Abstract
The MutL family of DNA mismatch repair proteins plays a critical role in excising and repairing misincorporation errors during DNA replication. In many eukaryotes, members of this family have evolved to modulate and resolve recombination intermediates into crossovers during meiosis. In these organisms, such functions promote the accurate segregation of chromosomes during the Meiosis I division. What alterations occurred in MutL homolog (MLH) family members that enabled them to acquire these new roles? In this review we present evidence that the yeast Mlh1-Mlh3 and Mlh1-Mlh2 complexes have evolved novel enzymatic and nonenzymatic activities and protein-protein interactions that are critical for their meiotic functions. Curiously, even with these changes, these complexes retain backup and accessory roles in DNA mismatch repair during vegetative growth.
Keywords: Mismatch repair, meiotic crossing over, MLH proteins, adaptation, endonuclease, Saccharomyces cerevisiae
1. INTRODUCTION
During meiosis a diploid cell undergoes successive reductional and equational divisions to form haploid gametes. In most organisms, proper chromosome disjunction during the reductional division requires crossing over between homolog pairs (Figure 1). Meiotic crossing over is thought to have evolved from DNA repair mechanisms present in vegetative (somatic) growth to allow for increased genetic recombination and a shift from sister chromatid to interhomolog repair (Barton & Charlesworth, 1998; Cavalier-Smith, 2002; Hunter, 2006; Kleckner, 1996; Marcon & Moens, 2005; Villeneuve & Hillers, 2001). Meiosis-specific functions needed to accomplish these tasks are thought to have emerged from the duplication of genes involved in cellular growth and maintenance, with such events and adaptive mechanisms permitting the acquisition of meiosis-specific gene expression and function. As described below, the MutL homolog (MLH) family of DNA mismatch repair (MMR) proteins provides a clear example of how genes that act in vegetative growth have acquired meiotic functions (Marcon & Moens, 2005).
FIGURE 1. A model indicating roles for MLH proteins in meiotic recombination.
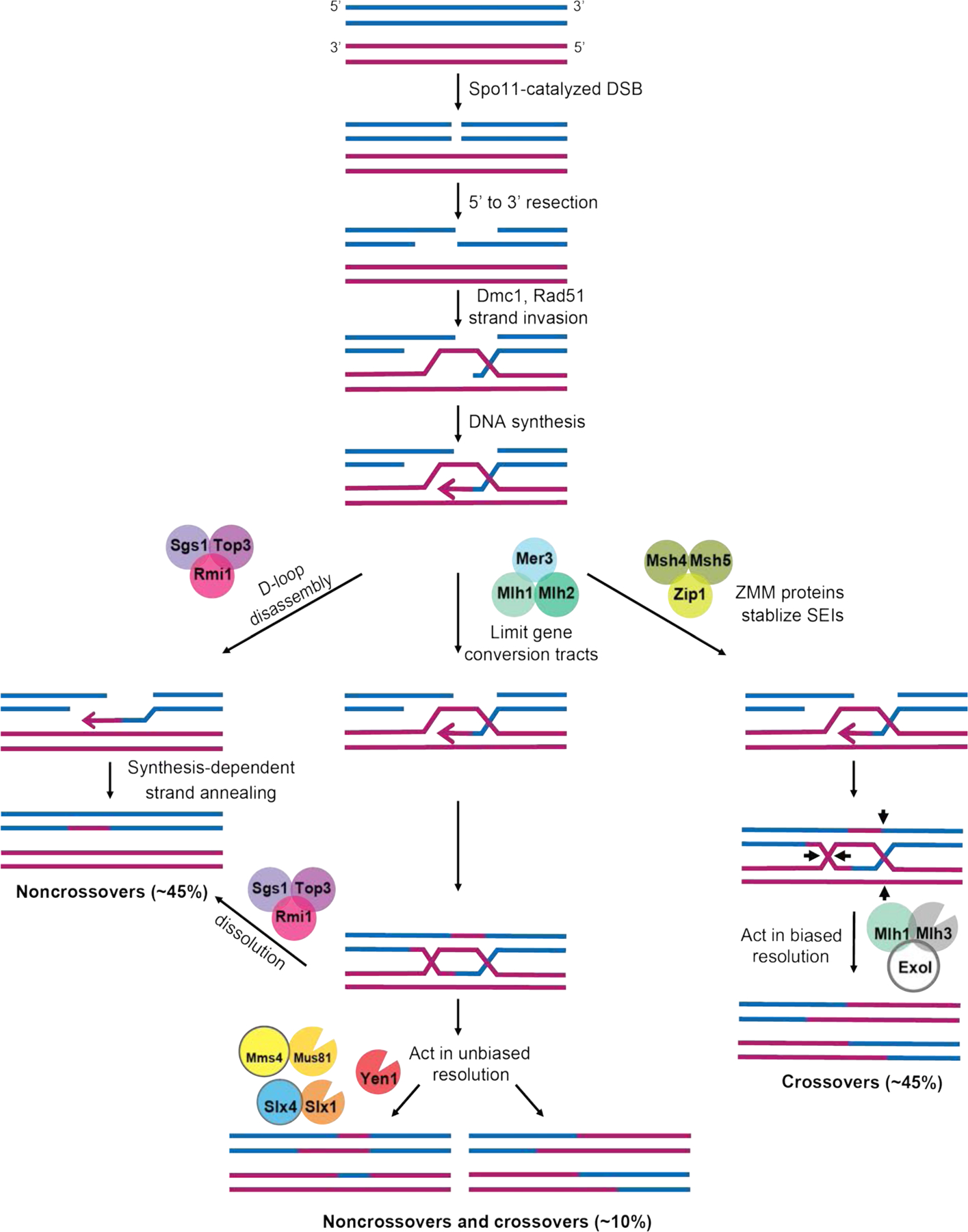
In diploid yeast induced to undergo meiosis, Spo11 catalyzes double-strand breaks (DSBs) throughout the genome, which undergo 5’ to 3’ resection. The resulting 3’ single stranded tail can invade the homologous chromosome to form a D-loop intermediate. In the ZMM-stabilized (Class I) pathway, the single end invasion intermediate (SEI) is stabilized by the actions of Msh4-Msh5 and Zip3 to promote D-loop extension through DNA repair synthesis. A double Holliday junction (dHJ) is then formed by second end capture that is asymmetrically cleaved in an Mlh1-Mlh3-dependent step to yield primarily crossover products. Note that in this model branch migration of Holliday junctions can occur but is not shown. Noncrossovers and a minority of crossovers (class II) are thought to occur through the other pathways shown (reviewed in Manhart & Alani, 2016; Wild & Matos, 2016; Zakharyevich et al., 2012). Roles for the Mlh1-Mlh2 complex in limiting gene conversion tract length, and Mlh1-Mlh3 in biased resolution of double-Holliday junctions to form crossovers, are shown. Mlh1-Pms1 acts to repair DNA mismatches that form in heteroduplex DNA in all pathways. The distribution of the types of meiotic recombination events in baker’s yeast is approximate and was calculated based on studies showing that Spo11 catalyzed DSBs produce equal numbers of crossovers and noncrossovers (Mancera, Bourgon, Brozzi, Huber, & Steinmetz, 2008; Martini, Diaz, Hunter, & Keeney, 2006; Marsolier-Kergoat et al., 2018). Of the crossovers, 85% are estimated to be class I, and 15% class II (Cooper et al., 2018; Jessop & Lichten, 2008; Oh, Lao, Taylor, Smith, & Hunter, 2008). Of the noncrossovers, 15% are estimated to result from the resolution of dHJs in the class II pathway (DeMuyt et al., 2012), with the remainder occurring through other mechanisms such as synthesis-dependent strand annealing. See text for details and Marsolier-Kergoat et al. (2018) and Pyatnitskaya, Borde, & De Muyt (2019) for more detailed models.
Yeast provides an ideal model to understand how gene duplication result in the expansion of functional roles for highly conserved proteins. The adaptation of such functions results from changes in gene expression and patterns of post-translational modifications, as well as the gain or loss of interacting protein partners and enzymatic functions. One such example in baker’s yeast involves the Rad51 and Dmc1 proteins, which promote early steps in homologous recombination by catalyzing the invasion of a 3’ single stranded end originating from a DNA double-strand break into a homologous template (Figure 1; Chan, Zhang, Weissman, & Bishop, 2019; Cloud, Chan, Grubb, Budke, & Bishop, 2012; Crickard, Kaniecki, Kwon, Sung, & Greene, 2018; Steinfeld et al., 2019). These proteins are thought to have evolved early in the evolution of eukaryotes from a single gene duplication of an ancestral archaea RadA recombinase. The duplication of an ancestral gene may have coincided with, or potentially enabled, meiosis (Lin, Kong, Nei, & Ma, 2006; Ramesh, Malik, & Logsdon, 2005). In support of these ideas, Rad51 is the only strand exchange protein present in vegetative growth, whereas both Rad51 and Dmc1 act in meiosis (Bishop, 2012; Bishop, Park, Xu, & Kleckner, 1992; Neale & Keeney, 2006). Rad51 and Dmc1 have biochemical and structural differences as well as unique subsets of protein interactors/regulators (Brown & Bishop, 2014; Chan et al., 2019; Lee et al., 2015; Steinfeld et al., 2019). In meiosis, Rad51 and Dmc1 are both needed to promote recombination between homologs, known as interhomolog bias (Figure 1). Interestingly, in a mutant background that lacks Dmc1 but is activated for Rad51 strand exchange activity, the bias towards interhomolog recombination is disrupted, and obligate crossovers are missing that are required for accurate chromosome segregation (Bishop 2012; Callender et al., 2016; Lao et al., 2013; Cloud et al., 2012). Additional studies showed that Rad51-mediated strand invasion activity is not required for meiotic recombination, but such an activity is critical for Dmc1 function (Cloud et al., 2012). Together, these studies identified a critical role for Dmc1 in strand exchange steps in meiosis. However, Rad51 meiotic functions are still essential in this process; Rad51 acts with the Mei5-Sae3 complex as a Dmc1 accessory factor to assist in the formation of the Dmc1 strand exchange filament (Chan et al., 2019; Cloud et al., 2012).
MutS provides another example of a single bacterial gene that duplicated and evolved early in eukaryogenesis to yield gene families with distinct vegetative growth and meiotic functions (Eisen, 1998; Lin, Nei, & Ma, 2007). In prokaryotic and eukaryotic MMR, MutS homolog (MSH) proteins recognize base-base and insertion/deletion mismatches that escape DNA polymerase proofreading, and transmit the recognition signal to downstream repair proteins such as the MLH family, which coordinate excision of the replication error and DNA resynthesis using the parental DNA strand as a repair template (Figures 2 and 3). Inactivation of MMR results in an increase in mutation rates by up to several orders of magnitude. Such an increase reduces fitness due to the accumulations of deleterious mutations yet can provide a source of beneficial mutations for adaptation to stressful environments (Kunkel & Erie, 2015; Raghavan, Aquadro & Alani, 2019). MutS is thought to have entered archaea and eukaryotes through horizontal gene transfer, followed by a series of gene duplications (Eisen, 1998; Lin et al., 2006). In baker’s yeast these events gave rise to MutS homologs (MSH) that function in the following pathways: 1. Msh2-Msh6 and Msh2-Msh3, which act to repair misincorporation errors during DNA replication and mismatches that form in heteroduplex DNA during genetic recombination; 2. Msh1, which functions in mitochondrial DNA metabolism; and 3. Msh4-Msh5, which promotes crossover formation in meiosis (Figures 1 and 3; Hollingsworth, Ponte, & Halsey, 1995; Kunkel & Erie, 2015; Reenan & Kolodner, 1992; Ross-Macdonald & Roeder, 1994; Snowden, Acharya, Butz, Berardini, & Fishel, 2004). Interestingly, Msh2-Msh6 (base-base and single nucleotide insertion/deletion mismatches) and Msh2-Msh3 (preference for insertion/deletion mismatches of up to 17 nucleotides in size) have evolved different mismatch repair specificities, and the Msh4-Msh5 complex lacks the amino-terminal mismatch recognition domain found in the other yeast MSH proteins, but recognizes Holliday junctions, a critical intermediate in meiotic recombination (Jensen, Jauert, & Kirkpatrick, 2005; Kunkel & Erie, 2015; Lahiri, Li, Hingorani & Mukerji, 2018; Pochart, Woltering, & Hollingsworth, 1997; Snowden et al., 2004).
FIGURE 2. Model for prokaryotic MMR.
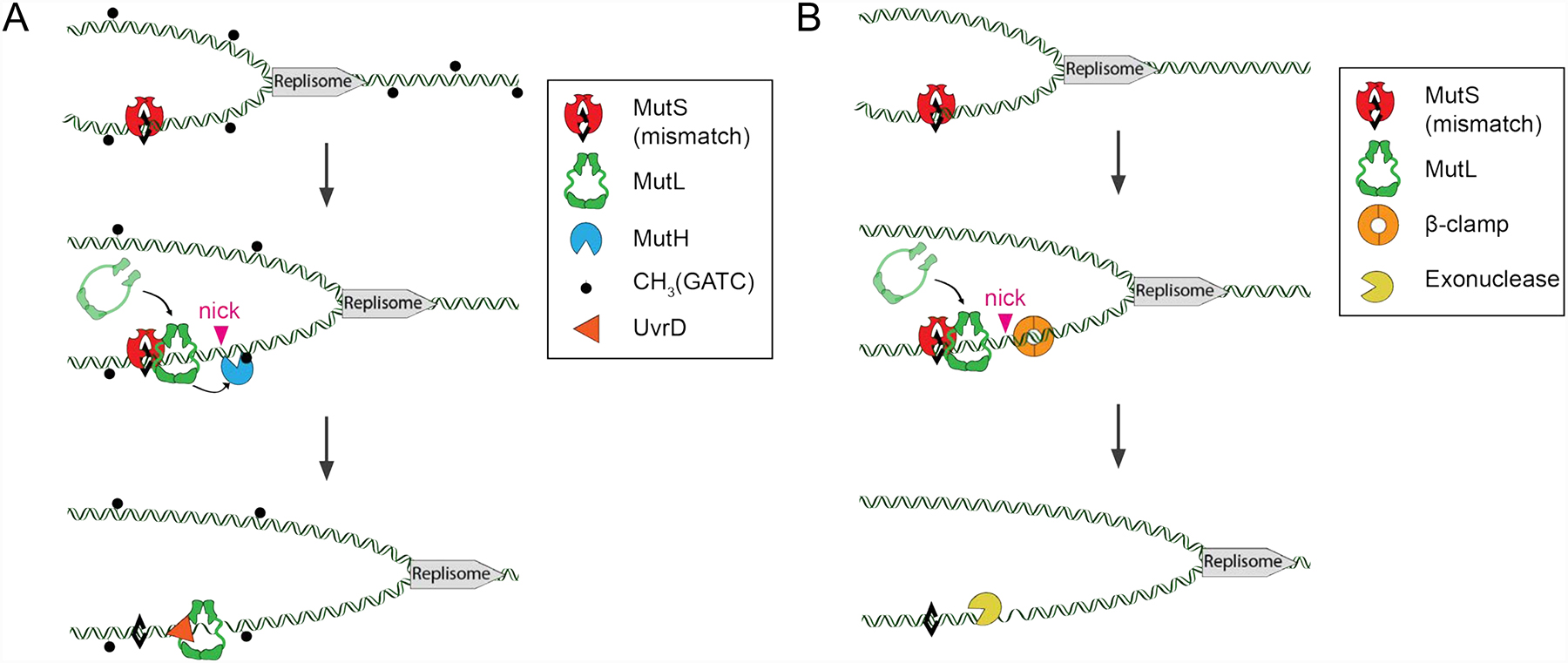
A model for mismatch repair in bacteria. A mismatch in DNA formed due to DNA polymerase misincorporation/DNA slippage is shown as a black diamond. MutS binds to the mismatch and undergoes an ATP-dependent conformational change to act as a sliding clamp and recruit MutL. (A) In E. coli, the MutS-MutL complex then acts as a sliding clamp complex that recruits MutH to nick the unmethylated (newly replicated) strand at hemi-methylated d(GATC) sites. These nicks allow for the helicase UvrD to unwind the DNA which is then excised, by a variety of nucleases, depending on the polarity of the nick relative to the mismatch. Alternatively, UvrD and MutL processively unwind the DNA between two d(GATC) sites (depicted). The resulting gap is then filled in by DNA polymerase. (B) In most other bacteria, MutL contains an intrinsic endonuclease activity that is stimulated by the DNA replication processivity factor β-clamp. These nicks act as entry sites for exonucleases to excise the mismatch, followed by re-synthesis of the gapped DNA by DNA polymerase. It is not clear if a UvrD helicase acts during MMR in bacteria that do not use MutH and Dam methylase. In such bacteria a recent model suggests that analogous to eukaryotic MMR, an exonuclease acts on double-stranded DNA to excise the mismatch, thus not requiring a UvrD helicase type activity (as shown, see Lenhart, Pillon, Guarne, Biteen, & Simmons, 2016).
FIGURE 3. Model for MMR in baker’s yeast.
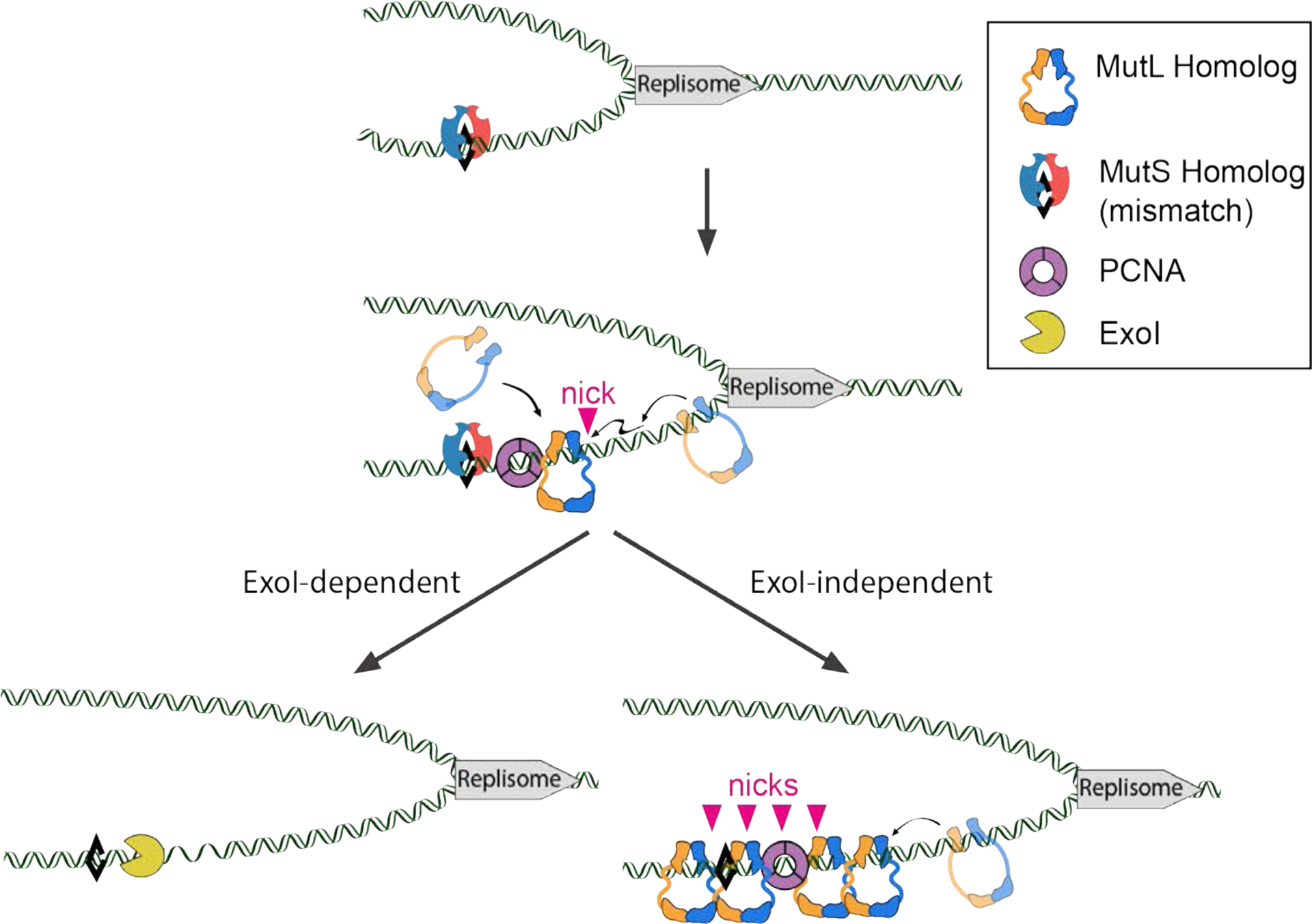
A mismatch in DNA formed due to DNA polymerase misincorporation is shown as black diamond. MSH complexes (Msh2-Msh3 or Msh2-Msh6) bind to the mismatch and undergo an ATP-dependent conformational change that allows them to act as a sliding clamp and recruit MLH complexes. Mlh1-Pms1 or Mlh1-Mlh3 undergo ATP-dependent conformational changes and orientation-specific interactions with PCNA (see text for a review of hypotheses for how these interactions with PCNA could be initiated), resulting in incision of the newly replicated DNA strand. This is followed by two potential pathways, ExoI-independent and -dependent, that can recruit downstream repair factors to excise and resynthesize the DNA and ligate the repaired strand. In the ExoI-dependent pathway, ExoI uses the nicks made by MLH complexes as an entry site to excise the mismatch containing DNA in a 5’−3’ direction. In the ExoI-independent pathway, multiple nicks made by MLH complexes in the vicinity of the mismatch promote DNA strand displacement and synthesis by DNA polymerase to repair the mismatch. Adapted from Kim et al., (2019).
This review focuses on how the MutL homolog (MLH) family of eukaryotic DNA MMR proteins has evolved distinct vegetative and meiotic life cycle functions; the causes of this specificity in function are not as well studied compared to those identified for the Rad51 and Dmc1 proteins. Most eukaryotes encode at least two MLH heterodimeric complexes that play critical roles in vegetative MMR and/or meiotic recombination (Kadyrov, Dzantiev, Constantin, & Modrich, 2006; Kaydrov et al., 2007; Räschle, Dufner, Marra, & Jiricny, 2002). In baker’s yeast three MLH complexes have been identified: Mlh1-Pms1, Mlh1-Mlh2, and Mlh1-Mlh3 (Wang, Kleckner, & Hunter, 1999). All three function during vegetative MMR, with Mlh1-Mlh3 and Mlh1-Mlh2 displaying minor and more specialized roles. (Romanova & Crouse, 2013; Harfe, Minesinger, & Jinks-Robertson, 2000). In meiosis, Mlh1-Mlh3 acts in the biased cleavage of double Holliday junctions to yield crossovers that are critical for the formation of gametes, Mlh1-Mlh2 regulates gene conversion tract lengths, and Mlh1-Pms1, analogous to its role in MMR, repairs mismatches that form in heteroduplex DNA during genetic recombination (Figure 1; Manhart & Alani, 2016; Zakharyevich, Tang, Ma, & Hunter., 2012; Abdullah, Hoffmann, Cotton, & Borts, 2004; Campbell et al., 2014; Duroc et al., 2017; Harfe et al., 2000; Hunter & Borts, 1997). These observations suggest that MLH complexes have evolved different roles during the vegetative and meiotic life cycles of a yeast cell. Such distinct roles have been conserved in higher eukaryotes, and in humans, defects in these factors have been linked to distinct diseases such as hereditary forms of colon cancer and infertility (Gray & Cohen, 2016; Lynch, Snyder, Shaw, Heinen, & Hitchens, 2015).
In addition to being functionally conserved, the MLH family of proteins are structurally conserved (Figure 4). Each subunit in the dimeric MLH complexes contain globular amino (N-) and carboxy (C-) terminal domains connected by linker arms. Structures of some of the N- and C-terminal domains have been solved for prokaryotic and eukaryotic proteins (Figure 4; Arana et al., 2010; Ban & Yang, 1998; Guarne et al., 2004; Gueneau et al., 2013). The N-terminal domain of these proteins contains an ATP-binding site similar in structure to the GHKL family of ATPases (DNA Gyrase, Hsp90, histidine kinases, MutL) (Dutta & Inouye, 2000). Upon nucleotide binding, the two ATP binding domains in the dimer physically interact, with the linker domains becoming more ordered, and the MLH complex primed for activation (Ban & Yang, 1998; Sacho, Kadryov, Modrich, Kunkel & Erie, 2008). The main dimerization interface for the two subunits is located at the C-terminal domain, and for most MLH complexes, a metal binding domain is also found at the C-terminus that is critical for endonuclease activity (Guarne et al., 2004; Kadyrov et al., 2006).
FIGURE 4. Structural homology of yeast Mlh1-Pms1, Mlh1-Mlh2, and Mlh1-Mlh3.
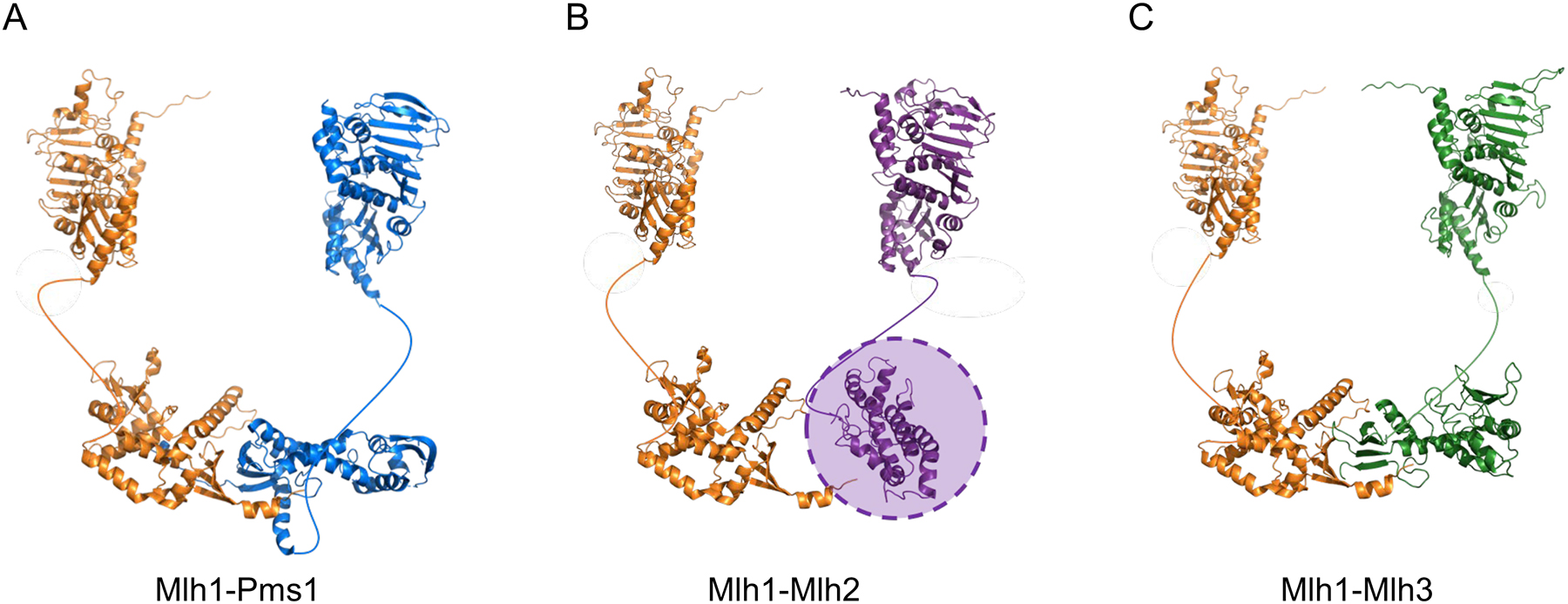
(A) For yeast Mlh1-Pms1, the N-terminal domain of Pms1 was obtained from PDB 3h4l, and the N-terminal domain of Mlh1 was modeled by a Phyre2 (amino acids 1–367 of Mlh1; http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index). The structure of the Mlh1-Pms1 C-terminus (PDB 4e4w) was solved by Gueneau et al. (2013). (B) For yeast Mlh1-Mlh2, the N-terminal domains of Mlh1 and Mlh2 were created using a Phyre2 homology model (amino acids 1–371 of Mlh2). The C-terminal domain of Mlh1 was obtained from PDB 4e4w. The C-terminal domain of Mlh2 (amino acids 503–695), created using a Phyre2 homology model, did not align well with the C-terminal domains of the other MLH proteins; for this reason, it is indicated by a shaded oval. (C) For yeast Mlh1-Mlh3, the N-terminal domains of Mlh1 and Mlh3 were created by a homology model presented in Al-Sweel et al. (2017). The C-terminal domain of Mlh1 was obtained from PDB 4e4w, and the C-terminal domain of Mlh3 (amino acids 491–715 of Mlh3) was created by a homology model presented in Al-Sweel et al. (2017). In all panels the unstructured linker domains are shown as curved lines.
What changes occurred in MLH family members that enabled them to acquire distinct vegetative and meiotic roles? As described below, there is strong conservation in the N- and C-terminal domains of the MLH proteins, but there are also amino acid sequences that are uniquely conserved for each MLH protein that likely confer different enzymatic functions and protein-protein interactions for the different MLH complexes. Also, the MLH proteins contain highly variable and non-conserved intrinsically disordered linker arms that range in size from about 100 amino acids (aa) in bacteria to 300 aa in yeast (Figure 5; Guarne et al., 2004; Sacho et al., 2008). In higher eukaryotes these linkers can approach 650 aa in length, as seen for the mouse and human Mlh3 linkers (Lipkin et al., 2000). The properties of these linkers have been proposed to facilitate conformational changes needed to activate latent MutL activities (Sacho et al., 2008), and recent studies in bacteria and yeast suggest that the linker arms are also important for MLH proteins to overcome barriers in the DNA landscape that would need to be traversed to locate the replication machinery and MSH proteins (Kim, Furman, Manhart, Alani, & Finkelstein, 2019; Mardenborough et al., 2019; Plys, Rogacheva, Greene, & Alani, 2012). Why linker lengths show such variability is not known, but they too could have evolved for different life cycle functions.
FIGURE 5. Composition of linker variation among MutL and MLH proteins.
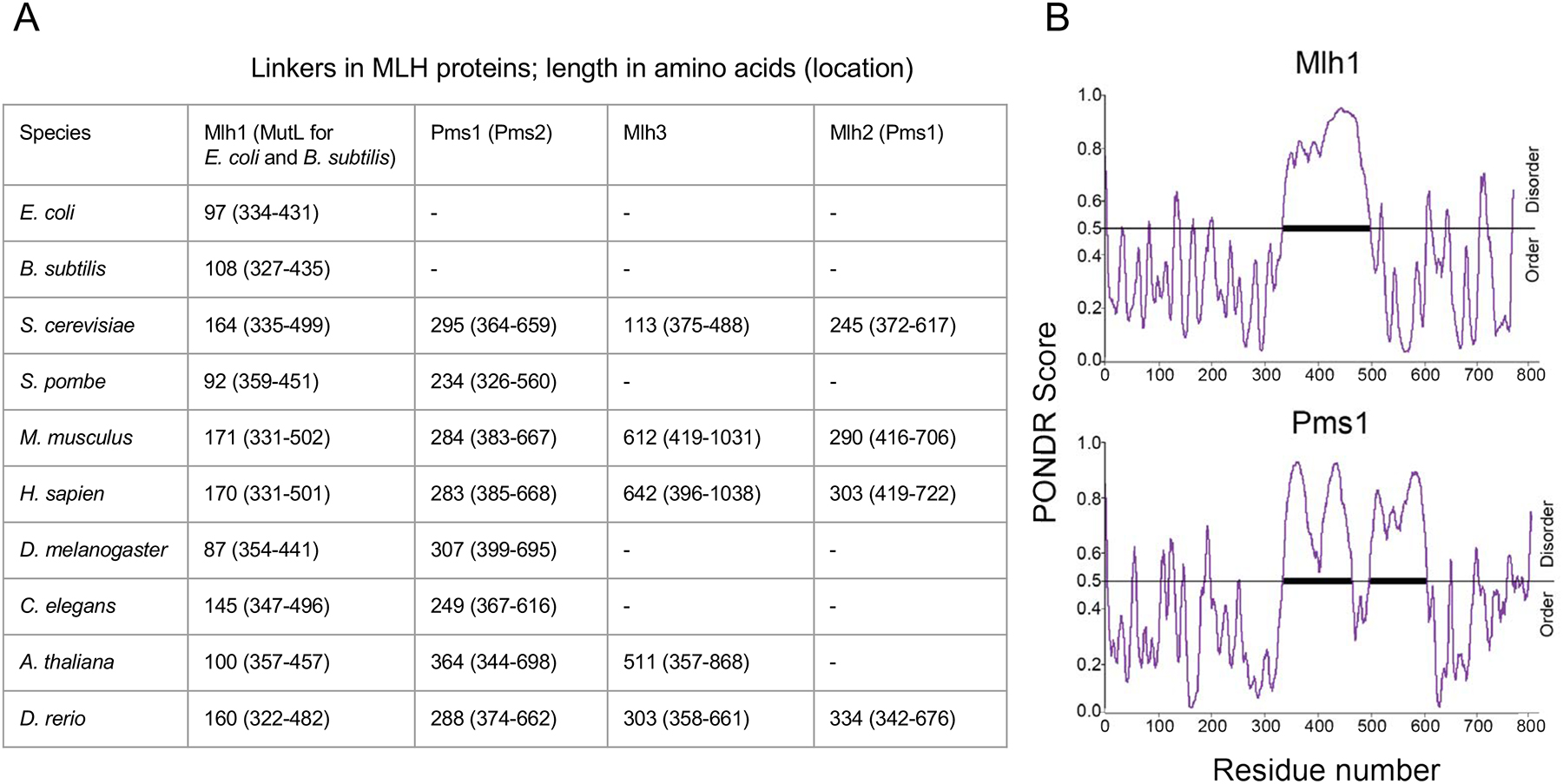
(A) Proposed linker lengths of various MutL and MLH proteins from bacteria to higher eukaryotes. Amino acid sequences were obtained from UniProt (https://www.uniprot.org), and linker lengths were determined using the Predictor of Natural Disorder (PONDR) VSL2 disorder prediction algorithm. (B) Shown is a PONDR analysis of S. cerevisiae Mlh1 and Pms1 (Kim et al., 2019; Obradovic et al., 2003).
2. OVERVIEW OF PROKARYOTIC MUTL PROTEINS AND THEIR RELATIONSHIP TO EUKARYOTIC MLH PROTEINS
Through genetic, in vitro reconstitution, and single-molecule analyses, scientists have obtained a thorough understanding of MMR in the Gram-negative bacteria E. coli. This work has provided detailed mechanistic information with respect to mismatch recognition, strand discrimination, and repair DNA synthesis steps (Figure 2; Jiricny 2013; Lahue, Au, & Modrich, 1989; Lamers et al., 2000; Liu et al., 2016; Modrich & Lahue, 1996; Obmolova, Ban, Hsieh, & Yang, 2000). In the model for E. coli shown in Figure 2, MMR is initiated by the MutS homodimer binding to a mismatch. This binding causes an ATP-dependent conformational change in MutS to form a sliding clamp that recruits the MutL homodimer (Acharya et al., 2003; Liu et al., 2016). The MutS-MutL complex then activates MutH, a restriction endonuclease-like protein, which nicks the unmethylated strand of hemimethylated DNA at d(GATC) sites. Importantly, E. coli uses the time interval needed for DNA adenine methyltransferase (Dam) to methylate newly replicated DNA at d(GATC) sites as a strand discrimination signal for MutH incision. After activating MutH incision, MutL recruits the UvrD helicase to unwind the newly replicated strand containing the mismatch, which in turns allows for single stranded exonucleases with different polarities (RecJ, ExoI, ExoVII, ExoX) to excise mismatches 5’ or 3’ to the nicked site (Burdett et al., 2001, but see Liu et al. (2019) for an exonuclease-independent UvrD unwinding model). MMR is completed upon DNA resynthesis by DNA Polymerase III, followed by sealing of nicks by DNA ligase.
In addition to roles in MMR, MutS and MutL act to prevent recombination between divergent homologous sequences, a process that is also thought to promote speciation. This heteroduplex rejection mechanism, which involves interactions with non-MMR proteins, acts to prevent deleterious recombination events, and is also seen in eukaryotes (Chakraborty & Alani, 2016; Rayssiguier, Thaler, & Radman, 1989; Worth, Clark, Radman, & Modrich, 1994).
E. coli is an outlier among prokaryotes in having acquired DNA methylation to distinguish template from nascent strands during replication fork passage. The acquisition of Dam and MutH nuclease functions is thought to be accompanied by the loss of MutL endonuclease activity (reviewed in Putnam 2016). In fact, most prokaryotes and all eukaryotes examined lack a MutH-Dam mechanism (Lenhart, Pillon, Guarné, Biteen, & Simmons, 2016), and in these organisms, MutL family proteins display an intrinsic endonuclease activity that is thought to be directed to nick the nascent strand through orientation-specific interactions with the DNA polymerase processivity clamp (β-clamp in prokaryotes, PCNA in eukaryotes; Kadyrov et al., 2006; Kadyrov et al., 2007; Pillon et al., 2010). While it is not known how this interaction occurs, recent studies have suggested that the discontinuity of Okazaki fragments on the lagging strand, processivity clamps remaining on DNA after replication, replication restart on the leading strand, and excision of ribonucleotides could provide the correct loading of the processivity clamp for strand-specific nicking by MLH endonucleases (Heller & Marians, 2006; Kawasoe, Tsurimoto, Nakagawa, Masukata, & Takahashi, 2016; Kunkel & Erie, 2015; Pluciennik et al., 2010).
3. BAKER’S YEAST MLH PROTEINS
3.1. MLH1-PMS1 PLAYS A MAJOR ROLE IN MMR
In current models for eukaryotic MMR (Figure 3; Kunkel & Erie, 2015), once an MSH complex binds to a mismatch, it undergoes, like MutS, an ADP-ATP exchange that alters the conformation of the complex to form a sliding clamp, allowing it to recruit the major MLH MMR complex, Mlh1-Pms1 (Gradia et al., 1999; Gorman et al., 2012). During this process, MSH heterodimers can interact with the replication processivity clamp PCNA that is loaded onto DNA by clamp loader (Clark, Valle, Drotschmann, Gary, & Kunkel, 2000; Johnson et al., 1996; Kleczkowska, Marra, Lettieri, & Jiricny, 2001). The MSH-PCNA interaction has been hypothesized to connect at least a portion of the MSH proteins to the replication fork and inhibit the unloading of PCNA during MMR, thus increasing the time interval for MMR (Hombauer, Campbell, Smith, Desai, & Kolodner, 2011; Kawasoe, Tsurimoto, Nakagawa, Masukata, & Takahashi, 2016; Kleczkowska, Marra, Lettieri, & Jiricny, 2001). PCNA then activates the endonuclease activity of Mlh1-Pms1 to nick the newly replicated daughter strand (Erdeniz, Dudley, Gealy, Jinks-Robertson, & Liskay, 2005; Kadyrov et al., 2006; Kawasoe, Tsurimoto, Nakagawa, Masukata, & Takahashi, 2016; Pluciennik et al., 2010). These nicks can facilitate two excision pathways (Figure 3). One is Exo1-dependent, where a nick 5′ to the mispair serves as an entry site for ExoI, a 5’ to 3’ exonuclease, to enter the DNA and excise DNA containing the mismatch. The resulting single-stranded DNA gap is coated by Replication Protein A (RPA), which facilitates DNA resynthesis steps. The second is Exo1-independent; Mlh1-Pms1 makes multiple nicks in the vicinity of the mismatch, allowing for strand displacement by Polymerase δ or ε. In both Exo1-dependent and independent mechanisms, nicks are sealed by DNA ligase, thus repairing the DNA and maintaining the original genetic information (reviewed in Goellner, Putnam, & Kolodner, 2015).
3.2. MLH1-MLH3 PLAYS A MAJOR ROLE IN MEIOTIC CROSSING OVER
Mlh1-Mlh3 plays a minor role in MMR; genetic studies have suggested that it is recruited by Msh2-Msh3 to repair primarily insertion/deletion mismatches (Flores-Rozas & Kolodner, 1998; reviewed in Manhart & Alani, 2016; Romanova & Crouse, 2013). However, Mlh1-Mlh3 plays a critical role in resolving recombination intermediates during meiotic crossing over, with mlh1 and mlh3 null mutants showing meiotic crossover defects (Hunter & Borts, 1997; Manhart & Alani, 2016).
Meiotic recombination is initiated by Spo11, which catalyzes 150–200 DNA double strand breaks genome-wide (Figure 1; Keeney, Giroux, & Kleckner, 1997; Pan et al., 2011; Pyatnitskaya, Borde, & De Muyt, 2019; Robine et al., 2007). The DSBs are resected in a 5’ to 3’ direction in steps involving Exo1 to form 3’ single stranded tails at each side of the double-strand break. The RecA family proteins Dmc1 and Rad51 form a filament on the tails and invade a homologous template, creating a displacement loop (D-loop; Brown, Grubb, Zhang, Rust, & Bishop, 2015). The D-loop intermediates serve as a key substrate for interconnected recombination pathways. In one pathway, the Sgs1-Top3-Rmi1 helicase/topoisomerase complex (STR), mediates the disassembly of the D-loop intermediate to form noncrossovers in a process known as synthesis-dependent strand annealing. In a second pathway the D-loop is stabilized by a class of ZMM (yeast Zip1/Zip2/Zip3/Zip4, Msh4/Msh5, Mer3) proteins to form a single-end invasion intermediate (SEI; Hunter and Kleckner, 2001; Pyatnitskaya, Borde, & De Muyt, 2019). Second-end capture of this intermediate, followed by branch migration, DNA synthesis, and ligation, results in the formation of a double-Holliday junction (dHJ) which is resolved in a biased manner to form crossovers (class I) that display interference (Allers & Lichten, 2001; Hunter, 2006; Zakharyevich et al., 2012). In a third pathway the D-loop progresses to form dHJs that are resolved by structure selective nucleases such as Mus81-Mms4 to form noncrossovers and crossovers (class II) that lack interference and are distributed randomly (rather than the tendency for crossovers to be evenly spaced; de los Santos et al., 2003; De Muyt et al., 2012; Zakharyevich, Tang, Ma, & Hunter, 2012).
Work in yeast demonstrated that the dHJs formed in the ZMM pathway are resolved through the actions of the Msh4-Msh5, Exo1, and the Mlh1-Mlh3 endonuclease. Specifically, Msh4-Msh5 acts to stabilize single-end invasion and Holliday junctions, after which Mlh1-Mlh3 and Exo1 promote biased resolution of double-Holliday junctions to form crossovers (Borner, Kleckner, & Hunter, 2004; Manhart et al., 2017; Ranjha, Anand, & Cejka, 2014; Rogacheva et al., 2014; Snowden et al., 2004; Zakharyevich et al., 2012). This model is supported by genetic and physical studies demonstrating that Mlh1-Mlh3 acts downstream of Msh4-Msh5 in meiosis (Kolas et al., 2005; Moens et al., 2002). Relevant to these studies is the work of Marsolier-Kergoat, Khan, Schott, Zhu, & Llorente (2018), who used a high-throughput DNA sequencing approach to analyze the meiotic progency of a hybrid yeast strain. In their study they analyzed heteroduplex DNA tracts associated with noncrossover and crossover events. Interestingly, they observed a biased pattern of crossover resolution events consistent with the resolution of double-Holliday junctions being directed towards strands containing newly replicated DNA near the junctions. These observations suggest that cleavage of double-Holliday junctions through the actions of the Mlh1-Mlh3 endonuclease might be similar to that seen for the Mlh1-Pms1 nuclease during MMR, rather than Mlh1-Mlh3 acting as a structure-specific nuclease that binds and symmetrically cleaves Holliday junctions (see Mlh1-Mlh3 polymerization models presented in Manhart et al., 2017).
It is important to note that the Mlh1-Mlh3 complex is likely to play similar roles in higher eukaryotes. For example, Mlh3−/− mice are viable, but sterile, and display dramatic decreases in meiotic crossovers. Mlh3−/− spermatocytes undergo apoptosis after metaphase, whereas oocytes fail to complete Meiosis I (Lipkin et al., 2002). These findings suggest that like in Baker’s yeast, Mlh3 plays a critical role in higher eukaryotes to direct the maturation of recombination intermediates into crossovers, promote accurate chromosome segregation, and generate viable gametes. In humans, there are few clinical studies linking hMLH3 polymorphisms to infertility; however, a detailed analysis of population variants in hMLH3 could provide mechanistic insights (Markandona et al., 2015).
Many questions remain for how Mlh1-Mlh3 resolves double-Holliday junctions into crossover products. It is not fully understood what kinds of chromosomal architecture or protein-protein interactions are required to stabilize Mlh1-Mlh3 or direct its function (Manhart et al., 2017). There is clear evidence for post-translational modifications regulating meiotic crossover formation (He et al., 2020; Hollingsworth & Gaglione, 2019; Sanchez et al., 2020; Sourirajan & Lichten, 2008; Wild & Matos, 2016; Wild et al., 2019). For example, the structure-specific endonucleases Mus81-Mms4 and Yen1 that act in the interference-independent crossover pathway are activated and inhibited, respectively, by phosphorylation (Figure 1; Matos, Blanco, Maslen, Skehel, & West, 2011; Wild & Matos, 2016). At present there is no evidence for phosphorylation directly regulating Mlh1-Mlh3 endonuclease activity, but it is likely that factors that interact with Mlh1-Mlh3 are regulated by phosphorylation and/or other modifications such as SUMOylation (Cheng et al., 2006; He et al., 2020; Manhart & Alani, 2016). It is also of interest to understand how the chromatin landscape is regulated to allow access of Mlh1-Mlh3 to recombination substrates (Wild et al., 2019).
Recent studies have also suggested roles for Mlh1-Mlh3 in chromosome disjunction and crossover interference (Chakraborty et al., 2017; Claeys-Bouuaert & Keeney, 2017). Crossover interference, seen in many organisms such humans, mice, Drosophila, and C. elegans, regulates the spatial distribution of crossovers; the formation of a crossover lowers the probability of a second crossover nearby (Wang et al., 2019). Widely spaced crossovers, exhibiting interference, help to ensure the fidelity of chromosome segregation. How Mlh1-Mlh3 participates in these roles is unclear.
Lastly, recent studies in yeast and higher eukaryotes have suggested a role for Mlh1-Mlh3 in mediating somatic CAG trinucleotide repeat instability (Pinto et al., 2013; Su & Freudenreich, 2017). In humans, CAG repeat expansions in exon 1 of the HTT gene results in Huntington’s disease, an inherited neurodegenerative disease, where age of disease onset depends on the length of the CAG repeat tract. It is unclear whether Mlh1-Mlh3 acts alone, or if this process also requires other MLH proteins (see Gomes-Pereira, Fortune, Ingram, McAbney, & Monckton, 2004). It could be that, analogous to MMR, there is some partial redundancy to Pms1 and Mlh3’s role in generating expansions. Curiously, Mlh1-Mlh3 has been shown to bind specifically to, but not cleave, DNA branch structures such as Holliday junctions (Ranjha, Anand, & Cejka, 2014; Rogacheva et al., 2014), substrates that could resemble intermediates that form during trinucleotide repeat expansion.
3.3. MLH1-MLH2 MODULATES MEIOTIC RECOMBINATION TRACT LENGTH
Unlike Mlh3 and Pms1, Mlh2 lacks an endonuclease motif, and although mlh2 mutants display sensitivity to DNA damaging agents, deletion of MLH2 confers only a mild mutator phenotype. Harfe et al. (2000) demonstrated that although not required for vegetative MMR, Mlh2 has a minor role in the repair of frameshift mutations. Additionally, Campbell et al. (2014) showed that Mlh1-Mlh2 plays a non-essential role as an accessory factor in DNA MMR, but whose function becomes more significant when essential MMR factors become limiting. More specifically, they showed through live cell imaging that Mlh2 forms foci in the presence of mispaired DNA, and that focus formation was dependent on MSH2, MSH6, and MLH1. Biochemical assays revealed that Mlh1-Mlh2 is recruited to DNA mismatches by both Msh2-Msh6 and Msh2-Msh3, and in vivo studies showed that deletion of MLH2 in backgrounds where essential MMR proteins were missing or reduced caused synergistic increases in mutation rate (Campbell et al., 2014). Taken together, these findings suggest that Mlh2 plays a minor role in MMR, potentially overlapping with Pms1 functions.
In contrast to minor roles in MMR, Mlh2 has been implicated through genetic analyses to interact with the Mer3 helicase to regulate meiotic gene conversion tract lengths (Figure 1; Abdullah et al., 2004; Duroc et al., 2017; Marsolier-Kergoat et al., 2018). Furthermore, Abdullah et al. (2004) suggested, in their analysis of meiotic recombination in mlh2 msh4 and mlh2 msh5 double mutants, that Mlh2 channels meiotic recombination intermediates into the Msh4-Msh5 interference-dependent meiotic crossover pathway. Consistent with regulating meiotic gene conversion tracts, Duroc et al. (2017) showed that the Mer3 helicase interacted with Mlh1-Mlh2 in vitro and in vivo, Mer3 and Mlh1-Mlh2 each preferentially bound to D-loop intermediates, Mlh2 was recruited to meiotic recombination hotspots through its interaction with Mer3, and Mlh2 recruitment did not require the presence of DNA mismatches or interactions with mismatch recognition factors. These studies suggested Mer3, initially in a structural role, binds to D-loop intermediates, and then recruits Mlh1-Mlh2 to prevent excessive D-loop extension and DNA synthesis. Such a mechanism was hypothesized to limit the exposure to DNA sequences that could participate in inappropriate meiotic recombination events that result in genome rearrangements, as well as to reduce the fraction of the genome converted at each meiosis (Duroc et al., 2017).
It is unclear whether Mlh2’s role in repairing frameshift mutations is mechanistically related to its meiotic recombination role, especially because its meiotic role appears distinct from MMR. A phylogenetic analysis of for Mlh2 in the unikont taxa revealed that S. cerevisiae MLH2 is the homolog to metazoan PMS1 (S. cerevisiae PMS1’s homolog is metazoan PMS2), which also lacks an endonuclease motif and exhibits a weak mutator phenotype (Campbell et al., 2014). MLH2 homologs were identified in species that diverged from S. cerevisiae prior to the whole genome duplication event, suggesting that the origin of MLH2 predated the yeast whole genome duplication event. Interestingly, a subset of unikont genomes contain a yeast MLH3 homolog but lack the yeast MLH2 homolog (the human homolog is PMS1), and other unikonts lack both yeast MLH3 and MLH2 homologs (see Vakirlis et al., 2016). Also, in a small number of fungal species, MLH2 homologs contained stop codons and frameshifts that could reflect inactivation of the MLH2 gene, providing support for accessory roles for Mlh2 (Campbell et al., 2014).
4. HOW DIFFERENT ARE THE ACTIVITIES OF MLH FAMILY PROTEINS?
There are many structural and functional similarities and differences between the Pms1, Mlh2, and Mlh3 proteins (Figure 4). Pms1 and Mlh3 share highly conserved functional domains: an ATP binding site in the N-terminus that drives the conformational changes observed in MLH proteins, an intrinsically disordered linker arm, and a C-terminus containing the endonuclease motif and Mlh1 dimerization site. All three proteins compete for the same dimerization/binding site on the Mlh1 protein, and this competition could regulate the levels of each heterodimer present at each stage of the yeast life cycle (Kondo, 2001). One interesting feature of these proteins is the variability in the lengths of their intrinsically disordered linkers (Figure 5). As mentioned earlier, the linker domains are poorly conserved, and in yeast the length of the Pms1 linker is twice that of Mlh1 and three-times that of Mlh3. Curiously, the Mlh3 linker has expanded significantly in higher eukaryotes. The mouse and human Mlh3 linkers are roughly six-times larger than the yeast linker. While the functions of these linkers are thought to primarily regulate ATP-driven conformational changes, a difference in length and sequence could facilitate physical interactions with other proteins or DNA, or serve as substrates for post-translational modifications (Claeys Bouuaert & Keeney, 2017; Sacho, Kadryov, Modrich, Kunkel & Erie, 2008; Kim et al., 2019).
4.1. MLH ENDONUCLEASE DOMAINS SHOW SUBTLE DIFFERENCES
Mlh1-Pms1 and Mlh1-Mlh3 display endonuclease activities dependent on highly conserved metal binding motifs present in both Pms1 and Mlh3 (Figure 6; Gueneau et al., 2013; Kadyrov et al., 2006; Rogacheva et al., 2014). Five conserved residues predicted to form the endonuclease active site in Pms1 were found in homologous positions in Mlh3. These proteins maintain the ancestral endonuclease motif, but also contain distinct residues that are likely required for their function and pathway specificity. The outlier is Mlh2, which has very limited conservation in its C-terminal domain, has lost the conserved endonuclease motif, and has no characterized enzymatic function. However, Mlh2 has adopted a novel function in regulating the length of gene conversion tract lengths, as well as playing a role as an accessory factor in MMR, but this was only seen when Pms1 levels were reduced (Abdullah et al., 2004; Campbell et al., 2014; Duroc et al., 2017). The endonuclease motif in Mlh3’s C-terminus also overlaps with the Mlh1 dimerization domain. Al Sweel et al. (2017) identified alleles in the endonuclease motif that disrupt Mlh1-Mlh3 interaction, but interestingly, these alleles are functional in meiotic crossing over and defective in MMR. These observations suggest the presence of other protein-protein interactions during meiosis that stabilize Mlh1-Mlh3 and promote crossover resolution. The identification and characterization of such interactions would also allow us to better understand how protein-protein interactions provide pathway specificity.
FIGURE 6. Alignment of the endonuclease domain of S. cerevisiae Pms1 (Gueneau et al., 2013) with Pms1 and Mlh3 homologs from 18 budding yeast (family Saccharomycetaceae) species (Clark, Alani, & Aquadro, 2012).

Saccharomyces cerevisiae (Scer) Pms1 and Mlh3 amino acid sequences are shown, followed by sequences from homologs from six of the 18 species. The metal binding site of Pms1 (green font), which forms the endonuclease active site, contains five residues (H703, E707, C817, C848, H850) that are highly conserved (100% identity in 18 Pms1 and 18 Mlh3 sequences; Clark, Alani, & Aquadro, 2012). The QXLXXP motif (blue font), important for interactions with PCNA (PIP), is highly conserved in the Pms1 sequences (>94% identity; Genschel et al., 2017), but is absent in Mlh3 sequences. Also shown are residues in red that are uniquely conserved (61 to 100% identity) within each Pms1 and Mlh3 homolog family. Homologs were obtained from Saccharomyces cerevisiae (Scer), Ashbya gossypii (Agos), Candida albicans (Calb), Candida dubliniensis (Cdub), Candida glabrata (Cgla), Candida guilliermondii (Cgui), Candida tropicalis (Ctro), Saccharomyces paradoxus, Saccharomyces mikatae, Saccharomyces bayanus, Saccharomyces kluyveri, Kluyveromyces thermotolerans, Kluyveromyces waltii, Kluyveromyces lactis, Kluyveromyces polysporus, Candida lusitaniae, Lodderomyces elongisporus, Debaryomyces hansenii, and Pichia stipitis. Because of incomplete sequence information, the Pms1 homolog for Debaryomyces hansenii, and the Mlh3 homolog of Candida lusitaniae were not included.
4.2. MLH PROTEINS INTERACT WITH SPECIFIC SETS OF PROTEINS
The MLH proteins do not act in isolated systems. The kinds of selective forces that drove the evolutionary adaptation of these genes also likely acted on other genes in the same pathway or molecular network (Clark, Alani, & Aquadro, 2013). For example, in MMR, the endonuclease activity of Mlh1-Pms1 is activated through its interaction with the DNA processivity factor PCNA. PCNA is a ring-shaped structure that acts to enhance the processivity of DNA polymerase during replication by creating a topological link with the DNA template and enabling sliding during chain elongation (Kelman, 1997; Pillon, Miller, & Guarné, 2011). Eukaryotic Msh2-Msh6 and Mlh1-Pms1 both interact with PCNA through the PCNA-interacting motif (PIP Box), which is defined as a six amino acid residue consisting of Qxφ[L/I]xP, where φ is a hydrophobic residue and x is any amino acid (Kelman, 1997; Pillon, Miller, & Guarné, 2011). The endonuclease activity of yeast Pms1 is highly stimulated by PCNA through the PIP-motif located in close proximity to the endonuclease motif (Genschel et al., 2017; Pluciennik et al., 2010). Mlh3, however, lacks any sort of motif at this specific site (Figure 6). Historically, in vitro studies have shown no requirement for PCNA to stimulate the endonuclease activity of Mlh1-Mlh3 (Ranjha, Anand, & Cejka, 2014; Rogacheva et al., 2014). However, studies (Cannavo et al., 2020; Kulkarni et al., 2020), have showed that PCNA stimulates the Mlh1-Mlh3 endonuclease activity in conjunction with Msh4-Msh5 and Exo1, and one group (Cannavo et al., 2020) characterize a potential PIP-motif in Mlh1, but whether this is a bonafide PCNA interaction site is not clear (Genschel et al., 2017; Lee & Alani, 2006). One explanation for these properties is that the MLH proteins have undergone changes in their ancestral catalytic motif to bind and resolve different substrates, and/or have shifted interactions with protein partners during MMR and meiosis.
4.3. DO MLH PROTEINS SHARE FUNCTIONS?
MLH proteins are likely to share many functions, with some of their specificities driven by recruitment to different substrates or different MSH proteins. For example, Mlh1-Mlh2 meiotic functions in limiting gene conversion tracts could be explained by its interactions with Mer3, and Mlh1-Mlh3’s functions actions in meiotic crossing over could be explained through its recruitment by Msh4-Msh5 to recombination intermediates (Duroc et al., 2017; Kolas et al., 2005; Moens et al., 2002). In support of this idea, work by Marsolier-Kergoat et al. (2018) showed that cleavage of double Holliday junctions, presumably through the actions of the Mlh1-Mlh3 endonuclease, was directed towards strands containing newly replicated DNA; this mechanism is analogous to Msh2-Msh6 or Msh2-Msh3 and PCNA directing the endonuclease activity of Mlh1-Pms1 to the newly replicated strand during MMR. Further support for this idea comes from studies which showed that Msh2-Msh3 recruits Mlh1-Mlh3 primarily for the repair of deletion mispairs (Flores-Rozas & Kolodner, 1998; Romanova & Crouse, 2013). Thus, the recruitment of MLH complexes to specific substrates by specialized MutS proteins could contribute to our understanding of how protein-protein interactions influence the difference in functionality. Further studies on Mlh1-Mlh3’s protein-protein interactions in meiosis would allow us to understand how DNA binding proteins and protein-protein interactions of MutL proteins evolved to adapt to meiotic processes. For example, if the residues that specify different MSH interactions were identified, and then introduced into the different MLH proteins, one could test if such interactions are sufficient for specificity, or if additional changes have occurred in the MLH proteins that specify their functions such as acquiring novel DNA binding activities.
4.4. BIOINFORMATIC AND STRUCTURAL APPROACHES
In addition to analyzing protein-protein interactions as descried above, bioinformatic strategies can be used to study the specificity of MLH complexes. Steinfeld et al. (2019) identified and analyzed amino acid residues that are well conserved within the Rad51 or Dmc1 lineage but differ between the two recombinases. This same methodology can be applied through computational approaches. For example, Multi-Harmony uses multiple sequence alignments between subfamilies of proteins, homology models, and multi-Relief and sequence-Harmony algorithms to identify amino acids that may suggest functional specificity (Brandt, Feenstra, & Heringa, 2010). By aligning Mlh3 and Pms1 amino acid sequences from 19 different budding yeast species, multi-Harmony alignments can identify residues that are well conserved within each subfamily but differ between Mlh3 and Pms1 (Figure 6). Having a three-dimensional structure of Mlh3 would allow for these residues to be mapped precisely.
4.5. REGULATION BY POST-TRANSLATIONAL MODIFICATIONS
At present there is little evidence in yeast to suggest that any of the MutL proteins are functionally regulated by phosphorylation. However, in mammalian cells, post translational modifications of MLH proteins have been proposed to regulate their functions. For example, recent studies have suggested that human Mlh1-Pms2 is phosphorylated by Casein Kinase II (CK2) at Mlh1’s S477 in vitro. Phosphorylated Mlh1 loses MMR activity and levels of phosphorylated Mlh1 vary during the cell cycle. In tumors, CK2 is overexpressed appears to inactivate MMR. This could be analogous to mechanisms of tumorigenesis where hyper-methylation of Mlh1 drives microsatellite instability (Webbecher & Brieger, 2018). The modification of MLH proteins in yeast is not well studied but could be an interesting avenue to explore with respect to understanding unique MLH functions.
5. CONCLUSIONS AND FUTURE DIRECTIONS
This review is focused on understanding how and why eukaryotes acquired multiple MLH complexes that act in the vegetative and meiotic stages of the yeast life cycle. In addition to the MLH, MSH, and Rad51/Dmc1 examples presented, many other components of the homologous recombination machinery have been adapted to function in meiosis (Marcon & Moens, 2005; Steinfeld et al., 2019). Thus, studies performed with MLH proteins will likely provide a model to understand how large classes of proteins have adapted novel roles. Such adaptations could involve new partner interactions (see Wild et al., 2019) or yield novel biochemical functions. We believe that a combination of structural (Figure 4) and computational evolutionary (e.g. using Multi-Harmony), analyses of MLH family proteins will encourage the creation of chimeric proteins that show how MLH proteins have evolved meiotic roles. We are optimistic that such an approach will be useful because aspects of this strategy have been successfully used by Steinfeld et al. (2019) to identify functional differences between Rad51 and Dmc1.
Molecular evolution provides another approach to study how proteins have adapted to new roles in the yeast life cycle. In work performed by Hsieh, Makrantoni, Robertson, Marston, & Murray (2020), the vegetative cohesin Scc1 was replaced with vegetative growth expression of the meiotic cohesin paralog Rec8. After passaging this strain through 1,750 vegetative growth generations, Hsieh et al. (2020) identified evolved fit populations that acquired mutations primarily in known and novel partner proteins of cohesins. Thus, this work provided a clear example of how one could “turn back the clock” to understand how proteins evolved to take on new roles. For example, a strain lacking Pms1 could be evolved to become fully functional in MMR. Such an experiment could be used to learn how the Mlh2, Mlh3, and Pms1 proteins diverged, with the goal of identifying novel protein domains and/or interacting partners.
ACKNOWLEDGMENTS
We thank Beata Mackenroth, Sara Liang, Michael Lichten, Nathan Clark, and anonymous reviewers for helpful comments on the manuscript, and Gianno Pannafino for help on the protein structure analysis of MLH proteins. C. M. F., R. E., and E.A. are supported by the National Institute of General Medical Sciences of the National Institutes of Health: R35GM134872. R. E. was also funded by the Cornell McNair Scholars Program, and the Frank L. and Lynnet Douglas Fellowship in Chemistry, Chemical Biology, and Biochemistry. The content of this review is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- aa
amino acid
- C-terminal
carboxy terminal
- D-loop
displacement loop
- Dam
DNA adenine methyltransferase
- HJ
double-Holliday junction
- HR
homologous recombination
- MLH
MutL homolog
- MMR
mismatch repair
- MSH
MutS homolog
- N-terminal
amino-terminal
- PCNA
proliferating cell nuclear antigen
- RPA
replication protein A
- SEI
single-end invasion
- ZMM
Zip1/Zip2/Zip3/Zip4, Msh4/Msh5, Mer3
Footnotes
CONFLICT OF INTEREST
Authors declare that they have no conflict of interests.
REFERENCES
- Abdullah MF, Hoffmann ER, Cotton VE, & Borts RH (2004). A role for the MutL homolog MLH2 in controlling heteroduplex formation and in regulating between two different crossover pathways in budding yeast. Cytogenetic Genome Research, 107, 180–190. [DOI] [PubMed] [Google Scholar]
- Acharya S, Foster PL, Brooks P, & Fishel R (2003). The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Molecular Cell, 12, 233–246. [DOI] [PubMed] [Google Scholar]
- Al-Sweel N, Raghavan V, Dutta A, Ajith VP, Di Vietro L, Khondakar N, Manhart CM, Surtees JA, Nishant KT, & Alani E (2017). mlh3 mutations in baker’s yeast alter meiotic recombination outcomes by increasing noncrossover events genome-wide. PLoS Genetics, 13, e1006974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, & Lichten M (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell, 106, 47–57. [DOI] [PubMed] [Google Scholar]
- Arana ME, Holmes SF, Fortune JM, Moon AF, Pedersen LC, & Kunkel TA (2010). Functional residues on the surface of the N-terminal domain of yeast Pms1. DNA Repair, 9, 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C, & Yang W (1998). Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell, 95, 541–552. [DOI] [PubMed] [Google Scholar]
- Barton NH & Charlesworth B (1998). Why sex and recombination? Science, 281, 1986–1990. [PubMed] [Google Scholar]
- Bishop DK (2012). Rad51, the lead in mitotic recombinational DNA repair, plays a supporting role in budding yeast meiosis. Cell Cycle, 11, 4105–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, & Kleckner N (1992). DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, & Hunter N (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell, 117, 29–45. [DOI] [PubMed] [Google Scholar]
- Brandt BW, Feenstra KA, & Heringa J (2010). Multi-Harmony: Detecting functional specificity from sequence alignment. Nucleic Acids Research, 38, W35–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, & Bishop DK (2014). DNA strand exchange and RecA homologs in meiosis. Cold Spring Harbor Perspectives in Biology, 7, a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Grubb J, Zhang A, Rust MJ, & Bishop DK (2015). Small Rad51 and Dmc1 complexes often co-occupy both ends of a meiotic DNA double strand break. PLoS Genetics, 11, e1005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett V, Baitinger C, Viswanathan M, Lovett ST, & Modrich P (2001). In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proceedings of the National Academy of Sciences, USA, 98, 6765–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callender TL, Laureau R, Wan L, Chen X, Sandhu R, Laljee S, Zhou S, Suhandynata RT, Prugar E, Gaines WA,Kwon Y, Börner GV,Nicolas A, Neiman AM, &Hollingsworth NM (2016). Mek1 Down Regulates Rad51 Activity during Yeast Meiosis by Phosphorylation of Hed1. PLoS Genetics, 12, e1006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CS, Hombauer H, Srivatsan A, Bowen N, Gries K, Desai A, Putnam CD, & Kolodner RD (2014). Mlh2 is an accessory factor for DNA mismatch repair in Saccharomyces cerevisiae. PLoS Genetics, 10, e1004327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Sanchez A, Anand R, Ranjha L, Hugener J, Adam C, Acharya A, Weyland N, Aran-Guiu X, Charbonnier J-B, Hoffmann ER, Borde V, Matos J, & Cejka P (2020). Regulation of the MLH1-MLH3 endonuclease in meiosis. BioRxiv, 2020.02.12.946293. 10.1101/2020.02.12.946293 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T (2002). Origins of the machinery of recombination and sex. Heredity, 88, 125–141. [DOI] [PubMed] [Google Scholar]
- Chakraborty U, & Alani E (2016). Understanding how mismatch repair proteins participate in the repair/anti-recombination decision. FEMS Yeast Research, 16, pii:fow071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, Pankajam AV, Lin G, Dutta A, Krishnaprasad GN, Tekkedil MM, Shinohara A, Steinmetz LM, & Nishant KT (2017). Modulating crossover frequency and interference for obligate crossovers in Saccharomyces cerevisiae meiosis. G3 (Bethesda), 7, 1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YL, Zhang A, Weissman BP, & Bishop DK (2019). RPA resolves conflicting activities of accessory proteins during reconstitution of Dmc1-mediated meiotic recombination. Nucleic Acids Research, 47, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, & Wang TF (2006). SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes and Development, 20, 2067–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys Bouuaert C, & Keeney S (2017). Distinct DNA-binding surfaces in the ATPase and linker domains of MutLγ determine its substrate specificities and exert separable functions in meiotic recombination and mismatch repair. PLoS Genetics, 13, e1006722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Alani E, & Aquadro CF (2012). Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome Research, 22, 714–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Alani E, & Aquadro CF (2013). Evolutionary rate covariation in meiotic proteins results from fluctuating evolutionary pressure in yeasts and mammals. Genetics, 193, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AB, Valle F, Drotschmann K, Gary RK, & Kunkel TA (2000). Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. Journal of Biological Chemistry, 275, 36498–36501. [DOI] [PubMed] [Google Scholar]
- Cloud V, Chan YL, Grubb J, Budke B, & Bishop DK (2012). Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science, 337, 1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TJ, Crawford MR, Hunt LJ, Marsolier-Kergoat MC, Llorente B, & Neale MJ (2018). Mismatch repair impedes meiotic crossover interference. BioRxiv, 480418 10.1101/480418. [DOI] [Google Scholar]
- Crickard JB, Kaniecki K, Kwon Y, Sung P, & Greene EC (2018). Spontaneous self-segregation of Rad51 and Dmc1 DNA recombinases within mixed recombinase filaments. Journal of Biological Chemistry, 293, 4191–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, & Hollingsworth NM (2003). The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics, 164, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, & Lichten M (2012). BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Molecular Cell, 46, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroc Y, Kumar R, Ranjha L, Adam C, Guérois R, Md Muntaz K, Marsolier-Kergoat MC, Dingli F, Laureau R, Loew D, Llorente B, Charbonnier JB, Cejka P, & Borde V (2017). Concerted action of the MutLβ heterodimer and Mer3 helicase regulates the global extent of meiotic gene conversion. eLife, 6, e21900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, & Inouye M (2000). GHKL, an emergent ATPase/kinase superfamily. Trends in Biochemical Sciences, 25, 24–28. [DOI] [PubMed] [Google Scholar]
- Eisen JA (1998). A phylogenomic study of the MutS family of proteins. Nucleic Acids Research, 26, 4291–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdeniz N, Dudley S, Gealy R, Jinks-Robertson S, & Liskay RM (2005). Novel PMS1 alleles preferentially affect the repair of primer strand loops during DNA replication. Molecular Cell Biology, 25, 9221–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H, & Kolodner RD (1998). The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proceedings of the National Academy of Sciences, USA, 95, 12404–12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschel J, Kadyrova LY, Iyer RR, Dahal BK, Kadyrov FA & Modrich P (2017). Interaction of proliferating cell nuclear antigen with PMS2 is required for MutLα activation and function in mismatch repair. Proceedings of the National Academy of Sciences, USA, 114, 4930–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goellner EM, Putman CD, & Kolodner RD (2015). Exonuclease 1-dependent and independent mismatch repair. DNA Repair, 32, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Pereira M, Fortune MT, Ingram L, McAbney JP, & Monckton DG (2004). Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Human Molecular Genetics, 13, 1815–1825. [DOI] [PubMed] [Google Scholar]
- Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, Alani EE, & Greene EC (2012). Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proceedings of the National Academy of Sciences, USA, 109, E3074–E3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, & Fishel R (1999). hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Molecular Cell, 3, 255–261. [DOI] [PubMed] [Google Scholar]
- Gray S, Cohen PE (2016). Control of meiotic crossovers: from double-strand break formation to designation. Annual Review of Genetics, 50, 175–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarné A, Ramon-Maiques S, Wolff EM, Ghirlando R, Hu X, Miller JH & Yang W (2004). Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO Journal, 23,4134–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueneau E, Dherin C, Legrand P, Tellier-Lebegue C, Gilquin B, Bonnesoeur P, Londino F, Quemener C, Le Du MH, Márquez JA, Moutiez M, Gondry M, Boiteux S, & Charbonnier JB (2013). Structure of the MutLα C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nature Structural and Molecular Biology, 20, 461–468. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Minesinger BK, & Jinks-Robertson S (2000). Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Current Biology, 10, 145–148. [DOI] [PubMed] [Google Scholar]
- He W, Rao HBDP, Tang S, Bhagwat N, Kulkarni DS, Ma Y, Chang MAW, Hall C, Bragg JW, Manasca HS, Baker C, Verhees GF, Ranjha L, Chen X, Hollingsworth NM, Cejka P, & Hunter N (2020). Regulated proteolysis of MutSg controls meiotic crossing over. Molecular Cell, 78, 168–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, & Marian KJ (2006). Replication fork reactivation downstream of a blocked nascent leading strand. Nature, 439, 557–562. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, & Gaglione R (2019). The meiotic-specific Mek1 kinase in budding yeast regulates interhomolog recombination and coordinates meiotic progression with double-strand break repair. Current Genetics, 65, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L, & Halsey C (1995). MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes and Development, 9,1728–1739. [DOI] [PubMed] [Google Scholar]
- Hombauer H, Campbell CS, Smith CE, Desai A & Kolodner RD (2011). Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell, 147, 1040–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y-YP, Makrantoni V, Robertson D, Marston AL, & Murray AW (2020). Evolutionary repair: changes in multiple functional modules allow meiotic cohesin to support mitosis. PLoS Biology, 18, e3000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N (2006). Meiotic recombination. Topics in Current Genetics, 17, 381–442. [Google Scholar]
- Hunter N, & Borts RH (1997). Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes and Development, 11, 1573–1582. [DOI] [PubMed] [Google Scholar]
- Hunter N, & Kleckner N (2001). The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell, 106, 59–70. [DOI] [PubMed] [Google Scholar]
- Jensen LE, Jauert PA, & Kirkpatrick DT (2005). The large loop repair and mismatch repair pathways of Saccharomyces cerevisiae act on distinct substrates during meiosis. Genetics, 170, 1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, & Lichten M (2008). Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Molecular Cell 31, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J (2013). Postreplicative mismatch repair. Cold Spring Harbor Perspectives in Biology, 5, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Kovvali GK, Guzder SN, Amin NS, Holm C, Habraken Y, Sung P, Prakash L, & Prakash S (1996). Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. Journal of Biological Chemistry, 271, 27987–27990. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, & Modrich P (2006). Endonucleolytic function of MutLalpha in human mismatch repair. Cell, 126, 297–308. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O’Donnell M, Kunkel TA & Modrich P (2007). Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. Journal of Biological Chemistry, 282, 37181–37190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasoe Y, Tsurimoto T, Nakagawa T, Masukata H & Takahashi TS (2016). MutSα maintains the mismatch repair capability by inhibiting PCNA unloading. eLife, 5, e15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, & Kleckner N (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell, 88, 375–84. [DOI] [PubMed] [Google Scholar]
- Kelman Z (1997). PCNA: Structure, functions and interactions. Oncogene, 14, 629–640. [DOI] [PubMed] [Google Scholar]
- Kim Y, Furman CM, Manhart CM, Alani E, & Finkelstein IJ (2019). Intrinsically disordered regions regulate both catalytic and non-catalytic activities of the MutLα mismatch repair complex. Nucleic Acids Research, 47, 1823–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N (1996). Meiosis: How could it work? Proceedings of the National Academy of Sciences, USA, 93, 8167–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowska HE, Marra G, Lettieri T, & Jiricny J (2001). hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes and Development, 15, 724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Svetlanov A, Lenzi ML, Macaluso FP, Lipkin SM, Liskay RM, Greally J, Edelmann W, & Cohen PE (2005). Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. Journal of Cell Biology, 171, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo E (2001). The interacting domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Research, 29, 1695–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni DS, Owens S, Honda M, Ito M, Yang Y, Corrigan MW, Chen L, Quan AL, & Hunter N (2020). PCNA activates the MutLγ endonuclease to promote meiotic crossing over. BioRxiv, 2020.02.12.946020. 10.1101/2020.02.12.946020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, & Erie DA (2015). Eukaryotic mismatch repair in relation to DNA replication. Annual Reviews of Genetics, 49, 291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S, Li Y, Hingorani MM, & Mukerji I (2018). MutSγ-induced DNA conformational changes provide insights into its role in meiotic recombination. Biophysical Journal, 115, 2087–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue RS, Au KG, & Modrich P (1989). DNA mismatch correction in a defined system. Science, 245, 160–164. [DOI] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, & Sixma TX (2000). The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature, 407, 711–717. [DOI] [PubMed] [Google Scholar]
- Lao JP, Cloud V, Huang C-C, Grubb J, Thacker D, Lee C-Y, Dresser ME, Hunter N, & Bishop DK (2013). Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genetics, 9, e1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SD, & Alani E (2006). Analysis of interactions between mismatch repair initiation factors and the replication processivity factor PCNA. Journal of Molecular Biology, 355, 175–184. [DOI] [PubMed] [Google Scholar]
- Lee JY, Terakawa T, Qi Z, Steinfeld JB, Redding S, Kwon Y, Gaines WA, Zhao W, Sung P, & Greene EC (2015). Base triplet stepping by the Rad51/RecA family of recombinases. Science, 349, 977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart JS, Pillon MC, Guarné A, Biteen JS, & Simmons LA (2016). Mismatch repair in Gram-positive bacteria. Research in Microbiology, 167, 4–12. [DOI] [PubMed] [Google Scholar]
- Lin Z, Kong H, Nei M, & Ma H (2006). Origins and evolution of the recA/RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer. Proceedings of the National Academy of Sciences, USA, 103, 10328–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Nei M, & Ma H (2007). The origins and early evolution of DNA mismatch repair genes—multiple horizontal gene transfers and co-evolution, Nucleic Acids Research, 35, 7591–7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin SM, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis AD, Lynch HT, Elliott RM, & Collins FS (2000). MLH3: A DNA mismatch repair gene associated with mammalian microsatellite instability. Nature Genetics, 24, 27–35. [DOI] [PubMed] [Google Scholar]
- Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, Schwartzberg P, Collins FS, & Cohen PE (2002). Meiotic arrest and aneuploidy in MLH3-deficient mice. Nature Genetics, 31, 385–90. [DOI] [PubMed] [Google Scholar]
- Liu J, Hanne J, Britton BM, Bennett J, Kim D, Lee JB & Fishel R (2016). Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature, 539, 583–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lee R, Britton BM, London JA, Yang K, Hanne J, Lee JB, & Fishel R (2019). MutL sliding clamps coordinate exonuclease-independent Escherichia coli mismatch repair. Nature Communications, 10, 5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HT, Snyder CL, Shaw TG, Heinen CD, & Hitchins MP(2015). Milestones of Lynch syndrome: 1895–2015. Nature Reviews Cancer, 15, 181–194. [DOI] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, & Steinmetz LM (2008). High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature, 454, 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhart CM, Alani E (2016). Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair 38, 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhart CM, Ni X, White MA, Ortega J, Surtees JA, & Alani E (2017). The mismatch repair and meiotic recombination endonuclease Mlh1-Mlh3 is activated by polymer formation and can cleave DNA substrates in trans. PLoS Biology, 15, e2001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon E, & Moens PB (2005). The evolution of meiosis: Recruitment and modification of somatic DNA-repair proteins. BioEssays, 27, 795–808. [DOI] [PubMed] [Google Scholar]
- Mardenborough Y, Nitsenko K, Laffeber C, Duboc C, Sahin E, Quessada-Vial A, Winterwerp H, Sixma TK, Kanaar R, Friedhoff P, Strick TR, & Lebbink J (2019). The unstructured linker arms of MutL enable GATC site incision beyond roadblocks during initiation of DNA mismatch repair. Nucleic Acids Research, 47, 11667–11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markandona O, Dafopoulos K, Anifandis G, Messini CI, Dimitraki M, Tsezou A, Georgoulias P, & Messinis IE (2015). Single-nucleotide polymorphism rs 175080 in the MLH3 gene and its relation to male infertility. Journal of Assisted Reproduction and Genetics, 32, 1795–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolier-Kergoat MC, Khan MM, Schott J, Zhu X, & Llorente B (2018). Mechanistic view and genetic control of DNA recombination during meiosis. Molecular Cell, 70, 9–20. [DOI] [PubMed] [Google Scholar]
- Martini E, Diaz RL, Hunter N, & Keeney S (2006). Crossover homeostasis in yeast meiosis. Cell, 126, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, & West SC (2011). Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell, 147, 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P, & Lahue R (1996). Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annual Reviews of Biochemistry, 65, 101–133. [DOI] [PubMed] [Google Scholar]
- Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, & Spyropoulos B (2002). The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. Journal of Cell Science, 115, 1611–22. [DOI] [PubMed] [Google Scholar]
- Neale MJ, & Keeney S (2006). Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature, 442, 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, & Yang W (2000). Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature, 407, 703–710. [DOI] [PubMed] [Google Scholar]
- Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, & Dunker AK (2003). Predicting intrinsic disorder from amino acid sequence. Proteins, 53, 566–572. [DOI] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Taylor AF, Smith GR, & Hunter N (2008). RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Molecular Cell, 31, 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, Hochwagen A, & Keeney S (2011). A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell, 144, 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon MC, Lorenowicz JJ, Uckelmann M, Klocko AD, Mitchell RR, Chung YS, Modrich P, Walker GC, Simmons LA, Friedhoff P, & Guarné A (2010). Structure of the endonuclease domain of MutL: unlicensed to cut. Molecular Cell, 39, 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon MC, Miller JH, & Guarné A (2011). The endonuclease domain of MutL interacts with the β sliding clamp. DNA Repair, 10, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto RM, Dragileva E, Kirby A, Lloret A, Lopez E, St Claire J, Panigrahi GB, Hou C, Holloway K, Gillis T, Guide JR, Cohen PE, Li GM, Pearson CE, Daly MJ, & Wheeler VC (2013). Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington’s disease mice: genome-wide and candidate approaches. PLoS Genetics, 9, e1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, & Modrich P (2010). PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proceedings of the National Academy of Sciences, USA, 107, 16066–16071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plys AJ, Rogacheva MV, Greene EC, Alani E (2012). The unstructured linker arms of Mlh1-Pms1 are important for interactions with DNA during mismatch repair. Journal of Molecular Biology, 422, 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochart P, Woltering D, & Hollingsworth NM (1997). Conserved properties between functionally distinct MutS homologs in yeast. Journal of Biological Chemistry, 272, 30345–30349. [DOI] [PubMed] [Google Scholar]
- Putnam CD (2016). Evolution of the methyl directed mismatch repair system in Escherichia coli. DNA Repair 38, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyatnitskaya A, Borde V, & De Muyt A (2019). Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma, 128, 181–198. [DOI] [PubMed] [Google Scholar]
- Raghavan V, Aquadro CF, & Alani E (2019). Baker’s yeast clinical isolates provide a model for how pathogenic yeasts adapt to stress. Trends in Genetics, 35, 804–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh MA, Malik SB, & Logsdon JM (2005). A phylogenomic inventory of meiotic genes: Evidence for sex in Giardia and an early eukaryotic origin of meiosis. Current Biology, 15, 185–191. [DOI] [PubMed] [Google Scholar]
- Ranjha L, Anand R, & Cejka P (2014). The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday Junctions. Journal of Biological Chemistry 289, 5674–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räschle M, Dufner P, Marra G & Jiricny J (2002). Mutations within the hMLH1 and hPMS2 subunits of the human MutLalpha mismatch repair factor affect its ATPase activity, but not its ability to interact with hMutSalpha. Journal of Biological Chemistry, 277, 21810–21820. [DOI] [PubMed] [Google Scholar]
- Rayssiguier C, Thaler DS, & Radman M (1989). The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature, 342, 396–401. [DOI] [PubMed] [Google Scholar]
- Reenan RA, & Kolodner RD (1992). Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics, 132, 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N, Uematsu N, Amiot F, Gidrol X, Barillot E, Nicolas A, & Borde V (2007). Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Molecular and Cellular Biology, 27, 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogacheva MV, Manhart CM, Chen C, Guarne A, Surtees J, & Alani E (2014). Mlh1-Mlh3, A meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. Journal of Biological Chemistry, 289, 5664–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova NV, & Crouse GF (2013). Different roles of eukaryotic MutS and MutL complexes in repair of small insertion and deletion loops in yeast. PLoS Genetics, 9, e1003920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P, & Roeder GS (1994). Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell, 79, 1069–1080. [DOI] [PubMed] [Google Scholar]
- Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, & Erie DA (2008). Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Molecular Cell, 29, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Adam C, Rauh F, Duroc Y, Ranjha L, Lombard B, Mu X, Loew D, Keeney S, Cejka P, Guérois R, Klein F, Charbonnier J-B, & Borde V (2020). Mechanism of in vivo activation of the MutLγ-Exo1 complex for meiotic crossover formation. BioRxiv, 2019.12.16.876623. 10.1101/2019.12.16.876623. [DOI] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, & Fishel R (2004). hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Molecular Cell, 15, 437–451. [DOI] [PubMed] [Google Scholar]
- Sourirajan A, & Lichten M (2008). Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes and Development, 22, 2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfeld JB, Beláň O, Kwon Y, Terakawa T, Al-Zain A, Smith MJ, Crickard JB, Qi Z, Zhao W, Rothstein R, Symington LS, Sung P, Boulton SJ, & Greene EC (2019). Defining the influence of Rad51 and Dmc1 lineage-specific amino acids on genetic recombination. Genes and Development, 33, 1191–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XA, & Freudenreich CH (2017). Cytosine deamination and base excision repair cause R-loop–induced CAG repeat fragility and instability in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences, USA, 114, E8392–E8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakirlis N, Sarilar V, Drillon G, Fleiss A, Agier N, Meyniel JP, Blanpain L, Carbone A, Devillers H, Dubois K, Gillet-Markowska A, Graziani S, Huu-Vang N, Poirel M, Reisser C, Schott J, Schacherer J, Lafontaine I, Llorente B, Neuvéglise C, & Fischer G (2016). Reconstruction of ancestral chromosome architecture and gene repertoire reveals principles of genome evolution in a model yeast genus. Genome Research, 26, 918–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve AM, & Hillers KJ (2001). Whence meiosis? Cell, 106, 647–650. [DOI] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, & Hunter N (1999). Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proceedings of the National Academy of Sciences, USA, 96, 13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liu Y, Shang Y, Zhai B, Yang X, Kleckner N, & Zhang L (2019). Crossover interference, crossover maturation, and human aneuploidy. Bioessays, 41, e1800221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßbecher IM, & Brieger A, (2018). Phosphorylation meets DNA mismatch repair. DNA Repair, 72,107–114. [DOI] [PubMed] [Google Scholar]
- Wild P, & Matos J (2016). Cell cycle control of DNA joint molecule resolution. Current Opinion in Cell Biology, 40, 74–80. [DOI] [PubMed] [Google Scholar]
- Wild P, Susperregui A, Piazza I, Dörig C, Oke A, Arter M, Yamaguchi M, Hilditch AT, Vuina K, Chan KC, Gromova T, Haber JE, Fung JC, Picotti P, & Matos J (2019) Network rewiring of homologous recombination enzymes during mitotic proliferation and meiosis. Molecular Cell, 75, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth L, Clark S, Radman M, & Modrich P (1994). Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proceedings of the National Academy of Sciences, USA, 91, 3238–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, & Hunter N (2012). Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell, 149, 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]


