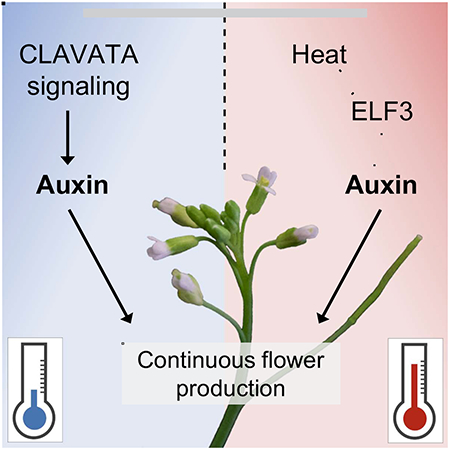
SUMMARY
The ability to thrive in diverse environments requires that species maintain development and reproduction despite dynamic conditions. Many developmental processes are stabilized through robust signaling pathways which cooperatively ensure proper development [1]. During reproduction, plants like Arabidopsis thaliana continuously generate flowers on growing indeterminate inflorescences [2]. Flower primordia initiation and outgrowth depends on the hormone auxin and is robust across diverse environments [3–6]. Here we show that reproductive development under different thermal conditions requires the integration of multiple pathways regulating auxin dependent flower production. In colder/ambient temperatures, the receptor complex CLAVATA2/CORYNE (CLV2/CRN) is necessary for continuous flower outgrowth during inflorescence development. CLV2/CRN signaling is independent of CLAVATA1 (CLV1)-related receptor signaling but involves the CLAVATA3 INSENSTIVE RECEPTOR KINASE (CIK) family co-receptors, with higher order cik mutant combinations phenocopying clv2/crn flower outgrowth defects. Developing crn inflorescences display reduced auxin signaling and restoration of auxin biosynthesis is sufficient to restore flower outgrowth in colder/ambient temperatures. In contrast, at higher temperatures both clv2/crn signaling and heat induced auxin biosynthesis via YUCCA family genes are synergistically required to maintain flower development. Our work reveals a novel mechanism integrating peptide hormone and auxin signaling in the regulation of flower development across diverse thermal environments.
Graphical Abstract
RESULTS AND DISCUSSION
Plants continually develop new organs throughout their life and do so across varied environmental conditions [7]. This indeterminate growth requires balanced cell proliferation and differentiation in stem cell niches, called meristems, at growing apices [8, 9]. During the reproductive phase of Arabidopsis thaliana, flower primordia are continuously produced from inflorescence meristems (IM) dependent on the hormone auxin [3–6]. Primordia then proliferate, forming flowers from secondary floral meristems [10, 11]. Cell recruitment into flower primordia is balanced by proliferation in the IM center. The conserved CLAVATA3 (CLV3) peptide signaling pathway dampens stem cell proliferation in shoot and floral meristems [12, 13]. CLV3 signals through a suite of receptors which repress the expression of WUSCHEL (WUS) in the center of the IM [8, 14]. WUS encodes a homeobox transcription factor that positively regulates stem cell proliferation [15]. Among these receptors is the atypical receptor pair CLAVATA2/CORYNE (CLV2/CRN), a leucine rich repeat (LRR) receptor-like protein and a transmembrane pseudokinase, respectively [16–19]. CLV2/CRN negatively regulate IM stem cell proliferation independent of other CLV3 receptors [17, 20]. Here we define a new role for CLV2/CRN in promoting auxin-dependent flower primordia outgrowth and show that signaling through this receptor complex contributes to an environmental buffering mechanism which ensures reproductive developmental stability.
The CLV2/CRN receptor complex promotes flower primordia outgrowth and development
After the production of 1-5 normal flowers in crn null mutants (crn-10; in the Col-0 ecotype), we noticed a novel phenotype in which flower primordia initiate but fail to develop further and inflorescence internode elongation stalls (Figures 1A–1B; the termination phase). After ~30-40 of these terminated primordia, flower development and inflorescence elongation resumes (recovery phase), indicating that continuous flower production requires CRN (Figures 1B and S1A; Video S1). clv2 null mutants (rlp10-1; in Col-0) displayed a similar phenotype to crn (Figure 1C), which was previously observed in clv2 in a survey of mutants in receptor-like protein genes, but not characterized [21]. To quantify primordia termination, we classified the first 30 attempts to make flowers along the primary inflorescence as normal (complete flowers; formation of all four flower organs [22]), terminated flower primordia (no flower organs develop), or terminated flowers (some flower organs develop, but no gynoecium). crn and clv2 single mutants displayed equivalent defects in flower production (Figure 1E), also observed in clv2 crn double mutants, consistent with the documented co-function of CLV2/CRN (Figures 1B–E). clv2/crn flower outgrowth defects, and floral meristem size (measured in carpels made per flower), were complemented by expressing fusion proteins from their native promoters (Figure S1B–S1C and S1I–S1J). Using standardized flower primordia staging, we found that crn primordia outgrowth deviated from WT (wild type) at flower primordia stage three (FP3), with little proliferation occurring afterwards (Figures 1F–1G; staging as in [23], or stage 2 using Smyth et. al. stages [22]). In crn, terminated primordia fail to develop floral organs, but occasionally produce bract-like structures, likely due to de-repression of cryptic bract outgrowth (Figure 1H) [24].
Figure 1. Flower primordia outgrowth is disrupted in clv2/crn.

(A-D) Inflorescences of Col-0, crn (crn-10), clv2 (rlp10-1), and crn clv2 double mutants. (A’-D’) Close up showing flower primordia termination in crn, clv2 and crn clv2.
(E) Quantification of flower termination, classifying the first 30 attempts to make a flower as: normal (grey), terminated primordia (blue) or terminated flowers (yellow) in Col-0 (n=28), crn (n=27), clv2 (n=25), and crn clv2 (n=25).
(F and G) 3-D reconstruction of inflorescence meristems of (F) Col-0 (n=4) and (G) crn (n=6). Axial view of the (F’, G’) third (FP3) and (F”, G”) fourth (FP4) flower primordia (labeled 3 and 4 in (F) and (G)) revealing developmental differences. FP3 and FP4 were determined by identifying the 3rd and 4th earliest detectable primordia along the IM, respectively. Staging similar to [23].
(H) Side view of a young inflorescence meristem of crn during the termination phase (n=5).
(I-K) Expression patterns of YPET-N7 reporter lines in the IM with XY view of L5 layer (I-K) and axial view (Z-axis) of same IM stack (I’-K’) shown for each: (I) CRNpro in Col-0 (n=6), (J) WUSpro in Col-0 (n=6), (K) WUSpro in crn (n=6). Tissue stained with propidium iodide (PI; magenta).
Statistical groupings based on significant differences found using Kruskal-Wallis and Dunn’s multiple comparison test correction (E). Scale bars, 50μm in (H), 20μm in (F-G) and (I-K).
Consistent with a role in flower primordia development, CRN expression was detected as early as incipient primordia (before primordia outgrowth) and remained throughout primordia formation (Figure 1I). Both CRN (CRN-GFP) and CLV2 (CLV2-Citrine) fusion proteins, expressed by native promoters, confirmed this expression pattern (Figures S1B and S1C). Supporting previous in situ results [15], WUS expression did not overlap with CRN spatially or temporally during primordia specification and outgrowth, when crn inflorescence phenotypes diverge from WT (Figures 1I and 1J). The WUS domain was expanded in crn IMs compared to WT but ectopic WUS expression was not detected in terminated crn primordia (Figures 1J and 1K) [17]. Additionally, other mutants known to have expanded WUS IM expression domains do not display clv2/crn primordia outgrowth defects (see below).
CLV2/CRN functions have been studied extensively in the Landsberg-erecta (Ler,) background [16, 17, 25]. We found no flower primordia outgrowth defects in null clv2-1 mutants in Ler, explaining why this phenotype has not been described in this ecotype (Figures S1D–S1E and S1I–S1J). Flower primordia termination was also not observed in crn-1 (Ler background [17]) or two CRISPR-derived null alleles of clv2 in Ler (clv2-10 and clv2-11; Figures S1F–S1H and S1I–S1K). F1 plants from an ecotype-hybrid clv2 null cross of rlp10-1 (Col-0) X clv2-1 (Ler) displayed mild termination (Figure S1L). Flower primordia outgrowth defects segregated in a digenic semi-dominant manner in the F2 population (Figure S1M), indicating that dominant modifiers in Col-0 underlie ecotype differences in clv2/crn flower outgrowth. Additionally, ecotypic differences were not attributable to the erecta (er) allele in Ler plants [26], as clv2 er double mutants in the Col-0 background had equivalent flower outgrowth defects to clv2 (Figures S1N–S1P). Double mutants between CLV2 and the floral identity gene LEAFY (LFY; clv2 lfy) displayed primordia outgrowth defects and lfy-like floral organ conversions (Figures S1Q–S1R), indicating that CLV2/CRN promote primordia formation independent of LFY-floral meristem specification [10, 27]. Collectively, these data show that CLV2/CRN signaling represents a novel ecotype-dependent process regulating primordia outgrowth following reproductive transition.
CLAVATA2/CRN-mediated flower outgrowth requires CIK co-receptors
The CLV2/CRN receptor complex lacks signaling capacity alone, as CRN is a transmembrane pseudokinase [19], suggesting CLV2/CRN require associated functional kinase(s) to signal. CLAVATA candidates with active kinase domains include the CLAVATA3 INSENSTIVE RECEPTOR KINASE1/2/3/4 (CIK1/2/3/4) family co-receptors, CLV1 and the CLV1-related BARELY ANY MERISTEM1/2/3 receptors (BAM1/2/3), which all regulate IM stem cell proliferation [28–31]. CLV1/BAM signals independent of CLV2/CRN in shoot and floral stem cell control [20]. Consistent with this, we found negligible amounts of flower primordia termination in clv1, bam1/2, or bam1/2/3 null mutants (in Col-0) and crn was additive in each higher order mutant combination (crn clv1, crn bam1/2, and crn bam1/2/3; Figures S2A–S2J). CIK1/2/3/4 are leucine-rich repeat (LRR)-II-receptor-like-kinase subfamily co-receptors with overlapping functions with CLAVATA primary receptors, several of which physically interact with CRN [31]. In a previous report we noticed a crn-like phenotype in specific cik mutant combinations [31]. To confirm this observation, we generated higher order CRISPR null alleles of CIK1/2/4 in Col-0 (cik1-3, cik2-3, cik4-3; Figure S2K). In contrast to clv1/bam mutants, cik1/2/4 displayed flower primordia termination equivalent to crn (Figures 2A–2C). Additionally, cik1/2/4 had enlarged floral meristems quantitatively similar to crn (Figure 2D). The protein phosphatase POLTERGEIST (POL), a downstream component of CLAVATA signaling, suppresses clv2/crn meristem size defects [17, 32]. pol restored flower outgrowth and internode elongation defects in crn (crn pol; Figures 2E–2H). Collectively, these data demonstrate that CLV2/CRN signal alongside CIK1/2/4 co-receptors to promote flower outgrowth through a POL-dependent pathway.
Figure 2. CLV2/CRN-mediated flower outgrowth requires CIK1/2/4 co-receptors and a downstream POL-dependent pathway.

(A-B) Inflorescence of crn and cik1/2/4 (cik1-3/cik2-3/cik4-3) mutant plants reveal similar flower termination defects. Quantification of (C) flower termination and (D) carpel number across Col-0 (n=16), crn (n=17), and cik1/2/4 (n=13).
(E-F) Inflorescence of crn and crn pol showing that flower termination is suppressed by pol (pol-6). Quantification of (G) flower termination and (H) carpel number across Col-0 (n=15), crn (n=15), pol (n=15), and crn pol (n=14).
Box and whisker plots show full range of data (min to max) with mean marked as “+”.
Statistical groupings based on significant differences found using Kruskal-Wallis and Dunn’s multiple comparison test correction (C-D and G-H).
See also Figure S2.
CLAVATA receptors respond to CLAVATA3(CLV3)/EMBRYO-SURROUNDING REGION (CLE) peptide ligand(s), and there are 32 CLEs in Arabidopsis [33]. CLV3 and a suite of redundant CLE peptides signal via CLV1 to repress IM stem cell proliferation parallel to CLV2/CRN [13, 17, 34, 35]. Flower primordia outgrowth is not impaired in clv3 or dodeca-cle higher order mutants, which combine clv3 with several redundant cle alleles [13], suggesting that additional unknown CLE peptides regulate flower outgrowth through CLV2/CRN/CIK (Figure S2L–S2M). Consistent with this, crn clv3 double mutants are additive with a clear disruption in flower primordial outgrowth and an enlarged disc-like IM (Figure S2N). The enlarged shoot and fasciated stem of crn clv3 made quantification of terminated primordia difficult; however, these data support previous work suggesting that CLV2/CRN can act independently of CLV3 [20].
Temperature and CLV2/CRN modulate auxin-dependent flower primordia outgrowth
Many developmental programs are robust, ensuring optimal morphology/function across varied conditions [13, 36]. Populations of A. thaliana can be found throughout the Northern Hemisphere thriving in diverse environments [7, 37]. Natural variation in traits like flowering time and freezing tolerance are influenced by and/or directly correlated with adaptations to local conditions [7]. While investigating clv2/crn we observed remarkable quantitative variability in flower primordia termination at different temperatures. Flower outgrowth defects in clv2/crn were suppressed when grown at higher temperatures (31°C) compared to colder/ambient temperatures (16°C/24°C; Figures 3A–3G). Previous work noted shoot defects in crn-1 mutants (Ler background) at high temperatures; however, we did not observe this under our conditions (Figures 3H–3I) [17]. Thermomorphogenic pathways regulate high temperature seedling growth by enhancing auxin biosynthesis [38, 39]. At higher temperatures, PHYTOCHROME INTERACTING FACTOR (PIF) family transcriptional regulators activate YUCCA (YUC) genes, which encode rate-limiting enzymes in auxin biosynthesis [40, 41]. Under colder/ambient temperatures, thermomorphogenesis is negatively regulated by the transcriptional repressor EARLY FLOWERING 3 (ELF3) [42]. As such, elf3 seedlings display constitutive thermomorphogenic responses and higher auxin production across temperatures. To test if the thermomorphogenesis pathway was sufficient to suppress clv2/crn flower outgrowth defects, we generated crn elf3 double mutants and grew them at colder/ambient temperatures. Consistent with high temperature mediated suppression of crn, flower primordia termination was suppressed in crn elf3 at colder temperatures (Figures 3J–3L). In contrast to the suppression of crn primordia outgrowth, elf3 slightly enhanced carpel numbers compared to crn (Figure 3M). This finding supports that CLV2/CRN-mediated outgrowth and CLV2/CRN-mediated meristem size regulation are separable with primordia outgrowth being highly sensitive to thermal conditions. Our data shows that CLV2/CRN/CIK signaling is critical for continuous flower production at colder/ambient temperatures but can be bypassed by thermomorphogenic responses to higher temperatures. As such, while Arabidopsis flower development is robust under various environmental conditions, distinct mechanisms maintain this stability across different temperatures.
Figure 3. High temperature responses modulate clv2/crn flower primordia outgrowth.
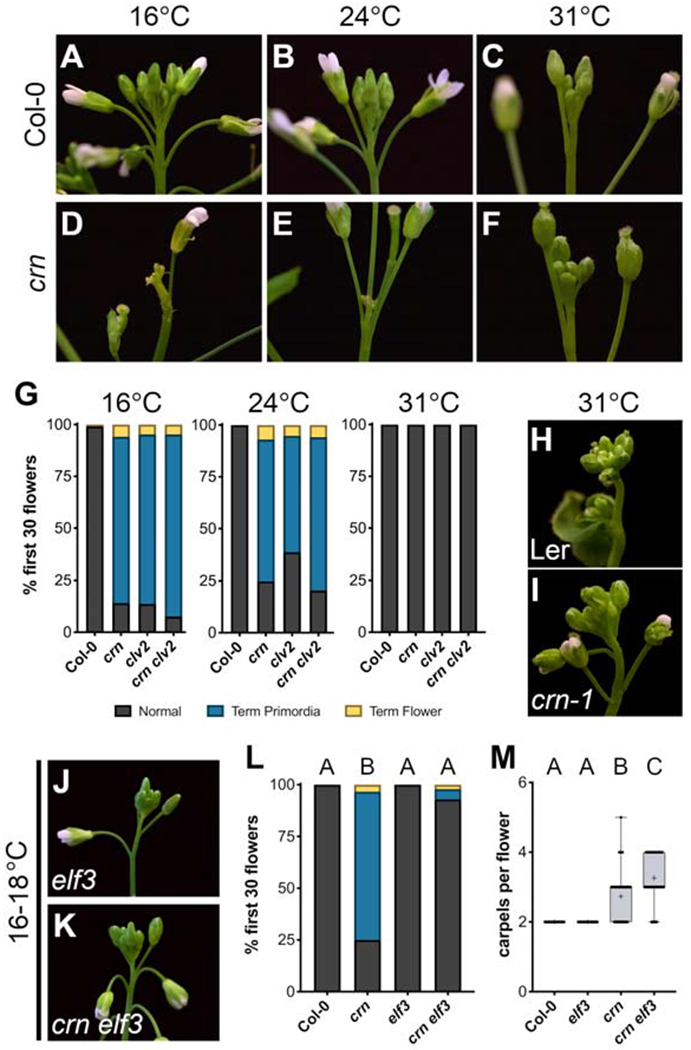
(A-F) Temperature-dependent inflorescence phenotypes of crn compared to Col-0. Flower primordia termination in crn is prevalent at (D) 16°C and (E) 24°C, but is suppressed at (F) 31°C.
(G) Quantification of flower primordia termination at 16°C: Col-0 (n=8), crn (n=9), clv2 (n=9), crn clv2 (n=7), 24°C: Col-0 (n=9), crn (n=9), clv2 (n=9), crn clv2 (n=9), and 31°C: Col-0 (n=8), crn (n=9), clv2 (n=9), crn clv2 (n=6).
(H-I) Inflorescences of Ler and crn-1 grown at 31°C.
(J-K) Inflorescences of elf3 (elf3-1) and crn elf3 grown at 16-18°C
(L-M) Quantification of (L) flower primordia termination and (M) carpel number in Col-0 (n=24), crn (n=18), elf3 (n=27), crn elf3 (n=26) grown at 16-18°C.
Box and whisker plots show full range of data (min to max) with mean marked as “+”.
Statistical groupings based on significant differences found using Kruskal-Wallis and Dunn’s multiple comparison test correction (L-M).
See also Figure S4.
To define the mechanisms underlying crn primordia termination, we used RNA-seq to identify differentially expressed genes (DEGs) in terminating crn IMs compared to WT. Using a strict cutoff (p-value < 0.001), we found 460 DEGs between crn and WT IMs, with 236 upregulated and 224 downregulated in crn (Figure 4A; Table S1). Enriched Gene Ontology (GO) terms among DEGs included meristem maintenance, flower development, and auxin function (Figure 4B; Table S2) [43–45]. The first two GO term groups are consistent with CRN’s role in meristem maintenance [17, 20], and flower development, documented in this study (Tables S1 and S2). The overrepresentation of auxin-associated genes in crn IM DEGs (Table S1) is complementary to the thermomorphogenic suppression of crn’s primordia termination (Figure 3), suggesting CLV2/CRN regulate auxin function during early flower primordia outgrowth. Therefore, we asked if clv2/crn were defective in auxin outputs at lower temperatures and if auxin biosynthesis was required for the high temperature suppression of clv2/crn. We visualized the auxin signaling reporter DR5::GFP (where GFP positively correlates with increased auxin signaling output) and the auxin perception reporter DII-Venus (where Venus negatively correlates with increased auxin perception) in terminating crn IMs [46, 47]. There was a significant reduction in DR5::GFP signal in crn IMs during termination, specifically in the L1 layer of incipient primordia (Figures 4C–4D and S3A–S3C). Consistently, DII::Venus accumulated in the L1 layer of terminated crn IMs, a pattern never observed in WT (Figures 4E–4F and S3D). DR5::GFP was restored to WT levels during crn’s recovery phase (Figures S3E–S3F). During flower development, the PIN-FORMED1 (PIN1) auxin efflux transporter concentrates auxin to the IM periphery, creating local maxima which trigger flower primordia initiation and subsequent outgrowth [4, 5]. PIN1 reporter levels (PIN1-GFP) [5] were decreased in terminating crn IMs (Figures S3G–S3H); consistent with PIN1 expression from our RNAseq DEG data. (Table S1). PIN1-GFP levels increased during crn’s recovery phase, but not to WT levels (Figure S3I). These data demonstrate an overall reduction in auxin signaling/perception within the IM and developing primordia of crn during the termination phase. This decrease is transient and corresponds with flower outgrowth defects, indicating that CLV2/CRN positively regulate auxin dependent flower primordia outgrowth in the IM.
Figure 4. Flower primordia outgrowth is maintained by separable auxin-dependent processes in different thermal conditions.

(A) Differentially expressed genes (DEG, p-value < 0.001) between crn and WT IMs; upregulated genes (red), downregulated genes (blue).
(B) Top GO terms enriched in crn DEGs, highlighting terms associated with flower development (blue), meristem maintenance (green) and auxin (red).
(C-D) Maximum intensity projection (MIP) of DR5::GFP (teal) in the IM of (C) Col-0 (n=14) and (D) crn (n=12). Stained with PI (magenta). Arrows point to primordia used for DR5 quantification/comparison.
(E-F) Axial view showing DII-Venus (green) expression pattern in (E) Col-0 (n=6) and (F) crn (n=8) IMs. Stained with PI (magenta). (E’-F’) L1 layer of IMs from (E’) Col-0 and (F’) crn used to quantify percent of L1 cells with DII marker. Arrows point to organ boundaries where DII reporter can be detected in the L1 of Col-0 and crn.
(G-H) Flower primordia termination across independent T1 lines of ANTpro::YUC1 in crn compared to Col-0 and crn when grown at (G) 16-18°C or (H) 22-24°C.
(I) clv2 yuc1/4 inflorescence grown at colder temperatures (16-18°C) and quantification of flower outgrowth defects in Col-0 (n=9), clv2 (n=9), yuc1/4 (n=8), clv2 yuc1/4 (n=10).
(J) clv2 yuc1/4 pin-like inflorescence grown at hot temperatures (28-31°C) and quantification of flower outgrowth defects, including instances of pins (pink bar; right y-axis) in Col-0 (n=29), clv2 (n=20), yuc1/4 (n=20), clv2 yuc1/4 (n=12).
Scale bars, 20μm in (C-D) and (E-F), 1mm in (I-J).
Bar plots show mean with SEM.
See also Figures S3 and S4, Tables S1–S3.
Several auxin biosynthetic genes had decreased expression in crn compared to WT, including YUC genes that regulate flower development (Figure S3J and Table S3) [48]. To test if low auxin levels contributes to crn’s flower outgrowth defects, we expressed YUC1 in developing primordia of crn using the AINTEGUMENTA promoter (ANTp::YUC1) [6, 49] and grew plants in colder (16-18°C) and ambient (22-24°C) temperatures. At 22-24°C, 9/13 T1 plants suppressed crn while only 2/18 plants partially suppressed crn at 16-18°C (Figures 4G–4H). This demonstrates that at ambient temperatures (where crn terminates), ectopic YUC1 can suppress flower outgrowth defects; however, the degree of suppression correlates with temperature. Higher order mutant combinations in IM-expressed YUC1/2/4/6 severely impair floral and vasculature development; however, yuc1/4 double mutants produce more typical inflorescences with identifiable flowers [48]. We generated clv2 yuc1/4 triple mutants to reduce YUC-dependent auxin in clv2 and test whether high temperature suppression of clv2/crn flower termination was dependent on YUC-mediated auxin biosynthesis. At 16-18°C, clv2 yuc1/4 triple mutants displayed rates of flower primordia termination comparable to clv2 (Figure 4I). As such, CLV2/CRN promote auxin mediated primordia outgrowth independent of YUC1/4 in colder temperatures. At 28-31 °C, some clv2 yuc1/4 triple mutant plants displayed clv2 flower primordia termination, indicating that at high temperatures YUC1/4 contribute to heat induced suppression (Figure 4J). Surprisingly though, the majority (~60%) of clv2 yuc1/4 plants had a synergistic response to high temperatures resulting in pin-like inflorescences completely lacking flower primordia (Figure 4J). These data suggest that high temperature suppression of clv2 is dependent on YUC1/4-mediated auxin biosynthesis and that under high temperatures, CLV2/CRN and YUC1/4 are synergistically required to maintain flower primordia initiation and outgrowth.
Ensuring robust development and reproduction across environments is a challenge all organisms face. Here we demonstrate that robust flower production in diverse thermal environments is achieved through the synergistic deployment of CLV2/CRN-signaling and ELF3-regulated auxin production via the thermomorphogenesis pathway in Arabidopsis. The relative contribution of each to flower development varies across thermal clines, with CLV2/CRN signaling being critical at colder/ambient temperatures and synergistic with heat-induced auxin production at higher temperatures (Figure S4). Arabidopsis seedlings respond to high temperatures by promoting auxin-dependent hypocotyl elongation, a process negatively regulated by ELF3 [38, 42]. Our work demonstrates that high temperatures and ELF3 also regulate auxin-dependent primordia production. Interestingly, clv2 yuc1/4 primordia outgrowth defects were strongly enhanced in warmer conditions. Higher temperatures also enhance penetrance of seedling defects in loss-of-function mutants in the TRANSPORT INHIBITOR RESPONSE1 (TIR)-family auxin receptors [50]. As such, heat might have an unappreciated negative impact on auxin function, with thermomorphogenesis-induced auxin playing a protective role rather than simply directing growth. How CLV2/CRN stimulate auxin-dependent flower initiation is unknown. Ectopic YUC1 expression or heat-induced auxin production is sufficient to restore primordia outgrowth to clv2/crn. This suggests that CLV2/CRN are not critically required for TIR-dependent auxin perception, ARF5/MONOPEROS dependent transcriptional activity, or the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA1)-mediated conversion of tryptophan to indole-3-pyruvate (IPA) step upstream of YUCCA in auxin biosynthesis [6, 51–53].
clv2/crn defects manifest early in inflorescence development and are transient. Recovery of primordia production is not linked to flower meristem identity or seed/fruit derived auxin production (clv2 Ify plants). This suggests that the transition from vegetative to reproductive meristem fate may be sensitized to CLV2/CRN signaling. Nevertheless, clv2 yuc1/4 plants reveals that CLV2/CRN signaling is required at later steps in inflorescence development as well. Heat stress is known to damage crops in ways that negatively impact yield, including the loss of flower production [54, 55]. As climate change increases global temperatures it will be necessary to mitigate heat impacts on crop yield. If the environmental buffering capacity of CLV2/CRN-signaling is conserved in crop species perhaps it could be deployed to help improve plant responses to climate change.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Information and resource/reagent requests should be directed to and will be fulfilled by the Lead Contact, Zachary Nimchuk (zackn@email.unc.edu).
Materials Availability
Plasmids and Arabidopsis lines made during this study are freely available to academic researchers through the Lead Contact.
Data and Code Availability
Raw RNAseq data described in this study has been deposited into the NCBI Short Read Archive (SRA) database under the BioProject PRJNA661065. Code used to analyze gene expression data can be found on the Nimchuk Lab GitHub page (https://github.com/NimchukLab). All other source data obtained throughout the course of this work have not been deposited to any public repository but are available upon request from the Lead Contact.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Arabidopsis thaliana accession Columbia (Col-0) was used as our primary model system throughout this work. Some phenotypic comparisons were also made with the accession Landsberg-erecta (Ler), as noted.
Plant growth conditions
Seeds were sterilized and plated on half-strength MS (Murashige-Skoog) media buffered with MES, pH 5.7. Plates were stratified in the dark at 4°C for 2 days and then moved to constant light in a custom-built grow room with environmental control (temperature maintained between 21-25°C), or in a Percival growth chamber (AR-75L3) when growing at a specified temperature. After 7-10 days, seedlings were transplanted to soil (Metro-Mix 360/sand/perlite supplemented with Marathon pesticide and Peter’s 20:20:20 [N:P:K] at recommended levels) and then placed back into the chamber they were germinated in and grown until flowering for phenotypic analysis.
METHOD DETAILS
Plant materials
Mutant alleles used in this study are all in the Col-0 background, unless otherwise noted, and information for each is as follows: crn (crn-10) [20], crn-1 in Ler [17], clv2 in Col-0 (rlp10-1) [21], clv2-1 in Ler [16], erecta (er-105) [56], Ify (lfy-1; ABRC CS6228) [27], clv1 (clv1-101) [57], baml (bam1-4) [30], bam2 (bam2-4) [30], bam3 (bam3-2) [30], pol (pol-6) [58], clv3 (clv3-9) [30], dodeca-cle (clv3-9; CRISPR alleles of: cle9, cle10, cle11, cle12, cle13, cle18, cle19, cle20, cle21, cle22, cle45) [13], elf3 (elf3-1; ABRC CS3787) [59], yucl (SALK_106293) [48], and yuc4 (SM_3_16128) [48]. Information for previously published transgenic complementation and reporter lines used are as follows: CRNpro::CRN-GFP in crn [20], CLV2pro::CLV2-CITRINE in clv2 [60], WUSpro::Ypet-N7 in Col-0 (gift from Paul Tarr – Caltech), DR5pro::GFP in Col-0 [61, 62], DII-Venus in Col-0 [63], PIN 1 pro::PIN 1-GFP in Col-0 [64]. The DR5pro::GFP, DII-Venus, and PIN1pro::PIN1-GFP lines were all crossed into crn (crn-10) for analysis of auxin signaling levels in crn shoots compared to WT.
CRISPR mutagenesis of CIK1/2/4 and CLV2
The pCUT vector system was used to simultaneously create cik1, cik2 and cik4 mutations in the Col-0 background (cik1-3, cik2-3, cik4-3), making the cik1/2/4 higher order mutant [65]. Similarly, the pCUT system was used to make multiple unique mutations in clv2 in the Ler background (clv2-10, clv2-11). A pENTR-D/TOPO entry vector was modified for golden gate cloning by TOPO cloning in Pmel and Bsal cut sites (pENTR-GG). pENTR-GG was used to clone tandem cassettes (cik1/2/4) or a single cassette (clv2) of the U6 promoter, 20 bp guide sequence, and the gRNA scaffold. These guide constructs were then gateway cloned into pCUT4GTWvector that expresses Cas9 from the UBIQUITIN10 promoter. Hygromycin resistant plants were selected in the T1 generation and sequenced to detect editing of the target genes; CIK1 (AT1G60800), CIK2 (AT2G23950), and CIK4 (AT5G45780) or CLV2 (AT1G65380). To make stable cik1/2/4, T2 seed from editing cik1/2/4 lines were grown for 2 weeks under standard conditions, heat shocked for 12-24 hours at 35°C [66], and screened for stable edits using dCAPS primers designed by the indCAPS webtool (http://indcaps.kieber.cloudapps.unc.edu/) [67]. Cas9 was segregated out of plants that had stable homozygous mutations in CIK1/2/4. To make stable clv2 alleles in Ler, T2 seed, collected from single branches of T1 plants that had the clv2 carpel phenotype, were grown in a Percival growth chamber at 16°C (see above), screened for stable mutations while segregating Cas9 out prior to phenotypic analysis.
Columbia-0 X Landsberg-erecfa hybrid clv2 population
Null clv2 lines in the Col-0 (r1p10-1) and Ler (clv2-1) backgrounds were crossed to generate a hybrid population with fixed clv2 mutations. Segregation of flower termination traits was assessed in the F2 generation. Phenotypic ratios were compared to expected values of a single causative locus (1:2:1) and digenic semi-dominant modifiers (7:6:3) using Chi-squared analysis (Microsoft Excel v.16.40) to determine the underlying genetic complexity of the ecotypic variability of clv2 termination.
Generation of binary vectors and transgenic lines
New transgenic lines were generated using floral-dip transformation of binary vectors into specified backgrounds [68]. pWUSpro::Ypet-N7 (gift from Paul Tarr – Caltech; cloning methods as in [69] but with Ypet-N7) was transformed directly into crn and the transgene was selected on ½ MS plates with Kanamycin. Four independent lines were selected for downstream analysis with the WUS expression domain being equivalent across all lines imaged (see imaging methods for data acquisition details). For pCRNpro::Ypet-N7, a pENTR-D Ypet-N7 (2xYpet-N7 fusion) entry vector was recombined by LR reaction into the pCRNpro::Gateway binary vector in the pMOA33 background [20]. For pANTpro::YUC1, the 4 kb 5’ ANT promoter was amplified from genomic DNA, and cloned up stream of a gateway::OCS terminator cassette in the pCR2.1 shuttle vector, and sequence verified. The resulting ANTpro::Gateway::OCS cassette was then mobilized as a Not1 fragment in to the pMOA33 binary vector backbone as previously described to create pANTpro::GTW [30]. The YUC1 CDS was amplified from Arabidopsis Col-0 cDNA and cloned into the pENTR-D topo vector and sequence verified. This vector was then recombined in an LR reaction into pANTpro::GTW to create the pANTpro::YUC1.
Photography and time-lapse imaging
Unless specified differently below, young inflorescences were staged at similar developmental timepoints and photographed using a Canon EOS Rebel T5 equipped with a Tokina 100mm f/2.8 AT-X M100 AF Pro D macro lens. Images were edited for brightness and contrast using Gimp v2.10.4 (https://www.gimp.org/). Young inflorescences from Col-0, crn, clv2, crn clv2, cik1/2/4, yuc1/4, and clv2 yuc1/4 (Figures 1A’–D’, 2B, 4E–F, and 4H–I) were imaged using a Zeiss Stemi 2000-C stereo microscope equipped with a Zeiss Axiocam 105-color digital camera and acquired using Zeiss ZEN software. Time-lapse imaging was done using the Lapse-it (http://www.lapseit.com/) app on an iPhone 6 operating iOS 12.3.1. Imaging began with ~3-week-old Col-0 (WT) and crn plants growing in our custom-built grow room. A single image was taken every 30 minutes over the course of 10-12 days. The final movie was compiled at 30 frames per sec and exported into iMovie where it was cropped to show only two plants for comparison of early flowering phenotypes.
Confocal microscopy
Live imaging of inflorescence meristems (IMs) was performed as previously reported [20, 70]. We used either an inverted Zeiss 710 (for: propidium iodide (PI) stained Col-0, crn, CRNpro:Ypet-N7, WUSpro::Ypet-N7, DR5pro::GFP, DII-Venus, and PIN1pro::PIN1-GFP) or a Zeiss 880 (for CRNpro::CRN-GFP and CLV2pro::CLV2-CITRINE) confocal laser scanning microscope equipped with an inverter (setup described in [71]). Young IMs were dissected immediately following floral transition in order to analyze expression patterns and reporter levels at the same developmental stage as clv2/crn flower primordia termination. When analyzing reporter status in recovered IMs of crn, shoots were dissected at a later timepoint after flower buds were visibly developing again. All IMs were briefly (~5 mins for WT and ~15 mins for crn) stained with PI (final concentration of 50μg/mL for WT shoots and 15μpg/mL for crn) on ice and placed into a petri dish with 2% agarose (w/v) and immersed in cold water, ensuring no bubbles formed around the IM [70]. IMs were imaged using a W Plan-APOCHROMAT 40X (NA = 1.0) water dipping objective. Laser excitation and detected emission ranges were as follows: PI only – laser 561nm diode, PI channel 600-750nm; Ypet/Venus markers with PI – 514nm argon laser, Ypet channel 520-581nm, PI channel 655-758nm; GFP markers (on Zeiss 710) with PI – 488nm argon laser, GFP channel 493-556nm, PI channel 598-642nm; GFP on Zeiss 880 with PI – 488nm argon laser, GFP channel (GaAsP detector) 490-550nm, PI channel (GaAsP detector) 565-610nm; Citrine on Zeiss 880 with PI – 514nm argon laser, Citrine channel (GaAsP detector) 519-550nm, PI channel (GaAsP detector) 565-610nm. Whole IMs were imaged as a z-stack series with a step size optimized for three-dimensional reconstruction of data. All images comparing reporter levels in different backgrounds were obtained with identical specifications: DR5pro::GFP, DII-Venus, and PIN1pro::PIN1-GFP in both WT and crn shoots. Live inflorescence micrographs were all post-processed using ZEN (Zeiss) for three-dimensional reconstructions of IMs and Fiji/ImageJ v.2.0.0-rc-69/1.52u (National Institutes of Health) [72] for single scan images as well as axial views of IMs. Channels corresponding to PI staining in crn IMs were almost always gamma corrected (0.8) as penetrance of this fluorescent dye in enlarged crn shoots was sometimes limited. DR5pro::GFP fluorescence quantification comparing WT and crn IMs was done as follows using Fiji/ImageJ v.2.0.0-rc-69/1.52u. Z-stacks were rendered as maximum intensity projections of the GFP channel only using data from the entire IM. A region of interest (ROI) was drawn around each of the first 3 developing primordia (identified as the 3 early primordia with the highest GFP intensities). GFP levels were quantified for each ROI, normalized to the final area of each ROI, and then averaged together to obtain a single value for WT and crn IMs. DII-Venus quantification was done using Fiji/ImageJ v.2.0.0-rc-69/1.52u. Due to rapid photobleaching of the Venus fluorescent reporter, single scan images were taken in the L1 layer of WT and crn IMs prior to z-stack scans and used for direct comparison. The percentage of L1 cells with the Venus reporter were determined across all imaged IMs, with WT IMs never having reporter in this layer.
Whole shoot reconstruction of crn during termination (Figure 1H) was done using fixed and cleared tissue, imaging structural autofluorescence (as in [13]). Young crn IM were fixed in FAA (2% formaldehyde, 5% acetic acid, 60% ethanol (w/v)) at 4°C overnight and then dehydrated in a graded ethanol series (70%, 80%, 95% and 100%) for 30 minutes each at room temperature. Tissue was then cleared overnight in methyl salicylate (catalog no. M6752; Sigma Aldrich) and placed in a small glass-bottom petri dish (catalog no. P35G-1.5-10-C; MatTek Corporation) and imaged on a Zeiss 710 CLSM using a Plan-APOCHROMAT 10X (NA = 0.45). Autofluorescence was detected using a 488nm argon laser for excitation and combining two channels for emission detection; Channel 1 – 504-597nm and channel 2 – 629-731nm. Data was gathered as a z-stack and three-dimensional reconstruction was done in Nikon NIS-Elements as a shaded render of both channels combined in grayscale.
RNA Sequencing and Data Analysis
Total RNA was isolated using the EZNA Plant RNA kit (Omega Bio-tek) from 45-50 inflorescence meristems for three biological replicates of both Col-0 and crn (crn-10) plants. RNA was treated with RNase-free DNase (Omega Bio-tek). Approximately 1.5 ug RNA was used as input material for library preparation, using the Stranded mRNA-Seq kit (Kapa Biosystems) at the High-throughput Sequencing Facility at UNC Chapel hill. 50bp paired-end reads were generated on the NovaSeq 6000 sequencer (illumina) with a read depth of 23-35 million reads per biological replicate. Trimmed raw data was aligned to the A. thaliana genome (TAIR10.1) using HISAT2 version 2.2.0 [73] and reads were counted using Subread version 1.5.1 [74]. Subsequent analysis was performed on RStudio with reads normalized using EDASeq version 2.22.0 [75] and RUVseq version 1.22.0 (upper quartile normalization) [76] and differentially expressed genes were identified with a p-value <0.001 using EdgeR version 3.33.0 [77]. These top 460 DEGs were used for GO term analysis from Panther [78]. To obtain the average TPM counts of auxin biosynthetic genes, reads were pseudoaligned to the Arabidopsis transcriptome (TAIR10 from plants.ensembl.org) using Kallisto version 0.44.0 [79] and quantified with Sleuth version 0.33.0 [80].
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative data from all experiments was compiled and analyzed in GraphPad Prism v.8.3.2. We performed at least two biological replicates for each experiment ensuring consistent results (sample sizes indicated in figure legends). For comparisons of flower termination across conditions, genotypes and/or transgenic lines (Figures 1E, 2C, 2G, 3G, 3L, 4G–H, S1I, S2C–D, S2I–J), only % flower primordia termination (as defined in the paper) was compare across samples using a non-parametric Kruskal-Wallis and a Dunn’s multiple comparison test correction where significance was defined as p-value < 0.05. For comparisons of carpel number across genotypes (Figures 2D, 2H, 3M, and S1J), 10 consecutive flowers on the primary inflorescence were counted, starting after the recovery phase in crn, clv2, and cik1/2/4 while starting at the 11th flower in genotypes that had no flower primordia termination or partially suppressed flower primordia termination (Col-0, Ler, clv2-1, crn-1, pol, crn pol, elf3, and crn elf3). Carpel number was compared statistically using a non-parametric Kruskal-Wallis and a Dunn’s multiple comparison test correction where significance was defined as p-value < 0.05. For comparison of DR5pro::GFP levels, WT and crn IM values were compared statistically using an unpaired t-test, where the p-value = 0.0007. Sample size (n) for all analyses can be found with each figure in the legend and refers to the number of individual plants analyzed.
Supplementary Material
Table S1. Normalized differentially expressed genes (DEG) list in crn IMs. Related to Figure 4 and Table S2
Genes with p-value < 0.001 showing their log2FC, p-value and false discovery rate (FDR). Red log2FC values indicate upregulation and blue log2FC values indicate downregulation. Red, green and blue highlights refer to GO processes indicating auxin, meristem and floral related genes, respectively, while purple highlights indicate the gene belongs to multiple of these groups.
Video S1 – Time-lapse of flower termination and recovery in crn. Related to Figure 1
Time-lapse of Col-0 (left) and crn (right) during early stages of flowering revealing both crn’s termination and recovery phases. Entire movie spans ~10 days. Movie rendered at 30 frames per second while each frame was captured in 30-minute intervals (see methods).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli strain DH5α | Widely distributed | N/A |
| E. coli strain DB3.1 | Widely distributed | N/A |
| Agrobacterium tumefaciens strain GV3101 | Widely distributed | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Propidium Iodide (PI) | Thermo Fisher | Cat#P1304MP |
| Murashige and Skoog (MS) Basal Salt Micronutrient Solution | Sigma-Aldrich | Cat#M0529 |
| Methyl Salicylate | Sigma-Aldrich | Cat#M6752 |
| Critical Commercial Assays | ||
| Gateway LR Clonase II Enzyme mix | Thermo Fisher | Cat#11791020 |
| Gateway BP Clonase II Enzyme mix | Thermo Fisher | Cat#11789020 |
| pENTR/D-TOPO Cloning Kit | Thermo Fisher | Cat#K240020 |
| Golden Gate Assembly Mix | NEB | Ca#E1601S |
| EZNA Plant RNA Kit | Omega Bio-tek | SKU:R6827-01 |
| RNase-free DNase | Omega Bio-tek | SKU:E1091-02 |
| Stranded mRNA-seq Kit | Kapa Biosystems | Cat#07962169001 |
| Deposited Data | ||
| Raw RNAseq data | This paper | SRA BioProject: PRJNA66106 |
| Code to analyze RNAseq data | This paper | https://github.com/NimchukLab |
| Arabidopsis thaliana TAIR10.1 reference genome | The Arabidopsis Information Resource (TAIR) | SRA BioSample: SAMN03081427 |
| Arabidopsis thaliana TAIR10 Transcriptome | EnsemblPlants | https://plants.ensembl.org/Arabidopsis_thaliana/Info/Index |
| Experimental Models: Organisms/Strains | ||
| Arabidopsis thaliana: Col-0 ecotype | N/A | N/A |
| Arabidopsis: crn-10 | [20] | N/A |
| Arabidopsis: rlp10-1 | [20] | GABI_686A09 |
| Arabidopsis: er-105 | [56] | N/A |
| Arabidopsis: lfy-1 | [27] | ABRC CS6228 |
| Arabidopsis: clv3-9 | [30] | N/A |
| Arabidopsis: dodeca-cle | [13] | N/A |
| Arabidopsis: clv1-101 | [57] | N/A |
| Arabidopsis: bam1-4 | [30] | N/A |
| Arabidopsis: bam2-4 | [30] | N/A |
| Arabidopsis: bam3-2 | [30] | N/A |
| Arabidopsis: pol-6 | [58] | N/A |
| Arabidopsis: elf3-1 | [59] | ABRC CS3787 |
| Arabidopsis: yuc1 | [48] | SALK_106293 |
| Arabidopsis: yuc4 | [48] | SM_3_16128 |
| Arabidopsis thaliana: Landsberg erecta (Ler) ecotype | N/A | N/A |
| Arabidopsis: crn-1 (Ler) | [17] | ABRC CS9853 |
| Arabidopsis: clv2-1 (Ler) | [16] | N/A |
| Arabidopsis: crn CRNpro::CRN-GFP | [20] | N/A |
| Arabidopsis: clv2 CLV2pro::CLV2-CITRINE | [60] | N/A |
| Arabidopsis: Col-0 WUSp:YN7 | gift from Paul Tarr - Caltech | N/A |
| Arabidopsis: Col-0 DR5-GFP | [61–62] | N/A |
| Arabidopsis: Col-0 DII-Venus | [63] | N/A |
| Arabidopsis: Col-0 PIN1pro::PIN1-GFP | [64] | N/A |
| Arabidopsis: crn WUSp:YN7 | This study | N/A |
| Arabidopsis: cik1-3 | This study | N/A |
| Arabidopsis: cik2-3 | This study | N/A |
| Arabidopsis: cik4-3 | This study | N/A |
| Arabidopsis: clv2-10 (Ler) | This study | N/A |
| Arabidopsis: clv2-11 (Ler) | This study | N/A |
| Oligonucleotides | ||
| cik1 BslI dCAPS F | dCAPs primer for cik1 genotyping | CTGGTTTTTGTATGGTTCTTTGATATCTCTTCTGCTACACTTCCTC |
| cik1 BslI dCAPS R | dCAPs primer for cik1 genotyping | GATAAATTCATAGAGAAAAAAATTGAGATTTCTCC |
| cik2 BslI dCAPS F | dCAPs primer for cik2 genotyping | CATCTTTGTTTTGTTTTTTCCCCCTTCCAGAG |
| cik2 BslI dCAPS R | dCAPs primer for cik2 genotyping | CAAACAAAATAATCTTACACTTGTCGAAG |
| cik4 BslI dCAPS F | dCAPs primer for cik4 genotyping | AACAAGATGAAAGATGAGAAAGAGGCCTT |
| cik4 BslI dCAPS R | dCAPs primer for cik4 genotyping | CTCACAGAGAAACCACAAAACCTTC |
| ANT Pro F | AINTEGUMENTA promoter for pANTpro:GTW | GGGAGCTCGTCGAGCGGCCGAAATTGGAATTATTCATATTAATGAGTTAGC |
| ANT Pro R | AINTEGUMENTA promoter for pANTpro:GTW | CCCCGGGGGTTTCTTTTTTTGGTTTCTGCTTCTCTTCTTTC |
| YUC1 F | YUCCA1 CDS for entry vector | CACCAAAAAATGGAGTCTCATCCTCACAACAAAACTGACCAG |
| YUC1 R | YUCCA1 CDS for entry vector | TTAGGATTTAGAGGTAAAGACAAAACGAGAACTGCACATATTCC |
| Recombinant DNA | ||
| Plasmid ANTp:YUC1 | This study | N/A |
| Plasmid CRNp:YN7 | This study | N/A |
| Plasmid pENTR-GG | This study | N/A |
| Software and Algorithms | ||
| Fiji/ImageJ v.2.0.0-rc-69/1.52u | National Institutes of Health | https://imagej.net/Fiji |
| Zen Microscope Imaging Software | Zeiss | https://www.zeiss.com/ |
| NIS-Elements Imaging Software | Nikon | https://www.microscope.healthcare.nikon.com/products/software |
| GIMP GNU image manipulation program 2.10.4 | GIMP | https://www.gimp.org/ |
| Lapse-it iOS 12.3.1 | Apple app store | http://www.lapseit.com/ |
| Prism v.8.3.2 | GraphPad | https://www.graphpad.com/ |
| indCAPS | [67] | http://indcaps.kieber.cloudapps.unc.edu/ |
| HISAT2 2.2.0 | [73] | https://github.com/DaehwanKimLab/hisat2 |
| Subread 1.5.1 | [74] | https://github.com/DeskGen/subread |
| EDASeq 2.22.0 | [75] | https://github.com/drisso/EDASeq |
| RUVseq 1.22.0 | [76] | https://github.com/drisso/RUVSeq |
| EdgeR 3.33.0 | [77] | https://gist.github.com/jdblischak/11384914 |
| Kallisto 0.44.0 | [79] | https://github.com/pachterlab/kallisto |
| Sleuth 0.33.0 | [80] | https://github.com/pachterlab/sleuth |
| GO term analysis Panther | [78] | http://geneontology.org/ |
| Other | ||
| Small glass-bottom petri dish | MatTek Corporation | Cat#P35G-1.5-10-C |
ACKNOWLEDGEMNTS
We thank Yunde Zhao, the ABRC stock center, Paul Tarr, Joe Kieber, and Elena Shpak for sharing seeds, vectors, and reporter lines. We thank Tony D. Perdue, director of the UNC Biology Microscopy Core, for assistance with imaging. We thank Jamie Winshell and James Garzoni for lab and plant growth facility support. We thank UNC’s High-Throughput Sequencing Facility for sequencing services. We thank members of the Nimchuk lab for critical feedback on this project. This research was supported by a NIGMS-MIRA award from National Institutes of Health (R35GM119614) and the National Science Foundation (NSF) Plant Genome Research Program (PGRP; IOS-1546837) to Z.L.N. D.S.J. is supported by an NSF Postdoctoral Research Fellowship in Biology through the PGRP (NSF# 1906389).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- 1.Hallgrimsson B, Green RM, Katz DC, Fish JL, Bernier FP, Roseman CC, Young NM, Cheverud JM, and Marcucio RS (2019). The developmental-genetics of canalization In Seminars in cell & developmental biology, Volume 88 (Elsevier; ), pp. 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradley D, Ratcliffe O, Vincent C, Carpenter R, and Coen E (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83. [DOI] [PubMed] [Google Scholar]
- 3.Reinhardt D, Mandel T, and Kuhlemeier C (2000). Auxin regulates the initiation and radial position of plant lateral organs. The Plant Cell 12, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, and Friml J (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- 5.Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, and Meyerowitz EM (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Current biology 15, 1899–1911. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi N, Wu M-F, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, and Wagner D (2013). A molecular framework for auxin-mediated initiation of flower primordia. Developmental cell 24, 271–282. [DOI] [PubMed] [Google Scholar]
- 7.Weigel D (2012). Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant physiology 158, 2–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somssich M, Je BI, Simon R, and Jackson D (2016). CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143, 3238–3248. [DOI] [PubMed] [Google Scholar]
- 9.Pierre-Jerome E, Drapek C, and Benfey PN (2018). Regulation of division and differentiation of plant stem cells. Annual review of cell and developmental biology 34, 289–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weigel D, Alvarez J, Smyth DR, Yanofsky MF, and Meyerowitz EM (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- 11.Pidkowich MS, Klenz JE, and Haughn GW (1999). The making of a flower: control of floral meristem identity in Arabidopsis. Trends in plant science 4, 64–70. [DOI] [PubMed] [Google Scholar]
- 12.Clark SE, Running MP, and Meyerowitz EM (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067. [Google Scholar]
- 13.Rodriguez-Leal D, Xu C, Kwon C-T, Soyars C, Demesa-Arevalo E, Man J, Liu L, Lemmon ZH, Jones DS, and Van Eck J (2019). Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nature genetics 51, 786–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soyars CL, James SR, and Nimchuk ZL (2016). Ready, aim, shoot: stem cell regulation of the shoot apical meristem. Current opinion in plant biology 29, 163–168. [DOI] [PubMed] [Google Scholar]
- 15.Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, and Laux T (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- 16.Jeong S, Trotochaud AE, and Clark SE (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. The Plant Cell 11, 1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller R, Bleckmann A, and Simon R (2008). The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. The Plant Cell 20, 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bleckmann A, Weidtkamp-Peters S, Seidel CA, and Simon R (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant physiology 152, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nimchuk ZL, Tarr PT, and Meyerowitz EM (2011). An evolutionarily conserved pseudokinase mediates stem cell production in plants. The Plant Cell 23, 851–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nimchuk ZL (2017). CLAVATA1 controls distinct signaling outputs that buffer shoot stem cell proliferation through a two-step transcriptional compensation loop. PLoS genetics 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, Mitchell K, Bastas K, Liu C-M, Woods-Tör A, and Zipfel C (2008). A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant physiology 147, 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth DR, Bowman JL, and Meyerowitz EM (1990). Early flower development in Arabidopsis. The Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy GV, Heisler MG, Ehrhardt DW, and Meyerowitz EM (2004). Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development 131, 4225–4237. [DOI] [PubMed] [Google Scholar]
- 24.Long J, and Barton MK (2000). Initiation of axillary and floral meristems in Arabidopsis. Developmental biology 218, 341–353. [DOI] [PubMed] [Google Scholar]
- 25.Kayes JM, and Clark SE (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843–3851. [DOI] [PubMed] [Google Scholar]
- 26.Rédei J (1992). A note on Columbia wild type and Landsberg erecta. Methods in Arabidopsis Research 3. [Google Scholar]
- 27.Schultz EA, and Haughn GW (1991). LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. The Plant Cell 3, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark SE, Williams RW, and Meyerowitz EM (1997). The CLAVATA1gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- 29.DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, and Clark SE (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. The Plant Journal 45, 1–16. [DOI] [PubMed] [Google Scholar]
- 30.Nimchuk ZL, Zhou Y, Tarr PT, Peterson BA, and Meyerowitz EM (2015). Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142, 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu C, Zhu Y, Cui Y, Cheng K, Liang W, Wei Z, Zhu M, Yin H, Zeng L, and Xiao Y (2018). A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nature plants 4, 205–211. [DOI] [PubMed] [Google Scholar]
- 32.Song S-K, Lee MM, and Clark SE (2006). POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 133, 4691–4698. [DOI] [PubMed] [Google Scholar]
- 33.Goad DM, Zhu C, and Kellogg EA (2017). Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function. New Phytologist 216, 605–616. [DOI] [PubMed] [Google Scholar]
- 34.Fletcher JC, Brand U, Running MP, Simon R, and Meyerowitz EM (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- 35.Ogawa M, Shinohara H, Sakagami Y, and Matsubayashi Y (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294–294. [DOI] [PubMed] [Google Scholar]
- 36.Lachowiec J, Queitsch C, and Kliebenstein DJ (2016). Molecular mechanisms governing differential robustness of development and environmental responses in plants. Annals of botany 117, 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alonso-Blanco C, El-Assal SE-D, Coupland G, and Koornneef M (1998). Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149, 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray WM, Östin A, Sandberg G, Romano CP, and Estelle M (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proceedings of the National Academy of Sciences 95, 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, and van Zanten M (2016). Molecular and genetic control of plant thermomorphogenesis. Nature plants 2, 1–9. [DOI] [PubMed] [Google Scholar]
- 40.Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, and Cohen JD (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proceedings of the National Academy of Sciences 108, 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, Qi L, Li Y, Chu J, and Li C (2012). PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS genetics 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, and Webb AA (2015). ELF3 controls thermoresponsive growth in Arabidopsis. Current biology 25, 194–199. [DOI] [PubMed] [Google Scholar]
- 43.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, and Eppig JT (2000). Gene ontology: tool for the unification of biology. Nature genetics 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Consortium GO (2019). The gene ontology resource: 20 years and still GOing strong. Nucleic acids research 47, D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, and Thomas PD (2019). Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v. 14.0). Nature protocols 14, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, and Weisbeek P (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472. [DOI] [PubMed] [Google Scholar]
- 47.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, and Jürgens G (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. [DOI] [PubMed] [Google Scholar]
- 48.Cheng Y, Dai X, and Zhao Y (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes & development 20, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krizek B (2009). AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant physiology 150, 1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prigge MJ, Platre M, Kadakia N, Zhang Y, Greenham K, Szutu W, Pandey BK, Bhosale RA, Bennett MJ, and Busch W (2020). Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, and Yao H (2011). The main auxin biosynthesis pathway in Arabidopsis. Proceedings of the National Academy of Sciences 108, 18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, and Zhao Y (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proceedings of the National Academy of Sciences 108, 18518–18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavy M, and Estelle M (2016). Mechanisms of auxin signaling. Development 143, 3226–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Björkman T, and Pearson KJ (1998). High temperature arrest of inflorescence development in broccoli (Brassica oleracea var. italica L.). Journal of Experimental Botany 49, 101–106. [Google Scholar]
- 55.Anderson R, Bayer PE, and Edwards D (2020). Climate change and the need for agricultural adaptation. Current Opinion in Plant Biology. [DOI] [PubMed] [Google Scholar]
- 56.Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, and Komeda Y (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. The Plant Cell 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, and Sawa S (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911–3920. [DOI] [PubMed] [Google Scholar]
- 58.Lita PY, Miller AK, and Clark SE (2003). POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Current Biology 13, 179–188. [DOI] [PubMed] [Google Scholar]
- 59.Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, Meeks-Wagner DR, and Kay SA (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274, 790–792. [DOI] [PubMed] [Google Scholar]
- 60.Hazak O, Brandt B, Cattaneo P, Santiago J, Rodriguez-Villalon A, Hothorn M, and Hardtke CS (2017). Perception of root-active CLE peptides requires CORYNE function in the phloem vasculature. EMBO reports 18, 1367–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, and Scheres B (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. [DOI] [PubMed] [Google Scholar]
- 62.Zhang W, To JP, Cheng CY, Eric Schaller G, and Kieber JJ (2011). Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. The Plant Journal 68, 1–10. [DOI] [PubMed] [Google Scholar]
- 63.Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, and Wells D (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Molecular systems biology 7, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiśniewska J, Xu J, Seifertová D, Brewer PB, Růžička K, Blilou I, Rouquié D, Benková E, Scheres B, and Friml J (2006). Polar PIN localization directs auxin flow in plants. Science 312, 883–883. [DOI] [PubMed] [Google Scholar]
- 65.Peterson BA, Haak DC, Nishimura MT, Teixeira PJ, James SR, Dangl JL, and Nimchuk ZL (2016). Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PloS one 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, and Jacob Y (2018). Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. The Plant Journal 93, 377–386. [DOI] [PubMed] [Google Scholar]
- 67.Hodgens C, Nimchuk ZL, and Kieber JJ (2017). indCAPS: A tool for designing screening primers for CRISPR/Cas9 mutagenesis events. PloS one 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clough SJ, and Bent AF (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The plant journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- 69.Ishihara H, Sugimoto K, Tarr PT, Temman H, Kadokura S, Inui Y, Sakamoto T, Sasaki T, Aida M, and Suzuki T (2019). Primed histone demethylation regulates shoot regenerative competency. Nature communications 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prunet N, Jack TP, and Meyerowitz EM (2016). Live confocal imaging of Arabidopsis flower buds. Developmental biology 419, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nimchuk ZL, and Perdue TD (2017). Live imaging of shoot meristems on an inverted confocal microscope using an objective lens inverter attachment. Frontiers in plant science 8, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, and Schmid B (2012). Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pertea M, Kim D, Pertea GM, Leek JT, and Salzberg SL (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11, 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liao Y, Smyth GK, and Shi W (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic acids research 41, e108–e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Risso D, Schwartz K, Sherlock G, and Dudoit S (2011). GC-Content Normalization for RNA-Seq Data. BMC Bioinformatics 12, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Risso D, Ngai J, Speed TP, and Dudoit S (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nature biotechnology 32, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, and Narechania A (2003). PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Research 13, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology 34, 525–527. [DOI] [PubMed] [Google Scholar]
- 80.Pimentel H, Bray NL, Puente S, Melsted P, and Pachter L (2017). Differential analysis of RNA-seq incorporating quantification uncertainty. Nature methods 14, 687–690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Normalized differentially expressed genes (DEG) list in crn IMs. Related to Figure 4 and Table S2
Genes with p-value < 0.001 showing their log2FC, p-value and false discovery rate (FDR). Red log2FC values indicate upregulation and blue log2FC values indicate downregulation. Red, green and blue highlights refer to GO processes indicating auxin, meristem and floral related genes, respectively, while purple highlights indicate the gene belongs to multiple of these groups.
Video S1 – Time-lapse of flower termination and recovery in crn. Related to Figure 1
Time-lapse of Col-0 (left) and crn (right) during early stages of flowering revealing both crn’s termination and recovery phases. Entire movie spans ~10 days. Movie rendered at 30 frames per second while each frame was captured in 30-minute intervals (see methods).
Data Availability Statement
Raw RNAseq data described in this study has been deposited into the NCBI Short Read Archive (SRA) database under the BioProject PRJNA661065. Code used to analyze gene expression data can be found on the Nimchuk Lab GitHub page (https://github.com/NimchukLab). All other source data obtained throughout the course of this work have not been deposited to any public repository but are available upon request from the Lead Contact.


