Abstract
Idiopathic or “unexplained” infertility represents as many as 30% of infertility cases worldwide. Conception, implantation, and term delivery of developmentally healthy infants require chromosomally normal (euploid) eggs and sperm. The crux of euploid egg production is error-free meiosis. Pathologic genetic variants dysregulate meiotic processes that occur during prophase I, meiotic resumption, chromosome segregation, and in cell-cycle regulation. This dysregulation can result in chromosomally abnormal (aneuploid) eggs. In turn, egg aneuploidy leads to a broad range of clinical infertility phenotypes, including primary ovarian insufficiency and early menopause, egg fertilization failure and embryonic developmental arrest, or recurrent pregnancy loss. Therefore, maternal genetic variants are emerging as infertility biomarkers, which could allow informed reproductive decision-making. Here, we select and deeply examine human genetic variants that likely cause dysregulation of critical meiotic processes in 14 female infertility-associated genes: SYCP3, SYCE1, TRIP13, PSMC3IP, DMC1, MCM8, MCM9, STAG3, PATL2, TUBB8, CEP120, AURKB, AURKC, and WEE2. We discuss the function of each gene in meiosis, explore genotype-phenotype relationships, and delineate the frequencies of infertility-associated variants.
Introduction
Healthy, chromosomally normal (euploid) eggs and sperm are essential to implantation, full-term pregnancy and healthy infant development. The contribution of low-quality eggs to female infertility is not fully understood, partly because human egg scarcity limits robust investigations. Egg quality decreases with increasing maternal age, but other causes of poor egg quality likely exist (Lindsay et al., 2015). Despite being of an optimal maternal age and the absence of reproductive pathology, many women have difficulty conceiving (Nandi et al., 2016) or endure recurrent idiopathic miscarriage (El Hachem et al., 2017). Why some women face exceptional difficulty conceiving and carrying a pregnancy to term is poorly understood. For these patients, the genetic determinants of egg quality may offer an answer.
During meiosis, oocytes must faithfully segregate their chromosomes in two rounds of division to create a euploid zygote upon fertilization. One-half of all spontaneous abortions are aneuploid; these comprise the largest fraction of chromosomally abnormal pregnancy losses (Hassold, 1986). A significant proportion of these pregnancy losses are attributed to errors in meiosis. Errors in meiotic processes, such as chromosome synapsis, crossing over and spindle building can impair chromosome segregation and cause aneuploidy. Aneuploidy is more frequently observed in oocytes than in spermatocytes (Hassold et al., 2001, Bernardini et al., 2000, Bell et al., 2019). Understanding the molecular and genetic underpinnings of how meiosis is regulated is key to unlocking the mystery of the origins of human aneuploidy.
Genomic approaches, including next-generation sequencing and cytogenetic arrays, have begun to identify relationships between clinical infertility phenotypes and maternal gene variants. The disease of infertility includes a spectrum of patient fecundity levels, including subfertility (i.e., any degree of reduced fecundity) (American College of Obstetrics and Gynecology, 2019). Identifying subfertility-associated literature is difficult because of the way cases and controls are typically defined: once a patient becomes pregnant and passes a threshold of gestation, the patient may be considered fertile. Alternative methods of defining subfertility have emerged, such as time-to-pregnancy and blastocyst aneuploidy measurements. Because these methods of subfertility assessment are not widely adopted, the clinical phenotypes associated with errors in meiosis encompass a list of diagnoses. After reviewing the literature using PubMed search terms such as “female infertility” and “fertility”, we identified the principal clinical phenotypes associated with aneuploid egg production and subfertility as: primary ovarian insufficiency1 (POI), oocyte arrest and embryonic arrest, fertilization failure, recurrent pregnancy loss and early menopause. Note that POI refers to secondary amenorrhea before 40 years of age, whereas early menopause is defined as secondary amenorrhea between the ages of 40 and 45 years old (Edmonds, 2012).
The genes associated with these clinical phenotypes span a wide range of meiotic processes. Here, we review selected human gene variants that may cause infertility or subfertility by impacting landmark cellular meiotic processes. We divided these processes into four groups: (i) prophase I; (ii) meiotic resumption; (iii) meiosis I (MI) chromosome segregation; and (iv) meiotic cell-cycle regulation. Recent reviews have discussed genetic drivers of female infertility through non-meiotic mechanisms such as altered ovarian reserve and ovarian function, defective follicle activation and growth (Yatsenko et al., 2019) and syndromic causes of infertility (Zorrilla et al., 2013, Jedidi et al., 2019). Another recent review discussed female infertility caused by DNA repair dysregulation (Veitia, 2020). To avoid redundancies, we took a deep dive to describe only a selection of gene variants that impact female gamete viability and/or quality by disrupting these four key meiotic processes. We discuss example genes and indicate the remainder of genes we identified in Figures 1–5. We review the genes by presenting them in a biologically-relevant temporal order in which they function (Fig. 1). To provide context, we first describe the biological function of these genes during the meiotic process often using findings from model organism studies, then consolidate findings from case studies, and, when possible, report functional assessments of the human gene variants.
Figure 1. Overview of steps in oocyte meiotic maturation vulnerable to infertility-causing genetic variants.
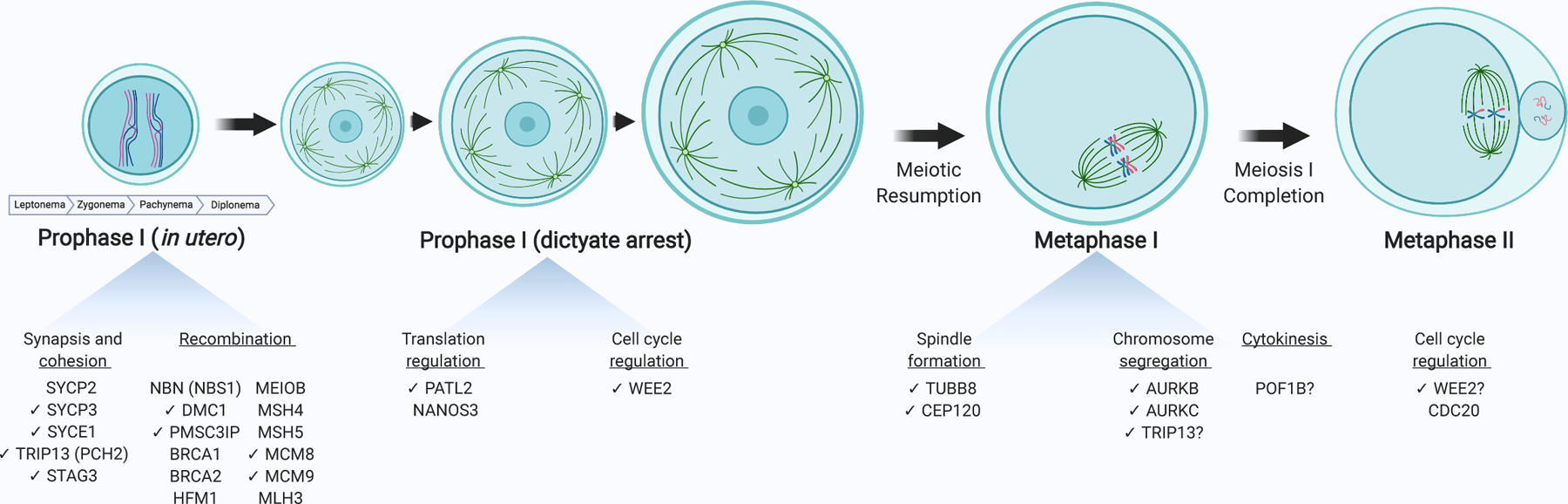
The oocyte matures through discrete meiotic phases, requiring genes that are susceptible to dysregulation by genetic variants; those associated with female infertility phenotypes are listed below each phase. Prophase I involves a program of synapsis, cohesion, and recombination. After the diplotene stage of prophase I, oocytes arrest in the dictyate stage of prophase I until puberty and while it grows in size. Upon ovulation, oocytes resume meiosis and initiate a burst of maternal RNA translation. To complete meiosis I, the spindle forms and, under the governance of the spindle assembly checkpoint, homologous chromosomes segregate. The spindle reflects one from a mouse oocyte. After an asymmetric cytokinesis, oocytes arrest at metaphase II awaiting fertilization. After fertilization, inhibition of CDK1 induces meiosis II completion. Exact stoichiometry is not depicted. Check marks (✔) indicate those genes discussed in this review. Created using BioRender.com.
Figure 5. Exits from meiosis I and II are targets for infertility-causing genetic variants.
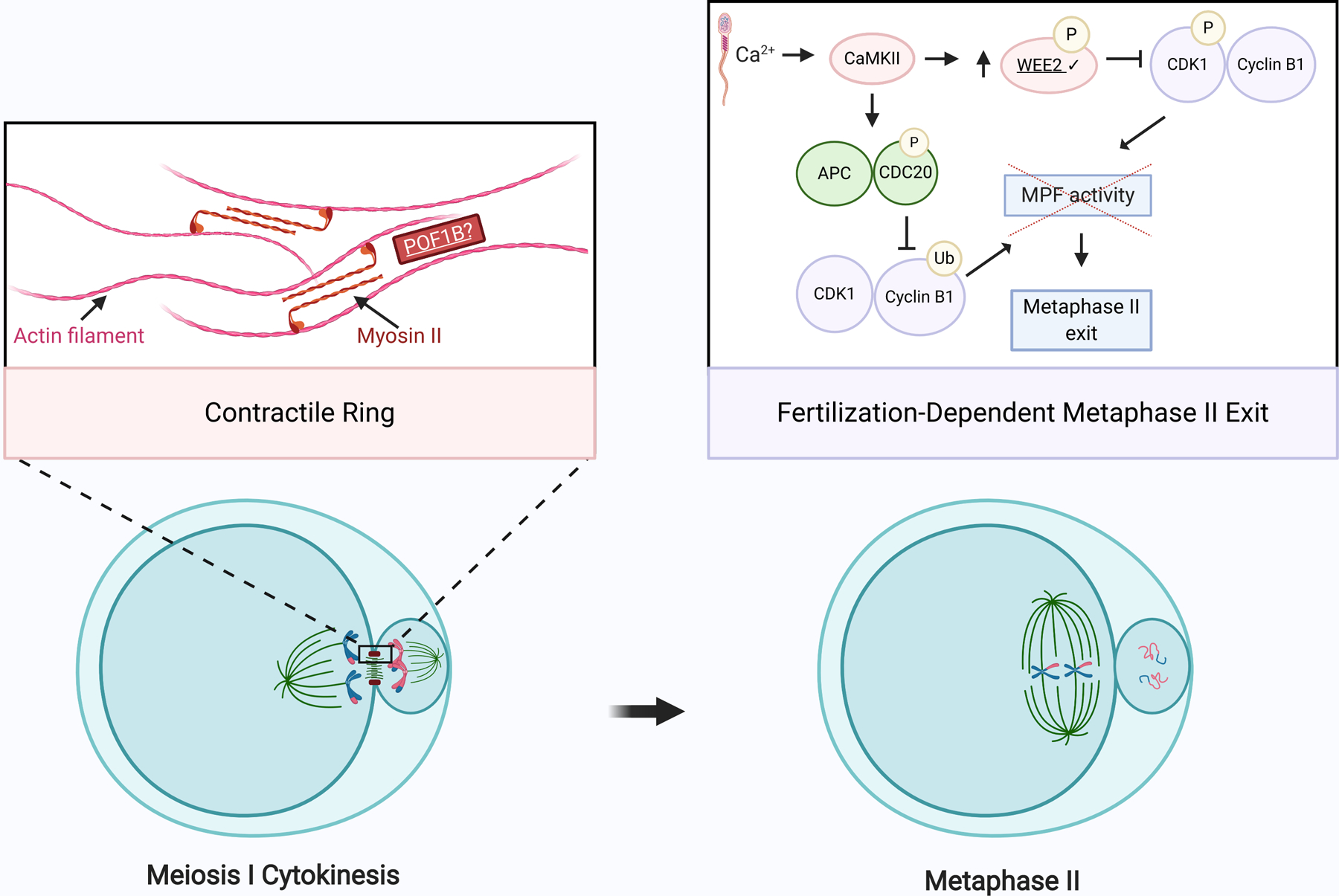
Schematic of meiosis I cytokinesis and meiosis II cell-cycle exit. During cytokinesis, actin filaments and myosin II form a contractile ring at the cleavage furrow. POF1B binds to non-muscle actin filaments and shares homology with myosin. Upon fertilization, an increase in intracellular calcium leads to activation of Ca2+/CaM-protein kinase II (CaMKII). CaMKII, in turn, activates WEE2 through phosphorylation. Activated WEE2 represses the maturation promoting factor (MPF), comprised of cyclin B1 and CDK1, by phosphorylating CDK1. This decrease in MPF activity, in concert with APC- and CDC20-mediated cyclin B1 decay, triggers exit from meiosis II. Exact stoichiometry is not depicted. Underlined gene products are implicated in infertility. Check marks (✔) indicate gene products discussed in this review. Created using BioRender.com.
1. Prophase I: making and repairing double-strand breaks
After pre-meiotic DNA replication, oocytes enter meiotic prophase I to prepare for homologous recombination (Fig. 1), an essential process for double-strand break (DSB) repair and successful MI chromosome segregation. Prophase I begins with chromosome DSBs initiated by the SPO11 topoisomerase (Keeney et al., 1995, Bergerat et al., 1997, Keeney et al., 1997, de Massy et al., 1995) and is followed by homologous chromosome pairing and synapsis through formation of the synaptonemal complex (SC). In parallel, DSB repair occurs. Repair can occur through homolog crossing over, leaving physical linkages (chiasmata) between the homologs. Chiasmata are essential to hold homologs together until anaphase I. Therefore, at least one crossover per homolog must occur, lest an aneuploid egg form after completion of MI. Notably, defects in prophase I can also give rise to POI because oocytes with persistent DNA damage undergo apoptosis and are culled from the ovarian reserve. Consistent with this hypothesis, most POI patients are subfertile: only 5 to 10% of POI patients eventually conceive and maintain a pregnancy (Welt, 2020). The following eight genes (Table 1) are essential regulators of distinct prophase I steps (Figs. 1, 2) and variants within these genes are associated with female infertility.
Table 1. Female infertility-associated genetic variants affecting prophase I and meiotic entry.
Genes are listed in order of appearance in text; their variants are listed in order of publication year. Frequencies of small (<1kb) genetic variants were identified in gnomAD v2.1.1 and ExAc v1.0 via ANNOVAR. Allele frequencies of structural variants reflect those of overlapping variants of the same type (i.e., deletion (DEL), duplication (DUP) etc.), rather than of exact matches; the gnomAD variant ID of the variant used is indicated.
| Gene | DNA Variant | Protein Variant | Key Phenotype | Reference | Allele Frequency | ||||
|---|---|---|---|---|---|---|---|---|---|
| gnomAD exomes1 | ExAc2 | gnomAD SVs3 | gnomAD genomes4 | ||||||
| DEL | DUP | ||||||||
| SYCP3 | NM_153694.1:c.553–16_19del | Intronic variant | Recurrent pregnancy loss | Bolor et al. (2009)† | NR | ||||
| SYCP3 | NM_153694.1:c.657T>C | Splice donor site variant | Recurrent pregnancy loss | Bolor et al. (2009)† | 5.63E-05 | 6.83E-05 | |||
| SYCP3 | NM_153694.1:c.657T>C | Splice donor site variant | Recurrent pregnancy loss | Mizutani et al. (2011)† | 5.63E-05 | 6.83E-05 | |||
| SYCP3 | NM_153694.1:c.657T>C | Splice donor site variant | Recurrent pregnancy loss | Sazegari et al. (2014)† | 5.63E-05 | 6.83E-05 | |||
| SYCE1 | NC_000010.10:g.135092227_135256027del | Structural variant | Primary ovarian insufficiency | McGuire et al. (2011)† | 1.52E-03 (10_115557) | ||||
| SYCE1 | NM_130784.2:c.613C>T | p.Q205* | Primary ovarian insufficiency | DeVries et al. (2014) | NR | ||||
| SYCE1 | NC_000010.10:g.135254039_135377532dup | Structural variant | Primary ovarian insufficiency | Jaillard et al. (2016)† | 4.97E-02 (10_31262) | ||||
| SYCE1 | NC_000010.10:g.135256762_135379710dup | Structural variant | Primary ovarian insufficiency | Tsuiko et al. (2016)† | 4.97E-02 (10_31262) | ||||
| SYCE1 | NC_000010.10:g.135281682_135377532dup | Structural variant | Primary ovarian insufficiency | Bestetti et al. (2019)† | 4.97E-02 (10_31262) | ||||
| SYCE1 | NC_000010.10:g.135281682_135378761dup | Structural variant | Primary ovarian insufficiency | Bestetti et al. (2019)† | 4.97E-02 (10_31262) | ||||
| SYCE1 | NC_000010.10:g.135252327_135378761dup | Structural variant | Primary ovarian insufficiency | Bestetti et al. (201)† | 4.97E-02 (10_31262) | ||||
| SYCE1 | NC_000010.10:g.135252327_135378761del | Structural variant | Primary ovarian insufficiency | Bestetti et al. (2019)† | 1.52E-03 (10_115557) | ||||
| SYCE1 | NC_000010.10:g.135281682_135404523dup | Structural variant | Primary ovarian insufficiency | Bestetti et al. (2019)† | 4.97E-02 (10_31262) | ||||
| TRIP13 | NM_004237.4: c.77A>G | p.H26R | Oocyte maturation arrest | Zhang et al. (2020b) | 9.19E-06 | 3.21E-05 | |||
| TRIP13 | NM_004237.4:c.518G>A | p.R173Q | Oocyte maturation arrest | Zhang et al. (2020b) | 7.95E-06 | 1.65E-05 | |||
| NM_004237.4:c.907G>A | p.E303K | Oocyte maturation arrest | Zhang et al. (2020b) | 1.60E-05 | 1.65E-05 | ||||
| TRIP13 | NM_004237.4:c.592A>G | p.I198V | Oocyte maturation arrest | Zhang et al. (2020b) | NR | ||||
| NM_004237.4:c.739G>A | pV247M | NR | |||||||
| STAG3 | NM_001282716:c.877_885del | p.293_295del | Primary ovarian insufficiency | Xiao et al. (2019)† | NR | ||||
| STAG3 | NM_001282716:c.891_893dup | p.297_298insAsp | Primary ovarian insufficiency | Xiao et al. (2019)† | NR | ||||
| STAG3 | NM_001282716:c.291dup; | p.N98Qfs*2 | Primary ovarian insufficiency | Franca et al. (2019)† | NR | ||||
| NM_001282716:c.1950C>A | p.Y650* | NR | |||||||
| STAG3 | NM_01282717:c.677C>G | p.S227* | Primary ovarian insufficiency | Colombo et al. (2017)† | 3.98E-06 | 8.24E-06 | |||
| STAG3 | NM_012447.2:c.1573+5G>A | p.L490Tfs*10 | Primary ovarian insufficiency | He et al. (2018) | 1.19E-05 | 3.19E-05 | |||
| STAG3 | NM_012447.3:c.1947_1948dup | p.Y650Sfs*22 | Primary ovarian insufficiency | Le Quesne Stabej et al. (2016) | NR | ||||
| STAG3 | NC_000007.13:g.99786485del | p.F187fs*7 | Primary ovarian insufficiency | Caburet et al. (2014) | NR | ||||
| STAG3 | NM_001282716.1:c.659T > G | p.L220R | Primary ovarian insufficiency | Heddar et al. (2019) | NR | ||||
| NM_001282716.1:c.3052del | p.R1018DfsTer14 | NR | |||||||
| STAG3 | NC_000007.13:g.99794799G>A | p.R321H | Primary ovarian insufficiency | Jaillard et al. (2020) | NR | ||||
| MCM8 | NC_000020.10:g.5948227G>A | p.E341K | Early menopause risk | He et al. (2009) | 0.0451 | 0.0451 | 0.0378 | ||
| MCM8 | NC_000020.10:g.5948227G>A | p.E341K | Early menopause risk | Murray et al. (2011) | 0.0378 | 0.0451 | 0.0451 | ||
| MCM8 | NC_000020.10:g.5948227G>A | p.E341K | Early menopause risk | Chen et al. (2011) | 0.0451 | 0.0451 | 0.0378 | ||
| MCM8 | NC_000020.11:g.5954739T>A | Intronic variant | Early menopause risk | Chen et al. (2012) | NR | ||||
| MCM8 | NM_001281520.1:c.1954–1G>A | p.V652Wfs*6, p.V652_Q664del, or p.V652_E721del | Primary ovarian insufficiency | Tenenbaum-Rakover et al. (2015) | 3.98E-06 | 8.25E-06 | |||
| MCM8 | NM_001281520.1:c.1469_1470insTA | p.L491Ifs*88 | Primary ovarian insufficiency | Tenenbaum-Rakover et al. (2015) | NR | ||||
| MCM8 | NM_032485:c.446C>G | p.P149R | Primary ovarian insufficiency | AlAsiri et al. (2015)† | NR | ||||
| MCM8 | NM_001281520.1:c.482A>C | p.H161P | Primary ovarian insufficiency | Bouali et al. (2017) | NR | ||||
| MCM8 | NC_000020.10:g.5948227G>A | p.E341K | Early menopause risk | Coignet et al. (2017) | 0.0451 | 0.0451 | 0.0378 | ||
| MCM8 | NC_000020.11:g.5954739T>A | Intronic variant | Early menopause risk | Coignet et al. (2017) | NR | ||||
| MCM8 | NM_001281522.1:c.925C>T | p.R309* | Growth retardation, recurrent pilomatricomas, and primary amenorrhea | Heddar et al. (2020) | 7.96E-06 | 8.24E-06 | |||
| MCM8 | NM_032485.4:c.351_354del | p.K118Efs*5 | Primary ovarian insufficiency | Zhang et al. (2020a) | NR | ||||
| MCM9 | NM_017696.2:c.1732+2T>C | Splice donor site variant | Primary ovarian insufficiency and short stature | Wood-Trageser et al. (2014) | 1.33E-05 | ||||
| MCM9 | NM_017696.2:c.394C>T | p.R132* | Primary ovarian insufficiency and short stature | Wood-Trageser et al. (2014) | 7.97E-06 | 8.24E-06 | |||
| MCM9 | NC_000006.11:c.672_673delinsC | p.Q225Kfs*4 | Primary ovarian insufficiency; mixed polyposis and colorectal cancer | Goldberg et al. (2015)† | NR | ||||
| MCM9 | NM_017696.2:c.1483G>T | p.E495* | Primary ovarian insufficiency and short stature | Fauchereau et al. (2016) | NR | ||||
| DMC1 | NM_007068.3: c.106G>A | p.D36N | Primary ovarian insufficiency | He et al. (2020b) | NR | ||||
| PSMC3IP | NM_016556.2:c.600_602del | p.E201del | 46,XX ovarian dysgenesis | Zangen et al. (2011) | NR | ||||
| PSMC3IP | NC_000017.10:c.489C>G | p.Y163* | Primary ovarian insufficiency | Al-Agha et al. (2018)† | NR | ||||
| PSMC3IP | NM_016556:c.496_497del | p.R166Afs | Primary ovarian insufficiency | Yang et al. (2019)† | NR | ||||
| PSMC3IP | NM_016556:c.430_431insGA | p.L144* | Primary ovarian insufficiency | Yang et al. (2019)† | NR | ||||
Reference sequence or reference sequence version number not reported
Frequency in gnomAD v.2.1.1 exomes
Frequency in ExAc v1.0
Frequency in gnomAD SVs v2.1
Frequency in gnomAD v2.1.1 genomes
NR, not reported
Figure 2. Meiotic prophase I and maturation are disrupted by female infertility-associated gene variants.
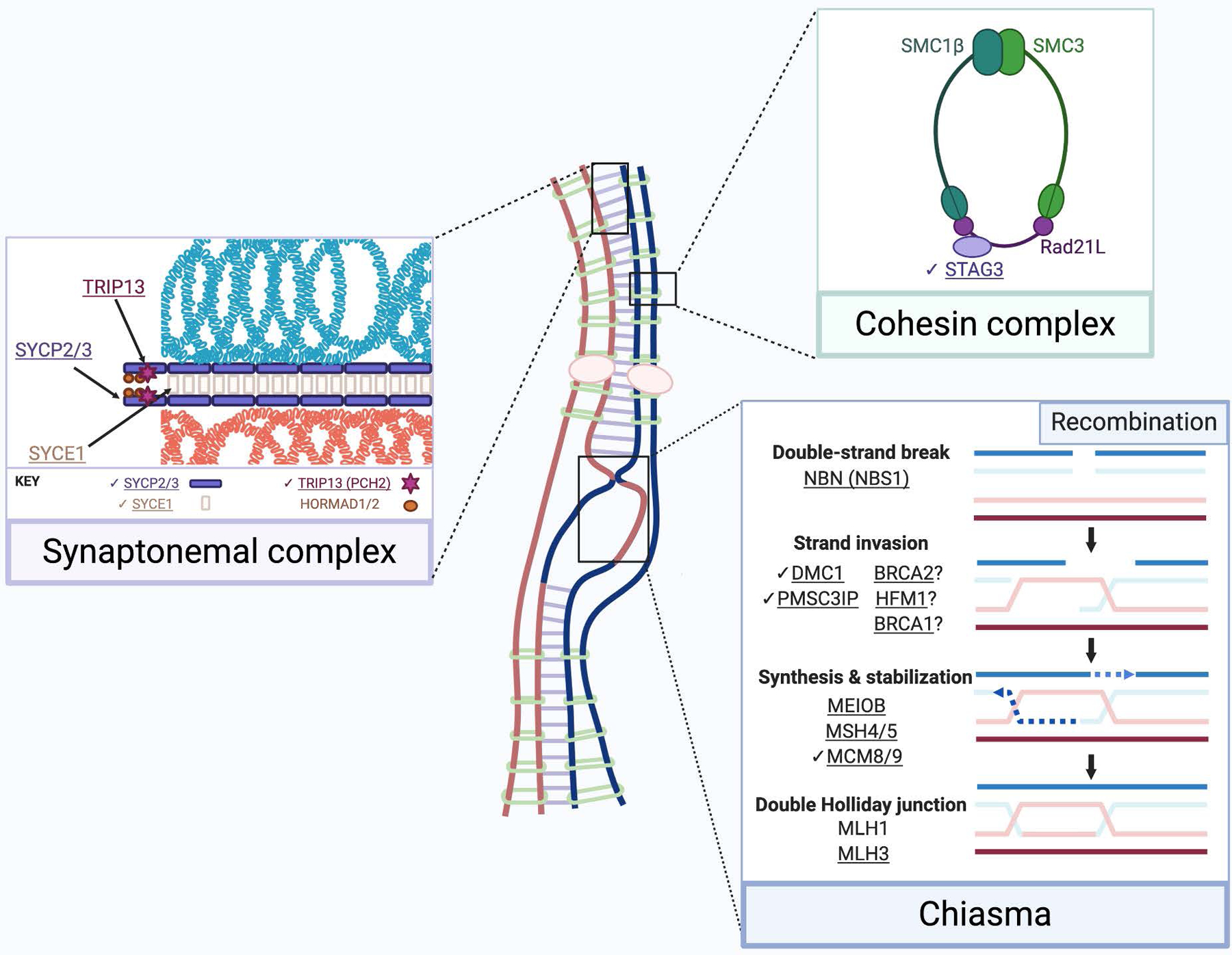
The synaptonemal complex (purple box) scaffolds homologous chromosomes and is composed of two lateral elements (SYCP2/3) and central elements SYCE1 (shown), SYCP1, SYCE2, SYCE3, and TEX12 (not shown). TRIP13 removes HORMA domain proteins (HORMADs), permitting synaptonemal complex assembly. The meiotic cohesin complex (green box) joins sister chromatids. In parallel with synaptonemal complex assembly, recombination (blue box) begins with double-strand breaks via SPO11. This requires the MRE11-RAD50-NBS1 (MRN) complex. Recombinases RAD51 (not shown) and DMC1 facilitate strand invasion into a chromosome’s homolog. HFM1 may also be essential for stable strand invasion (Guiraldelli et al., 2013). At double-strand breaks, the MCM8/9 helicase facilitates RAD51 recruitment and DNA synthesis. BRCA2 is proposed to localize to DSBs via MEILB2, and recruits RAD51 and DMC1 (Zhang et al., 2019a). A PMSC3IP-MND1 heterodimer stabilizes the RAD51 filament. BRCA1 may also stabilize the RAD51 filament in meiosis (Scully et al., 1997, Li et al., 2018). MEIOB is essential for maintenance of RAD51 and DMC1 foci (Souquet et al., 2013). The resulting double Holliday junction is stabilized by the MSH4/5 heterodimer. The MSH4/5 complex recruits the MLH1/MLH3 heterodimeric endonuclease, which processes most crossovers. Exact stoichiometry is not depicted. Underlined gene products are implicated in infertility. Check marks (✔) indicate gene products discussed in this review. Created using BioRender.com.
1.a. The synaptonemal complex and chromosome synapsis
The synaptonemal complex (SC), which forms during homologous chromosome synapsis, is a symmetrical, zipper-like protein complex that bridges two homologs (Fig. 2) (Fawcett, 1956, Moses, 1956). Three protein types comprise the SC: a central element, running parallel to the chromosome arms; transverse elements flanking the central element; and lateral elements coating the inner faces of the chromosome arms and framing the transverse elements (Dunne et al., 2019, Costa et al., 2005). The lateral elements of the SC assemble first. Lateral element proteins, SYCP2 and SYCP3, form a heterodimeric scaffold on chromosomes. The scaffold grows outward along the length of chromosomes, with chromatin forming perpendicular loops (Yang et al., 2006, Offenberg et al., 1998). Female mice deficient in the SYCP3 homolog, Scp3, have significantly more embryo death than their wildtype (WT) counterparts. Increased maternal age amplifies this phenotype: rates of embryo death in Scp3-deficient mice increase with age. As a result, Scp3-deficient female mice have a shorter reproductive lifespan than do WT female mice. At a cellular level, Scp3-deficient oocytes have greater proportions of chromosomes not linked by chiasmata, likely because chiasmata fail to form (Yuan et al., 2002). Therefore, SYCP3 dysfunction in humans could lead to infertility, specifically pregnancy loss and accelerated reproductive aging (reviewed in detail in (Geisinger et al., 2016)).
An SYCP3-specific sequencing screening of 26 women with three or more spontaneous miscarriages before ten weeks of gestation provided initial evidence for correlating SYCP3 malfunction with female infertility. The screening identified two women with heterozygous point mutations (NM_153694.1:c.IVS7–16_19delACTT and NM_153694.1:c.657T>C), both of which occur in putative splice sites of SYCP3. To ascertain their biological impact on meiosis, the putative splice-site variants were introduced into mouse testes. This analysis showed that the NM_153694.1:c.657T>C variant had a higher unspliced-to-spliced transcript ratio than the WT gene, while the NM_153694.1:c.IVS7–16_19delACTT variant did not change the splicing transcript ratio. The protein product of the NM_153694.1:c.657T>C variant was unable to form homopolymeric fibers in cultured somatic cells, whereas WT SYCP3 formed thick looped-shaped fibers (Bolor et al., 2009).
The association between SYCP3 NM_153694.1:c.657T>C and infertility was corroborated by targeted sequencing of 200 women, half of whom had recurrent pregnancy loss (RPL) of unknown cause and half of whom had successful pregnancies as controls (Sazegari et al., 2014). Of note, another study of 101 women with histories of at least three undiagnosable pregnancy losses did not identify the allele (Mizutani et al., 2011). These findings suggest that, although SYCP3 variants are linked to RPL in terms of both molecular characterization and patient genetic sequence data, SYCP3 variants are not the sole etiology of RPL.
After the lateral element assembles, the transverse filament and central element of the SC assemble between homologs. SYCP1, a central element protein, initiates this process by localizing between the lateral elements of two homologs and recruiting additional central element proteins, including synaptonemal complex central element protein 1, SYCE1 (Costa et al., 2005). SYCE1 is an essential component of the central element. For instance, mouse oocytes that lack SYCE1 fail to assemble the central element of the SC, a critical step in synapsis. Syce1−/− oocytes and spermatocytes have homologs that pair but do not form SCs. As a result, Syce1−/− spermatocytes arrest in meiotic prophase I, and the ovaries of Syce1−/− mice are small and depleted of follicles; both sexes are therefore sterile (Bolcun-Filas et al., 2009). The absence of follicles in Syce1−/− mice parallels POI (Welt, 2018).
In humans, SYCE1 variants are associated with POI. For example, a novel 0.16 MB microdeletion (hg19:g.135,092,227_135,256,027del) spanning SYCE1 and a cytochrome-encoding gene, CYP2E1, was identified via SNP array. One of 89 POI patients had this heterozygous microdeletion. This patient had menarche at age 14 but stopped menstruating at age 21, marking the onset of POI (McGuire et al., 2011). The association between POI and SYCE1 mutation was strengthened by whole-exome sequencing of a consanguineous family with multiple POI cases. In this study, two sisters harbored a single homozygous nonsense mutation in SYCE1 (NM_ 130784.2:c.613C>T), generating a truncated 205-amino acid protein. Both sisters (ages 16 and 17) had primary amenorrhea. Their heterozygote parents and brothers were fertile and healthy, suggesting autosomal recessive inheritance of the allele. A healthy, fertile sister and 90 ethnically-matched controls lacked the allele (de Vries et al., 2014).
Importantly, the biological impact of the SYCE1 c.613C>T variant has been assessed by creating a humanized mouse model. Female mice homozygous for the equivalent mutation lacked follicles and oocytes, and, similar to the family described, heterozygote mice were unaffected. Chromosomes in homozygous mutant gametes failed to synapse during prophase I (Hernandez-Lopez et al., 2020). This study demonstrates that this SYCE1 variant recapitulates the infertility phenotype in mouse and results in disrupted chromosome synapsis. Subsequent studies of POI patient cohorts that used SNP arrays also demonstrated enrichment of SYCE1 variants in POI patients (Zhen et al., 2013, Tsuiko et al., 2016, Jaillard et al., 2016). Additionally, whole-exome sequencing of a family containing two members with POI identified a gross deletion encompassing part of the SYCE1 locus (Zhe et al., 2020). The functional consequence of this deletion or of the truncating variant are unknown, but the generation of this humanized mouse model offers a promising avenue to interrogate the molecular relationship between SYCE1 function and subfertility.
1.b. Homologous recombination and genome integrity
HORMA domain (HORMAD) proteins accumulate on unsynapsed chromosomes and promote SPO11-mediated DSBs (Mao-Draayer et al., 1996, Woltering et al., 2000, Goodyer et al., 2008, Shin et al., 2010). SPO11-mediated DSB formation requires the MRE11-RAD50-NBS1 (MRN) complex (Garcia et al., 2011, Alani et al., 1990, Nairz et al., 1997, Cao et al., 1990, Girard et al., 2018). Thyroid hormone receptor interactor 13 (TRIP13 or PCH2) is an AAA+ ATPase that, in meiosis, catalyzes the removal of the meiosis-specific HORMADs, HORMAD1 and HORMAD2, from synapsed chromosome axes (Fig. 2) (Wojtasz et al., 2009, Chen et al., 2014a). The processes of DSB formation and HORMAD removal are susceptible to dysregulation by infertility-associated genetic variants. For example, variants in NBN (NBS1), a gene essential for DSB generation (Fig. 2), also lead to POI and Nijmegen breakage syndrome, a syndrome of microcephaly, recurrent cancer and infertility (Tucker et al., 2018, Fievet et al., 2020, Warcoin et al., 2009, Szeliga et al., 2019). In addition, several variants in TRIP13 have been linked with female infertility phenotypes both in mouse models and in clinical settings. TRIP13 (or PCH2) is an AAA+ ATPase that, in meiosis, catalyzes the removal of the meiosis-specific HORMADs, HORMAD1 and HORMAD2, from synapsed chromosome axes (Fig. 2) (Wojtasz et al., 2009, Chen et al., 2014a). Male and female TRIP13 knockout mice are sterile; female TRIP13 knockout mice undergo oocyte atresia resulting in POI (Li et al., 2007).
In contrast to the oocyte atresia phenotype in mouse, human TRIP13 variants are linked with oocyte maturation arrest (Table 1). The first is a homozygous missense variant in exon 1 (NM_004237.4: c.77A>G), found in three patients with infertility. Two other infertility patients had compound heterozygous variants. One patient’s compound heterozygous variants lie in exons 5 and 10 (NM_004237.4:c.518G>A and NM_004237.4:c.907G>A), the latter of which encodes the AAA+ ATPase domain. The other patient’s compound heterozygous variants lie in the AAA+ ATPase domain (NM_004237.4:c.592A>G; NM_004237.4:c.739G>A) (Zhang et al., 2020b). Each patient had primary infertility despite regular menstrual cycles. During IVF cycles, MI oocytes could be retrieved, but attempts to mature the oocytes failed. In the patients with the homozygous mutation and one of the compound heterozygous mutations (NM_004237.4:c.518G>A and NM_004237.4:c.907G>A), nearly all oocytes were arrested in metaphase I (Zhang et al., 2020b). However, in the patient with the other compound heterozygous mutations (NM_004237.4:c.592A>G; NM_004237.4:c.739G>A), 14 mature eggs were retrieved. Twelve of these eggs fertilized, but all subsequently arrested at the eight-cell stage. In vitro ATPase assay of the NM_004237.4:c.739G>A variant compared to WT TRIP13 showed significantly diminished ATPase activity; the other TRIP13 variants identified (Table 1) had no change in ATPase activity (Zhang et al., 2020b). Finally, the effects of these TRIP13 variants on HORMAD2 have been interrogated. Ectopic expression of WT TRIP13 in HeLa cells decreased total HORMAD2 protein content, but HORMAD2 levels were unchanged by ectopic expression of the NM_004237.4:c.739G>A, NM_004237.4:c.907G>A, and NM_004237.4: c.77A>G TRIP13 variants (Zhang et al., 2020b). These data suggest that these three variants may impair HORMAD2 removal. Interestingly, the clinical phenotype of patients harboring these variants is not POI, as it is in mouse, suggesting that the impact of TRIP13 mutation acts later during chromosome segregation (Fig. 1).
After DSBs are generated, DNA is resected and vulnerable single-stranded DNA remains. In mammals, two homologs of the bacterial recombinase RecA, RAD51 and DMC1, assemble on the single-stranded DNA, search for homologous DNA sequences and promote strand invasion. Proteasome 26S ATPase subunit 3-interacting protein (PSMC3IP), also called homologous-pairing protein 2 (HOP2) (Leu et al., 1998), heterodimerizes with MND1 to stimulate DMC1 and RAD51 activity. PSMC3IP/MND1 stabilizes DMC1 and RAD51 filaments on single-stranded DNA and catalyzes the capture of double-stranded DNA (Fig. 2) (Sansam et al., 2015). Hop2 and Dmc1-knockout mice lack ovarian follicles and have small ovaries, suggesting that loss of these DNA-repair proteins is associated with infertility phenotypes (Petukhova et al., 2003, Pittman et al., 1998).
Similar to the mouse knockout phenotypes, variants in both DMC1 and PSMC3IP are associated with POI (Table 1). A homozygous DMC1 missense variant (NM_007068.3:c.106G>A) was found in a consanguineous family with male and female infertility. At the time of the study, the female patient was 30 years old, had been infertile for at least 3 years and was diagnosed with POI at age 20 (He et al., 2018). The DMC1 variant in this family lies in the helix-3 turn-helix DNA-binding motif conserved between RAD51 and DMC1. In silico analysis predicts that the substitution alters hydrogen bonds within the structure, suggesting that the variant interrupts DNA interactions (He et al., 2018). Several other proteins essential in the process of strand invasion are implicated in infertility (Fig. 2), including BRCA1, BRCA2 and HFM1 (Caburet et al., 2020, Lambertini et al., 2018, Lin et al., 2017, Oktay et al., 2010, Oktay et al., 2020, Porcu et al., 2020, Pu et al., 2016, Turan et al., 2018, Turan et al., 2020, Valentini et al., 2013, Wang et al., 2014a, Wang et al., 2014b, Weinberg-Shukron et al., 2018, Zhe et al., 2019), suggesting that this process is particularly important for fertility.
Three studies have identified PSMC3IP variants unique to patients with subfertility. A homozygous 3-bp deletion in the conserved C-terminus of PSMC3IP (NM_016556.2:c.600_602del) was found in five patients with 46,XX ovarian dysgenesis. All affected patients had primary amenorrhea and their ovaries were undetectable on ultrasound (Zangen et al., 2011). Subsequently, a homozygous C-terminal truncation in exon 6 of PSMC3IP (NC_000017.10:c.489C>G) was identified. In this family, two sisters and one brother were homozygous for the mutation and had primary amenorrhea or azoospermia (Al-Agha et al., 2018). This sexually monomorphic phenotype mirrors that of the Hop2-knockout mice. The most recent detection of PSMC3IP POI-associated variants was in a woman who had two variant alleles of PSMC3IP predicted to be pathogenic (NM_016556:c.496_497del and NM_016556:c.430_431insGA). On ultrasound, ovaries and follicles were undetectable (Yang et al., 2019b). In contrast, a cohort of 50 Swedish women with POI did not have any pathogenic variants of PSMC3IP (Norling et al., 2014), highlighting the genetic complexities that give rise to this syndrome. The mechanism by which the mutations disrupt PSMC3IP is unknown.
DSBs are genotoxic and DNA repair is therefore critical for cell survival and genome integrity. Many DNA helicases are recruited during this process to protect the structural integrity of single-stranded DNA. Among these helicases is the complex formed by minichromosome maintenance proteins 8 and 9 (MCM8 and MCM9) (Yoshida, 2005, Gozuacik et al., 2003). MCM8 and MCM9 form a hexameric helicase, which promotes RAD51 recruitment and DNA synthesis at sites of DSBs (Fig. 2) (Park et al., 2013, Natsume et al., 2017, Hustedt et al., 2019). Both male and female Mcm8-knockout mice are sterile. The oocytes from Mcm8−/− mice cannot undergo DSB repair and their ovaries are small and adenoma-ridden. Upon histological examination, their primary follicles are shrunken and dysmorphic (Lutzmann et al., 2012). In contrast, the consequence of Mcm9 knockout is sexually dimorphic. Female knockouts are sterile and completely lack oocytes. However, male knockout mice are fertile, but have fewer germ cells than their WT counterparts (Hartford et al., 2011, Lutzmann et al., 2012). Primary follicles are completely absent in the ovaries of Mcm9 knockout mice (Lutzmann et al., 2012).
Reports of mixed reproductive-somatic syndromes associated with MCM8/9 variants indicate that this complex is essential in both human germ cells and somatic cells. One investigation identified two MCM9 variants, each in separate consanguineous families bearing daughters with POI and short stature. One variant alters a splice donor site (NM_017696.2:c.1732+2T>C), generating a truncated splice variant, while the other variant generates a premature stop codon (NM_017696.2:c.394C>T). When ectopically expressed in human HEK293T cells, truncated MCM9 failed to localize to sites of DNA damage (Wood-Trageser et al., 2014). A subsequent study identified a child of a consanguineous family with primary amenorrhea, short stature, and recurrent pilomatricomas (benign hair follicle tumors). This patient had a truncating mutation in exon 9 of MCM8 (NM_001281522.1:c.925C>T) (Heddar et al., 2020). Moreover, a homozygous MCM9 truncation (hg19:c.672_673delinsC) in a consanguineous family was linked with POI and gastrointestinal polyposis (Goldberg et al., 2015), a pathology classically associated with variants in genome stability genes (Grady et al., 2008). Heterozygotes in the family had no reproductive pathologies, but rather late-onset gastrointestinal tumors (Goldberg et al., 2015). Several studies have linked MCM8/9 variants with POI (Table 1), strengthening the association between the complex and female infertility.
Menopause represents the end of the female reproductive lifespan. Although there is a spectrum of causes of early menopause, one associated cause is depletion of the ovarian reserve, such as in POI (Edmonds, 2012). Analyses of several genome-wide association studies (GWAS) indicate that MCM8 variants are associated with early menopause. A SNP in exon 9 of MCM8 (NC_000020.10:g.5948227G>A) was first identified as associated with early natural menopause in the Nurse’s Health Study and the Women’s Genome Health Study (He et al., 2009). A subsequent study found that NC_000020.10:g.5948227G>A increased the risk of early menopause by 85% (Murray et al., 2011). An investigation of age-of-menopause-associated SNPs in Hispanic women corroborated NC_000020.10:g.5948227G>A and identified a novel variant, NC_000020.11:g.5954739T>A, as linked with early menopause (Chen et al., 2012). Additional studies using population-level data reinforced the association between NC_000020.10:g.5948227G>A and early menopause (Carty et al., 2013, Coignet et al., 2017). Two studies, a meta-analysis of 6,510 patients (Chen et al., 2014b) and a GWAS of 2,455 patients (Spencer et al., 2013), sought the age of menopause-associated alleles in African-American women. In contrast to the findings described above, neither of these studies found MCM8 alleles associated with early menopause. These studies collectively demonstrate an abundant range of fertility phenotypes associated with MCM8/9 helicase complex variants, which are complicated by ethnicity differences and a variety of alleles. Other work that identified infertility-linked variants in recombination, such as MEIOB, MSH4, MSH5 and MLH3 corroborate the essential role of recombination in fertility (Fig. 2) (Caburet et al., 2019, Carlosama et al., 2017, Guo et al., 2017, Mandon-Pepin et al., 2008, Ferras et al., 2012, Pashaiefar et al., 2013). Therefore, some genes associated with early menopause function in meiotic recombination, which illustrates unifying etiology of POI and some cases of early menopause.
In both mitosis and meiosis, cohesin complexes provide cohesion between sister chromatids and between homologous chromosomes; cohesion is essential for DSB repair by homologous recombination (Birkenbihl et al., 1992, Sjogren et al., 2001, Atienza et al., 2005, Sonoda et al., 2001, Schar et al., 2004). Together, two hinge-like SMC proteins, a kleisin and a stromalin form a ring-shaped cohesin complex (Fig. 2) (Chiu et al., 2004). During pre-meiotic DNA replication, cohesin complexes join homologous chromosomes along their arms and sister chromatids at their centromeres (Klein et al., 1999, Buonomo et al., 2000, Pasierbek et al., 2001). One subunit gene, stromal antigen protein 3 (STAG3) is expressed exclusively during meiosis (Prieto et al., 2001, Pezzi et al., 2000). Molecular and clinical studies of STAG3 indicate that it is essential for both mouse and human gametogenesis and fertility. Stag3 perturbation in mouse causes gametes to arrest in prophase I and Stag3 knockout mice of both sexes are sterile (Hopkins et al., 2014, Winters et al., 2014). At a molecular level, the chromosomes in Stag3 knockouts cannot crossover because intact cohesin is required for DSB repair (Hopkins et al., 2014). Additionally, STAG3 accumulates on microtubules in mouse oocytes and may be required for kinetochore-microtubule attachments to prevent aneuploidy (Zhang et al., 2017).
Similar to findings in mouse, STAG3 dysfunction in humans is associated with reduced oocyte viability. For example, a homozygous truncation mutation (NC_000007.13:g.99786485del) was identified in four sisters. Each presented with POI between ages 17 and 20. The premature stop codon interrupts the STAG domain in exon 7, thereby removing exons 8–34. Heterozygote family members did not have POI, indicating autosomal recessive inheritance. Notably, one patient homozygous for the NC_000007.13:g.99786485del allele developed bilateral ovarian cancer at age 19, but the relationship between this allele and ovarian cancer is unknown (Caburet et al., 2014). Subsequent studies have identified other STAG3 mutations associated with POI (Table 1). All of these cases share several features. First, the patient is either homozygous for a variant allele or is compound heterozygous for two variant alleles, suggesting that POI-causing STAG3 mutations are autosomal recessive. Second, all affected patients had primary amenorrhea. Finally, all affected patients examined via ultrasound had a structural gynecologic problem. Most often, patients had streak ovaries and, sometimes, they had small uteri. This clinical phenotype mimics Stag3 knockout in mouse, but defects in kinetochore-microtubule attachments have not been evaluated likely because of the severity of POI. Interestingly, other variants of STAG3 exist that have not been linked to POI and their clinical phenotype is unknown. For example, 569 missense variants are listed for this gene in the gnomAD database. Given STAG3’s critical role in meiosis and the large number of STAG3 genetic variants, there may be undescribed fertility phenotypes, such as embryonic death or aneuploidy, associated with other STAG3 variants.
2. Meiotic resumption: regulating translation and building a spindle
After completion of homologous recombination, oocytes arrest in the final stage of prophase I where they will eventually grow in size within follicles. During the growth phase, oocytes are transcriptionally active, but once they are fully grown, transcription ceases. Beginning at puberty and recurring in each menstrual cycle, a luteinizing hormone (LH) surge triggers ovulation and meiotic resumption. Meiotic resumption is the period between the exit from prophase I arrest and entry into metaphase I (Figs. 1, 3). It shares similar regulatory features of the mitotic G2/M cell-cycle transition, such as CDK1-dependent nuclear envelope breakdown and spindle building (Solc et al., 2010). To build a spindle, mouse oocytes use aggregated clusters of microtubule-organizing centers (MTOCs) to nucleate microtubules to form the meiotic spindles (Schuh et al., 2007). In human oocytes, however, a RAN-dependent chromatin-driven pathway nucleates microtubules (Holubcova et al., 2015).
Figure 3. Variants in mediators of meiotic arrest are associated with female infertility.
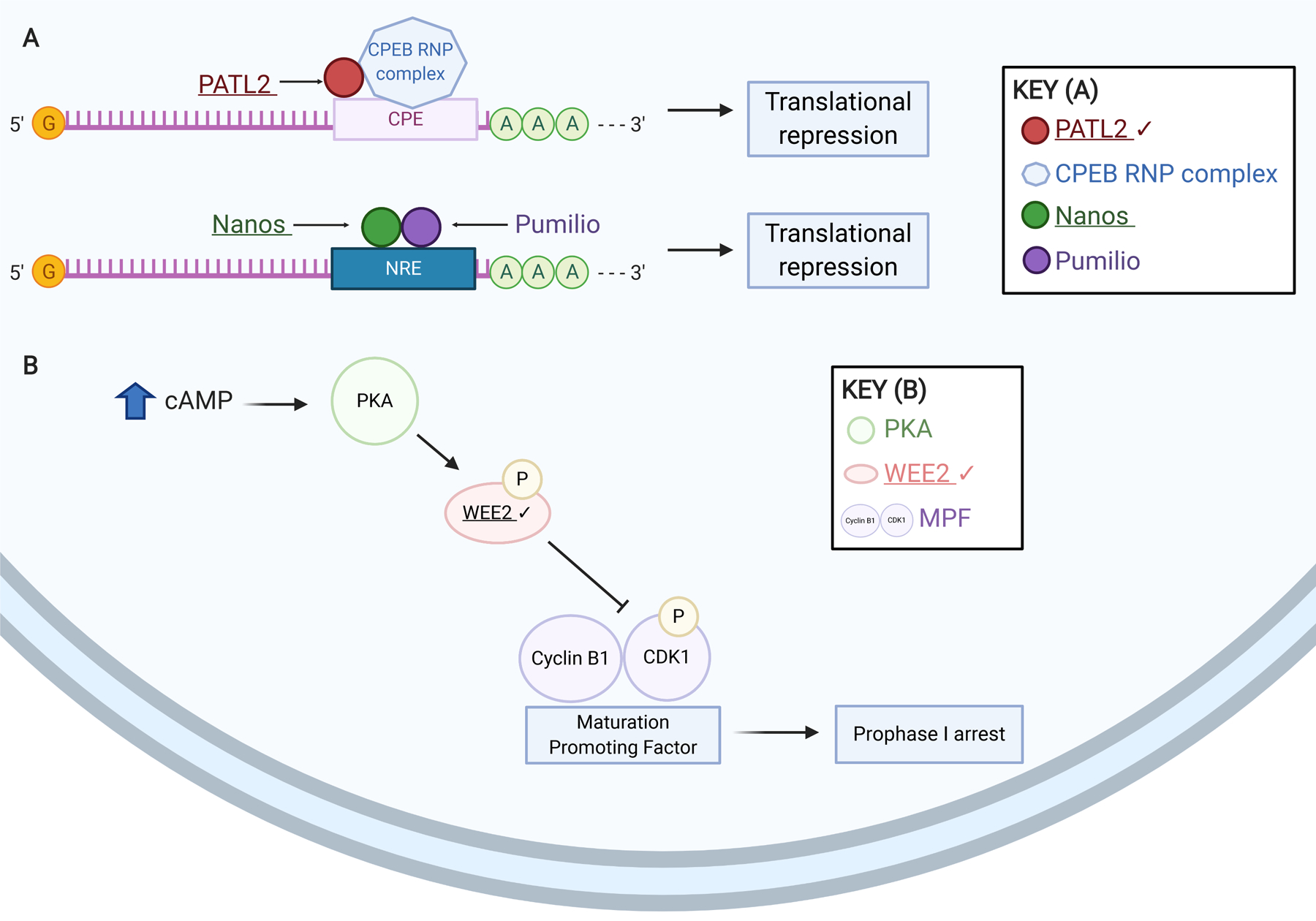
(A) Schematic of meiotic translational regulation mechanisms susceptible to infertility-causing genetic variants. Like PATL2, members of the Nanos family of proteins are recruited to a response element (NRE) in 3’ UTRs by Pumilio homolog(s) and repress translation of certain mRNAs. (B) Schematic of meiotic prophase I arrest mediators linked with female infertility. High cAMP levels trigger protein kinase A (PKA) phosphorylation of WEE2. Phosphorylated WEE2 in turn phosphorylates CDK1, which, with Cyclin B1, comprises the maturation promoting factor (MPF). This phosphorylation renders the MPF inactive and prophase I arrest is maintained. Exact stoichiometry is not depicted. Underlined gene products are implicated in infertility. Check marks (✔) indicate gene products discussed in this review. Created using BioRender.com.
2a. Translation repression
During oocyte growth and oogenesis, transcription and translation are tightly controlled. Oocyte maturation, fertilization and preimplantation embryonic development occur without transcription and rely on translation of stored maternal RNAs. Once oocytes resume meiosis, there is a regulated burst of maternal RNA translation. One negative regulator of oocyte translation is Protein Associated with Topoisomerase II Homolog 2 (PATL2). In Xenopus laevis eggs the PATL2 homolog, P100, complexes with the cytoplasmic polyadenylation element binding (CPEB) ribonucleoprotein complex and binds transcripts to repress translation (Fig. 3A) (Marnef et al., 2010). Loss of Patl2 caused decreased translational repression in mouse (Christou-Kent et al., 2018). P100 protein levels decrease rapidly after nuclear envelope breakdown, which marks meiotic resumption, thereby allowing translation of maternal RNAs (Nakamura et al., 2010, Marnef et al., 2010). Litter sizes from Patl2-knockout female, but not male, mice are reduced. Significantly more metaphase I-arrested oocytes were found in Patl2−/− mice than in WT, although some prophase I oocytes harvested from Patl2−/− mice reached meiosis II. Importantly, the arrested oocytes have significantly more spindle morphology defects and misaligned chromosomes. These data suggest that Patl2 loss reduces oocyte quality by changing regulation of mRNA transcripts, which makes it a strong candidate to cause human subfertility.
Two PATL2 variants were identified in two families with female infertility (Maddirevula et al., 2017). In both cases, the affected patients had oocyte maturation defects but had regular menstrual cycles. Neither family reported male infertility. In one family, two sisters presented with infertility at ages 27 and 35. Both tried to conceive for over six years and all IVF cycles failed because only prophase I-arrested oocytes were retrieved. In the second family, two sisters had several years of infertility characterized by retrieval of only prophase I-arrested oocytes. Positional mapping and whole-exome sequencing of these patients found two homozygous variants of PATL2, a truncation (NM_001145112.1:c.478C>T) in the first family and a missense mutation (NM_001145112.1:c.1108G>A) in the second family. Three fertile males in this study were homozygous for PATL2 variants (Maddirevula et al., 2017). Several other PATL2 mutations have been identified in female patients with infertility caused by oocyte maturation defects (Table 2). The phenotypic spectrum of patients with PATL2 variants sometimes includes early embryonic arrest and phenotypic heterogeneity. One hypothesis for the complexity in phenotypes is that differences in cis-regulatory elements cause variable penetrance (Wu et al., 2019). Other unidentified mutations in interacting proteins may also compound or dilute the phenotype. In aggregate, these results suggest that the most common phenotype of PATL2 variants is oocyte maturation defects. Although the structure of P100 is solved (Rother et al., 1992), the molecular biology of PATL2 variants is otherwise unexplored. However, infertility-linked variants in another translational regulator, NANOS3, suggest that translational regulation may be a key pathogenic mechanism in infertility (Santos et al., 2014, Sousa et al., 2016, Wu et al., 2013).
Table 2. Female infertility-associated genetic variants affecting meiotic resumption.
Genes are listed in order of appearance in text; their variants are listed in order of publication year. Frequencies of small (<1kb) genetic variants were identified in gnomAD v2.1.1 and ExAc v1.0 via ANNOVAR.
| Gene | DNA Variant | Protein Variant | Key Phenotype | Reference | Allele Frequency | ||
|---|---|---|---|---|---|---|---|
| gnomAD exomes1 | ExAc2 | gnomAD genomes3 | |||||
| PATL2 | NM_001145112.1:c.784C>T | p.R262* | Oocyte maturation arrest | Chen et al. (2017) | 0 | ||
| PATL2 | NM_001145112.1:c.558T>A; | p.Y186*; | Oocyte maturation arrest | Chen et al. (2017) | NF | ||
| NM_001145112.1:c. 223−14_223−2del | p.R75Vfs∗21 | 0.0002 | 5.55E-05 | 0.0003 | |||
| PATL2 | NM_001145112.1:c.953T>C; | p.I318T | Oocyte maturation arrest | Chen et al. (2017) | 6.50E-06 | 5.05E-05 | |
| NM_001145112.1:c.839G>A | p.R280Q | Chen et al. (2017) | 5.19E-05 | 3.19E-05 | |||
| PATL2 | NM_001145112.1:c.649T>A ; | p.Y217N; p.L189R | Oocyte maturation arrest | Chen et al. (2017) | 6.49E-06; | ||
| NM_001145112.1:c.566T>G | 6.49E-06 | ||||||
| PATL2 | NM_001145112.1:c.1224+2T>C; | p.Q411Wfs*3; | Oocyte maturation arrest | Chen et al. (2017) | 0.0002 | 5.55E-05 | 0.0003 |
| NM_001145112.1:c.223−14_223−2del | p.R75Vfs*21 | ||||||
| PATL2 | NM_001145112.1:c.478C>T | p.R160* | Oocyte maturation arrest | Maddirevula et al. (2017) | 3.25E-05 | ||
| PATL2 | NM_001145112.1:c.1108G>A | p.G370R | Oocyte maturation arrest | Maddirevula et al. (2017) | 6.60E-06 | ||
| PATL2 | ENST00000434130:c.478C>T | p.R160* | Oocyte maturation arrest | Christou-Kent et al. (2018)† | 3.25E-05 | ||
| PATL2 | c.778G>A, c.223–14_223–2del | p.V260M; | Oocyte maturation arrest | Wu et al. (2019)† | NF | ||
| p.R75Vfs*21 | 0.0002 | 5.55E-05 | 0.0003 | ||||
| PATL2 | c.1273A>C, c.898C>T | p.T425P; | Oocyte maturation arrest | Wu et al. (2019)† | NF | ||
| p.Q300* | NF | ||||||
| PATL2 | c.877G>T, c.223–14_223–2del | p.D293Y; | Embryonic arrest and oocyte maturation arrest | Wu et al. (2019)† | NF | ||
| p.R75Vfs*21 | 0.0002 | 5.55E-05 | 0.0003 | ||||
| PATL2 | c.716del, c.223–14_223–2del | p.N239fs*9; | Embryonic arrest and oocyte maturation arrest | Wu et al. (2019)† | NF | ||
| p.R75Vfs*21 | 0.0002 | 5.55E-05 | 0.0003 | ||||
| PATL2 | NC_000015.9:c.1528C>A | p.P510T | Oocyte maturation arrest | Liu et al. (2020)† | NF | ||
| PATL2 | NC_000015.9:c.1376C>A | p.S459Y | Oocyte maturation arrest | Liu et al. (2020)† | NF | ||
| TUBB8 | NM_177987:c.527C>T | p.S176L | Oocyte maturation arrest | Feng et al. (2016a)† | NF | ||
| TUBB8 | NM_177987:c.1249G>A | p.D417N | Oocyte maturation arrest | Feng et al. (2016a)† | NF | ||
| TUBB8 | NM_177987:c.5G>A | p.R2K | Oocyte maturation arrest | Feng et al. (2016a)† | NF | ||
| TUBB8 | NM_177987:c.785G>A | p.R262Q | Oocyte maturation arrest | Feng et al. (2016a)† | NF | ||
| TUBB8 | NM_177987:c.900G>A | p.M300I | Oocyte maturation arrest | Feng et al. (2016a)† | NF | ||
| TUBB8 | NM_177987:c.1088T>C | p.M363T | Oocyte maturation arrest | Feng et al. (2016a)† | NF | ||
| TUBB8 | NM_177987:c.686T>C | p.V229A | Oocyte maturation arrest | Feng et al. (2016a)† | NF | ||
| TUBB8 | c.426_427insG | p.T143Dfs*12 | Oocyte maturation arrest | Feng et al. (2016b)† | NP | ||
| TUBB8 | c.80_100del | p.E27_A33del | Oocyte maturation arrest | Feng et al. (2016b)† | NP | ||
| TUBB8 | c.1043A>G | p.N348S | Embryonic arrest | Feng et al. (2016b)† | NP | ||
| TUBB8 | c.628A>G | p.I210V | Embryonic arrest | Feng et al. (2016b)† | NP | ||
| TUBB8 | c.853A>C | p.T285P | Oocyte maturation arrest | Feng et al. (2016b)† | NP | ||
| TUBB8 | c.527C>T | p.S176L | Oocyte maturation arrest | Feng et al. (2016b)† | NP | ||
| TUBB8 | c.713C>T | p.T238M | Embryonic arrest | Feng et al. (2016b)† | NP | ||
| TUBB8 | c.784C>T | p.R262W | Oocyte maturation arrest | Feng et al. (2016b)† | NP | ||
| TUBB8 | c.763G>A | p.V255M | Oocyte maturation arrest | Feng et al. (2016b)† | 2.12E-05 | 6.77E-05 | 0.0005 |
| TUBB8 | ENST00000309812.4:c.10A>C | p.I4L | Oocyte maturation arrest | Chen et al. (2017) | 0 | 2.75E-05 | 3.19E-05 |
| TUBB8 | ENST00000309812.4:c.209C>T | p.P70L | Embryonic arrest | Chen et al. (2017) | 1.21E-05 | 8.44E-06 | |
| TUBB8 | ENST00000309812.4:c.1057G>A | p.V353I | Embryonic arrest | Chen et al. (2017) | NF | ||
| TUBB8 | ENST00000309812.4:c.35G>A | p.C12Y | Oocyte maturation arrest | Chen et al. (2017) | NF | ||
| TUBB8 | ENST00000309812.4:c.580G>A, | p.E194K; | Embryonic arrest | Chen et al. (2017) | NF | ||
| ENST00000309812.4:c.1245G>A | p.M415I | NF | |||||
| TUBB8 | ENST00000309812.4:c.5G>T | p.R2M | Oocyte maturation arrest | Chen et al. (2017) | NF | ||
| TUBB8 | ENST00000309812.4:c.613G>A | p.E205K | Embryonic arrest | Chen et al. (2017) | NF | ||
| TUBB8 | ENST00000309812.4:c.990G>A | p.M330I | Embryonic arrest | Chen et al. (2017) | NF | ||
| TUBB8 | Gross whole-gene deletion | N/A | Embryonic arrest | Chen et al. (2017) | NF | ||
| TUBB8 | NM_177987.2:c.292G>A | p.G98R | Oocyte maturation arrest | Wang et al. (2018) | NF | ||
| TUBB8 | NM_177987:c.322G>A | p.E108K | Oocyte maturation arrest | Yuan et al. (2018) | 3.58E-05 | 3.30E-05 | 3.18E-05 |
| TUBB8 | NM_177987.2:c.1205dup | p.M403Hfs*3 | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.426dup; | p.T143Dfs*12; | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| NM_177987.2:c.322G>A | p.E108K | 3.58E-05 | 3.30E-05 | 3.18E-05 | |||
| TUBB8 | NM_177987.2:c.10A>C | p.I4L | Embryonic arrest | Chen et al. (2019) | 0 | 2.75E-05 | 3.19E-05 |
| TUBB8 | NM_177987.2:c.1000C>G | p.Q334E | Embryonic arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.1072C>G | p.P358A | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.1073C>T | p.P358L | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.1171C>T | p.R391C | Embryonic arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.1270C>T | p.Q424* | Oocyte maturation arrest | Chen et al. (2019) | 8.16E-06 | 3.37E-05 | |
| TUBB8 | NM_177987.2:c.1286C>T; | p.T429M; | N/A | Chen et al. (2019) | 2.92E-05; | 2.69E-05 | 3.50E-05 |
| NM_177987.2:c.1301_1327del | p.434_442del | 6.87E-05 | 0.0002 | 3.40E-05 | |||
| TUBB8 | NM_177987.2:c.181C>A | p.P61T | Embryonic arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.527C>T | p.S176L | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.721C>T | p.R241C | Embryonic arrest | Chen et al. (2019) | 1.64E-05 | 0 | |
| TUBB8 | NM_177987.2:c.1057G>A | p.V353I | Embryonic arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.1061G>A | p.C354Y | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.1228G>A | p.E410K | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.1249G>T | p.D417Y | Embryonic arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.292G>A | p.G98R | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.523G>A | p.V175M | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.722G>A | p.R241H | Oocyte maturation arrest | Chen et al. (2019) | 8.90E-06 | ||
| TUBB8 | NM_177987.2:c.735G>C | p.Q245H | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.763G>A | p.V255M | Embryonic arrest | Chen et al. (2019) | 2.12E-05 | 6.77E-05 | 0.0005 |
| TUBB8 | NM_177987.2:c.763G>A | p.V255M | Oocyte maturation arrest | Chen et al. (2019) | 2.12E-05 | 6.77E-05 | 0.0005 |
| TUBB8 | NM_177987.2:c.883G>C | p.D295H | Embryonic arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.1099T>C | p.F367L | Oocyte maturation arrest | Chen et al. (2019) | NF | ||
| TUBB8 | NM_177987.2:c.600T>G | p.F200L | Embryonic arrest | Chen et al. (2019) | 0 | ||
| TUBB8 | NM_177987.2:c.735G>C | p.Q245H | Oocyte maturation arrest | Lanuza-Lopez et al. (2020) | NF | ||
| TUBB8 | NM_177987.2:c.845G>C | p.R282P | Oocyte maturation arrest | Lanuza-Lopez et al. (2020) | NF | ||
| TUBB8 | NM_177987.2:c.763G>A | p.V255M | Oocyte maturation arrest | Lanuza-Lopez et al. (2020) | 2.12E-05 | 6.77E-05 | 0.0005 |
| TUBB8 | NM_177987.2:c.178G>C | p.V60L | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.1130T>C | p.L377P | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.260C>T | P.P87L | Embryonic implantation failure; fertilization failure | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.1172G>A | p.R391H | Embryonic arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.608A>G | p.D203G | Embryonic implantation failure | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c. 728C>T | p.P243L | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.1103T>C; | p.I368T; | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| NM_177987.2:c.382dup | p.D128Gfs*27 | NF | |||||
| TUBB8 | NM_177987.2:c.80_100del; | p.E27_A33del; | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| NM_177987.2:c.1103T>C | p.I368T | NF | |||||
| TUBB8 | NM_177987.2:c.82C>T; | p.H28Y; | Embryonic implantation failure; embryonic arrest | Zhao et al. (2020a) | 2.48E-05; | 4.29E-05 | |
| NM_177987.2:c.148_154delinsCACCACCACGAGGCCAGCG GTGCGACCCCCGTCCTTCCC CCACCCAACGTGCACCACC | p.Y50_N52delinsHHHEASGATPVLPPPNVHHH | NF | |||||
| TUBB8 | NM_177987.2:c.400C>T; | p.Q134*; | Embryonic arrest | Zhao et al. (2020a) | 0 | 8.41E-06 | |
| NM_177987.2:c.353A>G | p.D118G | NF | |||||
| TUBB8 | NM_177987.2:c.716G>C | p.C239S | Embryonic implantation failure; fertilization failure | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.662C>T | p.T221I | Oocyte maturation arrest | Zhao et al. (2020a) | 0 | ||
| TUBB8 | NM_177987.2:c.1139G>A | p.R380H | Embryonic implantation failure; embryonic arrest | Zhao et al. (2020a) | 0 | ||
| TUBB8 | NM_177987.2:c.1163T>C | p.M388T | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.236G>A | p.G79E | Embryonic arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.293G>A | p.G98E | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.148T>C | p.Y50H | Oocyte maturation arrest | Zhao et al. (2020a) | 0 | 0 | |
| TUBB8 | NM_177987.2:c.743C>T | p.A248V | Embryonic arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.893A>G | p.N298S | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.940G>T | p.A314S | Embryonic arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.1187A>G | p.H396R | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.728C>G | p.P243R | Embryonic implantation failure; embryonic arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.497C>T | p.T166I | Embryonic arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.12C>G | p.I4M | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.1164G>A | p.M388I | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.49G>A | p.G17S | Embryonic implantation failure | Zhao et al. (2020a) | 5.24E-06 | ||
| TUBB8 | NM_177987.2:c.1076G>A | p.R359Q | Embryonic arrest | Zhao et al. (2020a) | 7.96E-06 | 1.66E-05 | 3.19E-05 |
| TUBB8 | NM_177987.2:c.736C>G | p.L246V | Embryonic arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.925C>T | p.R309C | Embryonic arrest | Zhao et al. (2020a) | 1.22E-05 | 8.58E-06 | 0.0002 |
| TUBB8 | NM_177987.2:c.535G>C | p.V179L | Oocyte maturation arrest | Zhao et al. (2020a) | NF | ||
| TUBB8 | NM_177987.2:c.82C>T; | p.H28Y; | Embryonic arrest | Zhao et al. (2020a) | 2.48E-05 | 4.29E-05 | |
| NM_177987.2:c.398T>C | p.F133S | NF | |||||
| CEP120 | NM_153223.3:c.2840G>A | p.R947H | Increased egg aneuploidy relative to maternal age | Tyc et al. (2020) | NF | ||
Reference sequence or reference sequence version number not reported
Frequency in gnomAD v.2.1.1 exomes
Frequency in ExAc v1.0
Frequency in gnomAD v2.1.1 genomes
NF, not found
NP, reference sequence not provided
2b. Spindle building
Because the spindle is essential for chromosome segregation, variants in genes encoding spindle components could cause infertility. Microtubules are composed of α- and β-tubulin heterodimers, and TUBB8 is the only primate-specific isotype of the β-tubulin family (Fig. 4). It contributes virtually all of the β-tubulin in primate oocytes (Feng et al., 2016a). TUBB8 was first identified as a genetic correlate for human infertility in a family with at least four generations of female infertility (Feng et al., 2016a). Two family members underwent IVF. In both cases, the retrieved oocytes were either morphologically abnormal or, more often, MI-arrested. Subsequently, six additional families with TUBB8 mutations and infertility were identified (Feng et al., 2016a) (Table 2). In each family, retrieved oocytes had either a morphologically abnormal or an absent spindle. When the mutant forms of TUBB8 were overexpressed in HeLa cells or microinjected into mouse oocytes, spindles were unipolar or absent (Feng et al., 2016a).
Figure 4. Spindle assembly and chromosome segregation are vulnerable to female infertility-associated genetic variants.
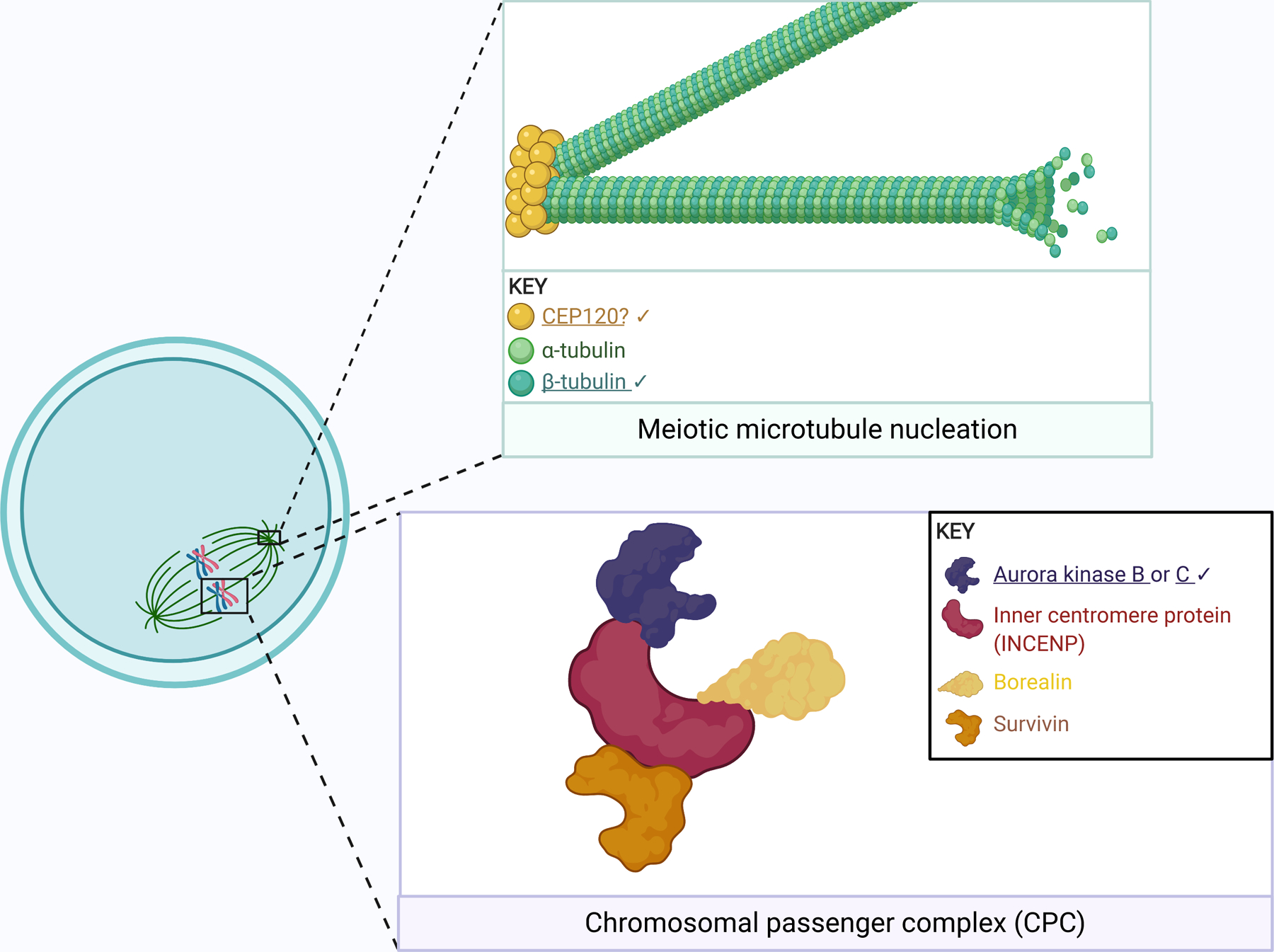
Schematic of microtubule nucleation (green box) and the chromosomal passenger complex (purple box). Microtubules are composed of tubulin, a heterodimeric polymer of α- and β-tubulin. TUBB8 encodes a β-tubulin. CEP120 may also act in meiotic spindle building, as it does in mitosis. The chromosomal passenger complex (purple box) is comprised of the illustrated components, with either Aurora kinase (AURK) B or C as the catalytic subunit. AURKC expression is exclusive to meiosis and participates in chromosome alignment, kinetochore-microtubule attachment correction, and spindle building; in meiosis, AURKB also supervises kinetochore-microtubule attachment. Note that the spindle reflects a mouse oocyte. Exact stoichiometry is not depicted. Underlined gene products are implicated in infertility. Check marks (✔) indicate gene products discussed in this review. Created using BioRender.com.
Until recently, TUBB8 function was poorly characterized, perhaps because its expression is limited to primate oocytes. New work shows that TUBB8’s numerous mutations give rise to a range of developmental arrest phenotypes (Table 2). Subsequent studies have identified several other TUBB8-variant phenotypes. One phenotype is fertilization failure (Feng et al., 2016b, Chen et al., 2019, Wang et al., 2018, Chen et al., 2017). Other phenotypes include, two-pronuclear zygotes with complete cleavage failure (Yuan et al., 2018), four-cell or eight-cell embryonic arrest (Chen et al., 2019, Feng et al., 2016b, Chen et al., 2017) and implantation failure (Chen et al., 2019). Perplexingly, there is no unifying genetic inheritance pattern of patients with TUBB8 variants; the cohorts include heterozygotes, homozygotes, and compound heterozygotes. The variants span all four exons of TUBB8 and its two functional domains.
Nearly all of the patients with TUBB8-deficient oocytes underwent years of repeated IVF procedures, despite bearing oocytes that could not complete development. However, women with TUBB8 mutations and oocyte maturation defects were able to successfully conceive with egg donation (Lanuza-Lopez et al., 2020). TUBB8 is thus a strong candidate for clinical validation as a predictive biomarker of IVF failure so that patients may consider egg donation earlier.
Centrosomal Protein 120 (CEP120) plays an essential role in mitotic microtubule nucleation and therefore is likely required for successful meiosis and fertility (Fig. 4) (Mahjoub et al., 2010). Whole-exome sequencing of infertile patients revealed enrichment of a Cep120 variant (NM_153223.3:c.2840G>A) in patients with disproportionately high rates of embryonic aneuploidy in their corresponding maternal age group (Tyc et al., 2020). When the identified variant was expressed in mouse oocytes, microtubule nucleation efficiency decreased and metaphase I chromosome misalignment increased, which resulted in increased frequency of egg aneuploidy at metaphase II.
Although mitotic cells form spindles using centriole-containing centrosomes, mammalian oocytes lack centrioles (Manandhar et al., 2005). Mouse oocytes contain acentriolar MTOCs. CEP120 localized to the forming MTOCs and ectopic expression of the variant reduced microtubule nucleation, which could contribute to unequal pulling forces on kinetochores upon anaphase I onset (Tyc et al., 2020). However, because human oocytes use a chromatin-based nucleation pathway, further evaluation of a role for CEP120 in human oocytes is warranted. Importantly, this study is the first whole-exome sequencing analysis in a cohort of subfertile patients grouped based on the proportion of in vitro fertilized embryos that are aneuploid. Such approaches help to develop biomarkers for age-independent risk of oocyte aneuploidy.
3. Chromosome segregation
Following meiotic resumption, oocytes continue to build the spindle and align chromosomes at the metaphase plate (Figs. 1, 4). The spindle is essential to segregate homologous chromosomes in anaphase I and, after fertilization, sister chromatids in anaphase II. To ensure euploid egg formation, spindle microtubules must correctly attach to kinetochore complexes that dock at centromeres. In meiosis I, sister chromatid kinetochores act as a unit and attach to microtubules that nucleate from the same spindle pole. However, in meiosis II, sister kinetochores will attach to microtubules from opposite poles. Gene variants appear to impact each of these steps and cause infertility.
The Aurora kinases (AURK) are serine/threonine kinases critical in mitosis and meiosis. Mammals have three isoforms: A, B, and C. AURKB and AURKC are members of the chromosomal passenger complex (CPC), essential for faithful chromosome transmission in meiosis by allowing correction of improperly attached kinetochore microtubules (Fig. 4) (Sharif et al., 2010, Balboula et al., 2014). Female Aurkc-knockout mice are subfertile and their oocytes frequently have chromosome misalignment (Schindler et al., 2012). Female Aurkb-knockout mice, however, are significantly less fertile than Aurkc-knockout mice and undergo premature age-related infertility (Nguyen et al., 2018).
Anaphase I onset in oocytes with chromosome misalignment often leads to aneuploidy. A protective AURKB mutation (hg19:g.8111091A>G) was identified in a patient who had low embryo aneuploidy relative to her age (39-years old, 33% aneuploid blastocysts) (Nguyen et al., 2017). Compared to mouse oocytes expressing WT AURKB, oocytes expressing the AURKB NC_000017.10:c.T116C variant more rapidly aligned their chromosomes when challenged with a microtubule re-growth assay. These data indicate that this gain-of-function AURKB variant protects against aneuploidy. Women who harbor this variant may have longer reproductive lifespans. Maternal Aurkb variants have also been associated with more complex aneuploid phenotypes, including a fetus with abnormal brain and skull development (anencephaly) with >10% aneuploid cells (Lopez-Carrasco et al., 2013). These results suggest that AURKB catalytic function is closely tied to egg ploidy where increased kinase function facilitates euploidy, while decreased kinase function can cause aneuploidy.
The difference in the impact of Aurkb and Aurkc loss on female mouse fertility is reflected in human fertility. AURKC expression is almost exclusive to germ cells (Fellmeth et al., 2015) and the first three embryonic division cycles (Avo Santos et al., 2011). AURKC variants are associated with spermatogenesis defects leading to male infertility (reviewed in Coutton et al., 2015, and Quartuccio et al., 2015), but AURKC dysfunction in female infertility is more sparsely described. An AURKC deletion has been detected in infertile men, but two fertile sisters also harbored the deletion (Dieterich et al., 2009). However, because the Aurkc knockout phenotype in female mice is smaller litter sizes, not sterility, a potential subfertility effect of AURKC defects in human females may be subtle and potentially overlooked.
4. Meiotic cell-cycle regulation: maintaining arrests and controlling exits
To maintain prophase I arrest and to trigger exit from the metaphase II arrest (Figs. 1, 5), the activity of maturation promoting factor complex (MPF), which consists of a heterodimer of CDK1 and CCNB1 (cyclin B) (Han et al., 2005), must be inhibited. This inhibition is accomplished by phosphorylation on a tyrosine residue in CDK1 and regulation of cyclin B stability. Aberrant regulation of CDK1 activity could alter the precision in timing required for fertilization. Indeed, several genes involved in the transition out of metaphase I and arrest at metaphase II are implicated in infertility, including POF1B and CDC20 (Fig. 5) (Huang et al., 2019, Lacombe et al., 2006, Ledig et al., 2015, Oral et al., 2019, Wang et al., 2020, Weinberg-Shukron et al., 2015, Zhao et al., 2020b).
WEE2 oocyte meiosis inhibiting kinase (WEE2 or WEE1B) is an oocyte-specific tyrosine kinase that phosphorylates and inhibits CDK1 (Figs. 3B, 5); in prophase I, this phosphorylation maintains arrest, but, after fertilization, it promotes meiotic exit (Han et al., 2005, Oh et al., 2011, Oh et al., 2010). In rhesus macaque and mouse oocytes, Wee2 knockdown causes some prophase I-arrested oocytes to resume meiosis prematurely (Hanna et al., 2010, Han et al., 2005). In humans, WEE2 inhibitors are being studied as hormonal contraception, suggesting that WEE2 may play a role in human reproduction (Hanna et al., 2019).
WEE2 variants are associated with fertilization failure after intracytoplasmic sperm injection (ICSI). In the first identified cases of WEE2-associated fertilization failure, metaphase II eggs were retrieved from four women ranging in age from 27 to 37. The WEE2 mutations were all homozygous (Table 3). In each case, the patients underwent menarche at an appropriate age, had regular menses, and generated polar-body-extruding oocytes. However, ICSI was unsuccessful and pronuclei failed to form. Surprisingly, the microinjection of WT-WEE2 cRNA into the four patients’ eggs rescued the fertilization failure phenotype, and two pronuclei formed. Two of the four oocytes developed into euploid blastocysts, while the others arrested (Sang et al., 2018). No further clinical follow-up was described. The homozygous inheritance of the mutation and the phenotype rescue with WT WEE2 suggests autosomal recessive inheritance.
Table 3. Female infertility-associated genetic variants affecting cell cycle regulation.
Variants are listed in order of publication year. Frequencies of small (<1kb) genetic variants were identified in gnomAD v2.1.1 and ExAc v1.0 via ANNOVAR.
| Gene | DNA Variant | Protein Variant | Clinical Phenotype | Reference | Frequency | ||
|---|---|---|---|---|---|---|---|
| gnomAD exome1 | ExAc2 | gnomAD genome3 | |||||
| WEE2 | NM_001105558.1:c.700G>C | p.D234H | Fertilization failure | Sang et al. (2018) | NF | ||
| WEE2 | NM_001105558.1:c.1473dup | p.T493Nfs*39 | Fertilization failure | Sang et al. (2018) | NF | ||
| WEE2 | NM_001105558.1:c.220_223del | p.E75Vfs*6 | Fertilization failure | Sang et al. (2018) | NF | ||
| WEE2 | NM_001105558.1:c.1006_1007insTA | p.H337Yfs*24 | Fertilization failure | Sang et al. (2018) | NF | ||
| WEE2 | NC_000007.13:c.598C>T | p.R200* | Fertilization failure | Zhou et al. (2019)† | 1.61E-0 | 2.50E-05 | 9.56E-05 |
| NC_000007.13:c.1319G>C | p.W440S | NF | |||||
| WEE2 |
NM_001105558:c.293_294ins CTGAGACACCAGCCCAACC |
p.P98P fsX2 | Fertilization failure | Zhao et al. (2019)† | NF | ||
| WEE2 | NM_001105558:c.1576T>G | p.Y526D | Fertilization failure | Zhao et al. (2019)† | NF | ||
| WEE2 | NM_001105558:c.991C>A | p.H331N | Fertilization failure | Zhao et al. (2019)† | NF | ||
| NM_001105558:c.1304_1307del | p.T435M fsX31 | NF | |||||
| WEE2 | NM_001105558:c.341_342del | p.K114N fsX20 | Fertilization failure | Zhao et al. (2019)† | NF | ||
| NM_001105558:c.864G>C | p.Q288H | NF | |||||
| WEE2 | NM_001105558:c.1A>G | p.0? | Fertilization failure | Zhao et al. (2019)† | NF | ||
| NM_001105558:c.1261G>A | p.G421R | NF | |||||
| WEE2 | NM_001105558.1:c.585G>C | p.K195N | Fertilization failure | Dai et al. (2019) | NF | ||
| WEE2 | NM_001105558.1:c.1228C>T | p.R410W | Blastocyst arrest after fertilization or fertilization failure | Dai et al. (2019) | 2.01E-05 | ||
| WEE2 | NM_001105558.1:c.1006_1007dup | p.H337Yfs*24 | Fertilization failure | Dai et al. (2019) | NF | ||
| WEE2 | NM_001105558.1:c.1006_1007dup | p.H337Yfs*24 | Fertilization failure | Dai et al. (2019) | NF | ||
| NM_001105558.1:c.1136–2A>G | p.G379Efs*6/ p.D380Lfs*39 | NF | |||||
| WEE2 | NM_001105558:c.619C>T | p.R207C | Fertilization failure | Yang et al. (2019) | NF | ||
| WEE2 | NC_000007.13:c.1228C>T | p.R410W | Fertilization failure | Zhang et al. (2019)† | 2.01E-05 | ||
| WEE2 | NC_000007.13:c.725G>C | p.R242P | Fertilization failure | Zhang et al. (2019)† | NF | ||
| NC_000007.13:c.997T>C | p.S333P | NF | |||||
| WEE2 | NC_000007.13:c.1221G>A | p.D408Vfs* | Fertilization failure | Zhang et al. (2019)† | NF | ||
| NC_000007.13:c.220_223del | p.E75Vfs*6 | NF | |||||
| WEE2 | NC_000007.13:c.1006_1007insTA | p.H337Yfs*24 | Fertilization failure | Zhang et al. (2019)† | NF | ||
| NC_000007.13:c.1286_1288del | p.G429del | NF | |||||
| WEE2 | NC_000007.13:c.220_223del | p.E75Vfs*6 | Fertilization failure | Zhang et al. (2019)† | NF | ||
| NC_000007.13:c.598C>T | p.R200* | 1.61E-05 | 9.50E-05 | 9.56E-05 | |||
| WEE2 | NC_000007.13:c.1184G>A | p.G395E | Fertilization failure | Zhang et al. (2019)† | 4.01E-06 | ||
Reference sequence or reference sequence version number not reported
Frequency in gnomAD v.2.1.1
Frequency in ExAc v1.0
Frequency in gnomAD v2.1.1 genomes. exomes.
When the identified mutants were expressed in mouse oocytes, polar body extrusion and phosphorylated CDK1 decreased significantly compared to controls (Sang et al., 2018). These data suggest that, in patients with these WEE2 variants, WEE2 is unable to sufficiently inactivate CDK1, resulting in impaired meiotic exit and pronucleus formation. Subsequently, WEE2 mutants were associated with patients who, despite having regular menses, had eggs that could not form pronuclei upon ICSI (Zhang et al., 2019b, Zhao et al., 2019, Zhou et al., 2019, Dai et al., 2019, Yang et al., 2019a). However, one patient with a WEE2 mutation (NM_001105558.1:c.1228C>T) demonstrated a different phenotype: poor-quality embryos. Of 30 eggs, three formed normal-looking 2-pronuclei zygotes and three formed abnormal 3-pronuclei zygotes. Of the 2-pronuclei zygotes, all later arrested after cleavage (Dai et al., 2019). The data from this patient show that WEE2 mutations may be associated with additional undiscovered oocyte quality phenotypes that impact embryonic development.
Conclusion
This review identified 251 reports of female patients with infertility-associated genotypes; some appeared frequently either within a given study, such as SYCE1 NC_000010.10:g.135254039_135377532dup in Bestetti et al. 2019, or across multiple studies, like MCM8 NC_000020.10:g.5948227G>A. Other reports, such as that of CEP120 NM_153223.3:c.2840G>A, were unique. In the gnomAD database, the population frequencies of the variants identified in this review range from not found to common (>2%) (Fig. 6). Most variants were either not found in the population database or were extremely rare, reflecting severe reproductive phenotypes associated with these variants that diminish evolutionary fitness, such as oocyte maturation arrest. In contrast, more common infertility variants tend to be associated with less severe reproductive phenotypes, such as increased risk of early menopause (MCM8) or increased egg aneuploidy (CEP120).
Figure 6. Population frequencies of infertility-associated variants.
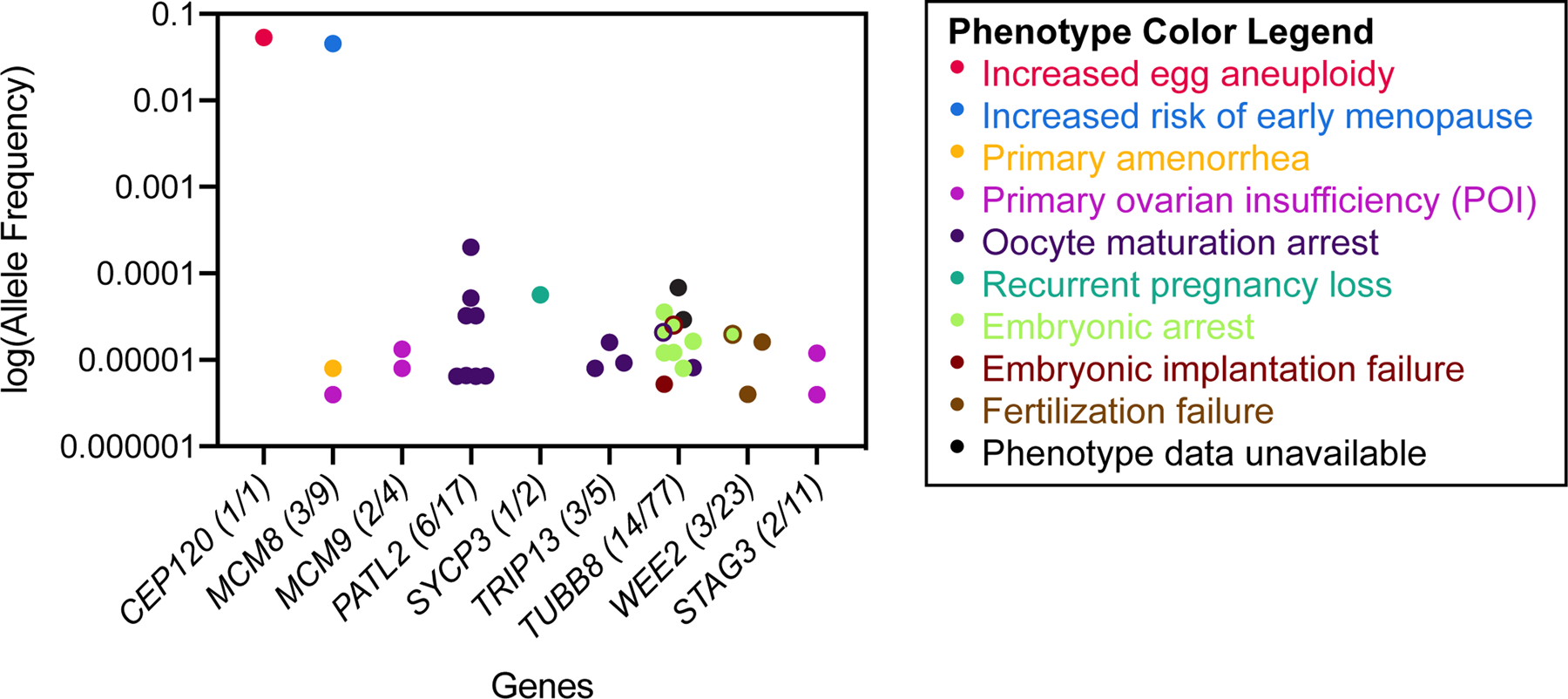
The population allele frequencies of the female infertility-associated variants listed in Tables 1 through 4 were identified in the gnomAD database (v2.1.1). The number of variants that had a non-zero allele frequency in gnomAD and the total number of variants in each gene is denoted in parentheses after the gene name. Genes for which no reported infertility-associated variants are present in the gnomAD database are not included in the figure. For each allele, dot color represents an associated reproductive phenotype (see key). Multiple phenotypes are represented by two-color dots. Frequencies are shown on log10 scale.
The range of reproductive phenotypes caused by meiotic dysregulation brings into focus the fundamental goal of fertility biomarker development: informed reproductive decision-making. Patients with pathologic variants in meiotic genes deserve realistic expectations of their fertility. For example, several patients with WEE2 variants that cause fertilization failure underwent a decade of fertility treatments with no success. It is critically important to identify and validate infertility-associated variants in meiotic genes so that clinicians can clearly assess future fertility.
This review shows that variants in meiotic genes can cause infertility. How do we bridge the gap between the literature reviewed here and the future of genetic fertility assessments? The most pressing problem in clearly identifying the molecular pathophysiology of oocyte-based infertility is the imprecision of the disease phenotype. The spectrum of subfertility is particularly problematic. A common treatment course in the United States is that, after having difficulty conceiving, a couple is referred to a fertility clinic. If conception occurs, the patient would leave the fertility clinic and continue prenatal care with an obstetrician. Patients such as these may be subfertile, rather than sterile, but there are very few studies conducted on such patients due to the transition of care. Clear metrics of subfertility and difficult conception are beginning to emerge. Increased use of metrics such as time to conception and blastocyst aneuploidy rate in assessing fertility status and the development of additional metrics will open the door to better understanding the etiology of subfertility.
The second opportunity for improvement in genetic infertility research is increasing the quality and standardization of genotype-phenotype associations. Currently, candidate variants are inconsistently experimentally validated; often, the initial study does not conduct further experiments after a candidate variant is identified. Increased collaboration between clinical researchers and basic scientists will produce a comprehensive picture of the genotype-phenotype association. In the absence of collaboration, study authors should actively facilitate future investigations by clearly reporting their results and depositing their data in public databases. This includes providing detailed clinical information when reporting an associated genotype. As such, we recommend that all researchers investigating genetic mechanisms of infertility follow two sets of standards: (i) the international nomenclature guidelines of the Human Genome Variation Society (den Dunnen et al., 2016) and (ii) the sequence variant causality guidelines published by a working group convened by the US National Human Genome Research Institute (MacArthur et al., 2014). Studies that adhere to these guidelines provide the foundation for follow-up research and can lead to strong clinical associations.
Given the apparent causal relationships between the meiotic genes that drive major meiotic processes described here and their clinical phenotypes, it is possible to envision a future in which maternal genetic biomarkers complement existing clinical approaches to infertility. Genetic evaluations could occur before a patient attempts to conceive or after an initial infertility diagnosis. In the interim, there are likely many undiscovered genotype-phenotype associations in meiotic-cause infertility. Together, the infertility and meiosis communities can develop new prognostic indicators of conception success.
Acknowledgements
The authors thank Siqi Sun for assistance with obtaining values for Figure 6. This work was supported by a grant from the National Institutes of Health to K.S. and J.X. (R01 HD091331).
Footnotes
Declaration of Interests
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported. Karen Schindler is an Associate Editor of Reproduction. Karen Schindler was not involved in the review or editorial process for this paper, on which she is listed as an author.
Primary ovarian insufficiency is the terminology preferred by the National Institutes of Health. Other works, including those cited in this review, may use the term “premature ovarian failure” (POF). Both terms refer to the same condition. (2014. Committee opinion no. 605: primary ovarian insufficiency in adolescents and young women. Obstet Gynecol, 124, 193–7.)
References
- 2014. Committee opinion no. 605: primary ovarian insufficiency in adolescents and young women. Obstet Gynecol, 124, 193–7. [DOI] [PubMed] [Google Scholar]
- AL-AGHA AE, AHMED IA, NUEBEL E, MORIWAKI M, MOORE B, PEACOCK KA, MOSBRUGER T, NEKLASON DW, JORDE LB, YANDELL M, et al. 2018. Primary Ovarian Insufficiency and Azoospermia in Carriers of a Homozygous PSMC3IP Stop Gain Mutation. J Clin Endocrinol Metab, 103, 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALANI E, PADMORE R & KLECKNER N 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell, 61, 419–36. [DOI] [PubMed] [Google Scholar]
- ALASIRI S, BASIT S, WOOD-TRAGESER MA, YATSENKO SA, JEFFRIES EP, SURTI U, KETTERER DM, AFZAL S, RAMZAN K, FAIYAZ-UL HAQUE M, et al. 2015. Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J Clin Invest, 125, 258–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMERICAN COLLEGE OF OBSTETRICS AND GYNECOLOGY 2019. Infertility Workup for the Women’s Health Specialist: ACOG Committee Opinion, Number 781. Obstet Gynecol, 133, e377–e384. [DOI] [PubMed] [Google Scholar]
- ATIENZA JM, ROTH RB, ROSETTE C, SMYLIE KJ, KAMMERER S, REHBOCK J, EKBLOM J & DENISSENKO MF 2005. Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol Cancer Ther, 4, 361–8. [DOI] [PubMed] [Google Scholar]
- AVO SANTOS M, VAN DE WERKEN C, DE VRIES M, JAHR H, VROMANS MJ, LAVEN JS, FAUSER BC, KOPS GJ, LENS SM & BAART EB 2011. A role for Aurora C in the chromosomal passenger complex during human preimplantation embryo development. Hum Reprod, 26, 1868–81. [DOI] [PubMed] [Google Scholar]
- BALBOULA AZ & SCHINDLER K 2014. Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet, 10, e1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELL AD, MELLO CJ, NEMESH J, BRUMBAUGH SA, WYSOKER A & MCCARROLL SA 2019. Insights about variation in meiosis from 31,228 human sperm genomes. bioRxiv, 625202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERGERAT A, DE MASSY B, GADELLE D, VAROUTAS PC, NICOLAS A & FORTERRE P 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature, 386, 414–7. [DOI] [PubMed] [Google Scholar]
- BERNARDINI L, GIANAROLI L, FORTINI D, CONTE N, MAGLI C, CAVANI S, GAGGERO G, TINDIGLIA C, RAGNI N & VENTURINI PL 2000. Frequency of hyper-, hypohaploidy and diploidy in ejaculate, epididymal and testicular germ cells of infertile patients. Hum Reprod, 15, 2165–72. [DOI] [PubMed] [Google Scholar]
- BIRKENBIHL RP & SUBRAMANI S 1992. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res, 20, 6605–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOLCUN-FILAS E, HALL E, SPEED R, TAGGART M, GREY C, DE MASSY B, BENAVENTE R & COOKE HJ 2009. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet, 5, e1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOLOR H, MORI T, NISHIYAMA S, ITO Y, HOSOBA E, INAGAKI H, KOGO H, OHYE T, TSUTSUMI M, KATO T, et al. 2009. Mutations of the SYCP3 gene in women with recurrent pregnancy loss. Am J Hum Genet, 84, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOUALI N, FRANCOU B, BOULIGAND J, IMANCI D, DIMASSI S, TOSCA L, ZAOUALI M, MOUGOU S, YOUNG J, SAAD A, et al. 2017. New MCM8 mutation associated with premature ovarian insufficiency and chromosomal instability in a highly consanguineous Tunisian family. Fertil Steril, 108, 694–702. [DOI] [PubMed] [Google Scholar]
- BUONOMO SB, CLYNE RK, FUCHS J, LOIDL J, UHLMANN F & NASMYTH K 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell, 103, 387–98. [DOI] [PubMed] [Google Scholar]
- CABURET S, ARBOLEDA VA, LLANO E, OVERBEEK PA, BARBERO JL, OKA K, HARRISON W, VAIMAN D, BEN-NERIAH Z, GARCIA-TUNON I, et al. 2014. Mutant cohesin in premature ovarian failure. N Engl J Med, 370, 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CABURET S, HEDDAR A, DARDILLAC E, CREUX H, LAMBERT M, MESSIAEN S, TOURPIN S, LIVERA G, LOPEZ BS & MISRAHI M 2020. Homozygous hypomorphic BRCA2 variant in primary ovarian insufficiency without cancer or Fanconi anaemia trait. J Med Genet [DOI] [PubMed] [Google Scholar]
- CABURET S, TODESCHINI AL, PETRILLO C, MARTINI E, FARRAN ND, LEGOIS B, LIVERA G, YOUNIS JS, SHALEV S & VEITIA RA 2019. A truncating MEIOB mutation responsible for familial primary ovarian insufficiency abolishes its interaction with its partner SPATA22 and their recruitment to DNA double-strand breaks. EBioMedicine, 42, 524–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAO L, ALANI E & KLECKNER N 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell, 61, 1089–101. [DOI] [PubMed] [Google Scholar]
- CARLOSAMA C, ELZAIAT M, PATINO LC, MATEUS HE, VEITIA RA & LAISSUE P 2017. A homozygous donor splice-site mutation in the meiotic gene MSH4 causes primary ovarian insufficiency. Hum Mol Genet, 26, 3161–3166. [DOI] [PubMed] [Google Scholar]
- CARTY CL, SPENCER KL, SETIAWAN VW, FERNANDEZ-RHODES L, MALINOWSKI J, BUYSKE S, YOUNG A, JORGENSEN NW, CHENG I, CARLSON CS, et al. 2013. Replication of genetic loci for ages at menarche and menopause in the multi-ethnic Population Architecture using Genomics and Epidemiology (PAGE) study. Hum Reprod, 28, 1695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN B, LI B, LI D, YAN Z, MAO X, XU Y, MU J, LI Q, JIN L, HE L, et al. 2017. Novel mutations and structural deletions in TUBB8: expanding mutational and phenotypic spectrum of patients with arrest in oocyte maturation, fertilization or early embryonic development. Hum Reprod, 32, 457–464. [DOI] [PubMed] [Google Scholar]
- CHEN B, WANG W, PENG X, JIANG H, ZHANG S, LI D, LI B, FU J, KUANG Y, SUN X, et al. 2019. The comprehensive mutational and phenotypic spectrum of TUBB8 in female infertility. Eur J Hum Genet, 27, 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN C, JOMAA A, ORTEGA J & ALANI EE 2014a. Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proc Natl Acad Sci U S A, 111, E44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN CT, FERNANDEZ-RHODES L, BRZYSKI RG, CARLSON CS, CHEN Z, HEISS G, NORTH KE, WOODS NF, RAJKOVIC A, KOOPERBERG C, et al. 2012. Replication of loci influencing ages at menarche and menopause in Hispanic women: the Women’s Health Initiative SHARe Study. Hum Mol Genet, 21, 1419–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN CT, LIU CT, CHEN GK, ANDREWS JS, ARNOLD AM, DREYFUS J, FRANCESCHINI N, GARCIA ME, KERR KF, LI G, et al. 2014b. Meta-analysis of loci associated with age at natural menopause in African-American women. Hum Mol Genet, 23, 3327–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIU A, REVENKOVA E & JESSBERGER R 2004. DNA interaction and dimerization of eukaryotic SMC hinge domains. J Biol Chem, 279, 26233–42. [DOI] [PubMed] [Google Scholar]
- CHRISTOU-KENT M, KHERRAF ZE, AMIRI-YEKTA A, LE BLEVEC E, KARAOUZENE T, CONNE B, ESCOFFIER J, ASSOU S, GUTTIN A, LAMBERT E, et al. 2018. PATL2 is a key actor of oocyte maturation whose invalidation causes infertility in women and mice. EMBO Mol Med, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COIGNET MV, ZIRPOLI GR, ROBERTS MR, KHOURY T, BANDERA EV, ZHU Q & YAO S 2017. Genetic variations, reproductive aging, and breast cancer risk in African American and European American women: The Women’s Circle of Health Study. PLoS One, 12, e0187205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLOMBO R, PONTOGLIO A & BINI M 2017. A STAG3 missense mutation in two sisters with primary ovarian insufficiency. Eur J Obstet Gynecol Reprod Biol, 216, 269–271. [DOI] [PubMed] [Google Scholar]
- COSTA Y, SPEED R, OLLINGER R, ALSHEIMER M, SEMPLE CA, GAUTIER P, MARATOU K, NOVAK I, HOOG C, BENAVENTE R, et al. 2005. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J Cell Sci, 118, 2755–62. [DOI] [PubMed] [Google Scholar]
- COUTTON C, ESCOFFIER J, MARTINEZ G, ARNOULT C & RAY PF 2015. Teratozoospermia: spotlight on the main genetic actors in the human. Hum Reprod Update, 21, 455–85. [DOI] [PubMed] [Google Scholar]
- DAI J, ZHENG W, DAI C, GUO J, LU C, GONG F, LI Y, ZHOU Q, LU G & LIN G 2019. New biallelic mutations in WEE2: expanding the spectrum of mutations that cause fertilization failure or poor fertilization. Fertil Steril, 111, 510–518. [DOI] [PubMed] [Google Scholar]
- DE MASSY B, ROCCO V & NICOLAS A 1995. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J, 14, 4589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE VRIES L, BEHAR DM, SMIRIN-YOSEF P, LAGOVSKY I, TZUR S & BASEL-VANAGAITE L 2014. Exome sequencing reveals SYCE1 mutation associated with autosomal recessive primary ovarian insufficiency. J Clin Endocrinol Metab, 99, E2129–32. [DOI] [PubMed] [Google Scholar]
- DEN DUNNEN JT, DALGLEISH R, MAGLOTT DR, HART RK, GREENBLATT MS, MCGOWAN-JORDAN J, ROUX AF, SMITH T, ANTONARAKIS SE & TASCHNER PE 2016. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum Mutat, 37, 564–9. [DOI] [PubMed] [Google Scholar]
- DIETERICH K, ZOUARI R, HARBUZ R, VIALARD F, MARTINEZ D, BELLAYOU H, PRISANT N, ZOGHMAR A, GUICHAOUA MR, KOSCINSKI I, et al. 2009. The Aurora Kinase C c.144delC mutation causes meiosis I arrest in men and is frequent in the North African population. Hum Mol Genet, 18, 1301–9. [DOI] [PubMed] [Google Scholar]
- DUNNE OM & DAVIES OR 2019. Molecular structure of human synaptonemal complex protein SYCE1. Chromosoma, 128, 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EDMONDS K 2012. Dewhurst’s Textbook of Obstetrics and Gynaecology, Hoboken, John Wiley & Sons, Inc. [Google Scholar]
- EL HACHEM H, CREPAUX V, MAY-PANLOUP P, DESCAMPS P, LEGENDRE G & BOUET PE 2017. Recurrent pregnancy loss: current perspectives. Int J Womens Health, 9, 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAUCHEREAU F, SHALEV S, CHERVINSKY E, BECK-FRUCHTER R, LEGOIS B, FELLOUS M, CABURET S & VEITIA RA 2016. A non-sense MCM9 mutation in a familial case of primary ovarian insufficiency. Clin Genet, 89, 603–7. [DOI] [PubMed] [Google Scholar]
- FAWCETT DW 1956. The fine structure of chromosomes in the meiotic prophase of vertebrate spermatocytes. J Biophys Biochem Cytol, 2, 403–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FELLMETH JE, GORDON D, ROBINS CE, SCOTT RT JR., TREFF NR & SCHINDLER K 2015. Expression and characterization of three Aurora kinase C splice variants found in human oocytes. Mol Hum Reprod, 21, 633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FENG R, SANG Q, KUANG Y, SUN X, YAN Z, ZHANG S, SHI J, TIAN G, LUCHNIAK A, FUKUDA Y, et al. 2016a. Mutations in TUBB8 and Human Oocyte Meiotic Arrest. N Engl J Med, 374, 223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FENG R, YAN Z, LI B, YU M, SANG Q, TIAN G, XU Y, CHEN B, QU R, SUN Z, et al. 2016b. Mutations in TUBB8 cause a multiplicity of phenotypes in human oocytes and early embryos. J Med Genet, 53, 662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERRAS C, FERNANDES S, SILVA J, BARROS A & SOUSA M 2012. Expression analysis of MLH3, MLH1, and MSH4 in maturation arrest. Reprod Sci, 19, 587–96. [DOI] [PubMed] [Google Scholar]
- FIEVET A, BELLANGER D, ZAHED L, BURGLEN L, DERRIEN AC, DUBOIS D’ENGHIEN C, LESPINASSE J, PARFAIT B, PEDESPAN JM, RIEUNIER G, et al. 2020. DNA repair functional analyses of NBN hypomorphic variants associated with NBN-related infertility. Hum Mutat, 41, 608–618. [DOI] [PubMed] [Google Scholar]
- FRANCA MM, NISHI MY, FUNARI MFA, LERARIO AM, BARACAT EC, HAYASHIDA SAY, MACIEL GAR, JORGE AAL & MENDONCA BB 2019. Two rare loss-of-function variants in the STAG3 gene leading to primary ovarian insufficiency. Eur J Med Genet, 62, 186–189. [DOI] [PubMed] [Google Scholar]
- GARCIA V, PHELPS SE, GRAY S & NEALE MJ 2011. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature, 479, 241–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEISINGER A & BENAVENTE R 2016. Mutations in Genes Coding for Synaptonemal Complex Proteins and Their Impact on Human Fertility. Cytogenetic and Genome Research, 150, 77–85. [DOI] [PubMed] [Google Scholar]
- GIRARD C, ROELENS B, ZAWADZKI KA & VILLENEUVE AM 2018. Interdependent and separable functions of Caenorhabditis elegans MRN-C complex members couple formation and repair of meiotic DSBs. Proc Natl Acad Sci U S A, 115, E4443–E4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOLDBERG Y, HALPERN N, HUBERT A, ADLER SN, COHEN S, PLESSER-DUVDEVANI M, PAPPO O, SHAAG A & MEINER V 2015. Mutated MCM9 is associated with predisposition to hereditary mixed polyposis and colorectal cancer in addition to primary ovarian failure. Cancer Genet, 208, 621–4. [DOI] [PubMed] [Google Scholar]
- GOODYER W, KAITNA S, COUTEAU F, WARD JD, BOULTON SJ & ZETKA M 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell, 14, 263–74. [DOI] [PubMed] [Google Scholar]
- GOZUACIK D, CHAMI M, LAGORCE D, FAIVRE J, MURAKAMI Y, POCH O, BIERMANN E, KNIPPERS R, BRECHOT C & PATERLINI-BRECHOT P 2003. Identification and functional characterization of a new member of the human Mcm protein family: hMcm8. Nucleic Acids Res, 31, 570–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRADY WM & CARETHERS JM 2008. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology, 135, 1079–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUIRALDELLI MF, EYSTER C, WILKERSON JL, DRESSER ME & PEZZA RJ 2013. Mouse HFM1/Mer3 is required for crossover formation and complete synapsis of homologous chromosomes during meiosis. PLoS Genet, 9, e1003383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO T, ZHAO S, ZHAO S, CHEN M, LI G, JIAO X, WANG Z, ZHAO Y, QIN Y, GAO F, et al. 2017. Mutations in MSH5 in primary ovarian insufficiency. Hum Mol Genet, 26, 1452–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN SJ, CHEN R, PARONETTO MP & CONTI M 2005. Wee1B is an oocyte-specific kinase involved in the control of meiotic arrest in the mouse. Curr Biol, 15, 1670–6. [DOI] [PubMed] [Google Scholar]
- HANNA CB, YAO S, MARTIN M, SCHONBRUNN E, GEORG GI, JENSEN JT & CUELLAR RAD 2019. Identification and Screening of Selective WEE2 Inhibitors to Develop Non-Hormonal Contraceptives that Specifically Target Meiosis. ChemistrySelect, 4, 13363–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANNA CB, YAO S, PATTA MC, JENSEN JT & WU X 2010. WEE2 is an oocyte-specific meiosis inhibitor in rhesus macaque monkeys. Biol Reprod, 82, 1190–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARTFORD SA, LUO Y, SOUTHARD TL, MIN IM, LIS JT & SCHIMENTI JC 2011. Minichromosome maintenance helicase paralog MCM9 is dispensible for DNA replication but functions in germ-line stem cells and tumor suppression. Proc Natl Acad Sci U S A, 108, 17702–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HASSOLD T & HUNT P 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet, 2, 280–91. [DOI] [PubMed] [Google Scholar]
- HASSOLD TJ 1986. Chromosome abnormalities in human reproductive wastage. Trends in Genetics, 2, 105–110. [Google Scholar]
- HE C, KRAFT P, CHEN C, BURING JE, PARE G, HANKINSON SE, CHANOCK SJ, RIDKER PM, HUNTER DJ & CHASMAN DI 2009. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet, 41, 724–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE WB, TU CF, LIU Q, MENG LL, YUAN SM, LUO AX, HE FS, SHEN J, LI W, DU J, et al. 2018. DMC1 mutation that causes human non-obstructive azoospermia and premature ovarian insufficiency identified by whole-exome sequencing. J Med Genet, 55, 198–204. [DOI] [PubMed] [Google Scholar]
- HEDDAR A, BECKERS D, FOUQUET B, ROLAND D & MISRAHI M 2020. A novel phenotype combining primary ovarian insufficiency growth retardation and pilomatricomas with MCM8 mutation. J Clin Endocrinol Metab [DOI] [PubMed] [Google Scholar]
- HEDDAR A, DESSEN P, FLATTERS D & MISRAHI M 2019. Novel STAG3 mutations in a Caucasian family with primary ovarian insufficiency. Mol Genet Genomics, 294, 1527–1534. [DOI] [PubMed] [Google Scholar]
- HERNANDEZ-LOPEZ D, GEISINGER A, TROVERO M, SANTINAQUE F, BRAUER M, FOLLE G, BENAVENTE R & RODRÍGUEZ-CASURIAGA R 2020. Familial primary ovarian insufficiency associated with a SYCE1 point mutation: Defective meiosis elucidated in humanized mice [DOI] [PubMed] [Google Scholar]
- HOLUBCOVA Z, BLAYNEY M, ELDER K & SCHUH M 2015. Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science, 348, 1143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOPKINS J, HWANG G, JACOB J, SAPP N, BEDIGIAN R, OKA K, OVERBEEK P, MURRAY S & JORDAN PW 2014. Meiosis-specific cohesin component, Stag3 is essential for maintaining centromere chromatid cohesion, and required for DNA repair and synapsis between homologous chromosomes. PLoS Genet, 10, e1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG W, WANG J, PANG M, ZHAO Q, KONG L, MAO Y, LI W & LIANG B 2019. Copy Number Variations in Female Infertility in China. Balkan J Med Genet, 22, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUSTEDT N, SAITO Y, ZIMMERMANN M, ALVAREZ-QUILON A, SETIAPUTRA D, ADAM S, MCEWAN A, YUAN JY, OLIVIERI M, ZHAO Y, et al. 2019. Control of homologous recombination by the HROB-MCM8-MCM9 pathway. Genes Dev, 33, 1397–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAILLARD S, AKLOUL L, BEAUMONT M, HAMDI-ROZE H, DUBOURG C, ODENT S, DUROS S, DEJUCQ-RAINSFORD N, BELAUD-ROTUREAU MA & RAVEL C 2016. Array-CGH diagnosis in ovarian failure: identification of new molecular actors for ovarian physiology. J Ovarian Res, 9, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAILLARD S, MCELREAVY K, ROBEVSKA G, AKLOUL L, GHIEH F, SREENIVASAN R, BEAUMONT M, BASHAMBOO A, BIGNON-TOPALOVIC J, NEYROUD AS, et al. 2020. STAG3 homozygous missense variant causes primary ovarian insufficiency and male non-obstructive azoospermia. Mol Hum Reprod [DOI] [PubMed] [Google Scholar]
- JEDIDI I, OUCHARI M & YIN Q 2019. Sex chromosomes-linked single-gene disorders involved in human infertility. Eur J Med Genet, 62, 103560. [DOI] [PubMed] [Google Scholar]
- KEENEY S, GIROUX CN & KLECKNER N 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell, 88, 375–84. [DOI] [PubMed] [Google Scholar]
- KEENEY S & KLECKNER N 1995. Covalent protein-DNA complexes at the 5’ strand termini of meiosis-specific double-strand breaks in yeast. Proc Natl Acad Sci U S A, 92, 11274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEIN F, MAHR P, GALOVA M, BUONOMO SB, MICHAELIS C, NAIRZ K & NASMYTH K 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell, 98, 91–103. [DOI] [PubMed] [Google Scholar]
- LACOMBE A, LEE H, ZAHED L, CHOUCAIR M, MULLER JM, NELSON SF, SALAMEH W & VILAIN E 2006. Disruption of POF1B binding to nonmuscle actin filaments is associated with premature ovarian failure. Am J Hum Genet, 79, 113–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMBERTINI M, GOLDRAT O, FERREIRA AR, DECHENE J, AZIM HA JR., DESIR J, DELBAERE A, T’KINT DE ROODENBEKE MD, DE AZAMBUJA E, IGNATIADIS M, et al. 2018. Reproductive potential and performance of fertility preservation strategies in BRCA-mutated breast cancer patients. Ann Oncol, 29, 237–243. [DOI] [PubMed] [Google Scholar]
- LANUZA-LOPEZ MC, MARTINEZ-GARZA SG, SOLORZANO-VAZQUEZ JF, PAZ-CERVANTES D, GONZALEZ-ORTEGA C, MALDONADO-ROSAS I, VILLEGAS-MORENO G, VILLAR-MUNOZ LG, ARROYO-MENDEZ FA, GUTIERREZ-GUTIERREZ AM, et al. 2020. Oocyte maturation arrest produced by TUBB8 mutations: impact of genetic disorders in infertility treatment. Gynecol Endocrinol, 1–6. [DOI] [PubMed] [Google Scholar]
- LE QUESNE STABEJ P, WILLIAMS HJ, JAMES C, TEKMAN M, STANESCU HC, KLETA R, OCAKA L, LESCAI F, STORR HL, BITNER-GLINDZICZ M, et al. 2016. STAG3 truncating variant as the cause of primary ovarian insufficiency. Eur J Hum Genet, 24, 135–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEDIG S, PREISLER-ADAMS S, MORLOT S, LIEHR T & WIEACKER P 2015. Premature ovarian failure caused by a heterozygous missense mutation in POF1B and a reciprocal translocation 46,X,t(X;3)(q21.1;q21.3). Sex Dev, 9, 86–90. [DOI] [PubMed] [Google Scholar]
- LEU JY, CHUA PR & ROEDER GS 1998. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell, 94, 375–86. [DOI] [PubMed] [Google Scholar]
- LI Q, SAITO TT, MARTINEZ-GARCIA M, DESHONG AJ, NADARAJAN S, LAWRENCE KS, CHECCHI PM, COLAIACOVO MP & ENGEBRECHT J 2018. The tumor suppressor BRCA1-BARD1 complex localizes to the synaptonemal complex and regulates recombination under meiotic dysfunction in Caenorhabditis elegans. PLoS Genet, 14, e1007701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI XC & SCHIMENTI JC 2007. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet, 3, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN W, TITUS S, MOY F, GINSBURG ES & OKTAY K 2017. Ovarian Aging in Women With BRCA Germline Mutations. J Clin Endocrinol Metab, 102, 3839–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDSAY TJ & VITRIKAS KR 2015. Evaluation and treatment of infertility. Am Fam Physician, 91, 308–14. [PubMed] [Google Scholar]
- LOPEZ-CARRASCO A, OLTRA S, MONFORT S, MAYO S, ROSELLO M, MARTINEZ F & ORELLANA C 2013. Mutation screening of AURKB and SYCP3 in patients with reproductive problems. Mol Hum Reprod, 19, 102–8. [DOI] [PubMed] [Google Scholar]
- LUTZMANN M, GREY C, TRAVER S, GANIER O, MAYA-MENDOZA A, RANISAVLJEVIC N, BERNEX F, NISHIYAMA A, MONTEL N, GAVOIS E, et al. 2012. MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol Cell, 47, 523–34. [DOI] [PubMed] [Google Scholar]
- MACARTHUR DG, MANOLIO TA, DIMMOCK DP, REHM HL, SHENDURE J, ABECASIS GR, ADAMS DR, ALTMAN RB, ANTONARAKIS SE, ASHLEY EA, et al. 2014. Guidelines for investigating causality of sequence variants in human disease. Nature, 508, 469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MADDIREVULA S, COSKUN S, ALHASSAN S, ELNOUR A, ALSAIF HS, IBRAHIM N, ABDULWAHAB F, AROLD ST & ALKURAYA FS 2017. Female Infertility Caused by Mutations in the Oocyte-Specific Translational Repressor PATL2. Am J Hum Genet, 101, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAHJOUB MR, XIE Z & STEARNS T 2010. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J Cell Biol, 191, 331–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANANDHAR G, SCHATTEN H & SUTOVSKY P 2005. Centrosome reduction during gametogenesis and its significance. Biol Reprod, 72, 2–13. [DOI] [PubMed] [Google Scholar]
- MANDON-PEPIN B, TOURAINE P, KUTTENN F, DERBOIS C, ROUXEL A, MATSUDA F, NICOLAS A, COTINOT C & FELLOUS M 2008. Genetic investigation of four meiotic genes in women with premature ovarian failure. Eur J Endocrinol, 158, 107–15. [DOI] [PubMed] [Google Scholar]
- MAO-DRAAYER Y, GALBRAITH AM, PITTMAN DL, COOL M & MALONE RE 1996. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics, 144, 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARNEF A, MALDONADO M, BUGAUT A, BALASUBRAMANIAN S, KRESS M, WEIL D & STANDART N 2010. Distinct functions of maternal and somatic Pat1 protein paralogs. RNA, 16, 2094–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCGUIRE MM, BOWDEN W, ENGEL NJ, AHN HW, KOVANCI E & RAJKOVIC A 2011. Genomic analysis using high-resolution single-nucleotide polymorphism arrays reveals novel microdeletions associated with premature ovarian failure. Fertil Steril, 95, 1595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIZUTANI E, SUZUMORI N, OZAKI Y, OSETO K, YAMADA-NAMIKAWA C, NAKANISHI M & SUGIURA-OGASAWARA M 2011. SYCP3 mutation may not be associated with recurrent miscarriage caused by aneuploidy. Hum Reprod, 26, 1259–66. [DOI] [PubMed] [Google Scholar]
- MOSES MJ 1956. Chromosomal structures in crayfish spermatocytes. J Biophys Biochem Cytol, 2, 215–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURRAY A, BENNETT CE, PERRY JR, WEEDON MN, JACOBS PA, MORRIS DH, ORR N, SCHOEMAKER MJ, JONES M, ASHWORTH A, et al. 2011. Common genetic variants are significant risk factors for early menopause: results from the Breakthrough Generations Study. Hum Mol Genet, 20, 186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAIRZ K & KLEIN F 1997. mre11S--a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev, 11, 2272–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAKAMURA Y, TANAKA KJ, MIYAUCHI M, HUANG L, TSUJIMOTO M & MATSUMOTO K 2010. Translational repression by the oocyte-specific protein P100 in Xenopus. Dev Biol, 344, 272–83. [DOI] [PubMed] [Google Scholar]
- NANDI A & HOMBURG R 2016. Unexplained subfertility: diagnosis and management. The Obstetrician & Gynaecologist, 18, 107–115. [Google Scholar]
- NATSUME T, NISHIMURA K, MINOCHERHOMJI S, BHOWMICK R, HICKSON ID & KANEMAKI MT 2017. Acute inactivation of the replicative helicase in human cells triggers MCM8–9-dependent DNA synthesis. Genes Dev, 31, 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NGUYEN AL, DRUTOVIC D, VAZQUEZ BN, EL YAKOUBI W, GENTILELLO AS, MALUMBRES M, SOLC P & SCHINDLER K 2018. Genetic Interactions between the Aurora Kinases Reveal New Requirements for AURKB and AURKC during Oocyte Meiosis. Curr Biol, 28, 3458–3468 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NGUYEN AL, MARIN D, ZHOU A, GENTILELLO AS, SMOAK EM, CAO Z, FEDICK A, WANG Y, TAYLOR D, SCOTT RT JR., et al. 2017. Identification and characterization of Aurora kinase B and C variants associated with maternal aneuploidy. Mol Hum Reprod, 23, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORLING A, HIRSCHBERG AL, KARLSSON L, RODRIGUEZ-WALLBERG KA, IWARSSON E, WEDELL A & BARBARO M 2014. No mutations in the PSMC3IP gene identified in a Swedish cohort of women with primary ovarian insufficiency. Sex Dev, 8, 146–50. [DOI] [PubMed] [Google Scholar]
- OFFENBERG HH, SCHALK JA, MEUWISSEN RL, VAN AALDEREN M, KESTER HA, DIETRICH AJ & HEYTING C 1998. SCP2: a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res, 26, 2572–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OH JS, HAN SJ & CONTI M 2010. Wee1B, Myt1, and Cdc25 function in distinct compartments of the mouse oocyte to control meiotic resumption. J Cell Biol, 188, 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OH JS, SUSOR A & CONTI M 2011. Protein tyrosine kinase Wee1B is essential for metaphase II exit in mouse oocytes. Science, 332, 462–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKTAY K, KIM JY, BARAD D & BABAYEV SN 2010. Association of BRCA1 mutations with occult primary ovarian insufficiency: a possible explanation for the link between infertility and breast/ovarian cancer risks. J Clin Oncol, 28, 240–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKTAY KH, BEDOSCHI G, GOLDFARB SB, TAYLAN E, TITUS S, PALOMAKI GE, CIGLER T, ROBSON M & DICKLER MN 2020. Increased chemotherapy-induced ovarian reserve loss in women with germline BRCA mutations due to oocyte deoxyribonucleic acid double strand break repair deficiency. Fertil Steril, 113, 1251–1260 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORAL E, TOKSOY G, SOFIYEVA N, CELIK HG, KARAMAN B, BASARAN S, AZAMI A & UYGUNER ZO 2019. Clinical and Genetic Investigation of Premature Ovarian Insufficiency Cases from Turkey. J Gynecol Obstet Hum Reprod, 48, 817–823. [DOI] [PubMed] [Google Scholar]
- PARK J, LONG DT, LEE KY, ABBAS T, SHIBATA E, NEGISHI M, LUO Y, SCHIMENTI JC, GAMBUS A, WALTER JC, et al. 2013. The MCM8-MCM9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination. Mol Cell Biol, 33, 1632–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASHAIEFAR H, SHEIKHHA MH, KALANTAR SM, JAHANINEJAD T, ZAIMY MA & GHASEMI N 2013. Analysis of MLH3 C2531T polymorphism in Iranian women with unexplained infertility. Iran J Reprod Med, 11, 19–24. [PMC free article] [PubMed] [Google Scholar]
- PASIERBEK P, JANTSCH M, MELCHER M, SCHLEIFFER A, SCHWEIZER D & LOIDL J 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev, 15, 1349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETUKHOVA GV, ROMANIENKO PJ & CAMERINI-OTERO RD 2003. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev Cell, 5, 927–36. [DOI] [PubMed] [Google Scholar]
- PEZZI N, PRIETO I, KREMER L, PEREZ JURADO LA, VALERO C, DEL MAZO J, MARTINEZ AC & BARBERO JL 2000. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams-Beuren syndrome deletion. FASEB J, 14, 581–92. [DOI] [PubMed] [Google Scholar]
- PITTMAN DL, COBB J, SCHIMENTI KJ, WILSON LA, COOPER DM, BRIGNULL E, HANDEL MA & SCHIMENTI JC 1998. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell, 1, 697–705. [DOI] [PubMed] [Google Scholar]
- PORCU E, CILLO GM, CIPRIANI L, SACILOTTO F, NOTARANGELO L, DAMIANO G, DIRODI M & RONCARATI I 2020. Impact of BRCA1 and BRCA2 mutations on ovarian reserve and fertility preservation outcomes in young women with breast cancer. J Assist Reprod Genet, 37, 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRIETO I, SUJA JA, PEZZI N, KREMER L, MARTINEZ AC, RUFAS JS & BARBERO JL 2001. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat Cell Biol, 3, 761–6. [DOI] [PubMed] [Google Scholar]
- PU D, WANG C, CAO J, SHEN Y, JIANG H, LIU J, WU BL, ZHANG W & WU J 2016. Association analysis between HFM1 variation and primary ovarian insufficiency in Chinese women. Clin Genet, 89, 597–602. [DOI] [PubMed] [Google Scholar]
- QUARTUCCIO SM & SCHINDLER K 2015. Functions of Aurora kinase C in meiosis and cancer. Front Cell Dev Biol, 3, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROTHER RP, FRANK MB & THOMAS PS 1992. Purification, primary structure, bacterial expression and subcellular distribution of an oocyte-specific protein in Xenopus. Eur J Biochem, 206, 673–83. [DOI] [PubMed] [Google Scholar]
- SANG Q, LI B, KUANG Y, WANG X, ZHANG Z, CHEN B, WU L, LYU Q, FU Y, YAN Z, et al. 2018. Homozygous Mutations in WEE2 Cause Fertilization Failure and Female Infertility. Am J Hum Genet, 102, 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANSAM CL & PEZZA RJ 2015. Connecting by breaking and repairing: mechanisms of DNA strand exchange in meiotic recombination. FEBS J, 282, 2444–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANTOS MG, MACHADO AZ, MARTINS CN, DOMENICE S, COSTA EM, NISHI MY, FERRAZ-DE-SOUZA B, JORGE SA, PEREIRA CA, SOARDI FC, et al. 2014. Homozygous inactivating mutation in NANOS3 in two sisters with primary ovarian insufficiency. Biomed Res Int, 2014, 787465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAZEGARI A, KALANTAR SM, PASHAIEFAR H, MOHTARAM S, HONARVAR N, FEIZOLLAHI Z & GHASEMI N 2014. The T657C polymorphism on the SYCP3 gene is associated with recurrent pregnancy loss. J Assist Reprod Genet, 31, 1377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHAR P, FASI M & JESSBERGER R 2004. SMC1 coordinates DNA double-strand break repair pathways. Nucleic Acids Res, 32, 3921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHINDLER K, DAVYDENKO O, FRAM B, LAMPSON MA & SCHULTZ RM 2012. Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proc Natl Acad Sci U S A, 109, E2215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHUH M & ELLENBERG J 2007. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell, 130, 484–98. [DOI] [PubMed] [Google Scholar]
- SCULLY R, CHEN J, PLUG A, XIAO Y, WEAVER D, FEUNTEUN J, ASHLEY T & LIVINGSTON DM 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell, 88, 265–75. [DOI] [PubMed] [Google Scholar]
- SHARIF B, NA J, LYKKE-HARTMANN K, MCLAUGHLIN SH, LAUE E, GLOVER DM & ZERNICKA-GOETZ M 2010. The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J Cell Sci, 123, 4292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIN YH, CHOI Y, ERDIN SU, YATSENKO SA, KLOC M, YANG F, WANG PJ, MEISTRICH ML & RAJKOVIC A 2010. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS Genet, 6, e1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SJOGREN C & NASMYTH K 2001. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol, 11, 991–5. [DOI] [PubMed] [Google Scholar]
- SOLC P, SCHULTZ RM & MOTLIK J 2010. Prophase I arrest and progression to metaphase I in mouse oocytes: comparison of resumption of meiosis and recovery from G2-arrest in somatic cells. Mol Hum Reprod, 16, 654–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SONODA E, MATSUSAKA T, MORRISON C, VAGNARELLI P, HOSHI O, USHIKI T, NOJIMA K, FUKAGAWA T, WAIZENEGGER IC, PETERS JM, et al. 2001. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell, 1, 759–70. [DOI] [PubMed] [Google Scholar]
- SOUQUET B, ABBY E, HERVE R, FINSTERBUSCH F, TOURPIN S, LE BOUFFANT R, DUQUENNE C, MESSIAEN S, MARTINI E, BERNARDINO-SGHERRI J, et al. 2013. MEIOB targets single-strand DNA and is necessary for meiotic recombination. PLoS Genet, 9, e1003784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOUSA BL, NISHI MY, SANTOS MG, BRITO VN, DOMENICE S & MENDONCA BB 2016. Mutation analysis of NANOS3 in Brazilian women with primary ovarian failure. Clinics (Sao Paulo), 71, 695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPENCER KL, MALINOWSKI J, CARTY CL, FRANCESCHINI N, FERNANDEZ-RHODES L, YOUNG A, CHENG I, RITCHIE MD, HAIMAN CA, WILKENS L, et al. 2013. Genetic variation and reproductive timing: African American women from the Population Architecture using Genomics and Epidemiology (PAGE) Study. PLoS One, 8, e55258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SZELIGA A, ZYSNARSKA A, SZKLARSKA Z, TRUSZKOWSKA E, PODFIGURNA A, CZYZYK A, GENAZZANI AR, CHRZANOWSKA K & MECZEKALSKI B 2019. A case of premature ovarian insufficiency in Nijmegen breakage syndrome patient and review of literature. From gene mutation to clinical management. Gynecol Endocrinol, 35, 999–1002. [DOI] [PubMed] [Google Scholar]
- TENENBAUM-RAKOVER Y, WEINBERG-SHUKRON A, RENBAUM P, LOBEL O, EIDEH H, GULSUNER S, DAHARY D, ABU-RAYYAN A, KANAAN M, LEVY-LAHAD E, et al. 2015. Minichromosome maintenance complex component 8 (MCM8) gene mutations result in primary gonadal failure. J Med Genet, 52, 391–9. [DOI] [PubMed] [Google Scholar]
- TSUIKO O, NOUKAS M, ZILINA O, HENSEN K, TAPANAINEN JS, MAGI R, KALS M, KIVISTIK PA, HALLER-KIKKATALO K, SALUMETS A, et al. 2016. Copy number variation analysis detects novel candidate genes involved in follicular growth and oocyte maturation in a cohort of premature ovarian failure cases. Hum Reprod, 31, 1913–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TUCKER EJ, GROVER SR, ROBEVSKA G, VAN DEN BERGEN J, HANNA C & SINCLAIR AH 2018. Identification of variants in pleiotropic genes causing “isolated” premature ovarian insufficiency: implications for medical practice. Eur J Hum Genet, 26, 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TURAN V, BEDOSCHI G, EMIRDAR V, MOY F & OKTAY K 2018. Ovarian Stimulation in Patients With Cancer: Impact of Letrozole and BRCA Mutations on Fertility Preservation Cycle Outcomes. Reprod Sci, 25, 26–32. [DOI] [PubMed] [Google Scholar]
- TURAN V & OKTAY K 2020. BRCA-related ATM-mediated DNA double-strand break repair and ovarian aging. Hum Reprod Update, 26, 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TYC KM, EL YAKOUBI W, BAG A, LANDIS J, ZHAN Y, TREFF NR, SCOTT RT, TAO X, SCHINDLER K & XING J 2020. Exome sequencing links CEP120 mutation to maternally derived aneuploid conception risk. Hum Reprod, 35, 2134–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VALENTINI A, FINCH A, LUBINSKI J, BYRSKI T, GHADIRIAN P, KIM-SING C, LYNCH HT, AINSWORTH PJ, NEUHAUSEN SL, GREENBLATT E, et al. 2013. Chemotherapy-induced amenorrhea in patients with breast cancer with a BRCA1 or BRCA2 mutation. J Clin Oncol, 31, 3914–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VEITIA RA 2020. Primary ovarian insufficiency, meiosis and DNA repair. Biomed J [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG AC, ZHANG YS, WANG BS, ZHAO XY, WU FX, ZHAI XH, SUN JX & MEI SY 2018. Mutation analysis of the TUBB8 gene in primary infertile women with arrest in oocyte maturation. Gynecol Endocrinol, 34, 900–904. [DOI] [PubMed] [Google Scholar]
- WANG ET, PISARSKA MD, BRESEE C, CHEN YD, LESTER J, AFSHAR Y, ALEXANDER C & KARLAN BY 2014a. BRCA1 germline mutations may be associated with reduced ovarian reserve. Fertil Steril, 102, 1723–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG F & LIU Y 2020. Identification of key genes, regulatory factors, and drug target genes of recurrent implantation failure (RIF). Gynecol Endocrinol, 36, 448–455. [DOI] [PubMed] [Google Scholar]
- WANG J, ZHANG W, JIANG H, WU BL & PRIMARY OVARIAN INSUFFICIENCY C 2014b. Mutations in HFM1 in recessive primary ovarian insufficiency. N Engl J Med, 370, 972–4. [DOI] [PubMed] [Google Scholar]
- WARCOIN M, LESPINASSE J, DESPOUY G, DUBOIS D’ENGHIEN C, LAUGE A, PORTNOI MF, CHRISTIN-MAITRE S, STOPPA-LYONNET D & STERN MH 2009. Fertility defects revealing germline biallelic nonsense NBN mutations. Hum Mutat, 30, 424–30. [DOI] [PubMed] [Google Scholar]
- WEINBERG-SHUKRON A, RACHMIEL M, RENBAUM P, GULSUNER S, WALSH T, LOBEL O, DREIFUSS A, BEN-MOSHE A, ZELIGSON S, SEGEL R, et al. 2018. Essential Role of BRCA2 in Ovarian Development and Function. N Engl J Med, 379, 1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEINBERG-SHUKRON A, RENBAUM P, KALIFA R, ZELIGSON S, BEN-NERIAH Z, DREIFUSS A, ABU-RAYYAN A, MAATUK N, FARDIAN N, REKLER D, et al. 2015. A mutation in the nucleoporin-107 gene causes XX gonadal dysgenesis. J Clin Invest, 125, 4295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WELT CK 2018. Pathogenesis and causes of spontaneous primary ovarian insufficiency (premature ovarian failure) In: BARBIERI RLC, WILLIAM F (ed.) UpToDate Waltham, MA: Wolters Kluwer. [Google Scholar]
- WELT CK 2020. Management of spontaneous primary ovarian insufficiency (premature ovarian failure) In: CROWLEY WFB, ROBERT L (ed.) UpToDate Wolters Kluwer. [Google Scholar]
- WINTERS T, MCNICOLL F & JESSBERGER R 2014. Meiotic cohesin STAG3 is required for chromosome axis formation and sister chromatid cohesion. EMBO J, 33, 1256–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOJTASZ L, DANIEL K, ROIG I, BOLCUN-FILAS E, XU H, BOONSANAY V, ECKMANN CR, COOKE HJ, JASIN M, KEENEY S, et al. 2009. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet, 5, e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOLTERING D, BAUMGARTNER B, BAGCHI S, LARKIN B, LOIDL J, DE LOS SANTOS T & HOLLINGSWORTH NM 2000. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol Cell Biol, 20, 6646–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOOD-TRAGESER MA, GURBUZ F, YATSENKO SA, JEFFRIES EP, KOTAN LD, SURTI U, KETTERER DM, MATIC J, CHIPKIN J, JIANG H, et al. 2014. MCM9 mutations are associated with ovarian failure, short stature, and chromosomal instability. Am J Hum Genet, 95, 754–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU L, CHEN H, LI D, SONG D, CHEN B, YAN Z, LYU Q, WANG L, KUANG Y, LI B, et al. 2019. Novel mutations in PATL2: expanding the mutational spectrum and corresponding phenotypic variability associated with female infertility. J Hum Genet, 64, 379–385. [DOI] [PubMed] [Google Scholar]
- WU X, WANG B, DONG Z, ZHOU S, LIU Z, SHI G, CAO Y & XU Y 2013. A NANOS3 mutation linked to protein degradation causes premature ovarian insufficiency. Cell Death Dis, 4, e825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIAO WJ, HE WB, ZHANG YX, MENG LL, LU GX, LIN G, TAN YQ & DU J 2019. In-Frame Variants in STAG3 Gene Cause Premature Ovarian Insufficiency. Front Genet, 10, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG F, DE LA FUENTE R, LEU NA, BAUMANN C, MCLAUGHLIN KJ & WANG PJ 2006. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol, 173, 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG X, SHU L, CAI L, SUN X, CUI Y & LIU J 2019a. Homozygous missense mutation Arg207Cys in the WEE2 gene causes female infertility and fertilization failure. J Assist Reprod Genet, 36, 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG X, TOURAINE P, DESAI S, HUMPHREYS G, JIANG H, YATSENKO A & RAJKOVIC A 2019b. Gene variants identified by whole-exome sequencing in 33 French women with premature ovarian insufficiency. J Assist Reprod Genet, 36, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YATSENKO SA & RAJKOVIC A 2019. Genetics of human female infertilitydagger. Biol Reprod, 101, 549–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOSHIDA K 2005. Identification of a novel cell-cycle-induced MCM family protein MCM9. Biochem Biophys Res Commun, 331, 669–74. [DOI] [PubMed] [Google Scholar]
- YUAN L, LIU JG, HOJA MR, WILBERTZ J, NORDQVIST K & HOOG C 2002. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science, 296, 1115–8. [DOI] [PubMed] [Google Scholar]
- YUAN P, ZHENG L, LIANG H, LI Y, ZHAO H, LI R, LAI L, ZHANG Q & WANG W 2018. A novel mutation in the TUBB8 gene is associated with complete cleavage failure in fertilized eggs. J Assist Reprod Genet, 35, 1349–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZANGEN D, KAUFMAN Y, ZELIGSON S, PERLBERG S, FRIDMAN H, KANAAN M, ABDULHADI-ATWAN M, ABU LIBDEH A, GUSSOW A, KISSLOV I, et al. 2011. XX ovarian dysgenesis is caused by a PSMC3IP/HOP2 mutation that abolishes coactivation of estrogen-driven transcription. Am J Hum Genet, 89, 572–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG J, FUJIWARA Y, YAMAMOTO S & SHIBUYA H 2019a. A meiosis-specific BRCA2 binding protein recruits recombinases to DNA double-strand breaks to ensure homologous recombination. Nat Commun, 10, 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG M, DAI X, SUN Y, LU Y, ZHOU C, MIAO Y, WANG Y & XIONG B 2017. Stag3 regulates microtubule stability to maintain euploidy during mouse oocyte meiotic maturation. Oncotarget, 8, 1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG YX, HE WB, XIAO WJ, MENG LL, TAN C, DU J, LU GX, LIN G & TAN YQ 2020a. Novel loss-of-function mutation in MCM8 causes premature ovarian insufficiency. Mol Genet Genomic Med, e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Z, LI B, FU J, LI R, DIAO F, LI C, CHEN B, DU J, ZHOU Z, MU J, et al. 2020b. Bi-allelic Missense Pathogenic Variants in TRIP13 Cause Female Infertility Characterized by Oocyte Maturation Arrest. Am J Hum Genet, 107, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Z, MU J, ZHAO J, ZHOU Z, CHEN B, WU L, YAN Z, WANG W, ZHAO L, DONG J, et al. 2019b. Novel mutations in WEE2: Expanding the spectrum of mutations responsible for human fertilization failure. Clin Genet, 95, 520–524. [DOI] [PubMed] [Google Scholar]
- ZHAO L, GUAN Y, WANG W, CHEN B, XU S, WU L, YAN Z, LI B, FU J, SHI R, et al. 2020a. Identification novel mutations in TUBB8 in female infertility and a novel phenotype of large polar body in oocytes with TUBB8 mutations. J Assist Reprod Genet [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO L, XUE S, YAO Z, SHI J, CHEN B, WU L, SUN L, XU Y, YAN Z, LI B, et al. 2020b. Biallelic mutations in CDC20 cause female infertility characterized by abnormalities in oocyte maturation and early embryonic development. Protein Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO S, CHEN T, YU M, BIAN Y, CAO Y, NING Y, SU S, ZHANG J & ZHAO S 2019. Novel WEE2 gene variants identified in patients with fertilization failure and female infertility. Fertil Steril, 111, 519–526. [DOI] [PubMed] [Google Scholar]
- ZHE J, CHEN S, CHEN X, LIU Y, LI Y, ZHOU X & ZHANG J 2019. A novel heterozygous splice-altering mutation in HFM1 may be a cause of premature ovarian insufficiency. J Ovarian Res, 12, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHE J, YE D, CHEN X, LIU Y, ZHOU X, LI Y, ZHANG J & CHEN S 2020. Consanguineous Chinese Familial Study Reveals that a Gross Deletion that Includes the SYCE1 Gene Region Is Associated with Premature Ovarian Insufficiency. Reprod Sci, 27, 461–467. [DOI] [PubMed] [Google Scholar]
- ZHEN XM, SUN YM, QIAO J, LI R, WANG LN & LIU P 2013. [Genome-wide copy number scan in Chinese patients with premature ovarian failure]. Beijing Da Xue Xue Bao Yi Xue Ban, 45, 841–7. [PubMed] [Google Scholar]
- ZHOU X, ZHU L, HOU M, WU Y, LI Z, WANG J, LIU Z, ZHANG D, JIN L & ZHANG X 2019. Novel compound heterozygous mutations in WEE2 causes female infertility and fertilization failure. J Assist Reprod Genet, 36, 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZORRILLA M & YATSENKO AN 2013. The Genetics of Infertility: Current Status of the Field. Curr Genet Med Rep, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]


