Abstract
Local spatial arrangement of nuclei in histopathology images of different cancer subtypes has been shown to have prognostic value. In order to capture localized nuclear architectural information, local cell cluster graph-based measurements have been proposed. However, conventional ways of cell graph construction only utilize nuclear spatial proximity, and do not differentiate between different cell types while constructing the graph. In this paper, we present feature-driven local cell cluster graph (FLocK), a new approach to constructing local cell graphs by simultaneously considering spatial proximity and attributes of the individual nuclei (e.g. shape, size, texture). In addition, we have designed a new set of quantitative graph-derived metrics to be extracted from FLocKs, in turn capturing the interplay between different proximally located clusters of nuclei. We have evaluated the efficacy of FLocK features extracted from H&E stained tissue images in two clinical applications: to classify short-term vs. long-term survival among patients of early stage non-small cell lung cancer (ES-NSCLC), and also to predict Human Papillomavirus (HPV) status of Oropharyngeal Squamous Cell Carcinoma (OP-SCCs).
In the classification of long-term vs. short-term survival among patients of ES-NSCLC (training cohort, n=434), the top 10 discriminative FLocK features related to the variation of FLocK sizes and intersected FLocK distance were identified, via the Minimum Redundancy and Maximum Relevance (MRMR), under 100 runs of 10-fold cross-validation, in conjunction with a linear discriminant classifier yielded a mean of AUC=0.68 for predicting survival in training cohort. This is better than when compared to other state-of-art histomorphometric and deep learning classifiers (cell cluster graphs (AUC=0.62), global cell graph (AUC=0.56), nuclear shape (AUC=0.54), nuclear orientation (AUC=0.61), AlexNet (AUC=0.55), ResNet (AUC=0.56)). The FLocK-based classifier yielded an AUC of 0.70 in an independent testing cohort (n=150). The patients identified as “high-risk” had significantly poorer overall survival in the independent testing cohort, with Hazard Ratio (95% Confident Interval) = 2.24 (1.24-4.05), p = 0.01144).
In the classification of HPV status of OP-SCC, the top three FLocK features pertaining to the portion of intersected FLocKs were identified to construct a classifier, which yielded an AUC of 0.80 in the training cohort (n=50), and an accuracy of 0.78 in an independent testing cohort (n=35). The combination of FLocK measurements with cell cluster graphs, nuclear orientation, and nuclear shape improved the training AUC to 0.87, 0.91 and 0.85, respectively. Deep learning approaches yield marginally better performance compared to the FLocK-based classifier in this application, with AUC=0.78 for AlexNet, AUC=0.81 for ResNet, and AUC=0.76 for FLocK-based classifier in the independent testing cohort. However, the combination of two hand-crafted features: FLocK and nuclear orientation yielded a better performance (with an AUC=0.84).
FLocK provides a unique and quantitative way to analyze histology image of solid tumor and interrogates tumor morphology from a different aspect compared to the existing histomorphometrics. The source code can be accessed at https://github.com/hacylu/FLocK.
Keywords: histology image analysis, feature extraction, oropharyngeal cancer, lung cancer
Graphical abstract
A. Introduction
Changes in distribution, appearance, size, morphology, and arrangement of histologic primitives, e.g., nuclei and glands, have been shown to predict cancer aggressiveness (Bera et al., 2019; Gurcan et al., 2009). For instance, in the context of lung cancer, it is known that lung cancer lethality can be characterized by the differences in nuclear shape, morphology and arrangement. For a number of different cancers, the hallmark of presence of disease is the disruption in the cohesion of architecture between cancer cells and other primitives belonging to the same family, e.g. lymphocytes. Conversely, more aggressive tumors tend to exhibit relatively lower degrees of structure and organization between the same class of primitives compared to less aggressive cancers.
There has been recent interest in developing computational graph-based approaches to characterize spatial arrangement of nuclei in histopathology images to be able to predict patient outcome (Ali et al., 2013a, 2013b; Bilgin et al., 2007; Lewis et al., 2014; Shin et al., 2015). Many of these approaches are based on capturing measurements relating to spatial architecture of histologic primitives. For instance, global graphs such as Voronoi and Delaunay triangulation strategies have been used to connect individual cell nuclei (representing graph vertices or nodes) and then extracting statistics relating to edge length and node density to predict disease outcome. Lewis et al. (Lewis et al., 2014) proposed cell cluster graphs (CCG) in which the nodes are defined on groups/clusters of nuclei rather than as individual nuclei, since there is a growing recognition that tumor aggressiveness might be driven more by the spatial inter-actions of proximally situated nuclei, compared to global interactions of distally located nuclei. While these approaches showed that attributes relating to spatial arrangement of proximal nuclei were prognostic (Ali et al., 2013a, 2013b; Bilgin et al., 2007; Lewis et al., 2014; Shin et al., 2015), the graph connections did not discriminate between different cell populations, e.g. whether the proximal cells were all cancer cells or belonged to other families such as immune cells.
Early-stage (Stage I, II) non-small cell lung carcinoma (ES-NSCLC) is typically treated with complete surgical excision followed by adjuvant chemotherapy, if required. However, 30 to 55 percent of patients still tend to have either local or distant recurrence even after systemic therapy. Currently, there is no clinically-used biomarker that is accurate and reproducible which can identify patients with a high risk of recurrence who might need early treatment intensification, while low-risk patients might be treated with surgery alone. Recent research has shown that computerized measurements related to the density of immune cells (Saltz et al., 2018) as well as the interplay between immune cells and cancer cells were able to risk stratify these patients (Corredor et al., 2019).
Human Papillomavirus (HPV) positive Oropharyngeal Squamous Cell Carcinoma (OP-SCC) accounts for 70 to 80 percent of all OP-SCC in North America and Europe (Chaturvedi et al., 2011). HPV status is critical for staging, treatment, and prognosis. Current standards to determine HPV status relies on immunohistochemical stain for p16 protein on tissue. However, there are just few works have been shown to determine the HPV status via the traditional H&E stain images (Kather et al., 2019).
In this work, we seek to go beyond the traditional approach of constructing cell graphs, which focus solely on cell proximity. We incorporate intrinsic nuclear morphologic features coupled with spatial distance to construct locally and morphologically distinct cell clusters. We introduce a new way of constructing a local cell graph called the Feature driven local cell cluster graph (FLocK), along with a corresponding new set of quantitative histomorphometric features. It is well established that cellular diversity is intrinsically linked to morphologic heterogeneity, and that increased morphologic heterogeneity is strongly associated with worse prognosis for a number of different cancer types (Almendro et al., 2013; Aum et al., 2014; Mroz and Rocco, 2016; Yuan et al., 2012). In fact, cellular diversity plays a critical role in a number of cancer-grading schemes, such as the Bloom-Richardson grading scheme for breast cancers (Genestie et al., 1998), where nuclear pleomorphism (which is focused on visual assessment of the diversity in nuclear shape, size and appearance) is the principal criteria. Similarly, pathologists have routinely observed spatial differences in arrangement of cancer cells and TILs in a variety of cancer types (Nawaz et al., 2015). However, unlike tasks relating to quantification of cell density, quantitating spatial architecture is more challenging. FLoCK allows for quantitatively capturing measurements relating to diversity in cellular appearance, morphology and architecture across the entirety of the whole slide image. FLoCK features are then used in conjunction with a machine learning classifier to prognosticate survival for early stage non-small cell lung carcinomas (ES-NSCLC), as also classifying HPV status for oropharyngeal cancers.
B. Previous Work and Novel Contributions
In traditional digital histopathology image analysis, quantitative imaging features derived from the histologic primitives have been used to quantify issue morphology (Bose et al., 2015; Gurcan et al., 2009; He et al., 2012). In non-small cell lung cancer, Yu et al. (Yu et al., 2016) and Wang. et al. (Wang et al., 2017) showed that the nuclear shape variations were associated with patient outcome. Similarly, a set of nuclear features that quantify the disorder of nuclear/gland polarity were found to be prognostic in prostate (Lee et al., 2014, 2013) and breast cancer (Lu et al., 2018) patients. In addition to the shape of nuclei, the textural intensity present on the surface of nuclei were also found to be correlated with patient outcomes (Bose et al., 2015; Noy et al., 2011). These above mentioned shape and texture features intended to quantify the nuclear morphology on a local and granular level, whereas the graph-based features aimed to capture the nuclear/glandular spatial arrangement (Ali et al., 2013a; Bilgin et al., 2007). Using nuclei or the glands as graph nodes, one can construct global graphs, such as Voronoi and Delaunay triangulation, and extract graph-based measurements and relate them to disease outcome (Lee et al., 2016; Nguyen et al., 2014). These graph measurements are mainly based on the density/distance of the graph nodes (i.e., the nuclei or gland locations), and do not take in to account the specific type of cells used to construct the graphs. Since a digitized histology image usually contains the entire tumoral landscape, which is informationally rich and complex, global graphs may dilute important diagnostic/prognostic signals that are only observed in local region. This has led to research which breaks down global graphs into local ones, and performed a more granular interrogation of local nuclear morphology (Ali et al., 2013a; Lewis et al., 2014). Ali et. al. (Ali et al., 2013b) proposed Cell Cluster Graph (CCG), in which the local cell graphs were constructed purely based on the proximity of nuclei. The graph complexity measurements (Ali et al., 2013b) were then extracted from the CCG to quantify the tumor morphology locally. While these approaches showed that attributes relating to spatial arrangement of proximal nuclei were prognostic (Ali et al., 2013a, 2013b; Bilgin et al., 2007; Lewis et al., 2014; Shin et al., 2015), the local graph connections again did not discriminate between different cell populations, e.g. whether the proximal cells were all cancer cells or belonged to other families such as immune cells (lymphocytes, neutrophils).
In this work, we have introduced a new family of quantitative histomorphometric features named FLocK, derived from local cell graphs that are driven by nuclear properties. The novel contributions of this work include:
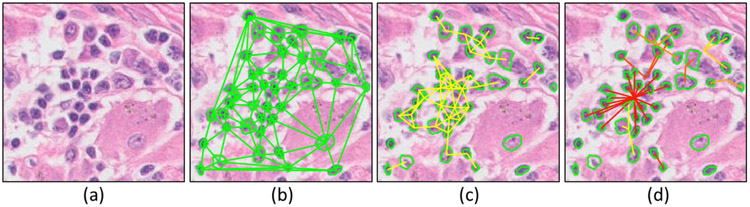
1) FLocK is a novel way to construct local cell cluster graphs which simultaneously consider nuclear properties as well proximities. This results in locally-packed cell graphs comprising nuclei with similar phenotype. Figure 1 illustrates the global cell graph (Bilgin et al., 2007), CCG (Lewis et al., 2014) and FLocK graph in the same local region contains lymphocytes and cancer cells in NSCLC. One may see that the global graph connects all the nuclei in the image and may not allow for capturing of local tumor morphology efficiently. Similarly, the CCG only considers nuclear locations, which results in connecting lymphocytes and cancer nuclei into a graph; important information involving local spatial interaction between different cellular clusters may be left unexploited. The FLocK incorporates nuclear morphologic feature (nuclear mean intensity in this case) into the graph constructing process, which enables us to interrogate the interaction between different groups of cell type and to reveal more sub-visual information from the underlying tissue image.
Figure 1:
Illustration of (a) original H&E stained color histology image of NSCLC, (b) global cell graph (Delaunay triangulation), (c) CCG, the cell clusters were created based on the proximity of nuclei, (d) FLocK driven by nuclear intensity and proximity.
2) In addition to constructing local cell graphs using FLocK, we designed a new set of quantitative histomorphometric features based on the constructed FLocKs. These feature sets are: Intersection between nearby FLocKs, size of FLocKs, disorder of nuclear morphology within FLocK, spatial arrangement of FLocK, and enrichment of the Nearest FLocKs. These features are different compared to standard features extracted from CCG and global graph methods, which only quantify the density of local/global graph, or the local/global distances between cells. The FLocK features attempt to capture the interactions between and within local cell clusters with similar morphological properties. Also, we have provided two methods for computation of FLocK features: one with predefined cluster type numbers, and the other one without pre-defined cluster type numbers. For the method with pre-defined cluster type numbers, the user is able to specify the cluster type number based on the domain specific knowledge. For example, for ES-NSCLC, one may want to set the cluster type number as 2, if they want to check if the interaction between tumor infiltrated lymphocytes and cancer cells are correlated for the prognosis. This allows the user to validate their own hypothesis using FLocK features. For the method without pre-defined cluster type numbers, we construct FLocK purely driven by the nuclear morphology and compute the features based on the constructed FLocK, which may reveal some important signatures.
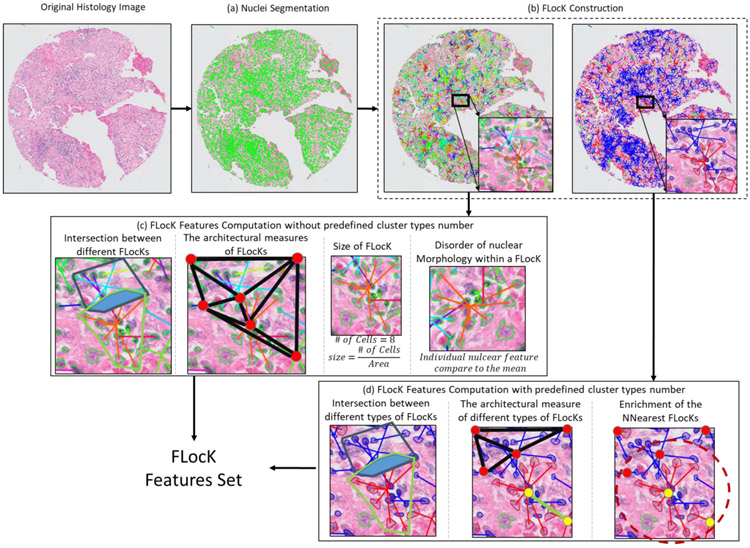
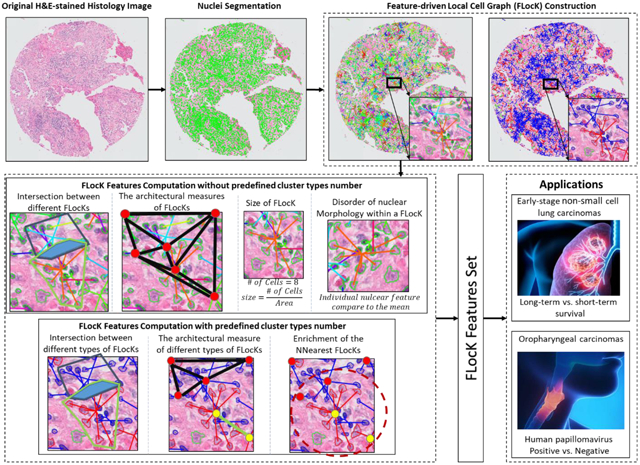
3) We employ the FLocK and associated Quantitative Histomorphometric (QH) features in conjunction with a linear machine learning classifier (Linear Discriminant Analysis) to predict risk of recurrence for ES-NSCLC and to classify the HPV status for OP-SCCs. For the ES-NSCLC, similar work has been reported by Yu et al.(Yu et al., 2016) and Wang et al. (Wang et al., 2017) for predicting recurrence in early-stage NSCLC patients, in which the global architecture and shape of nuclei features were found to be predictive. However, the interactions between different local cell clusters have not been explored. For the OP-SCC, to our best knowledge, there is no prior work using computer extracted hand-crafted features to classify the HPV status based on the H&E stained image. In the experiment, we compared the FLocK feature with other handcrafted nuclear features. Figure 2 shows the flowchart of FLocK construction and associated feature computation, which include nuclei segmentation, FLocK construction, and FLocK feature extraction modules.
Figure 2:
Flowchart of FLocK construction and associated feature computation. (a) Green contours indicate the nuclear boundaries (b) FLocK construction based on the nuclear features (used mean intensity of nuclei in this case). Nuclei that belong to the same FLocK have connecting edges with the same color. (c) FLocK feature computation without pre-defined cluster type number. (d) FLocK feature computation with pre-defined cluster type number.
C. Feature-driven Local Cell Graph
The schematic of FLocK feature computation is illustrated in Figure 2. Briefly, the steps comprise (a) nuclei segmentation, (b) FLocK construction, (c) and (d) FLocK feature computation without or with pre-defined cluster type number, respectively. In the experiment, computer-extracted FLocK features are then used in conjunction with a machine learning classifier to predict the overall survival in NSCLC (n=434 patients), and classification of HPV status in OP-SCC (n=85 patients) using digital pathology images. We also compare FLocK features with several states of the art histopathology image analysis and machine learning based approaches.
C.1. Nuclei segmentation and morphologic feature extraction
In order to efficiently segment nuclei, a multiple-pass adaptive voting method was employed to detect the cells (Lu et al., 2016) followed by a local optimal threshold method that segments nuclei based on analyzing the shape of these cells as well as the area occupied by them (Lu and Mandal, 2015). A set of 11 nuclear morphologic features that described the nuclear shape and texture were then computed based on these pre-segmented nuclei. There, nuclear features will be used in the downstream nuclear morphologic feature space analysis for constructing FLocK.
C.2. FLocK construction in nuclear morphologic feature space
In this step, spatial and morphological features of nuclei were used for feature space analysis to construct FLocK. Mean-shift clustering[25] was applied to perform the feature space analysis for sub-graph construction. It accomplishes this by first estimating the modes (i.e., stationary points of the density of nuclear morphologic feature) of the underlying density function of the nuclear morphologic feature. It then groups nuclei into different sub-graphs based on the corresponding modes.
We denote as N the total number of nuclei in the image, and each nucleus has a corresponding feature vector in d-dimensional Euclidean space Rd, so that we have a set of nuclear feature vectors X=x1, x2, …, xN, where xn∈Rd. For each feature vector xn ∈ X, there is a corresponding mode yi. In the beginning, the mode yi is initialized with the original feature vector xn, i.e., . The is then recursively updated, based on the neighborhood nuclear characteristics, using the following equation:
| (1) |
where is the updated version of . The vector is called the mean-shift vector and calculates the difference between the weighted mean and the center of the kernel. It has been previously shown that the mean-shift vector always points toward the direction of maximum increase in the underlying density function (Comaniciu and Meer, 2002). At the final step, each nuclear feature vector xn finds a corresponding mode yi which will be used for constructing the FLocK.
In this work, we explored a Q-dimensional feature space which includes 2-D spatial coordinates (i.e., centroid location) of nuclei in the image and Q-2 of the nuclear morphologic features. These features are chosen based on the observation that the same types of nuclei are usually located closely together and have a similar phenotype. The corresponding multivariate kernel is defined as the product of two radially symmetric kernels as follows:
| (2) |
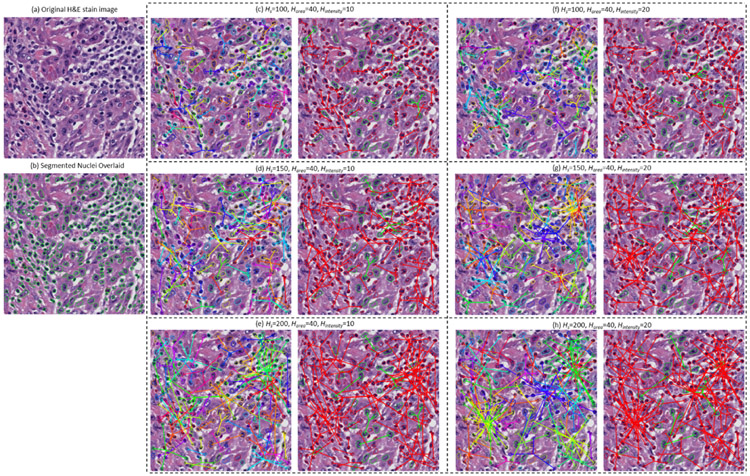
where k(·) is the profile of the kernel, xs is the spatial component, xm is the nuclear morphologic component, C is the normalization constant, and hs and hm are the kernel bandwidths controlling the size of the kernels. The higher value of the kernel bandwidths hs and hm correspond to more neighboring data points that are used to estimate the density in the Q-D feature space. This can be seen in Figure 1(d), in which the FLocKs were constructed in a 3-D feature space, i.e., the spatial x- and y-coordination, plus the nuclear intensity. Also Figure 6 illustrates how the bandwidths affect construction of FLocKs. In Figure 6, we illustrate FLocKs constructed in a 4-D feature space, i.e., comprising the spatial x and y coordinate frame, the nuclear area, and nuclear intensity. Appendix 2-B also provides quantitative analysis of the effect of kernel bandwidths (hs from 20 to 50 with an increment of 5, hintensity from 0 to 40 with an increment of 5, and harea from 0 to 50 with an increment of 5) and three selected FLocK feature values (portion of intersected FLocKs, mean size of FLocKs, and mean of variance of nuclear feature 1 with respect to the centroid).
Figure 6:
An example of FLocK configuration with different parameter settings. (a) and (b) show the original H&E stained image of OP-SCC and the segmented nuclei overlaid on the image, respectively. (c) to (e) show FLocKs constructed under the feature space consisted of nuclear co-ordinations (i.e., x and y locations of nuclei), nuclear Area and nuclear Mean Intensity, with spatial bandwidth Hs =100 to 200 (with an increasing step of 50), the nuclear Area and Mean Intensity bandwidth Harea=40, Hintensity=10. The left panel shows the FLocK configuration without predefined cluster types number (different FLocKs were highlighted with distinct colors), whereas the right panel shows the FLocK configuration with predefined cluster types number k=2. (f) to (h) show FLocKs with similar parameter settings compared to (c) to (e) with Hintensity=20.
Based on the description above, we are able to construct FLocKs without specifying the number of cluster types presented on the histology image (see Figure 2(b), left panel). This will allow us to interrogate the interactions between different cell families distributed locally on the image. On the other hand, we also provide another realization of FLocK, in which FLocKs are constructed by a pre-defined number of cluster types k, e.g., k = 2, one for tumor cell the other for tumor infiltrated lymphocytes (TILs) (see Figure 2(b) right panel and Figure 6).
C.3. FLocK Features Computation
Based on the FLocK created, several groups of features were derived as shown in Table 2 and Appendix 1 (see Figure 2 (c) and (d) for intuitive illustrations). These quantitative features are focused on capturing measurements relating to 1) the interaction between FLocKs, in which the relative portion, mean intersected region and number of intersected FLocKs are computed; 2) size of FLocK; 3) intrinsic variation of individual cells within each FLocK, in which variations are measured in terms of nuclear shape and appearance; 4) spatial arrangement of FLocKs, in which global graphs (Delaunay Triangulation, Voronoi Diagram, Minimum Spanning Trees) are constructed based on the centroids of FLocKs to quantify the architecture of FLocKs; 5) the enrichment of the K-nearest FLocKs, in which the diversity of neighboring FLocKs are captured.
Table 2.
Description of FLocK features. If the pre-defined cluster type number is set to zero, we have feature set A, these are the features that do not consider a pre-defined cluster types. If the predefined cluster number is set to >1, we have feature set B, these are the features that do consider a pre-defined cluster types. For more details on these feature names and the corresponding computation, we direct the reader to appendix 1 and to the code released on Github (Lu, 2020).
| A. Features that do not consider cluster types | |||
|---|---|---|---|
| Feature groups | # | Feature names | Explanation |
| Intersection between different FLocKs | 44 | Portion/ number of intersected FLocKs, mean intersected area | Quantify the interaction between FLocKs |
| Size of FLocK | 12 | Size of FLocK, nuclei number in FLocK | Quantify the size of FLocKs |
| Disorder of nuclear morphology | 18 | Variation in nuclear morphology within a FLocK | Quantify disorder of nuclear morphology locally |
| Spatial arrangement of FLocKs | 102 | Global graph measurements | Quantify global architecture of FLocKs |
| B. Features that consider cluster types (k is the number of cluster types). | |||
| Feature groups | # | Feature names | Explanation |
| Intersection between different types of FLocKs | 2xk | Portion of intersected FLocKs from different cluster types | Portion of intersected FLocKs, quantifying the intersection between different types (intertype) and within same type (intra-type) of FLocKs |
| Enrichment of the K-nearest FLocKs | 24 | Portion of other types of FLocKs within 5, 10, 15 nearest neighbors | Quantify the diversity of FLocKs, with pre-defined phenotype number, in terms of nearest neighbors |
| Spatial arrangement of a specific type of FLocK | 44xk | Global graph measurements based on a specific type of FLocK | Quantify the diversity of a specific type of FLocK, in terms of nearest neighbors |
D. Experimental Result and Discussion
D.1. Dataset Description
HPV classification for OP-SCC:
A training cohort of 50 patients, Otrain, with primary OP-SCC from Vanderbilt University Medical Center, including 25 HPV+ and 25 HPV−, were used to build the classifier. The H&E stained whole-mounted tissue slides from the primary tumors (biopsy or resection) were digitized at 40x magnification. Non-overlapped 2048 x 2048 pixels patches were cropped from the tumor region. FLocK features were then extracted from these patches, with nuclear Area and Mean Intensity as the feature space and spatial bandwidth Hs=100, nuclear morphology bandwidth Harea=40, Hintensity=10. The mean feature value for all patches from the same whole slide image was used to represent the signature of a patient. The top 3 FLocK features were selected to build the final classifier using 3-fold cross validation over 100 runs. To validate our classifier on an independent set Otest, we utilized another cohort of 35 patients with OP-SCC from Kaiser Permanente Medical System, of which 16 and 19 were HPV− and HPV+, respectively. One should note that the Kaiser cohort was not used in any way during the model training.
Predicting Patient Outcomes of ES-NSCLC:
The ES-NSCLC cohort comprises a total of 434 patients in the form of digitized TMA image (scanned at 20X magnification digitally). All of these patients are Stage I, II NSCLC cancers with long-term clinical survival follow-ups available for all patients in this cohort (collected between 2004 and 2014 from Cleveland Clinic), which ends up with 280 short-term survival patients (<5 years after surgery) and 154 long-term survival patients (>5 years after surgery). We used the same feature space as the case of OP-SCC, i.e., with nuclear Area and Mean Intensity as the feature space. Since the image was at 20X magnification, we divided the bandwidths that used in the OP-SCC case by two, i.e., the spatial bandwidth Hs=50, nuclear morphology bandwidth Harea=20, Hintensity=5. FLocK features were extracted from the TMA image and the top 10 features were selected to build the classifier using 10-fold cross validation over 100 runs. To validate our classifier on an independent test set Ltest, we utilized another cohort of 150 patients with Stage I, II NSCLC cancers collected from Yale Pathology between 2004 and 2014, the slides being scanned at 20x magnification. One should note that this independent test set Ltest was not used in any way during model training.
D.2. Classifier Construction and Performance Evaluation
Three different machine learning classifiers, Linear Discriminant Analysis (LDA), Quadratic Discriminant Analysis (QDA), and Random Forest (RF) were implemented and trained based on the patient ground truth labels, under k-fold cross-validation (CV) with 100 runs. Three different feature selection methods were implemented to select the most discriminative FLocK features within the training folds. The feature selection methods included minimum redundancy, maximum relevance (MRMR) feature selection method (Peng et al., 2005), Wilcoxon rank sum test (WRST), and T-test (TT).
We identified the top performing classifier-feature selection scheme combination based off the AUC values under k-fold CV with 100 runs. The best performing feature selection-classification combination was designated as the lock-down classifier, and the associated top features were further analyzed using box-plot and visualizing with feature map (Figure 3 and Figure 4). For both tasks, an independent testing cohort was used to validate the lock-down classifier. For prognosis on ES-NSCLC, the stratification of patients based on the predicted label obtained from the FLocK-based classier was evaluated using Kaplan-Meier (KM) product limit method.
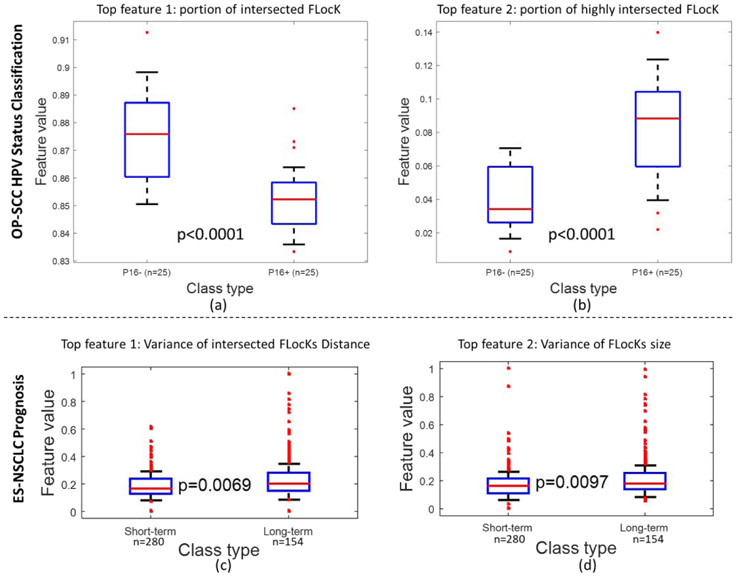
Figure 3:
The box plots of top two features for OP-SCC HPV status classification (upper panel (a) and (b)), and ES-NSCLC (lower panel, (c) to (d)). The p value was evaluated using WRST with all patients in training cohort. The separation between two groups of patients are statistically significant considering the significant level as 0.05.
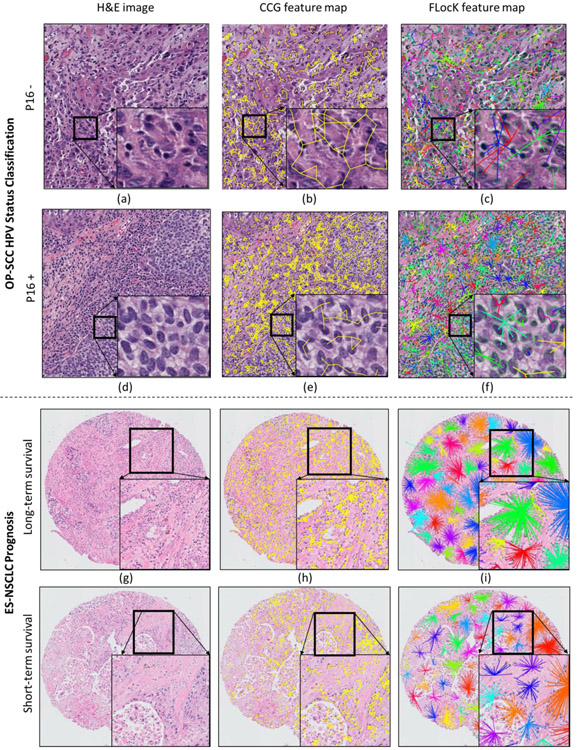
Figure 4:
Representative H&E stained images for OP-SCC HPV status classification (upper panel (a) to (f)), and ES-NSCLC (lower panel, (g) to (l)). The first column shows the original image, the second column shows the CCG configuration, and the third column shows the FLocK configuration, in which different FLocKs have different colors. The FLocKs shown in (c), (f), (i), and (l) reveal the grouping of different sub-type of nuclear as distinct clusters, unlike the CCG representation (shown in (b), (e), (h), and (k)) which does not distinguish between them.
D.3. Experimental Result
Experiment 1 - FLocK Classifier on HPV classification for OP-SCC:
The performance of the nine combinations of feature selection and classifier schemes in terms of AUC, sensitivity, specificity, and accuracy on the OP-SCC Cancer TMA cohort are summarized in Table 3. One may observe that the classification performance using FLocK features are relatively consistent across different feature selection and classifier combinations. Note that since the combination of WRST and LDA yielded the best performance in distinguishing two patient groups, AUC=0.80±0.04, we settled on the combination for WRST and LDA for constructing a lock-down classifier and comparative evaluation with other HQs using training cohort Otrain (Table 6). On the independent validation cohort (n=35 patients), the lock-down FLocK classifier achieved an AUC=0.76, accuracy=0.80, sensitivity=0.95, and specificity=0.63.
Table 3:
Performance of three different classifiers (LDA, QDA, and RF) with three different feature selection methods (WRST, TT, and MRMR) in the OP-SCC training cohort Otrain under 3-fold CV scheme.
| Classifier | Feature selection |
AUC | Accuracy | Specificity | Sensitivity |
|---|---|---|---|---|---|
| LDA | WRST | 0.80±0.04 | 0.76±0.03 | 0.82±0.11 | 0.71±0.10 |
| TT | 0.79±0.04 | 0.77±0.03 | 0.83±0.09 | 0.71±0.08 | |
| MRMR | 0.76±0.05 | 0.75±0.04 | 0.75±0.13 | 0.74±0.14 | |
| QDA | WRST | 0.77±0.04 | 0.75±0.03 | 0.78±0.12 | 0.72±0.12 |
| TT | 0.79±0.03 | 0.76±0.03 | 0.85±0.09 | 0.67±0.09 | |
| MRMR | 0.74±0.07 | 0.74±0.05 | 0.76±0.12 | 0.72±0.13 | |
| RF | WRST | 0.77±0.05 | 0.75±0.04 | 0.80±0.11 | 0.70±0.13 |
| TT | 0.78±0.04 | 0.77±0.04 | 0.85±0.10 | 0.68±0.11 | |
| MRMR | 0.76±0.05 | 0.75±0.05 | 0.76±0.11 | 0.73±0.14 |
Table 6:
Performance of five different histomorphometric feature families in the Vanderbilt OP-SCC training cohort Otrain under a 3-fold CV scheme.
| Model | Feature Family |
AUC | Accuracy | Specificity | Sensitivity |
|---|---|---|---|---|---|
| MCCG | CCG | 0.80±0.03 | 0.77±0.03 | 0.84±0.10 | 0.70±0.10 |
| MGlobalGraph | Global Cell Graph | 0.79±0.03 | 0.77±0.03 | 0.73±0.09 | 0.80±0.11 |
| MShape | Nuclear shape | 0.80±0.03 | 0.77±0.03 | 0.76±0.08 | 0.78±0.09 |
| MCOrE | COrE | 0.82±0.04 | 0.78±0.03 | 0.89±0.11 | 0.67±0.13 |
| MFLocK | FLocK | 0.80±0.04 | 0.77±0.03 | 0.83±0.09 | 0.71±0.08 |
Experiment 2 - FLocK Classifier for Predicting Patient Outcomes of ES-NSCLC:
The performance of the nine combinations of feature selection and classifier schemes in terms of AUC, sensitivity, specificity, and accuracy on the ES-NSCLC TMA cohort are summarized in Table 4. Since the combination of MRMR and LDA yielded the best performance in distinguishing two patient groups, AUC=0.68±0.03, we settled on the combination for MRMR and LDA for constructing the final classifier for survival analysis and comparative evaluation (Table 4). On the independent testing cohort Ltest (n=150 patients), the lock-down FLocK classifier achieved 0.70 AUC, with 0.72 accuracy, 0.75 sensitivity, and 0.70 specificity.
Table 4:
Performance of three different classifiers (LDA, QDA, and RF) with three different feature selection methods (WRST, TT, and MRMR) in the ES-NSCLC training cohort Ltrain, n=434, under a 10-fold CV scheme.
| Classifier | Feature selection |
AUC | Accuracy | Specificity | Sensitivity |
|---|---|---|---|---|---|
| LDA | WRST | 0.66±0.02 | 0.68±0.05 | 0.65±0.05 | 0.69±0.05 |
| MRMR | 0.68±0.03 | 0.67±0.06 | 0.71±0.10 | 0.65±0.09 | |
| TT | 0.65±0.03 | 0.70±0.07 | 0.57±0.04 | 0.76±0.03 | |
| QDA | WRST | 0.63±0.04 | 0.69±0.09 | 0.57±0.17 | 0.74±0.16 |
| MRMR | 0.67±0.04 | 0.71±0.05 | 0.63±0.09 | 0.75±0.17 | |
| TT | 0.63±0.07 | 0.68±0.08 | 0.66±0.21 | 0.68±0.13 | |
| RF | WRST | 0.67±0.02 | 0.66±0.06 | 0.77±0.14 | 0.61±0.16 |
| MRMR | 0.67±0.03 | 0.68±0.07 | 0.70±0.14 | 0.66±0.13 | |
| TT | 0.65±0.03 | 0.66±0.07 | 0.75±0.17 | 0.62±0.09 |
Experiment 3 - Comparison between FLocK and States of the Art Histomorphometric and Deep Learning Approaches
Description of the histomorphometric feature approaches
We compared the efficacy of FLocK features with four previously published histomorphometric feature approaches (Ali et al., 2013b; Kothari et al., 2013; Lee et al., 2016, 2013; Shin et al., 2015) describing both cell morphology and cellular architecture. In total, we investigated the performance of five hand-crafted feature families: (1) 100 features describing nuclear shape (Kothari et al., 2013), (2) 51 features describing global cell architectures (Shin et al., 2015), (3) 72 features describing cell orientation entropy by COrE (Lee et al., 2013) (24 features with three cell sub-graph setups), (4) 105 Cell Cluster Graph (CCG) features describing local cell cluster arrangement (Ali et al., 2013b) (35 features with three cell sub-graph setups), and (5) 235 FLocK features, characterizing the interaction between local cell clusters.
Description of the deep learning-based approaches
We also compared the FLocK based classifier MFLocK with the deep learning (DL)-based approaches: AlexNet (Janowczyk and Madabhushi, n.d.; Krizhevsky et al., 2012) MAlex and ResNet (He et al., 2016) MRes. The rationale for choosing AlexNet is based on our previous experience that a simple DL architecture with fewer parameters could achieve satisfactory or comparable performance compared to a more sophisticated one (Janowczyk and Madabhushi, n.d.). On the other hand, ResNet is a relatively deeper convolutional neural network (CNN) compared to AlexNet. We explored different configurations of MAlex and MRes, as well as the hyperparameters settings, and found that the following two CNNs achieved the best performance.
For the AlexNet, denoted as MAlex hence forth, a 10-layer CNN architecture comprising one input layer, five convolution layers, three fully connected layers and one output layer was constructed. The input layer accepts an image patch of 256 x 256 pixels, and the output layer is a soft-max function which outputs the class probability of being positive or negative (for the NSCLC use case, the positive label corresponds to the short-term survival patient, whereas the negative label corresponds to the long-term survival patient; for the OP-SCC use case, the positive label corresponds to the HPV+ patient, whereas the negative label corresponds to the HPV- patient). The batch size was set to 16 with initial learning rate at 0.001 and was divided by 10 when the error plateaued, and the weight decay was 0.004. Learning rate schedule was set to the adaptive gradient algorithm (Duchi et al., n.d.). The AlexNet has been reported to yield satisfactory performance for several pathology image analysis applications (Alom et al., 2019; Janowczyk and Madabhushi, n.d.; Nawaz et al., 2018).
For the ResNet network, denoted as MRes hence forth, a 34-layer architecture was chosen, with Stochastic Gradient Descent as optimizer, momentum being set to 0.9, binary crossentropy as the loss function, and the batch size being set to 16 with an initial learning rate at 0.01 and divided by 10 when the error plateaued. We used a weight decay of 0.0001 and dropout rate of 0.5.
For both CNN models, we split the training set (Otest for OP-SCC and Ltest for NSCLC) into train and validation sets with a ratio of 8:2. We performed the training and internal validation within the training cohort, all training and internal validation being done at the patient and not at the individual image patch-level. Once each of the individual image patches have been assigned a class label, majority voting is employed to aggregate all the individual predictions to generate a patient-level prediction. The model was trained for 100 epochs with the internal validation set being employed at the end of each epoch to track model convergence. The final model was selected in a way to minimize the error on the internal validation set and was subsequently locked down.
While we prepared the training data, we split each image into smaller patches of 256 x 256 pixels, the individual image patches from a single patient were all assigned the same class label, corresponding to the patient level diagnosis or prognosis. After filtering out unusable patches (not enough tissue, image patches with image markings, or other artifacts), the following number of image patches were left for evaluation in conjunction with the different use cases. For the OP-SCC training cohort Otrain, we have 115,968 patches at 40X magnification, each image patch of size 256 x 256 pixels, 30,848 patches from HPV+ patients, and 85,120 patches from HPV− patients. For the ES-NSCLC training cohort Ltrain, the average image size of the TMA spot was 3000x3000 pixels at 20x magnification, which in turn resulted in 43,872 patches for the short-term survival patients, and 24,129 patches for the long-term survival patients.
HPV classification for OP-SCC
While comparing FLocK feature family with other QH families, FLocK provided comparable classification AUC (Table 6). Since the FLocK feature family does not provide the best classification performance, we have investigated if it provides add-on information for separating HPV+ vs HPV− patients. We evaluated the performance of a classifier that utilized the combination of top two features from FLocK features and another feature family, i.e. FLocK + CCG (Ali et al., 2013b), FLocK + Nuclear shape, and FLocK + COrE (Lee et al., 2013), shown in Table 7. As one may observe that by combining the top features from different feature families, the classification performance improved significantly, especially in the case of COrE + FLocK. This suggests that the different ways of nuclear morphology quantification could help to stratify the HPV status for OP-SCC, and FLocK could provide a unique aspect to interrogate the local nuclear morphology. The boxplot of the top two FLocK features that were selected as most discriminative features during the training are shown in Figure 3 (a) and (b). It is interesting to observe that the feature portion of intersected FLocK has higher expression in HPV− image compared to that of the HPV+ image (representative FLocK configuration can be found in Figure 4(c) and (f) respectively). This suggests that in HPV+ cases, in general, there are less interactions between different FLocKs; in other words, the nuclear on the image are more homogeneous in terms of nuclear size and intensity. The other feature - portion of highly intersected FLocK (Figure 3(b)) shows that there are more highly overlapped FLocKs in HPV+ cases than that in HPV− cases, which suggests that locally there are more pleomorphism observed.
Table 7:
Performance of classifier based on top four features from the combination of two feature families in the OP-SCC training cohort under 3-fold CV scheme.
| Model | Feature Family | AUC | Accuracy | Specificity | Sensitivity |
|---|---|---|---|---|---|
| MFLocK+CCG | CCG + FLocK | 0.87±0.03 | 0.80±0.02 | 0.89±0.09 | 0.72±0.08 |
| MFLocK+COrE | COrE +FLocK | 0.91±0.02 | 0.84±0.02 | 0.83±0.11 | 0.86±0.10 |
| MFLocK+Shape | Nuclear shape+FLocK | 0.85±0.03 | 0.80±0.02 | 0.77±0.08 | 0.83±0.09 |
The performance of the FLocK feature-based classifier MFLock and CNN approaches MAlex and MRes, are shown in Table 9-A. In the independent test cohort Otrain, the CNN approaches MAlex and MRes outperformed the FLocK-based classifier MFLock as well. However, the combination of FLocK and COrE feature-based classifier MFlock+COrE yielded the best performance in the test cohort Otest.
Table 9:
Performance comparison between FLocK-based classifiers MFLocK and CNN approaches, AlexNet MAlex and ResNet MRes, for (a) OP-SCC HPV status classification and (b) prognosis of ES-NSCLC use cases. For the results on the training set Otrain and Ltrain, we reported the mean AUC under 3-fold CV for the OP-SCC use case and the mean AUC under 10-fold CV for the ES-NSCLC use case.
| OP-SCC HPV Status Classification | |||||
|---|---|---|---|---|---|
| Classifier | Cohort | AUC | Accuracy | Specificity | Sensitivity |
| MAlex | Otrain | 0.79±0.05 | 0.78±0.04 | 0.82±0.09 | 0.74±0.09 |
| MAlex | Otest | 0.78 | 0.77 | 0.72 | 0.80 |
| MRes | Otrain | 0.82±0.04 | 0.80±0.03 | 0.86±0.09 | 0.72±0.08 |
| MRes | Otest | 0.81 | 0.80 | 0.75 | 0.84 |
| MFLocK | Otrain | 0.80±0.04 | 0.77±0.03 | 0.83±0.09 | 0.71±0.08 |
| MFLocK | Otest | 0.76 | 0.80 | 0.63 | 0.95 |
| MFLocK+COrE | Otest | 0.84 | 0.85 | 0.82 | 0.88 |
| ES-NSCLC Prognosis | |||||
| Classifier | Cohort | AUC | Accuracy | Specificity | Sensitivity |
| MAlex | Ltrain | 0.55±0.04 | 0.54±0.04 | 0.57±0.13 | 0.53±0.15 |
| MAlex | Ltest | 0.56 | 0.57 | 0.55 | 0.58 |
| MRes | Ltrain | 0.56±0.03 | 0.55±0.04 | 0.58±0.12 | 0.54±0.13 |
| MRes | Ltest | 0.56 | 0.55 | 0.54 | 0.57 |
| MFLocK | Ltrain | 0.68±0.03 | 0.67±0.06 | 0.71±0.10 | 0.65±0.09 |
| MFLocK | Ltest | 0.70 | 0.72 | 0.70 | 0.75 |
Predicting Patient Outcomes of ES-NSCLC
While comparing FLocK feature family with other QH families, as shown in Table 8, the FLocK based classifier MFLock achieved the highest AUC of 0.68±0.02, outperforming the other handcrafted feature families. The Global graph, shape, COrE, and CCG based classifiers yielded AUCs of 0.56±0.02, 0.54±0.03, 0.61 ±0.02, and 0.62±0.03, respectively. The classification results suggested that the locally-extracted nuclear features provided better prognostic value than those associated with global architecture. Comparing the performance of CCG and FLocK based classifiers suggests that the organization of local cell clusters, where cluster membership was defined not solely based off spatial proximity but also on morphologic similarity, resulted in more prognostic signatures. Figure 4 shows two representative H&E stained TMA spot images for long-term and short-term survival NSCLC patients, along with the corresponding CCG and FLocK feature representations. The panel inset for FLocK reveals the grouping of the TIL and cancer nuclei as distinct clusters with the associated spatial interaction between these two cell families, unlike the CCG representation, which does not distinguish between the nuclei and TILs.
Table 8:
Performance of five different histomorphometric feature families in the ES-NSCLC training cohort under 10-fold CV scheme.
| Feature Family | AUC | Accuracy | Specificity | Sensitivity |
|---|---|---|---|---|
| CCG | 0.62±0.04 | 0.61±0.06 | 0.63±0.11 | 0.60±0.10 |
| Global Cell Graph | 0.56±0.07 | 0.58±0.09 | 0.60±0.12 | 0.54±0.15 |
| Nuclear shape | 0.54±0.09 | 0.55±0.08 | 0.58±0.14 | 0.52±0.16 |
| COrE | 0.61±0.05 | 0.60±0.07 | 0.65±0.11 | 0.57±0.11 |
| FLocK | 0.68±0.03 | 0.67±0.06 | 0.71±0.10 | 0.65±0.09 |
During feature discovery, we found that measures which reflect the degree of FLocKs intersection and the variance of FLocK graph sizes were the two most frequently selected features by WRST across 100 runs of 10-fold cross-validation (the boxplot of these two FLocK features are shown in Figure 3(c) and (d)). The top feature reflects the degree of interactions between the different local cell families. The boxplot in Figure 3(c) suggests that tumor outcome is improved with an increase in the total number of local cell cluster interactions. This may in turn reflect increase spatial interplay between tumor infiltrating lymphocytes (TIL) and cancer nuclei clusters. This is also reflected in the FLocK maps shown in Figure 4 (c) and (f), in which we observe a higher number of intersections between nearby FLocK graphs in the case of patient with long-term survival (Figure 4 (c)), compared to a short-term survivor (Figure 4 (f)).
The performance of the FLocK feature-based classifier MFLock and CNN approaches MAlex and MRes are shown in Table 9-B. One may observe that AlexNet MAlex and ResNet MRes resulted in mean AUCs of 0.55 and 0.56 respectively on Ltrain, lower compared to the FLocK-based classifier MFLock In the Ltest, MFLock outperformed the CNN approach. Considering the fact that in the ES-NSCLC case, we had a relatively small number of training samples (n=434), we also evaluated a transfer learning strategy (Litjens et al., 2017) in which a pre-trained ResNet on ImageNet dataset was fine-tuned using patches from the ES-NSCLC training cohort Ltrain. However, the performance of the transfer learning strategy was still found to be inferior to MFLocK (reported in Table 9). While CNN models have been reported good at low-level visual object detection and segmentation tasks, it is still unclear how to use models to summarize the sub-visual information extract from image patches in order to make prognostic predictions. Also, the deep learning approach we employed may be constrained by the fact that we had an unbalanced dataset, (280 short-term vs. 154 long-term survivals). It is likely that the relatively few long-term survival patients, coupled with the class imbalance resulted in a sub-optimally trained deep learning network. We have also implemented other CNN models, i.e., DenseNet (Huang et al., 2018); however, they suffered from the overfitting and did not provide better result compared to AlexNet MAlex.
Note that one limitation of the DL models implemented in this work is that we have not investigated how different resolutions of the patches used for the DL model training affect network performance. More specifically, we used the FLocK features from 3000x3000 pixels patches for the FLocK-based classifier, and we used 256 x 256 pixels patches for DL models training, which may have limited the spatial context and resolvability of the network.
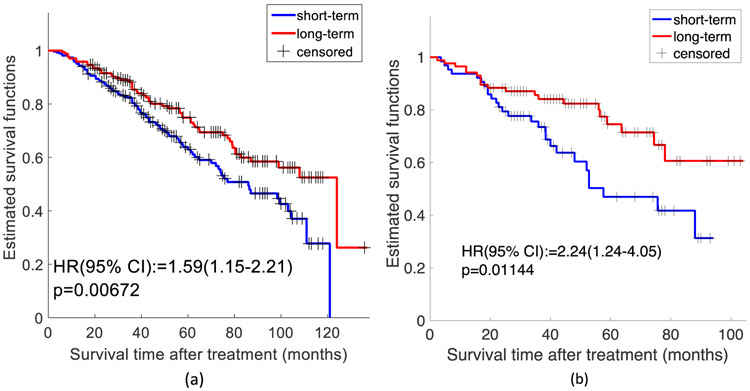
Since we have survival data in this application, we also performed survival analysis using a Cox proportional hazard model based on the predicted labels generated by the FLocK-based classifier. For each TMA spot image, the FLocK-based classifier MFLocK assigned a probability of risk. The Kaplan-Meier (KM) product limit method was used to estimate empirical survival probabilities as illustrated by KM curves. Log-rank tests were applied to examine survival differences, indicating the significance of a categorical variable being prognostic for a survival endpoint. Hazard ratios, associated 95 percent confidence intervals, and P-values from Log-rank tests were reported to evaluate their ability to predict patient survival, significance level being set at 0.05.
In univariable survival analysis, the Log-rank test was performed based on 1) the predicted labels generated by FLocK classifier in training cohort under a leave-one-out framework. The patients identified as high risk had significantly poorer overall survival, with Hazard Ratio (95% Confident Interval) =1.59 (1.15-2.21), p=0.00672; 2) the predicted labels generated by MFLocK in test set Ltest. The patients identified as high-risk had significantly poorer overall survival, with Hazard Ratio (95% Confidence Interval) = 2.24 (1.24-4.05), p=0.01144. The Kaplan-Meier (KM) curves for MFLocK applied to the training and testing sets, Ltrain and Ltest, are shown in Figures 5(a) and (b), respectively.
Figure 5:
(a) KM curves for the FLocK classifier associated with short-term and long-term survival under a leave-one-out framework in training cohort Ltrain. The patients identified as high-risk had significantly poorer overall survival, with Hazard Ratio (95% Confident Interval) =1.59 (1.15-2.21), p=0.00672. (b) KM curves for the FLocK classifier associated with short-term and long-term survival in testing cohort Ltest. The patients identified as high risk had significantly poorer overall survival, with Hazard Ratio (95% Confident Interval) = 2.24 (1.24-4.05), p = 0.01144.
Concluding Remarks
In this work, we presented a novel way to quantify nuclear morphology from histology image, Feature-driven Local Cell Graph (FLocK), and a new set of quantitative histomorphometrics that associated with FLocK which could interrogate the interactions between different local cell clusters presented in digitized tissue images. We applied the FLocK features in conjunction with linear classifiers on 1) classification of HPV status of OP-SCC patients and 2) prediction of patient survival of ES-NSCLC on traditionally used H&E stained histology images.
Although the results obtained via FLocK provide similar or outperformed other existing histomorphometric methods, we acknowledge limitations of our study. The FLocK construction required user to specify parameters for feature space analysis. For example, in our experiments for both of the two applications, the FLocK features were constructed under the feature space consisted of nuclear co-ordinations (i.e., x and y locations of nuclei), nuclear Area and nuclear Mean Intensity, and the spatial bandwidth Hs was set to 100 and the nuclear Area and Mean Intensity bandwidth were set to 40 and 10, respectively (this setting is for 40X magnification, for 20X magnification, the parameters should be divided by two). Since the combination of nuclear properties and bandwidth will affect the configuration of FLocKs, one should first check and visualize if the default parameters make sense in the underlying application (see Figure 6). However, in Appendix 2 and our released Matlab code at Github ((Lu, 2020) https://github.com/hacylu/FLocK/), we have provided a GUI to allow the user to check the FLocK configuration interactively, and also performed qualitative analysis on the effect of parameters on some final feature values with respect to feature space bandwidth (hs and hm).
In summary, FLocK is a unique and general quantitative methodology to analyze histology image of solid tumor and is able to provide nuclear morphology information from a different aspect compared to the existing histomorphometrics. With further larger scale clinical validation, we hope to be able to employ FLocK in different solid tumors for disease prognosis and predict to treatment response. The source code can be accessed at https://github.com/hacylu/FLocK.
Supplementary Material
Table 1:
Nuclear features considered in FLocK construction
| Feature Class | No. | Specific Attributes |
|---|---|---|
| Nuclear shape | 6 | Area, Eccentricity, Solidity, Circularity, Major/minor axis length of best fit eclipse |
| Nuclear appearance | 5 | Mean intensity, Intensity range, Mean inside/outside boundary intensity, Boundary Saliency |
Table 5:
Description of global cell graph, nuclear shape, COrE, CCG features in quantitative evaluation experiments.
| Feature set | # | Description |
|---|---|---|
| Global Cell Graph | 51 | Voronoi Diagram: Polygon area, perimeter, chord length; Delaunay Triangulation: Triangle side length, area; Minimum Spanning Tree: Edge length (compute mean, std. dev., range, skewness, kurtosis, disorder of each); Nearest Neighbors: Density of nuclei, distance to nearest nuclei |
| Nuclear Shape | 100 | Area, Mean Intensity/Intensity Range of Nuclei, Mean Intensity/Intensity Range Around Nuclei, Eccentricity, Perimeter, Smoothness, Invariant Moment 1-7, Fractal Dimension, Fourier Descriptor 1-10 (Mean, Std. Dev, Median, range, skewness, kurtosis of each) |
| Cell orientation entropy (CorE) | 72 | 4 Haralick measurements computed from nuclear orientation co-occurrence matrix (Mean, Std. Dev, median, range, skewness, kurtosis of each with three cell sub-graph setups) |
| Cell Cluster Graph (CCG) | 105 | Clustering Coeff C, Clustering Coeff D, Giant Connected Component, Average Eccentricity, Percent of Isolated Points, Number of Central Points, Skewness of Edge Lengths (Mean, Std. Dev, skewness, kurtosis, range of each with three cell sub-graph setups) |
| Feature-driven Local Cell Graph (FLocK) | 235 | See Table 2 and appendix 1 for more details. The total number of FLocK features depends on the pre-defined cell cluster type number k, and the feature space selected. In our experiment, we used the nuclear Centroid, Area and Mean Intensity as the feature space, and set Hs= 100, Area bandwidth Harea =40, Mean Intensity bandwidth Hintensity=10, and k=0 for feature calculation for 40x magnification image, which yielded 235 features. |
Highlights.
A new way to construct local cell graphs, named Feature-driven Local Cell Graph (FLocK), which simultaneously consider nuclear properties as well proximities, and enable us to interrogate the interaction between different groups of cell type and to reveal more sub-visual information from the underlying tissue image.
A new set of quantitative histomorphometric features based on the constructed FLocKs, which attempts to capture the interactions between and within local cell clusters with similar morphological properties.
Two ways for computation of FLocK features, one with pre-defined cluster type numbers, the other one without pre-defined cluster type numbers.
We validated the FLocK features in conjunction with a linear machine learning classifier (Linear Discriminant Analysis) to 1) predict risk of recurrence for early-stage lung cancers and 2) to classify the Human Papillomavirus status for oropharyngeal cancers.
Acknowledgements
The authors thank Autumn Watson, B.A., for her expert technical assistance with the tissue microarray construction. The authors also thank Donna M. Posey for her wonderful assistance with clerical support and spreadsheet data management for the various aspects of this study. The authors thank Tyler Smallman for his assistance with the digital image scanning and file management. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers U24CA199374, R01CA202752, R01CA208236, R21CA179327, R21CA195152, the National Institute of Diabetes and Digestive and Kidney Diseases under award number R01DK098503, National Center for Research Resources under award number C06 RR12463, the DOD Prostate Cancer Synergistic Idea Development Award (PC120857); the DOD Lung Cancer Idea Development New Investigator Award (LC130463), the DOD Prostate Cancer Idea Development Award; the DOD Peer Reviewed Cancer Research Program W81XWH-16-1-0329, the Case Comprehensive Cancer Center Pilot Grant, VelaSano Grant from the Cleveland Clinic, the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University. Dr. Lu is partially supported by the DoD Breast Cancer Research Program Breakthrough Level 1 Award W81XWH-19-1-0668 and NIH R21 grant CA253108-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest
Dr. Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata Inc.. He is also a scientific advisory consultant for Inspirata Inc. In addition he has served as a scientific advisory board member for Inspirata Inc, Astrazeneca, Bristol Meyers-Squibb and Merck. He also has sponsored research agreements with Philips and Inspirata Inc. His technology has been licensed to Elucid Bioimaging and Inspirata Inc. He is also involved in a NIH U24 grant with PathCore Inc, and 3 different R01 grants with Inspirata Inc.
Dr.’s Lu, Koyuncu, Bera, Velcheti, Wang, Prasanna, Corredor, and Janowczyk, Mr.’s Leo, declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The conflicts of interest for all the co-authors are stated below:
Dr. Janowczyk reports personal fees from Merck. Dr. Velcheti reports grants and personal fees from Merck, Bristol-Myers Squibb, Genentech, AstraZeneca, Celgene, Novartis, Amgen, Fulgent Genetics, Reddy Labs, Alkermes, Nektar Therapeutics, Novocure, and Foundation Medicine outside of the submitted work; advisory or consulting fees from Genentech, Merck, BMS, Astrazeneca, Foundation Medicine, Nektar Therapeutics, Alkermes, Reddy labs, and Millennium Pharma. Dr. Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata, Inc. He is also a scientific advisory consultant for Inspirata, Inc. and sits on its scientific advisory board. Additionally, his technology has been licensed to Elucid Bioimaging and Inspirata, Inc. He is also involved in an NIH U24 grant with PathCore Inc.
Dr.’s Lu, Koyuncu, Bera, Wang, Prasanna, and Corredor, and Mr. Leo, declare no competing financial interests.
References
- Ali S, Lewis J, Madabhushi A, 2013a. Spatially aware cell cluster(spACCI) graphs: predicting outcome in oropharyngeal pl6+ tumors. Med. Image Comput. Comput.-Assist. Interv. MICCAI Int. Conf. Med. Image Comput. Comput.-Assist. Interv 16, 412–419. [DOI] [PubMed] [Google Scholar]
- Ali S, Veltri R, Epstein JA, Christudass C, Madabhushi A, 2013b. Cell cluster graph for prediction of biochemical recurrence in prostate cancer patients from tissue microarrays, in: Gurcan MN, Madabhushi A (Eds.), . p. 86760H 10.1117/12.2008695 [DOI] [Google Scholar]
- Almendro V, Marusyk A, Polyak K, 2013. Cellular Heterogeneity and Molecular Evolution in Cancer. Annu. Rev. Pathol. Mech. Dis 8, 277–302. 10.1146/annurev-pathol-020712-163923 [DOI] [PubMed] [Google Scholar]
- Alom Z, Aspiras T, Taha T, Asari VK, 2019. Histopathological image classification with deep convolutional neural networks, in: Zelinski ME, Taha TM, Howe J, Awwal AA, Iftekharuddin KM (Eds.), Applications of Machine Learning. Presented at the Applications of Machine Learning, SPIE, San Diego, United States, p. 30 10.1117/12.2530291 [DOI] [Google Scholar]
- Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH, 2014. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg. Focus 37, E11 10.3171/2014.9.FOCUS14521 [DOI] [PubMed] [Google Scholar]
- Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A, 2019. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol 16, 703–715. 10.1038/s41571-019-0252-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin C, Demir C, Nagi C, Yener B, 2007. Cell-graph mining for breast tissue modeling and classification. Conf. Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. IEEE Eng. Med. Biol. Soc. Annu. Conf 2007, 5311–5314. 10.1109/IEMBS.2007.4353540 [DOI] [PubMed] [Google Scholar]
- Bose P, Brockton NT, Guggisberg K, Nakoneshny SC, Kornaga E, Klimowicz AC, Tambasco M, Dort JC, 2015. Fractal analysis of nuclear histology integrates tumor and stromal features into a single prognostic factor of the oral cancer microenvironment. BMC Cancer 15 10.1186/s12885-015-1380-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML, 2011. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 29, 4294–4301. 10.1200/JCO.2011.36.4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comaniciu D, Meer P, 2002. Mean shift: a robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell 24, 603–619. 10.1109/34.1000236 [DOI] [Google Scholar]
- Corredor G, Wang X, Zhou Y, Lu C, Fu P, Syrigos K, Rimm DL, Yang M, Romero E, Schalper KA, Velcheti V, Madabhushi A, 2019. Spatial Architecture and Arrangement of Tumor-Infiltrating Lymphocytes for Predicting Likelihood of Recurrence in Early-Stage Non-Small Cell Lung Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res 25, 1526–1534. 10.1158/1078-0432.CCR-18-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchi J, Hazan E, Singer Y, n.d. Adaptive Subgradient Methods for Online Learning and Stochastic Optimization 39. [Google Scholar]
- Genestie C, Zafrani B, Asselain B, Fourquet A, Rozan S, Validire P, Vincent-Salomon A, Sastre-Garau X, 1998. Comparison of the prognostic value of Scarff-Bloom-Richardson and Nottingham histological grades in a series of 825 cases of breast cancer: major importance of the mitotic count as a component of both grading systems. Anticancer Res. 18, 571–576. [PubMed] [Google Scholar]
- Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B, 2009. Histopathological image analysis: a review. IEEE Rev. Biomed. Eng 2, 147–171. 10.1109/RBME.2009.2034865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2016. Deep Residual Learning for Image Recognition, in: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Presented at the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778. 10.1109/CVPR.2016.90 [DOI] [Google Scholar]
- He L, Long LR, Antani S, Thoma GR, 2012. Histology image analysis for carcinoma detection and grading. Comput. Methods Programs Biomed 107, 538–556. 10.1016/j.cmpb.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Liu Z, van der Maaten L, Weinberger KQ, 2018. Densely Connected Convolutional Networks. ArXiv160806993 Cs. [Google Scholar]
- Janowczyk A, Madabhushi A, n.d. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use case. J. Pathol. Inform [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kather JN, Schulte J, Grabsch HI, Loeffler C, Muti H, Dolezal J, Srisuwananukorn A, Agrawal N, Kochanny S, von Stillfried S, Boor P, Yoshikawa T, Jaeger D, Trautwein C, Bankhead P, Cipriani NA, Luedde T, Pearson AT, 2019. Deep learning detects virus presence in cancer histology. bioRxiv 690206. 10.1101/690206 [DOI] [Google Scholar]
- Kothari S, Phan JH, Young AN, Wang MD, 2013. Histological image classification using biologically interpretable shape-based features. BMC Med. Imaging 13 10.1186/1471-2342-13-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhevsky A, Sutskever I, Hinton GE, 2012. ImageNet Classification with Deep Convolutional Neural Networks, in: Proceedings of the 25th International Conference on Neural Information Processing Systems - Volume 1, NIPS’12 Curran Associates Inc., USA, pp. 1097–1105. [Google Scholar]
- Lee G, Ali S, Veltri R, Epstein JI, Christudass C, Madabhushi A, 2013. Cell orientation entropy (COrE): predicting biochemical recurrence from prostate cancer tissue microarrays. Med. Image Comput. Comput.-Assist. Interv. MICCAI Int. Conf. Med. Image Comput. Comput.-Assist. Interv 16, 396–403. [DOI] [PubMed] [Google Scholar]
- Lee G, Sparks R, Ali S, Shih NNC, Feldman MD, Spangler E, Rebbeck T, Tomaszewski JE, Madabhushi A, 2014. Co-occurring gland angularity in localized subgraphs: predicting biochemical recurrence in intermediate-risk prostate cancer patients. PloS One 9, e97954 10.1371/journal.pone.0097954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Veltri RW, Zhu G, Ali S, Epstein JI, Madabhushi A, 2016. Nuclear Shape and Architecture in Benign Fields Predict Biochemical Recurrence in Prostate Cancer Patients Following Radical Prostatectomy: Preliminary Findings. Eur. Urol. Focus 10.1016/j.euf.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JS, Ali S, Luo J, Thorstad WL, Madabhushi A, 2014. A quantitative histomorphometric classifier (QuHbIC) identifies aggressive versus indolent p16-positive oropharyngeal squamous cell carcinoma. Am. J. Surg. Pathol 38, 128–137. 10.1097/PAS.0000000000000086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI, 2017. A survey on deep learning in medical image analysis. Med. Image Anal 42, 60–88. 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- Lu C, 2020. Matlab source code for FLocK at GitHub. [Google Scholar]
- Lu C, Mandal M, 2015. Automated analysis and diagnosis of skin melanoma on whole slide histopathological images. Pattern Recognit. 48, 2738–2750. 10.1016/j.patcog.2015.02.023 [DOI] [Google Scholar]
- Lu C, Romo-Bucheli D, Wang X, Janowczyk A, Ganesan S, Gilmore H, Rimm D, Madabhushi A, 2018. Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab. Invest 10.1038/S41374-018-0095-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Xu J, Gilmore H, Mandal M, Madabhushi A, 2016. Multi-Pass Adaptive Voting for Nuclei Detection in Histopathological Images. Sci. Rep 6, 33985 10.1038/srep33985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz EA, Rocco JW, 2016. Intra-tumor heterogeneity in head and neck cancer and its clinical implications. World J. Otorhinolaryngol.-Head Neck Surg 2, 60–67. 10.1016/j.wjorl.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz S, Heindl A, Koelble K, Yuan Y, 2015. Beyond immune density: critical role of spatial heterogeneity in estrogen receptor-negative breast cancer. Mod. Pathol 28, 766–777. 10.1038/modpathol.2015.37 [DOI] [PubMed] [Google Scholar]
- Nawaz W, Ahmed S, Tahir A, Khan HA, 2018. Classification Of Breast Cancer Histology Images Using ALEXNET, in: Campilho A, Karray F, ter Haar Romeny B (Eds.), Image Analysis and Recognition, Lecture Notes in Computer Science. Springer International Publishing, Cham, pp. 869–876. 10.1007/978-3-319-93000-8_99 [DOI] [Google Scholar]
- Nguyen K, Sarkar A, Jain AK, 2014. Prostate cancer grading: use of graph cut and spatial arrangement of nuclei. IEEE Trans. Med. Imaging 33, 2254–2270. 10.1109/TMI.2014.2336883 [DOI] [PubMed] [Google Scholar]
- Noy S, Vlodavsky E, Klorin G, Drumea K, Ben Izhak O, Shor E, Sabo E, 2011. Computerized morphometry as an aid in distinguishing recurrent versus nonrecurrent meningiomas. Anal. Quant. Cytol. Histol 33, 141–150. [PubMed] [Google Scholar]
- Peng H, Long F, Ding C, 2005. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell 27, 1226–1238. 10.1109/TPAMI.2005.159 [DOI] [PubMed] [Google Scholar]
- Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, Samaras D, Shroyer KR, Zhao T, Batiste R, Van Arnam J, Shmulevich I, Rao AUK, Lazar AJ, Sharma A, Thorsson V, 2018. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 23, 181–193.e7. 10.1016/j.celrep.2018.03.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Protano M-A, Polydorides AD, Dawsey SM, Pierce MC, Kim MK, Schwarz RA, Quang T, Parikh N, Bhutani MS, Zhang F, Wang G, Xue L, Wang X, Xu H, Anandasabapathy S, Richards-Kortum RR, 2015. Quantitative Analysis of High- Resolution Microendoscopic Images for Diagnosis of Esophageal Squamous Cell Carcinoma. Clin. Gastroenterol. Hepatol 13, 272–279.e2. 10.1016/j.cgh.2014.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Janowczyk A, Zhou Y, Thawani R, Fu P, Schalper K, Velcheti V, Madabhushi A, 2017. Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci. Rep 7 10.1038/S41598-017-13773-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K-H, Zhang C, Berry GJ, Altman RB, Ré C, Rubin DL, Snyder M, 2016. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun 7, 12474 10.1038/ncomms12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Failmezger H, Rueda OM, Ali HR, Graf S, Chin S-F, Schwarz RF, Curtis C, Dunning MJ, Bardwell H, Johnson N, Doyle S, Turashvili G, Provenzano E, Aparicio S, Caldas C, Markowetz F, 2012. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci. Transl. Med 4, 157ra143 10.1126/scitranslmed.3004330 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.