Abstract
Despite widespread concern for cytokine storms leading to severe morbidity in COVID-19, rapid cytokine assays are not routinely available for monitoring critically ill patients. We report the clinical application of a digital protein microarray platform for rapid multiplex quantification of cytokines from critically ill COVID-19 patients admitted to the intensive care unit (ICU) at the University of Michigan Hospital. The platform comprises two low-cost modules: (i) a semi-automated fluidic dispensing/mixing module that can be operated inside a biosafety cabinet to minimize the exposure of technician to the virus infection and (ii) a 12-12-15 inch compact fluorescence optical scanner for the potential near-bedside readout. The platform enabled daily cytokine analysis in clinical practice with high sensitivity (<0.4pg/mL), inter-assay repeatability (~10% CV), and rapid operation providing feedback on the progress of therapy within 4 hours. This test allowed us to perform serial monitoring of two critically ill patients with respiratory failure and to support the immunomodulatory therapy using the selective cytopheretic device (SCD). We also observed clear interleukin-6 (IL-6) elevations after receiving tocilizumab (IL-6 inhibitor) while significant cytokine profile variability exists across all critically ill COVID-19 patients and to discover a weak correlation between IL-6 to clinical biomarkers, such as ferritin and c-reactive protein (CRP). Our data revealed large subject-to-subject variability in patients’ response to COVID-19, reaffirming the need for a personalized strategy guided by rapid cytokine assays.
Graphical Abstract
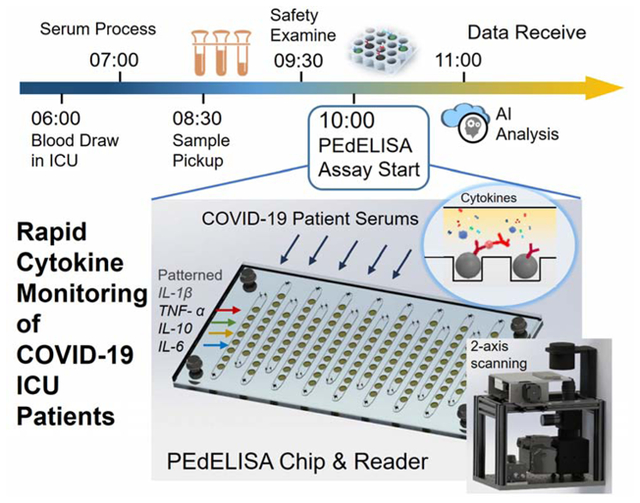
A digital microfluidic immunoassay platform enables rapid multiplex quantification of proinflammatory cytokines in serum for critically ill COVID-19 patients.
Introduction
With the global outbreak of the coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)1, accumulating evidence2–5 indicates that cytokine storm or cytokine release syndrome (CRS) is associated with severe illness. CRS is observed in several disease states associated with dysregulated immunity, including as a consequence of CAR-T cell immunotherapy6, a manifestation of hemophagocytic lymphohistiocytosis (HLH) in malignancy, macrophage activation syndrome in autoimmune disease7, or severe sepsis8. Selective cytokine blockade is a mainstay of care for CRS related cancer immunotherapy6, 9, 10, and macrophage activation syndrome11. In COVID-19, early translational studies suggest that high serum cytokines are a result of a complex interplay between lymphocytes and myeloid cells12. Modulation of cytokine signaling pathways is currently the subject of over 50 clinical trials worldwide13. However, most studies enroll based on clinical criteria without rapid assessment of specific cytokine levels, despite delivering therapies that are targeted to specific cytokines, such as interleukin (IL)-6. In our center, the current clinical practice is to use a variety of less specific surrogate markers, such as ferritin and CRP, to gauge a patient’s overall level of inflammation. While cytokine levels are being checked in patients with severe COVID-19, in practice, the results of these tests return in days, not hours. Ideally, treating physicians would understand the “real-time” level of a variety of cytokines in a particular patient before administering specific medications to blunt cytokine storm in critical illness, which urgently requires a low-cost multiplex cytokine profiling assay with a rapid assay turnaround.
Digital immunoassay14, 15 has been considered as the next generation protein detection method, which provides single-molecular sensitivity (aM-fM) detection by digitizing and amplifying enzymatic reaction in extremely confined volumes (fL-nL). The current commercial implementation of digital immunoassay is Quanterix Simoa HD-1/X Analyzer™16. Despite its full automation and ultra-high sensitivity, the high assay and instrumentation costs and large physical footprint prevent its application for timely near-bedside diagnosis. Several groups invented microfluidic platforms for lab-on-a-chip operation of digital assays17–20 and notably, Yelleswarapu et al21 demonstrated a mobile-phone-based, droplet microfluidic digital immunoassay for point-of-care (POC) settings. More recently, a few previous studies22, 23 have implemented a digital assay platform applicable to the clinical treatment of a COVID-19-induced cytokine storm. If continuous monitoring of the cytokine profiles of a COVID-19 patient is needed, the assay requires more than speed, sensitivity, and multiplex capacity. Other important but often overlooked requirements include (1) flexibility of running a small number of samples based on the demand of the physician with minimum preparation; (2) great inter-assay precision between multi-time point measurements, which is not an issue in conventional large batch-based retrospective tests; (3) a low-cost, compact, automated fluidic handling and readout instrumentation that can be operated inside the bio-safety cabinet with minimum user exposure to virus-contaminated blood samples.
Here, we report the development and application of an automated digital assay platform using a method termed the “pre-equilibrium digital enzyme-linked immunosorbent assay (PEdELISA) microarray” for rapid multiplex monitoring of cytokine: IL-6, TNF-α, IL-1β and IL-10 from COVID-19 patients admitted to the ICU in the University of Michigan hospital. The PEdELISA microarray analysis employs magnetic beads trapped into spatially registered microwell patterns on a microfluidic chip. The locations of the microwell patterns on the chip indicate which target analytes are detected. Combining single-molecular counting with early pre-equilibrium reaction quenching can achieve a rapid digital immunoassay with a clinically relevant fM-nM dynamic range without losing signal sensing linearity. The simple but effective bead spatial registration strategy coupled with machine learning-based image processing enables a multiplex digital immunoassay with high accuracy and eliminates bead loss, which has been experienced by a state-of-the-art commercial platform16. Our digital assay platform incorporates an automated fluidic dispensing and on-chip mixing system to precisely control the reaction time at the pre-equilibrium state after 9 min of incubation. This automated microfluidic operation leads to inter-assay precision with a coefficient of variation (CV) ~10%. Furthermore, the system using a low-cost compact fluorescence reader can potentially be adapted to bedside applications. Our digital assay demonstrated here works as a promising candidate for continuous cytokine profiling with the combination of speed and sensitivity, both greater than those of current analog24–27 and label-free POC diagnostic systems28–31.
Results and Discussion
Automated PEdELISA system
The PEdELISA microarray assay platform comprises a cartridge holding a disposable microfluidic chip with capture antibody (CapAb)-conjugated magnetic beads pre-settled in the designated microarray locations according to the antibody type, a parallel pipetting module controlled by Arduino for on-chip fluidic dispensing and mixing, and a 2-axis chip scanning and fluorescence imaging module (Fig. 1A–C, see Supplementary Information for system details). In this setup, each disposable microfluidic chip (Fig. 1A, inset) was designed to handle 16 samples with up to 8-plex maximum capacity. The chip contains two polymethyl methacrylate (PMMA) layers top (venting) and bottom layer (substrate) with countersink connectors that are seamlessly interfaced with fluidic dispensing tips, a thin polydimethylsiloxane (PDMS) layer (300 μm) which contains fL-sized microwell arrays for digital assay, and a polyethylene terephthalate (PET) thin (120 μm) film with microfluidic channels fabricated by laser cutting (see Fig. S1 for chip fabrication). A cluster of 66,724 arrayed microwells (each 3.4 μm in diameter and 8 μm in pitch) forms a circular biosensor pattern targeting a specific analyte species on the chip. Each microfluidic channel of the chip contains 8 biosensor patterns. The microwells are filled with pre-deposited CapAb-conjugated beads (see Methods and Fig. S2 for bead patterning). The use of these materials and processing methods significantly reduced the chip manufacturing cost (< $0.5/chip).
Figure 1.
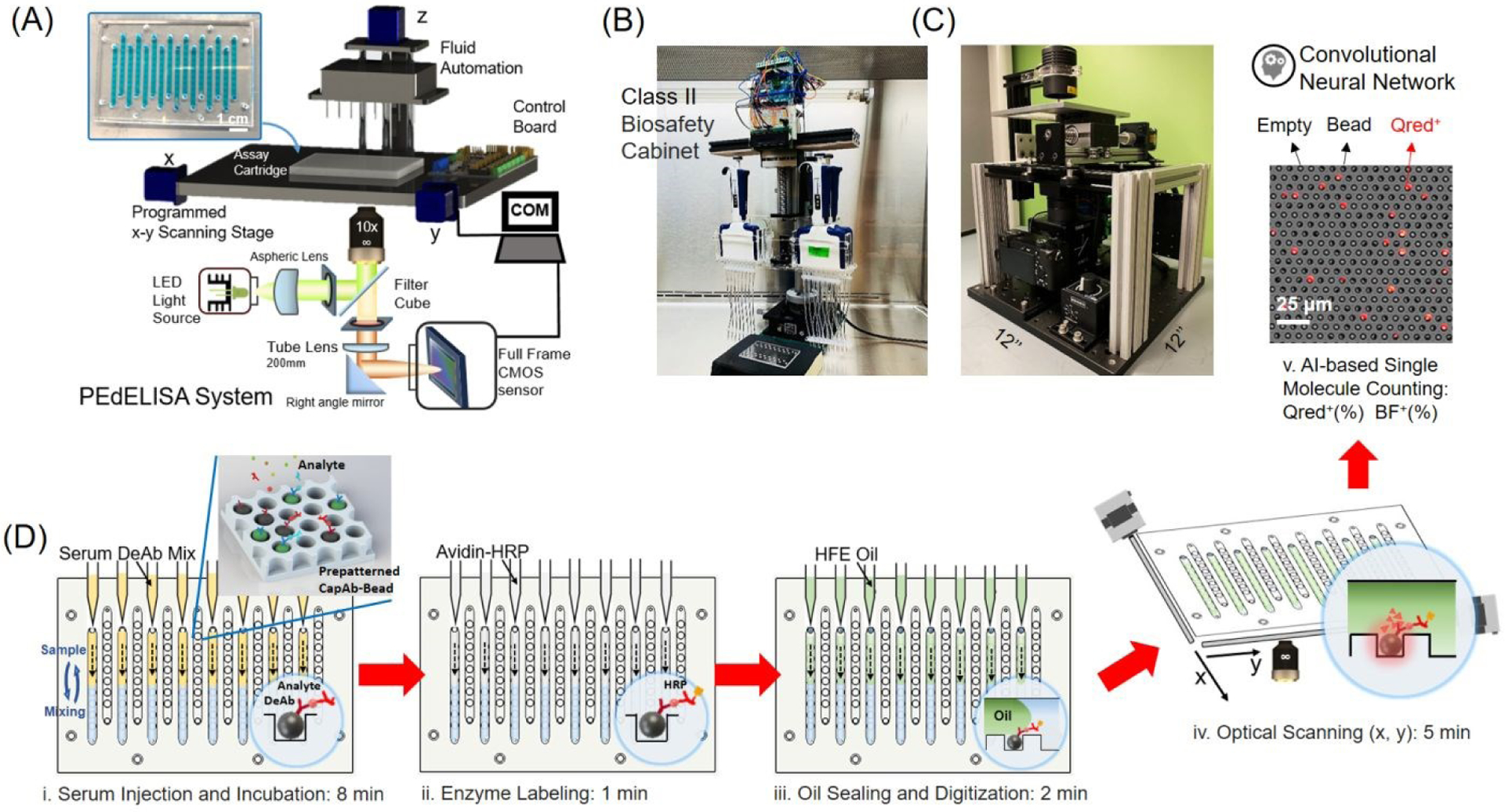
PEdELISA microarray assay platform for COVID-19 patient cytokine storm profiling. (A)-(C) Schematic and photo image of the assay system in a biosafety cabinet. The platform comprises a cartridge holding a disposable microfluidic chip (inset), an automated fluidic dispensing and mixing module, and a 2D inverted fluorescence scanning module. Each channel of the chip has 8 circular biosensor patterns formed by a cluster of 66,724 arrayed microwells. Each biosensor pattern detects one of four target analytes. (D) The 5-step assay procedure includes (i) automated injection and subsequent on-chip mixing of serum and a detection antibody solution with capture antibody-coated magnetic beads pre-deposited in microwell arrays, which is accompanied by a short incubation (9-min) and followed by washing (2-min), (ii) HRP enzyme labeling (1-min), followed by washing (5-min), (iii) fluorescence substrate loading and oil sealing (2-min), (iv) x-y optical scanning and imaging (5-min), and (v) Data analysis using a convolutional neural network-guided image processing algorithm (4sec/image) for high throughput and accurate single-molecule counting (5–7min). The total sample to answer time is around 30 min for 8 samples in 4-plex. Both the fluorescence substrate channel (Qred CH) and brightfield channel (BF CH) are analyzed to calculate the average number of immune-complexes formed on each bead surface.
The PEdELISA assay was carried out by the programmed pipetting module that allowed for microfluidic loading and handling in a consistent and repeatable manner (Fig. S3A). The module first mixed patient samples or assay standards with a detection antibody (DeAb) solution and then loaded them into the chip in parallel, followed by 50 automated cycles of on-chip mixing during incubation (9 min), washing (2 min), and enzyme labeling (1 min), washing (5 min), substrate loading, and oil sealing (Fig. 1D, see Methods for assay details). The chip was subsequently scanned and imaged by the compact and low-cost (<$5000) fluorescence imaging module using a consumer-grade CMOS camera (Fig. S4B), and the data was analyzed by a high-throughput in-house image processing algorithm based on machine learning and parallel computing. The PEdELISA data analysis algorithm (Fig. 2A) contains two convolutional neural networks (CNN) which were pre-trained to carry out two pathways: (i) Qred positive fluorescence microwell recognition and (ii) image defect recognition. The algorithm sequentially performed image pre-processing, category classification, and segmentation of image features, such as microwells, defects, and backgrounds, so that the digital assay counting results were generated without human supervision (see Methods for data analysis and algorithm details). The two networks executed in parallel computing were operated together to enhance the accuracy of image analysis while maintaining the high scanning speed. Fig. 2B and Fig. S4 show representative fluorescence images of Qred positive microwells on a biosensor pattern for various analyte concentrations. It generally took ~4s to analyze the whole biosensor pattern image of 66,724 microwells with 6000×4000 pixels using the Intel Core i7–8700 central processing unit (CPU) and the NVIDIA Quadro P1000 graphics processing unit (GPU). The assay also involved some manual work for assay reagent preparation and serial dilution, fluid waste collection, z-axis focusing, and origin/endpoint positioning to trigger the optical scanning.
Figure 2.
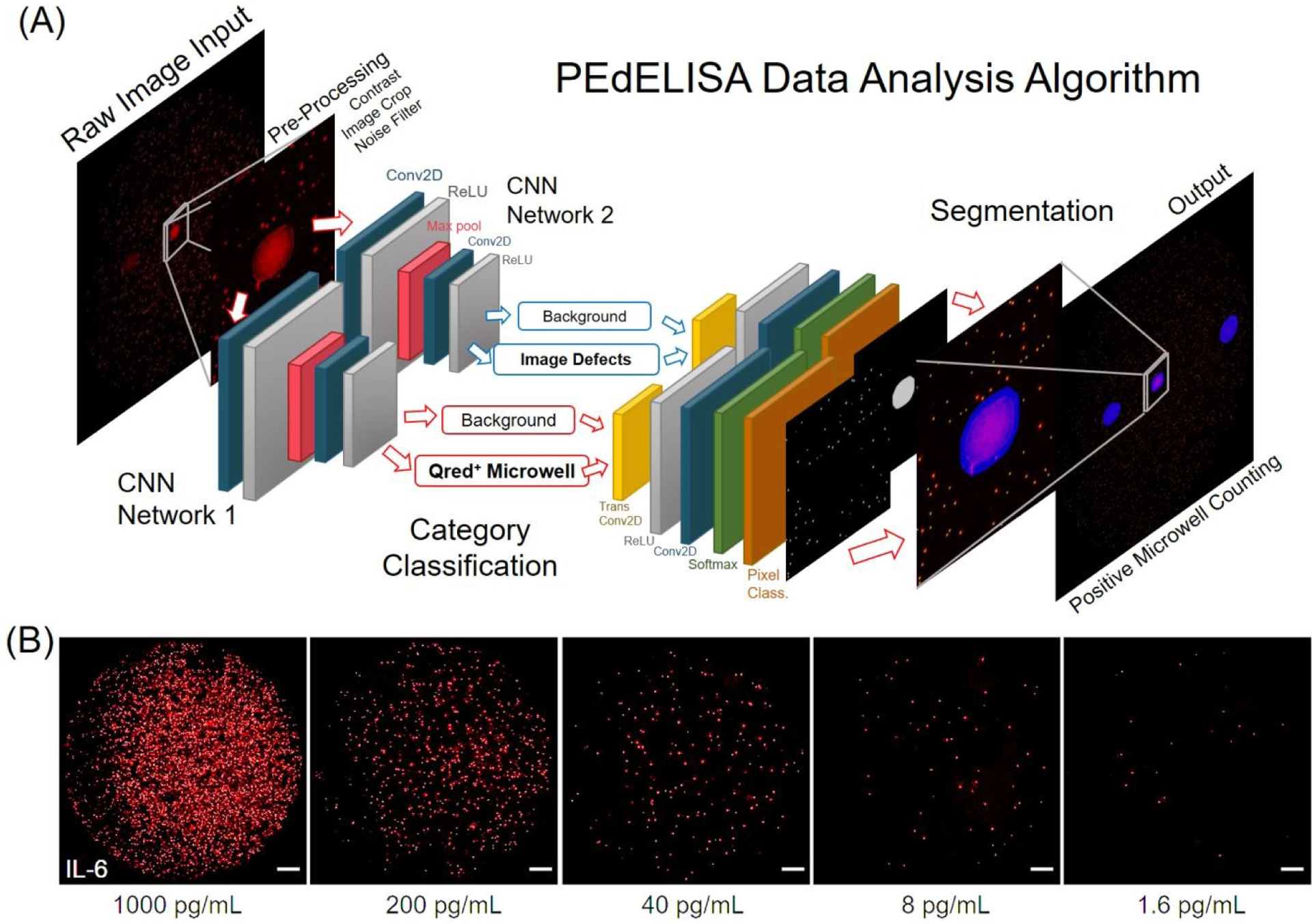
(A) Architecture of the PEdELISA data analysis algorithm using convolutional neural network (CNN) and parallel computing by MATLAB. Two CNN networks were trained and executed in parallel to read in the QuantaRedTM (Qred) fluorescence mirowell images, and sequentially performed image pre-processing (including image cropping, contrast enhancement, and noise filtering), category classification, image segmentation, and post-processing (image overlay, visualization, defect compensation, microwell counting). (B) Representative snapshot images of enzyme active “On” microwells on a biosensing pattern (66,724 wells/biosensor) for various analyte concentrations of IL-6. For clear visualization, images of 3600×3600 pixels were cropped from original raw images of 6000×4000 pixels with 80% brightness enhancement and 80% contrast enhancement. All of the scale bars are 200 μm. See Fig. S4 for images taken for IL-1β, TNF-α, IL-10, and IL-6.
PEdELISA performance optimization and characterization
We ensured the x-y optical scanning motion control accuracy each time by repetitively scanning and imaging the microarray structures on the chip. Post-image processing was used to calculate the x, y offset, which may be induced by the imperfection of system alignment, lead screw backlash, or motor step missing. We developed a mathematical algorithm to correct these offsets, and the scanning module was able to achieve less than 5 μm bidirectional repeatability and 0.31 μm minimum incremental movement (Fig. S5). Using the programmed fluidic dispensing system, we optimized the assay reaction parameters by performing a time-scanning assay standard test to evaluate the tradeoff between speed and sensitivity (see Fig. S6 and Supplementary Information). The optimization goal was to achieve the highest assay speed while maintaining the clinical dynamic range relevant to the cytokine storm (sub-pg/mL to ng/mL). We experimentally obtained the limit of detection (LOD) values of the cytokines for the varying sample reaction time (Fig. 3A). Observing that the LOD improvement tapered off with the sample reaction > 10 min, we selected 9 min as the optimal sample reaction time. The optimized assay conditions led to a limit of detection (LOD) < 0.4 pg/mL for all the measured cytokines (Table 1). We also characterized the typical filling rate of the bead array for cytokine antibody-conjugated magnetic beads, which was shown to be 50–80%, slightly varying based on the cytokine type (Fig. 3B). This filling rate ensures 36,000–54,000 beads to be interrogated for each assay condition.
Figure 3.
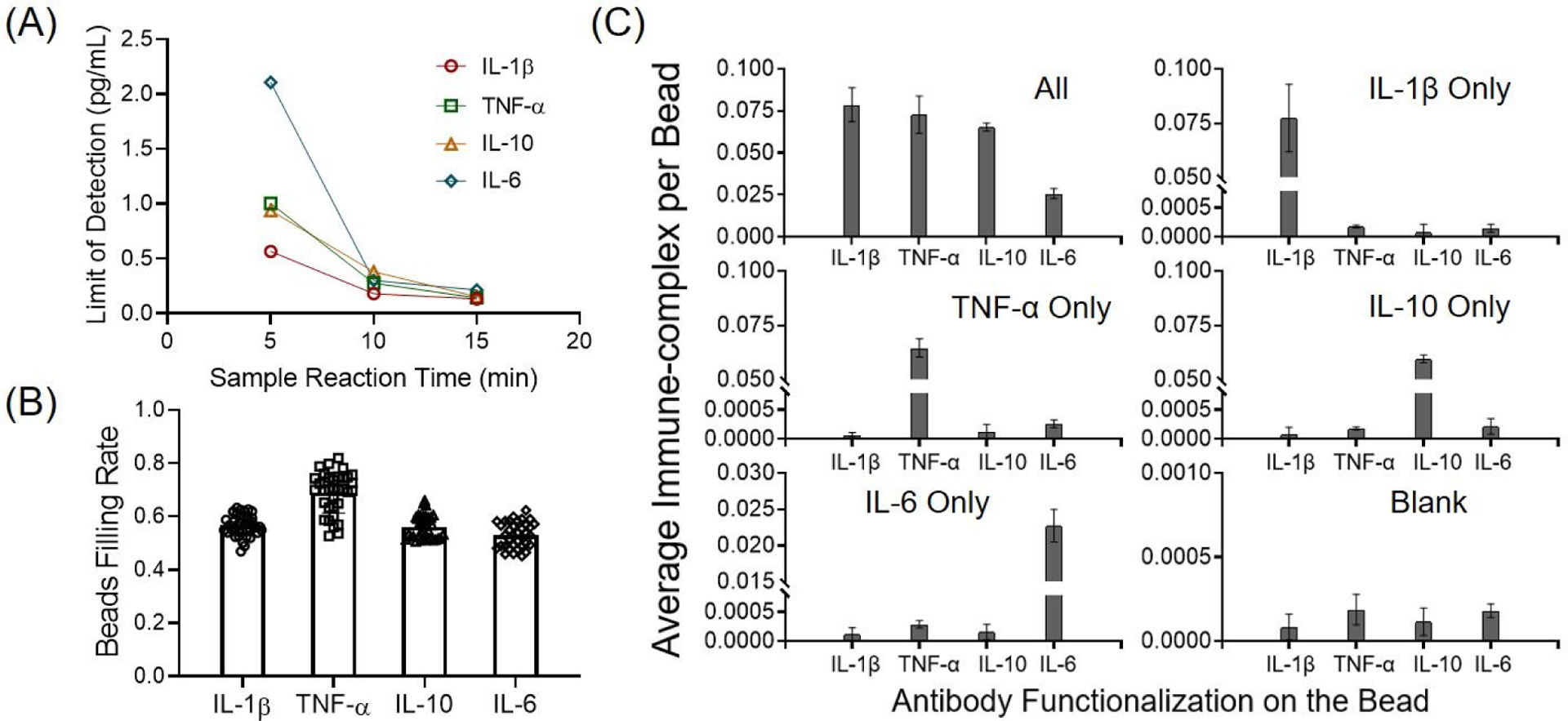
Assay characterization of the PEdELISA. (A) Limit of detection (LOD) of the assay as a function of sample/detection antibody reaction incubation time for the four cytokines. Each LOD value was estimated by taking the intercept of the fitted standard curve with the mean value of the negative control signal plus three times the standard deviation. (B) Statistical analysis of the bead array filling rate for cytokine antibody-conjugated magnetic beads. IL-1β: 56.7±3.9%; TNF-α: 68.7±7.3%; IL-10: 56.2±4.4%; and IL-6: 53.1±4.7%. Each data point represents the bead filling rate obtained across 66,724 microwells in each circular biosensor pattern on the PEdELISA chip. We examined 40 representative biosensor patterns from five different chips to obtain the statistically averaged value for each analyte. (C) Assay specificity test with “all-spike-in,” “single-spike-in,” and “no-spike-in” (negative) samples of recombinant cytokine marker(s) at 200 pg/mL in fetal bovine serum (FBS) buffer. The labels on the horizontal axis represent the cytokine analytes targeted by 4 biosensors in each detection channel on a 4-plex PEdELISA chip. The top label of each graph represents the recombinant cytokine marker(s) loaded to the chip.
Table 1.
Limit of detection (LOD), limit of Quantification (LOQ), and coefficient of variation (CV) of PEdELISA for a panel of 4 cytokines. Here, the LOD and LOQ values were determined from the blank signal + 3σ and the blank signal + 10σ, respectively. The intra-assay CV was determined by quadruplicate measurements of five COVID-19 patient samples at the range of 6–600 pg/mL in both rapid and retrospective assay modes. The inter-assay CV was determined by taking the root-mean-square average of signals from 40, 200, and 1000 pg/mL assay standard in 10-day continuous measurements of COVID-19 patients.
| Cytokine Type | Assay Blank (Average molecule per bead) | Assay Blank+3σ (Average molecule per bead) | LOD (pg/mL) | LOQ (pg/mL) | Intra-Assay CV (%) | Inter-Assay CV (%) |
|---|---|---|---|---|---|---|
| IL-1β | 0.000143 | 0.000347 | 0.191 | 1.188 | 4.68 | 9.98 |
| TNF-α | 0.000179 | 0.000379 | 0.198 | 1.889 | 8.77 | 11.66 |
| IL-10 | 0.000136 | 0.000378 | 0.350 | 1.552 | 5.70 | 9.63 |
| IL-6 | 0.000189 | 0.000309 | 0.377 | 2.378 | 4.55 | 10.80 |
We further assessed the 4-plex assay’s specificity and signal-to-noise ratio (SNR) by spiking-in each cytokine analyte in 100% fetal bovine serum buffer (FBS) to mimic the patient serum detection. Fig 3C shows the assay results of “all-spike-in,” “single-spike-in,” and “no-spike-in” samples using 200 pg/mL recombinant cytokine markers (a typical clinical threshold for cytokine storm). Negligible antibody cross-reactivity was observed between each cytokine analyte and SNR=488.0 was calculated on average (averaged assay signal over background signal).
Monitoring of cytokine storm for critically ill COVID-19 patients
In order to facilitate the care of patients with COVID-19 at the University of Michigan Hospital, we undertook a pragmatic study to rapidly return same-day cytokine levels to the clinical teams treating critically ill COVID-19 patients in ICU at the physicians’ request from April 9th to June 04th in 2020. Given the investigational nature of the assay, patients or their representatives provided informed consent for cytokine measurements to be provided for clinical use (UM IRB HUM00179668). COVID-19 patients with respiratory failure that required hospitalization in the ICU for mechanical ventilation were eligible for enrollment (see Methods for details). To ensure the accuracy of our data, the COVID-19 patient samples were run in quadruplicate with an assay standard curve calibrated every day.
The analytes IL-6, TNF-α, IL-1β, and IL-10 were selected based on our best understanding of clinical utility at the time of study design in March 2020. IL-6 had been reported as a likely mediator of COVID-19 severity and while several clinical trials of anti-IL6R agents were underway, off-label use was also common2. Likewise, TNF-α, IL-1β were selected due to the potential for off-label use of anti-cytokine antibodies against these mediators. IL-10 was selected due to the possible utility of monitoring a compensatory anti-inflammatory response32.
Fig. 4A shows assay standard curves that were accumulated for 10 different workdays of the patient cytokine monitoring period. Around three-order of linear dynamic range has been achieved. By quenching the reaction forming analyte-antibody complexes in the sample solution at the early pre-equilibrium state, the assay still retains the binary counting regime operation following the Poisson’s distribution. This approach allows the assay to cover an analyte concentration range in which the conventional digital assay cannot be operated15. The multiple assay standard curves yield excellent repeatability with the inter-assay coefficient of variation (CV) of ~ 10% due to the programmed fluidic handling and reaction (Table 1). We also characterized the intra-assay CV for five representative COVID-19 patient serum samples with cytokines at concentrations ranging from 6–600 pg/mL, each tested in quadruplicate measurements (Table 1). We compared assay data for these five patients resulting from rapid measurements of fresh samples drawn daily and retrospective measurements of stored samples after one freeze-thaw cycle. We observed a good linear correlation (R2=0.99) between the two measurement modes except for the TNF-α data yielding lower levels in the stored samples and for some of the IL-6 data showing slightly higher levels in the stored samples (Fig. 4B). This suggests that TNF-α in the stored serum could degrade by 20–40% after the freeze-and-thaw banking at −80 °C. Sample degradation is expected to yield lower signals for stored samples than for fresh samples. Therefore, the higher IL-6 signals of a fraction of the stored samples may potentially originate from sample handling error, not from a sample property change. Additionally, to validate our PEdELISA microarray assay, we compared the assay results with those of a conventional single-plex ELISA method that retrospectively measured 15 banked samples from identical patients. Because the ELISA test required a much larger sample volume (>200 μL for each measurement, in duplicate per analyte) than PEdELISA, it was practically difficult for us to manage the acquisition of a sufficiently large blood sample volume from critically ill COVID-19 patients. Therefore, we could only validate our assay against IL-6 detection results (Fig. 4C). Some discrepancy was observed at concentrations below 50 pg/mL (Fig. 4C inset), where PEdELISA yielded a slightly lower value than ELISA. We characterized the LOD of the ELISA test to be 12.03 pg/mL (Fig. S7), which is more than 30 times higher than that of PEdELISA (0.38 pg/mL). The discrepancy may derive from the poorer sensitivity of the ELISA assay. In addition, the limited sensitivity required the ELISA assay to be performed without dilution while 2x dilution was applied for the PEdELISA measurement. Background components in serum, such as carbohydrates, proteins, and phospholipids, can interfere with the ability of the antibody pairs to bind to their target, which is known as the “matrix effect”. The direct serum sample measurement with the ELISA assay potentially amplified the matrix effect at the lower analyte concentrations, which could be another source of the discrepancy. Nonetheless, the data between these two methods overall matched linearly (R2=0.95, P<0.0001). Especially, good agreement was obtained between them at the higher values of IL-6 for patients who received tocilizumab treatment.
Figure 4.
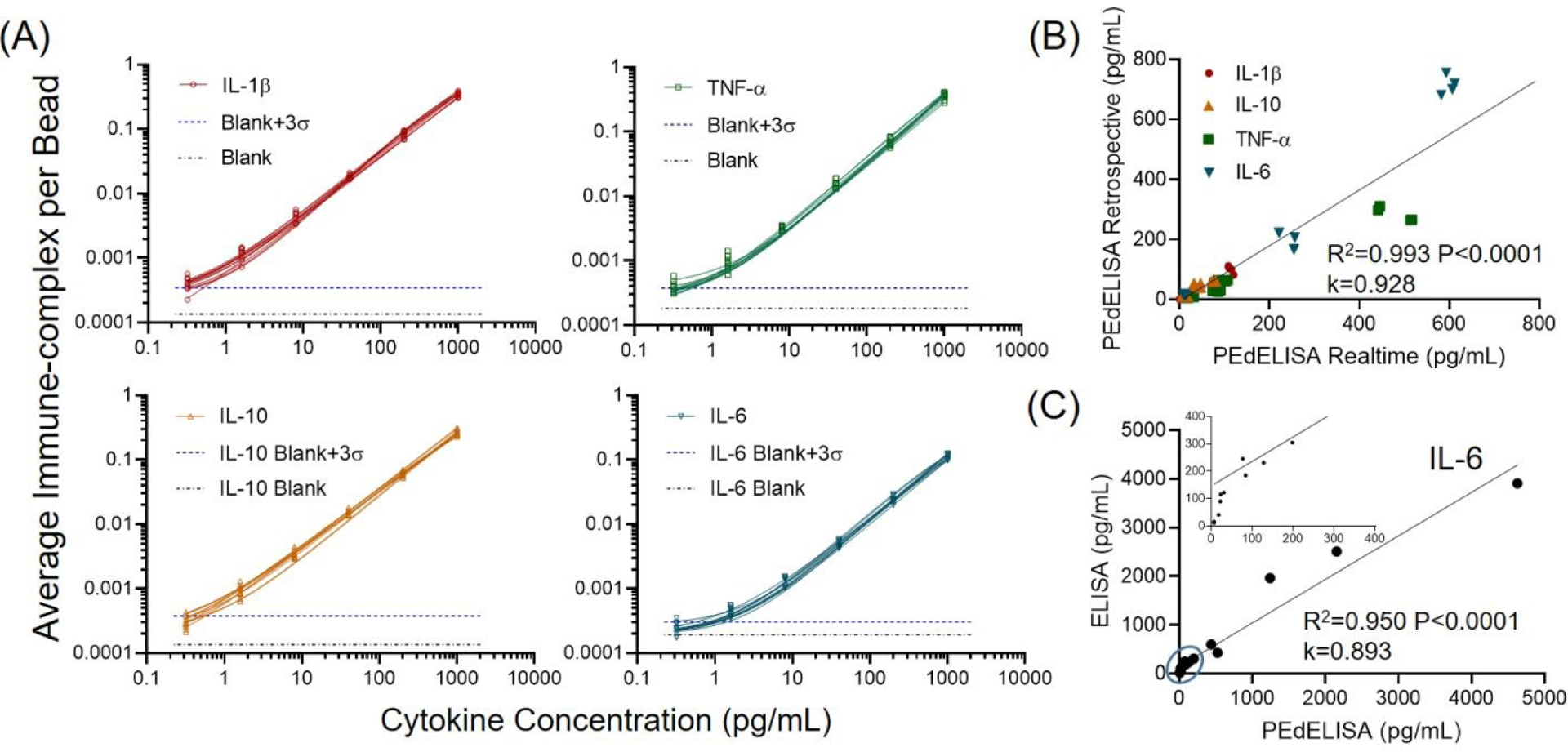
(A) Daily COVID-19 patient assay standard curves for four cytokines from 0.32 pg/mL to 1000 pg/mL in FBS (10 curves for each cytokine obtained over 10 workdays). The data points were fitted with four-parameter logistic (4PL) curves. The black dotted line represents the signal level from a blank solution. The blue dotted line shows 3σ above the blank signal, which is used to estimate the limit of detection (LOD) for each cytokine. (B) Linear correlation (R2=0.99, P<0.0001) between rapid measurements of fresh samples and retrospective measurements of samples stored for more than 30 days (1 freeze-and-thaw at −80 °C) in quadruplicate for 5 representative COVID-19 patients. (C) Good agreement was observed between single-plex IL-6 ELISA (R2=0.95, P<0.0001) and multiplex PEdELISA measurements for 15 COVID-19 patients. The inset shows the circled region and k is the slope of the linear regression for (B)-(C).
Fig. 5A shows a typical timeline of our daily cytokine profile measurement completed within 4 hours after the blood draws in the ICU. The assay itself was performed with a sample-to-answer time as short as 30 min for typical non-COVID-19 serum measurements. However, in the practical operation of our test starting from a patient blood draw and ending with data delivery to physicians, a larger amount of time was spent on sample processing, transport, and team coordination, as well as biosafety and disinfection protocol observation in handling COVID-19 samples. This resulted in a relatively long turnaround time for the entire clinically deployed test than the assay time itself. Further automation and miniaturization of the PEdELISA microarray platform permitting its near-patient operation could reduce the total test time to < 1 hour. The test allowed clinically meaningful time-course measurements for critically ill COVID-19 patients with respiratory failure during their stay at the ICU. Here, patients undergoing treatment with an expanded/emergency use of an immunomodulating selective cytophoretic device (SCD) were screened (first data point) and monitored with PEdELISA to provide quick feedback on the progress of therapy. The SCD therapy was applied to temper the cytokine storm by continuously processing circulating neutrophils and monocytes to gain a less proinflammatory phenotype using an extracorporeal membrane cartridge integrated into a renal replacement blood circuit33. Patients were selected if they were receiving extracorporeal membrane oxygenation and continuous renal replacement therapy and failing to clinically improve, and had evidence of active systemic inflammation with initial IL-6 >100 pg/mL. Treatment details and the response of the first two patients have been previously reported34. Here we present serial measurements from two additional patients for two-week continuous monitoring. Both patients experienced severe cytokine storm and acute respiratory distress syndrome (ARDS) on the screening time point, and demonstrated clinically improved treatment outcomes over the course of therapy. We observed a decreasing trend in both IL-6 and IL-6/IL-10 ratio (Fig. 5B, C) The IL-6/IL-10 ratio could be a positive prognostic indicator for their recovery from pneumonia and has been used to identify the “inflammatory pattern” by some studies32. With the successful implementation of this therapy monitored by PEdELISA, both of the patients came off from extracorporeal membrane oxygenation (ECMO) and mechanical ventilator support and were eventually discharged alive from the hospital.
Figure 5.
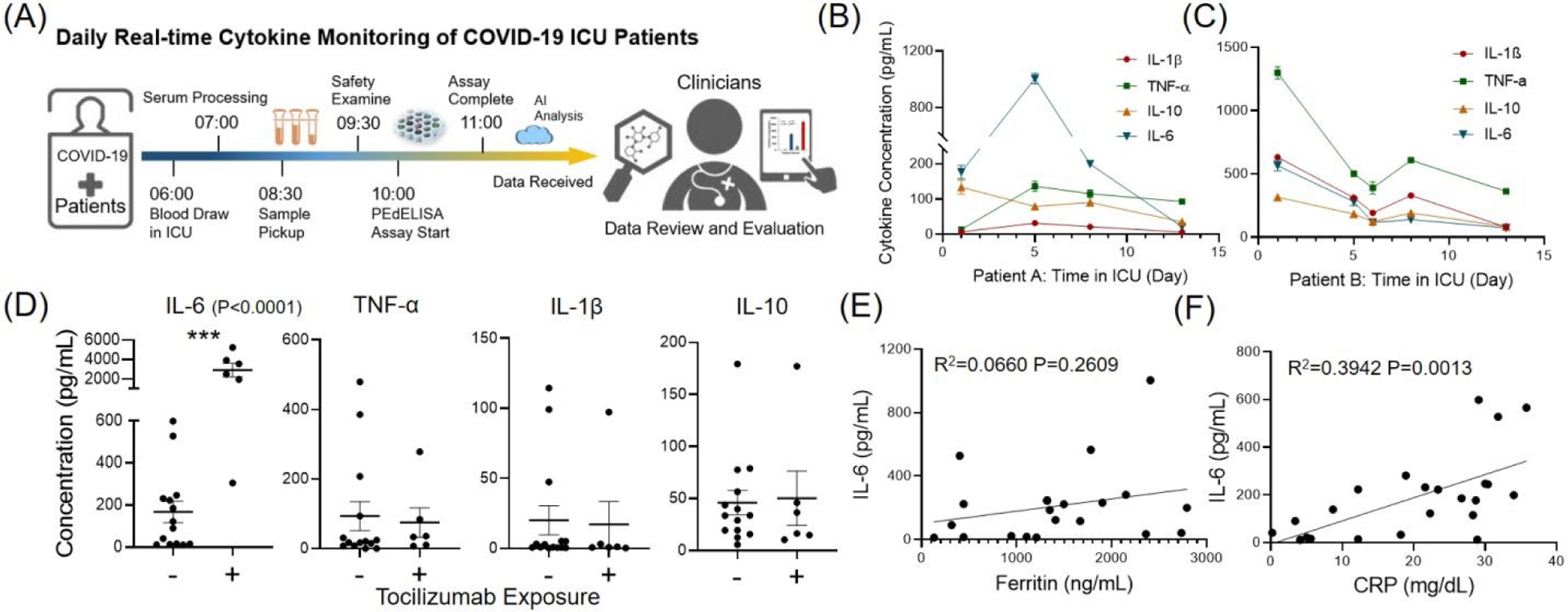
(A) Timeline of daily COVID-19 cytokine measurement. (B)-(C) Two-week serial monitoring of two critically ill COVID-19 patients with respiratory failure in the ICU. Both patients experienced severe cytokine storm and were under the emergency use of a selective cytophoretic device (SCD) by cytokine pre-screening (first data point). The patients demonstrated decreases in IL-6 and IL-6/IL-10 ratio and clinically improved treatment outcome over the course of therapy. (D) Statistical group analysis of patients that are dosed/undosed with Tocilizumab. Significant elevations of IL-6 levels were observed after the treatment of Tocilizumab (P<0.0001). (E)-(F) Correlation of cytokine IL-6 to Ferritin and C-Reactive Protein (CRP), standard clinical inflammatory biomarkers. Ferritin does not correlate well with IL-6 (R2 = 0.066, P=0.261). CRP correlates with IL-6 (R2=0.394, P=0.0013) better, but the IL-6 levels were widely distributed for patients with high levels of CRP.
Our rapid PEdELISA microarray cytokine measurement was also applied for a group of COVID-19 patents who were treated or untreated with tocilizumab. The test result revealed significant subject-to-subject heterogeneity despite all of these patients being critically ill. As expected, interruption of IL-6/IL-6R signaling in patients who received tocilizumab resulted in marked elevation of IL-6 levels in the setting of ongoing illness (p<0.0001, Fig. 5D)22, 35. Among patients who did not receive tocilizumab, we observed a large degree of variability in IL-6 levels, with a quarter of subjects having IL-6 <15 pg/mL, the median value of 106 pg/m, and the CV of 114%. Variability of TNF-α (CV 164%) and IL-1β (CV 193%) was driven by a small number of subjects with elevated levels. However, like IL-6, levels of IL-10 were also broadly distributed in patients who had not received tocilizumab (CV 93%).
Given the heterogeneity of cytokine levels in critically ill patients with COVID-19, we asked whether IL-6 levels were reflected in surrogate biomarkers. Direct measurement of cytokine levels has not been historically achievable in clinical practice. As a result, the presence of cytokine storm and risk of clinical deterioration is frequently judged by inflammatory markers, such as CRP and ferritin. Here, our data shows Ferritin did not predict IL-6 levels (Fig. 5E, R2 = 0.066, P=0.261). CRP was significantly associated with IL-6 (Fig. 5F, R2=0.394, P=0.0013). However, this association was driven by low IL-6 in subjects with low levels of CRP, while IL-6 values in subjects with high CRP were widely distributed. Therefore, while a normal CRP may identify low levels of IL-6, neither CRP nor ferritin is an accurate predictor of specific elevated IL-6 values. If a biomarker-driven strategy required only the identification of abnormal IL-6 levels, a CRP measurement would likely provide an adequate prediction. If differentiation among moderate and severe IL-6 elevations is needed, however, these data suggest that direct measurement of IL-6 may be required. For COVID-19 ARDS, there is a growing need to directly measure specific patient endotypes in order to effectively deploy the growing number of drugs targeting specific inflammatory pathways5. Understanding heterogeneity and patient-specific factors in critical illness has been identified as a priority by the National Institutes of Health36. Inflammatory markers such as CRP and ferritin, as we found here, may lie downstream of multiple inflammatory pathways, thus they are poor markers of specific cytokine activity.
Note that our pragmatic study of rapid cytokine measurements in patients with COVID-19 was designed to provide information to clinicians, rather than systematically study the biology of COVID-19. We, therefore, enrolled subjects without regard to time from the onset of infection. Furthermore, due to these subjects’ critical illness, many received empiric antibiotic therapy, limiting our ability to determine bacterial co-infection. These factors, as well as our small sample size, may have contributed to the heterogeneity of the cytokine response. Nevertheless, all subjects in this study were critically ill and had respiratory failure, underscoring the diversity of biological mechanisms that may lead to critical illness in COVID-19 and the importance of measuring, rather than inferring, cytokine storm.
Despite the likely benefits of rapid, clinically actionable cytokine measurement to the management of patients both with COVID19 and other forms of ARDS5, significant hurdles remain to implementation. In this study, we obtained informed consent for the potential clinical use of assay results. As of September 24, 2020, the US Food and Drug Administration had granted emergency use authorization (EUA) only to the Eclesys IL-6 assay (Roche) to be performed in clinical laboratories certified for moderate and high complexity testing37. While the manufacturer has reported unpublished data for the use of this assay in COVID-1938, we were unable to identify published reports of the use of this assay platform to provide rapid, clinically actionable cytokine levels. Rapid, clinically actionable use of the Ella assay platform (Biotechne) has been recently reported39 under EUA granted by the state of New York. This commercially available platform, as well as others, could potentially be validated as “laboratory-developed tests” for use in certified clinical laboratories even beyond emergency use authorization. However, significant barriers, including instrumentation and consumable supply costs, may limit their use in resource-constrained hospitals or in the setting of multisite clinical trials (Table 2).
Table 2.
Benchmarking of the PEdELISA microarray platform to commercial immunoassay platforms in cytokine detection.
| Diagnostic System | Cost/96 Assays | Instrument Cost | *Assay Time | LOD (pg/mL) | Sample Volume (μL) | Plexity | Ref |
|---|---|---|---|---|---|---|---|
| Colorimetric ELISA | $100–500 | $5,000 | > 4 hrs | 1–10 | 100 | 1 | (rndsystems.com) |
| Luminex | $1000–8000 | >$50,000 | > 4 hrs | 0.1–100 | 25–50 | 2–65 | (thermofisher.com) |
| Quanterix SIMOA | >$1500 | >$ 100,000 | ~2.5 hrs/96 plate | 0.002–1 | 100 | 1–6 | (quanterix.com) |
| Bio-Techne Ella | $2000–4500 | >$60,000 | ~1.5 hrs/16 samples | 0.2–2 | 25 | 1–8 | (proteinsimple.com/ella.html) |
| PEdELISA microarray | <$50 | <$5,000 | ~30 min/8 samples | 0.2–3 | 15 | 1–8 |
Assay Time refers to the sample-to-answer time, i.e., the total time between sample loading and delivery of analyzed data. Note that the assay and instrument cost of the commercial systems were based on their market prices and may not reflect the production cost. All the information was estimated in a range to the best of our knowledge based on the platforms’ website, online manual/brochure, quote, and do not count for marketing strategies, e.g. discount.
Conclusions
Timely intervention of cytokine storm guided by rapid cytokine measurement is critical for the management of severe COVID-19 infections resulting in respiratory failure. To this end, we have developed a microfluidic digital immunoassay platform that enables rapid 4-plex measurement of cytokines in COVID-19 patient serum. Our assay employed single-molecule counting for an antibody sandwich immune-complex formation quenched at an early pre-equilibrium state. The pre-equilibrium approach resulted in a detection limit < 0.4 pg/mL and a linear dynamic range of 103 while requiring an assay incubation time as short as 9 min. The platform incorporates a programmed fluidic dispensing and mixing module and a compact optical reader module for microfluidic analysis using low-cost disposable chips manufacturable at a large scale. Each chip contains spatially encoded microwell array patterns with capture antibody-coated magnetic beads pre-deposited for multiplex cytokine detection. The 4-plex on-chip measurement with a 15 μL sample volume showed negligible sensor cross-talk. The programmed fluidic handling and mixing module permitted high inter-assay repeatability (~10% CV). Our assay platform with the combination of high sensitivity, speed, and fidelity allowed us to characterize serum cytokines with a sample-to-answer time of 30 min. Using the platform, we were able to complete the entire cytokine storm monitoring test within 4 hours that involved blood draw, serum extraction, lab-to-lab sample transport, serum sample loading, biosensor signal measurements, serum cytokine data analysis, and feedback on the therapy outcome.
Multiple commercially available technologies are capable of multiplex cytokine measurement, and have been applied to cohorts of patients with COVID-19 disease39–41. Elevations in pro-inflammatory cytokines correlate with poor outcome39. These studies have also demonstrated significant heterogeneity in cytokine expression, underscoring the importance of timely measurement for prognostication, and eventually stratification in clinical trials. Anti-IL6 therapy in COVID-19 is under investigation in multiple clinical trials42–44 and has been employed in clinical practice as an off-label use. Results from trials of the anti-IL-6 receptor tocilizumab have not demonstrated efficacy in preventing intubation or death in moderately ill hospitalized patients with COVID-1942, 43. However, in a retrospective study of critically ill patients with COVID-19, early treatment with tocilizumab was associated with reduced mortality44. These results confirm that rapid, reliable, and repeatable direct cytokine measurement is needed to facilitate precision administration of anti-cytokine therapies only in patients who are experiencing severe cytokine storm. In agreement with other studies, our results here highlight the heterogeneity of cytokine response even among critically ill COVID-19 patients and the poor ability of surrogate inflammatory markers to predict an IL-6 response.
Our digital immunoassay platform may provide a promising means to enable such a precision medicine strategy in the pharmacotherapeutic management of life-threatening cytokine storm in COVID-19. The results presented here suggest that the basic characteristics of this assay platform allow it to deliver robust measurements with low sample volume, high sensitivity, low cost, and rapid answer times. Further miniaturization, automation, and casing of the platform may allow us to further decrease the need for operator supervision and device footprint, thus eventually enabling us to achieve near-bedside, near-real-time cytokine storm monitoring with the PEdELISA microarray technology.
Materials and Methods
Materials
We purchased human IL-6, TNF-α capture, and biotinylated detection antibody pairs from Invitrogen™, and IL-1β, IL-10 from BioLegend. We purchased the corresponding ELISA kits from R&D Systems (DuoSet®). We obtained Dynabeads, 2.7μm-diameter epoxy-linked superparamagnetic beads, avidin-HRP, QuantaRed™ enhanced chemifluorescent HRP substrate, bovine serum albumin (BSA), TBS StartingBlock T20 blocking buffer, and PBS SuperBlock blocking buffer from Thermo Fisher Scientific. We obtained Phosphate buffered saline (PBS) from Gibco™, Sylgard™ 184 clear polydimethylsiloxane (PDMS) from Dow Corning, and Fluorocarbon oil (Novec™ 7500) from 3M™. The automated PEdELISA system was mainly constructed by a micro-controller (Arduino Uno and MEGA 2560), stepper motors and shields (NEMA 17, 0.9 degree, 46 N·cm, TB6600 and DM542T motor shields), linear rail guide with ballscrews (5 mm/revolution), standard anodized aluminum profiles, clear acrylic boards and other supporting wheels, connectors and parts purchased from Amazon through various vendors. The optical scanning system mainly consists of a consumer-grade CMOS camera (SONY α6100), 10x Objective lens (Nikon, CFI Plan Achro), tube lens (200 mm), optical filter sets (Chroma), halogen light source, LED light source (560 nm), optical mountings and tubings (mainly from Thorlabs and Edmund Optics).
Antibody conjugation to magnetic beads
We conjugated human IL-6, TNF-α, IL-1β, IL-10 capture antibodies using the epoxy-linked Dynabeads (2.8 μm) with the capture antibody molecules at a mass ratio of 6 μg (antibody): 1 mg (bead) following the protocols provided by Invitrogen™ (Catalog number: 14311D). Based on the product datasheet, the estimated antibody coupling efficiency is 4–4.5 μg/mg (with each bead containing 2.4–2.7×105 antibodies approximately). The beads were then quenched (for unreacted epoxy groups) and blocked with TBS StartingBlock T20 blocking buffer. We stored the antibody-conjugated magnetic beads at 10 mg beads/mL in PBS (0.05% T20 + 0.1% BSA + 0.01% Sodium Azide) buffer sealed with Parafilm at 4 °C. No significant degradation of the beads was observed within the 3-month usage.
PEdELISA chip fabrication and patterning
The plastic-based disposable microfluidic chip used for PEdELISA assay was fabricated by laser cutting and PDMS molding. It has a transparent sandwich structure for optical imaging as shown in Fig. S1. The top and bottom layers were laser-cut using 3.175 mm (1/8 inch) and 1 mm thin clear polymethyl methacrylate (PMMA) boards which contain through-holes for venting and screw assembly purposes. The microfluidic channels (designed with AutoCAD software) were laser-cut through a 120 μm high definition transparency polyethylene terephthalate (PET) thin film (adopted from standard screen protector) which has a silicone gel layer to create a vacuum for securely sealing to the top acrylic layer without adhesives. The power and speed of the laser cutter were optimized to ensure a high-resolution smooth cut so that resistance difference or bubbles generation can be minimized during the fluidic handling process. The femtoliter-sized microwell array (3.4 μm diameter and 8 μm in pitch) layer (~300 μm) was made by polydimethylsiloxane (PDMS) through a standard SU-8 molding. First, we constructed SU-8 molds on oxygen plasma treated silicon wafers by standard photolithography which involved depositing negative photoresist (SU-8 2005 MicroChem) layers at 5000 rpm to form the desired thicknesses 3.8±0.1 μm. Subsequently, a precursor of PDMS was prepared at a 10:1 base-to-curing-agent ratio and deposited onto the SU-8 mold by spin coating (300 rpm) and baking overnight at 60 °C. We then transferred the fully cured PDMS thin film onto the bottom acrylic layer using a modified surface silanization bonding method based on a previous publication38. We also drilled 2 mm countersunk holes (60°) using a benchtop mini drill press (MicroLux®) on the top venting layer for guiding the multi-pin fluidic dispensing connector. Each layer was thoroughly cleaned through water bath sonication and the PET microchannel layer was carefully attached to the top venting layer for the later bead patterning process.
The PEdELISA bead patterning process (Fig. S2) first involved attaching the bead settling layer (containing long straight PDMS channels perpendicular to the PET microchannel layer) to the PDMS microwell array layer on the bottom PMMA substrate. Then, we prepared 4 sets of a 25 μL bead solution at the concentration of 1 mg/mL for IL-1β, TNF-α, IL-10, and IL-6 bead respectively. The bead solution was loaded into four different physically separated patterning channels in the bead settling layer. After waiting for beads to settle inside the microwells for 5 min, we washed the patterning channels with 200 uL PBS-T (0.1% Tween20) to remove the unstrapped beads. At this step, we imaged the microarray under the microscope to ensure that the microwells were filled with the beads at a sufficient rate (typically above 50%). If not, the bead mixture solution was reloaded and washed again. Finally, the bead settling layer was peeled off and replaced with the PET microchannel and top venting layer. Four layers of the cartridge were sandwiched together using M2 bolt screws. Note that the bonding between the PET layer and the PDMS layer was not permanent but achieved through pressure-based self-sealing, which can be later easily peel off and replaced. We then slowly primed each sample detection channel with Superblock buffer to passivate the cartridge surface and incubated the whole chip for at least 1 hour prior to the assay to avoid non-specific protein adsorption. The cartridge was typically prepared in batch and sealed in a moisture-controlled petri-dish at room temperature for up to a week with no significant degradation.
Programmed PEdELISA assay and imaging
The automated pipetting system was programmed to first draw 15 μL of the sample solution (patient serum and assay standard) and mix with 15 μL of detection antibody (DeAb) solution in the 96-well tube rack for 20 cycles (25-sec), and then draw 28 μL of the mixed solution and load them into the PEdELISA chip by two steps: Step 1. Load 14 μL of the sample-DeAb mix for channel buffer exchange, delay 10-sec for wiping away the original buffer solution inside the channel (1x PBS solution), Step 2. Load the rest of 14 μL, followed by 50-cycles of on-chip mixing (8-min). The mixing was performed by repeating a 10-sec cycle of pulling, holding, and pushing the plunger of a multi-channel pipette with a precisely programmed stepper motor. Then, the system loaded 200 μL of washing buffer (PBS-T 0.1% Tween20) into the channels of the chip with the pipette and slowly replaced the sample fluids with the PBS-T buffer solution for washing/quenching (2-min). Next, the system drew in 40 μL of the avidin-HRP solution (100 pM) and slowly loaded (micro-stepping) into the chip for enzyme labeling (1-min). The chip was washed again with the PBS-T solution for one cycle (200 μL) and 1x PBS solution for another cycle (200 μL), total to reduce the interference between Tween20 and the chemifluorescent HRP substrate later (total 5-min). Finally, the system drew and loaded 30 μL of the QuantaRed (Qred) substrate solution and then sealed with 35 μL of fluorinated oil (HFE-7500, 3M) for the digital counting process.
The scanning system was used to scan the image of the bead-filled microwell arrays on the PEdELISA chip right after the oil sealing step to detect the enzyme-substrate reaction activity. The imaging stage was pre-programmed to follow the designated path to scan the entire chip (64 microarrays) twice: 1. Scan the Qred channel (545nm/605nm, excitation/emission) 2. Scan the brightfield with the transmission light source on. It typically took ~ 5 min to scan the entire chip for 16 samples in 4-plex detection.
Data Analysis by the convolutional neural network
The collected images were analyzed by an in-house developed convolution neural network (CNN) algorithm. The CNN ran two signal recognition pathways in parallel, which were pre-trained to recognize enzyme active “On” microwells (Qred channel) versus defects and contaminations using >5,000 labeled images (see Supplementary Information for the training code). Briefly, the CNN method started from a pre-processing process, which included image cropping, contrast enhancement, and noise filtering. Then, the CNN classified each image pixel into the three pre-trained categories: Category 1: Enzyme Active Fluorescence Microwell (or “On” Microwell), Category 2: Image Defect, and Category 3: Background. The “On” microwells (Qred+) were segmented out as the output mask with defects being removed. The bright-field image was analyzed using the Sobel edge detection method to determine the overall beads filling rate. Finally, the fractional population of the Qred+ microwells with respect to the total bead-filled microwells, Pon, was calculated, and the Poisson’s distribution equation was used to calculate the mean expectation value: λ = −ln(1 − Pon), which represents the average number of analyte-antibody immune complexes per bead and the digital assay’s raw signal.
The architecture of the CNN contains a downsampling process for category classification and an upsampling process for image segmentation. The downsampling process consists of 5 layers, including two convolution 2D layers (Conv2D, in blue color, 6 filters, kernel of 3×3), two rectified linear unit layers (ReLU, in gray color), and one max-pooling layer (in red color, stride of 2). The upsampling process also consists of 5 layers, including one transposed convolution 2D layer (Trans Conv2D, in yellow color), one ReLu layer, one Conv2D layer, one softmax layer (in green color), one pixelClassificationLayer (in orange color, contains class weight balance).
Pragmatic study of rapid cytokine measurement in COVID-19
This study was approved by the University of Michigan Institutional Review Board (HUM00179668) and patients or their surrogates provided informed consent for the investigational use of this test. Patients with positive SARS-CoV-2 test via PCR and respiratory failure requiring hospitalization in the intensive care unit for heated high flow oxygen or mechanical ventilation were eligible for enrollment. Subjects were approached at the request of treating teams at any point in their disease course after intensive care unit admission. Due to restrictions on patient contact during the COVID-19 pandemic, samples were drawn by the subjects’ nurse with routine clinical labs in a serum separator tube and sent to the clinical specimen processing area, where they were centrifuged, aliquoted, and kept at 4°C until their transfer to the engineering lab at 9 am. Serum was then immediately analyzed to measure IL-6, TNF-α, IL-1β, and IL-10, and the results were posted to the patient’s chart on the same day. Ferritin and C-reactive protein (CRP) measurements made during routine clinical care were recorded if available from a sample within 24 hours of the cytokine measurement.
Live subject statement
All experiments were performed in compliance with the Office of Human Resarch Protections (OHRP) Guidance on COVID-19 of the U.S Department of Health & Human Services (DHH), U.S Food and Drug Administration (FDA) Guidance on Conduct of Clinical Trials of Medical Products During COVID-19 Public Health Emergency, and the guideliens of the University of Michigan’s Human Research Protection Program (HRPP). All experiments were approved by the University of Michgian Institutional Review Board (IRB), which aims to protect the rights and welfare of human research subjects recruited to participate in research activities condctued under the auspices of the University of Michigan. Informed consent was obtained for any experimentation with human subjects.
Statistics
Experiments with synthetic recombinant proteins were performed daily with 2 on-chip repeats. The duplicate measurement results were averaged to calculate the patient serum cytokine levels. Standard curves were obtained daily accumulated for 10 workdays, and the standard curve data was used to determine the inter-assay CV (CVinter-assay) for each analyte is listed in Table 1. The determination of CVinter-assay for a given analyte involved duplicate measurement of 100% fetal bovine serum (FBS) spiked with the target analyte (recombinant protein) at three (N = 3) representative concentrations of 40, 200, and 1000 pg/mL that fall into the linear dynamic range of our assay over 10 workdays. The CV value for a given analyte concentration was calculated as CV = STD/Sm, where STD is the standard deviation of the assay signal measured for spike-in FBS at the given concentration over 10 workdays, and Sm is the mean assay signal measured for spike-in FBS at the given concentration over 10 workdays. CVinter-assay was determined by taking the root mean square of the CV values for the three (N = 3) representative concentrations as .
In the daily cytokine profile monitoring test, we performed the measurement for each COVID-19 patient serum sample in quadruplicate and took an average value over the quadruplicate results for each data point. To evaluate the intra-assay CV (CVintra-assay) (see Table 1), we selected five (N = 5) COVID-19 patient serum samples with cytokines ranging from 6–600 pg/mL and measured the concentration in quadruplicate for the target analyte in each sample using the same chip on the same workday. The CV value for that target analyte in each sample was calculated as CV = STD/Cm, where STD is the standard deviation of the analyte concentration value measured for the given sample over 4 measurement repeats, and Cm is the mean analyte concentration value measured for the given sample over 4 measurement repeats. Similar to the procedure above, CVintra-assay was determined using the CV values for the five (N = 5) patient serum samples as .
A conventional ELISA test was conducted retrospectively for IL-6 in duplicate for selected banked patient samples. Here, Pearson’s R-value was used to quantify the PEdELISA to ELISA correlations and the t-test was used for the group analysis of IL-6 and TNF-α (normally distributed) between tocilizumab-treated and non-tocilizumab-treated subpopulations, and Mann-Whitney U test was used for IL-1β and IL-10 (not normally distributed). A p-value of < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgements
We thank the College of Engineering, Department of Internal Medicine at the University of Michigan (U-M) for their emergency approval of this COVID-19 related research. We specifically thank the UM Mechanical Engineering machine shop, Mr. Charles Bradley, Donald Wirkner, and Kent Pruss for the great help offered in designing and machining the automated system during the COVID-19 pandemic period. We also thank the personal protection equipment and the cleanroom fabrication support from the Lurie Nanofabrication Facility (LNF) at U-M. We acknowledge the funding support from the National Institute of Health K08NS101054 (B.H.S), the National Science Foundation CBET1931905, and CBET2030551 (K.K.), the University of Michigan COVID-19 Response Innovation Grant (B.H.S and K.K), and the University of Michigan Precision Health Scholars Grant (Y.S.).
Footnotes
Conflicts of interest
D.H. discloses the financial interest in SeaStar Medical, Inc. and Innovative Biotherapies, Inc, which has an exclusive license from the University of Michigan to develop the Selective Cytopheretic Device technology.
References
- 1.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J and Grp CS, Nat Microbiol, 2020, 5, 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J and Cao B, Lancet, 2020, 395, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ and Speciality HA, Lancet, 2020, 395, 1033–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore JB and June CH, Science, 2020, 368, 473–474. [DOI] [PubMed] [Google Scholar]
- 5.Sinha P, Matthay MA and Calfee CS, JAMA Internal Medicine, 2020, DOI: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 6.Neelapu SS, Tummala S, Kebriaei P, Wierda W, Gutierrez C, Locke FL, Komanduri KV, Lin Y, Jain N, Daver N, Westin J, Gulbis AM, Loghin ME, de Groot JF, Adkins S, Davis SE, Rezvani K, Hwu P and Shpall EJ, Nat Rev Clin Oncol Nat Rev Clin Oncol, 2018, 15, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin G, Shenoi S and Hughes GC, Best Pract Res Clin Rheumatol, 2020, DOI: 10.1016/j.berh.2020.101515,101515. [DOI] [PubMed] [Google Scholar]
- 8.Alcamo AM, Pang D, Bashir DA, Carcillo JA, Nguyen TC and Aneja RK, J Pediatr Intensive Care, 2019, 8, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotch C, Barrett D and Teachey DT, Expert Rev Clin Immunol, 2019, 15, 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen F, Teachey DT, Pequignot E, Frey N, Porter D, Maude SL, Grupp SA, June CH, Melenhorst JJ and Lacey SF, J Immunol Methods, 2016, 434, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehta P, Cron RQ, Hartwell J, Manson JJ and Tattersall RS, Lancet Rheumatol, 2020, DOI: 10.1016/S2665-9913(20)30096-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, Damoraki G, Gkavogianni T, Adami ME, Katsaounou P, Ntaganou M, Kyriakopoulou M, Dimopoulos G, Koutsodimitropoulos I, Velissaris D, Koufargyris P, Karageorgos A, Katrini K, Lekakis V, Lupse M, Kotsaki A, Renieris G, Theodoulou D, Panou V, Koukaki E, Koulouris N, Gogos C and Koutsoukou A, Cell Host Microbe, 2020, DOI: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. https://clinicaltrials.gov/ct2/results?cond=COVID-19&term=tocilizumab&cntry=&state=&city=&dist=&Search=Search).
- 14.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR and Duffy DC, Nat Biotechnol, 2010, 28, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y and Noji H, Anal Chem, 2017, 89, 13675–13675. [DOI] [PubMed] [Google Scholar]
- 16.Wilson DH, Rissin DM, Kan CW, Fournier DR, Piech T, Campbell TG, Meyer RE, Fishburn MW, Cabrera C, Patel PP, Frew E, Chen Y, Chang L, Ferrell EP, von Einem V, McGuigan W, Reinhardt M, Sayer H, Vielsack C and Duffy DC, Jala-J Lab Autom, 2016, 21, 533–547. [DOI] [PubMed] [Google Scholar]
- 17.Akama K, Iwanaga N, Yamawaki K, Okuda M, Jain K, Ueno H, Soga N, Minagawa Y and Noji H, Acs Nano, 2019, 13, 13116–13126. [DOI] [PubMed] [Google Scholar]
- 18.Sun J, Hu J, Gou T, Ding X, Song Q, Wu W, Wang G, Yin J and Mu Y, Biosens Bioelectron, 2019, 139, 111339. [DOI] [PubMed] [Google Scholar]
- 19.Piraino F, Volpetti F, Watson C and Maerkl SJ, Acs Nano, 2016, 10, 1699–1710. [DOI] [PubMed] [Google Scholar]
- 20.Araci IE, Robles M and Quake SR, Lab Chip, 2016, 16, 1573–1578. [DOI] [PubMed] [Google Scholar]
- 21.Yelleswarapu V, Buser JR, Haber M, Baron J, Inapuri E and Issadore D, Proc Natl Acad Sci U S A, 2019, DOI: 10.1073/pnas.1814110116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoiland RL, Stukas S, Cooper J, Thiara S, Chen LYC, Biggs CM, Hay K, Lee AYY, Shojania K, Abdulla A, Wellington CL and Sekhon MS, British Journal of Haematology, 2020, 190, e150–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, Breillat P, Carlier N, Gauzit R, Morbieu C, Pène F, Marin N, Roche N, Szwebel T-A, Merkling SH, Treluyer J-M, Veyer D, Mouthon L, Blanc C, Tharaux P-L, Rozenberg F, Fischer A, Duffy D, Rieux-Laucat F, Kernéis S and Terrier B, Science, 2020, 369, 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan R, Vermesh O, Srivastava A, Yen BKH, Qin LD, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L and Heath JR, Nat Biotechnol, 2008, 26, 1373–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan XT, Oo MKK, Gong Y, Li YX, Zhu HB and Fan XD, Analyst, 2017, 142, 2378–2385. [DOI] [PubMed] [Google Scholar]
- 26.McRae MP, Simmons G, Wong J and McDevitt JT, Accounts Chem Res Accounts Chem Res, 2016, 49, 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min J, Nothing M, Coble B, Zheng H, Park J, Im H, Weber GF, Castro CM, Swirski FK, Weissleder R and Lee H, Acs Nano, 2018, 12, 3378–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen PY, Huang NT, Chung MT, Cornell TT and Kurabayashi K, Adv Drug Deliver Rev, 2015, 95, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song YJ, Chen PY, Chung MT, Nidetz R, Park Y, Liu ZH, McHugh W, Cornell TT, Fu JP and Kurabayashi K, Nano Lett, 2017, 17, 2374–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park Y, Ryu B, Oh BR, Song YJ, Liang XG and Kurabayashi K, Acs Nano, 2017, 11, 5697–5705. [DOI] [PubMed] [Google Scholar]
- 31.Park Y, Ryu B, Deng QF, Pan BH, Song YJ, Tian YZ, Alam HB, Li YQ, Liang XG and Kurabayashi K, Small, 2020, 16. [DOI] [PubMed] [Google Scholar]
- 32.Lucena-Silva N, Torres LC, Luna CF, de Barros Correia J and da Silva GAP, Bmc Pulm Med, 2016, 16, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pino CJ, Westover AJ, Johnston KA, Buffington DA and Humes HD, Kidney Int Rep, 2018, 3, 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yessayan L, Szamosfalvi B, Napolitano L, Singer B, Kurabayashi K, Song Y, Westover A and Humes D, ASAIO Journal, 2020, DOI: 10.1097/MAT.0000000000001239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimoto N, Terao K, Mima T, Nakahara H, Takagi N and Kakehi T, Blood, 2008, 112, 3959–3964. [DOI] [PubMed] [Google Scholar]
- 36.Semler MW, Bernard GR, Aaron SD, Angus DC, Biros MH, Brower RG, Calfee CS, Colantuoni EA, Ferguson ND, Gong MN, Hopkins RO, Hough CL, Iwashyna TJ, Levy BD, Martin TR, Matthay MA, Mizgerd JP, Moss M, Needham DM, Self WH, Seymour CW, Stapleton RD, Thompson BT, Wunderink RG, Aggarwal NR and Reineck LA, Am J Respir Crit Care Med, 2020, DOI: 10.1164/rccm.201908-1595WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.In Vitro Diagnostics EUAs, https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas).
- 38.REGENERON AND SANOFI PROVIDE UPDATE ON U.S. PHASE 2/3 ADAPTIVE-DESIGNED TRIAL OF KEVZARA® (SARILUMAB) IN HOSPITALIZED COVID-19 PATIENTS, https://newsroom.regeneron.com/index.php/news-releases/news-release-details/regeneron-and-sanofi-provide-update-us-phase-23-adaptive).
- 39.Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D, Stock A, Marron TU, Xie H, Patel M, Tuballes K, Van Oekelen O, Rahman A, Kovatch P, Aberg JA, Schadt E, Jagannath S, Mazumdar M, Charney AW, Firpo-Betancourt A, Mendu DR, Jhang J, Reich D, Sigel K, Cordon-Cardo C, Feldmann M, Parekh S, Merad M and Gnjatic S, Nat Med, 2020, DOI: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed]
- 40.Kox M, Waalders NJB, Kooistra EJ, Gerretsen J and Pickkers P, JAMA, 2020, DOI: 10.1001/jama.2020.17052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Li S, Liu J, Liang B, Wang X, Wang H, Li W, Tong Q, Yi J, Zhao L, Xiong L, Guo C, Tian J, Luo J, Yao J, Pang R, Shen H, Peng C, Liu T, Zhang Q, Wu J, Xu L, Lu S, Wang B, Weng Z, Han C, Zhu H, Zhou R, Zhou H, Chen X, Ye P, Zhu B, Wang L, Zhou W, He S, He Y, Jie S, Wei P, Zhang J, Lu Y, Wang W, Zhang L, Li L, Zhou F, Wang J, Dittmer U, Lu M, Hu Y, Yang D and Zheng X, EBioMedicine, 2020, 55, 102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hermine O, Mariette X, Tharaux PL, Resche-Rigon M, Porcher R, Ravaud P and Group C-C, JAMA Intern Med, 2020, DOI: 10.1001/jamainternmed.2020.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stone JH, Frigault MJ, Serling-Boyd NJ, Fernandes AD, Harvey L, Foulkes AS, Horick NK, Healy BC, Shah R, Bensaci AM, Woolley AE, Nikiforow S, Lin N, Sagar M, Schrager H, Huckins DS, Axelrod M, Pincus MD, Fleisher J, Sacks CA, Dougan M, North CM, Halvorsen YD, Thurber TK, Dagher Z, Scherer A, Wallwork RS, Kim AY, Schoenfeld S, Sen P, Neilan TG, Perugino CA, Unizony SH, Collier DS, Matza MA, Yinh JM, Bowman KA, Meyerowitz E, Zafar A, Drobni ZD, Bolster MB, Kohler M, D’Silva KM, Dau J, Lockwood MM, Cubbison C, Weber BN, Mansour MK and Investigators BBTT, N Engl J Med, 2020, DOI: 10.1056/NEJMoa2028836. [DOI] [Google Scholar]
- 44.Gupta S, Wang W, Hayek SS, Chan L, Mathews KS, Melamed ML, Brenner SK, Leonberg-Yoo A, Schenck EJ, Radbel J, Reiser J, Bansal A, Srivastava A, Zhou Y, Finkel D, Green A, Mallappallil M, Faugno AJ, Zhang J, Velez JCQ, Shaefi S, Parikh CR, Charytan DM, Athavale AM, Friedman AN, Redfern RE, Short SAP, Correa S, Pokharel KK, Admon AJ, Donnelly JP, Gershengorn HB, Douin DJ, Semler MW, Hernán MA, Leaf DE and Investigators S-C, JAMA Internal Medicine, 2020, DOI: 10.1001/jamainternmed.2020.6252. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


