Abstract
The human gut microbiome is a collection of bacteria, protozoa, fungi, and viruses that coexist in our bodies and are essential in protective, metabolic, and physiologic functions of human health. Gut dysbiosis has traditionally been linked to increased risk of infection, but imbalances within the intestinal microbial community structure that correlate with untoward inflammatory responses are increasingly recognized as being involved in disease processes that affect many organ systems in the body. Furthermore, it is becoming more apparent that the connection between gut dysbiosis and age-related diseases may lie in how the gut microbiome communicates with both the intestinal mucosa and the systemic immune system, given that these networks have a common interconnection to frailty. We therefore discuss recent advances in our understanding of the important role the microbiome plays in aging and how this knowledge opens the door for potential novel therapeutics aimed at shaping a less dysbiotic microbiome to prevent or treat age-related diseases.
Keywords: Microbiome, Elderly, Age-related Diseases, Frailty, Inflammation
Microbiome Changes Occurring With Aging
Health care systems in the United States are experiencing increased and unsustainable burdens due to their aging populations. Improving elder health is essential, as the proportion of people older than 65 years is increasing in many countries. In fact, at the current rate of increase, it is projected that by 2030 more 1 in 5 Americans will be older than 65.1 Gut microbes occupy the interface between the external environment and the host, and interactions between the gut microbiota and humans occur at each stage of life; largely beginning soon after birth and continuing through old age (Figure 1). This sophisticated intestinal microbial ecosystem plays a pivotal role in an array of physiologic activities that are critical to human development and support health.2 This ecosystem is also finely tuned because when the cooperation between our own cells and the gut microbes falter, the microbial community within the gut can become a source of infection, and at times can lead to life-threating diseases.
Figure 1.
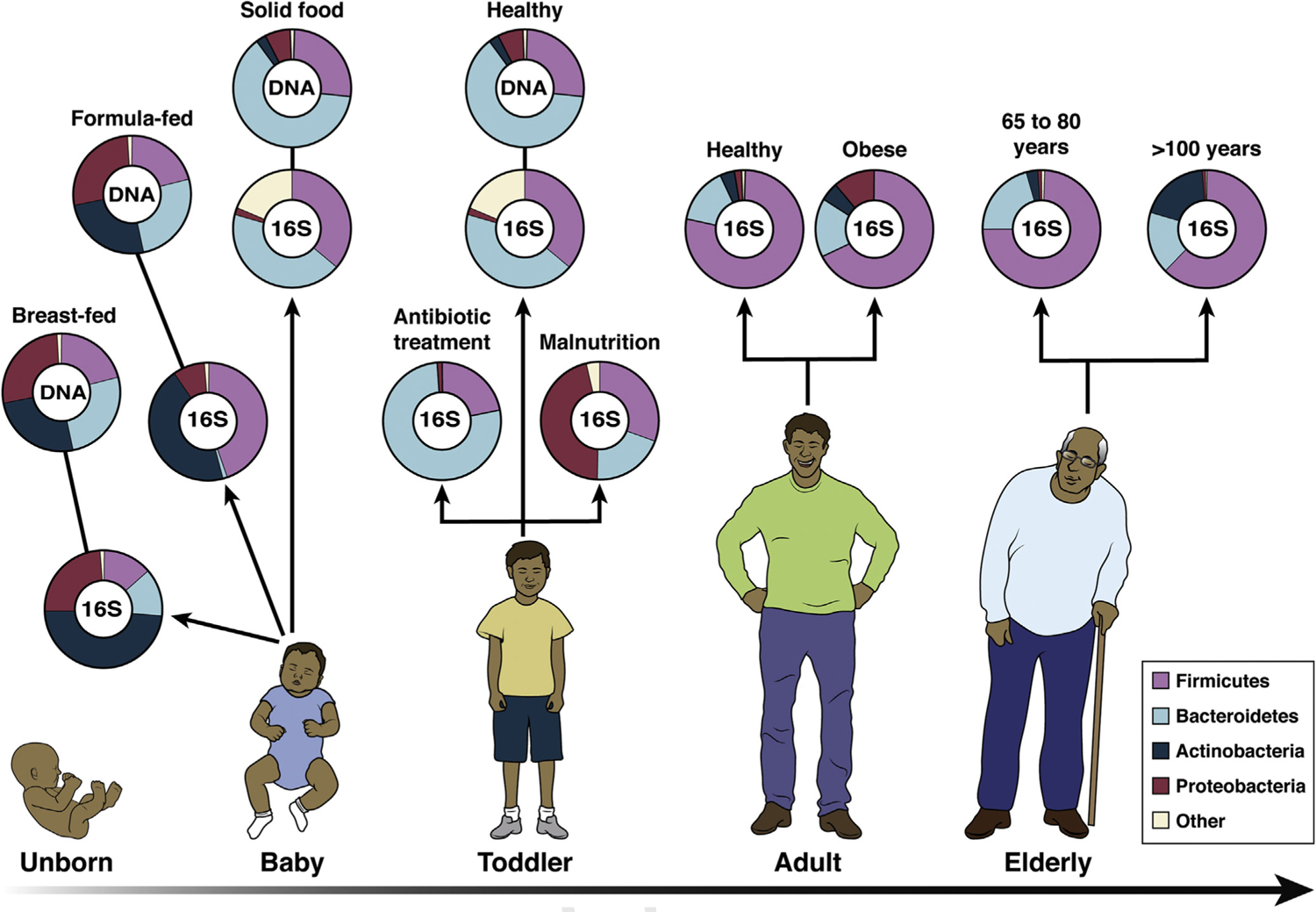
Human microbiota: onset and shaping through life stages. The graph provides a global overview of the relative abundance of key phyla of the human microbiota composition in different stages of life. Measured by either 16S RNA or metagenomic approaches (DNA). Data arriving from infants breast-and formula-fed (Schwartz et al, 2012), infant solid food (Koenig et al., 2011), toddler antibiotic treatment (Koenig et al., 2011), toddler healthy or malnourished (Monira et al., 2011), adult, elderly, and centenarian healthy (Biagi et al., 2010), and adult obese (Zhang et al., 2009).
Healthy individuals have many different types of microbes, whereas individuals with poor health, or older people (elders), will often have a less diverse and a higher proportion of disease-causing microbes. Therefore, as we age, our “aging microbiome” can undergo a number of compositional changes that can adversely affect digestive health and absorption,3,4 as well as immune function.5 Dysbiosis is a term describing a microbial imbalance or maladaptation on or inside the body and can be defined as either the loss or gain of bacteria that promote health or disease.6,7 A healthy non-dysbiotic microbiome works in a symbiotic fashion with its host to facilitate health by imparting critical protective functions (ie, pathogen displacement, nutrient competition, production of antimicrobials), structural functions (ie, barrier fortification, induction of immunoglobulin A, immune system development), and metabolic functions (ie, synthesis of biotin and folate, fermentation of nondigestible dietary products, energy salvation, ion adsorption, and control of intestinal epithelial cell differentiation and proliferation). Conversely, a dysbiotic, or maladaptive, microbiome has been associated with disease not only within the intestine8–10 but also several other organ systems with a few examples including but not limited to the cardiovascular,11,12 immune,13,14 neurological,15,16 and respiratory systems.17,18
Given the potential for the microbiome to influence a variety of dynamic disease processes, there is great interest to determine the composition of the gut microbiota of elders and to also characterize its variation as possible determinants of health.19–23 This is particularly germane to the elderly and aging individuals because increasing age is aligned with age-related morbidities that affect the quality and quantity of life (eg, heart disease, stroke, hypertension, cognitive impairment, and cancer). However, the changes that occur within the intestinal microbiome as we age are not completely understood.
Animal model systems have clearly demonstrated that the presence of certain gut-associated microbes have an influence over cellular aging; an excellent model being the fruit fly Drosophila melanogaster.24 Alterations in fruit fly microbiota composition have been linked to age-related intestinal barrier dysfunction, which also was found to lead to systemic immune activation, and eventually death.25 In addition, elimination of certain microbes, without causing detrimental side effects, has been shown to increase the fly’s life span.26 More recently, Smith et al27 used the short-lived African turquoise kill fish as another model to manipulate gut microbes in the study of longevity. Quite strikingly, this group found that when middle-aged fish were colonized with microbes transferred from younger fish, they lived longer and were significantly more active later in life than their control counterparts. It was also observed that middle-aged fish engrafted with the younger fish microbes retained a more diverse microbial community throughout their adulthood and shared key microbes with young fish; an observation inferred to also be associated with the improved health. Results from this study therefore suggest that the ability to control the composition of gut microbes can improve health and increase life span. Moreover, this model could be an important resource in providing new insights into how microbes can affect aging and to also delay the onset of age-related diseases (ARDs). Consistent with these findings, aging studies performed in other animal models and model organisms, such as in Caenorhabditis elegans8 and mice29,30 lend further support to the idea that microbiome modulation can lead to changes in the aging timeline with increasing evidence that such alterations can augment longevity. Collectively, these studies all point to the gut microbiome as playing a central role in the aging of the host.
Evidence of age-related changes in the gut microbiome are beginning to be described in different human aging populations. Such studies, however, have been limited by the paucity of elders as a research group on top of challenges that involve elders with dementia as a research group or elders who live in nursing home (NH) settings. Nevertheless, age-related microbiome changes are being uncovered that show a decline in bacterial diversity, shifts in dominant species, and changes in beneficial microorganisms and metabolic pathways.31–33 These changes are better resolved from a higher taxonomy approach where the major phyla of Bacteroidetes and Firmicutes switch in pre-dominance with older adults having higher abundances of Bacteroidetes as compared with higher Firmicutes abundances observed in younger counterparts.34 However, these observed shifts in composition do not stop at the phylum level; the species whose abundances are most prominently reduced in elders are the anaerobes,31,35 specifically with lower levels of Clostridium cluster XIVa, Faecalibacterium prausnitzii, and Actinobacteria (mainly among the bifido-bacterial genus).32,34–39 Other key metabolic species shown to decrease with increasing age include Akkermansia muciniphila,40 a mucin-degrading bacterium, Ruminococcus bromii,41 a keystone species in degradation of starch, as well as a prevalent gut commensal, Ruminococcus gnavus.32,33 Although these changes speak to an age-related dysbiotic microbiome, the variability in species abundance reported with age is likely due to external factors related to nationality, such as diet, environment, and life style.42
Apart from differences in nationality-based influences, elders living under different conditions also have been observed to differ in their microbiome structure. For instance, clear separations in microbiome signatures are noted between elders living in the community from those living in the NH setting.43,44 In fact, there is a distinct time-dependent manner in which the microbiome changes after an elder moves into a new NH environment with community structural changes taking approximately 1 year to occur.34 Although NHs in the United States provide services for elders that can be for custodial or skilled nursing in nature, these care settings also present an environment with frequent medication exposures, including antimicrobials, poorer diets, and increased pathogen prevalence, all which adversely affect the microbiome.36,45–48 Microbiota differences between NH and community-dwelling elders, in general, include higher proportions of Bacteroidetes and lower proportions of various other bacteria at the family and genus levels.36 Such changes in the bacterial populations among NH elders also represent loss of species that are associated with either a healthy or “youthful” microbiome.20 Curiously, and contrary to initial impressions, the elder gut microbiome exhibits temporal stability, outside of changes in medications, antimicrobial exposures, or major changes in health status.34,41 Each individual NH environment (such as floor/wing of residence) also plays a substantial role in shaping the microbiome,49 which may help to inform decisions and impart important consideration when grouping frail elders together to live.
In step with taxonomy differences, the metabolic potential of the microbiome also changes with age. Among elders (after accounting for nutrition and frailty) the metabolic dysbiosis associated with increasing age includes decreases in mucin and starch degradation, essential amino acid synthesis, and decreases in nitrogenous base and vitamin synthesis.41 Similarly, aging has been associated with a progressive loss of muscle mass (sarcopenia), which is linked to lower availability of essential amino acids.50,51 Moreover, among elder groups it has further been observed that intestinal microbiome alterations not only reveal a loss of genes for short-chain fatty acid production but also show an overall decrease in the saccharolytic potential, which correlates with the presence of opportunistic pathogens.52
Medications Influence Microbiome Composition
A key influencer of the aging microbiome structure is medications. Many medications commonly prescribed in elders (comprehensive of both NH and community settings) are well known to have specific effects on microbiota composition. The best example of this is with antibiotic exposures where there is a profound loss in diversity and shifts in microbial taxonomy abundances.48,53 Antibiotic exposures also lead to development of multidrug-resistant organisms (MDROs). This is a growing and significant health care problem among the elderly, especially those living in the NH environment. To date, there are an estimated 1.6 to 3.8 million infections per year in NHs 54,55 with as many as 400,000 resulting in death.56 Unfortunately, infections with MDROs continue to rise in NHs,56,57 and the mortality rate can be as high as 40% when an elder is hospitalized with an MDRO infection.58
Moreover, NHs in the United States have become the major reservoir for introduction of MDROs into other health care settings due to their uniquely high colonization prevalence.59–61 Because the microbiome plays a pivotal role that is central to human health,62 a healthy microbiome will, in turn, engage with the host immune system and contribute to pathogen resistance.63 Antibiotic therapies markedly decrease the intestinal microbiota diversity and richness. This creates a vulnerable immunodeficient environment that can be exploited by both antibiotic-resistant pathogenic and opportunistic bacteria that are frequently encountered nosocomially in the hospital as well as in the NH setting. The most clinically significant antibiotic-resistant intestinal pathogens include gram-positive C difficile and vancomycin-resistant Enterococcus faecium, along with gram-negative bacilli belonging to the Enterobacteriaceae family.64,65 Thus, the profound contributions made by commensal microbes toward resisting colonization and infection by pathogens are fundamental to host health and have long been observed. In spite of this, we are only now beginning to shed light on the molecular details underlying microbiome dysbiosis that occurs among elders and how this may be linked to pathogenic disease.9,41 Therefore, how we feed, treat, and group frail NH elders may offer new approaches to prevent MDRO spread.
Nonantibiotic medications have also been associated with changes in microbiome composition, and approximately 24% of marked drugs approved by the Food and Drug Administration have been shown to inhibit at least one common intestinal microbiome bacterial strain.66 The relative abundances of many microbiome members and changes noted during disease processes is extensive (Table 1). Furthermore, the effects of some nonantibiotic medications such as proton pump inhibitors,47,67,68 statins,69–71 nonsteroidal anti-inflammatory drugs,72 and atypical antipsychotics73–76 on the intestinal microbiome have been described only in healthy younger adults. Common among the elderly are the use of medications belonging to these classes as well as the mixture of these drugs in an individual. The combination of medications especially in excess, known as polypharmacy, is widespread in elders and has its own adverse effect on the microbiome and elder health.77,78 Indeed, polypharmacy is especially prevalent in the NH where more than half of the residents are on 5 or more daily medications.79
Table 1.
List of Microbiota Members by Genus or Species and the Published Disease Conditions With Which Each Has Been Shown to Have an Association
| Genus/Species | Related disease conditions | Abundances in disease | References |
|---|---|---|---|
| Butyrate producers | |||
| Anaerostipes | Alzheimer disease/ Cancer/ Colitis | Decreased | 1–3 |
| Butyricicoccus | Food allergy/ IBD | Decreased | 4,5 |
| Butyrivibrio | Age/ Alzheimer’s disease/ Amyotrophic lateral sclerosis | Decreased | 6–8 |
| Blautia hansenii | Alzheimer’s disease/ Autism/ Obesity | Decreased | 9–11 |
| Clostridial clusters IV and XIVa | Cystic Fibrosis/ IBD/ Multiple Sclerosis/ Parkinson’s | Decreased | 12–15 |
| Clostridium saccharolyticum | Alzheimer’s disease/ Parkinson’s disease | Decreased | 16,17 |
| Eubacterium species | Alzheimer’s disease/ Crohn’s disease/ Kidney stones | Decreased | 18,19–21 |
| Faecalibacterium prausnitzii | Alzheimer’s disease/ IBD/ Parkinson’s/ Psoriasis | Decreased | 2010,27,2815,22–24 |
| Roseburia hominis | Allergies/ Autoimmune diseases/ Diabetes Type 2/ Ulcerative Colitis | Decreased | 12,24,25 |
| Ruminococcus obeum | Age/ Liver Disease Obesity |
Decreased Increased |
26,27 28 |
| Ruminococcus bromii | Age/ Crohn’s disease/ Parkinson’s disease | Decreased | 29–31 |
| Lachnospiraceae bacterium | Diabetes / HIV/ Obesity/ | Increased | 32,33 |
| Frailty associated | |||
| Eggerthella lenta | Autoimmune/ Intestinal Infections | Increased | 34,35 |
| Eubacterium dolichum | Obesity | Increased | 36,37 |
| Methanobrevibacter | IBD | Decreased | 38,39 |
| Ruminococcus gnavus | Age/ Allergies/ Crohn’s disease/ Lupus | Increased | 31,40–42 |
| Malnutrition associated | |||
| Bifidobacterium | Antibiotic-associated diarrhea/ Cancer/ Eczema/ Ulcerative colitis | Decreased | 43–47 |
| Citrobacter freundii | Opportunistic pathogen | Increased | 31,48 |
| Enterococcus faecalis | Hospital-associated infections | Increased | 31,49 |
| Roseburia intestinalis | Anti-inflammatory/ Arthrosclerosis | Decreased | 50,51 |
| Inflammation /Autoimmune | |||
| Adlercreutzia equolifaciens | Multiple Sclerosis/ Primary sclerosing cholangitis | Decreased | 52,53 |
| Akkermansia muciniphila | Age/ Obesity/ Psoriasis/ Type 1 diabetes | Increased Decreased |
54–56 31,57 |
| Bacteroides dorei | Autoimmune Diseases/ Type 1 diabetes | Increased | 58,59 |
| Bacteroides vulgatus | Autism/ Autoimmune diabetes | Increased | 54,58,60,61 |
| Collinsella | Alzheimer’s disease/ Rheumatoid arthritis | Increased | 8,62 |
| Desulfovibrio fairfieldensis | IBD/ Obesity | Increased | 63,64 |
| Firmicutes bacterium | Obesity/ Type 2 diabetes | Decreased | 65,66 |
| Odoribacter splanchnicus | Hypertension/ IBD/ Lupus | Decreased | 67–69 |
| Parabacteroides distasonis | Multiple sclerosis/ Obesity/ Rheumatoid arthritis | Decreased | 70–72 |
| Pathogens | |||
| Bacteroides fragilis | Bacteroides fragilis | Increased | 73–76 |
| Campylobacter jejuni | Autoimmune/ Gastroenteritis/ Neurodegeneration | Increased | 77–79 |
| Cloacibacillus porcorum | Alzheimer’s disease/ Bacteremia | Decreased | 80–82 |
| Desulfovibrio fairfieldensis | Bacteremia/ IBD | Increased | 83,84 |
| Klebsiella pneumonia | Alzheimer’s disease/ Autoimmune/ Infections multiple sites | Increased | 85–89 |
| Peptostreptococcus anaerobius | Colorectal Cancer/ Multiple infections/ Septicemia | Increased | 90,91 |
NOTE. This list is based off of literature review of these microbiome members and organized into general categories that influence elder health. It is not intended to capture all of the published literature on each member but provide references to some of the relevant literature available.
HIV, human immunodeficiency virus; IBD, inflammatory bowel disease.
Role of Modulating the Microbiome to Improve Longevity
Taxonomy does inform and influence metabolic potential of the gut microbiome. Therefore, given that microbiome components change with age, it may offer an opportunity to intervene and slow or even reverse such age-related changes. In humans, centenarians have been used as a model for healthy aging studies because of their ability to delay, or even avoid, chronic diseases,80,81 and in addition their genetics have been extensively studied.82 However, to date only a few studies have interrogated the gut microbiome of this coveted population. To gain insight into which gut microbiome signatures are associated with longevity, Kong et al83 recently characterized the microbiota of a group of long-living (90 years of age or older) from the Dujiangyan region of China; 1 of 5 “longevous counties” in China. Comparing the gut microbiota in this long-living cohort with that of a younger adult group, Kong and colleagues83 found that the long-living group had a greater gut microbiome diversity than the younger adult group.
These results were not only validated using data from an independent Italian cohort that also included a group of long-living individuals32,84 but was also supported by 2 additional recent studies.37,85 Deeper characterization of the microbiota in the long-living cohort by Kong et al86 showed enrichment of several potentially beneficial bacterial taxa that are known to be short-chain fatty acid producers. Curiously, this result was coupled with a decrease of certain operational taxonomic units commonly associated with beneficial bacteria, such as Faecalibacterium, and an increase of some operational taxonomic units related to potential bacterial pathogens (eg, Escherichia and Shigella). Although it is too premature to draw any causal relationships between gut microbiota and healthy aging, this observational study does provide an important clue to suggest that maintaining a diverse and balanced gut microbiome may be a key contributor to healthy aging. All the same, whether one can modulate the intestinal microbiome to specifically target and promote healthy aging is an important question that needs to be carefully addressed. More specifically, because increasing age also engenders age-related morbidities that affect the quality and quantity of life (eg, heart disease, stroke, hypertension, cognitive impairment, and cancer), understanding how the aging microbiome affects these disease processes is critical to improving human health via the gut microbiome beyond just prevention of opportunistic pathogens.
The first association between microbes and healthy aging was made by Elie Metchnikoff, one of the founding fathers of modern microbiology and immunology. In 1908, he not only shared the Nobel Prize for Medicine with Paul Ehrlich but also published one of the most impactful books of that era entitled The Prolongation of Life (Metchnikoff, 1908). In this book Metchnikoff develops the concept that higher animals need an increasingly complex intestine to struggle for existence, and distinguished 2 types of metabolism for gut bacteria: (1) putrefaction that resulted in noxious metabolites as waste products, and (2) fermentation that resulted in beneficial metabolic end-products like lactic acid. To combat the process of putrefaction in the gut, he recommended improvements in diet and championed the notion that the fermentative metabolism of lactic acid bacteria would counterbalance putrefaction by the noxious gut bacteria and their toxic effect on our tissues. He backed these concepts by the observation that populations showing traditionally high yogurt consumption also showed increased longevity. More than 100 years later, modulation of the microbiome by either diet or probiotic interventions is evidenced in animal models supporting the tantalizing hypothesis that host longevity can be lengthened by shifting microbiome communities.87 Although this potential among humans is still relatively unexplored, and can be complicated by individual heterogeneity, it does tender a unique and potentially promising strategy to influence the aging process.
Inflammation and Age-related Diseases
One of the basic mechanisms shared in ARDs and geriatric syndromes is chronic low-grade inflammation called inflamm-aging.5,88 ARDs are diseases that increase in incidence exponentially with age and include disorders such as atherosclerosis, diabetes, hypertension, cancer, and Alzheimer disease (AD).89,90 Chronic upregulation of proinflammatory mediators (eg, tumor necrosis factor-a, interleukin (IL)-6) have been shown to be induced during the aging process. These proinflammatory mediators activate many signaling pathways91 that have a dramatic impact on immune function, which leads to a gradual deterioration of the immune system, called immunosenescence.14,92 Currently, both inflamm-aging and immunosenescence are thought to be responsible for most ARDs (and not just by the increased risk of bacterial infections) and are fertile ground for novel interventions to promote healthy aging.93 Dysbiosis of the gut microbiome can serve as a catalyst for fueling inflamm-aging (Figure 2).14 However, the contribution of dysbiosis in the context of the human microbiome interaction particularly regarding its impact on systemic immune functioning or deterioration of this function among the elderly as it relates to ARDs has not been rigorously studied.13
Figure 2.
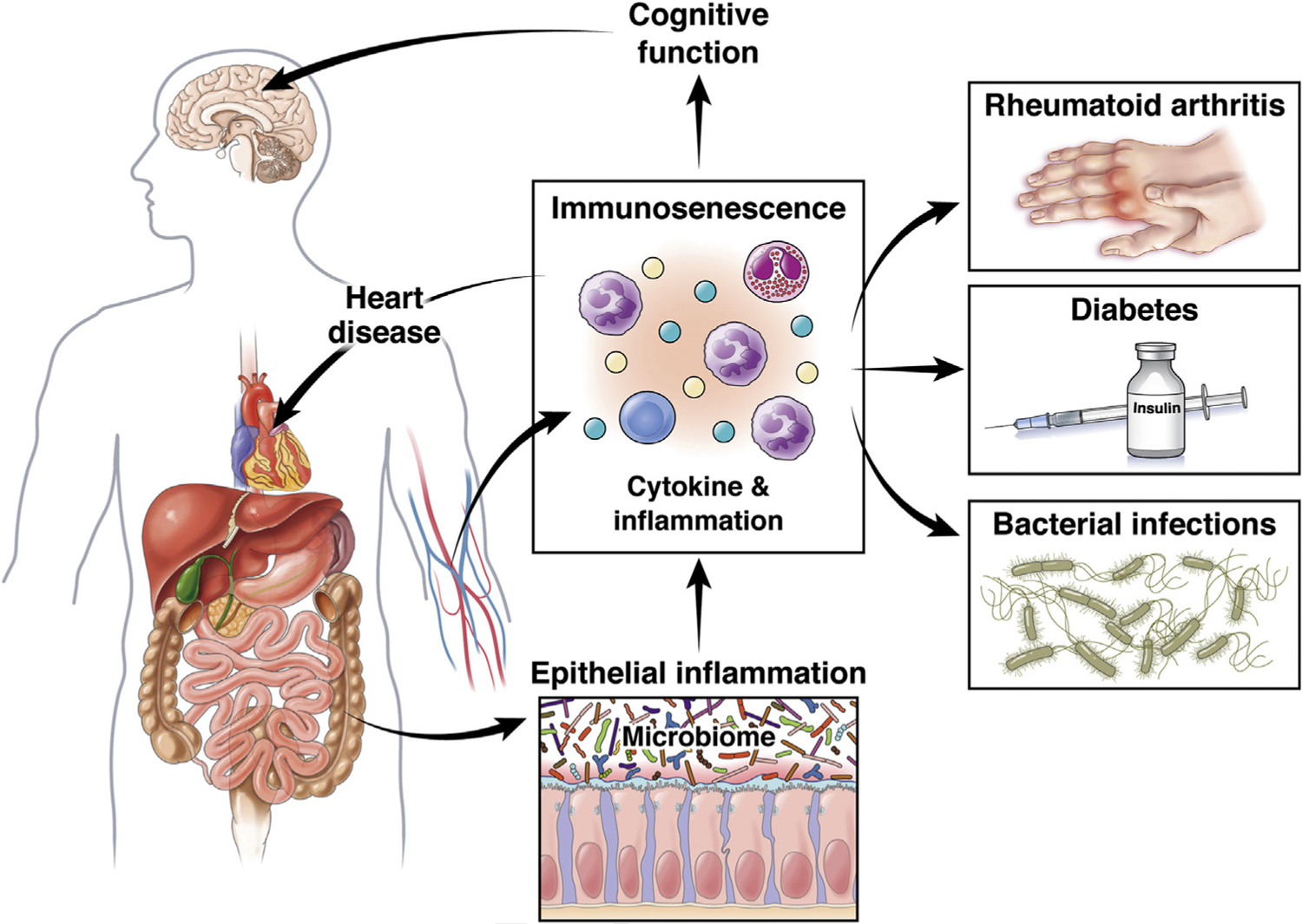
Inflamm-aging and related ARDs. The intestinal microbiome has been linked to disorders of the brain, heart, endocrine, musculoskeletal, and immune systems. This is an overview of the sections along the inflamm-aging to age-related disease pathways.
Nevertheless, there is a growing body of literature that implicates age-related dysbiosis of the gut microbiome as contributing to a global inflammatory state in the elderly.94,95 For example, neuroinflammation, one result of immunosenescence, has long been thought to promote progression of several neurological disorders, including AD and Parkinson disease (PD).96,97 Both acute and chronic systemic inflammation are associated with declining cognitive function in AD.98 To put this in perspective, more than 46 million people worldwide live with dementia, and this number is predicted to double in the next 20 years99 with an alarming projection of 3.3% of the US population being affected by AD.100 Both inflamm-aging and immunosenescence have been well described in patients with AD.101–104 The inflammatory response that accompanies AD pathology is hallmarked by higher peripheral concentrations of cytokines IL-6, tumor necrosis factor-α, IL-1β, transforming growth factor-β, IL-12, and IL-18.101 Moreover, both the innate and acquired immune systems have been shown to be altered in AD.105–107 For instance, patients with AD exhibit decreased levels of naïve T cells, along with elevated memory T-cell populations,108 and higher percentages of activated CD4+ CD25+ T cells.103 Such variances in T-cell populations, which are common in patients with AD, denote a heightened differentiated T-cell state. This is consistent with an adaptive immune system undergoing persistent antigen exposure and dysregulation of the naïve/memory T-cell balance.108
One area coming into focus as a potential driver of this proinflammatory state is the intestinal microbiome. The dysbiotic intestinal microbiome has been shown to induce systemic inflammation that triggers neuroinflammation leading to cognitive impairment.109 Glial cell phenotypes are known to be profoundly modulated by peripheral inflammatory stimuli, including those due to dysbiosis of the gut microbiota.110,111 Increased abundance of proinflammatory, with reduced abundance of anti-inflammatory, bacteria in the intestine also has been shown to be associated with systemic inflammatory states in patients with cognitive impairment and brain amyloidosis.112 With respect to the AD-intestinal microbiome interaction, AD pathogenesis has long been thought to be linked to chronic bacterial infections as a possible etiology.113 More recent 16S-based studies have found significant changes in the abundance of certain taxa in patients with AD compared with healthy controls,114,115 and one of these studies also linked microbiota composition back to AD cerebrospinal fluid biomarker levels.114 Thus, one prevailing theory is that AD pathogenesis is closely related to the imbalance of the gut microbiome and, in fact, may originate in the gut.
Although the role of microbes in promoting the inflammatory causal pathway of AD is becoming increasingly recognized,113,116,117 it is yet to be established. In taking a step toward addressing this goal, studies performed by Harach et al118 were among the first to report that gut microbes play a role in the development of cerebral Aβ amyloidosis in patients with AD. A key finding in this study was the dramatic shift in the gut microbiota of Aβ precursor protein (APP) transgenic mice as compared with non-transgenic wild-type mice. In addition, they also observed a profound reduction in cerebral Aβ amyloid pathology when APP transgenic mice were raised in a germ-free environment as compared with control mice, which harbored an intestinal microbiome. This observation was further supported by demonstrating that colonization of germ-free APP transgenic mice with microbiota from conventionally raised APP transgenic mice increased cerebral Aβ pathology, whereas colonization with microbiota from wild-type mice was much less effective. In summary, these findings reveal the potential of microbial involvement in the development of Aβ amyloid pathology, and more generally suggest that microbiota may contribute to the development of neurodegenerative diseases.
More recently, our group has reported findings among a cohort of NH elders that demonstrates a dysbiotic pattern is seen when comparing AD elders with those with no dementia.16 Such dysbiosis is characterized by a reduction in the proportion and prevalence of bacteria with the potential to synthesize butyrate, an essential metabolite in the human colon with anti-inflammatory properties, as well as an acquisition of taxa that are known to cause proinflammatory states. Consistent with these changes, we also demonstrated how the “AD microbiome” can adversely affect intestinal epithelial homeostasis via dysregulation of P-glycoprotein (P-gp). P-gp is a critical mediator of intestinal homeostasis,119 and when downregulated, can lead to a proinflammatory state (Figure 3). The bacterial species that differentiates the microbiome of AD from elders without dementia was also found to be predictive of lower P-gp expression levels among patients with AD. These species are key butyrate producers and include members of the Eubacterium, Clostridium, and Butyrivibrio genera,120 as well as bacteria known to associate with proinflammatory states in the intestines, such as Bacteroides dorei and Akkermansia glycaniphila.121 Therefore, the microbial members found to best predict the observed lower P-gp expression in patients with AD are all known to influence colonic inflammation in other pathological states. We are just beginning to disentangle the complex interplay involved in the gut-brain axis. Hence, a deeper understanding of the taxa and the role these microbial communities play in contributing to the progression of AD (as well as other neurodegenerative diseases) is needed to help advance our knowledge of causal relationship between dysbiosis and cognitive decline with the ultimate goal of preventing or halting disease.
Figure 3.
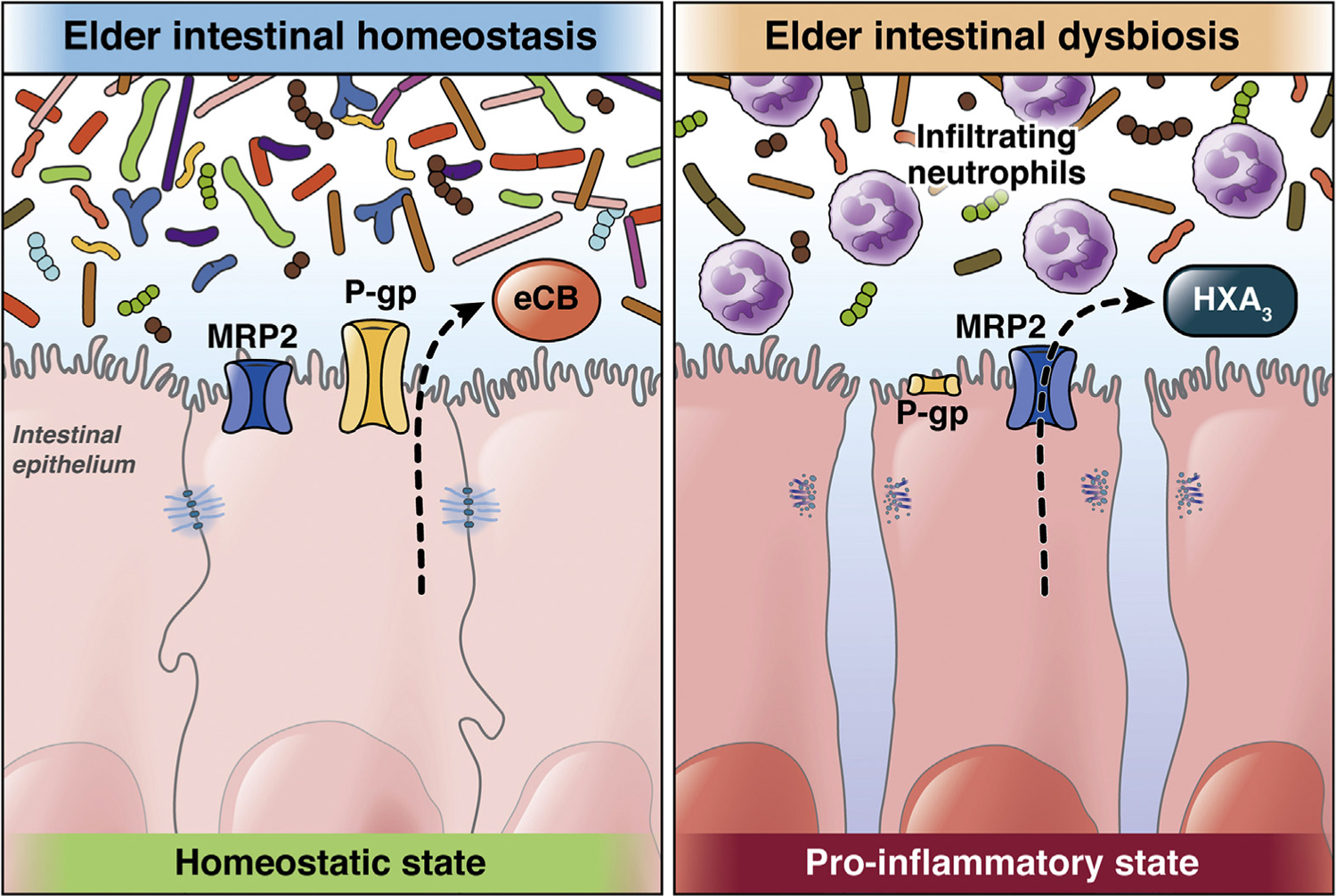
The MRP2/HXA3 (hepoxilin A3) axis forms the proinflammatory arm of a dynamically regulated system in which inflammatory pathways that activate responses to pathogens or aberrant signals are balanced against the anti-inflammatory P-glycoprotein (P-gp)/endocannabinoid (eCB) pathway that suppresses neutrophil responses in the context of normal commensal colonization. The 2 sets of lipid-based signaling molecules (eCB and HXA3) are released from the apical surface during periods of either tolerance or inflammation, which control the recruitment of neutrophils to the intestinal lumen. Dysregulation of this critical balance may contribute directly to inflammatory disorders of the intestine (108).
The gut microbiota also has been found to regulate motor deficits in neuroinflammation in a murine model of PD.122 Motor dysfunction in patients with PD is often characterized by aggregation of the protein alpha-synuclein (αSyn). Sampson et al122 used a mouse model that over-expresses αSyn to demonstrate that gut microbiota is required for motor deficits, microglia activation, and αSyn pathology. In this same study, colonization of αSyn-over-expressing mice with microbiota from PD-affected patients was also found to enhance physical deterioration as compared with microbiota engrafted from healthy human donors. Although the mechanism by which gut microbes affect the progression of PD is not well understood, the recent finding that αSyn is found in gut endocrine cells before appearing in the brain supports the contention that PD pathology originates first in the gut and then may spread to the central nervous system in a manner analogous to cell-to-cell prion-like propagation.123
Other ARDs share a similar inflamm-aging/immunosenescence profile that may have origins in the inflammatory type dysbiosis of the gut. For example, Fransen et al,29 when transferring aged microbiota to young germ-free mice, identified certain bacterial species within the aged microbiota that promote inflamm-aging. This effect was primarily associated with lower levels of Akkermansia and higher levels of TM7 bacteria and Proteobacteria in the aged microbiota after transfer. Such changes in the microbiota composition correlated with intestinal inflammation predominantly in the small intestine, leakage of inflammatory bacterial components into the circulation, and increased T-cell activation in the systemic compartment.29 In other examples, inoculation of mice with fecal samples from patients with rheumatoid arthritis promoted development of rheumatoid arthritis in the arthritis-prone mice via a Th17-dependent manner.124 Likewise, gut dysbiosis has been shown to contribute to systemic homeostasis disruption and subsequent proinflammatory pathways leading to obesity, B-cell decline, and type 2 diabetes.125 Lipopolysaccharides and other microbial factors promote inflammatory signaling and skeletal muscle changes that are also the hallmarks of the aging muscle phenotype.126 Finally, there is even an emerging, yet unproven, contributing role for the human microbiome in the cause and development of multiple different cancer types.127 Therefore, different forms of dysbiotic-induced inflammation, commencing locally and then exerting effects systemically, just might serve as the initiation and/or driver of many ARDs that pose significant burden to healthy human aging.
Is the Microbiome Frailty Connection a Linchpin to Other ARDs?
Frailty is a state of increased vulnerability and poor resolution of homeostasis following a stressful event to the elder.128,129 Frailty is highly prevalent in community-dwelling elders130 but is especially high in NH populations, with as many as 50% of elders being frail and an additional 40% meeting a prefrail definition.131 Fried et al128 provided one of the first operational definitions of frailty as meeting 3 of 5 phenotypic criteria indicating compromised energetics: low grip strength, low energy, slowed waking speed, low physical activity, and/or unintentional weight loss. Since then many other scoring systems have emerged that are easier to apply clinically.132,133 But even with all of these established parameters there remains a lack of a gold standard in defining an older adult as frail.134–136 Nevertheless, regardless of the instrument tool used to measure frailty, elders defined as frail have a clear increased risk of mortality among elders in the emergency department,137 admitted to the hospital,138 or living in either the community139,140 or NH141,142 settings.
This complex process, which is linked closely with aging, involves a decline in a constellation of physiological systems that leads to increased vulnerability and disproportionate changes in health status following even a minor stressor event.143 Outside of the relatively few medical causes (eg, medications, nutrition, or lack of exercise) the cause of frailty remains poorly understood.136,143,144 An emerging theory of a cause of frailty, however, ties back to inflamm-aging and the development of immunosenescence. As with other ARDs, frailty also has associations with immune dysfunction and inflammation, with a complex altered production of inflammatory cytokines.145,146 Nestled within this theory is an association to the gut microbiome; however, studies linking dysbiosis to frailty have been relatively unexplored.147 What is known is that frailty is hallmarked by a loss of microbiota diversity and specific taxonomic associations, such as increased abundances of Eubacterium dolichum and Eggerthella lenta and decreases in Faecalibacterium prausnitzii.21 In NH elders, this is associated with losses of community-dwelling associated microbiota36 that specifically involves a dysbiotic pattern where there is a loss of butyrate-producing organisms coupled with an increase in abundances of inflammation-associated organisms, as well as an increase in lipopolysaccharide biosynthesis and peptidoglycan biosynthesis metabolic pathways.41 Given the important connection to clinical outcomes and health-related quality of life in elders, combined with emerging theories and lack of any mechanism to treat frailty, the gut microbiome may hold a vital key to improving healthy aging.
Challenges and Opportunities to Improve the Elder Microbiome
The identification of gut microbiome associations with diseases in the elderly opens up the door for interventions to improve or prevent diseases. These microbiome-based interventions have lagged behind studies of younger-aged populations, but offer a great opportunity to improve human health given the increasing aging population and the burden of disease among elderly individuals. Microbiome-based interventions have traditionally focused on probiotics, typically lactobacilli and bifidobacterial, or prebiotics with nondigestible oligosaccharides.148 Clinical trials among elder participants have demonstrated not only the ability to manipulate the gut microbiome but also the safety among this population in doing so.149 However, clinical efficacy in this regard is yet to be well substantiated.
Advancing clinical trial work focused on manipulating the microbiome would avail new opportunities with the potential to address a wide range of disease processes. Lactobacilli, historically, have been one of the most chronicled probiotics studied. Since its first isolation from the feces of a normal healthy individual in 1987, Lactobacillis has been used for a wide variety of clinical indications. In healthy individuals it temporarily colonizes the distal gastrointestinal track and positively affects the resident microflora.150,151 In addition, Lactobacillis has been used in clinical trials addressing diarrhea from use of antibiotics152,153 to travelers’ diarrhea154 and diarrhea from autoimmune causes.155 Outside of gastrointestinal disorders, Lactobacillis has been used to prevent urinary tract infections,156 to treat rheumatoid arthritis,157 as an immune modulator for vaccine administration,158 and as preventive treatment in intensive care unit patients.159 Beyond Lactobacillis, other probiotic bacteria and bacterial combinations have been tested as a therapy to treat a multitude of human disease, many of which are age-related. Clearly, probiotics have a track record and potential to treat multiple disease processes that needs to be soundly tested in elder populations.
However, most probiotics are manufactured as food, which makes it challenging to ensure the quality and safety of these products as novel therapeutic agents. The basic issues of dosing, safety, and mechanism of action of these agents still need to be worked out because it is still unclear which bacterial strains hold benefit under different disease conditions. With the emergence of multistrain probiotics onto the market and the premise of engineered microorganisms for designer probiotics, it is even more crucial to move forward our understanding of how food and probiotics can influence exact mechanistic action on the microbiome and also necessitates a better understanding of the elder microbiome in health and disease.
Among elderly individuals, dietary interventions have shown promise in addressing some of the most devastating ARDs, such as AD. Large epidemiological studies have shown that healthy eating is protective against dementia and cognitive decline, which has been proven with diet interventions such as the MIND diet in AD.160 Large-scale interventions, such as the Finnish FINGER trial are under way and show promising results in preventing AD.161 Although the exact mechanism of these dietary interventions is not well known, there is mounting evidence that the gut microbiome alterations that occur during dietary intervention may be the driving force behind the improved outcomes in AD.95,162 In this regard, it is important for us to have a better understanding of how dietary interventions change the microbiome and if these changes are the primary drivers that improve AD symptomology. Whether the microbiome acts as a mediator or the primary agent in the causal pathway during a dietary intervention is still unclear; however, unraveling this mystery would greatly help us to understand the pathophysiology and treatment of cognition decline via the gut-brain axis.
The future of microbiome research is full of exciting possibilities. There is a wealth of evidence that links the gut microbiome to healthy human development and how dysbiosis of the microbiome leads to disease. It is now being increasingly recognized that it may not be the abundance of individual bacterial populations that drives a disease process, but the collective microbiome (ie, microbial consortia of functional genes and pathways) and its metabolites termed the “functional core microbiome” that may hold the key to understanding increased susceptibility to diseased states. Clinically, how we can alter the “functional core microbiome” in disease prevention or treatment still has a long way to go before it is put into practice.
Abbreviations used in this paper:
- AD
Alzheimer disease
- APP
Aβ precursor protein
- ARD
age-related disease
- IL
interleukin
- MDRO
multi-drug-resistant organism
- NH
nursing home
- PD
Parkinson disease
- P-gp
P-glycoprotein
- αSyn
alpha-synuclein
Footnotes
Conflict of interest
The authors disclose no conflicts.
References
- 1.National Population Projections. US Census Bureau 2017. Available at: https://www.census.gov/programs-surveys/popproj.html. Accessed August 4, 2020.
- 2.Kho ZY, Lal SK. The human gut microbiome - a potential controller of wellness and disease. Front Microbiol 2018; 9:1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camilleri M, Lee JS, Viramontes B, et al. Insights into the pathophysiology and mechanisms of constipation, irritable bowel syndrome, and diverticulosis in older people. J Am Geriatr Soc 2000;48:1142–1150. [DOI] [PubMed] [Google Scholar]
- 4.Kleessen B, Sykura B, Zunft HJ, et al. Effects of inulin and lactose on fecal microflora, microbial activity, and bowel habit in elderly constipated persons. Am J Clin Nutr 1997;65:1397–1402. [DOI] [PubMed] [Google Scholar]
- 5.Franceschi C, Bonafè M, Valensin S, et al. Inflammaging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci 2000;908:244–254. [DOI] [PubMed] [Google Scholar]
- 6.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol 2014; 16:1024–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkins LJ, Monga M, Miller AW. Defining dysbiosis for a cluster of chronic diseases. Sci Rep 2019;9:12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007;448:427–434. [DOI] [PubMed] [Google Scholar]
- 9.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015;517:205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y, Zhi F. Lower level of Bacteroides in the gut microbiota is associated with inflammatory bowel disease: a meta-analysis. Biomed Res Int 2016; 2016:5828959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013;368:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 2013;19:576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amsterdam D, Ostrov BE. The impact of the microbiome on immunosenescence. Immunol Invest 2018;47:801–811. [DOI] [PubMed] [Google Scholar]
- 14.Franceschi C, Garagnani P, Parini P, et al. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol 2018;14:576–590. [DOI] [PubMed] [Google Scholar]
- 15.Allen AP, Dinan TG, Clarke G, et al. A psychology of the human brain-gut-microbiome axis. Soc Personal Psychol Compass 2017;11:e12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haran JP, Bhattarai SK, Foley SE, et al. Alzheimer’s disease microbiome is associated with dysregulation of the anti-inflammatory P-glycoprotein pathway. mBio 2019;10:e00632–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bazett M, Bergeron ME, Haston CK. Streptomycin treatment alters the intestinal microbiome, pulmonary T cell profile and airway hyperresponsiveness in a cystic fibrosis mouse model. Sci Rep 2016;6:19189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoen AG, Li J, Moulton LA, et al. Associations between gut microbial colonization in early life and respiratory outcomes in cystic fibrosis. J Pediatr 2015;167:138–147. e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint HJ, O’Toole PW, Walker AW. Special issue: the human intestinal microbiota. Microbiology 2010; 156:3203–3204. [DOI] [PubMed] [Google Scholar]
- 20.Jeffery IB, Lynch DB, O’Toole PW. Composition and temporal stability of the gut microbiota in older persons. ISME J 2016;10:170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson MA, Jeffery IB, Beaumont M, et al. Signatures of early frailty in the gut microbiota. Genome Med 2016; 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Power SE, Jeffery IB, Ross RP, et al. Food and nutrient intake of Irish community-dwelling elderly subjects: who is at nutritional risk? J Nutr Health Aging 2014;18:561–572. [DOI] [PubMed] [Google Scholar]
- 23.Microbiota Brussow H. and healthy ageing: observational and nutritional intervention studies. Microb Biotechnol 2013;6:326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark RI, Salazar A, Yamada R, et al. Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality. Cell Rep 2015; 12:1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci U S A 2012;109:21528–21533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hye-Yeon L, Shin-Hae L, Ji-Hyeon L, et al. The role of commensal microbes in the lifespan of Drosophila melanogaster. Aging (Albany NY) 2019;11:4611–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith P, Willemsen D, Popkes M, et al. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. Elife 2017;6:e27014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han B, Sivaramakrishnan P, Lin CJ, et al. Microbial genetic composition tunes host longevity. Cell 2017; 169:1249–1262.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fransen F, van Beek AA, Borghuis T, et al. Aged gut microbiota contributes to systemical inflammaging after transfer to germ-free mice. Front Immunol 2017;8:1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison DE, Strong R, Allison DB, et al. Acarbose, 17-alpha-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell 2014; 13:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodmansey EJ, McMurdo ME, Macfarlane GT, et al. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl Environ Microbiol 2004;70:6113–6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biagi E, Nylund L, Candela M, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 2010;5:e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller S, Saunier K, Hanisch C, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol 2006; 72:1027–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claesson MJ, Cusack S, O’Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 2011; 108:4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hopkins MJ, Macfarlane GT. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J Med Microbiol 2002; 51:448–454. [DOI] [PubMed] [Google Scholar]
- 36.Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012;488:178–184. [DOI] [PubMed] [Google Scholar]
- 37.Odamaki T, Kato K, Sugahara H, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 2016;16:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hopkins MJ, Sharp R, Macfarlane GT. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 2001;48:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salazar N, López P, Valdés L, et al. Microbial targets for the development of functional foods accordingly with nutritional and immune parameters altered in the elderly. J Am Coll Nutr 2013;32:399–406. [DOI] [PubMed] [Google Scholar]
- 40.Collado MC, Derrien M, Isolauri E, et al. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 2007;73:7767–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haran JP, Bucci V, Dutta P, et al. The nursing home elder microbiome stability and associations with age, frailty, nutrition, and physical location. J Med Microbiol 2018; 67:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biagi E, Candela M, Fairweather-Tait S, et al. Aging of the human metaorganism: the microbial counterpart. Age (Dordr) 2012;34:247–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roghmann MC, Lydecker AD, Hittle L, et al. Comparison of the microbiota of older adults living in nursing homes and the community. mSphere 2017;2:e00210–e00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogawa T, Hirose Y, Honda-Ogawa M, et al. Composition of salivary microbiota in elderly subjects. Sci Rep 2018; 8:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saarela RK, Lindroos E, Soini H, et al. Dentition, nutritional status and adequacy of dietary intake among older residents in assisted living facilities. Gerodontology 2016;33:225–232. [DOI] [PubMed] [Google Scholar]
- 46.Rane PP, Guha S, Chatterjee S, et al. Prevalence and predictors of non-evidence based proton pump inhibitor use among elderly nursing home residents in the US. Res Social Adm Pharm 2017;13:358–363. [DOI] [PubMed] [Google Scholar]
- 47.Freedberg DE, Toussaint NC, Chen SP, et al. Proton pump inhibitors alter specific taxa in the human gastrointestinal microbiome: a crossover trial. Gastroenterology 2015;149:883–885.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dethlefsen L, Huse S, Sogin ML, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 2008;6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210–215. [DOI] [PubMed] [Google Scholar]
- 50.Volpi E, Kobayashi H, Sheffield-Moore M, et al. Essential amino acids are primarily responsible for the amino acid stimulation of muscle protein anabolism in healthy elderly adults. Am J Clin Nutr 2011;78:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Short KR, Vittone JL, Bigelow ML, et al. Age and aerobic exercise training effects on whole body and muscle protein metabolism. Am J Physiol Endocrinol Metab 2004;286:E92–E101. [DOI] [PubMed] [Google Scholar]
- 52.Rampelli S, Candela M, Turroni S, et al. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany NY) 2013;5:902–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 2011;108(Suppl 1):4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith PW, Bennett G, Bradley S, et al. SHEA/APIC guideline: infection prevention and control in the long-term care facility. Am J Infect Control 2008;36:504–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strausbaugh LJ, Joseph CL. The burden of infection in long-term care. Infect Control Hosp Epidemiol 2000; 21:674–679. [DOI] [PubMed] [Google Scholar]
- 56.Viray M, Linkin D, Maslow JN, et al. Longitudinal trends in antimicrobial susceptibilities across long-term-care facilities: emergence of fluoroquinolone resistance. Infect Control Hosp Epidemiol 2005;26:56–62. [DOI] [PubMed] [Google Scholar]
- 57.Vromen M, van der Ven AJ, Knols A, et al. Antimicrobial resistance patterns in urinary isolates from nursing home residents. Fifteen years of data reviewed. J Antimicrob Chemother 1999;44:113–116. [DOI] [PubMed] [Google Scholar]
- 58.CMS Manual System. Department of Health & Human Services (DHHS); Centers for Medicare & Medicaid Services (CMS) December 2, 2009; Pub. 100-07.
- 59.Trick WE, Weinstein RA, DeMarais PL, et al. Colonization of skilled-care facility residents with antimicrobial-resistant pathogens. J Am Geriatr Soc 2001;49:270–276. [DOI] [PubMed] [Google Scholar]
- 60.Pop-Vicas A, Mitchell SL, Kandel R, et al. Multidrug-resistant gram-negative bacteria in a long-term care facility: prevalence and risk factors. J Am Geriatr Soc 2008;56:1276–1280. [DOI] [PubMed] [Google Scholar]
- 61.Wang L, Lansing B, Symons K, et al. Infection rate and colonization with antibiotic-resistant organisms in skilled nursing facility residents with indwelling devices. Eur J Clin Microbiol Infect Dis 2012;31:1797–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morgan XC, Huttenhower C. Chapter 12: Human microbiome analysis. PLoS Comput Biol 2012;8: e1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 2013;13:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stecher B, Robbiani R, Walker AW, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 2007; 5:2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Endt K, Stecher B, Chaffron S, et al. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog 2010; 6:e1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018;555:623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imhann F, Bonder MJ, Vich Vila A, et al. Proton pump inhibitors affect the gut microbiome. Gut 2016;65:740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takagi T, Naito Y, Inoue R, et al. The influence of long-term use of proton pump inhibitors on the gut microbiota: an age-sex-matched case-control study. J Clin Biochem Nutr 2018;62:100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J, Lee H, An J, et al. Alterations in gut microbiota by statin therapy and possible intermediate effects on hyperglycemia and hyperlipidemia. Front Microbiol 2019; 10:1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y, Song X, Zhou H, et al. Gut microbiome associates with lipid-lowering effect of rosuvastatin in vivo. Front Microbiol 2018;9:530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nolan JA, Skuse P, Govindarajan K, et al. The influence of rosuvastatin on the gastrointestinal microbiota and host gene expression profiles. Am J Physiol Gastrointest Liver Physiol 2017;312:G488–G497. [DOI] [PubMed] [Google Scholar]
- 72.Rogers MAM, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect 2016;22:178.e1–178.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flowers SA, Evans SJ, Ward KM, et al. Interaction between atypical antipsychotics and the gut microbiome in a bipolar disease cohort. Pharmacotherapy 2017; 37:261–267. [DOI] [PubMed] [Google Scholar]
- 74.Flowers SA, Baxter NT, Ward KM, et al. Effects of atypical antipsychotic treatment and resistant starch supplementation on gut microbiome composition in a cohort of patients with bipolar disorder or schizophrenia. Pharmacotherapy 2019;39:161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morgan AP, Crowley JJ, Nonneman RJ, et al. The antipsychotic olanzapine interacts with the gut microbiome to cause weight gain in mouse. PLoS One 2014; 9:e115225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davey KJ, O’Mahony SM, Schellekens H, et al. Gender-dependent consequences of chronic olanzapine in the rat: effects on body weight, inflammatory, metabolic and microbiota parameters. Psychopharmacology 2012; 221:155–169. [DOI] [PubMed] [Google Scholar]
- 77.Ticinesi A, Milani C, Lauretani F, et al. Gut microbiota composition is associated with polypharmacy in elderly hospitalized patients. Sci Rep 2017;7:11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Masnoon N, Shakib S, Kalisch-Ellett L, et al. What is polypharmacy? A systematic review of definitions. BMC Geriatr 2017;17:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beloosesky Y, Nenaydenko O, Gross Nevo RF, et al. Rates, variability, and associated factors of polypharmacy in nursing home patients. Clin Interv Aging 2013;8:1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Engberg H, Oksuzyan A, Jeune B, et al. Centenarians–a useful model for healthy aging? A 29-year follow-up of hospitalizations among 40,000 Danes born in 1905. Aging Cell 2009;8:270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Medford A, Christensen K, Skytthe A, et al. A cohort comparison of lifespan after age 100 in Denmark and Sweden: are only the oldest getting older? Demography 2019;56:665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beekman M, Blanché H, Perola M, et al. Genome-wide linkage analysis for human longevity: Genetics of Healthy Aging Study. Aging Cell 2013;12:184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kong F, Hua Y, Zeng B, et al. Gut microbiota signatures of longevity. Curr Biol 2016;26:R832–R833. [DOI] [PubMed] [Google Scholar]
- 84.Biagi E, Franceschi C, Rampelli S, et al. Gut microbiota and extreme longevity. Curr Biol 2016;26:1480–1485. [DOI] [PubMed] [Google Scholar]
- 85.Bian G, Gloor GB, Gong A, et al. The gut microbiota of healthy aged Chinese is similar to that of the healthy young. mSphere 2017;2:e00327–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kong F, Deng F, Li Y, et al. Identification of gut microbiome signatures associated with longevity provides a promising modulation target for healthy aging. Gut Microbes 2019;10:210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Westfall S, Lomis N, Prakash S. Longevity extension in Drosophila through gut-brain communication. Sci Rep 2018;8:8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Franceschi C, Garagnani P, Morsiani C, et al. The continuum of aging and age-related diseases: common mechanisms but different rates. Front Med (Lausanne) 2018;5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ritchie K, Kildea D. Is senile dementia “age-related” or “ageing-related”?–evidence from meta-analysis of dementia prevalence in the oldest old. Lancet 1995; 346:931–934. [DOI] [PubMed] [Google Scholar]
- 90.Fransquet PD, Wrigglesworth J, Woods RL, et al. The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clin Epigenetics 2019;11:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chung HY, Cesari M, Anton S, et al. Molecular inflammation: underpinnings of aging and age-related diseases. Ageing Res Rev 2009;8:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jose SS, Bendickova K, Kepak T, et al. Chronic inflammation in immune aging: role of pattern recognition receptor crosstalk with the telomere complex? Front Immunol 2017;8:1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fulop T, Larbi A, Dupuis G, et al. Immunosenescence and inflamm-aging as two sides of the same coin: friends or foes? Front Immunol 2017;8:1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rehman T Role of the gut microbiota in age-related chronic inflammation. Endocr Metab Immune Disord Drug Targets 2012;12:361–367. [DOI] [PubMed] [Google Scholar]
- 95.Thevaranjan N, Puchta A, Schulz C, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe 2017;21:455–466.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Castillo E, Leon J, Mazzei G, et al. Comparative profiling of cortical gene expression in Alzheimer’s disease patients and mouse models demonstrates a link between amyloidosis and neuroinflammation. Sci Rep 2017; 7:17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Passamonti L, Tsvetanov KA, Jones PS, et al. Neuroinflammation and functional connectivity in Alzheimer’s disease: interactive influences on cognitive performance. J Neurosci 2019;39:7218–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Holmes C, Cunningham C, Zotova E, et al. Systemic inflammation and disease progression in Alzheimer disease. Neurology 2009;73:768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prince MJ. World Alzheimer Report 2015: The Global Impact of Dementia: an Analysis of Prevalence, Incidence, Cost and Trends; 2015.
- 100.Matthews KA, Xu W, Gaglioti AH, et al. Racial and ethnic estimates of Alzheimer’s disease and related dementias in the United States (2015–2060) in adults aged ≥65 years. Alzheimers Dement 2019;15:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Swardfager W, Lanctot K, Rothenburg L, et al. A meta-analysis of cytokines in Alzheimer’s disease. Biol Psychiatry 2010;68:930–941. [DOI] [PubMed] [Google Scholar]
- 102.Togo T, Akiyama H, Iseki E, et al. Occurrence of T cells in the brain of Alzheimer’s disease and other neurological diseases. J Neuroimmunol 2002;124:83–92. [DOI] [PubMed] [Google Scholar]
- 103.Pellicanò M, Larbi A, Goldeck D, et al. Immune profiling of Alzheimer patients. J Neuroimmunol 2012;242:52–59. [DOI] [PubMed] [Google Scholar]
- 104.Chen X, Hu Y, Cao Z, et al. Cerebrospinal fluid inflammatory cytokine aberrations in Alzheimer’s disease, Parkinson’s disease and amyotrophic lateral sclerosis: a systematic review and meta-analysis. Front Immunol 2018;9:2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pellicanò M, Bulati M, Buffa S, et al. Systemic immune responses in Alzheimer’s disease: in vitro mononuclear cell activation and cytokine production. J Alzheimers Dis 2010;21:181–192. [DOI] [PubMed] [Google Scholar]
- 106.Speciale L, Calabrese E, Saresella M, et al. Lymphocyte subset patterns and cytokine production in Alzheimer’s disease patients. Neurobiol Aging 2007;28:1163–1169. [DOI] [PubMed] [Google Scholar]
- 107.Xue SR, Xu DH, Yang XX, et al. Alterations in lymphocyte subset patterns and co-stimulatory molecules in patients with Alzheimer disease. Chin Med J (Engl) 2009; 122:1469–1472. [PubMed] [Google Scholar]
- 108.Larbi A, Pawelec G, Witkowski JM, et al. Dramatic shifts in circulating CD4 but not CD8 T cell subsets in mild Alzheimer’s disease. J Alzheimers Dis 2009;17:91–103. [DOI] [PubMed] [Google Scholar]
- 109.Daulatzai MA. Chronic functional bowel syndrome enhances gut-brain axis dysfunction, neuroinflammation, cognitive impairment, and vulnerability to dementia. Neurochem Res 2014;39:624–644. [DOI] [PubMed] [Google Scholar]
- 110.Erny D, Hrabe de Angelis AL, Jaitin D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 2015;18:965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rothhammer V, Mascanfroni ID, Bunse L, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med 2016;22:586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cattaneo A, Cattane N, Galluzzi S, et al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging 2017;49:60–68. [DOI] [PubMed] [Google Scholar]
- 113.Maheshwari P, Eslick GD. Bacterial infection and Alzheimer’s disease: a meta-analysis. J Alzheimers Dis 2015;43:957–966. [DOI] [PubMed] [Google Scholar]
- 114.Vogt NM, Kerby RL, Dill-McFarland KA, et al. Gut microbiome alterations in Alzheimer’s disease. Sci Rep 2017;7:13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhuang ZQ, Shen LL, Li WW, et al. Gut microbiota is altered in patients with Alzheimer’s disease. J Alzheimers Dis 2018;63:1337–1346. [DOI] [PubMed] [Google Scholar]
- 116.Lim C, Hammond CJ, Hingley ST, et al. Chlamydia pneumoniae infection of monocytes in vitro stimulates innate and adaptive immune responses relevant to those in Alzheimer’s disease. J Neuroinflammation 2014; 11:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ishida N, Ishihara Y, Ishida K, et al. Periodontitis induced by bacterial infection exacerbates features of Alzheimer’s disease in transgenic mice. NPJ Aging Mech Dis 2017;3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harach T, Marungruang N, Duthilleul N, et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci Rep 2017;7:41802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Szabady RL, Louissaint C, Lubben A, et al. Intestinal P-glycoprotein exports endocannabinoids to prevent inflammation and maintain homeostasis. J Clin Invest 2018;128:4044–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hippe B, Zwielehner J, Liszt K, et al. Quantification of butyryl CoA:acetate CoA-transferase genes reveals different butyrate production capacity in individuals according to diet and age. FEMS Microbiol Lett 2011; 316:130–135. [DOI] [PubMed] [Google Scholar]
- 121.Aaron L The anti-neo-epitopes tissue and microbial transglutaminases are new reliable serological markers in celiac disease diagnosis. Journal of Clinical & Cellular Immunology 2017;08. [Google Scholar]
- 122.Sampson TR, Debelius JW, Thron T, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 2016;167:1469–1480. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chandra R, Hiniker A, Kuo YM, et al. α-Synuclein in gut endocrine cells and its implications for Parkinson’s disease. JCI Insight 2017;2:e92295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maeda Y, Kurakawa T, Umemoto E, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol 2016;68:2646–2661. [DOI] [PubMed] [Google Scholar]
- 125.Lew KN, Starkweather A, Cong X, et al. A mechanistic model of gut-brain axis perturbation and high-fat diet pathways to gut microbiome homeostatic disruption, systemic inflammation, and type 2 diabetes. Biol Res Nurs 2019;21:384–399. [DOI] [PubMed] [Google Scholar]
- 126.Grosicki GJ, Fielding RA, Lustgarten MS. Gut microbiota contribute to age-related changes in skeletal muscle size, composition, and function: biological basis for a gut-muscle axis. Calcif Tissue Int 2018;102:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Scott AJ, Alexander JL, Merrifield CA, et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 2019;68:1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fried LP, Tangen CM, Walston J, et al. Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci 2001;56:146–156. [DOI] [PubMed] [Google Scholar]
- 129.Walston J, Hadley EC, Ferrucci L, et al. Research agenda for frailty in older adults: toward a better understanding of physiology and etiology: summary from the American Geriatrics Society/National Institute on Aging Research Conference on Frailty in Older Adults. J Am Geriatr Soc 2006;54:991–1001. [DOI] [PubMed] [Google Scholar]
- 130.Ofori-Asenso R, Chin KL, Mazidi M, et al. Global incidence of frailty and prefrailty among community-dwelling older adults: a systematic review and meta-analysis. JAMA Netw Open 2019;2:e198398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kojima G Prevalence of frailty in nursing homes: a systematic review and meta-analysis. J Am Med Dir Assoc 2015;16:940–945. [DOI] [PubMed] [Google Scholar]
- 132.Rockwood K, Song X, MacKnight C, et al. A global clinical measure of fitness and frailty in elderly people. CMAJ 2005;173:489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pijpers E, Ferreira I, Stehouwer CD, et al. The frailty dilemma. Review of the predictive accuracy of major frailty scores. Eur J Intern Med 2012;23:118–123. [DOI] [PubMed] [Google Scholar]
- 134.O’Caoimh R, Costello M, Small C, et al. Comparison of frailty screening instruments in the emergency department. Int J Environ Res Public Health 2019;16:3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Walston J, Buta B, Xue QL. Frailty screening and interventions: considerations for clinical practice. Clin Geriatr Med 2018;34:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dent E, Kowal P, Hoogendijk EO. Frailty measurement in research and clinical practice: a review. Eur J Intern Med 2016;31:3–10. [DOI] [PubMed] [Google Scholar]
- 137.Wallis SJ, Wall J, Biram RW, et al. Association of the clinical frailty scale with hospital outcomes. QJM 2015; 108:943–949. [DOI] [PubMed] [Google Scholar]
- 138.Hao Q, Zhou L, Dong B, et al. The role of frailty in predicting mortality and readmission in older adults in acute care wards: a prospective study. Sci Rep 2019;9:1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang MC, Li TC, Li CI, et al. Frailty, transition in frailty status and all-cause mortality in older adults of a Taichung community-based population. BMC Geriatr 2019;19:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Salminen M, Viljanen A, Eloranta S, et al. Frailty and mortality: an 18-year follow-up study among Finnish community-dwelling older people. Aging Clin Exp Res 2020;32:2013–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang X, Dou Q, Zhang W, et al. Frailty as a predictor of all-cause mortality among older nursing home residents: a systematic review and meta-analysis. J Am Med Dir Assoc 2019;20:657–663.e4. [DOI] [PubMed] [Google Scholar]
- 142.Luo H, Lum TY, Wong GH, et al. Predicting adverse health outcomes in nursing homes: a 9-year longitudinal study and development of the FRAIL-Minimum Data Set (MDS) Quick Screening Tool. J Am Med Dir Assoc 2015; 16:1042–1047. [DOI] [PubMed] [Google Scholar]
- 143.Clegg A, Young J, Iliffe S, et al. Frailty in elderly people. Lancet 2013;381:752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ho YY, Matteini AM, Beamer B, et al. Exploring biologically relevant pathways in frailty. J Gerontol A Biol Sci Med Sci 2011;66:975–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Di Iorio A Serum IL-1b levels in health and disease: a population-based study. ‘The InCHIANTI study’. Cytokine 2003;22:198–205. [DOI] [PubMed] [Google Scholar]
- 146.Sandmand M, Bruunsgaard H, Kemp K, et al. High circulating levels of tumor necrosis factor-alpha in centenarians are not associated with increased production in T lymphocytes. Gerontology 2003;49:155–160. [DOI] [PubMed] [Google Scholar]
- 147.Di Sabatino A, Lenti MV, Cammalleri L, et al. Frailty and the gut. Dig Liver Dis 2018;50:533–541. [DOI] [PubMed] [Google Scholar]
- 148.Hamilton-Miller JM. Probiotics and prebiotics in the elderly. Postgrad Med J 2004;80:447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rondanelli M, Giacosa A, Faliva MA, et al. Review on microbiota and effectiveness of probiotics use in older. World J Clin Cases 2015;3:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ling WH, Korpela R, Mykkanen H, et al. Lactobacillus strain GG supplementation decreases colonic hydrolytic and reductive enzyme activities in healthy female adults. J Nutr 1994;124:18–23. [DOI] [PubMed] [Google Scholar]
- 151.Hosoda M, He F, Kojima T, et al. Effect of administration of milk fermented with Lactobacillus acidophilus LA-2 on fecal mutagenicity and microflora in the human intestine. J Nutritional Food 1998;1:1–9. [DOI] [PubMed] [Google Scholar]
- 152.Cremonini F, Di Caro S, Nista EC, et al. Meta-analysis: the effect of probiotic administration on antibiotic-associated diarrhoea. Aliment Pharmacol Ther 2002; 16:1461–1467. [DOI] [PubMed] [Google Scholar]
- 153.McFarland LV. Meta-analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol 2006;101:812–822. [DOI] [PubMed] [Google Scholar]
- 154.Oksanen PJ, Salminen S, Saxelin M, et al. Prevention of travellers’ diarrhoea by Lactobacillus GG. Ann Med 1990;22:53–56. [DOI] [PubMed] [Google Scholar]
- 155.Frech TM, Khanna D, Maranian P, et al. Probiotics for the treatment of systemic sclerosis-associated gastrointestinal bloating/distention. Clin Exp Rheumatol 2011; 29:S22–S25. [PubMed] [Google Scholar]
- 156.Kontiokari T, Sundqvist K, Nuutinen M, et al. Randomised trial of cranberry-lingonberry juice and Lactobacillus GG drink for the prevention of urinary tract infections in women. BMJ 2001;322:1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.de los Angeles Pineda M, Thompson SF, Summers K, et al. A randomized, double-blinded, placebo-controlled pilot study of probiotics in active rheumatoid arthritis. Med Sci Monit 2011;17:CR347–CR354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Davidson LE, Fiorino AM, Snydman DR, et al. Lactobacillus GG as an immune adjuvant for live-attenuated influenza vaccine in healthy adults: a randomized double-blind placebo-controlled trial. Eur J Clin Nutr 2011;65:501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Morrow LE, Kollef MH, Casale TB. Probiotic prophylaxis of ventilator-associated pneumonia: a blinded, randomized, controlled trial. Am J Respir Crit Care Med 2010; 182:1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Morris MC, Tangney CC, Wang Y, et al. MIND diet associated with reduced incidence of Alzheimer’s disease. Alzheimers Dement 2015;11:1007–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kivipelto M, Solomon A, Ahtiluoto S, et al. The Finnish Geriatric Intervention Study to Prevent Cognitive Impairment and Disability (FINGER): study design and progress. Alzheimers Dement 2013;9:657–665. [DOI] [PubMed] [Google Scholar]
- 162.Xu R, Wang Q. Towards understanding brain-gut-microbiome connections in Alzheimer’s disease. BMC Syst Biol 2016;10(Suppl 3):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Table References
- 1.Kant R, Rasinkangas P, Satokari S, et al. Genome sequence of the butyrate-producing anaerobic bacterium Anaerostipes hadrus PEL 85. Genome Announc 2015;3 e00224–00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ai D, et al. Identifying gut microbiota associated with colorectal cancer using a zero-inflated lognormal model. Front Microbiol 2019;10:826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q, et al. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci Rep 2016;6:27572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eeckhaut V, et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut 2013;62:1745–1752. [DOI] [PubMed] [Google Scholar]
- 5.Ling Z, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol 2014;80:2546–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan SH, et al. Wheat bran promotes enrichment within the human colonic microbiota of butyrate-producing bacteria that release ferulic acid. Environ Microbiol 2016;18:2214–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu S, Yi J, Zhang YG, et al. Leaky intestine and impaired microbiome in an amyotrophic lateral sclerosis mouse model. Physiol Rep 2015;3:e12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haran JP, et al. Alzheimer’s disease microbiome is associated with dysregulation of the anti-inflammatory P-glycoprotein pathway. mBio 2019;10 e00632–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Finegold SM, Song Y, et al. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 2008;58:1896–1902. [DOI] [PubMed] [Google Scholar]
- 10.Ozato N, et al. Blautia genus associated with visceral fat accumulation in adults 20–76 years of age. NPJ Biofilms Microbiomes 2019;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue R, et al. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci Biotechnol Biochem 2016; 80:2450–2458. [DOI] [PubMed] [Google Scholar]
- 12.Duytschaever G, et al. Dysbiosis of bifidobacteria and Clostridium cluster XIVa in the cystic fibrosis fecal microbiota. J Cyst Fibros 2013;12:206–215. [DOI] [PubMed] [Google Scholar]
- 13.Miyake S, et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS One 2015;10:e0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labus JS, et al. Evidence for an association of gut microbial Clostridia with brain functional connectivity and gastrointestinal sensorimotor function in patients with irritable bowel syndrome, based on tripartite network analysis. Microbiome 2019;7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houser MC, Tansey MG. The gut-brain axis: is intestinal inflammation a silent driver of Parkinson’s disease pathogenesis? NPJ Parkinson Dis 2017;3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Zhu H, Zhang L, et al. The intestinal microbiome and Alzheimer’s disease: a review. Animal Model Exp Med 2018;1:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerhardt S, Mohajeri MH. Changes of colonic bacterial composition in Parkinson’s disease and other neurodegenerative diseases. Nutrients 2018;10:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eid HM, et al. Significance of microbiota in obesity and metabolic diseases and the modulatory potential by medicinal plant and food ingredients. Front Pharmacol 2017;8:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engels C, Ruscheweyh HJ, Beerenwinkel N, et al. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front Microbiol 2016;7:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi K, et al. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn’s disease. Digestion 2016;93:59–65. [DOI] [PubMed] [Google Scholar]
- 21.Stern JM, et al. Evidence for a distinct gut microbiome in kidney stone formers compared to non-stone formers. Urolithiasis 2016;44:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eppinga H, et al. Similar depletion of protective Faecalibacterium prausnitzii in psoriasis and inflammatory bowel disease, but not in hidradenitis suppurativa. J Crohns Colitis 2016;10:1067–1075. [DOI] [PubMed] [Google Scholar]
- 23.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014; 15:382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machiels K, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014;63:1275–1283. [DOI] [PubMed] [Google Scholar]
- 25.Tamanai-Shacoori Z, et al. Roseburia spp.: a marker of health? Future Microbiol 2017;12:157–170. [DOI] [PubMed] [Google Scholar]
- 26.Biagi E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 2010;5:e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guohong L, Qingxi Z, Hongyun W. Characteristics of intestinal bacteria with fatty liver diseases and cirrhosis. Ann Hepatol 2019;18:796–803. [DOI] [PubMed] [Google Scholar]
- 28.Castaner O, et al. The gut microbiome profile in obesity: a systematic review. Int J Endocrinol 2018;2018:4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrov VA, et al. Analysis of gut microbiota in patients with Parkinson’s disease. Bull Exp Biol Med 2017; 162:734–737. [DOI] [PubMed] [Google Scholar]
- 30.Nagao-Kitamoto H, Kamada N. Host-microbial crosstalk in inflammatory bowel disease. Immune Netw 2017;17:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haran JP, Bucci V, Dutta P, et al. The nursing home elder microbiome stability and associations with age, frailty, nutrition, and physical location. J Med Microbiol 2018;67:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kameyama K, Itoh K. Intestinal colonization by a Lachnospiraceae bacterium contributes to the development of diabetes in obese mice. Microbes Environ 2014;29:427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.San-Juan-Vergara H, et al. A Lachnospiraceae-dominated bacterial signature in the fecal microbiota of HIV-infected individuals from Colombia, South America. Sci Rep 2018;8:4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardiner BJ, et al. Clinical and microbiological characteristics of Eggerthella lenta bacteremia. J Clin Microbiol 2015;53:626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balakrishnan B, Luckey D, Taneja V. Autoimmunity-associated gut commensals modulate gut permeability and immunity in humanized mice. Mil Med 2019; 184:529–536. [DOI] [PubMed] [Google Scholar]
- 36.Jackson MA, et al. Signatures of early frailty in the gut microbiota. Genome Med 2016;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pallister T, et al. Untangling the relationship between diet and visceral fat mass through blood metabolomics and gut microbiome profiling. Int J Obes (Lond) 2017; 41:1106–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin HE, et al. Exercise, the gut microbiome, and frailty. Ann Geriatr Med Res 2019;23:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghavami SB, et al. Alterations of the human gut Methanobrevibacter smithii as a biomarker for inflammatory bowel diseases. Microb Pathog 2018;117:285–289. [DOI] [PubMed] [Google Scholar]
- 40.Hall AB, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 2017;9:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chua HH, et al. Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology 2018; 154:154–167. [DOI] [PubMed] [Google Scholar]
- 42.Kim JW, Kwok SK, Choe JY, et al. Recent advances in our understanding of the link between the intestinal microbiota and systemic lupus erythematosus. Int J Mol Sci 2019;20:4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zarrati M, et al. Effects of probiotic yogurt on fat distribution and gene expression of proinflammatory factors in peripheral blood mononuclear cells in overweight and obese people with or without weight-loss diet. J Am Coll Nutr 2014;33:417–425. [DOI] [PubMed] [Google Scholar]
- 44.Allen SJ, et al. Lactobacilli and bifidobacteria in the prevention of antibiotic-associated diarrhoea and Clostridium difficile diarrhoea in older inpatients (PLACIDE): a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2013;382:1249–1257. [DOI] [PubMed] [Google Scholar]
- 45.Wei H, et al. Antitumor mechanisms of bifidobacteria. Oncol Lett 2018;16:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makrgeorgou A, et al. Probiotics for treating eczema. Cochrane Database Syst Rev 2018;11:Cd006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duranti S, et al. Elucidating the gut microbiome of ulcerative colitis: bifidobacteria as novel microbial biomarkers. FEMS Microbiol Ecol 2016;92:fiw191. [DOI] [PubMed] [Google Scholar]
- 48.Whalen JG, Mully TW, English JC. Spontaneous Citrobacter freundii infection in an immunocompetent patient. Arch Dermaol 2007;143:124–125. [DOI] [PubMed] [Google Scholar]
- 49.Hidron AI, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol 2008;29:996–1011. [DOI] [PubMed] [Google Scholar]
- 50.Kasahara K, et al. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol 2018;3:1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo W, et al. Roseburia intestinalis supernatant ameliorates colitis induced in mice by regulating the immune response. Mol Med Rep 2019;20:1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep 2016;6:28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bajer L, et al. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol 2017;23:4548–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aaron L The anti-neo-epitopes tissue and microbial transglutaminases are new reliable serological markers in celiac disease diagnosis. J Clin Cellular Immunol 2017;08. [Google Scholar]
- 55.Xu Y, et al. Function of Akkermansia muciniphila in obesity: interactions with lipid metabolism, immune response and gut systems. Front Microbiol 2020; 11:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scher JU, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol 2015;67:128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collado MC, Derrien M, Isolauri E, et al. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 2007;73:7767–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis-Richardson AG, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol 2014;5:678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vatanen T, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 2016;165:842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finegold SM, et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis 2002;35:S6–S16. [DOI] [PubMed] [Google Scholar]
- 61.Bercik P, et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol Motil 2011; 23:1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horta-Baas G, et al. Intestinal dysbiosis and rheumatoid arthritis: a link between gut microbiota and the pathogenesis of rheumatoid arthritis. J Immunol Res 2017; 2017:4835189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao S, et al. A gut microbiota-targeted dietary intervention for amelioration of chronic inflammation underlying metabolic syndrome. FEMS Microbiol Ecol 2014;87:357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rooks MG, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J 2014;8:1403–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mathur R, Barlow GM. Obesity and the microbiome. Exp Rev Gastroenterol Hepatol 2015;9:1087–1099. [DOI] [PubMed] [Google Scholar]
- 66.Larsen N, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 2010;5:e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomez-Arango LF, et al. Increased systolic and diastolic blood pressure is associated with altered gut microbiota composition and butyrate production in early pregnancy. Hypertension 2016;68:974–981. [DOI] [PubMed] [Google Scholar]
- 68.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo XM, et al. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl Environ Microbiol 2018;84 e02288–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shahi SK, Freedman SN, Mangalam AK. Gut microbiome in multiple sclerosis: the players involved and the roles they play. Gut Microbes 2017;8:607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Picchianti-Diamanti A, et al. Analysis of gut microbiota in rheumatoid arthritis patients: disease-related dysbiosis and modifications induced by etanercept. Int J Mol Sci 2018;19:2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang K, et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep 2019; 26:222–235.e225. [DOI] [PubMed] [Google Scholar]
- 73.Wick EC, Sears CL. Bacteroides spp. and diarrhea. Curr Opin Infect Dis 2010;23:470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lukiw WJ. Bacteroides fragilis lipopolysaccharide and inflammatory signaling in Alzheimer’s disease. Front Microbiol 2016;7:1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwong TNY, et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology 2018;155:383–390.e388. [DOI] [PubMed] [Google Scholar]
- 76.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 2007;20:593–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Galanis E Campylobacter and bacterial gastroenteritis. CMAJ 2007;177:570–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nyati KK, Nyati R. Role of Campylobacter jejuni infection in the pathogenesis of Guillain-Barré syndrome: an update. BioMed Res Int 2013;2013:852195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phongsisay V The immunobiology of Campylobacter jejuni: innate immunity and autoimmune diseases. Immunobiology 2016;221:535–543. [DOI] [PubMed] [Google Scholar]
- 80.Levine UY, Looft T, Allen HK, et al. Butyrate-producing bacteria, including mucin degraders, from the swine intestinal tract. Appl Environ Microbiol 2013;79:3879–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Looft T, Levine UY, Stanton TB. Cloacibacillus porcorum sp. nov., a mucin-degrading bacterium from the swine intestinal tract and emended description of the genus Cloacibacillus. Int J Syst Evol Microbiol 2013; 63:1960–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Domingo MC, et al. Cloacibacillus sp., a potential human pathogen associated with bacteremia in Quebec and New Brunswick. J Clin Microbiol 2015;53:3380–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goldstein EJC, Citron DM, Peraino VA, et al. Desulfovibrio desulfuricans bacteremia and review of human desulfovibrio infections. J Clin Microbiol 2003;41:2752–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loubinoux J, Bronowicki JP, Pereira IA, et al. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol Ecol 2002;40:107–112. [DOI] [PubMed] [Google Scholar]
- 85.Shahnawaz M, Soto C. Microcin amyloid fibrils A are reservoir of toxic oligomeric species. J Biol Chem 2012; 287:11665–11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Friedland RP. Mechanisms of molecular mimicry involving the microbiota in neurodegeneration. J Alzheimer Dis 2015;45:349–362. [DOI] [PubMed] [Google Scholar]
- 87.Friedland RP, Chapman MR. The role of microbial amyloid in neurodegeneration. PLoS Pathogens 2017;13: e1006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vading M, Nauclér P, Kalin M, et al. Invasive infection caused by Klebsiella pneumoniae is a disease affecting patients with high comorbidity and associated with high long-term mortality. PLoS One 2018;13: e0195258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang L, et al. The association of HLA-B27 and Klebsiella pneumoniae in ankylosing spondylitis: a systematic review. Microb Pathog 2018;117:49–54. [DOI] [PubMed] [Google Scholar]
- 90.Tsoi H, et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology 2017;152:1419–1433.e1415. [DOI] [PubMed] [Google Scholar]
- 91.Murdoch DA. Gram-positive anaerobic cocci. Clin Microbiol Rev 1998;11:81–120. [DOI] [PMC free article] [PubMed] [Google Scholar]


