Abstract
Upon viral infection of a host cell, each virus starts a program to generate many progeny viruses. Although viruses interact with the host cell in numerous ways, one critical step of the virus life cycle is the expression of viral proteins, which are synthesized by the host ribosomes in conjunction with host translation factors. Here we review different mechanisms viruses have evolved to effectively seize host cell ribosomes, the roles of specific ribosomal proteins and their posttranslational modifications on viral RNA translation or the cellular response to infection. We will further highlight ribosomal proteins with extraribosomal function during viral infection and put the knowledge of ribosomal proteins during viral infection into the larger context of ribosome-related diseases, known as ribosomopathies.
Graphical Abstract
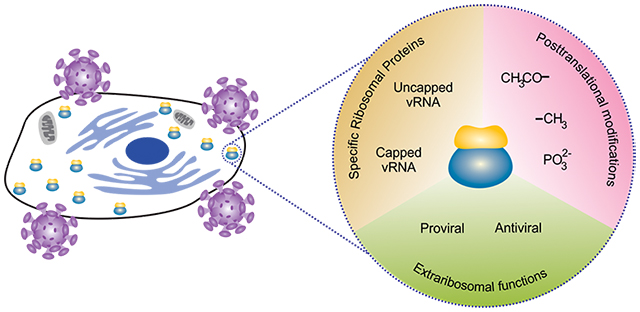
All viruses require host cell ribosomes for expression of viral proteins, starting a battle between the host cell and the virus for ribosomes. Viruses usurp specific ribosomal proteins to facilitate viral protein biosynthesis. Viruses directly or indirectly induce posttranslational modifications, or utilize extraribosomal functions in other steps of the virus life cycle. In contrast, cells try to control and limit viruses’ ribosome access, either by altering posttranslational modifications of ribosomes, by blocking viral replication, or by blocking other steps of the virus life cycle where ribosomal proteins function outside of the ribosome.
1. Introduction
Viruses extensively interact with the host cell to evade the host immune responses and to stimulate their own gene expression. Viral genomes are either composed of DNA or RNA, which is surrounded by a protein shell forming the capsid. Despite the kind of viral genome, whether DNA or RNA, whether positive-sense or negative-sense, viruses share certain steps of the virus life cycle. Specifically, all viruses must enter the host cell, they must replicate their genomes, synthesize viral proteins and leave the host cell after assembling new virions. The order of these steps can differ between viruses and depends on their genome and other virus characteristics. Although the genome size between viruses varies greatly, viruses require the host cells’ translation machinery, specifically ribosomes and translation factors, to synthesize viral proteins. Hence, viruses have evolved strategies to effectively compete for cellular ribosomes and translation factors.
1.1. Canonical cap-dependent translation
Translation of eukaryotic messenger RNAs (mRNAs) has been very well reviewed elsewhere and is only briefly recapped here (Hershey et al., 2018; Sonenberg & Hinnebusch, 2009). Cellular mRNAs are transcribed and processed in the nucleus, and contain a 7-methyl-guanosine (m7G) cap (Figure 1). This m7G cap not only stabilizes the mRNA and prevents its degradation, it also facilitates efficient translation by binding to the cap binding protein eukaryotic translation initiation factor eIF4E (Gingras et al., 1999). Next, other translation initiation factors, including eIF4G, eIF2 and the initiator tRNA, eIF3 and the 40S ribosomal subunit are recruited, and through ATP-dependent ribosome scanning the 40S ribosomal subunit reaches the AUG start codon. Following start codon recognition, the 60S subunit is then recruited to form the elongation competent 80S complex (Hershey et al., 2018; Sonenberg & Hinnebusch, 2009). However, there are exceptions to this canonical pathway, which are often referred to as cap-independent translation initiation mechanisms. As cellular ribosomes are critical for synthesis of viral proteins, viruses have evolved viral mRNA features and translation initiation mechanisms to seize these molecular protein synthesis machines.
Figure 1:
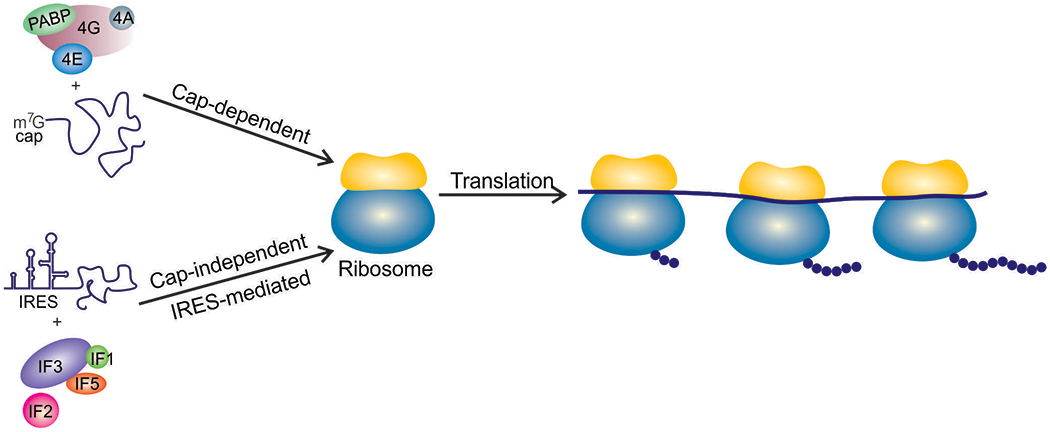
Canonical cap-dependent translation requires a m7G-capped mRNA and the eIF4F complex. A large group of viruses does not contain a cap and instead uses an IRES and a subset of translation factors for initiation.
1.2. The viral RNA 5′ end
Viral genomes are diverse and include positive and negative strand RNA genomes, single-stranded and double-stranded DNA genomes, non-segmented and segmented genomes. Hence, viral mRNAs also come in different appearances and use different 5′ ends and cap variants (Table 1). Viruses with a double-stranded DNA (dsDNA) genome such as herpesviruses use the cellular transcription machinery in the nucleus to synthesize m7G-capped viral mRNAs (Glaunsinger, 2015). Cytoplasmic DNA viruses such as poxviruses, and RNA viruses do not use the cellular transcription machinery, but encode their own viral transcription and capping proteins. Influenza virus, a member of the Orthomyxoviridae, which contains a segmented single-stranded negative RNA genome, replicates in the nucleus and uses a mechanism termed “cap-snatching” (De Vlugt et al., 2018). The cellular mRNA cap and a portion of RNA downstream of the cap is cleaved off and used as a template for viral RNA synthesis, yielding viral RNAs with a cellular m7G cap. In contrast, flaviviruses including Dengue virus and Zika virus encode their own methyltransferase enzyme that methylates the terminal nucleotide, creating a RNA cap-mimic (Issur et al., 2009).
Table 1:
Viruses use different cap structures and mechanisms for translation initiation
| Cap structure | Mechanism | Examples | References |
|---|---|---|---|
| m7G cap | Cellular transcription machinery | Herpesviruses (HSV, CMV, EBV) | (Glaunsinger, 2015) |
| m7G cap | Viral transcription and capping machinery | Poxviruses | (Meade et al., 2019) |
| m7G cap | Cap-snatching | Orthomyxoviridae (influenza virus) | (De Vlugt et al., 2018) |
| m7G cap | Encoded methyltransferase | Flaviviruses (Dengue virus, zika virus, West Nile virus) | (Issur et al., 2009) |
| VPg | VPg and IRES | Picornaviridae, Caliciviridae (poliovirus, norovirus) | (S. K. Jang et al., 1988; Pelletier & Sonenberg, 1989) |
| No cap | 5′ IRES | Hepatitis C virus | (Fraser & Doudna, 2007) |
| No cap | 3′ CITE | Barley yellow dwarf virus | (Allen et al., 1999) |
In contrast to these strategies that use either a cellular m7G cap or mimic the cap, many viral 5′ ends do not contain a m7G cap and instead are uncapped or contain a viral protein linked to the genome (VPg). The VPg acts as a proteinaceous cap-substitute and is found in viruses belonging to the Picornaviridae, Caliciviridae, and Potyviridae (Goodfellow, 2011). The VPg was shown to directly interact with eIF4E, eIF3, eIF4GI, and the 40S ribosomal subunit and to compete for m7G cap binding (Daughenbaugh et al., 2003, 2006; de Oliveira et al., 2019; Goodfellow et al., 2005).
1.3. IRESs and CITEs
Some viral RNAs contain untranslated genomic elements, which recruit the ribosome independent of the mRNA cap. RNA structural elements upstream of the open reading frame are referred to as internal ribosome entry sites (IRESs) (Figure 1). IRESs, which are found in several virus families including the Picornaviridae, Caliciviridae, Flaviviridae, and Dicistroviridae, are classified based on their overall secondary structure and the requirement for translation initiation factors (recently reviewed in (Mailliot & Martin, 2018; Stern-Ginossar et al., 2018)). While IRESs are typically found in the 5′ untranslated region (UTR), their position can vary, but they are always located upstream of the open reading frame they control. In contrast, many RNA plant viruses contain a cap-independent translation element (CITE) in the 3′ UTR to facilitate viral translation (Nicholson & White, 2011).
In addition to using cellular caps or cap-analogs and RNA structures, more recent evidence has shown that viruses have evolved to directly target ribosomes and translation factors. During viral infection, the availability of cellular translation factors and their posttranslational modifications change, which affects viral protein biosynthesis, and was recently reviewed elsewhere (Au & Jan, 2014; Stern-Ginossar et al., 2018). Since viruses critically rely on the host ribosomes, they have evolved unique methods of commandeering the cellular ribosomes and the ribosome biogenesis machinery. During infection with human cytomegalovirus (CMV) ribosome abundance increases. Bianco and Mohr showed that the increase in components of the translation machinery unexpectedly aided the host cell in efforts to elicit an immune response rather than benefiting the virus (Bianco & Mohr, 2019). The virally expressed protein pUL31, which reorganizes nucleolar proteins and is necessary to increase viral replication, may represent counterefforts by the virus (Westdorp et al., 2017). However, changes in rRNA expression, ribosome biogenesis, and their impact on viral infection is a complex topic that deserves a review on its own. Here we review how viruses utilize specific ribosomal proteins to enhance viral RNA translation, how posttranslational modifications of ribosomal proteins are altered in response to virus infection, and the evidence for extra-ribosomal functions of ribosomal proteins during other steps of the virus life cycle. Better understanding how interactions with ribosomal proteins contribute to ribosome recruitment during viral infection will offer insights into the molecular mechanisms through which ribosomes are usurped and paves the way for therapeutics against different types of viruses. In addition, this information can also be applied to research on ribosomopathies, diseases in which mutations in specific ribosomal proteins cause tissue-specific phenotypes.
2. Viruses exploit the function of specific ribosomal proteins
Concerning the function and impact of specific ribosomal proteins, two contrasting models are currently being discussed. In the “preference” model, certain RNAs have a higher dependency on specific ribosomal proteins. However, translation can still occur in the absence of the ribosomal protein, albeit less efficiently. In the “specialization” model, certain RNAs exclusively rely on a specific ribosomal protein. In the absence of such ribosomal protein, these mRNAs cannot be translated. In this section, we highlight the ribosomal proteins that support translation of viral RNAs and conclude by discussing evidence for preferential or specialized translation.
2.1. Translation of capped viral RNAs
Double stranded DNA viruses, such as herpesvirus 1 (HSV-1), cytomegalovirus (CMV), and Epstein Barr virus (EBV), use the cellular transcription machinery to synthesize m7G-capped viral mRNAs (Glaunsinger, 2015; Vincent et al., 2016). Several reports have implicated a role for the ribosomal protein Receptor for Activated C Kinase 1 (RACK1) during EBV infection (Baumann et al., 2000; Smith et al., 2000; Tardif et al., 2002). RACK1 interacts with various cellular signaling proteins, such as protein kinase C and Src tyrosine kinase, and is involved in many signaling events. Indirectly through these interactions but also directly as an integral part of the ribosome, RACK1 controls protein biosynthesis (Figure 2) (Adams et al., 2011; Calamita et al., 2018; Nielsen et al., 2017). Glutathione S-transferase (GST) pull-down of ubinuclein (Ubn-1) followed by proteomic analysis revealed an interaction of Ubn-1 with RACK1 in the cytoplasm. This interaction was further validated by co-immunoprecipitation and immunofluorescence assays in human epithelial HT29 cells (Lupo et al., 2012). In the context of EBV infection, Gruffat et al. proposed that Ubn-1 acts as a modulator of the EBV productive lifecycle in gastric carcinoma cell line AGS. Ubn-1 not only interacted with EBV transcription factor EB1, also referred to as BLZF1, Ubn-1 was relocalized to the tight junctions of nondividing AGS cells, possibly to allow for activation of the lytic cycle of EBV (Gruffat et al., 2011; Lupo et al., 2012). Based on these observations, it was suggested that the cytoplasmic interaction of Ubn-1 and RACK1 might shape the cellular organization and subcellular localization of proteins to aid EBV infection. Interestingly, an earlier study had suggested that RACK1 may act as a BZLF1 binding partner independent of its PKC-related functions (Baumann et al., 2000). Research by Smith et al. (2000) showed that the A73 protein, expressed from EBV viral RNA, co-precipitated with endogenous RACK1 in 293 cells and interacted with RACK1 in both EBV-infected cells and EBV-associated cancers. Through the A73-RACK1 interaction, A73 might modulate RACK1 function to support cell growth, possibly stimulating the unrestricted growth of EBV-associated cancers (Smith et al., 2000). Since members of the herpesvirus family, including EBV and CMV, have been associated with different types of cancers (Alibek et al., 2014) it is not surprising that related viruses utilize RACK1 in a pro-viral manner. Identifying viral binding partners of RACK1 and drug inhibitors of RACK1 in HSV-1-infected cells to reduce viral protein production, provides evidence that RACK1 inhibition could be employed as an antiviral therapy for the Herpesviridae (Baumann et al., 2000; Ullah et al., 2019).
Figure 2:
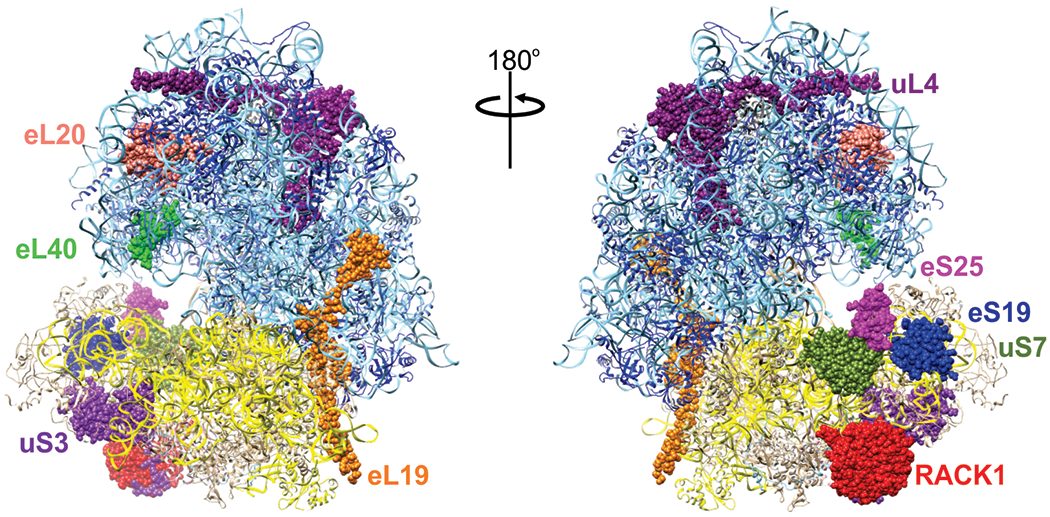
Front and back view of the human 80S ribosome highlighting ribosomal proteins directly targeted by viruses to facilitate translation of viral mRNAs. Human ribosome adapted from PDB 5T2C (X. Zhang et al., 2016)
Although vesicular stomatitis virus (VSV) is an ssRNA virus with a negative-strand genome its viral mRNAs are capped and polyadenylated like host mRNAs. In VSV, but also in other Mononegavirales viruses such as measles virus, rabies virus, and Ebola virus, the viral L protein caps the viral RNAs through a unique capping mechanism. Instead of using a GTP substrate like the host guanylyltransferase, the L protein attaches a GDP molecule to the 5’ terminal nucleotide (Gupta et al., 2002). Despite the production of viral capped and polyadenylated transcripts, VSV utilizes eL401 (RPL40) to ensure translation of viral mRNAs (Figure 2) (Lee et al., 2013). Because the stress-response gene DDR2 also relies on eL40 for translation VSV likely exploits an existing cellular translation pathway. The detailed mechanism, however, is unknown, and so it is possible that the viral mRNAs may directly bind to eL40 or that eL40 may allow for necessary conformational changes of the 60S subunit in order to permit preferential translation of the viral genome, but more investigations are required to determine how eL40 facilitates virus′ access to the ribosome during VSV infection.
Other negative strand ssRNA viruses such as Influenza virus and Hantavirus do not encode a viral protein to cap viral mRNAs. Instead, cellular mRNAs are cleaved downstream of the cap. The 3′ hydroxyl group of these short, capped mRNA pieces are then used to initiate viral transcription, generating hybrid RNA molecules of cellular and viral sequences. The hantavirus nucleocapsid protein (N) plays a critical role during viral replication and translation. Mass spectrometry experiments to identify interacting proteins suggested that the N protein may interact with uS15 (RPS13), eS17 (RPS17), and eS19 (RPS19). To determine the exact interacting proteins, the 18S rRNA of the 40S ribosomal subunit was nuclease-digested, followed by immunoprecipitation of the N protein. Using this approach, only eS19 was detected by immunoblot analysis (Cheng et al., 2011; Haque & Mir, 2010; Muyangwa et al., 2015). ES19 is a ribosomal protein located at the head of the 40S subunit (Figure 2), which rotates during mRNA binding (Spahn et al., 2001). Because of its ability to bind to both eS19 and to the viral 5′ UTR, the N protein has been suggested to be important for loading viral RNAs onto the ribosome (Cheng et al., 2011). In retroviruses, overexpression of uL4 (RPL4) (Figure 2) has been shown to cause a Gag protein processing defect in the Moloney murine leukemia virus (Green et al., 2012). Altering retroviral readthrough and frameshifting changed the ratio of Gag and Pol proteins, which ultimately decreased the number of correctly formed virus particles.
Since arboviruses such as Dengue virus (DV), Zika virus (ZIKV), West Nile virus (WNV), and Yellow Fever virus (YFV) pose a major health threat, recent research has focused on identifying host factors required by these viruses. Targeting these host factors might be one approach of treating an infected individual and to prevent further transmission. Several genome-wide screens identified certain ribosomal proteins as potential critical flaviviral host factors (Hafirassou et al., 2017; Marceau et al., 2016; Petrova et al., 2019). In addition to dolichyl-diphosphooligosaccharide-protein glycosyltransferase non-catalytic subunit (DDOST), which is part of the oligosaccharyltransferase (OST) complex, a gain-of-function screen identified uS3 (RPS3) and eL19 (RPL19) (Figure 2) as critical components for a chimeric yellow fever (YFV)/ West Nile (WNV) reporter genome (Petrova et al., 2019). Knockdown of uS3 and eL19 decreased translation of YFP and WNV viral proteins, while their overexpression slightly increased translation of a YFV/WNV chimeric GFP reporter genome by moderate two-fold; overexpression of DDOST increase translation by about five-fold. A haploid screen and a CRISPR genome-wide screen for DV host factors also identified and validated members of the OST complex, but only the haploid screen identified eS25 (RPS25) as a ribosomal protein required for DV (Marceau et al., 2016) (Figure 2). Interestingly, a siRNA screen for NS1-interacting proteins also revealed the OST complex, eS25, and RACK1 as binding partners (Hafirassou et al., 2017). Knockdown of eS25 decreased translation of YFV, DV, WNV, and ZIKV (Hafirassou et al., 2017; Petrova et al., 2019). Separation of early translation and translation during the replication phase further revealed that genome levels of a DV Renilla luciferase reporter virus were unaffected during early translation, but strongly decreased during the replication phase. In contrast, knockdown of RACK1 decreased DV, WNV, and ZIKV, but neither YFV nor HSV or VSV were affected (Hafirassou et al., 2017; Petrova et al., 2019). The observation that depletion of RACK1 did not affect HSV disagrees with a previously-mentioned study that used a RACK1 inhibitor (Ullah et al., 2019). Although the RACK1 inhibitor decreased HSV protein at a similar multiplicity of infection as used in the depletion studies, it cannot be excluded that the RACK1 inhibitor may bind and inhibit another cellular phosphoprotein and cause uncharacterized, secondary effects.
Both RACK1 and eS25 have been previously implicated in the translation of RNAs with IRESs (Hertz et al., 2013; LaFontaine et al., 2020; Landry et al., 2009; Majzoub et al., 2014). Flaviviruses were previously shown to contain a cap to stabilize the mRNA, to facilitate translation and to evade the host immune response (L. Liu et al., 2010; Saeedi & Geiss, 2013), but the need for RACK1 and eS25 suggested a possible IRES-mediated translation initiation mechanism. In contrast to an earlier report by Edgil et al. (Edgil et al., 2006), recent evidence supports the idea that the DV and ZIKV 5′ UTRs indeed contain an IRES (Song et al., 2019). The existence of an IRES in the DV and ZIKV 5′ UTRs plausibly explains why these viruses require RACK1 and eS25, however, neither the DV nor ZIKV IRES structures and translation initiation mechanisms have been characterized. We further do not know the significance of cap-dependent and cap-independent viral RNA translation on the virus life cycle. These important questions must be addressed to uncover the intricate translation regulation of these flaviviruses. Findings that a G protein-coupled receptor kinase 2 GRK2 (Le Sommer et al., 2012), an essential factor for Flaviviridae infection, phosphorylated ribosomal protein P2 (RPLP2) (Freeman et al., 2002), emphasized the role of the acidic ribosomal stalk proteins P1 (RPLP1) and P2 (RPLP2) during viral infection. Metabolic labeling confirmed earlier reports that loss of P1 or P2 may not affect global translation (Campos et al., 2017; M. et al., 1995; Martinez-Azorin et al., 2008). However, depletion of P1 and P2 prevented the early translation of a DENV-2 Renilla luciferase reporter genome. Because the ribosome stalk is close to the binding site of translation elongation factors, a potential involvement of the stalk in viral translation elongation has been proposed (Campos et al., 2017).
2.2. Translation of uncapped viral RNAs
Some viruses can bypass the canonical cap-dependent mode of translation initiation and use a cap-independent initiation pathway involving viral RNA structures. In this section, we will discuss examples of different viral IRESs and how certain ribosomal proteins facilitate IRES-mediated and cap-independent translation. IRESs are categorized due to their secondary structure and the requirement for translation initiation factors (Mailliot & Martin, 2018; Stern-Ginossar et al., 2018). Type 1 IRESs, which contain five core domains, are found in poliovirus and related enteroviruses and require all eIFs except for eIF4E. However, while the cap-binding function of eIF4E is not required for IRES function, eIF4E has recently been shown to stimulate eIF4A activity (Avanzino et al., 2017). Cardioviruses and Aphtoviruses including encephalomyocarditis virus (EMCV) and Foot-and-mouth disease virus (FMDV), respectively, use type 2 IRESs that forgo the scanning process. A type 3 IRES is not only found in hepatitis C virus (HCV), but also in many picornaviruses that contain an HCV-like IRES (Asnani et al., 2015; Hashem et al., 2013). These IRESs contain a compact fold and mostly require eIF2, eIF3, and eIF5 for translation initiation. Viruses that belong to the Dicistroviridae have been shown to contain two IRESs. The IRES at the 5′ end is a type 3 IRES (Gross et al., 2017), while the IRES in the intergenic region that separates the two open reading frames is a type 4 IRES. Type 4 IRESs, which include the Cricket Paralysis Virus (CrPV) IRES, are the smallest viral IRESs and function independently of any translation factors (Deniz et al., 2009; Jan & Sarnow, 2002). The compact IGR IRES contains three pseudoknots and mimics the initiator tRNA in the ribosomal P-site to initiate translation.
In an initial genome-wide screen 66 ribosomal proteins were identified as necessary host factors for viral IRES-mediated translation of Drosophila C virus (DCV) (Cherry et al., 2005). Due to recent advances in the field of cryo-EM, we now have high-resolution structures of mammalian ribosomes bound to the CrPV and HCV IRESs that support the observations of the genome-wide screen through structural evidence (Muhs et al., 2011; Nishiyama et al., 2007; Pisareva et al., 2018; Yamamoto et al., 2015). Based on these structures, certain ribosomal proteins may directly interact with IRESs. Specifically, ribosomal proteins eS25 and uS7 (Figure 2) are in the direct vicinity of domain II of the HCV IRES and eS28 is close to domain IV (Yamamoto et al., 2015). Of those proteins, uS7 has been crosslinked to the HCV IRES (Fukushi et al., 2001) and eS25 was shown not only to be critical for the translation of the CrPV and HCV IRES, but also for EMCV and PV IRESs (Hertz et al., 2013; Landry et al., 2009). Retroviruses such as human T-cell leukemiavirus type 1 (HTLV-1) utilize both cap-dependent and cap-independent translation. HTLV-1, which requires a ribosome scanning step to recognize the initiation codon by the 40S subunit, also showed dependency on eS25 for HTLV-1 IRES translation (Olivares et al., 2014). Within the family of retroviruses, human immunodeficiency virus (HIV) behaves in a similar fashion needing eS25 for efficient IRES-mediated translation. The HIV-1 IRES mediates the expression of the Gag protein, which is crucial for forming the viral core proteins. These retroviruses serve as examples for viruses that rely on eS25 for IRES-mediated translation but not for cap-dependent translation (Carvajal et al., 2016; Olivares et al., 2014)
The 40S ribosomal subunit protein RACK1 is involved in numerous signaling pathways (Adams et al., 2011; Gandin et al., 2013; Nielsen et al., 2017) (Figure 2). For an invading virus, RACK1 is hence an enticing target to commandeer during infection (Adams et al., 2011; Calamita et al., 2018; Dobrikov et al., 2018b, 2018a; Gallo et al., 2018). In addition to its potential role during EBV and HSV infection, RACK1 has been shown to be targeted by viruses initiating translation via an IRES. As such, RACK1 has been shown to be crucial for PV, HCV, DCV and CrPV 5′ IRES-mediated translation (LaFontaine et al., 2020; Majzoub et al., 2014). Although many viruses seize both eS25 and RACK1 for IRES-mediated translation, their roles in translation initiation appear to be distinct. Ribosomes lacking eS25 were unable to bind to the CrPV IGR IRES causing a loss in CrPV IGR IRES-mediated translation (Landry et al., 2009). In contrast, loss of RACK1 had no effect on CrPV IGR IRES-mediated translation (Johnson et al., 2019; Majzoub et al., 2014). The differences have been attributed to eIF3, which is not required by the CrPV IGR IRES. Indeed, in a cryoEM structure of a class III IRES in complex with the 40S ribosomal subunit and eIF3, domain III of the IRES interacted with the eIF3 complex (Hashem et al., 2013; Neupane et al., 2020; Siridechadilok et al., 2005). Further, Majzoub et al. showed that loss of the eIF3j subunit, which does not affect cell viability, decreased HCV IRES-mediated translation (Majzoub et al., 2014). In the context of a viral infection, RACK1 may live a double life by having both proviral and antiviral functions due to its wide array of functions inside the cell (Adams et al., 2011; Calamita et al., 2018; Nielsen et al., 2017). Based on currently available studies, cell-specific changes to cap-dependent translation of cellular mRNAs involved in the immune response cannot be entirely ruled out (Dobrikov et al., 2018b, 2018a; Gallo et al., 2018).
Most reported findings demonstrated binding of IRES structures to the small ribosomal subunit, which agrees with the early proposed translation initiation mechanism (Fraser et al., 2009; Fraser & Doudna, 2007). However, both CrPV IGR and HCV IRESs can capture 80S ribosomes, highlighting the importance of ribosomal proteins of the large subunit in CrPV IGR and HCV IRES-mediated translation (Petrov et al., 2016; Yokoyama et al., 2019). Hence, these IRESs might also directly interact with ribosomal proteins of the large subunit during IRES-mediated initiation. Indeed, the conserved L1.1 loop region within the CrPV IRES mimics E-site tRNA interactions with the uL1 (RPL10A) stalk protein of the large subunit to aid in 80S assembly (C. J. Jang et al., 2009). Dhar et al. were the first to show a requirement for a protein of the 60S ribosomal subunit in HCV IRES-mediated translation (Dhar et al., 2006). Through in vitro translation assays, they showed that eL20 (RPL18a) (Figure 2) not only interacted with the HCV IRES RNA but that addition of eL20 protein to in vitro translation extracts moderately stimulated HCV IRES activity (Dhar et al., 2006).
Although 3′ CITE elements serve a similar purpose as the 5′ IRESs, it is surprising that no specific ribosomal protein has been identified that facilitates 3′ CITE translation. Turnip crinkle virus (TCV) and Pea Enation Mosaic Virus (PEMV) represent two types of ribosome-binding 3’ CITEs that act as translational enhancers by either binding solely the 60S ribosomal subunit or the 40S, 60S, and 80S, respectively (Gao et al., 2013; Stupina et al., 2008). In the case of the PEMV 3′ CITE, a kissing loop hairpin and T-shaped structures mediate long range RNA interactions with the 5’ end, necessary for accumulation of PEMV in protoplasts (Gao et al., 2014). However, only some specific ribosomal proteins have been so far shown to be critical for plant virus translation. Cauliflower mosaic virus for example terminates and reinitiates translation of its viral RNA. The viral protein P6, critical for re-initiation, interacts with the plant ribosomal proteins eL13 (RPL13) (Bureau et al., 2004), eL18 (RPL18) (Bureau et al., 2004; Leh et al., 2000), and eL24 (RPL24) (Park et al., 2001). Several uncapped plant virus RNAs are covalently linked to the viral protein VPg at the 5’ end. The VPg not only protects the RNA from degradation, but also competes with capped cellular mRNAs for translation factor binding to promote translation of the viral RNA. Specifically, uL10 (RPLP0) acted synergistically with VPg to enhance translation of the plant virus Potato Virus A (PVA) (Hafren et al., 2013). Because the acidic P stalk proteins including RPLP0 are near the ribosomal A-site, it is possible that VPg and uL10 enhance aminoacyl-tRNA recruitment during early translation elongation (Inglis et al., 2019).
The mechanisms at which viruses have evolved to employ ribosomal proteins are diverse and extensive. Depending on the type of viral RNA circulating in a host cell during infection, unique ribosomal proteins can aid in translation initiation to express viral proteins. Identification of these ribosomal proteins and ribosome-associated proteins can provide further insights into viral translation initiation. Most cited studies use siRNAs or shRNAs to deplete ribosomal proteins. While depletion is often efficient, residual protein levels make it difficult to distinguish preferential from specialized translation. Studies using complete knockout cell lines provide evidence supporting the preferential translation model, as loss of eS25 or RACK1 decreased DV, WNV, ZIKV, and PV genome translation respectively (LaFontaine et al., 2020; Marceau et al., 2016). Based on the current evidence the preferential translation model appears to be supported, however, coexisting mechanisms cannot yet be ruled out. In addition to utilizing certain ribosomal proteins, viruses can manipulate the host cell machinery even further by posttranslationally modifying ribosomal proteins to their advantage.
3. Viruses induce changes in posttranslational modifications of ribosomal proteins
Posttranslational modifications are versatile in nature and lead to the expansion of the library of genetically encoded 20 amino acids. These posttranslational modifications either alter the functional groups of the amino acids or are different functional groups (or complex molecules or even small peptide chains). Enzymatic activity adds the modifications via a covalent linkage to the amino acids, including the N- and C-termini of the amino acids chain. Mammalian cells are capable of carrying out complex posttranslational modifications. Viral infection has been shown to alter the posttranslational modifications of many ribosomal proteins and translation factors in host cells (Figure 3). These modifications are often observed early during infection as they tend to target specific signaling pathways of the host and to regulate protein folding (Kaerlein & Horak, 1978). Since modifications of translation factors have been recently reviewed (Stern-Ginossar et al., 2018), we will focus on posttranslational modifications of ribosomal proteins during viral infection and their function.
Figure 3:
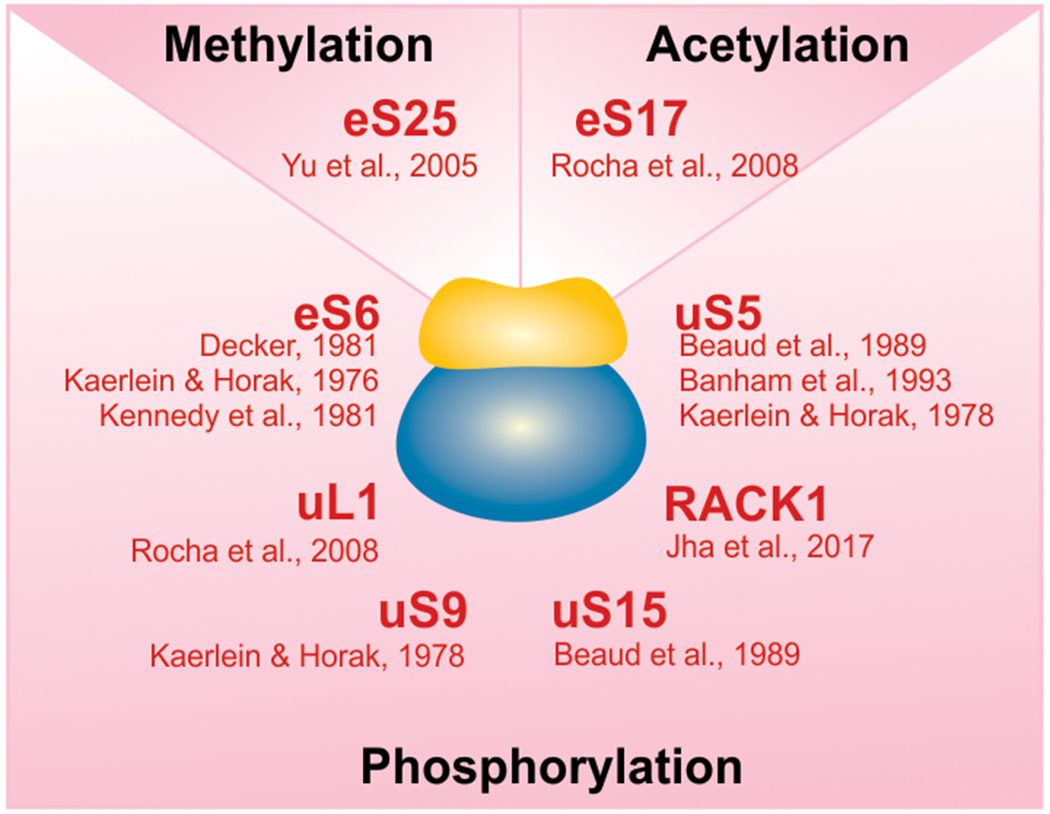
The PTM status of certain ribosomal proteins is altered in response to viral infection. Ribosomal proteins have been found to largely become phosphorylated, but changes in methylations and acetylations have also been reported.
Phosphorylation of ribosomal proteins is one of the most widely known posttranslational modifications in pathogen-infected cells (Figure 3) as well as uninfected cells. In addition to cellular kinases, protein kinases of viral origin can phosphorylate some ribosomal proteins. Phosphorylation due to virion-associated protein kinases tends to occur in the early stages of viral infection (Buendia et al., 1987; Kaerlein & Horak, 1978). The phosphorylation of the ribosomal protein eS6 is one of the most frequently observed posttranslational modifications in uninfected cells. Following stimulation of the mTOR pathway, S6 Kinase subsequently phosphorylates eS6 (Magnuson et al., 2012). However, the function of eS6 phosphorylation in cells has remained enigmatic, with some suggesting it function to finetune the cellular translation response (Meyuhas, 2008). In response to viral infection with vaccinia virus, herpes simplex virus-1, avian sarcoma virus, and pseudorabies virus, eS6 phosphorylation levels were elevated as compared to the uninfected cells (Decker, 1981; Kaerlein & Horak, 1976, 1978; Kennedy et al., 1981). The enhanced phosphorylation could have been caused by viral or host cellular kinases. To prove that virion-associated protein kinases were responsible for this phosphorylation, protein synthesis was inhibited in the host cell. Despite this inhibition phosphorylated eS6 was detected, suggesting that virion-associated protein kinases caused the phosphorylation (Kaerlein & Horak, 1976, 1978). However, the activity of virion-associated protein kinases was finally proven several years later. Upon addition of recombinantly expressed vaccinia virus B1R protein to purified 40S ribosomal subunits, several ribosomal proteins were phosphorylated in response, strongly suggesting the involvement of viral kinases (Banham et al., 1993). In addition to eS6 phosphorylation, vaccinia virus infection in HeLa cells induced the phosphorylation of ribosomal proteins uS5 (RPS2) (Banham et al., 1993; Beaud et al., 1989; Kaerlein & Horak, 1978), uS9 (RPS16) (Kaerlein & Horak, 1978), uS2 (RPSA) (Banham et al., 1993) and uS15 (Beaud et al., 1989) (Figure 3).
In contrast to the increase in eS6 phosphorylation observed during vaccinia virus infection, alphavirus infection drastically reduced eS6 phosphorylation (Montgomery et al., 2006). Of the two vaccinia-virus encoded kinases, the B1 kinase appears to be the main kinase responsible for phosphorylating ribosomal proteins. In addition to phosphorylating uS5 and uS2, B1 kinase also phosphorylates RACK1 (Jha et al., 2017). Through phosphorylation of a STSS motif in a loop in mammalian RACK1, the loop becomes negatively charged and mimics the negatively charged Asp/Glu residues in Arabidopsis thaliana RACK1. Interestingly, poxviruses and plant mRNAs both use polyA leader sequences. Mutagenesis of the STSS motif to a negatively charged EEEE sequence in the RACK1 loop increased translation of a luciferase reporter mRNA with a polyA leader, indicating that in the case of RACK1, phosphorylation specifically increases the translation of late vaccinia viral RNAs with polyA-leaders (Jha et al., 2017). Phosphorylation of ribosomal proteins also extends to plant viruses. NSP-interacting kinases (NIKs) mediate an antiviral response in plants and have been shown to specifically phosphorylate uL1 (RPL10A) in vitro (Rocha et al., 2008). Interestingly, loss of uL1 increased susceptibility to geminivirus infection suggesting that uL1 phosphorylation might be part of the plant’s antiviral response. ES6 (RPS6) phosphorylation is also targeted by plant viruses (Rajamäki et al., 2017; Yang et al., 2009). The VPg proteins of the potyviruses Turnip mosaic virus (TuMV) and Potato Virus A (PVA) interact with S6 Kinase (S6K) to avoid eS6 phosphorylation (Rajamäki et al., 2017). Silencing of eS6 in plants prevented TuMV RNA accumulation, while the Tobacco mosaic virus (TMV) RNA remained unaffected by loss of eS6. Although changes in phosphorylation of ribosomal protein during viral infection have been widely described, the function of these phosphorylated ribosomal proteins in infected cells remains mostly elusive. The ability to perform precise genome editing to mutate the phosphorylated amino acid residues provides new approaches to revisit these earlier findings and to dissect their molecular function.
In addition to phosphorylation by virus-encoded kinases, other posttranslational modifications on ribosomal proteins have been detected (Figure 3). Lysine acetylation is a reversible posttranslational modification, which neutralizes the positive charge of this amino acid and targets proteins of large macromolecular complexes, including ribosomes, for the regulation of gene expression (Choudhary et al., 2009). Inhibition of histone deacetylases increased acetylation of 15 different ribosomal proteins including acetylation of eL24 at K27, but not of eL24 at K93 (Wilson-Edell et al., 2014). Increased acetylation of eL24 K27 decreased association of the modified eL24 with polysomes and pointed towards acetylations functioning as regulatory posttranslational modifications in gene expression. Indeed, increased acetylation of microtubule proteins during viral infection with adenovirus, herpes simplex virus, and HIV1 supported the early post entry stages of infection (Sabo et al., 2013; Sadoul et al., 2011). Although ribosomal protein acetylation has not been described in the context of a viral infection, it is striking that a stretch of lysine residues within eS17 found inserted into a hepatitis E virus nonstructural protein increased the virus host range (Kenney & Meng, 2015). While unclear if these lysines are modified, it is possible that viruses also cause changes in ribosome acetylation.
Mass spectrometry is an important technique to identify posttranslational modifications other than phosphorylation. An earlier study by Yu et al. combined the top-down and bottom-up approach of mass spectrometry to identify multiple ribosomal proteins with their posttranslational modifications (Yu et al., 2005). They identified about 31 ribosomal proteins along with their posttranslational modifications and elucidated the variations in posttranslational modification in virus-infected and uninfected cells. Certain ribosomal proteins were observed to differ in their posttranslational modifications. HCV-IRES bound uS14 (RPS29) had no posttranslational modification whereas in the free 40S ribosomal subunit it contained a disulfide bond. Similarly, free 40S-bound eS25 was unmethylated, while eS25 in the HCV-IRES:40S complex contained mono- and dimethylated eS25. Since modifications like methylation are crucial for cellular localization and for regulating RNA binding proteins (Bedford & Richard, 2005), technological advances for the identification of PTMs are urgently needed. A recently used three-tiered mass spectrometry approach addressed ribosome heterogeneity and determined ribosome interacting factors (Van De Waterbeemd et al., 2018). Bottom-up LC-MS/MS focused on the posttranslational modifications of ribosomal proteins while the top-down LC-MS/MS revealed crosstalk between the multiple posttranslational modifications of ribosomal proteins. Finally, MS was also utilized to analyze the intact ribonucleoprotein complexes. Through this approach, cysteine methylation on human eS27 (RPS27) was observed. While combinatorial MS approaches can identify posttranslational modifications, several disadvantages remain. The use of trypsin on positively charged ribosomal proteins often generates very small peptides which do not have the optimal length for MS detection. Further, the presence of PTMs might affect the cleavage efficiency of the protease. To overcome these issues, novel techniques have been developed. PTMScan® Direct technology is an immunoaffinity based mass spectrometry approach to determine the post-translationally modified proteins (Stokes et al., 2012). The proteins are first digested into peptides, and peptides containing the PTM of interest are immunopurified and identified by MS. PTMselect is a recently developed software to optimize protein PTM and target PTM positions coverage (Perchey et al., 2019). In vitro translation systems may also prove to be useful. Zeenko et al. used a cell-free translation system to study protein PTMs (Zeenko et al., 2008). The in vitro system recapitulated the selectivity as compared with in vivo translation as the envelope protein gp120 of HIV type1 became glycosylated at multiple sites, while a signal peptide from β-lactamase was lost.
In conclusion, ribosome-modifications in response to viral infection have been observed, but are still limited to a small number of viruses. Unfortunately, the function of most modifications has remained largely unexplored. New mass spectrometry and gene editing technologies will allow researchers to investigate ribosomal PTMs to identify the nature and location of the modifications, the enzymes responsible for the modification, and to determine the molecular functions and mechanisms of these modifications.
4. Viruses exploit extra-ribosomal functions of ribosomal proteins
Many structural and non-structural viral proteins interact with cellular proteins to modulate cellular functions. These interactions are commonly identified by yeast-two-hybrid (Y2H) or by pull-down experiments combined with mass spectrometry analysis and confirmed via co-immunoprecipitation. Interactions of viral proteins with ribosomal proteins can either support or inhibit virus proliferation (Figure 4). However, because viral translation and replication are ultimately linked, determining whether a ribosomal protein truly functions outside of the ribosome has proven difficult. As such, depletion of a ribosomal protein that decreases virus proliferation could indicate an involvement in virus replication and virion assembly, but unless tested specifically, a role for the ribosomal protein in viral RNA translation cannot be excluded. In this section, we will discuss the evidence for ribosomal proteins with extra-ribosomal functions during viral infection.
Figure 4:
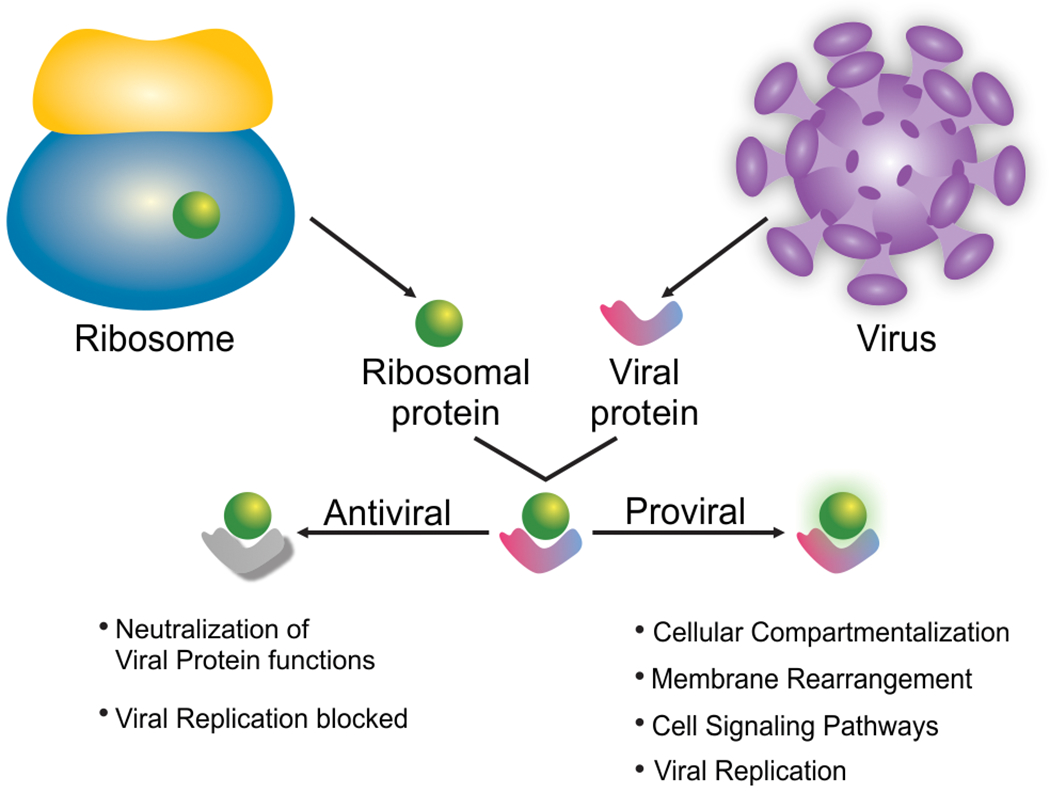
The binding of a ribosomal protein with extra-ribosomal function to a viral protein can either neutralize it and block virus proliferation, or it can help to activate cellular or viral pathways and functions to promote virus proliferation.
Several examples suggesting that ribosomal proteins act in an antiviral manner (Figure 4) support evidence for extra-ribosomal functions. FMDV structural protein VP1 has been shown to directly bind to ribosomal protein uS2 (RPSA) by Y2H and to have an antiviral function (Zhu et al., 2019). Zhu et al. showed further that FMDV upregulates the MAPK pathway and that overexpression of uS2 suppressed MAPK phosphorylation signaling and FMDV replication. Hence, binding of VP1 to uS2 likely blocks the inhibition of the MAPK pathway and boosts FMDV replication. While the antiviral function uS2 is blocked by FMDV VP1, in rabies virus (RABV) overexpression of uL6 (RPL9) decreased viral P protein expression and replication measured by a RABV-Renilla luciferase reporter construct (Y. Li et al., 2016). While uL6 overexpression caused a decrease in RABV RNA copy numbers in the absence or presence of the translation elongation inhibitor cycloheximide it had no effect on VSV RNA copy numbers, suggesting that the observed effect is independent of translation. Because the P protein has known roles in replication, these results are in agreement with a model that the interaction of uL6 with P could prevent P from interacting with other viral proteins during replication. However, whether RABV has evolved a specific mechanism to counteract inhibition through uL6 is currently not known. Another protein with antiviral function is uS10 (RPS20). During infection with classical swine fever virus (CSFV), viral protein Npro interacted with uS10 and targeted it for proteasomal degradation (Lv et al., 2017). Knockdown of uS10 decreased expression of Toll-like receptor 3 (TLR3) and promoted CSFV replication. Because TLR3 overexpression in conjunction with uS10 knockdown did not promote CSFV replication Lv. et al. concluded that uS10 likely indirectly modulates CSFV replication through TLR3. A better example of a ribosomal protein with extra-ribosomal function is ribosomal protein L22 (eL22). In Epstein-Barr virus (EBV)-infected cells, ribosomal protein eL22 is detached from the 40S ribosomal subunit and bound to the non-coding viral RNA EBER-1 (Fok et al., 2006; Houmani et al., 2009; Toczyski et al., 1994). Because EBER-1 also binds and inhibits PKR, Elia et al. determined that eL22 and PKR compete for a common EBER-1 binding site (Elia et al., 2004). By binding EBER-1 eL22 likely prevents PKR inhibition during viral infection. Further, higher expression of EBER-1 correlated with an increased cell growth, possibly by overcoming eL22 inhibition and PKR inactivation (Houmani et al., 2009). EBV infection has been shown to increase expression of another ribosomal protein uL4, which binds to the Epstein-Barr virus nuclear antigen 1 (EBNA1) (Shen et al., 2016). Depletion of uL4 decreased the ability of EBNA1 to bind an episomal vector, and specifically uL4 residues K380 and K393 are critical to mediate deposition of H3K4me2 modifications on the episomal vector to enable EBNA1-mediated transcription.
Evidence for extra-ribosomal function of proviral ribosomal proteins (Figure 4) mainly comes from findings that these ribosomal proteins interact with viral non-structural proteins. Because depletion of ribosomal proteins often decreases viral RNA translation it is difficult to distinguish if this effect is due to their general role in protein biosynthesis or due to the interaction with the non-structural viral proteins. TNF receptor-associated factor 2 (TRAF2) is a known regulator of cellular signaling pathways such as NF-κB and has previously been shown to promote vaccinia virus entry into the host cell (Haga et al., 2014).
RACK1, a ribosomal protein known to come off the ribosome (Johnson et al., 2019) and with links to cellular signaling, increased expression and phosphorylation of TRAF2, which in turn enhanced replication of porcine reproductive and respiratory syndrome virus (PRRSV) (X. Liu et al., 2019). Since RACK1 can stimulate translation in a PKC β II-dependent manner (Dobrikov et al., 2018a, 2018b), it is critical to show what triggers the increased expression and phosphorylation of TRAF2 to confirm that RACK1 functions as an extra-ribosomal protein in this case. While the evidence for an extra-ribosomal function of ribosomal protein uL30 (RPL7) only relies on the observation of its interaction with white spot syndrome virus (WSSV) VP51 protein and the finding that treatment with anti-uL30 antibody significantly delayed WSSV infection (Q. hui Liu et al., 2015), uL30 has also been shown to contain strong DNA/RNA chaperone activity. Together with the HIV-1 Gag protein, uL30 might drive rapid nucleic acid hybridization independent of its function as a ribosomal protein (Mekdad et al., 2016).
Surprisingly, a siRNA screen for proteins interacting with flavivirus NS1 (Hafirassou et al., 2017; Petrova et al., 2019) and a NS1 overexpression and co-immunoprecipitation approach followed by mass spectrometry (Cervantes-Salazar et al., 2015) yielded divergent results. While all three types of genetic screens (siRNA, CRISPR, and haploid screen) identified the OST complex as a critical complex for Dengue virus (DV) infection, the NS1 immunoprecipitation approach did not, possibly because the NS1 interaction with the OST complex is transient. The other major differences are the number of identified ribosomal proteins. Genetic screens only found eS25, RACK1, eL19, and uS3, indicating high stringency, while the NS1 interactome identified 33 ribosomal proteins, possibly due to interactions with cellular ribosomes. Since NS1 is a non-structural protein, Cervantes-Salazar et al. suggested that NS1 might interact with proteins of extra-ribosomal function such as eL18, uL30, eS1 (RPS3A), and eL20 (RPL18A). EL18 not only interacted with NS1, it also relocalized to the perinuclear region upon DV infection, where it co-localized with NS1 (Cervantes-Salazar et al., 2015). While knockdown of eL18 strongly inhibited DV in cells, depletion of eL18 had minimal effects on cellular translation as determined by metabolic labeling. Further, extra-ribosomal functions for uS3 and eL19 during YFV infection have also been suggested (Petrova et al., 2019). Flaviviruses cause major rearrangement in cellular membranes for replication (Apte-Sengupta et al., 2014; Gillespie et al., 2010), and the ZIKV NS1 crystal structure revealed a large hydrophobic surface to mediate membrane interactions (Brown et al., 2016). Thus, it is feasible that flaviviruses capture ribosomes in or near their replication complexes to increase viral protein synthesis. Similarly, influenza virus has been shown to capture and target ribosomes to autophagosomes (Becker et al., 2018).
Interactions of Chikungunya virus (CHIKV) nonstructural proteins 2 (nsP2), 3 (nsP3) and 4 (nsP4) with several ribosomal proteins of the 40S and 60S ribosomal subunits were identified by Y2H, and the interaction of eS6 with nsP2 was validated (Montgomery et al., 2006). Montgomery et al. observed that reduction of eS6 protein levels diminished expression from alphavirus subgenomic mRNAs, while the effect on cellular translation was minimal. Because expression of CHIKV nsP2 alone promoted mRNA translation (Bouraï et al., 2012) it is possible that the nsP2-eS6 interaction acts directly on translation, although an extra-ribosomal function cannot be excluded. Similarly, eL18 and uL4 directly interacted with the viral N protein of rice stripe virus (S. Li et al., 2018) and VP3 protein of infectious bursal disease virus (Chen et al., 2016), respectively. Although knockdown of these proteins inhibited viral replication, the evidence for extra-ribosomal function is inconclusive because the effects on viral and cellular translation have not been separately evaluated. Other interactions of viral non-structural proteins with ribosomal proteins have also been reported (Kim et al., 2016; Zhang et al., 2009), but the importance of the interaction to the virus life cycle have not been examined. Overall, there is strong evidence for interactions of ribosomal proteins with non-structural viral proteins, yet the molecular details of how extra-ribosomal proteins function on viruses are mostly vague. Few examples convincingly indicate antiviral effects due to extra-ribosomal protein functions. The majority of the observations of extra-ribosomal functions could be explained by changes in viral translation, changes in translation of specific antiviral cellular mRNAs, or changes in global translation. Further, many transformed cell lines overexpress ribosomal proteins, which in normal cells would be rapidly degraded (Guimaraes & Zavolan, 2016). These overexpressed ribosomal proteins can readily interact with viral proteins, possibly causing experimental artifacts and overall questioning the practice of using transformed cell lines in such interaction studies.
5. Conclusion
As diverse viruses are, the mechanisms they have evolved to seize cellular ribosomes for expression for viral proteins are equally diverse. Here we summarized several examples of viruses targeting specific ribosomal proteins for translation initiation. Ribosomal RNA is another intriguing target for viruses. A recent article by Netzband and Pager reviewed the RNA epitranscriptome during viral infection (Netzband & Pager, 2020). Although RNA modifying enzymes have been identified and viral RNA modifications clearly impact many viruses, the role of rRNA modifications during viral infection is largely unknown. Studies reporting differential expression of rRNA variants and functional importance of rRNA modifications were performed in the absence of a viral infection (Jack et al., 2011; Parks et al., 2018). Together, these studies highlight the importance to investigate rRNA modifications during infection, a field that has not yet received much attention.
The examples highlighted in this article span different virus families and various mechanisms of translation initiation. Numerous bacteriophages and Finkel–Biskis–Reilly murine sarcoma virus, a retrovirus, also appear to encode select ribosomal proteins (Mizuno et al., 2019), highlighting the evolutionary pressure to maintain the control over the host cell translation machinery. Further, although ribosomal proteins have been described to be posttranslationally modified during viral infection, the extent to which ribosomes become modified, whether these modifications are performed by the viral or cellular enzymes, and the functions of these modifications are largely unknown. New and improved mass spectrometry and genome editing techniques are required to address these questions and to characterize the molecular mechanism of these modifications on viral and cellular protein biosynthesis. Lastly, extra-ribosomal functions of ribosomal proteins are mostly based on interactions with viral nonstructural proteins. For many examples, an involvement in viral protein biosynthesis cannot yet be ruled out. Because viral translation and replication are interdependent, limiting the analysis to the step of viral protein biosynthesis is required to obtain conclusive results.
Although ribosomal proteins are thought to be expressed in equal stoichiometry, recent reports suggested that several ribosomal proteins are only expressed in substoichiometric quantities (Simsek et al., 2017). Further, loss or mutation of certain ribosomal proteins has been associated with a set of human diseases referred to as ribosomopathies (McCann & Baserga, 2013). Haploinsufficiency of the majority of ribosomal proteins causes bone marrow failure and Diamond–Blackfan anemia as well as other tissue-specific defects. Since many viruses target the same ribosomal proteins in different tissues, it is likely that viruses have co-opted already existing mechanisms for mRNA translation, and that the mechanistic insights into viral protein biosynthesis will contribute to our understanding of ribosome-related diseases.
Acknowledgements
We thank Dr. Cara Pager and Dr. Andrew Berglund for critically reading the manuscript and helpful discussions.
Funding information
This work was supported by a National Institutes of Health Grant R03 AI144839 to GF, and start-up funds from the University at Albany, State University of New York to GF.
Footnotes
A new nomenclature for ribosomal proteins has been established (Ban et al., 2014). In this review, we use the new nomenclature, but include the nomenclature as referred to in publications in parentheses.
Conflict of interest: The authors declare that they have no conflict of interest with the contents of this article. The content is solely the responsibility of the authors.
References
- Adams DR, Ron D, & Kiely PA (2011). RACK1, A multifaceted scaffolding protein: Structure and function. In Cell Communication and Signaling (Vol. 9). 10.1186/1478-811X-9-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibek K, Baiken Y, Kakpenova A, Mussabekova A, Zhussupbekova S, Akan M, & Sultankulov B (2014). Implication of human herpesviruses in oncogenesis through immune evasion and supression. In Infectious Agents and Cancer (Vol. 9, Issue 1). 10.1186/1750-9378-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Wang S, & Miller WA (1999). Barley yellow dwarf virus RNA requires a cap-independent translation sequence because it lacks a 5’ cap. Virology, 253(2), 139–144. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9918872 [DOI] [PubMed] [Google Scholar]
- Apte-Sengupta S, Sirohi D, & Kuhn RJ (2014). Coupling of replication and assembly in flaviviruses In Current Opinion in Virology (Vol. 9, pp. 134–142). Elsevier B.V; 10.1016/j.coviro.2014.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnani M, Kumar P, & Hellen CUT (2015). Widespread distribution and structural diversity of Type IV IRESs in members of Picornaviridae. Virology, 478, 61–74. 10.1016/j.virol.2015.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au HH, & Jan E (2014). Novel viral translation strategies. Wiley Interdisciplinary Reviews: RNA, 5(6), 779–801. 10.1002/wrna.1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanzino BC, Fuchs G, & Fraser CS (2017). Cellular cap-binding protein, eIF4E, promotes picornavirus genome restructuring and translation. Proceedings of the National Academy of Sciences of the United States of America, 114(36). 10.1073/pnas.1704390114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, McAlear MA, Moore PB, Noller HF, Ortega J, Panse VG, Ramakrishnan V, Spahn CM, Steitz TA, Tchorzewski M, … Yusupov M (2014). A new system for naming ribosomal proteins. Current Opinion in Structural Biology, 24, 165–169. 10.1016/j.sbi.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banham AH, Leader DP, & Smith GL (1993). Phosphorylation of ribosomal proteins by the vaccinia virus B1R protein kinase. FEBS Letters, 321(1), 27–31. 10.1016/0014-5793(93)80614-z [DOI] [PubMed] [Google Scholar]
- Baumann M, Gires O, Kolch W, Mischak H, Zeidler R, Pich D, & Hammerschmidt W (2000). The PKC targeting protein RACK1 interacts with the Epstein-Barr virus activator protein BZLF1. European Journal of Biochemistry, 267(12), 3891–3901. 10.1046/j.1432-1327.2000.01430.x [DOI] [PubMed] [Google Scholar]
- Beaud G, Masse T, Madjar J-J, & Leader DP (1989). Identification of induced protein kinase activities specific for the ribosomal proteins uniquely phosphorylated during infection of HeLa cells with vaccinia virus. FEBS Letters, 259(1), 10–14. 10.1016/0014-5793(89)81482-6 [DOI] [PubMed] [Google Scholar]
- Becker AC, Gannagé M, Giese S, Hu Z, Abou-Eid S, Roubaty C, Paul P, Bühler L, Gretzmeier C, Dumit VI, Kaeser-Pebernard S, Schwemmle M, Münz C, & Dengjel J (2018). Influenza a virus induces autophagosomal targeting of ribosomal proteins. Molecular and Cellular Proteomics, 17(10), 1909–1921. 10.1074/mcp.RA117.000364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, & Richard S (2005). Arginine methylation: An emerging regulator of protein function. In Molecular Cell (Vol. 18, Issue 3, pp. 263–272). 10.1016/j.molcel.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Bianco C, & Mohr I (2019). Ribosome biogenesis restricts innate immune responses to virus infection and DNA. ELife, 8 10.7554/eLife.49551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouraï M, Lucas-Hourani M, Gad HH, Drosten C, Jacob Y, Tafforeau L, Cassonnet P, Jones LM, Judith D, Couderc T, Lecuit M, André P, Kümmerer BM, Lotteau V, Desprès P, Tangy F, & Vidalain P-O (2012). Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component. Journal of Virology, 86(6), 3121–3134. 10.1128/JVI.06390-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WC, Akey DL, Konwerski JR, Tarrasch JT, Skiniotis G, Kuhn RJ, & Smith JL (2016). Extended surface for membrane association in Zika virus NS1 structure In Nature Structural and Molecular Biology (Vol. 23, Issue 9, pp. 865–867). Nature Publishing Group; 10.1038/nsmb.3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia B, PERSON-FERNANDEZ A, BEAUD G, & MADJAR J-J (1987). Ribosomal protein phosphorylation in vivo and in vitro by vaccinia virus. European Journal of Biochemistry, 162(1), 95–103. 10.1111/j.1432-1033.1987.tb10547.x [DOI] [PubMed] [Google Scholar]
- Bureau M, Leh V, Haas M, Geldreich A, Ryabova L, Yot P, & Keller M (2004). P6 protein of Cauliflower mosaic virus, a translation reinitiator, interacts with ribosomal protein L13 from Arabidopsis thaliana. Journal of General Virology, 85(12), 3765–3775. 10.1099/vir.0.80242-0 [DOI] [PubMed] [Google Scholar]
- Calamita P, Gatti G, Miluzio A, Scagliola A, & Biffo S (2018). Translating the Game: Ribosomes as Active Players. Frontiers in Genetics, 9 10.3389/fgene.2018.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos RK, Wong B, Xie X, Lu Y-F, Shi P-Y, Pompon J, Garcia-Blanco MA, & Bradrick SS (2017). RPLP1 and RPLP2 Are Essential Flavivirus Host Factors That Promote Early Viral Protein Accumulation. Journal of Virology, 91(4). 10.1128/JVI.01706-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal F, Vallejos M, Walters B, Contreras N, Hertz MI, Olivares E, Cáceres CJ, Pino K, Letelier A, Thompson SR, & López-Lastra M (2016). Structural domains within the HIV-1 mRNA and the ribosomal protein S25 influence cap-independent translation initiation. The FEBS Journal, 283(13), 2508–2527. 10.1111/febs.13756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Salazar M, Angel-Ambrocio AH, Soto-Acosta R, Bautista-Carbajal P, Hurtado-Monzon AM, Alcaraz-Estrada SL, Ludert JE, & Del Angel RM (2015). Dengue virus NS1 protein interacts with the ribosomal protein RPL18: This interaction is required for viral translation and replication in Huh-7 cells. Virology, 484, 113–126. 10.1016/j.virol.2015.05.017 [DOI] [PubMed] [Google Scholar]
- Chen Y, Lu Z, Zhang L, Gao L, Wang N, Gao X, Wang Y, Li K, Gao Y, Cui H, Gao H, Liu C, Zhang Y, Qi X, & Wang X (2016). Ribosomal protein L4 interacts with viral protein VP3 and regulates the replication of infectious bursal disease virus. Virus Research, 211, 73–78. 10.1016/j.virusres.2015.09.017 [DOI] [PubMed] [Google Scholar]
- Cheng E, Haque A, Rimmer MA, Hussein ITM, Sheema S, Little A, & Mir MA (2011). Characterization of the interaction between hantavirus nucleocapsid protein (N) and ribosomal protein S19 (RPS19). Journal of Biological Chemistry, 286(13), 11814–11824. 10.1074/jbc.M110.210179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, & Perrimon N (2005). Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes and Development, 19(4), 445–452. 10.1101/gad.1267905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, & Mann M (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science, 325(5942), 834–840. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19608861 [DOI] [PubMed] [Google Scholar]
- Daughenbaugh KF, Fraser CS, Hershey JWB, & Hardy ME (2003). The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment | The EMBO Journal. EMBO J. https://www.embopress.org/doi/10.1093/emboj/cdg251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh KF, Wobus CE, & Hardy ME (2006). VPg of murine norovirus binds translation initiation factors in infected cells. Virology Journal, 3 10.1186/1743-422X-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira LC, Volpon L, Rahardjo AK, Osborne MJ, Culjkovic-Kraljacic B, Trahan C, Oeffinger M, Kwok BH, & Borden KLB (2019). Structural studies of the eIF4E–VPg complex reveal a direct competition for capped RNA: Implications for translation. Proceedings of the National Academy of Sciences of the United States of America, 116(48), 24056–24065. 10.1073/pnas.1904752116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vlugt C, Sikora D, & Pelchat M (2018). Insight into influenza: A virus cap-snatching In Viruses (Vol. 10, Issue 11). MDPI AG; 10.3390/v10110641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker S (1981). Phosphorylation of ribosomal protein S6 in avian sarcoma virus-transformed chicken embryo fibroblasts. Proceedings of the National Academy of Sciences of the United States of America, 78(7 I), 4112–4115. 10.1073/pnas.78.7.4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz N, Lenarcic EM, Landry DM, & Thompson SR (2009). Translation initiation factors are not required for Dicistroviridae IRES function in vivo. RNA, 15(5), 932–946. 10.1261/rna.1315109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar D, Mapa K, Pudi R, Srinivasan P, Bodhinathan K, & Das S (2006). Human ribosomal protein L18a interacts with hepatitis C virus internal ribosome entry site. Archives of Virology, 151(3), 509–524. 10.1007/s00705-005-0642-6 [DOI] [PubMed] [Google Scholar]
- Dobrikov MI, Dobrikova EY, & Gromeier M (2018a). Ribosomal RACK1:Protein Kinase C βII Modulates Intramolecular Interactions between Unstructured Regions of Eukaryotic Initiation Factor 4G (eIF4G) That Control eIF4E and eIF3 Binding. Molecular and Cellular Biology, 38(19). 10.1128/MCB.00306-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikov MI, Dobrikova EY, & Gromeier M (2018b). Ribosomal RACK1:Protein Kinase C βII Phosphorylates Eukaryotic Initiation Factor 4G1 at S1093 To Modulate Cap-Dependent and -Independent Translation Initiation. Molecular and Cellular Biology, 38(19). 10.1128/MCB.00304-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgil D, Polacek C, & Harris E (2006). Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J Virol, 80(6), 2976–2986. 10.1128/JVI.80.6.2976-2986.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A, Vyas J, Laing KG, & Clemens MJ (2004). Ribosomal protein L22 inhibits regulation of cellular activities by the Epstein-Barr virus small RNA EBER-1. European Journal of Biochemistry, 271(10), 1895–1905. 10.1111/j.1432-1033.2004.04099.x [DOI] [PubMed] [Google Scholar]
- Fok V, Mitton-Fry RM, Grech A, & Steitz JA (2006). Multiple domains of EBER 1, an Epstein-Barr virus noncoding RNA, recruit human ribosomal protein L22. RNA, 12(5), 872–882. 10.1261/rna.2339606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, & Doudna JA (2007). Structural and mechanistic insights into hepatitis C viral translation initiation. In Nature Reviews Microbiology (Vol. 5, Issue 1, pp. 29–38). 10.1038/nrmicro1558 [DOI] [PubMed] [Google Scholar]
- Fraser CS, Hershey JWB, & Doudna JA (2009). The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nature Structural and Molecular Biology, 16(4), 397–404. 10.1038/nsmb.1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JLR, Gonzalo P, Pitcher JA, Claing A, Lavergne JP, Reboud JP, & Lefkowitz RJ (2002). β2-adrenergic receptor stimulated, G protein-coupled receptor kinase 2 mediated, phosphorylation of ribosomal protein P2. Biochemistry, 41(42), 12850–12857. 10.1021/bi020145d [DOI] [PubMed] [Google Scholar]
- Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, & Katayama K (2001). Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. The Journal of Biological Chemistry, 276(24), 20824–20826. 10.1074/jbc.C100206200 [DOI] [PubMed] [Google Scholar]
- Gallo S, Ricciardi S, Manfrini N, Pesce E, Oliveto S, Calamita P, Mancino M, Maffioli E, Moro M, Crosti M, Berno V, Bombaci M, Tedeschi G, & Biffo S (2018). RACK1 Specifically Regulates Translation through Its Binding to Ribosomes. Molecular and Cellular Biology, 38(23). 10.1128/MCB.00230-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Senft D, Topisirovic I, & Ronai ZA (2013). RACK1 Function in Cell Motility and Protein Synthesis. Genes & Cancer, 4(9–10), 369–377. 10.1177/1947601913486348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Gulay SP, Kasprzak W, Dinman JD, Shapiro B. a, & Simon AE (2013). The kl-TSS translational enhancer of PEMV can bind simultaneously to ribosomes and a 5’ proximal hairpin. Journal of Virology, August 10.1128/JVI.02005-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Kasprzak WK, Szarko C, Shapiro BA, & Simon AE (2014). The 3’ untranslated region of Pea Enation Mosaic Virus contains two T-shaped, ribosome-binding, cap-independent translation enhancers. Journal of Virology, 88(20), 11696–11712. 10.1128/JVI.01433-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie LK, Hoenen A, Morgan G, & Mackenzie JM (2010). The Endoplasmic Reticulum Provides the Membrane Platform for Biogenesis of the Flavivirus Replication Complex. Journal of Virology, 84(20), 10438–10447. 10.1128/jvi.00986-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A-C, Raught B, & Sonenberg N (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem, 68, 913–963. [DOI] [PubMed] [Google Scholar]
- Glaunsinger BA (2015). Modulation of the Translational Landscape During Herpesvirus Infection. Annual Review of Virology, 2(1), 311–333. 10.1146/annurev-virology-100114-054839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I (2011). The genome-linked protein VPg of vertebrate viruses - A multifaceted protein In Current Opinion in Virology (Vol. 1, Issue 5, pp. 355–362). Europe PMC Funders; 10.1016/j.coviro.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberté JF, & Roberts L (2005). Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Reports, 6(10), 968–972. 10.1038/sj.embor.7400510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L, Houck-Loomis B, Yueh A, & Goff SP (2012). Large Ribosomal Protein 4 Increases Efficiency of Viral Recoding Sequences. Journal of Virology, 86(17), 8949–8958. 10.1128/jvi.01053-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross L, Vicens Q, Einhorn E, Noireterre A, Schaeffer L, Kuhn L, Imler J-L, Eriani G, Meignin C, & Martin F (2017). The IRES5’UTR of the dicistrovirus cricket paralysis virus is a type III IRES containing an essential pseudoknot structure. Nucleic Acids Research, 45(15), 8993–9004. 10.1093/nar/gkx622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruffat H, Lupo J, Morand P, Boyer V, & Manet E (2011). The Nuclear and Adherent Junction Complex Component Protein Ubinuclein Negatively Regulates the Productive Cycle of Epstein-Barr Virus in Epithelial Cells. Journal of Virology, 85(2), 784–794. 10.1128/jvi.01397-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes JC, & Zavolan M (2016). Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biology, 17(1). 10.1186/s13059-016-1104-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta AK, Mathur M, & Banerjee AK (2002). Unique capping activity of the recombinant RNA polymerase (L) of vesicular stomatitis virus: Association of cellular capping enzyme with the L protein. Biochemical and Biophysical Research Communications, 293(1), 264–268. 10.1016/S0006-291X(02)00217-6 [DOI] [PubMed] [Google Scholar]
- Hafirassou ML, Meertens L, Umaña-Diaz C, Labeau A, Dejarnac O, Bonnet-Madin L, Kümmerer BM, Delaugerre C, Roingeard P, Vidalain P-O, & Amara A (2017). A Global Interactome Map of the Dengue Virus NS1 Identifies Virus Restriction and Dependency Host Factors. Cell Reports, 21(13), 3900–3913. 10.1016/j.celrep.2017.11.094 [DOI] [PubMed] [Google Scholar]
- Hafren A, Eskelin K, & Makinen K (2013). Ribosomal Protein P0 Promotes Potato Virus A Infection and Functions in Viral Translation Together with VPg and eIF(iso)4E. Journal of Virology, 87(8), 4302–4312. 10.1128/jvi.03198-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga IR, Pechenick Jowers T, Griffiths SJ, Haas J, & Beard PM (2014). TRAF2 Facilitates Vaccinia Virus Replication by Promoting Rapid Virus Entry. Journal of Virology, 88(7), 3664–3677. 10.1128/jvi.03013-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque A, & Mir MA (2010). Interaction of Hantavirus Nucleocapsid Protein with Ribosomal Protein S19. Journal of Virology, 84(23), 12450–12453. 10.1128/jvi.01388-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CUT, & Frank J (2013). Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature, 503(7477), 539–543. 10.1038/nature12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JWB, Sonenberg N, & Mathews MB (2018). Principles of Translational Control. Cold Spring Harbor Perspectives in Biology, a032607 10.1101/cshperspect.a032607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz MI, Landry DM, Willis AE, Luo G, & Thompson SR (2013). Ribosomal protein s25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Molecular and Cellular Biology, 33(5), 1016–1026. 10.1128/MCB.00879-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houmani JL, Davis CI, & Ruf IK (2009). Growth-Promoting Properties of Epstein-Barr Virus EBER-1 RNA Correlate with Ribosomal Protein L22 Binding. Journal of Virology, 83(19), 9844–9853. 10.1128/jvi.01014-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis AJ, Masson GR, Shao S, Perisic O, McLaughlin SH, Hegde RS, & Williams RL (2019). Activation of GCN2 by the ribosomal P-stalk. Proceedings of the National Academy of Sciences of the United States of America, 116(11), 4946–4954. 10.1073/pnas.1813352116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, & Bisaillon M (2009). The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA, 15(12), 2340–2350. 10.1261/rna.1609709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, Ruggero D, & Dinman JD (2011). RRNA Pseudouridylation Defects Affect Ribosomal Ligand Binding and Translational Fidelity from Yeast to Human Cells. Molecular Cell, 44(4), 660–666. 10.1016/j.molcel.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, & Sarnow P (2002). Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. Journal of Molecular Biology, 324(5), 889–902. http://www.ncbi.nlm.nih.gov/pubmed/12470947 [DOI] [PubMed] [Google Scholar]
- Jang CJ, Lo MCY, & Jan E (2009). Conserved Element of the Dicistrovirus IGR IRES that Mimics an E-site tRNA/Ribosome Interaction Mediates Multiple Functions. Journal of Molecular Biology, 387(1), 42–58. 10.1016/j.jmb.2009.01.042 [DOI] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJH, Duke GM, Palmenberg AC, & Wimmer E (1988). A segment of the 5’ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol, 62, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Rollins MG, Fuchs G, Procter DJ, Hall EA, Cozzolino K, Sarnow P, Savas JN, & Walsh D (2017). Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature, 546(7660). 10.1038/nature22814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AG, Lapointe CP, Wang J, Corsepius NC, Choi J, Fuchs G, & Puglisi JD (2019). RACK1 on and off the ribosome. 25(7). http://www.ncbi.nlm.nih.gov/pubmed/31023766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaerlein M, & Horak I (1976). Phosphorylation of ribosomal proteins in HeLa cells infected with vaccinia virus. Nature, 259(5539), 150–151. 10.1038/259150a0 [DOI] [PubMed] [Google Scholar]
- Kaerlein M, & Horak I (1978). Identification and Characterization of Ribosomal Proteins Phosphorylated in Vaccinia-Virus-Infected HeLa Cells. European Journal of Biochemistry, 90(3), 463–469. 10.1111/j.1432-1033.1978.tb12625.x [DOI] [PubMed] [Google Scholar]
- Kennedy IM, Stevely WS, & Leader DP (1981). Phosphorylation of ribosomal proteins in hamster fibroblasts infected with pseudorabies virus or herpes simplex virus. Journal of Virology, 39(2), 359–366. http://www.ncbi.nlm.nih.gov/pubmed/6268827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney SP, & Meng X-J (2015). The Lysine Residues within the Human Ribosomal Protein S17 Sequence Naturally Inserted into the Viral Nonstructural Protein of a Unique Strain of Hepatitis E Virus Are Important for Enhanced Virus Replication. Journal of Virology, 89(7), 3793–3803. 10.1128/jvi.03582-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Kim J, Bang B, Kim I, Lee HH, Park J, & Seo YS (2016). Comparative analyses of Tomato yellow leaf curl virus C4 protein-interacting host proteins in healthy and infected Tomato tissues. Plant Pathology Journal, 32(5), 377–387. 10.5423/PPJ.FT.08.2016.0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFontaine E, Miller CM, Permaul N, Martin ET, & Fuchs G (2020). Ribosomal protein RACK1 enhances translation of poliovirus and other viral IRESs. Virology, 545, 53–62. 10.1016/j.virol.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, & Thompson SR (2009). RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes & Development, 23(23), 2753–2764. 10.1101/gad.1832209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Sommer C, Barrows NJ, Bradrick SS, Pearson JL, & Garcia-Blanco MA (2012). G Protein-Coupled Receptor Kinase 2 Promotes Flaviviridae Entry and Replication. PLoS Neglected Tropical Diseases, 6(9). 10.1371/journal.pntd.0001820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS-Y, Burdeinick-Kerr R, & Whelan SPJ (2013). A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proceedings of the National Academy of Sciences of the United States of America, 110(1), 324–329. 10.1073/pnas.1216454109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leh V, Yot P, & Keller M (2000). The cauliflower mosaic virus translational transactivator interacts with the 60S ribosomal subunit protein L18 of Arabidopsis thaliana. Virology, 266(1), 1–7. 10.1006/viro.1999.0073 [DOI] [PubMed] [Google Scholar]
- Li S, Li X, & Zhou Y (2018). Ribosomal protein L18 is an essential factor that promote rice stripe virus accumulation in small brown planthopper. Virus Research, 247, 15–20. 10.1016/j.virusres.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Li Y, Dong W, Shi Y, Deng F, Chen X, Wan C, Zhou M, Zhao L, Fu ZF, & Peng G (2016). Rabies virus phosphoprotein interacts with ribosomal protein L9 and affects rabies virus replication. Virology, 488, 216–224. 10.1016/j.virol.2015.11.018 [DOI] [PubMed] [Google Scholar]
- Liu L, Dong H, Chen H, Zhang J, Ling H, Li Z, Shi PY, & Li H (2010). Flavivirus RNA cap methyltransferase: Structure, function, and inhibition. In Frontiers of Biology in China (Vol. 5, Issue 4, pp. 286–303). 10.1007/s11515-010-0660-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. hui, Ma F. fang, Guan GK, Wang XF, Li C, & Huang J (2015). White spot syndrome virus VP51 interact with ribosomal protein L7 ofLitopenaeus vannamei. Fish and Shellfish Immunology, 44(1), 382–388. 10.1016/j.fsi.2015.02.035 [DOI] [PubMed] [Google Scholar]
- Liu X, Bi J, Zhao Q, Li M, Zuo Q, Wang X, Lan R, Li X, Yang G, Liu J, & Yin G (2019). Overexpression of RACK1 enhanced the replication of porcine reproductive and respiratory syndrome virus in Marc-145 cells and promoted the NF-κB activation via upregulating the expression and phosphorylation of TRAF2. Gene, 709, 75–83. 10.1016/j.gene.2019.05.046 [DOI] [PubMed] [Google Scholar]
- Lupo J, Conti A, Sueur C, Coly PA, Couté Y, Hunziker W, Burmeister WP, Germi R, Manet E, Gruffat H, Morand P, & Boyer V (2012). Identification of new interacting partners of the shuttling protein ubinuclein (Ubn-1). Experimental Cell Research, 318(5), 509–520. 10.1016/j.yexcr.2011.12.020 [DOI] [PubMed] [Google Scholar]
- Lv H, Dong W, Qian G, Wang J, Li X, Cao Z, Lv Q, Wang C, Guo K, & Zhang Y (2017). uS10, a novel Npro-interacting protein, inhibits classical swine fever virus replication. Journal of General Virology, 98(7), 1679–1692. 10.1099/jgv.0.000867 [DOI] [PubMed] [Google Scholar]
- M. R, A. J-D, B. B, M.A. R-G, E. G, J.P.G. B, Remacha M, Jimenez-Diaz A, Bermejo B, Rodriguez-Gabriel MA, Guarinos E, & Ballesta JP (1995). Ribosomal acidic phosphoprotein P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae. Molecular and Cellular Biology, 15(9), 4754–4762. http://www.embase.com/search/results?subaction=viewrecord&from=export&id=L25259493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson B, Ekim B, & Fingar DC (2012). Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks In Biochemical Journal (Vol. 441, Issue 1, pp. 1–21). Portland Press; 10.1042/BJ20110892 [DOI] [PubMed] [Google Scholar]
- Mailliot J, & Martin F (2018). Viral internal ribosomal entry sites: four classes for one goal. Wiley Interdisciplinary Reviews: RNA, 9(2), e1458 10.1002/wrna.1458 [DOI] [PubMed] [Google Scholar]
- Majzoub K, Hafirassou ML, Meignin C, Goto A, Marzi S, Fedorova A, Verdier Y, Vinh J, Hoffmann JA, Martin F, Baumert TF, Schuster C, & Imler J-L (2014). RACK1 Controls IRES-Mediated Translation of Viruses. Cell, 159(5), 1086–1095. 10.1016/j.cell.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, & Carette JE (2016). Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature, 535(7610). 10.1038/nature18631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Azorin F, Remacha M, & Ballesta JPG (2008). Functional characterization of ribosomal P1/P2 proteins in human cells. Biochemical Journal, 413(3), 527–534. 10.1042/BJ20080049 [DOI] [PubMed] [Google Scholar]
- McCann KL, & Baserga SJ (2013). Mysterious Ribosomopathies. Science (New York, N.Y.), 341(6148), 849 10.1126/SCIENCE.1244156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade N, DiGiuseppe S, & Walsh D (2019). Translational control during poxvirus infection In Wiley Interdisciplinary Reviews: RNA (Vol. 10, Issue 2, p. e1515). Blackwell Publishing Ltd; 10.1002/wrna.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekdad H. El, Boutant E, Karnib H, Biedma ME, Sharma KK, Malytska I, Laumond G, Roy M, Réal E, Paillart J-C, Moog C, Darlix JL, Mély Y, & de Rocquigny H (2016). Characterization of the interaction between the HIV-1 Gag structural polyprotein and the cellular ribosomal protein L7 and its implication in viral nucleic acid remodeling. Retrovirology, 13(1), 54 10.1186/s12977-016-0287-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O (2008). Chapter 1 Physiological Roles of Ribosomal Protein S6: One of Its Kind. In International Review of Cell and Molecular Biology (Vol. 268, pp. 1–37). 10.1016/S1937-6448(08)00801-0 [DOI] [PubMed] [Google Scholar]
- Mizuno CM, Guyomar C, Roux S, Lavigne R, Rodriguez-Valera F, Sullivan MB, Gillet R, Forterre P, & Krupovic M (2019). Numerous cultivated and uncultivated viruses encode ribosomal proteins. Nature Communications, 10(1), 1–11. 10.1038/s41467-019-08672-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SA, Berglund P, Beard CW, & Johnston RE (2006). Ribosomal protein S6 associates with alphavirus nonstructural protein 2 and mediates expression from alphavirus messages. Journal of Virology, 80(15), 7729–7739. 10.1128/JVI.00425-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, & Spahn CM (2011). Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyangwa M, Martynova EV, Khaiboullina SF, Morzunov SP, & Rizvanov AA (2015). Hantaviral proteins: Structure, functions, and role in hantavirus infection In Frontiers in Microbiology (Vol. 6, Issue NOV). Frontiers Research Foundation; 10.3389/fmicb.2015.01326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzband R, & Pager CT (2020). Epitranscriptomic marks: Emerging modulators of RNA virus gene expression In Wiley Interdisciplinary Reviews: RNA (Vol. 11, Issue 3). Blackwell Publishing Ltd; 10.1002/wrna.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane R, Pisareva VP, Rodriguez CF, Pisarev AV, & Fernández IS (2020). A complex IRES at the 5’-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate. ELife, 9 10.7554/elife.54575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson BL, & White KA (2011). 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses In Current Opinion in Virology (Vol. 1, Issue 5, pp. 373–380). Elsevier B.V. 10.1016/j.coviro.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Nielsen MH, Flygaard RK, & Jenner LB (2017). Structural analysis of ribosomal RACK1 and its role in translational control. Cellular Signalling, 35, 272–281. 10.1016/j.cellsig.2017.01.026 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Yamamoto H, Uchiumi T, & Nakashima N (2007). Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Research, 35(5), 1514–1521. 10.1093/nar/gkl1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares E, Landry DM, Cáceres CJ, Pino K, Rossi F, Navarrete C, Huidobro-Toro JP, Thompson SR, & López-Lastra M (2014). The 5’ untranslated region of the human T-cell lymphotropic virus type 1 mRNA enables cap-independent translation initiation. Journal of Virology, 88(11), 5936–5955. 10.1128/JVI.00279-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Himmelbach A, Browning KS, Hohn T, & Ryabova LA (2001). A plant viral “reinitiation” factor interacts with the host translational machinery. Cell, 106(6), 723–733. 10.1016/S0092-8674(01)00487-1 [DOI] [PubMed] [Google Scholar]
- Parks MM, Kurylo CM, Dass RA, Bojmar L, Lyden D, Vincent CT, & Blanchard SC (2018). Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Science Advances, 4(2). 10.1126/sciadv.aao0665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, & Sonenberg N (1989). Internal binding of eucaryotic ribosomes on poliovirus RNA: translation in HeLa cell extracts. Journal of Virology, 63(1), 441–444. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=247704&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchey RT, Tonini L, Tosolini M, Fournié JJ, Lopez F, Besson A, & Pont F (2019). PTMselect: optimization of protein modifications discovery by mass spectrometry. Scientific Reports, 9(1). 10.1038/s41598-019-40873-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Grosely R, Chen J, O’Leary SE, & Puglisi JD (2016). Multiple Parallel Pathways of Translation Initiation on the CrPV IRES. Molecular Cell, 62(1), 92–103. 10.1016/j.molcel.2016.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova E, Gracias S, Beauclair G, Tangy F, & Jouvenet N (2019). Uncovering flavivirus host dependency factors through a genome-wide gain-of-function screen. Viruses, 11(1). 10.3390/v11010068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, & Fernández IS (2018). Dual tRNA mimicry in the cricket paralysis virus IRES uncovers an unexpected similarity with the hepatitis C virus IRES. ELife, 7 10.7554/eLife.34062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamäki M-L, Xi D, Sikorskaite-Gudziuniene S, Valkonen JPT, & Whitham SA (2017). Differential Requirement of the Ribosomal Protein S6 and Ribosomal Protein S6 Kinase for Plant-Virus Accumulation and Interaction of S6 Kinase with Potyviral VPg. Molecular Plant-Microbe Interactions, 30(5), 374–384. 10.1094/MPMI-06-16-0122-R [DOI] [PubMed] [Google Scholar]
- Rocha CS, Santos AA, Machado JPB, & Fontes EPB (2008). The ribosomal protein L10/QM-like protein is a component of the NIK-mediated antiviral signaling. Virology, 380(2), 165–169. 10.1016/j.virol.2008.08.005 [DOI] [PubMed] [Google Scholar]
- Sabo Y, Walsh D, Barry DS, Tinaztepe S, De Los Santos K, Goff SP, Gundersen GG, & Naghavi MH (2013). HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host and Microbe, 14(5), 535–546. 10.1016/j.chom.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoul K, Wang J, Diagouraga B, & Khochbin S (2011). The tale of protein lysine acetylation in the cytoplasm. Journal of Biomedicine & Biotechnology, 2011, 970382 10.1155/2011/970382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeedi BJ, & Geiss BJ (2013). Regulation of flavivirus RNA synthesis and capping. In Wiley Interdisciplinary Reviews: RNA (Vol. 4, Issue 6, pp. 723–735). 10.1002/wrna.1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CL, Liua C. Der, You RI, Ching YH, Liang J, Ke L, Chen YL, Chen HC, Hsu HJ, Liou JW, Kieff E, & Peng CW (2016). Ribosome Protein L4 is essential for Epstein-Barr Virus Nuclear Antigen 1 function. Proceedings of the National Academy of Sciences of the United States of America, 113(8), 2229–2234. 10.1073/pnas.1525444113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Tiu GC, Flynn RA, Byeon GW, Leppek K, Xu AF, Chang HY, & Barna M (2017). The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell, 169(6), 1051–1065.e18. 10.1016/j.cell.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, & Nogales E (2005). Structural Roles for Human Translation Factor eIF3 in Initiation of Protein Synthesis. Science, 310(5753), 1513–1515. 10.1126/science.1118977 [DOI] [PubMed] [Google Scholar]
- Smith PR, de Jesus O, Turner D, Hollyoake M, Karstegl CE, Griffin BE, Karran L, Wang Y, Hayward SD, & Farrell PJ (2000). Structure and Coding Content of CST (BART) Family RNAs of Epstein-Barr Virus. Journal of Virology, 74(7), 3082–3092. 10.1128/jvi.74.7.3082-3092.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, & Hinnebusch AG (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell, 136(4), 731–745. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19239892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Mugavero JA, Stauft CB, & Wimmer E (2019). Dengue and zika virus 5’ untranslated regions harbor internal ribosomal entry site functions. MBio, 10(2). 10.1128/mBio.00459-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, & Frank J (2001). Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science, 291(5510), 1959–1962. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Thompson SR, Mathews MB, & Mohr I (2018). Translational Control in Virus-Infected Cells. Cold Spring Harbor Perspectives in Biology, a033001 10.1101/cshperspect.a033001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Farnsworth CL, Moritz A, Silva JC, Jia X, Lee KA, Guo A, Polakiewicz RD, & Comb MJ (2012). PTMScan Direct: Identification and Quantification of Peptides from Critical Signaling Proteins by Immunoaffinity Enrichment Coupled with LC-MS/MS. Molecular & Cellular Proteomics, 11(5), 187–201. 10.1074/mcp.M111.015883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Meskauskas A, Mccormack JC, Yingling YG, Shapiro BA, Dinman JD, & Simon AE (2008). The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA, 14(11), 2379–2393. 10.1261/rna.1227808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif M, Savard M, Flamand L, & Gosselin J (2002). Impaired protein kinase C activation/translocation in Epstein-Barr virus-infected monocytes. Journal of Biological Chemistry, 277(27), 24148–24154. 10.1074/jbc.M109036200 [DOI] [PubMed] [Google Scholar]
- Toczyski DP, Matera AG, Ward DC, & Steitz JA (1994). The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proceedings of the National Academy of Sciences of the United States of America, 91(8), 3463–3467. 10.1073/pnas.91.8.3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Hou W, Dakshanamurthy S, & Tang Q (2019). Host targeted antiviral (HTA): Functional inhibitor compounds of scaffold protein RACK1 inhibit herpes simplex virus proliferation. Oncotarget, 10(35), 3209–3226. 10.18632/oncotarget.26907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Waterbeemd M, Tamara S, Fort KL, Damoc E, Franc V, Bieri P, Itten M, Makarov A, Ban N, & Heck AJR (2018). Dissecting ribosomal particles throughout the kingdoms of life using advanced hybrid mass spectrometry methods. Nature Communications, 9(1). 10.1038/s41467-018-04853-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Ziehr B, & Moorman NJ (2016). Human cytomegalovirus strategies to maintain and promote mRNA translation In Viruses (Vol. 8, Issue 4). MDPI AG; 10.3390/v8040097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westdorp KN, Sand A, Moorman NJ, & Terhune SS (2017). Cytomegalovirus Late Protein pUL31 Alters Pre-rRNA Expression and Nuclear Organization during Infection. Journal of Virology, 91(18). 10.1128/jvi.00593-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Edell KA, Kehasse A, Scott GK, Yau C, Rothschild DE, Schilling B, Gabriel BS, Yevtushenko MA, Hanson IM, Held JM, Gibson BW, & Benz CC (2014). RPL24: A potential therapeutic target whose depletion or acetylation inhibits polysome assembly and cancer cell growth. Oncotarget, 5(13), 5165–5176. 10.18632/oncotarget.2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Collier M, Loerke J, Ismer J, Schmidt A, Hilal T, Sprink T, Yamamoto K, Mielke T, Bürger J, Shaikh TR, Dabrowski M, Hildebrand PW, Scheerer P, & Spahn CMT (2015). Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA. The EMBO Journal, 34(24), 3042–3058. 10.15252/embj.201592469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Zhang C, Dittman JD, & Whitham SA (2009). Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology, 390(2), 163–173. 10.1016/j.virol.2009.05.018 [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Machida K, Iwasaki W, Shigeta T, Nishimoto M, Takahashi M, Sakamoto A, Yonemochi M, Harada Y, Shigematsu H, Shirouzu M, Tadakuma H, Imataka H, & Ito T (2019). HCV IRES Captures an Actively Translating 80S Ribosome. Molecular Cell, 74(6), 1205–1214.e8. 10.1016/j.molcel.2019.04.022 [DOI] [PubMed] [Google Scholar]
- Yu Y, Ji H, Doudna JA, & Leary JA (2005). Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci, 14(6), 1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeenko VV, Wang C, Majumder M, Komar AA, Snider MD, Merrick WC, Kaufman RJ, & Hatzoglou M (2008). An efficient in vitro translation system from mammalian cells lacking the translational inhibition caused by eIF2 phosphorylation. RNA, 14(3), 593–602. 10.1261/rna.825008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Villa NY, Rahman MM, Smallwood S, Shattuck D, Neff C, Dufford M, Lanchbury JS, LaBaer J, & McFadden G (2009). Analysis of vaccinia virus-host protein-protein interactions: Validations of yeast two-hybrid screenings. Journal of Proteome Research, 8(9), 4311–4318. 10.1021/pr900491n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Li W, Zhang X, Wang C, Gao L, Yang F, Cao W, Li K, Tian H, Liu X, Zhang K, & Zheng H (2019). Foot-and-mouth disease virus capsid protein VP1 interacts with host ribosomal protein SA to maintain the activation of MAPKs signal pathway and promote virus replication. Journal of Virology. 10.1128/JVI.01350-19 [DOI] [PMC free article] [PubMed] [Google Scholar]


