Abstract
Background and aims:
We sought to assess the performance of a comprehensive machine learning (ML) risk score integrating circulating biomarkers and computed tomography (CT) measures for the long-term prediction of hard cardiac events in asymptomatic subjects.
Methods:
We studied 1069 subjects (age 58.2±8.2 years, 54.0% males) from the prospective EISNER trial who underwent coronary artery calcium (CAC) scoring CT, serum biomarker assessment, and long-term follow-up. Epicardial adipose tissue (EAT) was quantified from CT using fully automated deep learning software. Forty-eight serum biomarkers, both established and novel, were assayed. A ML algorithm (XGBoost) was trained using clinical risk factors, CT measures (CAC score, number of coronary lesions, aortic valve calcium score, EAT volume and attenuation), and circulating biomarkers, and validated using repeated 10-fold cross validation.
Results:
At 14.5±2.0 years, there were 50 hard cardiac events (myocardial infarction or cardiac death). The ML risk score (area under the receiver operator characteristic curve [AUC] 0.81) outperformed the CAC score (0.75) and ASCVD risk score (0.74; both p=0.02) for the prediction of hard cardiac events. Serum biomarkers provided incremental prognostic value beyond clinical data and CT measures in the ML model (net reclassification index 0.53 [95% CI: 0.23–0.81], p<0.0001). Among novel biomarkers, MMP-9, pentraxin 3, PIGR, and GDF-15 had highest variable importance for ML and reflect the pathways of inflammation, extracellular matrix remodeling, and fibrosis.
Conclusions:
In this prospective study, ML integration of novel circulating biomarkers and noninvasive imaging measures provided superior long-term risk prediction for cardiac events compared to current risk assessment tools.
Keywords: Machine learning, artificial intelligence, serum biomarkers, cardiac computed tomography, cardiovascular risk stratification
INTRODUCTION
Atherosclerotic cardiovascular disease (ASCVD) causes significant morbidity and mortality in the United States, and early risk stratification of individuals for cardiovascular events is crucial in determining treatment strategies. Traditional risk assessment tools utilize demographic, anthropometric, and clinical patient characteristics1, 2. The ASCVD risk score is a reliable predictor of the 10-year risk of cardiac death, non-fatal myocardial infarction (MI), or stroke2. Coronary artery calcium (CAC) scoring using noncontrast cardiac computed tomography (CT) improves risk stratification over and above traditional risk assessment3. Further, CT-derived epicardial adipose tissue (EAT) volume has incremental prognostic value beyond the CAC score4. Circulating markers such as C-reactive protein (CRP) and low-density lipoprotein cholesterol (LDL) are established predictors of future CV events5, and novel serum biomarkers have recently been used to risk stratify individuals with ischemic heart disease6 and heart failure7. However, few studies have combined serum biomarker levels with clinical and imaging variables for prognostication. The EISNER (Early Identification of Subclinical Atherosclerosis by Noninvasive Imaging Research) trial8 consisted of asymptomatic subjects who underwent baseline CAC scoring CT scans and serum biomarker assessment, with 14-year follow-up. We previously trained a ML model using noncontrast CT parameters from this cohort for prognostication9. In the present EISNER substudy, we sought to determine if a comprehensive ML-based model integrating clinical risk factors, quantitative CT measures, and circulating biomarkers could outperform current risk assessment tools for the long-term prediction of hard cardiac events.
PATIENTS AND METHODS
Study population
The prospective EISNER trial8 comprised 1424 subjects who underwent baseline CAC scanning and blood sample collection at Cedars-Sinai Medical Center (CSMC) from May 2001-May 2005. Inclusion criteria for EISNER were: age 45–80 years and intermediate risk of CAD based on age (>55 years in men, >65 years in women) or the presence of ≥1 CAD risk factor in younger subjects (age 45–54 years in men or 55–64 years in women). Exclusion criteria were: history of cardiac disease or cerebrovascular disease or chest pain, prior CAC scanning or invasive coronary angiography, or significant medical comorbidity (including malignancy, infection, or a severe inflammatory condition). For the present substudy, we included 1069 patients who completed long-term follow-up and had CT images and comprehensive serum biomarker data available. Supplemental Figure 1 outlines the patient selection and study design. All subjects underwent clinical assessment at baseline, including measurements of blood pressure and body mass index (BMI). The ASCVD risk score was calculated using the Pooled Cohort Equation2.
Prognostic follow-up
Subjects were prospectively followed up during a mean of 14.5±2.0 years for hard cardiac events, defined as myocardial infarction (MI) or cardiac death. Follow-up was via clinical visits, detailed questionnaires sent by mail, or telephone contact. Reported event information was also verified by the National Death Index query and by comprehensive review of electronic medical, hospital, and death records by 2 independent cardiologists blinded to clinical data. The research was approved by the CSMC Review Board and all subjects provided written informed consent.
Biomarker analysis
Serum samples were collected at the time of CT, immediately centrifuged, and stored in a −80°C freezer until assayed. Fasting total cholesterol, high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglycerides, and glucose were measured using standard techniques. Serum biomarkers including adiponectin, angiotensinogen, creatine kinase MB (CKMB), interleukin 6 (IL-6), monocyte chemoattractant protein 1 (MCP-1), matrix metalloprotease 9 (MMP-9), myoglobin (MYO), endothelial plasminogen activator inhibitor 1 (PAI-1), soluble intercellular adhesion molecule 1 (sICAM-1), vascular cell adhesion molecule 1 (VCAM-1), neutrophil gelatinase-associated lipocalin (NGAL), macrophage inflammatory protein 3 (MIP3), chemokine (C-X-C motif) ligand 1 (CXCL1) and ligand 2 (CXCL2), peptidoglycan recognition proteins (PGRPs) and caspase 3 were measured by an independent and blinded laboratory (Biosite, San Diego, CA, USA) using sandwich enzyme-linked immunosorbent assays (ELISA) on a microtiter plate. Troponin I, brain natriuretic peptide (BNP), proBNP (3–108), high-sensitivity C-reactive protein (hs-CRP), D-dimer, myeloperoxidase (MPO), endothelial cell-selective adhesion molecule (ESAM), lymphotoxin beta receptor (LTBR), growth differentiation factor 15 (GDF-15), mesothelin, neuropilin 1 (NRP-1), atrial natriuretic peptide (ANP) propeptide, N-terminal pro C-type natriuretic peptide (NTProCNP), osteopontin, procalcitonin, pentraxin 3, periostin, polymeric immunoglobulin receptor (PIGR), pro-adrenomedullin, prosaposin B (PSAP-B), receptor for advanced glycation end products (RAGE), soluble ST-2, syndecan-1, tumor necrosis factor receptor 1 alpha (TNFR1A), vascular endothelial growth factor receptor 1 (VEGFR1), cystatin C and WAP four-disulfide core domain protein 4 (WAP4C) were measured using sandwich ELISAs on a Luminex® platform at a second independent laboratory (Alere™ Inc., San Diego, CA, USA).
Calcium assessment on CT
Noncontrast CT was performed using either an Electron Beam CT scanner (e-Speed, GE Healthcare, Milwaukee, WI, USA) or a 4-slice CT scanner (Somatom Volume zoom, Siemens Medical Solutions, Erlangen, Germany) with prospective ECG-triggering and a tube voltage of 120 kVp; slice thickness was either 2.5 or 3.0 mm. Each scan was analyzed by an experienced cardiologist using commercially available semi-automated software (ScImage Inc., Los Altos, CA, USA). The CAC score (Supplemental Figure 2A), number of calcified coronary lesions, and the aortic valve calcium score were calculated according to the Agatston method10.
Deep learning-based EAT quantification
EAT was defined as all adipose tissue within the visceral pericardium. EAT volume and attenuation were quantified from noncontrast CT using a fully automated deep learning (DL) algorithm11 incorporated into research software (QFAT version 2.0, CSMC, Los Angeles, CA) (Supplemental Figure 2B). This DL algorithm has been validated and tested in a large multicenter study, demonstrating high agreement with expert readers for quantification of EAT11. In this DL method, the pericardium was automatically segmented and the limits of the heart were automatically defined as the pulmonary artery bifurcation (superior limit) to the posterior descending artery (inferior limit). EAT volume (cm3) and attenuation (Hounsfield units [HU]) were automatically calculated from 3D fat voxels between the HU limits of (−190, −30 HU) within the visceral pericardium. The processing time for EAT quantification was approximately 25 s per case.
Machine learning model creation
In designing our predictive model for hard cardiac events, we aimed to integrate: i) clinical variables used in traditional risk scoring; ii) established prognostic parameters from CAC scoring CT; and iii) the most relevant serum biomarkers based on feature selection. Hence, our ML model included 12 individual components of the ASCVD risk score (age, gender, systolic and diastolic blood pressure, total cholesterol, LDL, HDL, diabetes, smoking, antihypertensive treatment, statin, aspirin), 5 quantitative CT measures (CAC score, number of calcified coronary lesions, aortic valve calcium score, EAT volume, EAT attenuation), and the top 15 serum biomarkers according to ML information gain. As a common feature selection method for decision tree algorithms, information gain is defined as a measure of the effectiveness of a feature in classifying the training data. It is calculated as the amount by which the entropy of the class decreases, and is hence a reflection of additional information about the class provided by the feature12.
Models were then built using XGBoost, a state-of-the-art ensemble boosting ML algorithm which has demonstrated high performance in cardiac CT-based risk stratification9, 13. The advantage of using a boosted ensemble algorithm is that it combines multiple weak classifiers (one-level decision trees) to produce a single strong classifier, which can improve prediction modeling. For each subject, the algorithm computed an individualized ML risk score according to the weighting of each variable.
Machine learning model training
To avoid reporting biased results and limit overfitting, we validated our ML algorithm using 10-fold cross-validation14. This involved stratifying and dividing the data into 10 folds of equal size: eight folds (80%) were used for training, one fold (10%) was used for tuning model parameters, and one fold (10%) was used for testing. Stratification was used to ensure a similar distribution of events across the 10 folds. This process was repeated 10 times, always using a different fold for model training, tuning, and testing. The ML risk scores were concatenated from all 10 testing data folds to allow assessment of model performance over the entire dataset. The set of hyperparameters that led to the highest overall area under the receiver-operating characteristic curve (AUC) was chosen for the final ML model.
Explainable individualized ML risk prediction
To demonstrate the clinical applicability of the ML model and explain how it provides accurate predictions for this cohort, we provide a detailed description of individualized risk prediction made by the ML algorithm. The model allows identification of important patient-specific variables and the role of the variable in the predicted score. We analyze the specific path a subject takes in the model; in each decision stump (or split) of the model, the individual lands in one of two leaves. Each leaf is associated with a weight: one leaf decreasing the risk of the event occurring, and the other one increasing the risk. These weights are associated with the variables used to generate the corresponding split. By cumulating all the weights used to refine per-patient prediction for each variable in the model, we can determine whether or not a parameter has a protective influence, depending on the weight sign. By considering the absolute values of cumulated weights, we can also obtain the global contribution of each parameter.
Statistical analysis
Continuous variables are presented as mean ± standard deviation or median (interquartile range), as appropriate. The two-sample t-test or Wilcoxon rank-sum test was used to compare continuous variables. A Chi-square or Fisher’s exact test was used to compare categorical variables. Distributions of CAC score and EAT volume were not normally distributed and hence normalized using logarithmic adjustment; base-2 logarithmic transformation was this represents doubling of the variable. Multivariable Cox regression with backward stepwise selection was used to identify independent predictors of cardiac events among the serum biomarkers which were statistically significant on univariable analysis, with adjustment for the ASCVD risk score and CAC score, and EAT volume. Receiver-operating characteristic curve analysis was used to assess the performance of the ML model, and AUC values were compared with the DeLong test15. The continuous net reclassification index (NRI)16 was used to measure the incremental prognostic value of adding serum biomarkers to a ML model with only clinical risk factors and CT measures. The highest Youden’s J index (J = sensitivity + specificity - 1) was used to identify an optimal cutoff for the ML score and stratify subjects into ‘high’ or low’ ML risk. Kaplan-Meier analysis was performed according to this threshold, and survival curves were compared with the log-rank test. Statistical analysis was performed using Stata/IC 15.1 (StataCorp LP, College Station, TX, USA), with SAS 9.4 (SAS 207 Institute, Cary, NC, USA) used for NRI computation. A p-value of <0.05 indicated statistical significance.
RESULTS
Patient characteristics
The final study population comprised 1069 subjects with mean age 58.2±8.2 years and 54.0% males. At mean follow-up of 14.5±2.0 years, 50 subjects experienced hard cardiac events. Subjects who experienced events were older and had higher mean systolic blood pressure and serum LDL cholesterol compared to subjects without events (both p <0.0001). CAC score and EAT volume were greater in subjects with events compared to those without events (both p <0.0001). Table 1 summarizes the baseline characteristics of subjects. There were no significant differences in clinical and CT parameters between the 1424 subjects who underwent baseline CAC scanning in the original EISNER trial and the 1069 subjects in the present substudy (all p>0.05).
Table 1.
Baseline characteristics of the study population
| Clinical characteristics | Original EISNER (n = 1424) | Current substudy (n = 1069) | Event (n = 50) | No event (n = 1019) | p-value |
|---|---|---|---|---|---|
| Age | 56.9±8.9 | 58.2±8.2 | 63.8±10.2 | 57.9±8.0 | <0.0001 |
| Male gender | 784 (55.1) | 558 (52.2) | 28 (56.0) | 530 (52.0) | 0.60 |
| BMI (kg/m2) | 27.1±5.1 | 27.5±5.3 | 28.5±5.9 | 27.5±5.3 | 0.30 |
| Hypertension | 751 (52.7) | 589 (55.1) | 39 (78.0) | 550 (54.0) | 0.001 |
| Hyperlipidemia | 978 (68.7) | 720 (67.4) | 37 (74.0) | 683 (67.0) | 0.30 |
| Diabetes mellitus | 94 (6.6) | 70 (6.5) | 3 (6.0) | 67 (6.6) | 0.87 |
| Smoking | 92 (6.5) | 69 (6.5) | 3 (6.0) | 66 (6.5) | 0.89 |
| Family history | 414 (29.1) | 301 (28.2) | 16 (32.0) | 285 (28.0) | 0.56 |
| Systolic blood pressure (mmHg) | 130.4±18.0 | 132.2±17.4 | 146.3±25.6 | 131.5±16.6 | <0.0001 |
| Diastolic blood pressure (mmHg) | 79.3±11.5 | 81.7±10.7 | 84.4±11.2 | 81.6±10.7 | 0.07 |
| Total cholesterol (mg/dL) | 212.4±41.2 | 215.4141.9 | 223.0135.9 | 215142.2 | 0.19 |
| HDL cholesterol (mg/dL) | 55.0±17.1 | 54.4±16.7 | 51.1±15.9 | 54.5±16.8 | 0.16 |
| LDL cholesterol (mg/dL) | 132.6±38.4 | 135.6±39.6 | 150.9±57.8 | 134.8±38.3 | <0.0001 |
| Triglycerides (mg/dL) | 106.0 (76.0–153.0) | 111.0 (79.0–156.8) | 136.0 (95.6–161.5) | 108.0 (78.0–156.3) | 0.19 |
| Glucose (mg/dL) | 95.8±17.4 | 94.6±16.1 | 98.2±14.9 | 94.4±16.1 | 0.10 |
| ASCVD risk score (%) | 6.1 (3.1–10.6) | 6.3 (3.4–11.4) | 15.0 (7.7–26.0) | 6.0 (3.3–11.0) | <0.0001 |
| Medications | |||||
| Beta blockers | 77 (5.4) | 70 (6.5) | 8 (16.0) | 62 (6.1) | 0.01 |
| ACE-I or ARB | 206 (14.5) | 167 (15.6) | 11 (22.0) | 157 (15.4) | 0.03 |
| Statin | 326 (22.9) | 255 (23.9) | 11 (22.0) | 244 (23.9) | 0.76 |
| Antihyperglycemic | 46 (3.2) | 31 (2.9) | 2 (4.5) | 29 (2.8) | 0.08 |
| Aspirin | 147 (10.3) | 131 (12.3) | 11 (21.3) | 120 (11.8) | 0.05 |
| Quantitative CT measures | |||||
| CAC score | 105.8±298.9 | 102.4±282.6 | 413.7±666.2 | 87.1±239.7 | <0.0001 |
| EAT volume (cm3)a | 87.5±40.0 | 90.4±40.9 | 111.4±43.8 | 89.3±40.5 | <0.0001 |
| EAT attenuation (HU)a | −74.2+4.8 | −74.7±4.9 | −76.8±4.6 | −74.6±4.9 | 0.002 |
Values are expressed as n (%), mean ± SD or median (IQR).
Data available in 1406 subjects from the original EISNER study.
ACE-I, angiotensin converting enzyme inhibitor; ARB, angiotensin receptor blocker; ASCVD, atherosclerotic cardiovascular disease; CAC, coronary artery calcium; EAT, epicardial adipose tissue; HDL, high-density lipoprotein; HU, Hounsfield units; LDL, low-density lipoprotein.
Serum biomarker levels
Serum levels of the traditional biomarkers hs-CRP, D-dimer, PAI-1, CKMB, myoglobin, and BNP were higher in subjects who experienced events compared to subjects without events. Similar results were observed for more novel biomarkers such as MPO, MMP-9, pentraxin 3, and PIGR (Supplemental Table 1).
Prediction of hard cardiac events
In multivariable Cox analysis, the serum biomarkers LDL (1.02 [95% CI: 1.01–1.03] per 1 mg/dL increase, p=0.01), MMP-9 (HR 1.04 [95% CI: 1.01–1.08] per 1 ng/mL increase, p=0.02), and MPO (HR 1.01 [95% CI: 1.01–1.02] per 1 pmol/L increase, p=0.01) were independently associated with MACE risk. The ASCVD risk score, CAC score, and EAT volume also had independent predictive value (Supplemental Table 2).
Using ML, the clinical and imaging parameters that had greatest variable importance for hard cardiac event prediction were age, CAC score, systolic blood pressure, number of calcified coronary lesions, and aortic valve calcium score (Figure 1). Established circulating markers of ASCVD risk such as LDL, D-dimer, and PAI-1 were highly-ranked variables in the ML model. Among novel serum biomarkers, MMP-9, pentraxin 3, PIGR and, GDF-15 had the greatest variable importance for ML risk prediction. The final ML score included other established inflammatory markers such as hs-CRP, MCP-1, and MPO.
Figure 1.
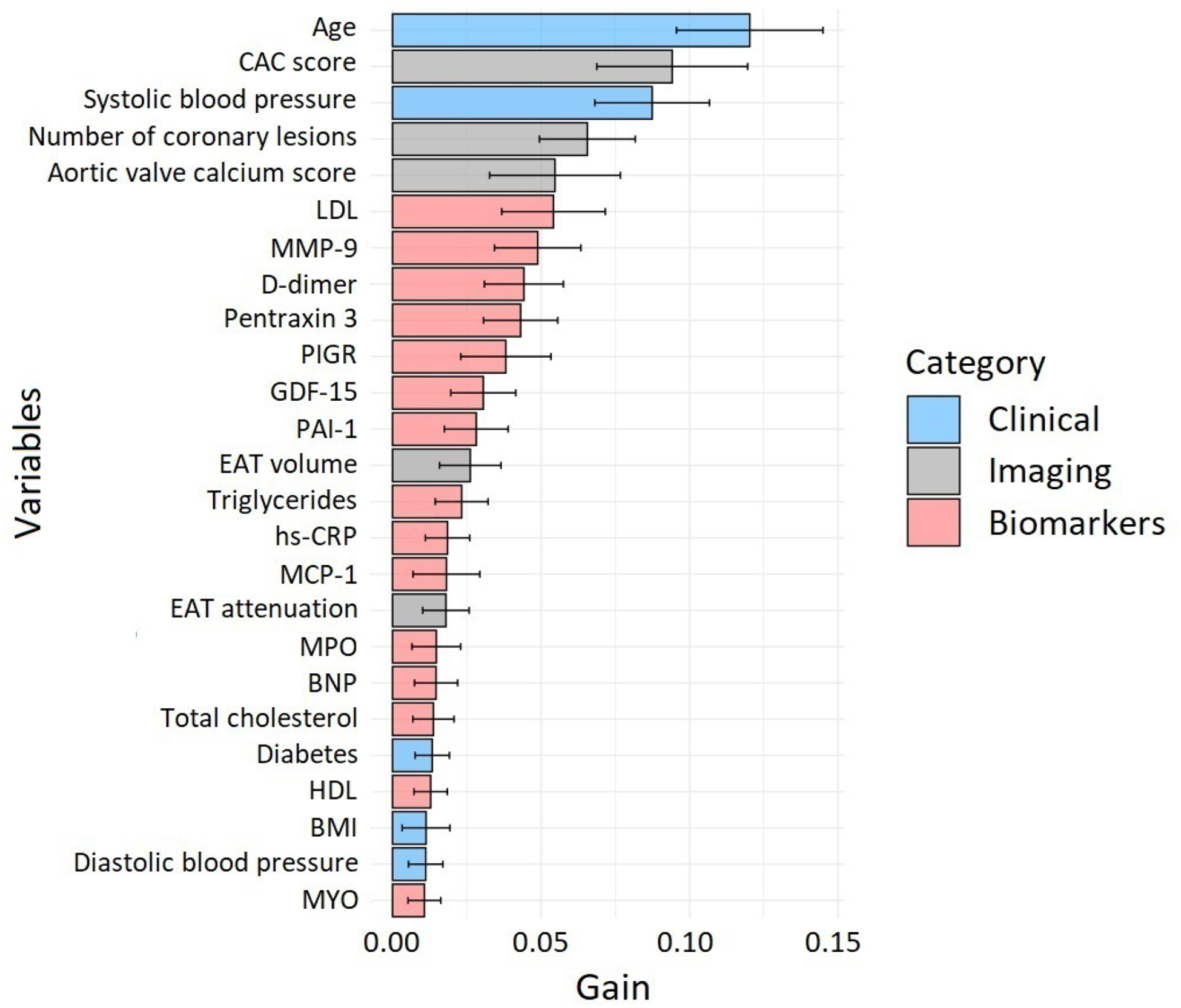
Variable importance for the classification of hard cardiac events.
The top 25 variables are displayed: clinical risk factors in blue, quantitative imaging measures in grey, and serum biomarkers in red. The “gain” denotes how much a variable contributes to the prediction made by the XGBoost algorithm.
Performance of the ML model
The comprehensive ML model integrating clinical risk factors, quantitative CT measures and circulating biomarkers had a significantly higher AUC (0.81 [95% CI: 0.75–0.87]) compared to CAC score (0.75 [95% CI: 0.68–0.81]) and ASCVD score (0.75 [95% CI 0.67–0.80]; both p=0.02) for long-term prediction of hard cardiac events (Figure 2). The comprehensive ML model also had a numerically higher AUC (0.81) than an ML model containing only clinical variables and CT measures (AUC 0.77), with trend towards statistical significance (p=0.10). The addition of serum biomarkers to a ML model with only clinical risk factors and CT measures resulted in substantial event risk reclassification (NRI 0.53 [95% CI: 0.23–0.81], p<0.0001). This was driven primarily by reclassification of non-events (45%, p<0.0001) over reclassification of events (8%, p=0.37).
Figure 2.
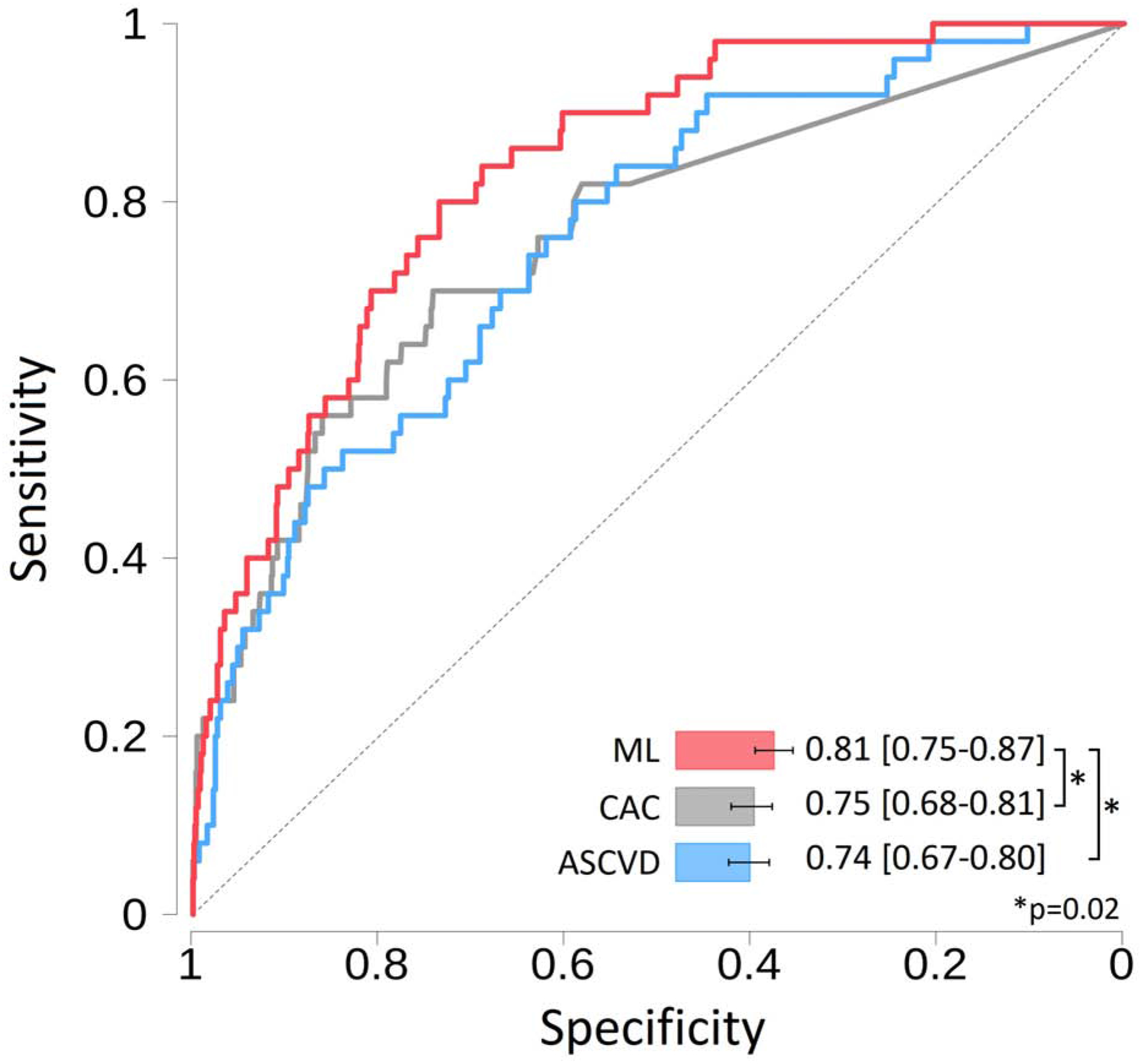
Receiver operator characteristic curves for the prediction of hard cardiac events.
The machine learning model with serum biomarkers performed significantly better than the ASCVD risk score and CAC score (both p=0.02).
Categorization of the ML risk score
The study population was stratified into ‘high’ and ‘low’ ML risk scores according to the optimal cutoff of 0.075 as determined by Youden’s index. At this value, the ML score was associated with sensitivity of 78.6% (95% CI: 61.8–86.9), specificity of 76.0% (95% CI: 73.1–78.7), and accuracy of 75.9% (95% CI: 73.1–78.6). Kaplan-Meier curves of hard events in subjects with high (≥0.075) versus low (<0.075) ML risk scores (log-rank p<0.0001) are shown in Figure 3.
Figure 3.
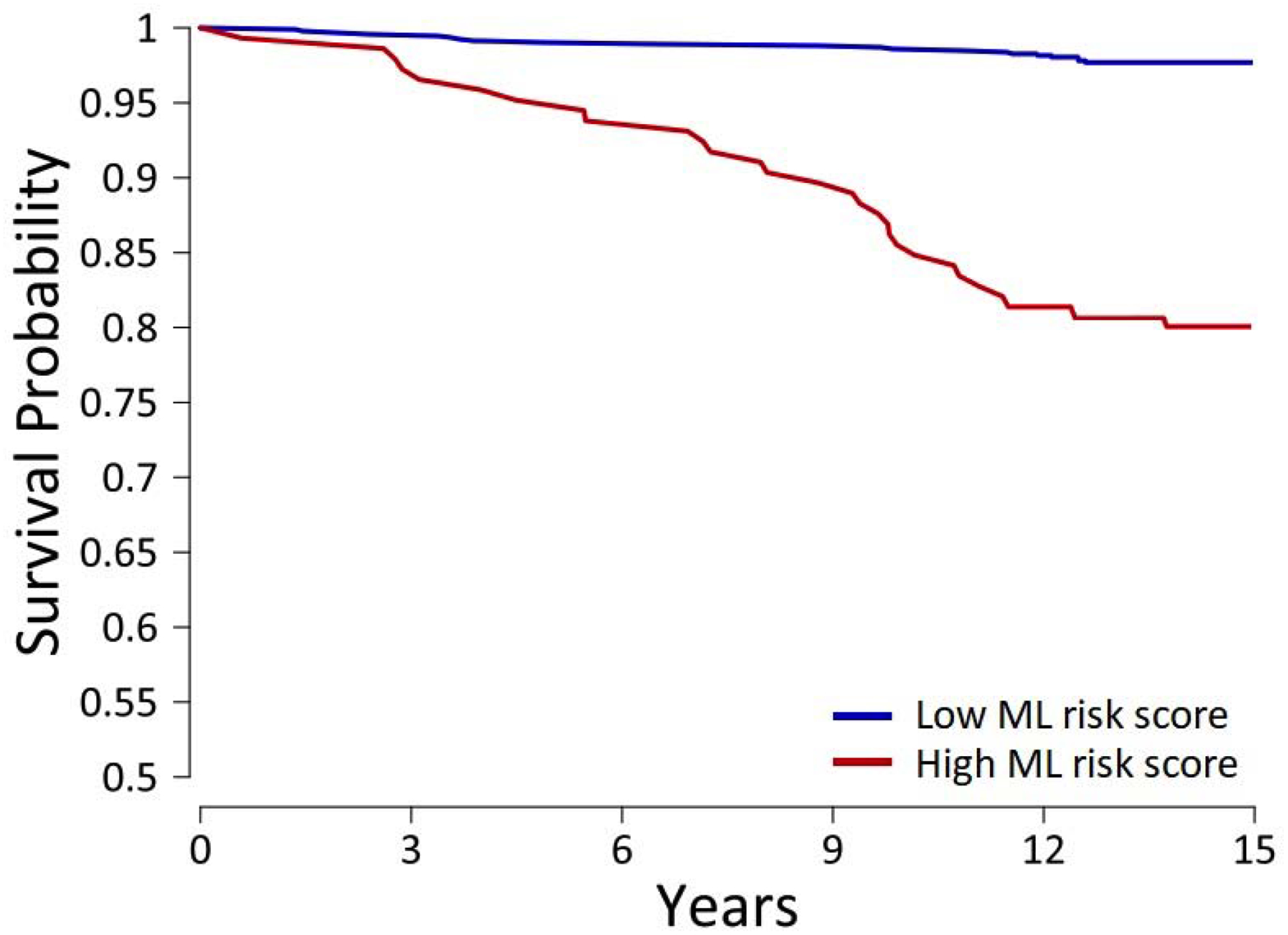
Kaplan-Meier curves of hard cardiac events with a high versus low ML risk score.
Cumulative probability of survival was worse in subjects with a high ML score (log-rank p<0.0001).
Explainable individualized ML risk prediction
Figure 5 demonstrates case examples of individualized ML risk score prediction: one for a male with no observed event over 14 years(Figure 4A), and one for a female with an observed event at 8.8 years(Figure 4B). The x-axis corresponds to the ML risk score. The arrows represent the influence of each variable on the overall prediction; blue and red arrows indicate whether the associated parameters decrease (blue) or increase (red) the risk of future events. The combination of all variables’ influence provides the final ML risk score. The blue and red backgrounds denote the separation between low versus high ML risk.
Figure 4.
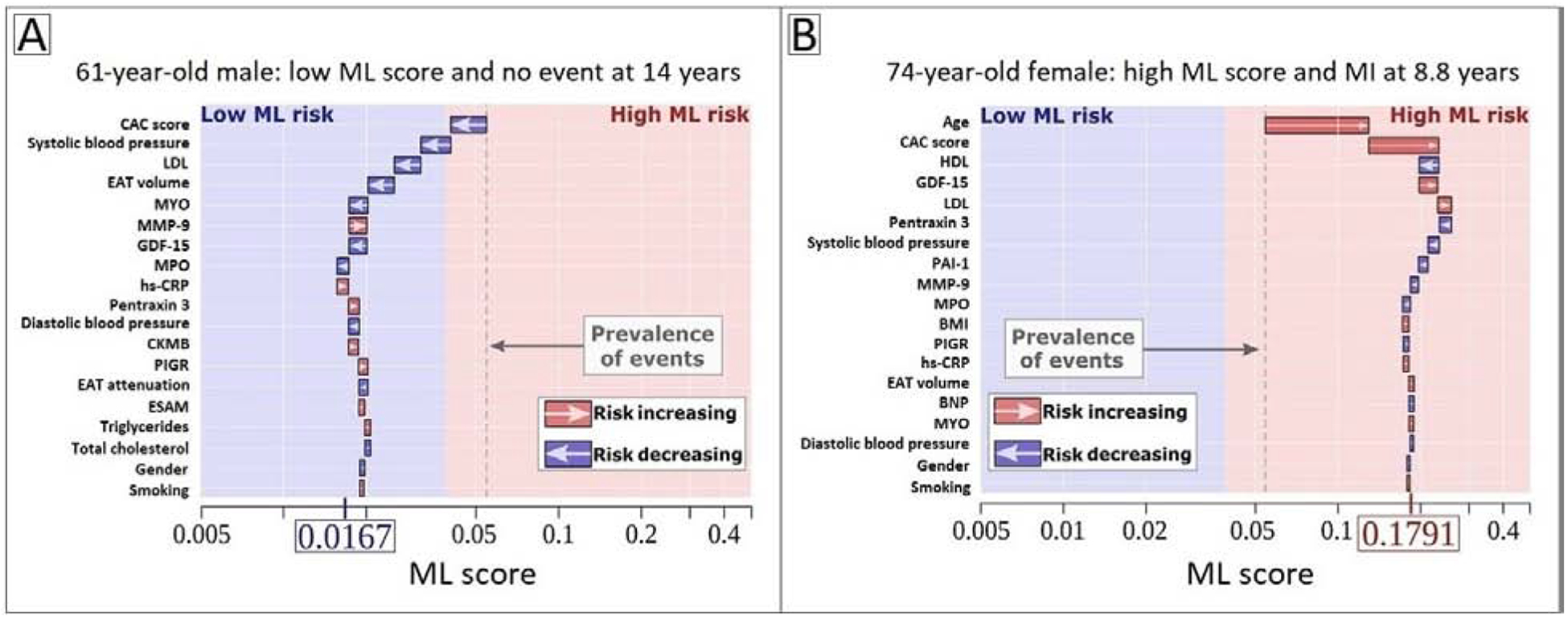
Individualized ML risk prediction with subject-specific variable importance.
(A) 62-year-old male with no event at 14 years and (B) 74-year-old female with an MI at 8.8 years. The X-axis denotes the ML risk score. The arrows represent the influence of each variable on the overall prediction; blue and red arrows indicate whether the associated parameters decrease (blue) or increase (red) the risk of future events. The combination of all variables’ influence provides the final ML risk score. The subject in (A) has a low ML risk score (0.0167), with an ASCVD risk score of 7.25% and a CAC score of 0. The subject in (B) has a high ML risk score (0.1791), with an ASCVD risk score of 30.4% and a CAC score of 324. The blue and red background colors indicate low versus high ML risk according to the Youden’s index cutoff of 0.075, and the gray dashed line corresponds to the base risk obtained from the prevalence of events in the population (4.7%).
ASCVD, atherosclerotic cardiovascular disease; BNP, brain natriuretic peptide; CAC, coronary artery calcium; CKMB, creatine kinase MB; CRP, C-reactive protein; EAT, epicardial adipose tissue; ESAM, endothelial cell-selective adhesion molecule; GDF-15, growth differentiation factor 15; HDL, high-density lipoprotein; LDL, low-density lipoprotein; MCP-1, monocyte chemoattractant protein 1; ML, machine learning; MMP-9, matrix metalloprotease 9; MPO: myeloperoxidase; PAI-1, plasminogen activator inhibitor 1; PIGR, polymeric immunoglobulin receptor.
DISCUSSION
In this study of asymptomatic subjects undergoing CAC scoring CT, our primary findings are: (1) an ML score integrating clinical risk factors, CT measures, and circulating biomarkers outperforms CAC score or ASCVD risk score for long-term prediction of hard cardiac events; (2) serum biomarkers provide incremental prognostic value over and above clinical and imaging variables in an ML model; (3) novel circulating markers of inflammation, extracellular matrix remodeling, and fibrosis have high variable importance for ML prediction.
There has been great interest in the identification of serum biomarkers, which can accurately risk stratify individuals for future major adverse cardiovascular events (MACE) and hence guide individualized medical therapy. Most studies have focused on cohorts with established CAD or heart failure6, 7, 17. Further, few have developed objective risk prediction scores from a comprehensive set of clinical variables, imaging metrics, and serum biomarkers. The present analysis is the one of the first to use ML to develop an integrated score that includes clinical and imaging parameters and novel circulating biomarkers for long-term prognostication in asymptomatic subjects.
CAC is a direct marker of coronary atherosclerosis and assessed noninvasively with low radiation dose noncontrast CT3. The CAC score is proportionally associated with increasing risk of MACE and outperforms traditional risk assessment tools such as the Framingham Risk Score (FRS) and ASCVD risk score3, 18. Beyond CAC scoring, the number of calcified coronary lesions19 and aortic valve calcium score20 are established predictors of all-cause and cardiovascular mortality, respectively, in asymptomatic individuals. Furthermore, EAT volume and attenuation quantified from CAC scoring CT have independent prognostic value for cardiac events21. With respect to circulating biomarkers, CRP, IL-6, MCP-1 fibrinogen, and MPO have been shown to predict MACE in asymptomatic individuals5, 22. A multiple biomarker strategy has also been applied for prediction of ASCVD morbidity and mortality in high-risk populations23. However, biomarkers used either individually or in combination have largely failed to improve MACE prediction over traditional risk factors in asymptomatic cohorts24–26. In a study of 1286 subjects from the EISNER study, Rana et al.24 showed that addition of multiple traditional biomarkers to FRS did not provide incremental predictive value for MACE at 4 years. By contrast, our ML technique integrating 48 traditional and novel blood biomarkers with comprehensive clinical and imaging data outperformed current risk assessment tools for predicting hard cardiac events at 14 years. While we have previously used XGBoost for risk prediction9, the present analysis is a unique EISNER substudy in which novel serum biomarkers were collected in addition to CAC scoring CT in asymptomatic subjects.
Among clinical risk factors, age and systolic blood pressure featured strongly in the ML score, consistent with traditional risk assessment tools. Notably, in our study, personalized clinical risk measures (such as systolic blood pressure, providing a more accurate indication of a subject’s hypertensive state) were more important than dichotomized risk factors (such as diabetes). As expected, LDL cholesterol, a causal risk factor for ASCVD27, was highly ranked in the ML algorithm. D-dimer and PAI-1, well-established inflammatory and thrombotic biomarkers of cardiac risk in asymptomatic populations28, had high variable importance in our ML model. The unique biomarkers with greatest contribution to ML risk prediction (MMP-9, pentraxin 3, PIGR and GDF-15) reflect the pathophysiological pathways of inflammation, extracellular matrix remodeling, and fibrosis. MMP-9 regulates pathological myocardial remodeling via degradation of the extracellular matrix and release of proinflammatory cytokines, and increased serum levels of this enzyme are observed in individuals with established CAD29 and heart failure30. Pentraxin 3, an acute phase inflammatory reactant, is expressed in coronary atherosclerotic lesions and associates with plaque vulnerability31. High serum levels of pentraxin 3 independently predict cardiac mortality in individuals with MI and heart failure32. PIGR is a transmembrane protein, which has been recently linked to CAC incidence and progression33. GDF-15, a stress-responsive member of the transforming growth factor-β cytokine superfamily, is an emerging prognostic biomarker in heart failure34. The present analysis is the first to apply these novel serum biomarkers to cardiac event prediction in asymptomatic subjects, and to demonstrate their prognostic value when combined with traditional risk markers such as CAC score and LDL. Our findings lend further mechanistic support to the role of inflammation and vascular remodeling in coronary atherosclerosis.
By objectively integrating clinical data, quantitative CT measures, and serum biomarkers, our ML score provides superior performance for MACE prediction compared to CAC score or ASCVD risk score alone. We also used repeated 10-fold cross-validation to provide a robust estimation of prediction accuracy with minimal bias; a powerful alternative when separate validation populations are not available35. Another advantage of using this ML model is the ability to explicitly describe the influence of each variable for individualized prediction. Such stratification of significant clinical parameters and serum biomarkers could potentially guide therapy targeted at the specific patient factors affecting cardiac outcomes. Further, combining different circulating biomarkers from distinct pathophysiological pathways enables the assessment of their interaction and independent contribution to risk. The recent advent of multiplex assays providing efficient and cost-saving multi-marker assessment will lead to more clinical studies of novel blood biomarkers, which may enhance outcome prediction and personalization of medical therapy. Finally, we used a rapid, fully automated deep learning method of EAT quantification from standard CAC scoring CT, which has the potential for integration into routine clinical practice, without additional radiation exposure to the patient or increased physician workload.
Limitations
Ours was a single center study of middle-aged asymptomatic subjects, hence limiting broad applicability of our findings to other populations. The limited number of hard cardiac events (50 events over 14.5±2.0 years) is typical of a low-risk patient group. We acknowledge the reduction in effective sample size and potential bias due to patients who were lost to follow-up or had missing CT image data. XGBoost, being a gradient boosting decision tree algorithm, requires longer training times and is prone to overfitting. Our ML model also requires external validation in an independent cohort. However, this would require an asymptomatic population with CAC scoring CT images, comprehensive data on novel serum biomarkers, and long-term follow-up; which was not available within the timeline of the present analysis.
CONCLUSION
In this prospective study, ML integration of novel circulating biomarkers and noninvasive imaging measures provided superior long-term risk prediction for cardiac events compared to current risk assessment tools. Serum biomarkers showed incremental prognostic value over and above clinical data and CT measures in the ML model.
Supplementary Material
HIGHLIGHTS.
We used machine learning (ML) to integrate clinical data, imaging measures, and serum biomarkers for cardiac prognostication.
The calculated ML risk score outperformed current risk assessment tools for the long-term prediction of hard cardiac events.
Serum biomarkers provided incremental prognostic value beyond clinical and imaging features in a ML model.
Novel biomarkers of inflammation, extracellular matrix remodeling, and fibrosis had high variable importance for ML prediction.
Our ML model can provide individualized, patient-specific explanations of its predictions.
FINANCIAL SUPPORT
This study was funded in part by NIH/NHLBI grants [1R01HL133616 and 1R01HL148787-01A1].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INEREST
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- 1.Pencina MJ, D’Agostino RB Sr., Larson MG, Massaro JM and Vasan RS. Predicting the 30-year risk of cardiovascular disease: the framingham heart study. Circulation. 2009;119:3078–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goff DC Jr., Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB, Gibbons R, Greenland P, Lackland DT, Levy D, O’Donnell CJ, Robinson JG, Schwartz JS, Shero ST, Smith SC Jr., Sorlie P, Stone NJ, Wilson PW, Jordan HS, Nevo L, Wnek J, Anderson JL, Halperin JL, Albert NM, Bozkurt B, Brindis RG, Curtis LH, DeMets D, Hochman JS, Kovacs RJ, Ohman EM, Pressler SJ, Sellke FW, Shen WK, Smith SC Jr. and Tomaselli GF. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129:S49–73. [DOI] [PubMed] [Google Scholar]
- 3.Kelkar AA, Schultz WM, Khosa F, Schulman-Marcus J, O’Hartaigh BW, Gransar H, Blaha MJ, Knapper JT, Berman DS, Quyyumi A, Budoff MJ, Callister TQ, Min JK and Shaw LJ. Long-Term Prognosis After Coronary Artery Calcium Scoring Among Low-Intermediate Risk Women and Men. Circ Cardiovasc Imaging. 2016;9:e003742. [DOI] [PubMed] [Google Scholar]
- 4.Mahabadi AA, Lehmann N, Mohlenkamp S, Pundt N, Dykun I, Roggenbuck U, Moebus S, Jockel KH, Erbel R, Kalsch H and Heinz Nixdorf Investigative G. Noncoronary Measures Enhance the Predictive Value of Cardiac CT Above Traditional Risk Factors and CAC Score in the General Population. JACC Cardiovasc Imaging. 2016;9:1177–1185. [DOI] [PubMed] [Google Scholar]
- 5.Kaptoge S, Di Angelantonio E, Lowe G, Pepys MB, Thompson SG, Collins R and Danesh J. C-reactive protein concentration and risk of coronary heart disease, stroke, and mortality: an individual participant meta-analysis. Lancet. 2010;375:132–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong YK, Cheung CYY, Tang CS, Au KW, Hai JSH, Lee CH, Lau KK, Cheung BMY, Sham PC, Xu A, Lam KSL and Tse HF. Age-Biomarkers-Clinical Risk Factors for Prediction of Cardiovascular Events in Patients With Coronary Artery Disease. Arterioscler Thromb Vasc Biol. 2018;38:2519–2527. [DOI] [PubMed] [Google Scholar]
- 7.Tromp J, Khan MA, Klip IT, Meyer S, de Boer RA, Jaarsma T, Hillege H, van Veldhuisen DJ, van der Meer P and Voors AA. Biomarker Profiles in Heart Failure Patients With Preserved and Reduced Ejection Fraction. J Am Heart Assoc. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rozanski A, Gransar H, Shaw LJ, Kim J, Miranda-Peats L, Wong ND, Rana JS, Orakzai R, Hayes SW, Friedman JD, Thomson LEJ, Polk D, Min J, Budoff MJ and Berman DS. Impact of Coronary Artery Calcium Scanning on Coronary Risk Factors and Downstream Testing. Journal of the American College of Cardiology. 2011;57:1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Commandeur F, Slomka PJ, Goeller M, Chen X, Cadet S, Razipour A, McElhinney P, Gransar H, Cantu S, Miller RJH, Rozanski A, Achenbach S, Tamarappoo BK, Berman DS and Dey D. Machine learning to predict the long-term risk of myocardial infarction and cardiac death based on clinical risk, coronary calcium, and epicardial adipose tissue: a prospective study. Cardiovascular Research. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agatston AS, Janowitz WR, Hildner FJ, Zusmer NR, Viamonte M Jr., and Detrano R Quantification of coronary artery calcium using ultrafast computed tomography. J Am Coll Cardiol. 1990;15:827–32. [DOI] [PubMed] [Google Scholar]
- 11.Commandeur F, Goeller M, Razipour A, Cadet S, Hell MM, Kwiecinski J, Chen X, Chang HJ, Marwan M, Achenbach S, Berman DS, Slomka PJ, Tamarappoo BK and Dey D. Fully Automated CT Quantification of Epicardial Adipose Tissue by Deep Learning: A Multicenter Study. Radiology: Artificial Intelligence. 2019;1:e190045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guyon I and Elisseeff A. An introduction to variable and feature selection. Journal of Machine Learning Research. 2003;3:1157–1182. [Google Scholar]
- 13.Al’Aref SJ, Singh G, Choi JW, Xu Z, Maliakal G, van Rosendael AR, Lee BC, Fatima Z, Andreini D, Bax JJ, Cademartiri F, Chinnaiyan K, Chow BJW, Conte E, Cury RC, Feuchtner G, Hadamitzky M, Kim Y-J, Lee S-E, Leipsic JA, Maffei E, Marques H, Plank F, Pontone G, Raff GL, Villines TC, Weirich HG, Cho I, Danad I, Han D, Heo R, Lee JH, Rizvi A, Stuijfzand WJ, Gransar H, Lu Y, Sung JM, Park H-B, Berman DS, Budoff MJ, Samady H, Stone PH, Virmani R, Narula J, Chang H-J, Lin FY, Baskaran L, Shaw LJ and Min JK. A Boosted Ensemble Algorithm for Determination of Plaque Stability in High-Risk Patients on Coronary CTA. JACC: Cardiovascular Imaging. 2020:3447. [DOI] [PubMed] [Google Scholar]
- 14.Molinaro AM, Simon R and Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics. 2005;21:3301–7. [DOI] [PubMed] [Google Scholar]
- 15.DeLong ER, DeLong DM and Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 16.Pencina MJ, D’Agostino RB Sr. and Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omland T and Kullo IJ. Biomarker-Based Risk Models to Risk Stratify Patients With Stable Coronary Heart Disease. J Am Coll Cardiol. 2017;70:827–829. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann N, Erbel R, Mahabadi AA, Rauwolf M, Mohlenkamp S, Moebus S, Kalsch H, Budde T, Schmermund A, Stang A, Fuhrer-Sakel D, Weimar C, Roggenbuck U, Dragano N and Jockel KH. Value of Progression of Coronary Artery Calcification for Risk Prediction of Coronary and Cardiovascular Events: Result of the HNR Study (Heinz Nixdorf Recall). Circulation. 2018;137:665–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams M, Shaw LJ, Raggi P, Morris D, Vaccarino V, Liu ST, Weinstein SR, Mosler TP, Tseng PH, Flores FR, Nasir K and Budoff M. Prognostic value of number and site of calcified coronary lesions compared with the total score. JACC Cardiovasc Imaging. 2008;1:61–9. [DOI] [PubMed] [Google Scholar]
- 20.Owens DS, Budoff MJ, Katz R, Takasu J, Shavelle DM, Carr JJ, Heckbert SR, Otto CM, Probstfield JL, Kronmal RA and O’Brien KD. Aortic Valve Calcium Independently Predicts Coronary and Cardiovascular Events in a Primary Prevention Population. JACC: Cardiovascular Imaging. 2012;5:619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goeller M, Achenbach S, Marwan M, Doris MK, Cadet S, Commandeur F, Chen X, Slomka PJ, Gransar H, Cao JJ, Wong ND, Albrecht MH, Rozanski A, Tamarappoo BK, Berman DS and Dey D. Epicardial adipose tissue density and volume are related to subclinical atherosclerosis, inflammation and major adverse cardiac events in asymptomatic subjects. J Cardiovasc Comput Tomogr. 2018;12:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnabel Renate B, Yin X, Larson Martin G, Yamamoto Jennifer F, Fontes João D, Kathiresan S, Rong J, Levy D, Keaney John F, Wang Thomas J, Murabito Joanne M, Vasan Ramachandran S and Benjamin Emelia J. Multiple Inflammatory Biomarkers in Relation to Cardiovascular Events and Mortality in the Community. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33:1728–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Holten TC, Waanders LF, de Groot PG, Vissers J, Hoefer IE, Pasterkamp G, Prins MWJ and Roest M. Circulating biomarkers for predicting cardiovascular disease risk; a systematic review and comprehensive overview of meta-analyses. PloS one. 2013;8:e62080–e62080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rana JS, Gransar H, Wong ND, Shaw L, Pencina M, Nasir K, Rozanski A, Hayes SW, Thomson LE, Friedman JD, Min JK and Berman DS. Comparative value of coronary artery calcium and multiple blood biomarkers for prognostication of cardiovascular events. Am J Cardiol. 2012;109:1449–53. [DOI] [PubMed] [Google Scholar]
- 25.Wang TJ, Gona P, Larson MG, Tofler GH, Levy D, Newton-Cheh C, Jacques PF, Rifai N, Selhub J, Robins SJ, Benjamin EJ, D’Agostino RB and Vasan RS. Multiple Biomarkers for the Prediction of First Major Cardiovascular Events and Death. New England Journal of Medicine. 2006;355:2631–2639. [DOI] [PubMed] [Google Scholar]
- 26.Folsom AR, Chambless LE, Ballantyne CM, Coresh J, Heiss G, Wu KK, Boerwinkle E, Mosley TH Jr., Sorlie P, Diao G and Sharrett AR. An assessment of incremental coronary risk prediction using C-reactive protein and other novel risk markers: the atherosclerosis risk in communities study. Arch Intern Med. 2006;166:1368–73. [DOI] [PubMed] [Google Scholar]
- 27.Ference BA, Ginsberg HN, Graham I, Ray KK, Packard CJ, Bruckert E, Hegele RA, Krauss RM, Raal FJ, Schunkert H, Watts GF, Borén J, Fazio S, Horton JD, Masana L, Nicholls SJ, Nordestgaard BG, van de Sluis B, Taskinen M-R, Tokgözoğlu L, Landmesser U, Laufs U, Wiklund O, Stock JK, Chapman MJ and Catapano AL. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. European Heart Journal. 2017;38:2459–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe GD, Yarnell JW, Sweetnam PM, Rumley A, Thomas HF and Elwood PC. Fibrin D-dimer, tissue plasminogen activator, plasminogen activator inhibitor, and the risk of major ischaemic heart disease in the Caerphilly Study. Thromb Haemost. 1998;79:129–33. [PubMed] [Google Scholar]
- 29.Blankenberg S, Rupprecht HJ, Poirier O, Bickel C, Smieja M, Hafner G, Meyer J, Cambien F, Tiret L and AtheroGene I. Plasma concentrations and genetic variation of matrix metalloproteinase 9 and prognosis of patients with cardiovascular disease. Circulation. 2003;107:1579–85. [DOI] [PubMed] [Google Scholar]
- 30.Morishita T, Uzui H, Mitsuke Y, Amaya N, Kaseno K, Ishida K, Fukuoka Y, Ikeda H, Tama N, Yamazaki T, Lee JD and Tada H. Association between matrix metalloproteinase-9 and worsening heart failure events in patients with chronic heart failure. ESC Heart Fail. 2017;4:321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soeki T, Niki T, Kusunose K, Bando S, Hirata Y, Tomita N, Yamaguchi K, Koshiba K, Yagi S, Taketani Y, Iwase T, Yamada H, Wakatsuki T, Akaike M and Sata M. Elevated concentrations of pentraxin 3 are associated with coronary plaque vulnerability. Journal of Cardiology. 2011;58:151–157. [DOI] [PubMed] [Google Scholar]
- 32.Latini R, Gullestad L, Masson S, Nymo SH, Ueland T, Cuccovillo I, Vårdal M, Bottazzi B, Mantovani A, Lucci D, Masuda N, Sudo Y, Wikstrand J, Tognoni G, Aukrust P, Tavazzi L, on behalf of the Investigators of the Controlled Rosuvastatin Multinational Trial in Heart F and trials GI-HF. Pentraxin-3 in chronic heart failure: the CORONA and GISSI-HF trials. European Journal of Heart Failure. 2012;14:992–999. [DOI] [PubMed] [Google Scholar]
- 33.Eisen A, Kornowski R, Hamdan A, Talmor-Barkan Y, Witberg G, Deshpande K, Paixao A, Ayers C, Joshi P, Rohatgi A, Khera A, Lemos J and Neeland I. NOVEL BIOMARKERS OF CORONARY ARTERY CALCIUM INCIDENCE OR PROGRESSION: INSIGHTS FROM THE DALLAS HEART STUDY. Journal of the American College of Cardiology. 2019;73:1783. [Google Scholar]
- 34.Cotter G, Voors AA, Prescott MF, Felker GM, Filippatos G, Greenberg BH, Pang PS, Ponikowski P, Milo O, Hua TA, Qian M, Severin TM, Teerlink JR, Metra M and Davison BA. Growth differentiation factor 15 (GDF-15) in patients admitted for acute heart failure: results from the RELAX-AHF study. Eur J Heart Fail. 2015;17:1133–43. [DOI] [PubMed] [Google Scholar]
- 35.Kim J-H. Estimating classification error rate: Repeated cross-validation, repeated hold-out and bootstrap. Computational Statistics & Data Analysis. 2009;53:3735–3745. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


