Abstract
Background
Susceptibility to Vibrio cholerae infection is affected by blood group, age, and preexisting immunity, but these factors only partially explain who becomes infected. A recent study used 16S ribosomal RNA amplicon sequencing to quantify the composition of the gut microbiome and identify predictive biomarkers of infection with limited taxonomic resolution.
Methods
To achieve increased resolution of gut microbial factors associated with V. cholerae susceptibility and identify predictors of symptomatic disease, we applied deep shotgun metagenomic sequencing to a cohort of household contacts of patients with cholera.
Results
Using machine learning, we resolved species, strains, gene families, and cellular pathways in the microbiome at the time of exposure to V. cholerae to identify markers that predict infection and symptoms. Use of metagenomic features improved the precision and accuracy of prediction relative to 16S sequencing. We also predicted disease severity, although with greater uncertainty than our infection prediction. Species within the genera Prevotella and Bifidobacterium predicted protection from infection, and genes involved in iron metabolism were also correlated with protection.
Conclusion
Our results highlight the power of metagenomics to predict disease outcomes and suggest specific species and genes for experimental testing to investigate mechanisms of microbiome-related protection from cholera.
Keywords: Vibrio cholera, cholera, microbiome, machine learning, metagenomics
Cholera infection and disease severity can be predicted using metagenomic sequencing of the gut microbiome before infection in a prospective cohort, and the results suggest potentially protective bacterial species and genes.
Cholera is an acute diarrheal disease caused by Vibrio cholerae. It is a major public health threat worldwide that continues to cause major outbreaks, such as in Yemen, where >1.7 million cases have been reported since 2016 [1, 2]. Transmission of V. cholerae between household members commonly occurs through shared sources of contaminated food or water or through fecal-oral spread [3, 4]. The clinical spectrum of disease ranges from asymptomatic infection to severe watery diarrhea that can lead to fatal dehydration [5]. Host factors such as age, innate immune factors, blood group, or prior acquired immunity partially explain why some people are more susceptible to V. cholerae infection than others, but a substantial amount of the variation remains unexplained [6].
The gut bacterial community can protect against enteropathogenic infections [7], and may explain some of the variation in V. cholerae susceptibility. Several studies have identified commensal bacteria and mechanisms that could be protective against V. cholerae. For instance, a species enriched in the gut microbiota of patients recovering from cholera, Blautia obeum, was found to interfere with V. cholerae pathogenicity through quorum-sensing inhibition in a mouse model [8]. Other experiments have demonstrated that alteration of commensal-derived metabolite levels influenced host susceptibility by affecting V. cholerae growth or colonization [9–13].
Studies of V. cholerae and the gut microbiota often focus on a few bacterial species or involve patients who already have symptomatic cholera [8, 14]. One study recently characterized the gut microbiome of healthy individuals exposed to V. cholerae. In that study, Midani et al [15] developed a machine learning model to predict susceptibility based on 16S ribosomal RNA (rRNA) gene amplicon sequencing of the gut microbiota in a group known to have high risk of infection: household contacts of confirmed cholera patients [4]. They showed that microbiome composition at the time V. cholerae exposure to can predict infection with similar or better accuracy as commonly measured host factors known to affect susceptibility. However, 16S rRNA sequencing has limited taxonomic resolution and does not identify the genetic mechanisms of protection.
In the current study we used shotgun metagenomics to analyze an expanded prospective cohort of persons exposed to V. cholerae in Bangladesh. Our metagenomic analysis yielded improved outcome predictions compared to 16S rRNA sequencing, and identified bacterial genes associated with remaining uninfected after exposure to V. cholerae. We are also able to predict disease severity among infected contacts, albeit with lower power and precision than susceptibility. Finally, we highlight several microbiome-encoded metabolic functions associated with protection against cholera.
METHODS
Sample Collection, Clinical Outcomes, and Metagenomic Sequencing
As described elsewhere (15), household contacts were enrolled within 6 hours of the presentation of an index cholera case at Dhaka Hospital, of the icddr,b (International Centre for Diarrhoeal Disease Research, Bangladesh). Index patients with severe acute diarrhea, a stool culture positive for V. cholerae, age 2–60 years, and no major comorbid conditions were recruited [4, 6]. A clinical assessment of symptoms in household contacts was conducted daily for the 10-day period after presentation of the index case, and repeated on day 30. We collected demographic information, rectal swab specimens, and blood samples for ABO typing and vibriocidal antibody titers as described in the Supplementary Methods.
During the observation period, contacts were determined to be infected if any rectal swab specimen culture was positive for V. cholerae and/or if the contact developed diarrhea and a 4-fold increase in vibriocidal titer during the follow-up period [4, 6]. Contacts with positive rectal swab specimens developing watery diarrhea were categorized as symptomatic, and those without diarrhea were considered asymptomatic (Figure 1). V. cholerae–positive contacts (by culture or deep 16S amplicon sequencing [15]) at the time of enrollment were excluded, in addition to contacts who reported antibiotic use or diarrhea during the week before enrollment. DNA extraction was performed for the selected samples and used for shotgun metagenomics sequencing. Details on cohorts, sequencing methods, and sample processing are described in the Supplementary Methods. The Ethical and Research Review Committees of the icddr,b and the Institutional Review Board of Massachusetts General Hospital reviewed the study. All adult subjects and parents/guardians of children provided written informed consent.
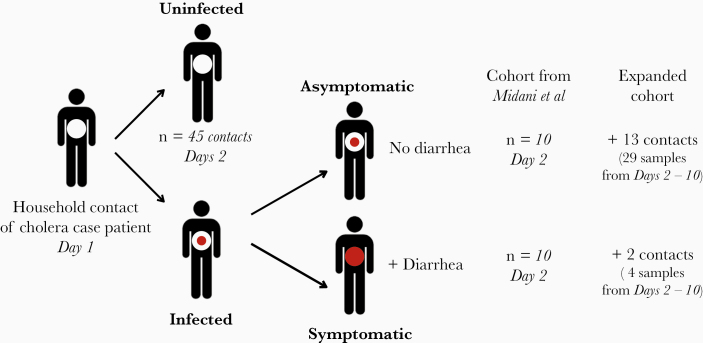
Figure 1.
Study cohort in Dhaka, Bangladesh. After presentation of a Vibrio cholerae culture-positive index case to the hospital on day 1, household contacts were enrolled on day 2. The expanded cohort includes the 2018 cohort from Midani et al [15], with an addition of 33 samples from infected individuals (13 asymptomatic and 2 symptomatic).
Taxonomic/Functional Profiling and Predictive Model Construction
We used MetaPhlAn2 software (version 2.9) [16] for taxonomic profiling and HUMAnN2 software [17] to profile cellular pathways (from the MetaCyc database) and gene families (identified using the Pfam database). To identify biomarkers of susceptibility and disease severity, we used MetAML software [18] to apply a random forest (RF) classifier on species, pathways, and gene family relative abundances, as well as the presence or absence of strain-specific markers. Models constructed using each of these features types were compared with a random data set with shuffled labels, and to a model constructed with clinical and demographic data, using 2-sample, 2-sided t tests over 20 replicate cross-validation [18].
We used a stratified 3-fold cross validation approach, splitting our data set into validation and training sets (one-third and two-thirds of samples, respectively) with the same infected-uninfected ratio. We used an embedded feature selection strategy to identify the most useful features and improve model accuracy. Feature relative importance was computed using the mean decrease in impurity strategy, which calculates the importance of each feature as the sum of the number of nodes (across all trees) that use the feature, proportional to the number of samples each of these nodes splits [18]. Further details are described in the Supplementary Methods.
Data availability
After removal of human reads, the sequence data has been deposited in NCBI under BioProject PRJNA608678.
RESULTS
Metagenomic Sequencing of the Gut Microbiome in Household Contacts Exposed to V. cholerae
We performed metagenomic sequencing of the gut microbiome in 65 contacts of cholera case patients from a cohort described by Midani et al [15], from which sufficient DNA remained. Of these 65 contacts, referred to as the Midani 2018 cohort, 20 experienced infection during the follow-up period, and 45 remained uninfected (Figure 1). Among the 20 contacts who became infected, 10 had no symptoms during the 30-day follow-up period and were classified as asymptomatic, and 10 experienced symptoms (Supplementary Methods). To increase our sample size, we surveyed an expanded cohort (Supplementary Table 1a; All supplementary Tables S1–S9 are available at: https://figshare.com/articles/Supplementary_Tables_-_Levade_et_al_2020/12440417.) by adding 33 samples, including 10 additional preinfection samples from time points for contacts in the Midani 2018 cohort, and 23 samples from 16 newly enrolled contacts from the same place and time (2012–2014; Dhaka, Bangladesh). We used preinfection samples to identify predictive features of disease outcomes in the Midani 2018 cohort, on which we base the majority of our analyses. We also performed exploratory analyses on the expanded cohort to determine the potential for predictive models to be generalized to larger samples.
We used the shotgun metagenomic DNA sequence reads from these samples to characterize 4 features of the microbiome: (1) relative abundances of microbial species, (2) the presence/absence of subspecies-level strains, 3) metabolic pathway relative abundances, and (4) gene family relative abundances (Table 1).
Table 1.
Assessment of Prediction Performance for a Random Forest Model Applied to the 2018 Cohort from Midani et al [15] and the Expanded Cohorta
| Mean Value (Margin of Error) Determined With Random Forest Model | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cohort from Midani et al [15] | Expanded Cohort | |||||||
| Prediction | Species Abundance | Strain Markers | Gene Families | Pathways | Species Abundance | Strain Markers | Gene Families | Pathways |
| Features, no. | 705 | 54 953 | 6810 | 443 | 807 | 62 965 | 7514 | 461 |
| Infected vs uninfected | ||||||||
| Accuracy | 0.73 (0.02) | 0.71 (0.02) | 0.76 (0.02) | 0.72 (0.02) | 0.76 (0.03) | 0.69 (0.03) | 0.80 (0.02) | 0.80 (0.03) |
| Precision | 0.71 (0.06) | 0.68 (0.06) | 0.77 (0.04) | 0.70 (0.05) | 0.76 (0.03) | 0.70 (0.03) | 0.81 (0.02) | 0.81 (0.03) |
| F1 score | 0.66 (0.02) | 0.64 (0.03) | 0.71 (0.03) | 0.66 (0.03) | 0. 75 (0.03) | 0.68 (0.03) | 0.80 (0.02) | 0.80 (0.03) |
| AUC | 0.61 (0.05) | 0.62 (0.04) | 0.74 (0.04) | 0.71 (0.04) | 0.83 (0.02) | 0.76 (0.03) | 0.87 (0.02) | 0.88 (0.02) |
| Shuffled | ||||||||
| F1 score | 0.55 (0.04) | 0.56 (0.04) | 0.56 (0.04) | 0.56 (0.05) | 0.40 (0.03) | 0.45 (0.03) | 0.48 (0.03) | 0.44 (0.03) |
| AUC | 0.40 (0.04) | 0.57 (0.04) | 0.50 (0.05) | 0.50 (0.04) | 0.39 (0.03) | 0.52 (0.03) | 0.51 (0.03) | 0.46 (0.03) |
| Asymptomatic vs symptomatic vs uninfected | ||||||||
| Accuracy | 0.70 (0.02) | 0.70 (0.02) | 0.69 (0.01) | 0.69 (0.01) | 0.68 (0.01) | 0.60 (0.03) | 0.69 (0.02) | 0.67 (0.03) |
| Precision | 0.53 (0.03) | 0.53 (0.03) | 0.60 (0.02) | 0.59 (0.02) | 0.60 (0.02) | 0.53 (0.03) | 0.61 (0.02) | 0.59 (0.02) |
| F1 score | 0.60 (0.02) | 0.59 (0.02) | 0.57 (0.02) | 0.57 (0.02) | 0.62 (0.02) | 0.55 (0.03) | 0.64 (0.02) | 0.62 (0.02) |
| AUC | NA | NA | NA | NA | NA | NA | NA | NA |
| Shuffled | ||||||||
| F1 score | 0.48 (0.04) | 0.49 (0.04) | 0.46 (0.03) | 0.55 (0.03) | 0.41 (0.03) | 0.35 (0.03) | 0.44 (0.04) | 0.37 (0.03) |
| AUC | NA | NA | NA | NA | NA | NA | NA | NA |
Abbreviations: AUC, area under the curve; NA, .
aSpecies abundances, presence or absence of strain-specific markers, relative abundance of gene families grouped according to the Pfam database, and pathways from the MetaCyc database were used as features. For each data set, we applied a binary (uninfected vs infected contacts) and a multiclass (asymptomatic vs symptomatic vs uninfected contacts) classifier and reported performance metrics for each data set. Metrics obtained by the same classifier applied to the same data sets with shuffled class labels (random assignment of labels to samples) are also reported (shuffled). Margins of error indicate 95% confidence intervals.
Predicting Susceptibility to V. cholerae Infection With an RF Model
We first used an RF model to predict V. cholerae susceptibility (developing infection or remaining uninfected) from baseline microbiome features (Figure 1). In the Midani 2018 cohort, functional pathways and gene families predicted infection significantly better than random (P < .05; 2-sample t tests comparing area under the curve [AUC] across 20 replicate 3 fold cross-validations) compared with data with shuffled (randomized) labels, and also predicted infection better than species or strain features (Table 1 and Supplementary Table 2). Pathways and gene families had significantly higher mean AUCs (0.71 and 0.74, respectively) than species or strains (0.61 and 0.62, respectively; P < .05) (Table 1, Supplementary Figure 1 and Supplementary Table 3).
To determine the minimum number of metagenomic features required for prediction, we repeated the analysis using smaller subsets of features. Using only 30 species, 60 gene families or pathways, or 200 strains achieved similar cross-validation AUC values (Supplementary Figure 2). We then trained an RF model on this reduced number of selected features, yielding improved predictions for all feature types (Supplementary Figure 1 and Supplementary Table 4). This suggests that only a limited number of strains, species, genes, and pathways in the gut microbiome at the time of exposure are sufficient to predict V. cholerae susceptibility. For example, prediction using strain-level markers after feature selection yielded an AUC of 0.95 (Supplementary Table 4). However, such high AUC values should be treated with caution because the models can be overfit when a supervised feature selection step is applied on the same data used to train the model [18].
Because we did not have a fully independent validation cohort (eg, from another continent) to test our model, we decided to use the features selected from the Midani cohort to make predictions on the expanded data set. Using the same features selected from the Midani 2018 training data set, we made predictions on the expanded cohort and achieved AUCs between 0.89 and 0.93 for prediction of infection using the 4 types of features (Supplementary Table 4). Again, because the expanded cohort partly overlaps with the Midani cohort and includes some repeated samples from the same individuals over time, these results could also be prone to overfitting, but they demonstrate the potential for generalized predictions.
Finally, we repeated the RF analysis using all features in the expanded data set, which increased predictive performance relative to the original Midani cohort (Supplementary Figure 1). Once again, genes and pathways outperformed species and strains according to all metrics, with AUC reaching approximately 0.88 using cellular pathways (Table 1). This improvement in the expanded cohort also highlights the importance of using larger, more balanced data sets as input to predictive models.
Improved Prediction Compared With Known Factors Affecting Susceptibility
To put the metagenomic predictions in context, we compared their predictive power and accuracy with clinical and demographic factors (Supplementary Table 1a). Three of these factors (age, baseline vibriocidal antibodies, and blood group) are known to affect susceptibility to V. cholerae infection [6, 15] and we used them to train RF models (Supplementary Table 5). As expected, contacts who became infected tended to be younger and have lower baseline antibody titers than those who remained uninfected (Supplementary Table 1b), but these small differences were not sufficient to train a significantly predictive model.
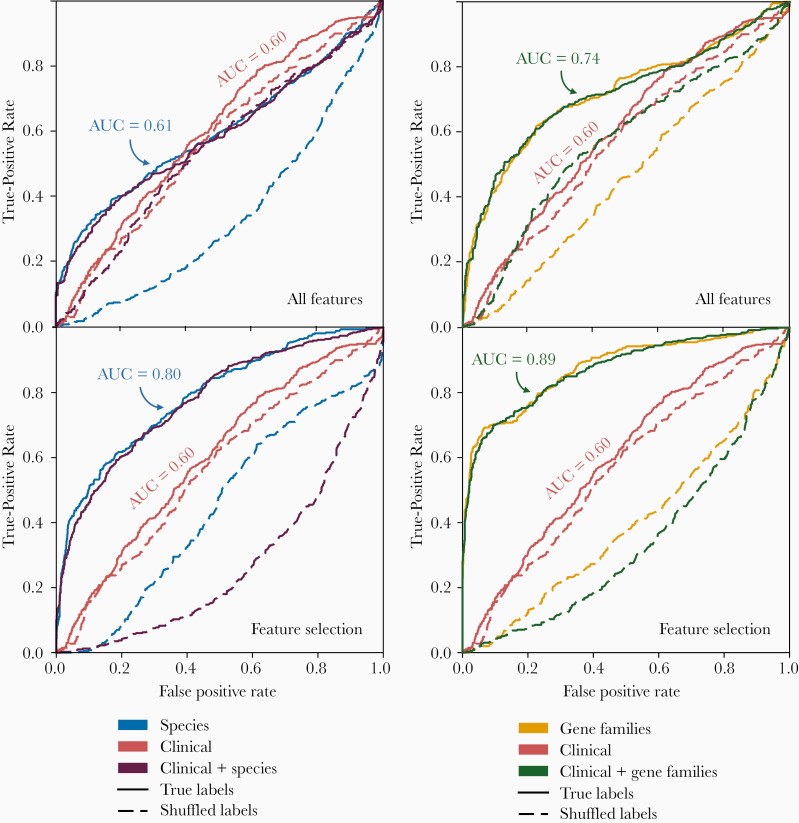
An RF model trained on the 7 clinical and demographic factors did not perform better than a random model with shuffled labels (AUC, 0.60; P = .66) (Figure 2). Predictions were not improved using all species-level metagenomic features present at the time of exposure to V. cholerae (AUC, 0.61), but they significantly improved with use of a selected number of species (AUC, 0.80; P < .001). The use of all gene families or a selected number of genes showed an increased predictive performance (AUC, 0.74 and 0.89, respectively; Figure 2) compared with species-level or clinical and demographic contact data (P < .001 for all comparisons).
Figure 2.
Metagenomic features predict Vibrio cholerae infection better than clinical and demographic features. Random forest prediction of infection status was applied to 7 clinical and demographic features, and compared with all species and all gene families (top row), as well as 30 selected species features from metagenomes and 60 selected gene family features (bottom row), or a combination of clinical, demographic, and metagenomic features. Plots show receiver operating characteristic (ROC) curves (average across cross-validations) for the 2018 data set from Midani et al [15]. Shuffled labels represent the prediction run on a data set with a random assignment of infection outcomes. Abbreviation: AUC, area under the curve.
We again note the caveat that models with selected features may be overfit and represent an upper bound for predictive power. Even without feature selection, we found that gene families clearly provide superior predictions, and adding clinical data did not improve the predictions based on microbiome features alone (Figure 2). Together, these results demonstrate that gene families present in the gut microbiome at the time of exposure contain more information about V. cholerae susceptibility than species-level or clinical and demographic contact data.
Difficulty of Predicting Disease Severity
To predict symptomatic disease among infected individuals (Figure 1), we divided samples into uninfected, symptomatic, and asymptomatic groups and again applied the RF approach. We used the F1 score as a performance metric because it is well suited for uneven class distributions in our uninfected/symptomatic/asymptomatic comparison. Applied to the Midani 2018 cohort, this model predicted outcomes significantly better than random (shuffled labels) using species, strains or pathway data, but not gene families (Table 1; see Supplementary Table 3 for P values). However, the F1 scores for the symptomatic/asymptomatic predictions were systematically lower (mean scores, 0.57–0.60) than for the infected/uninfected prediction (0.64 –0.71). In the expanded cohort, the scores were improved only slightly (Table 1). These results suggest that disease severity is predictable in principle, but with greater uncertainty than the infection outcome.
Taxonomic Biomarkers of Disease Susceptibility and Severity
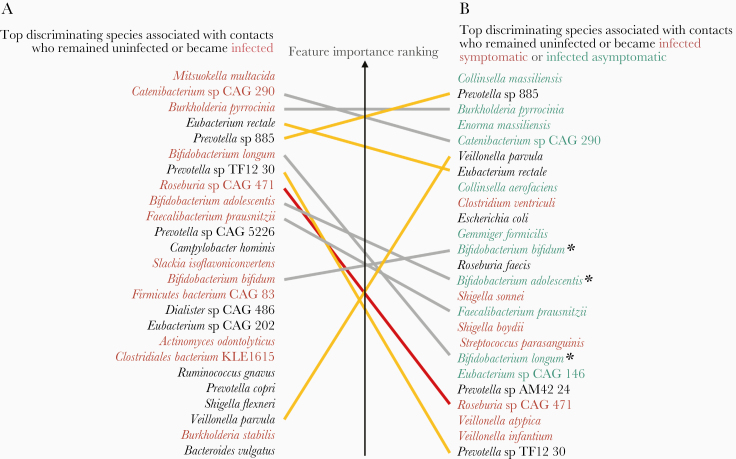
Predictive features in the gut microbiome identified to a species/strain or gene level allow the possibility of experimental follow-up to investigate mechanisms of the associations we observed. We characterized the most predictive species, pathways, and gene families (Supplementary Tables 6–9). The most common discriminating species in individuals that remained uninfected during the follow-up period were Eubacterium rectale, Campylobacter hominis, Ruminococcus gnavus, Bacteroides vulgatus, Veillonella parvula, and members of the Prevotella and Eubacterium genera (Figure 3A and Supplementary Figures3A and 4A). These species are ranked by their importance score, which is effectively their relative weighting in the RF model. Several species associated with contacts in whom V. cholerae infection developed belonged to the genera Bifidobacterium, Actinomyces, or Collinsella, and many of the species were also associated with asymptomatic infection (Figure 3B and Supplementary Figures 3B and 4B), including 3 species of Bifidobacterium.
Figure 3.
Most important discriminating species of the gut microbiome at the time of exposure to Vibrio cholerae identified in the 2018 data set from Midani et al [15], classified by clinical outcome. A, Species associated with contacts who became infected (red) or remained uninfected (black) during follow-up. B, Species associated with contacts who remained uninfected (black), or became infected asymptomatic (green), or symptomatic (red) during follow-up. The top 25 most important features for discriminating between classes in the random forest model are shown here; see Supplementary Table 6 for the full list. Yellow lines connect species associated with uninfected individuals in both A and B; red lines connect species associated with infection in A and symptomatic disease in B; gray lines connect species associated with infection in A but asymptomatic infection in B. Three species of Bifidobacterium are marked with asterisks.
Figure 4.
Most important discriminating gene families of the gut microbiome at the time of exposure to Vibrio cholerae identified in the 2018 data set from Midani et al [15], classified by clinical outcome. A, Genes families associated with contacts who became infected (red) or remained uninfected (black) during follow-up. B, Genes families associated with contacts who remained uninfected (black), or became infected asymptomatic (green) or symptomatic (red) during follow-up. The top 25 most important features for discriminating between classes in the random forest model are shown here; see Supplementary Table 8 for the full list. Yellow lines connect species associated with uninfected individuals in both A and B. Asterisks indicate genes involved in redox or iron metabolism. All PF gene name abbreviations can be found in the Pfam database at https://pfam.xfam.org/.
The top predictive species in contacts who developed symptomatic infection were Clostridium ventriculi (formerly Sarcina ventriculi), Streptococcus parasanguinis, and members of Veillonella. Shigella species were also associated with the gut microbiome of persons who developed symptomatic V. cholerae infection, although persons enrolled in this study had stool cultures negative for Shigella. Shigella identified by DNA presence in stool may be the result of recent or resolving infection or may be present at subclinical levels owing to ingestion of contaminated water. The features identified by the multivariate RF model were confirmed using univariate statistics for the uninfected/infected prediction (Supplementary Figure 5), but the overlap was poorer for the uninfected/symptomatic/asymptomatic prediction (Supplementary Figure 6). This is consistent with the difficulty of predicting disease severity.
In general, the most important species were selected by the model because of differences in relative abundance at baseline among uninfected, symptomatic, and asymptomatic outcomes (Supplementary Figures 7 and 8). In rare cases, species presence or absence was predictive. For example, R. gnavus is absent (near or below the limit of detection) in most of the individuals who become infected but present in many (but not all) of those who remained uninfected (Supplementary Figure 7). Thus, there is no single strong predictor of infection outcomes but rather a probabilistic combination of many species, each of relatively modest predictive value.
Identifying Functional Biomarkers of Disease Susceptibility and Severity
We also identified gene families in the gut microbiome of persons who remained uninfected during follow-up (Supplementary Figures 9 and 10), with some of the top gene families involved in DNA repair, transmembrane transporter activity, iron metabolism (indicated with asterisks in Figure 4), and genes of unknown function (Supplementary Table 8). Long-chain fatty acid biosynthesis pathways (eg, cis-vaccenate, gondoate, and stearate) were associated with individuals who remained uninfected, whereas amino acid biosynthesis and catabolic pathways were associated with individuals who became infected (Supplementary Figures 11 and 12 and Supplementary Table 9). We identified 3 iron-related genes associated with remaining uninfected [1]: the ferric uptake regulator (Fur), a major regulator of iron homeostasis [2]; thioredoxin, a redox protein involved in adaptation to oxidative and iron deficiency stress; and [3] the TonB/ExbD/TolQR system, a ferric chelate transporter [19–21]. In individuals who became infected but asymptomatic, 2 genes involved in the conversion of riboflavin into catalytically active cofactors, the riboflavin kinase and the flavin adenine dinucleotide (FAD) synthetase, were found to be the first and the third most discriminant features (Figure 4 and Supplementary Table 8).
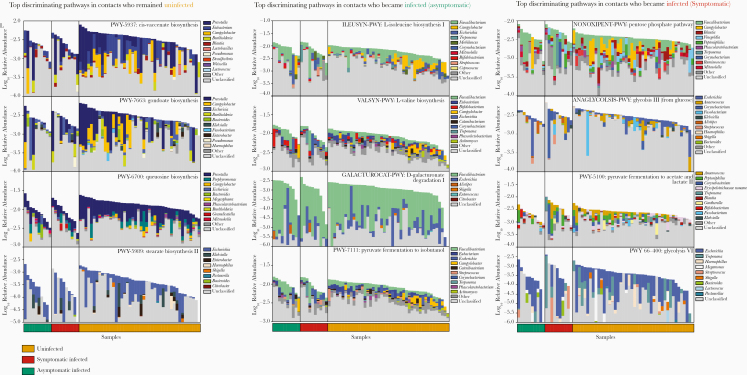
We next asked which taxa in the microbiome likely encoded these genes. In some cases, specific taxonomic groups corresponded to discrete gene functions. For example, several iron metabolism–related gene families tend to be encoded by Prevotella genomes (Supplementary Figure 13). In other cases, the major contributors to protective gene families were unclassified (Figures 5 and Supplementary Figure 14). These results partly explain why gene families or pathway features tend to outperform species-level features in predicting infection status—because predictive gene families are distributed across many species, including several with poor taxonomic annotation or families lacking representation in taxonomic databases.
Figure 5.
Top predictive cellular pathways of the gut microbiome at the time of exposure to Vibrio cholerae in the 2018 cohort from Midani et al [15], annotated by their taxonomic contributors. The 4 top-ranked pathways associated with uninfected contacts (left column), contacts who developed asymptomatic infection (middle column), and contacts who developed symptomatic infection (right column) are shown. Total bar height reflects log10-scaled community relative abundance of each pathway. The contributions of each genus to encoding these pathways are shown as stacked colors within each bar, linearly scaled within the total. See Supplementary Table 9 for the complete list of pathways.
DISCUSSION
The gut microbiome is a potentially modifiable host risk factor for cholera, and identification of specific genes and strains correlated with susceptibility is needed for experimental testing to understand the mechanisms of observed correlations. Compared with a previous study using a single marker gene, shotgun metagenomics provides this degree of resolution, potentially to the species and strain level, and to the level of individual genes and cellular functions. We found that gene families in the gut microbiome at the time of exposure to V. cholerae were more predictive of susceptibility compared with taxonomic or clinical and demographic information. Selecting a subset of the most informative features further improved predictions, but using these selected features may lead to overfitting. This suggests an upper limit to predictive power that requires validation in larger, independent cohorts.
Most of the top predictive biomarkers were associated with remaining uninfected after exposure to V. cholerae. An example is the genus Prevotella, including several strains within Prevotella sp. 885, identified only at the genus level in a previous study [15]. Prevotella species are hypothesized to be beneficial members of the microbiota in healthy individuals in non-Westernized countries, and this species is a potential candidate for follow-up experimental studies in V. cholerae susceptibility [14, 22, 23].
Several species known to ferment mucin glycans into short-chain fatty acids (SCFAs) are correlated with remaining uninfected, including E. rectale, R. gnavus, and B. vulgatus [24, 25]. This finding is consistent with experiments of SCFAs applied to animal models. B. vulgatus has been shown to inhibit V. cholerae colonization in mice, an effect that was dependent on SCFA production [13]. SCFAs are known to affect immune cell development and attenuate inflammation by inhibiting histone deacetylases and other mechanisms of altering gene expression [26–29].
All 3 Bifidobacterium species associated with contacts who developed infection were also associated with asymptomatic rather than symptomatic disease, and prior work on this genus supports several hypotheses for this relationship. First, Bifidobacterium species are known to produce the SCFA acetate that can protect against enteric infection in mice [30–32]. SCFAs are also known to inhibit cholera toxin–related chloride secretion in the mouse gut, reducing water and sodium loss and have been observed to increase cholera toxin–specific antibody responses [31, 33, 34]. Bifidobacterium species are also major producers of lactate, a metabolite that has been shown to impair V. cholerae biofilm formation, a function that can affect virulence [12]. Finally, Bifidobacterium bifidum and Bifidobacterium adolescentis are known to reduce the activity of V. cholerae type VI secretion systems through modification of bile acids [9].
Metagenomics also allowed us to identify bacterial functions that could affect the ability of V. cholerae to compete and colonize the gut. For example, several gene families involved in iron transport, iron regulation, and riboflavin conversion appeared among the top 20 features associated with uninfected and asymptomatic individuals, suggesting that competition for iron might be a protective mechanism of the gut microbiota against V. cholerae, as in other pathogens [7]. Iron is often a limiting redox cofactor in the gut, and bacteria have evolved strategies to solubilize and internalize iron [32, 35]. Riboflavin (another major redox cofactor in bacteria) and iron levels are reciprocally regulated in V. cholerae, and riboflavin may allow V. cholerae to overcome iron limitation in the gut [32, 36]. A gut microbiota more competitive for iron could therefore help resist V. cholerae colonization or reduce its virulence. Further work is thus needed to understand mechanisms whereby enrichment of these genes may protect people after exposure to V. cholerae.
Our results are currently not generalizable beyond the study cohort in Dhaka, Bangladesh, as a similar cohort in another geographic location is not available. As with any association-based study [37], it is unknown whether any of the metagenomic features correlated with protection from V. cholerae infection are causal, and many may be markers of clinical or environmental factors that themselves affect susceptibility. Despite our deep sequencing and collection of standard cholera risk factors, our study was unable to measure all potentially relevant environmental or clinical risk factors. In line with recent studies in Dhaka, we assume that V. cholerae transmission occurs mainly within households [3] and did not consider how the mode of transmission (eg, waterborne or not) might affect outcomes.
It has also been noted that microbiome-disease associations may be poorly portable across human populations [37]. For instance, we identified species of Prevotella as protective features in Bangladesh, but Prevotella is much less abundant and less diverse in Western countries [22]. It thus remains to be seen whether protective gene features (eg, iron metabolism) are encoded in other species of the microbiome outside endemic areas like Bangladesh, or if people outside these areas are simply at greater risk for cholera. Further experimental characterization of metagenomic features correlated with protection from infection or symptoms are needed to understand if factors we identified affect V. cholerae pathogenesis or host responses to infection. Ultimately, the strains and functionalities identified have the potential to inform microbiota-based therapeutics to ameliorate or prevent disease. Our results show the power of metagenomic data from the gut microbiome to predict health outcomes, such as susceptibility to infection and disease severity.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Tables S1–S9 are available at: https://figshare.com/articles/Supplementary_Tables_-_Levade_et_al_ 2020/12440417.
Notes
Acknowledgments. We thank Meti Debela for technical assistance. Finally, we are grateful to the people of Dhaka where our study was undertaken; to the field, laboratory, and data management staff who provided tremendous effort to make the study successful; and to the people who provided valuable support in our study. The icddr,b (International Centre for Diarrhoeal Disease Research, Bangladesh) gratefully acknowledges the government of the People’s Republic of Bangladesh, Global Affairs Canada, the Swedish International Development Cooperation Agency, and the Department for International Development.
Financial support. This work was supported by the Canadian Institutes of Health Research (B. J. S.) and the Canada Research Chairs program (B. J. S.), the icddr,b: Centre for Health and Population Research, the Alfred P. Sloan Fellowship (L. A. D.), the National Institutes of Health (grants AI099243 to J. B. H., AI103055 to J.B.H and F. Q, AI106878 to E. T. R and F. Q., AI058935 to E.T.R, S. B. C., and F. Q., and T32A1070611976 and K08AI123494 to A. A. W.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Ali M, Nelson AR, Lopez AL, Sack DA. Updated global burden of cholera in endemic countries. PLoS Negl Trop Dis 2015; 9:e0003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Camacho A, Bouhenia M, Alyusfi R, et al. Cholera epidemic in Yemen, 2016-18: an analysis of surveillance data. Lancet Glob Health 2018; 6:e680–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Domman D, Chowdhury F, Khan AI, et al. Defining endemic cholera at three levels of spatiotemporal resolution within Bangladesh. Nat Genet 2018; 50:951–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weil AA, Khan AI, Chowdhury F, et al. Clinical outcomes in household contacts of patients with cholera in Bangladesh. Clin Infect Dis 2009; 49:1473–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nelson EJ, Harris JB, Morris JG, Calderwood SB, Camilli A. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nature 2009; 7:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harris JB, LaRocque RC, Chowdhury F, et al. Susceptibility to Vibrio cholerae infection in a cohort of household contacts of patients with cholera in Bangladesh. PLoS Negl Trop Dis 2008; 2:e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ubeda C, Djukovic A, Isaac S. Roles of the intestinal microbiota in pathogen protection. Clin Transl Immunology 2017; 6:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hsiao A, Ahmed AM, Subramanian S, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 2014; 515:423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bachmann V, Kostiuk B, Unterweger D, Diaz-Satizabal L, Ogg S, Pukatzki S. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl Trop Dis 2015; 9:e0004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoon MY, Min KB, Lee K-M, et al. A single gene of a commensal microbe affects host susceptibility to enteric infection. Nat Commun 2016; 7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mao N, Cubillos-Ruiz A, Cameron DE, Collins JJ. Probiotic strains detect and suppress cholera in mice. Sci Transl Med 2018; 10:eaao2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaur S, Sharma P, Kalia N, Singh J, Kaur S. Anti-biofilm properties of the fecal probiotic lactobacilli against Vibrio spp. Front Cell Infect Microbiol 2018; 8:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. You JS, Yong JH, Kim GH, et al. Commensal-derived metabolites govern Vibrio cholerae pathogenesis in host intestine. Microbiome 2019; 7:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. David LA, Weil A, Ryan ET, et al. Gut microbial succession follows acute secretory diarrhea in humans. mBio 2015; 6:e00381–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Midani FS, Weil AA, Chowdhury F, et al. Human gut microbiota predicts susceptibility to Vibrio cholerae infection. J Infect Dis 2018; 218:645–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Truong DT, Franzosa EA, Tickle TL, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 2015; 12:902–3. [DOI] [PubMed] [Google Scholar]
- 17. Franzosa EA, McIver LJ, Rahnavard G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 2018; 15:962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pasolli E, Truong DT, Malik F, Waldron L, Segata N. Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput Biol 2016; 12:e1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fillat MF The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 2014; 546:41–52. [DOI] [PubMed] [Google Scholar]
- 20. Wang B- Y, Huang H- Q, Li S, et al. Thioredoxin H (TrxH) contributes to adversity adaptation and pathogenicity of Edwardsiella piscicida. Vet Res. 2019; 50:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 2010; 64:43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tett A, Huang KD, Asnicar F, et al. The Prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe 2019; 26:666–679.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kovatcheva-Datchary P, Nilsson A, Akrami R, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab 2015; 22:971–82. [DOI] [PubMed] [Google Scholar]
- 24. Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One 2013; 8:e76341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet 2015; 6:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun Y, O’Riordan MX. Regulation of bacterial pathogenesis by intestinal short-chain Fatty acids. Adv Appl Microbiol 2013; 85:93–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016; 165:1332–45. [DOI] [PubMed] [Google Scholar]
- 28. Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 2017; 19:29–41. [DOI] [PubMed] [Google Scholar]
- 29. Fachi JL, de Souza Felipe J, Pral LP, et al. Butyrate protects mice from Clostridium difficile-induced colitis through an HIF-1-dependent mechanism. Cell Rep 2019; 27:750–761.e7. [DOI] [PubMed] [Google Scholar]
- 30. Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011; 469:543–7. [DOI] [PubMed] [Google Scholar]
- 31. Rabbani GH, Albert MJ, Rahman H, Chowdhury AK. Short-chain fatty acids inhibit fluid and electrolyte loss induced by cholera toxin in proximal colon of rabbit in vivo. Dig Dis Sci 1999; 44:1547–53. [DOI] [PubMed] [Google Scholar]
- 32. Sepúlveda Cisternas I, Salazar JC, García-Angulo VA. Overview on the bacterial iron-riboflavin metabolic axis. Front Microbiol 2018; 9:1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol 2011; 17:1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang W, Xiao Y, Huang X, et al. Microbiota metabolite short-chain fatty acids facilitate mucosal adjuvant activity of cholera toxin through GPR43. J Immunol 2019; 203:282–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rivera-Chávez F, Mekalanos JJ. Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 2019; 572:244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sepúlveda Cisternas I, Aguirre LL, Flores AF, de Ovando IVS, García-Angulo VA. Transcriptomics reveals a cross-modulatory effect between riboflavin and iron and outlines responses to riboflavin biosynthesis and uptake in Vibrio cholerae. Sci Rep 2018; 8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmidt TSB, Raes J, Bork P. The human gut microbiome: from association to modulation. Cell 2018; 172:1198–215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
After removal of human reads, the sequence data has been deposited in NCBI under BioProject PRJNA608678.