Abstract
The gastrointestinal health of poultry can be impacted by a variety of factors including their environment. As egg production moves from conventional cage housing (CC) toward cage-free housing (CF), it is important to understand this impact on intestinal health. This study was conducted to determine if housing type impacted intestinal permeability, morphology, and microbial communities in commercial hens across housing systems. Hens were randomly selected from 2 rooms of CC (n = 25) and CF (n = 25) at a commercial facility. Birds were given fluorescein isothiocyanate dextran (FITC-D) by oral gavage to measure intestinal permeability. Jejunal and ileal samples were collected to evaluate villus height, crypt depth, and their ratio. Ileal contents were collected for bacterial DNA isolation and 16S rRNA gene sequencing. Serum FITC-D was similar between housing type (P = 0.709). Hens housed in the CF had increased jejunal villus height and crypt depth compared with hens from the CC (P < 0.002). Hens from the CC tended to have a greater villus height to crypt depth ratio in both the jejunum and ileum compared with the CF (P = 0.064; P = 0.091, respectively). Microbial community diversity measurements favored hens housed in the CC as ileal contents tended to have increased species richness (P = 0.059), had greater alpha diversity (P = 0.044), and had an increased number of over represented operational taxonomic units (46/64), including Romboutsia sp. (30.80%), Lactobacillus kitasatonis (17.16%), and Lactobacillus aviarius (11.15%). Correlations between microbial communities with intestinal traits identified significant association with the greatest number of correlations with FITC-D and ileal morphology. Many of these correlations identified microbial communities associated with expected traits; thus, providing limited functional data to microbial communities with limited information. The greater number of correlations of ileal morphology with ileal microbial communities suggesting local microbial communities contribute to the intestinal environment distant. In this limited study, several parameters favored hens from CC suggesting an advantage of this system for intestinal health. However, the lower intestinal health parameters observed in CF were not at levels to indicate detrimental effects.
Key words: fluorescein isothiocyanate dextran, jejunum, ileum, Lactobacillus, villus height to crypt depth ratio
Introduction
An increasing number of companies are pledging to no longer sell, serve, or utilize eggs from hens housed in conventional cages (CC). In addition to changes in general operations as farmers transition to cage-free systems (CF; Xin et al., 2012; Ward, 2014), much remains unclear regarding how these systems alter the physiology of the hens living in them in response to increased mobility and exposure to excreta. Understanding these differences will be critical for maximizing efficiencies and animal welfare.
The shift from CC increases the overall area a hen can move and is required to move as nest boxes, water lines, and feed lines are at increased distances compared with the CC system. Therefore, it was not surprising that numerical increases in energy requirements were observed in commercial hens raised in CF system compared with CC (Karcher et al., 2015). While this difference could be because of the increase in energy exerted in the form of movement, it is unclear if there are changes in intestinal morphology or permeability that can alter digestibility of consumed nutrients.
In addition to increasing the distance hens need to move for daily activities, the additional interaction with the environment including excreta may lead to chronic inflammation, dysbiosis or enteric disease. While no differences in eggshell contamination with Salmonella or Campylobacter were observed between housing systems, the observed increase in environmental contamination in the CF systems would suggest differences in how the hens interact with the environment (Jones et al., 2015). Therefore, these reduced hygienic conditions may put hens at an increased risk for colonization of microbial communities that compete for nutrients, secrete metabolites that suppress production, reduce nutrient digestion and absorption, increase subclinical infections, and/or allow for colonization of detrimental bacteria. A recent publication by van Goor et al. (2020), characterized microbial communities in the ceca of CF and CC hens. While they did not directly compare microbial communities between housing systems, microbial diversity of the ceca remained similar across stage of lay with CC but not with CF, suggesting the environment may alter stability of the microbiome which in turn may alter nutrient digestion and absorption (van Goor et al., 2020). To understand the effects of hen housing system on intestinal health which will contribute to nutrient digestion and absorption, it is critical to determine changes in bacterial communities and gastrointestinal health. Therefore, this study was designed to characterize bacterial communities, whole intestine permeability, and intestinal morphology between CF and CC systems and to determine associations between resident microbes and intestinal parameters.
Materials and methods
Animals
All procedures involving animals were approved by the Iowa State University's Institute of Animal Care and Usage Committee (IACUC number 18-231).
Twenty-five hens were randomly chosen and weighed from 2 different rooms of either a CF (n = 50) or CC (n = 50) housing system at a single commercial layer facility in Iowa. Hens within each room were the same age; however, the ages of the hens between rooms ranged from 26 to 70, and it is expected hens fed on performance/age appropriate diets. As the focus of this trial was to characterized parameters between commercial CF and CC systems, we chose to treat differences of management such as dietary formulation as a factor that is confounded with CF and CC systems.
Intestinal Permeability
Hens selected from CF and CC systems were orally inoculated with fluorescein isothiocyanate–dextran average molecular weight 3,000 to 5,000 (FITC-D; Sigma Aldrich, St. Louis, MO, FD4) at a rate of 16.64 mg/mL according to a previously described protocol by Baxter et al. (2017). Two hens per room were not inoculated and were used for control serum. One hour after hens were inoculated with FITC-D, hens were euthanized via cervical dislocation. Blood samples were collected from the femoral artery into serum blood collection tubes (BD367815; Fisher Scientific, Waltham, MA) and transported back to Iowa State University on ice for serum separation (10,000 × g for 15 min). Once the serum was separated, it was aliquoted and stored at −80°C in amber tubes to prevent break down of the fluorescence until analysis. All samples from hens given FITC-D were diluted at a ratio of 1:5 in saline. Using serum from control hens, a standard curve was generated for FITC-D. Diluted samples were plated in triplicate. Fluorescence was measured using a BioTek Cytation fluorescence spectrophotometer (BioTek US, Winooski, VT) with excitation and emission wavelengths of 485 and 528 nm, respectively. For data analysis, triplicates were averaged for each hen.
Jejunum and Ileum Morphometric Analysis
After euthanasia, a 2 cm section of the jejunum at Meckel's Diverticulum and of the ileum 5 cm proximal of the ileocecal junction was quickly excised, flushed with phosphate buffered saline, and placed in 10% formalin buffered saline. Formalin fixed samples were sectioned, embedded, and stained with hematoxylin and eosin stain by the Iowa State University Veterinary Histopathology Lab. Additionally, the ileal samples were stained with Alcian blue to determine goblet cell number. Images used for morphometric measurements (villus height and crypt depth) and cells counts were captured using an Olympus BX63 microscope and camera. Ten morphometric measurements per parameter were determined using the ImageJ software (Schindelin et al., 2012; Schnieder et al., 2012). Goblet cells were counted for the entire area of the image using color and shape filters in Image J. Data are expressed in counts per mm2. Three images per bird were used for analysis.
Characterization of Bacterial Communities and Sequence Analysis
Ileal luminal contents were aseptically removed from a 5 cm section adjacent and proximal to the section collected for morphometric analysis. Samples were transported on dry ice back to Iowa State University and stored at −80°C until DNA isolation. DNA from these ileal samples was extracted using the Qiagen Powerlyzer soil kit following the manufacturer's recommendations. After confirming DNA concentrations using a nanodrop (ND 2000; Fisher Scientific), 90 samples were found to contain DNA and were used to amplify bacterial and archaeal 16S rRNA genes. Samples were sequenced using 250 bp paired-end reads for each sample of the V4 region of the 16S rRNA gene (515F, 806R; Caporaso et al., 2011; Caporaso et al., 2012) at the Iowa State University DNA Facility using Illumina MiSeq sequencing technology.
Sequence analysis was done with mothur V1.40.4 following the mothur MiSeq Standard Operating Procedure (Kozich et al., 2013). Barcode sequences, primers, and low-quality sequences were trimmed using a minimum average quality score of 35, with a sliding window size of 50 bp. Chimeric sequences were removed with the “Chimera.uchime” command. For alignment and taxonomic classification of operational taxonomic units (OTU), the SILVA SSU NR reference database (V132) provided by the mothur website was used. Sequences were clustered into OTU with a cutoff of 99% 16S rRNA gene similarity (=0.01 distance).
To compare alpha diversity between experimental groups, reads were randomly subsampled to accommodate the sample with the lowest number of reads across data sets (20,000 sequences). Measurements of Chao species richness, Shannon diversity, and Simpson evenness were taken to compare community structures between experimental groups.
Average Bray-Curtis dissimilarity measures for each treatment group were compared using the analysis of similarity (ANOSIM) package provided by mothur (Clarke, 1993; Schloss et al., 2009). Bray-Curtis was selected as the dissimilarity coefficient because of its ability to compare closely related samples.
All plotting was completed using ggplot2, v2_3.1.1 graphing package (Wickham, 2016; R Core Team, 2019) in R 3.6.0. Overall variation in bacterial communities were visualized using principle coordinate analysis (PCoA). This information was generated with the Phyloseq (v1.28.0 [McMurdie and Holmes, 2013]) and Vegan (v2.5-5 [Oksanen et al., 2019]) packages using the shared and taxonomy file generated in mothur. Sequences were randomly subsampled to 20,000 sequences, and Bray-Curtis dissimilarity measures were used to generate distances between samples for the PCoA plot.
Statistical Analysis
Differences for intestinal permeability, morphology, microbial community parameters, and individual OTU were determined across housing type using PROC Glimmix in SAS (SAS Institute Inc., 2011) with housing type fit as a fixed effect and room fit as a random effect. Significance was set at a P < 0.05. To determine if specific bacterial OTU abundances were significantly different across housing type, data were normalized using the trimmed mean of the M-value (Robinson and Oshlack, 2010) for the top 200 OTU and had at least 2 reads in 45 of the 90 samples. Data were then analyzed using PROC Glimmix in SAS for each OTU following a negative binomial distribution and using housing type as a fixed effect (SAS Institute Inc., 2011). The q-values were used as a means to control for false discovery rate using the q-value package in R (Storey et al., 2004). For OTU, significance was set at a P < 0.05 and q < 0.05. To determine potential beneficial or detrimental bacterial communities, correlations were determined between bacterial communities and intestinal leakage or morphometric measurements using PROC CORR within housing type. Significance was set at a r2 > |0.35|.
Data Availability
The 16S rRNA gene sequences have been submitted to the NCBI Sequence Read Archive SRA and are available under the BioProject ID PRJNA647366.
Results AND discussion
Animal Parameters
All hens used for this study were apparently healthy at the time of selection. The average body weight of hens included in this study was 1.4 kg (1.42 ± 0.06 kg for CF; 1.41 ± 0.06 kg for CC P = 0.978).
Intestinal Parameters
Macromolecular flux of FITC-D, a nondigestible sugar, from the lumen of the intestine into circulation, was not altered by housing type (P = 0.348; See Table 1). Owing to the low levels of FITC-D in the serum, a large number of samples were not above the lowest standard. To ensure this was not bias to a single treatment, we also ran the samples based on fluorescence. Again, no difference was observed by housing type (P = 0.709; See Table 1). The lack of difference and low detection was expected as these birds were presumably healthy and on feed, 2 factors that are experimentally used to induce elevated intestinal permeability of FITC-D (Vicuña et al., 2015; Baxter et al., 2017). However, we observed high hen-to-hen variation across housing type, indicating that the individual hen interaction with the environment had more effects on intestinal permeability than housing system. While average hen weights were not significantly different across housing treatment, individual hen weights did vary; however, the inclusion of body weight as a covariate into the statistical model did not alter the results observed when weight was not included (P = 0.656).
Table 1.
Least squared means for intestinal parameters from hens housed in conventional cage and cage-free housing systems.
| Section | Parameter | Unit | CC | CF | SEM | P-value |
|---|---|---|---|---|---|---|
| Permeability | ng/mL | 114.23 | 101.31 | 9.8 | 0.348 | |
| Whole Intestine | Fluor | 971.69 | 866.68 | 199.16 | 0.709 | |
| Jejunum | Villus Height | μmol | 755.21 | 866.08 | 25.05 | 0.002 |
| Crypt Depth | μmol | 121.52 | 156.34 | 1.96 | <0.001 | |
| Ratio | μmol/μmol | 6.60 | 5.94 | 0.25 | 0.064 | |
| Ileum | Villus Height | μmol | 591.89 | 572.94 | 9.90 | 0.175 |
| Crypt Depth | μmol | 117.62 | 127.85 | 5.03 | 0.151 | |
| Ratio | μmol/μmol | 5.30 | 4.84 | 0.19 | 0.091 | |
| Goblet number | Count/mm21 | 1,170 | 686 | 74 | <0.001 |
Abbreviations: CC, conventional cage housing system; CF, cage-free housing system; Fluor, Fluorescence; SEM, standard error of the means.
Goblet cell count per area of each image.
Given the jejunum is the area of highest digestion and absorption, significant changes in this region could indicate changes in digestion and absorption across housing types. In this study, jejunal villus height and crypt depth were increased in hens from the CF system compared with hens from the CC system (P < 0.002; See Table 1). Villus height to crypt depth ratio tended to be greater in hens from the CC system compared with the CF system (P = 0.064; Table 1). Additionally, to observe changes where microbial populations increase and assist in the last of the small intestine digestion and absorption, ileal morphometric parameters were measured. Ileal villus height and crypt depth were not different between hens from the different housing systems. However, the villus height to crypt depth ratio tended to be greater in hens from CC systems (P = 0.091; Table 1), and number of goblet cells were increased in hens from CC (P < 0.001; Table 1).
Intestinal morphology is used as an indicator of intestinal health as values are often indicative of digestive and absorptive capacity. For both jejunum and ileum, villus height and crypt depth ratio were similar to previously reported length (Applegate et al., 2009; Deng et al., 2012; Pereira et al., 2019) indicating that intestinal absorption capacity was within normal ranges and not indicative of diseased states. In our study, hens from the CF system had increased villus height in the jejunum, an area of high nutrition absorption, as well as increased crypt depth, suggesting these hens may have higher intestinal absorption while continuing to proliferate new cells for the intestinal lining. This continued production of cells by the intestine is energetically unfavorable. Therefore, the ratio of the villus height to crypt depth is often used as a single measure of intestinal health. Surprisingly, the villus height to crypt depth ratio tended to be increased in the jejunum and ileum of CC hens, suggesting the intestine of these hens is more favorable (P < 0.092; Table 1). Extreme changes in villus height and crypt depth are observed during times of disease or toxin challenge with villus height decreasing as dying cells are removed and crypt depth increasing to support new cell growth (Yason et al., 1987). Extrapolation of these measures are often applied in nondisease challenges when changes are more subtle, as is the case in this study, and should be done cautiously.
Ileal Microbial Communities
Taxonomic Assignment
The 250 bp paired-end MiSeq sequencing of the 90 samples resulted in 8,570,879 raw sequences. After removing low-quality sequences, 6,474,777 sequences remained, which were clustered into 46,018 OTU. Both the SILVA SSU NR reference database (V132) provided by the mothur website and NCBI Blast on representative sequences were used to assign OTU a taxonomic classification and are provided in all tables where OTU data are present.
Alpha Diversity Measurements
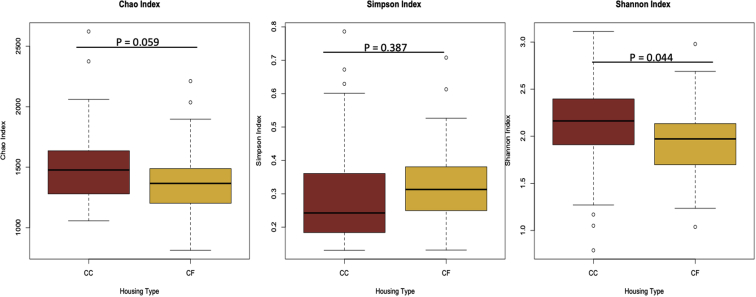
This is the first study to examine the changes in ileal microbial communities in laying hens across different housing environments. With the exception of evenness (Simpson index; P = 0.387), average ileal species richness (Chao index) tended to be higher (P = 0.059), and overall alpha diversity (Shannon index) was higher for hens housed in CC systems (P = 0.044; Figure 1). Results from this study suggest the species richness and alpha diversity of the microbial communities are more favorable in CC systems, which may provide greater plasticity of bacterial communities. However, the spread or evenness of microbial communities was similar. This evenness of microbial communities was, also, observed in cecal contents from hens housed in CF and CC systems (Hubert et al., 2019), potentially suggesting some structure or order to how microbial communities are allowed to flourish in the chicken intestine. Interestingly, Hubert et al. (2019) observed greater alpha diversity in cecal content of hens from CF systems, whereas van Goor et al. (2020) observed greater alpha diversity in cecal contents of hens from CC systems. While these communities were collected from different regions of the digestive system compared with this study, it should, also, be pointed out that hens housed in the CF environment in the Hubert et al. (2019) study had access to outdoor spaces, whereas hens in this study did not. While outdoor access was not mentioned in van Goor et al. (2020), the differences in alpha diversity measurements for these microbial communities may not only be a result of intestinal segment but access to outdoor microbes.
Figure 1.
Boxplots of alpha diversity measurements of ileal microbiota from hens in commercial conventional cage (CC) and cage-free (CF) systems. Goldenrod denotes the diversities from hens in CF systems, and red denotes the diversities from hens in CC.
Beta Diversity Measurements
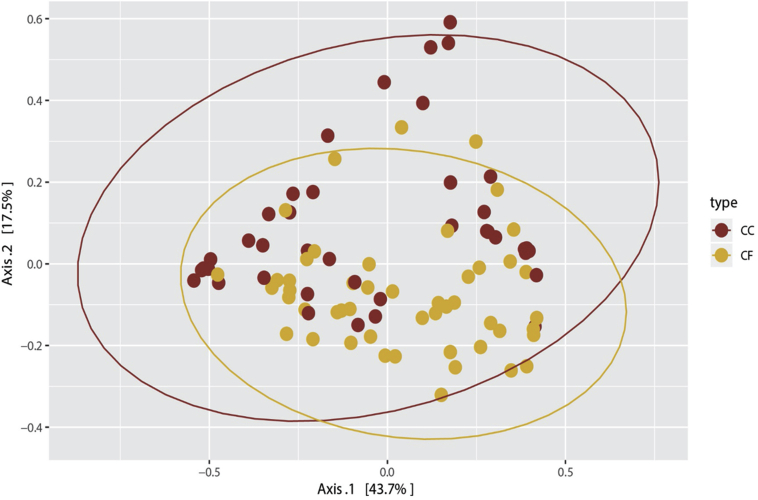
Whole community Beta diversity comparisons of CF and CC microbial community samples were made using ANOSIM and analysis of molecular variance comparing average Bray-Curtis distances per group and found significant differences in microbial communities between housing types (ANOSIM; P = 0.0003 and analysis of molecular variance; P = 0.004; Figure 2). However, PCoA plots revealed no clear clustering of the microbial communities based on housing type.
Figure 2.
Principle coordinate analysis comparing the ileal microbiota of hens in commercial conventional cage (CC) and cage-free (CF) systems. Goldenrod denotes hens in CF systems, and red denotes hens in CC systems.
Ileal Microbial Communities
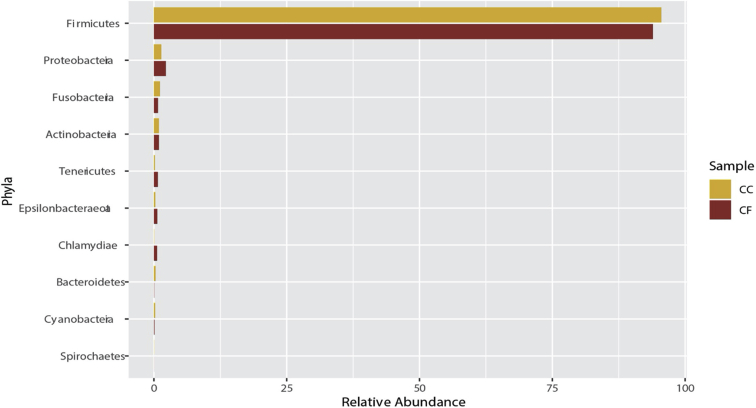
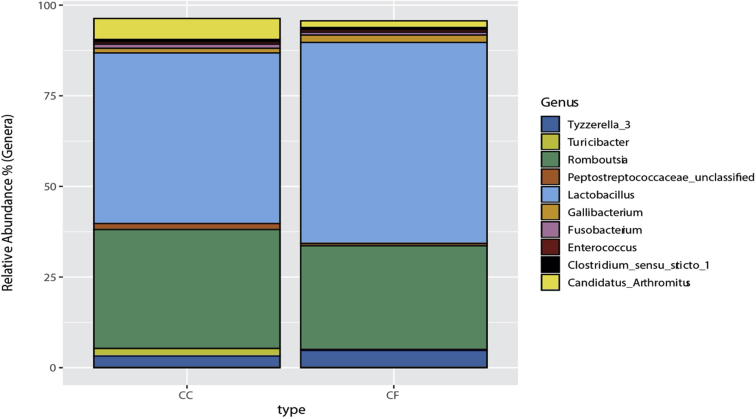
At the phylum level, 21 phyla were identified from samples between both housing types (Supplementary Figure 1). The majority of phyla were Firmicutes (91.5%), Proteobacteria (1.83%), Fusobacteria (0.85%), and Actinobacteria (0.63%). The major genera found in both housing types included mainly Lactobacillus (45.0%), Romboutsia (34.8%), Tyzzerella (3.74%), Candidatus Arthromitus (3.47%), Gallibacterium (1.76%), and Turicibacter (1.32%; Supplementary Figure 2). The percentage of phyla are similar to previously published ileal microbiome communities in laying hens (Ngunjiri et al., 2019; Wang et al., 2019).
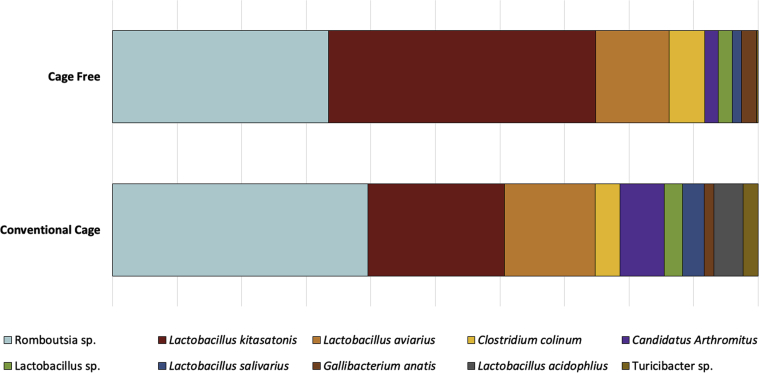
In hens from the CC system, Romboutsia was the most abundant genus (30.80%), followed by 2 Lactobacillus phylotypes: Lactobacillus kitasatonis (17.16%), and Lactobacillus aviarius (11.15%). In hens from CF system, L. kitasatonis was the most abundant genus (34.29%), followed by Romboutsia (27.68%) and L. aviarius (9.35%). The 10 most abundant genera and their relative abundances by housing system can be found in Figure 3.
Figure 3.
The 10 most abundant ileal microbiota operational taxonomic units (OTU) by commercial housing system. Percentages of the top 10 OTU are represented based on abundances for each commercial housing system. Each OTU genera or species identification can be found in the figure legend.
To determine specific OTU abundance differences across housing type, data were analyzed in SAS following abundance normalization which accounts for the number of sequence reads. Of the 200 OTU analyzed, 64 OTU were differentially abundant between housing types (Tables 2 and 3). Eighteen OTU were overrepresented in CF compared with CC systems (Table 2). The majority of these OTU were comprised of Lactobacillus sp. (5/18; 27.8%), Staphylococcus sp. (3/18; 16.7%), and Corynebacterium sp. (2/18; 11.1%) and did include over representation of OTU that aligned to the L. kitasatonis sequence at higher than 98% using BLAST (Altschul et al., 1990). This recently discovered bacterium has been isolated from the intestine, vagina, cloaca, and excreta of chickens (Mukai et al., 2003; Van Coillie et al., 2007; Yamazaki et al., 2012). While it has been studied for its ability to act as a probiotic and a competitive inhibitor of Salmonella enteritidis and typhimurium, it has not been shown to contribute significantly in either role (Van Coillie et al., 2007; Yamazaki et al., 2012).
Table 2.
Operational taxonomic units overrepresented in ileal digesta of hen housed in a commercial cage-free system.
| Group1 | Fold change2 | P-value | q-value | Taxonomy3 | Taxonomy based on NCBI BLASTn search4 |
|---|---|---|---|---|---|
| Otu00036 | 1.5374 | 0.0054 | 0.0183 | Streptococcus | Strep. alactolyticus; S. griseocameus; S. gallolyticus; S. macedonicus; S. pateurianus |
| Otu00004 | 1.5394 | 0.0169 | 0.0382 | Tyzzerella_3 | - |
| Otu00168 | 1.5625 | 0.0063 | 0.0200 | Lactobacillus | Lactobacillus kitasatonis |
| Otu00191 | 1.6654 | 0.0127 | 0.0310 | Nocardiopsis | Nocardiopsis alkaliphila; N. kunsanensis |
| Otu00110 | 1.7306 | 0.0041 | 0.0150 | Staphylococcus | Staphylococcus lentus; S. sciuri |
| Otu00055 | 1.7452 | 0.0002 | 0.0022 | Staphylococcus | Staphylococcus equorum |
| Otu00135 | 1.7470 | 0.0005 | 0.0032 | Lactobacillus | Lactobacillus acidophilus; L. crispatus |
| Otu00161 | 1.7958 | 0.0182 | 0.0400 | Bacteroides | Bacteroides salanitronis |
| Otu00116 | 1.8070 | 0.0007 | 0.0045 | Lactobacillus | Lactobacillus secaliphilus |
| Otu00157 | 1.8490 | 0.0074 | 0.0221 | Yaniella | Yaniella halotolerans |
| Otu00087 | 1.8546 | 0.0009 | 0.0048 | Staphylococcaceae_unclassified | Salinicoccus kekensis; S. gingdaonensis; S. alkaliphilus |
| Otu00166 | 1.9298 | 0.0046 | 0.0160 | Helicobacter | Helicobacter winghamenss; H. pametensi; H. macacae; H. brantae |
| Otu00096 | 2.0089 | 0.0003 | 0.0022 | Dietzia | Dietzia lutea; D. timorensis |
| Otu00072 | 2.0570 | 0.0008 | 0.0048 | Lactobacillus | Lactobacillus hayakitensis |
| Otu00035 | 2.4141 | 0.0074 | 0.0221 | Aeriscardovia | - |
| Otu00054 | 2.4993 | 0.0000 | 0.0001 | Corynebacterium_1 | Corynebacterium singular; C. sphenisorum; C. glyciniphilum; C. minutissimum |
| Otu00050 | 2.8528 | 0.0003 | 0.0024 | Corynebacterium_1 | Corynebacterium casei; C. ammoniagenes |
Group denotes the taxonomic group assigned to each unique sequence. This table only includes those significantly different from the top 200 taxonomic groups.
Fold change is expressed relative to CC system.
Taxonomic assignments are based the SILVA SSU NR reference database (v 132).
BLASTn search results were reported if the similarity was higher than 97%—indicates sequence alignments of less than 97%.
Table 3.
Operational taxonomic units overrepresented in ileal digesta of hen housed in a commercial conventional cage system.
| Group1 | Fold change2 | P-value | q-value | Taxonomy3 | Taxonomy based on NCBI BLASTn search4 |
|---|---|---|---|---|---|
| Otu00062 | 0.1670 | 0.0001 | 0.0012 | Clostridiaceae_1_unclassified | Clostridium fallax strain DSM 2631; C. chauvoei strain DSM 7528 |
| Otu00134 | 0.2049 | <0.0001 | 0.0001 | Peptostreptococcaceae_unclassified | Romboutsia timonensis strain Marseille-P326 |
| Otu00127 | 0.2130 | 0.0003 | 0.0022 | Romboutsia | Romboutsia timonensis strain Marseille-P326 |
| Otu00142 | 0.2242 | 0.0001 | 0.0016 | Turicibacter | Turicibacter sanguinis |
| Otu00148 | 0.2291 | 0.0010 | 0.0048 | Peptostreptococcaceae_unclassified | Clostridioides difficile |
| Otu00152 | 0.2418 | <0.0001 | 0.0004 | Peptostreptococcaceae_unclassified | Terrisporobacter othiniensis; Peptostreptococcaceae bacterium |
| Otu00197 | 0.2513 | 0.0004 | 0.0029 | Peptostreptococcaceae_unclassified | Clostridioides difficile |
| Otu00115 | 0.2622 | 0.0017 | 0.0075 | Tyzzerella_3 | T. sanguinis |
| Otu00080 | 0.2750 | 0.0003 | 0.0022 | Romboutsia | Romboutsia timonensis strain Marseille-P326 |
| Otu00108 | 0.2872 | 0.0002 | 0.0022 | Turicibacter | T. sanguinis |
| Otu00067 | 0.2906 | 0.0005 | 0.0034 | Romboutsia | Romboutsia timonensis strain Marseille-P326 |
| Otu00079 | 0.2939 | 0.0001 | 0.0017 | Clostridiales_unclassified | Corynebacterium atypicum; C. pseudogenitalium |
| Otu00154 | 0.2941 | 0.0017 | 0.0075 | Clostridiaceae_1_unclassified | Clostridium nigeriense strain Marseille-P2414 T |
| Otu00084 | 0.3151 | 0.0011 | 0.0053 | Turicibacter | T. sanguinis |
| Otu00170 | 0.3190 | <0.0001 | <0.0001 | Corynebacterium_1 | Corynebacterium glutamicum; C. efficiens |
| Otu00085 | 0.3260 | 0.0009 | 0.0048 | Turicibacter | T. sanguinis |
| Otu00189 | 0.3385 | 0.0001 | 0.0012 | Turicibacter | T. sanguinis |
| Otu00109 | 0.3819 | 0.0075 | 0.0221 | Bacteroides | - |
| Otu00178 | 0.3833 | 0.0110 | 0.0275 | Bacteroides | - |
| Otu00019 | 0.3919 | 0.0019 | 0.0081 | Peptostreptococcaceae_unclassified | Clostridioides difficile |
| Otu00039 | 0.4178 | 0.0038 | 0.0142 | Clostridiaceae_1_unclassified | Clostridium chauvoei; C. tertium; C. sartagoforme |
| Otu00183 | 0.4382 | 0.0171 | 0.0382 | Turicibacter | T. sanguinis |
| Otu00029 | 0.4463 | <0.0001 | 0.0008 | Terrisporobacter | Terrisporobacter othiniensis |
| Otu00100 | 0.4651 | 0.0002 | 0.0022 | Gallicola | uncultured bacterium |
| Otu00123 | 0.4713 | 0.0033 | 0.0133 | Romboutsia | Romboutsia timonensis strain Marseille-P326 |
| Otu00031 | 0.4997 | <0.0001 | 0.0007 | Lactobacillus | Lactobacillus ingluviei |
| Otu00010 | 0.5207 | 0.0012 | 0.0057 | Turicibacter | T. sanguinis |
| Otu00091 | 0.5290 | 0.0088 | 0.0240 | Lactobacillus | Lactobacillus aviarius |
| Otu00200 | 0.5319 | 0.0220 | 0.0474 | Ruminococcaceae_UCG-005 | - |
| Otu00175 | 0.5350 | 0.0009 | 0.0048 | Lactobacillus | L. aviarius |
| Otu00113 | 0.5452 | 0.0042 | 0.0150 | Aeriscardovia | Aeriscardovia aeriphila |
| Otu00009 | 0.5533 | 0.0095 | 0.0254 | Lactobacillus | Lactobacillus acidophilus; L. crispatus |
| Otu00195 | 0.5830 | 0.0105 | 0.0269 | Candidatus_Arthromitus | - |
| Otu00198 | 0.5884 | 0.0079 | 0.0229 | Romboutsia | Romboutsia timonensis strain Marseille-P326 |
| Otu00077 | 0.5951 | 0.0170 | 0.0382 | Lactobacillus | Lactobacillus ingluviei; L. senmaizukei |
| Otu00151 | 0.6035 | 0.0149 | 0.0351 | Lactobacillales_unclassified | Lactobacillus pobuzihii |
| Otu00027 | 0.6168 | <0.0001 | <0.0001 | Aeriscardovia | Aeriscardovia aeriphila |
| Otu00086 | 0.6299 | 0.0037 | 0.0142 | Lactobacillus | Lactobacillus agilis |
| Otu00094 | 0.6319 | 0.0063 | 0.0200 | Clostridiales_unclassified | Romboutsia timonensis strain Marseille-P326 |
| Otu00137 | 0.6434 | 0.0111 | 0.0275 | Lactobacillus | Lactobacillus mucosae |
| Otu00185 | 0.6456 | 0.0059 | 0.0197 | Romboutsia | Romboutsia timonensis strain Marseille-P326 |
| Otu00068 | 0.6604 | 0.0228 | 0.0486 | Candidatus_Arthromitus | - |
| Otu00130 | 0.6655 | 0.0082 | 0.0232 | Romboutsia | Romboutsia timonensis strain Marseille-P326 |
| Otu00114 | 0.6842 | 0.0083 | 0.0232 | Romboutsia | Romboutsia timonensis strain Marseille-P326 |
| Otu00025 | 0.7166 | 0.0099 | 0.0260 | Romboutsia | Romboutsia weinsteinii |
| Otu00028 | 0.7943 | 0.0147 | 0.0351 | Lactobacillus | Lactobacillus agilis |
Abbreviations: CC, Conventional cage.
Group denotes the taxonomic group assigned to each unique sequence. This table only includes those significantly different from the top 200 taxonomic groups.
Fold change is expressed relative to CC system.
Taxonomic assignments are based on sequence similarity to the SILVA SSU NR reference database (v 132).
BLASTn search results were reported if the similarity was higher than 97%—indicates sequence alignments of less than 97%.
The remaining 46 OTU were overrepresented in CC system (Table 3). The majority of these OTU were comprised of Romboutsia sp. (9/46; 19.6%), Lactobacillus sp. (8/46; 17.4%), Turicibacter sp. (7/46; 15%), Peptostreptococcaceae sp. (5/46; 10.9%), and Clostridiales sp. (5/46; 10.9%). As expected, many of the Romboutsia sp. were differently represented, with the closest BLAST aligned species being Romboutsia timonensis strain Marseille-P326. This strain was recently isolated in humans (Ricaboni et al., 2016). While it has been mentioned in poultry studies, it is largely unknown how this species is contributing to the chicken microbiota (Qiao et al, 2018, 2019). Turicibacter sp. have been identified with favorable feed conversion (low residual feed intake) in both broiler male and females (Siegerstetter et al., 2017). Unfortunately, the current study did not explore hen production parameters such as hen day egg production or egg weight across the housing systems and cannot speculate on this relationship in hens. Among the species of Clostridiales identified, Clostridioides difficile was the only microorganism to be identified as a potential human pathogen. It composed an average of 1.08% of the abundance and a median of 0.00175%. This small percentage and even lower median indicate a few birds had high abundance, whereas the majority had less than 0.002%.
Associations Between Intestinal Parameters and Microbial Communities
Correlation Summary
Overall, 48 correlations were identified for hens in CC systems, and 43 correlations were identified for hens in CF systems with r2 > |0.35|. For CC hens, 3 OTU were associated with body weight (1 negative and 2 positive); 11 OTU were associated with intestinal permeability (11 positive); 2 OTU were associated with jejunal villus height (1 positive and 1 negative); 1 OTU was negatively associated with jejunal crypt depth; 3 were positively associated with jejunal villus height to crypt depth ratio; 16 OTU were associated with ileal villus height (7 positive and 9 negative); 9 OTU were associated with ileal crypt depth (1 positive and 8 negative); and 3 were positively associated with the ileal villus height to crypt depth ratio. For hens housed in CF systems, 1 OTU was negatively associated with body weight; 25 OTU were associated with intestinal permeability (13 positive and 12 were negative); 2 OTU were positively associated with jejunal crypt depth; 2 OTU were negatively associated with ileal villus height; 9 OTU were negatively associated with ileal crypt depth; and 3 were associated with the ileal villus height to crypt depth ratio (2 negative and 1 positive). Correlations with r2 > |0.35| can be found in Table 4, Table 5, Table 6, Table 7.
Table 4.
Correlation of operational taxonomic units from ileal digesta in hen from conventional cage and cage-free systems for body weight.
| Housing | Group1 | Correlation | Taxonomy2 | Taxonomy based on NCBI BLASTn search3 |
|---|---|---|---|---|
| Conventional Cage | Otu00128 | −0.35 | Lactobacillus | Lactobacillus aviarius |
| Otu00136 | 0.38 | Peptostreptococcaceae | Romboutsia timonensis strain Marseille-P326 | |
| Otu00198 | 0.47 | Romboutsia | Romboutsia timonensis strain Marseille-P326 | |
| Cage-free | Otu00100 | −0.39 | Gallicola | uncultured bacterium |
Group denotes the taxonomic group assigned to each unique sequence. This table only includes those significantly different from the top 200 taxonomic groups.
Taxonomic assignments are based on sequence similarity to the SILVA SSU NR reference database (v 132).
BLASTn search results were reported if the similarity was higher than 97%—indicates sequence alignments of less than 97%.
Table 5.
Correlation of operational taxonomic units from ileal digesta in hen from conventional cage and cage-free systems for intestinal permeability1.
| Housing | Group2 | Correlation | Taxonomy3 | Taxonomy based on NCBI BLASTn search4 |
|---|---|---|---|---|
| Otu00126 | 0.36 | Lactobacillus | Lactobacillus kitasatonis | |
| Conventional Cage | Otu00137 | 0.39 | Lactobacillus | Lactobacillus mucosae |
| Otu00032 | 0.39 | Veillonellaceae | Veillonella magna | |
| Otu00082 | 0.40 | Turicibacter | Turicibacter sanguinis | |
| Otu00144 | 0.44 | Romboutsia | Romboutsia timonensis strain Marseille-P326 | |
| Otu00193 | 0.49 | Peptostreptococcaceae | Romboutsia timonensis strain Marseille-P326 | |
| Otu00174 | 0.50 | Lactobacillus | Lactobacillus aviarius | |
| Otu00077 | 0.50 | Lactobacillus | Lactobacillus ingluviei; senmaizukei | |
| Otu00064 | 0.53 | Tyzzerella_3 | - | |
| Otu00004 | 0.57 | Tyzzerella_3 | Natranaerovirga pectinivora strain DSM 24629 | |
| Otu00119 | 0.59 | Romboutsia | Romboutsia timonensis strain Marseille-P326 | |
| Otu00057 | 0.35 | Megamonas | Megamonas funiformis | |
| Otu00100 | 0.36 | Gallicola | uncultured bacterium | |
| Otu00054 | 0.36 | Corynebacterium_1 | Corynebacterium singular; C. sphenisorum; C. glyciniphilum; C. minutissimum | |
| Otu00055 | 0.37 | Staphylococcus | Staphylococcus equorum | |
| Cage-free | Otu00096 | 0.40 | Dietzia | Dietzia lutea |
| Otu00120 | 0.41 | Lactobacillus | Lactobacillus bacterium isolate MGYG-HGUT-01336 | |
| Otu00157 | 0.44 | Yaniella | Yaniela halotolerans | |
| Otu00011 | 0.44 | Lactobacillus | Lactobacillus vaginalis | |
| Otu00059 | 0.53 | Lactobacillus | Lactobacillus mucosae | |
| Otu00186 | 0.53 | Peptococcus | – | |
| Otu00101 | 0.59 | Lactobacillus | Lactobacillus oris; L. panis; L. antri; Lfrumenti; L. reuteri | |
| Otu00081 | 0.62 | Lactobacillus | L. kitasatonis | |
| Otu00023 | 0.67 | Lactobacillus | Lactobacillus mucosae | |
| Otu00117 | −0.50 | Aeriscardovia | ||
| Otu00187 | −0.45 | Lactobacillus | Lactobacillus kitasatonis; L. pasteurii | |
| Otu00035 | −0.41 | Aeriscardovia | – | |
| Otu00154 | −0.41 | Clostridiaceae_1_unclassified | Clostridium nigeriense | |
| Otu00134 | −0.41 | Peptostreptococcaceae_unclassified | Romboutsia timonensis | |
| Otu00153 | −0.40 | Gallibacterium | Gallibacterium anatis | |
| Otu00039 | −0.40 | Clostridiaceae_1_unclassified | Clostridium chauvoei | |
| Otu00147 | -0.39 | Lactobacillus | L. aviaries | |
| Otu00158 | −0.38 | Romboutsia | Romboutsia timonensis | |
| Otu00175 | −0.38 | Lactobacillus | L. aviaries | |
| Otu00021 | −0.38 | Lactobacillus | Lactobacillus collinoides; L. siliginis; L. paracollinoides | |
| Otu00082 | −0.35 | Turicibacter | T. sanguinis |
Permeability measured in fluorescence.
Group denotes the taxonomic group assigned to each unique sequence. This table only includes those significantly different from the top 200 taxonomic groups.
Taxonomic assignments are based on sequence similarity to the SILVA SSU NR reference database (v 132).
BLASTn search results were reported if the similarity was higher than 97%—indicates sequence alignments of less than 97%.
Table 6.
Correlation of operational taxonomic units from ileal digesta in hen from conventional cage and cage-free systems for jejunal morphology.
| Intestinal parameter | Housing | Group1 | Correlation | Taxonomy2 | BLAST search3 |
|---|---|---|---|---|---|
| Villus height | Conventional Cage | Otu00046 | −0.40 | Escherichia-Shigella | Clostridium cuniculli; Blastococcus litoris; E. coli O157H7; Escherichia albertii KF1; Shigella boydii |
| Otu00058 | 0.40 | Tyzzerella_3 | - | ||
| Cage-free | - | - | - | - | |
| Crypt depth | Conventional Cage | Otu00190 | −0.35 | Jeotgalicoccus | Jeotgalicoccus halotoleran; J. halophilus; J. saudimassiliensis |
| Cage-free | Otu00143 | 0.36 | Lactobacillus | L. acidophilus; L. crispatus | |
| Otu00082 | 0.43 | Turicibacter | Turicibacter sanguinis | ||
| Villus height to crypt depth ratio | Conventional Cage | Otu00173 | 0.36 | Streptococcus | Streptococcus hyovafinalis; S. acidominimus |
| Otu00088 | 0.47 | Lactobacillus | Lactobacillus acidophilus; L.s crispatus | ||
| Otu00058 | 0.48 | Tyzzerella_3 | - | ||
| Cage-free | - | - | - | - |
Group denotes the taxonomic group assigned to each unique sequence. This table only includes those significantly different from the top 200 taxonomic groups.
Taxonomic assignments are based on sequence similarity to the SILVA SSU NR reference database (v 132).
BLASTn search results were reported if the similarity was higher than 97%—indicates sequence alignments of less than 97%.
Table 7.
Correlation of operational taxonomic units from ileal digesta in hen from CC and CF systems for ileal morphology.
| Intestinal parameter | Housing | Group1 | Correlation | Taxonomy2 | Taxonomy based on NCBI BLASTn search3 |
|---|---|---|---|---|---|
| Villus Height | Conventional Cage | Otu00043 | −0.41 | Peptostreptococcaceae | Romboutsia timonensis strain Marseille-P326 |
| Otu00111 | −0.41 | Enterococcus | Enterococcus lactis; villorum, canis, cinnamoneus, canintestini; faecium | ||
| Otu00041 | −0.41 | Campylobacter | Campylobacter insulaenigrae, C. armoricus, C. helveticus | ||
| Otu00013 | −0.38 | Fusobacterium | Fusobacterium necrogenes, mortiferum, mvarium, ulcerans | ||
| Otu00008 | −0.38 | Gallibacterium | Gallibacterium anatis | ||
| Otu00175 | −0.37 | Lactobacillus | Lactobacillus aviarius | ||
| Otu00021 | −0.36 | Lactobacillus | Lactobacillus collinoides; L. siliginis; L. paracollinoides | ||
| Otu00016 | −0.36 | Clostridium_sensu_stricto_1 | Clostridium cuniculi; saudiense | ||
| Otu00150 | −0.35 | Lactobacillus | Lactobacillus aviarius; L. acidioiscis | ||
| Otu00131 | 0.36 | Lactobacillus | Lactobacillus kitasatonis | ||
| Otu00009 | 0.38 | Lactobacillus | Lactobacillus acidophilus; Lactobacillus crispatus | ||
| Otu00091 | 0.39 | Lactobacillus | L. aviarius | ||
| Otu00122 | 0.40 | Atopobium | - | ||
| Otu00018 | 0.41 | Lactobacillus | Lactobacillus gigerionum; amylolytics | ||
| Otu00118 | 0.42 | Bifidobacteriaceae | Bifidobacterium commune | ||
| Otu00156 | 0.46 | Lactobacillus | Lactobacillus kitasatonis; L. acidophilus | ||
| Cage-free | Otu00141 | −0.41 | Romboutsia | Romboutsia timonensis | |
| Otu00175 | −0.35 | Lactobacillus | L. aviaries | ||
| Crypt Depth | Conventional Cage | Otu00110 | −0.40 | Staphylococcus | Staphylococcus lentus; sciuri |
| Otu00150 | −0.38 | Lactobacillus | Lactobacillus aviarius; L. acidioiscis | ||
| Otu00166 | −0.38 | Helicobacter | Helicobacter winghamenss; pametensis, macacae, brantae | ||
| Otu00106 | −0.37 | Campylobacter | Campylobacter upsaliensis; coli, jejuni sp jejuni | ||
| Otu00146 | −0.36 | Phascolarctobacterium | Chryseobacterium oncorhynchi | ||
| Otu00179 | −0.36 | Parasutterella | - | ||
| Otu00157 | −0.36 | Yaniella | Yaniella halotolerans | ||
| Otu00148 | −0.35 | Peptostreptococcaceae | Clostridioides difficile | ||
| Otu00180 | 0.43 | Candidatus_Arthromitus | - | ||
| Cage-free | Otu00141 | −0.48 | Romboutsia | Romboutsia timonensis | |
| Otu00048 | −0.41 | Clostridium_sensu_stricto_1 | Clostridium disporicum | ||
| Otu00012 | −0.41 | Lactobacillus | L. taiwanensis; L.hominis; L. Paragasseri; L. gasseri; L. johnsonii | ||
| Otu00047 | −0.41 | Lactobacillus | Lactobacillus isolate MGYG-HGUT-01336 | ||
| Otu00069 | −0.41 | Romboutsia | Romboutsia timonensis | ||
| Otu00016 | −0.39 | Clostridium_sensu_stricto_1 | Clostridium cuniculi; saudiense | ||
| Otu00025 | −0.38 | Romboutsia | Romboutsia weinsteinii | ||
| Otu00038 | −0.36 | Romboutsia | Romboutsia timonensis | ||
| Otu00014 | −0.35 | Lactobacillus | L. delbrueckii bulgaricus | ||
| Villus Height to Crypt Depth Ratio | Conventional Cage | Otu00168 | 0.35 | Lactobacillus | L. kitasatonis |
| Otu00176 | 0.39 | Lactobacillus | L. aviarius | ||
| Otu00156 | 0.39 | Lactobacillus | Lactobacillus aviarius; L. acidioiscis | ||
| Cage-free | Otu00163 | −0.43 | Lactobacillus | L. aviaries | |
| Otu00154 | −0.37 | Clostridiaceae_1_unclassified | Clostridium nigeriense | ||
| Otu00059 | 0.35 | Lactobacillus | Lactobacillus mucosae |
Abbreviations: CC, Conventional cage; CF, cage-free.
Group denotes the taxonomic group assigned to each unique sequence. This table only includes those significantly different from the top 200 taxonomic groups.
Taxonomic assignments are based on sequence similarity to the SILVA SSU NR reference database (v 132).
BLASTn search results were reported if the similarity was higher than 97%—indicates sequence alignments of less than 97%.
Correlation of OTU With Body Weight
At the genus level, 1 Lactobacillus (L. aviarius) OTU was negatively associated with body weight, whereas 2 Romboutsia OTU (both showed highest similarity to Romboutsia timonensis strain Marseille-P326) were positively associated with body weight in the CC system. While no data has been reported regarding Romboutsia timonensis in chicken likely because of their recent identification (Ricaboni et al., 2016), ileal L. aviarius has been associated with high feed conversion rates in broiler chicken (Stanley et al., 2012). While feed conversion rates in broilers are a relationship between feed to body weight gain, laying hens convert feed to egg production, which is energetically taxing. Therefore, maintaining certain body conditioning or body weight is imperative. The presence of L. aviarius alone or in conjunction with Romboutsia timonensis may assist in this maintenance of body weight, but additional research is needed to understand this relationship. In CF, Gallicola sp. was the only microbe to be associated with body weight and was negatively associated with body weight. Unfortunately, our BLASTn search did not reveal a specific bacterium. Significant body weight correlations can be found in Table 4.
Correlation of OTU With Intestinal Permeability
Intestinal permeability as measured by the rate of FITC-D flux from the intestine into circulation was positively associated with 4 Lactobacillus, 3 Romboutsia, 2 Tyzzerella, 1 Turicibacter, and 1 Veillonellaceae OTU in hens from CC systems and 6 Lactobacillus, and 1 Megamonas, Gallicola, Corynebacterium, Staphylococcus, Dietzia, and Yaniella OTU in hens from CF systems. Significant intestinal permeability correlations can be found in Table 5.
While Lactobacillus sp. have been identified to have a protective nature in the intestine, the positive association between this genus and intestinal permeability would suggest that many species may have unfavorable impacts on intestinal health. Many of the positively associated species identified (L. kitasatonis, Lactobacillus mucosae, L. aviarius, and Lactobacillus ingluviei or Lactobacillus senmaizukei) have been identified in poultry but lack a described function (Qiao et al., 2019). The other genera have not been associated at the genus or species level to intestinal permeability. However, Veillonellaceae is a unique Firmicute in that it contains lipopolysaccharides incorporated into its cell membrane (Marchandin and Jumas-Bilak, 2014), which have been recognized to stimulate the immune response and increase intestinal permeability through decreasing tight junction proteins (Poltorak et al., 1998; Tanimura et al., 2008; Arce et al., 2010; Liu et al., 2012).
Interestingly, only negative correlations were identified in hens from CF systems and included the following OTU: 2 Clostridiaceae, 2 Aeriscardovia, 1 Romboutsia, 1 Turicibacter, and 1 Gallibacterium. Unlike with the positive correlations, 2 of these genera, Gallibacterium anatis, and 2 Clostridium (Clostridium nigeriense and Clostridium chauvoei) are associated directly with enteric disease or are potential pathogenic bacteria (Singh et al., 2016). Enteric disease in poultry has been associated with elevated intestinal permeability (Deng et al., 2012; Vicuña et al., 2015; Gilani et al., 2017a, 2017b). However, in this study, enteric disease was not identified, and our measure of intestinal permeability examines the whole intestinal tract and is not specific to the ileum where these microbial communities were isolated. Significant intestinal permeability correlations can be found in Table 5.
Correlation of OTU With Jejunal Intestinal Morphology
Jejunal morphology was associated with a limited number of ileal bacterial communities across both housing systems. In hens from CC systems, ileal microbial communities were associated with jejunal villus height (1 positive and 1 negative), jejunal crypt depth (1 negative; Jeotgalicoccus sp.), and jejunal villus height to crypt depth ratio (3 positive). In hens from CF systems, 2 ileal microbial phylotypes were positively associated with jejunal crypt depth (Turicibacter sanguinis and Lactobacillus acidophilus or Lactobacillus crispatus). While L. acidophilus has been used as a probiotic (De Cesare et al., 2017; Forte et al., 2018), T. sanguinis is an immunomodulating bacteria that may lead to secondary infections (Oh et al., 2017). Additionally, T. sanguinis has been associated with bile salt reabsorption and intestinal serotonin production; thus, it is unclear what role T. sanguinis has in regulating intestinal physiology. Significant jejunal morphology correlations can be found in Table 6.
Among these limited correlations, ileal Escherichia/Shigella was negatively associated with jejunal villus height in CC hens. This is not a surprising association as this taxonomic group has been shown to cause detachment of villus tips; thus, reducing the overall size (Shi et al., 2014). The other microbial phylotype associated with jejunal villus height, Tyzzerella sp., was positively associated with jejunal villus height and jejunal villus height to crypt depth ratio. In poultry, Tyzerella sp. abundance was elevated with probiotic supplementation (Gao et al., 2017), which has been associated with increasing villus height (Heak et al., 2017).
The 2 remaining species for jejunal villus height to crypt depth ratio were Streptococcus sp. and Lactobacillus sp. Interestingly, both genera have been used to formulate probiotics (Mallo et al., 2010; De Cesare et al., 2017; Forte et al., 2018; Hanchi et al., 2018), and the specific Lactobacillus sp. (L. acidophilus and L. crispatus) has been associated with increased jejunal villus height when administered in the feed (Chae et al., 2012; De Cesare et al., 2017; Forte et al., 2018). While this study identified correlations between ileal microbial communities and jejunal morphology, the number of correlations were limited suggesting that local or site-specific microbial communities likely play a larger role in shaping the intestinal physiology than presence in other areas of the intestinal tract. Additionally, this highlights the importance of characterizing site-specific communities and cautiously assigning interpretations across intestinal sections.
Correlation of OTU With Ileal Intestinal Morphology
As expected, the greatest number of correlations for ileal OTU were found with ileal intestinal morphology. See Table 7 for correlations between ileal microbial communities and ileal intestinal morphology with r2 > |0.35|. In hens from CC systems, 16 OTU associated with ileal villus height, 9 were negatively correlated, and 7 were positively correlated and 2 OTU were negatively associated with ileal villus height in hens from CF systems. The majority of the OTU associated with ileal villus height were Lactobacillus (4 negative, 5 positive; 8 CC and 1 CF). The negatively correlated Lactobacillus sp. included L. acidophilus, L. aviarius, and Lactobacillus collinoides. A single OTU, OTU 175, which aligned to L. aviarius, was negatively correlated across both housing types. Unfortunately, much remains unknown regarding the function of L. aviarius. The remaining negatively correlated OTU were Enterococcus, Campylobacter, Fusobacterium, Gallibacterium (G. anatis), and Clostridium (Clostridium cuniculi or C. saudinese). Campylobacter and G. anatis have been associated with either primary or secondary enteric diseases (Singh et al., 2016). Additionally, several Enterococcus sp. and Clostridium sp. have been associated with enteric diseases, but the particular phylotypes identified here have not been associated with enteric disease. The positively correlated OTU included the genera Atopobium and Bifidobacterium and of the positively associated OTU, the Lactobacillus species included L. acidophilus, L. aviarius, and L. kitasatonis. As previously mentioned, L. acidophilus is the only of the Lactobacillus species that have been associated with improved intestinal health (Brisbin et al., 2011). These correlations agree with published functional data suggesting that these analyses are correctly identifying known bacterial genera and species with local morphometric changes.
Ileal crypt depth was associated with 9 OTU for each of the 2 hen housing types. Interestingly in the CC system, 1 of the 9 OTU was positively correlated with crypt depth; whereas none of the 9 OTU were positively correlated with crypt depth in the CF system. The only positively correlated OTU was identified as a group of bacteria known as Candidatus Arthromitus or segmented filamentous bacteria. This group of bacteria are known to positively stimulate the gastrointestinal immune system (Bolotin et al., 2014), which could be through the expansion of the crypt (Schnupf et al., 2017; Flannigan and Denning, 2018). The 8 negatively associated OTU were associated with 8 different genera in hens from CC. The OTU of interest are known pathogenic bacteria such as Campylobacter jejuni, Helicobacter winghaensi, and potential pathogenic bacteria, Clostridium difficile, and Lactobacillus avarius. In the CF system, the 9 negatively associated OTU were grouped into 3 genera: Romboutsia (4 OTU); Lactobacillus. (3); and Clostridium (2). Of the Lactobacillus OTU, only 1 is a previously discussed in this manuscript, L. aviarius, and lacks previous research to agree or disagree with our finding. The remaining Lactobacillus phylotypes consist of Lactobacillus taiwanesis or Lactobacillus gasseri, or Lactobacillus johnsonii, Lactobacillus delbrueckii bulgaricus. L. taiwanesis or L. gasseri, or L. johnsonii have been identified in bobwhite quail and may serve a role in fatty acid and carbohydrate metabolism (Zhang et al., 2017), and L. delbrueckii bulgaricus has been associated with cheese and yogurt production (El Kafsi et al., 2014). While our correlations indicate the presence of these bacteria may decrease crypt depth, this may be a result of change in ileal digestibility that reduces the overall need to proliferate in the crypts.
A limited number of OTU were correlated with villus height to crypt depth ratio. Each system had 3 different OTU associated with villus height to crypt depth ratio. In the CC system, 3 OTU, all from Lactobacillus, were positively associated with ileal villus height to crypt depth ratio. Two of the OTU were associated with L. kitasatonis, and the other has high similarity to L. aviarius; both of which have limited functional data in previously published literature. In the CF system, 2 of the 3 OTU correlated with ileal villus height to crypt depth ratio were negatively associated. These species included L. aviarius and C. nigeriense, and based on limited research, it is unclear if either of the bacteria have been associated with intestinal health markers (Alou et al., 2017). L. mucosae was positively associated with ileal villus height to crypt depth ratio. Again, limited information is available regarding L. mucosae; however, this favorable correlation may provide some insight into the functionality.
The increased number of correlations between ileal OTU and ileal morphology compared with jejunal morphology would indicate the importance of using site specific microbial community data to assess influential communities. While this study identified several different phylotypes that were associated with ileal morphology, causation cannot be assessed from this study. Additional studies are needed to confirm these associations across these housing systems, and to define the roles, these phylotypes have in the intestinal physiology and overall performance of the hen.
Conclusions
This study investigated changes of intestinal health of commercial laying hens under optimal commercial conditions for each system. This is the first study to determine intestinal physiology, ileal communities, and association between intestinal physiology and ileal communities in hens across different commercial layer housing systems. In this study, we have identified greater changes in intestinal morphology in the jejunum compared with ileum. However, favorable villus height to crypt depth ratios in both the jejunum and ileum were observed in hens from CC systems, suggesting a balance in the production and sloughing of the intestinal epithelial lining in the CC hens. However, it should be noted in both groups, ranges for villus height, crypt depth, and their ratio were similar to previous reports, and these changes are likely only contributing to more efficient production. Additionally, the measurement of the macromolecule, FITC-D, was lowly detected and similar across housing types suggesting minimal intestinal permeability. Ileal bacterial community diversity measurements were different and favored hens housed in the CC types because of increased species richness, alpha diversity, and overrepresented OTU. Despite the increased overrepresented OTU in CC systems, neither housing type had a significant number of over-represented known pathogenic bacteria. Finally, we explored the correlation of bacterial communities with intestinal traits. A primary finding of this study was that a higher number of correlations were observed between ileal morphology and ileal microbial communities compared with jejunal morphology and ileal microbial communities. This suggests the site-specific microbial community contributes to the intestinal environment and comparisons across even segments of the small intestine should be limited. Additionally, this study identified several OTU that were associated with these traits as expected; thus, providing validation for correlations where OTU have limited information. For example, L. acidophilus phylotypes positively correlated with ileal villus height or an OTU associated with Escherichia/Shigella negatively correlated with jejunal villus height. In conclusion, these results were obtained from a commercial setting instead of a controlled research environment where one system is generally disadvantaged. Several parameters were found to be more favorable for hens housed in CC, suggesting an advantage of this system for intestinal health of these hens. However, it should be pointed out that the lower intestinal health parameters observed in CF were not at levels to indicate detrimental effects (e.g., similar macromolecular flux and pathogenic bacteria), but the differences may highlight known reduced efficiencies of the CF system (e.g., villus height to crypt depth ratio; microbial diversity). However, additional studies are needed to characterize these potentially beneficial bacterial interactions with the hen intestine across housing types to determine if these relationships can be obtained for both systems.
Acknowledgments
Funding for this project was provided by a grant from the Egg Industry Center. Any opinions, finding, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the Egg Industry Center and Iowa State University. Additionally, we would like to acknowledge the generous gift of hens used in the project by Hawkeye Pride Egg Farm.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.10.052.
Supplementary data
Supplementary Figure 1.
Supplementary Figure 2.
References
- Alou M.T., Ndongo S., Frégère L., Labas N., Andrieu C., Richez M., Couderc C., Baudoin J.-P., Abrahão J., Brah S., Diallo A., Sokhna C., Cassir N., La Scola B., Cadoret F., Raoult D. Taxonogenomic description of four new Clostridium species isolated from human gut: ‘Clostridium amazonitimonense’, ‘Clostridium merdae’, ‘Clostridium massilidielmoense’ and ‘Clostridium nigeriense. New Microbes. New Infect. 2017;21:128–139. doi: 10.1016/j.nmni.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Applegate T.J., Schatzmayr G., Pricket K., Troche C., Jiang Z. Effect of aflatoxin culture on intestinal function and nutrient loss in laying hens. Poult. Sci. 2009;88:1235–1241. doi: 10.3382/ps.2008-00494. [DOI] [PubMed] [Google Scholar]
- Arce C., Ramírez-Boo M., Lucena C., Garrido J.J. Innate immune activation of swine intestinal epithelial cell lines (IPEC-J2 and IPI-2I) in response to LPS from Salmonella typhimurium. Comp. Immunol. Microbiol. Infect. Dis. 2010;33:161–174. doi: 10.1016/j.cimid.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Baxter M.F.A., Merino-Guzman R., Latorre J.D., Mahaffey B.D., Yang Y., Teague K.D., Graham L.E., Wolfenden A.D., Hernandez-Velasco X., Bielke L.R., Hargis B.M., Tellez G. Optimizing fluorescein isothiocyanate dextran measurement as a Biomarker in a 24-h feed Restriction model to induce gut permeability in broiler chickens. Front. Vet. Sci. 2017;4:56. doi: 10.3389/fvets.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A., de Wouters T., Schnupf P., Bouchier C., Loux V., Rhimi M., Jamet A., Dervyn R., Boudebbouze S., Blottière H.M., Sorokin A., Snel J., Cerf-Bensussan N., Gaboriau-Routhiau V., van de Guchte M., Maguin E. Genome sequence of “Candidatus Arthromitus” sp. strain SFB-mouse-NL, a commensal bacterium with a key role in postnatal maturation of gut immune functions. Genome Announc. 2014;2:e00705–e00714. doi: 10.1128/genomeA.00705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisbin J.T., Gong J., Orouji S., Esufali J., Mallick A.I., Parvizi P., Shewen P.E., Sharif S. Oral treatment of chickens with lactobacilli influences elicitation of immune responses. Clin. Vaccine Immunol. 2011;18:1447–1455. doi: 10.1128/CVI.05100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J.A., Smith G., Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae B.-J., Ingale S.L., Kim J., Kim K.H., Sen S., Lee S.H., Khong C., Kim E.K., Kwon I.K. Effect of Dietary Supplementation of Probiotics on Performance, Caecal Microbiology and Small Intestinal Morphology of Broiler Chickens [WWW Document] Anim. Nutr. Technology. 2012;12:1–2. [Google Scholar]
- Clarke K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18:117–143. [Google Scholar]
- De Cesare A., Sirri F., Manfreda G., Moniaci P., Giardini A., Zampiga M., Meluzzi A. Effect of dietary supplementation with Lactobacillus acidophilus D2/CSL (CECT 4529) on caecum microbioma and productive performance in broiler chickens. PLoS One. 2017;12:e0176309. doi: 10.1371/journal.pone.0176309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Dong X.F., Tong J.M., Zhang Q. The probiotic Bacillus licheniformis ameliorates heat stress-induced impairment of egg production, gut morphology, and intestinal mucosal immunity in laying hens1. Poult. Sci. 2012;91:575–582. doi: 10.3382/ps.2010-01293. [DOI] [PubMed] [Google Scholar]
- El Kafsi H., Binesse J., Loux V., Buratti J., Boudebbouze S., Dervyn R., Kennedy S., Galleron N., Quinquis B., Batto J.-M., Moumen B., Maguin E., van de Guchte M. Lactobacillus delbrueckii ssp. lactis and ssp. bulgaricus: a chronicle of evolution in action. BMC Genomics. 2014;15:407. doi: 10.1186/1471-2164-15-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannigan K.L., Denning T.L. Segmented filamentous bacteria-induced immune responses: a balancing act between host protection and autoimmunity. Immunology. 2018;154:537–546. doi: 10.1111/imm.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte C., Manuali E., Abbate Y., Papa P., Vieceli L., Tentellini M., Trabalza-Marinucci M., Moscati L. Dietary Lactobacillus acidophilus positively influences growth performance, gut morphology, and gut microbiology in rurally reared chickens. Poult. Sci. 2018;97:930–936. doi: 10.3382/ps/pex396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Ma C., Sun Z., Wang L., Huang S., Su X., Xu J., Zhang H. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken. Microbiome. 2017;5:91. doi: 10.1186/s40168-017-0315-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilani S., Howarth G.S., Kitessa S.M., Tran C.D., Forder R.E.A., Hughes R.J. Intestinal permeability induced by lipopolysaccharide and measured by lactulose, rhamnose and mannitol sugars in chickens. Animal. 2017;11:1174–1179. doi: 10.1017/S1751731116002470. [DOI] [PubMed] [Google Scholar]
- Gilani S., Howarth G.S., Kitessa S.M., Tran C.D., Forder R.E.A., Hughes R.J. New biomarkers for increased intestinal permeability induced by dextran sodium sulphate and fasting in chickens. J. Anim. Physiol. Anim. Nutr. 2017;101:e237–e245. doi: 10.1111/jpn.12596. [DOI] [PubMed] [Google Scholar]
- Hanchi H., Mottawea W., Sebei K., Hammami R. The genus Enterococcus: between probiotic potential and Safety Concerns—an Update. Front. Microbiol. 2018;9:1791. doi: 10.3389/fmicb.2018.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heak C., Sukon P., Kongpechr S., Tengjaroen B., Chuachan K. Effect of Direct-fed microbials on intestinal villus height in broiler chickens: a Systematic review and Meta-analysis of controlled trials. Int. J. Poult. Sci. 2017;16:403–414. [Google Scholar]
- Hubert S.M., Al-Ajeeli M., Bailey C.A., Athrey G. The role of housing environment and dietary protein source on the gut microbiota of chicken. Anim. Open Access J. MDPI. 2019;9:1085. doi: 10.3390/ani9121085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.R., Cox N.A., Guard J., Fedorka-Cray P.J., Buhr R.J., Gast R.K., Abdo Z., Rigsby L.L., Plumblee J.R., Karcher D.M., Robison C.I., Blatchford R.A., Makagon M.M. Microbiological impact of three commercial laying hen housing systems. Poult. Sci. 2015;94:544–551. doi: 10.3382/ps/peu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher D.M., Jones D.R., Abdo Z., Zhao Y., Shepherd T.A., Xin H. Impact of commercial housing systems and nutrient and energy intake on laying hen performance and egg quality parameters. Poult. Sci. 2015;94:485–501. doi: 10.3382/ps/peu078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chen F., Odle J., Lin X., Jacobi S.K., Zhu H., Wu Z., Hou Y. Fish oil enhances intestinal integrity and inhibits TLR4 and NOD2 signaling pathways in weaned pigs after LPS challenge. J. Nutr. 2012;142:2017–2024. doi: 10.3945/jn.112.164947. [DOI] [PubMed] [Google Scholar]
- McMurdie P.J., Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo J.J., Rioperez J., Honrubia P. The addition of Enterococcus faecium to diet improves piglet’s intestinal microbiota and performance. Livest. Sci. 2010;133:176–178. [Google Scholar]
- Marchandin H., Jumas-Bilak E. The Family Veillonellaceae. In: Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. The Prokaryotes: Firmicutes and Tenericutes. Springer; Berlin, Heidelberg, Germany: 2014. pp. 433–453. [Google Scholar]
- Mukai T., Arihara K., Ikeda A., Nomura K., Suzuki F., Ohori H. Lactobacillus kitasatonis sp. nov., from chicken intestine. Int. J. Syst. Evol. Microbiol. 2003;53:2055–2059. doi: 10.1099/ijs.0.02815-0. [DOI] [PubMed] [Google Scholar]
- Ngunjiri J.M., Taylor K.J.M., Abundo M.C., Jang H., Elaish M., Kc M., Ghorbani A., Wijeratne S., Weber B.P., Johnson T.J., Lee C.-W. Farm stage, bird age, and body site dominantly affect the quantity, taxonomic composition, and dynamics of respiratory and gut microbiota of commercial layer chickens. Appl. Environ. Microbiol. 2019;85 doi: 10.1128/AEM.03137-18. e03137-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J.K., Pajarillo E.A.B., Chae J.P., Kim I.H., Yang D.S., Kang D.-K. Effects of Bacillus subtilis CSL2 on the composition and functional diversity of the faecal microbiota of broiler chickens challenged with Salmonella Gallinarum. J. Anim. Sci. Biotechnol. 2017;8:1. doi: 10.1186/s40104-016-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Kindt R., Legendre P., Hara B., Simpson G., Solymos P., Henry M., Stevens H., Maintainer H., Oksanen The vegan package. Community ecology package. 2009;10:631–637. [Google Scholar]
- Pereira A.P., Murakami A.E., Stefanello C., Iwaki L.C.V., Santos T.C. Productive performance, bone characteristics, and intestinal morphology of laying hens fed diets formulated with L-glutamic acid. Poult. Sci. 2019;98:2500–2508. doi: 10.3382/ps/pey595. [DOI] [PubMed] [Google Scholar]
- Poltorak A., He X., Smirnova I., Liu M.Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Qiao H., Shi H., Zhang L., Song Y., Zhang X., Bian C. Effect of Lactobacillus plantarum supplementation on production performance and fecal microbial composition in laying hens. Open Life Sci. 2019;14:69–79. doi: 10.1515/biol-2019-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Zhang L., Shi H., Song Y., Bian C. Astragalus affects fecal microbial composition of young hens as determined by 16S rRNA sequencing. AMB Express. 2018;8:70. doi: 10.1186/s13568-018-0600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2019. R: A language and environment for statistical computing. [Google Scholar]
- Ricaboni D., Mailhe M., Khelaifia S., Raoult D., Million M. Romboutsia timonensis, a new species isolated from human gut. New Microbes New Infect. 2016;12:6–7. doi: 10.1016/j.nmni.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc . SAS Institute; Cary, NC: 2011. SAS OnlineDoc 9.3. [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P., Westcottt S., Ryabin T., Hall J., Hartmann M., Hollister E., Lesniewski R., Oakley B., Parks D., Robinson C., Sahl J., Stres B., Thallinger G., Van Horn D., Weber C. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Env. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnieder C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P., Gaboriau-Routhiau V., Sansonetti P.J., Cerf-Bensussan N. Segmented filamentous bacteria, Th17 inducers and helpers in a hostile world. Curr. Opin. Microbiol. 2017;35:100–109. doi: 10.1016/j.mib.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Shi R., Yang X., Chen L., Chang H., Liu H., Zhao J., Wang X., Wang C. Pathogenicity of Shigella in chickens. PLoS One. 2014;9:e100264. doi: 10.1371/journal.pone.0100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegerstetter S.-C., Schmitz-Esser S., Magowan E., Wetzels S.U., Zebeli Q., Lawlor P.G., O’Connell N.E., Metzler-Zebeli B.U. Intestinal microbiota profiles associated with low and high residual feed intake in chickens across two geographical locations. PLoS One. 2017;12:e0187766. doi: 10.1371/journal.pone.0187766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D., Denman S.E., Hughes R.J., Geier M.S., Crowley T.M., Chen H., Haring V.R., Moore R.J. Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl. Microbiol. Biotechnol. 2012;96:1361–1369. doi: 10.1007/s00253-011-3847-5. [DOI] [PubMed] [Google Scholar]
- Singh S.V., Singh B.R., Sinha D.K., Vinodh K.O.R., Prasanna V.A., Bhardwaj M., Dubey S. Gallibacterium anatis: an emerging pathogen of poultry birds and domiciled birds. J. Veterinar. Sci. Techno. 2016;7:3. [Google Scholar]
- Storey J.D., Taylor J.E., Siegmund D. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J. R. Stat. Soc. Ser. B Stat. Methodol. 2004;66:187–205. [Google Scholar]
- Tanimura N., Saitoh S., Matsumoto F., Akashi-Takamura S., Miyake K. Roles for LPS-dependent interaction and relocation of TLR4 and TRAM in TRIF-signaling. Biochem. Biophys. Res. Commun. 2008;368:94–99. doi: 10.1016/j.bbrc.2008.01.061. [DOI] [PubMed] [Google Scholar]
- Van Coillie E., Goris J., Cleenwerck I., Grijspeerdt K., Botteldoorn N., Van Immerseel F., De Buck J., Vancanneyt M., Swings J., Herman L., Heyndrickx M. Identification of lactobacilli isolated from the cloaca and vagina of laying hens and characterization for potential use as probiotics to control Salmonella Enteritidis. J. Appl. Microbiol. 2007;102 doi: 10.1111/j.1365-2672.2006.03164.x. 1095-106. [DOI] [PubMed] [Google Scholar]
- Van Goor A., Redweik G.A.J., Stromberg Z.R., Treadwell C.G., Xin H., Mellata M. Microbiome and biological blood marker changes in hens at different laying stages in conventional and cage free housings. Poult. Sci. 2020;99:2362–2374. doi: 10.1016/j.psj.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicuña E.A., Kuttappan V.A., Tellez G., Hernandez-Velasco X., Seeber-Galarza R., Latorre J.D., Faulkner O.B., Wolfenden A.D., Hargis B.M., Bielke L.R. Dose titration of FITC-D for optimal measurement of enteric inflammation in broiler chicks. Poult. Sci. 2015;94:1353–1359. doi: 10.3382/ps/pev111. [DOI] [PubMed] [Google Scholar]
- Wang W.-W., Jia H.-J., Zhang H.-J., Wang J., Lv H.-Y., Wu S.-G., Qi G.-H. Supplemental plant Extracts from Flos lonicerae in Combination with Baikal skullcap Attenuate intestinal Disruption and Modulate gut microbiota in laying hens challenged by Salmonella pullorum. Front. Microbiol. 2019;10:1681. doi: 10.3389/fmicb.2019.01681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. Vol. 34. School of Public Policy Capstone; Amherst, MA: 2014. From Battery Cages to Barns: A Cost-Benefit Analysis of a National Standard for Cage-free Egg Production. [Google Scholar]
- Wickham H. Springer-Verlag; New York, NY: 2016. ggplot2: Elegant graphics for data analysis. [Google Scholar]
- Xin H., Hayes M., Ibarburu M.A., Millman S.T., Parsons R.L., Jung H. 2012. A Comprehensive Assessment of Aviary Laying- Hen Housing System for Egg Production in the Midwest. Agricultural and Biosystems Engineering Technical Reports and White Papers. 3. Accessed Nov. 2020. http://lib.dr.iastate.edu/abe_eng_reports/3. [Google Scholar]
- Yamazaki D.M., Ohtsu H., Yakabe Y., Kishima M., Abe H. In vitro screening of lactobacilli isolated from chicken excreta to control Salmonella Enteritidis and Typhimurium. Br. Poult. Sci. 2012;53:183–189. doi: 10.1080/00071668.2012.678814. [DOI] [PubMed] [Google Scholar]
- Yason C.V., Summers B.A., Schat K.A. Pathogenesis of rotavirus infection in various age groups of chickens and turkeys: pathology. Am. J. Vet. Res. 1987;48:927–938. [PubMed] [Google Scholar]
- Zhang M.Z., Yang M., Su H., Rollins D., Zhang S. Lactobacillus colini sp. nov., isolated from Northern Bobwhite (Colinus virginianus) Int. J. Syst. Evol. Microbiol. 2017;67:325–329. doi: 10.1099/ijsem.0.001624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The 16S rRNA gene sequences have been submitted to the NCBI Sequence Read Archive SRA and are available under the BioProject ID PRJNA647366.