Abstract
In recent months, Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread throughout the world. COVID-19 patients show mild, moderate or severe symptoms with the latter ones requiring access to specialized intensive care. SARS-CoV-2 infections, pathogenesis and progression have not been clearly elucidated yet, thus forcing the development of many complementary approaches to identify candidate cellular pathways involved in disease progression. Host lipids play a critical role in the virus life, being the double-membrane vesicles a key factor in coronavirus replication. Moreover, lipid biogenesis pathways affect receptor-mediated virus entry at the endosomal cell surface and modulate virus propagation. In this study, targeted lipidomic analysis coupled with proinflammatory cytokines and alarmins measurement were carried out in serum of COVID-19 patients characterized by different severity degree. Serum IL-26, a cytokine involved in IL-17 pathway, TSLP and adiponectin were measured and correlated to lipid COVID-19 patient profiles. These results could be important for the classification of the COVID-19 disease and the identification of therapeutic targets.
Subject terms: Biochemistry, Biotechnology, Diseases, Molecular medicine
Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread rapidly around the world in recent months and has become an overwhelming health threat globally. COVID-19 has affected more than 16 million individuals and caused nearly 650,000 deaths, resulting in an average mortality of 4% worldwide as of July 27th, 2020, compared with the influenza mortality rate of less than 1%. 80% of patients have mild or moderate symptoms and are therefore classified as mild or moderate COVID-191, whereas 20% of patients develop severe symptoms in time like respiratory distress and require immediate access to specialized intensive care and are therefore classified as clinically severe2–6.
Molecular bases of SARS-CoV-2 infections, pathogenesis and progression have not been clearly elucidated yet, thus forcing the development of many complementary approaches to identify candidate cellular pathways involved in disease progression. An incomplete understanding of the systemic response to SARS-CoV-2 is a major obstacle to the development of new therapeutics and a vaccine7. Double-membrane vesicles (DMVs)8 have been described to be optimal for coronavirus replication9 due to the unique lipid micro-environment supporting the hypothesis that host lipids play a critical role in replication cycle of viruses8. In addition, lipid biogenesis pathways affect receptor-mediated virus entry at the endosomal cell surface10,11 and modulate virus propagation12,13. The development of acute distress respiratory syndrome may also alter lipids implicated in inflammatory process.
In this study, we have combined targeted mass spectrometry (MS)-based lipidomics with the measurement of a subset of proinflammatory cytokines in blood serum of COVID-19 hospitalized patients. Circulating cytokines are crucially important in the systemic response to viruses. However, several studies have established that in severe COVID-19 cases massive cytokine released in the bloodstream may cause a cytokine storm, similar to the one occurring in the macrophage activation syndrome14,15. Such an hyperinflammatory response can contribute to systemic coagulation and fatality in infected patients. In severe cases of COVID-19 infection, an induction of reported increases in inflammatory monocytes and neutrophils and a sharp decrease in lymphocytes have been reported, with the pro- inflammatory cytokines include interleukins IL-1ß, IL-6 and IL-8, and TNF-α16–18.
Viral infection causes release of alarmins, also called damage- associated molecular patterns (DAMPs), by dying necrotic cells. Among the alarmins, thymic stromal lymphopoietin (TSLP) is a distant paralog of IL-7 mainly released by epithelial lining of the lungs, gastrointestinal tract and skin in response of an external insults, which when produced in high amount is released in the bloodstream19. In addition, it also participates to an adaptive immune response against viruses that target the respiratory mucosa20. Finally, in inflammatory processes triggered by viral infection adiponectin, selectively produced by adipose tissue, strongly correlates to severity of inflammation21–23.
In this paper, targeted metabolomic analysis was carried out to define the lipidomic serum profile of a cohort of COVID-19 patients characterized by different severity degree, following the hypothesis that SARS-CoV-2 induces metabolic changes in the body fluids, as reported very recently24,25. In all these patients, we previously found increased levels of serum interleukin (IL)-6 together with variable levels of tumor necrosis factor (TNF)-α, IL-17A and the serum receptor of IL-17A (IL-17RA)26. Here, we measured serum IL-26, a cytokine involved in IL-17 pathway, TSLP and adiponectin. Targeted lipidomics was correlated to inflammatory cytokines identifying significant correlations in infected patients. These findings may have important implications in the classification of the disease and in the identification of therapeutic targets.
Results
Serum lipidomics in COVID-19 patients
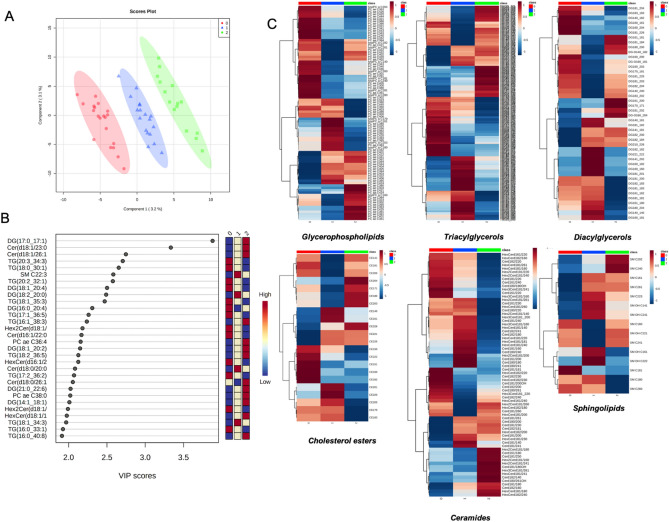
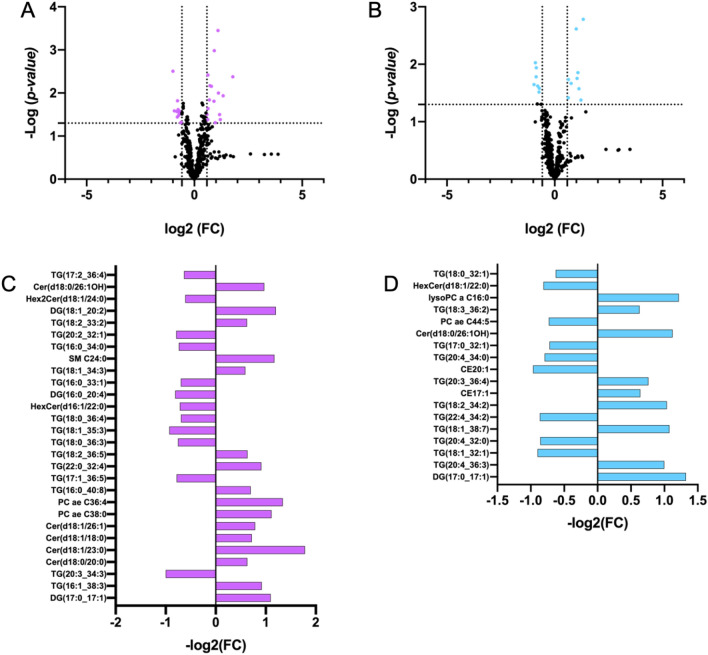
Six classes of lipids, as diacylglycerols (DG) (n = 44 molecules), triacylglycerols (TG) (n = 242 molecules), glycerophospholipids (LPC, PC aa, PC ae) (n = 90 molecules), cholesterol esters (CE) (n = 22), ceramides (Cer) (n = 70) and sphingolipids (SM) (n = 15 molecules), were dosed. A detailed list of the measured lipids is shown in Table S1 and their concentrations in Table S2. The concentrations of the lipids from serum COVID-19 patients were imputed to remove missing value, log(2) transformed and pareto scaled; the lipid datasets, corresponding to the 483 analyzed metabolites (Table S2), were processed by partial least squares-discriminant analysis (PLS-DA) to evaluate the difference rate and variance occurring between the groups according to pathology severity. These data points were clustered into three distinct groups in the plot map (Fig. 1A), separating the COVID-19 patients according to pathology-severity, (principal component, PC1 variance 3.2%, PC2 variance 3.1%). Outliers were not found in serum lipid datasets. The most discriminant lipidic molecules between the three groups of patients, estimated according to their Variable Importance in Projection (VIP) score (threshold > 2.0) included 28 metabolites (Fig. 1B); the levels of 12 lipidic molecules, including DG(17:0_17:1), DG(18:1_20:2), DG(21:0_22:6), DG(14:1_18:1), TG(16:1_38:3), TG(18:2_36:5), TG(16:0_40:8), PC ae C36:4, PC ae C38:0, Cer(d18:1/23:0), Cer(d18:1/26:1), HexCer(18:1/18:0), increased in severe COVID19 serum in respect to moderate and mild groups and 10, including DG(18:1_20:4), TG(20:3_34:3), TG(18:0_30:1), TG(18:1_35:3), TG(17:1_36:5), TG(17:2_36:2), TG(16:0_33:1), Cer(d16:1/22:0), Hex2Cer(d18:1/24:0), Hex2Cer(d18:1/22:0), decreased in severe COVID19 serum in respect to moderate and mild groups. In order to explore the details of the observed discrimination trend, a heatmap was constructed according to the degree of biochemical features of analyzed lipids. As indicated in Fig. 1C, changed lipid features affect all the analyzed lipidic classes, indicating that virus infection strongly perturbed lipid homeostasis. Univariate analysis allows to detect significant differences in lipidic profiles when severe-, medium- and mild-patients have been compared each other (Fig. 2A,B). Severe patients displayed altered levels of DG(17:0_17:1), DG(16:0_20:4), DG(18:1_20:2), TG(16:1_38:3), TG(20:3_34:3), TG(16:0_40:8), TG(17:1_36:5), TG(22:0_32:4), TG(18:2_36:5), TG(18:0_36:3), TG(18:1_35:3), TG(18:0_36:4), TG(16:0_33:1), TG(18:1_34:3), TG(16:0_34:0), TG(20:2_32:1), TG(18:2_33:2), TG(17:2_36:4), Cer(d18:0/20:0), Cer(d18:1/23:0), Cer(d18:1/18:0), Cer(d18:1/26:1), Cer(d18:0/26:1OH), HexCer(d16:1/22:0), Hex2Cer(d18:1/24:0), PC ae C38:0, PC ae C36:4, SM C24:0, in respect to mild patients (Table 1, Fig. 2C). In particular, serum concentrations of ceramides, as Cer(d18:0/20:0), Cer(d18:1/23:0), Cer(d18:1/18:0), Cer(d18:1/26:1), Cer(d18:0/26:1OH), were significantly increased while glycosylceramides as HexCer(d16:1/22:0), Hex2Cer(d18:1/24:0), were significantly decreased in severe condition versus mild condition. Moreover, severe patient serum concentrations of PC ae C38:0, PC ae C36:4, SM C24:0, resulted increased in respect to mild patients. When the severe condition was compared to moderate condition, significant alterations in serum level of DG(17:0_17:1), TG(20:4_36:3), TG(18:1_32:1), TG(20:4_32:0), TG(18:1_38:7), TG(22:4_34:2), TG(18:2_34:2), CE17:1, TG(20:3_36:4), CE20:1, TG(20:4_34:0), TG(17:0_32:1), Cer(d18:0/26:1OH), PC ae C44:5, TG(18:3_36:2), lysoPC a C16:0, HexCer(d18:1/22:0), TG(18:0_32:1) (Table 2, Fig. 2D). The alteration in ceramide levels was conserved also when the severe condition was compared to the moderate one. Indeed, serum concentrations of ceramide Cer(d18:0/26:1OH) were significantly increased, while glycosylceramide HexCer(d18:1/22:0) decreased in severe condition versus moderate condition. Lipid metabolites such as DG(17:0_17:1), TG(20:4_36:3), TG(18:1_38:7), TG(18:2_34:2), TG(20:3_36:4), TG(18:3_36:2), CE17:1, lysoPC a C16:0, have been observed as more abundant in severe versus moderate condition, and TG(20:4_36:3), TG(18:1_32:1), TG(22:4_34:2), CE20:1, TG(20:4_34:0), TG(17:0_32:1), PC ae C44:5 and TG(18:0_32:1), resulted less abundant.
Figure 1.
Lipid profiles in COVID-19 patient serum. (A) Supervised partial least squares-discriminant analysis (PLS-DA) score plot and (B) discriminant lipidic features according to the variable importance on projection (VIP) score of mild, moderate and severe COVID-19 patients. The top 30 important lipids (VIP score ≥ 2.0 were summarized according to their VIP score value. The intensity of the colored boxes indicates the relative lipids abundance in each group (0 mild, 1 moderate and 2 severe patients). (C) Heatmaps of the auto-scaled mean lipid concentrations of each patient groups (0 mild, 1 moderate and 2 severe patients) of significantly altered lipids (p < 0.05) in six different lipid classes. The heatmap color code represents the relative lipid abundance. The concentrations of the metabolites were imputed, log(2) transformed, and Pareto scaled.
Figure 2.
Targeted lipidomic analyses. Volcano plots of differential concentrations of 483 lipids in comparison of (A) severe patient serum to mild and (B) severe patient serum to moderate. The fold change of significant differentially abundant lipids (p < 0.05) in (C) severe condition versus mild and (D) severe condition versus moderate were plotted according to their significant p-value, reported in Tables 1 and 2, respectively.
Table 1.
Differentially abundant lipids in the severe condition versus mild condition.
| Lipid | FC | log2 (FC) | p value | − Log (p value) |
|---|---|---|---|---|
| DG(17:0_17:1) | 2.14 | 1.10 | 0.0004 | 3.45 |
| TG(16:1_38:3) | 1.89 | 0.92 | 0.0010 | 2.98 |
| TG(20:3_34:3) | 0.50 | − 1.00 | 0.0031 | 2.51 |
| Cer(d18:0/20:0) | 1.55 | 0.63 | 0.0039 | 2.41 |
| Cer(d18:1/23:0) | 3.44 | 1.78 | 0.0042 | 2.37 |
| Cer(d18:1/18:0) | 1.65 | 0.72 | 0.0068 | 2.17 |
| Cer(d18:1/26:1) | 1.73 | 0.79 | 0.0070 | 2.15 |
| PC ae C38:0 | 2.16 | 1.11 | 0.0100 | 2.00 |
| PC ae C36:4 | 2.53 | 1.34 | 0.0116 | 1.93 |
| TG(16:0_40:8) | 1.62 | 0.70 | 0.0145 | 1.84 |
| TG(17:1_36:5) | 0.58 | − 0.78 | 0.0151 | 1.82 |
| TG(22:0_32:4) | 1.88 | 0.91 | 0.0155 | 1.81 |
| TG(18:2_36:5) | 1.55 | 0.64 | 0.0230 | 1.64 |
| TG(18:0_36:3) | 0.59 | − 0.75 | 0.0242 | 1.62 |
| TG(18:1_35:3) | 0.53 | − 0.93 | 0.0261 | 1.58 |
| TG(18:0_36:4) | 0.62 | − 0.69 | 0.0265 | 1.58 |
| HexCer(d16:1/22:0) | 0.61 | − 0.72 | 0.0269 | 1.57 |
| DG(16:0_20:4) | 0.57 | − 0.81 | 0.0270 | 1.57 |
| TG(16:0_33:1) | 0.62 | − 0.70 | 0.0304 | 1.52 |
| TG(18:1_34:3) | 1.51 | 0.59 | 0.0314 | 1.50 |
| SM C24:0 | 2.25 | 1.17 | 0.0318 | 1.50 |
| TG(16:0_34:0) | 0.60 | − 0.74 | 0.0335 | 1.48 |
| TG(20:2_32:1) | 0.58 | − 0.79 | 0.0362 | 1.44 |
| TG(18:2_33:2) | 1.54 | 0.62 | 0.0414 | 1.38 |
| DG(18:1_20:2) | 2.30 | 1.20 | 0.0416 | 1.38 |
| Hex2Cer(d18:1/24:0) | 0.66 | − 0.61 | 0.0476 | 1.32 |
| Cer(d18:0/26:1OH) | 1.96 | 0.97 | 0.0488 | 1.31 |
| TG(17:2_36:4) | 0.64 | − 0.64 | 0.0493 | 1.31 |
Table 2.
Differentially abundant lipids in the severe condition versus medium condition.
| Lipid | FC | log2 (FC) | p value | − Log (p value) |
|---|---|---|---|---|
| DG(17:0_17:1) | 2.51 | 1.32 | 0.0017 | 2.78 |
| TG(20:4_36:3) | 2.00 | 1.00 | 0.0024 | 2.61 |
| TG(18:1_32:1) | 0.54 | − 0.90 | 0.0094 | 2.03 |
| TG(20:4_32:0) | 0.55 | − 0.86 | 0.0115 | 1.94 |
| TG(18:1_38:7) | 2.11 | 1.08 | 0.0140 | 1.85 |
| TG(22:4_34:2) | 0.55 | − 0.86 | 0.0165 | 1.78 |
| TG(18:2_34:2) | 2.05 | 1.04 | 0.0176 | 1.75 |
| CE17:1 | 1.56 | 0.64 | 0.0185 | 1.73 |
| TG(20:3_36:4) | 1.70 | 0.76 | 0.0216 | 1.67 |
| CE20:1 | 0.51 | − 0.97 | 0.0226 | 1.65 |
| TG(20:4_34:0) | 0.58 | − 0.79 | 0.0239 | 1.62 |
| TG(17:0_32:1) | 0.61 | − 0.72 | 0.0263 | 1.58 |
| Cer(d18:0/26:1OH) | 2.18 | 1.12 | 0.0266 | 1.57 |
| PC ae C44:5 | 0.60 | − 0.73 | 0.0305 | 1.52 |
| TG(18:3_36:2) | 1.55 | 0.63 | 0.0384 | 1.42 |
| lysoPC a C16:0 | 2.32 | 1.22 | 0.0421 | 1.38 |
| HexCer(d18:1/22:0) | 0.57 | − 0.81 | 0.0493 | 1.31 |
| TG(18:0_32:1) | 0.65 | − 0.63 | 0.0496 | 1.30 |
Serum levels of cytokines and their correlation with metabolic changes
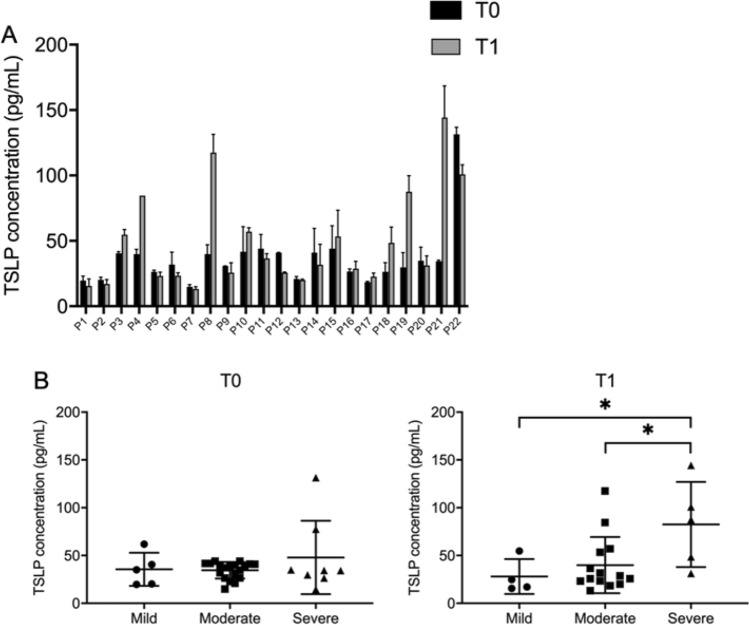
Being a lytic virus, SARS-CoV-2 causes cell death leading to local and systemic inflammation with the upregulation of several cytokines. Injured tissue releases alarmins such as TSLP that may be released in the bloodstream and have multiple effects. To test the possibility that SARS-CoV-2 infection leads to TSLP accumulation in the bloodstream, we measured the levels of serum TSLP in individuals affected by COVID-19 at two time points, namely at hospital admission and 7 days post admission. In most tested patients the TSLP serum levels remained similar or were elevated a week after admission, although this trend did not reach statistical significance (Fig. 3A). As for the other parameters, data were then clustered into three distinct groups according to pathology-severity. At admission (T0) no significant correlation in TSLP serum levels and clinical severity was observed, whereas, 7 days after hospitalization (T1) TSLP serum levels were significantly more elevated in severe versus moderate or mild cases (Fig. 3B and Table S3).
Figure 3.
TSLP protein levels in COVID-19 patient serum. (A) TSLP concentration expressed in pg/mL in the serum of COVID-19 patients at time of hospital admission (T0) or 1-week after admission (T1). (B) TSLP levels in severely affected individuals was significantly higher than in mild or moderate cases (adjusted p-value = 0.048 and 0.046 respectively).
IL-26 was not correlated to the severity of the disease in this cohort (Table S3), although we found that 5 moderate patients had IL-26 concentrations from fivefold to 200-fold higher than upper reference value (10 pg/mL) and only one mild patient showed a IL-26 level higher than reference value. At admission (T0) no significant correlations were found for IL-26. 7 days after hospitalization (T1), IL-26 positively correlated with Cer(d18:0/d18:0), whereas a negative correlation was observed between IL-6 and the cholesterol ester CE(22:1) (Table S3).
Serum level of adiponectin and its correlation with metabolic changes
Total adiponectin evaluation by ELISA-test showed that the COVID-19 patients subdivided in three sub-groups based have different adiponectin concentrations. In particular, in severe COVID-19 adiponectin levels are higher compared to moderate and mild COVID-19 patients. However, the difference between the three subgroups are not statistically significant (Table 3). Western blotting analysis showed the oligomeric profile of adiponectin in COVID-19 patients and confirmed the above described adiponectin differences (Supplementary Fig. 1). Full-length image of blot was reported in Supplementary Fig. 2. At T1 IL-6 serum levels positively correlated to adiponectin (Table S3). Furthermore, a positive correlation among adiponectin and ceramides and glycerophospholipids was found, at T0 (Table S3).
Table 3.
Adiponectin serum concentrations (µg/ml) in COVID− 19 patients.
| Patients | T0 | T1 | p-value |
|---|---|---|---|
| Covid-19 patients mild | 17.2 ± 3.14 | 15.6 ± 1.38 | ns |
| Covid-19 patients moderate | 19.3 ± 4.92 | 19.0 ± 0.57 | ns |
| Covid-19 patients severe | 21.4 ± 2.67 | 19.8 ± 2.57 | ns |
Discussion
MS-based lipidomic platform allowed to define the lipidic profiles according to infection-pathology severity by measuring 483 lipids, included in six different lipidic classes. MS targeted lipidomic approach coupled to multivariate and univariate statistical analyses identified alterations in serum concentrations of 46 lipids when severe patients were compered to mild and moderate patients. Among them, a group TGs resulted altered in severe conditions. This data seems to indicate the involvement of autophagy mechanism as observed in other virus infection as dengue virus infection27. It has been shown that cellular lipid metabolism is influenced by autophagy, using lipid droplets28, constituted by triglycerides (TGs) and cholesterol esters29. The release of free fatty acids30, from triglycerides hydrolysis by lipases enzyme, unbalances mitochondria homeostasis and subsequent β-oxidation and energy production. In addition, eleven TGs, as TG (20:3_34:3), TG(18:0_30:1), TG(20:2_32:1), TG(18:1_35:3), TG(17:1_36:5), TG(16:1_38:3), TG(18:2_36:5), TG(17:2_36:2), TG(18:1_34:3), TG(16:0_33:1), TG(16:0_40:8), were selected in the discriminant model to accurately differentiate severe conditions. According to plasma metabolomic panel differentiating COVID-19 patients from healthy controls in a recent study25, we defined two lipidic panels able to discriminate into severity degree of infective pathology. In Song et al.25 the SM sphingolipid class was demonstrated to display progressive increase with increasing of disease severity. Specifically, we found an increased level of SM C24:0 in severe versus mild condition.The membrane composition and fluidity alteration were described as the main biological process responsible of microdomains generation, able to affect the dynamic roles of membrane, as signal transduction and immune activation processes31.
It is interesting to note also the significant increased level of ceramides, Cer(d18:0/20:0), Cer(d18:1/23:0), Cer(d18:1/18:0), Cer(d18:1/26:1), in serum of severe patients. Recently it was proved the ability of ceramide to promote vasoconstriction32. Ceramides belong to sphingolipid family, known as essential constituents of organelle and cell membranes that play an additional role in signal transductions of crucial physiological processes such as growth, differentiation, proliferation, migration, apoptosis, and cell death33. Production of ceramide may be implicated in pathological processes including cardiovascular and lung diseases32. An increasing scientific evidence revealed the correlation between circulating ceramide levels with adverse cardiovascular events such as myocardial infarction and stroke34,35. In addition, the accumulation of ceramides with long chains resulted to be associated with deleterious outcomes but it was not correlated to traditional plasma lipid markers of cardiovascular risk factors36.
In severe cases of COVID-19, a dysfunctional immune response triggers a cytokine storm that mediates widespread lung inflammation, although the players of immunological misfiring are still incompletely understood. Here, we find that the alarmin TSLP is released into the bloodstream and is significantly associated to the severity of the disease specifically in advanced stages (i.e. one week after hospital admission), in contrast to TNF alpha, IL-6, IL-17 and IL-26 for which this correlation was not observed. Our results were in line with previous finding indicating a dysregulated immune response in COVID-19 patients, especially in the most severe cases25,26. Indeed a negative correlation was demonstrated between PCs and IL-625. In the present study a new negative correlation linking IL-6 and ceramides was observed, confirming the hypothesis of an inflammatory response as a consequence of the lipid alteration. In order to support these results, other clinical indices of systemic inflammation were investigated. Thymic stromal lymphopoietin (TSLP) is a mucosal tissue-associated cytokine that stimulates a T-helper type 2 (Th2) response in allergic diseases such as asthma and atopic dermatitis, and therefore plays a key role in the pathogenesis of inflammatory diseases19. In the human asthmatic lung TSLP is highly expressed and activates dendritic cells to strongly induce proallergic Th2 cell responses. Interestingly, a TSLP involvement in viral infection has been less investigated, although it is well established that its expression is elevated in respiratory syncytial virus-infected airway epithelial cells and plays a non-redundant role on anti-viral CD8 T cell responses37. In addition, it has been shown that epithelial cells exposed to HIV virus triggers DC-mediated mucosal infection of CD4 + T cells38. Therefore, if our findings will be extended in large number of COVID-19 patients around the world, they may provide evidence for TSLP beneficial role in antiviral immunity opening a window for therapeutic intervention, as human anti-TSLP antibody (Tezepelumab) have been proven effective for blocking the interaction between TSLP in asthmatic patients39.
In the present study, we found a correlation of adiponectin and IL-6 concentrations which could represent an additional attempt by adipose tissue to counteract inflammation mediated by the increase in IL-6. In addition, the correlation between adiponectin and ceramides confirmed the involvement of adiponectin in lipid metabolism40. These finding are suggestive of an active role of adipose tissue in COVID-19 onset.
Our data reinforce previous results that lipid metabolism play an important role in the viral infection cycle. Further work is necessary to investigate the role of lipids in the signaling pathway of the viral life cycle. These studies will establish new drug targets as well as the use of existing drugs.
Limitations of the study
Our results were based on a single Italian cohort of COVID-19 patients in Campania Region; future studies in different cohorts and in healthy controls will be needed for extending the understanding of lipid dysregulation and cytokine production in COVID-19 pathogenesis. The median age of severe patients is older than non-severe, so we did not check the influence of age on our data. However, other authors did not observe impact of age and sex on omics data in COVID-19 patients24. Severe patients also exhibit comorbidities like obesity which may influence the lipidomic profile. Lipidomic analysis was carried out in serum but not in lung tissues or in the BALF (bronchoalveolar lavage fluid) or sputum of COVID-19 and therefore we cannot establish a direct association between lipidomic and cytokine changes with SARS-CoV-2 infection in the lung. Furthermore, only a small number of cytokines has been measured and in particular, other alarmins beside TSLP, such as IL-25 and IL-33, have not been measured.
Materials and methods
Patients
A Regione-Campania-cohort of 53 patients with a diagnosis of COVID-19 (SARS-CoV-2 infection) admitted at one of the following hospitals was enrolled: Department of Clinical Medicine and Surgery—Section of Infectious Diseases, University Hospital Federico II, Naples; Department of Infectious Disease and Infectious Urgencies—Division of Respiratory Infectious Disease, Cotugno Hospital, AORN dei Colli, Naples, Pathophysiology and respiratory rehabilitation-1 utsir COVID. 53 patients were enrolled (37/53 males, 70% and 16/53 females, 30%), with median age of 58 years. Patients were classified using an seven-point ordinal scale which consists of following numeric scale and scale descriptors26: 1, not hospitalized with resumption of normal activities; 2, not hospitalized, but unable to resume normal activities; 3, hospitalized, not requiring supplemental oxygen; 4, hospitalized, requiring supplemental oxygen; 5, hospitalized, requiring nasal high-flow oxygen therapy, non-invasive mechanical ventilation, or both; 6, hospitalized, requiring ECMO, invasive mechanical ventilation, or both; and 7, death. 1–3 were subclassified as mild patients (n = 20), 4 was subclassified as moderate patients (n = 16), and 5–7 were subclassified as severe patients (n = 17). The aliquots of serum sample were collected at hospital admission and stored at − 80 °C for subsequent analysis. The study protocol was approved by Ethics Committee at The University of Naples Federico II n°191/20. All methods and experimental procedure were performed in accordance with the relevant guidelines and regulations included in n°191/20 protocol approved by Ethics Committee at The University of Naples Federico II. The present research was performed in accordance with the Declaration of Helsinki. The present research did not involve human participants under the age of 18 years. Each patient (and/or legal guardian) gave a fully informed consent to the participation to the study and to the use of their biological samples for research purpose.
Lipidomic analysis
Serum lipidome content was investigated by mass spectrometry-based targeted metabolomics41,42. The lipidomic platform was set to detect 483 lipids including 22 cholesterol esters, 34 glycosylceramides, 44 diacylglycerols, 90 glycerophospholipids, 15 sphingolipids, 242 triacylglycerols, 28 ceramides, 8 dihydroceramides according to Mxp Quant 500 protocols (Biocrates Life Sciences Innsbruck, Austria). For each analyzed sample, 10 µL of patient serum were processed and three technical replicates were carried out. Multiple reaction monitoring (MRM) was used to target and quantitate the molecular species from the different lipid classes by direct flow injection analysis (FIA), on a Triple Quad 5500 + System QTrap-Ready (AB Sciex) coupled with an Agilent 1260 Infinity II HPLC43–46. Briefly, 10 µL of each serum sample were added onto the wells of the 96-well extraction plate, with the exception of blank positions, PBS, calibrants, and quality controls (QC), and dried under nitrogen flow for 30 min. The samples were derivatized with 50 μL of 5% phenyl isothiocyanate at room temperature and then dried under N2 flow for 1 h. Metabolites were extracted with 300 µL of 5 mM ammonium acetate in methanol, shaked for 30 min (450 rpm) and eluted by centrifugation. Then, 10 µL of extract were diluted with 490 µl of FIA solvent. The injection volume into the system was 20 µL within the mobile phase (FIA solvent) at an initial flow rate of 0.03 mL/min until 1.6 min, followed by flow rates of 0.20 mL/min for 1.6 min and 0.02 mL/min for 0.20 min. The auto sampler was cooled at 10 °C. The ESI source operated in positive ion mode using the following parameters: spray voltage 55 kV, temperature of 450 °C, GS1 20, GS2 40, CUR 30, CAD 8. Data were acquired using Analyst software (version 1.7.1 Ab Sciex) and data analysis was performed used MetIDQ Oxygen 2976 (Biocrates Life Sciences Innsbruck, Austria)47,48.
Lipidomic feature selection
Serum concentrations of 483 lipids from COVID-19 patients were processed by multivariate analysis using MetaboAnalyst 4.0 (http://www.metaboanalyst.ca)49–51. The lipidic dataset was imputed to remove the missing values, log(2)-transformed and scaled according to the Pareto scaling method. The resulted dataset was used to define PLS-DA (Partial Least Squares—Discriminant Analysis), VIP (Variable Importance in Projection) score and heatmap. Finally, univariate statistics was carried out using GraphPad Prism 8.0, and the results presented as the mean standard error of the mean (SEM). The statistical significance of the difference in serum lipid concentrations was evaluated by parametric (unpaired t-test) or non-parametric (Mann–Whitney test) tests when data failed the Shapiro–Wilk normality test. Statistical significantly changed lipids were selected using criteria of p-value (p < 0.05) and log2 fold change (FC) larger than ± 0.58, corresponding to FC (defined as ratio between lipid non-null concentrations) value of 1.5.
Cytokine measurement
Concentrations of TSLP protein in blood serum were evaluated by enzyme-linked immunosorbent assay (ELISA) (Human TSLP Uncoated ELISA Kit; ThermoFisher) according to the manufacturer's instructions). Duplicate tests of each independent sample were conducted. 100 μL of serum samples was used for experiments. The results were expressed as protein concentration per mL of serum. Data of serum IL-17A, IL-17RA, IL-6 and TNF-α were already available from a previous study carried out on the same serum samples26. Serum IL-26 was measured by ELISA kit from Sigma-Aldrich (St. Louis, MO 63103 USA), in accordance with the manufacturer's instructions. The assay was performed using anti-human IL-26 precoated 96-well strip plates. Serum samples were analyzed using 100 μL of twofold diluted sample. All samples and standard solutions for calibration curves were analyzed in duplicate and mean concentrations were expressed as pg/mL.
Adiponectin measurement
The serum adiponectin concentrations were measured by enzyme-linked immunosorbent assay (ELISA) as previously reported21. Calibration curve was performed and quantified using human recombinant Adiponectin (Biovendor R&D, Brno, Czech Republic) as a standard. Each sample, diluted 1:5000, was assayed three times in duplicate. In addition, adiponectin profile was analyzed by western blot analysis as previously described21. The western-blot filter was scanned by using Chemidoc MP imaging system (BIORAD) able to provide automatic recognition adjustment of image scanning parameters.
Statistics
To calculate the statistical significance of changes in each cytokine in mild, moderate and severe patients a one-way ANOVA with Tukey’s multiple-comparison test was performed. To measure the strength of association between the 483 lipids and TNF-α, IL-6, IL-17A, IL-17RA, IL-26, TSLP and Adiponectin, the Spearman's rho method was used, where the value r = 1 means a perfect positive correlation and the value r = − 1 means a perfect negative correlation. The correlation was considered significant at the 0.01 level (2-tailed).
Supplementary Information
Acknowledgements
This study was supported by the project “CEINGE TASK-FORCE COVID19”, code D64I200003800 by Regione Campania for the fight against Covid-19 (DGR n.140 del 17 marzo2020).
Author contributions
R.P., M.D’A., F.M.F., C.dell’Isola, G.F., A.A., C.C., G.F. and I.G. contributed to study concept and design, clinical data collection and pathological analysis. M.C., R.F., M.C., E.M. and M.R. have contributed to study concept, design, lipidomic analysis and lipidomics data interpretation. M.G., M.C., R.R. and I.A. have contributed to statistical analysis, collection of the serum samples, and patient database establishment. S.S. and C.M. have contributed to TSLP results. A.D. and R.P. have contributed to Adiponectin results. M.R., C.M. and G.C. have contributed to the study supervision, interpretation of the results, and writing of the manuscript. All authors approved the final version.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Marianna Caterino, Monica Gelzo and Stefano Sol.
Contributor Information
Caterina Missero, Email: missero@ceinge.unina.it.
Margherita Ruoppolo, Email: margherita.ruoppolo@unina.it.
Giuseppe Castaldo, Email: giuseppe.castaldo@unina.it.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-82426-7.
References
- 1.Thevarajan I, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murthy S, Gomersall CD, Fowler RA. Care for critically Ill patients with COVID-19. JAMA. 2020 doi: 10.1001/jama.2020.3633. [DOI] [PubMed] [Google Scholar]
- 3.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease, (COVID-19) outbreak in China: summary of a report of 72314 cases from the chinese center for disease control and prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 4.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costanzo M, De Giglio MAR, Roviello GN. SARS-CoV-2: recent reports on antiviral therapies based on lopinavir/ritonavir, darunavir/umifenovir, hydroxychloroquine, remdesivir, favipiravir and other drugs for the treatment of the new coronavirus. Curr. Med. Chem. 2020 doi: 10.2174/0929867327666200416131117. [DOI] [PubMed] [Google Scholar]
- 8.Yan B, et al. Characterization of the lipidomic profile of human coronavirus-infected cells: implications for lipid metabolism remodeling upon coronavirus replication. Viruses. 2019 doi: 10.3390/v11010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knoops K, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taube S, Jiang M, Wobus CE. Glycosphingolipids as receptors for non-enveloped viruses. Viruses. 2010;2:1011–1049. doi: 10.3390/v2041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chazal N, Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 2003;67:226–237. doi: 10.1128/mmbr.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy PD, Strating JR, van Kuppeveld FJ. Building viral replication organelles: close encounters of the membrane types. PLoS Pathog. 2016;12:e1005912. doi: 10.1371/journal.ppat.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu NY, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta P, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, et al. Chemical inhibition of FBXO7 reduces inflammation and confers neuroprotection by stabilizing the mitochondrial kinase PINK1. JCI Insight. 2020 doi: 10.1172/jci.insight.131834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giamarellos-Bourboulis EJ, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000 e1003. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucas C, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020 doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roan F, Obata-Ninomiya K, Ziegler SF. Epithelial cell-derived cytokines: more than just signaling the alarm. J. Clin. Invest. 2019;129:1441–1451. doi: 10.1172/JCI124606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye L, et al. Interferon-lambda enhances adaptive mucosal immunity by boosting release of thymic stromal lymphopoietin. Nat. Immunol. 2019;20:593–601. doi: 10.1038/s41590-019-0345-x. [DOI] [PubMed] [Google Scholar]
- 21.Daniele A, et al. Adiponectin oligomerization state and adiponectin receptors airway expression in chronic obstructive pulmonary disease. Int. J. Biochem. Cell Biol. 2012;44:563–569. doi: 10.1016/j.biocel.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Lacedonia D, et al. Evaluation of adiponectin profile in Italian patients affected by obstructive sleep apnea syndrome. Pulm. Pharmacol. Ther. 2016;40:104–108. doi: 10.1016/j.pupt.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Signoriello E, et al. Adiponectin profile at baseline is correlated to progression and severity of multiple sclerosis. Eur. J. Neurol. 2019;26:348–355. doi: 10.1111/ene.13822. [DOI] [PubMed] [Google Scholar]
- 24.Shen B, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72 e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song JW, et al. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020 doi: 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polo A, et al. Identifying a panel of genes/proteins/miRNAs modulated by arsenicals in bladder, prostate, kidney cancers. Sci. Rep. 2018;8:10395. doi: 10.1038/s41598-018-28739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 30.Haemmerle G, Zimmermann R, Zechner R. Letting lipids go: hormone-sensitive lipase. Curr. Opin. Lipidol. 2003;14:289–297. doi: 10.1097/00041433-200306000-00009. [DOI] [PubMed] [Google Scholar]
- 31.Olsen ASB, Faergeman NJ. Sphingolipids: membrane microdomains in brain development, function and neurological diseases. Open Biol. 2017 doi: 10.1098/rsob.170069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cogolludo A, Villamor E, Perez-Vizcaino F, Moreno L. Ceramide and Regulation of vascular tone. Int. J. Mol. Sci. 2019;20:200. doi: 10.3390/ijms20020411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 34.Summers SA. Could ceramides become the new cholesterol? Cell Metab. 2018;27:276–280. doi: 10.1016/j.cmet.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Tippetts TS, Holland WL, Summers SA. The ceramide ratio: a predictor of cardiometabolic risk. J. Lipid Res. 2018;59:1549–1550. doi: 10.1194/jlr.C088377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meeusen JW, et al. Plasma ceramides. Arterioscler. Thromb Vasc. Biol. 2018;38:1933–1939. doi: 10.1161/ATVBAHA.118.311199. [DOI] [PubMed] [Google Scholar]
- 37.Shane HL, Klonowski KD. A direct and nonredundant role for thymic stromal lymphopoietin on antiviral CD8 T cell responses in the respiratory mucosa. J. Immunol. 2014;192:2261–2270. doi: 10.4049/jimmunol.1302085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fontenot D, et al. TSLP production by epithelial cells exposed to immunodeficiency virus triggers DC-mediated mucosal infection of CD4+ T cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16776–16781. doi: 10.1073/pnas.0907347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corren J, et al. Tezepelumab in adults with uncontrolled asthma. N. Engl. J. Med. 2017;377:936–946. doi: 10.1056/NEJMoa1704064. [DOI] [PubMed] [Google Scholar]
- 40.Qiao L, Zou C, van der Westhuyzen DR, Shao J. Adiponectin reduces plasma triglyceride by increasing VLDL triglyceride catabolism. Diabetes. 2008;57:1824–1833. doi: 10.2337/db07-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imperlini E, et al. Mass spectrometry-based metabolomic and proteomic strategies in organic acidemias. Biomed. Res. Int. 2016;2016:9210408. doi: 10.1155/2016/9210408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Auricchio R, et al. A phospholipid profile at 4 months predicts the onset of celiac disease in at-risk infants. Sci. Rep. 2019;9:14303. doi: 10.1038/s41598-019-50735-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruoppolo M, et al. Female and male human babies have distinct blood metabolomic patterns. Mol. Biosyst. 2015;11:2483–2492. doi: 10.1039/c5mb00297d. [DOI] [PubMed] [Google Scholar]
- 44.Scolamiero E, et al. Maternal vitamin B12 deficiency detected in expanded newborn screening. Clin. Biochem. 2014;47:312–317. doi: 10.1016/j.clinbiochem.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Scolamiero E, et al. Targeted metabolomics in the expanded newborn screening for inborn errors of metabolism. Mol. Biosyst. 2015;11:1525–1535. doi: 10.1039/c4mb00729h. [DOI] [PubMed] [Google Scholar]
- 46.Ruoppolo M, et al. Serum metabolomic profiles suggest influence of sex and oral contraceptive use. Am. J. Transl. Res. 2014;6:614–624. [PMC free article] [PubMed] [Google Scholar]
- 47.Ruoppolo M, et al. Targeted metabolomic profiling in rat tissues reveals sex differences. Sci. Rep. 2018;8:4663. doi: 10.1038/s41598-018-22869-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giacco A, et al. Exercise with food withdrawal at thermoneutrality impacts fuel use, the microbiome, AMPK phosphorylation, muscle fibers, and thyroid hormone levels in rats. Physiol. Rep. 2020;8:e14354. doi: 10.14814/phy2.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Pasquale V, et al. Targeted metabolomic analysis of a mucopolysaccharidosis IIIB mouse model reveals an imbalance of branched-chain amino acid and fatty acid metabolism. Int. J. Mol. Sci. 2020 doi: 10.3390/ijms21124211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caterino M, et al. Influence of sex on urinary organic acids: a cross-sectional study in children. Int. J. Mol. Sci. 2020 doi: 10.3390/ijms21020582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costanzo M, et al. Proteomics reveals that methylmalonyl-CoA mutase modulates cell architecture and increases susceptibility to stress. Int. J. Mol. Sci. 2020 doi: 10.3390/ijms21144998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.