Abstract
A combination of vaccination approaches will likely be necessary to fully control the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. Here, we show that modified vaccinia Ankara (MVA) vectors expressing membrane-anchored pre-fusion stabilized spike (MVA/S) but not secreted S1 induced strong neutralizing antibody responses against SARS-CoV-2 in mice. In macaques, the MVA/S vaccination induced strong neutralizing antibodies and CD8+ T cell responses, and conferred protection from SARS-CoV-2 infection and virus replication in the lungs as early as day 2 following intranasal and intratracheal challenge. Single-cell RNA sequencing analysis of lung cells on day 4 after infection revealed that MVA/S vaccination also protected macaques from infection-induced inflammation and B cell abnormalities and lowered induction of interferon-stimulated genes. These results demonstrate that MVA/S vaccination induces neutralizing antibodies and CD8+ T cells in the blood and lungs and is a potential vaccine candidate for SARS-CoV-2.
Keywords: Modified Vaccinia Ankara, Neutralizing antibody, CD8 T cells, Vaccine, Rhesus macaques, Protection, Spike, SARS-CoV-2, COVID-19
Graphical abstract
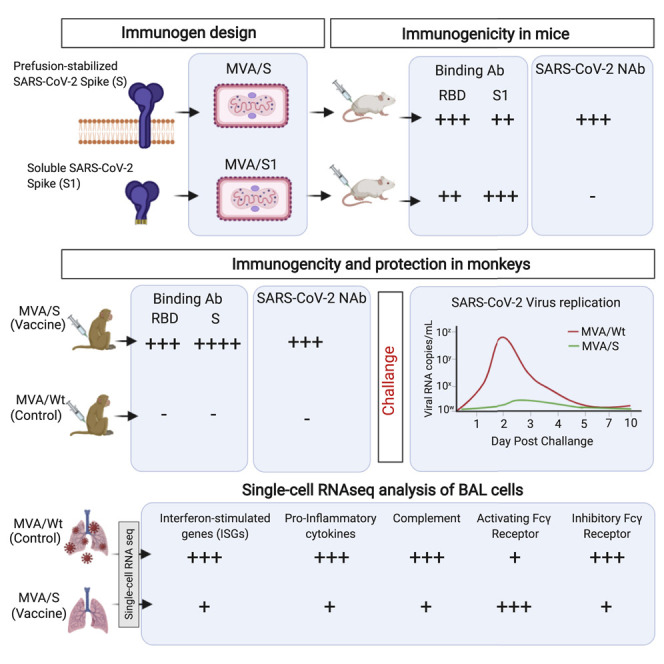
Modified vaccinia Ankara (MVA) vector-based vaccines are attractive because of their excellent safety and ability to induce long-lived humoral and cellular immunity in humans. Routhu et al. show that an MVA-based COVID-19 vaccine encoding prefusion-stabilized spike (MVA/S) induces strong neutralizing antibody and CD8+ T cell responses and protects macaques from SARS-CoV2 infection, immunopathology, and infection-induced B cell abnormalities in the lungs.
Introduction
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has emerged as a pandemic. As of January 21, 2021, SARS-CoV-2 has infected more than 95 million people, and over 2 million people have succumbed to coronavirus disease 2019 (COVID-19) disease. Thus, there is an urgent need for development of a vaccine that can rapidly induce immunity and prevent infection. Studies of closely related coronaviruses, such as SARS-associated coronavirus (SARS-CoV) and Middle East Respiratory Syndrome-related coronavirus (MERS-CoV), demonstrate that a strong neutralizing antibody response against the spike protein can effectively prevent infection (Chan et al., 2015; Haagmans et al., 2016; Xu et al., 2019; Yong et al., 2019). Building on these data, a number of vaccines are currently under development for SARS-CoV-2 using a variety of platforms, including mRNA, viral vectors, DNA, and proteins with different adjuvants (Brouwer et al., 2021; Corbett et al., 2020; Gao et al., 2020; Keech et al., 2020; Krammer, 2020; Laczkó et al., 2020; Mercado et al., 2020; Poland et al., 2020; Sahin et al., 2020; Smith et al., 2020; Tostanoski et al., 2020; van Doremalen et al., 2020; Walls et al., 2020; Yu et al., 2020). Phenomenal progress has already been made in a very short time. mRNA-based (Corbett et al., 2020; Laczkó et al., 2020; Sahin et al., 2020), adenoviral vector-based (Mercado et al., 2020; Tostanoski et al., 2020; van Doremalen et al., 2020), and spike trimer protein-based (Gao et al., 2020; Keech et al., 2020) vaccines are at the forefront and show induction of a strong neutralizing antibody response in macaques and humans. In addition, these vaccines confer protection from SARS-CoV-2 infection and replication in the lungs following intranasal and intratracheal challenge in macaques. Recent data from mRNA and chimpanzee adenovirus-based vaccines in human efficacy trials also show that these vaccines can protect humans from COVID-19 and have been approved for emergency use. Although remarkable progress has been made, many challenges still exist for SARS-CoV-2 vaccine development. These include lack of long-term safety and immunogenicity data for humans, poor induction of a CD8+ T cell response, lack of cross-reactive protective immunity against other human betacoronaviruses with pandemic potential, failure to provide protection against SARS-CoV-2 replication in the nasopharynx, and requirement of cold storage. A combination of vaccination approaches is needed to tackle these critical challenges. There is still a great need for development of SARS-CoV-2 vaccines using different platforms, especially ones that can synergize with current front runners to enhance the induction and durability of neutralizing antibodies and CD8 T cell responses with long-term safety.
Modified vaccinia Ankara (MVA) is a highly attenuated strain of vaccinia virus. The safety, immunogenicity, and protective capacity of replication-deficient MVA has been well established, and it is used widely to develop vaccines against infectious diseases and cancer in preclinical research and humans (Gilbert, 2013; Iyer and Amara, 2014). There are several advantages to MVA-based vaccines. (1) They are safe and well tolerated, including in HIV-infected individuals (Thompson et al., 2016). (2) They induce strong antibody responses after a single vaccination and can be boosted at least 10-fold with a second dose (Amara et al., 2002; Brault et al., 2017; Domi et al., 2018; Goepfert et al., 2011, 2014). (3) MVA vaccine-induced antibody responses in humans are durable, with little contraction over a 6-month time frame (Goepfert et al., 2014). (4) MVA can be delivered through multiple routes and can be used to generate a mucosal antibody response. (5) MVA can accommodate large inserts (>10 kb) that will allow expression of multiple antigens in a single vector. (6) MVA-vectored recombinants are stable and can be produced at high titer, facilitating vaccine manufacturing. (7) MVA vaccines can induce CD4 and CD8 T cell responses important for protection against some viral infections (Amara et al., 2002). (8) A lyophilized MVA vaccine can be stored at 37°C for 2 weeks and at 4°C for longer than a year (Zhang et al., 2007). (9) MVA-based vaccines have been shown to protect against SARS-CoV, MERS-CoV, Zika virus, and Ebola virus in animal models (Bisht et al., 2004; Brault et al., 2017; Domi et al., 2018; Haagmans et al., 2016). In addition, MVA-based vaccines can serve as excellent boosting agents for DNA-based and other viral vector-based vaccines, including chimpanzee adenovirus- and Ad26-based vaccines to boost cellular and humoral immunity (Amara et al., 2001; Barouch et al., 2012; Ewer et al., 2016).
In this study, we developed two MVA-based vaccines that express a membrane-anchored full-length spike protein (MVA/S) stabilized in a prefusion state or the soluble secreted trimeric S1 of the spike (MVA/S1). Both immunogens contained the receptor-binding domain (RBD), a known target of antibody-mediated neutralization in SARS-CoV-2-infected individuals (Suthar et al., 2020). MVA/S also incorporated two mutations that have been shown to maintain the spike protein in a prefusion confirmation (Pallesen et al., 2017; Wrapp et al., 2020). Using a mouse model, we selected the MVA/S vaccine based on its ability to induce a neutralizing antibody response. Vaccination of rhesus macaques followed by SARS-CoV-2 challenge demonstrated that the MVA/S vaccine induces neutralizing antibodies and CD8 T cells, protects from SARS-CoV-2 infection and replication in the lungs, and thus is a potential vaccine candidate for SARS-CoV-2.
Results
MVA vaccines express full-length stabilized spike and trimeric soluble S1 proteins
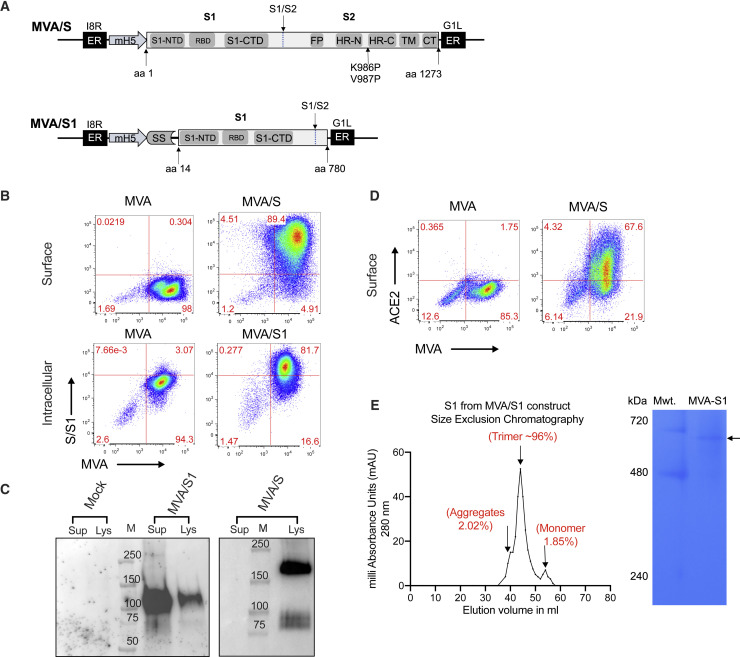
To develop the MVA recombinants, we synthesized the full-length spike gene (amino acids 1–1,273) with stabilizing mutations (K986P and V987P) or the S1 region with a small portion of the S2 region (amino acids 14–780). To promote active secretion of S1, we replaced amino acids 1–14 of the spike sequence with the signal sequence from Granulocyte macrophage-colony stimulating factor (GM-CSF) (Figure 1 A). Both sequences were optimized for MVA codon use, corrected for poxvirus transcription termination sequences, and cloned into the pLW73 vector, which allowed us to insert the recombinant sequences under the mH5 promoter into the essential region of MVA. The recombinants were selected as described previously (Chea et al., 2019) and characterized for protein expression by flow cytometry and western blotting. As expected, MVA/S expressed high amounts (based on mean fluorescence intensity) of spike on the cell surface (Figure 1B), and the expressed protein had a molecular mass of about 180 kDa (Figure 1C). Similarly, MVA/S1 was expressed intracellularly (Figure 1B), and a molecular mass of about 114 kDa was also secreted into the supernatants (Figure 1C). The spike protein expressed by MVA/S on the surface seemed to be folded correctly based on strong binding to ACE2 (Figure 1D). The S1 protein was found to form trimers based on the gel filtration profile and native PAGE analysis (Figure 1E).
Figure 1.
Construction and characterization of MVA/S and MVA/S1 recombinants
(A) Schematic representation of MVA/S and MVA/S1. MVA-SARS-CoV-2-S encodes for the SARS-CoV-2 S protein with the two indicated proline mutations for prefusion stabilization. MVA/S1 contains a GM-CSF signal sequence (SS) and replaces the first 14 amino acids of the S protein that contains the natural SS. Recombinant inserts were cloned in the essential region between the 18R and G1L genes under the mH5 promoter.
(B) Representative flow plots showing surface expression of membrane-anchored spike (MVA/S) and intracellular expression of secreted S1 (MVA/S1).
(C) Western blot analysis of expressed proteins in supernatants and lysates of MVA/S- and MVA/S1-infected cells.
(D) Binding of hACE2 to MVA/S-expressing cells.
(E) Size-exclusion chromatography (left) and blue native PAGE (right) analysis of S1 protein expressed by MVA/S1.
The experiments related to (B)–(E) were repeated twice, and representative data are shown.
MVA/S, but not MVA/S1, induces a strong neutralizing antibody response in mice
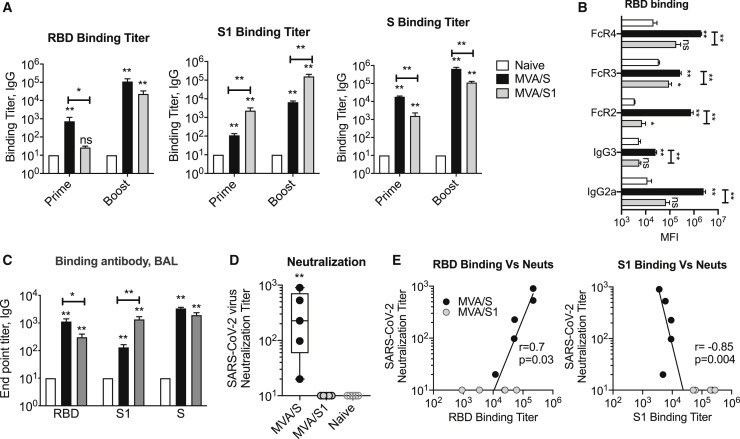
We immunized BALB/c mice with MVA/S or MVA/S1 at weeks 0 and 3 and measured binding antibody responses to total and different parts of spike (i.e., RBD, S1, and S) using ELISA 2 weeks after prime and boost (Figure 2 ). Although both vaccines induced a strong binding antibody response to S, they differentially targeted binding to RBD and S1 (Figure 2A; Figure S1A). The MVA/S sera showed higher binding to the RBD, whereas MVA/S1 sera showed higher binding to S1. This was interesting, considering that S1 protein includes the RBD region, and suggests that antibody binding in sera from MVA/S1-vaccinated mice may be targeting regions outside of the RBD. We further confirmed this differential targeting of antibody using a Luminex assay (Figure S1B). Analysis of immunoglobulin G (IgG) subclass and Fc-gamma receptor (FcγR) binding of an RBD-specific antibody showed strong IgG2a response and binding to all three FcγRs tested, with the strongest binding to FcγR2 and FcγR4 in the MVA/S group (Figure 2B). In contrast, poor binding of the RBD-specific antibody was observed in general with MVA/S1 sera. However, the S1-specific antibody showed similar results in both groups (Figure S2C). In addition, we also observed induction of spike-specific IgG responses in bronchoalveolar lavage (BAL) fluid (Figure 2C). Similar to serum, the RBD-specific IgG responses were higher in the MVA/S group compared with the MVA/S1 group. These results demonstrated differential targeting of a spike-specific antibody with a T helper 1 (Th1) profile induced by the MVA/S and MVA/S1 vaccines.
Figure 2.
Humoral responses in systemic and mucosal compartments of MVA/S- and MVA/S1-immunized mice
(A–E) Six-week-old female BALB/c mice (n = 5 per group) were immunized via the intramuscular route with MVA/S or MVA/S1 at weeks 0 and 3. The mouse immunization study was repeated twice, and representative data are shown.
(A) Endpoint IgG titers against the SARS-CoV-2 RBD, S1, and S, measured in serum collected at week 2 after prime and week 2 after boost immunization. Each sample was analyzed in duplicates.
(B) SARS-CoV-2 RBD-binding IgG subclass and soluble Fc receptor analysis in serum (week 3 after boost), performed using a Luminex assay. Data are from one experiment.
(C) Lung SARS-CoV-2 RBD-, S1-, and S-specific IgG responses measured in BAL (bronchoalveolar lavage) fluid collected 3 weeks after boost (at euthanasia) using ELISA. Each sample was analyzed in duplicates.
(D) Neutralization titer against the live mNeonGreen SARS-CoV-2 virus was performed in serum collected at week 2 after boost immunization. Each sample was analyzed in duplicates and repeated in two independent times.
(E) Correlations between neutralization titer and ELISA binding titers of RBD and S1 proteins.
Bars and columns show arithmetic mean values for each group ± SEM. Mann-Whitney test; ∗p < 0.05, and ∗∗p < 0.01. Spearman rank test was used to perform the correlation analysis. See Figure S1 for details.
We next evaluated the neutralization capacity of serum collected 2 weeks after boost from MVA-vaccinated mice using mNeonGreen SARS-CoV-2 virus and defined 50% reduction in focus reduction neutralization titer (FRNT) (Figure 2D). We observed a strong neutralizing antibody response in sera from mice vaccinated with MVA/S that ranged from 20–900 with a median of 200 (Figure 2D; Figure S1D). In contrast, we did not observe any detectable neutralization activity in sera from mice immunized with MVA/S1. This was despite the fact that MVA/S1 mice showed a higher binding antibody response to S1 and S proteins. The neutralization titer correlated directly with the RBD binding titer and negatively with the S1 binding titer (Figure 2E). In particular, it correlated with the RBD-specific IgG2a binding titer (Figure S1E). These results demonstrate that MVA/S immunogen can induce a strong neutralizing antibody response against SARS-CoV-2 and could be a potential vaccine for SARS-CoV-2. Importantly, they also reveal that MVA/S1 is not a good vaccine because it failed to induce a neutralizing antibody response.
To further understand the failure of the MVA/S1 vaccine to induce strong RBD binding antibodies and neutralizing antibodies, we purified the S1 trimer protein expressed by the MVA/S1 vaccine and determined its ability to bind to human ACE2 using biolayer interferometry (BLI) (Figure S2). We found that the affinity of S1 for ACE2 decreased by 5-fold when the protein was incubated at 25°C for 60 min, and this was not the case for the RBD. These data indicate the unstable nature of the RBD in S1 because the association with ACE2 protein is decreased upon prolonged incubation at room temperature.
The MVA/S and MVA/S1 vaccines induce strong T cell responses in the spleen and lungs of mice
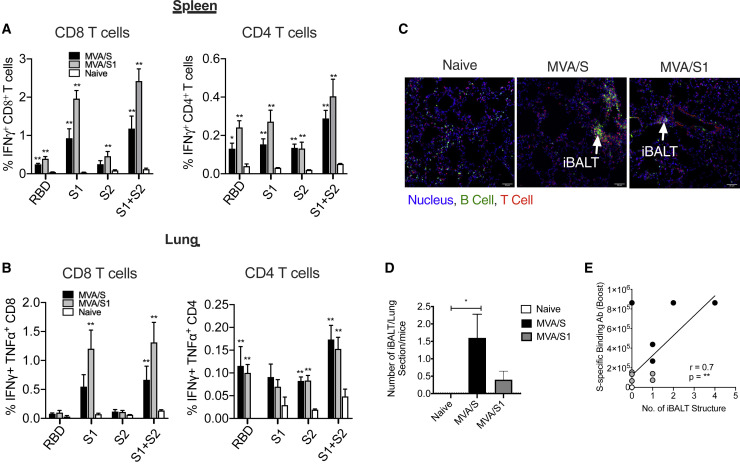
To determine T cell responses, we stimulated lymphocytes from the spleens and lungs of vaccinated mice with overlapping peptide pools specific to the RBD, S1, and S2 regions of the spike protein and measured the frequency of interferon γ (IFNγ)+ CD4 and CD8 T cells using the intracellular cytokine staining (ICS) assay. Both vaccines induced comparable IFNγ+ CD8 and CD4 T cell responses that primarily targeted the S1 region (including the RBD) in the spleen (Figure 3 A) and lungs (Figure 3B). We next studied whether vaccination induced formation of inducible bronchus-associated lymphoid tissue (iBALT), which has been shown to provide protection against influenza infection in the absence of peripheral lymphoid organs in mice (Moyron-Quiroz et al., 2004; Woodland and Randall, 2004) using immunohistochemistry 3 weeks after MVA boost by staining for B and T cells (Figures 3C and 3D). As expected, naive mice showed very little or no iBALT; however, MVA/S-vaccinated mice showed significant induction of iBALT, indicating generation of local lymphoid tissue. Formation of iBALT was also evident in the MVA/S1 group, but to a lesser extent. The relatively lower iBALT response in the MVA/S1 group could be due to the overall lower spike-specific antibody response induced in MVA/S1 mice compared with MVA/S mice. Although we do not know the longevity or persistence of these BALTs, they are expected to help with rapid expansion of immunity in the lungs following exposure to SARS-CoV-2 (Moyron-Quiroz et al., 2004; Wiley et al., 2009).
Figure 3.
The MVA/S and MVA/S1 vaccines induce strong T cell responses in the spleen and lungs of mice
(A and B) IFNγ+, CD8+, and CD4+ T cells in the spleen (A) and lungs (B) of MVA/S- and MVA/S1-immunized animals, analyzed 1 week after boost immunization, after re-stimulation with the indicated peptide pools. The mouse immunization study was repeated twice (n = 5 per group), except for the data presented in (C) and (D), and representative data are shown.
(C) Frozen lung sections from vaccinated mice were stained to analyze formation of iBALT aggregates to visualize the B cell- and T cell-forming follicle-like structure (iBALT) induced by MVA/S and MVA/S1 vaccination. The arrows in the immunofluorescence images indicate iBALT structures.
(D) The total number of iBALT-like structures, visualized in each section per mouse, was quantified and compared between the groups.
(E) Correlation between the total number of iBALT-like structures and ELISA binding antibody with S protein in serum.
Bars and columns show arithmetic mean values for each group ± SEM; Mann-Whitney test; ∗p < 0.05, and ∗∗p < 0.01. Spearman rank test was used to perform the correlation analysis.
The MVA/S vaccine induces a strong neutralizing antibody response with effector functions and CD8 T cell response in rhesus macaques
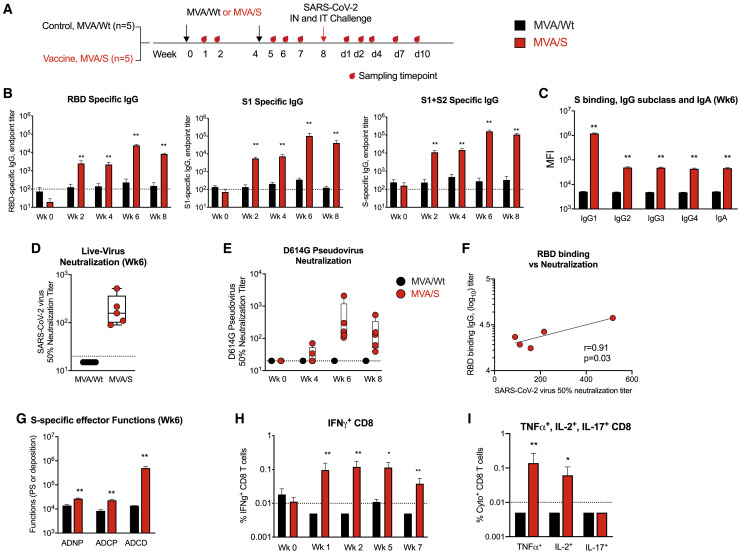
To test the immunogenicity and protective ability of the MVA/S vaccine, we immunized rhesus macaques (n = 5/group) with the MVA/S vaccine or MVA/wild type (WT) vaccine delivered intramuscularly at weeks 0 and 4 with a dose of 1 × 108 plaque-forming units (PFUs) (Figure 4 A). At week 8 (4 weeks after the boost), animals were challenged intranasally and intratracheally with live SARS-CoV-2 virus. Consistent with the data for mice, two MVA/S vaccinations induced a strong binding antibody against RBD (geometric mean titer of 2.4 × 104) and total S (geometric mean titer of 1.5 × 105) that persisted until 4 weeks after the boost (Figure 4B). IgG subclass analysis revealed the majority of the response as IgG1, indicating a Th1-dominant response (Figure 4C). We also detected a low titer of spike-specific IgA (Figure 4C). The MVA/S vaccine-induced antibody showed strong neutralization of live virus 2 weeks after boost, with a geometric mean titer of 177 (range, 90–516) (Figure 4D). A similar neutralization titer was also observed in a spike (having the D614G mutation) pseudotyped virus neutralization assay (Figure 4E). A low titer of neutralizing antibodies was detectable in 2 of the 5 animals after prime (Figure 4D). Antibody binding to the RBD correlated directly with live virus neutralization (Figure 4F). In addition to the neutralizing activity, the vaccine-induced sera showed strong antibody-dependent complement deposition (ADCD) activity and low antibody-dependent cellular phagocytosis (ADCP) and antibody-dependent neutrophil phagocytosis (ADNP) activity (Figure 4G).
Figure 4.
The MVA/S vaccine induces neutralizing antibodies with effector functions and CD8 T cell responses in rhesus macaques
10 macaques were divided into two groups (n = 5 per group). One group received MVA/Wt, and the other received MVA/S. The study was performed once.
(A) Schematic showing the timeline of vaccination, SARS-CoV-2 challenge, and sample collection.
(B) Endpoint IgG titers against the SARS-CoV-2 RBD, S1, and S measured in serum. Each sample was analyzed in duplicates.
(C) S protein-specific IgG subclass and IgA in serum. Each sample was analyzed in duplicates.
(D–E) 50% neutralization titer against mNeonGreen SARS-CoV-2 live virus at week 6 (D) and VRC7480.D614G pseudovirus at weeks 0, 4, 6, and 8 (E). Each sample was analyzed in duplicates, and the assay was repeated two independent times.
(F) Correlations between live virus neutralization titer and RBD-binding titer.
(G) S protein-specific antibody effector functions in serum.
(H) IFNγ+ CD8+ T cells specific to total S protein in blood.
(I) TNF-α+, IL-2+, and IL-17+ CD8+ T cells specific to total S protein in blood at week 2 after boost.
Bars and columns show mean responses in each group ± SEM. Mann-Whitney test was used to compare groups: ∗p < 0.05, and ∗∗p < 0.01. Spearman rank test was used to perform the correlation analysis. Dotted lines reflect the limit of detection.
The MVA/S vaccine also generated a strong spike-specific IFNγ+ CD8 T cell response that was evident as early as 1 week after the priming immunization (Figure 4H). The frequency of CD8 T cell responses was not boosted further following the second MVA/S immunization. Vaccine-induced CD8 T cells were also positive for tumor necrosis factor alpha (TNF-α) and interleukin-2 (IL-2) and negative for IL-17 (Figure 4I). The MVA/S vaccine induced very low frequencies of IFNγ+ CD4 T cells (data not shown). These data demonstrate that MVA/S vaccinations induce a poly-functional CD8 T cell response capable of producing IFNγ, IL-2, and TNF-α in macaques.
MVA/S vaccination provides protection from SARS-CoV2 infection and replication in the lungs
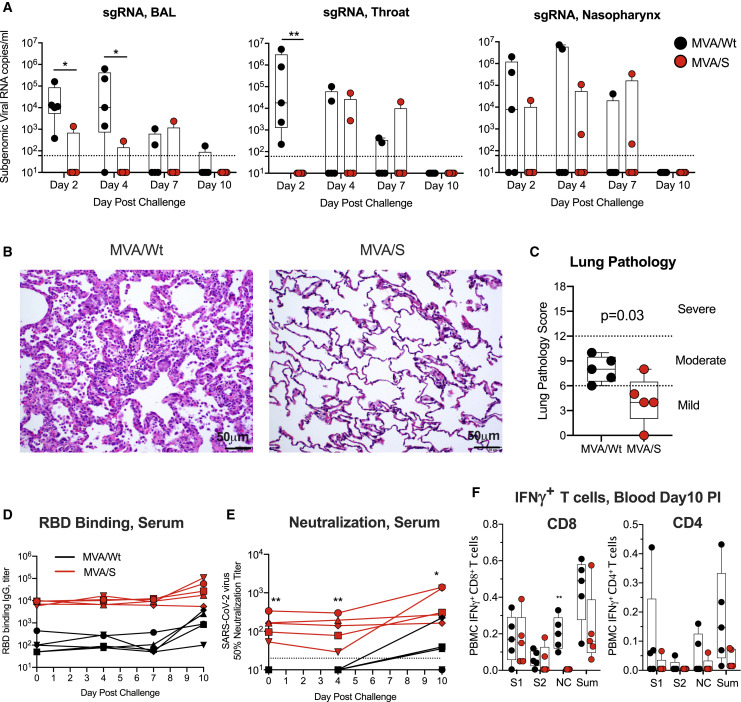
Following intranasal and intratracheal challenge, we monitored for subgenomic viral RNA to distinguish the replicating virus from the inoculum in the lungs (BAL fluid), nasopharynx, and throat on days 2, 4, 7, and 10 (day of euthanasia) (Figure 5 ). On day 2, all 5 controls (MVA/Wt vaccinated) tested positive for virus in BAL fluid and the throat, and 3 of 5 tested positive in the nasopharynx, indicating that all animals were infected productively (Figure 5A). The viral RNA titer was variable and ranged from 2 × 102–7 × 106 copies/mL. These titers were maintained in the BAL fluid until day 4 and then declined quickly with time in all animals (except in BAL fluid from one animal) and were below our detection limit by day 10. In contrast, MVA/S-vaccinated animals rapidly controlled SARS-CoV-2 replication in the lungs on day 2 (p < 0.05) and day 4 (p < 0.05) compared with controls (Figure 5A), with 4 of the 5 vaccinated animals being negative in BAL fluid. However, in the throat, all vaccinated animals tested negative on day 2 (p < 0.01), but a low titer of virus replication was evident in one or two vaccinated animals on days 4 and 7. Similarly, in the nasopharynx, one or two animals showed virus replication on days 2, 4, and 7, and virus replication was not significantly different between controls and vaccinated animals at all time points. By day 10, all control and vaccinated animals were negative in all 3 compartments. These results demonstrate that MVA/S vaccination provides protection from SARS-CoV2 infection and replication in the lower respiratory tract.
Figure 5.
The MVA/S vaccine protects from SARS-CoV-2 infection and replication and reduces lung pathology in rhesus macaques
Following vaccination, all macaques were challenged with SARS-CoV-2 at week 8 by intratracheal (IT) and intranasal (IN) route. Virus replication, lung pathology, binding and neutralizing antibody titers, and T cell responses were measured.
(A) SARS-CoV-2 subgenomic viral RNA in the lung (BAL fluid), throat, and nasopharynx on days 2, 4, 7, and 10 (day of euthanasia) after infection.
(B and C) Lung pathology on day 10 after infection. Shown is hematoxylin and eosin (H&E) staining of lung sections to analyze tissue structure and cell infiltration (B) and lung pathology score (C). See Table S1 for details.
(D and E) SARS-CoV-2 RBD-specific serum IgG responses (D) and live SARS-CoV-2 neutralization (E) in serum following challenge.
(F) IFNγ+ CD8+ and CD4+ T cells in blood on day 10 after infection after re-stimulation with a peptide pool specific to the indicated protein. S1, S1 region of spike; S2, S2 region of spike; NC, nucleocapsid; Sum, total response (S1+S2+NC).
Bars and columns show mean responses in each group ± SEM; Mann-Whitney test: ∗p < 0.05, and ∗∗p < 0.01. Dotted lines reflect the limit of detection. See also Figure S3 and Table S1 for details.
Consistent with early virus control in the lungs, vaccinated animals also showed lower lung pathology compared with control animals (Figure 5B). To assess lung pathology, we analyzed multiple regions of upper, middle, and lower lung lobes at euthanasia. Lung pathology analyses and scoring (considering severity and the number of affected lobes) were performed by two independent pathologists in a blinded fashion. The total pathology score was lower in the vaccinated group compared with the control group. Specifically, MVA/S-vaccinated animals showed decreased type 2 pneumocyte hyperplasia, peribronchiolar hyperplasia, alveolar septal thickening, and inflammatory cell infiltration (Figures 5B, Figure S3). Overall, these data support a beneficial role of MVA/S vaccination in reducing lung pathology. We also performed histopathological examination of various other tissue samples, including nasal turbinates, trachea, tonsils, hilar lymph nodes, spleen, heart, brain, gastrointestinal tract (stomach, jejunum, ileum, colon, and rectum), and testes. We did not observe significant histological lesions in upper respiratory tract tissues (nasal turbinates and trachea) and other examined tissues in control and vaccinated animals.
To understand the anamnestic expansion of antibodies and T cells after infection, we measured binding and neutralizing antibodies in serum on days 0, 4, 7, and 10 and T cells in the blood on day 10. Some of the control animals developed a low titer of binding and virus-neutralizing activity by day 10 that was markedly lower compared with the titers in vaccinated animals (Figures 5D and 5E). The neutralizing antibody titer in vaccinated animals was maintained in 2 animals and increased by 4-fold in 2 animals and by 25-fold in 1 animal. The control animals also showed IFNγ+ CD8 and CD4 T cells in the blood that were largely specific to S1 and nucleocapsid (NC) (Figure 5F). However, the vaccinated animals mainly showed IFNγ+ CD8 T cells against S1 but not NC. The S1-specific response in vaccinated animals after infection was likely due to the persisting vaccine-induced CD8 T cells. However, the lack of an NC-specific CD8 T cell response suggests that these animals were not exposed to a significant amount of the NC protein made by the replicating virus after infection. Thus, post-infection immune responses pointed to a less systemic spread of virus replication, at least in some of the vaccinated animals.
MVA/S vaccination protects from infection-induced immune abnormalities in the lungs
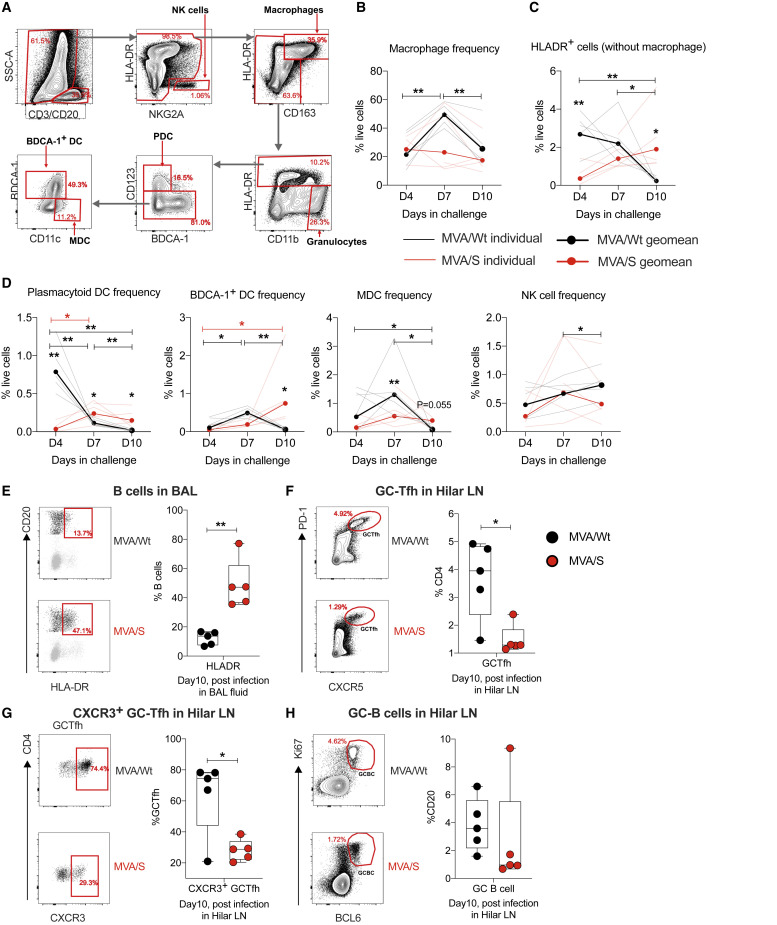
To further understand the influence of infection and vaccine protection on innate and adaptive immunity in the lungs early after infection, we monitored various innate cells and B cells in BAL fluid and Germinal Center - T follicular helper (GC-Tfh) and GC-B cell responses in lung draining hilar lymph nodes (LNs) on day 10 after infection (Figure 6 A). Longitudinal analysis of innate cells on days 4, 7, and 10 after infection in control (MVA/Wt) animals revealed a significant increase in the frequency of macrophages on day 7, followed by decline on day 10 (Figure 6B). In contrast to macrophages, the frequency of total Human Leukocyte Antigen-DR+ (HLA-DR+) dendritic cells (DCs) and plasmacytoid DCs (pDCs) on day 7 were higher in controls compared with MVA/S-vaccinated animals and decreased over time until day 10 (Figures 6C and 6D). The frequency of BDCA-1+ DCs and myeloid-derived DCs (MDCs) increased by day 7 and declined by day 10, and the frequency of natural killer (NK) cells increased gradually until day 10 (Figure 6D). However, the frequency of all of these cells stayed relatively stable in vaccinated (MVA/S) animals on days 4–10, except BDCA-1+ DCs, which showed a small increase on day 10 compared with day 4. On day 10, the control animals showed a lower frequency of HLA-DR+ DCs compared with vaccinated animals that was also reflected in the frequency of pDCs and BDCA-1+ DCs. Longitudinal analysis of innate cells revealed expansion and contraction of all studied subsets, but with different kinetics in controls but not in vaccinated animals, presumably because of high virus replication in controls. These data also suggest that there was a potential loss of some of the DCs in controls by day 10. It is difficult to determine this in the absence of data on pre-infection frequencies of innate cells in BAL fluid. In addition, B cells in the controls showed lower expression of HLA-DR, suggesting an impaired B cell response (Figure 6E; Titanji et al., 2010). Furthermore, the frequency of total (Figure 6F) and CXCR5+ GC-Tfh (Figure 6G) and GC-B cells (Figure 6H) in hilar LNs was higher in control animals compared with vaccinated animals, suggesting a higher antigen load and proinflammatory environment in the former. These results demonstrate distinct innate and adaptive immune profiles in the lungs between control and MVA/S-vaccinated animals that point to a lower antigen load and inflammation in vaccinated animals early after infection.
Figure 6.
Innate and adaptive immune responses after SARS-CoV-2 challenge in the lungs of rhesus macaques
Following SARS-CoV-2 infection, BAL fluid was collected on days 4, 7, and 10, and various innate cells were analyzed. On day 10 (at euthanasia), B cells were analyzed in BAL fluid, and GC-Tfh and GC-B cells were analyzed in hilar LNs. n =5/group, except for the MVA/S group on day 4, where data for one animal were not available. These analyses were performed only once because the macaque study was done only once.
(A) Gating strategy to identify innate cells in BAL fluid. Live cells were selected using a live/dead marker, and CD3+ and CD20+ cells were excluded. Then different innate cells were defined using the following combinations of markers: macrophages, HLA-DR+ CD163+; pDCs, HLA-DR+CD163−CD123+ BDCA1−; BDCA1+ DCs, HLA-DR+ CD163− CD123− BDCA1+; MDCs, HLA-DR+CD163−CD123−CD11C+; NK cells, HLA-DR− NKG2A+.
(B–D) Summary of changes in innate cell frequencies after infection.
(E) HLA-DR expression on B cells (CD20+) in BAL fluid on day 10 after infection. Left: representative flow plots. Right: summary of data for all animals.
(F–H) Total GC-Tfh (F), CXCR3+ GC-Tfh (G), and GC-B cells (H) on day10 after challenge in hilar LNs. Representative flow plots are shown on the right, and the data for all animals are shown on the left. GC-Tfh cells were CD3+CD4+CD8-CXCR5highPD-1high, and GC-B cells were CD3−CD20+BCL6+Ki67+.
NK, natural killer; pDC, plasmacytoid DC; MDC, myeloid-derived DC; LN, lymph node. Statistical differences between the groups and within the groups were analyzed using unpaired parametric t test and paired parametric t test, respectively. ∗p < 0.05, and ∗∗p < 0.005.
MVA/S vaccination reduces transcripts associated with inflammation and hyperimmune activation in lung macrophages
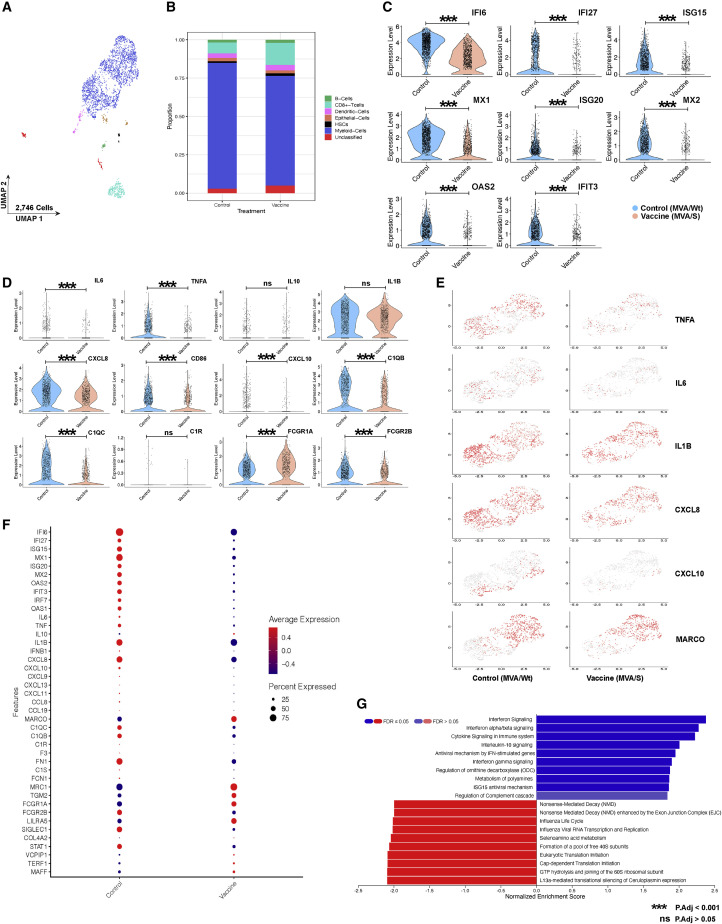
SARS-CoV-2 infection of non-human primates (NHPs), in most cases, does not recapitulate the full spectrum of clinical symptoms of severe COVID-19, with animals typically exhibiting a more moderate phenotype (Hartman et al., 2020; Munster et al., 2020). Recently, however, we and others have observed that SARS-CoV-2 infection of rhesus macaques (Aid et al., 2020; Hoang et al., 2021) and African green monkeys (AGMs) (Speranza et al., 2020). elicits many of the immunopathological events in the airway (elevated inflammatory cytokines and chemokines and recruitment of monocytes and neutrophils) that have been reported in the airways of individuals exhibiting severe COVID-19 symptoms (Liao et al., 2020). To assess the effect of our vaccine on these inflammatory sequelae in the airway, we performed single-cell RNA sequencing (RNA-seq) on the cellular fraction recovered from BAL fluid on day 4 after infection (Figure 7 ). The vast majority of cells identified in the BAL fluid were CD68+ myeloid cells (82% in controls and 71% in vaccinated animals), followed by CD8+ T cells (7% in controls and 14% in vaccinated animals) and minor populations (<5%) consisting of B cells, DCs, and epithelial cells (Figures 7A and 7B; Figure S4). Consistent with concurrent studies by us and others using NHPs, inoculation of NHPs with SARS-CoV-2 led to widespread induction of IFN-stimulated genes (ISGs) and related pathways as early as day 4 after infection, prevalently in the myeloid population (Aid et al., 2020; Hoang et al., 2021; Speranza et al., 2020; Figures 7C and 7G). However, we observed that expression of transcripts associated with ISGs was attenuated in vaccinated animals at the individual gene level and pathway level (Figures 7C and 7G). In human and NHP studies, SARS-CoV-2 has been observed to induce strong expression of inflammatory cytokines and chemokines in myeloid-origin cells in the BAL fluid and lungs (Hoang et al., 2021; Liao et al., 2020; Speranza et al., 2020). In the current study, control animals largely recapitulated these findings; we observed high expression of transcripts associated with IL-6, TNF-α, and IL-1β production (Figures 7D–7F). We also noted widespread expression of CXCL8, CXCL9, CXCL10, CXCL11, CCL8, and CCL19 (Figures 7D–7F). In vaccinated animals, expression of IL-6, TNF-α, IL-1b, CXCL8, and CXCL10 was reduced compared with controls (Figures 7D–7F). Recent studies in AGMs have reported that SARS-COV-2 infection elicits a population shift within the myeloid fraction in which interstitial macrophages/infiltrating monocytes (identified by lack of the macrophage receptor with collagenous structure (MARCO) cell marker) become predominant over MARCO+ resident alveolar macrophage (Speranza et al., 2020). Here we observed that vaccinated animals trended toward a higher proportion of MARCO+ cells relative to unvaccinated controls (70% versus 54%), indicating reduced recruitment of monocytes/interstitial macrophages (Figures 7E and 7F). In addition, expression of C1QB and C1QC, components of the C1q complement receptor, was also higher in control macrophages (Figures 7D and 7F). Furthermore, the transcripts associated with activating FcγR1A (required for the effector functions of IgG1 and IgG3) was lower and inhibitory FcγR2B was higher in controls (Figure 7D). These data demonstrate that MVA/S vaccination is able to reduce inflammatory events in the airway, including reduced infiltration of activated monocytes and lower production of inflammatory cytokines/chemokines. Additionally, our observation of attenuated expression of ISGs in the cellular fraction of BAL fluid of vaccinated animals provides orthogonal validation of the ability of MVA/S vaccination to reduce airway SARS-CoV-2 virus replication.
Figure 7.
MVA/S vaccination against SARS-CoV-2 protects from lower airway inflammation in BAL fluid on day 4
Single-cell suspensions from BAL fluid on day 4 after SARS-CoV-2 infection were collected from MVA/S-vaccinated (n = 4) and MVA/Wt control (n = 5) macaques and subjected to single-cell RNA seq. These analyses were performed only once because the macaque study was done only once.
A) UMAP showing major cell types in BAL fluid samples (n = 9; vaccinated, n = 4; control, n = 5).
(B) Proportions of cells clustering into identifiable clusters of B cells, CD8+ T cells, DCs, epithelial cells, hematopoietic stem cells, and myeloid cells and unclassified clusters.
(C and D) Violin plots showing expression of IFN-stimulated genes (ISGs) (C) and inflammatory mediators (D).
(E) Feature plots of reclustered myeloid cells of select inflammatory genes in MVA/S-vaccinated (right) and MVA/Wt control animals (left).
(F) Dot plots of selected genes, including inflammatory genes and ISGs. For each gene, the percentage of cells that expressed the transcript and the average expression per cell are shown.
(G) Gene set enrichment analyses of gene sets with higher expression in MVA/S-vaccinated versus MVA/Wt control animals. ∗p-value adjusted (p.adj) < 0.05. The p values were calculated using the differential expression method Model-based Analysis of Single-cell Transcriptomics (MAST), which uses a hurdle model to calculate the test statistics. See also Figures S4–S6 for details.
Discussion
There is an urgent global need for development of a safe and effective COVID-19 vaccine. Humoral and cell-mediated immune responses in systemic and mucosal compartments are crucial for preventing infection and counteracting virus replication. We focused our efforts on development of MVA-based vaccines for COVID-19 based on our nearly 20 years of experience with development of MVA-based vaccines for HIV (Chea and Amara, 2017; Robinson and Amara, 2005), which has demonstrated that MVA is safe in people (Goepfert et al., 2014), including HIV-infected individuals (Thompson et al., 2016), and induces strong humoral and cellular immunity that is long lasting (Goepfert et al., 2014; Nigam et al., 2007). Our results showed that two doses of recombinant MVA (rMVA) expressing the membrane-anchored prefusion stabilized full-spike protein can induce a strong neutralizing antibody against SARS-CoV-2 and the highly infectious D614G variant (Korber et al., 2020) and a CD8 T cell response and provides protection from SARS-CoV-2 replication in the lungs (lower respiratory tract). In addition, vaccination blunted virus replication very early on day 2 in the throat. Notably, hematoxylin and eosin staining analysis in individual vaccinated (MVA/S) animals showed that MVA/S vaccination retained healthy lung features, like thin alveolar septa, perivascular cuffing, peribronchiolar hyperplasia, and no inflammatory cells (except in one animal, few inflammatory cells were observed) in alveoli compared with control (MVA/Wt) animals. The effects of vaccination on protection in the nasopharynx was inconclusive because of the large variation of viral load in the control group and relatively small group sizes. Our results also showed that vaccination can provide protection from inflammation, Th1-biased hyperimmune activation, and immune dysfunction in the lung very early after infection.
We found that MVA expressing soluble trimeric S1 protein does not induce a neutralizing antibody response despite containing the RBD and inducing strong S1-binding antibodies. This suggests that the specificity of the antibody response induced by the S1 immunogen may be distinct from that induced by the S immunogen. This was also suggested by the distinct RBD- and S1-specific humoral immune profiles in the MVA/S and MVA/S1 groups, where the MVA/S immunization yielded a stronger RBD-specific IgG response, whereas MVA/S1 induced a stronger S1-specifc response. This was unexpected, given the fact that RBD is part of S1. It will be important to understand the mechanisms that lead to loss of neutralization when MVA/S1 is used as an immunogen. It will also be important to study whether the antibody response induced by the MVA/S1 vaccine possess any kind of antibody-dependent enhancement of infection activity. Our preliminary data in this direction point to the inability of the S1 protein to retain binding to ACE2 during prolonged incubation at room temperature. This could have contributed to induction of strong binding antibodies to regions in S1 other than the RBD following immunization. Further studies are needed to understand the binding specificity of antibodies induced by these two vaccines.
Our study demonstrated that vaccination can protect from infection-induced inflammation, Th1-biased hyperimmune activation, and humoral immune dysfunction in the lungs at the single-cell level when the virus is still actively replicating very early (day 4) after infection. Although a recent COVID-19 vaccine study demonstrated that vaccination can reduce inflammation in the lung by measuring selected cytokines, we were able to show these findings at the single-cell level in lung-resident macrophages using cutting-edge technologies. Single-cell RNA-seq allowed us study several hundred genes, including ISGs and immune function-associated genes, and specific cell subsets that contribute to inflammation and altered expression. SARS-CoV-2 infection, despite being largely an acute infection in this model, can cause significant inflammation, Th1-biased hyperimmune activation, and immune dysfunction, such as loss of HLA-DR expression on B cells and lower expression of activating (FcγR1A) and higher expression of inhibitory (FcγR2B) FcγRs, which can potentially lead to diminished effector functions of IgG1 and IgG3. These results are important because they demonstrate that systemic vaccination can provide protection from infection-induced immune abnormalities in the lungs very early (day 4) after infection. Although our preliminary analyses made important findings, future analyses will dig deeper into identification of individual clusters of cell populations within myeloid cells and see how they differ between control and vaccinated animals after infection.
It will be good to compare the immunogenicity and protective ability of the MVA/S vaccine with those of other vaccines that are based on mRNA, adenoviruses (Ad) (chimpanzee or Ad26), and proteins that have already been approved or are currently being tested in human trials (Brouwer et al., 2021; Corbett et al., 2020; Gao et al., 2020; Keech et al., 2020; Krammer, 2020; Laczkó et al., 2020; Mercado et al., 2020; Poland et al., 2020; Sahin et al., 2020; Smith et al., 2020; Tostanoski et al., 2020; van Doremalen et al., 2020; Walls et al., 2020; Yu et al., 2020). However, it is difficult to do so because different studies used different neutralization assays that can provide different results. In addition, some studies reported a 50% neutralization titer and others 80%–90%. Similarly, for protection studies, different stocks and doses of viruses and species of macaques are used. Because of these limitations, we refrain from making any comparisons between studies. However, it is clear that some of the protein-based vaccines with potent adjuvants (Brouwer et al., 2021; Walls et al., 2020) have induced much stronger neutralizing antibody responses compared with viral vector-based vaccines, including our MVA/S vaccine. In contrast to viral vector-based vaccines, protein-based vaccines induce a poor CD8 T cell response. In addition, it is important to compare the durability of these responses between different vaccines. Acknowledging the different conditions of viral challenges being used, the protection we observed in this study is quite comparable with most of the vaccines that have already been advanced into the clinic.
In conclusion, these results demonstrate that MVA/S vaccination can provide protection from intranasal and intratracheal SARS-CoV-2 challenge and support further development of this vaccine to test immunogenicity and efficacy in humans. They also suggest that the MVA/S vaccines can serve as an excellent boosting agent for COVID-19 vaccines based on mRNA, DNA, and chimpanzee adenovirus and Ad26 to boost cellular and humoral immunity.
Limitations of study
Although these results are encouraging, future studies will be necessary to determine the longevity of vaccine-induced immunity and durability of protection using larger groups. This is important because this vaccine is being developed for human use. It is encouraging to see that the MVA/S vaccine-induced antibody response contracted only minimally (less than 2-fold) between weeks 2 and 4 after boost, when maximum contraction of the antibody response happens (Kannanganat et al., 2016; Nigam et al., 2007). This, combined with our prior studies demonstrates the longevity of MVA vaccine-induced immunity in humans (Goepfert et al., 2014) and NHPs (Nigam et al., 2007). It is highly likely that the MVA/S-based vaccine will provide durable protection.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |
|---|---|---|---|
| Antibodies | |||
| APC Goat Anti-Mouse Ig (Multiple Adsorption) Clone Polyclonal (RUO) | BD PharMingen | Cat#550826; RRID:AB_398465 | |
| PE Donkey anti-rabbit IgG (minimal x-reactivity) Antibody | BioLegend | Cat#406421; RRID:AB_2563484 | |
| Alexa Fluor® 700 Rat Anti-Mouse CD4 | BD Biosciences | Cat#557956; RRID:AB_396956 | |
| PE Rat Anti-Mouse CD8a | BD Biosciences | Cat#553032; RRID:AB_394570 | |
| BV 711 anti-mouse TNF-α Antibody | BioLegend | Cat#506349; RRID:AB_2629800 | |
| FITC Rat Anti-Mouse IFN-γ | BD Biosciences | Cat#554411; RRID:AB_395375 | |
| APC Rat Anti-Mouse IL-2 | BD Biosciences | Cat#554429; RRID:AB_398555 | |
| SARS-CoV-2 (COVID-19) spike antibody | GenTex | Cat#GTX135356; RRID:AB_2887482 | |
| SARS-CoV / SARS-CoV-2 (COVID-19) spike antibody [1A9] | GenTex | Cat#GTX632604; RRID:AB_2864418 | |
| SARS-CoV-2 (2019-nCoV) Spike RBD Antibody, Rabbit PAb, Antigen Affinity Purified | Sino Biologicals | Cat#40592-T62 | |
| Monoclonal Anti-Vaccinia Virus E3L, Clone TW2.3 | BEI Resources | Cat#NR-4547 | |
| anti-CD28/CD49d antibodies | BD Biosciences | Cat#557393; RRID:AB_396676 | |
| anti-CD28 | BD Biosciences | Cat#555725; RRID:AB_396068 | |
| anti-CD49d | BD Biosciences | Cat#555501; RRID:AB_2130052 | |
| anti-CD3-PE-Texas Red | BD Biosciences | Cat#562406; RRID:AB_11154406 | |
| anti-CD4-BV650 | BD Biosciences | N/A - Custom reagent | |
| anti-CD8-BV510 | BD Biosciences | N/A - Custom reagent | |
| Live/Dead-APC-Cy7 | Invitrogen | Cat#L34976 | |
| anti-IFN-γ-V450 | BD Biosciences | Cat#560371; RRID:AB_1645594 | |
| anti-TNF-α -A700 | BD Biosciences | Cat#557996; RRID:AB_396978 | |
| anti-IL-2 -PE-Cy7 | BD Biosciences | Cat#560707; RRID:AB_1727542 | |
| anti-IL-17 -FITC | EBio | Cat#11-7179-42; RRID:AB_10805390 | |
| anti-NKG2A-APC | Milteny biotech | Cat#130-113-563; RRID:AB_2726170 | |
| anti-HLA-DR-PerCP | BD Biosciences | Cat#347364; RRID:AB_400292 | |
| anti-CD163-eflour 450 | Thermo scientific | Cat#48-1637-42; RRID:AB_2811826 | |
| anti-BDCA1-BV711 | BioLegend | Cat#331536; RRID:AB_2629760 | |
| anti-CD11C-BV650 | BioLegend | Cat#301638; RRID:AB_2563797 | |
| anti-CD123-PEcy7 | BD biosciences | Cat#560826; RRID:AB_10563898 | |
| anti-CD11b-PE/Dazzle 594 | BioLegend | Cat#301348; RRID:AB_2564081 | |
| anti-CD3-BV605 | BD biosciences | Cat#562994; RRID:AB_2737938 | |
| anti-CD20-BV605 | BioLegend | Cat#302334; RRID:AB_2563398 | |
| anti-Ki67 BV786 | BD Biosciences | Cat#563756; RRID:AB_2732007 | |
| anti- CD3-AF700 | BD Biosciences | Cat#557917; RRID:AB_396938 | |
| anti-CD21-BUV737 | BD Biosciences | Cat#612788 | |
| anti-CD3-PerCP | BD Biosciences | Cat#552851; RRID:AB_394492 | |
| anti-CD8-BV711 | BioLegend | Cat#344734; RRID:AB_2565243 | |
| anti-CXCR5-PE | Invitrogen | Cat#12-9185-42; RRID:AB_11219877 | |
| anti-PD1-BV421 | BioLegend | Cat#329920; RRID:AB_10900818 | |
| anti-Ki67-PEcy7 | BD Biosciences | Cat#561283; RRID:AB_10716060 | |
| anti-BCL6-PE-CF594 | BD Biosciences | Cat#562401; RRID:AB_11152084 | |
| anti-CD27-BUV496 | BD Biosciences | Cat#751678; RRID:AB_2875664 | |
| Bacterial and virus strains | |||
| NEB® Stable Competent E. coli (High Efficiency) | NEB | Cat#C3040H | |
| MVA/S (Modified Vaccinia virus Ankara) – Spike full-length with prefusion stabilized mutants | This paper | N/A | |
| MVA/S1 (Modified Vaccinia virus Ankara) – S1 | This paper | N/A | |
| MVA/Wt (Modified Vaccinia virus Ankara wild-type) | This paper | N/A | |
| SARS-CoV-2 (icSARS-CoV-2) | provided by Mehul Suthar, Emory University | N/A | |
| mNeonGreen SARS-CoV-2 (2019-nCoV/USA_WA1/2020) | Provided by Yong Shi, The University of Texas Medical Branch | N/A | |
| Biological samples | |||
| Serum samples from BALB/c mice | This paper | N/A | |
| Spleen from BALB/c mice | This paper | N/A | |
| Lungs from BALB/c mice | This paper | N/A | |
| BAL sample from BALB/c mice | This paper | N/A | |
| Serum samples from Rhesus monkey | This paper | N/A | |
| Plasma samples from Rhesus monkey | This paper | N/A | |
| Lungs from Rhesus monkey | This paper | N/A | |
| BAL samples from Rhesus monkey | This paper | N/A | |
| Swabs (nasal and throat) from Rhesus monkey | This paper | N/A | |
| PBMCs from Rhesus monkey | This paper | N/A | |
| Chemicals, peptides, and recombinant proteins | |||
| ExpiFectamineTM 293 transfection reagent | ThermoFisher, | Cat#A14524 | |
| SARS-CoV-2 (2019-nCoV) Spike S1+S2 ECD (R683A, R685A, F817P, A892P, A899P, A942P, K986P, V987P)-His Recombinant Protein | Sino Biologicals | Cat#40589-V08H4 | |
| ACE2 Protein, Human, Recombinant (His Tag), Biotinylated | Sino Biologicals | Cat#10108-H08H-B | |
| SARS-CoV-2 (2019-nCoV) Spike S1-His Recombinant Protein (HPLC-verified) | Sino Biologicals | Cat#40591-V08H | |
| Lipofectamine 2000 Transfection Reagent | Invitrogen/Thermo Fisher | Cat#11668019 | |
| Peptide Array, SARS-Related Coronavirus 2 Spike (S) Glycoprotein | BEI Resources | Cat#NR-52402 | |
| Peptide Array, SARS-Related Coronavirus 2 Nucleocapsid (N) Protein (Peptides and Peptide Arrays) | BEI Resources | Cat#NR-52404 | |
| HisPur Ni-NTA Resin | Thermo Scientific | Cat#88221 | |
| Galanthus Nivalis Lectin (GNL), Agarose bound | Vector Laboratories | Cat#AL-1243-5 | |
| Experimental models: cell lines | |||
| HEK293T | ATCC | ATCC® CRL-3216; RRID:CVCL_0063 | |
| DF-1 | ATCC | ATCC® CRL-12203; RRID:CVCL_0570 | |
| Vero | ATCC | ATCC® CCL-81; RRID:CVCL_0059 | |
| Experimental Models: organisms/strains | |||
| Mouse: BALB/c | Jackson Laboratory | Cat#000651 | |
| Monkey: Macaca mulatta | Yerkes primate breeding colony | N/A | |
| Oligonucleotides | |||
| 2019-nCoV_N2-F 5′-TTACAAACATTGGCCGCAAA-3′ | Waggoner et al., 2020 | N/A | |
| 2019-nCoV_N2-R 5′-GCGCGACATTCCGAAGAA-3′ | Waggoner et al., 2020 | N/A | |
| 2019-nCoV_N2-P 5′-FAM-ACAATTTGCCCCCAG CGCTTCAG-BHQ-3′ |
Waggoner et al., 2020 | N/A | |
| SGMRNA-E-F: 5′-CGATCTCTTGTAGATCTGTTCTC-3′ | Wölfel et al., 2020 | N/A | |
| SGMRNA-E-R 5′-ATATTGCAGCAGTACGCACACA-3′ | Wölfel et al., 2020 | N/A | |
| SGMRNA-E-Pr 5′-FAM-ACACTAGCCATCCTTACT GCGCTTCG-3′ |
Wölfel et al., 2020 | N/A | |
| RM-RPP30-F 5′-AGACTTGGACGTGCGAGCG-3′ | Waggoner et al., 2020 | N/A | |
| RM-RPP30-R 5′- GAGCCGCTGTCTCCACAAGT-3′ | Waggoner et al., 2020 | N/A | |
| RPP30-Pr 5′-FAM-TTCTGACCTGAAGGCTCTGC GCG-BHQ1-3′ |
Waggoner et al., 2020 | N/A | |
| Deposited data | |||
| BAL - scRNA-Seq | Gene Expression Omnibus | GEO: GSE165747 | |
| Recombinant DNA | |||
| pLW-73 | Provided by B. Moss and L. Wyatt, National Institute of Health (NIH) | N/A | |
| pLW-73 – SARS-CoV-2 S (PP) (Wuhan) | This paper | N/A | |
| pLW-73 – SARS-CoV-2 S1 (Wuhan) | This paper | N/A | |
| pCAGGS – SARS-CoV-2, RBD (C-ter His Tag) (Wuhan) | BEI Resources | Cat#NR-52309 | |
| pGA1 – SARS-CoV-2, S1 (Wuhan) | BEI Resources | Cat#NR-52309 | |
| Software and algorithms | |||
| FlowJo | FlowJo, LLC | v10.6.2 (https://www.flowjo.com/) | |
| GraphPad Prism | GraphPad | V8.4.3 (https://www.graphpad.com/) | |
Resource availability
Lead contact
Request for further information and for resources and reagents, should be directed to and will be fulfilled by the Lead Contact, Rama Rao Amara (ramara@emory.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact up on reasonable request after completion of a Materials Transfer Agreement.
Data and code availability
All data supporting the experimental findings of this study are available within the manuscript and are available from the corresponding author upon request. Data tables for expression counts for single-cell RNA-Seq for BAL are deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO accession GSE165747.
Experimental model and subject details
Ethics Statements
Mice and rhesus macaques were housed at the Yerkes National Primate Research Center and animal experiments were approved by the Emory University Institutional Animal Care and Use Committee (IACUC) using protocols PROTO201700014 and PROTO202000057. All animal experiments were carried out in accordance to USDA regulations and recommendations derived from the Guide for the Care and Use of Laboratory Animals.
Cells and Viruses
HEK (Human Embryonic Kidney)-293T cells, DF-1 (Chicken embryo fibroblasts), and Vero cells were obtained from ATCC. All MVA vaccines were produced in Amara’s laboratory at the Emory University. SARS-CoV-2 (icSARS-CoV-2) virus was obtained from BEI resources and grown in Suthar’s laboratory at the Emory University. mNeonGreen SARS-CoV-2 (2019-nCoV/USA_WA1/2020) virus was produced by Pei Yong Shi’s laboratory at the University of Texas. The infectious clone SARS-CoV-2 (icSARS-CoV-2) was propagated in VeroE6 cells (ATCC) and sequenced (Xie et al., 2020). The titer of MVA viruses was determined using DF-1 cells and SARS-CoV-2 viruses (icSARS-CoV-2 and 2019-nCoV/USA_WA1/2020) using VeroE6 cells. VeroE6 cells and DF-1 cells were cultured in complete DMEM medium consisting of 1x DMEM (Corning Cellgro), 10% Fetal bovine serum (FBS), 25 mM HEPES Buffer (Corning Cellgro), 2 mM L-glutamine, 1mM sodium pyruvate, 1x Non-essential Amino Acids, and 1x antibiotics. Viral stocks were stored at −80°C until further use.
Animal models
Specific-pathogen-free (SPF) 6–8-week-old female BALB/c mice (00065 strain) were obtained from Jackson Laboratories (Wilmington, MA, USA) and housed in the animal facility at the Yerkes National Primate Research Center of Emory University, Atlanta, GA. Male Indian rhesus macaques (Macaca mulatta), 3-4.5 years old, were housed in pairs in standard non-human primate cages and provided with both standard primate feed (Purina monkey chow) fresh fruit, and enrichment daily, as well free access to water. Immunizations, blood draws, and other sample collections were performed under anesthesia with ketamine (5-10 mg/kg) or telazol (3-5 mg/kg) performed by trained research and veterinary staff.
Method details
Construction and characterization of MVA vaccines
The MVA recombinants expressing the full-length spike (amino acids 1-1273) carrying the prefusion-stabilized mutations (MVA/S) or only S1 portion of spike (amino acids 14-780) (MVA/S1) were generated and confirmed by standard methods. SARS-CoV-2 (GenBank: MN996527.1; Wuhan strain) S ORF was codon optimized for vaccinia virus expression, synthesized using GenScript services, and cloned into pLW-73 (provided by L. Wyatt, National Institutes of Health) between the XmaI and BamHI sites under the control of the vaccinia virus modified H5 (mH5) early late promoter and adjacent to the gene encoding enhanced GFP (green fluorescent protein) (Wyatt et al., 2004). To promote active secretion of the S1, we replaced amino acids 1-14 of the spike sequence with the signal sequence from GM-CSF (WLQGLLLLGTVACSIS). Plaques were picked for 7 rounds to obtain GFP-negative recombinants and DNA sequenced to confirm lack of any mutations. Viral stocks were purified from lysates of infected DF-1 cells using a 36% sucrose cushion and titrated using DF-1 cells by counting pfu/ml. Absence of the wild-type MVA was confirmed by PCR using recombinant specific primers, flanking the inserts.
Flow staining for SARS-CoV-2 Spike protein expression
DF-1 cells were infected with MVA/S or MVA/S1 at an MOI of 1 and stained around 36hrs post-infection. MVA/S infected cells were harvested and stained with live-dead dye and anti- SARS-CoV-2 spike antibody (GTX135356, GeneTex), followed by donkey anti-rabbit IgG coupled to PE (406421, BioLegend). Cells were then fixed with Cytofix/cytoperm (BD PharMingen), permeabilized with permwash (BD PharMingen), and intracellularly stained for MVA protein using mouse monoclonal anti-vaccinia virus E3L Ab (NR-4547 BEI Resources) coupled to PacBlue. MVA/S1 infected cells were stained for live dead marker, fixed, permeabilized. and intracellularly stained for S1 protein using SARS-CoV-2 RBD Ab (40592-T62, SinoBiological), followed by staining with donkey anti-Rabbit IgG PE and anti -Vaccinia virus E3L-PacBlue. For detection of human ACE2 binding to surface expressed spike on MVA/S infected DF-1 cells, were incubated with biotinylated human ACE2 protein at 1:500 dilution (10108-H08H-B, Sino Biological) followed by streptavidin-PE (BD PharMingen). Cells were then stained intracellularly for MVA as described above.
Protein expression and purification
RBD-His and S1 proteins were produced in Amara laboratory by transfecting HEK293 cells using plasmids pCAGGS-RBD-His and pGA8-S1, respectively. The RBD-His plasmid was obtained from BEI resources (Cat# NR-52309). The pGA8-S1 plasmid was generated by cloning human codon-optimized S1 DNA sequence from amino acids 14-780 with GM-CSF signal sequence under the control of CMV promoter with intron A. Transfections were performed according to manufacturer’s instructions (Thermo Fisher). Briefly, HEK293F cells were seeded at a density of 2x106 cells/ml in Expi293 expression medium and incubated in an orbital shaking incubator at 37°C and 127 rpm with 8% CO2 overnight. Next day, 2.5x106 cells/ml were transfected using ExpiFectamine™ 293 transfection reagent (ThermoFisher, cat. no. A14524) as per manufacturer’s protocol. The cells were grown for 72h at 37°C,127 rpm, 8% CO2. The cells were harvested and collected by centrifugation at 2,000 g for 10 min at 4°C. The supernatant was filtered using a 0.22 μm stericup filter (ThermoFisher, cat.no. 290-4520) and loaded onto pre-equilibrated affinity column for protein purification. The SARS-CoV-2 RBD-His tag and S1 proteins were purified using Ni-NTA resin (ThermoFisher, cat.no. 88221) and Agarose bound Galanthus Nivalis Lectin (GNL) (Vector Labs, cat. no. AL-1243-5) respectively. Briefly, His-Pur Ni-NTA resin was washed with PBS by centrifugation at 2000 g for 10 min. The resin was resuspended with the supernatant and incubated for 2h on a shaker at RT. Polypropylene column was loaded on the supernatant-resin mixture and washed with wash Buffer (25mM Imidazole, 6.7mM NaH2PO4.H2O and 300mM NaCl in PBS) four times, after which the protein was eluted in elution buffer (235mM Imidazole, 6.7mM NaH2PO4.H2O and 300mM NaCl in PBS). S1 protein supernatants were mixed with GNL-resin overnight on rocker at 4°C. The supernatant-resin mix was loaded on to the column and washed 3 times with PBS and eluted using 1M methyl- α-D mannopyranoside (pH7.4). Eluted proteins were dialysed against PBS using Slide-A-lyzer Dialysis Cassette (ThermoScientific, Cat# 66030) and concentrated using either 10 kDa Amicon Centrifugal Filter Units (for RBD) or 50 kDa Amicon Centrifugal Filter Units (for S1), at 2000 g at 4°C. The concentrated protein elutes were run on a Superdex 200 Increase 10/300 GL (GE Healthcare) column on an Akta™Pure (GE Healthcare) system and collected the peak that is matching to corresponding protein. The quantity of the proteins were estimated by BCA Protein Assay Kit (Pierce) and quality by BN-PAGE (NuPAGE, 4%–12% Bis-Tris Protein Gels, ThermoScientific), SDS-PAGE and western blot.
Western Blotting
DF-1 cells were infected either with recombinant MVA/S or MVA/S1, at MOI of 1 for 36 h. Infected cells were lysed in ice-cold RIPA buffer and supernatants were collected. Lysates were kept on ice for 10 min, centrifuged, and resolved by SDS-PAGE using precast 4%–15% SDS polyacrylamide gels (BioRad). Proteins were transferred to a nitrocellulose membrane, blocked with 1% casein blocker overnight (Cat# 1610782 BioRad), and incubated for 1 h at room temperature with mouse monoclonal anti-SARS-CoV-2 spike antibody (Cat# GTX632604, GeneTex) for MVA/S, and rabbit SARS-CoV-2 RBD polyclonal antibody (Cat# 40592-T62, Sino Biological) for MVA/S1 diluted 1:2500 in blocking buffer, respectively. The membrane was washed in PBS containing Tween-20 (0.05%) and was incubated for 1 h with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody (Southern Biotech) diluted 1:20,000. The membranes were washed, and proteins were visualized using the ECL select chemiluminescence substrate (Cat# RPN2235 GEhealthcare).
Animal Vaccination
BALB/c mice of 6–8-week-old female were immunized with 10ˆ7 plaque-forming-units (pfu) of MVA/S or MVA/S1 vaccine in the thigh muscle (dose split equally into each thigh) using 25 guage needle on weeks 0 and 3. Following 2-weeks each immunizations, the blood samples were collected by facial vein puncture in BD Microtainer® Tube for analyzing SARS-CoV-2 S (RBD, S1 and S)-specific serum antibody responses. At three weeks after the boost, animals were euthanized using CO2, followed by cervical dislocation. Blood, lung tissue and bronchoalveolar lavage (BAL) were collected.
10 Indian-origin adult male rhesus macaques (Macaca mulatta), 4-5 years old, were randomly allocated into two groups; one group (n = 5) received MVA-empty vector (MVA-wt) and the second group (n = 5) received MVA-expressing prefusion stabilized (with proline mutations) SARS-CoV-2 full-length spike protein (MVA/S). Animals received 1x108 pfu in 1 mL vaccines at week 0 and week 4 by the intramuscular (IM) route.
SARS-CoV-2 Challenge of rhesus macaques
All macaques were challenged with a total of 5x104 pfu (2.5x104 pfu/ml) of SARS-CoV-2 at week 8. Virus was administered as 1 mL by intratracheal (IT) and 1ml by intranasal (IN) route (0.5 mL in each nostril). All the swab samples (nasal and throat) were collected, stored immediately in the viral transport media, and processed for viral RNA extraction the same day. The animal study was conducted at Yerkes National Primate Research Center, Emory University, and was approved by the Emory IACUC.
Binding antibody responses using ELISA
SARS-CoV-2 S (RBD, S1 and S) –specific IgG in serum and BAL was quantified by enzyme-linked immunosorbent assay (ELISA) as described previously (Chamcha et al., 2015). Briefly, Nunc high-binding ELISA plates were coated with 2 μg/ml of recombinant SARS-CoV-2 proteins (RBD, S1 and S) proteins in Dulbecco’s phosphate-buffered saline (DPBS) and incubated overnight at 4 °C. SARS-CoV-2 RBD and S1 proteins were produced in the lab whereas, S1 and S (S1 + S2 ECD) proteins were purchased. Plates were then blocked with 5% blotting-grade milk powder and 4% whey powder in DPBS with 0.05% Tween 20 for 2h at room temperature (RT). Plates were then incubated with serially diluted serum samples (starting from 100, 3 fold, 8x) and incubated for 2h at RT followed with 6 washes. Total SARS-CoV-2 S (RBD, S1 and S)–specific mouse IgG and monkey IgG antibodies were detected using HRP-conjugated anti-mouse (1:6000) (Southern Biotech; AL, USA) and goat anti-monkey IgG secondary antibody (1:10, 000), respectively for 1 h at RT. The plates were washed and developed using TMB (2-Component Microwell Peroxidase Substrate Kit) and the reaction was stopped using 1N phosphoric acid solution. Plates were read at 450 nm wavelength within 30 min using a plate reader (Molecular Devices; San Jose, CA, USA). ELISA endpoint titers were defined as the highest reciprocal serum dilution that yielded an absorbance > 2-fold over background values.
Live-virus neutralization
Live-virus SARS-CoV-2 neutralization antibodies were assessed using a full-length mNeonGreen SARS-CoV-2 (2019-nCoV/USA_WA1/2020), generated as previously described (Xie et al., 2020). Vaccinated mice, NHP and post-challenge sera were incubated at 56°C for 30 min and manually diluted in duplicate in serum-free Dulbecco’s Modified Eagle Medium (DMEM) and incubated with 750-1000 focus-forming units (FFU) of infectious clone derived SARS-CoV-2-mNG virus (Xie et al., 2020) at 37°C for 1 h. The virus/serum mixture was added to VeroE6 cell (C1008, ATCC, #CRL-1586) monolayers, seeded in 96-well blackout plates, and incubated at 37°C for 1 h. Post incubation, the inoculum was removed and replaced with pre-warmed complete DMEM containing 0.85% methylcellulose. Plates were incubated at 37°C for 24 h. After 24 h, the methylcellulose overlay was removed, cells were washed three times with phosphate-buffered saline (PBS), and fixed with 2% paraformaldehyde (PFA) in PBS for 30 min at room temperature. PFA is then removed and washed twice with PBS. The foci were visualized using an ELISPOT reader (CTL ImmunoSpot S6 Universal Analyzer) under a FITC channel and enumerated using Viridot (Katzelnick et al., 2018). The neutralization titers were calculated as follows: 1 - ratio of the (mean number of foci in the presence of sera: foci at the highest dilution of respective sera sample). Each specimen is tested in two independent assays performed at different times. The focus-reduction neutralization mNeonGreen live-virus 50% titers (FRNT-mNG50) were interpolated using a 4-parameter nonlinear regression in GraphPad Prism 8.4.3. Samples that did not neutralize at the limit of detection at 50% were plotted at 10 and were used for geometric mean calculations.
Pseudovirus neutralization assay
SARS-CoV-2 neutralization was assessed with Spike-pseudotyped virus in 293T/ACE2 cells as a function of reductions in luciferase (Luc) reporter activity. 293T/ACE2 cells were kindly provided by Drs. Mike Farzan and Huihui Mu at Scripps. Cells were maintained in DMEM containing 10% FBS and 3 μg/ml puromycin. An expression plasmid encoding codon-optimized full-length Spike of the Wuhan-1 strain (VRC7480), was provided by Drs. Barney Graham and Kizzmekia Corbett at the Vaccine Research Center, National Institutes of Health (USA). The D614G amino acid change was introduced into VRC7480 by site-directed mutagenesis using the QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent Technologies (Catalog # 210518). The mutation was confirmed by full-length Spike gene sequencing. Pseudovirions were produced in HEK293T/17 cells (ATCC cat. no. CRL-11268) by transfection using Fugene 6 (Promega Cat#E2692) and a combination of Spike plasmid, lentiviral backbone plasmid (pCMV ΔR8.2) and firefly Luc reporter gene plasmid (pHR’ CMV Luc) (Naldini et al., 1996) in a 1:17:17 ratio. Transfections were allowed to proceed for 16-20 h at 37°C. Medium was removed, monolayers rinsed with growth medium, and 15 mL of fresh growth medium added. Pseudovirus-containing culture medium was collected after an additional 2 days of incubation and was clarified of cells by low-speed centrifugation and 0.45 μm micron filtration and stored in aliquots at −80°C. TCID50 assays were performed on thawed aliquots to determine the infectious dose for neutralization assays (RLU 500-1000x background, background usually averages 50-100 RLU).For neutralization, a pre-titrated dose of virus was incubated with 8 serial 3-fold or 5-fold dilutions of serum samples or mAbs in duplicate in a total volume of 150 μl for 1 hr at 37°C in 96-well flat-bottom poly-L-lysine-coated culture plates (Corning Biocoat). Cells were suspended using TrypLE express enzyme solution (Thermo Fisher Scientific) and immediately added to all wells (10,000 cells in 100 μL of growth medium per well). One set of 8 control wells received cells + virus (virus control) and another set of 8 wells received cells only (background control). After 66-72 hr of incubation, medium was removed by gentle aspiration and 30 μL of Promega 1X lysis buffer was added to all wells. After a 10-minute incubation at room temperature, 100 μl of Bright-Glo luciferase reagent was added to all wells. After 1–2 min, 110 μl of the cell lysate was transferred to a black/white plate (Perkin-Elmer). Luminescence was measured using a PerkinElmer Life Sciences, Model Victor2 luminometer. Neutralization titers are the serum dilution (ID50/ID80) or mAb concentration (IC50/IC80) at which relative luminescence units (RLU) were reduced by 50% and 80% compared to virus control wells after subtraction of background RLUs. Maximum percent inhibition (MPI) is the % neutralization at the lowest serum dilution or highest mAb concentration tested. Serum samples were heat-inactivated for 30 min at 56°C prior to assay.
Antibody isotype, IgG subclass and FcR binding of mouse sera
For relative quantification of antigen-specific antibody titers, a customized multiplexed approach was applied, as previously described (Brown et al., 2012). Therefore, magnetic microspheres with a unique fluorescent signature (Luminex) were coupled with SARS-CoV-2 antigens including spike protein (S) (provided by Eric Fischer, Dana Farber), Receptor Binding Domain (RBD), and CoV HKU1 RBD (provided by Aaron Schmidt, Ragon Institute), CoV-2 S1 and S2 (Sino Biologicals) as well as influenza as control (Immune Tech). Coupling was performed using EDC (Thermo Scientific) and Sulfo-NHS (Thermo Scientific) to covalently couple antigens to the beads. 1.2x103 beads per region/ antigen were added to a 384-well plate (Greiner), and incubated with diluted plasma samples (1:90 for all readouts) for 16h while shaking at 900rmp at 4°C, to facilitate immune complex formation. The next day, immune complexed microspheres were washed three times in 0.1% BSA and 0.05% Tween-20 using an automated magnetic plate washer (Tecan). Anti-mouse IgG-, IgG2a-, IgG3-, IgA- and IgM-PE coupled (Southern Biotech) detection antibodies were diluted in Luminex assay buffer to 0.65ug/ml. Beads and detection antibodies were incubated for 1h at RT while shaking at 900rpm. Following washing of stained immune complexes, a tertiary goat anti-mouse IgG-PE antibody (Southern Biotech) was added and incubated for 1h at RT on a shaker. To assess Fc-receptor binding, mouse Fc-receptor FcγR2, FcγR3, FcγR4 (Duke Protein Production facility) were biotinylated (Thermo Scientific) and conjugated to Streptavidin-PE for 10 min (Southern Biotech) before adding to immune complexes and processed as described above. Finally, beads were washed and acquired on a flow cytometer, iQue (Intellicyt) with a robot arm (PAA). Events were gated on each bead region, median fluorescence of PE for bead positive events was reported. Samples were run in duplicate for each secondary detection agents.
Antibody isotype, IgG subclass and FcR binding of monkey sera
A Luminex assay was used to detect and quantify antigen-specific subclass, isotype and Fc-receptor (binding) factors (Brown et al., 2017). With this assay, we measured the antibody concentration against SARS-CoV-2 RBD (kindly provided by Aaron Schmidt, Ragon Institute) and SARS-CoV-2 S (Kindly provided by Erira Ollmann Saphire, La Jolla Institute). Carboxylate-modified microspheres (Luminex) were activated using EDC and Sulfo-NHS and antigens were covalently bound to the beads via NHS-ester linkages. Antigen-coupled beads were washed and blocked. Immune complexes were formed by mixing appropriately diluted plasma (1:100 for IgG1, IgG2, IgG3, IgG4, IgA, IgM, and 1:1000 for FcγRs) to antigen-coupled beads and incubating the complexes overnight at 4°C. Immune complexes were then washed with 0.1% BSA 0.02% Tween-20. PE-coupled secondary antibodies for each antibody isotype or subclass (Southern Biotech) was used to detect antigen-specific antibody titer. For FcRs, biotinylated FcRs were labeled with streptavidin-PE before addition to immune complexes. Fluorescence was measured with an iQue (Intellicyt) and analyzed using Forecyt software. Data are reported as median fluorescence intensity (MFI).
ADCP, ADNP, and ADCD Assays
Antibody-dependent cellular phagocytosis (ADCP), antibody-dependent neutrophil phagocytosis (ADNP) and antibody-dependent complement deposition (ADCD) was measured as previously described (Ackerman et al., 2011; Fischinger et al., 2019; Karsten et al., 2019). For ADCP and ADNP, yellow-green fluorescent neutravidin beads were coupled to biotinylated SARS-CoV-2 S or RBD. For ADCD, red fluorescent neutravidin beads were coupled to biotinylated SARS-CoV-2 S or RBD. Antigen-coupled beads were then incubated with appropriately diluted plasma (ADCP 1:100, ADNP 1:50, ADCD 1:10) for 2 h at 37°C to form immune complexes. For ADCP, THP-1 s (ATCC) were added at 1.25x105 cells/mL and incubated for 16 h at 37°C. For ADNP, leukocytes were isolated from fresh peripheral whole blood by lysing erythrocytes using ammonium-chloride potassium lysis. Leukocytes were added to immune complexes at 2.5x105 cells/mL and incubated for 1 h at 37°C. Neutrophils were detected using anti-human CD66b Pacblue (Biolegend). For ADCD, lyophilized guinea pig complement (Cedarlane) was resuspended, diluted in gelatin veronal buffer with calcium and magnesium (GVB++, Boston BioProducts) and added to immune complexes. The deposition of C3 was detected using an anti-C3 FITC antibody (Mpbio).
All functional assays were acquired with an iQue (Inellicyt) and analyzed using Forecyt software. For ADCP, event were gated on singlets and fluorescent cells. For ADNP, bead-positive neutrophils were defined as CD66b positive, fluorescent cells. For both ADCP and ADNP, a phagocytic score was defined as (percentage of bead-positive cells) x (MFI of bead-positive cells) divided by 10000. For ADCD, data were reported as median fluorescence of C3 deposition (MFI).
Cell processing
For mouse studies, spleens and lungs of vaccinated and control animals were removed and placed on ice in cold RPMI 1640 (1X) with 5% FBS (Company, state, USA). 1X β-Mercaptoethanol (Invitrogen, State, USA) was added to complete medium to isolate splenocytes. Whereas lungs were cut into small pieces and incubated at 37°C in RPMI (1X) medium containing Collagenase type IV and DNase I with gentle shaking for 30 min. After incubation, cells were isolated by forcing tissue suspensions through a 70 μM cell strainer. RBCs were removed by ACK lysis buffer and live cells counted by trypan blue exclusion. For macaques, PBMC from blood collected in sodium citrate CPT tubes were isolated using standard procedures. Post SARS-CoV-2 challenge, samples were processed and stained in BSL-3 facility.
To collect BAL fluids and processing, and single-cells isolation, up to 50 mL physiological saline was delivered through trachea to the lungs of anesthetized animals using a camera enabled fiberoptic bronchoscope. The flushed saline was re-aspirated 5 times before pulling out the bronchoscope. This collection was filtered through 70 μm cell strainer and centrifuged at 2200 rpm for 5 min. Pelleted cells were suspended in 1ml R10 medium (RPMI(1X), 10% FBS) and stained as described in sections below.
For processing lymph-node, lymph-node biopsies were dissociated using 70 μm cell strainer. The cell suspension was washed twice with R-10 media.
Intracellular Cytokine Staining (ICS)
Functional responses of SARS-CoV-2 RBD, S1 and S2 specific CD8+ and CD4+ T cells in vaccinated animals were measured using peptide pools and intracellular cytokine staining (ICS) assay. Overlapping peptides (13 or 17 mers overlapping by 10 amino acids) were obtained from BEI resources (NR-52402 for spike and NR-52419 for nucleocapsid) and different pools (S1, S2, RBD and NC) were made. The S1 pool contained peptides mixed from 1-97, S2 pool contained peptides mixed from 98-181, RBD pool contained peptides 46-76 and NC pool contained 57 peptides. Each peptide was used at 1 μg/ml concentration in the stimulation reaction. Two million cells suspended in 200 μL of RPMI 1640 medium with 10% FBS were stimulated with 1 μg/ml CD28 (BD Biosciences), 1 μg/ml CD49d (BD Biosciences) co-stimulatory antibodies and different peptide pools. These stimulated cells were incubated at 37°C in 5% CO2 conditioned incubator. After 2hrs of incubation, 1 μL Golgi-plug and 1 μL Golgi-stop/ml (both from BD Biosciences) were added and incubated for 4 more hours. After total 6 h of incubation, cells were transferred to 4°C overnight and were stained the next day. Cells were washed once with FACS wash (1XPBS, 2% FBS and 0.05% sodium azide) and surface stained with Live/Dead-APC-Cy7, anti-CD3, anti-CD4 and anti-CD8, each conjugated to a different fluorochrome for 30 min at RT. The stained cells were washed once with FACS wash and permeabilized with 200 μL of cytofix/cytoperm for 30 min at 4°C. Cells were washed once with perm wash and incubated with anti-cytokine antibodies for 30 min at 4°C. Finally, the samples were washed once with perm wash and once with FACS wash, and fixed in 4% paraformaldehyde solution for 20 min before acquiring on BD LSR Fortessa flow cytometer. Data were analyzed using FlowJo software.
Histopathological examination
For visualizing iBALT structures in mouse lungs by Immunohistochemistry, the lung tissues were fixed in 4% PFA for 12h followed by PBS wash. Fixed lungs tissues were kept in 30% sucrose overnight followed by freezing in OCT solution. Frozen blocks were cryosectioned, fixed, and immunostained for iBALT structure. Sections were incubated overnight at 4°C with primary antibodies containing rat anti-mouse B220 (Cat#130-042-401) and hamster anti-mouse CD3 (Cat#550277). Next day, primary antibodies were washed with chilled PBS thrice followed by incubation with secondary antibodies containing anti-rat IgG-Alexa 488 (Cat#ab150157) and anti-hamster IgG-Alexa 546 (Cat#A-21111). Sections were incubated with secondary antibodies at room temperature for 1h followed by wash with chilled PBS thrice. Washed sections were mounted with antifade mounting media with DAPI. Imaging was performed at Olympus FV1000 confocal microscope using 20X objective. Number of iBALT structures were quantified per image section and plotted using GraphPad prism version 8.
For histopathologic examination in macaques, the animals were euthanized due to the study end point, and a complete necropsy was performed. For histopathologic examination, various tissue samples including lung, nasal turbinates, trachea, tonsils, hilar lymph nodes, spleen, heart, brain, gastrointestinal tract (stomach, jejunum, ileum, colon, and rectum), testes were fixed in 10% neutral-buffered formalin for 24h at room temperature, routinely processed, paraffin-embedded, sectioned at 4 μm, and stained with hematoxylin and eosin (H & E). The H & E slides from all tissues were examined by two board certified veterinary pathologists. For each animal, all the lung lobes were used for analysis and affected microscopic fields were scored semiquantitatively as Grade 0 (None); Grade 1 (Mild); Grade 2 (Moderate) and Grade 3 (Severe). Scoring was performed based on these criteria: number of lung lobes affected, type 2 pneumocyte hyperplasia, alveolar septal thickening, fibrosis, perivascular cuffing, peribronchiolar hyperplasia, inflammatory infiltrates, hyaline membrane formation. An average lung lobe score was calculated by combining scores from each criterion. Digital images of H&E stained slides were captured at 100 × and 200 × magnification with an Olympus BX43 microscope equipped with a digital camera (DP27, Olympus) using Cellsens® Standard 2.3 digital imaging software (Olympus).
Immunophenotyping of BAL and LN cells
Briefly, the cells were stained with surface antibody cocktail and incubated at RT for 30 min. The stained cells were given a FACS wash and permeabilized with 1ml perm buffer (Invitrogen) for 30 min at RT. These cells were given a perm wash (Invitrogen) and stained with an intracellular antibody cocktail for 30 min at RT. Finally, the cells were washed once with perm wash and a FACS wash and fixed in 4% paraformaldehyde solution for 20 min before acquiring on BD LSR-II flow cytometer. Samples prior to challenge were acquired without 20-minute 4% paraformaldehyde fixation.
BAL innate cell surface antibody cocktail: live/dead stain-APC-cy7, anti-CD3-605, anti-CD20-605, anti-NKG2A-APC, anti-HLA-DR-PERCP, anti-cd11b-PE/Dazzle 594, anti-163-eflour-450, anti-CD123-PEcy7, anti-CD11c-BV655 and anti-BDCA1-BV711. BAL innate cell intracellular antibody: anti-Ki67-BV786. T cell phenotype surface antibody cocktail: live/dead stain-APC-cy7, anti-CD3-PERCP, anti-CD4-BV655, anti-CD8-BV711, anti-PD1-BV421, anti-CXCR5-PE and anti-CXCR3-BV605. T cell phenotype intracellular antibody: anti-Ki67-BV786. B Cell phenotype surface antibody cocktail: live/dead stain-APC-cy7, anti-CD3-AF700 and anti-CD20-BV605, B cell phenotype intracellular antibody: anti-BCL6-PE-CF594 and anti-Ki67-PEcy7.
Viral RNA extraction and quantification
SARS-CoV-2 genomic and subgenomic RNA was quantified in naso-pharyngeal (NP) swabs, throat swabs, and brocho-alveolar lavages (BAL). Swabs were placed in 1mL of Viral Transport Medium (VTM; Labscoop (VR2019-1L)). Viral RNA was extracted from NP swabs, throat swabs, and BAL on fresh specimens using the QiaAmp Viral RNA mini kit according to the manufacturer’s protocol. Quantitative PCR (qPCR) was performed on genomic viral RNA using the N2 primer and probe set designed by the CDC for their diagnostic algorithm: CoV2-N2-F: 5′-TTACAAACATTGGCCGCAAA-3′, CoV2-N2-R: 5′-GCGCGACATTCCGAAGAA-3′, and CoV2-N2-Pr: 5′-FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ-3′ (Waggoner et al., 2020). The primer and probe sequences for the subgenomic mRNA transcript of the E gene (Wölfel et al., 2020) are SGMRNA-E-F: 5′-CGATCTCTTGTAGATCTGTTCTC-3′, SGMRNA-E-R: 5′-ATATTGCAGCAGTACGCACACA-3′, and SGMRNA-E-Pr: 5′-FAM-ACACTAGCCATCCTTACTGCGCTTCG-3′. qPCR reactions were performed in duplicate with the Thermo-Fisher 1-Step Fast virus mastermix using the manufacturer’s cycling conditions, 200nM of each primer, and 125nM of the probe. The limit of detection in this assay was about 128 copies per mL of VTM/BAL depending on the volume of extracted RNA available for each assay. To verify sample quality the CDC RNase P p30 subunit qPCR was modified to account for rhesus macaque specific polymorphisms. The primer and probe sequences are RM-RPP30-F 5′-AGACTTGGACGTGCGAGCG-3′, RM-RPP30-R 5′- GAGCCGCTGTCTCCACAAGT-3′, and RPP30-Pr 5′-FAM-TTCTGACCTGAAGGCTCTGCGCG-BHQ1-3′ (Waggoner et al., 2020). A single well from each extraction was run as described above to verify RNA integrity and sample quality via detectable and consistent cycle threshold values (Ct between 25-32).
Single-cell RNA sequencing and analysis
BAL samples from 5 monkeys in each group were pooled together and two technical replicate 10X captures were performed. One replicate capture failed for the control group. The libraries were run on Nova Seq 6000 lanes and the resultant bcl files were converted to count matrices using Cell Ranger v3.1 (https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest?). Count matrices for each capture were processed using an in-house single-cell RNA-seq pipeline that uses Seurat v3.0 (Butler et al., 2018; Stuart et al., 2019). CITE-seq-Count (https://github.com/Hoohm/CITE-seq-Count) was used along with HTODemux function in Seurat to demultiplex samples. The cells expressing nFeature _RNA < 300 and > 10% mitochondrial genes, HBB, RPS or RPL genes were filtered along with doublets. One of the samples within the Vaccine group (Rcc18) was dropped due to 10-fold higher number of cells compared to other samples. Post filtration, cells from each capture were normalized using SCTransform normalization and then integrated in Seurat. After integration, Principal Component analysis was carried out. PCs 1-30 were chosen for clustering analysis, as there was very little additional variance beyond PC 30. Cell were clustered based on PC scores using the Louvain method. UMAP method (McInnes et al., 2018) was used to visualize the single cells in 2d embedding. We used Human primary cell atlas from SingleR (Aran et al., 2019) and knowledge of canonical markers to classify cells into different cell subtypes (Figures S4, Canonical, and S5, Top10Clusters). Differential gene expression between Control and Vaccine group was assessed by MAST (McDavid et al., 2017). Heatmaps, Dot plots, Violin plots and Feature plots were generated using seurat package in R. Additionally, we performed Gene set enrichment analysis using WebGestalt (Liao et al., 2019; Zhang et al., 2005).
Quantification and statistical analysis
The difference between any two groups at a time point was measured either using a two-tailed nonparametric Mann–Whitney rank-sum test or unpaired parametric t test depending on the distribution of the data. Comparisons between different time points within a group used paired parametric t test. P value of less than 0.05 was considered significant. The correlation analysis was performed using Spearman rank test. GraphPad Prism version 8.4.3 (471) (GraphPad Software) was used to perform data analysis and statistics.
Acknowledgments
We thank Traci H. Legere, Brenda Wehrle, and Zeba S. Momin for help with processing blood and tissue samples, Drs. Bernard Moss and Lynda Wyatt for providing the pLW73 transfer plasmid, the Yerkes Division of Pathology and Research Resources for outstanding animal care during the pandemic, and the Histology and Molecular Pathology Lab for help with tissue sectioning. The following reagent was produced under HHSN272201400008C and obtained through BEI Resources, NIAID, NIH: vector pCAGGS containing the SARS-related coronavirus 2, Wuhan-Hu-1 spike glycoprotein RBD, NR-52309. Imaging for this research project was supported in part by the Emory University Integrated Cellular Imaging Microscopy Core. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institute of Health. This work was supported in part by National Institutes of Health grant RO1 AI148378-01S1 and Fast Grants award 2166 (to R.R.A.) and NCRR/NIH base grant P51 OD011132 to YNPRC. Next-generation sequencing services were provided by the Yerkes NHP Genomics Core, which is supported in part by NIH P51 OD011132. Sequencing data were acquired on an Illumina NovaSeq6000 funded by NIH S10 OD026799 (to S.E.B.).
Author contributions
R.R.A. was responsible for the overall experimental design and supervision of laboratory studies and manuscript writing and editing. Sailaja Gangadhara developed, constructed, and characterized the rMVA/S and MVA/S1 vaccines and provided vaccine stocks for animal studies. N.K.R., Sailaja Gangadhara, N.C., Ayalnesh Shiferaw, V.S.B., A.K.B., S.A.R., Anusmita Sahoo, V.V.E., L.L., K.F., S.W., S.F., C.A., A.A.U., and K.P. were responsible for conducting experiments, data collection, and data analysis. M.S.S. conducted and supervised live virus neutralizing antibody assays. D.M. supervised pseudovirus neutralizing antibody assays. J.C., S.M.J., J.S.W., F.C.-S., and R.L.S. provided veterinary support in ABSL-3. G.A. supervised the Luminex assays. S.E.B. supervised the single-cell RNA-seq analyses. T.H.V. supervised the viral RNA measurements. Sanjeev Gumber and S.K. performed H&E staining and determined lung pathology scores. P.-Y.S. and V.D.M. provided virus for neutralization assays and challenge studies. All authors contributed to manuscript writing and editing.
Declaration of interests
R.R.A., Sailaja Gangadhara, and N.K.R. are co-inventors of the MVA/S vaccine technology. Emory University filed a patent on this technology. R.R.A. serves as a SAB member for Heat Biologics Inc.
Published: February 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.02.001.
Supplemental information
References
- Ackerman M.E., Moldt B., Wyatt R.T., Dugast A.-S., McAndrew E., Tsoukas S., Jost S., Berger C.T., Sciaranghella G., Liu Q., Irvine D.J., Burton D.R., Alter G. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J Immunol Methods. 2011;366:8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aid M., Busman-Sahay K., Vidal S.J., Maliga Z., Bondoc S., Starke C., Terry M., Jacobson C.A., Wrijil L., Ducat S., et al. Vascular Disease and Thrombosis in SARS-CoV-2-Infected Rhesus Macaques. Cell. 2020;183:1354–1366.e13. doi: 10.1016/j.cell.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara R.R., Villinger F., Altman J.D., Lydy S.L., O’Neil S.P., Staprans S.I., Montefiori D.C., Xu Y., Herndon J.G., Wyatt L.S., et al. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science. 2001;292:69–74. doi: 10.1126/science.1058915. [DOI] [PubMed] [Google Scholar]
- Amara R.R., Villinger F., Staprans S.I., Altman J.D., Montefiori D.C., Kozyr N.L., Xu Y., Wyatt L.S., Earl P.L., Herndon J.G., et al. Different patterns of immune responses but similar control of a simian-human immunodeficiency virus 89.6P mucosal challenge by modified vaccinia virus Ankara (MVA) and DNA/MVA vaccines. J. Virol. 2002;76:7625–7631. doi: 10.1128/JVI.76.15.7625-7631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D.H., Liu J., Li H., Maxfield L.F., Abbink P., Lynch D.M., Iampietro M.J., SanMiguel A., Seaman M.S., Ferrari G., et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. USA. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A.C., Domi A., McDonald E.M., Talmi-Frank D., McCurley N., Basu R., Robinson H.L., Hellerstein M., Duggal N.K., Bowen R.A., Guirakhoo F. A Zika Vaccine Targeting NS1 Protein Protects Immunocompetent Adult Mice in a Lethal Challenge Model. Sci. Rep. 2017;7:14769. doi: 10.1038/s41598-017-15039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Brinkkemper M., Maisonnasse P., Dereuddre-Bosquet N., Grobben M., Claireaux M., de Gast M., Marlin R., Chesnais V., Diry S., et al. Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection. Nature. 2021 doi: 10.1038/s41586-021-03275-y. Published online February 1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.P., Dowell K.G., Boesch A.W., Normandin E., Mahan A.E., Chu T., Barouch D.H., Bailey-Kellogg C., Alter G., Ackerman M.E. Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J Immunol Methods. 2017;443:33–44. doi: 10.1016/j.jim.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.P., Licht A.F., Dugast A.S., Choi I., Bailey-Kellogg C., Alter G., Ackerman M.E. High-throughput, multiplexed IgG subclassing of antigen-specific antibodies from clinical samples. J. Immunol. Methods. 2012;386:117–123. doi: 10.1016/j.jim.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamcha V., Jones A., Quigley B.R., Scott J.R., Amara R.R. Oral Immunization with a Recombinant Lactococcus lactis-Expressing HIV-1 Antigen on Group A Streptococcus Pilus Induces Strong Mucosal Immunity in the Gut. J. Immunol. 2015;195:5025–5034. doi: 10.4049/jimmunol.1501243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chea L.S., Amara R.R. Immunogenicity and efficacy of DNA/MVA HIV vaccines in rhesus macaque models. Expert Rev. Vaccines. 2017;16:973–985. doi: 10.1080/14760584.2017.1371594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chea L.S., Wyatt L.S., Gangadhara S., Moss B., Amara R.R. Novel Modified Vaccinia Virus Ankara Vector Expressing Anti-apoptotic Gene B13R Delays Apoptosis and Enhances Humoral Responses. J. Virol. 2019;93:93. doi: 10.1128/JVI.01648-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domi A., Feldmann F., Basu R., McCurley N., Shifflett K., Emanuel J., Hellerstein M.S., Guirakhoo F., Orlandi C., Flinko R., et al. A Single Dose of Modified Vaccinia Ankara expressing Ebola Virus Like Particles Protects Nonhuman Primates from Lethal Ebola Virus Challenge. Sci. Rep. 2018;8:864. doi: 10.1038/s41598-017-19041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer K., Rampling T., Venkatraman N., Bowyer G., Wright D., Lambe T., Imoukhuede E.B., Payne R., Fehling S.K., Strecker T., et al. A Monovalent Chimpanzee Adenovirus Ebola Vaccine Boosted with MVA. N. Engl. J. Med. 2016;374:1635–1646. doi: 10.1056/NEJMoa1411627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischinger S., Fallon J.K., Michell A.R., Broge T., Suscovich T.J., Streeck H., Alter G. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J Immunol Methods. 2019;473 doi: 10.1016/j.jim.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.C. Clinical development of Modified Vaccinia virus Ankara vaccines. Vaccine. 2013;31:4241–4246. doi: 10.1016/j.vaccine.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Goepfert P.A., Elizaga M.L., Sato A., Qin L., Cardinali M., Hay C.M., Hural J., DeRosa S.C., DeFawe O.D., Tomaras G.D., et al. National Institute of Allergy and Infectious Diseases HIV Vaccine Trials Network Phase 1 safety and immunogenicity testing of DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J. Infect. Dis. 2011;203:610–619. doi: 10.1093/infdis/jiq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert P.A., Elizaga M.L., Seaton K., Tomaras G.D., Montefiori D.C., Sato A., Hural J., DeRosa S.C., Kalams S.A., McElrath M.J., et al. HVTN 205 Study Group. National Institutes of Allergy and Infectious Diseases HIV Vaccines Trials Network Specificity and 6-month durability of immune responses induced by DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J. Infect. Dis. 2014;210:99–110. doi: 10.1093/infdis/jiu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., van den Brand J.M., Raj V.S., Volz A., Wohlsein P., Smits S.L., Schipper D., Bestebroer T.M., Okba N., Fux R., et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science. 2016;351:77–81. doi: 10.1126/science.aad1283. [DOI] [PubMed] [Google Scholar]
- Hartman A.L., Nambulli S., McMillen C.M., White A.G., Tilston-Lunel N.L., Albe J.R., Cottle E., Dunn M.D., Frye L.J., Gilliland T.H., et al. SARS-CoV-2 infection of African green monkeys results in mild respiratory disease discernible by PET/CT imaging and shedding of infectious virus from both respiratory and gastrointestinal tracts. PLoS Pathog. 2020;16:e1008903. doi: 10.1371/journal.ppat.1008903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T.N., Pino M., Boddapati A.K., Viox E.G., Starke C.E., Upadhyay A.A., Gumber S., Busman-Sahay K., Strongin Z., Harper J.L., et al. Baricitinib treatment resolves lower airway inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. Cell. 2021;184:460–475.e21. doi: 10.1016/j.cell.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S.S., Amara R.R. DNA/MVA Vaccines for HIV/AIDS. Vaccines (Basel) 2014;2:160–178. doi: 10.3390/vaccines2010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannanganat S., Wyatt L.S., Gangadhara S., Chamcha V., Chea L.S., Kozlowski P.A., LaBranche C.C., Chennareddi L., Lawson B., Reddy P.B., et al. High Doses of GM-CSF Inhibit Antibody Responses in Rectal Secretions and Diminish Modified Vaccinia Ankara/Simian Immunodeficiency Virus Vaccine Protection in TRIM5α-Restrictive Macaques. J. Immunol. 2016;197:3586–3596. doi: 10.4049/jimmunol.1600629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten C.B., Mehta N., Shin S.A., Diefenbach T.J., Slein M.D., Karpinski W., Irvine E.B., Broge T., Suscovich T.J., Alter G. A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. J Immunol Methods. 2019;471:46–56. doi: 10.1016/j.jim.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzelnick L.C., Coello Escoto A., McElvany B.D., Chávez C., Salje H., Luo W., Rodriguez-Barraquer I., Jarman R., Durbin A.P., Diehl S.A., et al. Viridot: An automated virus plaque (immunofocus) counter for the measurement of serological neutralizing responses with application to dengue virus. PLoS Negl. Trop. Dis. 2018;12:e0006862. doi: 10.1371/journal.pntd.0006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., 3rd, et al. A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses against SARS-CoV-2 in Mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDavid A., Finak G., Yajima M. MAST: Model-based Analysis of Single Cell Transcriptomics, R package version 1.2.1. 2017. http://rglab.github.io/MAST/
- McInnes L., Healy J., Melville J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv. 2018 https://arxiv.org/abs/1802.03426 arXiv:1802.03426. [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyron-Quiroz J.E., Rangel-Moreno J., Kusser K., Hartson L., Sprague F., Goodrich S., Woodland D.L., Lund F.E., Randall T.D. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat. Med. 2004;10:927–934. doi: 10.1038/nm1091. [DOI] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Nigam P., Earl P.L., Americo J.L., Sharma S., Wyatt L.S., Edghill-Spano Y., Chennareddi L.S., Silvera P., Moss B., Robinson H.L., Amara R.R. DNA/MVA HIV-1/AIDS vaccine elicits long-lived vaccinia virus-specific immunity and confers protection against a lethal monkeypox challenge. Virology. 2007;366:73–83. doi: 10.1016/j.virol.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson H.L., Amara R.R. T cell vaccines for microbial infections. Nat. Med. 2005;11(4, Suppl):S25–S32. doi: 10.1038/nm1212. [DOI] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speranza E., Williamson B.N., Feldmann F., Sturdevant G.L., Pérez-Pérez L., Mead-White K., Smith B.J., Lovaglio J., Martens C., Munster V.J., et al. SARS-CoV-2 infection dynamics in lungs of African green monkeys. bioRxiv. 2020 doi: 10.1101/2020.08.20.258087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar M.S., Zimmerman M., Kauffman R., Mantus G., Linderman S., Vanderheiden A., Nyhoff L., Davis C., Adekunle S., Affer M., et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep. Med. 2020;1:100040. doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M., Heath S.L., Sweeton B., Williams K., Cunningham P., Keele B.F., Sen S., Palmer B.E., Chomont N., Xu Y., et al. DNA/MVA Vaccination of HIV-1 Infected Participants with Viral Suppression on Antiretroviral Therapy, followed by Treatment Interruption: Elicitation of Immune Responses without Control of Re-Emergent Virus. PLoS ONE. 2016;11:e0163164. doi: 10.1371/journal.pone.0163164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titanji K., Velu V., Chennareddi L., Vijay-Kumar M., Gewirtz A.T., Freeman G.J., Amara R.R. Acute depletion of activated memory B cells involves the PD-1 pathway in rapidly progressing SIV-infected macaques. J. Clin. Invest. 2010;120:3878–3890. doi: 10.1172/JCI43271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., Yu J., Chan C.N., Bondoc S., Starke C.E., et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.2005.2013.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner J.J., Stittleburg V., Pond R., Saklawi Y., Sahoo M.K., Babiker A., Hussaini L., Kraft C.S., Pinsky B.A., Anderson E.J., Rouphael N. Triplex Real-Time RT-PCR for Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020;26:1633–1635. doi: 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O’Connor M.A., Chen C., et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e17. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley J.A., Richert L.E., Swain S.D., Harmsen A., Barnard D.L., Randall T.D., Jutila M., Douglas T., Broomell C., Young M., Harmsen A. Inducible bronchus-associated lymphoid tissue elicited by a protein cage nanoparticle enhances protection in mice against diverse respiratory viruses. PLoS ONE. 2009;4:e7142. doi: 10.1371/journal.pone.0007142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Woodland D.L., Randall T.D. Anatomical features of anti-viral immunity in the respiratory tract. Semin. Immunol. 2004;16:163–170. doi: 10.1016/j.smim.2004.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt L.S., Earl P.L., Liu J.Y., Smith J.M., Montefiori D.C., Robinson H.L., Moss B. Multiprotein HIV type 1 clade B DNA and MVA vaccines: construction, expression, and immunogenicity in rodents of the MVA component. AIDS Res. Hum. Retroviruses. 2004;20:645–653. doi: 10.1089/0889222041217428. [DOI] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Jia W., Wang P., Zhang S., Shi X., Wang X., Zhang L. Antibodies and vaccines against Middle East respiratory syndrome coronavirus. Emerg. Microbes Infect. 2019;8:841–856. doi: 10.1080/22221751.2019.1624482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent Advances in the Vaccine Development Against Middle East Respiratory Syndrome-Coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki475. W741-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.Z., Jiang C.L., Yu X.H., Lou C.P., Zhao D.H., Wu Y.G., Jin Y.H., Liu C.S., Kong W. Immunogenicity of lyophilized MVA vaccine for HIV-1 in mice model. Chem. Res. Chin. Univ. 2007;23:329–332. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the experimental findings of this study are available within the manuscript and are available from the corresponding author upon request. Data tables for expression counts for single-cell RNA-Seq for BAL are deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO accession GSE165747.