Abstract
SARS-CoV-2 is the etiological agent responsible for the ongoing pandemic of coronavirus disease 2019 (COVID-19). The main protease of SARS-CoV-2, 3CLpro, is an attractive target for antiviral inhibitors due to its indispensable role in viral replication and gene expression of viral proteins. The search of compounds that can effectively inhibit the crucial activity of 3CLpro, which results to interference of the virus life cycle, is now widely pursued. Here, we report that epigallocatechin-3-gallate (EGCG), an active ingredient of Chinese herbal medicine (CHM), is a potent inhibitor of 3CLpro with half-maximum inhibitory concentration (IC50) of 0.874 ± 0.005 μM. In the study, we retrospectively analyzed the clinical data of 123 cases of COVID-19 patients, and found three effective Traditional Chinese Medicines (TCM) prescriptions. Multiple strategies were performed to screen potent inhibitors of SARS-CoV-2 3CLpro from the active ingredients of TCMs, including network pharmacology, molecular docking, surface plasmon resonance (SPR) binding assay and fluorescence resonance energy transfer (FRET)-based inhibition assay. The SPR assay showed good interaction between EGCG and 3CLpro with KD ~6.17 μM, suggesting a relatively high affinity of EGCG with SARS-CoV-2 3CLpro. Our results provide critical insights into the mechanism of action of EGCG as a potential therapeutic agent against COVID-19.
Keywords: SARS-CoV-2, Traditional Chinese Medicine, Epigallocatechin-3-gallate
1. Introduction
The ongoing pandemic of coronavirus disease 2019 (COVID-19) has caused a major threat to public health and the global economy [1,2]. COVID-19 results from the infection of a severe acute respiratory syndrome (SARS)-like coronavirus named SARS-CoV-2 [3], whose RNA genome is closely similar to Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) [4]. SARS-CoV-2 is the seventh coronavirus that is able to infect humans with contagious and infective characteristics [5]. According to World Health Organization (WHO), the virus has infected thus far more than 53 million people worldwide, resulting the death of 1,308,975 people as of 15 November 2020 [6]. Effective pharmacological interventions are therefore urgently needed. Since the first report of COVID-19 in late 2019, relevant progress has been achieved in drug discovery and development within a remarkable timeline [[7], [8], [9], [10], [11]]. Several vaccines and drugs are undergoing clinical trials, but research on novel antivirals remains important. This is because of the growing clinical needs and also of the possible viral mutations that may render previous treatments ineffective.
In terms of drug discovery approaches against coronaviruses, the main protease (Mpro) has been identified as a highly promising drug target [12]. Mpro, also known as 3-chymotrypsin-like protease (3CLpro) or chymotrypsin-like protease, operates at no less than 11 cleavage sites on the large polyprotein to produce proteins necessary for the replication of virus [13]. Inhibiting the activity of 3CLpro will therefore block the viral replication. Since no human proteases are known with a similar cleavage specificity, targeting 3CLpro offers the advantage of virus-specific effect [14,15]. Moreover, 3CLpro is highly conserved in coronaviruses (CoVs). In fact, the sequence similarity between SARS-CoV and SARS-CoV-2 is as high as 96.08% [15,16]. Acting as a broad antiviral treatment option, drugs targeting 3CLpro can be crucial not only to inhibit SARS-CoV-2 but as well as other future coronavirus variants. For these reasons, 3CLpro has been well studied particularly following the SARS-CoV outbreak in 2002 [12,17,18]. The past experience with SARS-CoV is providing important foundational knowledge for the development of anti-SARS-CoV-2 drugs.
There are several ways to obtain small molecules that could be developed as viral target protein inhibitors. The first major option is in silico screening of large chemical libraries to select only the molecules that best fit with the target of interest for synthesis and experimental testing [19]. Other option is through structure-based drug design and chemical synthesis, such as the broad-spectrum inhibitor N3 that inhibits 3CLpro of SARS-CoV, MERS-CoV and SARS-CoV-2 [14,20,21]. An additional approach is bioprospecting in which potent compounds are identified and isolated from natural sources e.g. plant varieties. Over the past few decades, natural compounds of plant-based origin have been extensively studied as an exciting class of pharmacologically active molecules [[22], [23], [24]]. In particular, many natural products have demonstrated potent activity against CoVs [25]. Glycyrrhizic acid, baicalin, quercetin have been reported to inhibit the replication of SARS-CoV in vitro [[26], [27], [28]]. Also, natural products including betulinic acid, indigo, flavone amentoflavone, and luteolin have also been identified to inhibit the enzymatic activity of SARS-CoV 3CLpro [29,30]. Hence, the screening of inhibitors for SARS-CoV-2 3CLpro from natural products is a worthwhile direction.
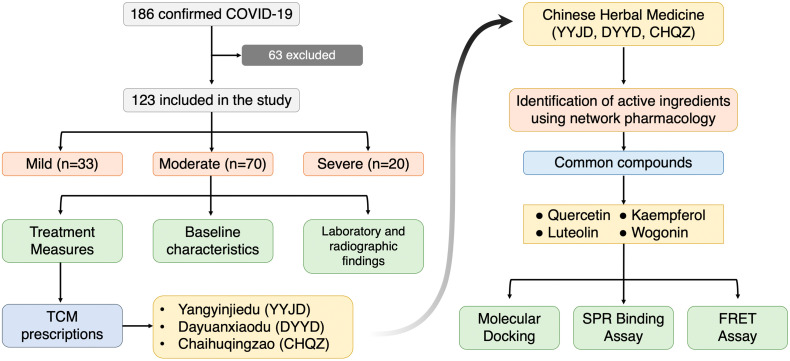
Chinese herbs are great sources of natural compounds. Many drugs that are used in the clinics were derived from Chinese herbs. Traditional Chinese Medicine has played an important role in the treatment of the past epidemics caused by viral infections. Notably, it has received renewed attention during the outbreak of COVID-19. Following reports of good clinical efficacy, several TCMs were officially endorsed for clinical use in China and were adopted as part of the treatment plan for COVID-19 [[31], [32], [33]]. An increasing number of clinical and experimental evidences have proved that TCMs inhibit viral replication owing to specific active ingredients [[34], [35], [36]]. Herein, we retrospectively analyzed 123 COVID-19 patients who received a combination of western and Traditional Chinese Medicines from the hospitals in Guizhou Province, China. Three TCM prescriptions were found to be effective. Multiple computational and experimental strategies were adopted to analyze the active components of the three TCMs. Seven active ingredients with high affinity with the SARS-CoV-2 3CLpro were identified (Fig. 1 ). Thereafter, in vitro binding and inhibition assays were performed.
Fig. 1.
A flow diagram illustrating the research design.
2. Materials and methods
2.1. Study design and participants
A total of 186 medical records of confirmed COVID-19 cases diagnosed between January 19, 2020 and March 10, 2020 in Mountain General Hospital of Guizhou, China were studied retrospectively. Incomplete records and children below the age of 18 were excluded, yielding 123 patients qualified for the study. The Mountain General Hospital in Guiyang City was designated by the provincial government as the COVID-19 referral hospital of Guizhou province. All patients included in this study were diagnosed according to the interim guideline of the World Health Organization. This study was approved by the Ethics Committee of Guizhou Provincial People's Hospital (Reference No.: 2020 504).
2.2. Collection of clinical data
The clinical, laboratory and radiological characteristics as well as treatment and clinical outcomes (recovered/expired, hospital stay) were obtained from electronic medical records and reviewed by a team of trained physicians. The data included gender, age, pre-existing medical conditions, date of onset of symptoms, date of admission, date of discharge, clinical diagnosis, laboratory tests, imaging data and treatment received. The treatment regimens included standard care, antiviral drugs, intravenous corticosteroids, intravenous γglobulin, blood profusion, respiratory supports, and the three TCM prescriptions [Yangyinjiedu (YYJD), Dayuanxiaodu (DYXX) and Chaihuqingzao (CHQZ)]. The TCM formulas were procured from Guizhou Province Diagnosis and Treatment Protocol for COVID-19.
2.3. Screening of active ingredients and their target genes
We retrieved the chemical composition of the three CHM formulas from Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://tcmspw.com/tcmsp.php), a unique system pharmacology platform of CHM that provides pharmacokinetic information for each compound such as oral bioavailability (OB), drug-like (DL) index and blood-brain barrier (BBB). OB is often used to evaluate the efficacy of oral drugs while DL index represents similarity between components and known chemical drugs. The DL index is a good assessment tool if a compound has a potential to become a drug. Thus, OB and DL index are considered here as important reference criteria for evaluating whether compounds can be used as drugs. In this study, OB ≥ 30% and DL ≥ 0.18 were used as screening threshold. Then, the targets of such compounds were obtained from the TCMSP database, in which the components not included in TCMSP were predicted through The Encyclopedia of Traditional Chinese Medicine database (ETCM; http://www.tcmip.cn/ETCM/index.php/Home/). Finally, the target proteins were identified using UniProt Database (https://www.uniprot.org/). The associated genes to COVID-19 were searched in GeneCard database (https://www.genecards.org) and NCBI (https://www.ncbi.nlm.nih.gov/) using the keyword “coronavirus pneumonia”.
2.4. Molecular docking of core compounds with 3CLpro of SARS-CoV-2
The seven most frequent compounds common among the three CHM formulas were docked with the key target SARS-CoV-2 3CLpro hydrolase. Chem3D software was used to draw the 3D structures of the core compounds and to optimize the energy. The protein crystal structure of 3CLpro (PDB: 6LU7) was downloaded from the Protein Data Bank (PDB) database (https://www.rcsb.org/). Pymol software (https://pymol.org/2/) was used to remove water molecules and heteromolecules while AutoDock Vina1.1.2 software (http://autodock.scripps.edu/resources/adt) was used for molecular docking. The compound with the lowest binding energy score conformation was selected.
2.5. SPR binding assay
SPR studies were performed on an Open SPR instrument (Nicoya Life Science, Inc., Kitchener, Canada) at room temperature. Briefly, a COOH sensor chip (Nicoya #SEN-AU-100-10-COOH) was installed according to the Open SPR standard operating procedures, and then, phosphate buffered saline (PBS, pH 7.4) was run at a maximum flow rate of 150 μL/min. After reaching the baseline of the signal, the air bubbles were evaporated with 200 μL of 80% isopropanol (IPA) and the sample loop was washed with buffer. Buffer flow rate was adjusted to 20 μL/min until the signal reached the baseline and 200 μL of 1-ethyl-3-(3-(dimethylamino)propyl) carbodiimide hydrochloride (EDC)/Nhydroxy succinimide (NHS) solution was loaded. Then the injection instrument of the 200 μL purified 3CLpro was operated for 4 min, diluted with activation buffer (sodium acetate pH = 4.5, 10 mM). After the 3CLpro protein-chip interaction ended, the upper sample mouth was rinsed with buffer and emptied by air. Thereafter, different concentrations of compounds were injected separately on the surface of the ligand chip (the concentration is detailed in the experimental results), and the analyte was sampled at 20 μL/min. The binding time of the analyte to the ligand was 240 s; and the natural dissociation 180 s was carried out. Finally, the kinetic parameters of the binding reactions were calculated and analyzed by Trace Drawer software (Ridgeview Instruments AB, The Kingdom of Sweden). The purity of SARS-CoV-2 3CLpro recombinant protein (Novoprotein #CR76) was over 95%.
2.6. Thermal shift assay
The thermal shift assay (TSA) was performed using CFX96 Touch Real-Time PCR Systems (Bio-Rad Laboratories, USA). The 3CLpro protein (Novoprotein #CR76) was equilibrated in sodium phosphate 50 mM, pH 8 and SYPRO Orange at a final concentration of 5× (Thermo Fisher Scientific), both with and without ligands. The samples were freshly prepared and dispensed into multiplate 96-well PCR plates (catalog #MLL9601) at a final volume of 50 μL. The initial thermal denaturation temperature was set at 25 °C for 2 min. The temperature was then systematically increased in 0.5–1.0 °C up to final temperature of 95 °C, with concomitant monitoring of fluorescence emission at the end of every 1 min hold at each temperature. Two methods were used to calculate the melting temperature (Tm) as previously described [37]. In brief, one method was used to determine the melting temperature from nonlinear fitting of the thermal denaturation data. The other method involved calculating the first derivative of fluorescence emission with respect to temperature. Since thermal stability of 3CLpro with the addition of quercetin was recently reported [16], quercetin was used as a positive control to test the feasibility of the PCR system. To assess the concentration dependence of the stability change induced by EGCG, 2-fold serial dilutions ranging from 0 to 250 μM were assayed by following the same protocol described above.
2.7. 3CLpro enzyme activity inhibition test
A fluorescence resonance energy transfer (FRET) protease assay was applied to measure the inhibitory activity of compounds against the SARS-CoV-2 3CLpro. The fluorogenic substrate (Dabcyl-TSAVLQ↓SGFRKMK-Edans) was synthesized by GenScript. The FRET-based protease assay was performed as follows. The recombinant SARS-CoV-2-3CLpro was mixed with serial dilutions of each compound, oral liquid or the dissolved lyophilized powder in 80 μL assay buffer (50 mM Tris–HCl, pH 7.3, 1 mM EDTA) and incubated. The reaction was initiated by adding 40 μL fluorogenic substrate with a final concentration of 20 μM. After that, the fluorescence signal at 360 nm (excitation)/490 nm (emission) was immediately measured every 30 s for 10 min with a BioTek Synergy4 plate reader. The Vmax of reactions added with compounds at various concentrations compared to the reaction added with DMSO were calculated and used to generate IC50 curves. For each compound, at least three independent experiments were performed for the determination of IC50 values.
2.8. Statistical analysis
Continuous variables were expressed as mean, median and interquartile (IQR). Categorical data were expressed as number (%). Categorical variables were compared by Chi-square (χ2) test. Line graphs were drawn to describe laboratory parameters. The distribution of normality was checked by Kolmogorov-Smirnov test. When data were not normally distributed, Mann-Whitney test was used to determine statistical significance. A p value of <0.05 was considered significant. Statistical analyses were performed using the SPSS software, version 23.
3. Results
3.1. Clinical features
Out of the 186 admitted patients between January 19 and March 10, 2020, 123 satisfied our inclusion criteria for this retrospective study (Table 1). Of these 123, 33 cases were classified as mild (26.8%), 70 were moderate (56.9%) and 20 were severe (16.3%). The median age was 36.0 years (IQR 24–48) with 71 (57.7%) being male. Age in the three groups (mild, moderate and severe) was statistically significant (IQR, 24–28; range, 18–72 years; p < 0.001), suggesting that age is associated with disease severity. The median length of hospital stay for all groups was 10 days (IQR, 6–15) whereas for mild patients it was 9 days (IQR, 6–15), 10 days (IQR, 6–14) for moderate patients, and 15 days (IQR, 4–20) for severe patients. There is statistical difference of hospital stay among the groups (p = 0.031). The common comorbidities in our patient cohort were hypertension (13 cases, 10.6%). Others included diabetes (9 cases, 7.5%), chronic liver disease (7 cases, 5.5%) cardiovascular disease (6 cases, 4.9%) and chronic kidney disease (1 case, 0.8%). The most common symptoms were fatigue (95 [77.2%]), fever (79 [64.2%]), headache (68 [55.3%]), dry cough (59 [48.0%]), and anorexia (59 [48.0%]). Among the rare symptoms included acute respiratory distress syndrome (5 [4.1%]), dyspnea (4 [3.3%]), diarrhea (12 [9.8%]), abdominal pain, nausea and vomiting. Compared with mild and moderate patients, severe patients had a longer hospital stay, a higher incidence of potential complications, and are more likely to have fever, headache, sore throat, cough, fatigue, anorexia, abdominal pain, diarrhea, and other symptoms.
Table 1.
Baseline characteristics of coronavirus disease 2019 (COVID-19) patients.
| Characteristics | Total (n = 123) | Disease severity |
p value | ||
|---|---|---|---|---|---|
| Mild (n = 33) | Moderate (n = 70) | Severe (n = 20) | |||
| Age, years, median (IQR) | 36 (24, 48) | 30 (18, 41) | 36 (24, 47) | 49 (35, 60) | <0.001 |
| Sex | |||||
| Male | 70 (57.0) | 17 (51.5) | 42 (60.0) | 11 (55.0) | |
| Female | 53 (43.1) | 16 (48.4) | 28 (40.0) | 9 (45.0) | |
| Hospitalization time/days | 10 (6, 15) | 9 (5, 15) | 10 (6, 14) | 15 (4, 20) | 0.031 |
| Comorbidities | |||||
| Hypertension | 13 (10.6) | 3 (9.1) | 6 (8.5) | 4 (20.0) | 0.256 |
| Cardiovascular disease | 6 (4.9) | 0 (0) | 5 (7.1) | 1 (5.0) | 0.473 |
| Diabetes | 8 (6.5) | 3 (9.1) | 4 (5.7) | 2 (10.0) | 0.544 |
| COPD | 4 (3.3) | 0 (0) | 2 (2.9) | 2 (10.0) | 0.143 |
| Chronic liver disease | 7 (5.5) | 2 (6.1) | 4 (5.7) | 1 (5.0) | 1.000 |
| Chronic kidney disease | 1 (0.8) | 0 (0) | 1 (1.4) | 0 (0) | 1.000 |
| Symptoms and signs | |||||
| Fever | 79 (64.2) | 5 (15.2) | 56 (80.0) | 18 (90.0) | <0.001 |
| Cough | 59 (48.0) | 8 (24.2) | 35 (50.0) | 18 (90.0) | <0.001 |
| Fatigue | 95 (77.2) | 20 (60.6) | 55 (78.5) | 20 (100) | 0.005 |
| Dyspnea | 4 (3.3) | 1 (3.0) | 3 (4.3) | 5 (25.0) | 0.001 |
| Anorexia | 59 (48.0) | 22 (66.7) | 37 (52.9) | 17 (85.0) | <0.001 |
| Headache | 68 (55.3) | 26 (78.8) | 24 (34.3) | 18 (90.0) | <0.001 |
| Myalgia | 47 (38.2) | 12 (36.4) | 17 (24.2) | 18 (90.0) | <0.001 |
| Pharyngalgia | 44 (35.8) | 6 (18.2) | 25 (35.7) | 13 (65.0) | <0.001 |
| Dizziness | 36 (29.3) | 5 (15.2) | 14 (20.0) | 17 (85.0) | <0.001 |
| Night sweats | 63 (51.2) | 18 (54.5) | 30 (42.8) | 15 (75.0) | 0.001 |
| Dry mouth | 51 (41.5) | 9 (27.2) | 34 (48.6) | 8 (40.0) | 0.001 |
| Diarrhea | 12 (9.8) | 0 (0) | 8 (11.4) | 4 (20.0) | 0.034 |
| Nausea | 18 (14.6) | 3 (9.0) | 9 (12.8) | 6 (30.0) | 0.053 |
| Vomiting | 23 (18.7) | 2 (6.0) | 14 (20.0) | 7 (35.0) | 0.016 |
| Abdominal pain | 22 (17.9) | 0 (0) | 17 (24.3) | 5 (25.0) | 0.010 |
| ARDS | 5 (4.1) | 0 (0) | 1 (1.4) | 4 (20.0) | 0.002 |
IQR = interquartile range. Data are presented as median (IQR), n (%). p values indicate differences of the different groups of varying disease severity (mild, moderate and severe). p < 0.05 was considered significant. COPD, chronic obstructive pulmonary disease; ARDS acute respiratory distress syndrome. One patient died of ARDS.
3.2. Laboratory tests and radiographic findings
Laboratory values and radiographic findings at the time of admission are presented in Table 2 . Regardless of the disease severity, all 123 patients had white blood cell, neutrophil, lymphocyte, and platelet counts within the normal ranges. At the time of admission, the median values for C-reactive protein, erythrocyte sedimentation rate, procalcitonin, creatine kinase, creatine kinase isoenzyme, alanine aminotransferase, aspartate aminotransferase, blood urea nitrogen, creatinine, and lactate dehydrogenase were also all within the normal ranges. There was no statistically significant difference among the groups (p > 0.05). Compared with mildly and moderately ill patients, severe patients had higher D-dimer levels (0.11 [0.07–0.42], p = 0.021). The number of neutrophils, C-reactive protein, GGT, and procalcitonin were also significantly higher in severely ill patients. The imaging data indicated that 74 [60.2%] patients showed bilateral distribution of patchy shadows in chest CT scans. There was no statistically significant difference between the groups (p > 0.05).
Table 2.
Laboratory values and radiographic findings of COVID-19 patients at the time of admission.
| Laboratory test | Normal range | Total (n = 123) | Disease severity |
p value | ||
|---|---|---|---|---|---|---|
| Mild (n = 33) | Moderate (n = 70) | Severe (n = 20) | ||||
| White blood cell count, ×109/L | 3.5–9.5 | 5.3 (4.4–6.2) | 5.1 (4.4–5.6) | 5.4 (4.5–6.6) | 5.0 (4.3–5.6) | 0.203 |
| Neutrophil count, ×109/L | 1.8–6.3 | 3.3 (2.2–5.6) | 2.6 (1.8–3.4) | 3.8 (2.5–6.3) | 3.9 (2.8–9.7) | 0.108 |
| Lymphocyte count, ×109/L | 1.1–3.2 | 1.6 (1.3–2.1) | 1.6 (1.4–1.7) | 1.6 (1.2–2.1) | 1.6 (1.4–1.9) | 0.214 |
| Platelet count, ×109/L | 125–350 | 206 (175–257) | 188 (168–226) | 213 (185–275) | 187 (165–222) | 0.064 |
| Procalcitonin ng/mL | <0.5 | 0.05 (0.01–0.20) | 0.02 (0.04–0.05) | 0.10 (0.05–0.20) | 0.16 (0.10–0.29) | 0.302 |
| Erythrocyte sedimentation rate, mm/h | 2–25.7 | 18 (11–32.2) | 17 (14.2–35) | 18 (11–30.2) | 17.5 (11–33.7) | 0.201 |
| Creatine kinase, U/L | 0–190 | 50 (35–72) | 56 (40,72) | 51 (32–70) | 43 (33.3–75.7) | 0.591 |
| Creatine kinase-MB, U/L | 0–24 | 11 (8–15) | 11.5 (8.3–13.0) | 11 (8–17) | 10 (8.3–12) | 0.629 |
| Alanine aminotransferase, U/L | 7–40 | 19 (16–26) | 21 (16–30.5) | 20 (15–26) | 18 (15.3–22.7) | 0.417 |
| Aspartate aminotransferase, U/L | 13–35 | 26 (17–42) | 24 (16.3–41.7) | 29 (18–45) | 24 (15.3–39) | 0.474 |
| Glutamyl transpeptidase | 11–49 | 16.4 (26.2–52.1) | 15.4 (22.2–31.6) | 28.8 (18.0–63.4) | 30.0 (15.0–47.7) | 0.427 |
| Blood urea nitrogen, mmol/L | 2.8–7.6 | 3.2 (2.8–4.0) | 3.2 (2.8–3.8) | 3.2 (2.8,4.1) | 3.2 (2.5–3.8) | 0.564 |
| Creatinine, μmol/L | 64–104 | 65.8 (53–77) | 68.9 (58.1–80.3) | 65.8 (50.5–78.2) | 60.6 (52.6–73.7) | 0.488 |
| Hypersensitive troponin I, pg/mL | <0.2 | 0.01 (0.01–0.01) | 0.01 (0.01–0.01) | 0.01 (0.01–0.01) | 0.01 (0.01–0.01) | 0.390 |
| Lactate dehydrogenase, U/L | 120–250 | 124 (112–153) | 118 (107–130) | 130 (114–156) | 122 (112–158) | 0.077 |
| C-reactive protein, mg/L | 0–5 | 0.6 (0.2–2.0) | 0.3 (0.1–0.8) | 0.5 (0.1–2.1) | 0.6 (0.3–2.2) | 0.065 |
| D-dimer, μg/mL | 0–1.5 | 0.09 (0.06–0.27) | 0.08 (0.06–0.09) | 0.09 (0.07–0.21) | 0.11 (0.07–0.42) | 0.021 |
| Bilateral distribution of patchy shadows or ground glass opacity, no. (%) | NA | 74 (60.2) | 11 (33.3) | 48 (68.5) | 15 (75) | 0.276 |
Data are presented as median (IQR) or n (%). p values indicate difference among groups (mild, moderate and severe). p < 0.05 was considered significant. NA, not available.
3.3. Treatment and outcomes
Drug treatment included commercially available drugs and TCMs including antivirals, antibiotics, antihypertensive drugs, lipid-lowering drugs, digestive system drugs, respiratory drugs, anti-hypoglycemics, antipyretics, analgesics and others. Antihypertensive drugs, lipid-lowering drugs, hypoglycemic drugs, and gastrointestinal medications were mainly used for the treatment of patients with underlying diseases, whereas respiratory drugs, antipyretics and analgesics were used for symptomatic treatment. Table 3 shows the antiviral drugs, antibacterial drugs, and Chinese medicines used to treat COVID-19. Most patients received antiviral therapy (lopinavir/ritonavir, 120 [97.6%]; abidol, 49 [39.8%]; interferon 115 [93.5%]), antibacterial therapy (moxifloxacin, 47 [38.2%]) and glucocorticoid therapy (34 [27.6%]). The most frequent TCM decoctions administered to patients as part of symptomatic treatment were YYJD (44 [35.8%]), DYXD (27 [22.0%]), and CHQZ (23 [18.7%]). The commonly used drugs-abidol, moxifloxacin, thymosin α1, corticosteroids- were statistically different among groups (p < 0.05). Of the 20 severe patients, 4 (3.3%) were admitted to the ICU and all received high-flow oxygen. Among them, one patient received non-invasive ventilation while others were treated with convalescent plasma therapy. There was no statistically significant difference in the application of other drugs among groups as well as the use of Chinese medicine.
Table 3.
Treatment regimens of coronavirus disease 2019 (COVID-19) patients.
| Antiviral drugs | Total (n = 123) | Mild (n = 33) | Moderate (n = 70) | Severe (n = 20) | p value |
|---|---|---|---|---|---|
| Abidol | 49 (39.8) | 6 (18.2) | 23 (32.9) | 20 (100) | 0.000 |
| Interferon | 115 (93.5) | 30 (90.9) | 65 (92.8) | 20 (100) | 0.397 |
| Lopinavir/ritonavir | 120 (97.6) | 30 (90.9) | 70 (100) | 20 (100) | 0.364 |
| Traditional Chinese Medicine | |||||
| Yangyinjiedu formula | 44 (35.8) | 8 (24.2) | 28 (40.0) | 8 (40.0) | 0.297 |
| Dayuanxiaodu formula | 27 (22.0) | 5 (15.2) | 18 (25.7) | 4 (20.0) | 0.816 |
| Chaihuqingzao formula | 23 (18.7) | 6 (18.2) | 10 (14.2) | 7 (35) | 0.004 |
| Sanren formula | 11 (8.9) | 2 (6.1) | 9 (12.8) | 0 (0) | 0.327 |
| Shenlingbaizhu san | 5 (4.1) | 2 (6.1) | 2 (2.9) | 1 (5.0) | 0.602 |
| Maimendong formula | 9 (7.3) | 1 (3.0) | 6 (8.6) | 2 (10.0) | 0.581 |
| Chaigechangyuan formula | 5 (4.1) | 0 (0) | 5 (7.1) | 0 (0) | 0.198 |
| Fuzhengjiedu powder | 5 (4.1) | 0 (0) | 3 (4.3) | 2 (10.0) | 0.173 |
| Belamcanda and Ephedre formula | 3 (2.4) | 0 (0) | 3 (4.3) | 0 (0) | 0.400 |
| Moxifloxacin | 47 (38.2) | 5 (15.2) | 26 (37.1) | 16 (80.0) | 0.000 |
| Thymosin alpha-1 | 87 (70.7) | 13 (39.4) | 54 (77.1) | 20 (100) | 0.000 |
| Corticosteroids | 34 (27.6) | 4 (12.1) | 12 (17.1) | 18 (90.0) | 0.000 |
Data are presented as n (%). p values indicate differences different groups (mild, moderate and severe). p < 0.05 was considered significant.
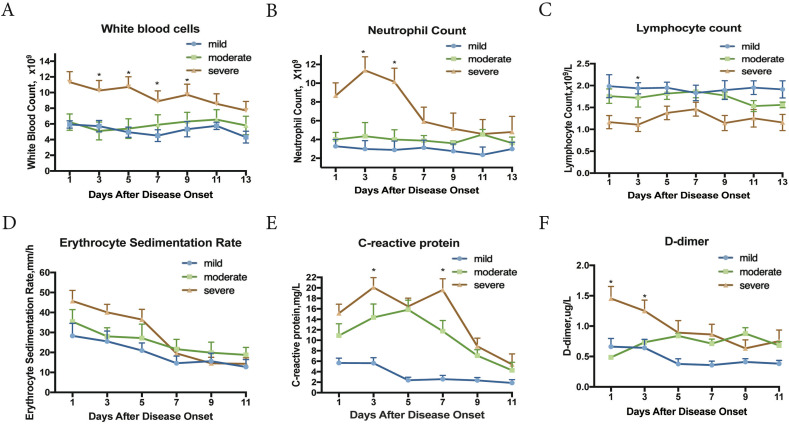
3.4. Dynamic profile of laboratory parameters in 60 patients with COVID-19
To determine the clinical features associated with COVID-19 progression, six clinical laboratory parameters were tracked in 60 patients every 2 days for a period of two weeks from the onset of symptoms (Fig. 2 ). Biochemical indicators (white blood cell, neutrophil, erythrocyte sedimentation rate, C-reactive protein, and D-dimer) in severe patients were higher than those of mild and moderate patients. But at the end of hospital admission, these biochemical parameters in severe patients decreased. After 1 week of hospitalization, all these laboratory findings in all our patient types subsided, which was related to faster recovery and shorter hospital stay. Taken the clinical data altogether, the use of TCMs seemed effective regardless of disease severity and appeared to modulate immune responses.
Fig. 2.
Dynamic profile of laboratory parameters in 60 patients (mild = 8, moderate = 32, and severe = 20 with COVID-19). Timeline charts illustrate the laboratory parameters taken every other day from the day after the onset of illness. p values indicate differences among different groups (mild, moderate and severe). *p < 0.05 was considered a statistically significant difference.
Our patient cohort received 9 different TCM prescriptions in total. Among these, three prescriptions, namely, Yangyinjiedu (YYJD), Dayuanxiaodu (DYXD) and Chaihuqingzao (CHQZ) were selected for further evaluation because they were used most frequently. A network pharmacology method was conducted to investigate their effectiveness for COVID-19. We screened the active ingredients of the three TCMs and predicted the component-target network (Fig. S1). Thereafter, we performed gene ontology (GO) enrichment analysis and KEGG pathway enrichment analysis to elucidate the diverse mechanisms of these TCMs at systematic level. Among the GO terms under molecular function were cytokine receptor binding, cytokine activity, and receptor ligand activity (Supplementary Fig. S2A, B, and C). In our KEGG analysis, the most enriched pathways were associated with immune and antiviral responses. These pathways included TNF signaling, IL-17 signaling, influenza A, C-type lectin receptor signaling pathway, and others (Supplementary Fig. S3). These GO, KEGG as well as the clinical data all showed the immunomodulatory effects of these three TCMs.
3.5. Acquisition of main active compounds and molecular docking
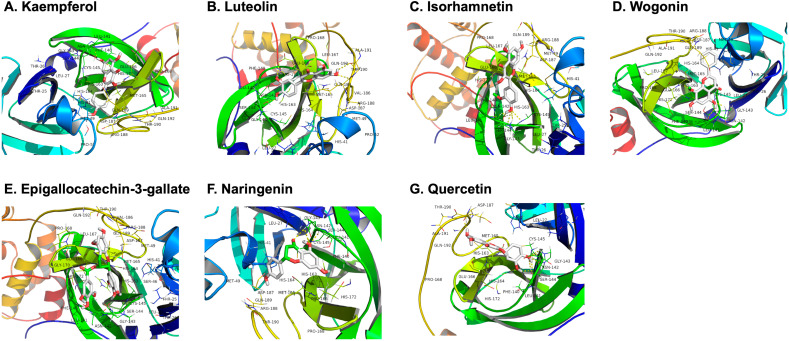
The active compounds of the three TCM prescriptions were retrieved from the TCMSP and ETCM databases (Table S1, Table S2, Table S3). Some active ingredients are common among all the three TCMs. Thereafter, we further determined the frequencies of these ingredients and noted their curated mechanism of actions. This screening strategy resulted to 7 compounds that are possibly responsible for the anti-COVID-19 properties of the TCMs. These compounds are quercetin, kaempferol, luteolin, isorhamnetin, epigallocatechin-3-gallate, naringenin, and wogonin. These compounds may underpin the antiviral activity of these TCMs, but the exact mechanism is unknown. While every viral enzyme of SARS-CoV-2 is a potential drug target, 3CLpro was selected to be the focus of our further analyses because of its indispensable function in the viral life cycle. The replication of SARS-CoV-2 is mediated by replicase polyprotein, consisting of pp1a and pp1ab. This polyprotein is then cleaved to form 16 functional polypeptides, also called non-structural protein, via extensive proteolytic processing predominantly by 3CLpro [38]. Thus, 3CLpro plays a critical function in the life cycle of SARS-CoV-2. More so, the cleavage sites of 3CLpro are highly conserved in CoVs with no known human proteases having similar cleavage specificity, providing added advantage of possible less toxicity of 3CLpro inhibitors [15]. With these reasons–functional importance in viral life cycle, high similarity of 3CLpro among CoVs, and cleavage specificity-, targeting 3CLpro of SARS-CoV-2 is an attractive antiviral strategy. We therefore docked the compounds with 3CLpro using AutoDock Vina1.1.2. It is generally believed that the lower energy of the conformational stability of the ligand and the receptor, the greater the possibility of interaction. When the binding energy ≤ − 5.0 kcal/mol is taken as the screening criterion, the results of molecular docking showed that all the compounds were all lower than −5 kcal/mol (Table 4). This indicates that they had a certain affinity for the protein crystal structure of 3CLpro (Fig. 3). Therefore, these compounds have good binding activity with 3CLpro. More so, the conformation of the conjugate was stable, indicating that these may indeed be the key components of the TCMs. Of all the 7 compounds, epigallocatechin-3-gallate (EGCG) has the lowest binding energy of −7.9 kcal/mol as well as highest DL index of 0.77.
Table 4.
Docking mode of the common active ingredients of Yangyinjiedu (YYJD), Dayuanxiaodu (DYXX) and Chaihuqingzao (CHQZ) with SARS-CoV-2 3CLpro. The oral bioavailability (OB) and drug-like (DL) index were retrieved from TCMSP database (http://tcmspw.com/tcmsp.php).
| Compound | Formula | MW (g/mol) | OB (%) | DL index | 3CLpro (kcal/mol) |
|---|---|---|---|---|---|
| Epigallocatechin-3-gallate | C22H18O11 | 458.40 | 55.09 | 0.77 | −7.9 |
| Kaempferol | C15H10O6 | 286.24 | 41.88 | 0.24 | −7.8 |
| Quercetin | C15H10O7 | 302.24 | 46.43 | 0.28 | −7.4 |
| Luteolin | C15H10O6 | 286.24 | 36.16 | 0.25 | −7.4 |
| Isorhamnetin | C16H12O7 | 316.26 | 49.6 | 0.31 | −7.1 |
| Naringenin | C15H12O5 | 272.25 | 59.29 | 0.21 | −7.1 |
| Wogonin | C16H12O5 | 284.26 | 30.68 | 0.23 | −7.0 |
Fig. 3.
Molecular models of the binding of SARS-CoV-2 3CLpro with kaempferol (A), luteolin (B), isorhamnetin (C) and wogonin (D), epigallocatechin-3-gallate (E), naringenin (F), and quercetin (G) shown as 3D diagrams.
The 3CLpro of SARS-CoV-2 is made up of 306 amino acid residues. The active site pocket consists of Thr25, Thr26, Leu27, His41, Ser46, Met49, Tyr54, Phe140, Leu141, Asn142, Gly143, Cys145, His163, Met165, Glu166, Leu167, Pro168, Phe185, Asp187, Gln189, Thr190, Ala191, and Gln192 [60]. In our molecular docking analysis, the key active sites were Met49, Phe140, Leu141, Ser144, His163, His164, His172, Glu-166, Met165, and Thr190. Interestingly, EGCG formed hydrogen bond with His41 and Cys145 (Fig. 3E). Besides these two key residues, several other amino acid residues in the active site were also involved in hydrogen bonding as well as different noncovalent interactions. Similarly, quercetin is also able to form various interactions with a number of residues of the binding pocket [16]. The key residue is Met165, which can form both hydrogen bonds and hydrophobic interactions with quercetin. Additional hydrogen bonds can be formed with the two residues Ser144 and Met165. Energetic contributions are also due to supplementary hydrophobic interactions with Met49, Phe140 and Leu141, and to electrostatic interaction formed with the polar residue His164 and the charged residue Glu166. All in all, EGCG and quercetin share certain similarity in terms of binding affinity with some key residues in the active site of 3CLpro. But since EGCG appears to interact with the catalytic residues of the 3CLpro protease and exhibited a more favorable binding affinity, it is likely that EGCG possibly inhibits the catalytic activity of 3CLpro. This therefore renders EGCG as a good candidate for drug development for the treatment of COVID-19.
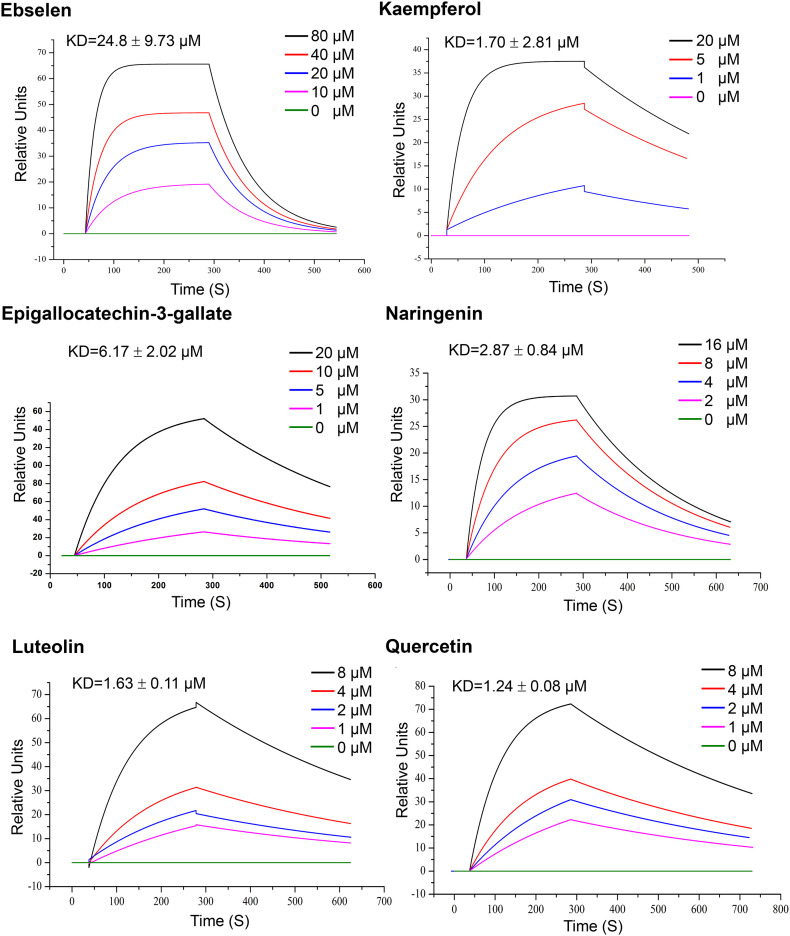
3.6. 3CLpro of SARS-CoV-2 binds with the active ingredients
Our previous analysis suggested that the 7 active compounds might underpin the antiviral activity of the 3 TCMs by binding with the 3CLpro of SARS-CoV-2. To gain deeper insights on the interactions between 3CLpro and the core compounds, an SPR biosensor-based assay was performed. SPR is an optical biosensor detection method that measures the interaction between ligand and analyte by monitoring the change in refractive index of the surface interface that occurs during the binding process. Following the immobilization of 3CLpro on the sensor COOH chip, the compounds were passed over the sensor's surface. The compound-protein binding reaction at different concentrations was continuously recorded, expressed in resonance units (RUs) and presented as a function of time (Fig. 4). The SPR analysis showed that all compounds increased the SPR sensorgram significantly in a dose-dependent manner. The dissociation constant KD (koff/kon) was calculated by globally fitting the kinetic data at various analyte concentrations to the Langmuir binding model. The calculation results show that the binding affinity of 3CLpro to quercetin, luteolin, kaempferol, naringenin, and epigallocatechin-3-gallate were 1.24 μM, 1.63 μM, and 1.70 μM, 2.87 μM, and 6.17 μM respectively. Ebselen served as positive control as it has been reported to inhibit 3CLpro activity with IC50 of 0.67 ± 0.09 μM and EC50 of 4.67 ± 0.80 μM [27]. Though wogonin and isorhamnetin were also predicted to have favorable affinity with 3CLpro, they were not analyzed by SPR due to its low water solubility.
Fig. 4.
SPR assay of specific binding affinities of several active compounds to immobilized 3CLpro of SARS-CoV-2. Different concentrations of the compounds (1–80 μM) were injected separately on the surface of the ligand chip, and the analyte was sampled at 20 μL/min. The binding time of the analyte to the ligand was 240 s; and the natural dissociation was carried out for 180 s. Data are representative of three independent experiments.
3.7. Enzyme inhibitory activity of active compounds against SARS-CoV-2
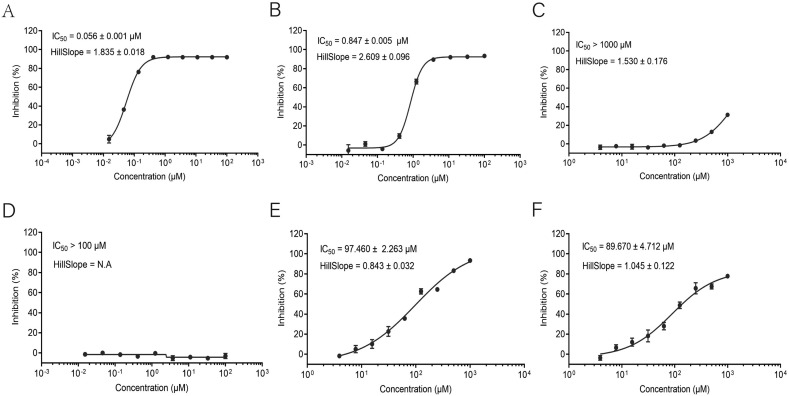
The in silico docking study and SPR binding assay both indicated that the active compounds have certain affinity with 3CLpro, the key protease during SARS-CoV-2 replication. The anti-COVID-19 property of these compounds is possibly due to its ability to inhibit the enzymatic activity of 3CLpro. In this regard, a fluorescence resonance energy transfer (FRET)–based cleavage assay was used to determine the median inhibitory concentration (IC50) values. As before, Ebselen was chosen as the positive control. A fluorescently-labeled substrate (Dabcyl-TSAVLQ↓SGFRKMK-Edans), derived from the N-terminal autocleavage sequence from the viral protease, was designed and synthesized for the enzymatic assay. Among the five soluble active ingredients, epigallocatechin-3-gallate has the best inhibitory effect on 3CLpro activity with IC50 of 0.847 μM, followed by quercetin and luteolin with IC50 of 97.460 μM and 89.670 μM, respectively (Fig. 5 ). Kaempferol and naringenin had IC50 of >100 μM.
Fig. 5.
Inhibitory screening of active compounds against SARS-CoV-2 3CLpro using FRET protease assay. (A) Ebselen. (B) Epigallocatechin-3-gallate. (C) Naringenin. (D) Kaempferol. (E) Quercetin. (F) Luteolin. Dose–response curves for IC50 values were determined by nonlinear regression. All data are shown as mean ± s.e.m., n = 3 biological replicates.
3.8. Thermal shift assay
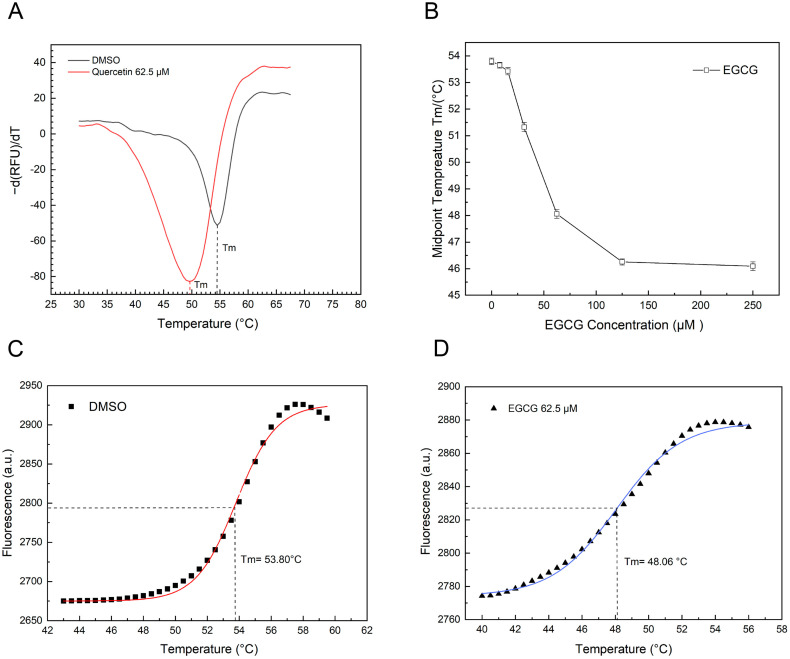
Generally, the stability of a protein can be affected by the interaction with a ligand [39]. Because of its easy use, thermal shift assay (TSA) has become an excellent platform for discovery of small molecule ligands [37]. To further characterize the potentials of EGCG as 3CLpro inhibitor, we performed TSA to determine if such compound alters thermal stability of 3CLpro. Since quercetin alters the thermal stability of 3CLpro by causing destabilization [16], we used quercetin as our positive control. To validate our own experimental system, we performed TSA on quercetin with 3CLpro. Compared with the control group (DMSO only), the addition of quercetin reduced the melting temperature (Tm) of 3CLpro (Fig. 6A), consistent with the previous report. This indicates that the experimental system is suitable. Similar to quercetin, EGCG also altered the thermal stability of SARS-CoV-2 3CLpro in a dose-dependent manner (Fig. 6B) with a melting temperature (Tm) of 53.80 °C at a concentration of 62.5 μM (Fig. 6D).
Fig. 6.
Thermal Shift Assay (TSA) of SARS-CoV-2 3CLpro stability and interaction with Epigallocatechin-3-gallate (EGCG). A. The experimental system for TSA was validated using quercetin, a compound that was previously reported to alter the thermal stability of 3CLpro by causing destabilization. The melting temperatures (Tm) was identified by plotting the first derivative of the fluorescence emission as a function of temperature (−dF/dT). Tm is represented as the lowest part of the curve. B. The melting temperature (Tm) of 3CLpro with various concentrations of EGCG, showing a dose-dependent trend. C and D. The sigmoidal curves showing the melting temperatures (Tm) of dimethyl sulfoxide (C; negative control) and 62.5 μM EGCG (D). The solid line represents the non-linear fit of the fluorescence curve to Boltzmann Equation.
4. Discussion
The lack of mass immunization programs and antiviral drugs continues to be the serious challenge to the current global effort to contain the COVID-19 epidemic. Several drugs were tested in randomized controlled clinical studies but unfortunately the vast majority of these drug interventions were not successful [[40], [41], [42]]. Therefore, different drug discovery strategies need to be considered in addition to the current widely used drug repurposing methods and structure-based drug design strategies. In this study, several natural compounds from Traditional Chinese Medicine were identified as SARS-CoV-2 3CLpro inhibitors. Here, we retrospectively evaluated medical records of patients who received a combination of Chinese and Western medicines. Three effective TCMs were identified, namely Yangyinjiedu, Dayuanxiaodu and Chaihuqingzao. Moreover, the active components of these three herbs were identified by network pharmacology. Additionally, the active compounds with potential therapeutic value for COVID-19 were screened through in vitro experiments.
In the past few years, medicinal plant-based natural compounds and Traditional Chinese Medicine have attracted much attention as important resources for the development of novel antiviral drugs. Plenty of natural products were reported to possess antiviral activities against various virus strains by in vivo and in vitro experiments, including coronaviruses, herpes simplex virus, influenza virus, human immunodeficiency virus [[43], [44], [45], [46], [47]]. In addition, TCM is widely used in clinical practice in the treatment of viral infectious diseases. For example, Lian-Hua-Qing-Wen Capsule (LHQWC), a commonly used Chinese medicine, was approved for the treatment of SARS and viral influenza [48,49]. In the height of SARS-CoV-2 outbreak in China, LHQWC was recommended again for clinical use to treat COVID-19. Subsequent clinical and experimental studies have confirmed the therapeutic benefits of LHWQC [35,50]. Its mode of action was by targeting virus replication and immunological regulation. In addition to LHQWC, several other TCMs have also been shown to be effective against COVID-19 and have been recommended for clinical use [51,52]. Therefore, it may be an economical and time-saving pursuit to screen inhibitors from effective TCM formulations.
Due to the critical function of 3CLpro in the life cycle of SARS-CoV-2, we screened several active ingredients against 3CLpro from TCMs. Our molecular docking analysis revealed that seven core compounds from the three effective TCMs are predicted to have good binding affinity with 3CLpro. Our SPR binding assay further confirmed the predicted affinity of the compounds with 3CLpro. This potential binding may directly inhibit its proteolytic activity. To test this hypothesis, we evaluated if the compounds can inhibit the enzymatic activity of 3CLpro using FRET assay. Of the soluble compounds, EGCG has the best inhibitory effect with IC50 of 0.874 ± 0.005 μM.
EGCG is a polyphenol catechin that is abundant in tea plants, especially green tea [53]. EGCG showed a wide range of antiviral activity against adenovirus, influenza virus, zika virus, herpesvirus, and hepatitis virus [54]. It has also been found to be a potential treatment option over synthetic chemical drugs. In addition, previous studies have reported that EGCG also has antiviral activity against coronavirus. In vitro studies showed that EGCG inhibited SARS-CoV 3CLpro with an IC50 value of 73 ± 2 μM [29]. Given the high similarity of 3CLpro in SARS-CoV and SARS-CoV-2, EGCG is a potential inhibitor of 3CLpro of SARS-CoV-2 and can be a lead candidate for drug development to treat COVID-19. Our molecular docking results showed that EGCG, with a docking score of −7.9, has the best in silico activity among all the compounds tested. This is consistent with the results of several recent reports [55,56]. Certain compounds bind to the allosteric site of an enzyme, resulting to conformational change of the active site of such enzyme. This results to non-competitive inhibition of its enzymatic action. However, a compound that binds to the active site of the enzyme and acts as a competitive inhibitor is a better choice. We thus explored if EGCG can bind directly with SARS-CoV-2 3CLpro. The molecular docking analysis provided an in silico evidence that EGCG binds with certain amino acid residues in the active site of 3CLpro. Furthermore, we employed SPR binding assay, a distinct biophysical technique that is routinely used to probe drug-protein engagement. The SPR results, combined with the results described above, confirmed the binding inhibitory activity of EGCG against 3CLpro.
Critically ill COVID-19 patients often show cytokine storm syndrome characterized by increased secretion of interleukin (IL)-6, IL-1β, IL-17, IL-8, tumour necrosis factor (TNF)-α, interferon (IFN)-γ, and other pro-inflammatory mediators [57]. The over-production of pro-inflammatory cytokines induces severe damage to the host. There is therefore a great clinical and research need for drugs to counteract COVID-19 hyper-inflammation. In this aspect, EGCG is potentially a promising agent for COVID-19 to revert hyperinflammation. Previous studies have demonstrated that EGCG can strongly inhibit the JAK/STAT pathway regulating the synthesis and secretion of several cytokines and chemokines [58,59]. Moreover, EGCG can suppress the canonical NF-κB pathway [60,61], which plays a key regulatory role in the expression of proinflammatory cytokines including IL-1, TNF-α, IL8, IL-6. All of these cytokines are induced in cytokine storm syndrome as well as in COVID-19. The KEGG analysis and the GO-based enrichment analyses also showed that a vast majority of the most enriched pathways of the components in the three TCMs were associated with immune response and inflammation-related signaling (Supplementary Figs. S2A, B, C and S3). As illustrated in the compound-target regulation network diagram (Fig. S1), several active components interact with a variety of targets. In particular, EGCG (ID: MOL006821) is predicted to target multiple genes. This prediction model provides a good insight but more research efforts are still needed to fully uncover the therapeutic benefits of EGCG.
In this work, we screened and identified several active ingredients of Traditional Chinese Medicine with inhibitory activity against SARS-CoV-2 3CLpro. Among them, EGCG is the most promising active compound due to its inhibitory activity. EGCG offers additional advantages conferred by its low toxicity and good absorptive capacity in human intestine [62]. Overall, EGCG warrants further study due to its promising potential in combating COVID-19 hyperinflammation and as lead compound for development of new antiviral drugs against COVID-19.
The following are the supplementary data related to this article.
Compound-target regulation network diagram. The red triangles represent targets. The surrounding ellipses represent the key active compounds.
A. Gene ontology terms of candidate targets of Yangyinjiedu (YYJD) against COVID-19. The top 20 GO functional categories with FDR < 0.05 are presented. B. Gene ontology terms of candidate targets of Dayuanxiaodu (DYXX) against COVID-19. The top 20 GO functional categories with p-adjust<0.05 are presented. C. Gene ontology terms of candidate targets of Chaihuqingzao (CHQZ) against COVID-19. The top 20 GO functional categories with p-adjust<0.05 are presented.
KEGG pathway enrichment analysis of candidate targets of A. Yangyinjiedu (YYJD), B. Dayuanxiaodu (DYXX) and C. Chaihuqingzao (CHQZ) against COVID-19. The bar graph shows the pathways with significant p-adjust <0.05.
Active ingredients of Yangyinjiedu (YYJD) herbal medicine.
Active ingredients of Dayuanxiaodu (DYXX) herbal medicine.
Active ingredients of Chaihuqingzao (CHQZ) herbal medicine.
CRediT authorship contribution statement
Ashuai Du: Resources, Data curation, Writing-original draft, Visualization. Rong Zheng: SPR analysis, FRET experiment, Writing-original draft, Software. Cyrollah Disoma, Shiqin Li, Zongpeng Chen, Sijia Li, Pinjia Liu, Yuzheng Zhou, Yilun Shen, Sixu Liu, Yongxing Zhang, Zijun Dong, Qinglong Yang, Moyed Alsaadawe, Aroona Razzaq, Yuyang Peng, Xuan Chen: Clinical data collection, Visualization Analysis, Investigation. Liqiang Hu, Jian Peng, Qianjun Zhang, Taijiao Jiang: Validation, Consultation. Long Mo: Writing-review & editing. Shanni Li, Zanxian Xia: Supervision, Methodology, Conceptualization, Writing reviewing and editing.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
Acknowledgement
We appreciate the members of Zanxian Xia's laboratory for their valuable comments and discussions. This work was supported by the National Key Research and Development Program of China [2016YFD0500300 and 2016YFC1200200 (Z. Xia)], the Key Research and Development Program of Hunan Province [2020SK2054 (Z. Xia)], the National Natural Science Foundation of China [82072293 (Z. Xia)], Zhejiang University special scientific research fund for COVID-19 prevention and control [2020XGZX033 (Z. Xia)], Changsha special scientific research fund for COVID-19 prevention and control [kq2001030 (Z. Xia)], the Central South University Graduate Research Innovation Project [2018zzts387 (Y. Zhou), 1053320184423 (S. Li), CX20200380 (S. Li), 1053320192850 (P. Liu), 1053320192068 (A. Du)], and the Central South University Innovation and Entrepreneurship Project [2018gczd016 (R. Zheng)].
References
- 1.Hui D.S., E. I.A., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., McHugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.V. Coronaviridae Study Group of the International Committee on Taxonomy of, The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2, Nat Microbiol 5(4) (2020) 536–544. [DOI] [PMC free article] [PubMed]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Q. Li, X. Guan, P. Wu, X. Wang, L. Zhou, Y. Tong, R. Ren, K.S.M. Leung, E.H.Y. Lau, J.Y. Wong, X. Xing, N. Xiang, Y. Wu, C. Li, Q. Chen, D. Li, T. Liu, J. Zhao, M. Liu, W. Tu, C. Chen, L. Jin, R. Yang, Q. Wang, S. Zhou, R. Wang, H. Liu, Y. Luo, Y. Liu, G. Shao, H. Li, Z. Tao, Y. Yang, Z. Deng, B. Liu, Z. Ma, Y. Zhang, G. Shi, T.T.Y. Lam, J.T. Wu, G.F. Gao, B.J. Cowling, B. Yang, G.M. Leung, Z. Feng, Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia, N Engl J Med 382(13) (2020) 1199–1207. [DOI] [PMC free article] [PubMed]
- 6.World Health Organization Coronavirus Disease 2019. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 7.Twomey J.D., Luo S., Dean A.Q., Bozza W.P., Nalli A., Zhang B. COVID-19 update: the race to therapeutic development. Drug Resist. Updat. 2020;53:100733. doi: 10.1016/j.drup.2020.100733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grobler J.A., Anderson A.S., Fernandes P., Diamond M.S., Colvis C.M., Menetski J.P., Alvarez R.M., Young J.A.T., Carter K.L. Accelerated preclinical paths to support rapid development of COVID-19 therapeutics. Cell Host Microbe. 2020;28(5):638–645. doi: 10.1016/j.chom.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Y. Chen, A. Zuiani, S. Fischinger, J. Mullur, C. Atyeo, M. Travers, F.J.N. Lelis, K.M. Pullen, H. Martin, P. Tong, A. Gautam, S. Habibi, J. Bensko, D. Gakpo, J. Feldman, B.M. Hauser, T.M. Caradonna, Y. Cai, J.S. Burke, J. Lin, J.A. Lederer, E.C. Lam, C.L. Lavine, M.S. Seaman, B. Chen, A.G. Schmidt, A.B. Balazs, D.A. Lauffenburger, G. Alter, D.R. Wesemann, Quick COVID-19 healers sustain anti-SARS-CoV-2 antibody production, Cell 183(6) (2020) 1496–1507.e16. [DOI] [PMC free article] [PubMed]
- 10.A.C. Walls, B. Fiala, A. Schafer, S. Wrenn, M.N. Pham, M. Murphy, L.V. Tse, L. Shehata, M.A. O'Connor, C. Chen, M.J. Navarro, M.C. Miranda, D. Pettie, R. Ravichandran, J.C. Kraft, C. Ogohara, A. Palser, S. Chalk, E.C. Lee, K. Guerriero, E. Kepl, C.M. Chow, C. Sydeman, E.A. Hodge, B. Brown, J.T. Fuller, K.H. Dinnon, 3rd, L.E. Gralinski, S.R. Leist, K.L. Gully, T.B. Lewis, M. Guttman, H.Y. Chu, K.K. Lee, D.H. Fuller, R.S. Baric, P. Kellam, L. Carter, M. Pepper, T.P. Sheahan, D. Veesler, N.P. King, Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2, Cell 183(5) (2020) 1367–1382.E17. [DOI] [PMC free article] [PubMed]
- 11.Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., Schneidman-Duhovny D., Zhang C., Shi Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370(6523):1479–1484. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 13.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10) doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T., Rizzuti B., Velazquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59(14):6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia B., Kang X. Activation and maturation of SARS-CoV main protease. Protein Cell. 2011;2(4):282–290. doi: 10.1007/s13238-011-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyu J., Wang S., Balius T.E., Singh I., Levit A., Moroz Y.S., O'Meara M.J., Che T., Algaa E., Tolmachova K., Tolmachev A.A., Shoichet B.K., Roth B.L., Irwin J.J. Ultra-large library docking for discovering new chemotypes. Nature. 2019;566(7743):224–229. doi: 10.1038/s41586-019-0917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82(5):2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren Z., Yan L., Zhang N., Guo Y., Yang C., Lou Z., Rao Z. The newly emerged SARS-like coronavirus HCoV-EMC also has an “Achilles’ heel”: current effective inhibitor targeting a 3C-like protease. Protein Cell. 2013;4(4):248–250. doi: 10.1007/s13238-013-2841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sferrazza G., Corti M., Brusotti G., Pierimarchi P., Temporini C., Serafino A., Calleri E. Nature-derived compounds modulating Wnt/beta-catenin pathway: a preventive and therapeutic opportunity in neoplastic diseases. Acta Pharm. Sin. B. 2020;10(10):1814–1834. doi: 10.1016/j.apsb.2019.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou H., Li Y., Liu X., Wu Z., Li J., Ma Z. Roles of plant-derived bioactive compounds and related microRNAs in cancer therapy. Phytother. Res. 2020:1–11. doi: 10.1002/ptr.6883. [DOI] [PubMed] [Google Scholar]
- 24.Xian Y., Zhang J., Bian Z., Zhou H., Zhang Z., Lin Z., Xu H. Bioactive natural compounds against human coronaviruses: a review and perspective. Acta Pharm. Sin. B. 2020;10(7):1163–1174. doi: 10.1016/j.apsb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J., Tao G., Liu J., Cai J., Huang Z., Chen J.X. Current prevention of COVID-19: natural products and herbal medicine. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.588508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoever G., Baltina L., Michaelis M., Kondratenko R., Baltina L., Tolstikov G.A., Doerr H.W., Cinatl J., Jr. Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J. Med. Chem. 2005;48(4):1256–1259. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- 27.Chen L., Li J., Luo C., Liu H., Xu W., Chen G., Liew O.W., Zhu W., Puah C.M., Shen X., Jiang H. Binding interaction of quercetin-3-beta-galactoside and its synthetic derivatives with SARS-CoV 3CL(pro): structure-activity relationship studies reveal salient pharmacophore features. Bioorg. Med. Chem. 2006;14(24):8295–8306. doi: 10.1016/j.bmc.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo M., Moccia S., Spagnuolo C., Tedesco I., Russo G.L. Roles of flavonoids against coronavirus infection. Chem. Biol. Interact. 2020;328:109211. doi: 10.1016/j.cbi.2020.109211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen T.T., Woo H.J., Kang H.K., Nguyen V.D., Kim Y.M., Kim D.W., Ahn S.A., Xia Y., Kim D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012;34(5):831–838. doi: 10.1007/s10529-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D., Nguyen T.T., Park S.J., Chang J.S., Park K.H., Rho M.C., Lee W.S. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg. Med. Chem. 2010;18(22):7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu P., Xing X., Yu K., Lv Z., Cui H., Shi Y., Chang T., Zhang D., Zhang Y., Wang K., Lu J., Huang Q., Li X., Cui Y., Shi L., Wang T., Niu J., Wang J. Profiles of COVID-19 clinical trials in the Chinese Clinical Trial Registry. Emerg. Microb. Infect. 2020;9(1):1695–1701. doi: 10.1080/22221751.2020.1791736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.China National Health Comission Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment (7 Edition). http://kjfy.meeting.so/msite/news/show/cn/3337.html.
- 33.Zhang L., Yu J., Zhou Y., Shen M., Sun L. Becoming a faithful defender: traditional Chinese medicine against coronavirus disease 2019 (COVID-19) Am. J. Chin. Med. 2020;48(4):763–777. doi: 10.1142/S0192415X2050038X. [DOI] [PubMed] [Google Scholar]
- 34.Xia L., Shi Y., Su J., Friedemann T., Tao Z., Lu Y., Ling Y., Lv Y., Zhao R., Geng Z., Cui X., Lu H., Schroder S. Shufeng Jiedu, a promising herbal therapy for moderate COVID-19: antiviral and anti-inflammatory properties, pathways of bioactive compounds, and a clinical real-world pragmatic study. Phytomedicine. 2020;153390 doi: 10.1016/j.phymed.2020.153390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Runfeng L., Yunlong H., Jicheng H., Weiqi P., Qinhai M., Yongxia S., Chufang L., Jin Z., Zhenhua J., Haiming J., Kui Z., Shuxiang H., Jun D., Xiaobo L., Xiaotao H., Lin W., Nanshan Z., Zifeng Y. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020;156:104761. doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X., Wu Y., Chen C., Gu Y., Zhu C., Wang S., Chen J., Zhang L., Lv L., Zhang G., Yuan Y., Chai Y., Zhu M., Wu C. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen capsule based on human exposure and ACE2 biochromatography screening. Acta Pharm. Sin. B. 2021;11(1):222–236. doi: 10.1016/j.apsb.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.K. Huynh, C.L. Partch, Analysis of protein stability and ligand interactions by thermal shift assay, Curr Protoc Protein Sci 79 (2015) 28 9 1–28 9 14. [DOI] [PMC free article] [PubMed]
- 38.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 39.Andreotti G., Monticelli M., Cubellis M.V. Looking for protein stabilizing drugs with thermal shift assay. Drug Test. Anal. 2015;7(9):831–834. doi: 10.1002/dta.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.B. Cao, Y. Wang, D. Wen, W. Liu, J. Wang, G. Fan, L. Ruan, B. Song, Y. Cai, M. Wei, X. Li, J. Xia, N. Chen, J. Xiang, T. Yu, T. Bai, X. Xie, L. Zhang, C. Li, Y. Yuan, H. Chen, H. Li, H. Huang, S. Tu, F. Gong, Y. Liu, Y. Wei, C. Dong, F. Zhou, X. Gu, J. Xu, Z. Liu, Y. Zhang, H. Li, L. Shang, K. Wang, K. Li, X. Zhou, X. Dong, Z. Qu, S. Lu, X. Hu, S. Ruan, S. Luo, J. Wu, L. Peng, F. Cheng, L. Pan, J. Zou, C. Jia, J. Wang, X. Liu, S. Wang, X. Wu, Q. Ge, J. He, H. Zhan, F. Qiu, L. Guo, C. Huang, T. Jaki, F.G. Hayden, P.W. Horby, D. Zhang, C. Wang, A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19, N Engl J Med 382(19) (2020) 1787–1799. [DOI] [PMC free article] [PubMed]
- 41.Lyngbakken M.N., Berdal J.E., Eskesen A., Kvale D., Olsen I.C., Rueegg C.S., Rangberg A., Jonassen C.M., Omland T., Rosjo H., Dalgard O. A pragmatic randomized controlled trial reports lack of efficacy of hydroxychloroquine on coronavirus disease 2019 viral kinetics. Nat. Commun. 2020;11(1):5284. doi: 10.1038/s41467-020-19056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R.H.M. Furtado, O. Berwanger, H.A. Fonseca, T.D. Correa, L.R. Ferraz, M.G. Lapa, F.G. Zampieri, V.C. Veiga, L.C.P. Azevedo, R.G. Rosa, R.D. Lopes, A. Avezum, A.L.O. Manoel, F.M.T. Piza, P.A. Martins, T.C. Lisboa, A.J. Pereira, G.B. Olivato, V.C.S. Dantas, E.P. Milan, O.C.E. Gebara, R.B. Amazonas, M.B. Oliveira, R.V.P. Soares, D.D.F. Moia, L.P.A. Piano, K. Castilho, R. Momesso, G.P.P. Schettino, L.V. Rizzo, A.S. Neto, F.R. Machado, A.B. Cavalcanti, C.C.-B.I. Investigators, Azithromycin in addition to standard of care versus standard of care alone in the treatment of patients admitted to the hospital with severe COVID-19 in Brazil (COALITION II): a randomised clinical trial, Lancet 396(10256) (2020) 959–967. [DOI] [PMC free article] [PubMed]
- 43.Kurokawa M., Ochiai H., Nagasaka K., Neki M., Xu H., Kadota S., Sutardjo S., Matsumoto T., Namba T., Shiraki K. Antiviral traditional medicines against herpes simplex virus (HSV-1), poliovirus, and measles virus in vitro and their therapeutic efficacies for HSV-1 infection in mice. Antivir. Res. 1993;22(2–3):175–188. doi: 10.1016/0166-3542(93)90094-y. [DOI] [PubMed] [Google Scholar]
- 44.Kannan S., Kolandaivel P. Antiviral potential of natural compounds against influenza virus hemagglutinin. Comput. Biol. Chem. 2017;71:207–218. doi: 10.1016/j.compbiolchem.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361(9374):2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin S.C., Ho C.T., Chuo W.H., Li S., Wang T.T., Lin C.C. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect. Dis. 2017;17(1):144. doi: 10.1186/s12879-017-2253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akram M., Tahir I.M., Shah S.M.A., Mahmood Z., Altaf A., Ahmad K., Munir N., Daniyal M., Nasir S., Mehboob H. Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: a systematic review. Phytother. Res. 2018;32(5):811–822. doi: 10.1002/ptr.6024. [DOI] [PubMed] [Google Scholar]
- 48.Dong L., Xia J.W., Gong Y., Chen Z., Yang H.H., Zhang J., He J., Chen X.D. Effect of lianhuaqingwen capsules on airway inflammation in patients with acute exacerbation of chronic obstructive pulmonary disease. Evid. Based Complement. Alternat. Med. 2014;2014 doi: 10.1155/2014/637969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding Y., Zeng L., Li R., Chen Q., Zhou B., Chen Q., Cheng P.L., Yutao W., Zheng J., Yang Z., Zhang F. The Chinese prescription lianhuaqingwen capsule exerts anti-influenza activity through the inhibition of viral propagation and impacts immune function. BMC Complement. Altern. Med. 2017;17(1):130. doi: 10.1186/s12906-017-1585-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang J., Li H., Du W., Yu P., Guan Y.Y., Ma S.Y., Liu D., Chen W., Shi G.C., Bian X.L. Efficacy of early combination therapy with lianhuaqingwen and arbidol in moderate and severe COVID-19 patients: a retrospective cohort study. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang R., Liu H., Bai C., Wang Y., Zhang X., Guo R., Wu S., Wang J., Leung E., Chang H., Li P., Liu T., Wang Y. Chemical composition and pharmacological mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction against Coronavirus Disease 2019 (COVID-19): in silico and experimental study. Pharmacol. Res. 2020;157:104820. doi: 10.1016/j.phrs.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang D., Zhang B., Lv J.T., Sa R.N., Zhang X.M., Lin Z.J. The clinical benefits of Chinese patent medicines against COVID-19 based on current evidence. Pharmacol. Res. 2020;157:104882. doi: 10.1016/j.phrs.2020.104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xing L., Zhang H., Qi R., Tsao R., Mine Y. Recent advances in the understanding of the health benefits and molecular mechanisms associated with green tea polyphenols. J. Agric. Food Chem. 2019;67(4):1029–1043. doi: 10.1021/acs.jafc.8b06146. [DOI] [PubMed] [Google Scholar]
- 54.Jang M., Park Y.I., Cha Y.E., Park R., Namkoong S., Lee J.I., Park J. Tea polyphenols EGCG and theaflavin inhibit the activity of SARS-CoV-2 3CL-protease in vitro. Evid. Based Complement. Alternat. Med. 2020;2020 doi: 10.1155/2020/5630838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peele K.A., Potla Durthi C., Srihansa T., Krupanidhi S., Ayyagari V.S., Babu D.J., Indira M., Reddy A.R., Venkateswarulu T.C. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Inform. Med. Unlocked. 2020;19:100345. doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leisman D.E., Ronner L., Pinotti R., Taylor M.D., Sinha P., Calfee C.S., Hirayama A.V., Mastroiani F., Turtle C.J., Harhay M.O., Legrand M., Deutschman C.S. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020;8(12):1233–1244. doi: 10.1016/S2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Menegazzi M., Mariotto S., Dal Bosco M., Darra E., Vaiana N., Shoji K., Safwat A.A., Marechal J.D., Perahia D., Suzuki H., Romeo S. Direct interaction of natural and synthetic catechins with signal transducer activator of transcription 1 affects both its phosphorylation and activity. FEBS J. 2014;281(3):724–738. doi: 10.1111/febs.12618. [DOI] [PubMed] [Google Scholar]
- 59.Menegazzi M., Campagnari R., Bertoldi M., Crupi R., Di Paola R., Cuzzocrea S. Protective effect of epigallocatechin-3-gallate (EGCG) in diseases with uncontrolled immune activation: could such a scenario be helpful to counteract COVID-19? Int. J. Mol. Sci. 2020;21(14):5171. doi: 10.3390/ijms21145171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.L. Wang, W. Lee, Y.R. Cui, G. Ahn, Y.J. Jeon, Protective effect of green tea catechin against urban fine dust particle-induced skin aging by regulation of NF-kappaB, AP-1, and MAPKs signaling pathways, Environ Pollut 252(Pt B) (2019) 1318–1324. [DOI] [PubMed]
- 61.Lakshmi S.P., Reddy A.T., Kodidhela L.D., Varadacharyulu N.C. The tea catechin epigallocatechin gallate inhibits NF-kappaB-mediated transcriptional activation by covalent modification. Arch. Biochem. Biophys. 2020;695:108620. doi: 10.1016/j.abb.2020.108620. [DOI] [PubMed] [Google Scholar]
- 62.Almatroodi S.A., Almatroudi A., Khan A.A., Alhumaydhi F.A., Alsahli M.A., Rahmani A.H. Potential therapeutic targets of epigallocatechin gallate (EGCG), the most abundant catechin in green tea, and its role in the therapy of various types of cancer. Molecules. 2020;25(14):3146. doi: 10.3390/molecules25143146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Compound-target regulation network diagram. The red triangles represent targets. The surrounding ellipses represent the key active compounds.
A. Gene ontology terms of candidate targets of Yangyinjiedu (YYJD) against COVID-19. The top 20 GO functional categories with FDR < 0.05 are presented. B. Gene ontology terms of candidate targets of Dayuanxiaodu (DYXX) against COVID-19. The top 20 GO functional categories with p-adjust<0.05 are presented. C. Gene ontology terms of candidate targets of Chaihuqingzao (CHQZ) against COVID-19. The top 20 GO functional categories with p-adjust<0.05 are presented.
KEGG pathway enrichment analysis of candidate targets of A. Yangyinjiedu (YYJD), B. Dayuanxiaodu (DYXX) and C. Chaihuqingzao (CHQZ) against COVID-19. The bar graph shows the pathways with significant p-adjust <0.05.
Active ingredients of Yangyinjiedu (YYJD) herbal medicine.
Active ingredients of Dayuanxiaodu (DYXX) herbal medicine.
Active ingredients of Chaihuqingzao (CHQZ) herbal medicine.