Abstract
The ongoing pandemic has demonstrated the utility of widespread surveillance and diagnostic detection of the novel SARS-CoV-2. Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) has enabled broader testing, but current LAMP tests only detect single targets and require separate reactions for controls. With flu season in the Northern Hemisphere, the ability to screen for multiple targets will be increasingly important, and the ability to include internal controls in RT-LAMP allows for improved efficiency. Here we describe multiplexed RT-LAMP with four targets (SARS-CoV-2, influenza A, influenza B, human RNA) in a single reaction using real-time and end point fluorescence detection. Such increased functionality of RT-LAMP will enable even broader adoption of this molecular testing approach and aid in the fight against this public health threat.
Keywords: : COVID-19 detection, fluorescence, LAMP, molecular diagnostics, multiplexing, sensors and probes
METHOD SUMMARY
This study describes enhancing loop-mediated isothermal amplification through multiplexed real-time and end point detection of SARS-CoV-2 combined with influenza and control targets. By enabling multiple target detection, loop-mediated isothermal amplification can be even more widely used for diagnostics in settings where multiple viral targets are potential infectious agents and where higher-throughput testing is advantageous.
Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) was developed as a nucleic acid amplification technique requiring only a single reaction temperature without the need for sophisticated thermal cycling equipment [1]. It is particularly well suited for simple molecular diagnostics as it is rapid and sensitive, with numerous and versatile detection methods for determining positive or negative reactions [2]. In the recent pandemic of COVID-19, LAMP has been successfully applied for the detection of SARS-CoV-2 using: colorimetric readouts in both simple, point-of-need assays and higher-throughput diagnostic platforms [3–11]; fluorescence [12–14] or turbidity [15] detection using simple instruments; Cas enzyme detection for lateral flow or fluorescence [9,16–18]; and next-generation sequencing, offering high levels of sample pooling and multiplexing [19,20]. Many of these tests have received Emergency Use Authorization from the US FDA or have been CE marked in Europe, and are used daily for COVID testing worldwide.
However, these initial LAMP tests are generally used for the detection of a single target per reaction, with samples split across multiple reactions for confirmatory detection or controls. A beneficial feature of many RT-qPCR assays, particularly in the widely-used TaqMan® method, is the ability to perform multiplex detection, whereby tests for numerous analyte targets and internal controls are conducted simultaneously in one reaction. In addition to reducing reagent usage and increasing testing throughput, multiplexing gives a reliable internal control, indicating successful amplification as well as validating sample extraction. Detecting multiple targets from the same pathogen of interest (e.g., SARS-CoV-2 genes N and E) increases confidence when calling a sample positive, or allows multiple different targets to be combined (e.g., SARS-CoV-2 and influenza) for more useful diagnostic tests. A number of methods exist for multiplexing LAMP, and we have previously presented a scheme called Detection of Amplification by Releasing of Quenching (DARQ) that supplements a standard LAMP primer set with a pair of oligonucleotides in duplex form for detection. This duplex consists of a 5′-modified version of the forward inner primer (FIP), Q-FIP, annealed to a 3′-modified oligonucleotide representing the F1 region (Fd), complementary to the FIP in its 5′ section [21]. The two modifications are selected to be dark quencher–fluorophore pairs; when the quenched FIP is incorporated into the LAMP product, subsequent amplification displaces the Fd oligo, releasing quenching and producing a fluorescent signal specific to the label and target of interest. Other versions of this approach have been used, either moving the quencher and fluorophore to a loop primer [22,23] or incorporating them into the amplification products and detecting after the reaction [24]. A detection approach based on molecular beacon hybridization has also been developed for LAMP amplification detection [25]. Several of these detection approaches have been applied to the detection of SARS-CoV-2 [23,25] enabling the detection of more than one target in the same reaction but as yet requiring additional probe design and without the inclusion of internal control assays.
As the COVID-19 pandemic continues into flu season, the ability to distinguish which causative agent is responsible for what can manifest as very similar symptoms will be of great importance for diagnostics and disease surveillance. A patient presenting with respiratory symptoms may have one of a number of viral infections (e.g., SARS-CoV-2, influenza A or B, respiratory syncytial virus, a rhinovirus), so using one test to identify multiple infectious agents in the same procedure will save time and cost. Most importantly, it gives a more definitive diagnostic identification. Syndromic panel tests (e.g., Genmark, Biofire) are the ultimate example of this principle, providing a simultaneous test for dozens of respiratory pathogens, but these tests are expensive and not suited to the need for expanded use of molecular testing that has arisen during COVID-19. Here we demonstrate a simple LAMP assay for detection of four targets (SARS-CoV-2, influenza A, influenza B and an internal control) that provides high sensitivity in a single reaction, expanding the utility of the widely used LAMP chemistry to a multiplex diagnostic setting.
DARQ LAMP primer sets include all conventional LAMP primers (outer primers F3 and B3, inner primers FIP and BIP, and Loop primers LF and LB) and a duplex oligonucleotide consisting of the FIP modified at its 5′ end with a dark quencher (Q-FIP) annealed to a complementary F1c oligonucleotide with 3′ fluorophore (Fd) (Table 1) [21]. E1 and N2 LAMP primers targeting SARS-CoV-2 sequence (GenBank accession number MN908947) were from our previous screening [11]. Primer sets for targeting multiple influenza A and B strains (IAV and IBV primer sets) were designed and validated previously [26]. Oligonucleotides were synthesized at Integrated DNA Technologies (IA, USA) with standard desalting for conventional LAMP primers and HPLC purification for Q-FIP and Fd oligonucleotides. Synthetic SARS-CoV-2 RNA containing an equal ratio of the viral genome regions was purchased from Twist Bioscience (Twist Synthetic SARS-CoV-2 RNA Control 2 #102024, MN908947.3; Twist Bioscience, CA, USA). RNAs for influenza A and B were purchased from ATCC (VA, USA): H1N1/2009pdm (VR-1737D), H1N1 A/PR/8/34 (VR-1469DQ), H3N2 A/Virginia/ATCC6/2012 (VR-1811D), H3N2 A/Aichi/2/68 (VR-1680D), B/Wisconsin/1/2010 BX-41A (VR-1885DQ) and B/Lee/40 (VR-1535D). Viral RNA was diluted to lower concentrations in 10 ng/μl Jurkat total RNA (Biochain, CA, USA) based on quantification provided by the manufacturers. For the 24 repeat reactions, the amount of RNA used was 50 copies of SARS-CoV-2 RNA, 1 μl of 1:10000 diluted influenza A RNA (VR-1737D) and approximately 21 copies of influenza B RNA (VR-1885DQ). This amount of viral RNA was sufficient for more than half of, but not all, the 24 reactions to show positive amplification, thus allowing detection of sensitivity change under different conditions.
Table 1. Loop-mediated isothermal amplification primer sequences.
| Primer set | Sequence | Ref. |
|---|---|---|
| E1 | [11] | |
| E1-F3 | TGAGTACGAACTTATGTACTCAT | |
| E1-B3 | TTCAGATTTTTAACACGAGAGT | |
| E1-FIP | ACCACGAAAGCAAGAAAAAGAAGTTCGTTTCGGAAGAGACAG | |
| E1-BIP | TTGCTAGTTACACTAGCCATCCTTAGGTTTTACAAGACTCACGT | |
| E1-LF | CGCTATTAACTATTAACG | |
| E1-LB | GCGCTTCGATTGTGTGCGT | |
| E1-QFIP | /5IABkFQ/ACCACGAAAGCAAGAAAAAGAAGTTCGTTTCGGAAGAGACAG | |
| E1-FD | ACTTCTTTTTCTTGCTTTCGTGGT/3Joe_N/ | |
| IAV | [26] | |
| IAV-F3-1 | GACTTGAAGATGTCTTTGC | |
| IAV-F3-2 | GACTGGAAAGTGTCTTTGC | |
| IAV-B3-1 | TRTTATTTGGGTCTCCATT | |
| IAV-B3-2 | TRTTGTTTGGGTCCCCATT | |
| IAV-FIP | TTAGTCAGAGGTGACARRATTGCAGATCTTGAGGCTCTC | |
| IAV-BIP | TTGTKTTCACGCTCACCGTGTTTGGACAAAGCGTCTACG | |
| IAV-LF | GTCTTGTCTTTAGCCA | |
| IAV-LB | CMAGTGAGCGAGGACTG | |
| IAV-QFIP | /5IAbRQ/TTAGTCAGAGGTGACARRATTGCAGATCTTGAGGCTCTC | |
| IAV-Fd | CAATYYTGTCACCTCTGACTAA/3Cy5Sp/ | |
| IBV | [26] | |
| IBV-F3 | GCAACCAATGCCACCATA | |
| IBV-B3 | TTCTCTCTTCAAGRGACATC | |
| IBV-FIP | TAGTCAAGGGCYCTTTGCCACTTTGAAGCAGGAATTCTGGA | |
| IBV-BIP | CAAGACCGCCTAAACAGACTAAACTTTTACTTTCAGGCTCACTT | |
| IBV-LF | TGAAAGYCTTTCATAGCAC | |
| IBV-LB | CAAGAATAAAGACTCACAAC | |
| IBV-QFIP | /5IABkFQ/TAGTCAAGGGCYCTTTGCCACTTTGAAGCAGGAATTCTGGA | |
| IBV-Fd | TGGCAAAGRGCCCTTGACTA/36-FAM/ | |
| ACTB | ||
| ACTB-F3 | AGTACCCCATCGAGCACG | |
| ACTB-B3 | AGCCTGGATAGCAACGTACA | |
| ACTB-FIP | GAGCCACACGCAGCTCATTGTATCACCAACTGGGACGACA | |
| ACTB-BIP | CTGAACCCCAAGGCCAACCGGCTGGGGTGTTGAAGGTC | |
| ACTB-LF | TGTGGTGCCAGATTTTCTCCA | |
| ACTB-LB | CGAGAAGATGACCCAGATCATGT | |
| ACTB-QFIP | /5IAbRQ/GAGCCACACGCAGCTCATTGTATCACCAACTGGGACGACA | |
| ACTB-Fd | TACAATGAGCTGCGTGTGGCTC/3Rox_N/ | |
| N2 | [11] | |
| N2-F3 | ACCAGGAACTAATCAGACAAG | |
| N2-B3 | GACTTGATCTTTGAAATTTGGATCT | |
| N2-FIP | TTCCGAAGAACGCTGAAGCGGAACTGATTACAAACATTGGCC | |
| N2-BIP | CGCATTGGCATGGAAGTCACAATTTGATGGCACCTGTGTA | |
| N2-LF | GGGGGCAAATTGTGCAATTTG | |
| N2-LB | CTTCGGGAACGTGGTTGACC | |
| N2-QFIP | /5IABkFQ/TTCCGAAGAACGCTGAAGCGGAACTGATTACAAACATTGGCC | |
| N2-FD22 | TCCGCTTCAGCGTTCTTCGGAA/36-FAM/ | |
Modifications as noted in Integrated DNA Technologies’ nomenclature.
BIP: Backward inner primer; E: SARS-CoV-2 E gene; FIP: Forward inner primer; IAV: Influenza A virus primer; IBV: Influenza B virus primer; N: SARS-CoV-2 N gene; QFIP: Quencher-modified forward inner primer.
All influenza primers were initially screened for performance using WarmStart® Colorimetric LAMP 2X Master Mix (DNA and RNA; New England Biolabs, M1800) and with WarmStart LAMP Kit (DNA and RNA) (E1700) supplemented with 1μM SYTO®-9 double-stranded DNA binding dye (Thermo Fisher S34854, MA, USA). DARQ LAMP reactions contained 1 × E1700, with an additional 0.32 U/μl Bst 2.0 WarmStart DNA Polymerase (M0538), 40 mM guanidine hydrochloride (Sigma-Aldrich RDD001, MO, USA) and standard concentrations of LAMP primers (0.2 μM F3, 0.2 μM B3, 1.6 μM FIP, 1.6 μM BIP, 0.4 μM Loop F, 0.4 μM Loop B) supplemented with DARQ FIP duplex (0.22μM QFIP:0.18 μM Fd, preannealed as 55 μM QFIP:45 μM Fd by heating to 95°C and slowly cooling to room temperature). The concentration for ACTB primers was reduced to 0.25 × the standard concentrations for LAMP primers (0.05 μM F3, 0.05 μM B3, 0.4 μM FIP, 0.4 μM BIP, 0.1 μM Loop F, 0.1 μM Loop B) with DARQ FIP duplex added as 0.066μM Q-FIP and 0.054 μM Fd. The reactions were incubated at 60°C in half-skirted plates (Bio-Rad HSP9601, CA, USA) on a real-time qPCR machine (Bio-Rad CFX96). Real-time LAMP signal was acquired every 15 s for 108 cycles (total incubation time ~40 min for single-channel or 49 min for four-channel acquisition). Completed LAMP reactions were then scanned on a BioTek (VT, USA) Synergy Neo2 microplate reader for fluorescence signal for 5-FAM (excitation 484/20 nm, emission 530/25 nm, signal gain 75), HEX (excitation 524/20 nm, emission 565/20 nm, signal gain 75), 5-ROX (excitation 569/20 nm, emission 615/25 nm, signal gain 85) and Cy5 (excitation 640/20 nm, emission 682/20 nm, signal gain 75). The threshold for the positive signal was set as above the sum of average raw fluorescence units of eight no-template control (NTC) reactions plus 10 × their standard deviation.
We had previously established sensitive LAMP primer sets for SARS-CoV-2 detection [11] and synthesized DARQ-compatible labeled FIP and Fd oligonucleotides with different detection fluorophores for Gene E (E1, JOE) and Gene N (N2, FAM), as well as our internal control primer set (ACTB, ROX) (Table 1). Both E1 and N2 primer sets were tested and found to be suitable for DARQ LAMP in multiplexing with other primers, but only data for E1 are shown here. For influenza detection we designed a variety of LAMP primer sets and pulled existing primers from literature reports [26–30] and evaluated them for speed, sensitivity and compatibility with our SARS-CoV-2 and control primers. From this comparison we selected the IAV [26,27] and IBV [26] primer sets (Table 1) as they were among the most sensitive primer sets in our screening and were designed for suitability with various strains. The IAV set targets the RNA sequence for the matrix (M) protein and has been validated to detect many avian and human influenza A strains from H1N1 to H15N8, including multiple H1N1/2009pdm and H7N9 strains [26,27]. In our testing, we found it amplified well with RNAs from the 2009 H1N1 (H1N1/2009pdm) and 1934 H1N1 (A/PR/8/34) strains, as well as 2012 H3N2 (A/Virginia/ATCC6/2012) and 1968 H3N2 (A/Aichi/2/68). The IBV set detects the RNA for the NS protein and has been tested with RNA from an influenza B virus isolated in 2012 [26]; we found it amplified well with RNA from strain B/Wisconsin/1/2010 BX-41A, but not with RNA from the B/Lee/40 strain isolated 80 years ago. We also performed sequence alignments of IAV and IBV target regions; these primer sets are expected to detect strains isolated in recent years. With this information, IAV and IBV primer sets were chosen for multiplexing with our SARS-CoV-2 primer set using Cy5 and FAM fluorophores, respectively.
First we compared DARQ LAMP with conventional intercalating dye detection of tenfold dilutions of viral RNAs for each primer set. The results for SARS-CoV-2 RNA using the E1 primer set, either with or without the ACTB control primer set, are shown in Figure 1. The speed by DARQ LAMP showed a target dosage response, just as in the conventional LAMP monitored by SYTO-9 dsDNA-binding dye (Figure 1A & B). Reaction speed decreased in the presence of the DARQ duplex and slowed slightly more with the addition of a second primer set (ACTB; Figure 1C & E). Similarly, in the duplex LAMP reactions, ACTB amplified slightly more slowly than in reactions containing only ACTB and Jurkat RNA (Figure 1D), but the signal level was sufficient for ACTB to serve as an internal positive and loading control. Using 0.25 × primer concentration for the ACTB internal control allowed for even amplification in single versus duplex reactions while still providing sufficient fluorescent signal for detection. Similar results were observed with IAV and IBV primer sets, indicating that these primer sets work well in DARQ LAMP and are compatible with ACTB primers.
Figure 1. Comparison between conventional and Detection of Amplification by Releasing of Quenching loop-mediated isothermal amplification.
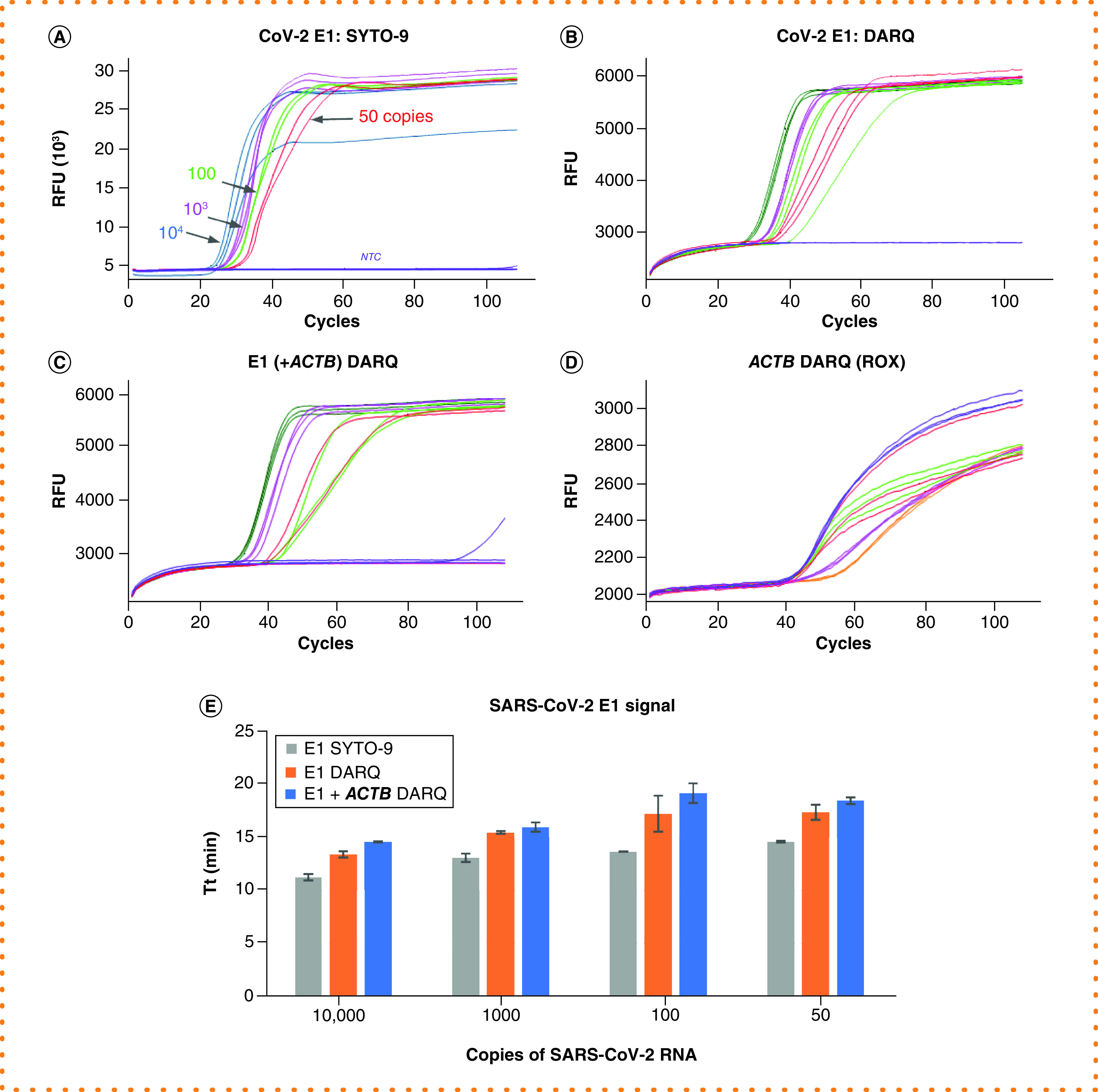
All reactions were performed using E1 primer targeting SARS-CoV-2 RNA at 104, 103 102 and 50 copies, and NTC in triplicate. (A) Conventional LAMP monitored by SYTO-9. (B) Single-plex DARQ LAMP. (C) Duplex DARQ LAMP including both E1 and ACTB primers. (D) ACTB detection from duplex DARQ LAMP. (E) Comparison of SARS-CoV-2 amplification speed in reactions with SYTO-9, single-plex and duplex DARQ LAMP.
DARQ: Detection of Amplification by Releasing of Quenching; LAMP: Loop-mediated isothermal amplification; NTC: No-template control.
Next we sought to evaluate whether DARQ LAMP has a detection sensitivity similar to that of standard LAMP. Figure 2 shows the results of 24 LAMP reactions, each with 50 copies of SARS-CoV-2 RNA, and 8 NTCs. In all configurations (standard intercalating LAMP, Figure 2A; single-plex DARQ LAMP, Figure 2B; duplex E1+ACTB DARQ LAMP, Figure 2C) we measured a similar number of positive LAMP reactions and negative NTCs. This indicates that DARQ LAMP has similar detection sensitivity to conventional LAMP reactions, and that the addition of a second primer set does not increase the rate of nontemplate amplification with our reaction conditions.
Figure 2. Maintaining detection sensitivity in Detection of Amplification by Releasing of Quenching loop-mediated isothermal amplification.
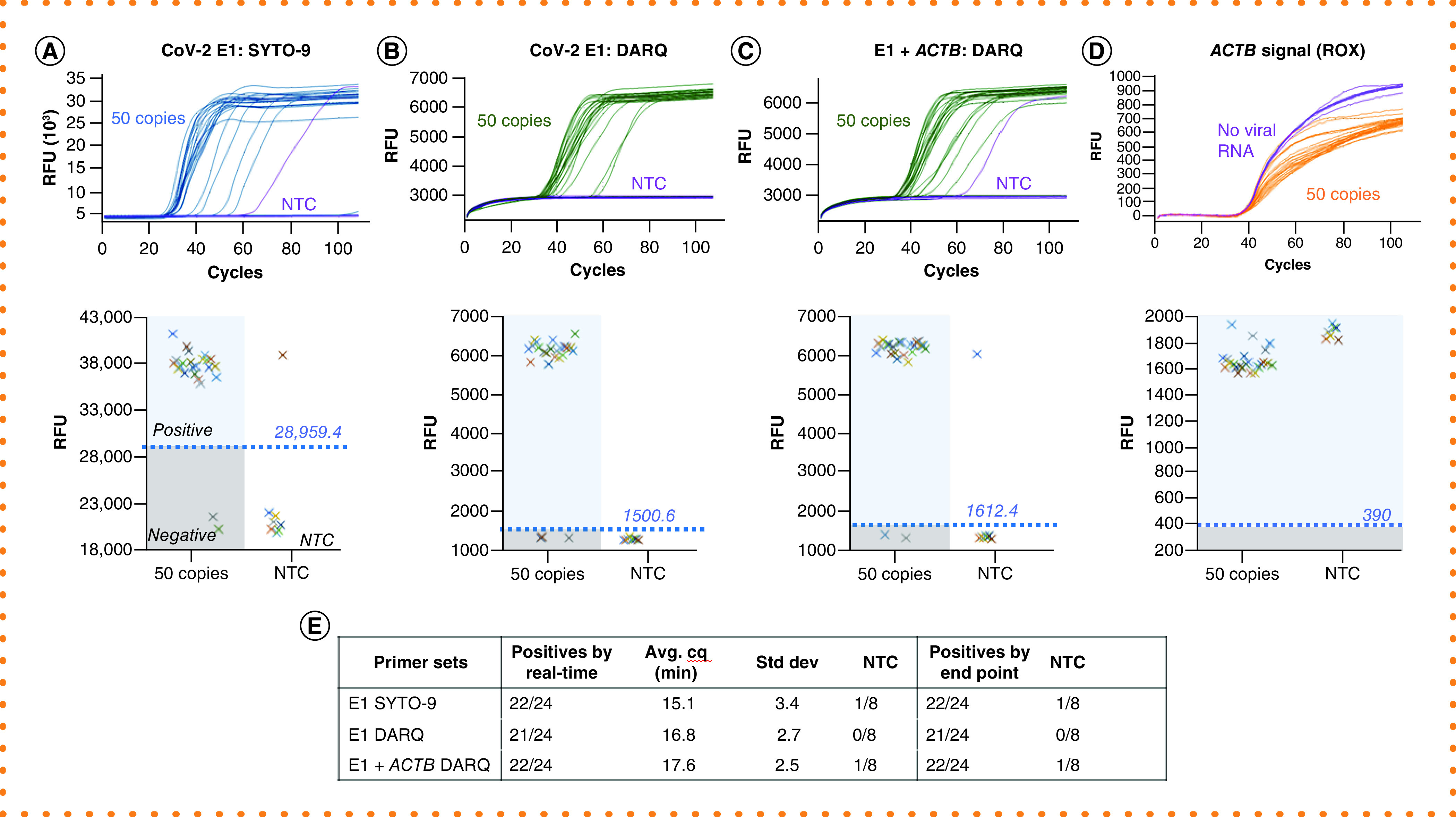
Amplifications were performed with 24 repeats, each containing 50 copies of SARS-CoV-2 RNA, or 8 repeats of NTC, using E1 or both E1 and ACTB primer sets. Two methods were used to determine positive amplification, and their results are arranged in the same panel: real-time curves (top) and plots of RFU obtained from end point scanning (bottom). In the RFU plotting, the threshold (value shown above the line) for positive reactions and a separation from NTC reactions divide the plot area into four quadrants as illustrated in panel A: positives, negatives, false positives and NTC. (A) Detection by SYTO-9. (B) Single-plex DARQ LAMP. (C) Duplex DARQ LAMP including both E1 and ACTB primers. (D) ACTB detection from duplex DARQ LAMP. (E) Summary of detection. Total number of positives, amplification speed (Cq) and correlation of results by real-time curve and end point scanning.
DARQ: Detection of Amplification by Releasing of Quenching; LAMP: Loop-mediated isothermal amplification; NTC: No-template control; RFU: Raw fluorescence unit.
Although real-time monitoring of LAMP provides some level of quantitative information, heating to 60°C with multiplex fluorescence requires a qPCR instrument or similar, and reactions must be incubated on the detection instrument for the entire reaction time. An alternative approach compatible with more instruments is to use end point fluorescence detection. We investigated the possibility of using end point fluorescence scanning to detect signal in DARQ LAMP reactions. After incubating at 60°C, the same plate used for real-time monitoring was scanned for fluorescence levels in a BioTek Synergy Neo2 plate reader and the raw fluorescence unit values were plotted (Figure 2A–C, lower panel). The difference in signal levels between positive and negative LAMP or NTC reactions was easily distinguished using a threshold value set based on negative (background value), and calling positives accordingly matched the results 100% with those obtained by real-time monitoring (Figure 2E). Similarly, the ACTB signal in duplex LAMP reactions was determined by scanning and matched the real-time results (Figure 2D). This result demonstrates the compatibility of end point plate reader measurements with multiplex DARQ LAMP, enabling its use on a wider range of instrument types and increasing the potential test throughput.
Next we extended the number and range of targets for DARQ LAMP for the detection of a single target and an internal control in the presence of up to four sets of LAMP primers. Figure 3 shows the results of 24 replicate LAMP reactions, each with 50 copies of SARS-CoV-2 RNA, and 8 NTCs using: E1 and ACTB primers (duplex; Figure 3A); E1, ACTB and IAV (three-plex, Figure 3B); or E1, ACTB, IAV and IBV (four-plex, Figure 3C). In all three cases, the SARS-CoV-2 RNA was successfully detected with equivalent sensitivity. We did observe a slowing of amplification with increasing primer amounts, but sensitivity was not significantly affected. Again, the real-time results matched exactly with the end point measurements on a plate reader (Figure 3A–C, lower half panel) for all primer combinations. The ACTB signal also appeared as expected and could be easily called as positive by the real-time data or end point fluorescence scanning in all reaction configurations (Figure 3D & E). A similar evaluation was performed with the IAV/influenza A RNA and IBV/influenza B RNA; equivalent performance was observed with these targets and combinations of primers and templates (Supplementary Figures 1 & 2). While not measuring a strict limit of detection, we used low-copy inputs (SARS-CoV-2 ~50 copies; influenza A 1:10,000 dilution; influenza B ~21 copies) for which <100% of reactions would indicate positive, in order to maximize our sensitivity to altered reaction performance. The results demonstrate that DARQ LAMP detection sensitivity is not significantly affected with up to four primer sets present in the reaction, though in testing 24 replicates at very low copy inputs we saw approximately 1 fewer positive result in reactions containing all four primer sets. Importantly, the rate of nontemplate amplification, which would present a false positive result, did not increase when adding multiple primer sets in the reaction.
Figure 3. Sensitive detection of SARS-CoV-2 and an internal control in the presence of multiple primer sets.
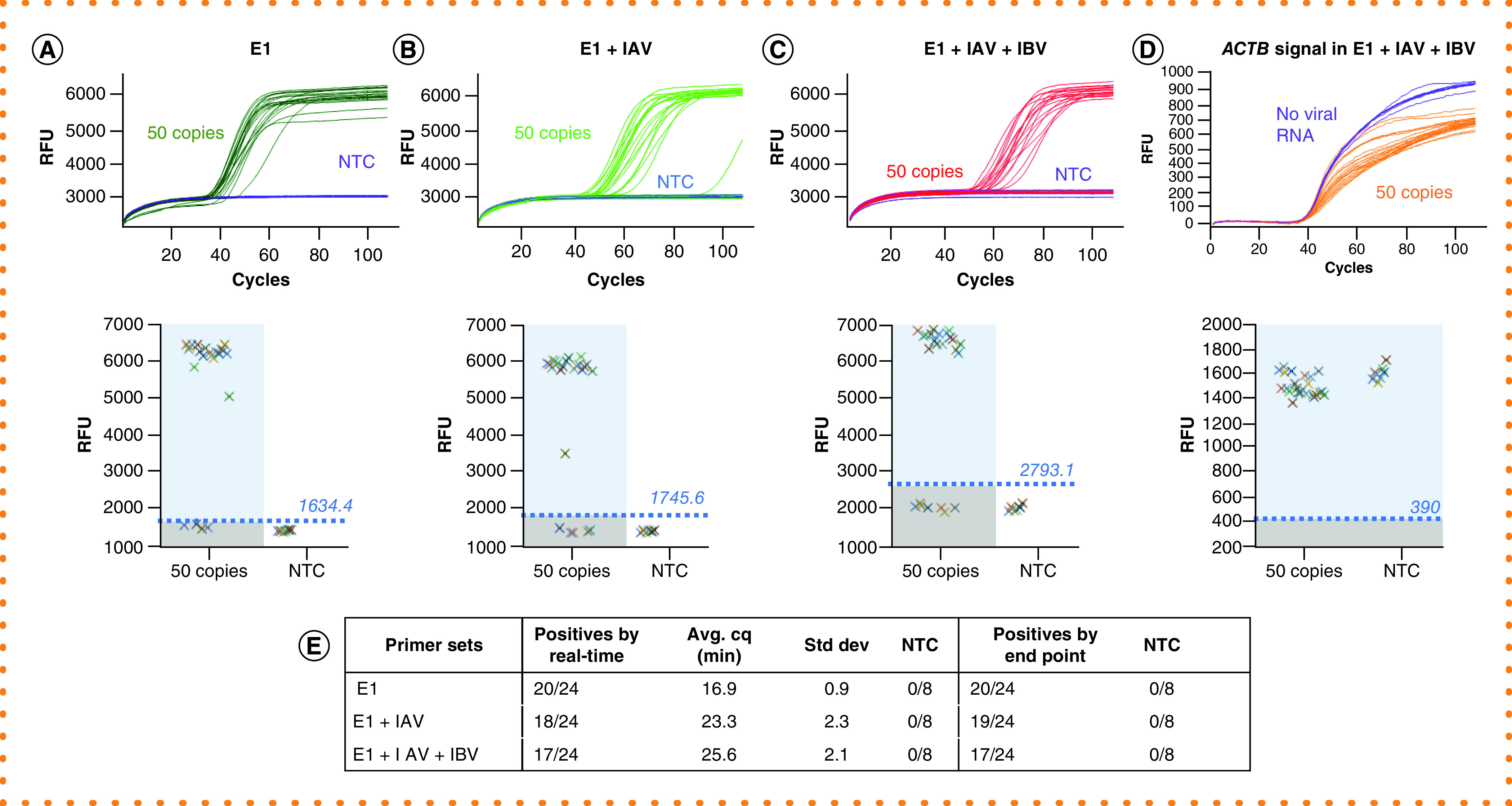
Amplifications were performed with 24 repeats, each containing 50 copies of SARS-CoV-2 RNA, or 8 repeats as NTC using E1 and ACTB primer sets or with additional primer sets. (A) Dual (E1 + ACTB) DARQ LAMP amplification. (B) Dual (E1 + ACTB) DARQ LAMP amplification in the presence of an unrelated primer set (IAV and its QFIP:Fd duplex). (C) Dual (E1 + ACTB) DARQ LAMP in the presence of two unrelated primer sets (IAV and IBV and their QFIP:Fd duplexes). (D) ACTB detection from conditions in (C). (E) Summary of detection: total number of positives, amplification speed (Cq) and correlation of results by real-time curve and end point scanning.
DARQ: Detection of Amplification by Releasing of Quenching; IAV: Influenza A virus primer; IBV: Influenza B virus primer; LAMP: Loop-mediated isothermal amplification: NTC: No-template control; QFIP: Quencher-modified forward inner primer.
With testing in real-world settings, the vast majority of samples will contain zero or one of the viral targets detectable in the multiplex assay. However, coinfections are possible and the assay must maintain performance in the presence of multiple target sequences. To study this scenario, we examined amplification of two DARQ LAMP targets and an internal control in the same reaction. We added two viral RNA targets to 24 DARQ LAMP reactions that all contained E1, IAV, IBV and ACTB primers (and Jurkat RNA for internal control) and monitored amplification by real-time and end point fluorescence scanning. In all three triplex combinations (SARS-CoV-2 + influenza A + ACTB; SARS-CoV-2 + influenza B + ACTB; influenza A + influenza B + ACTB), the positive rate of each target was equivalent to that of the single-plex or duplex reactions described above, indicating that sensitivity was unaffected (Figure 4). The majority of reactions showed detection of both viral targets, with a minority detecting only one of the targets (Figure 4). The shape of the real-time curves in these triplex LAMP reactions (two targets + ACTB) appeared slightly flatter and slower, but distinction above background was easily seen; by combining the real-time data with the end point fluorescence level (data not shown), positives could be differentiated from negatives in the same manner as described above.
Figure 4. Simultaneous detection of two viral targets and an internal control.
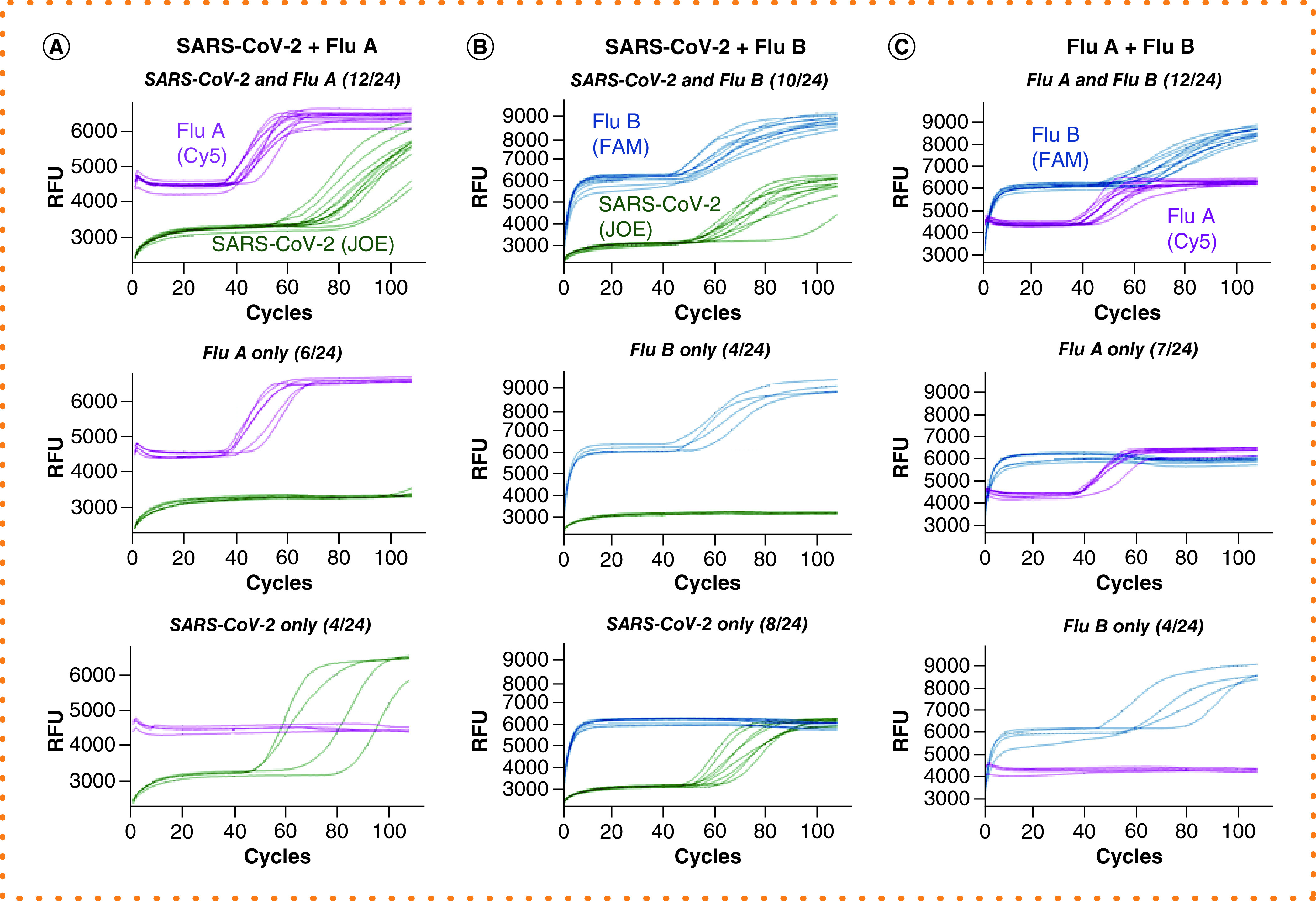
Reactions were performed in the presence of E1, IAV, IBV and ACTB primer sets and their QFIP:Fd duplexes with 24 repeats (for simplicity the ROX signal from ACTB is not shown). The targets are pairs of SARS-CoV-2, influenza A and influenza B RNAs. For each target pair, the reactions that had both targets amplified are shown in the top, and reactions that only had one target amplified are shown in the middle and bottom. The fractional numbers in brackets indicate the number of reactions in each category. (A) SARS-CoV-2, influenza A and Jurkat RNAs. (B) SARS-CoV-2, influenza B and Jurkat RNAs. (C) Influenza A, influenza B and Jurkat RNAs.
IAV: Influenza A virus primer; IBV: Influenza B virus primer; QFIP: Quencher-modified forward inner primer.
Detection of multiple viral targets in a molecular diagnostic reaction is hardly our original idea and is already widely used worldwide for multiplex PCR tests and syndromic panels. But as the COVID-19 pandemic continues into late 2020 and beyond, the need for more and varied testing remains an urgent priority worldwide. LAMP has found a use as a COVID testing modality due to its flexibility and simplicity, but as cold and flu season arrives, more functionality will be needed in LAMP tests to keep pace with diagnostic testing requirements. While fieldable colorimetric LAMP is extremely useful for screening and surveillance, the ability to multiplex influenza and potentially other targets will supplement the supply-constrained RT-qPCR workflows. Here we demonstrate multiplex fluorescent DARQ LAMP with detection of three primary respiratory viral RNA targets and an internal control in a single reaction. While not as simple as visual colorimetric LAMP, the DARQ approach is rapid (<40 min) and is compatible with standard qPCR instruments. We also showed that positive signals can be reliably detected by end point scanning of DARQ fluorescence levels in a plate reader, and thus the reactions could be performed on heat blocks, regular PCR cyclers or even simple incubators followed by quick scanning. The work presented here utilized only extracted and synthetic control RNA materials rather than real clinical samples, which would of course be needed for a real diagnostic test, but we hope that by demonstrating this multiplex approach we can help enable broader usage of LAMP testing and further support the continuing need for diagnostics to combat COVID-19. Ideally, as powerful molecular testing gains wider adoption, we will continue to utilize these approaches, with LAMP and other diagnostic methods tracking and fighting the further threats to public health that will inevitably arise in the future.
Supplementary Material
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.future-science.com/doi/suppl/10.2144/btn-2020-0157
Author contributions
Y Zhang conducted experiments, collected and analyzed data. Y Zhang and N Tanner conceived the study and wrote the manuscript.
Financial & competing interests disclosure
The authors are employees of, and received funding from, New England Biolabs, the manufacturer of reagents described in the paper. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
- 1.Notomi T, Okayama H, Masubuchi H et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28(12), E63 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, Lowe SB, Gooding JJ Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens. Bioelectron. 61, 491–499 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Anahtar MN, Mcgrath GEG, Rabe BA et al. Clinical assessment and validation of a rapid and sensitive SARS-CoV-2 test using reverse-transcription loop-mediated isothermal amplification without the need for RNA extraction. Open Forum Infect. Dis. (2020) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baek YH, Um J, Antigua KJC et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microbes Infect. 9(1), 998–1007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler DJ, Mozsary C, Meydan C et al. Shotgun transcriptome and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. bioRxiv (2020) (Preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dao Thi VL, Herbst K, Boerner K et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 12(556), eabc7075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lalli MA, Chen X, Langmade SJ et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric reverse-transcription loop-mediated isothermal amplification. Clin. Chem. (2020). (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabe BA, Cepko C SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl Acad. Sci. USA 117(39), 24450–24458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schermer B, Fabretti F, Damagnez M et al. Rapid SARS-CoV-2 testing in primary material based on a novel multiplex RT-LAMP assay. PLoS ONE 15(11), e0238612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Odiwuor N, Xiong J et al.Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv (2020) (Preprint). [Google Scholar]
- 11.Zhang Y, Ren G, Buss J, Barry AJ, Patton GC, Tanner NA Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. BioTechniques 69(3), 178–185 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Ganguli A, Mostafa A, Berger J et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl Acad. Sci. USA 117(37), 22727–22735 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JYH, Best N, Mcauley J et al. Validation of a single-step, single-tube reverse transcription loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA. J. Med. Microbiol. 69(9), 1169–1178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohon AN, Oberding L, Hundt J et al. Optimization and clinical validation of dual-target RT-LAMP for SARS-CoV-2. J. Virol. Methods 286, 113972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagura-Ikeda M, Imai K, Tabata S et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J. Clin. Microbiol. 58 9), e01438-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ali Z, Aman R, Mahas A et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 288, 198129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broughton JP, Deng X, Yu G et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38(7), 870–874 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joung J, Ladha A, Saito M et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 383(15), 1492–1494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James P, Stoddart D, Harrington ED et al. LamPORE: rapid, accurate and highly scalable molecular screening for SARS-CoV-2 infection, based on nanopore sequencing. medRxiv (2020) (Preprint). [Google Scholar]
- 20.Schmid-Burgk JL, Schmithausen RM, Li D et al. LAMP-Seq: population-scale COVID-19 diagnostics using combinatorial barcoding. bioRxiv (2020) (Preprint). [Google Scholar]
- 21.Tanner NA, Zhang Y, Evans TC Jr Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. BioTechniques 53(2), 81–89 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Curtis KA, Morrison D, Rudolph DL et al. A multiplexed RT-LAMP assay for detection of group M HIV-1 in plasma or whole blood. J. Virol. Methods 255, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Yaren O, Mccarter J, Phadke N et al. Ultra-rapid detection of SARS-CoV-2 in public workspace environments. medRxiv (2020) (Preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ball CS, Light YK, Koh CY, Wheeler SS, Coffey LL, Meagher RJ Quenching of unincorporated amplification signal reporters in reverse-transcription loop-mediated isothermal amplification enabling bright, single-step, closed-tube, and multiplexed detection of RNA viruses. Anal. Chem. 88(7), 3562–3568 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Sherrill-Mix S, Hwang Y, Roche AM et al. LAMP-BEAC: detection of SARS-CoV-2 RNA using RT-LAMP and molecular beacons. medRxiv (2020) (Preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takayama I, Nakauchi M, Takahashi H et al. Development of real-time fluorescent reverse transcription loop-mediated isothermal amplification assay with quenching primer for influenza virus and respiratory syncytial virus. J. Virol. Methods 267, 53–58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakauchi M, Yoshikawa T, Nakai H et al. Evaluation of reverse transcription loop-mediated isothermal amplification assays for rapid diagnosis of pandemic influenza A/H1N1 2009 virus. J. Med. Virol. 83(1), 10–15 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Parida M, Shukla J, Sharma S et al. Development and evaluation of reverse transcription loop-mediated isothermal amplification assay for rapid and real-time detection of the swine-origin influenza A H1N1 virus. J. Mol. Diagn. 13(1), 100–107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahony J, Chong S, Bulir D, Ruyter A, Mwawasi K, Waltho D Multiplex loop-mediated isothermal amplification (M-LAMP) assay for the detection of influenza A/H1, A/H3 and influenza B can provide a specimen-to-result diagnosis in 40 min with single genome copy sensitivity. J. Clin. Virol. 58(1), 127–131 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Ahn SJ, Baek YH, Lloren KKS et al. Rapid and simple colorimetric detection of multiple influenza viruses infecting humans using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. BMC Infect. Dis. 19(1), 676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


