B. pseudomallei causes significant morbidity and mortality, but its geographic prevalence and genetic diversity are not well characterized, especially in the Western Hemisphere. A better understanding of the genetic relationships among clinical and environmental isolates will improve knowledge of the population structure of this bacterium as well as the ability to conduct epidemiological investigations of cases of melioidosis.
KEYWORDS: melioidosis, infectious disease, molecular epidemiology, environmental, genome analysis
ABSTRACT
Burkholderia pseudomallei is a Gram-negative bacterium that causes the sapronotic disease melioidosis. An outbreak in 2003 in the state of Ceara, Brazil, resulted in subsequent surveillance and environmental sampling which led to the recognition of B. pseudomallei as an endemic pathogen in that area. From 2003 to 2015, 24 clinical and 12 environmental isolates were collected across Ceara along with one from the state of Alagoas. Using next-generation sequencing, multilocus sequence typing, and single nucleotide polymorphism analysis, we characterized the genomic diversity of this collection to better understand the population structure of B. pseudomallei associated with Ceara. We found that the isolates in this collection form a distinct subclade compared to other examples from the Western Hemisphere. Substantial genetic diversity among the clinical and environmental isolates was observed, with 14 sequence types (STs) identified among the 37 isolates. Of the 31,594 core single-nucleotide polymorphisms (SNPs) identified, a high proportion (59%) were due to recombination. Because recombination events do not follow a molecular clock, the observation of high occurrence underscores the importance of identifying and removing recombination SNPs prior to evolutionary reconstructions and inferences in public health responses to B. pseudomallei outbreaks. Our results suggest long-term B. pseudomallei prevalence in this recently recognized region of melioidosis endemicity.
IMPORTANCE B. pseudomallei causes significant morbidity and mortality, but its geographic prevalence and genetic diversity are not well characterized, especially in the Western Hemisphere. A better understanding of the genetic relationships among clinical and environmental isolates will improve knowledge of the population structure of this bacterium as well as the ability to conduct epidemiological investigations of cases of melioidosis.
INTRODUCTION
Melioidosis is a disease caused by Burkholderia pseudomallei, a bacterium that traditionally has been associated with Southeast Asia and northern Australia. It is difficult to diagnose, since it can present with symptoms that are nonspecific. Also, clinical laboratory staff often are not familiar with the bacterium and can confuse it with other bacteria (1, 2). The bacterium naturally occurs in the soil and water in habitats, typically in tropical and subtropical regions. An analysis of its predicted global distribution indicates that a large swath of South America, including a substantial portion of Brazil, has an environment suitable for survival of this pathogen (3). However, melioidosis had been noted only sporadically in South America, until an outbreak in the municipality of Tejucuoca in the state of Ceara, Brazil, in 2003, caused substantial concern, since it caused disease in four siblings who swam in a water reservoir near their residence, killing three of them in a short period of time (1, 4). This outbreak and subsequent cases of the disease in Ceara prompted improved surveillance for melioidosis in this state. The cases also prompted environmental sampling to identify potential sites of exposure to the bacterium, especially for the four siblings infected in 2003. Melioidosis was determined to be endemic in Ceara based on the cases found during improved surveillance as well as isolates recovered during environmental sampling (4–7).
Previous work to genetically characterize the isolates recovered in Ceara relied on pulsed-field gel electrophoresis (PFGE), ribotyping, and randomly amplified polymorphic DNA (4, 8). Although they provide some insight, these methods lack the high level of resolution now available with whole-genome sequencing (WGS), which can provide near-complete genomic sequences. Analysis of genomic data using single nucleotide polymorphisms (SNPs) has also been shown to be superior to multilocus sequence typing (MLST), which is currently the most common method to genetically subtype B. pseudomallei (9, 10). MLST relies on the analysis of portions of 7 housekeeping genes as a proxy for genome relatedness. Occasionally, the same MLST sequence types (STs) have been observed in divergent strains of B. pseudomallei due to ST homoplasy or within ST polyclonality due to recombination events. The appearance of relatedness by MLST in these instances can be overcome by analysis of the whole-genome data (11, 12).
Since 2005, melioidosis has been a reportable disease in Ceara, Brazil. Isolates from the original 2003 outbreak were combined with both clinical and environmental isolates collected until 2015 to make a panel representing geographic diversity across Ceara, except for one clinical isolate from the state of Alagoas, which does not share a border with Ceara but is approximately 300 km away at the closest point. To assess whether the pathogen came from a local or outside region, we performed WGS, MLST, and SNP analysis on this collection. We present our findings on the genomic diversity of isolates in this panel.
RESULTS
MLST performed on all 37 isolates gave a total of 14 STs, which consisted of four previously described STs (ST 92, ST 95, ST 297, and ST 1355) along with 10 new ones (ST 1454 to ST 1463) (Table 1). The most frequently observed sequence type in northeastern Brazil was ST 95 (38%; 14 of 37) and included isolates of clinical and environmental origin from 7 different municipalities.
TABLE 1.
Samples of B. pseudomallei collected throughout northeastern Brazila
| Isolate CEMM no. | Locationb | Sample type | Collection date | MLST sequence type | WGS SNP cluster |
|---|---|---|---|---|---|
| 03-6-033 | Tejucuoca, Ceara | Clinical | 2003 | 95 | B |
| 03-6-034 | Tejucuoca, Ceara | Clinical | 2003 | 1355 | A |
| 03-6-035 | Tejucuoca, Ceara | Clinical | 2003 | 1355 | A |
| 03-6-036 | Ipu, Ceara | Clinical | 2008 | 95 | C |
| 03-6-037 | Aracoiaba, Ceara | Clinical | 2005 | 1462 | G |
| 03-6-038 | Granja, Ceara | Clinical | 2009 | 95 | B |
| 03-6-039 | Tejucuoca, Ceara | Environmental | 2007 | 1460 | None |
| 03-6-040 | Tejucuoca, Ceara | Environmental | 2007 | 95 | B |
| 03-6-041 | Tejucuoca, Ceara | Environmental | 2007 | 95 | C |
| 03-6-042 | Tejucuoca, Ceara | Environmental | 2007 | 1463 | D |
| 03-6-043 | Tejucuoca, Ceara | Environmental | 2007 | 1463 | D |
| 03-6-044 | Tejucuoca, Ceara | Environmental | 2007 | 95 | A |
| 03-6-045 | Tejucuoca, Ceara | Environmental | 2007 | 95 | B |
| 03-6-046 | Tejucuoca, Ceara | Environmental | 2007 | 1463 | D |
| 03-6-047 | Tejucuoca, Ceara | Environmental | 2007 | 1463 | D |
| 03-6-048 | Tejucuoca, Ceara | Environmental | 2007 | 95 | C |
| 05-2-059 | Ipu, Ceara | Clinical | 2015 | 297 | None |
| 05-3-008 | Itapaje, Ceara | Clinical | 2009 | 1355 | A |
| 05-3-009 | Ipu, Ceara | Clinical | 2010 | 1454 | F |
| 05-3-010 | Pacoti, Ceara | Clinical | 2010 | 95 | F |
| 05-3-011 | Ocara, Ceara | Clinical | 2010 | 95 | B |
| 05-5-065 | Sao Joao do Jaguaribe, Ceara | Clinical | 2011 | 92 | E |
| 05-5-066 | Taua, Ceara | Clinical | 2012 | 1455 | None |
| 05-5-096 | Caridade, Ceara | Clinical | 2011 | 92 | E |
| 05-6-089 | Sao Gonçalo do Amarante, Ceara | Clinical | 2012 | 95 | G |
| 05-6-090 | Solonopole, Ceara | Clinical | 2014 | 1456 | A |
| 05-6-091 | Fortaleza, Ceara | Clinical | 2014 | 92 | E |
| 05-6-092 | Amontada, Ceara | Clinical | 2014 | 1457 | None |
| 05-6-093 | Granja, Ceara | Clinical | 2014 | 92 | E |
| 05-6-100 | Maceio, Alagoas | Clinical | 2010 | 95 | F |
| 05-6-101 | Banabuiu, Ceara | Clinical | 2004 | 1458 | None |
| 05-6-102 | Pacoti, Ceara | Clinical | 2010 | 95 | F |
| 05-6-103 | Pacoti, Ceara | Clinical | 2010 | 95 | F |
| 05-6-104 | Unknown, Ceara | Clinical | NA | 1459 | None |
| 05-6-105 | Unknown, Ceara | Clinical | 2009 | 1462 | G |
| 05-6-106 | Banabuiu, Ceara | Environmental | 2004 | 1461 | A |
| 05-6-107 | Banabuiu, Ceara | Environmental | 2004 | 1461 | A |
Clinical samples were derived from culture-confirmed cases of melioidosis, and environmental samples were directly collected from topsoil or inland waters. NA, not available.
For clinical isolates, the location refers to where the patient is believed to have been infected, sought treatment, or resided.
A phylogenetic tree based on SNP analysis of the draft genome sequence of each isolate indicates that all in our panel occur in the Western Hemisphere within a clade containing isolate BCC 215. BCC 215 was recovered during the original 2003 outbreak in Tejucuoca (Ceara), and its genome sequence was released to NCBI in 2007 (Fig. 1) (4, 13). These 38 isolates were further evaluated as a discrete group to identify clusters within the northeastern Brazilian clade and to further identify closely related isolates which may have epidemiological linkages.
FIG 1.
Maximum-likelihood phylogenetic tree of selected B. pseudomallei isolates collected throughout the world. Each leaf is labeled with its strain (or isolate) designation as the prefix, and geographic locations of isolation origin are suffixes. Bootstrap values above 70% are shown for more internal nodes, but none are shown for those near leaves. The term “ex” indicates the putative location where infection occurred. N.E., northeastern. The substitution bar indicates the number of substitutions per SNP site.
Extensive recombination.
With draft genome sizes ranging from 6.8 to 7.4 Mbp, the Brazilian clade’s whole-genome alignment captured 5.9 Mbp of the core genome, and 31,594 positions were variable (SNPs). Recombination assessment of the Brazilian clade genomes revealed that 12,976 (41%) of the SNPs were due to mutation. ClonalFrameML also estimated that 3 SNPs occurred for each recombination event, and the relative effect on recombination and mutation (r/m) was 6.53, which suggests that recombination introduced much more SNP changes than mutations did. A complementary approach with HomoplasyFinder gave a consistency index average of 62.3% (35.4% standard deviation) and reported 14,220 (45%) consistent sites, indicating that ClonalFrameML classified 3% more positions as recombination events. Such recombination events with no homoplasy, illustrated with a white background highlight and dark blue centered position in Fig. 2, were rare, but three large segments were inferred from an ancestral node that 35 of 38 isolates share. Here, “SNPs” refers to mutational SNPs with recombination sites removed unless otherwise specified.
FIG 2.
Maximum-likelihood phylogenetic tree of genome assemblies from northeast Brazil. Core SNP positions (n = 31,594) for each sample and each reconstructed ancestral node were labeled as mutational (light blue) or due to recombination (dark blue) on a per-site basis. Dotted horizontal lines connect samples and internal ancestral sequences to the illustration of inferred recombination positions. Cluster membership is indicated with color-coded circles and their corresponding-colored leaves, which contain MLST assignments for comparison. Isolate quantities per cluster are noted in parentheses in the key.
Outbreak diversity.
The outbreak had three clinical isolates and 10 environmental isolates from the city of Tejucuoca. Consistent with their MLST profiles, three clinical isolates from the original Tejucuoca (Ceara) outbreak (03-6-033 [ST 95], 03-6-034 [ST 1355], and 03-6-035 [ST 1355]) were found to belong to two different SNP clusters according to the genome analysis (Table 1; Fig. 2). The ST 1355 isolates reside in cluster A with an environmental (soil) isolate from Tejucuoca, a more recent clinical isolate from Itapaje, and isolate BCC 215 (4, 13). Isolates 03-6-035 and 05-3-008 are each separated from BCC 215 by just 7 SNPs, with 4 SNPs separating 03-6-035 and 05-3-008. The Tejucuoca ST 95 isolate, 03-6-33, resides in cluster B with four environmental isolates from Tejucuoca, suggesting that distinct genotypes (by MLST and SNPs) existed within the outbreak in the same city.
ST 95 diversity and patient-environment relatedness.
Isolates that 03-6-033 groups closely with are 10 to 18 SNPs apart (clusters B and C) and include four environmental isolates from Tejucuoca and four clinical isolates from other cities in Ceara. All of its closely related isolates are ST 95; however, several other isolates collected from the outbreak are also ST 95 and are more distant (Fig. 2). Such diversity of isolates that are ST 95 with distinct branches shows that more expansive SNP analysis was required to disentangle relatedness.
Three clinical isolates (all with ST 95) from a single patient were recovered in 2010 (05-6-102, 05-6-103, and 05-3-010) from Pacoti, although the exact time frame among isolation events is unknown. In this clade, 05-6-102 and 05-3-010 are separated by only 10 SNPs, whereas 05-6-103 and 05-3-010 are the most divergent, with 19 SNPs between them. These three isolates served as an internal control (with analysts initially unaware of their relationship) and show that while the ST did not change, the genomic mutations of B. pseudomallei in the infected patient varied by as much as 19 SNPs. From the observation that 19 SNPs occurred in the same patient, it seems plausible other ST 95 isolates in clusters B and C could be related closely enough to suggest that a transmission network was captured (Fig. 2).
Novel STs unrelated to patient isolates.
Four isolates from Tejucuoca were collected from environmental sources and formed a distinct cluster (D) with a more distantly related patient isolate from Fortaleza. These soil isolates possessed a unique profile of previously identified MLST alleles, which was assigned a new ST, ST 1463. According to the SNP phylogeny, the closest patient isolate to these ST 1463 environmental isolates is 05-6-091 from Fortaleza; however, the relatively large (4,971 to 4,977) SNP distances do not suggest a recent transmission or connection.
Two B. pseudomallei isolates collected in Banabuiu from soil (05-6-106) and from water (05-6-107) also appear clonal and have only 8 SNPs between them. The clinical isolate 05-6-101 associated with Banabuiu has 5,086 SNPs compared to 05-6-106, thus indicating that this clinical isolate is not closely related to either environmental isolate from Banabuiu. The MLST data corroborate this lack of a close relationship, because 05-6-101 occurs as its own unique ST, ST 1458, and the environmental samples were both ST 1461. The closest patient isolate to 05-6-107 differs by 4,805 SNPs, and therefore no patients sampled in the outbreak were infected from these Banabuiu sources.
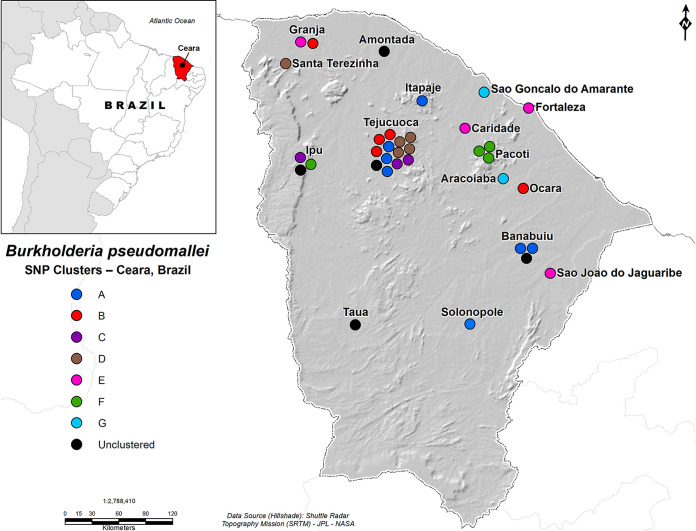
To geographically illustrate the genetic diversity of the isolates recovered from across Ceara, clusters are indicated on a map of the state (Fig. 3). A given color represents a cluster type, with each dot representing a genome from an isolate.
FIG 3.
Geographic distribution of SNP-based cluster types associated with isolates from across the state of Ceara. Each circle represents one genome of a given cluster and is placed adjacent to the name of the municipality with which it is associated. The dots do not directly correspond to a specific map coordinate. Three isolates are not shown: one from the state of Alagoas and two others known to be from Ceara but not associated with a specific municipality.
DISCUSSION
MLST limitations.
MLST is the most common method of genetically subtyping B. pseudomallei, so there is a large database available for comparisons to previous findings. The total number of genotypes exceeds 1,700 and generally captures diversity for new isolates of interest with a moderate resolution level, but in some cases the large geographic range of an ST limits epidemiological investigations. Among the previously defined STs that we also found in our data set from Ceara, ST 92 was previously noted in isolates associated with Puerto Rico (USA), Martinique (France), Brazil, and Mexico. ST 95 has been previously associated with isolates from Puerto Rico, from Mexico, and from a patient in Arizona originally from Central America. ST 297 was previously seen in isolates associated with Puerto Rico, Trinidad, and Mexico. ST 297 was also associated with an isolate originally thought to have been acquired by a World War II veteran in Southeast Asia while he was a prisoner of war, though it is now believed that he acquired his infection in the Americas (9). ST 1355 was previously noted for clinical isolate BCC 215, which was obtained during the original 2003 outbreak in Ceara (4, 9). In all of these cases, MLST points to a Western Hemisphere origin, but even establishing a continent of origin is not always possible.
Analysis of B. pseudomallei STs is not as useful for determining geographic provenance, since it provides only a moderate level of resolution. Because an ST can be found across different continents, countries, or regions, MLST lacks resolution to identify potential routes of transmission for epidemiological investigations. However, analysis of genome-wide SNPs using WGS has been shown to be useful (9–11, 14).
Diversity in Brazil.
In this study, all genomes (36 from Ceara and one from Alagoas) form a cohesive group within the previously established clade for genomes with origins in the Western Hemisphere (Fig. 1). This suggests that the Ceara and Alagoas isolates are members of a geographically defined subpopulation of B. pseudomallei, albeit with a high number of SNPs (diversity) among many of its members. The largest SNP distance within the clade is 5,390 between clinical isolates 05-5-066 (Taua) and 05-6-091 (Fortaleza), which were both recovered in 2012 (Table 1). Also noteworthy is the genome for isolate 4900CF from a patient in the Brazilian state of Mato Grosso do Sul in the midwestern part of the country (15). It is within the Western Hemisphere clade but not within the Ceara subgroup, suggesting an even higher level of diversity for B. pseudomallei in Brazil.
Recombination in B. pseudomallei.
We observed that 59% of the SNPs in these Brazilian isolates were due to recombination rather than mutation, and with a genome size (7.3 Mbp) larger than most bacterial pathogens, the total number of sites subject to mutation and recombination is high. Taken together with the fact that recombination does not occur at a molecular clock rate, these observations indicate that identifying and discarding all of these sites is important for making accurate phylogenetic inferences, which otherwise has shown incorrect branch lengths and topologies (16). The first report on a B. pseudomallei genome reported recombination as a possible mechanism for rearrangements throughout both chromosomes compared to other related Burkholderia spp. (17). Recombination positions are dispersed throughout the draft contigs in our isolates, and while one sample set previously reported more recombination in the larger chromosome, another study found that recombination is more frequent in the smaller chromosome (16, 18). Pearson et al. (16) reported that 6,331 of 14,544 (44%) SNPs were homoplasic, which is lower than the value for isolates from Brazil; however, Nandi et al. (18) reported a 7.2 r/m value, which was higher than what we observed. Such a high recombination rate is one of the highest among pathogenic bacteria (e.g., Streptococcus pneumoniae) (19).
Outbreak samples.
Considering first the clinical isolates in our panel from the original 2003 outbreak in Tejucuoca, 03-6-034 and 03-6-035 are in cluster A with 05-3-008 (Itapaje; clinical)—all ST 1355—as well as 03-6-044 (Tejucuoca; from an environmental sample from the area believed to be the source of infection for the 4 siblings), which is ST 95, and clinical isolate BCC215. Isolates 03-6-034 and 03-6-035 have 265 SNPs between them, indicating a modest level of relatedness. Although BCC215 is known to be from the original 2003 outbreak, it was originally sequenced in 2007, and we were unable to determine which isolate in the current panel it corresponds to. In our analysis, it matches 03-6-035 and 05-3-008 equally, with only 7 SNPs separating them, indicating clonality. Interestingly, 05-3-008, a clinical isolate from Itapaje, has only 4 SNP differences with 03-6-035 and 8 SNP differences with BCC215. This potentially indicates that a clonal lineage of B. pseudomallei has been disbursed in this region or that a common source of infection is involved in these cases. However, the possibility that a mix-up or mislabeling in the laboratory occurred is also plausible.
Cluster B consists of isolates that are all ST 95. The cluster consists of clinical isolate 03-6-033, also associated with the 2003 outbreak affecting the 4 siblings in Tejucuoca, along with four environmental isolates, 03-6-040, 03-6-041, 03-6-045, and 03-6-048, recovered during sampling around the siblings’ home and the nearby water reservoir believed to be the source of infection. This cluster also contains three other clinical isolates: 03-6-036 (Ipu), 03-6-038 (Granja), and 05-3-011 (Ocara). Using 03-6-033 as a reference, we see that it has 0 SNPs relative to 03-6-040, 03-6-041, 03-6-048, and 03-6-038; 1 SNP relative to 0306-045 and 05-3-011; and 3 SNPs relative to 03-6-036. Therefore, all members of cluster B appear clonal and were strong candidates for transmission analysis, especially given the potential environmental linkage to infected individuals, indicating the possibility that a clonal lineage of B. pseudomallei has been disbursed in this region or a common source of infection is involved in these cases. Again, the possibility that a mix-up or mislabeling in the laboratory occurred is also plausible.
It was previously documented that the four children who developed melioidosis in the 2003 Tejucuoca outbreak were exposed by playing in a water reservoir (4, 5). Our genome-wide analysis indicates a high level of genomic diversity among the isolates from this municipality, including those recovered from the environment in and near the water reservoir where the children were infected. The isolates are distributed in many disparate clusters, indicating high diversity, but at least some clinical isolates appear tightly linked to environmental isolates. Previous investigations by others have shown the utility of genomic analysis for matching probable sources of infection by comparison of clinical isolates to environmental samples. For example, investigations in Australia indicated that human clinical cases of melioidosis were probably due to exposure through the patients’ water supplies (20, 21), or potentially aerosolized bacteria near the patient’s residence (22). In another investigation, the likely source of infection for baby crocodiles was found to be an incubator used for their hatching (23).
The fact that the clinical isolates associated with municipalities that are distant from Tejucuoca but appear clonal with isolates from Tejucuoca (e.g., Ocara [05-3-011] and Granja [03-6-038], which are approximately 100 and 200 km away from Tejucuoca, respectively) is noteworthy. These findings suggest that some strains are widely dispersed throughout Ceara, causing relatively widespread infections, although we cannot rule out the possibility that patients were exposed to material from Tejucuoca. Other studies, such as that by Chapple et al. (24), found a geographically restricted zone for B. pseudomallei. They found that the maximum distance in their panel was 45 km for examples of ST 109 (24). However, another study indicated that examples of a truly clonal strain were found 460 km apart (25).
The clinical isolate 03-6-036 with only 15 SNPs different from other isolates, such as 03-6-040 from Tejucuoca or 03-6-038 from Granja, is also noteworthy, as it was recovered from a male from Fortaleza who traveled with his father to Ipu in 2008. The patient passed away 1 week after disease onset from pneumonia and sepsis. In 2010, his father, also from Fortaleza, developed melioidosis, which yielded isolate 05-3-009. The two isolates from son (03-6-036) and father (05-3-009) have 7,247 SNPs between them and are thus unrelated. Unfortunately, this genome distance information cannot help determine whether the father and son were exposed at different sites or were both exposed to a diverse population of bacteria at a single site. These results do support the hypothesis of widespread dispersal of clonal members of B. pseudomallei throughout Ceara. Without environmental isolates from Ocara, Granja, Ipu, and Itapaje for comparison, the population structure of B. pseudomallei in those municipalities remains unknown.
Another possibility to explain why isolates from disparate municipalities appear clonal would be a mislabeling or a mix-up in samples in the laboratory. To address this concern, we note that 13 isolates are associated with Tejucuoca either from the original 2003 outbreak or from environmental sampling in or near the area where the children are believed to have been infected. If isolates from Tejucuoca were clonal or even moderately related, we would expect to see a cluster of at least 13 members. The largest cluster in our dendrogram with near-intracluster genomic distances (cluster A) has only 8 members. Also, B. pseudomallei BCC215 served as an important internal control, because it was recovered from the original 2003 outbreak and was originally sequenced in 2007 before most members of this panel were isolated (4, 13). Its relation to clinical isolates 03-6-034 and 03-6-035 and environmental isolate 03-6-044 is supportive of their being from Tejucuoca. Another study of B. pseudomallei distribution in a discrete area in Western Australia addressed similar questions about the clonality observed in that collection. Their work indicated that B. pseudomallei can persist in the environment for decades with little evolution (26).
In summary, based on SNP analysis of whole-genome sequences, B. pseudomallei isolates in the state of Ceara form a phylogenetically discrete subclade within the Western Hemisphere clade. However, the isolates from Ceara do not all appear to be clonal based on the diversity of SNPs (by quantity and positions) observed among them, which is also apparent from the variety of STs identified. Based on our genome comparisons, almost a third of isolates had no close match, suggesting that it is necessary to isolate and sequence more samples from the area to aid in epidemiological investigations. Even within a single municipality, such as in Tejucuoca, a genetically diverse population of B. pseudomallei is present with distinct genotypes mixed together in the native environment. Therefore, it is probable that B. pseudomallei was introduced to Ceara sufficiently long ago for genetic diversity to have developed among some lineages. It appears that dispersion of clonal strains and mixing of the bacterial populations occurred within this region, resulting in some areas containing unrelated B. pseudomallei strains, which has been noted previously in other regions where the bacterium is endemic, such as in Southeast Asia. Further recovery and genetic characterization of the bacterium from Ceara and other states in Brazil will result in a better understanding of its population structure in this region of South America.
MATERIALS AND METHODS
B. pseudomallei isolates were collected in the state of Ceara, Brazil, from 2003 to 2015 from both clinical cases (n = 24) and environmental sampling (n = 12), as well as one from a clinical case associated with the Brazilian state of Alagoas. The collection and characterization of both clinical and environmental samples were reviewed under COMEPE no. 16/2005 (CEP-HUWC/UFC, Ethics Committee of the University Hospital Ceara Federal University) and project identification no. 434.786 (CEP-UNIFOR, University of Fortaleza). Environmental samples were collected on private property with permission of landowners. A review at the U.S. CDC also determined that this study was not human subject research. Municipalities recorded as sources for these isolates in Table 1 represent the location where the bacterium was recovered in the case of environmental sampling, whereas for clinical cases, they indicate where the patient was believed to have been infected, sought treatment, or resided. These data were mapped with the geographic information system (GIS) software ArcGIS (ArcMap) version 10.8.1 (https://www.esri.com/en-us/home). Genomic DNA was extracted using a High Pure PCR template preparation kit (Roche), according to the manufacturer’s recommendations, with some modifications. Briefly, after the cell lysis step, using specific buffers and proteinase K, the samples were treated with RNase at 37°C, for 30 min, to remove contaminating RNA and improve DNA quantification. Afterward, DNA was quantified, by using NanoDrop, yielding a minimum of 6.5 μg of DNA and a minimum A260/A280 ratio of 1.81, followed by electrophoretic analysis by agarose gel to evaluate the integrity of the DNA. Sequencing was performed on an Illumina MiSeq instrument (2 × 250 bp) as previously described (Table 1) (9).
Reads were filtered for PhiX using a 31-mer search query allowing for a single SNP with BBDUK version 35.92. Illumina adapter sequences were clipped from reads with Trimmomatic version 0.35 (27), which also quality trimmed sequences below 99.9% fidelity. Paired reads with at least 80% of their length overlapping due to small fragment size were assembled with PEAR version 0.9.10 (28) using “–keep-original –p- value 0.01” options. Paired and singleton reads at least 50 bp in length were used for assembly in SPAdes version 3.13.0 using the “–assembler-only” option (29). Contiguous sequences at least 1 kbp in length with at least 5-fold coverage were retained. Three sequential rounds of read mapping (with bwa mem version 0.7.17 [“-x intractg”] and samtools version 1.8) and consensus filtering (Pilon version 1.22 [30]) were used to correct SNPs and indels with the “–mindepth 0.5” option. After three sequential polishing rounds, no new errors were identified to be fixed.
SNPs were identified from genome assemblies using Parsnp version 1.3 (31). Both whole-genome alignment and core SNPs were extracted to evaluate effects on recombination. SNP sites were extracted from Parsnp's binary gingr file with harvest-tools version 1.3 (31). The FASTA format was converted to Phylip format with Biopython version 1.74 (32). PhyML version 20120412 was used to apply the general time-reversible (GTR) substitution model with 100 bootstrap replicates, branch length and rate parameter optimization, and “–search BEST” options to calculate nucleotide frequencies, transition mutations relative to transversion events (kappa), the shape of a gamma distribution, and branch dispersion (33). These values and the FASTA of SNPs were used in ClonalFrameML version 1.12 for 20 uncertainty estimate simulations of the Baum-Welch expectation maximization algorithm, which gave estimates of recombination-to-mutation per site, average length of recombined fragments, and divergence rate of recombination sites (34). Calculated values from PhyML, estimated values from the initial Baum-Welch algorithm, and the SNP sequences were all used in ClonalFrameML a second time to apply the expectation maximization algorithm for each branch individually. All reconstructed recombination events from ancestral nodes and samples along with their level of homoplasy were visualized in R version 3.3.2 with the APE version 4.1 and phangorn version 2.2.0 libraries (35–37). SNP distances between samples were used for OPTICS clustering with Scikit-learn version 0.23.1 (38) and NumPy version 1.13.1 (39). Graphical editing was performed with Inkscape version 1.0 (40). HomoplasyFinder version 0.0.999999 was used with default parameters (41).
Multilocus sequence typing (MLST) is based on sequencing a portion of seven single-copy housekeeping genes to assign alleles. The combination of alleles is used to designate a sequence type (ST). MLST was performed by either traditional Sanger sequencing or by in silico BLASTn analysis of the draft whole-genome sequence with the reference sequences available on the PubMLST website for B. pseudomallei (http://pubmlst.org/bpseudomallei/) (14, 42).
Data availability.
Sequences are available under NCBI BioProject no. PRJNA523188.
ACKNOWLEDGMENTS
This work was supported by grants from the National Council for Scientific and Technological Development (CNPq, Brazil). This publication made use of the Burkholderia pseudomallei MLST website (http://pubmlst.org/bpseudomallei/) sited at the University of Oxford; the development of this site was funded by the Wellcome Trust.
The conclusions, findings, and opinions expressed by the authors do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
REFERENCES
- 1.Benoit TJ, Blaney DD, Doker TJ, Gee JE, Elrod MG, Rolim DB, Inglis TJ, Hoffmaster AR, Bower WA, Walke HT. 2015. A review of melioidosis cases in the Americas. Am J Trop Med Hyg 93:1134–1139. doi: 10.4269/ajtmh.15-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiersinga WJ, Virk HS, Torres AG, Currie BJ, Peacock SJ, Dance DAB, Limmathurotsakul D. 2018. Melioidosis. Nat Rev Dis Primers 4:17107. doi: 10.1038/nrdp.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Limmathurotsakul D, Golding N, Dance DA, Messina JP, Pigott DM, Moyes CL, Rolim DB, Bertherat E, Day NP, Peacock SJ, Hay SI. 2016. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol 1:15008. doi: 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 4.Rolim DB, Vilar DC, Sousa AQ, Miralles IS, de Oliveira DC, Harnett G, O'Reilly L, Howard K, Sampson I, Inglis TJ. 2005. Melioidosis, northeastern Brazil. Emerg Infect Dis 11:1458–1460. doi: 10.3201/eid1109.050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rolim DB, Rocha MF, Brilhante RS, Cordeiro RA, Leitao NP, Jr, Inglis TJ, Sidrim JJ. 2009. Environmental isolates of Burkholderia pseudomallei in Ceara State, northeastern Brazil. Appl Environ Microbiol 75:1215–1218. doi: 10.1128/AEM.01953-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rolim DB, Vilar DC, de Goes Cavalcanti LP, Freitas LB, Inglis TJ, Nobre Rodrigues JL, Nagao-Dias AT. 2011. Burkholderia pseudomallei antibodies in individuals living in endemic regions in northeastern Brazil. Am J Trop Med Hyg 84:302–305. doi: 10.4269/ajtmh.2011.10-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rolim DB, Lima RXR, Ribeiro AKC, Colares RM, Lima LDQ, Rodriguez-Morales AJ, Montufar FE, Dance DAB. 2018. Melioidosis in South America. Trop Med Infect Dis 3:60. doi: 10.3390/tropicalmed3020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandeira T, Castelo-Branco D, Rocha MFG, Cordeiro RA, Ocadaque CJ, Paiva MAN, Brilhante RSN, Sidrim JJC. 2017. Clinical and environmental isolates of Burkholderia pseudomallei from Brazil: genotyping and detection of virulence gene. Asian Pac J Trop Med 10:945–951. doi: 10.1016/j.apjtm.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Gee JE, Gulvik CA, Elrod MG, Batra D, Rowe LA, Sheth M, Hoffmaster AR. 2017. Phylogeography of Burkholderia pseudomallei isolates, Western Hemisphere. Emerg Infect Dis 23:1133–1138. doi: 10.3201/eid2307.161978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsang AKL, Lee HH, Yiu SM, Lau SKP, Woo PCY. 2017. Failure of phylogeny inferred from multilocus sequence typing to represent bacterial phylogeny. Sci Rep 7:4536. doi: 10.1038/s41598-017-04707-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Smet B, Sarovich DS, Price EP, Mayo M, Theobald V, Kham C, Heng S, Thong P, Holden MT, Parkhill J, Peacock SJ, Spratt BG, Jacobs JA, Vandamme P, Currie BJ. 2015. Whole-genome sequencing confirms that Burkholderia pseudomallei multilocus sequence types common to both Cambodia and Australia are due to homoplasy. J Clin Microbiol 53:323–326. doi: 10.1128/JCM.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price EP, Sarovich DS, Viberg L, Mayo M, Kaestli M, Tuanyok A, Foster JT, Keim P, Pearson T, Currie BJ. 2015. Whole-genome sequencing of Burkholderia pseudomallei isolates from an unusual melioidosis case identifies a polyclonal infection with the same multilocus sequence type. J Clin Microbiol 53:282–286. doi: 10.1128/JCM.02560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukhopadhyay S, Thomason MK, Lentz S, Nolan N, Willner K, Gee JE, Glass MB, Inglis TJ, Merritt A, Levy A, Sozhamannan S, Mateczun A, Read TD. 2010. High-redundancy draft sequencing of 15 clinical and environmental Burkholderia strains. J Bacteriol 192:6313–6314. doi: 10.1128/JB.00991-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Spratt BG. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol 41:2068–2079. doi: 10.1128/jcm.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barth AL, de Abreu E Silva FA, Hoffmann A, Vieira MI, Zavascki AP, Ferreira AG, da Cunha LG, Albano RM, de Andrade Marques E. 2007. Cystic fibrosis patient with Burkholderia pseudomallei infection acquired in Brazil. J Clin Microbiol 45:4077–4080. doi: 10.1128/JCM.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, Price EP, Glass MB, Leadem B, Beckstrom-Sternberg JS, Allan GJ, Foster JT, Wagner DM, Okinaka RT, Sim SH, Pearson O, Wu Z, Chang J, Kaul R, Hoffmaster AR, Brettin TS, Robison RA, Mayo M, Gee JE, Tan P, Currie BJ, Keim P. 2009. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol 7:78. doi: 10.1186/1741-7007-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A 101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nandi T, Holden MT, Didelot X, Mehershahi K, Boddey JA, Beacham I, Peak I, Harting J, Baybayan P, Guo Y, Wang S, How LC, Sim B, Essex-Lopresti A, Sarkar-Tyson M, Nelson M, Smither S, Ong C, Aw LT, Hoon CH, Michell S, Studholme DJ, Titball R, Chen SL, Parkhill J, Tan P. 2015. Burkholderia pseudomallei sequencing identifies genomic clades with distinct recombination, accessory, and epigenetic profiles. Genome Res 25:608. [PMC free article] [PubMed] [Google Scholar]
- 19.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McRobb E, Sarovich DS, Price EP, Kaestli M, Mayo M, Keim P, Currie BJ. 2015. Tracing melioidosis back to the source: using whole-genome sequencing to investigate an outbreak originating from a contaminated domestic water supply. J Clin Microbiol 53:1144–1148. doi: 10.1128/JCM.03453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarovich DS, Chapple SNJ, Price EP, Mayo M, Holden MTG, Peacock SJ, Currie BJ. 2017. Whole-genome sequencing to investigate a non-clonal melioidosis cluster on a remote Australian island. Microb Genom 3:e000117. doi: 10.1099/mgen.0.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Currie BJ, Price EP, Mayo M, Kaestli M, Theobald V, Harrington I, Harrington G, Sarovich DS. 2015. Use of whole-genome sequencing to link Burkholderia pseudomallei from air sampling to mediastinal melioidosis, Australia. Emerg Infect Dis 21:2052–2054. doi: 10.3201/eid2111.141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rachlin A, Kleinecke M, Kaestli M, Mayo M, Webb JR, Rigas V, Shilton C, Benedict S, Dyrting K, Currie BJ. 2019. A cluster of melioidosis infections in hatchling saltwater crocodiles (Crocodylus porosus) resolved using genome-wide comparison of a common north Australian strain of Burkholderia pseudomallei. Microb Genom 5:e000288. doi: 10.1099/mgen.0.000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapple SN, Price EP, Sarovich DS, McRobb E, Mayo M, Kaestli M, Spratt BG, Currie BJ. 2016. Burkholderia pseudomallei genotype distribution in the Northern Territory, Australia. Am J Trop Med Hyg 94:68–72. doi: 10.4269/ajtmh.15-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aziz A, Sarovich DS, Harris TM, Kaestli M, McRobb E, Mayo M, Currie BJ, Price EP. 2017. Suspected cases of intracontinental Burkholderia pseudomallei sequence type homoplasy resolved using whole-genome sequencing. Microb Genom 3:e000139. doi: 10.1099/mgen.0.000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapple SNJ, Sarovich DS, Holden MTG, Peacock SJ, Buller N, Golledge C, Mayo M, Currie BJ, Price EP. 2016. Whole-genome sequencing of a quarter-century melioidosis outbreak in temperate Australia uncovers a region of low-prevalence endemicity. Microb Genom 2:e000067. doi: 10.1099/mgen.0.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 34.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paradis E, Claude J, Strimmer K. 2004. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 36.Schliep KP 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 38.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Dushesnay E. 2011. Scikit-learn: machine learning in Python. J Machine Learning Res 12:2825–2830. [Google Scholar]
- 39.Oliphant TE 2007. Python for scientific computing. Comput Sci Eng 9:90. [Google Scholar]
- 40.Contributors I 2020. Inkscape, version 1.0. https://inkscape.org/.
- 41.Crispell J, Balaz D, Gordon SV. 2019. HomoplasyFinder: a simple tool to identify homoplasies on a phylogeny. Microb Genom 5:e000245. doi: 10.1099/mgen.0.000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences are available under NCBI BioProject no. PRJNA523188.