Abstract
Bromodomain-containing proteins are often part of chromatin-modifying complexes, and their activity can lead to altered expression of genes that drive cancer, inflammation and neurological disorders in humans. Bromodomain-PHD finger protein 1 (BRPF1) is part of the MOZ (monocytic leukemic zinc-finger protein) HAT (histone acetyltransferase) complex, which is associated with chromosomal translocations known to contribute to the development of acute myeloid leukemia (AML). BRPF1 contains a unique combination of chromatin reader domains including two plant homeodomain (PHD) fingers separated by a zinc knuckle (PZP domain), a bromodomain, and a proline-tryptophan-tryptophan-proline (PWWP) domain. BRPF1 is known to recruit the MOZ HAT complex to chromatin by recognizing acetylated lysine residues on the N-terminal histone tail region through its bromodomain. However, histone proteins can contain several acetylation modifications on their N-terminus, and it is unknown how additional marks influence bromodomain recruitment to chromatin. Here, we identify the BRPF1 bromodomain as a selective reader of di-acetyllysine modifications on histone H4. We used ITC assays to characterize the binding of di-acetylated histone ligands to the BRPF1 bromodomain and found that the domain binds preferentially to histone peptides H4K5acK8ac and H4K5acK12ac. Analytical ultracentrifugation (AUC) experiments revealed that the monomeric state of the BRPF1 bromodomain coordinates di-acetylated histone ligands. NMR chemical shift perturbation studies, along with binding and mutational analyses, revealed non-canonical regions of the bromodomain-binding pocket that are important for histone tail recognition. Together, our findings provide critical information on how the combinatorial action of post-translational modifications can modulate BRPF1 bromodomain binding and specificity.
Keywords: Bromodomain, Di-acetyllysine, Epigenetics, Histone, Post-translational modification
Abbreviations: AML, acute myeloid leukemia; AUC, analytical ultracentrifugation; BRPF1, bromodomain-PHD finger protein 1; CD, circular dichroism; DTT, dithiothreitol; HAT, histone acetyltransferase; MEAF6, MYST/Esa1-associated factor 6; ING5, inhibitor of growth 5; ITC, isothermal titration calorimetry; MOZ, monocytic leukemic zinc-finger; NMR, nuclear magnetic resonance; PCR, polymerase chain reaction; PHD, plant homeodomain; PTMs, post-translational modifications; PWWP, proline-tryptophan-tryptophan-proline; SV, sedimentation velocity; TCEP, tris(2-carboxyethyl)phosphine
Graphical abstract
Introduction
In the human genome, DNA accessibility and transcriptional activation are often regulated by post-translational modifications (PTMs), among which acetylation of lysine residues on histones plays a major role. Histone acetylation enhances DNA accessibility by loosening the compact conformation of chromatin and by recruiting transcription and chromatin remodeling factors to increase transcriptional activity (Sanchez et al., 2014, Galvani and Thiriet, 2015). Bromodomains, which are epigenetic reader domains that specifically recognize ε-N-acetylated lysine residues on histone tails, are known to be important regulators of chromatin remodeling and transcriptional activation (Dhalluin et al., 1999). A total of 61 unique bromodomains are known to exist in 46 different bromodomain-containing proteins, and these bromodomains have been grouped into eight major families on the basis of sequence and structural homology (Filippakopoulos et al., 2012). Mutation and aberrant expression of bromodomain-containing proteins have been associated with multiple types of cancer, inflammation, neurological disorders and metabolic diseases. Importantly, bromodomains are often found in chromatin-modifying complexes, whose activity can lead to the altered expression of genes that drive these diseases in humans (Arrowsmith et al., 2012).
Bromodomain-PHD finger protein 1 (BRPF1) is known to play a role in maintaining the expression of genes involved in several developmental processes and hematopoiesis (Hibiya et al., 2009, Yan et al., 2017, You et al., 2016). The BPRF1 protein is a unique, multivalent chromatin reader that contains an N-terminal plant homeodomain (PHD)-zinc-knuckle-PHD (PZP) module, a bromodomain for acetyllysine recognition, and a C-terminal proline-tryptophan-tryptophan-proline (PWWP) domain (Laue et al., 2008). The N-terminal motif I of BRPF1 interacts with monocytic leukemic zinc finger (MOZ) and MOZ-related factor (MORF), while the downstream motif II interacts with inhibitor of growth 5 (ING5) and the MYST/Esa1-associated factor 6 (MEAF6) (Ullah et al., 2008). Thus, an important function of BRPF1 is to bridge the association of MOZ and MORF with ING5 and MEAF6, where it is an important regulator of many cellular processes including the self-renewal of leukemic stem cells (You et al., 2016), embryo development and cell proliferation (You et al., 2015), and osteoclast differentiation (Meier et al., 2017) through its activity with MOZ and MORF (You et al., 2016). BRPF1 enhances the acetyltransferase activity of the MOZ/MORF HATs via interaction with the histone acetyltransferase domain (Ullah et al., 2008), and also bridges the enzymatic complex that carries out specific PTMs in chromatin (Carlson and Glass, 2014).
PTMs on histones are known to be crucial for the regulation of chromatin and DNA accessibility (Li et al., 2007). PTMs modulate signal transduction pathways to recruit effector molecules, such as readers of histone modifications to chromatin (Yun et al., 2011). These histone reader domains direct biological outcomes based on the cellular context of the modifications (Berger, 2007). In humans, proteins are known to undergo a number of PTMs, including but not limited to phosphorylation, methylation, acetylation and ubiquitination, that greatly influence their function in cellular events (Vermeulen et al., 2010). Specifically, histone acetylation is an important component of epigenetic signaling, in that it weakens the interactions of histones with DNA by decreasing nucleosome compaction (Workman and Kingston, 1998), regulates enzymatic activity, and mediates the localization of macromolecular complexes to histones through bromodomain-containing proteins (Flynn et al., 2015). Therefore, the recognition of acetylated lysine by bromodomains has broad implications for cellular events including chromatin remodeling and transcriptional activation (Dyson et al., 2001).
The postulation that aberrant chromatin acetylation by MOZ may be a contributing factor to the development of cancer and other diseases was first demonstrated by cloning the breakpoint-associated genes of the recurrent t (8; 16) (p11; p13) translocation in patients with a subtype of acute myeloid leukemia (AML) (Borrow et al., 1996). This translocation, which results in a novel fusion that combines the PHD finger motifs and MYST acetyltransferase domain of MOZ with an intact CREB-binding protein (CBP), suggested that the MOZ-CBP fusion protein gives rise to leukemogenesis through chromatin hyperacetylation (Borrow et al., 1996). Since BRPF1 plays a crucial role in the MOZ/MORF complex, and the interaction domains of this complex remain intact after the translocation, interaction with BRPF1 may be implicated in other diseases in which MOZ and MORF are known to play a role. The MOZ gene has been reported to be mutated in esophageal adenocarcinoma (Dulak et al., 2013), while the MORF gene has been reported to be disrupted in leiomyomata (Moore et al., 2004, Panagopoulos et al., 2015), mutated in familial breast cancer (Lynch et al., 2013), and altered in castration-resistant prostate cancer (Grasso et al., 2012). Deletion of the mouse BRPF1 gene showed that BRPF1 is crucial for the growth and proliferation of embryonic fibroblasts and hematopoietic progenitors (You et al., 2015). Supporting evidence that BRPF1 acts through MOZ and MORF to modulate chromatin modification states comes from the report that multiple mutations in the BRPF1 gene have been reported to give rise to neurodevelopmental disorders associated with a deficiency in histone H3K23 acetylation (Yan et al., 2017).
The BRPF1 bromodomain belongs to family IV bromodomains, which also include the BRPF2, BRPF3, BRD7, BRD9, ATAD2 and ATAD2B bromodomains based on their sequence and structural homology (Filippakopoulos et al., 2012). Bromodomains share a conserved globular fold and are composed of a left-handed bundle of four α-helices αZ, αA, αB and αC. The helices are linked by loops of variable length (ZA and BC loops), which form the hydrophobic acetyllysine (Kac) binding pocket (Filippakopoulos et al., 2012). The difference in the acetyllysine recognition of these bromodomains is attributed mainly to the variability of their ZA and BC loops (Filippakopoulos et al., 2012). These differences have also permitted the development of small molecule inhibitors with selectivity for specific bromodomains within the same family (Bamborough et al., 2016, Bouche et al., 2017, Clark et al., 2015, Fernandez-Montalvan et al., 2017).
An earlier study characterized the interaction of the BRPF1 bromodomain with mono-acetylated ligands on the N-terminal of histone tails and showed selectivity for multiple acetylation modifications including H2AK5ac, H4K12ac and H3K14ac (Poplawski et al., 2014). However, each of these four histone proteins can be post-translationally modified at multiple locations, and with several different chemical marks (Huang et al., 2014). Proteomic studies have demonstrated that each histone tail carries a unique set of modifications (Young et al., 2009), and that the abundance and availability of specific histone proteoforms is dynamic (Wang et al., 2018, Jiang et al., 2018). The BET bromodomains BRD2, BRD4 and BRDT are all able to bind di-acetyllysine modifications including H4K5acK8ac, H4K5acK12ac, and H4K12acK16ac (Filippakopoulos et al., 2012; Moriniere et al., 2009). Other family IV bromodomains, including the BRD9 and ATAD2 bromodomain, are able to interact with di-acetylated histone ligands such as H4K5acK8ac and H4K5acK12ac, respectively (Flynn et al., 2015, Koo et al., 2016). However, the interaction of the BRPF1 bromodomain with di-acetyllysine modifications has not been systematically characterized, and it is unknown how multiple histone marks modulate acetyllysine recognition.
Previously published data on the BRPF1 bromodomain revealed that it recognizes several acetylated lysine residues on N-terminal histone tails (Poplawski et al., 2014). Related bromodomain proteins have been observed to cooperatively bind histone peptides with di-acetyllysine marks, through accommodation of the two acetyllysine residues in the same bromodomain binding pocket. For example, the BRDT bromodomain BD1 in the bromodomain and extra-terminal domain (BET) family, recognizes the di-acetylated histone ligand H4K5acK8ac (Moriniere et al., 2009). Other bromodomain proteins in the BET family including BRD2 and BRD4 are also known to recognize di-acetylated histone ligands, mainly through their enlarged binding pockets (Filippakopoulos et al., 2012). Three-dimensional X-ray crystal structures of the BAZ2A and BAZ2B bromodomains in family V have also been solved in complex with H4K16acK20ac and H4K8acK12ac di-acetylated histone ligands (Tallant et al., 2015). Family IV bromodomains possess a “keyhole style” pocket due to a larger aromatic gatekeeper residue that can only accommodate one acetyllysine group in the canonical binding site (Flynn et al., 2015, Moriniere et al., 2009). In ITC binding assays and crystallization studies of BRD9, the bromodomain interacts with the di-acetylated histone ligand by coordinating the H4K5ac group in one bromodomain with the H4K8ac site bound to a second bromodomain module (PDBID: 4YYI) (Flynn et al., 2015). However, the binding and recognition of di-acetylated histone ligands by the BRPF1 bromodomain has yet to be fully elucidated.
In this study, we characterized the binding affinity of the BRPF1 bromodomain with multiple di-acetylated histone ligands by isothermal titration calorimetry (ITC). We show that the BRPF1 bromodomain binds preferentially to histone peptides H4K5acK8ac and H4K5acK12ac. We utilized nuclear magnetic resonance (NMR) chemical shift perturbation techniques to map the BRPF1 bromodomain binding pocket and identify important residues involved in di-acetyllysine recognition. We performed site-directed mutagenesis experiments, together with ITC and analytical ultracentrifugation (AUC) experiments, to evaluate the relative contributions of specific amino acid residues in di-acetyllysine coordination. We also tested how adjacent PTMs impact acetyllysine recognition. The results provide new insights on how the BRPF1 bromodomain selects for its di-acetyllysine histone ligands at the molecular level, and how the combinatorial action of post-translational modifications can influence BRPF1 bromodomain binding and specificity. Our study further supports current research efforts geared toward the development of novel therapeutics targeting the BRPF1 bromodomain in acute myeloid leukemia.
Results
The BRPF1 bromodomain recognizes di-acetyllysine histone modifications
We postulated that the BRPF1 bromodomain may be able to recognize di-acetylated histone ligands in analogy to the closely related family IV bromodomain BRD9, which has been shown to recognize multiple diacyl modifications on histone H4K5/K8 (Flynn et al., 2015). To investigate our hypothesis, we sought to further characterize the histone ligands of this bromodomain by performing ITC experiments with mono- and di-acetylated lysine residues on histones H3 and H4. The BRPF1 bromodomain is able to recognize multiple acetyllysine modifications and binds to several mono-acetylated histone peptides, showing the highest affinity toward the H4K5ac peptides (residues 1–10 and 1–15) (Table 1 and Supplementary Fig. S1). Furthermore, when the ITC experiment was performed with the di-acetylated H4K5acK8ac (1–10) peptide, the BRPF1 bromodomain bound with a dissociation constant about two-fold stronger than that of the mono-acetylated H4K5ac peptide (27.7 ± 5.2 μM versus 57.6 ± 6.0). Similarly, we found that the combination of H4K5acK12ac marks increased the BRPF1 bromodomain binding affinity (KD of 49.7 ± 6.5 μM) six-fold over that for the singly acetylated H4K12ac mark, and also has a higher affinity than the single H4K5ac modification. Other combinations of di-acetylation marks on the histone tail resulted in lower binding affinities with the BRPF1 bromodomain, and the H4K8acK12ac mark completely abrogated the binding interaction.
Table 1.
Sequences of the mono- and di-acetylated histone peptides studied, and the dissociation constants of their complexes with the BRPF1 bromodomain as measured by ITC.
| Histone peptide | Sequence | BRPF1 KD (μM) | N value | ΔH (cal/Mol) | ΔS (cal/Mol/deg) |
|---|---|---|---|---|---|
| Histone H3 peptides | |||||
| H3 unmodified (1–24) | ARTKQTARKSTGGKAPRKQLATKA | No binding | – | – | – |
| H3K9ac (1–24) | ARTKQTAR (Kac)STGGKAPRKQLATKA | 718.0 ± 55.8 | 1.0 ± 0.0 | −918.8 ± 151.13 | 11.1 ± 0.4 |
| H3K14ac (1–24) | ARTKQTARKSTGG (Kac)APRKQLATKA | 183.7 ± 3.9 | 1.0 ± 0.0 | −1338.5 ± 48.7 | 12.3 ± 0.2 |
| H3K14ac (8–19)a | RKSTGG (Kac)APRKQ | 626 | – | – | – |
| H3K9acK14ac (1–24) | ARTKQTAR (Kac)STGG (Kac)APRKQLATKA | 233.8 ± 5.8 | 1.0 ± 0.0 | −1249.3 ± 136.7 | 12.1 ± 0.5 |
| Histone H4 peptides | |||||
| Mono-acetylated histone H4 peptides | |||||
| H4 unmodified (1–15) | SGRGKGGKGLGKGGA | No binding | – | – | – |
| H4K5ac (1–10) | SGRG (Kac)GGKGL | 57.6 ± 6.0 | 0.9 ± 0.1 | −3823.0 ± 220.0 | 5.7 ± 0.7 |
| H4K5ac (1–15) | SGRG (Kac)GGKGLGKGGA | 78.7 ± 15.4 | 1.0 ± 0.1 | −3487.3 ± 450.1 | 6.3 ± 1.3 |
| H4K5ac (4–17)a | G (Kac)GGKGLGKGGAKR | 1236b | – | – | – |
| H4K8ac (1–10) | SGRGKGG (Kac)GL | 189.1 ± 36.6 | 1.1 ± 0.2 | −622.5 ± 110.1 | 14.8 ± 0.1 |
| H4K8ac (4–17)a | GKGG (Kac)GLGKGGAKR | 697b | – | – | – |
| H4K12ac (1–15) | SGRGKGGKGLG (Kac)GGA | 100.0 ± 4.6 | 1.0 ± 0.1 | −2544.7 ± 430.0 | 9.0 ± 1.3 |
| H4K12ac (4–17)a | GKGGKGLG (Kac)GGAKR | 86.5 ± 9.1 | – | – | – |
| H4K16ac (10–20) | LGKGGA (Kac)RHRK | 517.9 ± 92.1 | 1.1 ± 0.1 | −3862.7 ± 112.0 | 1.2 ± 0.1 |
| Di-acetylated histone H4 peptides | |||||
| H4K5acK8ac (1–10) | SGRG (Kac)GG (Kac)GL | 27.7 ± 5.2 | 0.8 ± 0.0 | −1186 ± 35.3 | 16.6 ± 0.3 |
| H4K5acK12ac (1–15) | SGRG (Kac)GGKGLG (Kac)GGA | 49.7 ± 6.5 | 1.0 ± 0.0 | −7216.0 ± 378.5 | −6.2 ± 1.2 |
| H4K5acK16ac (1–24) | SGRG (Kac)GGKGLGKGGA (Kac)RHRKVLRD | 981 ± 11.3 | 1.0 ± 0.1 | −4560.0 ± 148.8 | 2.0 ± 0.3 |
| H4K8acK12ac (6–15) | GG (Kac)GLG (Kac)GGA | No binding | – | – | – |
| H4K8acK16ac (1–24) | SGRGKGG (Kac)GLGKGGA (Kac)RHRKVLRD | 444.7 ± 12.5 | 1.0 ± 0.0 | −3844.3 ± 56.0 | 1.5 ± 0.2 |
| H4K12acK16ac (10–20) | LG (Kac)GGA (Kac)RHRK | 644.2 ± 102.3 | 1.0 ± 0.0 | −4577.7 ± 179.4 | 1.8 ± 0.3 |
| Other PTMs on histone H4 peptides | |||||
| H4S1phosK5ac (1–15) | (Sp)GRG (Kac)GGKGLGKGGA | 220.3 ± 25.2 | 1.1 ± 0.1 | −3523.3 ± 195.3 | 4.1 ± 0.5 |
| H4K5me3K12ac (1–15) | SGRG (Kme3)GGKGLG (Kac)GGA | 179.8 ± 13.4 | 1.0 ± 0.0 | −4294.0 ± 64.3 | 1.7 ± 0.1 |
| H4K5acK12me3 (1–15) | SGRG (Kac)GGKGLG (Kme3)GGA | 70.3 ± 4.8 | 1.1 ± 0.0 | −6357.0 ± 205.9 | −3.8 ± 0.6 |
Previously published BRPF1 binding data (Poplawski et al., 2014).
Indicates dissociation constants measured by NMR titration experiments.
Interestingly, in some cases, the inclusion of residues 1–3 on the histone tail improved the binding interaction of histone H4 peptides that contained the K5 or K8 acetyllysine modification. This is the case for H4K5ac peptides (comparison of residues 1–10 and 4–17), and H4K8ac peptides (comparison of residues 1–10 and 4–17), which bind with significantly different affinities (Table 1) (Poplawski et al., 2014). However, in peptides with the H4K12ac modification, sequences 1–15 and 4–17 had similar binding constants. These data indicate that residues 1–3 on the N-terminus of histone H4 tail are important for recognition of acetyllysine modifications near the N-terminus of this histone, whereas the presence of N-terminal residues has little effect on the affinity of modifications further downstream. Furthermore, this is the first direct evidence that the BRPF1 bromodomain can read a span of 8–10 amino acid residues on the histone tail.
Since previous studies have shown that the BRPF1 bromodomain recognizes the histone H3K14ac peptide (Poplawski et al., 2014), we also tested the binding affinity of this bromodomain with mono- and di-acetylated peptides containing the first 24 amino acid residues of human histone H3. Our ITC experiments revealed that the BRPF1 bromodomain preferentially selects for the H3K14ac (1–24) modification over H3K9ac (1–24). A dissociation constant of 233.8 μM was obtained for the H3K9acK14ac (1–24) di-acetylated histone peptide, which indicates that the acetylation mark at K9 impairs the ability of the BRPF1 bromodomain to bind H3K14ac (Table 1, Supplementary Fig. S1).
We calculated ΔH and ΔS values for the interaction of BRPF1 with various mono- and di-acetylated histone ligands (Table 1). The bromodomain-histone binding interaction appears to be predominantly enthalpy driven as demonstrated by the large, negative changes in enthalpy (ΔH) observed upon ligand binding. This is in opposition to the smaller, generally positive changes in entropy (ΔS), which may reflect the displacement of water molecules in the bromodomain binding pocket upon ligand binding (Du et al., 2016). Taken together, our results show that the BRPF1 bromodomain preferentially selects for di-acetyllysine modifications on histone H4.
Crosstalk between PTMs modulate acetyllysine recognition by the BRPF1 bromodomain
To determine how the combinatorial action of PTMs on the histone tail modulates acetyllysine recognition, we performed ITC experiments to measure how the in vitro binding affinity of the BRPF1 bromodomain for acetylated histone peptides changes as a function of modifications located adjacent to the acetyllysine (Kac) recognition site (Table 1 and Supplementary Fig. S1). Our data show that recognition of the H4K5ac (1–15) mono-acetylated histone peptide decreased by approximately three-fold (KD = 228.5 μM) when serine 1 of histone H4 was phosphorylated. Additionally, the affinity of the H4K12ac (1–15) interaction decreased by ~1.5-fold (KD = 169.6 μM) when lysine 5 was tri-methylated. Interestingly, tri-methylation of lysine 12 in combination with acetylation of lysine 5 did not significantly alter the affinity of the BRPF1 bromodomain for H4K5ac (1–15), indicating that the H4K12me3 mark is permissive for H4K5ac recognition. Together, our results show that crosstalk between adjacent PTMs on the histone tail modulate how acetyllysine is recognized by the BRPF1 bromodomain.
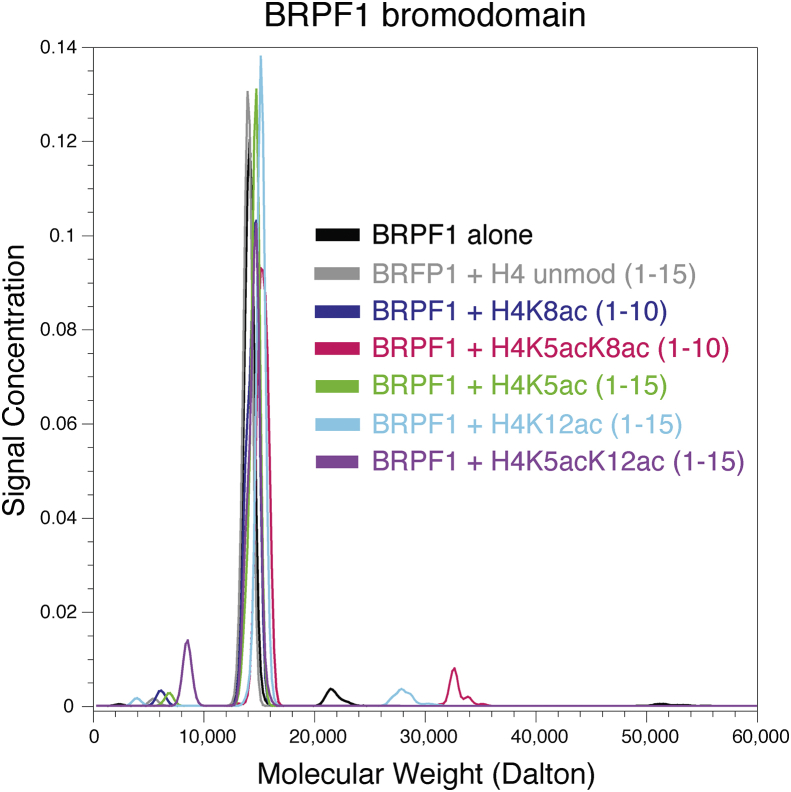
The BRPF1 bromodomain recognizes di-acetylated histone ligands as a monomer
In a recent crystal structure, the complex formed between the BRPF1 bromodomain and histone H4K5acK8ac is modeled as an apparent dimer (PDBID: 5FFW) (Meier et al., 2017). However, we hypothesized that BRPF1 likely functions as a monomer in its biologically active form when it associates with the MOZ/MORF HATs. As shown in Table 1, the N value observed in all our ITC data indicates that the BRPF1 bromodomain interacts with modified histone H3 and H4 tails as a monomer. To confirm this, we also performed analytical ultracentrifugation sedimentation velocity experiments to determine the molecular masses of the BRPF1 bromodomain in complexes with mono- and di-acetylated histone ligands. The molecular weight of the apo BRPF1 bromodomain is 13,703 Da, while the molecular weights of the histone ligands tested ranged from 958 to 1371 Da. Strikingly, the signal peaks of the BRPF1 bromodomain alone, or in complex with its mono- and di-acetylated histone ligands, overlapped at approximately 14–15 kDa (Fig. 1). These results support our ITC data showing that the BRPF1 bromodomain recognizes both mono- and di-acetylated histone ligands as a monomer in solution.
Fig. 1.
Molecular weight distributions of the BRPF1 bromodomain alone and in complex with mono- and di-acetylated histone ligands, determined by analytical ultracentrifugation. The distribution shows the molecular weights of the BRPF1 bromodomain alone, and in a 1:1 ratio with H4K5ac (1–15) (green), H4K5acK12ac (1–15) (purple), H4K12ac (1–15) (cyan), H4 unmodified (1–15) (grey), H4K5acK8ac (1–10) (magenta), and H4K8ac (1–10) (blue). All molar masses are consistent with a 1:1 ratio of BRPF1 bromodomain and histone peptide.
Characterization of the BRPF1 bromodomain binding pocket by NMR spectroscopy
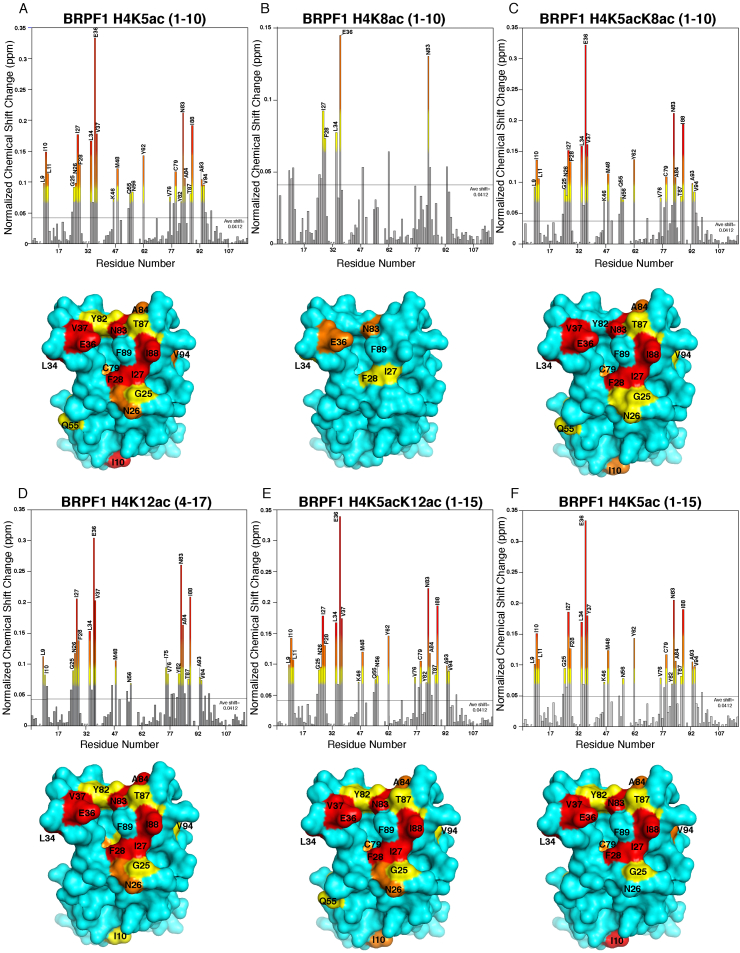
To outline specific amino acid interactions in the BRPF1 bromodomain binding pocket with the mono- and di-acetylated histone peptides, two-dimensional, 1H–15N heteronuclear single quantum coherence (HSQC) NMR experiments were performed. We previously reported the backbone assignments for the BRPF1 bromodomain accomplished by means of multidimensional NMR spectroscopy (Poplawski et al., 2014). Here, the amide-NH chemical shift perturbations induced in residues of the BRPF1 bromodomain sample were monitored upon addition of the acetylated histone peptides H4K5ac (1–10), H4K8ac (1–10), H4K12ac (4–17), H4K5ac (1–15), as well as the di-acetylated histone ligands H4K5acK8ac (1–10), and H4K5acK12ac (1–15), at increasing concentrations until the saturation point was observed. The bar graphs in Fig. 2A–F show the normalized chemical shift changes of individual residues in the BRPF1 bromodomain upon addition of the respective mono- and di-acetylated histone peptides, in a 1:5 M ratio. The affected amino acids were then mapped onto the surface of the apo BRPF1 bromodomain crystal structure (PDB ID: 2D9E), and it was observed that the majority of the altered resonances with large chemical shifts correspond to residues clustered around the acetyllysine binding pocket (Fig. 2A–F). Large chemical shift perturbations for residue N83 in the BRPF1 bromodomain were consistently observed in our NMR titration studies confirming the importance of this residue in the coordination of acetylated histone H4 peptides. Residues I27, L34, E36, V37, N83, and I88 also showed large chemical shift perturbations upon the addition of mono- and/or di-acetylated peptides. A few of the chemical shift perturbations corresponded to residues located more peripherally to the binding pocket. Amino acids including L9, I10, L11, Q55, and N56, located distal to the acetyllysine binding pocket, demonstrated chemical shift perturbations due to conformational changes within the bromodomain that occur in response to ligand binding.
Fig. 2.
Mapping of the mono- and di-acetylated histone ligand binding interfaces on the BRPF1 bromodomain. (A–F). Histograms show the normalized 1H–15N chemical shift changes in the backbone amides of the BRPF1 bromodomain in the presence of H4K5ac (1–10) (A), H4K8ac (1–10) (B), H4K5acK8ac (1–10) (C), H4K12ac (4–17) (D), H4K5acK12ac (1–15) (E), and H4K5ac (1–15) (F) in a 1:5 M ratio. Chemical shift changes 0.5, 1, and 2 standard deviations away from the average shift change are colored yellow, orange, and red, respectively. Amino acid residues with significant chemical shift perturbations upon addition of the histone ligands are mapped onto the surface of the apo structure of the BRPF1 bromodomain (PDBID: 2D9E).
Minimal chemical shift perturbations were seen with the H4K8ac (1–10) histone ligand with only E36 and N83 showing significant perturbations; this is consistent with the weak binding of the H4K8ac ligand to the BRPF1 bromodomain (KD = 168.4 ± 4.7 μM) found by our ITC experiment (Table 1 and Supplementary Fig. S1). Interestingly, in our NMR titration analysis, the mono- versus the di-acetylated histone ligands displayed a similar trend in their chemical shift perturbation patterns. The reason for this could be two-fold. Firstly, in the present in-vitro condition, the BRPF1 bromodomain shows a similar binding pocket for the mono- versus the di-acetylated histone H4 ligands, thus indicating that there may be other mechanisms involved in more specific interactions of the BRPF1 bromodomain with the di-acetylated histone H4 tail in-vivo. BRD7, which shows high structural similarity to BRPF1, has also been reported to lack specificity toward histone ligands under in-vitro conditions (Sun et al., 2007). Secondly, the second acetyllysine is likely making transient side chain interactions with certain residues surrounding the canonical BRPF1 bromodomain binding pocket not readily detected by our backbone amide NMR titrations. A similar observation was made for the BRDT BD1 bromodomain, which interacts with the di-acetylated H4K5acK8ac histone ligand via the methyl groups of the hydrophobic residues lining the bromodomain pocket (Moriniere et al., 2009).
Mutational analysis identifies residues that may coordinate the second acetyllysine moiety
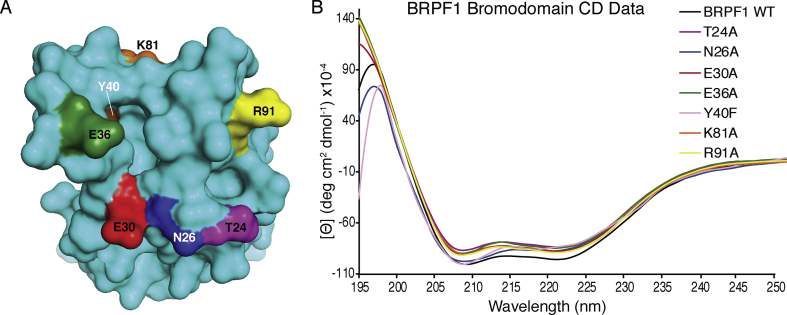
To obtain more information about coordination of di-acetyllysine by the BRPF1 bromodomain we used site-directed mutagenesis coupled to ITC binding assays (Table 2 and Supplementary Fig. S2). To investigate whether both acetyllysine groups in the di-acetylated histone ligands are accommodated in the canonical BRPF1 bromodomain binding pocket, or if secondary binding contacts exist on the BRPF1 bromodomain, we selected six amino acid residues (T24, N26, E30, E36, K81, R91) to mutate to alanine. These experiments were designed to test our hypothesis that if the second acetyllysine group is coordinated via specific side chain interactions on the BRPF1 bromodomain, then mutating it should decrease the binding affinity to what is seen for the singly acetylated histone ligands. We performed circular dichroism (CD) experiments on the mutant BRPF1 bromodomain proteins (Table 3 and Fig. 3) to ensure they were properly folded, and we ran ITC binding experiments to test the binding affinities of the BRPF1 bromodomain mutants in combination with H4K5ac (1–10), H4K12ac (1–15), H4K5acK8ac (1–10) and H4K5acK12ac (1–15) histone ligands. The binding affinities with observed stoichiometries of the mutant BRPF1 bromodomains for the histone ligands tested are shown in Table 2. The ITC traces of the mutant BRPF1 bromodomains with their histone ligands are shown in Supplemental Fig. S2. Interestingly, all of the alanine mutations to the BRPF1 bromodomain caused an increase in binding affinity for the H4K5acK12ac ligand, while most of these mutations caused at least a slight decrease in binding affinity for H4K5acK8ac. Our mutational study did not cause the binding affinity of di-acetylated ligands to revert to singly acetylated ligand values, but the T24A, E36A, and K81A mutations did show a weakened binding interaction between the BRPF1 bromodomain and the H4K5acK8ac ligand. The N26A mutation decreased the affinity for the H4K12ac (1–15) ligand, and increased the binding affinity for the H4K5acK12ac (1–15) and H4K5ac (1–10) ligands, but it did not have a large effect on binding of the H4K5acK8ac (1–10) ligand. The remaining mutations either caused no significant difference in the binding interaction or resulted in increased binding affinities with the BRPF1 bromodomain. The inconsistent binding affinity of BRPF1 bromodomain mutants suggests that multiple residues are involved in coordinating the second acetyllysine group.
Table 2.
Dissociation constants of the wild-type and mutant BRPF1 bromodomain proteins with histone ligands as measured by ITC experiments.
| BRPF1 | H4K5ac (1–10), N | H4K12ac (1–15), N | H4K5K8ac (1–10), N | H4K5K12ac (1–15), N |
|---|---|---|---|---|
| Wild type | 60.1 ± 6.8, 0.9 ± 0.1 | 97.3 ± 4.0, 1.0 ± 0.1 | 26.7 ± 1.8, 1.0 ± 0.0 | 52.0 ± 2.9, 1.0 ± 0.0 |
| T24A | 28.9 ± 1.0, 1.0 ± 0.0 | 105.7 ± 8.3, 0.9 ± 0.1 | 99.4 ± 5.3, 1.0 ± 0.1 | 31.2 ± 4.6, 1.0 ± 0.0 |
| E30A | 79.4 ± 5.7, 1.1 ± 0.1 | 104.5 ± 4.0, 1.1 ± 0.0 | 27.2 ± 0.7, 1.0 ± 0.0 | 39.0 ± 1.6, 1 ± 0.0 |
| E36A | 51.4 ± 1.9, 1.0 ± 0.0 | 54.7 ± 5.5, 1.0 ± 0.0 | 43.1 ± 1.4, 1 ± 0.0 | 31.0 ± 2.6, 1.0 ± 0.0 |
| K81A | 59.9 ± 3.8, 1.0 ± 0.0 | 45.0 ± 3.0, 1.0 ± 0.0 | 45.7 ± 5.6, 1 ± 0.0 | 24.6 ± 0.8, 1.0 ± 0.0 |
| R91A | 37.3 ± 1.3, 1.0 ± 0.0 | 79.7 ± 5.1, 1.0 ± 0.0 | 21.5 ± 1.9, 0.9 ± 0.0 | 23.0 ± 3.8, 1.0 ± 0.0 |
| N26A | 44.1 ± 0.9, 1.0 ± 0.0 | 122.3 ± 2.9, 1.2 ± 0.1 | 34.5 ± 1.3, 1.0 ± 0.0 | 34.2 ± 6.8, 1.0 ± 0.0 |
Table 3.
Percentages of α-helix and β-strand composition in wild-type (WT) and mutant BRPF1 bromodomain proteins calculated from circular dichroism experiments.
| BRPF1 bromodomain | % alpha helix | % beta strand |
|---|---|---|
| WT | 93.12% | 0.21% |
| T24A | 93.71% | 0.44% |
| E30A | 93.69% | 0.43% |
| E36A | 93.83% | 0.43% |
| K81A | 93.90% | 0.41% |
| R91A | 93.71% | 0.44% |
| N26A | 93.73% | 0.44% |
| Y40F | 93.12% | 0.19% |
Fig. 3.
Circular dichroism spectra of the wild-type and mutant BRPF1 bromodomains. (A). Surface representation of the apo structure (cyan) of the BRPF1 bromodomain showing mutated residues. The amino acid residues N26, T24, E30, E36, Y40, K81, and R91 mutated to alanine are indicated as shown. (B). Secondary structure of the BRPF1 bromodomain wild-type and mutant bromodomain proteins measured by circular dichroism. Circular dichroism molar ellipticity plots of the BRPF1 wild type and mutant bromodomains are shown. Each protein is represented by a different color and all show a clear α-helical structure to match the wild type BRPF1 bromodomain.
The role of ordered water in bromodomain structure and histone ligand coordination
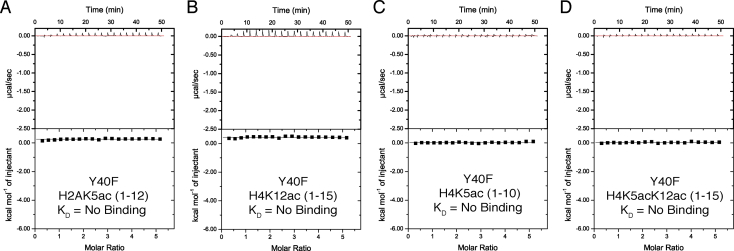
We made a conservative change to Y40 by mutating the residue to phenylalanine in order to investigate how loss of water coordination would impact acetyllysine recognition. The BRPF1 Y40F bromodomain variant failed to bind mono- or di-acetylated histone ligands (Table 2 and Fig. 4). Thus, the ordered water in the bromodomain binding pocket is important for the recognition of acetylated histone ligands.
Fig. 4.
ITC measurements of the Y40F mutant bromodomain involved in water coordination. No binding was detected for each of the four acetylated histone peptides tested with the Y40F mutant bromodomain, which highlights the important role of water coordination in the bromodomain binding pocket.
Discussion
Previous studies on the BRPF1 bromodomain revealed that it recognizes several acetylated lysine residues on the N-terminus of histone tails (Poplawski et al., 2014). The BRPF1 bromodomain predominantly selects for acetylation modifications on the histone H4 tail; however, it is also able to recognize the histone H3 and H2A ligands H3K14ac and H2AK5ac (Poplawski et al., 2014). More recently it has been discovered that bromodomain-containing proteins such as BRD2, BRD3, and BRD4 can bind to histone ligands with di-acetyllysine marks (Ferri et al., 2016). Another example of this is the first bromodomain (BD1) in the bromodomain testis-specific protein (BRDT), which is a member of the bromodomain and extra-terminal domain (BET) subfamily. BRDT BD1 failed to recognize mono-acetylated histone H4 tails, but was able to bind multiple di-acetylated histone H4 peptides as well as tetra-acetylated histone H3 peptides (Moriniere et al., 2009). BRDT BRD1 recognizes the di-acetylated histone ligand H4K5acK8ac with a low micromolar affinity, and the crystal structure of this complex demonstrated that the ‘wide pocket’ of the BRDT BD1 bromodomain is able to accommodate both acetylated lysine residues within the canonical bromodomain binding pocket (Moriniere et al., 2009). By contrast, the ‘keyhole pocket’ of the yeast GCN5 bromodomain is able to accommodate only one acetyllysine group inside the binding pocket at a time. Coordination of di-acetyllysine by the first bromodomain in the BET subfamily appears to be conserved as demonstrated by ligand binding assays with BRD1 and BRD2 (Ladurner et al., 2003, Kanno et al., 2004), as well as by structural models of BRD4 BD1 with H4K5acK8ac and H4K12acK16ac (PDBIDs 3UVW and 3UVX, respectively) (Filippakopoulos et al., 2012). It has been suggested that other bromodomain protein families can recognize di-acetylated histone ligands as well, possibly through alternative binding modes. The structures of the BAZ2A and BAZ2B bromodomains complexed, respectively, with the di-acetylated histone H4 ligands H4K16acK20ac and H4K8acK12ac have been determined (PDBIDs 4QBM and 4QC3) (Tallant et al., 2015). In these structures, BAZ2A can coordinate the di-acetylated ligand as a monomer, much like the BET family bromodomains, while the BAZ2B bromodomain forms a dimeric complex to coordinate the H4K8acK12ac histone ligand.
In this study, we show that the BRPF1 bromodomain can selectively recognize multiple histone mark combinations and that it binds preferentially to di-acetylated ligands on the N-terminus of histone H4. The results from our ITC binding assays demonstrate that this bromodomain has the highest affinity for the histone H4K5acK8ac ligand, followed by the H4K5acK12ac ligand. Histone H4 acetylation occurs progressively from mono-acetylated H4K16ac, followed by acetylation at the K12, K8, and K5 sites (Thorne et al., 1990, Zhang et al., 2002). Therefore the presence of K5 and K8 acetylation marks are characteristic of chromatin hyperacetylation in-vivo (Sasaki et al., 2009). Previous studies also reported that newly synthesized histone H4 has transient acetylation marks on the K5 and K12 positions (Koo et al., 2016, Sobel et al., 1995). Given the fact that BRPF1 recruits MOZ to particular sites on active chromatin and remains at chromosomes during mitosis (Laue et al., 2008), it is reasonable to assume that the BRPF1 bromodomain plays an important role in the recognition of hyperacetylated chromatin during cell division. BRPF1 binding would further drive the acetylation of the histones via the acetyl transferase activity of MOZ, which is activated by BRPF1 (Carlson et al., 2014). Since BRPF1 has been shown to play an important role in cell proliferation (You et al., 2015), it is possible that the recognition of these di-acetylation marks, characteristic of dividing cells, is driving cancer progression.
We also tested the histone H4K5acK16ac and H4K8acK16ac ligands and found that these di-acetyllysine combinations weakened the BRPF1 bromodomain interaction with the histone tail. Examination of the histone H3/H4 proteoforms found in human cancer cell lines revealed that in promyelocytic leukemia cells (HL60) the H4K5acK16ac mark is down-regulated, while it is up-regulated in acute promyelocytic (NB4) cells (Leroy et al., 2013). Additionally, the H4K8acK16ac mark was down-regulated in all leukemia cell lines tested. Thus, as histone PTMs are altered in disease states, the availability of particular acetyllysine combinations will enhance or inhibit the interaction of BRPF1 with chromatin and influence recruitment of the MOZ/MORF HAT complex.
Results from our previous study demonstrated that the BRPF1 bromodomain recognizes the singly-acetylated histone ligands H2AK5ac, H4K12ac, H4K8ac, H4K5ac, and H3K14ac respectively (Poplawski et al., 2014). Our current results demonstrate that of the singly acetylated histones, the BRPF1 bromodomain binds preferentially to H4K5ac, which is likely the predominant interaction driving recognition of di-acetylated ligands. Interestingly, the ITC binding data show that the BRPF1 bromodomain functions as a di-acetyllysine reader, with a binding pocket able to recognize PTMs over a span of 8–10 residues on each histone tail. Furthermore, our results show that the interaction of the BRPF1 bromodomain with histone ligands is enthalpy driven. A similar result was also reported for the BRPF1 bromodomain with small-molecule ligands and with ATAD2 bromodomain-ligand complexes (Gay et al., 2019, Zhu et al., 2018). Additionally, the BRPF1 bromodomain interacts with mono- and di-acetylated histone ligands as a monomer in solution based on the binding stoichiometry (N: peptide-to-bromodomain ratio) observed in the ITC binding assays and confirmed in AUC experiments.
This stoichiometry differs from that observed for family IV BRD9 bromodomain, which binds to the histone H4K5acK8ac ligand with an ITC N-value of 0.44, suggesting a dimeric interaction with two bromodomains bound to one histone peptide (Flynn et al., 2015). Family IV bromodomains possess a “keyhole style” pocket that, owing to the presence of a large aromatic gatekeeper residue, can only accommodate one acetyllysine group in the canonical binding site (Flynn et al., 2015, Moriniere et al., 2009). Thus, in ITC binding assays and crystallization studies the BRD9 bromodomain interacts with the di-acetylated histone ligand by coordinating the H4K5ac group in one bromodomain with the H4K8ac site bound to a second bromodomain module (PDBID: 4YYI) (Flynn et al., 2015). A related structure shows that the BRPF1 bromodomain binds to the histone H4K5acK8ac in an identical manner as an apparent dimer (PDBID: 5FFW) (Meier et al., 2017). However, this is likely due to crystal packing effects caused by the crystallization conditions used for solving the crystal structure, since our ITC binding data and analytical ultracentrifugation analysis of the BRPF1 bromodomain support monomeric coordination of both mono- and di-acetylated histone ligands in solution. Also, unlike the BET proteins, which contain two adjacent bromodomain modules (Shang et al., 2007), the BRPF1 protein has only one bromodomain, and this protein functions as a single subunit within the MOZ/MORF HAT complexes (Filippakopoulos et al., 2012, Ullah et al., 2008). Thus, the BRPF1 bromodomain is likely to be monomeric in its biologically active form.
Histone H4 can be post-translationally modified at multiple positions and is frequently acetylated at lysine 5, 8, 12, and 16. It also often carries a phosphorylation mark on serine 1, as well as methyl groups on arginine 3 and lysine 20 (Zhang and Reinberg, 2001). The SET-domain methyltransferases performsmethylation of lysine on the histone tails (Dillon et al., 2005), which can be mono-, di- or tri-methylated. Lysine at position 5 and 12 of histone H4 can also be methylated (Huang et al., 2014, Murray, 1964). Since acetylation at lysine 5 and 12 on histone H4 are the positions the BRPF1 bromodomain binds with highest affinity, we tested the effect tri-methylation at these sites would have on bromodomain binding. Our results demonstrated that tri-methylation of H4K5 reduced the binding affinity of the BRPF1 bromodomain for the H4K12ac ligand, while tri-methylation of H4K12 did not have a significant impact on the recognition of H4K5ac. We also characterized how phosphorylation of serine 1 modulates the acetyllysine binding activity of the BRPF1 bromodomain and showed that this mark has an inhibitory effect on recognition of H4K5ac. Histone phosphorylation has previously been linked to DNA damage repair mechanisms (Rossetto et al., 2012), and H4S1phos is known to be enriched 20- to 25-fold in nucleosomes that are located adjacent to double-strand breaks (Cheung et al., 2005). The impact of modifications adjacent to acetyllysine recognition sites may be biologically relevant to BRPF1's function in recruiting the MOZ HAT tetrameric complex to distinct sites on chromatin depending on the epigenetic landscape. For example, phosphorylation of histone H4S1 at sites of DNA damage could function to prevent or delay the transcriptional activation signaling by acetylation modifications until the genetic information can be repaired. Therefore, our study represents an important first step in understanding how the BRPF1 bromodomain is able to recognize and bind di-acetyllysine on the histone tail, and how cross-talk among adjacent marks influences its activity.
To delineate the acetyllysine binding site on the BRPF1 bromodomain, we used NMR chemical shift perturbation data to identify the residues involved in di-acetyllysine recognition, and to observe whether residues outside the canonical acetyllysine binding pocket are important for coordinating the second acetyllysine modification. We introduced additional mutations into the BRPF1 bromodomain binding pocket based on previous studies of acetyllysine coordination by other related bromodomains (Poncet-Montange et al., 2015, Zhu and Caflisch, 2016, Lubula et al., 2014, Langini et al., 2017). I27 is found in the BRPF1 “NIF Shelf”, which is important for ligand selectivity (Romero et al., 2016). L34, E36, and V37 are all located in the flexible ZA loop adjacent to the acetyllysine binding site, which is predicted to be important for increasing the on-rate of histone tail binding (Langini et al., 2017). N83 is the universally conserved asparagine residue involved in the coordination of the acetyllysine modification on histones (Romero et al., 2016). I88 is adjacent to the ‘gatekeeper’ residue F89 (unassigned) that lines the acetyllysine binding pocket. Residue E36 in the BRPF1 bromodomain showed the largest chemical shift perturbation upon the addition of either mono- or di-acetylated histones. Alignment of the apo crystal structure of the BRPF1 bromodomain (PDBID: 2D9E) with BRPF1 bromodomain structures in combination with the H2AK5ac and H4K12ac ligands (PDBIDs: 4QYL and 4QYD) (Lubula et al., 2014), demonstrate that the E36 residue does not contact the histone ligands directly, but undergoes a large conformational change as part of the ZA loop to accommodate peptide binding. Mutation of E36 to alanine did not significantly alter the binding affinity of the BRPF1 bromodomain with the H4K5ac (1–10) ligand, but it decreased the interaction of the BRPF1 bromodomain with the di-acetylated histone H4K5acK8ac (1–10) ligand (Table 2), suggesting it may be an important contact for the K8ac moiety. A recent study investigating the intermolecular contacts between the related ATAD2 family IV bromodomain and the H4K5acK12ac ligand found that residue E1017, which is equivalent to E36 in the BRPF1 bromodomain, comes within 5 Å of H4K8 in molecular dynamic simulations of the holo complex (Langini et al., 2017).
A prior study on the ATAD2 bromodomain found the H3K14ac ligand bound to a secondary site distal from the canonical bromodomain binding site (Poncet-Montange et al., 2015). In this ATAD2 structure (PDBID: 4TT4), two residues were involved in coordination of the acetyllysine moiety, R1005 and H1076. H1076 is equivalent to T24 in the BRPF1 bromodomain. The T24A mutant showed a two-fold stronger binding affinity for the H4K5ac (1–10) ligand than the wild-type BRPF1 bromodomain, while also having a weaker interaction with the H4K5acK8ac (1–10) ligand. This suggests that residue T24 plays some role in coordinating either the K5ac or the K8ac in the di-acetylated histone ligand. Notably, mutation of K81 to alanine significantly weakened the interaction of the BRPF1 bromodomain for the H4K5acK8ac ligand, also indicating a role for this residue in coordination of this di-acetylated histone ligand. However, the N26A and E30A mutations resulted in modest changes in the binding affinity of the BRPF1 bromodomain for the histone ligands, suggestive of a neutral role for these residues in acetyllysine binding. Similarly, the R91A mutation resulted in slightly stronger binding interactions of the BRPF1 bromodomain with both the mono- and di-acetyllysine ligands. When the histone peptides contain two acetyllysine groups, it is possible for either group to be inserted into the canonical bromodomain binding pocket. One potential reason we observed an increase in binding affinity of the mutant BRPF1 bromodomain proteins with the di-acetyllysine ligands could be because their binding is now stabilized in one orientation over the other. In a di-acetylated ligand, not only is each acetyllysine group able to insert into the canonical binding pocket, but the peptide can bind in different orientations (e.g. from N- to C-terminal or vice versa) across the surface of the bromodomain binding pocket. Thus, by altering the BRPF1 bromodomain surface contacts with histone residues surrounding the primary acetyllysine inserted into the canonical binding pocket, some of our mutations may be preferentially selecting for one orientation over another, thereby stabilizing the ligand binding interaction with both mono- and di-acetylated histone peptides. Overall, our NMR titration and mutagenesis data demonstrated that the acetyllysine binding pocket in BRPF1 bromodomain binds a variety of acetylated histone ligands with a conserved binding mechanism, and that increased side chain and/or hydrophobic interactions are involved in the coordination of the second acetyllysine group, thus resulting in stronger binding affinities for the di-acetylated histone ligands.
Previous studies have identified ordered waters in the bromodomain binding pocket that coordinate the acetyllysine modification (Lubula et al., 2014, Crawford et al., 2016). One of these waters (w1) connects the carbonyl oxygen of the acetyllysine moiety to the hydroxyl group of Y40. Y40 is located in the αAZ loop of the bromodomain module, and a multiple sequence alignment shows it is conserved in all but two of the human bromodomain proteins (Filippakopoulos et al., 2012). Thus, this residue is even more highly conserved than the binding site asparagine residue that contacts the acetyllysine group directly. Our conservative mutation of this residue to phenylalanine demonstrated that Y40 is crucial for water coordination and sheds light on why Y40 is the most highly conserved amino acid residue found in human bromodomains.
In summary, our results demonstrated that the BRPF1 bromodomain can recognize multiple acetyllysine combinations on the histone tail. The BRPF1 bromodomain preferentially selects for the di-acetyllysine modifications H4K5acK8ac and H4K5acK12ac, which are associated with hyperacetylation and cell division, respectively (Koo et al., 2016, Sasaki et al., 2009, Sobel et al., 1995). Our results also confirm that cross-talk between PTMs on histone tail peptides can modulate acetyllysine recognition by the BRPF1 bromodomain. This research provides further insights into how the BRPF1 bromodomain recruits its partners in the MOZ/MORF HAT complexes to chromatin, with different combinations of PTMs either promoting or inhibiting the binding interaction. Chromosomal translocations of the MOZ HAT are associated with acute myeloid leukemia, and fusion of MOZ with TIF2, CBP and p300 are thought to drive leukemogenesis through aberrant acetylation (Carapeti et al., 1998, Crowley et al., 2005, Kitabayashi et al., 2001). Understanding how cross-talk influences bromodomain binding specificity and affinity is an important first step to connect how epigenetic changes contribute to the etiology of disease and may provide new mechanisms for therapeutic intervention in the treatment of AML and other diseases.
Materials and methods
Plasmid construction
The cDNA coding for human BRPF1 was kindly provided to us by Dr. Xiang-Jiao Yang at McGill University. Residues 629–742 encoding the BRPF1 bromodomain region were amplified using PCR and subsequently cloned into the pDEST15 vector encoding an N-terminal glutathione S-transferase (GST) tag. The Gateway cloning technology (Invitrogen) was used for cloning the BRPF1 bromodomain as described previously (Poplawski et al., 2014). BRPF1 bromodomain mutants T24A, N26A, E30A, E36A, Y40F, K81A, and R91A were generated using the QuikChange® mutagenesis procedure (Stratagene) as described previously (Champagne et al., 2006). All of the mutants were also cloned into the pDEST15 vector and the DNA sequences of the wild-type (WT) and mutant proteins were verified by Eurofins Genomics. The WT and mutant plasmids were then transformed into Escherichia coli Rosetta™ 2 (DE3)pLysS competent cells (Novagen) for protein expression.
BRPF1 bromodomain expression and purification
E. coli Rosetta™ 2 (DE3)pLysS cells expressing the WT and mutant GST-tagged BRPF1 bromodomain were grown in terrific broth (TB) at 37 °C until an OD600 of 1.0 was reached. The temperature was reduced to 16 °C for 1 h, and the cultures were induced by 0.25 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and incubated overnight for over 16 h at 16 °C. The cells were harvested by centrifugation and sonicated after resuspension in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.05% Nonidet P-40, and 1 mM DTT) containing 0.1 mg/mL lysozyme (Thermo Scientific). After centrifugation at 10,000 RPM, the cell supernatant was mixed with glutathione agarose resin (Thermo Scientific) and incubated on ice (4 °C) for 2 h while agitating. The BRPF1 bromodomain protein was purified on glutathione agarose resin in a 25 mL Econo-Column® (Bio-Rad) by first washing the beads extensively with wash buffer containing 20 mM Tris pH 8.0, 150 mM NaCl, and 1 mM DTT. The GST tag was cleaved overnight at 4 °C by addition of PreScission Protease (GE Healthcare), and the BRPF1 bromodomain was eluted with wash buffer. The eluted fractions were pooled, concentrated to approximately 3 mL, and dialyzed into appropriate buffers. For NMR experiments the 15N-labeled BRPF1 bromodomain was expressed and purified as previously described (Poplawski et al., 2014). The uniformly 15N-labeled BRPF1 bromodomain was prepared at a concentration of 0.4 mM in 20 mM Tris–HCl pH 6.8, 150 mM NaCl, and 10 mM DTT with 10% D2O added to the sample. For ITC and circular dichroism (CD) experiments, the purified BRPF1 bromodomain was dialyzed, respectively, into ITC buffer (20 mM NaH2PO4 pH 7.0, 150 mM NaCl, and 1 mM TCEP), or CD buffer (50 mM NaH2PO4 pH 7.0 and 50 mM NaCl). For analytical ultracentrifugation (AUC) experiments, the protein was further purified over a gel filtration column (HiPrep 16/60 Sephacryl S-100 High Resolution, GE Healthcare) equilibrated with AUC buffer (50 mM NaH2PO4 pH 7.0, 50 mM NaCl, and 1 mM TCEP) via fast-protein liquid chromatography. The concentration of the BRPF1 bromodomain was calculated from optical absorbance measurements with the A280 = 7450 M-1 cm-1. The purity of the BRPF1 bromodomain was verified by SDS-polyacrylamide gel electrophoresis stained with GelCode Blue Safe protein stain (Thermo Scientific).
Histone peptide synthesis
Histone peptides containing unique acetylated and/or methylated lysine, as well as phosphorylated serine modifications were synthesized by GenScript and purified by HPLC to 98% purity. The correct composition of each peptide was confirmed by mass spectrometry.
Isothermal titration calorimetry
ITC measurements were conducted using a MicroCal iTC200 instrument (Malvern Panalytical). All experiments were performed at 5 °C. The BRPF1 WT and mutant bromodomain proteins were dialyzed into ITC buffer containing 20 mM NaH2PO4 pH 7.0, 150 mM NaCl, and 1 mM TCEP for 48 h before experiments were performed. All titration experiments were performed as follows: 200 μM of the BRPF1 bromodomain protein was injected into the sample cell using a loading syringe, with 5 mM of histone peptide loaded into the injection syringe. The sample cell received one preliminary injection of 0.5 μL of the histone peptide followed by 19 injections of 2 μL histone peptide. For all titration experiments, each injection was spaced at 150 s time intervals after an initial delay of 60 s, and the injection syringe was stirred continuously at 1000 RPM. The preliminary injection was excluded prior to integration of the binding curve and calculation of the dissociation constants (KDs). First, the raw data were corrected for non-specific heats of dilution determined by the magnitude of the peaks appearing after the system reaches complete saturation and normalized for concentration. The raw data were then integrated using a Marquadt nonlinear least-squares analysis according to a 1:1 binding model, normalized for concentration, and analyzed assuming a single set of identical binding sites to calculate the binding affinity, stoichiometry (N), and the thermodynamic values. Binding data for all experiments were analyzed using the ORIGIN 7.0 software (OriginLab Corporation). All experiments where binding occurred were performed in triplicate (with the exception of H3K14ac (1–24), which was performed in duplicate), while nonbinding experiments were performed in duplicate and standard errors were reported as standard deviations.
Circular dichroism spectroscopy
Circular Dichroism (CD) spectra were recorded on a JASCO J-815 CD Spectrometer (JASCO, Japan) at 25 °C in a 1.6 cm cell. BRPF1 bromodomain WT and mutant proteins were dialyzed in CD buffer containing 50 mM NaH2PO4 pH 7.0 and 50 mM NaCl and then diluted to between 0.1 μM and 0.5 μM concentration. CD spectra were measured from 199 nm to 260 nm. Two spectra were measured and averaged for each WT and mutant bromodomain protein sample. The spectra were analyzed using the K2D3 structure prediction software (Louis-Jeune et al., 2012, Andrade et al., 1993) to determine the percent α-helical and β-sheet content for the WT and each of the mutant BRPF1 bromodomain proteins.
NMR spectroscopy
Chemical shift perturbation experiments were performed using 0.4 mM of uniformly 15N-labeled BRPF1 bromodomain in NMR buffer containing 20 mM Tris–HCl pH 6.8, 150 mM NaCl, 10 mM DTT and 10% D2O. Titration mixtures of the BRPF1 bromodomain protein and each of the modified histone peptides (H4K5acK8ac residues 1–10, H4K5acK12ac (1–15), H4K5ac (1–10) or 1–15 and H4K8ac (1–10)) were prepared at concentration ratios of 1:0, 1:0.6, 1:1.2, 1:3 and 1:6 in a volume of 150 μL. The sample mixtures were then transferred into 3-mm NMR tubes (Bruker). Two-dimensional 15N HSQC (heteronuclear single quantum coherence) experiments for all samples were run at 25 °C and NMR data collected as described previously (Poplawski et al., 2014). Normalized chemical shift changes were calculated as described in (Williamson, 2013) using the equation:
where = 0.14 for all residues except glycine, which has = 0.2, and ΔδH and ΔδN, respectively, are the changes in proton and nitrogen chemical shift in ppm. To map the chemical shift perturbations onto the BRPF1 bromodomain surface, we calculated the standard deviation (σ) of the Euclidean chemical shift change over all residues of all bromodomain:peptide combinations. Residues whose chemical shift change was greater than 0.5σ were colored yellow, while residues with chemical shift changes greater than 1σ and 2σ were colored orange and red, respectively. The backbone resonance assignments for the BRPF1 bromodomain were completed as described previously (Poplawski et al., 2014).
Analytical ultracentrifugation
For all AUC experiments, samples of the WT and mutant BRPF1 bromodomain proteins were prepared at an OD280 of 0.6 (80.5 μM) in AUC buffer containing 50 mM NaH2PO4 pH 7.0, 50 mM NaCl, and 1 mM TCEP. The BRPF1 bromodomain protein alone, and in combination with unmodified histone H4 peptide residues 1–15 mixed in a 1:1 M ratio, were included as controls. All other acetylated histone peptides (H4K5ac 1–15, H4K12ac 1–15, H4K8ac 1–10, H4K5acK8ac 1–10, and H4K5acK12ac 1–15) tested with the BRPF1 bromodomain were also mixed in a 1:1 ratio. AUC measurements were performed at 20 °C by UV intensity detection in a Beckman Optima AUC at the Center for Analytical Ultracentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center at San Antonio and at the Canadian Center for Hydrodynamics at the University of Lethbridge, Alberta, Canada, using an 8-hole AN50Ti rotor and standard 2-channel epon centerpieces (Beckman–Coulter). All the samples were measured in the same run and at multiple wavelengths (278 nm, 280 nm, and 282 nm), and the concentrations measured were reported in 280 nm absorbance units. All the AUC data were analyzed with UltraScan-III ver. 4.0, release 2480 (Demeler et al., 2016). Hydrodynamic corrections for buffer density and viscosity were estimated by UltraScan to be 1.006 g/mL and 1.0239 cP. The partial specific volume of the BRPF1 bromodomain was estimated by UltraScan from the protein sequence. Sedimentation Velocity (SV) data were analyzed according to a published method (Demeler, 2010). Optimization was performed by two-dimensional spectrum analysis (2DSA) (Brookes et al., 2010) with simultaneous removal of time- and radially-invariant noise contributions (Schuck and Demeler, 1999). After inspection of the 2DSA solutions, the fits were further refined using the parametrically constrained spectrum analysis (PCSA) (Gorbet et al., 2014), coupled with Monte Carlo analysis to measure confidence limits for the determined parameters (Demeler and Brookes, 2008). The calculations are computationally intensive and were carried out on high performance computing platforms (Brookes and Demeler, 2008). All calculations were performed on the Lonestar cluster at the Texas Advanced Computing Center (TACC) at the University of Texas at Austin or on XSEDE clusters at TACC or at the San Diego Supercomputing Center.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Funding sources
This study was supported by the National Institutes of Health, National Institute of General Medical Sciences award number R15GM104865 to KCG. JO was the recipient of an ACPHS graduate research assistantship from 2018-2019. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399. NMRFAM equipment was purchased with funds from the University of Wisconsin-Madison, the National Institutes of Health (P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), and the NSF (National Science Foundation) (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA (United States Department of Agriculture). BD acknowledges support from NIH grant GM120600 and NSF grant NSF-ACI-1339649 supporting the development of the UltraScan software. Supercomputer calculations were performed on Comet at the San Diego Supercomputing Center (support through NSF/XSEDE (Extreme Science and Engineering Discovery Environment) grant TG-MCB070039N to BD) and on Lonestar-5 at the Texas Advanced Computing Center (supported through UT (University of Texas) grant TG457201 to BD).
Declaration of Competing Interest
The authors declare that they have no conflict with the contents of this article.
Acknowledgments
We are thankful to Dr. X.J. Yang at McGill University for providing us with the isolated cDNA from human BRPF1 (Gene ID:7862).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2020.05.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Andrade M.A., Chacon P., Merelo J.J., Moran F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- Arrowsmith C.H., Bountra C., Fish P.V., Lee K., Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- Bamborough P., Barnett H.A., Becher I., Bird M.J., Chung C.W., Craggs P.D., Demont E.H., Diallo H., Fallon D.J., Gordon L.J., Grandi P., Hobbs C.I., Hooper-Greenhill E., Jones E.J., Law R.P., Le Gall A., Lugo D., Michon A.M., Mitchell D.J., Prinjha R.K., Sheppard R.J., Watson A.J., Watson R.J. GSK6853, a chemical probe for inhibition of the BRPF1 bromodomain. ACS Med. Chem. Lett. 2016;7:552–557. doi: 10.1021/acsmedchemlett.6b00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Borrow J., Stanton V.P., Jr., Andresen J.M., Becher R., Behm F.G., Chaganti R.S., Civin C.I., Disteche C., Dube I., Frischauf A.M., Horsman D., Mitelman F., Volinia S., Watmore A.E., Housman D.E. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat. Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- Bouche L., Christ C.D., Siegel S., Fernandez-Montalvan A.E., Holton S.J., Fedorov O., Ter Laak A., Sugawara T., Stockigt D., Tallant C., Bennett J., Monteiro O., Diaz-Saez L., Siejka P., Meier J., Putter V., Weiske J., Muller S., Huber K.V.M., Hartung I.V., Haendler B. Benzoisoquinolinediones as potent and selective inhibitors of BRPF2 and TAF1/TAF1L bromodomains. J. Med. Chem. 2017;60:4002–4022. doi: 10.1021/acs.jmedchem.7b00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E., Demeler B. Parallel computational techniques for the analysis of sedimentation velocity experiments in UltraScan. Colloid Polym. Sci. 2008;286:139–148. [Google Scholar]
- Brookes E., Cao W., Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur. Biophys. J. 2010;39:405–414. doi: 10.1007/s00249-009-0413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapeti M., Aguiar R.C., Goldman J.M., Cross N.C. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–3133. [PubMed] [Google Scholar]
- Carlson S., Glass K.C. The MOZ histone acetyltransferase in epigenetic signaling and disease. J. Cell. Physiol. 2014;229:1571–1574. doi: 10.1002/jcp.24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne K.S., Piscitelli E., Francklyn C.S. Substrate recognition by the hetero-octameric ATP phosphoribosyltransferase from Lactococcus lactis. Biochem. 2006;45:14933–14943. doi: 10.1021/bi061802v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W.L., Turner F.B., Krishnamoorthy T., Wolner B., Ahn S.H., Foley M., Dorsey J.A., Peterson C.L., Berger S.L., Allis C.D. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 2005;15:656–660. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- Clark P.G., Vieira L.C., Tallant C., Fedorov O., Singleton D.C., Rogers C.M., Monteiro O.P., Bennett J.M., Baronio R., Muller S., Daniels D.L., Mendez J., Knapp S., Brennan P.E., Dixon D.J. LP99: discovery and synthesis of the first selective BRD7/9 bromodomain inhibitor. Angew Chem. Int. Ed. Engl. 2015;54:6217–6221. doi: 10.1002/anie.201501394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford T.D., Tsui V., Flynn E.M., Wang S.M., Taylor A.M., Cote A., Audia J.E., Beresini M.H., Burdick D.J., Cummings R., Dakin L.A., Duplessis M., Good A.C., Hewitt M.C., Huang H.R., Jayaram H., Kiefer J.R., Jiang Y., Murray J., Nasveschuk C.G., Pardo E., Poy F., Romero F.A., Tang Y., Wang J., Xu Z.W., Zawadzke L.E., Zhu X.Y., Albrecht B.K., Magnuson S.R., Bellon S., Cochran A.G. Diving into the water: inducible binding conformations for BRD4, TAF1(2), BRD9, and CECR2 bromodomains. J. Med. Chem. 2016;59:5391–5402. doi: 10.1021/acs.jmedchem.6b00264. [DOI] [PubMed] [Google Scholar]
- Crowley J.A., Wang Y., Rapoport A.P., Ning Y. Detection of MOZ-CBP fusion in acute myeloid leukemia with 8;16 translocation. Leukemia. 2005;19:2344–2345. doi: 10.1038/sj.leu.2403971. [DOI] [PubMed] [Google Scholar]
- Demeler B. Methods for the design and analysis of sedimentation velocity and sedimentation equilibrium experiments with proteins. Curr. Protoc. Protein Sci. 2010;7 doi: 10.1002/0471140864.ps0713s60. Unit 7 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B., Brookes E. Monte Carlo analysis of sedimentation experiments. Colloid Polym. Sci. 2008;286:129–137. [Google Scholar]
- Demeler B., Gorbet G.E. Analytical ultracentrifugation data analysis with UltraScan-III. In: Uchiyama S., Arisaka F., Stafford W.F., Laue T., editors. Analytical Ultracentrifugation: Instrumentation, Software, and Applications. Springer Japan; Tokyo: 2016. pp. 119–143. [Google Scholar]
- Dhalluin C., Carlson J.E., Zeng L., He C., Aggarwal A.K., Zhou M.M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dillon S.C., Zhang X., Trievel R.C., Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Li Y., Xia Y.L., Ai S.M., Liang J., Sang P., Ji X.L., Liu S.Q. Insights into protein-ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak A.M., Stojanov P., Peng S., Lawrence M.S., Fox C., Stewart C., Bandla S., Imamura Y., Schumacher S.E., Shefler E., McKenna A., Carter S.L., Cibulskis K., Sivachenko A., Saksena G., Voet D., Ramos A.H., Auclair D., Thompson K., Sougnez C., Onofrio R.C., Guiducci C., Beroukhim R., Zhou Z., Lin L., Lin J., Reddy R., Chang A., Landrenau R., Pennathur A., Ogino S., Luketich J.D., Golub T.R., Gabriel S.B., Lander E.S., Beer D.G., Godfrey T.E., Getz G., Bass A.J. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson M.H., Rose S., Mahadevan L.C. Acetyllysine-binding and function of bromodomain-containing proteins in chromatin. Front. Biosci. 2001;6:D853–D865. doi: 10.2741/dyson. [DOI] [PubMed] [Google Scholar]
- Fernandez-Montalvan A.E., Berger M., Kuropka B., Koo S.J., Badock V., Weiske J., Puetter V., Holton S.J., Stockigt D., Ter Laak A., Centrella P.A., Clark M.A., Dumelin C.E., Sigel E.A., Soutter H.H., Troast D.M., Zhang Y., Cuozzo J.W., Keefe A.D., Roche D., Rodeschini V., Chaikuad A., Diaz-Saez L., Bennett J.M., Fedorov O., Huber K.V.M., Hubner J., Weinmann H., Hartung I.V., Gorjanacz M. Isoform-selective ATAD2 chemical probe with novel chemical structure and unusual mode of action. ACS Chem. Biol. 2017;12(11):2730–2736. doi: 10.1021/acschembio.7b00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri E., Petosa C., McKenna C.E. Bromodomains: structure, function and pharmacology of inhibition. Biochem. Pharmacol. 2016;106:1–18. doi: 10.1016/j.bcp.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J.P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Muller S., Pawson T., Gingras A.C., Arrowsmith C.H., Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn E.M., Huang O.W., Poy F., Oppikofer M., Bellon S.F., Tang Y., Cochran A.G. A subset of human bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure. 2015;23:1801–1814. doi: 10.1016/j.str.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Galvani A., Thiriet C. Nucleosome dancing at the tempo of histone tail acetylation. Genes. 2015;6:607–621. doi: 10.3390/genes6030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay J.C., Eckenroth B.E., Evans C.M., Langini C., Carlson S., Lloyd J.T., Caflisch A., Glass K.C. Disulfide bridge formation influences ligand recognition by the ATAD2 bromodomain. Proteins. 2019;87:157–167. doi: 10.1002/prot.25636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbet G., Devlin T., Hernandez Uribe B.I., Demeler A.K., Lindsey Z.L., Ganji S., Breton S., Weise-Cross L., Lafer E.M., Brookes E.H., Demeler B. A parametrically constrained optimization method for fitting sedimentation velocity experiments. Biophys. J. 2014;106:1741–1750. doi: 10.1016/j.bpj.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C., Asangani I.A., Ateeq B., Chun S.Y., Siddiqui J., Sam L., Anstett M., Mehra R., Prensner J.R., Palanisamy N., Ryslik G.A., Vandin F., Raphael B.J., Kunju L.P., Rhodes D.R., Pienta K.J., Chinnaiyan A.M., Tomlins S.A. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibiya K., Katsumoto T., Kondo T., Kitabayashi I., Kudo A. Brpf1, a subunit of the MOZ histone acetyl transferase complex, maintains expression of anterior and posterior Hox genes for proper patterning of craniofacial and caudal skeletons. Dev. Biol. 2009;329:176–190. doi: 10.1016/j.ydbio.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Huang H., Sabari B.R., Garcia B.A., Allis C.D., Zhao Y. SnapShot: histone modifications. Cell. 2014;159:458–458 e1. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Hoover M.E., Holt M.V., Freitas M.A., Marshall A.G., Young N.L. Middle-down characterization of the cell cycle dependence of histone H4 posttranslational modifications and proteoforms. Proteomics. 2018;18 doi: 10.1002/pmic.201700442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Kanno Y., Siegel R.M., Jang M.K., Lenardo M.J., Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- Kitabayashi I., Aikawa Y., Yokoyama A., Hosoda F., Nagai M., Kakazu N., Abe T., Ohki M. Fusion of MOZ and p300 histone acetyltransferases in acute monocytic leukemia with a t(8;22)(p11;q13) chromosome translocation. Leukemia. 2001;15:89–94. doi: 10.1038/sj.leu.2401983. [DOI] [PubMed] [Google Scholar]
- Koo S.J., Fernandez-Montalvan A.E., Badock V., Ott C.J., Holton S.J., von Ahsen O., Toedling J., Vittori S., Bradner J.E., Gorjanacz M. ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget. 2016;7:70323–70335. doi: 10.18632/oncotarget.11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladurner A.G., Inouye C., Jain R., Tjian R. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell. 2003;11:365–376. doi: 10.1016/s1097-2765(03)00035-2. [DOI] [PubMed] [Google Scholar]
- Langini C., Caflisch A., Vitalis A. The ATAD2 bromodomain binds different acetylation marks on the histone H4 in similar fuzzy complexes. J. Biol. Chem. 2017;292:16734–16745. doi: 10.1074/jbc.M117.786350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue K., Daujat S., Crump J.G., Plaster N., Roehl H.H., Kimmel C.B., Schneider R., Hammerschmidt M. The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development. 2008;135:1935–1946. doi: 10.1242/dev.017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy G., Dimaggio P.A., Chan E.Y., Zee B.M., Blanco M.A., Bryant B., Flaniken I.Z., Liu S., Kang Y., Trojer P., Garcia B.A. A quantitative atlas of histone modification signatures from human cancer cells. Epigenet. Chromatin. 2013;6:20. doi: 10.1186/1756-8935-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Louis-Jeune C., Andrade-Navarro M.A., Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. 2012;80:374–381. doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- Lubula M.Y., Eckenroth B.E., Carlson S., Poplawski A., Chruszcz M., Glass K.C. Structural insights into recognition of acetylated histone ligands by the BRPF1 bromodomain. FEBS Lett. 2014;588:3844–3854. doi: 10.1016/j.febslet.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch H., Wen H., Kim Y.C., Snyder C., Kinarsky Y., Chen P.X., Xiao F., Goldgar D., Cowan K.H., Wang S.M. Can unknown predisposition in familial breast cancer be family-specific? Breast J. 2013;19:520–528. doi: 10.1111/tbj.12145. [DOI] [PubMed] [Google Scholar]
- Meier J.C., Tallant C., Fedorov O., Witwicka H., Hwang S.Y., van Stiphout R.G., Lambert J.P., Rogers C., Yapp C., Gerstenberger B.S., Fedele V., Savitsky P., Heidenreich D., Daniels D.L., Owen D.R., Fish P.V., Igoe N.M., Bayle E.D., Haendler B., Oppermann U.C.T., Buffa F., Brennan P.E., Muller S., Gingras A.C., Odgren P.R., Birnbaum M.J., Knapp S. Selective targeting of bromodomains of the bromodomain-PHD fingers family impairs osteoclast differentiation. ACS Chem. Biol. 2017;12:2619–2630. doi: 10.1021/acschembio.7b00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S.D., Herrick S.R., Ince T.A., Kleinman M.S., Dal Cin P., Morton C.C., Quade B.J. Uterine leiomyomata with t(10;17) disrupt the histone acetyltransferase MORF. Can. Res. 2004;64:5570–5577. doi: 10.1158/0008-5472.CAN-04-0050. [DOI] [PubMed] [Google Scholar]
- Moriniere J., Rousseaux S., Steuerwald U., Soler-Lopez M., Curtet S., Vitte A.L., Govin J., Gaucher J., Sadoul K., Hart D.J., Krijgsveld J., Khochbin S., Muller C.W., Petosa C. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- Murray K. The occurrence of epsilon-N-methyl lysine in histones. Biochem. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I., Gorunova L., Bjerkehagen B., Heim S. Novel KAT6B-KANSL1 fusion gene identified by RNA sequencing in retroperitoneal leiomyoma with t(10;17)(q22;q21) PloS One. 2015;10 doi: 10.1371/journal.pone.0117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet-Montange G., Zhan Y., Bardenhagen J.P., Petrocchi A., Leo E., Shi X., Lee G.R.t., Leonard P.G., Geck Do M.K., Cardozo M.G., Andersen J.N., Palmer W.S., Jones P., Ladbury J.E. Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2. Biochem. J. 2015;466:337–346. doi: 10.1042/BJ20140933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplawski A., Hu K., Lee W., Natesan S., Peng D., Carlson S., Shi X., Balaz S., Markley J.L., Glass K.C. Molecular insights into the recognition of N-terminal histone modifications by the BRPF1 bromodomain. J. Mol. Biol. 2014;426:1661–1676. doi: 10.1016/j.jmb.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero F.A., Taylor A.M., Crawford T.D., Tsui V., Cote A., Magnuson S. Disrupting acetyl-lysine recognition: progress in the development of bromodomain inhibitors. J. Med. Chem. 2016;59:1271–1298. doi: 10.1021/acs.jmedchem.5b01514. [DOI] [PubMed] [Google Scholar]
- Rossetto D., Avvakumov N., Cote J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7:1098–1108. doi: 10.4161/epi.21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R., Meslamani J., Zhou M.M. The bromodomain: from epigenome reader to druggable target. Biochim. Biophys. Acta. 2014;1839:676–685. doi: 10.1016/j.bbagrm.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Ito T., Nishino N., Khochbin S., Yoshida M. Real-time imaging of histone H4 hyperacetylation in living cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P., Demeler B. Direct sedimentation analysis of interference optical data in analytical ultracentrifugation. Biophys. J. 1999;76:2288–2296. doi: 10.1016/S0006-3495(99)77384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang E., Nickerson H.D., Wen D., Wang X., Wolgemuth D.J. The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development. 2007;134:3507–3515. doi: 10.1242/dev.004481. [DOI] [PubMed] [Google Scholar]
- Sobel R.E., Cook R.G., Perry C.A., Annunziato A.T., Allis C.D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Liu J., Zhang J., Shen W., Huang H., Xu C., Dai H., Wu J., Shi Y. Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem. Biophys. Res. Commun. 2007;358:435–441. doi: 10.1016/j.bbrc.2007.04.139. [DOI] [PubMed] [Google Scholar]
- Tallant C., Valentini E., Fedorov O., Overvoorde L., Ferguson F.M., Filippakopoulos P., Svergun D.I., Knapp S., Ciulli A. Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC. Structure. 2015;23:80–92. doi: 10.1016/j.str.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne A.W., Kmiciek D., Mitchelson K., Sautiere P., Crane-Robinson C. Patterns of histone acetylation. Eur. J. Biochem. 1990;193:701–713. doi: 10.1111/j.1432-1033.1990.tb19390.x. [DOI] [PubMed] [Google Scholar]
- Ullah M., Pelletier N., Xiao L., Zhao S.P., Wang K., Degerny C., Tahmasebi S., Cayrou C., Doyon Y., Goh S.L., Champagne N., Cote J., Yang X.J. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol. Cell Biol. 2008;28:6828–6843. doi: 10.1128/MCB.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M., Eberl H.C., Matarese F., Marks H., Denissov S., Butter F., Lee K.K., Olsen J.V., Hyman A.A., Stunnenberg H.G., Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Wang T., Holt M.V., Young N.L. The histone H4 proteoform dynamics in response to SUV4-20 inhibition reveals single molecule mechanisms of inhibitor resistance. Epigenet. Chromatin. 2018;11:29. doi: 10.1186/s13072-018-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Workman J.L., Kingston R.E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Yan K., Rousseau J., Littlejohn R.O., Kiss C., Lehman A., Rosenfeld J.A., Stumpel C.T.R., Stegmann A.P.A., Robak L., Scaglia F., Nguyen T.T.M., Fu H., Ajeawung N.F., Camurri M.V., Li L., Gardham A., Panis B., Almannai M., Sacoto M.J.G., Baskin B., Ruivenkamp C., Xia F., Bi W., Study D.D.D., Study C., Cho M.T., Potjer T.P., Santen G.W.E., Parker M.J., Canham N., McKinnon M., Potocki L., MacKenzie J.J., Roeder E.R., Campeau P.M., Yang X.J. Mutations in the chromatin regulator gene BRPF1 cause syndromic intellectual disability and deficient histone acetylation. Am. J. Hum. Genet. 2017;100:91–104. doi: 10.1016/j.ajhg.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L., Yan K., Zou J., Zhao H., Bertos N.R., Park M., Wang E., Yang X.J. The chromatin regulator Brpf1 regulates embryo development and cell proliferation. J. Biol. Chem. 2015;290:11349–11364. doi: 10.1074/jbc.M115.643189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L., Li L., Zou J., Yan K., Belle J., Nijnik A., Wang E., Yang X.J. BRPF1 is essential for development of fetal hematopoietic stem cells. J. Clin. Invest. 2016;126:3247–3262. doi: 10.1172/JCI80711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N.L., DiMaggio P.A., Plazas-Mayorca M.D., Baliban R.C., Floudas C.A., Garcia B.A. High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M., Wu J., Workman J.L., Li B. Readers of histone modifications. Cell Res. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Zhang K., Williams K.E., Huang L., Yau P., Siino J.S., Bradbury E.M., Jones P.R., Minch M.J., Burlingame A.L. Histone acetylation and deacetylation: identification of acetylation and methylation sites of HeLa histone H4 by mass spectrometry. Mol. Cell. Proteomics. 2002;1:500–508. doi: 10.1074/mcp.m200031-mcp200. [DOI] [PubMed] [Google Scholar]
- Zhu J., Caflisch A. Twenty crystal structures of bromodomain and PHD finger containing protein 1 (BRPF1)/Ligand complexes reveal conserved binding motifs and rare interactions. J. Med. Chem. 2016;59:5555–5561. doi: 10.1021/acs.jmedchem.6b00215. [DOI] [PubMed] [Google Scholar]
- Zhu J., Zhou C., Caflisch A. Structure-based discovery of selective BRPF1 bromodomain inhibitors. Eur. J. Med. Chem. 2018 doi: 10.1016/j.ejmech.2018.05.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.