Abstract
Despite extensive clinical research and management protocols applied in the field of coronary artery diseases (CAD), it still holds the number 1 position in mortality worldwide. This indicates that we need to work on precision medicine to discover the diagnostic, therapeutic, and prognostic targets to improve the outcome of CAD. In precision medicine, epigenetic changes play a vital role in disease onset and progression. Epigenetics is the study of heritable changes that do not affect the alterations of DNA sequence in the genome. It comprises various covalent modifications that occur in DNA or histone proteins affecting the spatial arrangement of the DNA and histones. These multiple modifications include DNA/histone methylation, acetylation, phosphorylation, and SUMOylation. Besides these covalent modifications, non-coding RNAs—viz. miRNA, lncRNA, and circRNA are also involved in epigenetics. Smoking, alcohol, diet, environmental pollutants, obesity, and lifestyle are some of the prime factors affecting epigenetic alterations. Novel molecular techniques such as next-generation sequencing, chromatin immunoprecipitation, and mass spectrometry have been developed to identify important cross points in the epigenetic web in relation to various diseases. The studies regarding exploration of epigenetics, have led researchers to identify multiple diagnostic markers and therapeutic targets that are being used in different disease diagnosis and management. Here in this review, we will discuss various ground-breaking contributions of past and recent studies in the epigenetic field in concert with coronary artery diseases. Future prospects of epigenetics and its implication in CAD personalized medicine will also be discussed in brief.
Keywords: Epigenetics, coronary artery disease (CAD), acute myocardial infraction, precision medicine, therapeutic targets
Introduction
Epigenetics is a term coined from Greek prefix, “epi,” meaning “over,” “external,” or “around,” and the traditional word “genetics” that is inheritance. Hence, epigenetics can be described as the changes affecting heritable phenotype variations without altering the DNA sequence of a genome. Epigenetic change is associated with the covalent modifications in the structural components of chromatin, nitrogenous bases of DNA, and expression of various non-coding RNA species, which decide the expression pattern of genes. They ultimately help in keeping the cell identity, which is censoriously important for normal development and causation of various diseases. During recent 2 decades, epigenetics has seen an extensive surge in the field of precision medicine, and epigenetic changes have been associated with almost all disease states, including malignant and non-malignant, especially lifestyle disorders. They mostly occur due to changes in the microenvironment of cells, which in turn is decided by various modifiable and nonmodifiable factors.1 Based on these facts, epigenetics plays an inevitable role in disease pathogenesis of coronary artery disease (CAD). Further, exploration of epigenetics underlying various diseases has paved way for the development of novel therapeutic agents. Several drugs targeted to epigenetic factors are under trial.1
Acute Myocardial Infarction (AMI) is the foremost cause of death worldwide.2 The role of epigenetic changes in patients with CAD is comparatively novel concept with interesting findings. Studies have shown that epigenetic alterations significantly contribute to the development of acute myocardial infarction. Endothelial dysfunction, imbalance in cholesterol metabolism, and atherosclerotic plaques are the primary pathophysiological modification leading to CAD. These changes, along with the risk factors of CAD-like obesity, diabetes mellitus (DM), hypertension, and insulin resistance, are interrelated to environmental risk factors associated to increased rate of CAD.3 The development of atherosclerotic plaque follows epigenetic changes of vascular smooth muscle cells (VSMCs), endothelial cells (ECs), and macrophages. Inflammation, cholesterol metabolism, and homocysteine homeostasis are associated with the development of atherosclerotic plaque, the main pathologic change resulting in CAD.4
A number of reports regarding influence of epigenetics on CAD are being added to the literature on an exponential rate, paving the way to explore the underlying epigenetic mechanisms of the disease. In this review, we will discuss the latest discoveries related to epigenetic modifications in fetal and adult stages that contribute to the CAD development and its progression. Also, how the environment coordinates with epigenetics of an individual towards the initiation and development of CAD, as well as the role of epigenetics in personalized medicine will also be discussed to provide a comprehensive view to readers.
Pathophysiology of CAD
Atherosclerosis is a chronic inflammatory condition which is characterized by thickening of vascular wall leading to luminal obstruction.5 It is the basic pathogenic lesion in vaso-occlusive conditions of different organ systems like coronary artery disease and peripheral artery disease. Atherosclerotic plaque is composed of an accumulation of lipids, T lymphocytes, macrophages and fibrous tissues in arterial walls. This theory is known as the “response to injury” hypothesis, implicating endothelial disorder as the primary step in the cascade of different pathological stages of this chronic inflammatory condition, which precedes any structural alterations in the arterial wall occur.6 The result of the process is the endorsed expression of intercellular and vascular cell adhesion molecules due to low-density lipoprotein (LDL) cholesterol oxidation in subendothelial space, which ultimately leads to monocyte adhesion/ migration to the subendothelial area. In the intima monocytes differentiate into macrophage which engulf oxidized LDL via scavenger receptor promoting foam cell formation and exhibit proinflammatory functions by secreting cytokines—viz. interleukins and tumor necrosis factor.
The end step of this process is the formation of atherosclerotic lesion I (fatty streak). Atherosclerotic build up results in plaque formation, vascular remodeling, acute and/or chronic luminal obstruction, abnormalities of blood flow and diminished oxygen/ nutrients supply to myocardium. By impairing or obstructing normal coronary blood flow, atherosclerotic build up causes myocardial ischemia.7 In this review we will discuss the epigenetics modifications played a significant role in the pathogenesis of CAD.
Risk factors for CAD have been classified into demographic, biochemical parameters, lifestyle and genetic factors. Demographic factors include advancing age and male sex, biochemical parameters include variables such as raised cholesterol levels, lifestyle factors include sedentary working habits, high levels of stress and depression, unbalanced dietary habits, socioeconomic status and alcohol or smoking/tobacco abuse and genetic factors include positive family history for CAD and inherited disorders of lipid metabolism.5
In demographic factor, age play a pivotal role in development of CAD.8,9 A study by Ravi et al demonstrated that increasing age is an independent risk factor for CAD.10 Considering gender, men of all ages have higher mortality from CAD than women before menopause.11 Several studies marked that; the incidence of CAD is similar in post-menopausal woman in age matched men.12-14 Several studies have reported high risk lifestyle choices are associated to epigenetic alterations such as histone modifications and DNA methylations are strongly associated with obesity leading to coronary artery disease.15 A recent study indicated that different candidate gene methylations are associated to lipid metabolism and inflammation in subjects with obesity.16 Also, a recently published study demonstrates that tobacco smoking is associated with DNA methylation at several loci of atherosclerotic carotid lesions.17 Moreover, a study by Rebecca et al. reveals that specific DNA methylation patterns have been found in tobacco smoking CAD patients.18 Those who smoke may have a vulnerability to illness with a heritable (genetic) component for CAD, T2DM, or hypertension, yet once the patient quits smoking, the likelihood of having/developing the disease phenotype starts to remit.19 Also, Lohoff et al revealed that inducement of chronic alcohol modifies PCSK9 methylation leads to impairment of lipid metabolism and CAD.20 Epidemiological studies propose that early life, mainly prenatal, environmental exposure can induce metabolic and physiological changes in fetus by modifying epigenetic profiles leading to predisposition of multiple chronic diseases such as diabetes, obesity, cardiovascular event, and cancer.21 Accumulating reports from numerous studies suggest that epigenetics might be one of the unique molecular mechanisms with different contributing factors like fetal programming, environmental stimulus, and childhood phenotype. Owing to the reversible nature of these factors, epigenetic modifications are becoming an attractive therapeutic agent.22
Inception of epigenetics in response to environmental variances
Intrauterine exposure
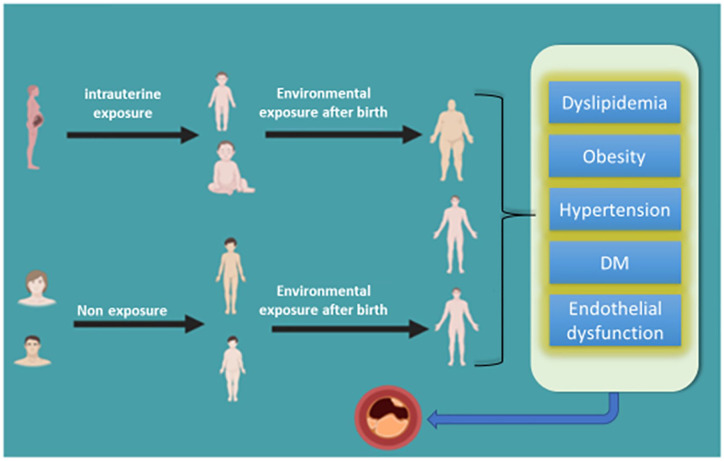
External stress modulates cellular and tissue level maturity. Severity varies depending time of exposure within narrow window period of genetic variation or specific organogenesis. Intrauterine development is highly sensitive for modifications in response to environmental stress in order to fine tune genotypic and phonotypic traits. This high level of developmental plasticity is not only adaptive but also may result in maladaptive change paving way for pathological processes.21,23 Epigenetic modulations can be inheritable through subsequent generations resulting in transgenerational epigenetic inheritance. Available micro milieu imparts most drastic effect on epigenetic status and modulation of transcriptome in early few weeks of pregnancy.22 Diet, pathological conditions like hypertension, DM, smoking, chemical pollutants, and psychological stress have been proven to have important role in determining epigenetic landscape for future disease processes (Figure 1)24,25.
Figure 1.
Increased risk of developing CAD with the effect of environmental inducements through intrauterine and after birth exposure.
Exposure after birth
Exposure to environmental pollution has been associated with cardiovascular risk factors. It is an established fact that smoking is the strongest independent risk predictor for premature coronary heart disease; moreover, second-hand smoke (SHS) has been characterized as an environmental pollutant contributing approximately to 80% death due to CAD and only <5% death from lung cancer.26 This suggests coronary atherosclerotic tissues are susceptible to environmental chemical pollutants. A meta-analysis of genome-wide DNA methylation conducted by Roby Joehanes et al in 2017 revealed that changes in DNA methylation sites are associated with cigarette smoking highlighting thousands of differentially methylated CpGs.27 Dietary influence on CAD has been studied widely suggesting both Mediterranean and DASH (Dietary Approaches to Stop Hypertension) diet, that is filled with vegetables, fruit, and nuts are associated with decreased risk of AMI and improved effects on blood pressure, lipid profile, inflammation, endothelial function, and thrombosis.28 Specific research to elucidate the effect of dietary determinants on CAD risk modifying epigenetic modifications is lacking, although literature suggests diet may influence factors such as DNA hyper and hypomethylation.29 Future research strategies might prove to be a worthwhile investment of resources to describe diet—epigenetics interactions leading to atherosclerotic disorders.
Another important factor resulting in epigenetic aberrations includes a sedentary lifestyle, which possesses a significant role in deciding the pattern of expression of different genes. This is achieved through changes in the microenvironment of chromatin affecting its spatial states; hetero/euchromatin ultimately changes the ability of transcription machinery to DNA accessibility despite the absence of DNA alterations.30 The epigenetic changes lead to gain and loss of function, as observed in animal models.31-35 In this article, we aimed to review Epigenetic factors, including DNA methylation, histone alteration, and noncoding RNA control, as a risk factor of CAD by altering the relationship between genes and the environment.
Epigenetic changes can present as:
(1) Covalent modifications including methylation, acetylation, phosphorylation, SUMOylating, ubiquitination.
(2) Non-coding RNAs—viz. miRNA, lncRNA, and circRNA (Figure 2).
Figure 2.
Environmental factors affecting epigenetic upbringing such as covalent modifications and different expression of non-coding RNAs leading to the development of Coronary Artery Disease over the years.
Covalent Modifications
Methylation of DNA
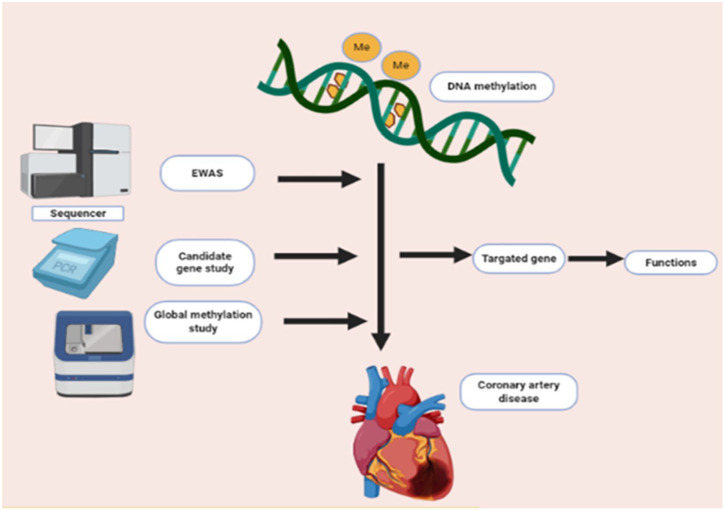
Amongst the covalent modifications, DNA methylation, which is a pre-transcriptional modification, has been extensively studied. It is the major epigenetic modification in mammals. DNA methylation shows a covalent transfer at CpG dinucleotide regions where methyl group (–CH3) attached to the fifth carbon of cytosine base. This process is catalyzed by particular enzymes called DNA methyltransferases (DNMTs).36 DNA methylation is involved in multiple factors such as basic biological activities of the cell, gene expression, aging, malignancies and non-malignancies such as CAD.36,37 It is estimated that a total of 70% to 80% of CpG dinucleotides exist in a state of methylation in the human genome’s dinucleotides may cluster as CpG islands and serve as functional units to regulate gene expression by deciding the binding of transcription factors.38 The frequency of CpG dinucleotide is higher in promoter regions as compared to intronic and exonic region.39 Hypermethylation of promoter regions in somatic cells inhibits their activity. On the other hand, promoters with strong activation are largely unmethylated.40 There are 3 angles that are being used to study the association between DNA methylation and CAD, as shown in Figure 3.
Figure 3.
Different approaches to study the association between DNA methylation and CAD.
Role of DNA methylation in CAD
Lipid metabolism plays a pivotal role in atherosclerosis. It is affected by genetic and environmental factors both. Hypermethylation of certain genes like insulin like growth factor-2 (IGF2) and leptin genes is associated with risk factors of CAD, namely obesity and insulin receptor (IR).41,42 Hypermethylation of ATP Binding Cassette Subfamily G Member (ABCG1)43 and hypomethylation of Carnitine palmitoyltransferase 1A (CPTIA)44 has been shown to be associated with postprandial lipemia. Differential patterns of methylation are observed in genes involved in the pathogenesis of CAD and lipid homeostasis.45
Global DNA methylation
The environmental and lifestyle factors which contribute to the development of CAD involve the epigenetic changes in the genome. Epigenetic modifications start in utero itself, which is a vulnerable period that can program the individual for risk of disease in adulthood.46 Later the early and midlife environmental exposures to smoke, environmental contaminants and pollution lead to methylation of DNA, laying the foundation of non-genetic mechanisms by which CAD might develop and manifest later in life. Also, a protein-restricted diet and diet deficient in vitamin B12, folate, methionine in pregnant animals, and a low-fat flavonoid-rich diet affects the methylation patterns in specific genes contributing to atherogenesis. Smoking, environmental toxins, and pollution contribute to atherogenesis and have been observed to alter the methylation in specific genes.47-49
The various reports have elucidated that modifications in DNA methylation sites contribute to the regulation of different biological processes, including coronary atherosclerosis, hypertension, and inflammation, that play a significant role in the development of CAD.50-52 For instance, mice with hypomethylated DNA had overexpression of inflammatory markers, which leads to the formation of aortic fatty streaks.53 In CAD prone mice lacking ApoE, modified DNA methylation sites are found in both blood leukocytes and aorta before the formation of vascular lesions.54 These modifications contribute to inflammation and atherosclerotic plaque.
Different studies have reported that both hypomethylation and hypermethylation status in CAD patients. Although global hypomethylation has been reported in DNA from atherosclerotic tissues,25,55,56 CAD associated genes have been found to be hypermethylated in promoter regions more commonly51 (Table 1). These differences could either be due to the presence of epigenetically/genetically non-identical cells in atherosclerotic tissue. Moreover, different methylation patterns may be associated with heterogeneous cell types of an individual’s tissue. While hypermethylation looks to be a greater contributing factor for CAD development, compared to hypomethylation, emerging single-cell sequencing technologies could overcome these discrepancies in the future.
Table 1.
Global DNA methylation pattern in CAD.
| References | Sample source and sample size (n) | Methods | Global DNA methylation status |
|---|---|---|---|
| Zaina et al57 | Post mortem donor: | Bisulfite treatment and DNA methylation microarray | Hypermethylation |
| Human atherosclerotic tissue (n = 1) | |||
| Human normal aortic tissue. (n = 1) | |||
| Nazarenko et al58 | Human atherosclerotic tissue obtained from right coronary arteries and carotid in an advanced stage (n = 12) | The Illumina HumanMethylation27 BeadChip microarrays | Hypomethylation |
| Human atherosclerotic tissue from resistant internal mammary arteries of men (n = 8) | |||
| Selected CpG sites quantified in vascular tissue (n = 80) | Bisulfite pyrosequencing | ||
| del Pilar Valencia-Morales et al59 | Human atherosclerotic aortic tissue samples ranging from histological grade III to VIA (n = 15) | DNA methylation microarray | Hypermethylation |
| Human normal aortic tissue samples (n = 15) | |||
| Hiltunen et al60 | Human normal arterial tissue samples (n = 3) | HPLC bisulfite sequencing | Hypomethylation |
| Tissue samples from fatty streaks (n = 23) | |||
| Tissue samples from advanced lesions (n = 29) | |||
| Sharma et al61 | Angiographically confirmed CAD patients (n = 18) | 12 k human CpG Island microarray | Hypermethylated |
| Healthy control (n = 18) | |||
| Sharma et al62 | Peripheral lymphocyte of angiographically confirmed CAD (n = 137) | Methylation-sensitive restriction enzymes (MSRE) | Hypermethylated |
| Control (n = 150) |
Candidate gene DNA methylation
An alternative way to investigate the methylation patterns is to analyze methylation at CpG dinucleotides of the targeted genes. Some reports from current studies associated to the pathophysiology of atherogenesis are compiled and depicted in Table 2. Modifications in the methylation status of a specific gene of interest can affect the functional pathways involved in atherogenesis such as LDL metabolism, lipid homeostasis, cholesterol biosynthesis, homocysteine metabolism, endothelial dysfunction and, inflammation.
Table 2.
Candidate gene DNA methylation status in CAD.
| Reference | Candidate gene | Technique | Sample source and sample size (n) | Methylation pattern | Methylation site | Expression pattern |
|---|---|---|---|---|---|---|
| Zhu et al63 | MTCT3 | Bisulfite sequencing | Aortas of heart donors | Hypermethylation | Exon2 | Downregulation |
| Aortic tissue (n = 23) | ||||||
| Normal aortic tissue (n = 10) | ||||||
| Huang et al64 | ES α | Nested methylation specific polymerase chain reaction (MS PCR) | Human Atherosclerosis blood (n = 54) | Hypermethylation | Promoter | Not measured |
| Healthy control blood (n = 24) | ||||||
| Kim et al65 | Esβ | Methylation-specific PCR (MS PCR) | Human atherosclerotic tissue (n = 2) | Hypermethylation | Promoter | Downregulation |
| Human normal tissue (n = 2) | ||||||
| Zuo et al66 | IL6 | Bisulfite treatment and pyrosequencing | CHD blood sample (n = 212) | Hypomethylation | Promoter | Not measured |
| CHD free blood sample (n = 218) | ||||||
| Bakshi et al67 | STAT1, IL12b, MHC2, iNOS, JAK1, JAK2 | Methylation specific high resolution melting curve (MS-HRM) assay | PBMCs of CAD patients (n = 25) | Hypomethylation | Promoter | Not measured |
| Control (n = 25) | ||||||
| Ghosh et al68 | GLANT2, HMGCR, CGK eNOS3 | Targeted bisulfite sequencing | Healthy subjects (n = 42) | Hypermethylated | Prompter | |
| LDLR | CAD patients (n = 33) | Hypomethylated |
Therefore, exploring candidate gene methylation pattern in CAD using novel cutting-edge techniques help us to confirm the contribution of new genes after global methylation studies. This will open up a new therapeutic window for targeted therapies and prognostic markers after validation.
Epigenome-wide association studies
Different epigenome-wide association studies (EWAS) have been conducted in coronary atherosclerosis linked to DNA methylation at the CpG site to elucidate its role at the molecular level.69,70 The results of recent studies are shown in Table 3. EWAS study reveals that patients with CAD have a different pattern of DNA methylation sites at several genomic loci.
Table 3.
Epigenome-wide association studies.
| Reference | Sample source and sample size (n) | Methods | Ethnicity | Methylation status | ||
|---|---|---|---|---|---|---|
| Total CpG sites | Hypomethylation | Hypermethylation | ||||
| Castillo-Díaz et al73 | Human atherosclerotic tissue (n = 45) | Microarray | Hong Kong | 10367 CpG sites | NA | 151 CpGs in the control site |
| Human non atherosclerotic tissue (n = 16) | 141 sites were partially methylated | |||||
| Yamada et al74 | Post mortem aortic tissue (n = 64) | DNA methylation-specific microarray (Infinium Methylation EPIC Bead Chip, Illumina, Inc.) | Japan | 853307CpG sites.2679 CpGs were significantly associated | 407 CpGs | 2272 CpGs |
| Shen et al75 | Blood sample of atherosclerotic stroke patients (n = 12) | Illumina Infinium Human Methylation 450K Bead chip | China | 482360 CpG site | 438 CpGs | 574 CpGs |
| Blood samples from matched control (n = 12) | ||||||
| Nakatochi et al69 | Blood sample of MI patients (n = 192) | Illumina Infinium Human Methylation 450 K Bead chip | Japan | 348595 CpG site | Not studied | Not studied |
| Blood sample matched control (n = 192) | ||||||
| Rask-Andersen et al70 | Blood sample (n = 729) | Illumina Infinium Human Methylation 450 K Bead chip | Sweden | 470789 Sites | Not studied | Not studied |
| Banerjee et al76 | Angiographically positive male (n = 6) | Illumina Infinium Human Methylation 450 K Bead chip | India | 429DMRs | 222 | 207 |
| Angiographically negative male (n = 6) | ||||||
Some investigators have found different methylated CpG sites in a specific gene; while others have observed a CpG island with increased methylation sites.71 Correspondingly, EWAS scrutinize gene-specific DNA methylation. Gradually, EWAS has substituted the candidate gene method as it is useful to find novel CpG sites linked to disease phenotype.72 Although a large proportion of previous EWAS confirmed their results on replication, the smaller sample size in these studies asks for further validation of the obtained results. A total of 84 translating loci with varied amounts of methylation were identified in at least more than 1 EWAS which suggests an investigation of epigenetic regulation of the identified genes in coronary atherosclerotic statuses. The results revealed a group of genes highlighting the significance of various conditions including carbohydrate and lipid metabolism along with obesity, inflammation and many others in CAD.71 Still, these patterns are mostly inconclusive, warranting future research with a larger sample cohort.
Histone modifications
Histones are alkaline proteins around which DNA is wrapped to form bead-like structures called nucleosomes. Histones are characterized by 5 families: H1/H5, H2A, H2B, H3, and H4.77 Histones are a fundamental unit of chromatin.78 These proteins are extremely unstable. They modify rapidly in response to any external signal. Histone molecules undergo several post-translational modifications due to different molecular mechanism at chromatin and amino acid levels.79 Amongst the multiple changes, histone acetylation, methylation, and phosphorylation are found to be associated with transcription regulation and gene expression. Post-translational modifications of histones can interfere with chromatin structure, organization, and function, which in turn alter mechanisms such as gene expression, DNA repair, DNA damage, and ultimately affecting the proliferative capacity of the cells.80 Moreover, histone methylation and demethylation have been studied in coronary atherosclerosis widely.81
Histone methylation and demethylation
Histone methylation is directly linked to activation or inactivation of a specific gene. Owing to the modified segment of histone a gene decides whether to activate/inactivate.82 In 1960s, methylation pattern on histone was first identified. The methylation enzyme was unknown until the event of the lysine methyltransferase (KMT) family was found.83 Histone methylation was considered to be the ultimate stone post-translational modification until the detection of first histone lysine de-methylase (KDM).84-86 Methylation of histone can occur both in arginine and lysine residues. At histone H3 and H4, the following methylation residues are documented: K4, K9, K27, K36, K79, and K20. Methylation of lysine residues can be found in multiple degrees, such as mono(me1)-, di (me2), or trimethyl (me3) groups.
Histone-lysine methyltransferase acts as a catalyst. It transfers a methyl group from S-adenosylmethionine (SAM) to amino groups on lysine residues to histone tails.87
Discovery of lysine demethylating enzymes altered the primitive concept of irreversible and stable nature of histone methylation. Now, things are changed, and 2 critical classes of histone-lysine dimethyl enzymes are known. It is reported that flavin adenosine dinucleotide is being used as a cofactor of lysine-specific de-methylases (LSDs). The most studied LSD includeLSD1, which interacts with protein complexes like CoREST (resting corepressor) and other protein complexes catalyzing reversible methylation of H3K4 and H3K9.85,88 A recent study has shown that JmjC domain-containing proteins are the largest class of histone de-methylases. They act as a catalyst and take part in histone demethylation by using Fe (II) and alpha-ketoglutarate as a cofactor.86 Though arginine demethylating enzymes have been studied extensively, still histone lysine demethylation is found to possess a predominant role in the process of gene transcription.87 A number of studies have been widely demonstrated the association between histone methylation and atherosclerotic plaque.88-90 In 2015, Greibel et al. established that H3K9me2 and H3K27me2 were significantly reduced in atherosclerotic plaque. They also found the H3K4me2 level in the atherosclerotic and healthy carotid artery. Simultaneously, immunohistochemistry results showed elevation of H3K4me2 in VSMCs, while H3K9me2 showed reduction. Likewise, H3K9me2 and H3k27me2 both the levels are reduced in inflammatory cells. Interestingly, the expression of the corresponding histone methyltransferases MLL2 and G9a was augmented in an advanced stage of atherosclerosis as compared to its early stage.88 Further, in 2015, Wierda et al observed that the global H3K27me3 level was less expressed in vessels with advanced atherosclerotic plaques. Though, this result does not affect the equivalent histone methyltransferase (EZH2) or de-methylase JMJD3, suggesting H3K9 and H3K27 demethylation were significantly associated with atherosclerotic plaque formation.89
Histone methylation of H3K4 is linked with the development and progression of coronary atherosclerosis.91 However, increased demethylation of H3K27 has been detected in the advanced stage of CAD.89
The significant impact of histone proteins is due to their intimate association with DNA, which makes them vital in a variety of regulatory processes. Hence histones are the main components that are exceptionally important in the onset and progression of CAD phenotype. However, their exact role is substantially diverse, and any specific pattern is yet to be established.
Acetylation of histone tails
Histone acetylation at lysine residues was discovered approximately 50 years ago.93,94 After the discovery of histone acetylation, researchers have identified some enzymes that catalyze reversible acetylation. The protein lysine acetyltransferase (KATs), initially known as histone acetyltransferase (HATs) and histone deacetylases (HDACs), also act on many non-histone substrates. Most acetylation substrates are localized in the nucleus and act as transcription factors and coregulators.95,96 Various studies have established a significant role of acetylation in gene expression and regulation of chromatin structure.97,98 Furthermore, acetylation and deacetylation are the fundamental natures of histone modifications that play a major role in many biological processes.99 Overall, transcriptional activation induced by HATs results from an addition of acetyl group. In contrast, the transcription inhibition induced by HDACs results from removal of the acetyl group from H3 and H4 in the N-terminal tails at lysine residues.60 In 2016, Greibel et al. revealed that H3K4 methylation and H3K9 acetylation were significantly correlated with the severity of coronary atherosclerosis.92 Different expression patterns of class II histone de-acetylase releases acetyl group from histone molecule and found to be associated with coronary atherosclerosis. Also, HDAC2 is less expressed due to oxidized low-density lipoprotein (ex-LDL), which shows increased oxidative stress. Overexpression HDAC2 cDNA in human aortic endothelial cell downregulate Arg2 expression, which in turn block the oxidized low-density lipoprotein mediated vascular dysfunction.100
HATs and HDACs are the primary enzymes for the regulation of lysine acetylation levels, suggesting these can be used as a therapeutic target in the management of coronary atherosclerosis.
Phosphorylation
Phosphorylation of histone proteins occurs in the hydroxyl groups of serine, threonine, and tyrosine residues. Kinases act as a catalyst and take part in the addition of phosphate group from ATP, whereas phosphatases catalyze the removal of the phosphate group. The phosphate group converses a negative charge in phosphorylation, which is associated with the chromatin model and leads to gene transcription.101 However, the calcium/calmodulin-dependent protein kinase II δ (CAMII δ) exhibit a significant role in the pathogenesis of cardiac hypertrophy.99 It is reported that chromatin remodeling is facilitated by nuclear CAMII δ through phosphorylation of H3Ser10, which endorse gene transcription and leads to coronary artery diseases.102,103
SUMOylation and ubiquitination
SUMOylation and ubiquitination are important mechanisms responsible for histone post-translational modification that comprises of addition or removal of large bulky groups.104 These 2 mechanisms show their resemblances in terms of 3D structures of SUMO and ubiquitin proteins.105 SUMOylation regulates miscellaneous cellular processes with cell cycle, transcription, protein stability, DNA replication/repair, signal transduction, apoptosis.106 SUMOylation is essential for cardiac function under pathological and physiological condition.107 It has been shown that failing heart encourages SUMO2/3 conjugation. Additionally, an elevated level of SUMO2/3-dependent modification has been observed to effect in congestive heart disease (CHD) such as cardiac hypertrophy by promoting cardiac cell death.108 Similarly, the role of SUMOylation in cardiac protein degradation has also been established.109 In contradiction, over-expression of SENP5, SUMO2/3-specific deconjugation enzyme has been detected to result in dilated cardiomyopathy or cardiac failure.110 The role of SUMOylation and ubiquitination has not been specifically explored in atherosclerotic conditions, in spite of the fact that these may prove to be attractive candidates to elucidate the molecular mechanisms of post-AMI cardiac remodeling. This could create a future area of research.
Non-coding RNAs
NIH conducted “ENCODE” project reported that only 3% of the human genome is translated to proteins, where 86% is transcribed to non-coding RNAs (nc RNAs).111,112 In the year 1950, worldwide published data demonstrated that there was no association between the genome and the developmental biology of organisms, and the non-coding genome was considered as a garbage gene.113-115
However, recent studies revealed the significant role of non-coding genome in the mechanism of gene expression, regulation followed by protein expression, and also in the pathogenesis of many diseases, both malignant and non-malignant, including coronary atherosclerosis.116 nc RNAs are transcribed from DNA and perform important regulatory and structural functions.117 Mainly, nc RNAs are divided into 2 classes (a) small ncRNAs containing <200 nucleotides and (b) long nc RNAs with a length more than 200 nucleotides. Nowadays, non-coding RNAs are being implicated in the pathogenesis of coronary atherosclerosis and hence act as important biomarkers of cardiovascular injury.118
miRNAs
Discovery of micro RNAs (miRNAs) is an innovative domain in translational research. These miRNAs can instantly regulate expression of different genes through various molecular mechanism. The nucleotide sequence of miRNAs is extremely preserved across, act as a modulator of gene expression and gene regulation.119 Till date, approximately 2500 miRNAs have been discovered in the human genome and these are responsible for the regulation of 62% of human genes.120 The role of miRNAs in coronary artery disease has been widely studied over the years. Thus, miRNAs are found to be major regulator in cellular gene expression for the initiation and progression of CAD.121 List of reported studies where miRNA played a substantial role in CAD are depicted in Table 4. Also, literature supports that miRNAs such as miRNA1 and miRNA133a can be used as better diagnostic and prognostic biomarkers for AMI in comparison with cardiac troponins.138
Table 4.
List of miRNA associated with Coronary artery disease.
| References | miRNAs | Sample source | Findings |
|---|---|---|---|
| Fichtlscherer et al124 | miRNA155, miRNA145, miRNA17a and miRNA92a, miRNA133a and miRNA208a | Human plasma | Reduced in patients with CAD compared healthy control |
| Overexpressed in patients with CAD compared to healthy control | |||
| Vindis et al125 | miRNA155, miRNA145 | Human plasma | Significantly under-expressed in stable coronary atherosclerosis patients as compared to healthy subjects |
| Wang et al126 | miRNA133 | Human plasma | Significantly higher in AMI patients as compared to control group |
| Positively correlated with the severities of the coronary artery stenosis | |||
| Liu et al127 | miRNA370 and miRNA208a | Human plasma | Both the miRNAs are significantly higher in CAD patients as compared to control |
| miRNA208a differentially expressed in heart muscle | |||
| miRNA370 associated with lipid metabolism | |||
| Zhang et al128 | miRNA208a | Human plasma | Associated with the severity of CAD |
| Wang et al129 | miRNA31 | Human plasma | Higher in the CAD patients with restenosis as compared to those without restenosis |
| Sayed et al130 | miR208,iR499, miR133 and miR1 | Human plasma | Upregulated in AMI patients compared to healthy control |
| Sondermeijer et al131 | miR340, miR451, miR624 | Human platelet | Upregulated in CAD patients compared to healthy control |
| Corsten et al132 | miRNA208a | Human plasma | 1550 fold higher in AMI patients as compared to healthy subjects |
| Xin et al133 | miRNA499-5p | Human plasma | 10 fold higher in AMI patients as compared to healthy subjects |
| Adachi et al134 | miRNA499 | Human plasma | It has higher sensitivity compared to troponin in diagnosing AMI (5%) |
| Increased in AMI patients in comparison to congestive HF | |||
| Olivieri et al135 | miRNA499-5p | Human plasma | This can be correlated better with AMI with no ST elevation as compared to troponin in aged AMI patients |
| Hoekstra et al136 | miRNA147 | PBMCs | 4 fold downregulated in CAD compared to control |
| Cheng et al137 | miRNA1 | Human plasma | 200 fold higher in patients with AMI at 6 h of the onset of symptoms and returned to normal levels after three days of the initial date of the symptoms |
| Ai et al138 | miRNA1 | Human plasma | Significantly increased in AMI patients compared to atherosclerosis without AMI or healthy controls |
| D’Alessandra et al139 | miRNA133 | Human and Mice plasma | Significantly upregulated in human AMI subjects and AMI animal models |
| Widera et al140 | miRNA1, miRNA133a | Human plasma | Significantly increased in AMI patients as compared to patients with unstable angina or any other coronary heart disease |
Long non-coding RNA (lncRNA)
In 2002, Okazaki and his team first identified the long non-coding RNAs (>200 bp) while performing a wide range sequencing in a mouse model.139 Subsequent studies found that long non-coding RNA (lncRNA) can be expressed in the cytoplasm or nucleus. The high stability of lncRNAs makes them interesting entities and an essential candidate for research. These long non coding RNAs may exist in extracellular fluids (ECF) like plasma with ultra-stability exhibiting a complex behavior on different disease states.140-142 These RNAs are packed in exosomes/extracellular vesicles and can reach to the bloodstream when it released from the dying or apoptotic cells.143,144 Packaging of lncRNAs into exosomes increases their stability and half-life. In addition, these large RNAs are found to be more stable when they linked to RNA binding proteins keeping them resistant against RNases. Due to the longer half-life and tremendously increased stability of lncRNAs make them easily detectible entities and hence may act as suitable and novel non-invasive diagnostic and prognostic biomarkers.
Like miRNAs, lncRNAs also do not encode protein though they exhibit similar structures like mRNA.145 The ncRNAs play a crucial role in gene regulation processes and epigenetic changes.146,147 The Long intergenic non-coding RNAs (lincRNAs) have been reported to play significant roles in a number of diseases, including coronary atherosclerosis,148 immune disease,149 and different type of malignancies.150 Over the years, many experimental findings have demonstrated that lincRNAs have significant roles in the pathogenesis of cardiovascular disease.151-153 Subsequently, the alterations in specific lncRNA are being proposed to be used as novel diagnostic and prognostic biomarkers of coronary atherosclerosis.154-156 A large-scale single nucleotide polymorphism (SNP) association study conducted in the Japanese population found a long non-coding myocardial infarction association transcript (MIAT). They elucidated 6 SNPs in the MIAT that were statistically increased in patients of AMI.157 In 2017 Qui and colleagues performed in vivo experiment and described MIAT as pro-fibrotic lncRNA in AMI.158 Recent studies conducted in different in vitro and vivo model demonstrated that, the long nc RNA named super enhancer associated RNA called Wisper (Wisp2) has been widely expressed in cardiac fibroblast (CF).They also observed that Whisper was significantly overexpressed in mice with AMI.159 Subsequent reports have shown that lncRNAs play a significant role in apoptotic cell death in patients with AMI, viz lncRNAs cardiac apoptosis-related lncRNA (Carl), and mitochondrial dynamic related lncRNA (Mdrl) were down-regulated AMI.160,161
The lncRNA called long intergenic non-coding RNA predicting cardiac remodeling (LIPCAR) is significantly associated with the remodeling of left ventricular (LV) in patients of AMI, which leads to heart failure.162 They demonstrated that circulating LIPCAR levels were higher in AMI patients with repeated heart failure. Moreover, the LIPCAR was also found to be significantly associated with chronic heart failure patients with a severe coronary atherosclerosis condition. The plasma levels of LIPCAR was recognized as an important risk predictor for diastolic dysfunction in patients with type 2 diabetes.163 It is also reported that circulating lncRNA, LIPCAR, and H19 were significantly associated with CAD and increased risk of chronic heart failure.164,165 In 2017, another study showed that raised levels of circulating ANRIL was significantly associated with stent restenosis.164 Therefore, detecting lncRNA in extracellular body fluids can be used as a prognostic and diagnostic biomarker repository for the pathology of coronary atherosclerosis and other diseases.142,167
Circular RNA (circRNA)
Circular RNAs (circRNAs) are an addition to the advanced group of non-coding RNAs with an exciting finding in molecular research.168 These circRNA are produced by the event of back-splicing method with a ring structure lacking 5′ cap and 3′ tail, which makes them more stable against RNAse degradation compared to linear nc RNA such as mRNA, miRNA, lncRNA etc. In the year of 1970s, scientists have first discovered circRNA in RNA viruses by the help of electron microscope.169 Till the development of high throughput RNA sequencing circRNAs were considered as junk RNAs for last 2 decade.170
Circular RNAs have added the worth of non-coding RNAs as a biomarker.171 Though the research about circRNAs is in its infancy, it has a role in various diseases. Studies have shown that circRNAs are significantly associated with Coronary Artery Disease. They are found ubiquitously in the eukaryotic world, most predominantly in the cytoplasm. Circular RNAs are found in different kind of species, comprising fungi, archaea, plants, insects, fish, and mammals.170,172
Circular RNAs have been designated to have multiple roles such as regulator of transcription and translation, splicing participants, protein binding, miRNA sponges, etc.170 Also, some circRNAs have been reported in protein synthesis using the cap-independent translational mechanism.169 They exhibit a tissue-specific and developmental stage-specific pattern and serve as gene expression regulators in mammalian cells. Moreover, circRNAs have been reported to play diverse roles in the initiation and progression of neurological disorders, atherosclerotic disease risk, and a variety of cancers.173
The research reports that have implicated the potential role of circRNAs in the initiation, development, and progression of Coronary artery disease are shown in Table 5. Most importantly, circRNAs have been found to be indispensable participants in the occurrence and progression of CAD.
Table 5.
List of circRNA associated with coronary artery disease.
| Reference | Sample source | Sample size | Methods | Type of circRNA | Expression pattern |
|---|---|---|---|---|---|
| Zhao et al176 | Blood | Discovery (case n = 12, control n = 12) | Microarray Real-time q PCR | Hsa_circ0124644 | Upregulation |
| Validation (cases n = 115, control n = 137) | Hsa_circ0098964 | ||||
| Vausort et al177 | Blood | Cases (n = 642) | Real time q PCR | MIRCA | Down |
| Li et al178 | Blood | (cases CAD = 6, T2DM = 6 CAD + T2DM = 6 CONTROL = 20) | Microarray Real time q PCR | Hsa_circ11783 | Downregulation |
| Vilades et al179 | Plasma | (n = 200) | Real time q PCR | Hsa_circ000145 | Down |
Here we summarized the recent studies related to the discovery and effect of circRNAs in CAD pathogenesis (Table 5) and discussed some of research articles commenting on the feasibility of circRNAs to serve as biomarkers for cardiovascular diseases.
circRNAs as miRNA Sponge in Coronary Artery Disease
Studies on circRNAs have evidence that some of these RNA species exert their control in the expression of a specific gene through miRNA sponging effect. Due to the interaction with RNA binding proteins, circRNAs are involved in disease progression in CAD and are summarized in Table 6.
Table 6.
circRNA with targeted miRNA in CAD.
| Reference | CircRNA | Target miRNA | Function |
|---|---|---|---|
| Lee et al180 | CDR1as | miR-7a | Overexpression of Poly (ADP-ribose) polymerase (PARP) and specificity protein 1 (SP1) acts as a miRNA sponge leads to apoptosis |
| Wang et al181 | Mitochondrial Fission and apoptosis-related circRNA MFACR | miRNA-652-3p | Upregulation of apoptosis and mitochondrial fission |
| Wang et al182 | circ-081881 | miRNA-548 | Regulates PRPyI |
| Zhou and Yu183 | circRNA-010567 | miRNA-141 | Mediate fibrosis-associated protein resection |
Epigenetic Biomarkers of CAD
Epigenetic biomarker defines any epigenetic mechanism which exposes the information about pathology of disease, detection of disease, predicts the risk of the disease development in future, predicts the significance of the disease, monitors to medication and therapy.182 Many epigenetics biomarkers have been reported for CAD and its progression.183,184 Epigenetic modifications can present in both the form of covalent modifications in DNA/ Histone and pattern nc RNA expressions. Evidence suggests that candidate gene methylation patterns can serve as a potential epigenetic biomarker for diagnosis and prognosis of CAD (Table 2). The role of miRNAs in coronary artery disease has been widely studied over the years. Therefore, miRNAs are found to be major regulators in cellular gene expression for the initiation and progression of CAD. As discussed above altered levels of different circulating miRNAs could be used as diagnostic and prognostic markers of CAD (Table 4). Moreover, lncRNAs (sec b) and circRNAs have been reported to be potential biomarkers of CAD (Table 5).
Epigenetic Diversity Among CAD Patients
In contrast to the conventional principle, 1 individual’s genome does not differ from another. Scientists have established by their extensive research work that abundant differences do exist in nature for an individual’s epigenome. Thus, epigenetic modifications come from the diversity of nature comprising environmental exposure and pollutants, which lead to the development of deleterious diseases like CAD. Keeping this in consideration, scientists and physicians are trying to evolve novel therapeutic strategies based on these differences.185 Furnishing the Human Genome project is the keystone of the inherent blueprint and backbone of genomics.186,187 Within a decade of declaring the human genome project, researchers have reached to next-generation technology to understand the mechanism behind structural DNA to crucial activities in genome biology. With the advent of next-generation sequencing, scientists have been able to understand the role played by candidate genes and alterations in their nucleotide sequence towards the development of different diseases. Despite these approaches, the epigenetic world remains unexplored about precision medicine in CAD as of now. Worldwide researchers have already initiated efforts to comprehend epigenomics related to coronary artery disease.188-190
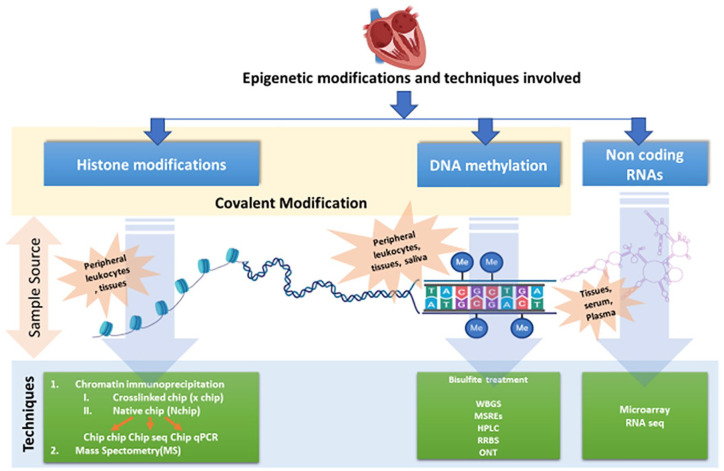
Nowadays, several epigenome-based techniques have come up to explore the epigenetics related to CAD (Figure 4).
Figure 4.
Epigenetic modifications and techniques involved with different kind of sample sources: WGBS, whole-genome bisulfite sequencing; MSRE, methylation-sensitive restriction enzymes; HPLC, high-performance liquid chromatography; RRBS, reduced representation bisulfite sequencing; ONT, Oxford nanopore technologies; Pyrosequencing—attractive alternative to the conventional bisulfite sequencing PCR (BSP); HRM assay; RT-PCR based high resolution melting curve analysis; ChIP, chromatin immunoprecipitation; MS, mass spectrometry; microarray; RNA seq, RNA sequencing.
Epigenetic Therapeutic Targets in CAD
The veracity of altered environmental exposure could be an epigenetic marker for upcoming treatment. According to researchers, these modifications can be inverted using pharmaceutical agents and nutraceuticals. For example, DNA methyltransferase inhibitors (DNMTis), histone acetyltransferase inhibitors, histone deacetylase inhibitors, histone methylase inhibitors, and bromodomain and extra terminal motif (BET) inhibitors have been extensively studied as conceivable therapeutics in inflammation and CAD.191 Although DNMTs are present in various food items naturally, many synthetic man-made ones are being used widely as well.192
Epigenetics Toward Personalized Medicine
Since the remote past, our health care professionals have been striving to offer the best care and effective treatment to their patients, as already discussed above. They have been doing this by experimenting with different treatment strategies, selecting the better successful ones based on their observational analysis, sharing their inferences with an expectation of improving the strategy of their predecessors. The ultimate goal of the efforts was to provide more accurate, precise, and effective treatment for each individual no matter how necessary the tools were at their disposal. However, nowadays, the advent of modern innovative techniques of genetic testing, data analysis, supercomputing devices, and electronic data storage has enabled physicians and scientists to take the area of precision medicine to a level which their historical predecessors could not even think of.
Precision medicine can take the management of a disease to its ultimate goal, i.e., complete cure of the disease. For example, Alexis and Noah, the famous twins who were misdiagnosed and mistreated, for about 14 years, for a condition called palsy. They received the treatment for palsy until human genome sequencing, and DNA sequencing made it possible to correctly diagnose their actual disease condition, that is, dystonia.193 The sequencing of the human genome took an international collaboration of 13 years and amounted to a financial costing of US$3 billion. However, the advancement in techniques involved, human genome sequencing, can be completed in 24 hours for just US$1000. This has made us capable of tailoring the prevention to an individual patient or group of patients using their genomic information in combination with their environment and lifestyles.
Although human genome sequencing has opened up the window for the interesting discipline of genetic medicine, which then leads the path to develop personalized medicine, it has become clear now, after 17 years, that our genetic material (genes) alone is not able to thoroughly explain the fundamental aspects of development and aging. Moreover, our genes are not enough to predict our susceptibility to different types of diseases. Epigenetics has already answered a lot of curiosity questions that bothered physicians and scientists for a large amount of time in the past. Epigenetics and factors affecting epigenetics mechanisms are attracting the attention of researchers and are being used to elucidate the underlying causes of the onset and progression of numerous life-threatening diseases. Innovative techniques that are harnessed to study epigenetics and epigenetic mechanisms are revolutionizing the approaches to personalized medicine (Figure 4). Innovative technologies to study epigenetics, such as bisulfite sequencing, have the ability to accurately quantify and map epigenetic marks underlying the causation of a particular disease phenotype, including CAD. This was not possible using traditionally used genetic research techniques. Epigenetic modifications determine the cell identity and make a skin cell different from a liver cell or heart cell. In many disease phenotypes like cardiac diseases, neurological diseases, or cancer, epigenetic changes are much more frequent than genetic variants, therefore, providing more vital insights into understanding the development and progression of these disease phenotypes.
Accurate mapping and quantitation of epigenetic markers help us in improving clinical practices and approaches by improving the diagnosis of diseases at the early stages of disease development, even prior to the happening of genetic changes.194 Research reports are demonstrating that epigenetic variations precede frequent genetic alterations in disease development.195 Therefore, detection of early disease signals by exploiting epigenetic based diagnostics offers opportunities for clinical intervention even before the appearance or progression of symptoms impact life quality when the patient is relatively fit. This favors our treatment process and success to treat the condition even before it appears. In this way, management of disease and prognosis is performed in more accurate manner.196-198
The concept of personalization of medicine in the context of coronary atherosclerosis could take the management of CAD to even more refinement than other diseases. Environmental factors are potential influencers of CADs. These environmental factors induce epigenetic changes, which in turn affect the genetic expression of an individual in the course of disease development. Therefore, taken together, the characterization of epigenetic aberrations in CAD precision medicine requires careful consideration.
Conclusion
This review summarized recent findings linked to epigenetic study in CAD and illustrated the impact of the epigenetic mechanism in CAD. Epigenetics has significantly influenced our understanding of gene regulation in CAD, although consensus regarding the magnitude of effect size is lacking. Epigenome intensely responds to variations in the environment during an individual’s lifetime contributions in the regulation of critical biological mechanisms and provides a significant association among life experiences, phenotypic expression, and risk of disease development. Since environmental inducements associated with epigenetics are inconsistent with the ethnicity of the population, results are difficult to generalize, and regional studies are imperative. Epigenetic aspects of in-utero exposure and after birth exposure, like dietary factors, have been inadequately explored. Future strategy for further research must incorporate elucidation of diagnostic biomarker potential and therapeutic targets for atherosclerotic disorders in order to translate emerging epigenetic data in bedside care. Moreover, with the help of epigenetics and techniques to study epigenetic factors, the present-day physician and researchers could take personalized medicine to a level unreachable to their ancestors.
Acknowledgments
Facilities provided by Govind Ballabh Pant Postgraduate Institute of Medical Education and Research (GIPMER), New Delhi, are acknowledged. Authors are grateful to the Department of Biotechnology Government of India for financial support provided under the head of “Establishment of Central Molecular Lab in GIPMER to study the diagnosis for precision medicine in order to understand the disease process and utilize it as Clinical Research Facility” with Grant No. BT/INF/22/SP33063/2019.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work is funded by the Department of Biotechnology (DBT), Govt. of India, India (BT/INF/22/SP33063/2019).
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: MPS and BM were involved in planning, designing the manuscript. MPS was involved in the writing of the manuscript. RSAS, N, A, AK, AKS, EA, and AA were involved in reviewing the manuscript. Finally, the manuscript was approved by BM and SSS.
List of Abbreviation: CAD: Coronary Artery Disease
AMI: Acute Myocardial Infarction
DM: Diabetes Mellitus
VSMCs: Vascular Smooth Muscle Cells
ECs: Endothelial Cells
SHS: Second Hand Smoke
DASH: Dietary Approaches to Stop Hypertension
DNMTs: DNA methyltransferases
IGF2: Insulin Like Growth Factor-2
IR: Insulin Receptor
ABCG1: ATP Binding Cassette Subfamily G Member
CPTIA: Carnitine palmitoyl transferase 1A
HPLC: High Performance Liquid Chromatography
MSRE: Methylation-Sensitive Restriction Enzymes
LDL: Low Density Lipoprotein
MS PCR: Methylation-specific Polymerase Chain Reaction
MS-HRM: Methylation Specific High Resolution Melting Curve
EWAS: Epigenome-Wide Association Studies
KMT: Lysine Methyltransferase
KDM: Lysine De-Methylase
SAM: S-adenosylmethionine
LSDs: Lysine-specific de-methylases
EZH2: Equivalent Histone Methyltransferase
KATs: Lysine Acetyltransferase initially
HATs: Histone Acetyltransferase
HDACs: Histone Deacetylases
ex-LDL: Oxidized Low-Density Lipoprotein
CAMII δ: Calcium/ calmodulin-Dependent Protein Kinase II δ
nc RNAs: non-coding RNAs
miRNAs: micro RNAs
lncRNA: Long non-coding RNA
ECF: Extracellular Fluids
SNP: Single Nucleotide Polymorphism
MIAT: Myocardial Infarction Association Transcript
LIPCAR: Long Intergenic non-coding RNA Predicting Cardiac Remodelling
ANRIL:
circRNA: circular RNA
WGBS: Whole Genome Bisulphite Sequencing,
RRBS: Reduced Representation Bisulphite Sequencing,
ONT: Oxford Nanopore Technologies
ChIP: Chromatin immunoprecipitation,
MS: Mass Spectrometry
RNA seq: RNA sequencing
DNMTis: DNA methyltransferase inhibitors
References
- 1. Liu C-F, Tang WHW. Epigenetics in cardiac hypertrophy and heart failure. JACC Basic Transl Sci. 2019;4:976–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roth GA, Johnson C, Abajobir A, et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol. 2017;70:1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Majnik AV, Lane RH. Epigenetics: where environment, society and genetics meet. Epigenomics. 2014;6:1-4. [DOI] [PubMed] [Google Scholar]
- 4. Grimaldi V, Vietri MT, Schiano C, et al. Epigenetic reprogramming in atherosclerosis. Curr Atheroscler Rep. 2015;17:476. [DOI] [PubMed] [Google Scholar]
- 5. Ross R. The pathogenesis of atherosclerosis—an update. N Engl J Med. 1986;314:488-500. [DOI] [PubMed] [Google Scholar]
- 6. Ross R. Atherosclerosis–an inflammatory disease. N Engl J Med. 1999;340:115-126. [DOI] [PubMed] [Google Scholar]
- 7. Sayols-Baixeras S, Lluís-Ganella C, Lucas G, Elosua R. Pathogenesis of coronary artery disease: focus on genetic risk factors and identification of genetic variants. Appl Clin Genet. 2014;7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanchis-Gomar F, Perez-Quilis C, Leischik R, Lucia A. Epidemiology of coronary heart disease and acute coronary syndrome. Ann Transl Med. 2016;4:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Varma PK, Kundan S, Ananthanarayanan C, et al. Demographic profile, clinical characteristics and outcomes of patients undergoing coronary artery bypass grafting—retrospective analysis of 4,024 patients. Indian J Thorac Cardiovasc Surg. 2014;30:272-277. [Google Scholar]
- 10. Dhingra R, Vasan RS. Age as a risk factor. Med Clin North Am. 2012;96:87-91. doi: 10.1016/j.mcna.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rossi R, Grimaldi T, Origliani G, Fantini G, Coppi F, Modena MG. Menopause and cardiovascular risk. Pathophysiol Haemost Thromb. 2002;32:325-328. [DOI] [PubMed] [Google Scholar]
- 12. Maas AH, Appelman YE. Gender differences in coronary heart disease. Neth Heart J. 2010;18:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grady D, Chaput L, Kristof M. Results of Systematic Review of Research on Diagnosis and Treatment of Coronary Heart Disease in Women: Summary. InAHRQ Evidence Report Summaries; 2003. Agency for Healthcare Research and Quality (US). [PMC free article] [PubMed] [Google Scholar]
- 14. Mosca L, Barrett-Connor E, Wenger NK. Sex/gender differences in cardiovascular disease prevention: what a difference a decade makes. Circulation. 2011;124:2145-2154. doi: 10.1161/CIRCULATIONAHA.110.968792ss [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smail HO. The epigenetics of diabetes, obesity, overweight and cardiovascular disease. AIMS Genetics. 2019;6:36-45. doi: 10.3934/genet.2019.3.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gallardo-Escribano C, Buonaiuto V, Ruiz-Moreno MI, et al. Epigenetic approach in obesity: DNA methylation in a prepubertal population which underwent a lifestyle modification. Clin Epigenetics. 2020;12:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Siemelink MA, van der Laan SW, Haitjema S, et al. Smoking is associated to DNA methylation in atherosclerotic carotid lesions. Circ Genom Precis Med. 2018;11:e002030. [DOI] [PubMed] [Google Scholar]
- 18. Steenaard RV, Ligthart S, Stolk L, et al. Tobacco smoking is associated with methylation of genes related to coronary artery disease. Clin Epigenetics. 2015;7:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Athyros VG, Katsiki N, Doumas M, Karagiannis A, Mikhailidis DP. Effect of tobacco smoking and smoking cessation on plasma lipoproteins and associated major cardiovascular risk factors: a narrative review. Curr Med Res Opin. 2013;29:1263-1274. [DOI] [PubMed] [Google Scholar]
- 20. Lohoff FW, Sorcher JL, Rosen AD, et al. Methylomic profiling and replication implicates deregulation of PCSK9 in alcohol use disorder. Mol Psychiatry. 2018;23:1900-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mendler L, Braun T, Müller S. The ubiquitin-like SUMO system and heart function: from development to disease. Circ Res. 2016;118:132-144. [DOI] [PubMed] [Google Scholar]
- 22. Barker DJ. Early growth and cardiovascular disease. Arch Dis Child.1999;80:305-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. Weight in infancy and death from ischaemic heart disease. Lancet. 1989;2:577-580. [DOI] [PubMed] [Google Scholar]
- 24. Agarwal P, Morriseau TS, Kereliuk SM, Doucette CA, Wicklow BA, Dolinsky VW. Maternal obesity, diabetes during pregnancy and epigenetic mechanisms that influence the developmental origins of cardiometabolic disease in the offspring. Crit Rev Clin Lab Sci. 2018;55:71-101. [DOI] [PubMed] [Google Scholar]
- 25. Ordovás J, Smith C. Epigenetics and cardiovascular disease. Nat Rev Cardiol. 2010;7:510-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bhatnagar A. Environmental cardiology: studying mechanistic links between pollution and heart disease. Circ Res. 2006;29;99:692-705. [DOI] [PubMed] [Google Scholar]
- 27. Joehanes R, Just AC, Marioni RE, et al. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet. 2016;9:436-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mozaffarian D. Dietary and policy priorities for cardiovascular disease, diabetes, and obesity: a comprehensive review. Circulation. 2016;133:187-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y, Kutateladze TG. Diet and the epigenome. Nat Commun. 2018;9:3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alegría-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. 2011;3:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Phillips NLH, Roth TL. Animal models and their contribution to our understanding of the relationship between environments, epigenetic modifications, and behavior. Genes (Basel). 2019;10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horii T, Morita S, Hino S, et al. Successful generation of epigenetic disease model mice by targeted demethylation of the epigenome. Genome Biol. 2020;21:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bennett AJ. Gene environment interplay: nonhuman primate models in the study of resilience and vulnerability. Dev Psychobiol. 2008;50:48-59. [DOI] [PubMed] [Google Scholar]
- 34. Rosenfeld CS. Animal models to study environmental epigenetics. Biol Reprod. 2010;82:473-488. [DOI] [PubMed] [Google Scholar]
- 35. Lloyd KC, Robinson PN, MacRae CA. Animal-based studies will be essential for precision medicine. Sci Transl Med. 2016;8:352ed12. [DOI] [PubMed] [Google Scholar]
- 36. Duan L, Liu Y, Wang J, Liao J, Hu J. The dynamic changes of DNA methylation in primordial germ cell differentiation. Gene. 2016;591:305-312. [DOI] [PubMed] [Google Scholar]
- 37. Smith ZD, Chan MM, Mikkelsen TS, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wong KY, Yu L, Chim CS. DNA methylation of tumor suppressor miRNA genes: a lesson from the miR-34 family. Epigenomics. 2011;3:83-92. [DOI] [PubMed] [Google Scholar]
- 39. Baccarelli A, Ghosh S. Environmental exposures, epigenetics and cardiovascular disease. Curr Opin Clin Nutr Metab Care. 2012;15:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457-466. [DOI] [PubMed] [Google Scholar]
- 41. Chaudhary N, Nakka KK, Maulik N, Chattopadhyay S. Epigenetic manifestation of metabolic syndrome and dietary management. Antioxid Redox Signal. 2012;17:254-281. [DOI] [PubMed] [Google Scholar]
- 42. Lewitt MS, Dent MS, Hall K. The insulin-like growth factor system in obesity, insulin resistance and type 2 diabetes mellitus. Journal of clinical medicine. 2014,3:1561-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hardy LM, Frisdal E, Le Goff W. Critical role of the human ATP-binding cassette G1 transporter in cardiometabolic diseases. Int J Mol Sci. 2017;18:1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frazier-Wood AC, Aslibekyan S, Absher DM, et al. Methylation at CPT1A locus is associated with lipoprotein subfraction profiles. J Lipid Res. 2014;55:1324-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hai Z, Zuo W. Aberrant DNA methylation in the pathogenesis of atherosclerosis. Clinica Chim Acta. 2016;456:69-74. [DOI] [PubMed] [Google Scholar]
- 46. Zhong J, Agha G, Baccarelli AA. The role of DNA methylation in cardiovascular risk and disease. Circ Res. 2016;118:119-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kalhan SC. One carbon metabolism in pregnancy: impact on maternal, fetal and neonatal health. Mol Cell Endocrinol. 2016;5;435:48-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Erdman JW, Jr, Balentine D, Arab L, et al. Flavonoids and Heart Health: proceedings of the ILSI North America flavonoids workshop, May 31–June 1, 2005, Washington, DC. J Nutr 2007;137(suppl 1):718S-737S. [DOI] [PubMed] [Google Scholar]
- 49. Sabogal C, Su S, Tingen M, Kapuku G, Wang X. Cigarette smoking related DNA methylation in peripheral leukocytes and cardiovascular risk in young adults. Int J Cardiol. 2020;306:203-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ Cardiovasc Genet. 2010;3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Turunen MP, Aavik E, Yla-Herttuala S. Epigenetics and atherosclerosis. Biochim Biophys Acta. 2009;1790:886-891. [DOI] [PubMed] [Google Scholar]
- 52. Makar KW, Wilson CB. DNA methylation is a nonredundant repressor of the Th2 effector program. J Immunol. 2004;173:4402-4406. [DOI] [PubMed] [Google Scholar]
- 53. Chen Z, Karaplis AC, Ackerman SL, et al. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum Mol Genet. 2001;10:433-443. [DOI] [PubMed] [Google Scholar]
- 54. Lund G, Andersson L, Lauria M, et al. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004;279:29147-29154. [DOI] [PubMed] [Google Scholar]
- 55. Baccarelli A, Wright R, Bollati V, et al. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21:819-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim M, Long TI, Arakawa K, Wang R, Mimi CY, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5:e9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zaina S, Heyn H, Carmona FJ, et al. DNA methylation map of human atherosclerosis. Circ Cardiovasc Genet. 2014;7:692-700. [DOI] [PubMed] [Google Scholar]
- 58. Nazarenko MS, Markov AV, Sleptcov AA, et al. Genome-wide profiling of DNA methylation in human atherosclerotic plaques. Atherosclerosis. 2015;241:e84-e85. [Google Scholar]
- 59. del Pilar Valencia-Morales M, Zaina S, Heyn H, et al. The DNA methylation drift of the atherosclerotic aorta increases with lesion progression. BMC Med Genomics. 2015;8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hiltunen MO, Turunen MP, Häkkinen TP, et al. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc Med. 2002;7:5-11. [DOI] [PubMed] [Google Scholar]
- 61. Sharma P, Garg G, Kumar A, et al. Genome wide DNA methylation profiling for epigenetic alteration in coronary artery disease patients. Gene. 2014;541:31-40. [DOI] [PubMed] [Google Scholar]
- 62. Sharma P, Kumar J, Garg G, et al. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. 2008;27:357-365. [DOI] [PubMed] [Google Scholar]
- 63. Zhu S, Goldschmidt-Clermont PJ, Dong C. Inactivation of monocarboxylate transporter MCT3 by DNA methylation in atherosclerosis. Circulation. 2005;112:1353-1361. [DOI] [PubMed] [Google Scholar]
- 64. Huang Y-S, Zhi Y-F, Wang S-R. Hypermethylation of estrogen receptor-alpha gene in atheromatosis patients and its correlation with homocysteine. Pathophysiology. 2009;16:259-265. [DOI] [PubMed] [Google Scholar]
- 65. Kim J, Kim JY, Song KS, et al. Epigenetic changes in estrogen receptor β gene in atherosclerotic cardiovascular tissues and in-vitro vascular senescence. Biochim Biophys Acta. 2007;1772:72-80. [DOI] [PubMed] [Google Scholar]
- 66. Zuo H-P, Guo Y-Y, Che L, Wu X-Z. Hypomethylation of interleukin-6 promoter is associated with the risk of coronary heart disease. Arq Bras Cardiol. 2016;107:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bakshi C, Vijayvergiya R, Dhawan V. Aberrant DNA methylation of M1-macrophage genes in coronary artery disease. Sci Rep. 2019;9:1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ghose S, Ghosh S, Tanwar VS, et al. Investigating coronary artery disease methylome through targeted bisulfite sequencing. Gene. 2019;721:144107. [DOI] [PubMed] [Google Scholar]
- 69. Nakatochi M, Ichihara S, Yamamoto K, et al. Epigenome-wide association of myocardial infarction with DNA methylation sites at loci related to cardiovascular disease. Clin Epigenetics. 2017;9:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rask-Andersen M, Martinsson D, Ahsan M, et al. Epigenome-wide association study reveals differential DNA methylation in individuals with a history of myocardial infarction. Hum Mol Genet. 2016;25:4739-4748. [DOI] [PubMed] [Google Scholar]
- 71. Fernández-Sanlés A, Sayols-Baixeras S, Subirana I, Degano IR, Elosua R. Association between DNA methylation and coronary heart disease or other atherosclerotic events: a systematic review. Atherosclerosis. 2017;263:325-333. [DOI] [PubMed] [Google Scholar]
- 72. Michels KB, Binder AM, Dedeurwaerder S, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10:949-955. [DOI] [PubMed] [Google Scholar]
- 73. Castillo-Díaz SA, Garay-Sevilla ME, Hernández-González MA, Solís-Martínez MO, Zaina S. Extensive demethylation of normally hypermethylated CpG islands occurs in human atherosclerotic arteries. Int J Mol Med. 2010;26:691-700. [DOI] [PubMed] [Google Scholar]
- 74. Yamada Y, Horibe H, Oguri M, et al. Identification of novel hyper- or hypomethylated CpG sites and genes associated with atherosclerotic plaque using an epigenome-wide association study. Int J Mol Med. 2018;41:2724-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shen Y, Peng C, Bai Q, et al. Epigenome-Wide Association Study Indicates Hypomethylation of MTRNR2L8 in Large-Artery Atherosclerosis Stroke. Stroke. 2019;50:1330-1338. [DOI] [PubMed] [Google Scholar]
- 76. Banerjee S, Ponde CK, Rajani RM, Ashavaid TF. Differential methylation pattern in patients with coronary artery disease: pilot study. Mol Biol Rep. 2019;46:541-550. [DOI] [PubMed] [Google Scholar]
- 77. Draizen EJ, Shaytan AK, Mariño-Ramírez L, Talbert PB, Landsman D, Panchenko AR. HistoneDB 2.0: a histone database with variants–an integrated resource to explore histones and their variants. Database (Oxford). 2016;2016:baw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407-412. [DOI] [PubMed] [Google Scholar]
- 79. Natsume-Kitatani Y, Shiga M, Mamitsuka H. Genome-wide integration on transcription factors, histone acetylation and gene expression reveals genes co-regulated by histone modification patterns. PLoS One. 2011;6:e22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074. [DOI] [PubMed] [Google Scholar]
- 81. Wei X, Yi X, Zhu X-H, Jiang D-S. Histone methylation and vascular biology. Clin Epigenetics. 2020;12:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta. 2014;1839:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rea S, Eisenhaber F, O’Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593-599. [DOI] [PubMed] [Google Scholar]
- 84. Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116(Pt 11):2117-2124. [DOI] [PubMed] [Google Scholar]
- 85. Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941-953. [DOI] [PubMed] [Google Scholar]
- 86. Shi YG, Tsukada Y. The discovery of histone demethylases. Cold Spring Harb Perspect Biol. 2013;5:a017947 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wolf SS. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell Mol Life Sci. 2009;66:2109-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432-435. [DOI] [PubMed] [Google Scholar]
- 89. Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811-816. [DOI] [PubMed] [Google Scholar]
- 90. Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444-447. [DOI] [PubMed] [Google Scholar]
- 91. Greißel A, Culmes M, Napieralski R, et al. Alternation of histone and DNA methylation in human atherosclerotic carotid plaques. Thromb Haemost. 2015;114:390-402. [DOI] [PubMed] [Google Scholar]
- 92. Greißel A, Culmes M, Burgkart R, et al. Histone acetylation and methylation significantly change with severity of atherosclerosis in human carotid plaques. Cardiovasc Pathol. 2016;25:79-86. [DOI] [PubMed] [Google Scholar]
- 93. Phillips DM. The presence of acetyl groups of histones. Biochem J. 1963;87:258-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Allfrey VG, Pogo BG, Littau VC, Gershey EL, Mirsky AE. Histone acetylation in insect chromosomes. Science. 1968;159:314-316. [DOI] [PubMed] [Google Scholar]
- 95. Yang X-J, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417-435. [DOI] [PubMed] [Google Scholar]
- 97. Cheung WL, Briggs SD, Allis CD. Acetylation and chromosomal functions. Curr Opin Cell Biol. 2000;12:326-333. [DOI] [PubMed] [Google Scholar]
- 98. Yang X-J, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310-5318. [DOI] [PubMed] [Google Scholar]
- 99. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pandey D, Sikka G, Bergman Y, et al. Transcriptional regulation of endothelial arginase 2 by histone deacetylase 2. Arterioscler Thromb Vasc Biol. 2014;34:1556-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Oki M, Aihara H, Ito T. Role of histone phosphorylation in chromatin dynamics and its implications in diseases. Subcell Biochem. 2007;41:319-336. [PubMed] [Google Scholar]
- 102. Backs J, Backs T, Neef S, et al. The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc Natl Acad Sci U S A. 2009;106:2342-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Awad S, Al-Haffar KMA, Marashly Q, et al. Control of histone H3 phosphorylation by CaMKIIδ in response to haemodynamic cardiac stress. J Pathol. 2015;235:606-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Awad S, Kunhi M, Little GH, et al. Nuclear CaMKII enhances histone H3 phosphorylation and remodels chromatin during cardiac hypertrophy. Nucleic Acids Res. 2013;41:7656-7672. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105. Song L, Luo Z-Q. Post-translational regulation of ubiquitin signaling. J Cell Biol. 2019;218:1776-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang J, Schwartz RJ. Sumoylation and regulation of cardiac gene expression. Circ Res. 2010;107:19-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Han Z-J, Feng Y-H, Gu B-H, Li Y-M, Chen H. The post-translational modification, SUMOylation, and cancer (Review). Int J Oncol. 2018;52:1081-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kang X, Qi Y, Zuo Y, et al. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell. 2010;38:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Li X-C, Zeng Y, Sun R-R, Liu M, Chen S, Zhang P-Y. SUMOylation in cardiac disorders - a review. Eur Rev Med Pharmacol Sci. 2017;21:1583-1587. [PubMed] [Google Scholar]
- 110. Yang Y, He Y, Wang X, et al. Protein SUMOylation modification and its associations with disease. Open Biol. 2017;7:170167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636-640. [DOI] [PubMed] [Google Scholar]
- 112. Bertone P, Stolc V, Royce TE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242-2246. [DOI] [PubMed] [Google Scholar]
- 113. Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kung JTY, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Thomas CA. The genetic organization of chromosomes. Annu Rev Genet. 1971;5:237-256. [DOI] [PubMed] [Google Scholar]
- 116. Mirsky AE, Ris H. The desoxyribonucleic acid content of animal cells and its evolutionary significance. J Gen Physiol. 1951;34:451-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Poller W, Dimmeler S, Heymans S, et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J. 2018;39:2704-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Boon RA, Jaé N, Holdt L, Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol. 2016;67:1214-1226. [DOI] [PubMed] [Google Scholar]
- 119. Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zhang Y, Liu T, Meyer CA, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Abi Khalil C. The emerging role of epigenetics in cardiovascular disease. Ther Adv Chronic Dis. 2014;5:178-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Fichtlscherer S, De Rosa S, Fox H, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677-684. [DOI] [PubMed] [Google Scholar]
- 123. Vindis C, Faccini J, Ruidavets JB, et al. Circulating miR-155, miR-145 and let-7c as diagnostic biomarkers of the coronary artery disease. Atherosclerosis. 2017;263:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wang F, Long G, Zhao C, et al. Plasma microRNA-133a is a new marker for both acute myocardial infarction and underlying coronary artery stenosis. J Transl Med. 2013;11:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Liu H, Yang N, Fei Z, et al. Analysis of plasma miR-208a and miR-370 expression levels for early diagnosis of coronary artery disease. Biomed Rep. 2016;5:332-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zhang Y, Li H-H, Yang R, Yang B-J, Gao Z-Y. Association between circulating microRNA-208a and severity of coronary heart disease. Scand J Clin Lab Invest. 2017;77:379-384. [DOI] [PubMed] [Google Scholar]
- 127. Wang J, Yan C-H, Li Y, et al. MicroRNA-31 controls phenotypic modulation of human vascular smooth muscle cells by regulating its target gene cellular repressor of E1A-stimulated genes. Exp Cell Res. 2013;319:1165-1175. [DOI] [PubMed] [Google Scholar]
- 128. Sayed ASM, Xia K, Yang T-L, Peng J. Circulating microRNAs: a potential role in diagnosis and prognosis of acute myocardial infarction. Dis Markers. 2013;35:561-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Sondermeijer BM, Bakker A, Halliani A, et al. Platelets in patients with premature coronary artery disease exhibit upregulation of miRNA340* and miRNA624. PLoS One. 2011;6:e25946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Corsten MF, Dennert R, Jochems S, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499-506. [DOI] [PubMed] [Google Scholar]
- 131. Xin Y, Yang C, Han Z. Circulating miR-499 as a potential biomarker for acute myocardial infarction. Ann Transl Med. 2016;4:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Adachi T, Nakanishi M, Otsuka Y, et al. Plasma MicroRNA 499 as a biomarker of acute myocardial infarction. Clin Chem. 2010;56:1183-1185. [DOI] [PubMed] [Google Scholar]
- 133. Olivieri F, Antonicelli R, Lorenzi M, et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int J Cardiol. 2013;167:531-536. [DOI] [PubMed] [Google Scholar]
- 134. Hoekstra M, van der Lans CA, Halvorsen B, et al. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem Biophys Res Commun. 2010;394:792-797. [DOI] [PubMed] [Google Scholar]
- 135. Cheng Y, Tan N, Yang J, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci. 2010;119:87-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ai J, Zhang R, Li Y, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391:73-77. [DOI] [PubMed] [Google Scholar]
- 137. D’Alessandra Y, Devanna P, Limana F, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Widera C, Gupta SK, Lorenzen JM, et al. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol. 2011;51:872-875. [DOI] [PubMed] [Google Scholar]
- 139. Okazaki Y, Furuno M, Kasukawa T, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563-573. [DOI] [PubMed] [Google Scholar]
- 140. Zhou X, Zhang W, Jin M, Chen J, Xu W, Kong X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017;8:e2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Martignano F, Rossi L, Maugeri A, et al. Urinary RNA-based biomarkers for prostate cancer detection. Clinica Chimica Acta. 2017;473:96-105. [DOI] [PubMed] [Google Scholar]
- 142. Terracciano D, Ferro M, Terreri S, et al. Urinary long noncoding RNAs in nonmuscle-invasive bladder cancer: new architects in cancer prognostic biomarkers. Transl Res. 2017;184:108-117. [DOI] [PubMed] [Google Scholar]
- 143. Li Q, Shao Y, Zhang X, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumor Biol. 2015;36:2007-2012. [DOI] [PubMed] [Google Scholar]
- 144. Viereck J, Thum T. Circulating Noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res. 2017;120:381-399. [DOI] [PubMed] [Google Scholar]
- 145. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62. [DOI] [PubMed] [Google Scholar]
- 146. Laham-Karam N, Laitinen P, Turunen TA, Ylä-Herttuala S. Activating the chromatin by noncoding RNAs. Antioxid Redox Signal. 2018;29:813-831. [DOI] [PubMed] [Google Scholar]
- 147. Butler AA, Webb WM, Lubin FD. Regulatory RNAs and control of epigenetic mechanisms: expectations for cognition and cognitive dysfunction. Epigenomics. 2016;8:135-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Haemmig S, Simion V, Yang D, Deng Y, Feinberg MW. Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr Opin Cardiol. 2017;32:776-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Mumtaz PT, Bhat SA, Ahmad SM, et al. LncRNAs and immunity: watchdogs for host pathogen interactions. Biol Proced Online. 2017;19:1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Su M, Wang H, Wang W, et al. LncRNAs in DNA damage response and repair in cancer cells. Acta Biochim Biophys Sin (Shanghai). 2018;50:433-439. [DOI] [PubMed] [Google Scholar]
- 151. Grote P, Wittler L, Hendrix D, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lin Z, Ge J, Wang Z, et al. Let-7e modulates the inflammatory response in vascular endothelial cells through ceRNA crosstalk. Sci Rep. 2017;7:42498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Viereck J, Kumarswamy R, Foinquinos A, et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med. 2016;8:326ra22. [DOI] [PubMed] [Google Scholar]
- 154. Ono K, Kuwabara Y, Horie T, Kimura T. Long non-coding RNAs as key regulators of cardiovascular diseases. Circ J. 2018;82:1231-1236. [DOI] [PubMed] [Google Scholar]
- 155. Arslan S, Berkan Ö, Lalem T, et al. Long non-coding RNAs in the atherosclerotic plaque. Atherosclerosis. 2017;266:176-181. [DOI] [PubMed] [Google Scholar]
- 156. Li H, Zhu H, Ge J. Long noncoding RNA: recent updates in atherosclerosis. Int J Biol Sci. 2016;12:898-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Ishii N, Ozaki K, Sato H, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 2006;51:1087-1099. [DOI] [PubMed] [Google Scholar]
- 158. Qu X, Du Y, Shu Y, et al. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci Rep. 2017;7:42657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Micheletti R, Plaisance I, Abraham BJ, et al. The long noncoding RNA <em>Wisper</em> controls cardiac fibrosis and remodeling. Sci Transl Med. 2017;9:eaai9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wang K, Long B, Zhou LY, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:1-3. [DOI] [PubMed] [Google Scholar]
- 161. Wang Z, Zhang XJ, Ji YX, et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22:1131-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Kumarswamy R, Bauters C, Volkmann I, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114:1569-1575. [DOI] [PubMed] [Google Scholar]
- 163. de Gonzalo-Calvo D, Kenneweg F, Bang C, et al. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep. 2016;6:37354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Hobuß L, Bär C, Thum T. Long non-coding RNAs: at the heart of cardiac dysfunction? Front Physiol. 2019;10:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Xuan L, Sun L, Zhang Y, et al. Circulating long non-coding RNA s NRON and MHRT as novel predictive biomarkers of heart failure. J Cell Mol Med. 2017;21:1803-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wang H, Liu Y, Zhong J, et al. Long noncoding RNA ANRIL as a novel biomarker of lymph node metastasis and prognosis in human cancer: a meta-analysis. Oncotarget. 2017;9:14608-14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Gutschner T, Richtig G, Haemmerle M, Pichler M. From biomarkers to therapeutic targets—the promises and perils of long non-coding RNAs in cancer. Cancer Metastasis Rev.2018;37:83-105. [DOI] [PubMed] [Google Scholar]
- 168. Haddad G, Lorenzen JM. Biogenesis and function of circular RNAs in health and in disease. Front Pharmacol. 2019;10:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [DOI] [PubMed] [Google Scholar]
- 171. Bayoumi AS, Aonuma T, Teoh J, Tang Y, Kim I. Circular noncoding RNAs as potential therapies and circulating biomarkers for cardiovascular diseases. Acta Pharmacol Sin. 2018;39:1100-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Wang PL, Bao Y, Yee M-C, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16:292-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Zhao Z, Li X, Gao C, et al. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7:39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Vausort M, Salgado-Somoza A, Zhang L, et al. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol. 2016;68:1247-1248. [DOI] [PubMed] [Google Scholar]
- 176. Li X, Zhao Z, Jian D, Li W, Tang H, Li M. Hsa-circRNA11783-2 in peripheral blood is correlated with coronary artery disease and type 2 diabetes mellitus. Diab Vasc Dis Res. 2017;14:510-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Vilades D, Martínez-Camblor P, Ferrero-Gregori A, et al. Plasma circular RNA hsa_circ_0001445 and coronary artery disease: performance as a biomarker. FASEB J. 2020;34:4403-4414. [DOI] [PubMed] [Google Scholar]
- 178. Lee ECS, Elhassan SAM, Lim GPL, et al. The roles of circular RNAs in human development and diseases. Biomed Pharmacother. 2019;111:198-208. [DOI] [PubMed] [Google Scholar]
- 179. Wang K, Gan T-Y, Li N, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Wang W, Wang Y, Piao H, et al. Circular RNAs as potential biomarkers and therapeutics for cardiovascular disease. PeerJ. 2019;7:e6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun. 2017;487:769-775. [DOI] [PubMed] [Google Scholar]
- 182. García-Giménez JL, Seco-Cervera M, Tollefsbol TO, et al. Epigenetic biomarkers: current strategies and future challenges for their use in the clinical laboratory. Crit Rev Clin Lab Sci. 2017;54:529-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Cardona-Monzonís A, Beltrán-García J, Ibañez-Cabellos JS, et al. Epigenetic biomarkers in cardiovascular disease. J Lab Precis Med. 2018;3:24. [Google Scholar]
- 184. Metzinger L, de Franciscis S, Serra R. The management of cardiovascular risk through epigenetic biomarkers. BioMed Res Int. 2017;2017:9158572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Fox CS, Hall JL, Arnett DK, et al. Future translational applications from the contemporary genomics era: a scientific statement from the American Heart Association. Circulation. 2015;131:1715-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860-921. [DOI] [PubMed] [Google Scholar]
- 187. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931-945. [DOI] [PubMed] [Google Scholar]
- 188. Green ED, Guyer MS. National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204-213. [DOI] [PubMed] [Google Scholar]
- 189. Altman RB, Ashley EA. Using “big data” to dissect clinical heterogeneity. Circulation. 2015;131:232-233. [DOI] [PubMed] [Google Scholar]
- 190. Leopold JA, Loscalzo J. Emerging role of precision medicine in cardiovascular disease. Circ Res. 2018;122:1302-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Nicorescu I, Dallinga GM, de Winther MPJ, Stroes ESG, Bahjat M. Potential epigenetic therapeutics for atherosclerosis treatment. Atherosclerosis. 2019;281:189-197. [DOI] [PubMed] [Google Scholar]
- 192. Chistiakov DA, Orekhov AN, Bobryshev YV. Treatment of cardiovascular pathology with epigenetically active agents: focus on natural and synthetic inhibitors of DNA methylation and histone deacetylation. Int J Cardiol. 2017;227:66-82. [DOI] [PubMed] [Google Scholar]
- 193. Yang YB. The role of genetics in medicine: a future of precision medicine. Br Col Med J. 2019;61:388-399. [Google Scholar]
- 194. Raiber E-A, Beraldi D, Martínez Cuesta S, et al. Base resolution maps reveal the importance of 5-hydroxymethylcytosine in a human glioblastoma. NPJ Genomic Med. 2017;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Vaz M, Hwang SY, Kagiampakis I, et al. Chronic cigarette smoke-induced epigenomic changes precede sensitization of bronchial epithelial cells to single-step transformation by KRAS mutations. Cancer Cell. 2017;32:360-376.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Van Neste L, Groskopf J, Grizzle WE, et al. Epigenetic risk score improves prostate cancer risk assessment. Prostate. 2017;77:1259-1264. [DOI] [PubMed] [Google Scholar]
- 197. Joosten SC, Deckers IA, Aarts MJ, et al. Prognostic DNA methylation markers for renal cell carcinoma: a systematic review. Epigenomics. 2017;9:1243-1257. [DOI] [PubMed] [Google Scholar]
- 198. Koschmieder S, Vetrie D. Epigenetic dysregulation in chronic myeloid leukaemia: A myriad of mechanisms and therapeutic options. Semin Cancer Biol. 2017;51:180-197. [DOI] [PubMed] [Google Scholar]