Dendritic cell (DC) actin dynamics at the immunological synapse with T cells is partially regulated by the WAVE regulatory complex. Absence of DC WAVE leads to reduced actin dynamics, prolonged cell–cell contacts, and impaired T cell priming.
Abstract
Dendritic cells (DCs) are crucial for the priming of naive T cells and the initiation of adaptive immunity. Priming is initiated at a heterologous cell–cell contact, the immunological synapse (IS). While it is established that F-actin dynamics regulates signaling at the T cell side of the contact, little is known about the cytoskeletal contribution on the DC side. Here, we show that the DC actin cytoskeleton is decisive for the formation of a multifocal synaptic structure, which correlates with T cell priming efficiency. DC actin at the IS appears in transient foci that are dynamized by the WAVE regulatory complex (WRC). The absence of the WRC in DCs leads to stabilized contacts with T cells, caused by an increase in ICAM1-integrin–mediated cell–cell adhesion. This results in lower numbers of activated and proliferating T cells, demonstrating an important role for DC actin in the regulation of immune synapse functionality.
Introduction
Dendritic cells (DCs) are the most potent antigen-presenting cells (APCs), with the unique ability to prime naive T cells in vivo (Banchereau and Steinman, 1998). T cell activation requires the formation of a transient cell–cell contact between T cell and APC. Within this immunological synapse (IS; Monks et al., 1998; Dustin et al., 1998), the T cell receptor (TCR), together with co-stimulatory molecules and adhesion receptors, engage their ligands on the APC surface to trigger downstream signaling, resulting in T cell activation and proliferation. The T cell face of the IS has been extensively studied in experimental setups where the APC is replaced by supported lipid bilayers (SLBs), which allow for imaging with high spatiotemporal resolution (Grakoui et al., 1999). On SLBs, the T cell IS is organized in the classic monofocal configuration, comprising three concentric domains: the central, peripheral, and distal supramolecular activation cluster (SMAC). While the central SMAC was initially thought to be the site of TCR signaling, it turned out to be the center of TCR recycling (Varma et al., 2006; Das et al., 2004). Instead, signaling occurs en route, when TCR microclusters that arise in the distal SMAC travel with a centripetal actin flow toward the central SMAC (Babich et al., 2012; Yi et al., 2012). The peripheral SMAC forms an adhesive ring that stabilizes the IS, mediated by the integrin lymphocyte function–associated antigen 1 (LFA-1) that binds its ligand intercellular adhesion molecule 1 (ICAM1) on the APC surface.
While the IS between B cells, which have weak priming capacity (Gunzer et al., 2004), and T cells resembles the aforementioned monofocal configuration found on SLBs, the IS between DCs and T cells is more complex and, despite its crucial physiological capacity, poorly understood (Fisher et al., 2008). T cell–DC ISs were described as multifocal, exhibiting multiple local ensembles of TCR, co-receptors, and adhesion molecules (Brossard et al., 2005; Tseng et al., 2008). In line with the idea that this multifocal structure determines the superior T cell priming capacity of DCs, engagement of the co-stimulatory molecule CD28 in multiple peripheral clusters, instead of one central cluster, correlates with stronger T cell activation (Shen et al., 2008).
The molecular driver of differential IS patterning is currently unknown. In the monofocal IS, formed between T and B cells and natural killer cells or cytotoxic T cells and target cells, the cytoskeleton seems largely cleared from the presynaptic face (Friedl et al., 2005). Therefore, SLBs, where ligands freely float in the bilayer, appear to be a valid model for such passive APCs. This is different in DCs, where the F-actin cytoskeleton polarizes toward the IS (Al-Alwan and Rowden, 2001; Benvenuti et al., 2004), supporting the idea that DCs actively pattern the IS via their cytoskeleton in order to support optimal T cell priming (Dustin et al., 2006a; Comrie and Burkhardt, 2016). As the presynaptic face of the IS has received little attention, direct evidence for this idea is missing. DC F-actin might form a stable scaffold, but could potentially also have a more dynamic role in structuring the IS.
Here, we use a multimodal imaging approach, in combination with functional in vitro and in vivo assays, to investigate the role of presynaptic actin dynamics in DCs and its impact on the priming of naive T cells.
Results
Depolymerization of DC actin converts multifocal into monofocal immune synapses
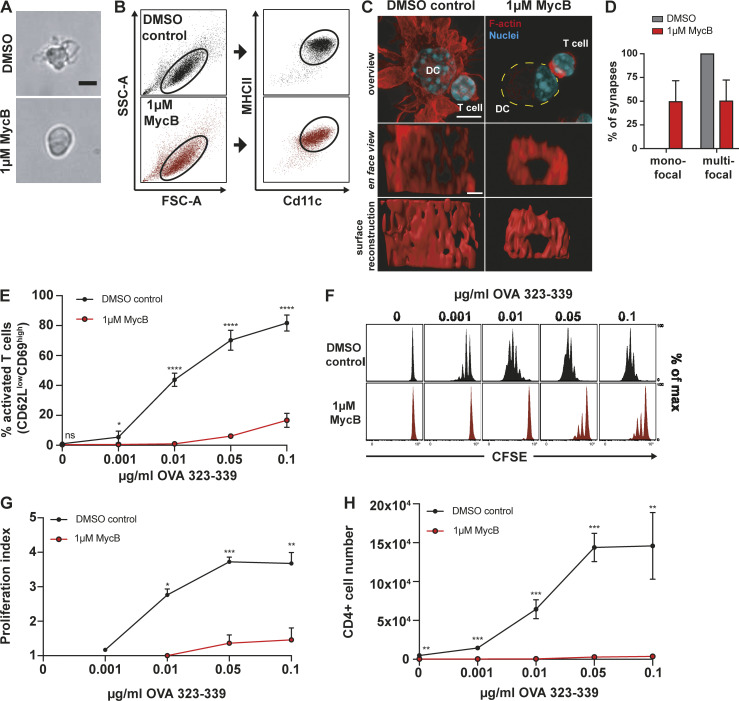
To explore how the structure of the T cell IS is influenced by the DC actin cytoskeleton, we depolymerized F-actin in DCs and characterized synapses formed with CD4+ T cells freshly isolated from OT-II transgenic mice whose T cells carry a TCR specific for an Ovalbumin (OVA)-derived peptide (Barnden et al., 1998). To selectively target actin in DCs and leave T cells unperturbed, we pretreated DCs with the drug mycalolide B (MycB). MycB causes actin depolymerization by severing F-actin filaments and irreversible sequestration of G-actin (Hori et al., 1993; Saito et al., 1994; Vaahtomeri et al., 2017). In contrast to other drugs, like cytochalasin D, which has been used in earlier studies (Al-Alwan and Rowden, 2001), MycB leads to covalent modification and therefore cannot be washed out. MycB-treated DCs lost their F-actin–rich veils, adopted a spherical shape, and lost the bulk signal in phalloidin stainings (Fig. 1, A and C). Pretreated DCs showed a shift toward slightly lower surface levels of the DC markers, Cd11c and major histocompatibility complex II (MHC II; Fig. 1 B), with no apparent effect on cell viability (Fig. S1, A and B). Synapses between MycB-treated or control DCs and T cells were fixed, stained for F-actin, and imaged via confocal microscopy. Around 50% of synapses between treated DCs and untreated T cells exhibited a monofocal structure with a pronounced ring of F-actin surrounding a central region devoid of F-actin. In contrast, all synapses in control samples showed a multifocal structure (Fig. 1, C and D; and Video 1). To exclude the possibility that this was an artifact caused by blurring of the pre- and postsynaptic side in fixed and stained samples, we performed live imaging of Lifeact-eGFP–expressing T cells and found a similar picture (Fig. S2, A–C; and Video 2). This confirms the notion of a multifocal T cell synapse with DCs (Brossard et al., 2005; Fisher et al., 2008) and suggests that the DC actin cytoskeleton prevents the formation of a monofocal IS, potentially by providing barriers that block or perturb centripetal flow of T cell actin (Dustin et al., 2006a). However, we also note that, upon DC F-actin depolymerization, only ∼50% of synapses become unambiguously monofocal, indicating that other F-actin–independent DC properties may contribute to IS patterning.
Figure 1.
DC F-actin depolymerization affects immune synapse structure and T cell priming efficiency. (A) Bright field images of mature DCs treated with 1 µM MycB (right) or DMSO (left). Scale bar: 10 µm. (B) Flow cytometry profiles of DMSO and 1 µM MycB-treated mature DCs. Cd11c/MHC II plots are pregated on FSC-A/SSC-A population defined by black oval on the left, representative example of three biological replicates. (C) Immunofluorescence images of synapses formed between DMSO or 1 µM MycB-treated mature DCs and T cells. Upper panel: overview pictures, yellow dotted line outlines DC. Scale bar: 5 µm. Middle panel: en face view on the synaptic interface. Scale bar: 1 µm. Lower panel: surface reconstruction of the synaptic interface. (D) Percentages of mono- and multifocal synapses formed between T cells and 1 µM MycB-treated mature DCs, n = 20 cell duplets for each condition, three biological replicates, mean ± SD. (E) Percentages of activated T cells assessed by CD62L/CD69 surface expression after 16 h of coculture with DMSO or 1 µM MycB-treated mature DCs at indicated OVA323-339 peptide concentrations, three biological replicates, mean ± SD. (F) CFSE dilution profile of T cells after 96 h of coculture with DMSO or 1 µM MycB-treated mature DCs at indicated OVA323-339 peptide concentrations, representative example of three biological replicates. (G) Proliferation indices of CFSE-labeled T cells after 96 h of coculture with DMSO or 1 µM MycB-treated mature DCs at indicated OVA323-339 peptide concentrations, three biological replicates, mean ± SD. (H) Absolute T cell numbers after 96 h of coincubation with DMSO or 1 µM MycB-treated mature DCs at indicated OVA323-339 peptide concentrations, three biological replicates, mean ± SD. Data in E, G, and H were tested for normal distribution, transformed if necessary, and tested by using Student’s t test. FSC-A, forward scatter–A; ns, not significant; SSC-A, side scatter–A. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
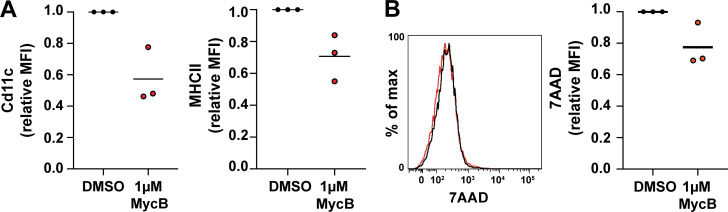
Figure S1.
Further characterization of MycB treated DCs. (A) Flow cytometry experiment displaying relative mean fluorescence intensities (MFIs) for Cd11c (left) and MHC II (right) of DMSO- or 1 µM MycB–treated mature DCs, three biological replicates. (B) 7–Actinomycin D (7-AAD) life/dead stain flow cytometry histograms (left) and relative mean fluorescence intensities (right) of DMSO- or 1 µM MycB–treated mature DCs, three biological replicates.
Video 1.
Immunological synapses of T cells and actin-depolymerized or control DCs. 3D reconstructions of mature OVA peptide–loaded DMSO control (left) or 1 µM MycB–treated (right) DCs, forming synapses with OT-II T cells. Cells were fixed and stained for F-actin with phalloidin (red) and NucBlue (blue). Frame rate, 12 frames per second.
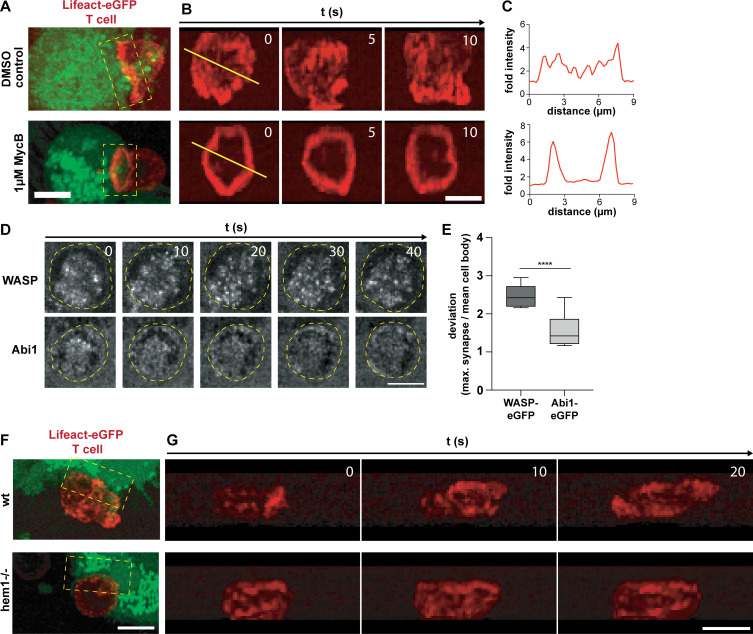
Figure S2.
Characterization of the structure and dynamics of the DC and T cell actin cytoskeleton at the immunological synapse. (A) 3D projection of Lifeact-eGFP–expressing OT-II T cells (red), interacting with DMSO control (upper) or 1 µM MycB–treated (lower) and 5-Carboxytetramethylrhodamine (TAMRA)-stained mature DC (green). (B) En face time-lapse reconstruction of T cell synapses shown in A (yellow boxes). (C) Relative Lifeact-eGFP intensities along yellow lines shown in B. (D) Time-lapse series of synapses formed between WASP-eGFP (upper) or eGFP-Abi1 (lower) expressing mature WT DCs and OT-II T cells in confiner setup. Yellow ovals demarcate the outline of the T cell. Scale bar: 5 µm. (E) Deviation of maximum WASP-eGFP or Abi1-eGFP signal at the synapse from mean intensity in the cell body, n = 7 synapses for each reporter construct, t test. (F) 3D projection of Lifeact-eGFP–expressing OT-II T cells (red) interacting with WT (top) or hem1−/− (bottom) TAMRA-stained mature DCs (green). (G) En face time-lapse reconstruction of T cell synapses shown in F (yellow boxes). All scale bars: 5 µm. ****, P ≤ 0.0001.
Video 2.
Live imaging of T cell immunological synapses with actin-depolymerized and control DCs. 3D reconstruction of live imaging of Lifeact-eGFP–expressing OT-II T cells (red), forming synapses with mature OVA peptide–loaded DMSO control (left) or 1 µM MycB–treated (right) DCs stained with TAMRA (green); time stamp: s:ms. Frame rate, 20 frames per second.
Next, we determined how actin depolymerization in DCs affects T cell priming. The capacity of MycB-pretreated DCs to activate T cells was dramatically decreased as measured by surface levels of early activation markers, CD62L and CD69 (Fig. 1 E). Impaired activation resulted in strongly reduced T cell proliferation, with few T cells entering division even at very high peptide concentrations (Fig. 1, F–H). Taken together, these results suggest that the DC actin cytoskeleton shapes the structure of the synapse, which is intricately linked to the strength of T cell activation. Importantly, these results are in line with observations in SLB systems where the addition of artificial barriers that prevent monofocal synapse formation enhances TCR signaling (Mossman et al., 2005).
Synaptic DC actin appears in foci that are dynamized by the WAVE regulatory complex
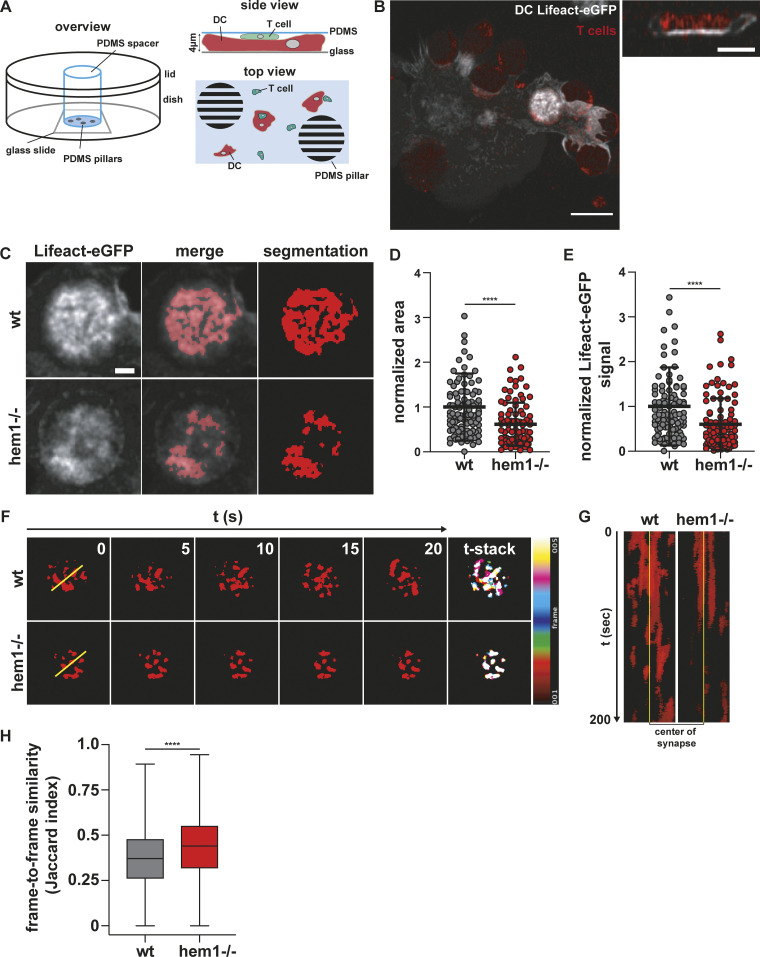
Next, we were interested in how DC actin at the IS might support T cell activation. Besides the fact that DC actin accumulates at the IS, little is known about its structure, dynamics, and molecular regulation. This is mainly due to the difficulty of imaging dynamic cell–cell contacts en face. To circumvent this problem, we developed a setup where antigen-loaded, Lifeact-eGFP expressing DCs and fluorophore-labeled T cells are confined between a coverglass and a layer of polydimethylsiloxane (PDMS; Le Berre et al., 2014). In this narrow space, DCs and T cells frequently position on top of each other and form synapses within the horizontal imaging plane (Fig. 2 A). Z-plane reconstruction of fast-spinning disc confocal movies showed that DC actin accumulates at the cell–cell interface as suggested previously by side views of fixed samples (Fig. 2 B, right). However, visualization of the synaptic plane revealed that DC actin dynamically appears in small foci and larger patches, which are separated by regions with low actin signal. Notably, there was no indication of any centripetal actin flow (Fig. 2, B, C, F, and G; Video 3; and Video 7).
Figure 2.
Dynamic F-actin foci at the DC immune synapse. (A) Schematic overview of PDMS confiner setup. (B) Left: Maximum intensity projection of Lifeact-eGFP–expressing mature WT DC interacting with TAMRA-stained T cells. Scale bar: 10 µm. Right: Z-stack of bright Lifeact-eGFP signal region on the left. Scale bar: 5 µm. (C) Examples of mature WT and hem1−/− DC–T cell immune synapses and machine learning–based segmentation of synaptic DC Lifeact-eGFP signal. Scale bar: 2 µm. (D) Normalized area of synaptic DC Lifeact-eGFP signal, n = 100 synapses each, Mann-Whitney test, mean ± SD, three biological replicates. (E) Normalized synaptic DC Lifeact-eGFP signal, n = 100 synapses each, Mann-Whitney test, mean ± SD, three biological replicates. (F) Left: Time-lapse series of synaptic DC Lifeact-eGFP signal. Right: Stack through time-lapse series. (G) Kymograph of yellow line in F. (H) Frame-to-frame similarity of synaptic DC Lifeact-eGFP signal, n ≪ 6,000 frame comparisons each, Mann-Whitney test, mean ± min/max, two biological replicates. ****, P ≤ 0.0001.
Video 3.
WT DC actin at the immunological synapse with T cells. 3D reconstruction of mature OVA peptide–loaded, Lifeact-eGFP–expressing WT DC (green), forming synapses with TAMRA-stained OT-II T cells (red) in confiner setup. Frame rate, 12 frames per second.
Video 7.
Actin dynamics of WT and hem1−/− DCs at the immunological synapse. Lifeact-eGFP signal of mature OVA peptide–loaded WT or hem1−/− DCs forming immune synapses with OT-II T cells in confined setup; Left: Raw data. Right: Segmentation. Frame interval: 5 s; time stamp: min:s; scale bar: 5 µm. Frame rate, 10 frames per second.
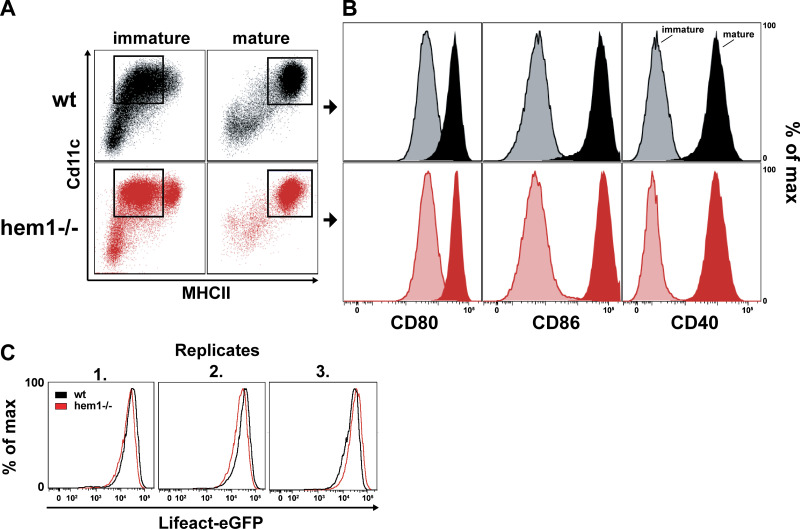
We next addressed the molecular regulation of synaptic actin in DCs. Wiskott-Aldrich-Syndrome-Protein (WASP), together with the WASP-family verprolin-homologous protein (WAVE) regulatory complex (WRC), are the main nucleation-promoting factors for the Arp2/3 complex that nucleates branched actin networks at the plasma membrane (Takenawa and Suetsugu, 2007; Alekhina et al., 2017). At the T cell face of the IS, the WRC and WASP have critical and divergent functions. WASP polymerizes F-actin foci that are associated with TCR microclusters that recruit PLCɣ1, leading to calcium influx and nuclear factor of activated T cells (NFAT) signaling. In contrast, the WRC creates and reorganizes the synaptic actin background that mediates integrin-dependent adhesion and PLCɣ1-independent signaling (Nolz et al., 2006; Kumari et al., 2015). By expressing fluorescent reporter constructs, we found a similar situation in DCs. While WASP localizes to distinct foci, the WRC localized throughout the actin cortex, including the synaptic interface (Fig. S2, D and E; and Videos 4 and 5). In DCs, WASP was shown to contribute to T cell–DC contact formation, maintenance, and IS stability (Bouma et al., 2011; Pulecio et al., 2008; Malinova et al., 2016). As the role of DC WAVE at the IS was unknown, we generated WRC-deficient DCs from the bone marrow of mice lacking hem1, an essential subunit of the pentameric WRC that is specific for the hematopoietic system (Park et al., 2008). Hem1−/− DCs contain 50% less F-actin, while surface levels of MHC II and co-stimulatory ligands are equal to control cells (Fig. S3, A and B; Leithner et al., 2016). Deletion of WRC did not lead to an overall qualitative change in the structure of the T cell synapse that remained multifocal (Fig. S2, F and G). We used machine learning–based image segmentation (Berg et al., 2019) to perform a quantitative and unbiased comparison of actin dynamics in the synapses of WT vs. hem1−/− DCs. Actin occupied a significantly reduced area fraction of the contact surface in hem1−/− compared with control DCs (Fig. 2, C–E; and Videos 3 and 6). This effect was not due to altered levels of the Lifeact-eGFP probe that was expressed at similar levels in WT and hem1−/− DCs (Fig. S3 C). When comparing time-lapse movies of synaptic Lifeact-eGFP in WT and hem1−/− DCs, we found that actin foci of WT DCs frequently appeared and disappeared throughout the whole synaptic interface. In contrast, comparing frame-to-frame similarities of Lifeact-eGFP signals revealed that hem1−/− DC actin foci display significantly reduced lateral dynamics (Fig. 2, F and G; and Video 7). Together, these data show that, in DCs, WRC-dependent actin polymerization contributes to the overall actin content at the synaptic interface and mediates lateral movement of actin foci.
Video 4.
DC WASP dynamics at the immunological synapse. 3D reconstruction of live imaging of mature OVA peptide–loaded, WASP-eGFP–expressing WT DC (green), forming synapses with TAMRA-stained OT-II T cells (red) in confiner setup; time stamp: s:ms. Frame rate, 24 frames per second.
Video 5.
DC WAVE dynamics at the immunological synapse. 3D reconstruction of live imaging of mature OVA peptide–loaded, eGFP-Abi1–expressing WT DC (green), forming synapses with TAMRA-stained OT-II T cells (red) in confiner setup; time stamp: s:ms. Frame rate, 24 frames per second.
Figure S3.
Flow cytometry analysis of immature and mature wt and hem1−/− DCs. (A) Flow cytometry profiles of MHC II– and Cd11c-stained immature (left) or mature (right) WT or hem1−/− DCs. (B) Flow cytometry histograms of immature (light colors) and mature (dark colors) of WT (top) or hem1−/− (bottom) DCs stained for CD80, CD86, or CD40, respectively. (C) Flow cytometry histograms of three biological replicates for Lifeact-eGFP–expressing mature WT and hem1−/− DCs.
Video 6.
Hem1−/− DC actin at the immunological synapse with T cells. 3D reconstruction of mature OVA peptide–loaded, Lifeact-eGFP–expressing hem1−/− DC (green), forming synapses with TAMRA-stained OT-II T cells (red) in confiner setup. Frame rate, 12 frames per second.
WRC-deficient DCs have reduced T cell priming capacity
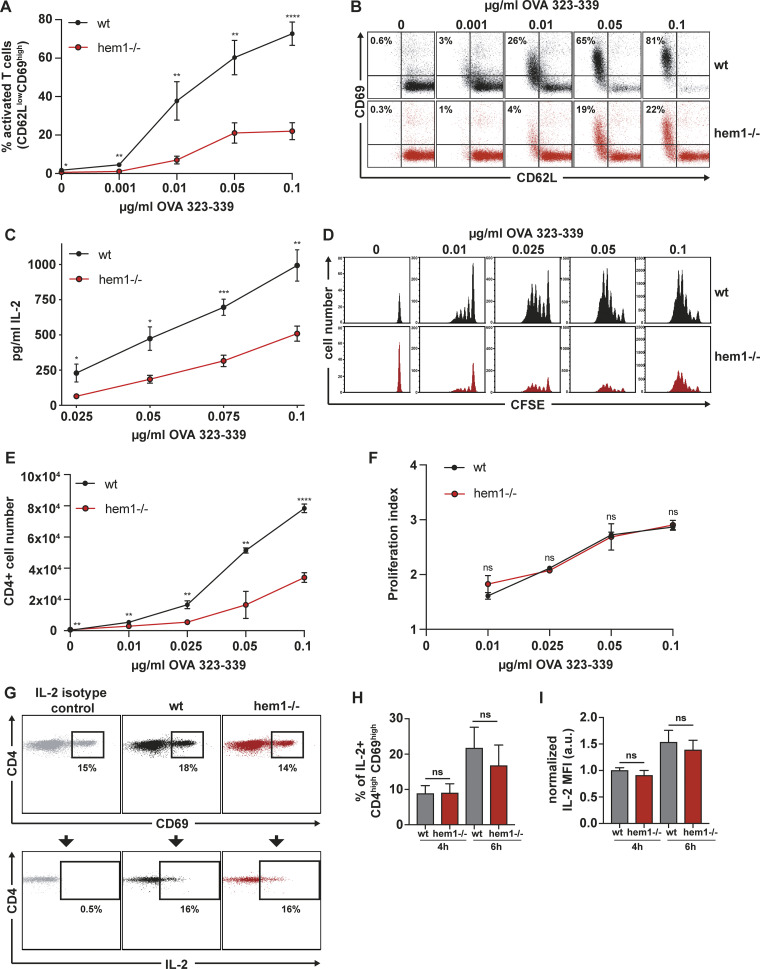
Next, we asked how WRC deficiency in DCs affects their T cell priming capacity. We found that, compared with control cells, hem1−/− DCs were only able to activate a small fraction of T cells at early time points, as measured by the modulation of early activation markers (Fig. 3, A and B). This deficiency in T cell priming was prominent even at highest peptide concentrations, accompanied by reduced interleukin 2 (IL-2) levels in the supernatant (Fig. 3 C), and resulted in significantly fewer T cells after 4 d of DC–T cell coculture (Fig. 3, D and E). However, in dye dilution assays, which measure proliferation at the single-cell level, we could not detect any differences in the proliferative indices of T cells stimulated by WT or hem1−/− DCs (Fig. 3 F). This might indicate that, although hem1−/− DCs activate fewer T cells, the ones that pass an activation threshold proliferate normally. To explore this further, we determined the capability of WT and hem1−/− DCs to induce TCR signaling. To this end, we measured the intracellular production of IL-2, the downstream product of TCR-triggered signaling cascades (Spolski et al., 2018), on the single-cell level. IL-2 production was measured at very early time points, before any auto- or paracrine loops take effect (Sojka et al., 2004). DCs and T cells were cocultured for 1 or 3 h, secretion was blocked with brefeldin A, and intracellular staining was performed after another 3 h to allow for cytokine accumulation. Comparable numbers of T cells activated by WT vs. hem1−/− DCs stained positive for IL-2 at these time points, and we could not detect any difference in the amount of produced cytokine (Fig. 3, G–I). These data suggest that, although they activate fewer T cells, hem1−/− DCs are otherwise fully capable of triggering TCR signaling, cytokine production, and effector T cell differentiation.
Figure 3.
Hem1−/− DCs are impaired in T cell activation. (A) Percentage of activated T cells assessed by CD62L/CD69 surface expression at indicated OVA323-339 peptide concentrations, three biological replicates, mean ± SD. (B) Exemplary CD62L/CD69 flow cytometry profile of T cells after 16 h of coculture with mature WT or hem1−/− DCs. (C) IL-2 ELISA after 16 h of T cell mature WT or hem1−/− DC coculture at indicated OVA323-339 peptide concentrations, three biological replicates, mean ± SD. (D) CFSE dilution profile of T cells after 96 h of coculture with mature WT or hem1−/− DCs at indicated OVA323-339 peptide concentrations, representative example of three biological replicates. (E) Absolute T cell numbers after 96 h of coincubation with mature WT or hem1−/− DCs at indicated OVA323-339 peptide concentrations, three biological replicates, mean ± SD. (F) Proliferation indices of CFSE-labeled T cells after 96 h of coculture with mature WT or hem1−/− DCs at indicated OVA323-339 peptide concentrations, three biological replicates, mean ± SD. (G) Exemplary CD4/CD69 (top) and CD4/IL-2 (bottom) flow cytometry profile of T cells after 6 h coculture with WT or hem1−/− DCs. (H) Fraction of IL-2–positive T cells after 4 or 6 h of coculture with mature WT or hem1−/− DCs, pregated on CD4high/CD69high, two biological replicates, one-way ANOVA, mean + SD. (I) Normalized IL-2 mean fluorescence intensity (MFI) of CD4high T cells after 4 or 6 h of coculture with mature WT or hem1−/− DCs, two biological replicates, one-way ANOVA, mean + SD. Data were normalized to WT 4 h. 10 µg/ml brefeldin A was added for the last 3 h of the cocultures in G, H, and I. Data in A, C, E, and F were tested for normal distribution, transformed if necessary, and tested by using Student’s t test. ns, not significant. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
WRC-deficient DCs show prolonged interaction times and altered synaptic structure
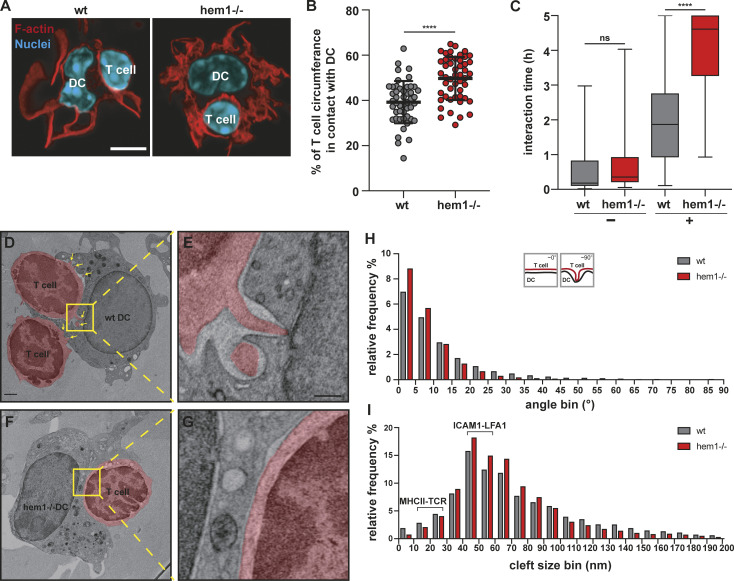
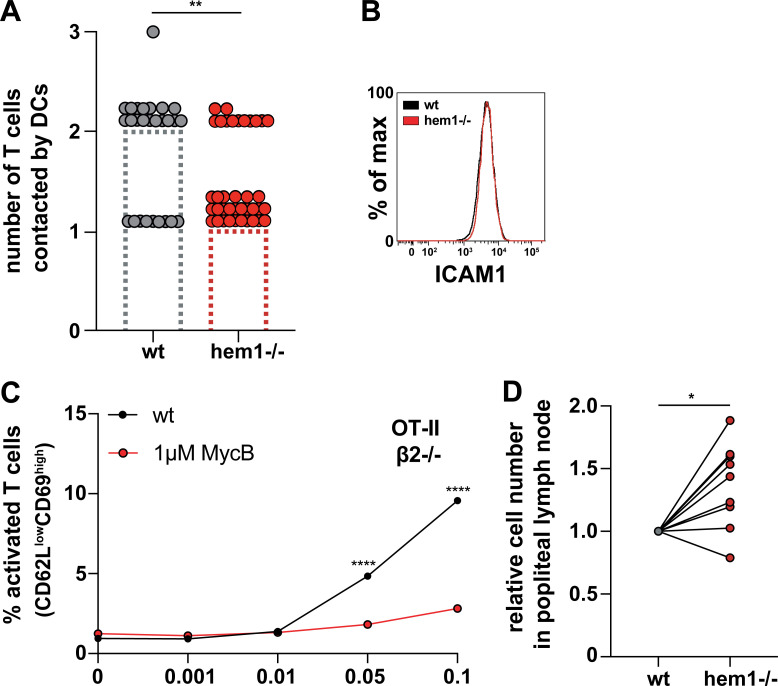
As these data indicated that changes in contact strength or frequency might reduce the capacity of DCs to prime multiple T cells, we analyzed the relative contact area of T cells with DCs in fixed duplets. Morphometry revealed that hem1−/− DCs formed significantly larger contacts with T cells compared with WT DCs, and that hem1−/− DCs often wrap around T cells, covering a substantial part of their surface (Fig. 4, A and B). These findings led us to speculate that reduced presynaptic actin dynamics in WRC-deficient DCs translate into an overall reduction in dynamism of cell–cell contacts. To test this idea, we seeded WT or hem1−/− DCs and T cells on glass slides and quantified their interactions by using live cell microscopy. Contact times between OVA peptide-loaded hem1−/− DCs and T cells were substantially increased compared with the WT control, while in the absence of peptide, interaction times were equally low (Fig. 4 C and Video 8). Consequently, hem1−/− DCs contacted significantly fewer T cells during the imaging period (Fig. S4 A).
Figure 4.
Hem1−/− DCs alter synapse structure and dynamics. (A) Fluorescence microscopy images of synapses formed between mature WT or hem1−/− DCs and T cells stained with phalloidin and DAPI. Scale bar: 5 µm. (B) Percentages of T cell surface area in contact with DC, ∼50 cells each, t test, mean ± SD, three biological replicates. (C) Interaction times of mature WT or hem1−/− DCs with T cells in the absence (−) or presence (+) of OVA323-339 peptide, n = 50 contacts each for (+) or n = 30 contacts each for (−), Mann-Whitney test, mean ± min/max, three biological replicates. (D and E) EM of WT DC–T cell synapse, T cells are colored in red, yellow box and dotted lines denote region magnified in E, yellow arrows highlight T cell protrusions. (F and G) EM of hem1−/− DC–T cell synapse, T cell is colored in red, yellow box and dotted lines denote region magnified in G. Scale bars: 1 µm in D and F, 300 nm in E and G. (H) Frequency histograms in percent of the angles found between DC and T cell membranes, n = 4 synapses each, two biological replicates, Kolmogorov-Smirnov test, P ≤ 0.0001. (I) Frequency histograms in percent of the cleft size found between DC and T cell membranes from H, Kolmogorov-Smirnov test, P ≤ 0.0001. ns, not significant. ****, P ≤ 0.0001.
Video 8.
In vitro contact times of WT or hem1−/− DCs with T cells. Interactions of mature OVA peptide–loaded WT or hem1−/− DCs with OT-II T cells on poly-L-lysine–coated glass. Frame interval: 30 s; time stamp: h:min; scale bar: 10 µm. Frame rate, 12 frames per second.
Figure S4.
The dependency of T cell priming defects on the pERM-ICAM1-LFA-1 axis are specific to hem1−/− DCs. (A) Quantification of the number of T cells contacted by mature WT and hem1−/− DCs. Bars represent the median. Mann-Whitney test, three biological replicates. (B) Flow cytometry histogram for ICAM1 in mature WT and hem1−/− DCs. (C) Percentages of WT or 1 µM MycB–treated mature DCs activating β2-integrin–deficient T cells, as assessed by CD62L/CD69 surface expression at indicated OVA323-339 peptide concentrations. (D) Normalized relative number of WT and hem1−/− DCs in the popliteal lymph node 24 h after coinjection in a 1:2 ratio. t test. Data are pooled from nine different mice. *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001.
To understand the mechanistic cause of these stabilized contacts, we resolved the ultrastructure of the whole synaptic interface by performing EM of high pressure–frozen and serially sectioned DC–T cell duplets. The WT DC–T cell synapse was characterized by multiple finger-like protrusions emanating from T cells and protruding into the DC cell body. At these focal protrusions, the plasma membranes of both cells came into direct contact (Fig. 4, D and E; and Video 9). Similar protrusions have been observed between T cells and other cell types (Ueda et al., 2011; Sage et al., 2012) and might correspond to T cell microvilli that have been recently observed on artificial substrates (Cai et al., 2017; Jung et al., 2016). In contrast, hem1−/− DC–T cell contacts were characterized by a complete lack of T cell protrusions, giving the T cell membrane at the synapse a smooth and stretched appearance (Fig. 4, F and G; and Video 9). To quantify this difference, we traced the plasma membranes of DCs and T cells and determined at which angle they are positioned relative to each other. Morphometry revealed a shift toward lower angles for hem1−/− DC–T cell synapses demonstrating parallel alignment of both membranes. WT DC–T cell synapses exhibited higher angles as membranes positioned orthogonal in regions of T cell protrusions (Fig. 4 H).
Video 9.
The ultrastructure of WT or hem1−/− DC and T cell synapses. Z-stack through high pressure–frozen and serially sectioned synapse formed between mature OVA peptide–loaded WT (left) or hem1−/− DC (right) and OT-II T cell (red); z-interval: 30 nm; scale bar: 1 µm. Frame rate, 10 frames per second.
The synaptic cleft was previously shown to be differentially spaced depending on the functional zone of the synapse. Sites of integrin-mediated adhesion are separated by 35–55 nm, matching the large extracellular parts of LFA-1 and ICAM1. The shorter TCR–MHC pair translates into closer contacts below 25 nm (Shaw and Dustin, 1997). Thus, we determined the distances between DC and T cell membranes and found an overrepresentation of 35–55-nm distances and an underrepresentation of below 25-nm distances in hem1−/− compared with control synapses (Fig. 4 I). Together, these data suggested that ICAM1–LFA-1 interactions were more frequent in hem1−/− DC–T cell synapses.
Increased interaction times are mediated by the pERM–ICAM1–LFA-1 axis
Integrin adhesiveness is regulated by clustering (valency) and ligand affinity (Kinashi, 2005). Integrins engage in catch bonds where application of pulling force on the integrin–ligand pair shifts them into a high-affinity conformation (Kong et al., 2009; Chen et al., 2010; Zhu et al., 2008). Consequently, it has been suggested that forces are provided by the T cell actin cytoskeleton (Zhu et al., 2008; Schürpf and Springer, 2011) and that these forces have to be balanced by opposing retention forces. In support of this idea, only immobilized, but not soluble, ICAM1 triggers conversion of LFA-1 to the high-affinity conformation (Perez et al., 2003; Feigelson et al., 2010).
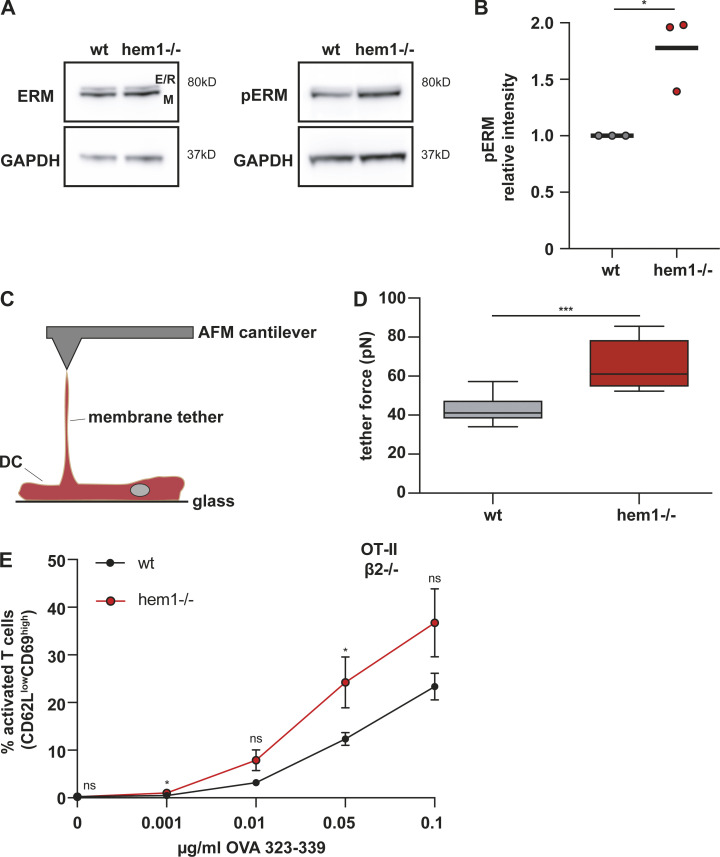
As surface ICAM1 levels of hem1−/− DCs were unchanged (Fig. S4 B), we turned our attention to the adaptor proteins that connect ICAM1 to the underlying actin cytoskeleton. In DCs, surface ICAM1 is immobilized by anchorage to F-actin via the phosphorylated, and thus active form, of the ezrin-radixin-moesin (ERM) protein, moesin. This enables DCs to oppose forces that are exerted on LFA-1 through the T cell actin cytoskeleton in a cellular tug of war, making them well suited to facilitate the transition of LFA-1 into its high-affinity conformation and thus support full T cell activation (Comrie et al., 2015). We found that pERM levels were elevated in hem1−/− compared with WT DCs, while total ERM levels were unchanged (Fig. 5, A and B). The functional correlate of elevated pERM levels is an increase in transmembrane protein-mediated membrane-to-cortex attachment. Thus, we employed atomic force microscopy (AFM) and found that significantly higher forces are required to pull membrane tethers from hem1−/− compared with WT DCs (Fig. 5, C and D), which is in line with increased membrane-to-cortex attachment. Taken together, our data suggest that elevated pERM levels immobilize ICAM1, potentially causing an increase in LFA-1 valency at the hem1−/− DC–T cell synapse. This might stabilize the synapse, counter its resolution, and thereby lead to increased interaction times.
Figure 5.
The hem1−/− DC–T cell priming defect is mediated by the pERM–ICAM1–LFA-1 axis. (A) Western blots for ERM, pERM, and GAPDH in mature WT and hem1−/− DCs, representative example of three biological replicates. (B) Relative intensity of pERM signal in mature WT and hem1−/− DCs, t test, three biological replicates. (C) Schematic overview of AFM setup for tether pulling. (D) Static tether force for mature WT and hem1−/− DCs, two biological replicates, t test. (E) Percentages of activated β2-integrin–deficient T cells assessed by CD62L/CD69 surface expression at indicated OVA323-339 peptide concentrations, three biological replicates, mean ± SD. Data were tested for normal distribution, transformed if necessary, and tested by using Student’s t test. ns, not significant. *, P ≤ 0.05; ***, P ≤ 0.001.
If the T cell priming deficiency of hem1−/− DCs was caused by an increase in adhesiveness, disruption of the ICAM1–LFA-1 axis should rescue this effect. To test this idea, we cocultured WT and hem1−/− DCs together with naive T cells of integrin β2-deficient, OT-II transgenic mice and monitored T cell activation. Removal of ICAM1–LFA-1 interactions led to an overall reduction of T cell activation (compare Fig. 3 A and Fig. 5 E). Notably, under these conditions, hem1−/− DCs were no more inferior in their priming capacity. In contrast, they even triggered slightly enhanced activation of integrin-deficient T cells compared with their WT counterparts. This was a hem1−/− DC–specific effect, as removal of the ICAM1–LFA-1 axis did not rescue the T cell priming deficiency of actin depolymerized DCs (Fig. S4 C). Taken together, these data suggest that stabilized cell–cell adhesion via the ICAM1–LFA-1 axis is the main cause of the T cell priming deficiency observed in hem1−/− DCs.
WRC-deficient DCs show impaired T cell priming capacity in vivo
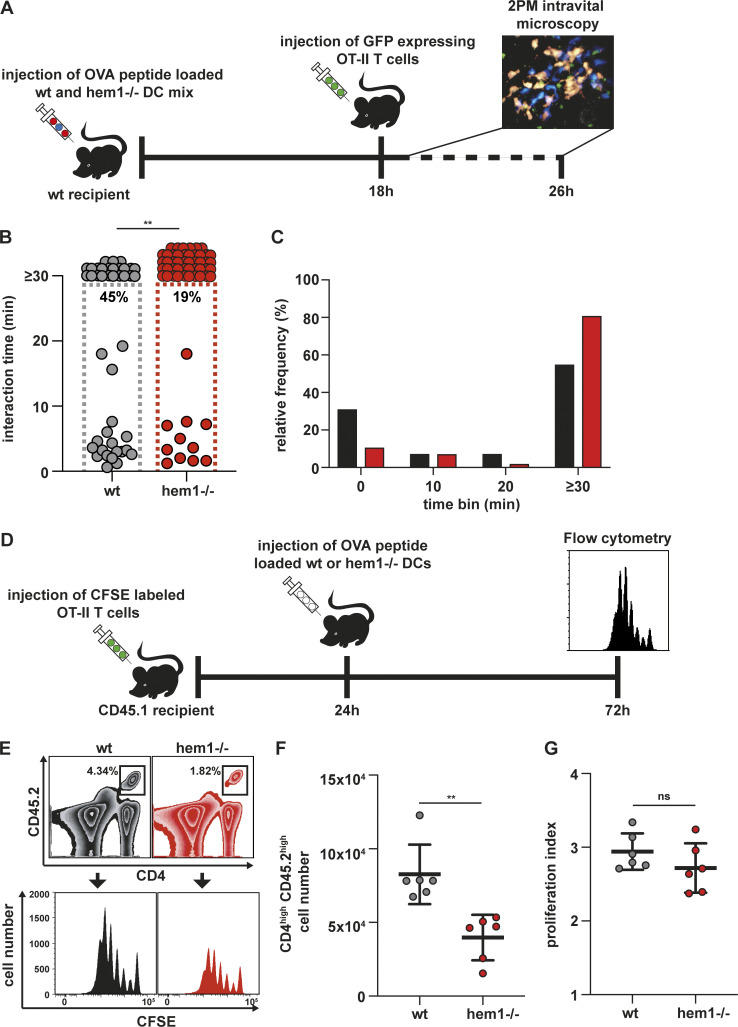
While the importance of the ICAM1–LFA-1 axis for in vitro T cell priming has been established (Abraham et al., 1999), its in vivo role remains controversial. Deletion of LFA-1 or talin in T cells leads to strong in vivo T cell proliferation defects (Kandula and Abraham, 2004; Wernimont et al., 2011); however, these results have to be interpreted with care due to the severe homing defects of these T cells. In contrast, deletion of ICAM1 on lymph node–resident (Feigelson et al., 2018) and migratory DCs (Kozlovski et al., 2019) appears to have no effect on initial T cell activation and proliferation, but is crucial for the development of immunological memory (Scholer et al., 2008). To address these points, we performed hock co-injections of differently labeled and peptide-loaded WT and hem1−/− DCs, which were followed by i.v. injection of labeled OT-II T cells (Fig. 6 A). This procedure mimics an infectious setting and causes DCs to home into the lymph node via the lymphatic route, while T cells enter the node via the blood circulation. To compensate for the inferior migratory capacity of hem1−/− DCs (Leithner et al., 2016), we injected two-fold higher numbers of hem1−/− DCs (Pulecio et al., 2008), which led to comparable or even slightly increased numbers of hem1−/− DCs in the draining lymph node (Fig. S4 D). We then performed two-photon intravital microscopy of the popliteal lymph node and quantified cellular interactions. Contact times between DCs and T cells were significantly increased for hem1−/− DC compared with WT DC, thus recapitulating the in vitro assays (Fig. 6, B and C; and Video 10). Finally, to determine if this change in contact times has an effect on T cell proliferation, we performed in vivo carboxyfluorescein diacetate succinimidyl ester (CFSE) dilution assays (Fig. 6 D). While activated T cells underwent the same number of divisions, significantly fewer T cells accumulated with hem1−/− DCs, again mirroring our in vitro findings (Fig. 6, E–G). Taken together, these data show that increased interaction times of hem1−/− DCs with T cells are also relevant for in vivo T cell priming.
Figure 6.
Hem1−/− DCs have T cell priming defects in vivo. (A) Schematic overview of experimental setup for two-photon intravital microscopy. (B) In vivo interaction times of mature WT or hem1−/− DCs and T cells, n = 45 cell–cell interactions each, Mann-Whitney test. Percentages refer to cell–cell interactions that start and end within the 30-min time window, two biological replicates. (C) Frequency histograms in percent of DC–T cell interaction times from B. (D) Schematic overview of experimental setup for in vivo T cell proliferation assays. (E) Gating strategy to determine the CFSE proliferation profiles of CD45.2/CD4 T cells in CD45.1 mice after WT or hem1−/− DC T cell priming. (F) Absolute CD45.2/CD4 T cell numbers in the popliteal lymph nodes of CD45.1 mice 72 h after DC injections, n = 6 lymph nodes each, Mann-Whitney test, two biological replicates. (G) Proliferation indices of CFSE-labeled T cells from F, t test. ns, not significant. **, P ≤ 0.01.
Video 10.
In vivo contact times of WT or hem1−/− DCs and T cells. Intravital microscopy of interactions between mature OVA peptide–loaded WT (blue, left) or hem1−/− (red, right) and OT-II T cells (green). Frame interval: 20 s; time stamp: min:s; scale bar: 5 µm. Frame rate, 12 frames per second.
Discussion
We show that an intact, dynamic DC actin cytoskeleton is a crucial prerequisite for effective T cell priming. F-actin depolymerization in DCs led to conversion of the multifocal to a monofocal T cell IS, associated with a drastic reduction in T cell priming efficiency. These results are in line with observations in SLB systems where the introduction of physical barriers prevented monofocal synapse formation and improved the T cell signaling response (Mossman et al., 2005). This underscores the importance of the DC F-actin cytoskeleton in actively structuring and organizing the IS in a way that supports optimal T cell activation. How DC actin orchestrates TCR signaling within the multifocal synapse remains to be studied at the molecular level. Here, biophysical parameters, like the lateral movement of MHC, and co-stimulatory molecules, but also mechanical properties, like cortical stiffness, might be crucial (Blumenthal et al., 2020; Comrie et al., 2015; Comrie and Burkhardt, 2016; Doty and Clark, 1998; Girard et al., 2012). Another open question of fundamental immunological importance is to what extent the mono- vs. multifocal synapse organization that different antigen-presenting cells promote determines their T cell priming capacity. Is it indeed their peculiar actin dynamics that make DCs the most potent activators of adaptive immunity?
In our study, we demonstrate that synaptic actin not only forms a structured scaffold to organize a multifocal synapse, but that its dynamism is an important driver of the interaction turnover between DCs and T cells, which is a decisive quantitative parameter of immune responses (Bousso, 2008). We show that synaptic actin on the DC side appears in highly dynamic foci with no sign of coherent centripetal flow. Upon depletion of the WAVE complex, total synaptic F-actin levels as well as lateral dynamics of actin foci decreased, and we found that this reduced dynamism led to an overall increase in T cell–DC contact times, effectively reducing contact turnover. Causative for this increase in contact times were elevated pERM levels that led to an overrepresentation of synapse-stabilizing ICAM1–LFA-1 contacts.
What is the molecular link between WRC loss of function and increased pERM levels? One possibility is that, in the absence of WRC, Arp2/3-mediated branched nucleation is compensated by formin-mediated actin polymerization. Such a shift, which we previously observed in WRC-deficient DCs, causes F-actin filaments to align and run parallel to the cell membrane (Leithner et al., 2016). It was shown in macrophages that formin-nucleated filaments are a preferential substrate for ERM proteins and thus restrict transmembrane receptor mobility. In contrast, Arp2/3-branched filaments localize to regions of increased receptor mobility and thus decrease ERM engagement (Freeman et al., 2018). A second, non-exclusive possibility is that WRC and WASP serve directly opposing functions at the DC IS. We show that the WRC localizes to the synaptic background, while WASP assembles in discrete foci. These dot-like WASP assemblies are in line with a study that used an inverted SLB system where the T cell side of the IS was mimicked by the bilayer. When DCs were placed on these artificial T cells, they developed synaptic WASP-dependent actin foci. These podosome-like structures were enhanced by integrin–ICAM1 interactions, indicating that WASP leads to adhesion stabilization, while the WRC dynamizes these adhesions. In line with this scenario, WASP-deficient DCs show defective ICAM1 organization, form smaller and less stable contacts with T cells, and, importantly, show an increase in F-actin turnover at the synapse (Malinova et al., 2016; Pulecio et al., 2008; Bouma et al., 2011). Taken together, this suggests that, while WASP has a role in stabilizing ICAM1–LFA-1-mediated DC–T cell interactions, the WRC has a role in adhesion dynamics and turnover that resolves this type of heterologous cell–cell contacts.
How does increased adhesion at the hem1−/− DC–T cell IS lead to a reduction in T cell priming? While we do not rule out the possibility that increased ICAM1–LFA-1 interaction might lead to suboptimal spacing of the DC and T cell membranes, thereby subtly affecting TCR signaling (Comrie and Burkhardt, 2016), the dominant effect we observed was at the level of DC–T cell interaction dynamics. Early studies suggested that the formation of a stable IS is a requirement for full T cell activation (Dustin et al., 1997); however, it later turned out that especially when T cells are activated by DCs, phases of short, sequential contacts and a dynamic synaptic interface (kinapse) are the prevalent pattern (Bousso, 2008; Gunzer et al., 2000; Dustin, 2008). We suggest that balancing cell–cell adhesion and de-adhesion, regulated by the WRC and WASP on the DC side, is an important factor in achieving full T cell expansion. DC–T cell contacts have to be long enough for T cells to become fully activated, but interaction time has to be regulated in a way that allows for contact with as many T cells as possible (Fig. 7). These results are in line with previous studies where DC adhesion to T cells was artificially increased by activation of normally inactive DC macrophage-1 antigen (MAC-1) or LFA-1, leading to impaired T cell priming (Varga et al., 2007; Balkow et al., 2010). Priming of T cells by hem1−/− DCs leads to a strong reduction in resulting T cell numbers, while the number of divisions that an individual T cell undergoes once it is activated is unchanged. Additionally, T cells activated by WT vs. hem1−/− DC produce the same amount of IL-2, which serves as a proxy for TCR signaling. This, together with the fact that their T cell priming deficiency can be rescued by removal of the ICAM1–LFA-1 axis, suggests that hem1−/− DCs are otherwise capable of activating T cells.
Figure 7.
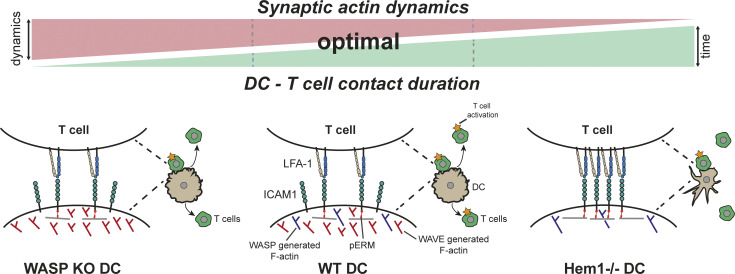
Working model showing how DC actin dynamics regulates DC–T cell contact time and priming efficiency. Center: Actin dynamics in WT DCs is regulated by WAVE and WASP in a way that allows for transiently stable DC–T cell contacts, mediated by the pERM–ICAM1–LFA-1 axis, which leads to the activation of a large number of T cells. Right: In the absence of DC WAVE, lateral actin dynamics is reduced, and the altered actin network serves as an ideal substrate for ERM proteins. This leads to an increase in pERM/ICAM1/LFA-1–mediated cell–cell adhesion and allows for the activation of only few T cells compared with WT. Left: in the absence of WASP, actin dynamics at the synapse increases, leading to destabilization of the cell–cell contact and a decrease in interaction time that results in insufficient T cell activation. KO, knockout.
Together, we show that DC F-actin actively structures the multifocal organization of the synaptic interface between DC and T cell, and that the dynamics at this interface promotes T cell expansion by increasing the turnover rather than stabilizing the structure of DC–T cell interactions.
Materials and methods
Cell culture
R10 medium, consisting of RPMI 1640 supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, and 50 µM 2-mercaptoethanol (all Thermo Fisher Scientific) was used as basic cell culture medium.
DCs were differentiated from bone marrow of male or female 6–12-wk-old WT or hem1−/− C57BL/6J mice according to established protocols (Lutz et al., 1999). F-actin reporter mice were obtained by breeding WT or hem1+/− mice to Lifeact-eGFP animals (Riedl et al., 2010), followed by backcrosses to hem1+/− mice. 2 × 106 WT or 1.25 × 106 hem1−/− bone marrow cells were seeded in 9 ml R10 medium, supplied with 1 ml of in-house–generated granulocyte-macrophage colony-stimulating factor (GM-CSF) in 94-mm Petri dishes (Greiner, 632180). At day 3, 8 ml R10 medium, supplied with 2 ml GM-CSF, was added to the dishes. At day 6, 10 ml of medium were removed and replaced with 8 ml R10 and 2 ml GM-CSF. On day 8 or 9, nonadherent cells from two dishes were collected and cryo-preserved in 1 ml 90% DMSO and 10% FCS. For maturation, two vials were thawed and cells were seeded overnight onto 150-mm cell culture dishes (VWR, 734–2322) in 18 ml R10 and 2 ml GM-CSF, supplied with 200 ng/ml LPS from Escherichia coli 0127:B8 (Merck, L4516). Only cells from the supernatant were used in experiments. For some experiments, cells were treated with 1 µM MycB (Enzo Lifesciences, BML-T123) for 15 min and then washed three times with R10 medium.
T cells were isolated from spleens and lymph nodes of C57BL/6J OTII transgenic mice (Barnden et al., 1998) by negative CD4+ selection (Stemcell, 19852). LFA-1–deficient T cells were obtained by breeding CD18−/− (Wilson et al., 1993) to OT-II mice. For these mice, negative selection for naive CD4+ was performed (Stemcell, 19765). T cells were directly used for experiments after isolation.
DC surface marker stainings
0.5 × 106 DCs/tube were spun down (300 g, 5 min), washed once, and then resuspended in FACS buffer (1× PBS, 5 mM EDTA, 1% BSA). Cells were first stained with Fc-block (1:100; 93) and then with a mixture of anti-mouse Cd11c-APC (1:300; N418) and anti-mouse MHC II–eFluor450 (1:800; M5/114.15.2; all eBioscience). All stainings were performed at 4°C for 15 min. Cells were washed once with 1 ml FACS buffer, resuspended, and analyzed on a FACS Canto II machine (BD Biosciences).
Imaging of live and fixed DC–T cell duplets
Glass-bottom dishes (35 mm; glass diameter 14-mm, glass thickness 0; Mattek) were plasma cleaned (pdc-002 plasma cleaner; Harrick) and coated with 1× poly-L-lysine (Merck, P8920) in H2O for 10 min. Dishes were washed twice with H2O and then dried for at least 4 h at RT. DCs were preloaded with 0.1 µg/ml OVA323-339 in R10 (vac-isq; Invivogen) for 2 h at 37°C/5% CO2. 1.5 × 105 DCs were mixed with 3 × 105 T cells and then pipetted onto coated dishes in a total volume of 300 µl. Cells were either followed by live imaging for 5 h or allowed to interact for 30 min at 37°C/5% CO2, followed by fixation with 300 µl 6% PFA in R10 medium. Cells were then washed twice with 1× TBS and permeabilized with 0.2% Triton X-100 in 1× TBS for 10 min. Samples were washed twice and then blocked with 3% BSA in 1× TBS for 1 h at RT. F-actin was stained with phalloidin-Alexa 488 (1:40; Thermo Fisher Scientific, A12379) for 30 min, washed twice with 1× TBS, and then embedded in mounting medium with DAPI (Fluoromount-G with DAPI, Thermo Fisher Scientific, 00–4959-52). Samples were imaged on an LSM880 with Airyscan (Zeiss).
Live imaging of T cell actin dynamics was performed on an Andor Dragonfly 505 Spinning Disc setup. To determine the fraction of monofocal and multifocal T cell synapses in fixed or live samples, the fluorescence intensity profile of a line through the center of each synapse was plotted. Monofocal synapses were characterized by two distinct peaks, separated by a valley of very low fluorescence. Synapses with at least one additional peak in the valley with an intensity of at least 25% of the flanking peaks were assigned as multifocal.
In vitro T cell activation, proliferation, and IL-2 assays
The assays were performed in 96-well, round-bottom plates (TPP). 10,000 DCs and 50,000 T cells per well were mixed in 200 µl R10 medium in the presence of different concentrations of OVA323-339 peptide.
To assess T cell activation, 16–18 h after beginning the coculture, plates were spun down (350 g, 10 min, 4°C) and the supernatants were removed and snap-frozen. Cells were stained with Fc block and then with a mixture of anti-mouse CD62L-PE (1:1500; MEL-14), anti-mouse CD69-APC-eFluor780 (1:200; H1.2F3), and anti-mouse CD4-eFluor450 (1:800; GK1.5, all eBioscience) in FACS buffer.
To measure IL-2 production, cells were allowed to interact for 1 or 3 h, after which 10 µg/ml brefeldin A was added. After another 3 h to allow for cytokine production and accumulation, cells were stained with Fc block and a mixture of anti-mouse CD69-PE (1:200; H1.2F3) and anti-mouse CD4-eFluor450. Cells were then fixed, permeabilized, and stained for intracellular CD69 and IL-2 (anti-mouse IL-2-APC, 1:100; eB149/10H5) by using the BD Fix/Perm kit (554714) following the manufacturer’s instructions.
To measure T cell proliferation, T cells were stained with 5 µM CFSE (Thermo Fisher Scientific, C1157), as described previously (Quah et al., 2007). 96 h after beginning the coculture, cells were spun down as described above and stained for CD4. 7-Actinomycin D (7AAD) was used to distinguish live cells from dead cells. Data were recorded with a FACS Canto II machine (BD Biosciences), equipped with an automated high-throughput sampler. FlowJo (http://www.flowjo.com) was used for data analysis.
The frozen supernatants were used for IL-2 ELISAs according to the manufacturer’s protocol (Thermo Fisher Scientific, KMC0021).
In vivo T cell proliferation
CD45.1 “pepboy” mice were injected i.v. with 106 freshly isolated CD4/CD45.2 OTII T cells and stained with CFSE as described above. 24 h later, DCs preloaded with 1 µg/ml OVA323-339 peptide were injected into the hind footpad. As hem1−/− DCs have a disadvantage in reaching the draining lymph node compared with WT DCs (Leithner et al., 2016), we injected double the amount of hem1−/− DCs (4 × 105), which led to comparable cell numbers in the lymph node. 72 h after DC injection, mice were sacrificed and popliteal lymph nodes were isolated. Lymph nodes were torn open and digested for 30 min at 37°C in digestion buffer (DMEM + 2% FCS, 3 mg/ml collagenase IV, 40 µg/ml DNaseI, 3 mM CaCl2). Cells were filtered through a cell strainer and washed and stained in FACS buffer with anti-mouse CD4-eFluor450 (1:300; eBioscience, GK1.5) and anti-mouse CD45.2-APC (1:300; eBioscience, 104).
Intravital microscopy
DCs were preloaded with 2 µM OVA323-339 peptide for 45 min at 37°C/5% CO2. In the last 20 min of loading, hem1−/− DCs were labeled with 25 µM CMTMR (Thermo Fisher Scientific, C2927) and WT DCs with 20 µM CMAC (Thermo Fisher Scientific, C2110). Cells were washed once, mixed in a 2:1 ratio (hem1−/−:WT), and 1.5 × 106 cells in 20 µl were injected into the hock. At 18 h postinjection, 3 × 106 GFP-expressing OT-II T cells were injected i.v. in a volume of 100 µl. Reactive popliteal lymph nodes were imaged by intravital two-photon microscopy in 30-min intervals, 2–8 h after T cell transfer.
EM
Sapphire discs were coated with carbon land markings and incubated with 1× poly-L-lysine (Merck, P8920) in H2O overnight at 4°C. On the next day, discs were placed in a 12-well dish, rinsed twice with H2O, and dried for 4 h at RT. WT or hem1−/− DCs preloaded with 0.1 µg/ml OVA323-339 peptide for 2 h together with T cells were seeded in R10 medium and allowed to interact for 2 h. Samples were mildly prefixed with 1% PFA for 10 min and then immediately transferred to a Baltec HPM010 machine for high-pressure freezing. Freeze substitution was performed in a Leica automated freeze substitution machine according to the following protocol: 23.5 h at −82°C with acetone and 0.1% tannic acid, followed by a change to acetone, 1% osmium tetroxide, and 0.2% uranyl acetate, and incubation for another 7 h. In the next 1.5 h, the temperature was then raised to −60°C, held for 3 h, and then raised to −30°C over the course of 2 h. The temperature was then raised to 0°C. Embedding was started with acetone washes and infiltration of the samples with resin/propylene oxide mixtures with increasing amounts of resin. Samples were infiltrated with pure resin overnight and then allowed to polymerize at 60°C for 2–3 d. Serial sections of 70 nm were cut with an automated tape collecting ultra-microtome (ATUMtome, RMC) and placed on a waiver for semiautomated image collection with a Merlin compact VP field-emission scanning EM (Zeiss). The Fiji plugin TrakEM2 (Cardona et al., 2012) was used for image alignment. To quantify the distances between DC and T cell membranes, the T cell volume was segmented manually by using Microscopy Image Browser (Belevich et al., 2016). The DC membrane was drawn as a region of interest (ROI). A simple Fiji script was then used to calculate the binary distance map from the T cell and to evaluate the distances at the location of the DC membrane (ROI). The angle between DC and T cell membranes was quantified by using a custom script. To this end, the manually drawn ROI of the DC membrane, as well the manual segmentation of the T cell, was used. Contour lines of the T cell outline are constructed as the normal of the gradient of the distance map of the T cell segmentation. The freehand line of the ROI that marks the DC membrane is cleaned up and a cubic smoothing spline is fitted to it. For the entire length of the membrane, the angle between the contour line and the membrane was calculated at steps of 100 nm.
Western blots
106 wt and hem1−/− DCs were spun down and lysed in 50 µl 1× lysis buffer (1 ml 10× radioimmunoprecipitation assay (RIPA) buffer, New England Biolabs; 9 ml H2O, 1 tablet PhosStop, and 1 tablet protease inhibitor mini, both Merck). Lysates were spun at full speed for 10 min at 4°C and supernatants were transferred to new tubes. Equal volumes of 2× loading buffer (4× lithium dodecyl sulfate (LDS) buffer, 10× sample-reducing agent) were added. Two lanes were run for each WT or hem1−/− sample. Per lane, 15 µl were denatured at 99°C for 5 min. Samples were loaded on gels (NuPage 4–12% Bis-Tris Protein gels, Thermo Fisher Scientific) and run with 180 V. After protein transfer, membranes were cut in a way to allow for separate incubation with anti-ERM (1:1,000; Cell Signaling Technology, #3142), anti-pERM (1:1,000; Cell Signaling Technology, #3141), and anti-GAPDH (1:10,000; Abcam, ab125247) antibodies. Membranes were blocked with 5% BSA in 1× TBS with Tween 20 (TBS-T) for 1 h at RT, followed by incubation with the respective antibodies overnight at 4°C. Membranes were washed three times for 10 min with 1× TBS-T and then incubated with HRP-coupled secondary antibodies for 1 h at RT. Membranes were washed again three times with 1× TBS-T, incubated with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific), and imaged on an Amersham imager 600 (GE Healthcare).
Manufacturing and imaging in PDMS confiner
In brief, the imaging chamber consists of two glass surfaces that are spaced by PDMS micropillars. The two surfaces are pressed together by a PDMS piston that is glued onto the lid of a glass-bottom dish. The photomask design for the PDMS micropillars was drawn with Coreldraw X8 (Corel) and printed on an emulsion film transparency with a resolution of 8 mm (JD Photo Data and Photo Tools). The mold was produced by using photolithography on a silicon wafer. In brief, the wafer was coated with SU8-GM1050 (Gersteltec) at 2.120 rpm for 40 s to achieve a height of 4 µm. The wafer was soft-baked for 1 min at 120°C and for 5 min at 95°C, and then exposed to UV light at 1 mJ/cm2 for 10 min by using a beam-expanded 365-nm UV LED (Thorlabs, M365L2-C1-UV). After UV exposure, the wafer was postbaked for 1 min at 65°C and 5 min at 95°C. The wafer was developed in an SU8 developer for 17 s and then silanized with trichloro (1H, 1H, 2H, 2H-perfluorooctyl) silane in a vacuum desiccator for 1 h. Micropillars were then produced by mixing silicone elastomer and curing reagent (PDMS Sylgard 184 Elastomere Kit, Dow Corning) in a 7:1 ratio. The mixture was then degassed by using a planetary centrifugal mixer (ARE250, Thinky) and carefully poured onto the wafer. Round cover glasses (#1, 12-mm diameter, Mentzel, Thermo Fisher Scientific) were plasma activated for 2 min (Plasma Cleaner, Harrick Plasma) and placed on the wafer with the activated surface facing the elastomere/curing agent mixture. The wafer was cured on a heating plate for 15 min at 95°C, and the micropillar-coated cover glasses were carefully removed with a sharp razor blade and isopropanol.
To produce the PDMS piston, silicone elastomer and curing reagent were mixed in a 30:1 ratio, degassed as described before, and poured into an aluminum mold with the needed dimensions. The PDMS pistons were cured for 6 h at 80°C, removed with isopropanol, and then glued into the middle of a 60 × 15-mm nonpyrogenic polystyrene tissue culture dish (Falcon) by using aquarium sealant (Marina). A hole with a 17-mm diameter was drilled into the center bottom of a 60 × 15-mm tissue culture dish (Falcon), and a glass slide (#2, 22 × 22 mm, Mentzel, Thermo Fisher Scientific) was glued onto the hole with aquarium sealant.
To assemble the device, a micropillar-bearing cover glass was mounted onto the PDMS piston with the micropillars facing upward. DCs and T cells were mixed in a volume of 5 µl that was then carefully pipetted onto the micropillar. The PDMS piston with the micropillar and cell mixture was then pressed onto the glass slide in the tissue culture dish that was then sealed with strong tape. Dishes were incubated for 1 h at 37°C/5% CO2 before imaging was performed on an inverted confocal microscope (Zeiss) equipped with a spinning-disc system (Andor, iXon897), a Plan-Apochromat 100×/1.4 oil objective (Zeiss), and 488 and 561 lasers.
Still images of the synapse were obtained by z-stacking in 27 ∼300-nm intervals. The Huygens Deconvolution Software package (https://svi.nl/Huygens-Deconvolution) was used for deconvolution according to the manufacturer’s manual. The synaptic plane was then identified as the one with the highest Lifeact-eGFP signal and, together with one plane above and below, was used to obtain maximum intensity projections that were used for further analysis. Live cell imaging was performed by recording three, ∼133-nm-spaced z-planes at the synaptic interface in 5-s intervals for 3 min. Maximum intensity projections were used for further analysis. To quantify F-actin and its dynamics, we first segmented the actin-rich area in the synapse by using ilastik (Berg et al., 2019). The segmentation was manually corrected and used to extract area and intensity measures from the raw data. All intensity measures were background corrected. Area and intensity measures were normalized to the respective mean of WT DCs. To quantify F-actin dynamics, single pixels were automatically removed. As a measure of how much the actin segmentation changes from frame to frame, we calculated the Jaccard similarity coefficient. It is given by the overlap of the segmentation area of the current frame with the segmentation of the previous frame divided by the area of the union of both. Hence, a completely static synapse would lead to a Jaccard coefficient of 1 and a lower number indicates a more dynamic synapse. To determine the clustering of Abi1 and WASP, their respective maximum intensity at the synapse was divided by the mean intensity in the cell body. The resulting deviation was used as proxy for synaptic clustering.
Membrane tension measurements
For membrane tension measurements, custom-made chambers were coated with 1× poly-L-lysine (Merck, P8920) in H2O. Cells were plated on each dish in R10 media and allowed to adhere for 10 min at 37°C. Cells were washed and probed in RPMI with 2% FBS at 30°C. Olympus BioLevers (k = 60 pN/nm) were calibrated by using the thermal noise method and incubated in 2.5 mg/ml concanavalin A (Sigma-Aldrich, C5275) for 1 h at RT. Before the measurements, cantilevers were rinsed in Dulbecco’s phosphate-buffered saline (DPBS). Cells were located by brightfield imaging, and the cantilever was positioned at any location over the cell for tether measurement. Cells were not used longer than 1 h for data acquisition. Tethers were pulled by using a Bruker Catalyst AFM controlled by custom-made LabVIEW software mounted on an inverted Zeiss fluorescent microscope. Approach velocity was set to 1 µm/s, contact force to 100 pN, contact time to 5–10 s, and retraction speed to 10 µm/s. We used the signature of tether breaking in the AFM traces to identify cells that have multiple tethers by visualizing multiple steps in the force trace—these cells with multiple tethers were excluded from analysis. After a 10-µm tether was pulled, the cantilever position was held constant until it broke. Only tethers that broke in less than 15 s were considered as actin polymerized inside longer-lived tethers. Resulting force–time curves were analyzed with the Kerssemakers algorithm (Kerssemakers et al., 2006) kindly provided by Jacob Kerssemakers.
Statistics and data analysis
All data were analyzed by using GraphPad Prism 8.
Online supplemental material
Fig. S1 shows supporting data for Fig. 1 B. Fig. S2 shows supporting data for Fig. 1, C and D and Fig. 2. Fig. S3 shows supporting data for Fig. 2 as well as the Cd11c/MHC II profile of immature and mature WT and hem1−/− DCs. Fig. S4 shows supporting data for Figs. 4, 5, and 6. Video 1 corresponds to Fig. 1 C and shows phalloidin stainings in DC–T cell conjugates under the indicated treatment. Video 2 corresponds to Fig. S1, A–C, and shows Lifeact-eGFP expression in T cells that formed conjugates with DCs under the indicated treatment. Videos 3 and 6 correspond to Fig. 2, A–E, and show Lifeact-eGFP expression in WT and hem1−/− DCs that formed synapses with T cells under confinement. Videos 4 and 5 correspond to Fig. S2, D–E, and show WASP- and Abi1-eGFP expression in DCs that formed synapses with T cells under confinement. Video 7 corresponds to Fig. 2, F–H, and shows live imaging of Lifeact-eGFP–expressing DCs forming synapses with T cells under confinement. Video 8 corresponds to Fig. 4 C and shows live imaging of WT and hem1−/− DCs forming conjugates with T cells. Video 9 corresponds to Fig. 4, D–G, and shows z-stacks through EM samples of WT and hem1−/− DCs that formed synapses with T cells. Video 10 corresponds to Fig. 6, A–C, and shows intravital microscopy of WT and hem1−/− DCs that form conjugates with T cells.
Acknowledgments
We thank the Electron microscopy facility, the Bioimaging facility, the Pre-clinical facility, and the Life Science facility of Institute of Science and Technology Austria for excellent support.
This work was supported by European Research Council grant ERC CoG 724373 and Austrian Science Fund grant FWF P29911 (to M. Sixt). K. Rottner and T.E.B. Stradal are funded by the Deutsche Forschungsgemeinschaft (PROCOMPAS GRK2223/1) and the Helmholtz Association (HGF impulse fund W2/W3-066).
The authors declare no competing financial interests.
Author contributions: A. Leithner: conceptualization, data curation, formal analysis, investigation, methodology, project administration, validation, visualization, writing—original draft, review, and editing; L.M. Altenburger: formal analysis, investigation; R. Hauschild: data curation, formal analysis, methodology, software; F.P. Assen: investigation; K. Rottner: writing—review and editing; T.E.B. Stradal: resources, writing—review and editing; A. Diz-Munoz: formal analysis, investigation; J.V. Stein: resources, writing—review and editing; M. Sixt: conceptualization, funding acquisition, project administration, supervision, writing—original draft, review, and editing.
References
- Abraham, C., Griffith J., and Miller J.. 1999. The dependence for leukocyte function-associated antigen-1/ICAM-1 interactions in T cell activation cannot be overcome by expression of high density TCR ligand. J. Immunol. 162:4399–4405. [PubMed] [Google Scholar]
- Al-Alwan, M., and Rowden G.. 2001. Cutting edge: the dendritic cell cytoskeleton is critical for the formation of the immunological synapse. J. Immunol. 166:1452–1456. 10.4049/jimmunol.166.3.1452 [DOI] [PubMed] [Google Scholar]
- Alekhina, O., Burstein E., and Billadeau D.D.. 2017. Cellular functions of WASP family proteins at a glance. J. Cell Sci. 130:2235–2241. 10.1242/jcs.199570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babich, A., Li S., O’Connor R.S., Milone M.C., Freedman B.D., and Burkhardt J.K.. 2012. F-actin polymerization and retrograde flow drive sustained PLCγ1 signaling during T cell activation. J. Cell Biol. 197:775–787. 10.1083/jcb.201201018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkow, S., Heinz S., Schmidbauer P., Kolanus W., Holzmann B., Grabbe S., and Laschinger M.. 2010. LFA-1 activity state on dendritic cells regulates contact duration with T cells and promotes T-cell priming. Blood. 116:1885–1894. 10.1182/blood-2009-05-224428 [DOI] [PubMed] [Google Scholar]
- Banchereau, J., and Steinman R.M.. 1998. Dendritic cells and the control of immunity. Nature. 392:245–252. 10.1038/32588 [DOI] [PubMed] [Google Scholar]
- Barnden, M.J., Allison J., Heath W.R., and Carbone F.R.. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34–40. 10.1046/j.1440-1711.1998.00709.x [DOI] [PubMed] [Google Scholar]
- Belevich, I., Joensuu M., Kumar D., Vihinen H., and Jokitalo E.. 2016. Microscopy Image Browser: A Platform for Segmentation and Analysis of Multidimensional Datasets. PLoS Biol. 14:e1002340. . 10.1371/journal.pbio.1002340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuti, F., Hugues S., Walmsley M., Ruf S., Fetler L., Popoff M., and Tybulewicz V.L.J.. 2004. Requirement of Rac1 and Rac2 Expression by Mature Dendritic Cells for T Cell Priming. Science. 305:1150–1153. [DOI] [PubMed] [Google Scholar]
- Berg, S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M., et al. 2019. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 16:1226–1232. 10.1038/s41592-019-0582-9 [DOI] [PubMed] [Google Scholar]
- Blumenthal, D., Chandra V., Avery L., and Burkhardt J.K.. 2020. Mouse T cell priming is enhanced by maturation-dependent stiffening of the dendritic cell cortex. eLife. 9:e55995. 10.7554/eLife.55995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma, G., Mendoza-Naranjo A., Blundell M.P., de Falco E., Parsley K.L., Burns S.O., and Thrasher A.J.. 2011. Cytoskeletal remodeling mediated by WASp in dendritic cells is necessary for normal immune synapse formation and T-cell priming. Blood. 118:2492–2501. 10.1182/blood-2011-03-340265 [DOI] [PubMed] [Google Scholar]
- Bousso, P. 2008. T-cell activation by dendritic cells in the lymph node: lessons from the movies. Nat. Rev. Immunol. 8:675–684. 10.1038/nri2379 [DOI] [PubMed] [Google Scholar]
- Brossard, C., Feuillet V., Schmitt A., Randriamampita C., Romao M., Raposo G., and Trautmann A.. 2005. Multifocal structure of the T cell - dendritic cell synapse. Eur. J. Immunol. 35:1741–1753. 10.1002/eji.200425857 [DOI] [PubMed] [Google Scholar]
- Cai, E., Marchuk K., Beemiller P., Beppler C., Rubashkin M.G., Weaver V.M., Gérard A., Liu T.-L., Chen B.-C., Betzig E., et al. 2017. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science. 356:eaal3118. 10.1126/science.aal3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona, A., Saalfeld S., Schindelin J., Arganda-Carreras ., Preibisch S., Longair M., Tomancak P., Hartenstein V., and Douglas R.J.. 2012. TrakEM2 software for neural circuit reconstruction. PLoS One. 7:e38011. . 10.1371/journal.pone.0038011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Lou J., and Zhu C.. 2010. Forcing switch from short- to intermediate- and long-lived states of the alphaA domain generates LFA-1/ICAM-1 catch bonds. J. Biol. Chem. 285:35967–35978. 10.1074/jbc.M110.155770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comrie, W.A., and Burkhardt J.K.. 2016. Action and traction: Cytoskeletal control of receptor triggering at the immunological synapse. Front. Immunol. 7:68. 10.3389/fimmu.2016.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comrie, W.A., Li S., Boyle S., and Burkhardt J.K.. 2015. The dendritic cell cytoskeleton promotes T cell adhesion and activation by constraining ICAM-1 mobility. J. Cell Biol. 208:457–473. 10.1083/jcb.201406120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, V., Nal B., Dujeancourt A., Thoulouze M.I., Galli T., Roux P., Dautry-Varsat A., and Alcover A.. 2004. Activation-induced polarized recycling targets T cell antigen receptors to the immunological synapse; involvement of SNARE complexes. Immunity. 20:577–588. 10.1016/S1074-7613(04)00106-2 [DOI] [PubMed] [Google Scholar]
- Doty, R.T., and Clark E.A.. 1998. Two regions in the CD80 cytoplasmic tail regulate CD80 redistribution and T cell costimulation. J. Immunol. 161:2700–2707. [PubMed] [Google Scholar]
- Dustin, M.L. 2008. Hunter to gatherer and back: immunological synapses and kinapses as variations on the theme of amoeboid locomotion. Annu. Rev. Cell Dev. Biol. 24:577–596. 10.1146/annurev.cellbio.24.110707.175226 [DOI] [PubMed] [Google Scholar]
- Dustin, M.L., Bromley S.K., Kan Z., Peterson D.A., and Unanue E.R.. 1997. Antigen receptor engagement delivers a stop signal to migrating T lymphocytes. Proc. Natl. Acad. Sci. USA. 94:3909–3913. 10.1073/pnas.94.8.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin, M.L., Olszowy M.W., Holdorf A.D., Li J., Bromley S., Desai N., Widder P., Rosenberger F., van der Merwe P.A., Allen P.M., and Shaw A.S.. 1998. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 94:667–677. 10.1016/S0092-8674(00)81608-6 [DOI] [PubMed] [Google Scholar]
- Dustin, M.L., Tseng S.-Y., Varma R., and Campi G.. 2006a. T cell-dendritic cell immunological synapses. Curr. Opin. Immunol. 18:512–516. 10.1016/j.coi.2006.05.017 [DOI] [PubMed] [Google Scholar]
- Feigelson, S.W., Pasvolsky R., Cemerski S., Shulman Z., Grabovsky V., Ilani T., Sagiv A., Lemaitre F., Laudanna C., Shaw A.S., and Alon R.. 2010. Occupancy of lymphocyte LFA-1 by surface-immobilized ICAM-1 is critical for TCR- but not for chemokine-triggered LFA-1 conversion to an open headpiece high-affinity state. J. Immunol. 185:7394–7404. 10.4049/jimmunol.1002246 [DOI] [PubMed] [Google Scholar]
- Feigelson, S.W., Solomon A., Biram A., Hatzav M., Lichtenstein M., Regev O., Kozlovski S., Varol D., Curato C., Leshkowitz D., et al. 2018. ICAMs Are Not Obligatory for Functional Immune Synapses between Naive CD4 T Cells and Lymph Node DCs. Cell Rep. 22:849–859. 10.1016/j.celrep.2017.12.103 [DOI] [PubMed] [Google Scholar]
- Fisher, P.J., Bulur P.A., Vuk-Pavlovic S., Prendergast F.G., and Dietz A.B.. 2008. Dendritic cell microvilli: a novel membrane structure associated with the multifocal synapse and T-cell clustering. Blood. 112:5037–5045. 10.1182/blood-2008-04-149526 [DOI] [PubMed] [Google Scholar]
- Freeman, S.A., Vega A., Riedl M., Collins R.F., Ostrowski P.P., Woods E.C., Bertozzi C.R., Tammi M.I., Lidke D.S., Johnson P., et al. 2018. Transmembrane Pickets Connect Cyto- and Pericellular Skeletons Forming Barriers to Receptor Engagement. Cell. 172:305–317.e10. 10.1016/j.cell.2017.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl, P., den Boer A.T., and Gunzer M.. 2005. Tuning immune responses: diversity and adaptation of the immunological synapse. Nat. Rev. Immunol. 5:532–545. 10.1038/nri1647 [DOI] [PubMed] [Google Scholar]
- Girard, T., El-Far M., Gaucher D., Acuto O., Beaulé G., Michel F., Mourad W., and Sékaly R.P.. 2012. A conserved polylysine motif in CD86 cytoplasmic tail is necessary for cytoskeletal association and effective co-stimulation. Biochem. Biophys. Res. Commun. 423:301–307. 10.1016/j.bbrc.2012.05.116 [DOI] [PubMed] [Google Scholar]
- Grakoui, A., Bromley S.K., Sumen C., Davis M.M., Shaw A.S., Allen P.M., and Dustin M.L.. 1999. The immunological synapse: a molecular machine controlling T cell activation. Science . 285:221–227. 10.1126/science.285.5425.221 [DOI] [PubMed] [Google Scholar]
- Gunzer, M., Schäfer A., Borgmann S., Grabbe S., Zänker K.S., Bröcker E.B., Kämpgen E., and Friedl P.. 2000. Antigen presentation in extracellular matrix: interactions of T cells with dendritic cells are dynamic, short lived, and sequential. Immunity. 13:323–332. 10.1016/S1074-7613(00)00032-7 [DOI] [PubMed] [Google Scholar]
- Gunzer, M., Weishaupt C., Hillmer A., Basoglu Y., Friedl P., Dittmar K.E., Kolanus W., Varga G., and Grabbe S.. 2004. A spectrum of biophysical interaction modes between T cells and different antigen-presenting cells during priming in 3-D collagen and in vivo. Blood. 104:2801–2809. 10.1182/blood-2004-03-1193 [DOI] [PubMed] [Google Scholar]
- Hori, M., Saito S., Shin Y.Z., Ozaki H., Fusetani N., and Karaki H.. 1993. Mycalolide-B, a novel and specific inhibitor of actomyosin ATPase isolated from marine sponge. FEBS Lett. 322:151–154. 10.1016/0014-5793(93)81557-G [DOI] [PubMed] [Google Scholar]
- Jung, Y., Riven I., Feigelson S.W., Kartvelishvily E., Tohya K., Miyasaka M., Alon R., and Haran G.. 2016. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc. Natl. Acad. Sci. USA. 113:E5916–E5924. 10.1073/pnas.1605399113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandula, S., and Abraham C.. 2004. LFA-1 on CD4+ T cells is required for optimal antigen-dependent activation in vivo. J. Immunol. 173:4443–4451. 10.4049/jimmunol.173.7.4443 [DOI] [PubMed] [Google Scholar]
- Kerssemakers, J.W.J., Munteanu E.L., Laan L., Noetzel T.L., Janson M.E., and Dogterom M.. 2006. Assembly dynamics of microtubules at molecular resolution. Nature. 442:709–712. . 10.1038/nature04928 [DOI] [PubMed] [Google Scholar]
- Kinashi, T. 2005. Intracellular signalling controlling integrin activation in lymphocytes. Nat. Rev. Immunol. 5:546–559. 10.1038/nri1646 [DOI] [PubMed] [Google Scholar]
- Kong, F., García A.J., Mould A.P., Humphries M.J., and Zhu C.. 2009. Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 185:1275–1284. 10.1083/jcb.200810002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovski, S., Atrakchi O., Feigelson S.W., Shulman Z., and Alon R.. 2019. Stable contacts of naïve CD4 T cells with migratory dendritic cells are ICAM-1-dependent but dispensable for proliferation in vivo. Cell Adhes. Migr. 13:314–321. 10.1080/19336918.2019.1644857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, S., Depoil D., Martinelli R., Judokusumo E., Carmona G., Gertler F.B., Kam L.C., Carman C.V., Burkhardt J.K., Irvine D.J., and Dustin M.L.. 2015. Actin foci facilitate activation of the phospholipase C-γ in primary T lymphocytes via the WASP pathway. eLife. 4:e04953. 10.7554/eLife.04953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Berre, M., Zlotek-Zlotkiewicz E., Bonazzi D., Lautenschlaeger F., and Piel M.. 2014. Methods for two-dimensional cell confinement. Methods Cell Biol. 121:213-229. [DOI] [PubMed] [Google Scholar]
- Leithner, A., Eichner A., Müller J., Reversat A., Brown M., Schwarz J., Merrin J., de Gorter D.J.J., Schur F., Bayerl J., et al. 2016. Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nat. Cell Biol. 18:1253–1259. 10.1038/ncb3426 [DOI] [PubMed] [Google Scholar]
- Lutz, M.B., Kukutsch N., Ogilvie A.L., Rössner S., Koch F., Romani N., and Schuler G.. 1999. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 223:77–92. . 10.1016/s0022-1759(98)00204-x [DOI] [PubMed] [Google Scholar]
- Malinova, D., Fritzsche M., Nowosad C.R., Armer H., Munro P.M.G., Blundell M.P., Charras G., Tolar P., Bouma G., and Thrasher A.J.. 2016. WASp-dependent actin cytoskeleton stability at the dendritic cell immunological synapse is required for extensive, functional T cell contacts. J. Leukoc. Biol. 99:699–710. 10.1189/jlb.2A0215-050RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks, C.R.F., Freiberg B.A., Kupfer H., Sciaky N., and Kupfer A.. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 395:82–86. 10.1038/25764 [DOI] [PubMed] [Google Scholar]
- Mossman, K.D., Campi G., Groves J.T., and Dustin M.L.. 2005. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 310:1191–1193. 10.1126/science.1119238 [DOI] [PubMed] [Google Scholar]
- Nolz, J.C., Gomez T.S., Zhu P., Li S., Medeiros R.B., Shimizu Y., Burkhardt J.K., Freedman B.D., and Billadeau D.D.. 2006. The WAVE2 complex regulates actin cytoskeletal reorganization and CRAC-mediated calcium entry during T cell activation. Curr. Biol. 16:24–34. 10.1016/j.cub.2005.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H., Staehling-Hampton K., Appleby M.W., Brunkow M.E., Habib T., Zhang Y., Ramsdell F., Liggitt H.D., Freie B., Tsang M., et al. 2008. A point mutation in the murine Hem1 gene reveals an essential role for Hematopoietic protein 1 in lymphopoiesis and innate immunity. J. Exp. Med. 205:2899–2913. 10.1084/jem.20080340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, O.D., Mitchell D., Jager G.C., South S., Murriel C., McBride J., Herzenberg L.A., Kinoshita S., and Nolan G.P.. 2003. Leukocyte functional antigen 1 lowers T cell activation thresholds and signaling through cytohesin-1 and Jun-activating binding protein 1. Nat. Immunol. 4:1083–1092. 10.1038/ni984 [DOI] [PubMed] [Google Scholar]
- Pulecio, J., Tagliani E., Scholer A., Prete F., Fetler L., Burrone O.R., and Benvenuti F.. 2008. Expression of Wiskott-Aldrich syndrome protein in dendritic cells regulates synapse formation and activation of naive CD8+ T cells. J. Immunol. 181:1135–1142. [DOI] [PubMed] [Google Scholar]
- Quah, B.J.C., Warren H.S., and Parish C.R.. 2007. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nat Protoc. 2:2049–2056. . 10.1038/nprot.2007.296 [DOI] [PubMed] [Google Scholar]
- Riedl, J., Flynn K.C., Raducanu A., Gärtner F., Beck G., Bösl M., Bradke F., Massberg S., Aszodi A., Sixt M., et al. 2010. Lifeact mice for studying F-actin dynamics. Nat Methods. 7:168–169. . 10.1038/nmeth0310-168 [DOI] [PubMed] [Google Scholar]
- Sage, P.T., Varghese L.M., Martinelli R., Sciuto T.E., Kamei M., Dvorak A.M., Springer T.A., Sharpe A.H., and Carman C.V.. 2012. Antigen recognition is facilitated by invadosome-like protrusions formed by memory/effector T cells. J. Immunol. 188:3686–3699. 10.4049/jimmunol.1102594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, S., Watabe S., Ozaki H., Fusetani N., and Karaki H.. 1994. Mycalolide B, a novel actin depolymerizing agent. J. Biol. Chem. 269:29710–29714. 10.1016/S0021-9258(18)43938-5 [DOI] [PubMed] [Google Scholar]
- Scholer, A., Hugues S., Boissonnas A., Fetler L., and Amigorena S.. 2008. Intercellular adhesion molecule-1-dependent stable interactions between T cells and dendritic cells determine CD8+ T cell memory. Immunity. 28:258–270. 10.1016/j.immuni.2007.12.016 [DOI] [PubMed] [Google Scholar]
- Schürpf, T., and Springer T.A.. 2011. Regulation of integrin affinity on cell surfaces. EMBO J. 30:4712–4727. 10.1038/emboj.2011.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, A.S., and Dustin M.L.. 1997. Making the T cell receptor go the distance: a topological view of T cell activation. Immunity. 6:361–369. 10.1016/S1074-7613(00)80279-4 [DOI] [PubMed] [Google Scholar]
- Shen, K., Thomas V.K., Dustin M.L., and Kam L.C.. 2008. Micropatterning of costimulatory ligands enhances CD4+ T cell function. Proc. Natl. Acad. Sci. USA. 105:7791–7796. 10.1073/pnas.0710295105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka, D.K., Bruniquel D., Schwartz R.H., and Singh N.J.. 2004. IL-2 secretion by CD4+ T cells in vivo is rapid, transient, and influenced by TCR-specific competition. J. Immunol. 172:6136–6143. 10.4049/jimmunol.172.10.6136 [DOI] [PubMed] [Google Scholar]
- Spolski, R., Li P., and Leonard W.J.. 2018. Biology and regulation of IL-2: from molecular mechanisms to human therapy. Nat. Rev. Immunol. 18:648–659. 10.1038/s41577-018-0046-y [DOI] [PubMed] [Google Scholar]
- Takenawa, T., and Suetsugu S.. 2007. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8:37–48. 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Tseng, S.-Y., Waite J.C., Liu M., Vardhana S., and Dustin M.L.. 2008. T cell-dendritic cell immunological synapses contain TCR-dependent CD28-CD80 clusters that recruit protein kinase C θ. J. Immunol. 181:4852–4863. 10.4049/jimmunol.181.7.4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H., Morphew M.K., McIntosh J.R., and Davis M.M.. 2011. CD4+ T-cell synapses involve multiple distinct stages. Proc. Natl. Acad. Sci. USA. 108:17099–17104. 10.1073/pnas.1113703108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaahtomeri, K., Brown M., Hauschild R., De Vries I., Leithner A.F., Mehling M., Kaufmann W.A., and Sixt M.. 2017. Locally Triggered Release of the Chemokine CCL21 Promotes Dendritic Cell Transmigration across Lymphatic Endothelia. Cell Rep. 19:902–909. 10.1016/j.celrep.2017.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, G., Balkow S., Wild M.K., Stadtbaeumer A., Krummen M., Rothoeft T., Higuchi T., Beissert S., Wethmar K., Scharffetter-Kochanek K., et al. 2007. Active MAC-1 (CD11b/CD18) on DCs inhibits full T-cell activation. Blood. 109:661–669. 10.1182/blood-2005-12-023044 [DOI] [PubMed] [Google Scholar]
- Varma, R., Campi G., Yokosuka T., Saito T., and Dustin M.L.. 2006. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 25:117–127. 10.1016/j.immuni.2006.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernimont, S.A., Wiemer A.J., Bennin D.A., Monkley S.J., Ludwig T., Critchley D.R., and Huttenlocher A.. 2011. Contact-dependent T cell activation and T cell stopping require talin1. J. Immunol. 187:6256–6267. 10.4049/jimmunol.1102028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.W., Ballantyne C.M., Smith C.W., Montgomery C., Bradley A., O’Brien W.E., and Beaudet A.L.. 1993. Gene targeting yields a CD18-mutant mouse for study of inflammation. J Immunol. 151:1571–1578. . [PubMed] [Google Scholar]
- Yi, J., Wu X.S., Crites T., and Hammer J.A. III. 2012. Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Mol. Biol. Cell. 23:834–852. 10.1091/mbc.e11-08-0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Luo B.H., Xiao T., Zhang C., Nishida N., and Springer T.A.. 2008. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. 32:849–861. 10.1016/j.molcel.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]