Abstract
Cercospora beticola is a hemibiotrophic fungus that causes cercospora leaf spot disease of sugar beet (Beta vulgaris). After an initial symptomless biotrophic phase of colonization, necrotic lesions appear on host leaves as the fungus switches to a necrotrophic lifestyle. The phytotoxic secondary metabolite cercosporin has been shown to facilitate fungal virulence for several Cercospora spp. However, because cercosporin production and subsequent cercosporin‐initiated formation of reactive oxygen species is light‐dependent, cell death evocation by this toxin is only fully ensured during a period of light. Here, we report the discovery of the effector protein CbNip1 secreted by C. beticola that causes enhanced necrosis in the absence of light and, therefore, may complement light‐dependent necrosis formation by cercosporin. Infiltration of CbNip1 protein into sugar beet leaves revealed that darkness is essential for full CbNip1‐triggered necrosis, as light exposure delayed CbNip1‐triggered host cell death. Gene expression analysis during host infection shows that CbNip1 expression is correlated with symptom development in planta. Targeted gene replacement of CbNip1 leads to a significant reduction in virulence, indicating the importance of CbNip1 during colonization. Analysis of 89 C. beticola genomes revealed that CbNip1 resides in a region that recently underwent a selective sweep, suggesting selection pressure exists to maintain a beneficial variant of the gene. Taken together, CbNip1 is a crucial effector during the C. beticola–sugar beet disease process.
Keywords: Cercospora beticola, necrosis‐inducing effector, virulence factor
The fungus Cercospora beticola secretes necrosis‐inducing molecules during infection of sugar beet that are optimized to be fully functional during the day or night.

1. INTRODUCTION
Cercospora leaf spot (CLS) disease is considered one of the most destructive foliar diseases of sugar beet worldwide (Rangel et al., 2020). The causal agent of CLS is the hemibiotrophic fungus Cercospora beticola, which belongs to the Dothideomycete class (Bolton et al., 2012). In the field, C. beticola overwinters as stromata, which serves as the primary inoculum in the following growing season (Khan & Khan, 2010; Rangel et al., 2020; Solel & Minz, 1971). Once infection is established, airborne conidia can be quickly dispersed throughout the field by wind, rain, and insect transfer (Khan & Khan, 2010; Rangel et al., 2020). On landing on a sugar beet leaf, spores germinate and grow towards stomata, where they form appressoria (Feindt et al., 1981; Rangel et al., 2020; Rathaiah, 1977). These hyphal structures enable the fungus to penetrate and enter the apoplast (Steinkamp et al., 1979). Once inside the host, C. beticola grows intercellularly and colonizes the mesophyll for up to 10 days (Rangel et al., 2020). During these early stages of infection, C. beticola lives a biotrophic lifestyle. However, unknown conditions trigger hemibiotrophic fungi to switch from a biotrophic to a necrotrophic lifestyle in which host cell death occurs to complete their life cycle (Horbach et al., 2011; Steinkamp et al., 1979).
Necrosis‐inducing molecules come in many forms and with various modes of actions. For example, necrotrophic effectors, also known as proteinaceous host‐selective toxins, depend on the presence of a corresponding target encoded by a susceptibility gene in their host to elicit host cell death (Faris et al., 2010; Friesen et al., 2007, 2008; Shi et al., 2016). This interaction is essentially the classic gene‐for‐gene interaction (Flor, 1942), but instead of providing resistance to the fungus, host cell death serves the necrotrophic needs of the fungus. Therefore, this interaction is also referred to as an inverse gene‐for‐gene interaction with host and pathogen traits that coevolve to avoid infection and obtain nutrients, respectively (Friesen et al., 2007). For example, the necrotrophic effector SnTox1 of the wheat pathogen Parastagonospora nodorum interacts with Snn1 encoded by a wheat receptor kinase gene, which activates programmed cell death in the host and facilitates a compatible interaction (Liu et al., 2004, 2012; Shi et al., 2016). However, not all necrosis‐inducing effectors are dependent on a host receptor to provoke host cell death. A family of Nep1‐like proteins (NLPs) has been identified in several oomycetes, fungi, and bacteria that elicit a hypersensitive response‐like host necrosis (Gijzen & Nürnberger, 2006; Pemberton & Salmond, 2004). The first family member discovered was Nep1 (necrosis‐ and ethylene‐inducing protein 1), a protein secreted by Fusarium oxysporum that was shown to trigger necrosis and ethylene production in Erythroxylum coca (coca plant) (Bailey, 1995). Besides high sequence homology, NLPs share a common necrosis‐inducing Phytophthora protein (NPP1) domain (Fellbrich et al., 2002). Motteram et al. (2009) reported an NPP1 domain‐carrying phytotoxic effector called MgNlp that is expressed during infection of the hemibiotrophic pathogen Zymoseptoria tritici, the causal agent of septoria tritici blotch on wheat. Furthermore, necrosis‐inducing activity was described as selective because MgNlp induced cell death in Arabidopsis and tobacco but not in wheat. Interestingly, targeted gene replacement of MgNlp did not affect fungal virulence in inoculation studies of susceptible wheat lines (Motteram et al, 2009). Additionally, analysis of Z. tritici culture filtrates led to the discovery of two light‐dependent phytotoxic proteins, ZtNip1 and ZtNip2, whose activities resemble those of host‐specific toxins (Ben M’Barek et al., 2015). While ZtNip1 displays homology to the Cladosporium fulvum effector protein Ecp2, which is known to elicit cell death in tomato and tobacco harbouring the Cf‐Ecp2 resistance gene (Laugé et al., 1998), ZtNip2 was identified to contain a putative MD‐2‐related lipid‐recognition domain, hinting at the ability to bind lipids that may have a potential role in innate immunity (Inohara & Nuñez, 2002; Mullen et al., 2003). Furthermore, the onset of ZtNip1 expression during infection matched with necrotic symptom development in planta (Ben M’Barek et al., 2015). Recently, the functional ribonuclease Zt6 was discovered in Z. tritici that targets not only plant but also mammalian ribosomal RNA for cleavage in vitro, a feature that makes it highly toxic to wheat, tobacco, bacterial, and yeast cells (Kettles et al., 2018). Intriguingly, the gene expression pattern of Zt6 during infection is marked by a double expression peak. The first boost in expression occurs at 1 day postinfection, followed by down‐regulation during the biotrophic life cycle phase. With onset of the necrotrophic phase at 14 days postinoculation, however, Zt6 gene expression increases again (Kettles et al., 2018).
Besides proteinaceous necrosis‐inducing agents, secondary metabolite effectors have also been reported to elicit cell death in their host. C. beticola is a producer of cercosporin and beticolin, two well‐known phytotoxic secondary metabolite effectors. Both toxins are only active in the presence of light and show no host specificity (Daub & Ehrenshaft, 2000; de Jonge et al., 2018; Schlösser, 1962). In multiple Cercospora species, targeted gene disruption mutants are unable to produce cercosporin and display reduced virulence, which underlines the importance of necrosis induction for the infection process in this genus (Callahan et al., 1999; Choquer et al., 2005). However, no proteinaceous phytotoxin has been reported for C. beticola to our knowledge. In this study, we describe the identification of the first proteinaceous C. beticola virulence factor. This protein can induce host cell death in the dark and therefore can complement the light‐dependent phytotoxins cercosporin and beticolin. Finally, we find evidence that the gene encoding this virulence factor has evolved under positive selection, possibly reflecting a role in an evolutionary host–pathogen arms race.
2. RESULTS
2.1. Necrosis‐inducing activity of C. beticola culture filtrate
Due to the hemibiotrophic lifestyle of C. beticola, we hypothesized that the fungus secretes effector proteins during infection that facilitate disease by causing necrosis and at least a portion of these are produced during in vitro growth. Therefore, we cultured C. beticola in different media (potato dextrose broth [PDB] and Fries medium) and under different conditions (e.g., shaking at 120 rpm or still cultures that were grown without movement, and sampling time points at 3, 5, 7, 12, and 14 days after medium inoculation) in attempts to identify an in vitro condition in which effector proteins were produced (Figure 1a,b). All culture conditions were tested for the presence of necrosis‐inducing activity by infiltrating culture filtrate into sugar beet leaves (Figure 1a) Ultimately, infiltration of culture filtrate of C. beticola grown in Fries medium for 7 days, shaking at 120 rpm, caused clear and repeatable necrosis of the host tissue (Figure 1a). Within 24 hr, the host cells within the infiltration zone had entirely collapsed while cells outside the area remained apparently unharmed. Because C. beticola is known to produce cercosporin and beticolin, which are phytotoxic (Daub & Ehrenshaft, 2000; Schlösser, 1962), culture filtrate was treated with a protease mixture to rule out the involvement of phytotoxic secondary metabolites for this necrosis formation. Treatment with proteases abolished necrosis, confirming that the necrosis‐inducing activity can be attributed to a proteinaceous component of the culture filtrate (Figure 1a). To single out the protein responsible for the necrotic phenotype, the active culture filtrate was fractionated using ion exchange chromatography and single fractions were screened for necrosis‐inducing activity by individual infiltration into sugar beet leaves (Figure 1b). The fraction that reproducibly caused necrosis was selected for protein identification using tandem mass spectrometry (MS/MS) analysis (Figure S1).
FIGURE 1.
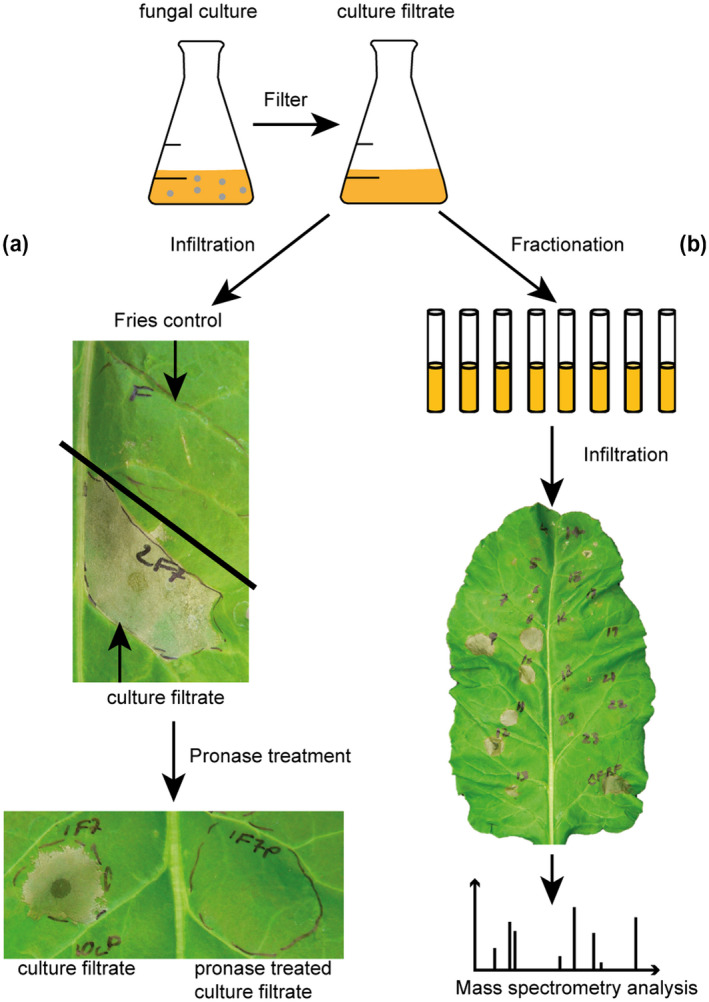
Scheme of the necrosis‐inducing effector identification pipeline. A 7‐day‐old Cercospora beticola 09‐40 wild‐type strain grown in Fries medium was filtered to remove fungal mycelia. (a) When the culture filtrate was infiltrated into 7‐week‐old sugar beet leaves, a clear necrotic phenotype was observed after 24 hr. Proteolysis treatment eliminated necrosis‐inducing activity of the culture filtrate. (b) Culture filtrate was fractionated using ion exchange chromatography and necrosis‐inducing activity of individual fractions was assayed by infiltration into sugar beet leaves. All infiltration experiments were repeated at least three times using different sugar beet plants
2.2. Effector protein candidate identification
Based on the analysis of MS/MS data and subsequent protein identity searches in the genome of C. beticola strain 09‐40 (de Jonge et al., 2018), three candidate proteins were identified: CB0940_03921, CB0940_10646, and CB0940_04765. While the mature CB0940_03921 peptide sequence was picked up in full in the MS/MS sample analysis, CB0940_10646 MS/MS coverage starts with the fifth amino acid of the mature peptide sequence and covers the peptide sequence until the 48th amino acid, totalling 75%. CB0940_04765 displayed peptide coverage to a total of 73% of the predicted protein sequence. Of these three proteins, only CB0940_03921 and CB0940_10646 displayed classic effector characteristics, including secretion signals, high cysteine content (Figure S2), and low molecular weight (9.2 and 6.6 kDa, respectively). In contrast, CB0940_04765 lacked a signal peptide and contained no cysteines. Analysis of the three candidates with the effector prediction software EffectorP v. 2.0 (Sperschneider et al., 2018) yielded an effector probability score for CB0940_03921 and CB0940_10646 of 0.945 and 0.9, respectively. CB0940_04765 was also predicted to be an effector, with a lower score of 0.557 compared to the other two candidates. Due to the lack of classical effector characteristics and a low predicted effector probability by EffectorP, CB0940_04765 was therefore excluded from further analysis.
The signal peptide cleavage sites were predicted to be between residues 18 and 19 for CB0940_03921 and between 16 and 17 for CB0940_10646 (Figure S2). Furthermore, the six cysteine residues found in the 85 amino acid sequence of the mature CB0940_03921 protein were predicted to form three disulphide bridges (Figure S2). Although CB0940_10646 is a rather small protein with 59 amino acids, it is predicted to have four disulphide bonds (Figure S2). On the nucleotide level, each of the two candidate genes had one intron, resulting in a coding sequence of 312 bp for CB0940_03921 and 228 bp for CB0940_10646. While no motifs were detectable for CB0940_03921, CB0940_10646 contains an AxxxG motif that may be involved in dimerization (Kairys et al., 2004). Additionally, a SxxV(K/R) motif associated with monocation specificity (Cu+, Ag+, and Au+) was also detected (Bird et al., 2013; Changela et al., 2003) (Figure S2). While a SxxV(K/R) motif was previously reported to occur in combination with a CxGxxxxDCP metal‐binding loop, CB0940_10646 appears to only be harbouring the monocation specificity domain without the metal‐binding loop motif. Notably, CB0940_03921, located on chromosome 4, is found on the edge of a contiguous sequence stretch and is thus flanked by a large sequence gap, a phenomenon typically observed for effector genes as a consequence of close association with transposable elements that are notoriously more difficult to assemble (Figure S3) (Thomma et al., 2016).
2.3. Protein sequence homology
While searching for potential homologs of our two effector candidates, two homologous hypothetical proteins from Colletotrichum spp. (CNYM01_08238 and CSIM01_12439) were identified for CB0940_10646 when queried by NCBI BLASTp (E < 10−6) against the nonredundant (NR) database (Table S1). No homologs were identified in the UniprotKB/Swiss‐Prot database consisting of functionally characterized proteins. NCBI tBLASTn analysis (E < 10−6) against fungal genome sequences also did not yield additional candidate homologous proteins. For CB0940_03921, we were able to identify 26 homologs via NCBI BLASTp (E < 10−6) against the NR database in several fungi in addition to C. beticola, specifically Alternaria spp., Fusarium spp., Colletotrichum spp., Stemphylium lycopersici, Didymella exigua, and Plenodomus tracheiphilus (Figure 2 and Table S2). In addition, we identified multiple candidate CB0940_03921 homologs in unannotated fungal genome sequences, specifically those belonging to Cercospora and Alternaria spp. and in S. lycopersici and Zymoseptoria passerinii (Table S2). CB0940_03921 (hereafter described as CbNip1 for necrosis‐inducing protein 1) homologs found in C. beticola strains clustered together (Figure 2) and showed high sequence conservation (Figure S4). One C. beticola strain (ICMP21690) appears to encode two CbNip1 copies with identical amino acid sequences (amino acid sequences of CbNip1 homologs can be accessed in Dataset S1), but detailed inspection of the ICMP21690 genome suggests this is most likely the result of erroneous genome assembly as it appears that the encoding contig (PDUI01000001.1) is represented by a near‐perfect head‐to‐head palindrome. Notably, we identified two, probably genuine, CbNip1 copies in Cercospora nicotianae and in Cercospora cf. sigesbeckiae (Dataset S1). In this case one paralog (g2604.t1 for C. nicotianae and g2944.t1 for C. cf. sigesbeckiae) clusters with C. beticola CbNip1 and several other Cercospora spp. CbNip1 homologs and the only remotely similar homolog in the barley pathogen Z. passerinii (Figure 2). The other paralogs (g2594.t1 for C. nicotianae and g2952.t1 for C. cf. sigesbeckiae, further referred to as CbNip1‐like) form a separate cluster and are more distant from CbNip1 (Figure 2). The two paralogs are spatially close on both the C. nicotianae (POSS01000015.1: 305,923‐306,288; POSS01000015.1: 279,073‐279,458) and the C. cf. sigesbeckiae genomes (NKQR01000009.1: 123,018‐123,382; NKQR01000009.1: 147,878‐ 148,249) at c.25 kb, suggesting a potentially ancestral organization that is further supported by the identification of a similar arrangement in Cercospora kikuchii strain ARG_18_001 and in Cercospora citrullina strain Cerl39‐09 following tBLASTn analysis using the CbNip1 and CbNip1‐like protein as queries. Careful analysis of the C. beticola genome revealed a predicted pseudogenized CbNip1‐like copy (i.e., with several in‐frame stop codons) distant from CbNip1 on chromosome 4. All homologs, except those in Fusarium fujikuroi (KLO92134.1, KLO99503.1, and XP_023427398.1) and in Colletotrichum asianum (KAF0315840.1), encode a predicted signal for secretion and encompass the previously mentioned six, similarly spaced, conserved cysteine residues (Figure S5).
FIGURE 2.
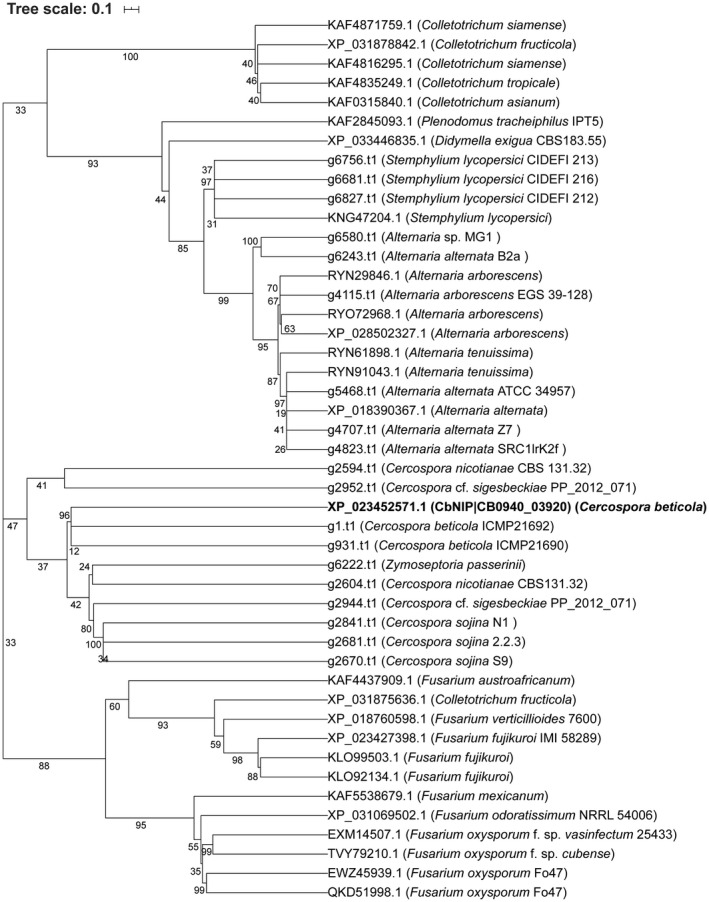
Phylogeny of CbNip1 and CbNip1 homologs found in Alternaria spp., Stemphylium lycopersici, Zymoseptoria passerinii, and different Cercospora species. The tree was constructed using the 46 proteins homologous to CbNip1. Peptide sequences were aligned using MAFFT (with default options) and subsequently filtered for phylogenetically relevant columns with trimAI. A maximum‐likelihood tree was built using automated protein model selection and 100 rapid bootstraps. The rooted tree was visualized in iTOL
2.4. CbNip1 protein structure homology
To gain insight into the biological function of CbNip1, we used the I‐TASSER server (Roy et al., 2010; Yang et al., 2015; Zhang, 2008) to predict the three‐dimensional structure of identified Nip1 proteins in C. beticola (XP_023452571.1), C. nicotianae (g2604.t1), C. cf. sigesbeckiae (g2944.t1), and Cercospora sojina (g2841.t1). When evaluating putative secondary structure, the most similar structure to C. beticola CbNip1 was C. nicotiniae with two highly conserved predicted α‐helices (not shown). However, the predicted β‐strand regions were more conserved between C. nicotiniae, C. cf. sigesbeckiae, and C. sojina proteins. The highest confidence protein model amongst the Cercospora spp. was for C. beticola Nip1 (confidence score [C‐score] = −1.7; Table S3 and Figure S6). The top protein model for each species, except C. sojina, showed closest structural homology to the β subunit of the KP6 killer toxin encoded by Ustilago maydis virus P6 (Table S3). C. sojina Nip1 had the lowest confidence protein model (C‐score = −3.84). Low C‐scores and disparate predictions between Nip1 proteins were obtained for ligand binding and enzymatic activity, making any inference of function difficult.
2.5. CbNip1 gene variation
We next investigated the CbNip1 gene sequence within 190 North American C. beticola isolates primarily sampled from North Dakota and Minnesota to assess the extent of within‐species variation in the gene (Spanner et al., unpublished data). In total, three different CbNip1 coding region haplotypes were identified. Most isolates (n = 189) had an identical CbNip1 sequence to the reference isolate 09‐40. Six strains harboured a synonymous mutation at codon D21 and the amino acid change N94H when compared to the reference haplotype. One strain had a single synonymous mutation at codon P41.
2.6. Genomic region encoding CbNip1 recently underwent a selective sweep
A selective sweep occurs when a beneficial mutation increases in frequency and becomes fixed in the population. The genomic signature of such positively selected mutations is a genomic region deprived of variation and with a high linkage disequilibrium. The “selective sweep map” of C. beticola was generated from a population genomic data set using 89 DMI‐resistant North American sugar beet‐infecting C. beticola isolates (Spanner et al., unpublished data). We correlated the genomic positions of the genes encoding CbNip1 homologs in C. beticola with the selective sweep map. In brief, the analysis used the site frequency spectrum (SFS) of genetic variants in the population genomic data sets with demographic simulations to assess the significance of outlier loci with deviations from the expected SFS under neutrality. Two independent approaches were used to detect outlier loci deviating from the expected distribution under neutrality: OmegaPlus and RAiSD (Alachiotis & Pavlidis, 2018; Alachiotis et al., 2012). Both programs detected an outlier locus at the end of chromosome 4 that spans the CbNip1 gene (Figure 3). This locus showed significantly higher values of both the two statistics ω (extent to which average linkage disequilibrium is increased on each side of the selective sweep) and μ (mutation rate per site per individual) as computed by OmegaPlus and RAiSD, respectively, when compared to values obtained from simulations under a neutral demographic scenario. The regions detected with the two methods include 10,750 bp (OmegaPlus) and 65,192 bp (RAiSD), and comprise 2,754 and 6,118 singe nucleotide polymorphisms (SNPs), respectively (Figure 3).
FIGURE 3.
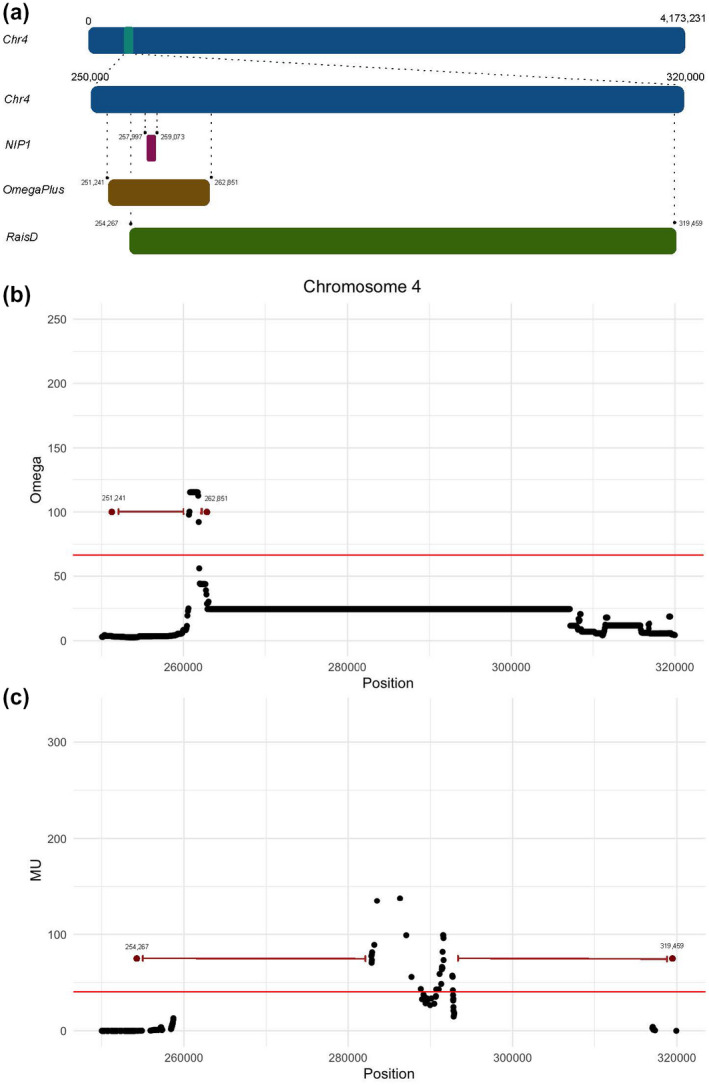
The genomic region encoding CbNip1 colocalizes with a selective sweep. Genome‐wide screen of selective sweeps in Cercospora beticola revealed a candidate region on chromosome 4 that spans the CbNip1 locus. (a) Position of the CbNip1 gene on chromosome 4 and the regions that were identified by the selective sweep analyses using demographic modelling and methods implemented in the programs Omega and RAiSD. (b) OmegaPlus and (c) RAiSD candidate regions on the chromosome arm of chromosome 4. The significance thresholds of the μ and ω statistics were determined with demographic simulations. Successive windows that were identified as significant were merged. The points in red represent the most left and the most right variants of the significant successive windows of the two statistics ω and μ computed by OmegaPlus and RAiSD, respectively
2.7. Heterologous expression of effector protein candidates and phenotype upon infiltration
To further characterize the candidate necrosis‐inducing effectors, the mature CbNip1 protein was produced heterologously in Escherichia coli and infiltrated into sugar beet leaves that were subsequently kept in a growth chamber with a 10‐hr light cycle. Unlike the response from the culture filtrate, no phenotype was observed for CbNip1 at 1 day postinfiltration (DPInf) (Figure 4a). However, after 2 DPInf the infiltration area of CbNip1 started to appear slightly chlorotic while the empty vector control remained unchanged (Figure 4a). Chlorosis of the CbNip1‐infiltrated area increased over time until it turned necrotic (Figure 4a). Because light is critical for the activity of C. beticola secondary metabolite effectors cercosporin and beticolin and has also been shown to be essential for functionality of other necrosis‐inducing factors (Ben M’Barek et al., 2015), we questioned whether light may play a role in the activity of CbNip1. To evaluate this, we infiltrated CbNip1 into sugar beet leaves that were subsequently placed in a growth chamber with different light/dark conditions (24 hr light, 10 hr light/14 hr dark cycle, 24 hr darkness). Of these growth chamber regimes, only incubation of CbNip1 infiltrated leaves incubated in 24 hr darkness resulted in clear necrosis of the complete infiltration area by 3 DPInf (Figure 4b). To assess the stability of CbNip1, we incubated the protein and empty vector control at 50 °C or 100 °C for 30 min, after which proteins were infiltrated into sugar beet leaves and subsequently shielded from light exposure. While exposure to 100 °C abolished necrosis formation, samples treated with 50 °C were still able to cause necrosis (Figure 4c). Furthermore, infiltrations of CbNip1 into Nicotiana benthamiana led to the same necrotic phenotype, indicating that the CbNip1 mode of action is not host‐specific (Figure 4d).
FIGURE 4.
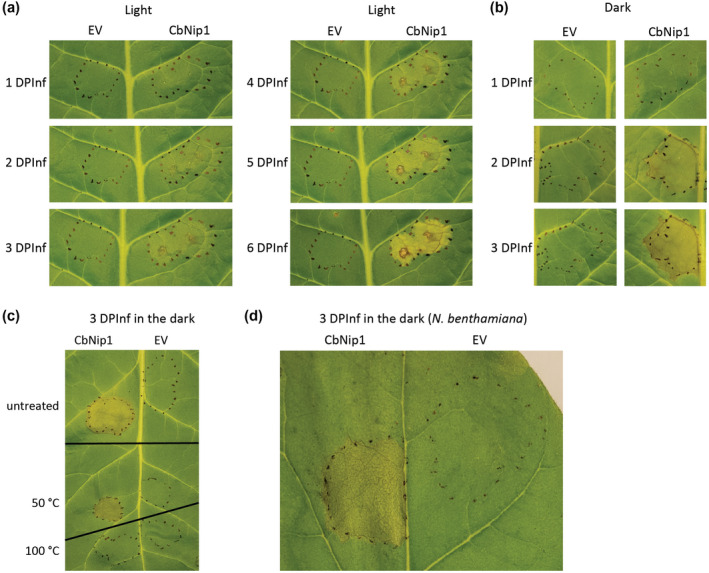
Necrosis‐inducing phenotype of CbNip1 protein. (a) Chlorosis/necrosis development after infiltration of CbNip1 into sugar beet leaf exposed to a 10 hr/14 hr light/dark cycle for up to 6 days postinfiltration (DPInf) and empty vector sample (EV) infiltration served as a control. (b) Necrosis development after infiltration of CbNip1 into a sugar beet leaf kept in 24 hr darkness and an EV infiltration that served as a control. (c) Treatment of CbNip1 and EV exposed to 50 °C for 30 min did not affect necrosis‐inducing activity of CbNip1 while treatment of both samples at 100 °C for 30 min abolished necrosis induction. Untreated samples served as controls. (d) Necrosis formation after infiltration of CbNip1 into a Nicotiana benthamiana leaf. An EV control sample served a control. All infiltration experiments were repeated at least three times using different sugar beet plants
We were unable to produce CB0940_10646 in sufficient amounts in either Pichia pastoris or E. coli. Therefore, chemically synthesized CB0940_10646 protein was used for infiltration into sugar beet leaves. In contrast to CbNip1, no phenotype was visible for the conditions tested, which included light/dark exposure, refolding of the protein, and the supplementation of trace elements (Figure S7). Consequently, CB0940_10646 was excluded from further analysis.
2.8. In planta gene expression profile of CbNip1 matches necrotic lesion development
To determine the expression pattern of CbNip1 during C. beticola colonization, we inoculated sugar beet plants with a C. beticola wild‐type strain and harvested leaf samples at 3, 6, 9, 12, 15, 18, and 21 days postinoculation (DPInoc). Gene expression analysis revealed that CbNip1 is minimally expressed at early time points (Figure 5). However, from 12 DPInoc onwards CbNip1 expression increased until peaking at 15 DPInoc. Interestingly, onset of CbNip1 up‐regulation at 12 DPInoc matched symptom development on the sugar beet leaves (Figure S8). At 15 DPInoc, many single necrotic spots were visible while CbNip1 expression reached its peak. However, with progressing necrosis expansion in planta, CbNip1 experienced a steady down‐regulation again from 18 DPInoc onwards (Figure 5).
FIGURE 5.
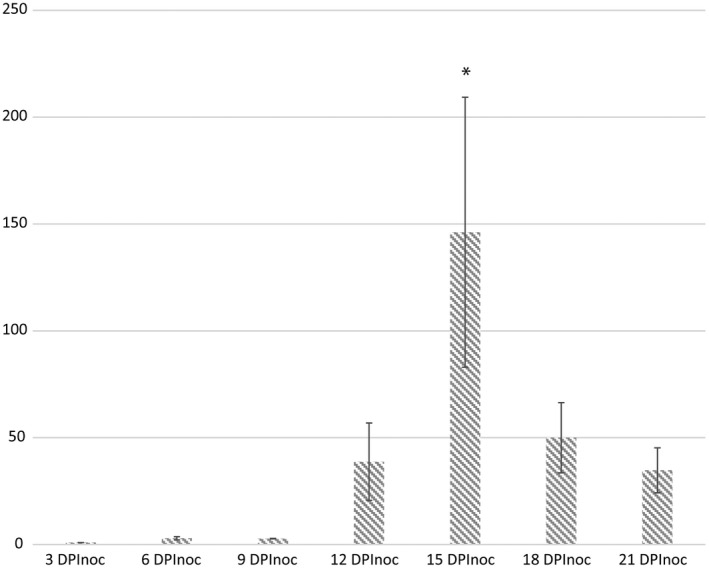
CbNip1 gene expression during Cercospora beticola infection on sugar beet. Gene expression profile of CbNip1 during C. beticola wild‐type strain 09‐40 infection course at 3, 6, 9, 12, 15, 18, and 21 days postinoculation (DPInoc). CbNip1 gene expression was normalized to C. beticola actin gene expression. The relative gene expression (displayed on the y axis) of three biological repetitions was calculated in comparison to the earliest measured time point using the Pfaffl method (Pfaffl, 2001). Error bars indicate the standard error of three biological replicates. By performing a one‐way analysis of variance (ANOVA) (p < .05) with a post hoc Tukey test, we tested for differences between the gene expression levels of different time points. A significant difference can be observed between 3, 6, and 9 DPInoc when compared to 15 DPInoc (p < .05, indicated by an asterisk). However, between 12 and 15 DPInoc there is no significant difference and also later time points such as 18 and 21 DPInoc do not show a significant difference to 15 DPInoc. This suggests an increase in CbNip1 gene expression at 12 DPInoc compared to earlier time points (3–9 DPInoc), but with an expression lower than the gene expression at 15 DPInoc. Furthermore, the loss of significance after 15 DPInoc suggests CbNip1 expression decreases again at later time points (18 and 21 DPInoc)
2.9. CbNip1 is a virulence factor
To investigate whether CbNip1 is required for full C. beticola virulence, we generated ΔCbNip1 mutants and inoculated sugar beet plants with wild‐type C. beticola, three individual ΔCbNip1 mutants (Figure S9), and an ectopic mutant (Figure S10). To ensure that the generated ΔCbNip1 mutants did not suffer any fitness penalties, the growth of the 09‐40 wild type strain and the three ΔCbNip1 mutants was assessed under in vitro growth conditions (Figure S11). No growth deficiency was detected between the wild‐type and mutant strains when grown on potato dextrose agar (PDA) and PDA amended with charcoal (0.01 g/ml), different cell wall (Calcofluor 100 µg/ml or Congo red 45 µg/ml), osmotic (1 M sorbitol or 1 M NaCl), or oxidative stressors (1 mM H2O2) (Figure S11). In addition to visible symptom assessment in planta, fungal biomass was measured using quantitative PCR (qPCR) for each treatment individually to determine the level of fungal colonization of the host plants. While clear infection symptoms were displayed by sugar beet plants inoculated with wild‐type C. beticola or the CbNip1 ectopic mutant, highly reduced symptom formation was observed for plants inoculated with any of the three individual ∆CbNip1 strains (Figure 6). In agreement with the noticeable difference in the in planta phenotype of wild‐type/ectopic mutant and ∆CbNip1 strains, evaluation of fungal biomass showed reduced fungal colonization in plants infected with ∆CbNip1 compared to high levels of fungal biomass found in sugar beet plants inoculated with the wild‐type C. beticola strain and the ectopic mutant (Figure 6).
FIGURE 6.
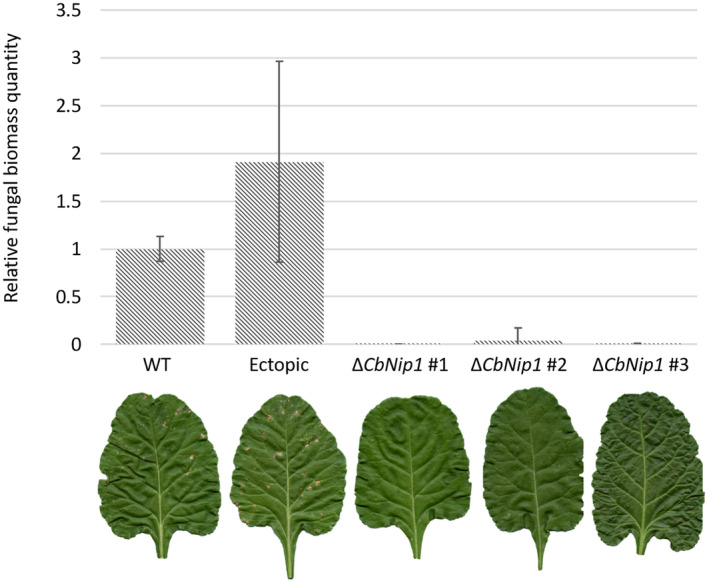
Fungal biomass quantification of Cercospora. beticola 09‐40 wild‐type strain, three individual ∆CbNip1 mutants, and one ectopic transformant. Sugar beet plants inoculated with C. beticola strains at 21 days postinoculation with photographs showing respective disease severity below. Gene expression was quantified with SbEc1‐F/SbEc1‐R quantitative PCR (qPCR) primers for sugar beet and CbActin Fp/CbActin Rp qPCR primers for C. beticola. Error bars represent standard error of three biological replicates. Fungal biomass was calculated using the ∆∆C t method relative to the average value of the wild‐type inoculated sugar beet plants
3. DISCUSSION
C. beticola is a hemibiotrophic fungus that is dependent on necrosis formation during infection (Weltmeier et al., 2011) and known to use the secondary metabolite effector cercosporin to cause host cell death (Daub & Ehrenshaft, 2000). Here, we report the identification of the C. beticola necrosis‐inducing effector CbNip1. By searching for in vitro parameters that trigger C. beticola to secrete effector proteins, we found growth conditions under which C. beticola produces proteinaceous effectors that cause necrosis on infiltration into sugar beet leaves within 24 hr. While infiltration of pure CbNip1 into sugar beet leaves took 48 hr to lead to visible necrosis, the timing difference in necrosis formation is probably due to the presence of multiple necrosis‐inducing effectors besides CbNip1 in the culture filtrate. Besides CbNip1, fractionation of the culture filtrate with subsequent mass spectrometry analysis of the necrosis‐inducing fraction identified the presence of two other proteins, CB0940_10646 and CB0940_04765, of which CB0940_04765 was excluded for further analysis due to the lack of typical effector characteristics.
For functional analysis, CbNip1 was heterologously produced and infiltrated into sugar beet leaves. As mentioned above, we found that the full potential of CbNip1 to induce host cell death was dependent on the absence of light. Light is known to influence Cercospora zeae‐maydis infection capability as the ability to find stomata and form appressoria is abolished in the dark (Kim et al., 2011). Plants are also impacted by light in various ways, including alteration of leaf physiology (Kami et al., 2010; Roberts & Paul, 2006). Furthermore, studies on host resistance responses have demonstrated that light is required for the full cascade of plant resistance responses (Guo et al., 1993; Roberts & Paul, 2006; Roden & Ingle, 2009). Based on microarray expression profiling of the C. beticola–sugar beet interaction, 571 sugar beet genes were induced, including pathogenesis‐related (PR) genes and genes involved in lignin and alkaloid biosynthesis at the onset of necrotic symptom formation (Weltmeier et al., 2011). While the products of these defence‐associated genes could potentially impede CbNip1 function in the presence of light, PR genes have been shown to be repressed in the dark (Guo et al., 1993; Roberts & Paul, 2006; Roden & Ingle, 2009). However, a recent paper on effector‐triggered immunity in a bacteria–plant interaction mentions that hypersensitive responses (HRs) caused by effector recognition via a nucleotide‐binding leucine‐rich repeat (NB‐LRR) immune receptor were more pronounced in the dark (Qi et al., 2018). While necrotrophic effectors such as SnTox1 have been reported to exploit interactions with defence‐associated genes such as specific plant receptors to induce cell death in the host specifically (also known as reverse gene‐for‐gene interaction) (Liu et al., 2004; Shi et al., 2016), in the case of CbNip1 necrosis is not only induced in sugar beet but also in the nonhost N. benthamiana. Therefore, CbNip1‐triggered induction of necrosis via interaction with a corresponding receptor protein would require that this receptor is conserved among various plant species. Given that ample predicted secondary structure similarity was seen between CbNip1 and C. nicotianae CnNip1 through I‐TASSER protein modelling (Roy et al., 2010; Yang et al., 2015; Zhang, 2008), the protein structure may be conserved enough between CbNip1 and CnNip1 such that CbNip1 can interact with the same target in the nonhost plant N. benthamiana.
Besides cell death through recognition, CbNip1 function may be the result of general toxicity to plants. For example, some effectors modulate targets in their host but potentially also in other plants for necrosis induction. This mode‐of‐action has been observed for the small sRNase Zt6 of Z. tritici, which displays universal cytotoxicity by cleaving plant and mammalian ribosomal RNA (Kettles et al., 2018). Nevertheless, necrosis formation could also be the result of cell wall‐degrading enzyme activity as cell wall‐degrading enzymes of various fungal pathogens have been found to be essential for fungal virulence (Bailey et al., 1990; Brito et al., 2006; Kars et al., 2005).
Protein modelling of Nip1 revealed that the closest structural analog to Nip1 from C. beticola, C. nicotianae, and C. cf. sigesbeckiae is the β subunit of the KP6 killer toxin produced by Ustilago maydis virus P6 (KP6β). Double‐stranded RNA totiviruses can infect certain strains of the corn smut fungus Ustilago maydis (Allen et al., 2013). These viruses produce potent antifungal proteins called “killer toxins” that are secreted by the host and kill competing uninfected strains of U. maydis (Koltin & Day, 1975). The KP6 toxin is one such killer toxin and is a heterodimer composed of two subunits: KP6α and KP6β (Allen et al., 2013). The KP6α and KP6β subunits have high structural similarity, but KP6β is responsible for cytotoxic activity via an unknown mechanism (Finkler et al., 1992). It is possible that Nip1 has similar cytotoxic activity, which causes the necrosis seen in leaf infiltrations of sugar beet and N. benthamiana.
Using population genomic data from a North American population of C. beticola isolates, we found evidence that the CbNip1 locus resides in a genomic region that has recently experienced a selective sweep. A selective sweep occurs when a favourable mutation appears in a population and becomes fixed as a result of its fitness advantage. Genetic variants in the same genomic background as the beneficial mutation will “hitchhike” with the selected mutation. This leaves a footprint at the genomic locus characterized by reduced variation and high linkage disequilibium compared to distal genomic regions (Smith & Haigh, 1974). The identification of CbNip1 within such a selective sweep region underscores the importance of this effector in the population. However, the nature of the selection pressure to retain CbNip1 remains unclear. The selection of this gene variant within this population could be the result of a conserved and specific host target that was widely introduced in sugar beet germplasm. For example, given the necrotic phenotype of CbNip1, it is tempting to speculate that this effector specifically interacts with a recently introgressed host immune receptor in a reverse gene‐for‐gene interaction, as known from other systems (Faris et al., 2010; Friesen et al., 2007, 2008; Shi et al., 2016). It is possible that the beneficial mutation that conferred a selective sweep in the CbNip1 region relates to regulation of gene expression. We speculate that the late onset of CbNip1 gene expression could be an adaptation to the necrotrophic stage of the pathogen. For example, there may have been directed evolution for CbNip1 to be expressed and recognized by sugar beet solely during its necrotrophic phase as a means of facilitating fungal growth. This may also explain cell death in nonhost plants upon infiltration with the CbNip1 protein where the necrotic response could be due to a defence‐inducing activity as seen in other necrotrophic fungal pathogens on nonhosts (Kettles et al., 2017; Raffaello & Asiegbu, 2017). Alternatively, CbNip1 may not have a host target and therefore its nonhost specificity is due to the general cytotoxic nature of this protein. Further investigation into nonhost responses to CbNip1 would be necessary to reveal a mechanism for the CbNip1‐induced necrotic phenotype.
While a necrotic phenotype is observed for CbNip1, the other effector candidate CB0940_10646 failed to induce any phenotype under tested conditions. Because CB0940_10646 has domains associated with dimerization and monocation specificity, the inability to induce necrosis may be due to the absence of a cognate cofactor. As CB0940_10646 was chemically synthesized, it is possible that due to its monocation specificity‐associated domain, the addition of trace elements (including copper) as present in the Fries medium of the initial culture filtrate might activate CB0940_10646 function. However, the supplementation of metal ions to CB0940_10646 did not lead to phenotype formation in sugar beet leaves. Moreover, infiltration of CB0940_10646 with CbNip1 did not obviously enhance CbNip1‐induced necrosis. Further research is required to identify the allied cofactors for CB0940_10646, if any.
In accordance with CbNip1 necrosis‐inducing ability, we have found that necrotic symptom development in planta correlates with up‐regulation of CbNip1 expression (Figure 5 and Figure S8). Induction of host necrosis during the biotrophic phase is not likely beneficial for the fungus, therefore it is not surprising that CbNip1 is minimally expressed at early infection time points (3–9 DPInoc). An increase in CbNip1 expression and the development of necrotic lesions occurred simultaneously, suggesting that CbNip1 is linked to the switch from biotrophic to necrotrophic lifestyle of the fungus. Once necrosis formation is ongoing and existing necrotic lesions start to fuse, CbNip1 expression is reduced again to a similar level as observed in the initial cell death induction phase at 12 DPInoc, indicating that necrosis induction by CbNip1 may still be important at later time points. Interestingly, the CbNip1 expression pattern is similar to expression patterns of other necrosis‐inducing effectors from different protein families found in the hemibiotroph Z. tritici. ZtNip1 showed an expression pattern where gene up‐regulation matched onset of symptom development in planta (Ben M’Barek et al., 2015). Similarly, the Z. tritici Nep‐1‐like protein gene MgNlp peaked towards the end of the biotrophic phase before necrotic lesions were visible (Motteram et al., 2009). However, there are also examples of contrasting expression patterns to CbNip1. For example, the expression of Zt6 in planta is characterized by a double peak probably attributed to a double functionality (Kettles et al., 2018).
Because C. beticola requires necrotic plant tissue to complete its life cycle (Weltmeier et al., 2011), we determined whether CbNip1 was also essential for fungal virulence. We found that site‐directed ΔCbNip1 mutants are impeded in virulence compared to the wild‐type C. beticola strain and an ectopic mutant strain. Not only did plants inoculated with ΔCbNip1 mutants develop fewer C. beticola‐specific lesions, biomass determination revealed there was less fungal biomass in plant tissue compared to the progenitor wild type (Figure 6). Taken together, this indicates that CbNip1 plays an important role in C. beticola virulence. As mentioned earlier, C. beticola produces the secondary metabolite cercosporin and a family of phytotoxins called beticolins, both of which are able to cause cell death in the presence of light (Daub & Ehrenshaft, 2000; Jalal et al., 1992; Schlösser, 1962; Yamazaki et al., 1975) and cercosporin was shown to be a virulence factor for several Cercospora species (Callahan et al., 1999; Choquer et al., 2005). Because light activation is essential for cercosporin and beticolin functionality, they are probably not active in the dark. With the secretion of CbNip1, however, C. beticola may be defying this light‐associated limitation using additional necrosis‐inducing agents to cover both light and dark conditions to achieve maximal host cell death to complete its life cycle.
In conclusion, we have shown that C. beticola secretes the effector protein CbNip1 during infection that in the absence of light has the ability to cause necrosis on infiltration into sugar beet leaves within 48 hr. Furthermore, CbNip1 expression in planta correlates with necrotic symptom appearance during C. beticola sugar beet infection. Targeted gene replacement of CbNip1 led to a reduction in virulence, indicating that CbNip1 is a virulence factor for C. beticola. As CbNip1 has no obvious homology to other characterized proteins in public databases, future studies will be directed to identify CbNip1 mode of action. Studies on pathogen–host plant interactions often focus on processes that occur in light conditions, but it may be interesting to understand how this interaction is altered in the dark, a vital condition for unhampered CbNip1 function. Consequently, CbNip1 is a fungal virulence factor that is hypothesized to take advantage of the reduced host plant defence response level due to the absence of light. Further analysis of yet unknown functional motifs of CbNip1 as well as localization studies will help to shed light on the biology of CbNip1.
4. EXPERIMENTAL PROCEDURES
4.1. Fungal strains
C. beticola wild‐type strain 09‐40 was isolated from leaf material collected from a sugar beet field in the Red River Valley, USA in 2009. The fungus was kept at 22 °C on PDA (Difco) and fungal site‐directed gene deletion mutants in a 09‐40 background on PDA amended with 150 µM hygromycin B (Duchefa).
4.2. Culture filtrate preparation and infiltration
A 5 mm plug was taken from the actively growing zone of C. beticola wild‐type strain 09‐40 on PDA and used to inoculate a 250 ml conical flask filled with 100 ml of Fries medium (Friesen & Faris, 2012). After 7 days of incubation at 120 rpm under 24‐hr light conditions at 21 °C, the liquid culture was run through two layers of Miracloth (EMS Millipore Corp.) to filter out fungal mycelia and subsequently filter‐sterilized with a 0.45 µm Filtropur membrane (Sarstedt). Approximately 30–50 µl of sterile culture filtrate was infiltrated into the leaves of 7‐week‐old sugar beet plants of the variety C093 (formerly 86RR66) using a 1 ml needleless syringe. Infiltration experiments were repeated at least three times with multiple individually produced culture filtrates. Plants were kept in a greenhouse chamber with an average temperature of 26 °C during the day and approximately 17 °C during the night. Chambers were equipped with additional lighting to ensure 16 hr of light a day. To confirm the proteinaceous nature of the necrosis‐inducing agent, 50 μl of 3‐(N‐morpholino)propanesulfonic acid (MOPS) buffer (1 M, pH 7.5) and 25 μl of pronase (1 mg/ml) (Sigma) or water as control was added to 425 μl of culture filtrate and incubated at 22 °C for 4 hr. Subsequently, samples were infiltrated into sugar beet leaves as described above.
4.3. Culture filtrate fractionation
Culture filtrate was partially purified as described in Liu et al. (2009). In short, 100 ml of 7‐day‐old C. beticola wild‐type strain 09‐40 grown in Fries medium was first filter‐sterilized and then dialysed against water using a 3.5 kDa molecular weight cut‐off dialysis membrane (Fisher Scientific). The next day, the dialysed culture filtrate was loaded onto a HiPrep SPXL 16/10 cation exchange column (GE Healthcare) using the ÄKTA prime plus (GE Healthcare) liquid chromatography system. After a washing step with 50 ml of 20 mM sodium acetate buffer pH 5.0, 5 ml fractions were collected during gradient elution of 0–300 mM NaCl plus 20 mM sodium acetate pH 5.0 at a flow rate of 5 ml/min over 20 min. Collected fractions were individually infiltrated into 7‐week‐old sugar beet plants of the variety C093 and screened for necrotic phenotype. Fractionation and infiltration experiments were repeated at least three times.
4.4. Preparation of necrosis‐inducing protein fraction for MS/MS analysis
The fraction that repeatedly caused necrosis was loaded onto a precast 16.5% Tris–Tricine polyacrylamide gel (Bio‐Rad). Protein spots were excised and sent to the Center for Mass Spectrometry and Proteomics at the University of Minnesota for trypsin digestion and subsequent LC‐MS analysis. Peptide mass fingerprints and peptide sequence information were used to search for protein identity using the annotated C. beticola 09‐40 genome (de Jonge et al., 2018).
4.5. gDNA extraction, RNA extraction, and cDNA synthesis
Genomic DNA was extracted using a modified version of the microprep protocol published by Fulton et al. (1995), replacing chloroform:isoamyl alcohol (24:1 vol/vol) with phenol:chloroform:isoamyl alcohol (25:24:1 vol/vol/vol).
RNA extraction followed the TRIzol method (Ambion) according to the manufacturer's protocol and subsequently cleaned up three times using the RNase‐free DNase set (Qiagen) according to Appendix E of the RNase Mini Handbook 06/2012. For cDNA synthesis, 1 µg of total RNA was used with the SuperScript III reverse transcriptase kit (Invitrogen) following the manufacturer's protocol.
4.6. Sequence analysis
CbNip1 homologs were identified online via NCBI BLASTP and tBLASTn analysis of annotated and unannotated whole‐genome sequences, respectively. Selected homologous sequences identified via tBLASTn analyses were subsequently extracted via local ab initio gene prediction of the corresponding genome sequence using the Augustus training parameters previously prepared for C. beticola (de Jonge et al., 2018). For phylogenetic analysis of CbNip1 and CbNip1 homologs (Table S2) and phylogenetic tree analysis we aligned all CbNip1 homologs by MAFFT v. 7.310 (Katoh & Standley, 2013) (setting: ‐‐auto), then filtered the alignment using trimAl v. 1.4.rev22 (Capella‐Gutiérrez et al., 2009) (settings: ‐gt 0.9 ‐cons 60 [remove all positions with gaps in 10% or more sequences, unless it leaves less than 60% of original alignment in which case the 60% best is reported]). The maximum‐likelihood (ML) phylogenetic tree including bootstrapping was calculated in RAxML v. 8.2.11 (Stamatakis, 2014) (settings: ‐f a [rapid bootstrap analysis and search for best‐scoring ML tree in one program run], ‐m PROTGAMMAAUTO [automatic protein substitution model selection with and without empirical base frequencies], ‐N 100 [100 rapid bootstraps]). The tree was rooted by running RAxML with setting “if I” which simply balances subtree lengths. InterProScan (https://www.ebi.ac.uk/interpro/) was used for protein sequence analysis.
Signal peptides (if present) were determined with the SignalP v. 5.0 online tool (ttp://www.cbs.dtu.dk/services/SignalP) while disulphide bonds were predicted using DISULFIND (http://disulfind.dsi.unifi.it/).
The detection of selective sweeps was conducted using a population genomics data set of 89 DMI‐resistant C. beticola isolates collected in sugar beet fields near Fargo, North Dakota in 2016 (Spanner et al., unpublished data). The selective sweep analysis used OmegaPlus v. 3.0.3 (Alachiotis et al., 2012) and RAiSD v. 2.9. These methods detect outlier regions based on two statistics, ω (OmegaPlus) and μ (RAiSD) (Alachiotis & Pavlidis, 2018; Alachiotis et al., 2012; Excoffier et al., 2013). To control for the effect of demographic history of the population on the site frequency spectrum, linkage disequilibrium, and genetic diversity along the genome when setting the significance threshold (Nielsen et al., 2005; Pavlidis et al., 2013), we simulated data sets using the ms coalescent simulator (Hudson, 2002) under the best neutral demographic model, as inferred with fastsimcoal2 (Excoffier et al., 2013).
4.7. Protein structure prediction
Mature protein sequences were submitted to the I‐TASSER online server for protein structure and function prediction (Roy et al., 2010; Yang et al., 2015; Zhang, 2008,). The query sequence was initially threaded through a nonredundant structure library (protein data bank, PDB) to identify structural templates, which were then used in an iterative Monte Carlo process to simulate protein structure. COACH analysis provided functional insight on ligand binding sites (LBS), Enzyme Commission (EC), and Gene Ontology (GO).
4.8. Quantitative reverse transcription PCR of CbNip1
Quantitative reverse transcription PCR (RT‐qPCR) was performed in triplicate using the SensiMix SYBR Hi‐Rox kit (Bioline) with an ABI 7300 PCR machine (Applied Biosystems) with cDNA of each time point for gene expression analysis and using the GoTaq qPCR Master Mix (Promega) with a CFX96 Real‐Time System (Bio‐Rad) with gDNA of each treatment for fungal biomass quantification. All reactions were done in triplicate and primers are listed in Table S4. Real‐time PCR conditions started with a denaturation step of 10 min at 95 °C; followed by denaturation for 15 s at 95 °C, annealing for 30 s at 60 °C, and extension for 30 s at 72 °C, for 30 cycles. Water as template control was included for all qPCR runs. With C. beticola actin as a reference gene for the gene expression study, relative gene expression of three biological repetitions was calculated in comparison to the earliest measured time point using the Pfaffl method (Pfaffl, 2001). Variation in gene expression was calculated using the standard error of the means of three biological replicates. A one‐way analysis of variance (ANOVA) (p < .05) with a post hoc Tukey test using GNU PSPP (2015) was performed to determine statistical differences in gene expression level means between time points and subsequently determine differences by pairwise comparing gene expression levels of different time points. Biomass was determined by quantifying the expression with SbEc1‐F/SbEc1‐R primers (De Coninck et al., 2012) for sugar beet (Table S4) and CbActin Fp/CbActin Rp primer for C. beticola (Table S4) and using the ∆∆C t method (Livak & Schmittgen, 2001) relative to the average value of the wild‐type inoculated sugar beet plants. Error bars indicate standard error of variation between three individual biological replicates.
4.9. Vector construction and protein production in E. coli
For heterologous protein expression in E. coli, the CbNip1 and CB0940_10646 coding sequence was amplified with GoTaq Long PCR Master Mix (Promega) from C. beticola 09‐40 wild‐type cDNA using primers MKE‐78/77 and MKE‐76/79 (Table S4), respectively. Amplicons and pET SUMO vector (Invitrogen) were digested with EcoRI and NotI and followed by ligation of the fragments into the double‐digested pET SUMO vector with T4 DNA ligase (NEB) and cloned into E. coli DH5α. Plasmids carrying the correct CbNip1 or CB0940_10646 coding sequence were verified by sequencing (Eurofins Genomics, Ebersberg, Germany) as well as an empty pET SUMO vector subsequently cloned into the E. coli Origami (DE3) strain.
For heterologous protein expression, 1 L of Luria‐Bertani (LB) broth was inoculated with 20 ml of a culture that was grown overnight in LB plus kanamycin 50 µg/ml with either CbNip1 pET expression construct or the empty vector control and grown at 37 °C, shaking at 200 rpm until an OD600 of 0.6–0.8 was reached. Protein production was induced with 0.05 mM isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) final concentration and kept growing at 20 °C and shaking at 200 rpm for 24 hr. Cells were pelleted, snap frozen with liquid nitrogen and then lysed with 20 ml of lysis buffer containing 50 mM Tris.HCl pH 8.5 (Invitrogen) and 150 mM NaCl (Sigma), 10% glycerol (Amresco), 6 mg/ml lysozyme from chicken egg white (Sigma), 2 mg/ml sodium deoxycholate (Sigma), 0.625 mg/ml deoxyribunuclease I from bovine pancreas (Sigma), and one cOmplete protease inhibitor pill (Sigma). After the cultures were kept on ice for 1.5 hr, cells debris was spun down for 1 hr at 20,000 × g at 4 °C and the soluble protein fraction was processed for protein purification.
4.10. Protein purification
In E. coli heterologously produced protein samples were loaded at 1 ml/min onto a column packed with 2 ml of Ni Superflow resin (Clontech) for purification. After a washing step with wash buffer (50 mM Na2HPO4, 300 mM NaCl, 40 mM imidazole [Merck]) at 2 ml/min to wash out contaminative E. coli native proteins, small ubiquitin‐like modifier (SUMO)‐tagged CbNip1 or the SUMO tag alone obtained from the empty vector sample were eluted with elution buffer (50 mM Na2HPO4, 300 mM NaCl, 250 mM imidazole). Elution samples were dialysed with a Spectra/Por dialysis membrane with molecular weight cut‐off (MWCO) of 3,500 (Spectrum Laboratories) against 200 mM NaCl containing ULP‐1 enzyme to cleave off the SUMO tag at 4 °C overnight without agitation. The next day samples were run through the nickle bead column with the same setup as before at 1 ml/min to allow cleaved off SUMO tags to bind to the nickel beads. Flow‐through was collected and again dialysed for 24 hr against 200 mM NaCl. Samples were concentrated with Amicon Ultra‐15 centrifugal filter unit with an Ultracel‐3 membrane (Millipore) with a 3 kDa cut‐off. For visualization, 5 µl of protein sample were loaded on Mini‐PROTEAN TGX stain‐free precast gels (Bio‐Rad).
4.11. Refolding and preparation of CB0940_10646 for sugar beet leaf infiltration
Synthesized mature CB0940_10646 purchased from GeneScript was dissolved in Milli‐Q water to 3 mg/ml. For refolding, oxidized glutathione (Sigma) and reduced glutathione (Sigma) were added to 1 mg/ml CB0940_10646 to an end ratio of 5:1 mM and incubated overnight. Milli‐Q water treated with the same glutathione ratio served as a control. To see whether the addition of trace elements leads to activation of necrosis‐inducing activity of CB0940_10646, 1 μl of trace element stock was added to 0.5 ml of protein sample or water control to a trace element end concentration as found in Fries medium used for the culture filtrate experiment.
4.12. Protein infiltration
Sugar beet plants of the variety C093 were grown in the climate chamber at 21 °C with 10 hr light with 10 lux and 70% humidity. After 7 weeks approximately 30–50 µl of purified protein (c.2 mg/ml) or empty vector (Figure S12) was infiltrated into the leaves using a 1 ml needleless syringe and the infiltration area was marked with a marker pen. Dark‐treated leaves were wrapped in aluminium foil to prevent light exposure. For this experiment, CB0940_10646 and three individually produced and purified CbNip1 samples were infiltrated at least three times.
4.13. CbNip1 deletion and ectopic mutants
Individual CbNip1 deletion mutants were generated following the split‐marker polyethylene glycol‐protocol described in Bolton et al. (2016). Primers are listed in Table S4. Three individual C. beticola 09‐40 CbNip1 deletion mutants were confirmed by full genome sequencing using Oxford Nanopore MinION technology with the Rapid Sequencing Kit (Oxford Nanopore Technologies) to determine a single hygromycin B phosphotransferase insertion replaced the CB0940_03921 gene as shown in Figure S9. The ectopic mutant was confirmed by PCR by absence of amplicon for site‐directed hygromycin integration, and presence of amplicons for hygromycin and CbNip1 gene amplification (Figure S10).
4.14. C. beticola in vitro fitness assay
A 5 mm plug was taken from the actively growing zone of 4‐day‐old cultures of the wild type and three individual ΔCbNip1 mutants and used to inoculate PDA plates supplemented with sorbitol (1 M), Calcofluor (100 μg/ml), charcoal (0.01 g/ml), Congo red (45 μg/ml), H2O2 (1 mM), or NaCl (1 M). Plates were incubated for 7 days at room temperature to observe fungal fitness and to take photographs.
4.15. Inoculation assay
Spore formation of C. beticola wild type, three individual ΔCbNip1 mutants, and one ectopic transformant was induced on CV8 agar plates as previously described (Secor & Rivera, 2012). Spores were harvested and adjusted to a concentration of 105 spores/ml and spore suspension was equally sprayed on the leaves of 5‐week‐old and 7‐week‐old sugar beet plants of the variety C093 for fungal biomass analysis and gene expression analysis, respectively. Inoculated plants were kept in a humidity chamber with about 27 °C and 90% humidity for 5 days after which plants were exposed to 22 °C with a 16‐hr/8‐hr day/night cycle. For fungal biomass analysis three leaves of three plants for three repetitions were harvested at 21 DPInoc and instantly snap frozen while plants for gene expression analysis were harvested at 3, 6, 9, 12, 15, 18, and 21 DPInoc using three leaves of two plants in three repetitions.
Supporting information
FIGURE S1 SDS‐PAGE gel displaying the protein content of active fraction #9 and crude culture filtrate. Protein samples of active fraction #9 and crude culture filtrate (indicated by arrows) were run on SDS‐PAGE. Subsequently, the SDS‐PAGE gel was stained with MS/MS compatible Coomassie Blue stain and band incisions are depicted by red boxes. Excised samples were sent for MS/MS analysis
FIGURE S2 Illustration of effector characteristics found in peptide sequences of CB0940_03921 (CbNIP1) and CB0940_10646. Both candidates display classic effector characteristics such as signal peptides (highlighted in pink) and predicted disulphide bonds (each individual disulphide bond is indicated by a differently coloured line) while the mature sequences are highlighted in blue. Underlined in the CB0940_10646 sequence are peptide sequence snippets SDAVRK, a conserved SxxV(K/R) motif associated with monocation specificity, and AYYYG, an AxxxG motif shown to be involved in dimerization
FIGURE S3 Genomic context CB0940_03920. The flanking genomic region of CbNip1 (CB0940_03920) highlighting a large sequence gap just upstream as well as several smaller repetitive elements. Prepared at https://www.ncbi.nlm.nih.gov/gene/?term=CB0940_03921
FIGURE S4 Alignment of CbNIP1 sequences found in different Cercospora species. Homologous protein sequences display high sequence conservation. Nucleotides of CbNip1 homologs that are identical to the nucleotide sequence of CbNip1 are indicated with a period, while variations are highlighted by displaying the corresponding nucleotide
FIGURE S5 Alignment of selected homologous CbNIP1 peptide sequences. The sequence alignment was constructed by mafft and the phylogenetically relevant positions are identified and shown here using trimAl. Sequence positions selected for phylogenetic analysis are listed below the alignment in dark grey “Selected Cols”. Asterisks indicate conserved cysteines. Signal peptides of CbNIP1 homologs are indicated with purple boxes. CbNIP homologs that lack a predicted signal peptide for secretion are depicted in red
FIGURE S6 The I‐TASSER server was used to predict the 3D structure of the identified NIP1 protein in Cercospora beticola (XP_023452571.1). The highest confidence model (C‐score = −1.7) for the CbNIP1 protein is displayed
FIGURE S7 No visible phenotype upon inoculation of CB0940_10646 protein. (a) No phenotype after CB0940_10646 infiltration into sugar beet leaf exposed to a 10 hr/14 hr light/dark cycle for 5 and 7 DPInf, while CbNIP1 served as a positive control to display expected chlorosis after 5 DPInf and beginning of necrosis after 7 DPInf. (b) No visible phenotype was identified after CB0940_10646 was infiltrated into a sugar beet leaf that was kept in 24 hr darkness for 2 and 3 DPInf. CT served as a control for CB0940_10646 infiltration and consisted of an equal amount of trace elements to what was supplied in the CB0940_10646 sample. As a positive control, we observe necrosis development after CbNIP1 inoculation, and an empty vector (EV) infiltration served as a control for CbNIP1 and showed no phenotype
FIGURE S8 Symptom development during Cercospora beticola 09‐40 wild type strain infection of sugar beet. No symptoms are detectable at early time points (3, 6, and 9 DPInoc). Red arrows at 12 and 15 DPInoc indicate formation of necrotic lesions. At 18 DPInoc, necrotic lesion formation intensifies, accompanied by some chlorosis around the necrotic lesions. At 21 DPInoc, single necrotic lesions are fused to form necrotic patches on the sugar beet leaf
FIGURE S9 Confirmation of site‐directed integration using Oxford Nanopore MinION reads. Mapping of Oxford Nanopore MinIon reads to Cercospora beticola 09‐40 genome. Genome browser view of the single hygromycin resistance gene insertion found in CbNip1 deletion mutant. Reads map to flanking regions of CbNip1 gene (CB0940_03921) with a gap representing the 2.8 kb hygromycin B phosphotransferase insertion is depicted above. Schematic representation of the hygromycin cassette insertion in the context of a 14 kb section of chromosome 4 encompassing CbNip1. Image is representative of genome sequences of three CbNip1 deletion mutants having a 21× genome coverage where 98% of the 847 hygromycin B phosphotransferase reads were mapped to CB0940_03921
FIGURE S10 CbNip1 ectopic transformant confirmation. The CbNip1 ectopic mutant was verified by PCR using three different primer pairs. All PCRs were run on gDNA of Cercospora beticola 09‐40 wild type (1), C. beticola ectopic transformant (2), and ∆CbNip1 #1. C. beticola 09‐40 wild type and ∆CbNip1 #1 served as controls. (a) In primer pair MDB‐1145/MDB‐1541, the forward primer amplifies 5′ of the upstream flanking region used for CbNip1 knockout construct generation and is combined with a hygromycin reverse primer. As expected, no amplicon is present in wild‐type and ectopic mutant, while PCR on the site‐directed ∆CbNip1 #1 yields the amplicon at the expected size (912 bp). (b) Primer pair MDB‐1628/MDB‐1629 anneals to the coding sequence of CbNip1 (expected size 540 bp). We are able to obtain an amplicon at the expected size with this primer pair when running on gDNA of C. beticola 09‐40 wild‐type and C. beticola ectopic transformant, but not of ∆CbNip1 #1. (c) Primer pair MDB‐258/MDB‐259 targets the hygromycin cassette. As expected, we amplified hygromycin from the C. beticola ectopic transformant and ∆CbNip1 #1, but not C. beticola 09‐40 wild type
FIGURE S11 Growth of Cercospora beticola 09‐40 under different stress conditions. Four‐day‐old cultures of wild type (labelled as WT) and three individual ΔCbNip1 mutants (labelled as #1, #2, and #3) were inoculated on to potato dextrose agar plates supplemented with sorbitol (1 M), calcofluor (100 μg/mL), charcoal (0.01 g/ml), Congo red (45 μg/ml), H2O2 (1 mM), or NaCl (1M). After incubation for 7 days at room temperature, fungal growth was assessed and photographs were taken
FIGURE S12 Gel visualization of infiltrated CbNIP1 protein and empty vector (EV) samples. SDS‐PAGE gel loaded with 5 µl of heterologously expressed protein sample used for infiltration studies. NIP1 I, NIP1 II, and NIP1 III are three individually produced and purified CbNIP1 protein samples (arrow indicates CbNIP1 protein). EVI is the individually produced and purified empty vector sample used as control
TABLE S1 NCBI hit table for homologs search results using CB0940_10646 as query
TABLE S2 NCBI hit table for homolog search results using CbNIP1 as query
TABLE S3 Summary of top results for 3D protein structure and function prediction for NIP1 proteins in Cercospora spp. using the I‐TASSER server (Roy et al., 2010; Yang et al., 2015; Zhang, 2008)
TABLE S4 Primers used in this study
DATASET S1 Amino acid sequences of CbNIP1 homologs identified via de novo sequence assembly from genome data available at the NCBI database
ACKNOWLEDGMENTS
M.D.B. is supported by USDA–ARS CRIS project 3060‐22000‐044 and grants from Beet Sugar Development Foundation and the Sugar Beet Research and Education Board of MN and ND. M.K.E. was supported by NWO grant 833.13.007.
Ebert MK, Rangel LI, Spanner RE, et al. Identification and characterization of Cercospora beticola necrosis‐inducing effector CbNip1. Mol Plant Pathol. 2021;22:301–316. 10.1111/mpp.13026
Malaika K. Ebert and Lorena I. Rangel contributed equally to this work.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Alachiotis, N. & Pavlidis, P. (2018) RAiSD detects positive selection based on multiple signatures of a selective sweep and SNP vectors. Communications Biology, 1, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alachiotis, N. , Stamatakis, A. & Pavlidis, P. (2012) OmegaPlus: a scalable tool for rapid detection of selective sweeps in whole‐genome datasets. Bioinformatics, 28, 2274–2275. [DOI] [PubMed] [Google Scholar]
- Allen, A. , Chatt, E. & Smith, T.J. (2013) The Atomic structure of the virally encoded antifungal protein, KP6. Journal of Molecular Biology, 425, 609–621. [DOI] [PubMed] [Google Scholar]
- Bailey, B.A. (1995) Purification of a protein from culture filtrates of Fusarium oxysporum that induces ethylene and necrosis in leaves of Erythroxylum coca . Phytopathology, 85, 1250–1255. [Google Scholar]
- Bailey, B.A. , Dean, J.F.D. & Anderson, J.D. (1990) An ethylene biosynthesis‐inducing endoxylanase elicits electrolyte leakage and necrosis in Nicotiana tabacum cv. Xanthi leaves. Plant Physiology, 94, 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben M’Barek, S. , Cordewener, J.H. , Tabib Ghaffary, S.M. , van der Lee, T.A. , Liu, Z. , Mirzadi Gohari, A. et al. (2015) FPLC and liquid‐chromatography mass spectrometry identify candidate necrosis‐inducing proteins from culture filtrates of the fungal wheat pathogen Zymoseptoria tritici . Fungal Genetics and Biology, 79, 54–62. [DOI] [PubMed] [Google Scholar]
- Bird, L.J. , Coleman, M.L. & Newman, D.K. (2013) Iron and copper act synergistically to delay anaerobic growth of bacteria. Applied and Environmental Microbiology, 79, 3619–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, M.D. , Ebert, M.K. , Faino, L. , Rivera‐Varas, V. , de Jonge, R. , Van de Peer, Y. et al. (2016) RNA‐sequencing of Cercospora beticola DMI‐sensitive and ‐resistant isolates after treatment with tetraconazole identifies common and contrasting pathway induction. Fungal Genetics and Biology, 92, 1–13. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , Secor, G.A. , Rivera, V. , Weiland, J.J. , Rudolph, K. , Birla, K. et al. (2012) Evaluation of the potential for sexual reproduction in field populations of Cercospora beticola from USA. Fungal Biology, 116, 511–521. [DOI] [PubMed] [Google Scholar]
- Brito, N. , Espino, J.J. & González, C. (2006) The endo‐β‐1,4‐xylanase xyn11A is required for virulence in Botrytis cinerea . Molecular Plant‐Microbe Interactions, 19, 25–32. [DOI] [PubMed] [Google Scholar]
- Callahan, T.M. , Rose, M.S. , Meade, M.J. , Ehrenshaft, M. & Upchurch, R.G. (1999) CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild type cercosporin production, resistance, and virulence on soybean. Molecular Plant‐Microbe Interactions, 12, 901–910. [DOI] [PubMed] [Google Scholar]
- Capella‐Gutiérrez, S. , Silla‐Martínez, J.M. & Gabaldón, T. (2009) trimAl: a tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics, 25, 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changela, A. , Chen, K. , Xue, Y. , Holschen, J. , Outten, C.E. , O’Halloran, T.V. et al. (2003) Molecular basis of metal‐ion selectivity and zeptomolar sensitivity by CueR. Science, 301, 1383–1387. [DOI] [PubMed] [Google Scholar]
- Choquer, M. , Dekkers, K.L. , Chen, H.‐Q. , Cao, L. , Ueng, P.P. , Daub, M.E. et al. (2005) The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin biosynthesis and fungal virulence of Cercospora nicotianae . Molecular Plant‐Microbe Interactions, 18, 468–476. [DOI] [PubMed] [Google Scholar]
- Daub, M.E. & Ehrenshaft, M. (2000) The photoactivated Cercospora toxin cercosporin: contributions to plant disease and fundamental biology. Annual Review of Phytopathology, 38, 461–490. [DOI] [PubMed] [Google Scholar]
- De Coninck, B.M.A. , Amand, O. , Delauré, S.L. , Lucas, S. , Hias, N. , Weyens, G. et al. (2012) The use of digital image analysis and real‐time PCR fine‐tunes bioassays for quantification of cercospora leaf spot disease in sugar beet breeding. Plant Pathology, 61, 76–84. [Google Scholar]
- Excoffier, L. , Dupanloup, I. , Huerta‐Sánchez, E. , Sousa, V.C. & Foll, M. (2013) Robust demographic inference from genomic and SNP data. PLoS Genetics, 9, e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris, J.D. , Zhang, Z. , Lu, H. , Lu, S. , Reddy, L. , Cloutier, S. et al. (2010) A unique wheat disease resistance‐like gene governs effector‐triggered susceptibility to necrotrophic pathogens. Proceedings of the National Academy of Sciences of the United States of America, 107, 13544–13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feindt, F. , Mendgen, K. & Heitefuss, R. (1981) Feinstruktur unterschiedlicher Zellwandreaktionen im Blattparenchym anfälliger und resistenter Rüben (Beta vulgaris L.) nach Infektion durch Cercospora beticola Sacc. Journal of Phytopathology, 101, 248–264. [Google Scholar]
- Fellbrich, G. , Romanski, A. , Varet, A. , Blume, B. , Brunner, F. , Engelhardt, S. et al. (2002) NPP1, a Phytophthora‐associated trigger of plant defense in parsley and Arabidopsis . The Plant Journal, 32, 375–390. [DOI] [PubMed] [Google Scholar]
- Finkler, A. , Peery, T. , Tao, J. , Bruenn, J. & Koltin, I. (1992) Immunity and resistance to the KP6 toxin of Ustilago maydis . Molecular and General Genetics, 233, 395–403. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1942) Inheritance of pathogenicity in Melampsora lini . Phytopathology, 32, 653–669. [Google Scholar]
- Friesen, T.L. & Faris, J.D. (2012) Characterization of plant–fungal interactions involving necrotrophic effector‐producing plant pathogens In: Bolton, M.D. and Thomma, B.P.H.J. (Eds.) Plant fungal pathogens: methods and protocols. Totowa, NJ: Humana Press, pp. 191–207. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Faris, J.D. , Solomon, P.S. & Oliver, R.P. (2008) Host‐specific toxins: effectors of necrotrophic pathogenicity. Cellular Microbiology, 10, 1421–1428. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Meinhardt, S.W. & Faris, J.D. (2007) The Stagonospora nodorum–wheat pathosystem involves multiple proteinaceous host‐selective toxins and corresponding host sensitivity genes that interact in an inverse gene‐for‐gene manner. The Plant Journal, 51, 681–692. [DOI] [PubMed] [Google Scholar]
- Fulton T.M., Chunwongse J., Tanksley S.D. (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Molecular Biology Reporter, 13, 207–209. [Google Scholar]
- Gijzen, M. & Nürnberger, T. (2006) Nep1‐like proteins from plant pathogens: Recruitment and diversification of the NPP1 domain across taxa. Phytochemistry, 67, 1800–1807. [DOI] [PubMed] [Google Scholar]
- GNU Project (2015) GNU PSPP for GNU/Linux (Version 0.8.5) [Computer Software]. Boston, MA: Free Software Foundation; Available from: https://www.gnu.org/software/pspp/ [Accessed 24 July 2020]. [Google Scholar]
- Guo, A. , Reimers, P.J. & Leach, J.E. (1993) Effect of light on incompatible interactions between Xanthomonas oryzae pv. oryzae and rice. Physiological and Molecular Plant Pathology, 42, 413–425. [Google Scholar]
- Horbach, R. , Navarro‐Quesada, A.R. , Knogge, W. & Deising, H.B. (2011) When and how to kill a plant cell: infection strategies of plant pathogenic fungi. Journal of Plant Physiology, 168, 51–62. [DOI] [PubMed] [Google Scholar]
- Hudson R.R. (2002) Generating samples under a Wright‐Fisher neutral model of genetic variation. Bioinformatics, 18, 337–338. [DOI] [PubMed] [Google Scholar]
- Inohara, N. & Nuñez, G. (2002) ML—A conserved domain involved in innate immunity and lipid metabolism. Trends in Biochemical Sciences, 27, 219–221. [DOI] [PubMed] [Google Scholar]
- Jalal, M.A.F. , Hossain, M.B. , Robeson, D.J. & van der Helm, D. (1992) Cercospora beticola phytotoxins: cebetins that are photoactive, magnesium ion‐binding, chlorinated anthraquinone–xanthone conjugates. Journal of the American Chemical Society, 114, 5967–5971. [Google Scholar]
- de Jonge, R. , Ebert, M.K. , Huitt‐Roehl, C.R. , Pal, P. , Suttle, J.C. , Spanner, R.E. et al. (2018) Gene cluster conservation provides insight into cercosporin biosynthesis and extends production to the genus Colletotrichum . Proceedings of the National Academy of Sciences of the United States of America, 115, E5459–E5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kairys, V. , Gilson, M.K. & Luy, B. (2004) Structural model for an AxxxG‐mediated dimer of surfactant‐associated protein C. European Journal of Biochemistry, 271, 2086–2092. [DOI] [PubMed] [Google Scholar]
- Kami, C. , Lorrain, S. , Hornitschek, P. & Fankhauser, C. (2010) Light‐regulated plant growth and development. Current Topics in Developmental Biology, 91, 29–66. [DOI] [PubMed] [Google Scholar]
- Kars, I. , Krooshof, G.H. , Wagemakers, L. , Joosten, R. , Benen, J.A.E. & Kan, J.A.L.V. (2005) Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris . The Plant Journal, 43, 213–225. [DOI] [PubMed] [Google Scholar]
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettles, G.J. , Bayon, C. , Canning, G. , Rudd, J.J. & Kanyuka, K. (2017) Apoplastic recognition of multiple candidate effectors from the wheat pathogen Zymoseptoria tritici in the nonhost plant Nicotiana benthamiana . New Phytologist, 213, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettles, G.J. , Bayon, C. , Sparks, C.A. , Canning, G. , Kanyuka, K. & Rudd, J.J. (2018) Characterization of an antimicrobial and phytotoxic ribonuclease secreted by the fungal wheat pathogen Zymoseptoria tritici . New Phytologist, 217, 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M.F.R. & Khan, J. (2010) Survival, spore trapping, dispersal, and primary infection site for Cercospora beticola in sugar beet In: Lartey, R.T. , Weiland, J.J. , Panella, L. , Crous, P.W. & Windels, C.E. (Eds.) Cercospora leaf spot of sugar beet and related species. St Paul, MN: APS Press; , pp. 67–75. [Google Scholar]
- Kim, H. , Ridenour, J.B. , Dunkle, L.D. & Bluhm, B.H. (2011) Regulation of stomatal tropism and infection by light in Cercospora zeae‐maydis: evidence for coordinated host/pathogen responses to photoperiod? PLoS Pathogens, 7, e1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltin, Y. & Day, P. (1975) Specificity of Ustilago maydis killer proteins. Applied Microbiology, 30, 694–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugé, R. , Joosten, M.H.A.J. , Haanstra, J.P.W. , Goodwin, P.H. , Lindhout, P. & De Wit, P.J.G.M. (1998) Successful search for a resistance gene in tomato targeted against a virulence factor of a fungal pathogen. Proceedings of the National Academy of Sciences of the United States of America, 95, 9014–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Faris, J. , Meinhardt, S. , Ali, S. , Rasmussen, J. & Friesen, T. (2004) Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host‐selective toxin produced by Stagonospora nodorum . Phytopathology, 94, 1056–1060. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Faris, J.D. , Oliver, R.P. , Tan, K.‐C. , Solomon, P.S. , McDonald, M.C. et al. (2009) SnTox3 acts in effector triggered susceptibility to induce disease on wheat carrying the Snn3 gene. PLoS Pathogens, 5, e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Zhang, Z. , Faris, J.D. , Oliver, R.P. , Syme, R. , McDonald, M.C. et al. (2012) The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1 . PLoS Pathogens, 8, e1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. & Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCt method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Motteram, J. , Küfner, I. , Deller, S. , Brunner, F. , Hammond‐Kosack, K.E. , Nürnberger, T. et al. (2009) Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain‐containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola . Molecular Plant‐Microbe Interactions, 22, 790–799. [DOI] [PubMed] [Google Scholar]
- Mullen, G.E.D. , Kennedy, M.N. , Visintin, A. , Mazzoni, A. , Leifer, C.A. , Davies, D.R. et al. (2003) The role of disulfide bonds in the assembly and function of MD‐2. Proceedings of the National Academy of Sciences of the United States of America, 100, 3919–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Williamson, S. , Kim, Y. , Hubisz, M.J. , Clark, A.G. & Bustamante, C. (2005) Genomic scans for selective sweeps using SNP data. Genome Research, 15, 1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P., Živković D., Stamatakis A., Alachiotis N. (2013) SweeD: Likelihood‐based detection of selective sweeps in thousands of genomes. Molecular Biology and Evolution, 30, 2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton, C.L. & Salmond, G.P.C. (2004) The Nep1‐like proteins – a growing family of microbial elicitors of plant necrosis. Molecular Plant Pathology, 5, 353–359. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Research, 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, T. , Seong, K. , Thomazella, D.P.T. , Kim, J.R. , Pham, J. , Seo, E. et al. (2018) NRG1 functions downstream of EDS1 to regulate TIR‐NLR‐mediated plant immunity in Nicotiana benthamiana . Proceedings of the National Academy of Sciences of the United States of America, 115, E10979–E10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaello, T. & Asiegbu, F.O. (2017) Small secreted proteins from the necrotrophic conifer pathogen Heterobasidion annosum s.l. (HaSSPs) induce cell death in Nicotiana benthamiana . Scientific Reports, 7, 8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel, L.I. , Spanner, R.E. , Ebert, M.K. , Pethybridge, S.J. , Stukenbrock, E.H. , de Jonge, R. et al. (2020) Cercospora beticola: The intoxicating lifestyle of the leaf spot pathogen of sugar beet. Molecular Plant Pathology, 21, 1020–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathaiah, Y. (1977) Stomatal tropism of Cercospora beticola in sugarbeet. Phytopathology, 67, 358–362. [Google Scholar]
- Roberts, M.R. & Paul, N.D. (2006) Seduced by the dark side: integrating molecular and ecological perspectives on the influence of light on plant defence against pests and pathogens. New Phytologist, 170, 677–699. [DOI] [PubMed] [Google Scholar]
- Roden, L.C. & Ingle, R.A. (2009) Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant–pathogen interactions. The Plant Cell, 21, 2546–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, A. , Kucukural, A. & Zhang, Y. (2010) I‐TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols, 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlösser, E. (1962) Über eine biologisch aktive Substanz aus Cercospora beticola . Journal of Phytopathology, 44, 295–312. [Google Scholar]
- Secor, G.A. & Rivera, V.V. (2012) Fungicide resistance assays for fungal plant pathogens. Plant Fungal Pathogens: Methods and Protocols, 385–392. [DOI] [PubMed] [Google Scholar]
- Shi, G. , Zhang, Z. , Friesen, T.L. , Raats, D. , Fahima, T. , Brueggeman, R.S. et al. (2016) The hijacking of a receptor kinase–driven pathway by a wheat fungal pathogen leads to disease. Science Advances, 2, e1600822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.M. & Haigh, J. (1974) The hitch‐hiking effect of a favourable gene. Genetics Research, 23, 23–35. [PubMed] [Google Scholar]
- Solel, Z. & Minz, G. (1971) Infection process of Cercospora beticola in sugarbeet in relation to susceptibility. Phytopathology, 61, 463–466. [Google Scholar]
- Sperschneider, J. , Dodds, P.N. , Gardiner, D.M. , Singh, K.B. & Taylor, J.M. (2018) Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Molecular Plant Pathology, 19, 2094–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkamp, M.P. , Martin, S.S. , Hoefert, L.L. & Ruppel, E.G. (1979) Ultrastructure of lesions produced by Cercospora beticola in leaves of Beta vulgaris . Physiological Plant Pathology, 15, 13–26. [Google Scholar]
- Thomma, B.P.H.J. , Seidl, M.F. , Shi‐Kunne, X. , Cook, D.E. , Bolton, M.D. , van Kan, J.A.L. et al. (2016) Mind the gap; seven reasons to close fragmented genome assemblies. Fungal Genetics and Biology, 90, 24–30. [DOI] [PubMed] [Google Scholar]
- Weltmeier, F. , Mäser, A. , Menze, A. , Hennig, S. , Schad, M. , Breuer, F. et al. (2011) Transcript profiles in sugar beet genotypes uncover timing and strength of defense reactions to Cercospora beticola infection. Molecular Plant‐Microbe Interactions, 24, 758–772. [DOI] [PubMed] [Google Scholar]
- Yamazaki, S. , Okubo, A. , Akiyama, Y. & Fuwa, K. (1975) Cercosporin, a novel photodynamic pigment isolated from Cercospora kikuchii . Agricultural and Biological Chemistry, 39, 287–288. [Google Scholar]
- Yang, J. , Yan, R. , Roy, A. , Xu, D. , Poisson, J. & Zhang, Y. (2015) The I‐TASSER suite: protein structure and function prediction. Nature Methods, 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. (2008) I‐TASSER server for protein 3D structure prediction. BMC Bioinformatics, 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 SDS‐PAGE gel displaying the protein content of active fraction #9 and crude culture filtrate. Protein samples of active fraction #9 and crude culture filtrate (indicated by arrows) were run on SDS‐PAGE. Subsequently, the SDS‐PAGE gel was stained with MS/MS compatible Coomassie Blue stain and band incisions are depicted by red boxes. Excised samples were sent for MS/MS analysis
FIGURE S2 Illustration of effector characteristics found in peptide sequences of CB0940_03921 (CbNIP1) and CB0940_10646. Both candidates display classic effector characteristics such as signal peptides (highlighted in pink) and predicted disulphide bonds (each individual disulphide bond is indicated by a differently coloured line) while the mature sequences are highlighted in blue. Underlined in the CB0940_10646 sequence are peptide sequence snippets SDAVRK, a conserved SxxV(K/R) motif associated with monocation specificity, and AYYYG, an AxxxG motif shown to be involved in dimerization
FIGURE S3 Genomic context CB0940_03920. The flanking genomic region of CbNip1 (CB0940_03920) highlighting a large sequence gap just upstream as well as several smaller repetitive elements. Prepared at https://www.ncbi.nlm.nih.gov/gene/?term=CB0940_03921
FIGURE S4 Alignment of CbNIP1 sequences found in different Cercospora species. Homologous protein sequences display high sequence conservation. Nucleotides of CbNip1 homologs that are identical to the nucleotide sequence of CbNip1 are indicated with a period, while variations are highlighted by displaying the corresponding nucleotide
FIGURE S5 Alignment of selected homologous CbNIP1 peptide sequences. The sequence alignment was constructed by mafft and the phylogenetically relevant positions are identified and shown here using trimAl. Sequence positions selected for phylogenetic analysis are listed below the alignment in dark grey “Selected Cols”. Asterisks indicate conserved cysteines. Signal peptides of CbNIP1 homologs are indicated with purple boxes. CbNIP homologs that lack a predicted signal peptide for secretion are depicted in red
FIGURE S6 The I‐TASSER server was used to predict the 3D structure of the identified NIP1 protein in Cercospora beticola (XP_023452571.1). The highest confidence model (C‐score = −1.7) for the CbNIP1 protein is displayed
FIGURE S7 No visible phenotype upon inoculation of CB0940_10646 protein. (a) No phenotype after CB0940_10646 infiltration into sugar beet leaf exposed to a 10 hr/14 hr light/dark cycle for 5 and 7 DPInf, while CbNIP1 served as a positive control to display expected chlorosis after 5 DPInf and beginning of necrosis after 7 DPInf. (b) No visible phenotype was identified after CB0940_10646 was infiltrated into a sugar beet leaf that was kept in 24 hr darkness for 2 and 3 DPInf. CT served as a control for CB0940_10646 infiltration and consisted of an equal amount of trace elements to what was supplied in the CB0940_10646 sample. As a positive control, we observe necrosis development after CbNIP1 inoculation, and an empty vector (EV) infiltration served as a control for CbNIP1 and showed no phenotype
FIGURE S8 Symptom development during Cercospora beticola 09‐40 wild type strain infection of sugar beet. No symptoms are detectable at early time points (3, 6, and 9 DPInoc). Red arrows at 12 and 15 DPInoc indicate formation of necrotic lesions. At 18 DPInoc, necrotic lesion formation intensifies, accompanied by some chlorosis around the necrotic lesions. At 21 DPInoc, single necrotic lesions are fused to form necrotic patches on the sugar beet leaf
FIGURE S9 Confirmation of site‐directed integration using Oxford Nanopore MinION reads. Mapping of Oxford Nanopore MinIon reads to Cercospora beticola 09‐40 genome. Genome browser view of the single hygromycin resistance gene insertion found in CbNip1 deletion mutant. Reads map to flanking regions of CbNip1 gene (CB0940_03921) with a gap representing the 2.8 kb hygromycin B phosphotransferase insertion is depicted above. Schematic representation of the hygromycin cassette insertion in the context of a 14 kb section of chromosome 4 encompassing CbNip1. Image is representative of genome sequences of three CbNip1 deletion mutants having a 21× genome coverage where 98% of the 847 hygromycin B phosphotransferase reads were mapped to CB0940_03921
FIGURE S10 CbNip1 ectopic transformant confirmation. The CbNip1 ectopic mutant was verified by PCR using three different primer pairs. All PCRs were run on gDNA of Cercospora beticola 09‐40 wild type (1), C. beticola ectopic transformant (2), and ∆CbNip1 #1. C. beticola 09‐40 wild type and ∆CbNip1 #1 served as controls. (a) In primer pair MDB‐1145/MDB‐1541, the forward primer amplifies 5′ of the upstream flanking region used for CbNip1 knockout construct generation and is combined with a hygromycin reverse primer. As expected, no amplicon is present in wild‐type and ectopic mutant, while PCR on the site‐directed ∆CbNip1 #1 yields the amplicon at the expected size (912 bp). (b) Primer pair MDB‐1628/MDB‐1629 anneals to the coding sequence of CbNip1 (expected size 540 bp). We are able to obtain an amplicon at the expected size with this primer pair when running on gDNA of C. beticola 09‐40 wild‐type and C. beticola ectopic transformant, but not of ∆CbNip1 #1. (c) Primer pair MDB‐258/MDB‐259 targets the hygromycin cassette. As expected, we amplified hygromycin from the C. beticola ectopic transformant and ∆CbNip1 #1, but not C. beticola 09‐40 wild type
FIGURE S11 Growth of Cercospora beticola 09‐40 under different stress conditions. Four‐day‐old cultures of wild type (labelled as WT) and three individual ΔCbNip1 mutants (labelled as #1, #2, and #3) were inoculated on to potato dextrose agar plates supplemented with sorbitol (1 M), calcofluor (100 μg/mL), charcoal (0.01 g/ml), Congo red (45 μg/ml), H2O2 (1 mM), or NaCl (1M). After incubation for 7 days at room temperature, fungal growth was assessed and photographs were taken
FIGURE S12 Gel visualization of infiltrated CbNIP1 protein and empty vector (EV) samples. SDS‐PAGE gel loaded with 5 µl of heterologously expressed protein sample used for infiltration studies. NIP1 I, NIP1 II, and NIP1 III are three individually produced and purified CbNIP1 protein samples (arrow indicates CbNIP1 protein). EVI is the individually produced and purified empty vector sample used as control
TABLE S1 NCBI hit table for homologs search results using CB0940_10646 as query
TABLE S2 NCBI hit table for homolog search results using CbNIP1 as query
TABLE S3 Summary of top results for 3D protein structure and function prediction for NIP1 proteins in Cercospora spp. using the I‐TASSER server (Roy et al., 2010; Yang et al., 2015; Zhang, 2008)
TABLE S4 Primers used in this study
DATASET S1 Amino acid sequences of CbNIP1 homologs identified via de novo sequence assembly from genome data available at the NCBI database
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


