Abstract
Simple Summary
Cancer cells in solid tumors often experience lack of oxygen (hypoxia), which they overcome with the help of hypoxia inducible transcription factors (HIFs). When HIFs are activated, they stimulate the expression of many genes and cause the production of proteins that help cancer cells grow and migrate even in the presence of very little oxygen. Many experiments have shown that agents that block the activity of HIFs (HIF inhibitors) can prevent growth of cancer cells under hypoxia and, subsequently, hinder formation of malignant tumors or metastases. Most small chemical HIF inhibitors lack the selectivity required for development of safe anticancer drugs. On the other hand, peptides derived from HIFs themselves can be very selective HIF inhibitors by disrupting specific associations of HIFs with cellular components that are essential for HIF activation. This review discusses the nature of available peptide HIF inhibitors and their prospects as effective pharmaceuticals against cancer.
Abstract
Reduced oxygen availability (hypoxia) is a characteristic of many disorders including cancer. Central components of the systemic and cellular response to hypoxia are the Hypoxia Inducible Factors (HIFs), a small family of heterodimeric transcription factors that directly or indirectly regulate the expression of hundreds of genes, the products of which mediate adaptive changes in processes that include metabolism, erythropoiesis, and angiogenesis. The overexpression of HIFs has been linked to the pathogenesis and progression of cancer. Moreover, evidence from cellular and animal models have convincingly shown that targeting HIFs represents a valid approach to treat hypoxia-related disorders. However, targeting transcription factors with small molecules is a very demanding task and development of HIF inhibitors with specificity and therapeutic potential has largely remained an unattainable challenge. Another promising approach to inhibit HIFs is to use peptides modelled after HIF subunit domains known to be involved in protein–protein interactions that are critical for HIF function. Introduction of these peptides into cells can inhibit, through competition, the activity of endogenous HIFs in a sequence and, therefore also isoform, specific manner. This review summarizes the involvement of HIFs in cancer and the approaches for targeting them, with a special focus on the development of peptide HIF inhibitors and their prospects as highly-specific pharmacological agents.
Keywords: hypoxia, HIF, HIF-1α, HIF inhibition, peptide inhibitors
1. Introduction
Oxygen is indispensable for cellular metabolism, as it is essential for aerobic respiration and energy production. Moreover, oxygen has a vital role as a substrate in many enzymatic reactions that regulate diverse biological processes. Mammals use oxygen sensing and delivery mechanisms to match available oxygen to their tissue demands so as to fulfill their metabolic needs but also prevent toxicity caused by excess oxygen [1]. Lack of sufficient oxygen or hypoxia can arise from an imbalance between oxygen delivery and consumption, that can occur under either physiological or pathological conditions [2]. A frequent example is ischemia, in which infarctions impair oxygen delivery via the circulation and form hypoxic areas. Oxygen delivery is also impaired in solid tumors because of the irregular vascularization caused by an imbalance between pro and anti-angiogenic signals. In addition, the high oxygen consumption rates of the rapidly proliferating cancer cell intensify the formation of hypoxic niches within solid tumors [3].
At the cellular level, hypoxia triggers the stabilization of the hypoxia inducible factors (known as HIFs) [2]. HIFs are transcription factors that initiate a cascade of events such as metabolic reprogramming, induction of angiogenesis and erythropoiesis that in healthy tissues facilitate adaptation to low oxygen conditions. Especially in cancer, HIFs, in addition to upregulating genes involved in glucose and lipid metabolism or vascularization, can also promote cell proliferation, resistance to apoptosis, evasion of the immune response, genomic instability, invasion, and metastasis [4,5]. Considering the pivotal role of HIFs and hypoxia in cancer, it is of no surprise that HIFs have been long targeted as means of anti-cancer therapy [6,7].
Here, we review the different strategies that directly or indirectly target control points of the hypoxic signaling pathway and their application in disease models. We specifically highlight the use of peptides as effectors of HIF activity and discuss their future perspectives and clinical significance.
2. The HIF-Dependent Response to Hypoxia
2.1. The HIF Family
The HIF family of heterodimeric transcription factors consists of 3 HIFα members (namely HIF-1α, HIF-2α, and HIF-3α) and one HIFβ member (HIF-1β, best known as ARNT) [8]. HIF-1α is the most well studied member of the family and the first to be discovered by Semenza and coworkers [9,10,11] by its ability to bind to a hypoxia response element (HRE) in the 3′ enhancer of the human EPO gene. Unlike HIF-1α that can be expressed in all types of cells, HIF-2α, encoded by the EPAS1 gene, is expressed in a few tissues such as placenta, lungs, liver, and heart, and holds a central role in angiogenesis and erythropoiesis [12,13]. HIF-3α, the less studied isoform, has many spliced variants with distinct expression pattern [14] and diverging functionalities ranging from HIF-1 inhibition to transcriptional activation of HIF targets [15,16].
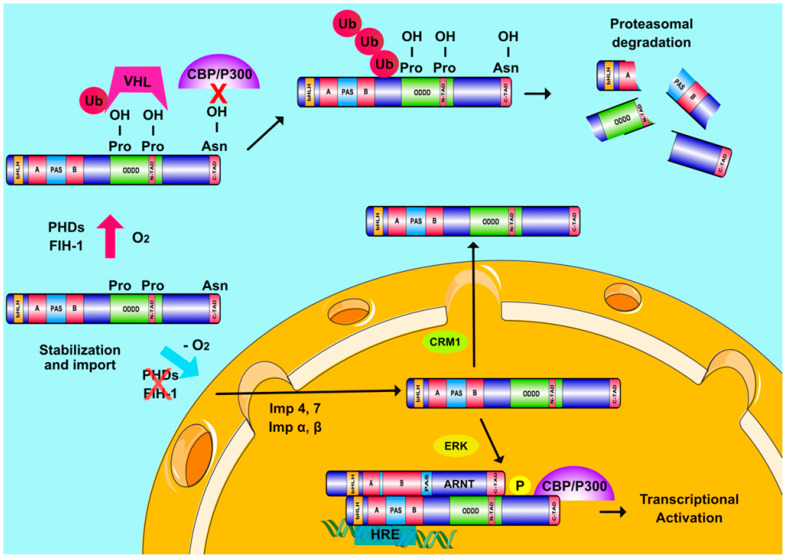
Detailed studies have shown that HIF-1α, as well as the other HIFα forms, are stabilized under hypoxia by an oxygen dependent mechanism (Figure 1) [17]. ARNT, which is constitutively expressed in cells regardless of oxygen concentration, associates with the HIFα subunits within the nucleus to form a functional heterodimer (HIF) that can bind to the HREs of hypoxia target genes and initiate the transcriptional hypoxic response [18]. Heterodimerization and DNA binding are mediated by the Per-Arnt-Sim (PAS) homology and basic helix-loop-helix (bHLH) domains, respectively, which are present at the N-terminal parts of all HIF subunits [18]. Structural data indicate that both HIF isoforms bind HRE sequences in an identical fashion. The α-helices of their bHLH domains associate with the major groove of the recognition motif and their PAS-A domains cooperate with the bHLH domains of the heterodimer to establish binding to DNA [19]. Furthermore, the PAS domains of both HIFα isoforms possess cavities, which can accommodate small ligands, but their size and distribution differs between isoforms [19,20].
Figure 1.
Regulation of HIFα subunits and Hypoxia Inducible Factors (HIF) transcriptional activity. When oxygen is abundant, PHDs and FIH hydroxylate two proline and one asparagine residue inside the ODDD and C-TAD regions of HIFα, respectively. Prolyl-hydroxylation leads to pVHL-mediated ubiquitination of HIFα subunits and their destruction in the proteasome while asparaginyl hydroxylation also inhibits HIFα interaction with CBP/P300. When oxygen levels drop, hydroxylases become inactive and HIFα subunits are stabilized and transported into the nucleus with the help of multiple importins (Imp α, β, 4, and 7). ERK-mediated phosphorylation of HIFα subunits ensures their nuclear accumulation by abrogating the association of HIFα with exportin CRM1. Nuclear HIFα subunits form a functional complex with ARNT, which binds to HREs on DNA and coactivators, such as CBP/p300 to induce transcription of hypoxia-regulated genes.
The C-terminal parts of both HIF-1α and HIF-2α are also critical for function as they contain their oxygen dependent degradation domain (ODDD) as well as two distinct transactivation domains, N-TAD (overlapping with ODDD) and C-TAD (at the very C-terminus), which is responsible for the interaction between HIFα and the transcriptional coactivator proteins CBP/p300 (Figure 1) [21]. HIF-1 and HIF-2 activate a great number of genes (more than 1000) which can either be specific for each factor or common for both of them [6,8]. Domain-swapping experiments have suggested that HIF target gene specificity may be conferred by the N-TAD through its interaction with additional transcriptional co-regulators [22].
2.2. HIFs and Cellular Oxygen Sensing
The cellular oxygen sensing mechanism has been characterized in the previous decade mainly by the work of G. Semenza, Sir P. Ratcliffe and W. Kaelin (2019 Nobel prize in Physiology or Medicine). Their breakthrough experiments revealed that the HIFα subunits are subjected to hydroxylation in two specific prolyl residues when cells are grown under atmospheric oxygen concentrations (normoxia) [21]. This post-translational modification is essential for interaction with the Von Hippel-Lindau (pVHL) tumor suppressor protein that is part of a ubiquitin E3 ligase complex. As a result of this interaction, HIFα subunits are polyubiquitinated and targeted to the proteasome for destruction [23,24,25]. The enzymes catalyzing this hydroxylation are prolyl-hydroxylases PHD1, 2, and 3 in humans, also known as EGLN2,1 and 3. PHDs belong to the 2-oxoglutarate-dependent dioxygenase family and are thought to act as oxygen sensors since they use molecular oxygen as substrate. PHD2 is the most abundant and best studied isoform in cells [1,26,27,28]. A second oxygen dependent hydroxylation occurs at an asparagine residue in the C-TAD of HIFα and is catalyzed by a different oxygenase, called FIH (Factor Inhibiting HIF) [29,30]. FIH downregulates HIF transcriptional activity by impairing HIF binding to CBP/p300. According to all the above, HIFαs are constitutively produced but in the presence of normal oxygen concentrations both their expression and activity remain minimal. Under hypoxia, low O2 levels as well as the production of ROS by oxygen-starving mitochondria inhibit hydroxylation and HIFαs escape degradation, accumulate, and translocate inside the nucleus where they assemble with ARNT into transcriptionally active heterodimers (Figure 1) [31].
2.3. Oxygen-Independent Regulation of HIFs
Over the past few years, it has become clear that HIFs can also be regulated by mechanisms not directly affected by oxygen concentration. This regulation can occur at multiple levels including transcription, translation, post-translational modification, stabilization, nuclear translocation and activation of HIFαs [2,31,32,33]. Post-translational modifications of HIFαs probably hold the most important role in their regulation [34]. HIFαs are subjected to acetylation, s-nitrosylation and sumoylation, but the significance of these modifications for HIF activity is still a matter of debate. On the other hand, HIFα phosphorylation is much better characterized with clearly demonstrated importance in various cellular models. Both HIF-1α and HIF-2α can be directly phosphorylated by several kinases including GSK3, PLK3, ATM, PKA, CDK1, and the extent of their modification depends on cell type and extracellular signals [34,35]. Previous work from our lab has also shown that HIF-1α is a direct target of kinases ERK1/2 and CK1δ, modification by which has distinct outcomes on HIF-1 activity [36,37,38,39]. More specifically, while import of HIF-1α inside the nucleus is constitutive, mediated by the importin α/β as well as importins 4/7 [40,41], nuclear export of HIF-1α is regulated in an ERK1/2- and CRM1-dependent manner (Figure 1) [38,39]. Phosphorylation of HIF-1α by ERK1/2 at Ser641/643, which lay inside a small domain termed ETD (ERK Targeted Domain; amino acids 616–658), masks a nearby CRM1-dependent nuclear export signal (NES), thus inhibiting HIF-1α nuclear export and increasing HIF-1α nuclear concentration and HIF-1 transcriptional activity [38,39]. Lack of this phosphorylation allows CRM1 binding to HIF-1α and its subsequent translocation to the cytoplasm, where, interestingly, HIF-1α interacts with mortalin and takes part in the assembly of anti-apoptotic complex on the surface of the mitochondria [36,42]. This mode of HIF-1α regulation by ERK1/2 has been exploited for the development of peptide HIF-1 inhibitors modelled after the ETD amino acid sequence (see below). On the other hand, phosphorylation of HIF-1α by Ck1δ at Ser247 inside the PAS domain has a negative effect by inhibiting the ability of HIF-1α to associate with ARNT [37,38,39,43,44,45]. Regulation of the HIF-1 heterodimer assembly by phosphorylation as well as by MgcRacGAP [46,47] highlights the HIF-1α/ARNT interaction as a target for peptide inhibitors modelled after the PAS domain, an approach that has indeed been successfully tried (see also below). ERK1/2 and CK1δ also modify HIF-2α at distinct sites, and in this case, they both appear to regulate HIF-2 activity by affecting the distribution of HIF-2α between nucleus and cytoplasm [44,45].
In addition to direct phosphorylation, signaling pathways involving PI3K, ERK1/2 or p38 MAPK when activated by non-hypoxic stimuli such as growth factors (e.g., PDGF, TGF-β, IGF-1, and EGF), cytokines or hormones can also affect HIF activation by indirectly modulating its expression or stability [34,35,48,49,50,51,52,53,54]. As an example, heregulin (a member of the EGF family of growth factors) induces HIF-1 by activating the PI3K/Akt/mTOR pathway and increasing the rate of HIF-1α translation [55]. Other exemplary modes of HIF regulation include the involvement of ROS signaling (reviewed in [56]), transcription factors such as NF-kb [57] and STAT3 [58] that upregulate transcription of the gene encoding HIF-1α and many interacting proteins such as HSP90 and RACK1, which stabilize or destabilize HIF-1α, respectively [59,60,61]. HIFα stability is also affected by CO2 concentration as both in vivo and in vitro hypercapnia decrease HIFα protein levels independently of the PHD/pVHL-mediated degradation pathway and, most likely, via lysosomal proteolysis [62].
3. The Involvement of HIFs in Cancer
HIFs and especially HIF-1 influence several hallmarks of cancer such as genomic instability, tumor cell invasion, metastasis, and angiogenesis as well as suppression of the anti-tumor immune response [5,63]. Most importantly, HIF-1 holds a prominent role as mediator of the metabolic reprogramming that characterizes many types of cancer cells [8,64]. HIF-1 upregulates expression of most enzymes of glycolysis as well as expression of pyruvate dehydrogenase kinase 1 (PDK1). PDK1 phosphorylates and inactivates pyruvate dehydrogenase (PDH), which catalyzes conversion of pyruvate into acetyl-CoA. Thus, HIF-1 drives pyruvate, the product of glycolysis away from the TCA cycle and towards production of lactate, even in the presence of oxygen, a phenomenon known as Warburg effect [8,64]. Lipid metabolism is also influenced by a HIF-1 and several HIF-1 gene targets are involved in lipogenesis, which is generally favored in cancer via an increase in fatty acid uptake or synthesis and storage and simultaneous downregulation of fatty acid oxidation (reviewed in [4]). This often leads to accumulation of lipid droplets [37,65,66,67] and protects cancer cells from lipotoxicity [37,65,66]. HIF-dependent gene expression is also important for the adaptation and metabolism of cells surrounding a tumor (e.g., stromal cells), which are known to play an important role for cancer development [64,68].
There are numerous studies in which HIF-α proteins are found overexpressed in malignant tumors [5]. In principle, overexpression of HIFα isoforms is associated with poor clinical outcomes in patients with solid tumors [5,6]. Interestingly, HIF-1α was also found elevated and correlated with bad prognosis in hematological malignancies (reviewed in [69]). However, there are few reports indicating that HIF-1α overexpression may be connected to a positive outcome in certain cancer types including head and neck [70], non-small cell lung [71] and neuroblastoma [72].
Another feature that gives HIFs a special role in cancer progression is their ability to promote epithelial to mesenchymal transition (EMT) as well as resistance to chemo- or radio-therapy. HIFs facilitate EMT mainly by enhancing the expression of genes such as TCF3, ZFHX1A/B, and TWIST, which repress E-cadherin and epithelial type promoting factors, while, at the same time, the expression of mesenchymal type genes is increased [73,74]. Furthermore, HIF-mediated gene expression drives extracellular matrix remodeling, resistance to anoikis-related cell death and establishment of new cancer colonies, all of which facilitate the metastatic phenotype of hypoxic tumors [75]. Moreover, HIF-1 mediates chemoresistance by inducing expression of proteins that enhance drug efflux such as multidrug resistance 1 (MDR1) [76,77] and MRP2 [78] or anti-apoptotic proteins that promote drug resistance such as survivin [79,80]. HIF-1 is also implicated in resistance to radiation therapy since it counteracts the cytotoxic effects of radiation such as DNA damage and production of reactive oxygen species (ROS) [81,82].
Despite the unequivocal involvement of HIFs in the adaptation of cancer cells in the hypoxic tumor microenvironment, which promotes tumor progression, metastasis and resistance to therapy, it is still questionable whether HIFs by themselves are pro-oncogenic in a normal genetic background [83]. HIFs are active under physiological conditions such as embryonic development, immune system development, high-altitude adaptation and exercise and play an essential role in the maintenance of normal tissue homeostasis. HIF activation under these conditions does not trigger oncogenesis. Even when HIFα is constitutively activated due to pVHL function loss in renal cells, development of renal carcinoma requires additional mutations [84]. These issues have become especially important due the recent licensing and wide clinical administration of PHD inhibitors as HIF activators, and subsequent erythropoiesis inducers, for the treatment of patients suffering from renal anemia [85]. Long-term administration of these PHD inhibitors has not demonstrated tumor-initiating or tumor-promoting effects in either animal models or phase III clinical trials, possibly because competitive inhibition of PHD catalytic activity cannot cause permanent and irreversible HIF activation or pharmacological HIF induction is graded and cannot exceed a physiologically acceptable threshold. Nevertheless, as discussed below, the proof-of-principle for HIF inhibition as a valid anticancer strategy has been demonstrated in several cases and in both animal and human studies.
4. HIF Inhibitors as Therapeutics
Activation of the HIF pathway is a common final aspect of many cancers arising from a variety of events, including intratumoral hypoxia, signaling molecules (reactive oxygen species, cytokines, and growth factors) and oncogenic transformation (e.g., VHL loss of function in renal carcinoma) [86]. As already discussed, HIF activation facilitates cancer progression and has been correlated with increased patient mortality [5]. Consequently, there has been great interest for the discovery of chemical agents and drugs that impair HIF activity and there is a fast growing list of such agents that act via various molecular mechanisms in cancer cells and tested for inhibition of tumor growth in animal models (comprehensibly reviewed in [7]).
4.1. Chemical Agents as HIF Inhibitors
Transcription factors such as HIFs have been long considered undruggable because they do not contain a single targetable catalytic site (as enzymes do) but their activity relies on protein–DNA or protein–protein interactions, specific inhibition of which by small molecules is very challenging [87], especially when no detailed structural information is available. Nevertheless, numerous attempts have been made to identify HIF inhibiting agents, mainly by screening chemical libraries for their effect on cellular systems that facilitate detection of HIF transcriptional activity. The well-characterized agents discussed in this section (Table 1) are only used as examples in order to highlight the constituents and/or processes of the HIF activation pathway that when targeted lead to inhibition of HIF activity. In brief, different compounds have been shown to interfere with HIFα mRNA production, protein synthesis, stability, nuclear transport, heterodimerization, DNA binding, and transcriptional activity.
Table 1.
Chemical and peptide or peptidomimetic inhibitors of HIFs discussed in this article.
| Inhibited Process |
Nature of Inhibitor | Inhibitor | Active Concentr.* | Ref |
|---|---|---|---|---|
| HIFα synthesis | Chemical | Aminoflavone | 0.25–0.5 μM | [88] |
| GL331 | 10 μM | [89] | ||
| Idarubicin | 0.625 μM | [90] | ||
| Digoxin | 0.1 μM | [91] | ||
| Topotecan | 0.05–0.1 μM | [92] | ||
| Calcitriol | 0.1 μM | [93] | ||
| Sorafenib | 10 μM | [94] | ||
| YC1 | 10–25 μM | [95] | ||
| EF24 | 1 μM | [96] | ||
| HIFα stability | Chemical | 17-AAG | 0.5 μM | [60] |
| NAC | 10 mM | [97] | ||
| HIFα binding
to ARNT |
Chemical | Acriflavine | 1–5 μM | [98] |
| PT2399 | 2 μM | [99] | ||
| Peptide | TAT-Ainp1 | 1–2 μM | [100] | |
| TAT-cyclo-CLLFVY | 10–50 μM | [101] | ||
| HIFα nuclear accumulation and activity | Chemical | PD98057 | 50 μM | [39] |
| U0126 | 5 μM | [42,44] | ||
| Kaempferol | 5–10 μM | [102] | ||
| Echinomycin | 1–5 nM | [103] | ||
| Doxorubicin | 0.2–1 μM | [104] | ||
| Chetomin | 10 nM | [105] | ||
| Peptide | TAT-EDT | 0.4 μM | [36] | |
| Peptido-mimetic | HBS 2 (C-TAD helix 2 mimic) |
1 μM | [106] | |
| HBS 1 (C-TAD helix 3 mimic) |
50 μM | [107] | ||
| OHM 1 (C-TAD helix 3 mimic) |
10 μM | [108] |
* Concentration of inhibitor that causes significant reduction (close to or more than 50%) in HIFα expression and/or HIF transcriptional activity when administered to HIF-expressing cultured cells.
4.1.1. Inhibition of HIFα mRNA Expression and Protein Synthesis
Although few in number, there are certain compounds that impair the transcription of HIF1A or EPAS1 genes to mRNA. These include agents such as aminoflavone shown to affect both HIF1A transcription and translation in breast cancer cell lines and xenograft models [88], GL331 that interferes with the activation of HIF1A promoter in a lung cancer cell model [89] and anthracyclines (such as idarubicin) that inhibit both HIF1A and EPAS1 transcription in pheochromocytoma cells and xenograft models [90]. A much larger group comprises compounds that decrease the translation rate of HIFα mRNA in several different ways. This group includes cardiac glycosides (e.g., digoxin) [91], topoisomerase inhibitors (e.g., topotecan) [92], agents that target components of the mTOR pathway (e.g., rapamycin) [109], steroids such as calcitriol that influence HIFα translation rates [93] or compounds that inhibit major signaling pathways such as sorafenib [94] or YC-1 [95]. This list also includes microtubule binding agents such as EF24 [96]. However, despite of the ability of this category of agents to interfere with the production of HIFα protein, most of these compounds do not specifically target the HIF-pathway nor can they differentially target HIF-1 or HIF-2, with few exceptions such as YC-1, which can specifically inhibit HIF-1 (but not HIF-2) expression in macrophages [110,111].
4.1.2. Inhibition of HIFα Stability
A distinguishing feature of HIFαs is that they can be quickly destroyed in response to oxygen or other stimuli in proteasomal or lysosomal compartments [24,112]. So, there have been numerous studies investigating agents that can promote HIFα degradation even under hypoxia or tumor-related stabilizing conditions. Archetypical examples of this group are HSP90 inhibitors (e.g., 17-AAG) that lead to RACK1-dependent and VHL-independent ubiquitination of HIFα [60]. Another example are antioxidant agents (e.g., N-acetyl cysteine) that destabilize HIFα subunits in a PHD/VHL-mediated manner [97].
4.1.3. Inhibition of HIF Heterodimerization
Agents that belong to this group target the PAS domains of HIF-1α and -2α thus interfering with HIFα heterodimerization with ARNT. For example, acriflavine (an antibacterial drug) can bind to the PAS-B domain of both HIFα subunits, which share significant homology in this part [98]. However, recent structural data of the HIFα PAS domains have shown that, despite the similarity between the HIF-1α and -2α PAS domains, the HIF-2α PAS-B contains a ligand-fitting cavity that is absent from the HIF-1α PAS-B [19,113]. Following these studies, subunit-specific inhibition of HIF-2α/ARNT dimerization was shown using a prototype chemical agent designed to bind to this cavity. More importantly, these compounds (PT2399, and PT2385) were efficient inhibitors of HIF-2 activity, exhibited anticancer properties in both cellular and animal renal carcinoma models and PT2385 administration to a patient with metastatic renal carcinoma (ccRCC) showed very promising results [99,114]. Furthermore, in a Phase I clinical trial with a population of patients with highly pretreated advanced ccRCC, PT2385 showed promising efficacy and favorable tolerability as a monotherapy, raising hopes for even better results in combination studies [115].
4.1.4. Inhibition of HIFα Intranuclear Localization
Other chemicals such as PD98059 and U0126 or naturally occurring substances such as Kaempferol that inhibit the activation of ERK1/2 pathway, impair the phosphorylation of both HIFα subunits, and thus promote CRM1-dependent nuclear export of HIFα, subsequent impeding HIF transcriptional activity and proliferation of cancer cells [39,42,44,102].
4.1.5. Inhibition of HIF DNA-Binding and Transcriptional Activity
DNA intercalating agents such as echinomycin or doxorubicin impair HIF-1 and HIF-2 binding to chromatin and, thus, inhibit HIF transcriptional activity [103,104]. On the other hand, chetomin targets the transcriptional activity of HIFs by influencing its ability to form a functional complex with transcriptional coactivators CBP/P300 [105].
Taken together, this growing number of preclinical data on chemical agents that inhibit HIF-pathway activation suggests that HIF inhibitors can improve the current treatment in many human cancer types. However, apart from the selective HIF-2 inhibitor PT2385 administered to patients with ccRCC in phase I trials and showing good tolerance [99,115], all other types of inhibitors discussed in this section represent either agents with tolerance limitations or chemicals that lack the selectivity for the hypoxic machinery or are not HIF-isoform specific.
4.2. Peptides as HIF Modulators
An approach that may lead to selective and isoform-specific inhibition or activation of HIFs is the disruption of protein–protein or protein–DNA interactions, using peptides modelled after the amino acid sequences of the HIFα isoforms. These peptides, when delivered inside cells, may compete with the endogenous HIFαs for critical interactions, thereby inhibiting HIF activity or impair their association with inhibitory proteins, thereby stimulating HIF activity. Of course, development of such peptide agents requires very detailed mapping and functional characterization of the specific domains they are modeled after (Figure 2) as well as practical ways for their administration and intracellular or intranuclear delivery. In the case of HIFαs, the peptide inhibition/activation approach is facilitated by their modular structure, having well separated DNA-binding and Transactivation/Regulatory domains, which makes targeting individual domains and activation steps more feasible. Concerning delivery of peptides, they can be overexpressed in cells using transfection with suitable vectors, but this is not a practical method, especially when it is intended to be developed for therapeutic purposes. An emerging approach to deliver peptides that modulate protein–protein interactions as means of treatment relies on protein transduction technology. This method involves small amino acid sequences of various origins (listed in detail in [116]) that are able to cross cellular membranes and deliver associated cargo intracellularly. Among these cell permeable peptides, HIV-1 Transactivator of Transcription (TAT) peptide has been extensively studied and has been frequently used as means of internalizing biomolecules with therapeutic potential [117]. Concerning HIFs, protein transduction technology was initially used in order to target interactors of the ODDD of HIF-1α and enhance HIF activity as potential therapeutic approach in ischemic diseases. Peptides derived from both the N-terminal (residues 343–417) and C-terminal (residues 549–582) part of the HIF-1α ODD and fused to the TAT sequence could efficiently penetrate into cells, stabilize HIF-1α even under normoxia, by competing for its PHD-dependent hydroxylation, and promote angiogenesis in both in vitro and animal models [118]. Moreover, a smaller ODD peptide fused to TAT could induce HIF-1α and promote VEGF production in rat cortical neurons possibly by the same degradation-protective mechanism [119].
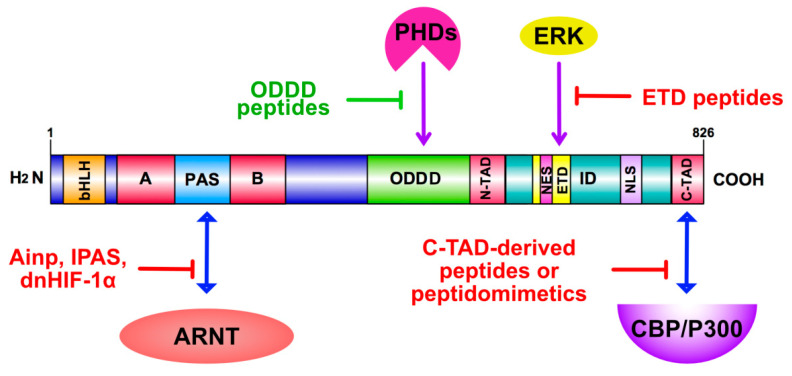
Figure 2.
Peptide-mediated HIFα targeting. Peptides target the association of distinct HIF-1α domains (as indicated) with components of the hypoxia signaling or transcription machinery, such as ARNT, PHDs and CBP/P300 (activating peptides in green, inhibiting in red). Modification of HIF-1α by ERK and its phosphorylation-dependent nuclear interactions can also be inhibited by ERK Targeted Domain (ETD)-derived peptides.
4.2.1. Peptide Inhibition of HIF Heterodimerization
The feasibility of inhibiting HIF heterodimerization using peptides stems from early studies which demonstrated that shorter splice variants of HIF-3α (IPAS) are expressed in human and mouse tissues and downregulate HIF activity [15,120]. These polypeptides comprise either the bHLH, PAS, and N-TAD or the bHLH and PAS domains with a length of 667 and 307 amino acid residues (aa), respectively, and can negatively regulate HIFs by inhibiting HRE-mediated transcription [15,121]. The various HIF-3α forms act as dominant negative regulators by forming transcriptionally inactive complexes with HIF-1α or HIF-2α that prohibit effective interaction with ARNT [15,120]. Along the same line, when a HIF-1α-derived polypeptide (dnHIF-1α; aa 30-389) containing the bHLH and PAS domain was expressed in pancreatic adenocarcinoma cell lines it acted as a dominant negative mutant (apparently by forming an DNA-binding but inactive complex with ARNT) as it reduced binding of endogenous HIF-1 to its target promoters and, subsequently, inhibited its activity. The inhibitory effectiveness of dnHIF-1α was demonstrated by causing reduced glucose uptake and cancer cell growth in both pancreatic cancer cells and xenograft animal models [122]. ARNT interacting peptide (Ainp1) is a 59 aa long peptide of unknown function, the mRNA of which is expressed in many human tissues. Ainp1 interacts with the bHLH domain of ARNT. Transient introduction of Ainp1 into cells led to ARNT sequestration to the cytoplasm, lower HIF-1α/ARNT complex formation, and reduced HIF-1 activity in Hep3B cells [123]. Furthermore, TAT-fused Ainp1 could localize inside the cell nucleus and suppress HIF-1 activity in three different cell lines by interfering with the bHLH domain of ARNT (Table 1) [100].
Another cell permeable peptide that targets the HIF-1α/ARNT association is the cyclic hexapeptide cyclo-CLLFVY isolated from a 3.2 million plasmid library coding for hexapeptides [101]. The synthetic hexapeptide could efficiently bind to the PAS-B domain of HIF-1α (but not HIF-2α) in vitro. MCF-7 and U2OS cells cultured in the presence of this peptide fused to the TAT epitope exhibited lower HIF-1α/ARNT association and decreased HIF-1 transcriptional activity (Table 1). Following these results, the same group engineered human cells conditionally expressing this HIF-1 inhibitory cyclic peptide. Inhibition of HIF-1 dimerization and hypoxia-response signaling was also verified in these engineered cells. Moreover, inhibition of HIF-1 by the cyclic peptide sensitized cells to the glycolysis inhibitor 2-deoxyglucose leading to reduced cell viability, showing synthetic lethality upon both inhibition of HIF-1 and glycolysis in cells under hypoxia [124].
4.2.2. Peptide Inhibition of HIF-Dependent Transactivation
Another target of peptide-mediated HIF inhibition is the HIFα/p300 binding interface. Initial studies used truncation mutants to map the minimum amino acid sequences that are required for the association between HIF-1α and p300. It was shown that 116 residues of p300 (aa 302–418) comprising its CH1 domain and 41 C-terminal HIF-1α residues (aa 786–826; C-TAD) were necessary for efficient HIF-1α/p300 binding [125]. Peptides that derive from HIF-1α C-TAD (aa 766–826 and aa 786–826) or p300 CH1 peptides reduced EPO promoter activity when overexpressed in HCT-116 colon and MDA-MB435 breast carcinoma cell lines [125]. Importantly, the expression of the HIF-1α C-TAD polypeptides could effectively suppress hypoxic signaling and tumor growth in a xenograft model. However, the same peptides also reduced STAT2 activity that requires the same CH1 domain of p300 for its transactivation, suggesting that binding of the HIF-1α peptides to p300 could affect the activity of other transcription factors in addition to HIF-1, although signaling pathways that relied on transcription factor binding with p300 domains other than CH1 were not impaired [125]. Recent studies have performed a more detailed structural investigation of the binding interface between the HIF-1α C-TAD region (aa 776–826) and the p300 CH1 domain (aa 330–420) [126]. NMR data revealed that the HIF-1α C-TAD region consists of three helical subdomains and wraps around the p300 CH1 domain with helices 2 and 3 being more important for HIF-1α/p300 association. Structural information from different HIF-1 C-TAD truncation and p300 point mutants revealed that helix 3 is essential for HIF-1α binding to a p300 binding patch. In this context, different peptide and phage display libraries as well as synthetic HIF-1α (aa 812–826)-derived peptides were examined for their binding to p300 and their adopted conformation when bound to the CH1 domain. Designed synthetic and constrained peptides that were able to adopt α-helical conformation in their bound state to p300 were more efficient competitors of HIF-1α C-TAD and may represent lead compounds for the development of peptidomimetic HIF-1α/p300 inhibitors (see also below) [126,127].
4.2.3. Inhibition of HIF-1α Nuclear Accumulation and Nuclear Interactions
As already mentioned, phosphorylation of Ser641/643 in the ETD domain of HIF-1α by p42/44 MAPK (ERK1/2), which are often activated in human cancers [128], is essential for HIF-1α nuclear accumulation and activity because it masks a CRM1-dependent NES [38]. Initial studies from our group showed that overexpression of ETD, as a 43-amino acid long free peptide, competed with endogenous HIF-1α and inhibited its activity in HeLa and Huh7 cells [36]. The specificity of this inhibition was shown by the fact that a mutant form of the ETD that could not be phosphorylated by ERKs (ETD-SA form, in which Ser641/643 were converted into Ala) left HIF-1 activity unaffected. In contrast, other mutant variants of the ETD that either mimic its phosphorylation by ERK (ETD-SE form, in which Ser641 was converted into Glu) or are constitutively nuclear (ETD-IA form in which the NES was mutated) were as efficient as their wild-type form in inhibiting HIF-1 activity, suggesting that the ETD peptides could act in either or both of two ways: as competitors of HIF-1α phosphorylation, being themselves ERK substrates, and/or as competitors of a phosphorylation-dependent essential nuclear interaction of the endogenous HIF-1α ETD. A better analysis of the effects of these inhibitory peptides became possible when they were delivered directly to the cells as cell-penetrating TAT-fusion recombinant moieties (Table 1) [36]. In this way, they were shown to efficiently disable the ERK-dependent transcriptional activity of HIF-1 without interfering with HIF-1α stabilization or affecting HIF-2 activity, as the ETD sequence is unique in HIF-1α. Moreover, administration of the inhibitory TAT-ETD peptides resulted in severe phenotypic defects, exclusively under hypoxia. These defects included inability of the cancer cells to metabolically adapt, migrate or form single cell colonies under low oxygen conditions. Furthermore, even though these peptides did not affect cell viability under normoxia, they significantly increased cell death rate by inducing apoptosis under hypoxia. Overall, these results underlined the significance of ERK-dependent phosphorylation of HIF-1α for the transcriptional response to hypoxia as well as the prospect of TAT-ETD peptides or their peptidomimetics as highly selective and isoform-specific inhibitors of the HIF-1 pathway in cancer cells.
5. Perspectives–Conclusions
Increased levels of HIF activity are a common feature of many cancer types and cancer cells that depend on HIF activity may be vulnerable to HIF inhibition alone or in combination with other more traditional treatments. However, most of the chemical HIF inhibitors identified so far suffer from poor selectivity against HIFs or limited patient tolerance as they target a broad range of cellular functions and potentially also interfere with the function of healthy cells. HIF activation and regulation involves many protein–protein interactions (PPIs) [59], targeting of which may have more selective and less toxic effects. PPI binding interfaces are usually large, shallow, and hydrophobic, which makes them largely intractable to small chemicals that can normally target well-defined cavities of enzymes or receptors. In contrast, the larger size and flexible backbones of peptides make them capable of binding and blocking the larger grooves or clefts of interacting interfaces [129]. Therefore, peptides that mimic endogenous interacting partners can very selectively and potently inhibit their association. Along this line, carefully characterized peptide sequences that target specific PPIs essential for HIF activation steps can be much more effective than small molecule inhibitors. However, clinical application of peptides presents major limitations due to their poor oral bioavailability, low cell permeability, and short plasma half-life. It is, therefore, a challenge to create peptide inhibitors that can share, at least some of, the advantages of small chemical inhibitors such as manufacturing cost, shelf life, oral delivery, metabolic stability, pharmacokinetics, bio-distribution, and permeability [130].
Once peptide–protein recognition mechanisms have been deciphered and characterized, a promising strategy is to improve the pharmaceutical properties of peptides through stabilization, fusion with cell-penetrating moieties, or even conversion into non-natural scaffolds, creating, thus, novel compounds often referred to as peptidomimetics [116,131]. Stabilization of peptides and improvement of their pharmacokinetic properties in vivo may include C-terminal amidation or N-terminal acetylation, conjugation with lipids or pegylated side chains or covalent "stapling" of helical regions along the peptide backbone [132]. More advanced strategies include construction of macrocycles (molecules containing a twelve or more-membered ring) through peptide cyclization, introduction of non-natural amino acid analogs or enantiomer (D-)amino acids and creation of peptide-inspired foldamers (chain molecules or oligomers that mimic the ability of peptides to fold into well-defined conformations, such as helices and β-sheets) [133,134]. These peptidomimetics may have small molecule drug properties like long in vivo stability, while maintaining robust affinity, specificity and minimal toxicity.
Such an approach has already been tried in the case of HIFs with promising results. Based on the sequence of HIF-1α C-TAD helix 2 (that is important for HIF-1α/p300 interaction) a hydrogen bond surrogate (HBS) α-helical peptide was synthesized that had better affinity for p300 and was effective in down-regulating HIF-1 activity in HeLa cells almost as efficiently as chetomin but without its toxic effects (Table 1) [106]. In an HBS peptide, a carbon–carbon bond replaces the intramolecular hydrogen bond, further stabilizing the α-helical conformation and promoting resistance to proteolysis. The same research group subsequently produced an HBS mimic of HIF-1α C-TAD helix 3, which, in addition to inhibiting HIF-1 activity in cancer-cell-based assays (Table 1), also impaired tumor growth when administered to a mouse xenograft tumor model, exhibiting, thus, much better in vitro and in vivo efficacy than the respective unconstrained peptide [107]. Furthermore, an oxopiperazine helix mimetic (OHM) of HIF-1α C-TAD helix 3, in which nitrogen atoms of neighboring backbone amides in a tetrapeptide are constrained with ethylene bridges, was able, despite its small size, to inhibit both HIF-1 activity and tumor growth rate in vitro and in vivo, respectively [108]. These results suggest that certain PPIs can depend on few protein side chains, so-called “hot-spot” residues, and mimicking the relative positioning and disposition of these important residues on synthetic scaffolds may lead to the production of small molecules that combine the advantages of both chemical and peptide inhibitors.
In conclusion, peptides as modulators of PPIs involving HIFs are not only important for in vitro experimentation and the elucidation of regulatory mechanisms but can also lead the way for the design and development of novel peptide analogues or synthetic peptidomimetics that can target exclusively HIF-overexpressing cells with great selectivity, low toxicity and pharmacological properties similar to those of small molecule chemical inhibitors. The effectiveness and advantages of HIF peptide inhibitors summarized herein, certainly provide proof-of-principle for the development of novel therapeutic options based on the interruption of PPIs that are essential for the adaptation of cancer cells to the hypoxic tumor micro-environment and urge for better and high resolution structural characterization of the corresponding HIFα PPI domains.
Author Contributions
Writing—original draft preparation and visualization: I.M. and G.C.; writing—review and editing G.S. All authors have read and agreed to the published version of the manuscript.
Funding
I.M. was funded by Fondation Santé (Project Code: 5360); The research work was supported by the Hellenic Foundation for Research and Innovation (H.F.R.I.) under the “First Call for H.F.R.I. Research Projects to support Faculty members and Researchers and the procurement of high-cost research equipment grant” (Project Numbers: HFRI-FM17-2132 to I.M and HFRI-FM17-1460 to G.C.); Research by G.S. has been co-financed by the European Regional Development Fund of the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH – CREATE – INNOVATE (project code:T1EDK-00204).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wilson J.W., Shakir D., Batie M., Frost M., Rocha S. Oxygen-sensing mechanisms in cells. FEBS J. 2020 doi: 10.1111/febs.15374. [DOI] [PubMed] [Google Scholar]
- 2.Semenza G.L. The Genomics and Genetics of Oxygen Homeostasis. Annu Rev. Genom. Hum. Genet. 2020;21:183–204. doi: 10.1146/annurev-genom-111119-073356. [DOI] [PubMed] [Google Scholar]
- 3.Rankin E.B., Nam J.M., Giaccia A.J. Hypoxia: Signaling the Metastatic Cascade. Trends Cancer. 2016;2:295–304. doi: 10.1016/j.trecan.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mylonis I., Simos G., Paraskeva E. Hypoxia-Inducible Factors and the Regulation of Lipid Metabolism. Cells. 2019;8:214. doi: 10.3390/cells8030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schito L., Semenza G.L. Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Albadari N., Deng S., Li W. The transcriptional factors HIF-1 and HIF-2 and their novel inhibitors in cancer therapy. Expert Opin. Drug Discov. 2019;14:667–682. doi: 10.1080/17460441.2019.1613370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenza G.L. Pharmacologic Targeting of Hypoxia-Inducible Factors. Annu. Rev. Pharm. Toxicol. 2019;59:379–403. doi: 10.1146/annurev-pharmtox-010818-021637. [DOI] [PubMed] [Google Scholar]
- 8.Dengler V.L., Galbraith M., Espinosa J.M. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2014;49:1–15. doi: 10.3109/10409238.2013.838205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenza G.L., Wang G.L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell Biol. 1992;12:5447–5454. doi: 10.1128/MCB.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G.L., Semenza G.L. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc. Natl. Acad. Sci. USA. 1993;90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G.L., Semenza G.L. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 12.Tian H., McKnight S.L., Russell D.W. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 13.Skuli N., Majmundar A.J., Krock B.L., Mesquita R.C., Mathew L.K., Quinn Z.L., Runge A., Liu L., Kim M.N., Liang J., et al. Endothelial HIF-2alpha regulates murine pathological angiogenesis and revascularization processes. J. Clin. Investig. 2012;122:1427–1443. doi: 10.1172/JCI57322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heikkila M., Pasanen A., Kivirikko K.I., Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3alpha variants in the hypoxia response. Cell Mol. Life Sci. 2011;68:3885–3901. doi: 10.1007/s00018-011-0679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makino Y., Cao R., Svensson K., Bertilsson G., Asman M., Tanaka H., Cao Y., Berkenstam A., Poellinger L. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 16.Tolonen J.P., Heikkila M., Malinen M., Lee H.M., Palvimo J.J., Wei G.H., Myllyharju J. A long hypoxia-inducible factor 3 isoform 2 is a transcription activator that regulates erythropoietin. Cell Mol. Life Sci. 2020;77:3627–3642. doi: 10.1007/s00018-019-03387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pugh C.W., Ratcliffe P.J. New horizons in hypoxia signaling pathways. Exp. Cell Res. 2017;356:116–121. doi: 10.1016/j.yexcr.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G.L., Jiang B.H., Rue E.A., Semenza G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu D., Potluri N., Lu J., Kim Y., Rastinejad F. Structural integration in hypoxia-inducible factors. Nature. 2015;524:303–308. doi: 10.1038/nature14883. [DOI] [PubMed] [Google Scholar]
- 20.Key J., Scheuermann T.H., Anderson P.C., Daggett V., Gardner K.H. Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B. J. Am. Chem. Soc. 2009;131:17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lando D., Gorman J.J., Whitelaw M.L., Peet D.J. Oxygen-dependent regulation of hypoxia-inducible factors by prolyl and asparaginyl hydroxylation. Eur. J. Biochem. 2003;270:781–790. doi: 10.1046/j.1432-1033.2003.03445.x. [DOI] [PubMed] [Google Scholar]
- 22.Hu C.-J., Sataur A., Wang L., Chen A., Simon M.C. The N-Terminal Transactivation Domain Confers Target Gene Specificity of Hypoxia-inducible Factors HIF-1α and HIF-2α. Mol. Biol. Cell. 2007;18:4528–4552. doi: 10.1091/mbc.e06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxwell P.H., Wiesener M.S., Chang G.W., Clifford S.C., Vaux E.C., Cockman M.E., Wykoff C.C., Pugh C.W., Maher E.R., Ratcliffe P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 24.Jaakkola P., Mole D.R., Tian Y.M., Wilson M.I., Gielbert J., Gaskell S.J., von Kriegsheim A., Hebestreit H.F., Mukherji M., Schofield C.J., et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 25.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J.M., Lane W.S., Kaelin W.G., Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 26.Epstein A.C., Gleadle J.M., McNeill L.A., Hewitson K.S., O’Rourke J., Mole D.R., Mukherji M., Metzen E., Wilson M.I., Dhanda A., et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/S0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 27.Bruick R.K., McKnight S.L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 28.Berra E., Benizri E., Ginouves A., Volmat V., Roux D., Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahon P.C., Hirota K., Semenza G.L. FIH-1: A novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lando D., Peet D.J., Gorman J.J., Whelan D.A., Whitelaw M.L., Bruick R.K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schödel J., Ratcliffe P.J. Mechanisms of hypoxia signalling: New implications for nephrology. Nat. Rev. Nephrol. 2019;15:641–659. doi: 10.1038/s41581-019-0182-z. [DOI] [PubMed] [Google Scholar]
- 32.Ivanova I.G., Park C.V., Kenneth N.S. Translating the Hypoxic Response-the Role of HIF Protein Translation in the Cellular Response to Low Oxygen. Cells. 2019;8:114. doi: 10.3390/cells8020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masoud G.N., Li W. HIF-1alpha pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B. 2015;5:378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albanese A., Daly L.A., Mennerich D., Kietzmann T., See V. The Role of Hypoxia-Inducible Factor Post-Translational Modifications in Regulating Its Localisation, Stability, and Activity. Int. J. Mol. Sci. 2020;22:268. doi: 10.3390/ijms22010268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kietzmann T., Mennerich D., Dimova E.Y. Hypoxia-Inducible Factors (HIFs) and Phosphorylation: Impact on Stability, Localization, and Transactivity. Front. Cell Dev. Biol. 2016;4:11. doi: 10.3389/fcell.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karagiota A., Kourti M., Simos G., Mylonis I. HIF-1alpha-derived cell-penetrating peptides inhibit ERK-dependent activation of HIF-1 and trigger apoptosis of cancer cells under hypoxia. Cell Mol. Life Sci. 2019;76:809–825. doi: 10.1007/s00018-018-2985-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kourti M., Ikonomou G., Giakoumakis N.N., Rapsomaniki M.A., Landegren U., Siniossoglou S., Lygerou Z., Simos G., Mylonis I. CK1delta restrains lipin-1 induction, lipid droplet formation and cell proliferation under hypoxia by reducing HIF-1alpha/ARNT complex formation. Cell Signal. 2015;27:1129–1140. doi: 10.1016/j.cellsig.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mylonis I., Chachami G., Paraskeva E., Simos G. Atypical CRM1-dependent nuclear export signal mediates regulation of hypoxia-inducible factor-1alpha by MAPK. J. Biol. Chem. 2008;283:27620–27627. doi: 10.1074/jbc.M803081200. [DOI] [PubMed] [Google Scholar]
- 39.Mylonis I., Chachami G., Samiotaki M., Panayotou G., Paraskeva E., Kalousi A., Georgatsou E., Bonanou S., Simos G. Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxia-inducible factor-1alpha. J. Biol. Chem. 2006;281:33095–33106. doi: 10.1074/jbc.M605058200. [DOI] [PubMed] [Google Scholar]
- 40.Chachami G., Paraskeva E., Mingot J.M., Braliou G.G., Gorlich D., Simos G. Transport of hypoxia-inducible factor HIF-1alpha into the nucleus involves importins 4 and 7. Biochem. Biophys. Res. Commun. 2009;390:235–240. doi: 10.1016/j.bbrc.2009.09.093. [DOI] [PubMed] [Google Scholar]
- 41.Depping R., Steinhoff A., Schindler S.G., Friedrich B., Fagerlund R., Metzen E., Hartmann E., Kohler M. Nuclear translocation of hypoxia-inducible factors (HIFs): Involvement of the classical importin alpha/beta pathway. Biochim. Biophys. Acta. 2008;1783:394–404. doi: 10.1016/j.bbamcr.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Mylonis I., Kourti M., Samiotaki M., Panayotou G., Simos G. Mortalin-mediated and ERK-controlled targeting of HIF-1alpha to mitochondria confers resistance to apoptosis under hypoxia. J. Cell Sci. 2017;130:466–479. doi: 10.1242/jcs.195339. [DOI] [PubMed] [Google Scholar]
- 43.Kalousi A., Mylonis I., Politou A.S., Chachami G., Paraskeva E., Simos G. Casein kinase 1 regulates human hypoxia-inducible factor HIF-1. J. Cell Sci. 2010;123:2976–2986. doi: 10.1242/jcs.068122. [DOI] [PubMed] [Google Scholar]
- 44.Gkotinakou I.M., Befani C., Simos G., Liakos P. ERK1/2 phosphorylates HIF-2alpha and regulates its activity by controlling its CRM1-dependent nuclear shuttling. J. Cell Sci. 2019;132 doi: 10.1242/jcs.225698. [DOI] [PubMed] [Google Scholar]
- 45.Pangou E., Befani C., Mylonis I., Samiotaki M., Panayotou G., Simos G., Liakos P. HIF-2alpha phosphorylation by CK1delta promotes erythropoietin secretion in liver cancer cells under hypoxia. J. Cell Sci. 2016;129:4213–4226. doi: 10.1242/jcs.191395. [DOI] [PubMed] [Google Scholar]
- 46.Lyberopoulou A., Mylonis I., Papachristos G., Sagris D., Kalousi A., Befani C., Liakos P., Simos G., Georgatsou E. MgcRacGAP, a cytoskeleton regulator, inhibits HIF-1 transcriptional activity by blocking its dimerization. Biochim. Biophys. Acta. 2013;1833:1378–1387. doi: 10.1016/j.bbamcr.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 47.Lyberopoulou A., Venieris E., Mylonis I., Chachami G., Pappas I., Simos G., Bonanou S., Georgatsou E. MgcRacGAP interacts with HIF-1alpha and regulates its transcriptional activity. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2007;20:995–1006. doi: 10.1159/000110460. [DOI] [PubMed] [Google Scholar]
- 48.Zhong H., Chiles K., Feldser D., Laughner E., Hanrahan C., Georgescu M.M., Simons J.W., Semenza G.L. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: Implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60:1541–1545. [PubMed] [Google Scholar]
- 49.Stiehl D.P., Jelkmann W., Wenger R.H., Hellwig-Burgel T. Normoxic induction of the hypoxia-inducible factor 1alpha by insulin and interleukin-1beta involves the phosphatidylinositol 3-kinase pathway. FEBS Lett. 2002;512:157–162. doi: 10.1016/S0014-5793(02)02247-0. [DOI] [PubMed] [Google Scholar]
- 50.Beppu K., Nakamura K., Linehan W.M., Rapisarda A., Thiele C.J. Topotecan blocks hypoxia-inducible factor-1alpha and vascular endothelial growth factor expression induced by insulin-like growth factor-I in neuroblastoma cells. Cancer Res. 2005;65:4775–4781. doi: 10.1158/0008-5472.CAN-04-3332. [DOI] [PubMed] [Google Scholar]
- 51.Calvani M., Trisciuoglio D., Bergamaschi C., Shoemaker R.H., Melillo G. Differential involvement of vascular endothelial growth factor in the survival of hypoxic colon cancer cells. Cancer Res. 2008;68:285–291. doi: 10.1158/0008-5472.CAN-07-5564. [DOI] [PubMed] [Google Scholar]
- 52.Mohlin S., Hamidian A., von Stedingk K., Bridges E., Wigerup C., Bexell D., Pahlman S. PI3K-mTORC2 but not PI3K-mTORC1 regulates transcription of HIF2A/EPAS1 and vascularization in neuroblastoma. Cancer Res. 2015;75:4617–4628. doi: 10.1158/0008-5472.CAN-15-0708. [DOI] [PubMed] [Google Scholar]
- 53.Gorlach A., Diebold I., Schini-Kerth V.B., Berchner-Pfannschmidt U., Roth U., Brandes R.P., Kietzmann T., Busse R. Thrombin activates the hypoxia-inducible factor-1 signaling pathway in vascular smooth muscle cells: Role of the p22(phox)-containing NADPH oxidase. Circ. Res. 2001;89:47–54. doi: 10.1161/hh1301.092678. [DOI] [PubMed] [Google Scholar]
- 54.Richard D.E., Berra E., Pouyssegur J. Nonhypoxic pathway mediates the induction of hypoxia-inducible factor 1alpha in vascular smooth muscle cells. J. Biol. Chem. 2000;275:26765–26771. doi: 10.1016/S0021-9258(19)61441-9. [DOI] [PubMed] [Google Scholar]
- 55.Laughner E., Taghavi P., Chiles K., Mahon P.C., Semenza G.L. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: Novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol. Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gorlach A., Kietzmann T. Superoxide and derived reactive oxygen species in the regulation of hypoxia-inducible factors. Methods Enzym. 2007;435:421–446. doi: 10.1016/S0076-6879(07)35022-2. [DOI] [PubMed] [Google Scholar]
- 57.Rius J., Guma M., Schachtrup C., Akassoglou K., Zinkernagel A.S., Nizet V., Johnson R.S., Haddad G.G., Karin M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papadakis A.I., Paraskeva E., Peidis P., Muaddi H., Li S., Raptis L., Pantopoulos K., Simos G., Koromilas A.E. eIF2alpha Kinase PKR modulates the hypoxic response by Stat3-dependent transcriptional suppression of HIF-1alpha. Cancer Res. 2010;70:7820–7829. doi: 10.1158/0008-5472.CAN-10-0215. [DOI] [PubMed] [Google Scholar]
- 59.Amir S., Wang R., Simons J.W., Mabjeesh N.J. SEPT9_v1 up-regulates hypoxia-inducible factor 1 by preventing its RACK1-mediated degradation. J. Biol. Chem. 2009;284:11142–11151. doi: 10.1074/jbc.M808348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y.V., Baek J.H., Zhang H., Diez R., Cole R.N., Semenza G.L. RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha. Mol. Cell. 2007;25:207–217. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Semenza G.L. A compendium of proteins that interact with HIF-1alpha. Exp. Cell Res. 2017;356:128–135. doi: 10.1016/j.yexcr.2017.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Selfridge A.C., Cavadas M.A., Scholz C.C., Campbell E.L., Welch L.C., Lecuona E., Colgan S.P., Barrett K.E., Sporn P.H., Sznajder J.I., et al. Hypercapnia Suppresses the HIF-dependent Adaptive Response to Hypoxia. J. Biol. Chem. 2016;291:11800–11808. doi: 10.1074/jbc.M116.713941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noman M.Z., Desantis G., Janji B., Hasmim M., Karray S., Dessen P., Bronte V., Chouaib S. PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J. Exp. Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakazawa M.S., Keith B., Simon M.C. Oxygen availability and metabolic adaptations. Nat. Rev. Cancer. 2016;16:663–673. doi: 10.1038/nrc.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mylonis I., Sembongi H., Befani C., Liakos P., Siniossoglou S., Simos G. Hypoxia causes triglyceride accumulation by HIF-1-mediated stimulation of lipin 1 expression. J. Cell Sci. 2012;125:3485–3493. doi: 10.1242/jcs.106682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoo W., Noh K.H., Ahn J.H., Yu J.H., Seo J.A., Kim S.G., Choi K.M., Baik S.H., Choi D.S., Kim T.W., et al. HIF-1alpha expression as a protective strategy of HepG2 cells against fatty acid-induced toxicity. J. Cell. Biochem. 2014;115:1147–1158. doi: 10.1002/jcb.24757. [DOI] [PubMed] [Google Scholar]
- 67.Triantafyllou E.A., Georgatsou E., Mylonis I., Simos G., Paraskeva E. Expression of AGPAT2, an enzyme involved in the glycerophospholipid/triacylglycerol biosynthesis pathway, is directly regulated by HIF-1 and promotes survival and etoposide resistance of cancer cells under hypoxia. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:1142–1152. doi: 10.1016/j.bbalip.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 68.Corbet C., Feron O. Tumour acidosis: From the passenger to the driver’s seat. Nat. Rev. Cancer. 2017;17:577–593. doi: 10.1038/nrc.2017.77. [DOI] [PubMed] [Google Scholar]
- 69.Deynoux M., Sunter N., Herault O., Mazurier F. Hypoxia and Hypoxia-Inducible Factors in Leukemias. Front. Oncol. 2016;6:41. doi: 10.3389/fonc.2016.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beasley N.J., Leek R., Alam M., Turley H., Cox G.J., Gatter K., Millard P., Fuggle S., Harris A.L. Hypoxia-inducible factors HIF-1alpha and HIF-2alpha in head and neck cancer: Relationship to tumor biology and treatment outcome in surgically resected patients. Cancer Res. 2002;62:2493–2497. [PubMed] [Google Scholar]
- 71.Volm M., Koomagi R. Hypoxia-inducible factor (HIF-1) and its relationship to apoptosis and proliferation in lung cancer. Anticancer Res. 2000;20:1527–1533. [PubMed] [Google Scholar]
- 72.Noguera R., Fredlund E., Piqueras M., Pietras A., Beckman S., Navarro S., Pahlman S. HIF-1alpha and HIF-2alpha are differentially regulated in vivo in neuroblastoma: High HIF-1alpha correlates negatively to advanced clinical stage and tumor vascularization. Clin. Cancer Res. 2009;15:7130–7136. doi: 10.1158/1078-0432.CCR-09-0223. [DOI] [PubMed] [Google Scholar]
- 73.Krishnamachary B., Zagzag D., Nagasawa H., Rainey K., Okuyama H., Baek J.H., Semenza G.L. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res. 2006;66:2725–2731. doi: 10.1158/0008-5472.CAN-05-3719. [DOI] [PubMed] [Google Scholar]
- 74.Yang M.-H., Wu M.-Z., Chiou S.-H., Chen P.-M., Chang S.-Y., Liu C.-J., Teng S.-C., Wu K.-J. Direct regulation of TWIST by HIF-1α promotes metastasis. Nat. Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 75.Schito L., Rey S. Hypoxic pathobiology of breast cancer metastasis. Biochim. Biophys. Acta Rev. Cancer. 2017;1868:239–245. doi: 10.1016/j.bbcan.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 76.Comerford K.M., Wallace T.J., Karhausen J., Louis N.A., Montalto M.C., Colgan S.P. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Res. 2002;62:3387–3394. [PubMed] [Google Scholar]
- 77.Ding Z., Yang L., Xie X., Xie F., Pan F., Li J., He J., Liang H. Expression and significance of hypoxia-inducible factor-1 alpha and MDR1/P-glycoprotein in human colon carcinoma tissue and cells. J. Cancer Res. Clin. Oncol. 2010;136:1697–1707. doi: 10.1007/s00432-010-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krishnamurthy P., Schuetz J.D. The ABC transporter Abcg2/Bcrp: Role in hypoxia mediated survival. Biometals. 2005;18:349–358. doi: 10.1007/s10534-005-3709-7. [DOI] [PubMed] [Google Scholar]
- 79.Chen Y.Q., Zhao C.L., Li W. Effect of hypoxia-inducible factor-1alpha on transcription of survivin in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2009;28:29. doi: 10.1186/1756-9966-28-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Faversani A., Vaira V., Moro G.P., Tosi D., Lopergolo A., Schultz D.C., Rivadeneira D., Altieri D.C., Bosari S. Survivin family proteins as novel molecular determinants of doxorubicin resistance in organotypic human breast tumors. Breast Cancer Res. 2014;16:R55. doi: 10.1186/bcr3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Befani C., Liakos P. The role of hypoxia-inducible factor-2 alpha in angiogenesis. J. Cell Physiol. 2018 doi: 10.1002/jcp.26805. [DOI] [PubMed] [Google Scholar]
- 82.Karakashev S.V., Reginato M.J. Progress toward overcoming hypoxia-induced resistance to solid tumor therapy. Cancer Manag. Res. 2015;7:253–264. doi: 10.2147/CMAR.S58285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cummins E.P., Strowitzki M.J., Taylor C.T. Mechanisms and Consequences of Oxygen and Carbon Dioxide Sensing in Mammals. Physiol. Rev. 2020;100:463–488. doi: 10.1152/physrev.00003.2019. [DOI] [PubMed] [Google Scholar]
- 84.Jonasch E., Walker C.L., Rathmell W.K. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat. Rev. Nephrol. 2020 doi: 10.1038/s41581-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanghani N.S., Haase V.H. Hypoxia-Inducible Factor Activators in Renal Anemia: Current Clinical Experience. Adv. Chronic Kidney Dis. 2019;26:253–266. doi: 10.1053/j.ackd.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Semenza G.L. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koehler A.N. A complex task? Direct modulation of transcription factors with small molecules. Curr. Opin. Chem. Biol. 2010;14:331–340. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Terzuoli E., Puppo M., Rapisarda A., Uranchimeg B., Cao L., Burger A.M., Ziche M., Melillo G. Aminoflavone, a ligand of the aryl hydrocarbon receptor, inhibits HIF-1alpha expression in an AhR-independent fashion. Cancer Res. 2010;70:6837–6848. doi: 10.1158/0008-5472.CAN-10-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang H., Shyu K.G., Lee C.C., Tsai S.C., Wang B.W., Hsien Lee Y., Lin S. GL331 inhibits HIF-1alpha expression in a lung cancer model. Biochem. Biophys. Res. Commun. 2003;302:95–100. doi: 10.1016/S0006-291X(03)00111-6. [DOI] [PubMed] [Google Scholar]
- 90.Pang Y., Yang C., Schovanek J., Wang H., Bullova P., Caisova V., Gupta G., Wolf K.I., Semenza G.L., Zhuang Z., et al. Anthracyclines suppress pheochromocytoma cell characteristics, including metastasis, through inhibition of the hypoxia signaling pathway. Oncotarget. 2017;8:22313–22324. doi: 10.18632/oncotarget.16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang H., Qian D.Z., Tan Y.S., Lee K., Gao P., Ren Y.R., Rey S., Hammers H., Chang D., Pili R., et al. Digoxin and other cardiac glycosides inhibit HIF-1alpha synthesis and block tumor growth. Proc. Natl. Acad. Sci. USA. 2008;105:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rapisarda A., Zalek J., Hollingshead M., Braunschweig T., Uranchimeg B., Bonomi C.A., Borgel S.D., Carter J.P., Hewitt S.M., Shoemaker R.H., et al. Schedule-dependent inhibition of hypoxia-inducible factor-1alpha protein accumulation, angiogenesis, and tumor growth by topotecan in U251-HRE glioblastoma xenografts. Cancer Res. 2004;64:6845–6848. doi: 10.1158/0008-5472.CAN-04-2116. [DOI] [PubMed] [Google Scholar]
- 93.Gkotinakou I.M., Kechagia E., Pazaitou-Panayiotou K., Mylonis I., Liakos P., Tsakalof A. Calcitriol Suppresses HIF-1 and HIF-2 Transcriptional Activity by Reducing HIF-1/2α Protein Levels via a VDR-Independent Mechanism. Cells. 2020;9:2440. doi: 10.3390/cells9112440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kumar S.M., Yu H., Edwards R., Chen L., Kazianis S., Brafford P., Acs G., Herlyn M., Xu X. Mutant V600E BRAF increases hypoxia inducible factor-1alpha expression in melanoma. Cancer Res. 2007;67:3177–3184. doi: 10.1158/0008-5472.CAN-06-3312. [DOI] [PubMed] [Google Scholar]
- 95.Hu H., Miao X.K., Li J.Y., Zhang X.W., Xu J.J., Zhang J.Y., Zhou T.X., Hu M.N., Yang W.L., Mou L.Y. YC-1 potentiates the antitumor activity of gefitinib by inhibiting HIF-1alpha and promoting the endocytic trafficking and degradation of EGFR in gefitinib-resistant non-small-cell lung cancer cells. Eur. J. Pharm. 2020;874:172961. doi: 10.1016/j.ejphar.2020.172961. [DOI] [PubMed] [Google Scholar]
- 96.Thomas S.L., Zhong D., Zhou W., Malik S., Liotta D., Snyder J.P., Hamel E., Giannakakou P. EF24, a novel curcumin analog, disrupts the microtubule cytoskeleton and inhibits HIF-1. Cell Cycle. 2008;7:2409–2417. doi: 10.4161/cc.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gao P., Zhang H., Dinavahi R., Li F., Xiang Y., Raman V., Bhujwalla Z.M., Felsher D.W., Cheng L., Pevsner J., et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee K., Zhang H., Qian D.Z., Rey S., Liu J.O., Semenza G.L. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc. Natl. Acad. Sci. USA. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99.Chen W., Hill H., Christie A., Kim M.S., Holloman E., Pavia-Jimenez A., Homayoun F., Ma Y., Patel N., Yell P., et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–117. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Y., Thompson J.D., Chan W.K. A cell-penetrating peptide suppresses the hypoxia inducible factor-1 function by binding to the helix-loop-helix domain of the aryl hydrocarbon receptor nuclear translocator. Chem. Biol. Interact. 2013;203:401–411. doi: 10.1016/j.cbi.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miranda E., Nordgren I.K., Male A.L., Lawrence C.E., Hoakwie F., Cuda F., Court W., Fox K.R., Townsend P.A., Packham G.K., et al. A cyclic peptide inhibitor of HIF-1 heterodimerization that inhibits hypoxia signaling in cancer cells. J. Am. Chem. Soc. 2013;135:10418–10425. doi: 10.1021/ja402993u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mylonis I., Lakka A., Tsakalof A., Simos G. The dietary flavonoid kaempferol effectively inhibits HIF-1 activity and hepatoma cancer cell viability under hypoxic conditions. Biochem. Biophys. Res. Commun. 2010;398:74–78. doi: 10.1016/j.bbrc.2010.06.038. [DOI] [PubMed] [Google Scholar]
- 103.Kong D., Park E.J., Stephen A.G., Calvani M., Cardellina J.H., Monks A., Fisher R.J., Shoemaker R.H., Melillo G. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res. 2005;65:9047–9055. doi: 10.1158/0008-5472.CAN-05-1235. [DOI] [PubMed] [Google Scholar]
- 104.Lee K., Qian D.Z., Rey S., Wei H., Liu J.O., Semenza G.L. Anthracycline chemotherapy inhibits HIF-1 transcriptional activity and tumor-induced mobilization of circulating angiogenic cells. Proc. Natl. Acad. Sci. USA. 2009;106:2353–2358. doi: 10.1073/pnas.0812801106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Kung A.L., Zabludoff S.D., France D.S., Freedman S.J., Tanner E.A., Vieira A., Cornell-Kennon S., Lee J., Wang B., Wang J., et al. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell. 2004;6:33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 106.Henchey L.K., Kushal S., Dubey R., Chapman R.N., Olenyuk B.Z., Arora P.S. Inhibition of hypoxia inducible factor 1-transcription coactivator interaction by a hydrogen bond surrogate alpha-helix. J. Am. Chem. Soc. 2010;132:941–943. doi: 10.1021/ja9082864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kushal S., Lao B.B., Henchey L.K., Dubey R., Mesallati H., Traaseth N.J., Olenyuk B.Z., Arora P.S. Protein domain mimetics as in vivo modulators of hypoxia-inducible factor signaling. Proc. Natl. Acad. Sci. USA. 2013;110:15602–15607. doi: 10.1073/pnas.1312473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lao B.B., Grishagin I., Mesallati H., Brewer T.F., Olenyuk B.Z., Arora P.S. In vivo modulation of hypoxia-inducible signaling by topographical helix mimetics. Proc. Natl. Acad. Sci. USA. 2014;111:7531–7536. doi: 10.1073/pnas.1402393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang W., Jia W.D., Xu G.L., Wang Z.H., Li J.S., Ma J.L., Ge Y.S., Xie S.X., Yu J.H. Antitumoral activity of rapamycin mediated through inhibition of HIF-1alpha and VEGF in hepatocellular carcinoma. Dig. Dis. Sci. 2009;54:2128–2136. doi: 10.1007/s10620-008-0605-3. [DOI] [PubMed] [Google Scholar]
- 110.Strowitzki M.J., Ritter A.S., Kimmer G., Schneider M. Hypoxia-adaptive pathways: A pharmacological target in fibrotic disease? Pharm. Res. 2019;147:104364. doi: 10.1016/j.phrs.2019.104364. [DOI] [PubMed] [Google Scholar]
- 111.Strowitzki M.J., Ritter A.S., Radhakrishnan P., Harnoss J.M., Opitz V.M., Biller M., Wehrmann J., Keppler U., Scheer J., Wallwiener M., et al. Pharmacological HIF-inhibition attenuates postoperative adhesion formation. Sci. Rep. 2017;7:13151. doi: 10.1038/s41598-017-13638-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hubbi M.E., Hu H., Ahmed I., Levchenko A., Semenza G.L. Chaperone-mediated autophagy targets hypoxia-inducible factor-1alpha (HIF-1alpha) for lysosomal degradation. J. Biol. Chem. 2013;288:10703–10714. doi: 10.1074/jbc.M112.414771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scheuermann T.H., Tomchick D.R., Machius M., Guo Y., Bruick R.K., Gardner K.H. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc. Natl. Acad. Sci. USA. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cho H., Du X., Rizzi J.P., Liberzon E., Chakraborty A.A., Gao W., Carvo I., Signoretti S., Bruick R.K., Josey J.A., et al. On-target efficacy of a HIF-2alpha antagonist in preclinical kidney cancer models. Nature. 2016;539:107–111. doi: 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Courtney K.D., Infante J.R., Lam E.T., Figlin R.A., Rini B.I., Brugarolas J., Zojwalla N.J., Lowe A.M., Wang K., Wallace E.M., et al. Phase I Dose-Escalation Trial of PT2385, a First-in-Class Hypoxia-Inducible Factor-2alpha Antagonist in Patients With Previously Treated Advanced Clear Cell Renal Cell Carcinoma. J. Clin. Oncol. 2018;36:867–874. doi: 10.1200/JCO.2017.74.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Habault J., Poyet J.L. Recent Advances in Cell Penetrating Peptide-Based Anticancer Therapies. Molecules. 2019;24:927. doi: 10.3390/molecules24050927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rizzuti M., Nizzardo M., Zanetta C., Ramirez A., Corti S. Therapeutic applications of the cell-penetrating HIV-1 Tat peptide. Drug Discov. Today. 2015;20:76–85. doi: 10.1016/j.drudis.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 118.Willam C., Masson N., Tian Y.M., Mahmood S.A., Wilson M.I., Bicknell R., Eckardt K.U., Maxwell P.H., Ratcliffe P.J., Pugh C.W. Peptide blockade of HIFalpha degradation modulates cellular metabolism and angiogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:10423–10428. doi: 10.1073/pnas.162119399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu X., Du J., Xi Q. HIF-1α ODD polypeptides increased the expression of HIF1 and VEGF in hypoxic rat cortical neuron. Neurol. Sci. 2011;32:1029–1033. doi: 10.1007/s10072-011-0574-8. [DOI] [PubMed] [Google Scholar]
- 120.Yamashita T., Ohneda O., Nagano M., Iemitsu M., Makino Y., Tanaka H., Miyauchi T., Goto K., Ohneda K., Fujii-Kuriyama Y., et al. Abnormal heart development and lung remodeling in mice lacking the hypoxia-inducible factor-related basic helix-loop-helix PAS protein NEPAS. Mol. Cell Biol. 2008;28:1285–1297. doi: 10.1128/MCB.01332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hara S., Hamada J., Kobayashi C., Kondo Y., Imura N. Expression and characterization of hypoxia-inducible factor (HIF)-3alpha in human kidney: Suppression of HIF-mediated gene expression by HIF-3alpha. Biochem. Biophys. Res. Commun. 2001;287:808–813. doi: 10.1006/bbrc.2001.5659. [DOI] [PubMed] [Google Scholar]
- 122.Chen J., Zhao S., Nakada K., Kuge Y., Tamaki N., Okada F., Wang J., Shindo M., Higashino F., Takeda K., et al. Dominant-negative hypoxia-inducible factor-1 alpha reduces tumorigenicity of pancreatic cancer cells through the suppression of glucose metabolism. Am. J. Pathol. 2003;162:1283–1291. doi: 10.1016/S0002-9440(10)63924-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Y., Li Y., Wang D., Li Y., Chang A., Chan W.K. Suppression of the hypoxia inducible factor-1 function by redistributing the aryl hydrocarbon receptor nuclear translocator from nucleus to cytoplasm. Cancer Lett. 2012;320:111–121. doi: 10.1016/j.canlet.2012.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mistry I.N., Tavassoli A. Reprogramming the Transcriptional Response to Hypoxia with a Chromosomally Encoded Cyclic Peptide HIF-1 Inhibitor. ACS Synth. Biol. 2017;6:518–527. doi: 10.1021/acssynbio.6b00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kung A.L., Wang S., Klco J.M., Kaelin W.G., Livingston D.M. Suppression of tumor growth through disruption of hypoxia-inducible transcription. Nat. Med. 2000;6:1335–1340. doi: 10.1038/82146. [DOI] [PubMed] [Google Scholar]
- 126.Kyle H.F., Wickson K.F., Stott J., Burslem G.M., Breeze A.L., Tiede C., Tomlinson D.C., Warriner S.L., Nelson A., Wilson A.J., et al. Exploration of the HIF-1alpha/p300 interface using peptide and Adhiron phage display technologies. Mol. Biosyst. 2015;11:2738–2749. doi: 10.1039/C5MB00284B. [DOI] [PubMed] [Google Scholar]
- 127.Hetherington K., Hegedus Z., Edwards T.A., Sessions R.B., Nelson A., Wilson A.J. Stapled Peptides as HIF-1alpha/p300 Inhibitors: Helicity Enhancement in the Bound State Increases Inhibitory Potency. Chemistry. 2020;26:7638–7646. doi: 10.1002/chem.202000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lavoie H., Gagnon J., Therrien M. ERK signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 2020;21:607–632. doi: 10.1038/s41580-020-0255-7. [DOI] [PubMed] [Google Scholar]
- 129.Lau J.L., Dunn M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018;26:2700–2707. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 130.Valeur E., Gueret S.M., Adihou H., Gopalakrishnan R., Lemurell M., Waldmann H., Grossmann T.N., Plowright A.T. New Modalities for Challenging Targets in Drug Discovery. Angew. Chem. Int. Ed. Engl. 2017;56:10294–10323. doi: 10.1002/anie.201611914. [DOI] [PubMed] [Google Scholar]
- 131.Joo S.H. Cyclic peptides as therapeutic agents and biochemical tools. Biomol. Ther. 2012;20:19–26. doi: 10.4062/biomolther.2012.20.1.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee A.C., Harris J.L., Khanna K.K., Hong J.H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019;20:2383. doi: 10.3390/ijms20102383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pelay-Gimeno M., Glas A., Koch O., Grossmann T.N. Structure-Based Design of Inhibitors of Protein-Protein Interactions: Mimicking Peptide Binding Epitopes. Angew. Chem. Int. Ed. Engl. 2015;54:8896–8927. doi: 10.1002/anie.201412070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ran X., Gestwicki J.E. Inhibitors of protein-protein interactions (PPIs): An analysis of scaffold choices and buried surface area. Curr. Opin. Chem. Biol. 2018;44:75–86. doi: 10.1016/j.cbpa.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]