SUMMARY
Glutamate receptor-like channels (GLRs) play important roles in numerous plant physiological processes. GLRs are homologous to ionotropic glutamate receptors (iGluRs) that mediate neurotransmission in vertebrates. Here we determine crystal structures of Arabidopsis thaliana GLR3.2 ligand-binding domain (LBD) in complex with glycine and methionine to 1.58 and 1.75 Å resolution, respectively. Our structures show a fold similar to iGluRs, but with several secondary structure elements either missing or different. The closed clamshell conformation of GLR3.2 LBD suggests that both glycine and methionine act as agonists. The mutation R133A strongly increases the constitutive activity of the channel, suggesting that the LBD mutated at the residue critical for agonist binding produces a more stable closed clamshell conformation. Furthermore, our structures explain the promiscuity of GLRs activation by different amino acids, confirm evolutionary conservation of structure between GLRs and iGluRs and predict common molecular principles of their gating mechanisms driven by bilobed clamshell-like LBDs.
Keywords: Glutamate receptor-like channels (GLR), plant, ionotropic glutamate receptor (iGluR), X-ray crystallography, Ca2+ channels
eTOC blurb
Glutamate receptor-like channels (GLRs) play important roles in plant physiology but their structural features have just begun unraveling. Gangwar et al. report structures of Arabidopsis thaliana GLR3.2 ligand-binding domain in complex with agonists glycine and methionine and discuss structural and functional relationships between GLRs and ionotropic glutamate receptors (iGluRs).
Graphical Abstract

INTRODUCTION
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that mediate excitatory neurotransmission throughout the vertebrate central nervous system (CNS) (Kumar and Mayer, 2013; Traynelis et al., 2010). iGluRs are assemblies of four subunits, each containing four main domains: the amino-terminal domain (ATD) implicated in receptor assembly, trafficking, and regulation; the ligand-binding domain (LBD or S1S2) that harbors binding sites for agonists, antagonists, and allosteric modulators; the transmembrane domain (TMD) forming an ion channel; and the cytosolic carboxy-terminal domain (CTD), which is involved in receptor localization and regulation (Sobolevsky, 2015; Twomey and Sobolevsky, 2018). Glutamate and other amino acids that function as neurotransmitters activate iGluRs by binding to the LBD and inducing conformational changes that lead to the opening of the ion channel (Armstrong and Gouaux, 2000; Twomey and Sobolevsky, 2018). Homologs of mammalian iGluRs have been identified in both vascular and non-vascular plants, known as glutamate receptor-like channels or GLRs, and are predicted to share the structural domain organization (Lam et al., 1998; Wudick et al., 2018a).
Recent studies revealed vital roles of GLRs in various physiological processes in plants, including wound response, stomatal aperture, seed germination, root development, innate immunity, and pollen tube growth (Kong et al., 2016; Kong et al., 2015; Li et al., 2013; Michard et al., 2011; Mousavi et al., 2013; Singh et al., 2016). GLRs are conserved along the plant lineage (2 in mosses, 4 in the lycophyte Sellaginella, 9 in Gingko) but went through an enormous expansion in the higher plants (40 in Pinus) and dramatic diversification into different clades in some angiosperms (Aouini et al., 2012; De Bortoli et al., 2016; Ortiz-Ramirez et al., 2017; Price et al., 2012; Wudick et al., 2018b). Arabidopsis thaliana has 20 AtGLRs phylogenetically divided into 3 clades (Chiu et al., 2002; Lacombe et al., 2001; Wudick et al., 2018a). AtGLR3.2, a representative of the third clade, is widely expressed in the plant, and displays highest expression in root cells where it localizes in the plasma membrane (Vincill et al., 2013). Overexpression of AtGLR3.2 in transgenic plants resulted in Ca2+ deprivation that was rescued by exogenous Ca2+ application, demonstrating ion channel functionality (Kim et al., 2001). While the structure of the LBD of AtGLR3.3 has been recently solved and predicted to accommodate various amino acids (Alfieri et al., 2020), there is no experimental confirmation that the predicted ligand promiscuity bears any functional consequence, namely in terms of activity elicitation, or other physiological consequences. Intriguingly, the sequence divergence of the ‘gate’ domain (the equivalent of the SYTANLAAF motif in iGluRs (Wollmuth and Sobolevsky, 2004) has led to the hypothesis that some GLRs might function without ligand-induced activation (Wudick et al., 2018a). This prediction is partially supported by patch-clamp recordings from plant protoplasts where constitutive currents are abolished in glr knock out (KO) lines (Mou et al., 2020). When expressed in the mammalian system, three channels (PpGLR1, AtGLR3.2, and AtGLR3.3) display constitutive currents in the absence of canonical ligands but are strongly activated by CORNICHON-homologue proteins (CNIHs) (Ortíz-Ramirez et al., 2017; Wudick et al., 2018b). Despite the constitutive activity reported for some GLRs, they remain to be gated by ligands, and screens designed to measure the effects of all proteinogenic amino acids showed an almost continuous gradient of AtGLR1.4 activation/inhibition (Tapken et al., 2013). A subsequent screen, using a different assay, showed a similar pattern for PpGLR1, but with the strongest activity inducer being the important plant hormone-like non-proteinogenic 1-aminocyclopropane-1-carboxylic acid (Mou et al., 2020). The apparent unique gating properties of GLRs, characterized by background ion channel activity and the amino acid stimulation requires structural and functional data to enlighten their possible physiological meaning.
While GLRs, including AtGLR3.2, govern a broad range of physiological and pathophysiological processes in plants, fundamental molecular mechanisms underlying their function remain elusive. To gain insight into how AtGLR3.2 binds to its activating ligands, we embarked on structural studies of its LBD. We found that the LBD of AtGLR3.2 binds to methionine (Met) and glycine (Gly), but the binding pocket is predicted to accommodate other amino acids as well. The LBD clamshell is closed in both structures, suggesting that they represent an active state of AtGLR3.2 that favors channel opening. Furthermore, we show that a point mutation of a residue critical for ligand binding increases the channel’s constitutive activity in the absence of either ligands or CNIHs.
RESULTS AND DISCUSSION
Structure determination
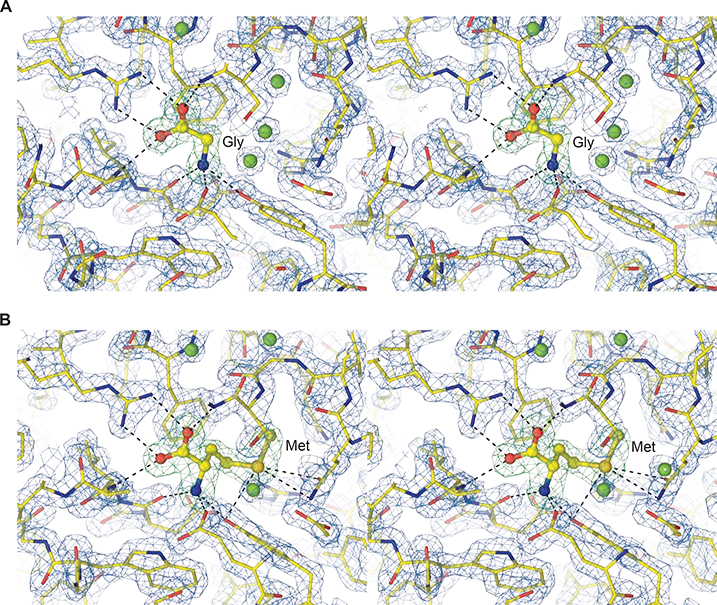
To determine the LBD structure, we used Arabidopsis thaliana GLR3.2 (AtGLR3.2) DNA to make a crystallizing construct, GLR3.2-S1S2. The boundaries of the two segments, S1 and S2 that assemble into the ligand-binding domain were determined based on the amino acid sequence alignment of AtGLR3.2 with mammalian iGluRs (Figure S1). At the beginning of S1 in the GLR3.2-S1S2 construct there are 46 N-terminal residues that have not been resolved in our crystal structures and presumably remain disordered. We expressed the GLR3.2-S1S2 construct in bacteria and purified the protein using affinity and ion-exchange chromatography (see Methods). Crystals of GLR3.2-S1S2 grew in the presence of methionine and glycine in sitting and hanging drops of vapor diffusion crystallization trays and were cryoprotected using glycerol for diffraction data collection at the synchrotron. Crystals of GLR3.2-S1S2 grown in the presence of glycine and methionine belonged to the P212121 space group, contained one S1S2 protomer in the asymmetric unit and diffracted to 1.58 and 1.75 Å resolution, respectively (Table 1). We solved the GLR3.2-S1S2Gly and GLR3.2-S1S2Met structures by molecular replacement, initially using a homology modeled search probe (see Methods). The clarity of the resulting electron density maps was sufficient (Figure 1) for the de novo building the structural models that included residues G47 to N286, with a 108 residue-long S1 GT-linked to a 130 residue-long S2.
Table 1.
Crystallographic statistics.
| GLR3.2-S1S2Gly | GLR3.2-S1S2Met | |
|---|---|---|
| Beamline | NE-CAT 24-ID-C | NE-CAT 24-ID-C |
| Wavelength (Å) | 0.97910 | 0.97910 |
| Space group | P212121 | P212121 |
| Cell parameters (a, b, c, Å) | 47.39, 64.37, 75.93 | 47.65, 65.47, 72.19 |
| Cell parameters (α, β, γ, °) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 47.39–1.58 (1.61–1.58) | 72.19–1.75 (1.78–1.75) |
| Number of Monomers in AU | 1 | 1 |
| Total observation | 146995 (5783) | 124336 (3896) |
| Unique observations | 32133 (1553) | 23419 (1258) |
| Rmerge | 0.06 (0.61) | 0.078 (0.67) |
| Rmease | 0.06 (0.67) | 0.87 (0.80) |
| Rpim | 0.03 (0.35) | 0.03 (0.43) |
| Mean (I)/sigma (I) | 14.9 (2.1) | 13.3 (1.8) |
| Completeness (%) | 99.2 (98.7) | 99.8 (99.1) |
| Multiplicity | 4.6 (3.7) | 5.3 (3.1) |
| CC (1/2) | 0.99 (0.69) | 0.99 (0.65) |
| Wilson B-factors (Å2) | 17.33 | 19.7 |
| Refinement | ||
| Resolution | 48.23 −1.58 | 48.50–1.75 |
| Reflections used in refinement | 32086 (3190) | 23364 (2295) |
| Rwork | 0.157 | 0.165 |
| Rfree | 0.183 | 0.199 |
| Number of non-hydrogen atoms | 2052 | 1962 |
| Macromolecule | 1852 | 1839 |
| Ligands | 9 | 11 |
| Average B factor | 21.13 | 23.87 |
| Macromolecule | 20.13 | 23.40 |
| Protein Residues | 240 | 238 |
| Number of water molecules | 202 | 112 |
| RMSD bond lengths (Å) | 0.01 | 0.01 |
| RMSD angles (°) | 1.89 | 1.90 |
| Ramachandran plot | ||
| Preferred regions (%) | 97.90 | 99.15 |
| Allowed regions (%) | 2.10 | 0.85 |
| Outliers (%) | 0 | 0 |
| PBD entry | 6VEA | 6VE8 |
Values in parentheses are for the highest-resolution shell.
Figure 1. AtGLR3.2-LBD electron density.
A-B, Close-up stereo view of AtGLR3.2 LBD (S1S2) in complex with (A) glycine and (B) methionine. Mesh shows a 2Fo-Fc electron density map contoured at 2 σ (blue) and Fo-Fc map contoured at 4 σ (green) when ligands were not present in the model.
See also Figure S2.
The structures of approximately 57×37×35 Å3 in dimension have a bilobed clamshell architecture (Figure 2A–B), with the ligand-binding site between the upper D1 lobe and the lower D2 lobe, similar to iGluR LBDs (Gouaux, 2004; Mollerud et al., 2017; Pohlsgaard et al., 2011). The GLR3.2-S1S2Gly and GLR3.2-S1S2Met structures superpose very well with the root mean square deviation (RMSD) of 0.275 Å for Cα atoms. For the ligand-binding pocket, even side-chain orientations are very similar between GLR3.2-S1S2Gly and GLR3.2-S1S2Met.
Figure 2. AtGLR3.2 ligand-binding domain structure.
A-B, Structures of isolated AtGLR3.2 LBD (S1S2) in complex with glycine (A) and methionine (B). The ligands are in ball-and-stick representation. Highly conserved cysteines, C230 and C284, are connected by disulfide bonds and shown in sticks. C-D, Close-up views of the ligand-binding pocket with bound glycine (C) and methionine (D). Residues involved in ligand binding are shown in sticks. Interactions between the ligands and the binding pocket residues are indicated by dashed lines.
See also Figures S1 and S2.
Ligand binding
The ligand-binding pocket of GLR3.2-S1S2 resembles the ligand-binding pocket of iGluR LBDs (Figure 2C–D), with the key interactions and binding residues conserved (Figure S1). The ligand glycine forms hydrogen bonds with Asp126, Ala128, Arg133 and Tyr178 and non-bonded contacts with Phe108, Asp126, Ile127, Ala128, Arg133, Ser177, Tyr178, Glu218 and Tyr221 (Figure S2A). Similarly, the ligand methionine establishes hydrogen bonds with Asp126, Ala 128, Arg133 and Tyr221 and forms non-bonded contacts with Arg57, Phe108, Asp126, Ile127, Ala 128, Arg133, Gln174, Val175, Gly176, Ser177, Tyr178, Glu218 and Tyr221 (Figure S2B).
For both glycine and methionine, the guanidinium group of Arg133 and the backbone amines of Ala128 and Tyr178 are hydrogen bonded to the carboxyl group of the ligand, while the backbone carbonyl oxygen of Asp126, the carboxyl group of Glu218 and the hydroxyl group of Tyr221 coordinate the amino group of the ligand. The thioether group of methionine is additionally coordinated by the hydroxyl group of Tyr221, guanidinium group of Arg57, and the amide group of Gln174. These interactions are specific to methionine and are missing in the case of glycine, which lacks the bulky side chain. Instead, two water molecules occupy the space that in the case of methionine is occupied by the thioether group. These two water molecules are stabilized by hydrogen bonds with Ser177 and Arg57.
Overall, the ligand-binding pocket of GLR3.2-S1S2 is shaped to bind differently sized amino acids (for example, glycine versus methionine) by exploiting the same interactions for binding the conserved amino acid core and adjusting the fit of the side chains into the corresponding binding pocket cavity with water. This explains a diverse range of ligand specificity previously observed for GLRs, with at least 12 of the 20 proteinogenic amino acids and D-Serine serving as agonists for the most studied AtGLR1.2, AtGLR1.4, AtGLR3.3, AtGLR3.4, and AtGLR3.5 (Forde and Roberts, 2014; Kong et al., 2016; Michard et al., 2011; Tapken et al., 2013; Vincill et al., 2012; Vincill et al., 2013; Wudick et al., 2018a). In agreement with our results, the recently determined structures of the AtGLR3.3-S1S2 (Alfieri et al., 2020) revealed similar ligand-binding promiscuity. The binding pocket and the mode of ligand binding, however, might be somewhat different among GLRs. For example, Trp, Phe, and Tyr can serve as agonists of AtGLR1.4 but not AtGLR3.3 or AtGLR3.4 (Tapken et al., 2013; Vincill et al., 2012; Vincill et al., 2013) suggesting that the ligand-binding pocket in AtGLR1.4 is likely larger to accommodate bulkier hydrophobic side chains. In part, differences in ligand binding among GLRs can originate from residues directly interacting with the ligand. For example, among eight GLR3.2 residues interacting with the ligand, six are conserved between clade 3 GLRs (Arg57, Asp126, Arg133, Tyr178, Glu218 and Tyr221) but two are not (Figure S1). Ala128 is Thr in GLR3.6, GLR3.4 and GLR3.7, while Gln174 is Pro in GLR3.6. Ligand binding can also be allosterically influenced by ATDs, which are much more variable in sequence compared to LBDs. In addition, GLR ligands may bind sites distinct from the site inside the LBD clamshell. For example, a bulky tripeptide glutathione that acts as an agonist of many GLRs is unlikely to fit the pocket accommodating Gly and Met (Figure 2) in the GLR3.2 LBD but it might bind somewhere else on the full-length protein.
Effect of a point mutation on gating
Given the structural determinants of ligand binding, we investigated the effects of possible disruption of ligand binding by mutating critical amino acids. We focused on the highly conserved Arg133 since the guanidinium group of this arginine coordinates the carboxyl group of both bound ligands and is critical for their binding. The possible effects of this point mutation were assayed by the transfection of mammalian COS-7 cells expressing the Ca2+ indicator Yellow CaMeleon 3.6 (YC3.6). To assay Ca2+ influx, COS-7 cells were first placed in a Ca2+- free solution containing EGTA, and subsequently subjected to 14.5 mM Ca2+ (see the top bar in Figure 3A). In the absence of ligand (Figure 3A, black dots), cytosolic Ca2+ showed a slight increase, revealing some basal conductance. When the experiment was repeated in the presence of 0.5 mM Gly, this elevation peaked at the same [Ca2+]cyt level and timing. Yet, while [Ca2+]cyt dropped immediately after peaking without the ligand, in the presence of 0.5mM Gly, [Ca2+]cyt levels went sustained for longer, producing a statistically detectable difference between essays (p<0.01). However, in the presence of 1 mM Gly, the elevation of cytosolic Ca2+ was more pronounced and statistically significant when compared to the other two experiments (p<10−6 to control and p=0.01 to 0.5 mM Gly). These elevations suggest that the wild-type AtGLR3.2 alone is moderately gated by 1 mM Gly. We then tested the effect of CNIHs that were previously shown to strongly promote ligand-independent activation of AtGLR3.2 currents (Wudick et al., 2018b). Expression of AtCNIH4 alone in COS-7 cells induces an increased Ca2+ influx (Figure S3). Given the conservation of CNIHs in plants and their capacity to complement other CNIH homologues, namely in yeast (Wudick et al., 2018b), we interpret this increase as a reflection of non-specific activation of COS-7 endogenous transport proteins. The effect of AtCNIH4 was insensitive to ligand addition (Figure S3). Yet, simultaneous expression of AtGLR3.2 and AtCNIH4 (Figure 3B) rendered much larger and robust Ca2+ elevations induced by both Met (red) and Gly (green) at 0.5 mM concentrations in comparison to the control (p<0.01 for both).
Figure 3. Effect of point mutations in ligand gating.
The possible effects of point mutations in the LBD gating of AtGLR3.2 were assayed by the transfection of mammalian COS-7 cells expressing a Ca2+ indicator (YC3.6). A, Expression of wild-type channel alone, shows its Ca2+ conductance to be gated by Glycine (Gly) at 1.0 mM. The experimental sequence is shown on the top black/yellows bar. Cells are Ca2+-starved with EGTA and then perfused with 14.5 mM Ca2+. In the absence of ligand (black dots) a slight increase occurs in cytosolic Ca2+. When the experiment is done in the presence 0.5 mM Gly, this elevation is slightly, but significantly, prolonged (p<0.01), but in the presence of 1.0 mM Gly there is a visible and statistically significant elevation of cytosolic Ca2+ (p<10−6 to control and p=0.01 to 0.5 mM). B, Simultaneous expression of AtGLR3.2 and AtCNIH4 renders the channel gated by both Met (red) and Gly (green) at 0.5 mM in comparison to the control (p<0.01 for all comparisons). However, when the critical residue 133 is substituted from Arginine to Alanine (C) the channel behaves as being constitutively open (black; compare with black control in B). Data are represented as mean ± SEM. All statistics obtained by two-way ANOVA with TukeyHSD.
See also Figures S3 and S4.
Finally, we tested the Ca2+ uptake by AtGLR3.2 with R133A mutation in the LBD, which was predicted to disrupt ligand binding (Figure 3C). Our Ca2+ uptake traces suggest that AtGLR3.2-R133A behaved as a constitutively open channel (compare black traces in Figure 3B and 3C), reaching the peak values of Ca2+ influx similar or higher than in the non-mutated channel in the presence of 0.5 mM Gly (green; p>0.1) or 0.5 mM Met (red; p<0.01 to the others). This apparent constitutive activation of the channel is independent of the presence of AtCNIH4 (Figure S4), which reached a similar level of Ca2+ flux in the presence or absence of AtCNIH4. Remarkably, the presence of AtCNIH4 affects the ligand binding properties, unveiling an apparent inhibitory effect of Gly (see Figures 3A and C). R133A mutation likely produces an alteration in the clamshell structure similar to ligand binding, i.e. clamshell closure, resulting in a similar effect on the pore. This result is hard to reconcile with no full-length GLR structure available, but it highlights the importance of the ligand binding domain for GLR gating. Mutations in the iGluR LBD have been shown to make AMPA receptors more responsive to kainate and less responsive to AMPA (Armstrong et al., 2003), to increase the efficacy of kainate receptor agonists (Meyerson et al., 2014), and to render NMDA receptors constitutively active (Blanke and VanDongen, 2008).
The strong increase in ligand-induced AtGLR3.2 activation caused by the presence of CNIH4 is consistent with the open state-stabilizing effects of HsCNIH2 and HsCNIH3 on AMPA receptors, where CNIHs slow down the deactivation and desensitization kinetics (Gill et al., 2011; Kato et al., 2010; Schwenk et al., 2009; Shi et al., 2010) and increase single-channel conductance (Coombs et al., 2012). While AMPA receptors are activated by ligands in the absence of CNIHs, the AtCNIH4 presence appears to always result in significant additional activation of AtGLR3.2. In the presence of AtCNIH4, glycine and methionine appear to act as an agonist and partial agonist on wild type AtGLR3.2 (Figure 3B). Methionine, however, acts like an inverse agonist on the R133A mutant. Indeed, strong activation of AtGLR3.2 by R133A in the presence of AtCNIH4 is not altered by glycine but suppressed to the level of partial activation in the presence of methionine (Figure 3C). Why these ligands, which cause the same clamshell closure in wild type LBD (Figure 2), behave so differently is currently unclear and may require full-length AtGLR3.2 structures to be understood.
Comparison of GLR and iGluR LBD structures
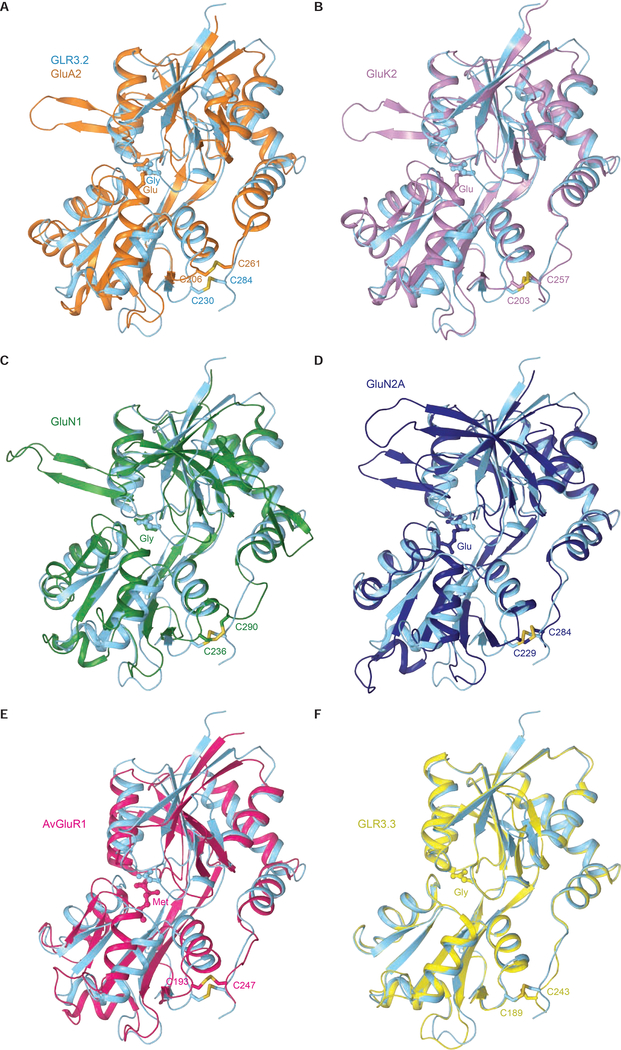
The ligand-binding domain, which binds agonists, competitive antagonists, and positive allosteric modulators, adopts a similar bilobed D1-D2 clamshell architecture in vertebrate, invertebrate, and plant glutamate receptors (Figure 4A–F). We compared the AtGLR3.2 LBD with the LBDs of three dominant mammalian iGluRs (AMPA, kainate and NMDA subtypes), rotifer Adienta vaga subunit 1 (AvGluR1), and Arabidopsis thaliana GLR3.3. These species are separated by millions of years of evolution and their LBD sequences share poor sequence identity. In Figure 4, we superimposed the GLR3.2-S1S2 with the previously solved agonist-bound S1S2 structures of GluA2 (PDB:1FTJ) (Armstrong and Gouaux, 2000), GluK2 (PDB:1S50) (Mayer, 2005), GluN1 (PDB:1PB7) (Furukawa and Gouaux, 2003), GluN2A (PDB:2A5S) (Furukawa et al., 2005), AvGluR1 (PDB:4IO2) (Lomash et al., 2013) and AtGLR3.3 (PDB:6R88) (Alfieri et al., 2020). The RMSD values calculated for all Cα atoms in each superposition with GLR3.2-S1S2 are 1.9 Å for GluA2, 1.5 Å for GluK2, 1.8 Å for GluN1, 4.5 Å for GluN2, 3 Å for AvGluR1, and 0.77 Å for AtGLR3.3. Structures of AtGLR3.3 and AtGLR3.2 LBDs are very similar, consistent with their sequence similarity. The amino acid sequences of AtGLR3.2 and AtGLR3.3 LBDs share 61.6% identity and all eight residues that interact with the agonist are 100% conserved, including Arg in the β1-β2 loop, Asp and Ala in the β5-αD loop, Arg in αD, Gln in β9, Tyr in αF, Glu in β10, and Tyr in αI (Figures S1 and S2). The extent of clamshell closure in AtGLR3.3 and AtGLR3.2 is also nearly identical and greatly resembles the one in AvGluR1 of the rotifer Adineta vaga (Lomash et al., 2013). More significant differences were observed in superpositions of GLR3.2-S1S2 with S1S2 of AMPA, kainate and NMDA receptors. The main regions of distinction are the β1-αB loop that is extended in GLRs compared to iGluRs, as well as the sticking out β hairpin loop β2-αC and the helices αA and αG, which are present in iGluRs but absent in GLRs. Instead of the helix G, GLRs have a short β strand that we named 9a. In addition, NMDA receptor LBDs have a large hairpin loop between β1 and αB, which is missing in GLRs, AMPA, and kainate receptors. Apart from these regions, the secondary structure organization of LBD is conserved between mammalian, rotifer, and plant receptors. The arginine in the αD helix (R133 in GLR3.2-S1S2 and R551 in the full-length GLR3.2), which forms bidentate hydrogen bonds with the ligand’s carboxyl group is highly conserved across all species (Lomash et al., 2013; Mayer, 2020). Other conserved residues include cysteines that form a disulfide bond between the C-terminal ends of the helices I and K (Cys230 and Cys284 in GLR3.2-S1S2), which are only missing in prokaryotic receptors (Lee et al., 2008; Mayer et al., 2001).
Figure 4. Comparison of AtGLR3.2 and iGluR LBDs.
A-F, Structural superpositions of isolated LBDs from AtGLR3.2 (cyan) in complex with glycine and (A) rat GluA2 (PDB ID: 1FTJ, orange) in complex with glutamate, (B) rat GluK2 (PDB ID: 1S50, purple) in complex with glutamate, (C) rat GluN1 (PDB ID: 1PB7, green) in complex with glycine and (D) rat GluN2A (PDB ID: 2A5S, blue) in complex with glutamate (E) rotifer AvGluR1 (PDB ID: 4IO2, magenta) in complex with Met (F) Arabidopsis GLR3.3 (PDB ID:6R88, yellow) in complex with Gly. The ligands are in ball-and-stick representation. Highly conserved cysteines connected by disulfide bonds are shown in sticks.
Compared to iGluRs that are selectively activated by certain amino acids, AtGLRs and AvGluR1 can be activated by different amino acids. Such promiscuity in amino acid ligand binding is supported by structures of S1S2 that were solved for AvGluR1 in complex with Glu, Asp, Ser, Ala, Met and Phe (Lomash et al., 2013), AtGLR3.3 in complex with Met, Glu, Ala, and Gly (Alfieri et al., 2020) and AtGLR3.2 in complex with Met and Gly (this study). This promiscuity is likely due to unique features of the LBDs in these receptors compared to mammalian iGluRs. The AvGluR1 requires a Cl− ion in the binding pocket for Ala, Ser, and Met complex. AtGLR3.3 did not require ions to interact with their ligand and not a trace of ion density was found in its binding pocket (Alfieri et al., 2020; Lomash et al., 2013). Moreover, only GLR3.2-S1S2Gly has two water molecules in the ligand binding pocket but GLR3.2-S1S2Met complex does not have any, unlike AvGluR1 and iGluRs. Interestingly, the AvGluR1 and AtGLR LBDs bound to different amino acid ligands have the same extent of the clamshell closure, which is also similar to agonist-bound iGluR LBDs. Since these AvGluR1 and AtGLRs ligands have different affinities and full versus partial agonistic character (Alfieri et al., 2020; Lomash et al., 2013), the extent of the LBD clamshell closure seems to be independent of these two characteristics. In some iGluR studies, the extent of the LBD clamshell closure was postulated as a measure of the ligand partial agonistic character (Jin et al., 2003), while other studies argued that it is rather the fraction of time that the clamshell spends in the fully closed conformation that matters (Ramaswamy et al., 2012; Salazar et al., 2017; Twomey and Sobolevsky, 2018). For example, based on the higher Ca2+ signal observed for glycine versus methionine, we hypothesize that methionine is rather a partial agonist compared to glycine. This difference in agonistic character is consistent with the previous reports on AtGLR3.1/3.5, where Met-activated Ca2+ currents were shown to be responsible for maintaining cytosolic Ca2+ (Kong et al., 2016). However, the structural basis for such differences are unclear until the structures of full-length GLRs are available as well as more detailed analysis of their kinetics and energetics.
In summary, the overall architecture of our GLR3.2-S1S2Gly and GLR3.2-S1S2Met structures as well as the type of ligand binding suggest that similar to iGluRs, the clamshell-like closure of LBDs in GLRs might provide a driving force to gate the GLR-associated ion channel (Armstrong and Gouaux, 2000; Twomey and Sobolevsky, 2018). To test this hypothesis, one would need to capture the full-length structure of GLR. The observed similarity in the LBD clamshell architecture, ligand binding, and predicted gating mechanism also suggests that plant GLRs and iGluRs originate from a common ancestor to function in different kingdoms of life yet utilize similar molecular mechanisms. Our structures of AtGLR3.2 LBD in complex with two different amino acid ligands along with the role of CNIH in Ca2+ uptake indicate that both ligand and auxiliary protein binding are necessary for AtGLR3.2 function.
STAR Methods text
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alexander Sobolevsky (as4005@cumc.columbia.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Coordinates and structure factors for the GLR3.2-S1S2Gly and GLR3.2-S1S2Met structures have been deposited to the PDB with the accession codes 6VEA and 6VE8, respectively. This study did not generate new code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Protein expression was performed in Escherichia coli Origami B (DE3) cells. Cells were cultured in LB media at 37°C until OD600 reached the value of 1.0–1.2, then cooled down to 20°C, induced with 250 μM IPTG and incubated for another 20 hours at 20°C.
COS-7 cells for calcium imaging experiments were maintained at 37°C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium, supplemented with 5 % fetal bovine serum and 1 % penicillin/streptomycin.
METHOD DETAILS
Cloning and mutagenesis
RNA was isolated from col-0 leaf tissue using Bioline ISOLATE II RNA Plant Kit. The Bioline SensiFAST cDNA Synthesis kit was used to generate cDNA from the col-0 RNA. The CDS for AtGLR3.2 was amplified from cDNA using the primers: 5’- gtaacggccgccagtgtgctggaattcA TGTTTTGGGTTTTGGTTCTGT-3’, 5’- atagggccctctagatgcatgctcgaGTCATATTGGTCTAGAAGGT-3’. The glr3.2 CDS PCR fragment was cloned into EcoRI/XhoI digested pCDNA3 via Gibson Isothermal Assembly to yield pCDNA3-AtGLR3.2(cDNA). The final construct was verified by Sanger Sequencing. The point mutant was amplified from pCDNA3-AtGLR3.2(cDNA) by two PCRs using overlapping mutagenic oligonucleotide primers. Primers were as follows, PCR one: 5’- TGATACTGTCTGGATCATTGC TCGAGCTGTTAAGAGACTTCTAG −3’; 5’- GAAATCCACAA TCCTTGTTGC TTTCGTAACAATAGCTATGTCTCC-3’. PCR two: 5’- GAGACATAGCTATT GTTACGAAAGC AACAAGGATTGTGGATTTCACTCAGC-3’; 5’- atagggccctctagatgcatgctcgaG TCA TATTGGTCTAGAAGGCT-3’. Inserts were ligated with a backbone of pCDNA3-AtGLR3.2 linearized at XhoI restriction sites to construct the final mutant vector by Gibson Assembly (Gibson et al., 2009).
Protein expression and purification
The boundaries of the GLR3.2 ligand-binding domain (S1S2) were determined based on the sequence alignment with GluA2 (Armstrong et al., 1998; Sobolevsky et al., 2009). The DNA encoding AtGLR3.2 residues, S420-V572 (S1) and P682-N811 (S2), were amplified using gene-specific primers and subcloned into the pET22b vector (Novagen) between NcoI and XhoI sites with a GT linker between S1 and S2 (Armstrong and Gouaux, 2000). For purification purposes, an 8xHis affinity tag followed by a thrombin cleavage site (LVPRG) was introduced at N-terminal.
The construct pET22b carrying GLR3.2-S1S2 was transformed into Escherichia coli Origami B (DE3) cells and grown in LB media supplemented with 100 μg/ml ampicillin, 15 μg/ml kanamycin and 12.5 μg/ml tetracycline. The freshly inoculated culture was grown at 37°C until OD600 reached the value of 1.0–1.2. Then cells were cooled down to 20°C, induced with 250 μM IPTG, and incubated in the orbital shaker for another 20 hours at 20°C. Cells were harvested by centrifugation at 5488 g for 15 min at 4°C and the cell pellet was washed with the buffer containing 20 mM Tris pH 8.0 and 150 mM NaCl. For protein extraction, cells were resuspended in lysis buffer consisting of 20 mM Tris pH 8.0, 200 mM NaCl, 1 mM glutamate, 5 mM methionine, 1 mM βME, 1 mM PMSF, 100 μg/ml lysozyme, 5 mM MgSO4 and DNAse. All purification steps were carried out in buffers supplemented with 1 mM glutamate and 5 mM methionine. The cells were disrupted by sonication and centrifuged at 18600 g in the Ti45 rotor for 1 hour at 4°C. The supernatant was mixed with His60 Ni superflow resin (Takara) and rotated for 2 hours at 4°C. The protein-bound resin was washed with the buffer containing 15 mM imidazole and the protein was eluted in 20 mM Tris pH 8.0, 150 mM NaCl, 1 mM glutamate, 5 mM methionine, 1 mM βME, and 200 mM imidazole. The protein was dialyzed overnight in the buffer containing 20 mM Tris pH 8.0, 75 mM NaCl, 1 mM glutamate, 5 mM methionine, 1 mM BME, and 4% (v/v) glycerol. After thrombin digest (1:500 w/w) at 22°C for 1-hour, the protein was further purified using ion-exchange Hi-Trap Q HP- (GE Healthcare). The protein quality was assessed by SDSPAGE and analytical size-exclusion chromatography using the Superpose 10/300 column (GE Healthcare).
Crystallization and structure determination
Crystallization screening was performed with GLR3.2-S1S2 protein at a concentration of ~7 mg/ml using Mosquito robot (TTP Labtech) and sitting drop vapor diffusion in 96-well crystallization plates. Small needle-shaped crystals, which appeared after two weeks of incubating crystallization trays at 4°C and 20°C, were further optimized using the hanging drop method and 24-well crystallization plates. The best-diffracting long needle-shaped crystals of methionine-bound GLR3.2-S1S2 grew at 20°C in 0.1 M MES pH 6.5, 18% PEG MME 2K and 0.1 M ammonium sulfate. Crystals of glycine-bound GLR3.2-S1S2 grew in a similar condition but in the presence of 0.3 μl of 1M glycine that supplemented the 4 μl crystallization drop as an additive. The best-diffracting needle-shaped crystals of glycine-bound GLR3.2-S1S2 grew at 4°C in 22 % PEG 4K, 0.1 M ammonium acetate, and 0.1 M sodium acetate pH 4.6. All crystals were cryoprotected using 25% glycerol and flash-frozen in liquid nitrogen for data collection. Crystal diffraction data were collected at the beamline 24-ID-C of the Advanced Photon Source and processed using XDS (Kabsch, 2010) and Aimless as a part of the CCP4 suite (Winn et al., 2011).
The structure of methionine-bound GLR3.2-S1S2 was solved by molecular replacement using Phaser (McCoy, 2007) and a search probe generated by SWISS-MODEL homology modeling (Waterhouse et al., 2018) from the ligand-binding domain of NMDA receptor (PDB ID: 6MMS) (Jalali-Yazdi et al., 2018). The initial partial solution was used again as a search probe for subsequent rounds of molecular replacement, which ultimately resulted in a complete GLR3.2-S1S2 model. The model was refined by alternating cycles of building in COOT (Emsley and Cowtan, 2004) and automatic refinement in Phenix (Adams et al., 2010). The structure of glycine-bound GLR3.2-S1S2 was solved by molecular replacement using the methionine-bound GLR3.2-S1S2 structure as a search probe. Water molecules were added in Coot and Phenix refine. All structural figures were prepared in PyMol (DeLano, 2002). The protein-ligand interaction plot was created using the Ligplot server (Wallace et al., 1995; Laskowski et al., 2018).
COS-7 cells transfection and calcium imaging
Protocols for COS-7 cells transfection and Ca2+ imaging were adapted from Ortiz-Ramirez et al. (2017). COS-7 cells (Sigma-Aldrich) were maintained at 37°C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium, supplemented with 5 % fetal bovine serum and 1 % penicillin/streptomycin (Gibco), and transfected at low passage (P < 7). Cells were plated at a density at 50% confluence in 35-mm diameter dishes and transfected using FugeneHD (Promega) as specified by the supplier. Cells were co-transfected with three plasmids: pCI-AtCNIH4 or empty pCI (0.3 μg) plus pcDNA3-AtGLR3.2 or empty pcDNA3 (0.9 μg) were co-transfected with pEF1-YC3.6 (0.5 μg). The co-transfection with pCI-AtCNIH4 was an experimental stratagem used to enhance functional expression of GLRs on the plasma membrane (Wudick et al., 2018b). Cells were used for imaging 38 to 41 hours after transfection. They were washed in a Ca2+-free solution (1 mM EGTA, 10 mM Bis-Tris propane buffered to pH 7.3 with HEPES and set to 350 mosmol.kg−1 with D-mannitol). Cells were imaged in the Ca2+-free solution for 1.5 min before the addition of Ca2+ to a final concentration of 14.5 mM (using Ca-Gluconate). The ligands (Met or Gly, 0.5 or 1.0 mM) are added at the beginning (even before calcium is added). Time-lapse acquisition was performed with a sampling interval of 30 secs. 8 to 12 cells were imaged in each dish using the stage position recording tool of the microscope system. Imaging was performed at room temperature using a DeltaVision Elite Deconvolution/TIRF microscope system (Olympus inverted IX-71) under a 60X lens (1.2NA UPLSAPO water /WD 0.28 mm). A xenon lamp from the DeltaVision system was used with a CFP excitation filter (438–424 nm). Two simultaneous emission records were captured: YFP emission (548–522 nm) and CFP emission (475–424 nm). To minimize bleaching, the laser was set to 2%. YFP and CFP imaging were recorded with 0.6 sec exposure time. Images were processed using ImageJ. Ratios were obtained after background subtraction and signal clipping using the “Ratio-plus” plug-in for ImageJ. The signal of each channel was averaged in a circle in the middle of the cell (with 100–200 pixel diameter, depending on the size of the cell). The YFP/CFP ratio was obtained by dividing the emission recorded for YFP (548–522 nm) by the one recorded for CFP (475–424 nm). No significant bleaching or ratio drift was observed in our experimental conditions.
QUANTIFICATION AND STATISTICAL ANALYSIS
The X-ray structures of GLR3.2-S1S2 were determined using software listed in the Key Resources Table. Statistics generated from the data processing, refinement and validation are displayed in Table 1.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ampicillin | Sigma | Cat# A8351 |
| Kanamycin | Fisher scientific | Cat# BP906–5 |
| Tetracycline | Fisher scientific | Cat# BP912 |
| IPTG | Zymo Research | Cat# I1001–5 |
| Tris | Fisher scientific | Cat# BP152–1 |
| NaCl | Fisher scientific | Cat# BP358–212 |
| L-Glutamate | Sigma | Cat# 49621 |
| L-Methionine | Sigma | Cat# M9625 |
| MgSO4 | Fluka | Cat# 13143 |
| DNAse | Sigma | Cat# DN25–1 |
| PMSF | Acros Organics | Cat# 215740500 |
| 2-Mercaptoethanol (BME) | Acros Organics | Cat# 125470100 |
| Ni-Affinity Resin | Takara | Cat# 635660 |
| Imidazole | Acros Organics | Cat# 301870025 |
| Thrombin | Haematologic Technologies | Cat# HCT-0020 |
| Glycerol | Fisher scientific | Cat# BP229–4 |
| MES buffer | Sigma | Cat# M2933 |
| PEG 2000 MME | Fluka | Cat# 81321 |
| Ammonium Sulfate | Fisher scientific | Cat# A702–500 |
| Glycine | Jena Biosciences | Cat# CS-507L |
| Ammonium acetate | Fisher scientific | Cat# BP326–500 |
| Sodium Acetate | Fisher scientific | Cat# S209–500 |
| Dulbecco’s Modified Eagle’s Medium | Gibco | Cat# 10566024 |
| Fetal bovine serum | Gibco | Cat# 16140071 |
| Penicillin Streptomycin Fungizone | Cytiva HyClone | Cat# SV3007901 |
| FugeneHD | Promega | Cat# E2311 |
| EGTA | Sigma | Cat# E4378 |
| Bis-Tris Propane | RPI | Cat# B78000100.0 |
| HEPES | Sigma | Cat# H3375–250G |
| D-mannitol | Fisher | Cat# M120–500 |
| Ca-Gluconate | Sigma | Cat# C8231–100G |
| Deposited Data | ||
| Coordinates of GLR3.2-S1S2-Glycine | This paper | PDB: 6VEA |
| Coordinates of GLR3.2-S1S2-Methionine | This paper | PDB: 6VE8 |
| S1S2 of GluA2 | (Armstrong and Gouaux, 2000) | PDB: 1FTJ |
| S1S2 of GluK2 | (Mayer, 2005) | PDB: 1S50 |
| S1S2 of GluN1 | (Furukawa and Gouaux, 2003) | PDB: 1PB7 |
| S1S2 of GluN2A | (Furukawa et al., 2005) | PDB: 2A5S |
| S1S2 of AvGluR1 | (Lomash et al., 2013) | PDB: 4IO2 |
| S1S2 of AtGLR3.3 | (Alfieri et al., 2020) | PDB: 6R88 |
| Ligand-binding domain of NMDA receptor | (Jalali-Yazdi et al., 2018) | PDB ID: 6MMS |
| Experimental Models: Cell Lines | ||
| COS-7 | ATTC | CRL-1651 |
| E. coli Origami B (DE3) | Novagen | Cat# 70837 |
| Recombinant DNA | ||
| pEF1-YC3.6 | Dr. Jörg Kudla lab, Univ. Muenster, Germany | N/A |
| Oligonucleotides | ||
| AtGLR3.2 amplification primer: 5’-gtaacggccgccagtgtgctggaattcA TGTTTTGGGTTTTGGTTCTGT-3’ | This paper | N/A |
| AtGLR3.2 amplification primer: 5’- atagggccctctagatgcatgctcgaGTCATATTGGTCTAGAAGGT-3’ | This paper | N/A |
| pcDNA3 | Invitrogen | N/A |
| pCI-AtCNIH4 | Wudick et al., 2018b | Genebank: NC_003070.9; At1g12390; Salk_145991 |
| pcDNA3-AtGLR3.2 | This paper | GeneBank: NC_003075; Araprot: At4G35290 |
| pET22b-GLR3.2-S1S2 | This paper | GeneBank: NC_003075; Araprot: At4G35290 |
| Software and Algorithms | ||
| Pymol (Schrödinger) | DeLano, 2002 | http://www.pymol.org |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org/ |
| CCP4 | Winn et al., 2011 | http://www.ccp4.ac.uk/ |
| COOT | Emsley et al., 2004 | http://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot |
| XDS | Kabsch, 2010 | http://xds.mpimf-heidelberg.mpg.de/ |
| Swiss-Model | Waterhouse et al., 2018 | https://swissmodel.expasy.org/ |
| PDBsum | Laskowski et al., 2018 | https://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html |
| SigmaPlot 11.0 | Systat Software Inc. | Systatsoftware.com |
| Other | ||
| DeltaVision Elite Deconvolution/TIRF microscope system | GE Healthcare | Part # 53–851206-001 |
| Ion Exchange Hi-Trap Q HP column | GE Healthcare | Cat# 17–1154-01 |
| Size Exclusion Superose 10/300 column | GE Healthcare | Cat# 17–5172-01 |
Statistical significance in calcium imaging experiments was calculated by two-way ANOVA with TukeyHSD using an R custom script or SigmaPlot 11.0 (Systat Software Inc).
Supplementary Material
Highlights.
AtGLR3.2 LBD structures were solved in complex with agonists glycine and methionine
AtGLR3.2 LBD structures show clamshell architecture typical for vertebrate iGluRs
Mutation of R133 that is critical for agonist binding increases channel’s activity
Structural conservation between GLRs and iGluRs predicts common gating principles
ACKNOWLEDGMENTS
We thank Dr. Surajit Banerjee for assistance with the data collection, Dr. Jesse Yoder for help with the molecular replacement, Dr. Appu K. Singh for advice in the crystallographic data processing and Drs. Maria Yelshanskaya and Kirill Nadezhdin for comments on the manuscript and for helpful discussions. pCI-YC3.6 construct was kindly supplied by Dr. Jorg Kudla (Univ. Muenster). We thank Dr. Daniel Damineli (Univ. São Paulo) for help with statistical analysis. A.I.S. is supported by the NIH (R01 CA206573, R01 NS083660, R01 NS107253), NSF (1818086), and the Irma T. Hirschl Career Scientist Award. Data were collected at the beamline 24-ID-C of the Advanced Photon Source. 24-ID-C is one of the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Pilatus 6M detector on the 24-ID-C beamline is funded by an NIH-ORIP HEI grant (S10 RR029205). M.N.G. received support from the Institute of Human Nutrition (IHN) training grant, Graduate Training in Nutrition (5T32DK007647-30). J.A.F. was supported by the NIH (R01 GM131043) and the NSF (MCB1616437, MCB1714993 and MCB1930165).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfieri A, Doccula FG, Pederzoli R, Grenzi M, Bonza MC, Luoni L, Candeo A, Romano Armada N, Barbiroli A, Valentini G, et al. (2020). The structural bases for agonist diversity in an Arabidopsis thaliana glutamate receptor-like channel. Proc Natl Acad Sci U S A 117, 752–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouini A, Matsukura C, Ezura H, and Asamizu E (2012). Characterisation of 13 glutamate receptor-like genes encoded in the tomato genome by structure, phylogeny and expression profiles. Gene 493, 36–43. [DOI] [PubMed] [Google Scholar]
- Armstrong N, and Gouaux E (2000). Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Mayer M, and Gouaux E (2003). Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic adjustment of agonist-induced conformational changes. Proc Natl Acad Sci U S A 100, 5736–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Sun Y, Chen GQ, and Gouaux E (1998). Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature 395, 913–917. [DOI] [PubMed] [Google Scholar]
- Blanke ML, and VanDongen AM (2008). Constitutive activation of the N-methyl-D-aspartate receptor via cleft-spanning disulfide bonds. J Biol Chem 283, 21519–21529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Brenner ED, DeSalle R, Nitabach MN, Holmes TC, and Coruzzi GM (2002). Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Mol Biol Evol 19, 1066–1082. [DOI] [PubMed] [Google Scholar]
- Coombs ID, Soto D, Zonouzi M, Renzi M, Shelley C, Farrant M, and Cull-Candy SG (2012). Cornichons modify channel properties of recombinant and glial AMPA receptors. J Neurosci 32, 9796–9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bortoli S, Teardo E, Szabo I, Morosinotto T, and Alboresi A (2016). Evolutionary insight into the ionotropic glutamate receptor superfamily of photosynthetic organisms. Biophys Chem 218, 14–26. [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002). The PyMOL Molecular Graphics System (San Carlos, CA, USA, DeLano Scientific; ). [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Forde BG, and Roberts MR (2014). Glutamate receptor-like channels in plants: a role as amino acid sensors in plant defence? F1000Prime Rep 6, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, and Gouaux E (2003). Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J 22, 2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, and Gouaux E (2005). Subunit arrangement and function in NMDA receptors. Nature 438, 185–192. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Gill MB, Kato AS, Roberts MF, Yu H, Wang H, Tomita S, and Bredt DS (2011). Cornichon-2 modulates AMPA receptor-transmembrane AMPA receptor regulatory protein assembly to dictate gating and pharmacology. J Neurosci 31, 6928–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouaux E (2004). Structure and function of AMPA receptors. The Journal of physiology. 554, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali-Yazdi F, Chowdhury S, Yoshioka C, and Gouaux E (2018). Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor. Cell 175, 1520–1532 e1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Banke TG, Mayer ML, Traynelis SF, and Gouaux E (2003). Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci 6, 803–810. [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010). Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Ho MT, Yu H, Tu Y, Siuda ER, Wang H, Qian YW, Nisenbaum ES, Tomita S, et al. (2010). Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron 68, 1082–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Kwak JM, Jae SK, Wang MH, and Nam HG (2001). Overexpression of the AtGluR2 gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol 42, 74–84. [DOI] [PubMed] [Google Scholar]
- Kong D, Hu HC, Okuma E, Lee Y, Lee HS, Munemasa S, Cho D, Ju C, Pedoeim L, Rodriguez B, et al. (2016). L-Met Activates Arabidopsis GLR Ca(2+) Channels Upstream of ROS Production and Regulates Stomatal Movement. Cell Rep 17, 2553–2561. [DOI] [PubMed] [Google Scholar]
- Kong D, Ju C, Parihar A, Kim S, Cho D, and Kwak JM (2015). Arabidopsis glutamate receptor homolog3.5 modulates cytosolic Ca2+ level to counteract effect of abscisic acid in seed germination. Plant Physiol 167, 1630–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, and Mayer ML (2013). Functional insights from glutamate receptor ion channel structures. Annu Rev Physiol 75, 313–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe B, Becker D, Hedrich R, DeSalle R, Hollmann M, Kwak JM, Schroeder JI, Le Novere N, Nam HG, Spalding EP, et al. (2001). The identity of plant glutamate receptors. Science 292, 1486–1487. [DOI] [PubMed] [Google Scholar]
- Lam HM, Chiu J, Hsieh MH, Meisel L, Oliveira IC, Shin M, and Coruzzi G (1998). Glutamate-receptor genes in plants. Nature 396, 125–126. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Jabłońska J, Pravda L, Vařeková RS, and Thornton JM (2018). PDBsum: Structural summaries of PDB entries. Prot. Sci 27, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kang GB, Lim HH, Jin KS, Kim SH, Ree M, Park CS, Kim SJ, and Eom SH (2008). Crystal structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with L-glutamate: structural dissection of the ligand interaction and subunit interface. J Mol Biol 376, 308–316. [DOI] [PubMed] [Google Scholar]
- Li F, Wang J, Ma C, Zhao Y, Wang Y, Hasi A, and Qi Z (2013). Glutamate receptor-like channel3.3 is involved in mediating glutathione-triggered cytosolic calcium transients, transcriptional changes, and innate immunity responses in Arabidopsis. Plant Physiol 162, 1497–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomash S, Chittori S, Brown P, and Mayer ML (2013). Anions mediate ligand binding in Adineta vaga glutamate receptor ion channels. Structure 21, 414–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML (2005). Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron 45, 539–552. [DOI] [PubMed] [Google Scholar]
- Mayer ML (2020). Glutamate receptors from diverse animal species exhibit unexpected structural and functional diversity. J Physiol. 10.1113/JP279026 [DOI] [PubMed] [Google Scholar]
- Mayer ML, Olson R, and Gouaux E (2001). Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state. J Mol Biol 311, 815–836. [DOI] [PubMed] [Google Scholar]
- McCoy AJ (2007). Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D 63, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, and Subramaniam S (2014). Structural mechanism of glutamate receptor activation and desensitization. Nature 514, 328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham M, Liu LH, Obermeyer G, and Feijo JA (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332, 434–437. [DOI] [PubMed] [Google Scholar]
- Mou W, Michard E, Sittman J, Simon A, Dong-Dong A, Feijo JA, Chang C, (2020). Ethylene-independent signalling by the ethylene precursor ACC in Arabidopsis ovular pollen tube attraction. Nat Commun 11, 4082. doi.org/ 10.1038/s41467-020-17819-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollerud S, Frydenvang K, Pickering DS, and Kastrup JS (2017). Lessons from crystal structures of kainate receptors. Neuropharmacology 112, 16–28. [DOI] [PubMed] [Google Scholar]
- Mousavi SA, Chauvin A, Pascaud F, Kellenberger S, and Farmer EE (2013). GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500, 422–426. [DOI] [PubMed] [Google Scholar]
- Ortiz-Ramirez C, Michard E, Simon AA, Damineli DSC, Hernandez-Coronado M, Becker JD, and Feijo JA (2017). GLUTAMATE RECEPTOR-LIKE channels are essential for chemotaxis and reproduction in mosses. Nature 549, 91–95. [DOI] [PubMed] [Google Scholar]
- Pohlsgaard J, Frydenvang K, Madsen U, and Kastrup JS (2011). Lessons from more than 80 structures of the GluA2 ligand-binding domain in complex with agonists, antagonists and allosteric modulators. Neuropharmacology 60, 135–150. [DOI] [PubMed] [Google Scholar]
- Price MB, Jelesko J, and Okumoto S (2012). Glutamate receptor homologs in plants: functions and evolutionary origins. Front Plant Sci 3, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Cooper D, Poddar N, MacLean DM, Rambhadran A, Taylor JN, Uhm H, Landes CF, and Jayaraman V (2012). Role of conformational dynamics in alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptor partial agonism. J Biol Chem 287, 43557–43564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar H, Eibl C, Chebli M, and Plested A (2017). Mechanism of partial agonism in AMPA-type glutamate receptors. Nat Commun 8, 14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, et al. (2009). Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323, 1313–1319. [DOI] [PubMed] [Google Scholar]
- Shi Y, Suh YH, Milstein AD, Isozaki K, Schmid SM, Roche KW, and Nicoll RA (2010). Functional comparison of the effects of TARPs and cornichons on AMPA receptor trafficking and gating. Proc Natl Acad Sci U S A 107, 16315–16319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Chien CT, and Chang IF (2016). The Arabidopsis glutamate receptor-like gene GLR3.6 controls root development by repressing the Kip-related protein gene KRP4. J Exp Bot 67, 1853–1869. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI (2015). Structure and gating of tetrameric glutamate receptors. J Physiol 593, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, and Gouaux E (2009). X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapken D, Anschutz U, Liu LH, Huelsken T, Seebohm G, Becker D, and Hollmann M (2013). A plant homolog of animal glutamate receptors is an ion channel gated by multiple hydrophobic amino acids. Sci Signal 6, ra47. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, and Dingledine R (2010). Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62, 405–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, and Sobolevsky AI (2018). Structural Mechanisms of Gating in Ionotropic Glutamate Receptors. Biochemistry 57, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincill ED, Bieck AM, and Spalding EP (2012). Ca(2+) conduction by an amino acid-gated ion channel related to glutamate receptors. Plant Physiol 159, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincill ED, Clarin AE, Molenda JN, and Spalding EP (2013). Interacting glutamate receptor-like proteins in Phloem regulate lateral root initiation in Arabidopsis. Plant Cell 25, 1304–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, and Thornton JM (1995). LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Engineering, Design and Selection 8, 127–134. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP, and Sobolevsky AI (2004). Structure and gating of the glutamate receptor ion channel. Trends Neurosci 27, 321–328. [DOI] [PubMed] [Google Scholar]
- Wudick MM, Michard E, Oliveira Nunes C, and Feijo JA (2018a). Comparing Plant and Animal Glutamate Receptors: Common Traits but Different Fates? J Exp Bot, 10. doi: 10.1093/jxb/ery153 [DOI] [PubMed] [Google Scholar]
- Wudick MM, Portes MT, Michard E, Rosas-Santiago P, Lizzio MA, Nunes CO, Campos C, Santa Cruz Damineli D, Carvalho JC, Lima PT, et al. (2018b). CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca(2+) homeostasis. Science 360, 533–536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the GLR3.2-S1S2Gly and GLR3.2-S1S2Met structures have been deposited to the PDB with the accession codes 6VEA and 6VE8, respectively. This study did not generate new code.