Abstract
PARP inhibitor monotherapy (olaparib) was recently FDA-approved for the treatment of BRCA1/2 mutant, HR (homologous recombination repair)-deficient pancreatic cancer. Most pancreatic cancers, however, are HR-proficient and thus resistant to PARP inhibitor monotherapy. We tested the hypothesis that combined therapy with radiation and ATR inhibitor (AZD6738) would extend the therapeutic indication of olaparib to HR-proficient pancreatic cancers. We show that olaparib combined with AZD6738 significantly reduced radiation survival relative to either agent alone, regardless of HR status. While catalytic inhibition of PARP with low concentrations of olaparib radiosensitized HR-deficient models, maximal sensitization in HR-proficient models required concentrations of olaparib that induce formation of PARP1-DNA complexes. Furthermore, CRISPR-Cas9-mediated PARP1 deletion failed to recapitulate the effects of olaparib on radiosensitivity and negated the combinatorial efficacy of olaparib and AZD6738 on radiosensitization, suggesting that PARP1-DNA complexes, rather than PARP catalytic inhibition, were responsible for radiosensitization. Mechanistically, therapeutic concentrations of olaparib in combination with radiation and AZD6738 increased DNA double strand breaks. DNA fiber combing revealed that high concentrations of olaparib did not stall replication forks but instead accelerated replication fork progression in association with an ATR-mediated replication stress response that was antagonized by AZD6738. Finally, in HR-proficient tumor xenografts, the combination of olaparib, radiation and AZD6738 significantly delayed tumor growth compared to all other treatments. These findings suggest that PARP1-DNA complexes are required for the therapeutic activity of olaparib combined with radiation and ATR inhibitor in HR-proficient pancreatic cancer and support the clinical development of this combination for tumors intrinsically resistant to PARP inhibitors.
INTRODUCTION
Pancreatic cancer is notoriously resistant to most cytotoxic chemotherapies, targeted therapies and immunotherapy (1,2). Recently, however, the POLO (Pancreas Cancer Olaparib Ongoing) trial demonstrated the activity of the PARP (polyADP-ribose polymerase) inhibitor olaparib in germline BRCA1/2 mutant pancreatic cancer patients leading to its FDA approval (3). While the findings of this trial are a major advancement in therapy for these patients, BRCA1/2 mutations or HR (homologous recombination repair) deficiencies only occur in about 10% of all pancreatic cancers (4). Combination strategies, however, may extend the therapeutic indications of PARP inhibitors to patients with BRCA1/2 wild-type, HR-proficient tumors.
The combinatorial efficacy of PARP inhibitors with radiation is well-established in HR-proficient cancers (5–7). Early studies demonstrated that this synergy between radiation and PARP inhibition requires DNA replication (7). In addition to radiation, agents that target the DDR (DNA damage response), in particular those which target replication fork progression, stability and/or restart, have been shown to overcome PARP inhibitor resistance and improve therapeutic efficacy (8–11). Given that both radiation and agents that target the DDR (such as CHK1 or WEE1 inhibitors) interact with olaparib by exacerbating replication stress and DNA damage, subsequent studies assessed olaparib-radiation therapy in combination with DDR inhibitors and found these therapies are highly efficacious in HR-proficient pancreatic cancer (12,13). Furthermore, mechanistic studies confirmed the importance of replication stress for sensitization to combined olaparib-radiation therapy by DDR inhibition (e.g., WEE1)(14). Therefore, we hypothesized that more direct targeting of DNA replication fork initiation and stability with an ATR inhibitor would further improve the efficacy of olaparib-radiation therapy.
The primary therapeutic target of olaparib is PARP1, a DDR protein that promotes the repair of both DNA single strand (SSB) and double strand breaks (DSB). After binding to sites of DNA damage, PARP1 acts primarily through the poly(ADP-ribosyl)ation (PARylation) of several proteins, including PARP1 itself. This PARylation signals the subsequent recruitment of DDR proteins such as XRCC1 to the damage site and facilities release of the PARP1 protein from DNA (15). Although early preclinical work defined the role of PARP in base excision repair, more recent studies have focused on the ability of PARP1 to regulate both DNA replication-associated repair, including stabilization and restart of stalled DNA replication forks (16–18), as well as DNA replication fork speed (19,20).
Although the replication-associated stress that results from PARP inhibition may determine the combinatorial efficacy of PARP inhibitors with a variety of DNA damaging agents, the cytotoxic activity of PARP inhibitor monotherapy has been attributed to a distinct process termed PARP trapping (21). PARP trapping is hypothesized to result from both inhibition of PARP catalytic activity, and thus the autoPARylation required for dissociation of PARP from DNA, as well as drug-induced allosteric changes in the PARP1 protein (22–24). We and others have reported that persistent PARP1-DNA binding, or trapping, is a key mechanism of radiosensitization by PARP inhibitors (5,14). Furthermore, it has been posited that trapped PARP1 interferes with DNA replication, causing fork stalling, which, if left unresolved due to PARP catalytic inhibition or impaired HR, ultimately leads to replication fork collapse and lethal DSBs (5,14,25). While more recent work has called this specific hypothesis into question (19), there is strong evidence that DNA replication is mechanistically important for olaparib-mediated radiosensitization (26,27).
The importance of replication and replication stress to the efficacy of DDR inhibitors such as AZD1775 (adavosertib) in combination with PARP inhibitors and radiation (14,28), suggests that ATR (Ataxia telangiectasia and Rad3-related) would also be a promising target for further improvement of therapeutic efficacy. ATR is a central mediator of the DNA replication stress response that is activated by Replication Protein A (RPA)-coated single stranded DNA at stalled replication forks (29,30). In cooperation with ATRIP and CHK1, ATR promotes the stabilization and restart of these stalled forks and pauses cell cycle progression to allow for the completion of DNA repair. Additionally, ATR regulates and maintains the timely firing of DNA replication origins (31,32). The ability of ATR inhibitors to overcome either acquired (11) or innate (33) resistance to PARP inhibitors further supports the hypothesis that ATR inhibitors sensitize HR-proficient cancers to olaparib-radiation combination therapies (10,11,33).
In this study we set out to extend the efficacy of PARP inhibitors to otherwise resistant HR-proficient pancreatic cancers through their combination with radiation and ATR inhibitor. We investigated the efficacy of olaparib combined with radiation and AZD6738 (ceralasertib)(34) in both HR-proficient and deficient pancreatic cancer cells as well as the role of PARP1-DNA binding as a mechanism of therapeutic activity. Given the importance of PARP and ATR in coordinating the cellular response to replication stress, we then used DNA fiber combing and a neutral comet assay to investigate the contributions of replication stress and DSBs to the efficacy of this combination therapy. Finally, we confirmed the tolerability and therapeutic efficacy of combined olaparib with radiation and AZD6738 in animal tumor models. Our results show that PARP1 protein and its association with DNA in response to trapping concentrations of olaparib are required for the therapeutic activity of olaparib combined with radiation and ATR inhibitor in HR-proficient pancreatic cancer and support the clinical development of this combination as a strategy to overcome the intrinsic resistance of HR-proficient pancreatic cancer to PARP inhibitor monotherapy
MATERIALS AND METHODS
Cell Culture and drug solutions
MiaPaCa2 and Panc1 cells were obtained from and authenticated by the American Type Culture Collection. Capan1.NEO, a clonal cell line expressing the neomycin resistance gene, was a gift from S. Powell (Memorial Sloan Kettering Cancer Center, New York, NY) (35). Cells were grown in either DMEM (MiaPaCa2 and Panc1; Invitrogen), or IMDM medium (Capan1.NEO; Invitrogen) supplemented with 10% fetal bovine serum (Premium Select; Atlanta Biologicals). These cell lines have been previously characterized as HR proficient (MiaPaCa2, Panc1) and HR deficient (Capan1)(35–37). To establish a conditional HR deficient model system, MiaPaCa2 cells were stably transduced with the Tet-pLKO-puro vector containing shRNA targeting RAD51 (sense 5’-CCGGAAGCTGAAGCTATGTTCGCCACTCGAGTGGCGAACATAGCTTCAGCTTTTTTTG; antisense 5’-AATTCAAAAAAAGCTGAAGCTATGTTC GCCACTCGAGTGGCGAACATAGCTTCAGCTT). Tet-pLKO-puro was a gift from Dmitri Wiederschain (Addgene plasmid 21915) (38). Stable clones were selected with puromycin (1 μg/ml; Sigma) and screened for RAD51 depletion following doxycycline treatment (Suppl. Fig. 1B). To generate stable PARP1-knockout clones (PARP1KO), MiaPaCa2 cells were transfected with a PARP1 CRISPR/Cas9 KO Plasmid (sc-419018, Santa Cruz) using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) per manufacturer’s protocol. Stable clones were selected with puromycin and isolated by limited dilution in 96-well plates. Wild-type (wt) PARP1 complemented clones (PARP1KO + wtPARP1) were generated by transfecting two different PARP1KO clones with a pcDNA4/V5-HisA plasmid expressing full-length wtPARP1 (1–1014) (39). Stable clones were selected with zeocin (100 μg/mL; Thermo Fisher) and screened by immunoblotting for expression of PARP1 protein and PAR. For in vitro experiments, AZD6738 (34) and olaparib (AstraZeneca) were each dissolved in dimethyl sulfoxide (Sigma) and stored in aliquots at −20°C. For in vivo studies, AZD6738 and olaparib were dissolved in DMSO (final concentration 10%) and then further diluted in 40% propylene glycol (Sigma) or 10% 2-hydroxypropyl-β-cyclodextrin (Cayman Chemical), respectively, and stored at room temperature for up to 3 days.
Clonogenic survival assays
Cells treated with drugs and/or radiation were processed for clonogenic survival as previously described (12,40). Unless otherwise indicated, AZD6738 and olaparib were given for 25 hours, beginning 1 hour prior to radiation. Radiation survival curves were normalized for drug toxicity and the radiation enhancement ratio (RER) was calculated as the ratio of the mean inactivation dose under control conditions divided by the mean inactivation dose after drug exposure (41). A value significantly greater than 1 indicates radiosensitization. Cytotoxicity in the absence of radiation treatment was calculated by normalizing the plating efficiencies of drug treated-cells to non-drug treated cells.
Immunoblotting
Whole cell lysates were prepared in ice cold RIPA buffer (1M NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 7.4) supplemented with both PhosSTOP phosphatase inhibitor and Complete protease inhibitor cocktails (Roche) (42). To assess PARP1 bound to chromatin, cells were fractionated using a series of salt stringency buffers as previously described (43). Briefly, nuclei from approximately 3×106 irradiated and drug-treated cells were isolated by gentle lysis in 100 μL ice-cold hypotonic buffer (50 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 0.1% Triton X-100, protease inhibitors), followed by slow-speed centrifugation (1,300 × g at 4°C for 4 minutes). Washed nuclei were then lysed in ice-cold buffer B (50 mM HEPES, pH 7.9, 100 mM KCl, 2.5 mM MgCl2, 0.05% Triton X-100, protease inhibitors) and insoluble chromatin was collected by centrifugation (15,000 × g at 4°C for 10 minutes) and washed once in buffer C (50 mM HEPES, pH 7.9, 250 mM KCl, 2.5 mM MgCl2, 0.05% Triton X-100, protease inhibitors) and once in buffer D (50 mM HEPES, pH 7.9, 500 mM KCl, 2.5 mM MgCl2, 0.1% Triton X-100, protease inhibitors). The chromatin pellet was then resuspended with Buffer B supplemented with 5 mM CaCl2 and incubated at 37°C for 10 minutes with 3 units of micrococcal nuclease, centrifuged, and processed for Western blot analysis (44).
Antibodies
The following antibodies were used: rabbit polyclonal anti-PARP1 (#9542), mouse monoclonal anti-Histone H3 (96C10; #3638) and rabbit monoclonal anti-GAPDH (14C10; #2118) from Cell Signaling Technology; mouse monoclonal anti-PAR (10H; ab14459) from Abcam. Densitometric analyses of immunoblots were carried out using Image J software (NIH).
Neutral comet assay
Cells were treated with AZD6738 and/or olaparib for 1 hour prior to and 24 hours post-8 Gy radiation. Neutral comet assay was performed according to the manufacturer’s protocol (Trevigen, USA). Briefly, cells were scraped, mixed 1:10 with 1% molten LMAgarose, pipetted onto CometSlides and submerged in neutral lysis buffer overnight at 4˚C. Slides were rinsed 3 times with TBE (90 mM Tris base, 90 mM boric acid, 2 mM EDTA, pH 8.0) and subjected to neutral electrophoresis for 35 minutes at 25V. Slides were then incubated in 2.5 μg/ml propidium iodide for 20 minutes, rinsed in 70% ethanol and allowed to dry overnight. Images were acquired using an Olympus ix73 inverted microscope with a 10x objective. The Olive tail moment (OTM) from at least 75 cells for each experimental condition was determined using Comet Assay IV software (Instem). Individual OTM measurements were first normalized to the mean OTM value from an internal control for each independent experiment (cells collected immediately following irradiation with 8 Gy on ice) and then to the mean normalized OTM for the radiation alone sample.
DNA fiber spreading
MiaPaCa2 cells were pulse-labeled with 25 μM 5-iododeoxyuridine (IdU) for 30 minutes, followed by two gentle washes with pre-warmed PBS and a second pulse with 250 μM 5-chloro-2’-deoxyuridine (CldU) for 30 minutes. Labelled cells were collected and fibers spread on silane-coated slides (Lab Scientific 7801B) as previously described (45) and consistent with the methodology with modifications noted below. Two slides were spread and stained for each experimental condition. IdU was detected with a mouse anti-bromodeoxyuridine (BrdU) antibody (Becton Dickinson, 347580) and CldU with a rat anti-BrdU antibody (BIORAD, OBT0030G). Secondary antibodies were Alexa Fluor 594 anti-mouse (Invitrogen, A11062) used at a 1:1000 dilution and Alexa Fluor 488 anti-rat (Invitrogen, A21470) used at a 1:200 dilution. Images of well-spread fibers were acquired using an Olympus ix73 inverted microscope with a 60x objective. For each experimental condition, double-labelled replication forks from 3–8 fields per slide were analyzed manually using ImageJ software (NIH), consistent with previously described methodology (46). Fork measurements from two independent experiments were pooled and the total number of DNA fibers from both experiments is presented.
Flow cytometry
Cells were trypsinized, washed with ice-cold PBS, and fixed at a concentration of 2 × 106 cells/mL in ice-cold 70% ethanol. For γH2AX analysis, samples were incubated with a mouse anti-γH2AX-specific antibody (clone JBW301; Millipore) overnight at 4°C followed by incubation with a FITC-conjugated secondary antibody (Sigma) as previously described (47). Samples were stained with propidium iodide to measure total DNA content and analyzed on a FACScan flow cytometer (Becton Dickinson) with FlowJo software (Tree Star). γH2AX positivity was quantified by setting a gate on the control, untreated sample to define a region of positive staining for γH2AX of 5–10%. This gate was then overlaid on the drug/radiation-treated samples.
Irradiation
Irradiations were performed using a Philips RT250 (Kimtron Medical) at a dose rate of ~2 Gy/min at the University of Michigan Comprehensive Rogel Cancer Center Experimental Irradiation Core. Dosimetry was performed using an ionization chamber connected to an electrometer system that is directly traceable to a National Institute of Standards and Technology calibration. For tumor irradiation, animals were anesthetized with isoflurane and positioned such that the apex of each flank tumor was at the center of a 2.4 cm aperture in the secondary collimator, with the rest of the mouse shielded from radiation.
Tumor growth studies
Animals were handled according to a protocol approved by the University of Michigan Committee for Use and Care of animals. MiaPaCa-2 cells (5×106) were suspended in a 1:1 mixture of DMEM+10%FBS:Matrigel (BD Biosciences) and injected subcutaneously, bilaterally into the flanks of 3–5 week old, male, athymic nude mice (Envigo). Treatment was initiated when the average tumor volume reached 100 mm3 and consisted of AZD6738 (25 mg/kg) and/or olaparib (50 mg/kg), once daily, 1 hour pre-radiation (1.8 Gy/fraction) Monday–Friday for 1 cycle. AZD6738 and olaparib were administered via oral gavage. Tumor size was measured two times per week. Tumor volume (TV) was calculated according to the equation: TV = π/6 (ab2), where a and b are the longer and shorter dimensions of the tumor, respectively. Measurements were made until day 80 or until the tumor volume increased by approximately a factor of 4.
Statistics
Statistically significant differences for the clonogenic survival and flow cytometry assays were determined by one-way ANOVA with the Tukey post-comparison test in GraphPad PRISM version 8.0 (GraphPad software). Comet assay and fiber combing data were also analyzed in PRISM with the Kruskal-Wallis one-way ANOVA. For tumor growth experiments, the time required for tumor volume tripling was determined for each xenograft by identifying the earliest day on which it was at least three times as large as on the first day of treatment. The Kaplan-Meier method was used to analyze the tripling times and the log-rank test was used to compare any two treatment groups in PRISM.
RESULTS
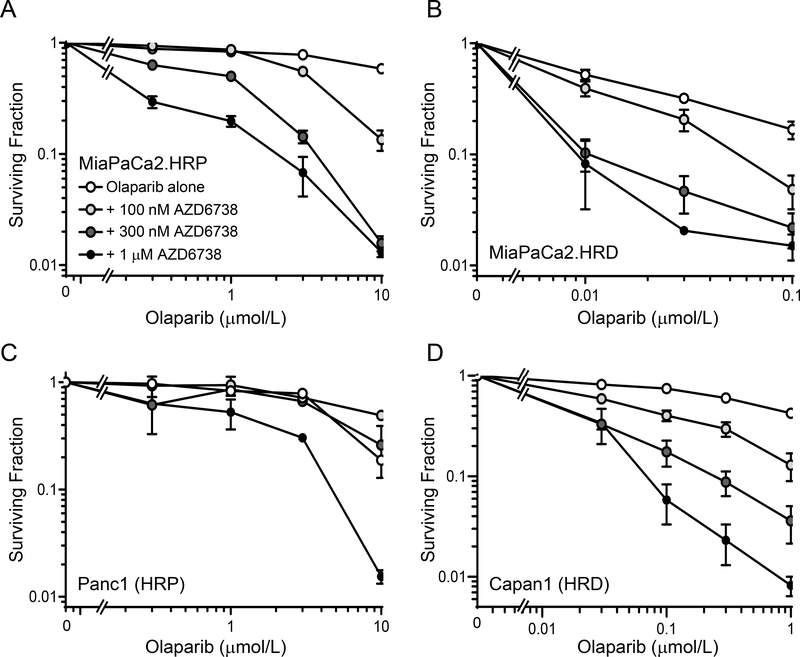
Before assessing the efficacy PARP inhibitors in combination with radiation and ATR inhibitor, we first determined the cytotoxicity of olaparib and AZD6738 in the absence of radiation. We hypothesized that this combination would be preferentially cytotoxic to HR-deficient cells due to the synthetic lethality between HR defects and PARP inhibition (48,49). Since silencing of RAD51 leads to functional inhibition of HR, we constructed a doxycycline-inducible shRAD51MiaPaCa2 cell line to test this hypothesis (Suppl. Fig. S1). As expected, RAD51-depleted, HR-deficient MiaPaCa2 cells (MiaPaCa2.HRD) were sensitive to olaparib-mediated loss of clonogenicity compared to HR-proficient MiaPaCa2 cells (MiaPaCa2.HRP) with IC50 values of 13.2 ± 0.04 nM and > 10 μM, respectively (Fig. 1A, B). We found however, that both HR-proficient and -deficient models were sensitized to olaparib by AZD6738, though much higher, albeit clinically relevant (50) concentrations of olaparib were required to achieve sensitization in HR-proficient cells. These findings extended to other HR-proficient (Panc1 cells) and -deficient (BRCA2 mutant; Capan1) pancreatic cancer cell lines (Fig. 1C, D) and support the hypothesis that ATR inhibition can overcome PARP inhibitor resistance, regardless of HR status (10).
Figure 1. Clonogenic survival of HR proficient and HR deficient pancreatic cancer cells treated with olaparib and AZD6738.
Cells treated with varying concentrations of olaparib and AZD6738 for 72 hours were trypsinized and assayed for drug-induced loss of clonogenicity. Data are the mean clonogenic survival ± SE of n, 2 (Panc1 cells) or 4 independent experiments for each drug combination and are normalized to the corresponding non-olaparib plating efficiency, either control, AZD6738 alone or doxycycline ± AZD6738.
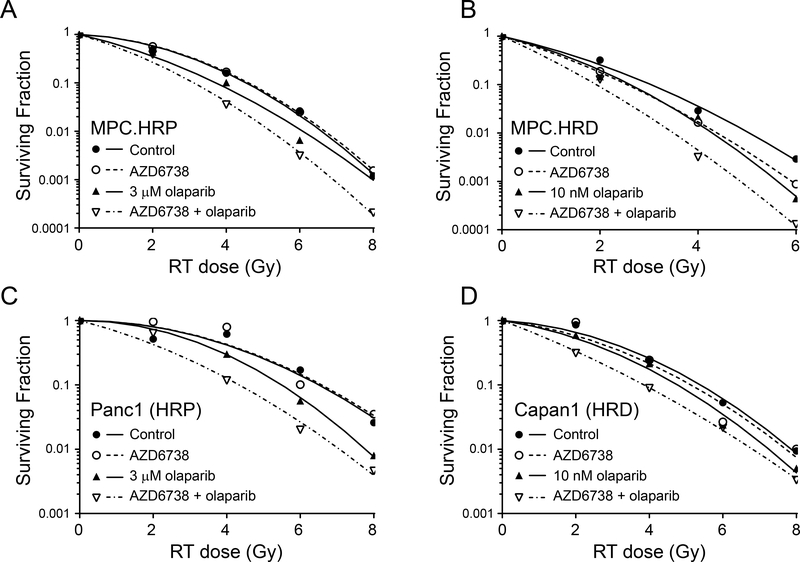
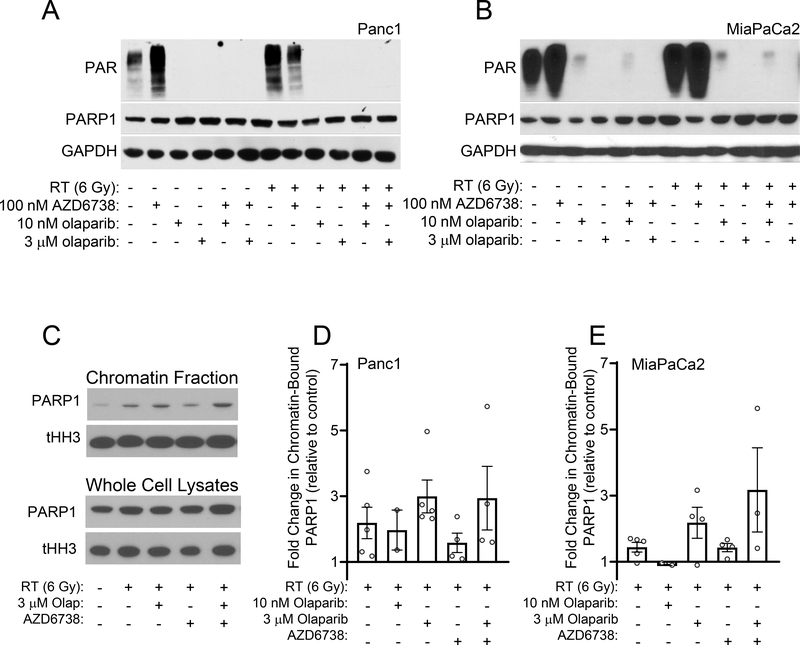
Using concentrations of olaparib chosen to inhibit PARP catalytic activity or induce PARP trapping (10 nM or 3 μM, respectively) (25), we next assessed the combinatorial efficacy of olaparib with radiation and AZD6738 in HR-deficient and HR-proficient cells. Although 10 nM olaparib in combination with AZD6738 was sufficient to radiosensitize HR-deficient cells, it had no effect on the radiosensitivity of the HR-proficient cell lines (Suppl. Fig. S2, Table 1A). In those cells, radiosensitization required the higher concentrations of olaparib, either alone or in combination AZD6738 (Fig. 2, Table 1A). Furthermore, the combined efficacy of 3 μM olaparib with AZD6738 in HR-proficient cells was significantly greater than the radiosensitizing activity of either drug alone. These data demonstrate the marked differences in the olaparib concentration required for radiosensitization in HR-proficient and HR-deficient cells, especially in the context of combined therapy with ATR inhibitor. Furthermore, these data suggested that while catalytic inhibition of PARP1 with low concentrations of olaparib (in combination with AZD6738) was sufficient to radiosensitize HR-deficient pancreatic cancer cells, radiosensitization in the HR-proficient cell lines required higher concentrations of inhibitor that would cause PARP trapping at sites of radiation-induced DNA damage. To further test the hypothesis that PARP trapping is required for the combinatorial efficacy of olaparib with radiation and AZD6738 in HR-proficient pancreatic cancer cells, we first compared the abilities of low (10 nM) and high (3 μM) concentrations of olaparib to inhibit PARP activity. In both Panc1 and MiaPaCa2 cells, both concentrations of olaparib, alone or in combination with AZD6738, effectively inhibited PAR formation, even in the presence of radiation-induced DNA damage (Fig. 3A, B). This result suggests comparable inhibition of PARP catalytic activity by either concentration of olaparib.
Table 1. Radiosensitization and cytotoxicity by combined ATR and PARP inhibition.
(A) MiaPaCa2.HRP, MiaPaCa2.HRD, Panc1, and Capan1 cells were treated with AZD6738 and/or olaparib beginning one hour pre- and continuing 24 hours post-RT (0–8 Gy). After drug treatment cells were processed for clonogenic survival. Data are the mean radiation enhancement ratio (RER) ± SE or the mean clonogenic survival ± SE for n, 3–6 independent experiments. Statistical significance (P < 0.05) is indicated versus controla, AZD6738b and olaparibc. (B) MiaPaCa2 Cas9 control, PARP1KO and PARP1KO + wtPARP1 cells were treated as described in (A). Data are the mean RER ± SE or the mean clonogenic survival ± SE for n, 3–4 independent experiments, or the mean ± SD for n, 2 independent experiments (PARP1KO + wtPARP1 clones). Statistical significance (P < 0.05) is indicated versus Cas9 controld, Cas9 AZD6738e and PARP1KO controlf. PARP1KO cells were not significantly radiosensitive relative to control Cas9 cells (RER 1.24 ± 0.1; P = 0.12). The mean RERs for the PARP1KO + wtPARP1 clones relative to the Cas9 control were 1.0 ± 0.18 and 0.96 ± 0.17 for clones 1 and 2, respectively.
| Condition | RER | Cytotoxicity | |
|---|---|---|---|
| A | MiaPaCa2 - RAD51 shRNA | ||
| HRP (no dox) | 1.0 | 1.0 | |
| AZD6738 (100 nmol/L) | 1.14 ± 0.05 | 0.93 ± 0.07 | |
| Olaparib (10 nmol/L) | 1.11 ± 0.04 | 0.88 ± 0.04 | |
| Olaparib (3 μmol/L) | 1.43 ± 0.08a | 0.80 ± 0.04 | |
| AZD6738 + olaparib (10 nmol/L) | 1.40 ± 0.10a,b | 0.84 ± 0.07 | |
| AZD6738 + olaparib (3 μmol/L) | 1.83 ± 0.14a,b,c | 0.66 ± 0.03 | |
| HRD (+ dox) | 1.0 | 1.0 | |
| AZD6738 (100 nmol/L) | 1.16 ± 0.06 | 0.98 ± 0.04 | |
| Olaparib (10 nmol/L) | 1.17 ± 0.03 | 0.80 ± 0.05 | |
| AZD6738 + olaparib | 1.43 ± 0.11a | 0.63 ± 0.02 | |
| Panc-1 (HRP) | |||
| AZD6738 (100 nmol/L) | 1.02 ± 0.03 | 0.96 ± 0.08 | |
| Olaparib (10 nmol/L) | 1.03 ± 0.07 | 0.83 ± 0.09 | |
| Olaparib (3 μmol/L) | 1.29 ± 0.06a | 0.80 ± 0.12 | |
| AZD6738 + olaparib (10 nmol/L) | 1.06 ± 0.04 | 0.87 ± 0.03 | |
| AZD6738 + olaparib (3 μmol/L) | 1.67 ± 0.07a,b,c | 0.94 ± 0.08 | |
| Capan-1 (HRD) | |||
| AZD6738 (100 nmol/L) | 1.14 ± 0.06 | 0.92 ± 0.04 | |
| Olaparib (10 nmol/L) | 1.26 ± 0.06 | 0.92 ± 0.12 | |
| AZD6738 + olaparib | 1.47 ± 0.13a | 0.87 ± 0.12 | |
| B | MiaPaCa2 – Cas 9 | 1.0 | 1.0 |
| AZD6738 (100 nmol/L) | 1.24 ± 0.06 | 0.88 ± 0.03 | |
| Olaparib (3 μmol/L) | 1.63 ± 0.10d | 0.92 ± 0.09 | |
| AZD6738 + olaparib | 2.04 ± 0.06d,e | 0.56 ± 0.07 | |
| MiaPaCa2 – PARP1KO | 1.0 | 1.0 | |
| AZD6738 (100 nmol/L) | 1.18 ± 0.04 | 1.03± 0.06 | |
| Olaparib (3 μmol/L) | 1.07 ± 0.06 | 1.00 ± 0.08 | |
| AZD6738 + olaparib | 1.26 ± 0.07f | 1.00 ± 0.07 | |
| PARP1KO + wtPARP1 (clone #1) | 1.0 | 1.0 | |
| AZD6738 (100 nmol/L) | 1.19 ± 0.02 | 1.01 ± 0.01 | |
| Olaparib (3 μmol/L) | 1.43 ± 0.01 | 0.98 ± 0.03 | |
| AZD6738 + olaparib | 1.48 ± 0.08 | 0.96 ± 0.05 | |
| PARP1KO + wtPARP1 (clone #2) | 1.0 | 1.0 | |
| AZD6738 (100 nmol/L) | 1.08 ± 0.02 | 0.99 ± 0.01 | |
| Olaparib (3 μmol/L) | 1.58 ± 0.08 | 0.85 ± 0.11 | |
| AZD6738 + olaparib | 1.61 ± 0.10 | 0.86 ± 0.02 | |
Figure 2. Radiosensitization by olaparib and AZD6738 in HR proficient and HR deficient pancreatic cancer cells.
Representative radiation survival curves for MiaPaCa2 shRAD51-expressing cells in the absence (A) or presence (B) of dox, Panc1 (C) or Capan1 (D) cells treated with 100 nM AZD6738 and/or olaparib beginning one hour pre- and continuing 24 hours post-radiation (RT). For mean radiation enhancement ratios (RER), survival data and statistical analyses, see Table 1A.
Figure 3. The effects of olaparib and AZD6738 on PARP catalytic inhibition and PARP1-DNA binding.
MiaPaCa2 and Panc1 cells were treated with either 10 nM or 3 μM olaparib and 100 nM AZD6738 beginning 1 hour pre- and continuing 24 hours post-RT (6 Gy). At the end of drug treatment, cells were analyzed by immunoblotting for the indicated proteins. Images from representative experiments are shown (A, B). Representative images of PARP1 in chromatin fractions or whole-cell lysates from Panc1 cells prepared 24 hours post-RT and analyzed by immunoblotting (C). Quantification of the fold change in chromatin-bound PARP1 relative to control, untreated cells. Data bars are the mean ± SE of n, 2–5 independent experiments. Individual data measurements are also shown (D, E).
We next evaluated the effects of olaparib alone or in combination with AZD6738 on PARP1-DNA binding in response to radiation-induced DNA damage. Relative to untreated control cells, we found that radiation alone caused a modest increase in chromatin associated PARP1, an effect which likely reflects the transient association of PARP1 with sites of DNA damage (Fig. 3C–E). Furthermore, 3uM olaparib, but not 10nM olaparib, either alone or in combination with AZD6738, increased radiation-induced PARP1-DNA binding with minimal change in the amount of total PARP1 in whole cell lysates, a finding that is consistent with prior studies demonstrating a lack of PARP1 trapping with lower (<100nM) concentrations of olaparib (25)(Fig. 3C–E, Suppl. Fig. S3). These data suggest that the therapeutic activity of olaparib in combination with radiation and/or AZD6738, is not only a function of PARP catalytic inhibition but is also associated with increased PARP1-DNA binding.
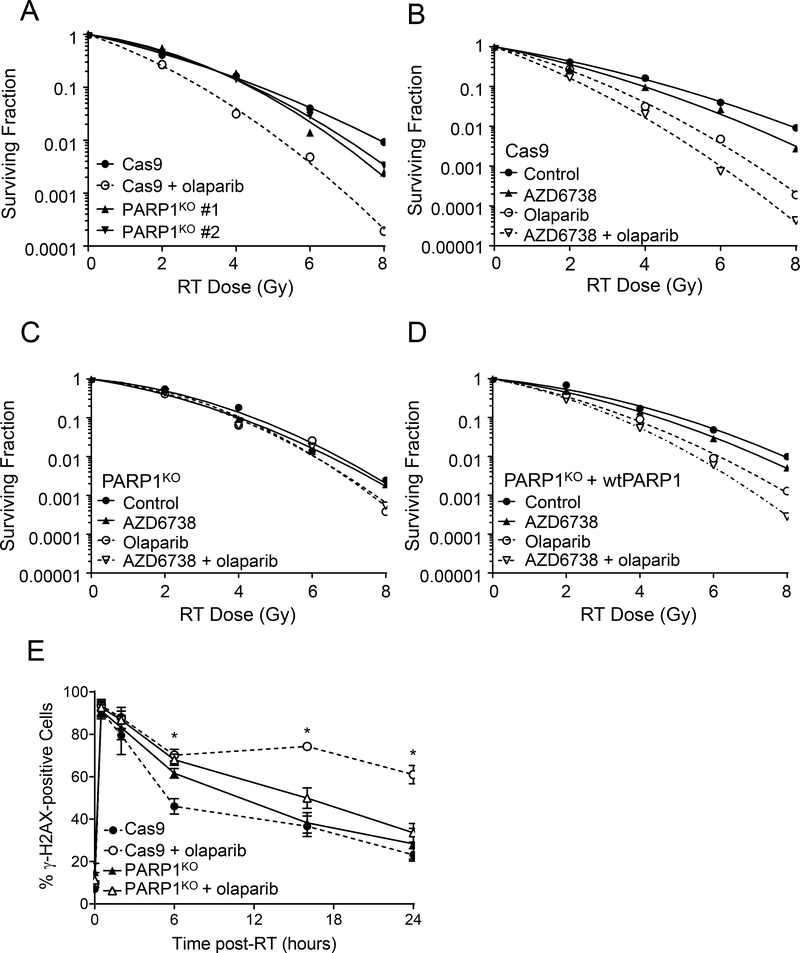
Much of the data in support of the PARP trapping model is derived from PARP1 knock-out studies which have demonstrated not only that deletion of PARP1 protein does not replicate the cytotoxic effects of pharmacological PARP inhibition, but additionally that PARP1 protein is required for the therapeutic efficacy of PARP inhibitors (25,51). In order to test the requirement for PARP1 protein for the activity of olaparib in combination with radiation and AZD6738, we used CRISPR-Cas9 to establish MiaPaCa2 PARP1 null cells (PARP1KO). As expected, PARP1KO cells displayed reduced levels of PAR comparable to that observed following treatment of Cas9 control cells with olaparib (Suppl. Fig. S4A). Furthermore, PARP1KO cells were resistant to the cytotoxic effects of the potent PARP trapping agent talazoparib (Suppl. Fig. S4B), and to the cytotoxicity of combined olaparib and AZD6738 (Suppl. Fig. S4C), sensitivities that were partially rescued by reintroduction of wild-type PARP1 (wtPARP1). To test the hypothesis that PARP trapping is required for the combinatorial efficacy of olaparib and radiation in HR proficient cells, we assessed the radiosensitivity of two independent MiaPaCa2 PARP1KO clones (#1 and #2) and found that PARP1 deletion did not replicate the sensitizing effects of 3 μM olaparib (Fig. 4A). This result is consistent with the lack of radiosensitization by 10 nM olaparib in parental MiaPaCa2 cells (Suppl. Fig. S2) and our prior study demonstrating minimal radiosensitization by PARP1 depletion with siRNA (14). Furthermore, while MiaPaCa2 Cas9 cells were sensitized to radiation by olaparib alone or in combination with AZD6738 (Fig. 4B; Table 1B), these effects were lost in PARP1KO cells (Fig. 4C) and partially restored by complementation with wtPARP1 (Fig. 4D). Taken together, these data demonstrate that PARP1 protein is required for the activity of olaparib in combination with radiation and AZD6738, and support the hypothesis that the combinatorial efficacy of PARP and ATR inhibitors with radiation is mediated by PARP1-DNA binding rather than PARP catalytic inhibition.
Figure 4. PARP1 protein is required for olaparib-mediated radiosensitization and DNA damage signaling.
Representative radiation survival curves for irradiated MiaPaCa2 Cas9 control cells (A, B), PARP1KO cells (A, C) or PARP1KO + wtPARP1 cells (D) treated with 3 μM olaparib and/or 100 nM AZD6738. For mean radiation enhancement ratios, survival data and statistical analysis, see Table 1B. To assess DNA damage signaling, irradiated cells treated ± olaparib were collected 0.5, 2, 6, 16, or 24 hours post-RT and assayed for γH2AX staining by flow cytometry (E). Data plotted are either the mean ± SD of n, 2 independent experiments (30 min post-RT) or the mean ± SE of n, 3–4 independent experiments. Statistical significance for Cas9 cells treated with radiation + olaparib versus radiation alone is indicated (*P <0.05, unpaired t test).
To begin to understand the mechanisms of, and the contribution of PARP1 protein to, the effects of olaparib in combination with radiation, we first assessed γH2AX, a marker of DSBs. Given that ATR is a direct kinase for H2AX (52) and that AZD6738 attenuates radiation-induced γH2AX signaling in parental MiaPaCa2 cells (Suppl. Fig. S5A), AZD6738 was not included in these experiments. Despite the lack of PARP catalytic activity in PARP1KO cells, PARP1 deletion had little effect on γH2AX induction or resolution following radiation (Fig. 4E), a result consistent with the inability of 10 nM olaparib to delay resolution of radiation-induced γH2AX signaling in parental MiaPaCa2 or Panc1 cells (Suppl. Fig. S5A, B). Furthermore, while 3 μM olaparib significantly increased the levels of γH2AX following radiation in Cas9 control cells, it did not affect radiation-induced γH2AX signaling in PARP1KO cells (Fig. 4E). These data suggest that the loss of PARP catalytic activity itself neither inhibits the repair of radiation-induced DSBs nor promotes the formation of additional DSBs as cells reenter S-phase and initiate DNA replication. Furthermore, these data support the hypothesis that the physical interaction of PARP1 protein with damaged DNA contributes to the olaparib-mediated increase in γH2AX signaling.
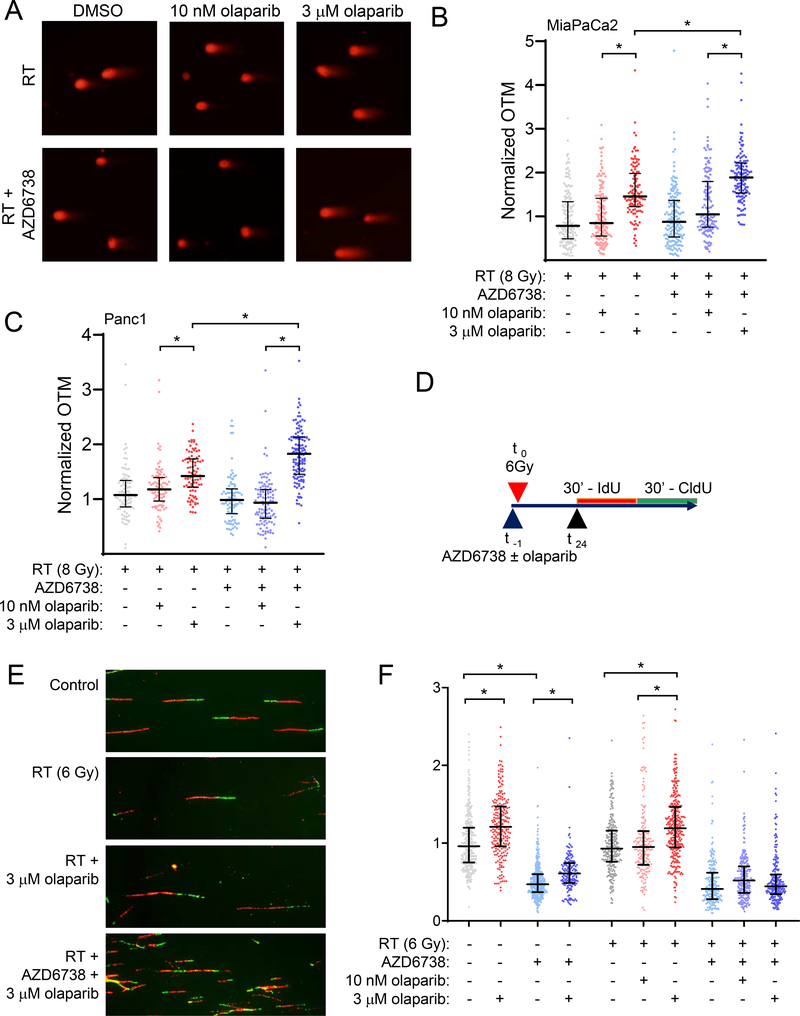
To more directly assess the effects of combined ATR and PARP inhibition on radiation-induced DNA damage, we next used a neutral comet assay to measure DSBs 24 hours post-radiation. In the absence of radiation, treatment with trapping concentrations of olaparib, alone or in combination with AZD6738, induced a small but significant level of DSBs relative to control (Suppl. Fig. S5C, D). Consistent with the γH2AX signaling patterns described above, 10 nM olaparib also had no effect on the resolution of radiation-induced DSBs while 3 μM olaparib significantly increased the extent of DSBs 24 hours post-radiation (Fig. 5A–C; Suppl. Fig. S5E). Furthermore, while AZD6738 in combination with 10 nM olaparib did not significantly affect repair of radiation-induced DNA damage, AZD6738 in combination with 3 μM olaparib caused a significant increase in DSBs. This damage may reflect persistent, unrepaired DSBs or possibly de novo DSBs generated when the DNA replication machinery encounters unrepaired, previously stalled forks (53). Taken together, these data demonstrate that the elevated DSBs mechanistically associated with therapeutic efficacy require concentrations of olaparib that cause persistent PARP1-DNA binding and are potentiated by an ATR inhibition.
Figure 5. Olaparib-mediated DNA damage and replication stress are concentration dependent.
MiaPaCa2 and Panc1 cells were treated with 100 nM AZD6738 and/or olaparib beginning one hour pre- and continuing 24 hours post-RT (8 Gy). At the end of drug treatment, DSBs were measured by neutral comet assay. Representative images of nuclei from MiaPaCa2 treated as indicated (A). Olive tail moment (OTM) measurements were normalized to an internal control of cells collected immediately post-RT. Error bars represent the median normalized OTM ± interquartile range from a single, representative experiment with individual measurements shown (B, C). Schedule for drug treatment and labeling of DNA fiber samples (D). Representative DNA fiber images from MiaPaCa2 cells treated as indicated (E). The effects of AZD6738 and olaparib on DNA replication rate (F). Replication fork speed was calculated for individual forks labelled with an IdU track (red) flanked by a single CldU track (green). Errors bars represent the median fork speed ± interquartile range of data pooled from 2 independent experiments. Scored forks: control = 354; 3 μM olaparib = 227; AZD6738 = 349; 3 μM olaparib + AZD6738 = 162; 6 Gy = 242; RT + 10 nM olaparib = 177; RT + 3 μM olaparib = 309; RT + AZD6738 = 201; RT + 10 nM olaparib + AZD6738 = 238; RT + 3 μM olaparib + AZD6738 = 163. Statistical significance between distributions was determined with the nonparametric Kruskal-Wallis test (P < 0.05) (B, C, F).
Based on our previous study which identified replication stress as a key factor in olaparib and AZD1775-mediated radiosensitization (14), and the well-established role of ATR in regulating origin firing and replication fork stability (31,54), we hypothesized that persistent PARP1-DNA binding creates an obstacle for active DNA replication forks that is further exacerbated by inhibition of ATR, ultimately resulting in fork stalling/collapse and lethal DSBs (25,33). To test this hypothesis, we used DNA fiber combing to directly visualize and measure DNA replication fork progression in cells treated with olaparib, radiation and AZD6738. In contrast to our initial hypothesis, combination treatment did not result in DNA replication fork stalling or collapse, which would be reflected in the accumulation of highly asymmetric replication forks (55)(Suppl. Fig. S6A, B). Instead, the higher concentrations of olaparib associated with PARP1-DNA binding significantly increased the rate of replication fork progression (Fig. 5E, F; Suppl. Fig. S6C). Furthermore, we found that canonical PARP1 trapping by treatment with temozolomide and talazoparib also failed to slow DNA replication rates (Suppl. Fig. S7). This olaparib-mediated increase in replication rate was associated with activation of an ATR-mediated replication stress response marked by increased ATR (T1989) and CHK1 (S345) phosphorylation (Suppl. Fig. S8) and γH2AX (Suppl. Fig. S5A, B). In contrast, ATR inhibition slowed DNA replication fork speed, a finding consistent with the role of ATR in suppressing aberrant origin firing to preserve replication machinery and maintain normal DNA replication rates (32,54,56). Furthermore, ATR inhibition negated the accelerating effects of olaparib on DNA replication fork rates, perhaps due to the effects of AZD6738 on origin firing limiting the capacity of the cell to accelerate replication fork progression (31). These data suggest the DNA damage response to olaparib-mediated replication stress is ATR-dependent, and, furthermore, that inhibition of ATR exacerbates this replication stress.
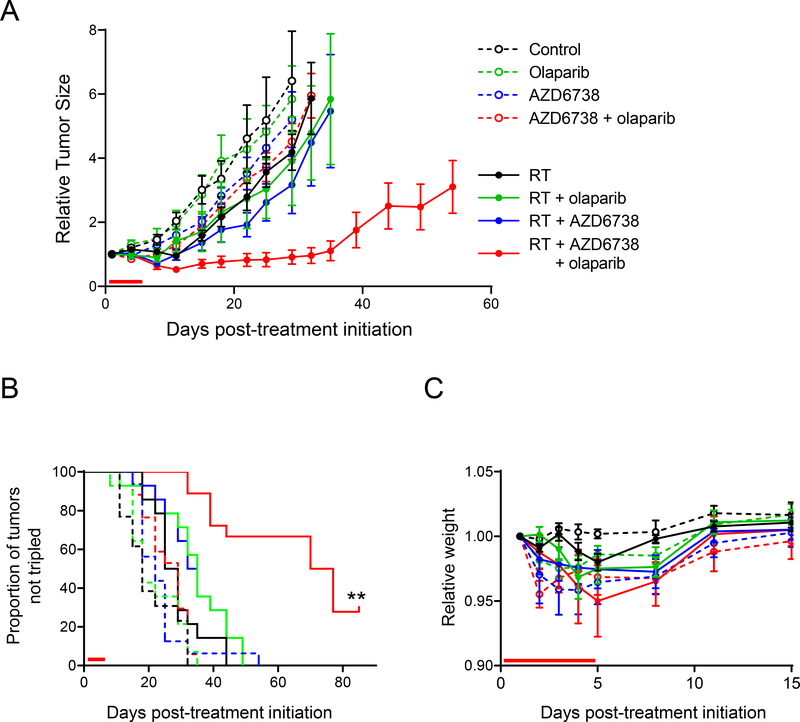
Based on the combined efficacy of PARP inhibitors with radiation and ATR inhibitor in vitro, we next tested the therapeutic efficacy and tolerability of combined olaparib, radiation and AZD6738 in animal tumor models. While radiation alone or in combination with olaparib or AZD6738 had a modest effect on tumor growth, the triple combination of olaparib, radiation and AZD6738 had a profound anti-tumor effect (Fig. 6A). Tumors treated with this combination displayed a significantly increased time to tumor volume tripling relative to all other groups including radiation combined with either single agent (Fig. 6B). Noteworthy, 28% of the tumors treated with the combination of olaparib, radiation and AZD6738 had complete responses that lasted for the duration of the study (80 days). This treatment regimen was well-tolerated (Fig. 6C). In summary, these data validate the further development of PARP inhibitors in combination with radiation and ATR inhibitor as a therapeutic strategy for overcoming the intrinsic resistance of HR-proficient pancreatic cancers to PARP inhibitor monotherapy
Figure 6. Radiosensitization of pancreatic tumor xenografts by AZD6738 and olaparib.
Athymic nude mice bearing bilateral, flank MiaPaCa2 xenografts were treated with AZD6738 (25 mg/kg) and/or olaparib (50 mg/kg) one hour pre-RT (1.8 Gy/fraction), daily Monday through Friday for one cycle as indicated by the red bars on the graphs above. Tumor volumes were normalized to the first day of treatment (day 1) and are plotted as the mean tumor volume ± SE of n, 13–18 tumors per treatment group (A). The Kaplan-Meier plot (B) illustrates the proportion of tumors not tripled in volume within the full 80-day monitoring period. Statistical significance (P < 0.001) is indicated versus all other treatment groups. Treatment caused no obvious systemic toxicity as assessed by weight loss (C).
DISCUSSION
In this study, we show that the intrinsic resistance of HR-proficient pancreatic cancer cells to PARP inhibitor monotherapy can be overcome by combination with radiation and ATR inhibitor. Using both pharmacological and genetic models, we further demonstrate that, while catalytic inhibition of PARP with low concentrations of olaparib is sufficient to radiosensitize HR-deficient models, radiosensitization in HR-proficient models requires both PARP1 protein and concentrations of olaparib that stabilize PARP1-DNA complexes. We also show that high concentrations of olaparib induce replication stress in the form of accelerated replication fork progression, and, in combination with radiation and ATR inhibitor, significantly increase DSBs in a PARP1 protein-dependent manner. While the effects of olaparib on replication fork progression are antagonized by AZD6738, olaparib-mediated DNA damage is amplified by ATR inhibition and correlates with radiosensitization. Finally, we show that combined treatment with olaparib, radiation and AZD6738 has a profound anti-tumor effect in vivo with minimal toxicity.
The combinatorial efficacy of radiation with a variety of different PARP inhibitors (talazoparib, olaparib, veliparib) is well-established in HR-proficient cancers where PARP inhibitor monotherapy has proven ineffective (57). Additionally, both intrinsic (33) and acquired PARP inhibitor resistance (11) may be overcome by targeting the ATR-mediated DDR. The therapeutic potential of PARP and ATR inhibitors with radiation is further supported by a study from Carruthers et al (28) which demonstrated that combined PARP and ATR inhibitors overcome the amplified DDR and intrinsic radioresistance of glioblastoma stem cells. While this study highlighted the potential of combined PARP and ATR inhibitors to enhance radiation induced DNA damage, the contributions of PARP trapping and its consequences on DNA replication and radiosensitization were not investigated.
Although the precise biochemical mechanism of PARP trapping is incompletely understood, it is hypothesized to result from both inhibition of PARP catalytic activity, and thus autoPARylation, as well as drug-induced allosteric changes in the PARP1 protein (22–24). Our data, however, demonstrate an increase in chromatin-associated PARP1 in the absence of olaparib, post-radiation (Fig. 3, Suppl. Fig. S7). Given that radiation alone does not inhibit PARP1 auto-PARylation, (Fig. 3), this specific interaction does not reflect PARP1 trapping. With this conclusion in mind, we cannot definitively state whether or not the chromatin-associated PARP1 in the presence of radiation plus olaparib is trapped PARP, only that there is increased PARP1 associated with chromatin under these conditions. It is possible that this interaction reflects enhanced chromatin associated PARP1 and not the bona fide PARP1 trapping that has been described in response to methyl methanesulfonate (MMS) or temozolomide-induced DNA lesions (25,43). Regardless, our studies suggest that the maximal therapeutic activity of olaparib in combination with radiation and ATR inhibitor requires the formation of these PARP1-DNA complexes.
Early studies on PARP inhibitor-radiation combination schedules identified DNA replication as a key factor in determining overall cytotoxicity (26). This finding suggested that the DSBs associated with therapeutic efficacy result from replication-associated events rather than directly from radiation-induced DNA damage. Furthermore, the kinetics of this model are consistent with our finding that radiation-induced PARP1-DNA complexes are a relatively late (24 hours post-radiation) event compared to the rapid recruitment of PARP to SSBs caused directly by radiation (58). Taken together, these observations support the hypothesis that replication is a key component of olaparib-mediated radiosensitization, suggesting a more nuanced view of how PARP is recruited to and retained on chromatin in response to ionizing radiation that may have clinical ramifications for the scheduling of these combination therapies.
In contrast to the posited inhibitory effect of PARP1-DNA binding on ongoing replication forks (5,14,25), we found that DNA replication rates accelerate in cells treated with concentrations of olaparib that cause PARP1-DNA binding, even in the presence of radiation-induced DNA damage. This finding is consistent with a previously published study on the replication stress caused by olaparib monotherapy (19) and supports their finding that PARP1-DNA complexes do not impede DNA replication. Of note, however, is that this replication stress is unlikely to result from p53-dependent p21 processes as MiaPaCa2 cells lack wt-p53 function. We propose that, combined with radiation, these competing stresses on replication (olaparib-mediated fork acceleration and ATR inhibitor-mediated suppression of replication fork progression), ultimately lead to unrepaired DSBs and reduced survival, as well as profound inhibition of tumor growth.
At the time of writing, eight active and recruiting trials are investigating the combination of olaparib and AZD6738, including one in pancreatic cancer. Many of these trials are founded on work by Kim et al (9) which demonstrated the preferential activity of combined ATR and PARP inhibition in BRCA1/2-mutant, HR-defective ovarian cancer. This strong emphasis on HR status is reflected in the patient stratification (NCT03330847) and inclusion criteria (NCT02576444) for subsequent trials investigating the combination of PARP and ATR inhibitors. The remarkable response we have seen in HR-proficient pancreatic tumors treated with olaparib in combination with radiation and ATR inhibitor however, suggests this combination strategy may be extended to resistant HR-proficient cancers. In addition, given that PARP and ATR inhibitors as well as radiation influence tumor immunity, their combination has the potential to enhance anti-tumor immune responses (59–61). We anticipate that the mechanisms highlighted in this study will help guide the choice of agents as well as their dose and schedule for future clinical trials combining PARP inhibitors with radiation and ATR inhibition.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge AstraZeneca, AZD6738 and olaparib teams for their support and thank Alan Lau and Mark O’Connor for their helpful comments and review of this manuscript.
Grant support: This work was funded by NIH Grants R01CA163895 (M.A.M.), R01CA240515 (M.A.M), P50CA130810 (T.S.L.), U01CA216449 (T.S.L.), R50CA251960 (L.A.P.), Cancer Center Support Grant P30CA46592, and a research grant from AstraZeneca (M.A.M.).
Disclosure of potential conflicts of interest: M, Morgan and T. Lawrence have received research funding (M.A.M.) and honoraria from AstraZeneca (M.A.M.; T.S.L.).
Abbreviations:
- ATR
Ataxia Telangiectasia and Rad3-related
- CldU
5-chloro-2’-deoxyuridine
- DDR
DNA Damage Response
- DSB
DNA double strand break
- HR
homologous recombination repair
- HRD
homologous recombination repair deficient
- HRP
homologous recombination repair proficient
- IdU 5
iododeoxyuridine
- OTM
Olive tail moment
- PAR
polyADP-ribose
- RER
radiation enhancement ratio
- RT
radiation therapy
REFERENCES
- 1.Diab M, Azmi A, Mohammad R, Philip PA. Pharmacotherapeutic strategies for treating pancreatic cancer: advances and challenges. Expert Opin Pharmacother 2019;20:535–46 [DOI] [PubMed] [Google Scholar]
- 2.Johnson BA 3rd, Yarchoan M, Lee V, Laheru DA, Jaffee EM. Strategies for Increasing Pancreatic Tumor Immunogenicity. Clin Cancer Res 2017;23:1656–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N Engl J Med 2019;381:317–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laird JH, Lok BH, Ma J, Bell A, de Stanchina E, Poirier JT, et al. Talazoparib Is a Potent Radiosensitizer in Small Cell Lung Cancer Cell Lines and Xenografts. Clin Cancer Res 2018;24:5143–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senra JM, Telfer BA, Cherry KE, McCrudden CM, Hirst DG, O’Connor MJ, et al. Inhibition of PARP-1 by olaparib (AZD2281) increases the radiosensitivity of a lung tumor xenograft. Mol Cancer Ther 2011;10:1949–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dungey FA, Caldecott KW, Chalmers AJ. Enhanced radiosensitization of human glioma cells by combining inhibition of poly(ADP-ribose) polymerase with inhibition of heat shock protein 90. Mol Cancer Ther 2009;8:2243–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lallo A, Frese KK, Morrow CJ, Sloane R, Gulati S, Schenk MW, et al. The Combination of the PARP Inhibitor Olaparib and the WEE1 Inhibitor AZD1775 as a New Therapeutic Option for Small Cell Lung Cancer. Clin Cancer Res 2018;24:5153–64 [DOI] [PubMed] [Google Scholar]
- 9.Kim H, George E, Ragland RL, Rafail S, Zhang R, Krepler C, et al. Targeting the ATR/CHK1 axis with PARP inhibition results in tumor regression in BRCA mutant ovarian cancer models. Clin Cancer Res 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Andrea AD. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair (Amst) 2018;71:172–6 [DOI] [PubMed] [Google Scholar]
- 11.Yazinski SA, Comaills V, Buisson R, Genois MM, Nguyen HD, Ho CK, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev 2017;31:318–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karnak D, Engelke CG, Parsels LA, Kausar T, Wei D, Robertson JR, et al. Combined Inhibition of Wee1 and PARP1/2 for Radiosensitization in Pancreatic Cancer. Clin Cancer Res 2014;20:5085–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vance S, Liu E, Zhao L, Parsels JD, Parsels LA, Brown JL, et al. Selective radiosensitization of p53 mutant pancreatic cancer cells by combined inhibition of Chk1 and PARP1. Cell Cycle 2011;10:4321–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsels LA, Karnak D, Parsels JD, Zhang Q, Velez-Padilla J, Reichert ZR, et al. PARP1 Trapping and DNA Replication Stress Enhance Radiosensitization with Combined WEE1 and PARP Inhibitors. Mol Cancer Res 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javle M, Curtin NJ. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer 2011;105:1114–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 2009;28:2601–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res 2012;72:2814–21 [DOI] [PubMed] [Google Scholar]
- 18.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 2010;37:492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maya-Mendoza A, Moudry P, Merchut-Maya JM, Lee M, Strauss R, Bartek J. High speed of fork progression induces DNA replication stress and genomic instability. Nature 2018;559:279–84 [DOI] [PubMed] [Google Scholar]
- 20.Merchut-Maya JM, Bartek J, Maya-Mendoza A. Regulation of replication fork speed: Mechanisms and impact on genomic stability. DNA Repair (Amst) 2019:102654. [DOI] [PubMed] [Google Scholar]
- 21.Murai J, Pommier Y. PARP Trapping Beyond Homologous Recombination and Platinum Sensitivity in Cancers. Annual Review of Cancer Biology 2019;3:131–50 [Google Scholar]
- 22.Hopkins TA, Shi Y, Rodriguez LE, Solomon LR, Donawho CK, DiGiammarino EL, et al. Mechanistic Dissection of PARP1 Trapping and the Impact on In Vivo Tolerability and Efficacy of PARP Inhibitors. Mol Cancer Res 2015;13:1465–77 [DOI] [PubMed] [Google Scholar]
- 23.Langelier MF, Zandarashvili L, Aguiar PM, Black BE, Pascal JM. NAD(+) analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat Commun 2018;9:844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zandarashvili L, Langelier MF, Velagapudi UK, Hancock MA, Steffen JD, Billur R, et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science 2020;368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res 2012;72:5588–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dungey FA, Loser DA, Chalmers AJ. Replication-dependent radiosensitization of human glioma cells by inhibition of poly(ADP-Ribose) polymerase: mechanisms and therapeutic potential. Int J Radiat Oncol Biol Phys 2008;72:1188–97 [DOI] [PubMed] [Google Scholar]
- 27.Loser DA, Shibata A, Shibata AK, Woodbine LJ, Jeggo PA, Chalmers AJ. Sensitization to radiation and alkylating agents by inhibitors of poly(ADP-ribose) polymerase is enhanced in cells deficient in DNA double-strand break repair. Mol Cancer Ther 2010;9:1775–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carruthers RD, Ahmed SU, Ramachandran S, Strathdee K, Kurian KM, Hedley A, et al. Replication Stress Drives Constitutive Activation of the DNA Damage Response and Radioresistance in Glioblastoma Stem-like Cells. Cancer Res 2018;78:5060–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnitz LM, Zou L. Molecular Pathways: Targeting ATR in Cancer Therapy. Clin Cancer Res 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol 2014;16:2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 2013;155:1088–103 [DOI] [PubMed] [Google Scholar]
- 32.Sorensen CS, Syljuasen RG. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res 2012;40:477–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murai J, Feng Y, Yu GK, Ru Y, Tang SW, Shen Y, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 2016;7:76534–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foote KM, Nissink JWM, McGuire T, Turner P, Guichard S, Yates JWT, et al. Discovery and Characterization of AZD6738, a Potent Inhibitor of Ataxia Telangiectasia Mutated and Rad3 Related (ATR) Kinase with Application as an Anticancer Agent. J Med Chem 2018;61:9889–907 [DOI] [PubMed] [Google Scholar]
- 35.Xia F, Taghian DG, DeFrank JS, Zeng ZC, Willers H, Iliakis G, et al. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci U S A 2001;98:8644–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, Hassan MC, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res 2010;70:4972–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsels LA, Morgan MA, Tanska DM, Parsels JD, Palmer BD, Booth RJ, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther 2009;8:45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiederschain D, Wee S, Chen L, Loo A, Yang G, Huang A, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 2009;8:498–504 [DOI] [PubMed] [Google Scholar]
- 39.Steffen JD, Tholey RM, Langelier MF, Planck JL, Schiewer MJ, Lal S, et al. Targeting PARP-1 allosteric regulation offers therapeutic potential against cancer. Cancer Res 2014;74:31–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence TS. Ouabain sensitizes tumor cells but not normal cells to radiation. Int J Radiat Oncol Biol Phys 1988;15:953–8 [DOI] [PubMed] [Google Scholar]
- 41.Fertil B, Dertinger H, Courdi A, Malaise EP. Mean inactivation dose: a useful concept for intercomparison of human cell survival curves. Radiat Res 1984;99:73–84 [PubMed] [Google Scholar]
- 42.Morgan MA, Parsels LA, Kollar LE, Normolle DP, Maybaum J, Lawrence TS. The combination of epidermal growth factor receptor inhibitors with gemcitabine and radiation in pancreatic cancer. Clin Cancer Res 2008;14:5142–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murai J, Zhang Y, Morris J, Ji J, Takeda S, Doroshow JH, et al. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J Pharmacol Exp Ther 2014;349:408–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 2000;20:8602–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foskolou IP, Biasoli D, Olcina MM, Hammond EM. Measuring DNA Replication in Hypoxic Conditions. Adv Exp Med Biol 2016;899:11–25 [DOI] [PubMed] [Google Scholar]
- 46.Techer H, Koundrioukoff S, Azar D, Wilhelm T, Carignon S, Brison O, et al. Replication dynamics: biases and robustness of DNA fiber analysis. J Mol Biol 2013;425:4845–55 [DOI] [PubMed] [Google Scholar]
- 47.Huang X, Halicka HD, Darzynkiewicz Z. Detection of histone H2AX phosphorylation on Ser-139 as an indicator of DNA damage (DNA double-strand breaks). Curr Protoc Cytom 2004;Chapter 7:Unit 7 27 [DOI] [PubMed] [Google Scholar]
- 48.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21 [DOI] [PubMed] [Google Scholar]
- 49.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913–7 [DOI] [PubMed] [Google Scholar]
- 50.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009;361:123–34 [DOI] [PubMed] [Google Scholar]
- 51.Murai J, Huang SY, Renaud A, Zhang Y, Ji J, Takeda S, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther 2014;13:433–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 2001;276:47759–62 [DOI] [PubMed] [Google Scholar]
- 53.Groth P, Orta ML, Elvers I, Majumder MM, Lagerqvist A, Helleday T. Homologous recombination repairs secondary replication induced DNA double-strand breaks after ionizing radiation. Nucleic Acids Res 2012;40:6585–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moiseeva T, Hood B, Schamus S, O’Connor MJ, Conrads TP, Bakkenist CJ. ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and And-1. Nat Commun 2017;8:1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinet A, Carvajal-Maldonado D, Lemacon D, Vindigni A. DNA Fiber Analysis: Mind the Gap! Methods Enzymol 2017;591:55–82 [DOI] [PubMed] [Google Scholar]
- 56.Zhong Y, Nellimoottil T, Peace JM, Knott SR, Villwock SK, Yee JM, et al. The level of origin firing inversely affects the rate of replication fork progression. J Cell Biol 2013;201:373–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lesueur P, Chevalier F, Austry JB, Waissi W, Burckel H, Noel G, et al. Poly-(ADP-ribose)-polymerase inhibitors as radiosensitizers: a systematic review of pre-clinical and clinical human studies. Oncotarget 2017;8:69105–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 2010;10:293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vendetti FP, Karukonda P, Clump DA, Teo T, Lalonde R, Nugent K, et al. ATR kinase inhibitor AZD6738 potentiates CD8+ T cell-dependent antitumor activity following radiation. J Clin Invest 2018;128:3926–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanpouille-Box C, Alard A, Aryankalayil MJ, Sarfraz Y, Diamond JM, Schneider RJ, et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun 2017;8:15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding L, Kim HJ, Wang Q, Kearns M, Jiang T, Ohlson CE, et al. PARP Inhibition Elicits STING-Dependent Antitumor Immunity in Brca1-Deficient Ovarian Cancer. Cell Rep 2018;25:2972–80 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.