Abstract
Spinal muscular atrophy (SMA) is one of the major genetic disorders associated with infant mortality. More than 90% of cases of SMA result from deletions of or mutations in the Survival Motor Neuron 1 (SMN1) gene. SMN2, a nearly identical copy of SMN1, does not compensate for the loss of SMN1 due to predominant skipping of exon 7. The spectrum of SMA is broad, ranging from prenatal death to infant mortality to survival into adulthood. All tissues, including brain, spinal cord, bone, skeletal muscle, heart, lung, liver, pancreas, gastrointestinal tract, kidney, spleen, ovary and testis, are directly and/or indirectly affected in SMA. Accumulating evidence on impaired mitochondrial biogenesis and defects in X chromosome-linked modifying factors, coupled with the sexual dimorphic nature of many tissues, point to sex-specific vulnerabilities in SMA. Here we review the role of sex in the pathogenesis of SMA.
Keywords: Spinal Muscular Atrophy (SMA), Survival Motor Neuron (SMN), X chromosome, mitochondria, male infertility, Intronic splicing silencer N1 (ISS-N1)
1. Introduction
Spinal muscular atrophy (SMA) is the leading genetic disease of children and infants affecting one in ~8,000 to 10,000 live births [1,2]. In more than 95% of incidences, SMA is caused by low levels of Survival Motor Neuron (SMN) protein due to deletions of or mutations in the SMN1 gene [3]. SMN2, a nearly identical copy of SMN1 universally present in humans, fails to prevent SMA since one of the SMN2 coding exons, namely exon 7, is predominantly skipped, resulting in the production of an unstable truncated protein SMNΔ7 [4]. The broad spectrum of SMA pathologies has been categorized into five types: 0, 1, 2, 3 and 4 [5]. The extremely severe type 0 is characterized by death at or shortly after birth [6]. Type 1 (also called Werdnig-Hoffmann disease) age of onset is at birth; type 1 patients are never able to sit or walk and generally succumb to death before their 2nd birthday [7]. In the case of type 2 (also called Dubowitz disease) patients show symptoms before 18 months; they survive beyond 2 years, they can sit but cannot walk [7]. Type 3 (also called Kugelberg-Welander disease) is manifested after18 months of age and patients can sit, walk and survive into adulthood [7,8]. Type 4, the mildest form of the disease, arises during early adulthood and patients may survive well into their fifties and sixties [9]. While degeneration of motor neurons and abnormalities of neuromuscular junctions are typical manifestations in severe SMA [10], in mild SMA impairments of peripheral tissues, such as muscle, as well as cardiac defects and male infertility problems can emerge prior to neuromuscular phenotypes [11]. Thus, these impairments could become the primary concerns in mild form of the disease. Of note, substantial heterogeneity exists between patients within each SMA type. Part of this heterogeneity could be attributed to the disease modifying factors, the numbers of which continues to grow. Additional heterogeneity could be due to the specific effects of epigenetic factors, the role of which remains largely unknown. Remaining heterogeneity could come from the sex-specific attributes that are the focus of this review.
Mouse models have been extensively employed to understand the pathogenesis of SMA [12-14]. Unlike humans, mice carry a single copy of the Smn gene termed Smn1. Consistent with the prenatal death in type 0 SMA, deletion of Smn1 in mouse is embryonic lethal [15,16]. Introduction of human SMN2 rescues Smn1 knockout mouse from the embryonic lethality and leads to the phenotype resembling that of the severe SMA [16,17]. A high copy number of SMN2 decreases disease severity in both humans and mice [17-20]. A growing number of proteins, including plastin (PLS3), neuritin 1 (NRN1), neurocalcin delta (NCALD), TIA1 cytotoxic granule associated RNA binding protein (TIA1), Ubiquitin Like Modifier Activating Enzyme 1 (UBA1), Ubiquitin Specific Peptidase 9 X-Linked (USP9X), Stathmin-1 (STMN1), Myostatin (MSTN) and ZPR1 zinc protein (ZPR1), have been suggested to modify SMA severity [21-30]. Several of these modifying factors, namely PLS3, USP9X and UBA1, are encoded by the genes located on the X chromosome; hence they have the intrinsic capability to affect SMA severity in a sex-specific manner [21,26,27].
SMN is implicated in multiple cellular processes, including DNA repair, transcription, pre-mRNA splicing, translation, stress-granule formation, macromolecular trafficking, cytoskeletal dynamics and cell signaling [31]. While most functions of SMN require its interactions with other proteins [31], there is also evidence to support a direct interaction of SMN with cellular transcripts [32]. The SMN genes generate a diverse repertoire of transcripts, including multiple alternatively spliced mRNAs, circular RNAs and long non-coding antisense RNAs [33-42]. Generation of these transcripts is likely to further expand the functions of SMN. Studies conducted in SMA patients and mouse models underscore the cell autonomous role of SMN in all tissues, including brain, spinal cord, liver, lung, heart, skeletal muscles, ovary and testis [Table 1; 43-137]. The tissue-specific requirement for SMN appears to vary depending on the developmental stage and the expression of the disease modifying factors [138]. In line with the distinct cell-specific roles, SMN impacts the expression of different genes in different tissues [139-141]. While not yet fully acknowledged, cumulative evidence supports the effect of sex on SMN functions as well as on the pathogenesis of SMA. Several recent reviews underscore the role of sex in genetic and epigenetic regulation as well as energy metabolism in mammals [142-145]. Sex also impacts the outcome of therapeutic interventions in several illnesses, including neurodegenerative, metabolic, and cardiovascular disorders [146-148]. Here we review the current literature to glean the sex-specific impact of SMN reduction that causes the disease. This review is inspired by independent reports supporting the adverse impact of low SMN on various tissues of males with mild SMA. SMN is particularly essential for maintaining male reproductive health [11], which deserves attention considering that male fertility remains a major global concern [149]. The less severe phenotype of SMA in females could be attributed to multiple sex-specific variables, including X chromosome-linked modifying factors, mitochondria and the action of the sex hormones.
Table 1.
Tissue-specific phenotypes of SMA.
| Tissue/organ | Organism | Pathology/Symptoms | Refs. |
|---|---|---|---|
| Body composition | Human (T1,2) | Higher body fat mass, lower fat-free mass/lean body mass | 43-45 |
| Blood vessels | Mouse (SV) | Decreased capillary bed density in skeletal muscles, increased capillary calibre, reduced capillary ramification into muscle | 46,47 |
| Bone | Human (T1-3) | Low bone mineral density, congenital bone fractures, extremely thin ribs, generalized osteopenia, impaired cartilage formation | 45,48-55 |
| Mouse (SV) | Impaired bone growth, pelvic bone fractures, decrease in total bone area, bone mineral content, bone mineral density | 55,56 | |
| Mouse (NSD) | Kyphosis of the spine | 57 | |
| Brain | Human (T3) | Epilepsy, atrophy of parahippocampal gyrus | 58 |
| Mouse (SV) | Impaired brain development | 59 | |
| GI tract | Human (T2) | Gastroesophageal reflux, constipation, delayed gastric emptying | 60,61 |
| Mouse (SV) | Morphological changes of intestine, increased inflamation, constipation, delayed colonic transit | 61-63 | |
| Heart | Human (T0) | Congenital septal defect | 64 |
| Human (T1-3) | Structural defects, bradycardia, ECG abnormalities, myocardial fibrosis | 49,65-68 | |
| Mouse (SV) | Structural defects, bradycardia, swollen and disorganized mitochondria, ECG abnormalities, blood clot accumulations | 63,69-72 | |
| Lung | Human (T1-3) | Respiratory problems and need for ventillation | 73 |
| Mouse (SV) | Pulmonary infarctions, lung emphysema, enlarged alveolar spaces | 63 | |
| Liver | Mouse (SV,IN) | Abnormal liver development, atrophy, steatosis | 72,74 |
| Mouse (LSD,EL) | Liver atrophy, severe liver dysfunction, iron overload, lack of regeneration | 75 | |
| Skeleton | Human (T1-3) | Hip dislocation/subluxation, colapsing spine, scoliosis | 11,65,76 |
| Mouse (SV) | Skeletal abnormalities in the lower body, reduced caudal vertebra | 77 | |
| Skeletal Muscle | Human (T0) | Immature muscle fibers | 78 |
| Human (T1-3) | Delayed muscle development, hypertrophy, impaired mitochondrial biogenesis, muscle weakness, lack of coordination | 11,79-82 | |
| Human (T4) | Abnormal gait | 11 | |
| Pig (NKD) | Muscle weakness, atrophy and fasciculations, decreased CMAP | 83 | |
| Mouse (SV,IN) | Morphological and functional deffects of muscle, impaired maturation of myofibers, macrophage depletion, muscle fatigue | 16,63,84-88 | |
| Mouse (MSD) | Severe degeneration, progressive myopathy, functional deficits | 89-91 | |
| Mouse (NSD) | Muscle atrophy, muscle fibre loss | 57 | |
| Drosophila | Reduced muscle growth, defective locomotion | 92 | |
| Motor nerons | Mouse (SV) | Reduced axon growth, molecular and functional abnormalities | 93,94 |
| Mouse (NSD) | Motor neuron degeneration | 57 | |
| Central and peripheral nervous system | Human (T1,2) | Loss of motor neurons, abnormal sensory conduction, neuroinflamation | 79,95-99 |
| Pig (NKD) | Loss of motor neurons | 83 | |
| Mouse (SV) | Myelination defects, sensory neuron developmental deffects, loss of synapse, functional deficits, neuroinflamation | 100-106 | |
| Mouse (NSD) | Reduction in lumbar and cervical motor neurons | 57 | |
| Zebrafish | Defects in motor neuron axonal outgrowth, abnormal branching | 107,108 | |
| Drosophila | Disfunctional sensory-motor network | 92 | |
| Autonomic nervous system | Human (T1-3) | Sympathetic-vagal imbalance, changes in blood pressure, digital necrosis, sympathetic nerve hyperactivity | 65,109-112 |
| Human (T2) | Gastroesophageal reflux and constipation | 60 | |
| Mouse (SV) | Decreased cardiac output, decreased contractility | 69-71 | |
| Mouse (SV) | Disrupted neuron signaling to colonic smooth muscles, diarrhea | 61-63 | |
| NMJ | Human (T1) | Arrested/delayed NMJ development, functional abnormalities of NMJ | 79 |
| Mouse (NSD) | Structural abnormalities of NMJs | 57 | |
| Mouse (SV,IN) | Structural and functionaly abnormalities of NMJs, decreased density and release of presynaptic vasicles | 63,86,88,113-117 | |
| Central synapse | Mouse (SV) | Abnormalities in central synapse connectivity and function | 101,102 |
| Mouse (NSD) | Defects in sensory-motor synapses | 57 | |
| Ovary | Mouse (SV,MD) | Decreased weight of ovary comapred to body weight | 118,119 |
| Pancreas | Human (T1) | Abnormalities morphology of pancreatic islet cells, pancreatitis | 65,120 |
| Mouse (Het) | Abnormalities of pancreatic islets | 121 | |
| Kideny | Human (T1,2) | Impaired kidney structure and function, nephrocalcinosis | 122,123 |
| Spleen | Human (T1) | Abnormal development, abnormal function | 124 |
| Mouse (SV,IN) | Reduced spleen weight, structural and functional defects | 74,86,124-126 | |
| Testis | Human (T1,2) | Cryptorchidism | 65,127 |
| Mouse (SV,MD) | Degeneration of seminiferous tubes, impaired spermatogenesis | 118,119 | |
| Thymus | Mouse (SV,IN) | Lymphocite apoptosis, misregulated T-cell development | 86,128 |
| Urinary tract | Human (T1,2) | Urinary incontinence, enuresis, hypercalciuria | 123,129 |
| Human (T3,4) | Nephrolithiasis,urinary retention problems | 130 | |
| Mouse (MD) | Reduced bladder and smooth muscle size | 131 | |
| Vascular | Human (T1) | Digital necrosis, generalized inflammation of vasculature | 110,111,132 |
| Mouse (SV) | Reduction vascular density in heart and small intestine | 47,62,132 | |
| Metabolic | Human (T1-4) | Abnormal glucose and lipid metabolism, dicarboxylic aciduria, chronic hypercalcaemia, precocious pubarche, hyperlipoproteinemia | 74,120,123, 133-137 |
| Mouse (IN) | Non-alcoholic fatty liver disease, hyperglycemia, hyperglucagonemia, increased insulin sensitivity, glucagon sensitivity | 74,120 | |
| Mouse (Het) | Abnormal glucose metabolsim, fasting hyperglycemia, hyperinsulinemia, hepatic glucagon sensitivity | 121 |
Abbreviations: EL, late embryonic lethality; Het, heterologous for Smn1 allele; GI, gastrointestinal; IN, intermediate phenotype; LSD, liver-specific depletion; MD, mild phenotype; MSD, muscle-specific depletion; NKD, motor-neuron-specific postnatal knockdown; NSD motor neuron progenitor-specific depletion; SV, severe phenotype; T0, SMA type 0; T1, SMA type 1; T2, SMA type 2; T3, SMA type 3; T4, SMA type 4;
The purpose of this review is to provide an assessment of SMN-dependent effects within cohorts of males and females rather than merely describe the differences between cohorts of males and females. Given the prominent role of SMN in gene regulation, including expression of factors with potential to be “overrepresented” in females such as those expressed from the X-chromosome and the mitochondrial genome, we expect SMN-linked sex-specific differences at all developmental stages. Further, due to changes in hormonal levels and the cumulative impact of the modifying factors, we anticipate a widening gap in SMN-dependent effects within cohorts of post-puberty patients in each sex category. While heterogeneity of the SMA patient population combined with the lack of sex-specific data in most reported studies may not allow sweeping claims of the gender-related effects, recent reports are beginning to capture differences between males and females caused by the loss of SMN. This review is aimed at motivating future studies in which sex-specific distinctions, regardless of their magnitude, are accurately captured. Understanding the effect of gender in SMA is critical for not only uncovering novel SMN functions but also for deciphering the role of a growing number of disease modifying factors and developing more efficient SMA therapies.
2. Sex and the prevalence of SMA
Major human diseases including cardiovascular diseases, Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS) show sex-specific bias in their prevalence [145,150-152]. While women are preferentially impacted by AD and other dementias [150], more men than women are affected by PD and ALS [151,152]. Further, women show more resistance to oxidative stress-associated diseases such as ischemic heart disease and ischemic stroke [145]. However, when it comes to muscular dystrophies, no clear generalizations can be made with respect to sex-specific differences, with Duchenne muscular dystrophy (DMD) and its milder form, Becker muscular dystrophy (BMD), being somewhat an exception. Both dystrophies are X-linked conditions that usually affect males. It should be noted, however, that some female dystrophin mutation carriers do present similar symptoms, particularly cardiac phenotypes, albeit in milder form [153-156]. At the same time, while men suffering from limb girdle muscular dystrophies (LGMD) show higher muscle fiber atrophy [157], women are more susceptible to developing musculoskeletal disorders [158]. Furthermore, in Facioscapulohumeral muscular dystrophy (FSHD), an autosomal dominant neuromuscular disorder with extremely variable symptoms [159], less females are affected than males [160]. A recent literature survey suggests differential interaction between a number of muscle pathologies and biological sex [161]. For instance, males and females display differences in muscle physiology, such as muscle size, muscle fiber composition, anabolic and catabolic factors as well as differences in circulating hormones and in mitochondrial content, biogenesis and activity [161-163].
Despite the fact that severe SMA is more prevalent than all other types of SMA combined [1], limited attention has been given to the sex-specific impact/differences in this type of the disease. The sex-specific statistics available for mild SMA are somewhat different. For instance, a study published in 1995 indicated that in the case of mild SMA, more males than females were affected by the disease [7]. In particular, this study found that in females the onset of mild SMA was delayed by about three years as compared to males [7]. While the study mostly analyzed Caucasian patients, findings are relevant to other ethnicities. For example, a recent study conducted on Japanese patients found a higher proportion of male cases of mild SMA [164]. Furthermore, an insurance claim study that analyzed the data from January 1, 2008 to October 1, 2015 found that adult patients with initial male infertility concerns were later diagnosed with mild SMA [11]. Of note, roughly equal number of males and females were represented in this insurance claim study [11]. Recently published results of the GTEx (Genotype-Tissue Expression) project, which investigated the impact of sex on gene expression among 44 different human tissues, found that ~37% of all genes show sex-biased expression [165]. Thus, strong evidence of the influence of sex on gene expression speaks to the SMA phenotype exhibiting sex-specific characteristics.
3. Role of mitochondria in sex-specific differences in SMA phenotype
Mitochondria play an important role in creating sex-specific pathologies including metabolic diseases, cardiovascular diseases, muscular dystrophies and neurodegenerative diseases [144]. The sex-specific effect of mitochondria is exerted through the activity of estrogens and estrogen receptors coupled with the asymmetric inheritance of the mitochondrial genome and the X chromosome. Throughout the evolution, mitochondrial DNA and X chromosomes spent more time under selective pressure in females than in males. Hence, genes located within mitochondrial DNA and on X chromosomes are better optimized to work in females than in males [166]. Mitochondria also impact the crosstalk between sex chromosomes and autosomes [167]. Studies in rodents and humans confirm sexual dimorphism in mitochondrial functions for multiple tissues including adipose tissue, skeletal muscle, cardiomyocytes and the brain. For example, in white and brown adipose tissue as well as in skeletal muscle of rodents, females possess more functional mitochondria than males [168-170]. In humans, female adipose tissue has higher expression of genes involved in mitochondrial function than male adipose tissue [171]. In rodent cardiomyocytes, mitochondria are more efficient in females than in males [172].
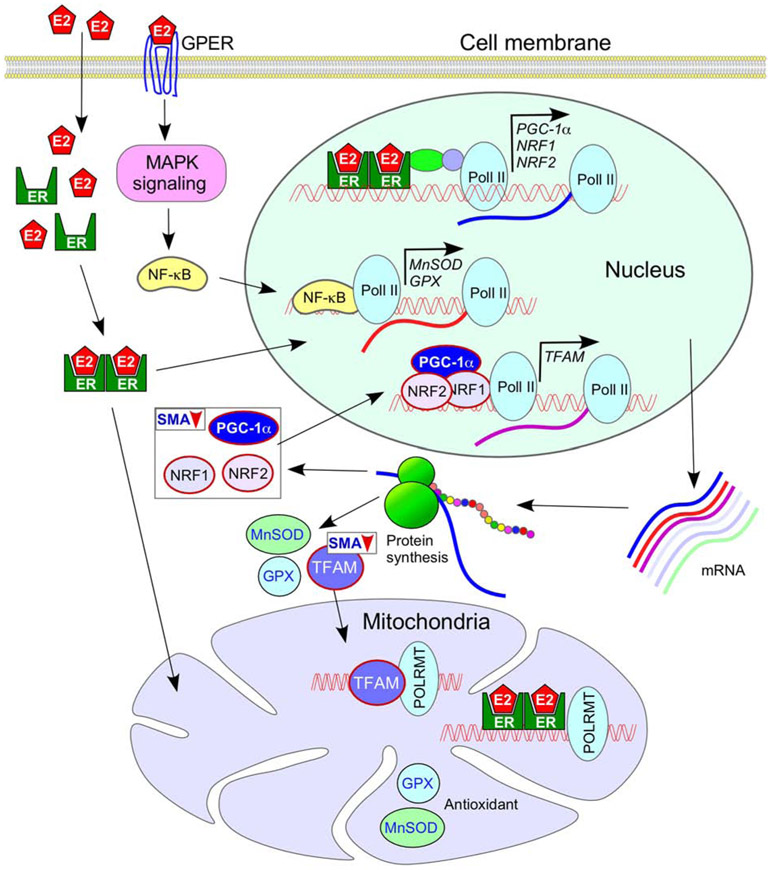
Impaired mitochondrial biogenesis has been recorded in muscle of SMA patients as well as in motor neurons of SMA mice [82,173]. Interestingly, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), one of the key factors involved in the biogenesis of mitochondria, is downregulated in skeletal muscle of SMA patients and in SMA mice (Fig. 1) [82,174]. Numerous signaling pathways central to the adaptation of muscle to endurance training converge at PGC-1α [175]. Furthermore, expression of the transcription factor A, mitochondrial (TFAM) as well as the nuclear respiratory factors, NRF1 and NRF2, is downregulated in muscle of SMA patients (Fig. 1) [82]. Relevant to sex-specific effects, mitochondria regulate the biosynthesis of sex hormones (estrogen, progesterone and testosterone), which in turn regulate mitochondrial function [167]. Mitochondria are the major source of the energy supply. They also modulate many cellular processes including calcium and ionic homeostasis, reactive oxygen species generation, pH maintenance, steroid hormone production, lipid and carbohydrate metabolism and cell death. Levels of multiple respiratory chain subunits associated with the mitochondrial energy production are substantially downregulated in the skeletal muscle of SMA patients [82]. Hence, functional defects associated with the mitochondrial respiratory chain subunits could be also expected in other high-energy-consuming tissues, such as cardiomyocytes, neurons, liver and kidney, all of which are impacted in SMA (Table 1). A recent study conducted on a mild SMA mouse model captured sex-specific differences, as females displayed better endurance than males in the rotarod performance test [176]. The authors attributed these differences to better motor neuron function in females. Another study conducted using the mouse brain showed lower oxidative stress in mitochondria of females as compared to males [177]. Hence, it is likely that the cumulative effect of oxidative stress brings an additional sex-specific difference during the lifespan of SMA patients.
Figure 1.
Sex-specific effect of mitochondrial functions in tissues with high energy demand. Interactions of pol II and POLRMT with the nuclear and the mitochondrial DNA, respectively, have been shown. The female sex hormone estrogen (E2) regulates expression of transcription factors PGC-1α, NRF1 and NRF2 that are important for the expression of TFAM, a transcription factor. TFAM is essential for mitochondrial biogenesis. Both TFAM and E2 regulate expression of genes located on the mitochondrial genome. Through the MAPK signaling pathway, E2 also regulates expression of antioxidants GPX and MnSOD that localize to the mitochondria. PGC-1α, NRF1 and NRF2 are known to be downregulated in SMA, although it is not yet known if the downregulation is more pronounced in males. Details of pathways depicted here are described in the body of the main text and the accompanying references (see section 3). Abbreviations: E2, estrogen; ER, estrogen receptor; MnSOD, manganese superoxide dismutase; GPER, G protein-coupled estrogen receptor; GPX, glutathione peroxidase; MAPK, mitogen associated protein kinase; NF-κB, nuclear factor kappa B; NRF, nuclear respiratory factor; PGC-1α, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha; pol II, RNA polymerase II; POLRMT, mitochondrial DNA-dependent RNA polymerase; TFAM, transcription factor A, mitochondrial.
4. Sex-specific effect of X chromosome inactivation
In order to maintain appropriate dosage of X-chromosomal genes between males and females, one of the two female X chromosomes undergoes random and permanent transcriptional inactivation called X-inactivation or Xi. However, several X-linked genes, including those involved in protection against oxidative stress and proteolytic stress, escape inactivation, creating a dosage imbalance between the two sexes [145]. Three X-linked genes, PLS3, USP9X and UBA1, have been suggested to be positive modifiers of SMA [21,26,27]. USP9X codes for a deubiquitinase (USP9X), which deubiquitinates SMN and consequently stabilizes it against degradation by the proteasome machinery [26]. UBA1 codes for Ubiquitin-like modifier activating enzyme 1 (UBA1), which directly interacts with SMN and mediates ubiquitin homeostasis and subsequent β-catenin signaling [178]. It has been shown that low levels of SMN cause downregulation of UBA1, leading to neuromuscular pathology due to disruption of ubiquitin homeostasis and β-catenin accumulation [178,179]. Systemic restoration of UBA1 ameliorates SMA phenotype in a mouse model of the disease [27]. The expression of both USP9X and UBA1 is female-biased due to Xi escape [145]. PLS3 codes for an actin-binding protein, PLS3, which is another positive modifier of SMA [21]. It should be noted that the correlation between PLS3 expression and SMA severity is not absolute as other disease modifying factors may confound the effect of PLS3 [22,180]. Although PLS3 is located within the X-chromosome evolutionary strata with the least chance of X-inactivation (Xi) escape, a published report showed that the gene was able to escape Xi in 4 out of 9 cases [181]. It also suggested that females would be variable in regard to PLS1 expression [181]. According to the same study, USP9X and UBA1 escaped from Xi completely [181]. Similarly, expression of UBA1 from the inactive X chromosome was demonstrated using female fibroblast cell line WI-38 [182]. One recent report found UBA1 to be expressed at higher levels in female cells as compared to male cells of dizygotic twins [183]. USP9X happens to be one of the six X-linked homologue of Y-linked genes. Interestingly, expression of USP9X has been shown to be significantly higher in female mouse brain as compared to male brain irrespective of its Xi status, potentially inducing sex-specific differences in neuronal function [184]. This finding supports a point of view that high expression of an X-inked gene could be achieved without Xi escape.
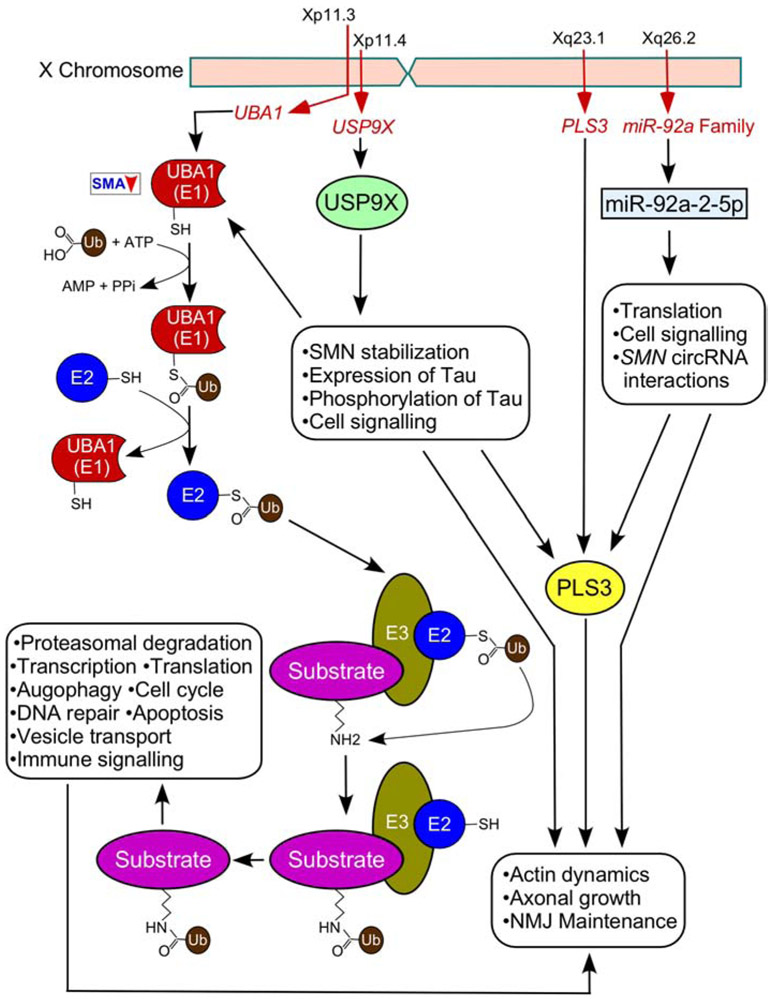
Other X-linked genes that may have sex-specific effects on SMA phenotype include the androgen receptor gene, cell-signaling receptor genes and microRNA genes [185]. In particular, location of the androgen receptor gene on the X-chromosome is suggestive of the impact this chromosome has on controlling the functions of the male sex hormone, testosterone. Furthermore, several X-linked genes are associated with enhanced female immunity against infections and with sexual dimorphism of the immune inflammatory response [185,186]. Impaired immune system and neuroinflammation are accompanying pathologies in SMA [126,128,187-189]. Future studies will determine if female SMA patients display milder inflammatory responses. About 10% of human microRNA genes are located on the X-chromosome and have the potential to exert substantial sex-specific effects on RNA metabolism in all tissues [190]. One of the X-linked microRNAs, miR-92a-2-5p, is predicted to bind circular RNAs generated by human SMN genes (Fig. 2) [41]. The role of miR-92a-2-5p has recently been implicated in mitochondrial translation [191]. Therefore, it is tempting to hypothesize that the sexual dimorphism in SMA could be partly regulated by X-linked microRNAs such as miR-92a-2-5p, availability of which is predicted to be modulated by SMN circular RNAs.
Figure 2.
Genes located on the X-chromosome modulate the severity of SMA. USP9X codes for USP9X, deubiquitinates SMN, protecting it from proteasomal degradation. UBA1 codes for UBA1 that mediates ubiquitin homeostasis through direct interaction with SMN. PLS3 codes for PLS3 and is a known positive modifier of SMA. PLS3 plays an important role in the maintenance of actin dynamics. X-linked coded miR-92a-2-5p is predicted to bind circular RNAs generated by the human SMN genes. Locations of the genes on the X-chromosome are indicated. Cellular functions of each protein and miR-92a-2-5p are listed in boxes. Details of pathways depicted here are described in the body of the main text and the accompanying references (see section 4).
5. Importance of sex hormones
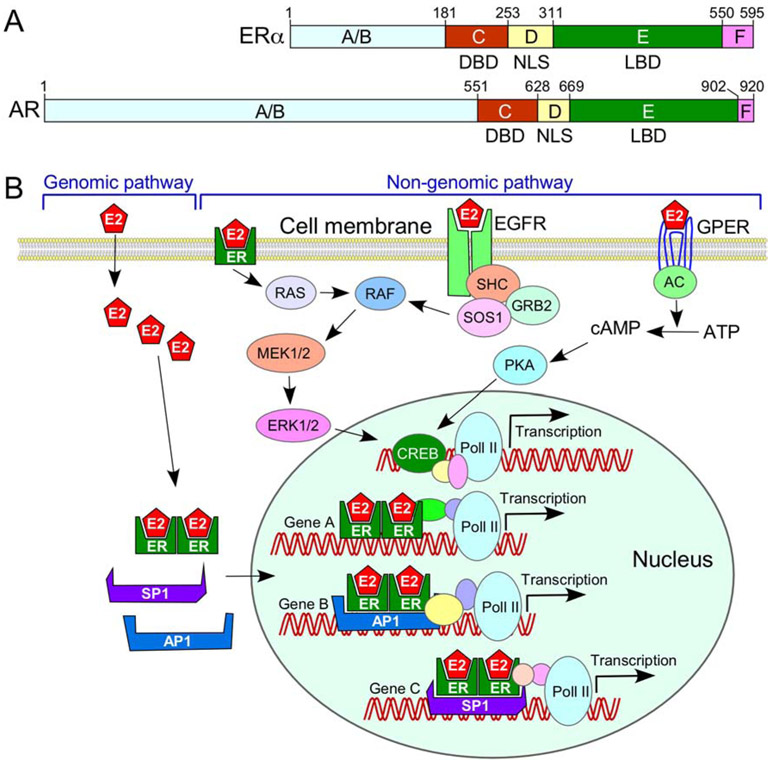
Differential exposure to sex hormones throughout embryonic development and adulthood is one of the driving forces behind sex-specific differences in the transcriptome and proteome of different mammalian tissues. Ovaries are the major source of estrogens, including 17β-Estradiol (E2), which plays an important role in both genomic (classical or canonical) and non-genomic (non-classical or non-canonical) regulatory mechanisms (Fig. 3). While the genomic mechanism refers to regulation of gene expression through chromatin remodeling and transcription, the non-genomic mechanism refers to regulation through posttranslational modifications, including phosphorylation of key intermediaries of signal transduction pathways. Among a large number of receptors that interact with E2, ERα plays an important role in cellular metabolism. ERα is considered to be a master regulator of transcription and the key factor of sexual differentiation in the rodent brain [143]. For the purposes of clarity, we use ER and ERα interchangeably hereafter. ERα encompasses nuclear localization signal (NLS), ligand binding domain (LBD) and the DNA binding domain (DBD) (Fig. 3A). Binding of E2 to LBD triggers a structural change favoring a stable ER dimerization, co-regulator interactions and nuclear localization. Once in the nucleus, ERs regulate transcription (genomic regulatory mechanism) by directly interacting with the estrogen response elements (EREs) through their DBD (Fig. 3B). Alternatively, ERs can regulate transcription through interactions with other transcription factors, such as stimulatory protein 1 (SP1) and activator protein 1 (AP1) that are bound to their cognate recognition sites on DNA [192,193] (Fig. 3B). E2 and ERα play an important role in neuroprotection [194]. Interestingly, ERα also interacts with PGC-1α to upregulate transcription of NRFs that are associated with biogenesis of mitochondria [144]. In addition, ERs localize to the mitochondrion and regulate mitochondrial transcription (Fig. 1) [195]. Independent of transcription, ligand-bound ERs can regulate signaling pathways, including ERK/MAPK and PI3K/AKT signaling cascades (non-genomic regulatory mechanism), modulating cellular metabolism.
Figure 3.
Role of sex hormones and their receptors in the sex-specific effect on cellular metabolism. (A) Diagrammatic representation of the domain structure of estrogen (E2) receptor ER (ERα) and androgen receptor AR. Both ERα and AR contain A/B, C, D E and F motifs and possess a DNA binding domain (DBD), nuclear localization signal (NLS) and a ligand binding domain (LBD). (B) Diagrammatic representation of two E2-mediated pathways. The genomic pathway refers to the role of E2 in transcription regulation through direct interaction with DNA via ER and/or ER-associated transcription factors, such as AP1 and SP1. The non-genomic pathway refers to the role of E2 executed through signaling cascades initiated/triggered within the cytosol. Details of pathways depicted here are described in the body of the main text and the accompanying references (see section 5).
The testes are the primary source of testosterone, which mediates its functions through the androgen receptor (AR). AR encompasses the same domain structures as ER (Fig. 3A). Similar to ER, AR acts through both genomic and non-genomic regulatory mechanisms [196]. At birth, male mice are exposed to exceptionally high levels of testosterone, which is converted into estradiol (E2) in the brain leading to the male-specific changes of transcription through ER receptors [197]. These changes, coupled with the relative levels of sex hormones, contribute to dimorphic immune response in mammals [185,186]. The compromised immune response in SMA is likely to add to sex hormone-dependent dimorphism. The SMN promoter encompasses motifs of transcription factors such as SP1, ELK1 and CREB that are targets of the genomic and non-genomic regulatory pathways of ERα/ERβ and AR [38,198]. Hence, the relative expression of sex hormones could potentially bring sex-specific bias in the regulation of SMN transcription at least in some tissues.
6. Sex and cell-signaling pathways
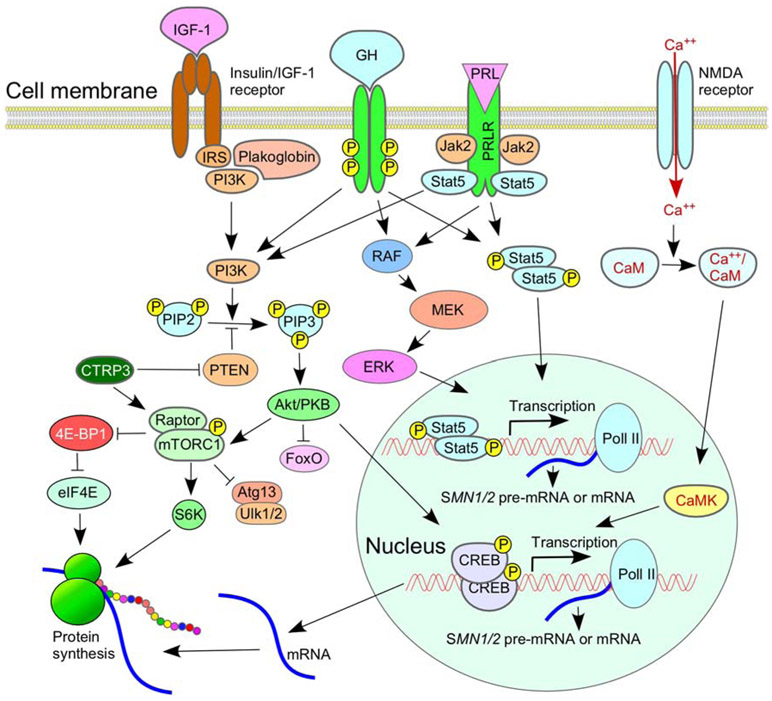
Factors linked to cell-signaling pathways elevate levels of SMN [199]. For example, prolactin (PRL) and human growth hormone (GH) enhance SMN2 transcription by activating the JAK/STAT pathway (Fig. 4) [200,201]. PRL and GH act through their cognate cell membrane receptors PRLR and GHR, respectively. PRLR and GHR share structural homology, including a cytoplasmic domain that provides a docking site for STAT5 [202]. It has been shown that STAT5 expression is affected by sex and that STAT5 knockout mice show sex-dependent pathologies of peripheral tissues [203,204]. A recent study uncovered a female-specific bias in the mechanism of prolactin action as well as its localization in the mouse brain [205]. The canonical mechanism of PRL/GR action involves binding to their cognate receptors with subsequent STAT5 phosphorylation catalyzed by cytosolic tyrosine kinase, Janus kinase 2 (JAK2). Phosphorylated STAT5 is then dimerized and translocated to the nucleus where it regulates the expression of its target genes [202]. PRL/GR also participates in other signaling pathways including mitogen-activated protein kinase (MAPK), AKT1/mTOR and NF-κB pathways that are known modulators of SMN2 transcription and/or SMN2 translation (Fig. 4) [174,206-209].
Figure 4.
Role of signaling pathways in SMA. Multiple signaling cascades are impacted by insulin-like growth factor 1 (IGF-1) and its receptor, both are aberrantly expressed in SMA. Prolactin (PRL) and human growth hormone (GH) enhance SMN2 transcription by activating JAK/STAT pathway. Ca2+/calmodulin-dependent protein kinase (CaMK) can also enhance SMN2 transcription through recruitment of cyclic AMP response element binding protein (CREB) to the SMN2 promoter. Details of pathways depicted here are described in the body of the main text and the accompanying references (see section 6).
Insulin-like growth factor-1 (IGF-1) is a signaling molecule that serves as a neuroprotective factor in mammals (Fig. 4) [210]. While the level of IGF-1 in circulation is markedly reduced in SMA, expression of IGF-1 receptor (IGF-1R) is found to be increased in tissues of SMA patients as well as in mouse models of SMA [211-213]. SMA mice lacking one of the Igf1r alleles show extended life expectancy and improved motor behavior [213]. IGF-1R encoded by Igf1r directly affects PI3/AKT and MAPK/ERK pathways and impacts several other signaling pathways, including N-methyl-D-aspartate (NMDA) pathway, in an indirect way [210,214]. The sex-specific effect of IGF-1R depletion has been observed in several mouse models [215-217]. In particular, heterozygous female mice (harboring a single Igf1r allele) lived longer and showed better resistance to oxidative stress than males [215]. Future studies will reveal if IGF-R1 contributes towards SMA pathogenesis in a sex-specific manner.
7. Effect of sex on metabolic and tissue-specific defects in SMA
7.1. Effect of sex on metabolic defects
Metabolic defects in mammals impact multiple tissues, including liver, pancreas, gastrointestinal tract, kidney, reproductive organs and cardiovascular system. These tissues are likely to display sexual dimorphism due to disparities found between males and females in gene expression, immune response and xenobiotic metabolism as well as in sex hormone effects. SMA patients and SMA mouse models display abnormal glucose, lipid and amino acid metabolisms, although the magnitude of abnormalities differs with respect to the type of SMA and the patient’s age [74,120,134,136,187,218,219]. Abnormal metabolism leads to reduced body weight in mouse models of SMA [12]. The reduction in body weight appears to be more prominent in females than males when compared to age- and sex-matched control littermates [13,119]. In other words, the effects of reduced SMN on body weight of female mice was greater than the one observed for male mice. These results may suggest that the loss of SMN differentially impacts the sex-specific network of interactions that account for the maintenance of body weight. In general, because of the extreme heterogeneity of SMA patients’ phenotypes and a small cohort size employed in most studies, it is not feasible to capture sex-specific differences with high confidence. Yet, several of the metabolic disorders observed in SMA patients are known to have sex-specific bias. For instance, SMA patients have increased susceptibility to dyslipidemia, liver steatosis and non-alcoholic fatty acid disease (NAFLD) [74]. While NAFLD data for SMA patients were not analyzed to draw sex-specific inference [74], a recent report highlighted that the prevalence and severity of NAFLD were higher in men than in women of reproductive age [220]. Defective functioning of other tissues described below could also be due to aberrant metabolic processes with sex-specific impact in SMA.
7.2. Effect of sex on skeletal muscle defects
Multiple factors contribute towards sex-specific differences in the functionality of different muscle types [161]. Skeletal muscle is one of the first tissues to manifest structural and functional defects in SMA (Table 1) [221,222]. While all types of skeletal muscles are affected by the disease [174], tibialis anterior (TA) muscle appears to be one of the most vulnerable [209]. Transcriptome and proteome analysis of muscles collected from an SMA mouse model revealed downregulation of PGC-1α, one of the key factors involved in biogenesis of mitochondria [174]. Among proteins secreted by muscle cells, C1q-TNF-related protein-3 (CTRP3) was shown to be downregulated in SMA [209]. This downregulation is consequential since CTRP3 was reported to promote SMN translation in motor neurons through activation of mTOR pathway [209]. Consistently, skeletal-muscle-specific depletion of SMN in mice leads to muscle fiber defects, neuromuscular junction abnormalities, compromised motor performance, and premature death [89]. Interestingly, female mice appear to be impacted more adversely than males [89]. These findings contrast a recent study showing higher susceptibility of skeletal muscle in males than females in a mild SMA mouse model [176]. The above discrepancy could be attributed to the fact that two different mouse models, one with selective depletion of SMN in skeletal muscle and the other with body-wide decrease in SMN levels, were used [89,176]. It is possible then that male mice are impacted more severely by body-wide depletion of SMN, while female mice are affected more when SMN depletion is restricted to skeletal muscle.
7.3. Effect of sex on neuromuscular junction and motor defects
Sex-specific effects on neuromuscular junction (NMJ) structure and motor functions in severe SMA are not easily captured as patients succumb to death before puberty. Recently, age and gender-specific motor defects were reported in a mild SMA mouse model [176]. In particular, male mice performed poorly on a rotarod test at 8-month compared to female mice of a similar age. The defects were attributed to the neurogenic atrophy and not to the loss of motor neurons. Analysis of TA muscle showed more atrophic fibers in males than in females [176]. An electrophysiological study performed on nerve-soleus muscle preparations of males showed less force generated upon stimulation of nerves than direct stimulation of muscle [176]. In contrast, a similar study on female samples showed no difference in force generated between nerve and muscle stimulation. These findings suggest possible disruption of signal transmission through NMJs in males but not in females. In addition, electrophysiological data pointed to possible deficiencies in NMJ/neuronal transmission in males as compared to females [176]. Blood circulation levels of CTRP3, which as described earlier promotes SMN translation, was shown to be higher in females than in males [209,223]. Thus, in addition to the protective effects against oxidative stress and favorable mitochondrial biogenesis in female neurons, high CTRP3 levels in blood may also benefit female neurons.
7.4. Sex and peripheral necrosis
Severe SMA patients, both males and females, occasionally display digital necrosis associated with thrombotic occlusions of small vessels [110,111]. However, due to a limited number of SMA patients with digital necrosis, no sex-specific inferences could be drawn. While human patients with mild SMA do not manifest digital necrosis, mouse models of mild SMA display tail and ear necrosis as hallmark features [13,16]. A sex-specific difference has been captured in the progression of peripheral necrosis in allele C mice that show mild SMA phenotype [224]. While the age of the peripheral necrosis onset was the same in both sexes, male animals showed a faster progression of tail necrosis [224]. Autonomic nervous system abnormalities and vascular perfusion defects have been proposed as the potential causes of peripheral necrosis in SMA [110,111]. Interestingly, blood vessels were shown to manifest sexual dimorphism due to the role of ERα in flow-mediated remodeling of resistance arteries [225]. In addition, the production of reactive oxygen by mitochondria could impact vascular lesions in a sex-specific manner [226]. Future studies will determine if sex plays an important role in determining the severity of peripheral symptoms in SMA patients.
7.5. Effect of sex on pulmonary functions
Inadequate pulmonary function is one of the major causes of death in type 1 SMA patients [227]. Weakness of respiratory muscles coupled with respiratory tract infections also contribute towards the mortality associated with SMA. While sex-specific defects in pulmonary function have not yet been captured in type 1 SMA patients, the observational study published in 2011 indicated that among types 2 and 3 SMA patients, females may be associated with a greater decline in pulmonary function [228]. This could be due to female anatomical features such as small size of the rib cage and short diaphragm [229]. In addition, differential exposure to sex hormones and sensory feedbacks through vagal and spinal pathways could contribute to sex dimorphism in pulmonary function of SMA patients [229,230]. The severity of the disease is enhanced by hypoxia [231]. Age and sex differences in the ventilatory response to hypoxia have been reported in rats [232]. Hence, it is possible that the sex-specific defects in pulmonary function in SMA are in part due to the ventilatory response to hypoxia.
7.6. Effect of sex on renal injury
The kidney happens to be one of the affected organs in SMA [122,233,234]. Type 1 SMA patients display varying degrees of renal pathology such as tubular injuries, including loss of brush borders, flattened epithelium with detachment, and occasional protein casts [135]. Consistent with these findings, mouse models of severe SMA show developmental defects of kidney and substantial reduction in nephron counts [235]. Recently, a type 3 SMA patient was found to have mild renal dysfunction and proteinuria [234]. Intriguingly, patients suffering from acute kidney injury (AKI) show reduction of SMN levels in renal tubular cells, suggesting a crosstalk between kidney function and SMN [233]. Compared to wild type mice, heterozygous mice with a single Smn allele showed declined renal function and tubular injury upon ischemia-reperfusion injury (IRI) [233]. Of note, IRI-induced AKI is a significant clinical problem associated with prolonged hospital stay and morbidity [236]. Kidneys are known to have sexual dimorphism with regard to their anatomy and physiology [237,238]. For example, a study conducted on rats found higher renovascular resistance, lower absolute glomerular filtration rate (GFR) and lower renal plasma flow in females than males [239]. Men are known to have a higher risk of AKI at all levels of estimated GFR (eGFR) and albumin-to-creatinine ratio [238], although this observation has not been tested in SMA yet. The male-specific vulnerability to AKI has been linked to enhanced apoptosis in renal tubular cells triggered by testosterone-induced upregulation of Fas and Fas ligands [240,241]. At the same time, the female-specific protection against AKI has been connected to the estrogen-induced sustained activation of cell survival pathways, including the Akt pathway [241,242].
7.8. Defects of reproductive organs
Abnormal development of the male reproductive system is one of the most consequential outcomes of low SMN in SMA. Independent studies conducted on SMA patients recorded cryptorchidism, a condition in which the testis fails to descend into the scrotum [65,127]. In several instances, cryptorchidism served as a good predictive marker of male infertility [243]. Young SMA patients (types 1 and 2) are not ideally suited to examine the effect of low SMN on male fertility; however, in adult SMA patients, male reproductive system dysfunctions have been documented [11]. Consistent with this, mild SMA mice (allele C model) show widespread degeneration of seminiferous tubules, loss of post-meiotic cells, and a drastic reduction in male fertility [118]. In contrast, female reproductive organs of allele C mice appear normal, although the uterus/ovary mass was reduced [118]. A recent study conducted in a severe mouse model of SMA found abnormalities in both ovary and testis [119]. This study confirmed the critical role of SMN in the survival of male germ cells and the maintenance of spermatogonia.
Testis shows the highest SMN expression as compared to all other tissues examined, suggesting the need for a sustainably higher level of SMN for various aspects of testis development and function [118]. Interestingly, unlike in other tissues, SMN2 exon 7 is predominantly included in the testis. It appears that the SMN2 exon 7 splicing switch from predominant skipping to significant inclusion fulfills, at least in part, the need for high SMN expression in human testes [118]. Additional mechanisms such as enhanced transcription and translation may also contribute towards the high expression of SMN in mammalian testes. Several cis-elements and transacting factors, including hnRNP A1/A2, hnRNP C, hnRNP Q, Tra2-β1, SRp30cPSF, Sam68, SRSF1, TDP-43 and TIA1, have been implicated in regulation of SMN exon 7 splicing [198,244-246]. Among them, hnRNP Q and Tra2-β1 have been linked to the promotion of SMN2 exon 7 inclusion in testes [247,248]. However, the mechanism of the testis-specific splicing switch of SMN2 exon 7 is not yet fully understood. Uncovering it has broad implication for understanding the tissue-specific splicing regulation as well as for finding novel therapeutic targets for manipulating SMN2 exon 7 splicing.
In addition to testosterone production, another main function of testes is to produce sperm. Multiple signaling pathways (in both somatic and germ cells) cooperate to regulate spermatogenesis. In fetal testes, TGFβ signaling through somatic cells plays an important role in the development of fetal testicular germ cells [249]. Proliferation of Sertoli cells that feed the germ cells is regulated by cAMP/PKA, ERK1/2, PI3K/Akt and mTORC1/p70SK6 signaling cascades [250]. Self-renewal of spermatogonial stem cells is controlled by glial cell line-derived neurotrophic factor, GDNF, and RET Tyrosine Kinase pathway [251]. In postnatal testes, the retinoic acid signaling pathway regulates the differentiation of spermatogonia and entry into meiosis [252]. The quality and quantity of spermatogenesis is controlled by the FSH signaling pathway, which acts through the FSH receptor located on Sertoli cells [253]. Finally, withdrawal of testosterone leads to an arrest of spermatogenesis in the post meiotic haploid phase [254]. Studies conducted using muscles and neurons of mouse models of SMA revealed dysregulation of several signaling cascades, including the most critical cAMP/PKA, ERK1/2, PI3K/Akt and mTORC1/p70SK6 cascades [174,199,209]. Future studies will uncover if relevant signaling pathways are particularly vulnerable to low SMN in specific cell types found in testes. In addition, it remains to be seen whether the aberrant expression of sex hormones by the reproductive organs causes a sex-specific impact on the development and function of other organs and tissues in SMA.
8. Sex-specific outcomes of SMA therapy
Considering that the overwhelming majority of SMA patients carry SMN2, correction of SMN2 exon 7 splicing emerged as the most attractive therapeutic option for the disease [255-258]. The discovery of intronic splicing silencer N1 (ISS-N1) revealed how an intronic site could be effectively targeted by an antisense oligonucleotide (ASO) to fully restore SMN2 exon 7 inclusion [258-262]. A number of in vivo studies independently validated the therapeutic efficacy of an ISS-N1-targeting ASO and confirmed the highest increase in the life expectancy in severe SMA mouse models [263-266]. Nusinersen (Spinraza), an ISS-N1-targeting ASO, became the first FDA approved therapy for SMA [260,261]. Gene therapy is the second approved therapy for SMA [267]. Both nusinersen and gene therapy have shown laudable outcomes in preventing death and ameliorating symptoms of the disease in SMA patients [268-270]. Recently approved Evrysdi (risdiplam) is the first orally deliverable small molecule for SMA therapy [271]. Similar to nusinersen, risdiplam promotes SMN2 exon 7 inclusion through targeting the sequence and/or structural elements at the 5′ splice site of exon 7 [258,272,273]. Additional orally deliverable small molecules currently in clinical trial for the treatment of SMA target either the SMN2 gene or downstream events [5,258]. While both sexes are benefitting from the currently approved drugs, no sex-specific outcome measures have been reported from preclinical and clinical studies of these drugs. However, a study conducted with an ISS-N2-targeting ASO showed sex-specific response in a mild SMA mouse model [224,274]. Of note, both ISS-N1- and ISS-N2-targeting ASOs abrogate the inhibitory context that sequesters the 5′ splice site of SMN2 exon 7 [232,258,260]. In particular, the study indicated that SMA female mice displayed better therapeutic outcomes than male mice upon treatment with ISS-N2-targeting ASO [224]. This finding is consistent with a recent report showing a better functional performance in a rotarod test by females than males of a mild SMA mouse model [176].
9. Conclusions
The housekeeping protein SMN is mechanistically linked to important functions in most cellular compartments, including mitochondria, which are known to have female sex-specific adaptations due to maternal inheritance. Reduced levels of SMN cause SMA characterized by a broad spectrum of pathologies (Table 1). SMA severity could be modified by multiple protein factors, some of which are encoded by genes located on the X chromosome. The development of ovaries and testes, organs that produce sex hormones (estrogen and testosterone), are differentially impacted by low levels of SMN. Given the important role of sex hormones in cellular metabolism coupled with the sex-specific bias brought in by mitochondria and X chromosomes, sex is likely to impact the structure and function of tissues expressing low levels of SMN in humans and animals. Unfortunately, the majority of SMA studies to date, either on animal models or patients, have used males and females together and interchangeably, without reporting any sex-dependent effects on disease progression, pathophysiology or response to treatment. Considering varied requirements of SMN in tissues at different developmental stages, capturing sex-specific variations during the early stages of SMA pathogenesis would be extremely valuable. However, this would require an arduous task of tissue-specific analysis of anatomical, physiological and biochemical parameters of sex- and age-matched patients coming from a similar genetic background. Recent studies employing animal models of SMA reveal sex-specific differences that increase with age [176]. Higher male susceptibility to the cumulative effects of oxidative stress [145] could be responsible for this observation. Testes appear to be one of the most and the earliest affected organs in all types of SMA. The adverse effect on testicular development with even a slight reduction of SMN provides a strong indication of the sexually dimorphic nature of SMA. Hence, deciphering the SMN-linked chain of molecular events during the testicular development has broad implications for uncovering novel transcriptional and posttranscriptional regulatory mechanisms.
Evaluating the sex-specific effects in model organisms and humans is of utmost importance for developing effective therapies [275,276]. There is a general concern about the immune response against the viral vector used for gene therapy for SMA [277]. Similar concerns could be raised for the antisense treatment as well as other potential SMA therapies [257,258,278]. Considering sex is an important determinant of immune response, the availability of sex-specific outcome measures would allow appropriate amendments to the existing treatment regimens or call for an altogether alternative treatment option. Emerging new data supporting sex-specific effects in motor neurons and other tissues in SMA warrant renewed efforts to discover novel therapeutic avenues for ameliorating the sex-specific pathologies in SMA.
Highlights:
Spinal muscular atrophy (SMA) is a disease of broad spectrum affecting infants, children and adults
Most tissues and organs are intrinsically (and independently) affected in SMA
Sex plays a role in determining the overall prevalence and severity of SMA
Male reproductive organ development and male fertility are adversely impacted in SMA
Acknowledgements
Authors acknowledge members of the Singh lab for critical reading. Authors have attempted to include relevant contributions on SMA and sex-specific studies, and they regret not being able to include several related references due to the lack of space.
Funding
This work was supported by grants from the National Institutes of Health (NIH) to RNS (R01NS055925) and PPR (R01HD094546). SH was supported by a NIH training grant (T35OD027967).
Footnotes
Declarations and competing interests
ISS-N1 target (US patent # 7,838,657) mentioned in this review was discovered in the Singh lab at UMASS Medical School (Worcester, MA, USA). Inventors, including RNS and UMASS Medical School, are currently benefiting from licensing of ISS-N1 target to IONIS Pharmaceuticals/Biogen, which is marketing Spinraza™ (Nusinersen), the FDA-approved drug, based on ISS-N1 target. RNS is co-founder of RNACorrect, Inc., an Iowa-based small business engaged in research and development.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lally C, Jones C, Farwell W, Reyna SP, Cook SF, Flanders WD, Indirect estimation of the prevalence of spinal muscular atrophy Type I, II, and III in the United States, Orphanet J Rare Dis 12(1) (2017) 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jha NN, Kim JK, Monani UR, Motor neuron biology and disease: A current perspective on infantile-onset spinal muscular atrophy, Future Neurol 13(3) (2018) 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Identification and characterization of a spinal muscular atrophy-determining gene, Cell 80(1) (1995) 155–65. [DOI] [PubMed] [Google Scholar]
- [4].Cho S, Dreyfuss G, A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity, Genes Dev 24(5) (2010) 438–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wirth B, Karakaya M, Kye MJ, Mendoza-Ferreira N, Twenty-Five Years of Spinal Muscular Atrophy Research: From Phenotype to Genotype to Therapy, and What Comes Next, Annu Rev Genomics Hum Genet 21 (2020) 231–261. [DOI] [PubMed] [Google Scholar]
- [6].Dubowitz V, Very severe spinal muscular atrophy (SMA type 0): an expanding clinical phenotype, Eur J Paediatr Neurol 3(2) (1999) 49–51. [DOI] [PubMed] [Google Scholar]
- [7].Zerres K, Rudnik-Schönebom S, Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications, Arch Neurol 52(5) (1995) 518–23. [DOI] [PubMed] [Google Scholar]
- [8].Wijngaarde CA, Stam M, Otto LAM, van Eijk RPA, Cuppen I, Veldhoen ES, van den Berg LH, Wadman RI, van der Pol WL, Population-based analysis of survival in spinal muscular atrophy, Neurology 94(15) (2020) e1634–e1644. [DOI] [PubMed] [Google Scholar]
- [9].Zerres K, Rudnik-Schönebom S, Forkert R, Wirth B, Genetic basis of adult-onset spinal muscular atrophy, Lancet 346(8983) (1995) 1162. [DOI] [PubMed] [Google Scholar]
- [10].Harding BN, Kariya S, Monani UR, Chung WK, Benton M, Yum SW, Tennekoon G, Finkel RS, Spectrum of neuropathophysiology in spinal muscular atrophy type I, J Neuropathol Exp Neurol 74(1) (2015) 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lipnick SL, Agniel DM, Aggarwal R, Makhortova NR, Finlayson SG, Brocato A, Palmer N, Darras BT, Kohane I, Rubin LL, Systemic nature of spinal muscular atrophy revealed by studying insurance claims, PLoS One 14(3) (2019) e0213680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bebee TW, Dominguez CE, Chandler DS, Mouse models of SMA: tools for disease characterization and therapeutic development, Hum Genet 131(8) (2012) 1277–93. [DOI] [PubMed] [Google Scholar]
- [13].Osborne M, Gomez D, Feng Z, McEwen C, Beltran J, Cirillo K, El-Khodor B, Lin MY, Li Y, Knowlton WM, McKemy DD, Bogdanik L, Butts-Dehm K, Martens K, Davis C, Doty R, Wardwell K, Ghavami A, Kobayashi D, Ko CP, Ramboz S, Lutz C, Characterization of behavioral and neuromuscular junction phenotypes in a novel allelic series of SMA mouse models, Hum Mol Genet 21(20) (2012) 4431–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bogdanik LP, Osborne MA, Davis C, Martin WP, Austin A, Rigo F, Bennett CF, Lutz CM, Systemic, postsymptomatic antisense oligonucleotide rescues motor unit maturation delay in a new mouse model for type II/III spinal muscular atrophy, Proc Natl Acad Sci U S A 112(43) (2015) E5863–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schrank B, Götz R, Gunnersen JM, Ure JM, Toyka KV, Smith AG, Sendtner M, Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos, Proc Natl Acad Sci U S A 94(18) (1997) 9920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H, A mouse model for spinal muscular atrophy, Nat Genet 24(1) (2000) 66–70. [DOI] [PubMed] [Google Scholar]
- [17].Monani UR, Sendtner M, Coovert DD, Parsons DW, Andreassi C, Le TT, Jablonka S, Schrank B, Rossoll W, Rossol W, Prior TW, Morris GE, Burghes AH, The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy, Hum Mol Genet 9(3) (2000) 333–9. [DOI] [PubMed] [Google Scholar]
- [18].Swoboda KJ, Prior TW, Scott CB, McNaught TP, Wride MC, Reyna SP, M B. Bromberg, Natural history of denervation in SMA: relation to age, SMN2 copy number, and function, Ann Neurol 57(5) (2005) 704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wirth B, Brichta L, Schrank B, Lochmüller H, Blick S, Baasner A, Heller R, Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number, Hum Genet 119(4) (2006) 422–8. [DOI] [PubMed] [Google Scholar]
- [20].Prior TW, Krainer AR, Hua Y, Swoboda KJ, Snyder PC, Bridgeman SJ, Burghes AH, Kissel JT, A positive modifier of spinal muscular atrophy in the SMN2 gene, Am J Hum Genet 85(3) (2009) 408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Oprea GE, Kröber S, McWhorter ML, Rossoll W, Müller S, Krawczak M, Bassell GJ, Beattie CE, Wirth B, Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy, Science 320(5875) (2008) 524–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yener İ, Topaloglu H, Erdem-Özdamar S, Dayangac-Erden D, Transcript levels of plastin 3 and neuritin 1 modifier genes in spinal muscular atrophy siblings, Pediatr Int 59(1) (2017) 53–56. [DOI] [PubMed] [Google Scholar]
- [23].Riessland M, Kaczmarek A, Schneider S, Swoboda KJ, Löhr H, Bradler C, Grysko V, Dimitriadi M, Hosseinibarkooie S, Torres-Benito L, Peters M, Upadhyay A, Biglari N, Kröber S, Hölker I, Garbes L, Gilissen C, Hoischen A, Nürnberg G, Nürnberg P, Walter M, Rigo F, Bennett CF, Kye MJ, Hart AC, Hammerschmidt M, Kloppenburg P, Wirth B, Neurocalcin Delta Suppression Protects against Spinal Muscular Atrophy in Humans and across Species by Restoring Impaired Endocytosis, Am J Hum Genet 100(2) (2017) 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Singh NN, Seo J, Ottesen EW, Shishimorova M, Bhattacharya D, Singh RN, TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy, Mol Cell Biol 31(5) (2011) 935–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Howell MD, Ottesen EW, Singh NN, Anderson RL, Seo J, Sivanesan S, Whitley EM, Singh RN, TIA1 is a gender-specific disease modifier of a mild mouse model of spinal muscular atrophy, Sci Rep 7(1) (2017) 7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Han KJ, Foster DG, Zhang NY, Kanisha K, Dzieciatkowska M, Sclafani RA, Hansen KC, Peng J, Liu CW, Ubiquitin-specific protease 9x deubiquitinates and stabilizes the spinal muscular atrophy protein-survival motor neuron, J Biol Chem 287(52) (2012) 43741–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Powis RA, Karyka E, Boyd P, Côme J, Jones RA, Zheng Y, Szunyogova E, Groen EJ, Hunter G, Thomson D, Wishart TM, Becker CG, Parson SH, Martinat C, Azzouz M, Gillingwater TH, Systemic restoration of UBA1 ameliorates disease in spinal muscular atrophy, JCI Insight 1(11) (2016) e87908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Villalón E, Kline RA, Smith CE, Lorson ZC, Osman EY, O'Day S, Murray LM, Lorson CL, AAV9-Stathmin1 gene delivery improves disease phenotype in an intermediate mouse model of spinal muscular atrophy, Hum Mol Genet 28(22) (2019) 3742–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhou H, Meng J, Malerba A, Catapano F, Sintusek P, Jarmin S, Feng L, Lu-Nguyen N, Sun L, Mariot V, Dumonceaux J, Morgan JE, Gissen P, Dickson G, Muntoni F, Myostatin inhibition in combination with antisense oligonucleotide therapy improves outcomes in spinal muscular atrophy, J Cachexia Sarcopenia Muscle 11(3) (2020) 768–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kannan A, Jiang X, He L, Ahmad S, Gangwani L, ZPR1 prevents R-loop accumulation, upregulates SMN2 expression and rescues spinal muscular atrophy, Brain 143(1) (2020) 69–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Singh RN, Howell MD, Ottesen EW, Singh NN, Diverse role of survival motor neuron protein, Biochim Biophys Acta Gene Regul Mech 1860(3) (2017) 299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ottesen EW, Singh NN, Luo D, Singh RN, High-affinity RNA targets of the Survival Motor Neuron protein reveal diverse preferences for sequence and structural motifs, Nucleic Acids Res 46(20) (2018) 10983–11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Singh NN, Seo J, Rahn SJ, Singh RN, A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes, PLoS One 7(11) (2012) e49595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Seo J, Singh NN, Ottesen EW, Lee BM, Singh RN, A novel human-specific splice isoform alters the critical C-terminus of Survival Motor Neuron protein, Sci Rep 6 (2016) 30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Seo J, Singh NN, Ottesen EW, Sivanesan S, Shishimorova M, Singh RN, Oxidative Stress Triggers Body-Wide Skipping of Multiple Exons of the Spinal Muscular Atrophy Gene, PLoS One 11(4) (2016) e0154390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].d'Ydewalle C, Ramos DM, Pyles NJ, Ng SY, Gorz M, Pilato CM, Ling K, Kong L, Ward AJ, Rubin LL, Rigo F, Bennett CF, Sumner CJ, The Antisense Transcript SMN-AS1 Regulates SMN Expression and Is a Novel Therapeutic Target for Spinal Muscular Atrophy, Neuron 93(1) (2017) 66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Woo CJ, Maier VK, Davey R, Brennan J, Li G, Brothers J, Schwartz B, Gordo S, Kasper A, Okamoto TR, Johansson HE, Mandefro B, Sareen D, Bialek P, Chau BN, Bhat B, Bullough D, Barsoum J, Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy, Proc Natl Acad Sci U S A 114(8) (2017) E1509–E1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ottesen EW, Seo J, Singh NN, Singh RN, A multilayered control of the human Survival Motor Neuron gene expression by Alu elements, Front Microbiol 8 (2017) 2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ottesen EW, Luo D, Seo J, Singh NN, Singh RN, Human Survival Motor Neuron genes generate a vast repertoire of circular RNAs, Nucleic Acids Res 47(6) (2019) 2884–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pagliarini V, Jolly A, Bielli P, Di Rosa V, De la Grange P, Sette C, Sam68 binds Alu-rich introns in SMN and promotes pre-mRNA circularization, Nucleic Acids Res 48(2) (2020) 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ottesen EW, Singh RN, Characteristics of circular RNAs generated by human Survival Motor Neuron genes, Cell Signal 73 (2020) 109696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Singh NN, Ottesen EW, Singh RN, A survey of transcripts generated by spinal muscular atrophy genes, Biochim Biophys Acta Gene Regul Mech 1863(8) (2020) 194562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bertoli S, De Amicis R, Mastella C, Pieri G, Giaquinto E, Battezzati A, Leone A, Baranello G, Spinal Muscular Atrophy, types I and II: What are the differences in body composition and resting energy expenditure?, Clin Nutr 36(6) (2017) 1674–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Baranello G, De Amicis R, Arnoldi MT, Zanin R, Mastella C, Masson R, Leone A, Alberti K, Foppiani A, Battezzati A, Bertoli S, Evaluation of body composition as a potential biomarker in spinal muscular atrophy, Muscle Nerve 61(4) (2020) 530–534. [DOI] [PubMed] [Google Scholar]
- [45].Martinez EE, Quinn N, Arouchon K, Anzaldi R, Tarrant S, Ma NS, Griffin J, Darras BT, Graham RJ, Mehta NM, Comprehensive nutritional and metabolic assessment in patients with spinal muscular atrophy: Opportunity for an individualized approach, Neuromuscul Disord 28(6) (2018) 512–519. [DOI] [PubMed] [Google Scholar]
- [46].Somers E, Stencel Z, Wishart TM, Gillingwater TH, Parson SH, Density, calibre and ramification of muscle capillaries are altered in a mouse model of severe spinal muscular atrophy, Neuromuscul Disord 22(5) (2012) 435–42. [DOI] [PubMed] [Google Scholar]
- [47].Shababi M, Habibi J, Ma L, Glascock JJ, Sowers JR, Lorson CL, Partial restoration of cardio-vascular defects in a rescued severe model of spinal muscular atrophy, J Mol Cell Cardiol 52(5) (2012) 1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Felderhoff-Mueser U, Grohmann K, Harder A, Stadelmann C, Zerres K, Bührer C, Obladen M, Severe spinal muscular atrophy variant associated with congenital bone fractures, J Child Neurol 17(9) (2002) 718–21. [DOI] [PubMed] [Google Scholar]
- [49].Vaidla E, Talvik I, Kulla A, Sibul H, Maasalu K, Metsvaht T, Piirsoo A, Talvik T, Neonatal spinal muscular atrophy type 1 with bone fractures and heart defect, J Child Neurol 22(1) (2007) 67–70. [DOI] [PubMed] [Google Scholar]
- [50].Khatri IA, Chaudhry US, Seikaly MG, Browne RH, Iannaccone ST, Low bone mineral density in spinal muscular atrophy, J Clin Neuromuscul Dis 10(1) (2008) 11–7. [DOI] [PubMed] [Google Scholar]
- [51].Vestergaard P, Glerup H, Steffensen BF, Rejnmark L, Rahbek J, Moseklide L, Fracture risk in patients with muscular dystrophy and spinal muscular atrophy, J Rehabil Med 33(4) (2001) 150–5. [PubMed] [Google Scholar]
- [52].Kinali M, Banks LM, Mercuri E, Manzur AY, Muntoni F, Bone mineral density in a pediatric spinal muscular atrophy population, Neuropediatrics 35(6) (2004) 325–8. [DOI] [PubMed] [Google Scholar]
- [53].Vai S, Bianchi ML, Moroni I, Mastella C, Broggi F, Morandi L, Arnoldi MT, Bussolino C, Baranello G, Bone and Spinal Muscular Atrophy, Bone 79 (2015) 116–20. [DOI] [PubMed] [Google Scholar]
- [54].Wasserman HM, Hornung LN, Stenger PJ, Rutter MM, Wong BL, Rybalsky I, Khoury JC, Kalkwarf HJ, Low bone mineral density and fractures are highly prevalent in pediatric patients with spinal muscular atrophy regardless of disease severity, Neuromuscul Disord 27(4) (2017) 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hensel N, Brickwedde H, Tsaknakis K, Grages A, Braunschweig L, Lüders KA, Lorenz HM, Lippross S, Walter LM, Tavassol F, Lienenklaus S, Neunaber C, Claus P, Hell AK, Altered bone development with impaired cartilage formation precedes neuromuscular symptoms in spinal muscular atrophy, Hum Mol Genet 29(16) (2020) 2662–2673. [DOI] [PubMed] [Google Scholar]
- [56].Shanmugarajan S, Tsuruga E, Swoboda KJ, Maria BL, Ries WL, Reddy SV, Bone loss in survival motor neuron (Smn(−/−) SMN2) genetic mouse model of spinal muscular atrophy, J Pathol 219(1) (2009) 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Park GH, Maeno-Hikichi Y, Awano T, Landmesser LT, Monani UR, Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene, J Neurosci 30(36) (2010) 12005–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Higashi K, Nakagawa M, Higuchi I, Saito K, Osame M, [Genetically confirmed spinal muscular atrophy type III with epilepsy, cerebral hypoperfusion, and parahippocampal gyrus atrophy], Rinsho Shinkeigaku 40(4) (2000) 334–8. [PubMed] [Google Scholar]
- [59].Wishart TM, Huang JP, Murray LM, Lamont DJ, Mutsaers CA, Ross J, Geldsetzer P, Ansorge O, Talbot K, Parson SH, Gillingwater TH, SMN deficiency disrupts brain development in a mouse model of severe spinal muscular atrophy, Hum Mol Genet 19(21) (2010) 4216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Messina S, Pane M, De Rose P, Vasta I, Sorleti D, Aloysius A, Sciarra F, Mangiola F, Kinali M, Bertini E, Mercuri E, Feeding problems and malnutrition in spinal muscular atrophy type II, Neuromuscul Disord 18(5) (2008) 389–93. [DOI] [PubMed] [Google Scholar]
- [61].Gombash SE, Cowley CJ, Fitzgerald JA, Iyer CC, Fried D, McGovern VL, Williams KC, Burghes AH, Christofi FL, Gulbransen BD, Foust KD, SMN deficiency disrupts gastrointestinal and enteric nervous system function in mice, Hum Mol Genet 24(19) (2015) 5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sintusek P, Catapano F, Angkathunkayul N, Marrosu E, Parson SH, Morgan JE, Muntoni F, Zhou H, Histopathological Defects in Intestine in Severe Spinal Muscular Atrophy Mice Are Improved by Systemic Antisense Oligonucleotide Treatment, PLoS One 11(5) (2016) e0155032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schreml J, Riessland M, Paterno M, Garbes L, Roßbach K, Ackermann B, Krämer J, Somers E, Parson SH, Heller R, Berkessel A, Sterner-Kock A, Wirth B, Severe SMA mice show organ impairment that cannot be rescued by therapy with the HDACi JNJ-26481585, Eur J Hum Genet 21(6) (2013) 643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Rudnik-Schöneborn S, Heller R, Berg C, Betzler C, Grimm T, Eggermann T, Eggermann K, Wirth R, Wirth B, Zerres K, Congenital heart disease is a feature of severe infantile spinal muscular atrophy, J Med Genet 45(10) (2008) 635–8. [DOI] [PubMed] [Google Scholar]
- [65].Bach JR, Medical considerations of long-term survival of Werdnig-Hoffmann disease, Am J Phys Med Rehabil 86(5) (2007) 349–55. [DOI] [PubMed] [Google Scholar]
- [66].Wijngaarde CA, Blank AC, Stam M, Wadman RI, van den Berg LH, van der Pol WL, Cardiac pathology in spinal muscular atrophy: a systematic review, Orphanet J Rare Dis 12(1) (2017) 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Menke LA, Poll-The BT, Clur SA, Bilardo CM, van der Wal AC, Lemmink HH, Cobben JM, Congenital heart defects in spinal muscular atrophy type I: a clinical report of two siblings and a review of the literature, Am J Med Genet A 146A(6) (2008) 740–4. [DOI] [PubMed] [Google Scholar]
- [68].Finsterer J, Stöllberger C, Cardiac involvement in Werdnig-Hoffmann's spinal muscular atrophy, Cardiology 92(3) (1999) 178–82. [DOI] [PubMed] [Google Scholar]
- [69].Bevan AK, Hutchinson KR, Foust KD, Braun L, McGovern VL, Schmelzer L, Ward JG, Petruska JC, Lucchesi PA, Burghes AH, Kaspar BK, Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery, Hum Mol Genet 19(20) (2010) 3895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Heier CR, Satta R, Lutz C, DiDonato CJ, Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice, Hum Mol Genet 19(20) (2010) 3906–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Shababi M, Habibi J, Yang HT, Vale SM, Sewell WA, Lorson CL, Cardiac defects contribute to the pathology of spinal muscular atrophy models, Hum Mol Genet 19(20) (2010) 4059–71. [DOI] [PubMed] [Google Scholar]
- [72].Szunyogova E, Zhou H, Maxwell GK, Powis RA, Muntoni F, Gillingwater TH, Parson SH, Survival Motor Neuron (SMN) protein is required for normal mouse liver development, Sci Rep 6 (2016) 34635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kapur N, Deegan S, Parakh A, Gauld L, Relationship between respiratory function and need for NIV in childhood SMA, Pediatr Pulmonol 54(11) (2019) 1774–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Deguise MO, Baranello G, Mastella C, Beauvais A, Michaud J, Leone A, De Amicis R, Battezzati A, Dunham C, Selby K, Warman Chardon J, McMillan HJ, Huang YT, Courtney NL, Mole AJ, Kubinski S, Claus P, Murray LM, Bowerman M, Gillingwater TH, Bertoli S, Parson SH, Kothary R, Abnormal fatty acid metabolism is a core component of spinal muscular atrophy, Ann Clin Transl Neurol 6(8) (2019) 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Vitte JM, Davoult B, Roblot N, Mayer M, Joshi V, Courageot S, Tronche F, Vadrot J, Moreau MH, Kemeny F, Melki J, Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload, Am J Pathol 165(5) (2004) 1731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Fujak A, Ingenhorst A, Heuser K, Forst R, Forst J, Treatment of scoliosis in intermediate spinal muscular atrophy (SMA type II) in childhood, Ortop Traumatol Rehabil 7(2) (2005) 175–9. [PubMed] [Google Scholar]
- [77].Shanmugarajan S, Tsuruga E, Swoboda KJ, Maria BL, Ries WL, Reddy SV, Bone loss in survival motor neuron (Smn(−/−) SMN2) genetic mouse model of spinal muscular atrophy, J Pathol 219(1) (2009) 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Nadeau A, D'Anjou G, Debray G, Robitaille Y, Simard LR, Vanasse M, A newborn with spinal muscular atrophy type 0 presenting with a clinicopathological picture suggestive of myotubular myopathy, J Child Neurol 22(11) (2007) 1301–4. [DOI] [PubMed] [Google Scholar]
- [79].Harding BN, Kariya S, Monani UR, Chung WK, Benton M, Yum SW, Tennekoon G, Finkel RS, Spectrum of neuropathophysiology in spinal muscular atrophy type I, J Neuropathol Exp Neurol 74(1) (2015) 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Martínez-Hernández R, Soler-Botija C, Also E, Alias L, Caselles L, Gich I, Bernal S, Tizzano EF, The developmental pattern of myotubes in spinal muscular atrophy indicates prenatal delay of muscle maturation, J Neuropathol Exp Neurol 68(5) (2009) 474–81. [DOI] [PubMed] [Google Scholar]
- [81].Martínez-Hernández R, Bernal S, Alias L, Tizzano EF, Abnormalities in early markers of muscle involvement support a delay in myogenesis in spinal muscular atrophy, J Neuropathol Exp Neurol 73(6) (2014) 559–67. [DOI] [PubMed] [Google Scholar]
- [82].Ripolone M, Ronchi D, Violano R, Vallejo D, Fagiolari G, Barca E, Lucchini V, Colombo I, Villa L, Berardinelli A, Balottin U, Morandi L, Mora M, Bordoni A, Fortunato F, Corti S, Parisi D, Toscano A, Sciacco M, DiMauro S, Comi GP, Moggio M, Impaired Muscle Mitochondrial Biogenesis and Myogenesis in Spinal Muscular Atrophy, JAMA Neurol 72(6) (2015) 666–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Duque SI, Arnold WD, Odermatt P, Li X, Porensky PN, Schmelzer L, Meyer K, Kolb SJ, Schümperli D, Kaspar BK, Burghes AH, A large animal model of spinal muscular atrophy and correction of phenotype, Ann Neurol 77(3) (2015) 399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Walker MP, Rajendra TK, Saieva L, Fuentes JL, Pellizzoni L, Matera AG, SMN complex localizes to the sarcomeric Z-disc and is a proteolytic target of calpain, Hum Mol Genet 17(21) (2008) 3399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hayhurst M, Wagner AK, Cerletti M, Wagers AJ, Rubin LL, A cell-autonomous defect in skeletal muscle satellite cells expressing low levels of survival of motor neuron protein, Dev Biol 368(2) (2012) 323–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Dachs E, Hereu M, Piedrafita L, Casanovas A, Calderó J, Esquerda JE, Defective neuromuscular junction organization and postnatal myogenesis in mice with severe spinal muscular atrophy, J Neuropathol Exp Neurol 70(6) (2011) 444–61. [DOI] [PubMed] [Google Scholar]
- [87].Boyer JG, Murray LM, Scott K, De Repentigny Y, Renaud JM, Kothary R, Early onset muscle weakness and disruption of muscle proteins in mouse models of spinal muscular atrophy, Skelet Muscle 3(1) (2013) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Lee YI, Mikesh M, Smith I, Rimer M, Thompson W, Muscles in a mouse model of spinal muscular atrophy show profound defects in neuromuscular development even in the absence of failure in neuromuscular transmission or loss of motor neurons, Dev Biol 356(2) (2011) 432–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kim JK, Jha NN, Feng Z, Faleiro MR, Chiriboga CA, Wei-Lapierre L, Dirksen RT, Ko CP, Monani UR, Muscle-specific SMN reduction reveals motor neuron-independent disease in spinal muscular atrophy models, J Clin Invest 130(3) (2020) 1271–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Cifuentes-Diaz C, Frugier T, Tiziano FD, Lacène E, Roblot N, Joshi V, Moreau MH, Melki J, Deletion of murine SMN exon 7 directed to skeletal muscle leads to severe muscular dystrophy, J Cell Biol 152(5) (2001) 1107–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Nicole S, Desforges B, Millet G, Lesbordes J, Cifuentes-Diaz C, Vertes D, Cao ML, De Backer E, Languille L, Roblot N, Joshi V, Gillis JM, Melki J, Intact satellite cells lead to remarkable protection against Smn gene defect in differentiated skeletal muscle, J Cell Biol 161(3) (2003) 571–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Imlach WL, Beck ES, Choi BJ, Lotti F, Pellizzoni L, McCabe BD, SMN is required for sensory-motor circuit function in Drosophila, Cell 151(2) (2012) 427–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Rossoll W, Jablonka S, Andreassi C, Kröning AK, Karle K, Monani UR, Sendtner M, Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons, J Cell Biol 163(4) (2003) 801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Jablonka S, Beck M, Lechner BD, Mayer C, Sendtner M, Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy, J Cell Biol 179(1) (2007) 139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Soler-Botija C, Ferrer I, Gich I, Baiget M, Tizzano EF, Neuronal death is enhanced and begins during foetal development in type I spinal muscular atrophy spinal cord, Brain 125(Pt 7) (2002) 1624–34. [DOI] [PubMed] [Google Scholar]
- [96].Rudnik-Schöneborn S, Goebel HH, Schlote W, Molaian S, Omran H, Ketelsen U, Korinthenberg R, Wenzel D, Lauffer H, Kreiss-Nachtsheim M, Wirth B, Zerres K, Classical infantile spinal muscular atrophy with SMN deficiency causes sensory neuronopathy, Neurology 60(6) (2003) 983–7. [DOI] [PubMed] [Google Scholar]
- [97].Anagnostou E, Miller SP, Guiot MC, Karpati G, Simard L, Dilenge ME, Shevell MI, Type I spinal muscular atrophy can mimic sensory-motor axonal neuropathy, J Child Neurol 20(2) (2005) 147–50. [DOI] [PubMed] [Google Scholar]
- [98].Rindt H, Feng Z, Mazzasette C, Glascock JJ, Valdivia D, Pyles N, Crawford TO, Swoboda KJ, Patitucci TN, Ebert AD, Sumner CJ, Ko CP, Lorson CL, Astrocytes influence the severity of spinal muscular atrophy, Hum Mol Genet 24(14) (2015) 4094–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Simic G, Mladinov M, Seso Simic D, Jovanov Milosevic N, Islam A, Pajtak A, Barisic N, Sertic J, Lucassen PJ, Hof PR, Kruslin B, Abnormal motoneuron migration, differentiation, and axon outgrowth in spinal muscular atrophy, Acta Neuropathol 115(3) (2008) 313–26. [DOI] [PubMed] [Google Scholar]
- [100].Jablonka S, Karle K, Sandner B, Andreassi C, von Au K, Sendtner M, Distinct and overlapping alterations in motor and sensory neurons in a mouse model of spinal muscular atrophy, Hum Mol Genet 15(3) (2006) 511–8. [DOI] [PubMed] [Google Scholar]
- [101].Ling KK, Lin MY, Zingg B, Feng Z, Ko CP, Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy, PLoS One 5(11) (2010) e15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Mentis GZ, Blivis D, Liu W, Drobac E, Crowder ME, Kong L, Alvarez FJ, Sumner CJ, O'Donovan MJ, Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy, Neuron 69(3) (2011) 453–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].McGivern JV, Patitucci TN, Nord JA, Barabas MA, Stucky CL, Ebert AD, Spinal muscular atrophy astrocytes exhibit abnormal calcium regulation and reduced growth factor production, Glia 61(9) (2013) 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Tarabal O, Caraballo-Miralles V, Cardona-Rossinyol A, Correa FJ, Olmos G, Lladó J, Esquerda JE, Calderó J, Mechanisms involved in spinal cord central synapse loss in a mouse model of spinal muscular atrophy, J Neuropathol Exp Neurol 73(6) (2014) 519–35. [DOI] [PubMed] [Google Scholar]
- [105].Gogliotti RG, Quinlan KA, Barlow CB, Heier CR, Heckman CJ, Didonato CJ, Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction, J Neurosci 32(11) (2012) 3818–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Hunter G, Aghamaleky Sarvestany A, Roche SL, Symes RC, Gillingwater TH, SMN-dependent intrinsic defects in Schwann cells in mouse models of spinal muscular atrophy, Hum Mol Genet 23(9) (2014) 2235–50. [DOI] [PubMed] [Google Scholar]
- [107].McWhorter ML, Monani UR, Burghes AH, Beattie CE, Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding, J Cell Biol 162(5) (2003) 919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Carrel TL, McWhorter ML, Workman E, Zhang H, Wolstencroft EC, Lorson C, Bassell EJ, Burghes AH, Beattie CE, Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis, J Neurosci 26(43) (2006) 11014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Hachiya Y, Arai H, Hayashi M, Kumada S, Furushima W, Ohtsuka E, Ito Y, Uchiyama A, Kurata K, Autonomic dysfunction in cases of spinal muscular atrophy type 1 with long survival, Brain Dev 27(8) (2005) 574–8. [DOI] [PubMed] [Google Scholar]
- [110].Araujo A, Araujo M, Swoboda KJ, Vascular perfusion abnormalities in infants with spinal muscular atrophy, J Pediatr 155(2) (2009) 292–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Rudnik-Schöneborn S, Vogelgesang S, Armbrust S, Graul-Neumann L, Fusch C, Zerres K, Digital necroses and vascular thrombosis in severe spinal muscular atrophy, Muscle Nerve 42(1) (2010) 144–7. [DOI] [PubMed] [Google Scholar]
- [112].Arai H, Tanabe Y, Hachiya Y, Otsuka E, Kumada S, Furushima W, Kohyama J, Yamashita S, Takanashi J, Kohno Y, Finger cold-induced vasodilatation, sympathetic skin response, and R-R interval variation in patients with progressive spinal muscular atrophy, J Child Neurol 20(11) (2005) 871–5. [DOI] [PubMed] [Google Scholar]
- [113].Murray LM, Comley LH, Thomson D, Parkinson N, Talbot K, Gillingwater TH, Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy, Hum Mol Genet 17(7) (2008) 949–62. [DOI] [PubMed] [Google Scholar]
- [114].Kariya S, Park GH, Maeno-Hikichi Y, Leykekhman O, Lutz C, Arkovitz MS, Landmesser LT, Monani UR, Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy, Hum Mol Genet 17(16) (2008) 2552–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kong L, Wang X, Choe DW, Polley M, Burnett BG, Bosch-Marcé M, Griffin JW, Rich MM, Sumner CJ, Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice, J Neurosci 29(3) (2009) 842–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Ling KK, Gibbs RM, Feng Z, Ko CP, Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy, Hum Mol Genet 21(1) (2012) 185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Cifuentes-Diaz C, Nicole S, Velasco ME, Borra-Cebrian C, Panozzo C, Frugier T, Millet G, Roblot N, Joshi V, Melki J, Neurofilament accumulation at the motor endplate and lack of axonal sprouting in a spinal muscular atrophy mouse model, Hum Mol Genet 11(12) (2002) 1439–47. [DOI] [PubMed] [Google Scholar]
- [118].Ottesen EW, Howell MD, Singh NN, Seo J, Whitley EM, Singh RN, Severe impairment of male reproductive organ development in a low SMN expressing mouse model of spinal muscular atrophy, Sci Rep 6 (2016) 20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Chang WF, Xu J, Lin TY, Hsu J, Hsieh-Li HM, Hwu YM, Liu JL, Lu CH, Sung LY, Survival Motor Neuron Protein Participates in Mouse Germ Cell Development and Spermatogonium Maintenance, Int J Mol Sci 21(3) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Bowerman M, Swoboda KJ, Michalski JP, Wang GS, Reeks C, Beauvais A, Murphy K, Woulfe J, Screaton RA, Scott FW, Kothary R, Glucose metabolism and pancreatic defects in spinal muscular atrophy, Ann Neurol 72(2) (2012) 256–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Bowerman M, Michalski JP, Beauvais A, Murray LM, DeRepentigny Y, Kothary R, Defects in pancreatic development and glucose metabolism in SMN-depleted mice independent of canonical spinal muscular atrophy neuromuscular pathology, Hum Mol Genet 23(13) (2014) 3432–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Nery FC, Siranosian JJ, Rosales I, Deguise MO, Sharma A, Muhtaseb AW, Nwe P, Johnstone AJ, Zhang R, Fatouraei M, Huemer N, Alves CRR, Kothary R, Swoboda KJ, Impaired kidney structure and function in spinal muscular atrophy, Neurol Genet 5(5) (2019) e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Khawaja K, Houlsby WT, Watson S, Bushby K, Cheetham T, Hypercalcaemia in infancy; a presenting feature of spinal muscular atrophy, Arch Dis Child 89(4) (2004) 384–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Thomson AK, Somers E, Powis RA, Shorrock HK, Murphy K, Swoboda KJ, Gillingwater TH, Parson SH, Survival of motor neurone protein is required for normal postnatal development of the spleen, J Anat 230(2) (2017) 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Deguise MO, Beauvais A, Schneider BL, Kothary R, Blood Flow to the Spleen is Altered in a Mouse Model of Spinal Muscular Atrophy, J Neuromuscul Dis 7(3) (2020) 315–322. [DOI] [PubMed] [Google Scholar]
- [126].Khairallah MT, Astroski J, Custer SK, Androphy EJ, Franklin CL, Lorson CL, SMN deficiency negatively impacts red pulp macrophages and spleen development in mouse models of spinal muscular atrophy, Hum Mol Genet 26(5) (2017) 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Brener A, Lebenthal Y, Shtamler A, Levy S, Stein R, Fattal-Valevski A, Sagi L, The endocrine manifestations of spinal muscular atrophy, a real-life observational study, Neuromuscul Disord 30(4) (2020) 270–276. [DOI] [PubMed] [Google Scholar]
- [128].Deguise MO, De Repentigny Y, McFall E, Auclair N, Sad S, Kothary R, Immune dysregulation may contribute to disease pathogenesis in spinal muscular atrophy mice, Hum Mol Genet 26(4) (2017) 801–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].von Gontard A, Laufersweiler-Plass C, Backes M, Zerres K, Rudnik-Schöneborn S, Enuresis and urinary incontinence in children and adolescents with spinal muscular atrophy, BJU Int 88(4) (2001) 409–13. [DOI] [PubMed] [Google Scholar]
- [130].Roth JD, Pariser JJ, Stout TE, Misseri R, Elliott SP, Presentation and Management Patterns of Lower Urinary Tract Symptoms in Adults Due to Rare Inherited Neuromuscular Diseases, Urology 135 (2020) 165–170. [DOI] [PubMed] [Google Scholar]
- [131].Yang Y, Vassilakos G, Hammers DW, Yang Z, Barton ER, Sweeney HL, Smooth muscle atrophy and colon pathology in SMN deficient mice, Am J Transl Res 11(3) (2019) 1789–1799. [PMC free article] [PubMed] [Google Scholar]
- [132].Somers E, Lees RD, Hoban K, Sleigh JN, Zhou H, Muntoni F, Talbot K, Gillingwater TH, Parson SH, Vascular Defects and Spinal Cord Hypoxia in Spinal Muscular Atrophy, Ann Neurol 79(2) (2016) 217–30. [DOI] [PubMed] [Google Scholar]
- [133].Kelley RI, Sladky JT, Dicarboxylic aciduria in an infant with spinal muscular atrophy, Ann Neurol 20(6) (1986) 734–6. [DOI] [PubMed] [Google Scholar]
- [134].Crawford TO, Sladky JT, Hurko O, Besner-Johnston A, Kelley RI, Abnormal fatty acid metabolism in childhood spinal muscular atrophy, Ann Neurol 45(3) (1999) 337–43. [DOI] [PubMed] [Google Scholar]
- [135].Zolkipli Z, Sherlock M, Biggar WD, Taylor G, Hutchison JS, Peliowski A, Alman BA, Ling SC, Tein I, Abnormal fatty acid metabolism in spinal muscular atrophy may predispose to perioperative risks, Eur J Paediatr Neurol 16(5) (2012) 549–53. [DOI] [PubMed] [Google Scholar]
- [136].Tein I, Sloane AE, Donner EJ, Lehotay DC, Millington DS, Kelley RI, Fatty acid oxidation abnormalities in childhood-onset spinal muscular atrophy: primary or secondary defect(s)?, Pediatr Neurol 12(1) (1995) 21–30. [DOI] [PubMed] [Google Scholar]
- [137].Kölbel H, Hauffa BP, Wudy SA, Bouikidis A, Della Marina A, Schara U, Hyperleptinemia in children with autosomal recessive spinal muscular atrophy type I-III, PLoS One 12(3) (2017) e0173144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Ramos DM, d'Ydewalle C, Gabbeta V, Dakka A, Klein SK, Norris DA, Matson J, Taylor SJ, Zaworski PG, Prior TW, Snyder PJ, Valdivia D, Hatem CL, Waters I, Gupte N, Swoboda KJ, Rigo F, Bennett CF, Naryshkin N, Paushkin S, Crawford TO, Sumner CJ, Age-dependent SMN expression in disease-relevant tissue and implications for SMA treatment, J Clin Invest 129(11) (2019) 4817–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G, SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing, Cell 133(4) (2008) 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Zhang Z, Pinto AM, Wan L, Wang W, Berg MG, Oliva I, Singh LN, Dengler C, Wei Z, Dreyfuss G, Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy, Proc Natl Acad Sci U S A 110(48) (2013) 19348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Motyl AAL, Faller KME, Groen EJN, Kline RA, Eaton SL, Ledahawsky LM, Chaytow H, Lamont DJ, Wishart TM, Huang YT, Gillingwater TH, Pre-natal manifestation of systemic developmental abnormalities in spinal muscular atrophy, Hum Mol Genet 29(16) (2020) 2674–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Deegan DF, Engel N, Sexual Dimorphism in the Age of Genomics: How, When, Where, Front Cell Dev Biol 7 (2019) 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Gegenhuber B, Tollkuhn J, Signatures of sex: Sex differences in gene expression in the vertebrate brain, Wiley Interdiscip Rev Dev Biol 9(1) (2020) e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Ventura-Clapier R, Moulin M, Piquereau J, Lemaire C, Mericskay M, Veksler V, Garnier A, Mitochondria: a central target for sex differences in pathologies, Clin Sci (Lond) 131(9) (2017) 803–822. [DOI] [PubMed] [Google Scholar]
- [145].Tower J, Pomatto LCD, Davies KJA, Sex differences in the response to oxidative and proteolytic stress, Redox Biol 31 (2020) 101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Gillies GE, McArthur S, Estrogen actions in the brain and the basis for differential action in men and women: a case for sex-specific medicines, Pharmacol Rev 62(2) (2010) 155–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Buzzetti E, Parikh PM, Gerussi A, Tsochatzis E, Gender differences in liver disease and the drug-dose gender gap, Pharmacol Res 120 (2017) 97–108. [DOI] [PubMed] [Google Scholar]
- [148].Mohadjer A, Brown G, Shah SR, Nallapati C, Waheed N, Bavry AA, Park K, Sex-Based Differences in Coronary and Structural Percutaneous Interventions, Cardiol Ther (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Xavier MJ, Salas-Huetos A, Oud MS, Aston KI, Veltman JA, Disease gene discovery in male infertility: past, present and future, Hum Genet (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Mazure CM, Swendsen J, Sex differences in Alzheimer's disease and other dementias, Lancet Neurol 15(5) (2016) 451–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Cerri S, Mus L, Blandini F, Parkinson's Disease in Women and Men: What's the Difference?, J Parkinsons Dis 9(3) (2019) 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Li R, Strykowski R, Meyer M, Mulcrone P, Krakora D, Suzuki M, Male-specific differences in proliferation, neurogenesis, and sensitivity to oxidative stress in neural progenitor cells derived from a rat model of ALS, PLoS One 7(11) (2012) e48581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Lim KRQ, Sheri N, Nguye Q, Yokota T. Cardiac involvement in Dystrophin-Deficient Females: Current Understanding and Implications for the Treatment of Dystrophinopathies, Genes (Basel), 2020, 11, 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Viggiano E, Ergoli M, Picillo E, Politano L. Determining the role of skewed X-chromosome inactivation in developing muscle symptoms in carriers of Duchenne muscular dystrophy. Hum. Genet 2016, 135, 685–698. [DOI] [PubMed] [Google Scholar]
- [155].Ishizaki M, Kobayashi M, Adachi K, Matsumura T, Kimura E. Female dystrophinopathy: Review of current literature. Neuromuscul. Disord 2018;28:572–581. [DOI] [PubMed] [Google Scholar]
- [156].Finsterer J, Stollberger C. Muscle, cardiac, and cerebral manifestations in female carriers of dystrophin variants. J. Neurol. Sci 2018, 388, 107–108. [DOI] [PubMed] [Google Scholar]
- [157].Fanin M, Nascimbeni AC, Angelini C, Gender difference in limb-girdle muscular dystrophy: a muscle fiber morphometric study in 101 patients, Clin Neuropathol 33(3) (2014) 179–85. [DOI] [PubMed] [Google Scholar]
- [158].Luger T, Seibt R, Rieger MA, Steinhilber B, Sex differences in muscle activity and motor variability in response to a non-fatiguing repetitive screwing task, Biol Sex Differ 11(1) (2020) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Tawil R, Van Der Maarel SM. Facioscapulohumeral muscular dystrophy. Muscle Nerve. 34 (2006) 1–15., [DOI] [PubMed] [Google Scholar]
- [160].Zatz M, Marie SK, Cerqueira A, Vainzof M, Pavanello RC, Passos-Bueno MR. The facioscapulohumeral muscular dystrophy (FSHD1) gene affects males more severely and more frequently than females. Am J Med Genet. 1998, 77, 155–161 [PubMed] [Google Scholar]
- [161].Rosa-Caldwell ME, Greene NP. Muscle metabolism and atrophy let’s talk about sex. Biol Sex Differ., 2019, 10, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Spangenburg EE, Geiger PC, Leinwand LA, Lowe DA. Regulation of physiological and metabolic function of muscle by female sex steroids. Med Sci Sports Exerc. 44(9) (2012) 1653–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Gouspillou G, Hepple RT. Editorial: Mitochondria in skeletal muscle health, aging and diseases. Frontiers in physiology, 7 (2016) 446). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Ar Rochmah M, Shima A, Harahap NIF, Niba ETE, Morisada N, Yanagisawa S, Saito T, Kaneko K, Saito K, Morioka I, Iijima K, Lai PS, Bouike Y, Nishio H, Shinohara M, Gender Effects on the Clinical Phenotype in Japanese Patients with Spinal Muscular Atrophy, Kobe J Med Sci 63(2) (2017) E41–E44. [PMC free article] [PubMed] [Google Scholar]
- [165].Oliva M, Muñoz-Aguirre M, Kim-Hellmuth S, Wucher V, Gewirtz ADH, Cotter DJ, Parsana P, Kasela S, Balliu B, Viñuela A, Castel SE, Mohammadi P, Aguet F, Zou Y, Khramtsova EA, Skol AD, Garrido-Martín D, Reverter F, Brown A, Evans P, Gamazon ER, Payne A, Bonazzola R, Barbeira AN, Hamel AR, Martinez-Perez A, Soria JM, Pierce BL, Stephens M, Eskin E, Dermitzakis ET, Segrè AV, Im HK, Engelhardt BE, Ardlie KG, Montgomery SB, Battle AJ, Lappalainen T, Guigó R, Stranger BE, Consortium G, The impact of sex on gene expression across human tissues, Science 369(6509) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Tower J, Sex-specific regulation of aging and apoptosis, Mech Ageing Dev 127(9) (2006) 705–18. [DOI] [PubMed] [Google Scholar]
- [167].Silkaitis K, Lemos B, Sex-biased chromatin and regulatory cross-talk between sex chromosomes, autosomes, and mitochondria, Biol Sex Differ 5(1) (2014) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Capllonch-Amer G, Lladó I, Proenza AM, García-Palmer FJ, Gianotti M, Opposite effects of 17-β estradiol and testosterone on mitochondrial biogenesis and adiponectin synthesis in white adipocytes, J Mol Endocrinol 52(2) (2014) 203–14. [DOI] [PubMed] [Google Scholar]
- [169].Nadal-Casellas A, Proenza AM, Lladó I, Gianotti M, Effects of ovariectomy and 17-β estradiol replacement on rat brown adipose tissue mitochondrial function, Steroids 76(10-11) (2011) 1051–6. [DOI] [PubMed] [Google Scholar]
- [170].Gómez-Pérez Y, Capllonch-Amer G, Gianotti M, Lladó I, Proenza AM, Long-term high-fat-diet feeding induces skeletal muscle mitochondrial biogenesis in rats in a sex-dependent and muscle-type specific manner, Nutr Metab (Lond) 9 (2012) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Nookaew I, Svensson PA, Jacobson P, Jemås M, Taube M, Larsson I, Andersson-Assarsson JC, Sjöström L, Froguel P, Walley A, Nielsen J, Carlsson LM, Adipose tissue resting energy expenditure and expression of genes involved in mitochondrial function are higher in women than in men, J Clin Endocrinol Metab 98(2) (2013) E370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Colom B, Oliver J, Roca P, Garcia-Palmer FJ, Caloric restriction and gender modulate cardiac muscle mitochondrial H2O2 production and oxidative damage, Cardiovasc Res 74(3) (2007) 456–65. [DOI] [PubMed] [Google Scholar]
- [173].Miller N, Shi H, Zelikovich AS, Ma YC, Motor neuron mitochondrial dysfunction in spinal muscular atrophy, Hum Mol Genet 25(16) (2016) 3395–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Ng SY, Mikhail A, Ljubicic V, Mechanisms of exercise-induced survival motor neuron expression in the skeletal muscle of spinal muscular atrophy-like mice, J Physiol 597(18) (2019) 4757–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Kupr B, Handschin C, Complex Coordination of Cell Plasticity by a PGC-1α-controlled Transcriptional Network in Skeletal Muscle, Front Physiol 6 (2015) 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Deguise MO, De Repentigny Y, Tierney A, Beauvais A, Michaud J, Chehade L, Thabet M, Paul B, Reilly A, Gagnon S, Renaud JM, Kothary R, Motor transmission defects with sex differences in a new mouse model of mild spinal muscular atrophy, EBioMedicine 55 (2020) 102750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Gaignard P, Savouroux S, Liere P, Pianos A, Thérond P, Schumacher M, Slama A, Guennoun R, Effect of Sex Differences on Brain Mitochondrial Function and Its Suppression by Ovariectomy and in Aged Mice, Endocrinology 156(8) (2015) 2893–904. [DOI] [PubMed] [Google Scholar]
- [178].Wishart TM, Mutsaers CA, Riessland M, Reimer MM, Hunter G, Hannam ML, Eaton SL, Fuller HR, Roche SL, Somers E, Morse R, Young PJ, Lamont DJ, Hammerschmidt M, Joshi A, Hohenstein P, Morris GE, Parson SH, Skehel PA, Becker T, Robinson IM, Becker CG, Wirth B, Gillingwater TH, Dysregulation of ubiquitin homeostasis and β-catenin signaling promote spinal muscular atrophy, J Clin Invest 124(4) (2014) 1821–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Groen EJN, Gillingwater TH, UBA1: At the Crossroads of Ubiquitin Homeostasis and Neurodegeneration, Trends Mol Med 21(10) (2015) 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Pane M, Lapenta L, Abiusi E, de Sanctis R, Luigetti M, Palermo C, Ranalli D, Fiori S, Tiziano FD, Mercuri E. Longitudinal assessments in discordant twins with SMA. Neuromuscul Disord. 27(10) (2017) 890–893. [DOI] [PubMed] [Google Scholar]
- [181].Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature, 434 (2005) 400–404. [DOI] [PubMed] [Google Scholar]
- [182].Carrel L, Clemson CM, Dunn JM, Miller AP, Hunt PA, Lawrence JB, Willard HF. X inactivation analysis and DNA methylation studies of the ubiquitin activating enzyme E1 and PCTAIRE-1 genes in human and mouse. Hum. Mol. Genet 5 (1996) 391–401 [DOI] [PubMed] [Google Scholar]
- [183].Witt E, Lorenz M, Völker U, Stangl K, Hammer E, Stangl VJ. Sex-specific differences in the intracellular proteome of human endothelial cells from dizygotic twins. Proteomics. 201 (2019) 48–56 [DOI] [PubMed] [Google Scholar]
- [184].Xu J, Burgoyne PS, Arnold AP. Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet, 11 (12) (2002) 1409–1419. [DOI] [PubMed] [Google Scholar]
- [185].Schurz H, Salie M, Tromp G, Hoal EG, Kinnear CJ, Möller M, The X chromosome and sex-specific effects in infectious disease susceptibility, Hum Genomics 13(1) (2019) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Klein SL, Marriott I, Fish EN, Sex-based differences in immune function and responses to vaccination, Trans R Soc Trop Med Hyg 109(1) (2015) 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Deguise MO, Kothary R, New insights into SMA pathogenesis: immune dysfunction and neuroinflammation, Ann Clin Transl Neurol 4(7) (2017) 522–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Wan B, Feng P, Guan Z, Sheng L, Liu Z, Hua Y, A severe mouse model of spinal muscular atrophy develops early systemic inflammation, Hum Mol Genet 27(23) (2018) 4061–4076. [DOI] [PubMed] [Google Scholar]
- [189].Abati E, Citterio G, Bresolin N, Comi GP, Corti S, Glial cells involvement in spinal muscular atrophy: Could SMA be a neuroinflammatory disease?, Neurobiol Dis 140 (2020) 104870. [DOI] [PubMed] [Google Scholar]
- [190].Di Palo A, Siniscalchi C, Salerno M, Russo A, Gravholt CH, Potenza N, What microRNAs could tell us about the human X chromosome, Cell Mol Life Sci (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Li H, Dai B, Fan J, Chen C, Nie X, Yin Z, Zhao Y, Zhang X, Wang DW, The Different Roles of miRNA-92a-2-5p and let-7b-5p in Mitochondrial Translation in db/db Mice, Mol Ther Nucleic Acids 17 (2019) 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Björnström L, Sjöberg M, Mutations in the estrogen receptor DNA-binding domain discriminate between the classical mechanism of action and cross-talk with Stat5b and activating protein 1 (AP-1), J Biol Chem 277(50) (2002) 48479–83. [DOI] [PubMed] [Google Scholar]
- [193].Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P, Estrogen receptor pathways to AP-1, J Steroid Biochem Mol Biol 74(5) (2000) 311–7. [DOI] [PubMed] [Google Scholar]
- [194].Arevalo MA, Azcoitia I, Garcia-Segura LM, The neuroprotective actions of oestradiol and oestrogen receptors, Nat Rev Neurosci 16(1) (2015) 17–29. [DOI] [PubMed] [Google Scholar]
- [195].Razandi M, Pedram A, Park ST, Levin ER, Proximal events in signaling by plasma membrane estrogen receptors, J Biol Chem 278(4) (2003) 2701–12. [DOI] [PubMed] [Google Scholar]
- [196].Davey RA, Grossmann M, Androgen Receptor Structure, Function and Biology: From Bench to Bedside, Clin Biochem Rev 37(1) (2016) 3–15. [PMC free article] [PubMed] [Google Scholar]
- [197].Clarkson J, Herbison AE, Hypothalamic control of the male neonatal testosterone surge, Philos Trans R Soc Lond B Biol Sci 371(1688) (2016) 20150115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Singh NN, Howell MD, Singh RN, Transcriptional and Splicing Regulation of Spinal Muscular Atrophy Genes, in: Charlotte SJ, Paushkin S, Ko C-P (Eds.), Spinal Muscular Atrophy: Disease Mechanisms and Therapy, Elsevier Inc. 2016. [Google Scholar]
- [199].Howell MD, Singh NN, Singh RN, Advances in therapeutic development for spinal muscular atrophy, Future Med Chem 6(9) (2014) 1081–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Farooq F, Molina FA, Hadwen J, MacKenzie D, Witherspoon L, Osmond M, Holcik M, MacKenzie A, Prolactin increases SMN expression and survival in a mouse model of severe spinal muscular atrophy via the STAT5 pathway, J Clin Invest 121(8) (2011) 3042–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].MacKenzie D, Shamim F, Mongeon K, Trivedi A, MacKenzie A, Farooq F, Human Growth Hormone Increases SMN Expression and Survival in Severe Spinal Muscular Atrophy Mouse Model, J Neuromuscul Dis 1(1) (2014) 65–74. [PubMed] [Google Scholar]
- [202].Waters MJ, The growth hormone receptor, Growth Horm IGF Res 28 (2016) 6–10. [DOI] [PubMed] [Google Scholar]
- [203].Sehgal PB, Yang YM, Yuan H, Miller EJ, STAT5a/b contribute to sex bias in vascular disease: A neuroendocrine perspective, JAKSTAT 4(3) (2015) 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Paul RG, Hennebry AS, Elston MS, Conaglen JV, McMahon CD, Regulation of murine skeletal muscle growth by STAT5B is age- and sex-specific, Skelet Muscle 9(1) (2019) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Salais-López H, Agustín-Pavón C, Lanuza E, Martínez-García F, The maternal hormone in the male brain: Sexually dimorphic distribution of prolactin signalling in the mouse brain, PLoS One 13(12) (2018) e0208960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Radhakrishnan A, Raju R, Tuladhar N, Subbannayya T, Thomas JK, Goel R, Telikicherla D, Palapetta SM, Rahiman BA, Venkatesh DD, Urmila KK, Harsha HC, Mathur PP, Prasad TS, Pandey A, Shemanko C, Chatterjee A, A pathway map of prolactin signaling, J Cell Commun Signal 6(3) (2012) 169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Branchu J, Biondi O, Chali F, Collin T, Leroy F, Mamchaoui K, Makoukji J, Pariset C, Lopes P, Massaad C, Chanoine C, Charbonnier F, Shift from extracellular signal-regulated kinase to AKT/cAMP response element-binding protein pathway increases survival-motor-neuron expression in spinal-muscular-atrophy-like mice and patient cells, J Neurosci 33(10) (2013) 4280–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Arumugam S, Mincheva-Tasheva S, Periyakaruppiah A, de la Fuente S, Soler RM, Garcera A, Regulation of Survival Motor Neuron Protein by the Nuclear Factor-Kappa B Pathway in Mouse Spinal Cord Motoneurons, Mol Neurobiol 55(6) (2018) 5019–5030. [DOI] [PubMed] [Google Scholar]
- [209].Rehorst WA, Thelen MP, Nolte H, Türk C, Cirak S, Peterson JM, Wong GW, Wirth B, Krüger M, Winter D, Kye MJ, Muscle regulates mTOR dependent axonal local translation in motor neurons via CTRP3 secretion: implications for a neuromuscular disorder, spinal muscular atrophy, Acta Neuropathol Commun 7(1) (2019) 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Dyer AH, Vahdatpour C, Sanfeliu A, Tropea D, The role of Insulin-Like Growth Factor 1 (IGF-1) in brain development, maturation and neuroplasticity, Neuroscience 325 (2016) 89–99. [DOI] [PubMed] [Google Scholar]
- [211].Millino C, Fanin M, Vettori A, Laveder P, Mostacciuolo ML, Angelini C, Lanfranchi G, Different atrophy-hypertrophy transcription pathways in muscles affected by severe and mild spinal muscular atrophy, BMC Med 7 (2009) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR, Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model, Nature 478(7367) (2011) 123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Biondi O, Branchu J, Ben Salah A, Houdebine L, Bertin L, Chali F, Desseille C, Weill L, Sanchez G, Lancelin C, Aïd S, Lopes P, Pariset C, Lécolle S, Côté J, Holzenberger M, Chanoine C, Massaad C, Charbonnier F, IGF-1R Reduction Triggers Neuroprotective Signaling Pathways in Spinal Muscular Atrophy Mice, J Neurosci 35(34) (2015) 12063–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Wang Y, Wang W, Li D, Li M, Wang P, Wen J, Liang M, Su B, Yin Y, IGF-1 alleviates NMDA-induced excitotoxicity in cultured hippocampal neurons against autophagy via the NR2B/PI3K-AKT-mTOR pathway, J Cell Physiol 229(11) (2014) 1618–29. [DOI] [PubMed] [Google Scholar]
- [215].Holzenberger M, Dupont J, Ducos B, Leneuve P, Géloën A, Even PC, Cervera P, Le Bouc Y, IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice, Nature 421(6919) (2003) 182–7. [DOI] [PubMed] [Google Scholar]
- [216].Garg N, Thakur S, McMahan CA, Adamo ML, High fat diet induced insulin resistance and glucose intolerance are gender-specific in IGF-1R heterozygous mice, Biochem Biophys Res Commun 413(3) (2011) 476–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Cintron-Colon R, Sanchez-Alavez M, Nguyen W, Mori S, Gonzalez-Rivera R, Lien T, Bartfai T, Aïd S, Françis JC, Holzenberger M, Conti B, Insulin-like growth factor 1 receptor regulates hypothermia during calorie restriction, Proc Natl Acad Sci U S A 114(36) (2017) 9731–9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Davis RH, Miller EA, Zhang RZ, Swoboda KJ, Responses to Fasting and Glucose Loading in a Cohort of Well Children with Spinal Muscular Atrophy Type II, J Pediatr 167(6) (2015) 1362–8.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Walter LM, Deguise MO, Meijboom KE, Betts CA, Ahlskog N, van Westering TLE, Hazell G, McFall E, Kordala A, Hammond SM, Abendroth F, Murray LM, Shorrock HK, Prosdocimo DA, Haldar SM, Jain MK, Gillingwater TH, Claus P, Kothary R, Wood MJA, Bowerman M, Interventions Targeting Glucocorticoid-Krüppel-like Factor 15-Branched-Chain Amino Acid Signaling Improve Disease Phenotypes in Spinal Muscular Atrophy Mice, EBioMedicine 31 (2018) 226–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Lonardo A, Nascimbeni F, Ballestri S, Fairweather D, Win S, Than TA, Abdelmalek MF, Suzuki A, Sex Differences in Nonalcoholic Fatty Liver Disease: State of the Art and Identification of Research Gaps, Hepatology 70(4) (2019) 1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Mutsaers CA, Wishart TM, Lamont DJ, Riessland M, Schreml J, Comley LH, Murray LM, Parson SH, Lochmüller H, Wirth B, Talbot K, Gillingwater TH, Reversible molecular pathology of skeletal muscle in spinal muscular atrophy, Hum Mol Genet 20(22) (2011) 4334–44. [DOI] [PubMed] [Google Scholar]
- [222].Fayzullina S, Martin LJ, Skeletal muscle DNA damage precedes spinal motor neuron DNA damage in a mouse model of Spinal Muscular Atrophy (SMA), PLoS One 9(3) (2014) e93329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Moradi N, Fadaei R, Khamseh ME, Nobakht A, Rezaei MJ, Aliakbary F, Vatannejad A, Hosseini J, Serum levels of CTRP3 in diabetic nephropathy and its relationship with insulin resistance and kidney function, PLoS One 14(4) (2019) e0215617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Howell MD, Ottesen EW, Singh NN, Anderson RL, Singh RN, Gender-Specific Amelioration of SMA Phenotype upon Disruption of a Deep Intronic Structure by an Oligonucleotide, Mol Ther 25(6) (2017) 1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Tarhouni K, Guihot AL, Freidja ML, Toutain B, Henrion B, Baufreton C, Pinaud F, Procaccio V, Grimaud L, Ayer A, Loufrani L, Lenfant F, Arnal JF, Henrion D, Key role of estrogens and endothelial estrogen receptor α in blood flow-mediated remodeling of resistance arteries, Arterioscler Thromb Vasc Biol 33(3) (2013) 605–11. [DOI] [PubMed] [Google Scholar]
- [226].Straface E, Vona R, Campesi I, Franconi F, Mitochondria can orchestrate sex differences in cell fate of vascular smooth muscle cells from rats, Biol Sex Differ 6 (2015) 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Thomas NH, Dubowitz V, The natural history of type I (severe) spinal muscular atrophy, Neuromuscul Disord 4(5-6) (1994) 497–502. [DOI] [PubMed] [Google Scholar]
- [228].Kaufmann P, McDermott MP, Darras BT, Finkel R, Kang P, Oskoui M, Constantinescu A, Sproule DM, Foley AR, Yang M, Tawil R, Chung W, Martens B, Montes J, O'Hagen J, Dunaway S, Flickinger JM, Quigley J, Riley S, Glanzman AM, Benton M, Ryan PA, Irvine C, Annis CL, Butler H, Caracciolo J, Montgomery M, Marra J, Koo B, De Vivo DC, M.S. Group, P.N.C.R.N.f.S.M. Atrophy, Observational study of spinal muscular atrophy type 2 and 3: functional outcomes over 1 year, Arch Neurol 68(6) (2011) 779–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Huff A, Reed MD, Iceman KE, Howland DR, Pitts T, Sex-specific vagal and spinal modulation of breathing with chest compression, PLoS One 15(6) (2020) e0234193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Behan M, Wenninger JM, Sex steroidal hormones and respiratory control, Respir Physiol Neurobiol 164(1-2) (2008) 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Bebee TW, Dominguez CE, Samadzadeh-Tarighat S, Akehurst KL, Chandler DS, Hypoxia is a modifier of SMN2 splicing and disease severity in a severe SMA mouse model, Hum Mol Genet 21(19) (2012) 4301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Holley HS, Behan M, Wenninger JM, Age and sex differences in the ventilatory response to hypoxia and hypercapnia in awake neonatal, pre-pubertal and young adult rats, Respir Physiol Neurobiol 180(1) (2012) 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Qian X, Du Y, Jiang G, Lin F, Yao L, Survival Motor Neuron (SMN) Protein Insufficiency Exacerbates Renal Ischemia/Reperfusion Injury, Front Physiol 10 (2019) 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].West M, Nash MM, Rapi L, Krizova A, Prasad GVR, Monitoring Kidney Dysfunction in Kugelberg-Welander Syndrome, Am J Case Rep 20 (2019) 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Allardyce H, Kuhn D, Hernandez-Gerez E, Hensel N, Huang YT, Faller K, Gillingwater TH, Quondamatteo F, Claus P, Parson SH, Renal pathology in a mouse model of severe Spinal Muscular Atrophy is associated with downregulation of Glial Cell-Line Derived Neurotrophic Factor (GDNF), Hum Mol Genet 29(14) (2020) 2365–2378. [DOI] [PubMed] [Google Scholar]
- [236].Fujii T, Uchino S, Takinami M, Bellomo R, Validation of the Kidney Disease Improving Global Outcomes criteria for AKI and comparison of three criteria in hospitalized patients, Clin J Am Soc Nephrol 9(5) (2014) 848–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Sabolić I, Asif AR, Budach WE, Wanke C, Bahn A, Burckhardt G, Gender differences in kidney function, Pflugers Arch 455(3) (2007) 397–429. [DOI] [PubMed] [Google Scholar]
- [238].Hosszu A, Fekete A, Szabo AJ, Sex differences in renal ischemia-reperfusion injury, Am J Physiol Renal Physiol 319(2) (2020) F149–F154. [DOI] [PubMed] [Google Scholar]
- [239].Munger K, Baylis C, Sex differences in renal hemodynamics in rats, Am J Physiol 254(2 Pt 2) (1988) F223–31. [DOI] [PubMed] [Google Scholar]
- [240].Verzola D, Gandolfo MT, Salvatore F, Villaggio B, Gianiorio F, Traverso P, Deferrari G, Garibotto G, Testosterone promotes apoptotic damage in human renal tubular cells, Kidney Int 65(4) (2004) 1252–61. [DOI] [PubMed] [Google Scholar]
- [241].Park KM, Kim JI, Ahn Y, Bonventre AJ, Bonventre JV, Testosterone is responsible for enhanced susceptibility of males to ischemic renal injury, J Biol Chem 279(50) (2004) 52282–92. [DOI] [PubMed] [Google Scholar]
- [242].Hosszu A, Antal Z, Veres-Szekely A, Lenart L, Balogh DB, Szkibinszkij E, Illesy L, Hodrea J, Banki NF, Wagner L, Vannay A, Szabo AJ, Fekete A, The role of Sigma-1 receptor in sex-specific heat shock response in an experimental rat model of renal ischaemia/reperfusion injury, Transpl Int 31(11) (2018) 1268–1278. [DOI] [PubMed] [Google Scholar]
- [243].Punjani N, Lamb DJ, Male infertility and genitourinary birth defects: there is more than meets the eye, Fertil Steril 114(2) (2020) 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Singh NN, Singh RN, Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model, RNA Biol 8(4) (2011) 600–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Singh NN, Lee BM, Singh RN, Splicing regulation in spinal muscular atrophy by an RNA structure formed by long-distance interactions, Ann N Y Acad Sci 1341 (2015) 176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Singh RN, Singh NN, Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes, Adv Neurobiol 20 (2018) 31–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Chen HH, Chang JG, Lu RM, Peng TY, Tam WY, The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene, Mol Cell Biol 28(22) (2008) 6929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Chen YC, Chang JG, Jong YJ, Liu TY, Yuo CY, High expression level of Tra2-β1 is responsible for increased SMN2 exon 7 inclusion in the testis of SMA mice, PLoS One 10(3) (2015) e0120721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Spiller C, Bumet G, Bowles J, Regulation of fetal male germ cell development by members of the TGFβ superfamily, Stem Cell Res 24 (2017) 174–180. [DOI] [PubMed] [Google Scholar]
- [250].Meroni SB, Galardo MN, Rindone G, Gorga A, Riera MF, Cigorraga SB, Molecular Mechanisms and Signaling Pathways Involved in Sertoli Cell Proliferation, Front Endocrinol (Lausanne) 10 (2019) 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Hofmann MC, Gdnf signaling pathways within the mammalian spermatogonial stem cell niche, Mol Cell Endocrinol 288(1-2) (2008) 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Griswold MD, Hogarth C, Beyond stem cells: Commitment of progenitor cells to meiosis, Stem Cell Res 27 (2018) 169–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Kumar TR, Fshb Knockout Mouse Model, Two Decades Later and Into the Future, Endocrinology 159(5) (2018) 1941–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Kerr JB, Millar M, Maddocks S, Sharpe RM, Stage-dependent changes in spermatogenesis and Sertoli cells in relation to the onset of spermatogenic failure following withdrawal of testosterone, Anat Rec 235(4) (1993) 547–59. [DOI] [PubMed] [Google Scholar]
- [255].Singh NN, Androphy EJ, Singh RN, The regulation and regulatory activities of alternative splicing of the SMN gene, Crit Rev Eukaryot Gene Expr 14(4) (2004) 271–85. [DOI] [PubMed] [Google Scholar]
- [256].Seo J, Howell MD, Singh NN, Singh RN, Spinal muscular atrophy: an update on therapeutic progress, Biochim Biophys Acta 1832(12) (2013) 2180–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Singh RN, More is needed to complement the available therapies of spinal muscular atrophy, Future Med Chem 11(22) (2019) 2873–2876. [DOI] [PubMed] [Google Scholar]
- [258].Singh RN, Seo J, Singh NN, RNA in spinal muscular atrophy: therapeutic implications of targeting, Expert Opin Ther Targets (2020) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Singh NK, Singh NN, Androphy EJ, Singh RN, Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron, Mol Cell Biol 26(4) (2006) 1333–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Ottesen EW, ISS-N1 makes the First FDA-approved Drug for Spinal Muscular Atrophy, Transl Neurosci 8 (2017) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Singh NN, Howell MD, Androphy EJ, Singh RN, How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy, Gene Ther 24(9) (2017) 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Singh NN, Luo D, Singh RN. Pre-mRNA Splicing Modulation by Antisense Oligonucleotides. Methods Mol Biol. 2018;1828:415–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Porensky PN, Mitrpant C, McGovern VL, Bevan AK, Foust KD, Kaspar BK, Wilton SD, Burghes AH, A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse, Hum Mol Genet 21(7) (2012) 1625–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Zhou H, Janghra N, Mitrpant C, Dickinson RL, Anthony K, Price L, Eperon IC, Wilton SD, Morgan J, Muntoni F, A novel morpholino oligomer targeting ISS-N1 improves rescue of severe spinal muscular atrophy transgenic mice, Hum Gene Ther 24(3) (2013) 331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Mitrpant C, Porensky P, Zhou H, Price L, Muntoni F, Fletcher S, Wilton SD, Burghes AH, Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy, PLoS One 8(4) (2013) e62114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Sivanesan S, Howell MD, Didonato CJ, Singh RN, Antisense oligonucleotide mediated therapy of spinal muscular atrophy, Transl Neurosci 4(1) (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Al-Zaidy SA, Mendell JR, From Clinical Trials to Clinical Practice: Practical Considerations for Gene Replacement Therapy in SMA Type 1, Pediatr Neurol 100 (2019) 3–11. [DOI] [PubMed] [Google Scholar]
- [268].Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, Yamashita M, Rigo F, Hung G, Schneider E, Norris DA, Xia S, Bennett CF, Bishop KM, Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study, Lancet 388(10063) (2016) 3017–3026. [DOI] [PubMed] [Google Scholar]
- [269].Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, Chiriboga CA, Saito K, Servais L, Tizzano E, Topaloglu H, Tulinius M, Montes J, Glanzman AM, Bishop K, Zhong ZJ, Gheuens S, Bennett CF, Schneider E, Farwell W, De Vivo DC, E.S. Group, Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy, N Engl J Med 377(18) (2017) 1723–1732. [DOI] [PubMed] [Google Scholar]
- [270].Al-Zaidy SA, Kolb SJ, Lowes L, Alfano LN, Shell R, Church KR, Nagendran S, Sproule DM, Feltner DE, Wells C, Ogrinc F, Menier M, L'Italien J, Arnold WD, Kissel JT, Kaspar BK, Mendell JR, AVXS-101 (Onasemnogene Abeparvovec) for SMA1: Comparative Study with a Prospective Natural History Cohort, J Neuromuscul Dis 6(3) (2019) 307–317. [DOI] [PubMed] [Google Scholar]
- [271].Singh RN, Ottesen EW, Singh NN. The First Orally Deliverable Small Molecule for the Treatment of Spinal Muscular Atrophy Neuroscience Insights 15, 2633105520973985; 10.1177/2633105520973985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Singh NN, Lee BM, DiDonato CJ, Singh RN, Mechanistic principles of antisense targets for the treatment of spinal muscular atrophy, Future Med Chem 7(13) (2015) 1793–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Singh NN, Singh RN, How RNA structure dictates the usage of a critical exon of spinal muscular atrophy gene, Biochim Biophys Acta Gene Regul Mech 1862(11-12) (2019) 194403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN, An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy, Nucleic Acids Res 41(17) (2013) 8144–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Perrin S, Preclinical research: Make mouse studies work, Nature 507(7493) (2014) 423–5. [DOI] [PubMed] [Google Scholar]
- [276].Clayton JA, Collins FS, Policy: NIH to balance sex in cell and animal studies, Nature 509(7500) (2014) 282–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Nidetz NF, McGee MC, Tse LV, Li C, Cong L, Li Y, Huang W, Adeno-associated viral vector-mediated immune responses: Understanding barriers to gene delivery, Pharmacol Ther 207 (2020) 107453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Alharbi AS, Garcin AJ, Lennox KA, Pradeloux S, Wong C, Straub S, Valentin R, Pépin G, Li HM, Nold MF, Nold-Petry CA, Behlke MA, Gantier MP, Rational design of antisense oligonucleotides modulating the activity of TLR7/8 agonists, Nucleic Acids Res 48(13) (2020) 7052–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]