Abstract
In this study, the spores and vegetative cells of B. clausii were independently evaluated for probiotic properties such as acid, gastric juice, bile, and intestinal fluid tolerance, adhesion to solvents/mucin and zeta potential. In addition, in silico identification of genome features contributing to probiotic properties were investigated. The results showed that spores were highly stable at gastric acidity and capable to germinate and multiply under intestinal conditions as compared to vegetative cells. The higher hydrophobicity of spores, compared to vegetative cells, is advantageous for colonization and persistence in the intestine. Furthermore, the presence of F0F1 ATP synthase, amino acid decarboxylase, bile acid symporter, mucin/collagen/fibronectin-binding proteins, heat/cold shock proteins, and universal stress proteins suggests that the strain is able to survive stress. In conclusion, the results demonstrate that B. clausii UBBC07 spores show significantly higher survival and adhesion in in vitro gastrointestinal conditions as compared to vegetative cells. Besides, this study provides a comparative analysis of the in vitro probiotic properties of spores and vegetative cells of Bacillus clausii UBBC07.
Keywords: Bacillus clausii UBBC07, Spores, Vegetative cells, Probiotics properties, Zeta potential
Introduction
Probiotics are defined as live microorganisms that, when administered in adequate amounts, confer a health benefit on the host (Hill et al. 2014). To date, Lactobacillus and Bifidobacteria are the most investigated probiotic cultures as compared to Bacillus (Bhushan et al. 2019). As per the recommendations of health institutions of Canada and Italy, the use of 1 × 109 colony forming units (cfu) of these bacteria were permitted per serving for nonstrain-specific claims (Hill et al. 2014). There are several probiotic products on the market, but only a few fulfill criteria of labelled concentration claims at the end of the shelf-life (Vecchione et al. 2018). Besides this, the ability of bacteria to tolerate gastric and intestinal conditions is imperative for the delivery of health benefits to the host (Bhushan et al. 2020). In comparison to vegetative cells, spores are highly stable at various industrial, environmental, and gastro-intestinal conditions, thus ensuring the delivery of recommended probiotic dose to the gut (Patel et al. 2009; Ahire et al. 2020c).
Comparative probiotic properties of spores and vegetative cells of spore-formers have rarely been investigated (Bernardeau et al. 2017). Cenci et al. (2006) showed the ability of Bacillus clausii spores to germinate during gastrointestinal transit and the possibility for vegetative cells to survive in the intestinal tract. Patel et al. (2009) evaluated probiotic properties of a mixture of spores and vegetative cells of B. megaterium. Recently, Sharma et al. (2020) characterized the probiotic properties of the ambiguous biotype i.e. spores or vegetative cells of Bacillus spp., isolated from fermented food. Moreover, probiotic properties of spore-formers were either reported for spores or vegetative cells or mixture.
Bacillus clausii is a Gram-positive, aerobic, spore-forming, motile, rod-shaped, facultative alkaliphilic soil bacterium (Cenci et al. 2006). It is one of the human probiotics, which is able to survive gastrointestinal transit and colonize the gut even in the presence of antibiotics (Duc et al. 2004; Ianiro et al. 2018). Preclinical and clinical studies suggest that B. clausii probiotic is effective in the treatment of diarrhea, recurrent respiratory infections and acute gastroenteritis (Marseglia et al. 2007; Ianiro et al. 2018; Paparo et al. 2020). Currently tested strain, Bacillus clausii UBBC07 (MTCC 5472) is a non-toxic, spore-forming probiotic bacterium available on the Indian market since 2005 (Upadrasta et al. 2016; Lakshmi et al. 2017). A daily dose of 4 billion cfu of UBBC07 spores is recommended to alleviate diarrhea in children and adults (Sudha et al. 2013, 2019). Recently, B. clausii UBBC07 has been reported for the production of lantibiotic clausin and reduction of uremic toxins in acetaminophen-induced uremic rats (Patel et al. 2019; Ahire et al. 2020b). In this study, for the first time, we describe the comparative probiotic properties of spores and vegetative cells of Bacillus clausii UBBC07 and in silico identification of genome features contributing to probiotic properties.
Materials and methods
Preparation of vegetative cells and spores
Bacillus clausii UBBC07 (MTCC 5472) was obtained from the Unique Biotech culture collection, Hyderabad, India. The strain was cultivated aerobically in BHI broth (HiMedia, India) at 37 °C for 24 h and purity confirmed by plating on BHI agar. A single colony was inoculated in 10 ml BHI broth and incubated for 24 h at 37 °C with shaking (180 rpm). Vegetative cells were harvested by centrifugation at 11,000 × g for 10 min at 4 °C (Sorvall Legend XTR, Thermo Scientific, USA) and washed twice with phosphate buffer saline (PBS, pH 7.3). The cell pellet obtained was resuspended in PBS and investigated for probiotic properties. Simultaneously, the strain was cultivated aerobically in BHI broth at 37 °C for 96 h to sporulate. Spores were harvested by centrifugation, washed twice with PBS, and heat-treated at 80 °C for 20 min to kill vegetative cells. The resultant spore suspension was evaluated for probiotic properties.
Survival of spores and vegetative cells under in vitro GIT conditions
Acid tolerance
The 100 µl of B. clausii UBBC07 vegetative cells and spore suspension was inoculated separately in 900 µl PBS pH (1.0, 2.0 and 3.0) and incubated aerobically at 37 °C for 0, 1, 2 and 3 h (Ahire 2012). Survivability was determined by plating on BHI agar.
Synthetic gastric juice tolerance
Spores or vegetative cells of UBBC07 were diluted 1:10 in filter sterilized (0.2 µm cellulose acetate; Sartorius, Germany) synthetic gastric juice [g l−1: pepsin (≥ 3000 NFU mg−1), 0.0133; lysozyme (≥ 40,000 U mg−1), 0.1; bile, 0.05; proteose peptone, 8.3; glucose, 3.5; KCl, 0.37; NaCl, 2.05; CaCl2, 0.11; KH2PO4, 0.6; pH 2.5] and incubated aerobically at 37 °C for 3 h (Pedersen et al. 2004). Survival was determined at 0, 30 and 180 min time intervals by plating appropriate dilutions on BHI agar plates.
Bile salt tolerance
The UBBC07 suspension (vegetative cells or spores) was inoculated 1:10 in BHI broth supplemented with 0.1, 0.3, 0.5, 1.0 and 2.0% (w/v) bile (HiMedia, India). The tubes were incubated aerobically at 37 °C for 24 h. Tolerance was evaluated by determining optical density at 600 nm (Ahire 2012).
Intestinal fluid tolerance
The UBBC07 suspension (vegetative cells or spores) was diluted 1:10 in filter sterilized intestinal fluid [1 mg ml−1 pancreatin (amylase 100 U mg−1; lipase 8 U mg−1; protease 100 U mg−1: Sisco Research Laboratory, India) prepared in 0.85% (w/v) NaCl supplemented with 0.3% bile (w/v); pH 8.0], and incubated aerobically at 37 °C for 6 h. Survivability was determined at 0 and 6 h by plating on BHI agar.
Microbial adhesion to solvents
The cell pellet or spores obtained from B. clausii UBBC07 were washed twice with PBS (pH7.3) and dissolved in 50 ml 0.1 mol l−1 KNO3 (pH 6.2). Absorbance of suspensions was measured at 600 as A0 using a UV–visible spectrophotometer (Thermo Scientific, USA). To every 3 ml of this suspension, 1 ml solvent (xylene, chloroform, and ethyl acetate) was added and left standing for 10 min at 37 °C. Thereafter, the two phases were mixed by vortexing for 2 min and incubated aerobically at 37 °C for 30 min. The aqueous phase was removed and the absorbance (600 nm) measured as A1 (Ahire et al. 2013). The percentage of microbial adhesion was calculated as (A0 – A1/A0) × 100.
Adhesion to porcine mucin
The 6-well tissue culture plates (Thermo Scientific, Denmark) were coated at 4 °C for 24 h with 100 µg ml−1 of porcine stomach mucin (Sigma Aldrich, USA) dissolved in 0.05 mol l−1 Na2CO3 (pH 9.7). After incubation, the coating solution was discarded and each well was treated with 2 ml PBS containing 1% (w/v) Tween 20 for 1 h. Finally, each well was washed with PBS containing 0.05% (w/v) Tween 20 and inoculated with 2 ml vegetative cells and or spore solution (0.5 OD) prepared in PBS (0.05% (w/v) Tween 20; pH 7.3) buffer. The plates were incubated overnight at 4 °C (Pedersen et al. 2004). After incubation, wells were washed with PBS containing 0.05% (w/v) Tween 20 and visualized using an inverted microscope (CKX53, Olympus, Japan). Adhesion was quantitatively determined by staining the wells with 0.1% (w/v) crystal violet (Ahire et al. 2014). Experiments were performed in triplicate.
Determination of zeta potential
The zeta potential of B. clausii UBBC07 (vegetative cells or spores) prepared in PBS (pH 7.3) was measured using the Zetasizer Nano-ZS (Malvern, UK). The DTS1070 capillary cell was used as per the procedure described by Ahire et al. (2020a).
In silico identification of genome features contributing to probiotic properties
Bacillus clausii UBBC07 whole genome (GenBank accession no. LATY00000000) was investigated for the presence of genes or specific domains involved in acid tolerance, bile salt tolerance, adhesion to gut mucosa and environmental stress resistance as described by Khatri et al. (2019). The RAST (Rapid Annotation using Subsystem Technology; Brettin et al. 2015) and SEED (Overbeek et al. 2014) viewer comparative blast search tool was used along with NCBI standard protein BLAST.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (USA). The statistical differences among means were determined using Tukey’s multiple comparison test and t-test. Data were presented as the mean and standard deviation. The p-value of less than 0.05 was considered significant.
Results
Acid tolerance
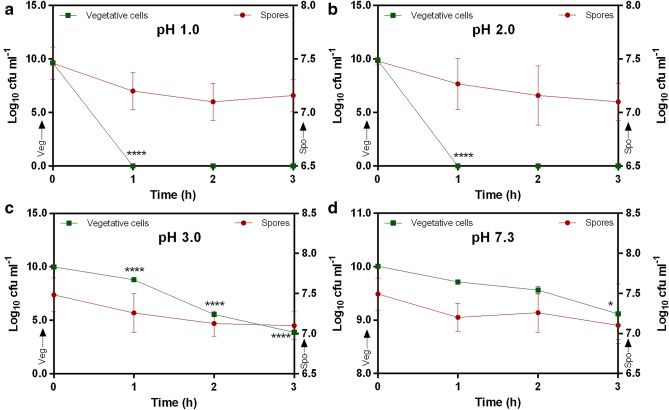
The exposure of spores (~ 7.48 log 10 cfu ml−1) to pH 1–3 for 3 h did not show any significant (p > 0.05) loss in survival (pH 1: 95.93 ± 2.02; pH 2: 94.92 ± 2.31; pH 3: 94.91 ± 2.30; pH 7.3: 94.79 ± 2.31%) as compared control (pH 7.3) (Fig. 1). On the contrary, the exposure of ~ 9.87 log 10 cfu ml−1 vegetative cells to pH 1 and 2 significantly (p < 0.0001) reduced survivability within an hour, with no vegetative cells surviving. At pH 3 the vegetative cells showed significant (p < 0.0001) reduction in survival up to 3 h (1 h: 88.02 ± 1.80; 2 h: 55.62 ± 0.75; 3 h: 38.80 ± 0.70%) (Fig. 1). Similar results were recorded when cells were exposed to pH 7.3, however, the decreased in survivability (1 h: 97.07 ± 0.30; 2 h: 95.57 ± 0.75; 3 h: 91.1 ± 5.56%) was less as compared to pH 3 (Fig. 1). The difference recorded in viability between 0 to 3 h were significant (p < 0.05).
Fig. 1.
Acid tolerance of spores and vegetative cells of Bacillus clausii UBBC07. Panel a pH 1.0; b pH 2.0; c pH 3.0; d pH 7.3 (control). The primary y-axis indicates vegetative cell count and secondary y-axis for spores. All data are represented as mean ± SD. *p < 0.05; ****p < 0.0001: significant difference compared to initial or 0 time point
Synthetic stomach juice tolerance
In synthetic gastric juice, the spore count was deceased significantly (p < 0.01) from 0 (7.62 ± 0.06 log 10 cfu ml−1) to 180 min (7.30 ± 0.07 log 10 cfu ml−1) of incubation (Fig. 2a). The percentage survival was determined as 95.75 ± 1.00%. Survival of vegetative cells were significantly (p < 0.0001) reduced to zero during the incubation (0 min: 9.7 ± 0.05; 30 min: 8.2 ± 0.09; 180 min: 0 log 10 cfu ml−1) in gastric juice (Fig. 2a).
Fig. 2.
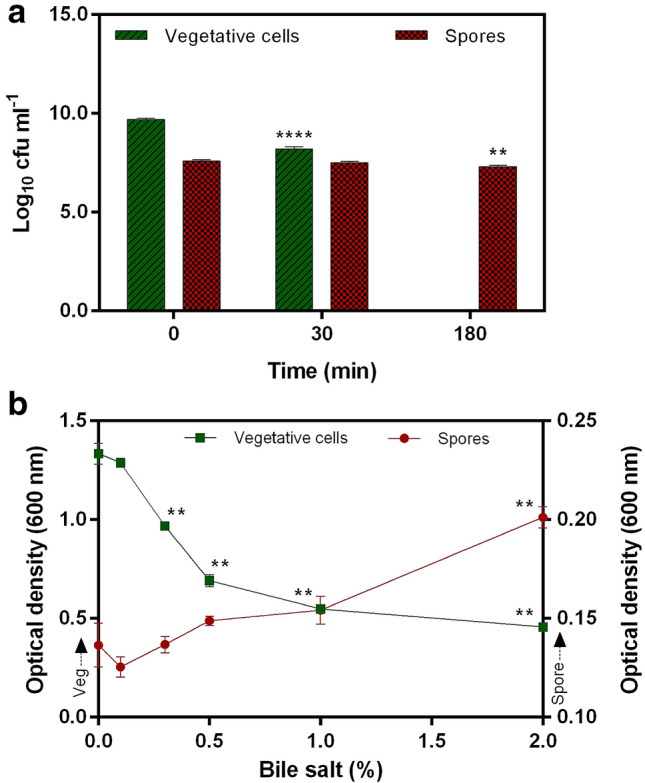
a Synthetic gastric juice; b bile salt tolerance of spores and vegetative cells of Bacillus clausii UBBC07. The primary y-axis indicates optical density for vegetative cell and secondary y-axis for spores. All data are represented as mean ± SD. **p < 0.01; ****p < 0.0001: significant difference compared to initial or 0 time point for gastric juice and 0% concentration for bile
Bile salt tolerance
Increasing concentrations of bile salts showed no adverse effects on the survivability of spores (bile 0.1%: 91.93 ± 3.76; 0.3%: 100.24 ± 3.05; 0.5%: 109.05 ± 1.70; 1.0%: 112.95 ± 5.13). In addition, 2.0% bile salt levels enhanced growth (147.43 ± 3.89%; p < 0.01) (Fig. 2b). Survivability of vegetative cells decreased (bile 0.3%: 72.56 ± 0.78; 0.5%: 51.80 ± 2.30; 1.0%: 41.04 ± 0.52) significantly (p < 0.01) when bile salt concentration was increased from 0.1% (Fig. 2b). No significant (p > 0.05) changes in survivability was recorded at 0.1% bile as compared with the control.
Intestinal fluid tolerance
In synthetic intestinal juice, the spore count increased significantly (p < 0.0001) from 0 (7.69 ± 0.08 log10 cfu ml−1) to 360 min (8.51 ± 0.07 log 10 cfu ml−1) of incubation (Fig. 3a). The survival was recorded as 110.66 ± 0.94%. On the contrary, the vegetative cell counts decreased significantly (p < 0.0001) from 7.59 ± 0.11 log 10 cfu ml−1 (0 min) to 5.77 ± 0.07 log10 cfu ml−1 (360 min) (Fig. 3a). Moreover, the vegetative cells showed 76.05 ± 0.96% survivability.
Fig. 3.
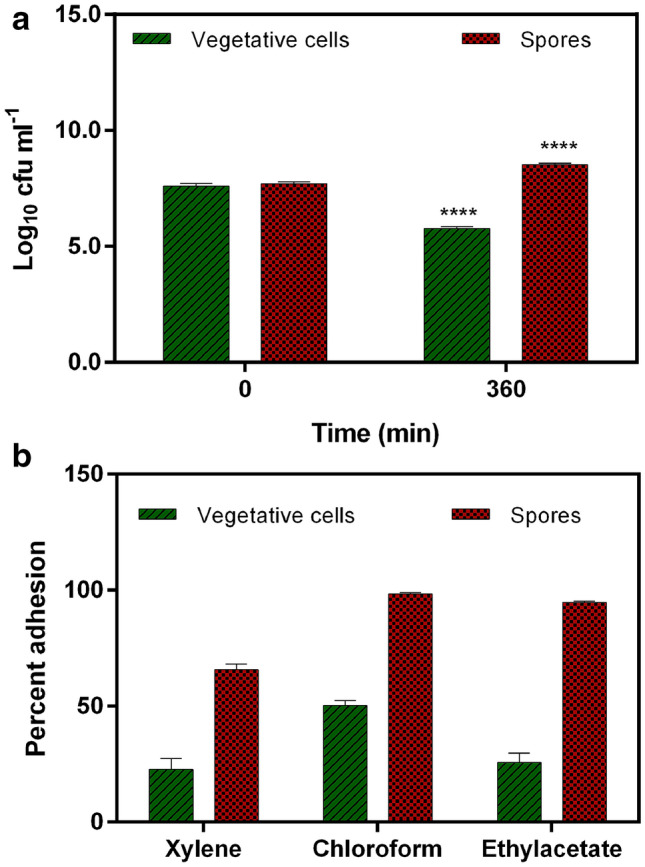
a Intestinal fluid tolerance; b Adhesion to solvents of spores and vegetative cells of Bacillus clausii UBBC07. All data are represented as mean ± SD. ****p < 0.0001: significant difference compared to initial or 0 time point
Microbial adhesion to solvents
Bacillus clausii UBBC07 spores had higher adhesion to chloroform (98.33 ± 0.57%) and ethyl acetate (94.66 ± 0.58%) as compared to xylene (65.66 ± 2.51%) (Fig. 3b). Whereas vegetative cells adhered greater to chloroform (50.33 ± 2.08%) as compared with xylene (22.66 ± 4.72%) and ethyl acetate (25.66 ± 4.16%) (Fig. 3b).
Adhesion to porcine mucin
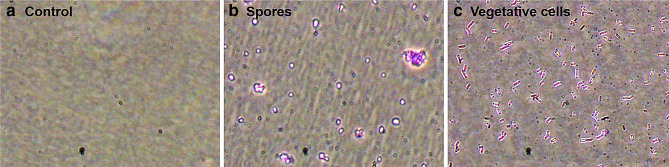
Spores had significantly (p < 0.01) higher crystal violet optical density readings (0.084 ± 0.004) as compared to vegetative cells (0.065 ± 0.005). Figure 4 describes the adhesion of spores and vegetative cells to mucin-coated wells.
Fig. 4.
Representative image of adhesion of spores and vegetative cells of Bacillus clausii UBBC07 to mucin. Panel a Control; b Spores; c Vegetative cells after crystal violet strain
Determination of zeta potential
Spores had significantly (p < 0.05) higher zeta potential (– 28.3 ± 1.04 mV; 7.95 ± 0.04 log10 cfu ml−1) as compared to vegetative cells (– 23.4 ± 2.23 mV; 8.01 ± 0.05 log10 cfu ml−1).
In silico identification of genome features contributing to probiotic properties
The in silico analysis of B. clausii UBBC07 genome revealed the presence of 10 domains for acid tolerance, three for bile tolerance, 11 for adhesion to gut mucosa, and 15 for environmental stress resistance (Table 1).
Table 1.
Distribution of proteins involved in probiotic properties in B. clausii UBCC07 genome
| Category | Probiotic feature | Accession numbers | Identified-domain |
|---|---|---|---|
| Acid tolerance | F0F1 ATP_synthase | KKI85936 | ATP synthase subunit A |
| KKI85898 | ATP synthase subunit C | ||
| KKI85899 | ATP synthase subunit B | ||
| KKI85900 | ATP synthase delta (OSCP) subunit | ||
| KKI85901 | ATP synthase subunit alpha | ||
| KKI85902 | ATP synthase subunit gamma | ||
| KKI85903 | ATP synthase subunit beta | ||
| KKI85904 | ATP synthase subunit epsilon | ||
| Amino acid decarboxylase | KKI85796 | Orn/Lys/Arg decarboxylase | |
| KKI84553 | Arginine/lysine/ornithine decarboxylase | ||
| Bile tolerance | Sodium bile acid symporter | KKI85025 | Bile acid sodium symporter |
| KKI85546 | Sodium transporter | ||
| KKI86848 | Sodium bile acid family transporter | ||
| Adhesion to gut mucosa | Mucus binding protein | * | Gram_pos_anchor |
| Collagen binding protein | † | Collagen_bind | |
| Fibronectin binding protein | KKI87777 | FbpA | |
| Sortase | KKI87126 | Sortase | |
| KKI87260 | Sortase | ||
| KKI84941 | Sortase | ||
| KKI86106 | Sortase | ||
| KKI84754 | Sortase | ||
| Flagellin | KKI85107 | Flagellin_C | |
| KKI87499 | Flagellin_N | ||
| Triosephosphate isomerase | KKI85379 | TIM | |
| Environmental stress resistance | Chaperonins GroEL | KKI84884 | Cpn60_TCP1 |
| Chaperonins GroES | KKI84885 | Cpn10 | |
| Clp protease | KKI84682 | CLP_protease | |
| KKI85387 | CLP_protease | ||
| KKI87878 | CLP_protease | ||
| Cold shock protein | KKI84482 | CspC | |
| KKI87492 | CSD | ||
| Heat shock protein | KKI85846 | HSP33 | |
| Heat resistance | KKI84985 | GrpE | |
| Hyperosmotic stress | KKI84983 | DnaJ | |
| Oxidative stress | KKI86288 | PMSR | |
| KKI88047 | PMSR | ||
| Universal Stress | KKI84787 | Usp | |
| KKI85810 | Usp | ||
| KKI87408 | Usp |
*Located on LATY01000009 with locus tag WZ76_RS06065
†Located on LATY01000017 with locus tag WZ76_RS12025
Discussion
The ability of probiotics to reach the gut in sufficient numbers is imperative in order for cells to confer health benefits. As per recommendations, most probiotic products contain billions of cells and the benefits they confer is dependent on the strains ability to survive transit through the gut. There are several factors which contribute to the success of probiotic, such as the stability at various industrial processes and tolerance to the gastrointestinal tract stress (Ahire 2012). The use of spore probiotic is advantageous over the vegetative cells since spore’s unique intrinsic makeup (dipicolinic acid, proteins, lipids, and carbohydrates) and extremely low permeability provides high tolerance to the stomach acidity, bile salt and intestinal conditions (Bernardeau et al. 2017). In this study, the Bacillus clausii UBBC07 spores demonstrated high resistance to acidic conditions (pH 1, 2, and 3) and synthetic gastric juice (pH 2.5) as compared to vegetative cells. These in vitro results suggests that spores are probably able to survive and deliver prerequisite quantities to the small intestine. Cenci et al. (2006) has shown that B. clausii spores tolerated pH 2 and vegetative cells pH ≤ 4. Recently, the in vitro investigation of B. clausii spore germination in the Simulator of Human Intestinal Microbial Ecosystem (SHIME) indicated the survival of spores and accompanied vegetative cells under SHIME-fed stomach simulations (Ahire et al. 2020b). Besides this, none of the studies evaluated the comparative probiotic properties of spore and vegetative cells.
Bile acid is the major component of bile, which acts as an emulsifier to facilitate the digestion of lipids and lipid-soluble-vitamins in the intestine. In higher concentrations, the bile acid is toxic to the bacterial cells by causing membrane damage, protein denaturation, and oxidative damage to the DNA (Prete et al. 2020). Therefore, the investigation of probiotic bacteria to survive bile acids is important to predict their persistence in the gut. In this study, the bile tolerance observed in UBBC07 spores was higher than the vegetative biotype, which is due to the intrinsic resistance of spores to the bile. However, the increased log10 cfu ml−1 from B. clausii spores at higher bile levels suggested bile-induced spore germination (Giel et al. 2010). The capabilities of B. clausii spores to germinate under fed and fasted in vitro intestinal-SHIME-conditions have recently been reported (Ahire et al. 2020b). Ghelardi et al. (2015) showed that the B. clausii spores germinate and undergoes multiplication under stimulated in vivo human intestinal environments. Moreover, bile tolerance is a strain-specific trait (Hyronimus et al. 2000).
The tolerance of probiotics to the intestinal fluid containing pancreatin and bile under alkaline conditions is a good model to estimate their survivability in the gut. In the present investigation, B. clausii spores replicated in simulated intestinal conditions as compared with vegetative cells. This finding indicates the germination and multiplication ability of B. clausii spores in the intestinal fluid. The 76% viability of vegetative cells to the intestinal fluid assures the persistence of the strain in the gut. Furthermore, it has been reported that B. clausii survival and persistence in alkaline conditions might be due to the alkaliphilic nature of this species (Nielsen et al. 1995; Vecchione et al. 2018). Overall, these results corroborate well with previous in-vitro and -vivo findings that B. clausii spores germinate and multiply in human intestinal conditions (Cenci et al. 2006; Ghelardi et al. 2015; Ahire et al. 2020b).
Like stomach and intestinal stress tolerance, the adhesion of probiotics is an important property for successful colonization in the gut. In the present study, we investigated the adhesion of spores and vegetative cells of B. clausii using adhesion to -solvents, -mucin and zeta potential. In adhesion to solvents, the adhesion to xylene is an indication of hydrophobic surface properties (Bellon-Fontaine et al. 1996). The high percent affinity of spores to xylene as compared with vegetative cells indicated higher surface hydrophobicity of spores. This may be due to the relative abundance of protein in the outer coat or exosporium of spore compared with peptidoglycan on the vegetative cell surface (Jindal and Anand 2018). High adhesion of spores to chloroform and ethyl acetate as compared with vegetative cells indicated the electron-donating and electron-accepting properties of biological surfaces (Bellon-Fontaine et al. 1996). The strong adhesion of spores to porcine mucin and significantly higher net negative zeta potential value over vegetative cells further confirmed the findings. Overall, these results show that spores are highly hydrophobic and more capable of adhering to gut epithelial lining as compared to vegetative cells.
In another investigation, we have analyzed the whole genome sequence of B. clausii UBBC07 to identify the genome features contributing to probiotic properties. The presence of F0F1 ATP synthase complex indicated the ability of bacteria to resist the acidic environment of the stomach by maintaining H+ homeostasis (Cotter and Hill 2003; Azcarate-Peril et al. 2004; Khatri et al. 2019). The ornithine/lysine/arginine decarboxylase family proteins catalyze the decarboxylation of amino acids resulting in the alkalinization of the cytosol and generation of a proton motive force, which can be exploited for acid resistance and/or the production of ATP (Romano et al. 2013). The sodium bile acid symporter family proteins contribute to bile resistance and adaptation to the gut environment (Price et al. 2006). Besides this, the proteins detected for mucus, collagen, and fibronectin-binding along with sortase, flagellin, and triosephosphate isomerase ensures adhesion to the intestinal mucosal layer and persistence of bacteria to the intestine. Furthermore, B. clausii UBBC07 harbors proteins for universal-, oxidative-, hyperosmotic-stress, heat resistance, cold and heat shock, Clp protease, and chaperonins (GroEL and GroES) for survival and growth under environmental stress. Overall, these results corroborate well with the previous reports on in silico analysis of proteins involved in probiotic properties of B. clausii Enterogermina® (Khatri et al. 2019).
In conclusion, Bacillus clausii UBBC07 spores demonstrated excellent gastro-intestinal resistance as compared with vegetative biotype. No loss in viability, good adhesion, and spore germination under simulated in vitro human intestinal conditions ensures the delivery of the recommended amount of probiotics to the gut. Moreover, in silico analysis revealed the presence of proteins involved in probiotic properties in B. clausii UBCC07 genome. Therefore, we recommend that spores of B. clausii UBBC07 be used to deliver probiotic to the human and or animal gut where they germinate and colonise to confer intended health benefits.
Author contributions
JJA contributed to the study conception, design, acquisition and analysis of data, drafting and critically revising the manuscript. MSK carried out the material preparation and data collection. RSM contributed to the study conception and review. All authors approved the final submitted manuscript.
Funding
No external funding was received. The financial support for the research described in the manuscript was provided by Unique Biotech Limited, Hyderabad, India.
Data availability
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Compliance with ethical standards
Conflict of interest
JJA, MSK and RSM are employed by Unique Biotech Limited, India, which is a manufacturer of probiotics. This does not alter our adherence to journal policies on sharing data and materials.
Ethics approval
The research conducted for this article did not involve studies on humans or animals.
References
- Ahire JJ (2012) Studies on probiotic microorganism(s) and its biogenic metabolite(s). Dissertation, North Maharashtra University, India.
- Ahire JJ, Dicks LM. 2, 3-Dihydroxybenzoic acid-containing nanofiber wound dressings inhibit biofilm formation by Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58:2098–2104. doi: 10.1128/AAC.02397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahire JJ, Mokashe NU, Patil HJ, Chaudhari BL. Antioxidative potential of folate producing probiotic Lactobacillus helveticus CD6. J Food Sci Tech. 2013;50:26–34. doi: 10.1007/s13197-011-0244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahire JJ, Kashikar MS, Lakshmi SG, Madempudi R. Identification and characterization of antimicrobial peptide produced by indigenously isolated Bacillus paralicheniformis UBBLi30 strain. 3 Biotech. 2020;10:112. doi: 10.1007/s13205-020-2109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahire JJ, Kashikar MS, Madempudi RS. Survival and germination of Bacillus clausii UBBC07 spores in in vitro human gastrointestinal tract simulation model and evaluation of clausin production. Front Microbiol. 2020;11:1010. doi: 10.3389/fmicb.2020.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahire JJ, Neelamraju J, Madempudi RS. Behavior of Bacillus coagulans Unique IS2 spores during passage through the simulator of human intestinal microbial ecosystem (SHIME) model. LWT-Food Sci Technol. 2020;124:109196. doi: 10.1016/j.lwt.2020.109196. [DOI] [Google Scholar]
- Azcarate-Peril MA, Altermann E, Hoover-Fitzula RL, Cano RJ, Klaenhammer TR. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl Environ Microbiol. 2004;70:5315–5322. doi: 10.1128/AEM.70.9.5315-5322.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon-Fontaine MN, Rault J, van Oss CJ. Microbial adhesion to solvents: a novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surf B. 1996;7:47–53. doi: 10.1016/0927-7765(96)01272-6. [DOI] [Google Scholar]
- Bernardeau M, Lehtinen MJ, Forssten SD, Nurminen P. Importance of the gastrointestinal life cycle of Bacillus for probiotic functionality. J Food Sci Technol. 2017;54:2570–2584. doi: 10.1007/s13197-017-2688-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan B, Singh BP, Saini K, Kumari M, Tomar SK, Mishra V. Role of microbes, metabolites and effector compounds in host–microbiota interaction: a pharmacological outlook. Environ Chem Lett. 2019;17:1801–1820. doi: 10.1007/s10311-019-00914-9. [DOI] [Google Scholar]
- Bhushan B, Sakhare SM, Narayan KS, Kumari M, Mishra V, Dicks LM. Characterization of riboflavin-producing strains of Lactobacillus plantarum as potential probiotic candidate through in vitro assessment and principal component analysis. Probiot Antimicro Prot. 2020 doi: 10.1007/s12602-020-09696-x. [DOI] [PubMed] [Google Scholar]
- Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, III, Stevens R, Vonstein V, Wattam AR, Xia F. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G, Caldini TF. Tolerance to challenges miming gastrointestinal transit by spores and vegetative cells of Bacillus clausii. J Appl Microbiol. 2006;101:1208–1215. doi: 10.1111/j.1365-2672.2006.03042.x. [DOI] [PubMed] [Google Scholar]
- Cotter PD, Hill C. Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol Mol Biol Rev. 2003;67:429–453. doi: 10.1128/mmbr.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc LH, Hong HA, Barbosa TM, Henriques AO, Cutting SM. Characterization of Bacillus probiotics available for human use. Appl Environ Microbiol. 2004;70:2161–2171. doi: 10.1128/AEM.70.4.2161-2171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghelardi E, Celandroni F, Salvetti S, Gueye SA, Lupetti A, Senesi S. Survival and persistence of Bacillus clausii in the human gastrointestinal tract following oral administration as spore-based probiotic formulation. J Appl Microbiol. 2015;119:552–559. doi: 10.1111/jam.12848. [DOI] [PubMed] [Google Scholar]
- Giel JL, Sorg JA, Sonenshein AL, Zhu J. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS ONE. 2010;5:e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- Hyronimus B, Le Marrec C, Sassi AH, Deschamps A. Acid and bile tolerance of spore-forming lactic acid bacteria. Int J Food Microbiol. 2000;61:193–197. doi: 10.1016/s0168-1605(00)00366-4. [DOI] [PubMed] [Google Scholar]
- Ianiro G, Rizzatti G, Plomer M, Lopetuso L, Scaldaferri F, Franceschi F, Cammarota G, Gasbarrini A. Bacillus clausii for the treatment of acute diarrhea in children: a systematic review and meta-analysis of randomized controlled trials. Nutrients. 2018;8:1074. doi: 10.3390/nu10081074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal S, Anand S. Comparison of adhesion characteristics of common dairy sporeformers and their spores on unmodified and modified stainless steel contact surfaces. J Dairy Sci. 2018;101:5799–5808. doi: 10.3168/jds.2017-14179. [DOI] [PubMed] [Google Scholar]
- Khatri I, Sharma G, Subramanian S. Composite genome sequence of Bacillus clausii, a probiotic commercially available as Enterogermina®, and insights into its probiotic properties. BMC Microbiol. 2019;19:307. doi: 10.1186/s12866-019-1680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmi SG, Jayanthi N, Saravanan M, Ratna MS. Safety assesment of Bacillus clausii UBBC07, a spore forming probiotic. Toxicol Rep. 2017;4:62–71. doi: 10.1016/j.toxrep.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marseglia GL, Tosca M, Cirillo I, Licari A, Leone M, Marseglia A, Castellazzi AM, Ciprandi G. Efficacy of Bacillus clausii spores in the prevention of recurrent respiratory infections in children: a pilot study. Ther Clin Risk Manag. 2007;1:13–17. doi: 10.2147/tcrm.2007.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P, Fritze D, Priest FG. Phenetic diversity of alkaliphilic Bacillus strains: proposal for nine new species. Microbiol. 1995;141:1745–1761. doi: 10.1099/13500872-141-7-1745. [DOI] [Google Scholar]
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparo L, Tripodi L, Bruno C, Pisapia L, Damiano C, Pastore L, Canani RB. Protective action of Bacillus clausii probiotic strains in an in vitro model of Rotavirus infection. Sci Rep. 2020;10:12636. doi: 10.1038/s41598-020-69533-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AK, Ahire JJ, Pawar SP, Chaudhari BL, Chincholkar SB. Comparative accounts of probiotic characteristics of Bacillus spp. isolated from food wastes. Food Res Int. 2009;42:505–510. doi: 10.1016/j.foodres.2009.01.013. [DOI] [Google Scholar]
- Patel C, Patel P, Acharya S. Therapeutic prospective of a spore forming probiotic-Bacillus clausii UBBC07 against acetaminophen-induced uremia in rats. Probiot Antimicro Prot. 2019;12:253–258. doi: 10.1007/s12602-019-09540-x. [DOI] [PubMed] [Google Scholar]
- Pedersen C, Jonsson H, Lindberg JE, Roos S. Microbiological characterization of wet wheat distillers grain, with focus on isolation of lactobacilli with potential as probiotics. Appl Environ Microbiol. 2004;70:1522–1527. doi: 10.1128/AEM.70.3.1522-1527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prete R, Long SL, Gallardo AL, Gahan CG, Corsetti A, Joyce SA. Beneficial bile acid metabolism from Lactobacillus plantarum of food origin. Sci Rep. 2020;10:1165. doi: 10.1038/s41598-020-58069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CE, Reid SJ, Driessen AJ, Abratt VR. The Bifidobacterium longum NCIMB 702259T ctr gene codes for a novel cholate transporter. Appl Environ Microbiol. 2006;72:923–926. doi: 10.1128/AEM.72.1.923-926.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A, Trip H, Lolkema JS, Lucas PM. Three-component lysine/ornithine decarboxylation system in Lactobacillus saerimneri 30a. J Bacteriol. 2013;195:1249–1254. doi: 10.1128/JB.02070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Devi PB, Ragul K, Shetty PH. Probiotic potential of Bacillus strains isolated from an acidic fermented food Idli. Probiot Antimicro Prot. 2020;12:1502–1513. doi: 10.1007/s12602-020-09650-x. [DOI] [PubMed] [Google Scholar]
- Sudha MR, Bhonagiri S, Kumar MA. Efficacy of Bacillus clausii strain UBBC-07 in the treatment of patients suffering from acute diarrhoea. Benef Microbes. 2013;4:211–216. doi: 10.3920/BM2012.0034. [DOI] [PubMed] [Google Scholar]
- Sudha MR, Jayanthi N, Pandey DC, Verma AK. Bacillus clausii UBBC-07 reduces severity of diarrhoea in children under 5 years of age: a double blind placebo controlled study. Benef Microbes. 2019;10:149–154. doi: 10.3920/BM2018.0094. [DOI] [PubMed] [Google Scholar]
- Upadrasta A, Pitta S, Madempudi RS. Draft genome sequence of Bacillus clausii UBBC07, a spore-forming probiotic strain. Genome Announc. 2016;4:e00235–e316. doi: 10.1128/genomeA.00235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchione A, Celandroni F, Mazzantini D, Senesi S, Lupetti A, Ghelardi E. Compositional quality and potential gastrointestinal behavior of probiotic products commercialized in Italy. Front Med. 2018;5:59. doi: 10.3389/fmed.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.