Abstract
Only 20% NSCLC patients benefit from immunotherapy with a durable response. Current biomarkers are limited by the availability of samples and do not accurately predict who will benefit from immunotherapy. To develop a unified deep learning model to integrate multimodal serial information from CT with laboratory and baseline clinical information. We retrospectively analyzed 1633 CT scans and 3414 blood samples from 200 advanced stage NSCLC patients who received single anti-PD-1/PD-L1 agent between April 2016 and December 2019. Multidimensional information, including serial radiomics, laboratory data and baseline clinical data, was used to develop and validate deep learning models to identify immunotherapy responders and nonresponders. A Simple Temporal Attention (SimTA) module was developed to process asynchronous time-series imaging and laboratory data. Using cross-validation, the 90-day deep learning-based predicting model showed a good performance in distinguishing responders from nonresponders, with an area under the curve (AUC) of 0.80 (95% CI: 0.74-0.86). Before immunotherapy, we stratified the patients into high- and low-risk nonresponders using the model. The low-risk group had significantly longer progression-free survival (PFS) (8.4 months, 95% CI: 5.49-11.31 vs. 1.5 months, 95% CI: 1.29-1.71; HR 3.14, 95% CI: 2.27-4.33; log-rank test, P<0.01) and overall survival (OS) (26.7 months, 95% CI: 18.76-34.64 vs. 8.6 months, 95% CI: 4.55-12.65; HR 2.46, 95% CI: 1.73-3.51; log-rank test, P<0.01) than the high-risk group. An exploratory analysis of 93 patients with stable disease (SD) [after first efficacy assessment according to the Response Evaluation Criteria in Solid Tumors (RECIST) 1.1] also showed that the 90-day model had a good prediction of survival and low-risk patients had significantly longer PFS (11.1 months, 95% CI: 10.24-11.96 vs. 3.3 months, 95% CI: 0.34-6.26; HR 2.93, 95% CI: 1.69-5.10; log-rank test, P<0.01) and OS (31.7 months, 95% CI: 23.64-39.76 vs. 17.2 months, 95% CI: 7.22-27.18; HR 2.22, 95% CI: 1.17-4.20; log-rank test, P=0.01) than high-risk patients. In conclusion, the SimTA-based multi-omics serial deep learning provides a promising methodology for predicting response of advanced NSCLC patients to anti-PD-1/PD-L1 monotherapy. Moreover, our model could better differentiate survival benefit among SD patients than the traditional RECIST evaluation method.
Keywords: NSCLC, SimTA, multi-omics serial deep learning
Introduction
Lung cancer is the leading cause of mortality globally. Non-small-cell lung cancer (NSCLC) accounts for 80%-85% of lung cancer cases [1]. In recent years, immunotherapy has significantly improved the overall survival (OS) and quality of life of NSCLC patients [2-7]. Nevertheless, only 20% of NSCLC patients benefit from immunotherapy with a durable response. Some biomarkers, such as tumor mutation burden [8,9], PD-L1 expression, tumor-infiltrating lymphocytes, and gut microbiome diversity have been reported to be associated with favorable responses to anti-PD-1/PD-L1 therapy [10]. However, the application of these potential biomarkers to a broader patient population in the clinical setting are limited by the availability of samples and the fact that none of the current biomarkers is applicable to all anti-PD-1/PD-L1 drugs [11]. Therefore, with rapid development of immunotherapy and its economic benefit analysis, the combination of biomarkers that can be easily obtained and followed up are urgently needed in the real-world clinical setting.
A growing body of evidence suggests that artificial intelligence (AI) could assess response to immunotherapy through recognition of radiomic biomarkers in NSCLC [12]. Radiomics, AI-enhanced high-dimensional radiological features, has a number of advantages such as noninvasiveness and easy and automated serial monitoring [13]. Radiographic images provide complete and comprehensive information of the entire tumor compared with biopsy. It could help identify early lung cancer and epithelial growth factor receptor (EGFR) and KRAS mutations in NSCLC [14,15]. It could reveal tumor heterogeneity and a possible heterogeneous treatment response [16]. Grossmann et al. reported two radiomics features: region dissimilarity and entropy, were associated with OS after second-line treatment with nivolumab [17]. The integration of radiomics data into conventional clinical and laboratory data could provide complementary patient- and tumor-specific information for the management and prognostic determination of patients with NSCLC [18,19]. Previous studies have demonstrated the value of radiomics in predicting the response of advanced NSCLC patient response to immunotherapy [17,20], but its survival benefit and effect on patients with stable disease (SD) are unclear. In addition, immunotherapy has introduced new response patterns such that long-term survival benefits for patients are not solely related to tumor shrinkage [21]. The value of multi-omics marker in predicting survival benefits in patients indistinguishable by conventional response criteria has not been explored.
The emerging deep learning technology [22] has dominated medical image analysis in a wide range of data modalities [e.g., ultrasound [23], computed tomography (CT) [24], and magnetic resonance imaging (MRI) [25] on several tasks [26-28]]. This study aimed to develop a unified deep learning model to integrate multimodal serial information from CT with laboratory and baseline clinical information. The asynchronism of clinical time series poses challenges to the design of deep learning models. Inspired by the recent advances in natural language processing [29], this study proposed a novel attention mechanism, named Simple Temporal Attention (SimTA), to process multimodal asynchronous time series from serial clinical data. The SimTA module was evaluated using serial radiographic and blood test information, as well as patient clinical data, for predicting clinical outcomes in patients with advanced NSCLC who received second-line and above anti-PD-1/PD-L1 monotherapy. The study also evaluated whether this multi-omics-based model could further detect patients with the same SD assessed by RECIST 1.1 who may derive a real long-term survival benefit from immunotherapy.
Patients and methods
Patients
This retrospective study included pathologically or cytologically proven stage IIIB and IV (2009 AJCC, 8th edition) NSCLC patients who received single-agent anti-PD-1/PD-L1 therapy between 2016 and 2019 at Shanghai Lung Cancer Center, Shanghai Chest Hospital, Jiao Tong University. Patients received 3 mg/kg nivolumab every 2 weeks, 200 mg pembrolizumab every 3 weeks, or 1200 mg atezolizumab every 3 weeks. We utilized 4 datasets in this study: 1) the datasets from the CheckMate-870 trial (ClinicalTrials.gov NCT03195491) with images acquired between January 2018 and July 2018, 2) the CheckMate-078 trial (Clinicaltrials.gov NCT02613507) with images acquired January 2016 and April 2016 [6], 3) the OAK (YO29232) trial (Clinicaltrials.gov NCT02008227) with images acquired between July 2016 and May 2017 [5], and 4) a real-world population with images acquired between June 2019 and December 2019.
The main inclusion criteria were 1) an Eastern Cooperative Oncology Group (ECOG) score of 0 or 1; 2) measurable lung lesions detected by CT scan as per the Response Evaluation Criteria in Solid Tumors (RECIST 1.1) [30]; 3) disease progression despite previous platinum-based chemotherapy for patients without EGFR mutation or disease progression during or after second-line or above platinum-based chemotherapy following first-line treatment with tyrosine kinase inhibitors for patients with EGFR mutation; 4) complete laboratory and radiological data on disease progression were available, and routine blood and biochemical tests were performed at baseline within 14 days of the first dose of anti-PD-1/PD-L1 and 3 days before each subsequent immunotherapy cycle, and chest CT scan was available at baseline and every 6 to 8 weeks after treatment initiation; 5) follow-up data were complete and endpoint events and status were clearly noted in the charts.
The study was approved by the ethics committee of Shanghai Chest Hospital (Approval No.: KS 1732) and informed consent was waived.
Data extraction
Patient demographics including age and sex, medical history including smoking history, and baseline data, routine laboratory investigations, and CT scans at baseline and follow ups were extracted from the hospital’s electronic medical records system. Baseline data included ECOG performance status, American Joint Committee on Cancer (AJCC) stage, histology, EGFR and KRAS mutational status, and concurrent therapy (chemotherapeutic regimen and line, and radiotherapy). Blood cell counts included white blood cell, neutrophil, lymphocyte, monocyte, eosinophil, and basophil counts and percentages; hemoglobin; red blood cell counts; hematocrit; mean corpuscular hemoglobin concentration; red blood cell distribution width; red blood cell distribution width standard deviation; platelet count; platelet-larger cell ratio; plateletcrit; mean platelet volume; platelet distribution width. Blood biochemistries included total protein, albumin, lactate dehydrogenase, alanine aminotransferase, aspartate aminotransferase, urea, and creatinine.
CT data
Chest CT scans were carried out according to standardized scanning protocols at our institution using a 64-layer LightSpeed Volume CT scanner (GE Healthcare, WI, USA). All imaging data were reconstructed using a medium-sharp reconstruction algorithm with a thickness of 3-5 mm. Tumor segmentation was performed to select the essential primary lesions of NSCLC cases after image acquisition. All nodules were manually delineated by two oncologists (Supplementary Methods). Target lesions were defined as any tumor that was well demarcated on both baseline and follow-up CT scans with a diameter ≥5 mm. Based on the evaluation of follow-up CT scans, patients with complete response (CR), partial response (PR) or SD were considered responders and patients with progressive disease (PD) were considered nonresponders according to RECIST 1.1 (Supplementary Figure 2). All CT scans were reviewed by two independent oncologists.
Follow up
Patients were followed up every 3 months by telephone, outpatient visit, or review of medical records. The date of diagnosis was the start date of follow-up, the first clinical efficacy assessment was between 6 and 8 weeks in the four datasets and the cutoff date was May 31, 2020.
Deep learning
The mean interval between baseline CT scan and the first dose of anti-PD-1/PD-L1 was 14 days (1-29 days). We collected 1633 CT scans of nodule markers and 3414 blood samples. Due to a limited number of CT scans, radiomics was used to represent the radiographic features instead of end-to-end convolutional neural networks. The radiomic features were extracted with PyRadiomics (Python 3.7.3, PyRadiomics 2.2.0) [31-35] with spatial normalization to 1 × 1 × 1 mm3. For each lesion, a radiomic feature of 107 dimensions was extracted from the CT scan as previously described [36]. The radiomic features were used as serial inputs to the module if CT scans were conducted more than once. Clinical and laboratory variables were categorized as serial and static. By filtering out fields with more than 10% missing information, the serial blood test features were of 22 dimensions and the static features were of 18 dimensions, where categorical features were one-hot encoded. Numeric features were normalized to enable more stable and better convergence for model training.
Both radiomic and blood test features were regarded as asynchronous time-series data with varying time intervals between neighboring time steps. Hence, a novel attention mechanism, named SimTA module, was developed to process the asynchronous clinical time series (Figure 1). The computation details of SimTA, model pipeline, and model training are provided in the following sections. Two SimTA modules were developed: 60- and 90-day response models. During the training phase, the models were trained with time-series data (radiomics and laboratory) 60 and 90 days prior to each efficacy assessment (responders/nonresponders, respectively). In the evaluation phase, only the serial data prior to baseline immunotherapy were used to predict response after 60 and 90 days. Deep learning models using only baseline PD-L1 expression, blood profile, radiomics, and recurrent neural network (RNN) [37,38] were also tested for comparison with SimTA modules.
Figure 1.
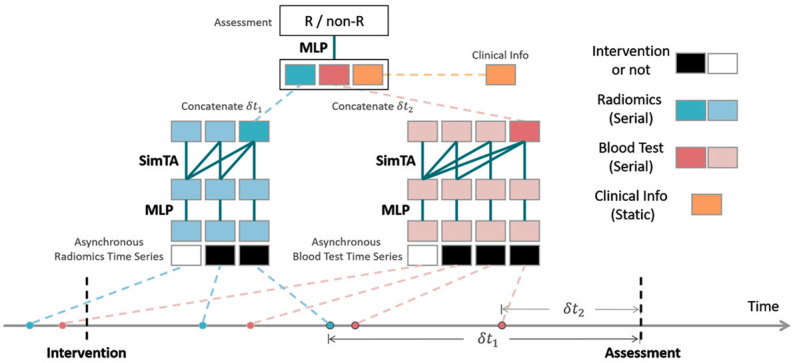
A deep learning model with Simple Temporal Attention (SimTA) modules. The SimTA modules process asynchronous clinical time series (i.e., the radiomics and blood tests) separately; the encoded features of these time series and static clinical information are then fused by a multiple-layer perceptron (MLP) to get the final output for the assessment prediction of responders/non-responders (R/non-R).
Furthermore, deep neural networks were implemented in PyTorch 1.2 [39]. We randomly split the dataset into 5 folds and performed 5-fold cross validation to evaluate the performance of the proposed method [40]. All hyper-parameters, including multiple layer perceptron (MLP) structure, learning rate, batch size, and regularization parameters, were selected with a random search via an additional cross-validation within the training dataset of each fold. The evaluation results were reported on all the validation sets on 5 folds.
Simple temporal attention (SimTA) module
The SimTA module learned representation from asynchronous time series of arbitrary length and interval and output a single feature vector as a summary of this time series. Given an asynchronous time series X 0 ∈ RT×c0, where T denoted the number of time steps in this series and c 0 denoted the input feature dimension of each time step, and τ ∈ R T-1 was denoted as the time intervals between neighboring time steps. For synchronous time series, τ was a constant vector. First, the feature vector X 0 was transformed into X 1 ∈ RT×c1 via MLP, where c 1 was its feature dimension. Inspired by the relative position encoding in attention transformers, the relations between time steps were learned via the time intervals [41]. Let X 1 (1), X 1 (2), ..., X 1 (T) ∈ Rc1 be the feature vectors from a time step, and τ(1),τ(2),...,τ(T-1) ∈ R be the time intervals, SimTA transformed feature vectors by (Equations 1, 2, 3 and 4):
Where λ and β were learnable scaling parameters. X 2=[X 2 (1), X 2 (2), ..., X 2 (T)] denoted the output by the SimTA module, which represented the transformed feature vectors for the time series. In the present study, X 2 (T) was taken as a summary vector of the time series to input into subsequent modules.
The relation learning formula -λτ (t)+β was based on the assumption that a measurement closer to assessment time was more important than a measurement further from the assessment time. Theoretically, a complex nonlinear temporal relation learning is of importance with MLP or self-attention. Besides, it is possible to stack more MLP and SimTA modules subsequently; however, a single MLP and a single SimTA module were used in this study to avoid the risk of overfitting. The softmax operation was defined as (Equation 5)
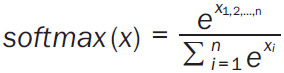 |
Model pipeline, training and tuning
The proposed SimTA module processed the input of asynchronous time series and output a summary vector. The whole model is illustrated in Figure 1. Multimodal information, including radiomics, blood tests and other clinical information, was processed separately. Two SimTA modules with MLPs processed the asynchronous radiomics time series (blue in Figure 1) and blood test time series (red in Figure 1), respectively, and integrated the information obtained with clinical information (yellow in Figure 1). A binary flag of 0/1, indicating whether a time step was before or after the intervention, was concatenated with each step of both time series, before inputting into the MLPs and SimTA modules. The summary vectors (of both radiomics series and blood test series) from SimTA modules, together with the static clinical information (yellow in Figure 1) and time intervals (i.e., δt 1 and δt 2 in Figure 1) between the last time steps and assessment time step, were fused with a final MLP to output the final prediction of R or non-R, with a final sigmoid activation function.
The neural network was trained on all assessment time steps. The optimization target was the assessment result of treatment response (responder versus nonresponder), while the network input was twofold: static and serial (i.e., the time series). Static input included patient clinical data. The time series input included all radiomic features and blood test data collected 30, 60, and 90 days prior to assessment. The neural network was optimized with an Adam optimizer [42], and the loss function was binary cross-entropy (Equation 6):
Where y and ŷ were ground-truth and predicted score, respectively.
When a neural network with limited data is trained, special care should be taken to prevent the network from overfitting. For time series model training, we applied data augmentation including random time step dropout, time scaling, and time shifting. Random time step dropout arbitrarily discarded features at certain time steps with a given dropout probability. Time scaling randomly scaled time interval, and time shifting added a random bias to it. The data augmentation rendered the model less prone to overfitting.
All hyper-parameters, including MLP structure, learning rate, batch size, and regularization parameters, were selected with a random search via an additional cross-validation within the training dataset of each fold. Crucially, this ensured that we did not violate the assumption of independence between the datasets used for developing and evaluating our method.
Statistical analysis
Data were analyzed using SPSS 22.0 (IBM, Armonk, NY, USA). Patient characteristics were analyzed using chi-square or Fisher’s exact test, where appropriate. To fairly evaluate the performance of SimTA, we conducted both receiver operating characteristic (ROC) and survival analyses using the proposed module. In the ROC analysis, module predictions of treatment response were compared with the ground truth (responders/nonresponders) in a binary classification fashion.
In survival analysis, patients were stratified into the low and high-risk groups for failure to respond to immunotherapy with a default threshold of 0.5 using model predictions, as we formalized the problem in this study as a binary classification of responders and nonresponders. Survival for each group was estimated using the Kaplan-Meier method and compared using log-rank test. Hazard ratios (HRs) and 95% confidence intervals (CIs) for the high and low risk groups were calculated using a Cox proportional-hazards model. OS was calculated from the start of immunotherapy to death from any cause or the final cutoff date. Progression-free survival (PFS) was defined as the time from the start of immunotherapy to disease progression per RECIST 1.1 or death. All tests were two sided, where applicable, and considered significant when P<0.05.
Results
Patient characteristics
The study flowchart is shown in Supplementary Figure 1. Totally 308 stage IIIB and IV patients received anti-PD-1/PD-L1 monotherapy. We excluded 108 patients with incomplete data; finally, 200 patients were included in the current analysis. Patient demographics, baseline and treatment characteristics are presented in Table 1. Their median age was 63 (range 23 to 82) years and 82.5% of them were male. Furthermore, 20.5% and 79.5% patients had stage IIIB and IV NSCLC, respectively. In addition, 57.5% patients were responders to anti-PD-1/PD-L1 monotherapy. Thirteen percent of the responders had an ECOG performance status score of 0 versus 0% in the nonresponders (P<0.001). The responders and nonresponders were comparable in the other demographic, baseline and treatment variables.
Table 1.
Patient demographics, baseline and treatment characteristics
| Variables | Responders* | Nonresponders* | P |
|---|---|---|---|
| N (%) | 115 (57.5) | 85 (42.5) | |
| Median age (95% CI), years | 62 (61-64) | 64 (61.5-66.5) | 0.56 |
| Male sex | 97 (84.3) | 68 (80.0) | 0.42 |
| ECOG performance status | |||
| 0 | 15 (13) | 0 | <0.01 |
| 1 | 100 (87) | 85 (100) | |
| Smoking Status# | |||
| Current or former smokers | 70 (60.9) | 49 (57.6) | 0.65 |
| Never smokers | 45 (39.1) | 36 (42.4) | |
| Tumor histology | |||
| Squamous | 40 (34.8) | 28 (32.9) | 0.79 |
| Adenocarcinoma/NOS | 75 (65.2) | 57 (67.1) | |
| AJCC stage | |||
| IIIB | 26 (22.6) | 15 (17.6) | 0.39 |
| IV | 89 (77.4) | 70 (82.4) | |
| EGFR mutation | |||
| Positive | 16 (13.9) | 14 (16.5) | 0.62 |
| Negative/NOS | 99 (86.1) | 71 (83.5) | |
| KRAS mutation | |||
| Positive | 9 (7.8) | 6 (7.1) | 0.84 |
| Negative/NOS | 106 (92.2) | 79 (92.9) | |
| Treatment | |||
| PD-1 | 93 (80.9) | 70 (82.4) | 0.79 |
| PD-L1 | 22 (19.1) | 15 (17.6) | |
| Line of therapy | |||
| 2nd line | 80 (69.6) | 61 (71.8) | 0.74 |
| >2nd line | 35 (30.4) | 24 (28.2) | |
| Radiation | |||
| Yes | 43 (37.4) | 32 (37.6) | 0.97 |
Data are presented as N (%) unless otherwise specified.
Best response was defined as complete or partial response or stable disease or progressive disease according to RECIST 1.1 throughout the course of nivolumab single drug immunotherapy.
Never smokers are defined as smoking <100 cigarettes/lifetime, and former smokers are defined as abstinence from smoking for at least 15 years on the day before the start of therapy.
EGFR, Epidermal growth factor receptor; NOS, not otherwise specified.
SimTA outperforms other models in distinguishing responders vs. nonresponders
In our study, we used the SimTA module, with pre-immunotherapy data input, for predicting response at 60 (SimTA60d) and 90 days (SimTA90d) post immunotherapy. The module showed a good performance in distinguishing responders from nonresponders, with an area under the curve (AUC) of 0.77 (95% CI: 0.70-0.84) and 0.80 (95% CI: 0.74-0.86), respectively (Figure 2A). At the same time, the proposed model was compared with other corresponding methods to demonstrate the effectiveness of the SimTA module. Both the deep learning model incorporating baseline blood test data and the model incorporating baseline radiomics performed poorly in distinguishing responders from nonresponders, with an AUC of 0.57 (95% CI: 0.49-0.65) and 0.64 (95% CI: 0.56-0.72), respectively (Figure 2B). Furthermore, the RNN model only fairly distinguished responders from nonresponders, with an AUC of 0.69 (95% CI: 0.62-0.76) (Figure 2B).
Figure 2.
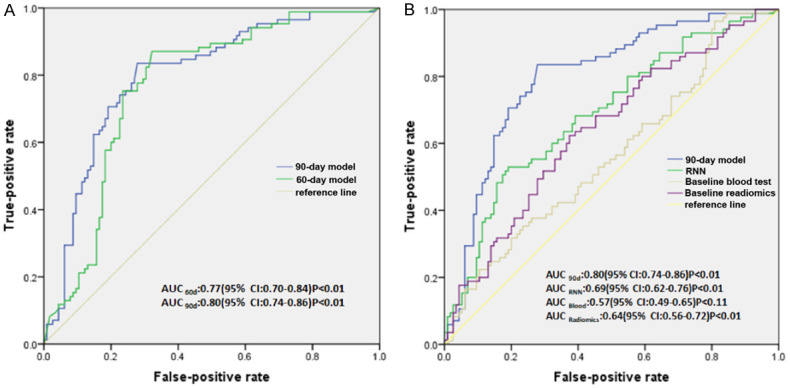
Model performance for response prediction in 200 patients. A. The AUC for the 60-day and 90-day response model. B. The AUC using the deep learning model incorporating baseline blood test data, baseline radiomics and using the RNN model. AUC, area under the ROC curve; ROC, receiver operating characteristic.
Immunohistochemical data on PD-L1 expression in tumor tissues was available in 102 patients (Supplementary Methods). PD-L1 performed poorly in distinguishing responders from nonresponders (Supplementary Figure 3A and 3B). SimTA60d had an AUC of 0.81 (95% CI: 0.73-0.90, P<0.001) versus an AUC of 0.45 (95% CI: 0.34-0.56, P=0.37) for baseline PD-L1 expression. SimTA90d had an AUC of 0.82 (95% CI: 0.73-0.90, P<0.001) versus an AUC of 0.38 (95% CI: 0.27-0.49, P=0.04).
Risk of nonresponding to immunotherapy and PFS and OS
The median duration of follow up was 12.7 (range 1.1 to 46.3) months. When SimTA60d was used for risk stratification, patients at low risk for nonresponding to immunotherapy were significantly more likely to have longer PFS and OS than patients at high risk [PFS: HR 2.49 (95% CI: 1.82-3.41); OS: HR 2.33 (95% CI: 1.61-3.37)] (Figure 3A and 3B). In addition, when SimTA90d was used for risk stratification, patients at low risk for nonresponding to immunotherapy were significantly more likely to have longer PFS and OS than patients at high risk [PFS: HR 3.14 (95% CI: 2.27-4.33); OS: HR 2.46 (95% CI: 1.73-3.51)] (Figure 3C and 3D). Patients at low risk had significantly longer median PFS than patients at high risk (8.4 months, 95% CI: 5.49-11.31) vs. 1.5 months, 95% CI: 1.29-1.71; log-rank test, P<0.01). They also had significantly longer median OS (26.7 months, 95% CI: 18.76-34.64 vs. 8.6 months, 95% CI: 4.55-12.65; log-rank test, P<0.01].
Figure 3.
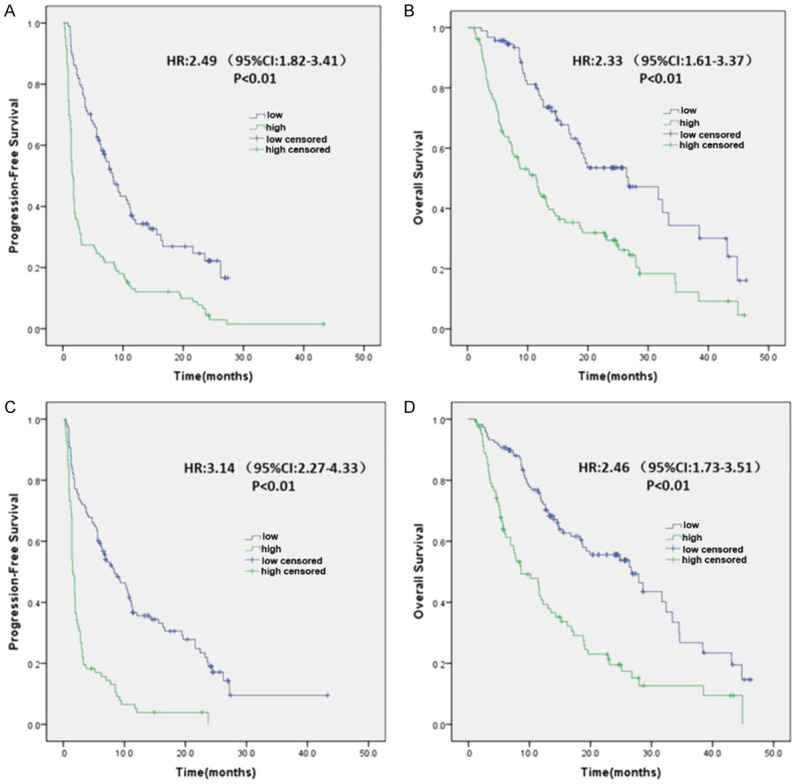
Deep learning prediction of PFS and OS in 200 patients. The prediction models stratify patients into high- and low-risk nonresponders using a default cutoff. A and B. PFS and OS according to risk stratification using the 60-day response model. C and D. PFS and OS according to risk stratification using the 90-day response model. HR, Hazards ratio; PFS, progression-free survival; OS, overall survival.
Ninety-three patients achieved SD. At the first efficacy assessment of anti-PD-1/PD-L1 therapy, 73 patients were at low risk and 20 patients were at high risk for nonresponding to immunotherapy. When SimTA60d was used for risk stratification, there was no statistical difference in both PFS and OS between patients at low risk and those at high risk (Supplementary Figure 4). When SimTA90d was used for risk stratification, patients at low risk had significantly longer median PFS than patients at high risk (11.1 months, 95% CI: 10.24-11.96 vs. 3.3 months, 95% CI: 0.34-6.26; HR 2.93, 95% CI: 1.69-5.10; log-rank test, P<0.01) (Figure 4A). Patients at low risk had significantly longer median OS than patients at high risk (31.7 months, 95% CI: 23.64-39.76 vs. 17.2 months, 95% CI: 7.22-27.18; HR 2.22, 95% CI: 1.17-4.20; log-rank test, P=0.01) (Figure 4B).
Figure 4.
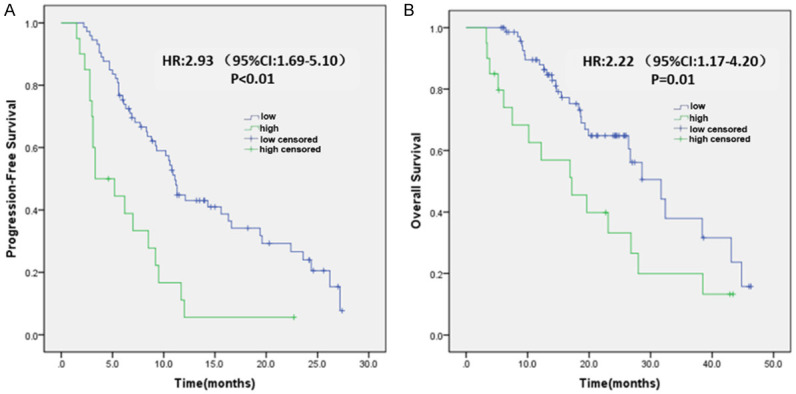
Deep learning prediction of survival in 93 patients with confirmed SD at the first efficacy assessment after anti-PD-1/PD-L1 treatment. The 90-day prediction model stratifies patients with SD into high- and low-risk nonresponders using a default cutoff. A. PFS in relation to risk stratification. B. OS in relation to risk stratification. Tumor response was evaluated according to RECIST 1.1. HR, Hazards ratio; SD, stable disease.
Discussion
In the present study, multidimensional serial information integrated with inputs from clinical data, laboratory tests and radiomics by deep learning could predict the response patterns of advanced stage NSCLC patients receiving anti-PD-1/PD-L1 monotherapy. Kaplan-Meier analysis further showed that when the 90-day model (SimTA90d) was used to stratify the risk of nonresponding to anti-PD-1/PD-L1 monotherapy, low risk patients had significantly longer median PFS and OS than high risk patients. Distinct from earlier similar studies, our study is novel in developing and validating a multi-omics-based serial deep learning approach for predicting clinical outcomes of advanced stage NSCLC patients receiving anti-PD-1/PD-L1 monotherapy, allowing more precise targeting of select advanced stage NSCLC patients who could truly benefit from anti-PD-1/PD-L1 immunotherapy. Particularly, the study is first in risk stratifying advanced NSCLC patients who have achieved SD and has successfully established a model that is capable of more precisely identifying which subset of SD patients could derive long term survival benefit from single-agent anti-PD-1/PD-L1 immunotherapy than the traditional RECIST evaluation method.
As a biomarker, radiomics is automated and noninvasive, and based on readily available imaging data. Some previous studies applied deep learning to predict treatment response of NSCLC patients using single-time-point or serial medical imaging [20,43,44]. Our study focused not only on the analysis of response patterns but also survival benefits. The previous deep learning models used only radiomics data and failed to integrate easily accessible clinical data and peripheral blood profiles which were also associated with clinical outcomes [18,19]. The present study showed that the multi-omics-based serial deep learning model integrating pretreatment patient data could predict survival benefits in patients with advanced NSCLC treated with anti-PD-1/PD-L1 monotherapy. The SimTA prediction model in this study is also more accurate than PD-L1 expression. Recently, a radiomic signature based on tumor-infiltrating CD8 cells has been demonstrated to predict OS of patients with solid tumors including lung cancer who received immune checkpoint inhibitors, showing a median OS of 24.3 months for patients with a high radiomic score and 11.5 months (HR=0.58, 95% CI: 0.39-0.87) for patients with a low radiomic score [36]. In our study, when the 90-day model was used to stratify risk of nonresponding to anti-PD-1/PD-L1 monotherapy, the low risk group had a median OS of 26.7 months versus 8.6 months for the high risk group (HR=2.46, 95% CI: 1.73-3.51). Some patients receiving immunotherapy experience severe toxicity or rapid progression. The rapid deterioration of clinical status prevents these patients from receiving a subsequent potentially effective treatment and results in a worse OS [45]. Indeed, 47 (23.5%) nonresponders in our dataset had an OS <6 months; of these, 18 (9%) could not receive later-line treatment due to the deterioration of their performance status. In contrast, durable disease control is often observed for responders to cancer immunotherapy [3,46]. In addition, immunotherapy could sensitize patients to subsequent chemotherapy [45], thereby contributing to the notable difference in survival outcome between responders and nonresponders.
In the present study, the 90-day response model better stratifies advanced stage NSCLC patients in survival outcomes than the 60-day response model. First of all, this is in line with the characteristics of immunotherapy, the effect of which is slow. Second, this could be attributed to the more accurate identification of nonresponders after 90 days with a confirmative follow-up CT assessment after the first efficacy evaluation. Response to immunotherapy involves infiltration of activated T cells into the tumor site, resulting in pseudoprogression in up to 10% patients with advanced cancer undergoing immunotherapy [45]. A total of 9 (4.5%) cases of pseudoprogression were documented in this study.
Traditional tumor temporal monitoring is often limited to size-based metrics due to the ease of clinical implementation (e.g., RECIST 1.1). Nevertheless, the emergence of targeted immunotherapy leads to novel patterns of tumor response, which might not be fully captured by changes in tumor diameter on imaging [13]. The interpretation of tumor size could be complicated by changes in the inflammatory, stromal, or fibrotic components of the tumor. Concerns are especially pertinent to patients with SD according to RECIST because this patient population has marked variability in prognosis. In the present study, 93 patients had SD according to RECIST at the first efficacy assessment. Among them, 20 patients were identified as high-risk nonresponders and 73 as low-risk nonresponders using the deep learning model. The lesion size increased in 8 (40%), remained unchanged in 8 (40%), and decreased in 4 (20%) cases in the 20 high-risk patients; further, it increased in 15 (20.54%), remained unchanged in 26 (35.62%), and decreased in 32 (43.84%) cases in the 73 low-risk patients. Interestingly, according to our 90-day prediction model, we found that among the patients who were also evaluated with the same SD, 20% of the shrinkable lesions had a poor prognosis and 20.54% of the enlarged lesions had a good prognosis, which cannot be achieved by the current traditional RECIST imaging assessment. These results suggested that long-term survival might not just be superior in patients with a moderate decrease (<30%) in the lesion size compared with patients with a moderate increase (<20%) in the lesion size after immunotherapy. AI-based radiomics could help overcome the limitation of RECIST with the analysis of multidimensional quantitative features, including tumor size, shape, density, and textural patterns [13]. Indeed, the use of radiomics [36] could more accurately determine the correlation between the heterogeneity of tumor-infiltrating lymphocytes and survival benefits for patients after immunotherapy. The risk stratification using the 90-day model was significantly associated with PFS (HR: 2.93, 95% CI: 1.69-5.10) and OS (HR: 2.22, 95% CI: 1.17-4.20). Our findings are consistent to the study by Korrahmi et al. who investigated changes in the radiomic texture of CT patterns of NSCLC patients and found that the radiomic texture could predict response to immune checkpoint inhibitors and OS of NSCLC patients [47]. However, the study by Korrahmi et al. did not take into consideration of such factors like clinicopathologic variables of the patients. A recent study by He et al. only incorporated tumor mutational burden and CT images for predicant response to immune checkpoint inhibitors in advanced stage NSCLC patients [48]. The present study is innovative in showing that the use of multidimensional serial information by deep learning could more accurately identify patients who can achieve the survival benefit from immunotherapy among patients with the same critical SD using RECIST evaluation. Despite the small sample size, the preliminary result of this study showed that the deep learning-based approach was superior to current assessment methods in accurately predicting patient prognosis, which needs further exploration.
The SimTA module developed in the present study combined serial imaging and laboratory data and the static clinical data through multimodal deep learning. It could process asynchronous, multisource sequential data. It is a common data format in clinical practice. This is the first method to process a complex multimodal asynchronous time series in clinical application with proven superiority over standard RNN. The SimTA module was flexible enough to encode multiple time series of arbitrary length and interval into a single unified representation, with no need for prespecified variables. In the present study, this attention module effectively and efficiently allowed the learning of temporal information. Nevertheless, the current formulation of SimTA encoded only linear temporal relations (by the attention weights). Although linear temporal relation learning performed well for this model, the model could still be refined by exploring nonlinear and complex temporal relations via introducing nonlinear temporal functions and self-attention. The findings need further investigation on more clinical applications and data modalities.
The present study has several limitations. First, the dataset had a limited number of patients. Although the sample size was small, deep learning-based multidimensional serial modeling in the training dataset was used to carry out responder versus nonresponder judgment learning at each evaluation point for every patient, thus virtually increasing the training dataset. Through the learning of the training datasets, a significant predictive value between responders and nonresponders was achieved in the validation sets. Additional studies with larger patient populations may help elucidate the role of SimTA-based multi-omics in predicting benefits of anti-PD-1/PD-L1 treatment in patients with advanced/metastatic NSCLC. Second, this is a retrospective study including patients only with intact blood tests, radiographic data, and complete follow-up information regarding assessment and survival after anti-PD-1/PD-L1 treatment. Although a proportion of patients in our dataset were from a real-world study, relatively strict inclusion criteria were applied. The usefulness of the model in a more prospective heterogeneous population should be examined.
Conclusions
In conclusion, our study has shown that the multidimensional serial information with inputs from pretreatment clinical data, laboratory tests, and radiomics combined with deep learning could help predict response to single-agent immunotherapy and offer a highly feasible, practical and operable model for risk stratification of advanced NSCLC patients. The proposed model provides a promising noninvasive biomarker that could be applied to a wide patient population in routine clinical practice, with standard-of-care clinical information.
Acknowledgements
The authors would like to thank Dr. Wei Zhao for insightful discussion and data support. The authors would also like to thank all the study patients and their families for their contributions to our scientific research. The study supported by the National Key R&D Program of China (2016YFC1303300), Young Scientists Fund of the National Natural Science Foundation of China (81802264), Shanghai Science and Technology Innovation Program (19411950500) and National Natural Science Foundation of China (82073152).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 3.Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, Castro G Jr, Srimuninnimit V, Laktionov KK, Bondarenko I, Kubota K, Lubiniecki GM, Zhang J, Kush D, Lopes G KEYNOTE-042 Investigators. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393:1819–1830. doi: 10.1016/S0140-6736(18)32409-7. [DOI] [PubMed] [Google Scholar]
- 4.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F, Kohlhäufl M, Arrieta O, Burgio MA, Fayette J, Lena H, Poddubskaya E, Gerber DE, Gettinger SN, Rudin CM, Rizvi N, Crinò L, Blumenschein GR Jr, Antonia SJ, Dorange C, Harbison CT, Graf Finckenstein F, Brahmer JR. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols MC, Cortinovis DL, Leach J, Polikoff J, Barrios C, Kabbinavar F, Frontera OA, De Marinis F, Turna H, Lee JS, Ballinger M, Kowanetz M, He P, Chen DS, Sandler A, Gandara DR. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389:255–265. doi: 10.1016/S0140-6736(16)32517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu YL, Lu S, Cheng Y, Zhou C, Wang J, Mok T, Zhang L, Tu HY, Wu L, Feng J, Zhang Y, Luft AV, Zhou J, Ma Z, Lu Y, Hu C, Shi Y, Baudelet C, Cai J, Chang J. Nivolumab versus docetaxel in a predominantly Chinese patient population with previously treated advanced NSCLC: checkmate 078 randomized phase III clinical trial. J Thorac Oncol. 2019;14:867–875. doi: 10.1016/j.jtho.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D, Ready N, Gainor J, Arén Frontera O, Havel L, Steins M, Garassino MC, Aerts JG, Domine M, Paz-Ares L, Reck M, Baudelet C, Harbison CT, Lestini B, Spigel DR. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai LL, Wang J. Liquid biopsy for lung cancer immunotherapy. Oncol Lett. 2019;17:4751–4760. doi: 10.3892/ol.2019.10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martens A, Wistuba-Hamprecht K, Yuan J, Postow MA, Wong P, Capone M, Madonna G, Khammari A, Schilling B, Sucker A, Schadendorf D, Martus P, Dreno B, Ascierto PA, Wolchok JD, Pawelec G, Garbe C, Weide B. Increases in absolute lymphocytes and circulating CD4+ and CD8+ T cells are associated with positive clinical outcome of melanoma patients treated with ipilimumab. Clin Cancer Res. 2016;22:4848–4858. doi: 10.1158/1078-0432.CCR-16-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen Y, Zheng H, Yao C, Wang Y, Lu S. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in chinese patients with NSCLC. J Thorac Oncol. 2019;14:1378–1389. doi: 10.1016/j.jtho.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17:e542–e551. doi: 10.1016/S1470-2045(16)30406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avanzo M, Stancanello J, Pirrone G, Sartor G. Radiomics and deep learning in lung cancer. Strahlenther Onkol. 2020;196:879–887. doi: 10.1007/s00066-020-01625-9. [DOI] [PubMed] [Google Scholar]
- 13.Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, Allison T, Arnaout O, Abbosh C, Dunn IF, Mak RH, Tamimi RM, Tempany CM, Swanton C, Hoffmann U, Schwartz LH, Gillies RJ, Huang RY, Aerts H. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin. 2019;69:127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Shi J, Ye Z, Dong D, Yu D, Zhou M, Liu Y, Gevaert O, Wang K, Zhu Y, Zhou H, Liu Z, Tian J. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53:11. doi: 10.1183/13993003.00986-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yip SS, Kim J, Coroller TP, Parmar C, Velazquez ER, Huynh E, Mak RH, Aerts HJ. Associations between somatic mutations and metabolic imaging phenotypes in non-small cell lung cancer. J Nucl Med. 2017;58:569–576. doi: 10.2967/jnumed.116.181826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts H. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grossmann P, Stringfield O, El-Hachem N, Bui MM, Rios Velazquez E, Parmar C, Leijenaar RT, Haibe-Kains B, Lambin P, Gillies RJ, Aerts HJ. Defining the biological basis of radiomic phenotypes in lung cancer. Elife. 2017;6:e23421. doi: 10.7554/eLife.23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martens A, Wistuba-Hamprecht K, Geukes Foppen M, Yuan J, Postow MA, Wong P, Romano E, Khammari A, Dreno B, Capone M, Ascierto PA, Di Giacomo AM, Maio M, Schilling B, Sucker A, Schadendorf D, Hassel JC, Eigentler TK, Martus P, Wolchok JD, Blank C, Pawelec G, Garbe C, Weide B. Baseline peripheral blood biomarkers associated with clinical outcome of advanced melanoma patients treated with ipilimumab. Clin Cancer Res. 2016;22:2908–2918. doi: 10.1158/1078-0432.CCR-15-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossmann P, Stringfield O, El-Hachem N, Bui MM, Rios Velazquez E, Parmar C, Leijenaar RT, Haibe-Kains B, Lambin P, Gillies RJ. Defining the biological basis of radiomic phenotypes in lung cancer. Elife. 2017;6:e23421. doi: 10.7554/eLife.23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trebeschi S, Drago SG, Birkbak NJ, Kurilova I, Calin AM, Delli Pizzi A, Lalezari F, Lambregts DMJ, Rohaan MW, Parmar C, Rozeman EA, Hartemink KJ, Swanton C, Haanen JBAG, Blank CU, Smit EF, Beets-Tan RGH, Aerts HJWL. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol. 2019;30:998–1004. doi: 10.1093/annonc/mdz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino M, Hatabu H, Hodi FS. Imaging of cancer immunotherapy: current approaches and future directions. Radiology. 2019;290:9–22. doi: 10.1148/radiol.2018181349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, Zheng Y, Park JH, Heng PA, Zhou SK. Iterative multi-domain regularized deep learning for anatomical structure detection and segmentation from ultrasound images. 2016 [Google Scholar]
- 24.Yan K, Wang X, Lu L, Zhang L, Harrison A, Bagheri M, Summers R. 2017. Deep Lesion Graphs in the Wild: Relationship Learning and Organization of Significant Radiology Image Findings in a Diverse Large-scale Lesion Database. Advances in Computer Vision and Pattern Recognition; pp. 413–435. [Google Scholar]
- 25.Bien N, Rajpurkar P, Ball RL, Irvin J, Park A, Jones E, Bereket M, Patel BN, Yeom KW, Shpanskaya K, Halabi S, Zucker E, Fanton G, Amanatullah DF, Beaulieu CF, Riley GM, Stewart RJ, Blankenberg FG, Larson DB, Jones RH, Langlotz CP, Ng AY, Lungren MP. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. PLoS Med. 2018;15:e1002699. doi: 10.1371/journal.pmed.1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 27.Tang H, Chen X, Liu Y, Lu Z, Xie X. Clinically applicable deep learning framework for organs at risk delineation in CT images. Nature Machine Intelligence. 2019;1:480–491. [Google Scholar]
- 28.Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging. 2019 doi: 10.1109/TMI.2019.2897538. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I. Attention is all you need. ArXiv. 2017 abs/1706.03762. [Google Scholar]
- 30.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 31.Chen D, She Y, Wang T, Xie H, Li J, Jiang G, Chen Y, Zhang L, Xie D, Chen C. Radiomics-based prediction for tumour spread through air spaces in stage I lung adenocarcinoma using machine learning. Eur J Cardiothorac Surg. 2020;58:51–58. doi: 10.1093/ejcts/ezaa011. [DOI] [PubMed] [Google Scholar]
- 32.Hepp T, Othman A, Liebgott A, Kim JH, Pfannenberg C, Gatidis S. Effects of simulated dose variation on contrast-enhanced CT-based radiomic analysis for non-small cell lung cancer. Eur J Radiol. 2020;124:108804. doi: 10.1016/j.ejrad.2019.108804. [DOI] [PubMed] [Google Scholar]
- 33.Yang M, Ren Y, She Y, Xie D, Sun X, Shi J, Zhao G, Chen C. Imaging phenotype using radiomics to predict dry pleural dissemination in non-small cell lung cancer. Ann Transl Med. 2019;7:259. doi: 10.21037/atm.2019.05.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang M, She Y, Deng J, Wang T, Ren Y, Su H, Wu J, Sun X, Jiang G, Fei K, Zhang L, Xie D, Chen C. CT-based radiomics signature for the stratification of N2 disease risk in clinical stage I lung adenocarcinoma. Transl Lung Cancer Res. 2019;8:876–885. doi: 10.21037/tlcr.2019.11.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Dong X, Wang J, Li W, Gu Z, Gao D, Zhong N, Guan Y. Computed tomography-based radiomics signature: a potential indicator of epidermal growth factor receptor mutation in pulmonary adenocarcinoma appearing as a subsolid nodule. Oncologist. 2019;24:e1156–e1164. doi: 10.1634/theoncologist.2018-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR, Verlingue L, Brandao D, Lancia A, Ammari S, Hollebecque A, Scoazec JY, Marabelle A, Massard C, Soria JC, Robert C, Paragios N, Deutsch E, Ferté C. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 37.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 38.Greff K, Srivastava RK, Koutnik J, Steunebrink BR, Schmidhuber J. LSTM: a search space odyssey. IEEE Trans Neural Netw Learn Syst. 2017;28:2222–2232. doi: 10.1109/TNNLS.2016.2582924. [DOI] [PubMed] [Google Scholar]
- 39.Adam Paszke SG, Soumith C, Gregory C, Edward Y, Zachary D, Lin ZM, Alban D, Luca A, Adam L. Automatic differentiation in PyTorch. 31st Conference oNeural Information Processing Systems. 2017 [Google Scholar]
- 40.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 41.Shaw P, Uszkoreit J, Vaswani A. Self-attention with relative position representations. Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 2 (Short Papers) 2018 [Google Scholar]
- 42.Kingma D, Ba J. Adam: a method for stochastic optimization. arVixorg. 2014 [Google Scholar]
- 43.Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, Bussink J, Gillies RJ, Mak RH, Aerts HJWL. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711. doi: 10.1371/journal.pmed.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y, Hosny A, Zeleznik R, Parmar C, Coroller T, Franco I, Mak RH, Aerts H. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin Cancer Res. 2019;25:3266–3275. doi: 10.1158/1078-0432.CCR-18-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borcoman E, Nandikolla A, Long G, Goel S, Le Tourneau C. Patterns of response and progression to immunotherapy. Am Soc Clin Oncol Educ Book. 2018;38:169–178. doi: 10.1200/EDBK_200643. [DOI] [PubMed] [Google Scholar]
- 46.Kojima T, Muro K, Francois E, Hsu CH, Metges JP. Pembrolizumab versus chemotherapy as second-line therapy for advanced esophageal cancer: phase III KEYNOTE-181 study. J. Clin. Oncol. 2019;37:2–2. [Google Scholar]
- 47.Khorrami M, Prasanna P, Gupta A, Patil P, Velu PD, Thawani R, Corredor G, Alilou M, Bera K, Fu P, Feldman M, Velcheti V, Madabhushi A. Changes in CT radiomic features associated with lymphocyte distribution predict overall survival and response to immunotherapy in non-small cell lung cancer. Cancer Immunol Res. 2020;8:108–119. doi: 10.1158/2326-6066.CIR-19-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He B, Dong D, She Y, Zhou C, Fang M, Zhu Y, Zhang H, Huang Z, Jiang T, Tian J, Chen C. Predicting response to immunotherapy in advanced non-small-cell lung cancer using tumor mutational burden radiomic biomarker. J Immunother Cancer. 2020;8:e000550. doi: 10.1136/jitc-2020-000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


