Abstract
Heavy metals cause considerable environmental pollution due to their extent and non-degradability in the environment. Analysis and trace levels of arsenic, lead, mercury, and cadmium as the most toxic heavy metals show that they can cause various hazards in humans’ health. To achieve rapid, high-sensitivity methods for analyzing ultra-trace amounts of heavy metals in different environmental and biological samples, novel biosensors have been designed with the participation of strategies applied in nanotechnology. This review attempted to investigate the novel, sensitive, efficient, cost-benefit, point of care, and user-friendly biosensors designed to detect these heavy metals based on functional mechanisms. The study’s search strategies included examining the primary databases from 2015 onwards and various keywords focusing on heavy metal biosensors’ performance and toxicity mechanisms. The use of aptamers and whole cells as two important bio-functional nanomaterials is remarkable in heavy metal diagnostic biosensors’ bioreceptor design. The application of hybridized nanomaterials containing a specific physicochemical function in the presence of a suitable transducer can improve the sensing performance to achieve an integrated detection system. Our study showed that in addition to both labeled and label-free detection strategies, a wide range of nanoparticles and nanocomposites were used to modify the biosensor surface platform in the detection of heavy metals. The detection limit and linear dynamic range as an essential characteristic of superior biosensors for the primary toxic metals are studied. Furthermore, the perspectives and challenges facing the design of heavy metal biosensors are outlined. The development of novel biosensors and the application of nanotechnology, especially in real samples, face challenges such as the capability to simultaneously detect multiple heavy metals, the interference process in complex matrices, the efficiency and stability of nanomaterials implemented in various laboratory conditions.
Keywords: biosensors, heavy metals, toxicity, nanomaterials, review
Introduction
Heavy metal pollution, especially various forms of arsenic, lead, mercury, and cadmium, is significant in most countries of the world and the United States. Of course, the situation is getting worse day by day. The upper crust of the earth contains many heavy metals due to agricultural pollution,1 mining, and industrial products.2 Heavy metals are absorbed via the gastrointestinal tract, inhalation of metal-containing fumes, and dermal exposure. Their transfer in the blood is through specific chaperons. Heavy metals do not cross lipid membranes, so they enter the cells by selective and nonselective channels.3 Arsenic is transported via aquaglyceroporins 7 and 9,4 whereas calcium channels mediate cadmium transport through cell;5 their entry is also through molecular Mimicry.6 Besides, lead can enter cells via calcium channels. Another route for entry of the lead into the cell is divalent metal transporter 1 (DMT1).7
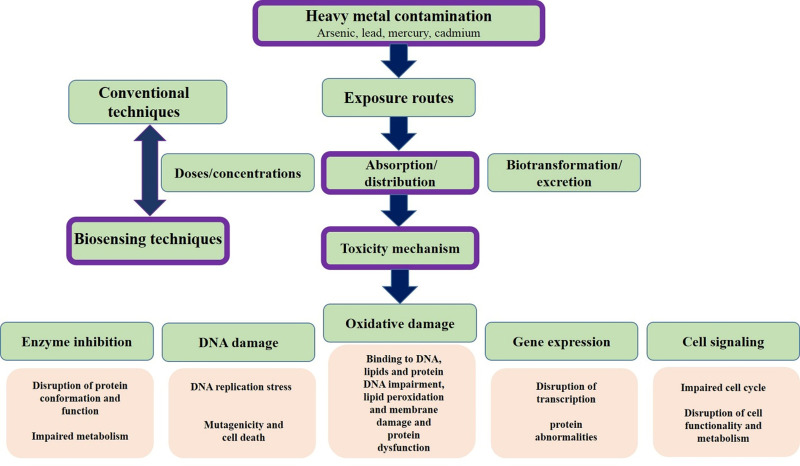
DMT1 also reuptakes mercury by cells.8 Mercury also enters the cell through amino acid transporters, organic anion transporters, and multidrug resistance-associated proteins (MRPs).9 Although these metals have no positive biological role in biological systems, they show their unfavorable effects after exposure.10,11 In addition to direct environmental impact, these metals are non-degradable and can accumulate in various organisms. Metals with toxic properties can lose one or more electrons and change to cations, so this exhibition of variable oxidation states enables them to react with biological organs.12 Thereby, there is a balance between the influx and efflux of these ions. The excessive amounts of these materials cause severe diseases in humans. The disease’s mechanism is the release of free radicals, cell injury, DNA destruction, enzyme inhibition, protein conformational change, promoting apoptosis, inhibition of neurotransmitters, and neuron damage.13,14 The manifestation of symptoms due to toxicity is common to other diseases, so toxicity is difficult unless there is an accident event (Figure 1).
Figure 1.
Heavy metals from the perspective of toxicity mechanisms and methods of analysis.
On the other hand, the techniques for diagnosing heavy metal toxicity are not available in all laboratories. At present, there are many various techniques for analyzing heavy metal toxicity such as atomic absorption spectrometry (AAS), electrothermal atomic absorption spectrometry (ET-AAS), flame atomic absorption spectrometry, hydride generation atomic absorption spectrometry, cold vapor atomic absorption spectrometry (HG-AAS, CV-AAS), inductively coupled plasma-optical emission spectrometry (ICP OES), inductively coupled plasma-mass spectrometry (ICP-MS), and HPLC-inductively coupled plasma-mass spectrometry (HPLC-ICP-MS).15 Although these techniques are susceptible and accurate, they require highly skilled technicians, complex instruments, and a long period for implementing the examination. Nowadays, new devices capable of performing quantitative and semi-quantitative analytic information tests by implementing a biological recognition element are used. These devices are biosensors that generate proportional signals to the analytes’ concentration and measure their physical or chemical reaction.16,17 The application of nanobiotechnology to design efficient biosensors in research and commercial fields allows the rapid and straightforward detection of biomarkers of various diseases, pathogens, pesticides, and other types of environmental pollutants.18–20
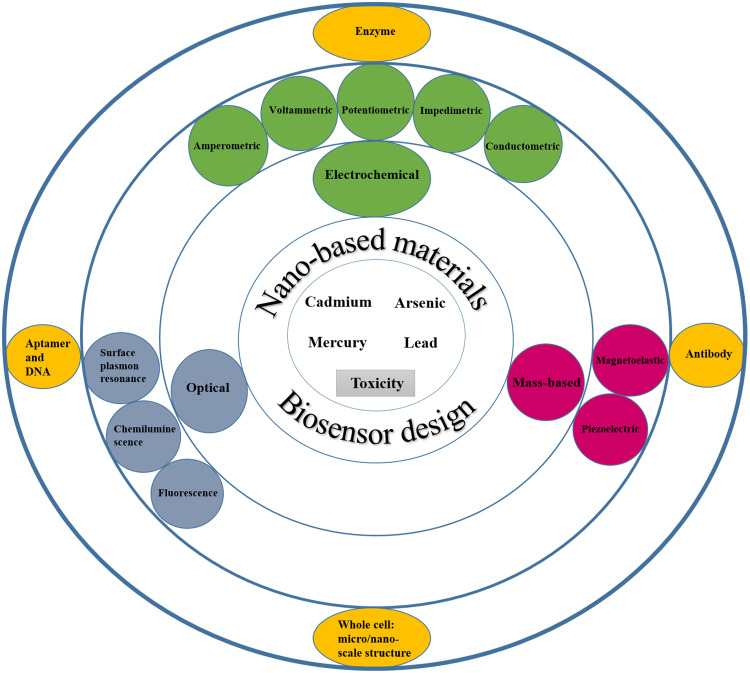
The biorecognition element, a particular transducer, and the signal processor form the main foundations of biosensors. Biosensors may be classified by their receptors or transducer21 (Figure 2). Some studies have been conducted to introduce nanomaterials utilized in the separation and detection of heavy metals based on nanotechnology in recent years.22–24 However, in addition to focusing on the toxicity mechanism, this study’s primary purpose is to investigate the designing and functionality of new biosensors from 2015 to 2020 to detect arsenic, lead, mercury, and cadmium, and the application of nanomaterials and nanotechnology methods in the construction of these biosensors. In this regard, keywords related to arsenic, lead, mercury, and cadmium biosensors, along with the functional characterization of biosensors and the mechanism of toxicity of these metals, were searched in PubMed, Science Direct, Web of Science, Scopus, and IEEE Xplore databases. The titles and abstracts of the English language articles were extracted and evaluated according to relevance to the topic. Then, the complete selected papers and related references were thoroughly studied.
Figure 2.
Schematic representation of the design of biosensors with the cooperation of nanotechnology for the detection of heavy metals.
Application of Nanomaterials in Biosensor Designing
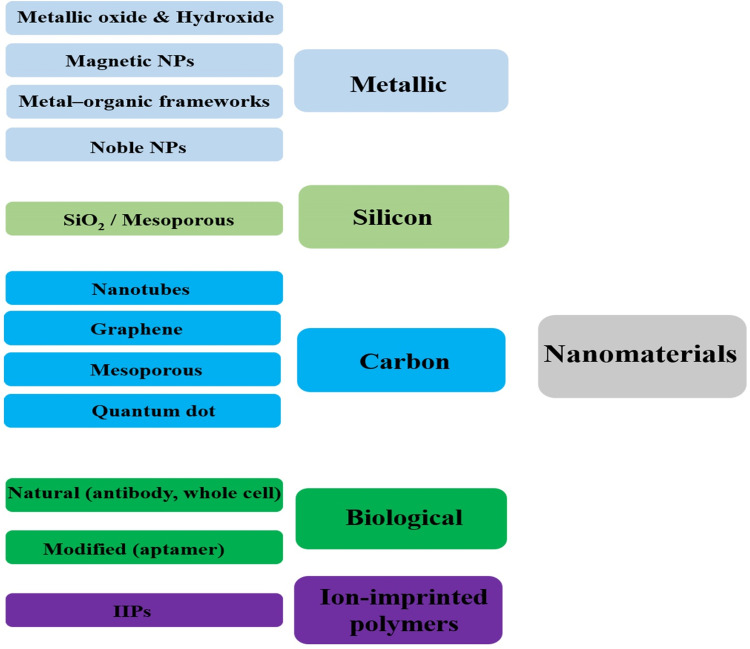
Having at least one external dimension in the range of 1 to 100 nm is the main characteristic of nanomaterials. The structures can have zero to three dimensions in cumulative, fusion, tubular, single, and irregular forms. A wide range of metal nanomaterials such as gold, copper, silver nanoparticles, non-metallic nanomaterials such as carbon-based compounds such as graphite, carbon nanotubes, and graphene is used for the immobilization process in the design of biosensors.23 Figure 3 depicts the nanomaterials used in sensing, preconcentration, and separation systems for heavy metals.23–26 The immobilization of nanoparticles on the surface increases the surface-to-volume ratio and more access to the active surface.27,28 Chemical and biochemical methods are used to synthesize nanomaterials employed to detect heavy metals. The advantage of the methods is producing smaller-sized nanomaterials with uniform size and increasing the surface to volume ratio to achieve better performance for detecting heavy metals. New nanomaterials such as nanoclusters, aptamers, antibodies, and quantum dots are used in heavy metal detection systems.25,29 Depending on the type of biosensor, it significantly increases sensitivity and specificity. Due to excellent resistance to oxidation and non-toxicity, gold nanoparticles are mostly used to manufacture biosensors applied in biotechnology and medicine. For instance, nanoparticles in electrochemical-based biosensors are often used as signal amplifiers and increase sensitivity and achieve lower detection limits. Nanoscale semiconductor materials such as quantum dots and nanocrystals such as iron oxide quantum dots and nanocrystals have optical and magnetic features. Magnetic nanoparticles implemented in biosensors’ design facilitate analyte purification and separation of interfering compounds in complex samples. Various studies have shown that metal nanoparticles and carbon nanotubes exhibit a massive increase in conductivity and improve electron transfer.30–32
Figure 3.
Schematic illustration of the possible nanomaterials used in biosensing system of heavy metal ions.
Biosensors Classification Based on Main Bioreceptors
Aptamer as Biological Nanomaterials
In addition to the double-stranded (ds) structures of DNA molecules, single-stranded DNA and RNA molecules are also utilized to design biosensor bioreceptors. Aptamers are fragments of single-stranded DNA and RNA selected by the in vitro evolution called systematic evolution of ligands by exponential enrichment (SELEX) technique to bound to the specific targets with high specificity and affinity. Aptamers consist of 30 to 100 nucleotides in size, consequently smaller than other biorecognition molecules such as antibodies and enzymes, resulting in efficient immobilization of high densities of aptamers on the surface.33–35 The binding of an aptamer to analyte changes the conformation in aptamers’ structure, which can be detected using the transducers’ output signal.36,37
The Antibody as Biological Nanomaterials
These specific proteins can detect analytes in other interfering agents due to their high binding affinity to the target.38 Antibodies used for the production of immunosensors are of two types: monoclonal and polyclonal. The sensitivity of polyclonal antibodies is more significant than their specificity, allowing them to detect different epitopes on the target antigen. In contrast, monoclonal antibodies are highly specific because they are made by one immune cell type and attached to the same corresponding antigen epitope.38,39
Enzyme
The properties and inherent catalytic activity of enzymes provide the advantage of achieving a lower detection limit than typical analyte binding methods. The enzyme recognizes a specific analyte similar to a key fitting a lock.40 The enzyme operating mechanisms in biosensors may include converting the analyte into a measurable sensor product without participating in the reaction detecting an analyte that performs as an enzyme activator or inhibitor and finally assessing the enzyme’s alteration properties after interaction with the analyte.41
Whole Cell-Based Biosensors as a Micro/Nano-Biosensor
A whole-cell microbial biosensor is an analytical method that detects target analytes by immobilizing microorganisms onto a transducer. The biosensors recognition elements for detecting particular molecules or the surrounding environment’s general condition could be microorganisms, such as bacteria or fungi.42 Most microbial biosensors are between 0.5 and 5 micrometers in size depending on the type of cell used in them, so they are in the nano/micro-biosensors category.43–45 One of the advantages of microbial-based biosensors is that they do not require purification, which is a time-consuming and costly process. Proteins and some intracellular components can be used as biorecognition elements to detect target analytes. Sensitive biosensors can be fabricated by integrating microorganisms with various electrochemical, optical, and mass-based transducers.46
Biosensors Classification Based on Primary Transducers
Electrochemical (EC)
EC biosensors measure the current resulting from the oxidation (ox) and reduction (red) reactions of the electrolyte (redox probe).47 EC transducers convert biological interactions into an electrochemical signal and rely on electrical parameters such as current, impedance, potential, and capacitance. As a result, these transducers’ main output is to convert information into measurable values using potentiometry, voltammetry, amperometry, and conductivity. The main elements of electrochemical studies are electrochemical data processing devices and electrochemical cells, including the working electrode, reference, and counter electrode in the presence of the electrolyte or redox probe.48,49 Since the expected redox reactions occur on the working electrode surface, the electrode must have good reproducibility and a high signal-to-noise ratio. A constant potential is generated in the electrochemical cell using a reference electrode. The counter-electrode is an electrode used to close the current circuit in an electrochemical cell, referred to as an auxiliary electrode. Researchers today highly regard electrochemical biosensors due to their simplicity and, at the same time, high processing speed, low cost, and miniaturization capability of electrodes (such as screen-printed electrode).50,51 Voltammetry is the most effective electrochemical procedure. The current and potential are monitored in voltammetric processing. The position and density of the obtained peak current are proportional to the chemical’s characteristics and individual concentration. Additionally, voltammetry can detect multiple chemicals with different peak potential in a single EC analysis, thereby simultaneously providing various analyte detection. The dedicated current- potential graph voltammetric technique is called a voltammogram and includes differential pulse voltammetry (DPV), cyclic voltammetry (CV), linear sweep voltammetry (LSV), and stripping voltammetry (SV) methods.52 Amperometric transducer performs electrochemical measurements of oxidation and reduction currents of electroactive species under a constant potential applied to the working electrode. The potential act as the driving force of the electrons’ transfer reaction causes a force to be applied to the electroactive species to gain or lose electrons. The output current obtained from this method is directly associated with the bioassay process and analyte concentration.53 The bio-recognition reaction triggers a modification of the transmembrane potential, a redox potential, or an ion’s activity in potentiometric biosensors. This technique implements the Nernst equation, so the electrode potential depends on the oxidized and reduced species’ concentration. A working electrode’s potential is measured compared to that reference electrode with a constant or null current flow by potentiometric instruments. The electrode-forming layers consist of a layer with a selectively permeable membrane and a membrane or surface with a high sensitivity to the analyte, such as a surface containing specific enzymes.54 Conductometric transducers permit to track of the biorecognition event, which changes the ionic concentration. Commonly, any chemical reaction is associated with a change in the concentration of ionic species present in the solution, which results in a change in electrical conductivity and current flow. In this method, two separate metal electrodes are used, spaced at a certain distance from each other. During the analysis process, a specific Ac voltage is applied to the electrode’s surface, which causes the current flow. The ionic composition changes with the reaction, and conductivity is measured between the metal electrodes.55 Electrochemical impedance spectroscopy (EIS) is a rapid and accurate procedure for characterizing biomaterial-functionalized electrodes’ structure and function. Immobilizing bio-materials on the electrodes’ surface alters interfacial electron transfer resistance and electrode capacitance changes and triggers impedance variations. This electrochemical technique can detect interfacial changes caused by biorecognition processes. An electrical circuit can model the measured impedance of the device. Electrochemical reactions on the electrode surface and the reaction solution are accompanied by the creation of impedance in the electrical circuit, including the electron transfer’s resistance between the electrode surface and the redox pair. Other impedance statements are the capacitance between the charged ions in solution and the electrode, the resistance due to the rate at which the redox couple penetrates the electrode, and the solution’s resistance present between the system electrodes.56
Optical-Based Biosensor
The utilization of optical biosensors technology is an essential approach for exploring and evaluating biomedical research, pharmaceuticals, environmental monitoring, national security, and warfare. Generally, optical transducers’ performance is based on changes in the amplitude, frequency of polarization, and phase of input light in response to physical and chemical changes caused by the reaction between the analyte and the bioreceptor.57 An optical biosensor’s main components include a light source, immobilized biorecognition elements, an optical analysis system, and, most importantly, an optical transmission device such as fiber.58 Surface plasmon resonance (SPR) is one of the label-free methods in producing optical biosensors. In the traditional SPR biosensor structure, a thin metallic layer is covered on one prism surface and distinguishes the prism and the sensing medium. The SPR sensing effect is responsive to target analyte bindings because the resulting increment in mass induces an expansion of the refractive index proportionally, which can be interpreted as a change in the resonance angle. When the analyte interacts with the sensor’s bioreceptor part, SPR biosensors utilize electromagnetic waves to detect changes.59 Luminescence is a widespread quantum phenomenon that occurs when specific molecules emit light after being excited to return to the ground state, and this transmission is recorded as luminescence. Chemiluminescence occurs by performing specific chemical reactions and generating an electronically excited state for a molecule by chemiexcitation.60 Chemiluminescence analysis consists of tracking the photon production rate, so light intensity depends on the photon production rate. The intensity of the output light is directly proportional to the limiting reactant’s concentration in the luminescence reaction. The purpose of designing biosensors based on luminescence chemistry is to combine the high sensitivity of light-emitting responses with the sensors’ structure, which results in the achievement of a highly sensitive and practical tool.61 Fluorescence is used frequently for signal transduction, particularly in the presence of antibodies and enzymes. In the fluorescence method, an external light source with a short wavelength is required to stimulate electrons’ transfer in an atom or molecule, producing luminescence with a longer wavelength.57 During the biorecognition process, a fluorescent biosensor embedded with fluorophore molecules is applied to generate light. Because most bio-recognition elements and target analytes lack intrinsic spectral properties, biosensing events are converted into optical signals using a combination of fluorescence-responsive optical reagents and sensing elements. Fluorescence-based biosensors such as FRET-biosensors are very sensitive and are used to detect analytes at low concentrations.62
Piezoelectric-Based Biosensor
Piezo means the application of pressure or squeeze. Due to mechanical stress in some materials, a potential difference is created, which is called piezoelectricity. The sensors are a subset of the mass-base and are divided into Bulk acoustic and Surface acoustic wave piezoelectric sensors. In piezoelectric-based biosensors, bioreceptors are coupled to piezoelectric materials such as ceramics and quartz, which produce a measurable signal in response to changes in mass-induced oscillations bonded to the piezoelectric crystal surface.63
Magnetoelastic-Based Biosensor
Magnetoelastic sensors are constructed from ribbons of amorphous ferromagnetic film. The ribbons are typically iron-rich alloys with a high magneto-elastic coupling coefficient and great mechanical tensile strength. Due to the magnetostriction property of ribbons, their shape changes in the presence of a magnetic field. This mechanism’s operation is similar to the quartz crystal microbalance method. The exception is that these types of biosensors work by magnetoelasticity instead of piezoelectricity.64,65
Field Effect Transistor-Based Biosensor
These semiconductor biosensors consist of three types of electrodes: source, drain, and gate. The part between the original and the drain acts as a biorecognition element. There is a nanowire channel between the discharge terminals and the source section in the FET biosensors’ configuration. The surface of nanowires is functionalized and modified by biorecognition elements. The measurable signal is generated based on the type of system, electrical activity, and analyte concentration.66 In the presence of a biorecognition element such as an antibody or aptamer, analyte concentration changes lead to a shift in charge near the sensor interface. The gate section’s voltage is changed, followed by a voltage difference, and the drain current changes. This analysis can be evaluated using I–V characterization. The basis of the detection method in the electric FET method is the effect of changes in permittivity and charge interactions.67
The General Strategy in the Accurate Design of Biosensors
The practical techniques implemented in biosensors detect labeled or label-free target analytes. Label-based detection relies primarily on the unique properties of label substances to detect target analytes. The performance of label-based biosensors is based on the basic concept that it is impossible to catch all analytes based on their inherent physical and chemical properties. Therefore, some radioactive labels, enzymes, or fluorescent molecules are attached to the target analyte to perform the detection process.68 Disadvantages of this method are its cost and time-consuming; besides, labeling may interfere with the bioreceptor’s active binding to the target analyte. In contrast, to facilitate measurements and time and cost savings, the label-free model is used in biosensors technology. In this method, analytes’ intrinsic physical and chemical properties such as size, electric charge, molecular weight, electrical impedance, refractive index, and dielectric properties are used. In recent years, label-free methods have found many medical science applications and environmental compound analysis due to the interdisciplinary convergence of electronics engineering, chemistry, and nanobiotechnology.69–71
Environmental Pollution, Toxicity Mechanisms, and Health Hazards of Heavy Metals
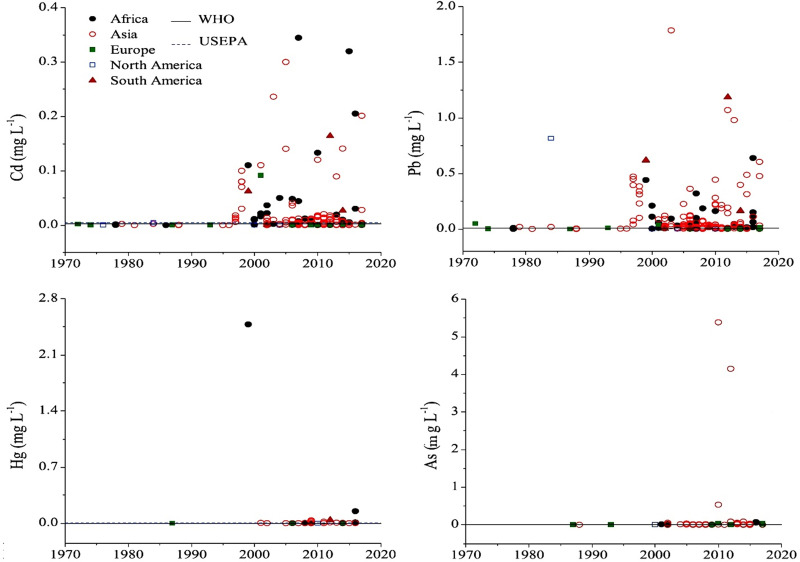
Natural and anthropogenic resources are two important sources of environmental pollution caused by heavy metals. According to recent reports from the World Health Organization, more than 1 billion people in the world do not have access to safe and clean water, and 1.6 million people pass away each year due to water contamination with biological and non-biological pollutants.72 Surface waters, including global rivers and lakes, are the best samples for monitoring the status of heavy metal pollution worldwide.73 Figure 4 shows the heavy metals’ total concentrations in global rivers and lakes on five continents from 1972 to 2017.74 The threshold concentration of these metals is also presented according to USEPA and WHO standards. Collectively, the average concentration of heavy metals has been increasing since the 1990s. Strict monitoring of health laws’ implementation to control global heavy metal pollution has led to relative control of these pollutants in developed countries.75 On the other hand, the level of this pollution is significantly increasing in developing countries. One of the practical aspects of establishing health legislation related to heavy metal pollution is continuous and accurate monitoring of these metals’ levels in the environment.
Figure 4.
Concentrations of heavy metals in global surface waters by continents from the 1970s to the present. Reprinted from Zhou Q, Yang N, Li Y, et al. Total concentrations and sources of heavy metal pollution in global river and lake water bodies from 1972 to 2017. Global Ecol Conserv. 2020;22:e00925. Copyright 2020, with permission from Elsevier.
Arsenic
Arsenic has caused many environmental concerns in personal and public health worldwide, as a toxic and carcinogenic metalloid with a wide distribution in the environment. Arsenic exists in organic and inorganic forms and is found in nature in three- and five-valent forms. The inorganic form of arsenic is more toxic than organic compounds. Because these compounds enter the human and animal food cycle and accumulate in biological systems.76 Occupational exposure to high levels of arsenic occurs in agricultural industries such as pesticide production. Also, metal smelting industries, especially copper, are essential sources of pollution with dust and arsenic fumes. Drinking water is the primary source of environmental exposure to arsenic.77–79 According to recent research, 150,000 people are exposed to arsenic-contaminated water (10 ppb) daily.80 Besides, a variety of foods, such as rice and seafood, contain arsenic. Arsenobetaine is an organic, less toxic form of arsenic found in seafood that causes a daily arsenic intake.81 Trivalent arsenic compounds react with and bind to sulfhydryl and thiol groups of enzymes and proteins, thereby altering their function and structure. One of the enzymes with a vital key role in the Krebs cycle inhibited by As (III) is pyruvate dehydrogenase, which eventually leads to disruption of cellular respiration and ATP production. On the other hand, the pentavalent form of As (V) is similar in structure to the phosphate group and is replaced in phosphate transfer reactions and can prevent the mitochondrial oxidative phosphorylation. Epidemiological and experimental studies have shown that chronic long-term exposure to low doses of arsenic is associated with developing crucial diseases such as cardiovascular diseases, hypertension, diabetes mellitus, hyperkeratosis, parenchymal cell damage, liver fibrosis, cirrhosis, gastroenteritis, peripheral neuropathy, encephalopathy, hepatocellular carcinoma, Pancytopenia, and a variety of cancers.12,82
Lead
As one of the most abundant elements on the earth, lead has long been considered by humans due to its desirable physical properties such as low melting point and high flexibility. The lead application is closely related to many industries, including smelting, mining, refining in mines, battery and glass production, rubber and plastics industries, and industries requiring lead soldering. Until the late 1980s, organo-lead compounds were widely used as additives to increase octane and gasoline combustion, which led to a sharp increase in the global atmospheric lead, the pollution of which is still debated in the environment. The widespread use of lead in the industry has led to environmental, soil, and groundwater pollution. As a non-essential metal for the human body, it is not biodegradable, and its ecotoxicity remains a concern despite warnings from international health organizations.82,83 The primary exposure to lead is water and food, and occupational exposure is due to inhalation and ingestion of lead-containing fumes and dust. The permissible limit for lead in drinking water is set by the World Health Organization at 0.01 µg/L. About 99% of blood lead in erythrocytes is bound to hemoglobin. Only 1% of circulating lead is present in serum, which is first distributed in soft tissues such as the liver and kidneys and then redistributed in the skeleton and hair. Blood lead levels (BLL) greater than 70 and 100 µg/dL indicate significant toxicity in children and adults, respectively.84 As a persistent pollutant in the environment and biological systems, lead covers a wide range from acute to chronic toxicity, resulting in a variety of abnormalities in the human body. Acute poisoning causes headaches, abdominal pain, hypertension, renal dysfunction, dizziness, hallucinations, insomnia and osteoarthritis, and chronic poisoning leading to congenital malformations, mental disability, allergies, autism, hyperactivity, kidney damage, severe muscle weakness, paralysis, and eventually leads to death.7,85 The known neurological, behavioral, and neurotoxic effects of lead can be explained by molecular, cellular mechanisms, including function as a substitute and mimic of calcium and its homeostasis, stimulation of protein kinase C, dysfunction of neurotransmitters in dopaminergic, cholinergic, and glutaminergic pathways. Lead has several hematological effects. The most important impact is interfering with the heme biosynthesis pathway by inhibiting key enzymes delta-aminolevulinic acid dehydratase and ferrochelatase. The bone effects of lead are notable, and its half-life in bones is very high compared to blood.3 Disorders of metabolic and homeostatic processes such as vitamin D, parathyroid hormone, calcitonin, replacement of lead with calcium in bone are among the adverse health effects of lead. According to various studies, lead carcinogenesis is due to inhibition of DNA synthesis and repair, replacement of lead with zinc in the structure of transcriptional regulators, production of ROS, oxidative stress and DNA damage, interference with DNA-binding proteins, and uncontrolled expression of genes.85,86
Mercury
Mercury is a heavy metal that is an excellent example to show metals’ movement in the environment. It is found in three forms: elemental, organic, and inorganic (Hg1+, Hg2+), each with its individual physical and chemical properties and toxicity profile. Atmospheric mercury is in mercury vapor (Hg0) at a specific temperature and pressure and is produced through many interactions in the Earth’s crust, oceans, and volcanic eruptions. Anthropogenic sources of extraction, smelting of metals in mines, coal combustion, various chlor-alkali industries, and waste incinerators are the primary atmospheric mercury sources. In rainwater, mercury returns to the earth’s surface in an inorganic form of mercury oxide. Mercury on the earth’s surface takes two paths: 1) the path of mercury vapor re-conversion and return to the atmosphere, 2) the conversion of mercury to methylmercury by microorganisms.3,87 The methylation pathway leads to mercury entering the aquatic food chain, and its cumulative levels in tissues are 1800 to 8000 times higher than the surrounding water. The process of bioconcentration and biomethylation is carried out, and human consumption of seafood eventually leads to methylmercury toxicity.82,88 Human exposure to various mercury forms occurs through accidental exposure, environmental and food contamination, dentistry and amalgam production, preservation of pharmaceutical products, occupational contamination in the electrical industry, precious metal processing, caustic soda, and antifungal production.89 Elemental mercury and methylmercury are highly absorbable. Mercury vapor is a lipophilic element and is quite effectively absorbed through the lungs and epithelial tissues. Upon entering the bloodstream, it is oxidized to Hg2+, which is highly reactive. The gastrointestinal tract also absorbs methylmercury accumulated in fish, and due to its high fat solubility, it crosses the maternal placental barrier and the blood-brain barrier. Unlike high absorption, mercury excretion is very low. A very high proportion of mercury is absorbed in the liver, kidneys, and nerve tissue, and its toxicity occurs in the gastrointestinal tract, nervous system, kidneys.90 Oxidative stress plays a crucial role in the development of mercury toxicity. This molecular mechanism indicates the reactivity of sulfhydryl-containing groups in the presence of mercury. Methylmercury and Hg2+ react covalently with cysteine residues to react proteins and enzymes and deplete cellular antioxidants. Accumulation of reactive oxygen species (ROS) occurs due to oxidative damage due to mercury compounds, which usually inhibit toxicity due to the cell’s high antioxidant capacity.91 Inorganic mercury increases ROS by disrupting oxidative phosphorylation in the ubiquinone cytochrome b5 step and electron transfer. The methylmercury form increases intracellular calcium by accelerating the influx of extracellular calcium and assembling intracellular stores, whereas inorganic mercury only affects the entry of extracellular calcium into the cell. Some studies have shown that mercury is genotoxic, but more studies are underway.92 Induced ROS due to mercury toxicity may initiate cancer-induced processes. On the other hand, the direct effect of free radicals on DNA and proteins responsible for DNA repair, chromosomal segregation, and the mitotic spindle has been observed in mercury toxicity mechanisms.93,94
Cadmium
As a ubiquitous heavy metal, cadmium has caused a great deal of concern in an occupational exposure and environmental pollution. The average concentration of cadmium in the earth’s crust is 0.1 mg/kg. The highest levels of cadmium compounds in the environment are found in sedimentary and marine rocks containing phosphate at a concentration of 15 mg/kg. Cadmium is used in a wide range of industries today. One of cadmium’s main applications in the industry is the production of alloys, pigments, and batteries.95 Commercial use of cadmium has been reduced as much as plausible due to environmental concerns. Daily cadmium intake in the United States does not exceed 0.4 µg/kg/day, half of the EPA’s reference dose.
Cadmium as a nonessential and toxic metal in biological organisms, although in minimal amounts, can accumulate in the body during life, and the effects of this accumulation can manifest years after. The main routes of exposure to cadmium are through food intake, passive smoking, or smoking. Circulation and blood vessels are essential routes of cadmium distribution to target organs.96,97 Chronic inhalation of cadmium particles causes pulmonary emphysema, which changes in pulmonary function are also seen on chest radiographs.98 Occupational exposure to airborne cadmium particles reduces olfactory function.99 Some epidemiological studies have confirmed an association between osteoporosis and decreased bone mineral density with chronic exposure to low cadmium levels.100,101 Cadmium acts as a potent irritant of the gastrointestinal tract and respiratory system, causing renal or hepatic injury and lung damage, along with symptoms such as abdominal pain, nausea and vomiting, muscle cramps, shock, seizures, and loss of consciousness that may eventually lead to coma and death.102 Chronic exposure to cadmium may even cause depression by affecting acetylcholine, norepinephrine, and serotonin.103 Molecular mechanisms of cadmium toxicity indicate cellular damage by inducing ROS production, leading to impaired synthesis of proteins and nucleic acids. Cadmium affects signaling pathways, including calcium channel blockade, increased cytosolic free calcium, and inositol polyphosphate. It also activates protein degradation processes, up-regulation of proto-oncogenes such as c-jun and c-fus and cytokines, and induces expression of the genes metallothioneins, glutathione transferases, heme oxygenases, and DNA polymerase β.104,105 Blood and urinary cadmium levels reflect recent and chronic cadmium exposure, respectively. The main sources of exposure and diseases caused by heavy metals are summarized in Table 1.
Table 1.
Heavy Metals and Their Source and Health Effect
| Major Items | As | Pb | Hg | Cd |
|---|---|---|---|---|
| Main sources | Metal smelters, fungicides, pesticides | Burning of coal, paint, automobile emission, pesticides, mining, smoking | Batteries, pesticides, paper, and the dental industry | Electroplating, smoking, welding, fertilizer, pesticides |
| Health effect | Dermatitis, bronchitis, cardiovascular disorders | Liver and renal failure, development delay, mental retardation, fatal neonatal encephalopathy, acute and chronic damage to nervous system | Gingivitis, tremors, protoplasm poisoning, spontaneous abortion, nervous system disorder | Pneumonitis and lung cancer, renal dysfunction, bone defects, bone marrow, kidney damage |
Biosensors Designing Based on Nanotechnology Approach to Heavy Metals Analysis
Arsenic
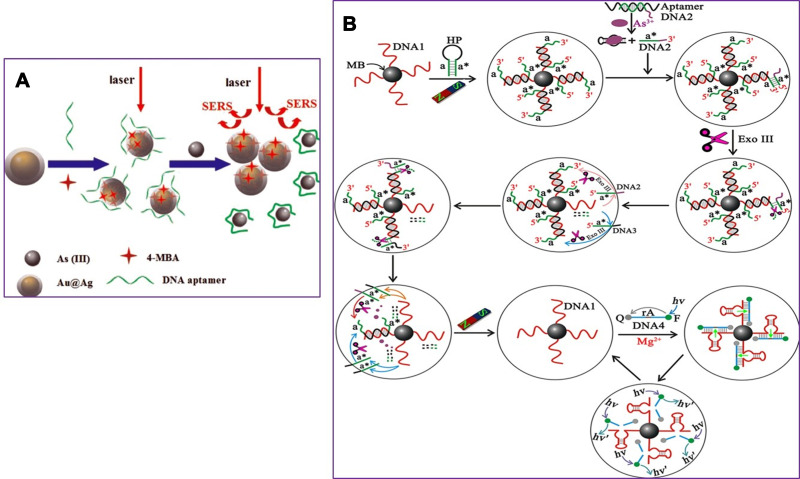
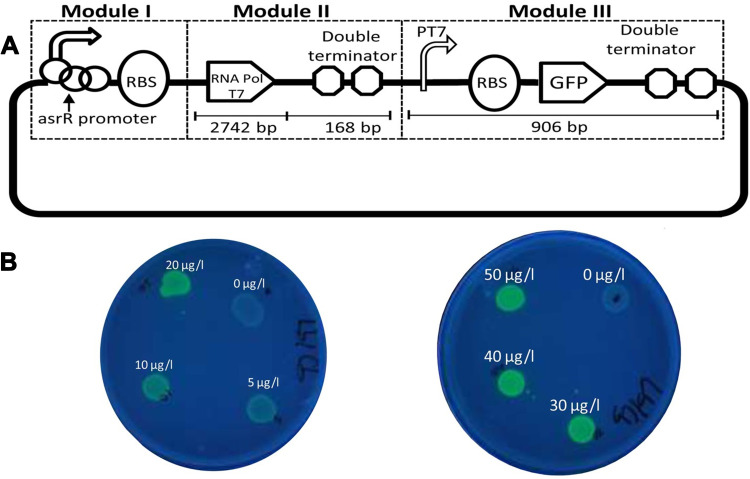
Given the importance of arsenic toxicity in biological systems and the environment, in recent years, in addition to conventional methods for measuring heavy metal contamination, several biosensors to analyze arsenic levels in complex matrices such as blood or various tissues and environmental samples are designed. An optical biosensor based on the Surface-enhanced Raman scattering (SERS) technique was developed to detect As (III). A combination of specific trivalent arsenic aptamer and Raman labeled Au@Ag core-shell nanoparticles were used. SERS performance increases in the presence of Au@Ag. Aptamer has been absorbed in 4-mercaptobenzoic (4-MBA) as a Raman reporter molecule and Au@Ag to act as a donor to SERS. In the presence of As (III), specific aptamers are separated from the surface of Au @ Ag and attached to As (III), resulting in the aggregation of nanoparticles. As a result, due to the formation of “hot spots” of SERS, the 4-MBA molecule signal of the Raman reporter was amplified. The detection limit and linear range of this novel SERS-based aptasensor were 0.1ppb and 0.5–10 ppb, respectively (Figure 5A).106 The structural similarity of the phosphate group with pentavalent arsenic is the basis for designing a high-sensitivity biosensor. In this new strategy, ss-DNA-MB is immobilized on CeO2 nanoparticles through its phosphate groups. Surface binding competition occurs in the presence of arsenic, and higher affinity of As (V), lead ss-DNA to separate from the surface, and As (V) is adsorbed to the nanoparticle surface. For analysis, the DPV electrochemical technique was used utilizing the ITO electrode, and the output signal was measured at each step. Methyl blue attached to ss-DNA as an electroactive compound facilitates electron transfer, and a significant DPV signal is obtained. The current signal decreases due to reduced diffusion to reach the electrode surface by increasing nanoparticles’ concentration and attaching ss-DNA. The competitive coordination-based immobilization-free electrochemical biosensor can detect pentavalent arsenic in the linear range from 400 ppb to 16 ppm and the detection limit of 164 ppb.107 Zeng et al108 demonstrated a high-sensitivity fluorescence platform for As (III) detection. The basis of this method is the amplification of the sequential signal in the presence of analytes. The specific As (III) -aptamer acts as a biorecognition element. In the analyte presence, the blocking DNA is released and stimulates the cascading signal process. By performing the digestion process of DNA recycling with Exo III’s cooperation, many mg-dependent DNAzyme enter the assay system. By performing magnetic separation, DNAzyme with multiple turnover numbers can catalyze the sequential cleavage of fluorophore- quencher-functionalized substrate strands. As a result, a powerful, amplified fluorescence signal is generated for analyte detection. The very low detection limit of 0.016 ppb results from further amplification of the Exo III and DNAzyme synergistic signal, which indicates the excessive sensitivity of the fluorescent biosensor (Figure 5B). Cell bioreporters are widely used to detect heavy metals. A new approach to the detection of arsenic has recently been suggested. The analysis results for a bacterial biosensor POLA (ABP), made from three genetic modules, include a promoter, reporter gene, and gene expression amplifier (Figure 6A and B). Pola-López et al109 designed a new vector that can exchange promoters, amplify gene expression, and green fluorescent protein (GFP), signaling at maturity and rapid growth. The basis of the method was a genetically modified bacterium from the IGEM 2012 series, conducted with the Bio parts of Escherichia coli MG1655k12. The fluorescence generated by the optical biosensor showed a linear response both in terms of time and arsenite concentration (ranged from 5 to 140 ppb). The fluorescent method for tracking inorganic arsenic levels in biosensor technology is based on the excitation or quenching of nanoparticle fluorescence in the presence of the target analyte. Zhang and co-workers used single-stranded DNA rich in guanine and thymine bases by the hydrothermal method to fabricate fluorescent DNA-containing quantum dots. A specific arsenic aptamer’s function is combined with the quantum dot’s optical properties in this probe. The binding of trivalent arsenic to QDs-DNA causes conformational changes in the ssDNA structure, ultimately increasing the output fluorescence signal. This method provided an appropriate linear range (1–150 ppb) and an acceptable detection limit (0.2 ppb) for arsenic detection.110 Aptasensors that use electrochemical techniques to detect arsenic are excellent examples of nanomaterials in biosensors’ fabrication, mainly when these nanomaterials modify the electrode’s surface. For instance, Cui et al111 applied a screen-printed carbon electrode modified with gold nanoparticles as the electrochemical reaction medium. The thiolated aptamer (Ars-3) was immobilized on the modified electrode’s surface by the self-assembly method. Polyatomic polymers such as PDDA are adsorbed to arsenic-specific aptamers by electrostatic interactions. The presence of arsenite in the reaction solution and its greater tendency to bind to aptamer alters the conformation of aptamer and the formation of the aptamer-arsenite complex, diminishing PDA’s adsorption. Under these conditions, the electrochemically active [Ru(NH3)6]3+ probe accumulates more and more on the electrode surface, which leads to an increase in electron transfer at the electrode surface, increasing DPV signal current. The linear range of the designed system was between 1.6 and 800 ppb, and the detection limit was 1.2 ppb. Divsar et al successfully designed an optical biosensor based on the colorimetric method for detecting. In this biosensor, thiolated aptamer conjugated with silver nanoparticles (Apt-SNPs) were employed. This biosensor’s function is based on the aggregation of aptamer-conjugated nanoparticles in the presence of arsenic ions. There are two binding sites on the aptamer for the binding of arsenic ions; arsenic ions in solution can act as a bridge between the dispersed Apt-SNPs and attach them. APT SNPs’ spectrophotometric characteristics in the As-containing solution show a significant decrease in the UV-VIS absorption peak at 430 nm. The optical biosensor’s linear dynamic range covers a wide range of arsenic ion concentrations from 50 to 700 ppb with a detection limit of 6 ppb.112 The operating mechanisms of other precision and important biosensors that have been developed in recent years are mentioned in Table 2.
Figure 5.
(A) Schematic illustration of the arsenic (III) SERS detection based on Au@Agcore–shell nanoparticles. SERS, Surface-enhanced Raman scattering. Reprinted from Song L, Mao K, Zhou X, Hu J. A novel biosensor based on Au@ Ag core–shell nanoparticles for SERS detection of arsenic (III). Talanta. 2016;146:285–290. Copyright 2016, with permission from Elsevier.106 (B) Representation of the Assay Principle for detection of As (III) based on target-triggered signal amplification processes, sequences a and a* are complementary, DNA1 corresponds to the Mg2+-dependent DNAzyme. Reprinted with permission from Zeng L, Zhou D, Gong J, Liu C, Chen J. Highly sensitive aptasensor for trace arsenic (III) detection using DNAzyme as the biocatalytic amplifier. Anal Chem. 2019;91(3):1724–1727. Copyright 2019 American Chemical Society.108
Abbreviations: HP, hairpin probe; MB, magnetic beads; F, fluorophore (FAM); Q, quencher; Exo III, exonuclease III.
Figure 6.
(A) Schematic representation of designs of whole-cell biosensors based on the total vector (POLA). (B) Fluorescence appearance of the bacterial biosensor exposed different concentrations of arsenite. Reprinted from Pola-López L, Camas-Anzueto J, Martínez-Antonio A, et al. Novel arsenic biosensor “POLA” obtained by a genetically modified E. coli bioreporter cell. Sens Actuators B Chem. 2018;254:1061–1068. Copyright 2018, with permission from Elsevier.109
Table 2.
Biosensing Characteristics for Arsenic and Lead Detection
| Analyte | Biosensing Main Components | Type of Transducer | LOD (ppb) | Linear Range (ppb) | References |
|---|---|---|---|---|---|
| Arsenic | Thiolated aptamer, glassy carbon electrode modified with Ag-Au alloy NPs. | Electrochemical (cyclic voltammetry and differential pulse voltammetry) | 0.003 × 10−3 | 0.01–10 | [119] |
| Label-free LC-based aptasensor, specific arsenic aptamer, CTAB as a cationic surfactant, micro-glass slides, OTS, 5CB. | Optical (polarized light microscope) | 400 | - | [120] | |
| Hairpin DNA restricted in PDANS, induction of SDA process by polymerase and endonuclease, ITO electrode in the presence of Ru (phen)32+ as ECL probe. | Optical (Electrochemiluminescence) | 1.2 × 10−3 | - | [121] | |
| Whole-cell (bacterial) biosensor based on arsenical-resistant genes, reporter gene carrying FGE, and sulfatase, which reacts with 4-MUS. | Optical (Fluorescence) | 5: As(III) 50: As(V) |
- | [122] | |
| Label-free aptasensor, gold electrode modified with a specific arsenic aptamer, [Fe(CN)6]3−/4−. | Electrochemical (Impedimetric) | - | 50–10000 | [123] | |
| Magnetotactic bacteria as a magnetic bacterial biosensor, magnetic bacterial strains host lux-based reporter plasmids. | Optical (Bioluminescent) | 4000 | 4000–40,000 | [124] | |
| FRET mechanism between (MPA)-capped CdTe QDs (energy donors) and R6G (energy acceptor), glutathione, FRET efficiency. | Optical (Fluorescence) | 48 | 160 −16,000 | [125] | |
| Aptamers: label-free and CSs labeled with FAM (donor fluorophore), BHQ-1 (acceptor dye), SNPs-Streptavidin, FRET reaction. |
Optical (Fluorescence) | 3.6 | 16 ppb-4000 | [126] | |
| Plasmonic biosensor, AuNPs as a signal probe, acid phosphatase, and AMP dephosphorylation, nucleoside. | Optical (Colorimetric) | 800 | up to 8000 | [127] | |
| Lead | Pb2+-dependent split DNAzyme, Au@Fe3O4, PDA+ as a redox probe, temple-free DNA extension reaction, electrodeposited gold nanoparticles on the surface of a glassy carbon electrode. |
Electrochemical (voltammetric) | 6.5×10−3 | 0.021–21.66 | [128] |
| SERS biosensor, thiolated 5ʹ-Cy3 DNA probe, DNAzyme-modified Fe3O4@Au@Ag NPs. | Optical | 1.08×10−3 | 0.0021–0.21 | [129] | |
| Pb2+-dependent GR-5 DNAzyme, H1 and H2 hairpin probes, catalyzed hairpin assembly, SRCHA reaction, ABTS2−. |
Optical (Colorimetric) | 0.56 | 4.33–21.66 | [130] | |
| Label-free GFET biosensor, graphene nanoelectronics, G-rich DNA single-strands with one-end confined on the graphene surface, alteration of G quadruplex conformation, alteration of charge-containing DNA strands, disruption of charge distribution near the graphene surface. | Field-effect transistor | 0.163 | - | [131] | |
| Whole-cell biosensor, violacein biosynthesis, T7 lac the promoter in E. coli, Pb(II)-dependent metalloregulator PbrR. | Optical (Colorimetric) | 40.62 | 40.62–325 | [132] | |
| C-PFBT Pdots as self-ECL emitter, Fc as Quencher, GCE, (Fc)-labeled DNA, DNAzyme-substrate hybrids. | Optical-electrochemical (Electrochemiluminescence) | 0.036×10−3 | 1.08×10−4-1.08×106 | [133] | |
| FFSSE, TLFC, flat screen-printed FFSSE, USB connection, finite element method FEM, ASV. | Electrochemical (voltammetric) | 0.2 | 0.5–100 | [134] | |
| Portable and label-free aptasensor, graphene FET, graphene surface functionalized by 8–17 DNAzyme, pyrene-NHS, pyrene-derivatized aptamer | Field-effect transistor | 0.037 | - | [135] | |
| Fungus as inactive biomass, GCE-SWCNT-CO-Biomass film electrodes, | Electrochemical (differential pulse anodic stripping voltammetry) | 2.16 | - | [136] | |
| Turn-on fluorescent aptasensor, C-PS2.M-DNA-templated Ag NCs, The template DNA is composed of the lead-specific aptamer. It contains two ends, including the AgNCs nucleus formation segment, Formation of G quadruplex, the proximity of DNA/Ag NCs at both ends fluorescence lighting. | Optical (fluorescence) | 0.65 | 1.08–10.83 | [137] | |
| Label-free peroxidase-like catalytic biosensor, GO–AuNP nanohybrids with peroxidase-like activity, peroxidase substrate TMB by H2O2, hairpin-like or quadruplex formation of ssDNA, and conformational changes. | Colorimetric | 108.33 | 0–10.83×106 | [138] |
Abbreviations: NPs, nanoparticles; LC, liquid crystal; CTAB, cetyltrimethylammonium bromide; OTS, octyltrichlorosilane; 5CB, nematic liquid crystal 4-cyano-4′-pentylbiphenyl; PDANS, polydopamine nanospheres; SDA, strand displacement amplification; FGE, formylglycine-generating enzyme; 4-MUS, 4-methylumbelliferyl sulfate; FRET, fluorescence resonance energy transfer; MPA, mercaptopropionic acid; R6G, rhodamine 6G; QDs, quantum dots; CSs, complementary strands of aptamer; FAM, fluorophores like fluorescein; BHQ-1, black hole quencher 1; SNPs, silica nanoparticles; AuNPs, gold nanoparticles; AMP, adenosine 5ʹ-monophosphate; PDA+, N,N′-bis(2-(trimethylammonium iodide)propylene)perylene-3,4,9,10-tetracarboxyldiimide; SERS, surface-enhanced Raman scattering; Ag NPs, Ag nanoparticles; SRCHA reaction, self-replicating catalyzed hairpin assembly; ABTS2−, 2,2ʹ-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid, GFET, fully integrated graphene field-effect transistor; PFBT, poly[(9,9-dioctylfuorenyl-2,7-diyl)-alt-co-(1,4-benzo-thiadiazole)]; C-PFBT Pdots, carboxyl-functionalized polymer dots; Fc, ferrocene; GCE, glassy carbon electrode; FFSSE, flow-field shaped solid electrode; TLFC, 3D-printed thin-layer flow cell; FEM, finite element method; ASV, square-wave anodic stripping voltammetry; SWCNT, single walled carbon nanotubes; Ag NCs, silver nanoclusters; GO–AuNP, graphene oxide–gold nanohybrids; TMB, 3.3,5,5-tetramethylbenzidine; ssDNA, single-stranded DNA; dsDNA, double stranded DNA.
Lead
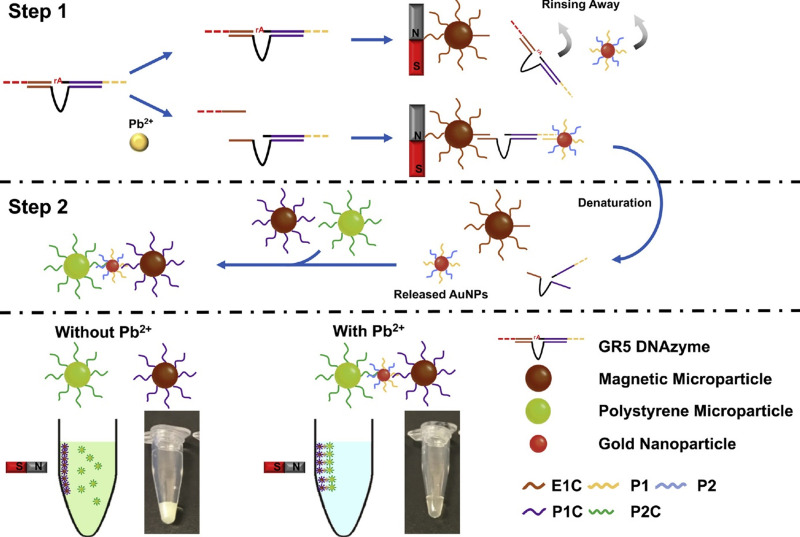
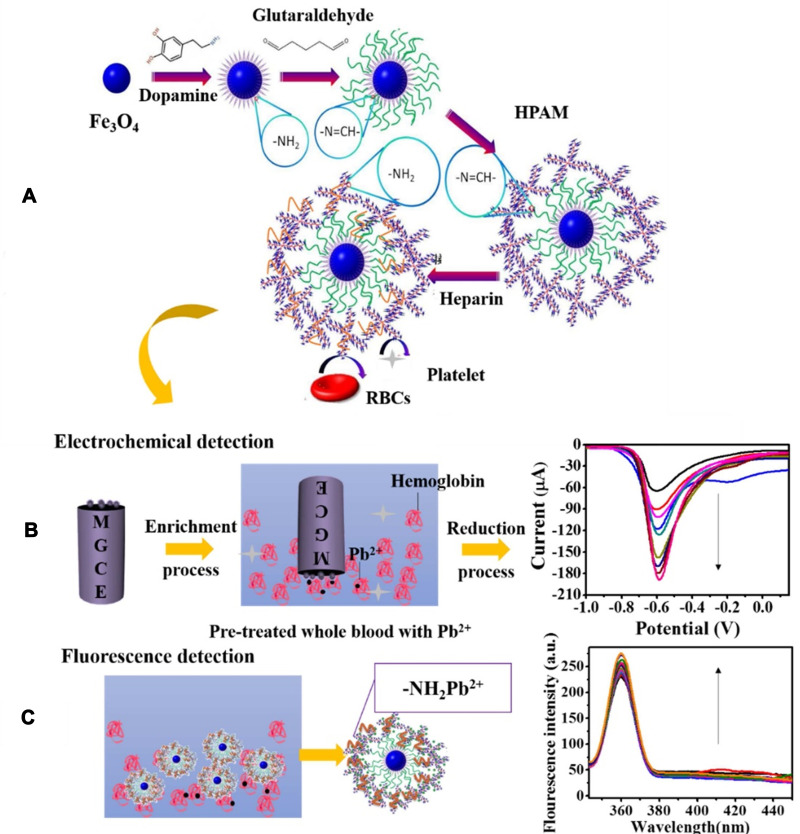
The lead level analysis is essential in environmental samples such as water, food, soil, and biological samples such as blood. In addition to conventional methods such as atomic absorption, spectrophotometry, and chromatography, various sensors and biosensors have been designed. Ran et al113 developed a simple, label-free, and highly sensitive electrochemical aptasensor based on polyadenine use for the first time as an assembly linker to bind aptamer to a screen-printed gold electrode surface. Polyadenine performs a crucial function in the fabrication of a single-stage aptasensor that no blocker compound is required for the electrode surface. DPV and CV techniques were used for electrochemical characterization. The presence of lead causes interaction with the aptamer. This change in the aptamer conformation increases the electrolyte’s access to the electrode surface, thus increasing the output current signal compared to the output signal in the absence of an analyte. The linear relationship of the fabricated aptasensor was obtained in the concentration range of 0.1–1000 ppb, and the detection limit of 0.03 ppb. DNAzyme has been utilized to design biosensors to detect heavy metal ions, as oligonucleotides can catalyze specific hydrolysis reactions. Visual detection of lead ions by Mie scattering in nanoparticle-amplified magnetophoresis is possible. Magnetic microparticles and gold nanoparticles are linked together via a fragment of DNA released through the reaction of lead ions with DNAzyme through hybridization. Then, with magnetic separation, AuNPs are separated, and this time used to connect the second pair, including magnetic microparticles and polystyrene microparticles. The reduction of solution turbidity from milky to smooth and clear is due to reducing Mie scattering property at the time of reduction of free polystyrene microparticles suspended in solution (Figure 7). The detection limit obtained from this method was reported to be 0.24 ppb.114 A biosensor was designed to measure blood lead levels to read in two different detectors, including fluorescence and electrochemical. The system was based on a heparin-modified hyperbranched magnetic polyamide (MHPAM-H). In the presence of magnetic Fe3O4, the integrated biosensor is rapidly immobilized on the surface of the magnetic electrode. Enriched amino groups in the structure of hyperbranched polyamides, along with the cavity structure and the appropriate fluorescence characterization, provide a reliable basis for the rapid accumulation of lead ions by forming a coordination bond. The electrochemical reaction method is based on current production due to enriched lead ions on the electrode surface. HPAM fluorescent substance has many advantages: comprehensive and continuous absorption band, good solubility in water, significant biological compatibility in comparison with conventional dyes. Amino groups in the HPAM structure are bonded to lead ions by coordination bonds, and the geometric mobility of the fluorescence centers is limited, increasing the output fluorescence intensity. Heparin allows the biosensor to function more accurately in the blood matrix. The proposed biosensor provided an excellent linear relationship (1.08×10−4-1.04 ppb and 3.25×10−4-1.04 ppb for fluorescent and electrochemical detection, respectively) and a low detection limit (2.16×10−4 ppb and 9.5×10−4 ppb for fluorescent and electrochemical techniques, respectively) for detecting lead in the blood (Figure 8).115 Xiong et al116 fabricated the electrochemiluminescence (ECL) biosensor using a glassy carbon electrode. The N doped carbon dots (N-CDs) were immobilized as a luminophore on the electrode surface using electropolymerization. Pd-Au hexoctahedron (Pd@Au HOHs) was used as an amplifier of the lead detection signal. In situ electropolymerization of o-phenylenediamine (OPD) was performed on the electrode surface to form N-CDs. Amino groups on the surface of N-CDs immobilize Ag nanoparticles on the electrode surface. The Ag-N band and the amino-modified capture DNA (T1) stabilize the Ag nanoparticles’ surface. Using the chain hybridization technique between T1 and complementary DNA (T2), ssDNA1 (S1) and T2 labeled with Pd @ AuHOHs are modified on the electrode. Simultaneously with this function, ssDNA2 (S2) labeled with Pd @ Au HOHs enters the system, and Pd @ Au HOHs DNA dendrimers are installed on the electrode surface. The Pd @ Au HOHs catalyst’s high activity increases N CDs’ ECL reaction, and as a result, significantly improves N CDs’ output signal. Repeated DNA sequences, including S1 and S2, are coupled with intracellular lead ions to form the G quadruplex structure containing Pb2+. The ECL intensity of the NCDs is quenched by pb2+-stabilized G-quadruplex (G4) structure. The linear range and detection limit of the desired ECL biosensor were 1–1375 ppb and 0.33 ppb, respectively. In another study, an electrochemical aptasensor based on reduced porous graphene oxide was fabricated. In this system, a graphene oxide substrate was employed to fix the aptamer, and gold-modified graphene oxide was used as a signal probe. Au modified the surface of the glassy carbon electrode @ p rGO. This compound’s very high loading capacity causes many gold nanoparticles to be loaded on its surface, enhancing electrical conductivity. Two types of aptamer strands called substrate aptamer 1 filament containing sulfhydryl group and aptamer 2 catalytic filament was developed in this technique to achieve a more accurate diagnosis of lead. Aptamer 1 binds to the surface of the Au @ p rGO nanocomposite, and Aptamer 2, which binds to Au NPs @ GO, is immobilized on the surface of the electrode based on the principle of DNA base pairing. The EIS technique was used for the electrochemical characterization of the fabricated aptasensor. In the lead ion’s presence as the analyte, the substrate strand is cleaved, the DNAzyme is activated, and Apt2 Au NPs @ GO is separated from the electrode surface, resulting in a decrease in electrochemical resistance. As the concentration of lead ions increases, the current response decreases due to the electrocatalytic state created by H2O2 in the presence of a DNAzyme. This complex biosensor revealed a linear range between 1.08×10−3 to 216.6 ppb with a detection limit of 0.36×10−3 ppb for lead ion detection.117 A new strategy based on the piezoelectric method was utilized to detect lead ions at low concentrations. In this study, a quartz crystal microbalance (QCM) biosensor was developed in which a highly specific GR-5 DNAzyme for lead ions was used. The two catalytic and substrate strands together form the GR 5 DNAzyme. Nanomagnetic beads (NMBs) were used to amplify the signal significantly due to their high mass, superparamagnetic properties, and amplifying the inductive effect. Nanomagnetic beads are combined with GR-5 DNAzyme and modified on the particular QCM electrode surface with the self-assembly procedure’s gold-sulfur bond. Under analytical conditions, when the modified electrode was exposed to the analyte, the DNAzyme split from the active RNA site into two parts and simultaneously released NMBs, resulting in an excessive increase in the frequency shifts of the QCM electrode. The linear correlation between Pb concentration logarithm and frequency changes in the proposed biosensor ranged from 0.21×10−3 to 10.83 ppb, and the detection limit was 0.06×10−3 ppb.118 Other remarkable biosensors designed for lead level analysis in recent years are listed in Table 2.
Figure 7.
Fabrication scheme of the visualized lead biosensor. Step 1. The decomposition of GR-5 DNAzyme by lead ions releases DNA fragments connecting magnetic particles and gold nanoparticles. Step 2. Subsequent magnetic separation, the connected AuNPs are released by denaturation and then used to attach a second pair of magnetic microparticles and polystyrene microparticles. Finally, the change of solution turbidity due to reduced Mie scattering. Reprinted from Chu LT, Leung HM, Lo PK, Chen T-H. Visual detection of lead ions based on nanoparticle-amplified magnetophoresis and Mie scattering. Sens Actuators B Chem. 2020;306:127564. Copyright 2020, with permission from Elsevier.114
Figure 8.
The synthesis scheme of the MHPAM-H NPs and their utilization in the electrochemical and fluorescence detection for blood lead. (A) Facilitate the biosensor’s immobilization on the magnetic electrode’s surface by magnetic Fe3O4 and Increased accumulation of blood lead in the presence of enriched amino groups with the formation of a coordination bond. (B) Electrochemical detection is based on the precipitation of enriched lead on the electrode surface. (C) Significant fluorescence detection with the participation of the expanded absorption band of HPAM. Reprinted from Chen H, Shao S, Yu Y, et al. A dual-responsive biosensor for blood lead detection. Anal Chim Acta. 2020;1093:131–141. Copyright 2020, with permission from Elsevier.115
Mercury
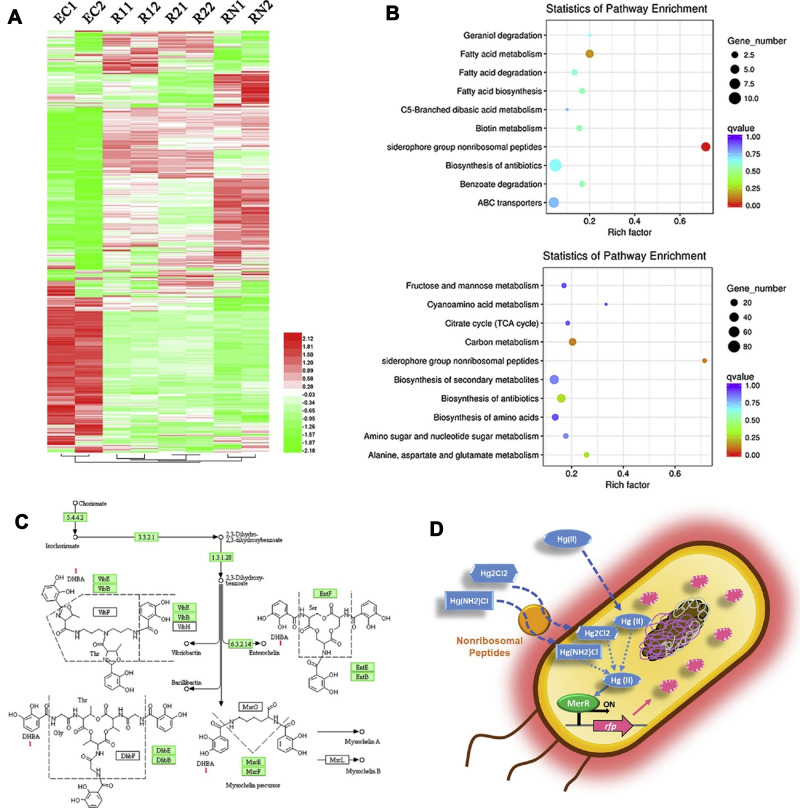
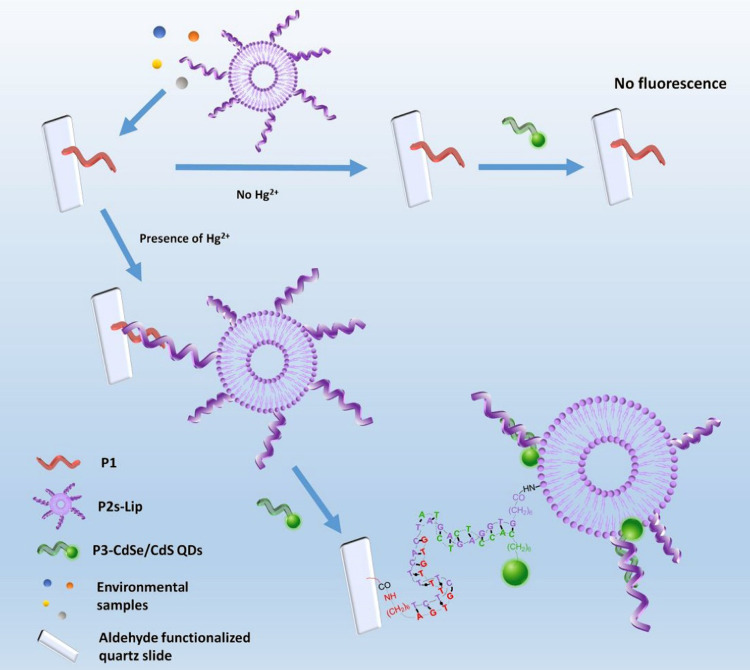
In addition to the general methods of analyzing the levels of various forms of mercury in environmental and biological samples, various biosensors have been designed to determine the levels of mercury. Guo et al designed a test strip platform based on a whole-cell microbial sensor for the simultaneous detection of inorganic mercury (HgCl2, Hg2Cl2, Hg (NH2) Cl) contaminants in cosmetic contents; the most crucial advantage is the lack of need for initial preparation and digestion of the sample. The genetic pathway contains MerR as the sensor protein, and the inducible red fluorescent protein as the reporting agent was located in Escherichia coli. The heatmap exhibits that the biosensors incubated with different mercury compounds show a similar gene expression profile based on the clustering. The results showed that insoluble (Hg2Cl2 and Hg(NH2)Cl) and soluble (HgCl2) mercury chlorinated compounds have a similar effect on the biosensor cell metabolism. It is also speculated that insoluble mercury compounds have been converted to divalent mercury by cell metabolism. Incubation with insoluble chlorinated mercury compounds significantly inhibited the expression of five genes among the seven genes present in the biosynthesis pathway of non-ribosomal peptides of the siderophore group. The production due to the mentioned biosynthesis can also include non-ribosomal enterochelin peptides, which participate in the absorption of iron from insoluble iron compounds. Evidence suggests that enterochelin and its analogs induce mercury uptake from insoluble mercury compounds, while this gene expression inhibition implies a cellular resistance mechanism to reduce mercury uptake. The sensor is installed on a paper filter and is useful in samples containing more than 1 mg/kg of mercury compounds. Fluorescence intensity showed an excellent linear relationship in the range of 50 nM to 10 μM mercury concentration (Figure 9).139 In combination with the inhibition of the hydrogen peroxide catalysis reaction, the amperometric technique was proposed for the indirect detection of divalent mercury ions. In this process, a glassy carbon electrode was applied as the biosensor’s functional substrate, the surface of which was modified by the catalase using cross-linking with glutaraldehyde and bovine serum albumin. In the application of enzymes in biosensors, optimization of conditions such as enzyme concentration, the enzyme loading, electrolyte pH, and applied potential is essential, which was performed in this study. The catalase enzyme’s irreversible inhibition in converting hydrogen peroxide by mercury ions is the biosensor’s functional basis. Electrochemical impedance spectroscopy was used to investigate the electrochemical events that occurred at the electrode surface. The linear inhibition signal was between 0.01 and 0.1 ppb, and the detection limit estimated based on 10% enzyme inhibition was 0.36×10−2 ppb.140 In 2020, Zhang and co-workers developed a highly sensitive fluorescence sensor whose components include integrated CdSe/CdS biosynthetic quantum dots with amplified liposome-carrying output signals. Three different oligonucleotide sequences, including a functional probe fixed on an aldehyde, functionalized quartz slide’s surface, an ssDNA labeled liposome carrier, and a CdSe/CdS QDs labeled index probe, are used in this biosensor. The presence of divalent mercury ions causes all three sequences’ participation, and the bond between the quartz slice and the functional liposome, and the fluorescence signal will be detectable. The binding of mercury ions follows the pattern of T-Hg2+ -T. The linear range of mercury concentration was determined between 0.05 to 20 ppb, and the biosensor detection limit was 0.002 ppb. In addition to the high specificity, the proposed biosensor showed high selectivity in the presence of high amounts of other ions. It also showed its efficiency in tap water, river water, and landfill leachate (Figure 10).141 Huang et al142 developed a signal-on label-free ECL biosensor using AuNPs-modified ITO electrodes with self-assembled thiol mercury-specific oligonucleotides. The oligonucleotide in the analyte’s presence undergoes a conformation change from linear to the hairpin related to mercury binding to the thymine-Hg2+-thymine form. The advantage of the suggested biosensor is that the oligonucleotide has a dual function. On the one hand, it acts as a probe for mercury ions, and from another side, it carries signaling molecules ([Ru(bpy)2(dppz)](BF4)2). Three types of oligonucleotide probes, including T-rich probe, A-rich probe, Hg2+ specific oligonucleotide probes with three different lengths, were used. The highest ECL signal intensity was observed using a longer dedicated probe. About eight molecules producing the ECL signal are loaded inside each oligonucleotide. The aim of a detection limit of 1.02×10−3 ppb was achieved by not utilizing costly and time-consuming processes of labeling oligonucleotides to transduce or amplify the signal. The electrochemical biosensor was produced according to the voltammetric technique based on the interaction of thymine-Hg (II)-thymine. A part of the biosensor consists of reduced graphene oxide modified by gold nanorods and loaded on this surface in two steps, thionine and streptavidin (RGO@AuNR-TH-SA). The other part contains a thiol-functionalized DNA fragment containing a thymine-rich sequence immobilized on the gold electrode surface. Another fragment is biotinylated T-rich DNA that binds to the first fragment in the presence of mercury by the interaction of T-Hg(II)-T. The joining of two strands of DNA in the presence of mercury ions binds RGO@AuNR-TH-SA containing streptavidin to a biotin-containing fragment at the electrode surface. The DPV technique exhibits an increased electrochemical signal, which depends on mercury ions concentration in the sample. Peak current in the concentration range between 0.2 to 40 ppb increased linearly, and the detection limit of this method was valued at 0.048 ppb.143 A novel and exciting method based on the SERS technique for detecting mercury ions was introduced, which depends on the binding coordination of T-Hg(II)-T and the structures of Au @ gap @ AuAg side by side nanorods. PEG-modified gold nanorods that bind to DNA1 via Au-S bond, as well as DNA2 bound to Cy5.5 dye, are used in this method. DNA1 and DNA2 were hybridized via A–T mismatches. Due to the proximity of these structures to gold nanorods, fluorescence is not observed. In the presence of mercury ions, by the interaction of T – Hg2+ – T, the nanorods are formed with regular side-by-side arrangement containing free DNA, resulting in increased fluorescence Cy5.5 dye and SERS signal. In the absence of Hg2+, a stable but dispersed mixture of AuNR-DNA1-DNA2-Cy5.5 is formed with deficient SERS activity and fluorescence intensity. The SERS amplified response was obtained by strong electromagnetic coupling of the Ag shell coating on the surface of assemblies and nanogap between Au @ Ag nanorod structures, which provides a cascade of SERS amplified signals to detect low concentrations of mercury ions. An acceptable linear relationship between peak intensity and target analyte concentration (0.005–1 ppb) and excellent detection limit (0.001 ppb) was obtained for this biosensor.144 Table 3 represents mercury detection biosensors.
Figure 9.
Schematic representation of the whole-cell (cMer-RFP) biosensor for the detection of mercury revealed by transcriptomics. (A) Gene expression heatmap. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. (C) Biosynthesis of siderophore group nonribosomal peptides metabolic pathway. (D) Potential mechanism of the whole-cell microbial biosensor. Reprinted from Guo M, Wang J, Du R, et al. A test strip platform based on a whole-cell microbial biosensor for simultaneous on-site detection of total inorganic mercury pollutants in cosmetics without the need for predigestion. Biosens Bioelectron. 2020;150:111899. Copyright 2020, with permission from Elsevier.139
Figure 10.
Schematic illustration of the fluorescent biosensing strategy for mercury detection. Reprinted with permission from Zhang Y, Xiao J-Y, Zhu Y, et al. Fluorescence Sensor Based on Biosynthetic CdSe/CdS Quantum Dots and Liposome Carrier Signal Amplification for Mercury Detection. Anal Chem. 2020;92(5):3990–3997. Copyright 2020 American Chemical Society.141
Table 3.
Biosensing Characteristics for Mercury and Cadmium Detection
| Analyte | Biosensing Main Components | Type of Transducer | LOD (ppb) | Linear Range (ppb) | References |
|---|---|---|---|---|---|
| Mercury | ExoIII and TdT dependent isothermal amplification, template sequence in the form of a hairpin, two thymine-rich ends, TMB, and H2O2. | Optical (colorimetric) | 0.082 | 0.2–5 | [150] |
| Label-free biosensor, Hg2+-induced cleavage of PS modified RNA, Hg-DPR linked with SWNTs/FET. | Electrochemical (conductometric) | 0.002 | 0.01–20, 20–2000 | [151] | |
| Off-On biosensor, GCE, ECL substrate: complex of Ruthenium(II) tris-(bipyridine)(Ru(bpy)3 2+)/CD-AuNps, ECL intensity switch: single hairpin DNA probe, ferrocene tag, | Optical (electrochemiluminescence) | 0.02 | 0.02–800 | [152] | |
| Reusable biosensor, AQDS as an anionic intercalator, glassy carbon electrode, ssDNA, AuNPs, OMC, SPAN nanofibers. | Electrochemical (voltammetric and impedimetric) | 0.12×10−6 | 1.2×10−6-200 | [153] | |
| A signal on the biosensor, Au electrode, Au screen-printed electrode, oligonucleotide consisting of polythymine modified in 5′ with a –SH functional group, and 3′ with methylene blue. | Electrochemical (square wave voltammetry) | 0.02 | 0.04–20 | [154] | |
| Fluorometric biosensor, CDs labeled thymine-rich22-mer oligonucleotide as the molecular recognition element and energy donor, GO as the FRET acceptor, ODN, NHS, EDC. | Optical (fluorescence) | 0.52 | 1–40 | [155] | |
| Signal-off and reusable biosensor, graphene nanosheets modified electrode, Fc-tagged T-rich ss-DNA, target DNA probe. | Electrochemical (differential pulse Voltammetry) | 0.001 | 0.005–2×10−6 | [156] | |
| Lateral flow biosensors, AC adsorbent, streptavidin-biotinylated DNA probes (Test and Control line probes), thiolated DNA probe modified with Au-NPs, diammonium hydrogen phosphate as an activating agent, quantification by smartphone reader. | Optical (colorimetric) | 0.5 | 0.4–1.4 | [157] | |
| δ-PGA-G-luminol composite, glassy carbon electrode, amino-modified DNA1, biotin-modified DNA2, streptavidin, | Optical (electrochemiluminescence) | 0.001 | 0.002–20 | [158] | |
| DNA-TWJ tagged with ruthenium (II) complex (5′-amino-ss-DNA (TW), complementary ss-DNA (J1) tagged with ruthenium at 5 terminal (Ru-J1)), glassy carbon electrode, 4-aminobenzoic acid. | Optical (electrochemiluminescence) | 0.008×10−3 | 2×10−5-0.002 | [159] | |
| Thymine-rich DNA, SYBR Green I, polyacrylamide hydrogel, 96-well microplates. | Optical (fluorescence) | 2 | - | [160] | |
| Cadmium | CPE, dsDNA: poly C DNA, poly G DNA, EG. | Electrochemical (differential pulse voltammetry) | 0.073×10−3 | 0.243×10−3-0.243 and 2.43–2438.5 | [161] |
| SPE, issAP08-Cd2+ aptamer, ITC, K3[Fe(CN)6] solution containing KCl. | Electrochemical (cyclic voltammetry, and differential pulse voltammetry) | 0.05 | 0.1–1000 | [162] | |
| Light-up biosensor, biotin, and streptavidin, CD, selective aptamers, FAM, Dabcyl (4-[4-(dimethylamino) phenylazo] benzoic acid. |
Optical (fluorescence) | 9.75 | 0–243.85 | [163] | |
| Label-free, regenerable biosensor, DNA probe, DPI, bare silicon oxynitride chip, PEI, T30 oligonucleotides. | Combination of CD, UV-vis, and FTIR techniques | 0.61 | 3.6–7280 | [164] | |
| LC biosensor, DNA containing –SH and –NH2 group in 5′-end and 3′ respectively, DMOAP, APTES, GA, 5CB, a glass slide. |
Optical (polarized light microscope) |
0.024 | - | [165] | |
| Enzyme biosensor, bare gold electrode, Aspergillus oryzae, beta-galactosidase, GA, K3Fe(CN)6, and K4Fe(CN)6 · 3H2O. |
Electrochemical (impedance spectroscopy, cyclic voltammetry, square wave voltammetry) | 6.95 by EIS, and 7.61× 10–3 by SWV | 2.36–2.36 × 107 by EIS, 2.36 × 10–3-2.94 × 107 by SWV | [166] | |
| Signal-on biosensor, gold electrode, thiolated ssDNA, methylene blue, K3Fe(CN)6 solution. | Electrochemical (cyclic voltammetry) | 0.3×10−3 | 10−3 - 20×10−3 | [167] | |
| Whole-Cell biosensor, Escherichia coli MG1655 (pBR-PzntA), green fluorescent protein: gfpuv, M9, and MOPS supplemented medium, Luria-Broth medium. |
Optical (fluorescence) | 2 | - | [146] | |
| Multiplex biosensor including E. coli DH5α, P. aeruginosa PAO1 (pBBcadRgfp-rfp), S. oneidensis MR-1 (pBBcadRgfprfp), Enterobacter sp. NCR3 (pBBcadRgfp-rfp), Enterobacter sp. LCR17 (pBBcadRgfp-rfp), cloning cadRgfp as a single-output cadmium biosensor element, mrfp1, broad-host range vector, red and green fluorescence proteins, slope agar plates. |
Optical (fluorescence) | DH5α: 500–2000 PAO1: 100 MR-1: 104 NCR3: 250 LCR17: 1000 |
- | [168] | |
| Portable biosensor, PGM as POU, aptamer, Exo III for Recycling signal amplification process, magnetic separation, SA-MBs-DNA1-DNA2, DNA3-DNA4 duplex, DNA5-invertase conjugates | Optical (colorimetric) | 1.219×10−3 | 4.877×10−3-48.77 | [169] | |
| Enzyme mimics biotinylated Cd2+ aptamers, thiolated csDNA, streptavidin-coated microplate, gold nanoparticles, MoS2 nanosheets, Au-MoS2 nanocomposites with peroxidase-like activity, TMB. | Optical (colorimetric) | 0.7 | 1–500 | [170] |
Abbreviations: ExoIII, exonuclease III; TdT, terminal deoxynucleotidyl transferase; TMB, 3,3′,5,5′-tetramethylbenzidine; PS, phosphorothioate; Hg-DPR, specific single stranded DNA embedded four PS-modified RNA; SWNTs/FET, single-walled carbon nanotube field effect transistor; ECL, electrochemiluminescence; GCE, glass carbon electrode; CD-AuNps, cyclodextrins-Au nanoparticles; ssDNA, single-stranded DNA; OMC, ordered mesoporous carbon; SPAN, self-doped polyaniline; AuNPs, gold nanoparticles; AQDS, disodium-anthraquinone-2, 6-disulfonate; CDs, carbon dots; GO, graphene oxide; ODN, oligodeoxyribonucleotide, NHS, N-hydroxysuccinimide; EDC, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide; Fc, ferrocene; AC, activated carbon; δ-PGA-G-luminol, gamma-polyglutamic acid-graphene-luminol; DNA-TWJ, DNA three-way junction structure; CPE, carbon paste electrode; dsDNA, double strand DNA; EG, ethyl green; SPE, screen-printed electrode; ITC, isothermal titration calorimetry; FAM, 6-carboxyl fluorescein; CD, circular dichroism; DPI, dual polarization interferometry, PEI, poly(ethyleneimine) solution, FTIR, Fourier transform infrared; LC, liquid crystal; DMOAP, N, N-dimethyl-N-octadecyl-3-aminopropyltrimethoxysilyl chloride; APTES, (3-aminopropyl) triethoxysilane; GA, glutaraldehyde; 5CB, LC 4-cyano-4ʹ-pentylbiphenyl; PGM, personal glucose meter; POU, point-of-use; SA-MBs, streptavidin-coated magnetic beads; TMB, 3,3ʹ,5,5ʹ-tetramethylbenzidine.
Cadmium
Analysis of cadmium levels in biological fluids and environmental samples is of great importance, and the design of high-sensitivity biosensors can be helpful in this field. Fakude and co-workers designed an aptamer-based electrochemical biosensor in which the surface of the screen-printed carbon electrode (SPCEs) was modified in a new approach. Surface modification was performed using carbon black (CB) and gold nanoparticles (AuNPs) by drop-casting and electrodeposition methods. The electrodeposition process was performed by the cyclic voltammetry technique in the range of 400 to 1100 mV with a scan rate of 50 mV/s. The Cd (II) specific thiolated aptamer is immobilized on the surface of SPCE CB AuNPs. Changes in aptamer conformation in interaction with the target analyte change the electrochemical current signal in the presence of the redox probe ([Fe (CN6)]3−/4−). In recent years, CB nanoparticles like other forms of carbon (carbon nanotubes, graphene) and gold nanoparticles have not been widely used; however, this combination has excellent conductivity and electrocatalytic properties, high porosity, and at the same time is cost-effective and can be considered as an excellent electrode surface modifier. SWV technique was used for analysis and concentration determination, and a linear range of 1 to 50 ppb with a detection limit of 0.14 ppb was observed for this biosensor (Figure 11A).145 Recently, a whole-cell bacterial biosensor was developed in which the strain of E. coli MG1655 (pBR-PzntA) was used as a fluorescent reporter to determine the concentration of inorganic cadmium. The transcriptional fusion of the zntA gene promoter with the less-promoter gene encoding the green fluorescent protein (gfpuv) causes the sensor plasmid. MOPS was used as a suitable assay medium to achieve the accurate operation of this biosensor. The detection limit of cadmium obtained in the optimized condition is 2 ppb.146 Another biosensor was designed based on an optical transducer, in which a multifunctional fluorescent aptamer probe was used to detect Cd (II). The system’s multifunctionality is due to the labeled aptamer’s simultaneous operation as a signal recognition and reporting element. Conformational change of a specific aptamer due to cadmium presence in the medium leads to a change in the aptamer’s structure from a random coil state to the stem-loop form. These changes bring the aptamer’s 5ʹ end, which contains G4, closer to the 3ʹ end, including 6- carboxyfluorescein (6-FAM), resulting in fluorescence quenched due to the photoinduced electron transfer (PET) phenomenon. The concentration of cadmium is proportional to the fluorescence response. The linear range and detection limit of the considered aptasensor were determined from 1.75 to 48.77 ppb and 0.52 ppb, respectively.147 Ruhan and co-workers designed a biosensor based on a carbon paste electrode (CPE) modified with dsDNA and a brilliant green (BG) indicator. BG is characterized by the ability to bind to ssDNA through interaction with the guanine base, and cadmium disrupts the structure of dsDNA and ssDNA generation. Oxidation of this indicator at the electrode surface causes changes in the output current signal. The reduction current of BG at the surface of the modified electrode increases significantly in the cadmium presence. In this study, the DPV technique was used to evaluate the electrode surface and determine cadmium concentration. The reduction peak current of BG increases linearly between 0.012 and 0.292 ppb of cadmium concentration, and the detection limit of the desired biosensor was 0.024× 10−3 ppb.148 Recently, an electrochemical aptasensor was designed using a modified SPGE platform with diazonium. The electrodeposition process first modifies the electrode’s surface with a diazonium salt film, in which 4-carboxymethyl aryl diazonium (CMA) is used. The carboxylic groups on the surface are then activated by the EDC/NHS. The Cd (II) aptamer containing the amino group is covalently bonded and immobilized on the surface by the reaction of carbodiimide with the active carboxylic groups present on the modified gold electrode’s surface. The step-by-step modification of the electrode surface was characterized by CV and EIS electrochemical techniques. Detection of cadmium and evaluation of the performance of the fabricated aptasensor was performed by the EIS method. In the presence of the analyte target, Rct increases due to the cadmium- aptamer complex’s formation, indicating a decrease in electron transfer between the electrode surface and the redox couple, which was also approved by the CV method. The designed aptasensor exhibits an excellent linear relationship between the impedance changes and the Cd (II) concentration logarithm in the range of 0.243×106 to 0.243 ppb. High sensitivity was achieved for the aptasensor with a detection limit of 0.670×10 −1 ppb (Figure 11B).149 Other noteworthy examples of cadmium detection biosensors are described in Table 3.
Figure 11.
(A) Procedure and electrochemical detection principle of the cadmium aptasensor based on carbon black (CB) and gold nanoparticles (AuNPs) application. Reprinted from Fakude CT, Arotiba OA, Mabuba N. Electrochemical aptasensing of cadmium (II) on a carbon black-gold nano-platform. J Electroanal Chem. 2020;858:113796. Copyright 2020, with permission from Elsevier.145 (B) Schematic illustration of cadmium detection by electrochemical aptasensor based on diazonium electrodeposition. Reprinted from Rabai S, Benounis M, Catanante G, et al. Development of a label-free electrochemical aptasensor based on diazonium electrodeposition: application to cadmium detection in water. Anal Biochem. 2020;612:113956. Copyright 2020, with permission from Elsevier.149
Conclusions and Outlook
The importance of investigating the toxicity of heavy metals and exploring the methods of analyzing these metals’ levels in different samples using integrated bio and nanotechnology techniques that have led to biosensors’ development in the last five years is the primary purpose of this study. In this study, we attempted to discuss the most sensitive and innovative biosensors designed over the previous five years from the functional mechanism, essential components, and detection of arsenic, lead, mercury, and cadmium as the most challenging toxic metals in the environment. The fabrication of nanomaterials with a high surface area such as porous materials or special functional groups on their surface is examined in the pre-treatment, absorbance, and separation stage of heavy metals. In addition to metal absorbers’ role, nanomaterials are used to design metal concentration tracking systems due to their unique electrical, optical, and magnetic properties. On the other hand, they have the role of amplifying electrical and optical signals. Our overview reveals that biosensor designers have often utilized optical and electrochemical transducers to design heavy metal detection biosensors. In recent years, aptamers and whole cells have been employed and developed more in detecting heavy metals than other biorecognition elements. The detection limit of heavy metals by various biosensors is in the micro to the femtomolar range, which indicates the acceptable sensitivity of biosensors. However, some of them had a limited linear dynamic range of concentration. Among the biosensors studied in this review, the best limit of detection for arsenic, lead, mercury, and cadmium were achieved 0.003×10−3, 2.16×10−4, 0.12×10−6, and 0.024×10−3 ppb, respectively. As mentioned above, the investigation of these biosensors’ structures revealed that the use of electrochemical and optical transducers of fluorescence type and surface modification with novel and composite nanomaterials leads to improved detection methods. The advent of biosensors and significant advances in this area have been made in recent years to achieve faster, cheaper, more sensitive, and more straightforward measurement methods than traditional analysis methods. Some biosensors provide a lower detection limit and a more sensitive method for measuring heavy metals while being simple, cost-effective, and time-consuming. However, most biosensors are not yet available for public and commercial applications; the main reason is that biosensors’ performance is suitable for research studies and laboratory conditions due to their components. Although, in addition to biosensors, other sensing mechanisms have been developed to assess the level of heavy metals, there is still a long way to develop miniaturized yet smart devices.171 Miniaturizing a device requires the use of materials that have several different functions at the same time. One of the most reliable methods to integrate nanomaterials’ performance into a system is to design multifunctional nanocomposites. An example of this aspect is flower-like nanocomposites such as ZnO - Ag2O,172 when used in sensing systems display two functions: high surface area and remarkable photocatalytic activity. Most heavy metals have several oxidation states that each of them exhibits varying degrees of toxicity. Biosensors are only able to detect one of these states in the sample. Another critical challenge in biosensors’ design is the simultaneous detection of different oxidation states of a metal. Therefore, it is suggested that the following items be considered in future studies (Figure 12): (1) Design of biosensors capable of simultaneously detecting a range of the most critical toxic heavy metals such as lead, mercury, cadmium, and arsenic. (2) Enhancing the functional stability of biosensors in different laboratory conditions. (3) Efforts to design more straightforward, more affordable, and user-friendly biosensors with a higher sensitivity approach, less sample volume, with fewer pre-treatment steps. (4) Focus on miniaturization such as microfluidic device fabrication and nanomaterials’ integration with other biosensors components for in-situ detection of heavy metals. (5) Utilization of nanotechnology in integrating artificial intelligence and biosensors to continue developing heavy metal detection processes towards scalability, high sensitivity, and multifunctionality.
Figure 12.
Futuristic options in the design of biosensors for the detection of heavy metals.
Funding Statement
The authors gratefully acknowledge the support from The Institute of Pharmaceutical Sciences (TIPS) and Iran Science Elites Federation (ISEF) for the first author postdoc program’s financial support and the general support of the INSF.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- 1.Xu J, Liu C, Hsu P-C, et al. Remediation of heavy metal contaminated soil by asymmetrical alternating current electrochemistry. Nat Commun. 2019;10(1):1–8. doi: 10.1038/s41467-018-07882-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masindi V, Muedi KL. Environmental contamination by heavy metals. Heavy Metals. 2018;10:115–132. [Google Scholar]
- 3.Jan AT, Azam M, Siddiqui K, Ali A, Choi I, Haq QM. Heavy metals and human health: mechanistic insight into toxicity and counter defense system of antioxidants. Int J Mol Sci. 2015;16(12):29592–29630. doi: 10.3390/ijms161226183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z. Roles of vertebrate aquaglyceroporins in arsenic transport and detoxification In: MIPs and Their Role in the Exchange of Metalloids. New York, NY: Springer; 2010:71–81. [DOI] [PubMed] [Google Scholar]
- 5.Yang H, Shu Y. Cadmium transporters in the kidney and cadmium-induced nephrotoxicity. Int J Mol Sci. 2015;16(1):1484–1494. doi: 10.3390/ijms16011484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chmielowska-Bąk J, Izbiańska K, Deckert J. The toxic Doppelganger: on the ionic and molecular mimicry of cadmium. Acta biochimica polonica. 2013;60:3. [PubMed] [Google Scholar]
- 7.Abadin H, Taylor J, Buser MC, et al. Toxicological profile for lead: draft for public comment. 2019.
- 8.Vázquez M, Vélez D, Devesa V, Puig S. Participation of divalent cation transporter DMT1 in the uptake of inorganic mercury. Toxicology. 2015;331:119–124. [DOI] [PubMed] [Google Scholar]
- 9.Hazelhoff MH, Trebucobich MS, Stoyanoff TR, Chevalier AA, Torres AM. Amelioration of mercury nephrotoxicity after pharmacological manipulation of organic anion transporter 1 (Oat1) and multidrug resistance-associated protein 2 (Mrp2) with furosemide. Toxicol Res (Camb). 2015;4(5):1324–1332. [Google Scholar]
- 10.Rana MN, Tangpong J, Rahman MM. Toxicodynamics of lead, cadmium, mercury and arsenic-induced kidney toxicity and treatment strategy: a mini review. Toxicol Rep. 2018;5:704–713. doi: 10.1016/j.toxrep.2018.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saadatzadeh A, Afzalan S, Zadehdabagh R, et al. Determination of heavy metals (lead, cadmium, arsenic, and mercury) in authorized and unauthorized cosmetics. Cutan Ocul Toxicol. 2019;38(3):207–211. doi: 10.1080/15569527.2019.1590389 [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Cobbina SJ, Mao G, Xu H, Zhang Z, Yang L. A review of toxicity and mechanisms of individual and mixtures of heavy metals in the environment. Environ Sci Pollut Res. 2016;23(9):8244–8259. doi: 10.1007/s11356-016-6333-x [DOI] [PubMed] [Google Scholar]
- 13.Mohod CV, Dhote J. Review of heavy metals in drinking water and their effect on human health. Int J Innovative Res Sci Eng Tech. 2013;2(7):2992–2996. [Google Scholar]
- 14.Sall ML, Diaw AKD, Gningue-Sall D, Aaron SE, Aaron -J-J. Toxic heavy metals: impact on the environment and human health, and treatment with conducting organic polymers, a review. Environ Sci Pollut Res Int. 2020;27(24):29927–29942. doi: 10.1007/s11356-020-09354-3 [DOI] [PubMed] [Google Scholar]
- 15.Arjomandi M, Shirkhanloo H. A review: analytical methods for heavy metals determination in environment and human samples. Analy Methods Environ Chem J. 2019;2(03):97–126. doi: 10.24200/amecj.v2.i03.73 [DOI] [Google Scholar]
- 16.Hassani S, Rezaei Akmal M, Salek Maghsoudi A, et al. High-Performance Voltammetric Aptasensing Platform for Ultrasensitive Detection of Bisphenol A as an Environmental Pollutant. Front Bioeng Biotech. 2020;8:1055. doi: 10.3389/fbioe.2020.574846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pandey CM, Malhotra BD. Biosensors: Fundamentals and Applications. Walter de Gruyter GmbH & Co KG; 2019. [Google Scholar]
- 18.Nehra M, Dilbaghi N, Marrazza G, et al. Emerging nanobiotechnology in agriculture for the management of pesticide residues. J Hazard Mater. 2020;401:123369. doi: 10.1016/j.jhazmat.2020.123369 [DOI] [PubMed] [Google Scholar]
- 19.Chakraborty U, Bhanjana G, Kaur G, Kaushik A, Chaudhary G. Electro-active silver oxide nanocubes for label free direct sensing of bisphenol A to assure water quality. Mater Today Chem. 2020;16:100267. doi: 10.1016/j.mtchem.2020.100267 [DOI] [Google Scholar]
- 20.MGd M, Martins VG, Steffens D, Pranke P, da Costa JAV. Biological applications of nanobiotechnology. J Nanosci Nanotechnol. 2014;14(1):1007–1017. doi: 10.1166/jnn.2014.8748 [DOI] [PubMed] [Google Scholar]
- 21.Salek-Maghsoudi A, Vakhshiteh F, Torabi R, et al. Recent advances in biosensor technology in assessment of early diabetes biomarkers. Biosens Bioelectron. 2018;99:122–135. doi: 10.1016/j.bios.2017.07.047 [DOI] [PubMed] [Google Scholar]
- 22.Aragay G, Merkoçi A. Nanomaterials application in electrochemical detection of heavy metals. Electrochim Acta. 2012;84:49–61. doi: 10.1016/j.electacta.2012.04.044 [DOI] [Google Scholar]
- 23.Kumar P, Kim K-H, Bansal V, Lazarides T, Kumar N. Progress in the sensing techniques for heavy metal ions using nanomaterials. J Industrial Eng Chem. 2017;54:30–43. doi: 10.1016/j.jiec.2017.06.010 [DOI] [Google Scholar]
- 24.Waheed A, Mansha M, Ullah N. Nanomaterials-based electrochemical detection of heavy metals in water: current status, challenges and future direction. TrAC Trends Anal Chem. 2018;105:37–51. doi: 10.1016/j.trac.2018.04.012 [DOI] [Google Scholar]
- 25.Liu Y, Deng Y, Dong H, Liu K, He N. Progress on sensors based on nanomaterials for rapid detection of heavy metal ions. Sci China Chem. 2017;60(3):329–337. [Google Scholar]
- 26.Yang J, Hou B, Wang J, et al. Nanomaterials for the removal of heavy metals from wastewater. Nanomaterials. 2019;9(3):424. doi: 10.3390/nano9030424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sepand MR, Ranjbar S, Kempson IM, et al. Targeting non-apoptotic cell death in cancer treatment by nanomaterials: recent advances and future outlook. Nanomedicine. 2020;102243. [DOI] [PubMed] [Google Scholar]
- 28.Vigneshvar S, Sudhakumari C, Senthilkumaran B, Prakash H. Recent advances in biosensor technology for potential applications–an overview. Front Bioeng Biotech. 2016;4:11. doi: 10.3389/fbioe.2016.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buledi JA, Amin S, Haider SI, Bhanger MI, Solangi AR. A review on detection of heavy metals from aqueous media using nanomaterial-based sensors. Environ Sci Pollut Res. 2020;1–9. [DOI] [PubMed] [Google Scholar]
- 30.Ahmad R, Wolfbeis OS, Hahn Y-B, Alshareef HN, Torsi L, Salama KN. Deposition of nanomaterials: a crucial step in biosensor fabrication. Mater Today Commun. 2018;17:289–321. doi: 10.1016/j.mtcomm.2018.09.024 [DOI] [Google Scholar]
- 31.Holzinger M, Le Goff A, Cosnier S. Nanomaterials for biosensing applications: a review. Front Chem. 2014;2:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Putzbach W, Ronkainen NJ. Immobilization techniques in the fabrication of nanomaterial-based electrochemical biosensors: A review. Sensors. 2013;13(4):4811–4840. doi: 10.3390/s130404811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn MR, Jimenez RM, Chaput JC. Analysis of aptamer discovery and technology. Nat Rev Chem. 2017;1(10):1–16. doi: 10.1038/s41570-017-0076 [DOI] [Google Scholar]
- 34.Hassani S, Akmal MR, Salek-Maghsoudi A, et al. Novel label-free electrochemical aptasensor for determination of Diazinon using gold nanoparticles-modified screen-printed gold electrode. Biosens Bioelectron. 2018;120:122–128. doi: 10.1016/j.bios.2018.08.041 [DOI] [PubMed] [Google Scholar]
- 35.Maghsoudi AS, Hassani S, Akmal MR, et al. An Electrochemical Aptasensor Platform Based on Flower-Like Gold Microstructure-Modified Screen-Printed Carbon Electrode for Detection of Serpin A12 as a Type 2 Diabetes Biomarker. Int J Nanomedicine. 2020;15:2219. doi: 10.2147/IJN.S244315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ali MH, Elsherbiny ME, Emara M. Updates on aptamer research. Int J Mol Sci. 2019;20(10):2511. doi: 10.3390/ijms20102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song K-M, Lee S, Ban C. Aptamers and their biological applications. Sensors. 2012;12(1):612–631. doi: 10.3390/s120100612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma S, Byrne H, O’Kennedy RJ. Antibodies and antibody-derived analytical biosensors. Essays Biochem. 2016;60(1):9–18. doi: 10.1042/EBC20150002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trilling AK, Beekwilder J, Zuilhof H. Antibody orientation on biosensor surfaces: a minireview. Analyst. 2013;138(6):1619–1627. doi: 10.1039/c2an36787d [DOI] [PubMed] [Google Scholar]
- 40.Zhu Y-C, Mei L-P, Ruan Y-F, et al. Enzyme-based biosensors and their applications In: Advances in Enzyme Technology. Elsevier; 2019:201–223. [Google Scholar]
- 41.Nguyen HH, Lee SH, Lee UJ, Fermin CD, Kim M. Immobilized enzymes in biosensor applications. Materials. 2019;12(1):121. doi: 10.3390/ma12010121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan NA, Wan J, Wong LS. Exploring the potential of whole cell biosensor: a review in environmental applications. Int J Chem Environ Biol Sci. 2014;2:1. [Google Scholar]
- 43.Lim JW, Ha D, Lee J, Lee SK, Kim T. Review of micro/nanotechnologies for microbial biosensors. Front Bioeng Biotech. 2015;3:61. doi: 10.3389/fbioe.2015.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai C, Choi S. Technology and applications of microbial biosensor. Open Journal of Applied Biosensor. 2013;02(03):83–93. doi: 10.4236/ojab.2013.23011 [DOI] [Google Scholar]
- 45.Ahmed A, Rushworth JV, Hirst NA, Millner PA. Biosensors for whole-cell bacterial detection. Clin Microbiol Rev. 2014;27(3):631–646. doi: 10.1128/CMR.00120-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gui Q, Lawson T, Shan S, Yan L, Liu Y. The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors. 2017;17(7):1623. doi: 10.3390/s17071623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Labuda J, Brett AMO, Evtugyn G, et al. Electrochemical nucleic acid-based biosensors: concepts, terms, and methodology (IUPAC Technical Report). Pure Applied Chem. 2010;82(5):1161–1187. doi: 10.1351/PAC-REP-09-08-16 [DOI] [Google Scholar]
- 48.Hassani S, Maghsoudi AS, Akmal MR, et al. A Sensitive Aptamer-Based Biosensor for Electrochemical Quantification of PSA as a Specific Diagnostic Marker of Prostate Cancer. J Pharm Pharm Sci. 2020;23:243–258. doi: 10.18433/jpps31171 [DOI] [PubMed] [Google Scholar]
- 49.Ronkainen NJ, Halsall HB, Heineman WR. Electrochemical biosensors. Chem Soc Rev. 2010;39(5):1747–1763. doi: 10.1039/b714449k [DOI] [PubMed] [Google Scholar]
- 50.Taleat Z, Khoshroo A, Mazloum-Ardakani M. Screen-printed electrodes for biosensing: a review (2008–2013). Microchimica Acta. 2014;181(9–10):865–891. [Google Scholar]
- 51.Ali MA, Dong L, Dhau J, Khosla A, Kaushik A. Perspective—electrochemical sensors for soil quality assessment. J Electrochem Soc. 2020;167(3):037550. doi: 10.1149/1945-7111/ab69fe [DOI] [Google Scholar]
- 52.Chillawar RR, Tadi KK, Motghare RV. Voltammetric techniques at chemically modified electrodes. J Anal Chem. 2015;70(4):399–418. doi: 10.1134/S1061934815040152 [DOI] [Google Scholar]
- 53.Farghaly OA, Hameed RA, Abu-Nawwas -A-AH. Analytical application using modern electrochemical techniques. Int J Electrochem Sci. 2014;9(1):3287–3318. [Google Scholar]
- 54.Ding J, Qin W. Recent advances in potentiometric biosensors. TrAC Trends Anal Chem. 2020;124:115803. doi: 10.1016/j.trac.2019.115803 [DOI] [Google Scholar]
- 55.Cesewski E, Johnson BN. Electrochemical biosensors for pathogen detection. Biosens Bioelectron. 2020;159:112214. doi: 10.1016/j.bios.2020.112214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bahadır EB, Sezgintürk MK. A review on impedimetric biosensors. Artif Cells, Nanomed Biotechnol. 2016;44(1):248–262. [DOI] [PubMed] [Google Scholar]
- 57.Chen C, Wang J. Optical biosensors: an exhaustive and comprehensive review. Analyst. 2020;145(5):1605–1628. [DOI] [PubMed] [Google Scholar]
- 58.Damborský P, Švitel J, Katrlík J. Optical biosensors. Essays Biochem. 2016;60(1):91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gridina N, Dorozinsky G, Khristosenko R, et al. Surface plasmon resonance biosensor. Sens Transducers. 2013;149(2):60. [Google Scholar]
- 60.Liu M, Lin Z, Lin J-M. A review on applications of chemiluminescence detection in food analysis. Anal Chim Acta. 2010;670(1–2):1–10. [DOI] [PubMed] [Google Scholar]
- 61.Bhand S, Kanungo L, Pal S. Chemiluminescence and Fluorescence Optical Biosensor for the Detection of Aflatoxins in Food. Food Biosensors. 2016;161–181. [Google Scholar]
- 62.Strianese M, Staiano M, Ruggiero G, Labella T, Pellecchia C, D’Auria S. Fluorescence-based biosensors In: Spectroscopic Methods of Analysis. Springer; 2012:193–216. [DOI] [PubMed] [Google Scholar]
- 63.Pohanka M. The piezoelectric biosensors: principles and applications. Int J Electrochem Sci. 2017;12:496–506. [Google Scholar]
- 64.Beltrami LV, Kunst SR, Birriel EJ, Malfatti C. Magnetoelastic biosensors: corrosion protection of an FeNiMoB alloy from alkoxide precursors. Thin Solid Films. 2017;624:83–94. [Google Scholar]
- 65.Guo X, Sang S, Guo J, et al. A magnetoelastic biosensor based on E2 glycoprotein for wireless detection of classical swine fever virus E2 antibody. Sci Rep. 2017;7(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wadhera T, Kakkar D, Wadhwa G, Raj B. Recent advances and progress in development of the field effect transistor biosensor: A review. J Electron Mater. 2019;48(12):7635–7646. [Google Scholar]
- 67.Vu C-A, Chen W-Y. Field-effect transistor biosensors for biomedical applications: recent advances and future prospects. Sensors. 2019;19(19):4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turner AP. Biosensors: sense and sensibility. Chem Soc Rev. 2013;42(8):3184–3196. [DOI] [PubMed] [Google Scholar]
- 69.Han K, Liang Z, Zhou N. Design strategies for aptamer-based biosensors. Sensors. 2010;10(5):4541–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sang S, Wang Y, Feng Q, Wei Y, Ji J, Zhang W. Progress of new label-free techniques for biosensors: a review. Crit Rev Biotechnol. 2016;36(3):465–481. [DOI] [PubMed] [Google Scholar]
- 71.Schöning MJ, Poghossian A. Label-Free Biosensing: Advanced Materials, Devices and Applications. Vol. 16 Springer; 2018. [Google Scholar]
- 72.Kumar S, Nehra M, Mehta J, Dilbaghi N, Marrazza G, Kaushik A. Point-of-care strategies for detection of waterborne pathogens. Sensors. 2019;19(20):4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y, Zhou Q, Ren B, et al. Trends and health risks of dissolved heavy metal pollution in global river and lake water from 1970 to 2017. Rev Environ Contam Toxicol. 2019;254:1–24. [DOI] [PubMed] [Google Scholar]
- 74.Zhou Q, Yang N, Li Y, et al. Total concentrations and sources of heavy metal pollution in global river and lake water bodies from 1972 to 2017. Global Ecol Conserv. 2020;22:e00925. [Google Scholar]
- 75.Kelessidis A, Stasinakis AS. Comparative study of the methods used for treatment and final disposal of sewage sludge in European countries. Waste Manage. 2012;32(6):1186–1195. [DOI] [PubMed] [Google Scholar]
- 76.Abdul KSM, Jayasinghe SS, Chandana EP, Jayasumana C, De Silva PMC. Arsenic and human health effects: A review. Environ Toxicol Pharmacol. 2015;40(3):828–846. [DOI] [PubMed] [Google Scholar]
- 77.Vimercati L, Gatti MF, Gagliardi T, et al. Environmental exposure to arsenic and chromium in an industrial area. Environ Sci Pollut Res. 2017;24(12):11528–11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hassani S, Maqbool F, Salek-Maghsoudi A, et al. Alteration of hepatocellular antioxidant gene expression pattern and biomarkers of oxidative damage in diazinon-induced acute toxicity in Wistar rat: a time-course mechanistic study. EXCLI j. 2018;17:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang -Y-Y, Chai L-Y, Yang W-C. Arsenic Distribution and Pollution Characteristics. Arsenic Pollution Control in Nonferrous Metallurgy. Springer; 2019:1–15. [Google Scholar]
- 80.Carlin DJ, Naujokas MF, Bradham KD, et al. Arsenic and environmental health: state of the science and future research opportunities. Environ Health Perspect. 2016;124(7):890–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh R, Singh S, Parihar P, Singh VP, Prasad SM. Arsenic contamination, consequences and remediation techniques: a review. Ecotoxicol Environ Saf. 2015;112:247–270. [DOI] [PubMed] [Google Scholar]
- 82.Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ. Heavy Metal Toxicity and the Environment. Molecular, clinical and environmental toxicology: Springer; 2012:133–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Karrari P, Mehrpour O, Abdollahi M. A systematic review on status of lead pollution and toxicity in Iran; Guidance for preventive measures. DARU J Pharm Sci. 2012;20(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pourmand A, Al-Tiae TK, Mazer-Amirshahi M. Perspective on lead toxicity, a comparison between the United States and Iran. DARU J Pharm Sci. 2012;20(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mitra P, Sharma S, Purohit P, Sharma P. Clinical and molecular aspects of lead toxicity: an update. Crit Rev Clin Lab Sci. 2017;54(7–8):506–528. [DOI] [PubMed] [Google Scholar]
- 86.Engwa GA, Ferdinand PU, Nwalo FN, Unachukwu MN. Mechanism and health effects of heavy metal toxicity in humans. Poisoning in the Modern World-New Tricks for an Old Dog? IntechOpen. 2019. [Google Scholar]
- 87.Mostafalou S, Abdollahi M. Environmental pollution by mercury and related health concerns: renotice of a silent threat. Arch Industrial Hygiene Toxicol. 2013;64(1):179–181. [DOI] [PubMed] [Google Scholar]
- 88.Rice KM, Jr EM W, Wu M, Gillette C, Blough ER. Environmental mercury and its toxic effects. J Preventive Med Public Health. 2014;47(2):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Drace K, Kiefer AM, Veiga MM. Cyanidation of mercury-contaminated tailings: potential health effects and environmental justice. Curr Environ Health Rep. 2016;3(4):443–449. [DOI] [PubMed] [Google Scholar]
- 90.Vardhan KH, Kumar PS, Panda RC. A review on heavy metal pollution, toxicity and remedial measures: current trends and future perspectives. J Mol Liq. 2019;290:111197. [Google Scholar]
- 91.Khalid M, Hassani S, Abdollahi M. Metals-induced oxidative stress: an evidence-based update of advantages and disadvantages. Curr Opinion Toxicol. 2020. [Google Scholar]
- 92.Dos Santos AA, Hort MA, Culbreth M, et al. Methylmercury and brain development: A review of recent literature. J Trace Elements Med Biol. 2016;38(99–107):254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ahmad S, Mahmood R. Mercury chloride toxicity in human erythrocytes: enhanced generation of ROS and RNS, hemoglobin oxidation, impaired antioxidant power, and inhibition of plasma membrane redox system. Environ Sci Pollut Res. 2019;26(6):5645–5657. [DOI] [PubMed] [Google Scholar]
- 94.Ynalvez R, Gutierrez J, Gonzalez-Cantu H. Mini-review: toxicity of mercury as a consequence of enzyme alteration. Biometals. 2016;29(5):781–788. [DOI] [PubMed] [Google Scholar]
- 95.Zhang H, Reynolds M. Cadmium exposure in living organisms: A short review. Sci Total Environ. 2019;678:761–767. [DOI] [PubMed] [Google Scholar]
- 96.Rahimzadeh MR, Rahimzadeh MR, Kazemi S. Cadmium toxicity and treatment: an update. Caspian j Internal Med. 2017;8(3):135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Faroon O, Ashizawa A, Wright S, et al. Toxicological profile for cadmium. 2012. [PubMed]
- 98.Hutchinson D, Müller J, McCarthy JE, et al. Cadmium nanoparticles citrullinate cytokeratins within lung epithelial cells: cadmium as a potential cause of citrullination in chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2018;13:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Branca JJV, Morucci G, Pacini A. Cadmium-induced neurotoxicity: still much ado. Neural Regener Res. 2018;13(11):1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li H, Wallin M, Barregard L, et al. Smoking-Induced Risk of Osteoporosis Is Partly Mediated by Cadmium From Tobacco Smoke: the MrOS Sweden Study. J Bone Mineral Res. 2020;35(8):1424–1429. doi: 10.1002/jbmr.4014 [DOI] [PubMed] [Google Scholar]
- 101.Organization WH. Preventing Disease Through Healthy Environments: Exposure to Cadmium: A Major Public Health Concern. World Health Organization; 2019. [Google Scholar]
- 102.Sharma H, Rawal N, Mathew BB. The characteristics, toxicity and effects of cadmium. Int j Nanotech Nanosci. 2015;3:1–9. [Google Scholar]
- 103.Scinicariello F, Buser MC, Cadmium B. Depressive Symptoms in Young Adults (20–39 years). Psychol Med. 2015;45(4):807. doi: 10.1017/S0033291714001883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rani A, Kumar A, Lal A, Pant M. Cellular mechanisms of cadmium-induced toxicity: a review. Int J Environ Health Res. 2014;24(4):378–399. doi: 10.1080/09603123.2013.835032 [DOI] [PubMed] [Google Scholar]
- 105.Sarkar A, Ravindran G, Krishnamurthy V. A brief review on the effect of cadmium toxicity: from cellular to organ level. Int J Biotechnol Res. 2013;3(1):17–36. [Google Scholar]
- 106.Song L, Mao K, Zhou X, Hu J. A novel biosensor based on Au@ Ag core–shell nanoparticles for SERS detection of arsenic (III). Talanta. 2016;146:285–290. doi: 10.1016/j.talanta.2015.08.052 [DOI] [PubMed] [Google Scholar]
- 107.Yang L, An B, Yin X, Li F. A competitive coordination-based immobilization-free electrochemical biosensor for highly sensitive detection of arsenic (v) using a CeO 2–DNA nanoprobe. Chem Commun. 2020;56(39):5311–5314. doi: 10.1039/D0CC01821J [DOI] [PubMed] [Google Scholar]
- 108.Zeng L, Zhou D, Gong J, Liu C, Chen J. Highly sensitive aptasensor for trace arsenic (III) detection using DNAzyme as the biocatalytic amplifier. Anal Chem. 2019;91(3):1724–1727. doi: 10.1021/acs.analchem.8b05466 [DOI] [PubMed] [Google Scholar]
- 109.Pola-López L, Camas-Anzueto J, Martínez-Antonio A, et al. Novel arsenic biosensor “POLA” obtained by a genetically modified E. coli bioreporter cell. Sens Actuators B Chem. 2018;254:1061–1068. doi: 10.1016/j.snb.2017.08.006 [DOI] [Google Scholar]
- 110.Zhang L, Cheng X-Z, Kuang L, Xu A-Z, Liang R-P, Qiu J-D. Simple and highly selective detection of arsenite based on the assembly-induced fluorescence enhancement of DNA quantum dots. Biosens Bioelectron. 2017;94:701–706. doi: 10.1016/j.bios.2017.03.057 [DOI] [PubMed] [Google Scholar]
- 111.Cui L, Wu J, Ju H. Label-free signal-on aptasensor for sensitive electrochemical detection of arsenite. Biosens Bioelectron. 2016;79:861–865. doi: 10.1016/j.bios.2016.01.010 [DOI] [PubMed] [Google Scholar]
- 112.Divsar F, Habibzadeh K, Shariati S, Shahriarinour M. Aptamer conjugated silver nanoparticles for the colorimetric detection of arsenic ions using response surface methodology. Anal Methods. 2015;7(11):4568–4576. doi: 10.1039/C4AY02914C [DOI] [Google Scholar]
- 113.Ran G, Wu F, Ni X, et al. A novel label-free electrochemical aptasensor with one-step assembly process for rapid detection of lead (II) ions. Sens Actuators B Chem. 2020;320:128326. doi: 10.1016/j.snb.2020.128326 [DOI] [Google Scholar]
- 114.Chu LT, Leung HM, Lo PK, Chen T-H. Visual detection of lead ions based on nanoparticle-amplified magnetophoresis and Mie scattering. Sens Actuators B Chem. 2020;306:127564. doi: 10.1016/j.snb.2019.127564 [DOI] [Google Scholar]
- 115.Chen H, Shao S, Yu Y, et al. A dual-responsive biosensor for blood lead detection. Anal Chim Acta. 2020;1093:131–141. doi: 10.1016/j.aca.2019.09.062 [DOI] [PubMed] [Google Scholar]
- 116.Xiong C, Liang W, Wang H, et al. In situ electro-polymerization of nitrogen doped carbon dots and their application in an electrochemiluminescence biosensor for the detection of intracellular lead ions. Chem Commun. 2016;52(32):5589–5592. doi: 10.1039/C6CC01078D [DOI] [PubMed] [Google Scholar]
- 117.Wang Y, Zhao G, Zhang Q, et al. Electrochemical aptasensor based on gold modified graphene nanocomposite with different morphologies for ultrasensitive detection of Pb2+. Sens Actuators B Chem. 2019;288:325–331. doi: 10.1016/j.snb.2019.03.010 [DOI] [Google Scholar]
- 118.Zhang Q, Cui H, Xiong X, et al. QCM-nanomagnetic beads biosensor for lead ion detection. Analyst. 2018;143(2):549–554. doi: 10.1039/C7AN01498H [DOI] [PubMed] [Google Scholar]
- 119.Yadav R, Kushwah V, Gaur M, et al. Electrochemical aptamer biosensor for As3+ based on apta deep trapped Ag-Au alloy nanoparticles-impregnated glassy carbon electrode. Int J Environ Anal Chem. 2020;100(6):623–634. doi: 10.1080/03067319.2019.1638371 [DOI] [Google Scholar]
- 120.Nguyen DK, Jang C-H. Label-free liquid crystal-based detection of As (III) ions using ssDNA as a recognition probe. Microchem J. 2020;156:104834. doi: 10.1016/j.microc.2020.104834 [DOI] [PubMed] [Google Scholar]
- 121.Zhu X, Zhang S, Li W, et al. Label-free and immobilization-free electrochemiluminescent sensing platform for highly sensitive detection of As (III) by combining target-induced strand displacement amplification with polydopamine nanospheres. Sens Actuators B Chem. 2020;311:127818. doi: 10.1016/j.snb.2020.127818 [DOI] [Google Scholar]
- 122.Sangkaew W, Sallabhan R, Ritcharoon B, Mongkolsuk S, Loprasert S. FGE‐sulfatase‐based bacterial biosensor with single copy evolved sensing cassette for arsenic detection. J Chem Technol Biotechnol. 2020;95(4):1173–1179. [Google Scholar]
- 123.Vega-Figueroa K, Santillan J, Ortiz-Gomez V, et al. Aptamer-based impedimetric assay of arsenite in water: interfacial properties and performance. ACS Omega. 2018;3(2):1437–1444. doi: 10.1021/acsomega.7b01710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dieudonne A, Preveral S, Pignol D. A sensitive magnetic arsenite-specific biosensor hosted in magnetotactic bacteria. Appl Environ Microbiol. 2020;86(14). doi: 10.1128/AEM.00803-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tang G, Wang J, Li Y, Su X. Determination of arsenic (III) based on the fluorescence resonance energy transfer between CdTe QDs and Rhodamine 6G. RSC Adv. 2015;5(23):17519–17525. doi: 10.1039/C4RA16789A [DOI] [Google Scholar]
- 126.Taghdisi SM, Danesh NM, Ramezani M, Emrani AS, Abnous K. A simple and rapid fluorescent aptasensor for ultrasensitive detection of arsenic based on target-induced conformational change of complementary strand of aptamer and silica nanoparticles. Sens Actuators B Chem. 2018;256:472–478. doi: 10.1016/j.snb.2017.10.129 [DOI] [Google Scholar]
- 127.Zhang J, Zhang C-L, Yu S-H. Tuning Gold Nanoparticle Aggregation through the Inhibition of Acid Phosphatase Bioactivity: A Plasmonic Sensor for Light-Up Visual Detection of Arsenate (As^ sup V^). ChemPlusChem. 2016;81(11):1147. doi: 10.1002/cplu.201600355 [DOI] [PubMed] [Google Scholar]
- 128.Zhang L, Deng H, Yuan R, Yuan Y. Electrochemical lead (II) biosensor by using an ion-dependent split DNAzyme and a template-free DNA extension reaction for signal amplification. Microchimica Acta. 2019;186(11):709. doi: 10.1007/s00604-019-3857-z [DOI] [PubMed] [Google Scholar]
- 129.Xu W, Zhao A, Zuo F, Khan R, Hussain HMJ, Li J. A highly sensitive DNAzyme-based SERS biosensor for quantitative detection of lead ions in human serum. Anal Bioanal Chem. 2020;412(19):4565–4574. doi: 10.1007/s00216-020-02709-2 [DOI] [PubMed] [Google Scholar]
- 130.Wang F, Dai J, Shi H, et al. A rapid and colorimetric biosensor based on GR-5 DNAzyme and self-replicating catalyzed hairpin assembly for lead detection. Anal Methods. 2020;12(17):2215–2220. doi: 10.1039/D0AY00091D [DOI] [Google Scholar]
- 131.Li Y, Wang C, Zhu Y, et al. Fully integrated graphene electronic biosensor for label-free detection of lead (II) ion based on G-quadruplex structure-switching. Biosens Bioelectron. 2017;89:758–763. doi: 10.1016/j.bios.2016.10.061 [DOI] [PubMed] [Google Scholar]
- 132.Hui C-Y, Guo Y, Liu L, et al. Genetic control of violacein biosynthesis to enable a pigment-based whole-cell lead biosensor. RSC Adv. 2020;10(47):28106–28113. doi: 10.1039/D0RA04815A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.He Y, Hu X, Gong Z, Chen S, Yuan R. A novel electrochemiluminescence biosensor based on the self-ECL emission of conjugated polymer dots for lead ion detection. Microchimica Acta. 2020;187(4):1–8. doi: 10.1007/s00604-020-4212-0 [DOI] [PubMed] [Google Scholar]
- 134.Sun Q, Wang J, Tang M, et al. A new electrochemical system based on a flow-field shaped solid electrode and 3D-printed thin-layer flow cell: detection of Pb2+ ions by continuous flow accumulation square-wave anodic stripping voltammetry. Anal Chem. 2017;89(9):5024–5029. doi: 10.1021/acs.analchem.7b00383 [DOI] [PubMed] [Google Scholar]
- 135.Wang C, Cui X, Li Y, et al. A label-free and portable graphene FET aptasensor for children blood lead detection. Sci Rep. 2016;6(1):21711. doi: 10.1038/srep21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dali M, Zinoubi K, Chrouda A, Abderrahmane S, Cherrad S, Jaffrezic-Renault N. A biosensor based on fungal soil biomass for electrochemical detection of lead (II) and cadmium (II) by differential pulse anodic stripping voltammetry. J Electroanal Chem. 2018;813:9–19. doi: 10.1016/j.jelechem.2018.02.009 [DOI] [Google Scholar]
- 137.Zhang B, Wei C. Highly sensitive and selective detection of Pb2+ using a turn-on fluorescent aptamer DNA silver nanoclusters sensor. Talanta. 2018;182:125–130. doi: 10.1016/j.talanta.2018.01.061 [DOI] [PubMed] [Google Scholar]
- 138.Chen X, Zhai N, Snyder JH, et al. Colorimetric detection of Hg 2+ and Pb 2+ based on peroxidase-like activity of graphene oxide–gold nanohybrids. Anal Methods. 2015;7(5):1951–1957. doi: 10.1039/C4AY02801E [DOI] [Google Scholar]
- 139.Guo M, Wang J, Du R, et al. A test strip platform based on a whole-cell microbial biosensor for simultaneous on-site detection of total inorganic mercury pollutants in cosmetics without the need for predigestion. Biosens Bioelectron. 2020;150:111899. doi: 10.1016/j.bios.2019.111899 [DOI] [PubMed] [Google Scholar]
- 140.Elsebai B, Ghica ME, Abbas MN, Brett CM. Catalase based hydrogen peroxide biosensor for mercury determination by inhibition measurements. J Hazard Mater. 2017;340:344–350. doi: 10.1016/j.jhazmat.2017.07.021 [DOI] [PubMed] [Google Scholar]
- 141.Zhang Y, Xiao J-Y, Zhu Y, et al. Fluorescence Sensor Based on Biosynthetic CdSe/CdS Quantum Dots and Liposome Carrier Signal Amplification for Mercury Detection. Anal Chem. 2020;92(5):3990–3997. doi: 10.1021/acs.analchem.9b05508 [DOI] [PubMed] [Google Scholar]
- 142.Huang R-F, Liu H-X, Gai -Q-Q, Liu G-J WZ, Wei Z. A facile and sensitive electrochemiluminescence biosensor for Hg2+ analysis based on a dual-function oligonucleotide probe. Biosens Bioelectron. 2015;71:194–199. doi: 10.1016/j.bios.2015.04.038 [DOI] [PubMed] [Google Scholar]
- 143.Jin H, Zhang M, Wei M, Cheng J-H. A voltammetric biosensor for mercury (II) using reduced graphene oxide@ gold nanorods and thymine-Hg (II)-thymine interaction. Microchimica Acta. 2019;186(4):264. doi: 10.1007/s00604-019-3372-2 [DOI] [PubMed] [Google Scholar]
- 144.Yuan A, Wu X, Li X, Hao C, Xu C, Kuang H. Au@ gap@ AuAg Nanorod Side‐by‐Side Assemblies for Ultrasensitive SERS Detection of Mercury and its Transformation. Small. 2019;15(27):1901958. doi: 10.1002/smll.201901958 [DOI] [PubMed] [Google Scholar]
- 145.Fakude CT, Arotiba OA, Mabuba N. Electrochemical aptasensing of cadmium (II) on a carbon black-gold nano-platform. J Electroanal Chem. 2020;858:113796. doi: 10.1016/j.jelechem.2019.113796 [DOI] [Google Scholar]
- 146.Elcin E, Öktem HA. Inorganic Cadmium Detection Using a Fluorescent Whole-Cell Bacterial Bioreporter. Anal Lett. 2020;1–19. [Google Scholar]
- 147.Zhu Y-F, Wang Y-S, Zhou B, et al. A multifunctional fluorescent aptamer probe for highly sensitive and selective detection of cadmium (II). Anal Bioanal Chem. 2017;409(21):4951–4958. doi: 10.1007/s00216-017-0436-1 [DOI] [PubMed] [Google Scholar]
- 148.Ruhan Q, Yinyin L, Dawei C, et al. Determination of Cadmium Ions Based on Electrochemical DNA Biosensors in Rat Tissues. Int J Electrochem Sci. 2020;15:7347–7358. doi: 10.20964/2020.08.70 [DOI] [Google Scholar]
- 149.Rabai S, Benounis M, Catanante G, et al. Development of a label-free electrochemical aptasensor based on diazonium electrodeposition: application to cadmium detection in water. Anal Biochem. 2020;612:113956. doi: 10.1016/j.ab.2020.113956 [DOI] [PubMed] [Google Scholar]
- 150.Li X, Du Z, Lin S, Tian J, Tian H, Xu W. ExoIII and TdT dependent isothermal amplification (ETDA) colorimetric biosensor for ultra-sensitive detection of Hg2+. Food Chem. 2020;316:126303. doi: 10.1016/j.foodchem.2020.126303 [DOI] [PubMed] [Google Scholar]
- 151.Wang H, Liu Y, Liu G. Electrochemical biosensor using DNA embedded phosphorothioate modified RNA for mercury ion determination. ACS sensors. 2018;3(3):624–631. doi: 10.1021/acssensors.7b00892 [DOI] [PubMed] [Google Scholar]
- 152.Cheng L, Wei B, He LL, et al. “Off-On” switching electrochemiluminescence biosensor for mercury (II) detection based on molecular recognition technology. Anal Biochem. 2017;518:46–52. doi: 10.1016/j.ab.2016.09.018 [DOI] [PubMed] [Google Scholar]
- 153.Tang L, Xie X, Zhou Y, et al. A reusable electrochemical biosensor for highly sensitive detection of mercury ions with an anionic intercalator supported on ordered mesoporous carbon/self-doped polyaniline nanofibers platform. Biochem Eng J. 2017;117:7–14. doi: 10.1016/j.bej.2016.09.011 [DOI] [Google Scholar]
- 154.Tortolini C, Bollella P, Antonelli ML, Antiochia R, Mazzei F, Favero G. DNA-based biosensors for Hg2+ determination by polythymine–methylene blue modified electrodes. Biosens Bioelectron. 2015;67:524–531. doi: 10.1016/j.bios.2014.09.031 [DOI] [PubMed] [Google Scholar]
- 155.Cui X, Zhu L, Wu J, et al. A fluorescent biosensor based on carbon dots-labeled oligodeoxyribonucleotide and graphene oxide for mercury (II) detection. Biosens Bioelectron. 2015;63:506–512. doi: 10.1016/j.bios.2014.07.085 [DOI] [PubMed] [Google Scholar]
- 156.Zhang Y, Xie J, Liu Y, et al. Simple and signal-off electrochemical biosensor for mercury (II) based on thymine-mercury-thymine hybridization directly on graphene. Electrochim Acta. 2015;170:210–217. doi: 10.1016/j.electacta.2015.04.152 [DOI] [Google Scholar]
- 157.Guo Z, Kang Y, Liang S, Zhang J. Detection of Hg (II) in adsorption experiment by a lateral flow biosensor based on streptavidin-biotinylated DNA probes modified gold nanoparticles and smartphone reader. Environ Pollut. 2020;266:115389. doi: 10.1016/j.envpol.2020.115389 [DOI] [PubMed] [Google Scholar]
- 158.Guo Z, Chen B, Wang Z, Jiang X. An electrochemiluminescence biosensor for mercury ion detection based on gamma-polyglutamic acid-graphene-luminol composite and oligonucleotides. Sens Actuators B Chem. 2015;209:579–585. doi: 10.1016/j.snb.2014.12.028 [DOI] [Google Scholar]
- 159.Ma F, Chen Y, Zhu Y, Liu J. Electrogenerated chemiluminescence biosensor for detection of mercury (II) ion via target-triggered manipulation of DNA three-way junctions. Talanta. 2019;194:114–118. doi: 10.1016/j.talanta.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 160.Pi K, Liu J, Van Cappellen P. A DNA-based biosensor for aqueous Hg (II): performance under variable pH, temperature and competing ligand composition. J Hazard Mater. 2020;385:121572. doi: 10.1016/j.jhazmat.2019.121572 [DOI] [PubMed] [Google Scholar]
- 161.Ebrahimi M, Raoof JB, Ojani R. Design of an electrochemical DNA-based biosensor for selective determination of cadmium ions using a DNA hybridization indicator. Int J Biol Macromol. 2018;108:1237–1241. doi: 10.1016/j.ijbiomac.2017.11.023 [DOI] [PubMed] [Google Scholar]
- 162.Li Y, Ran G, Lu G, et al. Highly sensitive label-free electrochemical aptasensor based on screen-printed electrode for detection of cadmium (II) ions. J Electrochem Soc. 2019;166(6):B449. doi: 10.1149/2.0991906jes [DOI] [Google Scholar]
- 163.Wang H, Cheng H, Wang J, Xu L, Chen H, Pei R. Selection and characterization of DNA aptamers for the development of light-up biosensor to detect Cd (II). Talanta. 2016;154:498–503. doi: 10.1016/j.talanta.2016.04.005 [DOI] [PubMed] [Google Scholar]
- 164.Xue Y, Wang Y, Wang S, Yan M, Huang J, Label-Free YX. Regenerable Aptasensor for Real-Time Detection of Cadmium (II) by Dual Polarization Interferometry. Anal Chem. 2020;92(14):10007–10015. doi: 10.1021/acs.analchem.0c01710 [DOI] [PubMed] [Google Scholar]
- 165.Deng S, Jiang Q, Zhang T, Xiong X, Chen P. Liquid Crystal Biosensor Based on Cd 2+ Inducing the Bending of PS-Oligo for the Detection of Cadmium. Health. 2015;7(08):986. doi: 10.4236/health.2015.78116 [DOI] [Google Scholar]
- 166.Fourou H, Zazoua A, Braiek M, Jaffrezic-Renault N. An enzyme biosensor based on beta-galactosidase inhibition for electrochemical detection of cadmium (II) and chromium (VI). Int J Environ Anal Chem. 2016;96(9):872–885. [Google Scholar]
- 167.Qu J, Wu L, Liu H, et al. A novel electrochemical biosensor based on DNA for rapid and selective detection of cadmium. Int J Electrochem Sci. 2015;10:4020–4028. [Google Scholar]
- 168.Bereza-Malcolm L, Aracic S, Kannan R, Mann G, Franks AE. Functional characterization of gram-negative bacteria from different genera as multiplex cadmium biosensors. Biosens Bioelectron. 2017;94:380–387. doi: 10.1016/j.bios.2017.03.029 [DOI] [PubMed] [Google Scholar]
- 169.Zeng L, Gong J, Rong P, Liu C, Chen J. A portable and quantitative biosensor for cadmium detection using glucometer as the point-of-use device. Talanta. 2019;198:412–416. doi: 10.1016/j.talanta.2019.02.045 [DOI] [PubMed] [Google Scholar]
- 170.Tao Z, Wei L, Wu S, Duan N, Li X, Wang Z. A colorimetric aptamer-based method for detection of cadmium using the enhanced peroxidase-like activity of Au–MoS2 nanocomposites. Anal Biochem. 2020;608:113844. doi: 10.1016/j.ab.2020.113844 [DOI] [PubMed] [Google Scholar]
- 171.Li Y, Chen Y, Yu H, Tian L, Wang Z. Portable and smart devices for monitoring heavy metal ions integrated with nanomaterials. TrAC Trends Anal Chem. 2018;98:190–200. doi: 10.1016/j.trac.2017.11.011 [DOI] [Google Scholar]
- 172.Chakraborty U, Bhanjana G, Adam J, et al. A flower-like ZnO–Ag 2 O nanocomposite for label and mediator free direct sensing of dinitrotoluene. RSC Adv. 2020;10(46):27764–27774. doi: 10.1039/D0RA02826F [DOI] [PMC free article] [PubMed] [Google Scholar]