Abstract
This study introduces an innovative methodological approach to identify potential drivers of structuring HIV-1 transmission clustering patterns between different subpopulations in the culturally and racially/ethnically diverse context of Houston, TX, the largest city in the Southern United States. Using 6332 HIV-1 pol sequences from persons newly diagnosed with HIV during the period 2010–2018, we reconstructed HIV-1 transmission clusters, using the HIV-TRAnsmission Cluster Engine (HIV-TRACE); inferred demographic and risk parameters on HIV-1 transmission dynamics by jointly estimating viral transmission rates across racial/ethnic, age, and transmission risk groups; and modeled the degree of network connectivity by using generalized estimating equations (GEE). Our results indicate that Hispanics/Latinos are most vulnerable to the structure of transmission clusters and serve as a bridge population, acting as recipients of transmissions from Whites (3.0 state changes/year) and from Blacks (2.6 state changes/year) as well as sources of transmissions to Whites (1.8 state changes/year) and to Blacks (1.2 state changes/year). There were high rates of transmission and high network connectivity between younger and older Hispanics/Latinos as well as between younger and older Blacks. Prevention and intervention efforts are needed for transmission clusters that involve younger racial/ethnic minorities, in particular Hispanic/Latino youth, to reduce onward transmission of HIV in Houston.
Subject terms: Bioinformatics, HIV infections, Population genetics, Risk factors, Statistics
Introduction
Although the rate of diagnosis of human immunodeficiency virus (HIV) in the United States (U.S.) decreased between 2011 and 2015, certain subpopulations bear an excess burden of new HIV diagnoses1. The highest HIV diagnosis rates were reported primarily for the subgroups of younger age, racial/ethnic minorities, men who have sex with men (MSM), and those who reside in the Southern region of the U.S.1. HIV-1 phylogenetic analysis has been increasingly used as a powerful methodological tool to reconstruct the HIV-1 transmission network by inferring putative HIV-1 transmission clusters formed by highly similar HIV strains. A number of HIV-1 molecular epidemiological studies have identified transmission clusters by assessing the viral genetic distance between these individual sequences2–7. Small genetic distances imply that individuals may have been infected from a common source in an overlapping time period. The shared common ancestry of a phylogenetic transmission cluster (or network) implies that members in that cluster share epidemiological characteristics of transmission. When combined with individuals’ demographic and transmission risk attributes, putative transmission clusters formed by highly similar HIV strains enable us to make inferences about clusters of epidemiologically connected individuals.
A number of HIV-1 molecular epidemiological studies have identified subpopulations in transmission networks that were characterized in terms of cluster membership, cluster size, and the degree of connectivity within a cluster. For instance, younger individuals, males, MSM, and heterosexuals tend to be members of a cluster6,8–10, while Black/African American (hereafter Black) individuals tend to cluster at lower frequencies than do White and Hispanic/Latino (hereafter Hispanic) individuals across the U.S.5,11–13. Within a transmission cluster, the degree of connectivity to other persons who are living with HIV increases for younger individuals, men, Blacks, and MSM4. Across three major U.S. metropolitan areas, Hispanic individuals cluster at high frequencies, whereas Black individuals cluster at the lowest frequencies and are more assortative than other race/ethnicity groups11.
Previous studies have identified and characterized emerging local transmission clusters and associated them with certain epidemiological profiles of co-clustered individuals in HIV-1 transmission networks. Little is known, however, about the drivers of HIV-1 transmission patterns between different vulnerable subpopulations who may fuel local HIV-1 sub-epidemics. As HIV-1 phylogenetic analysis is limited in its capability to establish directionality of transmission between individual members within these clusters14, it can provide only limited information in the absence of additional epidemiological data. As a consequence, there is scant knowledge of the epidemiological profiles of subpopulations who may, in fact, play a central role in the structuring of transmission clusters by acting as a recipient or source of HIV-1 infection.
In the present study, our primary goal was to identify potential drivers of structuring HIV-1 transmission clustering patterns between different subpopulations who act as sources and recipients of HIV-1 infection in the culturally and racially/ethnically diverse context of Houston/Harris County, TX. Our study took a hybrid heuristic and Bayesian approach15 and introduced an innovative methodology by synthesizing the HIV-TRAnsmission Cluster Engine (HIV-TRACE), Bayesian phylodynamics, and generalized estimating equations16 (GEE). Our approach enabled us to reconstruct HIV-1 transmission clusters; infer demographic and risk parameters of HIV-1 transmission dynamics by jointly estimating viral transmission rates across racial/ethnic, age, and transmission risk groups; and assess the effects of network mixing patterns based on racial/ethnic and age characteristics of the index person and partners on the degree of network connectivity. Our study is expected to inform the epidemiological profile of subpopulations with a disproportionate vulnerability and whose membership in clusters could be prioritized for urgent intervention efforts to eliminate the HIV epidemic in Houston.
Methods
Study setting and data
HIV molecular sequence data were reported from drug-resistance genotyping for people newly diagnosed with HIV while residing in Houston/Harris County or currently (as of January 1, 2019) residing in Houston/Harris County. Data were collected by the Houston Health Department as part of the routine HIV Surveillance Program. These samples were then stored in the Enhanced HIV/AIDS Reporting System. A total of 32,739 individuals older than 13 years and with an adult transmission risk were diagnosed in or currently residing in Houston/Harris County and entered into the surveillance system on or before December 31, 2018. Reporting of HIV genotypic testing has been required by law (Tex. Adm. Code Chapter 97, Subchapter F, §97.133) since January 1, 2010. Therefore, due to incompleteness of data prior to 2010, only individuals diagnosed in 2010 or later were included for analysis (N = 12,344).
The selection criteria for genotypes were: (1) had a reverse transcriptase (RT) or protease (PR)/RT HIV-1 pol sequence that was not ≤ 0.015 substitutions/site from the HXB2 reference sequence, and (2) had the month and year of sample date available. Further, PR and RT genotype were combined if they were reported at the same time but as separate sequences, and only the first PR/RT or RT genotype was used for constructing the network. Among 12,344 persons who were living with HIV diagnosed in or after 2010, 6516 (52.8%) had at least one HIV-1 sequence. Of the 6516 people with a sequence, 6332 had HIV-1 pol sequences that were used in the analysis.
This study was deemed to be exempt from institutional review by the Committee for the Protection of Human Subjects at the University of Texas Health Science Center at Houston because it was a retrospective analysis of surveillance data for the purposes of program evaluation. The data were analyzed in accordance with a Memorandum of Understanding for data sharing between the Houston Health Department and each institution. All methods were carried out in accordance with relevant guidelines and regulations.
The data that support the findings of this study are available from the corresponding author upon reasonable request. The data are not publicly available due to concerns regarding participant confidentiality.
Measures
We used the following study variables: age (Younger coded as individuals born after 1990, Older coded as individuals born in or before 1990); sex assigned at birth (Male, Female); race/ethnicity (Hispanics refers to Hispanics/Latinos of all races, Blacks refers to non-Hispanic Blacks, Whites refers to non-Hispanic Whites, Asians refers to non-Hispanic Asians, Others refers to non-Hispanic other racial categories, including American Indians and Alaskan Natives, Multi-race refers to non-Hispanic multi-race categories); HIV transmission risk (MSM refers to cisgender men who reported MSM, Cisgender men refers to other cisgender men who did not report injection drug use (IDU) or MSM, Cisgender women refers to cisgender women who did not report IDU, Transgender women refers to transgender women who did not report IDU, and PWID (people who inject drugs) refers to anyone who reported injection drug use, including MSM); first viral-load test result after HIV diagnosis (< 10,000, 10,000–100,000, > 100,000 copies/ml); and first CD4+ T-cell count test result after HIV diagnosis (< 50, 50–200, 201–350, > 350 cells/mm3). We defined age groups using a birth-year cutoff rather than age cohorts, as our data came from participants who were diagnosed over a range of years and aged through the life of our clusters.
Phylogenetic reconstruction and analysis
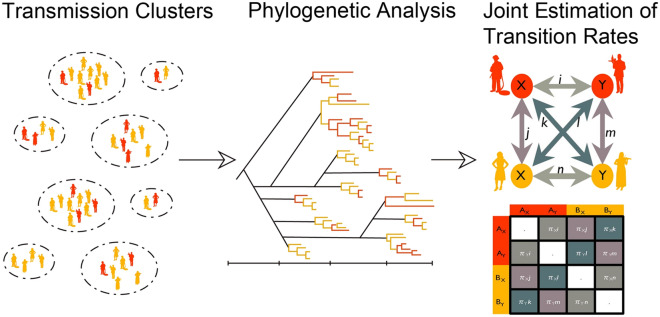
Testing hypotheses to identify underlying characteristics of HIV transmission clusters is challenging because most clusters contain few individuals, and, therefore, unbiased statistical inference is often not possible. To overcome this, we used a joint estimation procedure, whereby we investigated the relationships between transmission cluster characteristics and three epidemiological characteristics of race/ethnicity, age, and transmission risk to identify how these epidemiological characteristics may structure transmission clusters within Houston/Harris County. Our phylogenetic analysis design followed a three-step process: (1) identification of transmission clusters, (2) estimation of the time of the most recent common ancestor for each cluster, and (3) joint estimation of a single viral transition matrix for all clusters. For Steps 2 and 3, our phylogenetic analysis was based on the use of clusters with five or more members and exclusion of homogeneous clusters for computational tractability and the ability to infer rates that were meaningful and realistic across all identified clusters. Figure 1 provides a visual summary of our analytical design.
Figure 1.
Phylogenetic analytical design. The three-step process of estimating viral transmission between epidemiological characteristics that structure HIV transmission clusters within Houston/Harris County is summarized. In the first step, transmission clusters were identified. In the second step, a phylogenetic analysis was conducted to estimate the time of the most recent common ancestor for each cluster. In the third step, a joint estimation of transition rates of a single viral transition matrix for all clusters was conducted.
In Step 1, we constructed a genetic transmission network, using HIV-TRACE17,18. All sequences were aligned to an HXB2 reference sequence (positions 2253–3749). To identify genetically linked pairs, we used a pairwise genetic distances threshold of 0.015 substitutions/site and an ambiguity fraction of 1.5%.
In Step 2, we estimated a preliminary maximum likelihood phylogenetic tree, using a general time-reversible model with gamma distributed rate variation among sites (GTR Gamma), using RAxML v8.2.419. The “clocklikeness” of the tree was determined using TempEst v.1.5.3 with a root-to-tip linear regression20. Then, we conducted statistical phylodynamic analysis using BEAST v 1.10.121. All sequences were aligned to generate a rooted, time-measured phylogeny, inferred using a relaxed molecular clock model to estimate the time of the most recent common ancestor of each cluster. We used the GTR 4 substitution model that accounts for the proportion of invariant sites that does not undergo evolutionary change.
In Step 3, to assess the importance of epidemiological parameters in HIV transmission, we performed an ancestral state reconstruction22 of racial/ethnic and transmission risk characteristics using Bayesian Stochastic Search Variable Selection (BSSVS). For phylodynamic analysis, we used clusters constructed using a more conservative genetic distance threshold of 0.005 substitutions/site. This lower threshold identifies more recent potential transmission partners23 with fewer intermediate partners. Intermediate partners can confound estimates of frequencies of transmission among risk groups24. We estimated a single discrete state transition rate matrix jointly across all clusters independently instead of one matrix for a tree that contains all clusters under the assumption that underlying characteristics are shared by all clusters in Houston/Harris County25.
We jointly estimated a single phylodynamic discrete trait model to all clusters for each trait of race/ethnicity and transmission risk. We used this joint estimation approach because epidemiological characteristics of a population can determine the structure of outbreaks. Individual clusters, however, are linked on a phylogenetic tree by long branches. These branches represent unobserved members of an outbreak, viral migration, or a biological change that triggered the outbreak. Phylodynamic approaches to hypothesis testing, including ancestral state reconstruction of tip-associated discrete traits, assume that the rate of change is constant throughout the branches of the phylogenetic tree. Although this assumption is reasonable for individual clusters, it is unlikely to be valid for the entire tree. Therefore, a jointly fitted single discrete model should capture the signal of the population’s social structure that may determine patterns of HIV transmission.
The tip-associated traits that we analyzed included race/ethnicity, race/ethnicity plus year of birth (born in or before 1990 and after 1990), and transmission risk. We used an asymmetric continuous-time Markov chain model26 for discrete state reconstructions and performed three independent Markov chain Monte Carlo runs with a chain length of 10 million states, logging every 1000 for each model. We applied ancestral state reconstruction of a racial/ethnicity discrete state model to an empirical set of 1500 phylogenetic trees for each cluster.
Given the large number of states, BSSVS was employed to search parameter space and identify those parameters with significantly non-zero transition rates22,27. BSSVS explores and efficiently reduces the state space by employing a binary indicator (I)22,27. From the BSSVS results, we applied a Bayes Factor (BF) test to assess the support for individual transitions between discrete states. We removed a burn-in of 10% (100,000) and computed BF values using SpreaD327. We used the following scheme to assess the level of support: No support: BF < 3; substantial support: BF = 3–10; strong support: BF = 11–30; very strong support: BF = 31–100; decisive support: BF > 100. More details for Steps 2 and 3 are provided in the online supplemental materials.
Population averaged negative binomial models for connectivity
We conducted a supplementary statistical analysis to account for homogeneous clusters with regard to epidemiological characteristics in our phylogenetic analysis, as well as to control for clinical factors. We estimated GEE for our data clustered on identified phylogenetic clusters with five or more members. We conducted negative binomial regression analysis to model the connectivity level (i.e., network degree) as a function of the number of phylogenetically connected partners with matched or mismatched attributes of race/ethnicity (Blacks, Hispanics, Whites, and Asians/Others) and age (Younger and Older) with the index person. In this analysis, we created eight assortative mixing terms by computing the number of partners who are matched with an index on these subcategories as well as 42 disassortative mixing terms by computing the number of partners who are mismatched. Our model controlled for clinical factors (log-transformed viral load values, CD4+ T-cell count level), diagnosis year (2010–2018), and other sociodemographic and risk variables. We used an exchangeable correlation structure with empirical variance estimates to address potential mis-specifications on the correlation structure. We used the xtnbreg command implemented in Stata v16.0 (College Station, TX).
Results
Descriptive statistics
A majority of the 6332 individuals in our sample were older (born in or before 1990) (81%), males (80%), of a racial/ethnic minority (47% Blacks and 36% Hispanics), and MSM (54%). In addition, 42% had an initial viral load after HIV diagnosis greater than 10,000 copies/ml, and 88% had a CD4+ T-cell count of > 200. The detailed descriptive statistics for sample characteristics are presented in Supplemental Table 1S in the supplemental materials.
Phylogenetic analysis
Cluster identification
We identified 2675 (42%) samples with a cluster size of two or more out of 6332 samples, with a mean cluster size of 12 (SD = 14.2, min = 2, max = 58). Among these 2675 samples, 1529 (57%) belonged to 143 unique clusters, with five or more members with a mean cluster size of 18.5 (SE = 15.7, min = 5, max = 58). The categories of younger individuals (38%), males (27%), Whites (27%), MSM (31%), viral load of 10,000–100,000 (26%), and CD4+ T-cell count of > 350 (28%) showed a higher percentage of membership within a cluster that comprised ≥ 5 members (Supplemental Table 1S). An HIV-1 putative transmission network by racial/ethnic groups and risk categories is presented in Fig. 2.
Figure 2.
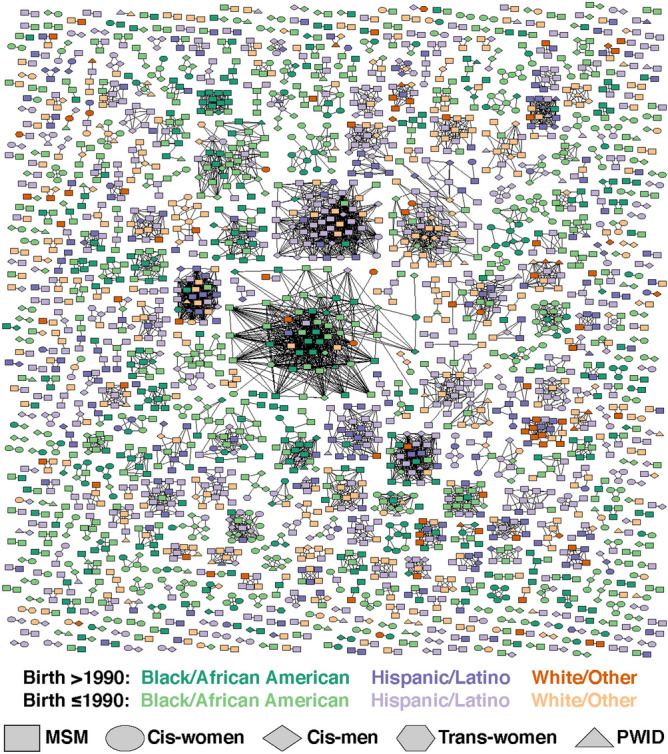
Visualization of HIV-1 transmission network in Houston/Harris County (2010–2018). MSM refers to cisgender men who report being MSM, cis-women refers to cisgender women who did not report injection drug use, trans-women refers to transgender women who did not report injection drug use, cis-men refers to cisgender men who did not report injection drug use or MSM, and PWID refers to anyone who reported injection drug use, including MSM. Hispanic/Latino refers to Hispanic/Latino of all races; Black/African American refers to non-Hispanic Black/African American; and White/Other refers to non-Hispanic Whites, non-Hispanic Asians, and non-Hispanic other racial categories, including American Indians, Alaskan Natives, and multi-race categories.
There were three large clusters, two of which were composed mainly of heterosexual males with minority racial/ethnic groups.
Comparative phylogenetic analysis
The meta-data of epidemiological profile variables associated with each cluster allowed us to estimate the transmission network that connects these different racial/ethnic groups. Out of 143 identified clusters with five or more members who were identified by our TRACE analysis, we identified 34 racially/ethnically non-homogeneous transmission clusters and 27 non-homogeneous clusters with respect to transmission risk categories, using a more conservative genetic distance threshold of 0.005 substitutions/site. It is important to note that, even though the majority of transmission is within racial/ethnic groups, the contact rates between groups is an important characteristic that can determine viral spread. We therefore excluded these homogeneous clusters from our analysis.
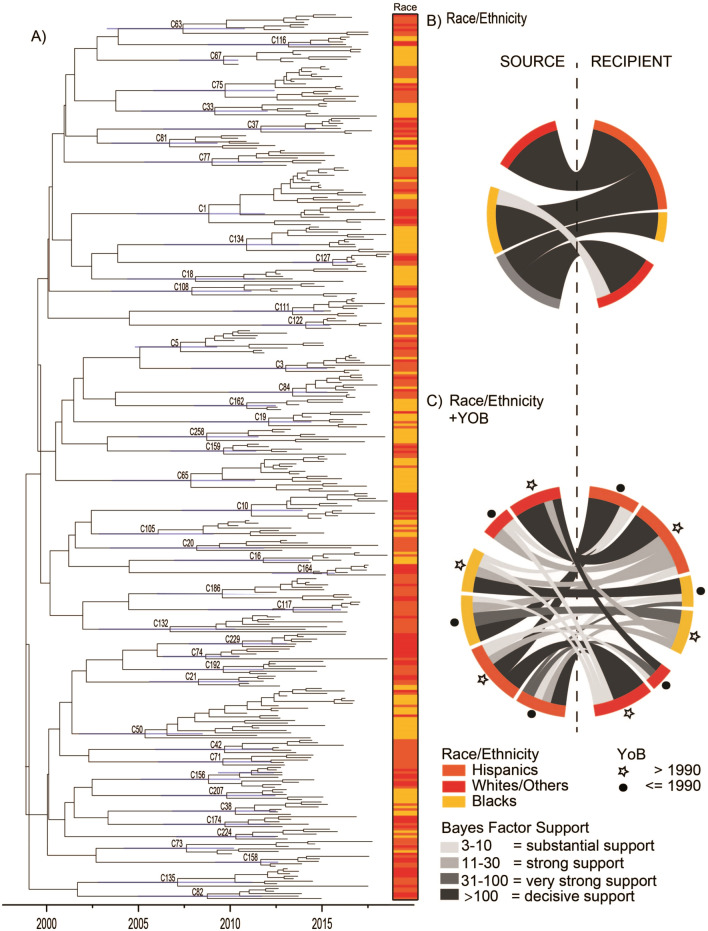
Members of these clusters were diagnosed between 2010 through the end of 2018. Phylogenetic analysis of the combined groups was conducted to determine the time of the most recent common ancestor for each cluster (Fig. 3A). Clusters circulated for 3.45 to 13.56 years, with larger clusters as circulating for longer periods. We estimated transmission between racial/ethnic groups, using an ancestral state reconstruction method. We assume that the underlying demographic characteristics are shared by all clusters, and, therefore, we jointly estimated a single transmission network shared among all clusters analyzed.
Figure 3.
(A) BEAST phylogeny of all clusters; (B) chord diagram for viral transmission between racial/ethnic groups (Hispanics, Blacks, Whites/Others); (C) chord diagram for viral transmission between racial/ethnic groups plus age group (born in or before 1990, born after 1990). (A) Shows a maximum clade credibility phylogenetic tree with all clusters that contain five or more cluster members. Tips are labeled with the race/ethnicity associated with the taxa at that tip. In (B) and (C), chord thickness indicates the magnitude of the transition rate, and color indicates Bayes Factor support. Only supported transition rates from discrete trait analysis are displayed. YoB Year of Birth.
A single discrete race/ethnicity model
Figure 3B shows a chord diagram that indicates viral transmission between different groups of races/ethnicities. We used an asymmetric model to allow for the directionality of transmission to be estimated. We observed decisively significant transitions (BF > 100) from Whites to Hispanics (2.99 state changes/year), from Blacks to Hispanics (2.55 state changes/year), and from Hispanics to Whites (1.84 state changes/year) (Fig. 3B, Supplemental Tables 3S). Further, Hispanics had decisively supported transitions (BF > 100) to Asians (0.50 state changes/year) and to Blacks (1.17 state changes/year) (Supplemental Table 3S). These results suggest that Hispanics are the most vulnerable population in structuring transmission clusters, acting as a recipient of viral transmission as well as a source. This result was further supported by our supplemental analysis of grouping Whites, Asians, and Multi-race categories into a single category in a single discrete race/ethnicity condensed model (Supplemental Table 4S).
A single discrete race/ethnicity plus age model
When the categories were broken down by age groups, we observed decisively supported (BF > 100) transitions from older (born in or before 1990) Hispanics and older Whites/Others (born in or before 1990) (Fig. 3C, Supplemental Table 5S). There was decisive support for both directions of transmission, with the actual rate as higher from older Whites/Others to older Hispanics than from older Hispanics to older Whites/Others (3.02 and 1.89, respectively). Decisively supported transition rates within a racial/ethnic group from older (born in or before 1990) to younger (born after 1990) groups were observed in Hispanic and Black racial/ethnic groups. The rate of transition from older Blacks to younger Blacks was 1.56. Other decisively supported transition rates were observed from younger Blacks to younger Hispanics (1.55 state transitions/year), younger Hispanics to older Whites/Others (1.50 state transitions/year), and older Whites/Others to younger Whites/Others (0.84 state transitions/year). More detailed findings on actual transition rates and corresponding BFs for a single discrete race/ethnicity model (Supplemental Table 3S), a single discrete race/ethnicity condensed model (Supplemental Table 4S), and a single discrete race/ethnicity plus age model (Supplemental Table 5S) are provided in the online supplemental materials.
A single discrete transmission risk model
We observed decisively significant (BF > 100) transitions from MSM to PWID (1.02 state transitions/year) and MSM to cisgender men (1.82 state transitions/year) (Supplemental Figure 1S and Supplemental Table 6S).
Generalized estimating equation (GEE) analysis
Table 1 presents a summary of estimated population averaged incident rate ratios (IRRs) for the assortative mixing (on-diagonals) and disassortative mixing (off-diagonals) terms based on racial-ethnic and age characteristics of the index person (rows) and partners (columns).
Table 1.
Summary of estimates of incident rate ratios, using the population averaged negative binomial model to predict network degree (connectivity level) for cluster size ≥ 5 (N = 1517 with number of clusters = 143), 2010–2018: phylogenetic links based on assortative mixing (diagonals) and disassortative mixing (off-diagonals).
| Population | Number of partners | |||||||
|---|---|---|---|---|---|---|---|---|
| Younger Blacks | Older Blacks | Younger Hispanics | Older Hispanics | Younger Whites | Older Whites | Younger Asians | Older Asians | |
| Younger Blacks | 1.14*** | 1.12*** | 1.14*** | 1.15*** | 1.02 | 1.14*** | 1.12 | 0.82 |
| Older Blacks | 1.11*** | 1.14*** | 1.18*** | 1.14*** | 0.93 | 1.14*** | 1.04 | 1.03 |
| Younger Hispanics | 1.11*** | 1.15*** | 1.15*** | 1.12*** | 1.20*** | 1.11*** | 1.14† | 1.12 |
| Older Hispanics (Index) | 1.06† | 1.15*** | 1.14*** | 1.11*** | 1.20*** | 1.14*** | 1.15 | 1.16** |
| Younger Whites | 1.01 | 1.30*** | 1.19*** | 1.21*** | 1.19*** | 1.19*** | 1.44*** | 1.13 |
| Older Whites | 1.09 | 1.11*** | 1.15*** | 1.10*** | 1.17*** | 1.18*** | 1.13 | 1.08 |
| Younger Asians | 1.04 | 1.16*** | 1.15*** | 1.11† | 1.23* | 1.17*** | 1.37 | 1.21 |
| Older Asians | 1.04 | 1.16*** | 1.15*** | 1.08*** | 1.19*** | 1.20*** | 1.45** | 0.77† |
Model controls for all study variables listed in Supplemental Table 1S and year of diagnosis. Complete estimated results are provided in Supplemental Table 7S in the online supplemental materials. Asians includes other racial categories of American Indians or Alaskan Natives and Multi-race. Younger is defined as born after 1990, and Older is defined as born in or before 1990. Exchangeable correlation structure and robust variance estimates were used.
†p < 0.1, *p < 0.05, **p < 0.01, ***p < 0.001.
A majority of disassortative mixing terms had significant positive associations with the connectivity level at p < 0.05. In relation to the earlier phylogenetic findings, all of the decisively supported patterns of transitions that we observed in the single discrete race/ethnicity plus age model also were statistically significant (p < 0.001) in their corresponding effects of the index-alter disassortative mixing patterns. The complete statistical analysis results are presented in Supplemental Table 7S in the supplemental materials.
Discussion
HIV-1 phylogenetic clusters in Houston/Harris County are characterized by subpopulations who are members of racial/ethnic minority groups. In particular, Hispanic and Latino populations are particularly vulnerable to HIV viral transmission dynamics, increasing the HIV infection disparities due to structural drivers (e.g., HIV stigma in Hispanic/Latino communities, treatment accessibility/availability) that underlie the increase in new HIV infections among these populations28. Our study indicates that Hispanic/Latino individuals often serve as the source and recipient of HIV-1 transmission across racial/ethnic groups in Houston. These viral transmission patterns are further characterized by younger Hispanic individuals’ being clustered with older Hispanic individuals at a high connectivity level. A similar pattern was observed among Black individuals. These findings are supported by a recent study based on the U.S. National HIV Surveillance System that reports the most substantial increases in linkage of older MSM to young MSM for Hispanic/Latinos, followed by Blacks, during 2009–201629.
Future research would benefit from an exploration of the effects of changes in the transmission clustering patterns in contexts where services differ in availability to these populations. Access to services, such as HIV testing, HIV medical care, and pre-exposure prophylaxis (PrEP) are of particular importance. Increased HIV testing can facilitate a linkage to HIV medical care for those who are positive and a linkage to PrEP for those who are negative, both of which can blunt the spread of HIV. Viral suppression through adherence to HIV treatment can effectively decrease onward sexual transmission to zero—often referred to as “treatment as prevention”30. PrEP is a highly effective pill that is associated with a decreased risk of acquiring HIV for populations at increased HIV infection risk, although the effectiveness level depends on the level of adherence31. A recent study demonstrated that increases in PrEP coverage, regardless of changes in viral suppression, are significantly associated with decreases in HIV diagnosis rates32, a finding that substantiates the need to increase service availability among those most vulnerable.
HIV-1 transmission cluster patterns that involve racial/ethnic minority groups are complex and multifaceted in terms of cluster membership, size, and connectivity within clusters, as they are inextricably associated with other demographic, transmission risk, and social/cultural factors. For instance, younger Black and Hispanic MSM tend to have other transmission partners similar in age33. Of note, the Hispanic and Latino culture in the U.S. is diverse, and regional differences exist in the predominant mode of transmission and associated HIV transmission risks34. Cluster membership and size also are further complicated by the way that other factors, such as immigration status and country of origin, interact in the transmission cluster35,36. For example, among Hispanic immigrants, cluster membership is associated with younger ages, more acculturation to the U.S., and being of Mexican origin35. Further, larger clusters that involve Hispanic members tend to contain fewer immigrants and more MSM, and Hispanic immigrants tend to be in clusters with other Hispanics35, indicating their having putative U.S.-born transmission partners36.
Given the racial/ethnic and cultural diversity in the Houston area, our study findings provide a unique perspective to investigating the HIV-1 clustering patterns in local HIV-1 transmission networks. Houston, in addition to being the largest city in the Southern U.S.37, is one of the fastest growing cities and has one of the youngest populations in the U.S., due mainly to the influx of immigrants38. In Harris County (which encompasses much of Houston), Hispanics are the plurality and, when combined with Black and other minority racial/ethnic groups, comprise 68.7% of the total population39.
Our study findings are constrained by some limitations. Our sample came from the first sequence test available for each person, which may exclude those who remained virally unsuppressed, which limits our ability to identify more of the clustered individuals when using multiple sequences per person in identifying clusters40. In addition, our sample accounts only for individuals who have been diagnosed and have had a sequence of good quality reported to the health department. Our study may have been limited in reconstructing closely related viral transmission clusters due to potential missing sequences from an intermediary or a common source of a pairwise, similarly-strained virus. A more complete reconstruction of the HIV-1 transmission networks could be realized by combining an epidemiological linkage via contact tracing of risk-sharing partners41, social network members42, or online or physical venue affiliation2,43. Future studies would likely benefit from combining molecular, epidemiological, and social network data to identify growing clusters and by considering the geographic proximity that may be associated with clustering patterns6,9,44. Further, HIV is known to be disproportionately transmitted by recently infected individuals45 and transmitted at an acute stage of HIV infection9,46. HIV-1 clustering has been reported to be associated with clinical factors, such as higher HIV viral load8,44,46, lack of antiretroviral use44, and higher CD4+ T-cell counts10,44. Future phylodynamic research also could consider these clinical factors in defining the epidemiological profile associated with the dynamics of HIV-1 transmission patterns. Finally, our statistical approach has limitations in dealing with potential autocorrelation of the error term and addressing network dependency of phylogenetic links. Future research could use exponential random graph models to estimate assortative mixing patterns with controls of the dependent nature of the HIV-1 transmission network by partitioning it into distinct neighborhoods47.
Despite these limitations, our study provides a unique opportunity to chart the dynamics of HIV-1 transmission between different groups by race/ethnicity, age, and transmission risk in the racially/ethnically diverse context of Houston, TX. The intensified transmission rates early in HIV infections have been attributed to the growth of larger clusters48,49. Our findings are consistent with the recommendation of the routine use of HIV sequence data to interrupt HIV transmission through prevention interventions, with the aim of preventing the onward transmission of HIV-1 among these vulnerable younger racial/ethnic minorities, in particular, younger Hispanic subpopulations.
Supplementary Information
Acknowledgements
We acknowledge R. Lubelchek, J. Xu, M. Imahashi, W. Zhou, N.-W. Sheung, M. Matsuda, A. Hochi, and individuals from the following areas of the Houston Health Department: Informatics Program, HIV Surveillance Program, and the Bureau of HIV/STD and Viral Hepatitis Prevention.
Author contributions
K.F., M.M., C.H., and L.Y.H. initiated the project. K.F. framed, designed, and analyzed statistical models and co-led the writing of the article with J.B. J.B. designed and conducted the phylogenetic analysis, reported results, and co-led the writing of the article. J.O.W. designed and conducted the phylogenetic analysis and reported results. N.D.V., R.M., and M.C. prepared and managed sequence data. N.D.V. computed measures, conducted preliminary statistical analysis, and reported results. J.T.H. and L.D. analyzed sequence data, conducted phylogenetic analysis, and reported results. M.M., C.H., and B.Y. designed the data collection, collected and managed the data, and interpreted the results. M.M., C.H., B.Y., J.O.W., J.A.S., R.L., and L.Y.H. assisted in interpretation and contributed to the implications of the results. All authors substantially contributed to the study, engaged in writing, and approved the final version.
Funding
This work was supported by the Centers for Disease Control and Prevention (CDC) (Grants 5 NU62PS924515-03-00 and 1 NU62PS924572-01-00 to M.M.) and the National Institutes of Health (NIH) (Grants 1R21GM113694 to K.F., 1R01MH100021 to K.F. and J.A.S., R01AI136056 to J.A.S., K01AI110181 to J.O.W., R01AI135992 to J.O.W., and 1R56AI150272-01A1 to K.F and J.A.S.).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kayo Fujimoto and Justin Bahl.
Change history
5/10/2021
A Correction to this paper has been published: 10.1038/s41598-021-89879-w
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-82673-8.
References
- 1.Centers for Disease Control and Prevention. HIV surveillance report: Diagnoses of HIV infection in the United States and dependent areas. Published November 2017. http://www.cdc.gov/hiv/library/reports/hiv-surveillance.html. Accessed October 4, 2020.
- 2.Chan PA, et al. Phylogenetic investigation of a statewide HIV-1 epidemic reveals ongoing and active transmission networks among men who have sex with men. J. Acquir. Immune Defic. Syndr. 2015;70(4):428–435. doi: 10.1097/QAI.0000000000000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassan AS, Pybus OG, Sanders EJ, Albert J, Esbjörnsson J. Defining HIV-1 transmission clusters based on sequence data. AIDS. 2017;31(9):1211–1222. doi: 10.1097/QAD.0000000000001470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan E, et al. HIV-1 Infection and transmission networks of younger people in Chicago, Illinois, 2005–2011. Public Health Rep. 2017;132(1):48–55. doi: 10.1177/0033354916679988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oster AM, et al. Using molecular HIV surveillance data to understand transmission between subpopulations in the United States. J. Acquir. Immune Defic. Syndr. 2015;70(4):444–451. doi: 10.1097/QAI.0000000000000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez-Losada M, et al. Characterization of HIV diversity, phylodynamics and drug resistance in Washington, DC. PLoS ONE. 2017 doi: 10.1371/journal.pone.0185644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wertheim JO, et al. Social and genetic networks of HIV-1 transmission in New York City. PLoS Pathog. 2017 doi: 10.1371/journal.ppat.1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lubelchek RJ, et al. Transmission clustering among newly diagnosed HIV patients in Chicago, 2008 to 2011: Using phylogenetics to expand knowledge of regional HIV transmission patterns. J. Acquir. Immune Defic. Syndr. 2015;68(1):46–54. doi: 10.1097/QAI.0000000000000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dennis AM, et al. Phylogenetic insights into regional HIV transmission. AIDS. 2012;26(14):1813–1822. doi: 10.1097/QAD.0b013e3283573244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf E, et al. Phylogenetic evidence of HIV-1 transmission between adult and adolescent men who have sex with men. AIDS Res. Hum. Retroviruses. 2017;33(4):318–322. doi: 10.1089/aid.2016.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ragonnet-Cronin, M. et al. Sorting by race/ethnicity across HIV genetic transmission networks in three major metropolitan areas in the United States. AIDS Res. Hum. Retroviruses (2020). [DOI] [PMC free article] [PubMed]
- 12.Balaban M, Moshiri N, Mai U, Jia X, Mirarab S. TreeCluster: Clustering biological sequences using phylogenetic trees. PLoS ONE. 2019 doi: 10.1371/journal.pone.0221068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragonnet-Cronin M, et al. HIV transmission networks among transgender women in Los Angeles County, CA, USA: A phylogenetic analysis of surveillance data. Lancet HIV. 2019;6(3):e164–e172. doi: 10.1016/S2352-3018(18)30359-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rose R, et al. Phylogenetic methods inconsistently predict the direction of HIV transmission among heterosexual pairs in the HPTN 052 cohort. J. Infect. Dis. 2019;220(9):1406–1413. doi: 10.1093/infdis/jiy734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong SL, Dellicour S, Vrancken B, et al. In search of covariates of HIV-1 subtype B spread in the United States—A cautionary tale of large-scale Bayesian phylogeography. Viruses. 2020;12(2):182. doi: 10.3390/v12020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harden JW, Hilbe JM. Generalized Estimating Equations. Boca Raton: Chapman and Hall/CRC; 2012. [Google Scholar]
- 17.Pond SLK, Weaver S, Leigh Brown AJ, Wertheim JO. HIV-TRACE (TRAnsmission Cluster Engine): A tool for large scale molecular epidemiology of HIV-1 and other rapidly evolving pathogens. Mol. Biol. Evol. 2018;35(7):1812–1819. doi: 10.1093/molbev/msy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weaver S, et al. Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018;35(3):773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rambaut A, Lam TT, Carvalho LM, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016 doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suchard MA, et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 110. Virus Evol. 2018 doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009 doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oster AM, et al. Identifying clusters of recent and rapid HIV transmission through analysis of molecular surveillance data. J. Acquir. Immune Defic. Syndr. 2018;79(5):543–550. doi: 10.1097/QAI.0000000000001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ragonnet-Cronin M, Hodcroft EB, Wertheim JO. Understanding disclosed and cryptic HIV transmission risk via genetic analysis: What are we missing and when does it matter? Curr. Opin. HIV AIDS. 2019;14(3):205–212. doi: 10.1097/COH.0000000000000537. [DOI] [PubMed] [Google Scholar]
- 25.Bahl J, et al. Influenza A virus migration and persistence in North American wild birds. PLoS Pathog. 2013 doi: 10.1371/journal.ppat.1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards CJ, et al. Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr. Biol. 2011;21(15):1251–1258. doi: 10.1016/j.cub.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P. SpreaD3: Interactive visualization of spatiotemporal history and trait evolutionary processes. Mol. Biol. Evol. 2016;33(8):2167–2169. doi: 10.1093/molbev/msw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guilamo-Ramos V, et al. The invisible US Hispanic/Latino HIV crisis: Addressing gaps in the national response. Am. J. Public Health. 2020;110(1):27–31. doi: 10.2105/AJPH.2019.305309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panneer N, France AM, Whiteside YO, Zhang T, Wertheim JO, Oster AM. Brief report: Temporal changes in HIV transmission patterns among young men who have sex with men, United States, 2009–2016. J. Acquir. Immune Defic. Syndr. 2020;84(1):1–4. doi: 10.1097/QAI.0000000000002305. [DOI] [PubMed] [Google Scholar]
- 30.Centers for Disease Control Prevention. Evidence of HIV treatment and viral suppression in preventing the sexual transmission of HIV. https://www.cdc.gov/hiv/pdf/risk/art/cdc-hiv-art-viral-suppression.pdf (2020).
- 31.Chou R, et al. Preexposure prophylaxis for the prevention of HIV infection: Evidence report and systematic review for the US Preventive Services Task Force. JAMA. 2019;321(22):2214–2230. doi: 10.1001/jama.2019.2591. [DOI] [PubMed] [Google Scholar]
- 32.Smith, D. K. et al. Evidence of an association of increases in pre-exposure prophylaxis coverage with decreases in Human Immunodeficiency Virus diagnosis rates in the United States, 2012–2016. Clin. Infect. Dis. (2020). [DOI] [PMC free article] [PubMed]
- 33.Whiteside YO, Ruiguang S, Wertheim JO, Oster AM. Molecular analysis allows inference into HIV transmission among young men who have sex with men in the United States. AIDS. 2015;29(18):2517–2522. doi: 10.1097/QAD.0000000000000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention. CDC fact sheet: HIV among Latinos. https://www.cdc.gov/nchhstp/newsroom/docs/factsheets/cdc-hiv-latinos-508.pdf (2017).
- 35.Dennis AM, et al. HIV transmission patterns among immigrant Latinos illuminated by the integration of phylogenetic and migration data. AIDS Res. Hum. Retroviruses. 2015;31(10):973–980. doi: 10.1089/aid.2015.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valverde EE, Oster AM, Xu S, Wertheim JO, Hernandez AL. HIV transmission dynamics among foreign-born persons in the United States. J. Acquir. Immune Defic. Syndr. 2017;76(5):445–452. doi: 10.1097/QAI.0000000000001541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.City of Houston. Houston facts and figures. https://www.visithoustontexas.com/about-houston/facts-and-figures/.
- 38.World Population Review. Houston population (demographics, maps, graphs). https://worldpopulationreview.com/us-cities/houston-population/.
- 39.Houston Health Department HIV Surveillance Program. HIV infection in Houston: An epidemiologic profile 2010–2014. http://www.houstontx.gov/health/HIV-STD/HI_%20Epi_Profile_20160506_this.pdf (2015).
- 40.Frost SD, Pillay D. Understanding drivers of phylogenetic clustering in molecular epidemiological studies of HIV. JID. 2015;211(6):856–858. doi: 10.1093/infdis/jiu563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith DM, et al. A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS. 2009;23(2):225–232. doi: 10.1097/QAD.0b013e32831d2a81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan E, Nyaku AN, D’Aquila RT, Schneider JA. Determinants of HIV phylogenetic clustering in Chicago among young black men who have sex with men from the uConnect cohort. J. Acquir. Immune Defic. Syndr. 2017;75(3):265–270. doi: 10.1097/QAI.0000000000001379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SS, et al. An exploratory study on the social and genotypic clustering of HIV infection in men having sex with men. AIDS. 2009;23(13):1755–1764. doi: 10.1097/QAD.0b013e32832dc025. [DOI] [PubMed] [Google Scholar]
- 44.Aldous JL, et al. Characterizing HIV transmission networks across the United States. Clin. Infect. Dis. 2012;55(8):1135–1143. doi: 10.1093/cid/cis612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fisher M, et al. Determinants of HIV-1 transmission in men who have sex with men: A combined clinical, epidemiological and phylogenetic approach. AIDS. 2010;24(11):1739–1747. doi: 10.1097/QAD.0b013e32833ac9e6. [DOI] [PubMed] [Google Scholar]
- 46.Poon AFY, et al. The impact of clinical, demographic and risk factors on rates of HIV transmission: A population-based phylogenetic analysis in British Columbia, Canada. J. Infect. Dis. 2015;211(6):926–935. doi: 10.1093/infdis/jiu560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweinberger M, Handcock MS. Local dependence in random graph models: characterization, properties and statistical inference. J. Am. Stat. Assoc. 2015;77(3):647–676. doi: 10.1111/rssb.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ragonnet-Cronin M, et al. Longitudinal phylogenetic surveillance identifies distinct patterns of cluster dynamics. J. Acquir. Immune Defic. Syndr. 2010;55(1):102–108. doi: 10.1097/QAI.0b013e3181e8c7b0. [DOI] [PubMed] [Google Scholar]
- 49.Volz EM, et al. HIV-1 transmission during early infection in men who have sex with men: A phylodynamic analysis. PLoS Med. 2013 doi: 10.1371/journal.pmed.1001568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.