Abstract
The advent of single-cell sequencing started a new era of transcriptomic and genomic research, advancing our knowledge of the cellular heterogeneity and dynamics. Cell type annotation is a crucial step in analyzing single-cell RNA sequencing data, yet manual annotation is time-consuming and partially subjective. As an alternative, tools have been developed for automatic cell type identification. Different strategies have emerged to ultimately associate gene expression profiles of single cells with a cell type either by using curated marker gene databases, correlating reference expression data, or transferring labels by supervised classification. In this review, we present an overview of the available tools and the underlying approaches to perform automated cell type annotations on scRNA-seq data.
Keywords: scRNA-seq, Cell type, Cell state, Automatic annotation
1. Introduction
Individual cells represent the basic building blocks of tissues and organisms [1]. In multicellular species, cells specialize to fulfill highly specific functions. This specialization occurs as a result of intrinsic and extrinsic cues, with spatial location and molecular profiles strongly modulating cell fate and function [2], [3]. In this context, the advent of robust and accessible single-cell sequencing technologies [4] has enormously advanced our capacity to resolve and understand the molecular mechanisms regulating cell behavior, including fate decisions, developmental transitions, and responses to injury and disease. Single-cell RNA sequencing (scRNA-seq), in particular, has revolutionized biological research, and enables the categorization of cell types across multiple species, tissues, and contexts.
From a biological perspective, classifying units into groups and categories is essential for their study, and makes it possible to draw parallels between analog units found either in different body compartments or in distinct species. However, while the human body is estimated to contain on average ~ 100 trillion cells, the number of distinct cell types remains unclear [5]. Moreover, to appropriately classify cells into different types, a fundamental question must be answered: what is a cell “type”? Defining cell identity is not a trivial endeavor, firstly because gene expression levels are not always binary, but can vary gradually over a spectrum. Secondly, and more important, transcriptional differences that would allow cells to be separated into different categories might not possess any biological relevance in terms of cellular function [6]. Considering that the human genome consists of approximately 20,000–25,000 genes, and that an average cell contains between 100,000 and 1,000,000 mRNA molecules [7], single-cell experiments require the use of amplification reactions to define the molecular profile of individual cells. Amplification, however, introduces technical variability, which increases the level of molecular noise and imposes additional difficulties in discerning between truly relevant changes in gene expression profiles and fluctuations in transcript levels inherent to cells. The transcriptional changes that occur during the cell cycle, for instance, remain challenging to isolate from simultaneous, albeit independent, cellular processes within the cell [8].
scRNA-seq is presently the dominant approach for defining cellular states at the molecular level [9]. To date, over 19,000 studies reporting the use of scRNA-seq in a variety of tissues, organisms, and contexts have been listed in Pubmed (search of term “single cell rna sequencing” on October 15th, 2020). Nonetheless, with novel sequencing methods being constantly developed [4], data standardization, curation, and integration have emerged as important challenges to be overcome for the precise and accurate categorization of cell types across species and developmental stages, as well as in injury and disease [10]. For many purposes, ensuring that a cell is significantly more similar to its in vivo counterpart than to other cell types might be sufficient [11]. Within the field of cellular engineering, profiling the transcriptional signatures of forward-programmed hiPSCs or of 3D organoids by scRNA-seq serves as a reliable quality-control measure by enabling the prompt and confident assessment of the capacity of diverse engineering strategies to drive cells into specific lineages [12], [13], [14]. For primary cells and tissues, however, the interpretation of scRNA-seq data requires caution and, when identifying novel cell types, validation by additional functional tests [15]. Starting from single-cell transcriptomes, numerous pipelines have been developed for studying cell heterogeneity [16], [17]. Manual annotation of cell types is often time-consuming and suffers from limited reproducibility. To overcome these limitations, computational methods have recently emerged for the automated annotation of cell clusters.
2. Automated cell type annotation of target scRNA-seq datasets
Analysis of scRNA-seq datasets generally starts with dimensionality reduction and clustering [16], [17]. Clusters represent groups of cells with relatively similar gene expression profiles. Hence, cells clustering together are likely to possess the same identity, although diverse cellular phenomena such as cell transitions might not be fully captured in scRNA-seq datasets. Consequently, cells might be assigned erroneous identities. Furthermore, the choice of clustering methods and granularity [18] yields different cluster numbers and compositions within the same dataset. Under-clustering, in particular, can result in insufficient resolution for identifying rare cell types or transition states. Thus, defining the appropriate granularity and assigning identities to the cells in each of the clusters generated, a process known as annotation, are both crucial steps in scRNA-seq data analysis. Here, we focus on the second of these steps. A straightforward approach for cluster annotation consists of the computation of differentially expressed genes (DEGs), or unbiased markers, that define the identity of each cluster. These are subsequently overlapped with specific marker-gene lists for the cell types expected in the dataset [19]. Alternatively, unbiased markers can be used as input for statistical tests or bioinformatic analysis tools, many of them originally developed to ascribe genotype-phenotype relations in bulk RNA-Seq datasets. The most widely used of these tools include over-representation analysis (ORA) and gene set enrichment analysis (GSEA), as well as AUCell, PROGENy and DoRothEA [20], [21].
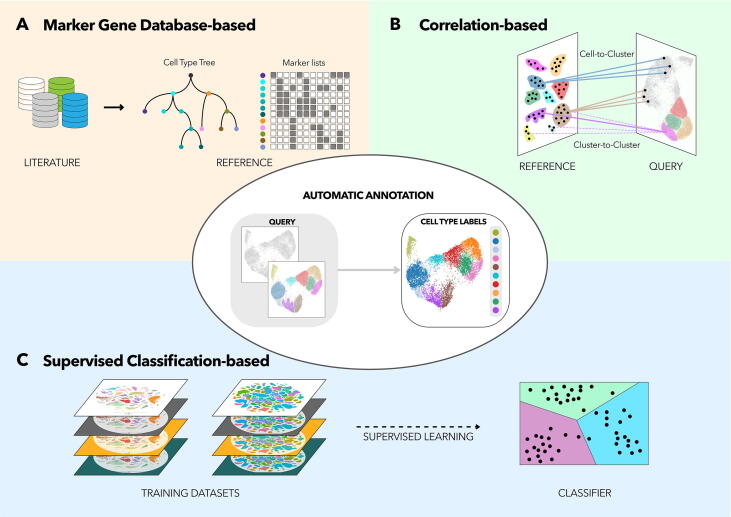
The task of cell type annotation is not trivial: multiple tools have been developed to automatically annotate single cells from their mRNA expression profiles. A reference cell type information is needed to label a query gene expression profile with its correspondent cell. First, marker genes related to cell types can be easily exploited. Lists of marker genes can be independently built by researchers or gathered from databases and ontologies. On the other hand, gene expression profiles of a reference dataset can be directly used for the annotation of a query. In particular, these tools have been designed either to annotate entire clusters or, to avoid clustering biases, to classify individual cells (reviewed in Wang et.al. [22]). Moreover, important characteristics of a tool for automated cell type annotation include: the capability to assemble multiple reference datasets to smooth batch effects; the possibility to classify cell types according to a hierarchical structure which can be given as input or learned from the data; the computation of a score of similarity between reference and query which can help identifying multiple cell types being harbored by the same cell; the ability of classifying cells as “unassigned” or “unknown” when they have an identity not represented in the reference. Beside such functionalities, three main methodological approaches can be identified (Table 1). The first approach relies on information from publicly available databases and ontologies describing cell type-specific markers (Fig. 1A). A second set of methods uses labeled scRNA-seq datasets as input for cell type identification, finding the best correlation between the reference and query datasets (Fig. 1B). Finally, a number of tools use a third alternative: supervised learning, which involves the training of a classifier with a labeled reference (Fig. 1C). Thereafter, the classifier is capable of determining cell types in unlabeled datasets. These methods, and the informatic tools employing them, are discussed in further detail below, with particular focus on their strengths and limitations.
Table 1.
Tools for automated cell type identification.
| Marker genes | Reference dataset | Tool name | Language | Computational approach | Unassigned | Multiple reference | Hierarchical classification | Additional features | Ref | |
|---|---|---|---|---|---|---|---|---|---|---|
| Marker gene database-based | • | scCATCH | R | Scoring system | – | [23] | ||||
| • | SCSA | Python | Scoring system | – | [24] | |||||
| • | SCINA | R | Bimodal distribution fitting to marker genes | ✓ | – | [25] | ||||
| • | CellAssign | R | Probabilistic Bayesian model | ✓ | – | [26] | ||||
| Correlation-based | • | scmap-cluster | R, web app | Cosine, Spearman, Pearson | ✓ | ✓ | [27] | |||
| • | scmap-cell | R, web app | Cosine distance based kNN | ✓ | ✓ | [27] | ||||
| • | SingleR | R | Spearman | score | ✓ | Harmonization of the labels allows for multiple reference datasets. | [28] | |||
| • | CHETAH | R, Shiny app | Spearman + confidence | ✓ | ✓ | [29] | ||||
| • | scMatch | Python | Spearman, Pearson | score | ✓ | Cell lineage is added as lower level of classification. | [30] | |||
| • | ClustifyR | R | Spearman, Pearson, Kendall, cosine | ✓ | Implements a consensus correlation score. | [31] | ||||
| • | CIPR | R, Shiny app | Dot product, Spearman, Pearson | score | Dot product implicitly involves feature selection. | [32] | ||||
| Supervised classification-based | • | CaSTLe | R | XGBoost classifier | ✓ | [33] | ||||
| • | Moana | Python | kNN-smoothing + SVM | [34] | ||||||
| • | LAmbDA | Python | Multiple ML techniques | ✓ | Training on multiple datasets to create a shared representation of the labels to smooth batch effects | [35] | ||||
| • | superCT | Web app | Artificial Neural Network | ✓ | Classifier trained on MCA with the possibility to add user defined datasets | [36] | ||||
| • | SingleCellNet | R | Random Forest | score | Similarity scores allow to find transition states and multiple identities in the same cell. | [37] | ||||
| • | Garnett | R | Elastic net regression | ✓ | ✓ | ✓ | Classification can be done using open chromatin information derived from scATAC-seq | [38] | ||
| • | scPred | R | SVM | ✓ | Allows to train different classifiers for defined labels | [39] | ||||
| • | ACTINN | Python | Artificial Neural Network | Robust against batch effects induced by sequencing technologies | [40] | |||||
| • | OnClass | Python | kNN and Bilinear Neural Network | ✓ | ✓ | ✓ | Use of a CellOntology to impute labels not present in the training data. | [41] | ||
| • | scClassify | R, Shiny app | Weighted kNN classifier | ✓ | ✓ | ✓ | Hierarchical cell type tree as reference. It combines six similarity matrices with five feature selection methods. | [42] | ||
| Others | • | scANVI | Python | kNN classifier | [43] | |||||
| • | Capybara | R | Quadratic programming | score | Cell engineering-oriented | [44] | ||||
| • | scID | R | Fisher’s Linear Discriminant Analysis | ✓ | [45] | |||||
| • | scNym | Python | Adversarial Neural Network | ✓ | ✓ | [46] | ||||
Fig 1.
Approaches for cell type annotation of scRNA-seq datasets. scRNA-seq datasets can be automatically annotated by tools implementing one of three approaches: annotation by marker gene databases; correlation-based methods; and annotation by supervised classification. The task of annotating a query scRNA-seq dataset consists of assigning a cell type identity to each one of the query single cells, or to a group of cells at once i.e. an unbiasedly calculated cluster. (A) Marker gene database-based annotation takes advantage of cell type atlases. Literature- and scRNA-seq analysis-derived markers have been assembled into reference cell type hierarchies and marker lists. In this approach, basic scoring systems are used to ascribe cell types at the cluster level in the query dataset. (B) Correlation-based methods make use of multiple correlation measures to compare gene expression profiles between a reference and a query dataset, at either single-cell or cluster level, by the use of centroids (pseudo-cells obtained by averaging the single-cell gene expression level of an entire cluster). Some of these tools assemble a reference of cell type gene-expression profiles from an ensemble of published studies and bulk RNA data repositories. The annotation step in this approach consists of finding the reference cell type that best correlates to the query cell or cluster, and every tool uses multiple steps for accurately finding the best match. (C) Annotation by supervised classification uses machine learning techniques for training a classifier on reference labeled scRNA-seq datasets. The classifier is subsequently applied to the query. Supervised learning is a powerful tool for building a model distribution of training labels as a function of features. Machine learning techniques offer a variety of alternatives in the training step and allow for hierarchical classification, which permits a more biologically-relevant identification of cell types.
2.1. Cluster annotation with marker gene databases
The widespread adoption of diverse scRNA-seq platforms has driven a rapid increase in the number of transcriptomic datasets published over the last years. Thousands of scRNA-seq datasets are now publicly available, with studies aiming not only to reveal the cellular heterogeneity of diverse tissues and organisms, but also to logically and accurately classify cells (Table 2) [47], [48], [49]. To unify results and organize information about cell types and states, thousands of publications have been manually curated and available datasets have been systematically re-analyzed, with results deposited in platforms such as CellMarker [50] and PanglaoDB [51]. In CellMarker, the cataloguing of manually-curated human and mouse cell type markers has allowed 13,605 genes to be mapped to 467 human cell types, and 9148 genes to 389 mouse cell types. For these analyses, gene-expression markers were gathered from over a thousand single-cell sequencing publications retrieved by specific PubMed queries, and collected from handbooks or company databases, such as those of BD biosciences and R&D Systems. From these datasets, CellMarker categorized cell types according to their tissue of origin, then hierarchically grouped them by localization, morphology, and functionality. PanglaoDB, is a cell type atlas in which information on gene expression and its relation to cell types is collected. To build PanglaoDB, an internal cell type marker database was assembled by automated abstract mining, followed by manual curation of the literature. Currently, PanglaoDB comprises 6631 marker genes mapping to 155 cell types. Similarly, CancerSEA provides markers, particularly protein-coding and long-non-coding transcripts, for 14 relevant functional cell states in cancer, including proliferative, invasive, and stemness states [52]. Altogether, these databases and online repositories offer an ample and ready-to-use source of cell type to marker gene relations derived from scRNA-seq experiments.
Table 2.
Publicly available repositories and datasets used by automated annotation tools.
| Data type | Species | Info | Tissues/cell types | Ref | |
|---|---|---|---|---|---|
| Human Primary Cell Atlas | Microarray | Human | Cell type profiles | Cell lines, tissues, primary cells | [53] |
| Blueprint | Bulk RNAseq | Human | Cell type profiles | Cell lines, tissues, primary cells | [54] |
| FANTOM5 | Bulk RNAseq | Human, Mouse, rat, dog and chicken | Cell type profiles | 15 cell types | [55] |
| Encode | Bulk RNAseq | Human, Mouse, Fly and Worm | Cell type profiles | Cell lines, tissues, primary cells | [56] |
| HCA | Single cell RNAseq | Human | Multi-organ datasets | 33 organs | [49] |
| MCA | Single cell RNAseq | Mouse | Multi-organ dataset | 98 major cell types | [48] |
| Tabula Muris | Single cell RNAseq | Mouse | Multi-organ datasets | 20 organs and tissues | [47] |
| Allen Brain Atlas | Single nuclei RNAseq | Human and Mouse | Brain datasets | 69 neuronal cell types | [57] |
| CellMaker | Marker genes | Human and Mouse | Marker Database | 467 (human), 389 (mouse) | [50] |
| PanglaoDB | Marker genes | Human | Marker Database | 155 cell types | [51] |
| CancerSEA | Marker genes | Human cancer | Marker Database | 14 cancer functional states | [52] |
Tools that have been natively developed to use the databases described previously for cell type inference include scCATCH [23] and SCSA [22]. In both cases, reference lists of markers were constructed by merging information from several sources. To use scCATCH, a tissue-specific cell taxonomy reference database known as CellMatch was assembled. Within it, markers were unified from CellMarker, the Mouse Cell Atlas project, CancerSEA, and the CD Marker Handbook. SCSA uses a collection of markers that were produced by merging CellMarker and CancerSEA. Additionally, SCSA allows users to add custom reference markers. Both scCATCH and SCSA calculate marker genes for the inputted clusters, and a scoring system subsequently assigns a cell type to each cluster. Additionally, SCSA provides an automatic GO term enrichment option, thereby adding information on the biological functions of the cells within each cluster.
As well as the tools above, more sophisticated statistical approaches have been used to transfer prior knowledge when trusted reference cell type markers are available, by performing a probabilistic assignment of the reference cell types. These include SCINA [25], which operates at the cluster level by fitting a bimodal distribution to marker genes; and CellAssign, which works at the single-cell level [26] using a Bayesian probabilistic model.
2.2. Correlation-based annotation
Correlation is the most straightforward statistical method for automatic comparison of gene expression data: it can easily use a reference dataset to reveal information about an unlabeled dataset. Moreover, correlating the expression levels of a set of genes, or of an entire transcriptomic profile, is a more refined way to find similarities between datasets than simply scoring the presence of marker genes in clusters. By combining the expression level of each gene with correlation methods, it is possible to evaluate both linear and non-linear interactions. Different strategies employing correlation have already been implemented in a variety of tools. These tools perform two main types of comparisons: either single cell-to-reference or cluster-to-reference. CIPR [32] and ClustifyR [31], for instance, employ a cluster-to-reference strategy. In particular, these tools cross-correlate unlabeled clusters to a reference of annotated clusters, with cell type labels assigned according to the best-correlating reference cell type. CIPR and ClustifyR represent clusters as centroids. Each centroid is a pseudo-cell whose expression level for each gene equals its averaged expression level in all cells of that cluster. After this step, both tools implement Spearman (default in ClustifyR) and Pearson correlation coefficients to determine the identity of each pseudo-cell (or cluster) in the query, with ClustifyR also integrating Kendall correlation and Cosine similarity, plus a consensus correlation score. In contrast to ClustifyR, CIPR recommends calculating the dot product of the logarithm-transformed fold change for each cluster, which implicitly involves feature selection.
In contrast, tools such as scmap [27], SingleR [28], and scMatch [30] correlate each cell of the query dataset to a reference collection of cell types or annotated clusters. SingleR and scMatch function in a similar way, as both use a collection of bulk datasets generated from human single cell types (Table 2). In particular, SingleR uses reference expression data from Blueprint [54], Encode [56], and the Human Primary Cell Atlas [53], while scMatch also uses FANTOM5 [58] and UCSC Xena Cancer Browser (https://xenabrowser.net) data, thereby also enabling the classification of cancer-related datasets. Moreover, since no assumptions can be made on the distribution of gene expression, both tools recommend the non-parametric Spearman rank correlation. To account for potential redundancies in the collection of bulk references, both SingleR and scMatch have an initial step for finding the top correlated cell types, and subsequent steps for refining these associations. Beyond this, the annotation strategy in scMatch groups cells by cell lineage or other ontological terms at a more general level than cell type. In contrast, SingleR, which was first published using only bulk references, has recently been updated to be used with single-cell references, and now incorporates a number of novel functionalities. For instance, there is now an option for using multiple reference datasets through label harmonization. Another tool for automated annotation, scmap, offers the scmap-cluster and scmap-cell options to annotate cells either to a reference cluster or to a reference cell. Thereby, it is possible to annotate single cells without requiring the user to define clusters a priori. To achieve this, scmap-cluster computes the similarity between each cell and the centroid of each reference cluster, while scmap-cell uses a fast-approximate k-nearest-neighbor search through product quantization, with an Euclidean distance algorithm adapted to incorporate cosine distance.
One requirement for the use of correlation methods, whether they map individual cells or entire clusters to a reference, is the selection of features. Feature selection consists of identifying and removing as many irrelevant or redundant features (genes in this context) from the data. Removing redundant genes is especially important when comparing datasets sequenced by different technologies, as the use of distinct sequencing parameters, i.e. variations in sequencing depth, may result in a different number of genes being detected in each cell. Of note, Kiselev and colleagues have reported that intra-dataset annotation performs poorly when unbiased gene selection methods such as highly variable genes (HVGs) or M3Drop are used instead of other feature-selection methods [59]. In their study, the best results were obtained by using random genes as features. In contrast, SingleR and ClustifyR utilize HVGs and DEGs, respectively, as default features. Feature-selection strategies were systematically tested during the development of CIPR and scmap. In CIPR, the performance of different correlation methods was evaluated when used either on all genes, or on a subset. Results suggested that methods using dot product operations on DEGs are best able to discriminate similar cell types, as they account for both down- and up-regulated genes. A distinct alternative was implemented in CHETAH [29], the only correlation-based tool implementing a method for hierarchical classification (see the next section). In CHETAH, candidate cells are compared to reference subsets in multiple rounds, with a different set of genes used to measure the similarity in each round. The best results were reported when the 200 genes with the largest absolute fold-change between a candidate cell and the averages of the sub-reference were used.
2.3. Annotation by supervised classification
Automatic cell type annotation methods attempt to identify similarities between scRNA-seq datasets, overcoming the intrinsic noise and variability of the data. Indeed, multiple confounding factors underlie the variability found across scRNA-seq datasets. Prominent drivers of variability include the sequencing platform used, the depth of sequencing chosen for the experiment, and the method of sample preparation. Such characteristic noise and the multidimensionality of scRNA-seq data have made machine-learning methods an outstanding resource for fulfilling a variety of tasks in analysis pipelines, including dimensionality-reduction operations [60], [61]. Supervised classification, i.e. the transferring of labels from labeled to unlabeled datasets, is a classic paradigm in machine learning, for which a wide range of techniques have been developed [62]. In the field of machine learning, the term ‘supervised learning’ is used to refer to the building of a model distribution of labels (cell types) in terms of a set of features (genes) which is trained on ground truth data (a previously annotated dataset). Thereafter, trained models are used to assign labels to instances of unlabeled datasets, according to their relative features. For automatic cell type annotation in scRNA-seq datasets, tools have already been developed which use supervised classification: here we highlight the main applications for scRNA-seq datasets.
Among the first classifiers for cell populations, CellNet was developed based on the Random Forest method [63]. Similarly, the same research group recently proposed a tool for single-cell classification named SingleCellNet [37]. Random Forest techniques are derived from decision trees, a class of logic-based algorithms [64], and have already proven useful in handling similarities within a scRNA-seq dataset [65]. By providing a quantitative score for the similarity between each cell class and each cell in the query dataset, SingleCellNet makes it possible to find multiple cell types associated with a single cell or with a group of cells: an extremely valuable functionality in the frame of cell type engineering, as cells in transition states can be identified.
In addition to Random Forest techniques, the k-nearest-neighbor (kNN) instance-based learning algorithm is also used for automatic cell type classification. This method is based on the principle that, in their feature-based representation, instances of the same class localize close to each other. Thus, kNN classifies cell type by representing labeled and unlabeled instances together in the same dimensions: this assigns unlabeled instances to the most-represented class in the neighborhood. OnClass is an example of a tool that takes advantage of the power of kNN classifiers. OnClass is able to impute labels not present in the training dataset by creating a low dimensional representation of the training set [41]. In this representation, a self-implemented CellOntology enables storage of information about numerous labels, even if they are unseen in the training set. Then, novel label imputation is carried out using a bilinear neural network. Similarly, a weighted kNN classification lies at the core of scClassify [42], a tool with the capacity to derive a hierarchical reference representation from multiple datasets. Thereafter, at each node of the reference cell type tree, scClassify trains 30 classifiers obtained by combining six similarity metrics with five feature selections.
Artificial neural networks (ANNs) are the basis for another class of supervised classifiers commonly referred to as perceptron-based. The great capacity of these techniques for solving non-linear relations between classes and features, together with advances in computation speed over recent years, has made ANN-based methods popular for tackling numerous tasks in the biomedical field [66], [67]. Examples of tools engaging ANNs for single-cell supervised classification are LAmbDA, SuperCT, and ACTINN [35], [36], [40]. LAmbDA is a framework that aims to perform multiple tasks on scRNA-seq data. ANNs perform the classification, interpreted as a transfer learning problem. By conducting the ANN training step on raw data from multiple datasets, LAmbDA creates a generalized representation of shared labels while correcting for potential batch effects. Another tool, SuperCT, was designed as a framework in which a supervised classifier is trained on all datasets within the Mouse Cell Atlas (MCA) [48], with the user able to expand this reference by submitting new datasets. In tests using the Tabula Muris Atlas as a training dataset [47], ACTINN was highly accurate in classifying strictly related cellular subtypes, and was robust against batch effects arising from the use of different sequencing techniques.
As with ANNs, Support Vector Machines (SVMs) have also been used in the context of scRNA-seq data analysis. SVMs allow multicollinearity and non-linear relationships to be harnessed within scRNA-seq data. Moana [34] and scPred [39] are two examples of tools which apply SVM-based classifiers on PCA-transformed gene expression matrices. Thereby, these tools prevent single genes from having an excessive impact on cell classification. More particularly, scPred uses SVMs with radial kernels as a standard, but allows the user to train other prediction models on specific labels as well (available in the R package caret [68]). Moana engages a hierarchical classification by recursively clustering and training a classifier over multiple iterations. Thereafter, Moana uses kNN to smooth the expression data minimally before training a SVM with a linear kernel to classify data in clusters in the two-PC dimension space. This operation is conducted for each cluster, until all labels in the reference dataset have been separated. Using this strategy for hierarchical classification allows Moana to maximize the number of cells it can analyze while minimizing the computation time required for the training step. The hierarchical classification approach is utilized not only for its efficiency in terms of computation time, but mostly because the classification it performs resembles the structured identity of cell types in tissues more closely. In fact, hierarchies between labels can be learned by the reference data (as in CHETAH (previous section), OnClass, and scClassify) or directly given as input by the user. The latter strategy was implemented in Garnett [38], a tool that allows cell type assignment according to a tree of cell types. Garnett creates a hierarchical model from the reference dataset by using cell type markers defined by the user. On these models, the software then trains an elastic net classifier. Notably, Garnett has been adapted to also be capable of classifying cell types according to their “gene activity score” as obtained from scATAC-seq data.
To harmonize cell counts between datasets while classifying unlabeled data with information from a labeled reference, semi-supervised learning techniques have also been implemented in the frame of scRNA-seq data analysis [43], [46]. Furthermore, Capybara [44] uses an unsupervised approach based on quadratic programming to score cells with a measure of cell identity which represents a linear combination of the cell types in the reference. Capybara can identify cells harboring characteristics of multiple cell types. The cell type classification task is performed in this tool by a statistical framework that makes it possible to find transition states between labels in the reference.
The power of scRNA-seq analysis tools lies primarily on their capacity to represent as many genes as possible in an unbiased manner. As computation tasks are presently feasible even when working with standard scRNA-seq dataset sizes, feature selection is not strictly required for supervised classification. Nonetheless, outliers and redundant features are detrimental to model training and classification in terms of computation speed and accuracy. Thus, feature selection and processing are key to enhancing the performance of supervised classification algorithms. Among the tools described in this section, many perform an initial data-processing step, while others select features. SingleCellNet, for example, selects features by pair transformation, keeping and binarizing only the most discriminant pairs before the training. CaSTLe also selects features, by using univariate methods such as selection by mean expression, mutual information, and correlation between genes, before splitting data into four bins according to expression levels [33]. ACTINN conducts a simple feature cleansing by considering as outlier genes whose mean expression level and standard deviation lie within the highest or lowest percentile. Instead of selecting features, Moana performs an initial kNN-smoothing step to remove unwanted noise. It is important to note that one main assumption of Moana (as well as superCT and ACTINN) is that all cell types existing in the query need to be present in the reference, to prevent that unseen cell types will be associated to the wrong labels. On the other hand, other tools implement a strategy to classify cells as “unassigned” or “unknown”, frequently by defining a cut off score for the annotation to be trusted. The ability of classify correctly the unseen cell types is key for these methods and it is benchmarked in scClassify, CHETAH, scPred and Garnett.
Reports suggest that all tools exploiting supervised clustering are reliable, efficient, and accurate. However, these conclusions might be the consequence of classification tests being relatively simple: classifying peripheral blood mononuclear cells or pancreatic cell types is relatively straightforward, given their high level of heterogeneity and the marked differences in the transcriptional profiles of the cells within each dataset. One task likely to be significantly more challenging is the identification of cell subtypes, for example within the neuronal classes, as only few genes may be crucial for their discrimination [57]. In case studies where the marker genes to be used are clearly defined, approaches like Garnett, SCINA, and CellAssign may outperform brute-force approaches. Similarly, if datasets with meaningful features and sufficient label representation are available, supervised learning methods might offer a powerful and flexible alternative for their analysis.
3. Summary and outlook
In the present review, we summarize the three main approaches used for automated cell type annotation on scRNA-seq data. A first category of tools relies on a set of trusted cell type-specific markers to ascribe the cell identity in the query. Such markers can be both database-derived or manually-curated lists. In the first case, the reference cell types we can use in the annotation is exhaustive, but the annotation can be uncertain if the query is not clean. On the other hand, manually-curated lists are usually limited in terms of cell type coverage, but allow for the use of sophisticated statistical approaches. Correlation-based methods require annotated bulk or single-cell RNA datasets as reference. These methods easily allow multiple references and large consortia data to be merged, making the annotation as comprehensive as possible. Ultimately, supervised classification methods represent a valid alternative when a meaningful reference dataset is available for the training step, being able to overcome characteristic scRNA-seq noise and batch effects given by different sequencing technologies. Automated cell type annotation tools have been assessed in a broad range of tissues, sample conditions and applications (Table 3). Notably, a benchmarking of supervised classification-based methods for automatic cell annotation was recently conducted by Abdelaal and colleagues [69], showing that each method possesses specific advantages over the others, and very good performances by using SVM with rejection option. Another benchmark study, comparing different classes of tools, shows that combining multiple tools is highly encouraged for improving the accuracy [70].
Table 3.
Classification challenges benchmarked in the original publication of each tool.
| Tool name | Pancreas | PBMC | BMDC | CBMC | Lung | Kidney | Liver | Retina | Brain | Differetiating/ transistioning cells | Whole organism | Tumor cells | Cross-species | Cross-platform | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| scCATCH | ✓ | ✓ | ✓ | ✓ | [23] | ||||||||||
| SCSA | ✓ | ✓ | ✓ | [24] | |||||||||||
| SCINA | ✓ | ✓ | ✓ | [25] | |||||||||||
| CellAssign | ✓ | ✓ | ✓ | ✓ | [26] | ||||||||||
| scmap-cluster | ✓ | ✓ | ✓ | ✓ | ✓ | [27] | |||||||||
| scmap-cell | ✓ | ✓ | ✓ | ✓ | ✓ | [27] | |||||||||
| SingleR | ✓ | ✓ | ✓ | [28] | |||||||||||
| CHETAH | ✓ | ✓ | ✓ | ✓ | [29] | ||||||||||
| scMatch | ✓ | ✓ | [30] | ||||||||||||
| ClustifyR | ✓ | ✓ | ✓ | ✓ | ✓ | [31] | |||||||||
| CIPR | ✓ | ✓ | [32] | ||||||||||||
| CaSTLe | ✓ | ✓ | ✓ | ✓ | ✓ | [33] | |||||||||
| Moana | ✓ | ✓ | ✓ | ✓ | [34] | ||||||||||
| LAmbDA | ✓ | ✓ | [35] | ||||||||||||
| superCT | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | [36] | ||||||||
| SingleCellNet | ✓ | ✓ | ✓ | ✓ | [37] | ||||||||||
| Garnett | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | [38] | ||||||||
| scPred | ✓ | ✓ | ✓ | ✓ | [39] | ||||||||||
| ACTINN | ✓ | ✓ | ✓ | [40] | |||||||||||
| OnClass | ✓ | [41] | |||||||||||||
| scClassify | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | [42] | ||||||||
| scANVI | [43] | ||||||||||||||
| Capybara | ✓ | ✓ | [44] | ||||||||||||
| scID | ✓ | ✓ | [45] | ||||||||||||
| scNym | ✓ | ✓ | ✓ | ✓ | [46] |
In the future, integrating the crucial role played by post-transcriptional regulatory mechanisms and epigenetic modifications on the genome with the in-depth knowledge currently being generated on the transcriptional profiles of a myriad of cell types across species and contexts will bring a better understanding of cellular identity. While the drop in the cost of sequencing technologies has allowed scRNA-seq technologies to become widely adopted, the implementation of strategies for the simultaneous extraction of transcriptomic, proteomic, and genomic regulatory information at the single-cell level will progressively allow for more refined cellular classifications [71], [72], [73], [74], [75], [76], [77]. The field will definitely benefit from a variety of computational tools for the efficient collection, standardization, and curation of discoveries related to cellular and molecular functions. Even in the absence of a final consensus in terms of what ultimately is entailed by the concept of cellular identity, sufficiently accurate approximations are expected to enable important advances in the field of cell and gene therapy.
CRediT authorship contribution statement
Giovanni Pasquini: Conceptualization, Writing - review & editing. Jesus Eduardo Rojo Arias: Writing - review & editing. Patrick Schäfer: Review & editing. Volker Busskamp: Review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Sara Oakeley for critical feedback on the manuscript. V.B. acknowledges funding by the European Research Council (ERC-StG-678071 – ProNeurons), by the Deutsche Forschungsgemeinschaft (SPP2127, EXC-2068-390729961 – Cluster of Excellence – Physics of Life at TU Dresden and EXC-2151-390873048 – Cluster of Excellence – ImmunoSensation2 at the University of Bonn) and the Volkswagen Foundation (Freigeist – A110720).
References
- 1.The T.W., Thoery C. Past and present. J Anat Physiol. 1890;24:253–287. [PMC free article] [PubMed] [Google Scholar]
- 2.Hosokawa H., Rothenberg E.V. How transcription factors drive choice of the T cell fate. Nat Rev Immunol. 2020 doi: 10.1038/s41577-020-00426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs E, Blau HM. Tissue Stem Cells: Architects of Their Niches. Cell Stem Cell 2020;27:532–56. DOI:10.1016/j.stem.2020.09.011. [DOI] [PMC free article] [PubMed]
- 4.Mereu E., Lafzi A., Moutinho C., Ziegenhain C., McCarthy D.J., Álvarez-Varela A. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat Biotechnol. 2020;38:747–755. doi: 10.1038/s41587-020-0469-4. [DOI] [PubMed] [Google Scholar]
- 5.Eberwine J., Sul J.Y., Bartfai T., Kim J. The promise of single-cell sequencing. Nat Methods. 2014;11:25–27. doi: 10.1038/nmeth.2769. [DOI] [PubMed] [Google Scholar]
- 6.Kolodziejczyk A.A., Kim J.K., Svensson V., Marioni J.C., Teichmann S.A. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Milo R., Jorgensen P., Moran U., Weber G., Springer M. BioNumbers the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2009;38:750–753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsiao C.J., Tung P.Y., Blischak J.D., Burnett J.E., Barr K.A., Dey K.K. Characterizing and inferring quantitative cell cycle phase in single-cell RNA-seq data analysis. Genome Res. 2020;30:611–621. doi: 10.1101/gr.247759.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tammela T., Sage J. Investigating tumor heterogeneity in mouse models. Annu Rev Cancer Biol. 2020;4:99–119. doi: 10.1146/annurev-cancerbio-030419-033413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuart T., Satija R. Integrative single-cell analysis. Nat Rev Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 11.Ng A.H.M., Khoshakhlagh P., Rojo Arias J.E., Pasquini G., Wang K., Swiersy A. A comprehensive library of human transcription factors for cell fate engineering. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Treutlein B., Lee Q.Y., Camp J.G., Mall M., Koh W., Shariati S.A.M. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature. 2016;534:391–395. doi: 10.1038/nature18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biddy B.A., Kong W., Kamimoto K., Guo C., Waye S.E., Sun T. Single-cell mapping of lineage and identity in direct reprogramming. Nature. 2018;564:219–224. doi: 10.1038/s41586-018-0744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowan C.S., Renner M., De Gennaro M., Gross-Scherf B., Goldblum D., Hou Y. Cell types of the human retina and its organoids at single-cell resolution. Cell. 2020;182(1623–1640) doi: 10.1016/j.cell.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J. Molecular architecture of the mouse nervous system. Cell. 2018;174(999–1014) doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15 doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiselev V.Y., Andrews T.S., Hemberg M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet. 2019;20:273–282. doi: 10.1038/s41576-018-0088-9. [DOI] [PubMed] [Google Scholar]
- 19.Pasquini G., Cora V., Swiersy A., Achberger K., Antkowiak L., Müller B. Using transcriptomic analysis to assess double- strand break repair activity: Towards precise in vivo genome editing. Int J Mol Sci. 2020:21. doi: 10.3390/ijms21041380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz-Mejia JJ, Meng EC, Pico AR, MacParland SA, Ketela T, Pugh TJ, et al. Evaluation of methods to assign cell type labels to cell clusters from single-cell RNA-sequencing data. F1000Research 2019;8:1–27. DOI:10.12688/f1000research.18490.3. [DOI] [PMC free article] [PubMed]
- 21.Holland C.H., Tanevski J., Perales-Patón J., Gleixner J., Kumar M.P., Mereu E. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 2020;21:1–19. doi: 10.1186/s13059-020-1949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Ding H., Zou Q. Identifying cell types to interpret scRNA-seq data: how, why and more possibilities. Brief Funct Genomics. 2020;19:286–291. doi: 10.1093/bfgp/elaa003. [DOI] [PubMed] [Google Scholar]
- 23.Shao X., Liao J., Lu X., Xue R., Ai N., Fan X. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. IScience. 2020:23. doi: 10.1016/j.isci.2020.100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Y., Wang X., Peng G. SCSA: a cell type annotation tool for single-cell RNA-seq data. Front Genet. 2020;11:1–8. doi: 10.3389/fgene.2020.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Luo D, Zhong X, Choi JH, Ma Y, Wang S, et al. Single Cells and Bulk Samples 2019. [DOI] [PMC free article] [PubMed]
- 26.Zhang A.W., O’Flanagan C., Chavez E.A., Lim J.L.P., Ceglia N., McPherson A. Probabilistic cell-type assignment of single-cell RNA-seq for tumor microenvironment profiling. Nat Methods. 2019;16:1007–1015. doi: 10.1038/s41592-019-0529-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiselev V.Y., Yiu A., Hemberg M. Scmap: projection of single-cell RNA-seq data across data sets. Nat Methods. 2018;15:359–362. doi: 10.1038/nmeth.4644. [DOI] [PubMed] [Google Scholar]
- 28.Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Kanter J.K., Lijnzaad P., Candelli T., Margaritis T., Holstege F.C.P. CHETAH: a selective, hierarchical cell type identification method for single-cell RNA sequencing. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou R., Denisenko E., Forrest A.R.R., Kelso J. ScMatch: a single-cell gene expression profile annotation tool using reference datasets. Bioinformatics. 2019;35:4688–4695. doi: 10.1093/bioinformatics/btz292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riemondy KA, Fu R, Gillen AE, Sheridan RM, Tian C, Daya M, et al. clustifyr: An R package for automated single-cell RNA sequencing cluster classification. F1000Research 2020;9:1–26. DOI:10.12688/f1000research.22969.2. [DOI] [PMC free article] [PubMed]
- 32.Ekiz H.A., Conley C.J., Stephens W.Z., O’Connell R.M. CIPR: a web-based R/shiny app and R package to annotate cell clusters in single cell RNA sequencing experiments. BMC Bioinf. 2020;21:191. doi: 10.1186/s12859-020-3538-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieberman Y, Rokach L, Shay T. Correction: CaSTLe - Classification of single cells by transfer learning: Harnessing the power of publicly available single cell RNA sequencing experiments to annotate new experiments(PLoS ONE (2018)13:10 (e0205499) DOI: 10.1371/journal.pone.0205499). PLoS One 2018;13:1–16. DOI:10.1371/journal.pone.0208349. [DOI] [PMC free article] [PubMed]
- 34.Wagner F, Yanai I. Moana: A robust and scalable cell type classification framework for single-cell RNA-Seq data. BioRxiv 2018:456129. DOI:10.1101/456129.
- 35.Johnson T.S., Wang T., Huang Z., Yu C.Y., Wu Y., Han Y. LAmbDA: label ambiguous domain adaptation dataset integration reduces batch effects and improves subtype detection. Bioinformatics. 2019;35:4696–4706. doi: 10.1093/bioinformatics/btz295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie P., Gao M., Wang C., Zhang J., Noel P., Yang C. SuperCT: a supervised-learning framework for enhanced characterization of single-cell transcriptomic profiles. Nucleic Acids Res. 2019;47:1–12. doi: 10.1093/nar/gkz116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan Y., Cahan P. SingleCellNet: a computational tool to classify single cell RNA-Seq data across platforms and across species. Cell Syst. 2019;9(207–213) doi: 10.1016/j.cels.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pliner H.A., Shendure J., Trapnell C. Supervised classification enables rapid annotation of cell atlases. Nat Methods. 2019;16:983–986. doi: 10.1038/s41592-019-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alquicira-Hernandez J., Sathe A., Ji H.P., Nguyen Q., Powell J.E. ScPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 2019;20:1–17. doi: 10.1186/s13059-019-1862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma F., Pellegrini M., Robinson M. ACTINN: automated identification of cell types in single cell RNA sequencing. Bioinformatics. 2020;36:533–538. doi: 10.1093/bioinformatics/btz592. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, Pisco AO, McGeever A, Brbic M, Zitnik M, Darmanis S, et al. Unifying single-cell annotations based on the Cell Ontology 2019. DOI:10.1101/810234.
- 42.Lin Y., Cao Y., Kim H.J., Salim A., Speed T.P., Lin D.M. scClassify: sample size estimation and multiscale classification of cells using single and multiple reference. Mol Syst Biol. 2020;16:1–16. doi: 10.15252/msb.20199389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu C, Lopez R, Mehlman E, Regier J, Jordan M, Yosef N. Probabilistic Harmonization and Annotation of Single-cell Transcriptomics Data with Deep Generative Models. BioRxiv 2019:532895. DOI:10.1101/532895. [DOI] [PMC free article] [PubMed]
- 44.Kong W, Fu Y, Morris S. Capybara: A computational tool to measure cell identity and fate transitions 2020. DOI:10.1101/2020.02.17.947390. [DOI] [PMC free article] [PubMed]
- 45.Boufea K., Seth S., Batada N.N. scID uses discriminant analysis to identify transcriptionally equivalent cell types across single-cell RNA-seq data with batch effect. IScience. 2020;23 doi: 10.1016/j.isci.2020.100914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimmel JC, Kelley DR. scNym: Semi-supervised adversarial neural networks for single cell classification. BioRxiv 2020:2020.06.04.132324.
- 47.Schaum N., Karkanias J., Neff N.F., May A.P., Quake S.R., Wyss-Coray T. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172(1091–1107) doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Regev A., Teichmann S., Lander E., Amit I., Benoist C., Birney E. Science forum: the human cell atlas. Elife. 2017:1–30. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C. Cell Marker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47:D721–D728. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franzén O., Gan L.M., Björkegren J.L.M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database. 2019;2019:1–9. doi: 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan H., Yan M., Zhang G., Liu W., Deng C., Liao G. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900–D908. doi: 10.1093/nar/gky939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mabbott N.A., Baillie J.K., Brown H., Freeman T.C., Hume D.A. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics. 2013;14:632. doi: 10.1186/1471-2164-14-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stunnenberg H.G., Hirst M. The international human epigenome consortium: a blueprint for scientific collaboration and discovery. Cell. 2016;167:1145–1149. doi: 10.1016/j.cell.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Alam T., Agrawal S., Severin J., Young R.S., Andersson R., Arner E. Comparative transcriptomics of primary cells in vertebrates. Genome Res. 2020;30:951–961. doi: 10.1101/gr.255679.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lizio M., Harshbarger J., Abugessaisa I., Noguchi S., Kondo A., Severin J. Update of the FANTOM web resource: high resolution transcriptome of diverse cell types in mammals. Nucleic Acids Res. 2017;45:D737–D743. doi: 10.1093/nar/gkw995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andrews T.S., Hemberg M. M3Drop: dropout-based feature selection for scRNASeq. Bioinformatics. 2019;35:2865–2867. doi: 10.1093/bioinformatics/bty1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van der Maaten L., Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 61.Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37:38–47. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 62.Kotsiantis S.B., Zaharakis I.D., Pintelas P.E. Machine learning: a review of classification and combining techniques. Artif Intell Rev. 2006;26:159–190. doi: 10.1007/s10462-007-9052-3. [DOI] [Google Scholar]
- 63.Cahan P., Li H., Morris S.A., Lummertz da Rocha E., Daley G.Q., Collins J.J. Cell net: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murthy S.K. Automatic construction of decision trees from data: a multi-disciplinary survey. Data Min Knowl Discov. 1998;2:345–389. doi: 10.1023/A:1009744630224. [DOI] [Google Scholar]
- 65.Pouyan M.B., Kostka D. Random forest based similarity learning for single cell RNA sequencing data. Bioinformatics. 2018;34:i79–i88. doi: 10.1093/bioinformatics/bty260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wainberg M., Merico D., Delong A., Frey B.J. Deep learning in biomedicine. Nat Biotechnol. 2018;36:829–838. doi: 10.1038/nbt.4233. [DOI] [PubMed] [Google Scholar]
- 67.Zemouri R., Zerhouni N., Racoceanu D. Deep learning in the biomedical applications: recent and future status. Appl Sci. 2019;9:1–40. doi: 10.3390/app9081526. [DOI] [Google Scholar]
- 68.caret: Classification and Regression Training 2020:https://CRAN.R-project.org/package=caret.
- 69.Abdelaal T., Michielsen L., Cats D., Hoogduin D., Mei H., Reinders M.J.T. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 2019;20:194. doi: 10.1186/s13059-019-1795-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao X., Wu S., Fang N., Sun X., Fan J. Evaluation of single-cell classifiers for single-cell RNA sequencing data sets. Brief Bioinform. 2020;21:1581–1595. doi: 10.1093/bib/bbz096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peterson V.M., Zhang K.X., Kumar N., Wong J., Li L., Wilson D.C. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 72.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Macaulay I.C., Haerty W., Kumar P., Li Y.I., Hu T.X., Teng M.J. G&T-seq: Parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 74.Clark S.J., Argelaguet R., Kapourani C.A., Stubbs T.M., Lee H.J., Alda-Catalinas C. ScNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells e. Nat Commun. 2018;9:1–9. doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu C., Yu M., Huang H., Juric I., Abnousi A., Hu R. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat Struct Mol Biol. 2019;26:1063–1070. doi: 10.1038/s41594-019-0323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen S., Lake B.B., Zhang K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat Biotechnol. 2019;37:1452–1457. doi: 10.1038/s41587-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu L., Liu C., Quintero A., Wu L., Yuan Y., Wang M. Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity. Nat Commun. 2019:10. doi: 10.1038/s41467-018-08205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]