Abstract
Phosphate-solubilizing bacteria (PSB) can convert insoluble rhizosphere phosphorus into forms that are absorbable by plants and thus enhance the growth of plants. Safflower is a cash crop that is a source of vegetable oils, food coloring and flavoring agents. This study sought to isolate PSB in safflower rhizosphere soil and investigate their effects on seedling growth. The isolated PSB were identified as belonging to the genera Pseudomonas, Sinorhizobium, Staphylococcus, Acinetobacter and Enterobacter using 16S rRNA gene sequence analysis. Acinetobacter sp RC04. showed the best performance in phosphate solubilization, with the efficiency of the process being influenced by carbon source, nitrogen source, cultivation temperature and initial culture pH. Acinetobacter sp. RC04 and Sinorhizobium sp. RC02 showed the ability to improve safflower seed germination and, when co-inoculated, improved seedling growth. Hence, we suggest that Acinetobacter sp. RC04 and Sinorhizobium sp. RC02 could be developed for field application to promote safflower growth. The results from this study will help drive novel biofertilizer discovery and could be included in integrated nutrient management regimes for safflower and other important economic crops.
Keywords: Acinetobacter spp, Sinorhizobium spp, Plant-growth promotion, Rhizobacteria, Co-inoculation
1. Introduction
Phosphorus (P) is an essential nutrient for crop growth and development [1, 2, 3]. In the soils of some agroecosystems, such as some arid and semi-arid regions, the total soil P concentration is adequate, but the content of soluble or plant-absorbable P (HPO42− or H2PO4−) is deficient [4,5]. In these soils, P tends to be limited because of immobilization and precipitation. For example, P is fixed by free oxides and hydroxides of aluminium and iron in acid soils and by calcium in alkaline soils, resulting in the low efficiency of soluble P fertilizers [6]. Solubilization of mineral phosphorus is beneficial to enhance plant growth.
Phosphate-solubilizing bacteria (PSB), associated with the plant rhizosphere, make mineral phosphorus more readily available for plant uptake by transforming insoluble P into forms that are available for the crop [7]. The effect of PSB is considered a mechanism for enhancing plant growth. Until now, previously known PSB belong to numerous genera including Pseudomonas, Bacillus, Rhizobium, Burkholderia, Achromobacter, Agrobacterium, Microccocus, Aereobacter, Flavobacterium and Erwinia [4]. Inoculation with these phosphate dissolvers as biofertilizers has been reported to increase P uptake and promote plant growth [8, 9, 10]. For example, treatment with PSB has increased the yield of wheat and promoted the growth of rice [11,12]. However, the ability of PSB to solubilize phosphate varies by bacterial species. The long-term stability and capability of phosphate dissolvers are key problems for their widespread application in promoting crop yields [13,14].
Safflower (Carthamus tinctorious L.) is a cash crop; its seeds are used for extraction of vegetable oil, and its petals are dried and used as food coloring and flavoring [15]. Safflower is suitable for cultivation in arid and semiarid regions in the Far East, central and northern Asia and the European Caucasus regions because of its drought tolerance and salt resistance [16]. As an important industrial and multipurpose crop, the global production of safflower exceeds 600 million tons per year [15]. However, PSB strains in the safflower rhizosphere still await investigation.
The present study aimed to isolate plant-growth-promoting PSB from safflower rhizosphere soil. Among those isolates, Acinetobacter sp RC04. showed the best performance in phosphate solubilization. To better understand its role in the rhizosphere, this study then surveyed the potential of the Acinetobacter sp RC04. It showed the ability to improve safflower seed germination and, when co-inoculated with Sinorhizobium sp. RC02, improve seedling growth. The results from this study could help drive novel biofertilizer discovery for safflower and other crops.
2. Materials and methods
2.1. Soil sample collection
This study was carried out at the agricultural experiment station of Shihezi University in Shihezi City, Xinjiang Province, China (44.27° N, 85.94° E), using the safflower cultivar Xinhong 4 as the model. The soil of the sampling sites is heavy loam with pH 8.0. According to the records of the station, the soil layer is approximately 15–21 cm thick and contains approximately 3.2 t ha−1 total P with 54 kg ha−1 available P, 1.6 t ha−1 total nitrogen with 88 kg ha−1 available nitrogen, 52 t ha−1 total potassium with 366 kg ha−1 available potassium and 30 t ha−1 soil organic matter. Samples were collected from soil sites not exposed to agrochemicals for several years. Sampling spots were selected by following an S pattern in the field. Rhizosphere samples (including roots and soil adhering to the roots) were collected at depths of 0–15 cm. Three biological replicates of plants were obtained at each spot. The samples were placed individually in sterile plastic bags and stored immediately in a cooler until arrival at the laboratory. All samples were stored at 4 °C until analysis and isolation.
2.2. PSB isolation
Excess soil was shaken from roots, leaving approximately 1 mm of soil still attached to the roots. About 1 g of the soil tightly adhering to the roots was separated from the roots by shaking in a sterile flask containing 50 ml of sterile phosphate-buffered saline (PBS) solution. The suspension was centrifuged twice for 1 min at 12,000 × g, serially diluted in PBS solution (10−2, 10−4, and 10−6) and plated on the P solubilization medium described below.
The P solubilization medium was modified from the National Botanical Research Institute’s phosphate-growth medium [17]. Tricalcium phosphate (TCP) was the sole P source. The medium contained (per liter) 10 g of glucose, 5 g of Ca3(PO4)2, 5 g of MgCl2·6H2O, 0.25 g of MgSO4·7H2O, 0.2 g of KCl, 0.5 g of (NH4)2SO4, 0.3 g of NaCl, 0.03 g of FeSO4·7H2O and 0.4 g of yeast extract in distilled water. Agar (20 g) was added to the medium for plate assays. The medium pH was adjusted to 7.0–7.5.
After 6 days of incubation on the P solubilization medium plates at 30 °C, colonies surrounded by a clear halo were considered phosphate dissolvers. Colonies were purified by restreaking on a plate. The capacity to dissolve phosphate on solid medium was measured, after 6 days of incubation at 30°C, as the ratio between the diameter of the phosphate solubilization halo around the colony and the diameter of the colony itself. The experiment was performed on a total of 9 plates, accounting for a total of 50 colonies.
2.3. Quantitative estimation of P solubilization in liquid culture
The potential PSB were grown in 100 mL of P solubilization medium and incubated with shaking at 300 rpm at 30 °C for 24 h. The approximate number of colony-forming units per milliliter (CFU mL−1) was determined by optical density measurement and serial dilutions with plate counts. One milliliter of culture (106 CFU mL−1) was transferred to a 500-mL Erlenmeyer flask containing 100 mL of P solubilization medium and incubated on a gyratory shaker (200 rpm) at 30 °C. The soluble P and optical density at 600 nm were measured every 12 h for 120 h using the phosphomolybdate blue colorimetric method [17]. Experiments were conducted five times per isolate. After the predefined incubation period, the cultures were harvested by centrifugation at 3000 × g for 15 min and the supernatant was filtered with 0.45-μm syringe filters for analysis of P concentration in the medium. Sterile, non-inoculated medium served as the control.
2.4. P solubilization assays
First, one-factor-at-a-time experiments were used to estimate whether a factor in PSB liquid cultivation had any effect on P solubilization and to seek to optimize the response. Carbon source, nitrogen source, initial pH, initial inoculum and cultivation temperature were separately tested. Each experimental factor was tested to determine optimum, keeping all other experimental factors constant as described above. After determination of the optimum of a given factor, the factor was subsequently held at the optimum throughout the remaining trials.
The carbon sources (20 g L−1) in liquid culture tested were fructose, glucose, sorbitol, sucrose and soluble starch. The nitrogen sources tested were (1.5 g L−1) (NH4)2SO4, NH4Cl, NH4NO3, urea and beef extract. Initial pH was 4, 5, 6, 7 or 8. For the inoculum assay, the initial bacterial cell suspension concentration was 3×104, 4×104, 5×104, 6×104 or 7×104 CFU mL−1. The cultivation temperature was 20, 25, 30, 35 or 40 °C. Each treatment had five replicates. After 6 days of cultivation, the bacterial cell suspension concentration was adjusted to 108 CFU mL−1 to estimate P solubilization.
To test possible factorial interactions influencing P solubilization, a full factorial experiment was performed. With five factors each taking two levels, the experiment had 32 treatment combinations (each having three replicates). It tested the effects of the five independent variables (NH4Cl concentration, glucose concentration, cultivation temperature, initial pH and inoculum amount) on the dependent variable (P solubilization) and possible interactions. The levels of the variables were designed according to the preceding one-factor-at-a-time experiments: [NH4Cl] had levels 1 g L−1 and 3 g L−1, [glucose] had levels 10 g L−1 and 30 g L−1, the temperature was 25 °C or 35 °C, the initial pH was 5.5 or 6.5 and the initial bacterial cell suspension concentration was 4×104 or 6×104 CFU mL−1. All other assay conditions were as described above.
2.5. PSB isolates identification
Bacterial genomic DNA was extracted by the phenol/chloroform method [18]. The 16S ribosomal DNA (rRNA) was amplified from extracted DNA by polymerase chain reaction with primers 5′-AGAGTTTGATCCTGGCTCAG-3′ and 5′-ACGGTTACCTTGTTACGACTT-3′ [19]. Amplification was performed by initial denaturation at 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 51 °C for 30 s and 72 °C for 1 min, with a final extension at 72 °C for 10 min. The 50-μL PCR mixtures contained 0.2 μM of each primer, 0.2 mM dNTPs, EasyTaq® buffer, 2.5 U EasyTaq® DNA polymerase (TRANSGEN, China) and 10 ng of template DNA. The PCR products were sequenced by Beijing Sun Biotech Co., Ltd. (Beijing, China). The sequences were then aligned to reference 16S rRNA sequences in the NCBI database using the BLAST program with default parameters [20]. The sequenced 16S rRNAs and reference 16S rRNAs were subjected to phylogenetic analysis using MEGA7 software [21]. The sequences were aligned by ClustalW to reconstruct a phylogenetic tree using the Kimura 2-parameter distance model and neighbor-joining method (1000 bootstrap replicates).
2.6. Germination assays
One milliliter of 24-h-old bacterial cultures was inoculated into 100 mL of Luria–Bertani (LB) medium (10 g L−1 tryptone, 5 g L−1 yeast extract and 10 g L−1 NaCl at pH 7.0), shaken for 72 h at 120 rpm at 30°C, and centrifuged for 10 min at 9,400 × g. The supernatant was discarded, and the pellet was resuspended in distilled water. Safflower seeds were surface sterilized with 5% sodium hypochlorite (commercial laundry bleach) for 15 min, and rinsed five times with sterile water.
The seed germination assay was based on a completely randomized design with three PSB inoculation treatments: (1) control without bacterial inoculation; (2) inoculation of one PSB strain (103 CFU mL−1); and (3) co-inoculation of two PSB strains (each with 103 CFU mL−1). Sterilized safflower seeds were placed on a filter paper in a plate (9 cm diameter). A five milliliter suspension of one treatment was added to one plate. Each treatment included at least three biological replicates. The seeds were germinated for 3 days at 28 °C in the dark.
2.7. Effects of PSB on plant growth
Sterilized seeds were sown in plastic pots (1 L) filled with autoclaved (121°C for 60 min) loamy soil and sand in 1:1 ratio (v/v) at 25 °C in a plant growth chamber (16-h light and 8-h dark, 40 ± 10 % relative humidity). Plants were watered with an equal volume of autoclaved sterilized water to keep the soil moist when needed. A pot with one seedling was considered an experimental unit, and three replicates per treatment were set up in a completely randomized design.
The PSB inoculation treatment for the greenhouse pot assay was designed with three treatments: (1) control without bacterial inoculation; (2) inoculation of one PSB strain (106 CFU mL−1); and (3) co-inoculation of two PSB strains (each with 106 CFU mL−1). Each treatment had three pots. A five milliliter suspension was inoculated on the top of the seed and the soil nearby at the time of planting. After 5 days, another 5 mL of suspension was added to the soil around the seeding area. Distilled water was used as a control. No other nutrients or bacterial inocula were supplied. Seedlings were harvested 4 weeks after sowing.
2.8. Data analysis
Differences were tested using ANOVA and groups were tested using Tukey’s HSD multiple comparisons procedure. Effects of factors in the P solubilization assay were evaluated using a GLM procedure (Gaussian error distribution).
Ethical approval: The conducted research is not related to either human or animals use.
3. Results and discussion
3.1. Characterization of PSB
On screening isolates from the safflower rhizosphere, six PSB strains were identified by their production of a halo around colonies on plates containing P solubilization medium. The 16S rRNA gene sequences of these strains were determined and deposited in the NCBI nucleotide sequence database (Table 1). They were clustered into the genera Pseudomonas, Sinorhizobium, Staphylococcus, Acinetobacter and Enterobacter by phylogenetic analysis (Figure 1). Their biochemical and physiological characteristics are shown in Table 1.
Table 1.
Characteristics of PSB.
| Strain | NCBI Accession | D/d | P (mg L−1) | Oxygen | Oxidase | Catalase | Starch | Methyl-Red | Indole Reduction |
|---|---|---|---|---|---|---|---|---|---|
| Pseudomonas sp. | KX387357 | 3.17±0.13bc | 131.6±1.79c | + | + | + | + | + | + |
| RC01 | |||||||||
| Sinorhizobium sp. | KX387354 | 2.32±0.18d | 90.9±1.56e | + | + | + | − | + | − |
| RC02 | |||||||||
| Staphylococcus sp. | KX387355 | 2.58±0.16cd | 112.5±1.91d | + | − | + | − | + | − |
| RC03 | |||||||||
| Acinetobacter sp. | KX387359 | 4.08±0.13a | 168.5±1.27a | + | + | + | − | − | + |
| RC04 | |||||||||
| Pseudomonas sp. | KX387358 | 3.33±0.12b | 157.2±1.21b | + | + | + | − | + | + |
| RC05 | |||||||||
| Enterobacter sp. | KX387356 | 2.35±0.19d | 92.4±0.75e | + | − | + | − | − | − |
| RC06 |
Six PSB strains were identified and their 16S rRNA gene sequences were deposited in the NCBI nucleotide sequence database. D, diameter of phosphate solubilization circle around the colony; d, diameter of the colony. P, phosphate solubilization. Values are the mean and standard error (n = 5). Means marked with different letters were significantly different (TukeyHSD, P < 0.05). The results of oxygen utilization, oxidase test, catalase test, starch utilization, methyl red test and indole reduction were given. +, tested positive or used as substrate; −, tested negative or not used as substrate.
Figure 1.
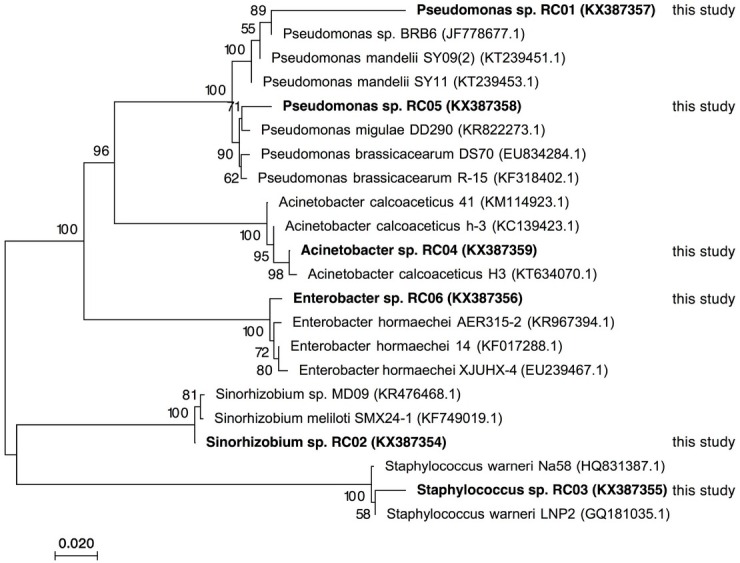
Evolutionary relationships of the phosphate-solubilizing bacteria (PSB) isolated in the present study and related strains in the NCBI database based on 16S rRNA gene sequences. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale with branch lengths similar to those of the evolutionary distances used to infer the phylogenetic tree. Bacterial strains in boldface indicate PSB included in the present study. Accession numbers are provided in parentheses.
The halo ratios of the six strains ranged between 2.32 and 4.08 after 6 days of incubation. The concentrations of soluble P they produced ranged between 90.9 and 168.5 mg L−1 in P solubilization medium. The strain Acinetobacter sp. RC04 showed the highest P solubilization among the strains (ANOVA, F(5, 24) = 504.4, P < 10−15; TukeyHSD, P < 0.05).
3.2. Growth profile of, and phosphate solubilization by, Acinetobacter sp. RC04
The growth profile of Acinetobacter sp. RC04 (judged by OD600) consisted of lag (0–12 h), exponential (12–36 h) and plateau (after 36 h) phases (Supplementary figure). The biomass (OD600) reached its maximum (1.25–1.29) at 48 h. The capacity to dissolve TCP also reached its maximum (182.77–184.31 mg L−1) at 48 h. The growth profile showed a similar trend to the phosphate solubility (Spearman’s rank correlation rho = 0.95, P < 10−16).
3.3. Effect of cultivation conditions on P solubilization by Acinetobacter sp. RC04
The cultivation conditions had a significant effect on the ability of Acinetobacter sp. RC04 to solubilize phosphate (Figure 2). The available P reached the maximum values with glucose as the carbon source, NH4Cl as the nitrogen source, initial pH = 6.0, initial inoculum of 4×104 CFU mL−1, and cultivation at 30 °C (Figure 2). Table 2 shows the effect of cultivation conditions ([NH4Cl], [glucose], cultivation temperature, initial pH and inoculum amount) on P solubilization and possible interactions between these factors. Interactions were observed between [glucose] and temperature, between [glucose] and [NH4Cl], and between pH and temperature (P < 0.001, GLM).
Figure 2.
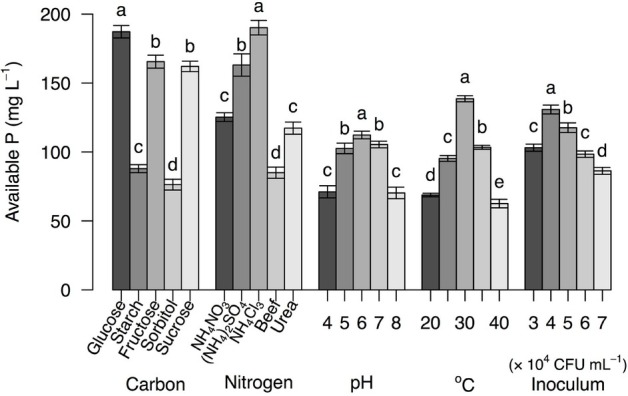
P solubilization assays of Acinetobacter sp. RC04. Bars represent means, and error bars show standard errors (n = 5). The means within one group marked with different lowercase letters were significantly different at P < 0.05 (TukeyHSD).
Table 2.
Generalized linear model.
| Terms | Generalized linear model |
||||
|---|---|---|---|---|---|
| Coefficients | t value | Pr (>|t|) | Sign | ||
| (Intercept) | 1499.94 | 7.08 | 5×10−10 | *** | |
| NH4Cl (g L−1) | −77.00 | −2.69 | 9×10−03 | ** | |
| Glucose (g L−1) | 11.40 | 3.99 | 1×10−04 | *** | |
| pH | −183.22 | −5.55 | 4×10−07 | *** | |
| Temp. (°C) | −28.59 | −5.43 | 6×10−07 | *** | |
| Inoculum (× 104 CFU mL | −1) | −102.24 | −3.77 | 3×10−04 | *** |
| NH4Cl : Glucose | −0.80 | −4.07 | 1×10-04 | *** | |
| NH4Cl : pH | 2.27 | 0.58 | 0.57 | ||
| NH4Cl : Temp. | 1.00 | 2.53 | 0.01 | * | |
| NH4Cl : Inoculum | 4.09 | 2.07 | 0.04 | * | |
| Glucose : pH | −0.71 | −1.79 | 0.08 | ||
| Glucose : Temp. | −0.15 | −3.92 | 2×10−04 | *** | |
| Glucose : Inoculum | 0.24 | 1.21 | 0.23 | ||
| pH : Temp. | 3.76 | 4.77 | 8×10−06 | *** | |
| pH : Inoculum | 8.09 | 2.05 | 0.04 | * | |
| Temp. : Inoculum | 0.71 | 1.81 | 0.07 | ||
| Residuals | |||||
Generalized linear model was used to test the effects of the five independent variables (NH4Cl concentration, glucose concentration, initial pH, cultivation temperature and initial inoculum amount) on the response variable (P solubilization) and possible interactions.
Sign, ‘***’ Pr < 0.001; ‘**’< 0.01; ‘*’< 0.05.
Compared with other carbon sources, glucose significantly promoted the P solubilizing capacity of Acinetobacter sp. RC04 (ANOVA, F(4, 20) = 468.1, P < 10−15, Figure 2). In addition, the amount of glucose played a role in P solubilization according to the GLM (Table 2). These results were consistent with the report that glucose could induce catabolite repression and affect the activity of acid and alkaline phosphatases in PSB [22]. Among the different nitrogen sources tested, NH4Cl was the best for Acinetobacter sp. RC04 to solubilize P (ANOVA, F(4, 20) = 243.8, P < 10−15). This result supports a previous suggestion that NH4Cl could be used as a nitrogen source to promote growth of PSB [17]. Moreover, a significantly negative interaction existed between NH4Cl and glucose concentrations (Table 2). The optimum P solubilization was obtained with the highest glucose concentration and the lowest NH4Cl concentration, consistent with previous reports that carbon and nitrogen concentrations modulate P solubilization efficiency [23, 24, 25].
Temperature influenced the efficiency of P solubilization by Acinetobacter sp. RC04 (ANOVA, F(4, 20) = 893.7, P < 10−15). Temperature plays a crucial role in influencing the activity of phytases [26]; the enzyme phytase releases P from phytate. Figure 2 shows that the P concentration increased with increasing temperature up to 30 °C, then decreased at higher temperatures. The trend is similar to that for Aspergillus oryzae and A. niger [26], although the optimal temperatures were different. The temperature of the highest activity of phytase varies widely for different microorganisms [27, 28, 29, 30, 31, 32]. In addition, pH also influenced the P solubilization (ANOVA, F(4, 20) = 169.4, P < 10−14). The P solubility was highest at pH 5–7 and lower at pH 4 and 8.
3.4. PSB effect on seedling growth
Inoculation of Acinetobacter sp. RC04 or Sinorhizobium sp. RC02 significantly promoted safflower seed germination (ANOVA, F(3, 8) = 97.43, P < 10−5, Figure 3). In addition, co-inoculation of the two isolates resulted in a significant increase in seedling length compared with single-strain treatment (TukeyHSD, P < 10−4). Pot trials showed that the co-inoculation had a positive effect on shoot length (ANOVA, F(3, 8) = 11.53, P = 0.003) and the number of secondary roots (ANOVA, F(3, 8) = 8.15, P = 0.008) compared with the control (Figure 4).
Figure 3.
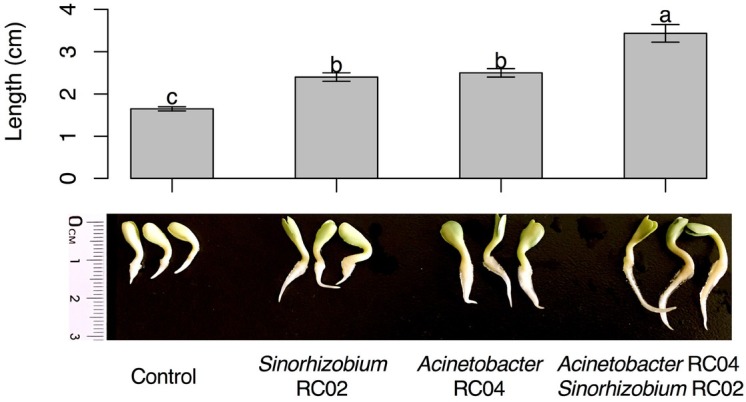
Promotion of safflower seed germination by PSB. Bars represent the mean, and error bars show the standard error (n = 3). Means marked with different lowercase letters were significantly different (TukeyHSD, P < 0.01).
Figure 4.
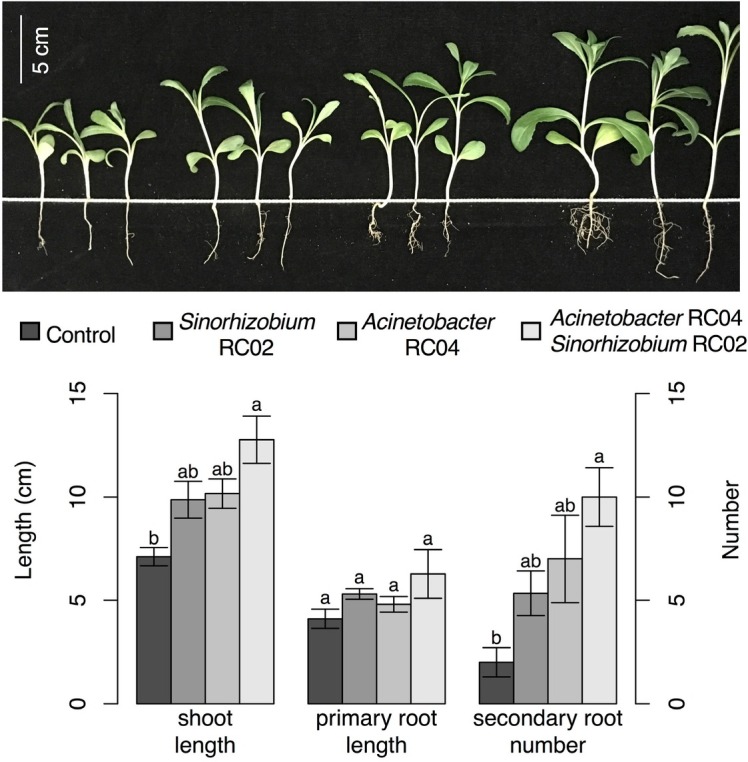
Growth promotion of safflower seedlings by PSB. Bars represent the mean, and error bars show the standard error (n = 3). Means within each group marked with different lowercase letters were significantly different (TukeyHSD, P < 0.01).
Acinetobacter sp. RC04 and Sinorhizobium sp. RC02 showed the ability to promote safflower seed germination and, when co-inoculated, improve seedling growth. These results indicate they might be incorporated into biofertilizers to increase safflower growth [33]. The improvement of plant growth and properties by PSB may be due to several possible mechanisms. PSB alter the plasticity of seeds and roots by changing the soil composition. For example, plant growth-promoting rhizobacteria may improve the solubility of mineral nutrients by releasing organic acids and thereby increasing the vegetative biomass and N and P accumulation in plant tissues, simulating plant growth [34]. This phenomenon, in turn, affects colonization and development of the bacteria [35]. Plant growth-promoting rhizobacteria can induce production of phytoalexin, antibiotics against pathogenic organisms, as well as siderophores, and they colonize root surfaces, thereby out-competing pathogens [34]. Inoculation with plant growth-promoting rhizobacteria can stimulate or inhibit functional community formation and growth in a given symbiotic relationship, depending upon the nature and concentration of secondary metabolites released by the partners in that plant–microbial relationship [34]. Thus, the interactive effect among rhizosphere microorganisms can influence P cycling and promote a sustainable nutrient supply to plants [34]. For instance, inoculation of mixed PSB or co-inoculation with other microorganisms can result in balanced nutrition for plants, such as providing P and N [36]. The present study confirmed the advantage of mixed PSB inoculation. The interactions among PSB, plants and other rhizobacteria create synergistic effects that improve the uptake of individual nutrients [37].
4. Conclusions
The present study screened phosphate dissolving bacteria from the rhizosphere soil of safflower. Among these strains, Acinetobacter sp. RC04 showed high performance in phosphate solubilization, and the efficiency of the process was influenced by cultivation conditions. Acinetobacter sp. RC04 and Sinorhizobium sp. RC02 showed the ability to improve safflower seed germination and, when co-inoculated, improve seedling growth. These results indicate the positive role of Acinetobacter sp. RC04 and Sinorhizobium sp. RC02 in enhancing the biomass of safflower seedlings. Their co-inoculation with plants will be highly beneficial in improving the growth of safflower. Further molecular and biochemical studies of these bacteria will provide efficient ways for incorporating these strains into biofertilizers to promote improved yield of agronomic crops and sustainable agriculture.
Acknowledgements
We thank Dave Baab for copyediting the manuscript and James Allen, DPhil, from Liwen Bianji, Edanz Group China, for English editing. We thank the reviewers for their helpful suggestions. The research was supported by the National Natural Science Foundation of China (31560310, 31760302, 31860308 and 31272416). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest: Authors state no conflict of interest
Supplemental Material: The online version of this article (DOI: 10.1515/biol-2019-0028) offers supplementary material.
Supplementary Material.
References
- [1].Chen YP, Rekha PD, Arun AB, Shen FT, Lai WA, Young CC. Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl Soil Ecol. 2006;34:33–41. [Google Scholar]
- [2].Díaz I, Torrent J. Changes in olsen P in relation to P balance in contrasting agricultural soils. Pedosphere. 2016;26:636–42. [Google Scholar]
- [3].Wei Y, Zhao Y, Fan Y, Lu Q, Li M, Wei Q. Impact of phosphate-solubilizing bacteria inoculation methods on phosphorus transformation and long-term utilization in composting. Bioresour Technol. 2017;241:134–41. doi: 10.1016/j.biortech.2017.05.099. [DOI] [PubMed] [Google Scholar]
- [4].Rodríguez H, Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv. 1999;17:319–39. doi: 10.1016/s0734-9750(99)00014-2. [DOI] [PubMed] [Google Scholar]
- [5].Goldstein AH. Recent Progress in Understanding the Molecular Genetics and Biochemistry of Calcium Phosphate Solubilization by Gram Negative Bacteria. Biol Agric Hortic. 1995;12:185–93. [Google Scholar]
- [6].Park JH, Bolan N, Megharaj M, Naidu R. Isolation of phosphate solubilizing bacteria and their potential for lead immobilization in soil. J Hazard Mater. 2011;185:829–36. doi: 10.1016/j.jhazmat.2010.09.095. [DOI] [PubMed] [Google Scholar]
- [7].Maroniche GA, Rubio EJ, Consiglio A, Perticari A. Plant-associated fluorescent Pseudomonas from red lateritic soil: Beneficial characteristics and their impact on lettuce growth. J Gen Appl Microbiol. 2016;62:248–57. doi: 10.2323/jgam.2016.04.006. [DOI] [PubMed] [Google Scholar]
- [8].Malboobi MA, Owlia P, Behbahani M, Sarokhani E, Moradi S, Yakhchali B. Solubilization of organic and inorganic phosphates by three highly efficient soil bacterial isolates. World J Microbiol Biotechnol. 2009;25:1471–7. [Google Scholar]
- [9].Wen Z, Shen J, Blackwell M, Li H, Zhao B, Yuan H. Combined applications of nitrogen and phosphorus fertilizers with manure increase maize yield and nutrient uptake via stimulating root growth in a long-term experiment. Pedosphere. 2016;26:62–73. [Google Scholar]
- [10].Eida AA, Hirt H, Saad MM. Challenges faced in field application of phosphate-solubilizing bacteria. Bioremediation, Springer; Singapore: 2017. p. 125–43. Rhizotrophs Plant Growth Promot. [Google Scholar]
- [11].Kloepper JW, Lifshitz R, Zablotowicz RM. Free-living bacterial inocula for enhancing crop productivity. Trends Biotechnol. 1989;7:39–44. [Google Scholar]
- [12].Panhwar QA, Naher UA, Jusop S, Othman R, Latif MA, Ismail MR. Biochemical and molecular characterization of potential phosphate-solubilizing bacteria in acid sulfate soils and their beneficial effects on rice growth. PLOS ONE. 2014;9:e97241. doi: 10.1371/journal.pone.0097241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ordoñez YM, Fernandez BR, Lara LS, Rodriguez A, Uribe-Vélez D, Sanders IR. Bacteria with phosphate solubilizing capacity alter mycorrhizal fungal growth both inside and outside the root and in the presence of native microbial communities. PLOS ONE. 2016;11:e0154438. doi: 10.1371/journal.pone.0154438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vaneeckaute C, Janda J, Vanrolleghem PA, Tack FMG, Meers E. Phosphorus use efficiency of bio-based fertilizers: bioavailability and fractionation. Pedosphere. 2016;26:310–25. [Google Scholar]
- [15].Hussain MI, Lyra D-A, Farooq M, Nikoloudakis N, Khalid N. Salt and drought stresses in safflower: a review. Agron Sustain Dev. 2015;36:1–31. [Google Scholar]
- [16].Nosheen A, Bano A, Ullah F, Farooq U, Yasmin H, Hussain I. Effect of plant growth promoting rhizobacteria on root morphology of Safflower (Carthamus tinctorius L.) Afr J Biotechnol. 2013;10:12638–49. [Google Scholar]
- [17].Nautiyal CS. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1999;170:265–70. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- [18].Sambrook J, Maniatis TE. Molecular cloning -a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- [19].Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol J Comput Mol Cell Biol. 2000;7:203–14. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- [21].Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim KY, Jordan D, McDonald GA. Enterobacter agglomerans, phosphate solubilizing bacteria, and microbial activity in soil: Effect of carbon sources. Soil Biol Biochem. 1998;30:995–1003. [Google Scholar]
- [23].Rodriguez H, Gonzalez T, Goire I, Bashan Y. Gluconic acid production and phosphate solubilization by the plant growth-promoting bacterium Azospirillum spp. Naturwissenschaften. 2004;91:552–5. doi: 10.1007/s00114-004-0566-0. [DOI] [PubMed] [Google Scholar]
- [24].Scervino JM, Mesa MP, Mónica ID, Recchi M, Moreno NS, Godeas A. Soil fungal isolates produce different organic acid patterns involved in phosphate salts solubilization. Biol Fertil Soils. 2010;46:755–63. [Google Scholar]
- [25].Lin T, Huang H, Shen F, Young C. The protons of gluconic acid are the major factor responsible for the dissolution of tricalcium phosphate by Burkholderia cepacia CC-Al74. Bioresour Technol. 2006;97:957–60. doi: 10.1016/j.biortech.2005.02.017. [DOI] [PubMed] [Google Scholar]
- [26].Naves L de P, Corrêa AD, Bertechini AG, Gomide EM, Santos C dos. Effect of ph and temperature on the activity of phytase products used in broiler nutrition. Braz J Poult Sci. 2012;14:181–5. [Google Scholar]
- [27].Igbasan FA, Männer K, Miksch G, Borriss R, Farouk A, Simon O. Comparative studies on the in vitro properties of phytases from various microbial origins. Arch Tierernahr. 2000;53:353–73. doi: 10.1080/17450390009381958. [DOI] [PubMed] [Google Scholar]
- [28].Greiner R, Konietzny U, Jany KD. Purification and characterization of two phytases from Escherichia coli. Arch Biochem Biophys. 1993;303:107–13. doi: 10.1006/abbi.1993.1261. [DOI] [PubMed] [Google Scholar]
- [29].Kim H, Kim Y, Lee J, Kim K, Kim Y. Isolation and characterization of a phytase with improved properties from Citrobacter braakii. Biotechnol Lett. 2003;25:1231–4. doi: 10.1023/a:1025020309596. [DOI] [PubMed] [Google Scholar]
- [30].Vats P, Banerjee UC. Production studies and catalytic properties of phytases (myo-inositolhexakisphosphate phosphohyd-rolases): an overview. Enzyme Microb Technol. 2004;35:3–14. [Google Scholar]
- [31].Wyss M, Brugger R, Kronenberger A, Rémy R, Fimbel R, Oesterhelt G. Biochemical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): catalytic properties. Appl Environ Microbiol. 1999;65:367–73. doi: 10.1128/aem.65.2.367-373.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wyss M, Pasamontes L, Rémy R, Kohler J, Kusznir E, Gadient M. Comparison of the thermostability properties of three acid phosphatases from molds: Aspergillus fumigatus phytase, A. niger phytase, and A. niger PH 2.5 acid phosphatase. Appl Environ Microbiol. 1998;64:4446–51. doi: 10.1128/aem.64.11.4446-4451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Naseri R, Mirzaei A. Response of yield and yield components of Safflower (Carthamus tinctorius L.) to seed inoculation with Azotobacter and Azospirillum and different nitrogen levels under dry land condition. Am-Eurasian J Agric Env Sci. 2010;9:445–9. [Google Scholar]
- [34].Zahir ZA, Arshad M, Frankenberger WT. Plant growth promoting rhizobacteria: Applications and perspectives In agriculture. Academic Press; 2003. p. 97–168. [Google Scholar]
- [35].Nosheen A, Bano A, Yasmin H, Keyani R, Habib R, Shah STA. Protein quantity and quality of safflower seed improved by NP fertilizer and rhizobacteria (Azospirillum and Azotobacter spp.) Front Plant Sci. 2016;7:1–12. doi: 10.3389/fpls.2016.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kushwaha DS. Growth and yield of different cultivars of sesame (Sesamum indicum L.) as influenced by seed applied Azotobacter and phosphate solubilizing bacteria. Indian J Agric Res. 2011;45:326–30. [Google Scholar]
- [37].Belimov AA, Kojemiakov AP, Chuvarliyeva CV. Interaction between barley and mixed cultures of nitrogen fixing and phosphate-solubilizing bacteria. Plant Soil. 1995;173:29–37. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


