Summary
Skeletal muscle insulin resistance is the earliest defect in type 2 diabetes (T2D), preceding and predicting disease development. To what extent this reflects a primary defect or is secondary to tissue crosstalk due to changes in hormones or circulating metabolites is unknown. To address this question, we have developed an in vitro disease-in-a-dish model using iPS cells from T2D patients differentiated into myoblasts (iMyo). We find that T2D iMyos in culture exhibit multiple defects mirroring human disease, including altered insulin signaling, decreased insulin-stimulated glucose uptake, and reduced mitochondrial oxidation. More striking, global phosphoproteomic analysis reveals a multi-dimensional network of signaling defects in T2D iMyos going beyond the canonical insulin signaling cascade and including proteins involved in regulation of Rho-GTPases, mRNA splicing/processing, vesicular trafficking, gene transcription and chromatin-remodeling. These cell-autonomous defects and dysregulated network of protein phosphorylation reveal a new dimension in the cellular mechanisms underlying the fundamental defects in T2D.
Introduction
Type 2 diabetes (T2D) and metabolic syndrome are major causes of increased morbidity and mortality worldwide. Both arise from a cluster of derangements in which insulin resistance stands as a central component (Boucher et al., 2014; Petersen and Shulman, 2018). Indeed, offspring of type 2 diabetic parents who are at high risk for developing T2D exhibit insulin resistance many years before diagnosis, and this predicts development of T2D in these individuals (Martin et al., 1992). The earliest site of insulin resistance is skeletal muscle (Rothman et al., 1995; Warram et al., 1990), which is of critical relevance since this tissue is the primary site of glucose disposal when insulin levels are high as in the post-prandial state or during an euglycemic-hyperinsulinemic clamp (DeFronzo and Tripathy, 2009; Kowalski and Bruce, 2014).
Despite the prominent role that skeletal muscle insulin resistance plays in T2D, the precise nature of the underlying mechanisms and whether it results from cell-autonomous signaling defects or from systemic changes due to tissue crosstalk remains debated. Multiple studies have shown that a variety of circulating factors, including increased levels of free fatty acids, ceramides, cytokines and other inflammatory mediators, as well as altered levels of branched chain amino acids and other metabolites may be increased in serum of T2D patients and contribute to the insulin resistance (Newgard, 2012; Petersen and Shulman, 2018; Saltiel and Olefsky, 2017). On the other hand, muscle biopsy studies have revealed multiple defects in glucose transport and utilization in T2D subjects. These include altered insulin-stimulated IRS protein phosphorylation and PI-kinase activation (Cusi et al., 2000), reduced expression of mitochondrial genes (Mootha et al., 2003; Patti et al., 2003) and hexokinase II (Ducluzeau et al., 2001), decreased GLUT4 translocation (Zierath et al., 1996), and impaired insulin-stimulated glucose transport, glycolytic flux, glycogen synthesis and mitochondrial ATP production (Cline et al., 1999; Petersen and Shulman, 2018; Stump et al., 2003). Likewise, primary skeletal muscle cultures from T2D patients show insulin resistance (Henry et al., 1995; Jiang et al., 2013). However, the molecular defects leading to these changes and to what extent they are primary or can be maintained in a cell-autonomous manner are unknown.
Induced pluripotent stem cells (iPSCs) have unlimited expansion potential and their ability to be differentiated in vitro provides a unique opportunity to study and model human disease (Shi et al., 2017). Using this technology, our group has demonstrated that severe insulin resistance due to mutations on the insulin receptor gene alters gene expression and glucose metabolism in iPSCs, as well as after their conversion to skeletal myocytes (Burkart et al., 2016; Iovino et al., 2014; Iovino et al., 2016). iPSCs have been successfully used to model other disorders involving muscle (Caron et al., 2016), as well as metabolic diseases including maturity onset diabetes of the young (MODY) (Teo et al., 2013), type 1 diabetes (Bhatt et al., 2015) and morbid obesity (Rajamani et al., 2018), but their application to the common polygenic T2D variety and its associated insulin resistance has been minimal. One advantage of this approach to reveal insight into these more complex disorders is that the impact of circulating factors and epigenetic factors in disease pathogenesis is minimized, since iPSC reprogramming normalizes the extracellular milieu and to a large extent resets the epigenome of the iPSCs and their derivatives compared to primary counterparts of the same lineage (Frobel et al., 2014).
To identify primary drivers of skeletal muscle insulin resistance in T2D without interference of systemic factors and other limitations posed by biopsy samples or primary myoblast cultures such as increased cellular heterogeneity and constant recruitment of new subjects, we have derived iPSC lines from people with T2D or normal glucose tolerant controls and have converted these into myoblasts (iMyos). We find that these cells in vitro closely mirror the impaired molecular responses to insulin observed in vivo, including defective insulin-stimulated glucose uptake and cellular respiration. Unexpectedly, global phosphoproteomics analysis demonstrates that, in addition to the alterations in insulin signaling, T2D is marked by a broad network of signaling changes in both the basal and insulin-stimulated states, including proteins involved in regulation of RNA processing, gene expression, cytoskeleton remodeling, and vesicular trafficking. Importantly, the regulatory events upstream of phosphoproteome rewiring in T2D imply dysfunction of a multiplicity of kinases, including mTOR, ROCK, novel PKCs and others, instead of a single kinase. These findings demonstrate a new, previously unrecognized layer in the cell-autonomous defects underlying skeletal muscle insulin resistance and open the opportunity for development of new therapeutic approaches to T2D.
Results
Derivation of Patient-Specific iPSCs and Conversion into Skeletal Myoblasts (iMyos)
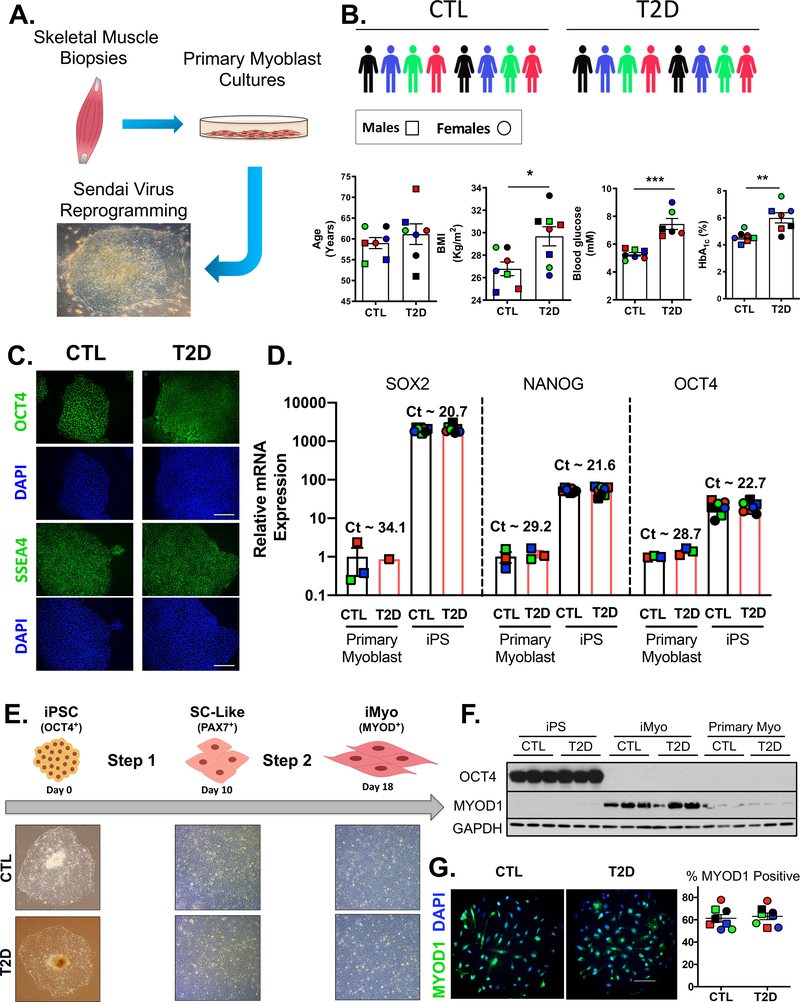
To identify primary determinants of insulin resistance associated with T2D in skeletal muscle, we derived iPSC lines from T2D patients and glucose tolerant healthy controls (CTL) and converted these into the myogenic lineage for metabolic and molecular studies. Because previous studies have demonstrated that there may be retention of developmental cues in iPSCs related to the tissue of origin (Quattrocelli et al., 2015), we derived iPSC lines from primary myoblasts cultures to maximize both the differentiation efficiency and the likelihood of capturing any cell-specific disease phenotype. The iPSC lines were generated by transducing primary myoblast cultures derived from skeletal muscle biopsies using Sendai viruses which do not integrate in the host cell genome (Figure 1A), as confirmed by RT-PCR (Figure S1A). The donor cohort was composed of 8 CTL and 8 T2D subjects equally divided between males and females, and matched for age. As expected, the T2D donor group had significantly higher BMIs, blood glucose levels and chronic hyperglycemia indicated by higher HbA1c levels (Figure 1B). Immunostaining, FACS analysis and qPCR confirmed the pluripotent state on the iPSCs. Thus, the iPSCs were positive for the transcription factor OCT4 and the pluripotency antigen SSEA4 (Figure 1C), which marked over 95% of the cells on FACS analysis (Figure S1B). Gene expression analysis showed prominent induction of pluripotency genes, such as SOX2, NANOG and OCT4, compared with primary myoblasts (Figure 1D). None of these parameters were affected by history of T2D.
Figure 1. Generation of Patient-Specific iPSCs and Differentiation into iMyos.
(A) Reprogramming strategy. (B) Cohort was composed of 8 non-diabetics (CTL) and 8 type 2 diabetic (T2D) subjects. Males are represented by □ symbols and females by ○ symbols. Biometric and biochemical features are shown. Data are means ± SEM, n = 6–8. * P < 0.05, ** P < 0.01, *** P < 0.001, Student’s t test. (C) Representative immunostaining of OCT4 and SSEA4, and DAPI in iPSCs (n = 3). Scale bar, 50 μm (D) Gene expression normalized to TBP of pluripotency markers in iPSCs (n = 8) and primary myoblasts (n = 3) expressed as fold-change over myoblast. Data are means ± SEM. (E) Two-step generation of iMyos in 18 days with chemically defined media. (F) OCT4, and MYOD1 western blot in donor-matched iPSCs, iMyos and primary myoblasts. GAPDH is used as loading control. (G) Representative MYOD1 immunostaining in iMyos and percentage of positive cells relative to DAPI (n = 8). Scale bar, 25 μm. See also Figure S1.
These human iPSCs were then differentiated into skeletal muscle myoblasts in a two-step process using a cocktail-based approach (Caron et al., 2016) that reproduces the hallmarks of myogenesis with early expression of PAX transcription factors followed by sequential expression of the myogenic regulators MYF5 and MYOD, and myogenin (MYOG) (Bentzinger et al., 2012). Step 1, conversion to muscle progenitors or satellite-like cells (SC-like) takes 10 days, and step 2 differentiation into myoblasts (iMyos) takes 8 days. There were no morphological distinctions between CTL and T2D cells (Figure 1E). As cells underwent differentiation, OCT4 protein fell to undetectable levels (Figure 1F), while PAX7 mRNA was highest in SC-like cells, and MYOD mRNA was highest in iMyos (Figure S1C). At the protein level, MYOD1 expression was 6-fold higher in iMyos than in primary myoblasts (Figure 1F and S1D) and over 60% of cell populations stained positive for this marker (Figure 1G). The differentiated state of iMyos was further confirmed by qPCR showing that, compared to primary myoblasts, iMyos had lower expression of MYF5 and higher levels of MYOG (Figure S1E). Similar trends were observed for myosin heavy chain isoforms (Figure S1F). iMyos also had higher mRNA levels of genes related to mitochondrial biogenesis (Figure S1G), electron transport chain and TCA cycle, while expression of glycolytic enzymes was lower than primary myoblasts (Figure S1H). Again, T2D status had no detectable influence on the differentiation efficiency of iPSCs towards the myogenic lineage.
Impaired Insulin Signaling, Glucose Uptake and Cellular Respiration in iMyos from T2D Subjects
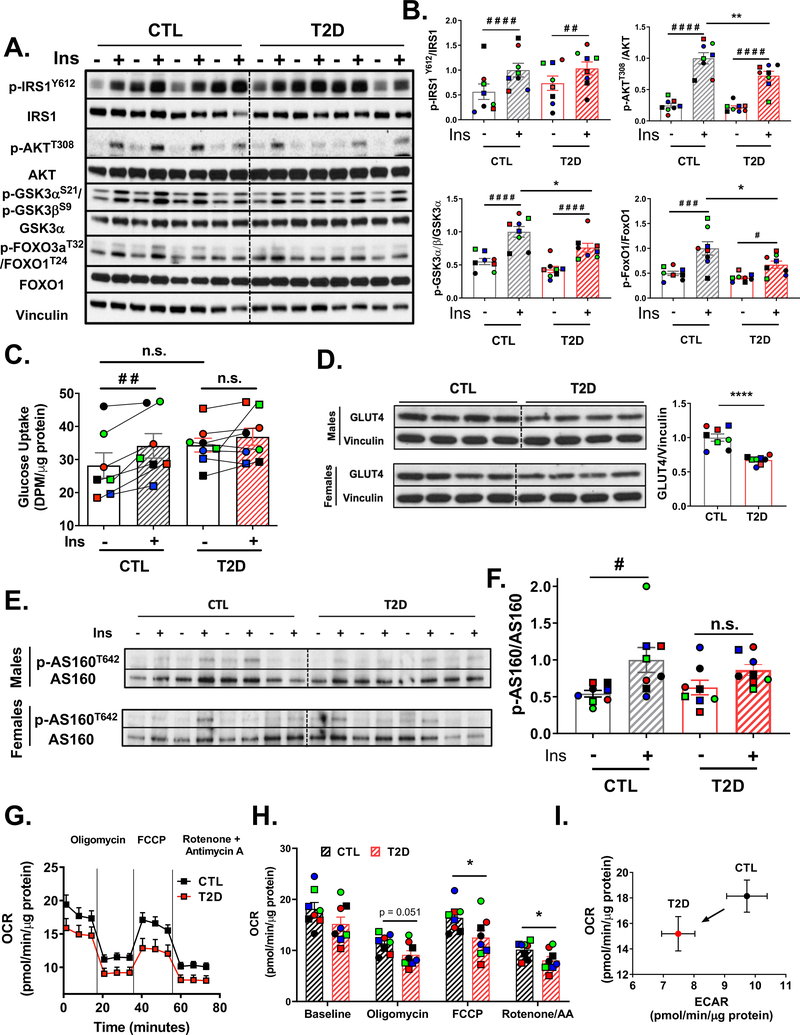
To investigate potential cell-autonomous defects related to T2D, we performed insulin signaling experiments in iMyos. Immunoblot assessment of total expression levels of key insulin signaling proteins in iMyos versus primary myoblasts revealed comparable GLUT4 and insulin receptor beta-subunit (IRβ), while protein levels of IRS-1, IRS-2, AKT2 and FOXO3a, normalized to GAPDH, were higher in general in iMyos than primary myoblasts (Figure S2A and S2B). In CTL iMyos, insulin stimulation produced a classical response inducing phosphorylation of multiple components of the insulin signaling cascade, including IRS-1, AKT, GSK3α, GSK3β, FOXO1 and FOXO3a (Figures 2A and S2C). Compared to iMyos from controls, T2D iMyos showed significantly lower insulin-induced phosphorylation of AKTT308, GSK3α S21/GSK3β S9 and FOXO1T24/FOXO3aT32 (Figure 2B). These occurred with no significant changes in protein levels. There was also a trend for decreased levels of IRS-1 protein (P = 0.059; Figure S2D), although the ratio of Y612-phosphorylated IRS-1 to total protein was unchanged in T2D iMyos. Insulin stimulation caused a significant increase in glucose uptake of 21% in CTL cells, as assessed using 2-[14C(U)]-deoxy-D-glucose (2-DOG), a magnitude typical of cultured myoblasts (Massart et al., 2017). With T2D iMyos there was a trend for elevated basal 2-DOG uptake and no significant stimulation by insulin (Figure 2C). There was also a 33% decrease in GLUT4 protein levels in T2D iMyos (P < 0.001, Figure 2D). Phosphorylation of AKT-substrate of 160 kDa (AS160, also known as TBC1D4), an important regulator of the translocation of glucose transporters to the plasma membrane, was increased by 1.9-fold upon insulin stimulation in CTL iMyos (P < 0.02), while in T2D iMyos this response was reduced to 1.4-fold and was not statistically significant (Figure 2E and 2F).
Figure 2. iMyos from T2D subjects Mirror Insulin Resistance.
(A) Insulin signaling in iMyos from male donors. (B) Quantification of insulin signaling experiments normalized by total protein. Data are means ± SEM, n = 8. # P < 0.05, ## P < 0.01, ### P < 0.001, #### P < 0.0001 basal vs insulin, * P <0.05, ** P <0.01 CTL vs T2D, n.s. = not significant, Two-way ANOVA. See also Figure S2. (C) 2-DOG uptake assay in iMyos. Data are means ± SEM, n = 7–8. ## P < 0.01 basal vs insulin, Two-Way ANOVA. (D) Total GLUT4 protein expression normalized to vinculin. Data are means ± SEM, n = 8. **** P < 0.0001, Student’s t test. (E-F) Immunoblot of AS160T642 normalized to total AS160. Data are means ± SEM, n = 8. # P < 0.05 basal vs insulin, Two Way ANOVA. (G-I) Seahorse Flux analysis showing (G) basal OCR profile or in response to oligomycin, FCCP or rotenone + antimycin A. (H) Average OCR at each condition during the assay. Data are means ± SEM, n = 8. * P < 0.05, Student’s t test. (I) Metabolic phenotyping plot of OCR vs ECAR.
An important metabolic function of insulin in muscle is to activate glycolytic and oxidative pathways for substrate utilization. Extracellular flux analysis using glucose as substrate demonstrated a trend of lower basal oxygen consumption rate (OCR) in T2D iMyos, which persisted following addition of oligomycin and was statistically significant after addition of FCCP to induce maximal respiration (Figure 2G and H). The reduced basal OCR also persisted after blockade of mitochondrial activity with a mixture of Rotenone/Antimycin A, indicating an impairment in both mitochondrial and non-mitochondrial respiration. Comparison of OCR to extracellular acidification rate (ECAR) revealed a shift downward and to the left, indicating that T2D iMyos were less metabolically active than controls with impaired glycolytic function (Figure 2I). Thus, the metabolic features of the iMyos from T2D patients closely mimic the changes observed in muscle of patients in vivo with impaired responses for insulin signaling, glucose uptake and signaling, and mitochondrial substrate oxidation.
Global Phosphoproteomics Reveals Disruption of IRS, AKT and mTORC1 Signaling in T2D
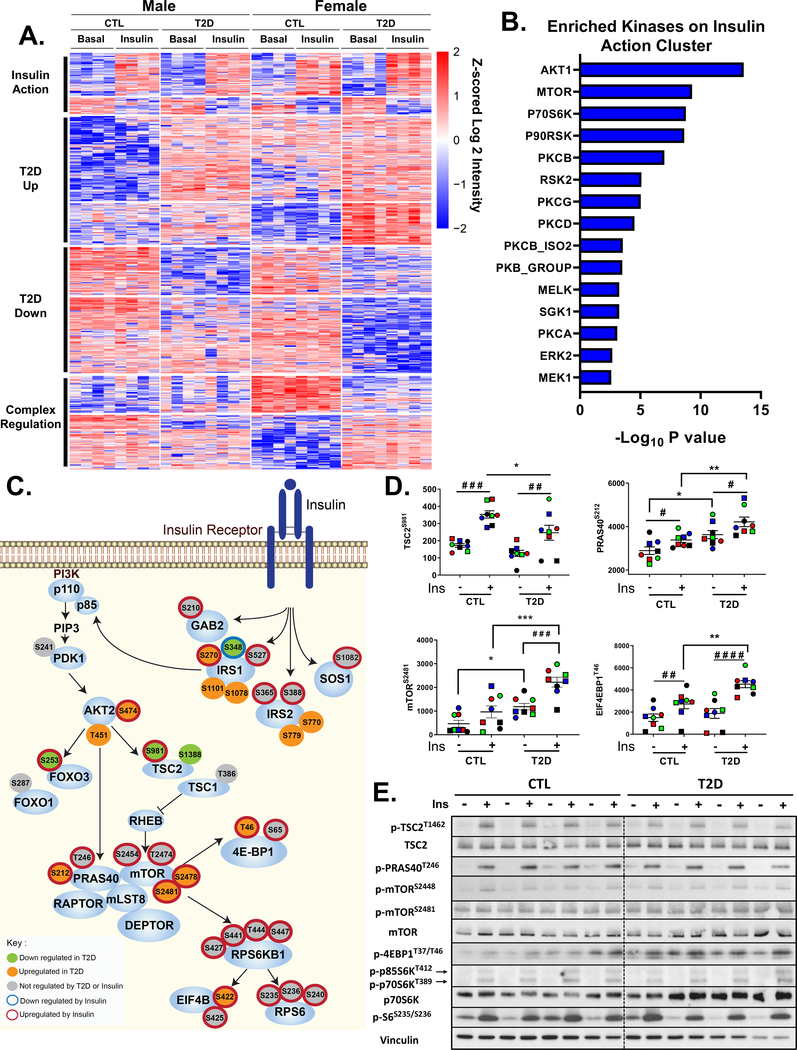
To identify the full extent of the signaling defects that contribute to T2D at the cellular level in the absence of systemic factors that might contribute to insulin resistance, we performed global phosphoproteomics analysis of all 16 CTL and T2D iMyo cell lines with or without insulin stimulation (10 min at 100 nM) in vitro (Figure S3A). Using this approach, a total of ~96,000 phosphosites were identified in all samples combined and of these 29,148 were considered class I (localization probability > 75%, average = 97%) and could be quantified (Figure S3B). The total number of phosphosites quantified per sample was similar among all cell lines (Figure S3C). Principal component analysis indicated clear separation of the phosophoproteome data with T2D as the dominant factor, followed by donor sex and insulin stimulation (Figure S3D). Heatmap analysis with hierarchical clustering revealed 1,172 significantly regulated phosphosites (FDR < 0.05). These formed four major clusters based on changes caused by insulin action and changes due to the effects of T2D to increase or decrease basal phosphorylation (Figure 3A, Table S1). In addition, one group of phosphosites affected by T2D showed regulation due to sexual dimorphism where differences were more pronounced in cells from female donors than male donors. The largest proportion of changes was observed in the diabetes-regulated clusters.
Figure 3. Phosphoproteomics Reveals Dysregulation of IRS/AKT/mTOR Signaling Nodes in T2D iMyos.
(A) Hierarchical clustering of the phosphopeptides showing the effects of insulin and T2D on the phosphoproteome. Rows are Z-scores of log2 transformed intensity of phosphosites for each sample (columns). See also Figure S3 and Table S1. (B) Overrepresented protein kinases within insulin action cluster (P < 0.01). (C) Representation of IR signaling pathway showing proximal and downstream phosphorylation events. Each of the phosphosites were color coded based on the effects of insulin or T2D on phosphorylation, Two-way ANOVA (P < 0.05). (D) Phosphosite quantification of mTORC1 signaling components. Data are means ± SEM of phosphosites intensity values (x105). # P < 0.05, ### P < 0.001, #### P < 0.0001 basal vs insulin, * P <0.05, ** P <0.01, *** P < 0.001 CTL vs T2D, Two-way ANOVA. (E) Validation of phosphoproteomics by immunoblot in iMyos from male subjects. See also Figure S4
Importantly, this phosphosite cluster distribution persisted even following correction by protein expression as estimated by MS/MS (Figure S3E, Table S2). Thus, out of the 831 protein-normalized phosphosites significantly regulated (FDR < 0.05), 613 (73.7%) had been found significantly regulated in the non-normalized analysis, and in the insulin action cluster this was even higher at 83% (Figure S3F). Other changes also persisted in the protein normalized data, but just lost statistical significance. Thus, changes in protein expression do not represent a major factor driving for the observed changes in the phosphoproteome. For all subsequent analysis, the non-normalized data were used to maximize the ability to identify key regulated pathways.
In the insulin action cluster, 125 phosphosites increased with insulin stimulation. These sites included many protein phosphorylations known to be involved in IR signaling, as well as a number of newly identified phosphorylation events. The former group included phosphorylation of IRS-1S527, GAB2S210, AKT2S474, mTORS2481, p70S6KT444, S447 and ERK2T185, Y187; the latter included phosphorylation of IRS-2S365, S388, SOS1S1082, mTORS2454, T2474, S2478, and p70S6KS427, S441 (Table S1). Also included in the insulin action cluster were many proteins outside the canonical insulin signaling pathway such as AfadinS1799 and MERIT40S29 (Figure S4A). Phosphorylation of these proteins is located within AKT consensus motifs (RxRxxS/T) and has been linked to negative feedback regulation of insulin action in adipocytes (Lundh et al., 2019) and response to DNA damage in breast epithelium (Brown et al., 2015). Insulin-induced phosphorylation of these proteins was confirmed in primary myoblasts with phosphosite-specific antibodies (Figure S4B), demonstrating the suitability of iMyos to uncover insulin responses not previously recognized in muscle. Kinase enrichment analysis (Hornbeck et al., 2015; Lee et al., 2011) of phosphosites up-regulated by insulin revealed strong enrichment for multiple kinases associated with insulin action including AKT, mTOR, the RSK kinases (p70S6K, p90RSK, RSK2) and many members of the PKC family (PKCβ, -γ, -δ, -α) (Figure 3B).
We identified 49 phosphosites in 39 proteins that were dephosphorylated within 10 minutes in response to insulin (Figure 3A and Table S1). These represented very different functional categories than those stimulated by insulin and included many nuclear proteins involved in cell cycle regulation (TOP2AS1471, TICRRS838), mitotic spindle formation and microtubule structures in the cell actomyosin cortex (CLASP1S687, S691, CLASP2S593) (Table S1). These could not be assigned to actions of a single kinase or phosphatase.
Interestingly, T2D did not cause disruption of the entire insulin action network but affected specific phosphorylation events in both the proximal (IRS-1, IRS-2) and downstream (AKT, mTOR) parts of the insulin signaling pathway. In Figure 3C we have represented these phosphosites individually and defined those that increased or decreased significantly in response to insulin stimulation (sites with red and blue halos) and those that increased or decreased significantly in T2D (sites with orange and green fills). For example, insulin significantly stimulated phosphorylation of IRS-1S270, and this effect was significantly higher in T2D iMyos. T2D also enhanced insulin-induced dephosphorylation of IRS-1S348, increased S474 phosphorylation on AKT2, impaired insulin-induced phosphorylation of AKT-consensus motifs on TSC2S981 and FOXO3S253, and increased insulin-induced phosphorylation of PRAS40S212, mTORS2475, S2481, 4EBP1T46 and EIF4BS422 in iMyos. Several of these are shown quantitated in Figure 3D. Many of these changes could be confirmed by western blot analysis using phosphosite-specific antibodies, which showed lower insulin-induced phosphorylation of TSC2 in the AKT consensus motif at T1462 and increased phosphorylation of 4EBP1T46 and mTORS2481 in T2D iMyos (Figure 3E, S4C and S4D). In addition, we identified several phosphosites on proteins involved in insulin signaling that were dysregulated in T2D iMyos in the basal state, i.e., independent of insulin action. These included increased phosphorylation of IRS-1S1101, IRS-1S1078, IRS-2S770 and IRS-2S779, and decreased phosphorylation of TSC2S1388 (Figure 3C and S4E).
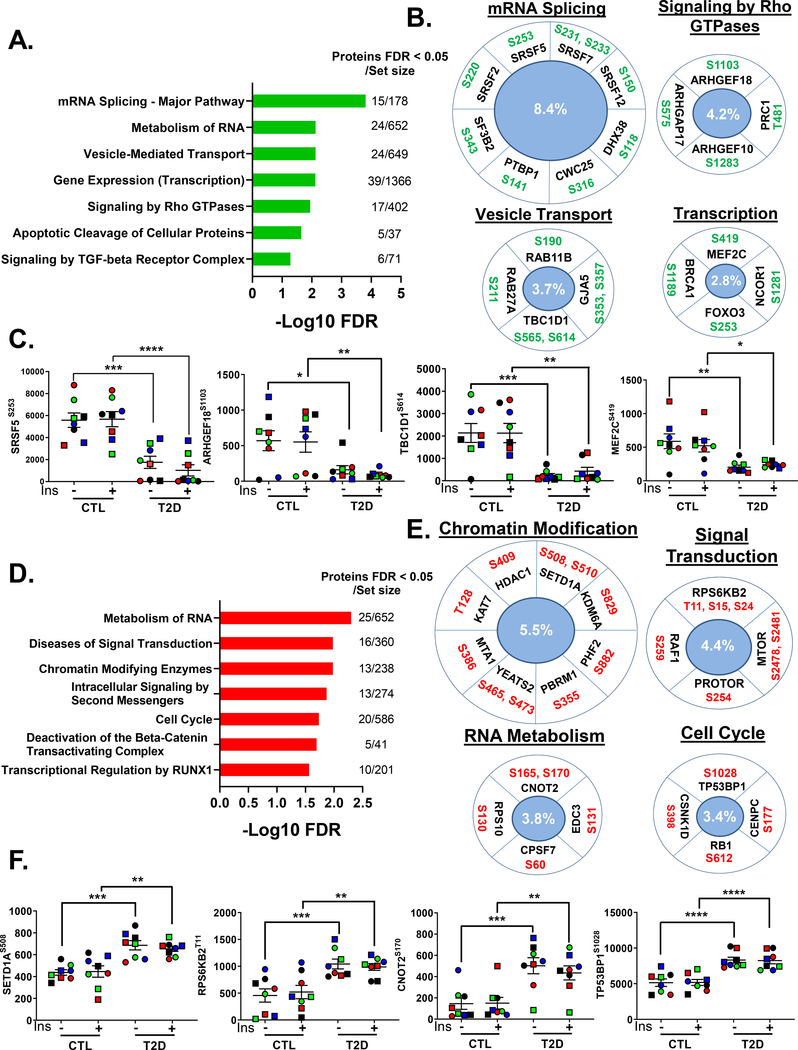
Disrupted Basal Phosphorylation of an Extensive Protein Network Underlies Diabetes Effects on Key Cellular Functions
Analysis of the full phosphoproteome revealed that in addition to changes in basal protein phosphorylation in proteins in the canonical insulin signaling pathway, there were a large number of changes in basal phosphorylation in T2D iMyos in other regulatory pathways (Figure 3A). Thus, there were 361 phosphosites on 290 proteins significantly down-regulated and 371 phosphosites on 271 proteins whose phosphorylation was increased in T2D iMyos. To gain more insight into the functions associated with this disruption of basal phosphorylation, we performed pathway and ontology analysis. For the down-regulated sites, the most enriched pathways shared by these phosphoproteins were related to mRNA splicing and mRNA metabolism, vesicle trafficking, gene transcription and signaling by Rho GTPases with up to 13% of proteins within some of these pathways affected (Figure 4A). Examples are shown and quantified in Figures 4B and Figure 4C. Proteins involved in mRNA splicing whose phosphorylation was down-regulated in T2D included core components of the spliceosome (SF3B2S343, CWC25S316) and a variety of serine/arginine (SR)-rich proteins (SRSF2S220, SRSF5S253) linked to alternative splicing regulation. There was also reduced phosphorylation of proteins involved in signaling by Rho GTPases, including multiple GEFs (ARHGEF18S1103, ARHGEF10S1283), which catalyze the exchange of GDP to GTP, and GAPs (ARHGAP17S575), which promote GTP hydrolysis rate and thus maintain GTPases in an inactive GDP-bound state. ARHGAP17 also regulates other GTPases such as RAC1, which has been implicated in insulin action (Sylow et al., 2013). Other proteins showing reduced phosphorylation included TBC1D1S565, S614 and MEF2CS419, which regulate GLUT4 trafficking and expression in skeletal muscle, respectively (Peck et al., 2009; Thai et al., 1998). These pathways were confirmed by molecular function analysis of the down-regulated phosphoproteins which showed strong enrichment for proteins with cytoskeleton and Rho GTPase binding properties and RNA, nucleic acid and transcription factor binding functions (Figure S5A).
Figure 4. Extensive Disruption of Basal Phosphorylation of Proteins Controlling Critical Cellular Functions in T2D.
(A and D) Enrichment analysis of overrepresented REACTOME pathways within (A) down-regulated or (D) up-regulated phosphosites in T2D iMyos (FDR < 0.05) (B and E) Selected phosphosites on significantly enriched pathways and (C and F) quantification of exemplary phosphosites that were (B and C) down-regulated or (E and F) up-regulated in T2D iMyos. Labels in the center indicate percentage of proteins regulated within each pathway. Data are means ± SEM of phosphosites intensity values (x105). * P <0.05, ** P <0.01, *** P < 0.001, **** P < 0.0001 CTL vs T2D, Two-way ANOVA. See also Figure S5 and Table S1.
Pathway analysis of the phosphosites whose phosphorylation was increased in the basal state in T2D iMyos showed overrepresentation of pathways related to RNA metabolism, signal transduction, chromatin modification and cell cycle (Figure 4D). Increased phosphorylation of chromatin regulators included a diverse set of proteins with acetyltransferase (KAT7T128) and methyltransferase (SETD1AS508, S510) activities, as well as deacetylases (HDAC1S409, PBRM1S355) and demethylases (KDM6AS829, PHF2S882) (Figure 4E and 4F). There was also increased phosphorylation of the epigenetic readers YEATS2S465 and PBRM1S355 in T2D iMyos, and up-regulation of phosphorylation of the mTORC2 component PRR5/PROTORS254 and the p70S6K family member RPS6KB2T11, S15, S24 (also known as S6K2), which has been linked to regulation of transcription, mRNA splicing and cell cycle. Additional nuclear pathways showing increased phosphorylation in T2D iMyos included regulators of mRNA processing, such as CCR4-NOT transcription complex subunit 2 (CNOT2S165, S170), which is involved in mRNA deadenylation, polyadenylation specific factor 7 (CPSF7S60), and the major cell cycle regulators RB1S612 and TP53BP1S1028 (Figure 4E and 4F). Interestingly, there was a strong enrichment of proteins with cytoskeleton and actin binding properties in this cluster (Figure S5B).
Analysis to identify potential kinases upstream of the phosphosites up-regulated in T2D iMyos in the basal state did not point to a single kinase, but were distributed over a variety of classes along the human kinome tree including AGC kinases (ROCK1, PKCs, PKA, P70S6K), TLK serine/threonine kinases (RAF1, DLK, ILK), CAMK-related kinases (SMMLCK, DAPK3), CMGC kinases (DYRK1A, CDC2) and STE (OSR1) kinases, which formed a network with potential targets involved in PI3K/AKT/mTOR signaling, cytoskeleton and cell cycle regulation (Figure S5C and S5D) indicating a multiplicity of mechanisms disrupted by diabetes. RNAseq analysis (Batista et al, manuscript in preparation) and western blotting for selected candidate kinases (Figure S5E) indicated no changes in the expression of any of these kinases in T2D iMyos.
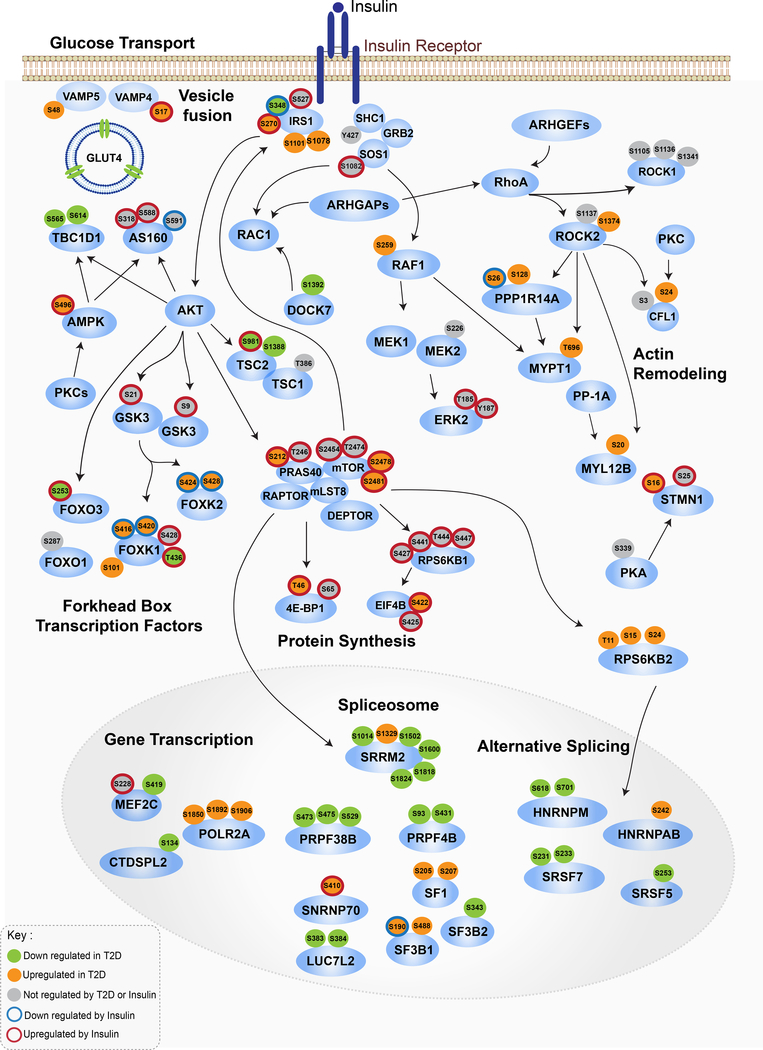
An Integrated Protein Phosphorylation Roadmap of Muscle Insulin Resistance in T2D
An integrated signaling map of the two major components in the altered phosphoproteome of T2D iMyos, namely 1) failure of insulin signaling downstream of IRS-1/2/AKT/mTOR pathways and 2) disruption of a broader network of basal phosphorylation is shown in Figure 5 with the phosphorylations which were increased or decreased in T2D shown as orange and green fills, and phosphorylations which were increased or decreased in response to insulin indicated by red and blue halos, respectively. The map reveals multiple instances whereby alterations at sites proximal (IRS-1) and downstream (AKT, mTOR) of IR signaling interact with mechanisms controlling GLUT4 vesicle trafficking and fusion, gene transcription, mRNA splicing and cytoskeleton organization. Thus, insulin-stimulated glucose transport is positively regulated by AKT and 5’-AMP-activated protein kinase (AMPK) through mechanisms that include inhibitory phosphorylation of the TBC1 domain-containing proteins TBC1D1 and AS160 (TBC1D4). Phosphoproteomics analysis revealed two phosphosites on TBC1D1 that were down-regulated in T2D, one of which, S565, is AMPK-specific (Peck et al., 2009). Phosphoproteomics also revealed that insulin-induced phosphorylation on AMPKS496 was enhanced in T2D iMyos, notably a site which inhibits enzyme activity (Heathcote et al., 2016).
Figure 5. Integrated Map of Signaling Networks Dysregulated in T2D.
Signaling cascade map showing integration between major IR signaling nodes and main pathways dysregulated in T2D identified by phosphoproteomics. Each of the phosphosites were color coded based on the effects of insulin or T2D on phosphorylation, Two-way ANOVA (P < 0.05). Arrows indicate protein-protein interactions and phosphorylation/dephosphorylation events curated from databases of experimentally defined kinase-substrate relationships (PhosphositePlus and RegPhos) and literature. See also Figure S5.
We also found a second component of the GLUT4 trafficking machinery that was defective in T2D, namely increased basal and insulin-stimulated phosphorylation of VAMP4S17 and VAMP5S48. These proteins are members of the soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptors (SNAREs) involved in fusion of intracellular vesicles with the plasma membrane. Silencing of VAMP4 and VAMP5 regulates GLUT4 and CD36 translocation implicating a role in the uptake of glucose and fatty acids (Schwenk et al., 2010). Although the role of phosphorylation on these specific sites has not been studied, serine and threonine phosphorylation on three phosphosites of VAMP8, one of which is homologous to VAMP5S48, has been shown to reduce vesicle fusion activity (Malmersjo et al., 2016).
Another factor contributing to lower glucose uptake in T2D iMyos is reduced GLUT4 protein levels. The myocyte enhancer factor 2 (MEF2) family of transcription factors control expression of several muscle metabolic genes including GLUT4, and the transcriptional activity of both MEF2A and MEF2C are reduced in muscle of insulin-deficient diabetic mice (Thai et al., 1998). Phosphorylation of MEF2C at S419 correlates with transcriptional activity (Han et al., 1997), and our data show that this phosphorylation was reduced in iMyos from T2D subjects.
Many proteins with dysregulated basal phosphorylation in T2D were related to cytoskeleton regulation (Figure S5A and S5B), and a subset of these phosphosites are potential targets of ROCK1, RAF1, SMMLCK, PKCs, and other kinases (Figure S5C and S5D). Given the central role of ROCK kinases in regulating cytoskeleton turnover and metabolism (Huang et al., 2013), these are potential drivers of a number of phosphorylation changes in the cytoskeleton seen in T2D iMyos. Consistent with this, we found changes in the phosphorylation of multiple RhoGEFs and RhoGAPs, both of which could regulate Rho-dependent ROCK activation (Figure 5 and S5F). Importantly, these ARHGAPs also modulate RAC1 activity, which together with hypophosphorylation of DOCK7S1392 and potential changes of its guanine exchange activity towards RAC1 (Watabe-Uchida et al., 2006), may impact on the cytoskeleton network controlling glucose transport.
ROCK activity is also regulated by serine phosphorylation (Lowery et al., 2007), and in the T2D iMyos there is increased phosphorylation of S1374 in ROCK2, which would be predicted to enhance its kinase activity. In T2D iMyos, increased ROCK activity is potentially linked to increased phosphorylation of non-sarcomeric myosin regulatory light chain 2 (MYL12BS20). ROCKs can also phosphorylate and inhibit the regulatory subunit of protein phosphatase 1A (PP-1A) MYPT1, which dephosphorylates MYL12B (Birukova et al., 2004). The AGC kinases ROCK, PKC and PKA further contribute to actin filament formation through inhibitory phosphorylation of the actin depolymerizing agent cofilin 1 (CFL1) (Sakuma et al., 2012) and phosphorylation of STMN1S16. Together, these phosphorylation changes in T2D are consistent with a state of impaired actin/microtubule filament turnover due to lower depolymerizing activity.
The effects of T2D may also modify gene transcription mediated by FOXO and FOXK families of forkhead box transcription factors. Insulin acts on these two families in reciprocal manner (Sakaguchi et al., 2019). Thus, insulin acting through AKT promotes phosphorylation of FOXO proteins, which results in nuclear exclusion and inhibition of their transcriptional activity (Nakae et al., 2000), whereas insulin promotes nuclear migration and increased activity of FOXK transcription factors through release from GSK3-mediated inhibitory phosphorylation (Sakaguchi et al., 2019). In T2D iMyos, insulin stimulation of FOXO3S253 phosphorylation via AKT was reduced while phosphorylation of FOXK1S416, S420/FOXK2S424/S428 on GSK3 motifs was enhanced in T2D iMyos. Collectively, these phosphorylation changes indicate that insulin resistance in T2D promotes higher transcriptional activity of FOXOs and impairs activity of FOXKs.
Altered basal phosphorylation in T2D might also affect post-transcriptional pathways, especially mRNA splicing factors. Decreases in phosphorylation were observed at multiple sites for the five small nuclear ribonucleoprotein particles (snRNPs), U1, U2 and the U4/U6.U5 tri-snRNP that compose the spliceosome, as well as phosphorylation of multiple serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs), which are also involved in modulation of alternative splicing (Figure 5). Our data indicate multiple ways by which insulin action could modulate this pathway through AKT/mTOR signaling. First, phosphorylation of SRRM2S1329 occurs on an mTOR motif (Hsu et al., 2011) suggesting direct regulation by this kinase. Second, we detected increased RPSKB2 (S6K2)T11, S15, S24 in T2D, and S6K2 has been shown to interact with transcription and splicing factors including SR proteins and hnRNPs (Pavan et al., 2016). Lastly, the phosphosites PRPF38BS475, S529 and SF3B2S343 occur within AKT consensus motifs. Thus, defects within the insulin signaling cascade are deeply integrated with the changes in basal phosphorylation of transcriptional regulators, splicing factors and cytoskeleton components observed in T2D iMyos.
Discussion
Skeletal muscle insulin resistance is the first defect detectable in individuals at high risk for T2D, and precedes and predicts disease development by many years (Warram et al., 1990). Nevertheless, the molecular defects underlying muscle insulin resistance and the extent to which it might be due to circulating factors, such as high free fatty acids or altered secretion of insulin, inflammatory cytokines or other hormones, remains unknown. Here, we show that myoblasts derived from iPSCs of T2D patients mirror many of the defects seen in skeletal muscle tissue of T2D patients, thus providing a unique disease-in-a-dish model to define the cell-autonomous molecular changes underlying these abnormalities. Using this system, we have uncovered a novel network of signaling defects in skeletal muscle underlying T2D. These defects occur both inside and outside the classical insulin signaling pathway, and in both the basal and insulin-stimulated states. Importantly, these defects occur in vitro in the absence of interference by changing levels of hormones, glucose and lipids, and thus define a unique signature of cell-autonomous changes in skeletal muscle underlying T2D pathogenesis.
The identification of the molecular defects of T2D requires an in vitro model that captures the fundamental defects associated with this condition. Physiological studies of T2D patients have revealed impaired glucose transport into muscle as a limiting factor causing downstream abnormalities of glucose utilization and glycogen formation (Cline et al., 1999; Rothman et al., 1995; Warram et al., 1990). Inefficient substrate utilization in skeletal muscle from T2D patients is also associated lower expression of oxidative metabolism genes (Mootha et al., 2003; Patti et al., 2003), and impaired ability of insulin to induce ATP production (Stump et al., 2003). All of these defects are reflected in iMyos from patients with T2D in vitro, far removed from the patient.
Metabolic actions of insulin are to a large extent downstream the PI3K/AKT branch of the insulin signaling network. Immunoblot analysis indicates that T2D iMyos exhibit defects at the level of GSK3αS21, GSK3βS9, FOXO1T24, and FOXO3aT32, all likely reflecting impaired AKT activation as a result of decreased T308 phosphorylation. Decreased AKT activity due to impaired phosphorylation of AKT on T308 has been described in skeletal muscle biopsies from T2D subjects (Karlsson et al., 2005; Krook et al., 1998). However, in our data and in studies using muscle biopsies, not all sites considered downstream of AKT are affected in T2D. Thus, despite reduced insulin-stimulated phosphorylation of FOXO1 at T24, we find no change in phosphorylation of FOXO1 at S287, although both are presumed AKT sites. Likewise, phosphorylation of FOXO1S256 was not significantly affected in muscle biopsies taken following stimulation during a euglycemic clamp (Tonks et al., 2013). Collectively, these findings suggest that T2D selectively impairs AKT action on its downstream targets.
Dysregulated serine phosphorylation of IRS-1 and IRS-2 represents a key mechanism to promote insulin resistance by interference of IRS-1/2 tyrosine phosphorylation and/or increased proteolytic degradation (Copps and White, 2012). Our analysis indicates that total IRS-1 protein tends to be reduced in T2D iMyos, and this is accompanied by increased serine phosphorylation at multiple sites. Increased serine phosphorylation of IRS-1/2 has been linked to overactivity of a variety of serine / threonine kinases including novel isoforms of PKCs (nPKCs), mTOR and S6K. Consistent with this notion, phosphorylation of IRS-1S270, S1101 is increased in T2D iMyos, and these are potential sites of S6K and nPKCs, such as PKCθ or PKCδ (Bezy et al., 2011; Li et al., 2004; Zhang et al., 2008). In addition, we find regulation of IRS-1 phosphorylation on S348 and S527 by insulin and increased phosphorylation on S1101 in T2D, mirroring findings from skeletal muscle biopsies of T2D subjects undergoing euglycemic-hyperinsulinemic clamps (Langlais et al., 2011) and amino acid-induced insulin resistance (Tremblay et al., 2007). We also observe increased phosphorylation of S770 and S779 of IRS-2 in T2D iMyos, sites not previously linked to T2D or insulin resistance. Since IRS-1 is dominant in skeletal muscle and IRS-2 is more important in liver (Previs et al., 2000), serine phosphorylation of IRS-1 likely constitutes an important factor underlying the disruption of proximal IR signaling and insulin resistance in T2D muscle.
Our phosphoproteomic approach greatly expands how insulin signaling nodes operate in T2D and points to dysregulation of several kinases, including mTOR, ROCK1/2 and the nPKCs, as parts of the signature of insulin resistance. Activation of mTORC1 results from a complex interplay between GTP-loading of RHEB, as a result of inhibition of TSC2 GAP activity by AKT (Manning and Toker, 2017), and relief of inhibitory activity on RAPTOR through AKT-mediated phosphorylation of PRAS40T246 (Sancak et al., 2007). Despite lower inhibitory phosphorylation of TSC2S981, T1462 by AKT in T2D cells, the increases of mTORS2478, S2481 autophosphorylation, 4EBP1T46 phosphorylation, and S6K-mediated EIF4BS422 phosphorylation, imply overactivity of mTORC1 in T2D. However, these increases are selective, since other targets of mTORC1, such as 4EBP1S65, S6K1S427, S441, T444, S447 and S6S235, S236, S240, are not altered by T2D. Thus, a complex interplay between PRAS40, mTOR, and potentially other kinases underlies the hyperphosphorylation of the proteins downstream of mTORC1.
The Rho-associated kinases ROCK1 and 2 are potential kinases accounting for some of the increased phosphorylation of proteins linked to actin cytoskeleton remodeling in T2D iMyos. However, their role in metabolism is complex with both positive and negative effects on glucose homeostasis, possibly related to effects of different isoforms. ROCK1 knockout or knockdown causes insulin resistance in vivo (Lee et al., 2009) and reduces glucose transport in vitro (Chun et al., 2012). However, in insulin resistant HFD-fed mice, expression of a dominant negative form of ROCK or treatment with a ROCK inhibitor ameliorates glucose intolerance (Noda et al., 2014). Haploinsufficiency of ROCK2 prevents insulin resistance induced by a high-fat diet in mice (Soliman et al., 2015). Our phosphorylation analysis indicates that there is increased phosphorylation of proteins downstream of ROCK1/2, accompanied by increased ROCK2S1374 phosphorylation in T2D iMyos. ROCK activity could also be increased by altered phosphorylation of one or more RhoGEFs or RhoGAPs. Given the important role of dynamic remodeling of the actin network in mediating metabolic functions of insulin in muscle (Klip et al., 2014), clearly more work investigating the role of Rho/ROCK pathway in T2D is warranted.
Protein kinase C (PKC) is a family of serine/threonine kinases that play important roles in cell growth, differentiation, apoptosis, and hormonal responses. PKCs are subclassified into conventional PKCs (α, βI, βII, γ), novel PKCs (δ, ε, ν, θ), and atypical PKCs (ζ, ι, λ). Several PKC isoforms have been implicated in both insulin action and insulin resistance (Farese et al., 2014). Activation of novel PKCs, especially PKCs δ, ε, and θ, by hormones, hyperglycemia, and lipids, especially diacylglycerol, has been shown to contribute to insulin resistance (Bezy et al., 2011; Li et al., 2004; Samuel et al., 2007). Importantly, to whatever extent PKCs contribute to the phenotypes observed in iMyos, this is occurring in the absence of these circulating factors.
The impaired OCR response to inhibitors of mitochondrial respiration and the lower ECAR activity, indicate that glycolytic and mitochondrial functions are also compromised in T2D iMyos. Two recent studies suggest that this may be due to alterations in the forkhead transcription factors FOXK1/K2 (Sakaguchi et al., 2019; Sukonina et al., 2019). In contrast to FOXOs, which are turned off by insulin, FOXKs translocate to the nucleus in response to insulin by release of inhibitory phosphorylation mediated by GSK3 on mouse FOXK1 at serines 402/406 and 454/458 (corresponding to human FOXK1S416/S420, S468/S472), and this may regulate expression of genes involved in cell cycle, apoptosis and lipid metabolism, at least in hepatocytes (Sakaguchi et al., 2019). Phosphorylation in some of these GSK3 candidate phosphosites on FOXK1 (S416/S420) and FOXK2 (S424/S428) is increased in T2D iMyos, consistent with inefficient inactivation of GSK3 by AKT. Moreover, FOXKs are important components regulating glucose uptake and lactate production in myocytes (Sukonina et al., 2019). Thus, impaired transcriptional activity of FOXKs due to enhanced inhibitory phosphorylation may contribute to the dysfunctional glucose uptake and cellular respiration in T2D.
A new dimension uncovered in our study is the extensive dysregulation in the phosphorylation of nuclear proteins, especially those involved in mRNA splicing. This extends our previous finding that insulin signaling can regulate a large number of genes involved in mRNA splicing in muscle in vivo (Batista et al., 2019). In recent years, abnormal spliceosome function has been linked to obesity (Vernia et al., 2016) and T2D (Pihlajamaki et al., 2011), however how these metabolic derangements influence spliceosome activity or vice versa is largely unknown. Dynamic cycles of phosphorylation are required for constitutive and alternative splicing, and often involve the activity of two families of splicing kinases – the cyclin-dependent-like kinases (CLKs) and SR-protein kinases (SRPKs). These kinases integrate signals from the external environment through interactions with other signaling effectors such as AKT and mTOR. AKT-dependent phosphorylation of CLK1 regulates glucose uptake in myocytes, and this signaling axis is dysregulated in diabetic mice (Jiang et al., 2009). Similarly, SRPK2 is phosphorylated by mTOR/S6K1, and this is required for SREBP-dependent expression of lipogenic genes in a cell model of cancer (Lee et al., 2017). Although we do not detect enrichment of these kinases by analysis of phosphosites in our dataset, they remain candidates for further investigation, since there is very limited knowledge of the substrates for this class of kinases (Needham et al., 2019).
While much remains to be learned about the fundamental defect(s) leading to the altered network of phosphorylation observed in this study, several points are worth considering. First, the defects in skeletal muscle insulin action and mitochondrial oxidation persist in vitro, thus indicating that changes in circulating hormones and metabolites may not be required to initiate the signaling defects leading to T2D. Second, the changes observed in iMyos in vitro occur after genetic reprogramming and dedifferentiation-redifferentiation of cells, suggesting that they are driven by either a genetic or persistent epigenetic change. However, the nature of this defect is difficult to link to the many polygenes currently associated to T2D, since the exact combination of genetic variants would differ from patient to patient and cell line to cell line. Clearly, we must think about new ways by which genetics and epigenetics can play a role in the cellular defects observed in T2D. Third, using existing analysis tools, no single kinase or phosphatase could be identified as the master regulator of the changes in phosphorylation observed in the T2D iMyos, pointing to cumulative signaling defects possibly driven by some factor that concertedly impacts on the activity of several kinases or phosphatases. Such factors could include, but are not limited to, changes in intracellular ions concentrations, redox state or ATP as it has been recently proposed (Su et al., 2019). Identifying this factor should be a high priority for future research, since it would be a novel target for therapy of T2D.
In summary, iMyos from T2D patients exhibit insulin resistance and mirror the key functional defects of glucose transport, mitochondrial respiration and insulin signaling in vitro removed from all systemic factors. These abnormalities are associated with a unique signature of altered protein phosphorylation, involving not only the canonical insulin signaling pathway but also regulatory loops involved in gene transcription, mRNA splicing, chromatin remodeling, vesicular trafficking, and cytoskeleton remodeling. Future work must be aimed at how genetic and potential residual epigenetic changes can create this complex cell-autonomous signature of T2D.
Limitations of the Study
Using a disease-in-a-dish model, we have uncovered a multidimensional network of signaling defects underlying skeletal muscle insulin resistance in T2D. While this represents a significant step forward towards elucidating the pathogenesis of the disease, there are limitations to consider. First, T2D is very heterogeneous, both clinically and genetically, and it is impossible for practical reasons to represent all potential subgroups in this study. Therefore, we chose to focus on a “typical” T2D population characterized by middle-aged people of European ancestry with higher BMI and chronic hyperglycemia. Second, by the nature of the study design, the iMyo system, while appropriate for defining cell type-specific molecular signatures, may not capture the cellular heterogeneity of whole muscle tissue or potential effects secondary to changes in blood flow or innervation which might occur in diabetic patients. Third, as with any in vitro model, iMyos are supported by an artificial culture media that may not reflect all aspects of in vivo physiology. However, it is a powerful system for studying cell-autonomous effects without interference of systemic factors associated with T2D as long as cells from non-diabetic controls are studied in parallel under identical conditions. Fourth and lastly, despite evidence from the literature that epigenetic effects are largely reset in iPSC-derivatives (Frobel et al., 2014), the role of epigenetics in regulating the changes observed in iMyos remains unclear and further investigation is warranted.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for reagents and resources should be directed to and will be fulfilled by the Lead Contact, C. Ronald Kahn (c.ronald.kahn@joslin.harvard.edu).
Materials Availability
Unique materials and resources generated in this study are available from the Lead Contact upon reasonable request.
Data and Code Availability
All mass spectrometry (MS) raw files acquired for this study are available at ProteomeXchange Consortium via the PRIDE partner repository under the identifier PXD015430.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study Subjects
All procedures were approved by the ethics committee at the Karolinska Institute. An informed consent was obtained from study participants. Color-coding, biometric and biochemical features are shown in Figure 1A. Biopsies from vastus lateralis muscle were collected from subjects with and without history of T2D under local anesthesia (5 mg/mL lidocaine hydrochloride) in chilled PBS containing a mixture of penicillin (100 U/mL) and streptomycin (100 μg/mL). Primary satellite cells were isolated and cultured as previously described (Al-Khalili et al., 2006).
iPSC Derivation and Maintenance
Primary myoblasts were seeded (4×104) in 6-well plates pre-coated with 0.1% gelatin for 2 hours and maintained on DMEM/F12 media supplemented with 20% FBS, 1% Pen Strep, 1% Glutamax. After 48 hours, cells were transduced using a CytoTune-iPS 2.0 Sendai Reprogramming Kit (MOI KOS:hc-Myc:hKLF4 = 5:5:3) (Thermo Fisher) and incubated overnight. Media was replaced every other day. On day 7, transduced myoblasts were treated with trypsin and transferred to 10 cm plates on top of irradiated mouse embryonic fibroblasts (MEFs) seeded at 1×106/plate the day before. From this point onwards, cells were grown on iPSC medium (DMEM/F12, 1% Glutamax, 20% KnockOut Serum Replacement, 1% MEM-NEAA, 55 μM β-mercaptoethanol, 10 μg/mL bFGF) with daily media changes. Colonies appeared within 2–4 weeks from transduction and were hand-picked within 1–2 weeks after appearance into plates coated with hESC-qualified Matrigel (Corning). Feeder-free iPSCs were cultured with mTeSR1 (StemCell Technologies) and passaged as aggregates at least once a week using ReLeSR (StemCell Technologies). Absence of Sendai virus genome was confirmed after 16–23 passages by RT-PCR following the instructions from the vendor (Thermo Fisher).
Myogenic Differentiation of iPSCs
iPSCs were differentiated into SC-Like cells and then into iMyos as previously described (Caron et al., 2016)1. First, approximately 7×103 iPSCs/cm2 were seeded onto collagen I-coated plates (Biocoat, Fisher) and grown for 10 days in skeletal muscle induction media containing 5 % horse serum (HS), 50 μg/mL fetuin, 3 μM CHIR99021, 2 μM Alk 5 inhibitor, 1 ng/mL bFGF, 10 ng/mL human recombinant epidermal growth factor (hr-EGF), 10 μg/mL insulin, 0.4 μg/mL dexamethasone, 10 μM Y27632 and 200 μM ascorbic acid with media change every other day, resulting in SC-like cells. Second, SC-like cells were treated with trypsin and approximately 7×103 iPSCs/cm2 were seeded onto collagen I-coated plates (Biocoat, Fisher) and grown for 8 days in skeletal myoblast media containing 5% HS, 50 μg/mL fetuin, 10 μg/mL insulin, 0.4 μg/mL dexamethasone, 10 μM Y27632, 10 ng/mL hr-EGF, 20 ng/mL hr-hepatocyte growth factor, 10 ng/mL hr-platelet-derived growth factor (PDGF-AB), 10 ng/mL oncostatin M, 20 ng/mL bFGF, 10 ng/mL insulin-like growth factor 1, 2 μM SB431542, and 200 μM ascorbic acid with media change every other day, resulting in iMyos.
METHOD DETAILS
Immunostaining and FACS analysis
For immunostaining, iPSCs from male subjects grown on 12-well plates or iMyos from all subjects grown on 24-well plates were fixed with 4% paraformaldehyde (Santa Cruz), permeabilized with 0.1% Triton-X and blocked with 10% normal goat serum for 10 min, with PBS washes in between steps. Plates were incubated with the primary antibodies SSEA4 (MAB4304, Millipore, 1:500 dilution), OCT4 (sc-9081, Santa Cruz, 1:100 dilution) or MYOD (sc-760, Santa Cruz, 1:100 dilution) overnight at 4 °C. Following incubation, cells were washed with PBS and incubated with appropriate secondary antibodies for 30 min and washed with PBS before imaging with an Olympus IX51 inverted fluorescence microscope.
For fluorescence activated cell sorting (FACS) analysis, iPSCs were dissociated to single cells using accutase (Stem Cell Tech). Unpermeabilized cells were washed in PBS with 2% FBS prior to fixation in 4% Paraformaldehyde and blocking is 0.5% BSA. Cells were stained with antibodies for SSEA4 - PerCP-Cy5.5 (#561565, BD Pharmingen) primary antibody and with a Propidium Iodide Staining Solution (eBiosciences) viability dye. Cells were analyzed on the BD LSR-II Flow Cytometer.
RNA Isolation and Semi-Quantitative qPCR
Total RNA from all cell types included in the study was isolated using TRIzol reagent (Thermo Fisher Scientific), followed by chloroform/isopropanol/ethanol extraction. Complementary DNA (cDNA) was synthesized from 400 ng RNA using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). qPCR reactions were prepared using iQ SybrGreen Supermix (Bio-Rad, catalog 1708884) and run on a C1000 Thermal Cycler (BioRad, catalog CFX384). TATA box binding protein (Tbp) or ribosomal subunit 18S were used to normalize gene expression. Primer sequences used are listed in Table S3.
Insulin Signaling and Immunoblotting
For insulin stimulation, iMyos were washed in PBS and incubated with starvation media (Ham’s F10 + 0.1% BSA) for 5 h before stimulation with 100 nM insulin for 10 min. Cells were then washed in ice cold PBS, snap frozen in liquid nitrogen and stored at −80 °C until analysis. Cells were homogenized in RIPA buffer (EMD Millipore) supplemented with protease and phosphatase inhibitors (Biotool). Equal protein ammounts (~ 10 μg) were resolved by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (EMD Millipore). Membranes were immunoblotted with the indicated antibodies: IRβ (sc-711, Santa Cruz), p-IRS-1Y612 (09–432, Millipore), IRS-1 (611394, BD), IRS-2 (sc-8299, Santa Cruz), p-AKTT308 (#4056, Cell Signaling), AKT pan (#4685, Cell Signaling), AKT2 (07–372, Millipore), p-GSK3αS21/βS9 (# 8566, Cell Signaling), GSK3α (# 9338, Cell Signaling), p-FOXO1T24/FOXO3aT32 (#9464, Cell Signaling), FOXO1 (#9454, Cell Signaling), FOXO3a (#12829, Cell Signaling), GLUT4 (ab654, Abcam), p-AS160T642 (44–1071G, Thermo Fisher Scientific), AS160 (# 2670, Cell Signaling), p-TSC2T1462 (# 3611, Cell Signaling), TSC2 (# 4308, Cell Signaling), p-PRAS40T246 (#13175, Cell Signaling), p-mTORS2448 (# 5536, Cell Signaling), p-mTORS2481 (# 2974, Cell Signaling), mTOR (# 2972, Cell Signaling), p-4EBP1T37, T46 (# 2855, Cell Signaling), p-p70S6KT389 (# 9205, Cell Signaling), p70S6K (# 9202, Cell Signaling), p-S6S235, S236 (# 2211, Cell Signaling), ROCK1 (# 4035, Cell Signaling), RAF1 (# 9422, Cell Signaling), p-PKAT197 (# 5661, Cell Signaling), PKCd (# 9616, Cell Signaling), DAPK3 (# 2928, Cell Signaling), OCT4 (sc-9081, Santa Cruz), MYOD (sc-760, Santa Cruz), AfadinS1799 (isoform 1)/AfadinS1718 (isoform 3) (# 5485, Cell Signaling), MERIT40S29 (#12110, Cell Signaling), H3 (#4499, Cell Signaling), H3K27me3 (07–449, Millipore), H3Ac (06–599, Millipore), GAPDH (#5174, Cell Signaling), Vinculin (#3574, Chemicon).
Glucose Uptake
iMyos grown in 12-well plates were serum starved (Ham’s F10 + 0.1% BSA) overnight, washed with PBS and incubated with Krebs-Ringer bicarbonate HEPEs (KRBH) buffer (120 mM NaCl, 10 mM NaHCO3, 4 mM KH2PO4, 1 mM MgSO4, 1 mM CaCl2, 30 mM HEPES) for 30 min at 37°C, then stimulated with 200 nM insulin for 40 min. During the last 10 min of insulin stimulation, 50 μL reaction buffer containing 0.1 μCi 2-[14C(U)]-deoxy-D-glucose (2-DOG, Perkin Elmer) and 200 μM non-radiolabeled 2-DOG (Sigma) was added to each well to a total reaction volume of 500 μL and incubated at room temperature. Glucose uptake reaction was stopped by addition of 50 μL 200 mM non-radiolabeled 2-DOG and immediate transfer of the plates to an ice bath. Cells were washed three times with PBS and lysed with 120 μL 0.1% SDS diluted in PBS followed by scintillation counting.
Assessment of Mitochondrial Function
The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a XFe96 Seahorse Extracellular Flux Analyzer (Seahorse Bioscience, MA, USA) according to the manufacturer’s instructions. One day prior to the assay, day 17 iMyos were treated with trypsin and seeded at 5×104 cells/well of a Seahorse XF96 well plate pre-coated with 0.1% gelatin (Stem Cell Technologies) for 2 h. Cells were washed twice and incubated with bicarbonate-free running buffer (DMEM containing 5.6 mM glucose, 1 mM sodium pyruvate, 1% Glutamax, pH=7.4) for 1 h at 37°C without CO2. Plate was transferred to the equipment and after a 15 min equilibration period, OCR and ECAR activities were recorded every 4 to 5 min (3 measurements per condition) at the basal state and after pneumatic injection of oligomycin (2 μM final concentration), carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP, 0.5 μM), and rotenone (0.1 μM) plus antimycin A (2.5 μM). After the assay, media was discarded, and cells were lysed with a 0.1% SDS solution followed by determination of protein concentration with a BCA kit (Thermo Fisher Scientific) used to normalize OCR and ECAR data.
Phosphoproteome Analysis of iMyos
Lysis and digestion
For phosphoproteomic analysis, serum-starved iMyos were stimulated with insulin as described above (100 nM, 10 min) and were processed according to the protocol described previously (Humphrey et al., 2018). In brief, cells were washed thoroughly in ice cold PBS to freeze insulin action, lysed immediately in SDC digestion buffer (4% SDC, 100 mM Tris pH8.5) and snap frozen. The samples were boiled at 95°C for 5 min, sonicated for 20 cycles in Biorupter plus (Diagenode), vortexed for 10 sec and protein concentration was determined by BCA assay. Per condition, 750 μg of protein lysate was used, alkylated with 10 mM CAA and reduced with 40 mM TCEP by incubating for 20 min on ice in dark. The samples were mixed with LysC and Trypsin (1:100 ratio) proteases and incubated overnight at 37°C, 1200 rpm in ThermoMixer.
Phosphopeptide enrichment
To the digested peptides 750 μl ACN and 250μl TK buffer (36% TFA and 3mM KH2PO4) were added and mixed in ThermoMixer for 30 seconds (1500 rpm). Debris was cleared by centrifugation at 13,000 rpm for 15 min and supernatant transferred to 2 ml Deep Well Plate (Eppendorf). For the phosphopeptide enrichment TiO2 beads (prepared in 80%ACN, 6%TFA buffer) were added (1:10 ratio protein/beads) and incubated at 40°C, 2000 rpm, 5 min in ThermoMixer. The TiO2 bound phosphopeptides were subsequently pelleted by centrifugation, transferred to clean tubes and washed 4 times in wash buffer (60% ACN, 1% TFA) to remove nonspecific or non-phosphorylated peptides. The beads were suspended in transfer buffer (80% ACN, 0.5% Acetic acid) and transferred on top of single layer C8 Stage Tips (stop-and-go-extraction tips) and centrifuged until dryness. The phosphopeptides were eluted with elution buffer (40% ACN, 20% NH4OH) and concentrated in a SpeedVac for 20 min at 45°C followed by phosphopeptides acidification by addition of 100 μl of 1% TFA. The acidified peptides were loaded on to equilibrated SDBRPS (styrenedivinylbenzene–reversed phase sulfonated, 3M Empore) Stage Tips for desalting and further clean up. The phosphopeptides containing SDBRPS StageTips were washed once in isopropanol/1% TFA and twice with 0.2% TFA. Finally, the desalted phosphopeptides were eluted with 60 μl of elution buffer (80%, 1.25% NH4OH). The dried elutes were resuspended in MS loading buffer (3% ACN, 0.3% TFA) and stored at −20°C until MS measurement.
LC-MS/MS measurement
The phosphopeptides were analyzed using Q Exactive HF-X Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fischer Scientific) coupled online to a nanoflow EASY-nLC1000 HPLC (Thermo Fisher Scientific). Briefly, the phosphopeptides were loaded onto an in house packed 50 cm C18 column with a 75 μM inner diameter (1.9 μM ReproSil particles, Dr. Maisch GmbH). The temperature of the column was maintained at 50°C by an in-house made column oven. The phosphopeptides were separated in a duration of 140-minute gradient with two mobile phase system buffer A (0.1% formic acid) and buffer B (60% ACN plus 0.1% formic acid) at a flow rate of 300 nl/min. The electro sprayed peptides were analyzed by the Q Exactive HF-X Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fischer Scientific) in a data dependent mode, with one survey scan at a target of 3×106 ions (300–1650 m/z, R=60,000 at 200 m/z), followed by Top10 MS/MS scans with HCD (high energy collisional dissociation) based fragmentation (target 1×105 ions, maximum filling time 120ms, Isolation window 1.6 m/z, and normalized collision energy 27%), detected in the Orbitrap (R=15,000 at 200 m/z). Apex trigger 4 to 7s, charge exclusion (unassigned, 1, 5, −8 & >8), and dynamic exclusion 40s were enabled.
QUANTIFICATION AND STATISTICAL ANALYSIS
Phosphoproteome Data analysis
The acquired raw files were processed using Maxquant (Cox and Mann, 2008) software environment (version 1.5.5.2) with the built in Andromeda search engine for identification and quantification of phosphopeptides. The data were searched using a target-decoy approach with a reverse database against Uniprot Human (August 2016 version) reference proteome fasta file with a false discovery rate of less than 1% at the level of proteins, peptides and modifications using minor changes to the default settings as follows: oxidized methionine (M), acetylation (protein N-term) and in case of phosphopetide search phospho (STY) was selected as variable modifications, and carbamidomethyl (C) as fixed modification. A maximum of 2 missed cleavages were allowed, a minimum peptide length of seven amino acids and enzyme specificity was set to Trypsin. In addition, the match between run algorithm was enabled. The Maxquant output phospho (STY) table was processed using Perseus (Tyanova et al., 2016) (version 1.5.2.11) software suite and prior to the analysis contaminants marked as potential contaminants and reverse hits were filtered out. Phosphopeptides that had more than 80% valid values in at least one group were selected for downstream analysis. Missing values were replaced by random numbers that were drawn from normal distributions with means that were down-shifted by 1.6 -fold of sample standard deviations from the sample means and standard deviations that were shanked to 0.6-fold of the sample standard deviations. Values were further normalized to make all samples to have the same median log intensity. Statistical significance of phosphopetides was assessed with empirical Bayesian linear modeling and moderated F-test using the limma package with default priors (Ritchie et al., 2015). P-values were corrected using the Benjamini-Hochberg false discovery rate (FDR), and FDR < 0.05 was considered statistically significant. Hierarchical cluster analysis was performed based on the Euclidean distance of the significant phosphopetides. Clusters were defined according to the hierarchical tree. Pathway and ontology analysis of protein clusters was done using STRING database (Szklarczyk et al., 2019). Protein sets based on human kinase substrates (PhosphositePlus and RegPhos) were tested using the Fisher exact test. Heatmaps were created with the pheatmap package. PCA plot was created with the ggplot2 package.
Statistical Analysis
Data are presented as means ± SEM. Comparisons between two groups was performed using two-tailed Student’s t test. Comparisons between two groups and two nominal variables (e.g. basal vs insulin) was performed using Two-way ANOVA matched for subject followed by Sidak’s correction. Significance level was set at P < 0.05.
Supplementary Material
Table S1. Hierarchical Clustering of Phosphoproteome. Average Intensity (Log2) and Hierarchical Clustering of Regulated Phosphosites, Related to Figure 3 and Figure 4.
Table S2. Hierarchical Clustering of Protein-Normalized Phosphoproteome. Average Intensity (Log2) and Hierarchical Clustering of Regulated Phosphosites Normalized by Total Protein Expression, Related to Figure 3.
Acknowledgements
We thank Andrew Minotti and Farzana Hakim from the iPS Core Facility of the Joslin Diabetes Center, and Dirk Wischnewski, Dr. Igor Paron, Christian Deiml, and Alex Strasse from the Proteomics and Signal Transduction Department at Max Planck Institute of Biochemistry for excellent technical assistance. This work was supported by NIH grants R01DK031036, R01DK033201 (to C.R.K.), P30DK036836 (to Joslin Diabetes Center), and the Mary K. Iacocca Professorship (to C.R.K.). Additional support was received from the Swedish Research Council (2015-00165) and the Novo Nordisk Foundation Center for Basic Metabolic Research at the University of Copenhagen (NNF18CC0034900) (to J.R.Z). T.M.B. was partially supported by grant from Sao Paulo Research Foundation (2014/25370-8).
Footnotes
Declaration of Interests
The authors declare no competing interests.
and personal communication from Dr. Leslie Caron, Ph.D.
References
- Al-Khalili L, Bouzakri K, Glund S, Lonnqvist F, Koistinen HA, and Krook A (2006). Signaling specificity of interleukin-6 action on glucose and lipid metabolism in skeletal muscle. Mol Endocrinol 20, 3364–3375. [DOI] [PubMed] [Google Scholar]
- Batista TM, Garcia-Martin R, Cai W, Konishi M, O’Neill BT, Sakaguchi M, Kim JH, Jung DY, Kim JK, and Kahn CR (2019). Multi-dimensional Transcriptional Remodeling by Physiological Insulin In Vivo. Cell Rep 26, 3429–3443 e3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF, Wang YX, and Rudnicki MA (2012). Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezy O, Tran TT, Pihlajamaki J, Suzuki R, Emanuelli B, Winnay J, Mori MA, Haas J, Biddinger SB, Leitges M, et al. (2011). PKCdelta regulates hepatic insulin sensitivity and hepatosteatosis in mice and humans. J Clin Invest 121, 2504–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gupta MK, Khamaisi M, Martinez R, Gritsenko MA, Wagner BK, Guye P, Busskamp V, Shirakawa J, Wu G, et al. (2015). Preserved DNA Damage Checkpoint Pathway Protects against Complications in Long-Standing Type 1 Diabetes. Cell metabolism 22, 239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birukova AA, Smurova K, Birukov KG, Usatyuk P, Liu F, Kaibuchi K, Ricks-Cord A, Natarajan V, Alieva I, Garcia JG, et al. (2004). Microtubule disassembly induces cytoskeletal remodeling and lung vascular barrier dysfunction: role of Rho-dependent mechanisms. J Cell Physiol 201, 55–70. [DOI] [PubMed] [Google Scholar]
- Boucher J, Kleinridders A, and Kahn CR (2014). Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KK, Montaser-Kouhsari L, Beck AH, and Toker A (2015). MERIT40 Is an Akt Substrate that Promotes Resolution of DNA Damage Induced by Chemotherapy. Cell Rep 11, 1358–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart AM, Tan K, Warren L, Iovino S, Hughes KJ, Kahn CR, and Patti ME (2016). Insulin Resistance in Human iPS Cells Reduces Mitochondrial Size and Function. Sci Rep 6, 22788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron L, Kher D, Lee KL, McKernan R, Dumevska B, Hidalgo A, Li J, Yang H, Main H, Ferri G, et al. (2016). A Human Pluripotent Stem Cell Model of Facioscapulohumeral Muscular Dystrophy-Affected Skeletal Muscles. Stem Cells Transl Med 5, 1145–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun KH, Araki K, Jee Y, Lee DH, Oh BC, Huang H, Park KS, Lee SW, Zabolotny JM, and Kim YB (2012). Regulation of glucose transport by ROCK1 differs from that of ROCK2 and is controlled by actin polymerization. Endocrinology 153, 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline GW, Petersen KF, Krssak M, Shen J, Hundal RS, Trajanoski Z, Inzucchi S, Dresner A, Rothman DL, and Shulman GI (1999). Impaired glucose transport as a cause of decreased insulin-stimulated muscle glycogen synthesis in type 2 diabetes. N Engl J Med 341, 240–246. [DOI] [PubMed] [Google Scholar]
- Copps KD, and White MF (2012). Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia 55, 2565–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cusi K, Maezono K, Osman A, Pendergrass M, Patti ME, Pratipanawatr T, DeFronzo RA, Kahn CR, and Mandarino LJ (2000). Insulin resistance differentially affects the PI 3-kinase- and MAP kinase-mediated signaling in human muscle. J Clin Invest 105, 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFronzo RA, and Tripathy D (2009). Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care 32 Suppl 2, S157–S163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducluzeau PH, Perretti N, Laville M, Andreelli F, Vega N, Riou JP, and Vidal H (2001). Regulation by insulin of gene expression in human skeletal muscle and adipose tissue. Evidence for specific defects in type 2 diabetes. Diabetes 50, 1134–1142. [DOI] [PubMed] [Google Scholar]
- Farese RV, Lee MC, and Sajan MP (2014). Atypical PKC: a target for treating insulin-resistant disorders of obesity, the metabolic syndrome and type 2 diabetes mellitus. Expert Opin Ther Targets 18, 1163–1175. [DOI] [PubMed] [Google Scholar]
- Frobel J, Hemeda H, Lenz M, Abagnale G, Joussen S, Denecke B, Saric T, Zenke M, and Wagner W (2014). Epigenetic rejuvenation of mesenchymal stromal cells derived from induced pluripotent stem cells. Stem Cell Reports 3, 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Jiang Y, Li Z, Kravchenko VV, and Ulevitch RJ (1997). Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386, 296–299. [DOI] [PubMed] [Google Scholar]
- Heathcote HR, Mancini SJ, Strembitska A, Jamal K, Reihill JA, Palmer TM, Gould GW, and Salt IP (2016). Protein kinase C phosphorylates AMP-activated protein kinase alpha1 Ser487. Biochem J 473, 4681–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RR, Abrams L, Nikoulina S, and Ciaraldi TP (1995). Insulin action and glucose metabolism in nondiabetic control and NIDDM subjects. Comparison using human skeletal muscle cell cultures. Diabetes 44, 936–946. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, and Skrzypek E (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43, D512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. (2011). The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Lee DH, Zabolotny JM, and Kim YB (2013). Metabolic actions of Rho-kinase in periphery and brain. Trends Endocrinol Metab 24, 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey SJ, Karayel O, James DE, and Mann M (2018). High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat Protoc 13, 1897–1916. [DOI] [PubMed] [Google Scholar]
- Iovino S, Burkart AM, Kriauciunas K, Warren L, Hughes KJ, Molla M, Lee YK, Patti ME, and Kahn CR (2014). Genetic insulin resistance is a potent regulator of gene expression and proliferation in human iPS cells. Diabetes 63, 4130–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovino S, Burkart AM, Warren L, Patti ME, and Kahn CR (2016). Myotubes derived from human-induced pluripotent stem cells mirror in vivo insulin resistance. Proc Natl Acad Sci U S A 113, 1889–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Patel NA, Watson JE, Apostolatos H, Kleiman E, Hanson O, Hagiwara M, and Cooper DR (2009). Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCbetaII messenger ribonucleic acid. Endocrinology 150, 2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang LQ, Duque-Guimaraes DE, Machado UF, Zierath JR, and Krook A (2013). Altered response of skeletal muscle to IL-6 in type 2 diabetic patients. Diabetes 62, 355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson HK, Zierath JR, Kane S, Krook A, Lienhard GE, and Wallberg-Henriksson H (2005). Insulin-stimulated phosphorylation of the Akt substrate AS160 is impaired in skeletal muscle of type 2 diabetic subjects. Diabetes 54, 1692–1697. [DOI] [PubMed] [Google Scholar]
- Klip A, Sun Y, Chiu TT, and Foley KP (2014). Signal transduction meets vesicle traffic: the software and hardware of GLUT4 translocation. Am J Physiol Cell Physiol 306, C879–886. [DOI] [PubMed] [Google Scholar]
- Kowalski GM, and Bruce CR (2014). The regulation of glucose metabolism: implications and considerations for the assessment of glucose homeostasis in rodents. Am J Physiol Endocrinol Metab 307, E859–871. [DOI] [PubMed] [Google Scholar]
- Krook A, Roth RA, Jiang XJ, Zierath JR, and Wallberg-Henriksson H (1998). Insulin-stimulated Akt kinase activity is reduced in skeletal muscle from NIDDM subjects. Diabetes 47, 1281–1286. [DOI] [PubMed] [Google Scholar]
- Langlais P, Yi Z, Finlayson J, Luo M, Mapes R, De Filippis E, Meyer C, Plummer E, Tongchinsub P, Mattern M, et al. (2011). Global IRS-1 phosphorylation analysis in insulin resistance. Diabetologia 54, 2878–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Shi J, Jeoung NH, Kim MS, Zabolotny JM, Lee SW, White MF, Wei L, and Kim YB (2009). Targeted disruption of ROCK1 causes insulin resistance in vivo. J Biol Chem 284, 11776–11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM, Yu Y, Liu X, He L, Cavaliere PM, et al. (2017). Post-transcriptional Regulation of De Novo Lipogenesis by mTORC1-S6K1-SRPK2 Signaling. Cell 171, 1545–1558 e1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TY, Bo-Kai Hsu J, Chang WC, and Huang HD (2011). RegPhos: a system to explore the protein kinase-substrate phosphorylation network in humans. Nucleic Acids Res 39, D777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Soos TJ, Li X, Wu J, Degennaro M, Sun X, Littman DR, Birnbaum MJ, and Polakiewicz RD (2004). Protein kinase C Theta inhibits insulin signaling by phosphorylating IRS1 at Ser(1101). J Biol Chem 279, 45304–45307. [DOI] [PubMed] [Google Scholar]
- Lowery DM, Clauser KR, Hjerrild M, Lim D, Alexander J, Kishi K, Ong SE, Gammeltoft S, Carr SA, and Yaffe MB (2007). Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J 26, 2262–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundh M, Petersen PS, Isidor MS, Kazoka-Sorensen DN, Plucinska K, Shamsi F, Orskov C, Tozzi M, Brown EL, Andersen E, et al. (2019). Afadin is a scaffold protein repressing insulin action via HDAC6 in adipose tissue. EMBO Rep 20, e48216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmersjo S, Di Palma S, Diao J, Lai Y, Pfuetzner RA, Wang AL, McMahon MA, Hayer A, Porteus M, Bodenmiller B, et al. (2016). Phosphorylation of residues inside the SNARE complex suppresses secretory vesicle fusion. EMBO J 35, 1810–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, and Toker A (2017). AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BC, Warram JH, Krolewski AS, Bergman RN, Soeldner JS, and Kahn CR (1992). Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study. Lancet 340, 925–929. [DOI] [PubMed] [Google Scholar]
- Massart J, Sjogren RJO, Lundell LS, Mudry JM, Franck N, O’Gorman DJ, Egan B, Zierath JR, and Krook A (2017). Altered miR-29 Expression in Type 2 Diabetes Influences Glucose and Lipid Metabolism in Skeletal Muscle. Diabetes 66, 1807–1818. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Nakae J, Barr V, and Accili D (2000). Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in teh forkhead transcription factor FKHR. EMBO J 19, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham EJ, Parker BL, Burykin T, James DE, and Humphrey SJ (2019). Illuminating the dark phosphoproteome. Sci Signal 12. [DOI] [PubMed] [Google Scholar]
- Newgard CB (2012). Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell metabolism 15, 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda K, Nakajima S, Godo S, Saito H, Ikeda S, Shimizu T, Enkhjargal B, Fukumoto Y, Tsukita S, Yamada T, et al. (2014). Rho-kinase inhibition ameliorates metabolic disorders through activation of AMPK pathway in mice. PLoS One 9, e110446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, et al. (2003). Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A 100, 8466–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan IC, Yokoo S, Granato DC, Meneguello L, Carnielli CM, Tavares MR, do Amaral CL, de Freitas LB, Paes Leme AF, Luchessi AD, et al. (2016). Different interactomes for p70-S6K1 and p54-S6K2 revealed by proteomic analysis. Proteomics 16, 2650–2666. [DOI] [PubMed] [Google Scholar]
- Peck GR, Chavez JA, Roach WG, Budnik BA, Lane WS, Karlsson HK, Zierath JR, and Lienhard GE (2009). Insulin-stimulated phosphorylation of the Rab GTPase-activating protein TBC1D1 regulates GLUT4 translocation. J Biol Chem 284, 30016–30023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen MC, and Shulman GI (2018). Mechanisms of Insulin Action and Insulin Resistance. Physiol Rev 98, 2133–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlajamaki J, Lerin C, Itkonen P, Boes T, Floss T, Schroeder J, Dearie F, Crunkhorn S, Burak F, Jimenez-Chillaron JC, et al. (2011). Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell metabolism 14, 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previs SF, Withers DJ, Ren JM, White MF, and Shulman GI (2000). Contrasting effects of IRS-1 versus IRS-2 gene disruption on carbohydrate and lipid metabolism in vivo. J Biol Chem 275, 38990–38994. [DOI] [PubMed] [Google Scholar]
- Quattrocelli M, Swinnen M, Giacomazzi G, Camps J, Barthelemy I, Ceccarelli G, Caluwe E, Grosemans H, Thorrez L, Pelizzo G, et al. (2015). Mesodermal iPSC-derived progenitor cells functionally regenerate cardiac and skeletal muscle. J Clin Invest 125, 4463–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamani U, Gross AR, Hjelm BE, Sequeira A, Vawter MP, Tang J, Gangalapudi V, Wang Y, Andres AM, Gottlieb RA, et al. (2018). Super-Obese Patient-Derived iPSC Hypothalamic Neurons Exhibit Obesogenic Signatures and Hormone Responses. Cell Stem Cell 22, 698–712 e699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman DL, Magnusson I, Cline G, Gerard D, Kahn CR, Shulman RG, and Shulman GI (1995). Decreased muscle glucose transport/phosphorylation is an early defect in the pathogenesis of non-insulin-dependent diabetes mellitus. Proc Natl Acad Sci U S A 92, 983–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M, Cai W, Wang CH, Cederquist CT, Damasio M, Homan EP, Batista T, Ramirez AK, Gupta MK, Steger M, et al. (2019). FoxK1 and FoxK2 in insulin regulation of cellular and mitochondrial metabolism. Nat Commun 10, 1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma M, Shirai Y, Yoshino K, Kuramasu M, Nakamura T, Yanagita T, Mizuno K, Hide I, Nakata Y, and Saito N (2012). Novel PKCalpha-mediated phosphorylation site(s) on cofilin and their potential role in terminating histamine release. Mol Biol Cell 23, 3707–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltiel AR, and Olefsky JM (2017). Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest 127, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel VT, Liu ZX, Wang A, Beddow SA, Geisler JG, Kahn M, Zhang XM, Monia BP, Bhanot S, and Shulman GI (2007). Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. J Clin Invest 117, 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, and Sabatini DM (2007). PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25, 903–915. [DOI] [PubMed] [Google Scholar]
- Schwenk RW, Dirkx E, Coumans WA, Bonen A, Klip A, Glatz JF, and Luiken JJ (2010). Requirement for distinct vesicle-associated membrane proteins in insulin- and AMP-activated protein kinase (AMPK)-induced translocation of GLUT4 and CD36 in cultured cardiomyocytes. Diabetologia 53, 2209–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Inoue H, Wu JC, and Yamanaka S (2017). Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 16, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman H, Nyamandi V, Garcia-Patino M, Varela JN, Bankar G, Lin G, Jia Z, and MacLeod KM (2015). Partial deletion of ROCK2 protects mice from high-fat diet-induced cardiac insulin resistance and contractile dysfunction. Am J Physiol Heart Circ Physiol 309, H70–81. [DOI] [PubMed] [Google Scholar]
- Stump CS, Short KR, Bigelow ML, Schimke JM, and Nair KS (2003). Effect of insulin on human skeletal muscle mitochondrial ATP production, protein synthesis, and mRNA transcripts. Proc Natl Acad Sci U S A 100, 7996–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Burchfield JG, Yang P, Humphrey SJ, Yang G, Francis D, Yasmin S, Shin SY, Norris DM, Kearney AL, et al. (2019). Global redox proteome and phosphoproteome analysis reveals redox switch in Akt. Nat Commun 10, 5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukonina V, Ma H, Zhang W, Bartesaghi S, Subhash S, Heglind M, Foyn H, Betz MJ, Nilsson D, Lidell ME, et al. (2019). FOXK1 and FOXK2 regulate aerobic glycolysis. Nature 566, 279–283. [DOI] [PubMed] [Google Scholar]
- Sylow L, Jensen TE, Kleinert M, Hojlund K, Kiens B, Wojtaszewski J, Prats C, Schjerling P, and Richter EA (2013). Rac1 signaling is required for insulin-stimulated glucose uptake and is dysregulated in insulin-resistant murine and human skeletal muscle. Diabetes 62, 1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47, D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo AK, Windmueller R, Johansson BB, Dirice E, Njolstad PR, Tjora E, Raeder H, and Kulkarni RN (2013). Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. J Biol Chem 288, 5353–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai MV, Guruswamy S, Cao KT, Pessin JE, and Olson AL (1998). Myocyte enhancer factor 2 (MEF2)-binding site is required for GLUT4 gene expression in transgenic mice. Regulation of MEF2 DNA binding activity in insulin-deficient diabetes. J Biol Chem 273, 14285–14292. [DOI] [PubMed] [Google Scholar]
- Tonks KT, Ng Y, Miller S, Coster AC, Samocha-Bonet D, Iseli TJ, Xu A, Patrick E, Yang JY, Junutula JR, et al. (2013). Impaired Akt phosphorylation in insulin-resistant human muscle is accompanied by selective and heterogeneous downstream defects. Diabetologia 56, 875–885. [DOI] [PubMed] [Google Scholar]
- Tremblay F, Brule S, Hee Um S, Li Y, Masuda K, Roden M, Sun XJ, Krebs M, Polakiewicz RD, Thomas G, et al. (2007). Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc Natl Acad Sci U S A 104, 14056–14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Vernia S, Edwards YJ, Han MS, Cavanagh-Kyros J, Barrett T, Kim JK, and Davis RJ (2016). An alternative splicing program promotes adipose tissue thermogenesis. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warram JH, Martin BC, Krolewski AS, Soeldner JS, and Kahn CR (1990). Slow glucose removal rate and hyperinsulinemia precede the development of type II diabetes in the offspring of diabetic parents. Ann Intern Med 113, 909–915. [DOI] [PubMed] [Google Scholar]
- Watabe-Uchida M, John KA, Janas JA, Newey SE, and Van Aelst L (2006). The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron 51, 727–739. [DOI] [PubMed] [Google Scholar]
- Zhang J, Gao Z, Yin J, Quon MJ, and Ye J (2008). S6K directly phosphorylates IRS-1 on Ser-270 to promote insulin resistance in response to TNF-(alpha) signaling through IKK2. Journal of Biological Chemistry 283, 35375–35382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierath JR, He L, Guma A, Odegoard Wahlstrom E, Klip A, and Wallberg-Henriksson H (1996). Insulin action on glucose transport and plasma membrane GLUT4 content in skeletal muscle from patients with NIDDM. Diabetologia 39, 1180–1189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Hierarchical Clustering of Phosphoproteome. Average Intensity (Log2) and Hierarchical Clustering of Regulated Phosphosites, Related to Figure 3 and Figure 4.
Table S2. Hierarchical Clustering of Protein-Normalized Phosphoproteome. Average Intensity (Log2) and Hierarchical Clustering of Regulated Phosphosites Normalized by Total Protein Expression, Related to Figure 3.
Data Availability Statement
All mass spectrometry (MS) raw files acquired for this study are available at ProteomeXchange Consortium via the PRIDE partner repository under the identifier PXD015430.