Abstract
Despite having a similar post-operative complication profile, cardiac valve operations are associated with a higher mortality rate compared to coronary artery bypass grafting (CABG) operations. For long-term mortality, few predictors are known. In this study, we applied an ensemble machine learning (ML) algorithm to 88 routinely collected peri-operative variables to predict 5-year mortality after different types of cardiac operations. The Super Learner algorithm was trained using prospectively collected peri-operative data from 8241 patients who underwent cardiac valve, CABG and combined operations. Model performance and calibration were determined for all models, and variable importance analysis was conducted for all peri-operative parameters. Results showed that the predictive accuracy was the highest for solitary mitral (0.846 [95% CI 0.812–0.880]) and solitary aortic (0.838 [0.813–0.864]) valve operations, confirming that ensemble ML using routine data collected perioperatively can predict 5-year mortality after cardiac operations with high accuracy. Additionally, post-operative urea was identified as a novel and strong predictor of mortality for several types of operation, having a seemingly additive effect to better known risk factors such as age and postoperative creatinine.
Subject terms: Biomarkers, Medical research, Risk factors, Mathematics and computing
Introduction
Whereas complications after cardiac operations are associated with increased risk of in-hospital mortality, only few predict long-term mortality. The best documented is post-operative acute kidney injury (AKI), a highly prevalent complication occurring in 15–30% of patients1,2 which is associated with both increased short- and long-term mortality1–4. The relation between postoperative AKI and mortality varies greatly per type of cardiac operation. Mortality risks related to AKI are well characterized for coronary artery bypass grafting (CABG), but less well studied in valve operations, despite these accounting for 24% of all cardiac operations and having higher mortality rates5,6. Recently, Bouma et al.5, showed post-operative AKI to be strongly associated with an increase in long-term mortality in patients with solitary valve and combined valve and CABG operations. Remarkably, even a mild impairment in renal function well below the threshold for AKI-1 (i.e., a mere 10% post-operative increase in serum creatinine) significantly increased long-term mortality risk in solitary valve operations5. Therefore, to date postoperative AKI represents the best studied organ injury related early marker of long-term mortality risk after cardiac operations.
Previously, we have demonstrated that machine learning (ML) predictive models proved superior to classical multivariable analysis in identifying patients at increased risk of long-term mortality after CABG operations7. Moreover, a unique property of ML is its ability to identify parameters predicting mortality and rank their importance by variable importance analysis. However, while ML analyses gain popularity in peri-operative care8, studies using ML techniques for long-term mortality analysis after cardiac valve operations are lacking. Several studies in different fields of healthcare have shown ensemble ML algorithms to be more accurate than individual algorithms in modelling complex outcomes such as mortality in critically ill patients9 and mortality following cardiac arrest10. In anesthesiology, recent studies showed that different machine learning algorithms could accurately predict acute hypotensive episodes 10 min in advance using patient characteristics and physiological variables11–13.
In this study, we combined multiple ML algorithms into an ensemble using the Super Learner (SL) algorithm14. This ensemble ML algorithm was trained to predict 5-year mortality in a large prospective cohort of patients undergoing cardiac valve, CABG, or combined operations using routinely collected peri-operative data in a single tertiary care hospital. We compared the accuracy of two SL training methodologies, using a targeted approach with patients split per operation type compared to the entire, unselected population. Furthermore, variable importance analysis was conducted to identify the strongest predictors of mortality.
Results
Patient characteristics and mortality per operation type
Patient characteristics, descriptives of all variables used in this study and mortality data per operation type are summarized in Table 1 (and Table 1 of the “Supplementary material”). Five years mortality rate of the full patient cohort was 16.5%. Operations involving valve procedures showed higher mortality amounting 16.9% for aortic valve alone, 19.7% for mitral valve alone, 21.0% for combined aortic valve/CABG and 28.9% for combined mitral valve/CABG (Table 1). Accordingly, mortality rate for CABG-only (13.8%) was lower than for the entire cohort.
Table 1.
Descriptives table per operation type.
| CABG | Aortic valve | Mitral valve | Aortic + coronary | Mitral + coronary | P value | |
|---|---|---|---|---|---|---|
| N = 4514 | N = 1663 | N = 884 | N = 813 | N = 367 | ||
| BMI | 27.34 | 27.01 | 26.03 | 27.40 | 26.56 | < 0.001 |
| Gender | < 0.001 | |||||
| Female | 1028 (22.77%) | 676 (40.65%) | 424 (47.96%) | 285 (35.06%) | 125 (34.06%) | |
| Male | 3486 (77.23%) | 987 (59.35%) | 460 (52.04%) | 528 (64.94%) | 242 (65.94%) | |
| Age | 66.26 | 64.80 (13.79) | 62.62 (13.62) | 72.25 (8.39) | 69.28 (8.57) | < 0.001 |
| Pre-operative eCCR | 71.42 | 72.32 (21.84) | 74.83 | 74.54 | 65.78 | 0.065 |
| Post-operative eCCR | 66.99 | 70.50 (39.87) | 67.43 (27.26) | 62.94 (24.22) | 59.03 (23.90) | < 0.001 |
| Per-operative eCCR decrease | 4.44 | 1.82 (32.85) | 7.40 (95.76) | 11.60 (106.61) | 6.75 (16.49) | 0.001 |
| Pre-operative eCCR ratio | 1.13 | 1.11 (0.38) | 1.27 (2.26) | 1.25 (1.18) | 1.23 (0.50) | 0.001 |
| Creatinine within 24 h before surgery (μmol/L) | 102.69 | 100.31 (78.26) | 98.75 (44.46) | 104.65 (79.01) | 107.90 (72.40) | 0.138 |
| Pre-operative creatinine | 101.95 | 99.21 (72.06) | 98.15 (40.11) | 102.89 (68.31) | 107.11 (70.80) | 0.110 |
| Creatinine 12–24 h after surgery | 91.83 | 89.10 (74.79) | 89.15 (45.57) | 96.46 (68.71) | 102.09 (65.22) | 0.002 |
| Creatinine 24 h after surgery | 92.84 | 90.71 (73.06) | 91.85 (45.48) | 98.40 (70.21) | 103.72 (65.53) | 0.002 |
| Creatinine at day 2 after surgery | 102.72 | 99.13 (73.60) | 96.19 (49.84) | 104.08 (66.63) | 107.76 (56.25) | 0.006 |
| Creatinine at day 4 after surgery | 98.61 | 94.35 (75.25) | 93.75 (57.22) | 100.73 (74.97) | 104.61 (69.26) | 0.007 |
| Maximum post-operative creatinine | 111.03 | 108.53 (86.88) | 110.96 (64.41) | 119.15 (86.85) | 126.22 (76.60) | < 0.001 |
| Absolute difference in creatinine | 9.08 | 9.32 | 12.81 (46.89) | 16.26 (43.28) | 19.11 (44.45) | < 0.001 |
| Relative difference in creatinine | 1.10 | 1.09 | 1.19 | 1.19 (0.79) | 1.19 (0.40) | < 0.001 |
| Percentual difference in creatinine | 10.20 | 8.71 | 19.27 | 19.45 (79.05) | 18.96 (39.91) | < 0.001 |
| Urea within 24 h before surgery (mmol/L) | 6.96 | 7.25 | 8.09 | 7.57 (3.27) | 8.20 (3.76) | < 0.001 |
| Pre-operative urea | 6.98 (3.29) | 7.34 (4.03) | 8.25 | 7.51 (3.12) | 8.52 (6.11) | < 0.001 |
| Urea 12–24 h after surgery | 7.24 (5.47) | 8.11 (16.73) | 8.55 | 8.19 (9.95) | 9.37 (8.95) | < 0.001 |
| Urea at day 2 after surgery | 10.13 (23.15) | 10.51 (25.40) | 11.89 | 14.51 (35.13) | 12.76 (24.00) | < 0.001 |
| Urea at day 4 after surgery | 8.49 (24.49) | 9.32 (29.78) | 10.74 | 14.14 (49.26) | 11.35 (16.44) | < 0.001 |
| Maximum CPB flow | 4.63 (1.47) | 4.12 (1.97) | 3.96 (2.05) | 4.02 (2.05) | 4.14 (2.01) | < 0.001 |
| Duration of perfusion | 100.07 (38.64) | 125.29 (48.12) | 169.76 (73.36) | 168.07 (50.43) | 214.06 (77.13) | 0.000 |
| Aortic cross-clamp time | 58.94 (25.42) | 83.36 (32.28) | 109.93 (53.19) | 110.25 (31.48) | 137.22 (52.95) | 0.080 |
| HR at start surgery | 62.22 (12.90) | 67.14 (14.00) | 70.80 (17.52) | 62.55 (13.90) | 66.17 (16.06) | 0.000 |
| HR during perfusion | 66.39 (57.83) | 61.24 (55.15) | 61.80 (53.20) | 60.09 (57.94) | 63.58 (61.49) | < 0.001 |
| SBP at start surgery (mmHg) | 113.54 (34.62) | 108.81 (31.98) | 102.25 (31.32) | 109.36 (33.49) | 105.28 (29.93) | 0.002 |
| SBP during perfusion | 61.76 (21.39) | 63.37 (22.60) | 63.17 (20.39) | 63.96 (20.10) | 62.83 (22.03) | < 0.001 |
| DBP at start surgery (mmHg) | 64.81 (31.81) | 61.87 (29.18) | 60.45 (27.90) | 60.51 (29.65) | 58.87 (22.32) | 0.012 |
| DBP during perfusion | 56.53 (18.09) | 58.52 (18.88) | 57.58 (17.00) | 59.16 (17.66) | 57.39 (17.37) | < 0.001 |
| CVP at start surgery (mmHg) | 12.58 (30.79) | 11.96 (28.43) | 13.88 (30.11) | 12.89 (32.66) | 12.32 (24.81) | < 0.001 |
| CVP during perfusion | 6.62 (8.31) | 5.03 (9.45) | 4.78 (15.07) | 5.65 (5.60) | 4.44 (7.75) | 0.653 |
| PaCO2 at start surgery (kPa) | 5.02 (0.63) | 5.08 (0.70) | 5.03 (0.69) | 5.07 (0.64) | 5.01 (0.72) | < 0.001 |
| PaCO2 during perfusion | 5.04 (0.54) | 5.17 (0.57) | 5.18 (0.62) | 5.09 (0.51) | 5.13 (0.57) | 0.010 |
| PaCO2 at end surgery | 4.84 (0.59) | 4.87 (0.63) | 4.99 (0.74) | 4.89 (0.62) | 5.04 (0.72) | < 0.001 |
| PaO2 at start surgery (kPa) | 21.49 (14.95) | 22.11 (14.43) | 22.03 (14.74) | 20.65 (12.85) | 19.81 (12.93) | < 0.001 |
| PaO2 during perfusion | 26.70 (10.88) | 25.59 (10.36) | 25.88 (9.82) | 25.87 (9.37) | 26.82 (10.52) | 0.018 |
| PaO2 at end surgery | 17.79 (11.58) | 22.27 (13.04) | 21.93 (12.82) | 21.30 (12.62) | 20.37 (11.37) | 0.001 |
| SaO2 at start surgery (%) | 0.98 (0.03) | 0.98 (0.03) | 0.98 (0.05) | 0.98 (0.03) | 0.98 (0.02) | < 0.001 |
| SaO2 during perfusion | 0.99 (0.03) | 0.99 (0.05) | 0.99 (0.05) | 0.99 (0.03) | 0.99 (0.06) | 0.206 |
| SaO2 end surgery | 0.98 (0.03) | 0.99 (0.04) | 0.98 (0.04) | 0.98 (0.04) | 0.98 (0.02) | 0.152 |
| ICU stay (hours) | 52.44 (163.21) | 47.51 (138.81) | 88.79 (216.86) | 88.72 (260.41) | 141.13 (267.53) | < 0.001 |
| ESR within 24 h before surgery (mm/h) | 20.61 (19.57) | 18.63 (19.96) | 20.21 (19.40) | 22.23 (20.25) | 23.01 (21.03) | < 0.001 |
| Pre-operative ESR | 20.85 (19.77) | 17.98 (19.27) | 19.22 (19.06) | 21.22 (19.82) | 23.15 (19.74) | < 0.001 |
| LDH within 24 h before surgery (U/L) | 227.71 (75.41) | 248.34 (115.16) | 259.90 (169.51) | 235.06 (70.41) | 228.79 (66.45) | < 0.001 |
| Pre-operative LDH | 228.65 (76.10) | 250.27 (142.33) | 273.05 (428.61) | 236.90 (74.54) | 230.82 (74.38) | < 0.001 |
| LDH 12- 24 h after surgery | 338.15 (273.89) | 396.80 (179.67) | 480.19 (484.17) | 456.26 (497.74) | 510.39 (662.83) | < 0.001 |
| LDH at day 2 after surgery | 338.30 (233.89) | 388.29 (252.01) | 461.39 (444.76) | 446.04 (312.62) | 474.68 (264.37) | < 0.001 |
| LDH at day 4 after surgery | 327.78 (882.49) | 382.96 (703.42) | 413.65 (329.29) | 424.52 (461.76) | 439.23 (340.88) | < 0.001 |
| Maximum post-operative LDH | 421.61 (896.25) | 461.39 (377.02) | 568.32 (731.73) | 558.72 (709.25) | 592.21 (543.21) | < 0.001 |
| Blood glucose 0–6 h after surgery (mmol/L) | 9.41 (2.46) | 8.41 (2.48) | 8.47 (2.84) | 8.48 (2.73) | 9.02 (2.70) | < 0.001 |
| Blood glucose 6–12 h after surgery | 10.22 (2.43) | 9.56 (2.00) | 9.49 (2.27) | 9.67 (2.15) | 9.57 (2.26) | < 0.001 |
| Blood glucose 12–24 h after surgery | 9.14 (2.48) | 8.39 (2.07) | 8.17 (2.21) | 8.27 (2.13) | 8.12 (2.08) | < 0.001 |
| Maximum post-operative glucose | 11.19 (4.37) | 10.38 (3.84) | 10.48 (2.58) | 10.53 (2.24) | 10.82 (2.69) | < 0.001 |
| Hb within 24 h before surgery (mmol/L) | 8.47 (1.09) | 8.45 (1.06) | 8.27 (1.20) | 8.35 (1.00) | 8.30 (1.11) | < 0.001 |
| Pre-operative Hb | 8.19 (1.36) | 8.24 (1.78) | 8.10 (1.65) | 8.26 (2.44) | 8.34 (2.90) | < 0.001 |
| Hb 0–6 h after surgery | 5.64 (0.73) | 5.69 (0.76) | 5.78 (0.82) | 5.52 (0.76) | 5.57 (0.84) | 0.135 |
| Hb 6–12 h after surgery | 6.02 (0.85) | 6.35 (1.16) | 6.22 (0.93) | 5.92 (0.84) | 5.80 (0.89) | < 0.001 |
| Hb 12–24 h after surgery | 6.18 (0.78) | 6.40 (0.85) | 6.25 (0.88) | 6.01 (0.77) | 5.92 (0.80) | < 0.001 |
| Hb at day 2 after surgery | 6.31 (0.78) | 6.26 (0.81) | 6.09 (0.86) | 6.01 (0.75) | 5.92 (0.76) | < 0.001 |
| Hb at day 4 after surgery | 6.52 (0.87) | 6.40 (1.33) | 6.22 (0.89) | 6.07 (0.81) | 5.97 (0.82) | < 0.001 |
| Minimum post-operative Hb | 5.31 (0.69) | 5.41 (0.73) | 5.29 (0.77) | 5.11 (0.65) | 5.01 (0.70) | < 0.001 |
| Leukocytes within 24 h before surgery (× 109/L) | 7.84 (2.73) | 7.44 (2.70) | 7.62 (3.32) | 7.77 (3.22) | 7.74 (2.18) | < 0.001 |
| Pre-operative leukocytes | 8.01 (2.96) | 7.53 (2.63) | 7.79 (3.12) | 7.88 (2.99) | 7.88 (2.46) | < 0.001 |
| Leukocytes 12–24 h after surgery | 13.95 (4.41) | 13.71 (4.36) | 13.79 (4.17) | 13.57 (4.93) | 13.49 (4.22) | < 0.001 |
| Leukocytes at day 2 after surgery | 17.08 (4.82) | 15.79 (4.81) | 15.99 (5.12) | 16.14 (4.74) | 16.49 (4.85) | 0.051 |
| Leukocytes at day 4 after surgery | 11.52 (4.15) | 10.00 (4.06) | 10.93 (9.74) | 10.99 (3.94) | 11.96 (4.78) | < 0.001 |
| Thrombocytes within 24 h before surgery (× 109/L) | 246.55 (73.47) | 231.91 (67.44) | 235.83 (72.42) | 234.86 (69.37) | 239.06 (72.70) | < 0.001 |
| Pre-operative thrombocytes | 238.69 (78.71) | 224.72 (71.67) | 230.34 (75.95) | 230.41 (73.07) | 233.71 (76.22) | < 0.001 |
| Thrombocytes 0–6 h after surgery | 152.85 (52.79) | 131.80 (44.10) | 132.02 (44.78) | 129.11 (46.63) | 131.97 (48.12) | < 0.001 |
| Thrombocytes 6–12 h after surgery | 171.17 (58.06) | 149.14 (49.54) | 141.39 (48.69) | 136.37 (47.14) | 140.20 (54.93) | < 0.001 |
| Thrombocytes 12–24 h after surgery | 174.48 (57.73) | 151.22 (50.74) | 141.85 (47.48) | 136.92 (46.77) | 138.73 (53.92) | < 0.001 |
| ALAT within 24 h before surgery (U/L) | 40.56 (35.46) | 28.54 (26.10) | 31.48 (29.11) | 30.31 (28.08) | 31.99 (26.70) | < 0.001 |
| Pre-operative ALAT | 40.80 (35.28) | 28.86 (27.93) | 33.58 (57.80) | 30.40 (26.64) | 33.25 (34.18) | < 0.001 |
| ALAT 12–24 h after surgery | 37.49 (79.01) | 29.24 (37.05) | 43.72 (160.67) | 35.59 (135.12) | 46.14 (198.66) | < 0.001 |
| ALAT at day 2 after surgery | 37.57 (146.05) | 31.20 (88.67) | 44.26 (123.93) | 40.56 (168.31) | 40.43 (105.37) | 0.002 |
| ASAT within 24 h before surgery (U/L) | 32.72 (20.36) | 29.50 (20.19) | 31.83 (24.10) | 28.95 (15.01) | 31.08 (28.86) | 0.169 |
| Pre-operative ASAT | 33.18 (24.15) | 30.11 (24.14) | 37.82 (166.12) | 29.45 (15.34) | 30.90 (18.47) | < 0.001 |
| ASAT 12–24 h after surgery | 59.82 (108.39) | 71.96 (83.88) | 112.66 (241.82) | 98.08 (206.81) | 121.96 (283.96) | < 0.001 |
| ASAT at day 2 after surgery | 53.36 (171.29) | 58.70 (113.12) | 92.51 (194.80) | 89.26 (347.04) | 90.90 (115.64) | 0.011 |
| ASAT at day 4 after surgery | 55.12 (422.01) | 54.44 (197.38) | 68.11 (217.54) | 72.37 (435.80) | 71.26 (248.91) | < 0.001 |
| Neutrophils 12–24 h after surgery (× 109/L) | 12.29 (3.86) | 12.07 (3.86) | 12.07 (3.80) | 11.86 (3.96) | 11.79 (3.82) | 0.584 |
| Monocytes 12–24 h after surgery (× 109/L) | 1.10 (1.73) | 1.32 (2.07) | 1.51 (2.25) | 1.42 (2.33) | 1.34 (2.20) | 0.004 |
| Lymphocytes 12–24 h after surgery (× 109/L) | 1.05 (2.05) | 1.12 (1.86) | 1.35 (2.60) | 1.15 (1.93) | 1.34 (3.11) | < 0.001 |
| 5-year mortality: | 0.001 | |||||
| Alive | 3890 (86.18%) | 1382 (83.10%) | 710 (80.32%) | 642 (78.97%) | 261 (71.12%) | < 0.001 |
| Deceased | 624 (13.82%) | 281 (16.90%) | 174 (19.68%) | 171 (21.03%) | 106 (28.88%) | |
| Minimum body temperature | 31.71 (1.82) | 31.20 (2.60) | 30.76 (2.36) | 31.23 (2.17) | 30.89 (1.86) | |
| AKI staging | < 0.001 | |||||
| No AKI | 3063 (67.86%) | 1142 (68.67%) | 584 (66.06%) | 462 (56.83%) | 199 (54.22%) | < 0.001# |
| Mild subclinical AKI | 841 (18.63%) | 268 (16.12%) | 133 (15.05%) | 145 (17.84%) | 62 (16.89%) | |
| Moderate subclinical AKI | 142 (3.15%) | 51 (3.07%) | 26 (2.94%) | 37 (4.55%) | 14 (3.81%) | |
| AKI 1–3 | 468 (10.37%) | 202 (12.15%) | 141 (15.95%) | 169 (20.79%) | 92 (25.07%) | |
| AKI 1 | 441 (9.77%) | 191 (11.49%) | 126 (14.25%) | 157 (19.31%) | 90 (24.52%) | |
| AKI 2 | 9 (0.20%) | 6 (0.36%) | 11 (1.24%) | 6 (0.74%) | 2 (0.54%) | |
| AKI 3 | 18 (0.40%) | 5 (0.30%) | 4 (0.45%) | 6 (0.74%) | 0 (0%) |
All values presented as mean (95% CI), and categorical variable with the percentage in parentheses.
BMI body mass index, eCCR estimated creatinine clearance, CPB cardio-pulmonary bypass, HR heart rate, SBP systolic blood pressure, DBP diastolic blood pressure, CVP central venous pressure, PaCO2 arterial CO2 pressure, PaO2 arterial oxygen pressure, SaO2 oxygen saturation, ICU intensive care unit, ESR erythrocyte sedimentation rate, LDH lactate dehydrogenase, Hb hemoglobin, ALAT alanine aminotransferase, ASAT aspartate aminotransferase, AKI acute kidney injury.
#Significance level presented is for AKI 1–3 combined, given that there are no patients in the mitral + coronary group with AKI 3.
Machine learning analysis
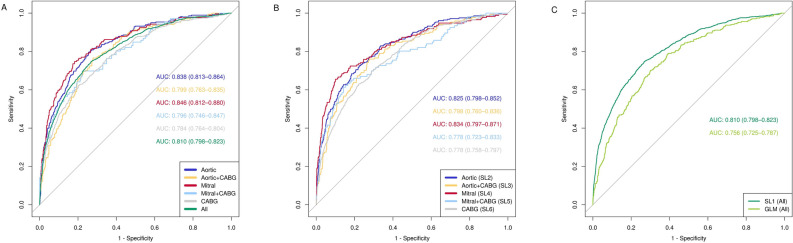
As a first step in the ML based prediction of long-term mortality, the ensemble was trained on the full cohort (SL1; Fig. 5, left part). ROC curves and their respective AUROCs were established for the full cohort and the different cardiac operation types (Fig. 1). SL1 achieved an AUROC of 0.810 [0.798–0.823]. When analyzed per operation type, the accuracy of SL1 was highest for solitary mitral valve (0.846) and solitary aortic valve operations (0.838), and lowest for solitary CABG (0.784) and mitral valve/CABG (0.796). In addition, the comparison between SL1 and the trained GLM showed that the SL1 significantly outperformed GLM (AUROC 0.756 [0.725–0.787]) for the full cohort (P = 0.0016; Fig. 1) as well as for solitary aortic valve and combined aortic valve and CABG (P < 0.01; Table 2 in the “Supplementary material”). Thus, SL1 produced sound long-term mortality prediction based on peri-operative routinely collected patient and operation data.
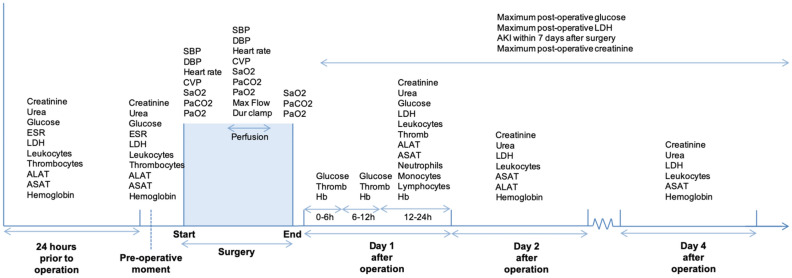
Figure 5.
Timeline of clinical measurements before, during, and after cardiac operation, in the intensive care unit (day 1 after operation), day 1 in the ward (day 2 after operation), and day 3 in the ward (day 4 after operation). Patient characteristics are not included here, but described in detail in Table 1. Dur CA duration of cardiac arrest, Dur clamp duration of aortic cross-clamp, Hb hemoglobin, ASAT aspartate aminotransferase, ALAT alanine aminotransferase, Thromb thrombocytes, ESR erythrocyte sedimentation rate, LDH lactate dehydrogenase, CVP central venous pressure, PaCO2 arterial carbon dioxide partial pressure, SaO2 oxygen saturation, PaO2 arterial oxygen partial pressure, SBP systolic blood pressure, DBP diastolic blood pressure.
Figure 1.
Plot of the receiver operating characteristic (ROC) curves and the respective areas under curve (AUCs) for the weighted Super Learner 1 for each of the 5 types of operation and for the whole cohort. Plot of the receiver operating characteristic (ROC) curves and the respective areas under curve (AUCs) for the weighted Super Learner and the generalized linear model (GLM) for the whole cohort. SL super learner, CABG coronary artery bypass grafting.
Next, we performed a similar analysis based on SL training per operation type, by making five training sets using 80% of the relevant patients to train five weighted ensembles (SL2–SL6). Comparison of AUROCs between SL1 versus SL2–6, showed identical ranking for specific operation subgroups. Predictive performance between the models generated by SL1 compared those from SL2 to SL6 did not differ (Fig. 1; Table 2 in the “Supplementary material”). SL3 and SL4 also outperformed GLM (P < 0.01; Table 4 in the “Supplementary material”). Lastly, because of its potential ability to identify patients at high risk prior to surgery, we examined the predictive performance when only pre-operative data are included. As expected, the model trained only on pre-operative data showed inferior performance to the full peri-operative model (AUROC 0.718 [0.687–0.749], P < 0.01, Fig. 12 in the “Supplementary material”).
Calibration, sensitivity analysis and adjusted risk thresholds based on predicted probability of mortality
Calibration of SL1 and SL2–6 was good for most models (Table 5 and Figs. 1–11 of the “Supplementary material”). Using the adjusted thresholds based on the Youden index and on a 50% increased risk of mortality lead to improved model sensitivity and specificity (Fig. 2). For all operations, the thresholds based on the Youden index approximated the baseline absolute mortality risk. Compared to the default threshold of 50% mortality risk, both the thresholds based on the Youden index and the thresholds defined by a 50% increased risk of mortality increased sensitivity substantially for all types of operation (Tables 6–15 of the “Supplementary material”). For the Youden index thresholds, this was paired with a steeper decrease in specificity than for the thresholds at 50% increased risk of mortality. As Table 2 shows, the threshold representing 50% increase in risk improved the number of patients correctly classified as “non-survivor” for all types of operation. The largest increase in correctly classified “non-survivors” was observed for aortic valve, CABG, combined aortic valve and CABG, and for all operations combined (3-, 4.7-, 2.2-, and 3-fold increase).
Figure 2.
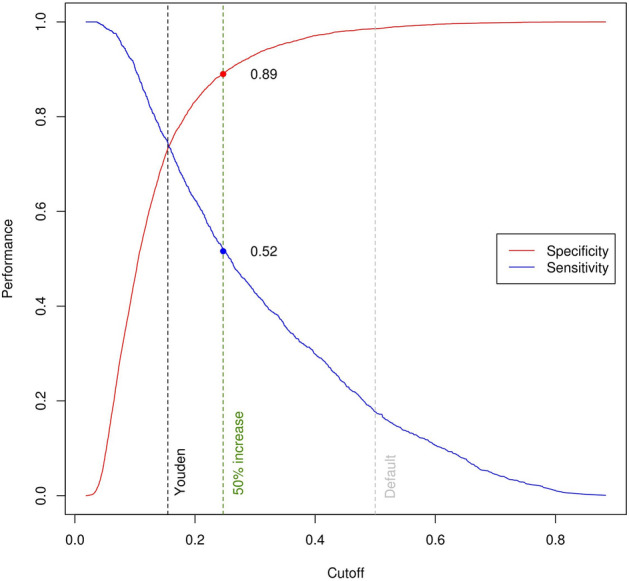
Specificity (blue) and sensitivity (red) values across all possible thresholds for all operations combined. The default 0.50 threshold is marked in grey, the threshold based on the maximized Youden index in black, and the threshold representing a 50% increase in mortality risk in green.
Table 2.
Percentage of correctly classified cases in survivors and non-survivors per operation type for SL1 predictions using the default and 50% increase in risk thresholds.
| Predictions matching actual patient outcome (%) | ||
|---|---|---|
| Survivors (%) | Non-survivors (%) | |
| Aortic valve | ||
| With default threshold | 98.8 | 18.1 |
| With 50% increased risk threshold | 90.5 | 53.0 |
| Difference | − 8.3 | + 34.9 |
| Mitral valve | ||
| With default threshold | 96.8 | 34.5 |
| With 50% increased risk threshold | 89.7 | 59.8 |
| Difference | − 7.1 | + 25.3 |
| CABG | ||
| With default threshold | 99.2 | 10.4 |
| With 50% increased risk threshold | 88.9 | 47.9 |
| Difference | − 9.3 | + 37.5 |
| Aortic + CABG | ||
| With default threshold | 97.0 | 19.9 |
| With 50% increased risk threshold | 88.8 | 43.3 |
| Difference | − 8.2 | + 23.4 |
| Mitral + CABG | ||
| With default threshold | 96.9 | 28.3 |
| With 50% increased risk threshold | 95.4 | 34.9 |
| Difference | − 1.5 | + 6.6 |
| All operations combined | ||
| With default threshold | 98.6 | 17.7 |
| With 50% increased risk threshold | 89.4 | 51.6 |
| Difference | − 9.2 | + 33.9 |
Variable importance analysis
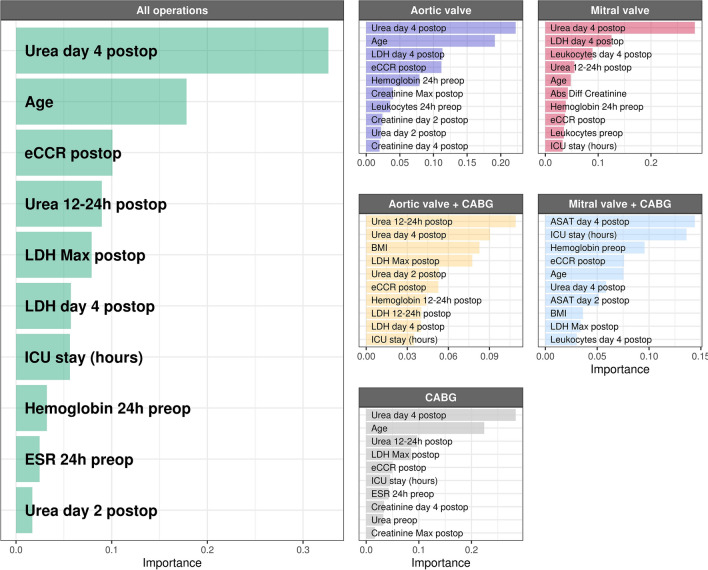
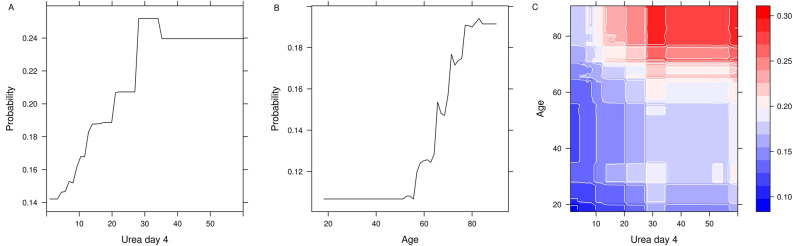
Unexpectedly, variable importance analysis of all operations combined (n = 8142) revealed serum urea at day 4 after operation as the top predictor variable for 5-year mortality (Fig. 3). Serum urea was also found the top predictor in all operation types, except for the smallest group (n = 367), combined mitral valve and CABG operations. Other important predictive variables included patient age, serum urea at other time points, indicators of kidney function, and serum markers for organ damage and inflammation. To better illustrate the impact of the changes in these variable and possible interactions, we constructed probability plots of the two highest ranking variables in all patients (Fig. 4). Mortality risk steeply increased from day 4 urea levels of 10 mmol/L, reaching a plateau at 30 mmol/L denoting a 50% increase in absolute risk compared to baseline. Likewise, mortality risk gradually increased between 60 and 80 years of age. Figure 4 illustrates the combined effect of serum urea day 4 and age on mortality risk.
Figure 3.
Top ten predictor variables for all types of operations combined. Variable coefficients indicate how much each parameter contributes to the outcome. eCCR estimated creatinine clearance, LDH lactate dehydrogenase, ESR erythrocyte sedimentation rate, ICU intensive care unit, ASAT aspartate transaminase, BMI body mass index.
Figure 4.
Partial dependence plots of urea at postoperative day 4 and age. Partial dependence plots of urea at postoperative day 4 against age. The vertical bar represents predicted risk (blue to red, low to high).
Discussion
This study shows that ensemble ML analysis achieves a high accuracy in predicting 5-year mortality in a cohort of 8241 patients with CABG and/or valve operations. Moreover, variable importance analysis revealed early postoperative urea as a novel and strong predictor of mortality in all types of cardiac operations. Furthermore, methodologically, a more targeted approach of training the algorithms on sub-groups instead of the full cohort did not significantly improve mortality prediction.
We demonstrated that using an ensemble algorithm with a combination of pre-operative, intra-operative, and first week post-operative data, achieves high accuracy in predicting 5-year mortality after different types of cardiac operations. These findings extend a previous study where we demonstrated the superiority of individual ML models compared to classical multivariable analysis in identifying patients at increased risk of long-term mortality after CABG7. Here, we reaffirm these findings using ensemble ML and data from different types of cardiac operations. Using peri-operative data, we achieved similar accuracy to a recently developed ML-based risk algorithm for prediction of 1- to 24-month mortality following major surgery15. Compared to other models that predict mortality specifically after cardiac surgery, the ensemble achieved superior performance8.
The application of algorithms such as the one we developed to pre-operative data would possibly predict patients at the highest risk of long-term complications prior to surgery. Expectedly, analysis of pre-operative data in the XGBoost model decreased performance significantly, which could be partly due to the limited set of pre-operative data available in our cohort, or to the lower frequency of the outcome (long-term mortality as opposed to short-term post-operative complications). Yet, it should be noted that the model’s performance using our restricted set of pre-operative data has comparable predictive power as currently used clinical scores8.
Methodologically, our study contributed to the discussion on the need of conducting predictive studies on operation-specific cohorts. Results from previous studies suggest that algorithms trained on pooled data from patients undergoing different types of surgeries were accurate in predicting outcomes for all these types of operations. In keeping, our findings show that both the model trained with the full cohort, and the models trained with the individual cardiac operation subgroups showed a good performance in predicting long-term mortality after aortic and mitral valve operations. This finding further questions the need to conduct ML analyses on operation-specific cohorts. Specifically, including full cohorts may lead to better model performance analyses due to the greater amount of data.
Additionally, by providing risk predictions at individual level, ML algorithms allow for the adjustment of the sensitivity and specificity of each model for different clinical settings15. Balancing sensitivity and specificity in the context of mortality risk predictions can be challenging. Lowering the prediction threshold may lead to excessive over-diagnosing and increase in healthcare costs. However, especially in populations with relatively low mortality rates such as cardiac surgery patients, a too high threshold would miss too many “non-survivors”. Here, we demonstrated that using a 50% increase in absolute risk of mortality as cut-off provides a favorable trade-off between false positives and true negatives, as previously shown in similar large studies predicting postoperative mortality and mortality in intensive care patients15,16. Validation of this approach merits further investigation, and may facilitate the translation of an algorithm’s good predictive performance into a clinically useful patient risk stratification tool17.
Variable importance analysis identified postoperative urea as the strongest predictor of 5-year mortality. This is consistent with our previous findings in a CABG-only population7. Yet, literature on the possible role of urea as a mortality predictor in cardiac operations is scarce7. Preoperative urea values above 10 mmol/L have been found to be associated with increased 30-day mortality risk after CABG and with increased risk of stroke in the 10 days after cardiac operations18,19. It should also be noted that, in heart failure patients, increased urea levels have been associated with derangements in cardiac output and renal perfusion20,21. These are, in turn, strongly related to patients’ overall performance status and prognosis, with both urea and the urea/creatinine ratio being known prognostic predictors22. In the context of this study, increased urea may originate from excess production and/or impaired excretion, yet mechanistic insight remains elusive. Possibly, urea production may be increased by mitochondrial dysfunction, caused by ischemia/reperfusion and increased systemic inflammatory response after cardiopulmonary bypass and surgical trauma23. Mitochondrial dysfunction may be amplified through excess reactive oxygen species (ROS) following accumulation of succinate during ischemia24,25. Additionally, recent evidence indicates that high urea levels generate ROS26. Furthermore, renal excretion of urea may decrease in response to kidney injury. Thus, urea likely reflects the compound pathological state of different organ systems, rather than just kidney function.
Lastly, this study also has some limitations to consider. Being a single center study, our findings need confirmation by external validation. Further, our analysis is limited to the variables in the CAROLA database. Detailed co-morbidity information, for instance, could help further improve model performance, especially for the CABG sub-group. Additionally, variable importance analysis as such does not provide directionality and assumptions about effect size between the variables and the outcome cannot be made directly. Finally, the current ensemble ML is not suited to use high-frequency, high-volume data, such as continuous intraoperative measurements of blood pressure, heart rate, oxygen saturation or temperature. Therefore, a study including algorithms suitable for such analysis, such as recurrent neural networks, is a logical follow-up.
In conclusion, ML analysis of 88 routinely collected peri-operative data achieved a high accuracy in predicting 5-year mortality after different cardiac operations in this large study of 8241 patients. A targeted approach of training the algorithms on sub-groups instead of the full cohort did not improve model performance. Moreover, variable importance analysis showed early postoperative urea as a novel and strong predictor of mortality in all types of cardiac operations. Similar studies enabling the identification of modifiable risk factors and providing individual patient predictions may form a first step towards facilitating personalized clinical interventions to improve patient care.
Methods
The electronic Cardiothoracic Anesthesiology Registry (CAROLA) comprises extensive prospective data of all adult patients who underwent first-time valve operation, CABG, or a combination of both between 1997 and 2017 in the University Medical Centre Groningen (UMCG), the Netherlands. The total number of patients is 11,286. This database study was approved by the Medical Ethical Committee of the UMCG, and the requirement to obtain informed consent was waived (waiver: METC#2010/118). All analyses were performed in accordance with relevant guidelines and regulations.
Patient population and outcome
Only patients who underwent valve operation, either solitary or combined with coronary artery bypass grafting (CABG), or solitary CABG, with cardiopulmonary bypass (CPB) were included (n = 8241). There were 1663 patients in the combined aortic and coronary group, 367 in the combined mitral and coronary group, 884 in the solitary mitral group, 813 in the solitary aortic group, and 4514 in the CABG-only group. Mortality data were obtained in November 2017 from the Dutch Municipal Personal Records Database comprising actual and reliable data of all citizens within the Netherlands.
Data selection and pre-processing
The dataset includes patient characteristics, peri-operative hemodynamic, CPB, respiratory and organ function data and blood values collected at different time points indicated in Fig. 5. Because for some patients referred from other hospitals the stay in our center was limited to the immediate peri-operative phase, a variable pattern of missing data was observed. Multivariate imputation by chained equations was performed on the set of variables with at least 50% non-missing data27. The final dataset without missing data consisted of 88 predictor variables and 5-year mortality as the outcome variable (Table 1). Baseline serum creatinine measurements was defined as the closest to the start of operation. Patients were classified for post-operative AKI 0–3 within the 7 days after operation according to the AKIN classification3.
Statistical analysis
The Super Learner, selected candidate algorithms, and hyper-parameter tuning
The Super Learner algorithm is a generalization of the stacking algorithms developed by Breiman28, which combines a set of candidate algorithms to make k-fold-cross-validated predictions9,29. In this process, the dataset is divided into k mutually exclusive and exhaustive subsets, with one set serving as a validation set, while the others are used for training each candidate algorithm14. This means that each patient is used only once in the validation set, and included in the training set for all other rounds. For each candidate learner, k risks are calculated and averaged into a “cross-validated risk”. Subsequently, the learners with the minimal risk are selected, applied to the entire dataset and included in the new weighted estimator (the SL), that attributes a relative coefficient to each of the learners. Those which reduce the calculated risk the most, will contribute to the final weighted prediction. Moreover, the SL presents individual patient predicted probabilities for 5-year mortality per ensemble. Five candidate algorithms were included in the SL: support Bayesian additive regression trees (BART), extremely randomized trees, elastic net, support vector machine, and extreme gradient boosted machine (XGBoost). Details of these five algorithms can be found in the “Supplementary material”. Since the performance of an algorithm varies greatly depending on its hyper-parameters and can be substantially improved by tuning, multiple hyper-parameter combinations were generated for each candidate algorithm. Details of each of these algorithms including the hyper-parameters, the tuning process, and final values are described in the “Supplementary material”. A 10-fold cross-validated generalized linear regression model (GLM) was trained on data from the full cohort for use as baseline comparison of the SL’s performance. Lastly, to test the performance of a model using only pre-operative data in predicting post-operative outcomes, a 10-fold cross-validated XGBoost model was trained on data from the full cohort.
Model training
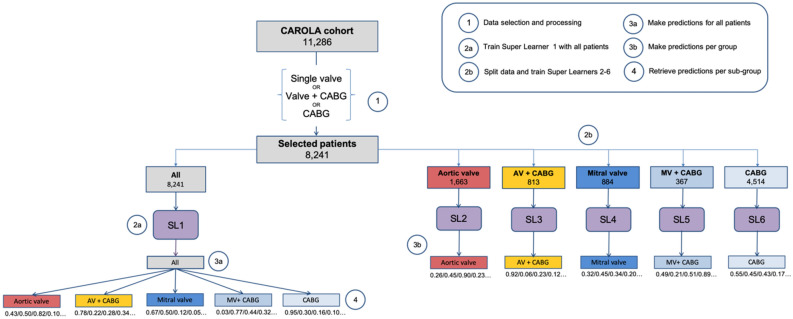
Two distinct training procedures for the SL were carried out (Fig. 6). First, one of the ensembles (SL1) was trained using the full cohort of 8241 patients. Secondly, the cohort was split into five different groups according to operation type, with one ensemble trained on data from each group (SL2–SL6). All six ensembles included the same candidate algorithms, and the same hyper-parameter configurations. Performance of two different approaches were assessed by comparison of the 10-fold cross-validated area under the receiver operated characteristic curve (AUROC), with a 95% confidence interval, for each of the weighted SL’s. Differences in the performance between SL’s and between SL1 and the GLM were assessed with DeLong’s nonparametric test for the difference in areas under the curve30.
Figure 6.
Diagram of the steps involved in data analysis: data split, algorithm training, and outcome prediction using different Super Learner ensembles. On the left, the process of training the single Super Learner on data of the whole cohort (n = 8241), obtaining the pooled predicted probabilities, and retrieving the group-specific probabilities to calculate the performance measures for each type of operation. On the right, the process of splitting the data into five groups, one per operation type, and training a different super learner on data from one type of operation only. SL super learner, AV aortic valve, MV mitral valve, CABG coronary artery bypass grafting.
Calibration, sensitivity analysis and adjusted risk thresholds based on predicted probability of mortality
Calibration plots and calibration indices (ECI)31 for all models are provided in the “Supplementary material”. Model performance metrics described above were obtained in a 2-step procedure: first using a default threshold to maximize the AUROC, and then using adjusted thresholds to optimize sensitivity and specificity. This process of tuning the operating points of the ROC using different risk thresholds depending on the requirements of a specific clinical setting has been previously shown to optimize model sensitivity and specificity for mortality prediction15. In the first step, a default threshold of 0.50 was used, where patients are classified as “non-survivors” if the predicted probability of mortality is greater than 50%. This is the standard threshold used to maximize algorithm performance during training. After this, a second and third risk thresholds were defined. The second one was calculated based on the maximized Youden index, which provides a balance between sensitivity and specificity15. The third one was based on the actual long-term mortality rate of each of the surgical sub-groups, and corresponds to a 50% increase in the absolute risk of mortality. We opted for this value as it represents a clinically relevant increase that could justify intervention. The confusion matrix, sensitivity, and specificity for each of the thresholds are reported in the “Supplementary material”.
Variable importance analysis
Variable importance measures aim at estimating the contribution of predictor variables to changes in the outcome32. The greater the association between each feature and the outcome, the greater the decrease in accuracy upon its removal, and the higher its reported importance32. We determined the variable importance of all routinely measured peri-operative clinical parameters in our cohort by training the best performing individual algorithm included in the ensemble—the XGBoost model—using the same hyper-parameter configurations as in the SL. The coefficients for the top ten features for each operation type, as well as for all operations combined, are presented.
All analyses were performed using R version 3.6.2 (The R Foundation for Statistical Computing; Vienna, Austria) for Ubuntu 16.04 LTS. Data are expressed as mean (95% confidence interval), and categorical as percentages. A P value < 0.05 was accepted as a statistically significant difference.
Supplementary Information
Author contributions
J.C.F., M.W., R.H., and A.E. designed and directed the study. J.C.F., M.W., and M.G. selected and implemented the machine learning algorithms. J.C.F., V.P., I.H., R.H., and A.E. drafted the paper. H.M., F.G., H.B., T.W.L.S., M.N., and M.M. contributed to data acquisition and revised the paper. All authors read and approved the manuscript.
Competing interests
TWLS received research grants and honoraria from Edwards Lifesciences (Irvine, CA, USA) and Masimo Inc. (Irvine, CA, USA) for consulting and lecturing and from Pulsion Medical Systems SE (Feldkirchen, Germany) for lecturing. All other authors have no competing interests to report.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-82403-0.
References
- 1.Loef BG, Epema AH, Smilde TD, Henning RH, Ebels T, et al. Immediate postoperative renal function deterioration in cardiac surgical patients predicts in-hospital mortality and long-term survival. J. Am. Soc. Nephrol. 2005;16(1):195–200. doi: 10.1681/ASN.2003100875. [DOI] [PubMed] [Google Scholar]
- 2.Loef BG, Epema AH, Navis G, Ebels T, Stegeman CA. Postoperative renal dysfunction and preoperative left ventricular dysfunction predispose patients to increased long-term mortality after coronary artery bypass graft surgery. Br. J. Anaesth. 2009;102(6):749–755. doi: 10.1093/bja/aep088. [DOI] [PubMed] [Google Scholar]
- 3.Mehta RL, Kellum JA, Shah SV, Molitoris BA, Ronco C, et al. Acute kidney injury network: Report of an initiative to improve outcomes in acute kidney injury. Crit. Care. 2007;11(2):R31. doi: 10.1186/cc5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lassnigg A, Schmidlin D, Mouhieddine M, Bachmann LM, Druml W, et al. Minimal changes of serum creatinine predict prognosis in patients after cardiothoracic surgery: A prospective cohort study. J. Am. Soc. Nephrol. 2004;15(6):1597–1605. doi: 10.1097/01.ASN.0000130340.93930.DD. [DOI] [PubMed] [Google Scholar]
- 5.Bouma HR, Mungroop HE, de Geus AF, Huisman DD, Nijsten MNW, et al. Acute kidney injury classification underestimates long-term mortality after cardiac valve operations. Ann. Thorac. Surg. 2018;106(1):92–98. doi: 10.1016/j.athoracsur.2018.01.066. [DOI] [PubMed] [Google Scholar]
- 6.D’Agostino RS, Jacobs JP, Badhwar V, Fernandez FG, Paone G, et al. The Society of Thoracic Surgeons Adult Cardiac Surgery Database: 2018 update on outcomes and quality. Ann. Thorac. Surg. 2018;105:15–23. doi: 10.1016/j.athoracsur.2017.10.035. [DOI] [PubMed] [Google Scholar]
- 7.Forte JNC, Wiering MA, Bouma HR, de Geus AG, Epema AH. Predicting long-term mortality with first week post-operative data after Coronary Artery Bypass Grafting using Machine Learning models. PMLR. 2017;68:39–58. [Google Scholar]
- 8.Allyn J, Allou N, Augustin P, Philip I, Martinet O, et al. A comparison of a machine learning model with euroscore II in predicting mortality after elective cardiac surgery: A decision curve analysis. PLoS ONE. 2017;12(1):e0169772. doi: 10.1371/journal.pone.0169772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pirracchio R, Petersen ML, Carone M, Rigon MR, Chevret S, et al. Mortality prediction in intensive care units with the Super ICU Learner Algorithm (SICULA): A population-based study. Lancet Respir. Med. 2015;3(1):42–52. doi: 10.1016/S2213-2600(14)70239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nanayakkara S, Fogarty S, Tremeer M, Ross K, Richards B, et al. Characterising risk of in-hospital mortality following cardiac arrest using machine learning: A retrospective international registry study. PLoS Med. 2018;15(11):e1002709. doi: 10.1371/journal.pmed.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherifa M, Blet A, Chambaz A, Gayat E, Resche-Rigon M, et al. Prediction of an acute hypotensive episode during an ICU hospitalization with a super learner machine-learning algorithm. Anesth. Analg. 2020;130(5):1157–1166. doi: 10.1213/ANE.0000000000004539. [DOI] [PubMed] [Google Scholar]
- 12.Hatib F, Jian Z, Buddi S, Lee C, Settels J, Sibert K, et al. Machine-learning algorithm to predict hypotension based on high-fidelity arterial pressure waveform analysis. Anesthesiology. 2018;129:663–674. doi: 10.1097/ALN.0000000000002300. [DOI] [PubMed] [Google Scholar]
- 13.Davies SJ, Vistisen ST, Jian Z, Hatib F, Scheeren TWL. Ability of an arterial waveform analysis-derived hypotension prediction index to predict future hypotensive events in surgical patients. Anesth. Analg. 2020;130:352–359. doi: 10.1213/ANE.0000000000004121. [DOI] [PubMed] [Google Scholar]
- 14.van der Laan MJ, Polley EC, Hubbard AE. Super learner. Stat. Appl. Genet. Mol. Biol. 2007;6(1):1544–6115. doi: 10.2202/1544-6115.1309. [DOI] [PubMed] [Google Scholar]
- 15.Bihorac A, Ozrazgat-Baslanti T, Ebadi A, Motaei A, Madkour M, et al. MySurgeryRisk: Development and validation of a machine-learning risk algorithm for major complications and death after surgery. Ann. Surg. 2019;269(4):652–662. doi: 10.1097/SLA.0000000000002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorsen-Meyer H-C, Nielsen AB, Nielsen AP, Kaas-Hansen BS, Toft P, et al. Dynamic and explainable machine learning prediction of mortality in patients in the intensive care unit: A retrospective study of high-frequency data in electronic patient records. Lancet Digital Health. 2020;2(4):e179–e191. doi: 10.1016/S2589-7500(20)30018-2. [DOI] [PubMed] [Google Scholar]
- 17.Gordon L, Austin P, Rudzicz F, Grantcharov T. MySurgeryRisk and machine learning: A promising start to real-time clinical decision support. Ann. Surg. 2019;269(1):e14–e15. doi: 10.1097/SLA.0000000000002877. [DOI] [PubMed] [Google Scholar]
- 18.Arnan MK, Hsieh TC, Yeboah J, Bertoni AG, Burke GL, et al. Postoperative blood urea nitrogen is associated with stroke in cardiac surgical patients. Ann. Thorac. Surg. 2015;99:1314–1320. doi: 10.1016/j.athoracsur.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 19.Chung PJ, Carter TI, Burack JH, Tam S, Alfonso A, et al. Predicting the risk of death following coronary artery bypass graft made simple: a retrospective study using the American College of Surgeons National Surgical Quality Improvement Program database. J. Cardiothorac. Surg. 2015;10:62. doi: 10.1186/s13019-015-0269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazory A. Emergence of blood urea nitrogen as a biomarker of neurohormonal activation in heart failure. Am. J. Cardiol. 2010;106:694–700. doi: 10.1016/j.amjcard.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 21.Gotsman E, Zwas D, Planer D, Admon D, Lotan C, Keren A. The significance of serum urea and renal function in patients with heart failure. Medicine. 2010;89(4):197–203. doi: 10.1097/MD.0b013e3181e893ee. [DOI] [PubMed] [Google Scholar]
- 22.Matsue Y, van der Meer P, Damman K, Metra M, O'Connor CM, et al. Blood urea nitrogen-to-creatinine ratio in the general population and in patients with acute heart failure. Heart. 2017;103(6):407–413. doi: 10.1136/heartjnl-2016-310112. [DOI] [PubMed] [Google Scholar]
- 23.Cherry AD. Mitochondrial dysfunction in cardiac surgery. Anesthesiol. Clin. 2019;37(4):769–785. doi: 10.1016/j.anclin.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chouchani ET, Pell VR, Gaude E, Aksentjevic D, Sundier SY, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J, Zhang J, Tian J, Virzì GM, Digvijay K, et al. Mitochondria in sepsis-induced AKI. J. Am. Soc. Nephrol. 2019;30(7):1151–1161. doi: 10.1681/ASN.2018111126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Apolito M, Du X, Pisanelli D, Pettoello-Mantovani M, Campanozzi A, et al. Urea-induced ROS cause endothelial dysfunction in chronic renal failure. Atherosclerosis. 2015;239(2):393–400. doi: 10.1016/j.atherosclerosis.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Buuren S, Groothuis-Oudshoorn K. Mice: Multivariate imputation by chained equations in r. J. Stat. Softw. 2011;45(3):1–67. doi: 10.18637/jss.v045.i03. [DOI] [Google Scholar]
- 28.Breiman L. Bagging predictors. Mach. Learn. 1996;24:123–140. [Google Scholar]
- 29.Dudoit, S., van der Laan, M.J. Asymptotics of cross-validated risk estimation in estimator selection and performance assessment. Accessed 1 June 2020; http://biostats.bepress.com/ucbbiostat/paper126/. (2006).
- 30.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988;44(3):837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 31.van Hoorde K, Van Huffel S, Timmerman D, Bourne T, Van Calster B. A spline-based tool to assess and visualize the calibration of multiclass risk predictions. J. Biomed. Inform. 2015;54:283–293. doi: 10.1016/j.jbi.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 32.Díaz I, Hubbard A, Decker A, Cohen M. Variable importance and prediction methods for longitudinal problems with missing variables. PLoS ONE. 2015;10(3):e0120031. doi: 10.1371/journal.pone.0120031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.