Abstract
Colorectal cancer, liver cancer, stomach cancer, pancreatic cancer, and esophageal cancer are leading causes of cancer related deaths worldwide. A fundamental trait of virtually all gastrointestinal cancers is genomic and epigenomic DNA alterations. Cancer cells acquire genetic and epigenetic alterations that drive the initiation and progression of the cancers by altering the molecular and cell biological processes of the cells. These alterations, as well as other host and microenvironment factors, ultimately mediate the clinical behavior of the pre-cancers and cancers and can be used as biomarkers for cancer risk determination, early detection of cancer and pre-cancer, determination of the prognosis of cancer and prediction of the response to therapy. Epigenetic alterations have emerged as one of most robust classes of biomarkers and are the basis for a growing number of clinical tests for cancer screening and surveillance.
Keywords: DNA methylation, noncoding RNA, chromatin, histone, colorectal cancer, esophageal cancer, gastric cancer, pancreatic cancer, Barretts esophagus, biomarkers, diagnosis, prognosis, predictive, treatment
INTRODUCTION
DNA Alterations In Cancer
Cancer develops as a consequence of disruption of the mechanisms that regulate fundamental processes, such as cellular proliferation, cell metabolism, angiogenesis, cell death, invasion, metastasis, as well as other hallmark behaviors of cancer1, 2. (Figure 1) These disruptions arise through an evolutionary process that stably encodes acquired oncogenic alterations in the genome and epigenome, which can then accumulate in clonal lineages. Genetic alterations, including gene sequence mutations, gene copy number alterations, insertions, deletions, and recombination events, were the first and most clearly demonstrated of these oncogenic alterations of DNA, leading to the prominence of the model of cancer genetics in cancer biology. More recently epigenetic alterations have been established as another fundamental oncogenic mechanism and appear to play a prominent role in the pathogenesis of cancer by inducing hallmark cancer cell behaviors. Genome-wide genomic and epigenomic analyses have revealed the widespread occurrence of mutations in epigenetic regulators as well as the breadth of alterations to the epigenome in cancer cells3, 4. It is clear that genetic and epigenetic mechanisms influence and cooperate with each other to enable the acquisition of the hallmarks of cancer2.
Figure 1:
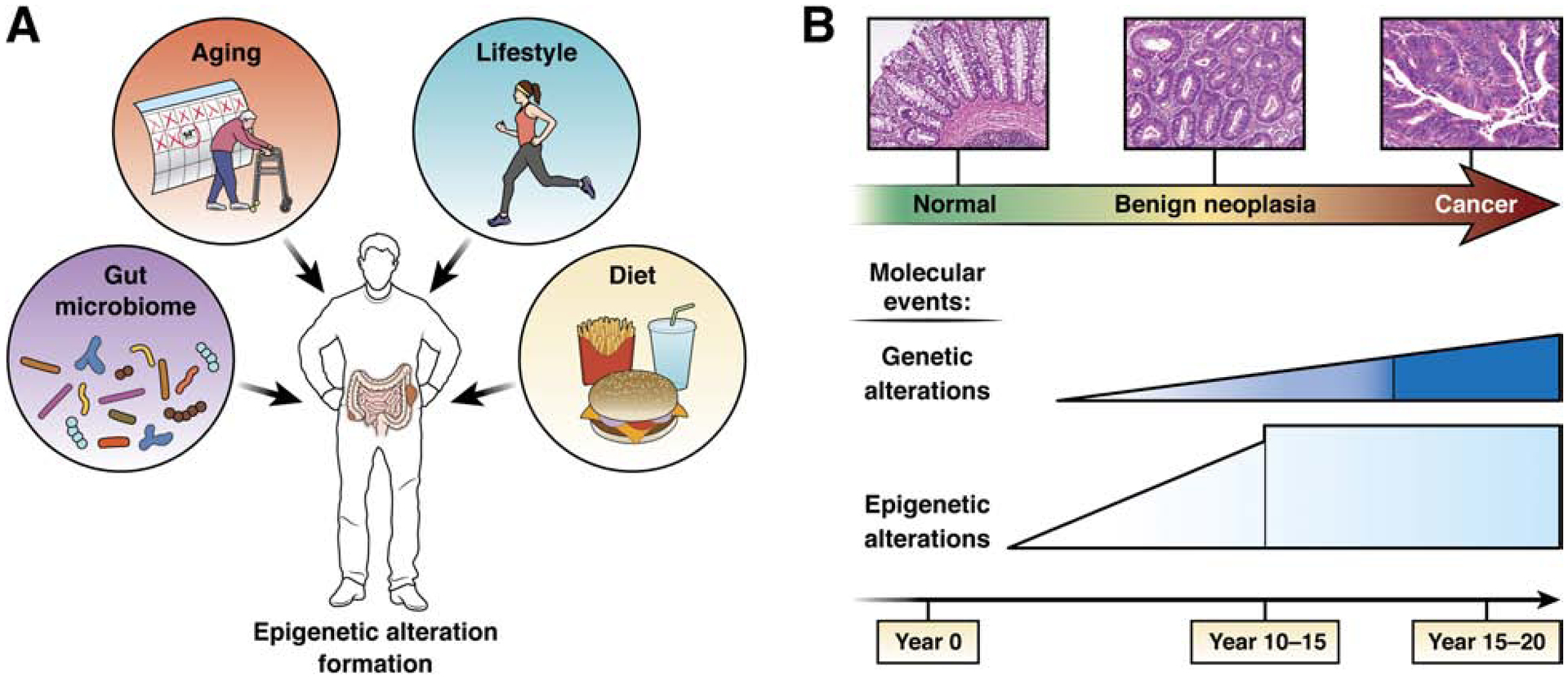
Schematic diagram showing factors that influence epigenetic alteration formation and the timing of epigenetic alteration formation in GI tract cancer formation.
The prevailing consensus suggests that epigenetic alterations in cancer occur early and are more common than genetic alterations. In addition, advances in genomic and epigenomic analysis technologies have led to the identification of a variety of specific epigenetic alterations that can be used as potential clinical biomarkers for gastrointestinal (GI) cancer patients. The use of these recently developed technologies has led to insights into cancer epidemiology and hereditary cancer syndromes as well as cancer biology. Because of the potential for environmental factors to modify epigenetic states, the association of cancer risk factors, such as tobacco exposure, on cancer epigenetics has also been assessed. There is modestly robust evidence to date suggesting such factors have significant effects in humans5, 6. It is also notable that GI cancer syndromes, such as Lynch syndrome, can arise from germline epigenetic alterations7, 8. However, familial epigenetic cancer syndromes are rare and appear to be rarely transmitted to offspring9. This review briefly outlines the fundamental basis of epigenetic alterations in cancer, and details the current state of the field regarding the promise and clinical usefulness of various epigenetic alterations as biomarkers for the early detection, diagnosis, prognosis and management of GI pre-cancer and cancer, with a primary focus of colorectal and esophageal cancer. The classes of cancer epigenetic alterations that will be the focus of this review will be DNA methylation and chromatin and histone structure and function. Noncoding RNAs and microRNAs are beyond the scope of this review and the interested reader is directed to recent reviews of this subject for more information10, 11.
Cancer Epigenetics
Epigenetic mechanisms stably regulate cell behavior by controlling the transcriptional availability of various parts of the genome through differential DNA methylation, chromatin marking and DNA packaging via histone modifications. The best studied of these mechanisms involve direct DNA modifications (primarily CpG cytosine-5’ methylation12, 13, as well as hydroxylation, formylation, and carboxylation14, 15); nucleosome occupancy and positioning16–18; nucleosome alterations (e.g. histone variants, different histone modifications)19; and noncoding RNAs20–22. Cancer related epigenetic alterations cooperate through a network of mutually reinforcing or counteracting signals. Genome-scale projects charting the human epigenome are rapidly extending our understanding of epigenetic marks and how they interact23. (The interested reader can visit the NIH Roadmap Epigenomics Project for more details at http://www.roadmapepigenomics.org/) Importantly, epigenetic mechanisms induce stable cell phenotypes but can adapt to changing developmental or environmental needs through the activities of proteins and noncoding RNAs (ncRNA), that regulate the epigenetic state of the genome and its effects on transcription. Recent studies of the aberrant regulation of gene enhancers and super-enhancers in cancer have further extended our understanding of the effects of epigenetic alterations on tumor formation and have shown how epigenetic alterations in noncoding DNA elements can promote cancer formation. The interested reader is directed to recent reviews of this topic24, 25 The epigenome regulators can be classified as: 1) initiators (e.g. long noncoding RNAs, transcription factors); 2) histone writers, which establish the epigenetic marks; 3) readers, which interpret the epigenetic marks; 4) erasers, which remove the epigenetic marks; 5) remodelers, which can alter chromatin states; and 6) insulators, which form boundaries between epigenetic domains. The establishment, maintenance, and modification of epigenetic marks are intricately regulated, with crosstalk among the marks and writers to shape and alter the epigenetic landscape23.
DNA Methylation
DNA methylation is an epigenetic DNA modification that can mediate a variety of processes in cells, such as maintenance of genome integrity, genomic imprinting, transcriptional regulation, and developmental processes26. DNA methylation in eukaryotic cells primarily occurs at the 5-prime position of the cytosine ring within CpG dinucleotides and regulates gene transcription via effects on promoters and noncoding DNA elements, such as enhancers. The modification at 5-methyl cytosine is catalyzed by a family of DNA methyltransferases (DNMTs) that include DNMT1, a DNA methylation maintenance enzyme, and DNMT3A and DNMT3B, which are de novo enzymes that target unmethylated CpGs to initiate methylation. DNMT3A and DNMT3B are highly expressed during embryogenesis and only minimally in adult tissues; whereas DNMT1 is constitutively active in all adult replicating tissues26. Another family member is DNMT-3L that lacks intrinsic methyltransferase activity and interacts with DNMT3A to facilitate DNA methylation27. In normal cells, DNA methylation occurs predominantly in repetitive genomic regions, including satellite DNA and parasitic elements such as long interspersed transposable elements (LINEs) and short interspersed transposable elements (SINEs), maintaining genomic integrity28, 29. (Figure 2A) Methylated cytosines account for approximately 1% of total nucleotides and are found in about 75% of all CpG dinucleotides in the human genome (approximately 28 million CpGs/genome). The CpG dinucleotides are unevenly distributed across the human genome and are concentrated in areas called CpG islands (CGIs). Roughly 50–60% of gene promoters lie within CpG islands, and it is estimated that the human genome contains approximately 29,000 CGI sequences30. Unlike the rest of the genome, CpG islands particularly those associated with promoters are generally unmethylated in normal cells, providing access to transcription factors and chromatin-associated proteins for the expression of most housekeeping genes and several other regulated genes, although some of them (~6%) become methylated in a tissue-specific manner during early development or in differentiated tissues26, 31. DNA methylation can inhibit gene expression directly, by inhibiting the binding of specific transcription factors, and indirectly, by recruiting methyl-CpG-binding domain (MBD) proteins. The associated MBD family members in turn recruit histone-modifying and chromatin-remodeling complexes to methylated sites32. To date, six methyl-CpG-binding proteins, including methylcytosine binding protein 2 (MECP2), MBD1, MBD2, MBD3, MBD4 and Kaiso, have been identified in mammals. Furthermore, it has been shown that nucleosome remodeling complex (NuRD) can methylate DNA by interacting with DNA methylation binding protein MBD2, which directs the NuRD complex to methylate DNA and that DNA can be actively demethylated by processes involving base excision repair (BER) enzymes33,34, 35. These and other recent findings have established that DNA cytosine methylation is a critical component of epigenetic gene regulation.
Figure 2A and B:
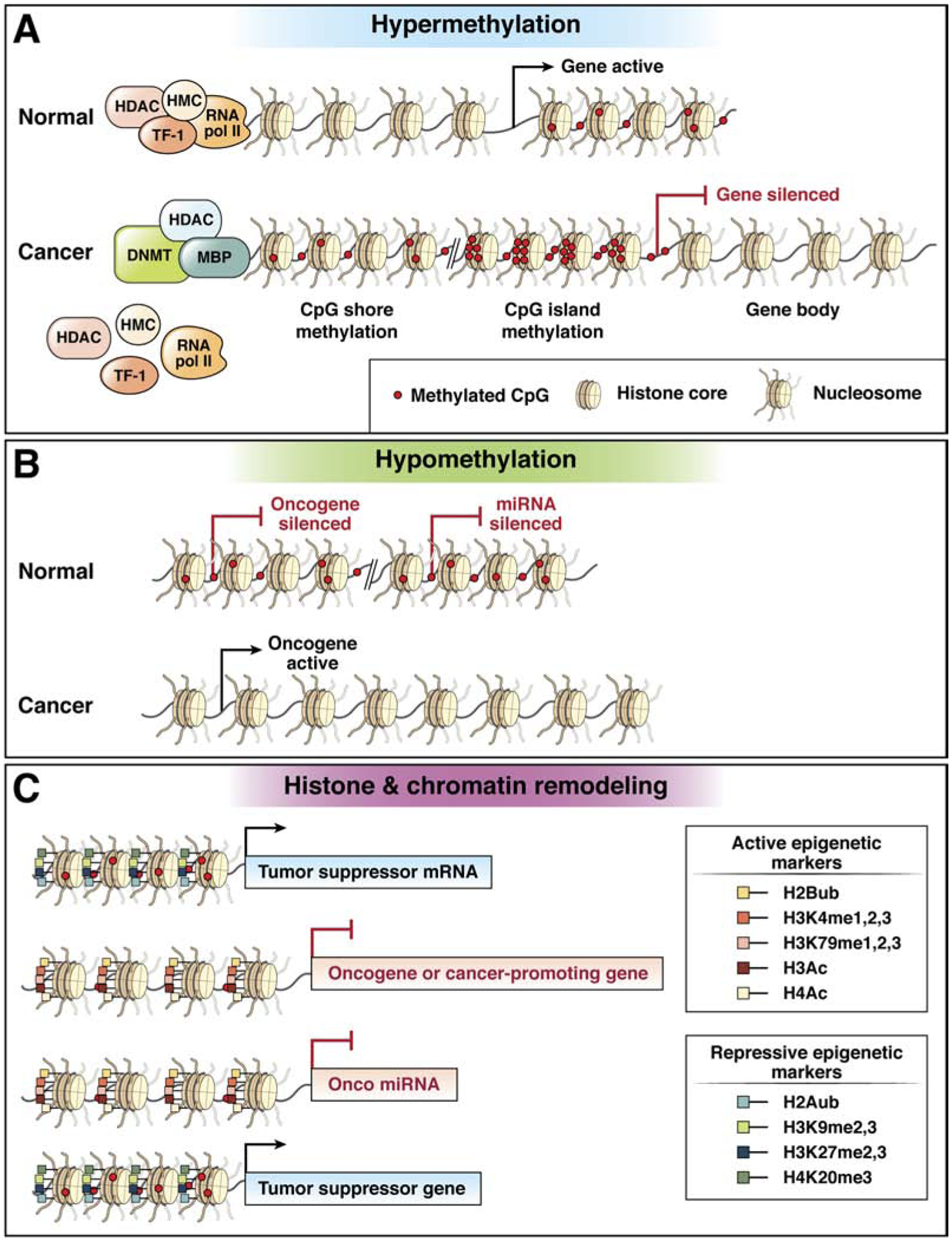
Schematic diagram of DNA hypermethylation and DNA hypomethylation in cancer. (TF-1=transcription factor 1, TF-2=transcription factor 2, TF-3=transcription factor 3, RNA pol II=RNA polymerase II, HDAC=histone deacetylase complex, DNMT=DNA methyltransferase, MBP=methyl binding protein, Onco miRNA=oncogenic microRNA. CpG shores are the regions immediately flanking CpG islands (CGI), and they can be methylated just as CpG islands can be. The consensus definition of a CpG shore is up to 2kbp away from the CpG island. miRNA silencing refers to hypermethylation of microRNA loci that silence miRNA expression, which can suppress tumor suppressor miRNAs or increase the expression of inactivated oncogenes that are the targets of the miRNAs
Figure 2C: Schematic diagram of DNA chromatin structure and histone modification states that affect conformation of chromatin. (H2A=histone 2A, H2B=histone 2B, H3=histone 3, H4=histone 4, ub=ubiquitylated, me=methylated, Ac=acetylated, K=lysine. Onco miR is an oncogenic microRNA.)
In cancer, including all gastrointestinal cancers, global DNA hypomethylation is observed as well as aberrant regional DNA hypermethylation. DNA hypermethylation has been extensively studied in virtually all cancers and is believed to contribute to cancer formation via repression of tumor suppressor gene expression36–38. DNA hypomethylation is also a hallmark feature of cancer, however, its functional role in cancer formation is less well understood. It has been suggested to contribute to cancer formation via the induction of genomic instability, inducing the expression of parasitic elements of DNA or by inducing the expression of oncogenes or cancer germline genes, but definitive evidence for an active causal (ie “driver”) role in cancer formation is lacking at this time39–42
Chromatin Alterations And Histone Modifications
Histones are a family of small basic proteins that include H2A, H2B, H3, and H4 and that have a globular domain and a flexible charged NH2 terminus known as the histone tail, which protrudes from a protein complex called a nucleosome. A nucleosome is an octomer of histone proteins and encompasses ~146 bp of DNA wrapped around this octomer. These octamers consist of two subunits of each of the core histone proteins43. Histone modifications influence chromatin structure, which plays an important role in gene regulation and carcinogenesis44, 45. In addition, there are histone variants that provide an additional layer of regulation, including H2A.Z, MacroH2A, H2A-Bbd, H2AvD, H2A.X, H3.3, CenH3, and H3.446.
Chromatin is a highly ordered B-form structure consisting of repeats of nucleosomes connected by linker DNA and exists in two distinct conformation states: heterochromatin, which is densely compacted and transcriptionally inert, and euchromatin, which is decondensed and transcriptionally active47.(Figure 2B). Chromatin consists of DNA, histones, and non-histone proteins condensed into nucleoprotein complexes and functions as the physiological template of all eukaryotic genetic information48. Regulation of gene expression occurs through posttranslational modifications of the histone tails provided by covalent modifications, such as acetylation, methylation, phosphorylation, ubiquitination, sumoylation, proline isomerization, and ADP ribosylation49–52. Posttranslational modifications to histone tails govern the structural status of chromatin and resulting transcriptional status of genes within a particular region of DNA. Euchromatin is characterized by high levels of acetylation and trimethylated H3K4, H3K36 and H3K79. On the other hand, heterochromatin is characterized by low levels of acetylation and high levels of H3K9, H3K27 and H4K20 methylation53, 54. These modifications are reversible and are controlled by a group of enzymes that include histone acetyltransferases (HATs) and deacetylases (HDACs), methyltransferases (HMTs) and demethylases (HDMs), kinases, phosphatases, ubiquitin ligases and deubiquitinases, SUMO ligases and proteases, which add and remove such modifications52, 55. These histone modifiers generally act in complexes, such as the repressive Polycomb (PcG) and activating Trithorax (TrxG) group complexes, which counterbalance each other in the regulation of genes important for development and have been implicated in cancer56.
ATP-dependent chromatin-remodeling complexes are responsible for sliding of the nucleosomes, as well as insertion and ejection of histone octamers, which are processes that are important for transcriptional repression and activation, and other important cellular functions such as DNA replication and repair. There are also remodeling complexes that can be divided into four families: SWI/SNF, CHD (chromodomain and helicase-like domain), ISWI, and INO80.
With regards to cancer, histone modification alterations, alterations in chromatin structure and deregulation of the genes that regulate histone modifications and chromatin structure have been found to occur commonly in many cancer types, including gastrointestinal cancers11. These alterations appear to have cancer specific signature patterns and have potential to be used as cancer biomarkers. There is also interest in developing epigenetic therapies for cancer, although these have not been shown to be particularly effective in the treatment of gastrointestinal cancers to date57, 58.
EPIGENETIC ALTERATIONS AND COLORECTAL CANCER
The current paradigm of the molecular pathogenesis of CRC is that colon neoplasms can arise through a variety of discrete molecular pathways that are driven predominantly by different primary mechanisms, including chromosomal instability (CIN), microsatellite instability (MSI), epigenetic instability (e.g. CpG Island Methylator Phenotype (CIMP) subclass of CRC), altered tumor microenvironments (e.g. CMS4 subclass of CRC) and altered metabolic states (e.g. CMS3 subclass of CRC)59–61.
With specific regards to the epigenetic alterations found in colorectal polyps and CRC, aberrant DNA methylation is the best understood class of epigenetic alterations in CRC and affects both the formation of colon polyps as well CRCs. The discovery of a molecular subclass of CRCs defined as having a “CpG island methylator phenotype (or CIMP)” in 1999 was a significant advance in our understanding of the molecular mechanisms that orchestrate colorectal tumor formation and revealed the role of aberrant DNA methylation, in particular, as an important epigenetic alteration in CRC62, 63. CIMP CRCs arise predominantly from sessile serrated lesions and exhibit unique clinicopathological and molecular features, including a predilection for proximal location in the colon, female gender, poor and mucinous histology, the presence of frequent KRAS and BRAF mutations and frequent MSI due to biallelic MLH1 methylation.62, 64–66,67, 68,37, 63.
DNA alterations in colon neoplasms include both hypermethylation and hypomethylation of DNA. DNA hypermethylation can silence tumor-suppressor genes via effects on promoters and noncoding DNA elements (e.g. enhancers, etc.)69, 70. Global DNA hypomethylation is commonly observed in most cancers, including CRC. It is believed to influence CRC development by inducing chromosomal instability, global loss of imprinting and super-enhancer activation70–73. Genome-wide hypomethylation generally occurs within repetitive transposable DNA elements such as the LINE-1 or short interspersed nucleotide elements (SINE, or Alu) sequences74–76. LINE-1 hypomethylation inversely associates with MSI and/or CIMP76, 77. Furthermore, a number of studies have demonstrated that a high degree of LINE-1 hypomethylation correlates with worse patient survival78–81. One hypothesis is that hypomethylation of LINE/SINE sequences may induce inadvertent activation of potential proto-oncogenes82, which implies that LINE-1 hypomethylation has a functional role in CRC formation83.
Aberrant DNA methylation in the “traditional” and “serrated” polyp pathways
As stated earlier, there are multiple pathways for the formation of colon polyps and CRC, and sessile serrated lesions (SSLs) as well as adenomas have malignant potential60, 84–86. It is of note that the terminology for serrated polyps has recently changed and that the term SSLs refers to lesions previously called sessile serrated adenomas or sessile serrated polyps87. There are at least 2 or 3 recognized serrated polyp pathways that begin with different precursor lesions: hyperplastic polyps, serrated polyps, and serrated adenomas, the malignant potential of these precursors varies significantly64. Notably, recent findings suggest that in addition to microenvironment factors, genetic and epigenetic alterations that occur at low frequency in the normal colon mucosa are a major factor involved with whether the initial lesion in CRC formation is an adenoma vs. SSL88–90, The bulk of epigenetic alterations appear to arise during polyp formation, which is unlike genetic alterations, which occur predominantly after the polyp initiation90.
The adenoma to CRC pathway is initiated by alterations in the WNT signaling pathway, most commonly APC mutations, and subsequent progression is associated with alterations that affect the MAPK and TP53 pathways (e.g. KRAS and TP53 mutations and methylation of genes that regulate these pathways). The SSL to CRC sequence is primarily characterized by activation of the mitogen activated protein kinase (MAPK) pathway by oncogenic mutations in BRAF and KRAS. Serrated polyps that carry mutant BRAF are also commonly CIMP and can progress to microsatellite stable or unstable CRC, depending on whether epigenetic inactivation of the mismatch repair protein MLH1 occurs84 In contrast, traditional serrated adenomas more commonly activate the MAPK pathway via KRAS mutations, carry RSPO fusion transcripts, and typically have a low CIMP status. Unlike classic adenomatous polyps, sessile serrated polyps and traditional serrated adenomas do not typically have genetic alterations in APC or CTNNB184,91, and appear to activate WNT signaling late in the polyp to CRC sequence via aberrant DNA methylation of SFRP family genes such as CDX2, MCC92, 93. SSLs also more commonly employ epigenetic alterations to disrupt other CRC pathways and processes, including the p53 signaling pathway (IGFBP7)94 and cell cycle control proteins (CDKN2A)95.
Clinical Applications Of DNA Methylation In The Prevention And Management Of CRC
There is considerable enthusiasm for the use of cancer related molecular alterations for the prevention and management of a variety of cancers, including the majority of GI cancers. Aberrantly methylated DNA biomarkers have proven the most robust and successful clinically used molecular markers to date. Another epigenetic feature, chromatin alterations, is an emerging class of promising biomarkers that will likely be in clinical use in the near future96. For the past two decades, we have witnessed a tremendous effort on the development of DNA-methylation based biomarkers in the prevention and management of CRC, and some of them are now available for clinical use (Figure 3, Table 1). The earliest application of methylated gene biomarkers in CRC clinical care occurred over a decade ago and was the use of methylated MLH1 for determining the likelihood of MSI CRC being sporadic vs. hereditary in origin97. Methylated MLH1 and BRAF mutations arise almost exclusively in sporadic MSI CRC and are used in the clinic to identify CRC patients who should be considered for genetic testing.
Figure 3:
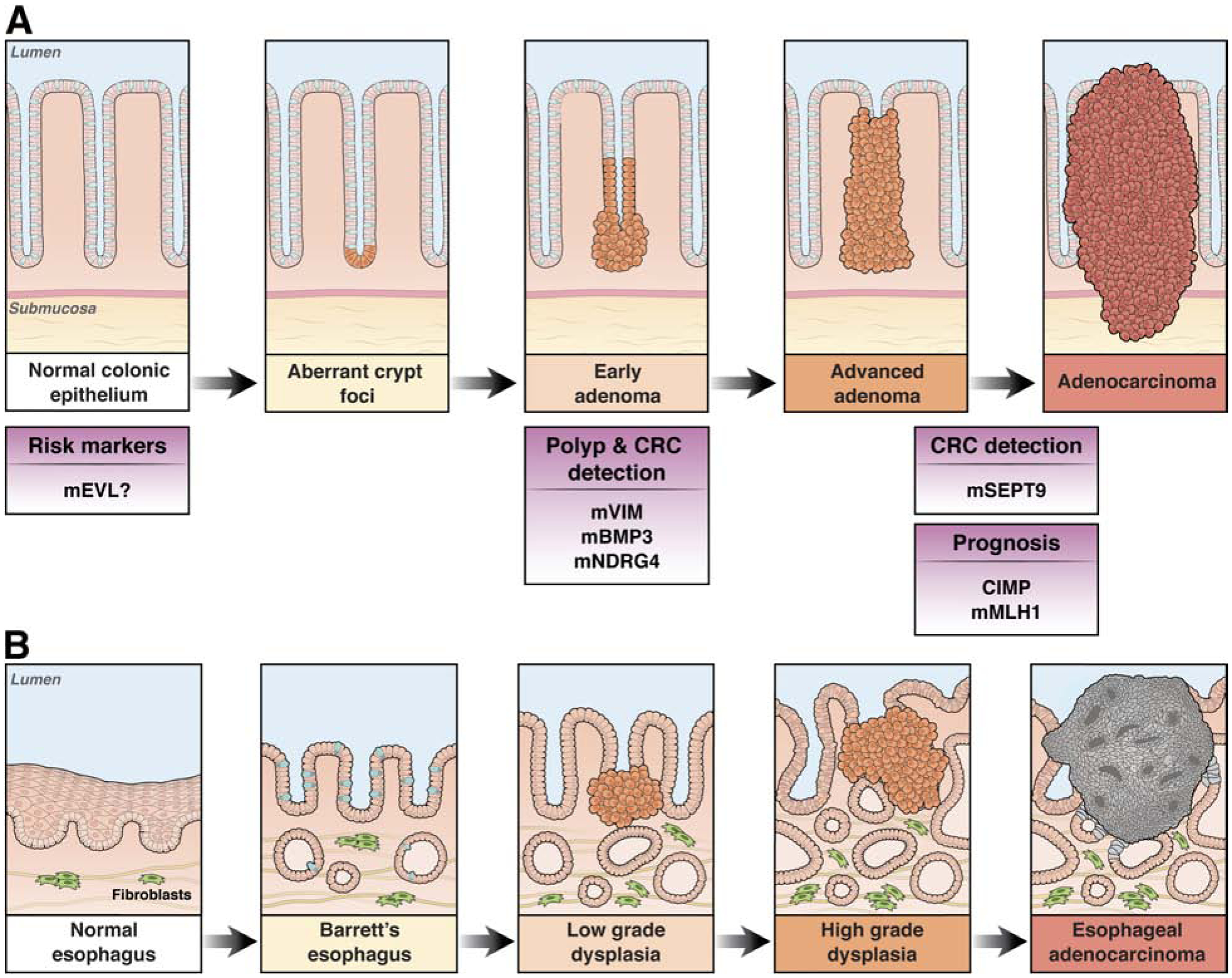
Diagram showing the polyp-to-CRC progression sequence and associated methylated gene alterations that have been shown to be potential or clinically used polyp and CRC detection markers or prognostic markers. The prefix “m” designates the methylated gene. mEVL and mMLH1 are used as tissue based biomarkers, whereas mVIM, mNDRG4, and mBMP3 are stool based biomarkers. mSEPT9 is a blood-based biomarker.
Table 1:
Validated DNA-methylation biomarkers for colorectal cancer
| Clinical Use | Biomarkers | Available Commercial Assays | Study Design of Major Trials with Assay | Ref. |
|---|---|---|---|---|
| Stool-based CRC screening | mVIM, | ColoSure™ | Case (N=42) control (N=241) study | 109 |
| mBMP3 and mNDRG4 | Cologuard® (detects mutant KRAS, and includes a FIT test) | Prospective cohort based clinical trial in screening population (N=9989) | 110 | |
| blood-based diagnostic marker | mSEPT9 | EpiproColon® 1.0; ColoVantage®; RealTime mS9 | Multiple trials: 1)Prospective cohort based clinical trial in screening population (N=7941) (Church);2) Case-Control study (N=269) (deVos); 3) Case-Control study (N=312) (Lofton-Day, 2008) | 125 135,138,139 |
| mBCATI mIKZFI | Colvera | Cross-sectional study (N=220) | 133 | |
| Tissue-based prognostic markers | CIMP panel | NA | Multiple trials:
|
140, 142, 159, 188 |
| Diagnostic tool to screen for Lynch syndrome | mMLHI | MLH1 Hypermethylation analysis |
|
67 |
Screening/Early detection
Because epigenetic alterations are much more frequent than genetic mutations in polyps98, they have greater potential as diagnostic biomarkers for the detection of colonic polyps and cancers, which is evident by the fact that the currently clinically approved screening assays for CRC, the ColoGuard assay (Exact Sciences) and EpiProcolon (Epigenomics) are based on methylated DNA. The greater potential for methylated DNA based alterations and perhaps other epigenetic alterations to detect colon polyp and early stage CRC in circulating DNA has also led to the development of blood-based assays for CRC screening that are currently under clinical investigation or that are available clinically. (See section below.)
Stool-based biomarkers
Specific biofluids, such as blood (plasma or serum) and feces are the most common analytes used in CRC screening tests. Since the initial discovery by Sidransky and colleagues of mutant KRAS in fecal specimens from patients with CRC99, numerous studies have supported using fecal DNA for potential screening assays for the early detection of CRC.
To date, a large number of hypermethylated genes including APC, ATM, BMP3, CDKN2A, SFRP2, GATA4, GSTP1, HLTF, MLH1, MGMT, NDRG4, RASSF2A, SFRP2, TFPI2, VIM, WIF1 as well as others, have been analyzed in fecal DNA for the early detection of CRC100–107. Of this large list of potential epigenetic based CRC screening markers, methylated VIM, BMP3, and NDRG4 have been shown to be robust and accurate enough to be approved for clinical use. Methylated VIM was the first stool-based epigenetic biomarker approved for the early detection of CRC and was marketed under the name ColoSure™, (Lab Corp, Burlington, NC)108, 109.
Subsequent to the ColoSure assay, a next generation stool based multi-target (MT) stool DNA assay was developed and FDA approved in 2014. This assay is a stool DNA based assay that detects methylated BMP3, methylated NDRG4, mutant KRAS, and occult hemoglobin (Cologuard® (Exact Sciences Corporation). In a large clinical trial of average risk individuals (the Deep C trial), this MT stool DNA assay was compared to the FIT assay and to colonoscopy (N=9989)110, and showed an overall sensitivity of 92% (95% CI, 83–97.5%) for CRC and 93% (95% CI 83.8–98.2%) for stage I-III CRC, compared to sensitivity of FIT at 74% (95% CI, 61.5–84%) and 73% (95% CI, 60.3–83.9%), respectively (p=0.002). For advanced adenomas and sessile serrated polyps, the sensitivity of the test increased proportionately with lesion size and grade. The molecular assay was significantly more sensitive than FIT for advanced adenomas: 42% (95% CI, 38.9–46%) vs. 24% (95% CI, 20.8–27%), respectively, for those ≥1 cm and 66% vs 43% for those ≥2 cm (p< 0.001). Sessile serrated polyps ≥1 cm were detected at a rate of 42% for the molecular assay compared 5% for FIT (p< 0.001); In this study the test specificity for detection of CRC and advanced pre-cancers was 87%110.
Since 2014, >3 million MT stool DNA assays have been conducted to date leading to a real-world use experience of this assay, which has similar performance to that seen in the Deep C trial111. In addition, recent studies have assessed the performance of the MT stool DNA assay in noncompliant patient populations and found improved compliance in this group112. The assay has been also assessed in African Americans, who have a higher risk for CRC than the general population, and was found to have equivalent performance to that seem in Caucasian patients113. Importantly, at its current price-point, cost-effectiveness studies have had conflicting results of the utility of the MT stool DNA assay compared to FIT tests114, 115.
Blood-based biomarkers
Due to accessibility and high patient acceptance, blood is invariably regarded to be the most ideal analyte for cancer biomarkers. The majority of blood-based biomarkers for cancer to date have been proteins or glycoproteins (e.g. PSA, CEA, CA-125, etc.). Recent studies have demonstrated the potential of circulating tumor DNA (ctDNA) for cancer detection and management, and there are now “liquid biopsy” assays being used in the clinical care of cancer patients116, 117. For CRC, somatic tumor-derived mutations in ctDNA are promising markers for the early detection of recurrent cancer, for monitoring treatment response, and for prognosis118–121. However, they have not been shown to be detected at a high frequency in patients with advanced polyps and early stage CRC using current technology, which likely reflects the very low frequency of these mutations in the plasma (6.6ng/ml blood, <0.1–0.01% ct DNA)98, 122–124. Methylated DNA and chromatin fragmentation patterns, in contrast can be found in patients with advanced polyps and early stage CRC more commonly. This is likely a consequence of the high frequency of epigenetic alterations in colon polyps and early CRC. Consequently, methylated DNA has been detected in higher proportions of patients with early stage CRC patients compared to DNA mutations, which has led to their assessment as blood-based early detection assays125, 126.
To date, several potential blood-based diagnostic methylation biomarkers have been identified for CRC detection, including ALX4127, APC107, CDKN2A105, HLTF128, HPP1129, MLH1128, MGMT107, NEUROG1130, NGFR, RASSF2A107, SFRP2, VIM107, and WIF1107, B4GAT1131, BCAT1, IKZF1132; SFRP1, SDC2, and PRIMA1126. The best studied methylated DNA blood-based biomarkers to date are mSEPT9, mSDC2, and a combination of mBCAT1 and mIKZF1, which is marketed under the name Colvera (Clinical Genomics). They have been shown to consistently detect CRC in serum or plasma samples and have high potential to eventually be used in the clinic for monitoring for recurrent CRC133,125,134,135. Other methylated DNA biomarkers are under active investigation for the early detection of recurrent CRC and for screening for CRC.
Currently, the most established methylated DNA blood biomarker is methylated Septin 9 (SEPT9), which belongs to the gene family that encodes a group of GTP-binding136 and filament-forming proteins137 involved in cytoskeletal formation. Lofton-Day and colleagues first identified methylated SEPT9 (mSEPT9) as a non-invasive diagnostic biomarker for CRC reporting a 69% sensitivity and 86% specificity138. However, a subsequent prospective CRC screening trial (PRESEPT) showed lower sensitivity for CRC (48.2% at 91.5% specificity)138. Subsequent studies validated the clinical significance of mSEPT9 as a potential biomarker for CRC screening, but the FDA stance on the test is that it is only recommended in people who refuse to undergo other CRC screening tests, because of its low sensitivity for CRC (52–72%) compared to other CRC screening assays125. It is commercially-offered as a blood-based screening test in various assays including EpiproColon® 1.0 (Epigenomics, Seattle, WA), ColoVantage® (Quest Diagnostics, Madison, NJ) and RealTime mS9 (Abbott Laboratories, Des Plaines, IL) and has been approved for CRC screening by the Chinese FDA. A major issue of mSEPT9 as a CRC screening assay, is its low sensitivity for the detection of advanced adenomas (11%), underscoring the need for further improvement of this test if it is to be used for population-based screening of colorectal neoplasia. A recent study demonstrated that the methylated SEPT9 assay was superior to fecal immunochemical (FIT) at detecting CRC neoplasms, but in this study both approaches were suboptimal for diagnosing patients with advanced adenomas139.
In summary, the data to support the clinical use of CRC screening biomarkers for the early detection of recurrent CRC suggests methylated DNA plasma-based markers have high potential to eventually be used in the clinic for the early detection of recurrent CRC. It is less clear if they will be effective CRC screening markers. The most promising CRC epigenetic alterations under evaluation at this time for CRC screening are based on a combination of biomarkers that include methylated DNA and chromatin fragmentation patterns. These combination biomarker assays are being assessed in ongoing CRC screening trials at this time [Circulating Cell-free Genome Atlas study (NCT02889978; GRAIL); Session VCTPLO2, CT021 - Prediction of cancer and tissue of origin in individuals with suspicion of cancer using a cell-free DNA multi-cancer early detection test, and the ECLIPSE trial of the Lunar-2 assay (Kim, AACR 2020 abstract #916; Guardant Health)]. There appears to be potential for a robust biomarker panel of methylated genes to be be developed into a clinically accurate CRC screening method in the near future.
Epigenetic Prognostic Biomarkers
Currently, the most accurate means for assessing CRC patient prognosis is based on the pathological staging and specific histologic features of the tumor. However, the heterogeneity of survival times in patients with the same stage of CRC is well known, which highlights the need for a more accurate system for determining CRC patient prognosis. Multiple large and sufficiently powered clinical studies with independent external validation cohorts have demonstrated the feasibility of using specific methylated DNA signatures for developing prognostic biomarkers in CRC11.
Among all epigenetic biomarker candidates, CIMP status is the most promising prognostic indicator for CRC patients to date. CIMP-positive cancers correlate with an overall unfavorable prognosis81, 140–143. Rijnsoever and colleagues showed in a cohort of 206 stage III CRC patients that CIMP-positive status associated with poor survival144. Another independent study analyzed more than 600 CRC patients and also found that CIMP associated with poor prognosis in MSS (microsatellite stable) CRC patients145. Some studies suggested that poor prognosis in CIMP-positive CRCs is from coexisting V600E BRAF mutations146, 147, however, in addition to CIMP, MSI status remains an important confounding factor that likely underlies the difference in prognosis of CIMP-positive MSS vs. MSI cancers148. These data highlight that the prognosis of patients with CIMP CRCs is affected by the MSI status of the tumor.
In addition to hypermethylation of various genes/loci, growing evidence suggests that DNA hypomethylation status associates with the prognosis of CRC patients. Ogino and colleagues have reported a correlation between LINE-1 hypomethylation and poor survival in prospective cohort studies of CRC patients78. Subsequent studies not only validated this association for LINE-1 hypomethylation and CRC prognosis79–81, but also identified other potential genes that correlate with adverse outcomes149–154.
In aggregate, these studies provide evidence that aberrantly methylated DNA loci have potential for use as prognostic biomarkers for CRC; however, further investigation is required to develop clinically reliable, standardized assays in order for these assays to be used in clinical care.
Epigenetic Predictive Biomarkers for Response to Treatment
Despite recent advances in the development of cancer therapeutics, the currently used chemotherapeutic drugs have modest efficacy for advanced CRCs, especially when used without consideration for molecular subtypes. It is now well recognized that molecular markers in CRC, such as mutant RAS (KRAS, NRAS) family genes and microsatellite instability, have clinical utility for targeting anti-EGFR (i.e. prediction of resistance to EGFR blockade) and immune checkpoint blockade therapies, respectively155. There is an unmet need for predictive biomarkers that can be used to target cytotoxic and targeted therapies to those CRC patients most likely to benefit.
Over the last decade, a number of aberrantly methylated genes have been assessed as predictive biomarkers for CRC patients undergoing various chemotherapeutic regimens. The majority of these studies have not progressed beyond Phase I/II discovery phase and thus will not be discussed further in this article. CIMP as a predictive marker has been intensively studied for more than a decade; however, the best done studies to date have yielded conflicting results on its use for predicting response to 5-fluorouracil (5-FU)144,156,157. Thus, at this time, CIMP is not used clinically for directing 5-FU based therapy. More recently, prospective studies assessing CIMP as a predictive marker for adjuvant irinotecan and oxalplatin have shown modest prognostic effects for overall survival in stage III, MSS, and CIMP-positive CRCs with the addition of irinotecan to adjuvant 5FU and leucovorin and in stage III CIMP-positive CRCs treated with 5FU, leucovorin, and oxaliplatin (FOLFOX-4)158,159. These studies suggest promise for the use of CIMP as a prognostic and possibly predictive marker and also highlight the need for additional studies of the interaction between CIMP status and therapeutic response to various treatments.
Although mostly still in the early phase of development, some promising single gene pharmaco-epigenetic biomarkers have been identified in various cancers with methylated MGMT for directing temozolamide treatment of gliomas being the best established to date160. A recent study showed the feasibility of using hypermethylated Transcription Factor AP-2 Epsilon (TFAP2E) as a predictive biomarker for response to 5-FU based chemotherapy in CRC patients161. Furthermore, DNA methylation microarray profiling of oxaliplatin sensitive vs. resistant CRC cell lines revealed that oxaliplatin-resistant cells exhibited hypermethylation of the BRCA1 interactor SRBC gene; which was subsequently shown to associate with poor progression free survival (PFS) in CRC cohorts treated with oxaliplatin although the results of the initial study have not been replicated to date162. The results of these early phase studies using methylated genes as predictive markers and the ability to develop reliable assays for methylated genes is expected to continue to drive the investigation of methylated genes as response predictors for CRC therapy.
Epigenetic CRC Risk Biomarkers: “Field Cancerization” and ‘Epigenetic Drift”:
The concept of “field cancerization” (or field effect) was first proposed in 1953 by Slaughter et al163. Field cancerization is characterized by the occurrence of genetic and epigenetic alterations in histologically normal-appearing tissues, and is believed to lead to an increased risk for synchronous or metachronous primary tumors. While genetic alterations are common in CRC cells, these are believed to be rare in normal cells. In contrast, some studies suggest somatic epigenetic dysregulation occurs not only in cancer tissues, but also in non-cancerous and pre-neoplastic tissues. Considering that epigenetic alterations could contribute to the early events predisposing to malignant transformation, these studies suggest epigenetic events are potentially more promising somatic CRC risk markers (aka field cancerization markers) than are gene mutations. Methylation changes in tumor-suppressor genes occurs more frequently in the normal colonic mucosa of CRC patients than healthy controls164, suggesting that they may be one of the earliest events that predispose normal mucosa to tumorigenic transformation in CRC165. Furthermore, loss of the insulin-like growth factor-II (IGF2) gene imprinting occurs at a higher frequency in the normal mucosa adjacent to cancer tissue, compared to normal mucosa in patients without CRC166, emphasizing the potential of loss of IGF2 imprinting to be a biomarker to identify patients at greater risk for CRC development. Other studies have revealed that both hypermethylation of tumor-suppressive genes such as SFRP, ESR1, MYOD, EVL, and MGMT, as well as LINE-1 hypomethylation in normal colonic mucosa correlates with an increased risk of CRC compared to patients without these traits167–171 (Grady, personal communication; Yu et al, under review)
It is of particular interest that accumulating evidence supports that the landscape of DNA methylation can be modified as a “function of age”. DNA methylation has been proposed to result from a gradual stochastic age-dependent dysregulation caused by a combination of external environmental factors and internal spontaneous random errors in the maintenance of methylation. This process of age-dependent alterations in methylation is defined as “epigenetic drift”172. Interestingly, such age-associated DNA methylation often targets the promoters of tumor-suppressive genes173, 174. In monozygotic twins, epigenetic divergence with age suggests the underlying epigenetic drift may in part help explain the disease discordance175–177. Since this new concept closely relates to the “field effect”, identification of biomarkers that overlap both the “epigenetic drift” and “field effect” in colorectal mucosa may allow development of next-generation biomarkers for determining risk for CRC development.
Histone modification alterations in CRC: potential for use as biomarkers.
Altered histone modifications are commonly found in CRCs178 and associate with altered chromatin activity states and gene expression through effects on gene promoters and noncoding regulatory elements179, 180. Some studies have demonstrated potential for histone modifications to be CRC biomarkers, however, due to the technical limitations of assays that assess the post-translational histone modification state, it has been difficult to reliably determine the histone modification state in primary cancer tissues and to develop tests that are sufficiently robust to be used in clinical care. Global alterations of specific-histones in primary tissues have been the focus for biomarker development in CRC. Studies of H3K4me2, H3K9ac, and H3K9me2 alterations detected by immunohistochemical staining in liver metastases suggest that low H3K4me2 expression levels correlate with poor prognosis181. Additionally, other studies in CRC suggest that histone modifications, such as acetylation of H3 lysine 56 and di- or tri-methylation of H3 lysine 9 and 27, have potential to be prognostic markers in CRC.178, 181–185. Similarly, studies of histone modifications in circulating nucleosomes have identified reduced levels of H3K9me3- and H4K20me3 as potential diagnostic biomarkers for CRC186, 187. However, thus far, these studies are all proof of concept Phase I biomarker studies. Further research is needed to determine whether any of these modifications will be clinically useful.
EPIGENETIC ALTERATIONS IN BARRETTS ESOPHAGUS AND ESOPHAGEAL ADENOCARCINOMA
Barretts Esophagus (BE) is specialized small intestinal metaplastic epithelium of the esophagus and is a precursor to esophageal adenocarcinoma (EAC). It has increased dramatically in the last 40 years and results in roughly 20,000 deaths/year in the U.S. Most, if not all, EAC originates in BE through a metaplasia-dysplasia-carcinoma sequence whereby BE progresses to low-grade dysplasia followed by high-grade dysplasia and then intramucosal carcinoma and invasive carcinoma189, 190
As with CRC, genetic and epigenetic alterations occur in BE and EAC and play an important role in the pathogenesis of EAC191. Epigenetic alterations identified in BE and EAC include DNA methylation alterations, histone alterations, aberrant expression of noncoding RNAs and chromatin alterations191–193. The best studied class of epigenetic alterations in BE and EAC is secondary to DNA methylation and found in the majority of BE and EAC cases194,192, 195, 196. Factors that influence DNA methylation in the esophagus include aging, smoking and obesity197, 198. Hypermethylated genes shown in BE and EAC include known tumor-suppressor genes, such as APC, CDKN2A (p16INK4a), RUNX3, MGMT, CDH1, and SFRP family members among others195. A subset of the hypermethylated genes are believed to play a causal (termed “driver”) role in driving the formation of EAC, while most appear to be BE and EAC specific passenger alterations, which do not functionally induce EAC formation distinguishing them from driver genes192, 195. Aberrant methylation of classic tumor suppressor genes such as CDKN2A and MGMT has been correlated with loss of mRNA and protein expression in the metaplasia-dysplasia-carcinoma sequence of BE to EAC199, 200.
Recently, Yu et al identified four methylation subtypes of EAC and BE through genome-wide DNA methylation profiling192. The four methylation subtypes were identified through the use of a recursively partitioned mixture model (RPMM) and included High Methylator (HM), Intermediate Methylator (IM), Low Methylator (LM), and Minimal Methylator (MM) subtypes, which were defined by the overall methylated CpG burden and pattern of methylation alterations. The high methylator subtype (HM) had more activating events in ERBB2 compared to the other subtypes, which suggested a unique dependence on epidermal growth factor (EGF) signaling in HM BE and HM EAC, and also a higher global mutation load. This EGF signaling pathway dependence of the HM subtype appears to arise from both oncogenic ERBB2 and through the epigenetic silencing of the tyrosine phosphatase non-receptor 13 (PTPN13), which is specific to the HM subtype. Subsequent studies have supported the observation of molecular subclasses of BE and EAC194.
Of relevance to biomarker discovery, a large number of genes and loci have been identified as high frequency targets of aberrant methylation in BE and EAC192. Although the functional significance of these methylated genes is still not clear, these DNA methylation events have proved to have potential as biomarkers of BE, as discussed below. The published studies to date suggest aberrant DNA methylation is a common molecular mechanism that mediates the development of esophageal cancer and that aberrantly methylated genes and loci are potential screening and surveillance biomarkers for BE and EAC.
METHYLATED DNA BIOMARKERS FOR BARRETTS ESOPHAGUS SCREENING AND SURVEILLANCE
Barretts Esophagus screening markers
Genetic and epigenetic alterations occurring in Barretts esophagus (BE) and early stage EAC are potential biomarkers for use in cancer care and prevention BE and EAC. Studies over the last 3 years have shown methylated DNA biomarkers to be the most promising class of BE and EAC biomarkers to date. Of particular interest is the recent development of a “molecular cytology” assay for methylated VIM in DNA samples from esophageal cytology brushings obtained during endoscopies of 322 individuals, divided into training and validation cohorts201. The assay showed 91% sensitivity for detecting BE, BE with dysplasia, and EAC at 93% specificity, with essentially identical results obtained in both the training and validation cohorts201.
This assay was further refined with the addition of a second methylated gene, CCNA1, and used on samples collected with an FDA approved swallowable balloon based device (the Esocheck device, Lucid Diagnostics) for obtaining targeted non-endoscopic brushings of the distal esophagus201. which improved the sensitivity to 95% (at 91% specificity) for BE, BE and dysplasia and EAC cases and for detecting 96% of BE with dysplasia and 96% of EAC201. A methylated DNA panel (tradename EsoGuard) based on these markers run on Esocheck collected samples is currently being further validated in a nationwide multi-center clinical trial and is undergoing commercial development.
Additional potential BE markers have been identified and validated by others include B3GAT2 and ZNF793, that are aberrantly methylated in BE. Clinical validation studies confirmed B3GAT2 and ZNF793 methylation levels were significantly higher in BE samples (median 32.5% and 33.1%, respectively) than in control tissues (median 2.29% and 2.52%, respectively; P < 0.0001 for both genes) and that gene-specific MethyLight assays could accurately detect BE (P < 0.0001 for both) in endoscopic brushing samples with mZNF793 having a sensitivity of 70% and specificity of 100% for BE202. These markers show promise to further improve the performance of a methylated gene panel for BE screening.
In addition to the Esocheck device, other swallowable cytology collection devices are being assessed and currently being evaluated for use in BE screening assays, such as the ‘Cytosponge’ and “Esophacap” devices, which are both swallowed capsules that degrade in the stomach to release a sponge tethered to a string203–205. Unlike the Esocheck device, these devices sample the entire esophagus and oropharynx, which increases the potential to impair biomarker performance. Using a Cytosponge based assay, Chettouh et al discovered and assessed hypermethylated TFPI2, TWIST1, ZNF345 and ZNF569 as potential BE screening markers. Methylated TFPI2 was shown to achieve the best sensitivity in both the pilot and validation Cytosponge cohorts (85% and 79%, respectively, AUC 0.88)206.
In summary, these studies have established that methylated DNA is a potential new biomarker class that will enable practical non-endoscopic screening and early detection of BE, an approach with potential to reduce the steadily increasing mortality from EAC. Table 2 below summarizes BE screening markers used for BE early detection that have been evaluated in clinical cohorts. (Table 2)
Table 2:
Validated Barretts Esophagus early detection markers
| BE early detection marker | Method | Study Design | AUC | Sensitivity | Specificity |
|---|---|---|---|---|---|
| mVIM and mCCNA1 | bsNSG (Esocheck device) | Case-control Validation cohort (N=86)201 | 90% | 92% | |
| mB3GAT2 | methyLight PCR (endoscopic brushings) | Case-control Validation cohort (N=66)202 | 0.95 | 80% | 86% |
| mZNF793 | methyLight PCR (endoscopic brushings) | Case-control validation cohort (N=66)202 | 0.96 | 80% | 93% |
| mTFPI2 | methyLight PCR (cytosponge) | Case-control validation cohort (N=278)206 | 0.88 (0.84–0.91 | 82% | 96% |
| mTWIST1 | methyLight PCR (cytosponge) | Case-control validation cohort (N=278)206 | 81% (0.77–0.86) | 70% | 93% |
IHC=immunohistochemistry, bsNSG=bisulfite next generation sequencing
Barretts Esophagus Surveillance And Risk Prediction Biomarkers
BE is associated with approximately 4X increased risk of EAC, which has led to the recommendation that patients with BE undergo regular endoscopic surveillance in order to prevent or detect EAC at its earliest stage207. However, only 0.1–0.3% of people/year with BE will progress to high-grade dysplasia or EAC, thus, a biomarker (or biomarker panel) that can accurately risk stratify high risk patients with BE who are likely to progress from those low risk BE patients who are unlikely to develop EAC is needed208. Such a marker could potentially spare the great majority of individuals with a diagnosis of BE from the cost, inconvenience, and risks of regular endoscopic surveillance. Being placed in a ‘low-risk’ group might also reduce the feelings of anxiety about developing EAC that have been shown to be associated with a diagnosis of BE209. The search for accurate risk stratification markers for BE is an area of intense investigation but has not yielded any markers have proven adequate to be used in the clinical setting. Immunostaining assays for p53 and aneuploidy appear to have the highest likelihood for eventual adoption into clinical care at this time207.
With regards to epigenetic risk prediction biomarkers, a retrospective study comparing BE patients who progressed to HGD or EAC to those who did not using hypermethylated CDKN2A (OR 1.74, 95% CI 1.33 – 2.20), RUNX3 (OR 1.80, 95% CI 1.08 – 2.81), and HPP1 (OR 1.77, 95% CI 1.06 – 2.81), has shown an association with an increased risk of progression. Age, BE segment length, and hypermethylation of other genes (TIMP3, APC, or CRBP1) were not found to be independent risk factors with this assay210. A follow-up study using these same epigenetic markers in combination with three clinical parameters (gender, BE segment length (SL), and pathologic assessment) demonstrated this multi-parameter method could stratify BE patients into high, intermediate, and low risk for progression to HGD or EAC. This tissue based assay has not been adopted into routine clinical use to date211. In a later iteration of this approach, this risk assessment tool was expanded to include additional genes previously shown to be hypermethylated in BE and/or EAC to generate an eight-marker risk-of-progression panel. In a retrospective analysis of 145 people with stable BE that did not progress to EAC vs. 50 people who did progress to EAC, this panel predicted progression with a sensitivity of ~50% when the specificity was set at 90%212. None of these candidates have advanced to phase III or IV biomarker trials. In summary, studies to date have demonstrated the potential for methylated DNA biomarkers to be risk prediction markers, but further studies are needed.
LIVER CANCER: EPIGENETIC ALTERATIONS
HCC arises secondary to a variety of etiologic factors, including predominately hepatitis B virus (HBV), hepatitis C virus (HCV), fatty liver disease (NAFLD), alcohol, and genotoxins. The HCCs that arise in the setting of these factors do so through a progressive pathway from premalignant cirrhosis-related premalignant nodular lesions, which include both regenerative and dysplastic nodules, to HCC213, 214. These factors induce genomic and epigenomic alterations as well as inflammatory cytokines that appear to mediate the formation and progression of HCC, with prominent genetic alterations including TERT promoter and TP53 mutations, and common epigenetic alterations including RASSF1A and SOCS1. The epigenetic alterations arise early in the pre-neoplastic cirrhotic phase of HCC carcinogenesis, precede many of the genetic alterations, and associate with a field effect in the cirrhotic liver215–217. Assessment of the clonal evolution of HCC has revealed that the epigenetic arise and evolve independent of the genetic alterations216. Furthermore, pre-malignant patterns of epigenetic alterations occur in some nodular lesions and are linked to the proliferative and dysplastic capacity of these nodules216.
Some of these genomic and epigenomic alterations are also known to occur preferentially or exclusively in the setting of specific causes, with distinct liver cancer epigenomes found in HCV and alcohol related HCC216. HBV-induced HCCs carry a high proportion of TP53 mutations and lack TERT promoter mutations. Furthermore, HBV integration sites appear to be common in HBV mediated HCC and to affect candidate oncogenes, such as TERT and MLL4 (most common), CCND1 and GLI2, as well as others218. In contrast, HCV mediated HCC display significantly increased frequencies of CDKN2A promoter silencing and TERT promoter mutations218. Some of the epigenetic alterations have been shown to affect the clinical behavior of the cancer as well as patient prognosis219, 220.
Methylated DNA Biomarkers For HCC Screening And Surveillance
As noted for CRC and EAC, methylated DNA (mDNA) alterations have properties that make them uniquely suited for development into molecular markers for HCC. In evidence of their potential as HCC biomarkers, a 6-marker circulating free methylated DNA (cf mDNA) marker panel yielded an AUROC of 0.96 with HCC detection sensitivity of 95% and specificity or 92% in a recent study221. Although selection and other biases almost certainly inflated the marker panel performance reported in this study, based on these encouraging results this panel is being further assessed and is undergoing external validation phase 2 studies in a variety of different patient populations.
GASTRIC CANCER EPIGENOMICS
Pangenomic analysis of gastric cancer (GCA) has revealed molecular subtypes of gastric cancer that vary based on mutation patterns, DNA methylation, and biological pathway dependence. Among several efforts to classify gastric cancer, The Cancer Genome Atlas (TCGA) and the Asian Cancer Research Group (ACRG) have proposed gastric cancer subtypes based on gene expression, microsatellite instability (MSI) and DNA methylation222. The TCGA identified four distinct subtypes: the Epstein Barr virus (EBV) subtype, which is enriched for tumors carrying this virus; the MSI subtype; the chromosomal instability (CIN) subtype; and the genome stable subtype. EBV subtype GCAs account for ~9% of GCA and exhibit distinct genome-wide increases in DNA methylation that is CIMP-like. EBV-CIMP gastric cancers possess the highest levels of DNA methylation seen among all cancer types222, 223. The MSI subtype (~22% of GCA) is defined by a classic microsatellite unstable genome, often is a consequence of aberrant methylation of the DNA mismatch repair gene, MLH1. MSI GCAs have a very high tumor mutation burden and have a hypermethylated CIMP epigenomic state, although to a lower extent than seen in the EBV subtype GCAs. The chromosomal instability (CIN) subtype cancers (~50% of GCAs) are defined by aneuploidy, and have relatively lower levels of methylation, as does the genome-stable subtype (~20% of GCAs), which are largely genetically stable. In the genome stable subtype, epigenetic alterations appear to be the primary mechanism for altering oncogene and tumor suppressor gene activity74.
Environmental factors such as infectious agents, chronic inflammation, diet, physical activity, age and smoking have been correlated with changes in the GCA methylome224. Helicobacter pylori mediated inflammation is associated with regional DNA hypermethylation and hypomethylation of Alu repeat elements in human gastric mucosa225, 226, and eradication of H. pylori has been associated with complete or partial reversal of methylation at cancer-related genes such as CDH1, MGMT and COX2227–229. Importantly, although H. pylori eradication results in decreased DNA methylation, the DNA methylation levels do not revert to those observed in uninfected states, suggesting long lasting effects from H pylori infection on the gastric mucosa, even after it is eliminated. By contrast, EBV-associated hypermethylation is thought to be caused by direct pathogen infection rather than inflammation-related intermediate pathways, possibly by EBV modulation of host DNMT1 activity and downregulation of the TET2 demethylase224, 230. Notably, CIMP can also occur in GCA that are EBV and MSI negative and account for ~26% of the total CIMP group in the TCGA cohort,222. The precise mechanism underlying the acquisition of CIMP in this subset of CIMP GCAs is not known at this time.
Epigenetic alterations in GCA can alter the activity of tumor suppressor genes and oncogenes. The most robust evidence to date with regards to the role of aberrantly methylated genes in GCA has been shown for CDKN2A/p16, which is methylated in all EBV subtype GCAs; MLH1, which is methylated in the majority of MSI GCAs and mediates the MSI phenotype; and CDH1, which is methylated commonly in diffuse GCAs and in GCAs arising in the Hereditary Diffuse Gastric Cancer syndrome36, 231 Notably, studies of the aberrant methylation of CDH1 provided some of the first evidence of the functional importance of DNA hypermethylation in cancer as a bona fide mechanism for biallelic inactivation of tumor suppressor genes36
Methylated DNA Biomarkers For GCA Prevention, Screening And Prognosis
The application of epigenetics to the management of gastric cancer has been assessed in early phase translational studies, which have demonstrated that aberrantly methylated genes may be used as risk markers, early detection markers, and prognostic markers for GCA. Ushijima and colleagues have carried out a comprehensive series of studies demonstrating that aberrantly methylated genes that arise in the normal stomach likely do so as a consequence of H pylori induced methylation and indicate a field cancerization effect associated with a 2–3X increased risk for GCA232, 233. DNA methylation markers to detect gastric cancer have also been identified in plasma, serum, gastric juice and fecal samples from patients with gastric cancer, albeit with varying specificity and sensitivity234–236. Patients with CIMP GCAs show superior survival in some cohort studies, suggesting the potential for CIMP to be used as a prognostic marker237. Regrettably, none of these assays have been subjected to clinical studies sufficient to determine whether they are suitable for use in the clinic.
PANCREATIC CANCER AND EPIGENETIC ALTERATIONS
The role of epigenetic alterations in pancreatic adenocarcinoma (PANCA) is less thoroughly understood compared to other GI cancers. Recent studies have begun to provide insight into the epigenomics of PANCA. Subtypes of PANCA have been identified though analysis of DNA methylation, copy number, lncRNA, miRNA, and aberrant protein expression238. In an assessment of 150 patients with PANCA in the TCGA, unsupervised clustering of DNA methylation data for high-purity samples revealed two major subgroups (termed H1 and H2). The H1 cluster (n = 41) had more extensive DNA hypermethylation than the H2 cluster (n = 35). Unlike with colorectal and gastric cancer, no CIMP subtype has been found to date in PANCA. Others have investigated the TDGC PANCA datasets and have identified three possible sub-groups based on methylation patterns, somatic mutations and copy number alterations, histologic features, and stage features, but not on gene expression239.
With regards to the functional consequence of epigenetic alterations in PANCA, in the TCGA study, integrated analysis of the DNA methylation and mRNA expression data revealed 98 genes that appear to be silenced by DNA methylation, including genes that have been implicated in the development of other cancers but not previously reported to be altered in PANCA. Presumably epigenetically silenced genes worth mention include ZFP82, a suspected tumor suppressor gene, PARP6, DNAJC15, BRCA1 and MGMT238. The application of aberrantly methylated genes to clinical care for PANCA patients, either as biomarkers or as therapeutic targets is in early development, and its translational potential is unclear at this time240–242.
CONCLUSIONS
In summary, epigenetic alterations are common in pre-malignant and malignant tumors of the gastrointestinal tract and have been found in all cancers arising in the gastrointestinal tract to date. These alterations not only effect the initiation and progression of gastrointestinal cancer but also appear to be robust biomarkers that can be used for the early detection of pre-malignant conditions, such as colon polyps and BE, as well as early stage cancers. Recent studies have led to FDA approved colon cancer screening assays based on methylated DNA and it is likely that additional screening and surveillance assays for CRC as well as BE and EAC will be available in the near future.
ACKNOWLEDGEMENTS
These studies were supported by funding from the NIH:UO1CA152756, RO1CA194663, P30CA015704, U54CA163060, UO1CA086402, UO1CA182940 to WMG, P50CA150964 and UO1CA152756 to SDM, R50CA233042 to MY. Funding is also provided by the Cottrell Family Fund, R.A.C.E. Charities and the Listwin Foundation to WMG.
We also wish to acknowledge the ColoCare study team (Fred Hutchinson Cancer Research Center) and GICaRes study team (University of Washington, Seattle, WA) for their efforts and support of biomarker assay development.
Grant support: NIH grants (P30CA15704, R01CA194663, RO1CA220004, RO1CA189184, U54CA143862, P01CA077852, R01CA207371, U01CA206110), R.A.C.E. Charities, Cottrell Family Fund, U01152756, Rodger Haggitt Endowed Chair, Listwin Family Foundation, Seattle Translational Tumor Research program, Fred Hutchinson Cancer Research Center to WMG; NCI (R50CA233042) to MY, and P50CA150964 and UO1CA152756 to SDM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interests: WM Grady is on the advisory boards for Freenome, Guardant Health, and SEngine and consults for Diacarta. He is also an investigator for a clinical trial sponsored by Janssen and receives services for investigator initiated research from Tempus.
REFERENCES
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57–70. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 2012;22:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epigenetics Herceg Z. and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis 2007;22:91–103. [DOI] [PubMed] [Google Scholar]
- 6.Holland N Future of environmental research in the age of epigenomics and exposomics. Rev Environ Health 2017;32:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hitchins MP. The role of epigenetics in Lynch syndrome. Fam Cancer 2013;12:189–205. [DOI] [PubMed] [Google Scholar]
- 8.Cerrato F, Sparago A, Ariani F, et al. DNA Methylation in the Diagnosis of Monogenic Diseases. Genes (Basel) 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morak M, Ibisler A, Keller G, et al. Comprehensive analysis of the MLH1 promoter region in 480 patients with colorectal cancer and 1150 controls reveals new variants including one with a heritable constitutional MLH1 epimutation. J Med Genet 2018;55:240–248. [DOI] [PubMed] [Google Scholar]
- 10.Dragomir MP, Kopetz S, Ajani JA, et al. Non-coding RNAs in GI cancers: from cancer hallmarks to clinical utility. Gut 2020;69:748–763. [DOI] [PubMed] [Google Scholar]
- 11.Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015;149:1204–1225 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet 2000;16:168–74. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Sethi NS, Hinoue T, et al. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell 2018;33:721–735 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paluch BE, Naqash AR, Brumberger Z, et al. Epigenetics: A primer for clinicians. Blood Rev 2016;30:285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meldi KM, Figueroa ME. Cytosine modifications in myeloid malignancies. Pharmacol Ther 2015;152:42–53. [DOI] [PubMed] [Google Scholar]
- 16.Brahma S, Henikoff S. Epigenome Regulation by Dynamic Nucleosome Unwrapping. Trends Biochem Sci 2020;45:13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaffney DJ, McVicker G, Pai AA, et al. Controls of nucleosome positioning in the human genome. PLoS Genet 2012;8:e1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valouev A, Johnson SM, Boyd SD, et al. Determinants of nucleosome organization in primary human cells. Nature 2011;474:516–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan S, Davey CA. Nucleosome structural studies. Curr Opin Struct Biol 2011;21:128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolin SL, Maquat LE. Cellular RNA surveillance in health and disease. Science 2019;366:822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabbri M, Calin GA. Beyond genomics: interpreting the 93% of the human genome that does not encode proteins. Curr Opin Drug Discov Devel 2010;13:350–8. [PubMed] [Google Scholar]
- 22.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857–66. [DOI] [PubMed] [Google Scholar]
- 23.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell 2013;153:38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer 2016;16:483–93. [DOI] [PubMed] [Google Scholar]
- 25.Sengupta S, George RE. Super-Enhancer-Driven Transcriptional Dependencies in Cancer. Trends Cancer 2017;3:269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanwal R, Gupta S. Epigenetics and cancer. J Appl Physiol (1985) 2010;109:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang ZM, Lu R, Wang P, et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 2018;554:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005;6:597–610. [DOI] [PubMed] [Google Scholar]
- 29.Ambrosi C, Manzo M, Baubec T. Dynamics and Context-Dependent Roles of DNA Methylation. J Mol Biol 2017;429:1459–1475. [DOI] [PubMed] [Google Scholar]
- 30.Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell Mol Life Sci 2009;66:596–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol 2010;28:1057–68. [DOI] [PubMed] [Google Scholar]
- 32.Leighton G, Williams DC, Jr. The Methyl-CpG-Binding Domain 2 and 3 Proteins and Formation of the Nucleosome Remodeling and Deacetylase Complex. J Mol Biol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013;502:472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dehan P, Kustermans G, Guenin S, et al. DNA methylation and cancer diagnosis: new methods and applications. Expert Rev Mol Diagn 2009;9:651–7. [DOI] [PubMed] [Google Scholar]
- 35.Torchy MP, Hamiche A, Klaholz BP. Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci 2015;72:2491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grady WM, Willis J, Guilford PJ, et al. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet 2000;26:16–7. [DOI] [PubMed] [Google Scholar]
- 37.Veigl ML, Kasturi L, Olechnowicz J, et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci U S A 1998;95:8698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klutstein M, Nejman D, Greenfield R, et al. DNA Methylation in Cancer and Aging. Cancer Res 2016;76:3446–50. [DOI] [PubMed] [Google Scholar]
- 39.Van Tongelen A, Loriot A, De Smet C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett 2017;396:130–137. [DOI] [PubMed] [Google Scholar]
- 40.Vilain A, Vogt N, Dutrillaux B, et al. DNA methylation and chromosome instability in breast cancer cell lines. FEBS Lett 1999;460:231–4. [DOI] [PubMed] [Google Scholar]
- 41.Dokun OY, Florl AR, Seifert HH, et al. Relationship of SNCG, S100A4, S100A9 and LCN2 gene expression and DNA methylation in bladder cancer. Int J Cancer 2008;123:2798–807. [DOI] [PubMed] [Google Scholar]
- 42.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta 2007;1775:138–62. [DOI] [PubMed] [Google Scholar]
- 43.Gilbert N Biophysical regulation of local chromatin structure. Curr Opin Genet Dev 2019;55:66–75. [DOI] [PubMed] [Google Scholar]
- 44.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009;10:295–304. [DOI] [PubMed] [Google Scholar]
- 45.Du Q, Luu PL, Stirzaker C, et al. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics 2015;7:1051–73. [DOI] [PubMed] [Google Scholar]
- 46.Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev 2005;19:295–310. [DOI] [PubMed] [Google Scholar]
- 47.Bersaglieri C, Santoro R. Genome Organization in and around the Nucleolus. Cells 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem 2011;80:473–99. [DOI] [PubMed] [Google Scholar]
- 49.Cohen I, Poreba E, Kamieniarz K, et al. Histone modifiers in cancer: friends or foes? Genes Cancer 2011;2:631–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kouzarides T Chromatin modifications and their function. Cell 2007;128:693–705. [DOI] [PubMed] [Google Scholar]
- 51.Bartke T, Kouzarides T. Decoding the chromatin modification landscape. Cell Cycle 2011;10:182. [DOI] [PubMed] [Google Scholar]
- 52.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol 2014;15:703–8. [DOI] [PubMed] [Google Scholar]
- 53.Hadley M, Noonepalle S, Banik D, et al. Functional Analysis of HDACs in Tumorigenesis. Methods Mol Biol 2019;1983:279–307. [DOI] [PubMed] [Google Scholar]
- 54.Bartova E, Krejci J, Harnicarova A, et al. Histone modifications and nuclear architecture: a review. J Histochem Cytochem 2008;56:711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kouzarides T SnapShot: Histone-modifying enzymes. Cell 2007;131:822. [DOI] [PubMed] [Google Scholar]
- 56.Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer 2010;10:669–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baretti M, Azad NS. The role of epigenetic therapies in colorectal cancer. Curr Probl Cancer 2018;42:530–547. [DOI] [PubMed] [Google Scholar]
- 58.Abdelfatah E, Kerner Z, Nanda N, et al. Epigenetic therapy in gastrointestinal cancer: the right combination. Therap Adv Gastroenterol 2016;9:560–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 2008;135:1079–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Komor MA, Bosch LJ, Bounova G, et al. Consensus molecular subtype classification of colorectal adenomas. J Pathol 2018;246:266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carethers JM, Jung BH. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology 2015;149:1177–1190 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 1999;96:8681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787–93. [DOI] [PubMed] [Google Scholar]
- 64.Pai RK, Bettington M, Srivastava A, et al. An update on the morphology and molecular pathology of serrated colorectal polyps and associated carcinomas. Mod Pathol 2019;32:1390–1415. [DOI] [PubMed] [Google Scholar]
- 65.Toyota M, Ohe-Toyota M, Ahuja N, et al. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A 2000;97:710–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 2005;129:837–45. [DOI] [PubMed] [Google Scholar]
- 67.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 2005;352:1851–60. [DOI] [PubMed] [Google Scholar]
- 68.Valle L, Serena-Acedo T, Liyanarachchi S, et al. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science 2008;321:1361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saghafinia S, Mina M, Riggi N, et al. Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell Rep 2018;25:1066–1080 e8. [DOI] [PubMed] [Google Scholar]
- 70.Heyn H, Vidal E, Ferreira HJ, et al. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol 2016;17:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science 2003;300:489–92. [DOI] [PubMed] [Google Scholar]
- 72.Eden A, Gaudet F, Waghmare A, et al. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 2003;300:455. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki K, Suzuki I, Leodolter A, et al. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 2006;9:199–207. [DOI] [PubMed] [Google Scholar]
- 74.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860–921. [DOI] [PubMed] [Google Scholar]
- 75.Yamamoto E, Toyota M, Suzuki H, et al. LINE-1 hypomethylation is associated with increased CpG island methylation in Helicobacter pylori-related enlarged-fold gastritis. Cancer Epidemiol Biomarkers Prev 2008;17:2555–64. [DOI] [PubMed] [Google Scholar]
- 76.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer 2008;122:2767–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Estecio MR, Gharibyan V, Shen L, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One 2007;2:e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst 2008;100:1734–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Antelo M, Balaguer F, Shia J, et al. A high degree of LINE-1 hypomethylation is a unique feature of early-onset colorectal cancer. PLoS One 2012;7:e45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahn JB, Chung WB, Maeda O, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer 2011;117:1847–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rhee YY, Kim MJ, Bae JM, et al. Clinical outcomes of patients with microsatellite-unstable colorectal carcinomas depend on L1 methylation level. Ann Surg Oncol 2012;19:3441–8. [DOI] [PubMed] [Google Scholar]
- 82.Xiao-Jie L, Hui-Ying X, Qi X, et al. LINE-1 in cancer: multifaceted functions and potential clinical implications. Genet Med 2016;18:431–9. [DOI] [PubMed] [Google Scholar]
- 83.Hur K, Cejas P, Feliu J, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2014;63:635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bettington M, Walker N, Clouston A, et al. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology 2013;62:367–86. [DOI] [PubMed] [Google Scholar]
- 85.Chang K, Willis JA, Reumers J, et al. Colorectal premalignancy is associated with consensus molecular subtypes 1 and 2. Ann Oncol 2018;29:2061–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bettington M, Rosty C, Whitehall V, et al. A morphological and molecular study of proposed early forms of traditional serrated adenoma. Histopathology 2018;73:1023–1029. [DOI] [PubMed] [Google Scholar]
- 87.Crockett SD, Nagtegaal ID. Terminology, Molecular Features, Epidemiology, and Management of Serrated Colorectal Neoplasia. Gastroenterology 2019;157:949–966 e4. [DOI] [PubMed] [Google Scholar]
- 88.Lee-Six H, Olafsson S, Ellis P, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 2019;574:532–537. [DOI] [PubMed] [Google Scholar]
- 89.Kaz AM, Wong CJ, Dzieciatkowski S, et al. Patterns of DNA methylation in the normal colon vary by anatomical location, gender, and age. Epigenetics 2014;9:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Luo Y, Wong CJ, Kaz AM, et al. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology 2014;147:418–29 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laiho P, Kokko A, Vanharanta S, et al. Serrated carcinomas form a subclass of colorectal cancer with distinct molecular basis. Oncogene 2007;26:312–20. [DOI] [PubMed] [Google Scholar]
- 92.Dhir M, Yachida S, Van Neste L, et al. Sessile serrated adenomas and classical adenomas: an epigenetic perspective on premalignant neoplastic lesions of the gastrointestinal tract. Int J Cancer 2011;129:1889–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kohonen-Corish MR, Sigglekow ND, Susanto J, et al. Promoter methylation of the mutated in colorectal cancer gene is a frequent early event in colorectal cancer. Oncogene 2007;26:4435–41. [DOI] [PubMed] [Google Scholar]
- 94.Suzuki H, Igarashi S, Nojima M, et al. IGFBP7 is a p53-responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis 2010;31:342–9. [DOI] [PubMed] [Google Scholar]
- 95.Kriegl L, Neumann J, Vieth M, et al. Up and downregulation of p16(Ink4a) expression in BRAF-mutated polyps/adenomas indicates a senescence barrier in the serrated route to colon cancer. Mod Pathol 2011;24:1015–22. [DOI] [PubMed] [Google Scholar]
- 96.Snyder MW, Kircher M, Hill AJ, et al. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016;164:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut;60:116–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Myint NNM, Verma AM, Fernandez-Garcia D, et al. Circulating tumor DNA in patients with colorectal adenomas: assessment of detectability and genetic heterogeneity. Cell Death Dis 2018;9:894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sidransky D, Tokino T, Hamilton SR, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science 1992;256:102–5. [DOI] [PubMed] [Google Scholar]
- 100.Leung WK, To KF, Man EP, et al. Detection of hypermethylated DNA or cyclooxygenase-2 messenger RNA in fecal samples of patients with colorectal cancer or polyps. Am J Gastroenterol 2007;102:1070–6. [DOI] [PubMed] [Google Scholar]
- 101.Chen WD, Han ZJ, Skoletsky J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst 2005;97:1124–1132. [DOI] [PubMed] [Google Scholar]
- 102.Wang DR, Tang D. Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J Gastroenterol 2008;14:524–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Glockner SC, Dhir M, Yi JM, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res 2009;69:4691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hellebrekers DM, Lentjes MH, van den Bosch SM, et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin Cancer Res 2009;15:3990–7. [DOI] [PubMed] [Google Scholar]
- 105.Petko Z, Ghiassi M, Shuber A, et al. Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clin Cancer Res 2005;11:1203–1209. [PubMed] [Google Scholar]
- 106.Lidgard GP, Domanico MJ, Bruinsma JJ, et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin Gastroenterol Hepatol 2013;11:1313–8. [DOI] [PubMed] [Google Scholar]
- 107.Lee BB, Lee EJ, Jung EH, et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res 2009;15:6185–91. [DOI] [PubMed] [Google Scholar]
- 108.Itzkowitz SH, Jandorf L, Brand R, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol 2007;5:111–7. [DOI] [PubMed] [Google Scholar]
- 109.Itzkowitz S, Brand R, Jandorf L, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol 2008;103:2862–70. [DOI] [PubMed] [Google Scholar]
- 110.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370:1287–97. [DOI] [PubMed] [Google Scholar]
- 111.Eckmann JD, Ebner DW, Kisiel JB. Multi-Target Stool DNA Testing for Colorectal Cancer Screening: Emerging Learning on Real-world Performance. Curr Treat Options Gastroenterol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Prince M, Lester L, Chiniwala R, et al. Multitarget stool DNA tests increases colorectal cancer screening among previously noncompliant Medicare patients. World J Gastroenterol 2017;23:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cooper GS, Markowitz SD, Chen Z, et al. Performance of multitarget stool DNA testing in African American patients. Cancer 2018;124:3876–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Force USPST, Bibbins-Domingo K, Grossman DC, et al. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2016;315:2564–2575. [DOI] [PubMed] [Google Scholar]
- 115.Sharma T Analysis of the effectiveness of two noninvasive fecal tests used to screen for colorectal cancer in average-risk adults. Public Health 2020;182:70–76. [DOI] [PubMed] [Google Scholar]
- 116.Mouliere F, Rosenfeld N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc Natl Acad Sci U S A 2015;112:3178–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thomas ML, Marcato P. Epigenetic Modifications as Biomarkers of Tumor Development, Therapy Response, and Recurrence across the Cancer Care Continuum. Cancers (Basel) 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bi F, Wang Q, Dong Q, et al. Circulating tumor DNA in colorectal cancer: opportunities and challenges. Am J Transl Res 2020;12:1044–1055. [PMC free article] [PubMed] [Google Scholar]
- 119.El Messaoudi S, Mouliere F, Du Manoir S, et al. Circulating DNA as a Strong Multimarker Prognostic Tool for Metastatic Colorectal Cancer Patient Management Care. Clin Cancer Res 2016;22:3067–77. [DOI] [PubMed] [Google Scholar]
- 120.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:827. [DOI] [PubMed] [Google Scholar]
- 121.Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016;65:625–34. [DOI] [PubMed] [Google Scholar]
- 122.Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tie J, Cohen JD, Wang Y, et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Y, Li L, Cohen JD, et al. Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nikolaou S, Qiu S, Fiorentino F, et al. Systematic review of blood diagnostic markers in colorectal cancer. Tech Coloproctol 2018;22:481–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bartak BK, Kalmar A, Peterfia B, et al. Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics 2017;12:751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ebert MP, Model F, Mooney S, et al. Aristaless-like homeobox-4 gene methylation is a potential marker for colorectal adenocarcinomas. Gastroenterology 2006;131:1418–30. [DOI] [PubMed] [Google Scholar]
- 128.Leung WK, To KF, Man EP, et al. Quantitative detection of promoter hypermethylation in multiple genes in the serum of patients with colorectal cancer. Am J Gastroenterol 2005;100:2274–9. [DOI] [PubMed] [Google Scholar]
- 129.Wallner M, Herbst A, Behrens A, et al. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin Cancer Res 2006;12:7347–52. [DOI] [PubMed] [Google Scholar]
- 130.Herbst A, Rahmig K, Stieber P, et al. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol 2011;106:1110–8. [DOI] [PubMed] [Google Scholar]
- 131.Picardo F, Romanelli A, Muinelo-Romay L, et al. Diagnostic and Prognostic Value of B4GALT1 Hypermethylation and Its Clinical Significance as a Novel Circulating Cell-Free DNA Biomarker in Colorectal Cancer. Cancers (Basel) 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Aad G, Abbott B, Abdallah J, et al. Combined Measurement of the Higgs Boson Mass in pp Collisions at sqrt[s]=7 and 8 TeV with the ATLAS and CMS Experiments. Phys Rev Lett 2015;114:191803. [DOI] [PubMed] [Google Scholar]
- 133.Young GP, Pedersen SK, Mansfield S, et al. A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer Med 2016;5:2763–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Oh T, Kim N, Moon Y, et al. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn 2013;15:498–507. [DOI] [PubMed] [Google Scholar]
- 135.Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014;63:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Finger FP. One ring to bind them. Septins and actin assembly. Dev Cell 2002;3:761–3. [DOI] [PubMed] [Google Scholar]
- 137.Sheffield PJ, Oliver CJ, Kremer BE, et al. Borg/septin interactions and the assembly of mammalian septin heterodimers, trimers, and filaments. J Biol Chem 2003;278:3483–8. [DOI] [PubMed] [Google Scholar]
- 138.Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008;54:414–23. [DOI] [PubMed] [Google Scholar]
- 139.Jin P, Kang Q, Wang X, et al. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 2015;30:830–3. [DOI] [PubMed] [Google Scholar]
- 140.Shen L, Catalano PJ, Benson AB 3rd, et al. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res 2007;13:6093–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev;29:181–206. [DOI] [PubMed] [Google Scholar]
- 142.Ogino S, Meyerhardt JA, Kawasaki T, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch 2007;450:529–537. [DOI] [PubMed] [Google Scholar]
- 143.Zlobec I, Kovac M, Erzberger P, et al. Combined analysis of specific KRAS mutation, BRAF and microsatellite instability identifies prognostic subgroups of sporadic and hereditary colorectal cancer. Int J Cancer. [DOI] [PubMed] [Google Scholar]
- 144.Van Rijnsoever M, Elsaleh H, Joseph D, et al. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res 2003;9:2898–903. [PubMed] [Google Scholar]
- 145.Lee S, Cho NY, Yoo EJ, et al. CpG island methylator phenotype in colorectal cancers: comparison of the new and classic CpG island methylator phenotype marker panels. Arch Pathol Lab Med 2008;132:1657–65. [DOI] [PubMed] [Google Scholar]
- 146.Pai RK, Jayachandran P, Koong AC, et al. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol 2012;36:744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst 2013;105:1151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Juo YY, Johnston FM, Zhang DY, et al. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis. Ann Oncol 2014;25:2314–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Baba Y, Nosho K, Shima K, et al. HIF1A overexpression is associated with poor prognosis in a cohort of 731 colorectal cancers. Am J Pathol 2010;176:2292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nilsson TK, Lof-Ohlin ZM, Sun XF. DNA methylation of the p14ARF, RASSF1A and APC1A genes as an independent prognostic factor in colorectal cancer patients. Int J Oncol 2013;42:127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nagasaka T, Sharp GB, Notohara K, et al. Hypermethylation of O6-methylguanine-DNA methyltransferase promoter may predict nonrecurrence after chemotherapy in colorectal cancer cases. Clin Cancer Res 2003;9:5306–12. [PubMed] [Google Scholar]
- 152.Draht MX, Smits KM, Tournier B, et al. Promoter CpG island methylation of RET predicts poor prognosis in stage II colorectal cancer patients. Mol Oncol 2014;8:679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang R, Kang KA, Piao MJ, et al. Epigenetic alterations are involved in the overexpression of glutathione S-transferase pi-1 in human colorectal cancers. Int J Oncol 2014;45:1275–83. [DOI] [PubMed] [Google Scholar]
- 154.Park SJ, Kim SM, Hong YS, et al. TFAP2E methylation status and prognosis of patients with radically resected colorectal cancer. Oncology 2015;88:122–32. [DOI] [PubMed] [Google Scholar]
- 155.Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol 2015;33:1787–96. [DOI] [PubMed] [Google Scholar]
- 156.Min BH, Bae JM, Lee EJ, et al. The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy. BMC Cancer;11:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jover R, Nguyen TP, Perez-Carbonell L, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology;140:1174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gallois C, Taieb J, Le Corre D, et al. Prognostic Value of Methylator Phenotype in Stage III Colon Cancer Treated with Oxaliplatin-based Adjuvant Chemotherapy. Clin Cancer Res 2018;24:4745–4753. [DOI] [PubMed] [Google Scholar]
- 159.Shiovitz S, Bertagnolli MM, Renfro LA, et al. CpG island methylator phenotype is associated with response to adjuvant irinotecan-based therapy for stage III colon cancer. Gastroenterology 2014;147:637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weller M, Stupp R. Prime time for molecular marker diagnostics in neuro-oncology. Curr Opin Neurol 2012;25:743–4. [DOI] [PubMed] [Google Scholar]
- 161.Ebert MP, Tanzer M, Balluff B, et al. TFAP2E-DKK4 and chemoresistance in colorectal cancer. N Engl J Med 2012;366:44–53. [DOI] [PubMed] [Google Scholar]
- 162.Moutinho C, Martinez-Cardus A, Santos C, et al. Epigenetic inactivation of the BRCA1 interactor SRBC and resistance to oxaliplatin in colorectal cancer. J Natl Cancer Inst 2014;106:djt322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 1953;6:963–8. [DOI] [PubMed] [Google Scholar]
- 164.Luo Y, Yu M, Grady WM. Field cancerization in the colon: a role for aberrant DNA methylation? Gastroenterol Rep (Oxf) 2014;2:16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Issa JP, Ottaviano YL, Celano P, et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 1994;7:536–40. [DOI] [PubMed] [Google Scholar]
- 166.Cui H, Onyango P, Brandenburg S, et al. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res 2002;62:6442–6. [PubMed] [Google Scholar]
- 167.Suzuki H, Watkins DN, Jair KW, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 2004;36:417–22. [DOI] [PubMed] [Google Scholar]
- 168.Kawakami K, Ruszkiewicz A, Bennett G, et al. DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer. Br J Cancer 2006;94:593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shen L, Kondo Y, Rosner GL, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst 2005;97:1330–8. [DOI] [PubMed] [Google Scholar]
- 170.Kamiyama H, Suzuki K, Maeda T, et al. DNA demethylation in normal colon tissue predicts predisposition to multiple cancers. Oncogene 2012;31:5029–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Grady WM, Parkin RK, Mitchell PS, et al. Epigenetic silencing of the intronic microRNA hsamiR-342 and its host gene EVL in colorectal cancer. Oncogene 2008;27:3880–8. [DOI] [PubMed] [Google Scholar]
- 172.Issa JP. Aging and epigenetic drift: a vicious cycle. J Clin Invest 2014;124:24–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ahuja N, Li Q, Mohan M, et al. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Research 1998;58:5489–5494. [PubMed] [Google Scholar]
- 174.Ahuja N, Issa JP. Aging, methylation and cancer. Histol Histopathol 2000;15:835–42. [DOI] [PubMed] [Google Scholar]
- 175.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 2005;102:10604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bell JT, Spector TD. DNA methylation studies using twins: what are they telling us? Genome Biol 2012;13:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bell JT, Saffery R. The value of twins in epigenetic epidemiology. Int J Epidemiol 2012;41:140–50. [DOI] [PubMed] [Google Scholar]
- 178.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 2005;37:391–400. [DOI] [PubMed] [Google Scholar]
- 179.Cohen AJ, Saiakhova A, Corradin O, et al. Hotspots of aberrant enhancer activity punctuate the colorectal cancer epigenome. Nat Commun 2017;8:14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harb Perspect Biol 2016;8:a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tamagawa H, Oshima T, Shiozawa M, et al. The global histone modification pattern correlates with overall survival in metachronous liver metastasis of colorectal cancer. Oncol Rep 2012;27:637–42. [DOI] [PubMed] [Google Scholar]
- 182.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Benard A, Goossens-Beumer IJ, van Hoesel AQ, et al. Histone trimethylation at H3K4, H3K9 and H4K20 correlates with patient survival and tumor recurrence in early-stage colon cancer. BMC Cancer 2014;14:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Benard A, Goossens-Beumer IJ, van Hoesel AQ, et al. Nuclear expression of histone deacetylases and their histone modifications predicts clinical outcome in colorectal cancer. Histopathology 2015;66:270–82. [DOI] [PubMed] [Google Scholar]
- 185.Tamagawa H, Oshima T, Numata M, et al. Global histone modification of H3K27 correlates with the outcomes in patients with metachronous liver metastasis of colorectal cancer. Eur J Surg Oncol 2013;39:655–61. [DOI] [PubMed] [Google Scholar]
- 186.Leszinski G, Gezer U, Siegele B, et al. Relevance of histone marks H3K9me3 and H4K20me3 in cancer. Anticancer Res 2012;32:2199–205. [PubMed] [Google Scholar]
- 187.Gezer U, Ustek D, Yoruker EE, et al. Characterization of H3K9me3- and H4K20me3-associated circulating nucleosomal DNA by high-throughput sequencing in colorectal cancer. Tumour Biol 2013;34:329–36. [DOI] [PubMed] [Google Scholar]
- 188.Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015;148:77–87 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sharma P Clinical practice. Barrett’s esophagus. N Engl J Med 2009;361:2548–56. [DOI] [PubMed] [Google Scholar]
- 190.Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet 2013;381:400–12. [DOI] [PubMed] [Google Scholar]
- 191.Kaz AM, Grady WM, Stachler MD, et al. Genetic and Epigenetic Alterations in Barrett’s Esophagus and Esophageal Adenocarcinoma. Gastroenterol Clin North Am 2015;44:473–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yu M, Maden SK, Stachler M, et al. Subtypes of Barrett’s oesophagus and oesophageal adenocarcinoma based on genome-wide methylation analysis. Gut 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Britton E, Rogerson C, Mehta S, et al. Open chromatin profiling identifies AP1 as a transcriptional regulator in oesophageal adenocarcinoma. PLoS Genet 2017;13:e1006879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jammula S, Katz-Summercorn AC, Li X, et al. Identification of Subtypes of Barrett’s Esophagus and Esophageal Adenocarcinoma Based on DNA Methylation Profiles and Integration of Transcriptome and Genome Data. Gastroenterology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kaz AM, Grady WM. Epigenetic biomarkers in esophageal cancer. Cancer Lett 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shah AK, Saunders NA, Barbour AP, et al. Early diagnostic biomarkers for esophageal adenocarcinoma--the current state of play. Cancer Epidemiol Biomarkers Prev 2013;22:1185–209. [DOI] [PubMed] [Google Scholar]
- 197.Kaz AM, Wong CJ, Varadan V, et al. Global DNA methylation patterns in Barrett’s esophagus, dysplastic Barrett’s, and esophageal adenocarcinoma are associated with BMI, gender, and tobacco use. Clin Epigenetics 2016;8:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Curtius K, Wong CJ, Hazelton WD, et al. A Molecular Clock Infers Heterogeneous Tissue Age Among Patients with Barrett’s Esophagus. PLoS Comput Biol 2016;12:e1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Salam I, Hussain S, Mir MM, et al. Aberrant promoter methylation and reduced expression of p16 gene in esophageal squamous cell carcinoma from Kashmir valley: a high-risk area. Mol Cell Biochem 2009;332:51–8. [DOI] [PubMed] [Google Scholar]
- 200.Kuester D, El-Rifai W, Peng D, et al. Silencing of MGMT expression by promoter hypermethylation in the metaplasia-dysplasia-carcinoma sequence of Barrett’s esophagus. Cancer Lett 2009;275:117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Moinova HR, LaFramboise T, Lutterbaugh JD, et al. Identifying DNA methylation biomarkers for non-endoscopic detection of Barrett’s esophagus. Sci Transl Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yu M, O’Leary RM, Kaz AM, et al. Methylated B3GAT2 and ZNF793 Are Potential Detection Biomarkers for Barrett’s Esophagus. Cancer Epidemiol Biomarkers Prev 2015;24:1890–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang Z, Kambhampati S, Cheng Y, et al. Methylation Biomarker Panel Performance in EsophaCap Cytology Samples for Diagnosing Barrett’s Esophagus: A Prospective Validation Study. Clin Cancer Res 2019;25:2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lao-Sirieix P, Bousioutas A, Kadri SR, et al. Non-endoscopic screening biomarkers for Barrett’s oesophagus: from microarray analysis to the clinic. Gut 2009;58:1451–9. [DOI] [PubMed] [Google Scholar]
- 205.Kadri SR, Lao-Sirieix P, O’Donovan M, et al. Acceptability and accuracy of a non-endoscopic screening test for Barrett’s oesophagus in primary care: cohort study. BMJ;341:c4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chettouh H, Mowforth O, Galeano-Dalmau N, et al. Methylation panel is a diagnostic biomarker for Barrett’s oesophagus in endoscopic biopsies and non-endoscopic cytology specimens. Gut 2018;67:1942–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Fitzgerald RC, di Pietro M, Ragunath K, et al. British Society of Gastroenterology guidelines on the diagnosis and management of Barrett’s oesophagus. Gut 2014;63:7–42. [DOI] [PubMed] [Google Scholar]
- 208.Spechler SJ. Barrett esophagus and risk of esophageal cancer: a clinical review. JAMA 2013;310:627–36. [DOI] [PubMed] [Google Scholar]
- 209.Chiba T, Marusawa H, Ushijima T. Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology 2012;143:550–63. [DOI] [PubMed] [Google Scholar]
- 210.Schulmann K, Sterian A, Berki A, et al. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett’s-associated neoplastic progression and predicts progression risk. Oncogene 2005;24:4138–48. [DOI] [PubMed] [Google Scholar]
- 211.Sato F, Jin Z, Schulmann K, et al. Three-Tiered Risk Stratification Model to Predict Progression in Barrett’s Esophagus Using Epigenetic and Clinical Features. PLoS One 2008;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jin Z, Cheng Y, Gu W, et al. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer Res 2009;69:4112–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kojiro M, Roskams T. Early hepatocellular carcinoma and dysplastic nodules. Semin Liver Dis 2005;25:133–42. [DOI] [PubMed] [Google Scholar]
- 214.Sato T, Kondo F, Ebara M, et al. Natural history of large regenerative nodules and dysplastic nodules in liver cirrhosis: 28-year follow-up study. Hepatol Int 2015;9:330–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hernandez-Vargas H, Lambert MP, Le Calvez-Kelm F, et al. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PLoS One 2010;5:e9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hlady RA, Tiedemann RL, Puszyk W, et al. Epigenetic signatures of alcohol abuse and hepatitis infection during human hepatocarcinogenesis. Oncotarget 2014;5:9425–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Schumacher FR, Olama AAA, Berndt SI, et al. Author Correction: Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet 2019;51:363. [DOI] [PubMed] [Google Scholar]
- 218.Ding X, He M, Chan AWH, et al. Genomic and Epigenomic Features of Primary and Recurrent Hepatocellular Carcinomas. Gastroenterology 2019;157:1630–1645 e6. [DOI] [PubMed] [Google Scholar]
- 219.Sung WK, Zheng H, Li S, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012;44:765–9. [DOI] [PubMed] [Google Scholar]
- 220.Sia D, Llovet JM. Liver cancer: Translating ‘-omics’ results into precision medicine for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2017;14:571–572. [DOI] [PubMed] [Google Scholar]
- 221.Kisiel JB, Dukek BA, VSRK R, et al. Hepatocellular Carcinoma Detection by Plasma Methylated DNA: Discovery, Phase I Pilot, and Phase II Clinical Validation. Hepatology 2019;69:1180–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Chang MS, Uozaki H, Chong JM, et al. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin Cancer Res 2006;12:2995–3002. [DOI] [PubMed] [Google Scholar]
- 224.Li Y, Liang J, Hou P. Hypermethylation in gastric cancer. Clin Chim Acta 2015;448:124–32. [DOI] [PubMed] [Google Scholar]
- 225.Maekita T, Nakazawa K, Mihara M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res 2006;12:989–95. [DOI] [PubMed] [Google Scholar]
- 226.Yoshida T, Yamashita S, Takamura-Enya T, et al. Alu and Satalpha hypomethylation in Helicobacter pylori-infected gastric mucosae. Int J Cancer 2011;128:33–9. [DOI] [PubMed] [Google Scholar]
- 227.Chan AO, Peng JZ, Lam SK, et al. Eradication of Helicobacter pylori infection reverses E-cadherin promoter hypermethylation. Gut 2006;55:463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sepulveda AR, Yao Y, Yan W, et al. CpG methylation and reduced expression of O6-methylguanine DNA methyltransferase is associated with Helicobacter pylori infection. Gastroenterology 2010;138:1836–44. [DOI] [PubMed] [Google Scholar]
- 229.Perri F, Cotugno R, Piepoli A, et al. Aberrant DNA methylation in non-neoplastic gastric mucosa of H. Pylori infected patients and effect of eradication. Am J Gastroenterol 2007;102:1361–71. [DOI] [PubMed] [Google Scholar]
- 230.Hino R, Uozaki H, Murakami N, et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res 2009;69:2766–74. [DOI] [PubMed] [Google Scholar]
- 231.Geddert H, Zur Hausen A, Gabbert HE, et al. EBV-infection in cardiac and non-cardiac gastric adenocarcinomas is associated with promoter methylation of p16, p14 and APC, but not hMLH1. Anal Cell Pathol (Amst) 2010;33:143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yamashita S, Kishino T, Takahashi T, et al. Genetic and epigenetic alterations in normal tissues have differential impacts on cancer risk among tissues. Proc Natl Acad Sci U S A 2018;115:1328–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Asada K, Nakajima T, Shimazu T, et al. Demonstration of the usefulness of epigenetic cancer risk prediction by a multicentre prospective cohort study. Gut 2015;64:388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun 2014;455:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Warton K, Mahon KL, Samimi G. Methylated circulating tumor DNA in blood: power in cancer prognosis and response. Endocr Relat Cancer 2016;23:R157–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Padmanabhan N, Ushijima T, Tan P. How to stomach an epigenetic insult: the gastric cancer epigenome. Nat Rev Gastroenterol Hepatol 2017;14:467–478. [DOI] [PubMed] [Google Scholar]
- 237.Kusano M, Toyota M, Suzuki H, et al. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer 2006;106:1467–79. [DOI] [PubMed] [Google Scholar]
- 238.Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research N. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017;32:185–203 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mishra NK, Guda C. Genome-wide DNA methylation analysis reveals molecular subtypes of pancreatic cancer. Oncotarget 2017;8:28990–29012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Byrne AT, Alferez DG, Amant F, et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer 2017. [DOI] [PubMed] [Google Scholar]
- 241.Eissa MAL, Lerner L, Abdelfatah E, et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin Epigenetics 2019;11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Brancaccio M, Natale F, Falco G, et al. Cell-Free DNA Methylation: The New Frontiers of Pancreatic Cancer Biomarkers’ Discovery. Genes (Basel) 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]


