Summary
Candida auris is an emerging multi-drug resistant human fungal pathogen. C. auris skin colonization results in environmental shedding, which underlies hospital transmissions, and predisposes patients to subsequent infections. We developed a murine skin topical exposure model for C. auris to dissect risk factors for colonization and to test interventions that might protect patients. We demonstrate that C. auris establishes long-term residence within the skin tissue compartment, which would elude clinical surveillance. The four clades of C. auris, with geographically distinct origins, differ in their abilities to colonize murine skin, mirroring epidemiologic findings. IL-17 receptor signaling and specific arms of immunity protect mice from long-term C. auris skin colonization. We further determine that commonly used chlorhexidine antiseptic serves as a protective and decolonizing agent against C. auris. This translational model facilitates an integrated approach to develop strategies to combat the unfolding global outbreaks of C. auris and other skin-associated microbial pathogens.
Keywords: Candida auris, fungal pathogen, skin colonization, epidemiology, translational mouse model, adaptive and innate immunity
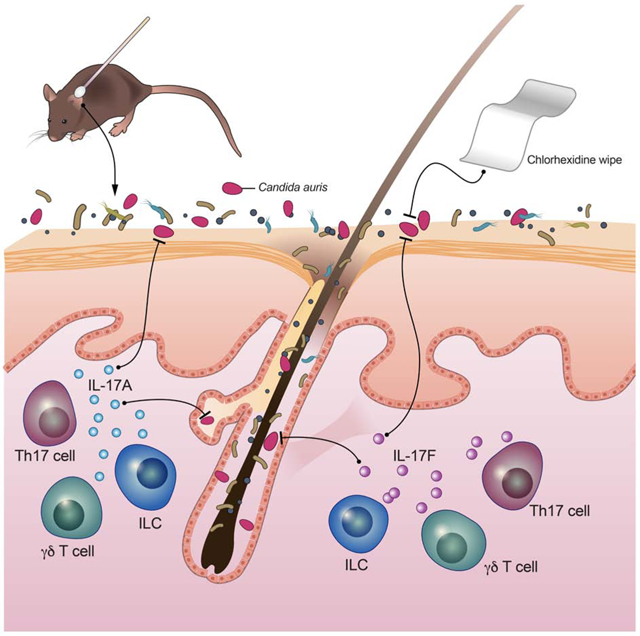
Graphical Abstract
In Brief
The mechanisms by which Candida auris, an emerging human fungal pathogen, colonizes skin remain largely uncharacterized. Using a translational model, Huang et al. demonstrate within tissue residence by C. auris, and reveal that IL-17 signaling provides protection against C. auris both on the skin surface and within skin tissue.
Introduction
The human-associated fungal pathogen Candida auris has emerged as a serious threat to global health because of its high rates of evolved antimicrobial resistance, high mortality associated with bloodstream infection and high transmission rates within healthcare facilities (https://www.cdc.gov/fungal/candida-auris) (Lockhart et al., 2017). C. auris was first identified in 2009 from a clinical culture of a Japanese patient’s external ear canal (Satoh et al., 2009) and in the ensuing decade, multiple outbreaks of skin colonization and/or associated bloodstream infection have occurred globally with an associated mortality rate of up to 60% (Forsberg et al., 2019). C. auris is one of only five microbial pathogens with the highest rating of urgent threat in the 2019 antibiotic resistance report released by the US Centers for Disease Control and Prevention (CDC) (CDC, 2019). Both the CDC (https://www.cdc.gov/fungal/candida-auris/candida-auris-alert.html) and Public Health England (https://www.gov.uk/government/publications/candida-auris-emergence-in-england/candida-auris-identified-in-england) issued multiple clinical alerts about C. auris. Within the United States, C. auris was made nationally notifiable in 2018 with over 1,000 cases reported to date. Over one million invasive fungal infections occur globally, increasingly from multi-drug resistant C. auris (Lionakis and Hohl, 2020). The advancement of medical technologies, such as organ and bone marrow transplantation, has also exposed more patients to nosocomial fungal infections
Many of the proposed factors for C. auris skin colonization and bloodstream infection co-occur in vulnerable hospitalized patients who are at risk for acquisition of nosocomial infections, thus, delineating primary drivers of patient susceptibility has been challenging (Vallabhaneni et al., 2019). Approximately half of the C. auris clinical cases in the United States have been in the bloodstream, but skin sites (axilla, groin) are considered the primary sites of asymptomatic colonization (Vallabhaneni et al., 2016). Colonization is distinct from infection as it typically results in asymptomatic carriage; however, skin colonization often precedes and is a risk factor for subsequent bloodstream infection. Hence, identifying C. auris colonized patients is a crucial component of infection control because these patients are at increased risk for developing a subsequent bloodstream infection, and pose a risk for nosocomial transmission to other patients in the ward.
C. auris has been demonstrated to colonize the skin of patients for months, and to persist as viable in the environment for weeks (Adams et al., 2018; Welsh et al., 2017), which both likely contribute to the nosocomial transmission of C. auris. Indeed, an investigation of a C. auris outbreak in an intensive care unit identified C. auris-bearing reusable skin-surface axillary temperature probes as a mode of nosocomial transmission (Eyre et al., 2018). Persistence on the skin and in the environment are unique features for C. auris which have not been observed with other Candida species that cause human infections, such as Candida albicans or Candida glabrata, which more typically reside within the human gastrointestinal tract.
Given the remarkable propensity of C. auris to colonize the human skin for prolonged periods of time, employing infection control measures for curtailing patient skin colonization by C. auris and preventing nosocomial transmission represents a major unmet medical need. To that end, bathing patient skin with the common antiseptic chlorhexidine gluconate (CHG) has increasingly become standard of care to reduce multi-drug resistant (MDR) bacterial transmission and infection (Huang et al., 2013; Huang et al., 2019a) and may represent a viable strategy to combat C. auris skin colonization in humans. However, this hospital practice has raised some concerns that CHG might deplete the commensal microbiota which provide colonization resistance and/or induce resistance among pathogens (Kampf, 2016), such as C. auris. Therefore, defining the efficacy of CHG in preventing or eliminating colonization by C. auris in preclinical models in vivo has important clinical implications.
Whole-genome sequencing and epidemiologic investigations have revealed near simultaneous emergence of genetically distinct clades of C. auris on different continents: East Asia, South Asia, South America, and Africa (Lockhart et al., 2017), with a potential fifth Iranian clade (Chow et al., 2019). The clades differ by tens to hundreds of thousands of DNA base pairs, but isolates are highly clonal within each clade (Chow et al., 2018). The simultaneous emergence of multiple phylogenetically distinct geographic clades presenting with high levels of antimicrobial resistance and causing disease among patients worldwide remains an epidemiologic enigma (Chow et al., 2020).
Previous studies examined the pathogenicity of systemically introduced C. auris infection in murine (Ben-Ami et al., 2017; Torres et al., 2019; Xin et al., 2019) and insect (Wurster et al., 2019) models. However, thus far, no animal model has been developed to examine fungal and host determinants of C. auris skin colonization. Here, we report a mouse model for C. auris skin colonization to facilitate translational research into features of human disease. We employ a series of studies to explore the underlying biology of C. auris as a human-associated pathogen, dissect risk factors for colonization and test preventive and therapeutic strategies which may be adopted to protect patients. This model provides the foundation for enhancing our understanding of fungal virulence traits, protective host immune responses, in vivo fitness of multi-drug resistant (MDR) strains, infection control interventions, and effective treatment modalities against C. auris skin colonization. This translational mouse model of skin colonization and evaluation of infection control measures could be adapted for other MDR microbes, such as Gram-negative and Gram-positive bacteria, which colonize the skin and cause life-threatening infections in hospitalized patients.
Results
Candida auris Persistently Colonizes Skin
To model epidemiologic features of nosocomial transmission, we extended our previously reported skin microbial colonization mouse model (Naik et al., 2015) and applied C. auris topically to the skin surface of the ear pinnae and shaved back (Figure 1A) of immunocompetent wild-type (WT) C57BL/6NTac mice. This model of skin colonization in the context of existing normal microbial communities has previously been used to demonstrate how tissue-resident immune cells are poised to sense and respond to alterations in microbial communities (Naik et al., 2015). In keeping with clinical relevance, the clinical isolate recovered from an NIH C. auris-infected patient (Vallabhaneni et al, 2016), which belongs to the South Asian clade, was used in most experiments in this study, unless noted otherwise.
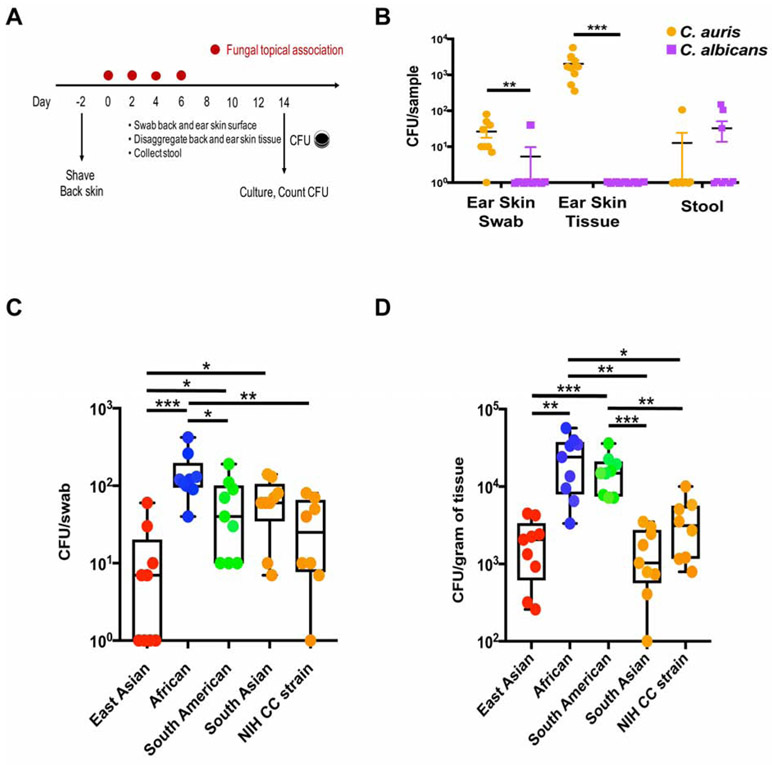
Figure 1. Candida auris colonizes skin surface and tissue compartment with clade specificity.
(A) Schematic for experimental design. Fungal cells were topically associated on ear pinna and dorsal back skin every other day four times. Skin swabs, skin tissue and stool were obtained on day 14 and processed for C. auris culturing.
(B) C. auris colonizes skin surface and tissue but not the gut compartment. Colony forming units (CFUs) per swab, gram of tissue or gram of stool from wild-type (WT) mice colonized with C. auris (orange) and C. albicans (purple) are plotted. Shown are mean values +/− SEM. Statistics were calculated using the Mann-Whitney U test. N=9 mice for each group, two independent experiments. See also Figure S1A and Table S1.
(C and D) C. auris colonization of WT mice with each of the four clades or the NIH Clinical Center strain (South Asian clade) was measured by culturing skin swabs (C) and disaggregated skin tissue (D). Statistics were calculated using one-way ANOVA with Tukey’s post-hoc tests or Games-Howell tests, as appropriate. N=8-9 mice for each group, two independent experiments. See also Figure S2.
*: p<0.05, **: p<0.01, ***: p<0.001.
To delineate body site tropism of fungi within the Candida genus, we compared fungal load on the skin and in the gut of mice colonized by C. auris and C. albicans over two weeks. Through mouse grooming behavior, fungal cells topically applied to mouse skin entered the gastrointestinal tract. At the 2-week endpoint, modeling the duration of a typical patient hospital stay, C. auris consistently colonized the skin surface of WT mouse ear pinnae (determined by swabbing), whereas C. albicans minimally colonized WT mouse ear skin (Figures 1A and 1B). Strikingly, higher concentrations of C. auris resided deeper within the skin (determined by disaggregation of tissue), whereas C. albicans did not reside within the skin tissue compartment (Figure 1B). Colonies, grown on selective media, were confirmed as C. auris with species-specific PCR primers (data not shown). As expected, C. albicans only sporadically colonized the WT mouse intestinal tract with very rare examples of C. auris intestinal colonization (Figure 1B).
Human patients are screened for C. auris colonization with non-invasive swabs of the skin surface. However, in the murine model following topical association, C. auris cells were observed both on the skin surface and deeper inside the tissue, resident within the hair follicle (Figure S1A). To test whether skin tissue could serve as an unrecognized reservoir for C. auris colonization, we examined persistence of C. auris residence on the surface and deeper within the skin tissue compartment. After topical association with C. auris, WT mice were examined for surface colonization on a weekly basis to model clinical epidemiologic surveillance. Skin surface swabs were negative 50 days after initial C. auris colonization in all 16 mice tested. At the same time, we recovered C. auris from deeper skin tissue in 3 of 4 mice tested (Table S1). With monthly surveillance, we continued to recover live C. auris colonies from deeper skin tissue for up to 4 months after initial colonization, which is two months after the mouse would have been considered ‘clear’ of C. auris skin colonization based on surface swabbing (Table S1). While C. auris colonization deeper in tissue has not been explored in human patients due to the risk of performing invasive sampling, it might potentially explain the clinical observation of skin surface surveillance cultures that grow out C. auris after multiple prior negative results.
Clinical reports have suggested that broad-spectrum antibiotic or azole antifungal pre-exposure and diabetes may be associated with C. auris colonization and/or infection (Khan et al., 2018; Lockhart et al., 2017; Parra-Giraldo et al., 2018). However, those features are common in patients with prolonged hospitalization and it is currently unknown if they increase the risk of developing C. auris colonization and infection. We utilized our model of C. auris skin association to examine the direct impact of broad-spectrum antibiotic pre-exposure (Kobayashi et al., 2015), fluconazole pre-exposure, diabetes (Grice et al., 2010), and high-fat diet (Ridaura et al., 2018), all of which are known to alter the skin microbiome and to affect topical host immune responses against microbes, on C. auris skin colonization. Notably, we found no increase in C. auris skin colonization in any of these conditions (Table S2), indicating that these factors may not directly predispose to skin C. auris colonization. Future studies will be needed to investigate whether the combination of these factors can contribute to higher C. auris burden on the skin.
In addition to their distinct genetic differences and geographic origins, the four clades of C. auris are reported as having differences in antimicrobial resistance, colonization and transmission rates (Chow et al., 2018). To assess if the clades feature intrinsic genetic differences that may account for their differential clinical manifestations, we tested the four clades in our murine skin colonization model. Mirroring reported epidemiologic features, the four clades of C. auris colonized WT skin at different levels. Specifically, two weeks after C. auris exposure, the African clade led to the highest topical skin colonization, with similar albeit lower rates of skin colonization observed for the South American, South Asian and East Asian clades (Figure 1C). In keeping with the paucity of outbreaks reported for the East Asian clade, this clade colonized WT mouse skin at lower levels (Figure 1C). Demonstrating the long-term risk for persistent and potentially undetected colonization, all C. auris clades became resident within the skin tissue, with the highest rate observed for the African clade and the lowest rate observed for the South Asian and East Asian clade (Figure 1D).
Previous studies that utilized a single clade suggested that C. auris was less virulent than C. albicans in a mouse model of disseminated candidiasis when introduced systemically into WT mice. Indeed, neutropenia and/or corticosteroid administration were essential in those studies to elicit lethality in C. auris-infected mice whereas C. albicans is known to cause lethality in immunocompetent WT mice upon intravenous inoculation (Ben-Ami et al., 2017; Lionakis and Netea, 2013; Torres et al., 2019; Wang et al., 2018). This inter-species difference may relate to the ability of C. albicans, and not C. auris, to filament in vivo, a critical determinant of renal injury and lethality in the mouse model of disseminated candidiasis (Ben-Ami et al., 2017; Lionakis and Netea, 2013). Because differential fungal strain and clade virulence have been documented for C. albicans in the systemic infection model (Marakalala et al., 2013), we explored whether any of the four clades of C. auris that effectively colonize the mouse skin would induce lethality upon systemic inoculation in WT mice, and used the virulent C. albicans SC5314 strain as a positive control. As expected, intravenous injection of 105 C. albicans cells resulted in mortality for all mice by day 30 post injection (Figure S2). By contrast, none of the four clade strains of C. auris resulted in similar rates of mortality, with only a single mouse, inoculated with the African clade, succumbing during the 30-day study. Taken together, these results demonstrate an intrinsic tropism for skin colonization by C. auris, with deeper skin residence as a potential reservoir for the pathogen and provide a model of experimental C. auris skin colonization with translational relevance.
Skin Association with C. auris Promotes a Protective IL-17A/IL-17F Response Derived from Innate and Adaptive Lymphoid Cells
As previously shown with commensal bacterial topical application (Naik et al., 2015; Naik et al., 2012), skin association with C. auris did not appear to cause overt skin inflammation or disruption of the skin architecture as evidenced by the absence of skin erythema or histological evidence of hyperkeratosis or spongiosis (Figure S1B-D). C. auris topical association resulted in the accumulation of CD4+ T cells and CD8+ T cells, but not γδ T cells or innate lymphoid cells (ILCs) (Figure 2A). Among T helper subsets, C. auris topical association led to a selective accumulation of CD4+ IL-17A+ and CD4+ IL-17F+ (Th17) cells within the skin compartment (Figures 2B, 2C and S3), without expansion in skin Th1 (CD4+IFN-g+), Th2 (CD4+IL-5+), regulatory T (CD4+Foxp3+) cells (Figures S3 and S4A-C). C. auris topical application also promoted the accumulation of IL-17A- and IL-17F-producing CD8+ T cells (Tc17), IL-17A-producing γδ T cells, and IL-17A-producing ILCs (Figures 2B, 2C and S3) within the skin.
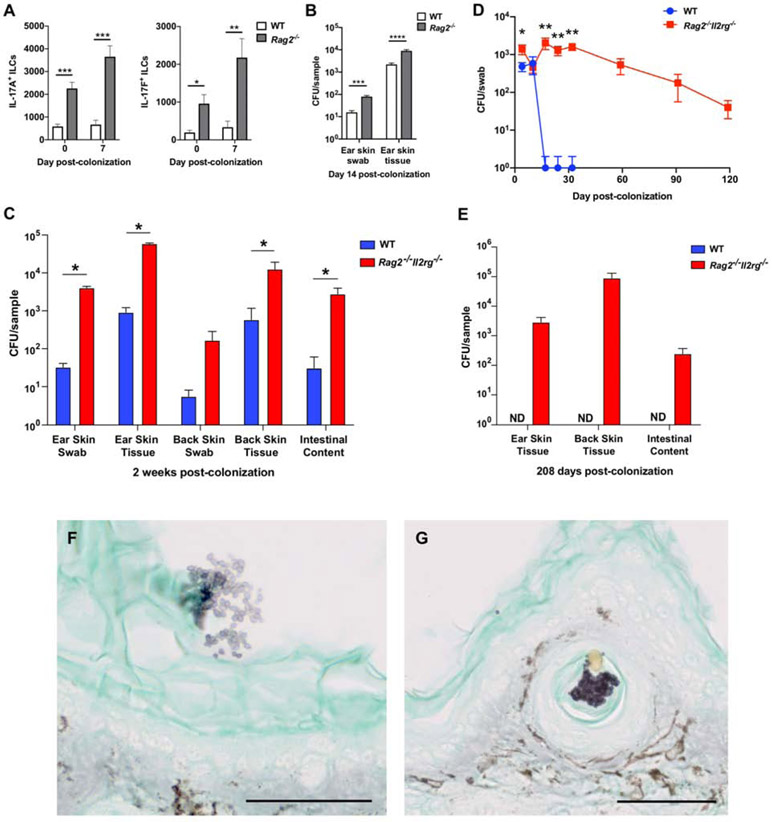
Figure 2. C. auris elicits a protective IL-17A/IL-17F response derived from innate and adaptive lymphoid cells.
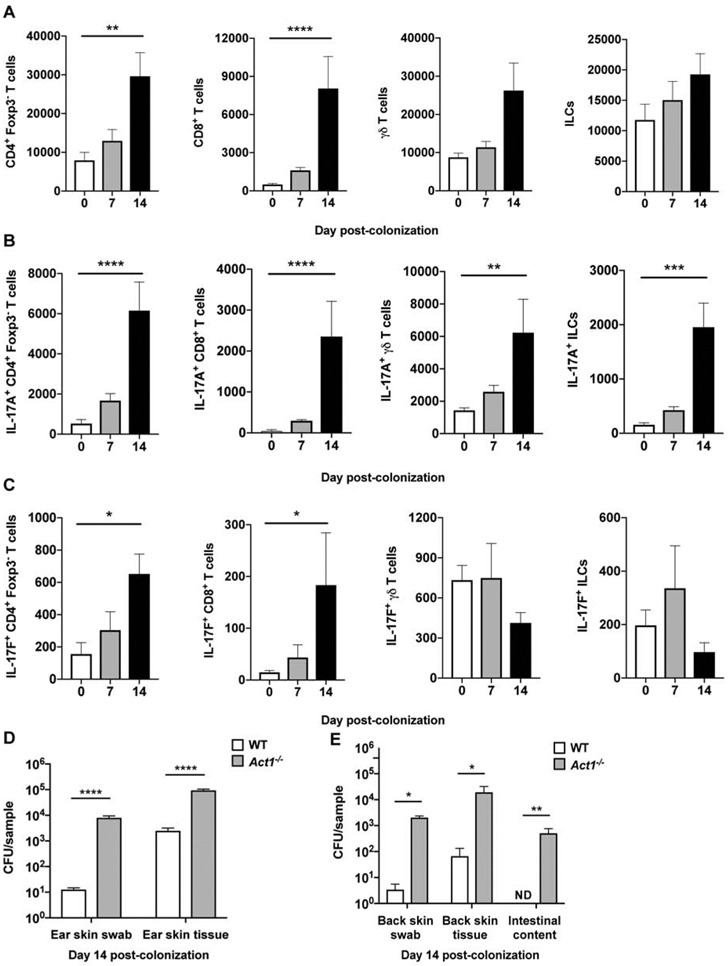
(A) Accumulation of CD4+ T cells, CD8+ T cells, γδ T cells, and innate lymphoid cells (ILCs) in the skin of WT mice 7 and 14 days after C. auris topical association as compared with naïve WT mice.
(B-C) Differences in IL-17A-producing (A) and IL-17F-producing (B) CD4+ T cells, CD8+ T cells, γδ T cells, and ILCs are shown for naïve WT mice, and WT mice 7 and 14 days after topical association with C. auris. (A-C) Total number of cells are shown for each cell type. Statistics were calculated using one-way ANOVA with Tukey’s HSD post-hoc test or Kruskal-Wallis test with Dunn’s multiple comparison test, as appropriate. N=6-9 mice for each group; two independent experiments. Data are shown with mean ± SEM. See also Figures S3, S4A-C and S5.
(D) Mice deficient in Act1, an essential adaptor of IL-17R family signaling, are defective in clearing C. auris from the ear skin surface (CFU/swab) and ear skin tissue (CFU/gram of tissue) relative to WT mice, measured 14 days after C. auris topical association. Statistics were calculated using the Mann-Whitney U test. N=10 mice for each group; two independent experiments. Data are shown with mean ± SEM.
(E) Compared with WT, mice deficient in Act1 have increased C. auris on the back skin surface (CFU/swab), back skin tissue (CFU/gram of tissue) and intestinal compartments (CFU/gram of intestinal content) on day 14 after topical C. auris association. Statistics were calculated using the Mann-Whitney U test. N=4-5 mice for each group. Data are shown with mean ± SEM.
*: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001, ND: not detected.
IL-17 receptor (IL-17R) signaling is well-recognized to play an indispensable role in mucocutaneous host defense against fungi (including Candida species) in mice and humans (Boisson et al., 2013; Conti et al., 2016; Lionakis and Levitz, 2018; Puel et al., 2011). Hence, we next asked whether the observed induction of an IL-17A/IL-17F response in C. auris-associated skin is protective against fungal skin colonization. We applied C. auris topically on the skin of mice deficient in the IL-17R-associated adaptor molecule Act1, which lack IL-17R-dependent cellular responses (Sonder et al., 2011), and recovered significantly increased amounts of C. auris from the Act1−/− mouse skin surface, skin tissue compartment, and intestinal luminal content (Figures 2D and 2E). Deficiency of the type 17-associated cytokine IL-22 was dispensable for the control of C. auris skin colonization as shown by experiments in both Il22−/− mice and Act1−/− mice in which IL-22 was neutralized using an antibody-based approach (Figure S4D-F). Collectively, these data indicate that skin association with C. auris drives an IL-17A-IL-17F/IL-17R response which promotes the control of C. auris skin colonization.
Previous studies have shown that induction of C. albicans-specific Th17 cells in the skin relies on Langerhans cells while induction of IL-17 at early time points post-C. albicans exposure is mediated by CD301b+ dermal dendritic cells (Igyarto et al. 2011; Kashem et al. 2015). Mice deficient in Langerhans cells (Lan-DTA) controlled C. auris skin colonization normally (Figure S4G). These data suggest that Langerhans cells may not be implicated in the overall cutaneous IL-17 response to C. auris, as it has been reported for C. albicans (Igyarto et al. 2011; Kashem et al. 2015). Moreover, the C-type lectin receptor adaptor molecule Card9 has been shown to promote the induction of Th17 cells upon systemic or oropharyngeal C. albicans infection and upon cutaneous Malassezia challenge, but it is dispensable for the development of innate type 17 responses in the setting of oropharyngeal candidiasis and skin Malassezia infection (LeibundGut-Landmann et al., 2007; Bishu et al., 2014; Kashem et al., 2015; Sparber et al. 2019). Mice deficient in Card9 controlled C. auris skin colonization in a similar manner to WT mice, suggesting that C-type lectin receptor signaling may not be implicated in the overall cutaneous IL-17 response to C. auris (Figures S4H).
To explore further the relative contribution of IL-17A/IL-17F derived from innate and/or adaptive lymphoid cells in protection against C. auris skin colonization, we next applied C. auris topically on the skin of Rag2−/− mice, which lack αβ and γδ T cells, and Rag2−/−Il2rg−/− mice, which also lack ILCs. With C. auris colonization, Rag2−/− mice exhibited enhanced accumulation of IL-17A and IL-17F producing ILCs in the skin relative to WT mice (Figure 3A). While Rag2−/− mice had greater C. auris skin colonization than WT mice, Act1−/− mice exhibited even greater C. auris skin colonization (Figures 2D and 3B). Of note, we found significantly increased C. auris colonization on the skin surface, deeper within the skin tissue, and in the intestinal luminal content of Rag2−/−Il2rg−/− mice (Figure 3C), which persisted for several months following the initial skin association with C. auris (Figures 3D and 3E). When comparing the degree of C. auris skin colonization in Rag2−/− versus Rag2−/−Il2rg−/− mice, Rag2−/−Il2rg−/− mice exhibited ~1 log greater CFU on the skin surface and deeper within the skin tissue (Figures 3B and 3C). Together these experiments suggest that production of IL-17A/IL-17F by a combination of αβ T cells, γδ T cells and ILCs contributes to the control of C. auris colonization. Future studies will be needed to define potential immune factors derived from innate and adaptive lymphoid cell subsets other than IL-17A/IL-17F that may confer protection against C. auris colonization in the model. Histological examination of the C. auris-associated skin of Rag2−/−Il2rg−/− mice using Grocott-Gomori's methenamine silver stain revealed clusters of C. auris on the skin surface as well as deeper in the skin compartment within the hair follicle structure (Figures 3F and 3G). Clusters of Grocott-Gomori's methenamine silver stained cells in naïve WT mice were not observed (data not shown).
Figure 3. Immunodeficient Rag2−/− and Rag2−/−Il2rg−/− mice exhibit greater propensity for C. auris skin colonization.
(A) IL-17A and IL-17F production by ILCs is increased in the skin of Rag2−/− mice. Shown are numbers of IL-17A-producing (left panel) and IL-17F-producing (right panel) ILCs in the skin of WT and Rag2-deficient mice at steady state and 7 days after C. auris colonization. The statistics were calculated using the t-test or Mann-Whitney U test, as appropriate. N=6-9 mice for each group; two independent experiments. Data are mean ± SEM.
(B) Rag2−/− mice are defective in controlling C. auris colonization on the ear skin surface (CFU/swab) and within the tissue compartment (CFU/gram of tissue). The statistics were calculated using the t-test or Mann-Whitney U test, as appropriate. N=9-10 mice for each group; two independent experiments.
(C) Immunodeficient Rag2−/−Il2rg−/− mice are defective in clearing C. auris from the ear and back skin surface (CFU/swab) and tissue (CFU/gram of tissue); and intestinal compartments (CFU/gram of intestinal content) as determined by culturing from WT and Rag2−/−Il2rg−/− mice two weeks after topical colonization. The statistics were calculated using the Mann-Whitney U test. N=5 mice for each group, representative of two independent experiments. Data are shown with mean ± SEM. See also Figure S4G-H.
(D) Longitudinally, WT and Rag2−/−Il2rg−/− mouse skin swabs were cultured for C. auris on days 4, 10, 17, 24, 32, and extended through days 59, 91 and 119 for Rag2−/−Il2rg−/− mice. The Mann-Whitney U test was used to compare between WT and Rag2−/−Il2rg−/− mice at each time point up to day 32. N=6 mice for each group, representative of three independent experiments. Data are shown with mean ± SEM.
(E) Immunodeficient Rag2−/−Il2rg−/− mice are defective in clearing C. auris from the ear and back skin tissue (CFU/gram of tissue); and intestinal compartments (CFU/gram of intestinal content) as measured by culturing from WT and Rag2−/−Il2rg−/− mice 208 days after topical association. No C. auris colonies were cultured from WT mice on day 208. N=6 mice for each group, representative of three independent experiments. Data are shown with mean ± SEM. ND: not detected.
(F and G) Grocott-Gomori's methenamine silver stain for fungi from ear pinna sections of a Rag2−/−Il2rg−/− mouse colonized by C. auris 7 days after topical association. Clusters of C. auris cells (purple) can be seen on the skin surface (stratum corneum, F) and located inside a hair follicle (G). 50 μm scale bar is displayed at the bottom right. The brown stain is endogenous mouse skin pigment also observed on control uncolonized mice. See also Figure S1A.
*: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001.
Chlorhexidine Gluconate Protects WT Mice from C. auris Colonization and Helps Decolonize Rag2−/−Il2rg−/− mice
Given C. auris’ high levels of antimicrobial resistance against multiple classes of antifungal drugs (i.e., azoles, polyenes, echinocandins), treatment options for C. auris bloodstream infection are limited (Larkin et al., 2017; Singh et al., 2019), especially with pan-resistant C. auris isolates recently reported in the United States (Ostrowsky et al., 2020). Limited therapeutic modalities underscore the essential role for effective infection control measures to prevent hospital transmission and bloodstream infections. The antiseptic CHG is commonly used to care for the skin of patients in hospitals and nursing homes as it has demonstrated effectiveness in reducing rates of MDR bacterial transmission and infection (Huang et al., 2019a). However, as C. auris outbreaks have occurred in facilities with established CHG bathing procedures, concern has been raised that CHG might be depleting the natural skin microbial shield allowing C. auris to persist on the skin and/or cause infections.
To disentangle CHG bathing from other concurrent epidemiologic risk factors seen in patients, we utilized our mouse model to explore the impact of CHG on C. auris skin colonization. CHG wipes, similar to those used on human skin, were used to ‘bathe’ the mouse skin daily for four days before C. auris topical association (Figure 4A), to mimic the effect of prophylactic use of CHG before patient exposure to C. auris. Next, to mimic the range of fungal exposures in healthcare facilities (Zhu et al., 2019), 107 and 109 CFUs of C. auris was topically applied to the skin of WT mice. CHG had a protective effect against C. auris skin colonization in both concentrations of applied C. auris, with complete eradication of skin colonization at the lower exposure concentration of 107 C. auris (Figure 4B). CHG treatment also prevented C. auris from establishing residence within the skin tissue compartment, again completely eliminating skin colonization at the lower exposure concentration of 107 C. auris (Figure 4C).
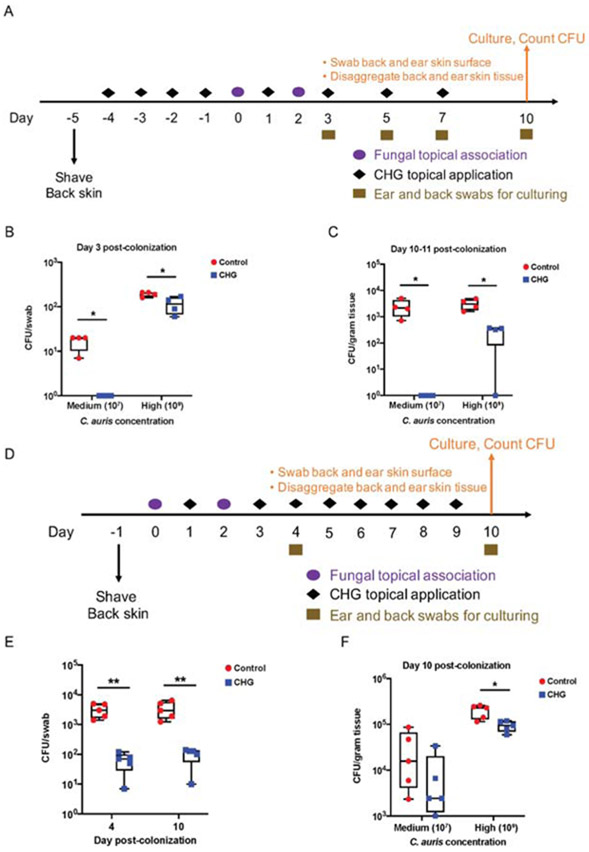
Figure 4. Skin antiseptic chlorhexidine protects WT mice from C. auris colonization and promotes C. auris decolonization of Rag2−/−Il2rg−/− mice.
(A) Schematic for experimental design to model chlorhexidine gluconate (CHG) skin bathing. Wipes impregnated with CHG were used daily for four days to ‘bathe’ the mouse skin before topical association. Cotton gauze saturated with sterile PBS was used as procedural control. C. auris cells were topically associated onto mouse ear pinna and dorsal back skin at two concentrations: 107 CFU as medium concentration, and 109 CFU as high concentration. After topical association, CHG was applied every other day for the duration of the experiment. Skin swabs were taken during the experiment and skin tissues were disaggregated at the end of the experiment and processed for C. auris culturing.
(B) CHG bathing protects WT mouse skin from C. auris topical exposure (medium and high concentrations) as measured by culturing skin swabs. N=4 mice for each group, representative of two independent experiments.
(C) CHG bathing protects WT mouse skin tissue from C. auris residence after topical exposure (medium and high concentrations). N=4 mice for each group, representative of two independent experiments.
(D) Schematic for experimental design to model CHG skin bathing to promote C. auris decolonization. After C. auris (medium and high concentrations) topical association, daily bathing with CHG was deployed for one week. Skin swabs were taken during the experiment and skin tissues were disaggregated at the end of the experiment for culturing.
(E) CHG bathing promotes C. auris decolonization on Rag2−/−Il2rg−/− ear skin swabs 4 and 10 days after high concentration topical exposure. The statistics were calculated using the t-test. N=5 mice for each group, representative of two independent experiments.
(F) CHG bathing promotes C. auris decolonization in Rag2−/−Il2rg−/− resident skin tissue after medium and high concentration topical exposure. N=5 mice for each group, representative of two independent experiments. See also Table S3. (B,C,E,F) The statistics were calculated using the t-test.
*: p<0.05, **: p<0.01.
Furthermore, to test whether CHG was effective at promoting decolonization of C. auris from colonized mouse skin, and to mimic the effect of CHG after patient exposure to C. auris, Rag2−/−Il2rg−/− mice (which were shown above to exhibit greater and persistent C. auris colonization in the skin) were first exposed to C. auris and then subsequently their skin was cleaned daily with CHG wipes (Figure 4D). Demonstrating its effectiveness, CHG reduced the C. auris load, both on the mouse skin surface and within the skin tissue compartment (Figures 4E and 4F). Together, these experiments demonstrate the effectiveness of CHG in preventing and ameliorating preexisting C. auris skin colonization and provide the basis for evaluating the efficacy of CHG in reducing the risk of subsequent infection and nosocomial transmission in the clinical setting.
Clinical microbiology laboratories are increasingly using quantitative PCR (qPCR)-based assays with C. auris specific primers to test surveillance skin swabs. A previous study demonstrated that CHG retains bacterial DNA on mouse skin (SanMiguel et al., 2018). We therefore sought to test whether CHG retained C. auris genomic DNA (gDNA) on mouse skin, which could have important implications for epidemiologic surveillance. Following an initial topical association with C. auris, the skin of mice was wiped every other day with either CHG or PBS as a control. During this time, skin swabs were taken every four days for both culture- and qPCR-based detection of C. auris. While C. auris load on the skin surface decreased over time on the skin of both CHG-exposed and control mice as determined by culture, the qPCR cycle threshold (Ct) values were consistently 2 cycles lower in the CHG group compared with control (Table S3). Since the levels of live (cultivable) C. auris were declining in CHG and control mice, the lower qPCR results in CHG mice likely represents PCR amplification from dead or compromised cells. Paradoxically, while CHG reduced the burden of live C. auris skin colonization, monitoring C. auris colonization levels solely with qPCR would have demonstrated an increased level of C. auris gDNA. These experiments underscore the need for clinical tests to benchmark molecular assays to culture based assays for hospital infection control.
Discussion
Over the last decade, C. auris has emerged simultaneously as four distinct genetic and geographic clades, with its origin still very much a mystery (Jackson et al., 2019). This MDR fungal pathogen is already causing global outbreaks that are difficult to control and are associated with significant healthcare-associated costs, as exemplified by one outbreak which required the facility to shut down for several months to eradicate the pathogen. Due to the paucity of antifungal treatment options for increasingly resistant C. auris bloodstream infections, effective infection control approaches are urgently needed to thwart the pathogen from spreading. Unlike other Candida species, C. auris’ unique ability to persist on human skin and in the hospital environment requires new biological insights into the host-pathogen life cycle. Our study presents a translational murine skin colonization model for C. auris which we employed to investigate risk factors for C. auris skin colonization, and to test infection control measures. Strikingly, we discover that C. auris is able to reside deep within the skin tissue compartment for months after skin surface swabs are repeatedly negative. Herein, we utilized the model to reveal a differential C. auris clade-specific propensity to persist in the skin, to define a clinically relevant immune deficiency state that predisposes to C. auris skin colonization, and to show that CHG can act as a protective and decolonizing agent against C. auris.
Previous studies have established various immunodeficient models for systemic C. auris infection in inbred mouse strains in the setting of neutropenia, corticosteroid use or chemotherapy administration (Ben-Ami et al., 2017; Torres et al., 2019; Xin et al., 2019). On the other hand, immunocompetent mice can survive high inocula of C. auris bloodstream challenge (Torres et al., 2019; Wang et al., 2018), with an exception of one study that used outbred ICR immunocompetent mice and showed lethality after C. auris challenge (Fakhim et al., 2018). Our results in immunocompetent C57BL/6NTac mice are consistent with most previous studies, in that intravenous challenge of a dose sufficient to kill C. albicans-infected mice does not pose a lethal threat to C. auris-challenged mice. Hence, C. auris appears to behave in this systemic model similar to C. glabrata, another medically important Candida species that does not result in in vivo filamentation or mouse lethality, yet it accounts for a significant and increasing proportion of human infections (Pappas et al., 2018). The fact that C. auris is primarily a skin colonizer underscores the difficulty in controlling the spread of the pathogen in healthcare settings (Ruiz-Gaitan et al., 2018). It is therefore likely that C. auris can colonize the patient skin long-term, and cause harm once it enters the bloodstream through breach of the mucocutaneous barriers in the setting of surgery or central venous catheters, and/or in the setting of underlying immune deficiency conditions of the patient.
Our surprising finding that C. auris resides within the mouse ear tissue long-term is indicative of potential major challenges in controlling C. auris in human populations. In our study, we occasionally observed re-emergence of C. auris on mouse skin that had been skin surface negative by culture several days prior. A previous clinical study had similarly observed the re-emergence of C. auris on patient skin after three consecutive negative culture screens (Eyre et al., 2018), suggesting that a skin tissue reservoir for C. auris may exist in humans, as in mice. Residence of C. auris within the skin tissue compartment might lead to bloodstream infection and its re-emergence on the skin surface could re-initiate hospital transmission, especially if infection control measures have meanwhile been withdrawn given prior consecutive negative skin surface culture screens. Our findings of a skin tissue reservoir of C. auris in our translational model and the early clinical reports are suggestive of such a reservoir in humans (Schelenz et al., 2016). However, directly testing humans for skin tissue residence of C. auris using punch biopsies is not advisable due to its invasive nature. Understanding how to deplete or deprive C. auris of this deeper tissue residence may prove critical for devising successful decolonization strategies in patients.
Our results that the four clades of C. auris differentially colonize the WT mouse skin are consistent with clinical observations that the East Asian clade is rarely implicated in outbreaks in human patients, unlike other clades (Jackson et al., 2019). As genetic tools are developed to manipulate the C. auris genome, future studies will explore the underlying differences that contribute to African, South Asian and South American clades’ proclivity for skin commensalism relative to the East Asian clade (Chow et al., 2020).
We found that C. auris skin colonization triggers an IL-17A+ and IL-17F+ response by several lymphoid cell populations, similar to the response elicited by C. albicans skin association (Kashem and Kaplan, 2018; Harrison et al., 2019). Interestingly, extending the well-recognized contribution of IL-17R signaling in mucocutaneous host defense in mice and humans (Boisson et al., 2013; Conti et al., 2016; Lionakis and Levitz, 2018; Puel et al., 2011), Act1 deleted mice, deficient in the IL-17 pathway, had a higher C. auris burden on the skin surface, skin tissue compartment and gut compared with WT mice. These results suggest that a competent IL-17R pathway contributes to keeping C. auris in check on the skin surface and tissue likely via facilitating the production of antifungal antimicrobial peptides by keratinocytes and promoting cutaneous barrier integrity and function. This finding has important translational implications because IL-17R-targeted biologics have been increasingly used in patients with autoimmune conditions such as psoriasis and inflammatory bowel disease (Frieder et al., 2018) and have been associated with mucocutaneous candidiasis in such patients (Saunte et al., 2017), which might also predispose them to greater and/or prolonged C. auris skin colonization. As well, other immunomodulatory agents such as corticosteroids can down-regulate IL-17R responses (Guggino et al., 2015), which could be a risk factor for greater and/or prolonged C. auris skin colonization in corticosteroid-treated patients. Of note, prior corticosteroid exposure has commonly been reported in patients with C. auris infection (Khan et al., 2018; Lockhart et al., 2017; Parra-Giraldo et al., 2018), making it plausible that corticosteroids may contribute to C. auris skin colonization via impaired IL-17A/IL-17F responses in the skin. Future studies will be needed to directly examine this hypothesis in the mouse model. Importantly, we show that both innate and adaptive lymphoid cellular sources of IL-17A/IL-17F appear to contribute in curtailing C. auris skin colonization as Rag2−/−Il2rg−/− mice, and to a lesser extent Rag2−/− mice, exhibited greater C. auris skin colonization. This finding supports the notion that deficiency of IL-17 production by a given cell population can be, at least partially, compensated at the mucocutaneous barrier by other IL-17-producing cells to promote IL-17R response competency. Taken together, these results suggest a specific clinically relevant immune deficiency as a risk factor for C. auris skin colonization. In addition, this work provides the basis for further investigations aiming to uncover other IL-17-independent immune factors produced by αβ T cells, γδ T cells, ILCs, and other immune and non-hematopoietic cells that may also regulate C. auris skin colonization.
CHG is typically used as an antiseptic in a wide range of healthcare settings. CHG has been tested extensively in humans and it is safe and effective against Gram-positive and Gram-negative bacteria and yeasts (Septimus and Schweizer, 2016). Increasingly, long-term acute care hospitals and skilled nursing facilities are adopting daily CHG bathing for their patients, although concerns have been raised about development of acquired resistance to CHG among bacterial pathogens (Kampf, 2016). A previous study indicated that CHG might disrupt the commensal bacterial communities on patient skin (Cassir et al., 2015). However, a more recent study appears to reach different conclusions (Kates et al., 2019). Importantly, topical CHG combined with nasal mupirocin has proven effective in reducing overall bloodstream infections for patients with medical devices (Huang et al., 2019a), and also in reducing postdischarge infection rates for methicillin-resistant Staphylococcus aureus (Huang et al., 2019b). Previous in vitro tests indicate at most 2 orders of magnitude reduction against C. auris growth by CHG (Rutala et al., 2019). Our in vivo study demonstrates the protective effect of CHG against C. auris colonization for both skin surface and skin tissue residence in mice by C. auris. This murine model supports the use of CHG bathing to protect patients from C. auris skin colonization, but this requires formal clinical trial testing.
Protocols to culture C. auris from surveillance or clinical samples require one-week growth, which is not practical for infection control in patients. Therefore, clinical microbiology laboratories are increasingly relying on qPCR-based results as the initial screen. We demonstrated that CHG trapped C. auris gDNA on the mouse skin, beyond the time point that live fungi could be recovered by culture. This discrepancy between molecular- and the culture-based assays might generate contradictory results regarding CHG’s ability to increase or decrease C. auris skin colonization, respectively. Future studies in C. auris-colonized patients will be required to directly compare the performance of qPCR- and culture-based assays in detecting C. auris biomass and viability on the human skin.
In summary, we have developed a murine model of C. auris skin colonization with translational relevance that demonstrates C. auris’ tropism for long-term skin colonization and provides a tractable in vivo system for the testing of C. auris virulence traits, host immune responses, the role of clinical risk factors (e.g., antibiotic pre-exposure, antifungal pre-exposure, diabetes), infection control measures and treatment approaches against this emerging MDR fungal pathogen of human patients.
STAR * Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents may be directed to and be fulfilled by the corresponding authors, Michail S. Lionakis (lionakism@niaid.nih.gov) and Julia A. Segre (jsegre@nhgri.nih.gov).
Materials Availability
Experimental models (organisms, strains) generated for use in this study will be made available on request, but we may require a completed Materials Transfer Agreement if there is potential for commercial application. These materials are available for distribution under the Uniform Biological Material Transfer Agreement, a master agreement that was developed by the NIH to simplify transfers of biological research materials.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study . This study did not generate any unique datasets or code
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal Studies
Wild-type (WT) and immunodeficient mice used in this study were female C57BL/6 mice at 8-10 weeks of age, unless specified below. Rag2−/− and Rag2−/−Il2rg−/− mice were on the C57BL/10 background, with age and sex matched WT C57BL/10 as controls. Male Il22−/− and Langerhans cell deficient (Lan-DTA) mice were used with appropriate controls. All mice were healthy before initiation of studies, were not subjected to previous procedures, and were naive to drugs. All mice were maintained in an SPF environment at an American Association for the Accreditation of Laboratory Animal Care (AAALAC)-accredited animal facility at the NIAID and housed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals. All experiments were performed at the NIAID under an animal study proposal approved by the NIAID Animal Care and Use Committee, in a BSL-2/3 room with necessary precaution for working with Candida auris. Mice were provided with water and a standard laboratory diet ad libitum except if noted otherwise. They were supplied with hardwood chips as bedding and housed in a temperature-controlled, air-conditioned room on a 12-hr light-dark cycle. All mice were co-housed at 5 mice maximum in each cage. Animals were assigned randomly to experimental groups.
Fungi
Stocks of Candida albicans (SC5314) and Candida auris (various clades) were streaked out on Sabouraud dextrose agar with chloramphenicol and incubated at 37°C for at least two days. Single colonies were grown with shaking overnight (220 rpm) at 30°C in Yeast Extract-Peptone-Dextrose (YPD).
METHOD DETAILS
Murine skin colonization model
One or two days before topical association, mouse dorsal hairs were shaved to provide easy access to the back skin. For skin colonization experiments, fungi were cultured, as described above, washed once with PBS, and resuspended at a concentration of 5x109 yeasts/mL of PBS unless otherwise noted. Individual mice were topically associated with 1x109 yeast cells on the shaved dorsal skin and pinna areas using a Puritan cotton swab every other day two (chlorhexidine experiments) or four times (all other experiments).
At defined time points, skin surface was swabbed and on day 14, skin swabs, skin tissue and intestinal content were taken, weighed and processed for culturing as described below. Experiments performed using this two-week model include: comparing C. auris with C. albicans for skin and gut colonization in WT mice; comparing skin colonization between different C. auris clades, i.e., East Asian (CDC # 0381), African (CDC # 0383), South American (CDC # 0385), South Asian (CDC # 0387) and NIH; comparing WT with genetically defined mouse models, including Rag2−/−, Rag2−/−Il2rg−/−, Card9−/−, Il22−/− and Lan-DTA mice.
Quantification of fungal load on mouse skin surface, within skin tissue and intestinal content
Skin surface swabs were taken periodically from dorsal and pinna skin using BD eSwab (moistened in sterile PBS prior to sampling) and placed in transport media. Skin swabs were processed as described in (Welsh et al., 2017) with both direct and enrichment cultures. Briefly, after 10 seconds of vigorous vortexing, 75 μl of the initial transport medium, plus the media on the cotton head, was plated on a CHORMagar Candida plate, and incubated at 37°C for up to one week. Separately, 150 μl of the transport medium was pipetted into a 14 ml round bottom tube with 2.5 ml of enrichment broth (Sabouraud dextrose broth with 10% Sodium Chloride and 50 mg/l chloramphenicol and gentamycin, pH 5.6±0.2). Tubes were incubated with shaking at 37°C for a week, before 100 μl of broth was plated on a CHORMagar Candida plate and incubated overnight at 37°C. For both direct and enrichment cultures, pink colonies, indicating C. auris, were verified with species specific PCR (Figure S1) and counted. To calculate the number of colony forming units (CFUs), colonies from direct plating were utilized. However, if direct plating demonstrated no growth, then the enrichment broth culture plate was examined for possible low level positivity and this number was utilized to calculate CFU.
For investigations of resident skin tissue colonization, ear sheets were separated and dorsal skin slices cut into 2-3 smaller pieces. Tissue was digested in RPMI containing 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 1x MEM nonessential amino acid solution, 20 μM HEPES, and 0.25 mg/ml Liberase TL or 2.5 mg/ml collagenase D by incubating for 1 hour and 45 minutes at 37°C and 5% CO2. Digested skin sheets were homogenized using a Medicon/Medimachine tissue homogenizer system. Liberated cells were suspended in approximately 2.5 ml (for pinna sheets) or 2 ml (for dorsal skin) RPMI media plus 3% fetal bovine serum. 250 μl of the suspension was plated on CHORMagar plates. Limit of detection is 40 CFU/g of skin tissue.
Cecal contents were obtained by making a cut along the cecal pouch and homogenizing contents in 3ml sterile PBS. Contents of small intestine were flushed out with PBS using a syringe and oral gavage tube and homogenized in 3ml sterile PBS. Colonic contents were extracted by pinching the colon with forceps and gently nudging out the contents, which were homogenized in 1.2ml sterile PBS. 250 μl of the suspension was plated on CHORMagar plates. Limit of detection is 20 CFU/g of gastrointestinal content.
PCR confirmation for selected colonies recovered from skin swabs, disaggregated skin tissue and intestinal content
To confirm the identity of the fungi cultured, select pink colonies from CHORMagar Candida plates were stored in YPD medium with 20% glycerol at −80°C. Each colony was tested using the following reaction: 10 μl of 2x Qiagen Hot Start Pre-mix, 1 μl of 10 μM C. auris species specific (Kordalewska et al., 2017) primers [CauF 5’-CGCACATTGCGCCTTGGGGTA-3’ and CauR 5’-GTAGTCCTACCTGATTTGAGGCGAC-3’], 4 μl of thawed out colony stock and 5 μl of PCR water. The PCR profile started with 5 minutes at 95°C, followed by 30 cycles of 30 seconds at 95°C, 30 seconds at 55°C and 1 minute at 72°C, ending with 10 minutes at 72°C. C. auris genomic DNA (gDNA) was included as a positive control and water as the negative control.
Long-term residence within ear skin tissue by C. auris in WT mice
Four cages, each with four WT mice, were set up in parallel and colonized four times on every other day. After all mice turned skin surface negative by swabbing (day 50), one mouse from each cage was sacrificed on a monthly basis (days 58, 98, 120, 153) to disaggregate the ear skin tissue and culture C. auris.
Antibiotic, antifungal treatments
To test whether antibiotic and/or antifungal receipt affected C. auris colonization, WT mice were pre-treated with an antibiotic cocktail [ampicillin (1 mg/ml), metronidazole (1 mg/ml), neomycin (1 mg/ml) and vancomycin (0.5 mg/ml)], fluconazole (0.25 mg/ml), or combination of antibiotics/fluconazole for four weeks. 1x109 C. auris cells were applied onto shaved mouse dorsal skin every other day four times. Only skin topical C. auris levels were tested at the end of the experiment.
In a separate experiment, WT mice were treated with skin-directed antibiotics [tetracycline (0.5 mg/ml), trimethoprim/sulfamethoxazole (0.13 mg/ml trimethoprim and 0.67 mg/ml sulfamethoxazole)] for four weeks prior to colonization with 1x109 C. auris. Skin tissue and small intestinal content were processed for culturing C. auris.
Diabetic, high-fat diet murine models tested
As epidemiological surveys point to diabetes as one potential clinical risk factor, we compared diabetic (db/db) mice with WT mice. Only skin topical C. auris levels were tested at the end of the experiment. We further tested whether high fat diet (HFD) could promote C. auris skin colonization. WT mice were fed with HFD or control diet for two weeks prior to colonization. Dorsal skin was colonized, and dorsal skin tissue and intestinal content processed for culturing.
Intravenous bloodstream infection
Candida albicans strains SC5314 (Lionakis et al., 2013), C. auris East Asian clade (CDC # 0381), C. auris African clade (CDC # 0383), C. auris South American clade (CDC # 0385), C. auris South Asian clade (CDC # 0387) and C. auris NIH strain were used in the present study. Fungi were grown on YPD (Yeast extract/Peptone/Dextrose) agar plates for 48 hours to obtain single colonies. A single colony was then used to inoculate YPD broth containing penicillin/streptomycin and grown for 16-24h at 30°C. The fungal cells were centrifuged, washed once with sterile PBS and reconstituted in PBS for infections. A standard model of invasive candidiasis was utilized (Lionakis et al., 2011; Spellberg et al., 2005), where 105 C. albicans or C. auris blastospores were injected per mouse via lateral tail vein in 200 μl volume of PBS. Survival was monitored for 28 days.
Histological staining of mouse skin tissue
Pinnae were cut with sterile scissors and fixed in 10% formalin. After 24 hours, fixed tissue specimens were washed in 70% ethanol and sent to Histoserv (Germantown, MD) for Grocott-Gomori's methenamine silver (GMS) and Hematoxylin and Eosin (H&E) staining. For the GMS staining, ear tissue specimens were cut parallel to the ear plane to maximize the visualization of fungal cells within the ear tissue. For H&E staining, ear tissue specimens were cut perpendicular to the ear plane.
IL-22 neutralizing antibody treatment in Act1−/− mice
Three treatment groups were established, each with five female mice: Act1−/− mice injected with IL-22 neutralizing antibody, Act1−/− mice injected with isotype and WT C57BL/6NTac mice injected with isotype. Injections started one day before topical association (day −1) and lasted 15 days daily, with each mouse receiving 150 μg of antibody in a total volume of 200 μl in sterile PBS. After dorsal shaving, approximately 1x109 C. auris cells were applied onto mouse dorsal and pinna skin every other day for four times (days 0, 2, 4, 6). Dorsal and pinna skin swabs were taken on days 4, 10 and 14. On day 14, mice were sacrificed, dorsal and ear skin tissue and intestinal content disaggregated, and CFUs from direct plating recorded.
Fluorescence activated cell sorting of immune cells isolated from disaggregated mouse ear skin tissue
Antibody staining was performed on tissue single-cell suspensions for 20-30 minutes at 4°C in Hanks' Balanced Salt Solution staining buffer and then washed twice with cold PBS. The sole exception was CD127, where staining was performed at room temperature for an hour. To measure cytokine production, cells were stimulated in vitro with Phorbol 12-myristate 13-acetate (PMA), ionomycin (Iono), and Brefeldin A (BFA) for 2 hours and 30 minutes. For intracellular staining, cells were fixed for 30 minutes with the eBioscience Fixation/Permeabilization kit as described by the manufacturer and stained intracellularly for at least 1 hour with anti-CD4, anti-CD8, anti-CD90.2, anti-TCRβ, anti-IFN-γ anti-IL-17A and anti-IL17F. Antibodies for flow cytometry were purchased from BD Biosciences, BioLegend, or eBiosciences. Antibodies used were conjugated to FITC, AF488, PE, PerCP-Cy5.5, PCP-eFluor 710, PeCy7, Alexa Fluor 780, Pacific Blue, BV605, BV650, BV 510, BV785, eFluor 450, APC, Alexa Flour 647, PE Texas Red, PE-CF594. DAPI or Live/Dead fixable stain (Life Technologies) was used to exclude dead cells in all experiments. Flow cytometric data was acquired on an LSR II or LSR Fortessa (BD Biosciences) and analyzed using the FlowJo software (Tree Star, Version 9).
Long-term colonization by C. auris in Rag2−/−Il2rg−/− mice
After dorsal shaving, approximately 1x109 C. auris cells were applied onto dorsal and pinna skin four times every other day of Rag2−/−Il2rg−/− and matched control mice. Skin swabs and stool samples were taken during topical association, then weekly for first month, and then monthly for up to 5 months. Experiments were repeated three times, both dorsal and pinna skin were colonized for the first and third experiments, and dorsal skin was colonized for the second experiment. For these three experiments, last skin surface and stool samples were taken on day 206, 183 and 180, and skin tissue and intestinal content processed on day 232, 204 and 208, respectively.
Chlorhexidine treatment
To test its protective effect against C. auris colonization, 2% chlorhexidine impregnated wipes (Sage Products) were applied daily onto mouse dorsal and pinna skin for four days. After C. auris topical colonization, chlorhexidine was applied every other day (Figure 4A). Skin swabs were taken throughout the experiment and skin tissue processed for culturing C. auris.
To test the decolonizing effect of chlorhexidine against C. auris, experiments were performed similar to above, except chlorhexidine impregnated wipes were applied daily onto mouse dorsal and pinna skin for seven days only after topical association (Figure 4D).
Chlorhexidine retention of C. auris gDNA
After dorsal shaving, approximately 5x109/ml C. auris cells were applied onto mouse dorsal and pinna skin every other day for two times. Chlorhexidine was then applied every other day to the dorsal and pinna skin. On days 3, 7, 11 and 15, skin swabs were taken from the left or right ears and left or right side of the dorsal skin for CFU plating or gDNA extraction/qPCR, respectively. DNA was extracted according to established protocols (Findley et al., 2013; Jo et al., 2016). After gDNA extraction, qPCR was performed according to (Leach et al., 2018), with 2.1 μl PCR water, 0.5 μl C. auris qPCR primer (10 μM), 5.0 μl TaqMan Fast Advanced Master Mix, 0.4 μl C. auris TaqMan probe and 2.0 μl gDNA, on a QuantStudio 6 Flex platform. Two technical replicates for each mouse gDNA sample were performed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample Sizes
N represents the number of mice as described in legends of each figure.
Statistical analysis
The Shapiro-Wilk test was first applied to test for normality. If normality was met, t-test or one-way analysis of variance (ANOVA) was used to prepare differences between two or more groups. If normality was not met, the non-parametric Mann-Whitney U test or Kruskal–Wallis one-way ANOVA was used to prepare differences between two or more groups. For parametric ANOVA tests, the Tukey’s honestly significant difference (HSD) test was used for comparing differences between two groups post ANOVA with equal variances, and the Games-Howell post-hoc test used with unequal variances. For non-parametric ANOVA tests, the Dunn’s test was used to compare between two groups post hoc. Statistical analyses were performed using R (v3.6.0) and Prism (v8).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse B220, PE-CF594 (RA3-6B2) | BD Biosciences | Cat # 562290 |
| Anti-mouse CCR6, PE (29-2L17) | Biolegend | Cat # 129804 |
| Anti-mouse CD4, AF700 (RM4-5) | eBioscience | Cat # 56-0042-82 |
| Anti-mouse CD4, BV510 (RM4-5) | BD Biosciences | Cat # 563106 |
| Anti-Mouse CD5, BV510 | Biolegend | Cat # 100627 |
| Anti-mouse CD8β, BV605 (H35-17.2) | BD Biosciences | Cat # 740387 |
| Anti-mouse CD8β, BV650 (H35-17.2) | eBioscience | Cat # 740552 |
| Anti-mouse CD8β, BUV395 (H35-17.2) | BD Biosciences | Cat #740278 |
| Anti-mouse CD11b, BV785 (M1/70) | Biolegend | Cat # 101243 |
| Anti-mouse CD11c, APC-eFluor 780 (N418) | eBioscience | Cat # 47-0114-82 |
| Anti-mouse CD44, AF700 (IM7) | eBioscience | Cat # 56-0441-82 |
| Anti-mouse CD44, PeCy7 (IM7) | eBioscience | Cat # 25-0441-82 |
| Anti-mouse CD45, APC-eFluor 780 (30-F11) | eBioscience | Cat # 47-0451-82 |
| Anti-mouse CD45, BV510 (30-F11) | Biolegend | Cat # 103138 |
| Anti-mouse CD90.2, BV605 (53-2.1) | Biolegend | Cat # 140318 |
| Anti-mouse CD90.2, BV785 (30-H12) | Biolegend | Cat # 105331 |
| Anti-mouse CD103, PerCP-eFluor 710 (2E7) | eBioscience | Cat # 46-1031-82 |
| anti-mouse CD127 (IL-7Rα), BV605 | Biolegend | Cat # 135041 |
| Anti-mouse FoxP3, APC (FJK-16s) | eBioscience | Cat #17-5773-82 |
| Anti-mouse FoxP3, FITC (FJK-16s) | eBioscience | Cat # 11-5773-82 |
| Anti-mouse Ly-6C, BV605 (HK1.4) | Biolegend | Cat # 128036 |
| Anti-mouse Ly-6G, PE-Cy7 (1A8) | BD Biosciences | Cat # 560601 |
| Anti-mouse MHC-II, AF700 (M5/114.15.2) | eBioscience | Cat # 56-5321-82 |
| Anti-mouse NK1.1, PE-CF594 (PK136) | BD Biosciences | Cat # 562864 |
| Anti-mouse RORγt, PE-CF594 (Q31-378) | BD Biosciences | Cat #562684 |
| Anti-mouse TCRβ, PerCP-Cy5.5 (H57-597) | eBioscience | Cat # 45-5961-82 |
| Anti-mouse TCRβ, BUV737 (H57-597) | BD Biosciences | Cat # 612821 |
| Anti-mouse TCRγδ, BV650 (GL3) | BD Biosciences | Cat # 563993 |
| Anti-mouse TCRγδ, PE-CF594 (GL3) | BD Biosciences | Cat # 563532 |
| Anti-mouse IFN-γ, eFluor450 (XMG1.2) | eBioscience | Cat # 48-7311-82 |
| Anti-mouse IFN-γ, Brilliant Violet 605 (XMG1.2) | Biolegend | Cat #505840 |
| Anti-mouse IL-5, APC | Biolegend | Cat # 504304 |
| Anti-mouse IL-13, PerCP-EF710 (eBio13A) | eBioscience | Cat # 46-7133-82 |
| Anti-mouse IL-17A, Pe-Cy7 (TC11-18H10.1) | Biolegend | Cat # 506922 |
| Anti-mouse IL-17F, PE (9D3.1C8) | Biolegend | Cat #517008 |
| IL-22 neutralizing antibody | Genentech (under a material transfer agreement) | Clone 8E11 |
| Mouse IgG1 isotype control | Bio X Cell | Clone MOPC-21 Cat # BE0083 |
| Fungal Strains | ||
| Candida auris, East Asia | CDC | 0381 |
| Candida auris, Africa | CDC | 0383 |
| Candida auris, South America | CDC | 0385 |
| Candida auris, South Asia | CDC | 0387 |
| Candida auris, NIH Clinical Center | NIH | N/A |
| Candida albicans, SC5314 | ATCC | Cat # ATCC MYA-2876 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 0.5 M EDTA pH 8 | Corning | Cat # MT-46034CI |
| Round-Bottom Polypropylene Tubes | Corning | Cat # 352059 |
| Ampicillin | NIH Veterinary Pharmacy | N/A |
| Metronidazole | NIH Veterinary Pharmacy | N/A |
| Neomycin | NIH Veterinary Pharmacy | N/A |
| Vancomycin | NIH Veterinary Pharmacy | N/A |
| Tetracycline | NIH Veterinary Pharmacy | N/A |
| Trimethoprim-Sulfamethoxazole | NIH Veterinary Pharmacy | N/A |
| BD Liquid Amies Elution Swab (Eswab) Collection/Transport System (BD 220245) | Fisher Scientific | Cat # 22-349-700 |
| Chloramphenicol | Sigma | Cat # C0378-5G |
| BD BBL™ Prepared Plated Media: CHROMagar™ Candida (BD 254093) | Fisher Scientific | Cat # B4354093 |
| Brefeldin A (GolgiPlug) | BD Biosciences | Cat #555029 |
| Collagenase D | Sigma Aldrich | Cat # 11088866001 |
| Control diet | Envigo | Cat # TD.08806 |
| Deoxyribonuclease I from bovine pancreas | Sigma Aldrich | Cat # DN25-5G |
| Ethyl Alcohol 200 Proof | Pharmco-Aaper | Cat # 111000200 |
| Fetal bovine serum | HyClone | Cat # C838R85 |
| Filcon, Sterile, Syringe-Type, 50 μm | BD Bioscience | Cat # 340601 |
| Fluconazole, ≥98% (HPLC), powder | Sigma Aldrich | Cat # F8929 |
| Formalin Solution, Neutral Buffered, 10% | Sigma Aldrich | Cat # HT501128-4L |
| Gentamycin | Fisher Scientific | Cat # 1405-41-0 |
| HEPES, Liquid | Corning | Cat # MT25060CI |
| High fat diet | Envigo | Cat # TD.06414 |
| Ionomycin | Sigma-Aldrich | Cat #I0634-5MG |
| Liberase™ TL Research Grade | Sigma Aldrich | Cat # 5401020001 |
| 50 μm Sterile Medicon | BD Bioscience | Cat # 340591 |
| MasterPure™ Yeast DNA Purification Kit | Lucigen | Cat # MPY80200 |
| MEM Nonessential Amino Acid Solution | Corning | Cat # MT25025CI |
| Monoject 20 mL Syringe | Covidien | Cat # 1182000777 |
| PCR water | Qiagen | Cat # 17000-10 |
| Penicillin and streptomycin, 100x | Corning | Cat # MT30002CI |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat #P8139-10MG |
| Puritan 6" Sterile Standard Foam Swab w/Polystyrene Handle | Harmony Lab & Safety Supplies | Cat # P25-1506PF-case |
| HotStarTaq Master Mix Kit | Qiagen | Cat # 203445 |
| RPMI 1640 medium 1X with L-Glutamine | Corning | Cat # MT10040CV |
| Sabouraud Dextrose Agar, Emmons w/Chloramphenicol | Thermo Scientific | Cat #R01770 |
| Sage 2% Chlorhexidine Gluconate (CHG) Cloths | Medline Industries | Cat # SGE9705 |
| Sabouraud Dextrose Broth | Fisher Scientific | Cat # DF0382-17-9 |
| Sodium Chloride | J. T. Baker | Cat # 3624-01 |
| Sodium Pyruvate 100 mM Solution | Corning | Cat # MT25000CI |
| TaqMan Fast Advanced Master Mix | Thermo Scientific | Cat # 4444557 |
| Yeast Extract-Peptone-Dextrose (YPD) Broth | BD Bioscience | Cat # 242820 |
| eBioscience Fixation/Permeabilization Concentrate | eBioscience | Cat #00-5123-43 |
| eBioscience Fixation/Permeabilization Diluent | eBioscience | Cat #00-5223-56 |
| eBioscience Permeabilization Buffer (10X) | eBioscience | Cat #00-8333-56 |
| Experimental Models: Organisms/Strains | ||
| Diabetic mice (db/db) | NIAID Taconic Contract | N/A |
| Rag2−/−Il2rg−/− | NIAID Taconic Contract | Line 111 |
| Rag2−/− | NIAID Taconic Contract | Line 103 |
| Il22−/− | Dr. Yasmine Belkaid | N/A |
| Card9−/− | Dr. Michail S. Lionakis | N/A |
| Act1−/− | NIAID Taconic Contract | Line 290 |
| Lan-DTA | Dr. Yasmine Belkaid | N/A |
| C57BL/6 mice | Taconic Biosciences | Cat # B6-F/B6-M |
| C57BL/10 mice | NIAID Taconic Contract | Line 8411 |
| Oligonucleotides | ||
| CauF 5’-CGCACATTGCGCCTTGGGGTA-3’ | Thermo Scientific | N/A |
| CauR 5’-GTAGTCCTACCTGATTTGAGGCGAC-3’ | Thermo Scientific | N/A |
| Software and Algorithms | ||
| FlowJo v9 | Tree Star | https://www.flowjo.com/solutions/flowjo/downloads |
| GraphPad Prism 4 | GraphPad Software | https://www.graphpad.com/scientificsoftware/prism/ |
| R v3.6.0 | The R Foundation | https://www.r-project.org/ |
| Other | ||
| Medimachine | BD Bioscience | Cat # 340588 |
| MediMachine II | Syntec International | Cat # 121600 |
| LSRFortessa | BD Biosciences | https://www.bd.com/en-us/offerings/brands/lsrfortessa |
| LSR II | BD Biosciences | https://www.bdbiosciences.com/en-us/go-campaign/lsr-ii-comp-cont |
| QuantStudio 6 Flex | Fisher Scientific | https://www.fishersci.com/shop/products/6-flx96wfast-instlptp-1-system/4485699 |
| GMS staining of mouse ear skin sections | Histoserv | N/A |
Highlights.
Fungal pathogen Candida auris colonizes mouse skin surface and tissue.
C. auris clades, of distinct geographic regions, differentially colonize mouse skin.
IL-17 receptor signaling protects mice from long-term C. auris colonization.
Chlorhexidine antiseptic protects against colonization of C. auris on mouse skin.
Acknowledgments
Research was funded by the Division of Intramural Research of the NHGRI and NIAID, NIH. The authors would like to thank Qiong Chen, ShihQueen Lee-Lin and Michael Abers for technical assistance with experiments, as well as Stephen Wincovitch for digitalization and visualization of histological slides. Darryl Leja crafted the Graphical Abstract. Helpful discussions and comments on the manuscript were provided by Ana Litvintseva, Tara Palmore, Heidi Kong and members of the Segre and Lionakis labs.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams E, Quinn M, Tsay S, Poirot E, Chaturvedi S, Southwick K, Greenko J, Fernandez R, Kallen A, Vallabhaneni S, et al. (2018). Candida auris in Healthcare Facilities, New York, USA, 2013-2017. Emerg Infect Dis. 24(10), 1816–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin S, Maor Y, Tarabia J, Schechner V, Adler A, et al. (2017). Multidrug-Resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis. 23(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishu S, Hernández-Santos N, Simpson-Abelson MR, Huppler AR, Conti HR, Ghilardi N, Mamo AJ and Gaffen SL (2014). The adaptor CARD9 is required for adaptive but not innate immunity to oral mucosal Candida albicans infections. Infect. Immun. 82(3), 1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B, Wang C, Pedergnana V, Wu L, Cypowyj S, Rybojad M, Belkadi A, Picard C, Abel L, Fieschi C, et al. (2013). An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity. 39(4), 676–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassir N, Papazian L, Fournier PE, Raoult D, and La Scola B (2015). Insights into bacterial colonization of intensive care patients' skin: the effect of chlorhexidine daily bathing. Eur J Clin Microbiol. 34(5), 999–1004. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States, 2019. Atlanta: CDC; https://www.cdc.gov/drugresistance/pdf/threats-report/2019-arthreats-report-508.pdf. [Google Scholar]
- Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, and Meis JF (2019). Potential Fifth Clade of Candida auris, Iran, 2018. Emerging Infectious Diseases. 25(9), 1780–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, Barrett PM, Kerins JL, Lockhart SR, Chiller TM, et al. (2018). Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis. 18(12), 1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow NA, Muñoz JF, Gade L, Berkow E, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, et al. (2020). Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. bioRxiv. 2020.2001.2006.896548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti HR, Bruno VM, Childs EE, Daugherty S, Hunter JP, Mengesha BG, Saevig DL, Hendricks MR, Coleman BM, Brane L, et al. (2016). IL-17 Receptor Signaling in Oral Epithelial Cells Is Critical for Protection against Oropharyngeal Candidiasis. Cell Host Microbe. 20(5), 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, Griffiths D, George S, Butcher L, Morgan M, et al. (2018). A Candida auris Outbreak and Its Control in an Intensive Care Setting. N Engl J Med. 379(14), 1322–1331. [DOI] [PubMed] [Google Scholar]
- Fakhim H, Vaezi A, Dannaoui E, Chowdhary A, Nasiry D, Faeli L, Meis JF, and Badali H (2018). Comparative virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a murine model. Mycoses. 61(6), 377–382. [DOI] [PubMed] [Google Scholar]
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Program NIHISCCS, et al. (2013). Topographic diversity of fungal and bacterial communities in human skin. Nature. 498(7454), 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg K, Woodworth K, Walters M, Berkow EL, Jackson B, Chiller T, and Vallabhaneni S (2019). Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med Mycol. 57(1), 1–12. [DOI] [PubMed] [Google Scholar]
- Frieder J, Kivelevitch D, Haugh I, Watson I, and Menter A (2018). Anti-IL-23 and Anti-IL-17 Biologic Agents for the T reatment of Immune-Mediated Inflammatory Conditions. Clin Pharmacol Ther. 103(1), 88–101. [DOI] [PubMed] [Google Scholar]
- Grice EA, Snitkin ES, Yockey LJ, Bermudez DM, Program NCS, Liechty KW, and Segre JA (2010). Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci U S A. 107(33), 14799–14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggino G, Giardina A, Ferrante A, Giardina G, Schinocca C, Sireci G, Dieli F, Ciccia F, and Triolo G (2015). The in vitro addition of methotrexate and/or methylprednisolone determines peripheral reduction in Th17 and expansion of conventional Treg and of IL-10 producing Th17 lymphocytes in patients with early rheumatoid arthritis. Rheumatol Int. 35(1), 171–175. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Linehan JL, Shih HY, Bouladoux N, Han SJ, Smelkinson M, Sen SK, Byrd AL, Enamorado M, Yao C, et al. (2019). Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science. 363(6422), eaat6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, Lankiewicz J, Gombosev A, Terpstra L, Hartford F, et al. (2013). Targeted versus universal decolonization to prevent ICU infection. N Engl J Med. 368(24), 2255–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Heim L, Gombosev A, Avery TR, Haffenreffer K, Shimelman L, et al. (2019a). Chlorhexidine versus routine bathing to prevent multidrug-resistant organisms and all-cause bloodstream infections in general medical and surgical units (ABATE Infection trial): a cluster-randomised trial. Lancet. 393(10177), 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SS, Singh R, McKinnell JA, Park S, Gombosev A, Eells SJ, Gillen DL, Kim D, Rashid S, Macias-Gil R, et al. (2019b). Decolonization to Reduce Postdischarge Infection Risk among MRSA Carriers. New Engl J Med. 380(7), 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igyarto BZ, Haley K, Ortner D, Bobr A, Gerami-Nejad M, Edelson BT, Zurawski SM, Malissen B, Zurawski G, Berman J, et al. (2011). Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity. 35(2), 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BR, Chow N, Forsberg K, Litvintseva AP, Lockhart SR, Welsh R, Vallabhaneni S, and Chiller T (2019). On the Origins of a Species: What Might Explain the Rise of Candida auris? J Fungi. 5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo JH, Deming C, Kennedy EA, Conlan S, Polley EC, Ng WI, Program NCS, Segre JA, and Kong HH (2016). Diverse Human Skin Fungal Communities in Children Converge in Adulthood. J Invest Dermatol. 136(12), 2356–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G (2016). Acquired resistance to chlorhexidine - is it time to establish an 'antiseptic stewardship' initiative? J Hosp Infect. 94(3), 213–227. [DOI] [PubMed] [Google Scholar]
- Kashem SW, Igyarto BZ, Gerami-Nejad M, Kumamoto Y, Mohammed JA, Jarrett E, Drummond RA, Zurawski SM, Zurawski G, Berman J, et al. (2015). Candida albicans morphology and dendritic cell subsets determine T helper cell differentiation. Immunity. 42(2), 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashem SW, and Kaplan DH (2016). Skin immunity to Candida albicans. Trends Immunol. 37(7), 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashem SW, Riedl MS, Yao C, Honda CN, Vulchanova L and Kaplan DH (2015). Nociceptive sensory fibers drive interleukin-23 production from CD301b+ dermal dendritic cells and drive protective cutaneous immunity. Immunity. 43(3), 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kates AE, Zimbric ML, Mitchell K, Skarlupka J, and Safdar N (2019). The impact of chlorhexidine gluconate on the skin microbiota of children and adults: A pilot study. Am J Infect Control. 47(8), 1014–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z, Ahmad S, Benwan K, Purohit P, Al-Obaid I, Bafna R, Emara M, Mokaddas E, Abdullah AA, Al-Obaid K, et al. (2018). Invasive Candida auris infections in Kuwait hospitals: epidemiology, antifungal treatment and outcome. Infection. 46(5), 641–650. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, Kong HH, Amagai M, and Nagao K (2015). Dysbiosis and Staphylococcus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity. 42(4), 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, and Perlin DS (2017). Rapid and Accurate Molecular Identification of the Emerging Multidrug-Resistant Pathogen Candida auris. J Clin Microbiol. 55(8), 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin E, Hager C, Chandra J, Mukherjee PK, Retuerto M, Salem I, Long L, Isham N, Kovanda L, Borroto-Esoda K, et al. (2017). The Emerging Pathogen Candida auris: Growth Phenotype, Virulence Factors, Activity of Antifungals, and Effect of SCY-078, a Novel Glucan Synthesis Inhibitor, on Growth Morphology and Biofilm Formation. Antimicrobial Agents and Chemotherapy. 61(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach L, Zhu Y, and Chaturvedi S (2018). Development and Validation of a Real-Time PCR Assay for Rapid Detection of Candida auris from Surveillance Samples. J Clin Microbiol. 56(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, Schweighoffer E, Tybulewicz V, Brown GD, Ruland J, et al. (2007). Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 8(6), 630–638. [DOI] [PubMed] [Google Scholar]
- Lionakis MS, and Hohl TM (2020). Call to Action: How to Tackle Emerging Nosocomial Fungal Infections. Cell Host Microbe. 27(6), 859–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, and Levitz SM (2018). Host Control of Fungal Infections: Lessons from Basic Studies and Human Cohorts. Annu Rev Immunol. 36, 157–191. [DOI] [PubMed] [Google Scholar]
- Lionakis MS, Lim JK, Lee CC, and Murphy PM (2011). Organ-specific innate immune responses in a mouse model of invasive candidiasis. J Innate Immun. 3(2), 180–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, and Netea MG (2013). Candida and host determinants of susceptibility to invasive candidiasis. Plos Pathog. 9(1), e1003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, Swamydas M, Fischer BG, Plantinga TS, Johnson MD, Jaeger M, Green NM, Masedunskas A, Weigert R, Mikelis C, et al. (2013). CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J Clin Invest. 123(12), 5035–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, et al. (2017). Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin Infect Dis. 64(2), 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marakalala MJ, Vautier S, Potrykus J, Walker LA, Shepardson KM, Hopke A, Mora-Montes HM, Kerrigan A, Netea MG, Murray GI, et al. (2013). Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. Plos Pathog. 9(4), e1003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, et al. (2015). Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 520(7545), 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. (2012). Compartmentalized control of skin immunity by resident commensals. Science. 337(6098), 1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowsky B, Greenko J, Adams E, Quinn M, O'Brien B, Chaturvedi V, Berkow E, Vallabhaneni S, Forsberg K, Chaturvedi S, et al. (2020). Candida auris Isolates Resistant to Three Classes of Antifungal Medications - New York, 2019. MMWR Morb Mortal Wkly Rep. 69(1), 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, and Kullberg BJ (2018). Invasive candidiasis. Nat Rev Dis Primers. 4, 18026. [DOI] [PubMed] [Google Scholar]
- Parra-Giraldo CM, Valderrama SL, Cortes-Fraile G, Garzon JR, Ariza BE, Morio F, Linares-Linares MY, Ceballos-Garzon A, de la Hoz A, Hernandez C, et al. (2018). First report of sporadic cases of Candida auris in Colombia. Int J Infect Dis. 69, 63–67. [DOI] [PubMed] [Google Scholar]
- Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, Migaud M, Israel L, Chrabieh M, Audry M, et al. (2011). Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 332(6025), 65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Bouladoux N, Claesen J, Chen YE, Byrd AL, Constantinides MG, Merrill ED, Tamoutounour S, Fischbach MA, and Belkaid Y (2018). Contextual control of skin immunity and inflammation by Corynebacterium. J Exp Med. 215(3), 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gaitan A, Moret AM, Tasias-Pitarch M, Aleixandre-Lopez AI, Martinez-Morel H, Calabuig E, Salavert-Lleti M, Ramirez P, Lopez-Hontangas JL, Hagen F, et al. (2018). An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 61(7), 498–505. [DOI] [PubMed] [Google Scholar]
- Rutala WA, Kanamori H, Gergen MF, Sickbert-Bennett EE, and Weber DJ (2019). Susceptibility of Candida auris and Candida albicans to 21 germicides used in healthcare facilities. Infect Cont Hosp Ep. 40(3), 380–382. [DOI] [PubMed] [Google Scholar]
- SanMiguel AJ, Meisel JS, Horwinski J, Zheng Q, Bradley CW, and Grice EA (2018). Antiseptic Agents Elicit Short-Term, Personalized, and Body Site-Specific Shifts in Resident Skin Bacterial Communities. J Invest Dermatol. 138(10), 2234–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, and Yamaguchi H (2009). Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 53(1), 41–44. [DOI] [PubMed] [Google Scholar]
- Saunte DM, Mrowietz U, Puig L, and Zachariae C (2017). Candida infections in patients with psoriasis and psoriatic arthritis treated with interleukin-17 inhibitors and their practical management. Brit J Dermatol. 177(1), 47–62. [DOI] [PubMed] [Google Scholar]
- Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF, et al. (2016). First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Septimus EJ, and Schweizer ML (2016). Decolonization in Prevention of Health Care-Associated Infections. Clin Microbiol Rev. 29(2), 201–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Uppuluri P, Mamouei Z, Alqarihi A, Elhassan H, French S, Lockhart SR, Chiller T, Edwards JE, and Ibrahim AS (2019). The NDV-3A vaccine protects mice from multidrug resistant Candida auris infection. Plos Pathog. 15(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonder SU, Saret S, Tang W, Sturdevant DE, Porcella SF, and Siebenlist U (2011). IL-17-induced NF-kappaB activation via CIKS/Act1: physiologic significance and signaling mechanisms. J Biol Chem. 286(15), 12881–12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparber F, De Gregorio C, Steckholzer S, Ferreira FM, Dolowschiak T, Ruchti F, Kirchner FR, Mertens S, Prinz I, Joller N, et al. (2019). The skin commensal yeast Malassezia triggers a type 17 response that coordinates anti-fungal immunity and exacerbates skin inflammation. Cell Host Microbe. 25(3), 389–403. [DOI] [PubMed] [Google Scholar]
- Spellberg B, Ibrahim AS, Edwards JE Jr., and Filler SG (2005). Mice with disseminated candidiasis die of progressive sepsis. J Infect Dis. 192(2), 336–343. [DOI] [PubMed] [Google Scholar]
- Torres SR, Pichowicz A, Torres-Velez F, Song R, Singh N, Lasek-Nesselquist E, and De Jesus M (2019). Impact of Candida auris infection in a neutropenic murine model. Antimicrobial Agents and Chemotherapy. AAC.01625–01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni S, Jackson BR, and Chiller TM (2019). Candida auris: An Emerging Antimicrobial Resistance Threat. Ann Intern Med. 171(6), 432–433. [DOI] [PubMed] [Google Scholar]
- Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, et al. (2016). Investigation of the First Seven Reported Cases of Candida auris, a Globally Emerging Invasive, Multidrug-Resistant Fungus - United States, May 2013-August 2016. MMWR Morb Mortal Wkly Rep. 65(44), 1234–1237. [DOI] [PubMed] [Google Scholar]
- Wang X, Bing J, Zheng Q, Zhang F, Liu J, Yue H, Tao L, Du H, Wang Y, Wang H, et al. (2018). The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infect. 7(1), 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, and Litvintseva AP (2017). Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J Clin Microbiol. 55(10), 2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurster S, Bandi A, Beyda ND, Albert ND, Raman NM, Raad II, and Kontoyiannis DP (2019). Drosophila melanogaster as a model to study virulence and azole treatment of the emerging pathogen Candida auris. J Antimicrob Chemoth. 74(7), 1904–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Mohiuddin F, Tran J, Adams A, and Eberle K (2019). Experimental Mouse Models of Disseminated Candida auris Infection. Msphere. 4(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, O'Brien B, Leach L, Clark A, Bates M, Adams E, Ostrowsky B, Quinn M, Dufort E, Southwick K, et al. (2019). Laboratory Analysis of an Outbreak of Candida auris in New York from 2016 to 2018-Impact and Lessons Learned. J Clin Microbiol. 58(4), e01503–01519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study . This study did not generate any unique datasets or code