Abstract
Physical exercise can produce changes in the microbiota, conferring health benefits through mechanisms that are not fully understood. We sought to determine the changes driven by exercise on the gut microbiota and on the serum and fecal metabolome using 16S rRNA gene analysis and untargeted metabolomics. A total of 85 serum and 12 fecal metabolites and six bacterial taxa (Romboutsia, Escherichia coli TOP498, Ruminococcaceae UCG-005, Blautia, Ruminiclostridium 9 and Clostridium phoceensis) were modified following a controlled acute exercise session. Among the bacterial taxa, Ruminiclostridium 9 was the most influenced by fecal and serum metabolites, as revealed by linear multivariate regression analysis. Exercise significantly increased the fecal ammonia content. Functional analysis revealed that alanine, aspartate and glutamate metabolism and the arginine and aminoacyl-tRNA biosynthesis pathways were the most relevant modified pathways in serum, whereas the phenylalanine, tyrosine and tryptophan biosynthesis pathway was the most relevant pathway modified in feces. Correlation analysis between fecal and serum metabolites suggested an exchange of metabolites between both compartments. Thus, the performance of a single exercise bout in cross-country non-professional athletes produces significant changes in the microbiota and in the serum and fecal metabolome, which may have health implications.
Subject terms: Microbiology, Physiology
Introduction
Physical exercise has numerous beneficial effects on health1. One of the mechanisms by which exercise can exert these effects is through changes to the gut microbiota2. While a clear effect of exercise on the gut microbiota has been demonstrated in animal models3–5, there is less robust evidence from human studies likely because the performance of physical exercise is associated with different feeding behaviors and with a healthier diet, which is a confounding factor when assessing the response to exercise6,7. Exercise is associated with an increase in the diversity of gut microbiota, including an increment in the number of health-promoting bacteria that produce short-chain fatty acids8,9. Indeed, acute physical exercise induces a series of metabolic changes both systemically and locally in specific tissues that are characterized by marked shifts in metabolism and oxygen consumption10, but its effects on the gut microbiota has not been studied in any great detail.
Advances in metabolomics have provided new opportunities for enriching our understanding of physical exercise-associated factors that stimulate metabolic responses11,12. In this line, a recent study by Zhao et al. found that the composition and functionality of the gut microbiota, as measured by fecal metabolomics, changed significantly in amateur runners after endurance exercise (completing a half marathon)13. The effect of an acute bout of exercise on the gut microbiota and its interaction with the serum and fecal metabolome has been scarcely investigated. We hypothesized that an acute exercise bout would impact on the serum and fecal metabolome and alter the gut microbiota. Here, using a combinatorial approach, we investigated serum and fecal metabolites and the microbiome in non-professional (cross-country) athletes before and after a session of moderate-intensity exercise to volitional exhaustion using untargeted metabolomics and 16S rRNA sequencing analysis, respectively. We also examined the potential associations between serum and fecal metabolites and the gut microbiota induced by exercise. This study is the first to our knowledge to reveal simultaneous changes in serum/fecal metabolic signatures and gut microbiota after a single acute bout of exercise.
Results
Characteristics of the study population, dietary habits and exercise performance
In total, 40 male endurance cross-country runners completed the study. Participants’ characteristics including age, weight, body mass index (BMI), dietary habits and sports performance data obtained in the exercise session are described in Table 1. The exercise session consisted of a treadmill test and running 1 km at maximum speed (see “Material and methods”
Table 1.
Participant characteristics.
| Age (y) | 35.79 ± 8.01 |
| Weight (kg) | 71.11 ± 8.24 |
| Height (cm) | 176.69 ± 6.01 |
| BMI (kg/m2) | 22.75 ± 2.12 |
| VO2maxREL (mL/kg/min) | 58.80 ± 3.24 |
| VT1 (km/h) | 13.28 ± 0.95 |
| VT2 (km/h) | 15.76 ± 1.01 |
| MAS (km/h) | 17.88 ± 1.25 |
| t1km (min) | 3.22 ± 0.26 |
| Energy (kcal) | 2229.32 ± 1118.74 |
| Carbohydrates (% of energy) | 45.41 ± 7.08 |
| Proteins (% of energy) | 19.18 ± 3.48 |
| Lipids (% of energy) | 35.17 ± 6.85 |
VO2maxREL relative maximum oxygen consumption, MAS maximal aerobic speed, VT1 first ventilatory threshold, VT2 second ventilator threshold, t1km time to run 1 km. Values are the mean ± standard deviation.
Fecal pH and ammonia
Fecal pH and ammonia were determined in samples collected before and after the exercise session. No changes in pH (pre = 7.59 ± 0.48 post = 7.54 ± 0.39; p = 0.337) were detected after the exercise session, but the concentration of fecal ammonia was significantly greater post-exercise (pre = 13.46 ± 7.25 mmol/L; post = 16.08 ± 8.37 mmol/L; p < 0.023).
Metabolomic serum profile
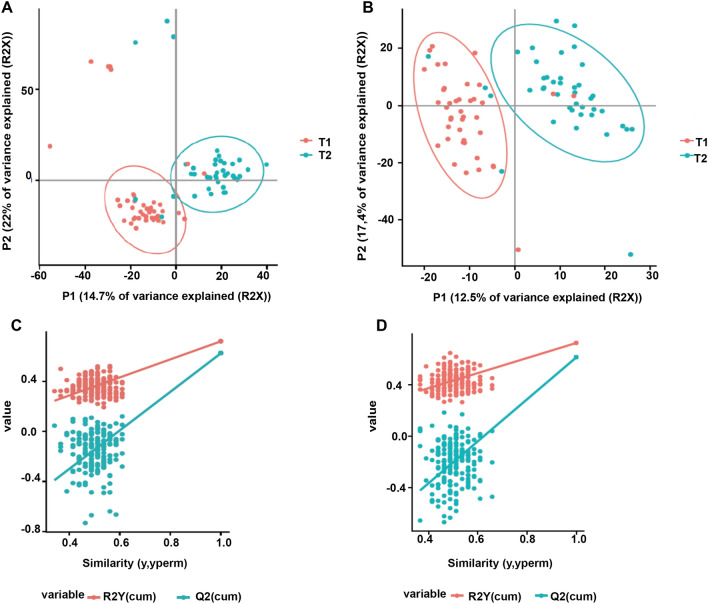
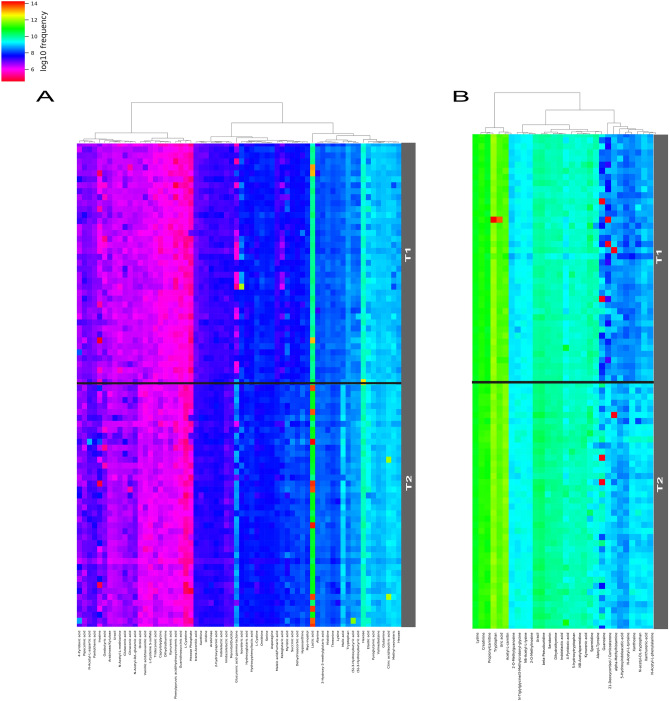
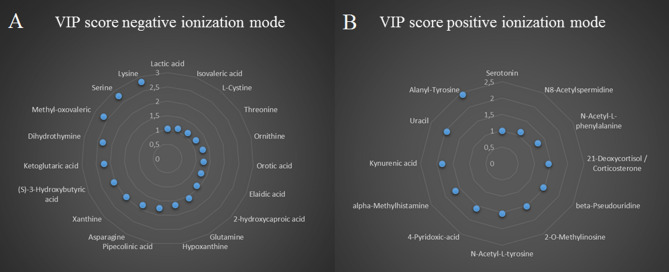
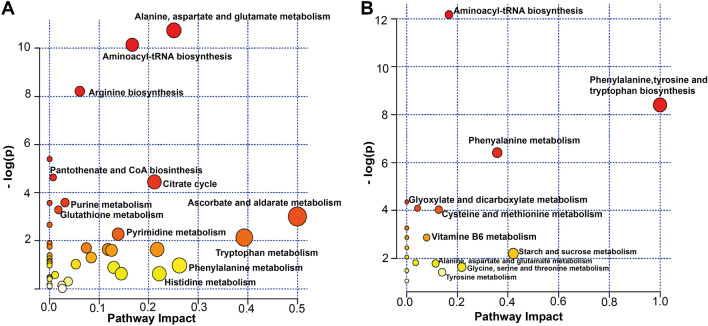
Metabolomics analyses were performed using liquid chromatography coupled to high-resolution mass spectrometry (LC-HRMS) with a combination of two complementary chromatographic methods: reversed-phase chromatography (C18 chromatographic column [C18(+)] and Hydrophilic Interaction Liquid Chromatography [HILIC(−)], for the analysis of hydrophobic and polar metabolites in the positive and the negative ionization modes, respectively). Using an untargeted approach, we detected a total of 3195 and 1600 metabolite features using C18(+) and HILIC(−) conditions, respectively. Among those metabolite features, 101 from the C18(+) and 159 from the HILIC(−) analysis accurately matched the mass and retention time of metabolites included in our chemical database (30 of which were common in both analyses). Differences between the two sampling times were investigated further using supervised partial least square discriminant analysis (PLS-DA). As shown in Fig. 1A,B, athletes’ samples before a session of acute exercise (T1) could be distinguished from those after a session of acute exercise (T2). The cross-validation parameters R2Y and Q2 indicated the variance and the predictive ability of the model. Permutation tests (200 times) were conducted to assess the robustness of the PLS-DA model when using a small sample size (Fig. 1C,D). Whole serum metabolomic profiles from both the negative and positive ionization modes following hierarchical clustering analysis are shown in Fig. 2A,B. Discriminant annotated metabolites between groups were ranked according to their variable importance in the projection (VIP) score, yielding a total of 31 features with a VIP score of > 1, including 13 amino acids, 4 lipids, 5 organic acids, 3 aromatic heterocyclic compounds, 1 aliphatic heterocycle compound, 3 aromatic heteropolycyclic compounds, and 2 nucleosides (Fig. 3A,B). Complementary to the panel of metabolites identified from the multivariate model, univariate analysis was applied using pairwise comparisons (T1 vs T2) of individual metabolites (Wilcoxon p-values with Benjamini–Hochberg correction). Under these conditions, up to 85 annotated metabolites were significantly different and were subjected to additional tandem mass spectrometry (MS/MS) experiments for identity confirmation (Supplementary Table S1). The 85 metabolites were then analyzed by MetaboAnalyst (with the KEGG database14) to identify potential discriminately perturbed metabolic pathways before and after the session of acute exercise (T1 vs T2). Results showed that alanine, aspartate and glutamate metabolism, aminoacyl-tRNA biosynthesis and arginine biosynthesis were the three most significantly modified pathways (Fig. 4A, Supplementary Table S3).
Figure 1.
Partial least square discriminant analysis (PLS-DA) of the metabolites in T1 (before exercise) and T2 (after exercise). The results are presented as principal component score plots, with each point in the plot representing an individual sample. (A,B) PLS-DA score plots obtained from LC-HRMS data in positive mode C18 and negative mode HILIC. (C,D) Statistical validation of the PLS-DA model (A,B), showing R2Y (pink dots) and the Q2 (light-blue dots) values from the permutated analysis (bottom left) lower than the corresponding original values (top right).
Figure 2.
Hierarchical clustering heatmaps representing annotated serum metabolites significantly (p < 0.05) different between T1 and T2. (A) Metabolites obtained from HILIC(−) LC-HRMS analysis. (B) Metabolites obtained from C18(+) LC-HRMS analysis.
Figure 3.
Variable important in projection (VIP) score from PLS-DA of serum metabolites. (A) VIP score from serum metabolites obtained in positive ionization mode. (B) VIP score of serum metabolites obtained in negative ionization mode.
Figure 4.
Summary of metabolic pathway enrichment analysis performed in MetaboAnalyst 4.0 using a panel of 85 metabolites found to be significantly altered by a session of acute exercise. All the matched pathways are displayed as circles. The node color is based on its p-value and the node radius is determined based on its pathway impact values. (A) Serum pathways modified by a session of acute exercise. (B) Fecal pathways modified by a session of acute exercise.
Metabolomic fecal profile
A similar experimental set-up was used for the fecal analysis. A total of 10,866 and 6795 metabolite features were detected under C18(+) and HILIC(−) LC-HRMS conditions, respectively. Firstly, lists of 176 and 335 metabolite features were obtained following annotation with our chemical database and using the C18(+) and HILIC(−) datasets, respectively (88 of which were common). The score plot of the principal component analysis (PCA) and PLS-DA did not reveal any distinction, showing that the two groups did not cluster separately (data not shown). However, 50 annotated metabolites were significantly different when comparing T1 and T2 groups using univariate statistical analysis, and were further confirmed by MS/MS analysis (Supplementary Table S2). Further correction of the p-values for multiple hypotheses using the Benjamini–Hochberg method narrowed down this list to 12 relevant metabolites (Table 2). Additional information for each metabolite is shown in the supplementary material (Supplementary Fig. S1). Metabolic pathway analysis showed that the top 3 affected metabolic pathways after the acute exercise session were the aminoacyl-tRNA biosynthesis, phenylalanine, tyrosine and tryptophan biosynthesis, and the phenylalanine pathway (Fig. 4B, Supplementary Table S4).
Table 2.
Annotated and top significant fecal metabolites.
| LC-HRMS conditions | Metabolite | p-corrected value | T1/T2 |
|---|---|---|---|
| C18(+) | Tryptophan | 0.0212 | 0.58 |
| C18(+) | Methionine | 0.0132 | 0.55 |
| C18(+) | S-ethyl-l-cysteine | 0.0143 | 0.57 |
| C18(+) | Phenol | 0.0145 | 0.63 |
| C18(+) | 4-Hydroxybenzyl-alcohol | 0.0234 | 0.64 |
| HILIC(−) | l/d-Glutamine | 0.0001 | 0.53 |
| HILIC(−) | Serine | 0.0212 | 0.60 |
| HILIC(−) | Phenylalanine | 0.0218 | 0.55 |
| HILIC(−) | Tyrosine | 0.0224 | 0.60 |
| HILIC(−) | Hexose alcohols | 0.0224 | 0.57 |
| HILIC(−) | Pyridoxamine | 0.0426 | 1.66 |
| HILIC(−) | Cinnamic acid | 0.0433 | 0.53 |
Changes originating in gut microbiota
Analysis of the gut microbiota before and after the acute exercise session revealed no difference in the composition of the bacterial community, and no changes were observed in any of the determined parameters of β-diversity (Bray–Curtis distance, Jaccard index and weighted and unweighted Unifrac; data not shown). Regarding α-diversity, no changes were observed in any of the parameters analyzed (observed operational taxonomic units, evenness, Shannon index and Faith's index) (Supplementary Fig. S2). To evaluate changes in bacterial taxa, we use an exploratory analysis of the bacterial taxa volatility—an approach that uses machine-learning regressors to establish the important bacterial taxa that predict the T2. The accuracy obtained for our model was significant (Mean squared error = 0.3194; R2 = 0.392; p = 0.0071). The bacterial taxa and those with the higher cumulative average change between T1 and T2 identified by the volatility plot were used to test whether the relative abundances of these features were impacted by exercise using a linear mixed effects analysis, in which time was included as a forced predictor (fixed effect) and subject identifier as a random effect. Six bacterial taxa were identified as significantly differentially abundant between T1 and T2 (Table 3). The exercise bout increased the abundance of Romboutsia genus, Ruminococcocaceae UCG-005, E. coli TOP498 and Blautia genus, and decreased the abundance of Ruminiclostridium 9 and Clostridium phoceensis.
Table 3.
Linear mixed-effects model results for bacterial taxa abundances modified by an acute session of physical exercise.
| Bacterial taxa | Estimate | SE | Z-score | P-value | Identity Confidence |
|---|---|---|---|---|---|
| D_5__Romboutsia;D_6__uncultured bacterium | 0.001 | 0.00 | 2.390 | 0.017 | 0.98558 |
| D_5__Escherichia-Shigella;D_6__Escherichia coli TOP498 | 0.001 | 0.001 | 2.534 | 0.011 | 0.99624 |
| D_5__Blautia;D_6__human gut metagenome | 0.000 | 0.000 | 3.107 | 0.002 | 0.96172 |
| D_5__Ruminococcaceae UCG-005;D_6__uncultured organism | 0.001 | 0.000 | 2.063 | 0.039 | 0.99940 |
| D_5__Ruminiclostridium9;D_6__uncultured Clostridia bacterium | − 0.000 | 0.000 | − 2.607 | 0.009 | 0.99772 |
| D_5__uncultured;D_6__Clostridium phoceensis | − 0.001 | 0.000 | − 2.413 | 0.016 | 0.99705 |
Parameter estimate (coefficient), standard error, Z-score, and P-value for each bacterial taxa. A positive coefficient indicates a higher relative abundance in T2.
Microbiota and metabolomic associations
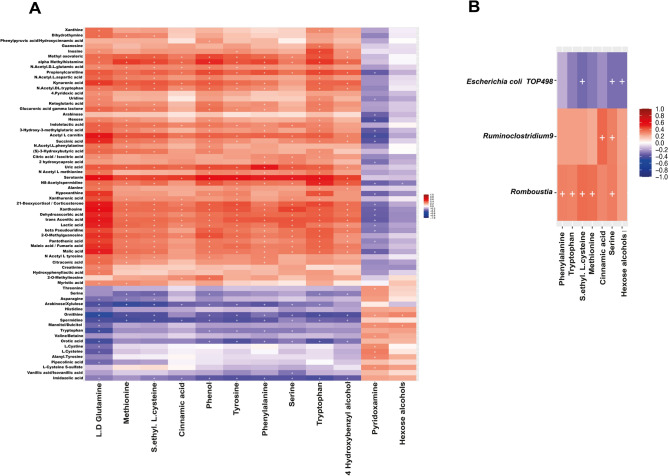
To examine for correlations between the metabolites detected in serum and feces and the changes in the gut microbiota, we performed several correlation analyses of the differentially detected metabolites and the 6 bacterial taxa that varied in the gut microbiota. There was a high number of plasma and feces metabolites that were significantly associated (Fig. 5A); however, none of the associations was strong (with ρ coefficient above 0.7). The fecal metabolites glutamine and tryptophan showed the highest number of moderate associations with serum metabolites (Fig. 5A). Some of the moderate positive associations were found between the fecal metabolite glutamine and serum xanthosine, hypoxanthine and deoxyadenosine, serotonin, acetyl-carnitine, 21-deoxycortisol, kynurenic acid, lactic, malic and succinic acid and pantothenic-acid (Fig. 5A). Serum phenylalanine was positively associated with fecal serotonin, formyl-methionine, alpha-methylhistamine and propionylcarnitine. Although some of the metabolites found in serum have a microbial origin, no clear associations between differential microbial taxa and serum metabolites were found. Correlation analysis between bacterial taxa and fecal metabolites showed 9 significant correlations, of which 5 were positive correlations between the Romboutsia genus and the metabolites S-ethyl-cysteine, methionine, serine, phenylalanine and tryptophan, two positive correlations between the Ruminiclostridium 9 genus and serine and cinnamic acid, and two negative correlations between E. coli TOP498 and the metabolites serine S-ethyl-cysteine and 4-hydroxybenzyl alcohol (Fig. 5B).
Figure 5.
Heatmaps showing significant statistical correlations values (r) between: (A) serum and fecal metabolites, and (B) microbial taxa and fecal metabolites. Significant correlations are marked with a cross (p-adjusted value < 0.05).
Multivariate association with linear models (MaAsLin) analysis was performed to analyze the associations between fecal and serum metabolites and significantly changed fecal microbiota taxa. Results showed that the presence of Ruminiclostridium 9 was partly predicted by fecal phenylalanine, serum tryptophan and uric acid metabolites (positive association), and negatively associated with the presence of succinic acid and 21-deoxycortisol serum metabolites (Table 4).
Table 4.
Regression analysis results using MaAsLin.
| Bacterial taxa | Metabolite | Regression coefficient | P-adj value |
|---|---|---|---|
| D_5__Ruminiclostridium9;D_6__uncultured Clostridia bacterium | Fecal phenylalanine | 0.3472 | 0.0067 |
| Serum tryptophan | 0.4493 | 0.0003 | |
| Serum uric acid | 0.36942 | 0.0019 | |
| Serum Succinic-acid | − 0.2486 | 0.0387 | |
| Serum 21-Deoxycortisol | − 0.4142 | 0.0387 |
Discussion
We previously showed that the performance of physical exercise in compliance with World Health Organization recommendations has a positive effect on the gut microbiota, increasing its diversity and complexity, and promoting the colonization of bacteria with potential health benefits2,15. To study how physical exercise modifies the gut microbiota, we designed a laboratory-based, moderate-intense exercise study to simultaneously investigate the potential changes that a single exercise bout induces in the serum and fecal metabolome and in the gut microbiota. During and immediately after exercise, there is a rapid upregulation of several metabolic pathways responsible for skeletal muscle substrate utilization, and a physiological response has been shown to occur in many organs12,16. Some of these changes do not seem to be completely reversed, as several of these metabolites can be considered markers of fitness status17. We conducted the present study with amateur athletes as it has been described that muscle signaling is more pronounced in trained people than in untrained people18. Our results reveal the activation of various metabolic pathways by exercise performance until volitional exhaustion. We found an increase in carbohydrate metabolites (hexoses, among others) and myo-inositol, reflective of gluconeogenic influx, which is typically induced after an initial depletion of glucose and glycogen stores19. Dysregulation of aminoacyl-tRNA biosynthesis reflects modifications in protein synthesis, with perturbations in proteinogenic amino acid levels. Consistent with a prior study using a maximal exercise cycling test12, we detected a decrease in serum tryptophan levels associated with an increase in indolelactic acid, a metabolite of the tryptophan-indole pathway20. Our results are also in accord with those reported after a single bout of endurance exercise21, with a decrease in serum tryptophan and an increase in kynurenic acid, increasing the kynurenic/tryptophan ratio. As previously reported by Lewis et al.17, we detected an increase in the abundance of metabolites associated with purine metabolism, including inosine, xanthine and hypoxanthine and uric acid. Of interest, hypoxanthine and xanthine accumulation in blood was also recently reported in response to a session of acute exercise22. The accumulation of intermediate metabolites of the tricarboxylic acid cycle, including aconitic acid, ketoglutaric acid, succinic acid, malic acid, citric acid and fumaric acid, and also lactic acid, was also evident, likely due to the activation of this pathway for ATP production, indicative of high energy demand. We also detected a slight but significant increase in hydroxybutyrate, a marker of ketone body production resulting from exhaustive exercise. Hydroxybutyrate is synthesized in the liver from fatty acids or ketogenic amino acids and can be used as an energy source by peripheral tissues when blood glucose is low23,24. The elevation of some acetylated amino acids and the decrease in pantothenic acid might also reflect an increased availability of acetyl-CoA by both pyruvate oxidation and beta-oxidation17,25. The involvement of the arginine biosynthesis pathway could be reflected by the observed decrease of spermidine and the increase of n(8)-acetylspermidine. Spermidine is a polyamine with anti-inflammatory effects, and it has been shown to enhance cell and organ function through autophagy and improve the bioavailability of arginine (not significant in the present study) required for nitric oxide biosynthesis26. The systemic levels of spermidine depend on oral intake, microbiota production and cellular metabolism26, but to our knowledge, this is the first time that its levels have been associated with exercise performance.
Exercise also induced changes in the fecal metabolome and in the gut microbiota. Metabolomic analysis revealed that the most relevant changes were in phenylalanine, tyrosine and tryptophan metabolism, reflected by an increase in tryptophan, tyrosine and phenylalanine metabolites. These essential amino acids are synthesized by gut microbiota, as humans (all animals) lack the shikimic pathway. Our results are similar to those of Zhao et al. in a half marathon running study, in which an increase of the phenylalanine, tyrosine and tryptophan biosynthesis pathway was also observed13. The analysis of the behavior of the microbiota regarding tryptophan synthesis during exercise is of great interest, as microbiota-contributed tryptophan in blood could counteract the decrease of tryptophan that is involved in the appearance of fatigue27, decreasing the effect of kynurenine—a metabolite that restricts exercise capacity28 and also related to depression and schizophrenia29. We detected kynurenine both in serum and feces, but it was not significantly modified by exercise in either compartment. While several studies have attempted to modify tryptophan levels in blood to delay central fatigue30–32, to the best of our knowledge no microbiota-based strategy has been proposed to increase circulating tryptophan levels. As mentioned, the microbiota plays an important role in tryptophan biosynthesis and catabolism. Because tryptophan is required for the survival of bacteria, they are able to synthesize this amino acid through a highly regulated tryptophan-biosynthesis operon, and the capacity of tryptophan synthesis is widely distributed in bacteria33. Bacteria also participate in tryptophan catabolism, which is directly or indirectly involved in the three major tryptophan metabolism pathways20. Furthermore, bacterial tryptophan metabolites influence host health, and contribute to the intestinal and systemic homeostasis in health and disease34, and our study suggests a strong cross-talk between microbiota and systemic tryptophan metabolism. Fecal tryptophan levels correlated negatively with the presence of E. coli TOP498 and positively with the presence of the Romboutsia genus. Also, we found changes in the abundance of E. coli and several bacterial taxa (Romboutsia, Ruminococcocaceae UCG-005, Blautia, Ruminiclostridium 9 and Clostridium phoceensis) belonging to the Clostridia class, which possess tryptophan synthesis machinery33.
We detected an increase in fecal ammonia after the exercise bout, which could be related to the metabolism of urea and glutamine that can occur during exercise, as both metabolites are physiological markers of exercise performance35. The main sources of ammonia in the gastrointestinal tract are from urea degradation and glutamine metabolism, and from dietary proteins36. Urea produced during exercise can permeate the intestinal tract and serve as a fuel for bacteria, enabling the growth of urease-containing bacteria36. The transfer of metabolites produced during exercise from the blood to the gastrointestinal system could be a mechanism by which exercise induces changes in the microbiota, as some of the bacteria detected in this study including Romboutsia, Ruminococcus and Clostridium exhibit urease activity37,38. In fact, a recent study demonstrated that the lactate produced during exercise can increase the abundance of the Veillonella genus in gut microbiota39.
The present study has some limitations that should be considered. The time of collection of the stool samples was not concurrent with the collection of serum samples. The timing for serum collection between T1 and T2 was clearly established; however, the collection of the stool samples was not as precise, although we limited the time to 4 h to avoid any other external influence. This could account for the lack of correlation between differential microbial taxa and serum metabolites.
In conclusion, we provide evidence that a single bout of acute physical exercise in amateur athletes triggers changes in serum and fecal metabolism and also in gut microbiota. The changes we have seen in the microbiota are subtle, as they have only been produced by an exercise bout. Despite being highly trained individuals the exercise they performed was very intense for them, reaching exhaustion, which can clearly be seen in the metabolomics approach. Further research is needed to better understand the interaction between the human body and the trillions of bacteria that inhabit it, as well as the changes induced by exercise on the gut microbiota and associated mechanisms. Exercise frequency, intensity, performing time, type of exercise, exercise volume and progression are all factors that influence physiological responses and exercise adaptations, and will need to be considered in future studies investigating the beneficial effect of exercise on the gut microbiota.
Materials and methods
Study design
The present study is a single-arm trial. Participants were recruited from different cross-country athletes' teams in Madrid, Spain. From the 68 participants screened, 40 male endurance cross-country athletes met the following inclusion criteria: 18–50 years of age, with a high physical condition (oxygen uptake VO2 ≥ 55 mL/kg/min), and body mass index 18–25 kg/m2 and treadmill experience. Exclusion criteria were antibiotics intake during 3 months prior to the study, smoking, nutritional or ergogenic supplements, prebiotics, probiotics, be vegetarian or vegan, chronic medication, gastrointestinal surgery, or any diagnosed disease. Participants were also excluded if they had any medical condition that could be exacerbated by exercise. The Ethics Committee for Clinical Research of the Community of Madrid Spain approved the study (Ref: 07/694487.9/17), and all procedures were in accordance with the 1964 Helsinki Declaration and its later amendments. Written informed consent was obtained from all the volunteers. The study was registered in Clinicaltrials.gov with the accession number NCT04244149.
Dietary habits
Dietary pattern characterization of the participants was carried out using a validated food frequency questionnaire with 93 food items40 and three 24-h dietary recalls (two weekdays and one weekend day). Data from the questionnaires were analyzed using Dietsource software 3.0 (Novartis, Barcelona, Spain) to obtain the total energy ingested (in kcals) of proteins, fat, carbohydrates and fiber.
Exercise protocol
Body weight and height were measured with a scale and a height meter (Asimed T2, Barcelona, Spain). BMI was calculated by dividing the weight by the square of the height. Participants refrained from any physical activity 24 h before the day of the physical test. All participants performed a standardized 10-min warm-up of continuous running on a treadmill (H/P/Cosmos Venus, Nussdorf-Traunstein, Germany) at 60% of their maximum heart rate. After the warm-up, they ran with a slope of 1% at a speed of 10 km/h, with increments of 0.3 km/h every 30 s until volitional exhaustion. Participants were verbally encouraged to give their maximal effort, particularly towards the end of the test. Oxygen consumption values were monitored during the test and the following variables were determined: VO2, pulmonary ventilation, ventilatory equivalents for oxygen and carbon dioxide, and end-tidal partial pressure of oxygen and carbon dioxide. These variables were used to calculate absolute maximal oxygen consumption (VO2maxABS), relative maximum oxygen consumption (VO2maxREL), maximal aerobic speed, and first and second ventilatory thresholds. After the exercise test, the participants performed a 1-km run on an athletics track at maximum speed. The time needed to complete the run was also measured.
Serum and stool sample collection
Venous blood samples were collected in vacutainer tubes immediately before and within 15 min of finishing the exercise test (T1 and T2, respectively). Serum was obtained after clotting and centrifugation at 760×g for 10 min at 4 °C; aliquots were immediately frozen and stored at − 80 °C. Stool samples were collected just before and after the exercise session. Participants were provided with a Fe–col stool collection device, an insulated bag and ice blocks to preserve the samples until they were delivered to the laboratory. The first stool sample was collected early in the morning, before physical exercise. The second sample was collected within four hours post-exercise to avoid diet interference. Exercise promotes bowel transit and normally, participants feel the need to defecate soon after intense exercise. Those participants who did not produce a stool sample within four hours after exercise were excluded from the study. On arrival at the laboratory, the samples were maintained at − 80 °C until processing. Stool samples for metabolomics analysis were lyophilized using the TFD5503 Bench-Top freezedryer (ilShin Biobase, Ede, Netherlands) and stored at − 80 °C.
Chemicals and reagents
All analytical grade reference compounds were from the Sigma Chemical Co. (Saint Quentin Fallavier, France). The standard mixtures used for the external calibration of the mass spectrometry (MS) instrument (Calmix-positive, for the positive ion mode, consisting of caffeine, l-methionyl-arginyl-phenylalanyl-alanine acetate; and Ultramark 1621, and Calmix-negative, for the negative ion mode, consisting of the same mixture plus sodium dodecyl sulfate and sodium taurocholate) were from Thermo Fisher Scientific (Courtaboeuf, France). Acetonitrile was from SDS (Peypin, France), formic acid from Merck (Briare-le-Canal, France), methanol from VWR Chemicals (Fontenay-sous-Bois, France) and deionized water from Biosolve chemicals (Dieuse, France).
Metabolite extraction from stool and serum and analytical measurements
Approximately 2 × 10 mg of lyophilized stool were weighed precisely in two distinct Precellys tubes (ref CK14-2 mL, Bertin Technologies, Montigny-le-Bretonneux, France), resuspended in 150 µL of pure water and thoroughly vortexed. Then, 600 µL of methanol were added. After vortexing, samples were lysed in a Precellys Device (Bertin Technologies) for 3 × 30 s at 6500 rpm and 4 °C, and were then left on ice for 90 min to facilitate complete protein precipitation.
Two 50-µL samples were withdrawn from each serum sample. Each sample was mixed with 200 µL of methanol and incubated on ice for 90 min to achieve protein precipitation.
After centrifugation at 20,000g for 15 min at 4 °C, 200 µL of the supernatant were withdrawn and evaporated to dryness under a nitrogen stream at 30 °C using a Turbovap instrument (Caliper Life Science Inc., Roissy, France). The resulting dried extracts were stored at − 80 °C until analysis. Dried aliquots were resuspended in either 100 µL of water/acetonitrile (95:5, v/v) with 0.1% formic acid for C18 analysis or 100 µL of a mixture of 10 mM ammonium carbonate buffer (pH 10.5) and acetonitrile (40:60) for ZIC-pHILIC analysis (see below). The tubes were then vortexed, incubated in an ultrasonic bath for 5 min, and centrifuged for a further 10 min. A volume of 95 μL of the supernatant was transferred to 0.2 mL-vials and mixed with 5 µL of a mixture of internal standards (13C-glucose, 15N-aspartate, ethylmalonic acid, amiloride, prednisone, metformin, atropine sulfate, colchicine, imipramine) in order to check for consistency of analytical results in terms of signal and retention time stability throughout the experiment. Quality control samples were obtained by pooling aliquots of each sample and these were injected every 5–10 samples throughout the analysis for further data normalization/standardization.
Untargeted metabolomics experiments were performed by liquid chromatography coupled to high-resolution mass spectrometry (LC-HRMS) using a combination of two complementary chromatographic methods41,42, consisting of reversed-phase chromatography (C18 chromatographic column) and hydrophilic interaction chromatography (HILIC) for the analysis of hydrophobic and polar metabolites, respectively. LC-HRMS experiments were conducted on an Ultimate 3000 chromatographic system (Thermo Fisher Scientific, Courtaboeuf, France) coupled to an Exactive mass spectrometer from Thermo Fisher Scientific fitted with an electrospray ionization source and operating in the positive and negative ion modes for C18 and HILIC separations, respectively (designated as C18(+) and HILIC(−), respectively). Metabolite separations were performed using a Hypersil GOLD C18 1.9 µm, 2.1 × 150 mm column maintained at 30 °C (Thermo Fisher Scientific) or a Sequant ZIC-pHILIC 5 µm, 2.1 × 150 mm column at 15 °C (Merck, Darmstadt, Germany), operated under gradient elution conditions as described41.
Metabolomic data processing, metabolite annotation and metabolic pathway analysis
The raw LC-HRMS data were first converted into mzXML files using MSConvert (ProteoWizard), and further data processing (e.g., normalization, scaling, filtering) and statistical treatments were performed using the Workflow4Metabolomics (W4M) platform43. Metabolite annotation was first realized using our spectral database according to accurately measured masses and chromatographic retention times41. Our chemical database initially included ~ 1000 metabolites mainly of human origin, but was then broadened by the addition of commercially available bacterial metabolites and other molecules described in the literature44,45. Confirmation of metabolite annotation was then accomplished by running additional LC–MS/MS experiments using a Dionex Ultimate chromatographic system combined with a Q-Exactive mass spectrometer (Thermo Fisher Scientific) under non-resonant collision-induced dissociation conditions using higher-energy C-trap dissociation. Obtained MS/MS spectra were both manually and automatically matched using MS-DIAL software46 to the spectra included in our in-house spectral database, as described47. To be identified, ions had to match at least 2 orthogonal criteria (accurately measured mass, isotopic pattern, MS/MS spectrum and retention time) to those of an authentic chemical standard analyzed under the same analytical conditions, as proposed by the Metabolomics Standards Initiative48. The identified metabolites were imported into the free online web-based platform MetaboAnalyst 4.049 for metabolic pathway enrichment. Thus, the annotated m/z features with a Wilcoxon p-value < 0.005 (BH-critical value) were imported as KEGG numbers using the appropriate Homo sapiens pathway library. The interpretation of the results was performed after considering data with a p-value < 0.05 and an impact value > 0.1.
pH and ammonia determination in stool samples
The pH was measured using a basic pH meter (Crisom, Hach Lange, Barcelona, Spain) according to the method described by Dai and Karring50. Ammonia content was determined using a high-performance ammonia-selective ion electrode (Orion, Thermofisher Scientific, Waltham, MA). Stool (100 mg) was dissolved in 5 mL of MilliQ water; the mixture was vortexed for 2 min and then sonicated (Ultrasons, Selecta, Barcelona, Spain) for 10 min. A volume of 100 µL 1 M NaOH was added to the sample and ammonia was immediately measured. A standard curve was made from serial dilutions of ammonium chloride from 0.1 M, according to the manufacturer's instructions.
Bacterial DNA extraction and bioinformatics
Bacterial DNA was extracted using the E.Z.N.A. Kit (Omega-Biotek, Norcross, GA) and a bead homogenizer (Bullet Blender Storm, Next Advance, Averill Park, NY). A DNA fragment comprising the V3 and V4 hypervariable regions of the 16 s rRNA gene was amplified for sequencing analysis6. Amplicons were sequenced on the MiSeq Illumina platform (Illumina, San Diego, CA). Sequence results were analyzed using Quantitative Insights into Microbial Ecology (QIIME2) v2019.751 and were processed with DADA2 for quality control52. The classify-sklearn method was used for taxonomy assignment53 with an in-house customized classifier based on the SILVA reference database54,55. To construct the customized reference database, we extracted the sequences according to our primers (forward primer sequence: CCTACGGGNGGCWGCAG, reverse primer sequence: GACTACHVGGGTATCTAATCC) from the SILVA 132 database clustered at 99% identity56. We trained the classifier using our tailored reference reads and SILVA 7-levels for reference taxonomy, including the species probability (weights) likely to be observed for human stool (downloaded from https://github.com/BenKaehler/readytowear)56,57.
Statistical analysis
For metabolomics data, multivariate analyses were used to identify molecular features that discriminate metabolite differences in athletes before and after a session of acute exercise. PCA and PLS-DA were performed using the W4M platform and were used to identify features with discriminative power and to maximize variation between the two groups (before and after the test). Also, permutation tests (200 cycles) were conducted to assess the robustness of the PLS-DA model when using a small sample size58. The significance of the discriminant metabolites from the two groups was defined by a statistically significant threshold of VIP, which was derived from the PLS-DA model. A value of VIP > 1.0 was considered sufficient for group discrimination59. The univariate data analyses included a Wilcoxon signed-rank test, corrected for multiple testing by the Benjamini–Hochberg procedure, to assess the statistical significance of each compound. The generation of the clustered heatmap was performed using the Canberra distance metric and normalizing data, adding a pseudocount of 1 before log10 conversion. To detect changes in microbiota, samples in T1 and T2 were compared using the q2-longitudinal method60. First, a feature volatility analysis was performed to explore the data. This type of analysis uses a supervised learning regressor to predict a continuous variable (time in this case) as a function of feature composition (bacterial taxa). Based on volatility analysis, some bacterial taxa were selected for subsequent analyses according to their importance and cumulative average change. Selected taxa were analyzed using a linear mixed-effects model to detect significant bacterial taxa. Associations between different variables were studied with Spearman´s correlation and multivariate association by linear models (MaAsLin), an additive general linear model to find associations between metadata and bacterial abundance61. The cumulative sum scaling normalization was used. All p-values were corrected for multiple testing using the Benjamini–Hochberg false discovery rate.
Supplementary Information
Acknowledgements
The authors acknowledge all the study participants. We thank Dr Kenneth McCreath for proofreading the manuscript.
Author contributions
M.T., F.F., D.M.P. and M.L. conceived and designed the experiments. J.A.G.M. and D.M.P. collected the samples and supervised the exercise session. J.A.G.M. and M.T. extracted DNA and prepared the metabolomic samples. M.T., E.C.V., F.A.C. and F.F. performed the acquisition and analysis of metabolomics data. C.B. and M.L. carried out the microbiota bioinformatics and statistical analysis. M.T. and M.L. wrote the manuscript. M.L. obtained the financial support for the project leading to this publication. All authors read and approved the final manuscript.
Funding
This research was funded by the Ministry of Economy and Competitiveness, Spain, project AGL2016-77288-R. MT received a short-term fellowship from the European Molecular Biology Organization (EMBO, STF 8131). JAGM holds a grant from the Ministry of Education, Culture and Sports Spain (FPU grant 2016/01110).
Data availability
The 16S rRNA dataset generated during the present study has been submitted to the NCBI Biosample database, https://submit.ncbi.nlm.nih.gov/subs/sra/SUB7780980/overview and will be available upon publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mariangela Tabone and Carlo Bressa.
These authors jointly supervised this work: François Fenaille and Mar Larrosa.
Contributor Information
François Fenaille, Email: francois.fenaille@cea.fr.
Mar Larrosa, Email: mar.larrosa@universidadeuropea.es.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-82947-1.
References
- 1.Fiuza-Luces C, et al. Exercise benefits in cardiovascular disease: Beyond attenuation of traditional risk factors. Nat. Rev. Cardiol. 2018;15:731–743. doi: 10.1038/s41569-018-0065-1. [DOI] [PubMed] [Google Scholar]
- 2.Cerdá B, et al. Gut microbiota modification: Another piece in the puzzle of the benefits of physical exercise in health? Front. Physiol. 2016;7:51. doi: 10.3389/fphys.2016.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert JE, et al. Exercise training modifies gut microbiota in normal and diabetic mice. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2015;40:749–752. doi: 10.1139/apnm-2014-0452. [DOI] [PubMed] [Google Scholar]
- 4.Campbell SC, et al. The effect of diet and exercise on intestinal integrity and microbial diversity in mice. PLoS ONE. 2016;11:e0150502. doi: 10.1371/journal.pone.0150502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denou E, Marcinko K, Surette MG, Steinberg GR, Schertzer JD. High-intensity exercise training increases the diversity and metabolic capacity of the mouse distal gut microbiota during diet-induced obesity. Am. J. Physiol. Endocrinol. Metab. 2016;310:E982–E993. doi: 10.1152/ajpendo.00537.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bressa C, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS ONE. 2017;12:e0171352. doi: 10.1371/journal.pone.0171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke SF, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63:1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- 8.Allen JM, et al. Exercise training-induced modification of the gut microbiota persists after microbiota colonization and attenuates the response to chemically-induced colitis in gnotobiotic mice. Gut Microbes. 2018;9:115–130. doi: 10.1080/19490976.2017.1372077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munukka E, et al. Six-week endurance exercise alters gut metagenome that is not reflected in systemic metabolism in over-weight women. Front. Microbiol. 2018;9:2323. doi: 10.3389/fmicb.2018.02323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinonen I, et al. Organ-specific physiological responses to acute physical exercise and long-term training in humans. Physiology. 2014;29:421–436. doi: 10.1152/physiol.00067.2013. [DOI] [PubMed] [Google Scholar]
- 11.Karl JP, et al. Military training elicits marked increases in plasma metabolomic signatures of energy metabolism, lipolysis, fatty acid oxidation, and ketogenesis. Physiol. Rep. 2017;5:1–14. doi: 10.14814/phy2.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manaf FA, et al. Characterizing the plasma metabolome during and following a maximal exercise cycling test. J. Appl. Physiol. 2018;125:1193–1203. doi: 10.1152/japplphysiol.00499.2018. [DOI] [PubMed] [Google Scholar]
- 13.Zhao X, et al. Response of gut microbiota to metabolite changes induced by endurance exercise. Front. Microbiol. 2018;9:1–11. doi: 10.3389/fmicb.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellanos N, et al. A critical mutualism—Competition interplay underlies the loss of microbial diversity in sedentary lifestyle. Front. Microbiol. 2019;10:3142. doi: 10.3389/fmicb.2019.03142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinonen I, et al. Organ-specific physiological responses to acute physical exercise and long-term training in humans. Physiology (Bethesda). 2014;29:421–436. doi: 10.1152/physiol.00067.2013. [DOI] [PubMed] [Google Scholar]
- 17.Lewis GD, et al. Metabolic signatures of exercise in human plasma. Sci. Transl. Med. 2010;2:33ra37. doi: 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lysenko EA, Popov DV, Vepkhvadze TF, Sharova AP, Vinogradova OL. Moderate-intensity strength exercise to exhaustion results in more pronounced signaling changes in skeletal muscles of strength-trained compared with untrained individuals. J. Strength Cond. Res. 2018 doi: 10.1519/JSC.0000000000002901. [DOI] [PubMed] [Google Scholar]
- 19.Maclaren, D. & Morton, J. Biochemistry for sport and exercise metabolism: Translate by gaeini a. Samt Publ. Tehran, Iran (2012).
- 20.Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Joisten N, et al. Exercise and the Kynurenine pathway: Current state of knowledge and results from a randomized cross-over study comparing acute effects of endurance and resistance training. Exerc. Immunol. Rev. 2020;26:24–42. [PubMed] [Google Scholar]
- 22.Contrepois K, et al. Molecular choreography of acute exercise. Cell. 2020;181:1112–1130.e16. doi: 10.1016/j.cell.2020.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman JC, Verdin E. beta-Hydroxybutyrate: A signaling metabolite. Annu. Rev. Nutr. 2017;37:51–76. doi: 10.1146/annurev-nutr-071816-064916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganeshan K, et al. Energetic trade-offs and hypometabolic states promote disease tolerance. Cell. 2019;177:399–413.e12. doi: 10.1016/j.cell.2019.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stander, Z. et al. The altered human serum metabolome induced by a marathon. Metabolomics14, 150 (2018). [DOI] [PubMed]
- 26.Madeo F, Eisenberg T, Pietrocola F, Kroemer G. Spermidine in health and disease. Science (80-). 2018;359:eaan2788. doi: 10.1126/science.aan2788. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Wu Y, Liu T, Li R, Xie M. Serotonin regulation in a rat model of exercise-induced chronic fatigue. Neuroscience. 2017;349:27–34. doi: 10.1016/j.neuroscience.2017.02.037. [DOI] [PubMed] [Google Scholar]
- 28.Yang F, et al. Liver CEBPbeta modulates the kynurenine metabolism and mediates the motility for hypoxia-induced central fatigue in mice. Front. Physiol. 2019;10:243. doi: 10.3389/fphys.2019.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cervenka, I., Agudelo, L. Z. & Ruas, J. L. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science357, eaaf9794 (2017). [DOI] [PubMed]
- 30.Khong TK, Selvanayagam VS, Hamzah SH, Yusof A. Effect of quantity and quality of pre-exercise carbohydrate meals on central fatigue. J. Appl. Physiol. 2018;125:1021–1029. doi: 10.1152/japplphysiol.00221.2018. [DOI] [PubMed] [Google Scholar]
- 31.Mikulski T, Dabrowski J, Hilgier W, Ziemba A, Krzeminski K. Effects of supplementation with branched chain amino acids and ornithine aspartate on plasma ammonia and central fatigue during exercise in healthy men. Folia Neuropathol. 2015;53:377–386. doi: 10.5114/fn.2015.56552. [DOI] [PubMed] [Google Scholar]
- 32.Strasser, B. et al. Probiotic supplements beneficially affect tryptophan-kynurenine metabolism and reduce the incidence of upper respiratory tract infections in trained athletes: A randomized, double-blinded, placebo-controlled trial. Nutrients8, (2016). [DOI] [PMC free article] [PubMed]
- 33.Merino E, Jensen RA, Yanofsky C. Evolution of bacterial trp operons and their regulation. Curr. Opin. Microbiol. 2008;11:78–86. doi: 10.1016/j.mib.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenham G, Buckley JD, Garrett J, Eston R, Norton K. Biomarkers of physiological responses to periods of intensified, non-resistance-based exercise training in well-trained male athletes: A systematic review and meta-analysis. Sports Med. 2018;48:2517–2548. doi: 10.1007/s40279-018-0969-2. [DOI] [PubMed] [Google Scholar]
- 36.Levitt DG, Levitt MD. A model of blood-ammonia homeostasis based on a quantitative analysis of nitrogen metabolism in the multiple organs involved in the production, catabolism, and excretion of ammonia in humans. Clin. Exp. Gastroenterol. 2018;11:193–215. doi: 10.2147/CEG.S160921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wozny MA, Bryant MP, Holdeman LV, Moore WE. Urease assay and urease-producing species of anaerobes in the bovine rumen and human feces. Appl. Environ. Microbiol. 1977;33:1097–1104. doi: 10.1128/AEM.33.5.1097-1104.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerritsen J, et al. Genomic and functional analysis of Romboutsia ilealis CRIB(T) reveals adaptation to the small intestine. PeerJ. 2017;5:e3698. doi: 10.7717/peerj.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheiman J, et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat. Med. 2019;25:1104–1109. doi: 10.1038/s41591-019-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vioque J, Gonzalez L. Validity of a food frequency questionnaire (preliminary results) Eur. J. Cancer Prev. 1991;1:19. doi: 10.1097/00008469-199110001-00029. [DOI] [Google Scholar]
- 41.Boudah S, et al. Annotation of the human serum metabolome by coupling three liquid chromatography methods to high-resolution mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014;966:34–47. doi: 10.1016/j.jchromb.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 42.Moreau R, et al. Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J. Hepatol. 2020 doi: 10.1016/j.jhep.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 43.Giacomoni F, et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics. 2015;31:1493–1495. doi: 10.1093/bioinformatics/btu813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aros-Calt S, et al. Metabolomic investigation of Staphylococcus aureus antibiotic susceptibility by liquid chromatography coupled to high-resolution mass spectrometry. Methods Mol. Biol. 2019;1871:279–293. doi: 10.1007/978-1-4939-8814-3_18. [DOI] [PubMed] [Google Scholar]
- 45.Aros-Calt S, et al. Annotation of the Staphylococcus aureus metabolome using liquid chromatography coupled to high-resolution mass spectrometry and application to the study of methicillin resistance. J. Proteome Res. 2015;14:4863–4875. doi: 10.1021/acs.jproteome.5b00697. [DOI] [PubMed] [Google Scholar]
- 46.Tsugawa H, et al. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 2015;12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbier Saint Hilaire P, et al. Comparative evaluation of data dependent and data independent acquisition workflows implemented on an orbitrap fusion for untargeted metabolomics. Metabolites. 2020;10:158. doi: 10.3390/metabo10040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chong J, et al. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai X, Karring H. A determination and comparison of urease activity in feces and fresh manure from pig and cattle in relation to ammonia production and pH changes. PLoS ONE. 2014;9:e110402. doi: 10.1371/journal.pone.0110402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolyen, E. et al. (2018) QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. 10.7287/peerj.preprints.27295v2. [DOI] [PMC free article] [PubMed]
- 52.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pedregosa F, et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 54.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yilmaz P, et al. The SILVA and ‘All-species Living Tree Project (LTP)’ taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bokulich NA, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2 ’ s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaehler, B. D., Bokulich, N. A., Caporaso, J. G. & Huttley, G. A. Species-level microbial sequence classification is improved by source-environment information. bioRxiv 406611 (2018).
- 58.Zhong F, et al. 1H NMR spectroscopy analysis of metabolites in the kidneys provides new insight into pathophysiological mechanisms: applications for treatment with Cordyceps sinensis. Nephrol. Dial. Transplant. 2012;27:556–565. doi: 10.1093/ndt/gfr368. [DOI] [PubMed] [Google Scholar]
- 59.Weljie AM, Bondareva A, Zang P, Jirik FR. (1)H NMR metabolomics identification of markers of hypoxia-induced metabolic shifts in a breast cancer model system. J. Biomol. NMR. 2011;49:185–193. doi: 10.1007/s10858-011-9486-4. [DOI] [PubMed] [Google Scholar]
- 60.Bokulich NA, et al. q2-longitudinal: Longitudinal and paired-sample analyses of microbiome data. mSystems. 2018;3:1–9. doi: 10.1128/mSystems.00219-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA dataset generated during the present study has been submitted to the NCBI Biosample database, https://submit.ncbi.nlm.nih.gov/subs/sra/SUB7780980/overview and will be available upon publication.