Abstract
There are extensive interactions between viruses and the host DNA damage response (DDR) machinery. The outcome of these interactions includes not only direct effects on viral nucleic acids and genome replication, but also the activation of host stress response signalling pathways that can have further, indirect effects on viral life cycles. The non-homologous end-joining (NHEJ) pathway is responsible for the rapid and imprecise repair of DNA double-stranded breaks in the nucleus that would otherwise be highly toxic. Whilst directly repairing DNA, components of the NHEJ machinery, in particular the DNA-dependent protein kinase (DNA-PK), can activate a raft of downstream signalling events that activate antiviral, cell cycle checkpoint and apoptosis pathways. This combination of possible outcomes results in NHEJ being pro- or antiviral depending on the infection. In this review we will describe the broad range of interactions between NHEJ components and viruses and their consequences for both host and pathogen.
Keywords: virus, DNA damage, NHEJ, double-stranded break, PRR sensing, DNA-PK
Introduction
Viruses hijack the machinery inside the hostile environment of a host cell in order to propagate. To make new infectious virions, the viral genome must be replicated and viral genes transcribed, necessitating extensive interactions between viruses and the host proteins that manipulate nucleic acids including DNA damage response (DDR) proteins. The DDR machinery is responsible for maintaining the integrity of the host genome by sensing and responding to damaged or mis-localized self-nucleic acids. The cellular DDR can promote or restrict virus growth by directly manipulating viral nucleic acids and by activating signalling pathways that can significantly impact viral life cycles. In return, viruses have evolved mechanisms to avoid or co-opt the DDR in order to maintain themselves successfully in the host cell.
One of the biggest threats to the integrity of the host genome are double-stranded DNA breaks (DSBs). Cells must repair DSBs or risk the rapid acquisition of potentially lethal mutations [1, 2]. Once a cell detects such a lesion it can be repaired by one of several complementary mechanisms including the rapid but error-prone non-homologous end-joining (NHEJ) pathway or the slower but more accurate homologous recombination (HR) pathway. Due to the cytotoxicity of DSBs, their detection leads to the activation of a broad range of stress-response pathways. The recruitment of the NHEJ or HR machinery to sites of DSBs results in downstream cell-cycle checkpoint activation by CHK2 and subsequent p53-driven senescence, or direct activation of p53-dependent apoptotic signalling [3]. The DDR machinery can also activate innate immune pathways to signal the presence of danger inside the cell, resulting in the activation of NF-κB and IFN regulatory factors (IRFs) that initiate an antiviral innate immune response [4]. In this way, the sensing of foreign, mislocalized or damaged self-DNA can be considered analogous. During infection, viruses produce nucleic acid structures in multiple intracellular compartments. These DNA structures can mimic damaged self-DNA in the nucleus, directly recruiting this DSB machinery and activating the same downstream signalling pathways as in cellular DSB repair (Fig. 1). Numerous studies have shown the complex cross-talk between viruses and components of the HR pathway [5, 6]. In this review we will focus solely on the interplay between viruses and components of the NHEJ pathway.
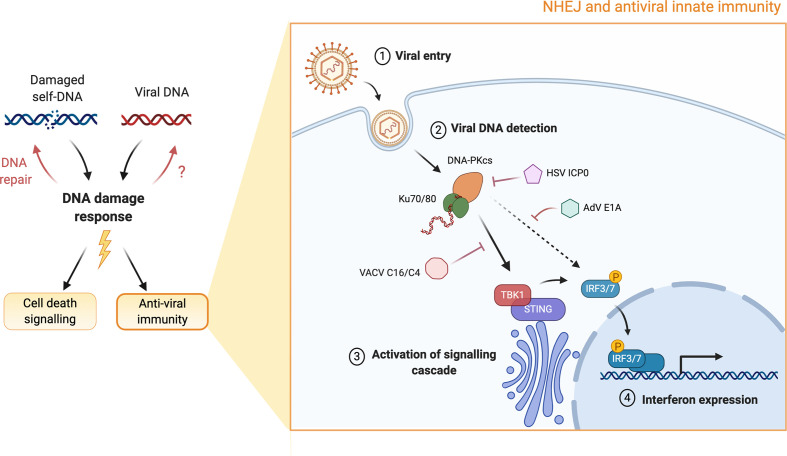
Fig. 1.
NHEJ and antiviral innate immunity. Damaged self-DNA and foreign viral DNA are both sensed by the DDR machinery (HR and NHEJ). The DDR machinery is responsible for repairing damaged DNA and ensuring survival of the cell. It also leads to the activation of a number of stress response pathways, which determine the fate of the cell, such as apoptosis/senescence, cellular antiviral state, etc. This graphic emphasizes the role of an NHEJ component, the DNA-PK complex, in antiviral immunity. DNA-PK recognizes intracellular viral DNA and activates the IFN innate immune response through activation of the STING adaptor protein or possibly via the direct phosphorylation of the IRF3 transcription factor. The indicated viral proteins suppress the innate immune response by blocking the signalling pathway at various stages. DDR, DNA damage response; HR, homologous recombination; NHEJ, non-homologous end-joining; DNA-PK, DNA-dependent protein kinase; STING, stimulator of IFN genes; IRF3, IFN regulatory factor 3; DNA-PKcs, DNA-dependent protein kinase catalytic subunit; HSV ICP0, herpes simplex virus infected cell polypeptide 0; VACV, vaccinia virus; AdV, adenovirus.
The non-homologous end-joining pathway
NHEJ rapidly repairs DNA double-stranded breaks, predominantly in the G1 phase of the cell cycle [7, 8]. Loss of NHEJ function results in cellular radiosensitivity via activation of p53-dependent apoptosis, an indication of the toxicity of unrepaired DSBs. The initial step of NHEJ is a sequence-independent recognition and binding of the Ku complex (consisting of the Ku70 and Ku80 subunits) to damaged DNA ends, which acts as a scaffold for recruitment of further NHEJ components, including the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), to form a heterotrimer with Ku70/80. This heterotrimer is sometimes referred to as DNA-PK. To complete a rapid repair, further factors are recruited, such as X-ray cross-complementing protein 4 (XRCC4), DNA Ligase IV (Lig4), XRCC4-like factor (XLF), and paralogue of XRCC4 and XLF (PAXX). This classical NHEJ complex results in re-ligation of the DNA sugar backbone by Lig4, which joins DNA ends using regions of microhomology and has therefore low fidelity (Fig. 2). Downstream of DNA repair, NHEJ recruitment to broken DNA can activate multiple stress responses, mainly via the PIKK family kinase, DNA-PKcs [3]. DNA-PKcs activity has a broad range of functions. It can activate apoptosis by direct phosphorylation of p53 and regulate transcription by phosphorylation of c-MYC and RNA-polymerase 1.
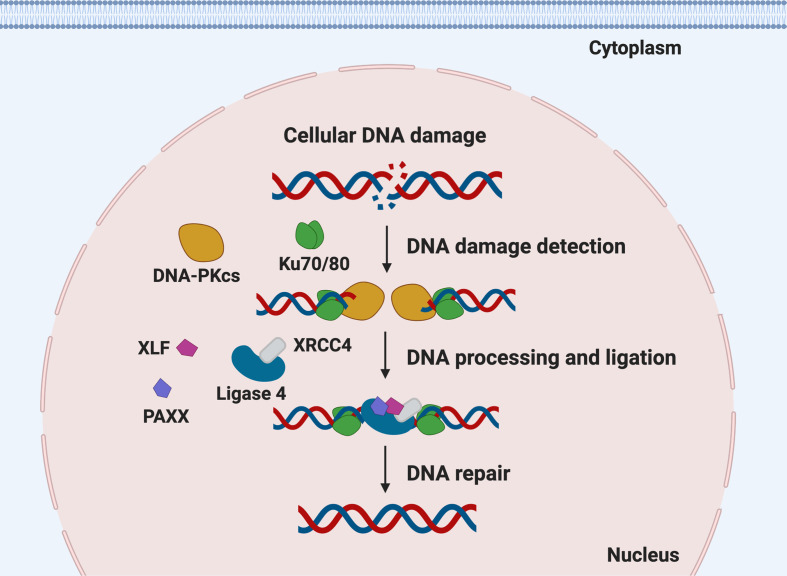
Fig. 2.
Schematic diagram of the NHEJ pathway. Ku70/80 recognize and bind to free ends of DNA at double-stranded breaks. The kinase DNA-PKcs is recruited and phosphorylates a number of downstream targets, contributing to further end-processing of the DNA. Ligase 4 in complex with XRCC4, XLF and PAXX re-ligate the broken DNA ends. Various virus types interact with proteins at multiple steps of the NHEJ pathway. DNA-PKcs, DNA-dependent protein kinase catalytic subunit; XRCC4, X-ray cross-complementing protein 4; XLF, XRCC4-like factor; PAXX, paralogue of XRCC4 and XLF.
NHEJ functions in the immune response
In addition to their role in protecting the genome from environmental stress, NHEJ components are an essential part of the mammalian immune system. The DNA repair function of NHEJ is required in the adaptive immune system for V(D)J recombination during T cell receptor rearrangement. Loss of NHEJ during development, for example through germline mutation of the prkdc gene that encodes DNA-PKcs, leads to severe combined immune disorder (SCID) due to an almost complete loss of T and B cells [9]. The commonly used scid mouse that phenocopies human SCID disease has a mutation that leads to expression of a C-terminally truncated, catalytically inactive DNA-PKcs protein [9]. Loss of DNA-PKcs activity results in reduced V(D)J recombination and hence very low levels of T and B cells in these mice. In addition to this function in adaptive immunity, DNA-PKcs and Ku70/80 act as sensors of DNA virus infection (pattern recognition receptors, PRRs) in the innate immune response [10, 11]. Upon infection with a virus, PRRs detect foreign DNA in the cytoplasm and possibly in the nucleus, which can trigger an antiviral response. The viral DNA-sensing DNA PRRs directly activate the production of type I IFN (IFN-I), mainly via the stimulator of IFN genes, tank-binding kinase 1, IFN regulatory factor 3 (STING-TBK1-IRF3) pathway [12] (Fig. 1). Ku70/80 and DNA-PKcs are essential for sensing multiple DNA virus infections in this way, as described in more detail in the following sections.
Interactions between NHEJ and viruses
Due to their ability to sense foreign or damaged self-DNA, NHEJ pathway components have extensive interactions with numerous different viruses (Table 1). These interactions can be antiviral, signalling the invasion into the cell and blocking viral replication, or pro-viral, being hijacked by viral processes for their own benefit to activate or enhance the viral life cycle. Below we will list the known interactions between NHEJ components and individual viruses and what is understood about the functional consequences of these interactions.
Table 1.
Known interactions between viruses and NHEJ components
|
Baltimore classification |
Virus |
Abbreviation |
Interaction with NHEJ components |
|---|---|---|---|
|
Group I: dsDNA viruses |
Adenovirus |
ADV |
DNA-PKcs [15, 18, 21], Lig4 [16], XRCC4 [17], Ku70 [20] |
|
Epstein Barr virus |
EBV |
||
|
Herpes simplex virus 1 |
HSV-1 |
DNA-PKcs [10, 34, 37, 39], Ku80 [38], Ku70 [39], Lig4/XRCC4 [40], PAXX [42], |
|
|
Human papillomavirus |
HPV |
Ku70/80 [45, 46] |
|
|
John Cunningham virus |
JC virus |
Ku70/80 [47] |
|
|
Kaposi sarcoma-associated herpesvirus |
KSHV |
||
|
Polyomavirus simian virus 40 |
SV40 |
DNA-PKcs [60] |
|
|
Vaccinia virus |
VACV |
||
|
Group II: ssDNA viruses |
Adeno-associated virus |
AAV |
|
|
Geminivirus |
– |
Ku80 [72] |
|
|
Parvovirus B19 |
B19 |
Ku80 [74] |
|
|
Group IV: (+) ssRNA viruses |
Hepatitis C virus |
HCV |
Ku70 [75] |
|
Group VI: ssRNA-RT viruses |
Avian sarcoma leukosis virus |
ASLV |
DNA-PKcs [76] |
|
Human immunodeficiency virus |
HIV |
DNA-PKcs [76, 79, 81–83, 85, 86], Ku80 [77, 78, 84, 85], Ku70 [80], Lig4 [81, 84, 85], XRCC4 [81, 85] |
|
|
Human T-cell leukaemia virus type |
HTLV |
||
|
Mouse mammary tumour virus |
MMTV |
Ku70/80 [94] |
|
|
Group VII: dsDNA-RT viruses |
Hepatitis B virus |
HBV |
Ku70/80 [96] |
|
Duck hepatitis B virus |
DHBV |
Ku80 [98] |
Double-stranded DNA viruses
Adenovirus
Adenoviruses are non-enveloped, dsDNA viruses. Various different serotypes of adenoviruses have been discovered in humans all having a linear non-segmented DNA genome [13]. Adenoviruses can cause a wide range of conditions, such as respiratory disease, conjunctivitis, adipogenesis and gastroenteritis in humans. The replication process is localized to the cellular nucleus and starts with the expression of mostly regulatory and non-structural proteins. The propagation of viral DNA is achieved via strand displacement and succeeded by the expression of structural proteins [14].
A number of studies have demonstrated a role for NHEJ components in interacting with adenovirus proteins. Adenovirus E4 34 k (early region 4) together with E4 11 k physically interact with DNA-PKcs [15] to inhibit DSB repair and prevent viral genome concatenation, thereby promoting viral DNA replication. Additionally, E4 34 k in combination with E1b 55 k induce the proteasomal degradation of Lig4 [16]. However, prior to initiation of the degradation process, E4 34 k contributes to the dissociation of the Lig4/XRCC4 complex independently of E1b 55 k [17]. E4orf4 is another adenovirus protein shown to interact with NHEJ proteins. More specifically, this protein physically binds to DNA-PK and uses it to reduce ATM/ATR activity and to inhibit the DDR response early during infection, whereas at a later stage the viral protein inhibits DNA-PK kinase properties, which enhances the viral replication [18].
After infection, DNA viruses can drive the cell cycle into S-phase to establish an environment conducive for viral replication. In this phase of the cell cycle the expression of many accessory factors on which the virus relies in its life cycle are upregulated. The key component for cell cycle switch is Early 1A proteins (E1A), which are expressed early post-infection [19]. In 2017, Frost et al. reported an interaction between the AdV E1A C terminus and the cellular Ku70 protein. They further showed that the diffusely dispersed Ku70 in the cell nucleus is localized at the viral genome during AdV infection but does not seem to interact directly with viral promoters. When Ku70 is depleted in cells, a consistent decrease of viral genome copy numbers per cell is observed at 72 h post-infection and viral yields are three-fold reduced in comparison to wild type cells during infection with AdV dl309. Controversially, Ku70 affects virus growth but has only a slight impact on viral genome replication and gene expression. A reduction in Ku70 levels seems to interfere with efficient induction of S-phase in arrested primary cells, which might explain the overall reduction of virus growth [20]. A recent study has also demonstrated a role for the E1A oncogene of human adenovirus 5 in the DNA-mediated antiviral immune response. The authors suggest that the oncogene acts downstream in the DNA sensing cascade, inhibits the complete phosphorylation of the IRF3 protein and blocks the activation of two antiviral pathways: the DNA-PK-mediated STING-independent (Fig. 1) and cGAS-mediated STING-dependent pathways [21].
Epstein Barr virus
Epstein Barr virus (EBV) is a human virus that infects mainly epithelial cells and B cells. The life cycle of EBV can be divided into two stages, a latent or dormant stage and the lytic, productive stage with S-phase-like cellular conditions. In the lytic stage, EBV is known to induce DNA damage and to inhibit DNA repair leading to instability of the host genome [22, 23]. One EBV component that has been associated with inhibition of NHEJ function in DSB repair is the transcription factor Zta, which is encoded by the BZLF1 gene and is crucial for lytic development of EBV [24]. Co-immunoprecipitation experiments have shown that the interaction of Ku80 with Zta can be mapped to the region associated with DNA binding and dimerization, between amino acids 168 and 245 of Zta. RNAP II seems to play a role in the replication process of many herpes viruses. RNAP II is a eukaryotic enzyme which catalyses DNA transcription to snRNA, mRNA and microRNA. Blockage of RNAP II, for example through oxidative DNA damage, increases double-stranded breaks [25]. In the case of EBV, it has been shown to support Zta-mediated transcription. By promoting the recruitment of the catalytic subunit of the kinase to RNAP II, Ku80 facilitates its phosphorylation, which is an important step for transcription [26, 27]. Zta has also been shown to interact with p53, which in turn is linked to transcriptional activation of RNAP II [28] and can be directly regulated by DNA-PKcs to drive cell death [29].
Han et al. showed in a co-immunoprecipitation experiment that another EBV protein, Epstein-Barr nuclear antigen (EBNA-LP), associates with DNA-PKcs in the C-terminal domain and leads to phosphorylation of EBNA-LP. When DNA-PKcs is inhibited by Wortmannin (an inhibitor of phosphoinositide 3-kinases) in vitro, no phosphorylation of EBNA-LP can be detected and hence can affect the role of EBNA-LP in viral transcription [30].
Herpes simplex virus 1
The herpes simplex virus 1 (HSV-1) genome is a large linear dsDNA molecule divided into a short and long region flanked by inverted repeats [31]. The expression of HSV-1 genes is regulated in a temporal cascade, starting with immediate early genes followed by the expression of early and late viral genes. One of the earliest genes expressed is the E3 ubiquitin-protein ligase ICP0, which prevents the antiviral function of nuclear bodies by degrading host promyelocytic leukaemia protein (PML), nuclear autoantigen SP-100 and gamma IFN-inducible protein 16 (IFI16) [32, 33].
ICP0 interferes with nuclear IRF3 signalling to prevent innate immune responses and IFN production by the infected cells [34]. Another cellular target of ICP0 is the NHEJ component DNA-PKcs, which is rapidly depleted as early as 2 h post-infection [35] (Fig. 1). In comparison, the levels of other NHEJ factors that have been shown to interact, such as Ku, seem unchanged by the virus. Lees-Miller et al. showed in 1996 that the half-life of DNA-PKcs is reduced by 50 % only 4 h after initial HSV infection and protein levels drop to 9 % in comparison to uninfected cells when using an HSV-1 KOS strain in human HeLa cells [35]. The authors suggest that the inhibition of DNA-PKcs activity counteracts its repressing effects on RNA polymerase II (RNAPII), paving the way for other viral proteins such as HSV-1 ICP27, which directly interacts with RNAPII to support viral transcription [36]. In cells lacking DNA-PKcs, HSV-1 showed increased replication efficiency, particularly after infection with low m.o.i. Accordingly, ICP0-null HSV strains exhibit poor growth, only rescued by infection with a high m.o.i. The exact mechanism of DNA-PKcs degradation is not understood in its entirety but the RING finger domain of ICP0 has been shown to be essential for the interaction [37]. Another explanation could be inferred by a study of Ferguson et al. who showed that HSV infection of murine embryonic cells (MEF) induces the STING-TBK1-IRF3 signalling pathway and type I IFN response to establish an antiviral state in the cell [4]. A novel role for DNA-PKcs has been described as a cytoplasmic innate immune sensor of viral DNA. For this reason, the inhibition of DNA-PKcs resulting in the prevention of a productive innate immune response would lead to a favourable environment for the virus. In neurons, HSV-1 also interacts with the NHEJ proteins Ku70 and Ku80, targeting these proteins for proteasomal degradation, leading to the accumulation of double-stranded breaks [38]. The authors speculate that this can result in the death of neuronal cells, implicating HSV-1 in the development of neurodegenerative disorders, such as Alzheimer’s disease. Another study demonstrated that in MEFs lacking Ku70, HSV-1 yields are 50-fold higher compared to control cells, implying an inhibitory role of Ku70 on viral replication [39].
The ubiquitously expressed DNA Lig4 is another core component of the NHEJ pathway that interacts with HSV-1. When Lig4 levels are reduced by RNA interference, the onset of virus production of HSV-1 is significantly impaired, reducing the number of infectious virions released from the cell. This effect is only overcome at later time points of infection with high m.o.i. It is suggested that Lig 4 plays a role early in HSV-1 replication to promote the formation of circular DNA molecules where the ligase is needed to conjoin linear DNA fragments producing the HSV-1 characteristic endless form [40].
One of the lesser studied components of the NHEJ machinery is the Paralogue of XRCC4 and XLF (PAXX), which was discovered in 2015 [41]. The exact function of PAXX remains somewhat elusive but it has a supportive role in NHEJ by stabilizing the complex consisting of DNA-PKcs, Ku70/80, Lig4, XRCC4 and XLF during end-joining. In the context of HSV-1 infection, Trigg et al. reported a role for PAXX in the viral life cycle. Although PAXX-knockout cells have an intact immune response, the virus seems to have a reproductive advantage and greater numbers of infectious virions are produced than in wild type cells. The authors suggest that PAXX acts to reduce the viral fitness by sequestering viral DNA in the cell leading to a reduced release of fully infectious HSV-1 particles [42].
Human papillomavirus
Human papillomavirus (HPV) is a member of the family Papillomaviridae, which consists of over 170 DNA viruses. Some HPV types can cause asymptomatic infections, form warts or lead to the formation of precancerous lesions [43]. HPV16 and HPV18 contribute to nearly 70 % of cervical cancer cases [44]. The integration process of HPV16 is poorly understood. Immunofluorescence studies have shown that Ku70/80 colocalize with HPV replication centres [45] and Winder et al. have demonstrated that in the absence of Ku70, an increased number of double-stranded breaks results in loss of HPV16 episome and de novo viral integration [46]. This effect is attributable to a combination of increased DSBs in Ku70-knockdown keratinocytes and alterations in the balance of linear versus circularized genomes.
Polyomaviruses
John Cunningham virus (JC) is a type of polyomavirus which can cause progressive multifocal leukoencephalopathy (PML) in immunocompromised patients [47]. Similar to other polyomaviruses [Simian virus 40 (SV40) and BK virus], a genome region of JC encodes for the small protein agnoprotein, found to be important for viral transcription and virion maturation [48]. A study by Darbinyan et al. showed that agnoprotein is able to prevent DSB repair after DNA damage. Agnoprotein associates with Ku70 in the perinuclear space by its N terminus. Moreover, the viral protein reduces the expression levels of Ku70/80 repair proteins [47]. The ability of agnoprotein to prevent DNA damage repair might be important for the virus life cycle and might contribute to malignancy formation.
SV40 is a polyomavirus that infects both humans and monkeys. SV40 is a tumour virus, which can suppress p53 via its T antigen (Tag) [49] but usually causes a long-term persistent infection. It has been proposed that following DNA damage DNA-PK plays a role in the regulation of SV40 replication in which DNA-PK phosphorylates and inactivates the viral Tag protein [50].
Kaposi sarcoma-associated herpesvirus
Kaposi sarcoma-associated herpesvirus (KSHV) is a human herpesvirus that causes tumours. The NHEJ components Ku70/80 localize to nuclear replication compartments during KSHV infection, suggesting that both proteins might have a role in the viral replication cycle [51]. Hollingworth et al. demonstrated that the viral KSHV DNA exhibits small double-stranded breaks which could act as potential targets for Ku recognition. Depletion of Ku80 in VK219-RTA cells infected with KSHV leads to an increase in the viral genome load and an increased release of infectious virions from the cells. Similar results have been obtained with the DNA-PK kinase inhibitor NU7441, suggesting that NHEJ components have a negative effect on KSHV replication and they might restrict the virus through inappropriate ligation of cleaved concatemeric viral DNA [51]. Additional evidence has been provided by Xiao et al., who showed that overexpression of KSHV processivity factor 8 (PF-8) can block the interaction between Ku70/80 and DNA-PKcs, which prevents the formation of the DNA-PK complex and the efficient repair of DNA DSBs [52]. The authors propose that this effect is a potential contributor to the KSHV-mediated carcinogenesis.
Recently, it has been demonstrated that KSHV DNA can be detected in the nucleus by a ribonuclear complex consisting of the DNA-PK holoenzyme and paraspeckle components. Detection of KSHV DNA activates cGAS and the STING-TBK1-IRF3 pathway in the same complex as DNA-PK, resulting in the induction of a type I IFN immune response and illustrating a role for DNA-PK in KSHV DNA sensing [53].
During latent infection KSHV genomes exist episomally and are replicated once per host cell cycle [54, 55]. In this phase of the viral life cycle several latency-associated genes are expressed, including latency-associated nuclear antigen (LANA), which acts in host and viral transcriptional regulation [56]. Cha et al. demonstrated that DNA-PK binds and phosphorylates LANA [57]. The authors suggest that the DNA-PK-mediated phosphorylation of LANA might drive the recruitment or inhibit the interaction with other cellular factors. Similar examples are PARP-1 poly(ADP-ribosyl)ation of LANA affecting viral replication [58], or Pim-1 and Pim-3 kinase which can phosphorylate LANA at serine 205 and 206 and inhibit its function [59, 60]. The overall conclusion is that DNA-PK activity negatively affects KSHV replication via multiple mechanisms.
Vaccinia virus
Vaccinia virus (VACV) is a large dsDNA virus from the family Poxviridae. VACV is revolutionary for its contribution to the eradication of the human disease smallpox. The unique replication cycle of poxviruses is limited only to the cytoplasm of the cell directly exposing their DNA to viral DNA sensors [61]. The role of the NHEJ component DNA-PKcs as a cytoplasmic DNA sensor driving type I antiviral innate immunity was discovered using VACV as an infection model [62]. DNA-PKcs, Ku70 and Ku80 are all essential for IRF-3 activation and type I IFN production following infection with VACV in murine fibroblasts and in mice. In turn, the poxvirus proteins C16 and C4 inhibit Ku70/80 binding to viral DNA and thus attenuate the antiviral response in cells and mice [63], suggesting the important role of DNA-PK in VACV infection (Fig. 1). Ku70 is also required for the production of type III IFNs in human cells via activation of the STING pathway [64], in a way that is likely to be relevant to sensing cytoplasmic DNA viruses such as VACV. Moreover, a role for Lig4 has been demonstrated during VACV infection. Introduction of DSBs in the poxvirus genome suppresses virus replication and DNA repair is performed by the cellular Lig4 acting outside of the nucleus [65].
Single-stranded DNA viruses
Adeno-associated virus
Adeno-associated virus (AAV) is a small DNA virus with a single-stranded genome representative of the family Parvoviridae. AAV is different from other DNA viruses because it depends on a helper virus (e.g. adenovirus) for productive infection and has the capacity to integrate at specific sites in the host genome. In cell culture, 65–90 % of AAV integration has been shown to be site-specific. An example is AAV2 which integrates into chromosome 19q13.4 in humans. Daya et al. showed that the process of integration, although not understood in its entirety, requires the NHEJ component DNA Lig4. Lig4-mutated cells exhibit a reduction of virus integration by 20 % [66].
The host response to AAV can be distinctly different depending on the associated helper virus. Co-infection with adenovirus activates cellular DNA damage response pathways in a different way compared to an infection with a single virus only. The former causes upregulation of DNA-PKcs levels and most signalling events are transmitted through it, as opposed to ATM/ATR-mediated signalling or the MRN complex, which play an essential role in HSV-1 infection [67]. Furthermore, during AAV infection DNA-Pkcs, Ku70 and Ku80 localize to viral replication centres, suggesting that the recognition of AAV is accomplished through the viral DNA hairpin loops known to activate DNA-PK [68].
DNA-PK has been confirmed to have a major role in the AAV life cycle in several studies. In vitro experiments in MO59 and 293 cells have shown a decrease in AAV replication when DNA-PK is inhibited. The same has been observed when Ku70 and Ku80 are depleted, confirming a positive role for DNA-PK in the viral life cycle [69]. In addition, it has been demonstrated that the lack of DNA-PKcs negatively affects site-specific integration of AAV and that DNA-PK contributes to the circularization of recombinant AAV episomes, particularly in skeletal muscle tissue [66, 70]. Several studies have focused on the cross-talk between AAV and DDR. However, the interaction between AAV and NHEJ proteins needs to be investigated in more detail in order to dissect the contradictory data. DNA-PK might differentially affect AAV integration or replication depending on the structure of the viral genome or the cellular context.
Geminivirus
Geminivirus is a single-stranded circular DNA virus that infects plants and causes leaf curling and stunting. Ku80 has been shown to impede geminivirus multiplication [71]. Southern blot analysis has shown greater viral DNA accumulation in Ku80 mutant plant lines (with abolished Ku80 transcription) compared to WT. Ku70 and Ku80 are found not only in the nucleus, but also in the cytoplasm in Arabidopsis thaliana and the authors propose a role for Ku80 as a novel viral DNA sensor in plants.
Parvovirus B19
Parvovirus B19 is a small ssDNA virus mainly infecting children via droplet transmission. An interesting characteristic of parvovirus B19 is its ability to infect red blood precursor cells in the bone marrow. The virus enters cells through receptor-mediated interaction with the blood group P antigen [72]. An important component to facilitate the binding is the NHEJ protein Ku80, which is normally localized in the cell nucleus but is expressed on the cell surface of erythroid progenitor cells. Munakata et al. showed that some cells with P antigen fail to bind B19 unless these cells express Ku80 on their surface. The authors suggest that Ku80 acts as a co-receptor for cell entry and they have further demonstrated that B19 infectivity could be inhibited by anti-Ku80 antibody or small interfering RNA (siRNA)-mediated knockdown [73].
Positive-sense single-stranded RNA viruses
Hepatitis C virus
Hepatitis C virus (HCV) is a small ssRNA virus that causes hepatitis C and some cancers, such as liver cancer. The HCV core protein has been found to ubiquitinate and reduce Ku70 protein levels in hepatocytes, attenuating the activity of DNA-PK [74].
Single-stranded RNA-RT viruses
Avian sarcoma leukosis virus
Avian sarcoma leukosis virus (ASLV) is an ssRNA retrovirus that productively infects chickens and other birds and causes a wide variety of tumours. Cell lines deficient in Ku and XRCC4 show a ten-fold lower integration rate of the virus [75]. Integration-competent ASV induces a three-fold increase in DNA-PK activity in HeLa cells compared to an infection with integration-deficient virus, suggesting that NHEJ plays a crucial role in viral integration. However, it is still unclear whether DNA-PK plays a direct or indirect role in completion of this integration process. Similar to human immunodeficiency virus (HIV), scid cells infected with ASLV undergo apoptosis, detected by TUNEL assay and increased caspase-3 activity. The results imply that scid cells are not able to repair the integration intermediate, resulting in a damaged DNA strand and ultimately death of the cell [75].
Human immunodeficiency virus
HIV is a member of the family Retroviridae that integrates into the host genome. HIV interacts with several components of the NHEJ machinery. For example, Jeanson et al. showed that Ku80 contributes to HIV genome circularization. Human CEM4fx cells (a human lymphoid cell line expressing high levels of CD4 antigen) with depleted Ku80 exhibit delayed HIV replication through inhibition of long terminal repeat (2-LTR) circle formation of viral cDNA in the nucleus [76]. This result has been further confirmed by siRNA-mediated knockdown of Ku80, which significantly reduces HIV-1 replication [77]. The regulatory HIV transactivator of transcription (Tat) protein, which significantly enhances viral transcription, decreases the expression of DNA-PKcs by binding to the core promoter region. Additionally, Tat interferes with the FAT domain that regulates the kinase activity of DNA-PKcs [78]. The third component of the DNA-PK heterotrimer, Ku70, interacts with HIV-1 integrase and protects it from the Lys [48]-linked polyubiquitination proteasomal pathway. Ku70 is able to downregulate the overall protein polyubiquitination level within the host cells and to specifically de-ubiquitinate HIV integrase, resulting in its stabilization [79]. There is conflicting information regarding the requirement of NHEJ components for HIV-1 integration. On the one hand, DNA-PK is reported as being required for efficient genome integration in MEFs [80], and DNA-PK-deficient scid cells and XRCC4-deficient Chinese hamster ovary (CHO) cells show substantially reduced levels of viral integration [75]. In these cases infected cells die by apoptosis, probably due to an inability to repair DNA damage following unsuccessful viral integration. On the other hand, DNA-PKcs-deficient MEF and MO59K cells do not exhibit defects when a vesicular stomatitis virus G protein (VSV-G)-pseudotyped HIV-1-derived lentiviral vector is transduced, suggesting that DNA-PK is not required for the retroviral integration process in all contexts [81, 82].
NHEJ further regulates cell death during HIV infection. Cells lacking Ku, ligase IV or XRCC4 exhibit virus-induced cell death independent of viral integration. It is hypothesized that the inability of the DNA repair pathway to enhance the formation of LTR circles results in the accumulation of retroviral DNA ends and the initiation of pro-apoptotic signals [83, 84]. Another study demonstrated that in primary human CD4 lymphocytes, which are the main target of HIV-1, viral integration induces cell death through DNA-PKcs-mediated phosphorylation of p53 and H2AX [85].
Human T-cell leukaemia virus type
Human T-cell leukaemia virus type (HTLV) is another member of the family Retroviridae. In humans, infection with HTLV is usually asymptomatic but this oncogenic retrovirus can cause adult T-cell leukaemia and demyelinating disease in approximately 2–5 % of infected individuals [86]. HTLV interacts with several members of the NHEJ complex. The HTLV-1 protein Tax can inhibit DNA double-stranded break repair leading to genomic instability of the host cell genome. The expression of Tax causes a decrease in protein and mRNA levels of Ku80 due to repressed transcription from the Ku80 promoter. Expression of Tax in Jurkat cells leads to a 60 % decrease in Ku80 mRNA levels while Ku70 is not affected [87]. In accordance with this study, the HTLV-1 Tax protein significantly increases genomic instability of the host cell, as demonstrated by an increasing number of DSBs and micronuclei (a sign of genotoxic stress), with unclear effects on the viral life cycle. These effects seem to be directly linked to the interaction of Tax with the NHEJ protein Ku80, but not with other proteins of the NHEJ pathway [88]. Another study has demonstrated that Ku80 decreases viral gene expression through targeting retroviral insertions to ‘silent’ chromatin areas (perinuclear regions of the genome) [89]. Overall, the consequences of Ku80/Tax interactions during HTLV infection are not fully understood. Tax can also exist in a stable complex with DNA-PKcs and Chk2, increasing DNA-PKcs activity leading to constitutive activation of the DNA damage response pathway [90]. The authors suggest that this could prevent the induction of host cell death and contribute to prolonged survival of infected cells.
There is evidence that other HTLV-1 proteins interact with NHEJ components. The viral oncoprotein basic leucine zipper (bZIP) associates with Ku70/80 and attenuates DNA-PK activity after topoisomerase inhibition with etoposide [91]. During the replication of retroviruses several nucleic acid structures are formed, one of which is the ssDNA intermediate. The expression of Ku70 is upregulated during HTLV-1 infection. Ku70 recognizes the ssDNA90 DNA and activates an antiviral innate immune response. Overexpression of Ku70 inhibits viral protein expression and increases the cellular immune response to HTLV-1 infection and ssDNA90 stimulation. An increased association of Ku70 with STING is observed in the cytoplasm of the host cell, demonstrating a role for Ku70 as a cytosolic DNA sensor of HTLV-1 infection that induces type III IFN. Ku80, however, does not appear to be involved in this process, indicating potential independent roles for members of the DNA-PK heterotrimer in regulating antiviral responses [92].
Mouse mammary tumour virus
Mouse mammary tumour virus (MMTV) is an obligatory integrating retrovirus that interacts with DNA-PK. The negative regulatory element (NRE1) of MMTV LTR is directly bound by DNA-PK, which prevents glucocorticoid-induced viral transcription [93]. This interaction suggests a mechanism for promoter/enhancer-driven modification of transcription factor activity and a repressed MMTV transcription.
Double-stranded DNA-RT viruses
Hepatitis B virus
Hepatitis B virus (HBV) is a DNA virus from the family Hepadnaviridae that infects mainly hepatocytes and can lead to the development of hepatitis, cirrhosis and hepatocellular carcinoma [94]. HBV interactions with NHEJ have only been studied in the context of viral DNA sensing and innate immunity. In liver cell lines the viral DNA is sensed by Ku70/80, which translocates to the cytoplasm and stimulates a STING-dependent type III IFN response, which leads to hepatitis-associated chemokine secretion and establishes an antiviral state in the cell [95].
Duck hepatitis B virus
Duck hepatitis B virus (DHBV) is a highly virulent Avihepadnavirus causing liver disease in ducks similar to other members of the family Hepadnaviridae, such as human BV (an orthohepadnavirus). Surprisingly, there are few sequence similarities between avi- and orthohepadnaviruses, except for highly conserved functional domains, such as the nucleocapsid [96]. DHBV is a dsDNA virus with a partially double-stranded circular genome and replicates in the nucleus via reverse transcription. In the nucleus, a minichromosome structure is formed, covalently closed circle DNA (cccDNA), which acts as a template for transcription of mRNA. Experiments in CHO cells have demonstrated that Ku80 is essential for cccDNA formation from double-stranded linear DNA [97]. In Xrs-5 Ku80 knockout cells cccDNA formation can be rescued with a Ku80-expressing plasmid.
Concluding remarks
The co-evolution of DNA viruses and their mammalian hosts has resulted in a complex network of interactions between DDR systems and the life cycle of these pathogens. Of all the DNA repair systems, NHEJ is the fastest and least accurate, functioning to sense broken ends of DNA and rejoin them as quickly as possible. NHEJ can therefore pose a threat to the integrity of DNA viral genomes, which must be accurately copied but often contain accessible DNA ends during replication. An additional threat posed by this machinery to the life cycle of DNA viruses is the PRR function of DNA-PKcs and Ku70/80. These proteins are targeted by most dsDNA viruses as an immune evasion mechanism to block the activation of IRF3 and type I IFN production. It is likely that the PRR function of DNA-PK has evolved concurrently with their role in DNA repair simply by plugging their sensing or receptor function into activation of alternative signalling outputs. The adenovirus E4 and E1A proteins bind DNA-PK to block both PRR and end-joining functions, thereby minimizing both unwanted genome concatenation and IFN-I production [15, 21].
On the other hand, NHEJ components are hijacked by some viruses to promote specific aspects of their life cycle, including generation of specific intermediates or genome structures that are required for efficient replication. For those viruses that have both circular and linear dsDNA genome structures as part of their replication cycle NHEJ has often been used to modulate the balance of linear (or monomeric) genomes to circular (or concatenated) viral genome structures by joining unbound genome ends. Indeed this has been shown for HSV-1, KSHV and HPV, which require different individual NHEJ components for efficient replication in the nucleus. HSV-1, for example, uses XRCC4 and Lig4 for the purpose of generating endless genomes [40], but also targets DNA-PKcs for degradation, presumably to evade the IFN-I response [4, 35]. Similarly retroviral genome circularization requires XRCC4, Lig4 and Ku80, but not DNA-PKcs to form 2-LTR circles [83, 84]. The absence of NHEJ proteins in retrovirus infection therefore often negatively impacts the viral life cycle and results in toxic build-up of non-integrated DNA. The direct role of NHEJ in retroviral integration is also well established but quite controversial in the case of DNA-PKcs and HIV specifically. As such there are several mechanisms by which NHEJ can assist in viral life cycles.
Overall, there is a mixture of positive and negative regulation of virus infection by NHEJ machinery with no specific pattern related to viruses of certain genome structures or life cycles. Mechanistically, when positively regulating virus replication, NHEJ components are used to assist in genome circularization, concatemerization, or integration in a broad variety of life cycles. In many cases, however, the precise molecular functions of NHEJ components in this context remain largely unexplored. In addition to direct manipulation of viral genome structure, NHEJ may also influence replication, transcription or packaging into newly formed capsids.
There are parallels with the other major double-stranded break repair pathway, homologous recombination. Cross-talk exists between HR and innate immune signalling. The PIKK ATM and the PARylation enzyme PARP-1 contribute to NF-κB activation in response to DNA damage by shuttling out of the nucleus and forming a complex with IKKκ/NEMO [98]. On the other hand, the DNA PRR cGAS negatively regulates HR via interaction with PARP-1 [99]. Both NHEJ and HR pathways can therefore carry out DNA repair processes and activation of stress responses and innate immune signalling, all of which have multiple potential consequences for virus infection.
Investigating the exact interactions between viruses and their host has become increasingly important with the introduction of viral vectors as delivery vehicles for gene therapy, or as vaccines. The choice of a specific vector which guarantees the best outcome for a patient or induces the most beneficial immune response in vaccine applications requires a deep understanding of the constituent components that are involved in the viral life cycle and are conducive to the desired outcome. Therefore, it is important to understand the mechanisms involved in guaranteeing integrity of the host cell, targeted insertion and productive expression systems. An example would be the above discussed AAV vector, which has been used to express FIX blood coagulation factor skeletal muscle of haemophilia patients [100]. As mentioned above, DNA-PK has a positive impact on the viral life cycle and viral fitness is reduced if inhibited in MO59 and 293 cells [69]. In the case of AAV this knowledge is key because it will influence the choice of helper system. A virus that downregulates DNA-PK activity or certain components of DNA-PK, such as HSV-1, would not present an ideal choice. Indeed, more such mechanistic information is required to understand precisely how NHEJ components regulate multiple aspects of the life cycles of viruses.
Funding information
This work received no specific grant from any funding agency.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AAV, adeno-associated virus; ASV, avian sarcoma leukosis virus; DDR, DNA damage response; DHBV, duck hepatitis B virus; DNA-PKcs, DNA-dependent protein kinase catalytic subunit; DSB, double strand break; EBV, Epstein Barr virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HPV, human papillomavirus; HR, homologous recombination; HSV, herpes simplex virus; HTLV, human T-cell leukaemia virus type; IRF, IFN regulatory factor; JC, John Cunningham virus; KSHV, Kaposi sarcoma-associated herpesvirus; LANA, latency-associated nuclear antigen; LTR, long terminal repeat; MMTV, mouse mammary tumour virus; NHEJ, non-homologous end-joining; PML, progressive multifocal leukoencephalopathy; PRR, pattern recognition receptor; SCID, severe combined immune disorder; siRNA, short interfering RNA; SV40, simian virus 40; VACV, vaccinia virus.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackford AN, Jackson SP. Atm, ATR, and DNA-PK: the Trinity at the heart of the DNA damage response. Mol Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Trigg BJ, Ferguson BJ. Functions of DNA damage machinery in the innate immune response to DNA virus infection. Curr Opin Virol. 2015;15:56–62. doi: 10.1016/j.coviro.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Weitzman MD, Lilley CE, Chaurushiya MS. Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol. 2010;64:61–81. doi: 10.1146/annurev.micro.112408.134016. [DOI] [PubMed] [Google Scholar]
- 6.Turnell AS, Grand RJ. Dna viruses and the cellular DNA-damage response. J Gen Virol. 2012;93:2076–2097. doi: 10.1099/vir.0.044412-0. [DOI] [PubMed] [Google Scholar]
- 7.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosma MJ, Carroll AM. The scid mouse mutant: definition, characterization, and potential uses. Annu Rev Immunol. 1991;9:323–350. doi: 10.1146/annurev.iy.09.040191.001543. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;2012 doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sui H, Zhou M, Imamichi H, Jiao X, Sherman BT, et al. Sting is an essential mediator of the Ku70-mediated production of IFN-λ1 in response to exogenous DNA. Sci Signal. 2017;10:eaah5054. doi: 10.1126/scisignal.aah5054. [DOI] [PubMed] [Google Scholar]
- 12.Mansur DS, Smith GL, Ferguson BJ. Intracellular sensing of viral DNA by the innate immune system. Microbes Infect. 2014;16:1002–1012. doi: 10.1016/j.micinf.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Nemerow GR, Stewart PL, Reddy VS. Structure of human adenovirus. Curr Opin Virol. 2012;2:115–121. doi: 10.1016/j.coviro.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guimet D, Hearing P. 3 - Adenovirus Replication. Curiel DTBT-AV for GT (Second E, editor. San Diego: Academic Press; 2016. pp. 59–84. [Google Scholar]
- 15.Boyer J, Rohleder K, Ketner G. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology. 1999;263:307–312. doi: 10.1006/viro.1999.9866. [DOI] [PubMed] [Google Scholar]
- 16.Baker A, Rohleder KJ, Hanakahi LA, Ketner G. Adenovirus E4 34k and E1B 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J Virol. 2007;81:7034–7040. doi: 10.1128/JVI.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayaram S, Gilson T, Ehrlich ES, Yu X-F, Ketner G, et al. E1B 55k-independent dissociation of the DNA ligase IV/XRCC4 complex by E4 34k during adenovirus infection. Virology. 2008;382:163–170. doi: 10.1016/j.virol.2008.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nebenzahl-Sharon K, Shalata H, Sharf R, Amer J, Khoury-Haddad H, et al. Biphasic functional interaction between the adenovirus E4orf4 protein and DNA-PK. banks L, editor. J Virol [Internet]. 2019;93:e01365–18. doi: 10.1128/JVI.01365-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lillie JW, Green MR. Transcription activation by the adenovirus E1A protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 20.Frost JR, Olanubi O, Cheng SK-H, Soriano A, Crisostomo L, et al. The interaction of adenovirus E1A with the mammalian protein Ku70/XRCC6. Virology. 2017;500:11–21. doi: 10.1016/j.virol.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Burleigh K, Maltbaek JH, Cambier S, Green R, Gale M, et al. Human DNA-PK activates a STING-independent DNA sensing pathway. Science Immunology. 2020;5:eaba4219. doi: 10.1126/sciimmunol.aba4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang C-Y, Lee C-H, Wu C-C, Chang Y-T, Yu S-L, et al. Recurrent chemical reactivations of EBV promotes genome instability and enhances tumor progression of nasopharyngeal carcinoma cells. Int J Cancer. 2009;124:2016–2025. doi: 10.1002/ijc.24179. [DOI] [PubMed] [Google Scholar]
- 23.McFadden K, Luftig MA. Interplay Between DNA Tumor Viruses and the Host DNA Damage Response BT - Intrinsic Immunity. In: Cullen BR, editor. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. pp. 229–257. editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C-C, Yang Y-C, Wang W-H, Chen C-S, Chang L-K. Enhancement of Zta-activated lytic transcription of Epstein-Barr virus by Ku80. J Virol. 2011;92:661–668. doi: 10.1099/vir.0.026302-0. [DOI] [PubMed] [Google Scholar]
- 25.Wei L, Levine AS, Lan L. Transcription-Coupled homologous recombination after oxidative damage. DNA Repair. 2016;44:76–80. doi: 10.1016/j.dnarep.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci U S A. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, et al. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 28.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill R, Madureira PA, Waisman DM, Lee PWK. DNA-PKCS binding to p53 on the p21WAF1/CIP1 promoter blocks transcription resulting in cell death. Oncotarget. 2011;2:1094–1108. doi: 10.18632/oncotarget.378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Han I, Harada S, Weaver D, Xue Y, Lane W, et al. EBNA-LP associates with cellular proteins including DNA-PK and HA95. J Virol. 2001;75:2475–2481. doi: 10.1128/JVI.75.5.2475-2481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacob RJ, Roizman B. Anatomy of herpes simplex virus DNA. VIII. Properties of the replicating DNA. J Virol. 1977;23:394–411. doi: 10.1128/JVI.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu H, Roizman B. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci U S A. 2003;100:8963–8968. doi: 10.1073/pnas.1533420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012;109:E3008-E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. The herpes simplex virus ICP0 ring finger domain inhibits IRF3- and IRF7-Mediated activation of interferon-stimulated genes. J Virol. 2004;78:1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lees-Miller SP, Long MC, Kilvert MA, Lam V, Rice SA, et al. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/JVI.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai-Ju JQ, Li L, Johnson LA, Sandri-Goldin RM. Icp27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J Virol. 2006;80:3567–3581. doi: 10.1128/JVI.80.7.3567-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkinson J, Lees-Miller SP, Everett RD. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650–657. doi: 10.1128/JVI.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Chiara G, Racaniello M, Mollinari C, Marcocci ME, Aversa G, et al. Herpes simplex Virus-Type1 (HSV-1) impairs DNA repair in cortical neurons. Front Aging Neurosci. 2016;8:242. doi: 10.3389/fnagi.2016.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor TJ, Knipe DM. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling Proteinswith ICP8. J Virol. 2004;78:5856–5866. doi: 10.1128/JVI.78.11.5856-5866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muylaert I, Elias P. Knockdown of DNA ligase IV/XRCC4 by RNA interference inhibits herpes simplex virus type I DNA replication. J Biol Chem. 2007;282:10865–10872. doi: 10.1074/jbc.M611834200. [DOI] [PubMed] [Google Scholar]
- 41.Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, et al. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015;347:185–188. doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trigg B, Lauer K, Fernandes dos Santos P, Coleman H, Balmus G, et al. The non-homologous end joining protein PAXX acts to restrict HSV-1 infection. Viruses. 2017;9:342. doi: 10.3390/v9110342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ljubojevic S, Skerlev M. Hpv-Associated diseases. Clin Dermatol. 2014;32:227–234. doi: 10.1016/j.clindermatol.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 45.Kadaja M, Isok-Paas H, Laos T, Ustav E, Ustav M. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog. 2009;5:e1000397. doi: 10.1371/journal.ppat.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winder DM, Pett MR, Foster N, Shivji MKK, Herdman MT, et al. An increase in DNA double-strand breaks, induced by Ku70 depletion, is associated with human papillomavirus 16 episome loss and de novo viral integration events. J Pathol. 2007;213:27–34. doi: 10.1002/path.2206. [DOI] [PubMed] [Google Scholar]
- 47.Darbinyan A, Siddiqui KM, Slonina D, Darbinian N, Amini S, et al. Role of JC virus agnoprotein in DNA repair. J Virol. 2004;78:8593–8600. doi: 10.1128/JVI.78.16.8593-8600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jay G, Nomura S, Anderson CW, Khoury G. Identification of the SV40 agnogene product: a DNA binding protein. Nature. 1981;291:346–349. doi: 10.1038/291346a0. [DOI] [PubMed] [Google Scholar]
- 49.Ahuja D, Sáenz-Robles MT, Pipas JM. Sv40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Zhou X-Y, Wang H, Huq MS, Iliakis G. Roles of replication protein A and DNA-dependent protein kinase in the regulation of DNA replication following DNA damage. J Biol Chem. 1999;274:22060–22064. doi: 10.1074/jbc.274.31.22060. [DOI] [PubMed] [Google Scholar]
- 51.Hollingworth R, Horniblow RD, Forrest C, Stewart GS, Grand RJ. Localization of double-strand break repair proteins to viral replication compartments following lytic reactivation of Kaposi's sarcoma-associated herpesvirus. J Virol. 2017;91:e00930-17. doi: 10.1128/JVI.00930-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao Y, Chen J, Liao Q, Wu Y, Peng C, et al. Lytic infection of Kaposi’s sarcoma-associated herpesvirus induces DNA double-strand breaks and impairs non-homologous end joining. J Gen Virol. 2013;2019:1870–1875. doi: 10.1099/vir.0.053033-0. [DOI] [PubMed] [Google Scholar]
- 53.Morchikh M, Cribier A, Raffel R, Amraoui S, Cau J, et al. Hexim1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol Cell. 2017;67:387–399. doi: 10.1016/j.molcel.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 54.Decker LL, Shankar P, Khan G, Freeman RB, Dezube BJ, et al. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in Ks tissue but replicates in the peripheral blood mononuclear cells of Ks patients. J Exp Med. 1996;184:283–288. doi: 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cesarman E, Moore PS, Rao PH, Inghirami G, Knowles DM, et al. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. doi: 10.1182/blood.V86.7.2708.2708. [DOI] [PubMed] [Google Scholar]
- 56.Renne R, Barry C, Dittmer D, Compitello N, Brown PO, et al. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Virol. 2001;75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cha S, Lim C, Lee JY, Song Y-J, Park J, et al. DNA-PK/Ku complex binds to latency-associated nuclear antigen and negatively regulates Kaposi’s sarcoma-associated herpesvirus latent replication. Biochem Biophys Res Commun. 2010;394:934–939. doi: 10.1016/j.bbrc.2010.03.086. [DOI] [PubMed] [Google Scholar]
- 58.Ohsaki E, Ueda K, Sakakibara S, Do E, Yada K, et al. Poly(ADP-Ribose) Polymerase 1 Binds to Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Terminal Repeat Sequence and Modulates KSHV Replication in Latency. J Virol. 2004;78:9936–9946. doi: 10.1128/JVI.78.18.9936-9946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bajaj BG, Verma SC, Lan K, Cotter MA, Woodman ZL, et al. Kshv encoded LANA upregulates Pim-1 and is a substrate for its kinase activity. Virology. 2006;351:18–28. doi: 10.1016/j.virol.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 60.Cheng F, Weidner-Glunde M, Varjosalo M, Rainio E-M, Lehtonen A, et al. Kshv reactivation from latency requires Pim-1 and Pim-3 kinases to inactivate the latency-associated nuclear antigen LANA. PLoS Pathog. 2009;5:e1000324. doi: 10.1371/journal.ppat.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith GL, Talbot-Cooper C, Lu Y. Chapter Fourteen - How Does Vaccinia Virus Interfere With Interferon? In: Kielian M, Mettenleiter TC, editors. Roossinck MJBT-A in VR. Academic Press; 2018. pp. 355–378. [DOI] [PubMed] [Google Scholar]
- 62.Peters NE, Ferguson BJ, Mazzon M, Fahy AS, Krysztofinska E, et al. A mechanism for the inhibition of DNA-PK-mediated DNA sensing by a virus. PLoS Pathog. 2013;9:e1003649. doi: 10.1371/journal.ppat.1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scutts SR, Ember SW, Ren H, Ye C, Lovejoy CA, et al. Dna-Pk is targeted by multiple vaccinia virus proteins to inhibit DNA sensing. Cell Rep. 2018;25:1953–1965. doi: 10.1016/j.celrep.2018.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J.i. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luteijn RD, Drexler I, Smith GL, Lebbink RJ, Wiertz EJHJ. Mutagenic repair of double-stranded DNA breaks in vaccinia virus genomes requires cellular DNA ligase IV activity in the cytosol. J Gen Virol. 2018;99:790–804. doi: 10.1099/jgv.0.001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daya S, Cortez N, Berns KI. Adeno-Associated virus site-specific integration is mediated by proteins of the nonhomologous end-joining pathway. J Virol. 2009;83:11655–11664. doi: 10.1128/JVI.01040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwartz RA, Carson CT, Schuberth C, Weitzman MD. Adeno-Associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase. J Virol. 2009;83:6269–6278. doi: 10.1128/JVI.00318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weitzman MD, Fisher KJ, Wilson JM. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J Virol. 1996;70:1845–1854. doi: 10.1128/JVI.70.3.1845-1854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi Y-K, Nash K, Byrne BJ, Muzyczka N, Song S. The effect of DNA-dependent protein kinase on adeno-associated virus replication. PLoS One. 2010;5:e15073. doi: 10.1371/journal.pone.0015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song S, Lu Y, Choi Y-K, Han Y, Tang Q, et al. Dna-Dependent pK inhibits adeno-associated virus DNA integration. Proc Natl Acad Sci U S A. 2004;101:2112–2116. doi: 10.1073/pnas.0307833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Richter KS, Jeske H. Ku80, a key factor for non-homologous end-joining, retards geminivirus multiplication. J Gen Virol [Internet] 2015;96:2913–2918. doi: 10.1099/jgv.0.000224. [DOI] [PubMed] [Google Scholar]
- 72.Brown K, Anderson S, Young N. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science. 1993;262:114–117. doi: 10.1126/science.8211117. [DOI] [PubMed] [Google Scholar]
- 73.Munakata Y, Saito-Ito T, Kumura-Ishii K, Huang J, Kodera T, et al. Ku80 autoantigen as a cellular coreceptor for human parvovirus B19 infection. Blood. 2005;106:3449–3456. doi: 10.1182/blood-2005-02-0536. [DOI] [PubMed] [Google Scholar]
- 74.Saitou K, Mizumoto K, Nishimura T, Kai C, Tsukiyama-Kohara K. Hepatitis C virus-core protein facilitates the degradation of Ku70 and reduces DNA-PK activity in hepatocytes. Virus Res. 2009;144:266–271. doi: 10.1016/j.virusres.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 75.Daniel R, Katz RA, Skalka AM. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 76.Jeanson L, Subra F, Vaganay S, Hervy M, Marangoni E, et al. Effect of Ku80 depletion on the Preintegrative steps of HIV-1 replication in human cells. Virology. 2002;300:100–108. doi: 10.1006/viro.2002.1515. [DOI] [PubMed] [Google Scholar]
- 77.Waninger S, Kuhen K, Hu X, Chatterton JE, Wong-Staal F, et al. Identification of cellular cofactors for human immunodeficiency virus replication via a Ribozyme-Based genomics approach. J Virol. 2004;78:12829–12837. doi: 10.1128/JVI.78.23.12829-12837.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang S-M, Zhang H, Yang T-Y, Ying T-Y, Yang P-X, et al. Interaction between HIV-1 Tat and DNA-PKcs modulates HIV transcription and class switch recombination. Int J Biol Sci. 2014;10:1138–1149. doi: 10.7150/ijbs.10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng Y, Ao Z, Wang B, Jayappa KD, Yao X. Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J Biol Chem. 2011;286:17722–17735. doi: 10.1074/jbc.M110.184739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daniel R, Greger JG, Katz RA, Taganov KD, Wu X, et al. Evidence that stable retroviral transduction and cell survival following DNA integration depend on components of the nonhomologous end joining repair pathway. J Virol [Internet]. 2004;78:8573 LP–8581. doi: 10.1128/JVI.78.16.8573-8581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ariumi Y, Turelli P, Masutani M, Trono D. Dna damage sensors ATM, ATR, DNA-PKcs, and PARP-1 are dispensable for human immunodeficiency virus type 1 integration. J Virol. 2005;79:2973–2978. doi: 10.1128/JVI.79.5.2973-2978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baekelandt V, Claeys A, Cherepanov P, De Clercq E, De Strooper B, et al. DNA-Dependent protein kinase is not required for efficient lentivirus integration. J Virol. 2000;74:11278–11285. doi: 10.1128/JVI.74.23.11278-11285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, et al. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. Embo J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kilzer JM, Stracker T, Beitzel B, Meek K, Weitzman M, et al. Roles of host cell factors in circularization of retroviral DNA. Virology. 2003;314:460–467. doi: 10.1016/S0042-6822(03)00455-0. [DOI] [PubMed] [Google Scholar]
- 85.Cooper A, García M, Petrovas C, Yamamoto T, Koup RA, et al. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 86.Gonçalves DU, Proietti FA, Ribas JGR, Araújo MG, Pinheiro SR, et al. Epidemiology, treatment, and prevention of human T-cell leukemia virus type 1-associated diseases. Clin Microbiol Rev. 2010;23:577–589. doi: 10.1128/CMR.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ducu RI, Dayaram T, Marriott SJ. The HTLV-1 Tax oncoprotein represses Ku80 gene expression. Virology. 2011;416:1–8. doi: 10.1016/j.virol.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Majone F, Jeang K-T. Unstabilized DNA breaks in HTLV-1 Tax expressing cells correlate with functional targeting of Ku80, not PKCs, XRCC4, or H2AX. Cell Biosci. 2012;2:15. doi: 10.1186/2045-3701-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Masson C, Bury-Moné Stéphanie, Guiot E, Saez-Cirion A, Schoëvaërt-Brossault D, et al. Ku80 participates in the targeting of retroviral transgenes to the chromatin of CHO cells. J Virol. 2007;81:7924–7932. doi: 10.1128/JVI.02015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Durkin SS, Guo X, Fryrear KA, Mihaylova VT, Gupta SK, et al. Htlv-1 Tax oncoprotein subverts the cellular DNA damage response via binding to DNA-dependent protein kinase. J. Biol. Chem. 2008;283:36311–36320. doi: 10.1074/jbc.M804931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rushing AW, Hoang K, Polakowski N, Lemasson I. The human T-cell leukemia virus type 1 basic leucine zipper factor attenuates repair of double-stranded DNA breaks via nonhomologous end joining. J Virol. 2018;92:e00672-18. doi: 10.1128/JVI.00672-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang J, Kang L, Song D, Liu L, Yang S, et al. Ku70 senses HTLV-1 DNA and modulates HTLV-1 replication. J Immun. 2017;199:2475–2482. doi: 10.4049/jimmunol.1700111. [DOI] [PubMed] [Google Scholar]
- 93.Giffin W, Torrance H, Rodda DJ, Préfontaine GG, Pope L, et al. Sequence-Specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 94.Schwalbe M, Ohlenschläger O, Marchanka A, Ramachandran R, Häfner S, et al. Solution structure of stem-loop α of the hepatitis B virus post-transcriptional regulatory element. Nucleic Acids Res. 2008;36:1681–1689. doi: 10.1093/nar/gkn006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y, Wu Y, Zheng X, Cong J, Liu Y, et al. Cytoplasm-Translocated Ku70/80 complex sensing of HBV DNA induces hepatitis-associated chemokine secretion. Front Immunol. 2016;7:569. doi: 10.3389/fimmu.2016.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ryu W-S. Ryu W-SBT-MV of HPV. Boston: Academic Press; 2017. Chapter 18 - Hepadnaviruses; pp. 247–260. [Google Scholar]
- 97.Guo H, Xu C, Zhou T, Block TM, Guo J-T. Characterization of the host factors required for hepadnavirus covalently closed circular (CCC) DNA formation. PLoS One. 2012;7:e43270. doi: 10.1371/journal.pone.0043270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Z-H W, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-κB signaling in response to genotoxic stimuli. Science. 2006;311:1141 LP–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 99.Liu H, Zhang H, Wu X, Ma D, Wu J, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–136. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 100.Buchlis G, Podsakoff GM, Radu A, Hawk SM, Flake AW, et al. Factor IX expression in skeletal muscle of a severe hemophilia B patient 10 years after AAV-mediated gene transfer. Blood. 2012;119:3038–3041. doi: 10.1182/blood-2011-09-382317. [DOI] [PMC free article] [PubMed] [Google Scholar]