Abstract
The present work summarizes the major research findings related to wastewater-based epidemiology (WBE) study of COVID-19 and puts forward a conceptual framework, termed as “Surveillance of Wastewater for Early Epidemic Prediction (SWEEP)” for implementation of WBE. SWEEP framework is likely to tackle few practical issues related to WBE and simultaneously proposes refinements to the approach for better outcome and efficiency to save precious lives around the globe. It is observed that the present pandemic offers an opportunity for SWEEP to get included in routine urban water management to put the humankind at front to stop such pandemic in future or at least be prepared to fight against it. With global collaboration, SWEEP can be fine-tuned to meet diverse needs, making the present and future generations resilient to future viral outbreaks. Recent WBE studies conducted to check for the presence of SARS-CoV-2 in wastewater revealed that raw sewage samples tested positive to PCR-based assays while the treated samples showed absence of viral titers. Moreover, the lockdown had a positive impact on decreasing the viral loading in sewage. The proposed SWEEP protocol has an advantage over testifying individuals for predicting the stage of pandemic.
Keywords: COVID-19, Early warning system, SARS-CoV-2, Surveillance of wastewater for early epidemic prediction (SWEEP), PCR-based assays
Graphical abstract
Authors' contributions
Satya Brat Tiwari: Conceptualization, Visualization, Data curation, writing, review & editing. Pallavi Gahlot: Conceptualization, Visualization, Data curation, writing, review & editing. Vinay Kumar Tyagi: Conceptualization, Visualization, Resources, Writing, review & editing, Supervision. Liang Zhang: Writing-review & editing. Yan Zhou: Writing-review & editing. A.A. Kazmi: Writing – review & editing. Manish Kumar: Conceptualization, Visualization, Writing, review & editing
1. Introduction
The occurrence of SARS-CoV-2 RNA in sewage offers an opportunity to use wastewater as a surveillance tool for the invasion, persistence, molecular epidemiology, fate and potential annihilation of the virus from the society. At present, there have been pieces of proof of gastrointestinal disorders triggered by SARS-CoV-2 infections in COVID-19 patients and the occurrence of viral RNA in feces and wastewater (Xiao et al., 2020). Hence there is a critical need to better understand WBE and its impact on human health. Table 1 summarizes the recent findings on wastewater surveillance related to SARS-CoV-2. Most papers related to WBE of SARS-CoV-2 are compiled in COVID-19 WBE Collaborative (https://www.covid19wbec.org/), a collaboration of researchers across the world investigating WBE for COVID-19 (Bivins et al., 2020b). The environmental monitoring and sewage surveillance might help determine whether the virus remains contagious in sewage or not (viable or non-viable) irrespective of symptomatic or asymptomatic patients.
Table 1.
Recent studies on SARS-CoV-2 related to wastewater surveillance.
| Country (State/city) | Sampling period | Sample type and sampling method | Take away | Reference |
|---|---|---|---|---|
| Brazil (Santa Catalina) | Oct 2019–Mar 2020 | Raw wastewater; six sampling events | SARS-CoV-2 detected in sewage samples collected and preserved can reveal the history of infecting pathogen collected in November 2019. Virus circulating much before first clinically confirmed case (late January 2020). | Fongaro et al., (2020) |
| Italy (Northern) | Oct 2019–Feb 2020 | Raw wastewater samples | Samples collected in December 2019 tested positive while the first documented indigenous COVID-19 case in Italy was in Feb 2020. | La Rosa et al., (2021) |
| Netherlands (many locations) | Three days in Feb. (Pre- outbreak), two days in early Mar. (outbreak started) and mid-Mar (outbreak spread) 2020 | Raw wastewater; composite sampling | First detection of SARS-CoV-2 fragments in wastewater, globally. WBE useful even in low prevalence regions. Predicted the contagion even before clinical reporting of cases. | Medema et al., (2020a) |
| China (Zhijiang) | Feb 2020 | Raw, partially disinfected, and disinfected sewage samples | All samples negative for viral culture of SARS-CoV-2, but viral RNA found in untreated samples. No RNA signals in disinfected samples. | Wang et al., (2020) |
| China (Wuhan) | Feb–Mar 2020 | Influent and effluent; 3 samplings | Current wastewater disinfection method recommended by WHO should be re-evaluated for decentralized disinfection system to effectively deactivate SARS-CoV-2 | Zhang et al., (2020a) |
| Germany | Mar 2020 | Raw wastewater | 10 out of 66 samples tested positive for SARS-CoV-2. Indirect viral transmission may play a minor role in disease spread. | Döhla et al., (2020) |
| USA (Massachusetts) | Mar 2020 | Raw wastewater | Viral concentration higher than clinical cases reported. Samples can be stored at 4 °C for more than a week without much loss of signal. | Wu et al., (2020a) |
| Australia (Brisbane) | Feb–Apr 2020 | Raw wastewater; composite and grab | Monte Carlo simulation can be used to predict the median range of affected individuals in the wastewater treatment plant (WWTP) catchment. Numbers predicted by WBE were similar to clinical observation. |
Albastaki et al., (2020) |
| China (Wuhan) | Mar–Apr 2020 | Raw and treated wastewater | TNO (The Netherlands Organization) SARS-CoV-2 detected in treated effluent are most often free from SARS-CoV-2 genes. but found in raw wastewater in some cases. Probable risk due to aerosolization of hospital wastewater poses transmission risk. |
Zhang et al., (2020c) |
| France (Paris) | Mar–Apr 2020; pre- and during lockdown period | Raw wastewater | RNA concentration in sewage increased concomitantly with reported cases. Lockdown had a positive impact on decreasing viral loading in sewage. | Wurtzer et al., (2020) |
| Israel | Mar–Apr 2020 | Raw wastewater | Proof-of-concept study on WBE. Presented a “dose-dependent” curve relating viral surveillance with number of infected persons. | Bar-Or et al., (2020) |
| Italy (Milano, Monza/Brianza) |
Apr 2020 (2 days) | Raw and treated wastewater and river samples; grab sampling | Raw sewage tested positive to PCR amplification while treated samples were negative. Decrease in RNA concentration followed clinical trend. Viral infectiveness was insignificant in treated sewage and rivers though some positive PCR signal was obtained in river water. | Rimoldi et al., (2020) |
| Japan (Ishikawa and Toyama Prefectures) | Mar–Apr 2020 | Raw wastewater; grab sampling | SARS-COV-2 RNA detected using different PCR based assays. Virus was detected even at low disease burden. | Hata et al., (2020) |
| Pakistan (many locations) | Mar–Apr 2020 | Raw wastewater | RT-qPCR assay useful for WBE but need to improve the sensitivity of detection of SARS-CoV-2 in wastewater. | Sharif et al., (2020) |
| Spain (Ourense) | Apr 2020 | Influent, primary sludge, secondary sludge, sludge thickener, digested sludge; grab samples | Sludge thickener is the suitable location for sample collection for detecting SARS-CoV-2 particles using WBE. | Balboa et al., (2020) |
| Spain (Valencia) | Feb–Apr 2020 | Raw and treated wastewater | Treated wastewater tested negative for viral titers. RNA fragments increased and plateaued in sewage faster than reported cases showing sensitivity of WBE to the contagion. | Randazzo et al., (2020a) |
| USA (Montana) | Mar–Apr 2020 | Raw wastewater; Manual and autosampler | Composite sampling is most reliable to get daily trend of viral concentration in sewage. Viral RNA concentration decreased with time suggesting efficacy social isolation measures. | Nemudryi et al., (2020) |
| India (Ahmedabad) | May 2020 | Influent and effluent samples | Samples tested positive for SARS-CoV-2 RNA in influent but not in treated wastewater. | Kumar et al., (2020) |
| Turkey (Istanbul) | May 2020 | Primary sludge and waste activated sludge (before dewatering); Grab sampling | First study globally on the fate of SARS-CoV-2 in sludge. Detection of SARS-CoV-2 in primary sludge like waste activated sludge. | Alpaslan Kocamemi et al., (2020) |
| USA (New York) | May 2020 | Raw wastewater; 24-h composite | Identify and optimize viral genome fragments from wastewater. Suggested ultracentrifugation with 50% sucrose cushion. | Green et al., (2020) |
| USA (Norfolk) | May 2020 | Raw wastewater; Grab and composite sampling | Grab sampling useful for determining SARS-CoV-2 concentrations but for additional calculations (like viral load), composite sampling is better. | Curtis et al., (2020) |
| Ecuador (Quito) | June 2020 | River water; grab sampling | First study quantifying SARS-CoV-2 in river water contaminated with untreated sewage. WBE useful for low sanitation countries as well. | Guerrero-Latorre et al., (2020) |
| India (Jaipur) | May–June 2020 | Untreated and treated wastewater | Viral genome detected at high ambient temperature (40–50 °C) validating WBE as potential tool for managing outbreaks. Some untreated samples tested positive while all treated samples were negative. | Arora et al., (2020) |
| Japan (Nigata and Kanagawa Prefectures, Tokyo) | May–June 2020 | Raw wastewater | Compare the various combinations of primary concentration and RNA extraction methods to find most suitable process recovery of Pseudomonas phage ϕ6 (a surrogate for enveloped viruses). Process recovery of non-enveloped surrogate (MS2) did not indicate that it is suitable for enveloped surrogate (ϕ6) as well. | Torii et al., (2020) |
| Mexico | Apr–July 2020 | Influent, effluent, and secondary sludge; grab sampling | Secondary sludge had higher RNA level than influent suggesting that total solids in wastewater may play a vital role in adsorbing RNA fragments. | Carrillo-Reyes et al., (2020) |
| Russia (Kazan) | Mar–July 2020 (2 days) | Raw wastewater; feces and urine samples of COVID-19 infected patients; model wastewater | Novel approach of WBE based on serial dilution of wastewater and prediction of number of infected persons through calibration curve obtained using feces and urine of COVID-19 patients. | Kuryntseva et al., (2020) |
| Argentina (Buenos Aires) | July- Sept 2020 | Raw surface water samples; composite sampling | WBE on surface water contaminated with sewage is useful for tracking COVID-19 in low-income areas (with suboptimal sewerage network) using surface water contaminated with sewage. | Iglesias et al., (2020) |
Sewage surveillance of such disease outbreaks has been extensively used as an early warning system to check for recurrence and magnitude of the viral disease in the community. Authentic and precise viral concentration and quantification methods are required for meticulous detection of virus in waste water. Centrifugal ultrafiltration, polyethylene glycol (PEG) precipitation and aluminium hydroxide (Al(OH)3) flocculation–precipitation are certain methods that are frequently used for concentration of viral fragments in wastewater (La Rosa et al., 2020b; Randazzo et al., 2020b; Zhang et al., 2020a). Evaluation of potential health risks of SARS-CoV-2 through wastewater (Amoah et al., 2020; Carducci et al., 2020; Foladori et al., 2020; Kitajima et al., 2020a; La Rosa et al., 2020a) and integrated use of WBE along with clinical investigation can prove to be economically beneficial for the community/country (Hart and Halden, 2020). In order to achieve sustainable development, the 2030 agenda for sustainable development emphasizes upon maintaining balance between economic growth, social inclusion and environmental protection (https://www.who.int/).
Regular wastewater surveillance could be applied as a non-invasive early-warning tool to aware people to up-to-the-minute COVID-19 infection (https://www.nature.com/articles/d41586-020-00973-x). According to the wastewater surveillance studies conducted in Netherlands, Sweden, and the US, SARS-CoV-2 can emerge in feces within three days of infection, much earlier than the time taken for people to appear visible symptoms required for official diagnosis i.e., 14 days incubation time (https://www.nature.com/articles/d41586-020-00973-x). Although measures are taken, such as social distancing or lockdowns, to suppress the epidemic by slowing down the spread of the virus, the virus might relapse once such measures are withdrawn. Wastewater monitoring might be used to evaluate the effectiveness of precautions like lockdowns and social distancing since early diagnosis can reduce the severity of the outbreak. In this paper, we have tried to embrace how wastewater surveillance can serve as an early warning system for COVID-19 outbreaks by analysing the presence of the virus or viral components in wastewater. The present work summarizes the major research findings related to WBE studies of COVID-19, a comparative account of SARS-CoV-2 wastewater surveillance in various countries, challenges of wastewater based epidemiological surveillance, and its effect on society and economy. This paper puts forward, a novel conceptual framework, termed as “Surveillance of Wastewater for Early Epidemic Prediction (SWEEP)” for implementation of WBE. By collaborating globally, SWEEP can be calibrated to meet diverse needs, making the present and future generations resilient to future viral outbreaks. Our aim through this paper is to promote SWEEP for routine urban water management to help mankind combat and mitigate the pandemic.
2. Surveillance of contaminated water for SARS-CoV-2
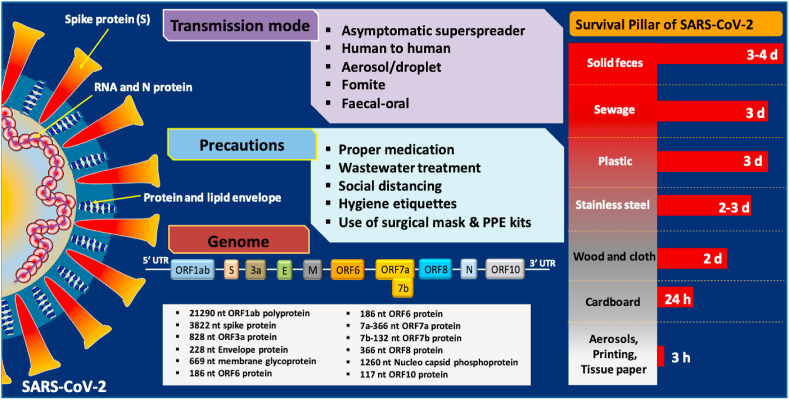
It is imperative to understand that we do not detect viable viruses in the water but the fragmented genes of SARS-CoV-2. Thus, we first need to understand the difference and similarities and its genome structure. SARS-CoV-2 is marked by the presence of single-stranded, positive-sense RNA genome, surrounded by spikes and protein envelope (Fig. 1 ). Four major structural proteins of SARS-CoV2 are the spike surface glycoprotein (S), a small envelope protein (E), matrix protein (M), and nucleocapsid protein (N), which are located in the 3′-terminus of the SARS-CoV-2 genome (Wu et al., 2020). The spike surface glycoprotein plays a vital role in binding to receptors on the host cell. The differences in amino acid substitutions could be used to study the functionality and pathogenesis of the SARS-CoV-2.
Fig. 1.
Transmission of SARS-CoV-2 and precautionary measures for COVID-19 along with its structure and genomic sequence. Also, survival of SARS-CoV-2 on different surfaces/medium (Chin et al., 2020; Nghiem et al., 2020).
Novel coronavirus or SARS-CoV-2 shows high similarity with SARS-CoV but has rapid human-to-human transmission in comparison with SARS-CoV and MERS-CoV (Chan et al., 2020). Enveloped viruses are surrounded by a lipid membrane and a capsid protein encapsulating the viral genome (either ss/dsDNA or ss/dsRNA). All the three viruses i.e. SARS-CoV, MERS-CoV and SARS-CoV-2 belong to Betacoronavirus (βCoV) and have emerged as highly contagious outbreaks with severe mortality rates.
3. Perspectives and challenges of wastewater-based epidemiology (WBE)
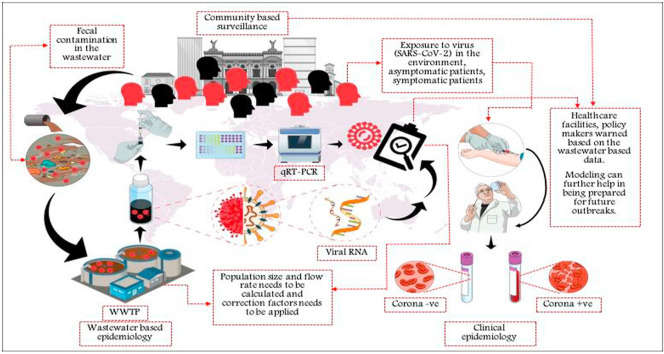
WBE can be a potent tool for mass and quick surveillance, especially during pandemic situations. Recent studies on SARS-CoV-2 highlighted WBE as a potential early warning system (refer Table 1). A schematic of the wastewater based epidemiological surveillance as an efficient early warning system (EWS) for COVID-19 and future outbreaks is represented in Fig. 2 , and discussed comprehensively in subsequent sections and subsections ( (see Fig. 3).
Fig. 2.
Schematic of wastewater based epidemiological surveillance (Source: Suthar et al., 2021; Randazzo et al., 2020b) Reprinted with permission from Elsevier.
Fig. 3.
SWEEP- Surveillance of wastewater for early epidemic prediction.
3.1. Perspectives
3.1.1. Tackling water and health security
Worldwide, 2.2 and 4.2 billion people face the lack of safe drinking water access, and basic sanitation facilities, respectively (Heller et al., 2020). Directly or indirectly, the water and wastewater sectors are bound to shape the current and future pandemics. As such, there is a need for further studies to understand the occurrence and behaviour of SARS-CoV-2 in water and wastewater. The spread of COVID-19 and other previous contagions like SARS (2002, China), Ebola (2013, Guinea, Liberia, and Sierra Leone) and MERS (2012, Saudi Arabia) highlight the need for a rigorous understanding of their behaviour, especially in water and wastewater. Using this understanding, subsequent preventive measures to control its spread can be taken.
Prior studies have indicated that coronaviruses originating from the fecal discharge of infected patients can exist in a viable state in sewage and hospital wastewater (Hung, 2003; Leung et al., 2003). Studies summarized in Table 1 have found SARS-CoV-2 fragments in sewage across the world with different positive rates. Coronaviruses have been reported to survive in sewage to some extent if it is not disinfected adequately (Naddeo and Liu, 2020). Viruses tend to colonize in the suspended solids in the water environment. Besides, bacteria colonizing the biofilm in the piping network can also provide viral stability. However, the survival and infectious nature of SARS-CoV-2 in the water environment are not yet clearly understood. Bivins et al. (2020a) found that time required for 1-log10 reduction (T90) of viable SARS-CoV-2 at room temperature is 1.5 and 1.7 days in wastewater and tap water, respectively. The average T90 of SARS-CoV-2 RNA varied from 8.04 to 27.8 days for untreated wastewater and 9.40–58.60 days for dechlorinated tap water, depending on storage temperature (Ahmed et al., 2020c). Its survival in/on different medium/surfaces based on recent studies are 3–4 days in solid feces, 3 days in sewage, 3 days on plastic, 2–3 days on stainless steel, 2 days on wood and cloth, and 3 h in aerosols, and on printing and tissue papers (Chin et al., 2020; Nghiem et al., 2020) (Fig. 1).
Coronaviruses can be transmissible in sewage up to several days and even longer in drinking water and thus the aerosolization of contaminated water and its exposure to humans can lead to infections (Casanova et al., 2009; Hung, 2003). For example, there was a community outbreak in a residential apartment in Hong Kong in 2003 due to aerosolization of infected water droplets from leaking sewage pipe (Hung, 2003). Aerosolization is also possible from shower heads (Naddeo and Liu, 2020). Thus aerosol-human transmission can be an important route for the spread of the disease.
The viral load in human excreta is not yet clearly understood but can surely provide idea regarding expected viral concentration in wastewater, fecal-oral transmission as well as correlation between wastewater viral load and rate of community infection. The viral load of coronaviruses in stool sample is much lower as compared to enteric viruses. According to studies conducted by Han et al. (2020)(Han et al., 2020) and Lescure et al. (2020) (Lescure et al., 2020), the viral load in stool samples of SARS-CoV-2 patients varied from 1.7 × 106–4.1 × 107 gc/mL and 6.3 × 106–1.26 × 108 gc/g, respectively. However, in case of anal swabs it was 105 gc/swab (Wölfel et al., 2020).
Disinfection is crucial for removing pathogens from wastewater. According to the new guidelines released by the US Occupational Safety and Health Administration (OSHA) in February 2020, the existing disinfection technology being used in WWTPs will likely protect wastewater workers and the public from COVID-19 (https://www.osha.gov/SLTC/covid-19/standards.html). Some studies listed in Table 1 also found the disinfection technology in WWTP in their research to be effective. However, additional research is needed to identify residual dosage and contact time for effective disinfection (Naddeo and Liu, 2020).
3.1.2. Early detection
WBE is useful to mitigate a disease outbreak by detecting it at the onset. In 2013, wild poliovirus type-1 (WPV1) was detected in the sewage from West Bank and the Gaza strip, while Israel or Palestine reported no paralytic polio cases (https://www.who.int/csr/don/2013_09_20_polio/en/). Based on this information WHO and regional authorities acted swiftly to contain the disease. Similarly, in countries like Egypt (Blomqvist et al., 2012) and Sweden (Hellmér et al., 2014), WBE has been recognized as an important early warning system. Medema and co-workers (2020) (Medema et al., 2020a) found that the SARS-CoV-2 virus RNA could be detected in the wastewater before the cases were clinically reported in the Netherlands. The RNA concentration increased in wastewater with the spread of the epidemic. Thus, WBE is important as a clinical surveillance tool, especially with asymptomatic behaviour of many viral diseases and their under-diagnosis (Johansson et al., 2014; Qi et al., 2018).
Further, some viruses like Saffold virus, Cosavirus and Salivirus/Klassevirus are seldom or never revealed by epidemiological monitoring methods (Bonanno Ferraro et al., 2020; Kitajima et al, 2014, 2015 bib_Kitajima_et_al_2015 bib_Kitajima_et_al_2014). SARS-CoV-2 can also cause asymptomatic infections (Lai et al., 2020; Mizumoto et al., 2020; Nishiura et al., 2020; Tang et al., 2020) and thus relying solely on clinical trial would be wastage of time (7–10 days) to control the epidemic locally (Mallapaty, 2020). Early detection is especially important for developing countries, which may lack necessary clinical testing resources and medical facilities to detect the epidemic at the onset and its control in later stages. WBE can also detect various strains of viruses through phylogenetic analysis, thus helping in the assessment of temporal and spatial evolution of SARS-CoV-2 (Nemudryi et al., 2020) from previously known enteric viruses (Bisseux et al., 2018; La Rosa et al., 2014; Lodder et al., 2013). It can even be used when a novel virus has entered a population (Savolainen-Kopra et al., 2011; Sinclair et al., 2008), making it useful for future outbreaks as well. SARS-CoV-2 has been observed in feces, sputum, blood and respiratory secretions of the symptomatic persons (Lescure et al., 2020; J. Wang et al., 2020; Zhang et al., 2020b) (https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf), though further research is required in the case of feces and urine (Wigginton and Boehm, 2020). All these eventually become part of wastewater and provides a “pooled sample” for detection.
3.1.3. Mass detection
The clinical trials for the mitigation of pandemic are of great concern globally. The global inequality and inadequate facilities in terms of labs, technicians, testing kits etc. might lead to further aggravation of COVID-19 crisis, especially in the poorer regions (Ahmed et al., 2020b; Wu et al., 2020b). Even the most advanced country cannot test every individual in the country repeatedly. WBE can supplement clinical surveillance. Testing wastewater from a single treatment plant can capture more than a million persons (Mallapaty, 2020). Hart and Halden (2020) (Hart and Halden, 2020) used computational simulation to suggest that theoretically, one symptomatic/asymptomatic infected case can be detected per 100 to 2 million non-infected persons, subject to the local variables. Ahmed and his co-workers (2020) (Ahmed et al., 2020b) used WBE and Monte Carlo simulation to arrive at a median range of infected persons, which was in “reasonable agreement with the clinical observation”. WBE can give us real-time status and trend of the outbreak spread, helping in informed decision making (Daughton, 2020) and soothing the nerves. Unlike the current situation where COVID-19 incidence is heavily biased towards the regions which are testing aggressively. In the other areas with insufficient testing, people are confused, and decision-makers are unsure about the next step. Mass detection can also reduce the stigma which infected persons and their families face due to individual diagnosis (Murakami et al., 2020). Stigmatization is related to racial profiling at the community level (Zhai and Du, 2020) and “finger-pointing” at the global level. WBE detours ethical and controversial issues like privacy, data sharing and tracking. Most importantly, promoting WBE inherently involves improving sanitation infrastructure, which is a seminal component for achieving most of the sustainable development goals (SDGs).
3.1.4. WBE, economy and society
Globally, the lockdown has been a powerful tool to curb the COVID-19 pandemic. However, the one-size-fits-all approach causes unnecessary social and economic hardship in the non-contagious areas. The estimated loss to the world economy due to the COVID-19 epidemic could be more than $10 trillion (Vos et al., 2020). International Labour Organization (ILO) estimated that full or partial lockdown affected 2.7 billion workers, that is, around 81% of the global workforce (https://www.ilo.org/wcmsp5/groups/public/---dgreports/---dcomm/documents/briefingnote/wcms_740877.pdf). Worldwide, poverty and hunger will exacerbate due to the pandemic (Vos et al., 2020), further accentuating the inequality.
Sustainable Development Goals (SDGs) seek to curb inequality around the world and create a better future. Goal 6: Clean water and sanitation requires robust sewerage network and efficient WWTPs, which is also very important for wastewater surveillance. WBE can help to formulate a “smart lockdown” or a graded response if the contagion can be pinned down to specific areas promptly. It is a much cheaper and faster tool compared to clinical surveillance. It is estimated that Germany, despite the highest testing capacity in Europe, will take approximately three months to test its entire population once (Hart and Halden, 2020). Conversely, Germany can use its around 9600 WWTPs and WBE to cover its entire population within 24–48 h, costing just 0.014% of the clinical surveillance (Hart and Halden, 2020). Studies in the Netherlands and France showed that RNA concentration in wastewater increased and decreased concomitantly with the number of infected persons (Medema et al., 2020b; Wurtzer et al., 2020). It will help in evaluating whether steps like lockdown and quarantine are effective or not (Kitajima et al., 2020a) and even predict the second wave of outbreak. Real-time monitoring will curb the public resentment against the lockdown and nudge them to behave responsibly as “seeing is believing” (https://www.sciencemag.org/news/2020/04/coronavirus-found-paris-sewage-points-early-warning-system).
Quantitative microbial risk assessment (QMRA), when integrated with WBE, can be useful in reducing the impact of COVID-19 pandemic (Haas, 2020; Wigginton and Boehm, 2020). QMRA predicts the chances of infection or illness based on the environmental concentration of an infectious agent and its exposure dose (Haas, 2020). Based on it, vital information like a gap for appropriate social distancing, indoor ventilation rate, and best disinfectant can be recommended (Kitajima et al., 2020b). Concerted global effort must be directed towards WBE research and its implementation. It will help tackle the current problems associated with WBE.
3.2. Challenges with WBE
Collecting samples and conducting research during a lockdown and ethical dilemma of diverting testing resources away from the health facilities are some basic issues (Mallapaty, 2020). Other problems are discussed in detail as follows:
3.2.1. Sample collection
A recent study showed that data variability between biological replicates was higher in the case of manual sampling than the autosampler (Nemudryi et al., 2020). Besides, there may be inconsistency due to different sampling methods (grab v/s composite), precipitation-induced dilution of wastewater, and stability of virus and genome fragments in sewage. SARS-CoV-2 may not be detected from the remote locations in the sewerage network due to a short lifespan in the wastewater. It will lead to false-negative results if the sampling is done only at the WWTP for a large sewerage network. However, the persistence of SARS-CoV-2 in wastewater is currently an open question; though, few studies have been done on coronaviruses regarding this (Chin et al., 2020; Kampf et al., 2020).
3.2.2. Sample processing and analysis
Coronaviruses are non-waterborne and hence not found abundantly in wastewater like water-borne viruses such as adenoviruses, astroviruses, hepatitis A and E viruses (Xagoraraki and O'Brien, 2020). In no or low infected regions, a large volume of water may have to be processed for data validity. Besides, there is no standard and optimized protocol for SARS-CoV-2 detection or quantification in sewage (Kitajima et al., 2020a). The group also attributed inconsistent results in isolating culturable virus from feces to the differences in protocols (W. Wang et al., 2020; Wölfel et al., 2020; Zhang et al., 2020). The viral concentration methods to extract enveloped viruses from water samples are inefficient (Haramoto et al., 2009; Ye et al., 2016), and most previous studies established these methods for non-enveloped enteric viruses (Haramoto et al., 2018). Even for enteric viruses, recovery using virus concentration methods would depend on the types of viruses and water (Haramoto et al., 2018). Moreover, SARS-CoV-2 has quite different structural and physical properties from enteric viruses (Kitajima et al., 2020a). We also need a better understanding of viral shedding in urine and feces as it varies from person-to-person and during the course of infection. Also, sample collection related problems and inadequate clinical testing (for cross-verification) make quantifying the number of infected persons difficult and uncertain.
The RT-qPCR assays being used for WBE are meant for clinical testing (Kitajima et al., 2020a) and thus not perfect for a heterogeneous sample like wastewater as it can lead to PCR inhibition (due to impurities or irrelevant genes). Often, serial dilution of the sample is used to reduce the inhibitions. For Biosafety Level (BSL)-3 and 4 microbes, lab facilities are limited and often used for other purposes like vaccine development and understanding the basic biology of organisms (Bibby et al., 2017). There are only about 54 upcoming or operational BSL-3+ or 4 globally, mostly in North America and Western Europe (https://www.who.int/ihr/publications/WHO-WHE-CPI-2018.40/en/). Thus, the researcher must rely on less pathogenic CoV strains like Murine Coronavirus (MHV) as a model for SARS-CoV-2 (Kitajima et al., 2020a; Naddeo and Liu, 2020). We also have a limited understanding of occurrence, persistence, and removal of SARS-CoV-2 in wastewater, partly due to the lack of previous studies on CoVs (Kitajima et al., 2020a). Further, animal studies, exposure pathways, dose-response curves, etc. are unavailable (or very less) for COVID-19 and conducting QMRA is very difficult at this stage.
4. Surveillance of Wastewater for Early Epidemic Prediction (SWEEP)
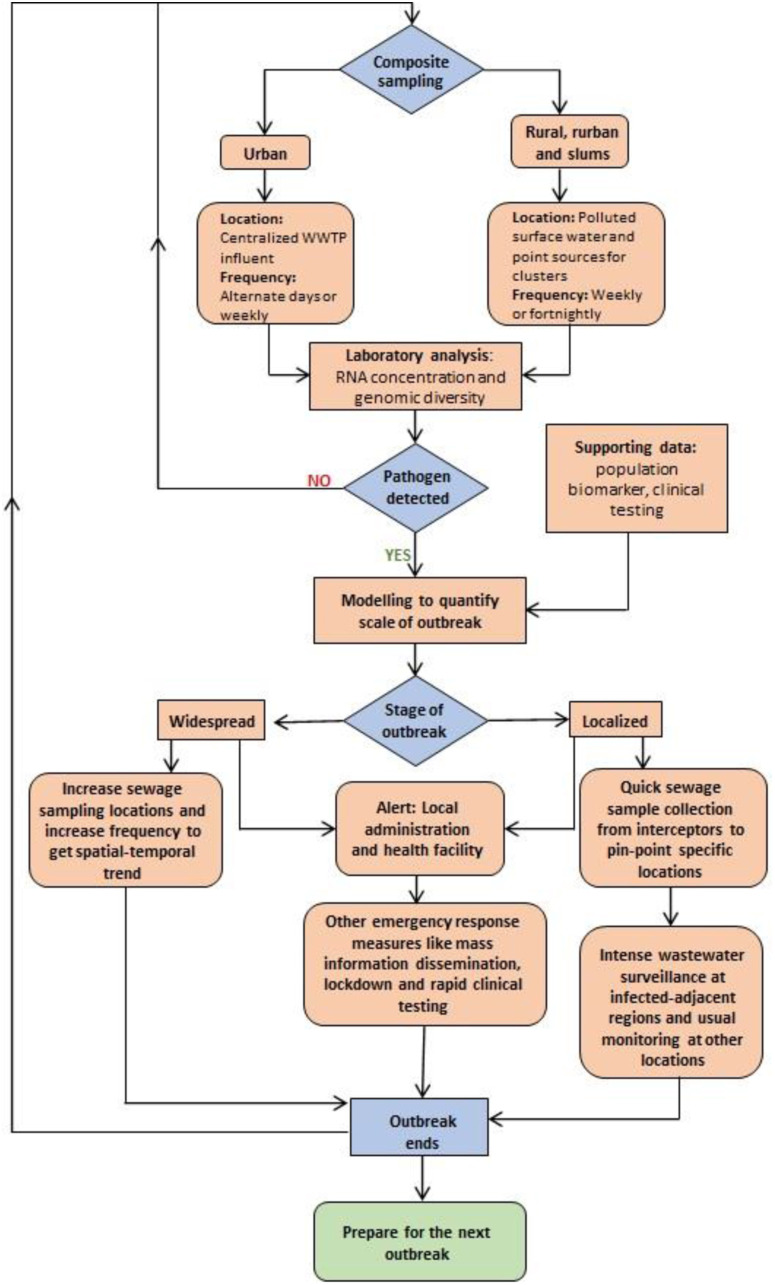
Through International Health Regulations (2005) (IHR), WHO requires countries to report events that are unusual, severe, likely to spread and prone to hamper international travel and trade (https://www.who.int/csr/disease/epidemic-focus/global-epidemic-response/en/). Such events can be promptly identified with a robust epidemic early warning system, which is direly needed as the events in the last two decades show. The next infectious disease could be more lethal and contagious, more so, as antimicrobial-resistant genes will become more common due to the greater use of disinfectant, mainly due to COVID-19 (Naddeo and Liu, 2020). United Nations International Strategy for Disaster Reduction (UNISDR) (2004) defines early warning systems (EWS) as the process of timely and useful information dissemination so that affected individuals can act to decrease or avoid the risk and prepare for the effective response. Four interdependent aspects of EWS are risk knowledge, monitoring and warning service, dissemination and communication, and response capability (https://www.undp.org/content/dam/rbec/docs/UNDP%20Brochure%20Early%20Warning%20Systems.pdf). These aspects are relevant throughout the continuum of pandemic phases (https://www.who.int/influenza/preparedness/pandemic/GIP_PandemicInfluenzaRiskManagementInterimGuidance_Jun2013.pdf?ua=1). Although the report discusses EWS in the context of natural hazards and climate risk, it broadly applies to epidemics as well. We hereby propose a further modified wastewater surveillance systems to incorporate the aspects of EWS and termed it as the Surveillance of Wastewater for Early Epidemic Prediction (SWEEP) (Fig. 3 ). It will have the following striking features:
-
(a)
Composite sampling: As mentioned in Table 1, composite sampling should be preferred over grab sampling. Grab sampling may suffice to find SARS-CoV-2 concentrations in wastewater, but it can give erroneous result for additional subsequent calculations (like viral load) (Curtis et al., 2020). A higher frequency of sampling is suggested for urban areas (especially tourist and commercial centres) because of the higher flux of the population. Rural, semi-urban, and slums often lack sewerage network, so surface water bodies are sampled where sewage is dumped. Sites with a higher probability of environment-human transmission should be included (like wet markets, slums, etc.). Further, during an outbreak/epidemic, frequency and sampling locations must be increased.
-
(b)
Laboratory analysis: It includes concentration, quantification, and identification of genome material as well as the infectiveness of the target agent. Current WBE studies on SARS-CoV-2 have used various concentration methods like ultrafiltration, ultracentrifugation, PEG precipitation, and electronegative membrane-direct RNA extraction (Kitajima et al., 2020a). A study conducted by Ahmed et al., 2020a, Ahmed et al., 2020c used murine hepatitis virus (MHV) as a control for SARS-CoV-2 surveillance in wastewater. The group used seven different virus concentration methods and found that out of centrifugal filter device methods, polyethylene glycol precipitation (PEG 8000) and ultracentrifugation method, adsorption-extraction with MgCl2 pre-treatment was most effective for viral concentration. They may be used for future outbreaks as well. Similarly, cell culture, PCR, qPCR, RT-qPCR, and advanced sequencing methods like nanopore sequencing may be used to quantify genomic diversity. RT-qPCR of heterogenous sample like wastewater is a challenge but serial dilution of the sample might prove beneficial to fulfil sensitivity of PCR based assays. Mao et al. (2020) (Mao et al., 2020) developed a portable paper-based device for on-site tracking of SARS-CoV-2 in wastewater. The group developed an analytical method to quantitate viral nucleic acids through agarose gel electrophoresis (AGE), a robust molecular method. This analytical method is fast, overcomes limitations of PCR, and might enable rapid real-time screening of potentially infected and asymptomatic patients. Thus, technology of laboratory analysis is constantly evolving, and the future looks optimistic.
-
(c)
Supporting data: It is useful for cross-verification of lab analysis data and as input parameters for modelling and inference. Flow rate, temperature, pH, biochemical oxygen demand (BOD) and chemical oxygen demand (COD) are some important parameters, which are already monitored in most WWTPs, and can be used as supporting data. Population biomarkers like creatinine and cholesterol can be quantified to estimate the population catered by the WWTPs. Other important data sources are hospitals (clinical testing, mortality rate, etc.) and government records (census data, immigration/outmigration, hydrological).
-
(d)
Modelling: It provides a “bigger picture” and predicts future outcomes based on the current trend. It helps to quantify the shedding rate and kinetic decay rate of the virus and its genome fragments (accounting to environmental conditions and virus type). Comparison of modelling data with clinical surveillance data can help to understand the trend in expected v/s actual infection rate and estimate the number of asymptomatic and/or pre-symptomatic cases.
-
(f)
Stage of outbreak: WBE is evolving and may not perfectly forecast the spread of infectious disease. If the outbreak is emergent/recent/localized, a different approach may be required compared to the widespread outbreak.
-
(g)
Other aspects of EWS: Complementary efforts like sensitizing the public about the outbreak and precautionary steps, instant message, and app-based display of WBE data from local WWTPs can be done. It also includes updating protocols for WWTP operators and waste handlers to keep them safe from infections.
-
(h)
Preparedness for the next epidemic: COVID-19 has shown that epidemics/pandemics can be far more damaging than other natural or anthropogenic disasters like earthquake, terrorism etc. Hence, global community must devote the attention to WBE that it deserves as an early warning system.
5. Conclusions
It is clear that COVID-19 is here to stay for the foreseeable future. Even after having vaccine and medicine for it, we will still need WBE as new strains are emerging. are . As a lot of research on WBE has already been done in the last 15 years. COVID-19 tailored WBE approach can be quickly made and implemented. It is heartening that funding on coronavirus research has jumped significantly (https://www.nature.com/articles/d41586-020-01314-8), but dedicated investment has to go into WBE and specifically into WBE as early warning system (EWS). United Nations Development Program (UNDP) has identified a lack of funding and ineffective institutions as a major roadblock to a functional EWS. We should not wait for the pandemic to act. The proposed SWEEP protocol has an advantage over testifying individuals for predicting the stage of pandemic. The SWEEP implementation requires suitable modelling at the core for effective prediction. Paper-based rapid testing kits could be used for on-site detection at wastewater treatment plants and track the source(s) as probable COVID-19 porters in local areas. Thus, regular wastewater investigation could be applied as a non-clinical early-warning tool to aware societies to new COVID-19 infections.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Authors are thankful to Department of Biotechnology-Government of India (GrantNo. BT/RLF/Re-entry/12/2016) for financial support to this research.
References
- Ahmed F., Ahmed N., Pissarides C., Stiglitz J. Why inequality could spread COVID-19. Lancet Public Heal. 2020;5 doi: 10.1016/S2468-2667(20)30085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191:110092. doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albastaki A., Naji M., Lootah R., Almeheiri R., Almulla H., Almarri I., Alreyami A., Aden A., Alghafri R. First confirmed detection of SARS-COV-2 in untreated municipal and aircraft wastewater in Dubai, UAE: the use of wastewater based epidemiology as an early warning tool to monitor the prevalence of COVID-19. Sci. Total Environ. 2020;143350 doi: 10.1016/j.scitotenv.2020.143350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpaslan Kocamemi B., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 detection in istanbul wastewater treatment plant sludges. medRxiv. 2020 doi: 10.1101/2020.05.12.20099358. [DOI] [Google Scholar]
- Amoah I.D., Kumari S., Bux F. Coronaviruses in wastewater processes: source, fate and potential risks. Environ. Int. 2020 doi: 10.1016/j.envint.2020.105962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S., Nag A., Rajpal A., Brat Tiwari S., Sethi J., Rajvanshi J., Saxena S., Srivastava S., Kazmi A.A., Kumar V., Kumar Tyagi V. Detection of SARS-CoV-2 RNA in fourteen wastewater treatment systems in Uttarakhand and Rajasthan States of North India. 2020. medRxiv. [DOI]
- Balboa S., Mauricio-Iglesias M., Rodríguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. 2020. The fate of SARS-CoV-2 in wastewater treatment plants points out the sludge line as a suitable spot for incidence monitoring. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O., Abu-Ali H., Ronen Z., Rinott E., Lewis Y.E., Friedler E., Bitkover E., Paitan Y., Berchenko Y., Kushmaro A. 2020. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burdelun in the population: a proof-of-concept for quantitative environmental surveillance. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Aquino de Carvalho N., Wigginton K. Research needs for wastewater handling in virus outbreak response. Environ. Sci. Technol. 2017 doi: 10.1021/acs.est.6b06492. [DOI] [PubMed] [Google Scholar]
- Bisseux M., Colombet J., Mirand A., Roque-Afonso A.-M., Abravanel F., Izopet J., Archimbaud C., Peigue-Lafeuille H., Debroas D., Bailly J.-L., Henquell C. Monitoring human enteric viruses in wastewater and relevance to infections encountered in the clinical setting: a one-year experiment in central France, 2014 to 2015. Euro Surveill. 2018;23 doi: 10.2807/1560-7917.ES.2018.23.7.17-00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., de los Reyes F.L., Delgado Vela J., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Blomqvist S., El Bassioni L., El Maamoon Nasr E.M., Paananen A., Kaijalainen S., Asghar H., de Gourville E., Roivainen M. Detection of imported wild polioviruses and of vaccine-derived polioviruses by environmental surveillance in Egypt. Appl. Environ. Microbiol. 2012;78:5406–5409. doi: 10.1128/AEM.00491-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanno Ferraro G., Mancini P., Veneri C., Iaconelli M., Suffredini E., Brandtner D., La Rosa G. Evidence of Saffold virus circulation in Italy provided through environmental surveillance. Lett. Appl. Microbiol. 2020;70:102–108. doi: 10.1111/lam.13249. [DOI] [PubMed] [Google Scholar]
- Carducci A., Federigi I., Dasheng L., Julian R.T., Marco V. Making waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020 doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Reyes J., Barragán-Trinidad M., Buitrón G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process Eng. 2020:101815. doi: 10.1016/j.jwpe.2020.101815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S., Tsoi H.-W., Lo S.K.-F., Chan K.-H., Poon V.K.-M., Chan W.-M., Ip J.D., Cai J.-P., Cheng V.C.-C., Chen H., Hui C.K.-M., Yuen K.-Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. The Lancet Microbe. 2020;1:e10. doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis K., Keeling D., Yetka K., Larson A., Gonzalez R. 2020. Wastewater SARS-CoV-2 concentration and loading variability from grab and 24-hour composite samples. medRxiv. [DOI] [Google Scholar]
- Daughton C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. Sci. Total Environ. 2020;726:138149. doi: 10.1016/j.scitotenv.2020.138149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhla M., Wilbring G., Schulte B., Kümmerer B.M., Diegmann C., Sib E., Richter E., Haag A., Engelhart S., Eis-Hübinger A.M., Exner M., Streeck H., Schmithausen R.M. 2020. SARS-CoV-2 in environmental samples of quarantined households. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fongaro G., Hermes Stoco P., Sobral Marques Souza D., Grisard E.C., Magri M.E., Rogovski P., Schorner M.A., Hartmann Barazzetti F., Christoff A.P., de Oliveira L.F.V., Bazzo M.L., Wagner G., Hernandez M., Rodriguez-Lazaro D. SARS-CoV-2 in human sewage in Santa Catalina, Brazil, November 2019. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- Green H., Wilder M., Middleton F., Collins M., Fenty A., Gentile K., Kmush B., Zeng T., Larsen D.A. Quantification of SARS-CoV-2 and cross-assembly phage (crAssphage) from wastewater to monitor coronavirus transmission within communities. 2020. medRxiv. [DOI]
- Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Granda M.G., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Total Environ. 2020;743:140832. doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas C.N. Coronavirus and environmental engineering science. Environ. Eng. Sci. 2020;37:233–234. doi: 10.1089/ees.2020.0096. [DOI] [Google Scholar]
- Han M.S., Seong M.W., Heo E.Y., Park J.H., Kim N., Shin S., Cho S.I., Park S.S., Choi E.H. Sequential analysis of viral load in a neonate and her mother infected with severe acute respiratory syndrome coronavirus 2. Clin. Infect. Dis. 2020;71:2236–2239. doi: 10.1093/cid/ciaa447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Katayama H., Ito T., Ohgaki S. Development of virus concentration methods for detection of koi herpesvirus in water. J. Fish. Dis. 2009;32:297–300. doi: 10.1111/j.1365-2761.2008.00977.x. [DOI] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.143578. 143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller L., Mota C.R., Greco D.B. COVID-19 faecal-oral transmission: are we asking the right questions? Sci. Total Environ. 2020;729:138919. doi: 10.1016/j.scitotenv.2020.138919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung L.S. The SARS epidemic in Hong Kong: what lessons have we learned? JRSM. 2003;96:374–378. doi: 10.1258/jrsm.96.8.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N.G., Gebhard L.G., Carballeda J.M., Aiello I., Recalde E., Terny G., Ambrosolio S., L'Arco G., Konfino J., Brardinelli J.I. SARS-CoV-2 surveillance in untreated wastewater: first detection in a low-resource community in Buenos Aires, Argentina. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- Johansson M.A., Vasconcelos P.F.C., Staples J.E. The whole iceberg: estimating the incidence of yellow fever virus infection from the number of severe cases. Trans. R. Soc. Trop. Med. Hyg. 2014;108:482–487. doi: 10.1093/trstmh/tru092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739:139076. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Rachmadi A.T., Haramoto E., Gerba C.P. Quantification and genetic analysis of salivirus/klassevirus in wastewater in Arizona, USA. Food Environ. Virol. 2014;6:213–216. doi: 10.1007/s12560-014-9148-2. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Rachmadi A.T., Iker B.C., Haramoto E., Pepper I.L., Gerba C.P. Occurrence and genetic diversity of human cosavirus in influent and effluent of wastewater treatment plants in Arizona, United States. Arch. Virol. 2015;160:1775–1779. doi: 10.1007/s00705-015-2435-x. [DOI] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746:141326. doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryntseva P., Karamova K., Fomin V., Selivanovskaya S., Galitskaya P. A simplified approach to monitoring the COVID-19 epidemiologic situation using waste water analysis 2 and its application in Russia Corresponding Author. 2020. medRxiv. [DOI]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Della Libera S., Iaconelli M., Ciccaglione A.R., Bruni R., Taffon S., Equestre M., Alfonsi V., Rizzo C., Tosti M.E., Chironna M., Romanò L., Zanetti A.R., Muscillo M. Surveillance of hepatitis A virus in urban sewages and comparison with cases notified in the course of an outbreak, Italy 2013. BMC Infect. Dis. 2014;14:419. doi: 10.1186/1471-2334-14-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., Lucentini L., Suffredini E. SARS-CoV-2 has been circulating in northern Italy since December 2019: evidence from environmental monitoring. Sci. Total Environ. 2021;750:141711. doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Liu Y.H., Wang C.-Y., Wang Y.-H., Hsueh S.-C., Yen M.-Y., Ko W.-C., Hsueh P.-R. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J. Microbiol. Immunol. Infect. 2020;53:404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V., Houhou-Fidouh N., Valette M., Mailles A., Lucet J.-C., Mentre F., Duval X., Descamps D., Malvy D., Timsit J.-F., Lina B., Van-der-Werf S., Yazdanpanah Y. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.K., To K., Chan P.K.S., Chan H.L.Y., Wu A.K.L., Lee N., Yuen K.Y., Sung J.J.Y. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection1 1The authors thank Man-yee Yung, Sara Fung, Dr. Bonnie Kwan, and Dr. Thomas Li for their help in retrieving patient information. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W.J., Rutjes S.A., Takumi K., Husman A.M. de R. Aichi virus in sewage and surface water, The Netherlands. Emerg. Infect. Dis. 2013;19:1222–1230. doi: 10.3201/eid1908.130312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. 2020. How Sewage Could Reveal True Scale of Coronavirus Outbreak. [DOI] [PubMed] [Google Scholar]
- Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. vol. 25. 2020. p. 2000180. (Estimating the Asymptomatic Proportion of Coronavirus Disease 2019 (COVID-19) Cases on Board the Diamond Princess Cruise Ship). Yokohama, Japan, 2020. Eurosurveillance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Hata A., Honda R., Watanabe T. Letter to the editor: wastewater-based epidemiology can overcome representativeness and stigma issues related to COVID-19. Environ. Sci. Technol. 2020;54:5311. doi: 10.1021/acs.est.0c02172. [DOI] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial Perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. Technol. 2020;6:1213–1216. doi: 10.1039/D0EW90015J. [DOI] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Reports Med. 2020;1:100098. doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem L.D., Morgan B., Donner E., Short M.D. The COVID-19 pandemic: considerations for the waste and wastewater services sector. Case Stud. Chem. Environ. Eng. 2020;1:100006. doi: 10.1016/j.cscee.2020.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Kobayashi T., Miyama T., Suzuki A., Jung S., Hayashi K., Kinoshita R., Yang Y., Yuan B., Akhmetzhanov A.R., Linton N.M. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int. J. Infect. Dis. 2020;94:154–155. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi R., Huang Y., Liu J., Sun Y., Sun X., Han H.-J., Qin X.-R., Zhao M., Wang L., Li W., Li J., Chen C., Yu X.-J. Global prevalence of asymptomatic norovirus infection: a meta-analysis. EClinicalMedicine. 2018;2–3:50–58. doi: 10.1016/j.eclinm.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. SSRN Electron. J. 2020 doi: 10.2139/ssrn.3586696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744:140911. doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen-Kopra C., Paananen A., Blomqvist S., Klemola P., Simonen M.-L., Lappalainen M., Vuorinen T., Kuusi M., Lemey P., Roivainen M. A large Finnish echovirus 30 outbreak was preceded by silent circulation of the same genotype. Virus Gene. 2011;42:28–36. doi: 10.1007/s11262-010-0536-x. [DOI] [PubMed] [Google Scholar]
- Sharif S., Ikram A., Khurshid A., Salman M., Mehmood N., Arshad Y., Ahmad J., Angez M., Alam M.M., Rehman L., Mujtaba G., Hussain J., Ali J., Akthar Ri, Malik M.W., Baig Z.I., Rana M.S., Usman M., Ali M.Q., Ahad A., Badar N., Umair M., Tamim S., Ashraf A., Tahir F., Ali N. Detection of SARS-Coronavirus-2 in wastewater, using the existing environmental surveillance network: an epidemiological gateway to an early warning for COVID-19 in communities. 2020. medRxiv. [DOI]
- Sinclair R.G., Choi C.Y., Riley M.R., Gerba C.P. Pathogen surveillance through monitoring of sewer systems. Adv. Appl. Microbiol. 2008;65:249–269. doi: 10.1016/S0065-2164(08)00609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar S., Das S., Nagpure A., Madhurantakam C., Tiwari S.B., Gahlot P., Tyagi V.K. Epidemiology and diagnosis, environmental resources quality and socio-economic perspectives for COVID-19 pandemic. J. Environ. Manag. 2021;280:111700. doi: 10.1016/j.jenvman.2020.111700. (Figure 5 Reprinted with permission from Elsevier) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z., Wang H., Dai Y., Li K., Liu J., Wu W., Yuan C., Yu M., Li P., Yan J. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. 2020;26:1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S., Furumai H., Katayama H. Applicability of polyethylene glycol precipitation followed by acid guanidinium thiocyanate-phenol-chloroform extraction for the detection of SARS-CoV-2 RNA from municipal wastewater. Sci. Total Environ. 2020;143067 doi: 10.1016/j.scitotenv.2020.143067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos R., Martin W., Laborde D. How much will global poverty increase because of COVID-19. Int. Food Policy Res. Inst. 2020 [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in wuhan, China. J. Am. Med. Assoc. 2020;323:1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y., Zhuo L., Zhong Z., Qu T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020;94:103–106. doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. J. Am. Med. Assoc. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigginton K.R., Boehm A.B. Environmental engineers and scientists have important roles to play in stemming outbreaks and pandemics caused by enveloped viruses. Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c01476. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V., Guggemos W., Seilmaier M., Zange S., Müller M., Niemeyer D., Kelly T., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized cases of coronavirus disease 2019. 2020. medRxiv. [DOI]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M., Bushman M., Chai P., Duvallet C., Erickson T., Foppe K., Ghaeli N., Gu X., Hanage W., Huang K., Lee W.L., Matus M., McElroy K., Nagler J., Rhode S., Santillana M., Tucker J., Wuertz S., Zhao S., Thompson J., Alm E. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. 2020. medRxiv. [DOI] [PMC free article] [PubMed]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. 2020. Evaluation of Lockdown Impact on SARS-CoV-2 Dynamics through Viral Genome Quantification in Paris Wastewaters. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I., O'Brien E. Women in Water Quality. Springer; 2020. Wastewater-based epidemiology for early detection of viral outbreaks; pp. 75–97. [DOI] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhai Y., Du X. Mental health care for international Chinese students affected by the COVID-19 outbreak. The Lancet Psychiatry. 2020;7:e22. doi: 10.1016/S2215-0366(20)30089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X., Wang X., Liu Y., Li G., Qu J. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020;741:140445. doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X., Wang X., Liu Y., Li G., Qu J. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020;741:140445. doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Yang Y., Huang X., Jiang J., Li M., Zhang X., Ling H., Li J., Liu Y., Li G., Li W., Yi C., Zhang T., Jiang Y., Xiong Y., He Z., Wang X., Deng S., Zhao P., Qu J. SARS-CoV-2 spillover into hospital outdoor environments. 2020. medRxiv. [DOI] [PMC free article] [PubMed]
- Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020;92:680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]