Abstract
Causes of death, in particular deaths due to infection, have not been widely studied in randomised trials in chronic lymphocytic leukaemia. With long-term follow-up (median 13 years) we examined the cause of death in 600/777 patients in the LRF CLL4 trial. Blood samples, taken at randomisation from 499 patients, were available for identifying gene mutations. Infection was a cause of death in 258 patients (43%). Patients dying of infection were more likely than those who died of other causes to have received ≥2 lines of treatment (194/258 [75%] versus 231/342 [68%], P = 0.04) and to have died in the winter months (149/258 [58%] versus 166/342 [49%], P = 0.03), respectively. In patients with mutation data, the factors significantly associated with death from infection versus all other deaths were 11q deletion (47/162 [29%] versus 40/209 [19%], P = 0.03) and mutations of the BRAF, FBXW7, NRAS and XPO1 genes. Death was caused by an infection in 46/67 assessable patients (69%) who had a mutation of one or more of these four genes versus only 129/333 patients (39%) without any of these mutations (odds ratio: 3.46 [95% CI 1.98–6.07] P < 0.0001). Careful management of infection risk, including prophylaxis against infection, may be important in patients who carry these mutations.
Subject terms: Chronic lymphocytic leukaemia, Cancer genetics, Infectious diseases
Introduction
Although overall survival (OS) has been an endpoint in many clinical trials in chronic lymphocytic leukaemia (CLL), the specific causes of patients’ deaths have not been widely studied in randomised trials. The LRF CLL4 trial is uniquely appropriate for this investigation for two reasons. First, it has long-term follow-up (median 13 years), which allowed us to examine the cause of death in 600 of the 777 patients randomised. Secondly, blood samples taken at randomisation from 499 patients are still available today for an analysis of baseline molecular data. Thus we have been able to examine the relationship between deaths from infection and a large panel of genes which have only relatively recently been found to be commonly mutated in CLL.
Infections are the leading cause of death in CLL, with around half of patients succumbing [1–3]. A study of 10,455 patients with CLL from the Danish Cancer Registry demonstrated a significant improvement in OS between 1978 and 2013, in parallel with the introduction of chemo-immunotherapeutic agents, but the risk of death from infections in CLL patients did not change over this time period. It remained 50% higher than for a matched cohort of 508,995 individuals without CLL [4]. Meanwhile, the organisms causing infections have changed with the introduction of purine analogues and monoclonal antibodies. Formerly mostly common bacterial organisms, they now include less common opportunistic pathogens such as listeria, candida, aspergillus, and pneumocystis jirovecii [2, 5, 6]. It has been suggested that the newer tyrosine kinase inhibitor ibrutinib might reduce the rate of infections [7–9].
The reasons why patients with CLL are susceptible to infections are multifactorial, including both therapy-related immunosuppression and defects of cellular and humoral immunity which are facets of the disease itself and which tend to increase over the course of the disease [3, 5, 10]. Factors may include defective T-cell and B-cell function, with low levels of serum immunoglobulins (hypogammaglobulinaemia) and defects in complement activity and neutrophil/monocyte function, all contributing to profound immune dysregulation [1, 2, 5, 6, 10, 11]. CLL treatment trials have identified various risk factors associated with the incidence of infections. These factors differ according to the treatments given and the patient demographics, but they include two or more previous lines of therapy, advanced disease stage, elevated LDH, low haemoglobin, low baseline IgG and IgA levels, renal insufficiency and older age [1, 5, 6, 8]. The Danish group have also identified factors associated with the severe infection prior to treatment in patients in the Danish National CLL Registry, notably an elevated β2 microglobulin level, low levels of IgA and a shorter time between previous infections [8, 12]. To our knowledge, risk factors in CLL for infections which result in death have not been separately studied, except recently in the setting of Covid-19 [13, 14].
Advances in identifying genes commonly mutated in CLL have led to the identification of several mutations which are linked to an earlier death. In the LRF CLL4 trial mutations of TP53 [15], SF3B1 [16], NOTCH1 (coding [16] and non-coding [17]) ATM with 11q deletion [18], EGR2 [19], and biallelic BIRC3 as well as MAPK-ERK genes [20] have all been shown to be predictive of shorter OS. In this study, we aimed to identify gene mutations which were specifically associated with death caused by infection.
Subjects and methods
LRF CLL4 was a large multi-centre randomised trial of 777 patients with previously untreated CLL. Patients were randomised between 1999 and 2004 to receive chlorambucil or fludarabine, with or without cyclophosphamide. Clinical follow-up was to October 2010 (median 7.1 years; range 6.0–11.7 years). Follow-up for OS was censored at October 2010 for 44 patients who were resident outside the UK but continued until September 2016 for UK-based patients (median 13.3 years follow-up for surviving patients; range 11.9–17.6 years). Causes of death were assessed centrally by the principal investigator, Daniel Catovsky, in his capacity as the single reviewer of all the death certificates and clinicians’ reports. For each patient up to two main causes of death were identified and classed simply as: CLL; Richter’s syndrome; infection; vascular; other cancer; other unclassified cause. Then, for this study, all the patients who had infection listed as one of their two causes of death were categorised as having died of infection. Thus this category did not include patients with incidental infections which were present at the time of death but which were not considered to be a main cause of death. For our primary analysis, the patients who died of infection were contrasted with all the other patients who died, i.e., those whose categorised causes of death did not include infection. A secondary analysis compared these same two groups within the subset of patients with available gene mutation data.
The following demographic, laboratory and molecular characteristics were recorded at randomisation, or assessed in stored blood samples collected at randomisation: age, sex, disease stage, size of the treatment centre, randomised treatment, haemoglobin and platelet levels, white blood count, lymphocyte and prolymphocyte counts, beta-2 microglobulin and lactate dehydrogenase levels, lymphadenopathy, IGHV mutation status, CD38 and Zap70 expression, telomere length, presence of trisomy 12 and deletions of chromosomes 13q, 11q and 17p. The cut-offs used for these variables have been previously published [21]. Not all the variables were assessable in all patients. Data on 623 high-confidence somatic mutations in ATM, BIRC3, BRAF, CHD2, DDX3X, EGR2, FBXW7, KRAS, MED12, MGA, MYD88, NFKBIE, NOTCH1, NRAS, POT1, RPS15, SAMHD1, SETD2, SF3B1, TP53 and XPO1 were available for analysis from our previous work, in 499 patients [20]. Cut-offs to define summer versus winter deaths were: 15th April (start of summer) and 15th October (start of winter)—reversed for patients in the southern hemisphere. World Health Organisation performance status (0 versus 1–4) was measured 6 months after the start of first-line treatment. The number of lines of treatment received and responses to treatments were also considered.
Statistics
Generalised linear modelling was used to identify which of the above variables were associated with deaths from infection versus patients whose causes of death did not include infection. Values of P ≤ 0.05 (two-sided) were considered significant and were included in a multivariate model using forward stepwise analysis. The Kaplan–Meier method was used to estimate OS. STATISTICA software from StatSoft, a subsidiary of Dell, Inc. (Tulsa, OK, USA) was used for the analyses. In addition, for the 21 genes with available data, Fisher’s exact test was used in R v3.3.0, followed up with multiple hypothesis testing.
LRF CLL4 is registered as an International Standard Randomised Controlled Trial, number NCT 58585610 and was approved by the UK multicentre research ethics committee. Informed consent was obtained from all subjects.
Results
In the LRF CLL4 trial, 614 of 777 patients (79%) died before the end of follow-up. The cause of death was known in 600 patients. Deaths tended to have more than one cause, but CLL was a cause in at least 520/600 patients (87%), including 258 deaths (43%) from infection (Fig. 1A).
Fig. 1. Mortality in the LRF CLL4 trial (n = 777).
A Causes of death. *‘Various’ included 21 cardiovascular causes and 46 other cancers. B Fatal infections. Note: this is a diagrammatic representation of the data. The areas of the circles correspond to the number of patients in each category, but the areas of the overlapping sections do not.
Fatal infections were pneumonia (67%), and/or sepsis (38%) and/or opportunistic infections (11%) (Fig. 1B). The latter group consisted of fungal infections (n = 9, 4%), mainly aspergillus, plus a range of other infections (n = 16, 7%) including adenoviral enteritis, clostridium difficile colitis, cryptosporidium colitis, cytomegalovirus, encephalitis, hepatitis, klebsiella, legionella pneumonia, listeria, necrotising fasciitis, parainfluenza and tuberculosis.
Of 137 patients who died from infection before the end of clinical follow-up in 2010, 59 (43%) died within 6 calendar months of the start of a line of treatment. Thus 13 died during first-line, 31 during second-line and 15 during third-line treatment. The remaining 57% died of infection when they were off treatment.
Comparing patients who died from infection versus those who died of all other causes
In the full dataset of 600 patients whose cause of death was known, the patients who died from infection were more likely than those whose death was not due to infection to have received two or more lines of treatment (Table 1A). Deaths from infection were more likely to occur during the winter, while deaths not due to infectious causes were evenly spread between the summer and winter months. Among the panel of 21 genes commonly mutated in CLL, univariate analysis showed that mutations of BRAF, FBXW7, NRAS and XPO1 were significantly associated with death from infection versus deaths not due to infection. However, NRAS was the only one of these recurrently mutated genes to remain significantly associated with death from infection after Benjamini and Hochberg false discovery adjustment (FDR, Q > P [P < 0.05]) (odds ratio: 17.51, 95% confidence interval (CI): 2.20–139.40, P = 0.0004); (data not shown). No other significant differences were found between patients who died of infection versus those who died from other causes, with respect to any other of the demographic, disease or treatment characteristics listed in the “Subjects and methods”. In particular, the rate of deaths from infection was not influenced by disease stage, the randomised treatment, the response to treatment, or the size/experience of the CLL treatment centre, as measured by the number of patients entered in the trial.
Table 1.
Univariate and multivariate analyses of baseline variables associated with deaths from infection versus deaths not due to infection.
| Variable | Deaths from infection (%) | Deaths not due to infection (%) | Odds ratio | 95% confidence levels | P-value |
|---|---|---|---|---|---|
| A. Univariate analysis: all trial patients with a known cause of death (n = 600) | |||||
| ≥2 lines of treatment | 194/258 (75) | 231/342 (68) | 1.46 | 1.01–2.09 | 0.04 |
| Winter season | 149/258 (58) | 166/342 (49) | 1.45 | 1.05–2.01 | 0.03 |
| BRAF mutation | 19/175 (11) | 9/225 (4) | 2.92 | 1.29–6.63 | 0.01 |
| FBXW7 mutation | 8/175 (5) | 2/225 (1) | 5.34 | 1.12–25.48 | 0.04 |
| NRAS mutation | 9/175 (5) | 1/225 (0.4) | 12.14 | 1.52–96.79 | 0.02 |
| XPO1 mutation | 17/175 (10) | 9/225 (4) | 2.58 | 1.12–5.94 | 0.03 |
| B. Univariate analysis: patients with a known cause of death and gene mutation data (n = 400) | |||||
| 11q deletion | 47/162 (29) | 40/209 (19) | 1.73 | 1.06–2.80 | 0.03 |
| BRAF mutation | 19/175 (11) | 9/225 (4) | 2.92 | 1.29–6.63 | 0.01 |
| FBXW7 mutation | 8/175 (5) | 2/225 (1) | 5.34 | 1.12–25.48 | 0.04 |
| NRAS mutation | 9/175 (5) | 1/225 (0.4) | 12.14 | 1.52–96.79 | 0.02 |
| XPO1 mutation | 17/175 (10) | 9/225 (4) | 2.58 | 1.12–5.94 | 0.03 |
| C. Multivariate analysis: patients with a known cause of death and gene mutation data (n = 400) | |||||
| 11q deletion | 1.79 | 1.09–2.94 | 0.02 | ||
| BRAF mutation | 3.22 | 1.33–7.80 | 0.01 | ||
| NRAS mutation | 10.78 | 1.31–88.43 | 0.03 | ||
| XPO1 mutation | 2.49 | 1.01–6.12 | 0.05 | ||
Of the 499 patients in the trial for whom gene mutation data were available, 411 died. Of these, 400 had a known cause of death. In the univariate analysis in this subset of 400 patients, 11q deletion was significantly associated with death from infection versus deaths not due to infection, along with mutations of the BRAF, FBXW7, NRAS and XPO1 genes (Table 1B). Multivariate analysis in these 400 patients showed that the factors most significantly associated with death from infection were 11q deletion and mutations of the BRAF, NRAS and XPO1 genes (Table 1C).
Of the 499 patients with gene mutation data, 73 (15%) carried one or more of the four gene mutations BRAF (6%), FBXW7 (2%), NRAS (2%) and XPO1 (6%). Of these, 67 (92%) died and only six (8%) remained alive (Fig. 2). Death was caused by an infection in 46 of the 67 who died (69%). In contrast, in the patients who died but did not carry any of the four mutations, an infection was a cause of death in only 129/333 (39%). Alternatively, of those who died of infection 46/175 assessable patients (26%) carried one of these four mutations versus only 21/225 (9%) of those who died of causes which did not include infection. (Odds ratio: 3.46 [95% CI: 1.98–6.07] P < 0.0001).
Fig. 2. Incidence of BRAF, FBXW7, NRAS and XPO1 mutations in deaths from infection, other deaths and patients remaining alive (n = 73).
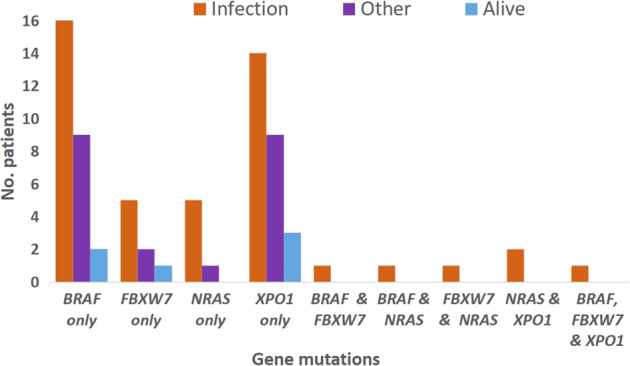
73/499 patients with gene mutation data carried one or more of these mutations.
Details of the patients with BRAF, FBXW7, NRAS and/or XPO1 mutations
Of the 30 BRAF-mutated patients in the trial 19 (63%) died of infection, nine of other causes and two survived (Fig. 2). Eleven patients had an FBXW7 mutation of whom eight (73%) died of infection, two died of other causes and one survived. There were ten NRAS-mutated patients of whom nine (90%) died of infection and one died of another cause. The single patient with an NRAS mutation who did not die of an infection nevertheless had an episode of severe and prolonged neutropenic sepsis after her first course of chlorambucil, followed by pneumocystis pneumonia 3 years later after completing second-line treatment with fludarabine. Of 29 trial patients with an XPO1 mutation, 17 (59%) died of infection, nine died of other causes and three survived. All six of the patients who carried more than one of these mutations died of infection.
The types of fatal infection were similar in these 46 patients to those for the trial as a whole (Fig. 1B), totalling 34 cases of pneumonia, 13 of sepsis, two of other infections and two with unspecified infections.
The demographics, clinical characteristics, and prognostic biomarkers of patients with versus without BRAF, FBXW7, NRAS, and/or XPO1 mutations are shown in Table 2. Patients with one or more of these mutations were significantly more likely than other patients to have unmutated IGHV genes, positive CD38 and ZAP70 expression, trisomy 12, absence of 13q deletion and a KRAS and/or NOTCH1 mutation. They were significantly less likely than others to have an SF3B1 mutation.
Table 2.
Demographics, clinical characteristics, and prognostic biomarkers in patients with versus without BRAF, FBXW7, NRAS, and/or XPO1 mutations (n = 499a).
| Patient/disease characteristic | BRAF, FBXW7, NRAS, and/or XPO1 mutation | P | |
|---|---|---|---|
| Yes % (n = 73) | No % (n = 426) | ||
| Aged >70 years | 34 | 30 | 0.5 |
| Male | 78 | 71 | 0.2 |
| Binet stage Cb | 36 | 31 | 0.4 |
| Randomised arm chlorambucil | 52 | 47 | 0.5 |
| Non-response to treatment | 21 | 21 | 1.0 |
| ≥2 lines of treatment | 73 | 66 | 0.3 |
| Unmutated IGHV genes (>98% homology)a | 84 | 59 | 0.0002 |
| β2 microglobulin ≥4 mg/La | 55 | 47 | 0.3 |
| CD38 positive (cut-off 7%)a | 78 | 61 | 0.02 |
| Zap-70 positive (cut-off 10%)a | 65 | 47 | 0.02 |
| Trisomy 12a | 29 | 14 | 0.003 |
| 11q deletiona | 24 | 20 | 0.5 |
| Absence of 13q deletiona | 62 | 37 | 0.0001 |
| 17p deletiona | 8 | 5 | 0.5 |
| ATM mutation | 8 | 7 | 0.8 |
| BIRC3 mutation | 12 | 6 | 0.07 |
| EGR2 mutation | 1 | 3 | 0.5 |
| KRAS mutation | 15 | 4 | 0.0003 |
| NOTCH1 mutation | 21 | 11 | 0.02 |
| SF3B1 mutation | 15 | 26 | 0.05 |
| TP53 mutation | 15 | 9 | 0.09 |
aEight variables had missing data, with the no. of cases ranging from 356 to 464.
bBinet stage C disease was not significant because, while patients with these gene mutations were more likely than others to have anaemia (haemoglobin < 100 g/L; 33% versus 18%; P = 0.003), they were less likely to have thrombocytopenia (platelets < 100 × 109/L; 10% versus 21%; P = 0.02).
Overall survival
Median OS from randomisation for the whole trial was 6 years, with surviving patients censored at their last follow-up date. OS was the same for patients who died of infection as it was for those who died of other causes: median 4 years 6 months versus 4 years 5 months, respectively. For the 67 patients carrying just one of the four mutations, there was no significant difference in OS between those who died from infection and those whose deaths were not due to infectious causes (median 4 years 1 month and 4 years 5 months, respectively). The median OS of the six patients who had more than one of the four mutations was 2 years 2 months.
Discussion
By 13 years median follow-up 79% of the patients in LRF CLL4 had died, the majority due to CLL or its complications, including infection, which was a cause in 43% of deaths. We do not have information on the particular pathogens involved, but two-thirds of deaths from infection had a respiratory origin.
Well-known adverse prognostic factors, associated with shorter OS in CLL, such as older age, male sex, TP53 or NOTCH1 abnormalities, or unmutated IGHV genes, were not significantly different between patients who died from infection and those who died from other causes. Many of these factors are also associated with poor response to treatment, as previously reported [21], but in this study patient who failed to respond to treatment succumbed to their CLL, either with or without infection in equal measure. Although stage C disease has been associated with a greater likelihood of severe infections [8], in this trial patients with stage C disease at randomisation were at no greater risk of death from infection than patients with stage A-progressive or stage B disease. Deletion of 11q was associated with death from infection in the subset of patients with mutation data, though not in the full trial dataset.
CLL is characterised by progressive immunodeficiency [2, 3, 8]. We found that patients who had undergone two or more lines of treatment were at greater risk of death from infection, consistent with previous reports associating longer disease duration with a greater likelihood of severe infections [5, 6, 22, 23], especially in heavily pre-treated patients with active disease [10]. In CLL severe infections are already common prior to treatment, with a five-year cumulative incidence of 31% among 2905 CLL patients [8], while documented infections may even predate CLL diagnosis by years, or decades, increasing in incidence over time [24]. This latter finding suggests that infections might contribute to CLL development through antigen stimulation, inducing somatic mutations, or epigenetic modifications in line with the antigenic drive in lymphoid malignancies [24]. Several of the gene polymorphisms implicated in CLL risk by GWAS suggest a key role for aberrant immune responses in CLL pathogenesis, suggesting that cumulative immune dysfunction is critical to CLL development [25].
With this in mind, our finding is intriguing but unsurprising that specific gene mutations, identified in samples obtained prior to initial treatment, namely those of the BRAF, FBXW7, NRAS and XPO1 genes, may, after further disease progression, be associated with infections so severe they result in death.
A possible mechanism for this has been reported with respect to NRAS, which is an oncogene involved in regulating cell division. When the human NRAS gene was activated in vitro, through point mutation in codon 61, in cells which were incubating the delNS1 H5N1 influenza virus, a significantly increased replication of the virus was observed in these cells compared with controls, while at the same time expression of IRF-3 and IFN-beta decreased [26]. The authors concluded that NRAS activated through point mutation, appeared to inhibit IFN antivirus defences and afford a more favourable environment for replication of the delNS1 influenza virus. In a similar experiment, Metzger et al. demonstrated that a mutated-NRAS recombinant virus was capable of transforming cells [27]. It should be noted that these findings apply to viral infections only and hence cannot be translated to bacterial or fungal infections.
BRAF mutations result in deregulation of B-cell receptor (BCR) intracellular signalling leading to disruption of hematopoietic and early B-cell differentiation [28]. FBXW7 is a potent tumour suppressor which is also involved in maintaining normal haematopoiesis. Mounting evidence has indicated the involvement of aberrant expression of FBXW7 for tumorigenesis [29]. An XPO1 mutation is a driving event in B cell malignancies through alteration of the nuclear trafficking of proteins involved in inflammatory signalling, DNA repair, RNA export and chromatin remodelling pathways [30]. Thus these three gene mutations may each be involved in CLL pathogenesis. BRAF, like NRAS, is a component in the MAPK/ERK signalling pathway, which may be usurped by DNA and RNA viruses to mediate multiple aspects of the virus infectious cycle and thus facilitate viral replication and subsequent pathogenesis [31]. But we are not aware of any studies, like that cited above for the NRAS gene, which might cast light on the possible mechanisms leading to a susceptibility to fatal infections in patients with a BRAF mutation, nor in those with an FBXW7 or XPO1 mutation.
The question arises whether patients who die of infection have a different disease course from those who die of other causes. IgA abnormalities at diagnosis have been shown to predict for both subsequent infections and a more aggressive disease course [9]. Another proposal is that infection may be the driver of more aggressive disease through tumour microenvironment interactions [12, 24]. Vendramini et al. showed a strong association between KRAS/NRAS/BRAF mutations and the presence of unmutated IGHV genes and trisomy 12 [32]. Similarly, we found that unmutated IGHV genes and trisomy 12, as well as KRAS mutations, were strongly associated with BRAF/FBXW7/NRAS/XPO1 mutations, suggesting a complex involvement of these mutations in CLL pathogenesis. We have previously shown an association of BRAF and NRAS mutations with shorter OS in this trial [20]. Our data in the present study (Fig. 2) suggest this may be explained by the tendency of patients with these mutations to succumb to fatal infections. Patients who died of other causes were less likely to present with these mutations.
It is hoped that an understanding of the molecular events underlying the susceptibility to infections in some patients may lead to improvements in infection control and targeted treatments. The main limitations of this study were: (1) the small number of patients with each of these gene mutations; (2) the ending of clinical follow-up in 2010, with the result that we have no data on the treatments received by these patients since then; and (3) the fact that the randomised treatments in LRF CLL4 are no longer in general use, raising the question as to whether deaths from infection may follow a different pattern in the era of BCR and BCL2 inhibitors. Other researchers may want to examine their databases to see if their findings match ours.
In conclusion, patients in LRF CLL4 were at some risk of death from infection, irrespective of their demographic characteristics, disease stage and treatment history. Nevertheless, those who had received two or more lines of treatment were particularly at risk, as were those who carried a BRAF, FBXW7, NRAS or XPO1 mutation. A meta-analysis of datasets from other trials could be important to assess the validity of the link between these gene mutations and deaths from infections in patients with CLL. Careful management of infection risk, including appropriate choice of CLL therapeutic agents and prophylaxis against infection, may be important in patients who carry one or more of these mutations.
Acknowledgements
The authors gratefully acknowledge all the patients and clinicians who contributed to this trial. We would also like to thank: David Gonzalez, Alison Morilla, Vasantha Brito-Babapulle, Zadie Davis, Ruth Clifford, Helen Parker, Matthew Rose-Zerilli and Jade Forster who performed the laboratory studies; Rachel Wade who gave statistical advice; Rachel Clack, Julie Burrett and Roger Else, who collated the data. The LRF CLL4 trial was funded by a core grant from Blood Cancer UK. ME was supported by the Arbib Charitable Fund. Laboratory studies were supported by the Kay Kendall Leukaemia Fund (KKL584), Blood Cancer UK (11052, 12036, 12050), Wessex Medical Research, Cancer Research UK (C328/A2738 and C328/A2737) and an educational grant from Schering Health Care (UK) and Schering AG (Germany).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Monica Else, Stuart J. Blakemore
References
- 1.Ravandi F, O’Brien S. Immune defects in patients with chronic lymphocytic leukemia. Cancer Immunol Immunother. 2006;55:197–209. doi: 10.1007/s00262-005-0015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamblin AD, Hamblin TJ. The immunodeficiency of chronic lymphocytic leukaemia. Br Med Bull. 2008;87:49–62. doi: 10.1093/bmb/ldn034. [DOI] [PubMed] [Google Scholar]
- 3.Forconi F, Moss P. Perturbation of the normal immune system in patients with CLL. Blood. 2015;126:573–81. doi: 10.1182/blood-2015-03-567388. [DOI] [PubMed] [Google Scholar]
- 4.da Cunha-Bang C, Simonsen J, Rostgaard K, Geisler C, Hjalgrim H, Niemann CU. Improved survival for patients diagnosed with chronic lymphocytic leukemia in the era of chemo-immunotherapy: a Danish population-based study of 10455 patients. Blood Cancer J. 2016;6:e499. doi: 10.1038/bcj.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wadhwa P, Morrison VA. Infectious complications of chronic lymphocytic leukemia. Semin Oncol. 2006;33:240–9. doi: 10.1053/j.seminoncol.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Morrison VA. Infectious complications of chronic lymphocytic leukaemia: pathogenesis, spectrum of infection, preventive approaches. Best Pr Res Clin Haematol. 2010;23:145–53. doi: 10.1016/j.beha.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Sun C, Tian X, Lee YS, Gunti S, Lipsky A, Herman SE, et al. Partial reconstitution of humoral immunity and fewer infections in patients with chronic lymphocytic leukemia treated with ibrutinib. Blood. 2015;126:2213–9. doi: 10.1182/blood-2015-04-639203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen MA, Eriksen CT, Brieghel C, Biccler JL, da Cunha-Bang C, Helleberg M, et al. Incidence and predictors of infection among patients prior to treatment of chronic lymphocytic leukemia: a Danish nationwide cohort study [letter] Haematologica. 2018;103:e300–e303. doi: 10.3324/haematol.2017.182006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishdorj G, Streu E, Lambert P, Dhaliwal HS, Mahmud SM, Gibson SB, et al. IgA levels at diagnosis predict for infections, time to treatment, and survival in chronic lymphocytic leukemia. Blood Adv. 2019;3:2188–98. doi: 10.1182/bloodadvances.2018026591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dearden C. Disease-specific complications of chronic lymphocytic leukemia. Hematol Am Soc Hematol Educ Program. 2008; 450–6. 10.1182/asheducation-2008.1.450. [DOI] [PubMed]
- 11.Morrison VA. Infectious complications in patients with chronic lymphocytic leukemia: pathogenesis, spectrum of infection, and approaches to prophylaxis. Clin Lymph Myeloma. 2009;9:365–70. doi: 10.3816/CLM.2009.n.071. [DOI] [PubMed] [Google Scholar]
- 12.Agius R, Brieghel C, Andersen MA, Pearson AT, Ledergerber B, Cozzi-Lepri A, et al. Machine learning can identify newly diagnosed patients with CLL at high risk of infection. Nat Commun. 2020;11:363. doi: 10.1038/s41467-019-14225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarfò L, Chatzikonstantinou T, Rigolin GM, Quaresmini G, Motta M, Vitale C, et al. COVID-19 severity and mortality in patients with chronic lymphocytic leukemia: a joint study by ERIC, the European Research Initiative on CLL, and CLL Campus. Leukemia. 2020;34:2354–63. doi: 10.1038/s41375-020-0959-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mato AR, Roeker LE, Lamanna N, Allan J, Leslie LA, Pagel JM, et al. Outcomes of COVID-19 in patients with CLL: a multicenter, international experience. Blood. 2020;136:1134–43. doi: 10.1182/blood.2020006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29:2223–9. doi: 10.1200/JCO.2010.32.0838. [DOI] [PubMed] [Google Scholar]
- 16.Oscier DG, Rose-Zerilli MJJ, Winkelmann N, Gonzalez de Castro D, Gomez B, Forster J, et al. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood. 2013;121:468–75. doi: 10.1182/blood-2012-05-429282. [DOI] [PubMed] [Google Scholar]
- 17.Larrayoz M, Rose-Zerilli MJ, Kadalayil L, Parker H, Blakemore S, Forster J, et al. Non-coding NOTCH1 mutations in chronic lymphocytic leukemia; their clinical impact in the UK CLL4 trial. Leukemia. 2017;31:510–4. doi: 10.1038/leu.2016.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rose-Zerilli MJ, Forster J, Parker H, Parker A, Rodri AÉ, Chaplin T, et al. ATM mutation rather than BIRC3 deletion and/or mutation predicts reduced survival in 11q-deleted chronic lymphocytic leukemia: data from the UK LRF CLL4 trial. Haematologica. 2014;99:736–42. doi: 10.3324/haematol.2013.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young E, Noerenberg D, Mansouri L, Ljungström V, Frick M, Sutton LA, et al. EGR2 mutations define a new clinically aggressive subgroup of chronic lymphocytic leukemia. Leukemia. 2017;31:1547–54. doi: 10.1038/leu.2016.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blakemore SJ, Clifford R, Parker H, Antoniou P, Stec-Dziedzic E, Larrayoz M, et al. Clinical significance of TP53, BIRC3, ATM and MAPK-ERK genes in chronic lymphocytic leukaemia: data from the randomised UK LRF CLL4 trial. Leukemia. 2020;34:1760–74. doi: 10.1038/s41375-020-0723-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oscier D, Wade R, Davis Z, Morilla A, Best G, Richards S, et al. Chronic Lymphocytic Leukaemia Working Group, UK National Cancer Research Institute. Prognostic factors identified three risk groups in the LRF CLL4 trial, independent of treatment allocation. Haematologica. 2010;95:1705–12. doi: 10.3324/haematol.2010.025338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molica S. Infections in chronic lymphocytic leukemia: risk factors, and impact on survival, and treatment. Leuk Lymph. 1994;13:203–14. doi: 10.3109/10428199409056283. [DOI] [PubMed] [Google Scholar]
- 23.Young JA. Epidemiology and management of infectious complications of contemporary management of chronic leukemias. Infect Disord Drug Targets. 2011;11:3–10. doi: 10.2174/187152611794407755. [DOI] [PubMed] [Google Scholar]
- 24.Andersen MA, Rostgaard K, Niemann CU, Hjalgrim H. Antimicrobial use before chronic lymphocytic leukemia: a retrospective cohort study. Leukemia. 2020; e-pub ahead of print 20 July 2020; 10.1038/s41375-020-0980-0. [DOI] [PubMed]
- 25.Law PJ, Berndt SI, Speedy HE, Camp NJ, Sava GP, Skibola CF, et al. Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukaemia. Nat Commun. 2017;8:14175. doi: 10.1038/ncomms14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, Zhou HB, Zou W, Jin ML. Effect of human activated NRAS on replication of delNS1 H5N1 influenza virus in MDCK cells. Virol J. 2011;8:240. doi: 10.1186/1743-422X-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metzger MJ, Miller AD. Acutely transforming retrovirus expressing NRAS generated from HT-1080 fibrosarcoma cells infected with the human retrovirus XMRV. J Virol. 2010;84:7908–10. doi: 10.1128/JVI.00389-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damm F, Mylonas E, Cosson A, Yoshida K, Della Valle V, Mouly E, et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 2014;4:1088–101. doi: 10.1158/2159-8290.CD-14-0104. [DOI] [PubMed] [Google Scholar]
- 29.Sailo BL, Banik K, Girisa S, Bordoloi D, Fan L, Halim CE, et al. FBXW7 in cancer: what has been unraveled thus far? Cancers. 2019;11:246. doi: 10.3390/cancers11020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tueur G, Lazarian G, Eclache V, Fleury C, Letestu R, Lévy V, et al. Prevalence, distribution and predictive value of XPO1 mutation in a real-life chronic lymphocytic leukaemia cohort [letter] Br J Haematol. 2020;191:e90–e94. doi: 10.1111/bjh.17046. [DOI] [PubMed] [Google Scholar]
- 31.DuShane JK, Maginnis MS. Human DNA virus exploitation of the MAPK-ERK cascade. Int J Mol Sci. 2019;20:3427. doi: 10.3390/ijms20143427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vendramini E, Bomben R, Pozzo F, Benedetti D, Bittolo T, Rossi FM, et al. KRAS, NRAS, and BRAF mutations are highly enriched in trisomy 12 chronic lymphocytic leukemia and are associated with shorter treatment-free survival. Leukemia. 2019;33:2111–5. doi: 10.1038/s41375-019-0444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]



