Abstract
Bone Morphogenetic Protein (BMP) signaling regulates diverse biological processes. Upon ligand binding, BMP receptors (BMPRs) phosphorylate SMAD1/5 and other non-canonical downstream effectors to induce transcription of downstream targets. However, the precise role of individual BMP receptors in this process remains largely unknown due to the complexity of downstream signaling and the innate promiscuity of ligand-receptor interaction. To delineate unique downstream effectors of individual BMPR1s, we analyzed the transcriptome of human umbilical endothelial cells (HUVECs) expressing three distinct constitutively active BMPR1s of which expression was detected in endothelial cells (ECs). From our analyses, we identified a number of novel downstream targets of BMPR1s in ECs. More importantly, we found that each BMPR1 possesses a distinctive set of downstream effectors, suggesting that each BMPR1 is likely to retain unique function in ECs. Taken together our analyses suggest that each BMPR1 regulates downstream targets non-redundantly in ECs to create context-dependent outcomes of the BMP signaling.
Keywords: BMP signaling, SMAD, BMPR1, Transcriptomics, Endothelial cell
1. INTRODUCTION
Bone Morphogenetic Proteins (BMPs), which are members of the Transforming Growth Factor-β (TGFβ) superfamily, function as essential regulators in diverse physiological processes (1–3). BMP signaling is transduced by two distinct sets of receptors; Type 1 receptors (BMPR1s) and Type 2 receptor (BMPR2). These two types of receptors form heterotetrameric complexes composed of two BMPR1s and two BMPR2s upon ligand binding (4–6). BMPR2, which is a constitutively active Receptor Ser/Thr Kinase (RSTK), phosphorylates BMPR1s within the complex and therefore, enables BMPR1s to function as RSTKs to induce activation of key downstream effectors, including SMAD1/5/8 (7). In the human genome, four BMPR1s, Activin A Receptor Like Type 1 (ACVLR1/ALK1), Activin A Receptor Type 1 (ACVR1/ALK2), BMP Receptor Type 1 A (BMPR1A/ALK3), and BMP Receptor Type 1 B (BMPR1B/ALK6), have been identified (8, 9). Considering that there is only one BMPR2 is present in human genome, it is possible that BMPR1s may be critical to elicit the specificity of signaling outcomes upon ligand binding.
In endothelial cells (ECs), BMP signaling modulates diverse behaviors of ECs in a context-dependent manner, by providing either pro-angiogenic or anti-angiogenic cues depending on the identity of the BMP ligand. In general, BMP2/4/6 generally appear to function as pro-angiogenic factors via ALK2, ALK3, or ALK6, while BMP9/10 act as anti-angiogenic factors via ALK1 (10–12). Consistent with this idea, endothelial specific deletion of Alk2 or Alk3 in mice has been reported to attenuate angiogenesis, while similar deletion of Alk1 in mice caused excessive angiogenesis (11). Interestingly, regardless of their nature as pro-angiogenic or anti-angiogenic cues, stimulation with BMP ligands or activation of cognate BMPR1s induces phosphorylation of canonical downstream effectors, SMAD1/5/8 and subsequent transcription of ID1/2/3 in ECs (7). Therefore, it is likely that the context-dependent outcomes of BMP signaling in ECs may not stem from activation of canonical downstream targets. Moreover, it seems plausible that each BMPR1 may possess a set of unique downstream targets in addition to these canonical downstream effectors, which could serve as a key factor to create context-dependent outcomes of BMP signaling in ECs. However, to date, relatively little is known about unique downstream targets of individual BMPR1s in ECs.
In this report, we analyzed the transcriptomic signature of human umbilical vein endothelial cells (HUVECs) expressing constitutively active ALK1, ALK2, or ALK3 to identify unique transcriptional changes induced by activation of individual BMPR1s. In addition, we identified a number of specific downstream targets for each BMPR1 in ECs. To validate our results, we examined a subset of newly identified transcriptional targets of ALK2 activation in detail in ECs. Taken together, our findings provide compelling evidence that each BMPR1 retains unique properties in activating downstream effectors, illustrating their functional divergence in ECs at a molecular level.
2. METHODS
Cell culture, treatment, and siRNA knockdown
Human Umbilical Vein Endothelial Cells (HUVEC) (Gibco), Human Dermal Lymphatic Endothelial Cells (HDLEC) (PromoCell), Human Pulmonary Artery Endothelial Cells (HPAEC) (PromoCell) and Human Aortic Endothelial Cells (HAEC) (PromoCell) were cultured in Endothelial Cell Growth Medium MV 2 supplemented with Growth Medium MV 2 SupplementMix (PromoCell) and penicillin/streptomycin (Gibco) at 37°C with 5% CO2 in a humidified incubator. Cells from passage 4 to 6 were used. For chemical treatment, HUVECs were treated with 5 μM DMH1 (Sigma-Aldrich), 10 μM U0126 (Sigma-Aldrich) for 12 hours.
For BMP ligand treatment, HUVECs were incubated with serum-free Basal Medium MV2 (PromoCell) for 6 hours. Subsequently, HUVECs were treated with 10 ng/mL BMP9 (R&D Systems), 50 ng/mL BMP6 (R&D Systems), 50 ng/mL BMP4 (R&D Systems), or 50 ng/mL BMP2 (R&D Systems) for 30 min or 2 hours. For knockdown, ACVRL1, ACVR1, BMPR1A, SMAD1/5, or control siRNA (GenePharma) were delivered to HUVECs using Lipofectamin RNAiMAX (Invitrogen) according to manufacturer’s instructions. For knockdown experiments, RNA or protein were extracted 48 hours after siRNA treatment.
Construction of adenoviral plasmids
pDONR223-ACVRL1 was a gift from William Hahn & David Root (Addgene plasmid # 23873; http://n2t.net/addgene:23873; RRID:Addgene_23873) (13). pCMV5-ALK2 Q207D was a gift from Jeff Wrana (Addgene plasmid # 11740; http://n2t.net/addgene:11740; RRID:Addgene_11740) (14). pCMV5-hBMPR-1A CA was a gift from Lee Niswander & Peter ten Dijke (Addgene plasmid # 49527; http://n2t.net/addgene:49527; RRID:Addgene_49527) (15). The constitutively activated (CA) ACVRL1 mutant (Q201D) construct was generated by site-directed mutagenesis using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). Adenoviral plasmids were constructed by AdEasy Adenoviral Vector System (Agilent Technologies). Each construct was inserted into pShuttle-CMV vector after restriction enzyme digestion using PmeI (NEB). Subsequently, linearized vectors were electroporated into BJ5183-AD-1 Electrocompetent Cells (Agilent Technologies). Recombinants were identified by PacI restriction enzyme (NEB) digestion.
Adenovirus transduction
Approximately 2.2 × 106 293A cells (Invitrogen) were plated in 60 mm dish 24 hours before transfection. 20μg of adenoviral vectors were digested with PacI, ethanol precipitated, and used for transfection with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. After six hours, culture medium was replaced with the growth medium. After one week of transfection, cells were collected and washed three times with Dulbecco’s Phosphate-Buffered Saline. Adenoviruses were subsequently released by three times of freezing-and-thawing, the soup was harvested and amplified in 293A cells. Amplified viruses were tittered using Adeno-X Rapid Titer Kit (Clontech Laboratories).
Single cell RNA-seq analysis
Single cell RNA-seq datasets were downloaded from EMBL-EBI ArrayExpress database (accession number: E-MTAB-8077) and re-analyzed using Seurat R package (v. 3.5.1) (16). Variable genes were identified based on variance stabilizing transformation using the FindVariableFeatures function with mean.cutoff = c(0.1, 8), dispersion.cutoff = c(1, Inf) parameters. Data was scaled and reduced by principal component analysis using the ScaleData and RunPCA (npcs = 100) functions, respectively. Subsequently, shared nearest neighbor-based clustering was performed to cluster cells using the FindNeighbors and FindClusters functions with resolution = 0.1, algorithm = 1 (Louvain algorithm) parameters. Data was then visualized using the RunUMAP function with min.dist = 0.3, dims = 1:100 parameters.
Analysis of SMAD1/5 ChIP-seq dataset
SMAD1/5 ChIP-seq datasets were downloaded from NCBI GEO database (accession number: GSE27661) and re-analyzed (17). Reads were aligned to hg18 human referenge genome using Bowtie2 and peaks were called using MACS2. Aligned reads were converted to bedgraph and visualized using IGV software (Broad Institute) (18).
Microarray Analysis
Microarray analysis was performed with GeneChip Human Genome U133 Arrays Plus 2.0 (Affymetrix) to analyze the mRNA expression levels. Total RNA of cells was isolated from the HUVECs transduced with adenovirus encoding constitutively active ALK1, ALK2, or ALK3 after 48-hours post-infection with RNeasy Mini Kit (Qiagen) and subjected to synthesize cRNA with 3’ IVT Express Kit (Affymetrix). The arrays were stained using the GeneChip Fluidics Station (Affymetrix) and scanned using the Affymetrix GeneChip Scanner (Affymetrix). Gene expression data were normalized and quality control-checked using Expression Console software (v. 1.4.1.46) (Affymetrix).
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis was performed using clusterProfiler R package (v. 3.14.3) (19). Hallmark gene sets from the Molecular Signatures Database (MSigDB v 7.1) was used to identify cellular processes and pathways. The parameters used were nPerm = 3000, minGSSize = 15, maxGSSize = 500 and pvalueCutoff = 0.1. The enrichment results were visualized using the enrichplot R package (v. 1.6.1).
Real-time RT-PCR
Total RNA of cells was isolated with RNeasy Mini Kit (Qiagen), and 1μg of RNA was subjected to reverse RT-PCR with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s instructions. PCR was performed with AccuPower PCR PreMix (Bioneer) using Mastercycler nexus X2 instrument (Eppendorf). Quantitative PCR was performed with TOPreal qPCR SYBR Green 2X premix (Enzynomics) using Rotor-Gene Q instrument (Qiagen). The quantitative PCR data were analyzed with Rotor-Gene Q Series Software (Qiagen). Primers used in this study are listed in Suppl. Table 1.
Western Blot
Cells were washed in cold Dulbecco’s Phosphate-Buffered Saline and harvested in a radio-immunoprecipitation assay buffer (Thermo Scientific) containing a protease and phosphatase inhibitor cocktail (Thermo Scientific). Whole cell extracts were fractionated by centrifugation. The amount of total protein was measured using BCA Protein Assay Kit (Thermo Scientific). Protein samples were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked with 5% non-fat milk in Tris-buffered saline with 0.1% Tween 20 (TBST) for one hour and incubated with antibodies against Smad1/5 (1:1000, Cell Signaling Technology), phospho-Smad1 (1:1000, Cell Signaling Technology), p44/42 MAPK (1:1000, Cell Signaling Technology), phospho-p44/42 MAPK (1:1000, Cell Signaling Technology), or actin (1:5000, Santa Cruz Biotechnology. Subsequently, membranes were washed three times in TBST and incubated with horseradish peroxidase-conjugated secondary antibodies. The protein bands were developed using chemiluminescent horseradish peroxidase substrate (Thermo Scientific). Images were acquired using ImageQuant LAS 500 (GE healthcare).
Statistical analysis
The data were analyzed using the Origin (v. 2020b) software (OriginLab Corporation). The differences between groups were compared using the independent Student’s t-test with a significance level of p < 0.05. Data are presented as means ± S.D.
3. RESULTS
3.1. Individual BMPR1s retain distinct expression pattern within ECs
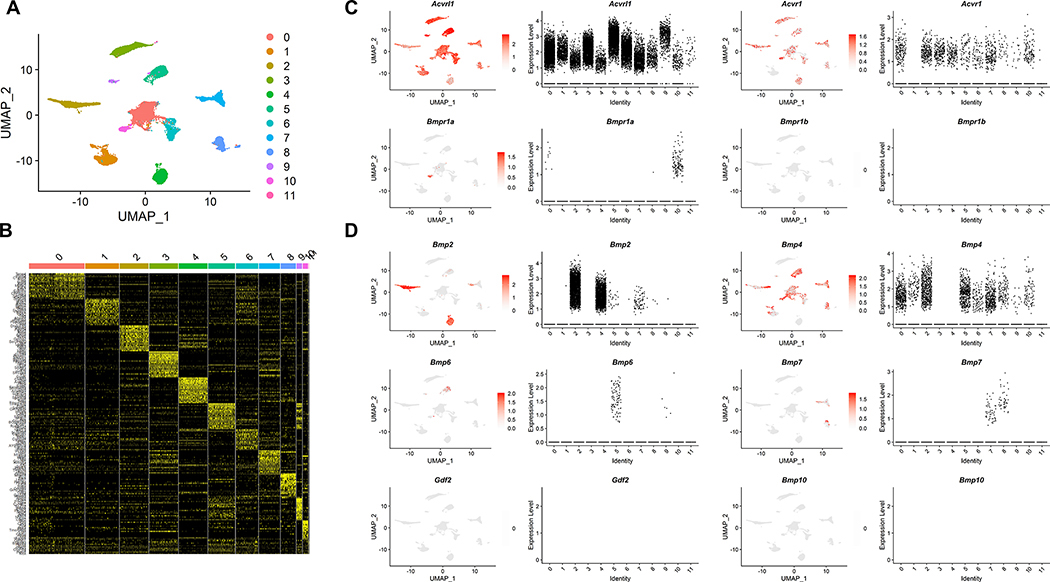
To investigate the distinct function of individual BMPR1s in ECs, we first determined the expression pattern and level of individual Bmpr1s by reanalyzing Pecam1 and Cdh5 expressing cells from 8-week-old male C57BL6/J mice, which are likely to be endothelial in nature, from a previously reported single cell transcriptomic data (20). UMAP visualization revealed that ECs could be further categorized into 12 distinct clusters (Figure 1A). Each cluster retains a unique transcriptomic profile, while expressing transcripts associated with endothelial identity (Figure 1B). Within ECs, Alk1 appears to be ubiquitously expressed. The expression of Alk2 was also detected in all but one clusters of ECs (Figure 1C), suggesting that these two receptors may be the most widely expressed Bmpr1s in ECs. In comparison, Alk3 appears to be expressed within a single subset of ECs only (Figure 1C), while Alk6 was not expressed at all in ECs in the dataset, suggesting that these BMPR1s may provide auxiliary function in ECs at adult stage (Figure 1C). In addition to BMPR1s, cognate ligands for Alk1, Alk2, and Alk3, appeared to be strongly expressed in ECs with the notable exception of Gdf2/Bmp9 and Bmp10 (Figure 1D), hinting that BMP ligands for the endothelial ALK2 and ALK3 could be provided by ECs themselves.
Figure 1: BMPR1s are expressed in a distinct pattern in ECs.
(A) UMAP visualization of CDH5 and PECAM1 expressing endothelial population from 8-week-old male C57BL6/J mice. Endothelial cells could further segregate into 12 distinct clusters. (B) Each cluster retained unique transcriptomic profiles while expressing key endothelial markers. (C) Distribution of cells expressing each BMPR1 within endothelial clusters was analyzed. Alk1/Acvrl1 and Alk2/Acvr1 were the most abundant and widely expressed BMPR1s, while Alk3/Bmpr1a expression appeared to be restricted to a specific cluster. Alk6/Bmpr1b expression was not detected in endothelial cells from our analyses. (D) Distribution of cells expressing each BMP ligand within endothelial clusters was analyzed. Bmp2 and Bmp4 were the most abundant and widely expressed within endothelial cells. Bmp6 and Bmp7 were expressed in endothelial cells, their expression appeared to be largely confined to a specific cluster of endothelial cells. In contrast, expression of Gdf2/Bmp9 or Bmp10 was largely absent in endothelial cells.
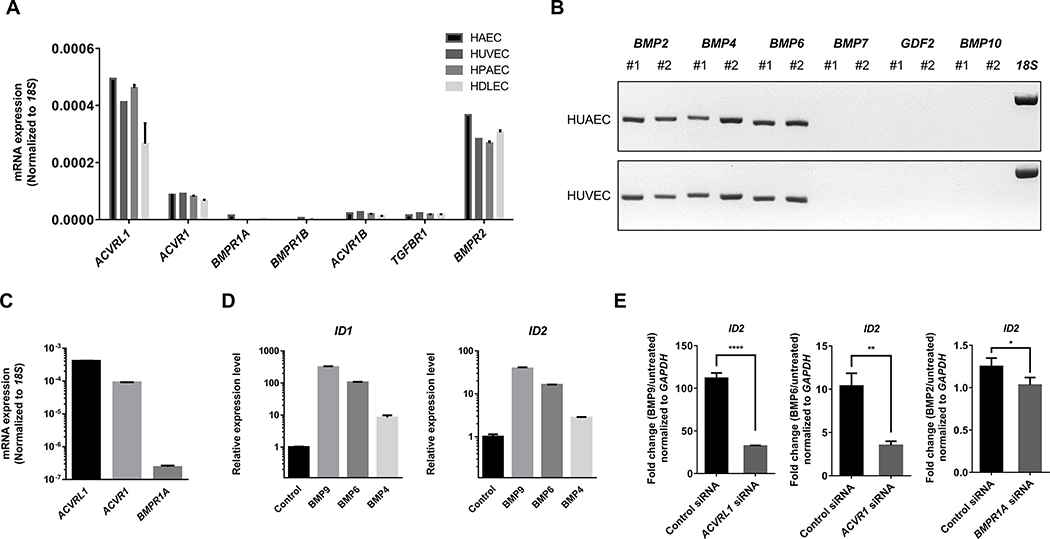
Next, we confirmed the expression of BMPR1s and BMP ligands in commercially available human ECs, including HUVECs, HAECs, HPAECs, and HDLECs to select ECs for further analyses. Consistent with single-cell transcriptomic data, we found that Alk1 and Alk2 were highly expressed in all ECs (Figure 2A and Figure 2B). In addition, a number of BMP ligands, including BMP2, BMP4, and BMP6, were expressed in ECs (Figure 2C). In HUVECs, stimulation with BMP9, BMP6, and BMP4, which are cognate ligands for ALK1, ALK2, and ALK3, respectively, induced transcription of canonical BMP targets such as ID1 and ID2 (Figure 2D). Conversely, inhibition of each BMPR1 significantly attenuated the upregulation of ID1/2 transcription and phosphorylation of SMAD1/5 by cognate ligand (Figure 2E and Supplementary Figure 1A and 1B), suggesting that these receptors are capable of relaying BMP stimulation and activating canonical downstream targets of BMP signaling in HUVECs.
Figure 2: A subset of BMPR1s is expressed and mediates BMP signaling in HUVECs.
(A) Expression of BMPR1s and TGFBR1s in diverse subtypes of human endothelial cells. Consistent with the UMAP visualization of mice endothelial cells, ALK1/ACVRL1 and ALK2/ACVR1 were two most abundantly expressed BMPR1s in human endothelial cells. (B) Expression of selected BMP ligands in HUAECs and HUVECs. Only three BMP ligands, BMP2, BMP4, and BMP6, but not GDF2/BMP9 and BMP10, were expressed within endothelial cells. (C) Relative expression of ALK1, ALK2, and ALK3 in HUVECs. (D) Stimulation with BMP9, BMP6, or BMP4, which are primary BMP ligand binding to ALK1, ALK2, and ALK3, respectively, induced robust phosphorylation of SMAD1/5 in HUVECs. (E) Stimulation with BMP9, BMP6, and BMP4 similarly promoted transcription of key downstream targets, including ID1 and ID2 in HUVECs.
3.2. Each BMPR1 regulates a unique set of downstream targets in ECs
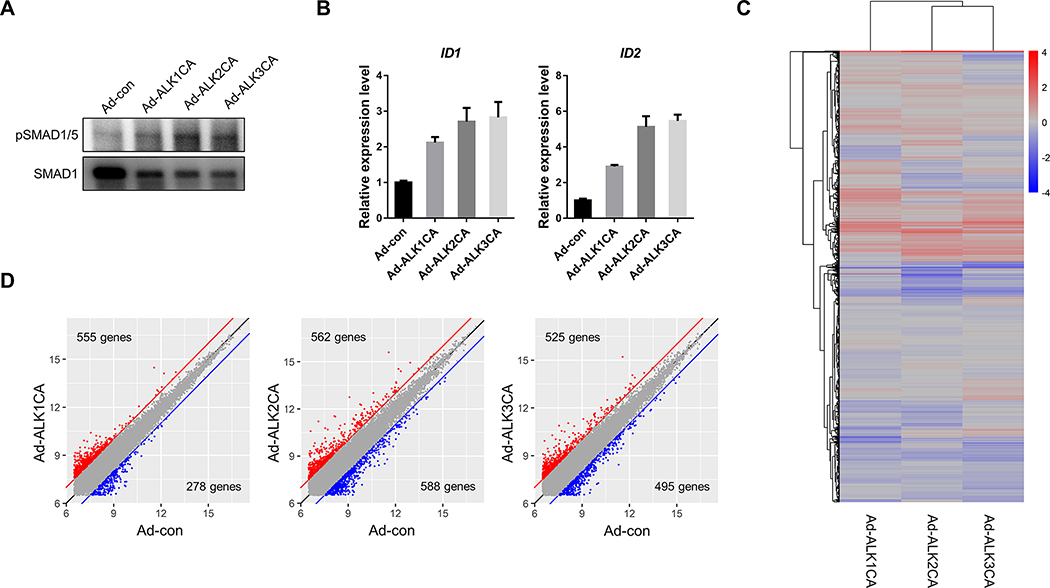
To identify unique downstream events of ALK1, ALK2, and ALK3 in ECs, HUVECs were transduced with previously reported constitutively active BMPR1 (henceforth BMPR1CA), including constitutively active ALK1/ACVRL1 (henceforth ALK1CA), constitutively active ALK2/ACVR1 (henceforth ALK2CA), and constitutively active ALK3/BMPR1A (henceforth ALK3CA). Overexpression of BMPR1CAs induced robust phosphorylation of SMAD1/5 even in the absence of exogenous BMP ligands (Figure 3A), and strongly upregulated transcription of canonical targets of BMP signaling, such as ID1 and ID2 at 48-hours post-infection (Figure 3B and Supplementary Figure 2), indicating that expression of BMPR1CAs in HUVECs could effectively activate downstream effectors. To comprehensively analyze changes in the transcriptomic profile induced by BMPR1CA, subsequently, respective transcriptomic signatures were assessed by microarray (Figure 3C). Comparison of 14040 transcripts showed that each BMPR1CA possesses distinct transcriptomic signatures (Figure 3D); 555 transcripts were upregulated and 278 transcripts were downregulated in HUVECs transduced with ALK1CA compared to mock transduced HUVECs, while expression of ALK2CA or ALK3CA upregulated 562 and 525 transcripts and downregulates 588 and 495 transcripts, respectively (Figure 3D and Suppl. Table 2).
Figure 3: Transcriptomic analyses reveals unique downstream targets of individual BMPR1s in ECs.
(A) Expression of a BMPR1CA in HUVECs induced phosphorylation of SMAD1/5 in the absence of BMP ligand. (B) Expression of BMPR1CA transcriptionally activated ID1 and ID2, canonical downstream effectors of BMP signaling. (C) Microarray analyses on HUVECs expressing ALK1CA, ALK2CA, or ALK3CA revealed that each BMPR1 may activate a unique set of downstream targets One of the sets from the experiments were shown as an example (n=3). Transcriptomic profiles of HUVECs expressing ALK1CA appears to be more divergent. (D) Pairwise comparison of transcriptome of HUVECs expressing BMPR1CA and control HUVECs identified transcripts of which expression was altered by BMPR1CA expression.
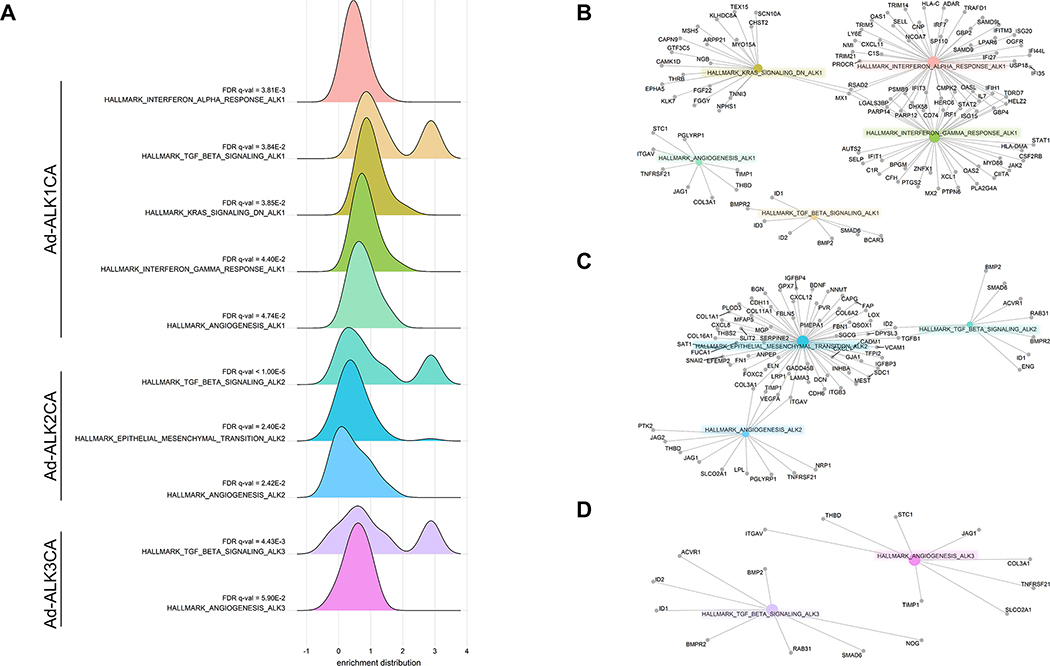
To better understand differences and similarities of downstream targets modulated by each BMPR1, we performed Gene Ontology (GO) enrichment analyses on transcriptomes of HUVECs expressing ALK1CA, ALK2Ca, or ALK3CA (Supple. Table 3). GO enrichment analyses showed that expression of any BMPR1CA led to upregulation of transcripts associated with Transforming Growth Factor (TGFβ) signaling and angiogenesis, indicating that these processes are shared targets of BMPR1s (Figure 4A). In addition, each BMPR1 appears to elicit unique responses. For instance, expression of transcripts associated with KRAS and Interferon γ (IFNγ) signaling was upregulated only in ALK1CA expressing HUVEC, while expression of transcripts associated with endothelial to mesenchymal transition was upregulated only in ALK2CA expressing HUVECs (Figure 4A). Similarly, gene network enrichment analyses in transcriptomes of BMPR1CA expressing HUVECs revealed upregulation of specific genes by individual BMPR1s, reiterating the possibility that each BMPR1 is likely to retain a unique set of downstream targets (Figure 4B to 4D).
Figure 4: Downstream targets of individual BMPR1s reflect functional divergence of BMPR1s in ECs.
(A) GO term analyses identified each BMPR1CA activates a unique set of transcripts in HUVECs. While expression of any BMPR1CA could induce transcriptional activation of genes associated with TGFβ signaling and angiogenesis in HUVECs, expression of ALK1CA additionally increased the transcription of IFNγ signaling or KRAS signaling associated genes, and expression of ALK2CA led to activation of genes implicated in epithelial to mesenchymal transition. (B-D) Gene network enrichment analyses showed ALK1CA (B), ALK2CA (C), or ALK3CA (D) upregulated a specific set of genes in HUVECs.
3.3. Transcriptomic analyses identified novel transcriptional targets for individual BMPR1s
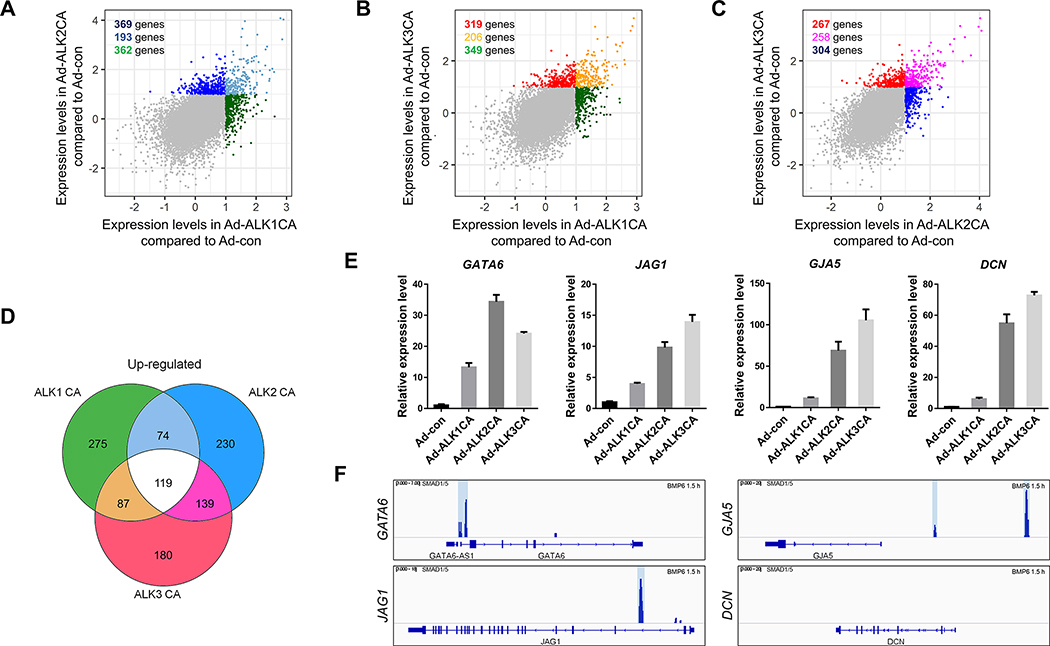
Next, we compared the transcriptomic signature of HUVECs expressing ALK1CA, ALK2CA, or ALK3CA to identify unique downstream targets for each BMPR1 in ECs. Pairwise comparisons show that ALK1 is the most functionally divergent BMPR1 expressed in ECs, consistent with its role as a mediator for anti-angiogenic cue in endothelial cells (10) (Figure 5A to 5C). Overall, 193 transcripts were upregulated and 40 transcripts were downregulated in both ALK1CA and ALK2CA expressing HUVECs (Figure 5A); 206 transcripts were upregulated and 66 transcripts were downregulated in both ALK1CA and ALK3CA expressing HUVECs (Figure 5B); 258 transcripts were upregulated and 190 transcripts were downregulated in both ALK2CA and ALK3CA expressing HUVECs (Figure 5C).
Figure 5: Transcriptomic analyses identify novel targets of BMP signaling in ECs.
(A-C) Pairwise comparison of transcripts of which expression was increased in HUVECs expressing BMPR1CA. Comparison of transcripts of which expression was elevated in ALK1CA or ALK2CA expressing HUVECs (A), in ALK1CA or ALK3CA expressing HUVECs (B), and in ALK2CA or ALK3CA expressing HUVECs (C) identified a number of transcripts of which expression was induced by activation of BMPR1s. (D) Venn diagram of transcripts of which expression was elevated in BMPR1CAs. 119 transcripts were identified as ‘shared’ downstream targets of BMPR1s in ECs. (E) Transcripts of which expression was increased by CA-BMPR1 were validated by quantitative RT-PCR in HUVECs expressing ALK1CA, ALK2CA, or ALK3CA. (F) SMAD1/5 binding to the 5′ regulatory region of the newly identified ‘shared’ downstream targets was assessed by previously reported chromatin immunoprecipitation.
To identify novel downstream targets which are shared by all BMPR1s, a Venn diagram was constructed. 119 transcripts, representing approximately 0.8 percent of the total transcripts, were significantly upregulated by the expression of either ALK1CA, ALK2CA, or ALK3CA (Figure 5D). We postulated that these transcripts are likely to represent the canonical BMP downstream targets since activation of any BMPR1 could induce upregulation of these transcripts. Consistent with this idea, we identified a number of well-characterized canonical targets of BMP signaling, including SMAD6, ID2, and ID1 among 119 transcripts (Figure 5D and Suppl. Table 4). In addition, we identified potential novel downstream effectors of BMP signaling, including surface receptors such as SSTR5, OR2H1, CRHBP, CSF2RB, and NRG4, transcription regulators such as GATA6 and HES4, as well as adhesion molecules such as MADCAM1 and PCDH7 (Suppl. Table 4). We validated a number of potential downstream targets of BMP signaling, including GATA6, JAG1, GJA5, and DCN, in HUVECs expressing BMPR1CAs (Figure 5E). Expression of BMPR1CA effectively elevated the level of these transcripts, suggesting that these transcripts are bona fide transcriptional targets of BMP signaling in ECs. In addition, we identified SMAD1/5 binding sites in the 5′ regulatory region of these novel downstream targets of BMP signaling (Figure 5F). Therefore, 119 transcripts of which expression was upregulated by activation of any BMPR1 may represent novel canonical downstream targets of BMP signaling in ECs. We also identified 23 transcripts which were downregulated by overexpression of any BMPR1CA, including MEOX2, DIAHG3, and DGCR2 (Suppl. Table 4). However, these transcripts were excluded from further analyses since downregulation induced by ectopic expression of BMPR1CA was not likely to have any physiological relevance.
3.4. Analyses on novel transcriptional targets of ALK2
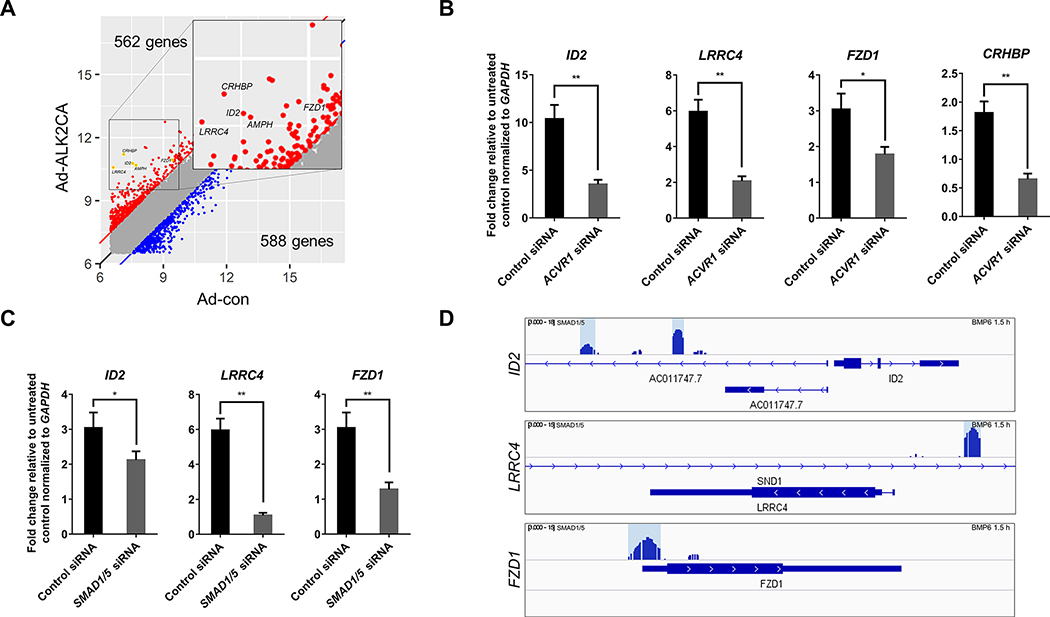
Next, we further validated our approach by analyzing the changes in the transcript level of newly identified BMPR1 targets upon BMP stimulation. In particular, we examined the transcriptional targets of ALK2, which functions as a primary BMPR1 to mediate pro-angiogenic BMP signaling in ECs (10, 11), since transcriptional targets of ALK1 have been previously reported (21), and the list of transcripts of which expression is regulated by ALK1 appear to largely overlap. We decided not to focus on ALK3 since the endogenous expression level of ALK3 appears to be much lower than other BMPR1s. For further analyses, a number of transcripts of which expression was highly upregulated by ALK2 activation were selected from 562 newly identified transcriptional targets of ALK2, including LRRC4, CRHBP, FZD1, and AMPH (Figure 6A). We found that the expression of these genes was highly upregulated by BMP6, a cognate ligand of ALK2. Moreover, transcriptional activation of these targets appeared to be ALK2 dependent, since abrogation of ALK2 substantially decreased the transcriptional upregulation of these genes in response to BMP6 stimulation (Figure 6B and 6C, and Supplementary Figure 3A). To exclude the possibility that alteration of the expression of other BMPR1s in the absence of ALK2 could contribute to the transcriptional regulation of these genes, we also examined the expression of ALK1 and ALK3 in ALK2 siRNA-treated HUVECs, and found that the expression of ALK1 and ALK3 remained largely unaffected by the lack of ALK2 (Supplementary Figure 3B and 3C).
Figure 6: A subset of newly identified ALK2 targets require SMAD1/5 for the transcriptional activation.
(A) Activation of ALK2 upregulates a set of previously unidentified transcriptional targets. Inset shows the location of each transcript shown in panel C and D, and Supplementary Figure 5. (B) Abrogation of ALK2 substantially attenuated the transcriptional activation of newly identified ALK2 targets in response to BMP6 stimulation. (C) Attenuation of SMAD1/5 expression decreased the expression of newly identified ALK2 targets in response to BMP6 stimulation. (D) Presence of SMAD1/5 binding sites in the 5′ regulatory region of newly identified SMAD1/5-dependent ALK2 targets was visualized.
Since ALK2 could transduce signaling from diverse ligands, including TGFβ1 and Activin A (22, 23), we examined whether these novel transcriptional targets of ALK2 activation would respond to activation of ALK2 by other ligands. While HUVECs stimulated with BMP9 or BMP6 could robustly induce transcription of two novel transcriptional targets of ALK2 we have identified, LRRC4 and FDZ1, Activin A or TGFβ1/2 stimulation did not elicit similar results (Supplementary Figure 4). Therefore, it appears that these novel transcription targets are likely to be endogenous targets of BMP signaling rather than related signaling transduced by ALK2 receptor. Since ALK1 and ALK2 share molecular similarities, we next assessed whether ALK1 could similarly activate these novel transcriptional targets of ALK2. We found that the expression of transcriptional targets of ALK2 such as CRHBP was not altered by the siRNA-mediated knockdown of ALK1 (Supplementary Figure 5A and 5B). However, in a subset of these targets, including LRRC4 and FDZ1, we found that siRNA-mediated knockdown of ALK1 decreased the expression although but not as much as ID2 (Supplementary Figure 5A and 5B). Therefore, it appears that at least a subset of novel transcriptional targets of ALK2 we have identified could be also regulated by ALK1 to a lesser degree.
Consistent with the idea that BMP signaling induces the transcription of downstream targets by SMAD1/5 activation, we found that the expression of a number of newly identified ALK2 targets was regulated by SMAD1/5. For instance, attenuation of SMAD1/5 substantially decreased the transcriptional activation of LRRC4 and FZD1 (Figure 6B and 6D, and Supplementary Figure 3A). Moreover, inhibition of ALK2 kinase activity by DMH1 abrogated the transcriptional activation of LRRC4 and FDZ1 (Supplementary Figure 6A and 6B), but chemical inhibition of ERK1/2 did not affect the expression of these genes (Supplementary Figure 6C and 6D), further corroborating that these novel transcriptional targets of ALK2 requires SMAD1/5 activity but not ERK1/2. Moreover, analyses on the previously reported SMAD1/5 Chromatin Immunoprecipitation seq (ChIP-seq) data in HUVECs revealed that phosphorylated SMAD1/5 could bind to the 5′ regulatory region of these genes in the presence of stimulated with BMP6 (17), validating that these genes are likely to be SMAD1/5-dependent ALK2 targets (Figure 6E).
4. DISCUSSION
In this report, we analyze downstream targets of individual BMPR1s in ECs by transcriptomic analyses, and show that each BMPR1 modulates a unique set of downstream targets. We show that the activation of each BMPR1 could induce robust activation of canonical downstream targets of BMP signaling, but also elicit transcriptional activation of a plethora of previously unidentified downstream targets. Our analyses reiterate that ALK1 and ALK2 are the most abundantly expressed BMPR1 in ECs. Considering that these two BMPR1s in ECs elicit opposite outcomes, with ALK1 being implicated as anti-angiogenic and ALK2 pro-angiogenic respectively, it is reasonable to assume that these BMPR1s possess unique downstream targets to manifest their respective outcomes. Consistent with this idea, we find that only one third of the downstream targets of ALK1 and ALK2 are shared with each other. Moreover, transcripts associated with KRAS and IFNγ signaling were upregulated by ALK1. In addition, those implicated in endothelial to mesenchymal transition were selectively upregulated by ALK2. Therefore, our data argue that each BMPR1 is likely to retain a specific function which helps to create context-dependent outcomes of BMP signaling in ECs. Considering that BMP signaling has been shown to intersect with diverse signaling pathways including IFNγ signaling to modulate endothelial behaviors (24), it is possible that signaling crosstalk may require activation of a specific BMPR1.
While our study provides a comprehensive overview of the transcriptomic changes elicited by individual BMPR1s, there are a number of innate limitations of the study; most importantly, our analyses primarily utilized HUVECs overexpressing BMPR1CA and examined the effects of BMPR1 activation at the maximum level, therefore, the subtle nuances which enable context-dependent outcomes of BMP signaling could not be precisely captured in our experimental setting. In addition, considering that BMPR1 could form homodimers as well as heterodimers which has been known to elicit distinct responses (25, 26), our experimental set-up which relied on the overexpression of each BMPR1, could not recapitulate the signaling outcomes induced by intricate balance between homodimeric and heterodimeric BMPR1s. For instance, while the majority of the newly identified transcriptional targets of ALK2 appear to be most sensitive to ALK2 activity by assays using BMPR1CAs, a number of these targets such as LRRC4 and FDZ1 appears to be modulated by ALK1 to a lesser degree in assays using siRNA. Therefore, our data should be interpreted as list of transcriptional targets of which transcriptional regulation is dependent on activity of ALK1, ALK2, or ALK3, rather than list of transcriptional targets exclusively regulated by specific BMPR1s.
Interestingly, we find that a significant portion of downstream targets activated by individual BMPR1s appears to be devoid of SMAD1/5 binding sites in their 5′ regulatory region, hinting the possibility of these targets being indirect targets of BMP signaling. While we cannot formally exclude the possibility, we believe that these targets represent bona fide direct targets of the BMP signaling, since our transcriptome analyses were performed at a time point when the expression of the canonical transcriptional targets, such as ID1/2/3, was initiated. The presence of the downstream targets, which SMAD1/5 do not bind to, alludes that non-canonical BMP signaling, such as mitogen-activated protein kinase (MAPK) signaling pathway, may be important to exert BMP signaling in ECs. Consistent with this notion, it has been previously proposed that BMP signaling could activate its downstream transcriptional targets in either SMAD1/5-dependent or SMAD1/5-independent manner (4). In particular, it has been previously proposed that BMP signaling can elicit distinct signaling outcomes based on the mode of receptor oligomerization (27–29), and a number of MAPKs could modulate transcription by altering the phosphorylation status or shifting subcellular localization of these transcription factors (30, 31). Therefore, it is tempting to speculate that a subset of novel transcriptional targets which do not interact with SMAD1/5 may be regulated by MAPK signaling. However, the majority of these novel transcriptional targets of BMPR1s are regulated in a SMAD1/5-dependent manner but not MAPK-dependent manner since chemical inhibition of SMAD1/5 activity but not ERK1/2 attenuated the expression of these genes upon BMP ligand stimulation. Therefore, while precise molecular mechanisms whereby these transcriptional targets are modulated are unknown, these targets are likely to be under the regulation of SMAD1/5 activity. Taken together, our analyses illustrate the complexity of BMP signaling and provide new insight into the molecular basis of context-dependent responses toward BMP signaling in ECs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the member of Jin laboratory for helpful discussion and critical reading of the manuscript. This study was supported by grants This work was supported by grants to SJ from the National Institutes of Health (HL114820) and National Research Foundation of Korea (NRF-2016R1A5A1007318, NRF-2017R1A2B2007211, and NRF-2019R1A2C2088125).
NONSTANDARD ABBREVIATIONS
- ACVR1
Activin A Receptor Type 1
- ACVRL1
Activin A Receptor Like Type 1
- BMP
Bone Morphogenetic Protein
- BMPR
Bone Morphogenetic Protein Receptor
- BMPR1
Bone Morphogenetic Protein Receptor Type 1
- BMPR1A: BMPR1:
Bone Morphogenetic Protein Receptor Type 1A
- BMPR2
Bone Morphogenetic Protein Receptor Type 2
- EC
Endothelial Cell
- ERK
Extracellular signal Regulated Kinase
- GO
Gene Ontology
- HAEC
Human Artery Endothelial Cell
- HDLEC
Human Dermal Lymphatic Endothelial Cell
- HUAEC
Human Umbilical Artery Endothelial Cell
- HUVEC
Human Umbilical Vein Endothelial Cell
- HPAEC
Human Pulmonary Artery Endothelial Cell
- IFNγ
Interferon γ
- MAPK
Mitogen-Activated Protein Kinase
- RSTK
Receptor Serine Threonine Kinase
- TGFβ
Transforming Growth Factor
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
REFERENCES
- 1.Lowery JW, and Rosen V (2018) Bone Morphogenetic Protein-Based Therapeutic Approaches. Cold Spring Harb Perspect Biol 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piek E, Heldin CH, and Ten Dijke P (1999) Specificity, diversity, and regulation in TGF-beta superfamily signaling. FASEB J 13, 2105–2124 [PubMed] [Google Scholar]
- 3.Gomez-Puerto MC, Iyengar PV, Garcia de Vinuesa A, Ten Dijke P, and Sanchez-Duffhues G (2019) Bone morphogenetic protein receptor signal transduction in human disease. J Pathol 247, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartung A, Bitton-Worms K, Rechtman MM, Wenzel V, Boergermann JH, Hassel S, Henis YI, and Knaus P (2006) Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol Cell Biol 26, 7791–7805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyer LA, Pi X, and Patterson C (2014) The role of BMPs in endothelial cell function and dysfunction. Trends Endocrinol Metab 25, 472–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai J, Pardali E, Sanchez-Duffhues G, and ten Dijke P (2012) BMP signaling in vascular diseases. FEBS Lett 586, 1993–2002 [DOI] [PubMed] [Google Scholar]
- 7.ten Dijke P, Miyazono K, and Heldin CH (2000) Signaling inputs converge on nuclear effectors in TGF-beta signaling. Trends Biochem Sci 25, 64–70 [DOI] [PubMed] [Google Scholar]
- 8.Wiley DM, and Jin SW (2011) Bone Morphogenetic Protein functions as a context-dependent angiogenic cue in vertebrates. Semin Cell Dev Biol 22, 1012–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JD, Lee HW, and Jin SW (2014) Diversity is in my veins: role of bone morphogenetic protein signaling during venous morphogenesis in zebrafish illustrates the heterogeneity within endothelial cells. Arterioscler Thromb Vasc Biol 34, 1838–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larrivee B, Prahst C, Gordon E, del Toro R, Mathivet T, Duarte A, Simons M, and Eichmann A (2012) ALK1 signaling inhibits angiogenesis by cooperating with the Notch pathway. Dev Cell 22, 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HW, Chong DC, Ola R, Dunworth WP, Meadows S, Ka J, Kaartinen VM, Qyang Y, Cleaver O, Bautch VL, Eichmann A, and Jin SW (2017) Alk2/ACVR1 and Alk3/BMPR1A Provide Essential Function for Bone Morphogenetic Protein-Induced Retinal Angiogenesis. Arterioscler Thromb Vasc Biol 37, 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David L, Mallet C, Mazerbourg S, Feige JJ, and Bailly S (2007) Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood 109, 1953–1961 [DOI] [PubMed] [Google Scholar]
- 13.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, Caponigro G, Hieronymus H, Murray RR, Salehi-Ashtiani K, Hill DE, Vidal M, Zhao JJ, Yang X, Alkan O, Kim S, Harris JL, Wilson CJ, Myer VE, Finan PM, Root DE, Roberts TM, Golub T, Flaherty KT, Dummer R, Weber BL, Sellers WR, Schlegel R, Wargo JA, Hahn WC, and Garraway LA (2010) COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468, 968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macias-Silva M, Hoodless PA, Tang SJ, Buchwald M, and Wrana JL (1998) Specific activation of Smad1 signaling pathways by the BMP7 type I receptor, ALK2. J Biol Chem 273, 25628–25636 [DOI] [PubMed] [Google Scholar]
- 15.Varley JE, McPherson CE, Zou H, Niswander L, and Maxwell GD (1998) Expression of a constitutively active type I BMP receptor using a retroviral vector promotes the development of adrenergic cells in neural crest cultures. Dev Biol 196, 107–118 [DOI] [PubMed] [Google Scholar]
- 16.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019) Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morikawa M, Koinuma D, Tsutsumi S, Vasilaki E, Kanki Y, Heldin CH, Aburatani H, and Miyazono K (2011) ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res 39, 8712–8727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29, 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G, Wang LG, Han Y, and He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalucka J, de Rooij L, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen LA, Veys K, Garcia-Caballero M, Khan S, Geldhof V, Sokol L, Chen R, Treps L, Borri M, de Zeeuw P, Dubois C, Karakach TK, Falkenberg KD, Parys M, Yin X, Vinckier S, Du Y, Fenton RA, Schoonjans L, Dewerchin M, Eelen G, Thienpont B, Lin L, Bolund L, Li X, Luo Y, and Carmeliet P (2020) Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 180, 764–779 e720 [DOI] [PubMed] [Google Scholar]
- 21.Ota T, Fujii M, Sugizaki T, Ishii M, Miyazawa K, Aburatani H, and Miyazono K (2002) Targets of transcriptional regulation by two distinct type I receptors for transforming growth factor-beta in human umbilical vein endothelial cells. J Cell Physiol 193, 299–318 [DOI] [PubMed] [Google Scholar]
- 22.Hatsell SJ, Idone V, Wolken DM, Huang L, Kim HJ, Wang L, Wen X, Nannuru KC, Jimenez J, Xie L, Das N, Makhoul G, Chernomorsky R, D’Ambrosio D, Corpina RA, Schoenherr CJ, Feeley K, Yu PB, Yancopoulos GD, Murphy AJ, and Economides AN (2015) ACVR1R206H receptor mutation causes fibrodysplasia ossificans progressiva by imparting responsiveness to activin A. Sci Transl Med 7, 303ra137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramachandran A, Vizan P, Das D, Chakravarty P, Vogt J, Rogers KW, Muller P, Hinck AP, Sapkota GP, and Hill CS (2018) TGF-beta uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo X, Liu Q, Wang G, Zhu S, Gao L, Hong W, Chen Y, Wu M, Liu H, Jiang C, and Kang J (2013) microRNA-29b is a novel mediator of Sox2 function in the regulation of somatic cell reprogramming. Cell Res 23, 142–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traeger L, Gallitz I, Sekhri R, Baumer N, Kuhlmann T, Kemming C, Holtkamp M, Muller JC, Karst U, Canonne-Hergaux F, Muckenthaler MU, Bloch DB, Olschewski A, Bartnikas TB, and Steinbicker AU (2018) ALK3 undergoes ligand-independent homodimerization and BMP-induced heterodimerization with ALK2. Free Radic Biol Med 129, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little SC, and Mullins MC (2009) Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nat Cell Biol 11, 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nohe A, Hassel S, Ehrlich M, Neubauer F, Sebald W, Henis YI, and Knaus P (2002) The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J Biol Chem 277, 5330–5338 [DOI] [PubMed] [Google Scholar]
- 28.Heinecke K, Seher A, Schmitz W, Mueller TD, Sebald W, and Nickel J (2009) Receptor oligomerization and beyond: a case study in bone morphogenetic proteins. BMC Biol 7, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilboa L, Nohe A, Geissendorfer T, Sebald W, Henis YI, and Knaus P (2000) Bone morphogenetic protein receptor complexes on the surface of live cells: a new oligomerization mode for serine/threonine kinase receptors. Mol Biol Cell 11, 1023–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitmarsh AJ (2007) Regulation of gene transcription by mitogen-activated protein kinase signaling pathways. Biochim Biophys Acta 1773, 1285–1298 [DOI] [PubMed] [Google Scholar]
- 31.Cargnello M, and Roux PP (2011) Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 75, 50–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.