Abstract
RNA has major regulatory roles in a wide range of biological processes and a surge of RNA research has led to the classification of numerous functional RNA species. One example is long non-coding RNAs (lncRNAs) that are structurally complex transcripts >200 nucleotides (nt) in length and lacking a canonical open reading frame (ORF). Despite a general lack of sequence conservation and low expression levels, many lncRNAs have been shown to have functionality in diverse biological processes as well as in mechanisms of disease. In parallel with the growing understanding of lncRNA functions, there is a growing subset of microsatellite expansion disorders in which the primary mechanism of pathogenesis is an RNA gain of function arising from RNA transcripts from the mutant allele. Microsatellite expansion disorders are caused by an expansion of short (3–10nt) repeats located within coding genes. Expanded repeat-containing RNA mediates toxicity through multiple mechanisms, the details of which remain only partially understood. The purpose of this review is to highlight the links between functional mechanisms of lncRNAs and the potential pathogenic mechanisms of expanded microsatellite RNA. These shared mechanisms include protein sequestration, peptide translation, micro-RNA (miRNA) processing, and miRNA sequestration. Recognizing the parallels between the normal functions of lncRNAs and the negative impact of expanded microsatellite RNA on biological processes can provide reciprocal understanding to the roles of both RNA species.
Graphical/Visual Abstract and Caption
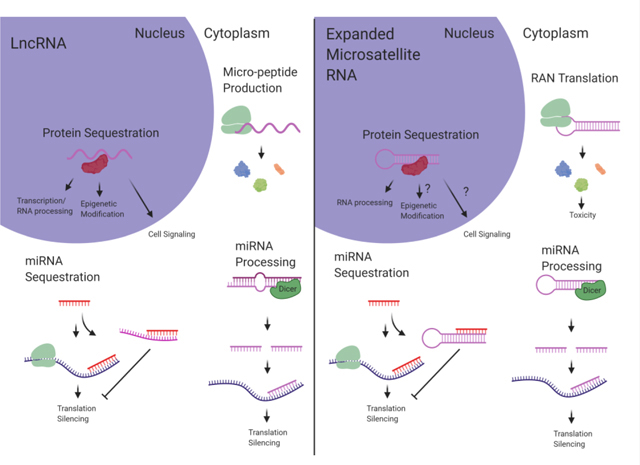
LncRNAs and expanded microsatellite RNA display extensive functional overlap. Shared mechanisms include protein sequestration, peptide translation, miRNA processing, and miRNA sequestration. Understanding parallel mechanisms shared between the two RNA species can reveal biological impact relevant to both lncRNAs and pathogenic microsatellite RNAs. (Created with BioRender.com)
1. INTRODUCTION
While only ~1% of the human genome is protein coding, 80% of the genome displays biochemical functionality and is annotated as non-protein-coding RNA according to data from the ENCODE Project Consortium (Dunham et al., 2012). This includes numerous categories such as micro-RNA (miRNA), small nucleolar RNA (snoRNA), and long noncoding RNA (lncRNA). Importantly, lncRNA lack further defined functional classification, making this an extremely broad category composed of all transcripts which are >200 nucleotides (nt) in length that have been annotated as having no protein-coding capacity (Ponting, Oliver, & Reik, 2009). Estimates of the number of lncRNA genes total >60,000 transcripts, of which 1,867 have been functionally characterized and experimentally supported (Ma et al., 2018). Although originally labeled as “junk RNA”, due to the lack of protein coding capacity, investigations over the past decades have shown lncRNAs to play roles in a wide range of biological processes including development, cellular reprogramming and disease (Kim et al., 2015; S. J. Liu et al., 2016; Luo et al., 2016; Beermann, Piccoli, Viereck, & Thum, 2016). A lack of sequence conservation creates difficulties in directly determining physiological relevance of many human lncRNAs in vivo, which has led to doubts of their functionality (Palazzo & Lee, 2015; Ruan et al., 2020). However, with the emergence of conceptual and technical developments came a hypothesis proposing multiple tiers of conservation: sequence, structure, function, and expression (Diederichs, 2014). Novel methodologies have provided evidence that lncRNAs can have highly complex secondary structure (Blythe, Fox, & Bond, 2016; Lu et al., 2016; Tavares, Pyle, & Somarowthu, 2019) that can impart functionality in multiple, wide-ranging processes from cardiovascular lineage determination to cell signaling (Xue et al., 2016; Uroda et al., 2019). While there is some debate regarding structural conservation of lncRNAs (Rivas, Clements, & Eddy, 2017), this complexity lends credence to the possibility of structural and subsequent functional conservation, even in the absence of strict sequence conservation (Ulitsky & Bartel, 2013; Lu et al., 2016; Tavares, Pyle, & Somarowthu, 2019). Another concern regarding the functionality of lncRNAs is that they are often expressed at a lower level compared to RNA from coding genes, though this can be somewhat offset by the fact that they display strong tissue, cell type, and temporal specificity and differential regulation throughout biological processes (Gloss & Dinger, 2016). The evidence of differential lncRNA expression levels supports their functionality and physiological relevance.
Microsatellite expansion disorders are an important class of genetic diseases that are caused by the expansion of microsatellites, 3–10nt in length, within coding genes. Microsatellite expansion disorders can be separated into two non-mutually exclusive categories based on toxicity mechanism: protein-mediated and RNA-mediated. For a portion of these disorders, disease pathology mainly arises from toxic protein production or loss of protein function of the expanded allele, while other disorders mediate pathology via a toxic RNA gain of function mechanism arising from the mutant RNA containing the expanded microsatellite. Microsatellite expansion disorders encompass a large number of pathologies and can cause disease by a combination of mechanisms. This review will focus on those with supported RNA toxicity including Myotonic Dystrophy type 1 (DM1) and type 2 (DM2), Fragile X Tremor and Ataxia Syndrome (FXTAS), and Amyotrophic Lateral Sclerosis (ALS) (Table 1).
Table 1.
Summary of expanded microsatellite disorders described in this review.
| Disease | Repeat Sequence | Repeat Length | RNA structure | Location of Repeat | Primary Toxicity (RNA vs Protein) |
|---|---|---|---|---|---|
| Myotonic Dystrophy Type 1 (DM1) | CTG | Unaffected: 5–38 Affected: 50–5000 |
Single or multi-hairpin with U/U loops (Mooers et al., 2005; Kumar et al., 2011; Cruchten, Wieringa, & Wansink, 2019) | 3’ UTR of DMPK gene (Brook et al.,1992) | RNA |
| Myotonic Dystrophy Type 2 (DM2) | CCTG | Unaffected: ~26 Affected: 75–11,000 |
Hairpin with CU/UC loops (Dere et al., 2004; Childs-Disney et al., 2013) | Intron 1 of CNBP gene (Liquori et al., 2001) | RNA |
| Fragile X-Associated Tremor/Ataxia Syndrome (FXTAS) | CGG | Unaffected: 6–54 Affected: 55–200 |
Hairpins with potential G-quadruplex (Handa et al., 2003; Blice-Baum & Mihailescu, 2014) | 5’ UTR of FMR1 gene (Kremer et al., 1991) | RNA |
| Amyotrophic Lateral Sclerosis (ALS) | GGGGCC | Unaffected: ≤11 Affected: >30 and up to thousands |
Hairpins and stable G-quadruplexes (Su et al., 2014; Balendra & Isaacs, 2018) | Intron 1 of C9orf72 gene (DeJesus-Hernandez et al., 2011) | Toxic protein production, RNA toxicity, and protein loss of function |
Similar to lncRNAs, expanded microsatellite RNA typically forms complex secondary structures. In FXTAS, the expanded CGG repeats in the FMR1 mRNA have been shown to form hairpin structures (Handa et al., 2003; Zumwalt, Ludwig, Hagarman, & Dieckmann, 2007) and the potential for G-quadruplex formation (Blice-Baum & Mihailescu, 2014). The expanded CUG repeat RNA within the DMPK mRNA, causing DM1, also forms stable hairpins (Tian et al., 2000; Mooers et al., 2005) and has recently been shown to form complex multi-branched hairpin structures (Cruchten, Wieringa, & Wansink, 2019). The GGGGCC RNA repeats expressed from the expanded C9orf72 gene responsible for ALS have been shown to form a complex G-quadruplex structure (Fratta et al., 2012) that can affect some aspects of downstream toxicity (Fay, Anderson, & Ivanov, 2017). Interestingly, lncRNA transcripts also often contain repeat tracts that mediate downstream functionality (Carrieri et al., 2012; Yang et al., 2015; Smola et al., 2016; Witkos, Krzyzosiak, Fiszer, & Koscianska, 2018). In terms of structural complexity, length, as well as the presence of repeat tracts, lncRNA and expanded microsatellite RNA are very similar RNA species. This similarity extends to functionality. This review will describe mechanistic similarities between lncRNA and expanded microsatellite RNA, in terms of protein sequestration, peptide translation, micro-RNA (miRNA) processing, and miRNA sequestration (Figure 1). The goal of this review is to establish reciprocal understanding between lncRNA and microsatellite expansion RNA studies to unite divergent fields and further discovery.
Figure 1.
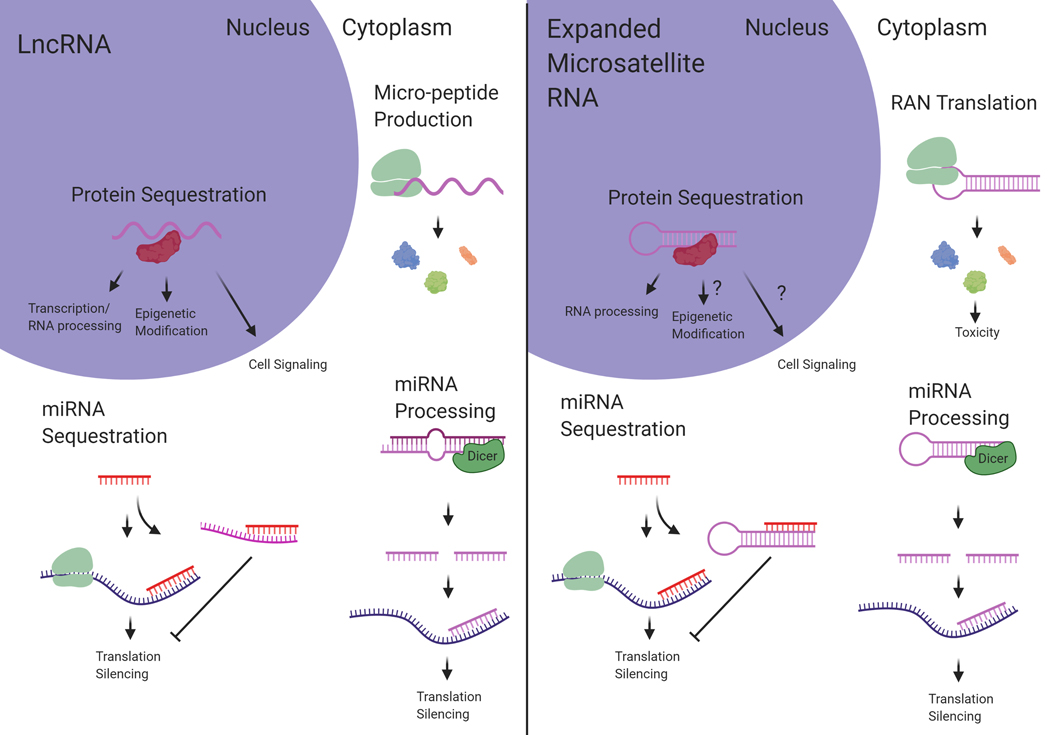
LncRNAs and expanded microsatellite RNA display extensive functional overlap. Shared mechanisms include protein sequestration, peptide translation, miRNA processing, and miRNA sequestration. Understanding parallel mechanisms shared between the two RNA species can reveal biological impact relevant to both lncRNAs and pathogenic microsatellite RNAs. (Created with BioRender.com)
PROTEIN SEQUESTRATION
Examples of protein sequestration mediated by lncRNA and expanded microsatellite RNA and the downstream functional consequences have been previously reviewed (Morris & Cooper, 2017). A myriad of lncRNAs have been shown to bind and sequester proteins (HafezQorani, Houdjedi, Arici, Said, & Kazan, 2019) and these lncRNA-protein interactions can be subdivided into different categories based on the proteins they sequester: transcription regulation, RNA processing regulation, epigenetic remodeling, and cell signaling.
Transcription and RNA processing regulation
Sequestration of transcription factors on lncRNAs has been demonstrated to result in downstream transcriptional regulation. For example, the Bvht lncRNA binds to the transcription factor CNBP and directly impacts regulation of cardiomyocyte differentiation via a transcriptional network (Xue et al., 2016). LncRNAs not only affect transcription, but also downstream RNA processing, including alternative splicing. Certain lncRNAs have been shown to bind to splicing factors resulting in splicing regulation (Wu et al., 2016). Similarly, microsatellite expansion disorders commonly display protein sequestration as a mechanism of pathogenesis. One such disorder, FXTAS, displays expansion of a CGG microsatellite in the 5’ untranslated region (UTR) of the FMR1 gene from 6–54 repeats in unaffected individuals to 55–200 repeats (Kremer et al., 1991) and displays RNA-mediated toxicity. In FXTAS, the RNA binding protein SAM68 is initially sequestered on the expanded CGG RNA repeats leading to a partial loss of SAM68-mediated splicing regulation in cell culture and in brain samples of affected individuals. In addition to the partial loss of SAM68 function, bound SAM68 was shown to recruit the RNA binding protein hnRNP G and the splicing regulator MBNL1 to the expanded RNA repeats, potentially impacting late-stage pathogenesis (Sellier et al., 2010). Another RNA binding protein and splicing regulator, TRA2, has been shown to colocalize with nuclear RNA foci of expanded CGG repeats in COS-7 cells transiently expressing 60 CGG repeats, in FXTAS mouse models and in post-mortem human brain samples. The same authors showed that the effect on TRA2 splicing targets was consistent with a loss of function (Cid-Samper et al., 2018).
The most common familial cause of ALS is an expanded repeat of GGGGCC within intron 1 of the C9orf72 gene leading to both toxic RNA gain of function, toxicity of peptides translated from the repeat region (DeJesus-Hernandez et al., 2011; Zhang et al., 2016), as well as loss of protein function (Burberry et al., 2020; McCauley et al., 2020). SRSF1 has been shown to colocalize with RNA foci in spinal cord tissue from individuals affected by C9orf72 ALS and in both cell culture and Drosophila models of the disease. SRSF1 sequestration leads to NFX1-mediated nuclear export of the expanded microsatellite RNA contributing to the production of toxic peptides via Repeat Associated Non-AUG (RAN) translation (Hautbergue et al., 2017) (see translation section below). RNA pulldown and RNA binding protein colocalization screens for expanded GGGGCC C9orf72 repeats have identified additional proteins that localize to RNA foci in cell models such as hnRNP A1, ALYREF, SRSF2, RanGap (Cooper-Knock et al., 2014; K. Zhang et al., 2015) and hnRNP H, that has also been identified in brain tissue from affected individuals (Lee et al., 2013).
DM1 is caused by an expansion of CTG repeats within the 3’UTR of the DMPK gene that ranges from 5–28 in unaffected individuals to as many as 5000 repeats in those affected (Brook et al., 1992). DM2 is caused by an expansion of CCTG repeats in the CNBP gene that ranges from 26 repeats in unaffected individuals up to 11,000 in individuals affected by the disease (Liquori et al., 2001). For both DM1 and DM2, the key mechanism of pathogenesis is RNA toxicity mediated primarily by sequestration of the MBNL family of RNA binding proteins on the RNA. The resulting loss of MBNL function leads to mis-regulated alternative splicing, polyadenylation and mRNA localization of MBNL pre-mRNA and mRNA targets (Miller et al., 2000; Ho, Bundman, Armstrong, & Cooper, 2005; Lin et al., 2006; Batra et al., 2014; Wang et al., 2015). The RNA helicase DDX5 also colocalizes with RNA foci in DM1 cell models, though the downstream functionality is uncertain (Laurent et al., 2012; Jones et al., 2015). Similar to its binding to the C9orf72 RNA repeat, the RNA binding protein hnRNP H has been shown to colocalize with the expanded CUG RNA in both DM1 cell models and tissue samples from affected individuals. Although hnRNP H does not colocalize as efficiently as MBNL, there is evidence that hnRNP H, as well as other nuclear speckle-associated proteins, localize to the repeats and may prevent nuclear export of the expanded CUG RNA (Jiang, Mankodi, Swanson, Moxley, & Thornton, 2004; D.-H. Kim et al., 2005; Paul et al., 2006; Holt et al., 2007).
Epigenetic Regulation
LncRNAs also sequester factors affecting epigenetic modifications leading to downstream regulation of gene expression. For example, the lncRNA HOTAIRM1 sequesters DNA methyltransferases and other epigenetic modifiers such as the histone methyltransferases G9A and EZH2, leading to increased HOXA1 expression due to a reduction in suppressive epigenetic modifications (Li, Dong, Cui, Wang, & Hong, 2018). LncRNAs can even function to guide proteins to particular chromatin locations, for example the lncRNA PAPAS guides the epigenetic regulators CDH4/NuRD and Suv4–20h2 to the rDNA promoter, encoding ribosomal RNA, impacting responses to heat shock and quiescence (Bierhoff et al., 2014; Zhao, Sentürk, Song, & Grummt, 2018). Several microsatellite expansion disorders display aberrant epigenetic modifications that arise at the mutant locus. For example, in DM1 the mutant DMPK locus is hypermethylated in a subset of affected individuals and is associated with larger repeat expansions and early disease onset (Santoro et al., 2015). In Fragile X Syndrome (FXS), expansion of the CGG microsatellite beyond 200 repeats leads to methylation-induced silencing of the FMR1 gene that causes pathogenesis due to loss of gene function (Kraan, Godler, & Amor, 2019). The specific mechanisms that form the basis for these epigenetic changes have yet to be fully elucidated.
Cell Signaling
LncRNAs have been shown to have regulatory roles in numerous signaling pathways including the p53, NFkB, AKT, and Notch pathways (Peng, Koirala, & Mo, 2017). Involvement in cell signaling partly stems from the ability of a subset of lncRNAs to influence a variety of post-translational modifications (PTMs) including SUMOylation (C. Liu et al., 2020), acetylation (Wan et al., 2013), ubiquitination (Yoon et al., 2013), and phosphorylation. For example, the lncRNA NBR2 directly interacts with the AMPKα subunit, promoting wide-spread AMPK-mediated phosphorylation during glucose starvation (X. Liu, Xiao, Han, et al., 2016). The lncRNA DANCR is a second example that binds to the RXRA nuclear receptor enhancing its phosphorylation via GSK3β (Tang et al., 2018). As an example of indirect phosphorylation regulation, the lncRNA Xist regulates cell signaling by inhibiting AKT phosphorylation. This inhibition occurs by Xist-mediated recruitment of the HDAC3 transcriptional repressor away from the promoter of the phosphatase PHLLP1 gene leading to increased PHLLP1 transcription and phosphatase activity. The increased PHLLP1 activity results in reduced phospho-AKT and subsequent reduced cell viability (Huang, Chang, Lee, Jou, & Shih, 2016). In addition to regulating PTMs, lncRNAs also impact signaling pathways directly by modulating the availability of cell signaling proteins. For example, the lncRNA p53RRA sequesters G3BP1, releasing bound p53 from G3BP1 and allowing for nuclear translocation of p53 and its downstream impact on cell cycle arrest, apoptosis, and ferroptosis (Mao et al., 2018).
A direct effect of expanded microsatellite RNAs on PTMs has not been demonstrated; however, multiple molecular phenotypes that are not fully explained by the current models imply changes in cell signaling in response to expanded microsatellite RNA. For example, the primary mechanism of DM1 pathogenesis is RNA toxicity via sequestration of MBNL proteins, as noted above. While some splicing defects have been directly linked to DM1 symptoms, splicing mis-regulation has not been linked to every feature of the disease brought about by the toxic RNA. Another molecular hallmark of DM1 is the upregulation of the RNA binding protein CELF1 that results in part from hyperphosphorylation by the kinases PKC and GSK3β (Kuyumcu-Martinez, Wang, & Cooper, 2007; Jones et al., 2012). The mechanism of RNA-mediated activation of PKC and GSK3β, remains to be identified. DM1 has also been shown to disrupt TWEAK/Fn14 signaling, but how the RNA initiates this disruption is unknown (Yadava et al., 2015). These molecular phenotypes arise from the toxic RNA but potentially come about from a mechanism other than splicing mis-regulation. These potential mechanistic gaps raise the possibility that the expanded microsatellite RNA could be exhibiting mechanisms similar to those utilized by lncRNAs.
TRANSLATION
Another commonality between lncRNA and pathogenic expanded microsatellite RNA is that their functionality was believed to be limited to the direct impact of the RNA. However, the discovery of translation of both lncRNAs and expanded microsatellite RNA resulted in recent shifts of thinking regarding mechanisms of function in both fields.
Coding transcripts were previously identified based on restrictions of ORFs greater than 200nt in length, AUG start codons, and sequence conservation (Fickett, 1982; Venter et al., 2001). Under these criteria, lncRNAs were classified as non-coding. LncRNA-encoded micro-peptides can also be difficult to identify via mass spectrometry due to their short length, reduced stability, degradation/loss during sample preparation and/or low expression levels (Starck et al., 2016; Yeasmin, Yada, & Akimitsu, 2018). However, further technological and computational developments have shown that some members of this broad class of genes contain ORFs that were previously not detected. Ribosome profiling and bioinformatics analyses led to suggestions that many lncRNA transcripts are translated to produce micro-peptides (Ingolia, Lareau, & Weissman, 2011; Ji et al., 2015). Additional reasoning for the initial mis-annotation of some lncRNAs as non-coding partly stems from the fact that some ORFs initiate from non-canonical start codons (non-AUG) that are typically one nucleotide different from the consensus (CUG, GUG, UUG). A study using RibORF, a technique of ribosome profiling analysis (Ji et al., 2015), has identified ORFs displaying similarities in translation criteria, including translation efficiency values and ribosome release scores, between 96 different lncRNAs and coding genes (Jackson et al., 2018). More than 50% of these lncRNA ORFs were initiated from non-AUG start codons (Jackson et al., 2018). The same study experimentally verified an 82 amino acid micro-peptide from the lncRNA AW112010 under a non-canonical ORF and discovered the peptide to be functional in mucosal immunity in mouse bone-marrow derived macrophages (Jackson et al., 2018). Ribosome profiling in mammalian cells has shown that ~50% of all translation events originate from non-AUG codons, with CUG being the most common near-cognate start codon (Cao & Slavoff, 2020). Although the fraction of lncRNAs that encode micro-peptides is up for debate, experimentally verified examples emphasize the presence and functionality of certain lncRNA-derived micro-peptides. A recent study performed CRISPR knockout of small ORFs identified by a combination of ribosome profiling and mass spectrometry and found 91 lncRNA-derived noncanonical micro-peptides that displayed physiological impact (Chen et al., 2020).
Further studies have led to the discovery and experimental validation of specific lncRNAs that express micro-peptides resulting in downstream functional impact (Anderson et al., 2015; Min et al., 2017; Jackson et al., 2018; Lu et al., 2019; Spencer et al., 2020; Niu et al., 2020). In one example the micro-peptide STORM, encoded by the lncRNA linc00689 via a previously unidentified AUG-initiated small ORF, was shown to mimic the signal recognition particle 19 (SRP19) that is required for proper assembly of the SRP complex in vitro, inhibiting SRP19 interaction with 7SL RNA. Inhibition of the SRP19–7SL interaction has been proposed to reduce SRP complex formation during TNF-α activation (Min et al., 2017). This same lncRNA has also been shown to sequester miRNAs in glioma and osteosarcoma (Xin Liu et al., 2019; Xing, Xu, Chang, Zhang, & Wang, 2020) (see miRNA sequestration mechanism described below), demonstrating an RNA-mediated mechanism in addition to micro-peptide expression. Another example is the micro-peptide SPAAR encoded within the lncRNA linc00961, in which a functional requirement was demonstrated in vivo. Knockout of the linc00961 locus in mice reduced the ability of muscle to recover from ischaemia due to a reduction in capillary density as a result of SPAAR’s role in endothelial network formation and angiogenesis (Spencer et al., 2020).
For microsatellite expansion disorders, such as DM1, DM2, FXTAS, and ALS, the discovery of proteins produced from the expanded repeats by RAN translation (Zu et al., 2011; Ash et al., 2013; Todd et al., 2013; Zhang et al., 2016) has demonstrated the potential for multiple pathogenic mechanisms. RAN translation leads to the production of large proteins encoded by the expanded microsatellite composed of repeats of single or multiple amino acids that can produce a toxic effect. The RAN translation mechanism in microsatellite repeat disorders has been previously reviewed (Green, Linsalata, & Todd, 2016). Since that review, RAN translation has been shown to require a 5’ cap, eIF4A, and eIF4E supporting the use of canonical translation machinery and a scanning mechanism at near-cognate initiation codons in a process that requires the formation of RNA secondary structure in both ALS and FXTAS (Green et al., 2017). RAN translation can occur in multiple reading frames generating multiple toxic protein elements, often from both the sense and antisense RNAs commonly transcribed from expanded microsatellites (Pearson, 2011; Zu et al., 2011; Todd et al., 2013). Evidence suggests that there may be a link between stress and the induction of RAN translation at near-cognate non-canonical ORFs (Green et al., 2017). RAN translation mechanisms are likely to differ between microsatellite expansion disorders (Zu et al., 2011; Todd et al., 2013; Kearse et al., 2016; Green et al., 2017). In FXTAS, the FMR1 CGG repeats undergo RAN translation of two reading frames to produce poly-glycine and poly-alanine peptides from both non-expanded and expanded repeats, though toxic aggregates only form when the repeats reach pathogenic lengths (Todd et al., 2013). Whether this toxicity is a result of the demonstrated increase in RAN translation with increasing CGG repeat length (Kearse et al., 2016), is unknown. The CCTG expansion causing DM2 is translated from sense and antisense RNA to produce tetrapeptides, a process that is partially regulated by MBNL1 sequestration at the RNA foci (Zu et al., 2017). While lncRNAs have not yet been shown to undergo RAN translation, the potential exists and the production of peptides from both RNA species highlights an additional parallel phenomenon.
MIRNA PROCESSING
Micro-RNAs (miRNAs) provide a wide-spread and potent mechanism of gene regulation. MiRNAs are processed from precursor transcripts within the nucleus by DROSHA to generate pre-miRNAs that are exported to the cytoplasm to undergo final processing to mature miRNAs by DICER. DICER recognizes dsRNA and cleaves it into ~22nt miRNAs that are subsequently loaded into the AGO2 protein complex where one miRNA strand is preferentially selected based on thermodynamic stability (Y. -K. Kim, Kim, & Kim, 2016). The miRNA within the complex binds to miRNA recognition elements (MREs) located primarily within the 3’UTR of coding transcripts. MiRNA binding recruits the RNA-induced silencing complex (RISC) and the mRNA transcript is translationally silenced and typically degraded.
The potential for lncRNAs of unknown function to produce miRNAs led to substantial efforts that identified numerous lncRNAs that are precursors for miRNAs with functional impact (Emmrich et al., 2014; Yuan, Liu, Han, & Zhao, 2019). One such case is the lncRNA RMRP that undergoes DICER-dependent production of two miRNAs, termed RMRP-S1 and RMRP-S2, (Maida et al., 2009) that exhibit wide-spread gene silencing activity related to hematopoiesis, splicing, and proliferation, among other processes (Rogler et al., 2014). Seventy-eight lncRNAs have been identified as potential host genes encoding miRNAs (HGNC database), such as mir675 derived from the lncRNA H19 (Cai & Cullen, 2007) and mir570 encoded by the lncRNA MIR570HG, previously named linc00969 (HGNC database). LncRNAs can also impact miRNA homeostasis through a mechanism that is reminiscent of their role in protein sequestration described above. In C. elegans the lncRNA rncs-1 is bound by DICER but the lncRNA is not cleaved leading to DICER sequestration and subsequent downregulation of overall miRNA production (Rybak-Wolf et al., 2014).
The process of miRNA production from lncRNA transcripts extends into the area of microsatellite expansion disorders. The CGG repeat hairpins, characteristic of FXTAS, have been shown to be cleaved by DICER into 22nt fragments, though inefficiently (Handa, Saha, & Usdin, 2003). Similarly, DICER has been shown to cleave the DM1 sense and antisense expanded repeats into CUG and CAG repeat-containing small RNAs, 21nt in length, in a cell-free system. The CUG repeat-containing small RNAs were shown to mediate downregulation of certain CAG-containing transcripts in cells (Krol et al., 2007) and in a DM1 Drosophila model (Yu, Teng, & Bonini, 2011). The extent to which miRNA production from expanded microsatellite RNA contributes to disease pathogenesis remains to be established, however, the potential for a miRNA-mediated mechanism is highlighted by the demonstrated roles for miRNAs generated from lncRNAs.
MIRNA Sequestration
The competitive endogenous RNA (ceRNA) hypothesis describes a transcriptome-wide RNA-mediated regulatory network in which lncRNAs harbor MREs that sequester specific miRNAs, leading to miRNA depletion and upregulation of the respective target mRNAs (Salmena, Poliseno, Tay, Kats, & Pandolfi, 2011). MiRNA sequestration is not limited to lncRNAs and can involve coding mRNAs, such as the ZEB2 mRNA that sequesters the miRNAs miR-181, miR-200b, miR-25, and miR-92a in melanoma cells decreasing their availability to the 3’UTR of the PTEN mRNA, leading to PTEN upregulation (Karreth et al., 2011). MiRNA sequestration has been studied extensively in relation to lncRNAs (Karreth et al., 2011; Guo et al., 2015); however, one concern, previously reviewed (Thomson & Dinger, 2016) is that lncRNAs are often not expressed at sufficiently high levels to sequester miRNAs and have a tangible titration effect, except for the lowest expressed miRNAs. Despite this valid concern, multiple examples of functional lncRNA-ceRNA relationships have been uncovered. LncRNA H19, for example, is upregulated in metastatic cancers and sequesters miR-196b leading to upregulation of the RNA binding protein LIN28B (Ren et al., 2018). H19 has also been shown to differentially sequester the miRNAs let-7b and mir-200b/c to initiate the EMT/MET switch in breast cancer. H19 sequesters let-7b leading to increased levels of the EMT-associated protein CYTH3. Following this, H19 sequesters mir-200b/c resulting in upregulation of the MET-associated protein GIT2 (Zhou et al., 2017). Another study has shown a ceRNA relationship in which upregulation of the lncRNA AK054386 during hepatic ischemia-reperfusion injury depletes functional miR-199 leading to sustained endoplasmic reticulum stress and subsequent cell death that was demonstrated in both cell culture and in mice (Dai et al., 2019). Numerous other studies have identified lncRNA-ceRNA relationships both in cells (Qu et al.,2016; Li Ma et al., 2019; Kong et al., 2019) and in vivo (Guo et al., 2015).
MiRNA sequestration on expanded microsatellite RNA has been documented for FXTAS, DM1 and DM2 (Witkos, Krzyzosiak, Fiszer, & Koscianska, 2018; C. Wang, Ge, Wu, Wang, & Yuan, 2018). In FXTAS, there is evidence of a network of genes commonly upregulated in this disease. A subset of the upregulated genes are targets of miRNAs that also bind to the FMR1 RNA (Zongaro et al., 2013). The presence of identical MREs on both mis-regulated mRNAs and the expanded microsatellite RNA raises the possibility that miRNA sequestration leads to this mis-regulation downstream. Further, bioinformatics studies have indicated that certain miRNAs may specifically bind to expanded CCUG and CUG microsatellite RNA leading to disruption of downstream mRNA silencing, highlighting the potential relevance to DM1 and DM2 pathogenesis (Koscianska, Witkos, Kozlowska, Wojciechowska, & Krzyzosiak, 2015; Witkos, Krzyzosiak, Fiszer, & Koscianska, 2018).
Sidebar: Phase Separation
Liquid-liquid phase separation (LLPS) is a process of cellular sub-compartmentalization that concentrates reaction components to increase reaction efficiency. Studies have shown that multivalent interactions, often mediated by protein domains, initiate this process (Erdel & Rippe, 2018). Although proteins are common mediators of LLPS, RNA has also been demonstrated to play a critical role in formation of several membrane-less organelles (Mitrea & Kriwacki, 2016). Certain lncRNAs have been shown to mediate LLPS leading to physiologically significant effects such as the lncRNA NEAT1 that forms phase-separated granules called paraspeckles that mediate a variety of RNA processing functions (Yamazaki, Nakagawa, & Hirose, 2020). Other lncRNAs have been suggested to form functional phase-separated droplets, recently reviewed (Krause, 2018; Yamazaki, Nakagawa, & Hirose, 2020), including the lncRNA Xist during X-chromosome inactivation (Cerase et al., 2019), omega speckles in Drosophila that require the lncRNA HSRω (Prasanth, Rajendra, Lal, & Lakhotia, 2000) and nuclear stress bodies formed from the lncRNA HSATIII (Aly, Ninomiya, Adachi, Natsume, & Hirose, 2019).
A role for expanded microsatellite RNA in LLPS has also been suggested. In one study the investigators postulated that the CUG and CAG microsatellite expansions result in multivalent electrostatic interactions that culminate in RNA-mediated LLPS for repeat lengths greater than 30 repeats (Jain & Vale, 2017). This study also demonstrated that chemical inhibition of RNA gelation dissolved the CUG RNA foci in patient-derived DM1 fibroblasts (Jain & Vale, 2017). Further studies will need to be performed to determine the extent to which microsatellite RNA exhibits LLPS, though the precedence of LLPS mediated by lncRNAs establishes this potential (Figure 2).
Figure 2.
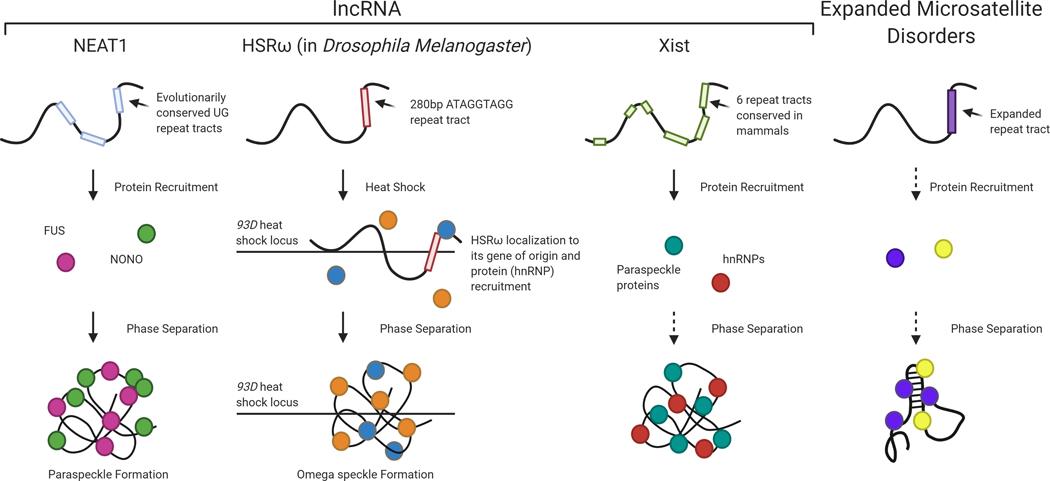
Certain lncRNAs have been shown to exhibit phase separation capacity. A commonality between these lncRNA is the presence of repeat tracts. The lncRNA NEAT1 contains evolutionarily conserved UG repeat tracts which recruit proteins including FUS and NONO leading to paraspeckle formation. The lncRNA HSRω in D. Melanogaster encodes a 280bp repeat tract. Upon heat shock, the lncRNA localizes to its locus of origin and recruits hnRNP proteins leading to omega speckle formation at the gene locus. The lncRNA Xist, containing six conserved repeat tracts (the sequence of the repeats varies), has also been hypothesized to exhibit phase separation during X-chromosome inactivation by recruiting hnRNPs and paraspeckle proteins. Evidence suggests that expanded repeat RNA, specifically CUG and CCUG (the repeat expansions exhibited in DM1 and DM2, respectively), phase separates to form the pathogenic RNA foci. Whether proteins are required for this process is yet to be fully understood as there is evidence for both instances. (Created with BioRender.com)
Conclusion
LncRNAs and the toxic RNA causing pathogenesis in some microsatellite expansion disorders share extensive length, complex higher order structure, a lack of canonical protein coding capacity and can share the presence of repeat tracts. From these similarities emerges the potential for further mechanistic overlap. The most extensively studied mechanism in common between lncRNAs and expanded microsatellite RNA is protein sequestration. The impact of protein sequestration on lncRNAs results not only in loss of protein function, but also protein scaffolding and guiding, leading to wide-spread regulation of RNA processing, epigenetic remodeling and cell signaling. While expanded microsatellite RNA directly impacts RNA processing through protein sequestration, there are additional epigenetic and cell signaling effects that occur through mechanisms that have yet to be fully elucidated. Expanding our knowledge of proteins that bind to the expanded microsatellite RNA could uncover links to these mechanisms. The potential for the expanded microsatellite RNA to perform scaffolding or guiding functions, as described for lncRNAs, also presents an interesting possibility to be explored. Meanwhile, RAN translation has yet to be discovered for lncRNAs and is an avenue for future studies, particularly in relation to lncRNAs containing repeat tracts such as Xist and NEAT1. MiRNA processing and miRNA sequestration of expanded microsatellite RNA has been explored in few expanded repeat disorders and the question remains as to the impact these processes may have in disease pathogenesis in vivo as well as whether these mechanisms transcend into other repeat disorders. Overall, these discoveries emphasize the potential for diverse mechanisms of functionality within microsatellite expansion disorders, reminiscent of phenomena exhibited within the lncRNA field.
Acknowledgments
Funding Information
Recent support for the Cooper lab has come from the NIH (R01HL045565, R01AR045653, R01AR060733 and R01HL147020) and the Muscular Dystrophy Association (MDA602529).
Footnotes
Research Resources
Notes
Contributor Information
Sara J. Johnson, Department of Molecular & Cellular Biology, Baylor College of Medicine.
Thomas A. Cooper, Department of Pathology & Immunology, Department of Molecular & Cellular Biology, Department of Physiology and Biophysics*.
References
- Aly MK, Ninomiya K, Adachi S, Natsume T, & Hirose T. (2019). Two distinct nuclear stress bodies containing different sets of RNA-binding proteins are formed with HSATIII architectural noncoding RNAs upon thermal stress exposure. Biochemical and Biophysical Research Communications, 516(2), 419–423. doi: 10.1016/j.bbrc.2019.06.061 [DOI] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang C-L, Makarewich CA, Nelson BR, McAnally JR, … Olson EN (2015). A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell, 160(4), 595–606. doi: 10.1016/j.cell.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin W-L, DeJesus-Hernandez M, … Petrucelli L. (2013). Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron, 77(4), 639–646. doi: 10.1016/j.neuron.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balendra R, & Isaacs AM (2018). C9orf72-mediated ALS and FTD: multiple pathways to disease. Nature Reviews Neurology, 14(9), 544–558. doi: 10.1038/s41582-018-0047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R, Charizanis K, Manchanda M, Mohan A, Li M, Finn DJ, … Swanson MS (2014). Loss of MBNL Leads to Disruption of Developmentally Regulated Alternative Polyadenylation in RNA-Mediated Disease. Molecular Cell, 56(2), 311–322. doi: 10.1016/j.molcel.2014.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beermann J, Piccoli M-T, Viereck J, & Thum T. (2016). Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiological Reviews, 96(4), 1297–1325. doi: 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, & Grummt I. (2014). Quiescence-Induced LncRNAs Trigger H4K20 Trimethylation and Transcriptional Silencing. Molecular Cell, 54(4), 675–682. doi: 10.1016/j.molcel.2014.03.032 [DOI] [PubMed] [Google Scholar]
- Blice-Baum AC, & Mihailescu M-R (2014). Biophysical characterization of G-quadruplex forming FMR1 mRNA and of its interactions with different fragile X mental retardation protein isoforms. RNA, 20(1), 103–114. doi: 10.1261/rna.041442.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe AJ, Fox AH, & Bond CS (2016). The ins and outs of lncRNA structure: How, why and what comes next? Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, 1859(1), 46–58. doi: 10.1016/j.bbagrm.2015.08.009 [DOI] [PubMed] [Google Scholar]
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, & Hudson T. (1992). Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell, 68(4), 799–808. 10.1016/0092-8674(92)90154-5 [DOI] [PubMed] [Google Scholar]
- Burberry A, Wells MF, Limone F, Couto A, Smith KS, Keaney J, … Eggan K. (2020). C9orf72 suppresses systemic and neural inflammation induced by gut bacteria. Nature, 582(7810), 89–94. doi: 10.1038/s41586-020-2288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, & Cullen BR (2007). The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA, 13(3), 313–316. doi: 10.1261/rna.351707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, & Slavoff SA (2020). Non-AUG start codons: Expanding and regulating the small and alternative ORFeome. Experimental Cell Research, 391(1), 111973. doi: 10.1016/j.yexcr.2020.111973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, … Gustincich S. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature, 491(7424), 454–457. doi: 10.1038/nature11508 [DOI] [PubMed] [Google Scholar]
- Cerase A, Armaos A, Neumayer C, Avner P, Guttman M, & Tartaglia GG (2019). Phase separation drives X-chromosome inactivation: a hypothesis. Nature Structural & Molecular Biology, 26(5), 331–334. doi: 10.1038/s41594-019-0223-0 [DOI] [PubMed] [Google Scholar]
- Chen J, Brunner A-D, Cogan JZ, Nuñez JK, Fields AP, Adamson B, … Weissman JS (2020). Pervasive functional translation of noncanonical human open reading frames. Science, 367(6482), 1140–1146. doi: 10.1126/science.aay0262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Yildirim I, Park H, Lohman JR, Guan L, Tran T, … Disney MD (2013). Structure of the Myotonic Dystrophy Type 2 RNA and Designed Small Molecules That Reduce Toxicity. ACS Chemical Biology, 9(2), 538–550. doi: 10.1021/cb4007387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid-Samper F, Gelabert-Baldrich M, Lang B, Lorenzo-Gotor N, Ponti RD, Severijnen L-AWFM, … Tartaglia GG (2018). An Integrative Study of Protein-RNA Condensates Identifies Scaffolding RNAs and Reveals Players in Fragile X-Associated Tremor/Ataxia Syndrome. Cell Reports, 25(12), 3422–3434.e7. doi: 10.1016/j.celrep.2018.11.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Walsh MJ, Higginbottom A, Highley JR, Dickman MJ, Edbauer D, … Shaw PJ (2014). Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain, 137(7), 2040–2051. doi: 10.1093/brain/awu120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchten RTP van, Wieringa B, & Wansink DG (2019). Expanded CUG repeats in DMPK transcripts adopt diverse hairpin conformations without influencing the structure of the flanking sequences. RNA, 25(4), 481–495. doi: 10.1261/rna.068940.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B, Qiao L, Zhang M, Cheng L, Zhang L, Geng L, … Yang J. (2019). lncRNA AK054386 Functions as a ceRNA to Sequester miR-199 and Induce Sustained Endoplasmic Reticulum Stress in Hepatic Reperfusion Injury. Oxidative Medicine and Cellular Longevity, 2019, 1–15. doi: 10.1155/2019/8189079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, … Rademakers R. (2011). Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron, 72(2), 245–256. doi: 10.1016/j.neuron.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere R, Napierala M, Ranum LPW, and Wells RD (2004). Hairpin Structure-forming Propensity of the (CCTG·CAGG) Tetranucleotide Repeats Contributes to the Genetic Instability Associated with Myotonic Dystrophy Type 2. Journal of Biological Chemistry, 279(40), 41715–41726. 10.1074/jbc.m406415200 [DOI] [PubMed] [Google Scholar]
- Diederichs S. (2014). The four dimensions of noncoding RNA conservation. Trends in Genetics, 30(4), 121–123. doi: 10.1016/j.tig.2014.01.004 [DOI] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, … Birney E. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature, 489(7414), 57–74. doi: 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmrich S, Streltsov A, Schmidt F, Thangapandi V, Reinhardt D, & Klusmann J-H (2014). LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Molecular Cancer, 13(1), 171. doi: 10.1186/1476-4598-13-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, & Rippe K. (2018). Formation of Chromatin Subcompartments by Phase Separation. Biophysical Journal, 114(10), 2262–2270. doi: 10.1016/j.bpj.2018.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MM, Anderson PJ, & Ivanov P. (2017). ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. Cell Reports, 21(12), 3573–3584. doi: 10.1016/j.celrep.2017.11.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett JW (1982). Recognition of protein coding regions in DNA sequences. Nucleic Acids Research, 10(17), 5303–5318. doi: 10.1093/nar/10.17.5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EMC, Parkinson G, & Isaacs AM (2012). C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Scientific Reports, 2(1), 1016. doi: 10.1038/srep01016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloss BS, & Dinger ME (2016). The specificity of long noncoding RNA expression. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, 1859(1), 16–22. doi: 10.1016/j.bbagrm.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, … Todd PK (2017). RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nature Communications, 8(1), 2005. doi: 10.1038/s41467-017-02200-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KM, Linsalata AE, & Todd PK (2016). RAN translation—What makes it run? Brain Research, 1647, 30–42. doi: 10.1016/j.brainres.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen Y, … Chen J-L (2015). A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene, 34(14), 1768–1779. doi: 10.1038/onc.2014.131 [DOI] [PubMed] [Google Scholar]
- HafezQorani S, Houdjedj A, Arici M, Said A, & Kazan H. (2019). RBPSponge: genome-wide identification of lncRNAs that sponge RBPs. Bioinformatics, 35(22), 4760–4763. doi: 10.1093/bioinformatics/btz448 [DOI] [PubMed] [Google Scholar]
- Handa V, Saha T, & Usdin K. (2003). The fragile X syndrome repeats form RNA hairpins that do not activate the interferon‐inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Research, 31(21), 6243–6248. doi: 10.1093/nar/gkg818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue GM, Castelli LM, Ferraiuolo L, Sanchez-Martinez A, Cooper-Knock J, Higginbottom A, … Shaw PJ (2017). SRSF1-dependent nuclear export inhibition of C9ORF72 repeat transcripts prevents neurodegeneration and associated motor deficits. Nature Communications, 8(1), 16063. doi: 10.1038/ncomms16063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HGNC Database, HUGO Gene Nomenclature Committee (HGNC), European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge CB10 1SD, United Kingdom: www.genenames.org. Retrieval date: 2020.08.02 https://www.genenames.org/data/genegroup/#!/group/1688 [Google Scholar]
- Ho TH, Bundman D, Armstrong DL, & Cooper TA (2005). Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Human Molecular Genetics, 14(11), 1539–1547. doi: 10.1093/hmg/ddi162 [DOI] [PubMed] [Google Scholar]
- Holt I, Mittal S, Furling D, Butler‐Browne GS, Brook JD, & Morris GE (2007). Defective mRNA in myotonic dystrophy accumulates at the periphery of nuclear splicing speckles. Genes to Cells, 12(9), 1035–1048. doi: 10.1111/j.1365-2443.2007.01112.x [DOI] [PubMed] [Google Scholar]
- Huang Y-S, Chang C-C, Lee S-S, Jou Y-S, & Shih H-M (2016). Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget, 7(28), 43256–43266. doi: 10.18632/oncotarget.9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, & Weissman JS (2011). Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell, 147(4), 789–802. doi: 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R, Kroehling L, Khitun A, Bailis W, Jarret A, York AG, … Flavell RA (2018). The translation of non-canonical open reading frames controls mucosal immunity. Nature, 564(7736), 434–438. doi: 10.1038/s41586-018-0794-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, & Vale RD (2017). RNA phase transitions in repeat expansion disorders. Nature, 546(7657), 243–247. doi: 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, & Struhl K. (2015). Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. ELife, 4, e08890. doi: 10.7554/elife.08890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, & Thornton CA (2004). Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Human Molecular Genetics, 13(24), 3079–3088. doi: 10.1093/hmg/ddh327 [DOI] [PubMed] [Google Scholar]
- Jones K, Wei C, Iakova P, Bugiardini E, Schneider-Gold C, Meola G, … Timchenko LT (2012). GSK3β mediates muscle pathology in myotonic dystrophy. Journal of Clinical Investigation, 122(12), 4461–4472. doi: 10.1172/jci64081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K, Wei C, Schoser B, Meola G, Timchenko N, & Timchenko L. (2015). Reduction of toxic RNAs in myotonic dystrophies type 1 and type 2 by the RNA helicase p68/DDX5. Proceedings of the National Academy of Sciences, 112(26), 8041–8045. doi: 10.1073/pnas.1422273112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, … Pandolfi PP (2011). In Vivo Identification of Tumor- Suppressive PTEN ceRNAs in an Oncogenic BRAF-Induced Mouse Model of Melanoma. Cell, 147(2), 382–395. doi: 10.1016/j.cell.2011.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, & Todd PK (2016). CGG Repeat-Associated Non-AUG Translation Utilizes a Cap-Dependent Scanning Mechanism of Initiation to Produce Toxic Proteins. Molecular Cell, 62(2), 314–322. doi: 10.1016/j.molcel.2016.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Marinov GK, Pepke S, Singer ZS, He P, Williams B, … Wold BJ (2015). Single-Cell Transcriptome Analysis Reveals Dynamic Changes in lncRNA Expression during Reprogramming. Cell Stem Cell, 16(1), 88–101. doi: 10.1016/j.stem.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Langlois M-A, Lee K-B, Riggs AD, Puymirat J, & Rossi JJ (2005). HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Research, 33(12), 3866–3874. doi: 10.1093/nar/gki698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-K, Kim B, & Kim VN (2016). Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proceedings of the National Academy of Sciences, 113(13), E1881–E1889. doi: 10.1073/pnas.1602532113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Duan Y, Sang Y, Li Y, Zhang H, Liang Y, … Yang Q. (2019). LncRNA–CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA‐215. Journal of Cellular Physiology, 234(6), 9105–9117. doi: 10.1002/jcp.27587 [DOI] [PubMed] [Google Scholar]
- Koscianska E, Witkos TM, Kozlowska E, Wojciechowska M, & Krzyzosiak WJ (2015). Cooperation meets competition in microRNA-mediated DMPK transcript regulation. Nucleic Acids Research, 43(19), 9500–9518. doi: 10.1093/nar/gkv849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraan CM, Godler DE, & Amor DJ (2019). Epigenetics of fragile X syndrome and fragile X‐related disorders. Developmental Medicine & Child Neurology, 61(2), 121–127. doi: 10.1111/dmcn.13985 [DOI] [PubMed] [Google Scholar]
- Krause HM (2018). New and Prospective Roles for lncRNAs in Organelle Formation and Function. Trends in Genetics, 34(10), 736–745. doi: 10.1016/j.tig.2018.06.005 [DOI] [PubMed] [Google Scholar]
- Kremer E, Pritchard M, Lynch M, Yu S, Holman K, Baker E, … Richards R. (1991). Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science, 252(5013), 1711. doi: 10.1126/science.1675488 [DOI] [PubMed] [Google Scholar]
- Krol J, Fiszer A, Mykowska A, Sobczak K, Mezer M. de, & Krzyzosiak WJ (2007). Ribonuclease Dicer Cleaves Triplet Repeat Hairpins into Shorter Repeats that Silence Specific Targets. Molecular Cell, 25(4), 575–586. doi: 10.1016/j.molcel.2007.01.031 [DOI] [PubMed] [Google Scholar]
- Kumar A, Park H, Fang P, Parkesh R, Guo M, Nettles KW, & Disney MD (2011). Myotonic Dystrophy Type 1 RNA Crystal Structures Reveal Heterogeneous 1 × 1 Nucleotide UU Internal Loop Conformations. Biochemistry, 50(45), 9928–9935. doi: 10.1021/bi2013068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang G-S, & Cooper TA (2007). Increased Steady-State Levels of CUGBP1 in Myotonic Dystrophy 1 Are Due to PKC-Mediated Hyperphosphorylation. Molecular Cell, 28(1), 68–78. doi: 10.1016/j.molcel.2007.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent F-X, Sureau A, Klein AF, Trouslard F, Gasnier E, Furling D, & Marie J. (2012). New function for the RNA helicase p68/DDX5 as a modifier of MBNL1 activity on expanded CUG repeats. Nucleic Acids Research, 40(7), 3159–3171. doi: 10.1093/nar/gkr1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-B, Chen H-J, Peres JN, Gomez-Deza J, Attig J, Štalekar M, … Shaw CE (2013). Hexanucleotide Repeats in ALS/FTD Form Length-Dependent RNA Foci, Sequester RNA Binding Proteins, and Are Neurotoxic. Cell Reports, 5(5), 1178–1186. doi: 10.1016/j.celrep.2013.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Dong C, Cui J, Wang Y, & Hong X. (2018). Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. Journal of Experimental & Clinical Cancer Research, 37(1), 265. doi: 10.1186/s13046-018-0941-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, … Thornton CA (2006). Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Human Molecular Genetics, 15(13), 2087–2097. doi: 10.1093/hmg/ddl132 [DOI] [PubMed] [Google Scholar]
- Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, … Ranum LPW (2001). Myotonic Dystrophy Type 2 Caused by a CCTG Expansion in Intron 1 of ZNF9. Science, 293(5531), 864. doi: 10.1126/science.1062125 [DOI] [PubMed] [Google Scholar]
- Liu C, Peng Z, Li P, Fu H, Feng J, Zhang Y, … Wu M. (2020). LncRNA RMST Suppressed GBM Cells Mitophagy through Enhancing FUS SUMOylation. Molecular Therapy - Nucleic Acids, 19, 1198–1208. doi: 10.1016/j.omtn.2020.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, … Lim DA (2016). Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biology, 17(1), 67. doi: 10.1186/s13059-016-0932-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Xiaowen, Xiao Z-D, Han L, Zhang J, Lee S-W, Wang W, … Gan B. (2016). LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nature Cell Biology, 18(4), 431–442. doi: 10.1038/ncb3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Xin, Zhu Q, Guo Y, Xiao Z, Hu L, & Xu Q. (2019). LncRNA LINC00689 promotes the growth, metastasis and glycolysis of glioma cells by targeting miR-338–3p/PKM2 axis. Biomedicine & Pharmacotherapy, 117, 109069. doi: 10.1016/j.biopha.2019.109069 [DOI] [PubMed] [Google Scholar]
- Lu S, Zhang J, Lian X, Sun L, Meng K, Chen Y, … He Q-Y (2019). A hidden human proteome encoded by ‘non-coding’ genes. Nucleic Acids Research, 47(15), 8111–8125. doi: 10.1093/nar/gkz646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, … Chang HY (2016). RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell, 165(5), 1267–1279. doi: 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, … Shen X. (2016). Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell, 18(5), 637–652. doi: 10.1016/j.stem.2016.01.024 [DOI] [PubMed] [Google Scholar]
- Ma Li, Sun X, Kuai W, Hu J, Yuan Y, Feng W, & Lu X. (2019). LncRNA SOX2 overlapping transcript acts as a miRNA sponge to promote the proliferation and invasion of Ewing’s sarcoma. American Journal of Translational Research, 11(6), 3841–3849. Retrieved from https://pubmed.ncbi.nlm.nih.gov/31312393 [PMC free article] [PubMed] [Google Scholar]
- Ma Lina, Cao J, Liu L, Du Q, Li Z, Zou D, … Zhang Z. (2018). LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Research, 47(D1), gky960-. doi: 10.1093/nar/gky960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, … Masutomi K. (2009). An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature, 461(7261), 230–235. doi: 10.1038/nature08283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y, … Tao Y. (2018). A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Research, canres.3454.2017. doi: 10.1158/0008-5472.can-17-3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley ME, O’Rourke JG, Yáñez A, Markman JL, Ho R, Wang X, Chen S, Lall D, Jin M, Muhammad A, Bell S, Landeros J, Valencia V, Harms M, Arditi M, Jefferies C, & Baloh RH (2020). C9orf72 in myeloid cells suppresses STING-induced inflammation. Nature, 585(7823), 96–101. 10.1038/s41586-020-2625-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng‐umnuay P, Stenberg MG, Byrne BJ, Thornton CA, & Swanson MS (2000). Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. The EMBO Journal, 19(17), 4439–4448. doi: 10.1093/emboj/19.17.4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K-W, Davila S, Zealy RW, Lloyd LT, Lee IY, Lee R, … Yoon J-H (2017). eIF4E phosphorylation by MST1 reduces translation of a subset of mRNAs, but increases lncRNA translation. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, 1860(7), 761–772. doi: 10.1016/j.bbagrm.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Mitrea DM, & Kriwacki RW (2016). Phase separation in biology; functional organization of a higher order. Cell Communication and Signaling, 14(1), 1. doi: 10.1186/s12964-015-0125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers BH, Logue JS, & Berglund JA (2005). The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proceedings of the National Academy of Sciences of the United States of America, 102(46), 16626–16631. 10.1073/pnas.0505873102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriss GR, & Cooper TA (2017). Protein sequestration as a normal function of long noncoding RNAs and a pathogenic mechanism of RNAs containing nucleotide repeat expansions. Human Genetics, 136(9), 1247–1263. doi: 10.1007/s00439-017-1807-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L, Lou F, Sun Y, Sun L, Cai X, Liu Z, … Wang H. (2020). A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Science Advances, 6(21), eaaz2059. doi: 10.1126/sciadv.aaz2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, & Lee ES (2015). Non-coding RNA: what is functional and what is junk? Frontiers in Genetics, 6, 2. doi: 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Dansithong W, Kim D, Rossi J, Webster NJ, Comai L, & Reddy S. (2006). Interaction of musleblind, CUG‐BP1 and hnRNP H proteins in DM1‐associated aberrant IR splicing. The EMBO Journal, 25(18), 4271–4283. doi: 10.1038/sj.emboj.7601296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE (2011). Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities! PLoS Genetics, 7(3), e1002018. doi: 10.1371/journal.pgen.1002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W-X, Koirala P, & Mo Y-Y (2017). LncRNA-mediated regulation of cell signaling in cancer. Oncogene, 36(41), 5661–5667. doi: 10.1038/onc.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, & Reik W. (2009). Evolution and Functions of Long Noncoding RNAs. Cell, 136(4), 629–641. doi: 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Rajendra TK, Lal AK, & Lakhotia SC (2000). Omega speckles - a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J. Cell Sci, 113(19), 3485 Retrieved from http://jcs.biologists.org/content/113/19/3485.abstract [DOI] [PubMed] [Google Scholar]
- Qu L, Ding J, Chen C, Wu Z-J, Liu B, Gao Y, … Wang L-H (2016). Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell, 29(5), 653–668. doi: 10.1016/j.ccell.2016.03.004 [DOI] [PubMed] [Google Scholar]
- Ren J, Fu J, Ma T, Yan B, Gao R, An Z, & Wang D. (2018). LncRNA H19-elevated LIN28B promotes lung cancer progression through sequestering miR-196b. Cell Cycle, 17(11), 1372–1380. doi: 10.1080/15384101.2018.1482137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Clements J, & Eddy SR (2017). A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nature Methods, 14(1), 45–48. doi: 10.1038/nmeth.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler LE, Kosmyna B, Moskowitz D, Bebawee R, Rahimzadeh J, Kutchko K, … Rogler CE (2014). Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage–hair hypoplasia. Human Molecular Genetics, 23(2), 368–382. doi: 10.1093/hmg/ddt427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan X, Li P, Chen Y, Shi Y, Pirooznia M, Seifuddin F, … Cao H. (2020). In vivo functional analysis of non-conserved human lncRNAs associated with cardiometabolic traits. Nature Communications, 11(1), 45. doi: 10.1038/s41467-019-13688-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Jens M, Murakawa Y, Herzog M, Landthaler M, & Rajewsky N. (2014). A Variety of Dicer Substrates in Human and C. elegans. Cell, 159(5), 1153–1167. doi: 10.1016/j.cell.2014.10.040 [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, & Pandolfi PP (2011). A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell, 146(3), 353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M, Fontana L, Masciullo M, Bianchi MLE, Rossi S, Leoncini E, … Silvestri G. (2015). Expansion size and presence of CCG/CTC/CGG sequence interruptions in the expanded CTG array are independently associated to hypermethylation at the DMPK locus in myotonic dystrophy type 1 (DM1). Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, 1852(12), 2645–2652. doi: 10.1016/j.bbadis.2015.09.007 [DOI] [PubMed] [Google Scholar]
- Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, … Charlet‐Berguerand N. (2010). Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. The EMBO Journal, 29(7), 1248–1261. doi: 10.1038/emboj.2010.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, … Weeks KM (2016). SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proceedings of the National Academy of Sciences, 113(37), 10322–10327. doi: 10.1073/pnas.1600008113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer HL, Sanders R, Boulberdaa M, Meloni M, Cochrane A, Spiroski A-M, … Baker AH (2020). The LINC00961 transcript and its encoded micropeptide SPAAR regulate endothelial cell function. Cardiovascular Research. doi: 10.1093/cvr/cvaa008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, … Walter P. (2016). Translation from the 5′ untranslated region shapes the integrated stress response. Science, 351(6272), aad3867. doi: 10.1126/science.aad3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, Johnston A, Overstreet K, Oh SY, Todd PK, Berry JD, Cudkowicz ME, Boeve BF, Dickson D, Floeter MK, Traynor BJ, … Disney MD (2014). Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron, 83(5), 1043–1050. 10.1016/j.neuron.2014.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Zhong G, Zhang H, Yu B, Wei F, Luo L, … Bi A. (2018). LncRNA DANCR upregulates PI3K/AKT signaling through activating serine phosphorylation of RXRA. Cell Death & Disease, 9(12), 1167. doi: 10.1038/s41419-018-1220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares RCA, Pyle AM, & Somarowthu S. (2019). Phylogenetic Analysis with Improved Parameters Reveals Conservation in lncRNA Structures. Journal of Molecular Biology, 431(8), 1592–1603. doi: 10.1016/j.jmb.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DW, & Dinger ME (2016). Endogenous microRNA sponges: evidence and controversy. Nature Reviews Genetics, 17(5), 272–283. doi: 10.1038/nrg.2016.20 [DOI] [PubMed] [Google Scholar]
- TIAN B, WHITE RJ, XIA T, WELLE S, TURNER DH, MATHEWS MB, & THORNTON CA (2000). Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA, 6(1), 79–87. doi: 10.1017/s1355838200991544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, … Paulson HL (2013). CGG Repeat-Associated Translation Mediates Neurodegeneration in Fragile X Tremor Ataxia Syndrome. Neuron, 78(3), 440–455. doi: 10.1016/j.neuron.2013.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, & Bartel DP (2013). lincRNAs: Genomics, Evolution, and Mechanisms. Cell, 154(1), 26–46. doi: 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uroda T, Anastasakou E, Rossi A, Teulon J-M, Pellequer J-L, Annibale P, … Marcia M. (2019). Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Molecular Cell, 75(5), 982–995.e9. doi: 10.1016/j.molcel.2019.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, … Zhu X. (2001). The Sequence of the Human Genome. Science, 291(5507), 1304. doi: 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- Wan G, Hu X, Liu Y, Han C, Sood AK, Calin GA, … Lu X. (2013). A novel non‐coding RNA lncRNA‐JADE connects DNA damage signalling to histone H4 acetylation. The EMBO Journal, 32(21), 2833–2847. doi: 10.1038/emboj.2013.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ge L, Wu J, Wang X, & Yuan L. (2018). MiR-219 represses expression of dFMR1 in Drosophila melanogaster. Life Sciences, 218, 31–37. doi: 10.1016/j.lfs.2018.12.008 [DOI] [PubMed] [Google Scholar]
- Wang ET, Ward AJ, Cherone JM, Giudice J, Wang TT, Treacy DJ, … Burge CB (2015). Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Research, 25(6), 858–871. doi: 10.1101/gr.184390.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkos TM, Krzyzosiak WJ, Fiszer A, & Koscianska E. (2018). A potential role of extended simple sequence repeats in competing endogenous RNA crosstalk. RNA Biology, 15(11), 1399–1409. doi: 10.1080/15476286.2018.1536593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Yin Q-F, Luo Z, Yao R-W, Zheng C-C, Zhang J, … Chen L-L (2016). Unusual Processing Generates SPA LncRNAs that Sequester Multiple RNA Binding Proteins. Molecular Cell, 64(3), 534–548. doi: 10.1016/j.molcel.2016.10.007 [DOI] [PubMed] [Google Scholar]
- Xing W, Xu W-Y, Chang L, Zhang K, & Wang S-R (2020). SP1-induced lncRNA LINC00689 overexpression contributes to osteosarcoma progression via the miR-655/SOX18 axis. European Review for Medical and Pharmacological Sciences, 24(5), 2205–2217. doi: 10.26355/eurrev_202003_20486 [DOI] [PubMed] [Google Scholar]
- Xue Z, Hennelly S, Doyle B, Gulati AA, Novikova IV, Sanbonmatsu KY, & Boyer LA (2016). A G-Rich Motif in the lncRNA Braveheart Interacts with a Zinc-Finger Transcription Factor to Specify the Cardiovascular Lineage. Molecular Cell, 64(1), 37–50. doi: 10.1016/j.molcel.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadava RS, Foff EP, Yu Q, Gladman JT, Kim YK, Bhatt KS, … Mahadevan MS (2015). TWEAK/Fn14, a pathway and novel therapeutic target in myotonic dystrophy. Human Molecular Genetics, 24(7), 2035–2048. doi: 10.1093/hmg/ddu617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Nakagawa S, & Hirose T. (2020). Architectural RNAs for Membraneless Nuclear Body Formation. Cold Spring Harbor Symposia on Quantitative Biology, 039404. doi: 10.1101/sqb.2019.84.039404 [DOI] [PubMed] [Google Scholar]
- Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, … Disteche CM (2015). The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biology, 16(1), 52. doi: 10.1186/s13059-015-0618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeasmin F, Yada T, & Akimitsu N. (2018). Micropeptides Encoded in Transcripts Previously Identified as Long Noncoding RNAs: A New Chapter in Transcriptomics and Proteomics. Frontiers in Genetics, 9, 144. doi: 10.3389/fgene.2018.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-H, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, … Gorospe M. (2013). Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nature Communications, 4(1), 2939. doi: 10.1038/ncomms3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Teng X, & Bonini NM (2011). Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy. PLoS Genetics, 7(3), e1001340. doi: 10.1371/journal.pgen.1001340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G, Liu B, Han W, & Zhao D. (2019). LncRNA-MIR17HG mediated upregulation of miR-17 and miR-18a promotes colon cancer progression via activating Wnt/β-catenin signaling. Translational Cancer Research, 8(4), 1097–1108. doi: 10.21037/tcr.2019.06.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, … Rothstein JD (2015). The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature, 525(7567), 56–61. doi: 10.1038/nature14973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-J, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu Y-F, … Petrucelli L. (2016). C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nature Neuroscience, 19(5), 668–677. doi: 10.1038/nn.4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Sentürk N, Song C, & Grummt I. (2018). lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes & Development, 32(11–12), 836–848. doi: 10.1101/gad.311688.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ye X, Xu J, Cao M-G, Fang Z-Y, Li L-Y, … Xie D. (2017). The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal, 10(483), eaak9557. doi: 10.1126/scisignal.aak9557 [DOI] [PubMed] [Google Scholar]
- Zongaro S, Hukema R, D’Antoni S, Davidovic L, Barbry P, Catania MV, … Bardoni B. (2013). The 3′ UTR of FMR1 mRNA is a target of miR-101, miR-129–5p and miR-221: implications for the molecular pathology of FXTAS at the synapse. Human Molecular Genetics, 22(10), 1971–1982. doi: 10.1093/hmg/ddt044 [DOI] [PubMed] [Google Scholar]
- Zu T, Cleary JD, Liu Y, Bañez-Coronel M, Bubenik JL, Ayhan F, … Ranum LPW (2017). RAN Translation Regulated by Muscleblind Proteins in Myotonic Dystrophy Type 2. Neuron, 95(6), 1292–1305.e5. doi: 10.1016/j.neuron.2017.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, … Ranum LPW (2011). Non-ATG–initiated translation directed by microsatellite expansions. Proceedings of the National Academy of Sciences, 108(1), 260–265. doi: 10.1073/pnas.1013343108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumwalt M, Ludwig A, Hagerman PJ, & Dieckmann T. (2007). Secondary Structure and Dynamics of the r(CGG) Repeat in the mRNA of the Fragile X Mental Retardation 1(FMR1)Gene. RNA Biology, 4(2), 93–100. doi: 10.4161/rna.4.2.5039 [DOI] [PubMed] [Google Scholar]


