Abstract
The retina experiences increased oxidative stress in diabetes, and the transcriptional activity of Nrf2, which is critical in regulating many antioxidant genes, is decreased. The nuclear movement/transcriptional activity of Nrf2 is mediated by its intracellular inhibitor Keap1, and retinal Keap1 levels are increased in diabetes. Gene expression is also regulated by long noncoding RNAs (LncRNAs). Our aim was to investigate the role of LncRNA MALAT1 in the regulation of Keap1-Nrf2-antioxidant defense in diabetic retinopathy. LncRNA MALAT1 expression (quantitative real-time PCR, immunofluorescence, and RNA sequencing), its interactions with Keap1 (FACS), Keap1-Nrf2 interactions, and transcription of the antioxidant response genes (immunofluorescence and nuclear RNA sequencing) were investigated in retinal endothelial cells exposed to high glucose. Glucose increased LncRNA MALAT1 levels by increasing Sp1 transcription factor binding at its promoter. Downregulation of LncRNA MALAT1 by its siRNA prevented glucose-induced increase in Keap1 and facilitated Nrf2 nuclear translocation and antioxidant gene transcription. Retinal microvessels from streptozotocin-induced diabetic mice and human donors with diabetic retinopathy also presented similar increases in LncRNA MALAT1 and its interactions with Keap1 and decreases in Nrf2-mediated antioxidant defense genes. Thus, LncRNA MALAT1, via Keap1-Nrf2, regulates antioxidant defense in diabetic retinopathy. Inhibition of LncRNA MALAT1 has potential to protect the retina from oxidative damage and to prevent or slow down diabetic retinopathy.
Introduction
Oxidative stress plays a major role in the pathogenesis of diabetic retinopathy (1). Retinal mitochondria become dysfunctional, and their microvasculature loses its integrity leading to accelerated apoptosis, a phenomenon that precedes the development of the histopathology characteristics of diabetic retinopathy (2–5). The antioxidant defense system to neutralize reactive oxygen species (ROS) is also compromised (6,7). Nuclear factor erythroid 2-related factor 2 (Nrf2), a master transcription factor, plays a crucial role in regulating the expression of many antioxidant defense genes (8). Nrf2 is sequestered in the cytoplasm by its specific repressor, Kelch-like ECH-associated protein 1 (Keap1), but under stress, it translocates in the nucleus to activate transcription of antioxidant defense genes, including hemeoxygenase 1 (HO1) and superoxide dismutase 2 (Sod2) (7,9). Keap1 levels are increased in the retina in diabetes, and despite no major change in the expression, nuclear translocation and transcriptional activity of Nrf2 are decreased (10). Decreased transcription of the antioxidant-response genes results in increased ROS accumulation and, ultimately, in mitochondrial damage (2–4,7). However, the mechanism by which diabetes increases Keap1 expression remains elusive.
Epigenetic modifications play an essential role in regulating gene expression, affecting signaling pathways and, ultimately, altering the cellular physiology. These modifications are controlled by many external factors including lifestyle and disease state (11,12). Diabetes is associated with several epigenetic modifications that result in transcriptional activation or repression of many genes implicated in the development of retinopathy (2,4). Methylation of lysine 4 of histones 3 at the Keap1 promoter in diabetes is associated with its transcriptional activation, resulting in increased tethering of Nrf2 (13). In addition to histones and DNA modifications, long noncoding RNA (LncRNA), a class of transcribed RNA molecules with more than 200 nucleotides, also regulate gene expression. These LncRNAs, by bringing the transcription factors to the transcription start site, or by interacting directly with the DNA at the gene promoter site, play an important role in transcription (14). Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is one of the highly conserved LncRNAs implicated in many pathological processes including inflammation, angiogenesis, and cell apoptosis (15,16). LncRNA MALAT1 is also shown to play a significant role in the pathogenesis of diabetic complications including cardiomyopathy and nephropathy (17). By directly interacting with the transcription factor Sp1 and facilitating its recruitment to the promoter, it regulates the expression of latent-transforming growth factor β-binding protein 3 gene (18). We have shown that Sp1 binding at the Keap1 promoter is increased in diabetes, resulting in its transcriptional activation (13). The role of LncRNA MALAT1 in regulation of Keap1 and its interactions with Nrf2 in diabetic retinopathy are unclear.
The goal of this study is to investigate the role of LncRNA MALAT1 in the development of diabetic retinopathy, specifically in the regulation oxidative stress, via Keap1-Nrf2 interactions. Using human retinal endothelial cells (HRECs), we investigated the effect of high glucose on LncRNA MALAT1 expression and the regulation of LncRNA MALAT1 on Keap1 transcription, Keap1-Nrf2 interactions, and antioxidant-response gene transcription. In vitro results are confirmed in the retinal microvessels from a mouse model of diabetic retinopathy and from human donors with documented diabetic retinopathy.
Research Design and Methods
Retinal Endothelial Cells
Endothelial cells from human retina (cat. no. ACBRI 181; Cell Systems Corp., Kirkland, WA) were cultured in DMEM (cat. no. D5523; Sigma-Aldrich, St. Louis, MO) supplemented with 12% heat-inactivated FBS, 15 μg/mL endothelial cell growth supplement, and 1% each insulin, transferrin, selenium, GlutaMAX, and antibiotic/antimitotic. Cells from the fifth to eighth passage were incubated in 5 mmol/L d-glucose (NG) or 20 mmol/L d-glucose (HG) for 96 h. l-glucose (L-Glu; 20 mmol/L), instead of 20 mmol/L d-glucose, was used as an osmotic/metabolic control (19,20).
HRECs from the fifth to sixth passage were transfected using two independent sequences of Silencer Select MALAT1-siRNA (MAL-si; no. 1, cat. no. 4392420, n272231; no. 2, cat. no. 4390771, n272237; Ambion, Carlsbad, CA) employing Lipofectamine RNAiMAX transfection reagent (cat. no. 13778–030; Invitrogen, Carlsbad, CA). Nontargeting scrambled RNA was used as a control. Each experiment had HRECs from the same batch and the same passage and was repeated three to four times (20). The transfection efficiency, evaluated by quantifying MALAT1 transcripts, was ∼50% for siRNA 1 and 40–45% for siRNA 2.
Mouse
For the experiment, 2-month-old C57BL/6J mice (either sex; The Jackson Laboratory, Bar Harbor, ME) were made diabetic by streptozotocin injection (55 mg/kg/mouse for four consecutive days) and maintained for 6 months, as described previously (19,20). Age-matched normal C57BL/6J mice were used as controls, and each group had 10–12 mice. The treatment of animals conformed to the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research, and the experiment was approved by the Wayne State University’s Institutional Animal Care and Use Committee.
Human Donors
Eye globes from human donors with documented diabetic retinopathy (four male and three female, 40–75 years of age and 14–41 years of diabetes), and their age-matched donors without diabetes (three male and four female), enucleated within 6–8 h of death, were obtained from the Eversight Eye Bank (Ann Arbor, MI). A small portion of the retina (one-sixth to one-eighth) was used to prepare the microvessels. Our exclusion criteria included donors with any other ocular diseases and chronic diseases (e.g., cancer, Alzheimer), HIV and other communicable diseases, and drug use during the past 3 years. Although retinopathy was documented in these donors with diabetes, information about the severity of their retinopathy was not available.
Retinal microvessels were isolated by incubating the retina (mouse/human) in 5–10 mL of deionized water for 60 min, as described previously (20,21).
Gene Expression
RNA was isolated by Trizol (cat. no. 15596018; Ambion), and gene transcripts were quantified by SYBR green–based, quantitative real-time (qRT)-PCR using gene- and species-specific primers (20) (Supplementary Table 1). Housekeeping genes included β-actin for human samples and 18S rRNA for mouse.
Keap1 protein expression was quantified by Western blot using Keap1 antibody (1:1,000 dilution, cat. no. sc-365626; Santa Cruz Biotechnology, Santa Cruz, CA) and β-actin (1:3,000 dilution, cat. no. A5441; Sigma-Aldrich) as a loading control.
The coimmunoprecipitation technique was used to examine Nrf2-Keap1 interactions (22). Cytosolic protein (150 μg) was incubated with 2-μg Keap1 antibody (Santa Cruz Biotechnology), followed by incubation with Protein A/G plus agarose immunoprecipitation beads. Proteins, extracted from the beads, were separated on an SDS-PAGE, and the membranes were immunoblotted with Nrf2 antibody (1:500 dilution, cat. no. ab92946; Abcam, Cambridge, MA).
Plasmid and Constructs
Keap1 promoter (−800 to +21) was PCR amplified and cloned into the promoter-less pGL3-Basic-IRES plasmid (plasmid no. 64784; Addgene, Watertown, MA) using pGL3-control vector (cat. no. E1741; Promega, Madison, WI) as a control (Supplementary Table 2 lists the primers). HRECs were transfected using Turbofect transfection reagent (cat. no. R0531; Thermo Fisher Scientific, Waltham, MA), as described previously (20).
Luciferase assay was performed by Pierce Firefly Luciferase Glow Assay Kit (cat. no. 16176; Thermo Fisher Scientific) (20). The results were normalized with the values obtained from pGL3-control vector–transfected cells.
Localization of Nrf2 and Keap1 was performed by the immunofluorescence technique using Nrf2 (1:200 dilution, cat. no. ab92946; Abcam) and Keap1 (1:100 dilution) antibodies. Secondary antibodies against Nrf2 and Keap1 included AlexaFluor 488-conjugated anti-rabbit and Texas red-conjugated anti-mouse (cat. no. A11008; Molecular Probes, Grand Island, NE, 1:500 dilution each, cat. no. TI-2000; Vector Laboratories, Burlingame, CA). The coverslips were mounted using DAPI-containing mounting medium (Vector Laboratories), and images were captured by Zeiss microscope at 40× magnification using the Apotome module. The intensity profile was determined by line region of interest across the cytosolic area, and the Pearson coefficient was evaluated using the Zeiss software module (20).
RNA Fluorescence In Situ Hybridization
A fuorescein-12-dUTP–incorporated probe was prepared from asymmetric PCR amplification, and the PCR products were gel purified. Briefly, HRECs fixed with 4% paraformaldehyde (PFA) were dehydrated with 70–100% ethanol, air dried, and incubated at 37°C for 2 h with a denatured probe in formamide-containing buffer. The cells were then washed in hybridization buffer, followed by saline sodium citrate buffer, and mounted using Vectashield. The hybridized probes were visualized by signals of fluorescein-12-dUTP–incorporated probe (23). The arithmetic mean intensity (AMI) was determined by region of interest using the Zeiss software module (20).
Dual-Stained Flow Cytometry
HRECs fixed in 70% ethanol were blocked in 5% BSA, and incubated with Keap1 antibody (1:100 dilution, cat. no. sc-365626; Santa Cruz Biotechnology). Texas red-conjugated anti-mouse secondary antibody (Vector Laboratories) was used as the secondary antibody. The cells were then washed with PBS and fixed with PFA, and fluorescein-12-dUTP incorporated LncRNA MALAT1 probe was hybridized with formamide-containing hybridization buffer. After washing with PBS, the suspended dual-stained HRECs were scanned under FL1 488 nm and FL3 640 nm wavelength excitation in BD Accuri C6 plus flow cytometer (BD Biosciences, San Jose, CA). Raw Flow Cytometry Standard files were analyzed by Winlist 3D v9 (Verity Software House, Topsham, ME) (24).
RNA Immunoprecipitation
Chromatin extract was prepared by sonication from the crosslinked samples, and uniform shearing was confirmed by agarose gel electrophoresis. Protein-DNA complex (24 µg DNA) was immunoprecipitated by Sp1 antibody (4 µg) (cat. no. sc-17824; Santa Cruz Biotechnology); normal rabbit IgG (4 µg IgG) (cat. no. ab172730; Abcam) was used as an antibody control. The antibody-chromatin complex was precipitated by Protein A/G PLUS-Agarose beads (cat. no. sc-2003; Santa Cruz Biotechnology), and the RNA protein complex was decrosslinked with proteinase K at 37°C for 30 min. RNA was then extracted using Trizol, cDNA was synthesized using high-capacity cDNA reverse transcription kit (cat. no. 4368814; Applied Biosystems), and the targeted RNA content was quantified by qRT-PCR (25).
Chromatin Isolation by RNA Purification
Samples were crosslinked with 1% PFA and sonicated to prepare chromatin extract. DNA (24 µg) from the sheared chromatin extract was used for chromatin isolation by RNA purification (ChIRP). The fluorescein-12-dUTP–incorporated LncRNA MALAT1 probe (100 pmol) (cat. no. FERR0101; Thermo Fisher Scientific) was hybridized, and the RNA-chromatin complex was immunoprecipitated with anti-fluorescein antibody–bound Protein A/G PLUS-Agarose beads (ref. no. 11426320001; Roche Diagnostics GmbH, Mannheim, Germany) (26). RNA immunoprecipitation-associated DNA fragments were quantified by qRT-PCR for the Keap1 promoter.
RNA Sequencing
Since cellular separation can significantly improve the analysis of complex transcriptomes (27), the nuclear fraction was isolated from HRECs by centrifugation using a kit from Thermo Fisher Scientific (cat. no. 89874). Nuclear RNA was purified, and outsourced for library preparation and Illumina next generation sequencing by GENEWIZ (South Plainfield, NJ). The raw FASTQ sequencing files were then uploaded in the server of www.usegalaxy.org and aligned against the hg38 human genome data set using the TopHat module. The aligned bam files were used to calculate the fold change and fragments per kilobase per million mapped reads (FPKM) by stringtie modules (28).
Chromatin Immunoprecipitation Sequencing
Protein-DNA complex (24 μg DNA) was immunoprecipitated by Sp1 antibody or IgG (4 μg each). The antibody-chromatin complex was precipitated by Protein A/G Agarose/Salmon Sperm DNA (cat. no. 16–157; Sigma-Aldrich Corp). DNA was isolated after overnight decrosslinking at 65°C and quantified by qRT-PCR targeting Keap1 promoter. The immunoprecipitated DNA was cleaned using NEBNext sample purification beads (cat. no. E7104S; New England Biolabs, Ipswich, MA), and ∼100 ng DNA was used for library preparation following the user manual protocol by NEBNext Ultra II DNA library prep kit for Illumina and NEBNext multiplex oligos index primers set 1 (cat. nos. E7645S and E7335S, respectively; New England Biolabs). The library was outsourced for Illumina Next Generation Sequencing by GENEWIZ.
The raw FASTQ sequencing files were uploaded on the www.usegalaxy.org server and aligned against the hg38 human genome data set using the bowtie 2 module. The aligned bam files were filtered with bamfilter and were compared using bamcompare and reads per kilobase per million mapped reads (RPKM), as the normalization criteria. The files in bigwig file format was analyzed in the UCSC Genome Browser (https://genome.ucsc.edu/) for promoter occupancy (28).
Micrococcal Nuclease Sequencing
Nucleosomal DNA was prepared from HRECs using the EZ Nucleosomal DNA preparation kit (cat. no. D5220; Zymo Research, Irvine, CA). About 100 ng nucleosomal DNA was processed for library preparation and 500 ng DNA was run on a 1.2% agarose gel to check for ∼150 base pair size uniformity (29).
Statistical Analysis
The statistical analysis was performed using Graph Pad Prism (San Diego, CA), and the results were expressed as mean ± SD. Significance of variance was analyzed using one-way ANOVA, and a P value <0.05 was considered as statistically significant.
Data and Resource Availability
The data sets generated during the current study are available from the corresponding author upon reasonable request.
Results
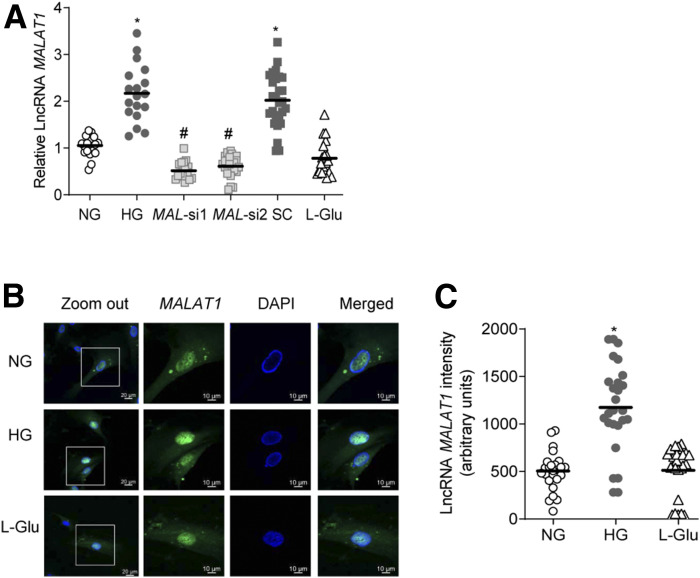
Gene transcripts of MALAT1 were elevated by over twofold in HRECs incubated in high glucose for 96 h, compared with HRECs in normal glucose (Fig. 1A). A similar increase in MALAT1 was observed as early as 12 h of high glucose exposure (data not shown). However, LncRNA MAL-si protected glucose-induced increases in MALAT1 transcripts. Compared with untransfected or scrambled RNA transfected cells in high glucose, siRNA-transfected cells (both MAL-si 1 and MAL-si 2) had significantly reduced MALAT1 (Fig. 1A). Consistent with the gene transcripts, RNA fluorescence in situ hybridization also showed significantly higher LncRNA MALAT1 labeling in glucose-exposed HRECs (Fig. 1B). Figure 1C represents the AMI of LncRNA MALAT1. Incubation of cells in 20 mmol/L l-glucose, instead of 20 mmol/L d-glucose, had no effect on MALAT1 expression.
Figure 1.
Effect of high glucose on LncRNA MALAT1 expression in retinal endothelial cells. A: HRECs, incubated in HG or NG, were analyzed for LncRNA MALAT1 expression of by qRT-PCR using β-actin as the housekeeping gene. Values obtained from the cells in NG were considered as 1. B: A representative image captured by Zeiss (original magnification ×40; objective magnification with Apotome module) to show localization and subcellular expression, determined by RNA-FISH technique using oligo probe PCR amplified with fluorescein dUTP (green), and counterstaining with DAPI (blue). C: AMI of LncRNA MALAT1, calculated by region of interest in the nuclear region from 30 or more cells in each group. The values in the graphs are represented as mean ± SD, obtained from 3–4 cell preparations, with each measurement made in duplicate. SC, scrambled control RNA. *P < 0.05 compared with NG and #P < 0.05 compared with HG.
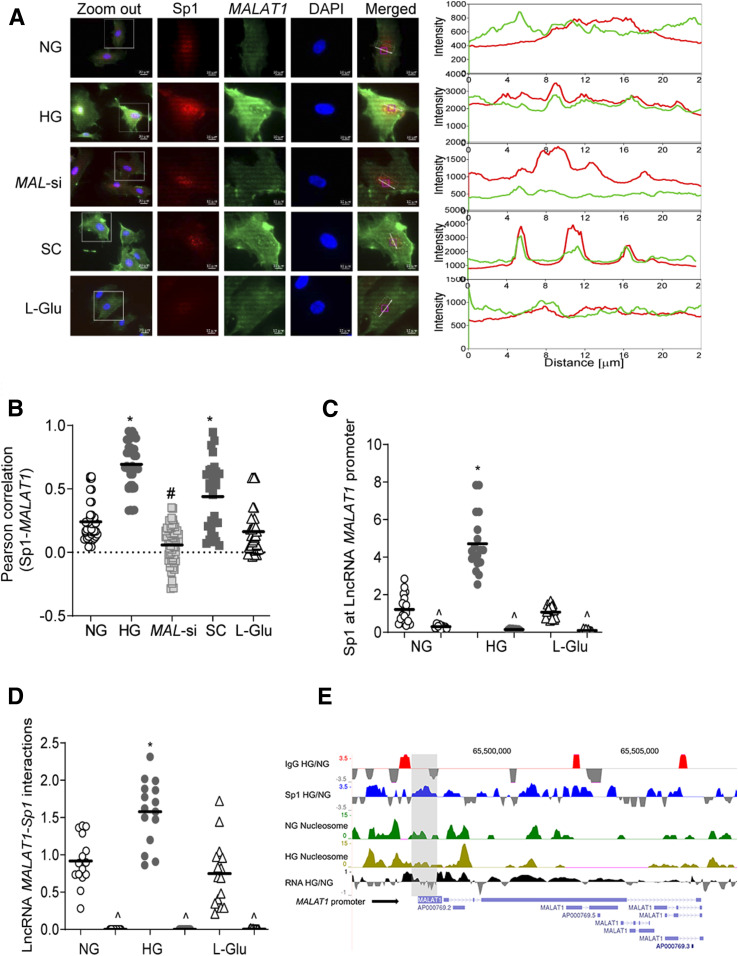
To understand the mechanism of LncRNA MALAT1 increase in the hyperglycemic milieu, its interactions with Sp1 were quantified. Cells in high glucose had significantly higher colocalization of LncRNA MALAT1 and Sp1, compared with normal glucose (Fig. 2A). The accompanying line intensity profile also demonstrated glucose-induced increased LncRNA MALAT1-Sp1 colocalization in the nucleus, which was further confirmed by the Pearson correlation graph (Fig. 2B). To investigate LncRNA MALAT1 transcriptional activation by Sp1, a chromatin immunoprecipitation (ChIP) assay was performed. Figure 2C shows over 2.5-fold increased Sp1 binding at LncRNA MALAT1 promoter by high glucose, and compared with Sp1 binding, IgG binding was <1% (indicated with ^). RNA immunoprecipitation further confirmed glucose-induced increased interactions of LncRNA MALAT1 and Sp1 (Fig. 2D). Consistent with the ChIP assay results, Sp1 ChIP-sequencing showed higher Sp1-LncRNA MALAT1 interactions and nucleosomal DNA remodeling in HRECs exposed to high glucose (Fig. 2E). This was accompanied by higher transcript RNA peaks of LncRNA MALAT1, as shown by the high glucose/normal glucose peak ratio (Fig. 2E). However, MAL-si attenuated glucose-induced increase in LncRNA MALAT1-Sp1 interactions and its decreased nuclear accumulation. Scrambled RNA had no effect on the glucose-induced increase in LncRNA MALAT1-Sp1 interactions (Fig. 2A and B).
Figure 2.
Effect of high glucose on the LncRNA MALAT1-Sp1 interactions. A: Colocalization of LncRNA MALAT1 and Sp1 was determined by RNA-FISH technique using fluorescein 12-dUTP–labeled LncRNA MALAT1 probe (green) and Texas Red (red) conjugated secondary antibody against Sp1. B: Pearson correlation showing the coefficient of interaction between LncRNA MALAT1 and Sp1, calculated using a random region of interest in the nuclear region from 30–40 cells in each group. C: Occupancy of Sp1 at LncRNA MALAT1 promoter was quantified by ChIP technique using IgG (^) as antibody control. D: LncRNA MALAT1 in Sp1-immunoprecipitated samples was quantified using the RNA immunoprecipitation technique. E: ChIP-sequencing showing comparative peaks in Sp1-immunoprecipitated chromatin and its occupancy patterns on LncRNA MALAT1 promoter in RPKM-normalized alignment. Nucleosome remodeling on LncRNA MALAT1 promoter at the Sp1 binding site was assessed by micrococcal nuclease–assisted nucleosomal DNA sequencing, and the figure shows the nucleosomal DNA remodeling patterns at the promoter site and the gene body. Values in the graphs are presented as mean ± SD, and NG values were considered as 1. *P < 0.05 vs. NG, #P < 0.05 vs. HG. SC, scrambled control RNA.
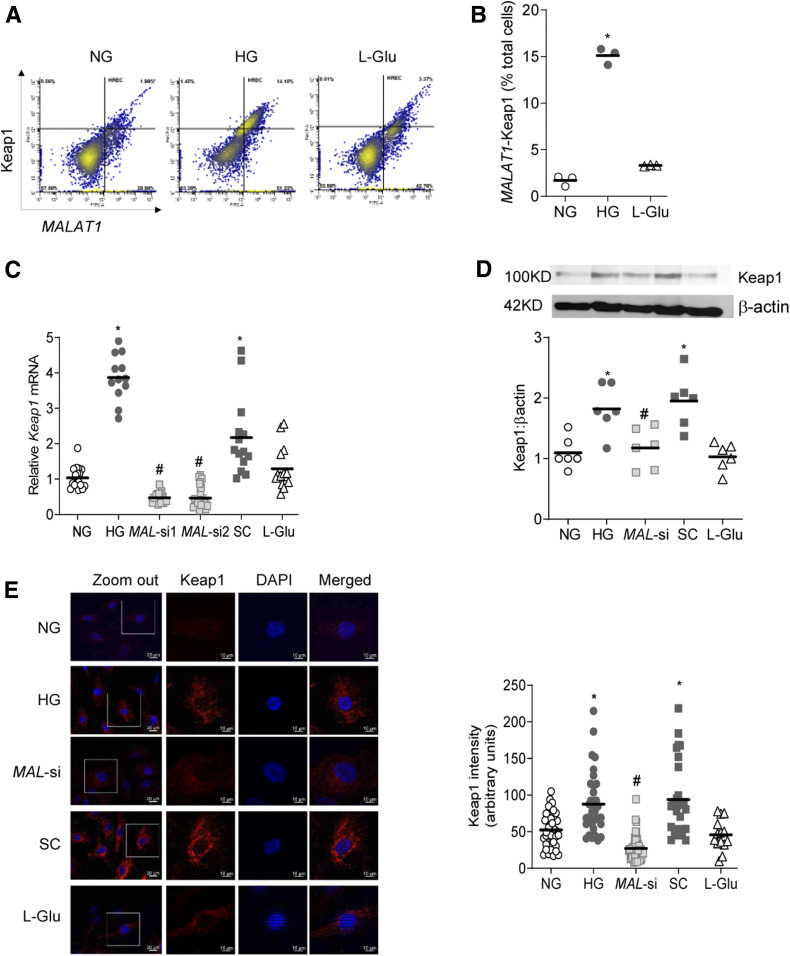
Hyperglycemia increases retinal Keap1 gene transcripts (13); to investigate LncRNA MALAT1 role in Keap1 regulation, dual dye-stained fluorescence flow cytometry was performed. Cells positive for Keap1 and LncRNA MALAT1, plotted from the values in the third quadrant, were significantly higher in high glucose compared with normal glucose (Fig. 3A and B). The role of LncRNA MALAT1 in regulation of Keap1 was further confirmed using MAL-si. Figure 3C shows significant attenuation of glucose-induced increase in Keap1 by both siRNAs 1 and 2, and siRNA 1 was used in the subsequent experiments. Consistent with gene transcripts, MAL-si also ameliorated glucose-induced increase in Keap1 protein expression, which was further confirmed by immunofluorescence staining and the accompanying AMI data (Fig. 3D and E). However, cells transfected with scrambled RNA failed to prevent glucose-induced increase in Keap1, and 20 mmol/L l-glucose, instead of 20 mmol/L d-glucose, did not increase Keap1 expression (transcripts or protein).
Figure 3.
Regulation of Keap1 by LncRNA MALAT1. A: HRECs incubated in high glucose or normal glucose were analyzed by FACS for LncRNA MALAT1 and Keap1 dual staining. B: LncRNA MALAT1- and Keap1-stained cells in the third quadrant. C: Keap1 gene expression was measured by qRT-PCR using β-actin as the housekeeping gene. The values obtained from cells in normal glucose were considered as 1. D and E: Keap1 expression was determined by Western blotting (D) using β-actin as the loading protein, and by immunofluorescence (E) using Texas Red conjugated secondary staining (red) and DAPI (blue)-containing Vectashield mounting reagent. AMI of Keap1 was calculated using the region of interest on the whole cell area. The values in the graphs are represented as mean ± SD, obtained from 3–4 cell preparations, with each measurement made in duplicate. NG values were considered as 1. *P < 0.05 vs. NG, #P < 0.05 vs. HG. SC, scrambled control RNA.
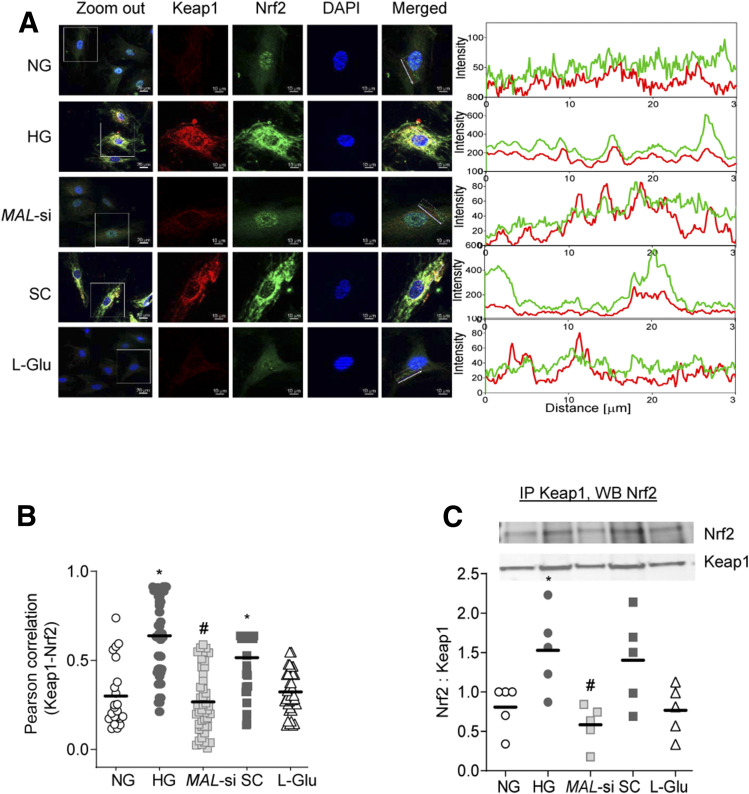
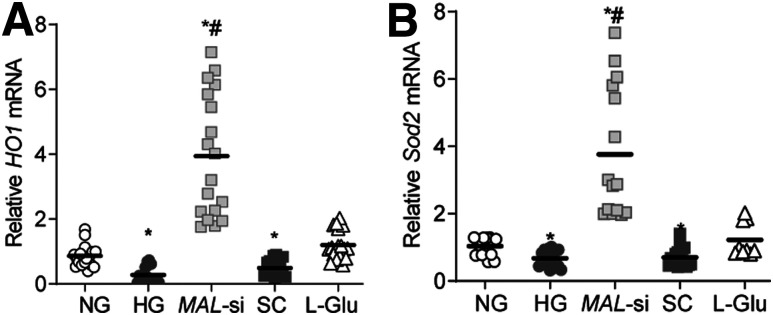
Keap1 quenches Nrf2 in the cytosol, and by blocking its translocation to the nucleus, inhibits the transcriptional activity of Nrf2 (13,30). As expected, glucose increased cytosolic accumulation of Nrf2 and its interactions with Keap1 and MAL-si prevented them (Fig. 4A). This was further confirmed by decreased Pearson correlation in siRNA-transfected cells compared with untransfected cells (Fig. 4B). However, the Pearson correlation was similar in untransfected cells and scrambled RNA transfected cells incubated in high glucose. Consistent with high glucose–induced increases in Keap-Nrf2 interactions, the transcription of Nrf2-responsive antioxidant defense genes HO1 and Sod2 was also decreased by high glucose, and MAL-si ameliorated such decreases (Fig. 5A and B).
Figure 4.
Effect of MAL-si on Keap1-Nrf2 subcellular localization. A: Colocalization of Keap1 and Nrf2 was determined by immunofluorescence using Texas red (red) and fluorescein-conjugated (green) secondary antibodies, respectively. The line represents the region of interest, used to determine the interaction of Keap1-Nrf2. B: Pearson correlation of Keap1 and Nrf2. C: Binding of Nrf2 with Keap1 was performed by immunoprecipitating cytosolic Keap1, followed by Western blot analysis for Nrf2, and Keap1 was used as a loading control. *P < 0.05 vs. NG, #P < 0.05 vs. HG. SC, scrambled control RNA.
Figure 5.
Effect of MAL-si on Nrf2-responsive genes. A and B: Gene expressions of HO1 (A) and Sod2 (B) were quantified by qRT-PCR using β-actin as a housekeeping gene. Values obtained from cells in normal glucose were considered as 1. *P < 0.05 vs. NG, #P < 0.05 vs. HG. SC, scrambled control RNA.
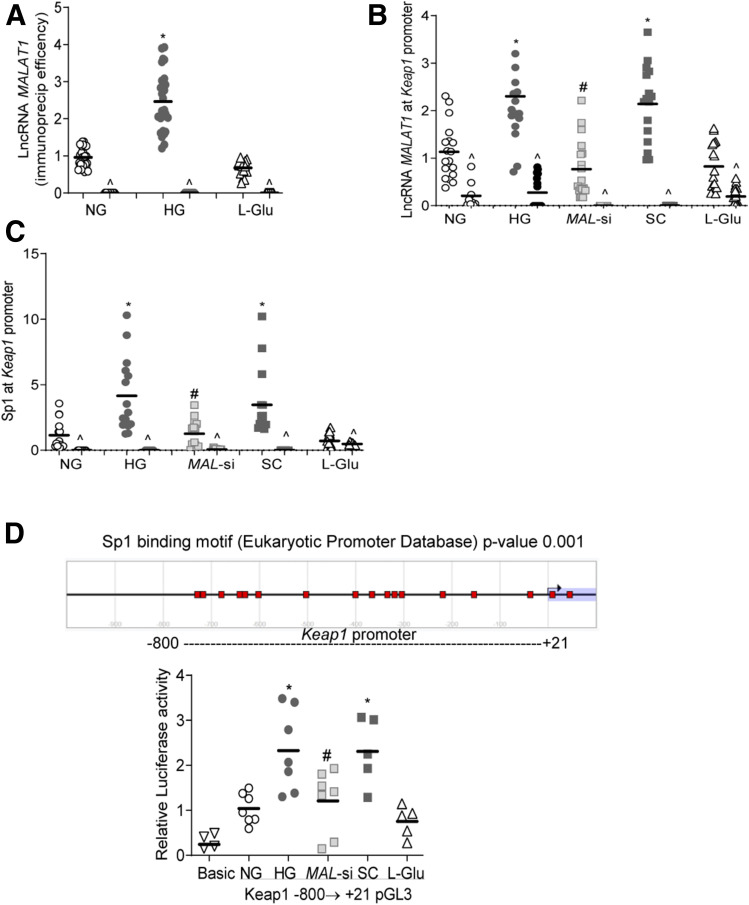
To understand the molecular mechanism of LncRNA MALAT1-mediated increase in Keap1, ChIRP was performed, and Fig. 6A shows twofold higher MALAT1 RNA immunoprecipitation efficiency in the cells incubated in high glucose compared with normal glucose or l-glucose. Similarly, cells in high glucose had two- to threefold higher LncRNA MALAT1 and Sp1 at the Keap1 promoter. While LncRNA MAL-si attenuated Sp1 occupancy at the Keap1 promoter, scrambled RNA had no effect (Fig. 6B and C). To confirm LncRNA MALAT1–mediated Sp1 recruitment at the Keap1 promoter, Keap1 promoter activity was quantified by luciferase assay. Consistent with the occupancy of Sp1 at Keap1, the luciferase assay also showed increased promoter activity in high glucose, which was attenuated by MAL-si (Fig. 6D), further confirming the importance of LncRNA MALAT1 in recruiting Sp1 at the Keap1 promoter.
Figure 6.
Regulation of Keap1 gene transcription by LncRNA MALAT1 and Sp1. A: LncRNA MALAT1 immunoprecipitation efficiency was determined using a DNA oligo probe against LncRNA MALAT1. B: LncRNA MALAT1 binding at the Keap1 promoter was quantified by ChIRP technique using fluorescein-12-dUTP incorporated oligo (LncRNA MALAT1) probe, and RNA-immunoprecipitated DNA fragments were quantified for LncRNA MALAT1 at the Keap1 promoter by qRT-PCR. C: The ChIP technique was used to determine Sp1 binding at the Keap1 promoter by performing qRT-PCR for Keap1 in Sp1-immunoprecipitated chromatin extract. IgG (^) was used as an antibody control. D: Keap1 promoter activity was quantified by luciferase assay using the cluster of Sp1-binding motif probable region −800 upstream of transcription start site to +21 downstream. Each graph has values represented as mean ± SD from 3–4 cell preparations. *P < 0.05 vs. NG, #P < 0.05 vs. HG. SC, scrambled control RNA.
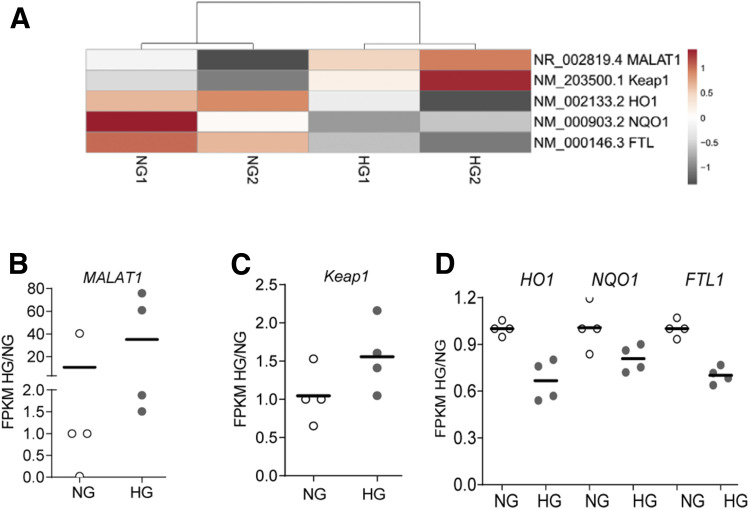
Consistent with the results detailed above, sequencing of the RNA isolated from the nuclear fraction of HRECs showed increased LncRNA MALAT1 in the cells incubated in high glucose, compared with normal glucose. The heatmap of genes generated by normalized FPKM showed an approximately twofold increase in LncRNA MALAT1 in the cells incubated in high glucose. Similarly, heatmap and scatter plots also confirmed increased expressions of Keap1 and decreased expression Nrf2-regulated HO1, NAD(P)H:quinone oxidoreductase-1 (NQO1), and ferritin-binding protein (light chain, FTL) in the same high glucose–treated cells (Fig. 7A–D).
Figure 7.
Expression and new synthesis of genes profiles by nuclear RNA sequencing. A: Heatmap of two different cell preparations each incubated in normal glucose (NG1 and NG2) and in high glucose (HG1 and HG2). B–D: FPKM ratio of HG/NG showing gene expression of LncRNA MALAT1 (B); Keap1 (C); and HO1, HQO1, and FTL1 (D) in the nuclear fraction.
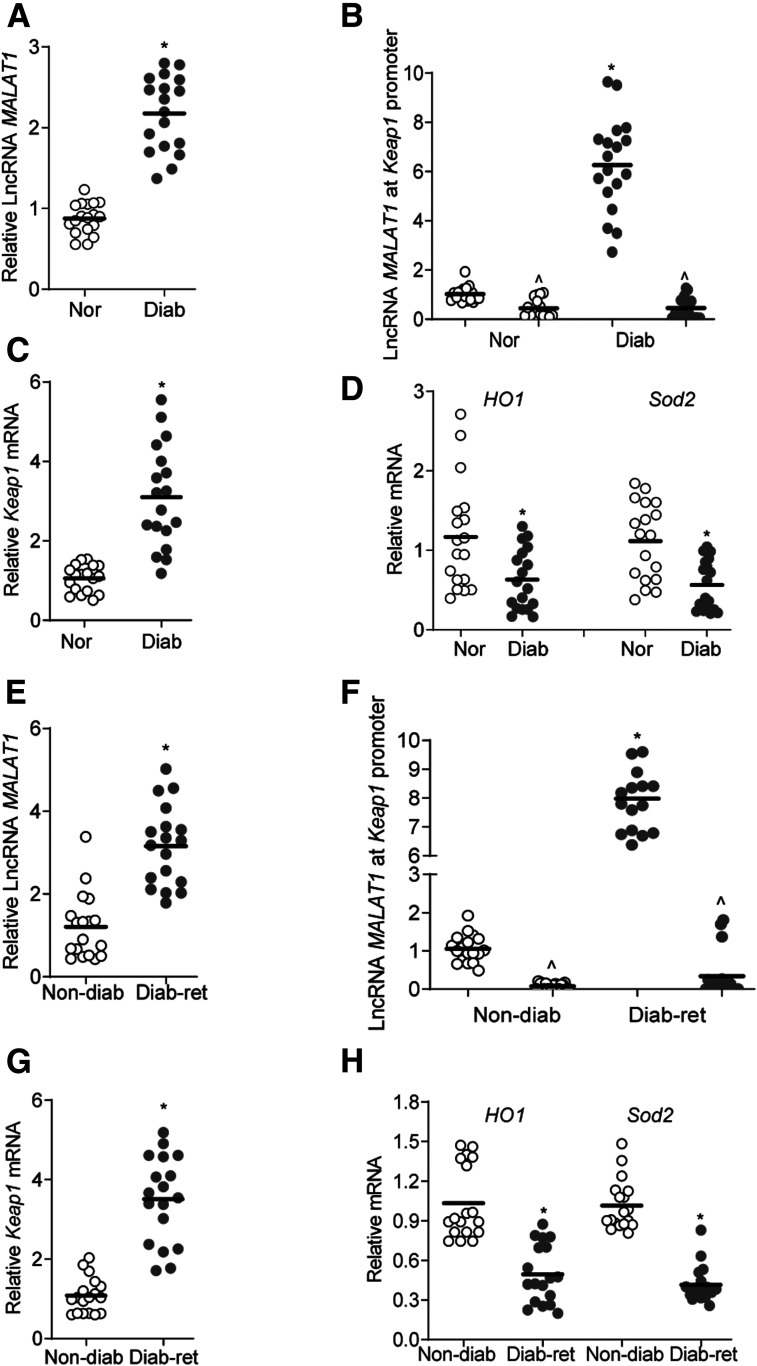
To confirm results from the in vitro model in an in vivo model, retinal microvessels from diabetic mice were analyzed. LncRNA MALAT1 transcripts were over twofold higher in diabetic mice, compared with normal mice, and LncRNA MALAT1 occupancy at the Keap1 promoter was greater than sevenfold higher (Fig. 8A and B). Furthermore, as expected (13), gene transcripts of Keap1 were elevated in diabetic mice, but HO1 and Sod2 were decreased by ∼50% (Fig. 8C and D).
Figure 8.
LncRNA MALAT1-Keap1 in retinal microvessels of diabetic mice and human donors with documented diabetic retinopathy. A–D: Retinal microvessels from diabetic mice (Diab) were analyzed for LncRNA MALAT1 expression (A) by qRT-PCR, LncRNA MALAT1 at the Keap1 promoter (B) was quantified employing ChIRP technique by Fluorescein dUTP-incorporated oligo (LncRNA MALAT1) immunoprecipitation, followed by quantification of Keap1 (C) and HO1 and Sod2 (D) gene transcripts by qRT-PCR using 18S rRNA as a housekeeping gene. Values obtained from age-matched normal mice (Nor) were considered as 1. E–H: Retinal microvessels from human donors with documented diabetic retinopathy (Diab Ret) were analyzed for LncRNA MALAT1 (qRT-PCR) (E), LncRNA MALAT1 at the Keap1 promoter (ChIRP) (F), and gene transcripts of Keap1 (G) and HO1 and Sod2 (H) by qRT-PCR, using β-actin as a housekeeping gene. Values from human donors without diabetes (Non-diab) were considered as 1. *P < 0.05 compared with normal mice, or to donors without diabetes. The values are represented as mean ± SD, and each measurement was made in duplicate in 7–9 mice per group or 6–7 human samples per group.
Consistent with the results from the experimental models, LncRNA MALAT1 transcripts and its occupancy at the Keap1 promoter were higher in retinal microvessels from human donors with diabetic retinopathy compared with their age-matched donors without diabetes (Fig. 8E and F). This was accompanied by over twofold increase in Keap1 gene transcripts and an over 50% decrease in HO1 and Sod2 gene transcripts (Fig. 8G and H). However, the limited number of samples (seven/group), did not allow us to establish the effect of the duration of diabetes or sex on the LncRNA MALAT1-Keap1-Nrf2 axis.
Discussion
The retina is a metabolically active tissue, and sustained hyperglycemic insult damages its vascular and neuronal structure, resulting in metabolic, molecular, functional, and structural abnormalities (31). Many metabolic abnormalities, including activation of polyol pathway and increased accumulation of inflammatory cytokines, are initiated by the diabetic environment (2,31). While production of free radicals is increased, their removal is decreased, and because of decreased transcriptional activity of Nrf2, the primary antioxidant defense mechanism is compromised (3,32). Although the role of LncRNA MALAT1 in the regulation of inflammatory cytokines and angiogenesis is reported by others (15,33), its role in maintaining oxidative stress is unclear. Here, our intriguing data show that LncRNA MALAT1 also has an important role in maintaining the antioxidant defense system. Hyperglycemia increases Sp1 binding at the MALAT1 promoter in retinal microvasculature, and elevated LncRNA MALAT1, by enhancing the binding of Sp1 at the Keap1 promoter, activates its transcription. High levels of Keap1 impede Nrf2 nuclear movement, which obstructs the transcription of the antioxidant response enzymes. Inhibition of LncRNA MALAT1 by its siRNA inhibits Keap1 upregulation, freeing Nrf2 to translocate in the nucleus and help the transcription of antioxidant genes (HO1 and Sod2). Consistent with the results from in vitro (HRECs) and in vivo (diabetic mice) models, retinal microvessels from human donors with diabetic retinopathy also have increases in LncRNA MALAT1 and its interactions with Keap1 and have increased levels of Keap1. Thus, LncRNA MALAT1, which is routinely implicated in inflammation, angiogenesis, and retinal neurodegeneration (34), also plays an important role in the regulation of the antioxidant defense system in the development of diabetic retinopathy.
LncRNAs are now considered to play important role in the regulation of various biological processes, including proliferation, differentiation, invasion, and apoptosis (14,35). LncRNA MALAT1 is a highly conserved LncRNA, which is linked to a variety of pathological processes including various malignancies and diabetes-related complications (15,16). Aqueous humor and fibrovascular membranes of patients with diabetes have increased LncRNA MALAT1 (36), and LncRNA MALAT1 expression is augmented by hypoxia contributing to the proliferative response in endothelial cells (37). Inhibition of LncRNA MALAT1 protects retinal photoreceptors and alleviates diabetic neurodegeneration (34), and LncRNA antisense noncoding RNA inhibits nuclear factor-kB–mediated signaling (38). Here, we show that interactions of LncRNA MALAT1 with Sp1 are increased in the nucleus in hyperglycemic milieu, resulting in its transcriptional activation. The Keap1 promoter also has a Sp1 binding site (−160/−153) (39), and in diabetic retinopathy, because of histone modifications of the Keap1 promoter, the binding of Sp1 is increased (13). Our results now show that MAL-si also inhibits diabetes-induced increases in Sp1 binding at the Keap1 promoter and attenuates Keap1 transcription and expression, suggesting a critical role of LncRNA MALAT1 in its expression in diabetic retinopathy.
Cysteine residues in Keap1 serve as oxidative stress sensors, and oxidation of thiol groups allows Nrf2 to detach from Keap1 (40,41). This facilitates Nrf2 to enter the nucleus and help transcribe the genes containing antioxidant response element (ARE), including mitochondrial and nonmitochondrial antioxidant proteins (9,41). In addition to cysteine modifications, Keap1 and Nrf2 also serve as substrates for the adaptor protein for Cullin3/Rbx1 ubiquitin ligase (42). In basal conditions, Keap1 tethers Nrf2 by continuously targeting its degradation by ubiquitin ligase; however, in stress conditions, ubiquitinylation is switched from Nrf2 to Keap1, and the proteasomal degradation releases Nrf2 to become transcriptionally active (42,43). In diabetes, overall cellular expression of retinal Nrf2 is not changed, but its binding with Keap1 is increased in the cytosol, resulting in decreased transcription of antioxidant defense genes (10,13). Here, we show that MAL-si decreases Keap1-Nrf2 interactions, and increases nuclear localization/transcription activity of Nrf2. LncRNA MALAT1 is shown to interact with ubiquitin 3 ligase in ovarian cancer cells (44), and the possibility of LncRNA MALAT1 assisting in the proteosomal degradation of Keap1-Nrf2 in diabetic retinopathy cannot be ruled out. MAL-si also prevents glucose-induced decreases in the ARE-containing genes HO1, an enzyme with endoplasmic reticulum localization, and Sod2, with mitochondrial localization, strongly suggesting that LncRNA MALAT1 upregulation in diabetes disrupts the overall cellular protective mechanisms of the retina. In support, LncRNA MALAT1, via upregulating endoplasmic reticulum stress, promotes glucose-induced angiogenesis and inflammation in retinal endothelial cells (45) and regulates Nrf2-mediated insulin action (46).
In vitro results are strongly supported by similar findings in the in vivo model. Retinal microvessels from diabetic mice have increased LncRNA MALAT1-Keap1 interactions and Keap1 expression. Furthermore, similar increases in LncRNA MALAT1-Keap1 interactions and Keap1 and decreases in ARE-containing genes in human donors with diabetic retinopathy further strengthen the role of LncRNA MALAT1 in the antioxidant defense system in diabetic retinopathy.
LncRNA MALAT1 is implicated in vascular growth and pathological angiogenesis and in regulation of VE-Cadherin (47). Others have shown that vascular leakage can be prevented by Nrf2 augmentation (48), and here, we show that LncRNA MALAT1 regulates the Keap1-Nrf2 axis. This raises a possibility that LncRNA could also be playing a role in retinal vascular leakage, which is commonly observed in diabetic retinopathy. Our study is focused on the role of LncRNA MALAT1 in regulating the Keap1-Nrf2 axis, but we recognize that it can regulate cell functions via other mechanisms, e.g., serving as a scaffold to bring two or more proteins into a functional ribonucleoprotein complex, or as an miRNA sponge to buffer their inhibitory function (49). A similar sponging role, or other modes of action, of LncRNA MALAT1 in diabetic retinopathy cannot be ruled out. Furthermore, at 2 months of diabetes in mice, the retinal LncRNA MALAT1 is upregulated (36) and the antioxidant system is compromised. When the duration is extended to ∼6 months, capillary cell apoptosis can be detected (2,3,50). However, even after 12–18 months of diabetes in the same mouse model, neovascularization cannot be detected, raising a possibility that LncRNA MALAT1 could be modulating cell death/capillary degeneration via mechanisms that could be different from those involved in angiogenesis.
In summary, LncRNA MALAT1 plays an important role in the regulation of the cellular antioxidant defense system in diabetic retinopathy. By increasing binding of the transcription factor at the Keap1 promoter, LncRNA MALAT1 facilitates its transcription. Elevated levels of Keap1 prevent the master regulator Nrf2 to move inside the nucleus to initiate transcription of the antioxidant response genes, and the retina fails to defend itself from the increased oxidative stress that is generated by the hyperglycemic milieu. Thus, regulation of LncRNA MALAT1 by pharmacological or molecular means, in addition to preventing angiogenesis during the later stages of diabetic retinopathy, has potential to protect the retina from increased oxidative stress and mitochondrial damage and to prevent further progression of diabetic retinopathy.
Article Information
Acknowledgments. The authors thank Gina Polsinelli for help with maintaining the animal colony.
Funding. The study was supported in parts by grants from the National Institutes of Health National Eye Institute (EY014370, EY017313, and EY022230) and from The Thomas Foundation to R.A.K., and an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology, Wayne State University.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. R.R. collected the data. R.A.K. wrote and edited the manuscript. All authors contributed to the experimental plan, data interpretation, and literature search. R.A.K. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 80th Scientific Sessions of the American Diabetes Association, 12–16 June 2020.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.13050809.
References
- 1.Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet 2010;376:124–136 [DOI] [PubMed] [Google Scholar]
- 2.Kowluru RA, Kowluru A, Mishra M, Kumar B. Oxidative stress and epigenetic modifications in the pathogenesis of diabetic retinopathy. Prog Retin Eye Res 2015;48:40–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kowluru RA, Mishra M. Oxidative stress, mitochondrial damage and diabetic retinopathy. Biochim Biophys Acta 2015;1852:2474–2483 [DOI] [PubMed] [Google Scholar]
- 4.Kowluru RA Mitochondrial stability in diabetic retinopathy: lessons learned from epigenetics. Diabetes 2019;68:241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrington DA, Fisher CR, Kowluru RA. Mitochondrial defects drive degenerative retinal diseases. Trends Mol Med 2020;26:105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowluru RA, Chan PS. Oxidative stress and diabetic retinopathy. Exp Diabetes Res 2007;2007:43603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowluru RA, Mishra M. Epigenetic regulation of redox signaling in diabetic retinopathy: role of Nrf2. Free Radic Biol Med 2017;103:155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh K, Wakabayashi N, Katoh Y, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 1999;13:76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Davies KJA, Forman HJ. Oxidative stress response and Nrf2 signaling in aging. Free Radic Biol Med 2015;88:314–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong Q, Mishra M, Kowluru RA. Transcription factor Nrf2-mediated antioxidant defense system in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci 2013;54:3941–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alegría-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics 2011;3:267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird A Perceptions of epigenetics. Nature 2007;447:396–398 [DOI] [PubMed] [Google Scholar]
- 13.Mishra M, Zhong Q, Kowluru RA. Epigenetic modifications of Keap1 regulate its interaction with the protective factor Nrf2 in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci 2014;55:7256–7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 2016;14:42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biswas S, Thomas AA, Chen S, et al. MALAT1: an epigenetic regulator of inflammation in diabetic retinopathy. Sci Rep 2018;8:6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmadi A, Kaviani S, Yaghmaie M, et al. Altered expression of MALAT1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings. Blood Res 2018;53:320–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon AD, Biswas S, Feng B, Chakrabarti S. MALAT1: a regulator of inflammatory cytokines in diabetic complications. Endocrinol Diabetes Metab 2018;1:e00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rickard JA, O’Donnell JA, Evans JM, et al. RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 2014;157:1175–1188 [DOI] [PubMed] [Google Scholar]
- 19.Duraisamy AJ, Mishra M, Kowluru A, Kowluru RA. Epigenetics and regulation of oxidative stress in diabetic retinopathy. Invest Ophthalmol Vis Sci 2018;59:4831–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammad G, Radhakrishnan R, Kowluru RA. Epigenetic modifications compromise mitochondrial DNA quality control in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci 2019;60:3943–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammad G, Kowluru RA. Homocysteine disrupts balance between MMP-9 and its tissue inhibitor in diabetic retinopathy: the role of DNA methylation. Int J Mol Sci 2020;21:1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos JM, Kowluru RA. Impaired transport of mitochondrial transcription factor A (TFAM) and the metabolic memory phenomenon associated with the progression of diabetic retinopathy. Diabetes Metab Res Rev 2013;29:204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Liu S, Zhou L, et al. Aberrant shuttling of long noncoding RNAs during the mitochondria-nuclear crosstalk in hepatocellular carcinoma cells. Am J Cancer Res 2019;9:999–1008 [PMC free article] [PubMed] [Google Scholar]
- 24.Arrigucci R, Bushkin Y, Radford F, et al. FISH-Flow, a protocol for the concurrent detection of mRNA and protein in single cells using fluorescence in situ hybridization and flow cytometry. Nat Protoc 2017;12:1245–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009;106:11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP). J Vis Exp 2012;61:3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaghlool A, Ameur A, Nyberg L, et al. Efficient cellular fractionation improves RNA sequencing analysis of mature and nascent transcripts from human tissues. BMC Biotechnol 2013;13:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afgan E, Baker D, Batut B, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 2018;46:W537–W544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chereji RV, Bryson TD, Henikoff S. Quantitative MNase-seq accurately maps nucleosome occupancy levels. Genome Biol 2019;20:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 2007;47:89–116 [DOI] [PubMed] [Google Scholar]
- 31.Frank RN Diabetic retinopathy. N Engl J Med 2004;350:48–58 [DOI] [PubMed] [Google Scholar]
- 32.Mishra M, Zhong Q, Kowluru RA. Epigenetic modifications of Nrf2-mediated glutamate-cysteine ligase: implications for the development of diabetic retinopathy and the metabolic memory phenomenon associated with its continued progression. Free Radic Biol Med 2014;75:129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martens CR, Bansal SS, Accornero F. Cardiovascular inflammation: RNA takes the lead. J Mol Cell Cardiol 2019;129:247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang YL, Hu HY, You ZP, Li BY, Shi K. Targeting long non-coding RNA MALAT1 alleviates retinal neurodegeneration in diabetic mice. Int J Ophthalmol 2020;13:213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 2016;96:1297–1325 [DOI] [PubMed] [Google Scholar]
- 36.Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci 2014;55:941–951 [DOI] [PubMed] [Google Scholar]
- 37.Kölling M, Genschel C, Kaucsar T, et al. Hypoxia-induced long non-coding RNA Malat1 is dispensable for renal ischemia/reperfusion-injury. Sci Rep 2018;8:3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei JC, Shi YL, Wang Q. LncRNA ANRIL knockdown ameliorates retinopathy in diabetic rats by inhibiting the NF-κB pathway. Eur Rev Med Pharmacol Sci 2019;23:7732–7739 [DOI] [PubMed] [Google Scholar]
- 39.Guo D, Wu B, Yan J, Li X, Sun H, Zhou D. A possible gene silencing mechanism: hypermethylation of the Keap1 promoter abrogates binding of the transcription factor Sp1 in lung cancer cells. Biochem Biophys Res Commun 2012;428:80–85 [DOI] [PubMed] [Google Scholar]
- 40.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells 2011;16:123–140 [DOI] [PubMed] [Google Scholar]
- 41.Baird L, Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol Cell Biol 2020;40:e00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci U S A 2005;102:10070–10075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med 2009;47:1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu J, Zhang L, Mei Z, et al. Interaction of E3 ubiquitin ligase MARCH7 with long noncoding RNA MALAT1 and autophagy-related protein ATG7 promotes autophagy and invasion in ovarian cancer. Cell Physiol Biochem 2018;47:654–666 [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Wang L, Guo H, et al. Knockdown of MALAT1 attenuates high-glucose-induced angiogenesis and inflammation via endoplasmic reticulum stress in human retinal vascular endothelial cells. Biomed Pharmacother 2020;124:109699. [DOI] [PubMed] [Google Scholar]
- 46.Chen J, Ke S, Zhong L, et al. Long noncoding RNA MALAT1 regulates generation of reactive oxygen species and the insulin responses in male mice. Biochem Pharmacol 2018;152:94–103 [DOI] [PubMed] [Google Scholar]
- 47.Liu P, Jia S-B, Shi J-M, et al. LncRNA-MALAT1 promotes neovascularization in diabetic retinopathy through regulating miR-125b/VE-cadherin axis. Biosci Rep 2019;39:BSR20181469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deliyanti D, Alrashdi SF, Tan SM, et al. Nrf2 activation is a potential therapeutic approach to attenuate diabetic retinopathy. Invest Ophthalmol Vis Sci 2018;59:815–825 [DOI] [PubMed] [Google Scholar]
- 49.Sun L, Lin JD. Function and mechanism of long noncoding RNAs in adipocyte biology. Diabetes 2019;68:887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kowluru RA, Kern TS, Engerman RL. Abnormalities of retinal metabolism in diabetes or experimental galactosemia. IV. Antioxidant defense system. Free Radic Biol Med 1997;22:587–592 [DOI] [PubMed] [Google Scholar]