Abstract
There is a limited understanding of how genetic loci associated with glycemic traits and type 2 diabetes (T2D) influence the response to antidiabetic medications. Polygenic scores provide increasing power to detect patterns of disease predisposition that might benefit from a targeted pharmacologic intervention. In the Study to Understand the Genetics of the Acute Response to Metformin and Glipizide in Humans (SUGAR-MGH), we constructed weighted polygenic scores using known genome-wide significant associations for T2D, fasting glucose, and fasting insulin, comprising 65, 43, and 13 single nucleotide polymorphisms, respectively. Multiple linear regression tested for associations between scores and glycemic traits as well as pharmacodynamic end points, adjusting for age, sex, race, and BMI. A higher T2D score was nominally associated with a shorter time to insulin peak, greater glucose area over the curve, shorter time to glucose trough, and steeper slope to glucose trough after glipizide. In replication, a higher T2D score was associated with a greater 1-year hemoglobin A1c reduction to sulfonylureas in the Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS) study (P = 0.02). Our findings suggest that individuals with a higher genetic burden for T2D experience a greater acute and sustained response to sulfonylureas.
Introduction
Metformin and sulfonylureas are widely prescribed medications for the treatment of type 2 diabetes (T2D). Metformin is the recommended first-line agent for T2D, owing to its high efficacy, low cost, and favorable side effect profile (1). Sulfonylureas are another commonly used agent due to their wide availability and glucose-lowering ability through stimulation of insulin secretion from pancreatic β-cells (2). Despite the recommendation that careful consideration of patient factors should inform the choice of therapy (3), clinicians typically do not account for the molecular target of each drug or integrate information about an individual’s genetic profile when prescribing a medication.
In the last decade, large-scale genome-wide association studies (GWAS) and high-throughput sequencing studies have identified >700 genetic signals influencing T2D risk and glycemic traits (4–10). The expanding list of genetic variants has resulted in a better understanding of the disease pathophysiology of T2D and the major processes that contribute to disease risk. However, the impact of these genetic loci on the response to pharmacological interventions for T2D has been less systematically studied.
With regards to metformin response, candidate gene studies have yielded initial findings in transporter gene variants (SLC22A1 and SLC47A1), but findings were not validated in subsequent large-scale meta-analyses (11). GWAS and meta-analyses have revealed additional loci, including single nucleotide polymorphisms (SNPs) in or near the gene encoding ataxia-telangiectasia mutated kinase (ATM) (12) and in an intron SNP of the GLUT2 (SLC2A2) (13). Pharmacogenetic studies of sulfonylurea response have been limited to candidate gene studies, and no GWAS for sulfonylurea response has been published to date (14–17).
The impact of T2D-associated genetic variants on drug response has been investigated as well. In particular, TCF7L2, the gene harboring common genetic variants with the largest effect on T2D susceptibility discovered to date, has been associated with drug response to sulfonylureas in those with established T2D (18) and in those at risk for T2D (19). For metformin, TCF7L2 has been associated with glycemic response in the early stages of disease (19,20). Because individual variants only have a modest effect, the field is now embracing the use of polygenic scores of aggregated variants, which offer increasing power and capture a greater proportion of the variance explaining a given trait (21).
As such, we examined whether polygenic scores derived from genome-wide significant loci for glycemic traits and T2D are associated with glycemic traits and the response to metformin and glipizide in the Study to Understand the Genetics of the Acute Response to Metformin and Glipizide in Humans (SUGAR-MGH). We hypothesized that polygenic scores constructed based on previously known genome-wide associations with fasting glucose (FG) and fasting insulin (FI) would be associated with these glycemic traits in SUGAR-MGH. Furthermore, we expected that a genetic predisposition to insulin secretion or action would influence the human response to glipizide or metformin, respectively. For findings that reached significance, we sought replication in the Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS) study, a longitudinal cohort study of T2D.
Research Design and Methods
Study Design and Participants
The study design of SUGAR-MGH has been previously described (22). Briefly, 1,000 participants were enrolled at three Boston academic medical centers between 2008 and 2015. Participants were preferentially enrolled in the study if they had risk factors for T2D (i.e., metabolic syndrome, obesity, polycystic ovarian syndrome, history of gestational diabetes, or positive family history) or lifestyle-controlled T2D. Some participants had previously unknown T2D, diagnosed at the time of study entry. All participants were naive to metformin and glipizide. Informed consent was obtained from all study participants, and the study protocol was approved by the Partners Human Research Committee (Partners HealthCare, Boston, MA).
After an overnight fast of at least 8 h, participants received a single dose of 5 mg glipizide if their fasting blood glucose was >4.4 mmol/L (visit 1). This threshold was chosen to minimize the risk of hypoglycemia. Glucose and insulin levels were subsequently measured at baseline, 30, 60, 90, 120, 180, and 240 min. The period of observation following glipizide administration was terminated early if the participant developed neuroglycopenic symptoms, a blood glucose ≤2.77 mmol/L with symptoms of hypoglycemia, blood glucose <2.50 mmol/L with or without symptoms of hypoglycemia, or at the discretion of study staff based on clinical assessment. Subjects who did not meet the threshold to receive glipizide or terminated the glipizide challenge early were excluded from analyses of glipizide response. Five days later, participants received a 2-day course of 500 mg metformin twice daily, followed by a 75-g oral glucose tolerance test at visit 2. Plasma glucose was measured by a hexokinase assay (Roche, Indianapolis, IN), and insulin was determined using a radioimmunoassay (Beckman Coulter, Fullerton, CA).
GoDARTS is a longitudinal case-control study that was established to study the genetics of T2D. Over 18,000 participants were enrolled between December 1998 and August 2012, of whom half were diagnosed with T2D and the remaining age- and sex-matched control subjects without diabetes were identified from general practice records in Tayside, Scotland. Details of the cohort have been previously described (23). The GoDARTS study was approved by the Tayside Committee for Medical Research Ethics (Tayside, Scotland, U.K.). Written informed consent was obtained from each participant.
For the replication analysis, we evaluated participants in GoDARTS who were diagnosed with T2D and were either on a sulfonylurea as monotherapy or as an add-on to metformin. Subjects with a history of insulin use, T2D diagnosed before 35 years of age, and a baseline hemoglobin A1c (HbA1c) <7% (53 mmol/mol) or >14% (130 mmol/mol) were excluded.
Genotyping
In SUGAR-MGH, DNA was extracted and genotyping was performed using the iPLEX Gold assay from Sequenom by allele-specific primer extension of amplified products with detection by mass spectroscopy (24). Hardy-Weinberg equilibrium was tested within each self-described ethnic group. SNPs with call rates <95% and samples with call rates <95% were excluded.
Genotyping and quality control of the GoDARTS data have been described previously (12,13). The SNPs included in the polygenic scores tested in this study were extracted from existing GWAS data. Imputed SNPs had an imputation score >0.9.
Polygenic Score Construction
Polygenic scores were constructed for T2D, FG, and FI by summing the number of risk alleles carried by each individual, weighted by the effect size estimates from well-established genome-wide significant associations derived from the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) or Europeans in the DIAbetes Meta-ANalysis of Trans-Ethnic association studies (DIAMANTE) Consortium (4,6,8). Due to the limited availability of SNPs on our genotyping platform in SUGAR-MGH, we were able to include only a subset of the known genome-wide significant loci for T2D and glycemic traits, resulting in a T2D polygenic score of 65 SNPs, FG score of 43 SNPs, and FI score of 13 SNPs. Supplementary Tables 1–3 list the genetic variants, corresponding genes, and original GWAS references for each score. Effect alleles were defined as T2D risk-raising, FG-raising, and FI-raising alleles. We used the 1000 Genomes database for global frequencies of the effect alleles because the individuals in SUGAR-MGH were largely without overt T2D, and we wanted to avoid using a reference database that included individuals from several T2D cohorts. If the lead SNP was not available, we used a proxy that had an r2 >0.8 for Europeans. In GoDARTS, polygenic scores were created in the same manner.
Statistical Analyses
In SUGAR-MGH, the area over the curve (AOC) for decreases in glucose during the glipizide challenge was calculated by subtracting glucose area under the curve (AUC) by the trapezoidal method from the baseline glucose value multiplied by total time for the glipizide challenge. The AUC for glucose and insulin following metformin administration was calculated by the trapezoidal method, which accounted for baseline glucose and insulin values, respectively. HOMA of insulin resistance (HOMA-IR) was calculated as previously described (25). Missing data were not imputed.
The mean ± SD or median (interquartile range) are reported for continuous normally or nonnormally distributed traits, respectively. Assessment of normality was performed using the Shapiro-Wilk test. Multiple linear regression with adjustments for age, sex, self-reported race/ethnicity, and BMI was used to test the association between each polygenic score and glycemic traits as well as pharmacodynamic end points. β Coefficients are presented as the incremental increase or decrease in the trait or end point per SD of the tested polygenic score. We assessed for both nominal significance (P < 0.05) and a more stringent P value of 0.008 for multiple comparisons (two drugs × three polygenic scores). Statistical analyses were performed using R 3.5.2 (26).
For the replication analyses in GoDARTS, multiple linear regression tested for the association between polygenic score and the outcome of HbA1c reduction, defined as baseline HbA1c (measured within 180 days prior to sulfonylurea initiation) minus on-treatment HbA1c at 1 year. Additional covariates included baseline HbA1c, age at diagnosis of diabetes, sex, BMI, average sulfonylurea dose, and mediation adherence as previously described (18).
Data and Resource Availability
The data sets analyzed during the current study are available from the corresponding author upon reasonable request. Data from SUGAR-MGH are also available at ClinicalTrials.gov.
Results
Subject Characteristics
The baseline characteristics of the 1,000 participants in SUGAR-MGH are summarized in Table 1. Approximately half of participants were female, the mean age was 47.2 years, and >35% of participants came from ethnic minority populations. The mean BMI was 30.2 kg/m2, and mean FG was 5.16 mmol/L, consistent with a population at risk for requiring future antidiabetic agents. Only 26 participants had a diagnosis of T2D (not treated pharmacologically) at the time of study entry. Of the 1,000 participants, 351 were either ineligible for the glipizide challenge due to low FG or terminated the challenge early in accordance with study protocol.
Table 1.
Demographic characteristics and baseline measurements of 1,000 participants in SUGAR-MGH
| All participants (n = 1,000) | |
|---|---|
| Female [n (%)] | 539 (54) |
| Age (years) | 47.2 ± 16.2 |
| BMI (kg/m2), n = 978 | 30.2 ± 7.1 |
| Self-reported race/ethnicity [n (%)] | |
| White, non-Hispanic | 639 (64) |
| Black, non-Hispanic | 209 (21) |
| Hispanic | 69 (6.9) |
| Asian, non-Hispanic | 59 (5.9) |
| Others | 24 (2.4) |
| Diagnosis of T2D | 26 (2.6) |
| FG (mmol/L) | 5.16 ± 0.93 |
| FI (pmol/L), n = 970 | 3.56 (3.03, 4.11) |
Age, BMI, and FG are mean ± SD. FI is median (interquartile range).
Construction of Polygenic Scores for T2D, FG, and FI
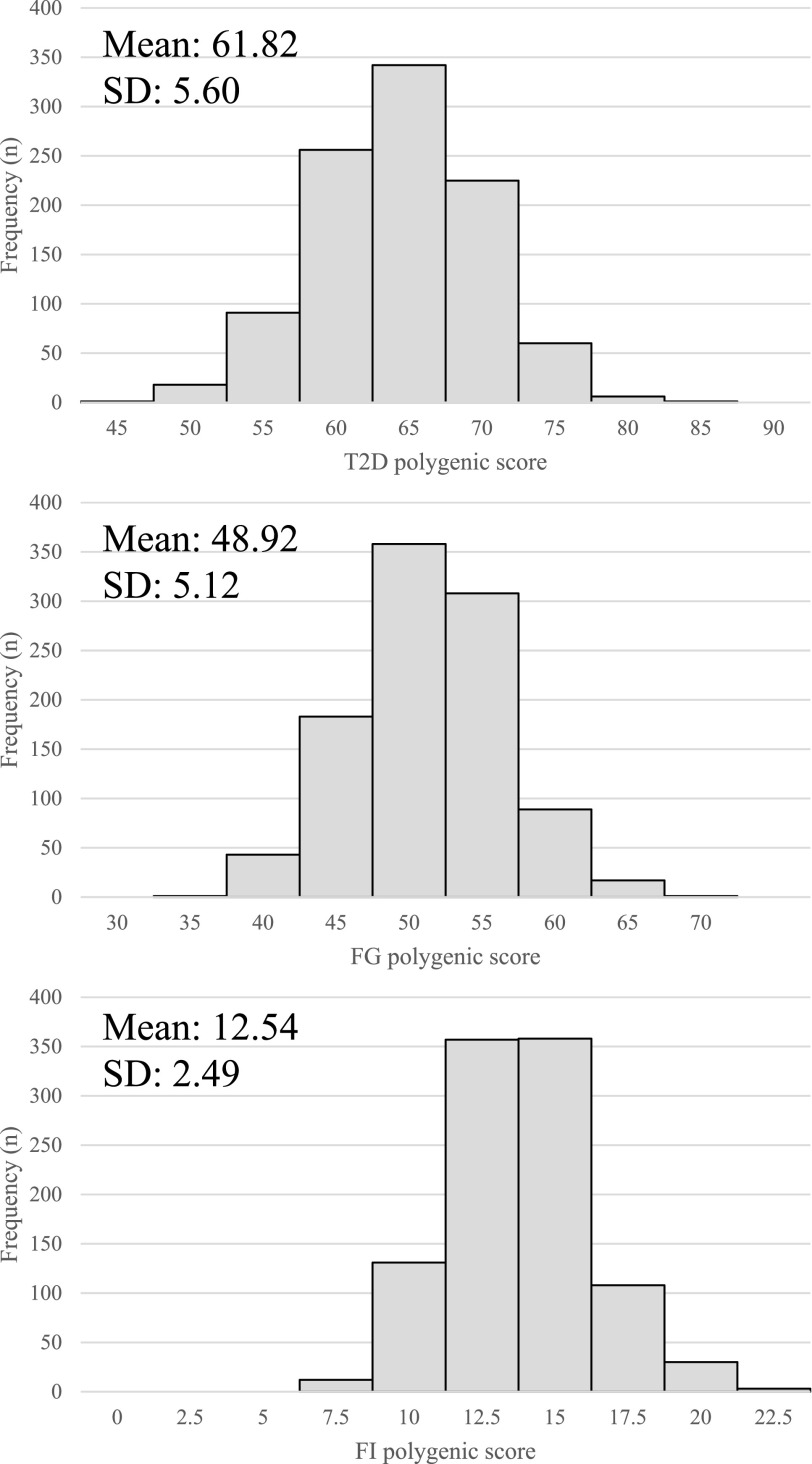
The distribution of all three polygenic scores is depicted in Fig. 1. The mean T2D polygenic score was 61.82 (range 44.49–80.93). The mean FG polygenic score was 48.92 (range 31.58–66.08). The mean FI polygenic score was 12.54 (range 5.11–22.74).
Figure 1.
Distribution of polygenic scores for T2D (A), FG (B), and FI (C) across 1,000 individuals in SUGAR-MGH.
Association Between Polygenic Scores and Baseline Glycemic Traits
Table 2 shows the associations between each polygenic score and either FG or FI at baseline in SUGAR-MGH. The FG polygenic score was strongly associated with FG in our cohort in multivariate analyses (P < 0.001), with each SD increase in score raising FG by 0.13 mmol/L. This association was present in stratified analyses of the non-Hispanic White (β = 0.09; P = 0.01) and non-Hispanic Black (β = 0.14; P = 0.007) individuals in SUGAR-MGH. Likewise, a higher FI polygenic score was associated with higher FI (P = 0.04); this finding was also present in stratified analyses of non-Hispanic Whites. A higher T2D polygenic score trended toward significance (P = 0.05) for the association with higher FG but was not associated with FI.
Table 2.
Association of polygenic scores with baseline glycemic traits in SUGAR-MGH
| Polygenic score | Trait | β (95% CI) | P |
|---|---|---|---|
| FG | FG (mmol/L) | 0.13 (0.07, 0.18) | <0.001 |
| FI | Ln FI (pmol/L) | 0.05 (0.003, 0.10) | 0.04 |
| T2D | FG (mmol/L) | 0.05 (−2.1e-5, 0.10) | 0.05 |
| T2D | Ln FI (pmol/L) | 0.009 (−0.04, 0.06) | 0.71 |
Linear regression model was adjusted for age, sex, race/ethnicity, and BMI. β values are reported per SD of polygenic score. P values of <0.05 are in bold and reflect significance. Ln, natural log transformation.
Association Between T2D Score and the Acute Response to Glipizide and Metformin
Table 3 summarizes the association between T2D polygenic score and select end points of glipizide and metformin response. A higher T2D polygenic score was associated with a greater glucose AOC, shorter time to glucose trough, steeper slope to glucose trough, and shorter time to insulin peak following glipizide administration at nominal significance (P < 0.05). When the more stringent P value of 0.008 was used to correct for multiple comparisons, the finding involving the insulin-based end point remained significant. We tested and did not find a significant association between T2D polygenic score and pharmacodynamic end points of metformin response (change in FG, change in FI, and change in HOMA-IR) (Table 3).
Table 3.
Association of T2D polygenic score with glipizide and metformin end points in SUGAR-MGH
| N | β (95% CI) | P§ | |
|---|---|---|---|
| Glipizide end point* | |||
| Glucose trough (mmol/L)† | 639 | −0.01 (−0.05, 0.02) | 0.50 |
| Glucose AOC (mmol/L ∗ min) | 633 | 10.05 (1.17, 18.93) | 0.03 |
| Time to glucose trough (min)† | 639 | −4.88 (−8.82, −0.94) | 0.02 |
| Slope to glucose trough (mmol/L/min)† | 638 | 7.6e-4 (1.2e-4, 1.4e-3) | 0.02 |
| Ln peak insulin (pmol/L)‡ | 615 | 0.04 (−0.009, 0.09) | 0.11 |
| Time to insulin peak (min)‡ | 615 | −5.83 (−9.91, −1.76) | 0.005 |
| Slope to insulin peak (pmol/L/min)‡ | 609 | −0.11 (−0.33, 0.12) | 0.35 |
| Metformin end point | |||
| FG V2-V1 (mmol/L)† | 924 | −0.009 (−0.04, 0.02) | 0.56 |
| Glucose AUC (mmol/L ∗ min) | 900 | 6.79 (−3.20, 16.77) | 0.18 |
| FI V2-V1 (pmol/L)‡ | 891 | −3.11 (−6.74, 0.52) | 0.09 |
| Insulin AUC (pmol/L ∗ min) | 831 | −66.27 (−2,561.34, 1,640.89) | 0.67 |
| Ln HOMA-IR V1 (mmol ∗ pmol/L2) | 915 | 0.02 (−0.03, 0.08) | 0.44 |
| Ln HOMA-IR V2 (mmol ∗ pmol/L2) | 914 | −0.01 (−0.07, 0.05) | 0.69 |
| HOMA-IR V2-V1 (mmol ∗ pmol/L2) | 914 | −0.84 (−1.73, 0.04) | 0.06 |
Ln, natural log transformation; V1, visit 1; V2, visit 2.
A total of 351 individuals did not meet the threshold to receive glipizide or terminated the glipizide challenge early and were excluded from analyses of glipizide response.
Adjusted for baseline glucose.
Adjusted for ln baseline insulin. Linear regression model was adjusted for age, sex, race/ethnicity, and BMI.
P values of <0.008 are in bold and reflect significance after adjustment for multiple testing.
Given that the T2D polygenic score was constructed using effect size estimates for European ancestries and proxies were selected based on linkage disequilibrium in Europeans, we performed stratified analyses for the non-Hispanic White and Black participants separately. In the non-Hispanic White subset of SUGAR-MGH (Supplementary Table 4), we observed that individuals with a higher T2D polygenic score trended toward having a greater glucose AOC and shorter time to insulin peak, though this did not reach our significance threshold after adjustment for multiple testing. Similarly in the non-Hispanic Black participants, a similar direction of association was seen between a higher T2D polygenic score and shorter time to glucose trough and insulin peak following glipizide (Supplementary Table 5). The relationship between higher T2D polygenic score and steeper slope to glucose trough trended toward but did not reach significance in both subgroups.
Association Between Glycemic Trait Polygenic Scores and the Acute Response to Glipizide and Metformin
Additionally, we observed associations between glycemic trait scores and end points of glipizide response, reaching only nominal significance but not meeting the more stringent significance threshold after adjustment for multiple testing. A higher FG polygenic score trended toward a higher glucose AOC (P = 0.02), with each SD increase in score raising the glucose AOC by 10.82 mmol/L ∗ min (Supplementary Table 6). Moreover, each SD increase in FI polygenic score trended toward a 0.05 mmol/L higher glucose trough following glipizide administration (P = 0.02) (Supplementary Table 7). No association was observed between glycemic trait polygenic score and select end points of metformin response (Supplementary Tables 6 and 7).
Replication in GoDARTS
The baseline characteristics of the 2,228 individuals in GoDARTS who underwent treatment with a sulfonylurea are summarized in Table 4. Approximately half of participants were female, the mean age was 59.7 years, and the baseline HbA1c was 8.97% (75 mmol/mol). All subjects were of European ancestry. To replicate our findings in SUGAR-MGH with respect to sulfonylurea response, we constructed a weighted T2D polygenic score for each individual in GoDARTS and tested for association with the HbA1c reduction over 1 year. The mean T2D polygenic score was 74.92 (range 53.29–93.08) with an SD of 5.90. In adjusted analyses, for each SD increase in T2D score, there was a 0.063% (0.07 mmol/mol) greater HbA1c reduction in response to sulfonylurea therapy (P = 0.02). Moreover, those in the top decile of T2D polygenic score had a 0.27 ± 0.12% greater HbA1c reduction compared with those in the bottom decile (P = 0.03).
Table 4.
Demographic characteristics and baseline measurements of 2,228 participants in GoDARTS
| All participants (n = 2,228) | |
|---|---|
| Age at diagnosis (years) | 59.7 ± 10.3 |
| Sex (% female) | 45 |
| Duration of diabetes (years) | 4.8 ± 4.4 |
| Baseline BMI (kg/m2) | 30.5 ± 5.4 |
| Baseline HbA1c (%) | 8.97 ± 1.47 |
| On-treatment HbA1c (%) | 7.64 ± 1.40 |
| Average HbA1c reduction (%) | 1.34 ± 1.69 |
| Sulfonylurea adherence (%) | 86 ± 20 |
| Sulfonylurea monotherapy (%) | 44 |
Age, BMI, and HbA1c values are mean ± SD.
Discussion
In SUGAR-MGH, we built polygenic scores for elevated T2D risk, FG, and FI using genome-wide significant variants discovered in GWAS for T2D and glycemic traits. We first assessed whether the three polygenic scores were associated with glycemic traits, which would indicate the generalizability of these scores to outcomes in this cohort. Subsequently, we tested the hypothesis that combining individual variants into a polygenic score may provide additional information on patterns of T2D disease predisposition that may benefit from tailored pharmacologic intervention.
We indeed demonstrated that sets of genome-wide significant genetic variants confirmed to be associated with glycemic traits were associated with FG and FI levels in SUGAR-MGH. Our findings were consistent in direction with and stronger in significance than previously reported findings in an interim analysis conducted for our design study at two-thirds study enrollment in SUGAR-MGH (22). Additionally, we examined whether a polygenic score for T2D risk would be associated with the same glycemic traits in our cohort. We found that there was a trend toward higher FG in those with a higher genetic burden for T2D, possibly related to the overlap of 14 SNPs between the T2D and FG scores. No association was seen between T2D polygenic score and FI, but this was not unexpected given that many of the genetic polymorphisms in the T2D score were those that directly or indirectly affect pancreatic β-cell function rather than insulin resistance.
We also tested the associations among each of the three polygenic scores and phenotypes of glipizide and metformin response. Individuals with a higher genetic burden for T2D were found to have a more robust response to glipizide, as indicated by a larger glucose AOC, representing a greater cumulative drop in glucose over time. Additionally, a higher T2D score was associated with a shorter time to glucose trough, steeper slope to glucose trough, and shorter time to insulin peak, all consistent with an enhanced glipizide response. We note that these findings were all at nominal significance (P < 0.05). Since the outcomes are correlated, we subsequently accounted for multiple comparisons, after which only the insulin-based outcome remained statistically significant. However, the presence of associations between T2D polygenic score and several glipizide challenge end points provides evidence for a true impact on glipizide response. These findings are additionally supported by the observation of a marginally higher glucose AOC in individuals with a higher FG polygenic score, again indicative of a greater glipizide response.
Since many of the SNPs comprising the T2D polygenic score influence β-cell function, it appears that treatment with glipizide, a sulfonylurea that stimulates insulin secretion from the β-cell, can overcome these genetic defects in the early stages of T2D pathogenesis. We speculate that perhaps those with a higher risk of T2D may have overly sensitized β-cells compared with those with a lower polygenic score, resulting in an accentuated response to glipizide. This is similar to what is observed in maturity-onset diabetes of the young type 3, which is characterized by HNF1A mutations causing decreased insulin secretion. Individuals with maturity-onset diabetes of the young type 3 demonstrate a heightened sensitivity to sulfonylureas (27) but require insulin as the secretory defect progresses. We hypothesize that individuals with a higher T2D risk score may behave in the same way, in which they initially have a sensitized β-cell early in the disease course but may achieve β-cell failure sooner.
Based on our findings, we sought replication in GoDARTS, a case-control study of T2D with longitudinal clinical and genetic data available. For a subset of 2,228 individuals who received a sulfonylurea, we tested whether a T2D polygenic score is associated with a clinical drug response. We found that the mean T2D polygenic score was higher in GoDARTS than in SUGAR-MGH, illustrating a higher burden of T2D risk variants. This was expected since GoDARTS participants have established T2D requiring sulfonylurea therapy. Moreover, we observed that a higher T2D score was again significantly associated with a greater sulfonylurea response, as measured by HbA1c reduction at 1 year. Thus, we demonstrated that the T2D score was not only associated with the physiologic response to an acute dose of glipizide, but also influenced the sustained glycemic response to sulfonylureas. We acknowledge that a 0.063% greater reduction in HbA1c per SD increase in T2D score is clinically small; however, this difference was as high as 0.27% when comparing the top and bottom deciles in T2D score. Therefore, the clinical utility of the T2D polygenic score may be limited in most of the population, but becomes more relevant in those at the extremes.
Interestingly, our findings appear to be in contrast with the candidate gene analysis of the TCF7L2 variant rs7903146 in GoDARTS, in which homozygotes for the T risk allele were less likely to respond to sulfonylureas (18). We have previously postulated that this genotype may have a differential effect in individuals with T2D who already have some degree of β-cell dysfunction compared with those without overt T2D (19). One might expect that similarly, those with a high T2D score and a predisposition to β-cell failure would benefit from sulfonylureas early in the disease course and have an attenuated response over time. However, our replication analyses in GoDARTS suggest otherwise, in that the association between a higher T2D score and greater response to sulfonylureas is observed even in those with established T2D and an average duration of disease of 4.8 years. Whether this effect would be observed for those with an even longer duration of T2D remains to be determined. If so, this could suggest that the T2D polygenic score captures additional mechanisms that remain to be elucidated.
We also demonstrated that individuals with a higher FI polygenic score trended toward a higher glucose trough, adjusted for baseline glucose, in response to glipizide. This finding might suggest that for the same 5-mg dose of glipizide resulting in the same amount of insulin secretion, individuals with a higher degree of insulin resistance respond worse and have a smaller glucose-lowering response. Notably, this observation was present after adjustment for BMI but did not meet the more stringent P value for multiple comparisons. We also did not observe an effect on other glipizide challenge end points.
With respect to metformin, we did not observe any significant associations between polygenic score and phenotypic end points of metformin response. This is not surprising, especially as T2D and FG polygenic scores comprising of predominantly β-cell function SNPs would not be expected to associate with metformin response. Similarly, in the Diabetes Prevention Program (DPP), a cohort with prediabetes, a genetic score of 34 T2D loci was associated with an increased risk of progression to diabetes and a lower probability of regression to normoglycemia, but there was no observed interaction effect of metformin on this association (28).
Prior pharmacogenetic studies of sulfonylurea response have been limited to candidate gene studies (14–17), and few have examined individual T2D-associated genes (18,19). Our study is the first, to our knowledge, to show a significant association between an aggregate score of T2D risk loci and drug response prospectively. One recently published study by Martono et al. (29) examined the added utility of genetic risk scores for insulin sensitivity, β-cell function, and T2D for prediction of the initial response to metformin or sulfonylureas in a primary care population with early T2D. They did not find an association between any of these scores and drug response, as measured by 6-month HbA1c, adjusted for baseline HbA1c. However, the study population was considerably smaller than ours (only 282 individuals initiating metformin and 89 individuals starting sulfonylureas) and may have been underpowered to detect significant effects. We note that our study also used weighted polygenic scores and data from the most recent GWAS for T2D (8).
Study strengths include the diverse population of our cohort, which allows for generalizability of our findings. Furthermore, SUGAR-MGH was conducted under fasting conditions, which limited the influence of dietary and lifestyle habits. While SUGAR-MGH had the advantage of examining a physiologic response to an acute perturbation in a controlled environment, the study design did not include an oral glucose tolerance test prior to metformin administration, which would have provided a dynamic glucose challenge for assessing metformin response.
Another shortcoming is that we were only able to assess a fraction of the known genomic loci for T2D and glycemic traits, due to the limited availability of SNPs on our genotyping platform. Genome-wide genotyping is currently underway in SUGAR-MGH, which will permit more extensive polygenic score construction in the future. This will include partitioned polygenic scores, which group variants by a common biological process and can provide insight into disease pathophysiology. The current study only analyzed restricted-to-significant polygenic scores, and future studies examining global extended polygenic scores, generated from large numbers of subthreshold significant variants, are needed as well. However, there appears to be limited improvement in predictive performance between a restricted polygenic score comprising 199 SNPs and a global polygenic score (21). These findings suggest that there may not be a significant step-up in power with increasing the number of variants included in the polygenic score. Finally, we note that the effect size estimates used in the polygenic score construction are for European ancestry, which does not take into consideration that risk variants can have different effect sizes in different populations. We also do not have ancestry information available on those individuals who self-reported as “Black” in our cohort. However, in our stratified analyses in non-Hispanic White and Black individuals (comprising 64% and 21% of SUGAR-MGH, respectively), we report findings that trend in the same direction as our primary analyses concerning the impact of the T2D polygenic score on glipizide response.
In summary, our findings suggest that there is some overlap between genes implicated in the risk of developing T2D and those associated with the response to treatment with sulfonylureas. We add to the growing body of literature on the potential utility of polygenic scores in understanding the response to T2D pharmacotherapy. Our study provides preliminary evidence that sulfonylureas could be more effective in T2D risk allele carriers, both in drug-naive individuals as well as those with established T2D. This finding is consistent with the recent results reported by Dennis et al. (30) in the A Diabetes Outcome Progression Trial (ADOPT) trial, showing that participants who cluster in the severely insulin-deficient diabetes phenotype (presumably enriched for β-cell deleterious alleles) experience a robust initial response to sulfonylureas, though it worsens over time. While genetic variation has been shown to alter the response to therapy in T2D, further confirmatory studies are necessary to clarify the role of polygenic scores in clinical decision-making.
Article Information
Funding. This work was conducted with support from National Institute of Diabetes and Digestive and Kidney Diseases awards R01 DK088214, R03 DK077675, and P30 DK036836, the Joslin Diabetes Center from its philanthropic donors, and the Harvard Catalyst: the Harvard University Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, awards M01-RR-01066, 1 UL1 RR025758-04, and 8UL1TR000170-05, and financial contributions from Harvard University and its affiliated academic health care centers). J.H.L. received individual support from National Institute of Diabetes and Digestive and Kidney Diseases grant T32DK007028. E.R.P. holds a Wellcome Trust New Investigator Award (102820/Z/13/Z).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.H.L., L.S., V.K., and J.C.F. conceived and designed the experiments in SUGAR-MGH. J.H.L., A.Y.D., E.R.P., and J.C.F. conceived and designed the replication analyses in GoDARTS. V.K. and J.C.F. recruited participants in SUGAR-MGH. J.H.L., L.S., A.Y.D., and V.K. analyzed the data. All authors took part in interpreting the data. J.H.L. and J.C.F. prepared the manuscript. All authors read and edited the manuscript. J.C.F. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in poster form at the 79th Scientific Sessions of the American Diabetes Association, San Francisco, CA, 7–11 June 2019.
Footnotes
L.S. is currently affiliated with Clinical Research Centre, Medical University of Bialystok, Bialystok, Poland.
J.N.T. is currently affiliated with Division of Endocrinology, Department of Pediatrics, University of Vermont Children’s Hospital, Burlington, VT.
This article contains supplementary material online at https://doi.org/10.2337/figshare.13135517.
References
- 1.Davies MJ, D’Alessio DA, Fradkin J, et al. . Management of hyperglycemia in type 2 diabetes, 2018. A consensus report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetes Care 2018;41:2669–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirst JA, Farmer AJ, Dyar A, Lung TW, Stevens RJ. Estimating the effect of sulfonylurea on HbA1c in diabetes: a systematic review and meta-analysis. Diabetologia 2013;56:973–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Diabetes Association 6. Glycemic targets: Standards of Medical Care in Diabetes—2019. Diabetes Care 2019;42(Suppl. 1):S61–S70 [DOI] [PubMed] [Google Scholar]
- 4.Dupuis J, Langenberg C, Prokopenko I, et al.; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk [published correction appears in Nat Genet 2010;42:464]. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris AP, Voight BF, Teslovich TM, et al.; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott RA, Lagou V, Welch RP, et al.; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012;44:991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manning AK, Hivert MF, Scott RA, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Multiple Tissue Human Expression Resource (MUTHER) Consortium . A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 2012;44:659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahajan A, Taliun D, Thurner M, et al. . Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flannick J, Mercader JM, Fuchsberger C, et al.; Broad Genomics Platform; DiscovEHR Collaboration; CHARGE; LuCamp; ProDiGY; GoT2D; ESP; SIGMA-T2D; T2D-GENES; AMP-T2D-GENES . Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature 2019;570:71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vujkovic M, Keaton JM, Lynch JA, et al.; HPAP Consortium; Regeneron Genetics Center; VA Million Veteran Program . Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet 2020;52:680–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dujic T, Zhou K, Yee SW, et al. . Variants in pharmacokinetic transporters and glycemic response to metformin: a metgen meta-analysis. Clin Pharmacol Ther 2017;101:763–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou K, Bellenguez C, Spencer CC, et al.; GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group; Wellcome Trust Case Control Consortium 2; MAGIC Investigators . Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet 2011;43:117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou K, Yee SW, Seiser EL, et al.; MetGen Investigators; DPP Investigators; ACCORD Investigators . Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat Genet 2016;48:1055–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sesti G, Laratta E, Cardellini M, et al. . The E23K variant of KCNJ11 encoding the pancreatic beta-cell adenosine 5′-triphosphate-sensitive potassium channel subunit Kir6.2 is associated with an increased risk of secondary failure to sulfonylurea in patients with type 2 diabetes. J Clin Endocrinol Metab 2006;91:2334–2339 [DOI] [PubMed] [Google Scholar]
- 15.Zhou K, Donnelly L, Burch L, et al. . Loss-of-function CYP2C9 variants improve therapeutic response to sulfonylureas in type 2 diabetes: a Go-DARTS study. Clin Pharmacol Ther 2010;87:52–56 [DOI] [PubMed] [Google Scholar]
- 16.Dujic T, Zhou K, Donnelly LA, Leese G, Palmer CNA, Pearson ER. Interaction between variants in the CYP2C9 and POR genes and the risk of sulfonylurea-induced hypoglycaemia: a GoDARTS Study. Diabetes Obes Metab 2018;20:211–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Li JH, Kaur V, et al. . The presence of two reduced function variants in CYP2C9 influences the acute response to glipizide. Diabet Med 2020;37:2124–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson ER, Donnelly LA, Kimber C, et al. . Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes 2007;56:2178–2182 [DOI] [PubMed] [Google Scholar]
- 19.Srinivasan S, Kaur V, Chamarthi B, et al. . TCF7L2 genetic variation augments incretin resistance and influences response to a sulfonylurea and metformin: the Study to Understand the Genetics of the Acute Response to Metformin and Glipizide in Humans (SUGAR-MGH). Diabetes Care 2018;41:554–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dujic T, Bego T, Malenica M, et al. . Effects of TCF7L2 rs7903146 variant on metformin response in patients with type 2 diabetes. Bosn J Basic Med Sci 2019;19:368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Udler MS, McCarthy MI, Florez JC, Mahajan A. Genetic risk scores for diabetes diagnosis and precision medicine. Endocr Rev 2019;40:1500–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walford GA, Colomo N, Todd JN, et al. . The Study to Understand the Genetics of the Acute Response to Metformin and Glipizide in Humans (SUGAR-MGH): design of a pharmacogenetic resource for type 2 diabetes. PLoS One 2015;10:e0121553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hébert HL, Shepherd B, Milburn K, et al. . Cohort profile: Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS). Int J Epidemiol 2018;47:380–381j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang K, Fu DJ, Julien D, Braun A, Cantor CR, Köster H. Chip-based genotyping by mass spectrometry. Proc Natl Acad Sci U S A 1999;96:10016–10020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- 26.R Development Core Team R: A Language and Environment for Statistical Computing. Vienna, Austria, R Foundation for Stastistical Computing, 2013 [Google Scholar]
- 27.Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet 2003;362:1275–1281 [DOI] [PubMed] [Google Scholar]
- 28.Hivert MF, Jablonski KA, Perreault L, et al.; DIAGRAM Consortium; Diabetes Prevention Program Research Group . Updated genetic score based on 34 confirmed type 2 diabetes loci is associated with diabetes incidence and regression to normoglycemia in the diabetes prevention program. Diabetes 2011;60:1340–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martono DP, Heerspink HJL, Hak E, Denig P, Wilffert B. No significant association of type 2 diabetes-related genetic risk scores with glycated haemoglobin levels after initiating metformin or sulphonylurea derivatives. Diabetes Obes Metab 2019;21:2267–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis JM, Shields BM, Henley WE, Jones AG, Hattersley AT. Disease progression and treatment response in data-driven subgroups of type 2 diabetes compared with models based on simple clinical features: an analysis using clinical trial data. Lancet Diabetes Endocrinol 2019;7:442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]