Abstract
Purpose:
Most prostate cancer in African American men lacks the ETS (E26 transforming specific) family fusion event (ETS–). We aimed to establish clinically relevant biomarkers in African American men by studying ETS dependent gene expression patterns to identified race specific genes predictive of outcomes.
Materials and Methods:
Two multicenter cohorts of a total of 1,427 men were used for the discovery and validation (635 and 792 men, respectively) of race specific predictive biomarkers. We used false discovery rate adjusted q values to identify race and ETS dependent genes which were differentially expressed in African American men who experienced biochemical recurrence within 5 years. Principal component modeling along with survival analysis was done to assess the accuracy of the gene panel in predicting recurrence.
Results:
We identified 3,047 genes which were differentially expressed based on ETS status. Of these genes 362 were differentially expressed in a race specific manner (false discovery rate 0.025 or less). A total of 81 genes were race specific and over expressed in African American men who experienced biochemical recurrence. The final gene panel included APOD, BCL6, EMP1, MYADM, SRGN and TIMP3. These genes were associated with 5-year biochemical recurrence (HR 1.97, 95% CI 1.27–3.06, p = 0.002) and they improved the predictive accuracy of clinicopathological variables only in African American men (60-month time dependent AUC 0.72).
Conclusions:
In an effort to elucidate biological features associated with prostate cancer aggressiveness in African American men we identified ETS dependent biomarkers predicting early onset biochemical recurrence only in African American men. Thus, these ETS dependent biomarkers representing ideal candidates for biomarkers of aggressive disease in this patient population.
Keywords: prostatic neoplasms, biomarkers, tumor, African Americans, gene expression regulation, neoplastic, neoplasm recurrence, local
PROSTATE cancer is currently the most common malignancy and the third leading cause of cancer associated death among men in the United States.1 Despite aggressive screening and advancements in treatment studies have shown that AAM continue to have worse outcomes than EAM.2 Although disparities in outcomes were previously attributed to socioeconomic factors such as access to health care,3 recent data suggest that biological and genetic factors may also contribute to a more aggressive cancer phenotype at presentation.4
The phenotypic cancer variations observed among AAM and EAM suggest that race specific factors have a role in prostate tumor biology and may in turn help identify clinically useful biomarkers. For example, some studies demonstrated variations in the androgen receptor signaling pathways between AAM and EAM, and detected substantially higher levels of androgen receptor protein expression among AAM with PCa.5,6 More recent studies indicated a correlation between specific biomarkers and PCa aggressiveness.7–9 Although these biomarkers have demonstrated the potential capability to predict disease aggressiveness, the clinical significance of these biomarkers is still lacking.
As targeted therapies come to the forefront of cancer treatment, understanding tumor biological differences based on ethnicity has become increasingly important. Previous data suggest that genomic mutations in ETS transcription factors are found in more than half of all prostate cancers.10 Gene fusion of the androgen responsive promoter TMPRSS2 to the ERG oncogene, a member of the ETS family, occurs early in the oncogenic process and was found to result in over expression of other prognostic biomarkers.11 Studies have shown that this genetic rearrangement behaves in a race dependent manner with ETS– tumors in most AAM.12
In an effort to identify clinically significant biomarkers in AAM we identified differences in ETS gene expression among AAM and EAM, and determined which genes predicted outcomes in a race specific manner.
METHODS AND MATERIALS
Patient Database and Tumor Tissue Selection
Previously published data sets from the Decipher® GRID were used to obtain microarray data. The Decipher GRID cohort included 635 CAPRA-S scores, a validated score of the BCR risk after surgery, in 127 and 508 matched AAM and EAM, respectively. With CAPRA-S matching AAM and EAM were at relatively similar risk for BCR. RNA was extracted from formalin fixed, paraffin embedded tissues, followed by amplification, labeling and hybridization to Affymetrix Human Exon 1.0 ST using Human Exon 1.0 ST Arrays to obtain expression profiling. All microarray data were obtained using the Decipher PCa classifier and performed in a CLIA (Clinical Laboratory Improvement Amendments) certified clinical laboratory. Patient profile preprocessing and normalization were completed with the SCAN algorithm13 and combat was used for debatching.14
Clinicopathological and Outcome Variables
Microarray data were obtained from radical prostatectomy samples from a previously published, retrospective GRID study consisting of a matched cohort of AAM and EAM.4 The 635 men were identified from a total of 4 institutions, including Thomas Jefferson University, Johns Hopkins University, Cleveland Clinic Foundation and Memorial Sloan Kettering Cancer Center. In addition to expression profiles, the discovery data set also contained patient baseline demographic and clinicopathological characteristics (table 1). Institutional Review Board approval was obtained before the formal analysis was performed (IRB No. MCC 18593).
Table 1.
Baseline clinicopathological characteristics by race in GRID discovery cohort of 635 African American and European American men
| AAM | EAM | p Value | |
|---|---|---|---|
| No. pts | 127 | 508 | – |
| No. ETS status (%): | |||
| Neg | 95 (74.8) | 214 (42.1) | <0.001 |
| Pos | 32 (25.2) | 294 (57.9) | |
| No. pathological Gleason Score (%): | |||
| 3 + 3 or Less | 13 (10.2) | 45 (8.9) | 0.03 |
| 3 + 4 | 70 (55.1) | 218 (42.9) | |
| 4 + 3 | 21 (16.5) | 94 (18.5) | |
| 8 or Greater | 23 (18.1) | 151 (29.7) | |
| No. ng/ml preop PSA (%): | |||
| 6 or Less | 35 (27.6) | 189 (37.2) | 0.08 |
| Greater than 6—10 | 47 (37.0) | 144 (28.3) | |
| Greater than 10—20 | 34 (26.8) | 145 (28.5) | |
| Greater than 20 | 11 (8.7) | 30 (5.9) | |
| No. surgical margins(%): | |||
| Present | 85 (66.9) | 239 (47.0) | <0.001 |
| Absent | 42 (33.0) | 269 (52.9) | |
| No. extracapsular extension (%): | |||
| Yes | 59 (46.5) | 343 (67.5) | <0.001 |
| No | 68 (53.5) | 165 (32.5) | |
| No. lymph node invasion (%): | |||
| Yes | 3 (2.4) | 37 (7.3) | 0.04 |
| No | 124 (97.6) | 471 (92.7) | |
| No. seminal vesical invasion (%): | |||
| Yes | 25(19.7) | 109 (21.5) | 0.66 |
| No | 102 (80.3) | 399 (78.5) | |
| No. CAPRA-S score (%): | |||
| 0–2 | 15 (11.8) | 64 (12.6) | 0.96 |
| 3–5 | 71 (55.9) | 280 (55.1) | |
| 6–12 | 41 (32.3) | 164 (32.3) | |
| No. age at diagnosis (%): | |||
| 50 or Less | 12 (9.4) | 24 (4.7) | 0.06 |
| Greater than 50—65 | 92 (72.4) | 363 (71.5) | |
| Greater than 65 | 23 (18.1) | 121 (28.8) |
Clinical End Points
BCR within 5 year of prostatectomy was a primary end point in the gene discovery work. BCR was defined as 2 consecutive postoperative PSA values of 0.2 ng/ml.15
ETS Expression Profiling and Molecular Characterization
ETS+ status was assigned to samples expressing high levels of ERG, and the ETS family members ETV1–5 and FLI1. All other tumors were assigned as ETS−. The ETS status of the GRID was determined as previously described for subtyping of microarray based ETS+ (ERG or ETV1/4/5).16
Genomic Resource Information Database Validation Cohort
A total of 792 prospectively collected radical prostatectomy tumor expression profiles served as validation data in this analysis (fig. 1). Microarray gene expression data and associated clinical information, including molecular subtypes, were retrieved from the Decipher GRID registry (ClinicalTrials.gov NCT02609269). The prospective GRID cohort (the GRID prospective) was obtained by clinical performance of the Decipher test. Samples from the GRID prospective cohorts are nonoverlapping and were analyzed with the same Affymetrix microarray platform.
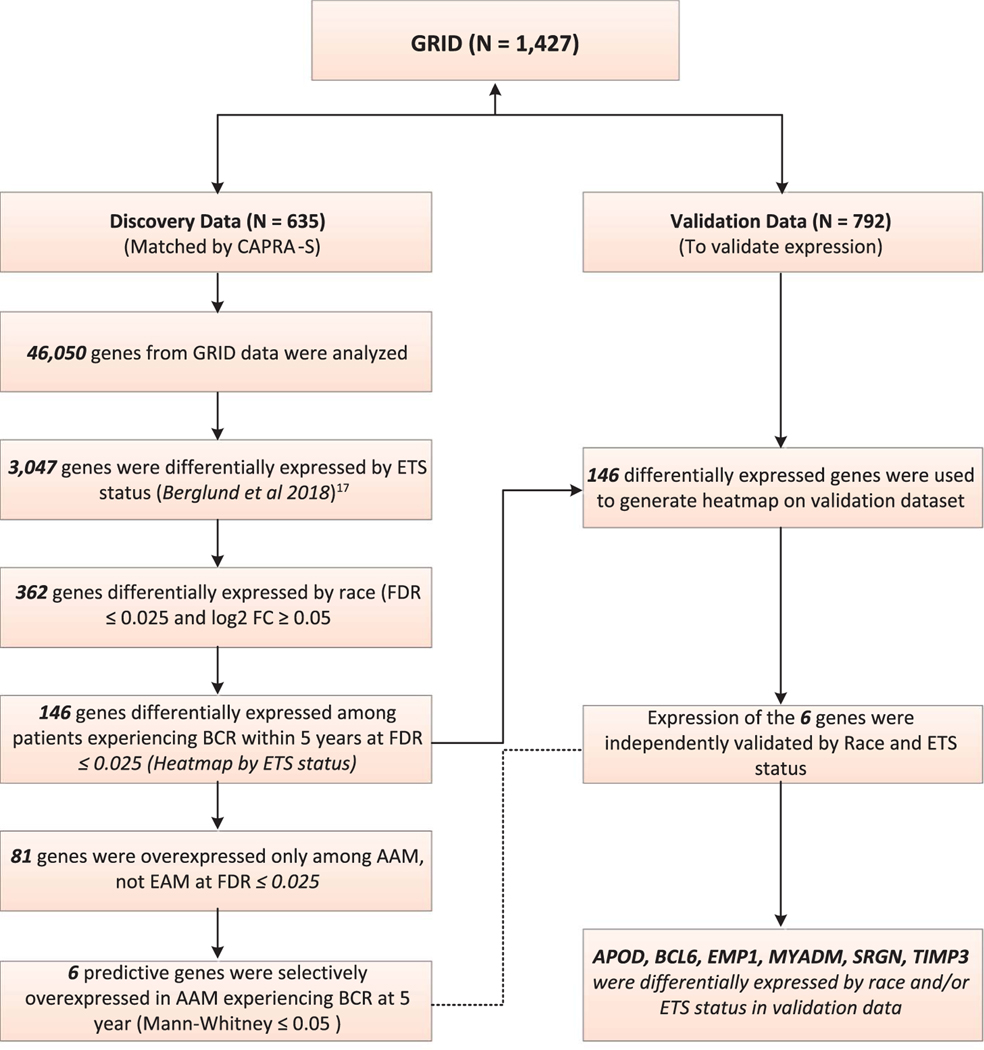
Figure 1.
CONSORT diagram of gene discovery and validation protocol using GRID data. FDR, false discovery rate.
Statistical Analysis
In a categorical data analysis of the GRID discovery and validation data we used the chi-square/exact method to assess race differences in ETS status along with clinicopathological and demographic characteristics (table 1 and supplementary table 1, https://www.jurology.com). The 2-tailed Mann-Whitney U test and the fold change cutoff (log2 fold change) were applied to assess differences in gene expression by ETS and race status. False discovery rate adjusted q values were used to account for multiple comparisons. The 3,047 ETS dependent genes identified by Berglund et al which were differentially expressed in a race specific manner (q ≤0.025 and log2 fold change ≥0.05)17 were selected to evaluate the association with PCa outcomes, which was BCR in 5 years.
PC analysis was run on the selected gene panel to obtain composite gene expression scores. Race stratified Kaplan-Meier curves were used to determine the association of the gene panel with 5-year BCR. Furthermore, the predictive accuracy of the final gene panel when added to the standard clinicopathological variables was determined by calculating the tAUC at a specific time point (60 months) by the methods described by Guo et al.18 Analysis was completed with the SAS® and R (https://www.r-project.org/foundation/) statistical packages.
RESULTS
Median followup in the discovery cohort was 108 months. Table 1 shows clinicopathological characteristics of the CAPRA-S matched (1:4 ratio) cohort of 635 patients, of whom 127 were AAM and 508 were EAM. In the discovery and validation data sets a significantly higher proportion of AAM were ETS negative compared to EAM. Table 1 and supplementary table 1 (https://www.jurology.com) show associations of race with ETS and clinicopathological variables for discovery and validation data, respectively.
Genomic Resource Information Database Gene Discovery
The CONSORT (Consolidated Standards of Reporting Trials) diagram details the gene discovery algorithm and the validation process (fig. 1). We interrogated the gene expression profile of the GRID discovery cohort, which contains the expression profile of 46,050 genes, to identify ETS dependent genes. We used 3,047 previously identified ETS dependent genes from the GRID according to the criteria of Berglund et al (q ≤0.05 and a ≥0.05 expression fold change).17 From the pool of 3,047 ETS dependent genes 362 were differentially expressed between AAM and EAM after adjusting for multiple comparisons using a stringent FDR correction of ≤0.025 with an expression fold change of ≥0.05. The 362 race dependent genes included previously identified biomarkers which have been shown to impact PCa biology, such as AMACR, AMD1, ANPEP, CACNA1D, CCL3, CRISP3, ERG, HPGD, INPP4B, LSAMP, PCGEM1 and SPINK1 (supplementary table 2, https://www.jurology.com).
To evaluate the clinical usefulness of these ETS dependent, race specific genes we then interrogated these 362 race specific genes for expression in patients who experienced BCR (supplementary table 3, https://www.jurology.com). A total of 146 genes were differentially expressed in patients with BCR. Supplementary figure 1 (https://www.jurology.com) shows a cluster analysis heat map of these 146 genes sorted based on ETS status, which were differentially expressed in a race and outcome specific manner. There was a cluster of AAM cases in the ETS− gene cluster compared to the ETS+ gene cluster (supplementary figs. 1 and 2, https://www.jurology.com).
Final Gene Panel
Next we sought to identify genes which could be further developed and assayed for clinical usefulness as predictive biomarkers in AAM. Using 146 race dependent and outcome specific genes we identified 81 genes which were over expressed only in AAM and not in EAM (supplementary table 2, https://www.jurology.com). These 81 AAM specific genes were then used to select the final gene panel. To ensure the robustness of our gene panel we further evaluated the expression of these genes in a race specific manner, comparing patients with vs without BCR (supplementary table 2, https://www.jurology.com).
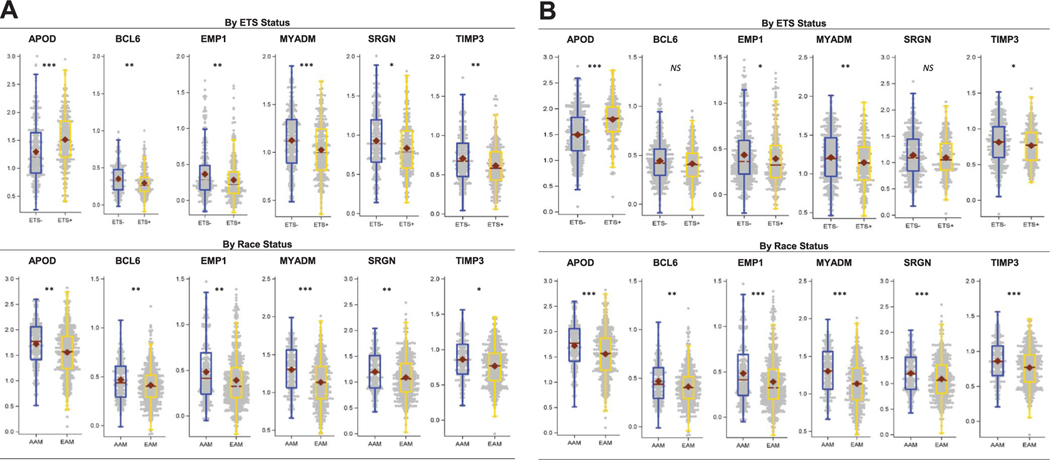
Six genes (APOD, BCL6, EMP1, MYADM, SRGN and TIMP3) were selectively over expressed in AAM who experienced BCR within 5 years after prostatectomy and were identified as putative biomarkers of early onset BCR in AAM. Figure 2, A shows median expression of these genes by ETS and race status. To further verify the observed differential expression in the discovery data set we validated these genes by race and ETS status in an AAM enriched validation data set of 792 patients (fig. 2, B). This 6-gene panel represents promising putative biomarkers of aggressive biology in AAM and it can be useful for the future development of AAM specific signatures (table 2).
Figure 2.
Expression pattern of AAM and 6 outcome specific ETS dependent genes by race and ETS status. A, discovery data set. Single asterisk indicates p <0.025, double asterisks indicate p <0.01 and triple asterisks indicate p <0.001. B, validation data set. NS, not significant. Single asterisk indicates p <0.05, double asterisks indicate p <0.01 and triple asterisks indicate p <0.001.
Table 2.
Differential expression of 6 putative biomarker genes by race and ETS status in discovery and validation data sets
| Gene Panel | Discovery Data Set False Discovery Rate Adjusted q Value* | Validation Data Set p Value (Mann-Whitney test) | ||
|---|---|---|---|---|
| AAM vs EAM | ETS− vs ETS+ | AAM vs EAM | ETS− vs ETS+ | |
| APOD | 0.008 | 2.12E-08 | 1.25E-06 | 1.79E-19 |
| BCL6 | 0.007 | 0.00117 | 0.001 | 0.11 |
| EMP1 | 0.005 | 0.00295 | 0.0005 | 0.03 |
| MYADM | 0.0005 | 0.000958 | 1.25E-11 | 0.007 |
| SRGN | 0.008 | 0.0111 | 0.0002 | 0.18 |
| TIMP3 | 0.02 | 0.00161 | 4.13E-05 | 0.01 |
Differential median expression.
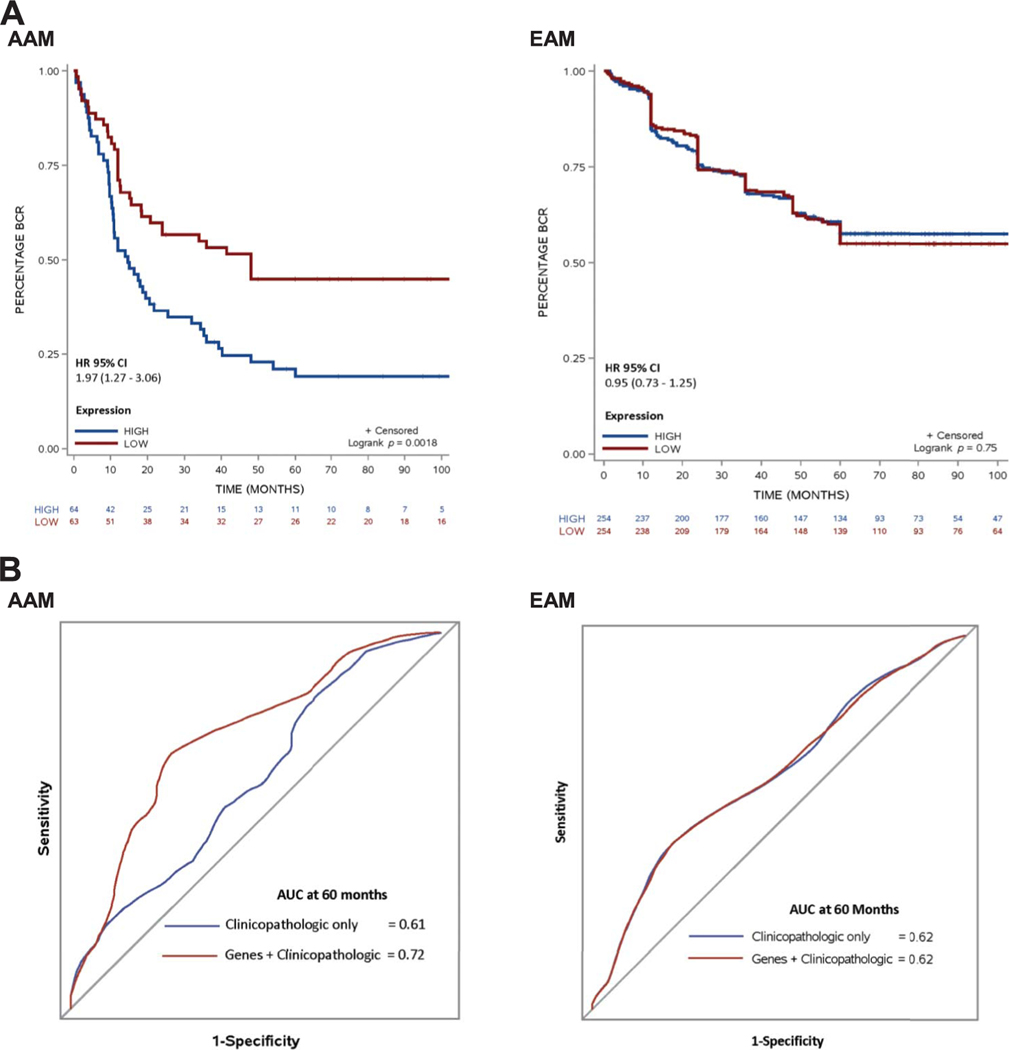
To establish the clinical usefulness of our gene panel in the prediction of 5-year BCR a final 6-gene panel was introduced in a PC model. The first PC (PC1) of the final gene panel explains 57.2% of the variation with a PC1-to-PC2 ratio of 3.5, making this model better than a random gene model.17 Based on the PC analysis model we identified a composite expression score for the gene panel, which was further categorized based on its median value. On Kaplan-Meier analysis higher expression of AAM specific genes was associated with a higher risk of BCR only among AAM but no difference was observed among EAM (fig. 3, A).
Figure 3.
A, race stratified Kaplan-Meier curves of 5-year BCR risk using AAM specific gene panel. B, race stratified, time dependent area operating curve of Cox proportional hazard model using discovery data with AUC showing predictive ability of ETS dependent AAM specific genes for 5-year BCR at 60 months. Clinicopathologic only, Cox proportional hazard model with preoperative PSA, pathological Gleason score and age at diagnosis. Genes + clinicopathologic, Cox proportional hazard model with AAM specific genes and preoperative PSA, pathological Gleason score and age at diagnosis.
To assess predictive accuracy the final gene panel was then introduced into a standard Cox model along with known clinicopathological variables. Using the methods described by Guo et al18 we derived the tAUC of the model at the specific time point of 60 months. Including our gene panel in the standard Cox model yielded high accuracy selectively in AAM (tAUC 0.72, fig. 3, B). No apparent benefit was observed among EAM.
DISCUSSION
Underrepresentation of an AAM study population in the development of most commercially available biomarkers and gene signatures often limits the effectiveness and reliability of these signatures in AAM.19 Given the differences in underlying biology, there is a need to further explore AAM specific biomarkers which can aid in the development of AAM specific signatures to better understand the PCa prognosis among AAM. Previous research has demonstrated that mutations in ETS genes are part of an early oncogenic process in PCa which behaves in a race specific manner.11,20,21
In the current study we found race differences in ETS expression and identified a race specific ETS dependent gene panel associated with clinically relevant outcomes such as BCR with increased predictive accuracy, particularly in AAM. While previous studies showed differentially expressed genes in prostate tumors of AAM and EAM, few studies have been able to link these genomic differences to clinically relevant outcomes. Currently identification of TMPRSS2-ERG gene fusion as an early process in PCa has led to the implementation of ETS over expression as a PCa biomarker.11 However, the clinical implications of ETS over expression may only be pertinent in EAM as studies have demonstrated that most AAM lack ETS fusion.4,12 Our study consistently showed that race was one of the strongest predictors of ETS status with ETS− findings in approximately 75% of AAM in our cohort. Additionally, we identified a subset of ETS dependent genes predicting clinical outcomes in a race dependent manner. In this gene subset we determined which genes were differentially expressed by race and, therefore, could serve as clinically relevant biomarkers among AAM.
Importantly 5 genes out of the final 6-gene panel, including BCL6, EMP1, MYADM, SRGN and TIMP3, were over expressed in ETS− tumors, making these genes extremely relevant to AAM tumor biology. Although to our knowledge the mechanistic interaction between race and the ETS dependent biology of our final 6-gene panel remains largely unknown, the role of these genes has been documented in cell proliferation and tumorigenesis.22−29 Interestingly, 2 of the 6 genes (EMP1 and MYADM) have been shown to interact with Rac1, a member of the Rho-GTPase family, which when activated results in progression and metastasis of various cancers, including PCa.22,23 Pathway dependence in the cellular functionality of EMP1 and MYADM may provide biological basis for the importance of Rho-GTPase protein in AAM and should be explored further.
SRGN has been shown to be markedly over expressed in malignant tumors and it has been implicated in other cancers.24,25 Another gene, BCL6, a suppressor of p53 genes, is known to modulate DNA damage via altered apoptotic response26 and is over expressed in metastatic PCa.27 TIMP3 is considered to be associated with advanced stage tumors, given its selectively higher expression in adjacent stromal cells in close proximity to invasive tumor cells.28 Lastly, APOD, the only gene in our 6-gene panel which is over expressed in ETS+ tumors, has androgen mediated action which impacts cell proliferation in PCa.29
Given the nature of race specific variations in ETS rearrangement in PCa, the development of AAM specific, ETS dependent gene signatures can be vital for future biomarker discovery work and targeted drug development. In the era of personalized medicine our biomarker signature may be useful to stratify patients and guide treatment decisions in AAM, potentially helping to close the disparities gap.
Although previous studies have described single genes which may predict prognosis, to our knowledge this study is the only one of its kind to identify a gene panel of clinically relevant ETS dependent biomarkers which is differentially expressed in a race specific manner. Enrichment of the traditionally underrepresented population of African American patients in our discovery and validation data sets significantly improved the generalizability of our results.
Our study also has some limitations, including the lack of clinically robust long-term end points, ie PCa specific survival and metastasis, in discovery data. Although BCR is a good surrogate for the treatment response, further investigation may be warranted to assess the clinical usefulness of our gene panel to predict metastasis and survival. Additionally, while our 6-gene panel was validated for race and ETS dependent differential expression in the GRID validation cohort, the lack of survival end points such as BCR in these data limited our ability to validate the predictive accuracy of our gene panel. Therefore, a future direction of this study would be to prospectively collect clinical data and validate our gene biomarker panel.
CONCLUSIONS
In an effort to elucidate biological features associated with PCa aggressiveness in AAM we identified ETS dependent biomarkers predicting early onset BCR in AAM. Our results show that differences in tumor biology based on ethnicity may help predict aggressive disease in AAM and may be used to stratify patients and direct treatment decisions. Although our results provide a promising predictive tool for AAM with PCa, future validation studies are still needed to adopt this ETS dependent biomarker signature into clinical practice.
Supplementary Material
Acknowledgments
Supported by the Prostate Cancer Foundation Young Investigator Award (KY), American Cancer Society Grant MRSG-17–108-01-TBG (KY), the V Foundation (KY, JYP) and the NCI Comprehensive Cancer Center P30 CA076292 Grant (Moffitt Cancer Center).
Abbreviations and Acronyms
- AAM
African American men
- APOD
apolipoprotein D
- BCL6
B cell CLL/lymphoma 6
- BCR
biochemical recurrence
- CAPRA-S
Cancer of the Prostate Risk Assessment–post-surgical
- EAM
European American men
- EMP1
epithelial membrane protein 1
- ETS
E26 transforming specific
- ETS−
ETS negative
- ETS+
ETS positive
- GRID
Genomic Resource Information Database
- MYADM
myeloid associated differentiation marker
- PC
principal component
- PCa
prostate cancer
- PSA
prostate specific antigen
- SRGN
secretory granule proteoglycan core protein
- tAUC
time dependent AUC
- TIMP3
tissue inhibitor of metalloproteinases 3
Footnotes
The corresponding author certifies that, when applicable, a statement(s) has been included in the manuscript documenting institutional review board, ethics committee or ethical review board study approval; principles of Helsinki Declaration were followed in lieu of formal ethics committee approval; institutional animal care and use committee approval; all human subjects provided written informed consent with guarantees of confidentiality; IRB approved protocol number; animal approved project number.
No direct or indirect commercial, personal, academic, political, religious or ethical incentive is associated with publishing this article.
REFERENCES
- 1.Siegel RL, Miller KD and Jemal A: Cancer statistics, 2017. CA Cancer J Clin 2017; 67: 7. [DOI] [PubMed] [Google Scholar]
- 2.Altekruse SF, Kosary CL, Krapcho M et al. : SEER Cancer Statistics Review. Available at http://seer.cancer.gov/csr/1975_2007/. Accessed September 18, 2017. [Google Scholar]
- 3.Williams VL, Awasthi S, Fink AK et al. : African-American men and prostate cancer-specific mortality: a competing risk analysis of a large institutional cohort, 1989–2015. Cancer Med 2018; 7: 2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamoah K, Johnson MH, Choeurng V et al. : Novel biomarker signature that may predict aggressive disease in African American men with prostate cancer. J Clin Oncol 2015; 33: 2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Platz EA, Rimm EB, Willett WC et al. : Racial variation in prostate cancer incidence and in hormonal system markers among male health professionals. J Natl Cancer Inst 2000; 92: 2009. [DOI] [PubMed] [Google Scholar]
- 6.Gaston KE, Kim D, Singh S et al. : Racial differences in androgen receptor protein expression in men with clinically localized prostate cancer. J Urol 2003; 170: 990. [DOI] [PubMed] [Google Scholar]
- 7.Perner S, Hofer MD, Kim R et al. : Prostate-specific membrane antigen expression as a predictor of prostate cancer progression. Hum Pathol 2007; 38: 696. [DOI] [PubMed] [Google Scholar]
- 8.Perner S, Mosquera JM, Demichelis F et al. : TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol 2007; 31: 882. [DOI] [PubMed] [Google Scholar]
- 9.Fine SW, Gopalan A, Leversha MA et al. : TMPRSS2-ERG gene fusion is associated with low Gleason scores and not with high-grade morphological features. Mod Pathol 2010; 23: 1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narod SA, Seth A and Nam R: Fusion in the ETS gene family and prostate cancer. Br J Cancer 2008; 99: 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlins SA, Rhodes DR, Perner S et al. : Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005; 310: 644. [DOI] [PubMed] [Google Scholar]
- 12.Magi-Galluzzi C, Tsusuki T, Elson P et al. : TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate 2011; 71: 489. [DOI] [PubMed] [Google Scholar]
- 13.Piccolo SR, Withers MR, Francis OE et al. : Multiplatform single-sample estimates of transcriptional activation. Proc Natl Acad Sci U S A 2013; 110: 17778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson WE, Li C and Rabinovic A: Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8: 118. [DOI] [PubMed] [Google Scholar]
- 15.Yamoah K, Deville C, Vapiwala N et al. : African American men with low-grade prostate cancer have increased disease recurrence after prostatectomy compared with Caucasian men. Urol Oncol 2015; 33: 70.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres A, Alshalalfa M, Tomlins SA et al. : Comprehensive determination of prostate tumor ETS gene status in clinical samples using the CLIA decipher assay. J Mol Diagn 2017; 19: 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berglund AE, Rounbehler RJ, Gerke T et al. : Distinct transcriptional repertoire of the androgen receptor in ETS fusion-negative prostate cancer. Prostate Cancer Prostatic Dis 2018; doi: 10.1038/s41391-018-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo C, So Y and Jang W: Paper SAS462–2017. Evaluating predictive accuracy of survival models with PROC PHREG. Available at https://pdfs.semanticscholar.org/0f63/7c13f7eac0dbbe-b1a691da46197593fa131b.pdf. Accessed February 4, 2019. [Google Scholar]
- 19.Pietro GD, Chornokur G, Kumar NB et al. : Racial differences in the diagnosis and treatment of prostate cancer. Int Neurourol J, suppl., 2016; 20: S112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrovics G, Liu A, Shaheduzzaman S et al. : Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene 2005; 24: 3847. [DOI] [PubMed] [Google Scholar]
- 21.Mosquera JM, Mehra R, Regan MM et al. : Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res 2009; 15: 4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aranda JF, Reglero-Real N, Kremer L et al. : MYADM regulates Rac1 targeting to ordered membranes required for cell spreading and migration. Mol Biol Cell 2011; 22: 1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmat Amin MK, Shimizu A, Zankov DP et al. : Epithelial membrane protein 1 promotes tumor metastasis by enhancing cell migration via copine-III and Rac1. Oncogene 2018; 37: 5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li XJ, Ong CK, Cao Y et al. : Serglycin is a theranostic target in nasopharyngeal carcinoma that promotes metastasis. Cancer Res 2011; 71: 3162. [DOI] [PubMed] [Google Scholar]
- 25.Korpetinou A, Papachristou DJ, Lampropoulou A et al. : Increased expression of serglycin in specific carcinomas and aggressive cancer cell lines. Biomed Res Intern 2015; 2015: 690721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phan RT and Dalla-Favera R: The BCL6 protooncogene suppresses p53 expression in germinal-centre B cells. Nature 2004; 432: 635. [DOI] [PubMed] [Google Scholar]
- 27.Kristedja TO, Park SE, Tiwari RK: Synthesis and antiproliferative activity of hybrid peptides for ovarian and prostate cancer. Int J Pept Res Ther 2018; doi: 10.1007/s10989-0189751-4. [DOI] [Google Scholar]
- 28.Kornfeld JW, Meder S, Wohlberg M et al. : Overexpression of TACE and TIMP3 mRNA in head and neck cancer: association with tumour development and progression. Br J Cancer 2011; 104: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Otın C and Diamandis EP: Breast and prostate cancer: an analysis of common epidemiological, genetic, and biochemical features. Endocr Rev 1998; 19: 365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.