Abstract
Adult stem cells must continuously fine‐tune their behavior to regenerate damaged organs and avoid tumors. While several signaling pathways are well known to regulate somatic stem cells, the underlying mechanisms remain largely unexplored. Here, we demonstrate a cell‐intrinsic role for the OvoL family transcription factor, Shavenbaby (Svb), in balancing self‐renewal and differentiation of Drosophila intestinal stem cells. We find that svb is a downstream target of Wnt and EGFR pathways, mediating their activity for stem cell survival and proliferation. This requires post‐translational processing of Svb into a transcriptional activator, whose upregulation induces tumor‐like stem cell hyperproliferation. In contrast, the unprocessed form of Svb acts as a repressor that imposes differentiation into enterocytes, and suppresses tumors induced by altered signaling. We show that the switch between Svb repressor and activator is triggered in response to systemic steroid hormone, which is produced by ovaries. Therefore, the Svb axis allows intrinsic integration of local signaling cues and inter‐organ communication to adjust stem cell proliferation versus differentiation, suggesting a broad role of OvoL/Svb in adult and cancer stem cells.
Keywords: Drosophila, enterocyte differentiation, intestinal stem cells, OvoL transcription factors, Wnt and EFGR pathways
Subject Categories: Cancer, Signal Transduction, Regenerative Medicine
Post‐translational processing of the transcription factor Shavenbaby defines its dichotomous function in fly midgut homeostasis.

Introduction
Living organisms are constantly exposed to aging and environmental challenges that disturb cell functions and ultimately lead to cell death. To maintain homeostasis, most adult organs are regenerated by self‐renewing stem cells, which differentiate to replace dead cells and replenish damaged tissues. The highly regenerative digestive system is kept intact during adulthood by the activity of resident intestinal stem cells. Drosophila intestinal stem cells have emerged as a powerful system to understand the signaling networks underlying stem cell biology and their implication in cancers (reviewed in (Li & Jasper, 2016; Perochon et al, 2018)).
The adult fly intestine consists of a compartmentalized epithelium (Buchon et al, 2013), which shares anatomical and physiological similarities with its mammalian counterpart. Drosophila intestinal stem cells (ISCs) are small diploid cells scattered along the basement membrane (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2006). In steady‐state conditions, ISC divide asymmetrically to generate a new stem cell and a transient post‐mitotic progenitor cell called enteroblast (EB) (Ohlstein & Spradling, 2007). ISCs and early EBs express Escargot (Esg), a transcription factor of the Snail/Slug family that maintains diploidy and prevents premature differentiation (Korzelius et al, 2014; Loza‐Coll et al, 2014). EBs progressively acquire characteristics of polyploid absorptive enterocytes (ECs), representing the main population of intestinal cells (Ohlstein & Spradling, 2007). The second type of differentiated intestinal cells is hormone‐secreting enteroendocrine cells (EEs). They emerge from a separate pool of progenitors (Biteau & Jasper, 2014; Zeng & Hou, 2015), called pre‐enteroendocrines (pre‐EEs), which express markers of both ISCs (Esg) and EEs (Prospero).
The evolutionarily conserved Notch pathway establishes the asymmetry between ISCs and EBs (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2007; Bardin et al, 2010; Perdigoto et al, 2011). ISCs express Delta, a ligand that activates the Notch receptor in daughter EBs, as seen by Su(H) expression. The EC fate requires high levels of Notch, whereas lower Notch activity induces the production of EEs that maintain Prospero expression. Gut homeostasis relies on a tight regulation of ISC division through cooperative activity of conserved developmental signaling pathways, such as the epidermal growth factor receptor (EGFR), Wnt, and JAK/STAT pathways (Jiang & Edgar, 2009; Biteau & Jasper, 2011; Jiang et al, 2011). Despite the wealth of knowledge accumulated on the role of signaling pathways in regulating ISC maintenance, division, and differentiation, the intrinsic mechanisms by which ISCs integrate these cues remain largely unknown.
During embryogenesis, the activity of Wnt and EGFR pathways in the epidermis is mediated by a common target gene, ovo/shavenbaby (svb), which encodes a transcription factor governing epidermal differentiation (Payre et al, 1999). The Svb factor undergoes post‐translational processing from a repressor (SvbREP) to an activator (SvbACT) via limited proteasome degradation (Zanet et al, 2015). Svb maturation is triggered by Polished rice (Pri) peptides (Kondo et al, 2010), which are founding members of a growing family of peptides translated from small open reading frames, called smORF peptides (Saghatelian & Couso, 2015; Plaza et al, 2017). ovo/svb is also critical for maintenance and differentiation of the germline (Mevel‐Ninio et al, 1995). There are two Svb germline‐specific isoforms, called OvoA and OvoB, which are insensitive to Pri peptides (Kondo et al, 2010) and act as constitutive repressor and activator (Andrews et al, 2000), respectively. Throughout development, the production of SvbACT in somatic tissues is triggered by periodic peaks of ecdysone, the main steroid hormone in insects. Upon hormone binding, the ecdysone receptor (EcR) directly activates the expression of pri, triggering, in turn, Svb processing (Chanut‐Delalande et al, 2014). The ecdysone signaling pathway has also wide‐ranging functions in adults, including regulation of stress resistance, nutritional state, and reproduction (Uryu et al, 2015).
Ovo/Svb defines a metazoan‐specific family of transcription factors, comprising three paralogs in vertebrates called OvoL1‐3, which are crucial regulators of epithelial lineage determination and differentiation. For example, human OvoL2 is required for the maintenance of corneal epithelium cells (Kitazawa et al, 2016) and its alteration is a major cause of inherited corneal dystrophies (Davidson et al, 2016). OvoL factors have been involved in the metastatic/stemness potential of various tumors, including in breast (Roca et al, 2013), prostate (Fu et al, 2016), lung (Wang et al, 2017), and colorectal (Ye et al, 2016) cancers. Moreover, OvoLs also act for the repair of epithelial tissues from stem/progenitor cells, e.g., for epidermal and mammary regeneration (Watanabe et al, 2014; Haensel et al, 2019). In the flatworm, OvoL/Svb is expressed in eye progenitors and required for eye regeneration from multipotent stem cells (Lapan & Reddien, 2012). Hence, a growing body of evidence suggests a role of OvoL/Svb in stem/progenitor cells across animals. Indeed, we recently found that Svb is required for the survival of renal nephric stem cells (RNSCs) in adult flies, via direct interaction with Yorkie (a.k.a. YAP/TAZ), the nuclear effector of the Hippo pathway (Bohere et al, 2018). RNSCs derive from progenitors that also produce intestinal stem cells (Xu et al, 2018), suggesting a broader function of Svb in adult stem cells.
Here, we demonstrate that the Shavenbaby transcription factor is essential to adult midgut homeostasis. Importantly, proteasome‐mediated processing allows Svb isoforms to exert antagonistic functions along the ISC lineage. Through clonal analysis of a null allele of svb, and cell type‐specific RNAi knockdown or overexpression, we conclude that the processed SvbACT is required to maintain ISCs and sufficient to induce their self‐renewal. In contrast, the unprocessed SvbREP directs differentiation into ECs, in which it is further required to maintain the differentiated state. svb expression in either ISC/EBs, or ECs, is driven by separate regulatory networks. Results from a large in vivo screen reveal that svb enhancers are directly regulated on the one hand by Wnt and EGFR local signaling for ISC/EB survival and self‐renewal, and, on the other hand, by intrinsic regulatory factor Pdm1 for EC differentiation. Moreover, recent studies show that the systemic steroid hormone ecdysone, which is produced in ovaries (Uryu et al, 2015), increases proliferation and regulates the fate of stem cells in the intestine (Ahmed et al, 2020; Zipper et al, 2020). Our data suggest that these effects of ecdysone are due, at least in part, to the activation of pri expression that triggers, in turn, Svb processing. Together, these results reveal the dual role of OvoL/Shavenbaby in stemness versus differentiation and provide a first molecular frame to explain how local and systemic regulatory signals, in coordination with intrinsic cues, are integrated within the adult stem cell lineage.
Results
Svb is required to maintain adult intestinal progenitors
svb expression is driven by a large array of enhancers, which collectively define at single‐cell resolution the pattern of epidermal differentiation in the embryo (Sucena et al, 2003; McGregor et al, 2007; Frankel et al, 2011; Preger‐Ben Noon et al, 2016). To monitor svb expression in the adult midgut epithelium, we tested the activity of main svb enhancers. While one svb enhancer (7) was active in terminally differentiated cells (see below), we found that the E enhancer (Fig 1A) drives specific expression in esg + progenitors (Fig 1B), i.e., in stem cells (ISCs) and enteroblasts (EBs). Dissection of the E enhancer (5kb) delineated two separate elements called E3N (292 bp) and E6 (1kb) that each drives similar expression in intestinal progenitors (Figs 1C and EV1A).
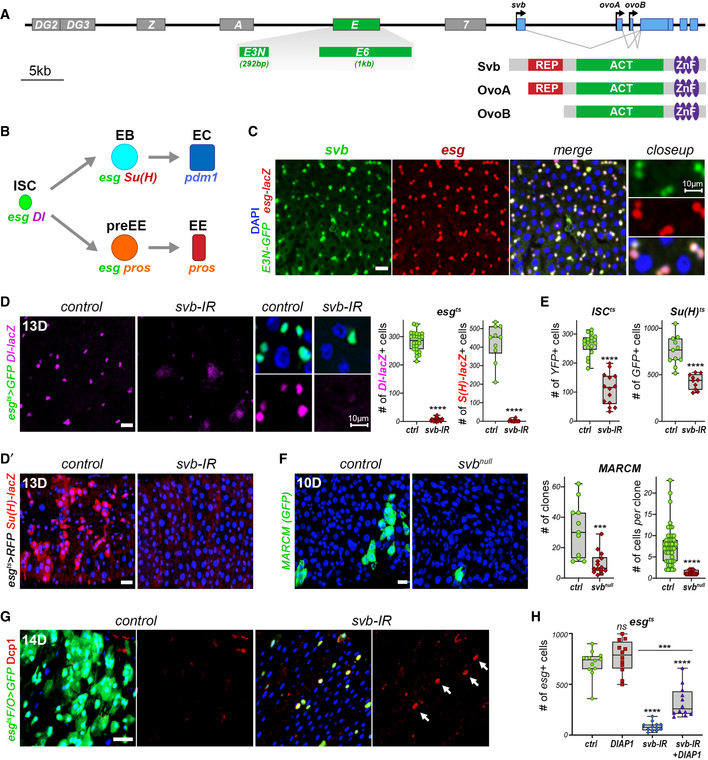
Figure 1. svb is expressed in ISC/EBs and is required for their maintenance.
-
ASchematic representation of the svb locus, showing location of enhancers as well as functional organization of somatic (Svb) and germline (OvoA, OvoB) protein isoforms. Red and green boxes represent the repressor and activator domains, respectively; purple ovals depict the DNA‐binding zinc fingers.
-
BThe adult intestinal stem cell lineage, with markers of stem cells (Dl), enteroblasts (Su(H)) and enterocytes (Pdm1). Esg is expressed in progenitor cells gathering stem cells (ISC), enteroblasts (EB), and pre‐enteroendocrines (preEE). Both pre‐EEs and mature enteroendocrine cells (EE) express Prospero (pros).
-
CPosterior midgut showing expression of the E3N svb enhancer (GFP, green) in ISC/EBs, as shown by co‐staining with esg‐lacZ (β‐Gal, red).
-
D, D′Staining for Dl‐lacZ (purple) or Su(H)‐lacZ (red) in esgts midguts expressing GFP alone (control), or expressing svb‐RNAi. Samples were stained for GFP (green) and β‐Gal. Close‐ups show separate channels for GFP and β‐Gal. The graphs show quantification of the number of ICS (Dl‐positive) and EBs (Su(H)‐positive).
-
EQuantification of the number of YFP‐positive cells (left) and GFP‐positive cells (right) in control and upon expression of svb‐RNAi driven by ISCts and Su(H)ts, respectively.
-
FPosterior midguts containing control or svbR9 MARCM clones (GFP, green), and quantification of the number of clones, and of the average number of cells per clone.
-
GesgtsF/O midguts expressing GFP alone (control) or expressing svb‐RNAi. Samples were stained for GFP (green) and the apoptotic marker cleaved Dcp1 (red). Arrows highlight GFP‐positive cells that are also positive for Dcp1.
-
HQuantification of GFP‐positive cells per posterior midgut in esgts expressing GFP alone (ctrl), or expressing DIAP1, svb‐RNAi, and svb‐RNAi+ DIAP1.
Data information: Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the horizontal line in each box is plotted at the median; data were collected from three independent replicates. P values from Mann–Whitney tests (D,E,F) and one‐way ANOVA (H) are ns > 0.05, * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001.
DAPI is blue, scale bars, 20 µm, except in close‐ups (10 µm).
Source data are available online for this figure.
Figure EV1. svb is required in ISCs/EBs and svb loss does not affect differentiation.

- Expression of E6 svb enhancer in the posterior midgut, as monitored by X‐Gal staining of E6‐LacZ reporter line.
- ISCts midguts expressing YFP alone (control), or expressing svb‐RNAi. Samples were stained for YFP (yellow) and DAPI (blue); the quantification is shown in Fig 1E.
- Su(H)ts midguts expressing GFP alone (control), or expressing svb‐RNAi. Samples were stained for GFP (green), Prospero (red), and DAPI (blue). The graph shows quantification of the number of Prospero‐positive cells (EEs). See also Fig 1E.
- Voilats midguts expressing GFP alone (control), or expressing svb‐RNAi. Samples were stained for GFP (green), Prospero (red), and DAPI (blue); lower panels show separate channels. The graph displays quantification of the average number of Prospero‐positive cells (EEs) in control conditions, or upon svb‐RNAi treatment.
- ActtsF/O midguts expressing GFP alone (control), or expressing svb‐RNAi. Samples were stained for GFP (green) and DAPI (blue).
- Schematic representation of the ReDDM lineage tracing system (Antonello et al, 2015) in which esgts drives expression of both mCD8::GFP (green) and H2B::RFP (red). esg + cells are labeled by cytoplasmic GFP and nuclear RFP, while cells of their differentiated progeny only maintain the very stable H2B::RFP. Pictures show posterior midguts in control conditions, or upon expression of svb‐RNAi. Samples were stained for GFP (green), RFP (red), and DAPI (blue). For each genotype, merge picture is show at left and the red channel at right. Bottom panels show magnified views; arrow show differentiated cells (RFP‐positive/GFP‐negative). Graphs show quantification of the number of GFP‐positive (precursors) and the percentage of GFP‐negative RFP‐positive (differentiated) cells.
Data information: Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the line in each box is plotted at the median; data were collected from three independent replicates. P values from Mann–Whitney tests are ns > 0.05, **** < 0.0001. Scale bars are 20 µm.
To investigate the function of Svb in adult intestinal stem cells, we used targeted RNAi depletion using conditional and temperature‐sensitive drivers, typically induced in 3‐day‐old mated females. As a first step, we used the esgts driver (Micchelli & Perrimon, 2006) to drive svb knockdown in adult progenitor cells (esg+). Knockdown of svb in the esg + population for 2 weeks led to almost complete disappearance of ISCs, as seen by loss of Delta‐lacZ + cells (Fig 1D), as well as loss of EBs marked by Su(H)‐GBE‐lacZ (Fig 1D′). Consistently, svb depletion specifically targeted either in stem cells by the ISCts system (Wang et al, 2014), or in enteroblasts by using Su(H)ts (Zeng et al, 2010), caused the loss of ISCs or EBs, respectively (Figs 1E and EV1B). In contrast, svb knockdown did not affect the enteroendocrine lineage (Fig EV1C and D). Hence, these data show that Svb is specifically required for the maintenance of ISCs and EBs.
The loss of stem/progenitor cells upon svb knockdown could be due to premature differentiation and/or cell death; we then performed a series of genetic experiments to discriminate between these possibilities. The acttsF/O system allowed random knockdown of svb in dividing intestinal cells and their progeny (marked by GFP), leading to a strong decrease in both the number and size of GFP+ clones (Fig EV1E). We next used the mosaic analysis with a repressible cell marker (MARCM) technique (Lee & Luo, 2001) to generate positively marked clones (GFP+) in the midgut epithelium for a null mutation in svb (Delon et al, 2003). Svb‐mutant clones were rare and far smaller than control clones, being often restricted to single cells (Fig 1F). Therefore, the loss of stem cells observed upon svb inactivation was likely resulting from their death, a conclusion we further tested by lineage tracing experiments. We used the repressible dual differential stability markers (ReDDM) approach (Antonello et al, 2015) in which esg + cells express both short (mCD8::GFP) and long (Histone::RFP) half‐lives proteins, the latter persisting in differentiated progeny (GFP negative) for several weeks (Fig EV1F). ReDDM results confirmed that ISC/EBs did not prematurely differentiate upon svb knockdown, since the loss of progenitors (GFP+/RFP+) was not paralleled by an increased number of differentiated cells (GFP−/RFP+). We also generated clones of intestinal cells using the esgtsF/O system (Jiang et al, 2009), which marks both ISC/EBs and their descendant progeny by GFP, and stained for the apoptotic marker cleaved‐Dcp1. Two weeks after induction, large GFP+ clones and only rare apoptotic cells were observed in control midguts. In contrast, svb knockdown led to sparse GFP+ cells, often positive for Dcp1, thus demonstrating that progenitors lacking svb underwent apoptosis (Fig 1G). Accordingly, expression of the apoptosis inhibitor DIAP1 was sufficient to significantly rescue the ISC/EB population following svb knockdown (Fig 1H).
Hence, loss of svb leads to a loss of stem/progenitor cell population, demonstrating that Svb is required for their maintenance and protection from apoptosis.
The Pri/Ubr3/proteasome axis controls Svb function in stem cells
Svb is translated as a large (1,354 aa) repressor (SvbREP) that is processed into a shorter (910 aa) transcriptional activator (SvbACT) (Kondo et al, 2010). This switch is gated by Pri peptides that bind to and activate the E3 ubiquitin ligase Ubr3, triggering Ubr3 binding to Svb (Zanet et al, 2015). The Ubr3/UbcD6 complex then ubiquitinates Svb, inducing in turn Svb processing via limited proteasome degradation of its N‐term repressor domain (see Fig 2C). Originally identified in the epidermis (Kondo et al, 2010; Chanut‐Delalande et al, 2014), there is growing evidence that Pri‐dependent processing underlies Svb function in other somatic tissues (Pueyo & Couso, 2011; Ray et al, 2019), including in adult stem cells (Bohere et al, 2018).
Figure 2. Pri/proteasome processing of Svb is required for ISC/EB maintenance.

- Schematic representation of the pri locus, with tested enhancers and location of the pri‐Gal4 gene trap insertion.
- Expression of priA, priJ, or priH enhancers in the posterior midgut as seen by lacZ reporters (X‐Gal staining, blue), and pri‐Gal4 gene trap expressing GFP (green).
- Schematic representation of Svb maturation by proteasome processing, which is triggered by EcR‐mediated expression of pri.
- esgts midguts expressing GFP alone (control), or expressing pri‐RNAi, and quantification of GFP‐positive cells (green). The graph also shows the number of YFP‐positive cells in ISCts midguts expressing YFP alone (ctrl), or pri‐RNAi (see Fig EV2A).
- ISCts midguts expressing YFP alone (control), or expressing two non‐overlapping EcR‐RNAi, and quantification of the number of YFP‐positive cells (yellow). The graph also plots the number of GFP‐positive cells in esgts midgut expressing GFP alone (ctrl), or expressing EcR‐RNAi#1, EcR‐RNAi#2, and mcherry‐RNAi as an additional negative control (see Fig EV2B).
- esgts midguts expressing GFP alone (control), or expressing EcR‐DN, and EcR‐DN+ pri. Samples were stained for GFP (green). The graph shows quantification of the number of GFP‐positive cells in the different genotypes.
- Posterior midguts containing control and Ubr3 null MARCM clones (GFP, green).
- esgts midguts expressing GFP alone (control), or expressing Ubr3‐RNAi, and Ubr3‐RNAi+ OvoB. Samples were stained for GFP (green). (H') quantification of GFP‐positive cells from H.
Data information: Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the horizontal line in each box is plotted at the median; data were collected from three independent replicates. P values from Mann–Whitney tests (D) and one‐way ANOVA (E,F,H′) are: ns:> 0.5, * < 0.05, ** < 0.01, **** < 0.0001. Blue is DAPI, scale bars, 20 µm.
To investigate whether Svb processing regulated stem cell fate, we first examined pri expression in the adult midgut. Profiling of reporter lines covering the entire pri locus (Chanut‐Delalande et al, 2014) showed that three pri enhancers (priA, priH, and priJ) were active in ISC/EBs (Fig 2A and B). We also monitored a Gal4 gene trap within pri gene that faithfully reflects the pattern of pri in many tissues (Galindo et al, 2007). This experiment confirmed pri expression in ISC/EBs, and not in large polyploid ECs (Fig 2B). Since pri was specifically expressed in stem/progenitor cells, we investigated its putative function by targeted knockdown. Upon 2 days of pri‐RNAi induction in esg + progenitors, the majority of GFP+ cells had disappeared from the midgut (Fig 2D). We also observed an acute loss of stem cells when pri‐RNAi was driven using ISCts (Figs 2D and EV2A). Hence, loss of pri leads to a loss of stem/progenitor cells, demonstrating that, like svb, pri is required for their maintenance.
Figure EV2. Pri and Ubr3 are required for the maintenance of progenitor cells.
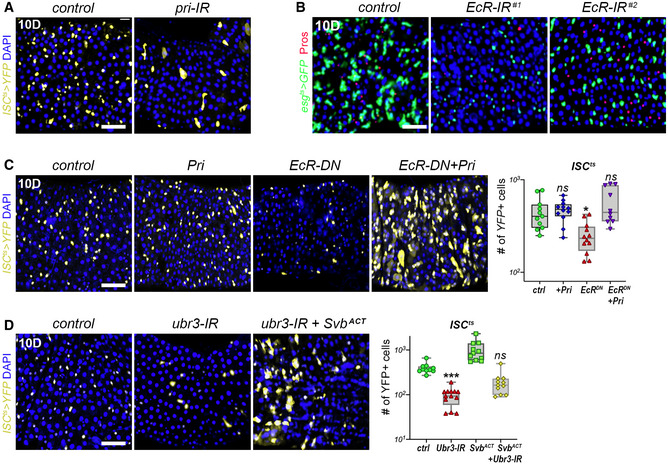
- ISCts midguts expressing YFP alone (control), or expressing pri‐RNAi. Samples were stained for YFP (yellow) and DAPI (blue); quantification is shown in Fig 1E.
- esgts midguts expressing GFP alone (control), or expressing two RNAi lines that target non‐overlapping regions of the Ecdysone receptor (EcR) mRNA. Samples were stained for GFP (green), Prospero (red), and DAPI (blue); quantification is shown in Fig 2E.
- ISCts midguts expressing YFP alone (control), or expressing UAS‐pri, EcR‐DN, and EcR‐DN+ pri. Samples were stained for YFP (yellow) and DAPI (blue). The graph shows quantification of the number of YFP‐positive cells for each genotype.
- ISCts midguts expressing YFP alone (control), or expressing Ubr3‐RNAi, and Ubr3‐RNAi+ OvoB. Samples were stained for YFP (yellow) and DAPI (blue). The graph shows quantification of the number of YFP‐positive cells for each genotype.
Data information: Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the line in each box is plotted at the median; data were collected from three independent replicates. P values from one‐way ANOVA are: ns > 0.05; * < 0.05, *** < 0.001. Graphs are drawn with a log (10) Y scale. Scale bars are 20 µm.
Throughout development, ecdysone signaling times pri expression through direct activation by EcR (Chanut‐Delalande et al, 2014). We reasoned that if this hormonal control of pri expression was occurring in the adult midgut, cell‐autonomous disruption of the ecdysone pathway should affect the behavior of ISCs. Consistent with this prediction, EcR knockdown—using two non‐overlapping RNAi driven by ISCts or esgts—led to a loss of ISCs and EBs (Figs 2E and EV2B). Similar results were obtained when driving EcR‐DN, a dominant negative form of the receptor, confirming that ecdysone signaling is required within ISC/EBs (Figs 2F and EV2C). Furthermore, expression of pri was able to rescue the loss of ISC/EBs caused by EcR‐DN (Figs 2F and EV2C). These data indicate that ecdysone signaling is required for ISC homeostasis and that pri is a main target of EcR within adult stem cells.
Our observations supported a model in which Pri peptides act in ISC/EBs to trigger Ubr3‐mediated processing of Svb. To test this model, we generated MARCM clones of intestinal cell homozygous mutant for a null allele of Ubr3 (Zanet et al, 2015). As observed for svb mutants, clones lacking Ubr3 were very rare and consisted of only a few cells (Fig 2G). Knockdown of Ubr3 in progenitors (esgts > Ubr3‐RNAi), or specifically in stem cells (ISCts > Ubr3‐RNAi), also strongly decreased their number (Figs 2H and H′ and EV2D). If this loss of stem/progenitor cells was due to impaired Svb processing, then expression of a constitutive activator form of Svb (Andrews et al, 2000; Kondo et al, 2010) should suppress this phenotype. As predicted, co‐expression of constitutive SvbACT with Ubr3 RNAi significantly restored the pool of ISCs and EBs (Figs 2H and EV2D). Similar results were obtained by expressing both SvbACT and EcR‐DN with the esgts driver, showing that SvbACT is also able to override pri downregulation (Fig 2F).
Taken together, these results demonstrate that ecdysone signaling is required within intestinal stem cells, in which it promotes the expression of pri. They further show that a main function of Pri and Ubr3 in the adult intestine is to trigger Svb maturation in order to maintain the pool of intestinal stem cells.
Svb activator promotes stem cell renewal and sustains stemness
Having shown that SvbACT is required to maintain stem/progenitor cells, we then asked whether elevated SvbACT activity could be sufficient to trigger stem cell hyperplasia in homeostatic conditions.
We used different means to increase SvbACT levels within ISC/EBs, i.e., expression of the constitutively active form (Kondo et al, 2010), co‐expression of Svb and Pri, or expression of a construct engineered to express the precise protein form normally resulting from Svb maturation (Ray et al, 2019). In all cases, we observed very similar results, with a strong increase in stem/progenitor population (Figs 3A and EV3A and B). For the sake of simplicity, the term SvbACT will be used in the following to collectively refer to these conditions. Examination of ISCs marked with Dl‐lacZ showed that stem cells reached up to fourfold the normal population upon 2 weeks of esgts‐driven SvbACT expression (Fig 3A). Similar increase in ISC population was also overserved when SvbACT was specifically targeted in ISCs (Fig EV3C). The expansion of stem cells resulted from over‐proliferation, as seen by increased number of mitotic cells marked by phosphorylated‐histone3 (PH3), while svb knockdown conversely reduced the mitotic index (Fig 3B). Taken together, these results thus demonstrate that high SvbACT is sufficient to trigger stem cell hyperproliferation.
Figure 3. Svb activator induces stem cell proliferation.

- esgts midguts expressing GFP alone (control), or expressing OvoB, and carrying a Dl‐lacZ transgene that marks ISCs. Samples were stained for GFP (green) and β‐Gal (red); the graph shows quantification of β‐Gal‐positive cells (ISCs).
- Quantification of number of mitotic PH3‐positive cells/midgut in esgts guts expressing GFP alone (control), or expressing OvoB and svb‐RNAi; y‐axis is drawn as log(10).
- esgts midguts expressing GFP alone (control) or expressing SvbACT. Samples were stained for GFP (green), and β‐catenin (purple), DE‐Cadherin (white) or Scribble (yellow). Top and bottom pictures show separate channels for a same region.
- esgts midguts expressing GFP alone (control), or expressing miR8, OvoB, and mir8+ OvoB. Samples were stained for GFP (green) and β‐catenin (purple). The graph shows quantification of GFP‐positive cells for each genotype. The y‐axis is drawn using a log(10) scale.
- esgts midguts expressing GFP alone (control), or expressing Notch Intra Cellular Domain (NICD), and NICD+ OvoB. In close‐ups (right), DAPI is shown in purple for improved contrast; the arrow highlights a cell with intermediate phenotype.
Data information: Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the line in each box is plotted at the median; data were collected from three independent replicates. P values from Mann–Whitney tests (A) and one‐way ANOVA (B,D) are: * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001. Blue is DAPI. Scale bars are 20 µm.
Figure EV3. Svb acts as a transcriptional activator in ISC/EB cells.

-
A, A′esgts midguts expressing GFP alone (control), or expressing OvoB, and SvbREP+ pri. Samples were stained for GFP (green) and DAPI (blue). The graph (A′) plots the number of GFP‐positive cells in each genotype.
-
Besgts midguts expressing GFP alone (control) or expressing SvbACT. Samples were stained for GFP (green) and DAPI (blue).
-
CISCts midguts expressing YFP alone (control), or expressing OvoB, and SvbREP+ pri. Samples were stained for YFP (yellow) and DAPI (blue). The graph shows quantification of the number of YFP‐positive cells in ISCts midguts expressing YFP alone (control), or expressing OvoB, SvbREP+ pri, and pri.
-
DSnapshot view of ChIPseq signal in embryonic cells (Menoret et al, 2013), showing in vivo binding of Svb on the singed (sn) locus that encodes Fascin. The snE1 enhancer (purple) contains two Svb‐binding sites and is directly activated by SvbACT. Pictures at right show expression in the posterior midgut of wild‐type snE1 (snE1‐wt), and a variant of it that contains mutation of the two Svb‐binding sites (snE1‐Svb‐mt). Samples were stained for β‐Gal activity (cyan blue).
Data information: Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the horizontal line is plotted at the median; data were collected from three independent replicates. P values from one‐way ANOVA are ns > 0.05, ** < 0.01; *** < 0.001; **** < 0.0001. Scale bars are 20 µm.
Epithelial to Mesenchymal Transition (EMT) is a key process for the acquisition of stemness for both normal and cancer stem cells, and OvoL emerge as epithelial stabilizing factors able to counteract EMT (Nieto et al, 2016). Besides overgrowth, we then investigated whether Svb could as well influence epithelial features. ISCs are characterized by prominent basolateral accumulation of β‐catenin (Ohlstein & Spradling, 2006), whereas β‐catenin is restricted to apical cell junctions in differentiated cells. Like wild‐type ISCs, SvbACT cell clusters displayed basolateral accumulation of β‐catenin (Fig 3C). The same was also true for DE‐Cadherin that is a hallmark of epithelial tissues (Nieto et al, 2016). In contrast, Scribble, a tumor suppressor that defines lateral domains, was reduced in both wild‐type ISCs and SvbACT clones (Fig 3C). Previous work has shown that EMT‐inducing factors Esg (Snail in mammals) and ZFh1 (Zeb1,2) are expressed in ISCs and required to maintain stemness and suppress differentiation (Korzelius et al, 2014). As in mammals, miR8 (miR200) downregulates Esg and Zfh1 levels in the fly midgut and miR8 upregulation (esgts > miR8) induces precocious differentiation, resulting to the loss of stem cells (Antonello et al, 2015). We found that SvbACT was sufficient to overcome downregulation of EMT factors (egsts > miR8 + SvbACT), restoring the population of stem cells, i.e., esg‐GFP+ cells with enriched β‐catenin in basolateral domains (Fig 3D). Notch promotes EMT and constitutive activation of Notch signaling in ISCs (esgts > NICD) enforces differentiation resulting in giant cells, with polyploid nuclei. Co‐expression of SvbACT with NICD in ISC/EBs largely suppressed these defects (Fig 3E), restoring esg + cells with normal‐looking morphology and nuclei. Of note, some cells yet displayed intermediate phenotypes (Fig 3E, see close‐ups), reinforcing the conclusion that SvbACT actively counteracts differentiation. Finally, singed that encodes Fascin, an actin‐bundling protein strongly upregulated in epithelial tumors, is a direct target of Svb in epidermal cells (Chanut‐Delalande et al, 2006) and the snE1 enhancer provides readout of SvbACT activity (Menoret et al, 2013). We found that snE1 was specifically expressed in ISC/EBs and mutations of Svb‐binding sites abrogated snE1 activity in the midgut (Fig EV3D). These results thus provide conclusive evidence that Svb behaves as an activator in intestinal stem cells, further suggesting that it regulates the expression of cytoskeleton and cell junction factors, as in embryonic epithelial cells (Chanut‐Delalande et al, 2006; Fernandes et al, 2010; Menoret et al, 2013).
Taken together, our data show that SvbACT is sufficient to induce characteristics of stem cells such as typical cellular architecture and proliferative capability, and to prevent differentiation.
Svb mediates Wnt and EGFR mitogenic pathways in intestinal stem cells
During embryogenesis, Svb mediates the activity of EGFR and Wnt signaling pathways for epidermal differentiation (Payre et al, 1999; Payre, 2004). Since these pathways are key regulators of normal and cancer stem cells (Li & Jasper, 2016; Perochon et al, 2018), we investigated their putative relationship with Svb in intestinal stem cells.
EGFR and Wnt are the main mitogenic pathways in the intestine under homeostatic conditions. Upregulation of EGFR (esgtsF/O > RasV12) or Wnt (esgts > ArmS10) leads to ISC proliferation (Lin et al, 2008) and svb knockdown was sufficient to suppress these phenotypes, resulting in a strongly decreased population of ISC/EBs (Fig 4A and B). Therefore, the mitogenic activity of Wnt and EGFR in stem cells requires svb function. Furthermore, we found that SvbACT was capable to induce ISC hyperproliferation even when these pathways were blocked. Inhibition of EGFR (esgts > EGFR‐DN) or Wnt (esgts > TCF‐DN) induces a marked loss of ISC/EBs (Lin et al, 2008) and, in both cases, the expression of SvbACT rescued these phenotypes, still leading to a twofold‐threefold increase in stem/progenitor population when compared to controls (Fig 4A and B). Hence, Svb is epistatic to, in other words is a downstream effector of, Wnt and EGFR pathways and mediates their activity for stem cell maintenance and self‐renewal.
Figure 4. Svb acts downstream of Wnt and EGFR mitogenic signaling pathways in the adult midgut.

- esgtsF/O midguts expressing GFP alone (control), or expressing RasV12, and RasV12+ svb‐RNAi (top panels). Bottom panels show esgts midguts expressing GFP alone (control), or expressing EGFR‐DN, and EGFR‐DN+ OvoB. Samples were stained for GFP (green). The graph shows quantification of the number of GFP‐positive cells in esgts midguts expressing GFP alone (ctrl), or expressing EGFR‐DN, EGFR‐DN+ OvoB, and EGFR‐DN+ svb‐RNAi. The y‐axis is plotted as log(10).
- esgts midguts expressing GFP alone (control), or expressing ArmS10, ArmS10+ svb‐RNAi, ArmS10+ OvoB, TCF‐DN, and TCF‐DN+ OvoB. Samples were stained for GFP (green). The graph shows quantification of the number of GFP‐positive cells in esgts midguts expressing GFP alone (ctrl), or expressing TCF‐DN, TCF‐DN+ OvoB, and TCF‐DN+ svb‐RNAi. The y‐axis is plotted as log(10).
- The drawing at left schematizes the svb locus, with position of the E3N enhancer. Close‐up shows E3N sequence that correspond to binding sites for Pnt (red) and TCF (green). Right subpanels are pictures of posterior midguts showing expression of wild‐type E3N‐lacZ (E3Nwt), or E3N‐Pnt‐mt, and E3N‐TCF‐mt, as seen from X‐Gal staining (cyan blue).
Data information: Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the line in each box is plotted at the median; data were collected from three independent replicates. P values from one‐way ANOVA are ** < 0.01, **** < 0.0001. In all pictures of (A and B) panels, blue is DAPI. Scale bars are 20 µm.
To get comprehensive insight into the mechanisms linking mitogenic pathways to the control of svb expression, we undertook an in vivo screen to identify transcription factors that regulate svb enhancers (see Materials and Methods). Following the individual inactivation of 220 candidates, the two top list factors were Pointed (Pnt), an ets effector of the EGFR pathway, and TCF, the nuclear effector of Wnt. The E3N sequence contains putative binding sites for both Pnt and TCF (Figs 4C and EV4A), suggesting that they directly activate E3N expression in ISC/EBs. To confirm this, we generated E3N variants‐bearing mutations that inactivate Pnt (E3N‐Pnt‐mt) or TCF (E3N‐TCF‐mt)‐binding sites. Both E3N‐Pnt‐mt and E3N‐TCF‐mt displayed strongly decreased activity (Fig EV4B and C), leading to barely detectable expression in ISC/EBs (Fig 4C). Therefore, the binding of Pnt and TCF appears critical for the function of the E3N enhancer that drives svb transcription in ISC/EBs.
Figure EV4. Identification of transcription factors required for the activity of E3N and 9CJ2 svb enhancers in the embryo.

- Drawing of the E3N svb enhancer, with position of putative binding sites for Pnt (red) and TCF (green) factors, and evolution of DNA sequences across Drosophila species. Nucleotides in red represent point mutations introduced to disrupt either Pnt‐ or TCF‐binding sites.
- Consequences of knocking out Pnt‐ or TCF‐binding sites on expression of the E3N svb enhancer in the embryonic epidermis. Pictures show ventral views of stage‐15 embryos. Scale bar is 50 µm.
- Trichome rescue assays (Crocker et al, 2015) showing the influence of TCF‐binding sites on E3N function. Picture show cuticle preparations of wild‐type and svb‐mutant embryos, focusing on the ventral region of A6 segments. svb mutants display strong reduction in the number of trichomes, remaining ones being highly abnormal. Consistent with its expression pattern, E3N driving svb cDNA (E3N‐wt::svb) rescues formation of the anterior‐most trichome row (arrow). Knocking out TCF‐binding sites (E3N‐TCF‐mt::svb) disrupts rescuing ability of the E3N enhancer. The graph plots the number of trichomes in the anterior‐most row. Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the horizontal line in each box is plotted at the median; data were collected from three independent replicates. P values from one‐way ANOVA are ns > 0.5, **** < 0.0001. Scale bar is 15 µm.
- Drawing of the 9CJ2 svb enhancer, with position of putative binding sites for Pdm‐1 (orange) and evolutionary conservation of the DNA sequence. Nucleotides in red show mutations that have been introduced to disrupt Pdm‐binding sites.
- Consequences of knocking down Pdm‐1‐binding sites on expression of the 9CJ2 svb enhancer in the embryonic epidermis. Pictures show lateral (left), ventral (middle), and dorsal (right) views of stage‐15 embryos. Scale bar are 50 µm.
Data information: Dmel, Drosophila melanogaster; Dsim, Drosophila simulans; Dyac, Drosophila yacuba; Dere, Drosophila erecta; Dfic Drosophila ficusphila; Dtak, Drosophila takahashii; Dana, Drosophila ananassae.
These results support the conclusion that Svb is a direct downstream target of Wnt and EGFR in adult ISC/EBs and integrates local signaling pathways to endorse renewal and stemness of intestinal progenitors.
Svb repressor promotes differentiation into enterocytes
In addition to SvbACT in ISC/EBs, we next explored the putative role of SvbREP and whether Svb was also active in later stages of the intestinal lineage.
In situ hybridization confirmed svb expression in the adult intestine. Basal views of the intestinal epithelium showed svb mRNA accumulating in ISC/EBs, which are seen as small doublet cells apposed to the basement membrane (Fig 5A). Apical views further revealed svb expression in ECs, characterized by their very large polyploid nuclei (Fig 5A). Svb expression in both ISC/EBS and ECs was also confirmed by analysis of a svb::GFP mini‐gene rescue construct (Menoret et al, 2013). The Svb::GFP protein was detected in ISC/EBs and in ECs (Fig 5B–B′), but not in EEs (that are not affected by svb loss of function, see Fig EV1). As aforementioned, svb expression in ECs is driven by a separate enhancer, called 7 (Fig 1A). Within svb enhancer 7, we delineated a minimal region, 9CJ2 (232 bp), that drives specific expression in ECs (Fig 5C). Therefore, svb expression in the intestinal lineage relies on two distinct enhancers: E3N in stem/progenitor cells and 9CJ2 in enterocytes.
Figure 5. Svb repressor promotes enterocyte differentiation.

- Drawing of apical–basal organization of the intestinal epithelium and expression of svb mRNA as revealed by in situ hybridization. The inlet shows an enlarged view.
- esg‐Gal4 midguts expressing mCherry and a svb::GFP rescue mini‐gene, consisting of svb cDNA tagged by GFP (green) and driven by E and 7 svb enhancers (see Fig 1). Samples were stained for GFP (green), mCherry (red), and Prospero (white). (B') shows close‐up views.
- The drawing at left schematizes the svb locus, with position of E3N and 9CJ2 enhancers. Close‐up shows 9CJ2 sequence with binding sites for Pdm1 (orange). The right subpanels are posterior midguts showing expression of wild‐type 9CJ2 svb enhancer, and expression of 9CJ2‐Pdm‐mt in which Pdm1‐binding sites have been mutated.
- esgts midguts expressing GFP alone (control), or expressing SvbREP. Samples were stained for GFP (green) and Prospero (red). Close‐ups correspond to boxed regions, with DAPI shown in purple and GFP‐positive cells outlined in yellow.
- esgtsF/O midguts expressing GFP alone (control), or expressing SvbREP. Samples were stained for GFP (green) and cleaved DCP1 (red). (E') shows quantification of the number of GFP‐positive cells in esgts guts expressing GFP alone (ctrl), or DIAP, SvbREP, and SvbREP+ DIAP.
- ReDDM lineage tracing in control midguts, or in midguts expressing SvbREP, and quantification of the percentage of progenitors (GFP‐positive, RFP‐positive) versus differentiated cells (GFP‐negative, RFP‐positive).
- esgts midguts expressing GFP alone (control), or expressing SvbREP. Samples were stained for GFP (green) and Pdm1 (red); arrows show enlarged GFP‐positive cells which are also positive for Pdm1.
Data information: Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the line in each box is plotted at the median; data were collected from three independent replicates. P values from one‐way ANOVA (E′) and Mann–Whitney tests (F) are: ns > 0.5, **** < 0.0001. Blue is DAPI, scale bars, 20 µm.
The differential expression of svb enhancers implied that they capture different regulatory inputs. We used our in vivo screen to identify factors responsible for 9CJ2 activity and found that Pdm1 (a.k.a. Nubbin) is critical for 9CJ2 function. Interestingly, Pdm1 is a conserved POU factor that is a hallmark of ECs (Jiang et al, 2009; Beebe et al, 2010). There are two putative Pdm1‐binding sites within the 9CJ2 svb enhancer (Fig EV4D), which we inactivated by point mutations (9CJ2‐Pdm‐mt). Knockout of Pdm1 sites disrupted 9CJ2 activity (Figs 5C and EV4E), supporting that svb expression in ECs is under direct control of the enterocyte factor Pdm1.
The switch in Svb transcriptional activity triggered by Pri peptides is associated with a marked change in Svb intranuclear distribution: Whereas SvbACT diffuses within the nucleoplasm, SvbREP accumulates in dense foci (Kondo et al, 2010; Zanet et al, 2015). We found that Svb is diffused in esg + cells that express pri, while displaying foci in ECs, which do not (Fig 5B′). Hence, unlike ISCs that rely on SvbACT, later stages of the intestinal lineage were likely to involve SvbREP. To test this, we assayed consequences of expressing SvbREP using the esgts system. The number of ISC/EBs was markedly reduced and remaining esg + cells displayed aberrant morphology (Fig 5D). These esg + cells were larger and their nuclei were significantly bigger than nuclei of wild‐type ISCs (see close‐ups Fig 5D). SvbREP also severely reduced the growth of esgtsF/O clones, which contained individual cells with large nuclei (Fig 5E). These results led us to hypothesize that SvbREP causes stem cell loss through precocious differentiation rather than cell death. Indeed, GFP+ cells of esgtsF/O > SvbREP intestines were negative for Dcp1 apoptotic staining (Fig 5E) and DIAP1 overexpression did not suppress SvbREP phenotypes (Fig 5E′). These data ruled out stem cell apoptosis and lineage tracing fully supported the notion that SvbREP induces massive differentiation. When SvbREP was expressed in esg + cells using the ReDDM system, the loss of ISC/EBs was accompanied by a strong increase in their differentiated progeny (Fig 5F). We also observed that enlarged SvbREP cells that still express low levels of esg‐GFP became positive for Pdm1 (Fig 5G), indicating that they engaged precocious differentiation. Thus, SvbREP is sufficient to trigger a loss of stem cell identity and results in the initiation of EC differentiation.
A main determinant of ISC differentiation is the activation of Notch. We thus assayed whether the differentiation potential of SvbREP relied on Notch and/or other regulatory pathways of intestinal stem cells. Inhibition of Notch (esgts > Notch‐RNAi) induces dramatic tumor‐like expansion of ISCs (Ohlstein & Spradling, 2007). Strikingly, co‐expression of SvbREP was sufficient to suppress Notch‐deficient tumors and enforce differentiation, as manifested by enlarged GFP+ cells with big nuclei (Fig 6A). SvbREP also suppressed ISC‐derived tumors resulting from the inactivation of JAK/STAT (Fig 6B), which also regulates differentiation of the intestinal lineage (Buchon et al, 2009; Jiang et al, 2009). Finally, SvbREP was able to suppress stem cell hyperplasia triggered by Wnt overactivation (Fig 6C). These results well illustrate that SvbREP forces tumor cells to differentiate, as seen by prominent changes in morphology and increased nuclear size. Of note, these phenotypes were strikingly different from those observed for svb loss of function (Fig 6C), which prevents stem cell overgrowth but cannot impose differentiation. Hence, SvbREP acts as a potent tumor suppressor, sufficient to impose differentiation and prevent stem cell proliferation triggered by altered signaling.
Figure 6. SvbREP suppresses stem cell tumors in the gut epithelium.

- esgts midguts expressing GFP alone (control), or expressing Notch‐RNAi, Notch‐RNAi+ svb‐RNAi, and Notch‐RNAi+ SvbREP. Samples were stained for GFP (green) and Prospero (red). The graph shows quantification of the number of GFP‐positive cells.
- esgts midguts expressing GFP alone (control), or expressing STAT‐RNAi, STAT‐RNAi+ svb‐RNAi and STAT‐RNAi+ SvbREP. Samples were stained for GFP (green) and Prospero (red). The graph shows quantification of the number of GFP‐positive cells.
- esgts midguts expressing GFP alone (control), or expressing Wg, Wg+ OvoB, Wg+ SvbREP, and Wg+ svb‐RNAi. In close‐up views, nuclei are in purple.
Data information: Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the line in each box is plotted at the median, data were collected from three independent replicates. P values from one‐way ANOVA are: ns P > 0.5, **** < 0.0001. Graphs are drawn using a log(10) y‐axis scale. DAPI is blue; scale bars, 20 µm.
In sum, svb enhancers directly integrate different regulatory inputs to drive specific expression either in stem cells or in enterocytes. While SvbACT promotes stemness and proliferation, these results demonstrate that SvbREP drives enterocyte differentiation, in both normal and tumorous contexts.
SvbREP is required to maintain enterocyte differentiation, and SvbACT triggers hallmarks of dedifferentiation
We further investigated the role of Svb isoforms in differentiated ECs. To avoid indirect consequences linked to expression in stem/progenitor cells, we used the temperature‐sensitive driver MyoIAts (Jiang et al, 2009). MyoIA encodes a gut‐specific myosin that is a component of the apical brush border and found only in differentiated enterocytes.
Knocking down svb in ECs (MyoIAts > svb‐RNAi) led to a gross alteration of the midgut, with a thinner epithelium and enlarged lumen (Fig 7A). The lack of svb also impaired EC differentiation, as MyoIA‐GFP expression was decreased (Fig 7B). Elevated Dcp1 levels were suggestive of increased apoptosis (Fig EV5A), as also supported by ultra‐structural analyses showing pyknotic nuclei and defective cell contacts (Fig 7C). As observed for ISC/EBs, svb also prevents apoptosis of mature ECs, in which svb function is further required to maintain differentiation.
Figure 7. Svb repressor is required to maintain enterocyte differentiation.

- Control MyoIAts midguts, and MyoIAts > svb‐RNAi or MyoIAts > OvoB midguts. Cyan dye stains the lumen.
- MyoIAts midguts expressing GFP alone (control), or expressing svb‐RNAi, and SvbACT. Samples were stained for GFP (green).
- Electron micrographs of MyoIAts control midguts, or expressing svb‐RNAi and SvbACT. Brush border microvilli are pseudo‐colored in green, and high magnification views are shown in inlets. Nuclei are pseudo‐colored in purple, and visceral muscles located above the basement membrane are in pink. Arrowheads point to impaired cell contacts, the arrow points to a pyknotic nucleus.
- Cross sections of control MyoIAts midguts (expressing GFP and mCherry‐RNAi), or expressing svb‐RNAi, SvbACT, and Pri. Samples were stained for F‐actin (white), GFP (green) and Tsp2a (yellow).
Data information: Blue is DAPI. Scale bars are 20 µm (A, B, D) and 5 µm in (C).
Figure EV5. Svb acts as a transcriptional repressor in differentiated enterocytes.
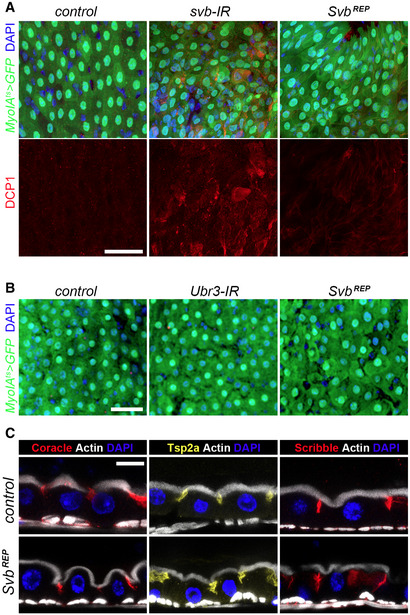
- Myo1Ats midguts expressing GFP alone (control), or expressing svb‐RNAi and SvbREP. Samples were stained for GFP (green) and DAPI (blue). Lower panel shows staining for cleaved DCP1 (red). Scale bar is 20 µm.
- Myo1Ats midguts expressing GFP alone (control), or expressing Ubr3‐RNAi and SvbREP. Samples were stained for GFP (green) and DAPI (blue). Scale bar is 20 µm.
- Cross sections of control MyoIAts midguts (expressing GFP and mCherry‐RNAi, top row), or SvbREP (bottom row). Samples were stained for F‐actin (white), DAPI (blue), and Coracle (red), Tsp2a (yellow) or Scribble (red). Scale bar is 5 µm.
The pattern of Svb::GFP intranuclear distribution suggested that Svb was acting as a repressor in ECs (see Fig 5). We tested this hypothesis through a series of complementary experiments. While MyoIAts > SvbREP intestines showed no detectable homeostatic or structural changes (Fig EV5A–C), forced expression of SvbACT in ECs had dramatic effects on the midgut, with abnormal multilayered intestinal epithelium and a reduced lumen (Fig 7A and D). It also caused loss of MyoIA‐GFP, indicating deeply compromised differentiation (Fig 7B), as also manifested by disruption of brush border microvilli (Fig 7C). The dramatic phenotypes observed upon 2 weeks of induction prompted us to use shorter treatments (6 days). Even in these milder conditions, SvbACT disrupted the intestinal epithelium, with multilayered cells displaying reduced apical actin and altered organization, as highlighted by staining for Tsp2a or Coracle (Fig 7D). The conclusion that svb function in ECs relies on the unprocessed SvbREP raised specific predictions, which we assayed directly. First, unlike in stem cells, Svb activity in ECs should not depend on factors that operate its processing into the activator, i.e., it should be insensitive to the loss of Ubr3. Accordingly, we did not detect defects upon Ubr3 knockdown in ECs, intestines exhibiting proper levels of GFP and organization (Fig EV5B). Second, since pri is normally absent from ECs (Fig 2), forced expression of pri should trigger processing of endogenous SvbREP into SvbACT. Indeed, MyoIAts‐driven expression of pri in ECs induced defects resembling those seen with SvbACT, albeit of weaker severity (Fig 7D). Hence, these data demonstrate that the repressor form of Svb is required to maintain differentiation of ECs.
We then investigated in more details the phenotypes caused by SvbACT in enterocytes, which were stronger than the loss of svb. As seen in stem cells (Fig 3), expression of SvbACT in ECs led to remodeling of the epithelial architecture, featured by basolateral accumulation of β‐Catenin, and decreased Scribble in lateral domains (Fig 8A). Close inspection revealed that large polyploid EC‐like cells with reduced or undetectable GFP levels remained in the gut following induction of SvbACT in ECs. Some cells displayed extreme phenotypes, with massive accumulation of β‐catenin and withdrawal of Scribble (Fig 8A′). SvbACT also induced over‐proliferation, with high increase in the number of PH3+ intestinal cells (Fig 8B). These mitotic cells were likely ISCs, since damaged or dying ECs produce short‐range signals, such as Upd1‐3 cytokines, which foster regenerative proliferation of neighbor stem cells (Buchon et al, 2009; Jiang et al, 2009). However, we observed some PH3+ cells that also express Myo1Ats‐GFP (Fig 8C), suggesting that SvbACT can force late EBs or ECs to reenter the cell cycle.
Figure 8. Ectopic Svb processing disrupts enterocyte differentiation.

- MyoIAts midguts expressing GFP alone (control), or expressing SvbACT. Samples were stained for GFP (green), Scribble (yellow), β‐catenin (purple), and DAPI (Blue). (A') pictures display cross sections of the regions shown in (A).
- MyoIAts midguts expressing GFP alone (control), or expressing SvbACT. Samples were stained for GFP (green) and PH3 (red). The graph plots number of mitotic PH3‐positive cells per midgut of MyoIAts guts expressing GFP alone (ctrl), or expressing SvbACT, and SvbREP+ pri.
- MyoIAts midguts expressing GFP and SvbACT (green), and stained for GFP (green) and PH3 (red). The picture is a single focal plane; the arrow shows a large GFP‐positive cell, which is also positive for mitotic PH3.
- Summary of the role of SvbACT and SvbREP in the control of intestinal stem cell maintenance, proliferation, and differentiation.
Data information: Boxes extend from the 25th to 75th percentiles, whiskers from min to max, the horizontal line in each box is plotted at the median; data were collected from three independent replicates. P values from one‐way ANOVA are: ** <0.01, **** <0.0001. Scale bars are 20 µm.
Therefore, our data show the importance of SvbREP to trigger and maintain enterocyte differentiation. Furthermore, the proper regulation of Svb processing is crucial, since ectopic production of SvbACT in ECs induces loss of differentiation markers and gain of features normally seen in stem cells.
Discussion
Our data show that the OvoL/Shavenbaby transcription factor is a key integrator of intrinsic, local, and systemic cues to control the behavior of adult intestinal stem cells and of their progeny. In stem cells, Svb is processed into the activator form (SvbACT) that mediates EGFR and Wnt activities for stem cell self‐renewal. Pdm1 then drives svb expression in later stages of the lineage, during which Svb behaves as a repressor (SvbREP) that direct differentiation into enterocytes. The balance between SvbACT and SvbREP is gated by Pri peptides, which allow conversion of Svb transcriptional activity in response to systemic ecdysone signaling. These results show the pivotal role of Svb in balancing stem cell renewal/proliferation versus differentiation, and further suggest that OvoL factors are evolutionarily conserved determinants of stemness.
svb integrates multiple regulatory cues for the homeostasis of adult stem cells
Numerous studies have demonstrated the role of Wnt and EGFR signaling pathways in somatic stem cells and cancers (Normanno et al, 2006; Zhan et al, 2017). In the Drosophila intestine, EGFR pathway acts in an autocrine/paracrine manner to promote homeostatic stem cell self‐renewal (Jiang & Edgar, 2009; Biteau & Jasper, 2011; Li & Jasper, 2016), whereas Wnt signals are mainly produced by visceral muscles that act as a niche (Perochon et al, 2018) (see Fig 8D). We find that ovo/svb is a common target of Wnt and EGFR in adult stem cells that mediates their activity to promote stem cell self‐renewal. Our data further indicate that the nuclear mediators of Wnt (TCF) and EGFR (Pointed) activate svb expression in stem cells, through direct regulation of an enhancer (E3N) driving ISC/EB‐specific expression. Although the precise register of Wnt activity in the midgut remains to be confirmed (Perochon et al, 2018), the main mitogenic pathway EGFR is high in ISC/EBs and strongly reduced in ECs (Jiang & Edgar, 2009; Jin et al, 2015), explaining specific expression of E3N in stem and progenitor cells (Fig 8D).
A separate regulatory module consisting of the 9CJ2 svb enhancer, under direct control of the POU transcription factor Pdm1 (Fig 8D), maintains svb expression in later stages of the lineage. Pdm1 is highly expressed in differentiated enterocytes (Jiang et al, 2009; Beebe et al, 2010) and is a main marker of mature ECs (Li & Jasper, 2016). Since there is evidence for mutual antagonism between Pdm1 and Escargot (Korzelius et al, 2014; Tang et al, 2018), Escargot might repress Pdm1 expression in stem/progenitor cells and, thereby, would restrict activity of the 9CJ2 svb enhancer to enterocytes.
E3N and 9CJ2 enhancers also drive svb expression in the embryo, supporting the notion that they harbor pleiotropic functions across the life‐cycle (Preger‐Ben Noon et al, 2018). Both svb enhancers capture additional regulatory cues during development, including Hox proteins and Exd/Hth cofactors (Crocker et al, 2015), as well as GATA and LIM homeodomain factors in the case of E3N (Preger‐Ben Noon et al, 2016), opening the possibility that these factors could as well regulate intestinal stem cells in the adult. svb expression in the adult midgut also involves the E6 svb enhancer, which like E3N is active in ISC/EBs. Apparently redundant svb enhancers may ensure robustness of intestinal homeostasis in the face of genetic or environmental variations, as shown for epidermal development (Frankel et al, 2010). Together, our data provide mechanistic information on how the svb cis‐regulatory landscape integrates multiple cues to drive stage‐specific expression in the intestinal stem cell lineage (Fig 8D).
SvbACT: a key factor for stemness and stem cell renewal
In addition to the control of svb expression, activity of the Svb factor is tightly regulated by post‐translational modification, which relies on proteasome‐mediated processing (Kondo et al, 2010; Zanet et al, 2015). We show that Svb processing into an activator is indispensable to maintain and prevent differentiation of intestinal stem cells, which otherwise undergo apoptosis, as recently reported for renal nephric stem cells (Bohere et al, 2018). Although renal stem cells are mostly quiescent (Bohere et al, 2018; Xu et al, 2018), intestinal stem cells self‐renew under homeostatic conditions and proliferate in response to various challenges (Li & Jasper, 2016). This high plasticity of the intestinal lineage further reveals that SvbACT is both required and sufficient to promote stem cell proliferation; high SvbACT in ISCs leading to hyperplasic overgrowth (Fig 8D). Supernumerary cells induced by SvbACT display typical features of stem cells, including redistribution of cell junction and apical‐basal polarity complexes. These data support a model in which SvbACT might be an intrinsic determinant of stemness. Furthermore, forced expression of SvbACT strongly alters ECs, which lose differentiation and in some cases engage mitosis. Similar epithelial dysplasia progressively appears when the gut experiences aging (Biteau et al, 2008; Biteau et al, 2010). Future studies will determine whether Svb mis‐regulation is involved in aging, and/or if SvbACT is capable to induce dedifferentiation.
SvbREP triggers enterocyte differentiation
Our results show the function of Shavenbaby in enterocytes, but not in enteroendocrine cells, consistent with an early separation between EC and EE lineages (Biteau & Jasper, 2014; Guo & Ohlstein, 2015; Zeng & Hou, 2015). In contrast to stem cells, Svb acts as a repressor within ECs in which it is required for their maintenance and differentiation. Ectopic processing of Svb in ECs disrupts epithelial organization, leading to multilayered cells that lose features of mature ECs, including brush border microvilli, as well as properly organized cell–cell junctions. Junctional complexes are progressively established during EB to EC maturation and they are essential for differentiation. For instance, the septate junction component Tsp2A is required for downregulation in ECs of Hippo and JAK/STAT signaling, which otherwise promote proliferation (Xu et al, 2019). Likewise, SvbREP promotes EC differentiation and is also a potent inhibitor of stem cell proliferation. This is the case under homeostatic conditions and, importantly, SvbREP can also suppress hyperproliferation of stem cells induced by altered Notch, STAT, Wnt, or EGFR signaling (Fig 8D). Of note, SvbREP enforced tumor cell differentiation, while svb loss of function prevents stem cell growth but does not induce differentiation. Therefore, SvbACT and SvbREP exert antagonistic functions within the adult intestinal lineage, SvbACT promoting stem cell survival and proliferation, while SvbREP later acts to induce and maintain enterocyte differentiation.
Ecdysone function in intestinal stem cells
Throughout development, the maturating processing of Svb is triggered by Pri peptides (Kondo et al, 2010; Chanut‐Delalande et al, 2014; Zanet et al, 2015; Bohere et al, 2018). In the adult intestine, pri is specifically expressed in ISC/EBs, and Pri peptides are required—with their target Ubr3 ubiquitin ligase—for stem cell maintenance. Previous findings have led us to propose that a key role of Pri is to mediate ecdysone signaling to implement systemic hormonal control within gene regulatory networks, as seen for developmental timing of epidermal derivatives (Chanut‐Delalande et al, 2014). Consistent with this view, we show that inactivation of the ecdysone receptor EcR within intestinal stem cells and enteroblasts strongly impacts their behavior, decreasing proliferation and promoting differentiation, i.e., as seen upon inhibition of Svb processing. These results were particularly surprising because ecdysone is not produced in the gut, ovaries being the major source of ecdysone in adult females after mating (Uryu et al, 2015; Ahmed et al, 2020). Thus, they imply the existence of sex‐specific inter‐organ communication that regulates the fate of somatic stem cells, a feature that has never been reported so far, to our best knowledge. Two contemporary studies confirm the role of ecdysone in sustaining stemness and undifferentiated state of Drosophila ISCs in the midgut. Both studies demonstrate that EcR and its cofactor Usp foster division and expansion of ISCs in response to a peak of steroids synthesized in ovaries upon mating (Ahmed et al, 2020; Zipper et al, 2020). These data provide compelling evidence for ovary‐to‐gut communication and show that sex hormones remodel stem cell fate to adjust organ size, as means to face elevated energetic costs imposed by reproduction. Because the expression of pri, or of SvbACT, can overcome EcR inactivation in ISC/EBs, our data suggest that the activation of pri to increase SvbACT levels is a nexus target of steroid action in intestinal stem cells.
OvoL/Svb transcriptional switch for stem cell control across animals
Mounting evidence suggests a wide role of OvoL/Svb factors in progenitor and stem cells across animals. Unlike Drosophila, most insects develop by sequential addition of posterior segments, from a group of embryonic precursors referred to as posterior growth zone. In such species, Svb is specifically expressed in these precursors and required for the formation of posterior structures, together with Pri and Ubr3 (Ray et al, 2019). OvoL factors also display evolutionarily conserved role in germ cell precursors (Hayashi et al, 2017). In flies, the germline‐specific OvoB activator and OvoA repressor are produced from two alternative promoters. OvoB is required for the maintenance of germ cells, while OvoA later acts for their differentiation (Andrews et al, 2000; Hayashi et al, 2017). Precocious expression of OvoA leads to germ line loss (Andrews et al, 2000) and other ovo mutations cause ovarian tumors (Oliver et al, 1993). Although relying on different mechanisms between soma (post‐translational processing) and germline (alternative promoters), the REP‐to‐ACT switch appears as a key feature of Ovo/Svb function in the control of stem/progenitor cells.
In mammals, OvoLs have been implicated in the reprogramming of mesenchymal fibroblasts toward induced pluripotent stem cells (Kagawa et al, 2019) and epithelial lineages (Watanabe et al, 2019). OvoLs are also associated with human cancers, in particular those of epithelial origin that often display deregulated Wnt and EGFR signaling (Normanno et al, 2006; Zhan et al, 2017). Our studies in flies demonstrate opposing effects of SvbACT versus SvbREP that promotes or suppresses stem cell‐derived tumors, respectively. Interestingly, individual OvoL2 isoforms in mice display strikingly different effects when expressed in patient‐derived xenografts, only the OvoL2 repressor can inhibit tumor progression (Watanabe et al, 2014). Therefore, OvoL/Svb repressors appear as evolutionarily conserved tumor suppressors, a finding that might open new paths for cancer diagnostic and treatment.
Several studies have shown that OvoL/Svb factors behave as epithelial gatekeepers (Nieto et al, 2016), which counteract Snail and Zeb1‐2 transcription factors to prevent epithelial to mesenchymal transition (EMT). In agreement with this antagonistic model, Drosophila Escargot (Snail) and ZFh1 (Zeb1,2) maintain stemness and prevent ISC differentiation (Korzelius et al, 2014; Loza‐Coll et al, 2014; Antonello et al, 2015), while SvbREP promotes EC differentiation. However, our results draw a more complex picture, where SvbACT contrariwise cooperate with EMT factors in early stages of the intestinal lineage for the maintenance of ISCs. Indeed, SvbACT can suppress the phenotypes resulting from downregulation of EMT regulators, restoring the pool of stem cells, which display proper cellular architecture. Recent studies show that EMT is not an all‐or‐none process and instead progresses through a series of reversible intermediate states between the epithelial (E) and mesenchymal (M) phenotypes (Nieto et al, 2016). Such hybrid E/M phenotypes are hallmarks of normal and cancer stem cells, and relative doses of EMT factors and OvoL/Svb may provide a tunable window of stemness (Jolly et al, 2015).
Taken together, these data show the importance of OvoL/Shavenbaby factors in the control of adult stem cell behavior, in both normal and tumorous conditions. We propose that OvoL/Shavenbaby epithelial factors are ancestral regulators of stemness in animals and their study would provide key insights into stem cell biology. Future work remains to determine how the intrinsic regulatory hub provided by Svb/Pri for intestinal stem cells in flies has evolved both across species and amid the distinct populations of stem cells that regenerate adult organs.
Materials and Methods
Reagents and Tools table
| Reagent/Resource | Reference or source | Identifier or catalog number |
|---|---|---|
| Experimental Models: D. melanogaster | ||
| esg‐LacZ: y1 w67c23; P{w+mC=lacW}esgk00606/CyO | BDSC | BDSC Cat# 10359, RRID:BDSC_10359 |
| Su(H)GBE‐LacZ | Furriols and Bray (2001) | N/A |
| DL‐LacZ: ry506 P{ry+t7.2=PZ}Dl05151/ TM3, ryRK Sb1 Ser1 | BDSC | BDSC Cat# 11651, RRID:BDSC_11651 |
| UAS‐w‐RNAi: y1 v1; P{y+t7.7 v+t1.8=TRiP.JF01545}attP2 | BDSC | BDSC Cat# 28980RRID:BDSC_28980 |
| UAS‐mCherry‐RNAi y1 sc* v1 sev21; P{y+t7.7 v+t1.8=VALIUM20‐mCherry}attP2 | BDSC | BDSC Cat# 35785 RRID:BDSC_35785 |
| UAS‐svb‐RNA: i w1118; P{GD9026}v41584 | VDRC | Cat# FBst0464178, RRID:FlyBase_FBst0464178 |
| UAS‐Ubr3‐RNAi: w1118; P{GD12698}v22901 | VDRC | Cat# FBst0454736, RRID:FlyBase_FBst0454736 |
| UAS‐Notch‐RNAi: w*; P{w+mC=UAS‐N.dsRNA.P}9G | BDSC | BDSC Cat# 7077, RRID:BDSC_7077 |
| UAS‐STAT92E‐RNAi: y1 sc* v1 sev21; P{y+t7.7 v+t1.8=TRiP.GL00437} attP40/ CyO | BDSC | BDSC Cat# 35600, RRID:BDSC_35600 |
| UAS‐pri‐RNAi: P{UAS‐tal.dsRNA} | Galindo et al (2007) | FBtp0072543 |
| UAS‐pri: P{UAS‐tal}/CyO | Galindo et al (2007) | N/A |
| UAS‐EcR‐DN: w*; P{w+mC=UAS‐EcR.B2.W650A}TP5 | BDSC | BDSC Cat# 9449, RRID:BDSC_9449 |
| UAS‐EcR‐RNAi#1: y1 v1; P{y+t7.7 v+t1.8=TRiP.HMJ22371}attP40 | BDSC | BDSC Cat# 58286, RRID:BDSC_58286 |
| UAS‐EcR‐RNAi#2: y1 v1; P{y+t7.7 v+t1.8=TRiP.JF02538}attP2 | BDSC | BDSC Cat# 29374, RRID:BDSC_29374 |
| UAS‐SvbACT(OvoB): w118;; P{UAS‐ovo.B2} | Payre et al (1999) | FBtp0012383 |
| UAS‐SvbREP w118;; P{UAS‐ovo.svb} | Delon et al (2003) | FBtp0017877 |
| UAS‐SvbACT: Y, w;; P{UAS‐SvbACT::GFP} | Ray et al (2019) | FBtp0134164 |
| UAS‐TCF‐DN: y1 w1118; P{w+mC=UAS‐pan.dTCFDeltaN}4 | BDSC | BDSC Cat# 4784, RRID:BDSC_4784 |
| UAS‐ArmS10: P{w+mC=UAS‐arm.S10}C, y1 w1118 | BDSC | BDSC Cat# 4782, RRID:BDSC_4782 |
| UAS‐EGFR‐DN: y1 w*; P{w+mC=UAS‐Egfr.DN.B}29‐77‐1; P{w+mC=UAS‐Egfr.DN.B}29‐8‐1 | BDSC | BDSC Cat# 5364, RRID:BDSC_5364 |
| UAS‐NICD: P{UAS‐N.icd} | Cooper and Bray (2000) | FBtp0013654 |
| UAS‐DIAP: w*; P{w+mC=UAS‐DIAP1.H}3 | BDSC | BDSC Cat# 6657 |
| UAS‐Wg: w*; P{UAS‐wg.h.t:HA1}6C | BDSC | BDSC Cat# 5918, RRID:BDSC_5918 |
| UAS‐RasV12: w1118; P{w+mC=UAS‐Ras85D.V12}TL1 | BDSC | BDSC Cat# 4847, RRID:BDSC_4847 |
| esgts: esg‐Gal4, UAS‐GFP, tubP‐Gal80ts | Jiang et al (2009) | N/A |
| NREts: Su(H)‐GBE‐Gal4, UAS‐GFP; tubP‐Gal80ts | Zeng et al (2010) | N/A |
| ISCts: esg‐Gal4, UAS‐GFP; Su(H)‐GBE‐GAL80, tubP‐Gal80ts | Wang et al (2014) | N/A |
| esg‐ReDDM: esg‐Gal4, UAS‐mCD8::GFP/Cyo; UAS‐H2B::RFP, tubP‐Gal80ts/TM2 | Antonello et al (2015) | N/A |
| MyoIAts: MyoIA‐Gal4, UAS‐GFP, tubP‐Gal80ts | Jiang et al (2009) | N/A |
| Voilats: tubP‐Gal80ts; Voila‐GAL4, UAS‐GFP.nls | Balakireva et al (1998) | N/A |
| ActtsF/O: hs‐FLP; actin < y+< Gal4; UAS–GFP | Chanut‐Delalande et al (2014) | N/A |
| esgtsF/O: w; esg‐Gal4, UAS‐GFP, tubP‐Gal80ts/CyO; UAS‐FLP, act > CD2>Gal4/TM6B | Jiang et al (2009) | N/A |
| Pri‐Gal4: P{GaWB}talKG/TM3, Sb | Galindo et al (2007) | N/A |
| MARCM‐19A: P{ry+t7.2=hsFLP}1, P{w+mC=tubP‐GAL80}LL1 w*, P{ry+t7.2=neoFRT}19A; P{w+mC=UAS‐mCD8::GFP.L}LL5/ Cyo; P{w+mC=tubP‐GAL4}LL7/TM6B,Tb | N. Tapon | N/A |
| svbR9: y* w1118 svbR9, P{ry+t7.2=neoFRT}19A/FM0 | Delon et al (2003) | FBal0151651 |
| Ubr3B: y1 w* Ubr3B P{ry+t7.2=neoFRT}19A/FM0 | Zanet et al (2015) | FBal0319860 |
| svbE6‐lacZ: w1118; DmE6‐lacZ | Frankel et al (2011) | FBtp0085021 |
| PriA‐LacZ | Chanut‐Delalande et al (2014) | N/A |
| PriH‐LacZ | Chanut‐Delalande et al (2014) | N/A |
| PriJ‐LacZ | Chanut‐Delalande et al (2014) | N/A |
| svbE3N‐GFP | This paper | N/A |
| svbE3N‐LacZ | Crocker et al (2015) | N/A |
| svbE3N‐Pnt‐mt‐LacZ | This paper | N/A |
| svbE3N‐TCF‐mt‐LacZ | This paper | N/A |
| svb::GFP: E+7‐svbP‐svb‐cDNA::GFP (pRSQ8) | Menoret et al (2013) | N/A |
| E3N‐svbP‐svb‐cDNA | This paper | N/A |
| E3N‐TFC‐mt‐svbP‐svb‐cDNA | This paper | N/A |
| 9CJ2‐LacZ | This paper | N/A |
| 9CJ2‐Pdm‐mt‐LacZ | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: placZAttB | DGRC | Cat# 1421 |
| Plasmid: pRSQsvb | Frankel et al (2011) | N/A |
| Antibodies | ||
| Mouse monoclonal anti‐GFP (1: 500) | Sigma‐Aldrich | Cat# 11814460001, RRID:AB_390913 |
| Rabbit anti‐GFP (1: 500) | Torrey Pines Biolabs | Cat# TP401 071519, RRID:AB_10013661 |
| Rabbit anti‐β‐galactosidase (1:500) | MP Biomedicals | Cat# 559761, RRID:AB_2687418 |
| Mouse monoclonal anti‐β‐galactosidase (1:1,000) | Promega | Cat# 53781, |
| Rat monoclonal antibody to Red Fluorescent Proteins (1:800) | Chromotek GmbH | Cat# 5f8‐100, RRID:AB_2336064 |
| Cleaved Drosophila Dcp‐1 (Asp216) antibody (1:100) | Cell Signaling Technology | Cat# 9578, RRID:AB_2721060 |
| Mouse anti‐Cora antibody (1:100) | DSHB | Cat# C615.16, RRID:AB_1161644 |
| Mouse anti‐Prospero antibody (1:100) | DSHB | Cat# MR1A, RRID:AB_528440 |
| Mouse anti‐Armadillo (β‐catenin) antibody (1:100) | DSHB | Cat# N2 7A1, RRID:AB_528089 |
| Rat anti‐DE‐Cadherin antibody (1:50) | DSHB | Cat# DCAD2, RRID:AB_528120 |
| Biotinylated goat anti‐rabbit IgG antibody (1:1,000) | Vector Laboratories | Cat# BA‐1000, RRID: AB_2313606 |
| Rabbit polyclonal anti‐Tsp2a antibody (1:1,000) | Izumi et al (2016) | N/A |
| Rabbit polyclonal anti‐Scribble antibody (1:1,000) | Chen et al (2018) | N/A |
| Rabbit polyclonal Anti‐phospho‐Histone H3 (Ser10) (1:1,000) | Millipore | Cat# 06‐570, RRID:AB_310177 |
| Sheep Anti‐Digoxigenin Fab fragments antibody, Alkaline Phosphatase conjugated (1:2,000) | Roche | Cat# 11093274910, RRID:AB_51449 |
| Goat anti‐Rabbit IgG (H+L) Secondary Antibody, AlexaFluor‐488 conjugate (1:500) | Thermo Fisher Scientific | Cat# A‐11034, RRID:AB_2576217 |
| Goat anti mouse IgG (H+L) secondary antibody, AlexaFluor‐488 (1:500) | Quantum Dot Corporation | Cat# 1100‐1, RRID:AB_346865 |
| Goat anti‐rabbit IgG (H+L) secondary antibody, AlexaFluor‐555 (1:500) | Molecular Probes | Cat# A‐21428, RRID:AB_141784 |
| Goat anti‐mouse IgG (H+L) secondary antibody, AlexaFluor‐555 (1:500) | Molecular Probes | Cat# A‐21422, RRID:AB_141822 |
| Goat anti‐rat IgG (H+L) secondary antibody, AlexaFluor‐555 (1:500) | Molecular Probes | Cat# A‐21434, RRID:AB_141733 |
| Oligonucleotides | ||
| Primer: Fwd_9CJ2: CGGTACCCCGCGGCCGCCATATGTCAACG | This paper | N/A |
| Primer: Rev_9CJ2: TCCGGCGCTCCTCGAGACTATTGGGATACC | This paper | N/A |
| Primer: Fwd_E3‐14: CGGTACCCCGCGGCCGCCATATGTCTTTTTTTTTATCC | This paper | N/A |
| Primer: Rev_E3‐14: CCGGCGCTCCTCGAGGTAGGTTAGG | This paper | N/A |
| Chemicals, enzymes and other reagents | ||
| Sucrose, BioXtra, >=99.5% (GC) | Sigma‐Aldrich | Cat# 57‐50‐1 |
| DAPI (4′,6‐diamidino‐2‐phenylindole) | Thermo Fisher Scientific | Cat# D1306 |
| X‐Gal (5‐bromo‐4‐chloro‐3‐indoyl‐β‐D‐Galactopyranoside) | Biosolve | Cat# 7240‐90‐6 |
| NBT/BCIP (C40H30Cl2N10O6 / C8H6NO4BrClP x C7H9N) | Sigma Aldrich | Cat# 11681451001 |
| 16% Paraformaldehyde, methanol free | Electron microscopy Sciences | Cat# 30525‐89‐4 |
| Formaldehyde | Electron microscopy Sciences | Cat# 50‐00‐0 |
| Blocking Reagent for nucleic acid hybridization and detection | Roche | Cat# 11096176001 |
| Phalloidin conjugated to Rhodamin (1:500) | Thermo Fisher Scientific | Cat# R415, RRID:AB_2572408 |
| VECTASHIELD Mounting Medium antibody | Vector Laboratories | Cat# H‐1000, RRID:AB_2336789 |
| VECTASHIELD Mounting Medium with DAPI antibody | Vector Laboratories | Cat# H‐1200, RRID:AB_2336790 |
| XhoI | New England Biolabs | Cat# R0146L |
| NotI | New England Biolabs | Cat# R0189L |
| Phusion High‐Fidelity PCR Master Mix with HF Buffer | Thermo Fisher Scientific | Cat# F531L |
| Phusion High‐Fidelity DNA Polymerase | Thermo Fisher Scientific | Cat# F530L |
| Software | ||
| ImageJ 1.52a | https://imagej.net/ | RRID:SCR_003070 |
| Fiji | http://fiji.sc | RRID:SCR_002285 |
| Prism 8 | GraphPad | RRID:SCR_002798 |
| Photoshop CC | Adobe | RRID:SCR_014199 |
| FlyBase | http://flybase.org/ | RRID:SCR_006549 |
| Clustal Omega | http://www.ebi.ac.uk/Tools/msa/clustalo/ | RRID:SCR_001591 |
| MUSCLE | http://www.ebi.ac.uk/Tools/msa/muscle/ | RRID:SCR_011812 |
| JASPAR | http://jaspar.genereg.net | RRID:SCR_003030 |
| Clone Manager Software | http://www.scied.com/pr_cmbas.htm | RRID:SCR_014521 |
| ZEN Digital Imaging for Light Microscopy | http://www.zeiss.com/microscopy/en_us/products/microscope‐software/zen.html | RRID:SCR_013672 |
| Leica Application Suite | https://www.nikoninstruments.com/Products/Software | RRID:SCR_016555 |
| NIS‐Elements | https://www.nikoninstruments.com/Products/Software | RRID:SCR_014329 |
| Other | ||
| VECTASTAIN ABC‐Peroxidase Kit | Vector Laboratories | Cat# PK‐4001, RRID:AB_2336810 |
| Qiaquick PCR Purification kit | Qiagen | Cat# 28104 |
| QIAmp DNA Micro Kit | Qiagen | Cat# 56304 |
| In‐Fusion® HD Cloning Plus | Takara | Cat# 638920 |
| DIG RNA Labeling Kit (SP6/T7) | ROCHE | Cat# 11 175 025 910 |
Methods and Protocols
Animal breeding and maintenance
Flies were kept at 25°C and grown on a standard cornmeal food medium (per liter: 17 g inactivated yeast powder, 80 g corn flour, 9 g agar, 45 g white sugar, and 17 ml of Moldex). Crosses involving targeted expression under the control of Gal4/Gal80ts were maintained at 18°C until 3–4 days post‐hatching, and mated females were shifted to 29°C for 10–14 days for optimal activity of the UAS/GAL4 system. Flies were transferred to fresh food vials daily. For flip‐out (F/O) and MARCM clonal analyses, 3‐ to 4‐day mated adult female flies of the indicated genotypes were heat shocked 1 h at 37°C and then shifted to 25°C for 10 days. The genotype of each Drosophila sample is detailed in the Appendix.
In vivo screening of transcription factors
To avoid indirect effects due to alteration of cell survival/proliferation, the screen was performed in late embryos, when signaling pathways and Svb do not impinge on cell survival and proliferation, as opposed to adult stem cells. Briefly, we knocked down every candidate factor and examined whether it affected the activity of individual svb enhancers. We selected transcription factors showing detectable expression in stage‐15 whole embryos (Menoret et al, 2013) and/or enriched in dorsal trichome cells (Preger‐Ben Noon et al, 2016), resulting in a list of 227 candidate factors. 273 representative UAS‐RNAi lines were obtained from Bloomington and VDRC stock centers, taken from the TRIP or VDRC collection, respectively. Males from each UAS::RNAi carrying line (Table EV1) were crossed with virgin females of stock w; ptc‐Gal4; E3N‐lacZ or w; ptc‐Gal4; 9CJ2‐lacZ and eggs were collected for 12 h at 28°C. Embryos were dechorionated, fixed, and stained using standard protocols (Fernandes et al, 2010), with mouse anti‐β‐galactosidase 1:500 (Promega) and biotinylated goat anti‐rabbit (1:1,000) antibodies, revealed using VECTASTAIN ABC Peroxidase Kit (Vector Laboratories). After washing, embryos were mounted in Glycerol/PBS (80/20%) and imaged using a Nikon Eclipse 90i microscope using NIS‐elements software (Nikon). Each experiment (typical 200 embryos per genotype) was performed at least three times and also included UAS‐w‐RNAi and w embryos as negative controls. Reporter patterns upon RNAi treatment were classified into “no change”, “reduced”, or “ectopic” expression; the two latter were kept for additional characterization (Table EV1). For rescuing assays, males carrying pRSQsvb constructs were crossed with females of stock w* btd1, svb1/FM7‐kr > GFP, allowing phenotypical identification of svb‐mutant embryos. First instar larva cuticles were prepared in Hoyer’s/lactic acid 1:1, imaged with phase‐contrast microscopy, and trichomes were counted in the ventral region of A6 segments.
DNA constructs and transgenic lines
DNA fragments from svb enhancers was cloned into placZAttB for reporter constructs and into pRSQsvb for rescue constructs (Frankel et al, 2011; Menoret et al, 2013; Crocker et al, 2015). All plasmids were integrated using the PhiC31 system into the same attP landing site (zh‐86F) by Bestgene (Chino Hills, CA, USA) or in the Payre laboratory. Putative binding sites for transcription factors were identified using JASPAR and their evolutionary conservation was assayed using multiple sequence alignments (Clustal Omega and MUSCLE) of orthologous regions from other species (http://flybase.org). Site‐specific mutations were introduced using DNA synthesis by Genscript, and modified enhancers were sub‐cloned in reporter and rescue constructs using ligation‐free cloning (In‐Fusion, Takara). All constructs were verified by sequencing.
Immunofluorescence
Stage‐15 embryos were processed using standard protocols, using mouse anti‐β‐galactosidase (1:500, Promega), rabbit anti‐Dyl at 1:400 (Fernandes et al, 2010), rabbit anti‐Min 1:200 (Chanut‐Delalande et al, 2006) antibodies, Alexa Fluor‐488, or Alexa Fluor‐555 secondary antibodies at 1:500 (Molecular Probes). Embryos were mounted in Vectashield mounting media (Vector Laboratories) and imaged using X20 and X40 objectives on a Leica Spe confocal laser scanning microscope with Leica Application Suite software, or a Zeiss 710 confocal microscope using the ZEN software (Zeiss).
Adult midguts were dissected in PBS and fixed for 1h at room temperature in a fresh 4% paraformaldehyde solution (Electron microscopy Science) prepared in PBS. Following three washes of 15 min each in 0.1% Triton‐PBS (PBST), samples were blocked in 1% BSA‐PBST for 30 min at room temperature, prior to overnight incubation with primary antibody at 4°C. The following antibodies were used: mouse and rabbit anti‐GFP at 1:500, rabbit anti‐β‐Galactosidase (MP Biomedicals) at 1:1,000, rat anti‐RFP (5F8) at 1:800, cleaved Dcp‐1 (Asp216) rabbit antibody at 1:100 (Cell signaling), mouse anti‐Prospero (DSHB) at 1:100, rabbit anti‐Phospho‐Histone 3 (Millipore) at 1:1,000, mouse anti‐Coracle at 1:100 (DSHB), mouse anti‐β‐catenin 1:100 (DSHB), rat anti‐DE‐Cadherin 1:50 (DSHB), rabbit anti‐Pdm1 1:100 (gift from F.J. Díaz‐Benjumea), rabbit anti‐Tsp2A, and anti‐Scribble (gifts from D. St Johnston) at 1:1,000 both. F‐Actin was stained with Rhodamine‐conjugated phalloidin (Invitrogen, at 1:500). The next day, samples were washed 3 times in PBST for 15 min each and next incubated with Alexa Fluor‐488 or Alexa Fluor‐555 secondary antibodies at 1:500 (Molecular Probes) for 2 h at room temperature. After three washes, tissues were mounted in Vectashield mounting media containing DAPI (Vector Laboratories) for nuclear staining. Images of posterior midgut were acquired on Leica SPE and SP8 confocal microscopes (X40 objective). 3 to 5 images were acquired for each posterior midgut to cover the 4a to 5 regions.
In situ hybridization
RNA probes were synthetized from svb cDNA (Delon et al, 2003) using a DIG RNA Labeling Kit (Roche). Guts were dissected in PBS and fixed in freshly prepared 4% formaldehyde 5 mM EGTA fix solution in PBS. After two washes in PBS, guts were dehydrated in successive baths of methanol (5 times), ethanol (5 times), followed by 1 h in 1:1 xylene/ethanol, and rinsed in methanol. Guts were post‐fixed for 30 min at room temperature in the same fix solution and washed in PBS‐0.1% Tween 20 (PBSTw). Samples were treated by proteinase‐K at room temperature, and the reaction was stopped by a 5‐min treatment in 2 mgl/ml glycine, followed by washes in cold PBSTw. Samples were incubated in the fix solution overnight at 4°C and washed in PBST. After 2 hrs. at 60°C in hybridization buffer (HB: 50% formamide, 2X SSC, 1 mg/ml Tortula RNA, 0.05 mg/ml Heparin, 2% blocking reagent (Roche), 0.1% CHAPS, 5 mM EDTA, 0.1% Tween 20), guts were incubated overnight with svb anti‐sense DIG‐labeled RNA probe diluted in HB, at 60°C. After several washes in HB, PBSTw, and PBSTw‐1% BSA (blocking solution), samples were incubated with anti‐DIG primary antibody conjugated to alkaline phosphatase (Roche), at 1:2,000 in blocking solution. After washes in PBSTw, in situ hybridization signals were developed by incubating samples in a fresh staining buffer containing NBT/BCIP stock solution (Sigma Aldrich) diluted at 1:50. Finally, samples were washed and mounted in 50% glycerol/PBS.
X‐Gal staining assays
Adult females were dissected in 1% PBS and guts were fixed in 1% glutaraldehyde‐PBS for 15 min at room temperature. Samples were washed three times for 15 min each. The staining buffer (10 mM Na2HPO4, 1.6 mM NaH2PO4, 150 mM NaCl, 1 mM MgCl2, 3.5 mM K3FeCN6, 3.5 mM K4FeCN6) was warmed up at 37°C for 10 min; an 8% X‐Gal solution (5‐bromo‐4‐chloro‐3‐indoyl‐β‐D‐Galactopyranoside, Sigma Aldrich) was added (final concentration 2.5% X‐Gal in SB) and kept for an additional 10 min at 37°C, before centrifugation (5′ at 12,000 g). Samples were incubated in staining solution overnight at 37°C, washed three times for 15 min with PBS, and mounted in 50% glycerol/PBS. Bright‐field pictures were acquired using a Nikon Eclipse 90i microscope.
Quantification and statistical analysis
Images were analyzed by using ImageJ, with macros we developed for quantification of indicated cell types (codes are available upon request). Briefly, images were acquired with same setting and transformed into multilayered TIFF files. To count the number of cells positive for a given marker (e.g., GFP), the corresponding channel was used to generate a ROI mask in which DAPI‐labeled nuclei were automatically segmented and the number, size, and morphometric features of each object were recorded. Similar analyses were performed for ReDDM assays, quantifying the number of nuclei in GFP+/RFP+ progenitors versus GFP−/RFP+ differentiated cells. Data of at least three independent experiments were combined. Statistical analyses were carried out with Prism 8 (GraphPad), using nonparametric unpaired two‐tailed Mann–Whitney tests for comparison of two samples, and one‐way ANOVA for three or more samples, using Welch’s ANOVA with Dunnet’s T3 correction for multiple comparisons between samples showing normal distribution (Shapiro–Wilk tests alpha = 0.01) and nonparametric Kruskal–Wallis tests with Dunn’s correction for multiple comparison tests when at least one sample did not passed normality test. In each figure, graphs show all individual points, boxes extend from the 25th to 75th percentiles, whiskers to min and max values, and the horizontal line in each box is plotted at the median. Images were processed and figures drawn using Adobe Photoshop CC.
Author contributions
Project setup: DO, SP, and FP; Work conceptualization and supervision: FP and DO; Experiment design: SAH, FP, and DO; Experiments on intestinal stem cells: SAH, DO, JPB, JB, and CI; in vivo screening of upstream TFs, transgenic enhancer constructs and genetic rescuing assays: AA; Expertise with microscopy and macros development for automated image analyses: BR; Key resource sharing and materials: ZK and BL; Manuscript writing: SAH, DO and FP. Manuscript edition and comments: All authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
Source Data for Figure 1
Acknowledgements
We would like to thank C. Polesello, all members of the FP laboratory and G. Kolahgar for invaluable help, as well as T. Reiff, S. Ahmed, and B. Edgar for kind sharing of unpublished results. We are also grateful to M. Bou Sleiman and Z. Zhai for discussions and critical reading of the manuscript. We thank P. Valenti for excellent assistance in the early stages of this work, O. Bohner, J. Favier, and A. Destenabes for help in transgenic lines, S. Bosch and the LITC platform (https://www‐litc.biotoul.fr/) for imaging. We thank A. Bardin, H. Bellen, D. Bilder, J. Chen, J.P. Couso, A. Debec, F.J. Díaz‐Benjumea, M. Dominguez, B. Edgar, N. Frankel, D. Stern, D. St Johnston, and N. Tapon for providing fly stocks and reagents, the Bloomington Drosophila Stock Center, Kyoto Drosophila Genomics and Genetic Resources, Vienna Drosophila Resource Center, Flybase and Developmental Studies Hybridoma Bank for key data and materials. SAH was supported by Fondation pour la Recherche Médicale and Communauté des Communes de Dannieh‐Université Libanaise. This work was supported by Agence Universitaire de la Francophonie (PCSI AUF‐BMO), École Doctorale des Sciences et de Technologie‐Université Libanaise, the Federation of European Biochemical Societies, American University of Beirut URB, Agence Nationale de la Recherche (ANR, ChronoNet), and Fondation pour la Recherche Médicale (DEQ20170336739). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The EMBO Journal (2021) 40: e104347.
Contributor Information
François Payre, Email: francois.payre@univ-tlse3.fr.
Dani Osman, Email: dani.osman@ul.edu.lb.
References
- Ahmed SMH, Maldera JA, Krunic D, Paiva‐Silva GO, Penalva C, Teleman AA, Edgar BA (2020) Fitness trade‐offs incurred by ovary‐to‐gut steroid signalling in Drosophila . Nature 584: 415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews J, Garcia‐Estefania D, Delon I, Lu J, Mevel‐Ninio M, Spierer A, Payre F, Pauli D, Oliver B (2000) OVO transcription factors function antagonistically in the Drosophila female germline. Development 127: 881–892 [DOI] [PubMed] [Google Scholar]
- Antonello ZA, Reiff T, Ballesta‐Illan E, Dominguez M (2015) Robust intestinal homeostasis relies on cellular plasticity in enteroblasts mediated by miR‐8‐Escargot switch. EMBO J 34: 2025–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakireva M, Stocker RF, Gendre N, Ferveur JF (1998) Voila, a new Drosophila courtship variant that affects the nervous system: behavioral, neural, and genetic characterization. J Neurosci 18: 4335–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Perdigoto CN, Southall TD, Brand AH, Schweisguth F (2010) Transcriptional control of stem cell maintenance in the Drosophila intestine. Development 137: 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe K, Lee WC, Micchelli CA (2010) JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev Biol 338: 28–37 [DOI] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H (2008) JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell 3: 442–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Jasper H (2011) EGF signaling regulates the proliferation of intestinal stem cells in Drosophila . Development 138: 1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Jasper H (2014) Slit/Robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila . Cell Rep 7: 1867–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H (2010) Lifespan extension by preserving proliferative homeostasis in Drosophila . PLoS Genet 6: e1001159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohere J, Mancheno‐Ferris A, Al Hayek S, Zanet J, Valenti P, Akino K, Yamabe Y, Inagaki S, Chanut‐Delalande H, Plaza S et al (2018) Shavenbaby and Yorkie mediate Hippo signaling to protect adult stem cells from apoptosis. Nat Commun 9: 5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Chakrabarti S, Lemaitre B (2009) Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila . Genes Dev 23: 2333–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Osman D, David FP, Fang HY, Boquete JP, Deplancke B, Lemaitre B (2013) Morphological and molecular characterization of adult midgut compartmentalization in Drosophila . Cell Rep 3: 1725–1738 [DOI] [PubMed] [Google Scholar]
- Chanut‐Delalande H, Fernandes I, Roch F, Payre F, Plaza S (2006) Shavenbaby couples patterning to epidermal cell shape control. PLoS Biol 4: e290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanut‐Delalande H, Hashimoto Y, Pelissier‐Monier A, Spokony R, Dib A, Kondo T, Bohere J, Niimi K, Latapie Y, Inagaki S et al (2014) Pri peptides are mediators of ecdysone for the temporal control of development. Nat Cell Biol 16: 1035–1044 [DOI] [PubMed] [Google Scholar]
- Chen J, Sayadian AC, Lowe N, Lovegrove HE, St Johnston D (2018) An alternative mode of epithelial polarity in the Drosophila midgut. PLoS Biol 16: e3000041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MT, Bray SJ (2000) R7 photoreceptor specification requires Notch activity. Curr Biol 10: 1507–1510 [DOI] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F et al (2015) Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160: 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AE, Liskova P, Evans CJ, Dudakova L, Noskova L, Pontikos N, Hartmannova H, Hodanova K, Stranecky V, Kozmik Z et al (2016) Autosomal‐dominant corneal endothelial dystrophies CHED1 and PPCD1 are allelic disorders caused by non‐coding mutations in the promoter of OVOL2. Am J Hum Genet 98: 75–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delon I, Chanut‐Delalande H, Payre F (2003) The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila . Mech Dev 120: 747–758 [DOI] [PubMed] [Google Scholar]
- Fernandes I, Chanut‐Delalande H, Ferrer P, Latapie Y, Waltzer L, Affolter M, Payre F, Plaza S (2010) Zona pellucida domain proteins remodel the apical compartment for localized cell shape changes. Dev Cell 18: 64–76 [DOI] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL (2010) Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL (2011) Morphological evolution caused by many subtle‐effect substitutions in regulatory DNA. Nature 474: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Qi L, Chen L, He Y, Zhang N, Guo H (2016) Expression of Ovol2 is related to epithelial characteristics and shows a favorable clinical outcome in hepatocellular carcinoma. Onco Targets Ther 9: 5963–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols M, Bray S (2001) A model Notch response element detects Suppressor of Hairless‐dependent molecular switch. Curr Biol 11: 60‐64 [DOI] [PubMed] [Google Scholar]
- Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5: e106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Ohlstein B (2015) Stem cell regulation. Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency. Science 350: aab0988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haensel D, Sun P, MacLean AL, Ma X, Zhou Y, Stemmler MP, Brabletz S, Berx G, Plikus MV, Nie Q et al (2019) An Ovol2‐Zeb1 transcriptional circuit regulates epithelial directional migration and proliferation. EMBO Rep 20: e46273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Shinozuka Y, Shigenobu S, Sato M, Sugimoto M, Ito S, Abe K, Kobayashi S (2017) Conserved role of Ovo in germline development in mouse and Drosophila . Sci Rep 7: 40056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Motoishi M, Furuse K, Furuse M (2016) A tetraspanin regulates septate junction formation in Drosophila midgut. J Cell Sci 129: 1155–1164 [DOI] [PubMed] [Google Scholar]
- Jiang H, Edgar BA (2009) EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development 136: 483–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA (2011) EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila . Cell Stem Cell 8: 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA (2009) Cytokine/Jak/Stat Signaling Mediates Regeneration and Homeostasis in the Drosophila Midgut. Cell 137: 1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Ha N, Fores M, Xiang J, Glasser C, Maldera J, Jimenez G, Edgar BA (2015) EGFR/Ras signaling controls Drosophila intestinal stem cell proliferation via capicua‐regulated genes. PLoS Genet 11: e1005634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly MK, Jia D, Boareto M, Mani SA, Pienta KJ, Ben‐Jacob E, Levine H (2015) Coupling the modules of EMT and stemness: a tunable “stemness window” model. Oncotarget 6: 25161–25174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa H, Shimamoto R, Kim SI, Oceguera‐Yanez F, Yamamoto T, Schroeder T, Woltjen K (2019) OVOL1 Influences the determination and expansion of iPSC reprogramming intermediates. Stem Cell Rep 12: 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa K, Hikichi T, Nakamura T, Mitsunaga K, Tanaka A, Nakamura M, Yamakawa T, Furukawa S, Takasaka M, Goshima N et al (2016) OVOL2 maintains the transcriptional program of human corneal epithelium by suppressing epithelial‐to‐mesenchymal transition. Cell Rep 15: 1359–1368 [DOI] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y (2010) Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329: 336–339 [DOI] [PubMed] [Google Scholar]
- Korzelius J, Naumann SK, Loza‐Coll MA, Chan JS, Dutta D, Oberheim J, Glasser C, Southall TD, Brand AH, Jones DL et al (2014) Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J 33: 2967–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW (2012) Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell Rep 2: 294–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L (2001) Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24: 251–254 [DOI] [PubMed] [Google Scholar]
- Li H, Jasper H (2016) Gastrointestinal stem cells in health and disease: from flies to humans. Dis Model Mech 9: 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Xu N, Xi R (2008) Paracrine Wingless signalling controls self‐renewal of Drosophila intestinal stem cells. Nature 455: 1119–1123 [DOI] [PubMed] [Google Scholar]
- Loza‐Coll MA, Southall TD, Sandall SL, Brand AH, Jones DL (2014) Regulation of Drosophila intestinal stem cell maintenance and differentiation by the transcription factor Escargot. EMBO J 33: 2983–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL (2007) Morphological evolution through multiple cis‐regulatory mutations at a single gene. Nature 448: 587–590 [DOI] [PubMed] [Google Scholar]
- Menoret D, Santolini M, Fernandes I, Spokony R, Zanet J, Gonzalez I, Latapie Y, Ferrer P, Rouault H, White KP et al (2013) Genome‐wide analyses of Shavenbaby target genes reveals distinct features of enhancer organization. Genome Biol 14: R86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel‐Ninio M, Terracol R, Salles C, Vincent A, Payre F (1995) ovo, a Drosophila gene required for ovarian development, is specifically expressed in the germline and shares most of its coding sequences with shavenbaby, a gene involved in embryo patterning. Mech Dev 49: 83–95 [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N (2006) Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439: 475–479 [DOI] [PubMed] [Google Scholar]
- Nieto MA, Huang RY, Jackson RA, Thiery JP (2016) Emt: 2016. Cell 166: 21–45 [DOI] [PubMed] [Google Scholar]
- Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS (2006) Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366: 2–16 [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A (2006) The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439: 470–474 [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A (2007) Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315: 988–992 [DOI] [PubMed] [Google Scholar]
- Oliver B, Kim YJ, Baker BS (1993) Sex‐lethal, master and slave: a hierarchy of germ‐line sex determination in Drosophila . Development 119: 897–908 [DOI] [PubMed] [Google Scholar]
- Payre F (2004) Genetic control of epidermis differentiation in Drosophila . Int J Dev Biol 48: 207–215 [PubMed] [Google Scholar]
- Payre F, Vincent A, Carreno S (1999) ovo/svb integrates Wingless and DER pathways to control epidermis differentiation. Nature 400: 271–275 [DOI] [PubMed] [Google Scholar]
- Perdigoto CN, Schweisguth F, Bardin AJ (2011) Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development 138: 4585–4595 [DOI] [PubMed] [Google Scholar]
- Perochon J, Carroll LR, Cordero JB (2018) Wnt signalling in intestinal stem cells: lessons from mice and flies. Genes 9: 138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza S, Menschaert G, Payre F (2017) In search of lost small peptides. Annu Rev Cell Dev Biol 33: 391–416 [DOI] [PubMed] [Google Scholar]
- Preger‐Ben Noon E, Davis FP, Stern DL (2016) Evolved Repression Overcomes Enhancer Robustness. Dev Cell 39: 572–584 [DOI] [PubMed] [Google Scholar]
- Preger‐Ben Noon E, Sabaris G, Ortiz DM, Sager J, Liebowitz A, Stern DL, Frankel N (2018) Comprehensive analysis of a cis‐regulatory region reveals pleiotropy in enhancer function. Cell Rep 22: 3021–3031 [DOI] [PubMed] [Google Scholar]
- Pueyo JI, Couso JP (2011) Tarsal‐less peptides control Notch signalling through the Shavenbaby transcription factor. Dev Biol 355: 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Rosenberg MI, Chanut‐Delalande H, Decaras A, Schwertner B, Toubiana W, Auman T, Schnellhammer I, Teuscher M, Valenti P et al (2019) The mlpt/Ubr3/Svb module comprises an ancient developmental switch for embryonic patterning. Elife 8: e39748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca H, Hernandez J, Weidner S, McEachin RC, Fuller D, Sud S, Schumann T, Wilkinson JE, Zaslavsky A, Li H et al (2013) Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS One 8: e76773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelian A, Couso JP (2015) Discovery and characterization of smORF‐encoded bioactive polypeptides. Nat Chem Biol 11: 909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena E, Delon I, Jones I, Payre F, Stern DL (2003) Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424: 935–938 [DOI] [PubMed] [Google Scholar]
- Tang X, Zhao Y, Buchon N, Engstrom Y (2018) The POU/Oct transcription factor nubbin controls the balance of intestinal stem cell maintenance and differentiation by isoform‐specific regulation. Stem Cell Rep 10: 1565–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uryu O, Ameku T, Niwa R (2015) Recent progress in understanding the role of ecdysteroids in adult insects: Germline development and circadian clock in the fruit fly Drosophila melanogaster . Zoological Lett 1: 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Li Z, Hu M, Yang QJ, Yan S, Wu RS, Li BA, Guo M (2017) Ovol2 gene inhibits the Epithelial‐to‐Mesenchymal Transition in lung adenocarcinoma by transcriptionally repressing Twist1. Gene 600: 1–8 [DOI] [PubMed] [Google Scholar]
- Wang L, Zeng X, Ryoo HD, Jasper H (2014) Integration of UPRER and oxidative stress signaling in the control of intestinal stem cell proliferation. PLoS Genet 10: e1004568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Liu Y, Noguchi S, Murray M, Chang JC, Kishima M, Nishimura H, Hashimoto K, Minoda A, Suzuki H (2019) OVOL2 induces mesenchymal‐to‐epithelial transition in fibroblasts and enhances cell‐state reprogramming towards epithelial lineages. Sci Rep 9: 6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Villarreal‐Ponce A, Sun P, Salmans ML, Fallahi M, Andersen B, Dai X (2014) Mammary morphogenesis and regeneration require the inhibition of EMT at terminal end buds by Ovol2 transcriptional repressor. Dev Cell 29: 59–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Tang HW, Hung RJ, Hu Y, Ni X, Housden BE, Perrimon N (2019) The septate junction protein Tsp2A restricts intestinal stem cell activity via endocytic regulation of aPKC and hippo signaling. Cell Rep 26: 670–688 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Liu X, Wang Y, Wong C, Song Y (2018) Temporospatial induction of homeodomain gene cut dictates natural lineage reprogramming. Elife 7: e33934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye GD, Sun GB, Jiao P, Chen C, Liu QF, Huang XL, Zhang R, Cai WY, Li SN, Wu JF et al (2016) OVOL2, an inhibitor of WNT signaling, reduces invasive activities of human and mouse cancer cells and is down‐regulated in human colorectal tumors. Gastroenterology 150: 659–671 [DOI] [PubMed] [Google Scholar]
- Zanet J, Benrabah E, Li T, Pelissier‐Monier A, Chanut‐Delalande H, Ronsin B, Bellen HJ, Payre F, Plaza S (2015) Pri sORF peptides induce selective proteasome‐mediated protein processing. Science 349: 1356–1358 [DOI] [PubMed] [Google Scholar]
- Zeng X, Chauhan C, Hou SX (2010) Characterization of midgut stem cell‐ and enteroblast‐specific Gal4 lines in Drosophila . Genesis 48: 607–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Hou SX (2015) Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development 142: 644–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T, Rindtorff N, Boutros M (2017) Wnt signaling in cancer. Oncogene 36: 1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipper L, Jassmann D, Burgmer S, Gorlich B, Reiff T (2020) Ecdysone steroid hormone remote controls intestinal stem cell fate decisions via the PPARgamma‐homolog Eip75B in Drosophila . Elife 9: e55795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
Source Data for Figure 1


