Nitrate-reducing bacteria (NRB) and sulfate-reducing bacteria (SRB) colonize diverse anoxic environments, including soil subsurface, groundwater, and wastewater. NRB and SRB compete for resources, and their interplay has major implications on the global cycling of nitrogen and sulfur species, with undesirable outcomes in some contexts.
KEYWORDS: cysteine, denitrification, environmental microbiology, hydrogen sulfide, metabolomics, microbial ecology, nitrate-reducing bacteria, systems biology, transcriptomics
ABSTRACT
Competition between nitrate-reducing bacteria (NRB) and sulfate-reducing bacteria (SRB) for resources in anoxic environments is generally thought to be governed largely by thermodynamics. It is now recognized that intermediates of nitrogen and sulfur cycling (e.g., hydrogen sulfide, nitrite, etc.) can also directly impact NRB and SRB activities in freshwater, wastewater, and sediment and therefore may play important roles in competitive interactions. Here, through comparative transcriptomic and metabolomic analyses, we have uncovered mechanisms of hydrogen sulfide- and cysteine-mediated inhibition of nitrate respiratory growth for the NRB Intrasporangium calvum C5. Specifically, the systems analysis predicted that cysteine and hydrogen sulfide inhibit growth of I. calvum C5 by disrupting distinct steps across multiple pathways, including branched-chain amino acid (BCAA) biosynthesis, utilization of specific carbon sources, and cofactor metabolism. We have validated these predictions by demonstrating that complementation with BCAAs and specific carbon sources relieves the growth inhibitory effects of cysteine and hydrogen sulfide. We discuss how these mechanistic insights give new context to the interplay and stratification of NRB and SRB in diverse environments.
IMPORTANCE Nitrate-reducing bacteria (NRB) and sulfate-reducing bacteria (SRB) colonize diverse anoxic environments, including soil subsurface, groundwater, and wastewater. NRB and SRB compete for resources, and their interplay has major implications on the global cycling of nitrogen and sulfur species, with undesirable outcomes in some contexts. For instance, the removal of reactive nitrogen species by NRB is desirable for wastewater treatment, but in agricultural soils, NRB can drive the conversion of nitrates from fertilizers into nitrous oxide, a potent greenhouse gas. Similarly, the hydrogen sulfide produced by SRB can help sequester and immobilize toxic heavy metals but is undesirable in oil wells where competition between SRB and NRB has been exploited to suppress hydrogen sulfide production. By characterizing how reduced sulfur compounds inhibit growth and activity of NRB, we have gained systems-level and mechanistic insight into the interplay of these two important groups of organisms and drivers of their stratification in diverse environments.
INTRODUCTION
Over the past century, the nitrogen cycle has become increasingly perturbed by anthropogenic inputs of fixed nitrogen from synthetic fertilizers, industrial activity, and wastewater. This perturbation has global effects, contributing to increased groundwater contamination, eutrophication, and emissions of the greenhouse gas nitrous oxide (1). In the environment, microbially driven redox reactions are mainly responsible for transforming nitrogen-containing compounds (2). Microbial activities are impacted by factors that include resource concentration, pH, metal availability, and interactions between microorganisms in the environment (3–5). Knowing how these factors differentially impact subsurface microbial nitrogen cycling is essential for developing a predictive understanding of the fate of different nitrogen species in natural and engineered systems. This has been a specific focus at the Oak Ridge Field Research Center (FRC), a Department of Energy legacy nuclear waste site with high levels of heavy metals and radionuclides (6). Nitrate is a major cocontaminant at the site and has been shown to negatively impact the remediation potential of uranium (U) by hindering U(VI) reduction by sulfate-reducing bacteria (SRB) and iron-reducing bacteria (7). Recent work has also shown that groundwater nitrate levels are associated with nitrous oxide production at the FRC and enrichment of denitrification pathway genes (6, 8). Taken together, these observations suggest that dissimilatory nitrate reduction by nitrate-reducing bacteria (NRB) is a major metabolic process in nitrate-contaminated regions of the subsurface.
Both SRB and NRB play a variety of important roles in subsurface environments such as the FRC. In addition to reducing U(VI), SRB can stabilize heavy metals such as lead, zinc, and cadmium through the formation of insoluble metal sulfides, thus providing a means to sustainably sequester and immobilize these metals (9). SRB also play important roles in marine systems, where they mineralize an estimated 29% of the organic matter deposited to the seafloor (10). Meanwhile, in sediments, NRB can facilitate the retention of nitrogen via dissimilatory nitrate reduction to ammonia (DNRA), converting mobile nitrate to ammonium retained by the soil matrix (11). The activity of denitrifiers can also lead to significant losses of soil nitrogen through reduction of nitrate to gaseous forms, including nitrous oxide or dinitrogen gas (12).
Previous studies have shown that intermediates derived from nitrogen and sulfur cycling (e.g., hydrogen sulfide, nitrite, etc.) can impact the activities of SRB and NRB in oxygen-limited environments, such as sediments and the terrestrial subsurface. For example, as a consequence of the higher energy yield of nitrate respiration relative to that of sulfate respiration and NRB production of toxic nitrite, SRB are generally less competitive for carbon when nitrate is available (13). For this reason, nitrate is often used to prevent the souring of oil reservoirs by stimulating NRB activity and depressing SRB activity (14). However, the production of hydrogen sulfide by SRB is also known to impact transformation of different nitrogen species. Heterotrophic denitrification has been shown to be inhibited by hydrogen sulfide in Pseudomonas, and inhibition of two key enzymes in the pathway (nitric oxide and nitrous oxide reductase) was thought to be involved (15). However, there has not been much progress on further characterizing the mechanism of sulfide-induced inhibition of NRB. Prior studies have also demonstrated that hydrogen sulfide can cause NRB to shift from denitrification to DNRA (16). Additionally, inhibiting the activity of ammonia oxidizers indirectly influences both the NRB and SRB by depressing the conversion of ammonia to nitrite (17). Despite much empirical evidence regarding the influences of SRB and NRB on one another, the specific mechanism(s) by which reduced sulfur species act to impact the activity of NRB remains unresolved. Here, we have used a systems approach to characterize the mechanisms by which reduced sulfur compounds modulate the growth and activity of NRB. We have characterized the effects of two naturally occurring reduced sulfur compounds, hydrogen sulfide (here referred to as sulfide) and cysteine, on Intrasporangium calvum C5. Whereas sulfide is encountered as an end product of sulfate respiration by SRB, cysteine is typically used as a reducing agent to achieve anoxic conditions in laboratory growth medium (3). Interestingly, we observed that cysteine significantly lowered the growth rate, growth yield, and denitrification activity of I. calvum C5. Therefore, we sought to understand and contrast the mechanistic underpinnings of cysteine- and sulfide-induced inhibition of denitrification and growth of I. calvum C5, which has the genome-encoded metabolic capabilities to perform both denitrification and DNRA (Fig. 1; see also Fig. S1 in the supplemental material) (3). Also, as an isolate recovered from the FRC, we anticipate its activities are representative of processes occurring at this contaminated field site. We profiled changes in the transcriptome and metabolome at a global level to characterize the physiological response of I. calvum to cysteine and sulfide (18). Cysteine and sulfide had significant and distinct impacts on the physiology and growth characteristics of I. calvum that were correlated with dysregulation (i.e., abnormal changes in the activity) of multiple pathways, including branched-chain amino acid (BCAA) biosynthesis, carbon utilization, and cofactor metabolism. This work expands the scope of a previously established mechanism for growth inhibition of NRB by cysteine and provides new evidence for the mechanism of sulfide inhibition (19).
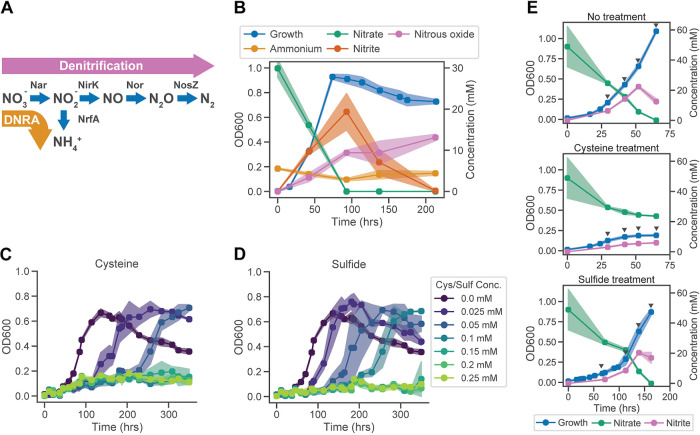
FIG 1.
Nitrate-reducing phenotype of I. calvum and the growth inhibitory effects of cysteine and sulfide. (A) The genome sequence of I. calvum encodes two nitrate reduction pathways: partial denitrification to nitrous oxide and dissimilatory reduction to ammonia (DNRA). (B) Growth characteristics and dynamics of nitrate reduction by I. calvum with 30 mM nitrate and 20 mM lactate. Growth characteristics of I. calvum in the presence of increasing concentrations of cysteine (C) and sulfide (D). (E) Growth characteristics of cultures sampled for transcriptomic and metabolomic profiling. Profiling timelines varied across conditions based on growth characteristics. Cysteine treatment cultures contained 0.25 mM cysteine. Sulfide treatment cultures contained 0.25 mM sulfide. Triangles indicate points at which samples were collected. Samples for transcriptomics were collected as biological triplicates at all four time points, whereas samples for metabolomics were collected as five biological replicates at the first two time points for each condition. Shaded regions in all plots represent standard deviation across biological replicates (n ≥ 3).
Carbon-to-nitrogen ratio determines dissimilatory nitrate reduction end products in continuous flow reactors. Growth and metabolic activity of I. calvum in chemostat reactors with various C:N ratios. Dashed lines indicate the point at which chemostat flow was initiated, with a constant dilution rate of 650 ml/day. Download FIG S1, TIF file, 0.6 MB (567.9KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RESULTS
Phenotypic characterization of I. calvum during nitrate reduction and the impact of cysteine and sulfide on growth.
To characterize growth effects of reduced sulfur compounds, we first investigated the ability of I. calvum to grow anaerobically in the absence of cysteine, which is typically used as a reducing agent in anoxic culture medium (3, 20). Consistent with its genome-encoded capabilities (Fig. 1A), in the absence of cysteine, I. calvum utilized incomplete denitrification to grow on nitrate. Within the first 50 h, ∼50% of the total nitrate in the growth medium was reduced to nitrite, which accumulated transiently, before complete reduction to nitrous oxide after ∼100 h (Fig. 1B). We also found that varying the carbon-to-nitrogen ratio in the medium led to increased DNRA activity when nitrate was limited (see Fig. S1 in the supplemental material). Subsequently, we subjected I. cavum to growth assays in minimal medium with increasing concentrations of cysteine or sulfide. Both compounds inhibited the growth of I. calvum in a dose-dependent manner, with complete growth inhibition at >0.05 mM cysteine and >0.1 mM sulfide (Fig. 1C and D). Notably, the inhibitory effect of cysteine and sulfide was observed only under anoxic conditions, when the primary mode of growth was by nitrate reduction (see Fig. S2B). Furthermore, we confirmed that recovery from inhibition at lower concentrations of cysteine and sulfide was not due to oxygen leakage, suggesting potential physiological adaptation, including plausible activation of a tolerance mechanism(s) (Fig. S2C).
Treatment of sulfide batch effects and the role of oxygen during cysteine and sulfide treatment. (A) Log recovery time differences are predictive of effective sulfide concentration across batches and account for batch effects. Shaded regions represent model uncertainty estimated from propagation of parameter error. (B) Aerobically grown cultures (uncapped tubes shaken at 250 rpm) in minimal medium (20 mM lactate, 0 mM nitrate) with and without the addition of 0.25 mM cysteine or 0.25 mM sulfide and monitored over a period of 40 h. (C) Monitoring of in situ oxygen levels using oxygen sensors attached to the interior of sealed Balch tubes confirm recovery is not due to oxygen leakage. Growth conditions included standard anoxic growth medium as well as supplementation with cysteine (0.05 mM), sulfide (0.025 mM), or aerated medium. Shaded regions depict the standard deviations from biological replicates (n ≥ 3). Download FIG S2, TIF file, 0.7 MB (739.8KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Transcriptome-wide changes induced by cysteine and sulfide treatment.
To elucidate mechanisms of growth inhibition and recovery, we profiled temporal changes in both the transcriptome and metabolome of I. calvum cultured with and without cysteine or sulfide (Fig. 1E). Cysteine revealed a substantial inhibitory effect, with ∼20% of the growth and ∼50% of the nitrate reduced compared to that of the control, while cultures treated with sulfide showed a similar growth and nitrate reduction/nitrite production extent but at a lower rate, requiring ∼160 h compared to ∼70 h in the control culture. Differentially expressed genes (DEGs) across treatments and regular growth conditions were identified using DESeq2-normalized transcriptome sequencing (RNA-seq) data and Boruta (21, 22). Altogether, a total of 362 of 3,687 genes in the I. calvum genome were differentially regulated in response to cysteine or sulfide treatment (see Fig. S3), of which, 162 DEGs were associated with the response to cysteine, with 82 genes downregulated and 80 genes upregulated. A similar number of DEGs (139 in total) were associated with response to sulfide treatment; 90 of these genes were significantly downregulated and 49 were significantly upregulated. Finally, a total of 146 DEGs were different between cysteine and sulfide treatments; of these, 90 were downregulated by cysteine relative to sulfide treatment, and 56 were upregulated. We performed k-means clustering and pathway ontology enrichment analysis to elucidate global patterns in pathway regulation associated with growth inhibition and recovery in response to cysteine and sulfide treatment (Fig. 2). We present the results of this analysis in the context of treatment type, highlighting similarities and differences across responses to cysteine and sulfide.
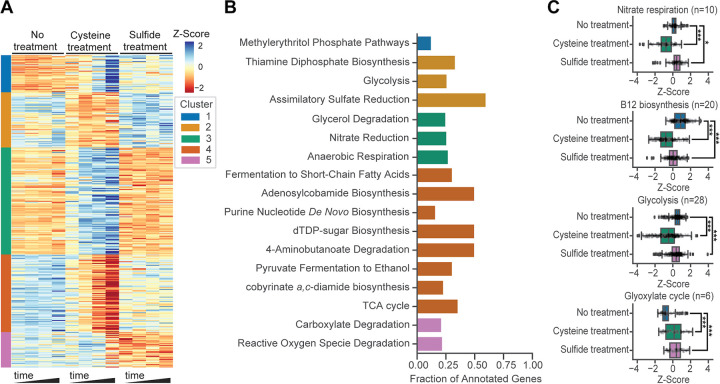
FIG 2.
I. calvum’s transcriptional responses to cysteine and sulfide treatment. (A) Differentially expressed genes (DEGs) clustered into five groups using k-means. Expression levels displayed were normalized using the Z-score. MetaCyc pathway terms enriched in each cluster (B) and average Z-score normalized expression of select pathways and processes (C). Bars indicate comparisons for which differences were significant. *, P < 0.05; ***, P < 0.001. Boxplots display estimates of data minimum (left whisker), median (line within box), maximum (right whisker), and interquartile range (box dimensions) as well as possible outliers (points beyond whiskers). Data are overlaid as scatter points.
Differentially expressed genes identified by Boruta and global expression patterns. (A) Pairwise comparisons of condition-specific normalized gene expression data using Boruta-identified differentially expressed gene sets which best predicted condition labels. For each comparison, the total number of genes is indicated as well as the set of genes unique to that comparison (e.g., not shared across comparisons). The total number of DEGs identified is noted in the center as well as the number of genes shared across all comparisons. (B) Z-score-normalized expression for select pathways identified by enrichment of DEG clusters. Bars indicate comparisons for which differences were significant. *, P < 0.05; ***, P < 0.001. Boxplots display estimates of data minimum (left whisker), median (line within box), maximum (right whisker), and interquartile range (box dimensions) as well as possible outliers (points beyond whiskers). Data are overlaid as scatter points. Global patterns in normalized gene expression data for all genes (C) and differentially expressed genes identified by Boruta (D). Download FIG S3, TIF file, 0.8 MB (807.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(i) Transcriptional response to cysteine treatment.
We discovered that cysteine treatment resulted in the differential regulation of 162 genes within 94 of 294 pathways annotated by MetaCyc, 55 of 108 pathways annotated by SEED, and 117 of 548 GO biological process terms, including those associated with amino acid, fatty acid, and cofactor biosynthesis, transport processes, sulfur and central carbon metabolism, and terminal cytochrome oxidase activity (Fig. 2; Fig. S3). Clustering and pathway enrichment analysis revealed that 28 MetaCyc pathways were significantly enriched in DEG clusters. While the differential regulation of 23 of 34 genes of sulfur metabolism was expected, there were distinct patterns of change that differed across related pathways and treatments. For instance, 19 of 25 total genes in the cysteine and methionine biosynthesis pathways and 1 of 2 predicted cysteine transporters were downregulated in response to cysteine but not sulfide treatment (Fig. S3B). Similarly, 3 of 5 genes involved in the biosynthesis of the glutathione analog mycothiol were also downregulated. In contrast, while the upregulation of sulfur oxidation (3 of 8 genes) in response to cysteine was consistent with concomitant downregulation of sulfate uptake (4 of 6 predicted transporters or uptake facilitators), there was a variable response of genes associated with disulfide bond reduction, with 1 upregulated and 1 downregulated among the 5 predicted to be involved in this process.
The dysregulation of cobalamin (vitamin B12) biosynthesis (17 of 20 genes were downregulated and 1 was upregulated) was consistent with the role of this cofactor in DNA metabolism and fatty acid biosynthesis (Fig. 2C; Fig. S3B), which were also downregulated (13 of 27 genes of fatty acid biosynthesis; 15 of 55 DNA metabolism genes). The upregulation of 7 of 18 genes associated with iron and copper uptake potentially reflects the importance of these metals as enzyme cofactors across metabolic pathways that were differentially regulated and the need to replenish copper(II) and iron(III) reduced or sequestered by cysteine (23, 24). Finally, cysteine treatment resulted in a shift in central carbon metabolism toward utilization of the glyoxylate cycle, based on the downregulation of 5 of 7 genes associated with the conversion of isocitrate to malate in the tricarboxylic acid (TCA) cycle and upregulation of 4 of 6 genes associated with the production of glyoxylate and its subsequent conversions (Fig. 2C; Fig. S3B).
(ii) Transcriptional response to sulfide treatment.
Sulfide treatment resulted in the differential regulation of 139 genes across metal ion transport (19 of 38 genes), RNA metabolism (19 of 63 genes), glycerol uptake and utilization (4 of 6 genes), vitamin B12 biosynthesis (11 of 20 genes), central carbon metabolism (28 of 64 genes with DEGs distributed among the TCA cycle, glycolysis, and pyruvate metabolism), sulfite oxidation (YedY-YedZ), and terminal cytochrome oxidase activity (6 of 11 genes) (Fig. 2C and S3B). Some of these responses triggered by sulfide treatment, such as downregulation of the TCA cycle (15 of 24 genes), were similar to the response to cysteine treatment (Fig. S3B). However, several responses differed between the two treatments, including the absence of differential regulation of cysteine, methionine, and mycothiol biosynthesis genes in response to sulfide treatment. The upregulation of thiol-oxidizing cytochrome oxidases by cysteine and sulfide implicates these genes in relieving oxidative stress resulting from the two treatments. However, the specific cytochrome oxidases upregulated differed across the two treatments, with Ical_3589, Ical_3588, and Ical_3156 (annotated as thioredoxin/protein-disulfide isomerases and a cytochrome c biogenesis protein CcdA) upregulated by cysteine and Ical_0754, Ical_0755, and Ical_0756 (all cytochrome c biogenesis proteins CcdA and CcdB) upregulated by sulfide (Fig. S3B). Similarly, the expression of many transporters that were downregulated by cysteine were less affected by sulfide (e.g., lactate and sulfate). The up- and downregulation patterns of specific metal ion transporters also varied across the two treatments (e.g., cobalt, nickel, zinc, and iron). Ferrous iron transport genes, for instance, were downregulated in the presence of sulfide but not by cysteine treatment, while cobalt and nickel transporters were unaffected by sulfide but were upregulated by cysteine treatment. This was unexpected given the ability of sulfide to sequester these metals and potentially induce stress via their limitation. Finally, the upregulation of glycerol-3-phosphate transporters by sulfide treatment, even though glycerol and glycerol-3-phosphate were not added to the growth medium, suggested that high levels of sulfide might be coupled to availability of these compounds in some niches occupied by I. calvum (Fig. S3B).
Metabolome changes during cysteine and sulfide treatment.
We investigated the physiological consequences of sulfide and cysteine treatment by performing untargeted metabolomics using liquid chromatography-electrospray ionization-quadrupole time of flight mass spectrometry (LC-ESI-QTOF-MS) for samples collected at early phases of growth (Fig. 1E). Clustering of metabolomes into distinct groups by principal-component analysis (PCA) demonstrated high reproducibility across replicate measurements and suggested that treatment conditions had distinct effects on feature abundances (see Fig. S4). The mass spectra were analyzed using XCMS Online to identify mass spectral features that were significantly and reproducibly different in relative abundance of total ion intensities across replicates in each of the two treatments (25, 26). Altogether, the untargeted metabolomics identified a total of 51 mass spectral features which mapped to 60 putative metabolites. Of these, 32 were differentially abundant in cysteine-treated cultures, and 39 were differentially abundant in sulfide-treated cultures. We mapped the putative metabolites to MetaCyc metabolic pathways encoded in the I. calvum genome using XCMS Online and identified 12 pathways, including isoleucine biosynthesis, glycerol degradation, and methionine biosynthesis, that were dysregulated in both treatments. Five pathways, including the glyoxylate cycle and TCA cycle, were dysregulated only in cysteine treatment, whereas 16 pathways, including valine biosynthesis, leucine biosynthesis, and alanine biosynthesis, were dysregulated in just sulfide treatment (Fig. 3A and B).
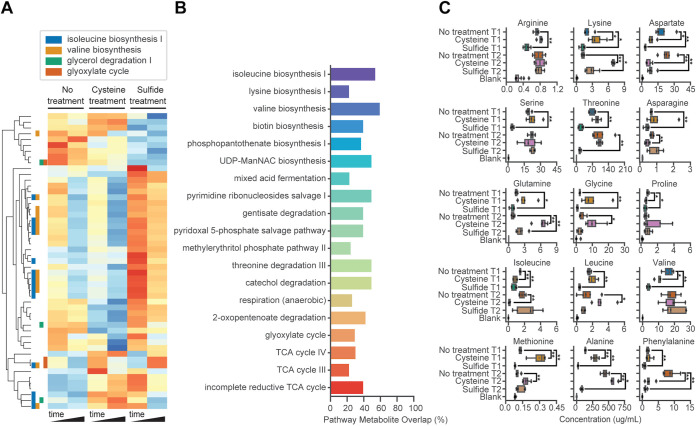
FIG 3.
Metabolome changes during physiological adaptation to cysteine and sulfide treatment. (A) Hierarchically clustered metabolomic feature abundances for those with putative identities scaled using natural logarithm and normalized with Z-score. (B) Pathways identified as significantly dysregulated in cysteine or sulfide treatment relative to no treatment based on enrichment of differentially abundant putative metabolites. (C) Concentrations of amino acids quantified using targeted metabolomics displayed. Bars indicate significance level of select comparisons. *, P < 0.05; **, P < 0.01. Boxplots display estimates of data minimum (left whisker), median (line within box), maximum (right whisker), and interquartile range (box dimensions) as well as possible outliers (points beyond whiskers). Data are overlaid as scatter points.
Differentially abundant metabolomic features and global patterns in feature abundance. (A) Pairwise comparisons of condition-specific feature relative abundances identified differentially abundant features for those with putative identities. For each comparison, the total number of features is indicated as well as the set of features unique to that comparison. The total number of features identified is noted in the center as well as the number of features shared across all comparisons. (B) Relative abundance of select features identified as differentially abundant. Bars indicate comparisons for which differences were significant. *, P < 0.05; ***, P < 0.001. Boxplots display estimates of data minimum (left whisker), median (line within box), maximum (right whisker), and interquartile range (box dimensions) as well as possible outliers (points beyond whiskers). Data are overlaid as scatter points. Global patterns in normalized log feature abundance data for all features (C) and for features with putative identities (D). Download FIG S4, TIF file, 0.6 MB (628.4KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To confirm the finding that amino acid metabolism was dysregulated across both treatments, we quantified the absolute abundance of 15 amino acids using targeted metabolomics and amino acid standards (Fig. 3C). This analysis revealed that with the exception of arginine and threonine, which changed in abundance only in sulfide treatment, the abundance of 9 amino acids, including 2 of 3 BCAAs (valine and isoleucine), was significantly reduced by both treatments but generally more so by sulfide treatment (except isoleucine). Dysregulation of the TCA cycle, glyoxylate cycle, mixed acid fermentation, and anaerobic respiration suggested that cysteine and sulfide treatment also resulted in a potential shift in the carbon metabolism of I. calvum.
Putative mechanisms for growth inhibition and recovery of I. calvum during cysteine and sulfide treatment.
The dysregulated pathways identified by metabolomics were consistent with the differential regulation of enzymes observed at the transcriptional level (e.g., mycothione biosynthesis, amino acid biosynthesis, glyoxylate cycle, and TCA cycle). For instance, consistent with transcriptional downregulation of mycothione synthase, the abundance of mycothione was decreased to an almost undetectable level in early phases of growth for cysteine treatment (see Fig. S5). This low-molecular-weight thiol is known to play a role in mitigating oxidative stress (27). However, it is unclear what role this compound may have played in the response to cysteine and sulfide treatment given the reduced levels of its oxidized form and downregulation of biosynthesis genes.
Differential expression and abundance of the cellular redox regulatory mycothiol. (A) Normalized expression of mycothiol biosynthesis genes identified using gene ontologies with genes listed in order of pathway steps. (B) Relative abundance of metabolite features identified as mycothione (the oxidized disulfide form of mycothiol) and precursor UDP-N-acetyl-α-d-glucosamine at time points T1 and T2 across all conditions. Boxplots display estimates of data minimum (lower whisker), median (line within box), maximum (upper whisker), and interquartile range (box dimensions) as well as possible outliers (points beyond whiskers). Data are overlaid as scatter points. Download FIG S5, TIF file, 0.6 MB (583.6KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
All enzymes of both denitrification and DNRA pathways were progressively downregulated by cysteine through all stages of growth (Fig. 4A). This pattern of downregulation was consistent with reduced nitrate and nitrite reductase activity as well as complete growth inhibition through the entire course of the experiment (Fig. 1E). In contrast, sulfide treatment resulted in transient downregulation of the nitric oxide-producing nitrite reductase (Nirk; Ical_2449) and nitric oxide reductase (Nor; Ical_0054), both of which were impacted during the early phase of growth but later recovered. Meanwhile, both nitrate reductase (Nar; Ical_1176, Ical_1188, and Ical_1210) and the ammonium-producing nitrite reductase (NrfA; Ical_0747 and Ical_0748) were upregulated by sulfide treatment, especially in mid-to-late growth phases, suggesting a shift from denitrification to DNRA (Fig. 4A). These expression patterns were consistent with growth inhibition and recovery patterns as well as changes in nitrate and nitrite levels during sulfide treatment (Fig. 1E).
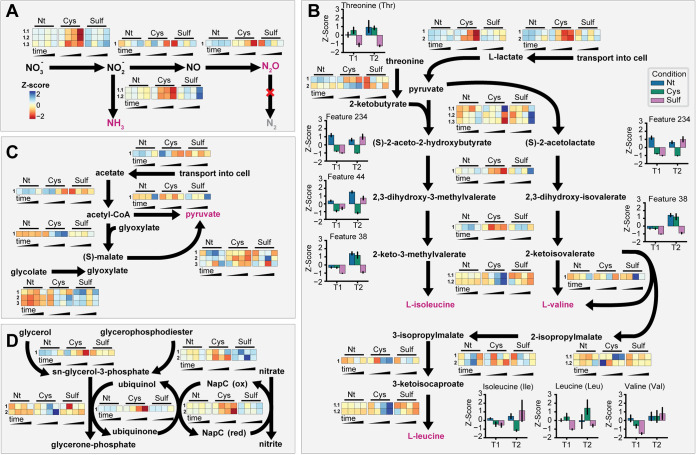
FIG 4.
Integrated analysis of transcriptional and metabolic changes. Expression profiles for each condition are displayed as a heat map for genes predicted to be involved in nitrate reduction (A), branched-chain amino acid biosynthesis (B), acetate utilization (C), and glycerol-3-phosphate coupled nitrate reduction (D). In each panel, the expression level normalized by Z-score is displayed. In each heat map, rows are genes associated with the pathway step (integer numbers note individual enzymes, while decimals indicate subunits), and columns are condition time points. Nt, no treatment control; Cys, cysteine treatment; Sulf, sulfide treatment. Additionally, for specific metabolites for which the absolute concentration was quantified (e.g., amino acids) or for which a metabolomic feature was associated, the Z-score of the metabolite abundance (or relative abundance) is displayed. The metabolomic feature number is indicated for metabolites with putative identification. Error bars represent the standard deviations from biological replicates (n ≥ 3).
The uptake and metabolism of acetate and lactate were repressed in both treatments, but the specific regulation patterns of genes associated with the transport and metabolism of these compounds varied somewhat between treatments. Lactate transport was downregulated more severely in the presence of cysteine than with sulfide treatment, while acetate transport was less affected by cysteine treatment and more so by sulfide treatment (Fig. 4B and C). These observations were consistent with perturbed abundance of intermediates of both the carbon and amino acid metabolism pathways (Fig. 4B and C). Cysteine and sulfide treatments also resulted in the dysregulation of 8 and 3 of 15 genes, respectively, for BCAA biosynthesis (e.g., threonine deaminase; Ical_2218 and Ical_2634) and 2-isopropylmalate synthase (Ical_1041 and Ical_1304) (19). The differential regulation of these pathways was consistent with the altered abundance of intermediates and end products of this pathway (Fig. 4B). For instance, the downregulation of threonine deaminase coincided with the reduced abundance of mass spectral features that matched end products of this enzyme (2-ketobutyrate and 2-iminobutanoate) as well as threonine (Fig. 4B). Additionally, 11 of 20 genes associated with l-valine degradation were upregulated initially (first time point) or later in growth during cysteine treatment (e.g., Ical_1011 and Ical_1143), which may have served as a source of carbon (via production of propanoyl coenzyme A [propanoyl-CoA]) in lieu of repressed carbon uptake (28).
The global transcriptomics and metabolomics analyses (Fig. 2 and 3) also suggested that glycerol degradation was dysregulated during cysteine and sulfide treatment. Exploring this possibility, we identified a pathway coupling glycerol and glycerophosphodiester oxidation to nitrate reduction (Fig. 4D). Supporting the relevance of this pathway, transport of glycerol-3-phosphate and oxidation of glycerophosphodiesters to glycerol-3-phosphate were upregulated in the presence of sulfide (e.g., Ical_0677, Ical_0177, and Ical_0523). Subsequent steps linking oxidation of glycerol-3-phosphate to nitrate reduction were unaffected by sulfide treatment (i.e., neither upregulated or downregulated). Meanwhile, all pathway steps except the oxidation of glycerol-3-phosphate to glycerone-phosphate were downregulated by cysteine treatment (Fig. 4D). Thus, glycerol or glycerophosphodiesters may provide an alternative or complementary means of producing reducing equivalents for nitrate reduction during sulfide-induced stress. In summary, the integrated analysis of changes at the levels of the transcriptome and the metabolome in the context of growth and phenotype characteristics identified a shift in carbon metabolism and the repression of branched-chain amino acid biosynthesis as potential mechanisms of growth inhibition by cysteine and sulfide.
Alternate carbon sources and BCAA supplementation rescue growth inhibition by cysteine and sulfide.
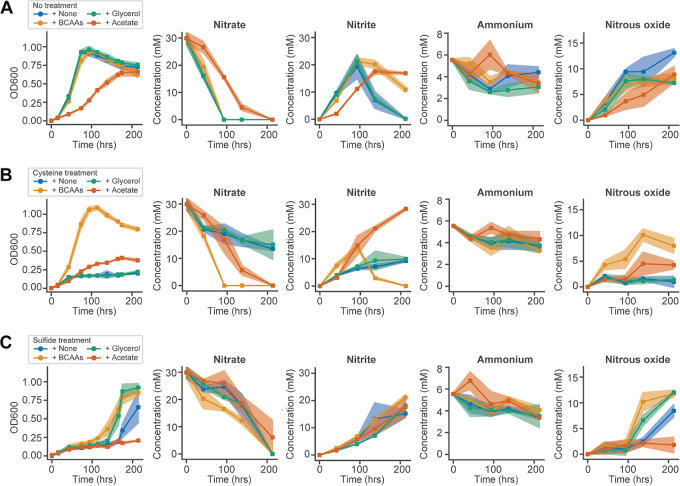
Based on the findings of the systems analysis, we designed experiments to test hypotheses regarding specific mechanisms underlying the growth inhibition effects of cysteine and sulfide. We hypothesized that providing an alternative carbon source or supplementing with BCAAs would rescue the growth-inhibitory effects based on dysregulation of both pathways at the transcriptional and metabolic levels. To test these hypotheses, we assayed growth characteristics and nitrate metabolism of I. calvum in media supplemented with various carbon sources (lactate, acetate, and glycerol) or a mixture of BCAAs (Fig. 5). Supplementation with 1 mM BCAAs (in the presence of lactate) relieved growth inhibition by both reduced sulfur compounds, with complete growth recovery of cultures treated with 0.2 mM cysteine. In the presence of BCAAs, cysteine-treated cultures also readily reduced nitrate, nitrite, and accumulated nitrous oxide. In the absence of supplementation, cysteine treatment impaired these activities, in agreement with previous observations. BCAAs also relieved growth inhibition by 0.2 mM sulfide, contributing to a significant reduction in recovery time (t test P = 4 × 10−4) compared to that of cultures with sulfide alone, which took an additional 60 h to recover on average. Sulfide-treated cultures supplemented with BCAAs also reduced nitrate at a greater rate within the first 100 h of growth than cultures treated with sulfide alone and accumulated greater quantities of nitrous oxide following recovery (Fig. 5).
FIG 5.
Supplementation experiments support the role of pathway dysregulation predicted by transcriptomics and metabolomics. Supplementation conditions consisted of a branched-chain amino acid mixture (Iso, Leu, and Val, each at 1 mM [+BCAAs]) plus 20 mM lactate, 30 mM glycerol (+Glycerol) plus 20 mM lactate, 20 mM lactate alone (+None), or 20 mM acetate alone (+Acetate). The effects of supplementation on growth and denitrification/DNRA for untreated (A), 0.2 mM cysteine treated (B), and 0.2 mM sulfide treated (C). Shaded regions depict the standard deviations from biological replicates (n ≥ 3).
Changing the carbon source from lactate to acetate resulted in slower growth and reduced carrying capacity (i.e., maximum optical density at 600 nm [OD600]). Acetate also mitigated the inhibitory effect of cysteine in the early and mid-growth phases but not in the stationary phase (Fig. 5B). Acetate worsened the growth-inhibitory effect of sulfide. Interestingly, the inhibitory effects of cysteine and sulfide mirrored the patterns of differential regulation of acetate uptake genes during growth with lactate (Fig. 4C). In contrast, glycerol by itself did not support growth of I. calvum (unpublished observation), but in combination with lactate, it reduced the inhibitory effects of sulfide but not cysteine. Sulfide-treated cultures supplemented with glycerol and lactate had significantly decreased recovery times relative to those of sulfide-treated cultures grown with lactate alone (t test P = 0.027), although the effect was less significant than for supplementation with BCAAs. Meanwhile, glycerol supplementation had no detectable effect on the recovery from cysteine inhibition in cultures grown with lactate (no recovery observed over 200 h), which was consistent with expression patterns associated with glycerol utilization (Fig. 4D). Altogether, these experiments corroborated our hypotheses for how dysregulation of carbon metabolism and BCAA biosynthesis mechanistically contribute to the inhibitory effects of cysteine and sulfide on the growth of I. calvum.
DISCUSSION
In this study, we characterized dose-dependent growth inhibition by cysteine and sulfide on a groundwater-associated nitrate reducer, I. calvum. Our systems analysis revealed that a shift in carbon metabolism and the repression of BCAA biosynthesis were two potential mechanisms of growth inhibition by reduced sulfur compounds, which we subsequently tested in a series of supplementation experiments. Growth inhibition by cysteine and sulfide in the presence of lactate was consistent with the downregulation patterns of lactate uptake and metabolism, which were severe in the presence of cysteine but were only observed in early stages of sulfide treatment (Fig. 4B and 5). Acetate, on the other hand, relieved growth inhibition by cysteine, which upregulated acetate uptake and metabolism during early stages of growth. However, acetate failed to relieve growth inhibition during sulfide treatment, likely due to the downregulation of both its uptake and metabolism (Fig. 4C and 5). In contrast, glycerol reduced the severity of growth inhibition by sulfide but not cysteine treatment, consistent with the expression patterns of the associated pathways and modest increase in the rate of nitrate reduction for sulfide-treated cultures supplemented with glycerol (Fig. 4D and 5). Collectively, these data demonstrate that the availability of an appropriate carbon source can play an important role in mediating the growth-inhibitory effects of reduced sulfur compounds and thus potentially also mediate the interplay between NRB and SRB.
Cysteine is known to have cytotoxic effects in various contexts, and studies in Escherichia coli established a mechanistic link between cysteine-induced growth inhibition and repression of BCAA biosynthesis (19, 29–31). While our findings on the mechanism by which cysteine inhibits growth of I. calvum are consistent with previous literature, we have discovered that growth inhibition by sulfide also acts through dysregulation of BCAA biosynthesis. However, our results demonstrated that cysteine and sulfide may act on different enzymatic steps in BCAA biosynthesis (Fig. 4B). Furthermore, our data showed that in addition to BCAA biosynthesis, carbon uptake played an important role in growth inhibition and that cysteine and sulfide treatment had broad and distinct impacts on the regulation of other pathway activities (e.g., TCA cycle and vitamin B12 and mycothiol biosynthesis). In addition, whereas growth inhibition of I. calvum by cysteine and sulfide treatments was observed only under anoxia, in E. coli, this phenomenon was observed under oxic conditions (30, 31). Thus, for I. calvum, repression of genes involved in nitrate respiration likely also underlies the observed phenomenon. Indeed, all genes in the denitrification pathway were severely repressed during cysteine treatment, and pathway activity was significantly reduced (Fig. 1E and 4A). Sulfide treatment also resulted in downregulation of the denitrification pathway prior to recovery, but interestingly, the DNRA pathway was upregulated (Fig. 4A). While significant accumulation of ammonium was not observed during sulfide treatment, the sulfide-induced shift from denitrification to DNRA is consistent with previous work on the effect of sulfide on the activities of nitrate-reducing communities in freshwater, wastewater, and sediment (16, 32–34).
I. calvum recovered from the inhibitory effects of cysteine and sulfide and achieved a normal growth rate within 300 h of treatment, with low-to-moderate doses of cysteine (<0.1 mM) and sulfide (<0.15 mM). The recovery from cysteine inhibition could have been mediated by the upregulation of a putative cystathionine gamma-lyase (EC 4.4.1.1, Ical_3515), which metabolizes cysteine into pyruvate, hydrogen sulfide, and ammonia. Overexpression of this enzyme, also known as cysteine desulfhydrase, confers resistance to cysteine in E. coli (35). We also discovered that genes associated with pyridoxidal-5′-phosphate biosynthesis (a cofactor of cystathionine gamma-lyase), iron transport, cytochrome c assembly, and sulfide:quinone oxidoreductase (EC 1.8.5.4, Ical_1214) were upregulated. Taken together, these observations suggest a recovery mechanism in which cysteine is first reduced to sulfide by cystathionine gamma-lyase and then oxidized to polysulfide by sulfide:quinone oxidoreductase (36). This putative cystathionine-mediated recovery mechanism might be utilized due to lack of availability of the mycothione-based primary redox stress response as a result of the downregulation of mycothiol synthase and other mycothiol-dependent enzymes during treatment with cysteine (see Fig. S5 in the supplemental material) (37). While cysteine is a reactant in mycothiol biosynthesis, the decrease in mycothione levels was likely due to the unavailability of precursors generated by BCAA biosynthesis, which was downregulated by cysteine treatment. It is also plausible that reducing conditions and increased thiol levels resulting from excess cysteine could have triggered sulfur relay pathways to decrease the synthesis of mycothione. Dysregulation of the sulfur pool can also alter protein translation via tRNA thiolation pathways and affect growth (38). Like cystathione and mycothione, the thiols of thioredoxins, which were upregulated by both cysteine and sulfide, could have also played a role in recovery from reduced sulfur stress by scavenging excess cysteine through the formation of intermolecular disulfide bonds (39).
Interestingly, sulfide:quinone oxidoreductase and the other genes implicated in recovery from cysteine were not upregulated during sulfide treatment, suggesting the mechanism of recovery from sulfide inhibition is different. Terminal cytochrome c oxidases that were upregulated by sulfide treatment (e.g., Ical_0754, Ical_0755, Ical_0756, Ical_0747, and Ical_0748) could have facilitated its oxidation via the reduction of heme (Fig. S3B) (40). Among these cytochrome c oxidases are thioredoxin/protein-disulfide isomerases, cytochrome c biogenesis protein CcdA, and those in the DNRA pathway, which suggests that sulfide oxidation may have been coupled to dissimilatory nitrate reduction to ammonia, as observed previously in freshwater nitrate-reducing communities exposed to sulfide (16). In stark contrast to the response to cysteine, mycothione biosynthesis and thioredoxin system genes were unaffected by sulfide treatment, suggesting their possible involvement in sulfide scavenging, clearing, or removal of free sulfide-induced disulfide bonds or nitroso groups (41). Thus, the systems analysis demonstrates that the recovery of growth of I. calvum from cysteine and sulfide inhibition likely occurs through distinct mechanisms.
Our results broaden the scope of knowledge regarding the inhibitory effects of reduced sulfur compounds and provide additional insights into the competition between SRB and NRB in the environment. It is a well-accepted principle that NRB can outcompete SRB for carbon based on the thermodynamics of denitrification and sulfate reduction, which is the basis for nitrate injections into oil wells to prevent souring from sulfide production (14, 42, 43). However, several studies have found exceptions to simple thermodynamic principles, which suggests that in order to better understand microbial community assembly and interactions in the environment, more research is needed (44–47). In line with this notion, the interactions between NRB, SRB, and their metabolites are complex and not fully characterized. For instance, certain SRB have been shown to perform DNRA (a higher energy-yielding metabolism than sulfate reduction), but this activity is inhibited by the presence of sulfide (48). Furthermore, previous work has shown that SRB that lack nitrite reductase are inhibited by nitrate and nitrite (13). Other studies have shown that certain NRB are inhibited by sulfide (15). Our findings, that growth and activity of the NRB I. calvum is inhibited by elevated levels of sulfide and cysteine, build on this prior work and provide a mechanistic explanation for mutual exclusion of SRB and NRB in certain environments. Specifically, thermodynamic considerations and the inhibitory effects of nitrite likely exclude SRB from niches occupied by NRB and, conversely, elevated levels of sulfide may prevent growth of NRB in niches where SRB are active. These mechanistic hypotheses are supported by observations of the effects of sulfide on denitrification activity in freshwater and wastewater communities (16, 32) and are also consistent with an observed stratification of SRB and NRB activities down the vertical transect of sediment cores sampled and analyzed from the FRC. Specifically, transects of the cores that had supposed sulfate reduction activity (based on sulfide detection) and denitrification activity (based on acetylene block activity assays) were spatially separated (K. De León, E. Majumder, and F. von Netzer, personal communication, December 2020). However, there are notable exceptions, such as certain bacteria that couple nitrate reduction to sulfide oxidation and thrive under conditions where both nitrate and sulfide are present (49). Additionally, the observation that I. calvum was capable of recovering from reduced sulfur stress suggests that NRB have developed mechanisms to overcome the inhibitory effects of moderate cysteine and sulfide levels and coexist with SRB in some environments. Functional genomic screens and laboratory evolution studies using mixed communities of NRB and SRB could help to better elucidate the biotic and abiotic constraints on competition between NRB and SRB (50).
MATERIALS AND METHODS
Strains and medium preparation.
I. calvum C5 was isolated from groundwater obtained from a well (GW 247) at the Oak Ridge Field Research Center (FRC) that is highly contaminated with nitrate (>200 mM). Growth studies were performed at 30°C in defined minimal medium containing 20 mM sodium lactate and 30 mM sodium nitrate at pH 7.2 with a 80:20 N2-CO2 headspace (see Text S1 in the supplemental material for additional details). For supplementation experiments, cysteine (l-cysteine; Sigma-Aldrich), hydrogen sulfide (sodium sulfide hydrated technical grade chips [Fisher Scientific] or sodium sulfide nonahydrate [Santa Cruz Biotechnology]), isoleucine (l-isoleucine; Sigma-Aldrich), leucine (l-leucine, Sigma-Aldrich), valine (l-valine; Sigma-Aldrich), sodium acetate (Sigma-Aldrich), and glycerol (molecular biology grade; Sigma-Aldrich) were added from sterile anoxic stocks. All growth assays were performed using cultures revived from freezer stocks, which were allowed at least one full growth cycle.
Additional details regarding experimental procedures which were omitted from the main text are presented. Included are details regarding the preparation of medium, isolation of I. calvum C5, treatment of sulfide batch effects, and the metabolomics analyses. Download Text S1, DOCX file, 0.1 MB (25.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Growth measurements.
For all experiments, cell concentration was monitored in Balch tubes (10-ml culture volume) with periodic measurements of the optical density at 600 nm (OD600) using a Spectronic 200 spectrophotometer (Thermo Fisher). Blank subtraction was performed using Balch tubes containing medium with no cells. Initial cell densities were normalized to ∼0.01 OD600 units for all experiments.
Measurement of nitrogen species.
Nitrite, total oxidized nitrogen (TON; nitrite plus nitrate), and total ammonia (ammonia and ammonium) content in media were quantified using the Gallery automated photometric analyzer (Thermo Fisher). Measurements were calibrated using sodium nitrite, sodium nitrate, and ammonium chloride. Nitrous oxide was measured by gas chromatography (model 8610; SRI Instruments) with nitrogen as the carrier gas, a 182.9-cm HayeSep D column (SRI Instruments), and an electron capture detector (ECD).
Measurement of oxygen concentration.
The concentration of dissolved oxygen was monitored in Balch tubes using a FireStingO2-Mini fiber optic oxygen meter with a temperature‐compensated 3-mm-diameter optode and oxygen sensor spots (Pyroscience). Sensor spots were attached to the interior of Balch tubes below the 10-ml liquid level using silicone glue and calibrated using sodium sulfite solution and oxygen-saturated growth medium. Both temperature and the ionic strength of the medium were taken into account for the purpose of calibration and subsequent measurements.
Transcriptomics profiling and analysis.
(i) Sample collection and sequencing. I. calvum C5 was cultured in biological triplicates, in medium containing either 0.25 mM cysteine or 0.25 mM sulfide (Fisher) (see Fig. S2A and Text S1 for treatment of batch effects). Aliquots (10 ml) were collected at four time points for transcriptome profiling (Fig. 1E), and cell pellets were harvested under anoxic conditions by centrifugation at 4,000 × g. Cell pellets were flash frozen in liquid nitrogen, and total RNA was extracted using hot phenol-chloroform (51). The Ribo-Zero bacterial kit (Illumina) was used for rRNA depletion, and the TruSeq Stranded mRNA library preparation kit (Illumina) was used for library preparation. Sequencing was performed using the NextSeq platform (2 by 75 bp, Illumina) with 10 to 15 million reads per sample.
(ii) Read processing. RNA sequencing reads were analyzed with FastQC according to Illumina’s default quality filtering process and then trimmed using base quality scores by Trimmomatic (52, 53). A quality score of 20 was used for read trimming and quality filtering. Reads were aligned to the genome of Intrasporangium calvum C5 (NCBI BioProject PRJNA475609) (3, 52) using Spliced Transcripts Alignment to a Reference (STAR) (54) followed by tabulation of transcript abundances by HTSeq-count (55).
(iii) Differential expression analysis, clustering, and functional enrichment. Normalized expression data obtained from DESeq2 (21) were filtered to remove genes with consistently low expression levels (among the lower 1% of normalized expression values across all conditions). Boruta, a random forest-based feature selection algorithm, was used to perform pairwise comparisons of transcriptional profiles across treatments in order to identify differentially expressed genes (DEGs) using Z-score-transformed normalized expression (22). Gene ontology (GO) and MetaCyc annotations were obtained by comparing features in the genomes of I. calvum C5 and the type strain I. calvum 7 KIP (for which these annotations were available) using reciprocal BLAST (56, 57). Reciprocal BLAST default parameters were used. Sequence matches were required to be at least 70% identical within the aligned region, and high-scoring segment pair alignment had to cover at least 50% of the query sequence. SEED annotations were obtained using DIAMOND to map transcript reads against the SEED database of annotated microbial protein sequences (58, 59). Default DIAMOND search parameters and the BLASTX option were used to query transcript sequences against the SEED protein database. DEGs identified by Boruta were clustered using the scikit-learn implementation of the k-means algorithm (60). Additional statistical analyses, including principal-component analysis (PCA), significance testing, and functional enrichment were performed using tools from the python scikit-learn, SciPy, and NumPy packages (60–62). The significance of GO, MetaCyc, and SEED term enrichment among k-means clusters was assessed by comparing term frequencies within each cluster to their frequencies in the genome using a hypergeometric test. Significantly enriched terms (P ≤ 0.05 and Benjamini-Hochberg false-discovery rate q < 0.01) were reported (see Table S1) (63). Gene level expression comparisons were performed for the pathways identified using SEED, GO, and MetaCyc process and pathway annotations (59, 64, 65). Expression level differences were assessed by Welch’s t test, and genes were reported as dysregulated if differences were significant (P ≤ 0.05 and Benjamini-Hochberg false discovery rate q < 0.01) (see Table S2).
Cellular process and pathway terms enriched among differentially expressed gene (DEG) clusters. This table provides a comprehensive list of GO, MetaCyc, and SEED ontologies enriched among k-means clustered DEGs along with enrichment P values (hypergeometric test, corrected using Benjamini-Hochberg false-discovery rate), and average process-level expression Z-scores. Download Table S1, DOCX file, 0.1 MB (48.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gene level expression comparisons for select pathways and processes. This table provides a comprehensive list of gene level expression comparisons for specific pathways. Included are gene pathway annotations, gene descriptions, average gene level expression Z-scores, and corrected P values (Welch’s t test, corrected using Benjamini-Hochberg false-discovery rate). Download Table S2, DOCX file, 0.1 MB (86.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Metabolomics profiling and analysis.
(i) Extraction. Metabolomic analysis was performed for five biological replicates over two time points during growth of I. calvum C5 in medium containing 0.25 mM cysteine or 0.25 mM sulfide (Fisher) (see Text S1 for treatment of batch effects). Cell culture aliquots (10 ml) were harvested under anoxic conditions by centrifugation at 4,000 × g, flash frozen in liquid nitrogen and stored at −80°C. Frozen cell pellets were thawed on ice and resuspended in a 2:2:1 solution of acetonitrile, methanol, and water. Metabolites were extracted according to a standard approach, dried, and reconstituted in volumes of acetonitrile-water solution (1:1 [vol/vol]) normalized to protein content in the sample. Following extraction, samples were transferred to liquid chromatography-mass spectrometry autosampler vials and were stored at −80°C until analysis. See Text S1 for additional details.
(ii) High-resolution mass spectrometry untargeted analysis. Metabolite mixtures were analyzed with liquid chromatography electrospray ionization quadrupole time-of-flight mass spectrometry (LC-ESI-QTOF-MS) (Bruker impact II) in both positive and negative electrospray ionization modes. Metabolites were separated by gradient elution with the mobile phase consisting of various ratios of water containing 0.1% formic acid and acetonitrile containing 0.1% formic acid. For the amide hydrophilic interaction chromatography analysis, metabolites were separated by gradient elution with the mobile phase composed of various ratios of water-acetonitrile (95:5 [vol/vol]) containing 20 mM ammonium acetate, 40 mM ammonium hydroxide, and water-acetonitrile (5:95 [vol/vol]). See Text S1 for additional details.
(iii) Targeted metabolomics analysis of amino acid concentration. Samples were analyzed on an Agilent 6495 triple quadrupole mass spectrometer coupled to an Agilent 1290 ultraperformance liquid chromatography stack. Separation was carried out using an Imtakt amino acid column. Targeted masses and retention times were selected based on previously established reference standards for each amino acid.
(iv) Data processing, metabolite annotation, and statistical analysis. Raw data were processed and analyzed using XCMS Online (25, 66–68). Metabolite features shared across all samples were identified using a multigroup analysis and compared for differential abundance. Differentially abundant metabolite features were identified with a total ion intensity of >10,000 and an associated abundance fold change of >3 in sulfide or cysteine-treated samples relative to the control sample (P ≤ 0.01). The filtered feature data table was annotated via an accurate mass search against METLIN using a 25-ppm window and manual inspection of putative identification hits for each feature (69–71). Dysregulated pathways were annotated by considering the enrichment of differentially abundant metabolites predicted by METLIN using the Systems Biology results feature of XCMS Online (25, 26). Pathways with a significant enrichment of differentially abundant metabolite features (P ≤ 0.01) and coverage of at least 20% were reported. Additional statistical analyses, including PCA, clustering, and significance testing, were performed using tools from the python scikit-learn, SciPy, and NumPy packages with Z-score-standardized log peak areas (60–62). Pathways and putative metabolites were compared to the results of the corresponding samples in the transcriptomic analysis.
Data availability.
Processed data tables as well as code to reproduce the main figures and key analyses in this study are available as part of a GitHub repository at https://github.com/baliga-lab/Ical_S_Inhibition. The untargeted metabolomics data reported in this paper are publicly available on XCMS Online (https://xcmsonline.scripps.edu, in the public shares section). The RNA-sequencing data reported in this paper have been deposited in the NCBI GEO database (accession no. GSE158822).
ACKNOWLEDGMENTS
A.V.C. and S.M.G. were supported by a Washington Research Foundation Distinguished Investigator Award and by startup funds from the Institute for Systems Biology. A.E.O., A.V.C., E.L.W.M., M.K.R., R.L.W., D.A.E., D.A.S., G.S., and N.S.B. were supported by Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a scientific focus area program at Lawrence Berkeley National Laboratory based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research under contract number DE-AC02-05CH11231. N.S.B. was supported by the National Science Foundation under grant no. OCE-1558924, CBET-1606206, MCB-1518261, DBI-1565166, and MCB-1616955.
We thank Kara De León, Frederick von Netzer, Lauren Lui, and Heidi Smith for discussions about analyses they performed on sediment cores from the Oak Ridge Field Research Center, which supported mutual exclusion of nitrate-reducing bacteria and sulfate-reducing bacteria.
REFERENCES
- 1.Fowler D, Coyle M, Skiba U, Sutton MA, Cape JN, Reis S, Sheppard LJ, Jenkins A, Grizzetti B, Galloway JN, Vitousek P, Leach A, Bouwman AF, Butterbach-Bahl K, Dentener F, Stevenson D, Amann M, Voss M. 2013. The global nitrogen cycle in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 368:20130164. doi: 10.1098/rstb.2013.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuypers MMM, Marchant HK, Kartal B. 2018. The microbial nitrogen-cycling network. Nat Rev Microbiol 16:263–276. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 3.Vuono DC, Read RW, Hemp J, Sullivan BW, Arnone JA, III, Neveux I, Blank RR, Loney E, Miceli D, Winkler M-KH, Chakraborty R, Stahl DA, Grzymski JJ. 2019. Resource concentration modulates the fate of dissimilated nitrogen in a dual-pathway actinobacterium. Front Microbiol 10:3. doi: 10.3389/fmicb.2019.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson HK, Price MN, Callaghan M, Aaring A, Chakraborty R, Liu H, Kuehl JV, Arkin AP, Deutschbauer AM. 2019. The selective pressures on the microbial community in a metal-contaminated aquifer. ISME J 13:937–949. doi: 10.1038/s41396-018-0328-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson HK, Lui LM, Price MN, Kazakov AE, Carr AV, Kuehl JV, Owens TK, Nielsen T, Arkin AP, Deutschbauer AM. 2020. Selective carbon sources influence the end products of microbial nitrate respiration. ISME J 14:2034–2045. doi: 10.1038/s41396-020-0666-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith MB, Rocha AM, Smillie CS, Olesen SW, Paradis C, Wu L, Campbell JH, Fortney JL, Mehlhorn TL, Lowe KA, Earles JE, Phillips J, Techtmann SM, Joyner DC, Elias DA, Bailey KL, Hurt RA, Jr, Preheim SP, Sanders MC, Yang J, Mueller MA, Brooks S, Watson DB, Zhang P, He Z, Dubinsky EA, Adams PD, Arkin AP, Fields MW, Zhou J, Alm EJ, Hazen TC. 2015. Natural bacterial communities serve as quantitative geochemical biosensors. mBio 6:e00326-15. doi: 10.1128/mBio.00326-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finneran KT, Housewright ME, Lovley DR. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ Microbiol 4:510–516. doi: 10.1046/j.1462-2920.2002.00317.x. [DOI] [PubMed] [Google Scholar]
- 8.Hemme CL, Tu Q, Shi Z, Qin Y, Gao W, Deng Y, Van Nostrand JD, Wu L, He Z, Chain PSG, Tringe SG, Fields MW, Rubin EM, Tiedje JM, Hazen TC, Arkin AP, Zhou J. 2015. Comparative metagenomics reveals impact of contaminants on groundwater microbiomes. Front Microbiol 6:1205. doi: 10.3389/fmicb.2015.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayangbenro AS, Olanrewaju OS, Babalola OO. 2018. Sulfate-reducing bacteria as an effective tool for sustainable acid mine bioremediation. Front Microbiol 9:1986. doi: 10.3389/fmicb.2018.01986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasmund K, Mußmann M, Loy A. 2017. The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ Microbiol Rep 9:323–344. doi: 10.1111/1758-2229.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Lan T, Müller C, Cai Z. 2015. Dissimilatory nitrate reduction to ammonium (DNRA) plays an important role in soil nitrogen conservation in neutral and alkaline but not acidic rice soil. J Soils Sediments 15:523–531. doi: 10.1007/s11368-014-1037-7. [DOI] [Google Scholar]
- 12.Smirnov PM, Kidin VV, Pedishyus RK. 1979. Loss of nitrogen by denitrification. Biol Bull Acad Sci USSR 6:450–459. [PubMed] [Google Scholar]
- 13.He Q, He Z, Joyner DC, Joachimiak M, Price MN, Yang ZK, Yen H-CB, Hemme CL, Chen W, Fields MM, Stahl DA, Keasling JD, Keller M, Arkin AP, Hazen TC, Wall JD, Zhou J. 2010. Impact of elevated nitrate on sulfate-reducing bacteria: a comparative study of Desulfovibrio vulgaris . ISME J 4:1386–1397. doi: 10.1038/ismej.2010.59. [DOI] [PubMed] [Google Scholar]
- 14.Xue Y, Voordouw G. 2015. Control of microbial sulfide production with biocides and nitrate in oil reservoir simulating bioreactors. Front Microbiol 6:1387. doi: 10.3389/fmicb.2015.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sørensen J, Tiedje JM, Firestone RB. 1980. Inhibition by sulfide of nitric and nitrous oxide reduction by denitrifying Pseudomonas fluorescens . Appl Environ Microbiol 39:105–108. doi: 10.1128/AEM.39.1.105-108.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunet RC, Garcia-Gil LJ. 1996. Sulfide-induced dissimilatory nitrate reduction to ammonia in anaerobic freshwater sediments. FEMS Microbiol Ecol 21:131–138. doi: 10.1111/j.1574-6941.1996.tb00340.x. [DOI] [Google Scholar]
- 17.Joye SB, Hollibaugh JT. 1995. Influence of sulfide inhibition of nitrification on nitrogen regeneration in sediments. Science 270:623–625. doi: 10.1126/science.270.5236.623. [DOI] [Google Scholar]
- 18.Zhang W, Li F, Nie L. 2010. Integrating multiple “omics” analysis for microbial biology: application and methodologies. Microbiology (Reading) 156:287–301. doi: 10.1099/mic.0.034793-0. [DOI] [PubMed] [Google Scholar]
- 19.Harris CL. 1981. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as the target enzyme. J Bacteriol 145:1031–1035. doi: 10.1128/JB.145.2.1031-1035.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukushima RS, Weimer PJ, Kunz DA. 2003. Use of photocatalytic reduction to hasten preparation of culture media for saccharolytic Clostridium species. Braz J Microbiol 34:22–26. doi: 10.1590/S1517-83822003000100006. [DOI] [Google Scholar]
- 21.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kursa MB, Jankowski A, Rudnicki WR. 2010. Boruta – a system for feature selection. Fundam Inform 101:271–285. doi: 10.3233/FI-2010-288. [DOI] [Google Scholar]
- 23.Folsom JP, Parker AE, Carlson RP. 2014. Physiological and proteomic analysis of Escherichia coli iron-limited chemostat growth. J Bacteriol 196:2748–2761. doi: 10.1128/JB.01606-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giles NM, Watts AB, Giles GI, Fry FH, Littlechild JA, Jacob C. 2003. Metal and redox modulation of cysteine protein function. Chem Biol 10:677–693. doi: 10.1016/s1074-5521(03)00174-1. [DOI] [PubMed] [Google Scholar]
- 25.Huan T, Forsberg EM, Rinehart D, Johnson CH, Ivanisevic J, Benton HP, Fang M, Aisporna A, Hilmers B, Poole FL, Thorgersen MP, Adams MWW, Krantz G, Fields MW, Robbins PD, Niedernhofer LJ, Ideker T, Majumder EL, Wall JD, Rattray NJW, Goodacre R, Lairson LL, Siuzdak G. 2017. Systems biology guided by XCMS Online metabolomics. Nat Methods 14:461–462. doi: 10.1038/nmeth.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsberg EM, Huan T, Rinehart D, Benton HP, Warth B, Hilmers B, Siuzdak G. 2018. Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS Online. Nat Protoc 13:633–651. doi: 10.1038/nprot.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ung KSE, Av-Gay Y. 2006. Mycothiol-dependent mycobacterial response to oxidative stress. FEBS Lett 580:2712–2716. doi: 10.1016/j.febslet.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 28.Massey LK, Sokatch JR, Conrad RS. 1976. Branched-chain amino acid catabolism in bacteria. Bacteriol Rev 40:42–54. doi: 10.1128/BR.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takumi K, Nonaka G. 2016. Bacterial cysteine-inducible cysteine resistance systems. J Bacteriol 198:1384–1392. doi: 10.1128/JB.01039-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korshunov S, Imlay KRC, Imlay JA. 2020. Cystine import is a valuable but risky process whose hazards Escherichia coli minimizes by inducing a cysteine exporter. Mol Microbiol 113:22–39. doi: 10.1111/mmi.14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chonoles Imlay KR, Korshunov S, Imlay JA. 2015. Physiological roles and adverse effects of the two cystine importers of Escherichia coli . J Bacteriol 197:3629–3644. doi: 10.1128/JB.00277-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan Y, Ye L, Yuan Z. 2013. Effect of H2S on N2O reduction and accumulation during denitrification by methanol utilizing denitrifiers. Environ Sci Technol 47:8408–8415. doi: 10.1021/es401632r. [DOI] [PubMed] [Google Scholar]
- 33.Bowles MW, Nigro LM, Teske AP, Joye SB. 2012. Denitrification and environmental factors influencing nitrate removal in Guaymas Basin hydrothermally altered sediments. Front Microbiol 3:377. doi: 10.3389/fmicb.2012.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones ZL, Jasper JT, Sedlak DL, Sharp JO. 2017. Sulfide-induced dissimilatory nitrate reduction to ammonium supports anaerobic ammonium oxidation (anammox) in an open-water unit process wetland. Appl Environ Microbiol 83:e00782-17. doi: 10.1128/AEM.00782-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soutourina J, Blanquet S, Plateau P. 2001. Role of d-cysteine desulfhydrase in the adaptation of Escherichia coli to d-cysteine. J Biol Chem 276:40864–40872. doi: 10.1074/jbc.M102375200. [DOI] [PubMed] [Google Scholar]
- 36.Chan L-K, Morgan-Kiss RM, Hanson TE. 2009. Functional analysis of three sulfide:quinone oxidoreductase homologs in Chlorobaculum tepidum . J Bacteriol 191:1026–1034. doi: 10.1128/JB.01154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fahey RC. 2013. Glutathione analogs in prokaryotes. Biochim Biophys Acta 1830:3182–3198. doi: 10.1016/j.bbagen.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Paritala H, Carroll KS. 2013. New targets and inhibitors of mycobacterial sulfur metabolism. Infect Disord Drug Targets 13:85–115. doi: 10.2174/18715265113139990022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Laer K, Hamilton CJ, Messens J. 2013. Low-molecular-weight thiols in thiol–disulfide exchange. Antioxid Redox Signal 18:1642–1653. doi: 10.1089/ars.2012.4964. [DOI] [PubMed] [Google Scholar]
- 40.Vitvitsky V, Miljkovic JL, Bostelaar T, Adhikari B, Yadav PK, Steiger AK, Torregrossa R, Pluth MD, Whiteman M, Banerjee R, Filipovic MR. 2018. Cytochrome c reduction by H2S potentiates sulfide signaling. ACS Chem Biol 13:2300–2307. doi: 10.1021/acschembio.8b00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng H, Shen J, Edmonds KA, Luebke JL, Hickey AK, Palmer LD, Chang F-MJ, Bruce KA, Kehl-Fie TE, Skaar EP, Giedroc DP. 2017. Sulfide homeostasis and nitroxyl intersect via formation of reactive sulfur species in Staphylococcus aureus . mSphere 2:e00082-17. doi: 10.1128/mSphere.00082-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolfing J, Hubert CRJ. 2017. Using thermodynamics to predict the outcomes of nitrate-based oil reservoir souring control interventions. Front Microbiol 8:2575. doi: 10.3389/fmicb.2017.02575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. 2011. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev 75:361–422. doi: 10.1128/MMBR.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Hanke A, Tegetmeyer HE, Kattelmann I, Sharma R, Hamann E, Hargesheimer T, Kraft B, Lenk S, Geelhoed JS, Hettich RL, Strous M. 2017. Impacts of chemical gradients on microbial community structure. ISME J 11:920–931. doi: 10.1038/ismej.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takaya N, Catalan-Sakairi MAB, Sakaguchi Y, Kato I, Zhou Z, Shoun H. 2003. Aerobic denitrifying bacteria that produce low levels of nitrous oxide. Appl Environ Microbiol 69:3152–3157. doi: 10.1128/aem.69.6.3152-3157.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao H, Schreiber F, Collins G, Jensen MM, Svitlica O, Kostka JE, Lavik G, de Beer D, Zhou H-Y, Kuypers MMM. 2010. Aerobic denitrification in permeable Wadden Sea sediments. ISME J 4:417–426. doi: 10.1038/ismej.2009.127. [DOI] [PubMed] [Google Scholar]
- 47.Canfield DE, Stewart FJ, Thamdrup B, De Brabandere L, Dalsgaard T, Delong EF, Revsbech NP, Ulloa O. 2010. A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science 330:1375–1378. doi: 10.1126/science.1196889. [DOI] [PubMed] [Google Scholar]
- 48.Dalsgaard T, Bak F. 1994. Nitrate reduction in a sulfate-reducing bacterium, Desulfovibrio desulfuricans, isolated from rice paddy soil: sulfide inhibition, kinetics, and regulation. Appl Environ Microbiol 60:291–297. doi: 10.1128/AEM.60.1.291-297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villahermosa D, Corzo A, Garcia-Robledo E, González JM, Papaspyrou S. 2016. Kinetics of indigenous nitrate reducing sulfide oxidizing activity in microaerophilic wastewater biofilms. PLoS One 11:e0149096. doi: 10.1371/journal.pone.0149096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. doi: 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 51.Khodursky AB, Bernstein JA, Peter BJ, Rhodius V, Wendisch VF, Zimmer DP. 2003. Escherichia coli spotted double-strand DNA microarrays: RNA extraction, labeling, hybridization, quality control, and data management. Methods Mol Biol 224:61–78. doi: 10.1385/1-59259-364-X:61. [DOI] [PubMed] [Google Scholar]
- 52.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. [Google Scholar]
- 54.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S, Pyl PT, Huber W. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cock PJA, Chilton JM, Grüning B, Johnson JE, Soranzo N. 2015. NCBI BLAST+ integrated into Galaxy. Gigascience 4:39. doi: 10.1186/s13742-015-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 59.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang H-Y, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. [Google Scholar]
- 61.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0 Contributors . 2020. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, Kern R, Picus M, Hoyer S, van Kerkwijk MH, Brett M, Haldane A, Del Río JF, Wiebe M, Peterson P, Gérard-Marchant P, Sheppard K, Reddy T, Weckesser W, Abbasi H, Gohlke C, Oliphant TE. 2020. Array programming with NumPy. Nature 585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57:289–300. [Google Scholar]
- 64.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. 2014. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gowda H, Ivanisevic J, Johnson CH, Kurczy ME, Benton HP, Rinehart D, Nguyen T, Ray J, Kuehl J, Arevalo B, Westenskow PD, Wang J, Arkin AP, Deutschbauer AM, Patti GJ, Siuzdak G. 2014. Interactive XCMS Online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal Chem 86:6931–6939. doi: 10.1021/ac500734c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. 2012. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal Chem 84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benton HP, Ivanisevic J, Mahieu NG, Kurczy ME, Johnson CH, Franco L, Rinehart D, Valentine E, Gowda H, Ubhi BK, Tautenhahn R, Gieschen A, Fields MW, Patti GJ, Siuzdak G. 2015. Autonomous metabolomics for rapid metabolite identification in global profiling. Anal Chem 87:884–891. doi: 10.1021/ac5025649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, Koellensperger G, Huan T, Uritboonthai W, Aisporna AE, Wolan DW, Spilker ME, Benton HP, Siuzdak G. 2018. METLIN: a technology platform for identifying knowns and unknowns. Anal Chem 90:3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xue J, Guijas C, Paul Benton H, Warth B, Siuzdak G. 2020. METLIN MS2 molecular standards database: a broad chemical and biological resource. Nat Methods 17:953–954. doi: 10.1038/s41592-020-0942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. 2012. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol 30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Carbon-to-nitrogen ratio determines dissimilatory nitrate reduction end products in continuous flow reactors. Growth and metabolic activity of I. calvum in chemostat reactors with various C:N ratios. Dashed lines indicate the point at which chemostat flow was initiated, with a constant dilution rate of 650 ml/day. Download FIG S1, TIF file, 0.6 MB (567.9KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Treatment of sulfide batch effects and the role of oxygen during cysteine and sulfide treatment. (A) Log recovery time differences are predictive of effective sulfide concentration across batches and account for batch effects. Shaded regions represent model uncertainty estimated from propagation of parameter error. (B) Aerobically grown cultures (uncapped tubes shaken at 250 rpm) in minimal medium (20 mM lactate, 0 mM nitrate) with and without the addition of 0.25 mM cysteine or 0.25 mM sulfide and monitored over a period of 40 h. (C) Monitoring of in situ oxygen levels using oxygen sensors attached to the interior of sealed Balch tubes confirm recovery is not due to oxygen leakage. Growth conditions included standard anoxic growth medium as well as supplementation with cysteine (0.05 mM), sulfide (0.025 mM), or aerated medium. Shaded regions depict the standard deviations from biological replicates (n ≥ 3). Download FIG S2, TIF file, 0.7 MB (739.8KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differentially expressed genes identified by Boruta and global expression patterns. (A) Pairwise comparisons of condition-specific normalized gene expression data using Boruta-identified differentially expressed gene sets which best predicted condition labels. For each comparison, the total number of genes is indicated as well as the set of genes unique to that comparison (e.g., not shared across comparisons). The total number of DEGs identified is noted in the center as well as the number of genes shared across all comparisons. (B) Z-score-normalized expression for select pathways identified by enrichment of DEG clusters. Bars indicate comparisons for which differences were significant. *, P < 0.05; ***, P < 0.001. Boxplots display estimates of data minimum (left whisker), median (line within box), maximum (right whisker), and interquartile range (box dimensions) as well as possible outliers (points beyond whiskers). Data are overlaid as scatter points. Global patterns in normalized gene expression data for all genes (C) and differentially expressed genes identified by Boruta (D). Download FIG S3, TIF file, 0.8 MB (807.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differentially abundant metabolomic features and global patterns in feature abundance. (A) Pairwise comparisons of condition-specific feature relative abundances identified differentially abundant features for those with putative identities. For each comparison, the total number of features is indicated as well as the set of features unique to that comparison. The total number of features identified is noted in the center as well as the number of features shared across all comparisons. (B) Relative abundance of select features identified as differentially abundant. Bars indicate comparisons for which differences were significant. *, P < 0.05; ***, P < 0.001. Boxplots display estimates of data minimum (left whisker), median (line within box), maximum (right whisker), and interquartile range (box dimensions) as well as possible outliers (points beyond whiskers). Data are overlaid as scatter points. Global patterns in normalized log feature abundance data for all features (C) and for features with putative identities (D). Download FIG S4, TIF file, 0.6 MB (628.4KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differential expression and abundance of the cellular redox regulatory mycothiol. (A) Normalized expression of mycothiol biosynthesis genes identified using gene ontologies with genes listed in order of pathway steps. (B) Relative abundance of metabolite features identified as mycothione (the oxidized disulfide form of mycothiol) and precursor UDP-N-acetyl-α-d-glucosamine at time points T1 and T2 across all conditions. Boxplots display estimates of data minimum (lower whisker), median (line within box), maximum (upper whisker), and interquartile range (box dimensions) as well as possible outliers (points beyond whiskers). Data are overlaid as scatter points. Download FIG S5, TIF file, 0.6 MB (583.6KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Additional details regarding experimental procedures which were omitted from the main text are presented. Included are details regarding the preparation of medium, isolation of I. calvum C5, treatment of sulfide batch effects, and the metabolomics analyses. Download Text S1, DOCX file, 0.1 MB (25.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cellular process and pathway terms enriched among differentially expressed gene (DEG) clusters. This table provides a comprehensive list of GO, MetaCyc, and SEED ontologies enriched among k-means clustered DEGs along with enrichment P values (hypergeometric test, corrected using Benjamini-Hochberg false-discovery rate), and average process-level expression Z-scores. Download Table S1, DOCX file, 0.1 MB (48.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gene level expression comparisons for select pathways and processes. This table provides a comprehensive list of gene level expression comparisons for specific pathways. Included are gene pathway annotations, gene descriptions, average gene level expression Z-scores, and corrected P values (Welch’s t test, corrected using Benjamini-Hochberg false-discovery rate). Download Table S2, DOCX file, 0.1 MB (86.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Processed data tables as well as code to reproduce the main figures and key analyses in this study are available as part of a GitHub repository at https://github.com/baliga-lab/Ical_S_Inhibition. The untargeted metabolomics data reported in this paper are publicly available on XCMS Online (https://xcmsonline.scripps.edu, in the public shares section). The RNA-sequencing data reported in this paper have been deposited in the NCBI GEO database (accession no. GSE158822).