Abstract
The genus Claviceps has been known for centuries as an economically important fungal genus for pharmacology and agricultural research. Only recently have researchers begun to unravel the evolutionary history of the genus, with origins in South America and classification of four distinct sections through ecological, morphological, and metabolic features (Claviceps sects. Citrinae, Paspalorum, Pusillae, and Claviceps). The first three sections are additionally characterized by narrow host range, whereas section Claviceps is considered evolutionarily more successful and adaptable as it has the largest host range and biogeographical distribution. However, the reasons for this success and adaptability remain unclear. Our study elucidates factors influencing adaptability by sequencing and annotating 50 Claviceps genomes, representing 21 species, for a comprehensive comparison of genome architecture and plasticity in relation to host range potential. Our results show the trajectory from specialized genomes (sects. Citrinae and Paspalorum) toward adaptive genomes (sects. Pusillae and Claviceps) through colocalization of transposable elements around predicted effectors and a putative loss of repeat-induced point mutation resulting in unconstrained tandem gene duplication coinciding with increased host range potential and speciation. Alterations of genomic architecture and plasticity can substantially influence and shape the evolutionary trajectory of fungal pathogens and their adaptability. Furthermore, our study provides a large increase in available genomic resources to propel future studies of Claviceps in pharmacology and agricultural research, as well as, research into deeper understanding of the evolution of adaptable plant pathogens.
Keywords: adaptive evolution, gene cluster expansion, fungal plant pathogens, RIP
Significance
Lack of genomic data for the Claviceps genus has hampered the ability to identify factors influencing the adaptation of Claviceps species and mechanisms associated with the broad host range of some species. Our analysis reveals the trajectory from specialized genomes toward adaptive genomes through a variety of genomic mechanisms which coincided with increases in host range potential. These results demonstrate a clear example of how genomic alterations can influence and shape the evolutionary trajectory of fungal pathogens in association with host range.
Introduction
Fungi, particularly phytopathogenic species, are increasingly being used to gain insight into the evolution of eukaryotic organisms, due to their adaptive nature and unique genome structures (Gladieux et al. 2014; Dong et al. 2015). Adaptation and diversification of fungal species can be mediated by changes in genome architecture and plasticity, such as genome size, transposable element (TE) content, localization of TEs to specific genes, genome compartmentalization, gene duplication rates, recombination rates, and presence/absence polymorphism of virulence factors (Dong et al. 2015; Möller and Stukenbrock 2017). The presence or absence of repeat-induced point (RIP) mutation is also an important mechanism for fungal genome evolution, as RIP works on a genomewide scale to silence TEs and duplicated genes, which can also “leak” onto neighboring genes (Galagan et al. 2003; Galagan and Selker 2004; Raffaele and Kamoun 2012; Urguhart et al. 2018; Möller and Stukenbrock 2017). It is becoming increasingly evident that variations in these factors can be used to classify genomes as a one speed (one compartment), such as the powdery mildew fungi Blumeria graminis f.sp. hordei and f.sp tritici, two speed (two compartments), such as the late blight pathogen Phytophthora infestans, or multispeed (multicompartment) such as the multihost pathogen Fusarium oxysporum (Dong et al. 2015; Frantzeskakis et al. 2019). These different “speeds” are characterized by their potential adaptability such that one-speed genomes are often considered less adaptable, whereas two-speed and multispeed genomes are often considered more adaptable (Dong et al. 2015; Frantzeskakis et al. 2019; Möller and Stukenbrock 2019).
The ergot fungi of the genus Claviceps (Ascomycota, Hypocreales) are biotrophic species that share a specialized ovarian-specific nonsystemic parasitic lifestyle with their grass hosts (Píchová et al. 2018). Infections are fully restricted to individual unpollinated ovaries (Tudzynski and Scheffer 2004), and the fungus actively manages to maintain host cell viability to obtain nutrients from living tissue through a complex cross-talk of genes related to pathogenesis, such as secreted effectors, secondary metabolites, or cytokinin production (Hinsch et al. 2015, 2016; Oeser et al. 2017; Kind, Schurack, et al. 2018; Kind, Hinsch, et al. 2018). Species of Claviceps are most notably known for their production of toxic alkaloids and secondary metabolites but are also known for their expansive host range and negative impact on global cereal crop production and livestock farming. These negative effects on human and livestock health are the primary reason Claviceps species are referred to as plant pathogens. However, under the light of coevolution with their grass hosts, some Claviceps species are considered conditional defensive mutualists with their hosts as they prevent herbivory and can improve host fitness (Raybould et al. 1998; Fisher et al. 2007; Wäli et al. 2013).
The genus Claviceps contains 59 species divided into four sections as follows: Claviceps, Pusillae, Citrinae, and Paspalorum (Píchová et al. 2018). It was postulated that sections Citrinae and Paspalorum originated in South America, whereas section Pusillae experienced speciation throughout the Eocene, Oligocene, and Miocene as these species encountered newly emergent PACMAD warm-season grasses (subfamilies Panicoideae, Aristidoideae, Chloridoideae, Micrairoideae, Arundinoideae, and Danthonioideae) when an ancestral strain was transferred from South America to Africa (Píchová et al. 2018). In contrast, the crown node of section Claviceps is estimated at 20.4 Ma and was followed by a radiation of the section corresponding to a host jump from ancestral sedges (Cyperaceae) to the Bamboo, Oryzoideae, Pooideae (BOP) clade (cool-season grasses; subfamilies Bambusoideae, Oryzoideae [syn: Ehrhartoideae]; Soreng et al. 2017, Pooideae) in North America (Bouchenak-Khelladi et al. 2010; Píchová et al. 2018). Section Claviceps has the largest host range with C. purpurea sensu stricto (s.s.) having been reported on up to 400 different species in clade BOP (Alderman et al. 2004, Píchová et al. 2018) across six tribes and retains the ability to infect sedges (Cyperaceae) (Jungehülsing and Tudzynski 1997). In contrast, section Pusillae is specialized to the tribes Paniceae and Andropogoneae, and sections Citrinae and Paspalorum only infect members of tribe Paspaleae and tribe Cynodonteae, respectively (Píchová et al. 2018). The shared specialized infection life cycle of the Claviceps genus, the drastic differences in host range potential of different species, and geographic distribution represent a unique system to study the evolution and host adaptation of eukaryotic organisms.
Despite their ecological and agriculture importance, little is known about the evolution and genomic architecture of these important fungal species in comparison with other cereal pathogens such as species in the genera Puccinia (Cantu et al. 2013; Kiran et al. 2016, 2017), Zymoseptoria (Estep et al. 2015; Grandaubert et al. 2015, 2019; Poppe et al. 2015; Testa, Oliver et al. 2015; Wu et al. 2017; Stukenbrock and Dutheil 2018), or Fusarium (Kvas et al. 2009; Ma et al. 2010; Rep and Kistler 2010; Watanabe et al. 2011; Sperschneider et al. 2015). Unfortunately, the lack of genome data for the Claviceps genus has hampered our ability to complete comparative analyses to identify factors that are influencing the adaptation of Claviceps species across the four sections in the genus, and the mechanisms by which species of section Claviceps have adapted to such a broad host range, in comparison with the other three sections. Here we present the sequences and annotations of 50 Claviceps genomes, representing 19 species, for a comprehensive comparison of the genus to understand evolution within the genus Claviceps by characterizing the genomic plasticity and architecture in relation to adaptive host potential. Our analysis reveals the trajectory from specialized one-speed genomes (sects. Citrinae and Paspalorum) toward adaptive two-speed genomes (sects. Pusillae and Claviceps) through colocalization of TEs around predicted effectors and a putative loss of RIP resulting in tandem gene duplication coinciding with increased host range potential.
Materials and Methods
Sample Acquisition
Field collected samples (Clav) were surfaced sterilized, allowed to grow as mycelia, and individual conidia transferred to make single spore cultures. Thirteen cultures were provided by Dr Miroslav Kolařík from the Culture Collection of Clavicipitaceae (CCC) at Institute of Microbiology, Academy of Sciences of the Czech Republic. Raw Illumina reads for samples (LM28, LM582, LM78, LM81, LM458, LM218, LM454, LM576, and LM583) were downloaded from NCBI SRA database. Raw Illumina reads from an additional 21 LM samples were generated by Dr Liu’s lab (AAFC), sequencing protocol of these 21 samples followed (Wingfield et al. 2018). Summarized information can be found in supplementary table S1, Supplementary Material online.
Preparation of Genomic DNA
Cultures grown on cellophane PDA plates were used for genomic DNA extraction from lyophilized mycelium following a modified CTAB method (Doyle JJ and Doyle JL 1987; Wingfield et al. 2018) without using the RNase Cocktail Enzyme Mix, only RNase A was used. DNA contamination was checked by running samples on a 1% agarose gel and a NanoDrop Onec (Thermo Fishcer Scientific). Twenty samples (7 Clav and 13 CCC) were sent to BGI-Hong Kong HGS Lab for 150-bp paired-end Illumina sequencing on an HiSeq 4000.
Genome Assembly
Preliminary data showed that raw reads of LM458 were contaminated with bacterial DNA but showed strong species similar to Clav32 and Clav50. To filter out the bacterial DNA sequences, reads of LM458 were mapped against the assembled Clav32 and Clav50 genomes using BBSplit v38.41 (Bushnell 2014). All forward and reverse reads mapped to each of the genomes were concatenated, respectively. Both sets were then interleaved to remove duplicates and used for further analysis. Reads for all 50 samples were checked for quality with FastQC v0.11.5 (Andrews 2010) and trimmed with Trimmomatic v0.36 (Bolger et al. 2014) using the commands (SLIDINGWINDOW: 4:20; MINLEN:36; HEADCROP:10) to remove poor quality data, only paired-end reads were used. To better standardize the comparative analysis, all 50 samples were subject to de novo genome assembly with Shovill v0.9.0 (https://github.com/tseemann/shovill; last accessed May 11, 2020) using SPAdes v3.11.1 (Nurk et al. 2013) with a minimum contig length of 1,000 bp.
The reference genomes of C. purpurea strain 20.1 (SAMEA2272775), C. fusiformis PRL 1980 (SAMN02981339), and C. paspali RRC 1481 (SAMN02981342) were downloaded from NCBI. Proteins for C. fusiformis and C. paspali were not available on NCBI so they were extracted from GFF3 files provided by Dr Chris Schardl and Dr Neil Moore, University of Kentucky, corresponding to the 2013 annotations (Schardl et al. 2013) available at http://www.endophyte.uky.edu (last accessed March 22, 2020). Reference genomes were standardized for comparative analysis with our 50 annotated genomes, by implementing a protein length cutoff of 50 aa and removal of alternatively spliced proteins in C. fusiformis and C. paspali, only the longest spliced protein for each locus remained.
Transposable Elements
TE fragments were identified following procedures for establishment of de novo comprehensive repeat libraries set forth in Coghlan et al. (2018), a brief summary is described below. The following steps were automated through construction of a custom script, TransposableELMT (https://github.com/PlantDr430/TransposableELMT). Each of the 53 Claviceps genome were used to create a respective repeat library using RepeatModeler v1.0.8 (Smit and Hubley 2015), TransposonPSI (Hass 2010), and long terminal repear (LTR) LTR_finder v1.07 (Xu and Wang 2007) on default settings. LTR_harvest v1.5.10 (Ellinghaus et al. 2008) was additionally run on default settings, and results were filtered with LTR_digest v1.5.10 (Steinbiss et al. 2009) with an HMM search for Pfam domains associated with TEs; only candidates with domain hits were kept. Repeat libraries from these four programs were concatenated with all curated TEs from RepBase (Bao et al. 2015) and redundant sequences were removed using Usearch v11.0.667 (Edgar 2010) with a percent identity cutoff of ≥80%. TEs for each of the nonredundant libraries were classified using RepeatClassifier v1.0.8 (Smit and Hubley 2015). RepeatMasker v4.0.7 (Smit et al. 2015) was then used, on default settings with each assemble genome and its respective repeat library, to soft mask the genomes and identify TE regions. TE content was represented as the proportion of the genome masked by TE regions determined by RepeatMasker, excluding simple and low complexity repeats.
The TE divergences, calculated from RepeatMasker for TEs in all 53 Claviceps genomes, were used to plot the divergence landscape using a custom script (https://github.com/PlantDr430/CSU_scripts/blob/master/TE_divergence_landscape.py). The RepeatMasker results were also used with the respective GFF3 file from each genome to calculate the average distance (kb) of each gene to the closest TE fragment on the 5′ and 3′ flanking side. Values were calculated for predicted effectors, noneffector secreted genes, nonsecreted metabolite genes, and all other genes using a custom script (https://github.com/PlantDr430/CSU_scripts/blob/master/TE_closeness.py).
Genome Annotation
AUGUSTUS v3.2.2 (Mario et al. 2008) was used to create pretrained parameters files using the reference C. purpurea strain 20.1, available expressed sequence tag (EST) data from NCBI, and wild-type RNAseq data (SRR4428945) created in Oeser et al. (2017). RNA-seq data was subject to quality check and trimming as above. All three data sets were also used to train parameter files for the ab initio gene model prediction software’s GeneID v1.4.4 (Blanco et al. 2007) and CodingQuarry v2.0 (Testa et al. 2015). GeneID training followed protocols available at http://genome.crg.es/software/geneid/training.html. For CodingQuarry training, RNA transcripts were created de novo using Trinity v2.8.4 (Grabherr et al. 2011) on default settings and EST coordinates were found by mapping the EST data to the reference genome using Minimap2 v2.1 (Li 2018).
Gene models for the 50 genomes were then predicted with GeneID and CodingQuarry using the trained C. purpruea parameter files. CodingQuarry prediction was also supplemented with transcript evidence by mapping the available EST and RNA-seq C. purpurea data to each genome using Minimap2. BUSCO v3 (Waterhouse et al. 2018) was run on all 50 genomes using the AUGUSTUS C. purpurea pretrained parameter files as the reference organism and the Sordariomyceta database. The resulting predicted proteins for each sample were used as training models for ab initio gene prediction using SNAP (Korf 2004) and GlimmerHMM v3.0.1 (Majoros et al. 2004). Last, GeMoMa v1.5.3 (Keilwagen et al. 2016) was used for ab initio gene prediction using the soft-masked genomes and the C. purpruea 20.1 reference files.
Funannotate v1.6.0 (Palmer and Stajich 2019) was then used as the primary software for genome annotation. Funannotate additionally uses AUGUSTUS and GeneMark-ES (Ter-Hovhannisyan et al. 2008) for ab initio gene model prediction, Exonerate for transcript and protein evidence alignment, and EVidenceModeler (Hass et al. 2008) for a final weighted consensus. All C. purpurea EST and RNAseq data were used as transcript evidence and the Uniport Swiss-Prot database and proteins from several closely related species (C. purpurea strain 20.1, C. fusiformis PRL1980, C. paspali RRC1481, Fusarium oxysporum f. sp. lycopersici 4287, Pochonia chlamydosporia 170, Ustilago maydis 521, and Epichloe festucae F1) were used as protein evidence. The AUGUSTUS pretrained C. purpurea files were used as BUSCO seed species along with the Sordariomyceta database and all five ab initio predictions were passed through the –other_gff flag with weights of 1. The following flags were also used in Funannotate “predict”: –repeats2evm, –optimize_augustus, –soft_mask 1000, –min_protlen 50. BUSCO was used to evaluate annotation completeness using the Dikarya and Sordariomyceta databases (odb9) with –prot on default settings.
Functional Annotation
Functional analysis was performed using Funannotate “annotate.” The following analyses were also performed on the three reference Claviceps genomes. Secondary metabolite clusters were predicted using antiSMASH v5 (Blin et al. 2019) with all features turned on. Functional domain annotations were conducted using eggNOG-mapper v5 (Huerta-Cepas et al. 2017, 2019) on default settings and InterProScan v5 (Jones et al. 2014) with the –goterms flag. Phobius v1.01 (Käll et al. 2007) was used to assist in prediction of secreted proteins. In addition to these analyses Funannotate also performed domain annotations through an HMMer search against the Pfam-A database and dbCAN CAZYmes database, a BlastP search against the MEROPS protease database, and secreted protein predictions with SignalP v4.1 (Nielsen 2017).
For downstream analysis, proteins were classified as secreted proteins if they had signal peptides detected by both Phobius and SignalP and did not possess a transmembrane domain as predicted by Phobius and an additional analysis of TMHMM v2.0 (Krogh et al. 2001). Effector proteins were identified by using EffectorP v2.0 (Sperschneider et al. 2018), with default settings, on the set of secreted proteins for each genome. Transmembrane proteins were identified if both Phobius and TMHMM detected transmembrane domains. Secondary metabolite proteins were identified if they resided within metabolite clusters predicted by antiSMASH. Proteins were classified as having conserved protein domains if they contained any Pfam or IPR domains.
Gene Family Identification and Classification
OrthoFinder v2.3.3 (Emms and Kelly 2019) was run on default settings using Diamond v0.9.25.126 (Buchfink et al. 2015) to infer groups of orthologous gene clusters (orthogroups) based on protein homology and Markov Cluster Algorithm (MCL) clustering. To more accurately place closely related genes into clusters an additional 78 fungal genomes (supplementary table S3, Supplementary Material online) with emphasis on plant associated fungi of the order Hypocreales were added. To standardize, all 78 additional genomes were subject to a protein length cutoff of 50 amino acids and genomes downloaded from http://www.endophyte.uky.edu had alternatively spliced proteins removed. For downstream analysis, orthogroups pertaining to the 53 Claviceps genomes were classified as secreted, predicted effectors, transmembrane, metabolite, and conserved domain orthogroups if ≥50% of the Claviceps strains present in a given cluster had at least one protein classified as such.
Phylogeny and Genome Fluidity
Phylogenetic relationship of all 53 Claviceps genomes, with Fusarium graminearum, F. verticillioides, Epichloe festucae, and E. typhina as outgroups, was derived from 2,002 single-copy orthologs obtained from our OrthoFinder defined gene clusters (described above). This resulted in a data set of 114,114 amino acids sequences that were concatenated to create a supermatrix and aligned using MAFFT v7.429 (Katoh and Standley 2013) on default settings. Uninformative sites were removed using Gblocks v0.91 (Castresana 2000) on default settings. Due to the large scale of the alignment maximum likelihood reconstruction was performed using FastTree v2.1.11 (Price et al. 2010) using the Whelan and Goldman matrix model of amino acid substitution with the –gamma, –spr 4, –mlacc 2, –slownni, and –slow flag with 1,000 bootstraps. MEGA X (Sudhir et al. 2018) was used for neighbor joining (NJ) reconstruction using the Jones, Taylor, and Thorton matrix model of amino acid substitution with gamma distribution and maximum parsimony (MP) reconstruction using the tree bisection reconstruction (TBR) algorithm with 100 repeated searches. Nodal support for both NJ and MP reconstructions were assessed with 1,000 bootstraps. In addition, an alignment and maximum likelihood (ML) reconstruction was performed on each of the 2,002 protein sequences following the procedure as above (MAFFT, Gblocks, FastTree). A density consensus phylogeny was created from all gene trees using the program DensiTree v2.2.5 (Bouckaert and Heled 2014). PhyBin v0.3-1 (Newton RR and Newton IL 2013) was used to cluster trees from three data sets (1: Claviceps genus without outgroups, 2: section Pusillae species, and 3: section Claviceps species) together to identify frequencies of concordant topologies using the –complete flag with –editdist = 2. To reduce noise, from abundant incomplete lineage sorting in section Claviceps, we implemented a –minbranchlen = 0.015 for our Claviceps genus data set.
Following methodologies established in Kislyuk et al. (2011) genomic fluidity, which estimates the dissimilarity between genomes by using ratios of the number of unique gene clusters to the total number of gene clusters in pairs of genomes averaged over randomly chosen genome pairs from within a group on N genomes, was used to assess gene cluster dissimilarity within the Claviceps genus. For a more detailed description refer to Kislyuk et al. (2011). Data sets containing gene clusters from representative members of section Pusillae, section Claviceps, Clavieps genus, and all C. purpurea strains were extracted from our OrthoFinder defined gene clusters. Additional species- and genus-wide gene cluster data sets from the additional 78 fungal genomes were extracted for comparative purposes. All section- and genus-wide data sets contained one representative isolate from each species to reduce phylogenetic bias. Each extracted data set was used to calculate the genomic fluidity using a custom script (https://github.com/PlantDr430/CSU_scripts/blob/master/pangenome_fluidity.py). The result files for each data set were then used for figure creation and two-sample two-sided z test statistics (Kislyuk et al. 2011) using a custom script (https://github.com/PlantDr430/CSU_scripts/blob/master/combine_fluidity.py).
Gene Density Compartmentalization
A custom script (https://github.com/PlantDr430/CSU_scripts/blob/master/genome_speed_hexbins.py) was used to calculate local gene density measured as 5′ and 3′ flanking distances between neighboring genes (intergenic regions). To statistically determine whether specific gene types had longer intergenic flanking regions than all other genes within the genome we randomly sampled 100 each group of genes (specific gene vs. other genes) 1,000 times for both the 5′ and 3′ flanking distances. Mann–Whitney U test was used to test for significance on all 2,000 subsets corrected with Benjamini–Hochberg. Corrected P values were averaged per flanking side and then together to get a final P value. Genes that appeared on a contig alone were excluded from analysis (supplementary table S4, Supplementary Material online). For graphical representation, genes that were located at the start of each contig (5′ end) were plotted along the x axis, whereas genes located at the end of each contig (3′ end) were plotted along the y axis.
RIP and Blast Analyses
For all 53 genomes a self-BlastP v2.9.0+ search was conducted to identify best hit orthologs within each genome with a cutoff e-value of 10−5 and removal of self-hits. This process was automated using a custom script (https://github.com/PlantDr430/CSU_scripts/blob/master/RIP_blast_analysis.py). We further examined if gene pairs with a pairwise identity of ≥80% were located next to each other and/or separated by five or fewer genes. Fifty-six important Claviceps genes (supplementary table S7, Supplementary Material online) including the rid-1 homolog (Freitag et al. 2002) were used in a BlastP analysis to identify the number of genes present that passed an e-value cutoff of 10−5, 50% coverage, and 35% identity. Genes that appeared as best hits for multiple query genes were only recorded once for their overall best match. In addition, the web-based tool The RIPper (Van Wyk et al. 2019) was used on default settings (1-kb windows in 500-bp increments) to scan whole genomes for presence of RIP and large RIP affected regions (LRARs).
Statistical Programs and Plotting
Statistics and figures were generated using Python3 modules SciPy v1.3.1, statsmodel v0.11.0, and Matplotlib v3.1.1. Heatmaps were generated using ComplexHeatmap v2.2.0 in R (Gu 2016).
Results
Genome Assembly and Annotation
To provide a comprehensive view of variability across Claviceps, we sequenced and annotated 50 genomes (19 Claviceps spp.), including C. citrina the single species of section Citrinae, six species belonging to section Pusillae, and 44 genomes (12 species) belonging to section Claviceps, of which 23 genomes belong to C. purpurea s.s. (table 1 and supplementary table S1, Supplementary Material online). The assemblies and annotations were of comparable quality to the reference strains (table 1). A more detailed representation of the assembly and annotation statistics can be seen in table 1 and supplementary figure S1 and table S2, Supplementary Material online.
Table 1.
Assembly and Annotations Statistics for the Three Reference Claviceps Genomes and the 50 Claviceps Genomes Used in This Study
| Organism | Strain | Section | Host of Origin |
Read Coverage | Genome size (Mb) | Contig (#) | N50 | Genomic GC (%) | TE Content (%) | Gene Count | BUSCO Completeness |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family/Tribe | Genus/Species | Dikarya (%) | Sordario-myceta (%) | ||||||||||
| References | |||||||||||||
| C. purpruea | 20.1 | Claviceps | Triticeae | Secale cereale | — | 32.1 | 1,442b | 46,498b | 51.6 | 10.9 | 8,703 | 95.30 | 94.70 |
| C. fusiformis | PRL1980 | Pusillae | Paniceae | Pennisetum typhoideum | — | 52.3 | 6,930 | 19,980 | 37.3 | 47.5 | 9,304 | 96.70 | 94.90 |
| C. paspali | RRC1481 | Paspalorum | Paspaleae | Paspalum sp. | — | 28.9 | 2,304 | 26,898 | 47.7 | 17.5 | 8,400 | 94.30 | 93.30 |
| This study | |||||||||||||
| C. purpruea | Clav04 | Claviceps | Bromeae | Bromus inermis | 46× | 31.8 | 3,288 | 21,051 | 51.7 | 10.1 | 8,824 | 95.50 | 94.10 |
| C. purpruea | Clav26 | Claviceps | Triticeae | Hordeum vulgare | 59× | 30.8 | 1,361 | 49,697 | 51.7 | 9.1 | 8,737 | 97.70 | 96.50 |
| C. purpruea | Clav46 | Claviceps | Triticeae | Secale cereale | 58× | 30.8 | 1,409 | 49,302 | 51.7 | 9.7 | 8,597 | 98.00 | 96.60 |
| C. purpruea | Clav55 | Claviceps | Poeae | Lolium perenne | 59× | 30.7 | 1,525 | 44,299 | 51.8 | 9.8 | 8,480 | 97.10 | 95.90 |
| C. purpruea | LM4 | Claviceps | Triticeae | Tricosecale | 64× | 30.6 | 1,296 | 47,441 | 51.8 | 10.0 | 8,470 | 97.00 | 95.80 |
| C. purpruea | LM5 | Claviceps | Triticeae | Hordeum vulgare | 67× | 30.5 | 1,258 | 51,505 | 51.8 | 9.0 | 8,508 | 96.90 | 95.50 |
| C. purpruea | LM14 | Claviceps | Triticeae | Hordeum vulgare | 49× | 30.6 | 1,297 | 49,955 | 51.8 | 10.0 | 8,422 | 97.40 | 95.60 |
| C. purpruea | LM28 | Claviceps | Triticeae | Triticum aestivum | 49× | 30.6 | 1,343 | 51,635 | 51.7 | 9.6 | 8,713 | 97.30 | 96.10 |
| C. purpruea | LM30 | Claviceps | Triticeae | Secale cereale | 64× | 30.6 | 1,224 | 51,374 | 51.8 | 9.4 | 8,526 | 97.00 | 95.50 |
| C. purpruea | LM33 | Claviceps | Triticeae | Secale cereale | 45× | 30.5 | 1,398 | 44,564 | 51.8 | 9.2 | 8,557 | 96.30 | 95.50 |
| C. purpruea | LM39 | Claviceps | Triticeae | Triticum turgidum subsp. durum | 81× | 30.5 | 1,282 | 48,443 | 51.8 | 10.1 | 8,591 | 97.10 | 96.10 |
| C. purpruea | LM46 | Claviceps | Triticeae | Triticum turgidum subsp. durum | 79× | 30.6 | 1,291 | 50,932 | 51.8 | 9.6 | 8,455 | 97.00 | 95.80 |
| C. purpruea | LM60 | Claviceps | Poeae | Avena sativa | 81× | 30.6 | 1,259 | 47,464 | 51.7 | 9.3 | 8,498 | 97.00 | 95.80 |
| C. purpruea | LM71 | Claviceps | Poeae | Alopercurus myosuroides | 168× | 30.5 | 1,400 | 45,114 | 51.8 | 9.6 | 8,472 | 97.10 | 95.60 |
| C. purpruea | LM207 | Claviceps | Triticeae | Elymus repens | 53× | 30.5 | 1,352 | 45,388 | 51.8 | 9.2 | 8,475 | 97.00 | 95.70 |
| C. purpruea | LM223 | Claviceps | Bromeae | Bromus riparius | 74× | 30.8 | 1,297 | 46,577 | 51.7 | 10.5 | 8,438 | 97.00 | 95.70 |
| C. purpruea | LM232 | Claviceps | Poeae | Phalaris canariensis | 53× | 30.7 | 1,348 | 49,571 | 51.7 | 9.4 | 8,512 | 96.60 | 95.70 |
| C. purpruea | LM233 | Claviceps | Poeae | Phalaris canariensis | 49× | 30.6 | 1,331 | 50,327 | 51.8 | 9.9 | 8,717 | 96.70 | 95.90 |
| C. purpruea | LM461 | Claviceps | Triticeae | Elymus repens | 37× | 30.5 | 1,440 | 44,216 | 51.8 | 8.4 | 8,656 | 96.60 | 95.20 |
| C. purpruea | LM469 | Claviceps | Triticeae | Triticum aestivum | 75× | 30.5 | 1,257 | 48,403 | 51.8 | 10.0 | 8,394 | 97.30 | 96.00 |
| C. purpruea | LM470 | Claviceps | Triticeae | Elymus repens | 26× | 30.5 | 1,797 | 32,579 | 51.8 | 9.0 | 8,591 | 96.50 | 95.30 |
| C. purpruea | LM474 | Claviceps | Triticeae | Hordeum vulgare | 64× | 30.6 | 1,354 | 47,245 | 51.8 | 9.4 | 8,500 | 96.80 | 95.70 |
| C. purpruea | LM582 | Claviceps | Triticeae | Secale cereale | 89× | 30.7 | 1,600 | 39,003 | 51.8 | 9.6 | 8,518 | 97.20 | 95.40 |
| C. aff. purpruea | Clav52 | Claviceps | Poeae | Poa pratensis | 60× | 29.6 | 1,334 | 48,893 | 51.8 | 8.2 | 8,316 | 96.80 | 96.20 |
| C. quebecensis a | Clav32 | Claviceps | Triticeae | Hordeum vulgare | 64× | 28.7 | 1,068 | 58,118 | 51.6 | 4.5 | 8,232 | 98.00 | 96.60 |
| C. quebecensis a | Clav50 | Claviceps | Triticeae | Elymus sp. | 59× | 28.8 | 1,075 | 66,795 | 51.6 | 6.9 | 8,046 | 97.50 | 96.30 |
| C. quebecensis a | LM458 | Claviceps | Poeae | Ammophila (plant) | 78× | 28.4 | 1,166 | 45,693 | 51.6 | 6.1 | 8,055 | 97.10 | 95.80 |
| C. occidentalis a | LM77 | Claviceps | Poeae | Phleum pratense | 58× | 28.7 | 1,728 | 29,222 | 51.4 | 6.0 | 8,162 | 96.10 | 94.70 |
| C. occidentalis a | LM78 | Claviceps | Bromeae | Bromus inermis | 64× | 28.8 | 1,689 | 29,608 | 51.4 | 6.0 | 8,231 | 95.80 | 94.70 |
| C. occidentalis a | LM84 | Claviceps | Bromeae | Bromus inermis | 164× | 28.9 | 1,404 | 36,685 | 51.4 | 6.0 | 8,221 | 97.00 | 95.40 |
| C. ripicola a | LM218 | Claviceps | Poeae | Phalaris arundinacea | 146× | 31.1 | 1,072 | 60,464 | 51.4 | 10.3 | 8,327 | 96.70 | 95.70 |
| C. ripicola a | LM219 | Claviceps | Poeae | Phalaris arundinacea | 55× | 30.8 | 1,239 | 55,312 | 51.4 | 9.5 | 8,381 | 96.80 | 95.80 |
| C. ripicola a | LM220 | Claviceps | Poeae | Phalaris arundinacea | 91× | 30.9 | 1,223 | 54,100 | 51.4 | 9.3 | 8,449 | 97.10 | 95.90 |
| C. ripicola a | LM454 | Claviceps | Poeae | Ammophila breviligulata | 156× | 31.2 | 1,508 | 40,844 | 51.4 | 8.4 | 8,562 | 97.10 | 96.10 |
| C. spartinae | CCC535 | Claviceps | Zoysieae | Sporobolus anglicus | 60× | 29.3 | 1,456 | 42,688 | 51.4 | 7.1 | 8,433 | 97.50 | 95.90 |
| C. arundinis | LM583 | Claviceps | Molinieae | Phragmites australis | 69× | 30.6 | 996 | 70,672 | 51.4 | 9.8 | 8,235 | 96.80 | 95.70 |
| C. arundinis | CCC1102 | Claviceps | Molinieae | Phragmites australis | 61× | 30.3 | 896 | 91,905 | 51.4 | 8.3 | 8,486 | 97.70 | 96.50 |
| C. humidiphila | LM576 | Claviceps | Poeae | Dactylis sp. | 77× | 31.2 | 1,236 | 55,717 | 51.5 | 9.9 | 8,440 | 97.00 | 95.90 |
| C. perihumidiphila a | LM81 | Claviceps | Triticeae | Elymus albicans | 140× | 31.2 | 1,003 | 67,487 | 51.5 | 11.0 | 8,291 | 97.10 | 95.90 |
| C. cyperi | CCC1219 | Claviceps | Cyperaceae (family) | Cyperus esculentus | 56× | 26.6 | 1,921 | 27,113 | 51.7 | 8.9 | 7,673 | 97.70 | 95.40 |
| C. capensis | CCC1504 | Claviceps | Ehrharteae | Ehrharta villosa | 66× | 27.7 | 1,136 | 59,777 | 51.7 | 6.2 | 8,037 | 97.60 | 95.70 |
| C. pazoutovae | CCC1485 | Claviceps | Stipeae | Stipa dregeana | 61× | 27.6 | 1,304 | 42,785 | 51.7 | 6.8 | 7,941 | 97.50 | 96.00 |
| C. monticola | CCC1483 | Claviceps | Brachypodieae | Brachypodium sp. | 58× | 27.8 | 1,144 | 56,619 | 51.6 | 7.0 | 7,977 | 98.10 | 96.50 |
| C. pusilla | CCC602 | Pusillae | Andropogoneae | Bothriochloa insculpta | 52× | 45.9 | 5,068 | 15,010 | 40.4 | 42.1 | 8,735 | 90.90 | 88.30 |
| C. lovelessii | CCC647 | Pusillae | Eragostidinae | Eragrostis sp. | 53× | 41.1 | 5,300 | 12,480 | 42.1 | 33.9 | 8,862 | 91.60 | 88.20 |
| C. digitariae | CCC659 | Pusillae | Paniceae | Digitaria eriantha | 57× | 33.4 | 1,773 | 32,638 | 44.8 | 20.0 | 8,285 | 95.90 | 94.70 |
| C. maximensis | CCC398 | Pusillae | Paniceae | Megathyrsus maximus | 58× | 33.0 | 829 | 81,956 | 44.9 | 19.8 | 7,943 | 98.30 | 96.50 |
| C. sorghi | CCC632 | Pusillae | Andropogoneae | Sorghum bicolor | 60× | 35.6 | 3,660 | 16,225 | 44.4 | 30.4 | 8,208 | 89.90 | 87.10 |
| C. africana | CCC489 | Pusillae | Andropogoneae | Sorghum bicolor | 56× | 37.7 | 1,781 | 37,639 | 42.5 | 34.0 | 8,119 | 95.00 | 91.50 |
| C. citrine | CCC265 | Citrinae | Cynodonteae | Distichlis spicata | 64× | 43.5 | 4,772 | 16,294 | 41.5 | 51.7 | 7,821 | 92.20 | 88.20 |
Note.—TE content represented as percent of the genome masked by TEs.
Newly identified species (Liu et al. 2020).
The reference strain C. purpurea 20.1 was additionally assembled into 191 scaffolds with a scaffold N50 of 433,221.
Overall, species of section Claviceps had better assemblies and annotations than species of other sections regarding contig numbers, N50’s, and BUSCO completeness scores (table 1). Nearly all species of section Claviceps showed higher BUSCO scores than the references, whereas species of sections Pusillae and Citrinae generally showed lower scores, likely due to their higher TE content (average 34.9 ± 11.0%, table 1). Exceptions to the low BUSCO scores were C. digitariae and C. maximensis (sect. Pusillae), which had lower TE content, 20.0% and 19.8%, respectively, than the rest of the species in section Pusillae (table 1). Although, C. africana (sect. Pusillae, TE content = 34.0%) also had comparable BUSCO scores, to the references, with a higher N50 and lower contig number, than the rest of the species in section Pusillae (table 1). Despite the differences in assembly quality between species of section Pusillae, the genomic findings reported in this study were found to be comparable between members of this section indicating that both higher quality and lower quality genomes of section Pusillae provided similar results.
Phylogenomics and Genome Fluidity
Orthologous gene clusters (orthogroups), which contain orthologs and paralogs, were inferred from protein homology and MCL clustering using OrthoFinder. Across the 53 Claviceps isolates and outgroups species Fusarium graminearum, F. verticillioides, Epichloe festucae, and E. typhina, we identified 2,002 single-copy orthologs. We utilized a supermatrix approach to infer an ML species tree, based on these protein sequences. Results showed statistical support for four sections of Claviceps with a near concordant topology to the Bayesian five-gene phylogeny in Píchová et al. (2018). In addition, our topology of section Claviceps is concordant with a larger multilocus phylogeny of the section (Liu et al. 2020). Our ML topology was also supported by NJ and maximum parsimony supermatrix analyses (supplementary fig. S2 and S3, Supplementary Material online). Notable exceptions were the placement of C. paspali (sect. Paspalorum) which grouped closer to C. citrina (sect. Citrinae) instead of section Claviceps, and C. pusilla which grouped closer to C. fusiformis instead of C. maximensis (fig. 1). We also found that section Claviceps diverged from a common ancestor with section Pusillae as opposed to section Paspalorum. Our results provide support for the deeply divergent lineages of sections Pusillae, Paspalorum, and Citrinae with a long divergent branch resulting in section Claviceps (fig. 1).
Fig. 1.
ML phylogenetic reconstruction of the Claviceps genus using amino acid sequences of 2,002 single copy orthologs with 1000 bootstrap replicates. Pink dots at branches represent bootstrap values ≥95. Arrows and descriptions indicate potential changes in genomic architecture between Claviceps sections identified in this study.
Each of the 2,002 single-copy orthologs were also independently aligned and analyzed in the same manner as our supermatrix phylogeny from representative isolates of each species. A density consensus tree of all 2,002 topologies was concordant with our supermatrix analysis but reveals evidence of incongruencies, particularly within section Claviceps (supplementary fig. S4, Supplementary Material online), which could be caused by biological, analytical, and sampling factors (Steenwyk et al. 2019). Although grouping of species generally held true to figure 1, variation was more related to the order of branches, with C. cyperi, C. arundinis, C. humidiphila, and C. perihumidiphila showing the most variability. These results indicate the presence of some incongruencies within section Claviceps, section Pusillae, and across the genus (supplementary fig. S5–S7, Supplementary Material online) but a consensus supporting our ML species tree (fig. 1 and supplementary fig. S4, Supplementary Material online). There are several potential causes of these incongruencies that are currently the focal point of an ongoing study.
To further elucidate trends of divergence within the genus, we examined genomic fluidity (Kislyuk et al. 2011) using all 82,267 orthogroups from our previous OrthoFinder analysis. Genomic fluidity estimates the dissimilarity between genomes by using ratios of the number of unique orthogroups to the total number of orthogroups in pairs of genomes averaged over randomly chosen genome pairs from within a group on N genomes. For example, a fluidity value of 0.05 indicates that randomly chosen pairs of genomes in a group will on average have 5% unique orthogroups and share 95% of their orthogroups (Kislyuk et al. 2011). Section Claviceps, which is composed of 12 different species, showed a relatively small genomic fluidity (0.0619 ± 0.0019) with limited variation, indicating pairwise orthogroup dissimilarity between randomly sampled genomes was quite low. The amount of variation between 12 different Claviceps species was similar to the variation between 24 C. purpurea s.s. isolates, however, the fluidities were significantly different (P < 0.0001; supplementary table S5, Supplementary Material online). In comparison, the fluidity of section Pusillae (0.126 ± 0.014; P < 0.0001; supplementary table S5, Supplementary Material online) was two times greater than the fluidity of section Claviceps and exhibited greater variation, indicating greater dissimilarities in orthogroups between randomly sampled species of section Pusillae.
Overall, our ML phylogeny (fig. 1) and genome fluidity analysis (fig. 2) indicate a large evolutionary divergence separating section Claviceps. Our subsequent analyses of the genomic architecture of all Claviceps species examine factors that could be associated with the evolutionary divergence of section Claviceps and those driving cryptic speciation.
Fig. 2.
Genomic fluidity (dashed lines) for specified groups within the order Hypocreales. Species level groups contain multiple isolates of a given species, whereas section and genus level groups contain one strain from representative species to remove phylogenetic bias. Shaded regions represent standard error and were determined from total variance, containing both the variance due to the limited number of samples genomes and the variance due to subsampling within the sample of genomes. Letters correspond to significant difference between fluidities determined through a two-sided two-sample z test (P < 0.05; supplementary table S4, Supplementary Material online). Legend is in descending order based on fluidity, and names are additionally appended to mean lines for clarity.
TE Divergences and Locations
Due to variation in sequencing platforms that generated the genome data, we examined the relationship of sequence quality with predicted TE content to test for potential biases. Results identified two clusters of genomes with differing sequence qualities, which was determined to be a result of the sequencer used. Although these differences existed, analysis of each cluster showed a lack of relationship between sequence quality and TE content (supplementary fig. S8, Supplementary Material online). In addition, section Claviceps samples were sequenced with both sequencers and results were highly comparable between these samples (reported below), indicating no sequence quality bias.
TE divergence landscapes revealed an overrepresentation of LTR elements in sections Pusillae, Citrinae, and Paspalorum. All three sections showed a similar large peak of LTRs with divergences between 5% and 10% (fig. 3 and supplementary fig. S9, Supplementary Material online), indicating a relatively recent expansion of TEs. The landscapes of sections Pusillae, Citrinae, and Paspalorum are in striking contrast to species of section Claviceps that showed more similar abundances of LTR, DNA, LINE, SINE, and RC (helitron) elements. Species of section Claviceps showed broader peaks of divergence between 5% and 30% but also showed an abundance of TEs with ∼0% divergence suggesting very recent TE expansion (fig. 3 and supplementary fig. S9, Supplementary Material online). The TE landscape of C. cyperi showed a more striking peak of divergence between 5% and 10% that more closely resembled the TE divergences of sections Pusillae, Paspalorum, and Citrinae. However, the content of the TE peak in C. cyperi largely contained DNA, LINE, and unclassified TEs as opposed to LTR’s (supplementary fig. S9, Supplementary Material online).
Fig. 3.
TE fragment divergence landscapes for representative species of each Claviceps section; C. purpurea 20.1 (sect. Claviceps), C. maximensis CCC398 (sect. Pusillae), C. paspali RRC1481 (sect. Paspalorum), and C. citrina (sect. Citrinae). Stacked bar graphs show the nonnormalized sequence length occupied in each genome (y axis) for each TE type based on their percent divergence (x axis) from their corresponding consensus sequence. Landscape for all remaining isolates can be seen in supplementary figure S8, Supplementary Material online.
To identify where genes were located in relation to TEs, we calculated the average distance (kb) of each gene to the closest TE fragment. This analysis was performed for predicted effectors, secreted (noneffector) genes, secondary metabolite (nonsecreted) genes, and all other genes. Secreted genes and predicted effectors of sections Claviceps and Pusillae species were found to be significantly closer to TEs compared with other genes within each respective section (fig. 4; P < 0.0001), suggesting that these genes could be located in more repeat-rich regions of the genome. It should be noted that we did observe a significant difference (P < 0.001, Welch’s test) in TE content between section Pusillae (32.5 ± 9.59%) and section Claviceps (8.79 ± 1.52%). In both sections Claviceps and Pusillae, secondary metabolite genes were located farther away from TEs (fig. 4; P < 0.0001), that is, repeat-poor regions of the genome. These trends hold true for individual isolates, with a notable exception of C. pusilla (sect. Pusillae) showing no significant differences in the proximity of TEs to specific gene types (P > 0.12; supplementary fig. S10, Supplementary Material online). Variation existed in whether particular isolates had significant differences between all other genes compared with secreted genes and secondary metabolite genes, but all species in sections Claviceps and Pusillae (aside from C. pusilla) had predicted effector genes located significantly closer to TEs (P < 0.003; supplementary fig. S10, Supplementary Material online). No significant differences in the proximity of TEs to specific gene types were observed in sections Citrinae and Paspalorum (fig. 4; P > 0.11), suggesting that TE’s are more randomly distributed throughout these genomes.
Fig. 4.
Boxplot distributions of predicted effectors, secreted (noneffectors), secondary metabolite (nonsecreted) genes, and other genes (i.e., genes that are not effectors, secreted, or secondary [2°] metabolite genes) in Claviceps sections showing the mean distance (kb) of each gene to the closest TE fragment (5′ and 3′ flanking distances were averaged together). Kruskal–Wallis (P value: *<0.05, **<0.01, ***<0.001, n.s. = not significant). Pairwise comparison was performed with Mann–Whitney U test with Benjamini–Hochberg multitest correction. Letters correspond to significant differences between gene categories within sections (P < 0.05). Plots for all individual isolates can been seen in supplementary figure S9, Supplementary Material online.
Gene Density Compartmentalization
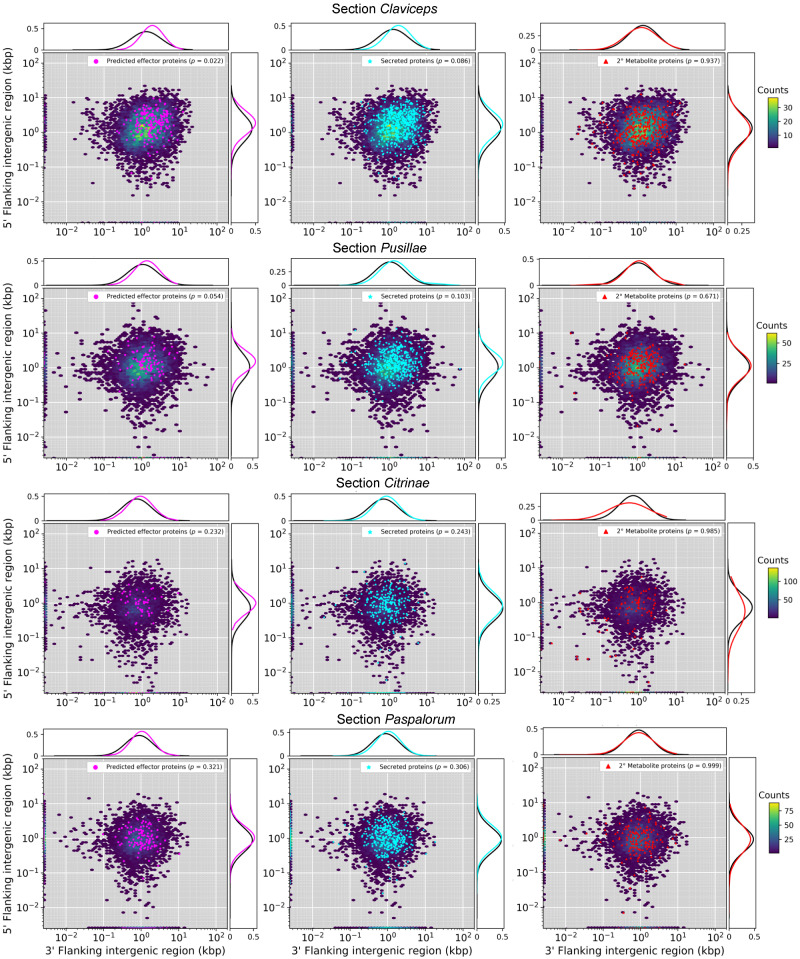
To further examine genome architecture, we analyzed local gene density measured as flanking distances between neighboring genes (intergenic regions) to examine evidence of gene density compartmentalization (i.e., clustering of genes with differences in intergenic lengths) within each genome. Results showed that all 53 Claviceps strains exhibited a one-compartment genome (lack of multiple compartments of genes with different intergenic lengths). Although, there was a tendency for more genes with larger intergenic regions in sections Claviceps and Pusillae compared with sections Citrinae and Paspalorum (fig. 5; supplementary fig. S11, Supplementary Material online).
Fig. 5.
Gene density as a function of flanking 5' and 3' intergenic region size (y- and x axis) of representative isolates of each of the four sections within the Claviceps genus; C. purpurea 20.1 (sect. Claviceps), C. maximensis CCC398 (sect. Pusillae), C. paspali RRC1481 (sect. Paspalorum), and C. citrina (sect. Citrinae). Colored hexbins indicate the intergenic lengths of all genes with color code indicating the frequency distribution (gene count) according to the legend on the right. Overlaid markers indicate specific gene types corresponding to legends in the top right within each plot. Line graphs (top and right of each plot) depict the frequency distributions of specific gene types (corresponding legend color) and all other genes not of the specific type (black). For visualization purposes, the first genes of contigs (5′ end) are plotted along the x axis and the last gene of each contig (3′ end) are plotted along the y axis. For information on statistical test, see Methods and for plots of all remaining isolates see supplementary figure S10, Supplementary Material online.
To further clarify evolutionary tendencies, we evaluated whether gene types showed a difference in their flanking intergenic lengths compared with other genes within their genomes. Results showed that predicted effector genes in section Claviceps had significantly larger intergenic flanking regions compared with other genes, indicating they may reside in more gene-sparse regions of the genome (P < 0.04, fig. 5, supplementary fig. S11, Supplementary Material online). Only C. digitariae and C. lovelessi (P < 0.01, P = 0.024, respectively; supplementary fig. S11, Supplementary Material online) of section Pusillae had predicted effector genes with significantly larger intergenic regions than other genes, although C. fusiformis and C. pusilla were near significant (fig. 5, P = 0.054, P = 0.056, respectively; supplementary fig. S11, Supplementary Material online). Flanking intergenic lengths of secreted genes also showed larger intergenic lengths and were often significantly larger than other genes in section Claviceps (fig. 5; supplementary fig. S11, Supplementary Material online). In contrast, secondary metabolite genes exhibited a widespread distribution of intergenic lengths that were not significantly different than other genes in all 53 Claviceps strains (P > 0.55, fig. 5; supplementary fig. S11, Supplementary Material online).
RIP Analysis
To test for effects of RIP-like signatures, we assessed the bidirectional similarity of genes against the second closest BlastP match within each isolate’s own genome (Galagan et al. 2003; Urguhart et al. 2018), supported by a BlastP analysis against the rid-1 RIP gene of Neurospora crassa, and calculations of RIP indexes in 1-kb windows (500 bp increments) using The RIPper (Van Wyk et al. 2019). Results showed that sections Pusillae, Citrinae, and Paspalorum had homologs of rid-1, fewer genes with close identity (≥80%), on average 27.4 ± 11.4% of their genomes affected by RIP, a mean RIP composite index of −0.03 ± 0.21, and 325 ± 138 LRARs covering 3,984 ± 2,144 kb of their genomes, indicating past or current activity of RIP-like mechanisms (fig. 6; supplementary tables S6–S8, Supplementary Material online). This is further supported by an average GC content of 42.84 ± 3.03% (table 1) in sections Pusillae, Citrinae, and Paspalorum, which is on average 8.81% lower than in section Claviceps that shows an absence of RIP (reported below). The presence of RIP-like mechanisms in sections Pusillae, Citrinae, and Paspalorum was unexpected, given the abundance of TEs within genomes of these sections (table 1, fig. 3, and supplementary fig. S9, Supplementary Material online) as RIP-like mechanisms should be working to silence and inactivate these TEs. Although we did not directly test the activity of TEs within our genomes, due to lack of RNAseq data, the peaks of low TE nucleotide divergence (<10%) in sections Pusillae, Citrinae, and Paspalorum (fig. 3, supplementary fig. S9, Supplementary Material online) suggest recent activity of TEs (Frantzeskakis et al. 2018).
Fig. 6.
Representative isolates of each Claviceps species showing the fraction of Blast hits at a given % identity (y axis) within each isolate (z axis) at a given percent identity (x axis) from the second closet BlastP match of proteins within each isolate’s own genome. Two C. purpruea s.s. isolates are shown to compare a newly sequenced genome versus the reference.
In comparison, species in section Claviceps lack rid-1 homologs, showed larger amounts of gene similarity, and a general lack of evidence of RIP-like signatures with only 0.13 ± 0.03% of their genomes putatively affected by RIP, and a mean RIP composite index of −0.59 ± 0.01 suggesting that RIP-like mechanisms are inactive (fig. 6 and supplementary tables S6–S8, Supplementary Material online). Gene pairs sharing a ≥80% identity to each other were often located near each other. On average 27.02 ± 5.91% of the pairs were separated by five or fewer genes, and 15.95 ± 3.50% of the pairs were located next to each other, indicating signs of tandem gene duplication within the section (supplementary table S6, Supplementary Material online). C. cyperi showed the smallest proportions of highly similar tandem genes (7.77% and 5.7%) compared with other species within section Claviceps. Additional variations in the proportions of highly similar tandem genes between other species of section Claviceps were not evident as these proportions appeared to vary more between isolate than species (supplementary table S6, Supplementary Material online).
Gene Cluster Expansion
The proteome of Claviceps genomes were used to infer orthologous gene clusters (orthogroups) through protein homology and MCL clustering using OrthoFinder. Our results revealed evidence of orthogroup expansion within section Claviceps as species contained more genes per orthogroup than species of the other three sections (supplementary fig. S12, Supplementary Material online). To identify the types of gene clusters that were showing putative expansion, we filtered our clusters by following two criteria: 1) at least one isolates had two or more genes in the orthogroup and 2) there was a significant difference in the mean number of genes per orthogroup between all 44 isolates in section Claviceps and the 9 isolates from sections Pusillae, Citrinae, and Paspalorum (α ≤ 0.01, Welch’s test).
Overall, we identified 863 (4.7%) orthogroups showing putative expansion. We observed extensive expansion (orthogroups with observations of greater than or equal to ten genes per isolate) present in many unclassified, predicted effectors, secreted (noneffector) orthogroups, and orthogroups encoding genes with conserved domains (fig. 7 and supplementary figs. S13 and S14, Supplementary Material online). Transmembrane orthogroups also showed evidence of expansion with several isolates having five to ten genes. Orthogroups with secondary metabolite genes showed the lowest amount of expansion (supplementary fig. S15, Supplementary Material online). Overall, section Claviceps showed expansion in a greater number of orthogroups than section Pusillae, Citrinae, and Paspalorum in all categories except transmembranes (supplementary fig. S15, Supplementary Material online). Orthogroups with an average greater than or equal to five genes per isolate, within section Claviceps, contained a variety of functional proteins, with generally more proteins encoding protein/serine/tyrosine kinase domains (supplementary table S9, Supplementary Material online). Additional details can be obtained from supplementary tables S10 (ordered orthogroups corresponding to heatmaps; fig. 7 and supplementary figs. S13 and S14, Supplementary Material online), S11-1, and S11-2, Supplementary Material online (orthogroups identification and functional annotation of all proteins).
Fig. 7.
Heatmap of gene counts in orthogroups for all 53 Claviceps strains ordered based on ML tree in figure 1 and separated by sections. Orthogroups are separated based on their classification and are only represented once (i.e., secondary [2°] metabolite orthogroups shown are those that are not already classified into the effector or secreted orthogroups) and are ordered based on hierarchical clustering, see supplementary table S9, Supplementary Material online, for list of orthogroups corresponding to the order shown in the heatmaps. The host spectrum (right) is generalized across species, as no literature has determined the existence of race specific isolates within species, is shown on the left side of the figure determined from literature review of field collected samples (Supplementary Material in Píchová et al. 2018) and previous inoculation tests Campbell (1957) and Liu et al. (2020). For heatmap of conserved domains, see supplementary figure S12, Supplementary Material online, and for unclassified gene families, see supplementary figure S13, Supplementary Material online.
Within section Claviceps patterns of gene counts per orthogroup appeared to break down and contain variations in the number of genes per orthogroups with some presence/absences occurring between isolates and species. Notably, C. cyperi (CCC1219) showed the lowest amount of expansion, across all taxa, in comparison with other species of section Claviceps. In addition, C. spartinae (CCC535), C. capensis (CCC1504), C. monticola (CCC1483), C. pazoutovae (CCC1485), C. occidentalis (LM77, 78, 84), and C. quebecensis (LM458, Clav32, 50) also showed lower expansion (fig. 7, supplementary figs. S13 and S14, Supplementary Material online). However, no patterns were observed linking the variation in expansions with the literature determined host range of different species within section Claviceps.
Discussion
Our comparative study of 50 newly annotated genomes from four sections of Claviceps has provided us with an enhanced understanding of evolution in the genus through knowledge of factors associated with its diversification. Our results have revealed that despite having nearly identical life strategies, these closely related species have substantially altered genomic architecture and plasticity, which may drive genome adaptation. One key difference we observe is a shift from aspects that are characteristic of a one-speed genome (i.e., less adaptable) in narrow host-range Claviceps species (sects. Citrinae and Paspalorum) toward aspects that are characteristic of a two-speed genome (i.e., more adaptable) in broader host-range lineages of sections Pusillae and Claviceps (fig. 1; Dong et al. 2015; Frantzeskakis et al. 2019).
The oldest divergent species of the genus (Píchová et al. 2018), C. citrina (sect. Citrinae) and C. paspali (sect. Paspalorum), are characterized by a proliferation of TEs, particularly LTRs, which do not appear to be colocalized around particular gene types (fig. 4). Coupled with a lack of large-scale genome compartmentalization (fig. 5), these two species can be considered to fit with aspects of a one-speed genome which are often considered to be less adaptable and potentially more prone to being purged from the biota (Dong et al. 2015; Frantzeskakis et al. 2019). This could help explain the paucity of section lineages and restricted host range to one grass tribe, as similar patterns of large genome size, abundant TE content, and equal distribution of TEs has been observed in the specialized barley pathogen Blumeria graminis f.sp. hordei (Frantzeskakis et al. 2018). Although, rapid adaptive evolution within B. graminis f.sp. hordei, has been suggested to occur through copy-number variation and/or heterozygosity of effector loci (Dong et al. 2015; Frantzeskakis et al. 2018, 2019). Our results show a lack of gene duplication occurring in sections Citrinae and Paspalorum likely due to the presence of RIP-like mechanisms. However, even with the presence of RIP-like mechanisms, there was a high LTR content in these species (fig. 3). This suggests that these LTR elements have found a way to avoid RIP-like mechanisms or indicate that these species harbor a less active version of an RIP-like mechanisms as is found in several fungal species (Kachroo et al. 1994; Nakayashiki et al. 1999; Graïa et al. 2001; Ikeda et al. 2002; Chalvet et al. 2003; Kito et al. 2003). Nonetheless, due to the high abundance of TEs (fig. 4) and presence of RIP (fig. 6 and supplementary tables S6 and S7, Supplementary Material online), we hypothesize that aspects of RIP-like “leakage” could be a likely mechanism for evolution in C. citrina and C. paspali (and similarly sect. Pusillae) as has been shown to occur in other fungi (Fudal et al. 2009; Van de Wouw et al. 2010; Hane et al. 2015). It should be noted that since the estimated divergence of section Citrinae 60.5 Ma (Píchová et al. 2018), it has remained monotypic. It was only recently that unknown lineages of section Paspalorum were identified (Oberti et al. 2020), although these lineages were found on the same genera of host as C. paspali (Paspalum spp.) supporting our hypothesis that species within section Paspalorum have restricted host ranges. These recent findings further suggest that lack of additional lineages within these sections could be due to limited records of Claviceps species in South America, where the genus is thought to have originated (Píchová et al. 2018). Further research into South American populations of Claviceps will provide significant insight into the evolution of these two sections.
Members of section Pusillae also exhibited a proliferation of TEs, however, as this section diverged from sections Citrinae and Paspalorum, the genomic architecture evolved such that TEs colocalized around predicted effector genes (fig. 4). This proximity of TEs to effectors persisted in section Pusillae species (except C. pusilla; supplementary fig. S10, Supplementary Material online) and section Claviceps species potentially resulting in the large intergenic regions flanking predicted effector genes (fig. 5, supplementary fig. S11, Supplementary Material online). Together, these genomic alterations indicate aspects of a two-speed genome (Dong et al. 2015; Möller and Stukenbrock 2017). These observed genomic changes may have influenced the divergence and adaptability of sections Pusillae and Claviceps (fig. 1) similar to what has been observed in other fungi (Raffaele and Kamoun 2012; Stukenbrock 2013; Möller and Stukenbrock 2017) and has been proposed to promote genomic flexibility and drive accelerated evolution of these genome compartments (Raffaele et al. 2010; Rouxel et al. 2011; de Jonge et al. 2013; Faino et al 2015, 2016; Seidl et al. 2015). Despite the number of studies that suggest this role of TEs in genome evolution, there has been limited evidence for the mechanism by which TEs drive evolution in filamentous pathogens. However, studies incorporating improved genome assemblies of multiple individuals of a species along with transcriptome data have been able to demonstrate that transcriptionally active TEs were observed in lineage-specific regions of the plant pathogen Verticillium dahliae (Amyotte et al. 2012; Faino et al. 2016), resulting in genomic diversity through large scale duplications in these lineage-specific regions (Faino et al. 2016). This also lead to the frequent loss of the effector Ave1 in populations of V. dahliae, which is located in a TE-rich lineage-specific region (de Jonge et al. 2012).
Although we did not have transcriptome data to determine how many of the TEs are transcriptionally active, our data do show that most of the repetitive elements in section Claviceps species have very low nucleotide divergence (<1%) compared with TEs in sections Pusillae, Paspalorum, and Citrinae (5–20% nucleotide divergence; fig 3), suggesting a recent section specific expansion of TEs that are associated with a recent host range and geographic expansion and proliferation of recently described cryptic species (Liu et al. 2020) within section Claviceps. Similar observations placing TE bursts around speciation times have been reported in the plant pathogen Leptosphaeria maculans (Rouxel et al. 2011; Grandaubert et al. 2014), and the grass-infecting (Blumeria spp.) and dicot-infecting (Erysiphe spp.) powdery mildews (Frantzeskakis et al. 2018). Theoretical models have proposed that repeated changes in phenotypic optimum in a dynamic fitness landscape may induce explosive bursts of transposon activity associated with faster adaptation (Startek et al. 2013). However, long-term maintenance of transposon activity is unlikely, and this may contribute to significant variation in the TE copy number among closely related species. Our findings that the variation in TE copy number between species in the genus Claviceps fits this pattern and call for future studies to clarify the relationship between TE expansion and changes in host range, geographic distribution, and cryptic speciation.
Furthermore, our analyses revealed that a key difference between section Claviceps and section Pusillae is a putative loss of RIP-like mechanisms (figs. 1, 6 and supplementary table S7, Supplementary Material online). In the absence of RIP-like mechanisms, the gene-sparse regions rich in TEs, and effectors could be hot spots for duplication, deletion, and recombination (Galagan et al. 2003; Galagan and Selker 2004; Raffaele and Kamoun 2012; Dong et al. 2015; Faino et al. 2016; Möller and Stukenbrock 2017; Frantzeskakis et al. 2018, 2019). This would explain the observations of tandem gene duplication within the section (figs. 6, 7 and supplementary table S6, figs. S12–S15, Supplementary Material online), which may facilitate rapid speciation, as has been postulated in several smut fungi (Kämper et al. 2006; Schirawski et al. 2010; Dutheil et al. 2016). In fact, C. cyperi, a species of section Claviceps and thought to be ancestral from ancestral state reconstructions of host range (Píchová et al. 2018), showed the least amount of gene cluster expansion and tandem duplication (fig. 7 and supplementary table S6, figs. S13 and S14, Supplementary Material online), indicating that gene duplication may be contributing to the divergence of new species, as other species in section Claviceps have increased genome size, gene count, and number of closely related gene pairs (≥80% identity) (table 1 and supplementary table S6, Supplementary Material online). It is unclear if these changes in gene duplication rate are a selective or neutral mutational process. Because the increased occurrence of gene duplication within section Claviceps is likely a result of a loss of RIP-like mechanisms, it is more plausible to suggest that the change in propensity for gene duplication was a neutral process. However, our evidence of effector duplications suggests that this change in propensity may have allowed an increase chance for future adaptive events. Within section Claviceps gene duplication is likely facilitated by recombination events during annual sexual reproduction (Esser and Tudzynski 1978). Future studies on recombination will be critical to our understanding of the mechanisms driving gene duplication and elucidating factors associated with the observations of potential incomplete lineage sorting (Pease and Hahn 2013) within the section.
Substantially altered genomic architecture and plasticity between Claviceps sections was observed in this study, yet it is unclear whether the evolution of these genomes were caused by contact with new hosts and different climates as ancestral lineages migrated out of South America (Píchová et al. 2018) or if the evolution toward aspects of a two-speed genome provided an advantage in adapting to new hosts or environments. Further research is needed to clarify this point. As sections Pusillae and Claviceps have larger host ranges (5 tribes and 13 tribes, respectively) and increased levels of speciation (Píchová et al. 2018), they represent ideal systems to test this hypothesis. It is postulated that section Pusillae was transferred to Africa (ca. 50.3 Ma), whereas section Claviceps originated in North America (ca. 20.7 Ma), and it is likely that the common ancestor shared between these sections (fig. 1) had strains that were transferred to Africa likely due to insect vectors via transatlantic long-distance dispersal (Píchová et al. 2018). The strains that remained, in South America, likely persisted but appeared to not speciate for roughly 30 Ma (Píchová et al. 2018), despite having aspects of a more adaptable two-speed genome (figs. 4, 5). Limited sampling records could be a factor contributing to this lack of speciation during this 30 Myr period, but it could also be suggested that the ancestral species of sections Claviceps did not diverge due to a lack of diversification of host species (Píchová et al. 2018). It is well known that Claviceps species share a rather unique relationship with their hosts (strict ovarian parasites). The evolution of the Claviceps genus appears to be primarily driven by the evolution and diversification of the host species (Píchová et al. 2018). This can be inferred from divergence time estimates which show that the crown node of section Pusillae aligns with the crown node of PACMAD grasses (ca. 45 Ma) (Bouchenak-Khelladi et al. 2010; Píchová et al. 2018), suggesting that these two organisms radiated in tandem after ancestral strains of section Pusillae were transferred to Africa. Similarly, the estimated crown node of section Claviceps corresponds with the origin of the core Pooideae (Poeae, Triticeae, Bromeae, and Littledaleae), which occurred in North America (ca. 33–26 Ma) (Bouchenak-Khelladi et al. 2010; Sandve and Fjellheim 2010).
Such a large difference between the estimate divergence age (∼30 Myr) and long divergence branch (fig. 1) between section Clavcieps and the other three sections (Píchová et al. 2018) could suggest that a sudden event sparked the adaptive radiation within this section (fig. 1). Under an assumption that ancestral strains of section Claviceps were infecting sedges (Cyperaceae), as is seen in the ancestral C. cyperi (Píchová et al. 2018), a host jump to BOP grasses could have ignited the rapid speciation of section Claviceps, similar to the suggested tandem radiation of section Pusillae with the PACMAD grasses in Africa. However, unknown factors might be responsible for the drastic genomic changes (i.e., putative loss of RIP-like mechanisms) observed in section Claviceps, as no such changes were observed in section Pusillae. The radiation of the core Pooideae occurred after a global supercooling period (ca. 33–26 Ma) in North America. During this period, Pooideae experienced a stress response gene family expansion that enabled adaptation and diversification to cooler, more open, habitats (Kellogg 2001; Sandve and Fjellheim 2010). As gene cluster expansion was observed in section Claviceps (the only section to infect BOP grasses), it suggests that the same environmental factors that caused the radiation of Pooideae could have similarly affected section Claviceps (Kondrashov 2012) and might have resulted in the host jump to Pooideae, and potentially other BOP tribes. Interestingly, one of the orthogroups significantly expanded in section Claviceps (OG0000016) contains proteins associated with a cold-adapted (Alias et al. 2014) serine peptidase S8 subtilase (MER0047718; S08.139) (supplementary table S9, Supplementary Material online). Although the crown node of section Claviceps is estimated at ∼5–10 Myr before the radiation of the core Pooideae, the 95% highest posterior density determined in Píchová et al. (2018) could indicate both radiation events occurred at similar times.
Further examination of Claviceps species in South and Central America needs to be conducted to better elucidate the evolution and dispersal of the genus (Píchová et al. 2018). Efforts should focus on the elusive C. junci, a pathogen of Juncaceae (rushes), which is thought to reside in section Claviceps based on morphological and geographic characteristics (Langdon 1952; Píchová et al. 2018). This species, and potentially others, will provide further insight into the early evolution of section Claviceps and could bridge the current gap between the environmental factors that sparked the radiation of the core Pooideae and section Claviceps. Last, it would be interesting to examine if other phytopathogenic fungal species that diverged in North America ∼20 Ma experienced similar genomic alterations and host range expansions.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr Miroslav Kolařík for providing Claviceps isolates from the Culture Collection of Clavicipitaceae at Institute of Microbiology, Academy of Sciences of the Czech Republic (CCC samples); Parivash Shoukouhi, Dr Jim Menzies, and Zlatko Popovic for collection, isolation, maintenance, and DNA extraction of LM samples; Dr Chris Schardl and Dr Neil Moore, University of Kentucky for providing the 2013 GFF3 files for C. paspali and C. fusiformis; Dr Joshua Weitz and the Franklin Graybill Statistical Laboratory at Colorado State University for their help in data analysis of genomic fluidity; Molecular Technologies Laboratory (MTL) at the Ottawa Research & Development Centre, Agriculture and Agri-Food Canada, especially Kasia Dadej for technical assistance. For genomes downloaded from JGI, these sequence data were produced by the US Department of Energy Joint Genome Institute https://www.jgi.doe.gov/ in collaboration with the user community. This work was supported by the Agriculture and Food Research Initiative (AFRI) National Institute of Food and Agriculture (NIFA) (Fellowships Grant Program: Predoctoral Fellowships Grant No. 2019-67011-29502/Project Accession No. 1019134) from the United States Department of Agriculture (USDA) and by the American Malting Barley Association (Grant No. 17037621). Dr Broders was supported by the Simon’s Foundation (Grant No. 429440) to the Smithsonian Tropical Research Institute. Whole-genome sequencing of LM samples was supported, in part, by funding provided to Dr Jeremy Dettman from Agriculture and Agri-Food Canada’s Biological Collections Data Mobilization Initiative (BioMob, Work Package 2, project J-001564).
Author Contributions
The project was conceived and designed by S.A.W., S.J.M., and K.B.; S.A.W. performed the research, annotations, bioinformatic workflows, and analyzed the data with technical troubleshooting from S.J.M.; M.L. and J.R.D initiated whole-genome sequencing of LM samples; M.L., V.N., and K.B. provided management, research advice, and editorial contributions; S.A.W. wrote the paper with contributions from all other authors.
Data Availability
Data sets and scripts are available on Dryad: Stephen et al. (2020), whole-genome comparisons of ergot fungi reveals the divergence and evolution of species within the genus Claviceps are the result of varying mechanisms driving genome evolution and host range expansion, v4, Dryad, Data set, https://doi.org/10.5061/dryad.18931zcsk (submitted upon publication). Genomes and Illumina raw reads were deposited to NCBI under the BioProject PRJNA528707 (supplementary table S1, Supplementary Material online). Scripts are maintained within the GitHub repository of the primary author’s, https://github.com/PlantDr430/CSU_scripts. TransposableELMT can be found at Zenodo doi: 105281/zenodo3469661. All phylogenetic trees were made available at TreeBase (ID: TB2:S26278).
Literature Cited
- Alderman SC, Halse RR, White JF. 2004. A reevaluation of the host range and geographical distribution of Claviceps species in the United States. Plant Disease 88(1):63–81. [DOI] [PubMed] [Google Scholar]
- Alias N, et al. 2014. Molecular cloning and optimization for high level expression of cold-adapted serine protease from Antarctic yeast Glaciozyma antarctica PI12. Enzyme Res. 2014:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyotte SG, et al. 2012. Transposable elements in phystopathogenic Verticillium spp.: insights into genome evolution and inter- and intra-specific diversification. BMC Genomics 13(1):314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S 2010. FastQC: a quality control tool for high throughput sequence data. Available from: http://wwwbioinformaticsbabrahamacuk/projects/fastqc.
- Bao W, Kojima KK, Kohany O. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E, Parra G, Guigo R. 2007. Using geneid to identify genes. Curr Protoc Bioinformatics. Chapter 4:Unit 4.3. [DOI] [PubMed] [Google Scholar]
- Blin K, et al. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47(W1):W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchenak-Khelladi Y, Verboom GA, Savolainen V, Hodkinson TR. 2010. Biogeography of the grasses Poaceae: a phylogenetic approach to reveal evolutionary history in geographical space and geological time. Bot J Linn Soc. 162(4):543–557. [Google Scholar]
- Bouckaert R, Heled J. 2014. DensiTree 2: seeing trees through the forest. bioRXiv. doi: 10.1101/012401. [DOI]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. [DOI] [PubMed] [Google Scholar]
- Bushnell B 2014. BBMap: a fast, accurate, splice-aware aligner. Available from: https://sourceforgenet/projects/bbmap/.
- Campbell WP 1957. Studies on ergot infection in gramineous hosts. Can J Bot. 35(3):315–320. [Google Scholar]
- Cantu D, et al. 2013. Genome analyses of the wheat yellow stripe rust pathogen Puccinia striiformis f sp. triticireveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics 14(1):270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Chalvet F, Grimaldi C, Kaper F, Langin T, Dabousii MJ. 2003. Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Molecular Biology and Evolution. 20(8):1362–1375. [DOI] [PubMed] [Google Scholar]
- Coghlan A, Coghlan A, Tsai IJ, Berriman M. 2018. Creation of a comprehensive repeat library for newly sequenced parasitic worm genome. Protocol Exchange. doi: 101038/protex2018054. [Google Scholar]
- de Jonge R, et al. 2012. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci USA. 109(13):5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, et al. 2013. Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 23(8):1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Raffaele S, Kamoun S. 2015. The two-speed genomes of filamentous pathogens: waltz with plants. Curr Opin Genet Dev. 35:57–65. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Dutheil JY, et al. 2016. A tale of genome compartmentalization: the evolution of virulence clusters in smut fungi. Genome Biol Evol. 8(3):681–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser K, Tudzynski P. 1978. Genetics of the ergot fungus Claviceps purpurea. Theor Appl Genet. 53(4):145–149. [DOI] [PubMed] [Google Scholar]
- Estep LK, et al. 2015. Emergence and early evolution of fungicide resistance in North American populations of Zymoseptoria tritici. Plant Pathol. 64(4):961–971. [Google Scholar]
- Faino L, et al. 2015. Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genomes. mBio 6(4):pe00936-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino L, et al. 2016. Transposons passively and actively contribute to evolution of the two-speed genome of a fungal pathogen. Genome Res. 26(8):1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AJ, DiTomaso JM, Gordon TR, Aegerter BJ, Ayres DR. 2007. Salt marsh Claviceps purpurea in native and invaded Spartina marshes in Northern California. Plant Disease 91(4):380–386. [DOI] [PubMed] [Google Scholar]
- Frantzeskakis L, et al. 2018. Signatures of host specialization and a recent transposable element burst in the dynamic one-speed genome of the fungal barley powdery mildew pathogen. BMC Genomics. 19(1):381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantzeskakis L, Kusch S, Panstruga R. 2019. The need for speed: compartmentalized genome evolution in filamentous phytopathogens. Mol Plant Pathol. 20(1):3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M, Williams RL, Kothe GO, Selker EU. 2002. A cytosine methyltransferase homologue is essential for repeat_induced point mutation in Neurospora crassa. Proc Natl Acad Sci U S A. 99(13):8802–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudal I, et al. 2009. Repeat-induced point mutation RIP as an alternative mechanism of evolution towards virulence in Leptosphaeria maculans. Mol Plant Microbe Interact. 22(8):932–941. [DOI] [PubMed] [Google Scholar]
- Galagan JE, et al. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422(6934):859–868. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Selker EU. 2004. RIP: the evolutionary cost of genome defense. Trends Genet. 20(9):417–423. [DOI] [PubMed] [Google Scholar]
- Gladieux P, et al. 2014. Fungal evolutionary genomics provides insight into the mechanisms of adaptive divergence in eukaryotes. Mol Ecol. 23(4):753–773. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graïa F, et al. 2001. Genome quality control: RIP repeat-induced point mutation comes to Podospora. Mol Microbiol. 40(3):586–595. [DOI] [PubMed] [Google Scholar]
- Grandaubert J, Bhattacharyya A, Stukenbrock EH. 2015. RNA-seq-based gene annotation and comparative genomics of four fungal grass pathogens in the genus Zymoseptoria identify novel orphan genes and species-specific invasions of transposable elements. G3 (Bethesda) 5:1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandaubert J, Dutheil JY, Stukenbrock EH. 2019. The genomic determinants of adaptive evolution in a fungal pathogen. Evol Lett. 3(3):299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandaubert J, et al. 2014. Transposable element-assisted evolution and adaptation to host plant within the Leptosphaeria maculans-Leptosphaeria biglobosa species complex of fungal pathogens. BMC Genomics 15(1):891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32(18):2847–2849. [DOI] [PubMed] [Google Scholar]
- Hane JK, Williams AH, Taranto AP, Solomon PS, Oliver RP. 2015. Repeat-induced point mutation: a fungal-specific, endogenous mutagenesis process In: van den Berg MA, Maruthachalam K, editors. Genetic transformation systems in fungi. Vol. 2 Springer International Publishing, p. 55–68. [Google Scholar]
- Hass BJ 2010. TransposonPSI. Available from: http://transposonpsisourceforgenet.
- Hinsch J, Galuszka P, Tudzynski P. 2016. Functional characterization of the first filamentous fungal tRNA-isopentenyltransferase and its role in the virulence of Claviceps purpurea. New Phytol. 211(3):980–992. [DOI] [PubMed] [Google Scholar]
- Hinsch J, et al. 2015. De novo biosynthesis of cytokinins in the biotrophic fungus Claviceps purpurea. Environ Microbiol. 17(8):2935–2951. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2017. Fast genome-wide functional annotation through orthology assignment by eggNog-Mapper. Mol Biol Evol. 34(8):2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D, et al. 2019. eggNog 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47(D1):D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K-I, et al. 2002. Repeat-induced point mutation RIP in Magnaporthe grisea: implications for its sexual cycle in the natural field context. Mol Microbiol. 45(5):1355–1364. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungehülsing U, Tudzynski P. 1997. Analysis of genetic diversity in Claviceps purpurea by RAPD markers. Mycol Res. 101(1):1–6. [Google Scholar]
- Kachroo P, Leong SA, Chatto BB. 1994. Pot2, an inverted repeat transposon from the rice blast fungus Magnaporthe grisea. Mol Gen Genet. 245(3):339–348. [DOI] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer EL. 2007. Advantages of combined transmembrane topology and signal peptide prediction: the Phobius web server. Nucleic Acids Res. 35(Web Server):W429–W32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper J, et al. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444(7115):97–101. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J, et al. 2016. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 44(9):e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA 2001. Evolutionary history of the grasses. Plant Physiol. 125(3):1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind S, Schurack S, Hinsch J, Tudzynski P. 2018. Brachypodium distachyon as alternative model host system for the ergot fungus Claviceps purpurea. Molecular Plant Pathology. 19(4):1005–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind S, Hinsch J, et al. 2018. Manipulation of cytokinin level in the ergot fungus Claviceps purpurea emphasizes its contribution to virulence. Curr Genet. 64(6):1303–1319. [DOI] [PubMed] [Google Scholar]
- Kiran K, et al. 2017. Dissection of genomic features and variations of three pathotypes of Puccinia striiformis through whole genome sequencing. Sci Rep. 7(1):42419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran K, et al. 2016. Draft genome of the wheat rust pathogen Puccinia triticina unravels genome-wide structural variations during evolution. Genome Biol Evol. 8(9):2702–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislyuk AO, Haegeman B, Bergman NH, Weitz JS. 2011. Genomic fluidity: an integrative view of gene diversity within microbial populations. BMC Genomics 12(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kito H, et al. 2003. Occan, a novel transposon in the Fot1 family, is ubiquitously found in several Magnaporthe grisea isolates. Curr Genet. 42(6):322–331. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc R Soc B. 279(1749):5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I 2004. Gene finding in novel genomes. BMC Bioinformatics 5(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kvas M, Marasas WFO, Wingfield BD, Wingfield MJ, Steenkamp ET. 2009. Diversity and evolution of Fusarium species in the Gibberella fujikuroi complex. Fungal Divers. 34:1–21. [Google Scholar]
- Langdon RFN 1952. Studies on ergot [PhD thesis]. [ Brisbane (Australia)]: Queensland University. [Google Scholar]
- Li H 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, et al. 2020. Four new ergot species based on morphology, alkaloid production, pathogenicity and DNA sequences analyses. Mycologia 112(5):974–988. [DOI] [PubMed] [Google Scholar]
- Ma L-J, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464(7287):367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros WH, Pertea M, Salzberg SL. 2004. TigrScan and GlimmerHMM: two open-source ab initio eukaryotic gene-finders. Bioinformatics 20(16):2878–2879. [DOI] [PubMed] [Google Scholar]
- Mario S, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24(5):637–644. [DOI] [PubMed] [Google Scholar]
- Möller M, Stukenbrock EH. 2017. Evolution and genome architecture in fungal plant pathogens. Nat Rev Microbiol. 15(12):756–771. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H, Nishimoto N, Ikeda K, Tosa Y, Mayama S. 1999. Degenerate MAGGY elements in a subgroup of Pyricularia grisea: a possible example of successful capture of a genetic invader by a fungal genome. Mol Gen Genet. 261(6):958–966. [DOI] [PubMed] [Google Scholar]
- Newton RR, Newton IL. 2013. PhyBin: binning trees by topology. Peer J. 1:e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H 2017. Predicting secretory proteins with SignalP In: Kihara D, editor. Protein function prediction. Methods in Molecular Biology. Vol. 1611 New York (NY): Humana Press. [DOI] [PubMed] [Google Scholar]
- Nurk S, et al. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads In: Deng M, Jiang R, Sun F, Zhang X, editors. Research in computational molecular biology RECOMB 2013. Lecture Notes in Computer Science, vol. 7821 Berlin (Heidelberg): Springer. [Google Scholar]
- Oberti H, et al. 2020. Diversity of Claviceps paspali reveals unknown lineages and unique alkaloid genotypes. Mycologia 112(2):230–214. [DOI] [PubMed] [Google Scholar]
- Oeser B, et al. 2017. Cross-talk of the biotrophic pathogen Claviceps purpurea and its host Secale cereale. BMC Genomics. 18(1):273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J, Stajich J. 2019. nextgenusfs/funannotate: funannotate. Version 1.6.0. Zenodo. doi: 105281/zenodo3354704.
- Pease JB, Hahn MW. 2013. More accurate phylogenies inferred from low-recombination regions in the presence of incomplete lineage sorting. Evolution 67(8):2376–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Píchová K, et al. 2018. Evolutionary history of ergot with a new infrageneric classification (Hypocreales: Clavicipitaceae: Claviceps). Mol Phylogenet Evol. 123:73–87. [DOI] [PubMed] [Google Scholar]
- Poppe S, Dorcheimer L, Happel P, Stukenbrock EH. 2015. Rapidly evolving genes are key players in host specialization and virulence of the fungal wheat pathogen Zymoseptoria tritici Mycosphaerella graminicola. PLoS Pathog. 11(7):e1005055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Farrer RA, Cano LM, et al. 2010. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330(6010):1540–1543. [DOI] [PubMed] [Google Scholar]
- Raffaele S, Kamoun S. 2012. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol. 10(6):417–430. [DOI] [PubMed] [Google Scholar]
- Raybould AF, Gray AJ, Clarke RT. 1998. The long-term epidemic of Claviceps purpurea on Spartina anglica in Poole Harbour: pattern of infection, effects on seed production and the role of Fusarium heterosporum. New Phytol. 138(3):497–505. [Google Scholar]
- Rep M, Kistler HC. 2010. The genomic organization of plant pathogenicity in Fusarium species. Curr Opin Plant Biol. 13(4):420–426. [DOI] [PubMed] [Google Scholar]
- Rouxel T, Grandaubert J, Hane JK, et al. 2011. Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat-induced point mutations. Nat Commun. 2:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandve SR, Fjellheim S. 2010. Did gene family expansions during the Eocene-Oligocene boundary climate cooling play a role in Pooideae adaptation to cool climates? Mol Ecol. 19(10):2075–2088. [DOI] [PubMed] [Google Scholar]
- Schardl CL, Young CA, Hesse U, et al. 2013. Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 9(2):e1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski J, Mannhaupt G, Münch K, et al. 2010. Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330(6010):1546–1548. [DOI] [PubMed] [Google Scholar]
- Seidl MF, et al. 2015. The genome of the saprophytic fungus Verticillium tricorpus reveals a complex effector repertoire resembling that of its pathogenic relatives. Mol Plant Microbe Interact. 28(3):362–345. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R. 2015. RepeatModeler Open-10. Available from: http://wwwrepeatmaskerorg.
- Smit AFA, Hubley R, Green P. 2015. RepeatMasker Open-40. Available from: http://wwwrepeatmaskerorg.
- Soreng RJ, et al. 2017. A worldwide phylogenetic classification of the Poaceae Gramineae II: an update and a comparison of two 2015 classifications. J Syst Evol. 55(4):259–290. [Google Scholar]
- Sperschneider J, Dodds PN, Gardiner DM, Singh KB, Taylor JM. 2018. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol Plant Pathol. 19(9):2094–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J, et al. 2015. Genome-wide analysis in three Fusarium pathogens identifies rapidly evolving chromosomes and genes associated with pathogenicity. Genome Biol Evol. 7(6):1613–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Startek M, Le Rouzic A, Capy P, Grzebelus D, Gambin A. 2013. Genomic parasites or symbionts? Modeling the effects of environmental pressure on transposition activity in asexual populations. Theor Popul Biol. 90:145–151. [DOI] [PubMed] [Google Scholar]
- Steenwyk JL, Shen X-X, Lind AL, Goldman GH, Rokas A. 2019. A robust phylogenomic time tree for biotechnologically and medically important fungi in the genera Aspergillus and Penicillium. mBio 10(4):e00925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbiss S, Willhoeft U, Gremme G, Kurtz S. 2009. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37(21):7002–7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH 2013. Evolution, selection and isolation: a genomic view of speciation in fungal plant pathogens. New Phytol. 199(4):895–907. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH, Dutheil JY. 2018. Fine-scale recombination maps of fungal plant pathogens reveal dynamic recombination landscape and intragenic hotspots. Genetics 208(3):1209–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhir K, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18(12):1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa A, Hane JK, Ellwood SR, Oliver RP. 2015. CodingQuarry: highly accurate hidden Markov model gene prediction in fungal genomes using RNA-seq transcripts. BMC Genomics. 16(1):170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa A, Oliver R, Hane J. 2015. Overview of genomic and bioinformatic resources for Zymoseptoria tritici. Fungal Genet Biol. 79:13–16. [DOI] [PubMed] [Google Scholar]
- Tudzynski P, Scheffer JAN. 2004. Claviceps purpurea: molecular aspects of a unique pathogenic lifestyle. Mol Plant Pathol. 5(5):377–388. [DOI] [PubMed] [Google Scholar]
- Urguhart AS, Mondo SJ, Makela MR. 2018. Genomic and genetic insights into a cosmopolitan fungus, Paecilomyces variotii Eurotiales. Front Microbiol. 9:3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wouw AP, Cozijnsen AJ, et al. 2010. Evolution of linked avirulence effectors in Leptosphaeria maculans is affected by genomic environment and exposure to resistance genes in host plants. PLoS Pathog. 6(11):e1001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wyk S, et al. 2019. The RIPper, a web-based tool for genome-wide quantification of repeat-induced point RIP mutations. PeerJ. 7:e7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wäli PP, Wäli PR, Saikkonen K, Tuomi J. 2013. Is the pathogenic ergot fungus a conditional defensive mutualist for its host grass? PLoS One 8(7):e69249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, et al. 2011. Molecular phylogeny of the higher and lower taxonomy of the Fusarium genus and differences in the evolutionary histories of multiple genes. BMC Evol Biol. 11(1):322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, et al. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield BD, et al. 2018. Nine draft genome sequences of Claviceps purpurea s.lat., including C. arundinis, C. humidiphila, and C. cf. spartinae, pseudomolecules for the pitch canker pathogen Fusarium circinatum, draft genome of Davidsoniella eucalypti, Grosmannia galeiformis, Quambalaria eucalypti, and Teratosphaeria destructans. IMA Fungus 9(2):401–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Macielog AI, Hao W. 2017. Origin and spread of spliceosomal introns: insights from the fungal clade Zymoseptoria. Genome Biol Evol. 9(10):2658–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang H. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35(Web Server):W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sets and scripts are available on Dryad: Stephen et al. (2020), whole-genome comparisons of ergot fungi reveals the divergence and evolution of species within the genus Claviceps are the result of varying mechanisms driving genome evolution and host range expansion, v4, Dryad, Data set, https://doi.org/10.5061/dryad.18931zcsk (submitted upon publication). Genomes and Illumina raw reads were deposited to NCBI under the BioProject PRJNA528707 (supplementary table S1, Supplementary Material online). Scripts are maintained within the GitHub repository of the primary author’s, https://github.com/PlantDr430/CSU_scripts. TransposableELMT can be found at Zenodo doi: 105281/zenodo3469661. All phylogenetic trees were made available at TreeBase (ID: TB2:S26278).