Abstract
The novel coronavirus (SARS-CoV-2) has become a pandemic and is threatening human health globally. Here, we report nine newly evolved SARS-CoV-2 single nucleotide polymorphism (SNP) alleles those underwent a rapid increase (seven cases) or decrease (two cases) in their frequency for 30–80% in the initial four months, which are further confirmed by intrahost single nucleotide variation analysis using raw sequence data including 8,217 samples. The nine SNPs are mostly (8/9) located in the coding region and are mainly (6/9) nonsynonymous substitutions. The nine SNPs show a complete linkage in SNP pairs and belong to three different linkage groups, named LG_1 to LG_3. Analyses in population genetics show signatures of adaptive selection toward the mutants in LG_1, but no signal of selection for LG_2. Population genetic analysis results on LG_3 show geological differentiation. Analyses on geographic COVID-19 cases and published clinical data provide evidence that the mutants in LG_1 and LG_3 benefit virus replication and those in LG_1 have a positive correlation with the disease severity in COVID-19-infected patients. The mutants in LG_2 show a bias toward mildness of the disease based on available public clinical data. Our findings may be instructive for epidemiological surveys and disease control of COVID-19 in the future.
Keywords: COVID-19, SARS-CoV-2, SNP, iSNV, mutants
Significance
Understanding the evolutionary dynamics of SARS-CoV-2 is important in the control of COVID-19 already pandemic worldwide. Through comprehensive analyses on global SARS-CoV-2 genomes at intra- and inter-host levels, we found nine mutant alleles showing rapid increase or decrease in frequency, possibly driven by directional selection. Based on statistics on global clinical data, we predicted that the fast increasing mutants are capable of promoting viral replication and enhancing the severity of the disease, while the fast decreasing mutants are milder than the original alleles. These reflect a potential evolutionary trend of SARS-CoV-2 and possible subsequent clinical outcomes resulted from the trend.
Introduction
SARS-CoV-2, also named 2019-nCoV, is a novel coronavirus that causes novel coronavirus pneumonia (COVID-19). The rapid spread of SARS-CoV-2 has become a global threat, since its first identification in Wuhan City, China, last December (Ralph et al. 2020). To date, there have been more than 43 million confirmed COVID-19 cases and more than one million cases have resulted in deaths around the world. To control the COVID-19 epidemic, the research on the genomic epidemiology of the virus is important for the prediction of global evolutionary trends. Furthermore, the identification of the diversification in patterns and selection signatures in the SARS-CoV-2 genome (Lu et al. 2020) during the evolution of the virus is essential for the early diagnosis and control of this disease (Ayres 2020).
Although the origin of the virus is still a mystery, SARS-CoV-2 has displayed higher divergence in genomic sequence to its possible origins (Bat-CoV-RaTG13 or Pangolin-CoV-2019) (Lam et al. 2020) than previously anticipated (Tang et al. 2020). The genome of SARS-CoV-2 is undergoing continuous evolution. Just two months after the virus was first reported (Lu et al. 2020; Ralph et al. 2020), there have been more than 100 polymorphic sites identified in SARS-CoV-2’s protein coding region. Most of these mutations are located in the coding region of polyprotein 1ab (pp1ab, ORF1) (Namy et al. 2006) and structural proteins (Fehr and Perlman 2015). It has also been reported that SARS-CoV-2 is undergoing recombination (Yi 2020), which is a common event among RNA viruses. Meanwhile, there are reports saying that there is no detectable genetic recombination in SARS-CoV-2 strains (Richard et al. 2020) (virological.org/t/testing-recombination-in-the-pandemic-sars-cov-2-strains/492). A previous study has suggested that the virus has evolved into two subtypes (L and S) classified by two complete linked single nucleotide polymorphisms (SNPs) at genome locations 8792 and 28144 (Tang et al. 2020). SNP 29144 leads to an amino acid (AA) change from LEU (L) to SER (S) in ORF8, which is supposed to be related to viral replication (Muth et al. 2018). In the study Tang et al. (2020), it was predicted that S is less aggressive but more adaptive than L and may increase in frequency in the future. With more SARS-CoV-2 genomes sequenced and deposited, we are now able to reevaluate the performance of the polymorphic alleles.
Materials and Methods
Identification of Rapidly Changing Mutants
We collected genomic sequences and related information of coronavirus from GISAID (www.gisaid.org), NCBI, CoVdb (Zhu et al. 2020) and ViralZone (Hulo et al. 2011). The whole genome alignments were calculated using Clustal Omega (Sievers and Higgins 2014). To assess genomic differences between months, we grouped coronavirus genomes into three groups according to their collection dates. We combined the genomes collected in December, 2019 and January, 2020, considering that there are few samples in 2019 (16 cases). Based on this approach, the number of the samples in December–January, February, March, and April were 344, 661, 16,270, and 10,042, respectively. Using libraries in BioPerl, we wrote scripts to extract mutations with a change in frequency by near or more than 30% between January and April. Finally, we performed a manual check and identified nine cases complying with the request. The significance of the change in frequency was evaluated using the chi square test by R.
Linkage Disequilibrium Analysis
According to the published algorithms (Morton 1955; Lewontin 1964; Slatkin 2008), we counted the possibility of coupling and repulsion gametes. The coupling gametes are alleles on the same chromosome that remain together, while the repulsion gametes are alleles on the same chromosome that are repulsed by each other and pair with alleles on the opposite strand. Then we wrote Perl scripts to calculate D’, ρ2, and logarithm of the odds (LOD), which refer to a normalized basic linkage disequilibrium parameter, a squared correlation coefficient (Lewontin 1964), and a statistical test to infer the likelihood of obtaining the test data if the two loci are indeed linked (Morton 1955), respectively. They all positively correlate with the degree of genetic linkage.
Intrahost Single Nucleotide Variation Analysis
Following previous efforts (Rueca et al. 2020; Wang et al. 2020; Zhou et al. 2020), we performed intrahost single nucleotide variation (iSNV) analyses at different scales. First, we tried to calculate the variation of identified mutants in one single host. We analyzed 13 consensus genomes of SARS-CoV-2 extracted from six patients with COVID-19 (Rueca et al. 2020). We used the genome of the strain MN908947 as the reference genome in this study. We performed pairwise sequence alignment between the reference genome and each of the 13 genomes by LASTZ (Harris 2007) and wrote Perl scripts to identify the allele in the nine SNP sites. In the same way, we performed analysis on 43 consensus genomes collected at different time points from eight pneumonia patients with COVID-19 (Project accession CNP0001004, CNGB, https://db.cngb.org/) (Wang et al. 2020). We performed iSNV analysis of the raw sequence data sets (including 91 samples which were also collected from the same eight patients) (Wang et al. 2020) generated by metatranscriptomic sequencing with hybrid capture methods. A previously reported pipeline (Zhou et al. 2020) was applied. We used Bowtie2 (Langdon 2015) to map reads onto the reference genome (MN908947) and mark duplicated reads by GATK MarkDuplicates (McKenna et al. 2010; DePristo et al. 2011). Both the mapping quality and the base quality were required to be better than 20 (Li 2011). Perl scripts were written to count the frequency of target alleles based on the SAMtools (Li et al. 2009) and view results through the BAM files. During the calculation of allele frequency, we required the sample size to be higher than 20 in general. Using the same method, we performed iSNV analyses on the raw sequence data sets of SARS-CoV-2 including 112 samples from China (NCBI Project ID: PRJNA627662), 1,938 samples from Australia (PRJNA613958), 1,542 samples from USA (PRJNA614995), and 4,521 samples from United Kingdom (PRJEB37886). We visualized the distribution of allele frequencies at different time points by ggplot2 and tested the significance in the change using Wilcoxon test by R. For allele frequency calculation for UK samples, we required that the sample size higher than 10 because of the difference in sequencing depth.
Evolutionary Analysis
Using LASTZ (Harris 2007), we performed genome–genome alignments between any two coronavirus strains and outputted the results in AXT format. From the results, we retrieved the corresponding sequences of other strains of one coronavirus gene, and realigned these sequences using MUSCLE (Edgar 2004a, 2004b). To detect selection signals, the sliding widow analysis was used with a window size of 200 bp and a step size of 50 bp. For each sliding window, we calculated the scores of Pi (Tajima 1993) and Tajima’s D (Tajima 1989) by VariScan 2.0 (Vilella et al. 2005; Hutter et al. 2006). The fixation index (Fst) was calculated according to published algorithms (Fumagalli et al. 2013). We further calculated the allele frequencies and used SweepFineder2 (DeGiorgio et al. 2016) to calculate the composite likelihood ratio (CLR, step size = 50) (Nielsen et al. 2005; Zhu and Bustamante 2005). The figures were generated using the R libraries “gdata” and “ggplot2”.
To test whether a negative Tajima’s D is biased caused because of a genetic bottleneck, a simple model was built assuming that the population size of COVID-19 shrunk from N1 = 2,558 to N2 = 450 and then expanded to N3 = 12,196, based on the number of daily confirmed infections in February 15, 2020, in Asia, in February 24, 2020, in Asia, and in March 16, 2020, in Europe. We assume that on February 29, 2020, the infected population started to expand. We used the software ms (Hudson 2002) to generate simulation data according to the model with the parameter “-G 11.189 -eG 0.3 0.0 -eN 0.5 0.2”. We tested the significance by ranking in distribution.
Function Prediction Based on Statistical Analysis
Countries with more than 20 sequenced COVID-19 strains are used to do analysis in statistics. The case fatality rates (CFR) in countries were calculated based on the data provided by the European Centre for Disease Prevention and Control from March to May. The daily exponential growth rates (λ) were calculated as λ = ln[Y(t)]/t (Lipsitch et al. 2003). The calculation periods for CFR and λ are 10 days. A two-sided test was used to assess the significance of Pearson’s product-moment correlation in the correlation analysis.
We obtained the sequence data set with annotation on patients’ status from GISAID and developed a local database. We performed pairwise alignment between each of these sequences and the reference genome (MN908947) and the identification of alleles in the nine SNPs’ positions. For the identification of the possible clinical effects resulted from the mutations in the nine SNPs, we marked records according to pairs of related patient status. We used a series of keywords to search for records and then marked them, for example, we marked a record “Outpatient” if it is annotated as “Outpatient”, “without hospitalization”, or “Not hospitalized.” We grouped records according to these marks and performed poststatistical analyses by R.
Results
SNP and iSNV Frequency, Linkage Disequilibrium, and Haplotype Analyses
Inspired by a previous effort (Tang et al. 2020), we performed comprehensive analyses on 27,388 full-length COVID-19 genomes (collected from December, 2019 to May, 2020, supplementary table S1, Supplementary Material online) with a focus on evolutionary dynamics, selection, and gene function. We searched for SNP mutants with a monthly identification frequency (IF) variation in past four months higher than 0.3. A total of 1,710 polymorphic sites were identified, and nine of those SNPs were highlighted in such cases (fig. 1A and B). From January to April, seven mutants increased from a monthly frequency of 0–2% to 35–82%, while two decreased from a monthly frequency of 33.92% and 34.02% to 3.12% and 3.14% (table 1). These changes in frequency are statistically significant (Chisq-Test, P value < 2.2e-16). Linkage disequilibrium analyses show that the nine SNPs can be clustered into three linkage groups, in which SNPs show significant complete linkage (D’ > 0.97, ρ2 > 0.96) (fig. 1C and D) with a median LOD score of 146.62 (supplementary table S2, Supplementary Material online). For convenience, we named SNPs using the format “SNP_Location”, where the genome of the strain MN908947 is used as the reference. For example, we use SNP_241 to represent the SNP at location 241. Linkage groups are named using the format of “LG_” plus a serial number (table 1), from LG_1 to LG_3.
Fig. 1.
A is the density curve showing the distribution of frequency changes in 1710 SNPs. B is the change in frequency of SNPs (y-axis) along different chromosome positions (x-axis), with the nine new SNPs are marked by red dots. C shows the LOD score of each pair of SNPs (y-axis) against the squared correlation coefficient ρ2 between that pair (x-axis). D shows ρ2 of each pair of SNPs (y-axis) against the genomic distance between that pair. In C and D, the pairs of the new nine alleles showing complete or near complete linkages are marked in red. The orange dotted lines in B–D are the top 5% positions in the corresponded axes.
Table 1.
Overview of the Nine New SNPs. “Loc”, “Mutat”, “Chg”, and “AA” Are Simplified Expressions for Location, Mutation, Change, and Amino Acids, Respectively
| Loc | Mutat | Dec–Jan |
Feb |
Mar |
Apr |
Chg in Perc. |
Chisq-Test | Protein | Pos in Codon |
AA Mutat | Linkage Group |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Perc (a) | a/A | Perc (a) | a/A | Perc (a) | a/A | Perc (a) | a/A | ||||||||
| 241 | C->T | 0.88% | 3/337 | 14.93% | 96/547 | 66.08% | 10,397/5,337 | 82.87% | 8,211/1,697 | 81.99% | < 2.2e-16 | LG_1 | |||
| 3037 | C->T | 1.46% | 5/337 | 14.88% | 97/555 | 65.56% | 10,573/5,553 | 82.86% | 8,295/1,716 | 81.40% | < 2.2e-16 | nsp3 | 3 | Synonymous | LG_1 |
| 8782 | C->T | 33.92% | 116/226 | 26.03% | 170/483 | 10.67% | 1,713/14,335 | 3.12% | 308/9,556 | −30.80% | < 2.2e-16 | nsp4 | 3 | Synonymous | LG_2 |
| 14408 | C->T | 0.58% | 2/340 | 14.48% | 95/561 | 65.47% | 10,605/5,594 | 82.95% | 8,308/1,708 | 82.36% | < 2.2e-16 | RdRp | 2 | PRO->LEU | LG_1 |
| 23403 | A->G | 1.45% | 5/339 | 15.05% | 99/559 | 65.60% | 10,624/5,571 | 82.92% | 8,301/1,710 | 81.47% | < 2.2e-16 | S | 2 | ASP->GLY | LG_1 |
| 28144 | T->C | 34.02% | 116/225 | 24.88% | 161/486 | 10.63% | 1,723/14,493 | 3.14% | 315/9,703 | −30.87% | < 2.2e-16 | ORF8 | 2 | LEU->SER | LG_2 |
| 28881 | G->A | 0.00% | 0/343 | 6.42% | 42/612 | 20.06% | 3,234/12,889 | 35.52% | 3,546/6,436 | 35.52% | < 2.2e-16 | N | 2 | ARG->LYS | LG_3 |
| 28882 | G->A | 0.00% | 0/343 | 6.41% | 42/613 | 19.99% | 3,226/12,909 | 35.48% | 3,541/6,438 | 35.48% | < 2.2e-16 | N | 3 | LG_3 | |
| 28883 | G->C | 0.00% | 0/343 | 6.41% | 42/613 | 19.99% | 3226/12910 | 35.48% | 3542/6440 | 35.48% | < 2.2e-16 | N | 1 | GLY->ARG | LG_3 |
Note.—The locations of SNPs are according to MN908947. “Perc” refers to the percentage of minor alleles. “a/A” refers to the number of minor alleles (a) and the number of major alleles (A) in January, February, March, and April. “Pos in Codon” refers to the position of the mutation in the codon.
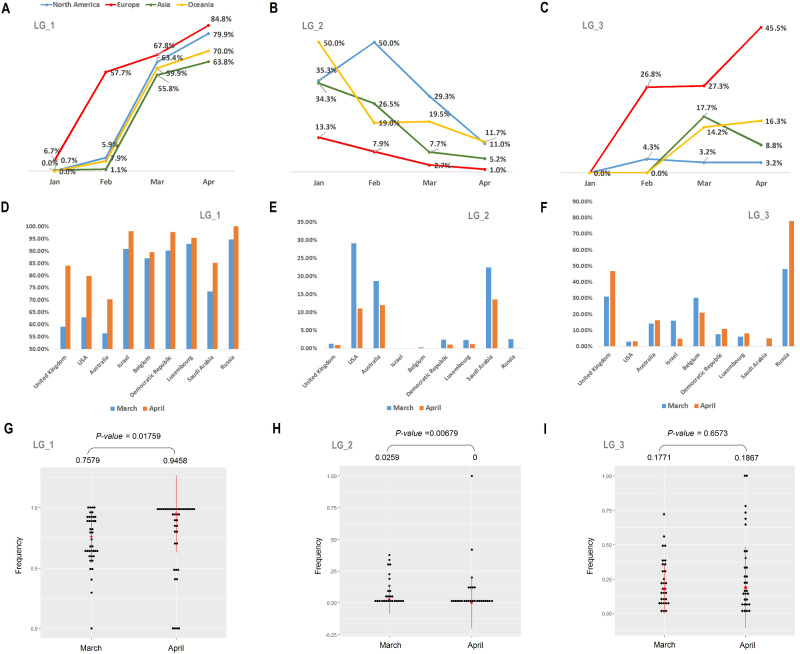
To understand whether the increase of the mutants is universal or only happened in some specific region, we calculated statistics for their IF in different continents. We found that the IF of the mutants clustered in LG_1 were arising in all continents, while the IF of those in LG_2 were decreasing (fig. 2A and B). The IF of the mutants in LG_3 does not show converged trend across all continents. Their IF increased in Europe and Oceania from January to April, but decreased in Asia from March to April (fig. 2C). We also counted the independent IF of mutants among different countries, and evaluated the overall changes of IF among countries from March to April. The results show that the overall IF of LG_1 are significantly higher in April than in March (fig. 2D and G) while those of LG_2 are significantly lower in April than in March (fig. 2E and H). We did not observe a significance in statistics for those of LG_3 (fig. 2F and I).
Fig. 2.
Changes in frequencies of the mutants in LG_1 to LG_3 in different continents/countries. A–C is the change in frequency in four continents, North America, Europe, Asia, and Oceania. D–F is the change in frequency from March (blue) to April (Orange) in nine countries with high quantities of sequenced COVID-19 strains. G–I is tests of the frequencies of mutants in countries with sample size > 10 between March and April. The medians and the P values calculated by Wilcox test are list at the top of each figure.
In addition to interhost SNP analysis, we performed iSNVs analysis on published sequencing data (for specifics, see “Materials and Methods”) to trace the evolutionary dynamics of the mutants. We performed analyses of the SARS-CoV-2 sequencing data (supplementary table S3, Supplementary Material online) collected from six patients (Rueca et al. 2020), but observed no allele change event in a single patient. However, we observed that the mutants in LG_1 emerged and fixed in four patients tested later. The cycle threshold for positive signal in E gene-based RT-PCR (Ct) is a surrogate for relative viral loads. Low Ct values are always related to high viral loads (Corman et al. 2020). The strains with LG_1 mutants have a lower Ct (20.7 in median) than others (21.5 in median) but the difference is not significant (P value = 0.4633).
Then we have performed analyses on the raw sequence data of 91 SARS-CoV-2 samples collected at different time points and from eight patients with COVID-19 in Guangzhou, China (Wang et al. 2020) and correlated assembled consensus genomes. We found the mutants in LG_1 have begun to emerge, for example from A to G in SNP_23403, in February (supplementary table S4, figs. S1A and S2A, Supplementary Material online). There is a significant change in the allele frequency of LG_2. In the consensus genomes, we found the ratio of the mutants to the original alleles (T/C) has changed from 18/5 (counted from January 27 to February 7 in 2020) to 6/14 (counted from February 8 to February 26 in 2020, Chisq-Test, P value = 0.00148, supplementary table S4, Supplementary Material online). We observed a significant increase (Wilcox test, P value = 0.02518) of the IF for the mutants in LG_2, from 0.0037 (median, counted from January 27 to February 7 in 2020) to 0.99285 (median, counted from February 8–26 in 2020) based on the metatranscriptomic data (supplementary fig S1B, Supplementary Material online). We also observed a significant increase of the IF for the mutants in LG_2 based on the hybrid capture data (P value = 0.02452, supplementary fig. S2B, Supplementary Material online). This is in accordance to a previous observation (Tang et al. 2020). The IF of the mutants in LG_3 is low compared with LG_1 or LG_2 (supplementary figs. S1 and S2, Supplementary Material online) in the Guangzhou data set. At the meantime, we analyzed the raw sequence data sets of SARS-CoV-2 strains collected in January and February of this year and from 112 patients in Shanghai, China (supplementary table S5, Supplementary Material online). We found that some mutants in LG_1 have high IF and most mutants in LG_3 have low IF (supplementary fig. S3A and C, Supplementary Material online). We did not observe a significant change in IF for the mutants in LG_2 (supplementary fig. S3B, Supplementary Material online) from the Shanghai data set.
Furthermore, we performed iSNV analysis using the raw sequence data including: 1,938 SARS-CoV-2 samples from Australia patients, 4,521 samples from UK, and 1,542 samples from USA (supplementary tables S6–S8, Supplementary Material online). The results show that the IF of the mutants in LG_1 are in a steady increase from January to June, in Australia, UK, and USA (supplementary figs. S4–S6, Supplementary Material online). There is also a significant IF increase of the mutants in LG_3 from January to June in Australia and UK but USA. The overall trend of mutants; IF in LG_2 is decreasing. These findings based on iSNV analysis are generally in consistent with the inter-host SNP analysis results based on global consensus genomes.
In order to avoid the possible bias caused by geographical difference and experimental methods, we have done further test by selecting out samples collected by the same institution and collected in the same place. The 1,938 Australia samples are all collected in a state, Victoria. In the 1,938 samples, we selected out 1,755 that were collected and sequenced by the Victorian Infectious Diseases Reference Laboratory. We performed the same analysis onto the 1,755 samples (fig. 3). The results are in agreement with our previous findings. For the 4,521 SARS-CoV-2 genome sequences collected in England, UK, we performed iSNV analysis of 335 samples collected and sequenced by Northumbria University et al and the 4,186 samples collected and sequenced by Public Health England (Colindale), separately. We obtained consistent results too (supplementary figs. S7 and S8, Supplementary Material online). The 1,542 genome sequences collected in USA are all produced by the Utah Public Health Laboratory.
Fig. 3.
Changes in the distribution of IF along time for the mutants in LG_1 to LG_3 based on the analyses of the raw-sequence data of SARS-CoV-2 including 1,755 samples collected from patients in Victoria, Australia and by VIDRL. X-axis is the IF, from 0 to 1 (mutants are fixed), and “Frequency” (y-axis) refer to the counts of samples with some IF value, from 0 to 1. We used Wilcoxon test to test the difference between two distributions.
We classified all COVID-19 genomes available publicly into 30 haplotypes based on the nine SNPs. From a network view, we observed that there are only two major haplotypes in January but four major haplotypes in April (supplementary fig. S9, Supplementary Material online). This suggests that two newly evolved haplotypes became majority in the last three months. This change is rapid, and is reflected by a significantly high Tajima’s D (3.07169, P value = 0.00857) of all haplotypes in April. The two major haplotypes in January correspond to the two subtypes L and S referred to previously (Tang et al. 2020), which are now clustered into LG_2 in this study.
Population Genetics and Evolutionary Patterns
We performed sliding window analyses in population genetics to address recent evolutionary patterns in the nine SNP sites (fig. 4A). To avoid oscillation caused by time scale, we only targeted the COVID-19 strains collected in April, 2020 (10,042 samples). The results show that SNP_3037 (LG_1) and SNP_23403 (LG_1) both have CLR (Nielsen et al. 2005; Zhu and Bustamante 2005) peaks (fig. 4B and C). SNP_14408 (LG_1) is adjacent to a CLR peak (fig. 4D). These indicated that the increase of the mutants in LG_1 may be resulted from directional selection. A parallel simulation test was performed to assess effects caused by genetic bottleneck on mutation sites showing directional selection signals (for details, see Materials and Methods and supplementary fig. S10, Supplementary Material online). The results show that SNP_3037 (LG_1) and SNP_23403 (LG_1) both have a negative Tajima’s D with significance in statistics, while SNP_14408 (LG_1) has a negative Tajima’s D with weak significance (supplementary table S9, Supplementary Material online).
Fig. 4.
Population genetic analyses of the nine new SNPs. A is the distributions of CLR and Tajima’s D of 200 bp windows with 50 bp steps for all strains. B–D are sliding window analysis views of CLR at three SNP sites, indicated by red arrows.
Mutants in LG_2 and LG_3 do not have CLR peaks, although they all are adjacent to a CLR peak (supplementary fig. S11A and B, Supplementary Material online). The decrease of the minor haplotype (S) in LG_2 may be resulted from genetic drift. In other words, the initial higher percentage of the major haplotype (L) may lead to the successive increase of its frequency and the decrease of the S. The change of mutants’ frequencies in LG_3 is different. The frequency increased more rapidly in Europe than in North America (Chisq-Test, P value < 3.968e-11, fig. 2C). In accordance, LG_3 has a high Fst (Holsinger and Weir 2009) in comparison with North American and European strains (supplementary fig. S11C, Supplementary Material online). We detected a CLR peak at LG_3 for England strains (supplementary fig. S11D, Supplementary Material online). Even though, the Tajima’s D of LG_3 is not significantly negative (–1.636, test by simulation, P value = 0.2474). The observed increase in frequency of this mutant in England may be resulted from positive selection.
Potential Functional Consequences of the Rapid Spread Mutant Alleles
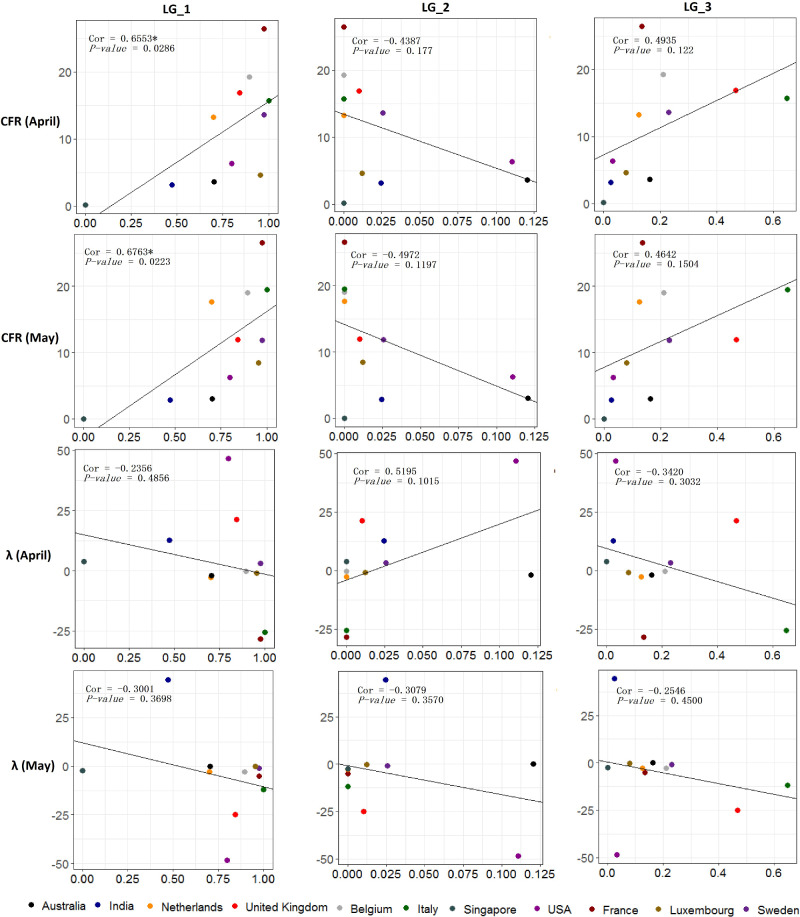
Based on the global COVID-19 cases reported (supplementary table S10, Supplementary Material online) by the European Centre for Disease Prevention and Control (www.ecdc.europa.eu), we calculated the daily CFR and the daily exponential growth rate (λ) (Lipsitch et al. 2003) of COVID-19 in different countries (supplementary tables S11–S13, Supplementary Material online). We also calculated the percentages of minor alleles in different countries in April and May (supplementary table S13, Supplementary Material online). With these parameters, correlation analysis was performed to deduce the possible phenotype of the nine alleles. We performed analysis with the data in 11 countries, for the adequacy of sample size (> 20). We calculated the median of the daily CFR or λ for postcorrelation coefficiency calculation. The result (fig. 5) shows that the mutants in LG_1 is positively correlated with the CFR (both in April, P value = 0.0286, and May, P value = 0.0223), which suggests that the virus possessing mutants in LG_1 is more aggressive than others. Mutants in LG_2 and LG_3 show negative and positive correlations with the CFR respectively, but with no statistical significance. Different from previous predictions (Tang et al. 2020), the mutant in LG_2 (S) is negatively correlated with λ but not significant (P value > 0.1), indicating that S may not have advantages in phenotypes comparing to L.
Fig. 5.
Correlation analysis results between the ratios of mutants (x-axis) and the median case fatality rate (CFR) or the median daily exponential growth rate (λ) of COVID-19 in one month (y-axis). The P value was calculated by Spearman’s test. Coefficients with significance are marked by “*”. Analyses were performed on data collected in April as well as in May.
To further validate the possibility of mutants’ clinical feature, we performed statistics analyses on total 7,692 sequenced strains with information of coordinated patient status (supplementary table S14, Supplementary Material online, from GISAID). We remarked the records based on four pairs of keywords, “Deceased” and “Released”, “Hospitalization” and “Outpatient”, “Symptomatic” and “Asymptomatic”, “Severe” and “Mild”, for convenience in comparison (supplementary table S15, Supplementary Material online, for specifics, see Materials and Methods).
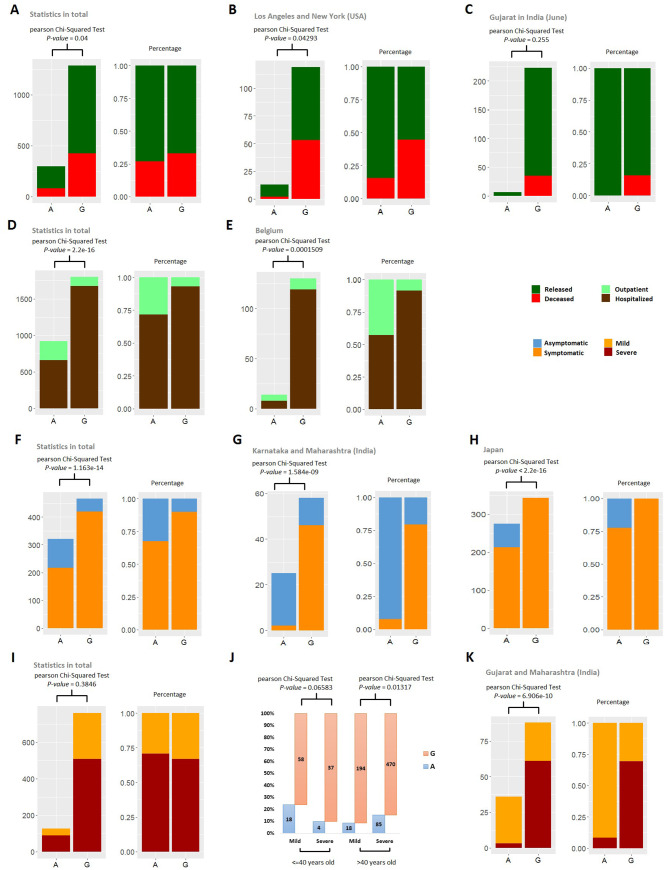
Consistently with the results from correlation analysis based on CFR, mutants in LG_1 show a significant increase in the ratio of deceased patients compared with released patients (fig. 6A). We tried to avoid the bias toward specific geographic areas and performed statistics on the data collected in a small scale, for example, a city or a country/region within a small area. Based on reports on surveys (Czeisler et al. 2020), we have selected Los Angeles and New York for the test. We assumed that the pandemic of COVID-19 and hospital capacity are the same in these two cities. The analysis was repeated using the data of the two cities and obtained a consistent result (fig. 6B). The data collected in June from a third region, Gujarat of India, was used for further validation of the test. These results show bias but not significantly different (fig. 6C).
Fig. 6.
Potential clinical outcomes of the mutants in SNP_23403, LG_1. The mutant is G and the original allele is A. A–C compare two clinical status, deceased and released. D and E, outpatient and hospitalized. F–H, asymptomatic and symptomatic. I–K, severe (hospitalized) and mild (hospitalized). For display, we used different pairs of colors to represent these four pairs of opposite clinical status. In A–K, excluding J, the left subdiagram shows the count and the right shows the percentage. J shows the ratio of the mutant and the original allele (G/A) in patients with mild or severe disease and young (age ≤ 40 years) or old (age > 40 years). J and I are based on the same data. A, D, F, I, and J are based on all available global data and others are based on selected data on small scale. Related description is at the head of each subfigure. For sufficient data in statistics, we assume that the two cities, Los Angeles and New York, in USA are with similar situation, for example, the pandemic of COVID-19 and hospital capacity. We also assumed that those in all area of Japan are similar, those in Karnataka and Maharashtra (India) are similar, and those in Gujarat and Maharashtra (India) are similar.
We also observed that the strains holding mutants in LG_1 have bias toward hospitalization compared with outpatient (fig. 6D and E), and more likely to make patients symptomatic (fig. 6F–H), which is supported by analysis both on global and small scales (including Belgium, two Indian cities Karnataka and Maharashtra, and Japan).
The keyword pair “Severe” and “Mild” describe the severity of the disease for hospitalized patients. In total statistics analysis for this issue, we did not find a significant difference in ratio of severe patients compared with mild ones between the mutant and the original allele in SNP_23403, LG_1 (fig. 6I). However, our results show that young patients (age < 40 years) with SNP_23403 mutant may have a disease more severe than old patients (age > 40 years) with SNP_23403 mutant (fig. 6J). We observed a bias in severity correlated to SNP_23403 of LG_1 in two Indian states (Gujarat and Maharashtra), with the assumption that the two states are in the similar situation facing COVID-19 (fig. 6K).
In the UK, raw sequence data set used for iSNV analysis referred includes 335 records with Ct value. These records are all deposited by the same institution. Based on the records, we performed correlation analysis between Ct and mutants’ IF. The calculation also reveals the relationship between the high efficiency in replication/transmission of virus and the mutant SNP_23403, LG_1 (D614G) (supplementary fig. S12, Supplementary Material online).
Taking together, results from the analyses on patients’ status provide statistical supports that the mutants in LG_1 have correlations with more severe disease development in human. These are predictions based on statistical analysis. It is far from conclusion because of the possible bias caused by several unavoidable factors, such as spatiotemporal changes, how viral genomes got sequenced and deposited in the databases. Until now, parts of our prediction are supported by virology experiments using hamster as animal model, which focused on the clinical influence of SNP_23403, LG_1 (D614G). This experiment indicate that D614G SARS-CoV-2 mutant strains cause more severe pathological changes in the lung tissues when compared with unmutated isolates (Mok et al. 2020). The mutation of D614G has been further confirmed to have correlation with the increase infectivity (Korber et al. 2020).
Our calculation also suggested that the mutants in LG_2 may increase the ratio of symptomatic patients compared with asymptomatic ones if performing statistics in total (supplementary fig. S13A, Supplementary Material online). We observed consistent results based on Japanese data but inconsistent results based on Indian data (supplementary fig. S13B and C, Supplementary Material online). This may be due to the small sample size of the mutants in India, considering the LG_2 mutant is decreasing in IF from January to April (fig. 2). In the comparison of “Hospitalized” and “Outpatient” and “Released” and “Deceased”, the mutants in LG_2 showed a bias toward mildness (supplementary fig. S13D–H, Supplementary Material online). For hospitalized samples, there is no significant difference in statistics in total but a bias toward mildness based on Indian data (supplementary fig. S13I and J, Supplementary Material online). Our calculation suggested that the mutants in LG_2 may result in the mildness of the virus, as proposed by other research group (Tang et al. 2020).
The analysis on the mutants of LG_3 reveal a negative correlation between the mutants and Ct (supplementary fig. S12, Supplementary Material online), suggesting that the LG_3 mutants benefits the increase of viral load. This is supported by another experiment showing an unprecedented capacity of replication of the SARS-CoV-2 GZ69 strain, including the mutant of LG_3, in Vero E6 cells (Caccuri et al. 2020). LG_3 showed a bias toward patients symptomatic (supplementary fig. S14A–C, Supplementary Material online) and patients hospitalized (supplementary fig. S14D and E, Supplementary Material online), but no significant differences in the ration of severity ratio and death ratio (supplementary fig. S14F–J, Supplementary Material online). One explanation is that the mutants in LG_3 may promote viral replication, which is referred in our study described above and previous reports (Caccuri et al. 2020).
Potential Structural Variations Led by the SNPs
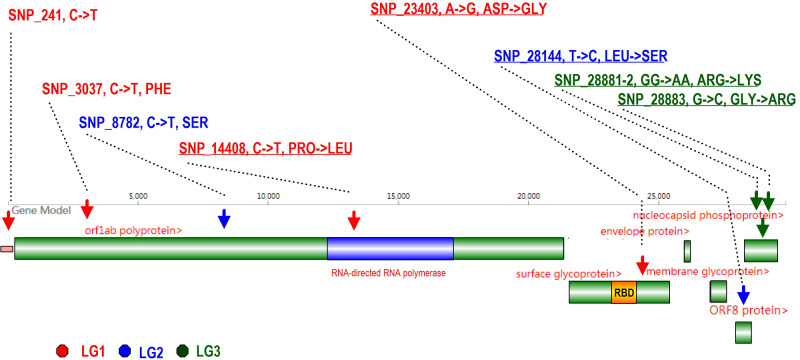
As shown in figure 7, most SNPs (8/9) are located in the coding region. SNP_251 (LG_1) is the only SNP located in the noncoding region. It is located at the leader sequence (Sawicki et al. 2007) in front of ORF1 and may affect the protein-to-RNA interaction (Pasternak et al. 2004, 2006; Sawicki et al. 2007) in the assembly into membrane-bound replication–transcription complexes (Curtis et al. 2004; Zúñiga et al. 2004; Sola et al. 2005; Enjuanes et al. 2006; Yount et al. 2006). Three SNPs are located in ORF1ab. Two are synonymous mutations. The three all are transitions from “C” to “T”. This is consistent with coronavirus codon usage bias toward U-ending (Castells et al. 2017). SNP_14408 (LG_1) is located at the RNA-directed RNA polymerase and the AA mutation from PRO to LEU (P4715L) may lead to a change in protein flexibility and may further influence viral replication. The mutation SNP_23403 (LG_1) located in the surface glycoprotein (S) is 60 AA to the 3' end of the receptor-binding domain. This mutation changes AA side chain polarity, from a negative ASP to a neutral GLY (D614G), which may reduce protein rigidity and increase the membrane fusion efficiency during the virus infection (Korber et al. 2020). SNP_28881, SNP_28882, and SNP_28883 (in LG_3) are three consecutive mutations located in the nucleoprotein (N). They make two AA changes from ARG to LYS (R203K) and from GLY to ARG (G204R). These changes will modify the protein charge distribution, which may affect the recognition of genomic RNA during virus replication and assembly. SNP_28144 (L84S, LG_2) is located in ORF8, which is also related to viral replication (Muth et al. 2018).
Fig. 7.
A diagram showing an overview of the nine SNPs in the virus genome. SNP sites are marked by upside-down arrows and colored according to the linkage groups to which they belong. “RBD” denotes the receptor binding domain, marked in yellow. A pink rectangle ahead of ORF1 denotes the leader sequence for transcription.
Discussion
With the purpose of predicting the evolutionary pattern of SARS-CoV-2 causing the pandemic COVID-19, we performed a thorough analysis of the virus’ genome sequences. Our results show that the IF of LG_1 is increasing continuously from January to April and the mutants are nearly fixed in the end (∼83%). A strain may be sequenced multiple times just because it happens to find its way into a city/country with good sequencing facility, whereas another strain landing in a city with poor sequencing facility may have 0 or low representation in the SARS-CoV-2 database, even if the strain has infected many people. For avoiding the situation that the strains sequenced are not a random sampling over time, we made statistics of mutants’ IF in different countries and test the overall trend. The results show that the rapid increase of the IF of LG_1 is convincing and universal. From population genetic analyses, we also observed corresponding positive selection signatures in some SNP sites of LG_1. Moreover, we confirmed those findings in intrahost level, which also dynamically displayed the process of mutants’ spread (fig. 3 and supplementary figs. S4–S8, Supplementary Material online).
The increments of most mutants are rapid, linked, and simultaneous. This may be resulted from the potential benefit of the mutant in new environments and a successive increasing population size. Beneficial mutants are common in a large population and can dramatically alter the genetic diversity at linked sites (Desai and Fisher 2007). Moreover, the viral population easily experiences evolution via multiple concurrent mutations (Desai and Fisher 2007). Considering the nearby sequences around SNP_3037 and SNP_23403 show the strongest selection signals in LG_1, they may be the causative mutation in these linkage groups. The increments of other two mutants may be driven by hitchhiking effects (Smith and Haigh 2007). Thus, our results suggest paying more attention to these causative SNP sites in further epidemiological investigations of COVID-19.
The newly evolved mutants in LG_1 show to be more effective in replication and may be more aggressive from our analysis in the correlation with CFR, patients’ status, and Ct. Experiments on virus by other groups confirmed those feature of LG_1 (Korber et al. 2020; Mok et al. 2020). In accordance, we observed that the CFR is increasing from March to April in most countries (supplementary fig. S15, Supplementary Material online). The frequency increase of the mutants in LG_1 may be one of the contributor. The S type of LG_2 shows no evolutionary advantages from past four months statistics. Its IF is diminishing continuously from January to April. This indicates that the L type may be more adaptive than S type. S type is at least not positive enough to replace L type in global COVID-19 populations, indicated by our study and other work (Xu et al. 2020). The intrahost dynamics (fig. 3) and the predicted contribution to viral replication of the mutants in LG_3 is a surprise, which may tract more attention for its possible clinical influences. Functions of LG_1 to LG_3 are only predictions from statistics and correlation analyses in this study. More animal or clinical experiments are needed to test these linked mutants’ function (Caccuri et al. 2020; Korber et al. 2020; Mok et al. 2020; Xu et al. 2020). They are potential targets for medicine and clinical researches.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We gratefully acknowledge the submitting and the originating laboratories where genetic sequence data were generated and shared via NCBI and the GISAID Initiative. This work was supported by grants from the National Key Research and Development Program (2019YFC1604600), the National Natural Science Foundation of China (31200941, 31660713), the Fundamental Research Funds for the Central Universities (106112016CDJXY290002), and the National Natural Science Foundation of HeBei province (19226631D).
Author Contributions
Z.Z. collected, analyzed, and compiled the data. G.L., K.M., L.Y., and D.L. took part in the analysis. Z.Z. and G.M. conceived the idea, coordinated the project, and wrote the manuscript.
Data Availability
The consensus sequences of SARS-CoV-2 strains and related information are available in GISAID (gisaid.org, IDs are in supplementary tables S1 and S14, Supplementary Material online), NCBI GenBank (MN908947 and the 13 consensus genomes used in iSNV analysis), CNGB (db.cngb.org, Accession is CNP0001004), and CoVdb (covdb.popgenetics.net). The raw sequencing data sets used for iSNV analysis are available in CNGB (CNP0000997) and NCBI SRA database (accessions are PRJNA627662, PRJNA613958, PRJNA614995, and PRJEB37886). The geographic distribution of COVID-19 cases worldwide is available in an online file provided by the European Centre for Disease Prevention and Control (https://www.ecdc.europa.eu/sites/default/files/documents/COVID-19-geographic-disbtribution-worldwide.xlsx). The Perl scripts used in the research referred in this manuscript are available in Github (https://github.com/alahunan/GBE200707).
Literature Cited
- Ayres JS 2020. Surviving COVID-19: a disease tolerance perspective. Sci Adv 6(18):eabc1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccuri F, et al. 2020. A persistently replicating SARS-CoV-2 variant derived from an asymptomatic individual. J Transl Med 18(1):362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells M, Victoria M, Colina R, Musto H, Cristina J. 2017. Genome-wide analysis of codon usage bias in Bovine Coronavirus. Virol J 14(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, et al. 2020. Detection of 2019 Novel Coronavirus (2019-nCoV) by Real-Time RT-PCR. Euro Surveill. 25(36):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis KM, Yount B, Sims AC, Baric RS. 2004. Reverse genetic analysis of the transcription regulatory sequence of the coronavirus transmissible gastroenteritis virus. JVI 78(11):6061–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeisler ME, et al. 2020. Public attitudes, behaviors, and beliefs related to COVID-19, stay-at-home orders, nonessential business closures, and public health guidance – United States, New York City, and Los Angeles, May 5–12, 2020. MMWR Morb Mortal Wkly Rep 69(24):751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGiorgio M, Huber CD, Hubisz MJ, Hellmann I, Nielsen R. 2016. SweepFinder2: increased sensitivity, robustness and flexibility. Bioinformatics 32(12):1895–1897. [DOI] [PubMed] [Google Scholar]
- DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MM, Fisher DS. 2007. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics 176(3):1759–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2004a. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2004b. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L, Almazan F, Sola I, Zuniga S. 2006. Biochemical aspects of coronavirus replication and virus-host interaction. Annu Rev Microbiol 60(1):211–230. [DOI] [PubMed] [Google Scholar]
- Fehr AR, Perlman S. 2015. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 1282:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, et al. 2013. Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195(3):979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS 2007. Improved pairwise alignment of genomic DNA. [Ph.D. thesis]: [State College (PA)]: Pennsylvania State University.
- Holsinger KE, Weir BS. 2009. Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat Rev Genet 10(9):639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18(2):337–338. [DOI] [PubMed] [Google Scholar]
- Hulo C, et al. 2011. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res 39(suppl_1):D576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter S, Vilella AJ, Rozas J. 2006. Genome-wide DNA polymorphism analyses using VariScan. BMC Bioinformatics 7(1):409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, et al. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182(4):812–827. e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT, et al. 2020. Identifying SARS-CoV-2. Related Coronaviruses in Malayan Pangolins. Nature 583(7815):282–285. [DOI] [PubMed] [Google Scholar]
- Langdon WB 2015. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min 8(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC 1964. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49(1):49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H Genome Project Data Processing S, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M, et al. 2003. Transmission dynamics and control of severe acute respiratory syndrome. Science 300(5627):1966–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, et al. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10224):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok BW-Y, et al. Forthcoming 2020. SARS-CoV-2 spike D614G variant exhibits highly efficient replication and transmission in hamsters. bioRxiv.
- Morton NE 1955. Sequential tests for the detection of linkage. Am J Hum Genet 7(3):277–318. [PMC free article] [PubMed] [Google Scholar]
- Muth D, et al. 2018. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci Rep 8(1):15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. 2006. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441(7090):244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, et al. 2005. Genomic scans for selective sweeps using SNP data. Genome Res 15(11):1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak AO, Spaan WJ, Snijder EJ. 2004. Regulation of relative abundance of arterivirus subgenomic mRNAs. JVI 78(15):8102–8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak AO, Spaan WJ, Snijder EJ. 2006. Nidovirus transcription: how to make sense…? J Gen Virol 87(6):1403–1421. [DOI] [PubMed] [Google Scholar]
- Ralph R, et al. 2020. 2019-nCoV (Wuhan virus), a novel Coronavirus: human-to-human transmission, travel-related cases, and vaccine readiness. J Infect Dev Ctries 14(1):3–17. [DOI] [PubMed] [Google Scholar]
- Richard D Owen CJ van Dorp L Balloux F. 2020. No detectable signal for ongoing genetic recombination in SARS-CoV-2. bioRxiv. [Google Scholar]
- Rueca M, et al. 2020. Compartmentalized replication of SARS-Cov-2 in upper vs. lower respiratory tract assessed by whole genome quasispecies analysis. Microorganisms 8(9):1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki SG, Sawicki DL, Siddell SG. 2007. A contemporary view of coronavirus transcription. JVI 81(1):20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Higgins DG. 2014. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol 1079:105–116. [DOI] [PubMed] [Google Scholar]
- Slatkin M 2008. Linkage disequilibrium – understanding the evolutionary past and mapping the medical future. Nat Rev Genet 9(6):477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Haigh J. 2007. The hitch-hiking effect of a favourable gene. Genet Res 89(5–6):391–403. [DOI] [PubMed] [Google Scholar]
- Sola I, Moreno JL, Zúñiga S, Alonso S, Enjuanes L, 2005. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. JVI 79(4):2506–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F 1993. Measurement of DNA polymorphism. In: Takahata N and Clark AG, editors. Mechanisms of molecular evolution. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Tang X, et al. 2020. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev 7(6):1012–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, Blanco-Garcia A, Hutter S, Rozas J. 2005. VariScan: analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics 21(11):2791–2793. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Forthcoming 2020. Intra-host variation and evolutionary dynamics of SARS-CoV-2 population in COVID-19 Patients. bioRxiv. [DOI] [PMC free article] [PubMed]
- Xu Y, et al. Forthcoming 2020. Hybrid capture-based sequencing enables unbiased recovery of SAR-CoV-2 genomes from fecal samples and characterization of the dynamics of intra-host variants. bioRxiv.
- Yi H 2020. 2019 novel coronavirus is undergoing active recombination. Clin Infect Dis 71(15):884–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B, Roberts RS, Lindesmith L, Baric RS. 2006. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc Natl Acad Sci U S A 103(33):12546–12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z-Y, et al. Forthcoming 2020. Worldwide tracing of mutations and the evolutionary dynamics of SARS-CoV-2. bioRxiv.
- Zhu L, Bustamante CD. 2005. A composite-likelihood approach for detecting directional selection from DNA sequence data. Genetics 170(3):1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Meng K, Liu G, Meng G Forthcoming 2020. A database resource and online analysis tools for coronaviruses on a historical and global scale. Database (Oxford). [DOI] [PMC free article] [PubMed]
- Zúñiga S, Sola I, Alonso S, Enjuanes L, 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. JVI 78(2):980–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The consensus sequences of SARS-CoV-2 strains and related information are available in GISAID (gisaid.org, IDs are in supplementary tables S1 and S14, Supplementary Material online), NCBI GenBank (MN908947 and the 13 consensus genomes used in iSNV analysis), CNGB (db.cngb.org, Accession is CNP0001004), and CoVdb (covdb.popgenetics.net). The raw sequencing data sets used for iSNV analysis are available in CNGB (CNP0000997) and NCBI SRA database (accessions are PRJNA627662, PRJNA613958, PRJNA614995, and PRJEB37886). The geographic distribution of COVID-19 cases worldwide is available in an online file provided by the European Centre for Disease Prevention and Control (https://www.ecdc.europa.eu/sites/default/files/documents/COVID-19-geographic-disbtribution-worldwide.xlsx). The Perl scripts used in the research referred in this manuscript are available in Github (https://github.com/alahunan/GBE200707).