Abstract
The COVID-19 pandemic has created unprecedented challenges worldwide. Strained healthcare providers make difficult decisions on patient triage, treatment and care management on a daily basis. Policy makers have imposed social distancing measures to slow the disease, at a steep economic price. We design analytical tools to support these decisions and combat the pandemic. Specifically, we propose a comprehensive data-driven approach to understand the clinical characteristics of COVID-19, predict its mortality, forecast its evolution, and ultimately alleviate its impact. By leveraging cohort-level clinical data, patient-level hospital data, and census-level epidemiological data, we develop an integrated four-step approach, combining descriptive, predictive and prescriptive analytics. First, we aggregate hundreds of clinical studies into the most comprehensive database on COVID-19 to paint a new macroscopic picture of the disease. Second, we build personalized calculators to predict the risk of infection and mortality as a function of demographics, symptoms, comorbidities, and lab values. Third, we develop a novel epidemiological model to project the pandemic’s spread and inform social distancing policies. Fourth, we propose an optimization model to re-allocate ventilators and alleviate shortages. Our results have been used at the clinical level by several hospitals to triage patients, guide care management, plan ICU capacity, and re-distribute ventilators. At the policy level, they are currently supporting safe back-to-work policies at a major institution and vaccine trial location planning at Janssen Pharmaceuticals, and have been integrated into the US Center for Disease Control’s pandemic forecast.
Electronic supplementary material
The online version of this article (10.1007/s10729-020-09542-0) contains supplementary material, which is available to authorized users.
Supplementary Information
The online version contains supplementary material available at (10.1007/s10729-020-09542-0)
Keywords: COVID-19, Epidemiological modeling, Machine learning, Optimization
Highlights
This paper introduces a comprehensive data-driven approach for the COVID-19 pandemic with goals to inform the overall scientific community, to estimate the epidemiological spread of the virus, to provide clinical insights, and to support ventilator allocation decisions for policy makers.
To consolidate medical insights, a clinical database on the disease is aggregated from available scientific literature.
To assess the risk of infection and mortality, we provide personalized risk prediction models from the electronic health records by leveraging machine learning algorithms.
To forecast the progression of COVID-19 and evaluate the impact of various social distancing policies, we develop a dynamic epidemiological model.
To inform operational decisions for government officials, our optimization model addresses surges in ventilator demand through state-level and federal reallocation.
Introduction
In just a few weeks, the whole world was upended by the outbreak of COVID-19, an acute respiratory disease caused by a new coronavirus called SARS-CoV-2. The virus is highly contagious: it is easily transmitted from person to person via respiratory droplet nuclei and can persist on surfaces for days [22, 43]. As a result, COVID-19 has spread rapidly—classified by the World Health Organization as a public health emergency on January 30, 2020 and as a pandemic on March 11. As of November 2020, over 51 million cases and 1.2 million deaths have been reported globally [20].
Given the uncertainty surrounding the disease and its treatment, healthcare providers and policy makers have wrestled with unprecedented challenges. Hospitals and other care facilities have faced shortages of beds, ventilators and personal protective equipment—raising hard questions on how to treat COVID-19 patients with scarce supplies and how to allocate resources to prevent further shortages. At the policy level, most countries have imposed “social distancing” measures and other non-pharmaceutical interventions to slow the spread of the pandemic. These measures allow strained healthcare systems to cope with the disease by “flattening the curve” [2] but also come at a steep economic price [11, 32]. This trade-off has prompted difficult decisions balancing public health and socio-economic outcomes.
This paper proposes a comprehensive data-driven approach to combat the COVID-19 pandemic. We leverage a broad range of data sources, which include (i) our own cohort-level data aggregating hundreds of clinical studies, (ii) patient-level data obtained from electronic health records, and (iii) census reports on the scale of the pandemic. We develop an integrated approach spanning descriptive analytics (to derive a macroscopic understanding of the disease), predictive analytics (to forecast the near-term impact and longer-term dynamics of the pandemic), and prescriptive analytics (to support healthcare and policy decision-making).
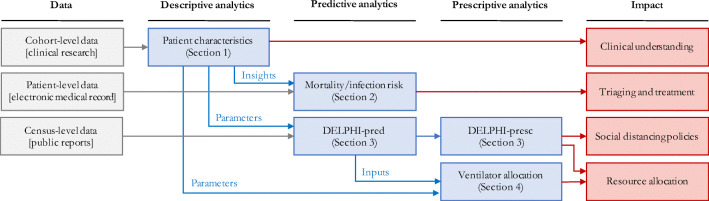
Our approach comprises four steps (Fig. 1):
Aggregating and visualizing the most comprehensive clinical database on COVID-19 as of May 2020 (Section 1). We aggregate cohort-level data on demographics, comorbidities, symptoms and lab values from 160 clinical studies. These data paint a broad picture of the disease: identifying common symptoms, disparities between mild and severe patients, and geographic disparities—insights that are hard to derive from any single study and can orient future clinical research on COVID-19, its mutations, and its disparate effects across ethnic groups.
Providing personalized indicators to assess the risk of mortality and infection (Section 2). Using patient- level data, we develop machine learning models to predict mortality and infection risk, as a function of demographics, symptoms, comorbidities, and lab values. Using gradient boosting methods, the models achieve strong predictive performance—with an out-of-sample area under the curve above 90%. These models yield personalized calculators that can (i) guide triage, treatment, and care management decisions for strained healthcare systems, and (ii) serve as pre-screening tools for patients before they visit healthcare or testing facilities.
Developing a novel epidemiological model to forecast the evolution of the disease and assess the effects of social distancing (Section 3). We propose a new compartmental model called DELPHI, which accounts for COVID-19 features such as underdetection and government response. The model estimates the disease’s spread with high accuracy; notably, its projections from as early as April 3 have matched the number of cases observed in the United States up to mid-May and outperforms comparable methods during such period. We also provide a data-driven assessment of social distancing policies, showing that the pandemic’s spread is highly sensitive to the stringency and timing of mitigating measures.
Proposing an optimization model to support ventilator allocation in response to the pandemic (Section 4). We formulate a mixed-integer optimization model to allocate ventilators efficiently in a semi-collaborative setting where resources can be shared both between healthcare facilities or through a central authority. In the United States, this allows us to study the trade-offs of managing the federal ventilator stockpile in conjunction with inter-state transfers. Results show that limited ventilator transfers could have eliminated shortages in April 2020.
Fig. 1.
Overview of our end-to-end analytics approach. We leverage diverse data sources to inform a family of descriptive, predictive and prescriptive tools for clinical and policy decision-making support
This work makes two key contributions. First, we derive data-driven insights about the early stages of the COVID-19 pandemic. Although some of the results should be treated with caution when extrapolated beyond the period spanning March to May 2020, these insights help understand the clinical characteristics of the disease, predict its mortality, forecast its evolution, and ultimately alleviate its impact. Second, we provide a comprehensive roadmap to guide short-term responses to new, and unforeseen epidemics. The proposed approach involves four steps: (i) gathering meta-data from early small-scale clinical studies to derive a fast and broad understanding of the disease, (ii) applying predictive analytics based on patient-level data to identify the drivers of the disease and its mortality, (iii) using population-level data on cases, hospitalizations and deaths to predict the macroscopic evolution of the disease, and (iv) leveraging these models for resource allocation optimization to alleviate the near-term damage of the disease. A major feature of this approach is to treat these different questions as interdependent challenges, as opposed to a series of isolated problems. Indeed, clinical decision-making depends directly on patient inflows and available supplies, while resource planning and government responses react to patient-level outcomes. By combining various data sources into descriptive, predictive and prescriptive methods, this paper proposes an end-to-end approach to design a comprehensive and cohesive response to the COVID-19 pandemic and future epidemics.
Ultimately, this paper develops analytical tools to inform clinical and policy responses to the COVID-19 pandemic. These tools are available to the public on a dedicated website. 1 They have also been deployed in practice to combat the spread of COVID-19 globally. Several hospitals in Europe have used our risk calculators to support pre-triage and post-triage decisions, and a major financial institution in South America is applying our infection risk calculator to determine how employees can safely return to work. A major hospital system in the United States, Hartford Healthcare, planned its intensive care unit (ICU) capacity based on our forecasts, and leveraged our optimization results to allocate ventilators across hospitals when the number of cases was rising. Our epidemiological predictions are used by one of the largest pharmaceutical companies, Janssen Pharmaceuticals, to design a vaccine trial location selection strategy . They have also been one of the top 5 models that are consistently incorporated into the US Center for Disease Control’s forecasts [42] and its core ensemble model.
Descriptive analytics: clinical outcomes database
Early responses to the COVID-19 pandemic have been inhibited by the lack of available data on patient outcomes. Individual centers released reports summarizing patient characteristics. Yet, this decentralized effort makes it difficult to construct a cohesive picture of the pandemic.
To address this problem, we construct a database that aggregates demographics, comorbidities, symptoms, laboratory blood test results (“lab values”, henceforth) and clinical outcomes from 160 clinical studies released between December 2019 and May 2020—made available on our website for broader use. The database contains information on 133,600 COVID-19 patients (3.13% of the global COVID-19 patients as of May 12, 2020), spanning mainly Europe (81,207 patients), Asia (19,418 patients) and North America (23,279 patients). To our knowledge, this is the largest dataset on COVID-19 that contains detailed clinical outcomes (as of May 2020).
Data aggregation
Each study was read by a researcher, who transcribed numerical data from the manuscript, and subsequently checked by a second researcher, to verify correctness. The Appendix reports the main transcription assumptions.
Each row in the database corresponds to a cohort of patients—some papers study a single cohort, whereas others study several cohorts or sub-cohorts. Each column reports cohort-level statistics on demographics (e.g., average age, gender breakdown), comorbidities (e.g., prevalence of diabetes, hypertension), symptoms (e.g., prevalence of fever, cough), treatments (e.g., prevalence of antibiotics, intubation), lab values (e.g., average lymphocyte count), and clinical outcomes (e.g., average hospital length of stay, mortality rate). We also track whether the cohort comprises “mild” or “severe” patients (mild and severe cohorts are only a subset of the data).
We should note that, as the pandemic has progressed, clinical knowledge of COVID-19 has improved beyond what was knowable when this study was performed. As a result, the prevalence of the symptoms and the mortality rates reported here may not accurately reflect the current status of the pandemic. For instance, Anosmia (loss of sense of taste/smell) is now recognized as one of the main symptoms of COVID-19 but is not mentioned in this section. Similarly, mortality rates have dropped significantly since March/April due, in particular, to advances by the medical community.
Due to the pandemic’s urgency, many papers were published before all patients in a cohort were discharged or deceased. Accordingly, we estimate the mortality rate from discharged and deceased patients only (referred to as “Projected Mortality”).
Objectives
Our main goal is to leverage this database to derive a macroscopic understanding of the disease. We break it down into the following questions:
Which symptoms are most prevalent?
How do “mild” and “severe” patients differ in terms of symptoms, comorbidities, and lab values?
Can we identify epidemiological differences in different parts of the world?
Descriptive statistics
Table 1 depicts the prevalence of COVID-19 symptoms, in aggregate, classified into “mild” or “severe” patients, and classified per geographic region. Our key observations are that:
Cough, fever, shortness of breath, and fatigue are the most prevalent symptoms of COVID-19.
COVID-19 symptoms are much more diverse than those listed by public health agencies. COVID-19 patients can experience at least 15 different symptoms. In contrast, the US Center for Disease Control and Prevention lists seven symptoms (cough, shortness of breath, fever, chills, myalgia, sore throat, and loss of taste/smell) [47]; the World Health Organization lists three symptoms (fever, cough, and fatigue) [52]; and the UK National Health Service lists two main symptoms (fever and cough) [35]. This suggests a lack of consensus among the medical community, and opportunities to revisit public health guidelines to capture the breadth of observed symptoms.
Shortness of breath and elevated respiratory rates are much more prevalent in cases diagnosed as severe.
Symptoms are quite different in Asia vs. Europe or North America. In particular, more than 75% of Asian patients experience fever, as compared to less than half in Europe and North America. Conversely, shortness of breath is much more prevalent in Europe and North America.
Table 1.
Count and prevalence of symptoms among COVID-19 patients, in aggregate, broken down into mild/severe patients, and broken down per continent (Asia, Europe, North America)
| Symptom | All patients | Mild | Severe | Asia | Europe | North America | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | (%) | Count | (%) | Count | (%) | Count | (%) | Count | (%) | Count | (%) | ||||||
| Cough | 94,950 | 52.8% | 6,833 | 63.0% | 5,803 | 50.4% | 14,034 | 56.2% | 78,430 | 52.2% | 1,113 | 63.6% | |||||
| Fever | 95,870 | 48.1% | 6,864 | 79.3% | 6,077 | 76.7% | 14,750 | 76.6% | 78,450 | 43.5% | 1,481 | 41.3% | |||||
| Short Breath | 17,290 | 33.7% | 6,006 | 16.1% | 5,373 | 60.7% | 11,330 | 19.7% | 3,512 | 69.9% | 1,111 | 49.2% | |||||
| Fatigue | 11,560 | 31.4% | 5,313 | 35.3% | 1,989 | 40.6% | 11,320 | 30.8% | 226 | 64.2% | − | − | |||||
| Sputum | 7,613 | 26.3% | 4,995 | 29.2% | 1,216 | 34.2% | 7,395 | 26.7% | − | − | 176 | 10.9% | |||||
| Sore Throat | 83,170 | 22.2% | 3,513 | 14.2% | 921 | 8.2% | 6,013 | 10.4% | 75,235 | 22.9% | 550 | 9.8% | |||||
| Myalgia | 12,150 | 17.5% | 4,455 | 16.4% | 1,643 | 19.1% | 8,517 | 15.5% | 1,633 | 33.5% | 755 | 25.3% | |||||
| Elev. Resp. Rate | 7,376 | 16.4% | 527 | 9.7% | 642 | 38.4% | 1,257 | 14.6% | − | − | 6,117 | 16.8% | |||||
| Anorexia | 3,928 | 15.8% | 1,641 | 14.2% | 808 | 15.4% | 3,566 | 13.8% | 312 | 40.5% | − | − | |||||
| Headache | 11,430 | 15.7% | 5,068 | 12.2% | 1,541 | 8.6% | 7,929 | 9.9% | 1,633 | 27.2% | 551 | 8.7% | |||||
| Nausea | 10,070 | 12.4% | 4,238 | 6.5% | 1,798 | 5.6% | 8,262 | 8.2% | 312 | 22.4% | 259 | 9.0% | |||||
| Chest Pain | 3,303 | 11.3% | 767 | 12.2% | 588 | 19.6% | 2,984 | 12.2% | − | − | − | − | |||||
| Diarrhea | 16,520 | 11.1% | 5,687 | 9.7% | 5,369 | 9.0% | 11,470 | 10.8% | 3,512 | 10.4% | 1,066 | 15.4% | |||||
| Cong. Airway | 1,639 | 8.7% | 2,176 | 6.5% | 234 | 14.1% | 1,369 | 8.9% | − | − | 258 | 7.4% | |||||
| Chills | 3,116 | 8.7% | 2,751 | 9.9% | 520 | 9.4% | 2,794 | 8.2% | − | − | 268 | 11.5% | |||||
| Proj. Mortality | 111,700 | 11.7% | 7,428 | 0.4% | 9,146 | 74.0% | 12,820 | 16.7% | 79,750 | 9.9% | 19,060 | 15.8% | |||||
Mild and severe patients only form a subset of the data, and so do patients from Asia, Europe and North America. A “-” indicates that fewer than 100 patients in a subpopulation reported on this symptom
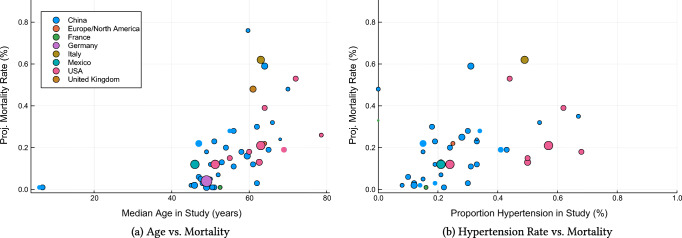
Using a similar nomenclature, Table 2 reports comorbidities, demographics, average lab values and average clinical outcomes among all patients, mild patients and severe patients. In terms of demographics, severe populations of patients have a higher incidence of male subjects and are older on average. Severe patients also have elevated comorbidity rates. Figure 2 visually confirms the impact of age and hypertension rates on population-level mortality—consistent with [15, 16, 40]. In terms of lab values, CRP, AST, BUN, IL-6 and Protocalcitonin are highly elevated among severe patients.
Table 2.
Comorbidities, demographics, average lab values, average length of stay and projected mortality among COVID-19 patients, in aggregate and broken down into mild/severe patients
| Feature | All Patients | Mild Patients | Severe Patients | |||||
|---|---|---|---|---|---|---|---|---|
| No. Report | Avg. | No. Report. | Avg. | No. Report. | Avg. | |||
| Demographics | ||||||||
| Male (%) | 131,200 | 53.0% | 9,570 | 48.8% | 10,120 | 68.7% | ||
| Age (years) | 119,000 | 51.3 | 8,022 | 46.1 | 9,685 | 68.2 | ||
| White/European (%) | 55,490 | 22.2% | 10,120 | 9.7% | 9,887 | 63.9% | ||
| African American (%) | 55,490 | 5.4% | 10,120 | 3.5% | 9,887 | 2.5% | ||
| Asian (%) | 55,320 | 51.3% | 10,300 | 80.2% | 9,933 | 31.2% | ||
| Hispanic/Latino | 50,630 | 19.9% | 8,017 | 0% | 9,107 | 0% | ||
| Multiple ethnicities/other | 55,190 | 3.6% | 10,120 | 6.9% | 9,887 | 2.7% | ||
| Comorbidities | ||||||||
| Smoking history | 27,900 | 16.1% | 6,080 | 12.2% | 1,973 | 16.6% | ||
| Hypertension | 38,390 | 35.9% | 8,252 | 15.2% | 8,449 | 54.4% | ||
| Diabetes | 39,790 | 20.8% | 8,396 | 6.8% | 8,818 | 26.1% | ||
| Cardio Disease | 40,030 | 12.4% | 8,028 | 3.0% | 9,540 | 20.3% | ||
| COPD | 34,150 | 6.0% | 6,297 | 2.8% | 8,727 | 10.0% | ||
| Cancer | 29,170 | 7.2% | 6,259 | 3.2% | 8,355 | 12.9% | ||
| Liver Disease | 18,300 | 2.8% | 1,875 | 2.3% | 6,832 | 3.5% | ||
| Cebrovascular | 6,830 | 9.8% | 3,245 | 2.7% | 1,360 | 24.8% | ||
| Kidney Disease | 35,500 | 5.7% | 6,152 | 1.2% | 8,139 | 10.8% | ||
| Lab values | ||||||||
| WBC Count (109/L) | 19,970 | 6.41 | 5,403 | 5.07 | 2,305 | 6.80 | ||
| Neutrophil Count (109/L) | 12,500 | 4.72 | 2,236 | 5.12 | 1,410 | 5.78 | ||
| Platelet Count (109/L) | 12,125 | 195.7 | 5,165 | 184.0 | 2,105 | 170.4 | ||
| ALT (U/L) | 14,467 | 29.0 | 2,840 | 24.6 | 2,428 | 31.1 | ||
| AST (U/L) | 14,214 | 37.3 | 2,766 | 27.1 | 2,366 | 45.7 | ||
| BUN (mmol/L) | 4,822 | 5.22 | 1,700 | 4.18 | 1,138 | 6.86 | ||
| Creatinine (μ mol/L) | 8,504 | 63.08 | 2,529 | 66.0 | 2,454 | 56.4 | ||
| CRP Count (mg/L) | 17,090 | 76.5 | 2,573 | 18.9 | 2,339 | 94.1 | ||
| Interleukin-6 (pg/mL) | 2,582 | 24.57 | 1,127 | 4.17 | 552 | 38.63 | ||
| Procalcitonin (ng/mL) | 14,750 | 2.26 | 1,468 | 1.85 | 1,969 | 4.81 | ||
| D-Dimer (mg/L) | 13,330 | 38.81 | 2,478 | 8.04 | 2,401 | 165.9 | ||
| Length of Stay (days) | 16,010 | 10.7 | 4,131 | 14.0 | 5,642 | 7.97 | ||
| Proj. Mortality (%) | 111,700 | 11.7% | 7,428 | 0.4% | 9,146 | 74.0% | ||
Fig. 2.
Impact of cohort characteristics on projected mortality, assessed at a cohort level. The size of each dot represents the number of patients in the cohort, and its color represents the nation the study was performed in. We only include studies reporting both discharged and deceased patients
Discussion and impact
Our database is the largest available source of clinical information on COVID-19 assembled to date (as of May 2020). As such, it provides new insights on common symptoms and the drivers of the disease’s severity. Ultimately, this database can support guidelines from health organizations, and contribute to ongoing clinical research on the disease.
Another benefit of this database is its geographical reach. Results highlight disparities in patients’ symptoms across regions. These disparities may stem from (i) different reporting criteria; (ii) different treatments; (iii) disparate impacts across different ethnic groups; and (iv) mutations of the virus since it first appeared in China. This information contributes to early evidence on COVID-19 mutations [12, 17] and on its disparate effects on different ethnic groups [13, 48].
Finally, the database provides average values of key parameters into our epidemiological model of the disease’s spread and our optimization model of resource allocation (e.g., average length of stay of hospitalizations, average fraction of hospitalized patients put on a ventilator).
The insights derived from this descriptive analysis highlight the need for personalized data-driven clinical indicators. Yet, our population-level database cannot be leveraged directly to support decision-making at the patient level. We have therefore initiated a multi-institution collaboration to collect electronic medical records from COVID-19 patients and develop clinical risk calculators. These calculators, presented in the next section, are informed by several of our descriptive insights. Notably, the disparities between severe patients and the rest of the patient population inform the choice of the features included in our mortality risk calculator. Moreover, the geographic disparities suggest that data from Asia may be less predictive when building infection or mortality risk calculators designed for patients in Europe or North America—motivating our use of data from Europe.
Predictive analytics: mortality and infection risk
Throughout the COVID-19 crisis, physicians have made difficult triage and care management decisions on a daily basis. These decisions initially relied on small-scale clinical tests; each clinical test requires significant time, personnel and equipment and thus cannot be easily replicated. As the burden on “hot spots” ebbed in late spring, hospitals began to aggregate rich data on COVID-19 patients. This data offers opportunities to develop algorithmic risk calculators for large-scale decision support, facilitating a more proactive and data-driven strategy to combat the disease globally.
We have established a patient-level database of thousands of COVID-19 hospital admissions. Using state-of-the-art machine learning methods, we develop a mortality risk calculator and an infection risk calculator. The resultant models enable the rapid identification of risk factors and clinical insights which were lacking at the pandemic’s onset. Machine learning is particularly useful in such a setting, where we seek to analyze diverse datasets in a timely and scalable fashion. These personalized risk assessment tools can support critical care management decisions, spanning hospital triage to testing prioritization.
Methods
This investigation constitutes a multi-center study from healthcare institutions in Spain and Italy, two countries severely impacted by COVID-19. Specifically, we collected data from (i) Azienda Socio-Sanitaria Territoriale di Cremona (ASST Cremona), the main hospital network in the Province of Cremona, and (ii) HM Hospitals, a leading hospital group in Spain with 15 general hospitals and 21 clinical centers spanning the regions of Madrid, Galicia, and León. We applied the following inclusion criteria to the calculators:
Mortality Risk: We include adult patients diagnosed with COVID-19 and hospitalized. We consider patients who were either discharged from the hospital or deceased within the visit—excluding active patients. We include only lab values and vital values collected on the first day in the emergency department to match the clinical decision setting—predicting prognosis at the time of admission.
Infection Risk: We include adult patients who underwent a polymerase chain reaction test for detecting COVID-19 infection at the ASST Cremona hospital [26].2 We include all patients, regardless of their clinical outcome. Each patient was subject to a blood test. We omit comorbidities since they are derived from the discharge diagnoses, hence not available for all patients.
We train two models for each calculator: one with lab values and one without lab values. Missing values are imputed using k-nearest neighbors imputation [46]. We exclude features missing for more than 40% of patients. We train binary classification models for both risk calculators, using the XGBoost algorithm [8]. We use SHapley Additive exPlanations (SHAP) [30, 31] to generate importance plots that identify risk drivers and provide transparency on the model predictions. All statistical analyses have been conducted using Python 3.7 [38].
To evaluate predictive performance, we use 40 random data partitions into training and test sets. We compute the average Area Under the Curve (AUC), sensitivity, specificity, precision, negative predictive value, and positive predictive value. We calculate 95% confidence intervals using bootstrapping.
Results
Study population
The mortality study population comprises 2,831 patients, 711 (25.1%) of whom died during hospitalization while the remaining ones were discharged. Table 3 summarizes the clinical characteristics of the cohort, both in aggregate and broken down by survival status. The reported features are those used in the final model, that is age, gender, 3 vitals values, 13 lab results, and 4 comorbidities.
Table 3.
Characteristics of study population for mortality prediction model
| All (N = 2, 831) | Survivors (N = 2, 120) | Non-Survivors (N = 711) | P-Value | |
|---|---|---|---|---|
| Age | 68.0 (57.0-79.0) | 63.0 (54.0-74.0) | 81.0 (73.2-86.0) | 1.28E-185 |
| Female a | 1095.0 (38.7%) | 868.0 (40.9%) | 227.0 (31.9%) | 1.18E-05 |
| Heart Rate | 89.0 (79.0-101.0) | 90.0 (80.0-102.0) | 87.0 (78.0-100.0) | 1.29E-03 |
| Oxygen Saturation | 94.0 (90.0-96.0) | 94.4 (92.0-96.0) | 88.5 (80.0-93.6) | 3.16E-37 |
| Temperature (F) | 98.4 (97.5-99.7) | 98.4 (97.5-99.6) | 98.8 (97.7-100.0) | 2.42E-04 |
| Alanine Aminotransferase | 27.0 (17.0-44.0) | 27.8 (17.5-45.0) | 25.5 (16.0-41.0) | 3.77E-02 |
| Aspartate Aminotransferase | 36.0 (25.0-55.0) | 34.0 (24.4-51.0) | 45.0 (30.0-69.0) | 1.55E-11 |
| Blood Glucose | 118.0 (105.0-141.0) | 115.0 (103.4-133.0) | 134.0 (113.0-171.0) | 1.12E-22 |
| Blood Urea Nitrogen | 17.0 (12.6-25.2) | 15.0 (11.5-20.0) | 29.5 (20.3-47.2) | 1.02E-65 |
| C-Reactive Protein | 74.2 (29.1-149.5) | 58.6 (22.7-119.3) | 141.1 (72.0-223.1) | 4.76E-50 |
| Creatinine | 1.0 (0.8-1.2) | 0.9 (0.7-1.1) | 1.3 (1.0-1.8) | 2.84E-36 |
| Hemoglobin | 13.9 (12.7-15.0) | 14.0 (12.9-15.0) | 13.5 (12.0-14.7) | 9.11E-10 |
| Mean Corpsular Volume | 87.8 (84.9-91.0) | 87.5 (84.7-90.4) | 89.3 (85.8-92.7) | 2.80E-08 |
| Platelets | 201.0 (156.0-263.0) | 206.0 (160.0-266.5) | 185.0 (141.0-246.8) | 6.62E-08 |
| Potassium | 4.1 (3.7-4.4) | 4.0 (3.7-4.4) | 4.1 (3.7-4.6) | 1.43E-04 |
| Prothrombin Time (INR) | 1.1 (1.0-1.2) | 1.1 (1.0-1.2) | 1.1 (1.0-1.3) | 3.20E-05 |
| Sodium | 137.1 (135.0-140.0) | 137.0 (135.0-139.4) | 138.0 (135.0-141.0) | 5.65E-08 |
| White Blood Cell Count | 6.8 (5.2-9.2) | 6.5 (5.0-8.7) | 8.0 (5.7-11.4) | 3.00E-15 |
| Cardiac dysrhythmias a | 200.0 (7.1%) | 127.0 (6.0%) | 73.0 (10.3%) | 6.50E-04 |
| Chronic kidney disease a | 65.0 (2.3%) | 33.0 (1.6%) | 32.0 (4.5%) | 3.67E-04 |
| Heart disease a | 125.0 (4.4%) | 80.0 (3.8%) | 45.0 (6.3%) | 1.10E-02 |
| Diabetes a | 345.0 (12.2%) | 234.0 (11.0%) | 111.0 (15.6%) | 2.73E-03 |
| Mortality a | 711 (25.1%) | 0 (0%) | 711 (100%) | – |
a Count (proportion) is reported for binary variables
Our infection cohort comprises 3,135 patients, 1,661 (53.0%) of whom tested positive for COVID-19. This cohort only includes patients from ASST Cremona, as negative tests results were not available from HM Hospitals. Table 4 summarizes the clinical characteristics of the cohort, both in aggregate and broken down by COVID-19 test result. Again, the reported features are those used in the final model, that is age, gender, 4 vitals values, and 14 lab values.
Table 4.
Characteristics of study population for infection test prediction model
| All (N = 3, 135) | No Infection (N = 1, 474) | Infection (N = 1, 661) | P-Value | |
|---|---|---|---|---|
| Age | 63.0 (49.0-78.0) | 58.0 (42.0-78.0) | 66.0 (55.0-78.5) | 9.00E-28 |
| Female a | 1444.0 (46.1%) | 777.0 (52.7%) | 667.0 (40.2%) | 1.70E-12 |
| Heart Rate | 88.5 (78.0-100.5) | 89.0 (78.0-101.2) | 88.0 (78.2-100.0) | 6.13E-01 |
| Oxygen Saturation | 95.4 (91.8-97.0) | 96.5 (94.8-97.5) | 94.2 (89.6-96.4) | 1.68E-31 |
| Respiratory Frequency | 18.0 (16.0-19.0) | 18.0 (16.0-18.0) | 18.0 (16.0-20.0) | 9.64E-21 |
| Temperature | 98.3 (97.5-99.5) | 97.7 (97.2-98.7) | 99.0 (97.9-100.0) | 4.51E-80 |
| Alanine Aminotransferase | 22.0 (15.0-37.0) | 19.0 (13.0-30.0) | 27.0 (18.0-43.0) | 6.59E-09 |
| Aspartate Aminotransferase | 29.0 (21.0-47.0) | 23.0 (19.0-31.0) | 37.0 (26.0-57.0) | 1.20E-20 |
| Blood Urea Nitrogen | 17.0 (13.0-25.0) | 16.0 (12.0-22.0) | 18.0 (13.0-27.0) | 3.78E-05 |
| Calcium | 9.3 (8.9-9.7) | 9.6 (9.2-9.9) | 9.0 (8.7-9.4) | 1.90E-96 |
| C-Reactive Protein | 31.0 (3.4-107.6) | 4.7 (1.1-35.4) | 69.8 (23.2-152.3) | 1.28E-83 |
| Creatinine | 0.9 (0.8-1.2) | 0.9 (0.7-1.1) | 1.0 (0.8-1.2) | 2.19E-05 |
| Hemoglobin | 13.5 (12.3-14.7) | 13.4 (12.1-14.6) | 13.6 (12.5-14.8) | 5.70E-05 |
| Mean Corpsular Volume | 87.2 (84.0-90.3) | 87.7 (84.2-90.7) | 86.8 (83.9-90.0) | 1.43E-01 |
| Platelets | 223.0 (174.0-285.0) | 241.0 (198.0-297.0) | 202.0 (156.0-266.0) | 1.41E-18 |
| Red Cell Distrbution Width | 13.2 (12.5-14.3) | 13.2 (12.5-14.5) | 13.1 (12.4-14.0) | 1.59E-06 |
| Sodium | 139.0 (137.0-141.0) | 140.0 (138.0-142.0) | 139.0 (136.0-141.0) | 1.12E-14 |
| Prothrombin Time (INR) | 1.0 (1.0-1.1) | 1.1 (1.0-1.1) | 1.0 (1.0-1.1) | 8.96E-01 |
| Total Bilirubin | 0.6 (0.5-0.8) | 0.6 (0.4-0.9) | 0.6 (0.5-0.8) | 8.83E-03 |
| White Blood Cell Count | 7.6 (5.8-10.1) | 8.7 (7.0-11.1) | 6.6 (5.1-8.7) | 7.59E-38 |
| COVID-19 Positive Test a | 1661 (53.0%) | 0 (0%) | 1661 (100%) | – |
Count (proportion) is reported for binary variables
Performance evaluation
All models achieve strong out-of-sample performance. Our mortality risk calculator has an AUC of 93.8% with lab values and 90.5% without lab values. Our infection risk calculator has an AUC of 91.8% with lab values and 83.1% without lab values. These values suggest a strong discriminative ability of the proposed models. We report average results across all random data partitions in the Appendix.
We also report threshold-based metrics in the Appendix, which evaluate the discriminative ability of the calculators at a fixed cutoff. With the threshold set to ensure a sensitivity of at least 90% (motivated by the high costs of false negatives), we obtain accuracies spanning 65%–80%.
The mortality model achieves better overall predictive performance than the infection model. As expected, both models have better predictive performance with lab values than without lab values. Yet, the models without lab values still achieve strong predictive performance.
Model interpretation
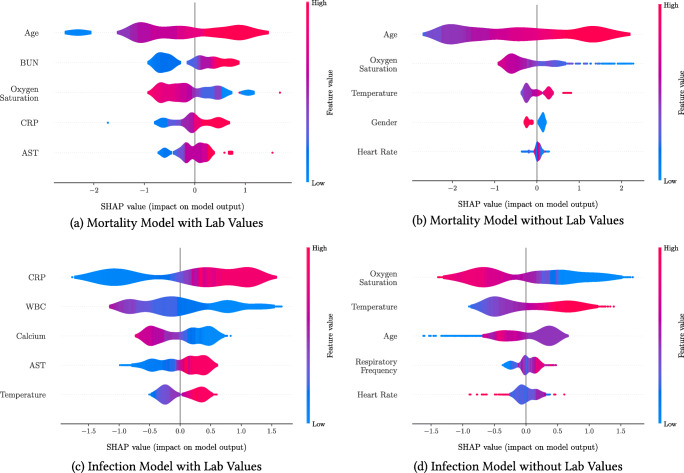
Figure 3 plots the SHAP importance plots for all models. The figures sort the features by decreasing significance. For each one, the row represents its impact on the SHAP value, as the feature ranges from low (blue) to high (red). Higher SHAP values correspond to increased likelihood of a positive outcome (i.e. mortality or infection). Features with the color scale oriented blue to red (resp. red to blue) from left to right have increasing (resp. decreasing) risk as the feature increases. For example, “Age” is the most important feature of the mortality score with lab values (Fig. 3a), and older patients have higher predicted mortality.
Fig. 3.
SHapley Additive exPlanations (SHAP) importance plots for the mortality and infection risk calculators. The five most important features are shown for each model. Gender is a binary feature (female is equal to 1, shown in red; male is equal to 0, shown in blue). Each row represents the impact of a feature on the outcome, with higher SHAP values indicating higher likelihood of a positive outcome
Discussion and impact
The proposed models provide algorithmic screening tools that deliver COVID-19 risk predictions using common clinical features. In a constrained healthcare system or in a clinic without access to advanced diagnostics, clinicians can use these models to rapidly identify high-risk patients to support triage and treatment decisions. The models without lab values offer an even simpler tool that could be used outside of a clinical setting. In strained healthcare systems, it can be difficult for patients to obtain direct advice from providers, and so this tool could serve as a pre-screening step. While the exclusion of lab values reduces the AUC (especially for infection), these calculators still perform strongly.
Our models provide insights into risk factors and biomarkers related to COVID-19 infection and mortality. Our results suggest that the main indicators of mortality risk are age, BUN, CRP, AST, and low oxygen saturation. These findings validate several population-level insights from Section 2 and are in agreement with clinical studies and public health guidance. Age is widely recognized as a primary risk factor [55]. Studies have found shortness of breath as a common symptom of severe cases [50, 53], and low oxygen saturation has been further identified as a risk factor even without respiratory symptoms [51]. BUN, CRP, and AST have been identified as key biomarkers in severe COVID-19 cases: elevated BUN as an indicator of kidney dysfunction [9], elevated levels of CRP as an inflammatory marker [7, 49, 54], and elevated AST levels related to liver dysfunction [16, 18].
Turning to infection risk, the main indicators are CRP, WBC, Calcium, AST, and temperature. These findings are also in agreement with clinical reports: an elevated CRP generally indicates an early sign of infection and implies lung lesions from COVID-19 [28], elevated levels of leukocytes suggest cytokine release syndrome caused by SARS-CoV-2 virus [45], and lowered levels of serum calcium signal higher rate of organ injury and septic shock [44]. The agreement between our findings and clinical observations offers credibility for the use of our calculators to support clinical decision-making—although they are not intended to substitute clinical diagnostic or medical expertise.
When lab values are not available, the widely accepted risk factors of age, oxygen saturation, temperature, and heart rate become the key indicators for both risk calculators. We observe that mortality risk is higher for male patients (blue in Fig. 3b) than for female patients (red), confirming clinical reports [21, 29]. An elevated respiratory frequency becomes an important predictor of infection, as reported in [55]. These findings suggest that demographics and vitals provide valuable information in the absence of lab values. However, when lab values are available, these other features become secondary.
The timing of the clinical data, obtained from March through May 2020, is relevant in considering the generalizability of the models. Various factors have affected COVID-19 severity as the pandemic has continued to progress. Treatment protocols, government policies, seasonal factors, and the evolution of the virus itself all potentially contribute to changes in risk determinants. In theory, the risk factors identified in this paper may not generalize to broader populations (e.g., non-European populations, populations in the Fall of 2020). However, our findings have been validated by more recent studies, such as the 4C Mortality Score, which also highlighted the importance of age, oxygen saturation, and CRP in the classification of severe COVID-19 infections [24]. As a result, our proposed infection and mortality models are relevant beyond the geographical limits of Italy and Spain and beyond the critical months of March and April 2020. More broadly, these recent results underscore the importance of prospective validation of risk calculators as we enter subsequent phases of the pandemic to assess their ongoing applicability.
A limitation of the current mortality model is that it does not take into account medication and treatments during hospitalization. This is mainly due to the fact that, during the first months of the COVID-19 pandemic, no systematic treatment protocol had yet been established, and it was therefore challenging to account for treatment effects in our model. Accordingly, our objective in this paper is to uncover the associations between patient characteristics and risk scores, without making claims related to causality. We have addressed this limitation in later research, by leveraging data on treatments to delineate the impact of medications at the patient level 〈REF〉.3
Overall, we have developed data-driven calculators that allow physicians and patients to assess mortality and infection risks in order to guide care management—especially with scarce healthcare resources. These tools were developed in the early stages of the pandemic, to support an initial pandemic response. As the pandemic has progressed, testing has become more widespread and affordable. Yet, reagent resources for Reverse Transcription Polymerase Chain Reaction (RT-PCR) testing remain relatively scarce, and the time frame to get RT-PCR results ranges between 24 to 48 hours. Therefore, molecular tests cannot be implemented at large scale in resource constrained systems, let alone applied on a daily basis. In such instances, our infection score can aid fast and effective decision-making in clinical settings. In addition, testing capacities remain even more limited in non-clinical settings. Notably, our infection calculator has been employed by the Banco de Credito del Peru, the largest bank in Peru, to guide safety protocols for employees and work from home policies. Finally, our mortality calculator remains useful as one of the first models to synthesize clinical features into a single risk score upon hospital admission. This calculator is being used by several hospitals within the ASST Cremona system to support triage and treatment decisions, ultimately alleviating the toll of the pandemic.
Predictive and prescriptive analytics: disease projections and government response
We develop a new epidemiological model, called DELPHI (Differential Equations Leads to Predictions of Hospitalizations and Infections). The model first provides a predictive tool to forecast the number of detected cases, hospitalizations and deaths—we refer to this model as “DELPHI-pred”. It then provides a prescriptive tool to simulate the effect of policy interventions and guide government response to the COVID-19 pandemic—we refer to this model as “DELPHI-presc”. All models are fit in each US state (plus the District of Columbia).
DELPHI-pred: projecting early spread of COVID-19
Model development
DELPHI is a compartmental model, with dynamics governed by ordinary differential equations. It extends the standard SEIR model by defining 11 core states (Fig. 4): susceptible (S), exposed (E), infectious (I), undetected people who will recover (UR) or decease (UD), detected hospitalized people who will recover (DHR) or decease (DHD), quarantined people who will recover (DQR) or decease (DQD), recovered (R) and deceased (D). The separation of the UR/UD, DQR/DQD and DHR/DHD states enables separate independence in fitting recoveries and deaths from the data. Further auxiliary states, including Total Detected Cases (DT) and Total Detected Deaths (DD), are defined relative to the core states in order to correspond to actual data.
Fig. 4.
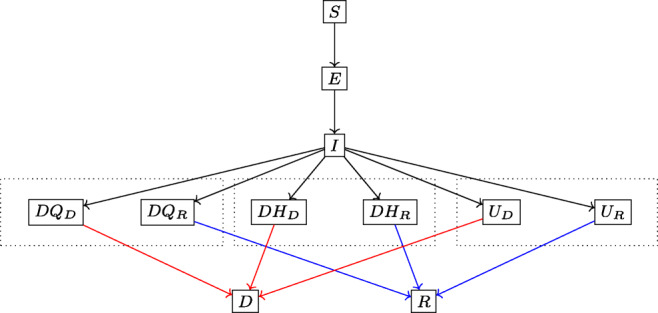
Simplified flow diagram of DELPHI
As opposed to many other COVID-19 models [?[, see, e.g.,]]kissler2020projecting, DELPHI captured two key elements of the pandemic explicitly since its inception in late March:
Underdetection: Many cases remain undetected due to limited testing, record failures, and detection errors. Ignoring them would underestimate the scale of the pandemic. We capture them through the UR and UD states.
-
Government Response: “Social distancing” policies limit the spread of the virus. Ignoring them would overestimate the spread of the pandemic. We model them through a decline in the infection rate over time. Specifically, we write: where α is a constant baseline rate and γ(t) is a time-dependent function characterizing each state’s policies, modeled as follows:
The inverse tangent function provides a concave-convex relationship, capturing three phases of government response. In Phase I, most activities continue normally as people adjust their behavior. In Phase II, the infection rate declines sharply as policies are implemented. In Phase III, the decline in the infection rate reaches saturation. The parameters t0 and k can be respectively thought of as the median date and the strength of the response.
Ultimately, DELPHI involves 13 parameters that define the transition rates between the 11 states (details in the Supplementary Materials). We calibrate six of the biological parameters from our clinical outcomes database (Section 2). The remaining 7 parameters are optimized using constrained Nelder-Mead optimization [25] and trust-region methods [6] by minimizing a weighted mean squared error on detected cases and deaths:
Where DT(t) is the historical detected cases for a region on day t, the predicted detected cases from DELPHI, and similarly for DD(t) and corresponding to deaths. The historical data for the number of cases and deaths per US county is taken from the New York Times database [36]. The lambda factor balances the fitting between detected cases and deaths; this re-scaling coefficient was obtained experimentally. Each state is included as soon as it records more than 100 cases so that isolated outbreaks are ignored. Every day as new data becomes available, we reoptimize the parameters while restricting the optimization range to be within 10% of the current fitted parameters to ensure a smooth drift. Further details on the fitting procedure are contained in the Appendix.
Validation
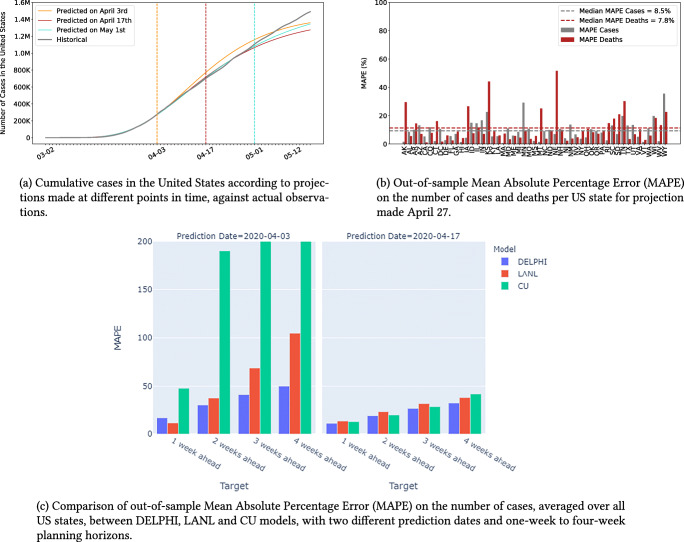
DELPHI was created in late March and has been continuously updated to reflect new observed data. Figure 5a shows our projections made on three different dates, and compares them against historical observations. This plot focuses on the number of cases, but a similar plot for the number of deaths is reported in the Appendix.
Fig. 5.
Projection accuracy for the United States
In addition to providing aggregate validation figures, we also evaluate the model’s out-of-sample performance quantitatively, using a backtesting procedure. To our knowledge, this represents the first attempt to assess the predictive performance of COVID-19 projections. Specifically, we fit the model’s parameters using data up to April 27, build projections from April 28 to May 12, and evaluate the resulting Mean Absolute Percentage Error (MAPE). Figure 5b reports the results in each US state.
Finally, we compare the predictions from the DELPHI model to the one reported by benchmark models. For obvious reasons, we restrict our attention to models that were available early on in the pandemic, and for which historical predictions are still publicly available. Specifically, we consider the models from the Los Alamos National Laboratory (LANL) [27] and Columbia University (CU) [39]. Figure 5c reports the Mean Absolute Percentage Error (MAPE) in the number of cases (averaged over all US states) from the projections made at two different points in time (April 03 and April 17, 2020) for different planning horizons (1, 2, 3, 4 weeks).
Discussion and impact
Results suggest that DELPHI-pred achieves strong predictive performance. The model has been consistently predicting, with high accuracy the overall spread of the disease for several weeks. Notably, DELPHI-pred was able to anticipate, as early as April 3rd, the dynamics of the pandemic in the United States up to mid-May. At a time where 200,000–300,000 cases were reported, the model was predicting 1.2M–1.4M cases by mid-May—a prediction that proved accurate 40 days later. Note that, at the time, the DELPHI model underpredicted the spread of the pandemic in May, as compared to ex-post data. This is due to the fact that early predictions were based on limited data and limited visibility into subsequent governmental policies. As the pandemic progressed, the DELPHI model has been able to address these limitations.
Our quantitative results confirm the visual evidence. The MAPE is small across US states. The median MAPE is 8.5% for the number of cases—the 10% and 90% percentiles are equal to 1.9% and 16.7%. The median MAPE is 7.8% for the number of deaths—the 10% and 90% percentiles are equal to 3.3% and 25.1%. Given the high level of uncertainty and variability in the disease’s spread, this level of accuracy is suggestive of excellent out-of-sample performance.
This behavior is further confirmed when we compare our MAPE to the two benchmark models. For almost all targets, DELPHI outperforms both models, resulting in lower MAPE—especially in the earliest phases of the pandemic when data remained scarcely available.
As Fig. 5b shows, a limitation of our model is that the relative error remains large for a small minority of US states. These discrepancies stem from two main reasons. First, errors are typically larger for states that have recorded few cases (WY) or few deaths (AK, KS, NE). Like all SEIR-derived models, DELPHI performs better on large populations. Moreover, the MAPE metric emphasizes errors on smaller population counts. Second, our model is fitted at the state level, implicitly assuming that the spread of the pandemic is independent from one state to another—thus ignoring inter-state travel. This limitation helps explain the above-median error in a few heartland states which were confronted with the pandemic in later stages (MN, TN, IA).
In summary, DELPHI-pred is a novel epidemiological model of the pandemic that has provided high-quality estimates of the daily number of cases and deaths per US state since the early pandemic period. This model is one of the top 5 consistent contributors to the forecasts used by the US Center for Disease Control to chart and anticipate the spread of the pandemic [42]. It has also been used by the Hartford Healthcare system—the major hospital system in Connecticut, US—to plan its ICU capacity, and by Janssen Pharmaceuticals to design a vaccine distribution strategy for their leading candidate Ad26.Cov2.S.
As the pandemic continued to spread throughout the world, governments have enacted further measures, including massive testing and contact-tracing efforts, while clinical treatments have substantially improved. Therefore, some of the assumptions included in the original DELPHI model (such as a time-invariant detection and mortality rates) have become increasingly untenable. Moreover, some of the quantities estimated through the clinical databases have become obsolete as the standard of care has improved. Finally, the behavior of the population in response to governments’ policies has also varied over time. In response, we have since significantly updated the structure of the DELPHI model and its empirical calibration, in order to incorporate these dynamics 〈REF〉.4
DELPHI-presc: toward re-opening society
To inform the relaxation of social distancing policies, we link policies to the infection rate using machine learning. Specifically, we predict the values of γ(t), obtained from the fitting procedure of DELPHI-pred. For simplicity and interpretability, we consider a simple model based on regression trees [5] and restrict the independent variables to the policies in place. We classify policies based on whether they restrict mass gatherings, school , travel and work activities. We group travel and work restrictions together as ”other” restrictions as they are most often implemented simultaneously. Then, using such grouping, we define a set of seven mutually exclusive and collectively exhaustive policies observed in the US data: (i) No measure; (ii) Restrict mass gatherings; (iii) Restrict others; (iv) Authorize schools, restrict mass gatherings and others; (v) Restrict mass gatherings and schools; (vi) Restrict mass gatherings, schools and others; and (vii) Stay-at-home. The remaining possible policies were not implemented. The classifier was trained with policy data up till end of April so that the assumption of increasing governmental intervention is still valid. The regression tree is in the Supplementary materials, obtained from state-level data in the United States (here states include District of Columbia). This model achieves an out-of-sample R2 of 0.8, suggesting a good fit to the data.
The key results for various policies are summarized in Table 5. Policy (ii) and Policy (iv) are not shown in the table as these policies were not collectively implemented sufficiently to derive meaningful statistical inference. “States” record the number of states that had implemented the policy and “State-Days” record the total implemented days across states. The residual infection rate is calculated by normalizing the γ(t) under no policy to be 100%, while the standard errors are given assuming each state as an independent sample. We observe that tighter restrictions do indeed generate larger reductions in the residual infection rates. Furthermore, the results also provide comparisons between various policies—for instance, although Other (travel and work) restrictions seems to be able to effectively reduce the infection rate by 33.2 ± 5.9% when applied alone, its incremental effect after mass gathering and school restrictions have been implemented is insignificant (5.7 ± 10.2%). This seems to imply a large overlap between the behavior change caused by mass gathering and school restrictions with Other restrictions, so that once mass gathering and school restrictions are implemented, the effect of further limiting travel and work is severely limited. These quantitative results allow us to predict the value of γ(t) as a function of the policies (see Appendix for details), and simulate the spread of the disease as states progressively loosen social distancing policies.
Table 5.
Implementation length and effect of each policy category as implemented in the US
| Restrictions | States | State-Days | Residual Infection Rate |
|---|---|---|---|
| None | 51 | 749 | 100% |
| Others | 25 | 218 | 66.8 ± 5.9% |
| Mass Gathering and Schools | 9 | 72 | 47.9 ± 9.0% |
| Mass Gathering, School, and Others | 37 | 805 | 42.2 ± 4.8% |
| Stay-at-Home Order | 39 | 1876 | 23.9 ± 4.0% |
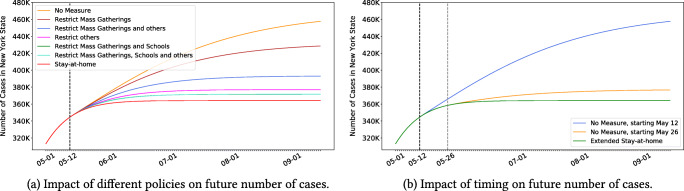
Figure 6a plots the projected case count in the state of New York (NY), for different policies (we report a similar plot for the death count in the Appendix). Note that the stringency of the policies has a significant impact on the pandemic’s spread and ultimate toll. For instance, relaxing all social distancing policies on May 12 can increase the cumulative number of cases in NY by up to 25% by September.
Fig. 6.
Reopening scenarios for New York
Using a similar nomenclature, Fig. 6b shows the case count if all social distancing policies are relaxed on May 12 vs. May 26. Note that the timing of the policies also has a strong impact: a two-week delay in re-opening society can greatly reduce a resurgence in NY.
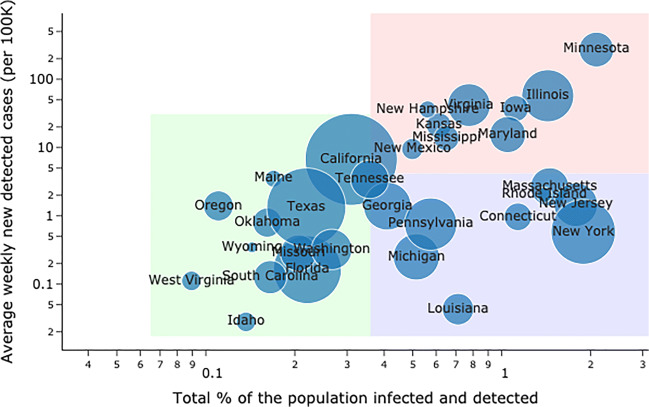
The road back to a new normal is not straightforward: results suggest that the disease’s spread is highly sensitive to both the intensity and the timing of social distancing policies. As governments grapple with an evolving pandemic, DELPHI-presc can be a useful tool to explore alternative scenarios and ensure that critical decisions are supported with data. To further understand the disparate impact of the policies across states, we made predictions for the situation across the US assuming a policy that involves mass gathering, travel, and work restrictions was implemented in all states on June 16th. Figure 7 represents, for many states in the US, the average weekly prevalence per 100K people in the first two weeks of July against the total percentage of detected cases among the population. One can observe three distinct clusters of states:
States with a small number of cumulative cases (relative to the population), and that are in a late stage of the pandemic with relatively few new cases, such as California, Florida, Texas or West Virginia.
States with a large number of cumulative cases, but that are in a late stage of the pandemic, with relatively few new cases, such as Connecticut, Louisiana, Massachusetts or New York.
States where the pandemic has had a large impact with a large number of cumulative cases, and where the situation will still be worsening at an alarming rate. These include states like Illinois, Minnesota, Iowa and Virginia. While the worst-case scenario shows a maximum of 2-3% of the population being infected, this suggests that for these particular states, such hypothetical policy could be inadequate for controlling the epidemic, and a stronger policy (such as Stay-at-Home orders) is needed for a little more time.
Fig. 7.
United States predictions for mid-July under mass gathering, travel and work restrictions
Prescriptive analytics: ventilator allocation
COVID-19 is primarily an acute respiratory disease. The World Health Organization recommends that patients with oxygen saturation levels below 93% receive respiratory support [52]. Following the standard Acute Respiratory Distress Syndrome protocol, COVID-19 patients are initially put in the prone position and then put in a drug-induced paralysis via a neuromuscular blockade to prevent lung injury [10]. Patients are then put on a ventilator, which delivers high concentrations of oxygen while removing carbon dioxide [3]. Early evidence suggests that ventilator intubation reduces the risk of hypoxia for COVID-19 patients [34].
As a result, hospitals have been facing ventilator shortages worldwide [41]. Still, local shortages do not necessarily imply global shortages. For instance, in April 2020, the total supply of ventilators in the United States exceeded the projected demand from COVID-19 patients. Ventilator shortages could thus be alleviated by pooling the supply, i.e., by strategically allocating the surge supply of ventilators from the federal government and facilitating inter-state transfers of ventilators.
We propose an optimization model to support the allocation of ventilators in a semi-collaborative setting where resources can be shared both between healthcare facilities or through a central authority. Based on its primary motivation, we formulate the model to support the management of the federal supply of ventilators and inter-state ventilator transfers in the United States. A similar model has also been used to support inter-hospital transfers of ventilators. This model leverages the demand projections from DELPHI-pred (Section 4) to prescribe resource allocation recommendations—with the ultimate goal of alleviating the health impact of the pandemic.
Model
Resource allocation is critical when clinical care depends on scarce equipment, and the COVID-19 has therefore sparked renewed interest in resource allocation problems [37]. In particular, several studies have used optimization to support ventilator pooling. In the context of influenza planning, [19] proposed a time-independent model for stockpiling ventilators. For COVID-19, [33] developed a time-dependent stochastic optimization model to support transfers to and from the federal government, given scenarios regarding the pandemic’s spread. Additionally, [1] and [4] proposed simple network optimization models to evaluate policy scenarios in ventilator sharing. Optimization can also be used to improve patient-level ventilator allocation decisions: for instance, [14] proposed a simulation model to compare the efficacy of respiratory medical interventions with different invasiveness levels. In this section, we propose a deterministic time-dependent ventilator sharing model. As compared to the earlier literature, our model optimizes both the management of the federal stockpile as well as inter-state transfers; allows each state’s fraction of pooled ventilators to vary continuously over time as a function of the underlying dynamics of the pandemic; allows ventilators to be shared proactively ahead of a state’s peak; and, most importantly, leverages the projections from DELPHI-pred as inputs, thus bridging the gap from predictive to prescriptive analytics.
We model ventilator pooling as a multi-period resource allocation over S states and D days. The model takes as input ventilator demand in state s and day d, denoted as vs,d, as well as parameters capturing the surge supply from the federal government and the extent of inter-state collaboration. We formulate an optimization problem where the key decisions are the number of ventilators transferred from state s to state on day d, and the number of ventilators allocated from the federal government to state s on day d. The problem has a multi-period network flow structure with lead times, with additional problem-specific constraints: for instance, we do not allow states currently facing shortages to ship out ventilators.
We propose a bi-objective formulation. The first objective is to minimize ventilator-day shortages; for robustness, we consider both projected shortages (based on demand forecasts) and worst-case shortages (including a buffer in the demand estimates). The second objective is to minimize inter-state transfers, to limit the operational and political costs of inter-state coordination. Mixed-integer optimization provides modeling flexibility to capture spatial-temporal dynamics and the trade-offs between these various objectives. We report the full mathematical formulation of the model, along with the key assumptions, in the Appendix.
Results
We implemented the model on April 15, a time of pressing ventilator need in the United States. We estimate the number of hospitalizations from DELPHI-pred as the sum of DHR and DHD. From our clinical outcomes database in Section 2, we estimate that 25% of hospitalized patients are put on a ventilator, which we use to estimate the demand for ventilators. We also obtain the average length of stay from our clinical outcomes database (Table 2).
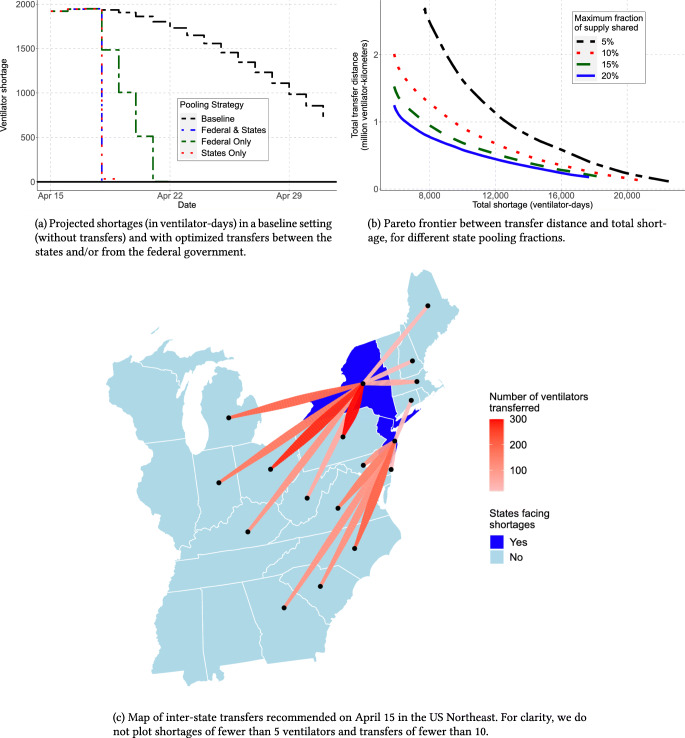
Figure 8a shows the evolution of ventilator shortages with and without ventilator transfers from the federal government and inter-state transfers. These results indicate that ventilator pooling can rapidly eliminate all ventilator shortages. Figure 8c shows ventilator transfers recommended in the US Northeast on April 15 (with inter-state transfers only), overlaid on a map displaying the predicted shortage without transfers.
Fig. 8.
The edge of optimization to eliminate ventilator shortages
There are different pathways toward eliminating ventilator shortages. Figure 8b shows the trade-off between shortages and transfer distance—each line corresponds to the maximal fraction of its own ventilators that each state can pool. Overall, states do not have to share more than 10% of their supply at any time to efficiently eliminate shortages. States can largely meet their needs with help from neighboring states, with cross-country transfers only used as a last resort. Broadly, results underscore trade-offs between ventilator shortages, the extent of inter-state transfers, the number of ventilators allocated from the federal government, and the robustness of the solution.
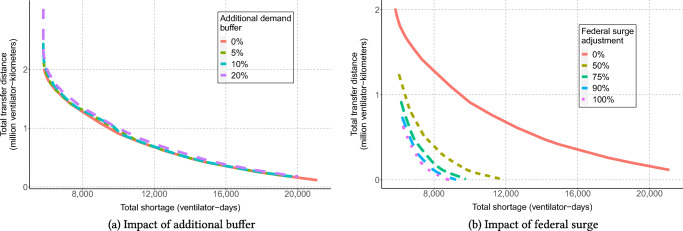
For instance, Fig. 9 shows the Pareto frontier between the model’s two objectives, inter-state transfer distance and ventilator shortage, as a function of two model parameters: α (capturing the demand buffer that states would like to plan for) and a surge supply multiplier (capturing by how much the federal government’s ventilator supply varies from our estimates). Note that the buffer α does not impact the number of inter-state transfers and the amount of ventilator shortages too significantly, suggesting that the solution can be made robust at a limited overall cost. In contrast, as the surge supply is decreased, the number of required ventilator transfers and the amount of ventilator shortages increase—highlighting the need for stronger cooperation between states as federal supply drops.
Fig. 9.
Influence of additional buffer and federal surge availability on ventilator shortages and transfers
Discussion and impact
Our main insight is that ventilator shortages could be eliminated altogether through inter-state transfers and strategic management of the federal supply. Results also underscore (i) the benefits of inter-state coordination and (ii) the benefits of early coordination. First, ventilator shortages can be eliminated through inter-state transfers alone: leveraging a surge supply from the federal government is not required, though it may reduce inter-state transfers. Under our recommendation, the most pronounced transfers occur from states facing no shortages (Ohio, Pennsylvania, and North Carolina) to states facing strong shortages (New York, New Jersey). Second, most transfers occur in the early stages of the pandemic. This underscores the benefits of leveraging a predictive model like DELPHI-pred to align the ventilator supply with demand projections as early as possible.
We have developed a similar model to support the re-distribution of ventilators across hospitals within the Hartford HealthCare system in Connecticut—using county-level forecasts of ventilator demand obtained from DELPHI-pred. This model was used by Hartford HealthCare to align ventilator supply with projected demand at a time where the pandemic was on the rise.
Looking ahead, the model provides a methodology to recommend allocations of critical resources in the next phases of the pandemic, from ventilators to drugs to personal protective equipment. Since epidemics do not peak in each state at the same time, states whose infection peak has already passed or lies weeks ahead can help other states facing immediate shortages at little costs to their constituents. Inter-state transfers of ventilators occurred in isolated fashion through April 2020; our model proposes an automated decision-making tool to support these decisions systematically. As our results show, proactive coordination and resource pooling can significantly reduce shortages—thus increasing the number of patients that can be treated without resorting to extreme clinical recourse with side effects (such as splitting ventilators). Other ventilator sharing studies, including [1], [4], and [33], come to similar conclusions despite differences in modeling assumptions. Though the exact nature of the best ventilator sharing policy can be debated, our results confirm that even simple collaboration policies can alleviate, or even eliminate, ventilator shortages.
Conclusion
This paper proposes a comprehensive data-driven approach to address several core challenges faced by healthcare providers and policy makers in the midst of the COVID-19 pandemic. We have gathered and aggregated data from hundreds of clinical studies, electronic health records, and census reports. We have developed descriptive, predictive and prescriptive models, combining methods from machine learning, epidemiology, and mixed-integer optimization. Results provide insights on the clinical aspects of the disease, on patients’ infection and mortality risks, on the dynamics of the pandemic, and on the levers that policy makers and healthcare providers can use to alleviate its toll. The models developed in this paper also yield decision support tools that have been deployed on our dedicated website and that are actively being used by several hospitals, companies and policy makers.
Supplementary Information
(PDF 896 KB)
Funding
HW and IP supported by the National Science Foundation Graduate Research Fellowship under Grant No. 174530. Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors(s) and do not necessarily reflect the views of the National Science Foundation. This work was partially supported by grants from c3.ai and IBM for COVID-19 related research; neither organization played any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. No further funding was provided for the study.
Data Availability
We have already released our aggregated clinical database on our website. Another of our data sources is the daily census report from the New York Times, which is publicly available. A third data source stems from patients’ electronic medical records, obtained from collaborations with several hospitals. Due to its highly sensitive nature, this data cannot be made available to the public directly.
Compliance with Ethical Standards
Ethics Approval
All independent organizationsapproved this 1006 protocol as minimal-risk research using data collected for standard 1007 clinical practice and waived the requirement for informed consent.
Conflict of interests
The authors have no competing interests to declare that may be relevant to the submitted work.
Footnotes
HM Hospitals patients were not included since no negative case data was available.
Reference redacted for double blind review.
Reference redacted for double blind review.
Code Availability
The code for this project is available at https://github.com/COVIDAnalytics/. The website is available at www.covidanalytics.io.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Adelman D (2020) Thousands of lives could be saved in the US during the COVID-19 pandemic if states exchanged ventilators: study examines how lives could be saved by allowing US states to exchange ventilators during the COVID-19 pandemic. Health Affairs, pp 10–1377 [DOI] [PubMed]
- 2.Anderson RM, Heesterbeek H, Klinkenberg D, Hollingsworth TD. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395(10228):931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bein T, Grasso S, Moerer O, Quintel M, Guerin C, Deja M, Brondani A, Mehta S. The standard of care of patients with ARDS: ventilatory settings and rescue therapies for refractory hypoxemia. Intensive Care Med. 2016;42(5):699–711. doi: 10.1007/s00134-016-4325-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billingham S, Widrick R, Edwards NJ, Klaus S (2020) COVID-19 (SARS-CoV-2) ventilator resource management using a network optimization model and predictive system demand. medRxiv
- 5.Breiman L, Friedman J, Stone CJ, Olshen RA . Classification and regression trees. Boca Raton: CRC press; 1984. [Google Scholar]
- 6.Byrd RH, Gilbert JC, Nocedal J. A trust region method based on interior point techniques for nonlinear programming. Math Program. 2000;89(1):149–185. doi: 10.1007/PL00011391. [DOI] [Google Scholar]
- 7.Caruso D, Zerunian M, Polici M, Pucciarelli F, Polidori T, Rucci C, Guido G, Bracci B, de Dominicis C, Laghi A (2020) Chest CT features of COVID-19 in Rome, Italy. Radiology 201237 [DOI] [PMC free article] [PubMed]
- 8.Chen T, Guestrin C (2016) Xgboost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, pp 785–794
- 9.Cheng Y, Luo R, Wang K, Zhang M, Wang Z, Dong L, Li J, Yao Y, Ge S, Xu G (2020) Kidney disease is associated with in-hospital death of patients with covid-19. Kidney Int [DOI] [PMC free article] [PubMed]
- 10.Cornejo RA, Díaz JC, Tobar EA, Bruhn AR, Ramos CA, González RA, Repetto CA, Romero CM, Gálvez LR, Llanos O, et al. Effects of prone positioning on lung protection in patients with acute respiratory distress syndrome. Am J Respir Crit Care Med. 2013;188(4):440–448. doi: 10.1164/rccm.201207-1279OC. [DOI] [PubMed] [Google Scholar]
- 11.Fernandes N (2020) Economic effects of coronavirus outbreak (COVID-19) on the world economy. Available at SSRN 3557504
- 12.Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci. 2020;117(17):9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garg S (2020) Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019—COVID-NET, 14 states, March 1–30, 2020. MMWR Morbidity and Mortality Weekly Report 69 [DOI] [PMC free article] [PubMed]
- 14.Gershengorn HB, Hu Y, Chen JT, Hsieh SJ, Dong J, Gong MN, Chan CW (2020) The impact of high-flow nasal cannula use on patient mortality and the availability of mechanical ventilators in covid-19. Ann Am Thorac Soc (ja) [DOI] [PMC free article] [PubMed]
- 15.Goyal P, Choi JJ, Pinheiro LC, Schenck EJ, Chen R, Jabri A, Satlin MJ, Campion TR Jr, Nahid M, Ringel JB et al (2020) Clinical characteristics of Covid-19 in New York City. N Engl J Med [DOI] [PMC free article] [PubMed]
- 16.Guan Wj, Zy Ni, Hu Y, Liang Wh, Ou Cq, He Jx, Liu L, Shan H, Cl Lei, Hui DS, et al. Clinical characteristics of coronavirus disease 2019 in china. New Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland LA, Kaelin EA, Maqsood R, Estifanos B, Wu LI, Varsani A, Halden RU, Hogue BG, Scotch M, Lim ES (2020) An 81 nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (Jan-Mar 2020). J Virol [DOI] [PMC free article] [PubMed]
- 18.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang HC, Araz OM, Morton DP, Johnson GP, Damien P, Clements B, Meyers LA. Stockpiling ventilators for influenza pandemics. Emerg Infect Dis. 2017;23(6):914. doi: 10.3201/eid2306.161417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johns Hopkins University (2020) Center for systems science and engineering. https://systems.jhu.edu/research/public-health/ncov/
- 21.Jordan RE, Adab P, Cheng K (2020) COVID-19: risk factors for severe disease and death [DOI] [PubMed]
- 22.Kampf G, Todt D, Pfaender S, Steinmann E (2020) Persistence of coronaviruses on inanimate surfaces and its inactivation with biocidal agents. J Hosp Infect [DOI] [PMC free article] [PubMed]
- 23.Kissler SM, Tedijanto C, Goldstein E, Grad YH, Lipsitch M (2020) Projecting the transmission dynamics of sars-cov-2 through the postpandemic period. Science [DOI] [PMC free article] [PubMed]
- 24.Knight SR, Ho A, Pius R, Buchan I, Carson G, Drake TM, Dunning J, Fairfield CJ, Gamble C, Green CA et al (2020) Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. bmj 370 [DOI] [PMC free article] [PubMed]
- 25.Lagarias JC, Reeds JA, Wright MH, Wright PE. Convergence properties of the Nelder–Mead simplex method in low dimensions. SIAM J Optim. 1998;9(1):112–147. doi: 10.1137/S1052623496303470. [DOI] [Google Scholar]
- 26.Lan L, Xu D, Ye G, Xia C, Wang S, Li Y, Xu H. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323(15):1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LANL (2020) LANL COVID-19 cases and deaths forecasts
- 28.Ling W (2020) C-reactive protein levels in the early stage of COVID-19. Medecine et Maladies Infectieuses [DOI] [PMC free article] [PubMed]
- 29.Liu Y, Du X, Chen J, Jin Y, Peng L, Wang HH, Luo M, Chen L, Zhao Y (2020) Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J Infect [DOI] [PMC free article] [PubMed]
- 30.Lundberg SM, Lee SI (2017) A unified approach to interpreting model predictions. In: Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, Garnett R (eds) Advances in neural information processing systems, vol 30. Curran Associates Inc., pp 4765–4774
- 31.Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, Nair B, Katz R, Himmelfarb J, Bansal N, Lee SI. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2(1):2522–5839. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKibbin WJ, Fernando R (2020) The global macroeconomic impacts of COVID-19: Seven scenarios. CAMA Working Paper
- 33.Mehrotra S, Rahimian H, Barah M, Luo F, Schantz K (2020) A model of supply-chain decisions for resource sharing with an application to ventilator allocation to combat COVID-19. Naval Research Logistics (NRL) [DOI] [PMC free article] [PubMed]
- 34.Meng L, Qiu H, Wan L, Ai Y, Xue Z, Guo Q, Deshpande R, Zhang L, Meng J, Tong C et al (2020) Intubation and ventilation amid the COVID-19 Outbreak Wuhan’s experience. Anesthesiology [DOI] [PMC free article] [PubMed]
- 35.(2020) Check if you have Coronavirus symptoms. https://www.nhs.uk/conditions/coronavirus-covid-19/check-if-you-have-coronavirus-symptomshttps://www.nhs.uk/conditions/coronavirus-covid-19/check-if-you-have-coronavirus-symptomshttps://www.nhs.uk/conditions/coronavirus-covid-19/check-if-you-have-coronavirus-symptoms, (Accessed 11 May 2020)
- 36.New York Times (2020) New York Times, Coronavirus in the U.S., Latest Map and Case Count. https://www.nytimes.com/interactive/2020/us/coronavirus-us-cases.html
- 37.Pathak PA, Sönmez T, Unver MU, Yenmez MB (2020) Leaving no ethical value behind: Triage protocol design for pandemic rationing. Working Paper 26951, National Bureau of Economic Research. 10.3386/w26951. http://www.nber.org/papers/w26951
- 38.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 39.Pei S, Shaman J (2020) Initial simulation of SARS-CoV2 spread and intervention effects in the continental US. medRxiv
- 40.Petrilli CM, Jones SA, Yang J, Rajagopalan H, O’Donnell LF, Chernyak Y, Tobin K, Cerfolio RJ, Francois F, Horwitz LI (2020) Factors associated with hospitalization and critical illness among 4,103 patients with COVID-19 disease in New York City. medRxiv [DOI] [PMC free article] [PubMed]
- 41.Ranney ML, Griffeth V, Jha AK (2020) Critical supply shortages—the need for ventilators and personal protective equipment during the COVID-19 pandemic. New Engl J Med [DOI] [PubMed]
- 42.Ray EL, Wattanachit N, Niemi J, Kanji AH, House K, Cramer EY, Bracher J, Zheng A, Yamana TK, Xiong X et al (2020) Ensemble forecasts of coronavirus disease 2019 (covid-19) in the us. MedRXiv
- 43.Sanchel S, Ting Lin Y, Xu C, Romero-Severson E, Hengartner N, Ke R (2020) High contagiousness and rapid spread of severe acute respiratory syndrome Coronavirus 2. Emerg Infect Dis 27(7) [DOI] [PMC free article] [PubMed]
- 44.Shi Q, Sun J, Zhang W, Zou L, Liu Y, Li J, Kan X, Dai L, Yuan S, Yu W et al (2020a) Serum calcium as a biomarker of clinical severity and prognosis in patients with coronavirus disease 2019: a retrospective cross-sectional study. Crit Care Emerg Med [DOI] [PMC free article] [PubMed]
- 45.Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, Bucci E, Piacentini M, Ippolito G, Melino G (2020b) COVID-19 infection: the perspectives on immune responses [DOI] [PMC free article] [PubMed]
- 46.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17(6):520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 47.US Center for Disease Control (2020) https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html, (Accessed 11 May 2020)
- 48.Vahidy FS, Nicolas JC, Meeks JR, Khan O, Jones SL, Masud F, Sostman HD, Phillips RA, Andrieni JD, Kash BA et al (2020) Racial and ethnic disparities in SARS-CoV-2 pandemic: Analysis of a COVID-19 observational registry for a diverse US metropolitan population. medRxiv [DOI] [PMC free article] [PubMed]
- 49.Velavan TP, Meyer CG. The COVID-19 epidemic. Trop Med Int Health. 2020;25(3):278. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. 2020;92(6):568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkerson RG, Adler JD, Shah NG, Brown R (2020) Silent hypoxia: A harbinger of clinical deterioration in patients with covid-19. Am J Emerg Med [DOI] [PMC free article] [PubMed]
- 52.World Health Organization (2020) Coronavirus. https://www.who.int/health-topics/coronavirus, (Accessed 11 May 2020)
- 53.Xie J, Covassin N, Fan Z, Singh P, Gao W, Li G, Kara T, Somers VK (2020) Association between hypoxemia and mortality in patients with COVID-19. Mayo Clin Proc. 10.1016/j.mayocp.2020.04.006. https://linkinghub.elsevier.com/retrieve/pii/S0025619620303670 [DOI] [PMC free article] [PubMed]
- 54.Yan L, Zhang HT, Goncalves J, Xiao Y, Wang M, Guo Y, Sun C, Tang X, Jing L, Zhang M et al (2020) An interpretable mortality prediction model for covid-19 patients. Nat Mach Intell, 1–6
- 55.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 896 KB)
Data Availability Statement
We have already released our aggregated clinical database on our website. Another of our data sources is the daily census report from the New York Times, which is publicly available. A third data source stems from patients’ electronic medical records, obtained from collaborations with several hospitals. Due to its highly sensitive nature, this data cannot be made available to the public directly.