Abstract
High-sensitivity detection of minute quantities or concentration variations of analytes of clinical importance is critical for biosensing to ensure accurate disease diagnostics and reliable health monitoring. A variety of sensitivity-improving concepts have been proposed from chemical, physical, and biological perspectives. In this review, we classify and discuss elements that are responsible for sensitivity enhancement in accordance with their operating steps in a typical biosensing workflow that runs through sampling, analyte recognition, and signal transduction. With a focus on optical biosensing, we introduce exemplary sensitivity-improving strategies, which can be developed into “plug-and-play” modules for many of current and future sensors, and discuss their mechanisms to enhance biosensing performance. We cover three major strategies: (i) amplification of signal transduction by polymerization and nanocatalysts, (ii) diffusion-limit-breaking systems for enhancing sensor–analyte contact and subsequent analyte recognition by fluid-mixing and analyte-concentrating, and (iii) combined approaches that utilize renal concentration at the sampling and recognition steps and chemical signal amplification at the signal transduction step.
Keywords: optical biosensing, sensitivity, signal amplification, analyte–sensor contact
Graphical Abstract
A variety of sensitivity-improving concepts has been proposed from chemical, physical, and biological perspectives. In this review, sensitivity-enhancing elements in accordance with their operating steps in a typical biosensing workflow that runs through sampling, analyte recognition, and signal transduction are identified. Exemplary sensitivity-improving strategies, which can be developed into “plug-and-play” modules for many of current and future sensors, are discussed.
1. Introduction
Biosensing, a technique capable of producing signals that are correlated to specific biomolecules, has long been one of the hot research topics in medical diagnostics,[1-8] biology,[9-12] environmental monitoring,[13-16] and food quality control.[17-19] Optical biosensing employs an optical signal as an indication and quantification of target analytes in samples. Along with developments of photonic materials, optical modalities such as colorimetry,[20-22] fluorescence,[23,24] chemiluminescence[25-29] and Raman scattering[30-32] have been successfully integrated into biosensing for their high sensitivity that enables concentration-dependent readouts. For example, colorimetric immunoassays are a standard of disease-associated antigen and immunogenic antibody testing, while a fluorescence modality is widely used in nucleic acid testing with specific molecular designs (e.g., cleavable fluorophore-quencher probes in real-time polymerase chain reaction) to analyze genetic information and to diagnose infectious diseases.[33-39] Considering that one of the major goals of biosensing is to identify the presence or concentration variations of relevant analytes in samples, sensitivity at a given time has been regarded as one of the key performance parameters. In the case of the recent COVID-19 pandemic, viral loads in clinical samples vary significantly among patients and the stage of infection.[40-43] In order to draw accurate diagnostic outcomes regardless of the viral load, high-sensitivity viral detection methods are desired. A fundamental element of biosensing is molecular recognition and binding events between targets and target-specific capture molecules, which enable the specific identification of analytes. Therefore, molecular designs with a high affinity to the target analytes are of great importance. Besides the selection and optimization of interacting molecules from the field of biochemistry,[44-50] multidisciplinary approaches from chemistry, mechanics, and optics have been introduced to enhance sensitivity.
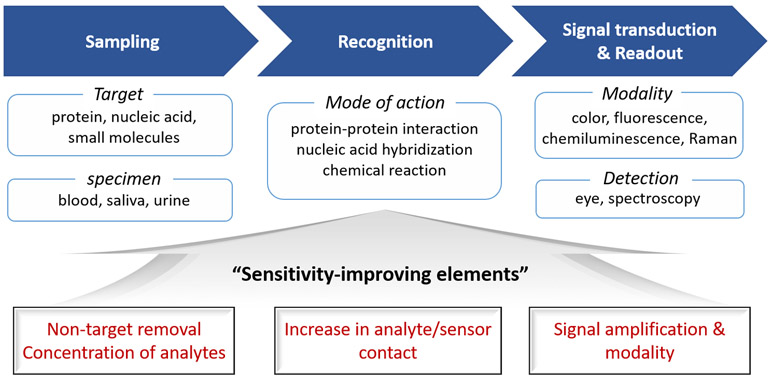
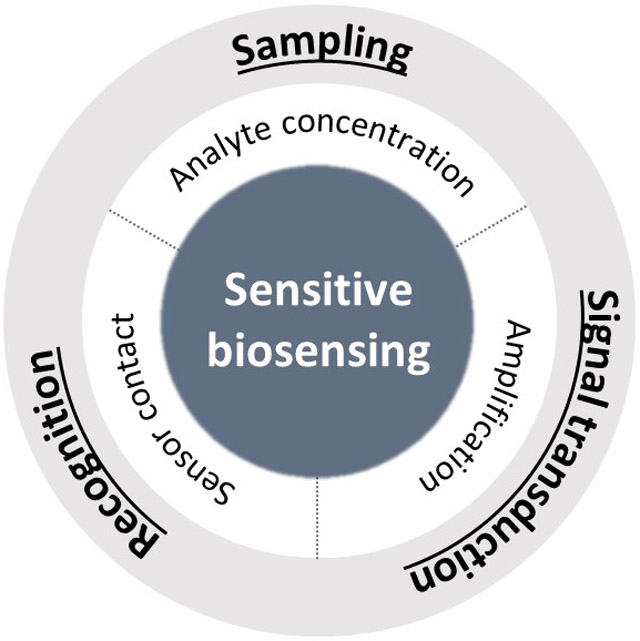
The scope of biosensing covers in vitro biochemical assays for the detection or quantification of biomarkers and in vivo imaging for the visualization of biomarkers. This review focuses on the in vitro assays. A general workflow of biosensing consists of collecting target-containing samples, recognizing targets by specific biomolecular interactions, and signal-transducing from recognition events to a detectable readout (Figure 1). Typical target analytes are small molecules (e.g., glucose, amino acids, and reactive oxygen species), proteins (e.g., enzymes and viral proteins) and nucleic acids (e.g., pathogen-specific DNA/RNA sequences), which are involved in metabolisms or disease development. In practice, biological fluids such as blood, saliva, and urine in intact or purified forms that contain analytes are collected from human bodies. In this sampling step, the removal of non-target materials to circumvent their potential interference with sensor operations and the preconcentration of analytes to maximize sensing performances can be identified as sensitivity-enhancing strategies. Classic examples are the centrifugation of blood samples to remove visible-light-absorbing red blood cells for improving signal detection and the filtering out of high-molecular-weight materials like proteins to minimize the autofluorescence background for the fluorescent detection of small molecules.
Figure 1.
A general workflow of optical biosensing along with demonstrated sensitivity-improving elements (at the bottom in red) at each step of the sensor operation (i.e., “Sampling”, “Recognition”, and “Signal transduction & Readout”).
At the recognition step, molecular interactions are employed to endow the sensor with specificity to target analytes. The mode of interaction depends on the type of targets, which includes protein–protein interaction in most of the immunoassays, nucleotide hybridization or polymerase-based gene amplification for nucleic acid detection, and chemical reactions for small molecules. Due to the facile separation of analytes from a sample solution to a substrate and substrate-confined readout, in vitro biochemical assays mostly adopt substrates such as solid supports on which capture molecules are immobilized. Therefore, sensitivity-enhancing strategies at this step are concerned with how to increase analyte–sensor contact probability. In the final step of biosensing, analyte-correlated signals are transduced from responsible probes. A variety of signal-transducing modalities such as color, fluorescence, chemiluminescence, and Raman scattering have been used with respect to readout methods (e.g., colorimetric modality for eye detection, and luminescence and scattering modalities with spectroscopic analysis). In recent decades, other optical phenomena based on surface plasmon resonance, waveguide resonance, whispering-gallery resonance, and interferometry have also been studied for the label-free sensing.[51-61] It is noteworthy that the signal transduction step is integrated with the biorecognition step in some sensor designs where signal amplification is one of the major approaches to enhance the sensitivity of biosensors.
In this review, exemplary strategies for sensitivity enhancement are classified into three sections based on the various steps of sensor operation. Firstly, chemical signal amplification is introduced as a sensitivity-enhancing strategy at the signal-transducing and readout step. Secondly, we discuss fluid mixing and analyte concentrating as physical approaches to increase analyte–sensor contact at the recognition step. Finally, we introduce combined approaches concerning sampling, target recognition, and signal transduction. At each section, basic concepts and designs of representative sensors are presented.
2. Chemical Signal Amplification
At the signal-transducing step of biosensing, signal intensity per a single biorecognition event determines the sensitivity of the biosensor. Accordingly, one of the effective ways to enhance sensitivity is to maximize the number or intensity of signals per event. In a basic approach, instead of single signaling molecule tagging, micro- or nano-carriers loaded with multiple signaling molecules are used as probes to improve the sensitivity.[62,63] Variable signal modalities have also been studied for sensitivity improvements. For example, near-infrared-emitting fluorophores have been suggested as a candidate for fluorescence modality to enhance a signal-to-noise ratio, especially in clinically relevant samples like serum where light absorbance autofluorescence in a visible range is inevitable.[64,65] Plasmon-enhanced fluorescence has also been successfully deployed in biosensing for sensitivity improvement.[63,66]
Beyond the selection of signaling modes, the introduction of chemical reactions at the signal-transducing step to amplify the signals is a promising way to enhance sensitivity. One example is the current standard of protein sensing, called enzyme-linked immunosorbent assay (ELISA). The capability of enzymes to repeatedly drive chemical reactions of specific substrates without permanent chemical change of themselves enables signal amplification. By conjugating enzymes to antibodies that are specific to target antigens and applying substrate molecules (chromogenic or fluorogenic) at the final incubation step, one can dramatically amplify the detection signals from biorecognition events for enhanced sensitivity. Inspired by the concept of enzymatic amplification, chemical amplification concepts for biosensing based on polymerization and nanocatalysts have been reported. Representative strategies of chemical signal amplification are summarized in Table 1.
Table 1.
Strategies of chemical signal amplification.
| Category | Strategy | Model system | Signal modality | Detection limit | Reference |
|---|---|---|---|---|---|
| Polymerization | Branched polymerization | Sandwich-type DNA hybridization | Opaqueness | 1 fM | [67] |
| Branched polymerization | Antigen-antibody binding | Surface plasmon resonance | 2.19 fmol/spot | [69] | |
| Photoinitiated polymerization with macroinitiator | Spotted oligonucleotide | Opaqueness | ~ amol/spot | [68] | |
| Nanocatalyst | Au/Pt NPs peroxidation | Sandwich-type lateral flow immunoassay | Chromogenesis | 3.1 pg/mL | [73] |
| Porous Pt NPs peroxidation | Sandwich-type lateral flow immunoassay | Chromogenesis | 0.8 pg/mL | [74] |
2.1. Signal Amplification by Polymerization at the Signal Transduction Step
Polymerization is a chemical reaction that produces a long-chain polymer from monomer molecules. Due to the propagating nature of polymerization at specified conditions (i.e., in the presence of reaction components), it can be employed as a tool to amplify biorecognition events through the growth of polymer bodies. In terms of reaction mechanisms, chain-growth polymerization, especially radical-based reactions, has an advantage over step-growth polymerization. In chain-growth polymerization, the reaction progresses through the addition of individual monomers to an active growing chain, enabling the controlled growth of a polymer with a narrow molecular weight distribution and concomitant target-proportional signal transduction. Given that polymerization occurs only if initiators are present, target-triggered polymerization is realized by labeling end biomolecules that recognize the target with initiators as enzyme-labeled antibodies in ELISA.
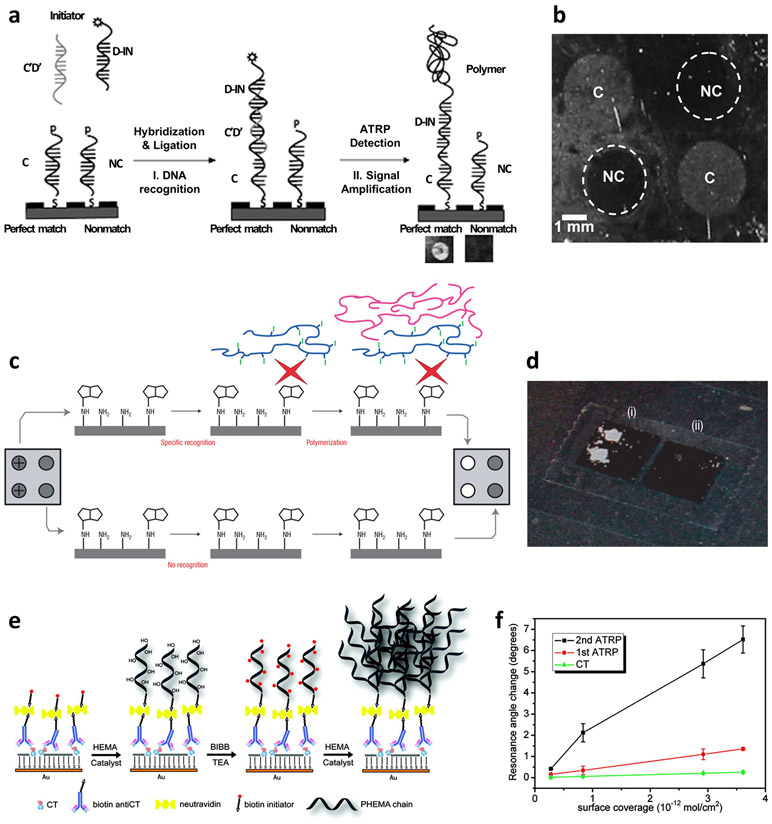
The concept of polymerization-based signal amplification was first demonstrated by Lou et al.[67] They have shown that the atom transfer radical polymerization reaction is an effective and robust reaction that produces well-controlled polymers by using alkyl halide radical initiators coupled with copper-based redox catalysts. Initiators (bromoisobutyrroyl bromide) were conjugated to probe DNA in a sandwich-type DNA hybridization scheme. After the completion of hybridization between surface capture, complementary target, and probe DNAs, the solution of monomer (2- hydroxyethyl methacrylate, HEMA) and catalyst mixture (CuCl, CuBr2 and 2,2’-bipyridyl) was applied to the substrate to grow a polymer (Figure 2a). Reaction conditions were optimized to enable quantitative correlation between the target and the polymer product. It was demonstrated that the growth of polyHEMA, indicated by the opaqueness of the film, was specific to the complementary sequence whereas the film remained transparent when the non-complementary sequence was applied (Figure 2b). In this work, a single mismatch in the DNA sequence was successfully distinguished.
Figure 2.
Signal amplification by polymerization. (a, b) Amplified DNA detection with atom transfer radical polymerization: (a) schematic description of the system and (b) photographs of target DNA detected by the growth of polymer film. Reproduced with permission.[67] Copyright 2005, American Chemical Society. (c, d) Amplification by macroinitiator-mediated photo-initiated polymerization: (c) scheme and (d) photographs of oligonucleotide-immobilized substrate after polymerization. Reproduced with permission.[68] Copyright 2008, Springer Nature. (e, f) Double amplification approach: (e) scheme and (f) comparison between double- and single-amplified methods. Reproduced with permission.[69] Copyright 2010, American Chemical Society.
Another approach of polymerization-based amplification was demonstrated with photoinitiated radical polymerization.[68] Photoinitiation can reduce reaction time by triggering initiators throughout the whole substrate with universal light radiation. An additional key design was macroinitiators that have the function of both biorecognition and initiation (Figure 2c,d). Compared to one-to-one conjugation of an initiator with a probe molecule, macroinitiators that carry multiple initiators can further facilitate polymerization. For this purpose, polymeric materials were used as a macroinitiator platform by which polymerization advances. In a direct comparison to classical enzymatic amplification (horseradish peroxidase and 3,3’,5,5’-tetramethylbenzidine (TMB)-dextran), the polymeric signal amplification of substrate-immobilized nucleotides showed sensitvity enhancement by orders of magnitude.
Efforts to maximize the amplification capacity were made from design approaches such as two-step polymerization wherein an additional polymerization step is introduced to further amplify the signal. This is accomplished by the branched growth of polymers from the linear polymers grown at the first polymerization step.[69] The main idea of this design is the activation of polyHEMA, a product of the first polymerization step, for the consequent polymerization. By treating the as-grown linear polyHEMA with 2-bromoisobutyryl bromide, hydroxyl groups were converted into initiators that enabled further polymerization (Figure 2e). Moreover, in this work, ascorbic acid was used to grow polymer in ambient conditions. With surface plasmon resonance as a signal modality and surface-immobilized bacterial cholera toxin as an analyte, the degree of amplification was enhanced by 5 times in the two-step polymerization compared to the single-step polymerization (Figure 2f). Further studies have been performed with combined design of polymerization and enzymatic amplification as well as the integration with colorimetric, electrochemiluminescent and electrochemical readouts.[70-72] Although polymerization-based approaches have shown potential for chemical signal amplification, requirements of multiple reagents and reaction conditions should be alleviated before widespread practical implementation is undertaken.
2.2. Signal Amplification by Nanocatalysts
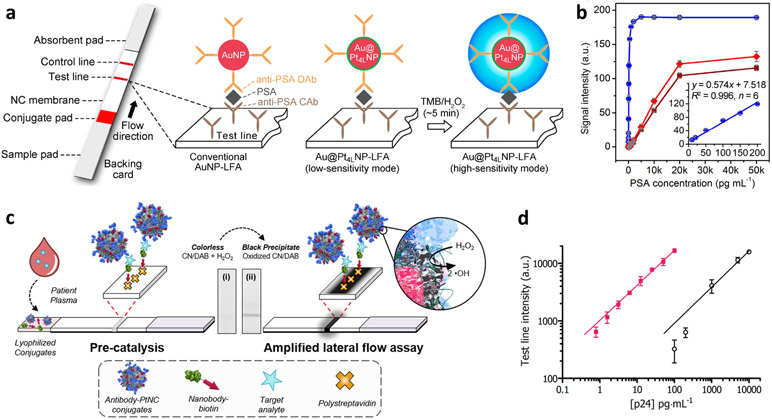
Another class of signal amplification is nanocatalyst-driven chemical reactions. Classically, enzymes have been driving signal-amplification strategies in biosensing. However, the structure and concomitant reaction activity of enzymes are subject to change by various parameters such as pH and temperature. In this regard, there have been efforts to replace enzymes with robust, stable, and cost-efficient inorganic catalysts such as noble-metal nanoparticles (NPs). For example, Gao et al. reported that platinum-decorated gold NPs (Au/Pt NPs) could be utilized as nanocatalysts for ultrasensitive colorimetric assay for in vitro diagnostics.[73] In a lateral flow assay (LFA) platform, Au/Pt NPs were introduced to the probe antibody, triggering peroxidase-like catalytic reactions for signal amplification (Figure 3a). An oxidative dye (TMB) with hydrogen peroxide was used as a chromophore. Human prostate-specific antigen (PSA) was studied as a model. Compared to the conventional LFA that employs intrinsic color of Au NPs as signal, catalytic amplification was capable of generating a color signal several orders of magnitude higher (Figure 3b). Further development of this concept was done through different NP designs. Loynachan et al. demonstrated that porous Pt NPs enabled strong catalytic amplification due to the larger catalytic surface area (Figure 3c).[74] In the LFA model for p24 (HIV biomarker) detection, signal enhancement by two orders of magnitude was achieved (Figure 3d). These studies have successfully showed that nanocatalyst-based signal amplification is able to improve the limited senstivity of the naked-eye-detectable LFA.
Figure 3.
Signal amplification by nanocatalysts. (a, b) Catalytic amplification by Au/Pt NPs: (a) schematic of Au/Pt NPs-based lateral flow assay and (b) calibration curves for PSA detection. Reproduced with permission.[73] Copyright 2017, American Chemical Society. (c, d) Catalytic amplification by Pt nanocrystals: (c) schematic of catalyst-amplified lateral flow assay and (d) tested results for p24 spiked into sera. Reproduced with permission.[74] Copyright 2018, American Chemical Society.
Within the scope of chemical signal amplification, color tonality has been suggested as an effective signal mode. Eye-detectable biosensing is of specific interest and is regarded as a core technology for point-of-care testing and home diagnostics. Most of the current commercial products are based on the generation of a colored signal as in LFA. However, the color signal is only detectable at a high concentration of analytes, limiting their usage in the detection of low-concentration, clinically relevant analytes. Rather than the on-off colorimetric mode, color tonality (i.e., distinguishable color change) has been suggested as a signal modality where signal amplification can be applied to enhance the sensitivity. Pertinent research has been motivated by the color-changing phenomenon of plasmonic NPs by growth and aggregation.[75,76]
3. Diffusion-Limit-Breaking Systems for Enhancing Sensor–Analyte Contact at the Biorecognition Step
Molecular recognition in biosensing is governed by collisions and subsequent binding reactions of interacting molecules. While the binding reactions rely on the binding affinity between two types of molecules determined by molecular structures and reaction mechanisms, the collisions are subject to the diffusive mass transport of molecules in solutions to surface-immobilized counterparts in surface-based sensor configurations. Given the concentration dependence of diffusion, the degree of recognition at any given time is determined by the concentration of analytes in a solution. In practice, substrates are often incubated with a solution for molecular binding. As an example, each incubation step in ELISA takes 30 minutes to one hour. When the concentration of analytes is diluted, it often takes too long for binding events to transduce detectable signals in the diffusion-based incubation. In this context, diffusion-limit-breaking elements that can enhance the contact probability between sensor surfaces and solutes have been proposed to improve the sensitivity of surface-based sensors. Most of the diffusion-limit-breaking approaches fit into two major categories: (i) effectively mixing solutions during incubation, and (ii) concentrating solutes near the substrates. Table 2 summarizes representative strategies for enhancing sensor–analyte contact.
Table 2.
Strategies for enhancing analyte–sensor contact.
| Category | Strategy | Model system | Signal modality | Sensitivity enhancementa) |
Reference |
|---|---|---|---|---|---|
| Fluid mixing | Acoustic microstreaming | DNA hybridization | Fluorescence | × 5 | [79] |
| Micromotor | Sandwich-type immunoassay | Fluorescence | × 3.5 | [84] | |
| Analyte concentrating | Solvent evaporation with surface nanostructuring | Direct DNA sensing | Fluorescence | 1 aM (detection limit) | [87] |
| Alternative current electrothermal effect | Antigen-antibody binding in microfluidics | Fluorescence | ×10 (time reduction) | [91] | |
| Biphasic bubble | Protein-protein interaction | Fluorescence | × 8.7 | [107] |
Enhancement factor compared to static incubation methods.
3.1. Enhanced Analyte–Sensor Contact by Fluid Mixing
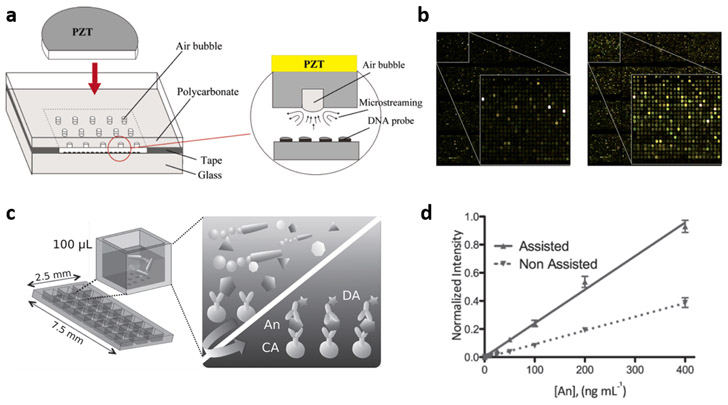
The purpose of mixing-enhanced biosensing is to break the diffusion limit of analytes by continuously agitating the fluid during incubation. Beyond simple approaches such as rotating the reactor and stirring the solution with magnetic bars,[77,78] efforts have been made to harness the physical phenomena of external-field-driven fluid dynamics. One candidate is acoustic streaming, which produces controlled fluid flows through acoustic oscillations. Several examples of this approach can be found in the conventional DNA microarray platform where static incubation methods typically require several hours for the sensing. Liu et al. reported that acoustic microstreaming can generate a steady, global convective flow throughout the DNA microarray, leading to the mixing-induced acceleration of DNA hybridization.[79] As shown in Figure 4a, air bubbles were introduced in the DNA biochip and microstreaming was generated by applying acoustic waves to actuate the bubbles through a piezoelectric disk. For a single hybridization pair of complementary DNAs with a fluorescence detection mode, 5-fold enhancement in signal intensity and signal homogeneity was observed for the mixing-enhanced hybridization in comparison to static hybridization. Several different designs of acoustic-microstreaming-based mixing have been studied to obtain optimal detection outputs, which include enhanced and homogenized DNA hybridization by multiple piezoelectric transducers (Figure 4b).[80]
Figure 4.
Enhanced analyte–sensor contact by fluid mixing. (a) Schematic of the acousitic microscreaming device. Reproduced with permission.[79] Copyright 2003, American Chemical Society. (b) Fluorescence images of DNA microarray with static (left) and acoustic-streaming-assisted dynamic (right) hybridization. Reproduced with permission.[80] Copyright 2012, Royal Society of Chemistry. (c, d) Micromotor-assisted immunosensing: (c) schematic illustration of microarray platform and working principle, and (d) calibration curves. Reproduced with permission.[84] Copyright 2014, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
An alternative approach was demonstrated by utilization of micromotors as a mixing element. Micromotors are generally referred to as artificial micromachines designed to convert stimuli such as chemical fuel and light into motion. There have been extensive studies on the design of micromotors and their utilization in biosensing.[81-84] While micromotors can serve as actively moving surfaces to which dispersed analytes bind effectively,[82] they can also be used as actuators that mix fluids. Both approaches contribute to sensitivity enhancement in biosensing. Morales-Narváez et al. studied the mixing effect of micromotors on sensing performance in the protein microarray platform.[84] Micromotors made of polyaniline and platinum with the capability of being self-propelled by chemical fuels (i.e., hydrogen peroxide) were used in this study. In the sandwich immunoassay scheme that consists of capture antibody (CA), target antigen (An), and detection (Ab) (Figure 4c), surface biorecognition (CA-An complexation) was enhanced by 3.5 times in magnitude compared to the diffusion-limited static incubation (Figure 4d).
3.2. Enhanced Analyte–Sensor Contact by Analyte Concentrating
Another concept to aid in overcoming the diffusion limit is to induce surface-localized concentration of analytes. This concept can significantly enhance sensing performance by directly exposing analytes to sensor surfaces in a concentrated form. One of the available approaches is evaporating a solution droplet to concentrate solutes. However, the evaporation of droplets typically causes a nonuniform and undesirable distribution of solutes deposited on the sensor surface.[85,86] De Angelis et al. proposed that the confinement of solutes to defined sensing areas was possible by utilizing a superhydrophobic surface.[87] On a superhydrophobic surface made of silicon micropillar arrays, high contact angles (> 150°) were observed, which enabled the steady concentration of solutes within a stably shrinking, quasi-spherical droplet (Figure 5a). By concentrating solutes on a sensing area of a few tens of square micrometers, the fluorescent detection of lambda DNA (labeled with YOYO-1 dye) was achieved at an initial concentration of 1 aM (Figure 5b). Although surface biorecognition events were not incorporated in this study, evaporation-based concentration with stable droplets, when combined with sensor configurations and controllable evaporation, may show promise as an effective sensitivity-enhancing element.
Figure 5.
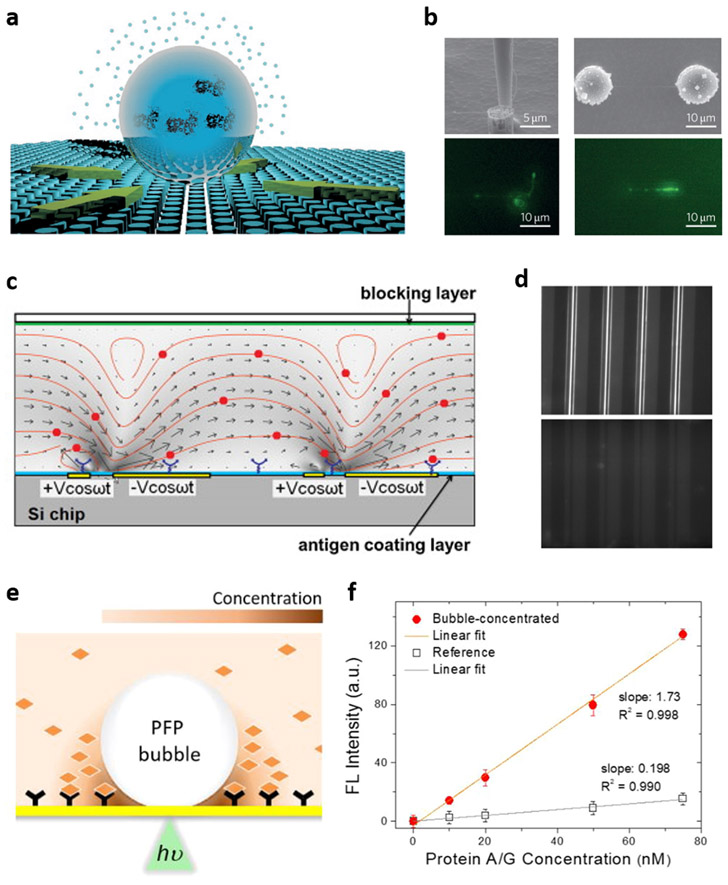
Surface concentration of analytes. (a, b) Evaporation-induced concentration at droplets with high contact angles: (a) conceptual illustration and (b) scanning electron micrographs and fluorescence images of localized DNA from 10 aM solution. Reproduced with permission.[87] Copyright 2011, Springer Nature. (c, d) ACET-based concentration: (c) simulation of ACET-driven convection in a microfluidic channel and (d) fluorescence images of antigen-antibody binding with (top) and without (bottom) ACET flow. Reproduced with permission.[91] Copyright 2011, Elsevier. (e, f) Concentration by an optothermally generated microbubble: (e) schematic of bubble-mediated molecular concentration in a perfluoropentane (PFP)-in-water biphasic liquid system and (f) surface binding profiles in the bubble-concentrated system and the reference of static incubation. Reproduced with permission.[107] Copyright 2020, American Chemical Society.
Another approach to overcoming the diffusion limit is phoretic manipulation and concentration of biomolecules at a sensor surface. Several studies based on thermo- and electrophoresis have been conducted,[88-90] but specific working conditions required for phoresis limit their applicability to certain types of biomolecules. Alternatively, the manipulation of fluid dynamics can be deployed to induce surface-localized concentration of solutes. Liu et al. reported an immunoassay system based on the alternative current electrothermal (ACET) effect.[91] By applying an alternative current to an electrode array in a microfluidic setting, swirl-like microflow patterns were generated toward the electrodes, concentrating the dispersed proteins on the substrate surface and enabling flow-driven mixing (Figure 5c). An immunoassay scheme for immunogenic antibody testing was adopted for the detection of immunoglobulin G against Johne’s disease in serum. With surface-immobilized disease-specific antigens as a capture and fluorescent secondary antibodies as a probe, the fluorescent detection was accelerated by ten times compared to the conventional microfluidic setup (Figure 5d). In comparison to other electrical concentration methods such as direct current electrophoresis that works only for charged biomolecules, ACET benefits from a high level of versatility due to the indirect manipulation of biomolecules by flow control.
Another exciting approach within the scope of analyte concentration is microbubble-based solute accumulation. It is well-known that dispersoids such as colloidal particles and bacteria can be favorably trapped at the air–water interface by surface tension. Spurred by this phenomenon, implementation of microbubble-mediated concentration in the field of biosensing has been conducted. Tantussi et al. demonstrated that an expanding microbubble generated by a pulse laser beam through a plasmonic nanoantenna was able to concentrate extracellular membrane vesicles (EVs) on the bubble surface.[92] On the other hand, optothermally generated microbubbles on a plasmonic surface have also been proposed as an effective concentrating method. These bubbles are capable of inducing strong Marangoni convective flow by utilizing a surface tension gradient along the bubble surface that drives solutes to the bubble–liquid–substrate interface.[93-95] Along with surface assembly of microbes, synthetic particles, and molecules, small-molecule sensing has been reported based on this phenomenon.[96-105] However, a high working temperature required to vaporize water has prevented the optothermally generated microbubbles from being applied to sensing of proteins whose activity is subject to thermal denaturation. [95,106]. Kim et al. reported a biphasic liquid system wherein volatile, water-immiscible perfluoropentane was emulsified into an aqueous medium as a bubble-generating liquid (Figure 5e).[107] With a single protein–protein interaction model (immunoglobulin G as a surface capture molecule and protein A/G as an analyte), one-order-of-magnitude enhancement of surface capture was achieved within 1 minute, compared to a static incubation method with incubation time of 30 minutes (Figure 5f).
4. Combined Approaches Utilizing Multiple Sensitivity-Enhancing Elements
The biological specimen of concern varies with respect to target analytes. For example, blood is drawn from the human body for assessing bloodborne pathogens, immunogenic antibodies, or metabolites. Respiratory fluids are collected for detecting viruses associated with respiratory illnesses. Among a variety of available biofluids, urine is especially intriguing in that endo- and exogenous substances are conditionally filtered out by the renal clearance system of our bodies. Upon the systemic administration, nanoscale molecules (e.g., short peptides and proteins) and nanoparticles have been found to undergo rapid renal clearance when their hydrodynamic size is less than 6 nm, also depending on their shape and surface chemistry.[108-110] Based on these unique characteristic, the urinary system can serve as a natural concentrator through which substances with rapid clearance kinetics are concentrated from the whole body (typically throughout the vascular system) to early post-administration or post-release urine samples. Especially, there exist cases where target biomarkers are not adequately available in body-circulating fluids due to limited vascular release and dilution in systemic circulation, such as biomarkers retained in tumor regions at the early stages of cancer.[111,112] In such cases, artificial probes are needed to transfer the information of hidden biomarkers to samplable biofluids. When target-correlated probes are constructed to undergo rapid renal clearance as a result of their interaction with targets in the body, like the working principle of prodrugs, the use of urinary samples takes advantage of the concentration effect at the sampling step for sensing unavailable or low-abundant targets in body fluids.
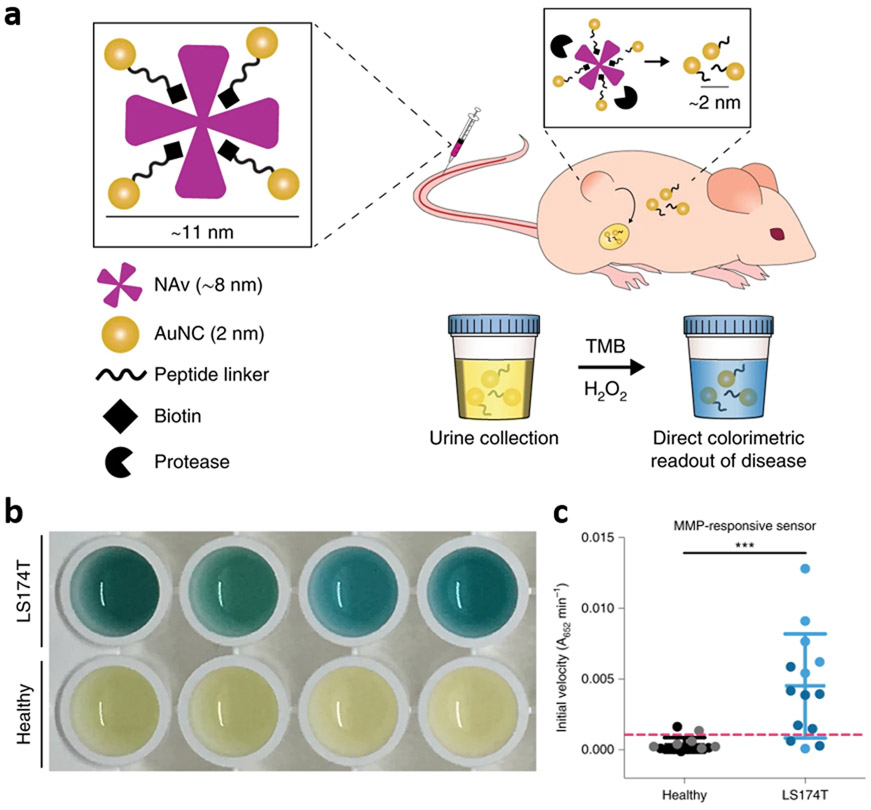
A strategy that combines multiple sensitivity-enhancing elements throughout sampling, target recognition, and signal transduction has been proposed by Loynachan et al.[112] This combined strategy capitalizes on the renal concentration at the sampling step by using synthetic biomarkers. Protein-based nanostructures in which 2 nm gold nanocrystals (AuNCs) were conjugated to 8 nm proteins through protease-cleavable peptide linkers were designed to enable the renal concentration of cleaved AuNCs in a size-dependent manner (Figure 6a). Upon the in vivo administration of the nanocomplexes, AuNCs were cleaved by target protease abundant in tumor regions (i.e., biorecognition event), and concentrated from blood to urine after systemic circulation (i.e., concentration event). The presence of AuNCs in urine, correlated with the tumor diagnosis, was detected by nanocatalytic signal amplification with the oxidative chromogenesis (TMB and hydrogenperoxide) enabled by the intrinsic catalytic activity of AuNCs (i.e., signal amplification) (Figure 6b and 6c). This work further implies that sensitivity-enhancing elements can be found in biological systems, and sensor designs can be expanded to harness these endogenous elements although case-specific designs of probes are required for succeessful implementation.
Figure 6.
Combination of renal concentration and nanocatalytic signal amplification. (a-c) Signal concentration and amplification enabled by renal-clearable gold-protein nanocomplexes: (a) schematic of nanocomplex and its working principle for in vivo signal concentration and in vitro signal amplification, (b) colorimetric urinary readout for tumor diagnosis, and (c) corresponding catalytic activity assay. Reproduced with permission.[112] Copyright 2019, Springer Nature.
5. Conclusion and Outlook
Various approaches have been developed to enhance sensitivity of optical biosensing. Chemical signal amplification strategies that utilize polymerization and nanocatalysts effectively enhance sensitivity at the signal transduction step by multiplying the signal-to-recognition ratio. Fluid mixing and analyte concentrating based on external-field-driven flow dynamics and evaporation enhance sensitivity by increasing the propensity of sensor–analyte contact at the biorecognition step. The concept of analyte concentration can be extended to the sampling step by utilizing the renal clearance system, which can be further combined with nanocatalyic signal amplification at the transduction step.
When combined with rational sensor design and optimization, the sensitivity-enhancing elements will contribute to the development of high-performance diagnostic platforms capable of detecting target analytes of a minute quantity or variation thereof. Although this review focuses on examples in conventional optical biosensing schemes, these elements are conceptually compatible with other sensing modes such as those based on electrical and electrochemical methods. One of the most prominent applications of biosensing is point-of-care testing (POCT) for managing infectious diseases and monitoring personal health, where a trade-off between sensitivity and processing time has often been inevitable. Proper implementation of the sensitivity-enhancing elements into POCT devices may lead to simplified procedures throughout sampling, recognition, and signal transduction. Another exciting direction is to develop the sensitivity-enhancing elements into “plug-and-play” modules, which can be readily integrated with many of current and future sensors to boost performance. It is also noteworthy that the incorporation of machine learning into biosensing can be a synergistic way of improving sensor performance. There have been successful applications of machine learning for the precise analysis of biological data such as genetic information[113-115] and cellular images.[116-120] With its capability of processing a large amount of data, machine learning enables the detection of complex and/or marginally-varying sensing signals in an accurate and rapid way.[121-123] Along with their contributions to developing autonomous systems and optimizing sensor designs, machine-learning-based approaches can fully benefit multiplexed and real-time sensing (e.g., wearable health monitoring systems) where complex and fluctuating signal matrices must be cross-interpreted to draw diagnostic outcomes. To conclude, high sensitivity combined with user-friendliness and facile readouts will eventually enable larger-scale uses of various sensors for self- and home-diagnosis and Internet of Things.
Acknowledgements
The authors acknowledge the financial support of the National Science Foundation (NSF-CMMI-1761743 and NSF-ECCS-2001650), the National Aeronautics and Space Administration Early Career Faculty Award (80NSSC17K0520), and the National Institute of General Medical Sciences of the National Institutes of Health (DP2GM128446).
References
- [1].Clarke JR, Expert Rev. Mol. Diagn 2002, 2, 233. [DOI] [PubMed] [Google Scholar]
- [2].Moody A, Clin. Microbiol. Rev 2002, 15, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nainan OV, Xia G, Vaughan G, Margolis HS, Clin. Microbiol. Rev 2006, 19, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NTN, Jones-López EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D, J. Clin. Microbiol 2010, 48, 229.19864480 [Google Scholar]
- [5].DiMaio MA, Sahoo MK, Waggoner J, Pinsky BA, Virol J. Methods 2012, 186, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bohunicky B, Mousa SA, Nanotechnol. Sci. Appl 2011, 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yoo E-H, Lee S-Y, Sensors 2010, 10, 4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sin ML, Mach KE, Wong PK, Liao JC, Expert Rev. Mol. Diagn 2014, 14, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Von Erlach TC, Bertazzo S, Wozniak MA, Horejs CM, Maynard SA, Attwood S, Robinson BK, Autefage H, Kallepitis C, Del Río Hernández A, Chen CS, Goldoni S, Stevens MM, Nat. Mater 2018, 17, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cheng X, Ferrell JE, Science 2018, 361, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS, Nat. Chem. Biol 2011, 8, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Miller EW, Tulyathan O, Isacoff EY, Chang CJ, Nat. Chem. Biol 2007, 3, 263. [DOI] [PubMed] [Google Scholar]
- [13].Horsburgh AM, Mardlin DP, Turner NL, Henkler R, Strachan N, Glover LA, Paton GI, Killham K, Biosens. Bioelectron. 2002, 17, 495. [DOI] [PubMed] [Google Scholar]
- [14].Ron EZ, Curr. Opin. Biotechnol 2007, 18, 252. [DOI] [PubMed] [Google Scholar]
- [15].Boonjob W, Miroó M, Segundo MA, Cerdaá V, Anal. Chem 2011, 83, 5237. [DOI] [PubMed] [Google Scholar]
- [16].Pérez-Fernández V, Mainero Rocca L, Tomai P, Fanali S, Gentili A, Anal. Chim. Acta 2017, 983, 9. [DOI] [PubMed] [Google Scholar]
- [17].Scognamiglio V, Arduini F, Palleschi G, Rea G, TrAC Trends Anal. Chem 2014, 62, 1. [Google Scholar]
- [18].Alves RC, Barroso MF, González-García MB, Oliveira MBPP, Delerue-Matos C, Crit. Rev. Food Sci. Nutr 2016, 56, 2304. [DOI] [PubMed] [Google Scholar]
- [19].Khuda S, Slate A, Pereira M, Al-Taher F, Jackson L, Diaz-Amigo C, Bigley EC, Whitaker T, Williams KM, Agric J. Food Chem. 2012, 60, 4195. [DOI] [PubMed] [Google Scholar]
- [20].Song Y, Wei W, Qu X, Adv. Mater 2011, 23, 4215. [DOI] [PubMed] [Google Scholar]
- [21].Yang C, Wang Y, Marty J-L, Yang X, Biosens. Bioelectron. 2011, 26, 2724. [DOI] [PubMed] [Google Scholar]
- [22].Wang C, Ma Z, Wang T, Su Z, Adv. Funct. Mater 2006, 16, 1673. [Google Scholar]
- [23].Zhong W, Anal. Bioanal. Chem 2009, 394, 47. [DOI] [PubMed] [Google Scholar]
- [24].Zhang C, Yuan Y, Zhang S, Wang Y, Liu Z, Angew. Chemie Int. Ed 2011, 50, 6851. [DOI] [PubMed] [Google Scholar]
- [25].Browne KA, Deheyn DD, El-Hiti GA, Smith K, Weeks I, J. Am. Chem. Soc 2011, 133, 14637. [DOI] [PubMed] [Google Scholar]
- [26].Tanaka T, Matsunaga T, Anal. Chem 2000, 72, 3518. [DOI] [PubMed] [Google Scholar]
- [27].Brabant G, von zur Mühlen A, Wüster C, Ranke MB, Kratzsch J, Kiess W, Ketelslegers J-M, Wilhelmsen L, Hulthén L, Saller B, Mattsson A, Wilde J, Schemer R, Kann P, Horm. Res. Paediatr 2003, 60, 53. [DOI] [PubMed] [Google Scholar]
- [28].Bi S, Yan Y, Yang X, Zhang S, Chem. - A Eur. J 2009, 15, 4704. [DOI] [PubMed] [Google Scholar]
- [29].Ge L, Wang S, Song X, Ge S, Yu J, Lab Chip 2012, 12, 3150. [DOI] [PubMed] [Google Scholar]
- [30].Tripp RA, Dluhy RA, Zhao Y, Nano Today 2008, 3, 31. [Google Scholar]
- [31].Bantz KC, Meyer AF, Wittenberg NJ, Im H, Kurtuluş Ö, Lee SH, Lindquist NC, Oh S-H, Haynes CL, Phys. Chem. Chem. Phys 2011, 13, 11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Laing S, Jamieson LE, Faulds K, Graham D, Nat. Rev. Chem 2017, 1, 0060. [Google Scholar]
- [33].Shen M, Zhou Y, Ye J, Abdullah AL-maskri AA, Kang Y, Zeng S, Cai S, J. Pharm. Anal 2020, 10, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Balboni A, Gallina L, Palladini A, Prosperi S, Battilani M, Sci. World J 2012, 2012, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Uhlenhaut C, Cohen JI, Pavletic S, Illei G, Gea-Banacloche JC, Abu-Asab M, Krogmann T, Gubareva L, McClenahan S, Krause PR, Transpl. Infect. Dis 2012, 14, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wan Z, Zhang Y, He Z, Liu J, Lan K, Hu Y, Zhang C, Int. J. Mol. Sci 2016, 17, 1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Corman VM, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, van Boheemen S, Gopal R, Ballhause M, Bestebroer TM, Muth D, Müller MA, Drexler JF, Zambon M, Osterhaus AD, Fouchier RM, Drosten C, Eurosurveillance 2012, 17, 1. [DOI] [PubMed] [Google Scholar]
- [38].Noh JY, Yoon S-W, Kim D-J, Lee M-S, Kim J-H, Na W, Song D, Jeong DG, Kim HK, Arch. Virol 2017, 162, 1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lu X, Whitaker B, Sakthivel SKK, Kamili S, Rose LE, Lowe L, Mohareb E, Elassal EM, Al-sanouri T, Haddadin A, Erdman DD, J. Clin. Microbiol 2014, 52, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C, Nature 2020, 581, 465. [DOI] [PubMed] [Google Scholar]
- [41].Yang Y, Yang M, Yuan J, Wang F, Wang Z, Li J, Zhang M, Xing L, Wei J, Peng L, Wong G, Zheng H, Wu W, Shen C, Liao M, Feng K, Li J, Yang Q, Zhao J, Liu L, Liu Y, Innov. 2020, 1, 100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pan Y, Zhang D, Yang P, Poon LLM, Wang Q, Lancet Infect. Dis 2020, 20, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Winichakoon P, Chaiwarith R, Liwsrisakun C, Salee P, Goonna A, Limsukon A, Kaewpoowat Q, J. Clin. Microbiol 2020, 58, e00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ellington AD, Szostak JW, Nature 1990, 346, 818. [DOI] [PubMed] [Google Scholar]
- [45].Zeng X, Shen Z, Mernaugh R, Anal. Bioanal. Chem 2012, 402, 3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nakatsuka N, Cao HH, Deshayes S, Melkonian AL, Kasko AM, Weiss PS, Andrews AM, ACS Appl. Mater. Interfaces 2018, 10, 23490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Union A, Meheus L, Humbel RL, Conrad K, Steiner G, Moereels H, Pottel H, Serre G, De Keyser F, Arthritis Rheum. 2002, 46, 1185. [DOI] [PubMed] [Google Scholar]
- [48].Brock I, Weldingh K, Leyten EMS, Arend SM, Ravn P, Andersen P, J. Clin. Microbiol 2004, 42, 2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Volland H, Pradelles P, Ronco P, Azizi M, Simon D, Créminon C, Grassi J, J. Immunol. Methods 1999, 228, 37. [DOI] [PubMed] [Google Scholar]
- [50].Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS, Int. J. Cancer 2009, 125, 1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yanik AA, Huang M, Kamohara O, Artar A, Geisbert TW, Connor JH, Altug H, Nano Lett. 2010, 10, 4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Riedel T, Rodriguez-Emmenegger C, de los Santos Pereira A, Bědajánková A, Jinoch P, Boltovets PM, Brynda E, Biosens. Bioelectron 2014, 55, 278. [DOI] [PubMed] [Google Scholar]
- [53].Pimková K, Bocková M, Hegnerová K, Suttnar J, Čermák J, Homola J, Dyr JE, Anal. Bioanal. Chem 2012, 402, 381. [DOI] [PubMed] [Google Scholar]
- [54].Yun J, Park YH, Bae TS, Lee S, Lee GH, ACS Appl. Mater. Interfaces 2013, 5, 164. [DOI] [PubMed] [Google Scholar]
- [55].Endo T, Kerman K, Nagatani N, Hiepa HM, Kim D-K, Yonezawa Y, Nakano K, Tamiya E, Anal. Chem 2006, 78, 6465. [DOI] [PubMed] [Google Scholar]
- [56].Armani AM, Kulkarni RP, Fraser SE, Flagan RC, Vahala KJ, Science 2007, 317, 783. [DOI] [PubMed] [Google Scholar]
- [57].He L, Özdemir ŞK, Zhu J, Kim W, Yang L, Nat. Nanotechnol 2011, 6, 428. [DOI] [PubMed] [Google Scholar]
- [58].Ymeti A, Greve J, Lambeck PV, Wink T, van Hövell, Beumer, Wijn RR, Heideman RG, Subramaniam V, Kanger JS, Nano Lett. 2007, 7, 394. [DOI] [PubMed] [Google Scholar]
- [59].Xu J, Suarez D, Gottfried DS, Anal. Bioanal. Chem 2007, 389, 1193. [DOI] [PubMed] [Google Scholar]
- [60].Zaytseva N, Miller W, Goral V, Hepburn J, Fang Y, Appl. Phys. Lett 2011, 98, 163703. [Google Scholar]
- [61].Zhao C, Xu X, Ferhan AR, Chiang N, Jackman JA, Yang Q, Liu W, Andrews AM, Cho N-J, Weiss PS, Nano Lett. 2020, 20, 1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chikkaveeraiah BV, Mani V, Patel V, Gutkind JS, Rusling JF, Biosens. Bioelectron. 2011, 26, 4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Luan J, Seth A, Gupta R, Wang Z, Rathi P, Cao S, Gholami Derami H, Tang R, Xu B, Achilefu S, Morrissey JJ, Singamaneni S, Nat. Biomed. Eng 2020, 4, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kim J, Kwon JH, Jang J, Lee H, Kim S, Hahn YK, Kim SK, Lee KH, Lee S, Pyo H, Song CS, Lee J, Biosens. Bioelectron 2018, 112, 209. [DOI] [PubMed] [Google Scholar]
- [65].Kang D, Lee S, Shin H, Pyun J, Lee J, Biosens. Bioelectron 2020, 150, 111921. [DOI] [PubMed] [Google Scholar]
- [66].Xu W, Wang L, Zhang R, Sun X, Huang L, Su H, Wei X, Chen CC, Lou J, Dai H, Qian K, Nat. Commun 2020, 11, 1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lou X, Lewis MS, Gorman CB, He L, Anal. Chem 2005, 77, 4698. [DOI] [PubMed] [Google Scholar]
- [68].Sikes HD, Hansen RR, Johnson LM, Jenison R, Birks JW, Rowlen KL, Bowman CN, Nat. Mater 2008, 7, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Liu Y, Dong Y, Jauw J, Linman MJ, Cheng Q, Anal. Chem 2010, 82, 3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yuan L, Xu L, Liu S, Anal. Chem 2012, 84, 10737. [DOI] [PubMed] [Google Scholar]
- [71].Zheng W, He L, Anal. Bioanal. Chem 2009, 393, 1305. [DOI] [PubMed] [Google Scholar]
- [72].Sun H, Kong J, Wang Q, Liu Q, Zhang X, ACS Appl. Mater. Interfaces 2019, 11, 27568. [DOI] [PubMed] [Google Scholar]
- [73].Gao Z, Ye H, Tang D, Tao J, Habibi S, Minerick A, Tang D, Xia X, Nano Lett. 2017, 17, 5572. [DOI] [PubMed] [Google Scholar]
- [74].Loynachan CN, Thomas MR, Gray ER, Richards DA, Kim J, Miller BS, Brookes JC, Agarwal S, Chudasama V, McKendry RA, Stevens MM, ACS Nano 2018, 12, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].De La Rica R, Stevens MM, Nat. Nanotechnol 2012, 7, 821. [DOI] [PubMed] [Google Scholar]
- [76].Qu W, Liu Y, Liu D, Wang Z, Jiang X, Angew. Chemie - Int. Ed 2011, 50, 3442. [DOI] [PubMed] [Google Scholar]
- [77].Yuen PK, Li G, Bao Y, Müller UR, Lab Chip 2003, 3, 46. [DOI] [PubMed] [Google Scholar]
- [78].Vanderhoeven J, Pappaert K, Dutta B, Van Hummelen P, Desmet G, Anal. Chem 2005, 77, 4474. [DOI] [PubMed] [Google Scholar]
- [79].Liu RH, Lenigk R, Druyor-Sanchez RL, Yang J, Grodzinski P, Anal. Chem 2003, 75, 1911. [DOI] [PubMed] [Google Scholar]
- [80].Maturos T, Pogfay T, Rodaree K, Chaotheing S, Jomphoak A, Wisitsoraat A, Suwanakitti N, Wongsombat C, Jaruwongrungsee K, Shaw P, Kamchonwongpaisan S, Tuantranont A, Lab Chip 2012, 12, 133. [DOI] [PubMed] [Google Scholar]
- [81].Kagan D, Calvo-Marzal P, Balasubramanian S, Sattayasamitsathit S, Manesh KM, Flechsig GU, Wang J, J. Am. Chem. Soc 2009, 131, 12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kagan D, Campuzano S, Balasubramanian S, Kuralay F, Flechsig GU, Wang J, Nano Lett. 2011, 11, 2083. [DOI] [PubMed] [Google Scholar]
- [83].Wu J, Balasubramanian S, Kagan D, Manesh KM, Campuzano S, Wang J, Nat. Commun 2010, 1, 1. [DOI] [PubMed] [Google Scholar]
- [84].Morales-Narváez E, Guix M, Medina-Sánchez M, Mayorga-Martinez CC, Merkoçi A, Small 2014, 10, 2542. [DOI] [PubMed] [Google Scholar]
- [85].Dash S, Garimella SV, Langmuir 2013, 29, 10785. [DOI] [PubMed] [Google Scholar]
- [86].Das S, Dey A, Reddy G, Sarma DD, J. Phys. Chem. Lett 2017, 8, 4704. [DOI] [PubMed] [Google Scholar]
- [87].De Angelis F, Gentile F, Mecarini F, Das G, Moretti M, Candeloro P, Coluccio ML, Cojoc G, Accardo A, Liberale C, Zaccaria RP, Perozziello G, Tirinato L, Toma A, Cuda G, Cingolani R, Di Fabrizio E, Nat. Photonics 2011, 5, 682. [Google Scholar]
- [88].Nicoli F, Verschueren D, Klein M, Dekker C, Jonsson MP, Nano Lett. 2014, 14, 6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Crick CR, Albella P, Kim HJ, Ivanov AP, Kim KB, Maier SA, Edel JB, ACS Photonics 2017, 4, 2835. [Google Scholar]
- [90].Shi X, Verschueren DV, Dekker C, Nano Lett. 2018, 18, 8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Liu X, Yang K, Wadhwa A, Eda S, Li S, Wu J, Sensors Actuators, A Phys. 2011, 171, 406. [Google Scholar]
- [92].Tantussi F, Messina GC, Capozza R, Dipalo M, Lovato L, De Angelis F, ACS Nano 2018, 12, 4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lin L, Peng X, Mao Z, Li W, Yogeesh MN, Rajeeva BB, Perillo EP, Dunn AK, Akinwande D, Zheng Y, Nano Lett. 2016, 16, 701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Rajeeva BB, Kunal P, Kollipara PS, Acharya PV, Joe M, Ide MS, Jarvis K, Liu Y, Bahadur V, Humphrey SM, Zheng Y, Matter 2019, 1, 1606. [Google Scholar]
- [95].Xie Y, Zhao C, Nanoscale 2017, 9, 6622. [DOI] [PubMed] [Google Scholar]
- [96].Tokonami S, Kurita S, Yoshikawa R, Sakurai K, Suehiro T, Yamamoto Y, Tamura M, Karthaus O, Iida T, Sci. Adv 2020, 6, eaaz5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Yamamoto Y, Tokonami S, Iida T, ACS Appl. Bio Mater 2019, 2, 1561. [DOI] [PubMed] [Google Scholar]
- [98].Karim F, Vasquez ES, Sun Y, Zhao C, Nanoscale 2019, 11, 20589. [DOI] [PubMed] [Google Scholar]
- [99].Kotnala A, Kollipara PS, Li J, Zheng Y, Nano Lett. 2020, 20, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Li J, Hill EH, Lin L, Zheng Y, ACS Nano 2019, 13, 3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lin L, Hill EH, Peng X, Zheng Y, Acc. Chem. Res 2018, 51, 1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Rajeeva BB, Wu Z, Briggs A, Acharya PV, Walker SB, Peng X, Bahadur V, Bank SR, Zheng Y, Adv. Opt. Mater 2018, 6, 1701213. [Google Scholar]
- [103].Rajeeva BB, Alabandi MA, Lin L, Perillo EP, Dunn AK, Zheng Y, J. Mater. Chem. C 2017, 5, 5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Bangalore Rajeeva B, Lin L, Perillo EP, Peng X, Yu WW, Dunn AK, Zheng Y, ACS Appl. Mater. Interfaces 2017, 9, 16725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Wu Z, Kelp G, Yogeesh MN, Li W, McNicholas KM, Briggs A, Rajeeva BB, Akinwande D, Bank SR, Shvets G, Zheng Y, Nanoscale 2016, 8, 18461. [DOI] [PubMed] [Google Scholar]
- [106].Moon S, Zhang Q, Huang D, Senapati S, Chang HC, Lee E, Luo T, Adv. Mater. Interfaces 2020, 2000597, 1. [Google Scholar]
- [107].Kim Y, Ding H, Zheng Y, Nano Lett. 2020, 20, 7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Longmire M, Choyke PL, Kobayashi H, Nanomedicine 2008, 3, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Liu J, Yu M, Zhou C, Zheng J, Mater. Today 2013, 16, 477. [Google Scholar]
- [110].Warren AD, Kwong GA, Wood DK, Lin KY, Bhatia SN, Proc. Natl. Acad. Sci. U. S. A 2014, 111, 3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Hori SS, Gambhir SS, Sci. Transl. Med 2011, 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Loynachan CN, Soleimany AP, Dudani JS, Lin Y, Najer A, Bekdemir A, Chen Q, Bhatia SN, Stevens MM, Nat. Nanotechnol 2019, 14, 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wylie D, Beaudenon-Huibregtse S, Haynes BC, Giordano TJ, Labourier E, J. Pathol. Clin. Res 2016, 2, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Salem H, Attiya G, El-Fishawy N, Appl. Soft Comput 2017, 50, 124. [Google Scholar]
- [115].Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velázquez Vega JE, Brat DJ, Cooper LAD, Proc. Natl. Acad. Sci 2018, 115, E2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Christiansen EM, Yang SJ, Ando DM, Javaherian A, Skibinski G, Lipnick S, Mount E, O’Neil A, Shah K, Lee AK, Goyal P, Fedus W, Poplin R, Esteva A, Berndl M, Rubin LL, Nelson P, Finkbeiner S, Cell 2018, 173, 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Wang H, Rivenson Y, Jin Y, Wei Z, Gao R, Günaydın H, Bentolila LA, Kural C, Ozcan A, Nat. Methods 2019, 16, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Mata G, Radojević M, Fernandez-Lozano C, Smal I, Werij N, Morales M, Meijering E, Rubio J, Neuroinformatics 2019, 17, 253. [DOI] [PubMed] [Google Scholar]
- [119].Sommer C, Gerlich DW, J. Cell Sci 2013, 126, 5529. [DOI] [PubMed] [Google Scholar]
- [120].Nawaz AA, Urbanska M, Herbig M, Nötzel M, Kräter M, Rosendahl P, Herold C, Toepfner N, Kubánková M, Goswami R, Abuhattum S, Reichel F, Müller P, Taubenberger A, Girardo S, Jacobi A, Guck J, Nat. Methods 2020, 17, 595. [DOI] [PubMed] [Google Scholar]
- [121].Jin X, Liu C, Xu T, Su L, Zhang X, Biosens. Bioelectron. 2020, 165, 112412. [DOI] [PubMed] [Google Scholar]
- [122].Cui F, Yue Y, Zhang Y, Zhang Z, Zhou HS, ACS Sensors 2020, 5, 3346. [DOI] [PubMed] [Google Scholar]
- [123].Oldfrey B, Jackson R, Smitham P, Miodownik M, Front. Robot. AI 2019, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]