Abstract
Cancer stem cells (CSCs) show a self-renewal capacity and differentiation potential that contribute to tumor progression and therapy resistance. However, the underlying processes are still unclear. Elucidation of the key hallmarks and resistance mechanisms of CSCs may help improve patient outcomes and reduce relapse by altering therapeutic regimens. Here, we reviewed the identification of CSCs, the intrinsic and extrinsic mechanisms of therapy resistance in CSCs, the signaling pathways of CSCs that mediate treatment failure, and potential CSC-targeting agents in various tumors from the clinical perspective. Targeting the mechanisms and pathways described here might contribute to further drug discovery and therapy.
Subject terms: Cancer stem cells, Cancer therapy
Introduction
Therapy resistance is becoming a major problem in medicine; while patients initially respond to treatment, sustained administration frequently results in therapy resistance along with a poor prognosis. Both genetic and nongenetic mechanisms enable cancer cells to resist treatment.1 Traditionally, cancer is viewed as a homogenous mass of rapidly proliferating cells. Over the last decades, a more complex model, in which cancer tissue is composed of heterogeneous cell populations with a hierarchical organization has replaced the previous one-dimensional view. Cancer stem cells (CSCs) are on top of this hierarchical structure.2 Eppert and colleagues3 published their pioneering work showing that a defined subset of leukemia cells (CD38+CD34-) was solely responsible for propagating acute myeloid leukemia (AML). A decade later, Al-Hajj’s team showed that as few as 100 cells with a CD44+CD24- phenotype could form tumors in mice, whereas tens of thousands of cells with alternate phenotypes failed to form tumors.4 Many other teams have also found that only a few fractions of cancer cells can reform secondary tumors after transplantation into immunodeficient mice. This cluster of cells are commonly described using special terms such as CSCs, tumor propagating cells, tumor progenitor cells (TPCs) and cancer-initiating cells (CICs). In many adult tissues, stem cells (SCs) are responsible for tissue homeostasis and regeneration, and they can give rise to transit-amplifying (TA) cell populations.5 Similar to normal tissue SCs, CSCs show self-renewal and are defined by their ability to (i) generate a xenograft that histologically resembles the parent tumor from which it was derived, (ii) be serially transplanted in a xenograft assay thereby showing self-renewal (regenerate), and (iii) generate daughter cells that possess some proliferative capabilities but are unable to initiate or maintain the cancer because they lack intrinsic regenerative potential6 (Fig. 1).
Fig. 1.
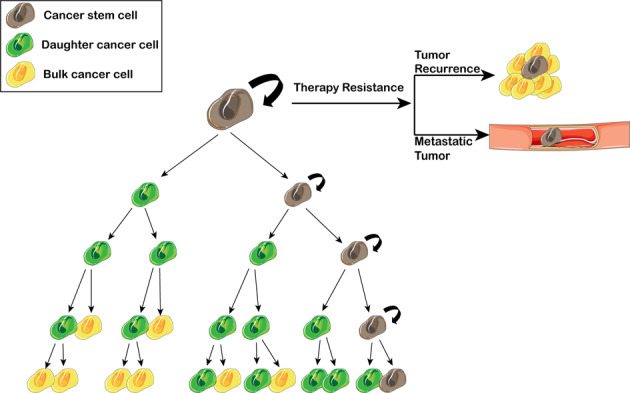
Poor response to therapy due to CSCs. In heterogeneous tumors that contain CSCs, though non-CSCs are ablated, CSC will sustain tumor growth for its ability of self-renewing, then no long-term tumor recurrence or metastatic tumor will be observed
The mechanisms of therapy resistance in cancer can be categorized as intrinsic and acquired. Intrinsic mechanisms are due to preexisting factors of the cancer that are present prior to any treatment, thus rendering certain treatments useless. Acquired drug resistance develops during treatment. Accumulating evidence has shown that the expression of markers related to stemness is crucial for tumor maintenance and that these molecule also mediate resistance. In most cases, tumor recurrence is the result of a resistant CSC (intrinsic or acquired) in the primary tumor and its sphere formation7 and self-renewing abilities8 (Fig. 1). Based on the “seed and soil” theory, at a distant site, a resistant CSC can drive metastasis and then form a metastatic tumor9 (Fig. 1); thus, an increase in the CSC signature in tumors is associated with a worse prognosis.3
Based on the functional CSC concept, CSCs are naturally resistant to chemo- or radiotherapy, indicating they can survive after chemoradiotherapy and develop into a new cancer. However, understanding the properties of CSCs is the first step. The eventual goal is to investigate why CSCs can escape treatment, be retained, and form a new carcinoma.
Identification of CSCs
The classical definition of CSCs is a rare subpopulation of cells endowed with the capacity for self-renew and tumor-generating potential. Therefore, different methods have been developed and are currently exploited to isolate CSCs from patient-derived tumors or cancer cell lines in vitro. In this context, specific patterns of biomarkers that identify CSCs have been determined for some solid tumors such as CD44+CD24- for breast cancer (Table 1). However, increasing findings have showed that the previously defined CSC population is still heterogeneous, and thus, researchers must further enrich these cells by additional differentially expressed markers.
Table 1.
Identification of cancer stem cells in human cancer
| Classification | Markers | Function and role |
|---|---|---|
| Isolation markers | CD133 |
A common CSC marker in various cancers. CD133+Nestin+ is better for CSCs in glioma.15,16 CD133+CD44+ is better for CSCs in HCC19 and colon tumor.18 |
| CD44 |
A common CSC marker in various cancers. CD44+CD133+28/CD44+EpCAM+23/CD44+ALDH+19,29 is better for CSCs in CRC. CD44+c-Met+30 is better for CSCs in pancreatic cancer; |
|
| EpCAM |
A common CSC marker in various cancers. EpCAM+CD44+ CD166+23 is better for CSCs in CRC. EpCAM+ CD133+9 is better for CSCs in HCC |
|
| ALDH |
An enzyme that is identified as a common CSC marker in various cancers. ALDHhigh CD44+CD24-/ALDHhigh CD133+CD44+41 is better for CSCs in breast cancer. ALDH+CD133+42 is better for CSCs in HCC. |
|
| CD90 |
A common CSC marker in various cancers. CD90+CD44+49 is better for CSCs in lung cancer cell. |
|
| Intracellular markers | Oct4 |
Oct4 is a homeodomain transcription factor by binding to octamers, and regulates the expression of many genes.52 |
| Nanog |
Nanog is a homeobox transcription factor, And plays a crucial role in the second embryonic cell-fate specification.62 |
|
| Sox2 |
Sox2 has an important function in the early development and maintenance of undifferentiated ESCs. |
|
| SP fraction |
Hoechst 33342- negative population |
SP cells can be separated by fluorescence screening after the outflow of Hoechst 33342. And SP cells have high homology, self-renewal and multidirectional differentiation potential.74 |
| Noncoding RNAs | Circ008913 | Regulate CSC phenotype in nasopharyngeal carcinoma cell line.91 |
| CirGprc5a | Regulate CSC in bladder tumor.92 | |
| Circ001680 | Promote CSC in CRC and induce irinotecan resistance.93 | |
| CircLgr4 | Regulate CSC in CRC.94 | |
| LncTCF7 | Promote CSC by Wnt signaling pathway.95 | |
| Lnc-β-Catm | Promote CSC together with Wnt.96 | |
| H19 | Regulate CSC in breast cancer97 and HCC.98 |
Isolation markers
CSCs can be isolated through different biomarkers on the cell surface by fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting (MACS). Classical surface markers, such as CD133, CD44, epithelial cell adhesion molecule (EpCAM) and CD90 are extensively applied. However, some CSC surface markers are shared with normal stem cells. Therefore, multiple markers must be utilized for the accurate targeting of CSCs.
CD133
CD133 is a membrane-bound pentaspan glycoprotein first identified in neuroepithelial SCs in mice and later found in human tissues.10 CD133 was used as a CSC marker in a series of tumors. In 2004, Singh and colleagues11 identified CD133 as a CSC marker in brain TICs because injection of as few as 100 CD133+ cells produced a tumor that could be serially transplanted and was a phenocopy of the patient’s original tumor, whereas injection of 105 CD133− cells engrafted but did not result in a tumor. Then, CD133 was identified as a CSC marker in hepatocellular carcinoma (HCC),12 glioblastoma,13 colon tumors7 and ovarian cancers (OCs).14 However, CD133 alone cannot always indicate the CSC phenotype. Researchers have thus focused on investigating combined signatures. CD133 combined with Nestin may be an optimal CSC-specific marker in glioma patients.15,16 The combination of CD133 and CD44 was used to define a novel HCC subpopulation. CD133+CD44high xenografts, but not CD133+CD44−/low, CD133−CD44high or CD133−CD44−/low xenografts, produced intrahepatic or lung metastasis in nude mice.17 Similarly, Naotsugu Haraguchi’s team showed that the CD133+CD44+ population may identify TICs in human colon cancer.18 In 2009, aldehyde dehydrogenase (ALDH) was reported to contribute to the stemness of the CD133+CD44+ fraction in colon tumors.19 Using ALDH in combination with CD133 to analyze OC cell lines, Ines A Silva observed greater growth in ALDH+CD133+ cells than ALDH+CD133− cells, suggesting a further enrichment of ovarian CSCs in ALDH+CD133+ cells.20 More recently, research showed that double-positive L1 cell adhesion molecule (L1CAM)+CD133+ cells displayed higher spherogenic and clonogenic properties than L1CAM−CD133− cells in OC and indicated radiotherapy resistance.21
CD44
CD44, a nonkinase transmembrane glycoprotein, is thought to play a role in CSCs.22 As mentioned previously, CD44 was first used as a CSC marker in breast cancer.4 Then CD44 was identified as a CSC marker in colorectal cancer (CRC),23,24 pancreatic cancer,25 OC,26 gastric cancer,27 and others. In CRC, a recent meta-analysis suggested that the combination of CD44 and CD133 indicated an approximately sevenfold increase in the tumorigenic potential, while CD133 alone indicated 1.45-foldchange, and CD44 alone indicated twofold increase.28 Moreover, the ability to engraft in vivo in immunodeficient mice was restricted to a minority subpopulation of EpCAMhigh/CD44+ epithelial cells in CRC;23 furthermore, ALDH combined with either CD44 or CD133 could increase the tumor-initiating ability.19,29
Chenwei Li’s team30 showed that cells that expressed CD44 (0.5–5%) and c-Met showed a capacity for self-renewal and had the highest tumorigenic potential of all cell populations studied in pancreatic cancer.
In gastric CSCs, CD44+ gastric cancer cells showed self-renewal and the ability to form differentiated progeny and gave rise to CD44− cells.31 Phu Hung Nguyen showed that CD44 and ALDH are the most specific biomarkers to detect and isolate tumorigenic and chemoresistant gastric CSCs in non-cardia gastric carcinomas independent of the histologic classification of the tumor.32 CD44 is a cell surface transmembrane glycoprotein encoded by the CD44 gene, a 20-exon DNA segment,33 of which exons 1–5 and 6–20 are spliced together to form CD44s, the standard isoform. In addition, variant exons 6–15 can be alternatively spliced and assembled in different combinations with the standard exons to generate other variant (CD44v) isoforms. From this perspective, further research on gastric cancer found that CD44v8-10 but not CD44s increased the frequency of tumor initiation,34 which suggests a strategy to target CSCs in gastric cancer.
EpCAM
EpCAM is a transmembrane glycoprotein expressed on the surface of healthy epithelial cells.35 EpCAM is increasingly recognized as a specific CSC marker for various tumors such as breast cancer,4 colon cancer,36 HCC37 and pancreatic cancer.38 CRC originating from EpCAMhigh/CD44+ cells maintained a differentiated phenotype and reproduced the full morphologic and phenotypic heterogeneity of their parental lesions; moreover, CD166 could be an additional differentially expressed marker, for CSC isolation in CRC.23 Compared with EpCAM−/CD133−HCC cells, EpCAM+/CD133+ cells appear to be a CSC subpopulation in HCC.9
ALDH
ALDH detoxifies intracellular aldehydes through oxidation and may have a role in the differentiation of SCs through the oxidation of retinoic acid.39 In head and neck squamous cell carcinoma (HNSC), ALDH can be used as a single marker of CSCs.40 More often, ALDH is combined with other CSC markers: ALDHhighCD44+CD24- and ALDHhighCD44+CD133+ cells may be important mediators of breast CSCs.41 Further research was conducted and the results revealed the existence of a hierarchical organization in HCC cells with tumorigenic potential as follows: CD133+ALDH+ > CD133+ALDH− > CD133−ALDH−.42
CD90
CD90, a glycoprotein, also known as thymocyte differentiation antigen-1 (Thy-1), is a cell adhesion molecule and the smallest member of the immunoglobulin superfamily.43 CD90+ cells were found to be CSCs in HCC,44 as well as prostate cancer,45 insulinomas,46 OC,47 and could predict the response to sorafenib in patients.48 Co-expression with the additional surface marker, CD44, produced an even more aggressive phenotype, including a higher metastatic and self-renewal capacity, than that of the CD90+CD44− counterparts.49
Intracellular markers
The most important properties of CSCs are self-renewal and the ability to differentiate into one or two more specialized cell types.50 Oct4, Nanog and SOX2 are transcription factors that play essential roles during early embryonic development.50 On this basis, Shinya Yamanaka’s group found that several transcription factors (Oct4, Sox2, Klf4 and c-Myc) can convert a differentiated cell back to a pluripotent phenotype over the course of a few weeks, thus reprogramming the cells into induced pluripotent stem (iPS) cells.51 Upon expression of the reprogramming factors, some cells start to rapidly divide and quickly lose their differentiated cell characteristics with robust downregulation of somatic genes. These transcription factors can be re-expressed or reactivated in CSCs with the ability to self-renew and differentiate.50 In theory, the ideal CSC markers are those that are required to maintain their stemness features. Therefore, Oct4, Sox2 and Nanog can be considered as CSC markers.
Oct4
Oct4 functions as a homeodomain transcription factor by binding to octamers,52 which regulate the expression of many genes. Oct4 was expressed early in the preimplantation embryo and thus regulated early events of murine development.53,54 Oct4 was first shown to be associated with cancer by M Monk in 2001.55 In 2005, based on CSCs in breast cancer, Ponti and colleagues56 found that the CD44+CD24- fraction expressed Oct4 and gave rise to new tumors. Oct4high cells have more SC-like traits, such as self-renewal, chemoresistance and xenograft tumorigenicity, than Oct4low cells.57 To date, Oct4 has been used to isolate CSCs by Oct4 promoter-mediated activity in breast cancer,58 non-small cell lung cancer (NSCLC),59 gastric cancer60 and HCC.61
Nanog
Nanog, a homeobox transcription factor, plays a crucial role in the second embryonic cell-fate specification.62 This molecule is required for the maintenance of pluripotency but absent from differentiated cells.63,64 In 2004, Kristian Almstrup and colleagues65 revealed embryonic SC-like features of testicular carcinoma in situ by genome-wide expression profiling: Nanog was upregulated during progression to embryonic carcinoma. In 2010, the expression of Nanog was directly correlated with CSCs (CD133high/CD44high),66 and Nanog+ NSCLC cells were shown to exihibit CSC properties.58 Furthermore, the status of Nanog determines the switch between cancer cells and CSCs.67 Moreover, Nanog expression was associated with enhanced ALDH activity and cellular radioresistance68 and chemoreistance.69
Sox2
Sox2 belongs to the family of high-mobility group transcription factors and has an important function in the early development and maintenance of undifferentiated ESCs. Sox2 is commonly used as a stemness-associated marker in CSC research. Increased expression of Sox2 was observed in CD133+ NSCLC cells70 and ALDHhigh cells.71 Zhu and colleagues72 showed that Sox2 is a marker for CSCs in bladder cancer. In a reporter system (SORE6), which allows the monitoring of viable cells expressing Sox2 and/or Oct4, SORE6+ cells were found to be significantly more tumorigenic than SORE6- cells.60,73
Side population (SP) cells
SP cells were discovered in 1996 by M A Goodell in hematopoietic stem cells (HSCs): SP cells were not stained by Hoechst 33342.74 SP fractions were shown to protect recipients from lethal irradiation at low cell doses, and to contribute to both lymphoid and myeloid lineages.74 SP cells exhibit a low Hoechst 33342 staining pattern because of the high expression of ATP-binding cassette transporters (ABC transporters), especially ABCG2.75 Therefore, the ABCG2 transporter is an efficient Hoechst 33342 efflux pump. Moreover, ABCG2 is preferentially expressed by immature human hematopoietic progenitors.76 Transplantable HSCs in human fetal liver have an SP phenotype.77 Therefore, the SP fraction might indicate a stemness phenotype. Research on AML showed that SP identifies a CD34+CD38- progenitor cells.78 Then, the SP fraction was used to identify CSCs in solid tumors: Lubna Patrswala’s team first identified the SP fraction in human cancer cells and SP cells were shown to possess some intrinsic SC properties.79
Another more recent study showed that P-gp pump function was required for amplification of both phenotypically defined SP cells and functionally defined repopulating cells.80 In 2006, SP cells were detected in HCC cells, and the SP fraction presented a CSC phenotype.81 Then, the SP fraction was identified in a human nasopharyngeal carcinoma cell line,82 OC,83 brain tumor,84 lung cancer,85 especially for CSCs with unknown cell surface markers. Currently, SP analysis is increasingly applied in CSCs research as an indication of stemness86–88 and therapy resistance.89,90
Noncoding RNAs
In recent years, research on noncoding RNAs has become increasingly prominent. Many studies have indicated noncoding RNA can be a CSC marker. Circ008913 was reported to be involved in CSC-like properties.91 CircGprc5a regulated CSCs in bladder tumors.92 Circ001680 could enhance the CSC population in CRC and induce therapeutic resistance to irinotecan.93 Moreover, CircLgr4 knockdown inhibited colorectal CSC self-renewal, while CircLgr4 overexpression had the opposite effects.94 In addition, several long noncoding RNA (lncRNA)-based regulatory circuits that promote CSCs formation highlight the importance of lncRNAs in CSCs: lncTCF7 promotes CSCs through activation of Wnt signaling,95 lnc-β-Catm together with Wnt is required for self-renewal of CSCs,96 H19 is associated with CSCs in breast cancer97 as well as HCC,98 and H19 facilitates angiogenesis by an exosome-mediated mechanism in CSC-like cells.98
Overall, although surface markers, SC-specific transcription factors, the SP fraction and noncoding RNAs can be used as CSC markers, the identification and isolation of CSCs in clinical specimens is challenging. The extent to which these marker-identified populations are actual CSCs remains unclear. Moreover, CSC-defining molecules have functions in addition to the roles as markers. Accumulating evidence has shown that these molecules could have specific biological functions in tumor initiation and progression. CD44 is preferentially involved in invasion, adhesion and metastasis, and CD133 tends to be involved in the maintenance of the CSC population. Therefore, instead of solely examing the marker expression in CSCs, we hope to specifically clarify the biological roles played by these markers and the regulatory mechanisms through novel technologies such as live-cell RNA detection and single-cell DNA and RNA sequencing methods.
Resistance mechanisms of CSCs
Accumulating evidence shows that the CSCs are critically associated with drug resistance: ionizing radiation induces the upregulation of CD133+ CSCs in glioblastoma xenografts99 and CSCs are enriched in breast cancer after radiation therapy.100 Moreover, increasing evidence has demonstrated that acquired resistance to one specific drug can result in cross-resistance to other chemotherapeutics.101–103 CSCs can resist therapy mainly because they express multidrug resistance (MDR) transporters and display a more active DNA repair capacity and induce more apoptotic arrest than other cells.104 Therefore, researches should focus on CSCs to reverse therapy resistance (Fig. 2).
Fig. 2.
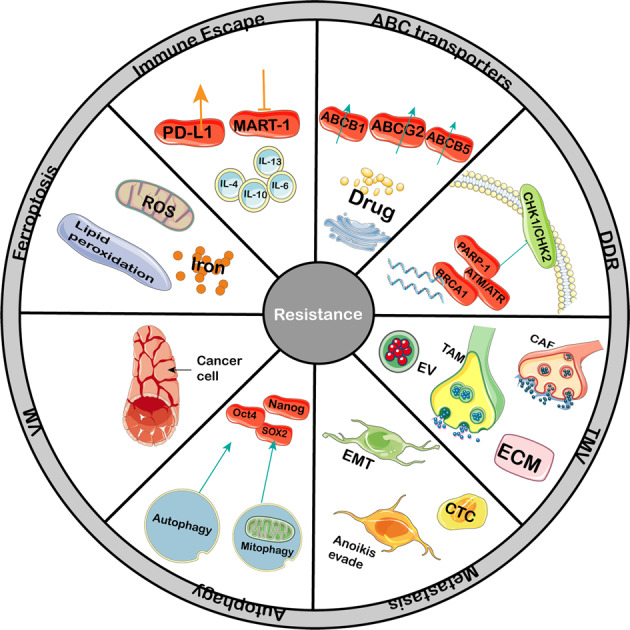
Mechanisms mediating the resistance of CSCs to scores of therapy. Multiple intrinsic and extrinsic resistant mechanisms controlling CSC respond to traditional or novel therapy. In the process or after therapy, CSCs display several properties: higher degree of drug efflux activity, active DNA repair, high ROS levels, the tendency of VM; moreover, non-CSCs may reacquire CSC properties by EMT, microenvironment, autophagy and extracellular vesicles also contribute to tumor relapse
Increased drug efflux activity of CSCs
ABC transporters can export wide range of toxic substrates from cells105 and thus directly contribute to the acquisition of resistance and CSCs exhibit increased ABC transporter expression.106 CSC-mediated drug resistance is supported by the hypothesis that the SP fraction can be identified as CSCs. The number of ABC transporters was shown to be correlated with maturation state: cells that exhibit the greatest efflux activity are the most primitive.76,107 ABCG2 was the first ABC transporter reported to determine the SP phenotype.75 A more comprehensive study was conducted in 2001 and the results indicated that ABCG2 was a determinant of the SP phenotype and could be a marker for SCs from various sources.75 Various factors such as glutamine,108 DNA methyltransferase activity109 and hypoxia-inducible factor (HIF)110 can regulate the SP population by controlling ABCG2. However, the role of ABCG2 in the SP fraction is controversial: the ABCG2+ population did not show significant drug resistance compared with the ABCG2- population. Moreover, ABCG2- cells exhibited higher sphere formation than ABCG2+ cells,111 which is consistent with the findings in a study published more than 10 years ago.112 Patrawala indicated that ABCG2+ cancer cells can generate ABCG2- cells and ABCG2- cancer cells can also generate ABCG2+ cells.112 Another study showed that ABCG2 activity was not responsible for the stem-like phenotypes of CSCs.113 At present, the more pertinent conclusion is that the SP fraction is composed of heterogeneous cell populations. ABCG2 expression mainly identifies fast-cycling tumor progenitors, and the ABCG2- population contains primitive stem-like cancer cells in the SP fraction. Other subtypes of the ABC transporter family also contributed to CSC-mediated chemoresistance: ABCB5 was colabeled with CD133114 and CD44115 and clinically correlated with chemoresistance.115 Furthermore, ABCB5 controls chemoresistant and ABCB5 blockade-induced cellular differentiation,116 which is possibly mediated by a cell cycle checkpoint mechanism.117 ABCB1 is another important ABC transporter contributing to the chemoresistance-phenotype of CSCs118 by PKC/PI3K/Akt.119
Although specific ABC transporters are inhibited, cancer cells display an MDR phenotype. Tepotinib significantly reversed ABCB1-mediated MDR but not ABCC1-or ABCG2-mediated MDR.120 While this phenotype can protect cells from cytotoxic agents, MDR genes are sensitive to drugs such as doxorubicin,121 motixantrone.122 Currently, (i) the expression of multiple ABC transporters in CSCs can compensate the effect of the inhibition of a single ABC transporter, (ii) the key regulatory SP-related pathways that modulate ABC transporter expression are poorly understood, and (iii) limited research on and comprehension of the characteristic of the SP fraction are the main reasons for clinical failure.
Enhanced capability of DNA repair in CSCs
The hypothesis that therapy resistance is due to increased levels of ABC transporters cannot explain the enhanced therapy resistance of CSCs. Cancer cells show decreased DNA repair and thus display many mutations and genomic instability, ultimately resulting in apoptosis after multiple therapies. CSCs have highly active DNA repair mechanism, which results in effective DNA protection.123–125
In addition to chemotherapy drugs causing DNA damage, radiotherapy can induce DNA damage. In short, radiotherapy induced DNA damage, directly or indirectly, via the production of water-derived radicals and reactive oxygen species (ROS), which then interact with macromolecules, including DNA, lipids and proteins. Then, the DNA repair machinery is induced. Distinct from normal cancer cells, CSCs have both lower ROS levels and enhanced DNA damage repair.
CSCs have low levels of ROS due to increases in ROS scavengers to reduce ROS-induced DNA damage and apoptosis,126,127 and the ROS scavenger N-acetylcysteine (NAC) restored the CSC phenotypes.128 Salinomycin, a compound that can selectively eradicate CSCs, could target the CD44+CD24- fraction and upregulate ROS levels.129 NRF2 silencing reversed the ability of CD44+ cells to retain high levels of ROS and the sensitivity to anticancer drugs.130 Depletion of glutamine decreased the proportion of SP cells by increasing the intracellular ROS levels;131 glycolysis promoted the expression of doublecortin-like kinase 1 (DCLK1) and maintained the CSC phenotypes via maintenance of low ROS levels in gemcitabine-resistant pancreatic cancer cells.128 However, Lee et al.132 revealed that myelocytomatosis oncogene (MYC) and myeloid cell leukemia-1 (MCL1) cooperate to maintain chemotherapy resistance of CSCs by increasing ROS production and HIF-1α expression, which might be explained by the independence of the apoptosis domain. Inhibition of HIF-1α blocked CSC expansion and restored the sensitivity to chemotherapy.
The key sensors of DNA damage are the ataxia telangiectasia mutated (ATM) and ataxia telangiectasia mutated-RAD3-related (ATR) protein kinases. Upon DNA damage, ATM and ATR kinases form complexes with poly ADP-ribose polymerase (PARP-1) and breast cancer 1 (BRCA1) to phosphorylate checkpoint kinase 1 (CHK1) and CHK2, which subsequently drive the activation of targeted proteins, inducing DNA repair. CHK-associated contributors lead to therapy resistance of CSCs: a clinical cohort indicated that CHK1 phosphorylated at serine 345 is a predictor of radioresistance in breast cancer.133 Wang demonstrated that the c-MYC-CHK1/CHK2 axis regulates the DNA damage-checkpoint response and CSC characteristics, resulting in radiotherapy resistance.134 Srivastava found that enhanced expression of DNA polymerase contributes to cisplatin resistance in ovarian CSCs.135 In contrast, pharmacological inhibition of the DNA damage checkpoints CHK1 and CHK2 sensitized CSCs to radiotherapy.99 Furthermore, CSCs can develop high drug resistance through regulating their cell cycle. During chemotherapy, the cell cycle of CSCs slows down and the cells fall into a “quiescent” state. In this state, protective mechanisms can be initiated by the DNA damage repair system. When the DNA damage repair finished, CSCs resume tumorigenesis and thereby escape apoptosis.
Despite the interest and investment of pharmaceutical companies in the development of treatments that prevent DNA repair in cancer cells, the results tend to be worse than expected. The novel targets proposed herein proposed provide inspiration.
Strong correlation od CSCs with metastasis
Metastasis is a complex cascade of events including tumor cell conversion into mobile tumor cells, invasion into blood vessels, survival in circulation, attachment to endothelial cells in vessels, extravasation and finally colonization and growth in the host organ. During epithelial mesenchymal transition (EMT), epithelial cancer cells lose their polarity and cell-cell contacts, generating a mesenchymal phenotype with migratory and invasive characteristics. Although EMT is present in most cancer cells and not specific to CSCs, mobile CSCs might derive from stationary CSCs through the acquisition of a transient EMT phenotype except stemness. Paget compared metastatic cancer cells to “seeds” that, once released from the plant (primary tumor), can spread, survive and proliferate when on “congenial soil.”136 Although the metastatic process is considered highly inefficient because only a cluster of cancer cells can drive metastasis following their transplantation into immunodeficient mice,137 which is consistent with the low percentage of CSCs in cancer, the self-renewal and differentiation of CSCs play an important role: genome sequencing showed metastatic clones are genetically evolved from the original cells.138
Therefore, acquiring metastatic characteristics is the first step that may be mediated by EMT in CSCs. A clinical analysis of β-catenin+ cancer cells (strong indication of stemness) that coexpressed E-cadherin and vimentin in core-needle biopsies from patients with various advanced metastatic carcinomas showed a significant association among CSCs, EMT and metastasis.139 Recent studies have shown that, several signaling pathways and molecules play an important role in this process. SOX8 bound to the promoter region of Frizzled 7 (FZD7) and mediated EMT processes in chemoresistant tongue squamous cell carcinoma (TSCC) via the FZD7/Wnt/β-catenin pathway.140 Moreover, extracellular matrix 1 (ECM1) regulated Wnt-mediated EMT by increasing the association between β-catenin and MUC1 cytoplasmic tail.141 Twist1 is a basic helix-loop-helix transcription factor that potently drives the EMT process. Metadherin (MTDH) indirectly activates Twist1 expression by facilitating histone H3 acetylation on the Twist1 promoter, a process mediated by the histone acetyltransferase cAMP response element-binding protein-binding protein (CBP), resulting in CSC traits and drug resistance.142 Moreover, S-phase protein kinase 2 (Skp2) regulates castration-resistant prostate cancer through Twist-mediated EMT and CSC acquisition. Skp2 interacts with Twist and promotes the nondegradative ubiquitination of Twist. Consequently, Skp2 stabilizes Twist protein expression by preventing proteasomal degradation of Twist by β-TrCP.143 Zinc-finger E-box-binding homeobox-2 (EZB2) is an EMT inducing transcription factor. The FBXW7-ZEB2 axis links EMT and the tumor microenvironment (TMV) to promote colorectal CSCs and chemoresistance.144
The EMT provides a basis for CSCs with unique tendencies, which allows them to be better leveraged for treatments that are more strategic than treatments employing non-CSCs. So we expect that a successful CSCs therapy might be achieved by preventing the induction of EMT, selectively killing CSCs during the EMT process or pharmacologically inducing the reversal process (EMT to MET).
After metastasizing and infiltrating into the surrounding parenchyma, tumor cells enter the blood circulation, where they can induce anoikis.145 CSCs can evade anoikis and enter the circulation to reach distant target organs. Grillet and colleagues146 reported that circulating tumor cells (CTCs) from patients with CRC displayed CSC hallmarks in ex vivo culture. Moreover, a transgenic mouse model demonstrated that CTCs returned to the primary tumor and generated new tumors with enhanced tumorigenic capacity.147 Mechanistic investigations demonstrated that overexpression of stromal-derived factor-1γ (SDF-1γ or CXCL12γ) induced CSC phenotypes in prostate cancer cells through CXCR4-mediated PKCα/NFκB signaling148 and Wnt signaling,147 which promoted tumor outgrowth, metastasis and chemoresistance in vivo. For invasion of anoikis, androgen receptor (AR) may be important in CSCs: AR maintains a CSC-like tumor-initiating population and serves as an antiapoptotic factor, facilitating anchorage independence and metastasis149 and constant ubiquitination and degradation of AR by MDM2 conserves the CSC integrity.150
Finally, a single CSC could not survive alone after anoikis; other CSCs need to be preserved in the niche. A key factor that modulates the microenvironment and CSCs resulting in drug resistance is hypoxia.151–153 Hypoxia activates multiple signaling pathways by activating hypoxia-inducible factor-1α and 2α (HIF1α, HIF2α) or phosphatidylinositol 3-kinase (PI3K/AKT), which bind to promoters containing the hypoxia-response element (HRE) and then regulate gene expression. As a feedback loop, activation of the PI3K/ATK pathway promotes CSCs by activating HIF1α and HIF2α.154 The cascade of activation leads to the induction of stemness and self-renewal, which results in secondary tumors.
The development of new drugs targeting EMT program could have a significant impact on the CSC therapy field. However, effective targeting of CSCs still faces a variety of challenges, as the mechanism that regulates the retention or induction of EMT programs in CSCs remains unclear. Overcoming these challenges will require that the risk of resistance be minimized, but a successful therapeutic strategy will eventually open the door for curing cancer by targeting CSCs.
Vasculogenic mimicry (VM) in CSCs
VM is a newly defined pattern of tumor microvascularization that is different from angiogenesis and vasculogenesis and lacks the participation of endothelial cells, by which highly aggressive tumor cells can form vessel-like structures due to their high plasticity. VM channels provide a functional blood supply in malignant tumors and mediate therapy resistance. Increasing studies have found that CSCs directly line VM channels and provide VM-related molecules to enhance VM formation.155 Rates of CD133, ALDH, and VM were positively associated with lymph node metastasis, distant metastasis, Enneking stages, and overall survival of patients.156 A similar phenomenon was found: VM formation was associated with altered CSC-associated proteins,157 and CSCs directly line VM channels. Additionally, CSCs provide VM-related molecules to promote VM formation.155 CSCs can form VM-mediated resistance and acquire resistance to antiangiogenic therapy.158,159
Increased autophagic activity in CSCs
Autophagy is an evolutionarily conserved physiological process in cells that generates intracellular nutrients, growth factors and energy to support cell survival and cellular activities during stress, such as nutrition deprivation, hypoxia or ischemia.160,161 Autophagy was upregulated in CD133+ cells, and promoted resistance to photodynamic therapy (PDT).162 Moreover, CD44+CD117+ ovarian CSCs presented higher basal autophagy than their nonstem cell counterparts. Inhibiting autophagy could reduce chemoresistance in CSCs.163 The differential regulation of autophagy is a molecular link between the differing chemosensitivity of CSCs and differentiated cancer cells,164 consistent with the fact that ATG7 or ATG12 KD could decrease the pluripotency and promote the differentiation and/or senescence of CSCs.165 Some studies have revealed the mechanism by which autophagy facilitates the degradation of Sox2.166 Moreover, mitophagy could regulate the binding of the Nanog promoter to PINK1 via p53.167 Furthermore, ATG7 facilitated the transcription of Oct4 via β-catenin, which binds to the Oct4 promoter.168 TARBP2 was reported to be destabilized through autophagic-lysosomal proteolysis, thereby stabilizing the expression of Nanog.69
Overall, autophagy is an important mechanism activated by CSCs to increase their resistance to therapy. Autophagic inhibitors might decrease the stemness properties and reverse therapy resistance.
Decreased ferroptosis in CSCs
Ferroptosis is a recently described form of cell death that is distinct from other known cell death pathways.169 However, the precise mechanism of ferroptotic cell death is still unclear. Iron, ROS and lipid peroxidation are critical mediators of ferroptosis.170,171 The ability of iron to cycle between oxidized and reduced forms contributes to the formation of free radicals, and an excess of free radicals leads to lipid peroxidation, increased ROS and oxidative stress, and DNA damage.
CSCs are generally characterized by a high intracellular iron content.172 Iron addiction could be a therapeutic target in CSCs and could reverse therapy resistance.173 A forced reduction in intracellular iron reduced the proliferation of CSCs in OC. Moreover, CD44 expression suppressed ferroptosis in cancer cells, which indicated a correlation between CSCs and ferroptosis.174 Inhibition of autophagy increased the susceptibility of glioblastoma SCs to temozolomide by initiating ferroptosis.175 Furthermore, inducing ferroptosis could sensitize CSCs to chemotherapy in OC.176 In addition, as we mentioned before, salinomycin is a selective agent against CSCs and triggers ferritin degradation and ROS-mediated ferroptosis in CSCs,177 which may reverse the radiotherapy resistance caused by low ROS levels and enhanced DNA repair in CSCs.
However, few studies directly focusing on ferroptosis and CSC-mediated therapy resistance are available. Based on the association among ferroptosis and CSCs, CSC renewal and therapy resistance, further research should investigate the regulatory mechanisms of Oct4, Nanog and Sox2 by iron, ROS and lipid peroxidation, at both the transcriptional and translational levels.
Favorable TMV in the CSC niche
Accumulating evidence suggests that the TMV plays a crucial role in CSC development and is a potential target for therapy resistance. Extracellular vesicles (EVs), carcinoma-associated fibroblasts (CAFs), tumor-associated macrophages (TAMs) and chemokines in the CSC niche have important roles. Chemotherapy-induced EVs promote CSC traits and therapy resistance.178 The presence of Cav-1 in EVs acts as a potent driver to induce CSC phenotypes and can induce radio- and chemoresistance in recipient cells.179 Unfortunately, exosomes derived from gemcitabine-resistant pancreatic CSCs mediate the horizontal transfer of drug-resistant traits to gemcitabine-sensitive pancreatic cancer cells.180 Lysine-specific demethylase 1 (LSD1) expression was increased in CAFs as an upstream driver of Notch3-mediated CSC self-renewal.181 ZEB2-mediated induction of EMT was associated with stromal factors secreted from CAFs, which induced chemotherapy resistance.144 Furthermore, a specific subset of CAFs, CD10+GPR77+ CAFs promotes tumor formation and chemoresistance by providing a niche for survival of CSCs. Mechanistically, CD10+GPR77+ CAFs are driven by persistent NF-kB activation via p65 phosphorylation and acetylation.182 Although studies on TAMs are limited, Masahisa reported that TAMs produce milk fat globule epidermal growth factor 8 (MFG-E8), and MFG-E8 mainly activates Shh and further amplifies its anticancer drug resistance.183 Interestingly, the extracellular matrix (ECM) is also an integral part of the CSC niche that mediates therapy resistance. Hyaluronic acid in the ECM is a ligand for the CD44 receptor and can affect CSC stemness along with the response to differentiation therapy.184 Another component of the ECM, laminin-332, could reduce cell mitosis, present resistance to doxorubicin and sorafenib treatment, and increase the SP fraction.185
Therefore, crosstalk occurring in the TMV can expedite and confer resistance of CSCs to radiotherapy and chemotherapy. Elucidation of the content of the CSC niche would provide us with valuable information to design therapeutic targets, e.g., exosome-like nanovesicles could be used to specifically target CSCs.
Immune escape in CSCs
Immunotherapy has recently attracted global attention and has emerged as the “new hope” for cancer treatment. However, CSCs have developed many strategies to circumvent immune attack and maintain the immune-resistant phenotype.
PD-L1, a T-cell inhibitor expressed on T cells, B cells, and natural killer cells, can eliminate tumor cells.186,187 PD-L1 expression was substantially increased in chemoresistant CRC through the PI3K/Akt and MEK/ERK pathways;188 conversely, CSCs showed a low-immunogenic profile: ABCB5+ melanoma cells did not express the immunogenic differentiation antigen MART-1 at significant levels,189 nor did they express cancer testis antigens.190 Moreover, loss of the tumor suppressor PTEN in CSCs led to reduced expression of neoantigens that demonstrate strong immune reactivity and was thus associated with resistance to anti-PD-1 checkpoint blockade therapy.191 Furthermore, STAT3 signaling can functionally render CSCs immunosuppressive as inhibition of STAT3 can restore T-cell function.192 In addition, CSCs isolated from various solid tumors have been shown to release various cytokines and soluble immunosuppressive factors such as IL-4, IL-6, IL-10, and IL-13.193
Signaling pathways of therapy resistance in CSCs
Various signaling pathways are involved in therapy resistance of CSCs. Some of the most important and well- characterized signaling pathways include Hedgehog(Shh), Wnt/β-catenin, Notch and NF-κB pathways.
Sonic Hedgehog pathway
The Sonic Hedgehog (Shh) pathway was initially identified in the fruit fly and has an important role in embryonic development. Binding of the Hh ligand to its receptor Patched (PTCH) enables Smoothened (SMO)-mediated translocation of glioma-associated protein 1/2(Gli1/2) to the cell nucleus to drive the transcription of Shh target genes194 (Fig. 3). Shh regulates the proliferation, differentiation and migration of target cells in a spatial, temporal and concentration- dependent manner.195 Enhanced Hedgehog activation can increase proliferation-associated genes: cyclin D1, cyclin D2, N-Myc, Hes1 and Igf-2.195 Shh is related to chemoresistance.9 Shh signaling regulates the ABCG2 efflux pump196 along with ALDH activity144,197 and reverses epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) resistance. Furthermore, simultaneously inhibiting the Shh pathway could kill imatinib-sensitive or -resistant BCR-ABL+ cells.198 Moreover, Shh signaling maintained CSC phenotypes and malignant transformation phenotypes in CD44+ gastric cancer cells, and Shh inhibition could reverse chemotherapy resistance in CD44+ cells.199 LncRNA-cCSC1 activates the Shh signaling pathway and regulates the expression of CD44 and CD133.200 In contrast, miR-200b and let-7c significantly diminished Shh-mediated-erlotinib resistance in CSCs.201
Fig. 3.
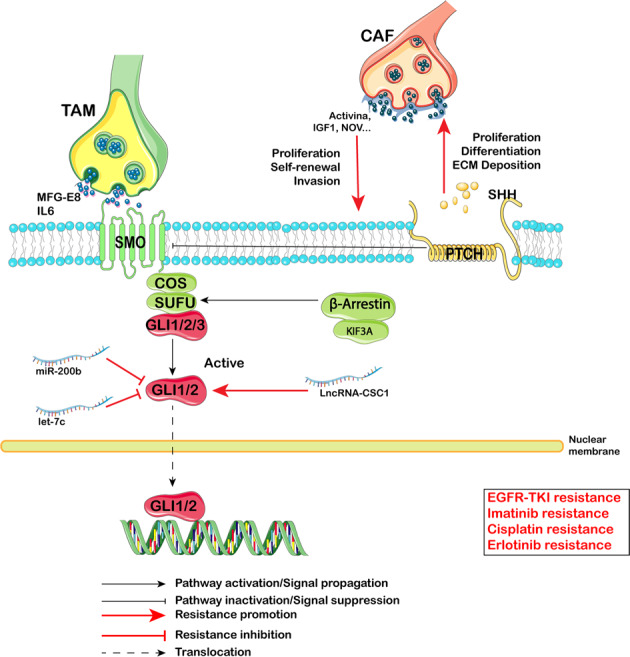
Hedgehog (Hh) signaling pathway-mediated therapy resistance in CSCs. Binding of the Hh ligand to its receptor Patched (PTCH) enables Smoothened (SMO)-mediated translocation of glioma-associated protein 1/2(Gli1/2) to the cell nucleus to drive the transcription of Shh target genes. During activation of the Hh pathway, some proteins (IL-6, MFG-E8), microRNA (miR-200b, let-7c) and the long noncoding RNA LncRNA-CSC1 are involved in the Hedgehog pathway to regulate EGFR-TKI resistance, Imatinib resistance, Cisplatin resistance, and Erlotinib resistance
From the perspective of the TMV, TAMs and CAFs contribute to Shh-mediated therapy resistance: TAMs produce MFG-E8, and MFG-E8 mainly activates Shh and further amplifies its anticancer drug resistance.183 Moreover, Shh participates in an intracellular signaling module that synergistically regulates CAFs and CSCs to mediate therapy resisitance.202
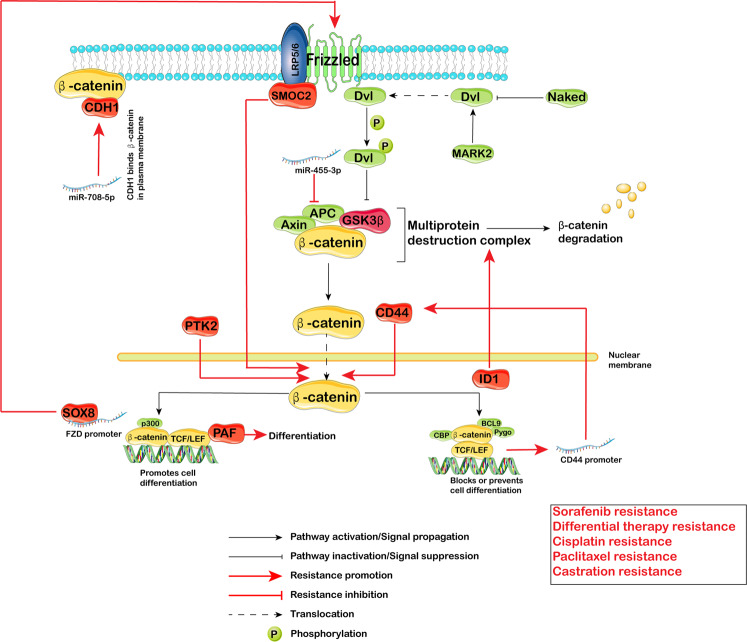
Wnt/β-catenin pathway
Wnt/β-catenin signaling plays a crucial role during embryogenesis. In general, the Wnt signaling pathway can be divided into canonical Wnt signaling (through the FZD-LRP5/6 receptor complex, leading to depression of β-catenin) and noncanonical Wnt signaling. Canonical Wnt signaling is the best-known (Fig. 4). In the absence of Wnt signaling, β-catenin is bound to the Axin complex, which contains APC and GSK3β and is, phosphorylated, leading to ubiquitination and proteasomal degradation through the β-Trcp pathway. In the presence of Wnt signaling, the binding of LRP5/6 and FZD inhibits the activity of the Axin complex and the phosphorylation of β-catenin, allowing β-catenin to enter the nucleus, and then bind to TEF/TCF to form a complex, which then recruits cofactors to initiate downstream gene expression.
Fig. 4.
Wnt/β-Catenin signaling pathway-mediated therapy resistance in CSCs. In the absence of Wnt signaling, β-catenin is bound to the Axin complex, which contains APC and GSK3β and is, phosphorylated, leading to ubiquitination and proteasomal degradation through the β-Trcp pathway. In the presence of Wnt signaling, the binding of LRP5/6 and FZD inhibits the activity of the Axin complex and the phosphorylation of β-catenin, allowing β-catenin to enter the nucleus, and then bind to TEF/TCF to form a complex, which then recruits cofactors to initiate downstream gene expression. Several proteins (CDH1, SMOC2, SOX8, PAF, PTK2, CD44 and ID1) along with miR-708-5p regulate Sorafenib resistance, Differential therapy resistance, Cisplatin resistance, Paclitaxel resistance and Castration resistance
The Wnt/β-catenin pathway regulates CSC-mediated therapy resistance: PTK2 promoter hypomethylation induces PTK2 overexpression and activates Wnt signaling, leading to the CSC phenotype and sorafenib resistance in HCC.203 MiR-708-5p could inhibit CSCs by repressing the Wnt pathway through promotion of CDH1 to bind β-catenin in the plasma membrane, resulting in loss of the release of β-catenin.204 PAF could induce the differentiation and lose of stemness of CSCs by binding β-catenin in a chemoresistance model.205 A more direct association between differentiation and therapy resistance was identified by Xiong Jin: ID1, which is important in lineage differentiation, could sensitize glioma CSCs to differentiation therapy by inhibiting β-catenin degradation,206 similar to the function of miR-455-3p.207 Moreover, SMOC-2 could activate Wnt by binding FZD6 and LRP6, resulting in paclitaxel resistance and cisplatin resistance.208 Furthermore, Wnt signaling is a key pathway regulating the well-known SC marker CD44 by directly interacting with the promoter and presenting castration resistance.209 Combined with the research conducted by Souvick Roy,210 these findings indicate a positive feedback mechanism between CD44 and Wnt: CD44 binds to β-catenin and activates Wnt, resulting in cispatin resistance. In addition, the transcription factor Sox8 was reported to promote the Wnt/β-catenin pathway by binding to the promoter of FZD7, eventually leading to cisplatin resistance.140
Notch pathway
DLL1, DLL3 and DLL4, and Jagged ligands (JAG1 and JAG2) expressed on the cell surface can induce signaling in adjacent cells expressing their cognate receptors Notch1–4. Ligand binding promotes sequential cleavage of the Notch receptors by ADAM/TACE enzymes (S2 cleavage) and then γ-secretase (S3 cleavage), resulting in release the NICD, which interacts with transcriptional regulators in the nucleus to induce a Notch gene-expression profile (Fig. 5). Notch target genes, in turn, regulate pivotal cell-fate choices, including differentiation, cell cycle progression, and survival.211
Fig. 5.
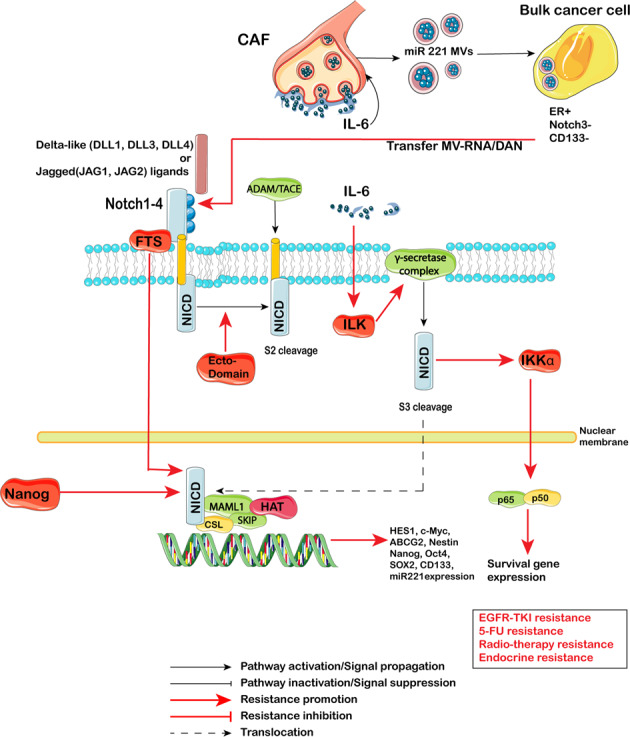
Notch signaling pathway-mediated therapy resistance in CSCs. DLL1, DLL3 and DLL4, and Jagged ligands (JAG1 and JAG2) expressed on the cell surface can induce signaling in adjacent cells expressing their cognate receptors Notch1–4. Ligand binding promotes sequential cleavage of the Notch receptors by ADAM/TACE enzymes (S2 cleavage) and then γ-secretase (S3 cleavage), resulting in release the NICD, which interacts with transcriptional regulators in the nucleus to induce a Notch gene-expression profile. Some proteins (FTS, ILK, Nanog) along with CAF and EVs regulate EGFR-TKI resistance, 5-FU resistance, Radio-therapy resistance and Endocrine resistance
CSCs activate the Notch pathway to promote of resistance to chemotherapy212 and radiation.213 Inhibiting Notch signaling could sensitize CSCs to cisplatin or carboplatin214 and radiation therapy.213 Nanog regulated Notch signaling along with ALDH activity and radiotherapy resistance in breast cancer.68 Moreover, crosstalk between Notch and NF-κB contributed to therapy resistance in triple-negative breast cancer (TNBC):215 Jagged1 triggers nuclear, NF-κB-dependent transcription of antiapoptotic gene cIAP-2. Furthermore, extracellular signals can regulate Notch. En-Chi Hsu reported the indispensable role of ILK in regulating IL-6-induced Notch1 activation and CSC expansion through γ-secretase assembly at the caveolae.216 In addition, inhibiting ADAM-17, a major component of Notch signaling, by Nectin-4, could partially reserve 5-FU resistance.217 FTS could bind with Notch1 and then activate Notch signaling and upregulate Nanog, Oct4 and Sox2 expression, which contributed to radiotherapy resistance.218 TMV also participated in regulation of the CSC phenotype regulation. Stroma microvesicles mediated CSC evolution in endocrine resistant metastatic breast cancer.219 Autocrine IL-6/Stat3 signaling induces the proliferation of CAFs and the biogenesis of onco-miR221/222+ MVs; these MVs are taken up by estrogen receptor+ (ER+) breast cancer cells and lead to the potent suppression of ER signaling, resulting in Notch3 upregulation, which in turn sustains the self-renewal of CD133+ CSCs in an ER-independent manner.
NF-κB pathway
The NF-κB pathway mediates acute and chronic inflammation in tumors through the association of inflammation with stemness;220 it plays a crucial role in tumor biology and regulates key processes during the initiation and progression of various carcinomas.221,222 The main physiological component of NF-κB is the p50-p65 dimer. The active p50-p65 dimer is further activated by post-translational modification and transported into the nucleus, inducing the expression of target genes in combination with other transcription factors (Fig. 6).
Fig. 6.
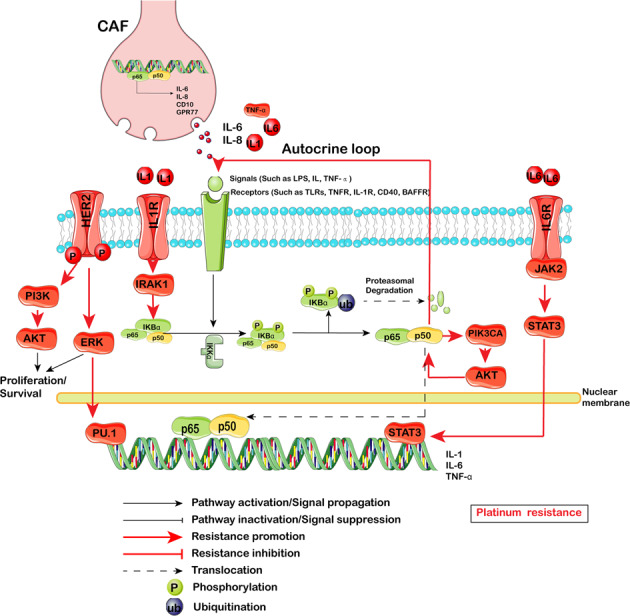
NF-κB signaling pathway-mediated therapy resistance in CSCs. The main physiological component of NF-κB is the p50-p65 dimer. The active p50-p65 dimer is further activated by post-translational modification and transported into the nucleus, inducing the expression of target genes in combination with other transcription factors. PI3K/Akt, ERK, IRAK1, Jak2/STAT3 and CAF regulate Platinum resistance
More recently, NF-κB signaling was found to be preferentially activated in CSCs.223,224 Salinomycin, an inhibitor of NF-κB, could induce apoptosis in cisplatin-resistant OC.225 Furthermore, NF-κB participates in the progression of EMT: Twist2 transcriptionally enhances NF-κB activation, and NF-κB upregulates Twist2 expression, thereby forming a positive feedback loop that activates EMT and enhances CSC-like properties.226 Moreover, NF-κB activated hypoxia related stemness signaling,227 and revertes ROS-induced apoptotic cell death in CSCs. In addition, a model of HER2-induced sequential activation of the IL-1α and IL-6 signaling pathways was supported by the following evidence: (i) HER2 upregulates IL-1α expression via MAPK-mediated activation of the PU.1 transcription factor; (ii) secreted IL-1α binds to its receptor and activates NF-κB, which subsequently binds to and activates the IL1A and IL6 promoters via a feedback mechanism; (iii) secreted IL-6 binds to its receptor and activates the downstream STAT3 transcription factor.228 The HER2/NF-κB model could promote tumorigenesis and chemotherapy resistance. Another autocrine loop was reported by Bhushan Thakur: cisplatin mediated NF-κB activation only in CSCs, which in turn activated the bimodal feedback loop of NF-κB-TNFα and NF-κB-PIK3CA. On the one hand, this mechanism promotes an autocrine loop by activating TNFα-NF-κB in CSCs, and on the other hand, it increases PIK3CA and PI3K/AKT signaling thus leading to NF-κB stabilization. Activated PI3K/AKT confers resistance against cisplatin through modulation of antiapoptotic (increase in cFLIP) and proapoptotic (decrease in Bax and PUMA) genes. A constant supply of NF-κB through the TNFα-NF-κB autocrine loop and enhanced stabilization of NF-κB by activated AKT maintains an antiapoptotic, quiescent CSC state that confers survival against chemotherapeutics in resistant cells.229 Similar to other signaling pathways, complement signaling maintains NF-κB activation in the TMV. CD10+GPR77+ CAFs promote tumor formation and chemoresistance by providing a niche for CSC survival. Mechanistically, CD10+GPR77+CAFs are driven by persistent NF-κB activation via p65 phosphorylation and acetylation, which is maintained by complement signaling via GPR77, a C5a receptor.182
RhoA/ROCK pathway
RhoA is the founding member of the Rho GTPase family, which also includes Cdc42 and Rac1.230 RhoA acts through Rho-associated, coiled-coil-containing protein kinase (ROCK) to control processes such as actin-myosin-dependent cell contractility, cell motility, and the cell cycle. Currently, a few groups have unveiled the significant role of RhoA/ROCK in CSC therapy resistance.231 In diffuse-type gastric adenocarcinoma (DGA), RhoA signaling promotes CSC phenotypes, which mediate cisplatin resistance.232 RhoA is involved in upregulating MDR1 in CSCs thus promoting drug resistance in CRC.233 Ephrin-B2 signaling also promoted tumorigenesis in a cell-autonomous manner, by mediating anchorage-independent cytokinesis via RhoA in glioblastoma stem-like cells (GSCs).234 The cyclin-dependent kinase 7/9 (CDK7/9) inhibitor SNS-032 repressed the transcription of the RhoA gene, and thereby decreased RhoA GTPase activity and actin polymerization, reducing the frequency of CSCs.235
Overcoming therapy resistance of CSCs by prospective agents: from experimental research to clinical evaluation
Although the ability to target these resistant cell populations is approaching fruition, the majority of currently available anti-CSC strategies target stemness-associated factors, which are shared between CSCs and normal SCs. The therapeutic window of these approaches remains unclear. A more comprehensive understanding of CSC-specific targets, optimization of dosing relative to biological function, and the use of rationally designed combination strategies based on data from relevant preclinical models will yield an improved therapeutic window and targeting efficacy. For the above signaling pathways, which may contribute to CSC-mediated therapy resistance, new strategies targeting CSCs and the results of anti-CSC clinical trials (Table 2) will be discussed in detail below. Several factors limit the interpretation of the results of these trials: (i): Most of these studies lack robust SC readouts to prove the efficacy of drugs that directly target CSCs. (ii): For ethical reasons, most clinical trials are conducted with combined treatment for efficiency and safety. Most of these studies were not designed to target only CSCs. Therefore, while providing a mechanistic view of anti-CSC therapeutics, we preferred to focus on trials that reported subanalyses showing that the actual CSC compartment was targeted. In addition, studies on the proficiency of protein kinase inhibitors (PKIs) have shown cutting-edge results in reversing therapy resistance. Multikinase inhibitors such as regorafenib, sorafenib and EGFR-TKIs are discussed as below.
Table 2.
Emerging agents targeting CSC-associated pathways
| Drug class/mechanism | Agent | Experimental research | Suggested patient population | Notes | Phase |
|---|---|---|---|---|---|
| Agents targeting the Sonic Hedgehog pathway | |||||
| SMO antagonists |
Vismodegib (GDC-0449) |
GDC-0449 could inhibit stemness209 and reverse erlotinib resistance, radiation and carboplatin resistance;258 | Multiple basel-cell carcinomas (MIKIE)239 | Good activity in long-term regimens of MIKIE | 2 |
| TNBC240 | Downregulates CSC markers expression and sensitizes tumors to docetaxel | 1 | |||
| Myelofibrosis241 | Not improved any of the efficacy outcome | 1b | |||
|
Sonidegib (LDE225) |
LDE225 could destroy CSCs niche and reverse docetaxel resistance.240 | TNBC242 | No drug-to-drug interactions between sonidegib and docetaxel were found in the PK assessment | 1b | |
| mBCC243 | Sonidegib continued to demonstrate long-term efficacy and safety in mBCC. | 2 | |||
| SMO inhibitors |
Glasdegib (PF-04449913) |
Myelofibrosis244 | Further study of glasdegib in combination with JAKi in a MF population may be warranted | 1b/2 | |
|
Taladegib (LY2940680) |
Advanced solid tumors245 | Taladegib doses of 100 mg and 200 mg were well tolerated in this population of Japanese patients with advanced solid tumors. | 1 | ||
| BCC246 | LY2940680 treatment resulted in an acceptable safety profile in patients with advanced/metastatic cancer | 1 | |||
|
Saridegib (IPI-926) |
Advanced Pancreatic Adenocarcinoma247 | The study closed early | 1 | ||
| Agents targeting Notch pathway | |||||
| γ-secretase inhibition (GSI) | MK-0752 | Pancreatic cancer257 | Tumor response evaluation was available in 19 of 33 | 1 | |
| RO4929097 | RO4929097 reverse antiandrogen resistance,259 radiation resistance,260 and tamoxifen resistance261 mediated by CSCs; | Recurrent Malignant Glioma263 | Combination of antiangiogenic and notch signaling inhibitors should be considered | 1 | |
| Glioma262 | A specific decrease in the CD133+ CSC population | 0/1 | |||
| PF-03084014 | PF-03084014 reverse docetaxel resistance in CSCs.265 | Advanced TNBC268 | 16% of 25 response-evaluable patients achieved a confirmed partial response | 1 | |
| Desmoid Fibromatosis269 | Objective response rate of 71.4% | 1 | |||
| Aggressive Fibromatosis270 | PF-03084014 was well tolerated and demonstrated promising clinical benefit in patients | 1 | |||
| DLL4 inhibitors |
Demcizumab (OMP-21M18) |
Metastatic Non-Squamous NSCLC271 | 50% had objective tumor responses | 1b | |
| Agents targeting Wnt/β-catenin pathway | |||||
| Ligand sequestration |
OMP-54F28 (FZD8-Fc) |
Advanced solid tumors249 | Agent was well tolerated | 1 | |
| Recurrent platinum-sensitive ovarian cancer250 | 75.7% of overall response rate | 1b | |||
| Inhibitors of β-catenin | PRI-724 |
PRI-724 could downregulate expression of SOX2, |
Hepatitis C Virus-related Cirrhosis255 | Liver injury may be a possible related serious adverse event | 1 |
| CWP232291 |
CWP232291 could reverse castration resistance in CSCs.256 |
NCT03055286 | Recommended Phase 2 dose | 1b | |
| Agents targeting NF-κB pathway | |||||
| Nuclear export protein exportin 1 inhibitor | Selinexor | Selinexor could reverse paclitaxel resistance mediated by CSCs.273 | Triple-class refractory multiple myeloma | Approved by FDA29 | |
Agents targeting the Shh pathway
As we noted previously, SMO activates a cascade. Vismodegib (GDC-0449) and sonidegib (LDE225) are oral SMO antagonists that have been approved by FDA.236,237
Ahmad and colleagues201 showed that erlotinib resistance was mediated by CSCs, and inhibition of the Shh signaling pathway by GDC-0449 resulted in the attenuation of CSC markers, leading to sensitization of EMT cells to drug treatment.201 Moreover, GDC-0449 could decrease stemness and both radiation and carboplatin resistance.238 Furthermore, in vivo treatment with GDC-0449 disrupted the intracellular signaling model mediated by Shh and reduced CAF and CSC expansion.202
A phase 2 trial has been conducted in patients with multiple basal-cell carcinomas treated with vismodegib,239 and both intermittent dosing schedules of vismodegib (group A and group B) seemed to show good activity in long-term regimens in patients with multiple basal-cell carcinomas. In the phase 1 clinical trial EDALINE, 3 of 12 patients with metastatic triple-negative breast cancer (TNBC) derived clinical benefit from combination therapy with a SMO inhibitor and docetaxel chemotherapy, with one patient experiencing a complete response.240 However, in the MYLIE study, which assessed the safety and efficacy of combining ruxolitinib with vismodegib in ruxolitinib-naive patients with myelofibrosis, no new safety concerns were reported, but the addition of vismodegib to ruxolitinib was not shown to improve the efficacy of the treatment.241
Sonidegib (LDE225) is another potent and selective SMO inhibitor. In mouse models of TNBC, Hedgehog ligand produced by neoplastic cells reprogrammed CAFs to provide a supportive niche for the acquisition of a chemoresistant, CSC phenotype via FGF5 expression and the production of fibrillar collagen. Stromal treatment of patient-derived xenografts with SMO inhibitors downregulated CSC marker expression and sensitizes tumors to docetaxel, leading to substantially improved survival and reduced metastatic burden.240
A phase 1 clinical study was designed to explore the combination of sonidegib plus docetaxel (fixed dose at 75 mg/ml) in advanced TNBC patients:242 no drug-to-drug interactions between sonidegib and docetaxel were found, and the combination showed antitumor activity in three of 10 patients with measurable disease. The median time to progression for the overall study was 42.5 days. 30-month analysis of the randomized phase 2 BOLT study was conducted to assess the long-term efficacy and safety of sonidegib in patients with locally advanced and metastatic basal-cell carcinoma. A possitive outcome was exhibited:243 patients treated with 200 mg sonidegib, had objective response rates of 56.1% (central) and 71.2% (investigator) in locally advanced basal-cell carcinoma and 7.7% (central) and 23.1% (investigator) in metastatic basal-cell carcinima.
Other selective SMO inhibitors, glasdegib (PF-04449913),244 taladegib (LY2940680),245,246 and saridegib (IPI-926)247 have entered clinical trials to be tested in various tumors. Since the safety profile of glasdegib monotherapy was manageable in patients with primary/secondary MF, further study of glasdegib in combination with JAK inhibitors in an MF population may be warranted. For taladegib, phase 1 dose escalation studies were designed and a low dose was tolerated in patients. The clinical efficacy of this drug should be further investigated. A phase 1 study of FOLFIRINOX plus IPI-926 for advanced pancreatic adenocarcinoma was closed early when a separate phase 2 trial of IPI-926 plus gemcitabine indicated detrimental effects of this combination.
Overall, despite the impressive preclinical activity and sheer number of trials with Hh inhibitors, the clinical efficacy of these agents has been modest.
Agents targeting the Wnt/β-catenin pathway
OMP-54F28 is a fusion protein that combines the cysteine-rich domain of Fzd8 with the immunoglobulin Fc domain that competes with the native Fzd8 receptor for its ligands and antagonizes Wnt signaling.248 Twenty-six patients were treated in a phase 1 study of the anticancer SC agent OMP-54F28249 and another phase 1b dose escalation study of OMP54F28 in combination with paclitaxel and carboplatin in patients with recurrent platinum-sensitive ovarian cancer. Further investigation is limited because of bone toxicity.250
PRI-724 is an inhibitor of β-catenin. PRI-724 reduced drug resistance and CSC phenotypes in TNBC251 and downregulated Sox2 and CD44 expression.252 Moreover, the combination of PRI-724 with cisplatin synergistically suppressed cell growth.252 While preclinical models showed that this treatment can reverse therapy resistance by targeting CSCs,253,254 its function in cancer has not been clinically determined, and liver injury may be a possible serious adverse event.255
CWP232291 is a small molecule Wnt/β-catenin inhibitor that blocks the growth of castration-resistant prostate cancer by activating the endoplasmic reticulum stress pathway.256 CWP232291 (NCT03055286) was evaluated in a phase 1b study of 45 patients with AML to determine the recommended phase 2 dose (RP2D) of CWP232291 in combination with cytarabine (ara-C) administered to subjects with relapsed or refractory AML. Published articles are currently unavailable.
Agents targeting the Notch pathway
γ-Secretase inhibitors (GSIs), such as MK-0752 and RO4929097, and the use of antibodies against the Notch receptor or ligand are the major clinical approaches targeting Notch signaling. A multicenter, nonrandom Bayesian adaptive design study of MK-0752 was performed to determine the safety of combination treatment and the recommended phase 2 dose (RP2D):257 tumor response evaluation was available in 19 patients; 13 achieved stable disease and 1 patient achieved a confirmed partial response. MK-0752 plus docetaxel could decrease CD44+CD24- and ALDH+ cell fractions.258
RO4929097, another GSI, could sensitize prostate cancer cells to antiandrogen therapy.259 Moreover, RO4929097 could reduce IDO1 expression in cervical CSCs and reduce the binding of NICD on the IDO1 promoter, as well as sensitize xenograft tumors to radiation treatment.260 Furthermore, RO4929097 could overcome acquired tamoxifen resistance in CSCs in human breast cancer.261 In a phase 0/1 trial, 21 patients with newly diagnosed glioblastoma or anaplastic astrocytoma received RO4929097 combined with temozolomide and radiotherapy,262 and a specific decrease in the CD133 CSC population was observed. Thirteen subjects were enrolled in a phase 1 study of RO4929097 with bevacizumab in patients with recurrent malignant glioma.263The median overall survival was 10.9 months with a median progression-free survival of 3.7 months.
Finally, PF-03084014 is also a GSI. PF-03084014 inhibited HCC growth via suppression of cancer stemness.264 Moreover, PF-03084014 was reported to enhance the antitumor effect of docetaxel in prostate cancer stem-like cells.265 In pancreatic ductal adenocarcinoma (PDA), a combination of PF-03084014 with gemcitabine reduced putative CSCs. Notably, in a highly aggressive orthotopic model, a PF-03084014 and gemcitabine combination was effective in inducing apoptosis, and inhibiting tumor cell proliferation and angiogenesis, resulting in the attenuation of primary tumor growth as well as controlling metastatic dissemination, compared to gemcitabine treatment.266 Furthermore, a synergistic effect of PF-03084014 with docetaxel through targeting of CSCs was observed in breast cancer.267 Studies of PF-03084014 on advanced TNBC,268 desmoid fibromatosis,269 and aggressive fibromatosis270 demonstrated efficacy in desmoid tumors in phase 1 studies.
Demcizumab (OMP-21M18) is a first-in-class humanized antiDLL4 antibody. Twenty of 40 evaluable patients (50%) had objective tumor responses in the phase 1b trial of anti-CSC therapy.271 Moreover, demcizumab in combination with paclitaxel has a manageable toxicity profile and showed activity in patients with heavily pretreated platinum-resistant ovarian cancer.272 However, a recent study in metastatic pancreatic cancer failed to demonstrate survival benefit when demcizumab was added to gemcitabine plus Abraxane (YOSEMITE study) (NCT02289898). Another phase 2 study investigating the addition of demcizumab to standard first-line treatment with carboplatin plus pemetrexed in lung cancer (DENALI study) is ongoing (NCT02259582).
Agents targeting the NF-κB pathway
Selinexor is an oral inhibitor of the nuclear export protein exportin 1, which inhibits NF-κB signaling. Selinexor compounds synergize with gemcitabine and nanoparticle albumin-bound (nab)-paclitaxel, leading to suppression of pancreatic ductal adenocarcinoma (PDAC) growth and CSC spheroid disintegration.273 Recently, the FDA granted accelerated approval to selinexor plus low-dose dexamethasone for triple-class refractory multiple myeloma,29 because selinexor-dexamethasone resulted in objective treatment responses in patients who displayed resistance to several proteasome inhibitors.274 As it is for relapsed patients, selinexor may play a role in reversing therapy resistance. Combined selinexor and gemcitabine could suppress CSC spheroids in a PDAC phase 1b trial,273 identifying selinexor as a promising agent targeting CSCs.
Effects of protein kinase inhibitors (PKIs)
Given that accumulating evidence has demonstrated that eventual treatment failure resultes from multiple defense mechanisms of CSCs, we should block the compensatory responses induced by mutual communication in these cells. Several multikinase inhibitors have been approved for patients who failed to respond to currently available chemotherapeutic agents.275 The potential efficacy of PKIs on CSCs has been investigated: multikinase inhibitors (such as regorafenib and sorafenib) and a group of EGFR-TKIs are being examined.
Regorafenib is an oral multikinase inhibitor that blocks the activity of protein kinases involved in angiogenesis, oncogenesis, metastasis, and tumor immunity.276,277 Limited studies have indicated that regorafenib can reverse drug resistance caused by CSCs: regorafenib treatment decreased the stemness phenotypes including tumor sphere formation and the SP fraction of HCT-116R and DLD-1R cells. The combination of regorafenib and 5-FU significantly suppressed the tumorigenesis and stemness markers of 5-FU resistant cells.278 In addition, regorafenib could decrease the expression of CSC markers in PDAC.279 Moreover, targeting the TMV with regorafenib altered the tumor cell-marrow-derived mesenchymal stem cells (MSCs) interaction, which in turn inhibited the growth and metastasis of colon cancer.280 Furthermore, in an AML model, leukemic SCs were sensitive to regorafenib treatment.281
Increasing studies have found that combinatorial administration could possibly reverse therapy resistance in CSCs. The poor efficacy of first‑generation EGFR‑TKIs for lung adenocarcinoma appears to be related to the increased expression of CSC markers.282 Overexpression of shisa3 inhibited CSC properties in lung adenocarcinoma cells and reversed resistance to gefitinib/osimertinib, which are EGFR-TKIs.283 The EGFR-TKI, brexpiprazole, combined with osimertinib, is a potential therapeutic strategy for glioblastoma by chemosensitizing glioma CSCs through the downregulation of survivin expression.284
Sorafenib is another oral multitargeted receptor tyrosine kinase inhibitor, and although it significantly prolonged progression-free survival,285 its long-term success is quite low due to the development of resistant cells286 and adverse drug reactions (ADRs).287 Efforts should also be made to explore other potent molecular targets that can improve the efficiency of sorafenib. Huang’s team reported that lysine-specific histone demethylase 1A (KDM1A) inhibitors dramatically suppressed the stem-like properties of sorafenib-resistant cells by regulating the Wnt signaling pathway.288 An inhibitor of cyclin-dependent kinase 1(CDK1), RO3306, combined with sorafenib could potently decrease tumor growth in patient-derived xenograft (PDX) models, and the combined administration synergistically downregulated CDK1/β-catenin signaling as well as the pluripotency proteins Oct4, Sox2, and Nanog.289 NF-κB signaling mediated sorafenib resistance,290 and cotreating cells with sorafenib and sulforaphane downregulated NF-κB and reversed sorafenib-induced NF-κB binding, which was associated with decreased clonogenicity, spheroid formation, ALDH activity and migratory capacity.291
Perspectives
Overall, CSCs are key players in tumorigenesis and, through multiple and different mechanisms, contribute to the therapy-resistant phenotype. Innovative treatments for CSC sensitization should include the combination of drugs targeting ABC transporters, DNA damage repair, metastasis, autophagic inhibition, ferroptosis and the TMV disruption and immunotherapies. The level of resistance may reflect the sum of alterations of different molecular pathways (in which resistance-related proteins are deregulated). From this point of view, the therapeutic approach needs to be extremely effective and efficient in space (tumor volume) and time (effective in the first-line of treatment). As noted previously, the number of patients in current clinical trials remains limited, which is not conducive to carrying out pivotal phase 3 trials. In addition, preclinical models of therapy resistant are usually conducted in normal cancer cell lines rather than CSC models. This condition could lead to errors: although normal cancer cell lines were killed by preclinical therapy, the CSC subpopulation still remained, which induces cancer recurrence. In addition, the extent to which the current marker-designated populations are actual CSCs remains to be answered. Improvement of preclinical models of CSC should be further undertaken, and identification of surrogate markers or functional assays to monitor biological activity and treatment responses are needed. The landscape of agents targeting CSC self-renewal pathways or TKIs is expanding, and combined treatments to avoid off-target effects is one notable strategy. Finally, the reversal of therapy resistance of CSCs is not straightforward and require the following: (i) improved understanding of the mechanisms regulating CSC resistance to therapy; (ii) a combination of pharmacology and pharmacology for design and structural modification of drugs; (iii) both experimental and molecular modeling simulations of crystal structures, properties and formation of targeted proteins and agents; (iv) some new strategies, e.g., targeting drug-efflux pumps, targeting the CSC niche and the quiescent state and induction of CSC apoptosis and ferroptosis.
Acknowledgements
This work was sponsored by the Interdisciplinary Program of Shanghai Jiao Tong University (No. YG2017MS29). The funding body has no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author contributions
Z.H.M. and Z.J.G. drafted the manuscript. Z.X. participated in interpretation. L.Q. participated in the design of the manuscript and helped modifying the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: He-Ming Zhou, Ji-Gang Zhang
References
- 1.Salgia R, Kulkarni P. The genetic/non-genetic duality of drug ‘resistance’ in cancer. Trends Cancer. 2018;4:110–118. doi: 10.1016/j.trecan.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo M, Brooks M, Wicha MS. Epithelial-mesenchymal plasticity of breast cancer stem cells: implications for metastasis and therapeutic resistance. Curr. Pharm. Des. 2015;21:1301–1310. doi: 10.2174/1381612821666141211120604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eppert K, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 4.Al-Hajj M, et al. Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanpain C, Fuchs E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science. 2014;344:1242281. doi: 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Brien CA, Kreso A, Jamieson CHM. Cancer stem cells and self-renewal. Clin. Cancer Res. 2010;16:3113–3120. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 7.Ricci-Vitiani L, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 8.Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat. Rev. Cancer. 2008;8:545–554. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, et al. Epithelial mesenchymal transition and hedgehog signaling activation are associated with chemoresistance and invasion of hepatoma subpopulations. J. Hepatol. 2011;55:838–845. doi: 10.1016/j.jhep.2010.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shmelkov SV, St Clair R, Lyden D, Rafii S. AC133/CD133/Prominin-1. Int. J. Biochem. Cell Biol. 2005;37:715–719. doi: 10.1016/j.biocel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 12.Ma S, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Beier D, et al. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 14.Baba T, et al. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene. 2009;28:209–218. doi: 10.1038/onc.2008.374. [DOI] [PubMed] [Google Scholar]
- 15.Zhang M, et al. Nestin and CD133: valuable stem cell-specific markers for determining clinical outcome of glioma patients. J. Exp. Clin. Cancer Res. : CR. 2008;27:85. doi: 10.1186/1756-9966-27-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colman H, et al. A multigene predictor of outcome in glioblastoma. Neuro-Oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou Y, et al. The critical role of CD133(+)CD44(+/high) tumor cells in hematogenous metastasis of liver cancers. Cell Res. 2012;22:259–272. doi: 10.1038/cr.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haraguchi N, et al. CD133+CD44+ population efficiently enriches colon cancer initiating cells. Ann. Surg. Oncol. 2008;15:2927–2933. doi: 10.1245/s10434-008-0074-0. [DOI] [PubMed] [Google Scholar]
- 19.Huang EH, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva IA, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71:3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terraneo, N. et al. L1 cell adhesion molecule confers radioresistance to ovarian cancer and defines a new cancer stem cell population. Cancers (Basel). 12, 217–234 (2020). [DOI] [PMC free article] [PubMed]
- 22.Chen C, Zhao S, Karnad A, Freeman JW. The biology and role of CD44 in cancer progression: therapeutic implications. J. Hematol. Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl Acad. Sci. USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du L, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2008;14:6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 25.Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. J. Clin. Oncol. 2008;26:2806–2812. doi: 10.1200/JCO.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- 26.Alvero AB, et al. Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle. 2009;8:158–166. doi: 10.4161/cc.8.1.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takaishi S, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells (Dayt., Ohio). 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbasian M, Mousavi E, Arab-Bafrani Z, Sahebkar A. The most reliable surface marker for the identification of colorectal cancer stem-like cells: a systematic review and meta-analysis. J. Cell Physiol. 2019;234:8192–8202. doi: 10.1002/jcp.27619. [DOI] [PubMed] [Google Scholar]
- 29.XPO1 Inhibitor Approved for Multiple Myeloma. Cancer Discov. 9, 1150–1151 (2019). [DOI] [PubMed]
- 30.Li, C. et al. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology141, 2218–2227 (2011). [DOI] [PubMed]
- 31.Takaishi S, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen PH, et al. Characterization of biomarkers of tumorigenic and chemoresistant cancer stem cells in human gastric carcinoma. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2017;23:1586–1597. doi: 10.1158/1078-0432.CCR-15-2157. [DOI] [PubMed] [Google Scholar]
- 33.Screaton GR, et al. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc. Natl Acad. Sci. USA. 1992;89:12160–12164. doi: 10.1073/pnas.89.24.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lau WM, et al. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014;74:2630–2641. doi: 10.1158/0008-5472.CAN-13-2309. [DOI] [PubMed] [Google Scholar]
- 35.Boesch M, Spizzo G, Seeber A. Concise review: aggressive colorectal cancer: role of epithelial cell adhesion molecule in cancer stem cells and epithelial-to-mesenchymal transition. Stem Cells Transl. Med. 2018;7:495–501. doi: 10.1002/sctm.17-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shmelkov SV, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J. Clin. Invest. 2008;118:2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita T, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 39.Chute JP, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc. Natl Acad. Sci. USA. 2006;103:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clay MR, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head. Neck. 2010;32:1195–1201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Croker AK, et al. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J. Cell. Mol. Med. 2009;13:2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma S, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol. Cancer Res. 2008;6:1146–1153. doi: 10.1158/1541-7786.MCR-08-0035. [DOI] [PubMed] [Google Scholar]
- 43.Kisselbach L, Merges M, Bossie A, Boyd A. CD90 Expression on human primary cells and elimination of contaminating fibroblasts from cell cultures. Cytotechnology. 2009;59:31–44. doi: 10.1007/s10616-009-9190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang ZF, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 45.True LD, et al. CD90/THY1 is overexpressed in prostate cancer-associated fibroblasts and could serve as a cancer biomarker. Mod. Pathol. 2010;23:1346–1356. doi: 10.1038/modpathol.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buishand FO, et al. Identification of CD90 as putative cancer stem cell marker and therapeutic target in insulinomas. Stem Cells Dev. 2016;25:826–835. doi: 10.1089/scd.2016.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W-C, et al. Cancer stem cell marker CD90 inhibits ovarian cancer formation via β3 integrin. Int. J. Oncol. 2016;49:1881–1889. doi: 10.3892/ijo.2016.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim BH, et al. Stem cell markers predict the response to sorafenib in patients with hepatocellular carcinoma. Gut Liver. 2019;13:342–348. doi: 10.5009/gnl18345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang P, et al. Identification and characterization of cells with cancer stem cell properties in human primary lung cancer cell lines. PLoS ONE. 2013;8:e57020. doi: 10.1371/journal.pone.0057020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Zhang H, Wang ZZ. Mechanisms that mediate stem cell self-renewal and differentiation. J. Cell Biochem. 2008;103:709–718. doi: 10.1002/jcb.21460. [DOI] [PubMed] [Google Scholar]
- 51.Papp B, Plath K. Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell Res. 2011;21:486–501. doi: 10.1038/cr.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schöler HR, et al. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 53.Schöler HR. Octamania: the POU factors in murine development. Trends Genet. 1991;7:323–329. doi: 10.1016/0168-9525(91)90422-M. [DOI] [PubMed] [Google Scholar]
- 54.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 55.Monk M, Holding C. Human embryonic genes re-expressed in cancer cells. Oncogene. 2001;20:8085–8091. doi: 10.1038/sj.onc.1205088. [DOI] [PubMed] [Google Scholar]
- 56.Ponti D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 57.Koo BS, et al. Oct4 is a critical regulator of stemness in head and neck squamous carcinoma cells. Oncogene. 2015;34:2317–2324. doi: 10.1038/onc.2014.174. [DOI] [PubMed] [Google Scholar]
- 58.Ghanei, Z., Jamshidizad, A., Joupari, M. D. & Shamsara, M. Isolation and characterization of breast cancer stem cell-like phenotype by Oct4 promoter-mediated activity. J Cell Physiol.235, 7840–7848 (2020). [DOI] [PubMed]
- 59.Liu L, et al. Inhibition of Wnt/β-catenin pathway reverses multi-drug resistance and EMT in Oct4/Nanog NSCLC cells. Biomed. Pharmacother. 2020;127:110225. doi: 10.1016/j.biopha.2020.110225. [DOI] [PubMed] [Google Scholar]
- 60.Pádua, D. et al. A SOX2 reporter system identifies gastric cancer stem-like cells sensitive to monensin. Cancers (Basel). 12, 495–515 (2020). [DOI] [PMC free article] [PubMed]
- 61.Gao W, et al. Development of a novel and economical agar-based non-adherent three-dimensional culture method for enrichment of cancer stem-like cells. Stem Cell Res. Ther. 2018;9:243. doi: 10.1186/s13287-018-0987-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cavaleri F, Schöler HR. Nanog: a new recruit to the embryonic stem cell orchestra. Cell. 2003;113:551–552. doi: 10.1016/S0092-8674(03)00394-5. [DOI] [PubMed] [Google Scholar]
- 63.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 64.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/S0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 65.Almstrup K, et al. Embryonic stem cell-like features of testicular carcinoma in situ revealed by genome-wide gene expression profiling. Cancer Res. 2004;64:4736–4743. doi: 10.1158/0008-5472.CAN-04-0679. [DOI] [PubMed] [Google Scholar]
- 66.Botchkina GI, et al. New-generation taxoid SB-T-1214 inhibits stem cell-related gene expression in 3D cancer spheroids induced by purified colon tumor-initiating cells. Mol. Cancer. 2010;9:192. doi: 10.1186/1476-4598-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu S, et al. Methylation status of the promoter determines the switch between cancer cells and cancer stem cells. Adv. Sci. (Weinh.) 2020;7:1903035. doi: 10.1002/advs.201903035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dehghan Harati, M., Rodemann, H. P. & Toulany, M. Nanog signaling mediates radioresistance in aldh-positive breast cancer cells. Int. J. Mol. Sci. 20, 1151–1165 (2019). [DOI] [PMC free article] [PubMed]
- 69.Lai H-H, et al. TARBP2-mediated destabilization of Nanog overcomes sorafenib resistance in hepatocellular carcinoma. Mol. Oncol. 2019;13:928–945. doi: 10.1002/1878-0261.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsu H-S, et al. Cucurbitacin I inhibits tumorigenic ability and enhances radiochemosensitivity in nonsmall cell lung cancer-derived CD133-positive cells. Cancer. 2011;117:2970–2985. doi: 10.1002/cncr.25869. [DOI] [PubMed] [Google Scholar]
- 71.Masciale V, et al. Isolation and identification of cancer stem-like cells in adenocarcinoma and squamous cell carcinoma of the lung: a pilot study. Front. Oncol. 2019;9:1394. doi: 10.3389/fonc.2019.01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu F, et al. SOX2 is a marker for stem-like tumor cells in bladder cancer. Stem Cell Rep. 2017;9:429–437. doi: 10.1016/j.stemcr.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Menendez, S. T. et al. SOX2 expression and transcriptional activity identifies a subpopulation of cancer stem cells in sarcoma with prognostic implications. Cancers (Basel). 12, 964–981 (2020). [DOI] [PMC free article] [PubMed]
- 74.Goodell MA, et al. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou S, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat. Med. 2001;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 76.Scharenberg CW, Harkey MA, Torok-Storb B. The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood. 2002;99:507–512. doi: 10.1182/blood.V99.2.507. [DOI] [PubMed] [Google Scholar]
- 77.Uchida N, Fujisaki T, Eaves AC, Eaves CJ. Transplantable hematopoietic stem cells in human fetal liver have a CD34(+) side population (SP) phenotype. J. Clin. Investig. 2001;108:1071–1077. doi: 10.1172/JCI13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feuring-Buske M, Hogge DE. Hoechst 33342 efflux identifies a subpopulation of cytogenetically normal CD34(+)CD38(-) progenitor cells from patients with acute myeloid leukemia. Blood. 2001;97:3882–3889. doi: 10.1182/blood.V97.12.3882. [DOI] [PubMed] [Google Scholar]
- 79.Patrawala L, et al. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 80.Bunting KD, Zhou S, Lu T, Sorrentino BP. Enforced P-glycoprotein pump function in murine bone marrow cells results in expansion of side population stem cells in vitro and repopulating cells in vivo. Blood. 2000;96:902–909. doi: 10.1182/blood.V96.3.902.015k40_902_909. [DOI] [PubMed] [Google Scholar]
- 81.Chiba T, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology (Baltim., Md.). 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 82.Wang J, et al. Identification of cancer stem cell-like side population cells in human nasopharyngeal carcinoma cell line. Cancer Res. 2007;67:3716–3724. doi: 10.1158/0008-5472.CAN-06-4343. [DOI] [PubMed] [Google Scholar]
- 83.Szotek PP, et al. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc. Natl Acad. Sci. USA. 2006;103:11154–11159. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fan X, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 85.Reynolds SD, et al. Molecular and functional properties of lung SP cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2007;292:L972–L983. doi: 10.1152/ajplung.00090.2006. [DOI] [PubMed] [Google Scholar]
- 86.Li H, et al. Nucleolar and spindle associated protein 1 promotes metastasis of cervical carcinoma cells by activating Wnt/β-catenin signaling. J. Exp. Clin. Cancer Res. : CR. 2019;38:33. doi: 10.1186/s13046-019-1037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cho Y, et al. Post-translational modification of OCT4 in breast cancer tumorigenesis. Cell Death Differ. 2018;25:1781–1795. doi: 10.1038/s41418-018-0079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhong Q, et al. The fusion gene induces cancer stem cell-like properties and therapeutic resistance in nasopharyngeal carcinoma. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2018;24:659–673. doi: 10.1158/1078-0432.CCR-17-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gao M, et al. Identification and characterization of tumor-initiating cells in multiple myeloma. J. Natl Cancer Inst. 2020;112:507–515. doi: 10.1093/jnci/djz159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Islam SS, Aboussekhra A. Sequential combination of cisplatin with eugenol targets ovarian cancer stem cells through the Notch-Hes1 signalling pathway. J. Exp. Clin. Cancer Res. : CR. 2019;38:382. doi: 10.1186/s13046-019-1360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiao T, et al. Circ008913, via miR-889 regulation of DAB2IP/ZEB1, is involved in the arsenite-induced acquisition of CSC-like properties by human keratinocytes in carcinogenesis. Metallomics. 2018;10:1328–1338. doi: 10.1039/C8MT00207J. [DOI] [PubMed] [Google Scholar]
- 92.Gu C, et al. circGprc5a promoted bladder oncogenesis and metastasis through Gprc5a-targeting peptide. Mol. Ther. Nucleic Acids. 2018;13:633–641. doi: 10.1016/j.omtn.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jian X, et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol. Cancer. 2020;19:20. doi: 10.1186/s12943-020-1134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhi, X. et al. circLgr4 drives colorectal tumorigenesis and invasion through Lgr4-targeting peptide. Int. J. Cancer. 10.1002/ijc.32549 (2019). [DOI] [PubMed]
- 95.Wang Y, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 96.Zhu P, et al. lnc-β-Catm elicits EZH2-dependent β-catenin stabilization and sustains liver CSC self-renewal. Nat. Struct. Mol. Biol. 2016;23:631–639. doi: 10.1038/nsmb.3235. [DOI] [PubMed] [Google Scholar]
- 97.Shima H, et al. Lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res. Treat. 2018;170:507–516. doi: 10.1007/s10549-018-4793-z. [DOI] [PubMed] [Google Scholar]
- 98.Conigliaro A, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 100.Diehn M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen J, et al. Characteristics of doxorubicin-selected multidrug-resistant human leukemia HL-60 cells with tolerance to arsenic trioxide and contribution of leukemia stem cells. Oncol. Lett. 2018;15:1255–1262. doi: 10.3892/ol.2017.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Luzhna L, Lykkesfeldt AE, Kovalchuk O. Altered radiation responses of breast cancer cells resistant to hormonal therapy. Oncotarget. 2015;6:1678–1694. doi: 10.18632/oncotarget.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alderton GK. Tumour evolution: Epigenetic and genetic heterogeneity in metastasis. Nat. Rev. Cancer. 2017;17:141. doi: 10.1038/nrc.2017.11. [DOI] [PubMed] [Google Scholar]
- 104.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat. Rev. Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 105.Hou Y, et al. The FOXM1-ABCC5 axis contributes to paclitaxel resistance in nasopharyngeal carcinoma cells. Cell Death Dis. 2017;8:e2659. doi: 10.1038/cddis.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ohashi R, et al. Expression of MRP1 and ABCG2 is associated with adverse clinical outcomes of papillary thyroid carcinoma with a solid component. Hum. Pathol. 2017;67:11–17. doi: 10.1016/j.humpath.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 107.Jiang Y, et al. Expressions of putative cancer stem cell markers ABCB1, ABCG2, and CD133 are correlated with the degree of differentiation of gastric cancer. Gastric Cancer. 2012;15:440–450. doi: 10.1007/s10120-012-0140-y. [DOI] [PubMed] [Google Scholar]
- 108.Liao, J. et al. Regulation of stem-like cancer cells by glutamine through beta-catenin pathway mediated by redox signaling. Mol. Cancer. 16, 51–64 (2017). [DOI] [PMC free article] [PubMed]
- 109.Wang X-k, et al. Afatinib enhances the efficacy of conventional chemotherapeutic agents by eradicating cancer stem-like cells. Cancer Res. 2014;74:4431–4445. doi: 10.1158/0008-5472.CAN-13-3553. [DOI] [PubMed] [Google Scholar]
- 110.Das B, et al. MYC regulates the HIF2 alpha stemness pathway via nanog and sox2 to maintain self-renewal in cancer stem cells versus non-stem cancer cells. Cancer Res. 2019;79:4015–4025. doi: 10.1158/0008-5472.CAN-18-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sasaki N, et al. Stemness and anti-cancer drug resistance in ATP-binding cassette subfamily G member 2 highly expressed pancreatic cancer is induced in 3D culture conditions. Cancer Sci. 2018;109:1135–1146. doi: 10.1111/cas.13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Patrawala L, et al. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2(+) and ABCG2(-) cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 113.Miranda-Lorenzo I, et al. Intracellular autofluorescence: a biomarker for epithelial cancer stem cells. Nat. Methods. 2014;11:1161–1169. doi: 10.1038/nmeth.3112. [DOI] [PubMed] [Google Scholar]
- 114.El-Khattouti A, et al. CD133(+) melanoma subpopulation acquired resistance to caffeic acid phenethyl ester-induced apoptosis is attributed to the elevated expression of ABCB5: Significance for melanoma treatment. Cancer Lett. 2015;357:83–104. doi: 10.1016/j.canlet.2014.10.043. [DOI] [PubMed] [Google Scholar]
- 115.Grimm M, et al. ABCB5 expression and cancer stem cell hypothesis in oral squamous cell carcinoma. Eur. J. Cancer. 2012;48:3186–3197. doi: 10.1016/j.ejca.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 116.Wilson BJ, et al. ABCB5 maintains melanoma-initiating cells through a proinflammatory cytokine signaling circuit. Cancer Res. 2014;74:4196–4207. doi: 10.1158/0008-5472.CAN-14-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee CAA, et al. Targeting the ABC transporter ABCB5 sensitizes glioblastoma to temozolomide-induced apoptosis through a cell-cycle checkpoint regulation mechanism. J. Biol. Chem. 2020;295:7774–7788. doi: 10.1074/jbc.RA120.013778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sugano T, et al. Inhibition of ABCB1 overcomes cancer stem cell-like properties and acquired resistance to MET inhibitors in non-small cell lung cancer. Mol. Cancer Ther. 2015;14:2433–2440. doi: 10.1158/1535-7163.MCT-15-0050. [DOI] [PubMed] [Google Scholar]
- 119.Huang B, et al. PKC epsilon inhibits isolation and sternness of side population cells via the suppression of ABCB1 transporter and PI3K/Akt, MAPK/ERK signaling in renal cell carcinoma cell line 769P. Cancer Lett. 2016;376:148–154. doi: 10.1016/j.canlet.2016.03.041. [DOI] [PubMed] [Google Scholar]
- 120.Wu Z-X, et al. Tepotinib reverses ABCB1-mediated multidrug resistance in cancer cells. Biochem. Pharm. 2019;166:120–127. doi: 10.1016/j.bcp.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 121.Tsou SH, Chen TM, Hsiao HT, Chen YH. A critical dose of doxorubicin is required to alter the gene expression profiles in MCF-7 cells acquiring multidrug resistance. PLoS ONE. 2015;10:24. doi: 10.1371/journal.pone.0116747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou S, Zong Y, Lu T, Sorrentino BP. Hematopoietic cells from mice that are deficient in both Bcrp1/Abcg2 and Mdr1a/1b develop normally but are sensitized to mitoxantrone. BioTechniques. 2003;35:1248–1252. doi: 10.2144/03356ss04. [DOI] [PubMed] [Google Scholar]
- 123.Yuan R, et al. Cyclin F-dependent degradation of E2F7 is critical for DNA repair and G2-phase progression. EMBO J. 2019;38:e101430. doi: 10.15252/embj.2018101430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schulz, A., Meyer, F., Dubrovska, A. & Borgmann, K. Cancer stem cells and radioresistance: DNA repair and beyond. Cancers (Basel). 11, 862–882 (2019). [DOI] [PMC free article] [PubMed]
- 125.Gold A, et al. Spironolactone inhibits the growth of cancer stem cells by impairing DNA damage response. Oncogene. 2019;38:3103–3118. doi: 10.1038/s41388-018-0654-9. [DOI] [PubMed] [Google Scholar]
- 126.Skvortsova I, Debbage P, Kumar V, Skvortsov S. Radiation resistance: Cancer stem cells (CSCs) and their enigmatic pro-survival signaling. Semin. Cancer Biol. 2015;35:39–44. doi: 10.1016/j.semcancer.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 127.Wang K, et al. Redox homeostasis: the linchpin in stem cell self-renewal and differentiation. Cell Death Dis. 2013;4:e537. doi: 10.1038/cddis.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao H, et al. Up-regulation of glycolysis promotes the stemness and EMT phenotypes in gemcitabine-resistant pancreatic cancer cells. J. Cell. Mol. Med. 2017;21:2055–2067. doi: 10.1111/jcmm.13126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Versini A, et al. Salinomycin derivatives kill breast cancer stem cells by lysosomal iron targeting. Chemistry. 2020;26:7416–7424. doi: 10.1002/chem.202000335. [DOI] [PubMed] [Google Scholar]
- 130.Ryoo I-G, Choi B-H, Ku S-K, Kwak M-K. High CD44 expression mediates p62-associated NFE2L2/NRF2 activation in breast cancer stem cell-like cells: Implications for cancer stem cell resistance. Redox Biol. 2018;17:246–258. doi: 10.1016/j.redox.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liao J, et al. Regulation of stem-like cancer cells by glutamine through β-catenin pathway mediated by redox signaling. Mol. Cancer. 2017;16:51. doi: 10.1186/s12943-017-0623-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee, K.-M. et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 26, 633–647 (2017). [DOI] [PMC free article] [PubMed]
- 133.Alsubhi N, et al. Chk1 phosphorylated at serine345 is a predictor of early local recurrence and radio-resistance in breast cancer. Mol. Oncol. 2016;10:213–223. doi: 10.1016/j.molonc.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang WJ, et al. MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res. 2013;73:1219–1231. doi: 10.1158/0008-5472.CAN-12-1408. [DOI] [PubMed] [Google Scholar]
- 135.Srivastava AK, et al. Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc. Natl Acad. Sci. USA. 2015;112:4411–4416. doi: 10.1073/pnas.1421365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Paget, S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 8, 98–101 (1989). [PubMed]
- 137.Dieter SM, et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 2011;9:357–365. doi: 10.1016/j.stem.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 138.Gundem G, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Navas T, et al. Clinical evolution of epithelial-mesenchymal transition in human carcinomas. Cancer Res. 2020;80:304–318. doi: 10.1158/0008-5472.CAN-18-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xie SL, et al. SOX8 regulates cancer stem-like properties and cisplatin-induced EMT in tongue squamous cell carcinoma by acting on the Wnt/β-catenin pathway. Int. J. Cancer. 2018;142:1252–1265. doi: 10.1002/ijc.31134. [DOI] [PubMed] [Google Scholar]
- 141.Lee KM, et al. ECM1 regulates tumor metastasis and CSC-like property through stabilization of β-catenin. Oncogene. 2015;34:6055–6065. doi: 10.1038/onc.2015.54. [DOI] [PubMed] [Google Scholar]
- 142.Liang Y, et al. Epigenetic activation of TWIST1 by MTDH promotes cancer stem-like cell traits in breast cancer. Cancer Res. 2015;75:3672–3680. doi: 10.1158/0008-5472.CAN-15-0930. [DOI] [PubMed] [Google Scholar]
- 143.Ruan D, et al. Skp2 deficiency restricts the progression and stem cell features of castration-resistant prostate cancer by destabilizing Twist. Oncogene. 2017;36:4299–4310. doi: 10.1038/onc.2017.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li NN, et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis. 2019;8:17. doi: 10.1038/s41389-019-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Douma S, et al. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430:1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- 146.Grillet F, et al. Circulating tumour cells from patients with colorectal cancer have cancer stem cell hallmarks in culture. Gut. 2017;66:1802–1810. doi: 10.1136/gutjnl-2016-311447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu T, et al. Circulating glioma cells exhibit stem cell-like properties. Cancer Res. 2018;78:6632–6642. doi: 10.1158/0008-5472.CAN-18-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jung Y, et al. CXCL12γ promotes metastatic castration-resistant prostate cancer by inducing cancer stem cell and neuroendocrine phenotypes. Cancer Res. 2018;78:2026–2039. doi: 10.1158/0008-5472.CAN-17-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barton VN, et al. Androgen receptor supports an anchorage-independent, cancer stem cell-like population in triple-negative breast cancer. Cancer Res. 2017;77:3455–3466. doi: 10.1158/0008-5472.CAN-16-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vummidi Giridhar P, et al. Constant degradation of the androgen receptor by MDM2 conserves prostate cancer stem cell integrity. Cancer Res. 2019;79:1124–1137. doi: 10.1158/0008-5472.CAN-18-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ghattass K, Assah R, El-Sabban M, Gali-Muhtasib H. Targeting hypoxia for sensitization of tumors to radio- and chemotherapy. Curr. Cancer Drug Targets. 2013;13:670–685. doi: 10.2174/15680096113139990004. [DOI] [PubMed] [Google Scholar]
- 152.Qian J, Rankin EB. Hypoxia-induced phenotypes that mediate tumor heterogeneity. Adv. Exp. Med. Biol. 2019;1136:43–55. doi: 10.1007/978-3-030-12734-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gao T, et al. The mechanism between epithelial mesenchymal transition in breast cancer and hypoxia microenvironment. Biomed. Pharmacother. 2016;80:393–405. doi: 10.1016/j.biopha.2016.02.044. [DOI] [PubMed] [Google Scholar]
- 154.O’Reilly D, Johnson P, Buchanan PJ. Hypoxia induced cancer stem cell enrichment promotes resistance to androgen deprivation therapy in prostate cancer. Steroids. 2019;152:108497. doi: 10.1016/j.steroids.2019.108497. [DOI] [PubMed] [Google Scholar]
- 155.Sun H, et al. Cancer stem-like cells directly participate in vasculogenic mimicry channels in triple-negative breast cancer. Cancer Biol. Med. 2019;16:299–311. doi: 10.20892/j.issn.2095-3941.2018.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bao Z, Cheng Z, Chai D. The expressions of CD133, ALDH1, and vasculogenic mimicry in osteosarcoma and their clinical significance. Int. J. Clin. Exp. Pathol. 2018;11:3656–3663. [PMC free article] [PubMed] [Google Scholar]
- 157.Wang, H et al. ZEB1-mediated vasculogenic mimicry formation associates with epithelial-mesenchymal transition and cancer stem cell phenotypes in prostate cancer. J. Cell. Mol. Med.22, 3768–3781 (2018). [DOI] [PMC free article] [PubMed]
- 158.Wu H-B, et al. Autophagy-induced KDR/VEGFR-2 activation promotes the formation of vasculogenic mimicry by glioma stem cells. Autophagy. 2017;13:1528–1542. doi: 10.1080/15548627.2017.1336277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schnegg CI, Yang MH, Ghosh SK, Hsu M-Y. Induction of vasculogenic mimicry overrides VEGF-A silencing and enriches stem-like cancer cells in melanoma. Cancer Res. 2015;75:1682–1690. doi: 10.1158/0008-5472.CAN-14-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wei M-F, et al. Autophagy promotes resistance to photodynamic therapy-induced apoptosis selectively in colorectal cancer stem-like cells. Autophagy. 2014;10:1179–1192. doi: 10.4161/auto.28679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pagotto A, et al. Autophagy inhibition reduces chemoresistance and tumorigenic potential of human ovarian cancer stem cells. Cell Death Dis. 2017;8:e2943. doi: 10.1038/cddis.2017.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sharif T, et al. HDAC6 differentially regulates autophagy in stem-like versus differentiated cancer cells. Autophagy. 2019;15:686–706. doi: 10.1080/15548627.2018.1548547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sharif T, et al. Autophagic homeostasis is required for the pluripotency of cancer stem cells. Autophagy. 2017;13:264–284. doi: 10.1080/15548627.2016.1260808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ji J, et al. XIAP limits autophagic degradation of Sox2 and is a therapeutic target in nasopharyngeal carcinoma stem cells. Theranostics. 2018;8:1494–1510. doi: 10.7150/thno.21717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu, K. et al. Mitophagy controls the activities of tumor suppressor p53 to regulate hepatic cancer stem cells. Mol Cell. 68, 281–292 (2017). [DOI] [PMC free article] [PubMed]
- 168.Huang H, et al. Reciprocal network between cancer stem-like cells and macrophages facilitates the progression and androgen deprivation therapy resistance of prostate cancer. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2018;24:4612–4626. doi: 10.1158/1078-0432.CCR-18-0461. [DOI] [PubMed] [Google Scholar]
- 169.Stockwell BR, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xu Y, et al. Recent progress in nanotechnology based ferroptotic therapies for clinical applications. Eur. J. Pharm. 2020;880:173198. doi: 10.1016/j.ejphar.2020.173198. [DOI] [PubMed] [Google Scholar]
- 171.El Hout M, Dos Santos L, Hamaï A, Mehrpour M. A promising new approach to cancer therapy: targeting iron metabolism in cancer stem cells. Semin. Cancer Biol. 2018;53:125–138. doi: 10.1016/j.semcancer.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 172.Recalcati S, Gammella E, Cairo G. Dysregulation of iron metabolism in cancer stem cells. Free Radic. Biol. Med. 2019;133:216–220. doi: 10.1016/j.freeradbiomed.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 173.Basuli D, et al. Iron addiction: a novel therapeutic target in ovarian cancer. Oncogene. 2017;36:4089–4099. doi: 10.1038/onc.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu T, Jiang L, Tavana O, Gu W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 2019;79:1913–1924. doi: 10.1158/0008-5472.CAN-18-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Buccarelli M, et al. Inhibition of autophagy increases susceptibility of glioblastoma stem cells to temozolomide by igniting ferroptosis. Cell Death Dis. 2018;9:841. doi: 10.1038/s41419-018-0864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tesfay L, et al. Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Res. 2019;79:5355–5366. doi: 10.1158/0008-5472.CAN-19-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mai TT, et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat. Chem. 2017;9:1025–1033. doi: 10.1038/nchem.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shen M, et al. Chemotherapy-Induced extracellular vesicle miRNAs promote breast cancer stemness by targeting. Cancer Res. 2019;79:3608–3621. doi: 10.1158/0008-5472.CAN-18-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lin C-J, et al. The paracrine induction of prostate cancer progression by caveolin-1. Cell Death Dis. 2019;10:834. doi: 10.1038/s41419-019-2066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang Z, et al. Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210. Cell Oncol. (Dordr.) 2020;43:123–136. doi: 10.1007/s13402-019-00476-6. [DOI] [PubMed] [Google Scholar]
- 181.Liu C, et al. LSD1 stimulates cancer-associated fibroblasts to drive notch3-dependent self-renewal of liver cancer stem-like cells. Cancer Res. 2018;78:938–949. doi: 10.1158/0008-5472.CAN-17-1236. [DOI] [PubMed] [Google Scholar]
- 182.Su, S et al. CD10GPR77 Cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell172, 841–856 (2018). [DOI] [PubMed]
- 183.Jinushi M, et al. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc. Natl Acad. Sci. USA. 2011;108:12425–12430. doi: 10.1073/pnas.1106645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lee IC, Fadera S, Liu H-L. Strategy of differentiation therapy: effect of dual-frequency ultrasound on the induction of liver cancer stem-like cells on a HA-based multilayer film system. J. Mater. Chem. B. 2019;7:5401–5411. doi: 10.1039/C9TB01120J. [DOI] [PubMed] [Google Scholar]
- 185.Govaere O, et al. Laminin-332 sustains chemoresistance and quiescence as part of the human hepatic cancer stem cell niche. J. Hepatol. 2016;64:609–617. doi: 10.1016/j.jhep.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 186.Agata Y, et al. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int. Immunol. 1996;8:765–772. doi: 10.1093/intimm/8.5.765. [DOI] [PubMed] [Google Scholar]
- 187.Chang W-S, et al. Cutting edge: Programmed death-1/programmed death ligand 1 interaction regulates the induction and maintenance of invariant NKT cell anergy. J. Immunol. 2008;181:6707–6710. doi: 10.4049/jimmunol.181.10.6707. [DOI] [PubMed] [Google Scholar]
- 188.Wei, F et al. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. 450, 1–13 (2019). [DOI] [PubMed]
- 189.Schatton T, Frank MH. Antitumor immunity and cancer stem cells. Ann. N. Y Acad. Sci. 2009;1176:154–169. doi: 10.1111/j.1749-6632.2009.04568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schatton T, et al. Modulation of T-cell activation by malignant melanoma initiating cells. Cancer Res. 2010;70:697–708. doi: 10.1158/0008-5472.CAN-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.George S, et al. Loss of PTEN is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46:197–204. doi: 10.1016/j.immuni.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wei J, et al. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol. Cancer Ther. 2010;9:67–78. doi: 10.1158/1535-7163.MCT-09-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Maccalli C, Parmiani G, Ferrone S. Immunomodulating and immunoresistance properties of cancer-initiating cells: implications for the clinical success of immunotherapy. Immunol. Invest. 2017;46:221–238. doi: 10.1080/08820139.2017.1280051. [DOI] [PubMed] [Google Scholar]
- 194.Hui M, et al. The Hedgehog signalling pathway in breast development, carcinogenesis and cancer therapy. Breast Cancer Res. : BCR. 2013;15:203. doi: 10.1186/bcr3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Merchant AA, Matsui W. Targeting Hedgehog-a cancer stem cell pathway. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2010;16:3130–3140. doi: 10.1158/1078-0432.CCR-09-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bai X-Y, et al. Blockade of hedgehog signaling synergistically increases sensitivity to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer cell lines. PLoS ONE. 2016;11:e0149370. doi: 10.1371/journal.pone.0149370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang M, et al. Mithramycin represses basal and cigarette smoke-induced expression of ABCG2 and inhibits stem cell signaling in lung and esophageal cancer cells. Cancer Res. 2012;72:4178–4192. doi: 10.1158/0008-5472.CAN-11-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zeng X, et al. Targeting Hedgehog signaling pathway and autophagy overcomes drug resistance of BCR-ABL-positive chronic myeloid leukemia. Autophagy. 2015;11:355–372. doi: 10.4161/15548627.2014.994368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yoon C, et al. CD44 expression denotes a subpopulation of gastric cancer cells in which Hedgehog signaling promotes chemotherapy resistance. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2014;20:3974–3988. doi: 10.1158/1078-0432.CCR-14-0011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 200.Zhou H, et al. LncRNA-cCSC1 modulates cancer stem cell properties in colorectal cancer via activation of the Hedgehog signaling pathway. J. Cell Biochem. 2020;121:2510–2524. doi: 10.1002/jcb.29473. [DOI] [PubMed] [Google Scholar]
- 201.Ahmad A, et al. Inhibition of Hedgehog signaling sensitizes NSCLC cells to standard therapies through modulation of EMT-regulating miRNAs. J. Hematol. Oncol. 2013;6:77. doi: 10.1186/1756-8722-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Valenti G, et al. Cancer stem cells regulate cancer-associated fibroblasts via activation of hedgehog signaling in mammary gland tumors. Cancer Res. 2017;77:2134–2147. doi: 10.1158/0008-5472.CAN-15-3490. [DOI] [PubMed] [Google Scholar]
- 203.Fan Z, et al. PTK2 promotes cancer stem cell traits in hepatocellular carcinoma by activating Wnt/β-catenin signaling. Cancer Lett. 2019;450:132–143. doi: 10.1016/j.canlet.2019.02.040. [DOI] [PubMed] [Google Scholar]
- 204.Liu T, et al. Downregulation of DNMT3A by miR-708-5p Inhibits Lung Cancer Stem Cell-like Phenotypes through Repressing Wnt/β-catenin Signaling. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2018;24:1748–1760. doi: 10.1158/1078-0432.CCR-17-1169. [DOI] [PubMed] [Google Scholar]
- 205.Wang X, et al. PAF-Wnt signaling-induced cell plasticity is required for maintenance of breast cancer cell stemness. Nat. Commun. 2016;7:10633. doi: 10.1038/ncomms10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jin X, et al. Inhibition of ID1-BMPR2 intrinsic signaling sensitizes glioma stem cells to differentiation therapy. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2018;24:383–394. doi: 10.1158/1078-0432.CCR-17-1529. [DOI] [PubMed] [Google Scholar]
- 207.Liu A, et al. Antagonizing miR-455-3p inhibits chemoresistance and aggressiveness in esophageal squamous cell carcinoma. Mol. Cancer. 2017;16:106. doi: 10.1186/s12943-017-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lu H, et al. Targeting cancer stem cell signature gene SMOC-2 Overcomes chemoresistance and inhibits cell proliferation of endometrial carcinoma. EBioMedicine. 2019;40:276–289. doi: 10.1016/j.ebiom.2018.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yun E-J, et al. Targeting cancer stem cells in castration-resistant prostate cancer. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2016;22:670–679. doi: 10.1158/1078-0432.CCR-15-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Roy S, et al. Inhibition of CD44 sensitizes cisplatin-resistance and affects Wnt/β-catenin signaling in HNSCC cells. Int. J. Biol. Macromol. 2020;149:501–512. doi: 10.1016/j.ijbiomac.2020.01.131. [DOI] [PubMed] [Google Scholar]
- 211.Takebe N, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Capodanno Y, et al. Notch pathway inhibition targets chemoresistant insulinoma cancer stem cells. Endocr. -Relat. Cancer. 2018;25:131–144. doi: 10.1530/ERC-17-0415. [DOI] [PubMed] [Google Scholar]
- 213.Panaccione A, et al. NOTCH1 and SOX10 are essential for proliferation and radiation resistance of cancer stem-like cells in adenoid cystic carcinoma. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 2016;22:2083–2095. doi: 10.1158/1078-0432.CCR-15-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Muñoz-Galván S, et al. New markers for human ovarian cancer that link platinum resistance to the cancer stem cell phenotype and define new therapeutic combinations and diagnostic tools. J. Exp. Clin. cancer Res. : CR. 2019;38:234. doi: 10.1186/s13046-019-1245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hossain F, et al. Notch signaling regulates mitochondrial metabolism and NF-κB activity in triple-negative breast cancer cells via IKKα-dependent non-canonical pathways. Front. Oncol. 2018;8:575. doi: 10.3389/fonc.2018.00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hsu E-C, et al. Function of integrin-linked kinase in modulating the stemness of IL-6-abundant breast cancer cells by regulating γ-secretase-mediated notch1 activation in caveolae. Neoplasia. 2015;17:497–508. doi: 10.1016/j.neo.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Nayak A, et al. Nanoquinacrine sensitizes 5-FU-resistant cervical cancer stem-like cells by down-regulating Nectin-4 via ADAM-17 mediated NOTCH deregulation. Cell Oncol. (Dordr.) 2019;42:157–171. doi: 10.1007/s13402-018-0417-1. [DOI] [PubMed] [Google Scholar]
- 218.Prabakaran DS, et al. Silencing of FTS increases radiosensitivity by blocking radiation-induced Notch1 activation and spheroid formation in cervical cancer cells. Int J. Biol. Macromol. 2019;126:1318–1325. doi: 10.1016/j.ijbiomac.2018.09.114. [DOI] [PubMed] [Google Scholar]
- 219.Sansone P, et al. Evolution of Cancer Stem-like Cells in Endocrine-Resistant Metastatic Breast Cancers Is Mediated by Stromal Microvesicles. Cancer Res. 2017;77:1927–1941. doi: 10.1158/0008-5472.CAN-16-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yeh DW, et al. Interplay between Inflammation and Stemness in Cancer Cells: The Role of Toll-Like Receptor Signaling. J. Immunol. Res. 2016;2016:4368101. doi: 10.1155/2016/4368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Naugler WE, Karin M. NF-kappaB and cancer-identifying targets and mechanisms. Curr. Opin. Genet. Dev. 2008;18:19–26. doi: 10.1016/j.gde.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Biswas DK, et al. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc. Natl Acad. Sci. USA. 2004;101:10137–10142. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Garner JM, et al. Constitutive activation of signal transducer and activator of transcription 3 (STAT3) and nuclear factor kappaB signaling in glioblastoma cancer stem cells regulates the Notch pathway. J. Biol. Chem. 2013;288:26167–26176. doi: 10.1074/jbc.M113.477950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zakaria N, et al. Inhibition of NF-κB Signaling Reduces the Stemness Characteristics of Lung Cancer Stem Cells. Front. Oncol. 2018;8:166. doi: 10.3389/fonc.2018.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Parajuli B, et al. Salinomycin inhibits Akt/NF-kappaB and induces apoptosis in cisplatin resistant ovarian cancer cells. Cancer Epidemiol. 2013;37:512–517. doi: 10.1016/j.canep.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 226.Wu X, et al. AKR1B1 promotes basal-like breast cancer progression by a positive feedback loop that activates the EMT program. J. Exp. Med. 2017;214:1065–1079. doi: 10.1084/jem.20160903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhu H, et al. Role of the Hypoxia-inducible factor-1 alpha induced autophagy in the conversion of non-stem pancreatic cancer cells into CD133+ pancreatic cancer stem-like cells. Cancer Cell Int. 2013;13:119. doi: 10.1186/1475-2867-13-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liu S, et al. HER2 overexpression triggers an IL1α proinflammatory circuit to drive tumorigenesis and promote chemotherapy resistance. Cancer Res. 2018;78:2040–2051. doi: 10.1158/0008-5472.CAN-17-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Thakur B, Ray P. Cisplatin triggers cancer stem cell enrichment in platinum-resistant cells through NF-κB-TNFα-PIK3CA loop. J. Exp. Clin. Cancer Res. : CR. 2017;36:164. doi: 10.1186/s13046-017-0636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Guan R, et al. Advances in the studies of roles of Rho/Rho-kinase in diseases and the development of its inhibitors. Eur. J. Med. Chem. 2013;70:613–622. doi: 10.1016/j.ejmech.2013.10.048. [DOI] [PubMed] [Google Scholar]
- 231.Pranatharthi A, Ross C, Srivastava S. Cancer stem cells and radioresistance: Rho/ROCK pathway plea attention. Stem Cells Int. 2016;2016:5785786. doi: 10.1155/2016/5785786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yoon C, et al. Chemotherapy resistance in diffuse-type gastric adenocarcinoma is mediated by RhoA activation in cancer stem-like cells. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2016;22:971–983. doi: 10.1158/1078-0432.CCR-15-1356. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 233.Zhang S, et al. GPR56 drives colorectal tumor growth and promotes drug resistance through upregulation of MDR1 expression via a RhoA-mediated mechanism. Mol. Cancer Res. 2019;17:2196–2207. doi: 10.1158/1541-7786.MCR-19-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Krusche, B et al. EphrinB2 drives perivascular invasion and proliferation of glioblastoma stem-like cells. Elife5, e14845 (2016). [DOI] [PMC free article] [PubMed]
- 235.Zhang J, Liu S, Ye Q, Pan J. Transcriptional inhibition by CDK7/9 inhibitor SNS-032 abrogates oncogene addiction and reduces liver metastasis in uveal melanoma. Mol. Cancer. 2019;18:140. doi: 10.1186/s12943-019-1070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Dummer, R. et al. The 12-month analysis from basal cell carcinoma outcomes with LDE225 treatment (BOLT): A phase II, randomized, double-blind study of sonidegib in patients with advanced basal cell carcinoma. J. Am. Acad. Dermatol. 75, 113–125 (2016). [DOI] [PubMed]
- 237.Axelson M, et al. U.S. Food and Drug Administration approval: vismodegib for recurrent, locally advanced, or metastatic basal cell carcinoma. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2013;19:2289–2293. doi: 10.1158/1078-0432.CCR-12-1956. [DOI] [PubMed] [Google Scholar]
- 238.Wang, D. et al. Hedgehog pathway as a potential intervention target in esophageal cancer. Cancers (Basel). 11, 821–834 (2019). [DOI] [PMC free article] [PubMed]
- 239.Dréno B, et al. Two intermittent vismodegib dosing regimens in patients with multiple basal-cell carcinomas (MIKIE): a randomised, regimen-controlled, double-blind, phase 2 trial. Lancet Oncol. 2017;18:404–412. doi: 10.1016/S1470-2045(17)30072-4. [DOI] [PubMed] [Google Scholar]
- 240.Cazet AS, et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat. Commun. 2018;9:2897. doi: 10.1038/s41467-018-05220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Couban S, et al. A phase Ib study to assess the efficacy and safety of vismodegib in combination with ruxolitinib in patients with intermediate- or high-risk myelofibrosis. J. Hematol. Oncol. 2018;11:122. doi: 10.1186/s13045-018-0661-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ruiz-Borrego, M. et al. A phase Ib study of sonidegib (LDE225), an oral small molecule inhibitor of smoothened or Hedgehog pathway, in combination with docetaxel in triple negative advanced breast cancer patients: GEICAM/2012-12 (EDALINE) study. Invest. New Drugs. 37, 98–108 (2019). [DOI] [PubMed]
- 243.Lear JT, et al. Long-term efficacy and safety of sonidegib in patients with locally advanced and metastatic basal cell carcinoma: 30-month analysis of the randomized phase 2 BOLT study. J. Eur. Acad. Dermatol. Venereol. 2018;32:372–381. doi: 10.1111/jdv.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Gerds AT, et al. Phase 1/2 trial of glasdegib in patients with primary or secondary myelofibrosis previously treated with ruxolitinib. Leuk. Res. 2019;79:38–44. doi: 10.1016/j.leukres.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ueno H, et al. A phase I and pharmacokinetic study of taladegib, a Smoothened inhibitor, in Japanese patients with advanced solid tumors. Invest. N. Drugs. 2018;36:647–656. doi: 10.1007/s10637-017-0544-y. [DOI] [PubMed] [Google Scholar]
- 246.Bendell J, et al. Phase I study of LY2940680, a smo antagonist, in patients with advanced cancer including treatment-naïve and previously treated basal cell carcinoma. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2018;24:2082–2091. doi: 10.1158/1078-0432.CCR-17-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ko AH, et al. A phase I study of FOLFIRINOX plus IPI-926, a hedgehog pathway inhibitor, for advanced pancreatic adenocarcinoma. Pancreas. 2016;45:370–375. doi: 10.1097/MPA.0000000000000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Le, P. N., McDermott, J. D. & Jimeno, A. Targeting the Wnt pathway in human cancers: therapeutic targeting with a focus on OMP-54F28. Pharmacol Ther. 146, 1–11 (2015). [DOI] [PMC free article] [PubMed]
- 249.Jimeno A, et al. A first-in-human phase i study of the anticancer stem cell agent ipafricept (OMP-54F28), a decoy receptor for wnt ligands, in patients with advanced solid tumors. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2017;23:7490–7497. doi: 10.1158/1078-0432.CCR-17-2157. [DOI] [PubMed] [Google Scholar]
- 250.Moore KN, et al. A phase 1b dose escalation study of ipafricept (OMP54F28) in combination with paclitaxel and carboplatin in patients with recurrent platinum-sensitive ovarian cancer. Gynecol. Oncol. 2019;154:294–301. doi: 10.1016/j.ygyno.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ring, A. et al. CBP/β-catenin/FOXM1 Is a novel therapeutic target in triple negative breast cancer. Cancers (Basel). 10, 525–544 (2018). [DOI] [PMC free article] [PubMed]
- 252.Chan KC, et al. Therapeutic targeting of CBP/β-catenin signaling reduces cancer stem-like population and synergistically suppresses growth of EBV-positive nasopharyngeal carcinoma cells with cisplatin. Sci. Rep. 2015;5:9979. doi: 10.1038/srep09979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zhang Z, et al. Inhibition of the Wnt/β-catenin pathway overcomes resistance to enzalutamide in castration-resistant prostate cancer. Cancer Res. 2018;78:3147–3162. doi: 10.1158/0008-5472.CAN-17-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kartha VK, et al. Functional and genomic analyses reveal therapeutic potential of targeting β-catenin/CBP activity in head and neck cancer. Genome Med. 2018;10:54. doi: 10.1186/s13073-018-0569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Kimura K, et al. Safety, tolerability, and preliminary efficacy of the anti-fibrotic small molecule PRI-724, a CBP/β-catenin inhibitor, in patients with hepatitis c virus-related cirrhosis: a single-center, open-label, dose escalation phase 1 trial. EBioMedicine. 2017;23:79–87. doi: 10.1016/j.ebiom.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Pak S, et al. The small molecule WNT/β-catenin inhibitor CWP232291 blocks the growth of castration-resistant prostate cancer by activating the endoplasmic reticulum stress pathway. J. Exp. Clin. Cancer Res. : CR. 2019;38:342. doi: 10.1186/s13046-019-1342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Cook N, et al. A phase I trial of the γ-secretase inhibitor MK-0752 in combination with gemcitabine in patients with pancreatic ductal adenocarcinoma. Br. J. Cancer. 2018;118:793–801. doi: 10.1038/bjc.2017.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Schott AF, et al. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2013;19:1512–1524. doi: 10.1158/1078-0432.CCR-11-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Rice MA, et al. Loss of Notch1 activity inhibits prostate cancer growth and metastasis and sensitizes prostate cancer cells to antiandrogen therapies. Mol. Cancer Ther. 2019;18:1230–1242. doi: 10.1158/1535-7163.MCT-18-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Low, H.-Y. et al. Reciprocal regulation between indoleamine 2,3-dioxigenase 1 and Notch1 involved in radiation response of cervical cancer stem cells. Cancers (Basel). 12, 1547–1563 (2020). [DOI] [PMC free article] [PubMed]
- 261.Simões BM, et al. Anti-estrogen resistance in human breast tumors is driven by JAG1-NOTCH4-dependent cancer stem cell activity. Cell Rep. 2015;12:1968–1977. doi: 10.1016/j.celrep.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Xu R, et al. Molecular and clinical effects of notch inhibition in glioma patients: a phase 0/I trial. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2016;22:4786–4796. doi: 10.1158/1078-0432.CCR-16-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Pan E, et al. Phase I study of RO4929097 with bevacizumab in patients with recurrent malignant glioma. J. Neurooncol. 2016;130:571–579. doi: 10.1007/s11060-016-2263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wu CX, et al. Notch inhibitor PF-03084014 inhibits hepatocellular carcinoma growth and metastasis via suppression of cancer stemness due to reduced activation of Notch1-Stat3. Mol. Cancer Ther. 2017;16:1531–1543. doi: 10.1158/1535-7163.MCT-17-0001. [DOI] [PubMed] [Google Scholar]
- 265.Wang L, et al. Inhibition of Notch pathway enhances the anti-tumor effect of docetaxel in prostate cancer stem-like cells. Stem Cell Res. Ther. 2020;11:258. doi: 10.1186/s13287-020-01773-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Yabuuchi S, et al. Notch signaling pathway targeted therapy suppresses tumor progression and metastatic spread in pancreatic cancer. Cancer Lett. 2013;335:41–51. doi: 10.1016/j.canlet.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zhang CC, et al. Synergistic effect of the γ-secretase inhibitor PF-03084014 and docetaxel in breast cancer models. Stem Cells Transl. Med. 2013;2:233–242. doi: 10.5966/sctm.2012-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Locatelli MA, et al. Phase I study of the gamma secretase inhibitor PF-03084014 in combination with docetaxel in patients with advanced triple-negative breast cancer. Oncotarget. 2017;8:2320–2328. doi: 10.18632/oncotarget.13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Villalobos VM, et al. Long-term follow-up of desmoid fibromatosis treated with PF-03084014, an oral gamma secretase inhibitor. Ann. Surg. Oncol. 2018;25:768–775. doi: 10.1245/s10434-017-6082-1. [DOI] [PubMed] [Google Scholar]
- 270.Kummar S, et al. Clinical activity of the γ-secretase inhibitor PF-03084014 in adults with desmoid tumors (aggressive fibromatosis) J. Clin. Oncol. 2017;35:1561–1569. doi: 10.1200/JCO.2016.71.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.McKeage MJ, et al. Phase IB trial of the anti-cancer stem cell DLL4-binding agent demcizumab with pemetrexed and carboplatin as first-line treatment of metastatic non-squamous NSCLC. Target Oncol. 2018;13:89–98. doi: 10.1007/s11523-017-0543-0. [DOI] [PubMed] [Google Scholar]
- 272.Coleman RL, et al. Demcizumab combined with paclitaxel for platinum-resistant ovarian, primary peritoneal, and fallopian tube cancer: The SIERRA open-label phase Ib trial. Gynecol. Oncol. 2020;157:386–391. doi: 10.1016/j.ygyno.2020.01.042. [DOI] [PubMed] [Google Scholar]
- 273.Azmi AS, et al. Preclinical assessment with clinical validation of selinexor with gemcitabine and nab-paclitaxel for the treatment of pancreatic ductal adenocarcinoma. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 2020;26:1338–1348. doi: 10.1158/1078-0432.CCR-19-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Chari A, et al. Oral selinexor-dexamethasone for triple-class refractory multiple myeloma. N. Engl. J. Med. 2019;381:727–738. doi: 10.1056/NEJMoa1903455. [DOI] [PubMed] [Google Scholar]
- 275.Bertocchi P, et al. Chemotherapy rechallenge after regorafenib treatment in metastatic colorectal cancer: still hope after the last hope? J. Chemother. (Florence, Italy) 2017;29:102–105. doi: 10.1080/1120009X.2016.1247205. [DOI] [PubMed] [Google Scholar]
- 276.Wilhelm SM, et al. Regorafenib (BAY 73-4506): a new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int. J. Cancer. 2011;129:245–255. doi: 10.1002/ijc.25864. [DOI] [PubMed] [Google Scholar]
- 277.Abou-Elkacem L, et al. Regorafenib inhibits growth, angiogenesis, and metastasis in a highly aggressive, orthotopic colon cancer model. Mol. Cancer Ther. 2013;12:1322–1331. doi: 10.1158/1535-7163.MCT-12-1162. [DOI] [PubMed] [Google Scholar]
- 278.Cai MH, et al. Regorafenib suppresses colon tumorigenesis and the generation of drug resistant cancer stem-like cells via modulation of miR-34a associated signaling. J. Exp. Clin. Cancer Res. : CR. 2018;37:151. doi: 10.1186/s13046-018-0836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Mayer B, et al. A marginal anticancer effect of regorafenib on pancreatic carcinoma cells in vitro, ex vivo, and in vivo. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2017;390:1125–1134. doi: 10.1007/s00210-017-1412-1. [DOI] [PubMed] [Google Scholar]
- 280.Takigawa H, et al. Multikinase inhibitor regorafenib inhibits the growth and metastasis of colon cancer with abundant stroma. Cancer Sci. 2016;107:601–608. doi: 10.1111/cas.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Her Z, et al. An improved pre-clinical patient-derived liquid xenograft mouse model for acute myeloid leukemia. J. Hematol. Oncol. 2017;10:162. doi: 10.1186/s13045-017-0532-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Hu F, et al. Lung adenocarcinoma resistance to therapy with EGFR‑tyrosine kinase inhibitors is related to increased expression of cancer stem cell markers SOX2, OCT4 and NANOG. Oncol. Rep. 2020;43:727–735. doi: 10.3892/or.2019.7454. [DOI] [PubMed] [Google Scholar]
- 283.Si J, et al. Shisa3 brakes resistance to EGFR-TKIs in lung adenocarcinoma by suppressing cancer stem cell properties. J. Exp. Clin. Cancer Res. : CR. 2019;38:481. doi: 10.1186/s13046-019-1486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Suzuki, S. et al. Brexpiprazole, a serotonin-dopamine activity modulator, can sensitize glioma stem cells to osimertinib, a third-generation EGFR-TKI, via survivin reduction. Cancers (Basel). 11, 947–960 (2019). [DOI] [PMC free article] [PubMed]
- 285.Gounder MM, et al. Sorafenib for advanced and refractory desmoid tumors. N. Engl. J. Med. 2018;379:2417–2428. doi: 10.1056/NEJMoa1805052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Mendez-Blanco C, et al. Sorafenib resistance in hepatocarcinoma: role of hypoxia-inducible factors. Exp. Mol. Med. 2018;50:134. doi: 10.1038/s12276-018-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Feldt S, et al. Incidence of thyroid hormone therapy in patients treated with sunitinib or sorafenib: a cohort study. Eur. J. Cancer (Oxf., Engl. : 1990). 2012;48:974–981. doi: 10.1016/j.ejca.2012.01.036. [DOI] [PubMed] [Google Scholar]
- 288.Huang M, et al. Targeting KDM1A attenuates Wnt/beta-catenin signaling pathway to eliminate sorafenib-resistant stem-like cells in hepatocellular carcinoma. Cancer Lett. 2017;398:12–21. doi: 10.1016/j.canlet.2017.03.038. [DOI] [PubMed] [Google Scholar]
- 289.Wu CX, et al. Blocking CDK1/PDK1/beta-Catenin signaling by CDK1 inhibitor RO3306 increased the efficacy of sorafenib treatment by targeting cancer stem cells in a preclinical model of hepatocellular carcinoma. Theranostics. 2018;8:3737–3750. doi: 10.7150/thno.25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Lo J, et al. Nuclear factor kappa B-mediated CD47 up-regulation promotes sorafenib resistance and its blockade synergizes the effect of sorafenib in hepatocellular carcinoma in mice. Hepatology (Baltim., Md.). 2015;62:534–545. doi: 10.1002/hep.27859. [DOI] [PubMed] [Google Scholar]
- 291.Rausch V, et al. Synergistic activity of sorafenib and sulforaphane abolishes pancreatic cancer stem cell characteristics. Cancer Res. 2010;70:5004–5013. doi: 10.1158/0008-5472.CAN-10-0066. [DOI] [PubMed] [Google Scholar]