Abstract
Rationale: The 17q12–21.1 locus is one of the most highly replicated genetic associations with asthma. Individuals of African descent have lower linkage disequilibrium in this region, which could facilitate identifying causal variants.
Objectives: To identify functional variants at 17q12–21.1 associated with early-onset asthma among African American individuals.
Methods: We evaluated African American participants from SAPPHIRE (Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race–Ethnicity) (n = 1,940), SAGE II (Study of African Americans, Asthma, Genes and Environment) (n = 885), and GCPD-A (Study of the Genetic Causes of Complex Pediatric Disorders–Asthma) (n = 2,805). Associations with asthma onset at ages under 5 years were meta-analyzed across cohorts. The lead signal was reevaluated considering haplotypes informed by genetic ancestry (i.e., African vs. European). Both an expression-quantitative trait locus analysis and a phenome-wide association study were performed on the lead variant.
Measurements and Main Results: The meta-analyzed results from SAPPHIRE, SAGE II, and the GCPD-A identified rs11078928 as the top association for early-onset asthma. A haplotype analysis suggested that the asthma association partitioned most closely with the rs11078928 genotype. Genetic ancestry did not appear to influence the effect of this variant. In the expression-quantitative trait locus analysis, rs11078928 was related to alternative splicing of GSDMB (gasdermin-B) transcripts. The phenome-wide association study of rs11078928 suggested that this variant was predominantly associated with asthma and asthma-associated symptoms.
Conclusions: A splice-acceptor polymorphism appears to be a causal variant for asthma at the 17q12–21.1 locus. This variant appears to have the same magnitude of effect in individuals of African and European descent.
Keywords: asthma, African Americans, chromosome 17, GSDMB, ORMDL3
At a Glance Commentary
Scientific Knowledge on the Subject
Attempts to identify causal variants for asthma at the 17q12–21.1 locus have been hindered by the high degree of linkage disequilibrium among individuals of European descent, who predominate most genetic studies of asthma. It has been postulated that lower linkage disequilibrium at 17q12–21.1 among individuals of African descent may assist in identifying causal asthma variants. Existing association studies in African Americans have relied on candidate genotyping or commercial arrays, thereby requiring tagging or imputation to characterize this region.
What This Study Adds to the Field
This is the first study using whole-genome sequence data to characterize the asthma association signal at 17q12–21.1 by meta-analyzing data from three large cohorts of African American individuals. Asthma risk was localized to a small intragenic region in GSDMB (gasdermin-B). An expression analysis using whole-blood transcriptome data from African Americans demonstrated that the top SNP association, rs11078928, was functionally related to alternative splicing of GSDMB transcripts. A phenome-wide association analysis provided further support for the selective role of this variant in asthma. This study provides strong evidence for a causal variant underlying the asthma association signal at 17q12–21.1 and thereby focuses the effort to develop targeted asthma treatments.
Asthma is a common condition affecting over 300 million individuals worldwide (1–3). Although asthma can occur at any age, epidemiologic studies suggest that new cases peak early in life and again in adulthood (4–6). Therefore, the pathologic mechanisms that contribute to asthma development likely differ by age (7, 8). A large twin study showed that the likelihood of asthma cooccurring in monozygotic twins decreased with age of disease onset, implying that genetics play a larger role in early-life asthma (9, 10). Similarly, London and colleagues showed that family history was significantly associated with asthma onset at any age with the largest effect sizes in early-onset persistent asthma (11).
Genome-wide association studies have repeatedly identified a relationship between chromosomal region 17q12–21.1 and asthma status (12–16). Subsequent work suggests that this area is associated with age of asthma onset (17), specifically childhood-onset asthma (18). However, fine mapping the 17q12–21.1 locus to determine the causative variant or gene has proven to be difficult, especially among individuals of European or East Asian descent in whom a large stretch (∼100–200 kb in length) of the region is in strong linkage disequilibrium (LD) (19). This LD structure results in a high degree of correlation between variants, such as SNPs. In contrast, LD between genetic variants at 17q12–21.1 among African Americans tends to be much lower, suggesting that association studies in this group may be more successful in uncovering causal variants (19). Here we used whole-genome sequencing (WGS) data and transcriptomic data from African American participants in three large cohort studies to more fully evaluate the 17q12–21.1 locus.
Methods
Study Populations
This study included cohorts participating in the Asthma Translational Genomics Collaborative. As part of the NHLBI’s Trans-Omics for Precision Medicine program, WGS data were generated on Asthma Translational Genomics Collaborative cohorts. The studies included in the current analysis are as follows: SAPPHIRE (Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race–Ethnicity), SAGE II (Study of African Americans, Asthma, Genes and Environment), and the GCPD-A (Study of the Genetic Causes of Complex Pediatric Disorders–Asthma). All of these studies were approved by their respective institutional review boards. Written informed consent (and written assent for minors) was obtained for each study participant before the collection and use of their data. A description of these cohorts can be found elsewhere and in the in the online supplement (20–22). Except when otherwise specified, individuals included in this analysis were African American by self-report. “European American” refers to individuals who identified as non-Hispanic white.
Population Structure and LD
DNA isolation, sequencing, read alignment, and variant calls are discussed in the online supplement. We used the R packages PC-AiR and PC-relate (R Foundation for Statistical Computing) to estimate underlying population structure in our study population (23, 24). Principal components (PCs) are axes of variation that reflect underlying population structure attributable to biogeographic ancestry and admixture (25). Local ancestry (i.e., African or European ancestry at each SNP location) was estimated using the program RFMix (26). The software package KING was used to estimate relatedness between participants (27); we randomly excluded one individual among pairs in which the coefficient of relationship was ≥0.088 (i.e., greater than or equal to a second degree relationship). Scree plots were used to determine the number of PCs characterizing underlying population structure in the study cohorts (28). These analyses suggested no additional population structure was captured after the first three PCs (data not shown); therefore, only three PCs were included in our analytic models. Figure E1 in the online supplement shows the plots of the first two PCs against the following reference populations from the 1,000 Genomes Project: the Yoruba population in Ibadan, Nigeria; Utah residents with Northern and Western European ancestry; and Han Chinese population in Beijing, China (29).
LD plots were created using the program Haploview 4.2 (Broad Institute) (30), and haplotype blocks were defined using the approach described by Gabriel and colleagues (31) To distinguish the different LD structures by ancestry, we generated plots for the following groups: African American individuals homozygous for African ancestry across the 17q12–21.1 locus, African American individuals homozygous for European ancestry across the 17q12–21.1 locus, and European American SAPPHIRE participants.
RNA-Sequence Pipeline
A portion of the blood collected from SAPPHIRE participants at the time of enrollment was stored in PAXgene Blood RNA tubes (BD Biosciences) for later transcriptomic analyses. Sequencing libraries were constructed using TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Globin (Illumina) on 417 African American individuals with asthma and 427 healthy African American control subjects. RNA libraries were sequenced on Illumina HiSeq machines with v4 kits. The software program HISAT2 2.1.0 was used to map reads to human-genome build GRCh38.p5 (32, 33), and mapped reads were quantified at the transcript level using StringTie v1.3.3 (34, 35). Gene and splice-variant expression were measured as transcripts per kilobase million. When using gene expression as an outcome for the regression models, transcripts-per-kilobase-million values were natural log–transformed. We limited our analysis to genes with one or more read counts in at least 10% of study individuals.
Statistical Analysis
We performed a case–control meta-analysis of the 17q12–21.1 region using case patients and unaffected control subjects from the SAPPHIRE, SAGE II, and GCPD-A cohorts. Each cohort was adjusted for sex and the first three PCs. Results were meta-analytically combined assuming a fixed-effect model and using the program METAL (36). We focused on a 1-Mb region bounded by positions 39,500,000 and 40,500,000 within the chromosomal region 17q12–21.1 (Genome Reference Consortium Human Build 38-17q12 positions 33,500,001–39,800,000 and 17q21.1 positions 39,800,001–40,200,000). We limited our evaluation to biallelic variants with a minor allele frequency ≥ 1% in control subjects. In total, 4,001 variants were retained for analysis. Analyses assumed an additive genetic model. Early asthma cases were defined as an age of onset <5 years. This age range was found to have the strongest association with asthma in an earlier candidate-variant study of the 17q21 region (37). Locus zoom plots were created using the program available at https://github.com/pgxcentre/region-plot and used elsewhere (38).
Functional variants in the haplotype block containing the lead variant from above meta-analysis were used to construct haplotypes that were assessed for their association with asthma onset at age <5 years. Haplotypes were assessed within each cohort, and the results were meta-analytically combined.
RNA-sequence (RNA-seq) data generated from whole blood RNA in SAPPHIRE participants were used to determine if rs11078928 was an expression-quantitative trait locus (eQTL) for regional genes and transcript isoforms. Linear regression was used to test the association between expression and genotype adjusting for patient age (at the time of sample collection), sex, RNA-seq batch, and probabilistic estimation of expression residuals (39).
The entire GCPD (Study of the Genetic Causes of Complex Pediatric Disorders) cohort was used for a phenome-wide association study (PheWAS). Logistic and linear regression was used to evaluate the association between codified clinical outcomes from the electronic medical record and rs11078928 genotype. Analyses were preformed within each population group (adjusted for patient age, sex, type of commercial array used, and the first 10 PCs) and were then meta-analytically combined.
Association analyses were performed using PLINK and R statistical software (40, 41). For single-variant association, we used a P value threshold of <2.86 × 10−5, which was derived using the genetic type I error calculator GEC (42). For the haplotype analysis, we used a P value threshold of 0.003 (0.05/16). The PheWAS meta-analysis was performed using the R package PheWAS. The P value thresholds among African Americans, Asians, European Americans, Latino individuals, and all groups meta-analyzed were 4.30 × 10−5, 1.16 × 10−4, 3.81 × 10−5, 1.14 × 10−4, and 3.67 × 10−5, respectively. Statistical significance for the eQTL and expression analyses was derived using the R package, fdrtool (43); associations with a false discovery rate (FDR)-adjusted P value < 0.05 were considered statistically significant.
Results
Sample Characteristics
Table 1 shows the characteristics of participants enrolled in the three study cohorts: SAPPHIRE (1,143 case patients, 797 control subjects), SAGE II (393 case patients, 492 control subjects), and GCPD-A (1,042 case patients, 1,763 control subjects). Case patients were those individuals with asthma onset at an age <5 years. SAPPHIRE participants were older at the time of study enrollment when compared with SAGE II and GCPD-A participants; the latter two studies almost exclusively enrolled children.
Table 1.
Characteristics of African American Study Participants Stratified by Cohort and Asthma Status
| Variable | SAPPHIRE Cohort |
SAGE II Cohort |
GCPD-A |
P Value for the Comparison of Cases across Cohorts* | |||
|---|---|---|---|---|---|---|---|
| Case Patients (n = 1,143) | Control Subjects (n = 797) | Case Patients (n = 393) | Control Subjects (n = 492) | Case Patients (n = 1,042) | Control Subjects (n = 1,763) | ||
| Age at enrollment, yr† | 25.57 ± 12.74 | 39.45 ± 12.59 | 13.28 ± 3.58 | 15.82 ± 3.73 | 5.29 ± 4.25 | 9.88 ± 5.48 | <0.001 |
| Sex, F | 622 (54.4) | 542 (68.0) | 180 (45.8) | 289 (57.3) | 588 (56.4) | 879 (49.4) | <0.001 |
| Proportion of African ancestry‡ | 0.80 ± 0.11 | 0.81 ± 0.10 | 0.77 ± 0.13 | 0.78 ± 0.12 | 0.70 ± 0.14 | 0.70 ± 0.15 | <0.001 |
| BMI, kg/m2† | 30.53 ± 9.45 | 31.78 ± 7.86 | 23.80 ± 6.56 | 24.81 ± 6.91 | 20.5 ± 6.18 | 20.9 ± 6.39 | <0.001 |
| BMI percentile§ | — | — | 76.28 ± 24.15 | 70.64 ± 26.98 | — | — | — |
| Percentage of predicted FEV1, %† | 87.05 ± 19.75 | 96.26 ± 15.34 | 98.86 ± 13.81 | 103.45 ± 13.16 | 89.45 ± 18.92 | 99.10 ± 8.75 | <0.001 |
| Age of asthma onset‖ | |||||||
| 0–1 yr | 740 (64.7) | — | 161 (41.0) | — | 219 (21) | — | <0.001 |
| 2–4 yr | 403 (35.3) | — | 232 (59.0) | — | 823 (79) | — | |
| Average age of asthma onset, yr | 1.45 ± 1.23 | — | 2.0 ± 1.30 | — | 2.12 ± 1.25 | — | |
| ACT score ≤ 19¶ | 574 (50.2) | — | — | — | — | — | — |
Definition of abbreviations: ACT = Asthma Control Test; BMI = body mass index; GCPD-A = Study of the Genetic Causes of Complex Pediatric Disorders–Asthma; SAGE II = Study of African Americans, Asthma, Genes and Environment; SAPPHIRE = Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race–Ethnicity.
Data are shown as mean ± SD or n (%).
Differences in patient characteristics among individuals with asthma from the three cohorts were assessed using a chi-squared test for categorical variables and ANOVA for continuous variables.
Measured at the time of study enrollment.
Proportion of African ancestry was estimated using genome-wide autosomal polymorphisms.
BMI percentile was estimated using growth charts specific for age and sex.
Available at http://www.cdc.gov/nccdphp/dnpao/growthcharts/resources/sas.htm. Age of asthma onset was based on patient or parent self-report.
A composite ACT score ≤ 19 indicates poorly controlled asthma.
Meta-analysis of 17q12–21.1 Variants Associated with Asthma Onset at <5 Years of Age
We assessed genetic variants on chromosome 17 between positions 39,500,000 and 40,500,000 for association with asthma onset at the age of <5 years (i.e., case patients vs. healthy control subjects); this region was selected because it encompasses the broad asthma signal first identified in the 17q region by Moffatt and colleagues (12). We evaluated associations in the SAPPHIRE, SAGE II, and GCPD-A cohorts separately and then meta-analytically combined these results (Table 2; for full results, see Table E1 in the online supplement). The quantile-quantile plots for associations at the 17q12–21.1 locus are shown in Figure E2; the observed deviation is consistent with the high degree of regional LD, as demonstrated after LD pruning. The top 78 early asthma associations at the 17q12–21.1 locus were common variants (minor allele frequency > 5%). The lead association from the meta-analysis was rs11078928, a purported intronic splice-acceptor variant. A locus zoom plot of the 17q12–21.1 locus of the meta-analysis results is shown in Figure 1A; associations in this region were markedly diminished after adjusting by rs11078928 genotype (Figure 1B and Table E1).
Table 2.
Meta-analysis of Associations with Early Asthma Age at the 17q12–21.1 Locus among Three Cohorts with African American Participants (n = 5,630)*
| Association Rank | Variant | Chromosome 17 Position† | Allele 1 | Allele 2 | Allele 1 Frequency‡ | Odds Ratio | P Value§ | R2 with rs11078928‖ | P Value, Adjusted¶ |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs11078928 | 39,908,216 | C | T | 0.147 | 0.734 | 1.47 × 10−7 | 1.000 | NA |
| 2 | rs34120102 | 39,869,782 | A | G | 0.151 | 0.737 | 1.75 × 10−7 | 0.954 | 7.62 × 10−1 |
| 3 | rs12949100 | 39,900,936 | A | G | 0.147 | 0.737 | 1.99 × 10−7 | 1.000 | 3.75 × 10−1 |
| 4 | rs12232497 | 39,883,866 | C | T | 0.152 | 0.739 | 2.18 × 10−7 | 0.964 | 9.88 × 10−1 |
| 5 | rs35736272 | 39,876,427 | C | T | 0.152 | 0.741 | 2.58 × 10−7 | 0.964 | 8.44 × 10−1 |
| 6 | rs12939832 | 39,908,623 | A | G | 0.146 | 0.739 | 2.67 × 10−7 | 0.995 | 3.40 × 10−1 |
| 7 | rs2305480 | 39,905,943 | A | G | 0.154 | 0.747 | 4.04 × 10−7 | 0.950 | 9.68 × 10−1 |
| 8 | rs4795398 | 39,881,926 | T | C | 0.154 | 0.747 | 4.33 × 10−7 | 0.951 | 7.59 × 10−1 |
| 9 | rs35569035 | 39,879,371 | T | C | 0.154 | 0.748 | 5.40 × 10−7 | 0.951 | 6.31 × 10−1 |
| 10 | rs12936409 | 39,887,396 | T | C | 0.154 | 0.749 | 5.62 × 10−7 | 0.951 | 6.01 × 10−1 |
| 11 | rs10852935 | 39,875,421 | T | C | 0.156 | 0.751 | 6.59 × 10−7 | 0.938 | 6.64 × 10−1 |
| 12 | rs34189114 | 39,876,207 | T | C | 0.164 | 0.762 | 1.41 × 10−6 | 0.885 | 7.17 × 10−1 |
| 13 | rs34074973 | 39,879,513 | G | GAGA | 0.164 | 0.763 | 1.60 × 10−6 | 0.885 | 6.64 × 10−1 |
| 14 | rs11557466 | 39,868,373 | T | C | 0.164 | 0.763 | 1.64 × 10−6 | 0.882 | 6.66 × 10−1 |
| 15 | rs4795400 | 39,910,767 | T | C | 0.169 | 0.770 | 1.64 × 10−6 | 0.856 | 9.00 × 10−1 |
| 16 | rs11078925 | 39,868,955 | C | T | 0.164 | 0.763 | 1.65 × 10−6 | 0.885 | 6.56 × 10−1 |
| 17 | rs5820308 | 39,913,111 | TCAAAA | T | 0.163 | 0.771 | 2.29 × 10−6 | 0.868 | 9.56 × 10−1 |
| 18 | rs62067029 | 39,882,138 | T | A | 0.154 | 0.686 | 2.58 × 10−6 | 0.951 | 3.38 × 10−1 |
| 19 | rs17608925 | 39,926,578 | C | T | 0.060 | 0.666 | 6.76 × 10−6 | 0.340 | 6.37 × 10−2 |
| 20 | rs59716545 | 39,875,604 | G | T | 0.164 | 0.706 | 9.04 × 10−6 | 0.885 | 7.63 × 10−1 |
| 21 | rs36000226 | 39,907,676 | C | T | 0.187 | 0.794 | 1.14 × 10−5 | 0.752 | 9.18 × 10−1 |
| 22 | rs2305479 | 39,905,964 | T | C | 0.187 | 0.795 | 1.20 × 10−5 | 0.752 | 8.92 × 10−1 |
| 23 | rs883770 | 39,907,128 | T | C | 0.187 | 0.795 | 1.26 × 10−5 | 0.752 | 8.97 × 10−1 |
| 24 | rs8076131 | 39,924,659 | G | A | 0.187 | 0.797 | 1.33 × 10−5 | 0.737 | 9.44 × 10−1 |
| 25 | rs56750287 | 39,906,691 | C | A | 0.179 | 0.794 | 1.42 × 10−5 | 0.794 | 6.25 × 10−1 |
| 26 | rs62067034 | 39,907,485 | T | C | 0.187 | 0.797 | 1.44 × 10−5 | 0.752 | 8.44 × 10−1 |
| 27 | rs11651596 | 39,899,863 | C | T | 0.255 | 0.818 | 1.49 × 10−5 | 0.525 | 3.77 × 10−1 |
| 28 | rs907092 | 39,766,006 | A | G | 0.174 | 0.792 | 1.53 × 10−5 | 0.716 | 9.62 × 10−1 |
| 29 | rs4795399 | 39,905,186 | C | T | 0.154 | 0.709 | 1.68 × 10−5 | 0.950 | 4.87 × 10−1 |
Definition of abbreviation: NA = not applicable.
Associations meta-analyzed across GCPD-A (Study of the Genetic Causes of Complex Pediatric Disorders–Asthma), SAGE II (Study of African Americans, Asthma, Genes and Environment), and SAPPHIRE (Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race–Ethnicity) using fixed-effect model. Case patients were individuals with reported asthma onset at an age of <5 years as compared with healthy control subjects. Associations below the threshold of P < 2.86 × 10−5 are listed.
Positions based on Genome Reference Consortium Human Build 38.
Allele frequencies are based on results from the participants without asthma in SAPPHIRE cohort.
Genotypes were analyzed using an additive model for the number of copies of allele 1 (coded as 0, 1, or 2); models were adjusted for patient sex and the first three principal components for population structure.
Linkage disequilibrium between given variant and rs11078928. Values of 1 imply perfect correlation between markers.
P value for the genotype association with asthma status after adjusting for rs11078928 genotype.
Figure 1.
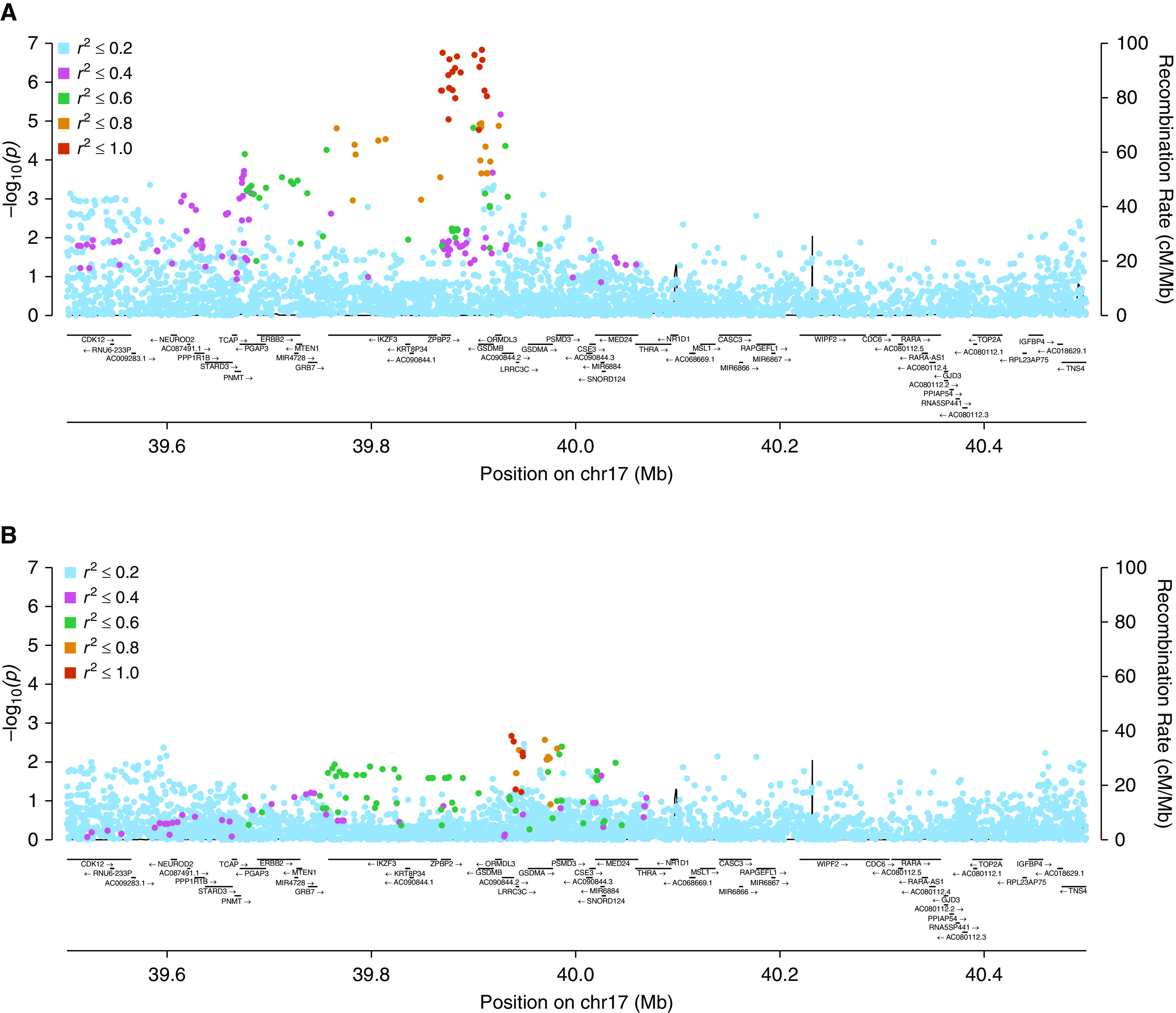
Locus zoom plots of variants in the chromosome 17q12–21.1 region and the association with asthma onset at age <5 years versus control subjects among African American participants from the (A) three-cohort meta-analysis and (B) after conditioning on the rs11078928 genotype. chr17 = chromosome 17; GSDMB = gasdermin-B; ZPBP2 = zona pellucida–binding protein 2.
Population Group Differences at 17q12–21.1
The LD pattern at 17q12–21.1 for common variants among African American SAPPHIRE participants homozygous for European ancestry (n = 174) is shown in Figure 2A (Figure E3); the LD pattern for African Americans homozygous for African ancestry (n = 507) is shown in Figure 2B (Figure E4). The LD pattern for European American SAPPHIRE participants (n = 132) is shown in Figure E5. Variants residing within the transcribed portions of GSDMB (gasdermin-B) and ORMDL3 are highlighted in blue and yellow, respectively, in Figures E3–E5. Differences in LD patterns between the groups are shown graphically in Figures E6–E8. These plots show the differences in LD structure between European and African ancestry (Figures E6 and E7) and show the near-complete LD concordance between individuals homozygous for European ancestry at this locus, regardless of race (Figure E8). The case–control associations for rs11078928 and early-onset asthma in these groups were as follows: odds ratio (OR), 0.59 (P = 0.163) in African Americans homozygous for European ancestry at 17q12–21.1; OR, 0.57 (P = 0.005) in African Americans homozygous for African ancestry at 17q12–21.1; and OR, 0.59 (P = 0.148) in European Americans.
Figure 2.
Linkage disequilibrium (LD) between variants at the 17q12–21.1 locus among African American SAPPHIRE (Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race–Ethnicity) participants without asthma but (A) homozygous for European ancestry and (B) homozygous for African ancestry. The haplotype block containing the most significant association for early-onset asthma is noted (black circle) and (C) shown in greater detail. The degree of pairwise LD (r2 × 100) between two markers is shown in each square; darker shaded squares and higher numbers (range, 0–100) denote higher LD.
The haplotype block including rs11078928 was similar for African America individuals homozygous for European ancestry and European American participants (>100 kb) (Figures E3 and E5). Among African American individuals, the haplotype block was much smaller (∼4 kb) (Figure 2C), suggesting that the asthma signal localized to a region spanning introns 3–10 of GSDMB. The 4-kb block included four potentially functional polymorphisms: spice-acceptor variant rs11078928, adjacent to exon 6, and three missense mutations in exon 9 (rs2305479, rs2305480, and rs16965388). Haplotypes with these four variants were evaluated for their association with asthma onset at the age of <5 years in SAPPHIRE, SAGE II, and GCPD-A (Table 3). Haplotypes containing the minor alleles for rs2305479, rs2305480, and rs16965388 without the minor allele for rs11078928 (i.e., haplotypes 2, 3, and 4 in Table 3) were not significantly associated with early-onset asthma when compared with the haplotype containing the major allele for all 4 variants. The only significant haplotype in all three cohorts individually and combined contained the minor C-allele of rs11078928 (SAPPHIRE: OR, 0.67; [P = 6.33 × 10−5]; SAGE II: OR, 0.71 [P = 0.017]; GCPD-A: OR, 0.78 [P = 4.78 × 10−4]; meta-analysis: OR, 0.73 [P = 8.94 × 10−8]).
Table 3.
Association between Early-Onset Asthma (Age < 5 yr) and Haplotypes among African American Study Participants
| Haplotype* | Meta-analysis |
SAPPHIRE |
SAGE II |
GCPD-A |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR† | P Value | Haplotype Frequency in Case Patients/Control Subjects | OR† | P Value | Haplotype Frequency in Case Patients/Control Subjects | OR† | P Value | Haplotype Frequency in Case Patients/Control Subjects | OR† | P Value | |
| 1: M-M-M-M (G-G-C-T) | Ref | — | 0.751/0.701 | Ref | — | 0.724/0.697 | Ref | — | 0.728/0.694 | Ref | — |
| 2: m-M-M-M (A-G-C-T) | 0.91 (0.81–1.03) | 0.151 | 0.098/0.112 | 0.86 (0.69–1.05) | 0.143 | 0.102/0.101 | 1.02 (0.75–1.41) | 0.887 | 0.113/0.124 | 0.92 (0.77–1.10) | 0.374 |
| 3: M-M-m-M (G-G-T-T) | 1.01 (0.82–1.24) | 0.948 | 0.032/0.033 | 0.90 (0.62–1.29) | 0.556 | 0.043/0.037 | 1.15 (0.71–1.85) | 0.564 | 0.032/0.032 | 1.04 (0.76–1.41) | 0.826 |
| 4: M-m-m-M (G-A-T-T) | 0.99 (0.63–1.57) | 0.978 | 0.009/0.007 | 1.01 (0.47–2.15) | 0.987 | 0.008/0.002 | 3.35 (0.59–18.93) | 0.172 | 0.008/0.008 | 0.85 (0.46–1.56) | 0.598 |
| 5: M-m-m-m (G-A-T-C) | 0.73 (0.65–0.82) | 8.94 × 10−8 | 0.110/0.147 | 0.67 (0.55–0.81) | 6.33 × 10−5 | 0.123/0.164 | 0.71 (0.54–0.94) | 0.017 | 0.116/0.142 | 0.78 (0.66–0.92) | 4.78 × 10−4 |
Definition of abbreviations: GCPD-A = Study of the Genetic Causes of Complex Pediatric Disorders–Asthma; OR = odds ratio; Ref = reference; SAGE II = Study of African Americans, Asthma, Genes and Environment; SAPPHIRE = Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race–Ethnicity.
Haplotypes were based on functional polymorphisms within the African 17q12–21.1 haplotype block containing the early-onset asthma signal. Haplotype order is as follows: rs16965388 (G is the M, and A is the m); rs2305480 (G is the M, and A is the m); rs2305479 (C is the M, and T is the m); and rs11078928 (T is the M, and C is the m). Of note, variants are ordered by mapped position (lowest to highest) in reference genome hg38. However, the GSDMB (gasdermin-B) gene is transcribed from the negative strand. Therefore, rs11078928 is 5′ to the rs16965388 transcribed gene. The GSDMB nucleotide polymorphisms are reported according to the positive (nontranscribed) strand.
ORs are in comparison with the reference haplotype (G-G-C-T).
PheWAS of rs11078928
Longitudinal electronic-medical-record data were available for children as part of the larger GCPD study, including 19,433 African Americans, 1,329 Asian individuals, 20,667 European Americans, and 1,102 Latino individuals (Tables E2A–E2D). “Asthma with exacerbation” was the diagnosis most strongly associated with rs11078928 genotype in African Americans (OR, 0.80, P = 1.78 × 10−8), whereas “asthma” was the top diagnosis in European Americans (OR, 0.85, P = 4.48 × 10−10). In Asian and Latino individuals, “asthma with exacerbation” was less significantly associated with the rs11078928 genotype (OR, 0.64 [P = 0.014] and OR, [P = 0.042], respectively). In the meta-analysis, the top five clinical associations were related to respiration, led by asthma with exacerbation (OR, 0.81; P = 7.81 × 10−15) and asthma (OR, 0.86; P = 1.08 × 10−14) (Table E2E).
Association between Variant rs11078928 and Gene Expression
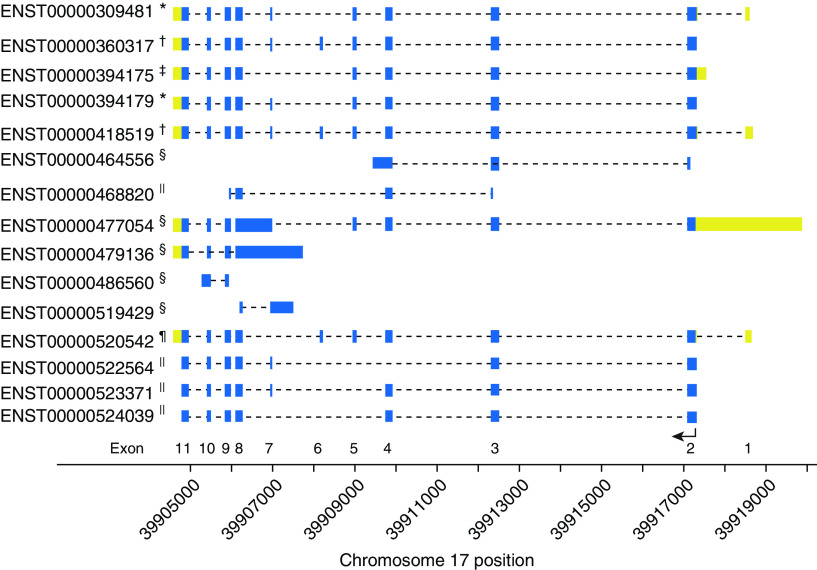
RNA-seq data was available on 844 SAPPHIRE participants. The rs11078928 C-allele dosage was negatively associated with overall expression of GSDMB (P = 3.60 × 10−12) and ORMDL3 (P = 7.00 × 10−5) (Table E3). Because rs11078928 immediately precedes exon 6 (44, 45), we evaluated the relationship between genotype and transcript isoforms (Table E4). The alternative transcripts of GSDMB are shown in Figure 3. Some of these spice isoforms correspond to the four protein-encoding isoforms: GSDMB-1 missing exon 6, GSDMB-2 missing exons 6 and 7, GSDMB-3 containing all 11 exons, and GSDMB-4 missing exon 7. The C-allele of the rs11078928 genotype was significantly associated with lower levels of Ensembl-spliced transcript 00000360317 (Figure 4 and Table E4) and the combination of all transcripts containing exon 6 (P = 6.86 × 10−15; Table E5). Only transcript Ensembl-spliced transcript 00000394175, encoding GSDMB-2 (i.e., missing exons 6 and 7), was significantly increased among rs11078928 C-allele carriers (P = 3.06 × 10−7). The rs11078928 C-allele was also associated with lower expression of transcripts encoding the full-length ORMDL3 protein (P = 8.32 × 10−4; Table E5). Similar transcript expression patterns were observed when the analytic set was restricted to individuals homozygous for African ancestry at 17q12–21.1, although the only significant relationships seen were for GSDMB and not ORMDL3 (Table E5).
Figure 3.
Shown are the various GSDMB (gasdermin-B) transcript isoforms and their relationship to their mapped genomic position on chromosome 17. GSDMB is transcribed from the reverse strand; hence, the exons (labeled at the bottom of the figure) are numbered in order from right to left. The thicker boxes denote regions that are transcribed: yellow signifies the untranslated regions, and blue signifies the translated regions. The bottom arrow shows the location of the start codon and the direction of translation. Dashed lines are intronic regions not included in the final transcript isoform. The footnote for each transcript isoform denotes the resulting product encoded: *protein with 403 amino acids (GSDMB-1), †protein with 416 amino acids (GSDMB-3), ‡protein with 394 amino acids (GSDMB-2), §no protein because of retained intron, ||nonsense-mediated decay, and ¶protein with 407 amino acids (GSDMB-4).
Figure 4.

The relationship between rs11078928 genotype and GSDMB (gasdermin-B) isoforms. The ENST identification numbers correspond to the alternatively spliced gene isoforms shown in Figure 3. The P value (Kruskal-Wallis one-way ANOVA) for the univariable relationship between genotype and transcript expression is shown at the top of each plot. Solid circles denote transcript read abundance within individuals; the boxes denote the interquartile range of transcript expression; and the middle line within each box is the median level of transcript expression. ENST = Ensembl-spiced transcript.
Assessing the relationship between asthma status and transcript expression (Table E5), we found asthma to be associated higher expression in blood of transcripts encoding GSDMB-3 and GSDMB transcripts containing exon 6 (FDR-adjusted P = 0.020 and P = 0.007, respectively). Asthma was also associated with higher expression of transcripts encoding full-length ORMDL3 (FDR-adjusted P = 0.020).
Discussion
Since it was first identified, the 17q12–21.1 locus has been considered a marker of childhood asthma (12). However, with some exceptions (16, 46), the studies evaluating this region and its relationship with asthma have almost exclusively included white individuals of European descent (13, 15).
Using UK Biobank data (47), Ferreira and colleagues showed that the genetic correlation between childhood-onset asthma and adult-onset asthma was 0.67, suggesting that there are both shared and independent genetic components in these phenotypes (48). In their meta-analysis of childhood-onset asthma among white individuals of European descent, marker rs4795399 within GSDMB was their strongest genome-wide association (P = 1 × 10−257). This variant is located at the edge of the 4-kb block that we defined as containing the early asthma signal using African ancestry. Other association studies for age of asthma onset have also used white individuals of European descent for their discovery populations (17, 49), and Sarnowski and colleagues identified rs9901146 between genes GSDMB and ZPBP2 (zona pellucida–binding protein 2) as their most significant association with time of asthma onset (49).
In large part, the high degree of LD between markers at 17q12–21.1 among European individuals precluded further fine mapping of the region for causal variants (19). This is the first study to employ WGS data to evaluate the 17q12–21.1 region in African American individuals. As a result, we did not need to rely on imputation or tagging SNPs to characterize this region. Although large panels with WGS data have markedly improved the quality of imputation (50, 51), imputation is still less effective in individuals of African descent, which is due in part to the greater genetic diversity in this group. We found that the greater genetic diversity among individuals of African ancestry permitted more precise localizing of the early asthma signal at 17q12–21.1. Specifically, the signal was largely restricted to a haplotype block extending from introns 3–10 of GSDMB. Further analysis of this haplotype block suggested that asthma risk most closely partitioned with rs11078928, a splice-acceptor variant located just before exon 6 (44).
Interestingly, we found that asthma risk associated with rs11078928 was similar for individuals of African and European descent. However, African American individuals are much more likely to carry the risk allele (T allele) when compared with European American individuals (i.e., 78.7% vs. 54.0% in the population with African ancestry in Southwest United States and population of Utah residents with Northern and Western European ancestry from the 1000 Genomes Project, respectively) (52). Allele frequency differences and LD differences may explain why earlier array-based genome-wide association studies and candidate-variant analyses of the 17q12–21.1 locus have generally showed different effect sizes by ancestry for alleles falling outside of the 4-kb haplotype block when compared with variants that are inside this block (14, 16).
Ober and colleagues recently performed an association analysis and eQTL study in nine longitudinal cohorts, which make up the Children’s Respiratory and Environmental Workgroup consortium (53). This group consisted of 1,613 European American children (296 [36%] with asthma) and 870 African American children (319 [45%] with asthma). They genotyped nine SNPs across the 17q12–21.1 locus for association with asthma and then evaluated their top associations for a relationship with gene expression in blood and epithelial cells. None of the nine SNPs were associated with asthma among African Americans in the Children’s Respiratory and Environmental Workgroup, but two SNPs, rs2305480 and rs80776131, were associated with asthma when meta-analyzed with samples from the EVE consortium (14). Variant rs2305480, a nonsynonymous variant in GSDMB exon 9, was associated with GSDMB expression in both peripheral blood mononuclear cells and upper airway epithelial cells; variant rs11078928 was not evaluated. In our study, we showed that rs2305480 was located in the haplotype block that appeared to contain the early asthma signal in African Americans. Although variant rs2305480 was in high LD with both rs11078928 and rs2305479 (another nonsynonymous variant in GSDMB exon 9), our haplotype-association analysis suggested that the early-onset asthma signal traveled most closely with the rs11078928 allele. Disentangling the relative importance of these variants on asthma development will require additional functional studies.
In strong support for role of rs11078928 in asthma development, Panganiban and colleagues also found the rs11078928 C-allele to have a protective association with asthma status (45). The effect estimate was similar across population groups (i.e., European Americans, Latino individuals, and African Americans), despite the different minor allele frequencies (0.45, 0.32, and 0.14, respectively). Importantly, these researchers also showed that inducing an aspartate-to-alanine missense mutation at amino acid 236 (D236A) located in exon 7 abolished caspase-1 cleavage of GSDBM into an N-terminal and C-terminal fragment. When expressed in human embryonic kidney 293T cell line cells, the N-terminal fragment of GSDMB alone could induce pyroptotic cell death, whereas the full-length protein and the C-terminal fragment did not. Coexpression of caspase-1 with wild-type GSDMB resulted in cell death, whereas coexpression of caspase-1 with the D236A mutated form of GSDMB did not. We did not observe, nor are we aware of, a commonly occurring missense mutation at residue 236. However, we did find that individuals with the protective rs11078928 C-allele had almost no expression of transcripts containing exon 6, and they had increased expression of a transcript missing both exons 6 and 7 (corresponding to missing amino acids 221–243).
Somewhat counter to the above findings, Das and colleagues showed that GSDMB-1, the isoform missing exon 6, was highly expressed in bronchial epithelial cells and that in vitro overexpression of GSDMB-1 resulted in increased production of other factors associated with airway remodeling and inflammation, such as transforming growth factor β-1, 5-lipoxygenase, and matrix metalloproteinase 9 (54). However, expression of full-length human GSDMB in mice, which do not have a naturally occurring GSDMB analog, resulted peribronchial smooth-muscle thickening and lung fibrosis. Increased expression of GSDMB-2 has been observed in other disease conditions. For example, Hergueta-Redondo and colleagues found GSDMB-2 expression in breast carcinomas to be associated with decreased survival and an increased likelihood of distant metastasis (55).
In addition to asthma (56–59), variants in or near GSDMB have also been associated with ulcerative colitis (60, 61), primary biliary cirrhosis (62, 63), type 1 diabetes (64), and cervical cancer (65, 66). However, in our PheWAS analysis in children, the rs11078928 variant was overwhelmingly associated with an asthma diagnosis when compared with other clinical diagnoses. DeBoever and colleagues examined the relationship between known protein-truncating variants and multiple clinical phenotypes among 337,205 unrelated individuals from the UK Biobank (67). A set of 3,724 protein-truncating variants was assessed for its relation to 135 phenotypes derived from surveys and clinical databases. Although the authors did not specify time of disease onset, the most significant relationship that they observed among all comparisons was between rs11078928 and asthma status (OR, 0.90; adjusted P = 9.1 × 10−45). The rs11078928 C-allele was also associated with a lower risk of bronchitis (OR, 0.91; adjusted P = 0.032); however, hay fever and/or allergic rhinitis (OR, 0.96), inflammatory bowel disease (OR, 1.09), female genital-tract cancer (OR, 1.07), and cervical cancer (OR, 1.09) were not significantly associated with the rs11078928 genotype. The above findings suggest that the strongest impact of the rs11078928 variant is through its effect on asthma.
Although rs11078928 has a functional impact on GSDMB isoform expression, this variant may also be an eQTL for ORMDL3. We found significant associations between rs11078928 genotype and both overall ORMDL3 gene expression and the expression of individual transcripts. GSDMB and ORMDL3 are adjacent to start codons located only 6.9 kb apart. In the initial genome-wide association study of asthma by Moffatt and colleagues, the most significant variant, rs7216389, was located in intron 2 of GSDMB—closer to ORMDL3, but well within the large haplotype block seen at 17q12–21.1 among Europeans (12). The investigators found rs7216389 to be strongly associated with ORMDL3 expression. The fact that we did not identify rs7216389 as the top association for early-onset asthma may relate to population group differences or our ability to more finely map this region among individuals of African descent. When we restricted the eQTL analysis to individuals homozygous for African ancestry at 17q12–21.1, we did not observe a significant association between rs11078928 genotype and ORMDL3 expression in blood. This lack of association may be the result of limited power in the smaller-sized group, or it could reflect ancestry-dependent differences in LD with ORMDL3 eQTL.
There have been multiple animal models and in vitro studies of ORMDL3 that support its functional role in asthma (68). These potential functions include regulating calcium signaling and contractility in airway smooth muscle (69), affecting ICAM-1 surface expression and sphingolipid metabolism in epithelial cells (70) and influencing inflammatory responses of type 2 T-helper cells (71). Because we studied gene expression in blood, we cannot comment on the effects of our lead association in other tissue types; however, some have speculated that 17q21 variants disproportionately affect lymphocyte populations (72). Schmiedel and colleagues identified two 17q12–21.1 variants, rs4065275 and rs12936231 (in the genes ORMDL3 and ZPBP2, respectively), that appear to affect CTCF (CCCTC-binding factor) motifs and ORMDL3 promoter–enhancer interactions (72). In our meta-analysis, neither variant was independently associated with early-onset asthma among African Americans (Table E1), and neither variant was in high LD with rs11078928 among individuals homozygous for African ancestry at 17q12–21,1 (Figure E4). However, these variants were in high LD among African Americans homozygous for European ancestry at 17q12–21.1 and European American individuals (Figures E3 and E5, respectively). Ancestry-based differences may explain why the 17q12–21.1 locus has been robustly associated with asthma in genomic studies of European ancestry populations in whom the large haplotype block may summarize the effects of multiple causal variants.
In summary, by studying a large population of individuals of African descent, we were able to localize an asthma association signal at 17q12–21.1. The top functional candidate, rs11078928, appeared to affect splicing of GSDMB transcripts. Expression levels of these transcript isoforms in blood were associated with asthma status. The PheWAS analysis provided additional support that asthma development and symptoms are the primary clinical consequence of this variant. Most importantly, the variant appeared to have the same magnitude of effect on asthma status in African American and European American individuals, suggesting that the mechanism and consequence of this polymorphism are not strongly influenced by ancestry. As a result, future therapies targeting this pathway are likely to prove equally beneficial to risk allele carriers regardless of racial or ethnic background.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank the studies and participants who provided biological samples and data for the Trans-Omics for Precision Medicine (TOPMed) program. They also thank all of those involved in the TOPMed Consortium, especially the members of the Lung and Asthma working groups. A complete list of TOPMed Consortium participants can be found at https://www.nhlbiwgs.org/topmed-banner-authorship. The authors thank SAGE II (Study of African Americans, Asthma, Genes and Environment) study collaborators Harold J. Farber, Emerita Brigino-Buenaventura, Michael A. LeNoir, Kelley Meade, Luisa N. Borrell, Adam Davis, and Fred Lurmann; SAGE II study coordinator Sandra Salazar; and SAGE II recruiters Lisa Caine, Elizabeth Castellanos, Brenda Lopez, and Shahdad Saeedi. The authors thank all of the participants, families, and clinic staff who made these studies possible.
Footnotes
Supported by the Fund for Henry Ford Hospital (H.G., A.M.L., D.E.L., and L.K.W.); the American Asthma Foundation (L.K.W. and E.G.B.); the Sandler Family Foundation (E.G.B.); the Robert Wood Johnson Foundation Amos Medical Faculty Development Program (E.G.B.); Harry W. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II (E.G.B.); and the following institutes of the NIH: National Institute of Allergy and Infectious Diseases (U19AI077439 to D.J.E. and R01AI079139 and R01AI061774 to L.K.W.), the NHLBI (R01HL128439, R01HL135156, R01HL117004, and X01HL134589 to E.G.B. and R01HL079055, R01HL118267, R01HL141845, and X01HL134589 to L.K.W.), the National Institute of Diabetes and Digestive and Kidney diseases (R01DK064695 and R01DK113003 to L.K.W.), the National Institute of Health and Environmental Sciences (R01ES015794 and R21ES024844 to E.G.B.), and the National Institute on Minority Health and Health Disparities (P60MD006902 and R01MD010443 to E.G.B.). Whole-genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the NHLBI. WGS for the NHLBI TOPMed SAGE (Study of African Americans, Asthma, Genes and Environment) study (phs000921.v1.p1) was performed at the New York Genome Center (3R01HL117004-02S3). WGS for the NHLBI TOPMed Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race–Ethnicity (phs001467.v1.p1) was performed at University of Washington’s Northwest Genome Center (HHSN268201600032I). WGS for the NHLBI TOPMed Genetics of Complex Pediatric Disorders—Asthma study (phs001661) was performed at Northwest Genomics Center (HHSN268201600032I). Centralized read-mapping and genotype-calling, together with variant quality metrics and filtering, were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Phenotype harmonization, data management, sample-identity quality control, and general study coordination were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1; contract HHSN268201800001I).
Author Contributions: H.G., A.M.L., D.H., P.S., S.X., A.C.Y.M., H.H., E.G.B., and L.K.W. contributed to the study conception and design. H.G., A.M.L., D.H., P.S., S.X., A.C.Y.M., M.Y., A.J.B., S. Huntsman, C.E., S. Hochstadt, K.W., S.S., W.C., S.T., G.A., T.W.B., H.M.K., D.A.N., S.G., D.J.E., H.H., E.G.B., and L.K.W. participated in the acquisition and/or analysis of the data. H.G., A.M.L., D.H., P.S., S.X., A.C.Y.M., M.Y., S. Huntsman, G.A., T.W.B., H.M.K., D.A.N., D.E.L., F.G., W.J.G., R.K., D.J.E., F.D.M., H.H., E.G.B., and L.K.W. were involved in the interpretation of the data. H.G., A.M.L., D.H., P.S., S.X., A.C.Y.M., E.Z., H.H., E.G.B., and L.K.W. drafted the work and/or substantially revised it. H.G., A.M.L., D.H., P.S., S.X., A.C.Y.M., M.Y., A.J.B., S. Huntsman, C.E., S. Hochstadt, E.Z., K.W., S.S., W.C., S.T., G.A., T.W.B., H.M.K., D.A.N., S.G., D.E.L., F.G., W.J.G., R.K., D.J.E., F.D.M., H.H., E.G.B., and L.K.W. approved of the submitted version and agree to be accountable for the accuracy and integrity of their contributions.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202006-2623OC on September 23, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Vos T, Flaxman AD, Naghavi M, Lozano R, Michaud C, Ezzati M, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010 Lancet 20123802163–2196.[Published erratum appears in Lancet 381:628.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asher MI, Montefort S, Björkstén B, Lai CK, Strachan DP, Weiland SK, et al. ISAAC Phase Three Study Group. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC phases one and three repeat multicountry cross-sectional surveys. Lancet. 2006;368:733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- 3.Papi A, Brightling C, Pedersen SE, Reddel HK. Asthma. Lancet. 2018;391:783–800. doi: 10.1016/S0140-6736(17)33311-1. [DOI] [PubMed] [Google Scholar]
- 4.Broder I, Higgins MW, Mathews KP, Keller JB. Epidemiology of asthma and allergic rhinitis in a total community, Tecumseh, Michigan: IV. Natural history. J Allergy Clin Immunol. 1974;54:100–110. doi: 10.1016/0091-6749(74)90038-4. [DOI] [PubMed] [Google Scholar]
- 5.Dodge RR, Burrows B. The prevalence and incidence of asthma and asthma-like symptoms in a general population sample. Am Rev Respir Dis. 1980;122:567–575. doi: 10.1164/arrd.1980.122.4.567. [DOI] [PubMed] [Google Scholar]
- 6.de Marco R, Locatelli F, Sunyer J, Burney P. Differences in incidence of reported asthma related to age in men and women: a retrospective analysis of the data of the European Respiratory Health Survey. Am J Respir Crit Care Med. 2000;162:68–74. doi: 10.1164/ajrccm.162.1.9907008. [DOI] [PubMed] [Google Scholar]
- 7.Strachan DP, Butland BK, Anderson HR. Incidence and prognosis of asthma and wheezing illness from early childhood to age 33 in a national British cohort. BMJ. 1996;312:1195–1199. doi: 10.1136/bmj.312.7040.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh RE, Cullinan P, Fishwick D, Hoyle J, Warburton CJ, Strachan DP, et al. Asthma and occupation in the 1958 birth cohort. Thorax. 2013;68:365–371. doi: 10.1136/thoraxjnl-2012-202151. [DOI] [PubMed] [Google Scholar]
- 9.Thomsen SF, Duffy DL, Kyvik KO, Backer V. Genetic influence on the age at onset of asthma: a twin study. J Allergy Clin Immunol. 2010;126:626–630. doi: 10.1016/j.jaci.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 10.Thomsen SF, van der Sluis S, Kyvik KO, Skytthe A, Backer V. Estimates of asthma heritability in a large twin sample. Clin Exp Allergy. 2010;40:1054–1061. doi: 10.1111/j.1365-2222.2010.03525.x. [DOI] [PubMed] [Google Scholar]
- 11.London SJ, James Gauderman W, Avol E, Rappaport EB, Peters JM. Family history and the risk of early-onset persistent, early-onset transient, and late-onset asthma. Epidemiology. 2001;12:577–583. doi: 10.1097/00001648-200109000-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 13.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. GABRIEL Consortium. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Mexico City Childhood Asthma Study (MCAAS); Children’s Health Study (CHS) and HARBORS study; Genetics of Asthma in Latino Americans (GALA) Study, Study of Genes-Environment and Admixture in Latino Americans (GALA2) and Study of African Americans, Asthma, Genes & Environments (SAGE); Childhood Asthma Research and Education (CARE) Network; Childhood Asthma Management Program (CAMP); Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-Ethnicity (SAPPHIRE); Genetic Research on Asthma in African Diaspora (GRAAD) Study. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmuller J, Ang W, et al. Australian Asthma Genetics Consortium (AAGC) Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50:42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daya M, Rafaels N, Brunetti TM, Chavan S, Levin AM, Shetty A, et al. CAAPA. Association study in African-admixed populations across the Americas recapitulates asthma risk loci in non-African populations. Nat Commun. 2019;10:880. doi: 10.1038/s41467-019-08469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forno E, Lasky-Su J, Himes B, Howrylak J, Ramsey C, Brehm J, et al. Genome-wide association study of the age of onset of childhood asthma. J Allergy Clin Immunol. 2012;130:83–90, e4. doi: 10.1016/j.jaci.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halapi E, Gudbjartsson DF, Jonsdottir GM, Bjornsdottir US, Thorleifsson G, Helgadottir H, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18:902–908. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein MM, Thompson EE, Schoettler N, Helling BA, Magnaye KM, Stanhope C, et al. A decade of research on the 17q12-21 asthma locus: piecing together the puzzle. J Allergy Clin Immunol. 2018;142:749–764, e3. doi: 10.1016/j.jaci.2017.12.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin AM, Gui H, Hernandez-Pacheco N, Yang M, Xiao S, Yang JJ, et al. Integrative approach identifies corticosteroid response variant in diverse populations with asthma. J Allergy Clin Immunol. 2019;143:1791–1802. doi: 10.1016/j.jaci.2018.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White MJ, Risse-Adams O, Goddard P, Contreras MG, Adams J, Hu D, et al. Novel genetic risk factors for asthma in African American children: precision medicine and the SAGE II Study. Immunogenetics. 2016;68:391–400. doi: 10.1007/s00251-016-0914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children N Engl J Med 201036236–44.[Published errata appear in N Engl J Med 363:994 and N Engl J Med 366:672.] [DOI] [PubMed] [Google Scholar]
- 23.Conomos MP, Miller MB, Thornton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free estimation of recent genetic relatedness. Am J Hum Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cattell RB. The scree test for the number of factors. Multivariate Behav Res. 1966;1:245–276. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
- 29.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 32.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–1994. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 38.Tardif JC, Rhéaume E, Lemieux Perreault LP, Grégoire JC, Feroz Zada Y, Asselin G, et al. Pharmacogenomic determinants of the cardiovascular effects of dalcetrapib. Circ Cardiovasc Genet. 2015;8:372–382. doi: 10.1161/CIRCGENETICS.114.000663. [DOI] [PubMed] [Google Scholar]
- 39.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 41.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant P-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 44.Morrison FS, Locke JM, Wood AR, Tuke M, Pasko D, Murray A, et al. The splice site variant rs11078928 may be associated with a genotype-dependent alteration in expression of GSDMB transcripts. BMC Genomics. 2013;14:627. doi: 10.1186/1471-2164-14-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panganiban RA, Sun M, Dahlin A, Park HR, Kan M, Himes BE, et al. A functional splice variant associated with decreased asthma risk abolishes the ability of gasdermin B to induce epithelial cell pyroptosis. J Allergy Clin Immunol. 2018;142:1469–1478, e2. doi: 10.1016/j.jaci.2017.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galanter J, Choudhry S, Eng C, Nazario S, Rodríguez-Santana JR, Casal J, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira MAR, Mathur R, Vonk JM, Szwajda A, Brumpton B, Granell R, et al. 23andMe Research Team; eQTLGen Consortium; BIOS Consortium. Genetic architectures of childhood- and adult-onset asthma are partly distinct. Am J Hum Genet. 2019;104:665–684. doi: 10.1016/j.ajhg.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarnowski C, Sugier PE, Granell R, Jarvis D, Dizier MH, Ege M, et al. Identification of a new locus at 16q12 associated with time to asthma onset. J Allergy Clin Immunol. 2016;138:1071–1080. doi: 10.1016/j.jaci.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 50.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowalski MH, Qian H, Hou Z, Rosen JD, Tapia AL, Shan Y, et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; TOPMed Hematology & Hemostasis Working Group. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 2019;15:e1008500. doi: 10.1371/journal.pgen.1008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ober C, McKennan CG, Magnaye KM, Altman MC, Washington C, III, Stanhope C, et al. Environmental Influences on Child Health Outcomes–Children’s Respiratory Research Workgroup. Expression quantitative trait locus fine mapping of the 17q12-21 asthma locus in African American children: a genetic association and gene expression study. Lancet Respir Med. 2020;8:482–492. doi: 10.1016/S2213-2600(20)30011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das S, Miller M, Beppu AK, Mueller J, McGeough MD, Vuong C, et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc Natl Acad Sci U S A. 2016;113:13132–13137. doi: 10.1073/pnas.1610433113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hergueta-Redondo M, Sarrió D, Molina-Crespo Á, Megias D, Mota A, Rojo-Sebastian A, et al. Gasdermin-B promotes invasion and metastasis in breast cancer cells. PLoS One. 2014;9:e90099. doi: 10.1371/journal.pone.0090099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu H, Romieu I, Sienra-Monge JJ, Li H, del Rio-Navarro BE, London SJ. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64:629–635. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bisgaard H, Bønnelykke K, Sleiman PM, Brasholt M, Chawes B, Kreiner-Møller E, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med. 2009;179:179–185. doi: 10.1164/rccm.200809-1436OC. [DOI] [PubMed] [Google Scholar]
- 58.Kang MJ, Yu HS, Seo JH, Kim HY, Jung YH, Kim YJ, et al. GSDMB/ORMDL3 variants contribute to asthma susceptibility and eosinophil-mediated bronchial hyperresponsiveness. Hum Immunol. 2012;73:954–959. doi: 10.1016/j.humimm.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 59.Leung TF, Sy HY, Ng MC, Chan IH, Wong GW, Tang NL, et al. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy. 2009;64:621–628. doi: 10.1111/j.1398-9995.2008.01873.x. [DOI] [PubMed] [Google Scholar]
- 60.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McGovern DP, Gardet A, Törkvist L, Goyette P, Essers J, Taylor KD, et al. NIDDK IBD Genetics Consortium. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X, Invernizzi P, Lu Y, Kosoy R, Lu Y, Bianchi I, et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet. 2010;42:658–660. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hitomi Y, Kojima K, Kawashima M, Kawai Y, Nishida N, Aiba Y, et al. Identification of the functional variant driving ORMDL3 and GSDMB expression in human chromosome 17q12-21 in primary biliary cholangitis. Sci Rep. 2017;7:2904. doi: 10.1038/s41598-017-03067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Type 1 Diabetes Genetics Consortium. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi Y, Li L, Hu Z, Li S, Wang S, Liu J, et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat Genet. 2013;45:918–922. doi: 10.1038/ng.2687. [DOI] [PubMed] [Google Scholar]
- 66.Lutkowska A, Roszak A, Lianeri M, Sowińska A, Sotiri E, Jagodziński PP. Analysis of rs8067378 polymorphism in the risk of uterine cervical cancer from a polish population and its impact on gasdermin B expression. Mol Diagn Ther. 2017;21:199–207. doi: 10.1007/s40291-017-0256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DeBoever C, Tanigawa Y, Lindholm ME, McInnes G, Lavertu A, Ingelsson E, et al. Medical relevance of protein-truncating variants across 337,205 individuals in the UK Biobank study. Nat Commun. 2018;9:1612. doi: 10.1038/s41467-018-03910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller M, Broide DH. Why is ORMDL3 on chromosome 17q21 highly linked to asthma? Am J Respir Crit Care Med. 2019;199:404–406. doi: 10.1164/rccm.201810-1941ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen J, Miller M, Unno H, Rosenthal P, Sanderson MJ, Broide DH. Orosomucoid-like 3 (ORMDL3) upregulates airway smooth muscle proliferation, contraction, and Ca 2+ oscillations in asthma. J Allergy Clin Immunol. 2018;142:207–218, e6. doi: 10.1016/j.jaci.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Willis-Owen SAG, Spiegel S, Lloyd CM, Moffatt MF, Cookson WOCM. The ORMDL3 asthma gene regulates ICAM1 and has multiple effects on cellular inflammation. Am J Respir Crit Care Med. 2019;199:478–488. doi: 10.1164/rccm.201803-0438OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller M, Rosenthal P, Beppu A, Mueller JL, Hoffman HM, Tam AB, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192:3475–3487. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmiedel BJ, Seumois G, Samaniego-Castruita D, Cayford J, Schulten V, Chavez L, et al. 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells. Nat Commun. 2016;7:13426. doi: 10.1038/ncomms13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.